- 1Key Laboratory of Sustainable Exploitation of Oceanic Fisheries Resources, Ministry of Education, College of Marine Sciences, Shanghai Ocean University, Shanghai, China
- 2Shanghai Engineering Research Center of Hadal Science & Technology, College of Marine Sciences, Shanghai Ocean University, Shanghai, China
- 3Key Laboratory of Aquaculture Resources and Utilization, Ministry of Education, College of Fisheries and Life Sciences, Shanghai Ocean University, Shanghai, China
- 4National Distant-Water Fisheries Engineering Research Center, Shanghai Ocean University, Shanghai, China
Hadal trenches are commonly referred to as the deepest areas in the ocean and are characterized by extreme environmental conditions such as high hydrostatic pressures and very limited food supplies. Amphipods are considered the dominant scavengers in the hadal food web. Alicella gigantea is the largest hadal amphipod and, as such, has attracted a lot of attention. However, the adaptive evolution and gigantism mechanisms of the hadal “supergiant” remain unknown. In this study, the whole-body transcriptome analysis was conducted regarding the two hadal amphipods, one being the largest sized species A. gigantea from the New Britain Trench and another the small-sized species Bathycallisoma schellenbergi from the Marceau Trench. The size and weight measurement of the two hadal amphipods revealed that the growth of A. gigantea was comparatively much faster than that of B. schellenbergi. Phylogenetic analyses showed that A. gigantea and B. schellenbergi were clustered into a Lysianassoidea clade, and were distinct from the Gammaroidea consisting of shallow-water Gammarus species. Codon substitution analyses revealed that “response to starvation,” “glycerolipid metabolism,” and “meiosis” pathways were enriched among the positively selected genes (PSGs) of the two hadal amphipods, suggesting that hadal amphipods are subjected to intense food shortage and the pathways are the main adaptation strategies to survive in the hadal environment. To elucidate the mechanisms underlying the gigantism of A. gigantea, small-sized amphipods were used as the background for evolutionary analysis, we found the seven PSGs that were ultimately related to growth and proliferation. In addition, the evolutionary rate of the gene ontology (GO) term “growth regulation” was significantly higher in A. gigantea than in small-sized amphipods. By combining, those points might be the possible gigantism mechanisms of the hadal “supergiant” A. gigantea.
Introduction
Hadal, the deepest area of the ocean (6,000–11,000 m), is a unique harsh environment characterized by extremely high hydrostatic pressures, low temperature, and limited food sources (Somero, 1992; Jamieson et al., 2010). Hadal is an extremely hostile environment for most organisms; however, some species seem to be adapted very well to the hadal environment (Somero, 1992; Bartlett, 2002; Yancey and Siebenaller, 2015). With increasing sampling efforts regarding the hadal trenches, a wide range of organisms was found to thrive within the extreme environment (Nunoura et al., 2015; Tarn et al., 2016). How those hadal creatures adapted to the extreme environment is a topic of great interest to researchers.
The organisms that are endemic to hadal zones have evolved unique physiological and biochemical functions necessary for growth and survival in this hadal habitat. Hadal creatures were found to have more fluidity of cell membranes due to an increase of less rigid unsaturated fatty acid chains (Cossins and Macdonald, 1984, 1989). For example, compared with the bathyal roundnose grenadier Coryphaenoides rupestris (400–1,200 m depth), the abyssal grenadier Coryphaenoides armatus (2,000–5,000 m depth) was found to have consistently higher fluidity in membranes (Cossins and Macdonald, 1989). Hadal creatures were also found to own some external types of “assistance” to improve protein stability and functional adaptability under high hydrostatic pressures. The types of “assistance” include stress proteins, phospholipids, and micromolecular counteractants, for example, trimethylamine N-oxide (TMAO), which was reported to be of great significance for the adaptation of hadal species (Yancey, 2020).
With the development of molecular technology, researchers tried to explore the molecular adaptation mechanisms of the hadal creatures at the genetic level. For example, the whole genome sequence of Mariana hadal snailfish (Pseudoliparis Swirei) was recently analyzed. The bone Gla protein (BGLAP) gene possessed a frameshift mutation in P. swirei, which resulted in an incomplete ossification of the hadal snailfish to adapt to a high-pressure environment. The hadal snailfish also lost several important photoreceptor genes to adapt to the lightless hadal environment (Wang et al., 2019).
Amphipods (Arthropoda: Crustacea: Amphipoda) have a long evolutionary history, wide variety of species, and wide distribution ranging from 0 to 11,000 m, and are considered to be the dominant scavengers in the hadal food web (Eustace et al., 2016; Lacey et al., 2016; Copilaş-Ciocianu et al., 2020). These animals are among the few fauna species that can be readily obtained in large numbers and diversities. Therefore, hadal amphipods are commonly used in the study of adaptability to an extreme environment. By comparing hadal and littoral amphipods, researchers have found that TMAO, scyllo-inositol, and other pressure counteractants increase with depth (Downing et al., 2018). The hadal Hirondellea gigas was found to have a unique enzyme component, cellulase (HGcel), which can digest the wooden debris buried in the deepest seafloor (Kobayashi et al., 2012).
Among the reported hadal amphipods so far, one unique hadal amphipod, Alicella gigantea, attracts wide attention due to its significant gigantism. A. gigantea, also called “supergiant,” inhabits in the deep abyssal plains in the Northern Hemisphere of the North Atlantic Ocean (off the Canaries, Cape Verde, and in the Demerara Basin) and near the Hawai'ian Islands of the North Pacific Ocean (Barnard and Ingram, 1986; Hasegawa et al., 1986; De Broyer and Thurston, 1987). It is the largest known amphipod, whose adult body length ranges from 240 to 340 mm (Harrison et al., 1983; Barnard and Ingram, 1986; Jamieson et al., 2013). Chapelle and Peck (2004) counted the body lengths of over 2,000 amphipod species and found that the body sizes of most amphipod species were <40 mm. It has also been suggested that, with reduced temperature and increased hydrostatic pressure, there should be an increase in cell size and life span (Timofeev, 2001), which might be an explanation for the gigantism of A. gigantea. However, until now there has not been a genetic mechanism study on the gigantism of A. gigantea.
Bathycallisoma schellenbergi is a geographically widely distributed hadal amphipod species. It is found in the North Pacific, the Puerto Rico Trench in the Southwest Pacific, and the Java Trench in the Indian Ocean (Lacey et al., 2013). Compared with the supergiant A. gigantea, B. schellenbergi is quite a small hadal amphipod. For example, B. schellenbergi collected from the Tonga Trench had an average size of 25 mm (Jamieson, 2015). Similar to most hadal amphipods, B. schellenbergi is an obligate scavenger, feeding on carrion falling to the deep-sea floor (Sainte-Marie, 1992; Britton and Morton, 1994; Kaiser and Moore, 1999). It has some hadal adaptations, such as the acute chemoreceptor organs used to detect carrion and the ability to resist chronic hunger, which is conducive to the scavenging foraging strategy (Smith and Baldwin, 1982; Sainte-Marie, 1992). Therefore, B. schellenbergi can be used as a fine reference species to explore the gigantism mechanism of A. gigantea.
Indeed, to explore the hadal adaptation mechanisms and the possible causes of gigantism in A. gigantea, its whole genome has to be analyzed. However, the estimated huge genome size of A. gigantea (34.02 Gb) makes such a task cumbersome (Ritchie et al., 2017). It has been also well-known that the hadal amphipods possess a striking large genome, ranging from 4.04 Gb in Paralicella caperesca to 34.02 Gb in A. gigantea (Ritchie et al., 2017). Therefore, transcriptome sequencing and analysis could be an effective option for genome-wide comparative adaptation studies of hadal species. Some researchers have identified a genome-wide positive selection by combining the next-generation sequencing of the transcriptome with branching site modeling to reveal the underlying mechanisms of molecular adaptation (Tsaur and Wu, 1997; Roux et al., 2014; Dungan et al., 2016), which provides technical support for the hadal species adaptation studies at the genetic level. As of now, only H. gigas from the Mariana Trench has been analyzed by transcriptome (Lan et al., 2017), and the transcriptome analysis of other hadal amphipod species is lacking.
In this study, two different sized amphipods, A. gigantea and B. schellenbergi, were selected as the research objects for the transcriptome and evolutionary analysis, which could improve the adaptation mechanisms of hadal amphipods at the genetic level. Meanwhile, the gigantism mechanism of the “supergiant” A. gigantea was also explored in this study.
Materials and Methods
Sample Collection
Two amphipod species, A. gigantea and B. schellenbergi, were collected from the New Britain Trench (8,824 m, 7.02°S 149.16°E) and Marceau Trench (6,690 m, 1.42°N 148.74°E) in the West Pacific Ocean (Figure 1). The autonomous deep-ocean lander vehicle, equipped with two cage traps baited with a suitable amount of mackerels, was launched from the “Zhang Jian” research vessel and deployed to the sea floor for up to 10 h. Detailed information about the lander vehicle and sampling was described in Chan et al. (2020, 2021). Upon the recovery of the lander, amphipods were preserved immediately at −80°C until processed for analysis. The body length and weight were measured in a land-based laboratory.
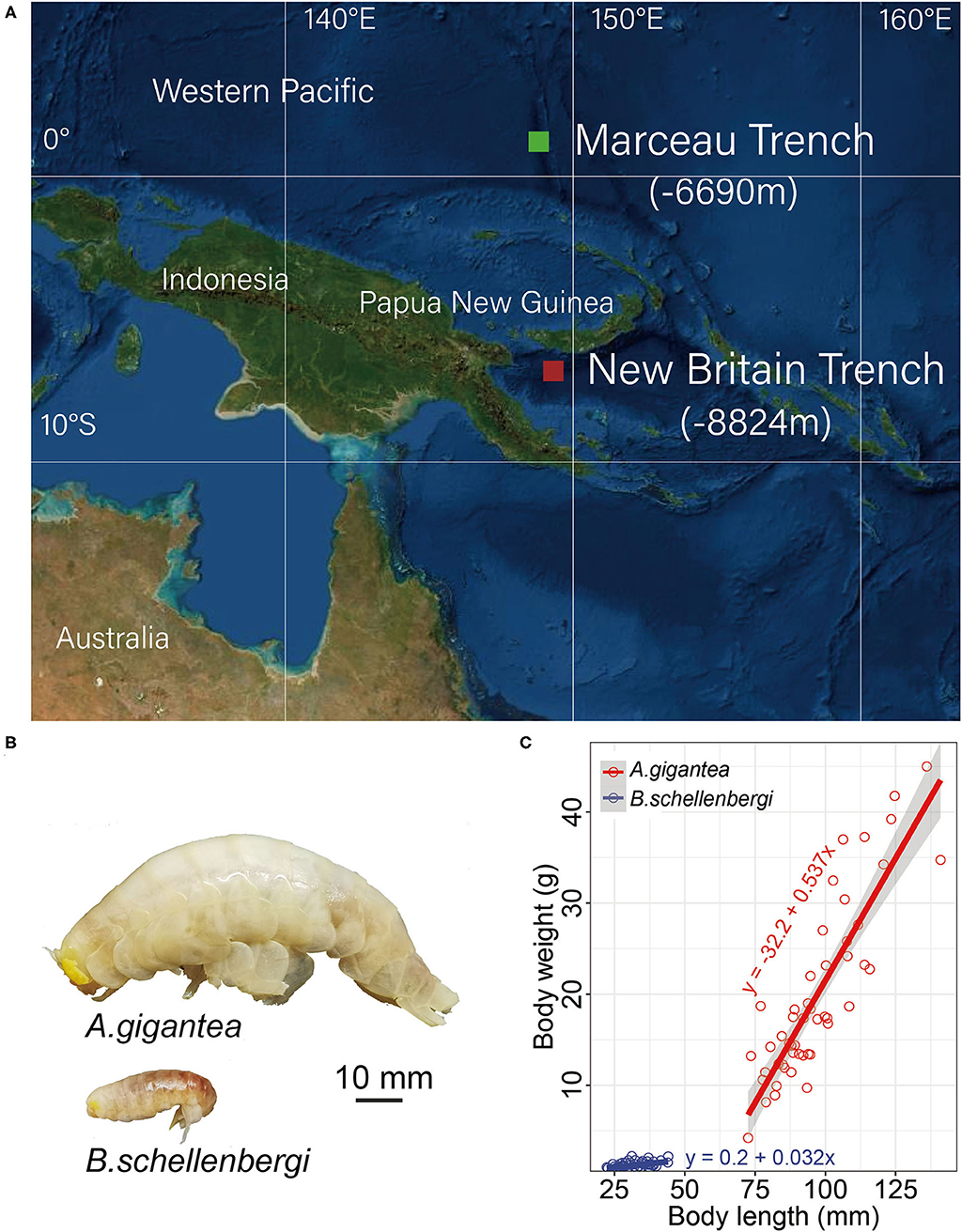
Figure 1. Sample collection and size and weight measurement. (A) The sampling sites of two amphipods were used in this study. Alicella gigantea and Bathycallisoma schellenbergi individuals were sampled from the New Britain Trench (8,224 m) (red square) and the Marceau Trench (6,990 m) (green square), respectively. (B) The pictures of A. gigantea (upper) and B. schellenbergi (down) exhibited a significant difference in their body size. (C) The growth fitting curves for A. gigantea (red) and B. schellenbergi (blue). About 50 individuals for each species were used for the body length (x-axis, mm) and body weight (y-axis, g) measurement. The linear correlation equations between the body lengths and weights for A. gigantea (red) and B. schellenbergi (blue) were also shown.
Transcriptome Sequencing, de novo Assembly, and Gene Prediction
Nine juvenile individuals of A. gigantea (5–7 cm) and B. schellenbergi (2–3 cm) were selected and divided into three subgroups (three individuals for each subgroup), which were assigned as three biological replicates. Total RNA from the whole body of A. gigantea and B. schellenbergi was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). The complementary DNA (cDNA) libraries were constructed by VAHTS Universal V6 RNA-seq Library Prep Kit Illumina (Vazyme Biotech, Nanjing, China, Cat: NR604-01/02) and sequenced on Illumina NovaSeq 6000/PE150 (Novogene, Beijing, China). The paired-end cleaned reads were obtained by trimmomatic (Bolger et al., 2014) (0.33) with the parameter: HEADCROP:3 AVGQUAL:30 TRAILING:30 SLIDINGWINDOW:4:20 MINLEN:50. All clean reads of the three biological replicates were merged and assembled by the Trinity (Haas et al., 2013) (v2.6.6) software, maintaining the length of over 200 bp transcript. To evaluate the quality of the assemblies, contigs number, N50 length, assembly size, and the predicted gene number were assessed. Then, we selected the longest isoform of the gene as the unigene had to do the following analysis. To assess the completeness of the assembled transcripts, single-copy marker genes were checked by the benchmarking universal single-copy orthologs (BUSCO) (3.0.2) software package (Simão et al., 2015; Seppey et al., 2019) using the Arthropoda subset (arthropoda_odb10). The TransDecoder (Haas et al., 2013) (5.2.0) software was used to predict the protein sequence with the annotation result of blastp (Altschul et al., 1997) (2.5.0+) and hmmscan (Finn et al., 2011) (3.1b2) from the UniProt and Pfam database.
Gene Functional Annotation
Following the de novo assembly and gene prediction, we annotated the predicted genes from different databases, including the National Center for Biotechnology Information non-redundant (NCBI-nr), KEGG (Kyoto Encyclopedia of Genes and Genomes) automatic annotation server (KAAS) (Moriya et al., 2007), and eggNOG (Jensen et al., 2008) database. The DIAMOND (Buchfink et al., 2015) software (v0.8.22.84) was used to mapping the NR database with an E-value cutoff of 1e-10 and a minimum match percentage identity of 50%. The gene ontology (GO) terms were extracted from the eggNOG and Swissprot results. KAAS (https://www.genome.jp/tools/kaas/) was used for an orthology assignment and a pathway mapping by using a bidirectional best hit (BBH) method.
Identification of Orthologs and Phylogenetic Analysis
The whole-body transcriptome and genome data for 11 arthropoda species were obtained from multiple sources. Gammarus fossarum was obtained from Bourbre River in central France and its body length was about 2 mm (Straub et al., 2017). Gammarus minus was collected from the Ward Spring in Greenbrier County, West Virginia, USA (Carlini and Fong, 2017). The male adults were about 6–9 mm in length (David et al., 2013). Gammarus chevreuxi was collected from the River Plym, Plymouth, UK (Collins et al., 2017). H. gigas was collected from the Mariana Trench with a depth of 10,929 m (Lan et al., 2017). The body length of H. gigas can reach more than 30 mm (Kobayashi et al., 2019). Echinogammarus marinus was from the sea coast (50.79°N 1.03°W) of the UK (Cogne et al., 2019). We downloaded the published Gammaridea species transcriptome raw data from SRA (https://www.ncbi.nlm.nih.gov/sra) (G. fossarum ERR386132, G. minus SRR5576331, SRR5576333, G. chevreuxi SRR5109803, SRR5109804, SRR5109805, H. gigas SRR3822238, and E. marinus SRR8089734, SRR8089735), and then cleaned and assembled the data by using the same pipeline like our A. gigantea and B. schellenbergi data.
Genome sequence and gene annotation files of Hyalella azteca (Poynton et al., 2018), Penaeus vannamei (Zhang et al., 2019), Eurytemora affinis (Eyun et al., 2017), Daphnia pulex (Colbourne et al., 2011), Cryptotermes secundus (Harrison et al., 2018) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/). The genome sequence of Parhyale hawaiensis (Kao et al., 2016) was downloaded from the NCBI, and the coding DNA sequence of P. hawaiensis was annotated by the gene model mapper (Keilwagen et al., 2018) (GeMoMa, 1.5.3) software using P. vannamei homology protein. OrthoMCL (Li et al., 2003) (v2.0.9) was employed to retrieve the groups of homologous genes from the longest protein sequences of each gene. Single-copy orthologous genes were extracted using a manual Perl script. These sequences were aligned using the MAFFT (Katoh et al., 2002) (v7.407) software. The aligned sequences were converged to a supergene. Gblocks (Castresana, 2000) (0.91b) software was used to get the conserved region. Using the best model according to the ProtTest (Darriba et al., 2011) (3.4) software: LG+I+G+F, the phylogenetic tree was reconstructed using the RAxML (Stamatakis et al., 2008) software (v8.1.24, raxmlHPC-PTHREADS-SSE3, bootstrap: 1,000 iterations). C. secundus was selected as the outgroup species. Figtree (v1.4.4) was used for the cladogram tree visualization. The classification of Arthropoda in this study was made according to the Taxonomicon (http://taxonomicon.taxonomy.nl/).
Determination of Positively Selected Genes
Single-copy orthologous genes were selected from A. gigantea, B. schellenbergi, G. minus, G. fossarum, G. chevreuxi, and E. marinus using the OrthoMCL software. PRANK was used to align the codon sequences by using a codon substitution matrix. Gblocks (0.91b) software was used to get the conserved region using the –t = c parameter. The signatures of positively selected genes (PSGs) along a specific branch can be detected by branch-site models implemented in the CodeML module of the phylogenetic analysis by maximum likelihood (PAML) package (Yang, 2007) version 4.9 g. To detect PSGs in the hadal amphipod, the A. gigantea lineage was designated as “foreground” phylogeny, and the other four shallow-water Gammarida lineages were assigned as “background” phylogeny. The tree file for the branch-site model is [((E. marinus, (G. chevreuxi, (G. minus, G. fossarum))), A. gigantea #1);] which the foreground species labeled with #1. In a similar way, the B. schellenbergi lineage was designated as “foreground” phylogeny to detect the PSGs in B. schellenbergi, and the H. gigas lineage was designated as “foreground” phylogeny to detect the PSGs in H. gigas.
To detect the PSGs that are unique to the “supergiant” amphipod, single-copy orthologous genes were selected among A. gigantea, B. schellenbergi, H. gigas, G. chevreuxi, and E. marinus using the OrthoMCL software, and A. gigantea was set as a foreground branch. The tree file for the branch-site model is [((E. marinus, G. chevreuxi), (H. gigas, (B. schellenbergi, A. gigantea #1)));]. The custom python script was used to implement the GO and KEGG enrichment analysis by using a hypergeometric test method.
Identifications of Rapidly Evolving GO Terms
To identify the rapidly evolving GO terms in A. gigantea or B. schellenbergi, we estimated the evolution rate by calculating the average dN/dS values for GO terms. We combined the GO annotation both from the eggNOG database and the Swissprot results. The GO categories, including at least 30 but <800 single-copy orthologous genes, were used for the following analysis. We concatenated the single-copy orthologous genes to a “supergene” according to the GO category. We calculated the dN/dS values of supergenes using a free-ratio model (M1) of the CodeML program, which is integrated into the PAML package. Supergenes with N*dN <1 or S*dS <1 or dS >1 were filtered.
Results
Size Measurement of the Two Hadal Amphipods
In this study, 50 individuals for each species were randomly selected in terms of their size and weight measurement. The body size of A. gigantea ranges from 72.5 to 141.0 mm and its weight ranges from 4.2 to 45 g, whereas the body size of B. schellenbergi ranges from 22.3 to 44.0 mm and its weight ranges from 0.5 to 2.2 g. From the perspective of range, body size, and weight have a much wider distribution in A. gigantea than in B. schellenbergi. It is also obvious that body size and weight are much larger in A. gigantea than in B. schellenbergi. From the growth trend, the growth fitting curves of both A. gigantea and B. schellenbergi are linear, but the slope of A. gigantea is much larger than that of B. schellenbergi, which might indicate that the weight of A. gigantea increases comparatively rapidly (Figure 1C).
Data Filtering and de novo Assembly
The transcriptome sequencing results for the two hadal amphipods, A. gigantea and B. schellenbergi, were summarized in Supplementary Table 1. A total of 18.9 and 16.6 Gb raw data were generated for A. gigantea and B. schellenbergi, respectively. The trimmed reads were then used in the transcriptome assembly. The final assembled transcriptome contained 244,665 and 204,636 contigs, which have a total length of 159.1 and 128.2 M and a contig N50 value of 986 and 917 bp, respectively, for A. gigantea and B. schellenbergi. About 26,559 and 21,879 contigs were annotated for A. gigantea and B. schellenbergi, respectively. The completeness of the assembled transcriptomes against the BUSCO database was 90.1% in A. gigantea and 86.3% in B. schellenbergi.
Gene Annotations
The predicted genes were blasted from the NCBI-nr protein database using the DIAMOND software. The percent ratio of the genes that have homologs to the genes from the NCBI-nr database for A. gigantea and B. schellenbergi is 55.2% (14,661/26,559) and 58.4% (12,774/21,879), respectively, suggesting that the two hadal amphipods contained a certain number of orphan genes. Most of the genes of A. gigantea and B. schellenbergi have homologs in the NCBI-nr database and are mapped to Malacostraca animals, 70.1 and 69.6% mapping to H. azteca (Amphipoda), 11.5 and 12.2% mapping to P. vannamei (Decapoda), and 1.5 and 1.4% mapping to Armadillidium vulgare (Isopoda), respectively (Figure 2). A total of 11,185 (42.1%) in A. gigantea and 9,466 (43.3%) in B. schellenbergi translated proteins had at least one significant hit on GO terms, and 10,771 (40.5%) in A. gigantea and 9,110 (41.6%) in B. schellenbergi were matched to the KEGG pathway database.
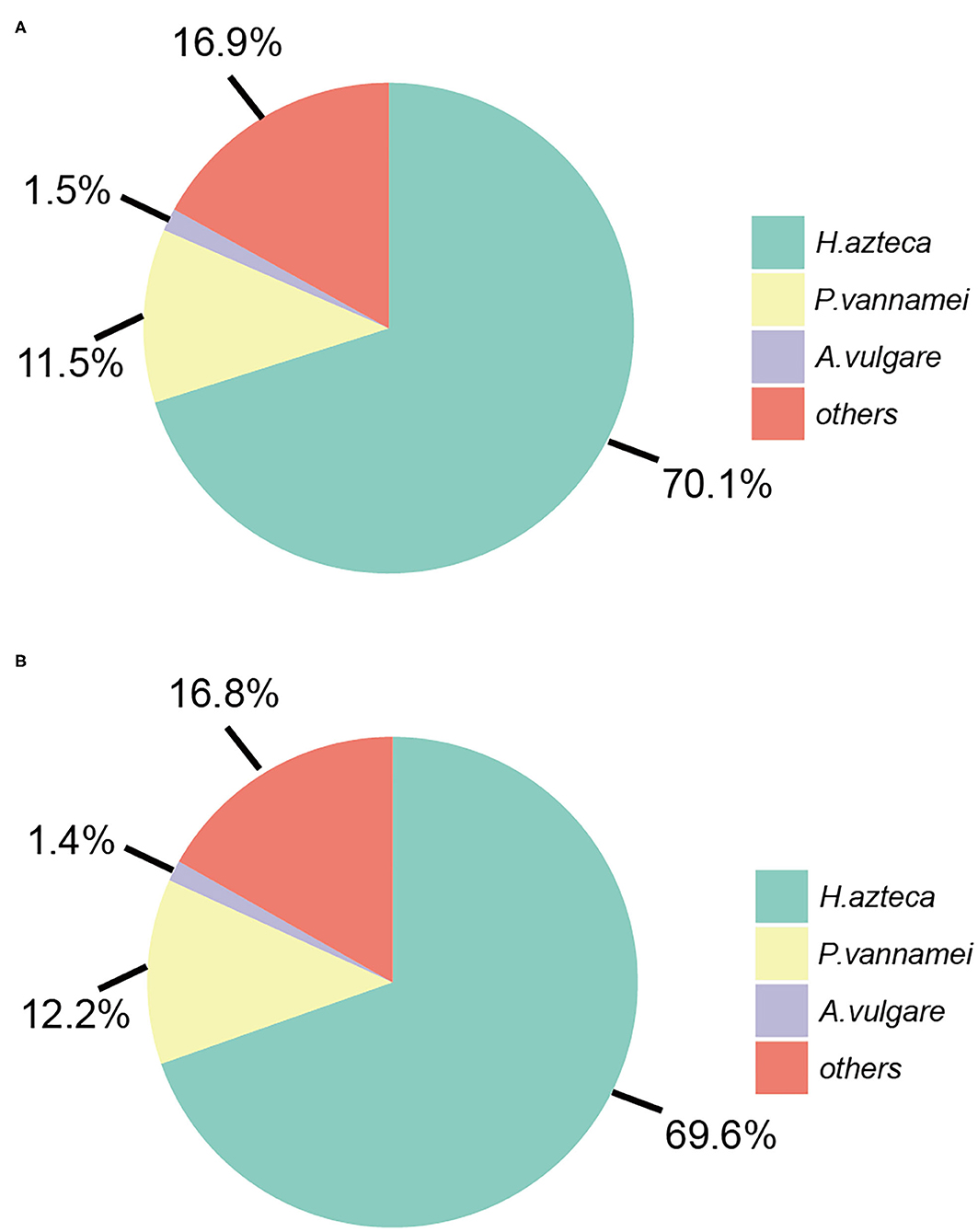
Figure 2. The distributions of the hit species for A. gigantea (A) and B. schellenbergi (B) based on BLASTP against non-redundant (Nr) databases.
Phylogenetic Relationships Between Hadal and Shallow-Water Arthropods
A total of 31,051 orthologous gene families were clustered using the OrthoMCL software with default parameters. To reveal the phylogenetic relationship between the hadal amphipods and shallow-water arthropods, a total of 512 single-copy orthologous genes among A. gigantea, B. schellenbergi, H. gigas, G. minus, G. fossarum, G. chevreuxi, E. marinus, P. hawaiensis, H. azteca, P. vannamei, E. affinis, D. pulex, and C. secundus (served as an outgroup) were identified and used in the tree construction. After the alignment and removal of poorly aligned positions and regions, 91,385 positions (19%) remained in 1,832 selected block(s) were used for the phylogenetic tree reconstruction (Figure 3A). All nodes were supported with the bootstrap values of 100, indicating a well-resolved relationship between the 11 species (Figure 3A). It is evident that the three hadal species, A. gigantea, B. schellenbergi, and H. gigas, are clustered into one clade (Lysianassoidea clade), and that the four shallow-water Gammarus species, G. minus, G. fossarum, G. chevreuxi, and E. marinus, are clustered into another clade (Gammaroidea clade) (Figure 3A).
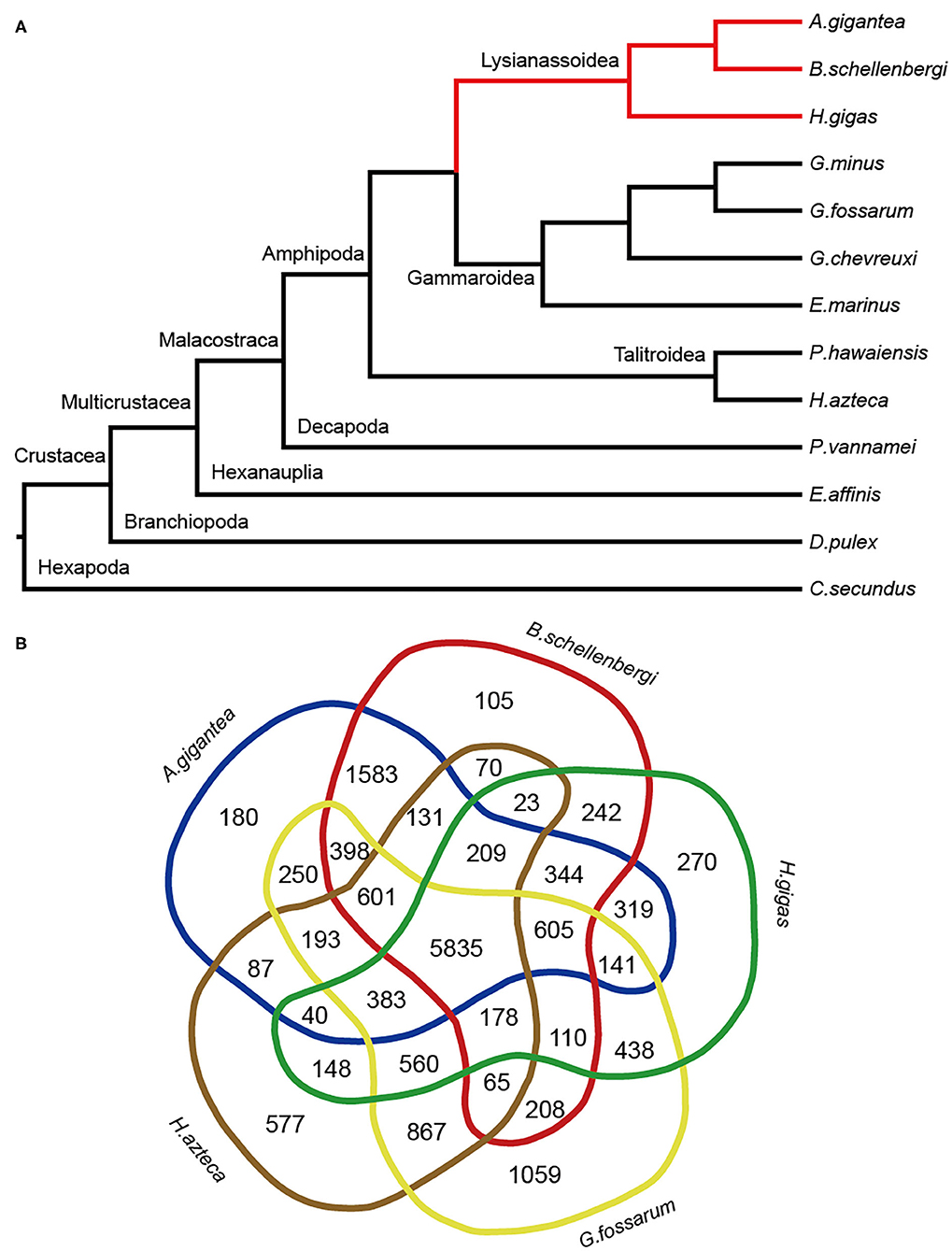
Figure 3. Evolution and homology analysis. (A) Phylogenetic analysis of A. gigantea, B. schellenbergi, and other arthropods. The hadal species were clustered in the clade Lysianassoidea. A. gigantea showed a close phylogenetic relationship with B. schellenbergi. All nodes received 100% bootstrap support. (B) Comparisons of the predictive genes between A. gigantea, B. schellenbergi, Hirondellea gigas, Hyalella Azteca, and Gammarus fossarum. A. gigantea and B. schellenbergi have more gene families in common.
When we compared the predicted genes of the five species, including three hadal amphipods (A. gigantea, B. schellenbergi, and H. gigas) and two shallow-water arthropods (G. fossarum and H. azteca), 5,835 gene families were shared by the five species (Figure 3B). Moreover, 1,583 gene families were particularly shared by A. gigantea and B. schellenbergi (Figure 3B).
Positive Selection Genes of the Hadal Amphipods
We detected PSGs using the branch-site models from CodeML and obtained 147 genes across the two hadal amphipod species. About 111 and 103 PSGs were identified in A. gigantea and B. schellenbergi, respectively. About 67 PSGs (45.6%) were shared between A. gigantea and B. schellenbergi (Figure 4A and Supplementary Tables 2, 3), which include carnitine O-palmitoyltransferase 2 (Cpt2), acylglycerol kinase, mitochondrial (AGK). Cpt2 is reported to be involved in lipid transport (Liu et al., 2018), and AGK, a mitochondrial membrane kinase, is reported to be involved in lipid and glycerolipid metabolism (Mårtensson and Becker, 2017).
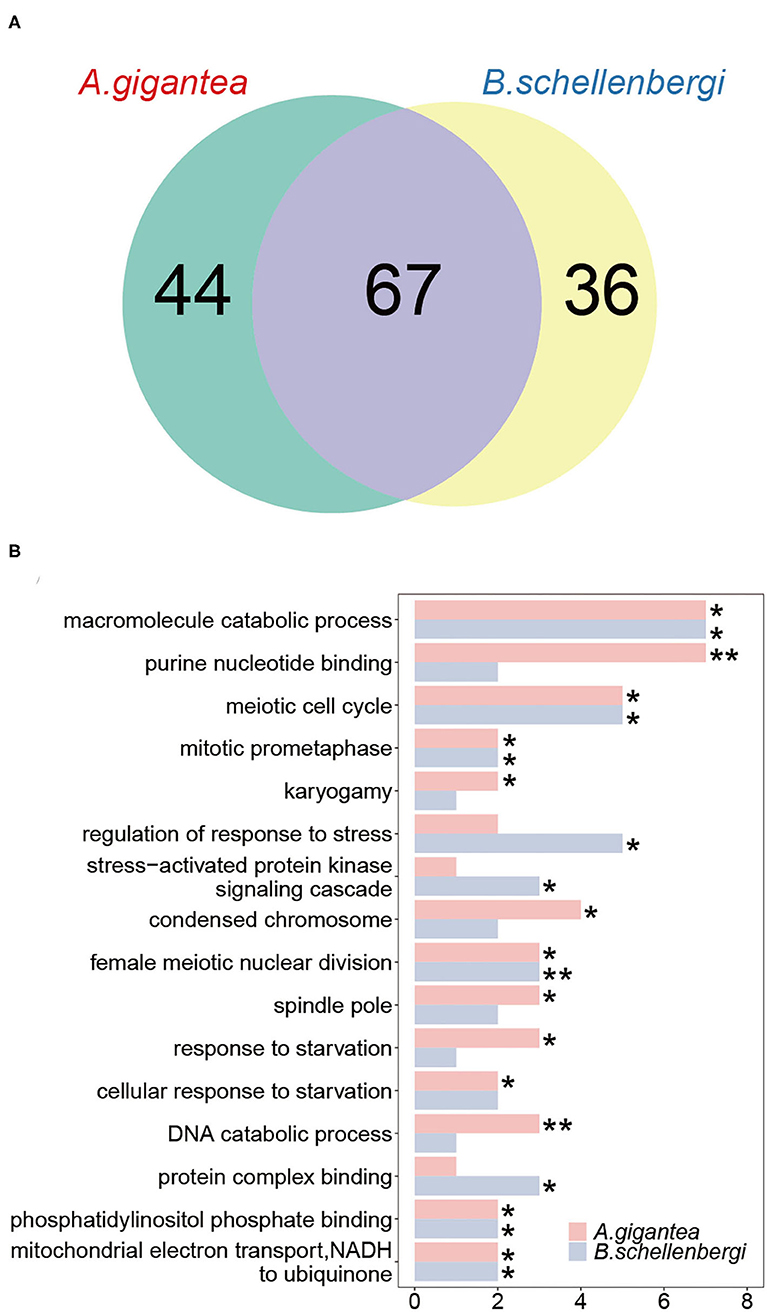
Figure 4. Gene ontology (GO) enrichment analysis of the positively selected genes (PSGs) in A. gigantea and B. schellenbergi. (A) The Venn diagram shows the numbers of the PSGs in A. gigantea and B. schellenbergi. The two hadal species have more shared PSGs (67) than they solely owned PSGs (44 for A. gigantea and 36 for B. schellenbergi). (B) GO enrichment results of PSGs in A. gigantea and B. schellenbergi. The abscissa indicates the number of annotated genes that were assigned to GO terms. These enriched genes were mainly related to “starvation response” and “meiosis.” Statistically significant enrichment results are indicated by asterisks (*indicates the value of p < 0.05, **indicates the value of p < 0.01).
About 20 PSGs in A. gigantea or B. schellenbergi were found to overlap with H. gigas using identical shallow-water species as the background. The shared PSGs included NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9 (NDUFA9), Leucine-rich pentatricopeptide repeat containing (LRPPRC), Microtubule-associated protein futsch (MAP1B), SprT-like N-terminal domain (SPRTN), Nucleolar protein 6 (NOL6), and Nucleolar pre-ribosomal-associated protein 1 (URB1), which indicated these hadal amphipods undergoing similar evolutionary pressures (Supplementary Figure 1 and Supplementary Tables 2–4).
As for the PSGs for both A. gigantea and B. schellenbergi, GO terms associated with “response to starvation,” “cellular response to starvation,” “mitochondrial electron transport, NADH to ubiquinone,” “meiotic cell cycle,” “spindle pole,” “karyogamy,” “condensed chromosome,” and “regulation of response to stress” were found to be shared by the two hadal amphipods (Figure 4B and Supplementary Tables 2, 3). Additionally, several relevant KEGG categories “glycerolipid metabolism,” “mitophagy-animal,” and “homologous recombination” were enriched in A. gigantea, whereas the KEGG categories “ECM-receptor interaction,” “glycerolipid metabolism,” “ribosome biogenesis in eukaryotes,” and “RNA transport” were found to be enriched in B. schellenbergi (Table 1).
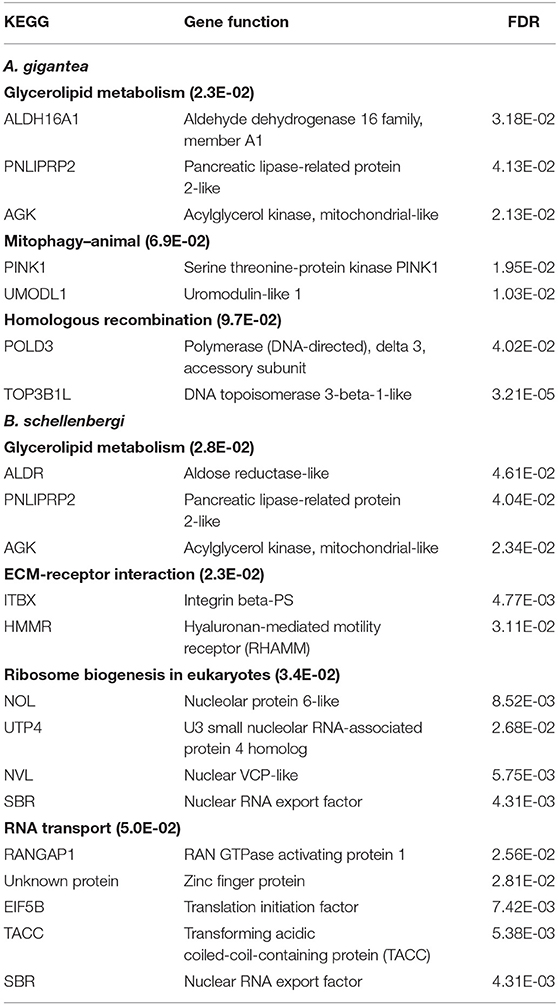
Table 1. KEGG enrichment analysis of the positively selected genes (PSGs) in the hadal amphipods Alicella gigantea and Bathycallisoma schellenbergi.
Positively Selected Genes of the “Supergiant” Amphipod
We further explored the PSGs in the “supergiant” amphipod within a total of 3,597 single-copy orthologs. About 58 genes were solely identified in the “supergiant” A. gigantea (value of p < 0.05; Supplementary Table 5), among which 14 PSG genes were shown with a false discovery rate (FDR <0.05; Table 2). Among these 14 critically PSGs (FDR <0.05; Table 2), we found 2 inositol-related genes, inositol-trisphosphate 3-kinase homolog (ITPK) and inositol monophosphatase 2 (IMPA2) (Figure 5A and Table 2), which are involved in the inositol biosynthetic process and inositol phosphate metabolism (Figure 5B). We also found that a gene-encoding rate-limiting enzyme, inosine-5′-monophosphate dehydrogenase 1 (IMPDH1) (Figure 5A and Table 2), which was involved in the de novo synthesis of guanine nucleotides and acts as a homotetramer to regulate cell growth (Slee and Bownes, 1995; Hossain et al., 2016), was under positive selection in A. gigantea (Figure 5A and Table 2).
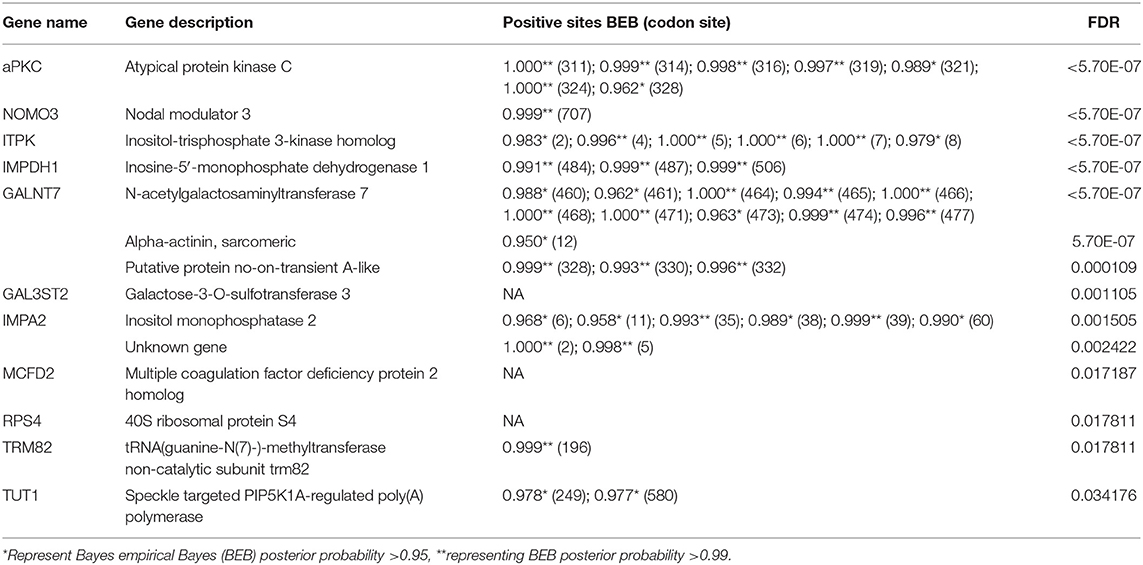
Table 2. A total of 14 PSGs in the supergiant amphipod A. gigantea was identified by a branch-site model in the CodeML Program.
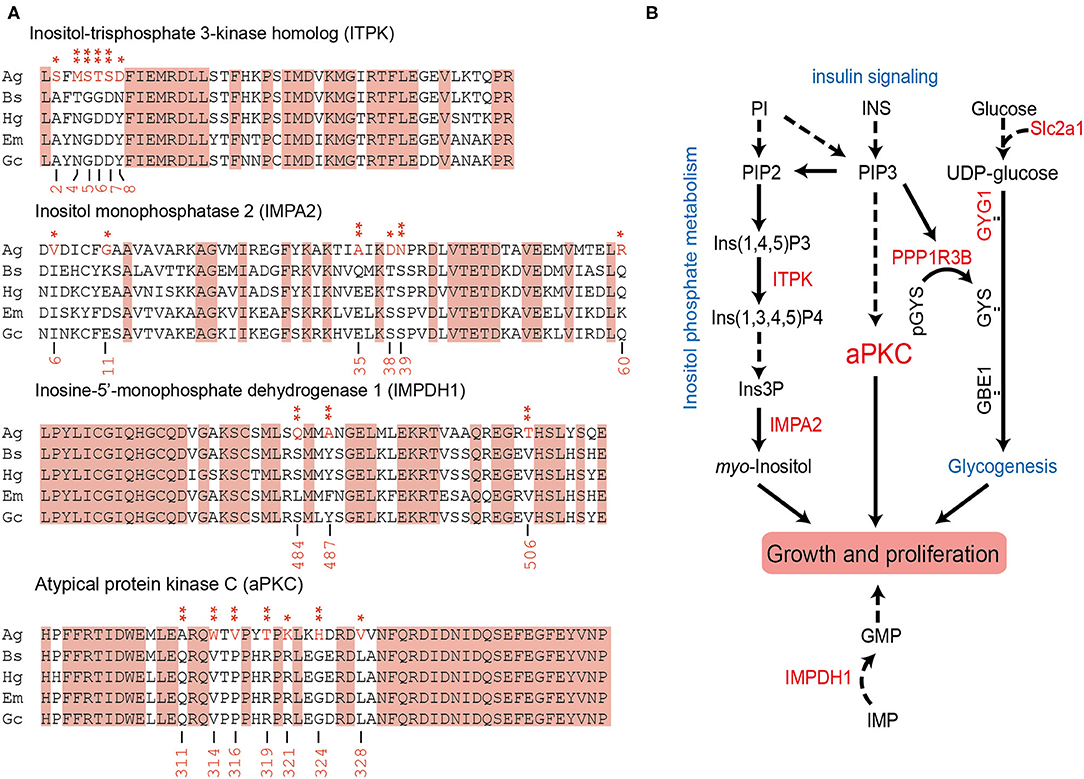
Figure 5. The possible signaling pathways involved by the PSGs solely identified in the “supergiant” A. gigantea. (A) The positively selected sites in the four PSGs (ITPK, IMPA2, IMPDH1, and aPKC) were solely identified in A. gigantea. To detect PSGs solely identified in the “supergiant” A. gigantea, we calculated by setting A. gigantea as the foreground and the other two small hadal amphipods (B. schellenbergi and H. gigas) as well as the two shallow-water amphipods (Gammarus chevreuxi and Echinogammarus marinus) as the background. The positively selected sites were marked in red asterisks. (B) The seven PSGs (ITPK, IMPA2, IMPDH1, aPKC, PPP1R3B, GYG1, and Slc2a1) were solely identified in A. gigantea were involved in inositol phosphate metabolism, insulin signaling, and glycogenesis signaling, which were related to growth and proliferation. PI, phosphatidylinositol; PIP2, phosphatidylinositol-4,5-bisphosphate; Ins(1,4,5)P3, inositol 1,4,5-trisphosphate; ITPK, inositol-triphosphate 3-kinase homolog; Ins(1,3,4,5)P4, inositol 1,3,4,5-tetrakisphosphate; Ins3P, inositol 3-monophosphate; IMPA2, inositol monophosphatase 2; INS, insulin; PIP3, phosphatidylinositol-3,4,5-trisphosphate; aPKC, atypical protein kinase C; PPP1R3B, protein phosphatase 1 regulatory subunit 3B; GYG1, glycogenin-1; GYS, glycogen synthase; GBE1, 1,4-alpha-glucan branching enzyme; Slc2a1, solute carrier family 2, facilitated glucose transporter member 1; IMPDH1, inosine-5′-monophosphatase dehydrogenase 1; IMP, inosine monophosphate; GMP, guanosine monophosphate.
Among the 14 PSGs solely identified in the “supergiant” A. gigantea, the most significant PSG (FDR <5.70E-07) is atypical protein kinase C (aPKC) (Figure 5A and Table 2), which encodes a member of the protein kinase C (PKC) family of serine/threonine protein kinases and plays an important role in the insulin signaling pathway (Figure 5B; Luna et al., 2006). Seven positively selected sites (sites 311, 314, 316, 319, 321, 324, and 328) were found in A. gigantea aPKC (Figure 5A and Table 2). In addition, protein phosphatase 1 regulatory subunit 3B (PPP1R3B, the value of p < 0.01), glycogenin-1 (GYG1), and Solute Carrier Family 2 (Slc2a1, facilitated glucose transporter member 1, the value of p < 0.01) were considered to be under positive selection (Supplementary Table 5). The three genes were reported to be involved in glycogenesis and glucose transport (Figure 5B; Zhao and Keating, 2007; Bilyard et al., 2018). Therefore, the PSGs solely identified in the “supergiant” A. gigantea were involved in inositol phosphate metabolism, insulin signaling, and glycogenesis signaling, which were ultimately related to growth and proliferation (Figure 5B).
Elevated dN/dS Ratios in the Lineages of Hadal Amphipods and the Supergiant Hadal Amphipod
To identify whether the GO categories were evolving faster in the hadal amphipods or the shallow-water amphipods, the dN/dS ratios of 3,380 single-copy orthologous genes among the four gammarideas (2 hadal species: A. gigantea and B. schellenbergi and two shallow-water species: E. marinus and G. fossarum) together with H. azteca (set as outgroup) were calculated and the mean dN/dS value of the genes associated with each GO term was calculated for each species. By comparing with E. marinus and G. fossarum, we screened for the GO categories that underwent rapid evolution in the hadals A. gigantea and B. schellenbergi. We identified 549 GO categories, which showed increased dN/dS ratios in the (A. gigantea or B. schellenbergi)/(E. marinus or G. fossarum) comparisons. The common GO categories included “regulation of response to stimulus,” “mitochondrial matrix,” “DNA repair,” “meiotic cell cycle,” “lipid biosynthetic process,” and “defense response,” indicating that the genes in these categories may be under higher evolutionary pressure than those in the shallow-water species (Figures 6A,B,D,E).
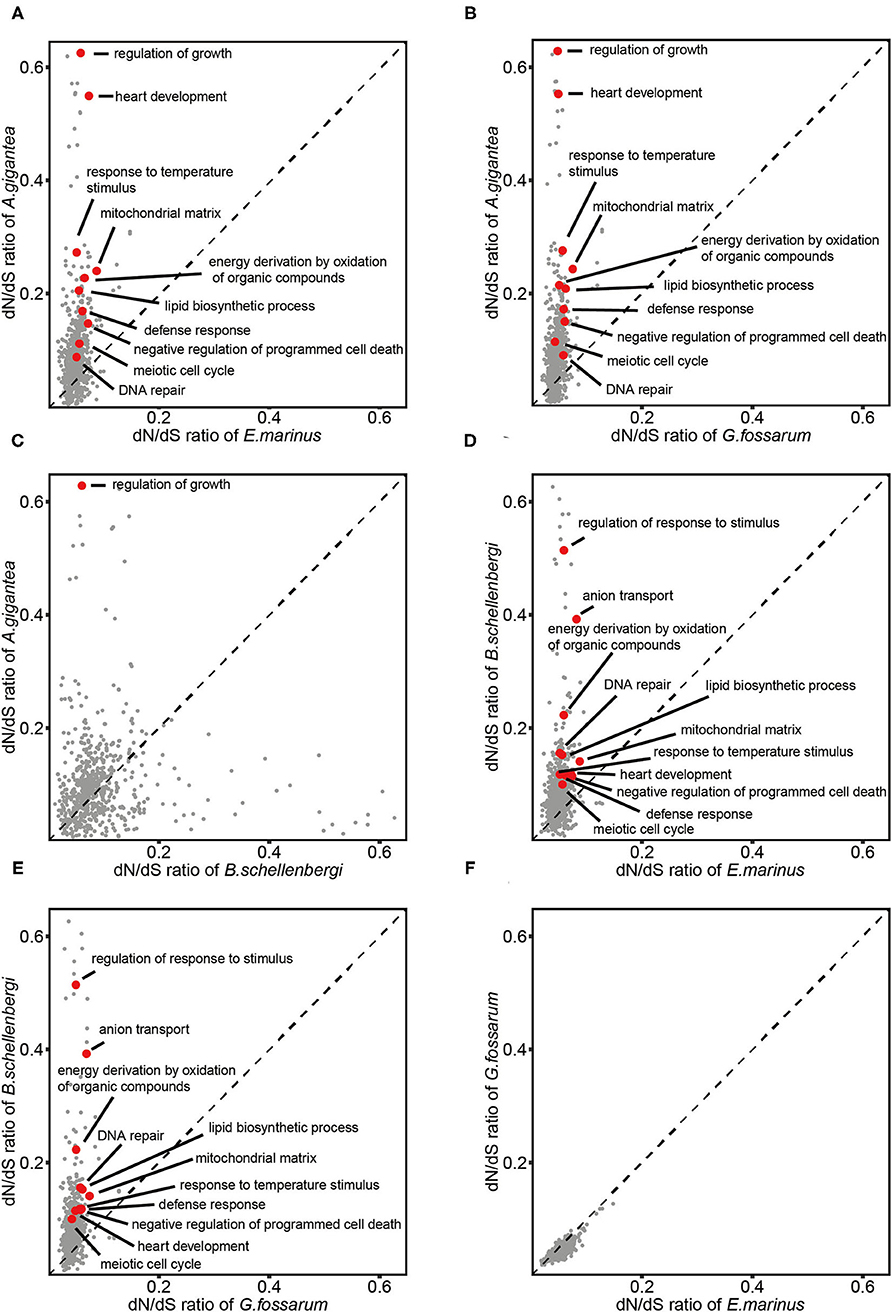
Figure 6. Comparisons of dN/dS of GO terms between the two hadal amphipods (A. gigantea and B. schellenbergi) and two shallow-water amphipods (G. fossarum and E. marinus). The evolution rates of A. gigantea (A–C) and B. schellenbergi (D,E) were much faster than those of G. fossarum and E. marinus (F) on the whole. Several representative GO terms including “regulation of response to stimulus,” “mitochondrial matrix,” “DNA repair,” “meiotic cell cycle,” “lipid biosynthetic process,” and “defense response” (A,B,D,E) were selected and marked in red. Compared with the small-sized amphipod species (B. schellenbergi, G. fossarum, and E. marinus), the dN/dS of “regulation of growth” in the “supergiant” A. gigantea was significantly higher (A–C). The comparison of shallow-water amphipods did not have these GO terms with significantly increased evolutionary rates (F).
To identify the GO categories that were evolving faster in the “supergiant” amphipods, by comparing with B. schellenbergi, E. marinus, and G. fossarum, we screened for the GO categories that underwent rapid evolution in A. gigantea (Figures 6A–C). The common GO category is “regulation of growth,” suggesting that the genes involved in the “regulation of growth” were evolving faster in the “supergiant” hadal amphipods than in other amphipods.
Discussion
Hadal zone is an extreme environment and is also the least known area. How the organisms endemic to hadal zones adapted to an extreme environment attract a wide interest of several researchers. Due to technical limitations, previous studies were conducted mainly at biochemical levels, such as the studies on the osmotic factors, lipids, or proteins (Yancey, 2020), and there are very few studies regarding the adaptive mechanisms of the hadal species at the genetic level. With the development of the next-generation sequencing technology, transcriptome studies of many species have been conducted. However, due to the restriction by the difficulties of the hadal species samplings, only one hadal amphipod species, H. gigas, has been analyzed by using transcriptome sequencing (Lan et al., 2017). In this study, the two hadal amphipods, A. gigantea and B. schellenbergi, were selected for transcriptome sequencing and comparative evolutionary analysis. Compared to the published H. gigas transcriptomes (Lan et al., 2017), similarly predicted gene number and much more contigs were obtained from the de novo assembly for A. gigantea and B. schellenbergi (Supplementary Table 1), which probably indicated comparably higher incomplete contigs produced in this study. The higher contig number obtained in our assembly might result from the biological variability (Smith-Unna et al., 2016). This study not only would improve the adaptation biology studies of hadal amphipods from the perspective of phylogeny but also could explore the underlying reasons for the gigantism of A. gigantea.
According to the phylogenetic tree constructed from the orthologous genes of the hadal and shallow-water arthropods, we found that A. gigantea, B. schellenbergi, and H. gigas all belong to the lysianassoidea clades (Figure 3A). The result is consistent with a recent study on a large-scale molecular phylogeny of amphipoda, in which ecologically diverse deep-sea species could be gathered together in a clade of lysianassoids (Copilaş-Ciocianu et al., 2020). It should be noticed that A. gigantea and B. schellenbergi have a close distance in the phylogenetic tree (Figure 3A) and also shared the more common gene families (Figure 3B), which indicated that A. gigantea and B. schellenbergi have a close relationship. However, it should be noticed that the number of single-copy orthologous genes used in the phylogeny tree construction was only 512, thus our results could not fully determine the species differentiation time among the three hadal amphipods, A. gigantea, B. schellenbergi, and H. gigas. However, this result could also provide the reference data for species phylogeny relationship identification studies.
A large-scale phylogeny study of amphipoda also indicates a close relationship between the deep-sea lysianassoids and shallow-water gammaroids (Copilaş-Ciocianu et al., 2020). Thus, the hadal species belonging to Lysianassoidea could be compared against shallow-water species belonging to Gammaroidea for a positive selection analysis. Among the taxa of shallow-water marine amphipods, which possess the available transcriptome data of Melita plumulosa (Hadzioids) (Hook et al., 2014), P. hawaiensis (Talitroids) (Kao et al., 2016), Grandidierella japonica (Corophioids) (Hiki et al., 2019), Gondogeneia Antarctica (Kang et al., 2015), E. marinus (Cogne et al., 2019), and Eogammarus possjeticus (Chen et al., 2019), three species (G. antarctica, E. marinus, and E. possjeticus) belong to the Gammaroidea taxa, which could be chosen as the reference species for a positive selection analysis. G. antarctica, inhabiting another extreme environment (the Antarctic Pole) evolved despite a constant cold environment (Kang et al., 2015), which might disturb our evolutionary analysis. E. possjeticus was widely distributed in the coastal and estuarine areas, and the transcriptome data of E. possjeticus were released in 2019 (Chen et al., 2019). However, muscle tissues were used for RNA sequencing (RNA-seq) in that study. Therefore, we finally chose the four Gammaroidean species, including a shallow-water marine species (E. marinus) and three freshwater species (G. minus, G. fossarum, and G. chevreuxi), as the reference species for evolutionary analysis.
The PSG detection results of A. gigantea and B. schellenbergi showed that the number of PSGs shared by the two hadal amphipods exceeded the number of unique PSGs possessed by each species (Figure 4A), which further indicated a close genetic relationship between the two species. The PSGs of A. gigantea and B. schellenbergi were enriched by the GO and KEGG analysis, and it was found that they were mainly related to “starvation response” and “meiosis” (Figure 4B and Table 1). Hadal is an environment with limited food supplies, and most of the falling organic matter will be decomposed by bacteria and consumed by animals (Jamieson et al., 2010). However, amphipods can survive long periods of starvation (Jamieson, 2015). In the transcriptome analysis of the hadal amphipod H. gigas, researchers found that the key genes directly involved in the “energy metabolism” pathway were positively selected, which were suggested to be a new genetic adaptation strategy for H. gigas to survive in the limited food supply environment (Lan et al., 2017). In our study, we found that the PSGs in the KEGG pathway “glycerolipid metabolism” were enriched both in A. gigantea and in B. schellenbergi (Table 1). Such a result is consistent with the findings of Lan et al. (2017). Meiosis is a biological process that depends on the cytoskeleton (Bourns et al., 1988), and high hydrostatic pressures in the hadal environment will affect the cytoskeleton, thus affecting meiosis (Ishii et al., 2004). The PSGs involved in the “meiosis” pathway would be related to the environmental adaptations of high hydrostatic pressure.
As mentioned earlier, A. gigantea and B. schellenbergi have a closer genetic relationship compared with another hadal amphipod, H. gigas (Figure 3A). However, their body sizes are greatly different from the H. gigas body size (Figure 1B). As shown in Figure 1C, the growth fitting curves of both A. gigantea and B. schellenbergi are linear, but the slope of A. gigantea is much steeper than that of B. schellenbergi. It is well-known that the body size and weight fitting curve of crustaceans are the common references for growth and development studies. Researchers studied the correlations between the body size and weight of a shallow-water crustacean, Pederson cleaner shrimp (Ancylomenes pedersoni), and found that the precisely measured total length increased linearly with the carapace length while the wet mass increased exponentially with the carapace length (Gilpin and Chadwick, 2017). This is not consistent with the linear body size and weight fitting curve of our hadal amphipods, which might be due to a harsh hadal environment. With the shortage of food supplies, the wet mass of hadal amphipods could not increase exponentially with respect to their body length. However, the growth fitting slope curve of A. gigantea is much steeper than that of the B. schellenbergi, indeed suggesting that, with the same body length, A. gigantea might gain more weight and grow much faster than B. schellenbergi.
In combination with the PSG detection using A. gigantea as the foreground and other small-sized amphipods as the background, the results showed that 58 genes were considered to be under positive selection in A. gigantea (value of p < 0.05; Supplementary Table 5) (14 PSGs with FDR <0.05, see Table 2). Two PSGs (ITPK and IMPA2) are involved in the Inositol phosphate metabolism pathway [process from phosphatidylinositol (PI) to myo-inositol], where ITPK converts inositol 1,4,5-trisphosphate [Ins (1,4,5) P3] to inositol 1,3,4,5-tetrakisphosphate [Ins (1,3,4,5) P4] (Nalaskowski et al., 2003), and IMPA2 converts inositol 3-phosphate (Ins3P) to myo-inositol (Figure 5B; McAllister et al., 1992). It is well-known that myo-inositol is often added to animal feeds to meet the metabolic needs of animals (Wang et al., 2020). Moreover, IMPDH1 (PSG as shown in Figure 5A and Table 2) is involved in nucleotide metabolism by converting IMP to XMP (Slee and Bownes, 1995). XMP can be further converted to GMP (Shivakumaraswamy et al., 2020), which can promote the growth performance of organisms (Hossain et al., 2016). Therefore, we can conclude that the two PSGs (ITPK and IMPA2) are involved in the inositol phosphate metabolism pathway, and together with IMPDH1 would ultimately be related to growth and proliferations in A. gigantea.
On the other hand, four PSGs (aPKc, PPP1R3B, GYG1, and Slc2a1), which are associated with insulin signaling and involved in the glycogenesis pathway, were identified in A. gigantea (Figure 5B). PPP1R3B, a key factor, connects the insulin signaling and glycogenesis signaling pathway (Luo et al., 2011). Slc2a1, an important factor, plays a role in glucose transport (Zhao and Keating, 2007), and together with GYG1 was involved in the glycogenesis process. Therefore, the PSGs involved in the glycogenesis suggested that A. gigantea might undergo active energy metabolism, which could help the “supergiant” to survive in the harsh hadal environment with scarce food sources. Similar to the previous studies on H. gigas (Lan et al., 2017), these PSGs involved in the “energy metabolism” pathway might be also related to starvation resistance in the hadal amphipods. In addition, researchers also found that larger animals showed better resistance to starvation compared to smaller animals (Cushman et al., 1993; Arnett and Gotelli, 2003). The insulin production-related PSGs shown in larger-sized A. gigantea (Figure 5) possibly explain the reason for larger animals possessing a better resistance to starvation.
To explore the gigantism of A. gigantea, aPKC, the most significant PSG (FDR <5.70E-07) identified in the “supergiant” A. gigantea (Figure 5A and Table 2), invokes our attentions. APKC, which encodes a member of the PKC family of serine/threonine protein kinases, was reported to affect insulin regulation (Zhao et al., 2017). PRKCI, an aPKC isoform (PRKC iota), was reported to be related to the gigantism of capybara (Hydrochoerus hydrochaeris), the world's largest living rodent (Herrera-Álvarez et al., 2021) as it is involved in cell survival, differentiation, and proliferation by accelerating G1/S transition (Ni et al., 2016). Therefore, we could expect that the most significant PSG, aPKC identified in A. gigantea might ultimately be related to cell proliferation and growth of the “supergiant” amphipod.
From the evolutionary rate comparisons between the hadal and shallow-water amphipods, it was clearly shown that the evolutionary rates of GO categories, such as “lipid synthesis,” “meiosis,” and “DNA repair,” increased in the hadal amphipods (Figures 6A,B,D,E). However, the evolutionary rates of “lipid synthesis” and “meiosis” are consistent with the abovementioned PSG enrichment results. The hadal environment has an extremely high hydrostatic pressure, which can lead to DNA damage (Abe et al., 1999; Rothschild and Mancinelli, 2001; Aertsen et al., 2004). Therefore, researchers suggested that hadal organisms might require a high frequency of DNA repair (Dixon et al., 2004), which is also consistent with our results. Notably, the most significantly enriched GO category in A. gigantea was “regulation of growth,” (Figure 6C) which indicated that size control or growth regulation mechanisms might hide under the growth of the “supergiant” amphipod, and this could explain why A. gigantea is so huge. In mammals, growth is regulated by growth hormones, excessive growth hormone secretion can cause gigantism (Lodish et al., 2016), and possibly, the existence of a similar regulatory mechanism in the “supergiant” amphipods.
Conclusion
In this research, a comparative evolutionary study regarding two different-sized hadal amphipods, A. gigantea and B. schellenbergi, was conducted. Many PSGs involved in “glycerolipid metabolism,” “response to starvation,” and “meiosis” were found in the two hadal species, suggesting that these pathways might be the most important adaptation mechanisms for the hadal creatures. Moreover, seven PSGs (especially the most significant PSG, aPKC) solely identified in the A. gigantea showed to be related to inositol phosphate metabolism, insulin signaling, and glycogenesis signaling. Together, the evolutionary rate of the GO term “growth regulation” was significantly higher in the “supergiant” A. gigantea than in B. schellenbergi and other small-sized amphipods. These points might be the possible gigantism mechanisms of A. gigantea.
Data Availability Statement
All sequencing data associated with this project were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database [BioProject Accession Numbers: PRJNA739006 (Alicella gigantea) and PRJNA739007 (Bathycallisoma schellenbergi)].
Author Contributions
QX conceived the experiments, led the whole project, and contributed to edits to the manuscript. WL and FW analyzed the data. BP designed the lander vehicle for sample collection. JC and BP collected the samples. JC extracted the RNA. SJ performed RNA-seq. WL, FW, and QX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Funding Project of the National Key Research and Development Program of China (2018YFC0310600), the National Key Research and Development Program of China (2018YFD0900601), the National Natural Science Foundation of China (Grant No. 31772826), and the Major Scientific Innovation Project from Shanghai Committee of Education (2017-01-07-00-10-E00060).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Shanghai Rainbowfish Ocean Technology Co., Ltd. for the sample collection. We also thank the research group members of Prof. Weicheng Cui and Prof. Jiasong Fang and other people for sample collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.743663/full#supplementary-material
Supplementary Figure 1. The Venn diagram shows the numbers of the positively selected genes (PSGs) in the three hadal amphipods, Alicella gigantea, Bathycallisoma Schellenbergi, and Hirondellea gigas.
Supplementary Table 1. Summary of the transcriptome sequencing results.
Supplementary Table 2. A list of positively selected genes (PSGs) of the hadal amphipod Alicella gigantea.
Supplementary Table 3. A list of the PSGs of the hadal amphipod Bathycallisoma schellenbergi.
Supplementary Table 4. A list of the PSGs of the hadal amphipod Hirondellea gigas.
Supplementary Table 5. The identification of 58 PSGs in the “supergiant” A. gigantea.
References
Abe, F., Kato, C., and Horikoshi, K. (1999). Pressure-regulated metabolism in microorganisms. Trends Microbiol. 7, 447–453. doi: 10.1016/s0966-842x(99)01608-x
Aertsen, A., Van Houdt, R., Vanoirbeek, K., and Michiels, C. W. (2004). An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 186, 6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arnett, A. E., and Gotelli, N. J. (2003). Bergmann's rule in larval ant lions: testing the starvation resistance hypothesis. Ecol. Entomol. 28, 645–650. doi: 10.1111/j.1365-2311.2003.00554.x
Barnard, J. L., and Ingram, C. L. (1986). The supergiant amphipod, Alicella gigantea chevreux from the north pacific gyre. J. Crustacean Biol. 6, 825–839. doi: 10.1163/193724086X00613
Bartlett, D. H. (2002). Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595, 367–381. doi: 10.1016/s0167-4838(01)00357-0
Bilyard, M. K., Bailey, H. J., Raich, L., Gafitescu, M. A., Machida, T., Iglésias-Fernández, J., et al. (2018). Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis. Nature 563, 235–240. doi: 10.1038/s41586-018-0644-7
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bourns, B., Franklin, S., Cassimeris, L., and Salmon, E. D. (1988). High hydrostatic pressure effects in vivo: changes in cell morphology, microtubule assembly, and actin organization. Cell Motil. Cytoskel. 10, 380–390. doi: 10.1002/cm.970100305
Britton, J. C., and Morton, B. (1994). Marine carrion and scavengers. Oceanogr. Mar. Biol. Ann. Rev. 32, 369–434.
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Carlini, D. B., and Fong, D. W. (2017). The transcriptomes of cave and surface populations of Gammarus minus (Crustacea: Amphipoda) provide evidence for positive selection on cave downregulated transcripts. PLoS ONE 12:e0186173. doi: 10.1371/journal.pone.0186173
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chan, J., Geng, D., Pan, B., Zhang, Q., and Xu, Q. (2021). Metagenomic insights into the Sructure and function of intestinal microbiota of the hadal amphipods. Front. Microbiol. 12:668989. doi: 10.3389/fmicb.2021.668989
Chan, J., Pan, B., Geng, D., Zhang, Q., Zhang, S., Guo, J., et al. (2020). Genetic diversity and population structure analysis of three deep-sea amphipod species from geographically isolated hadal trenches in the Pacific Ocean. Biochem. Genet. 58, 157–170. doi: 10.1007/s10528-019-09935-z
Chapelle, G., and Peck, L. S. (2004). Amphipod crustacean size spectra: new insights in the relationship between size and oxygen. Oikos 106, 167–175. doi: 10.1111/j.0030-1299.2004.12934.x
Chen, J., Liu, H., Cai, S., and Zhang, H. B. (2019). Comparative transcriptome analysis of Eogammarus possjeticus at diferent hydrostatic pressure and temperature exposures. Sci Rep. 9:3456. doi: 10.1038/s41598-019-39716-y
Cogne, Y., Degli-Esposti, D., Pible, O., Gouveia, D., François, A., Bouchez, O., et al. (2019). De novo transcriptomes of 14 gammarid individuals for proteogenomic analysis of seven taxonomic groups. Sci. Data. 6:184. doi: 10.1038/s41597-019-0192-5
Colbourne, J. K., Pfrender, M. E., Gilbert, D., Thomas, W. K., Tucker, A., Oakley, T. H., et al. (2011). The ecoresponsive genome of Daphnia pulex. Science 331, 555–561. doi: 10.1126/science.1197761
Collins, M., Tills, O., Spicer, J. I., and Truebano, M. (2017). De novo transcriptome assembly of the amphipod Gammarus chevreuxi exposed to chronic hypoxia. Mar. Genom. 33, 17–19. doi: 10.1016/j.margen.2017.01.006
Copilaş-Ciocianu, D., Borko, Š., and Fišer, C. (2020). The late blooming amphipods: global change promoted post-Jurassic ecological radiation despite Palaeozoic origin. Mol. Phylogenet. Evol. 143:106664. doi: 10.1016/j.ympev.2019.106664
Cossins, A. R., and Macdonald, A. G. (1984). Homeoviscous theory under pressure: II. The molecular order of membranes from deep-sea fish. Biochim. Biophys. Acta Biomembr. 776, 144–150. doi: 10.1016/0005-2736(84)90260-8
Cossins, A. R., and Macdonald, A. G. (1989). The adaptations of biological membranes to temperature and pressure: fish from the deep and cold. J. Bioenerg. Biomembr. 21, 115–135. doi: 10.1007/BF00762215
Cushman, J. H., Lawton, J. H., and Manly, B. F. (1993). Latitudinal patterns in European ant assemblages: variation in species richness and body size. Oecologia 95, 30–37. doi: 10.1007/BF00649503
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
David, B., Carlini, D. B., Satish, S., and Fong, D. W. (2013). Parallel reduction in expression, but no loss of functional constraint, in two opsin paralogs within cave populations of Gammarus minus (Crustacea: Amphipoda). BMC Evol. Biol. 13:89. doi: 10.1186/1471-2148-13-89
De Broyer, C., and Thurston, M. H. (1987). New Atlantic material and redescription of the type specimens of the giant abyssal amphipod Alicella gigantea Chevreux (Crustacea). Zool. Scr. 16, 335–350. doi: 10.1111/j.1463-6409.1987.tb00079.x
Dixon, D. R., Pruski, A. M., and Dixon, L. R. (2004). The effects of hydrostatic pressure change on DNA integrity in the hydrothermal-vent mussel Bathymodiolus azoricus: implications for future deep-sea mutagenicity studies. Mutat. Res. 552, 235–246. doi: 10.1016/j.mrfmmm.2004.06.026
Downing, A. B., Wallace, G. T., and Yancey, P. H. (2018). Organic osmolytes of amphipods from littoral to hadal zones: Increases with depth in trimethylamine N-oxide, scyllo-inositol and other potential pressure counteractants. Deep Sea Res. Part I 138, 1–10. doi: 10.1016/j.dsr.2018.05.008
Dungan, S. Z., Kosyakov, A., and Chang, B. S. (2016). Spectral tuning of killer whale (Orcinus orca) rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323–336. doi: 10.1093/molbev/msv217
Eustace, R. M., Kilgallen, N. M., Ritchie, H., Piertney, S. B., and Jamieson, A. J. (2016). Morphological and ontogenetic stratification of abyssal and hadal Eurythenes gryllus (Amphipoda: lysianassidae) from the Peru-Chile Trench. Deep-Sea Res. Part I 109, 91–98. doi: 10.1016/j.dsr.2015.11.005
Eyun, S. I., Soh, H. Y., Posavi, M., Munro, J. B., Hughes, D. S. T., Murali, S. C., et al. (2017). Evolutionary history of chemosensory-related gene families across the arthropoda. Mol. Biol. Evol. 34, 1838–1862. doi: 10.1093/molbev/msx147
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gilpin, J. A., and Chadwick, N. E. (2017). Life-history traits and population structure of pederson cleaner shrimps Ancylomenes pedersoni. Biol. Bull. 233, 190–205. doi: 10.1086/695802
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Harrison, C. S., Hida, T. S., and Seki, M. P. (1983). Hawaiin seabird feeding ecology. Wildlife Monogr. 85, 1–71.
Harrison, M. C., Jongepier, E., Robertson, H. M., Arning, N., Bitard-Feildel, T., Chao, H., et al. (2018). Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2, 557–566. doi: 10.1038/s41559-017-0459-1
Hasegawa, M., Kurohiji, Y., Takayanagi, S., Sawadaishi, S., and Yao, M. (1986). Collection of fish and amphipoda from abyssal sea-floor at 30°N-147°E using traps tied to 10000 m wire of research vessel. Bull. Tokai Reg. Fish. Res. Lab. 119, 65–75.
Herrera-Álvarez, S., Karlsson, E., Ryder, O. A., Lindblad-Toh, K., and Crawford, A. J. (2021). How to make a rodent giant: genomic basis and tradeoffs of gigantism in the capybara, the world's largest rodent. Mol. Biol. Evol. 38, 1715–1730. doi: 10.1093/molbev/msaa285
Hiki, K., Nakajima, F., Tobino, T., Watanabe, H., and Yamamoto, H. (2019). Whole transcriptome analysis of an estuarine amphipod exposed to highway road dust. Sci. Total Environ. 675, 141–150. doi: 10.1016/j.scitotenv.2019.04.201
Hook, S. E., Twine, N. A., Simpson, S. L., Spadaro, D. A., Moncuquet, P., and Wilkins, M. R. (2014). 454 pyrosequencing-based analysis of gene expression profiles in the amphipod Melita plumulosa transcriptome assembly and toxicant induced changes. Aquat. Toxicol. 153, 73–88. doi: 10.1016/j.aquatox.2013.11.022
Hossain, M. S., Koshio, S., Ishikawa, M., Yokoyama, S., and Sony, N. M. (2016). Effects of dietary administration of guanosine monophosphate on the growth, digestibility, innate immune responses and stress resistance of juvenile red sea bream, Pagrus major. Fish Shellfish Immun. 57, 96–106. doi: 10.1016/j.fsi.2016.08.026
Ishii, A., Sato, T., Wachi, M., Nagai, K., and Kato, C. (2004). Effects of high hydrostatic pressure on bacterial cytoskeleton FtsZ polymers in vivo and in vitro. Microbiology 150, 1965–1972. doi: 10.1099/mic.0.26962-0
Jamieson, A. J. (2015). The Hadal Zone: Life in the Deepest Oceans. Cambridge: Cambridge University Press.
Jamieson, A. J., Fujii, T., Mayor, D. J., Solan, M., and Priede, I. G. (2010). Hadal trenches: the ecology of the deepest places on Earth. Trends Ecol. Evol. 25, 190–197. doi: 10.1016/j.tree.2009.09.009
Jamieson, A. J., Lacey, N. C., Lörz, A., Rowden, A. A., and Piertney, S. B. (2013). The supergiant amphipod Alicella gigantea (Crustacea: Alicellidae) from hadal depths in the Kermadec Trench, SW Pacific Ocean. Deep-Sea Res. Part II 92, 107–113. doi: 10.1016/j.dsr2.2012.12.002
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. doi: 10.1093/nar/gkm796
Kaiser, M. J., and Moore, P. G. (1999). Obligate marine scavengers: do they exist? J. Nat. Hist. 33, 475–481. doi: 10.1080/002229399300191
Kang, S., Kim, S., and Park, H. (2015). Transcriptome of the Antarctic amphipod Gondogeneia antarctica and its response to pollutant exposure. Mar Genomics 24, 253–254. doi: 10.1016/j.margen.2015.07.012
Kao, D., Lai, A. G., Stamataki, E., Rosic, S., Konstantinides, N., Jarvis, E., et al. (2016). The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. Elife 5:e20062. doi: 10.7554/eLife.20062
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Keilwagen, J., Hartung, F., Paulini, M., Twardziok, S. O., and Grau, J. (2018). Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics 19:189. doi: 10.1186/s12859-018-2203-5
Kobayashi, H., Hatad, Y., Tsubouchi, T., Nagahama, T., and Takami, H. (2012). The hadal amphipod Hirondellea gigas possessing a unique cellulase for digesting wooden debris buried in the deepest seafloor. PLoS ONE 7:e42727. doi: 10.1371/journal.pone.0042727
Kobayashi, H., Shimoshige, H., Nakajima, Y., Arai, W., and Takami, H. (2019). An aluminum shield enables the amphipod Hirondellea gigas to inhabit deep-sea environments. PLoS ONE 14:e0206710. doi: 10.1371/journal.pone.0206710
Lacey, N. C., Jamieson, A. J., and Soreide, F. (2013). Successful capture of ultradeep sea animals from the Puerto Rico Trench. Sea Technol. 54, 19–21. doi: 10.2307/3548406
Lacey, N. C., Rowden, A. A., Clark, M. R., Kilgallen, N. M., Linley, T., Mayor, D. J., et al. (2016). Community structure and diversity of scavenging amphipods from bathyal to hadal depths in three South Pacific trenches. Deep Sea Res. Part I 111, 121–137. doi: 10.1016/j.dsr.2016.02.014
Lan, Y., Sun, J, Tian, R., Bartlett, D. H., Li, R., Wong, Y. H., et al. (2017). Molecular adaptation in the world's deepest-living animal: Insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Mol. Evol. 26, 3732–3743. doi: 10.1111/mec.14149
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Liu, L., Long, X., Deng, D., Cheng, Y., and Wu, X. (2018). Molecular characterization and tissue distribution of carnitine palmitoyltransferases in Chinese mitten crab Eriocheir sinensis and the effect of dietary fish oil replacement on their expression in the hepatopancreas. PLoS ONE 13:e0201324. doi: 10.1371/journal.pone.0201324
Lodish, M. B., Trivellin, G., and Stratakis, C. A. (2016). Pituitary gigantism: update on molecular biology and management. Curr. Opin. Endocrinol. Diabetes Obes. 23, 72–80. doi: 10.1097/MED.0000000000000212
Luna, V., Casauban, L., Sajan, M. P., Gomez-Daspet, J., Powe, J. L., Miura, A., et al. (2006). Metformin improves atypical protein kinase C activation by insulin and phosphatidylinositol-3,4,5-(PO4)3 in muscle of diabetic subjects. Diabetologia 49, 375–382. doi: 10.1007/s00125-005-0112-4
Luo, X., Zhang, Y., Ruan, X., Jiang, X., Zhu, L., Wang, X., et al. (2011). Fasting-induced protein phosphatase 1 regulatory subunit contributes to postprandial blood glucose homeostasis via regulation of hepatic glycogenesis. Diabetes 60, 1435–1445. doi: 10.2337/db10-1663
Mårtensson, C. U., and Becker, T. (2017). Acylglycerol kinase: mitochondrial protein transport meets lipid biosynthesis. Trends Cell Biol. 27, 700–702. doi: 10.1016/j.tcb.2017.08.006
McAllister, G., Whiting, P., Hammond, E. A., Knowles, M. R., Atack, J. R., Bailey, F. J., et al. (1992). cDNA cloning of human and rat brain myo-inositol monophosphatase. Expression and characterization of the human recombinant enzyme. Biochem. J. 284, 749–754. doi: 10.1042/bj2840749
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Nalaskowski, M. M., Bertsch, U., Fanick, W., Stockebrand, M. C., Schmale, H., and Mayr, G. W. (2003). Rat inositol 1,4,5-trisphosphate 3-kinase C is enzymatically specialized for basal cellular inositol trisphosphate phosphorylation and shuttles actively between nucleus and cytoplasm. J. Biol. Chem. 278, 19765–19776. doi: 10.1074/jbc.M211059200
Ni, S., Chen, L., Li, M., Zhao, W., Shan, X., Wu, M., et al. (2016). PKC iota promotes cellular proliferation by accelerated G1/S transition via interaction with CDK7 in esophageal squamous cell carcinoma. Tumor Biol. 37, 13799–13809. doi: 10.1007/s13277-016-5193-9
Nunoura, T., Takaki, Y., Hirai, M., Shimamura, S., Makabe, A., Koide, O., et al. (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. U.S.A. 112, E1230–E1236. doi: 10.1073/pnas.1421816112
Poynton, H. C., Hasenbein, S., Benoit, J. B., Sepulveda, M. S., Poelchau, M. F., Hughes, D. S. T., et al. (2018). The toxicogenome of Hyalella azteca: a model for sediment ecotoxicology and evolutionary toxicology. Environ. Sci. Technol. 52, 6009–6022. doi: 10.1021/acs.est.8b00837
Ritchie, H., Jamieson, A. J., and Piertney, S. B. (2017). Genome size variation in deep-sea amphipods. R. Soc. Open Sci. 4:170862. doi: 10.1098/rsos.170862
Rothschild, L. J., and Mancinelli, R. L. (2001). Life in extreme environments. Nature 409, 1092–1101. doi: 10.1038/35059215
Roux, J., Privman, E., Moretti, S., Daub, J. T., Robinson-Rechavi, M., and Keller, L. (2014). Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 31, 1661–1685. doi: 10.1093/molbev/msu141
Sainte-Marie, B. (1992). “Foraging of scavenging deep-sea lysianassoid amphipods,” in Deep-Sea Food Chains and the Global Carbon Cycle, eds G. T. Rowe and V. Pariente (Dordrecht: Kluwer Academic), 105–124.
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shivakumaraswamy, S, Pandey, N., Ballut, L., Violot, S., Aghajari, N., and Balaram, H. (2020). Helices on interdomain interface couple catalysis in the ATPPase domain with allostery in plasmodium falciparum GMP synthetase. Chembiochem 21, 2805–2817. doi: 10.1002/cbic.202000158
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Slee, R., and Bownes, M. (1995). The raspberry locus encodes Drosophila inosine monophosphate dehydrogenase. Mol. Gen. Genet. 248, 755–766. doi: 10.1007/BF02191716
Smith, K. L. Jr., and Baldwin, R. J. (1982). Scavenging deep-sea amphipods: effects of food odor on oxygen consumption and a proposed metabolic strategy. Mar. Biol. 68, 287–298. doi: 10.1007/BF00409595
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Somero, G. N. (1992). Adaptations to high hydrostatic pressure. Annu. Rev. Physiol. 54, 557–577. doi: 10.1146/annurev.ph.54.030192.003013
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Straub, S., Hirsch, P. E., and Burkhardt-Holm, P. (2017). Biodegradable and petroleum-based microplastics do not differ in their ingestion and excretion but in their biological effects in a freshwater invertebrate Gammarus fossarum. Int. J. Environ. Res. Public Health 14:774. doi: 10.3390/ijerph14070774
Tarn, J., Peoples, L. M., Hardy, K., Cameron, J., and Bartlett, D. H. (2016). Identification of free-living and particle-associated microbial communities. Front. Microbiol. 7:665. doi: 10.3389/fmicb.2016.00665
Timofeev, S. F. (2001). Bergmann's principle and deep-water gigantism in marine crustaceans. Biol. Bull. Russ. Acad. Sci. 28, 646–650. doi: 10.1023/A:1012336823275
Tsaur, S. C., and Wu, C. I. (1997). Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14, 544–549. doi: 10.1093/oxfordjournals.molbev.a025791
Wang, C., Lu, S., Li, J., Wang, L., Jiang, H., Liu, Y., et al. (2020). Effects of dietary myo-inositol on growth, antioxidative capacity, and nonspecific immunity in skin mucus of taimen Hucho taimen fry. Fish Physiol. Biochem. 46, 1011–1018. doi: 10.1007/s10695-020-00766-z
Wang, K., Shen, Y., Yang, Y., Gan, X., Liu, G., Hu, K., et al. (2019). Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat. Ecol. Evol. 3, 823–833. doi: 10.1038/s41559-019-0864-8
Yancey, P. H. (2020). Cellular responses in marine animals to hydrostatic pressure. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 398–420. doi: 10.1002/jez.2354
Yancey, P. H., and Siebenaller, J. F. (2015). Co-evolution of proteins and solutions: protein adaptation versus cytoprotective micromolecules and their roles in marine organisms. J. Exp. Biol. 218, 1880–1896. doi: 10.1242/jeb.114355
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, X., Yuan, J., Sun, Y., Li, S., Gao, Y., Yu, Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. commun. 10:356. doi: 10.1038/s41467-018-08197-4
Zhao, F. Q., and Keating, A. F. (2007). Functional properties and genomics of glucose transporters. Curr. Genomics 8, 113–128. doi: 10.2174/138920207780368187
Keywords: hadal, amphipod, Alicella gigantea, gigantism, positively selected gene, evolution rate
Citation: Li W, Wang F, Jiang S, Pan B, Chan J and Xu Q (2021) The Adaptive Evolution and Gigantism Mechanisms of the Hadal “Supergiant” Amphipod Alicella gigantea. Front. Mar. Sci. 8:743663. doi: 10.3389/fmars.2021.743663
Received: 19 July 2021; Accepted: 06 September 2021;
Published: 13 October 2021.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Denis Copilas-Ciocianu, Nature Research Centre, LithuaniaGuoyong Yan, Hong Kong University of Science and Technology, Hong Kong SAR, China
Copyright © 2021 Li, Wang, Jiang, Pan, Chan and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianghua Xu, cWh4dUBzaG91LmVkdS5jbg==
†These authors share first authorship
 Wenhao Li
Wenhao Li Faxiang Wang
Faxiang Wang Shouwen Jiang
Shouwen Jiang Binbin Pan
Binbin Pan Jiulin Chan1
Jiulin Chan1 Qianghua Xu
Qianghua Xu