- 1CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory of Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 4University of Chinese Academy of Sciences, Beijing, China
The endoparasitic dinoflagellate Amoebophrya infects a number of marine dinoflagellates, including toxic and harmful algal bloom-forming species. The parasite kills its host and has been proposed to be a determining factor in the demise of dinoflagellate blooms in restricted coastal waters. Previous studies have mainly focused on the occurrence, prevalence, and diversity of Amoebophrya, while the interactions between the parasite and its host have received limited attention. Herein, an Amoebophrya sp.-Akashiwo sanguinea co-culture was established from Chinese coastal waters, and morphological, physiological, and transcriptional changes throughout an infection cycle of the parasite were systemically studied. The parasitic dinoflagellate was very infectious, resulting in an infection rate up to 85.83% at a dinospore:host ratio of 10:1. Infected host cells died eventually and released approximately 370 dinospores/cell. The host nuclear structures were rapidly degraded by Amoebophrya infection, and the chloroplasts of parasitized host cells remained intact until the parasite filled the almost entire cell structure. Nevertheless, infected cultures showed sustained but lower levels of photosynthetic performance (∼64% of control cultures), and the photosynthesis-related genes were significantly down-regulated. These findings provide a better understanding of the biological basis of the complex parasite-host interactions, which will be helpful to further elucidate the ecological significance of parasitic dinoflagellates in marine ecosystems.
Introduction
Marine dinoflagellates represent a key component of marine ecosystems. Besides being major contributors of primary production, dinoflagellates possess a variety of other life and feeding strategies, including osmotrophy, phagotrophy, mixotrophy, symbiosis, and parasitism (Taylor, 1987; Fensome et al., 1996), thereby playing complicated ecological roles in the marine environment. In particular, parasitic dinoflagellates account for a large proportion of species richness in coastal and oceanic waters, and are critical for biomass transfer between trophic levels through impacting population dynamics, species succession, host biodiversity, and co-evolution of marine plankton organisms (Chambouvet et al., 2008, 2011a; Dougherty et al., 2016; Chen et al., 2020; Paseka et al., 2020). For example, the parasitic dinoflagellates of the family Amoebophryidae, the major component of the order Syndiniales or marine Alveolate group II, are known to infect a wide range of unicellular planktonic organisms, and have been associated with the demise of harmful algal blooms (Mazzillo et al., 2011; Lima-Mendez et al., 2015).
Species of Amoebophrya have a relatively simple life cycle consisting of a free-swimming infective dinospore that attaches to the host and penetrates the host cell membrane, an endoparasitic trophont grows inside the host cell by completely digesting the cell contents, and a multinucleate flagellated vermiform is released upon death of the host and then divides into hundreds of infectious dinospores (Cachon, 1964; Coats and Bockstahler, 1994; Chambouvet et al., 2008). The parasite completes its full life cycle in 2–3 days, while its dinospores can survive 3–15 days searching for a new host or dying in the water column (Cachon, 1964). The latest molecular approaches have revealed enormous diversity and abundance of Amoebophryidae organisms in the global plankton interactome (Guillou et al., 2008; Chambouvet et al., 2011b; de Vargas et al., 2015; Lima-Mendez et al., 2015; Chen et al., 2019; Cai et al., 2020), and stimulated attention on the biological characteristics and ecological significance of the parasitic dinoflagellates.
Previous studies have revealed the infectivity and host specificity of Amoebophrya. For instance, the parasites are more infectious in nutrient-enriched environments than those in nutrient-limited conditions, and newly formed dinospores lose their infectivity quickly in the water column in the absence of susceptible hosts (Yih and Coats, 2000; Chen et al., 2018). Different strains of Amoebophrya indicate various degrees of host specificity ranging from extremely species-specific (Coats and Park, 2002; Mazzillo et al., 2011; Chen et al., 2018), to moderately specific (Kim, 2006), to non-specific (Sengco et al., 2003; Chambouvet et al., 2011b). The parasitic infections also cause morphological and behavioral changes in host cells. Host cells at the late infection stage always exhibit as “giant cells,” with negative phototaxis and absence of diel/diurnal vertical migration (Park et al., 2002a; Kim et al., 2004). Interestingly, the infected dinoflagellate is able to maintain a degenerating, but steady swimming behavior until the host cell dies (Park et al., 2002b; Kayal et al., 2020; personal observations), suggesting the host still retains a certain degree of physiological function and is digested “alive.”
The intrinsic relationship between Amoebophrya and its dinoflagellate host implies a delicate balance. The parasite may subvert host metabolism to fulfill its needs while the host develops strategies to survive from the parasitic infection (Kim and Park, 2016; Scholz et al., 2017; Marc et al., 2021). The developments of Amoebophrya infection are different between the thecate and athecate dinoflagellates, and the two groups show various responses to the parasitic infection (Kim, 2006; Park et al., 2013). The parasitic waterborne cues are able to trigger series of physiological responses in host dinoflagellates, such as energy production, ATP synthesis, fatty acid production, and calcium-mediated signal transduction (Lu et al., 2016). Park et al. (2002b) noticed that Amoebophrya infections resulted in a decrease of host photosynthesis. The ultrastructure of Amoebophrya sp. and changes during the course of infection were elucidated in Miller et al. (2012). Most recently, Kayal et al. (2020) found that the chloroplast of the thecate dinoflagellate Scrippsiella acuminata remained transcriptomically functional during the course of Amoebophrya infection, and the photosynthetic activity of infected cultures was similar to that of uninfected cultures in continuous light conditions. Photosynthesis is strongly related to irradiation times in the marine environment where the key chlorophyll pigments and main phases of the process of photosynthesis localize in chloroplast of the eukaryotic photosynthetic cells (Blankenship, 2014; Sun et al., 2019). Yet, effects of Amoebophrya infections on chloroplast and photosynthetic activities of the athecate dinoflagellate in natural conditions still needs further study.
Herein, a strain of Amoebophrya known to infect Akashiwo sanguinea, an athecate dinoflagellate commonly observed in estuarine and coastal waters around the world (Botes et al., 2003; Jessup et al., 2009; Trainer et al., 2010; White et al., 2014; Kang et al., 2020), was successfully isolated and maintained in a monoclonal culture of A. sanguinea. The objective of this study was to investigate the variations in chloroplast ultrastructure and photosynthetic response of A. sanguinea during the course of Amoebophrya infection or exposed to parasitic waterborne cues. By analyzing a series of changes in A. sanguinea, we were able to identify the physiological characteristics and related molecular regulation mechanisms of the dinoflagellate during the Amoebophrya sp. infection. These findings will foster our understanding of complex host-parasite interactions and contribute to better understanding of the ecological roles of parasitic dinoflagellates in the marine ecosystem.
Materials and Methods
Host Dinoflagellate and Parasite Cultures
Both Akashiwo sanguinea and its corresponding strain of Amoebophrya sp. were isolated from Jiaozhou Bay (36°24′ N, 120°11′ E), China in June 2020 using single cell isolation methods, and established as continuous cultures as described in Kim et al. (2004). The identifications of A. sanguinea and Amoebophrya sp. were first made with microscopic observation (by visualization of the natural bright-green auto-fluorescence of the parasite, Coats and Bockstahler, 1994), and further confirmed with molecular characterization (18S rDNA sequencing, see details in Supplementary Text). In brief, A. sanguinea was maintained in f/2-Si medium (Guillard and Ryther, 1962) made with sterile natural seawater (salinity of 30 ± 0.1). The Amoebophrya sp. ex A. sanguinea cross was maintained in stock culture by sequentially transferring aliquots of infected host culture into uninfected host culture of exponentially growing A. sanguinea at approximately 2-day intervals. Stock cultures were incubated at 20 ± 1°C under a 14 h: 10 h light: dark cycle, with cool white fluorescent light providing 175 μE m–2 s–1.
Time-Course Infection Experiment
The time-course infection of Amoebophrya sp. in A. sanguinea was established following the methods of Coats and Park (2002), which covered a complete life cycle of the parasite. Briefly, newly formed dinospores (≤6 h old) were separated from the parasite cultures by gravity filtration using 47-mm polycarbonate membranes (Whatman Nuclepore, 10-μm pore size). The subsamples (1 ml) of newly formed dinospores (≤6 h old) were immediately fixed with buffered formaldehyde (final conc. 1%), and enumerated with hemacytometers using an Olympus IX71 epifluorescence microscope (excitation 450–480 nm, emission 500 nm; Coats and Bockstahler, 1994).
Batch cultures of exponentially growing A. sanguinea (800 ml) were prepared in 1-L culture flasks, with an initial density of approximately 3 × 103 cells ml–1. To investigate the responses of A. sanguinea to Amoebophrya sp., A. sanguinea cultures were inoculated with the recently formed dinospores (≤6 h old) at a dinospore: host ratio of 10:1 to insure infection of most host cells (Chen et al., 2018). To examine the potential effects of the waterborne cues from the parasite, A. sanguinea cultures were treated with the addition of an equivalent volume of dinospore filtrate (Millipore PC filters, 1-μm pore size). In addition, an equivalent volume of sterile f/2-Si medium was added in the separate culture as experimental control. All treatments were conducted in triplicate and incubated under the same condition as mentioned above.
Host abundance and parasite prevalence (i.e., percent cells infected) were assessed and further quantified at time points of 0, 12, 30, 54, and 70 h post inoculation. To determine host abundance, subsamples (1 ml) were collected and fixed with buffered formaldehyde (final conc. 1%), then enumerated using triplicate plankton counting chambers under an epifluorescence microscope (IX71, Olympus, Japan). The parasitized A. sanguinea were identified based on the characteristic green autofluorescence of the parasite and red fluorescence of the host chloroplasts under the blue light excitation (excitation 450–480 nm, emission 500 nm; Coats and Bockstahler, 1994). The prevalence was calculated as the percentage of infected cells when at least 100 host cells per sample were counted. Morphological variation, photosynthesis activity, pigment composition, and transcription of photosynthesis-related genes of host cells were also studied at the five time points (0, 12, 30, 54, and 70 h) and as described below.
Epifluorescence and Electron Microscopy
Epifluorescence microscopy, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) were used to characterize the morphology and development of Amoebophrya sp. ex A. sanguinea. Live observation of the algal culture (1 ml) was made at 400 × magnification and recorded with an Olympus DP73 digital camera attached to an Olympus IX71 epifluorescence microscope.
Aliquots of 30 ml of newly formed dinospores (≤ 6 h old) of Amoebophrya sp. ex A. sanguinea, with a density of ∼105 cells ml–1 were fixed with OsO4 (final conc. 1%, Sigma-Aldrich, Seoul, Korea) for 40–50 min at room temperature, and rinsed with deionized water. The solution was gently filtered using gravity through a 5-μm pore sized Millipore nylon membrane (Cork, Ireland) to capture the fixed cells. Then the sample was dehydrated in a graded acetone series (10, 30, 50, 70, 90%, 3 × 100%) for 15 min at each step, critical point dried with liquid CO2 (EM CPD300, Leica, Vienna, Austria), and sputter-coated with gold (EM ACE200, Leica, Vienna, Austria). Digital images were collected with an S-3400 N SEM (Hitachi, Hitachinaka, Japan) and processed using Adobe Photoshop (Adobe, San Jose, CA, United States). Cell dimensions of 50 cells were calculated from SEM micrographs.
Aliquots of experimental cultures (30 ml) were fixed with 2.5% glutaraldehyde (final conc.) for 3 h at 4°C, and post-fixed with OsO4 prepared with filtered seawater (final conc. 1%) overnight at 4°C. Cells were then centrifuged at 720 g for 5 min to form pellets and remove the supernatant. Pellets were rinsed three times (10 min each) in fresh f/2 medium. Dehydration was carried out in a graded acetone series (10, 30, 50, 7%, 90%) for 10 min each and then rinsed three times in absolute acetone for 15 min each. Finally, the pellets were infiltrated with 2:1, 1:1, and 1:2 mixtures of mixtures of acetone: Spurr’s resin (at least 8 h for each dilution) and embedded in 100% Spurr’s resin (Spurr Formula Kit I, SPI Supplies West Chester, Pennsylvania, United States; Spurr, 1969) overnight. The material was sectioned on an ultramicrotome (EM UC7, Leica, Vienna, Austria). Sections were mounted on Formvar-coated grids, stained with uranyl acetate (3%) and lead citrate (2%), and examined with an HT7700 TEM (Hitachi, Hitachinaka, Japan).
Photosynthesis Activity and Pigment Composition of Akashiwo sanguinea During the Course of Infection
Aliquots of 2 ml algal culture were used to determine the chlorophyll fluorescence of A. sanguinea using a FEM-2 pulse-modulated fluorometer (Hansatech, Norfolk, United Kingdom). The maximal photosynthetic efficiency (Fv/Fm) was calculated as:
Where Fv represents the variable fluorescence, Fm represents the maximum fluorescence, and Fo represents the minimum fluorescence. Samples were dark adapted for 30 min (Schreiber et al., 1995) before transfer to a cuvette for the measurement of Fo, and after a saturating pulse (0.3 s; 2,600 μmol photonsm–2 s–1), the measurement of Fm. The actual photochemical efficiency of PSII in the light (ΦPSII) was also measured at the same time.
Another 10 ml of algal culture were filtered through the Whatman GF/F filters (25-mm in diameter, Whatman Inc.) with a vacuum pump (<0.03 MPa). Filters were then folded and wrapped against light in aluminum foil, and immediately frozen at –80°C for further processing. Pigments were determined by an alliance High-Pressure Liquid Chromatograph (e2695, Waters, Milford, Massachusetts, United States) using a 100 μl sample injection according to Kong et al. (2012).
Transcript Analysis of Photosynthesis-Related Genes of Akashiwo sanguinea During the Course of Infection
Aliquots of 30 ml algal cultures were pipetted and concentrated at 4°C via gentle centrifugation (500 g, 5 min) to form pellets and remove the supernatant, then flash-frozen in liquid nitrogen, and stored at –80°C until RNA extraction. Total RNA was extracted using TRIzol reagent (TakaRa, Japan), and further treated with RNase-free DNase I (TakaRa, Japan). The quality of RNA samples was ascertained by the spectrophotometric method with an A260 nm/A280 nm ratio from 1.8 to 2.0, and further assessed with 1% agarose gel electrophoresis based on the integrity of 18S and 28S rRNA bands. The cDNA was synthesized from 1 μg of total RNA using the PrimeScriptTM RT reagent Kit (TaKaRa, Japan) with addition of gDNA Eraser to eliminate the genomic DNA contamination according to manufacture protocols. The cDNA templates were diluted 10 times with nuclease-free water for subsequent analyses.
The transcriptions of photosynthesis-related genes, including psbA, psbD, psaA, psaB, petA, and petB, were detected with the quantitative Real Time PCR (qRT-PCR) assay using the specific primers reported in Liu et al. (2020). To normalize the mRNA expression of the target genes, 18S rRNA, TUA and GAPDH were selected as endogenous control and their expression stability in Amoebophrya infected cultures was confirmed prior to the experiments based on the assays described in Pfaffl et al. (2004) and Deng et al. (2016). The qRT-PCR assay was performed with a Rotor-Gene Q 2plex HRM thermocycler (QIAGEN, Germany). Reactions (triplicate for each sample) were carried out in a total reaction volume of 20 μl, containing 10 μl SYBR Premix Ex Taq II (2 ×) (TaKaRa, Japan), 0.5 μl for each primer (10 μM), 1 μl template cDNA, and DNAse/RNAse free water (8 μl, Sigma-Aldrich, MO, United States). Amplification conditions were 95°C for 30s, followed by 40 cycles of denaturation at 95°C for 5 s, and annealing at 60°C for 30 s. Melt curve analysis was added at the end of each PCR thermal profile to assess the specificity of amplification. Amplification efficiency (E) was calculated from the given slopes in Rotor-gene Real-Time PCR System Manager software by a 10-fold diluted cDNA sample series, with six dilution points measured in triplicate. The relative mRNA levels were calculated by the 2–Δ Δ Ct method as described in Livak and Schmittgen (2001). The gene expression levels in control cultures were used as control.
Statistical Analysis
All data were presented as mean ± standard error of the mean (SE) in the text, and subjected to a t-test to determine the difference in the three treatments using the SPSS 18.0 for windows (SPSS, Chicago, IL, United States), with the significance level of P < 0.05.
Results
Development of Amoebophrya sp. Infections in Cultured Akashiwo sanguinea
In the filtrate-added and control groups, Akashiwo sanguinea showed a steady growth during the experiment, reaching 10.05 ± 0.73 × 103 and 10.35 ± 0.18 × 103 cells ml–1 by 70 h, respectively (Figure 1A). In the infected groups, the growth of A. sanguinea was significantly affected by introduction of Amoebophrya dinospores. The cell density of A. sanguinea decreased dramatically after 30 h post inoculation (p.i.), and the final density was 1.71 ± 0.74 × 103 cells ml–1, even though there was no significant difference in the cell densities among the three groups within the first 30 h (P > 0.05).
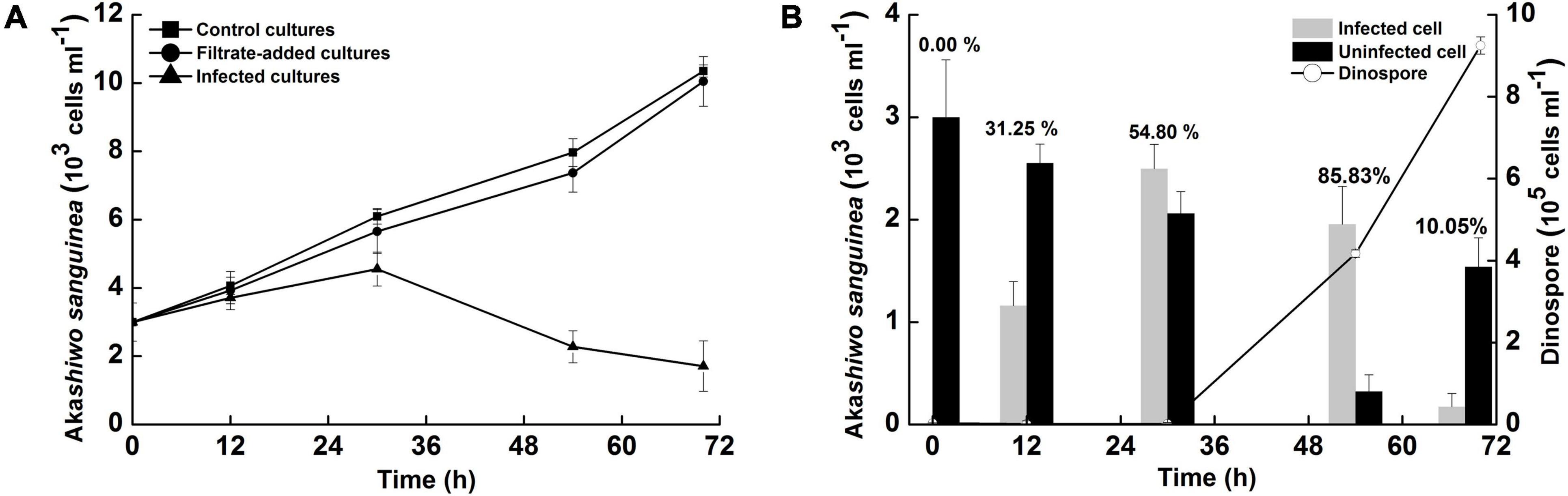
Figure 1. Time-course studies of Akashiwo sanguinea infected with Amoebophrya sp. at a dinospore: host ratio of 10: 1. (A) Total A. sanguinea cell concentrations in control, filtrate-added and infected cultures. (B) Estimation of numbers of dinospores, infected and uninfected hosts in infected cultures, with the prevalence (data on the column). No infected cells or dinospores were detected in the filtrate-added and control cultures. Error bars indicate the standard error.
Using fluorescence microscopy, we were able to detect Amoebophrya infection in A. sanguinea based on the presence of green autofluorescence signals inside host nuclei (Figure 2). In the early infection stage (12 h p.i.), the green autofluorescing parasite (approximately 18.20 μm length) was surrounded by red-fluorescing chloroplasts of host dinoflagellates (Figure 2C). By this time, the abundance of infected cells was 1.16 ± 0.24 × 103 cells ml–1 and the prevalence in the infected cultures was 31.25 ± 1.73% (Figure 1B). In the middle stage of infection (30 h p.i.), the presence of the beehive-like structure (approximately 40.01 μm length) could be easily discriminated in the Ameobophrya-infected host cells (Figure 2D), even though the green autofluorescence was mostly concealed by the red autofluorescence of host chloroplasts. The abundance of infected host cells increased to 2.50 ± 0.24 × 103 cells ml–1 and the prevalence of Amoebophrya infection was 54.80 ± 2.84% at the time point (Figure 1B). In the late stage of infection (54 h p.i.), the characteristic “beehive” structure of Amoebophrya multinucleate trophonts (approximately 65.80 μm length) was easily observed in the infected A. sanguinea cells (Figure 2E) where they appeared bright green with grooved and doted structure when examined with an epifluorescence microscope. The abundance of infected host cells was 1.95 ± 0.37 × 103 cells ml–1 and the prevalence increased to 85.83 ± 1.40% at 54 h p.i. By 70 h p.i., only 0.17 ± 0.03 × 103 cells ml–1 were infected and the prevalence of Amoebophrya infection dropped to 10.05 ± 0.73% (Figure 1B). Most of the infected host cells broke up at the time point, and the parasite emerged as a conspicuously motile vermiform (approximately 133.33 μm length) (Figure 2F). The vermiform persisted for approximately 3–10 min before suddenly breaking apart to release numerous dinospores, and the abundance of motile dinospores was 9.25 ± 0.41 × 105 cells ml–1 in the infected groups. In the case of A. sanguinea as host, each infected cell produced 370 ± 51 Amoebophrya dinospores. No infected cells or dinospores were detected in the filtrate-added and control groups.
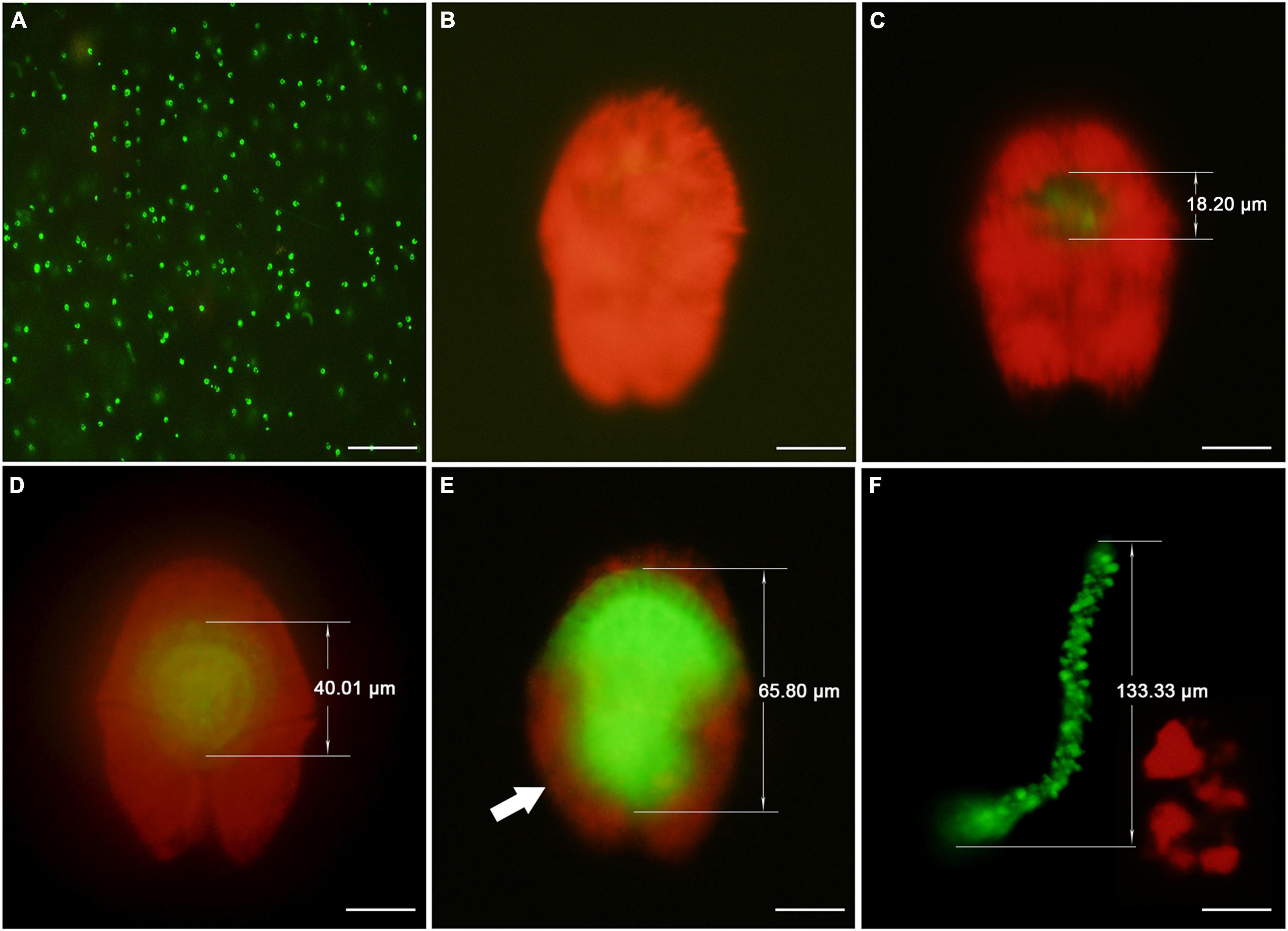
Figure 2. Epifluorescence micrographs of free-swimming dinospores and Amoebophrya sp. within Akashiwo sanguinea. Amoebophrya sp. exhibited green autofluorescence throughout its life cycle and A. sanguinea showed red fluorescence under the blue light excitation. (A) Free-living dinospores of Amoebophrya sp. (B) Uninfected A. sanguinea. (C) Twelve hours infection showing a very early trophont surrounded by the red fluorescing chloroplasts of A. sanguinea. (D) Thirty hours infection showing a mid trophont surrounded by the red fluorescing chloroplasts of A. sanguinea and took on a “beehive” appearance. (E) Fifty-four hour infection showing a mature trophont and the parasites filled almost the entire host cell; the remnants of the host chloroplasts (arrow) were also shown. (F) Seventy hours infection showing a free-swimming Amoebophrya sp. vermiform and the remaining host cell fragments. Scale bars = 20 μm.
Ultrastructure of Akashiwo sanguinea Infected by Amoebophrya sp.
The free-living stage (dinospore) of Amoebophrya sp. was a flagellated zooid with a large bulbous episome (4.01 ± 0.37 μm in diameter, n = 50, using SEM) and a narrow hyposome, which was characterized by the presence of two flagella (Figure 3A). The transverse flagellum exhibited flagellar hairs over its full length and was approximately 11.21 ± 1.05 μm long (n = 50, using SEM), whereas the longitudinal flagellum was bare (4.05 ± 0.53 μm long, n = 50, using SEM) and progressively thinner toward the end. As indicated by the TEM ultrastructure, there was an elongated, typically compact nucleus located in the cell center of uninfected A. sanguinea, with a distinct nucleus membrane condensed chromosomes (Figure 3B). Chloroplasts were ellipsoidal or elongated, rod-like or irregularly shaped, and located almost throughout the healthy host cells.
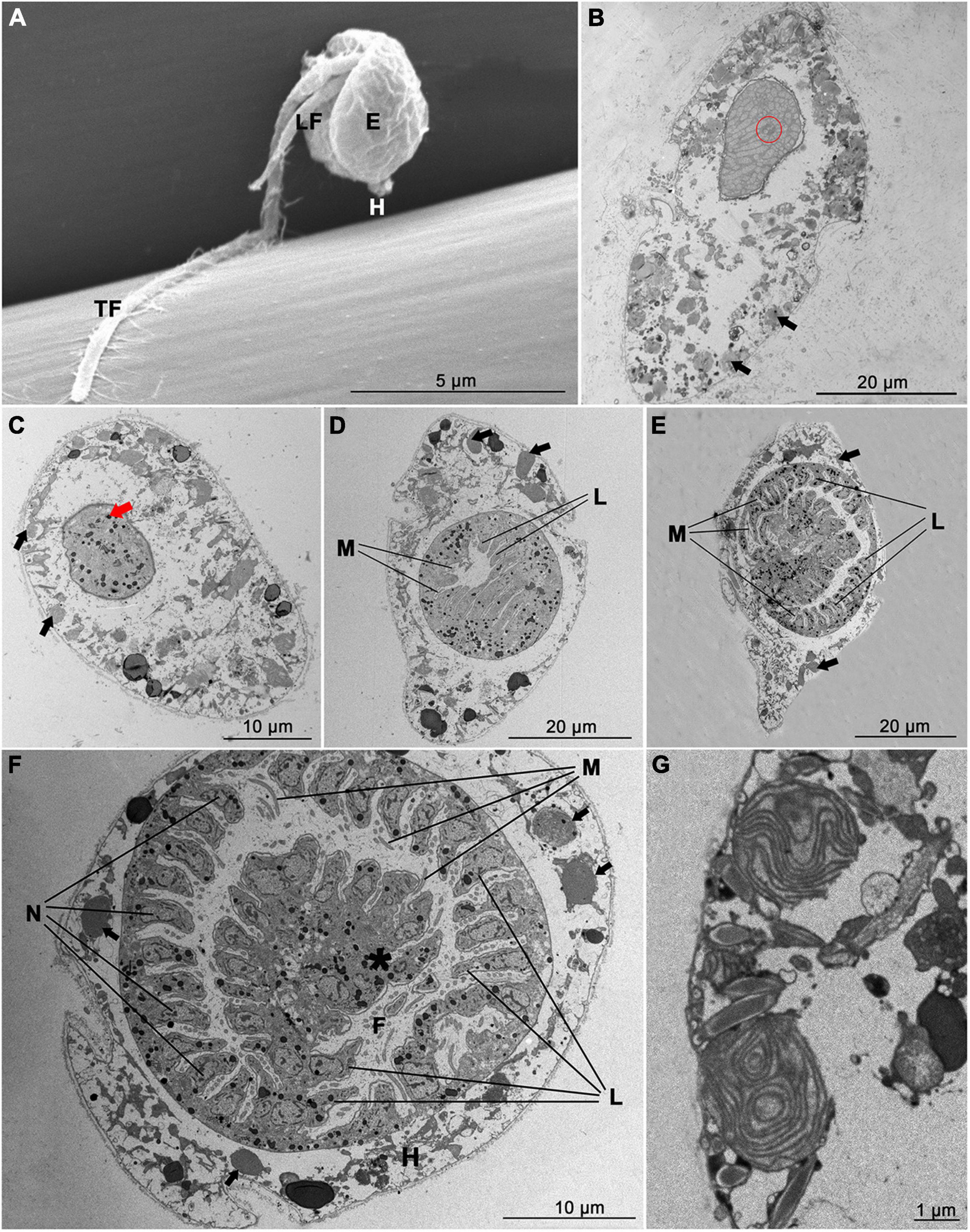
Figure 3. Electron micrographs of free-swimming dinospores and Amoebophrya sp. within Akashiwo sanguinea. (A) Scanning electron microscope image of Amoebophrya sp. dinospore showing the episome (E), hyposome (H), longitudinal flagellum (LF), transverse flagellum (TF), and flagellar hairs (black arrow). (B–G) Transmission electron microscope images of A. sanguinea infected with Amoebophrya sp. Black arrows indicated the chloroplast of A. sanguinea. (B) Uninfected A. sanguinea showing nucleus, nucleolus (red circle) and nucleus membrane. (C) 12 h infection with multiple parasite (red arrow) in host nucleus. (D) 30 h infection showing nascent mastigocoel (M) and multilobed cavity (L). (E) 54 h infection showing “beehive” with fully developed mastigocoel (M) and multilobed cavity (L). (F) Transverse section through 54 h Amoebophrya sp. trophont within the A. sanguinea nucleus. The host cytoplasm (H) was relegated by the parasite to the margin of the cell. The mastigocoel (M) contains numerous flagella (F), and the nucleus (N) line up in a continuous fashion along the folds of the “beehive.” Asterisk indicated the clusters of parasite trichocysts. (G) Detail of chloroplasts in the 54 h infected A. sanguinea.
At 12 h p.i., the condensed chromosomes of host cells were overtly disrupted and trophonts of Amoebophya sp. appeared as spherical cells showing electron-dense indentation at the surface (Figure 3C). By 30 h p.i., the parasite grew in size as they consumed the nucleonic contents of the host. The host nucleus was significantly enlarged and mostly occupied by the developing trophont. The parasite started to take on a “beehive” appearance (Figure 3D), resulting from helical extension of the parasitic cingulum and sinking of the parasitic apex during its growth. Subsequently, the development of the parasite largely consumed the nucleonic contents of the host and took up a large proportion of the host dinoflagellate. Further invagination of the developing parasite created a cavity structure (lobed cavity and mastigocoel) within the trophont cytoplasm, appearing as a narrow space associated with basal bodies. By 54 h p.i., cytoplasm was relegated to the margin of the host cells by the well-developed trophont of the parasite (Figure 3E), the chloroplasts of the host cell remained intact at the late stage of the parasitic infection (Figure 3G). Inside the giant trophont, multiple nuclei (will develop into the nuclei of dinospores in next few hours) lined up in a continuous fashion along the folds of the beehive, each section lobe contained one nucleus, with numerous flagella extended into the cavity (amphiesmalined mastigocoel) (Figure 3F). Ultimately, the multiflagellated and multinucleated trophont ruptured the host cell and emerged as a conspicuously motile vermiform, which split into fragments of variable sizes before suddenly break, releasing numerous individual dinospores (Supplementary Videos 1–3).
Variation of Host Photosynthetic Activity During the Course of Infection
Photosynthesis is an important determinant of the ability of marine microalgae to survive biotic stresses (Baker, 2008), and the Fv/Fm is widely used to indicate photosynthetic activity of microalgae (Maxwell and Johnson, 2000). The photosynthetic activities of A. sanguinea were overtly impacted by Amoebophrya sp. infection (Figure 4). The Fv/Fm of A. sanguinea remained steady in the filtrate-added and control cultures, with near-optimal quantum yield values (Fv/Fm = 0.6) over the experimental period. In the cultures inoculated with Amoebophrya dinospores, the Fv/Fm decreased continuously to 0.45 ± 0.03 at 54 h p.i., then recovered to 0.51 ± 0.02 by 70 h p.i., which was significantly different from that of control and filtrate-added groups (P < 0.05) (Figure 4A). The values of photochemical efficiency (ΦPSII) in the infected cultures ranged from 0.47 ± 0.03 to 0.54 ± 0.01, significantly lower than that of the control and filtrate-added cultures (0.47 ± 0.03 to 0.57 ± 0.04) (P < 0.05), but exhibited a similar pattern of variation (Figure 4B).
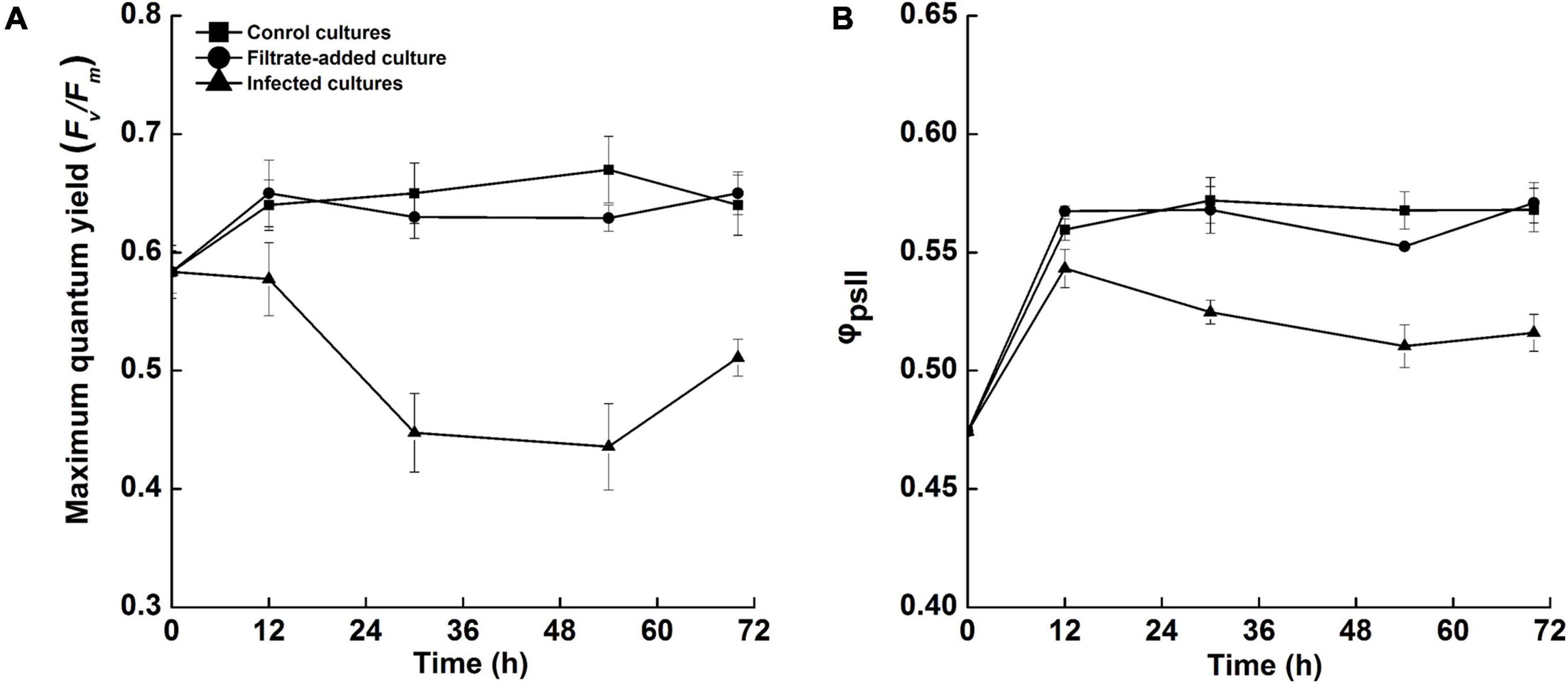
Figure 4. Variations in the photosynthetic efficiencies of Akashiwo sanguinea during the course of infection by Amoebophrya sp. at a dinospore: host ratio of 10: 1. (A) Fv/Fm. (B) ΦPSII. Error bars indicate the standard error.
Photosynthesis of marine dinoflagellates largely depends on the cellular photosynthetic pigments (Blankenship, 2014), thus variations in the contents of major pigments were further quantified to assess the effects of Amoebophrya infection on A. sanguinea. For the filtrate-added and control cultures, the cellular photosynthetic pigments showed a similar pattern of variation, and no significant difference was found between the two groups (P > 0.05). In the infected cultures, chlorophyll a (Chl a) exhibited a significant difference from the filtrate-added and control groups, which decreased sharply by 30 h p.i. (36.41 ± 2.66 pg cell–1) (P < 0.05), then recovered gradually to 48.86 ± 2.83 pg cell–1 at 70 h p.i. (Figure 5A). The light-harvesting pigment peridinin of the infected cultures ranged from 40.01 ± 0.03 to 48.22 ± 2.35 pg cell–1, Figure 5B). The photoprotective pigments diatoxanthin, β-carotene, and diadinoxanthin increased gradually in the infected cultures, with fluctuated variation during the time course (Figures 5C–E). It was noteworthy that the β-carotene of the infected cultures decreased overtly by 30 h p.i., then gradually increased by 70 h p.i. (Figure 5D).
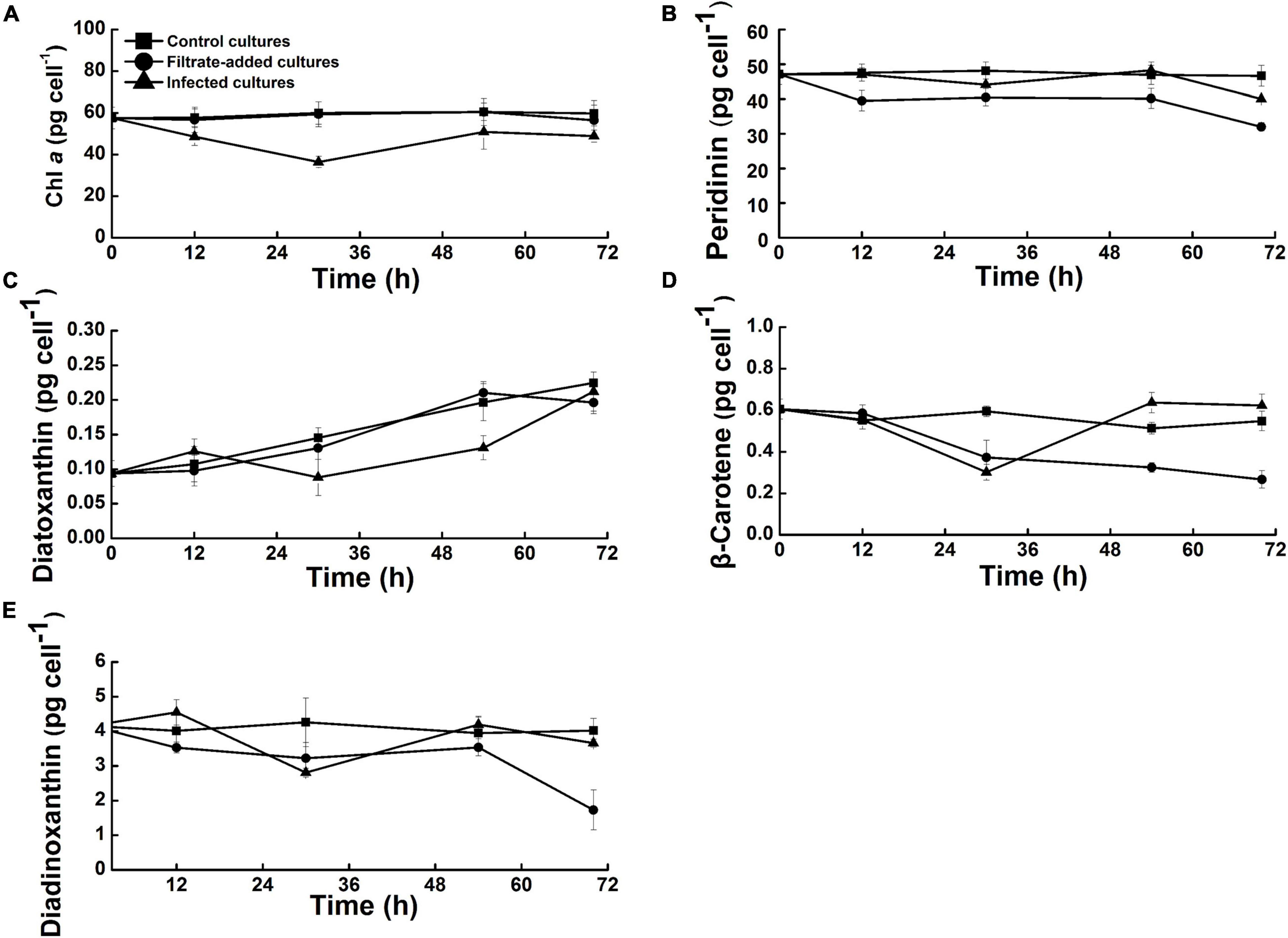
Figure 5. Variations in the pigments of Akashiwo sanguinea during the course of infection by Amoebophrya sp. at a dinospore: host ratio of 10: 1. (A) Chlorophyll a. (B) Peridinin. (C) Diatoxanthin. (D) β-Carotene. (E) Diadinoxanthin. Error bars indicate the standard error.
Variation in the Transcripts of Photosynthetic Genes
The two photosystem I-related genes—psaA and psaB—were down-regulated (< 1-fold) in the A. sanguinea cultures with Amoebophrya infection, but showed different patterns. The psaA transcripts were significantly down-regulated at 30 h p.i. and 70 h p.i. (P < 0.05), with the lowest level of transcription (0.06 ± 0.01-fold) at 30 h p.i. (Figure 6A). The psaB transcripts decreased significantly during the infection cycle (P < 0.05), with the lowest transcription (0.18 ± 0.05-fold) at 30 h p.i. (Figure 6B). The photosystem II-related genes—psbA and psbD—were significantly suppressed during the time course of infection (P < 0.05), with the lowest level of transcription at 70 and 30 h p.i., respectively (Figures 6C,D). In contrast, the psbA and psbD were almost up-regulated (> 1-fold) in the control cultures, with the highest level of transcription at 70 and 54 h p.i., respectively. The two cytochrome b6f complex-related genes—petA and petB showed various patterns in the infected cultures. The petA gene was down-regulated (< one fold) during the time course, with significant suppression at 12 and 30 h p.i. (P < 0.05) (Figure 6E). The petB was down-regulated significantly (P < 0.05), with the lowest transcription (0.13 ± 0.07-fold) at 30 h p.i. (Figure 6F). The petB transcripts in the control cultures increased during the time course, with the highest transcription (2.00 ± 0.04-fold) at 54 h p.i (P < 0.05). Compared to control cultures, all genes except psbA were down-regulated in the filtrate-added cultures (< one fold), and the psbD was significantly suppressed throughout the infection cycle (P < 0.05).
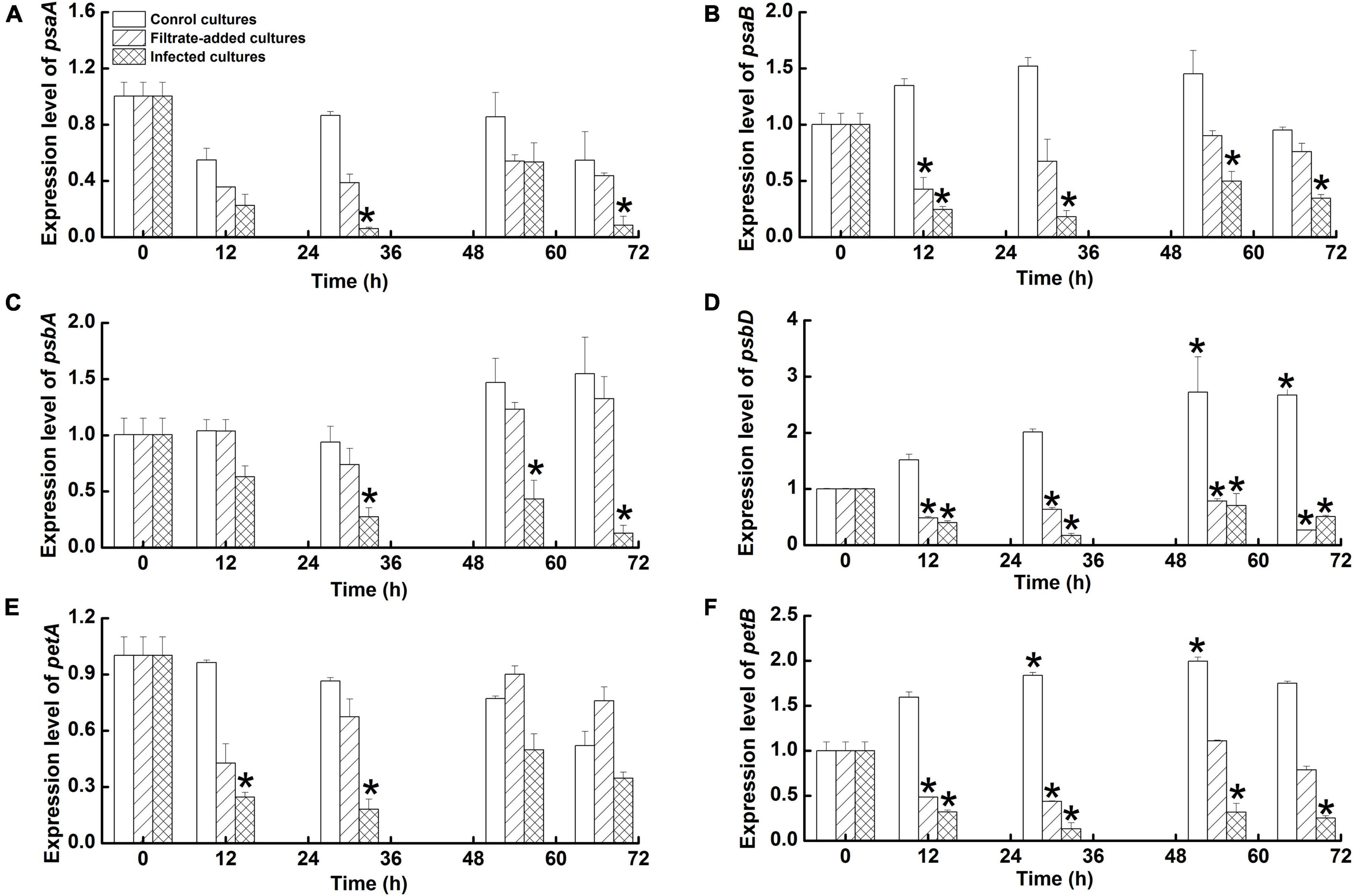
Figure 6. Relative mRNA expressions of photosynthesis gene in Akashiwo sanguinea during the course of infection by Amoebophrya sp. at a dinospore: host ratio of 10: 1. Temporal mRNA expression of psaA (A), psaB (B), psbA (C), psbD (D), petA (E), petB (F) were quantitatively assessed using the qRT-PCR assays. The transcriptional expression was calculated by the 2–Δ Δ Ct method using the 18S rRNA, TUA and GAPDH as the reference gene. Error bars indicate the standard error. Statistical significances across control in the same time of sampling were indicated with an asterisk (P < 0.05).
Discussion
The parasitic dinoflagellate Amoebophrya infects a wide range of marine dinoflagellates and is closely associated with succession of harmful algal blooms (Taylor, 1968; Coats, 1999; Chambouvet et al., 2008; Mazzillo et al., 2011), thus leading to growing interest between the parasite and its dinoflagellate hosts. Our results demonstrate that the athecate dinoflagellate Akashiwo sanguinea can trigger multiple responses to the parasitic infections, such as decreased cell density, impaired photosynthetic activities, and down-regulation of related gene expressions. Amoebophrya sp. targeted and digested the nucleus of A. sanguinea cells, the chloroplasts of infected cells remained intact throughout the infection cycle. These findings provide valuable insight to understanding the complex parasite-host relationship between Amoebophrya and its host dinoflagellate.
Amoebophrya is an aerobic phagotrophic endoparasite and relies solely upon their hosts to supply necessary resources for their own metabolism and reproduction (Cachon, 1964; Farhat et al., 2018). After settling inside its host cell, the parasite avoids killing its host and proliferates rapidly during the intracellular developmental stage (Miller et al., 2012; Kayal et al., 2020). Miller et al. (2012) described the ultrastructure of Amoebophrya sp. and its changes during the course of infection in A. sanguinea. Herein, we focused on the ultrastructure of parasitized host cells, particularly the chloroplast. No host nuclear structures (or remnants) were conclusively identified in any observed thin sections of infected A. sanguinea even in the early stage of infection, suggesting that the nucleus was attacked early on by the parasite. At the late stage of infection, the parasite occupied the entire host nucleus and the host cytoplasm was relegated by the parasites to the margin of the cell, the chloroplasts of A. sanguinea cells with Amoebophrya infections remained intact. This may partially be due to indirect contact of host cytoplasmic structures with the growing multinucleate of the parasite once the parasitic infection was established inside host nucleus (Park et al., 2002a).
The index Fv/Fm is an important to indicator of photosynthetic capacity of microalgae (Maxwell and Johnson, 2000). In the filtrate-added cultures (addition of an equivalent volume of dinospore filtrate), Fv/Fm did not show a significant difference from that of control cultures, suggesting that the parasitic waterborne cues had little effect on photosynthesis of A. sanguinea. Fv/Fm in the infected groups gradually decreased until 54 h p.i. due to the mortality of most infected host cells in the late stage of infection. It then increased dramatically when the continuous presence of uninfected host cells contributed more to the photosynthetic efficiency of the infected cultures. As shown by Park et al. (2002b), the Fv/Fm of infected Gymnodinium instriatum cultures was approximately 80% that of uninfected cultures, which was slightly higher than that of infected A. sanguinea cultures (∼64% of control cultures) in the present study. Most recently, Kayal et al. (2020) reported that the photosynthetic activities of the infected Scrippsiella acuminata by Amoebophrya sp. were similar to those of the uninfected cells in continuous light condition. It is likely that S. acuminata was capable of relying on the existing pool of nuclear-encoded photosynthetic proteins to maintain photosynthetic activities despite digestion of its nucleus in continuous light conditions (Kayal et al., 2020). Overall, the infected dinoflagellates were still able to maintain relatively high photosynthetic performance, and there may be various physiological strategies among different Amoebophrya strains.
Infection by Amoebophrya induced changes in the pigment composition of A. sanguinea, except for the peridinin content. Similar contents of Chl a were observed in the filtrate-added and control cultures; Chl a content in infected cultures was significantly lower than that of the other two cultures, indicating the direct impact of Amoebophrya infections on its host cells. The same effect was observed in the co-cultures of Amoebophrya sp.—Scrippsiella acuminata (Kayal et al., 2020). The Chl a content of infected G. instriatum cells (121.50 ± 4.19 pg cell–1) was comparable to that of uninfected cells (Park et al., 2002b), which was higher than that of A. sanguinea in the present study. Such difference in the variation of Chl a content might result from the different sites of infection in the three host species. The parasite mainly developed in the nucleus of A. sanguinea and S. acuminata, but in the cytoplasm of G. instriatum. The β-carotene content in the infected A. sanguinea cultures showed no significant difference from that of control groups before 30 h p.i., after which it increased dramatically, which was in agreement with previous studies on β-carotene in other microalgae (Zubia et al., 2014; Benavente-Valdés et al., 2016). With the rapid growth of Amoebophrya in host cell, the major host defense against parasites may thus be production of reactive oxygen species (ROS) (Imlay, 2003; Lu et al., 2016). The elevated ROS in host cells can cause strong oxidative damage, resulting in an increased level of photoprotective β-carotene to prevent photooxidation (Zubia et al., 2014; Lu et al., 2016). In the infected S. acuminata cultures, the contents of β-carotene were relatively stable during the parasitic infection cycle suggesting the parasite might not induce significant photophysiological stress under continuous light condition (Kayal et al., 2020).
The dinospore filtrate triggered none-significant down-regulation of most photosynthesis-related genes (except psbD), suggesting the host was capable of sensing the parasite, but the level of stimulation could not trigger prominent response of host cells. The encoding the PSII core subunit psbD (D2) can be directly correlated with the production of certain proteins essential for photosynthesis (Sharon et al., 2007), and the lower expressions of the psbD in the filtrate-added cultures might result from the stimulation of signal transduction chains in the adaptive responses of the host induced by the parasitic waterborne cues (Lu et al., 2016). A. sanguinea triggered a strong response to Amoebophrya infections along with progress of the parasitic infection, and there was a significant decline of photosynthetic activity in A. sanguinea cultures before 54 h p.i. The transcripts of all tested genes were significantly down-regulated during this period, but seemed to recover at 54 h p.i., which may be due to the continuous presence of uninfected host cells and growth after collapse of infected host cells. Despite the fact that parasites filled almost the entire host nucleus at the late stage, the expression of the nuclear-encoded petA gene was still slightly higher than that seen at 30 h, suggesting that the host nucleus remained functional and the host was digested “alive.” The petB levels can be directly correlated with the production of certain proteins essential for photosynthesis (e.g., light-harvesting chlorophyll proteins and components of cytochrome b6/f complex encoded in the nucleus), and the lower expression levels of petB also correspond to the lower efficiency of photosystem in the infected cultures (Falkowski and Raven, 1997). While the expressions of chloroplast genes (associated with complexes PSI, PSII, and Cytb6f) were significantly up-regulated in infected S. acuninata cultures (Kayal et al., 2020). This discrepancy may result from the difference in light conditions, and the infected host cell exposed to continuous light conditions may need enhanced energy and oxygen to function (Lu et al., 2014, 2016; Kayal et al., 2020).
The parasite-host interactions between Amoebophrya and host dinoflagellates are complicated and far from being fully understood. Several key features of the Amoebophrya infection inside athecate dinoflagellate were further identified in the present study. Amoebophrya sp. was highly infectious to A. sanguinea, had a short lifespan, and high reproductive outputs. Infection by Amoebophrya caused a series of changes to host cells, including cellular and nuclear degradation, impaired physiological activity, and down-regulation of key genes associated with photosynthesis; the chloroplasts of infected hosts remained intact and functional. These findings highlight the fundamental cellular processes involved in Amoebophrya development and preservation of chloroplast activity toward host cell collapsing. Further research on the physiological and underlying molecular processes should decode the complex parasite-host relationship between Amoebophrya and its dinoflagellate hosts.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
TC, YL, and CL designed and conducted the experiments. YL and ZH conceived and supervised the project. SS and CL managed the funding acquisition. TC and CL compiled and analyzed output data, designed, and wrote the first version of the manuscript. All authors edited and approved the final version of this manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (Grant Nos. 41876120 and 41906122), the Key Deployment Project of Centre for Ocean Mega-Science, Chinese Academy of Sciences (Grant No. COMS2020Q06), China Postdoctoral Science Foundation (Grant No. 2018M642709), and the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (Grant No. 2018SDKJ0504-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are highly grateful of the two reviewers for their patience, insights, critical comments and generous suggestions, which helped greatly the improvement of the manuscript. We thank Sandra E. Shumway from Department of Marine Science, University of Connecticut for her valuable help in the language. We also thank Drs. Yuanyuan Sun and Litao Zhang for their assistance in the preparations of electron microscope and chlorophyll fluorescence analysis, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.742498/full#supplementary-material
Supplementary Video 1 | The emergence process of Amoebophrya sp. (green fluorescence) from an Akashiwo sanguinea cell (red fluorescence) observed under epifluorescence.
Supplementary Video 2 | A free-swimming Amoebophrya sp. vermiform observed under epifluorescence.
Supplementary Video 3 | The process of a free-swimming Amoebophrya sp. vermiform split into dinospores.
References
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Benavente-Valdés, J. R., Aguilar, C., Contreras-Esquivel, J. C., Méndez-Zavala, A., and Montañez, J. (2016). Strategies to enhance the production of photosynthetic pigments and lipids in chlorophycae species. Biotechnol. Rep. 10, 117–125. doi: 10.1016/j.btre.2016.04.001
Blankenship, R. E. (2014). Molecular Mechanisms of Photosynthesis, 2nd Edn. Hoboken, NJ: Wiley Blackwell, 1–9.
Botes, L., Smit, A. J., and Cook, P. A. (2003). The potential threat of algal blooms to the abalone (Haliotis midae) mariculture industry situated around the South African coast. Harmful Algae 2, 247–259. doi: 10.1016/S1568-9883(03)00044-1
Cachon, J. (1964). Contribution à l′étude des péridiniens parasites. Cytologie, cycles évolutifs. Ann. Sci. Nat. Zool. 6, 1–158.
Cai, R., Kayal, E., Alves-de-Souza, C., Bigeard, E., Corre, E., Jeanthon, C., et al. (2020). Cryptic species in the parasitic Amoebophrya species complex revealed by a polyphasic approach. Sci. Rep. 10:2531. doi: 10.1038/s41598-020-59524-z
Chambouvet, A., Alves-de-Souza, C., Cueff, V., Marie, D., Karpov, S., and Guillou, L. (2011a). Interplay between the parasite Amoebophrya sp. (Alveolata) and the cyst formation of the red tide dinoflagellate Scrippsiella trochoidea. Protist 162, 637–649. doi: 10.1016/j.protis.2010.12.001
Chambouvet, A., Laabir, M., Sengco, M., Vaquer, A., and Guillou, L. (2011b). Genetic diversity of Amoebophryidae (Syndiniales) during Alexandrium catenella/tamarense (Dinophyceae) blooms in the Thau lagoon (Mediterranean Sea, France). Res. Microbiol. 162, 959–968. doi: 10.1016/j.resmic.2011.03.002
Chambouvet, A., Morin, P., Marie, D., and Guillou, L. (2008). Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science 322, 1254–1257. doi: 10.1126/science.1164387
Chen, T., Liu, Y., Song, S., and Li, C. (2018). Characterization of the parasitic dinoflagellate Amoebophrya sp. infecting Akashiwo sanguinea in coastal waters of China. J. Eukaryot. Microbiol. 65, 448–457. doi: 10.1111/jeu.12489
Chen, T., Liu, Y., Xu, S., Song, S., and Li, C. (2020). Variation of Amoebophrya community during bloom of Prorocentrum donghaiense Lu in coastal waters of the East China Sea. Estuar. Coast. Shelf Sci. 243:106887. doi: 10.1016/j.ecss.2020.106887
Chen, T., Xiao, J., Liu, Y., Song, S., and Li, C. (2019). Distribution and genetic diversity of the parasitic dinoflagellate Amoebophrya in coastal waters of China. Harmful Algae 89:101633. doi: 10.1016/j.hal.2019.101633
Coats, D. W. (1999). Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 46, 402–409. doi: 10.1111/j.1550-7408.1999.tb04620.x
Coats, D. W., and Bockstahler, K. R. (1994). Occurrence of the parasitic dinoflagellate Amoebophrya ceratii in Chesapeake Bay populations of Gymnodinium sanguinea. J. Eukaryot. Microbiol. 41, 586–593. doi: 10.1111/j.1550-7408.1994.tb01520.x
Coats, D. W., and Park, M. G. (2002). Parasitism of photosynthetic dinoflagellates by three strains of Amoebophrya (Dinophyta): parasite survival, infectivity, generation time, and host specificity. J. Phycol. 38, 520–528. doi: 10.1046/j.1529-8817.2002.01200.x
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi: 10.1126/science.1261605
Deng, Y., Hu, Z., Ma, Z., and Tang, Y. (2016). Validation of reference genes for gene expression studies in the dinoflagellate Akashiwo sanguinea by quantitative real-time RT-PCR. Acta Oceanol. Sin. 35, 106–113. doi: 10.1007/s13131-016-0887-9
Dougherty, E. R., Carlson, C. J., Bueno, V. M., Burgio, K. R., Cizauskas, C. A., Clements, C. F., et al. (2016). Paradigms for parasite conservation: parasite conservation. Conserv. Biol. 30, 724–733. doi: 10.1111/cobi.12634
Farhat, S., Florent, I., Noel, B., Kayal, E., Silva, C. D., Bigeard, E., et al. (2018). Comparative time-scale gene expression analysis highlights the infection processes of two Amoebophrya strains. Front. Microbiol. 9:2251. doi: 10.3389/fmicb.2018.02251
Fensome, R. A., Riding, J. B., and Taylor, F. J. R. (1996). “Dinoflagellates,” in Palynology: Principles and Applications, Vol. 1, eds J. Jansonius and D. C. McGregor (Salt Lake City, UT: AASP Foundation), 107–169.
Guillard, R. R. L., and Ryther, J. H. (1962). Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R. M., Kirkham, A. R., Massana, R., et al. (2008). Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10, 3349–3395. doi: 10.1111/j.1462-2920.2008.01731.x
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
Jessup, D. A., Miller, M. A., Ryan, J. P., Nevins, H. M., Kerkering, H. A., Mekebri, A., et al. (2009). Mass stranding of marine birds caused by a surfactant-producing red tide. PLoS One 4:e4550. doi: 10.1371/journal.pone.0004550
Kang, J., Park, J. S., Jung, S. W., Kim, H.-J., Joo, H. M., Kang, D., et al. (2020). Zooming on dynamics of marine microbial communities in the phycosphere of Akashiwo sanguinea (Dinophyta) blooms. Mol. Ecol. 30, 207–221. doi: 10.1111/mec.15714
Kayal, E., Alves-de-Souza, C., Farhat, S., Velo-Suarez, L., Monjol, J., Szymczak, J., et al. (2020). Dinoflagellate host chloroplasts and mitochondria remain functional during Amoebophrya infection. Front. Microbiol. 11:600823. doi: 10.3389/fmicb.2020.600823
Kim, S. (2006). Patterns in host range for two strains of Amoebophrya (Dinophyta) infecting thecate dinoflagellates: Amoebophrya spp. ex Alexandrium affine and ex Gonyaulax polygramma. J. Phycol. 42, 1170–1173. doi: 10.1111/j.1529-8817.2006.00277.x
Kim, S., and Park, M. G. (2016). Effect of the endoparasite Amoebophrya sp. on toxin content and composition in the paralytic shellfish poisoning dinoflagellate Alexandrium fundyense (Dinophyceae). Harmful Algae 51, 10–15. doi: 10.1016/j.hal.2015.10.016
Kim, S., Park, M. G., Yih, W., and Coats, D. W. (2004). Infection of the bloom-forming thecate dinoflagellates Alexandrium affine and Gonyaulax spinifera by two strains of Amoebophrya (Dinophyta). J. Phycol. 40, 815–822. doi: 10.1111/j.1529-8817.2004.04002.x
Kong, F., Yu, R., Zhang, Q., Yan, T., and Zhou, M. (2012). Pigment characterization for the 2011 bloom in Qinghuangdao implicated ‘brown tide’ events in China. Chin. J. Oceanol. Limnol. 30, 361–370. doi: 10.1007/s00343-012-1239-z
Lima-Mendez, G., Faust, K., Henry, N., Decelle, J., Colin, S., Carcillo, F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348:1262073. doi: 10.1126/science.1262073
Liu, Y., Chen, T., Wang, X., Song, S., and Li, C. (2020). Variation in the photosynthetic activities of the dinoflagellate Akashiwo sanguinea during formation of resting cysts. Mar. Biol. 167:158. doi: 10.1007/s00227-020-03774-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Y., Wohlrab, S., Glockner, G., Guillou, L., and John, U. (2014). Genomic insights into processes driving the infection of Alexandrium tamarense by the parasitoid Amoebophrya sp. Eukaryot. Cell 13, 1439–1449. doi: 10.1128/EC.00139-14
Lu, Y., Wohlrab, S., Groth, M., Glockner, G., Guillou, L., and John, U. (2016). Transcriptomic profiling of Alexandrium fundyense during physical interaction with or exposure to chemical signals from the parasite Amoebophrya. Mol. Ecol. 25, 1294–1307. doi: 10.1111/mec.13566
Marc, L., Dominique, M., Jeremy, S., Jordan, T., Estelle, B., Marc, S., et al. (2021). Dinophyceae use exudates as weapons against the parasite Amoebophrya sp. (Syndiniales). bioRxiv [preprint] doi: 10.1101/2021.01.05.425281
Maxwell, K., and Johnson, G. N. (2000). Chlorophyll fluorescence – a practical guide. J. Exp. Bot. 51, 659–668. doi: 10.1093/jexbot/51.345.659
Mazzillo, F. F. M., Ryan, J. P., and Silver, M. W. (2011). Parasitism as a biological control agent of dinoflagellate blooms in the California Current System. Harmful Algae 10, 763–773. doi: 10.1016/j.hal.2011.06.009
Miller, J. J., Delwiche, C. F., and Coats, D. W. (2012). Ultrastructure of Amoebophrya sp. and its changes during the course of infection. Protist 163, 720–745. doi: 10.1016/j.protis.2011.11.007
Park, M. G., Cooney, S. K., Kim, J. S., and Coats, D. W. (2002a). Effects of parasitism on diel vertical migration, phototaxis/geotaxis, and swimming speed of the bloom-forming dinoflagellate Akashiwo sanguinea. Aquat. Microb. Ecol. 29, 11–18. doi: 10.3354/ame029011
Park, M. G., Cooney, S. K., Yih, W., and Coats, D. W. (2002b). Effects of two strains of the parasitic dinoflagellate Amoebophrya on growth, photosynthesis, light absorption, and quantum yield of bloom-forming dinoflagellates. Mar. Ecol. Prog. Ser. 227, 281–292. doi: 10.3354/meps227281
Park, M. G., Kim, S., Shin, E.-Y., Yih, W., and Coats, D. W. (2013). Parasitism of harmful dinoflagellates in Korean coastal waters. Harmful Algae 30, S62–S74. doi: 10.1016/j.hal.2013.10.007
Paseka, R. E., White, L. A., Van de Waal, D. B., Strauss, A. T., González, A. L., Everett, R. A., et al. (2020). Disease-mediated ecosystem services: pathogens, plants, and people. Trends Ecol. Evol. 35, 731–743. doi: 10.1016/j.tree.2020.04.003
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Scholz, B., Küpper, F., Vyverman, W., Ólafsson, H., and Karsten, U. (2017). Chytridiomycosis of marine diatoms-the role of stress physiology and resistance in parasite-host recognition and accumulation of defense molecules. Mar. Drugs 15:26. doi: 10.3390/md15020026
Schreiber, U., Endo, T., Mi, H., and Asada, K. (1995). Quenching analysis of chlorophyll fluorescence by the saturation pulse method: particular aspects relating to the study of eukaryotic algae and cyanobacteria. Plant Cell Physiol. 36, 873–882. doi: 10.1093/oxfordjournals.pcp.a078833
Sengco, M. R., Coats, D. W., Popendorf, K. J., Erdner, D. L., Gribble, K. E., and Anderson, D. M. (2003). “Biological and phylogenetic characterization of Amoebophrya sp. ex Alexandrium tamarense. Abstract,” in Proceedings of the Second Symposium on Harmful Marine Algae in the U.S. Woods Hole, MA, 57.
Sharon, I., Tzahor, S., Williamson, S., Shmoish, M., Man-Aharonovich, D., Rusch, D. B., et al. (2007). Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 1, 492–501. doi: 10.1038/ismej.2007.67
Spurr, A. R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–43.
Sun, K., Xin, M., Li, Y., Li, R., Tang, X., Wang, Z., et al. (2019). Effects of dark-adaptation time, irradiation time and nutrient level on Fv/Fm in Prorocentrum donghaiense (Dinophyceae) during different growth phases. Phycologia 58, 18–25.
Taylor, F. J. R. (1968). Parasitism of the toxin-producing dinoflagellate Gonyaulax catenella by the endoparasitic dinoflagellate Amoebophrya ceratii. J. Fish. Res. Board Can. 25, 2241–2245. doi: 10.1139/f68-197
Taylor, F. J. R. (Ed.) (1987). Botanical Monographs, 21: The Biology of Dinoflagellates. Oxford: Blackwell Scientific, 785.
Trainer, V. L., Pitcher, G. C., Reguera, B., and Smayda, T. J. (2010). The distribution and impacts of harmful algal bloom species in eastern boundary upwelling systems. Prog. Oceanogr. 85, 33–52. doi: 10.1016/j.pocean.2010.02.003
White, A. E., Watkins-Brandt, K. S., Mckibben, S. M., Wood, A. M., Hunter, M., Forster, Z., et al. (2014). Large-scale bloom of Akashiwo sanguinea in the Northern California current system in 2009. Harmful Algae 37, 38–46. doi: 10.1016/j.hal.2014.05.004
Yih, W., and Coats, D. W. (2000). Infection of Gymnodinium sanguineum by the dinoflagellate Amoebophrya sp.: effect of nutrient environment on parasite generation time, reproduction, and infectivity. J. Eukaryot. Microbiol. 47, 504–510. doi: 10.1111/j.1550-7408.2000.tb00082.x
Keywords: Amoebophrya, dinoflagellate, parasitism, photophysiology, ultrastructure
Citation: Chen T, Liu Y, Hu Z, Song S and Li C (2021) Chloroplast Ultrastructure and Photosynthetic Response of the Dinoflagellate Akashiwo sanguinea Throughout Infection by Amoebophrya sp. Front. Mar. Sci. 8:742498. doi: 10.3389/fmars.2021.742498
Received: 16 July 2021; Accepted: 03 November 2021;
Published: 26 November 2021.
Edited by:
Angel Borja, Technological Center Expert in Marine and Food Innovation (AZTI), SpainReviewed by:
Catharina Alves-de-Souza, University of North Carolina at Wilmington, United StatesFlora Julie Vincent, Weizmann Institute of Science, Israel
Copyright © 2021 Chen, Liu, Hu, Song and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiwen Li, Y3dsaUBxZGlvLmFjLmNu
 Tiantian Chen
Tiantian Chen Yun Liu
Yun Liu Zhangxi Hu
Zhangxi Hu Shuqun Song
Shuqun Song Caiwen Li
Caiwen Li