- 1Department of Health, Life and Environmental Sciences, University of L’Aquila, L’Aquila, Italy
- 2Dipartimento di Scienze, Università Roma Tre, Rome, Italy
The recently published mitochondrial genome of the fingerprint oyster Alectryonella plicatula (Gmelin, 1791) with GenBank accession number MW143047 was resolved in an unexpected phylogenetic position, as sister to the Pacific cupped oyster Magallana gigas (Thunberg, 1793) and share with this species three typical gene duplications that represent robust synapomorphies of the Magallana clade. In this study, we verified the identity of MW143047 using direct comparisons of single gene sequences, DNA barcoding and phylogenetic analyses. BLAST searches using as query each of the 12 protein coding genes (PCGs) and rRNA genes extracted from MW143047 retrieved M. gigas as best hit with 100% sequence identity for all genes. MW143047 is nested within the clade formed by M. gigas sequences, with virtually zero-length terminal branch, both in the cox1 gene tree (based on 3639 sequences) and in the 16S gene tree (based on 1839 sequences), as well as in the Maximum Likelihood mitogenomic tree based on concatenated sequence of 12 PCGs. Our findings suggest that the original specimen used for mitogenome sequencing was misidentified and represents an individual of M. gigas. This study reinforces the notion that morphological shell analysis alone is not sufficient for oyster identification, not even at high taxonomic ranks such as subfamilies. While it is well established that morphological identification of oysters should be validated by molecular data, this study emphasizes that also molecular data should be taxonomically verified by means of DNA barcoding and phylogenetic analyses. The implications of the publication of taxonomically misidentified sequences and mitogenomes are discussed.
Introduction
Oysters are distributed worldwide in temperate and tropical waters and several of them have a great economic importance. Taxonomic identification of oysters based on morphological characters is challenging, even for species locally cultivated since centuries (e.g., Wang et al., 2004; Hsiao et al., 2016). Indeed, oysters’ shells show a high degree of phenotypic plasticity driven by environmental factors, therefore, shell morphology is often uninformative or misleading for taxonomic identification and classification. The use of molecular data has been fruitful for species identification and has resulted in a well-established phylogeny and systematics of oysters (Salvi et al., 2014; Salvi and Mariottini, 2017). The mitochondrial genome has been the most valuable source of molecular data for oyster species identification (DNA barcoding), phylogenetic reconstruction and classification (e.g., Wang et al., 2004; Liu et al., 2011; Salvi et al., 2014; Raith et al., 2016). Moreover, mitochondrial gene rearrangements, such as transpositions and duplications, has provided additional characters for phylogenetic inference, classification and diagnosis of oysters’ genera and subfamilies (Salvi and Mariottini, 2021). Molecular resources of oyster are continuously growing, and most studies currently implement these data for taxonomic identification. For this purpose, a reliable reference of taxonomically identified sequences and mitogenomes is necessary (Bortolus, 2008; Jin et al., 2020; Salvi et al., 2020).
Recently, the complete mitochondrial genome of the fingerprint oyster Alectryonella plicatula (Gmelin, 1791), with GenBank accession number MW143047, has been characterized (Wang et al., 2021) and resolved in an unexpected phylogenetic position, as sister to the Pacific cupped oyster Magallana gigas (Thunberg, 1793). Unfortunately, in this mitogenome announcement the phylogenetic position of MW143047 is described in a cladogram with arbitrary branch lengths (Wang et al., 2021), therefore masking the true evolutionary divergence between MW143047 and the mitogenome of M. gigas (see Botero-Castro et al., 2016). However, their sister relationship is surprising and in sharp contrast with all previous phylogenetic studies that have consistently established the placement of A. plicatula within the lophinae lineage, that is nested within the subfamily Ostreinae Rafinesque, 1815, whereas M. gigas belong to the well-defined clade of Indo-Pacific Crassostreinae Scarlato and Starobogatov, 1979 (O’Foighil and Taylor, 2000; Salvi et al., 2014; Crocetta et al., 2015; Salvi and Mariottini, 2017; Al-Kandari et al., 2021). Moreover, the newly published mitogenome MW143047 conforms to the mitochondrial gene arrangement of M. gigas, that is characterized by the duplication of trnK, trnQ, and rrnS genes that are exclusive of the Magallana clade (Ren et al., 2010) and represent robust synapomorphies of this clade (Salvi et al., 2014; Salvi and Mariottini, 2017, 2021). These intriguing points are urgent to clarify as MW143047 might become the mitogenomic reference of A. plicatula. In this study, we verified the taxonomic identification of Wang et al. (2021) using available quality control guidelines for taxonomic validation of new mitogenomes (Botero-Castro et al., 2016).
Materials and Methods
We verified the identity of MW143047 using DNA barcoding and phylogenetic analyses.
We extracted from the mitogenome MW143047 the two barcoding fragments commonly used for oysters, the cox1 and the 3′ half portion of the 16S rRNA (Liu et al., 2011; Crocetta et al., 2015), as well the remaining protein coding genes and rRNAs (12S and the 5′ half portion of the 16S) using Geneious Prime 2021 (Biomatters Ltd., Auckland, New Zealand). Sequence of each gene were used as query in BLAST searches using default settings. Sequences of the barcoding markers cox1 and the 16S were aligned with oysters’ sequences available from public database (BOLD and NCBI) assembled, dereplicated, and aligned following the procedure by Salvi et al. (2020). A Neighbor-Joining (NJ) tree was constructed based on uncorrected p-distance values in MEGA v. 7 (Kumar et al., 2016) with pairwise deletion and 100 replicates of bootstrap (BS).
We inferred a Maximum Likelihood (ML) tree based on the concatenated sequences of 12 protein-coding genes (PCGs) of the same oyster taxa analyzed by Wang et al. (2021) plus six additional mitogenome sequences of M. gigas, to further assess phylogenetic relationships and divergence between the latter and the mitogenome MW143047. ML analyses were performed in IQTREE v. 1.6.12 (Nguyen et al., 2015) using for each gene partition the best substitution model determined by the ModelFinder module (Kalyaanamoorthy et al., 2017) and 1000 replicates of ultrafast bootstrapping.
Results
Results of BLAST searches using as query the cox1 and the 16S sequences extracted from MW143047 retrieved as best hits sequences assigned to M. gigas with a sequence identity of 100% (sequence identity ranging from 99.85 to 100% among the best 10 hits for cox1 and of 100% for 16S; Table 1). The same result was obtained in BLAST searches using as query the other 11 protein coding genes and rRNAs extracted from MW143047, with 100% of nucleotides identical to multiple sequences of M. gigas.
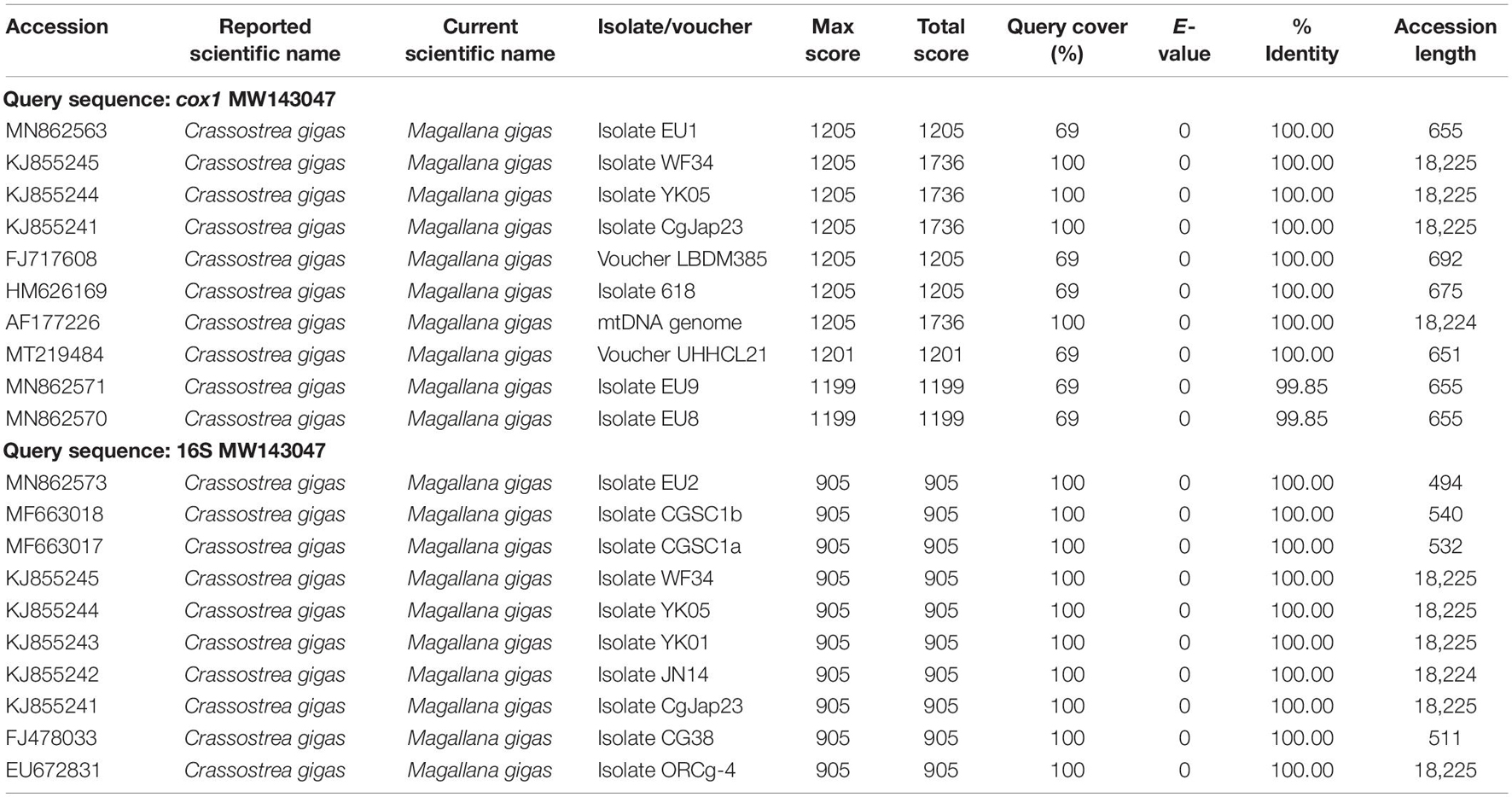
Table 1. Top 10 best hits of BLAST results using as query the sequences of the barcoding fragments cox1 (above) and 16S rRNA (below) extracted from the complete mitochondrial genome MW143047.
In the gene tree based on 3639 cox1 sequences (Figure 1A) and in the gene tree based on 1839 16S sequences (Figure 1B) MW143047 clustered with M. gigas with maximum bootstrap support (BS = 100%).
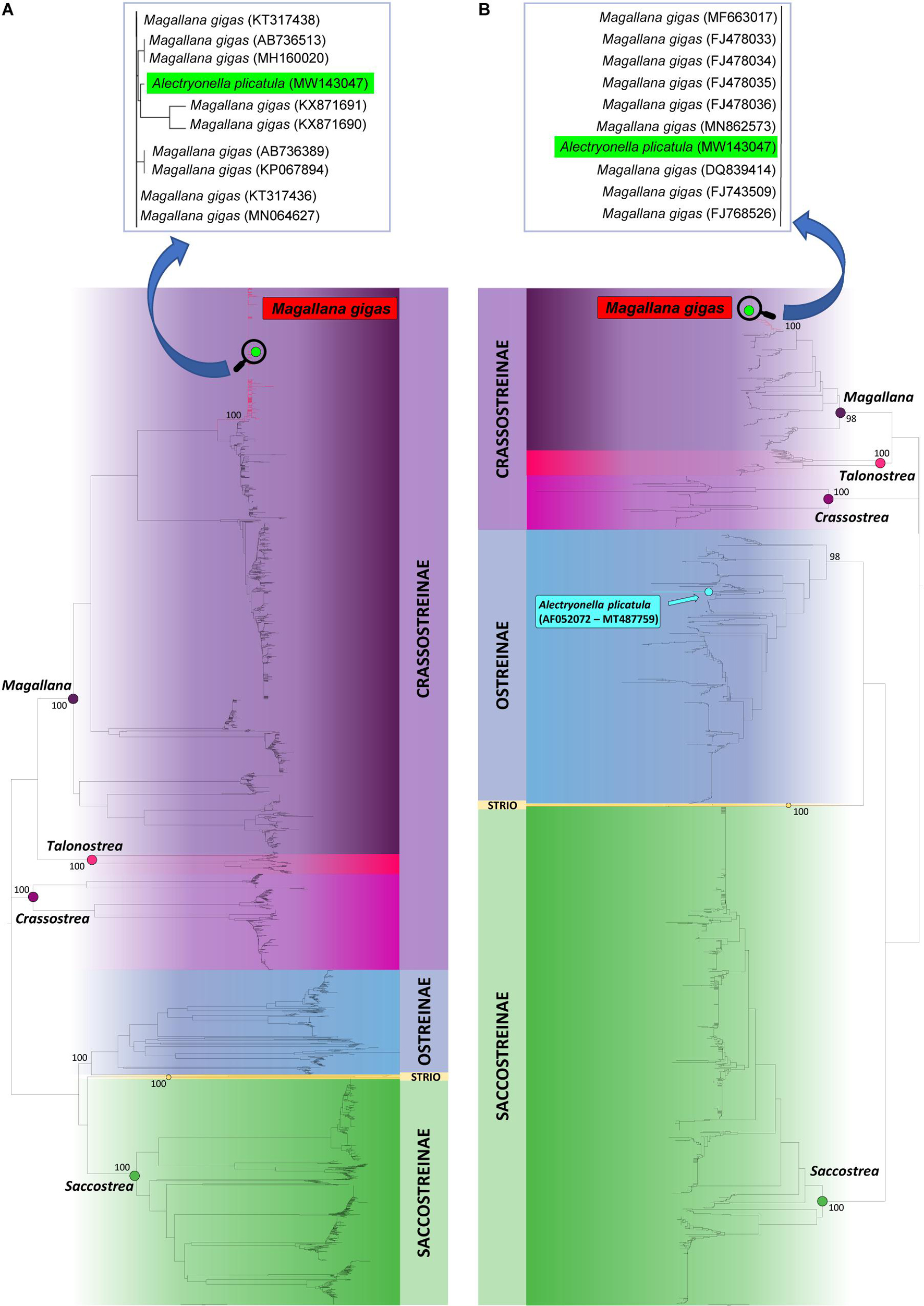
Figure 1. Neighbor-Joining trees based on 3639 cox1 sequences (A) and 1839 16S sequences (B) available from public databases. Bootstrap values are reported in correspondence of main nodes. In both trees MW143047 is nested within the clade formed by sequences of Magallana gigas within the Crassostreinae lineage. Instead, available 16S rRNA sequences of Alectryonella plicatula generated in previous studies cluster within the Ostreinae lineage.
In the ML mitogenomic tree (Figure 2) MW143047 is nested within the clade formed by M. gigas sequences, with virtually zero-length terminal branches. This clade was sister to the mitogenome sequence of Magallana angulata (BS = 100%) within the well supported clade formed by Magallana species (BS = 100%).
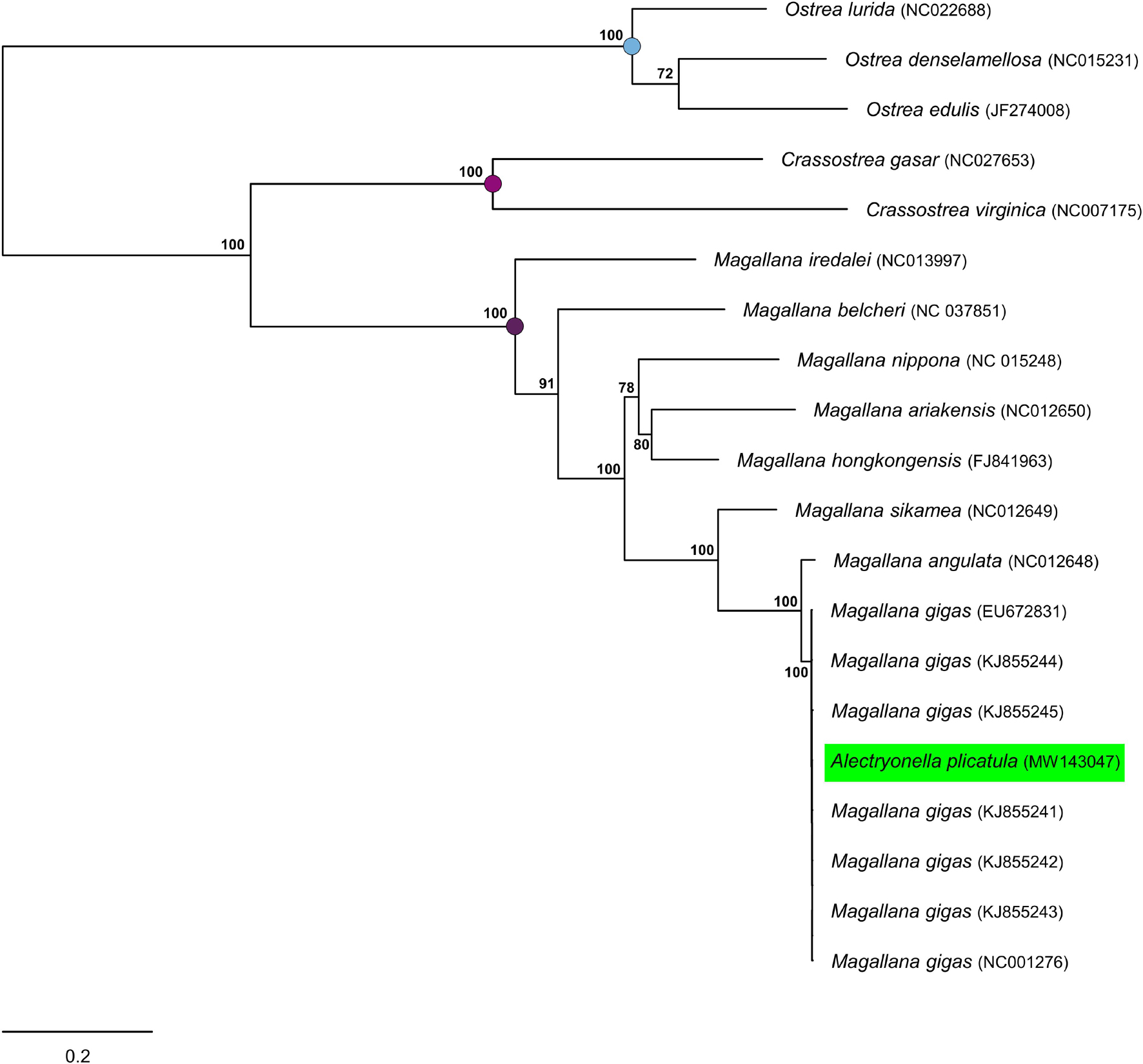
Figure 2. Maximum Likelihood tree based on the concatenated sequences of 12 protein-coding genes from complete mitochondrial genomes of the same oyster taxa analyzed by Wang et al. (2021) plus six additional mitogenome sequences of Magallana gigas. Bootstrap values are reported in correspondence of main nodes. The mitogenome MW143047 is nested within the clade formed by mitogenomes of M. gigas.
Discussion
Results of DNA barcoding, BLAST and phylogenetic analyses show that MW143047, attributed by Wang et al. (2021) to the fingerprint oyster A. plicatula, is identical to mitochondrial DNA sequences of the Pacific cupped oyster M. gigas (Table 1). The MW143047 sequences cluster within the clade of M. gigas both in the gene trees based on the barcoding markers cox1 and 16S and in the ML mitogenome tree based on concatenated sequence of 12 PCGs (Figures 1, 2). On the other hand, two mitochondrial 16S rRNA sequences of A. plicatula generated in previous studies (Jozefowicz and O’Foighil, 1998; Ardura et al., 2021), and available in GenBank under the accession numbers AF052072 and MT487759, show a high genetic divergence (p-distance: 19 and 18% respectively) with MW143047. The most likely explanation for these results is that the original specimen used for mitogenome sequencing was misidentified and represents an individual of M. gigas. A morphological re-assessment of this specimen (voucher no. CP-202005; Wang et al., 2021) was not possible despite our requests to the authors of the mitogenome MW143047.
The hypothesis of contamination by DNA of M. gigas, either prior to PCR amplification or as PCR product prior to sequencing, is unlikely. In these cases, often chimera sequence artifacts are observed (e.g., Sangster and Luksenburg, 2020), whereas all PCGs and rRNA genes of MW143047 are identical to sequences of M. gigas thus indicating that MW143047 is a bona fide mitogenome of M. gigas. Even less likely is the hypothesis of mitochondrial introgression of M. gigas in A. plicatula following hybridization. Indeed, while these two species might co-occur in the collection site of the original specimen used for sequencing (Shicheng Island, Dalian, China), their genetic divergence is very large (∼19% at the 16S rRNA) as they belong to distinct evolutionary lineages within Ostreidae Rafinesque, 1815: A. plicatula belongs to the Ostreinae lineage whereas M. gigas to the Crassostreinae lineage (e.g., O’Foighil and Taylor, 2000; Salvi et al., 2014; Crocetta et al., 2015; Salvi and Mariottini, 2017; Al-Kandari et al., 2021).
While M. gigas in A. plicatula are readily distinguishable using mitochondrial (Liu et al., 2011; Crocetta et al., 2015) or nuclear markers (O’Foighil and Taylor, 2000; Salvi et al., 2014; Mazón-Suástegui et al., 2016), morphological misidentification between the two might be easy according to Bishop et al. (2017) due the extensive degree of phenotypic plasticity of oysters. This example highlights the common difficulties encountered for identifying oysters based on shell morphology alone and provides one more demonstration that misidentification regards not only closely related species but also taxonomic ranks as high as subfamilies (discussed in Salvi and Mariottini, 2021; see Salvi et al., 2014 and Raith et al., 2016 for examples regarding the subfamilies Striostreinae Harry, 1985, Ostreinae Rafinesque, 1815, and Saccostreinae Salvi and Mariottini, 2016).
Previous studies on oyster systematics strongly advice that morphological identification of oysters should be validated by molecular data (e.g., Wang et al., 2004; Lam and Morton, 2006; Hamaguchi et al., 2017). This study also emphasizes that molecular data should be taxonomically verified by means of DNA barcoding and phylogenetic analyses. Taxonomic validation of mitogenomes is straightforward following the quality control guidelines of Botero-Castro et al. (2016) (see also Sangster and Luksenburg, 2020) and most of these recommendations can be applied also for a taxonomic verification of sequences from single gene fragments. The publication of taxonomically misidentified sequences and mitogenomes can have profound implications if few sequences are available for the species so that misidentified sequence ends up as the reference for the species in public databases. A great source of new mitochondrial DNA sequences is represented by the journal Mitochondrial DNA Part B that is specifically aimed at publishing whole mitochondrial genomes such as the mitogenome of A. plicatula (Wang et al., 2021). Unfortunately, this journal did not give us the possibility to publish the evidence that this mitogenome was based on a misidentification. Therefore, many misidentified mitochondrial genomes have the potential to enter public database without validation. In such cases misidentification errors can propagate in future studies that use the wrong reference-sequences in taxonomic and phylogenetic comparisons.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Table 1 and Figures 1, 2.
Author Contributions
DS contributed to the conception and design of the study and wrote the first draft of the manuscript. DS, EB, and MG organized the database and performed the statistical analysis. PM revised the analyses carried out. DS, EB, MG, and PM wrote the sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Kandari, M., Oliver, P. G., and Salvi, D. (2021). Molecular and morphological systematics of a new, reef forming, cupped oyster from the northern Arabian Gulf: Talonostrea salpinx new species. Zookeys 1043, 1–20. doi: 10.3897/zookeys.1043.66992
Ardura, A., Fernandez, S., Haguenauer, A., Planes, S., and Garcia-Vazquez, E. (2021). Ship-driven biopollution: how aliens transform the local ecosystem diversity in Pacifc islands. Mar. Pollut. Bull. 166:112251. doi: 10.1016/j.marpolbul.2021.112251
Bishop, M., Brumbaugh, R., Luckenbach, M., and Ruesink, J. (2017). Alectryonella plicatula. The IUCN Red List of Threatened Species 2017: e.T200866A2683909. Available Online at: https://dx.doi.org/10.2305/IUCN.UK.2017-3.RLTS.T200866A2683909.en (accessed April 5, 2021).
Bortolus, A. (2008). Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. AMBIO 37, 114–118. doi: 10.1579/0044-7447(2008)37[114:ecitbs]2.0.co;2
Botero-Castro, F., Delsuc, F., and Douzery, E. J. (2016). Thrice better than once: quality control guidelines to validate new mitogenomes. Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 449–454. doi: 10.3109/19401736.2014.900666
Crocetta, F., Mariottini, P., Salvi, D., and Oliverio, M. (2015). Does GenBank provide a reliable DNA barcode reference to identify small alien oysters invading the Mediterranean Sea?. Marine biological association of the United Kingdom. J. Mar. Biol. Assoc. U. K. 95, 111–122. doi: 10.1017/s0025315414001027
Hamaguchi, M., Manabe, M., Kajihara, N., Shimabukuro, H., Yamada, Y., and Nishi, E. (2017). DNA barcoding of flat oyster species reveals the presence of Ostrea stentina Payraudeau, 1826 (Bivalvia: Ostreidae) in Japan. Mar. Biodivers. Rec. 10:4.
Hsiao, S. T., Chuang, S. C., Chen, K. S., Ho, P. H., Wu, C. L., and Chen, C. A. (2016). DNA barcoding reveals that the common cupped oyster in taiwan is the portuguese oyster Crassostrea angulata (Ostreoida; Ostreidae), not C. gigas. Sci. Rep. 6:34057.
Jin, S., Kim, K. Y., Kim, M. S., and Park, C. (2020). An assessment of the taxonomic reliability of DNA barcode sequences in publicly available databases. Algae 35, 293–301. doi: 10.4490/algae.2020.35.9.4
Jozefowicz, C. J., and O’Foighil, D. (1998). Phylogenetic analysis of southern hemisphere flat oysters based on partial mitochondrial 16S rDNA gene sequences. Mol. Phylogenet. Evol. 10, 426–435. doi: 10.1006/mpev.1998.0529
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lam, K., and Morton, B. (2006). Morphological and mitochondrial DNA analysis of the Indo-West pacific rock oysters (Ostreidae: Saccostrea species). J. Molluscan Stud. 72, 235–245. doi: 10.1093/mollus/eyl002
Liu, J. U. N., Li, Q. I., Kong, L., Yu, H., and Zheng, X. (2011). Identifying the true oysters (Bivalvia: Ostreidae) with mitochondrial phylogeny and distance-based DNA barcoding. Mol. Ecol. Resour. 11, 820–830. doi: 10.1111/j.1755-0998.2011.03025.x
Mazón-Suástegui, J. M., Fernández, N. T., Valencia, I., Cruz-Hernández, P., and Latisnere-Barragán, H. (2016). 28S rDNA as an alternative marker for commercially important oyster identification. Food Control 66, 205–214. doi: 10.1016/j.foodcont.2016.02.006
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
O’Foighil, D., and Taylor, D. J. (2000). Evolution of parental care and ovulation behaviour in oysters. Mol. Phylogenet. Evol. 15, 301–313. doi: 10.1006/mpev.1999.0755
Raith, M., Zacherl, D. C., Pilgrim, E. M., and Eernisse, D. J. (2016). Phylogeny and species diversity of Gulf of California oysters (Ostreidae) inferred from mitochondrial DNA. Am. Malacol. Bull. 33, 263–283. doi: 10.4003/006.033.0206
Ren, J., Liu, X., Jiang, F., Guo, X., and Liu, B. (2010). Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. BMC Evol. Biol. 10:394. doi: 10.1186/1471-2148-10-394
Salvi, D., Berrilli, E., D’Alessandro, P., and Biondi, M. (2020). Sharpening the DNA barcoding tool through a posteriori taxonomic validation: the case of Longitarsus flea beetles (Coleoptera: Chrysomelidae). PLoS One 15:e0233573. doi: 10.1371/journal.pone.0233573
Salvi, D., Macali, A., and Mariottini, P. (2014). Molecular phylogenetics and systematics of the bivalve family Ostreidae based on rRNA sequence-structure models and multilocus species tree. PLoS One 9:e108696. doi: 10.1371/journal.pone.0108696
Salvi, D., and Mariottini, P. (2017). Molecular taxonomy in 2D: a novel ITS2 rRNA sequence-structure approach guides the description of the oysters’ subfamily Saccostreinae and the genus Magallana (Bivalvia: Ostreidae). Zoological J. Linn. Soc. 179, 263–276.
Salvi, D., and Mariottini, P. (2021). Revision shock in Pacific oysters taxonomy: the genus Magallana (formerly Crassostrea in part) is well-founded and necessary. Zool. J. Linn. Soc. 192, 43–58. doi: 10.1093/zoolinnean/zlaa112
Sangster, G., and Luksenburg, J. A. (2020). The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA B Resour. 5, 2661–2664. doi: 10.1080/23802359.2020.1785349
Wang, H., Guo, X., Zhang, G., and Zhang, F. (2004). Classification of jinjiang oysters Crassostrea rivularis (Gould, 1861) from China, based on morphology and phylogenetic analysis. Aquaculture 242, 137–155. doi: 10.1016/j.aquaculture.2004.09.014
Keywords: DNA barcoding, Magallana, misidentification, Ostreidae, oyster, phylogeny
Citation: Salvi D, Berrilli E, Garzia M and Mariottini P (2021) Yet Another Mitochondrial Genome of the Pacific Cupped Oyster: The Published Mitogenome of Alectryonella plicatula (Ostreinae) Is Based on a Misidentified Magallana gigas (Crassostreinae). Front. Mar. Sci. 8:741455. doi: 10.3389/fmars.2021.741455
Received: 14 July 2021; Accepted: 20 August 2021;
Published: 09 September 2021.
Edited by:
Christian Marcelo Ibáñez, Andrés Bello University, ChileReviewed by:
Felipe Aguilera, University of Concepción, ChileJuan E. Uribe, Smithsonian National Museum of Natural History (SI), United States
Copyright © 2021 Salvi, Berrilli, Garzia and Mariottini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniele Salvi, ZGFuaWVsZXNhbHZpLmJpb0BnbWFpbC5jb20=
†ORCID: Daniele Salvi, orcid.org/0000-0002-3804-2690; Emanuele Berrilli, orcid.org/0000-0001-8081-8600; Matteo Garzia, orcid.org/0000-0002-0918-9925; Paolo Mariottini, orcid.org/0000-0003-1044-7108
 Daniele Salvi
Daniele Salvi Emanuele Berrilli
Emanuele Berrilli Matteo Garzia
Matteo Garzia Paolo Mariottini
Paolo Mariottini