
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 03 November 2021
Sec. Marine Molecular Biology and Ecology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.742469
 Qikun Xing1†
Qikun Xing1† Miriam Bernard1†
Miriam Bernard1† Sylvie Rousvoal1
Sylvie Rousvoal1 Erwan Corre2
Erwan Corre2 Gabriel V. Markov1
Gabriel V. Markov1 Akira F. Peters3
Akira F. Peters3 Catherine Leblanc1*
Catherine Leblanc1*The filamentous algal endophyte Laminarionema elsbetiae is highly prevalent in European populations of the brown alga Saccharina latissima, but has also been found occasionally in the other kelp species Laminaria digitata. The presence of L. elsbetiae coincides with morphological changes in the hosts such as twisted stipes and deformed blades, however, little is known about the molecular bases of these algal host-endophyte interactions. Using a co-cultivation experiment, we showed that physiological and gene regulation responses, and later endophyte prevalences are different between the main and the occasional host. The contact with the endophyte L. elsbetiae induced a stronger and faster transcriptomic regulation in the occasional host L. digitata after 24 h, from which growth rate was later affected. During the first two days of co-cultivation, only 21 differentially expressed genes (DEGs) were common in both kelps, indicating a crucial difference between the molecular responses of the two hosts. By functional annotation, we identified DEGs related to host-endophyte recognition, defense response and cell wall modification. Our results suggest that expression pattern differences between the two kelps related to the recognition of the endophyte and later defense reactions could explain the variability of observed physiological responses and host-endophyte specificity in kelp natural populations.
Kelps – including large brown macroalgae of the order Laminariales – are major components of rocky intertidal and subtidal habitats (Wynne and Bold, 1985). They do not only serve as food source or habitats for animals, but also provide a substratum for smaller organisms growing on (epiphytes) or inside (endophytes) of their thalli, such as fungi, oomycetes or filamentous algae (Bartsch et al., 2008; Gachon et al., 2010). The prevalence of the latter can be very high, reaching up to 100% of infected individuals in natural kelp population (Lein et al., 1991; Ellertsdottir and Peters, 1997; Bernard M. et al., 2018). Furthermore, filamentous algal endophytes often coincide with disease symptoms in their hosts such as twisted stipes, crippled thalli or a reduced growth of the kelps (Peters, 1996; Gauna et al., 2009; Thomas et al., 2009), but the nature of endophytic relationships in different kelps species, from detrimental to neutral ones, is still an open question. As they have also been reported to lower the commercial value of infected kelps (Yoshida, 1979), endophytes represent a potential threat to the globally increasing seaweed aquaculture (Gachon et al., 2010).
Laminarionema elsbetiae is a filamentous brown alga, which is commonly found as an endophyte in the sugar kelp Saccharina latissima along European coasts (Ellertsdottir and Peters, 1997; Bernard M. et al., 2018; Bernard M.S. et al., 2018). Occasionally it also infects Laminaria digitata, although this kelp is more often infected by other endophyte species belonging to the genus Laminariocolax (Russell, 1964; Kornmann and Sahling, 1994). In Asia, L. elsbetiae has been described infecting the economically important Saccharina japonica, but none of the other kelp species in the direct vicinity, such as Costaria costata or Undaria pinnatifida (Kawai and Tokuyama, 1995). Similarly, kelps in Northern Brittany have shown significant variation in the prevalence of L. elsbetiae according to different host species (Bernard M. et al., 2018). It therefore seems that kelp-endophyte relationships underlie a certain specificity, but the molecular bases of the interaction between kelps and brown algal endophytes remain poorly understood.
In macroalgae, as for most eukaryotic organisms, the activation of defense responses and innate immunity relies on a successful recognition of the potential attacker. This may either involve the perception of exogenous elicitors, i.e., highly conserved patterns in the cell envelope or cell wall, which are found only on the attacker, but not on the host itself, or endogenous elicitors, such as oligosaccharides deriving from the host’s cell wall, which are released following an enzymatic degradation during a biotic attack (Weinberger, 2007). This non se recognition is followed by different inducible defense reactions. A fast and common eukaryotic stress response is the so-called oxidative burst, a release of reactive oxygen species (ROS). ROS do not only have direct cytotoxic effects on attackers, but are also involved in cell-wall strengthening and signaling processes (Hancock et al., 2001; Küpper et al., 2001, 2002). Other defense pathways in kelps that may be activated during biotic interactions through gene expression regulation (Cosse et al., 2009) involve the production of fatty acids and oxylipins and the emission of volatile halogenated organic compounds (Leblanc et al., 2006; La Barre et al., 2010).
A well-studied alga-endophyte pathosystem is the interaction between the red alga Chondrus crispus and the green algal endophyte Ulvella operculata. Sporophytes of C. crispus are regularly infected by U. operculata, but the endophyte cannot penetrate beyond the outer cell layers of the gametophyte of C. crispus (Correa and McLachlan, 1994). U. operculata expresses carrageenolytic activity to degrade and penetrate into the cell wall of C. crispus (Bouarab et al., 1999). Previous studies on C. crispus suggested that the oxidative burst and the oxylipin pathway play an important role in the natural resistance of C. crispus gametophytes against U. operculata (Bouarab et al., 1999, 2004). Electron microscopy observation suggested that the spores of L. elsbetiae penetrate the surface of S. latissima by locally dissolving the cell wall using alginolytic enzymes (Heesch and Peters, 1999). Oligosaccharides released during this interaction could act as endogenous elicitors that could be recognized by the kelp and trigger an activation of defense responses. However, further biochemical and molecular studies are necessary to confirm this hypothesis in kelps. Resistance of L. digitata against the endophyte Laminariocolax tomentosoides was increased after an oxidative burst elicited by endogenous oligoalginate elicitors or by a pre-treatment with arachidonic acid, a polyunsaturated fatty acid (Küpper et al., 2002, 2009). Thus, several different pathways may be involved in the inducible defense of kelps against algal endophytes. In situ surveys in Brittany have shown that S. latissima sporophytes are infected early in their life by L. elsbetiae (Bernard M. et al., 2018). They confirmed different prevalence according to host species, with S. latissima being the principal host and L. digitata being infected only occasionally.
This paper aims at exploring the importance of both early recognition of endophytic presence and inducible defense responses in two different kelp species, in relationship with specific infection patterns observed in natural kelp populations. For this purpose, we investigated and compared the physiological and molecular responses of young sporophytes of the main host S. latissima and the occasional host L. digitata to an infection with L. elsbetiae. We developed a co-cultivation bioassay to measure the kelps’ growth over 14 days in the presence of the endophyte and measured the production of H2O2 in kelp-endophyte co-cultures to follow the oxidative response of the kelps in the presence of endophytic algae. We tested pre-treatment with oligoalginates to further explore L. digitata physiological responses in presence of L. elsbetiae endophyte, using the co-cultivation bioassay. To understand the molecular bases of kelp-endophyte early interaction and its specificity, an RNA sequencing analysis was conducted to compare the regulation of gene expression of both kelp species during the first 2 days of contact with the endophyte L. elsbetiae in laboratory conditions.
Spores of fertile individuals of S. latissima and L. digitata, collected at Perharidy (near Roscoff, 48.73°N, 4.00°W) were released onto cover slips using the hanging-drop technique (Wynne, 1969). The cover slips with settled spores were cultivated at 14°C and exposed to 20 μmol photons s–1m–2 of white light, with a 12 h light/dark cycle. The developing sporophytes were raised in 100 mL Petri dishes with weekly changes of culture medium. For all cultures, filtered autoclaved natural seawater (FSW) was enriched with Provasoli solution (10 mL Provasoli solution/L seawater) (Provasoli, 1968). After 4 weeks, the sporophytes were detached from the cover slips and transferred to 10 L bottles connected to an aeration system. Culture medium in the 10 L bottles was changed weekly. The culture conditions were the same as for the production of sporophytes from settled spores.
Cultures of the filamentous brown algae L. elsbetiae, Laminariocolax tomentosoides, and Microspongium tenuissimum were obtained from the Bezhin Rosko culture collection. They were kept in Petri dishes at 14°C and 5 μmol photons s–1m–2 with monthly changes of culture medium.
Both S. latissima and L. digitata species were co-cultivated with the endophyte for 2 weeks as described below.
Fifteen 2 L bottles were filled with 1.5 L sterile Provasoli-enriched FSW and connected to an aeration system (Supplementary Figure 1A). One kelp sporophyte was transferred to each of the bottles. A hole was punched in the kelp sporophytes at 1 cm distance from the basal meristem using a pipet tip. In the following experiment, the longitudinal growth of the sporophyte blade was measured by monitoring the distance of the hole from the basal meristem with a ruler (Parke, 1948) (Supplementary Figure 1B). The first measurement was done after 3–5 days to ensure that the growth behavior of all sporophytes was similar. Subsequently, a filament of either L. elsbetiae or M. tenuissimum of similar size was added to 5 bottles each (N = 5). M. tenuissimum – a filamentous brown alga which is not endophytic in S. latissima and L. digitata - was used as a control to test a nutrient competition effect. Nothing was added to the remaining 5 bottles (control cultures of sporophytes without endophyte).
After the addition of the filaments (day 0), growth of S. latissima was measured on days 3, 6, 9, 11, and 14. Growth of L. digitata was measured on days 3, 6, 10, and 14. To ensure a sufficient nutrient supply, an amount of 0.5 mL of Provasoli solution per day of experiment was added after each measurement. All co-cultivation experiments were performed at 14°C and 20 μmol photons s–1m–2 with a 12 h light/dark cycle.
The maximum quantum yield of photosystem II (Fv/Fm) was measured on the same days using a JuniorPAM fluorometer (Walz, Germany). The sporophytes were dark-acclimated for 20 min prior to the measurement. After the last measurement, the kelp sporophytes were frozen in liquid nitrogen and kept at −80°C for the molecular detection of the endophyte in the kelp tissue.
The growth curves and Fv/Fm graphs were drawn with GraphPad prism (GraphPad Prism Software, Inc., United States) and SPSS was used for statistical analyses (IBM Corp. Released 2015. IBM SPSS Statistics for Windows, Version 23.0. Armonk, NY, United States: IBM Corp.). Normality of the data and homogeneity of variances were tested with the Shapiro-Wilk test and the Levene test, respectively. Subsequently, data were analyzed with one-way ANOVAs. Significant differences were evaluated with the Tukey post hoc test.
Direct microscopic observation was not adapted to the observation of endophyte spore settlements after 2 weeks, as first tested on two L. digitata plantlets, co-cultivated with endophytes. We therefore used a qPCR-based molecular approach: DNA extraction and qPCR quantification of endophytes in the hosts were performed as described previously (Bernard M. et al., 2018). In total, up to 15 individuals of S. latissima and 13 individuals of L. digitata from three independent co-cultivation bioassays with L. elsbetiae as well as 3 randomly chosen control sporophytes of each species were analyzed by qPCR for the detection of L. elsbetiae DNA using specific ITS primers. Cycle threshold (CT) values were calculated with the LightCycler 480 Software (Roche, Germany): endophyte DNA was amplified with a maximum of 33 cycles, and above 34 cycles, it was considered as undetectable.
Sixteen L. digitata sporophytes raised in laboratory culture were transferred to small glass beakers, filled with 50 mL autoclaved FSW. 150 μg/ml of oligoguluronate blocks (GG, prepared from L. hyperborea according to Haug et al. (1974) were added to 8 sporophytes. All beakers were placed on a shaker for 3 h (100 rpm) and the occurrence of an oxidative burst was measured as described below. After the incubation, the sporophytes were washed by transferring them to new beakers containing 50 mL autoclaved FSW and shaking for another 15 min. This washing step was repeated twice. A hole was punched in 1 cm distance of the meristem in the kelps and they were transferred to 2 L bottles. The first measurement was done after 3 days to assure that growth behavior of all sporophytes (control and GG-treated) was similar. Then, filaments of L. elsbetiae were added to 4 of the GG pre-treated and to 4 of the untreated L. digitata sporophytes. Growth was measured as described above on days 3, 7, 10, and 14. Statistical analysis were performed as described above.
The net production of H2O2 in seawater surrounding kelp-endophyte co-cultures was determined using a luminol chemiluminescence method (Küpper et al., 2001). After measuring the fresh weight of young sporophytes of S. latissima and L. digitata, they were transferred to glass beakers containing 50 mL seawater and placed on a shaker (100rpm). The experimental set-up consisted of a control (only S. latissima or L. digitata), both kelps co-cultivated with the endophytes L. elsbetiae or L. tomentosoides, and 50 mL of seawater containing only filaments of L. elsbetiae or L. tomentosoides. As a positive control, 150 μg/mL of GG were added to another glass beaker containing 50 mL of seawater and a sporophyte of either S. latissima or L. digitata.
150 μL of seawater were taken as sample for each measurement. Measurements were done before starting the treatment (t = 0), and 2, 4, 6, 8, 10, 15, 20, 25, and 30 min after the addition of the endophytes or GG. For each measurement, 50 μL of 20 U.mL–1 horseradish peroxidase, dissolved in pH 7.8 phosphate buffer, and 100 μL of 0.3 M luminol (5-amino-2,3-dihydro-1,4-phtalazinedione) were added automatically to the sample by two injectors of the GloMax 20/20 Luminometer (Promega, United States). Chemiluminescence was measured immediately after the injection with a signal time of 1 s. A standard calibration curve from 0.1 μM to 20 mM H2O2 was drawn to determine the concentration of H2O2 in the seawater samples. H2O2 production by the kelp per g fresh weight was estimated by integrating the total amount of H2O2 monitored over 30 min and expressed as log2-transformed fold changes between control and treatments. The experiment was repeated 3 times and a one-sample-t-test was used for statistical analysis.
For transcriptomic analysis, 16 bottles were filled with 1.5 L autoclaved Provasoli enriched FSW and adapted to an aeration system. One sporophyte of S. latissima (3–5 cm) was added to each bottle. After 24 h of acclimation time, filaments of L. elsbetiae were added to 8 of the bottles. The four individuals of the control group and 4 individuals co-cultivated with L. elsbetiae were taken after 24 and 48 h. The kelp sporophytes were blotted dry with tissue paper, frozen in liquid nitrogen and stored in −80°C until RNA extraction. The same experimental set-up was used for L. digitata sporophytes (Supplementary Figure 1C).
RNA was extracted as described by Heinrich et al. (2012) with a combination of a classical CTAB-based method and the RNeasy Mini kit (QIAGEN, Hilden, Germany) including an on-column DNA digestion. Quantity and purity of the extracted RNA were tested on a NanoDropTM spectrophotometer (Thermo Fisher Scientific Inc., Waltham, United States) and on a 2% agarose gel.
Based on the quality and concentration, three replicates of each condition and each kelp species were chosen for commercial library preparation and Illumina paired-end sequencing (HiSeq3000) at the Plateforme Génomique du Genopole Toulouse Midi-Pyrénées GeT (France).
The quality of the Illumina reads was checked using FastQC (Andrews, 2010). Reads were cleaned by removing adapters, low quality reads (Phred score < 33) and short reads (<50 nucleotides) with Trimmomatic (Bolger et al., 2014) and residual rRNA was removed with SortMeRNA (Kopylova et al., 2012). Another quality check was performed with FastQC on the processed reads to ensure that high quality reads were obtained through the cleaning steps. An additional cleaning step was done by removing reads of the endophyte by mapping to the transcriptome of L. elsbetiae, produced in the context of the Phaeoexplorer project1.
A de novo transcriptome assembly was created for both kelp species separately based on the pooled processed control reads using Trinity (Grabherr et al., 2011) with the default options. The quality of the assembly was assessed by re-mapping the cleaned reads using the bowtie2 aligner (Langmead and Salzberg, 2012). Transcript abundance was estimated in TPM with PERL scripts implemented in Trinity. The quality of the transcriptome assemblies was evaluated by using BUSCO v2.0 (Simão et al., 2015) with eukaryote dataset and redundancy further reduced according to the TPM value.
Gene annotation was performed with a Blastx search against the NCBI-NR (Release 239) and the Uniprot databases (Release 2020_02) with an E-value cut-off of 10–5. Furthermore, genes were assigned to 2nd level GO subcategories within the three root categories molecular function, cellular component and biological process using Blast2GO (Conesa et al., 2005).
Differential gene expression between the control and the co-cultivation treatments was determined separately for the 24 and 48 h samples using DESeq2 (Love et al., 2014). Log2 fold change values ≥ 0.7 and ≤ −0.7 with a p-value < 0.01 were considered to be up- and downregulated, respectively. Heat maps were plotted using the R package pheatmap.
The genes that were differentially expressed in both species were compared with Blastn (E-value cut-off of 10–5) against each other in order to identify common DEGs.
No significant differences in growth occurred within 2 weeks of co-cultivation of S. latissima with the non-endophytic M. tenuissimum and endophytic L. elsbetiae (Figure 1A and Supplementary Table 1).
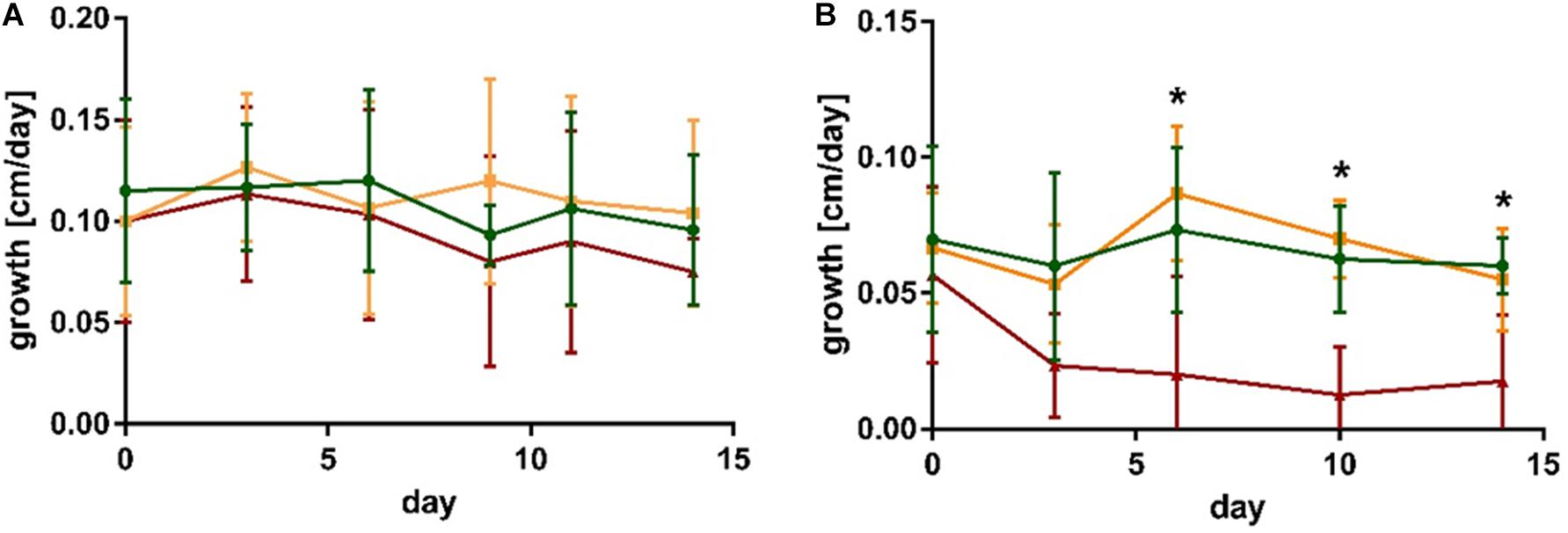
Figure 1. Growth of (A) S. latissima and (B) L. digitata in control conditions (green), in presence of the non-endophytic M. tenuissimum (yellow) and the endophyte L. elsbetiae (red). The presented values are mean values with standard deviation (N = 5). Significant differences are indicated by asterisk (see Supplementary Table 1).
In the case of L. digitata, a significant difference between the treatments occurred 6 days after the addition of L. elsbetiae (Figure 1B and Supplementary Table 1). The growth of L. digitata decreased significantly (Figure 1B) as compared to the other treatments after 6 days of co-cultivation (Supplementary Table 1, one-way ANOVA, p = 0.013) and the difference persisted until the end of the experiment (Supplementary Table 1). There was no significant effect of co-cultivation with M. tenuissimum on the growth of L. digitata.
No significant differences in Fv/Fm occurred within S. latissima or L. digitata alone or in co-cultivation with M. tenuissimum and L. elsbetiae (Supplementary Table 1).
At the end of the experiment, i.e., after 2 weeks of co-cultivation, DNA of the endophyte was detected in 7 out of the fifteen S. latissima DNA samples using qPCR Laminarionema specific primers, whereas it was only detected in 4 out of 13 samples of the L. digitata DNA samples. No L. elsbetiae DNA was detected by qPCR in any of the controls.
Co-cultivation of L. digitata sporophytes with the endophyte L. elsbetiae resulted in a significant decrease of growth from day 3 until day 14 (red line in Figure 2 and Supplementary Table 2), as already described before (control as green line in Figure 2). However, the addition of L. elsbetiae did not have any effect on the growth of L. digitata sporophytes that had been pre-treated with GG 3 days before the co-cultivation was started (golden line, Figure 2). There was no effect of the GG elicitation pre-treatment alone on the growth of L. digitata (gray line in Figure 2).
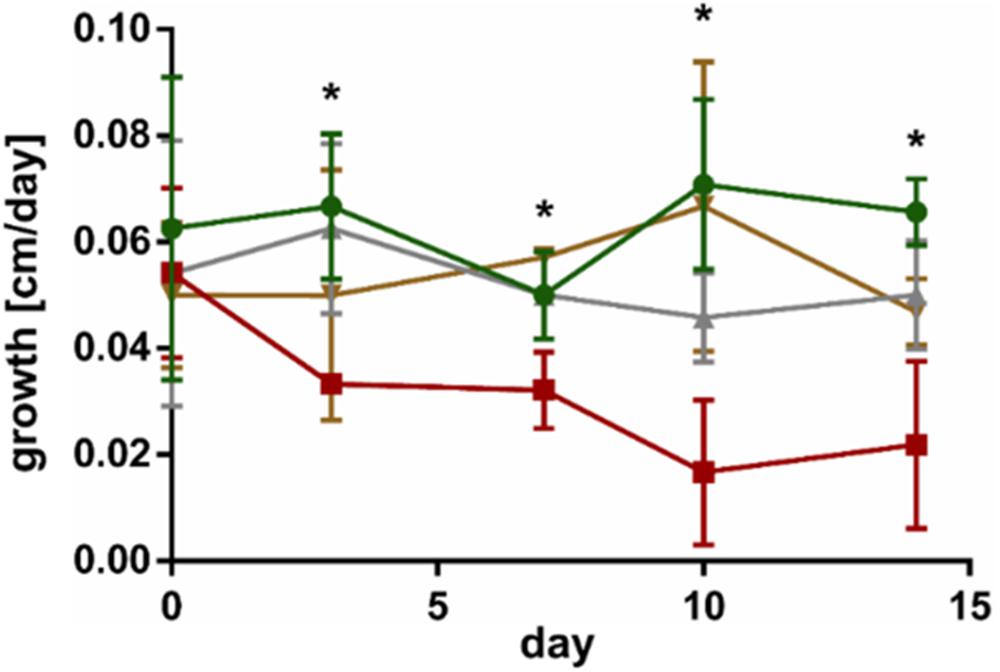
Figure 2. Growth of L. digitata without GG pre-treatment in control conditions (green) and in co-cultivation with L. elsbetiae (red) and of L. digitata with GG pre-treatment in control conditions (gray) and with L. elsbetiae (gold). Significant differences are indicated by asterisks (see Supplementary Table 2). The presented values are mean values with standard deviation (N = 4).
As previously demonstrated by Küpper et al. (2001), oligoguluronate (GG) blocks triggered an oxidative burst in both kelp species, which is indicated by a significant fold change of H2O2 release as compared to the control (Table 1).
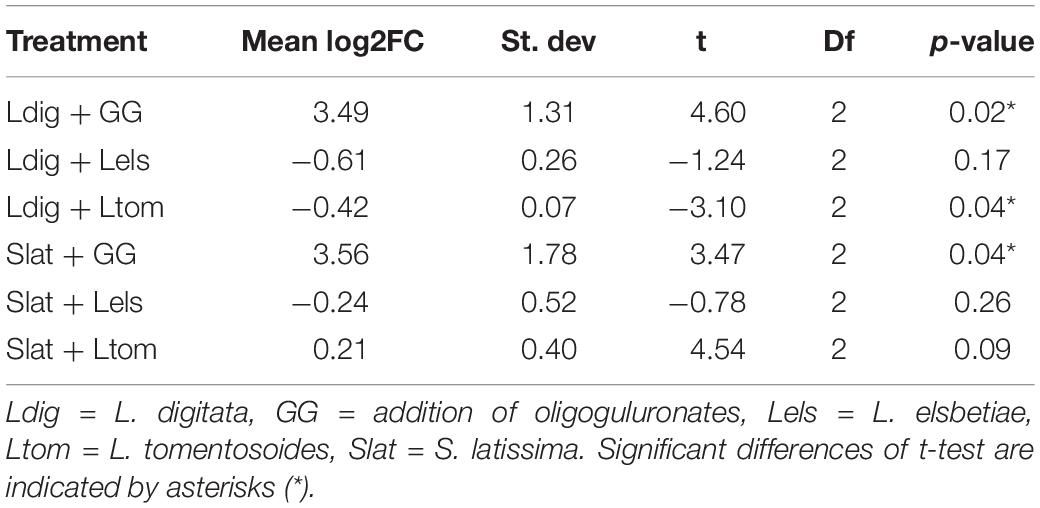
Table 1. Mean values of log2FC in H2O2 content monitored during 30 min in seawater surrounding the treated kelps as compared to the control (N = 3) and results of the statistical analysis using a one-sample t-test.
The addition of L. tomentosoides to S. latissima resulted in a slight increase of H2O2 concentration in the seawater (log2FC = 0.21, one-sample t-test, p = 0.09, Table 1). When added to L. digitata, on the other hand, L. tomentosoides caused a significant decrease of the H2O2 concentration in the seawater (log2FC = −0.42, one-sample t-test, p = 0.04, Table 1). No significant changes in the H2O2 concentration were observed after the addition of L. elsbetiae to both kelp species.
The cleaned RNA sequencing reads of S. latissima and L. digitata were de novo assembled by Trinity and the general features of these two de novo transcriptomes are presented in Supplementary Table 3. 90% of total expression was present in 24,733 and 34,251 transcripts, respectively. The results of the BUSCO analysis revealed a near-complete gene sequence information for two transcriptomes with 82.4 and 84.4% complete BUSCO matches with eukaryotic dataset in S. latissima and L. digitata, respectively.
Overall, the distribution of GO terms for the three root categories “Molecular Function,” “Cellular Component,” and “Biological process” were similar for the assembled transcriptomes of S. latissima and L. digitata (Supplementary Figure 2). Within the molecular function category, most hits were assigned to catalytic activity and binding. Within the biological process root, most genes belonged to metabolic and cellular processes (Supplementary Figure 2). Overall, 47.34% of the obtained genes of S. latissima could be annotated whereas the annotation rate was slightly lower in L. digitata (45.66%).
The comparison between control and co-cultivated conditions identified 107 and 155 differentially expressed genes (DEG) at both time points in S. latissima and L. digitata, respectively (Supplementary Figure 3). Only six DEG were detected in S. latissima in presence of L. elsbetiae after 24 h, whereas 46 genes were significantly differentially expressed in L. digitata between control condition and co-culture with the endophyte (Supplementary Table 4). At 24 h, the majority of DEGs were downregulated in both species (S. latissima, 5 genes, i.e., 83.33% of DEG, L. digitata: 29 DEG, i.e., 63%), with stronger repression for those identified in S. latissima and for 16 DEG of L. digitata (Figure 3A). Unlike S. latissima, where only one gene was moderated upregulated, 11 genes featured log2FC value above 5 compared to control in L. digitata (Figure 3A).
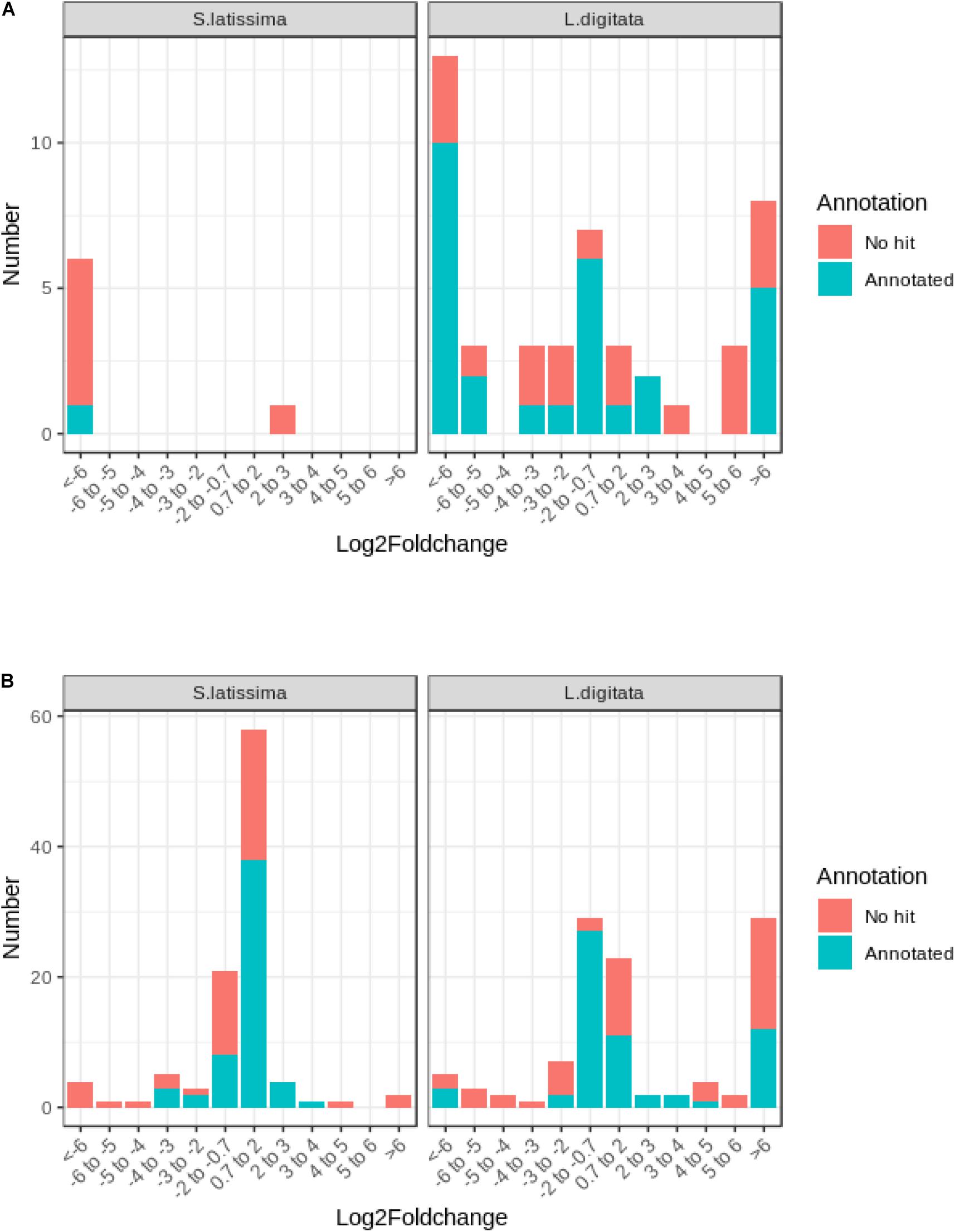
Figure 3. Frequency distribution of log2FC among differentially expressed genes in S. latissima and L. digitata after (A) 24 h and (B) 48 h of co-cultivation with L. elsbetiae.
After 48 h, more changes in gene expression occurred and most detected DEG were upregulated in both species (Figure 3B). In S. latissima, log2FC values of the 101 identified DEG ranged from 11.41 to −28.27 (Supplementary Table 4), but the majority (78.22%) showed moderate expression log2FC between 2 and −2 (Figure 3B and Supplementary Table 4). In L. digitata, log2FC values of the 109 DEG ranged from 16.51 to −14.28 (Supplementary Table 4): 52 DEGs (47.71%) showed moderate log2FC between 2 and −2 and 29 DEGs (26.61%) were strongly upregulated (log2FC > 6) (Figure 3B, Supplementary Table 4). Among 262 DEGs identified in S. latissima and L. digitata in the 2-days cocultures with the endophyte, only 21 homologous genes were shared by both kelps.
Similar to the whole transcriptome analysis, an important part of the differentially expressed genes of both kelp species (between 39.0 to 45.0%) did not have a match through Blastx search in the available protein databases, as represented on Figure 3. In the case of S. latissima, after 24 h of co-cultivation with the endophyte, only one gene had a match through Blastx search as a conserved unknown Ectocarpus protein (Figure 3A, Supplementary Table 4). In L. digitata, among the 23 DEGs annotated based on at least one database after 24 h of the addition of L. elsbetiae, one putative respiratory burst oxidase homolog protein was significantly strongly upregulated (Table 2), and 9 genes corresponded to conserved unknown or hypothetical proteins in Ectocarpus genome (Supplementary Table 4).
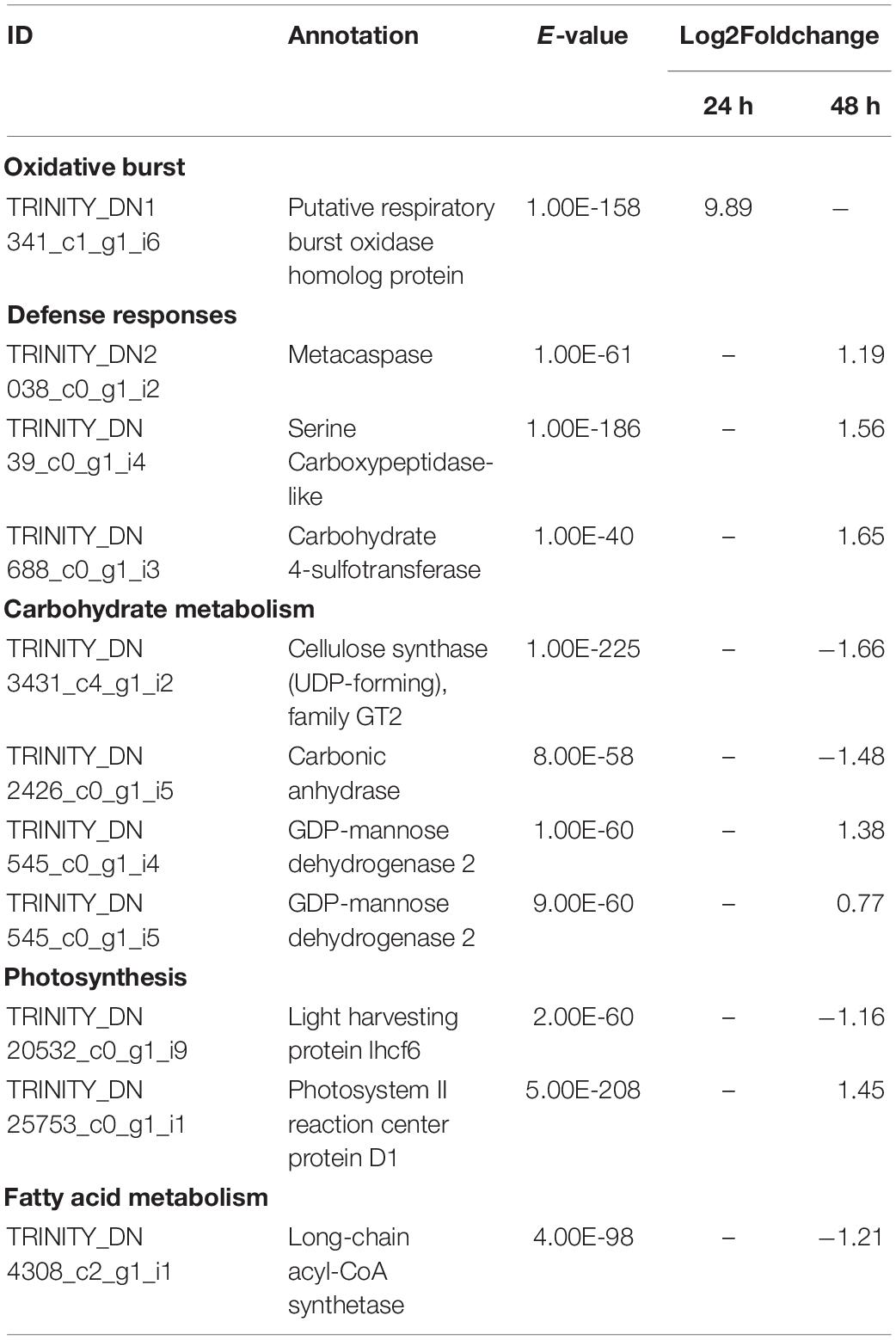
Table 2. Selected significant differentially expressed genes (DEGs) in L. digitata after co-cultivation with L. elsbetiae for 24 and 48 h vs. the control group (q ≤ 0.01, | log2 FC|≥ 0.7).
After 48 h of co-cultivation of S. latissima with the endophyte, 19 DEGs corresponded to conserved unknown or hypothetical Ectocarpus proteins. A functional putative annotation was retrieved for only 38 genes out of 101 DEG (see Supplementary Table 4). Among those DEGs, several genes related to cell wall metabolism were significantly up-regulated, including five mannuronan C-5-epimerases, an endo-1,3-beta-glucanase, and an alginate lyase (Table 3). Two vanadium-dependent bromoperoxidases and a PAP2/haloperoxidase-like protein were also up-regulated, which could be related to antioxidative responses. In addition, two defense-related genes were identified as DEGs. One upregulated gene encoding a putative respiratory burst oxidase homolog protein might be involved in the oxidative burst and another gene encoding a putative LRR receptor-like serine/threonine-protein kinase related to PAMPs recognition was downregulated in the presence of L. elsbetiae. Additionally, three genes that might be involved in oxylipin signaling pathways were up-regulated, including one gene encoding lipoxygenase and two genes encoding lipases (Table 3). In L. digitata, a putative annotation was obtained for 60 DEGs (Supplementary Table 4). 31 DEGs were annotated as conserved unknown or hypothetical Ectocarpus proteins whereas a putative functional annotation was retrieved for 25 DEGs (see Supplementary Table 4). Among those DEG putative annotations, there were three up-regulated genes that might be related to defense responses such as metacaspase, serine carboxypeptidase-like and carbohydrate 4-sulfotransferase genes (Table 2). The co-cultivation with endophyte repressed the expression of genes involved in photosynthesis and carbohydrate metabolism such as a light harvesting protein lhcf6, a carbonic anhydrase and a cellulose synthase. However, a photosystem II reaction center protein D1 gene and two GDP-mannose dehydrogenase genes were up-regulated during the co-cultivation. The co-cultivation also induced the down-regulation of a gene involved in fatty acid metabolism, a long-chain acyl-CoA synthetase putative gene. Furthermore, a gene coding for a protein homologous to one of those upregulated in the Ectocarpus imm-mutant and a gene with high similarity to the Ectocarpus virus gene (EsV 1–7), encoding for a cystein-rich protein, were strongly upregulated in the co-cultivation treatment with L. elsbetiae (Supplementary Table 4).
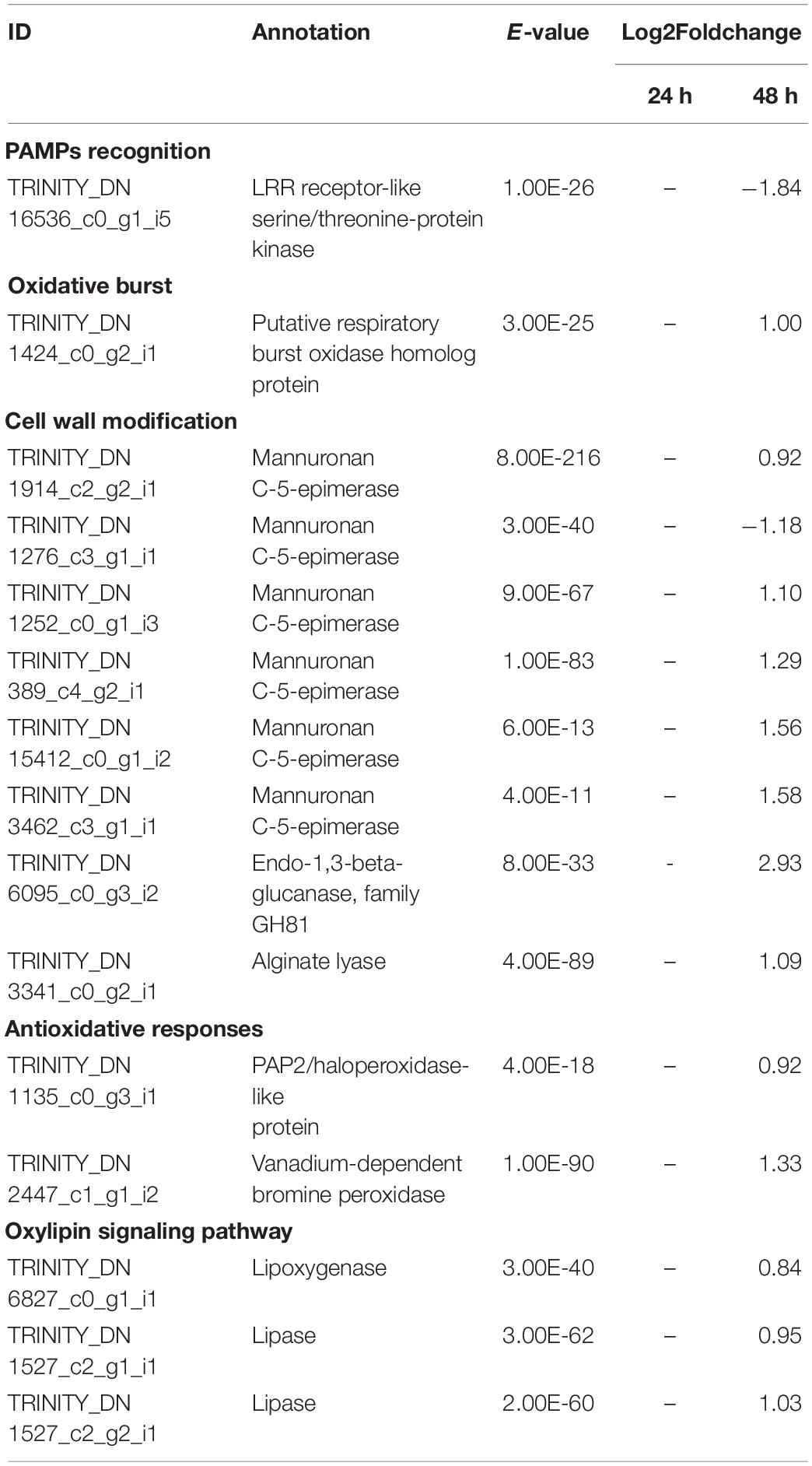
Table 3. Selected significant differentially expressed genes (DEGs) in S. latissima after co-cultivation with L. elsbetiae for 24 and 48 h vs. the control group (q ≤ 0.01, | log2 FC|≥ 0.7).
In Brittany, L. elsbetiae is mainly found in S. latissima, whereas L. digitata is not only infected less frequently, but also with lower severity (Bernard M. et al., 2018; Bernard M.S. et al., 2018). However, until now, these endophytic interactions have rarely been studied apart from epidemiological surveys in natural kelp populations. This study provides a first insight into the bases of kelp-endophyte interactions on both physiological and molecular level and highlights the complex and specific cross-talk occurring after the recognition of endophytes by kelps which could explain host specificity.
Previously, it was reported that algal endophytes can reduce the growth of their hosts by up to 70%, as it has been shown for the red algal endophyte Hypneocolax stellaris in its host, the rhodophyte Hypnea musciformis (Apt, 1984). Here we show that an effect on growth depends on the host species, as different physiological responses occurred between the two hosts S. latissima and L. digitata challenged by the endophyte L. elsbetiae.
The growth of the main host S. latissima was not affected by the endophyte during the 2 weeks of co-cultivation, and L. elsbetiae DNA was detected in half of the sporophytes at the end of the experiment. Although this detection does not prove that endophytic filaments were already growing inside of the kelp thallus, at least spores of the endophytes were likely to be attached to the kelp tissue by the end of the experiment. Thus, a direct and tiny contact between the endophyte and the kelp had been established, as already observed (Heesch and Peters, 1999), but without directly affecting the growth of S. latissima.
In contrast, the growth of the occasional host L. digitata was significantly reduced after a few days of co-cultivation, when only one third of the plantlets featured L. elsbetiae DNA contamination. As the filamentous brown alga, M. tenuissimum, did not have any effect on both kelps, nutrient competition could be excluded as a possible cause of the growth reduction. On the transcriptome level, the expression of several growth-related genes in L. digitata was impacted by the co-cultivation with L. elsbetiae. One gene encoding a putative cellulose synthase was significantly down-regulated 48 h after the addition of L. elsbetiae, and this might decrease cell elongation and division rate in young sporophytes. In Arabidopsis thaliana, lack of cellulose synthase resulted in a reduced cell growth rate of young seedlings (Hu et al., 2018). A gene coding for carbonic anhydrase, an essential enzyme for CO2 fixation, was also down-regulated in L. digitata, suggesting an impact on carbon uptake and photosynthesis. It has been reported that endophytic pathogens can impair the efficiency of energy transfer from the light harvesting complexes to the reaction center of PS II in land plants (Luque et al., 1999; Guidi et al., 2007) and in the seagrass Zostera marina (Ralph and Short, 2002). In L. digitata co-cultivated with the endophyte, we also observed differential expression of photosynthesis-related genes. The down-regulation of a light harvesting protein gene lhcf6 suggests photoinhibition, which may be counteracted by the up-regulation of a photosystem II reaction center protein D1 gene as a key step in the repair of photodamaged PSII (Murata et al., 2007). Indeed, PAM measurements did not indicate any impact on the performance of photosystem II of the two kelp species, during the 2 weeks of co-cultivation. According to our data, the presence of L. elsbetiae did not strongly affect its main host, while it had a significant impact on photosynthesis and growth physiology of L. digitata, its occasional host. The decreased growth rate could be due to the trade-offs between algal defense and growth metabolisms during biotic interactions, as already shown in land plants (Züst and Agrawal, 2017).
In our study, the endophyte-detection results showed that the association with its host tends to be lower for L. digitata than for S. latissima after 2 weeks of co-cultivation, indicating that the mechanism of early resistance to the infection with L. elsbetiae may differ between the two hosts. In addition to different regulations of growth or photosynthesis pathways, the RNA-seq data provide insights of the defense-related transcriptional mechanisms occurring during the first infection step. Only a small portion (0.4%) of both transcriptomes showed significant expression differences, as already observed during the early steps of biotic interactions such as grazing in brown algae (Flöthe et al., 2014; Ritter et al., 2017) or bacterial infection in red algae (de Oliveira et al., 2017). Distinct transcriptomic regulation patterns appeared between the two kelps species, according to time and intensity of gene regulation. After 24 h of co-cultivation with the endophyte, only few genes were differentially regulated in S. latissima, whereas L. digitata already featured a stronger transcriptomic response. Although most of the DEGs were down-regulated in both species, a small portion of genes in L. digitata were already highly up-regulated after 24 h. After 48 h, almost the same amount of DEGs were found in both kelp species. Our results therefore suggest that the first contacts with the endophyte induced a faster and stronger transcriptomic response in the occasional host, L. digitata, than in the main host, S. latissima. The majority of differentially expressed genes were unique in the two hosts and only twenty-one genes were commonly differentially expressed in both kelps. This confirms that overall the two kelps react differently to the contact with the endophyte.
The overall rate of functional annotations was very low, as it is usually the case for non-model organisms (Armengaud et al., 2014). It is therefore difficult to fully understand the molecular responses of the two kelps toward an infection with L. elsbetiae, and further investigations will be necessary to characterize the functions of some conserved or specific unknown genes. However, some interesting gene candidates were annotated that are related to defense responses. Putative respiratory burst oxidase homolog protein (rboh) genes were strongly upregulated in the L. digitata samples after 24 h of the addition of the endophyte (Figure 4A), and in S. latissima after 48 h, but with lower log2FC value (Figure 4B). Rbohs are involved in oxidative bursts of plants (Torres and Dangl, 2005) and have been shown to be induced by elicitors and other stress conditions in some macroalgae (Luo et al., 2015; Wang et al., 2018). The local oxidative burst is known to be the earliest conserved response following recognition of the attack(er) in the plant and macroalgal innate immunity systems and the reactive oxygen species produced by Rbohs act as toxic compounds and/or as defense signals (Potin et al., 2002). The comparison of the expression of the putative rboh genes in the two species suggests that the immunity responses in L. digitata could be much quicker and stronger than the one in S. latissima after the first contact with the endophyte. The 24-h delay of transcriptomic regulations in S. latissima might be due to the repression of the attack(er) recognition in this species. A gene encoding LRR receptor-like serine/threonine-protein kinase was indeed downregulated in S. latissima, and not in L. digitata, after 48 h of the co-cultivation with L. elsbetiae. LRR receptor-like serine/threonine-protein kinases play a central role in the signaling of pathogen recognition in land plants (Afzal et al., 2008). It has been shown that viruses and bacteria have developed mechanisms to suppress the expression of genes involved in pathogen recognition in plants before an infection (Stack et al., 2005; Akira et al., 2006; Boller and He, 2009). The downregulation of this gene in S. latissima could result in an incomplete or inaccurate recognition of L. elsbetiae as an external biotic attack, avoiding the induction of immunity defense responses and leading to stronger infection. In S. latissima, no other putative known defense-related genes were found to be upregulated. Based on transcriptomic regulations, a different scenario seems to occur in L. digitata, where several defense-related genes were upregulated. For instance, a putative carbohydrate sulfotransferase gene, upregulated after 48 h, might have an effect on the recognition between host and endophyte. Carbohydrate sulfotransferases act by adding sulfonyl groups from 3′phosphoadenosine-5′phosphosulfate (PAPS) to a glycoside receptor and the sulfated carbohydrate can serve as a source of extracellular biological information to cells, influencing recognition events and signaling pathway (Bowman and Bertozzi, 1999). Metacaspase belongs to a cysteine protease family and has an essential role in programmed cell death caused by the immunity response of plants (Spoel and Dong, 2012). The upregulation of these genes were also observed in terrestrial plants during the contact with pathogens (Hoeberichts et al., 2003; Jayaswall et al., 2016). Furthermore, a serine carboxypeptidase-like protein (SCPL) was upregulated in L. digitata. SCPLs comprise a large family of protein hydrolyzing enzymes that play roles in multiple cellular processes. In addition, some SCPLs are involved in the biosynthesis of antimicrobial compounds and disease resistance in land plants (Liu et al., 2008; Mugford et al., 2009). The SCPL we found might be also involved in the biosynthesis of defense-related compounds. Their strong upregulation in L. digitata suggests that - unlike S. latissima - L. digitata recognizes the endophyte as a thread and activates defense reactions and efficient immunity responses, which could explain the lower infection patterns in natural L. digitata populations (Bernard M. et al., 2018; Bernard M.S. et al., 2018). In young plantlets, the activation of defense reactions could consume energy, resulting in the decrease of growth rate during the first contacts with endophytes.
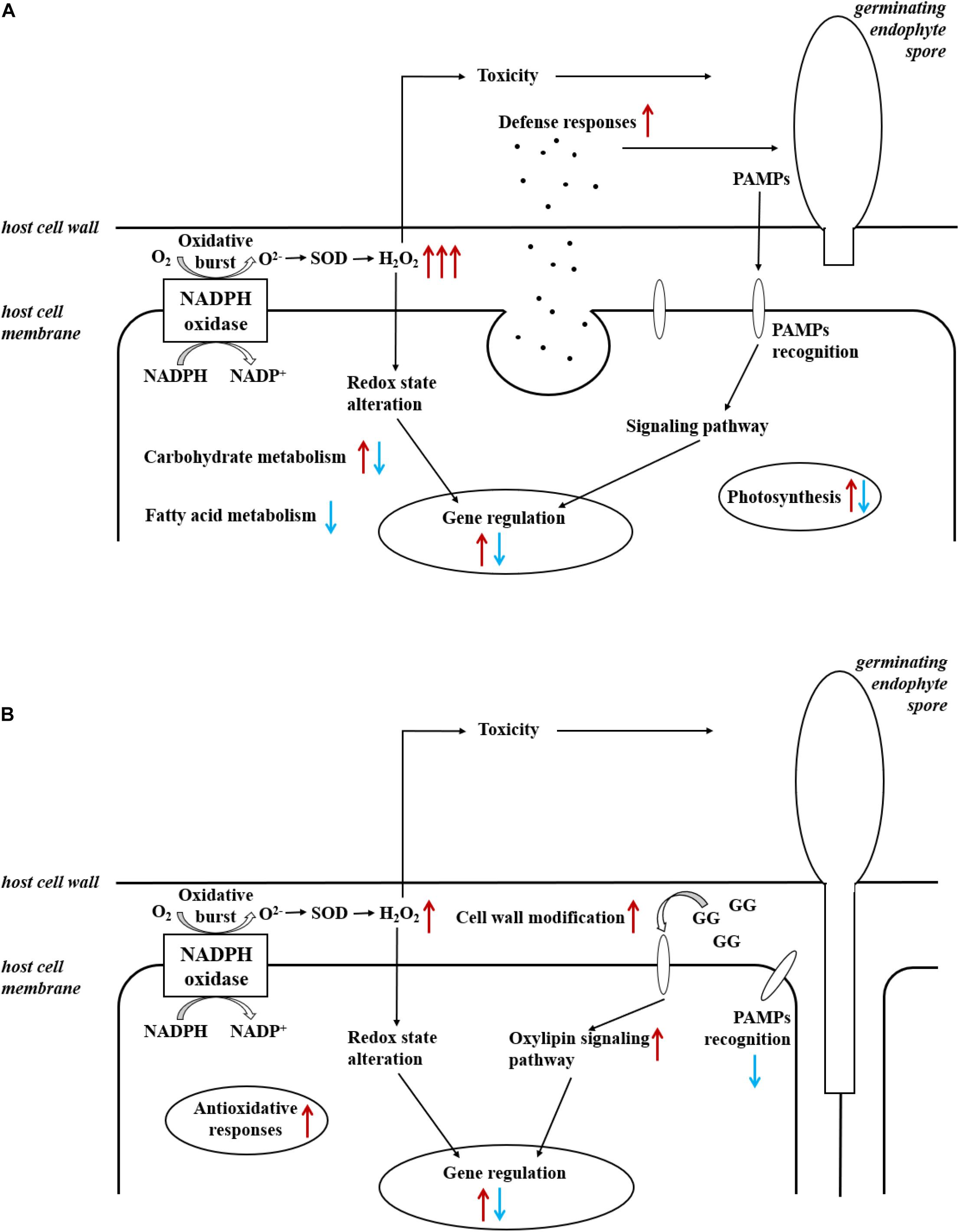
Figure 4. Summary of induced responses in (A) L. digitata and (B) S. latissima during the interactions with L. elsbetiae. Red and blue arrows indicated, respectively, up- and downregulation of differentially expressed genes, in relationships with main cellular metabolisms (see also Tables 2, 3).
Above we hypothesized that the growth of L. digitata in the presence of L. elsbetiae was slowed down due to an activation of energy-costing defense reactions. Growth behavior of L. digitata in co-cultivation with the endophyte after GG elicitation, however, was similar to the controls. An elicitation with GG has been shown to strongly induce defense-related genes in L. digitata (Cosse et al., 2009). The pre-treatment could therefore restore normal growth behavior of the kelp in co-culture with the endophyte, due to the activation of the kelp defense reactions prior to the co-cultivation. Previously, resistance of L. digitata sporophytes against the filamentous algal endophyte L. tomentosoides has been induced by a GG pre-treatment (Küpper et al., 2002). The authors suggested that the oxidative burst caused by the addition of GG activated secondary, long-term defense mechanisms in the kelps that in a second time lead to a strengthening of their cell wall, thereby building up a mechanical barrier against the endophyte. In the field, the protection of juvenile sporophytes by GG elicitation was also observed in S. latissima, which reduced the density of endobionts and the number of bacterial cells on sporophytes (Wang et al., 2019).
Our experimental set-up, on the other hand, was rather monitoring the initial steps of kelp-endophyte interactions, mainly during the spore settlement and germination. The results suggest rapid and direct defense mechanisms that may have been enhanced through a GG-induced priming effect, as already observed in L. digitata (Thomas et al., 2011). Future studies using the qPCR bioassay could test a potential effect on long-term resistance of the kelp against the endophyte.
In the co-cultivation with L. elsbetiae, many cell-wall modification-related genes were upregulated in S. latissima. Five mannuronan C-5 epimerase (MC5E) were upregulated in S. latissima. MC5E is catalyzing the last step of alginate biosynthesis, i.e., the conversion of mannuronic to guluronic acids (Michel et al., 2010). The upregulation of MC5E indicates activation of alginate synthesis and modification of the cell wall toward strengthening in S. latissima upon co-cultivation (Figure 4B). Other genes associated with cell-wall metabolism were endo-1,3-beta-glucanase and alginate lyase. Endo-1,3-beta-glucanase catalyzes the hydrolysis of 1,3-beta-D-glucosidic linkages in callose, laminarin, and various carbohydrates found in the cell wall of plants (Casu et al., 2007). Alginate lyase can degrade alginate by cleaving the glycosidic bond through a β-elimination reaction, generating an oligomer with a 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the non-reducing end (Kim et al., 2011). The functions of both genes are related to the degradation of the cell wall. The fact that these enzymes were upregulated 48 h after the infection with the endophyte suggests an enhanced activity of either decomposing the host cell wall or degrading the endophyte cell wall. Two DEGs in S. latissima, imm upregulated 3 and EsV 1–7, were previously identified as members of a putative regulatory cascade with their potential life-cycle-related roles in E. siliculosus (Peters et al., 2008; Macaisne et al., 2017). The strong up-regulation of these two genes might lead to the regulation of the cell-cycle in order to repair damaged cell wall, which was also observed in the wounding responses of land plants (Dombrowski et al., 2020). The other gene regulations occurring after 48 h could be related to the accumulation and perception of cell wall degradation products, such as oligoalginates (Figure 4B). Indeed, some stress-response related genes were upregulated in S. latissima such as vanadium dependent bromoperoxidases (vBPO) and vanadium dependent iodoperoxidases (vIPO) involved in halide metabolism of brown algae and upregulated during abiotic and biotic oxidative responses (Cosse et al., 2009; Strittmatter et al., 2016; Salavarría et al., 2018). Other upregulated genes were putatively involved in calcium and oxylipin signaling pathways, two key pathways activated upon biotic and abiotic stress (Knight, 1999; Eckardt, 2008; Zhang et al., 2014).
The results presented in this study demonstrate that the main host S. latissima and the occasional host L. digitata both react to the endophyte L. elsbetiae on physiological and transcriptomic levels. These biotic interactions were not neutral, but the reactions of the two hosts showed significant different patterns during the first hours of contact with the endophytes. Based on dynamics of gene regulations, we propose that differences in the early recognition and the subsequent induced defense reactions in both kelps could explain the important prevalence of L. elsbetiae in S. latissima, and the lower infection of L. digitata in natural kelps populations.
During biotic interactions, the fitness of the endophyte could be related to its ability to infect its host whereas the fitness of the kelp host could be linked to its capacity to resist or adapt to the infection. Therefore, both partners are underlying strong selective pressures which are driving and accelerating co-evolution. The comparison of physiological interactions between the brown algal endophyte L. elsbetiae and its different kelp hosts here provides an experimental system to study defense responses in kelps upon endophyte infection and further explore co-evolution of algal endophytes and hosts.
The RNA-seq datasets analyzed during the current study are available in the EMBL databases under the accession ID: PRJEB37483. All other data generated during this study are either included in this published article and its supplementary information files, or available from the corresponding author on request.
QX, MB, AP, and CL conceived the experiments. QX, MB, and SR performed the experiments, processed algal, and RNA samples. QX and EC performed the bioinformatic analyses. QX, MB, GM, and CL interpreted the data. QX and MB wrote the first draft of the manuscript. All authors contributed to the revisions, read and approved the final manuscript.
This work has benefited from the support of the European Union’s Horizon 2020 Research and Innovation Program under the grant agreement N°727892, GENIALG (GENetic diversity exploitation for Innovative macro-228 ALGal biorefinery, http://genialgproject.eu/) for data analysis and under the Marie Składowska-Curie grant agreement N° 642575 (MB’s Ph.D. work), from the Blue Granary company (QX’s Ph.D. work) and from the Centre National de la Recherche Scientifique (CNRS). It was also supported by the French National Research Agency via the ‘Investment for the Future’ project IDEALG (no. ANR-10-BTBR-04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank L. Dartevelle and E. Rolland from the LBI2M’s Algal Culture facilities and Nadia Collet for technical support during her internship. We are grateful to the Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr), and to the Institut Français de Bioinformatique (ANR-11-INBS-0013), and BioGenouest for providing help and computational resources.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.742469/full#supplementary-material
Supplementary Figure 1 | Experimental set-up of the co-cultivation bioassay and sampling. (A) 2 L bottles used for co-cultivation bioassay; (B) Punching hole method of kelp sporophyte growth measurement. The tip of the red arrow shows the position of the hole punched with a pipet tip in 1 cm distance from the basal meristem; (C) Treatment and sampling of algae for transcriptomic analysis.
Supplementary Figure 2 | Distribution of the functional categories derived from Gene Ontology terms obtained by Blast2GO hits of genes from the S. latissima and the L. digitata transcriptome.
Supplementary Figure 3 | Expression levels of unigenes in triplicate samples from (A) S. latissima and (B) L. digitata after 24 and 48 h of culture without (Control: C1–3) or with L. elsbetiae (Endophyte co-culture: E1–3).
Supplementary Table 1 | Statistical analysis of growth and Fv/Fm measurements during the co-cultivation bioassay with the two kelp species.
Supplementary Table 2 | Statistical analysis of growth measurement between the co-cultivation bioassay and GG pre-treatment using L. digitata.
Supplementary Table 3 | Summary of the Trinity assembly and annotation for S. latissima and L. digitata.
Supplementary Table 4 | Lists of differentially expressed genes (DEG) of L. digitata and S. latissima after 24 and 48 h of co-culture with L. elsbetiae.
Afzal, A. J., Wood, A. J., and Lightfoot, D. A. (2008). Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 21, 507–517. doi: 10.1094/MPMI-21-5-0507
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Institute.
Apt, K. E. (1984). Effects of the symbiotic red alga Hypneocolax stellaris on its host Hypnea musciformis (hypneaceae, gigartinales). J. Phycol. 20, 148–150. doi: 10.1111/j.0022-3646.1984.00148.x
Armengaud, J., Trapp, J., Pible, O., Geffard, O., Chaumot, A., and Hartmann, E. M. (2014). Non-model organisms, a species endangered by proteogenomics. J. Proteomics 105, 5–18. doi: 10.1016/j.jprot.2014.01.007
Bartsch, I., Wiencke, C., Bischof, K., Buchholz, C. M., Buck, B. H., Eggert, A., et al. (2008). The genus Laminaria sensu lato: recent insights and developments. Eur. J. Phycol. 43, 1–86. doi: 10.1080/09670260701711376
Bernard, M. S., Strittmatter, M., Murúa, P., Heesch, S., Cho, G. Y., Leblanc, C., et al. (2018). Diversity, biogeography and host specificity of kelp endophytes with a focus on the genera Laminarionema and Laminariocolax (Ectocarpales, Phaeophyceae). Eur. J. Phycol. 54, 39–51. doi: 10.1080/09670262.2018.1502816
Bernard, M., Rousvoal, S., Jacquemin, B., Ballenghien, M., Peters, A. F., and Leblanc, C. (2018). qPCR-based relative quantification of the brown algal endophyte Laminarionema elsbetiae in Saccharina latissima: variation and dynamics of host—endophyte interactions. J. Appl. Phycol. 30, 2901–2911. doi: 10.1007/s10811-017-1367-0
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Bouarab, K., Adas, F., Gaquerel, E., Kloareg, B., Salaün, J.-P., and Potin, P. (2004). The innate immunity of a marine red alga involves oxylipins from both the eicosanoid and octadecanoid pathways. Plant Physiol. 135, 1838–1848. doi: 10.1104/pp.103.037622
Bouarab, K., Potin, P., Correa, J., and Kloareg, B. (1999). Sulfated oligosaccharides mediate the interaction between a marine red alga and its green algal pathogenic endophyte. Plant Cell 11, 1635–1650. doi: 10.1105/tpc.11.9.1635
Bowman, K. G., and Bertozzi, C. R. (1999). Carbohydrate sulfotransferases: mediators of extracellular communication. Chem. Biol. 6, R9–R22. doi: 10.1016/S1074-5521(99)80014-3
Casu, R. E., Jarmey, J. M., Bonnett, G. D., and Manners, J. M. (2007). Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip sugarcane genome array expression profiling. Funct. Integr. Genomics 7, 153–167. doi: 10.1007/s10142-006-0038-z
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Correa, J. A., and McLachlan, J. (1994). Endophytic algae of Chondrus crispus (Rhodophyta). V. Fine structure of the infection by Acrochaete operculata (Chlorophyta). Eur. J. Phycol. 29, 33–47. doi: 10.1080/09670269400650461
Cosse, A., Potin, P., and Leblanc, C. (2009). Patterns of gene expression induced by oligoguluronates reveal conserved and environment-specific molecular defense responses in the brown alga Laminaria digitata. New Phytol. 182, 239–250. doi: 10.1111/j.1469-8137.2008.02745.x
de Oliveira, L. S., Tschoeke, D. A., Lopes, A. C. R. M., Sudatti, D. B., Meirelles, P. M., Thompson, C. C., et al. (2017). Molecular mechanisms for microbe recognition and defense by the red seaweed Laurencia dendroidea. Msphere 2:e00094-17. doi: 10.1128/mSphere.00094-17
Dombrowski, J. E., Kronmiller, B. A., Hollenbeck, V., and Martin, R. C. (2020). Transcriptome analysis of wounding in the model grass Lolium temulentum. Plants 9:780. doi: 10.3390/plants9060780
Eckardt, N. A. (2008). Oxylipin signaling in plant stress responses. Plant Cell 20, 495–497. doi: 10.1105/tpc.108.059485
Ellertsdottir, E., and Peters, A. F. (1997). High prevalence of infection by endophytic brown algae in populations of Laminaria spp. (Phaeophyceae). Mar. Ecol. Prog. Ser. 146, 135–143. doi: 10.3354/meps146135
Flöthe, C. R., Molis, M., and John, U. (2014). Induced resistance to periwinkle grazing in the brown seaweed Fucus vesiculosus (Phaeophyceae): molecular insights and seaweed-mediated effects on herbivore interactions. J. Phycol. 50, 564–576. doi: 10.1111/jpy.12186
Gachon, C. M., Sime-Ngando, T., Strittmatter, M., Chambouvet, A., and Kim, G. H. (2010). Algal diseases: spotlight on a black box. Trends Plant Sci. 15, 633–640. doi: 10.1016/j.tplants.2010.08.005
Gauna, M. C., Parodi, E. R., and Cáceres, E. J. (2009). Epi-endophytic symbiosis between Laminariocolax aecidioides (Ectocarpales, Phaeophyceae) and Undaria pinnatifida (Laminariales, Phaeophyceae) growing on Argentinian coasts. J. Appl. Phycol. 21, 11–18. doi: 10.1007/s10811-007-9298-9
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guidi, L., Mori, S., Degl’innocenti, E., and Pecchia, S. (2007). Effects of ozone exposure or fungal pathogen on white lupin leaves as determined by imaging of chlorophyll a fluorescence. Plant Physiol. Biochem. 45, 851–857. doi: 10.1016/j.plaphy.2007.07.001
Hancock, J., Desikan, R., and Neill, S. (2001). Role of reactive oxygen species in cell signalling pathways. Biochem. Soc. Trans. 29, 345–349. doi: 10.1042/bst0290345
Haug, A., Larsen, B., and Smidsrød, O. (1974). Uronic acid sequence in alginate from different sources. Carbohydr. Res. 32, 217–225. doi: 10.1016/S0008-6215(00)82100-X
Heesch, S., and Peters, A. F. (1999). Scanning electron microscopy observation of host entry by two brown algae endophytic in Laminaria saccharina (Laminariales, Phaeophyceae). Phycol. Res. 47, 1–5. doi: 10.1111/j.1440-1835.1999.tb00277.x
Heinrich, S., Valentin, K., Frickenhaus, S., John, U., and Wiencke, C. (2012). Transcriptomic analysis of acclimation to temperature and light stress in Saccharina latissima (Phaeophyceae). PLoS One 7:e44342. doi: 10.1371/journal.pone.0044342
Hoeberichts, F. A., Ten Have, A., and Woltering, E. J. (2003). A tomato metacaspase gene is upregulated during programmed cell death in Botrytis cinerea-infected leaves. Planta 217, 517–522. doi: 10.1007/s00425-003-1049-9
Hu, H., Zhang, R., Tao, Z., Li, X., Li, Y., Huang, J., et al. (2018). Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in Arabidopsis. Plant Cell Physiol. 59, 1144–1157. doi: 10.1093/pcp/pcy050
Jayaswall, K., Mahajan, P., Singh, G., Parmar, R., Seth, R., Raina, A., et al. (2016). Transcriptome analysis reveals candidate genes involved in blister blight defense in tea (Camellia sinensis (L) Kuntze). Sci. Rep. 6:30412. doi: 10.1038/srep30412
Kawai, H., and Tokuyama, M. (1995). Laminarionema elsbetiae gen. et sp. nov. (Ectocarpales, Phaeophyceae), a new endophyte in Laminaria sporophytes. Phycol. Res. 43, 185–190. doi: 10.1111/j.1440-1835.1995.tb00024.x
Kim, H. S., Lee, C.-G., and Lee, E. Y. (2011). Alginate lyase: structure, property, and application. Biotechnol. Bioprocess Eng. 16, 843–851. doi: 10.1007/s12257-011-0352-8
Knight, H. (1999). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–324. doi: 10.1016/S0074-7696(08)62707-2
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Kornmann, P., and Sahling, P.-H. (1994). Meeresalgen von Helgoland: Zweite Ergänzung. Helgoländer Meeresuntersuchungen 48, 365–406. doi: 10.1007/BF02366253
Küpper, F. C., Gaquerel, E., Cosse, A., Adas, F., Peters, A. F., Müller, D. G., et al. (2009). Free fatty acids and methyl jasmonate trigger defense reactions in Laminaria digitata. Plant Cell Physiol. 50, 789–800. doi: 10.1093/pcp/pcp023
Küpper, F. C., Kloareg, B., Guern, J., and Potin, P. (2001). Oligoguluronates elicit an oxidative burst in the brown algal kelp Laminaria digitata. Plant Physiol. 125, 278–291. doi: 10.1104/pp.125.1.278
Küpper, F. C., Müller, D. G., Peters, A. F., Kloareg, B., and Potin, P. (2002). Oligoalginate recognition and oxidative burst play a key role in natural and induced resistance of sporophytes of Laminariales. J. Chem. Ecol. 28, 2057–2081. doi: 10.1023/A:1020706129624
La Barre, S., Potin, P., Leblanc, C., and Delage, L. (2010). The halogenated metabolism of brown algae (Phaeophyta), its biological importance and its environmental significance. Mar. Drugs 8, 988–1010. doi: 10.3390/md8040988
Langmead, B., and Salzberg, S. (2012). Fast gapped-read alignment with Bowtie2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leblanc, C., Colin, C., Cosse, A., Delage, L., La Barre, S., Morin, P., et al. (2006). Iodine transfers in the coastal marine environment: the key role of brown algae and of their vanadium-dependent haloperoxidases. Biochimie 88, 1773–1785. doi: 10.1016/j.biochi.2006.09.001
Lein, T. E., Sjøtun, K., and Wakili, S. (1991). Mass-occurrence of a brown filamentous endophyte in the lamina of the kelp Laminaria hyperborea (Gunnerus) Foslie along the southwestern coast of Norway. Sarsia 76, 187–193. doi: 10.1080/00364827.1991.10413474
Liu, H., Wang, X., Zhang, H., Yang, Y., Ge, X., and Song, F. (2008). A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene 420, 57–65. doi: 10.1016/j.gene.2008.05.006
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Luo, Q., Zhu, Z., Yang, R., Qian, F., Yan, X., and Chen, H. (2015). Characterization of a respiratory burst oxidase homologue from Pyropia haitanensis with unique molecular phylogeny and rapid stress response. J. Appl. Phycol. 27, 945–955. doi: 10.1007/s10811-014-0391-6
Luque, J., Cohen, M., Savé, R., Biel, C., and Lvarez, I. F. (1999). Effects of three fungal pathogens on water relations, chlorophyll fluorescence and growth of Quercus suber L. Ann. For. Sci. 56, 19–26. doi: 10.1051/forest:19990103
Macaisne, N., Liu, F., Scornet, D., Peters, A. F., Lipinska, A., Perrineau, M.-M., et al. (2017). The Ectocarpus immediate upright gene encodes a member of a novel family of cysteine-rich proteins with an unusual distribution across the eukaryotes. Development 144, 409–418. doi: 10.1242/dev.141523
Michel, G., Tonon, T., Scornet, D., Cock, J. M., and Kloareg, B. (2010). The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 188, 82–97. doi: 10.1111/j.1469-8137.2010.03374.x
Mugford, S. T., Qi, X., Bakht, S., Hill, L., Wegel, E., Hughes, R. K., et al. (2009). A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484. doi: 10.1105/tpc.109.065870
Murata, N., Takahashi, S., Nishiyama, Y., and Allakhverdiev, S. I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767, 414–421. doi: 10.1016/j.bbabio.2006.11.019
Parke, M. (1948). Studies on British laminariaceae. I. Growth in Laminaria saccharina (l.) lamour. J. Mar. Biol. Assoc. U. K. 27, 651–709. doi: 10.1017/S0025315400056071
Peters, A. (1996). New record of the kelp endophyte Laminarionema elsbetiae (Phaeophyceae, Ectocarpales) at Helgoland and its life history in culture. Nova Hedwigia 62, 341–349.
Peters, A. F., Scornet, D., Ratin, M., Charrier, B., Monnier, A., Merrien, Y., et al. (2008). Life-cycle-generation-specific developmental processes are modified in the immediate upright mutant of the brown alga Ectocarpus siliculosus. Development 135, 1503–1512. doi: 10.1242/dev.016303
Potin, P., Bouarab, K., Salaün, J.-P., Pohnert, G., and Kloareg, B. (2002). Biotic interactions of marine algae. Curr. Opin. Plant Biol. 5, 308–317. doi: 10.1016/S1369-5266(02)00273-X
Provasoli, L. (1968). “Media and prospects for the cultivation of marine algae. Cultures and collections of algae,” in Proceedings of US-Japan Conference, September 1966. Japan Society of Plant Physiology, (Hakone).
Ralph, P. J., and Short, F. T. (2002). Impact of the wasting disease pathogen, Labyrinthula zosterae, on the photobiology of eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 226, 265–271. doi: 10.3354/meps226265
Ritter, A., Cabioch, L., Brillet-Guéguen, L., Corre, E., Cosse, A., Dartevelle, L., et al. (2017). Herbivore-induced chemical and molecular responses of the kelps Laminaria digitata and Lessonia spicata. PLoS One 12:e0173315. doi: 10.1371/journal.pone.0173315
Russell, G. (1964). Laminariocolax tomentosoides on the Isle of Man. J. Mar. Biol. Assoc. U. K. 44, 601–612. doi: 10.1017/S0025315400027806
Salavarría, E., Paul, S., Gil-Kodaka, P., and Villena, G. K. (2018). First global transcriptome analysis of brown algae Macrocystis integrifolia (Phaeophyceae) under marine intertidal conditions. 3 Biotech 8:185. doi: 10.1007/s13205-018-1204-4
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Spoel, S. H., and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Stack, J., Haga, I. R., SchröDer, M., Bartlett, N. W., Maloney, G., Reading, P. C., et al. (2005). Vaccinia virus protein A46R targets multiple Toll-like–interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201, 1007–1018. doi: 10.1084/jem.20041442
Strittmatter, M., Grenville-Briggs, L. J., Breithut, L., Van West, P., Gachon, C. M., and Küpper, F. C. (2016). Infection of the brown alga Ectocarpus siliculosus by the oomycete Eurychasma dicksonii induces oxidative stress and halogen metabolism. Plant Cell Environ. 39, 259–271. doi: 10.1111/pce.12533
Thomas, D., Beltrán, J., Flores, V., Contreras, L., Bollmann, E., and Correa, J. A. (2009). Laminariocolax sp.(phaeophyceae) associated with gall developments in Lessonia nigrescens (phaeophyceae). J. Phycol. 45, 1252–1258. doi: 10.1111/j.1529-8817.2009.00749.x
Thomas, F., Cosse, A., Goulitquer, S., Raimund, S., Morin, P., Valero, M., et al. (2011). Waterborne signaling primes the expression of elicitor-induced genes and buffers the oxidative responses in the brown alga Laminaria digitata. PLoS One 6:e21475. doi: 10.1371/journal.pone.0021475
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Wang, F., Lv, Y., Lin, L., Xu, N., Lu, K., and Sun, X. (2018). Characterization of a respiratory burst oxidase homolog from Gracilariopsis lemaneiformis (Rhodophyta) during stress and phytohormone treatments. Bot. Mar. 61, 511–519. doi: 10.1515/bot-2018-0013
Wang, G., Chang, L., Zhang, R., Wang, S., Wei, X., Rickert, E., et al. (2019). Can targeted defense elicitation improve seaweed aquaculture? J. Appl. Phycol. 31, 1845–1854. doi: 10.1007/s10811-018-1709-6
Weinberger, F. (2007). Pathogen-induced defense and innate immunity in macroalgae. Biol. Bull. 213, 290–302. doi: 10.2307/25066646
Wynne, M. (1969). Life history and systematic studies of some Pacific North American Phaeophyceae (brown algae). Univ. Cal. Publ. Bot. 50, 1–88.
Wynne, M. J., and Bold, H. (1985). Introduction to the Algae: Structure and Reproduction. Englewood Cliffs, NJ: Prentice-Hall, Incorporated.
Yoshida, T. (1979). Streblonema (Phaeophyceae) infection in the frond of cultivated Undaria (Phaeophyceae). Proc. Int. Seaweed Symp. 9, 219–223.
Zhang, L., Du, L., and Poovaiah, B. (2014). Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 9:e973818. doi: 10.4161/15592324.2014.973818
Keywords: biotic interactions, brown algae, defense responses, endophyte, Laminariales, physiology, transcriptomic (RNA-Seq)
Citation: Xing Q, Bernard M, Rousvoal S, Corre E, Markov GV, Peters AF and Leblanc C (2021) Different Early Responses of Laminariales to an Endophytic Infection Provide Insights About Kelp Host Specificity. Front. Mar. Sci. 8:742469. doi: 10.3389/fmars.2021.742469
Received: 16 July 2021; Accepted: 24 September 2021;
Published: 03 November 2021.
Edited by:
Jin Liu, Peking University, ChinaReviewed by:
Jianhua Fan, East China University of Science and Technology, ChinaCopyright © 2021 Xing, Bernard, Rousvoal, Corre, Markov, Peters and Leblanc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine Leblanc, Y2F0aGVyaW5lLmxlYmxhbmNAc2Itcm9zY29mZi5mcg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.