- 1Laboratory of Environmental Pollution and Integrative Omics, Guilin Medical University, Guilin, China
- 2Department of Chemistry, City University of Hong Kong, Kowloon, Hong Kong SAR, China
- 3State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon, Hong Kong SAR, China
- 4State Key Laboratory of Agrobiotechnology, School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong SAR, China
- 5Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 6Center for Promotion of International Education and Research, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
- 7Department of Biomedical Sciences, City University of Hong Kong, Kowloon, Hong Kong SAR, China
Recent studies have shown hypoxia to be an endocrine disruptor that impairs sex differentiation and reproductive function, leading to male-biased F1 populations in fish. However, the molecular mechanisms through which hypoxia alters fish sex differentiation and therefore sex ratios remain poorly understood. In order to understand the potential role of miRNAs in mediating hypoxia-altered sex determination and differentiation in fish, we conducted small RNA sequencing and transcriptome sequencing on marine medaka (Oryzias melastigma) embryos that were exposed to hypoxia (2.0 ± 0.2 mg O2 L–1) for 40 h (encompassing a critical window of sex determination). We identified dysregulated miRNAs and mRNAs in the hypoxia-exposed embryo, and bioinformatic analysis of the integrative small RNA sequencing and transcriptome sequencing results revealed hypoxia to cause alterations of genes related to embryonic development through miRNA regulation. Importantly, we have identified miRNA-mRNA pairs that were reported to play roles in gonad development (novel miR-145-col9a3 and novel miRNA-94- arid5b), in sex hormone response (novel miRNA-210-ca2, novel miRNA-106-nr2f2, nbr-miR-29c-nr4a1, and ola-miR-92b-akr1d1), and in sex characteristic development (novel miRNA-145-mns1, nle-miR-20-sord, and ipu-miR-219b-abcc8). Our findings highlighted the possible roles of miRNA–mRNA in regulation of embryonic development and sex determination in response to hypoxic stress.
Introduction
Hypoxia is a widespread and pressing environmental concern in aquatic habitats, causing severe habitat damage and major disruption to aquatic ecosystems worldwide. The occurrence of hypoxia in water bodies has increased globally over the past 30 years due to escalating eutrophication and organic pollution. Presently, over 500 hypoxic areas (<2 mg O2 L–1) spanning hundreds of thousands of square kilometers have been reported worldwide (Thrash et al., 2017), and is likely to worsen in the future, as a result of global warming and rapid coastal development, thereby threatening the sustainability of natural populations (Pörtner and Peck, 2010). Compared with mammals, fishes possess an exceptional range of sex determination and differentiation processes, which are modulated by a variety of environmental factors such as population density, social behaviors, pH, and dissolved oxygen (Baroiller et al., 2009; Reddon and Hurd, 2013; Yamamoto et al., 2014).
Previous studies from our group have shown that hypoxia is an endocrine disruptor that impairs reproductive activities and affects sexual differentiation in fish, leading to male-biased F1 generations (Shang et al., 2006; Cheung et al., 2014). Similar findings on sex ratio alterations have been observed in other studies which showed that the forkhead transcription factor (foxl2) and doublesex and mab3-related transcription factor 1 (dmrt1) were downregulated following exposure to hypoxia (Pelley, 2006; Baroiller et al., 2009). Importantly, a largescale field study from the Gulf of Mexico, one of the largest hypoxic dead zones in the world, reported similar endocrine disruption phenomena − extensive reproductive disruption, ovarian masculinization and male-biased Atlantic croaker sex ratios − occurring in natural fish populations (Thomas and Rahman, 2012). DNA methylation of the gonadal cyp19a (aromatase) gene has been linked to temperature-dependent sex ratio shifts in European sea bass (Navarro-Martín et al., 2011), suggesting that epigenetic modifications in response to environmental changes may also play a role in sex determination. Additionally, several recent studies have reported regulatory roles of miRNAs in sex differentiation. For example, let-7 and miR-21 have been shown to regulate egg development in rainbow trout (Ma et al., 2012), and miR-141 and miR-429 have been found to play crucial roles in regulating testis development and spermatogenesis in yellow catfish (Jing et al., 2014). Distinct subsets of miRNAs have also been observed in both male and female Nile tilapia embryos (Eshel et al., 2014) and gonads (Wang et al., 2016), indicating potential regulatory roles for miRNAs in the sexual differentiation of fish. Overall, the abovementioned studies strongly implicate miRNAs as a new and important class of effector molecules controlling gonadal development and sexual differentiation in vertebrates.
Marine medaka Oryzias melastigma (synonyms: Oryzias dancena) is a model marine fish species for ecotoxicological studies (Lau et al., 2014). The sex-determining gene DMY is absent in the O. melastigma (Juanchich et al., 2013). Genetic studies using a progeny test of sex-reversed individuals indicated the presence of XX/XY sex-determination system absent in the O. melastigma (Bouchareb et al., 2017). In addition, SRY-related HMG-Box gene 3 (sox3) has been co-opted as the male sex determination gene in the O. melastigma (Gay et al., 2018). Our group has previously identified several gonad-specific miRNAs in marine medaka, O. melastigma, as well as specific miRNAs from medaka embryos that are responsive to hypoxia (Feng et al., 2014). Likewise, differentially expressed miRNAs was found to play important roles during fish ovarian development (Kang et al., 2004). A genome-wide study identified novel ovarian-predominant miRNAs in the medaka fish (Mishima et al., 2008). And, miR-202 was reported to control female fecundity by regulating medaka oogenesis (Fagegaltier et al., 2014), suggesting that tissue-specific miRNAs may play specific roles in gonadal functions. Using marine medaka embryos as a model, together with integrative omics analysis, including small-RNA sequencing and transcriptome sequencing, we investigated the role of specific hypoxia-responsive miRNAs in directly targeting sex determination genes. Our results identify miRNA–mRNA pairs that could be important for controlling sexual differentiation, resulting in hypoxia-altered sex ratios in fish.
Materials and Methods
Fish Embryo Maintenance and Hypoxic Exposure
All animal research procedures were approved by the Animal Ethics Committee on Research Experiments involving Animal Subjects (A-0244) of the City University of Hong Kong. Oryzias melastigma embryos used in our experiments were obtained from State Key Laboratory of Marine Pollution, City University of Hong Kong. Embryos within 1 day old (< 24 h after fertilization) were collected from parental marine medakas that were maintained in seawater with salinity of 3.5% under optimal growth and breeding conditions (5.8 mg O2 L–1, 28 ± 2°C, pH 7.2 in a 14-h light: 10-h dark cycle). The collected embryos were randomly selected under a dissecting microscope. Filaments attached to the clustered embryos were carefully removed via constant rolling of the embryos on a net with fingertips and pulling of forceps until the embryo was fully separated and clean. Abnormal or unfertilized embryos were removed from the batch. The selected embryos were then distributed into 12 separate 50-mL beakers, each holding 30 embryos. Six beakers of embryos were exposed to normoxia (5.8 ± 0.2 mg O2 L–1; outside the hypoxic chamber) and six beakers of embryos were exposed to hypoxia (2.0 ± 0.2 mg O2 L–1; inside the hypoxic chamber) continuously for 40 h. The desired dissolved oxygen (DO) level was achieved by a constant flow of premixed air and nitrogen (0.5% oxygen) inside a hypoxic chamber. The DO level was measured and monitored using a DO meter (YSI model 580). After the exposure period, the DO level of the hypoxic water was measured again to ensure DO levels were maintained at 2.0 ± 0.2 mg O2 L–1. Water temperature was maintained at 26 ± 2°C by placing the hypoxic chamber inside an incubator.
Embryo Survivability and Hatchability
After the exposure to normoxic or hypoxic conditions, embryos from each condition were transferred back to normoxic condition. The number of hatched eggs and dead eggs was recorded daily for 4 weeks. Temperature was maintained at 26 ± 2°C under a 14:10 light:dark photoperiod. Statistical analyses were performed using the paired t-test in GraphPad Prism 9.1.0 (GraphPad Software).
RNA Sample Preparation and Small RNA Sequencing
After the normoxic or hypoxic exposure, one beaker of embryos from each condition was used as one biological replicate for the RNA preparation, and there were three replicates per condition. Total RNA was extracted immediately using the mirVanaTM miRNA isolation kit (Applied Biosystems) following the manufacturer’s instructions. RNA quality was assessed using the Agilent 2100 Bioanalyzer system and samples with an RNA Integrity Number (RIN) greater than 8 were used to construct the small RNA and complementary DNA (cDNA) libraries (Supplementary Table 1). Five μg of total RNA were used from each sample. Short RNA transcripts (18–30 nucleotides long) were resolved and isolated using polyacrylamide gel electrophoresis (PAGE) gels. The isolated small RNA molecules were ligated with 3′ and 5′ adapters, and single-stranded cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen). The cDNA was then amplified using indexing primers. Following quantitative real-time PCR (qRT-PCR) amplification, the library size was determined using the Bioanalyzer (Agilent Technologies 2100) and library concentrations were assessed using qRT-PCR (EvaGreen). The libraries were then processed by Novogene. Single-end reads [50 base-pair (bp) read length] were sequenced using the Illumina Novaseq 6000 sequencer to produce at least 20 M clean reads per sample. The adaptor sequences were trimmed and bases with a Phred quality score less than 20 were removed.
Prediction of Mature miRNA Sequences
Raw sequencing reads were trimmed to remove adapters using Cutadapt 2.8 software (Martin, 2011). Trimmed reads were processed using the mapper.pl function of miRDeep2 software (Friedländer et al., 2012) to remove reads shorter than 18 nucleotides and collapsed reads. The processed reads were then mapped onto the genome assembly of O. melastigma (Om_v0.7.RACA) with Bowtie 1.2.2 software (Langmead, 2010), using the following parameters: -n 1 -e 80 -l 18 -a –best –strata, producing mapped reads in BWT format. The BWT files were converted to mirDeep2 ARF format and parsed to remove unmatched nucleotides at the 3′ end, using the convert_bowtie_output.pl and parse_mappings.pl functions of mirDeep2 respectively. The parsed mapped reads were used to predict miRNA sequences using the miRDeep2.pl function of mirDeep2. The miRNA prediction results from all samples were merged and parsed into a single list of unique predicted mature miRNA sequences, which were then matched to known miRNAs by nucleotide BLAST to the miRBase database (Kozomara et al., 2018) using the blastn function of BLAST + 2.6.0 (Camacho et al., 2009) with the following parameters: -task blastn-short -strand plus -num_alignments 1. The BLAST hits were filtered using the following parameters: the first 2–7 nucleotides are identical between the predicted miRNA and known miRNA sequences, hit coverage was ≥90%, no indels present, and a maximum of two mismatches.
Expression Analysis of Small RNAs
Trimmed sequencing reads were remapped onto the mature miRNA sequences using Bowtie with the following parameters: -v 2 -a -m 1 –norc –best –strata, with mapped reads output in SAM format. The SAM files were converted to BAM format using samtools (Li et al., 2009). The number of reads mapped to each mature miRNA sequence was then counted, producing a read counts table. Differential expression analysis of the miRNA read counts was performed using DESeq2 (Love et al., 2014). Differentially expressed miRNAs (DEmiRNAs) were determined using cutoff values of | log2 (fold change: treatment/control)| > 1 and B&H corrected p-value < 0.05.
miRNA Target Genes Prediction
Target mRNAs of significant differentially expressed miRNAs were predicted using IntaRNA v2:3:1 (Camacho et al., 2009; Li et al., 2009; Langmead, 2010; Martin, 2011; Friedländer et al., 2012; Fagegaltier et al., 2014; Love et al., 2014; Wright et al., 2014; Mann et al., 2017; Busch et al., 2018; Kozomara et al., 2018; Raden et al., 2018)[20, 21] and miRanda v3:3a (John et al., 2005), with default parameters. Target mRNAs were treated as true targets when they met the following criteria: (1) targets were predicted by both tools; (2) interaction sites overlapped between predictions.
Transcriptome Sequencing and Bioinformatic Analysis
Total RNA extracted from the same set of embryos (three replicates each from normoxic and hypoxic conditions) were subjected to cDNA library construction. The libraries were processed by Novogene. Paired-end reads (150 bp × 2) were sequenced on the Illumina Novaseq 6000 sequencer to produce at least 23 M clean reads per sample. The raw sequencing reads were checked for quality using FastQC (Andrews, 2010). The sequence reads were then mapped onto the genome assembly of O. melastigma (Om_v0.7.RACA) using Spliced Transcripts Alignment to a Reference (STAR) 2.7.1a (Dobin et al., 2013), and aligned reads were output in BAM format. Aligned reads were annotated to exon features of the Om_v0.7.RACA gene annotation file using the htseq-count function of HTseq 0.11.2 (Anders et al., 2015), and the resulting read-count data were subjected to differential expression analysis using DESeq2. Genes with | log2 (fold change: treatment/control)| > 1 and B&H corrected p-value < 0.05 were considered differentially expressed genes (DEGs).
Assignment of Human Gene Symbols to Differentially Expressed Gene and Functional Analysis of Differentially Expressed Gene
Differentially expressed gene were assigned human gene symbols by reciprocal best hit (RBH) matching of the human and O. melastigma proteomes. Briefly, protein BLAST of the human (GRCh38.p12) protein sequences was performed against the medaka (Om_v0.7.RACA) protein sequences, and vice versa, using BLAST + 2.6.0 (Camacho et al., 2009). The top hit, with the lowest e-value, for each query sequence was extracted and compared between the two sets of BLAST queries; Protein sequences where the query and top hit matched in both directions were identified as RBHs. The DEGs were then subjected to gene ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using DAVID 6.81, enriched biological processes and KEGG pathways (p < 0.05) were identified.
Complementary DNA Synthesis and Quantitative PCR Analysis
One μg of total RNA was converted to cDNA by using SuperScript™ VILO™ cDNA Synthesis Kit (Invitrogen). Then qPCR analysis was performed on the cDNA from normoxia and hypoxia groups using StepOnePlus™ Real-Time PCR System (Thermofisher). The primer sequencing was shown as Supplementary Table 2.
Results
Exposure to Hypoxia Did Not Affect the Hatchability and Survival Rate of Medaka Embryos
Following normoxia or hypoxia exposure, the viability and hatchability of the embryos were measured for 4 weeks (28 days). Our results indicated that exposure to hypoxia did not affect the hatchability (Figure 1A) and survival rates (Figure 1B) of the medaka embryos, as compared to the normoxic group.
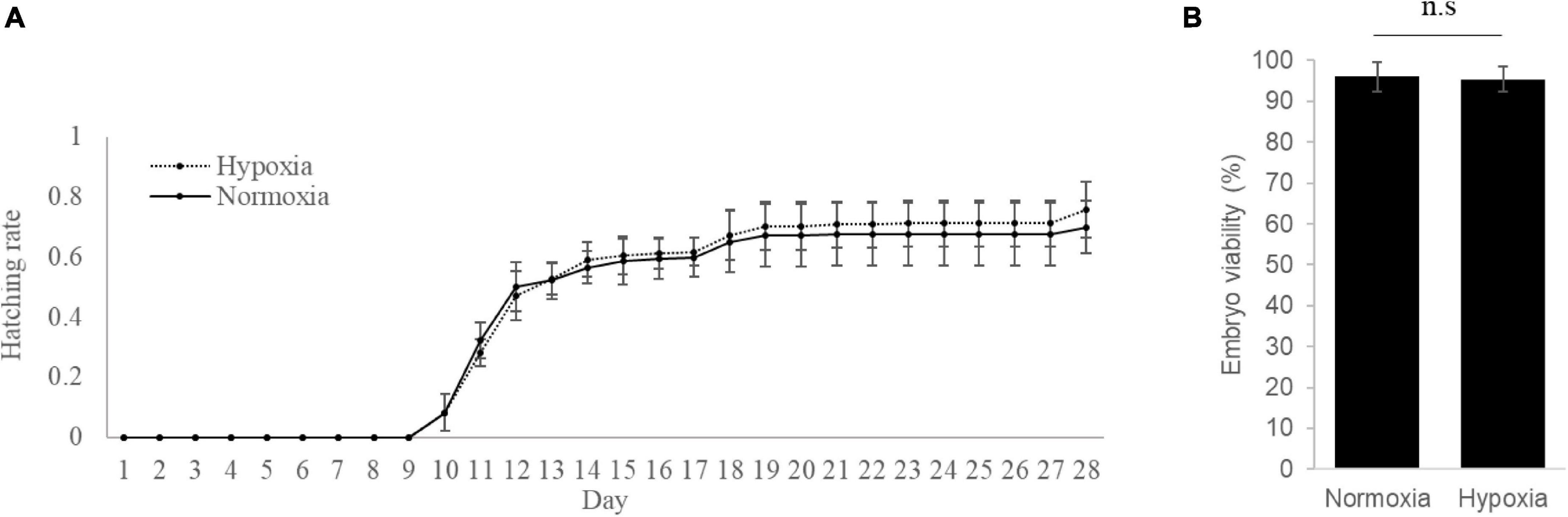
Figure 1. Hypoxia exposure has no effect on the hatchability and viability of medaka embryos. (A) Embryo hatchability was assessed continuously for 28 days after normoxia or hypoxia exposure. (B) Total embryo viability was determined at day 28 after normoxia or hypoxia exposure. n.s. represents not statistically significant (p > 0.05).
Hypoxia Was Predicted to Alter Embryonic Development Through miRNA Dysregulation
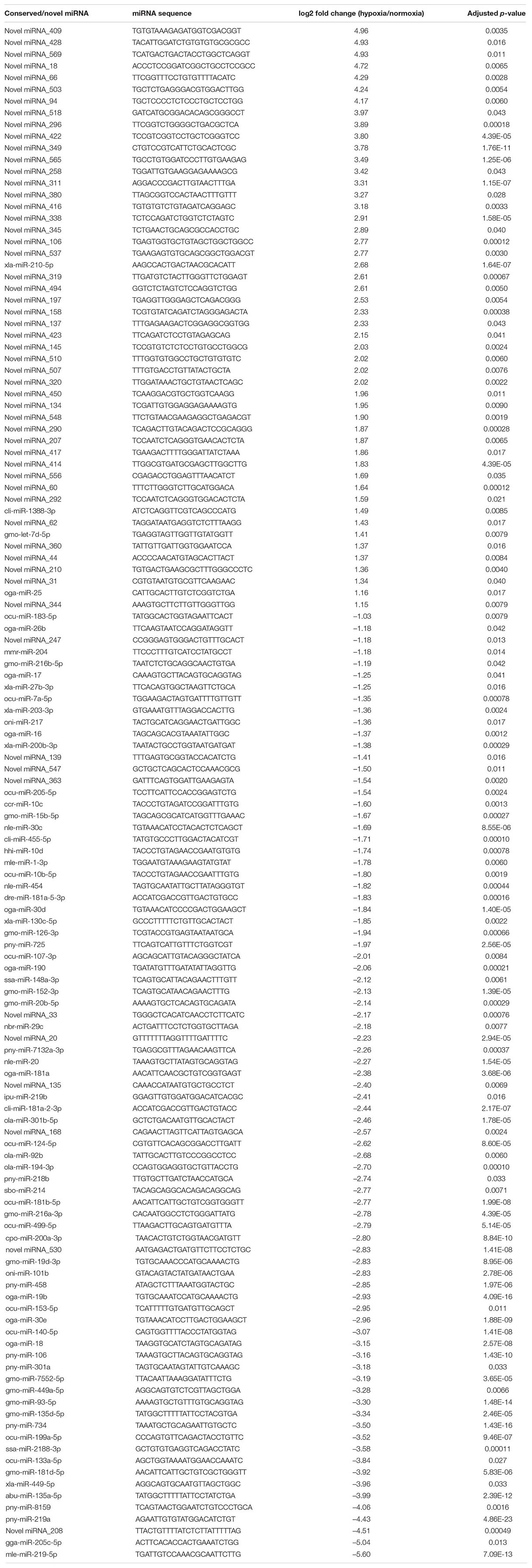
To identify changes to the miRNA profile in the hypoxia-exposed embryos, small RNA sequencing was employed. For this, we obtained 72.8 and 79.3 million quality-trimmed raw reads from normoxic and hypoxic-exposed embryos, respectively, translating to a total of 4.36 Gb of qualified data (Table 1). The clean sequencing reads were mapped to the O. melastigma genome to determine the miRNA content of marine medaka embryos. We identified 253 conserved miRNAs and 572 novel miRNAs (Supplementary Table 3) in the medaka embryo. When we compared the expression level of these miRNAs in the normoxic and hypoxic-exposed embryos, we found 131 miRNAs with altered regulation, including 50 upregulated and 81 downregulated miRNAs (Figure 2A and Table 2). Of these, 75 and 56 dysregulated miRNAs were conserved and novel miRNAs, respectively (Table 2).
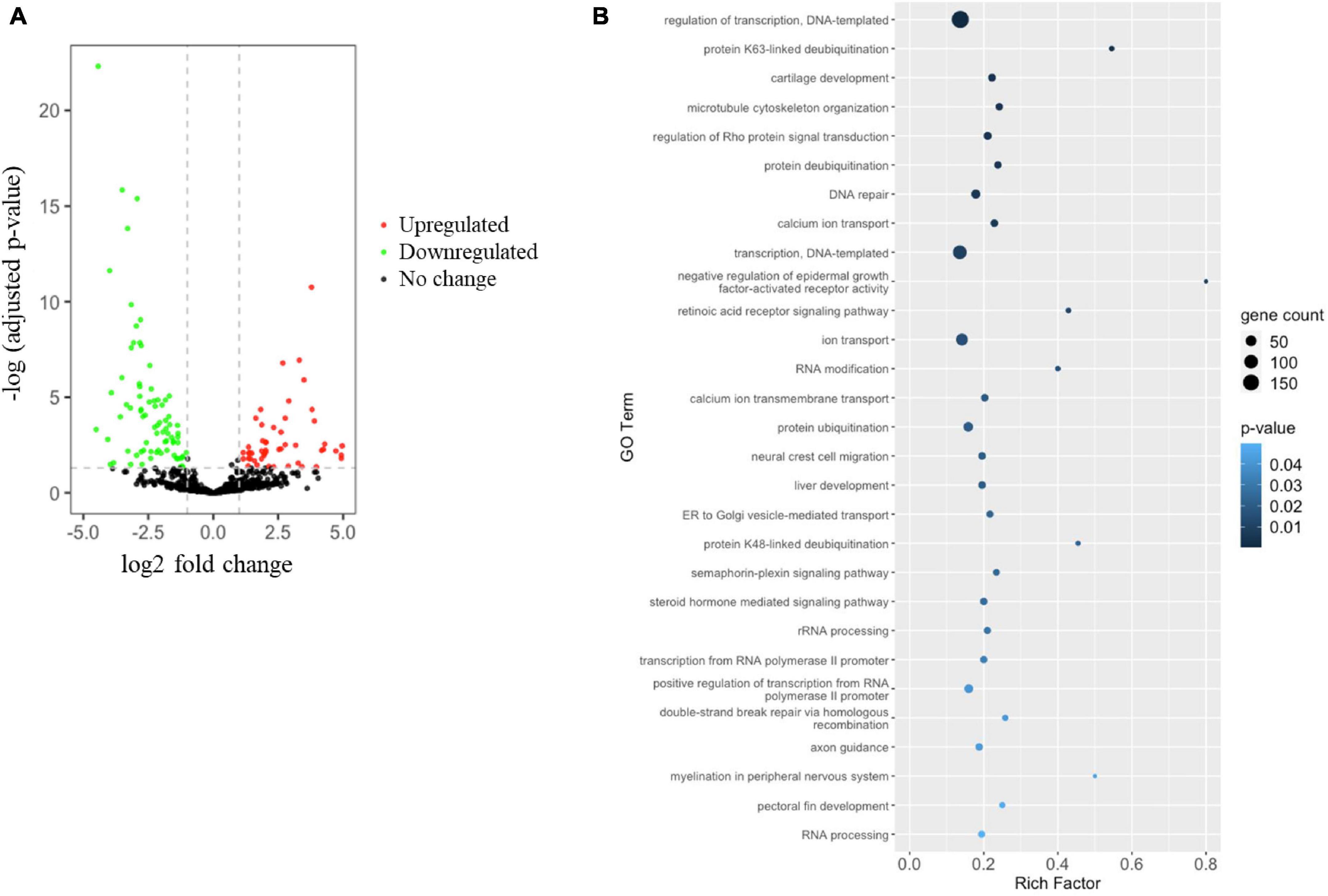
Figure 2. Hypoxia induces miRNA dysregulation, leading to altered medaka embryonic development. (A) Volcano plot indicating differential miRNA expression in medaka embryos after hypoxia or normoxia exposure. miRNA with | log2 (fold change: hypoxia/normoxia)| > 1 and –log B&H corrected p-value > 1.3 were considered differentially expressed. Red dots represent upregulated miRNA, green dots represent downregulated miRNA, and black dots represent miRNA with no significant change. (B) Rich factor plot showing the biological processes altered by the miRNAs target predicted by Gene Ontology (GO) enrichment analysis of Database for Annotation, Visualization and Integrated Discovery (DAVID). Dot size represents the number of target genes. The color intensity of each dot represents the significance of the biological process.
Bioinformatic analysis using the miRanda and IntaRNA algorithms was used to predict the dysregulated miRNA target genes. By combining the results of the 2 algorithms, we found that the hypoxia-dysregulated miRNAs were predicted to target 2466 genes in the embryo (Supplementary Table 4). The predicted target genes were subjected to Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 analysis to further understand the effect of hypoxia-dysregulated miRNA. Our results showed that the hypoxia exposure could alter the gene cluster involved in many biological processes related to embryo development, such as cartilage development, microtubule cytoskeleton organization, liver development, neural crest cell migration, myelination in the peripheral nervous system, and pectoral fin development (Figure 2B). More importantly, fertility and sex determination-related cell signaling including the retinoic acid receptor signaling pathway and the steroid hormone-mediated signaling pathway were highlighted in our analysis (Figure 2B). Although, the hypoxia exposure had no effect on the GSI and HSI indexes (Supplementary Figure 1).
Hypoxia Altered Embryonic Development and Sex Determination Through the Regulation of miRNA–mRNA Pairs
As miRNAs are one of the major mediators of gene expression, we applied comparative transcriptomic analysis to determine the differential gene expression occurring as a result of hypoxia exposure. A total of 163.6 million quality-trimmed raw reads, translating to 8.18 Gb of data were obtained from the RNA sequencing (Table 3). In the comparative transcriptome analysis, we found 3009 DEGs, including 1543 upregulated genes and 1466 downregulated genes in the embryos exposed to hypoxia (Figure 3A and Supplementary Table 5).
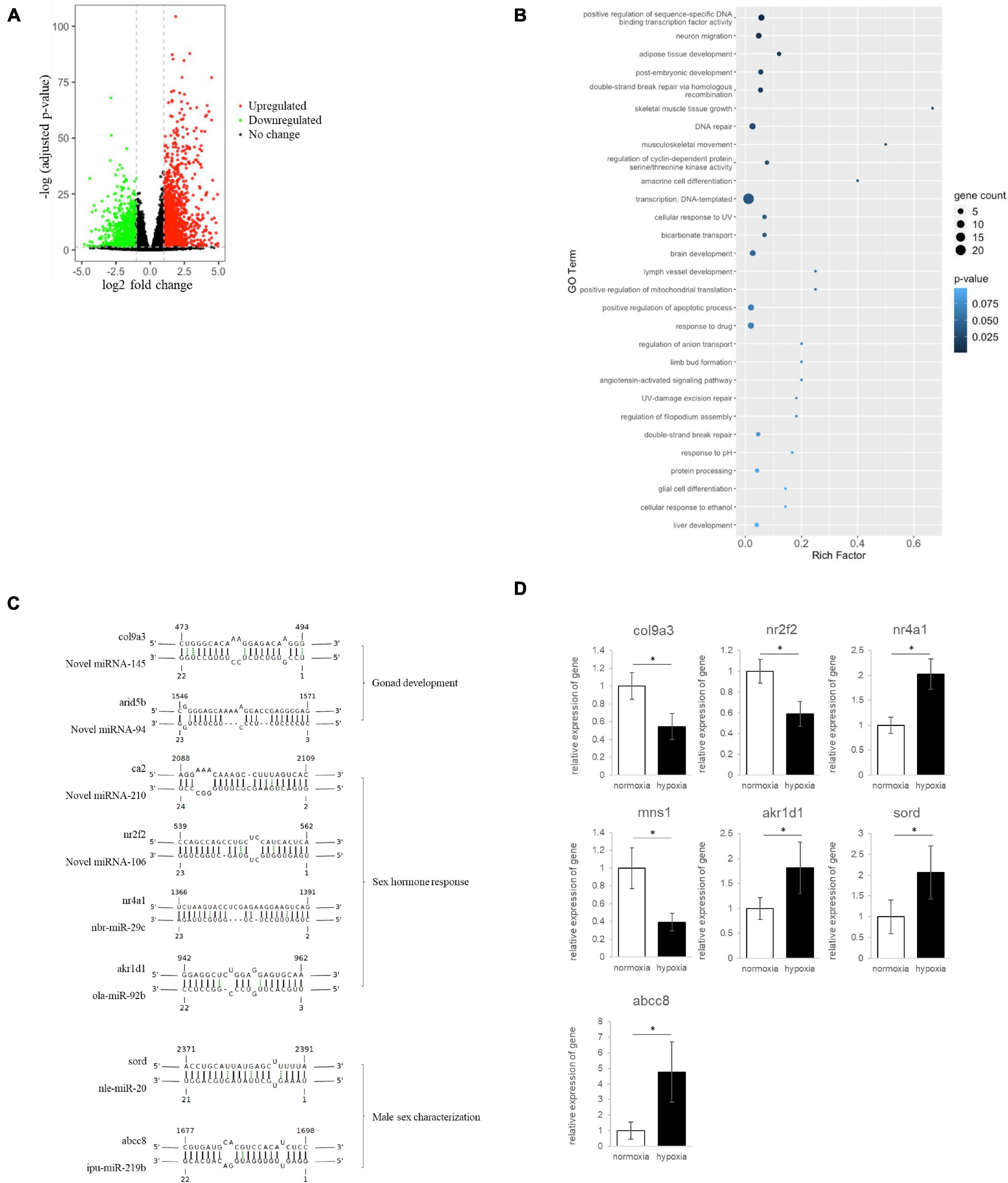
Figure 3. Hypoxia-induced miRNA–mRNA dysregulation, resulting in developmental defects and alteration of sex determination and differentiation in medaka embryos. (A) Volcano plot showing differential mRNA expression in embryos after hypoxia or normoxia exposure. mRNAs with | log2 (fold change: hypoxia/normoxia)| > 1 and –log B&H corrected p-value > 1.3 were considered differentially expressed. Red dots represent upregulated mRNA, green dots represent downregulated mRNA, and black dots represent mRNA with no significant change. (B) Rich factor plots showing the biological processes altered by the miRNA–mRNA pairs using Gene Ontology (GO) enrichment analysis of Database for Annotation, Visualization and Integrated Discovery (DAVID). Dot size represents the number of genes. The color intensity of each dot represents the significance of the biological process. (C) Hypoxia-dysregulated miRNA–mRNA pairs related to gonad development, sex hormone response, male sex characterization. The complementary sequence shows the interaction between miRNA and mRNA. (D) Quantitative PCR validated the differential gene expression in embryo after the hypoxia exposure. * Represented a statistically significant different of gene expression between normoxia and hypoxia groups.
We then integrated the miRNA and mRNA sequencing results to look at the reversed expression of miRNA and mRNA. Our results indicated that hypoxia could lead to the dysregulation of 167 miRNA–mRNA interaction pairs including 31 downregulated miRNA-upregulated mRNA pairs (Table 4) and 136 upregulated miRNA-downregulated mRNA pairs (Table 5). The miRNA-targeted mRNA was subjected to DAVID analysis to understand the effect of hypoxia-dysregulated miRNA–mRNA pairs in the embryo. Our results showed that hypoxia exposure could result in alterations to miRNA-mediated genes related to different developmental processes, such as adipose tissue development, skeletal muscle tissue growth, and post-embryonic development (Figure 3B). Additionally, hypoxia exposure altered the gene cluster related to neuron migration and amacrine cell differentiation, leading to the interference of brain development through the control of miRNA–mRNA interactions (Figure 3B). Finally, we further assessed the possible impact of hypoxia on sex determination and differentiation through miRNA-mRNA regulation. Using gene ontology analysis, we identified miRNA-mRNA pairs that may be involved in biological functions related to sex determination and differentiation including novel miR-145-col9a3 and novel miRNA-94- arid5b in gonad development, novel miRNA-210-ca2, novel miRNA-106-nr2f2, nbr-miR-29c-nr4a1, and ola-miR-92b-akr1d1 in sex hormone response, and novel miRNA-145-mns1, nle-miR-20-sord, and ipu-miR-219b-abcc8 in sex characteristic development (Figure 3C). The differential gene expression was further validated by using quantitative PCR (qPCR), our data showed that the result of qPCR matched with the finding of RNA sequencing (Figure 3D). Taken together, our data suggest that hypoxia may alter sex determination and differentiation through the regulation of miRNA-mRNA pairs.
Discussion
Using small-RNA sequencing analysis, we identified 253 conserved miRNA and 572 novel miRNAs in the medaka embryo. The larger number of novel miRNAs being identified in this study compared to conserved miRNAs suggests that many marine medaka miRNAs are yet to be identified. When we compared our results to the limited miRNAs previously identified in marine medaka which is approximately 200 novel miRNAs identified in different organs such as brain, liver, and gonads (Lai et al., 2016). Many novel miRNAs in embryos are important in fish embryonic development, such as bone and gonadal development (Hausser and Zavolan, 2014; Li et al., 2016).
We then looked at the miRNAs altered by hypoxia exposure by comparing the miRNA profile of normoxic and hypoxic exposed embryos. We observed some conserved miRNAs, which are reported to play important roles in embryonic development. For example, the downregulated miR-214 is a developmental regulator, which controls the polycomb protein, Ezh2, in skeletal muscle and embryonic stem cells (Gan et al., 2016). Additionally, an in vitro study demonstrated that miR-214 expression levels are controlled by the transcription factor Twist-1 in the development of specific neural cell populations (Presslauer et al., 2017). Another hypoxia-downregulated miRNA − miR-29c reportedly affects lateral development and cardiac circulation through the Wnt4/β-catenin signaling pathway in zebrafish (Juan et al., 2009). The involvement of miR-29c has also been implicated in bovine blastocyst development and embryo implantation of rats with endometriosis (Lee et al., 2009; Shen et al., 2020), suggesting the importance of hypoxia dysregulated miR-29c in embryonic development.
Our results also highlighted miR-19b, which was reduced following hypoxia exposure, and is reported to impair cardiac development in zebrafish by targeting ctnnb1(Goossens et al., 2013). Our results in medaka are also concordant with a bird study of miRNAs in great tits that found miRNA-19b to be a hypoxia-responsive miRNA, which regulated MAPK1 expression in embryonic fibroblasts (Cai et al., 2018). In addition, the expression level of miRNA-19b is also associated with embryo quality (Li et al., 2014). We also found hypoxia to suppress the expression of miR-204; this miRNA is reported to alter neuronal migration and cortical morphogenesis during embryonic development in mouse embryos (Chen et al., 2018), and both human and zebrafish studies have demonstrated miR-204-mediated control of developmental lymphangiogenesis (Abu-Halima et al., 2020). The reduction of these miRNAs by hypoxia is suggestive of the possible effects of hypoxia exposure on embryonic development through miRNA regulation. Following analysis of miRNAs for which expression was induced by hypoxia exposure, we found elevated expression levels in a large number of novel miRNAs, but not conserved miRNAs. This result made determining the possible effect of the novel miRNA profile change complex. Therefore, we applied two algorithms (MiRanda and IntaRNA) to predict miRNA target genes. To strengthen our findings, we also conducted comparative transcriptome sequencing followed by the miRNA and mRNA integrative analysis, to further determine miRNA-mRNA interaction pairs. Hundred and ninety-nine miRNA–mRNA pairs were identified, and DAVID analysis was performed on the miRNA target genes to determine the effect of hypoxia-dysregulated miRNA-mRNA pairs. For the data analysis, we focused primarily on the endpoint of embryonic development and sex determination and differentiation. In the functional characterization, novel miRNA-145-col9a3 was reported to be involved in gonad development, as Col9a3 encodes the α3 chain of type IX collagen, which is expressed during mouse testis development (Venø et al., 2017; Jung et al., 2019). Another mouse study demonstrated the specified expression of Col9a3 in testicular cords during the early stages of gonadal differentiation (Hanson-Kahn et al., 2018). More importantly, Col9a3 expression was markedly upregulated in the male, but remained very low in the female during embryo development (Hanson-Kahn et al., 2018), suggesting the possible role of Col9a3 in sex differentiation.
Our findings also highlighted some hypoxia-dysregulated miRNA-mRNA pairs such as novel miRNA-210-ca2, novel miRNA-106-nr2f2, and nbr-miR-29c-nr4a1, which could contribute to sex-steroid hormone response. The novel miRNA, miRNA-210, mediates carbonic anhydrase II (CA2), which is a metalloenzyme responsible for the maintenance of the acid-base balance in body systems (McClive and Sinclair, 2003). Rat studies have demonstrated an association between CA2 and sex-steroid hormones, for example, CA2 expressed in the rat lateral prostate and seminal vesicles is under testosterone regulation (Perera et al., 2001), and it is involved in bicarbonate production − a function that particularly characterizes the rat lateral prostate (Perera et al., 2001). The expression level of CA2 is also differentially regulated by testosterone in the dorsal and lateral prostate in rats (Sanyanga et al., 2019). Other than male sex hormones, CA2 is also regulated by female sex hormones. Hepatic carbonic anhydrase is reportedly induced by estrogen in rat models (Härkönen and Väänänen, 1988), and estrogen and progesterone differentially regulate CA2 in ovariectomized rat uteri (Härkönen et al., 1991). The other hypoxia-altered miRNA-mRNA pair, novel miRNA-106-nr2f2, was also found to be associated with the development of sex organs. Nuclear Receptor Subfamily 2 Group F Member 2 (nr2f2), encodes a ligand-inducible transcription factor and is expressed during early ovarian development of embryogenesis in ovarian somatic cells (Garg, 1975). A review of genetic disease studies showed that rare disease differences or disorders of sex development (DSD) are associated with mutations in NR2F2 (Karim et al., 2016). Furthermore, a study of human genetic disorders demonstrated that NR2F2 loss of function caused defects in testis development in individuals with DSD (Rastetter et al., 2014).
As well as novel miRNAs, our result also highlighted the conserved miR-29c-nr4a1, which is also involved in sex differentiation. Nr4a1, an orphan nuclear receptor, is important for hormone-induced steroidogenesis in Leydig cells, where it plays a pivotal role in regulating the expression of several genes involved in male sex differentiation (Kremen and Chan, 2019). Nr4a1 can be activated by many transcription factors such as myocyte enhancer factor 2 to control Leydig cell gene expression (Bashamboo et al., 2018). It has also been reported that Nr4a1 is a regulator of Insulin-like 3 (INSL3) transcription (Garg, 1975), which is a hormone produced by fetal Leydig cells of the testis to regulate testicular descent during fetal life. In adults, Nr4a1 acts as a germ cell survival factor (Abdou et al., 2016).
Finally, novel miR-145-mns1 and nle-miR-20-sord pairs are reported to play roles in male sex characteristics. Meiosis-specific nuclear structural protein 1 (Mns1) is necessary for spermiogenesis and motile cilia function, such as sperm flagella, through its interaction with ciliary proteins (Robert et al., 2006; Daems et al., 2014). Studies of Mns1-deficient mice demonstrate that homozygous loss-of-function mutations in Mns1 resulted in laterality defects and male infertility (Boschen et al., 2018). This finding was supported by a human population study combining genome-wide SNP mapping and whole-exome sequencing analysis that identified MNS1 variant as a cause of male infertility in humans (Zhou et al., 2012).
For nle-miR-20-sord, sorbitol dehydrogenase (Sord) expression is reported to be regulated by androgens in the human prostate, suggesting that this is the location of the physiological role of sord (Ta-Shma et al., 2018). Furthermore, Sord expression is induced to support epididymal sperm motility during spermatogenic differentiation in mouse spermatogenesis (Leslie et al., 2020). Accordingly, reduced expression of Sord in the mouse epididymis results in inhibition of sperm maturation through the impairment of secretory functions of the epididymis (Szabó et al., 2010).
Conclusion
Our integrative analysis of small RNA sequencing and transcriptome sequencing identified a cluster of miRNA–mRNA pairs that may involve in embryonic development. Our findings have also provided important insights into the molecular mechanisms through which miRNAs interact with different target genes that may regulate the reproductive axis and mediate hypoxia-altered plasticity of sex determination and differentiation in fish. But further studies are required to confirm the role of miRNA–mRNA pairs in physiological processes of hypoxia-altered sex biased in fish.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA706118.
Ethics Statement
The animal study was reviewed and approved by all animal research procedures were approved by the Animal Ethics Committee on Research Experiments involving Animal Subjects (A-0244) of the City University of Hong Kong.
Author Contributions
KL, SC, and RK contributed to conception and design of the study. NT, YK, and WT organized the database. CT, CL, and YK performed the experiments. YC, XL, and TC performed the bioinformatics and statistical analysis. KL and RK wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was fully supported by a grant from the General Research Fund (CityU 11102918) of the Research Grants Council of Hong Kong SAR, People’s Republic of China. KL was supported by the Hong Kong SAR, Macao SAR, and Taiwan Province Talented Young Scientist Program of Guangxi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.736362/full#supplementary-material
Footnotes
References
Abdou, H. S., Robert, N. M., and Tremblay, J. J. (2016). Calcium-dependent Nr4a1 expression in mouse Leydig cells requires distinct AP1/CRE and MEF2 elements. J. mol. Endocrinol. 56, 151–161. doi: 10.1530/JME-15-0202
Abu-Halima, M., Khaizaran, Z. A., Ayesh, B. M., Fischer, U., Khaizaran, S. A., Al-Battah, F., et al. (2020). MicroRNAs in combined spent culture media sterility. Fertil. Steril. 113, 970–980. doi: 10.1016/j.fertnstert.2019.12.028
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England). 31, 166–169. doi: 10.1093/bioinformatics/btu638
Andrews, S. (2010). A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Baroiller, J. F., D’Cotta, H., and Saillant, E. (2009). Environmental effects on fish sex determination and differentiation. Sex Dev. 3, 118–135. doi: 10.1159/000223077
Bashamboo, A., Eozenou, C., Jorgensen, A., Bignon-Topalovic, J., Siffroi, J. P., Hyon, C., et al. (2018). Loss of function of the nuclear receptor NR2F2, encoding COUP-TF2, causes testis development and cardiac defects in 46,XX children. Am. J. Hum. Genet. 102, 487–493. doi: 10.1016/j.ajhg.2018.01.021
Boschen, K. E., Gong, H., Murdaugh, L. B., and Parnell, S. E. (2018). Knockdown of Mns1 increases susceptibility to craniofacial defects following gastrulation-stage alcohol exposure in mice. Alcohol. Clin. Exp. Res. 42, 2136–2143. doi: 10.1111/acer.13876
Bouchareb, A., Le Cam, A., Montfort, J., Gay, S., Nguyen, T., Bobe, J., et al. (2017). Genome-wide identification of novel ovarian-predominant miRNAs: new insights from the medaka (Oryzias latipes). Sci. Rep. 7:40241. doi: 10.1038/srep40241
Busch, A., Andreas, S. A., and Backofen, R. (2018). IntaRNA: ecient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24, 2849–2856. doi: 10.1093/bioinformatics/btn544
Cai, H., Zhu, X. X., Li, Z. F., Zhu, Y. P., and Lang, J. H. (2018). MicroRNA dysregulation and steroid hormone receptor expression in uterine tissues of rats with endometriosis during the implantation window. Chin. Med. J. 131, 2193–2204. doi: 10.4103/0366-6999.240808
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chen, X., Qu, Y., Cheng, Y., Wang, J., Lei, X., Song, G., et al. (2018). MiR-19b-3p Regulates MAPK1 expression in embryonic fibroblasts from the great tit (parus major) under hypoxic conditions. Cell. Physiol. Biochem. 46, 546–560. doi: 10.1159/000488621
Cheung, C. H., Chiu, J. M., and Wu, R. S. (2014). Hypoxia turns genotypic female medaka fish into phenotypic males. Ecotoxicology 23, 1260–1269. doi: 10.1007/s10646-014-1269-8
Daems, C., Martin, L. J., Brousseau, C., and Tremblay, J. J. (2014). MEF2 is restricted to the male gonad and regulates expression of the orphan nuclear receptor NR4A1. Mol. Endocrinol. (Baltimore, Md.) 28, 886–898. doi: 10.1210/me.2013-1407
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. doi: 10.1093/bioinformatics/bts635
Eshel, O., Shirak, A., Dor, L., Band, M., Zak, T., Markovich-Gordon, M., et al. (2014). Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15:774. doi: 10.1186/1471-2164-15-774
Fagegaltier, D., König, A., Gordon, A., Lai, E. C., Gingeras, T. R., Hannon, G. J., et al. (2014). Genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics 198, 647–668. doi: 10.1534/genetics.114.169268
Feng, Y. P., Chen, J. F., Huang, P., Wang, X., Wang, J., Peng, X. L., et al. (2014). Expression analysis of differentially expressed miRNAs in male and female chicken embryos. Genet. Mol. Res. 13, 3060–3068.
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gan, S., Huang, Z., Liu, N., Su, R., Xie, G., Zhong, B., et al. (2016). MicroRNA-140-5p impairs zebrafish embryonic bone development via targeting BMP-2. FEBS Lett. 590, 1438–1446. doi: 10.1002/1873-3468.12190
Garg, L. C. (1975). Induction of hepatic carbonic anhydrase by estrogen. J. Pharmacol. Exp. Ther. 192, 297–302.
Gay, S., Bugeon, J., Bouchareb, A., Henry, L., Delahaye, C., Legeai, F., et al. (2018). MiR-202 controls female fecundity by regulating medaka oogenesis. PLoS Genet. 14:e1007593. doi: 10.1371/journal.pgen.1007593
Goossens, K., Mestdagh, P., Lefever, S., Van Poucke, M., Van Zeveren, A., Van Soom, A., et al. (2013). Regulatory microRNA network identification in bovine blastocyst development. Stem Cells Dev. 1, 1907–1920. doi: 10.1089/scd.2012.0708
Hanson-Kahn, A., Li, B., Cohn, D. H., Nickerson, D. A., Bamshad, M. J., University of Washington Center for Mendelian Genomics, et al. (2018). Autosomal recessive Stickler syndrome resulting from a COL9A3 mutation. Am. J. Med. Genet. 176, 2887–2891. doi: 10.1002/ajmg.a.40647
Härkönen, P. L., Mäkelä, S. I., Valve, E. M., Karhukorpi, E. K., and Väänänen, H. K. (1991). Differential regulation of carbonic anhydrase II by androgen and estrogen in dorsal and lateral prostate of the rat. Endocrinology 128, 3219–3232. doi: 10.1210/endo-128-6-3219
Härkönen, P. L., and Väänänen, H. K. (1988). Androgen regulation of carbonic anhydrase II, a major soluble protein in rat lateral prostate tissue. Biol. Reprod. 38, 377–384. doi: 10.1095/biolreprod38.2.377
Hausser, J., and Zavolan, M. (2014). Identification and consequences of miRNA-target interactions beyond repression of gene expression. Nat. Rev. Genet. 15, 599–612.
Jing, J., Wu, J. J., Liu, W., Xiong, S. T., Ma, W. G., Zhang, J., et al. (2014). Sexbiased miRNAs in gonad and their potential roles for testis development in yellow catfish. PLoS One 9:e107946. doi: 10.1371/journal.pone.0107946
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S. (2005). Human MicroRNA targets. PLoS Biol. 2:e363. doi: 10.1371/journal.pbio.0020363
Juan, A. H., Kumar, R. M., Marx, J. G., Young, R. A., and Sartorelli, V. (2009). Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 36, 61–74. doi: 10.1016/j.molcel.2009.08.008
Juanchich, A., Le Cam, A., Montfort, J., Guiguen, Y., and Bobe, J. (2013). Identification of differentially expressed miRNAs and their potential targets during fish ovarian development. Biol. Reprod. 88:128. doi: 10.1095/biolreprod.112.105361
Jung, H. M., Hu, C. T., Fister, A. M., Davis, A. E., Castranova, D., Pham, V. N., et al. (2019). MicroRNA-mediated control of developmental lymphangiogenesis. eLife 8:e46007. doi: 10.7554/eLife.46007
Kang, L., Cui, X., Zhang, Y., Yang, C., and Jiang, Y. (2004). Identification of miRNAs associated with sexual maturity in chicken ovary by Illumina small RNA deep sequencing. BMC Genomics 14:352. doi: 10.1186/1471-2164-14-352
Karim, K., Giribabu, N., Muniandy, S., and Salleh, N. (2016). Estrogen and progesterone differentially regulate carbonic anhydrase II, III, IX, XII, and XIII in ovariectomized rat uteri. Syst. Biol. Reprod. Med. 62, 57–68. doi: 10.3109/19396368.2015.1112699
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2018). Mirbase: from microrna sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kremen, J., and Chan, Y. M. (2019). Genetic evaluation of disorders of sex development: current practice and novel gene discovery. Curr. Opin. Endocrinol. Diabetes obes. 26, 54–59. doi: 10.1097/MED.0000000000000452
Langmead, B. (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformat. 2010:Chater11,Unit–11.7. doi: 10.1002/0471250953.bi1107s32
Lai, K. P., Li, J. W., Tse, A. C. K., Chan, T. F., and Wu, R. S. S. (2016). Hypoxia alters steroidogenesis in female marine medaka through miRNA regulation. Aquat. Toxicol. 172, 1–8.
Lau, K., Lai, K. P., Bao, J. Y., Zhang, N., Tse, A., Tong, A., et al. (2014). Identification and expression profiling of microRNAs in the brain, liver and gonads of marine medaka (Oryzias melastigma) and in response to hypoxia. PLos One 9:e110698. doi: 10.1371/journal.pone.0110698
Lee, Y. B., Bantounas, I., Lee, D. Y., Phylactou, L., Caldwell, M. A., and Uney, J. B. (2009). Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 37, 123–128. doi: 10.1093/nar/gkn920
Leslie, J. S., Rawlins, L. E., Chioza, B. A., Olubodun, O. R., Salter, C. G., Fasham, J., et al. (2020). MNS1 variant associated with situs inversus and male infertility. Eur. J. Hum. Genet. 28, 50–55. doi: 10.1038/s41431-019-0489-z
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, J. W., Lin, X., Tse, A., Cheung, A., Chan, T. F., Kong, R. Y., et al. (2016). Discovery and functional characterization of novel miRNAs in the marine medaka Oryzias melastigma. Aquat. Toxicol. (Amsterdam, Netherlands) 175, 106–116. doi: 10.1016/j.aquatox.2016.03.013
Li, M., Hu, X., Zhu, J., Zhu, C., Zhu, S., Liu, X., et al. (2014). Overexpression of miR-19b impairs cardiac development in zebrafish by targeting ctnnb1. Cell. Physiol. Biochem. 33, 1988–2002. doi: 10.1159/000362975
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, H., Hostuttler, M., Wei, H. R., Rexroad, C. E., and Yao, J. B. (2012). Characterization of the rainbow trout egg microRNA transcriptome. PLoS One 7:e39649. doi: 10.1371/journal.pone.0039649
Mann, M., Wright, P. R., and Backofen, R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45, W435–W439. doi: 10.1093/nar/gkx279
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10–12. doi: 10.1089/cmb.2017.0096
McClive, P. J., and Sinclair, A. H. (2003). Type II and type IX collagen transcript isoforms are expressed during mouse testis development. Biol. Reprod. 68, 1742–1747. doi: 10.1095/biolreprod.102.008235
Mishima, T., Takizawa, T., Luo, S. S., Ishibashi, O., Kawahigashi, Y., Mizuguchi, Y., et al. (2008). MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction 136, 811–822.
Navarro-Martín, L., Viñas, J., Ribas, L., Díaz, N., Gutiérrez, A., Di Croce, L., et al. (2011). DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7:e1002447. doi: 10.1371/journal.pgen.1002447
Pelley, J. (2006). Dead zones might masculinize fish. Environ. Sci. Technol. 40:2862. doi: 10.1021/es0630137
Perera, E. M., Martin, H., Seeherunvong, T., Kos, L., Hughes, I. A., Hawkins, J. R., et al. (2001). Tescalcin, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, Col9a3, and renin are expressed in the mouse testis during the early stages of gonadal differentiation. Endocrinology 142, 455–463. doi: 10.1210/endo.142.1.7882
Pörtner, H. O., and Peck, M. A. (2010). Climate change effects on fishes and fisheries: towards a cause-and-effect understanding. J. Fish. Biol. 77, 1745–1779. doi: 10.1111/j.1095-8649.2010.02783.x
Presslauer, C., Tilahun Bizuayehu, T., Kopp, M., Fernandes, J. M., and Babiak, I. (2017). Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 7:43850. doi: 10.1038/srep43850
Raden, M., Ali, S. M., Alkhnbashi, O. S., Busch, A., Costa, F., Davis, J. A., et al. (2018). Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Res. 46, W25–W29. doi: 10.1093/nar/gky329
Rastetter, R. H., Bernard, P., Palmer, J. S., Chassot, A. A., Chen, H., Western, P. S., et al. (2014). Marker genes identify three somatic cell types in the fetal mouse ovary. Dev. Biol. 394, 242–252. doi: 10.1016/j.ydbio.2014.08.013
Reddon, A. R., and Hurd, P. L. (2013). Water pH during early development influences sex ratio and male morphin a West African cichlid fish, Pelvicachromis pulcher. Zoology (Jena) 116, 139–143. doi: 10.1016/j.zool.2012.11.001
Robert, N. M., Martin, L. J., and Tremblay, J. J. (2006). The orphan nuclear receptor NR4A1 regulates insulin-like 3 gene transcription in Leydig cells. Biol. Reprod. 74, 322–330. doi: 10.1095/biolreprod.105.044560
Sanyanga, T. A., Nizami, B., and Bishop, ÖT. (2019). Mechanism of action of non-synonymous single nucleotide variations associated with α-carbonic anhydrase ii deficiency. Molecules (Basel, Switzerland). 24, 3987. doi: 10.3390/molecules24213987
Shang, E. H. H., Yu, R. M. K., and Wu, R. S. S. (2006). Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio). Environ. Sci. Technol. 40, 3118–3122. doi: 10.1021/es0522579
Shen, Y., Lu, H., Chen, R., Zhu, L., and Song, G. (2020). MicroRNA-29c aing Wnt4. Mol. Med. Rep. 22, 4675–4684. doi: 10.3892/mmr.2020.11584
Szabó, Z., Hämäläinen, J., Loikkanen, I., Moilanen, A. M., Hirvikoski, P., Väisänen, T., et al. (2010). Sorbitol dehydrogenase expression is regulated by androgens in the human prostate. Oncol. Rep. 23, 1233–1239. doi: 10.3892/or_00000755
Ta-Shma, A., Hjeij, R., Perles, Z., Dougherty, G. W., Abu Zahira, I., Letteboer, S. J. F., et al. (2018). Homozygous loss-of-function mutations in MNS1 cause laterality defects and likely male infertility. PLoS Genet. 14:e1007602. doi: 10.1371/journal.pgen.1007602
Thomas, P., and Rahman, M. S. (2012). Extensive reproductive disruption, ovarian masculinization and aromatase suppression in Atlantic croaker in the northern Gulf of Mexico hypoxic zone. Proc. Biol. Sci. 279, 28–38. doi: 10.1098/rspb.2011.0529
Thrash, J. C., Seitz, K. W., Baker, B. J., Temperton, B., Gillies, L. E., Rabalais, N. N., et al. (2017). Metabolic roles of uncultivated bacterioplankton lineages in the northern Gulf of Mexico “Dead Zone. mBio 8, e1017–e1017. doi: 10.1128/mBio.01017-17
Venø, M. T., Venø, S. T., Rehberg, K., van Asperen, J. V., Clausen, B. H., Holm, I. E., et al. (2017). Cortical morphogenesis during embryonic development is regulated by miR-34c and miR-204. Front. Mol. Neurosci. 10:31. doi: 10.3389/fnmol.2017.00031
Wang, W., Liu, W., Liu, Q., Li, B., An, L., Hao, R., et al. (2016). Coordinated microRNA and messenger RNA expression profiles for understanding sexual dimorphism of gonads and the potential roles of microRNA in the steroidogenesis pathway in Nile tilapia (Oreochromis niloticus). Theriogenology 85, 970–978. doi: 10.1016/j.theriogenology.2015.11.006
Wright, P. R., Georg, J., Mann, M., Sorescu, D. A., Richter, A. S., Lott, S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42, W119–W123. doi: 10.1093/nar/gku359
Yamamoto, Y., Zhang, Y., Sarida, M., Hattori, R. S., and Strüssmann, C. A. (2014). Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. PLoS One 9:e102574. doi: 10.1371/journal.pone.0102574
Keywords: hypoxia, fish, sequencing, miRNA, bioinformatics
Citation: Lai KP, Tam NYK, Chen Y, Leung CT, Lin X, Tsang CF, Kwok YC, Tse WKF, Cheng SH, Chan TF and Kong RYC (2022) miRNA–mRNA Integrative Analysis Reveals the Roles of miRNAs in Hypoxia-Altered Embryonic Development- and Sex Determination-Related Genes of Medaka Fish. Front. Mar. Sci. 8:736362. doi: 10.3389/fmars.2021.736362
Received: 08 July 2021; Accepted: 14 December 2021;
Published: 21 January 2022.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Jae-Sung Rhee, Incheon National University, South KoreaBindhu Paul, Amrita Vishwa Vidyapeetham, India
Copyright © 2022 Lai, Tam, Chen, Leung, Lin, Tsang, Kwok, Tse, Cheng, Chan and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keng Po Lai, a2VuZ3BsYWlAY2l0eXUuZWR1Lmhr; Richard Yuen Chong Kong, Ymhya29uZ0BjaXR5dS5lZHUuaGs=
 Keng Po Lai
Keng Po Lai Nathan Yi Kan Tam2
Nathan Yi Kan Tam2 Yuelong Chen
Yuelong Chen Xiao Lin
Xiao Lin William Ka Fai Tse
William Ka Fai Tse Ting Fung Chan
Ting Fung Chan Richard Yuen Chong Kong
Richard Yuen Chong Kong