
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 29 September 2021
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.731517
This article is part of the Research Topic Mollusk Breeding and Genetic Improvement View all 25 articles
 Xiaokai Bao1†
Xiaokai Bao1† Yan Li1†
Yan Li1† Jianbai Zhang2
Jianbai Zhang2 Xipan Chen1
Xipan Chen1 Xiaohui Xu1
Xiaohui Xu1 Yanwei Feng1
Yanwei Feng1 Guohua Sun1
Guohua Sun1 Xiumei Liu3
Xiumei Liu3 Bin Li1,4
Bin Li1,4 Weijun Wang1,5
Weijun Wang1,5 Zan Li1*
Zan Li1* Jianmin Yang1*
Jianmin Yang1*Immune defense systems are indispensable for living organisms. Within an immune network, problems with any given link can impact the normal life activities of an organism. Amphioctopus fangsiao is a cephalopod that exists widely throughout the world’s oceans. Because of its nervous system and locomotive organs, it has become increasingly studied in recent years. Vibrio anguillarum is one of the most common pathogenic bacteria in aquaculture organisms. It is highly infectious and can infect almost all aquaculture organisms. V. anguillarum infection can cause many adverse biological phenomena, including tissue bleeding. Study the immune response after V. anguillarum infection would help us to understand the molecular mechanisms of immune response in aquaculture organisms. In this research, we infected the primary incubation A. fangsiao with V. anguillarum for 24 h. We analyzed gene expression in A. fangsiao larvae via transcriptome profiles at 0, 4, 12, and 24 h after hatching, and 1,385, 734, and 6,109 differentially expressed genes (DEGs) were identified at these three time points. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were used to identify immune-related DEGs. Protein–protein interaction networks were constructed to examine interactions between immune-related genes. Twenty hub genes involved in multiple KEGG signaling pathways or with multiple protein–protein interaction relationships were identified, and their differential expression verified by quantitative RT-PCR. We first studied V. anguillarum infection of A. fangsiao larvae by means of protein–protein interaction networks. The results provide valuable genetic resources for understanding immunity in molluscan larvae. These data serve as a theoretical basis for the artificial breeding of A. fangsiao.
As a new research direction, mollusks have received extensive attention recently because of their complex immune responses and molecular mechanisms (Sokolova, 2009; Wang et al., 2019). organism growth is inseparable from immune response, and loss of immunity leads to disease and even death, suggesting that immune response is a core factor of the growth of organisms (Rowley and Powell, 2007; Barcia and Ramos-Martínez, 2011; Wang et al., 2019). The larval is a relatively vulnerable stage in life (Ginger et al., 2013; Kaplan et al., 2013). Growth quality among larvae directly affects the quality of life of the adult, impacting characteristics including vitality and physique. The immune functions of larvae are important to the growth process. It is well known that organisms have a higher ability to resist invasion by external pathogens when the numbers and activity of their immune cells are high. Therefore, immune function can effectively protect larvae of organisms, maintaining their normal growth (Wang et al., 2013; Matozzo, 2016).
As a marine mollusk, Amphioctopus fangsiao is widely distributed along the coasts of the Pacific, the Yellow Sea, and the Bohai Sea. Due to the difficulties associated with catching wild A. fangsiao, numbers of A. fangsiao bred in captivity have gradually risen in recent years. However, whether A. fangsiao is wild or bred the larvae are vulnerable to a variety of pathogens including bacteria and viruses (Budelmann, 1994; Wei et al., 2015, 2018). Vibrio anguillarum is a gram-positive bacterium that can cause many popular diseases in aquaculture animals. In recent years, multiple studies have confirmed V. anguillarum infections in many fish species, including Salmo salar and Paralichthys olivaceus (Hoel et al., 1997; Li et al., 2015; Bao et al., 2019). V. anguillarum can infect bivalve and gastropod such as Chlamys nobilis and Haliotis gigantea (Cong et al., 2008; Iehata et al., 2009; Zhang et al., 2016). When aquaculture organisms are infected by V. anguillarum, they display red gills, muscle bleeding, mucosal tissue decay, and other symptoms, ultimately leading to significant losses to the aquaculture industry. V. anguillarum infection requires certain conditions, and the incidence rate of V. anguillarum infection tends to be higher in poor environments or among injured or stimulated organisms (Vázquez et al., 2006; Zhou et al., 2010; Nie et al., 2017). Although there are widely studies on the infection of V. anguillarum in fish, bivalve, and gastropod, there are still few studies have been conducted on the infection of mollusks, especially cephalopods. The pathogenic mechanisms of V. anguillarum in cephalopods are still unclear, and require further study.
Transcriptome sequencing technology has recently become an important method for studying differences between individuals of the same species. To aid in assessing biological immune response mechanisms, transcriptome sequencing can analyze differences in gene expression between different individuals or tissues (Zhao et al., 2012; Silva et al., 2013; Zhang et al., 2018). It can help identify important immune genes and further study biological immune response mechanisms. Previous researches have carried out transcriptome analyses of aquatic mollusks such as Pinctada martensii (Zhao et al., 2012), Biomphalaria glabrata (Silva et al., 2013), Haliotis diversicolor (Zhang et al., 2018), Octopus vulgaris (Castellanos-Martínez et al., 2014), and Euprymna tasmanica (Salazar et al., 2015; Schultz and Adema, 2017). The transcriptome analyses of A. fangsiao is still lacking, which need a lot of experiments to explore.
In this research, we infected the primary incubation A. fangsiao larvae with V. anguillarum for 24 h. We then took A. fangsiao larvae at 0, 4, 12, and 24 h after hatching for transcriptome sequencing and bioinformatics analyses, including gene function annotation, differentially expressed genes (DEGs) analysis, Gene Ontology (GO) functional enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, and immune-related protein–protein interaction network studies. We identified and verified 20 differentially expressed hub genes using quantitative RT-PCR. These results revealed the immunity of A. fangsiao larvae after infection with V. anguillarum, providing a valuable resource for understanding immune response mechanisms in mollusks, and laying a foundation for exploring the immune response mechanisms of other mollusks.
Amphioctopus fangsiao samples were obtained from a commercial hatchery. This research was conducted in accordance with the protocols of the Institutional Animal Care and Use Committee of the Ludong University (protocol number LDU-IRB20210308NXY) and the China Government Principles for the Utilization and Care of Invertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No. 2, October 31, 19881).
The wild parent used in these experiments were obtained from the Rizhao Sea area. After spawning, the eggs were protected and incubated by female parent A. fangsiao. During the incubation process, the seawater temperature was maintained at 19–20.8°C, and the incubation lasted approximately 29 days. After hatching, larvae were temporarily cultured in floating seawater containing 1 × 107 CFU/mL V. anguillarum for 24 h. Primary incubation larvae were collected and stored for 4, 12, and 24 h in liquid nitrogen until RNA was extracted using the TRIzol method.
Nine larvae from primary incubation were randomly selected from each time point for RNA extraction: A. fangsiao larvae infected for 0 h (Oo-C), A. fangsiao larvae infected for 4 h (Oo-4h), A. fangsiao larvae infected for 12 h (Oo-12h), and A. fangsiao larvae infected for 24 h (Oo-24h). At each time point, equal molar masses of RNA from any three of nine larvae were pooled into one replicate to provide a template for the construction of the transcriptome library; any three of the remaining six larvae were pooled into the second replicate; and the final three larvae were pooled into the third replicate. Remaining RNA was retained for subsequent quantitative RT-PCR verification.
Library construction was carried out according to a method described by Li et al. (2017). Finally, we sequenced each sample using an Illumina HiSeq 4000 platform.
A reference sequence was obtained by splicing Trinity with RSEM, and clean reads were mapped to this reference sequence. We then obtained the number of read counts for each sample that mapped to each gene. The read count was transformed into fragments per kilobase of transcript per million mapped reads (FPKM) to analyze the gene expression and abundance, and expression levels of samples were positively correlated with FPKM. The unigenes were annotated by searching the sequences against the NR, NT, GO, SwissProt, and KOG databases using BLASTX with a cut-off of E value ≤ 1e-5. Meanwhile, we annotated unigenes into Pfam database with Hmmer 3.0 package (E value ≤ 0.01).
Differentially expressed genes were screened out using the DESeq2 package for R as a negative binomial distribution model. First of all, data were imported for building the dds model, and then the DESeq function was used to estimate the dispersion of the samples. After that, the difference in gene expressions was analyzed by this package. Finally, DEGs with q-value ≤ 0.05 and |log2 fold change| ≥ 1 were screened out (Love et al., 2014; Ho et al., 2017). Significant DEGs in each time point were screened by GO functional enrichment analysis to obtain GO terms and distribution of DEGs. Statistical analyses of DEGs were conducted. To further understand functions of DEGs, KEGG signaling pathway analysis was performed for up- and down-regulated genes, and we annotated immune pathways involving DEGs. Finally, we screened the signaling pathways in which DEGs were significantly enriched, and species and numbers of DEGs in these pathways were counted.
Through trend analysis, we understood the change trend of gene expression over time. STEM was used to analyze the expressing trend of DEGs, and cluster trends in gene expression quantity. The parameters were set as follows: the Maximum Unit Change in model profiles between time points is 1; maximum output profiles number is 20 (similar profiles will be merged); minimum ratio of fold change of DEGs is no less than 2.0; the criterion for screening trends with significant differences is p-value ≤ 0.05 (Ernst and Bar-Joseph, 2006). Trend analysis showed the expressing trend of genes in the process of larval growth after infection, and predicted the future trend, which can help us explore the relationship between immunity and larval growth of A. fangsiao after infection.
We used STRING v11.0 with default parameters to construct protein–protein interaction networks to further explore gene relationships in immune regulation pathways (Damian et al., 2018).
We verified the accuracy of our RNA-Seq results using quantitative RT-PCR to validate 20 selected genes (Supplementary Table 1). Three biological replicates of each time point were used in the experiment. Primer Premier 5.0 was used to design gene-specific primers. Table 1 lists the 20 selected genes and their corresponding primer sequences. We evaluated the stability of GAPDH, β-actin, and 18S genes in different tissues and embryo development stages of A. fangsiao. The expression level of A. fangsiao β-actin tended to be stable, and it was used as an endogenous control in this experiment. Quantitative RT-PCR was performed according to a method described by Li et al. (2019).
Samples at four time points were sequenced by RNA-Seq. The overview of sequencing results is provided in Table 2. Raw sequencing reads were submitted to Sequence Read Archive in NCBI; the SRA accession numbers were SRR15204591, SRR15204592, SRR15204593, SRR15204594, SRR15204595, SRR15204596, SRR15204597, SRR15204598, SRR15204599, SRR15204600, SRR15204601, and SRR152046022.
Through differential expression analysis, we identified 1,385, 734, and 6,109 DEGs at 4, 12, and 24 h after infecting compared with primary incubation larvae. Among these, 480 DEGs were up-regulated, and 905 DEGs were down-regulated at 4 h after infection; 338 DEGs were up-regulated, and 396 DEGs were down-regulated at 12 h after infection; 1,685 DEGs were up-regulated, and 4,424 DEGs were down-regulated at 24 h after infection (Figure 1 and Supplementary Tables 2–4). A Venn diagram shows all DEGs after infecting. Because DEGs at each of the three time points might be key genes affecting immunity in A. fangsiao larvae, their union set (7,019 DEGs) was selected for subsequent analysis (Figure 2 and Supplementary Table 5). The heatmap (Figure 3 and Supplementary Table 6) intuitively shows the clustering distribution of these DEGs.
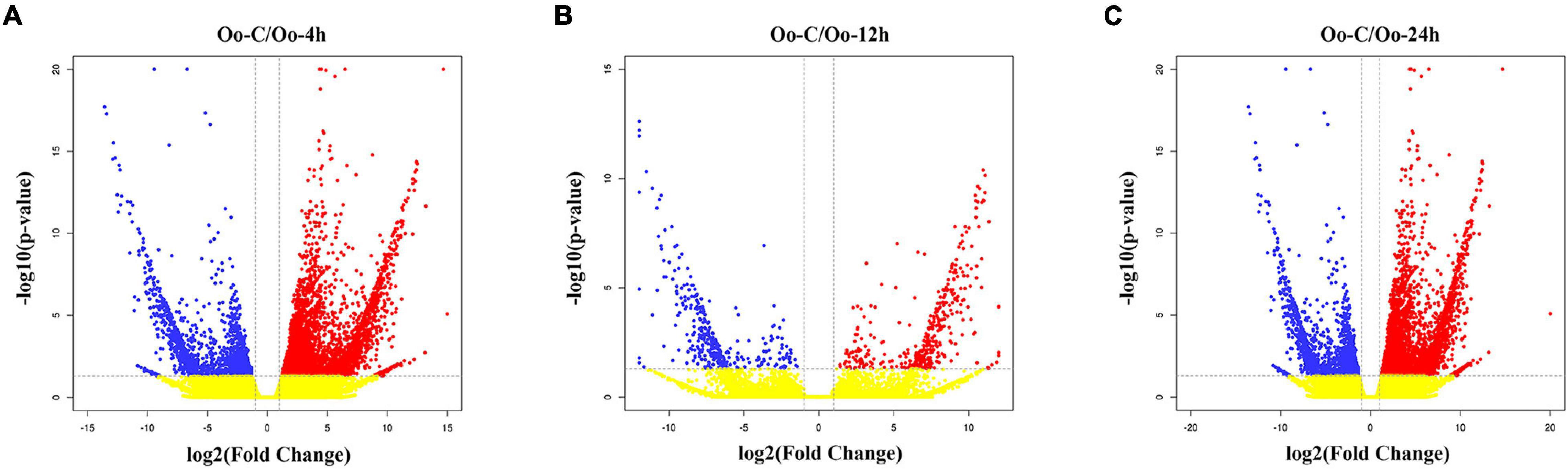
Figure 1. (A) Volcano plot of DEGs distribution trends between Oo-C and Oo-4h. Each dot stands for a gene. Red dots indicate up-regulated DEGs; blue dots are down-regulated DEGs; and yellow dots stand for the genes with no difference. (B) Volcano plot of DEGs distribution trends between Oo-C and Oo-12h. (C) Volcano plot of DEGs distribution trends between Oo-C and Oo-24h.
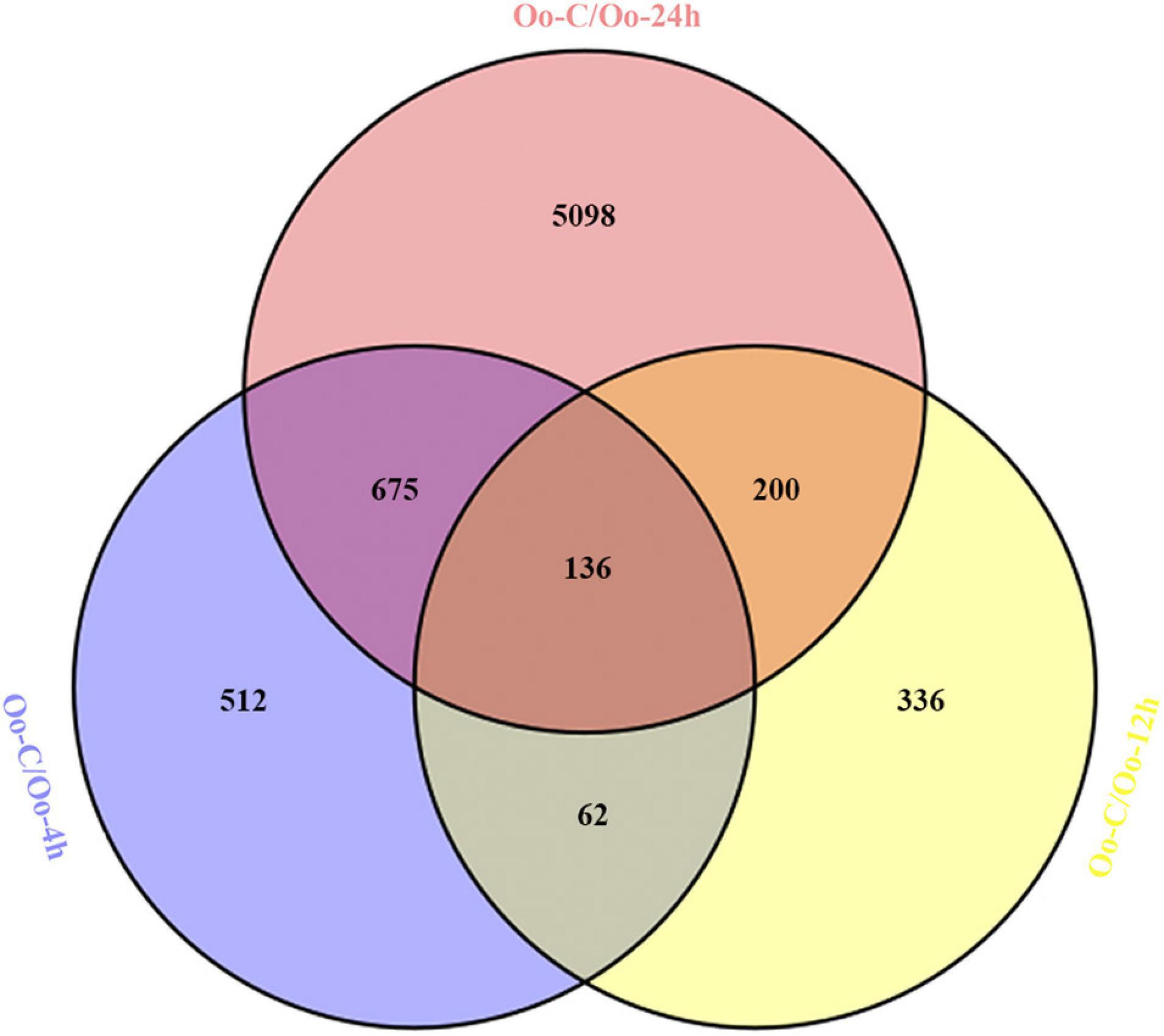
Figure 2. The Venn diagram shows the overlap of the DEGs at 4 h (blue), 12 h (yellow), and 24 h (red) after infecting.
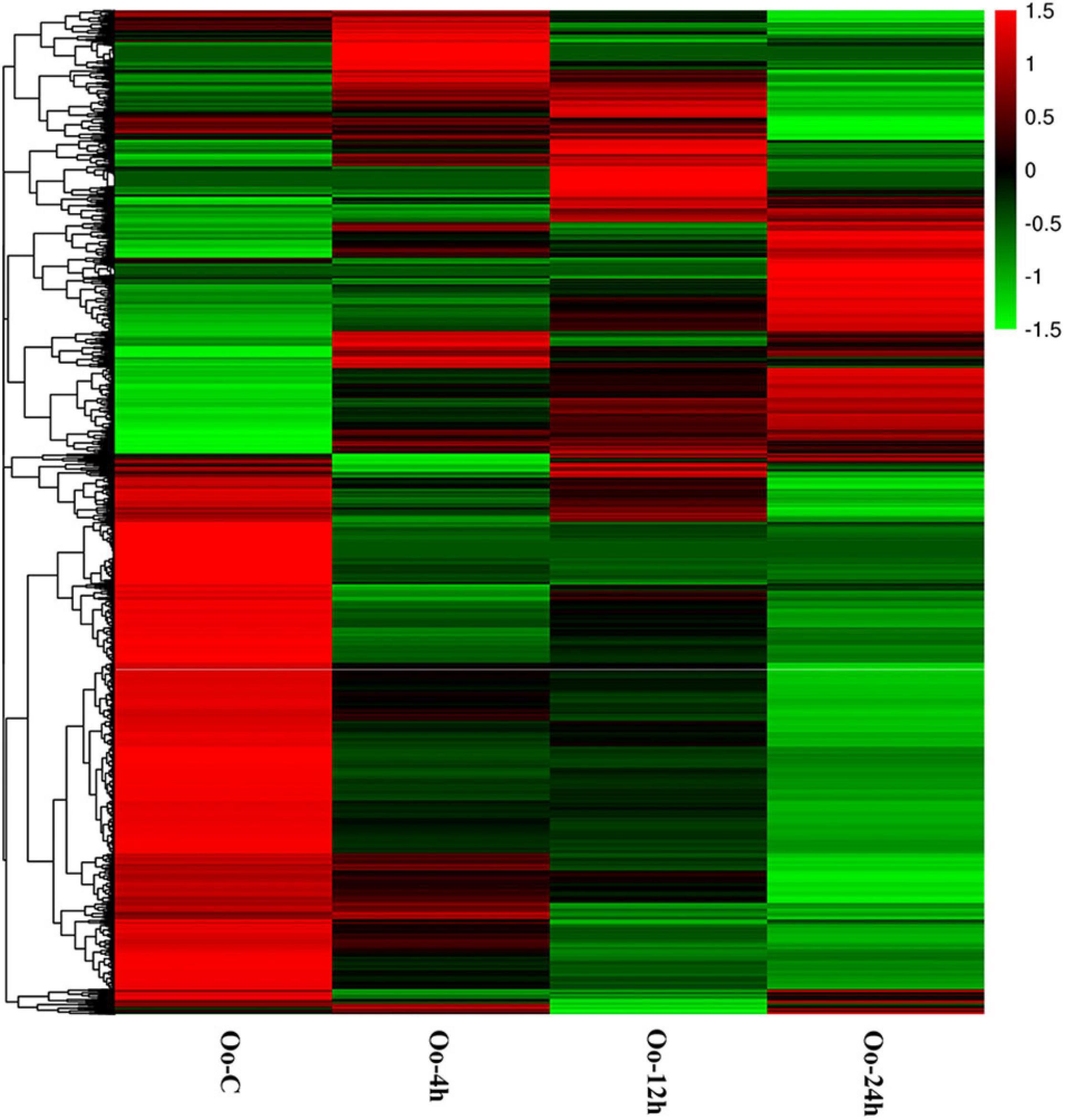
Figure 3. Heatmap analysis of hierarchical clustering of DEGs at four time points. Each row represents one gene, and each column stands for a time point. The colors range from green to red, indicating the level of expression from low to high.
STEM was used to analyze trends in 7,019 DEGs. The results showed that DEGs were divided into 20 expression trends, involving six significantly enriched ones (Figure 4). Their p-value were less than 0.05, and 4,940 DEGs were differentially expressed in these trends. Among them, profile 0 (Figure 4D) is the most significant trend, and profile 9 (Figure 4C) has the most genes expressed in this trend.
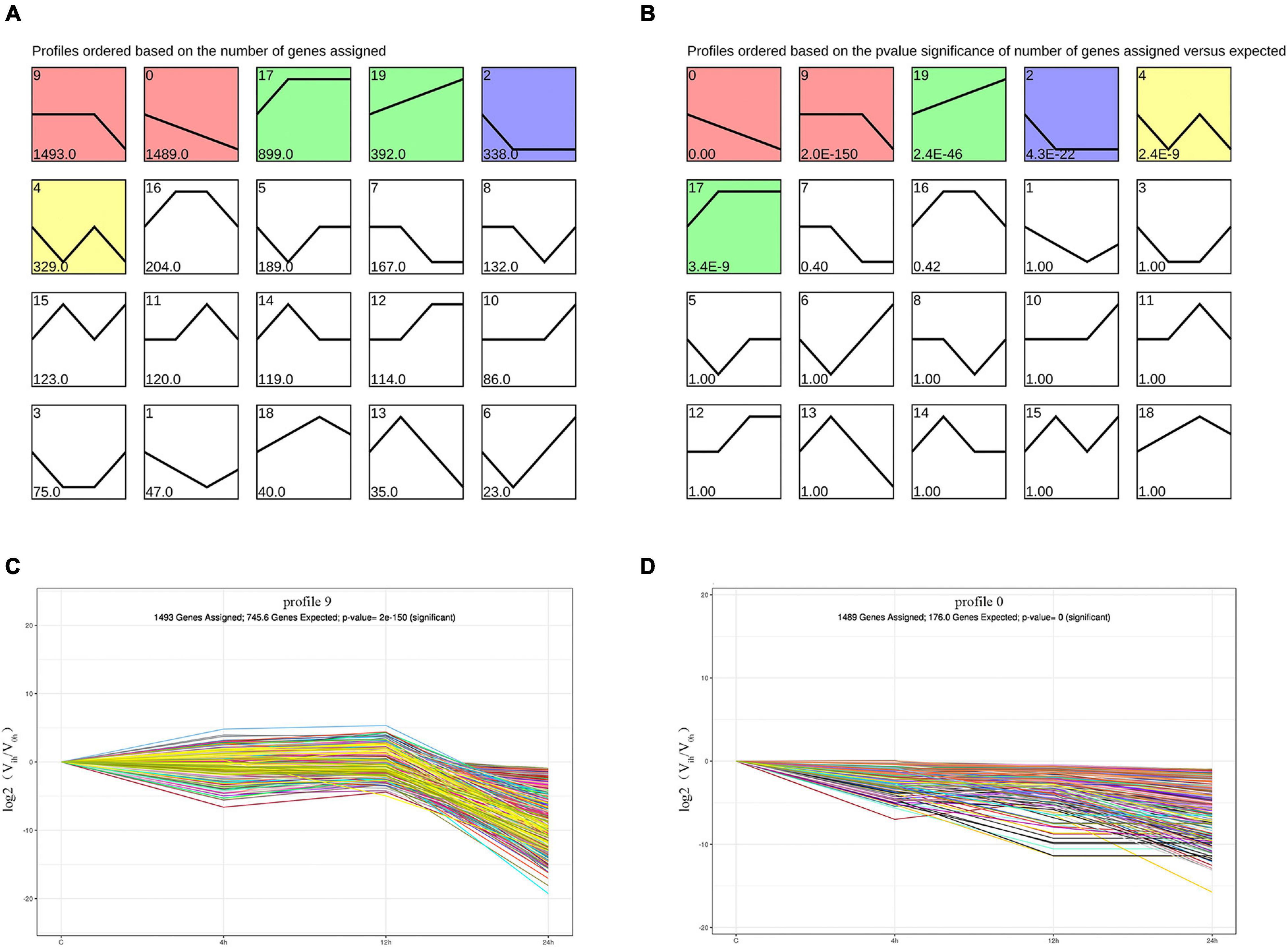
Figure 4. Trend analysis was used to analysis the trend changes of DEGs. (A) Trend analysis results sorted by the number of enriched DEGs. (B) Trend analysis results sorted by p-value. (C) The expression trend of DEGs in profile 9. The x-axis lists the time point after hatching; the y-axis indicates log2(fold change). (D) The expression trend of DEGs in profile 0.
We identified the top ten level-3 terms of the three categories (biological process: 352 level-3 subclasses, molecular function: 107 level-3 subclasses, and cellular component: 84 level-3 subclasses) (Figure 5 and Supplementary Table 7) by GO functional enrichment analysis. For instance, the signal transduction term and the cell differentiation term in biological process were closely related to immunity. Similarly, KEGG enriched DEGs in different pathways guided us to further understand the functions of these genes. Of 7,019 DEGs, 1,548 DEGs were enriched in 263 level-2 KEGG class pathways (Figure 6 and Supplementary Table 8). Table 3 showed the 20 immune-related pathways that were significantly enriched including biosynthesis of antibiotics signaling pathway, Wnt signaling pathway, and PI3K-Akt signaling pathway. Because some DEGs were involved in the regulation of multiple signal pathways, we finally obtained 208 genes enriched in 20 immune-related pathways for follow-up analyses.
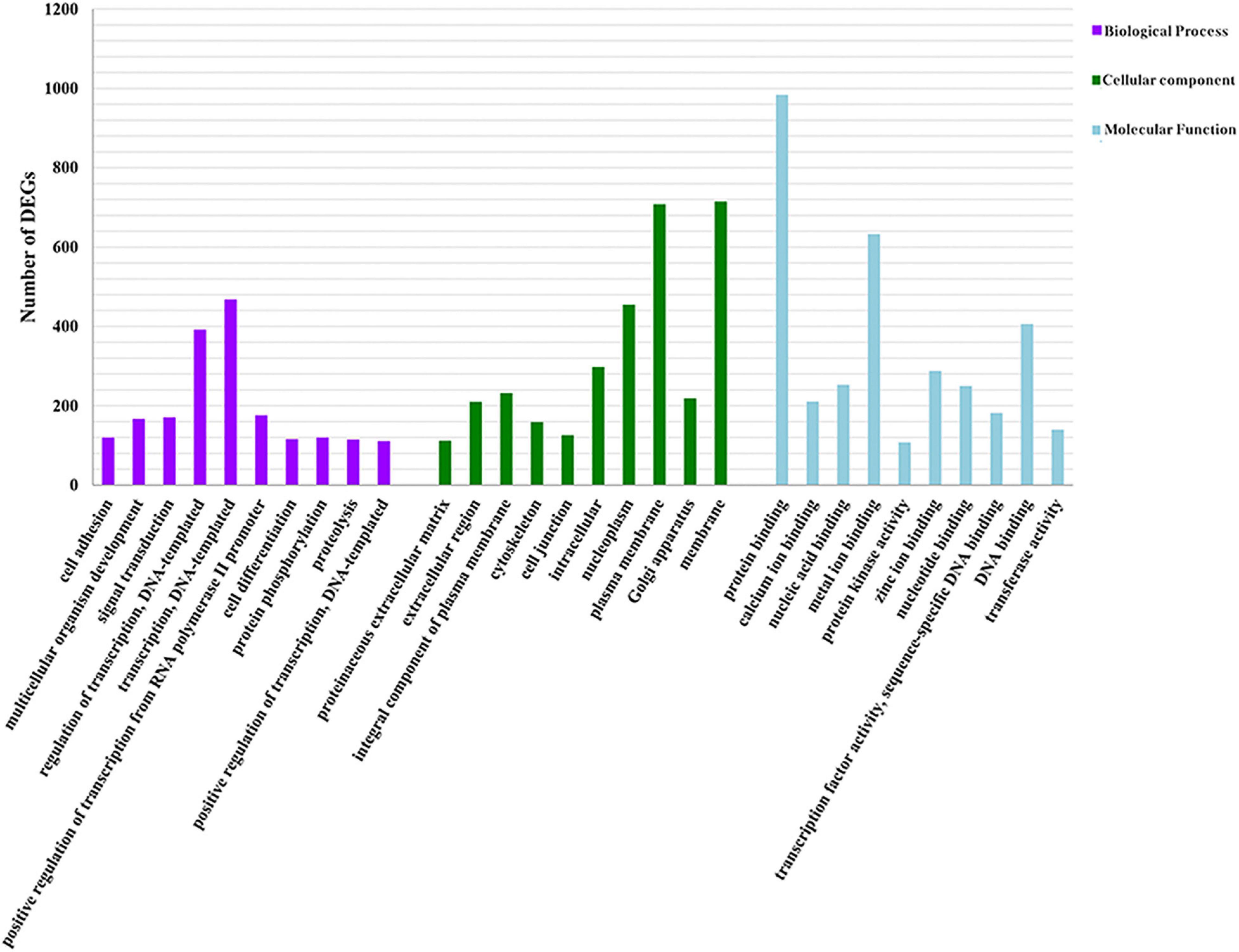
Figure 5. GO analysis of DEGs. Distribution of level-3 GO annotation among the three categories. The y-axis represents the corresponding number of DEGs; the x-axis stands for the gene function classification based on GO.
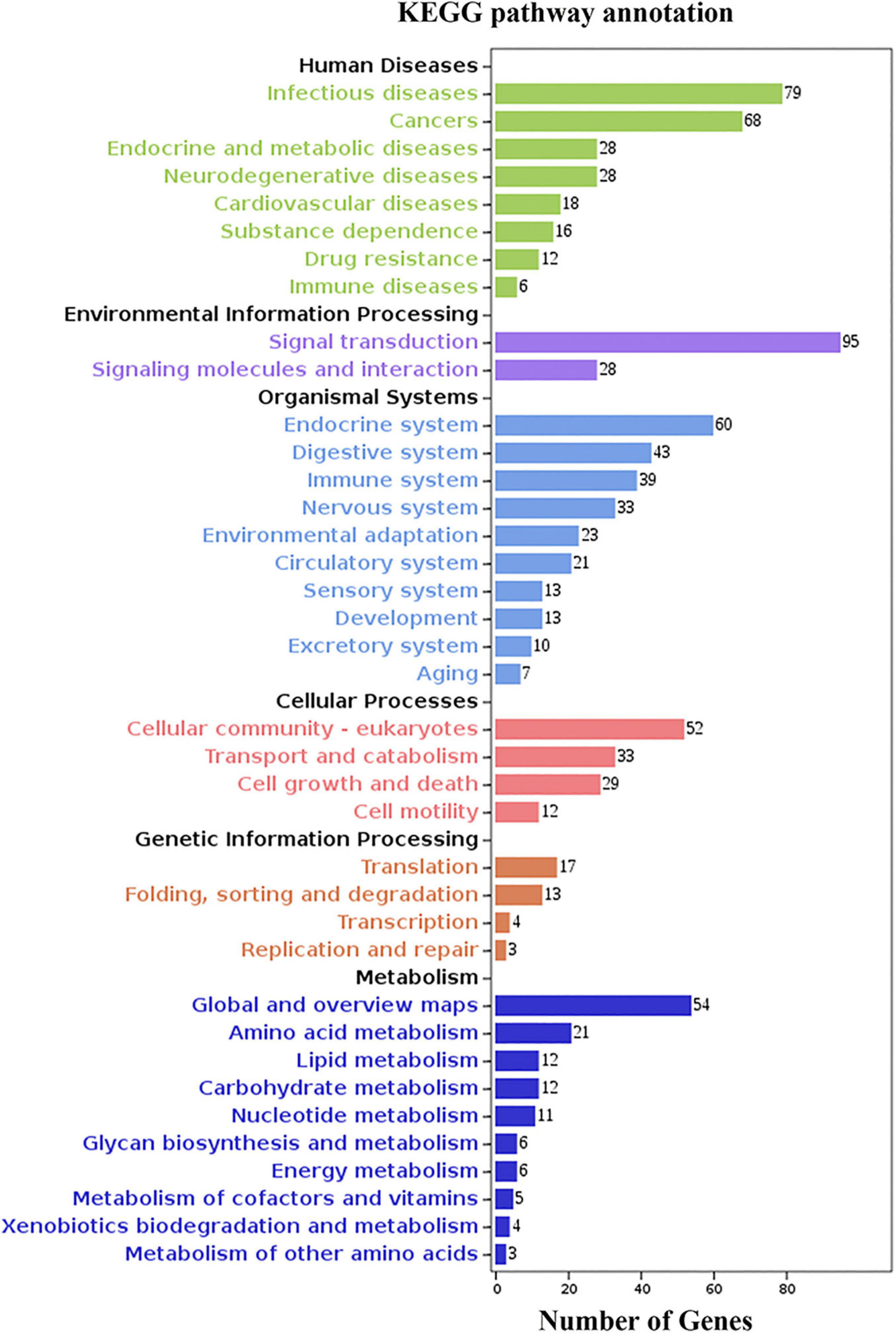
Figure 6. KEGG analysis of DEGs. The y-axis indicates level-2 KEGG classes; the x-axis represents the corresponding number of DEGs.
Proteins are important components of all cells and tissues in living organisms. All important parts of an organism require functional proteins, which are the main mediators of the activities of life. Construction of protein–protein interaction networks can help reveal the key genes involved in immune processes. We constructed protein–protein interaction networks using the protein sequences encoded by 208 genes in 20 significantly enriched immune-related signaling pathways. Figure 7 shows these networks. Among this figure, the CASP gene had the highest number of protein-protein interactions, and the other 5 genes including AXN1, CREBBP, MTOR, PTK2, and WNT5A were interacted with more than 20 proteins. Relevant parameter information of them was listed in Table 4. There were 148 nodes and 622 edges in the network, and each node was interacted with an average of 8.41 nodes. The clustering coefficient was 0.466, and the p-value of this network was ≤ 1.0e-16.
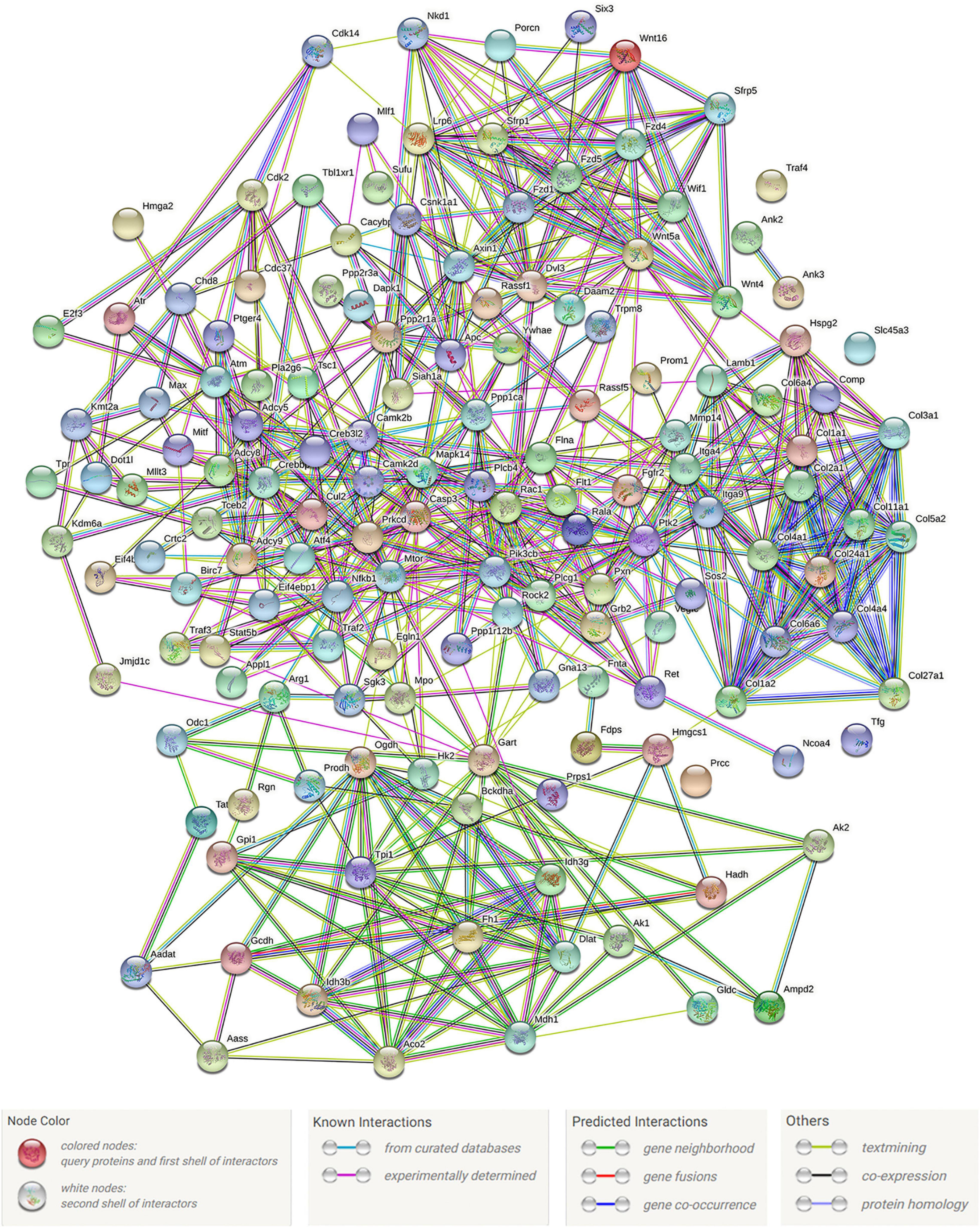
Figure 7. Immune-related protein–protein interaction networks. Network nodes stand for proteins. The legend represents the relationships between nodes.
This research focused on interactions between key immune-related genes. Twenty key DEGs (Table 5) involved in multiple signaling pathways or multiple interaction relationships were obtained from KEGG pathway analysis and protein–protein interaction networks. These were then used to explore interaction mechanisms between them. The 20 key genes were divided into five categories as follows: Collagen family, Wnt signaling pathway, biosynthesis of antibiotics signaling pathway, tumor necrosis factor (TNF) signaling pathway, and other important immune-related genes. To verify the accuracy of these key genes, we used quantitative RT-PCR to quantitatively assess the relative expression of 20 immune-related genes at each time point identified as DEGs by RNA-Seq. Quantitative RT-PCR results showed that all DEGs measured were single products. A comparison of gene expression profiles by RNA-Seq and quantitative RT-PCR (Figure 8) indicated that the quantitative RT-PCR results correlated significantly with the RNA-Seq results, and the two methods presented the same trend pattern.
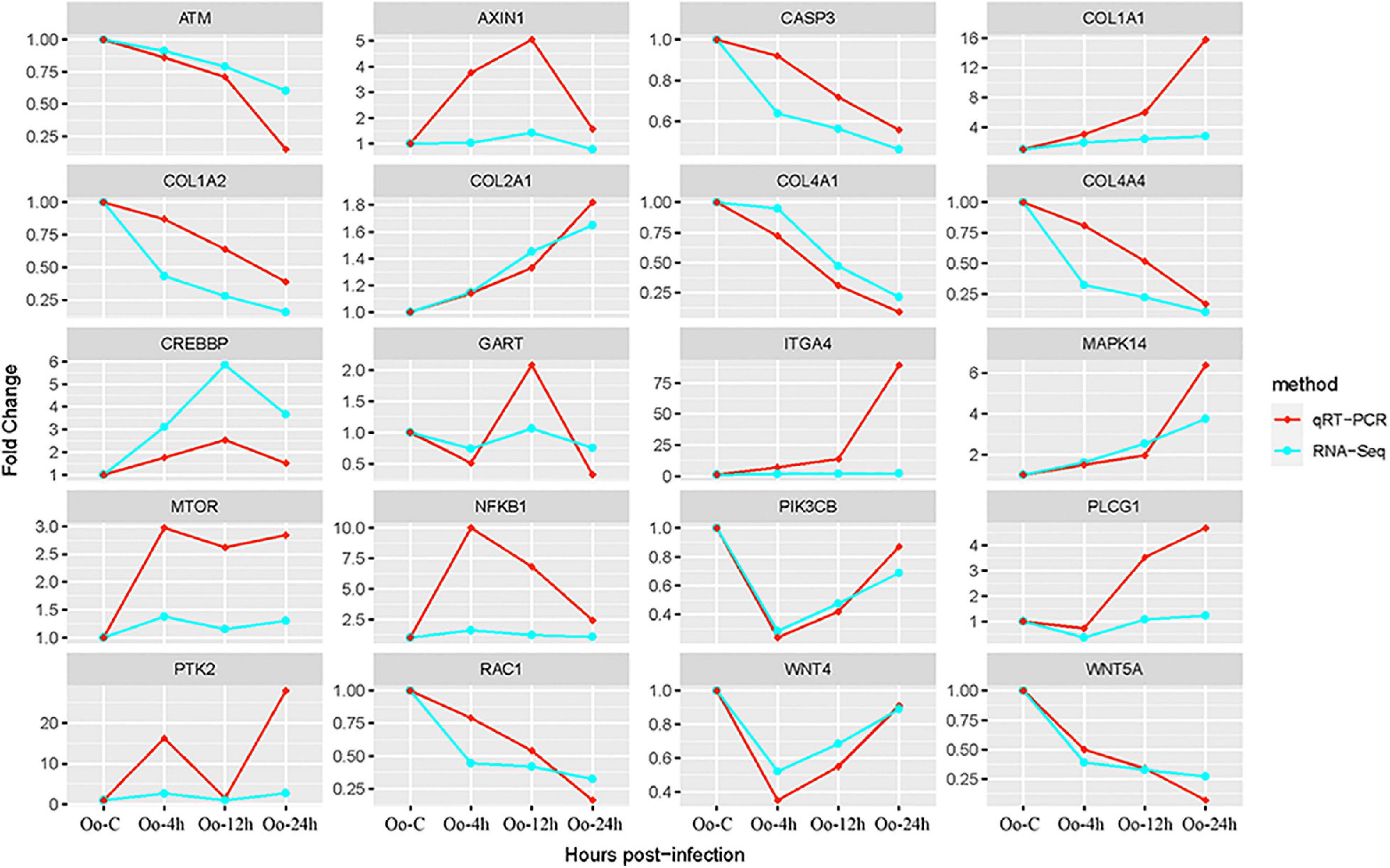
Figure 8. Comparison of expression of 20 hub genes between qRT-PCR and RNA-Seq results. The transcript expression levels of the selected DEGs were each normalized to that of the β-actin gene. The x-axis represents the growth time after hatching; the y-axis stands for Fold change.
Amphioctopus fangsiao has become increasingly popular because of its rich nutritional value and delicious meat. As an economically important species employed in mariculture, the artificial breeding of A. fangsiao has been well concerned (Iehata et al., 2009; Zhang et al., 2016; Li et al., 2019). The larval growth stage is very important for A. fangsiao, which determines whether the adult can grow healthily. The larvae of aquaculture organisms are easily infected by a variety of pathogens, which can seriously hinder aquaculture development (Barcia and Ramos-Martínez, 2011; Wang et al., 2019). Research into biological immune responses to bacterial infection will help breeders to prevent and mitigate bacterial infections. We infected primary incubation larvae for 24 h with V. anguillarum, and identified the changes of the biological immune response mechanisms. We identified 7,019 DEGs and believed that these genes have an important relationship with the immune response mechanisms of A. fangsiao larvae. A heatmap showed that there were significant differences in immunity with increase in A. fangsiao larvae infection time. The results of trend analysis showed that the six of the 20 trends were significantly enriched, and half of DEGs were expressed in these trends, indicating significant differences in the immunity after larval infection. KEGG enrichment analysis obtained 20 immune-related pathways enriched in DEGs, which were interacted with each other and shared biological immune functions. Finally, we used 208 genes in these pathways to construct protein–protein interaction networks to understand immune response mechanisms of A. fangsiao larvae.
The results of the trend analysis in Figure 4 indicated that profile 9 trend had the largest number of DEGs, and the significance of profile 0 trend was the highest. Among them, the gene expression trends showed continuously down-regulation and from stable to down, respectively, which were inconsistent with the expectation of our experiment. Therefore, we conducted GO and KEGG enrichment analyses of the genes involved to further explore the causes. In our analysis, we found that these genes were mainly involved in metabolic processes, including protein digestion and absorption signaling pathway, insulin signaling pathway, and cell adhesion term (Curtis et al., 1978; Lee and Pilch, 1994; Schepis et al., 2012; Brezas and Hardy, 2020). These results showed that the metabolic capacity of A. fangsiao larvae decreased gradually in the course of infection. Meanwhile, 1,291 DEGs in profile 17 and profile 19 were up-regulated. We used DEGs from these two profiles for GO and KEGG enrichment analyses, and identified a number of immune-related terms and pathways. For instance, positive regulation of JNK cascade term, biosynthesis of antibiotics signaling pathway, and NF-kappa B signaling pathway were significantly enriched. The JNK cascade is a key step in cell apoptosis mediated by the NF- kappa B signaling pathway, which is involved in the apoptosis process of diseased cells (Bubici et al., 2004; Lanna et al., 2017). NF-κB is involved in the regulation of immunity, lymphocyte development, and tumorigenesis (Bubici et al., 2004; Sun, 2017). Antibiotics can effectively resist the invasion of bacteria, and their synthesis can improve biological resistance (Bulfon et al., 2020). In conclusion, although the DEGs for the two most critical trends were down-regulated, they were highly correlated with metabolism. The results indicated that the metabolism of larvae was affected and had a downward trend after infection. The other DEGs in two up-regulated trends were more associated with immunity, suggesting that the immune response of A. fangsiao larvae increased gradually with the prolongation of infection. Above results revealed significant changes in the responses of A. fangsiao after infection. The complex mechanisms of A. fangsiao after infection need to be further studied and discussed in the future.
Many immune-related terms and pathways were yielded through GO and KEGG enrichment analyses, including signal transduction term, cell differentiation term, biosynthesis of antibiotics signaling pathway, TNF signaling pathway, and other key terms and pathways. These terms and pathways were vital in biological immunity, indicating that there were a large number of immune-related genes in the larvae infected with V. anguillarum, and thus resulted in complex immune responses in the larvae. A comprehensive analysis of the enriched KEGG pathways and GO terms contributes to investigate the molecular mechanisms and immune systems changes of A. fangsiao larvae after infection with V. anguillarum, thereby aiding our artificial breeding work.
Proteins are the basis of biological activities, and their interactions maintain the growth and development of organisms. We used 208 key DEGs identified in immune-related pathways to construct protein–protein interaction networks. The results suggested that interactions between these proteins was greater than expected relative to a randomly selected a group of proteins of similar size from the genome. This conclusion proves that above proteins cooperate in carrying out coordinated functions. We thus suggested that nodes with more edges were hub proteins in immune response. The genes corresponding to these hub proteins were identified as hub genes for further studies and verification.
Transcriptome profiling was analyzed to compare the changes of immune response mechanisms in A. fangsiao larvae infected with V. anguillarum within 24 h. This helps us to further understand the A. fangsiao larval immune response to V. anguillarum infection with the growth of them. Finally, we speculatively identified 20 hub genes involved in multiple protein–protein interaction relationships or KEGG signaling pathways, and these hub genes, KEGG signaling pathways, and immune differences at each time point were investigated.
Collagen is one of the most abundant proteins in the organisms. It is an important part of extracellular matrix, and plays a regulatory role in tissue structure. Its subunits directly affect cell phenotypes by mediating cell differentiation and apoptosis via relevant signaling pathways (Brondijk et al., 2010; Jürgensen et al., 2020). Collagen also acts as a major part in the immune system. When cells are stimulated by bacteria, collagen receptors are responsive to change the extracellular environment, thus indirectly regulating the activities of immune cells to resist the invasion of bacteria (Kresina et al., 1984; Aragona et al., 1998; Brondijk et al., 2010). In this study, five key genes involved in the immune response of collagen family (COL1A1, CoL1A2, COL2A1, COL4A1, and COL4A4) were identified. Among them, COL1A1 and COL2A1 presented the most obvious effect on enhancing the biological resistance to external invasion through the protection of bones, kidney, skin, and other organs. The lack of collagens in the organisms can lead to osteogenic and skin damages, which can reduce the immunity of organisms indirectly (Kuppevelt et al., 1995; Takai et al., 2001; Makino et al., 2010). In our study, the five genes in collagen family were enriched significantly, indicating an obvious bacteriological stimulation effect. Collagen family may be an important part to regulate the biological immune functions. In the above five genes, COL1A1 and COL2A1 were significantly up-regulated with the prolongation of infection time. This result indicated that A. fangsiao larvae had a strong immunity to effectively resist the invasion of pathogens. The other three genes were continuously down-regulated within 24 h of infection, the specific reasons need additional explorations.
Wnt is an important immune regulatory protein widely existing in various organisms. It participates in the immune response of multiple organs and acts a key role in maintaining tissue homeostasis (Famili et al., 2016). The Wnt signaling pathway can be divided into two subtypes: typical Wnt signaling pathway and atypical Wnt signaling pathway. The interaction between these two pathways regulates the growth, proliferation, differentiation, and apoptosis of immune cells, which can affect multiple immune responses of organisms (Naskar et al., 2014; Yu et al., 2014; Chae and Bothwell, 2018). Moreover, the Wnt signaling pathway is responsible for maintaining homeostasis through repairing damaged tissues and regulating the inflammatory response, which plays a role in the resistance and treatment of some immune diseases (Staal et al., 2008; Pai et al., 2017; Ruan et al., 2021). In this study, the Wnt signaling pathway was significantly enriched, and multiple DEGs including TBL1XR1, ROCK2, and CSNK1A1 were enriched in this pathway. They are the key factors in the immune system, and play a significant role in immune regulation during larval growth. Meanwhile, these genes are functional in the resistance of larvae against the invasion of pathogens and promote the normal growth of organisms (Puigvert et al., 2013; Nikola et al., 2017; Venturutti et al., 2020). Their up-regulation after larval infection suggested the improved immune ability of A. fangsiao larvae after being infected with bacteria.
Antibiotics are a class of metabolites produced in the life of higher animals, plants, and fungi, which have the ability to resist the invasion of pathogens. They can affect the growth of organisms by regulating cell growth and apoptosis (Pomorska-Mól and Pejsak, 2012; Herman, 2020). Biosynthesis of antibiotics can improve the ability of organisms to resist bacterial infection and increase the resistance of larvae. When organisms are invaded by external bacteria, antibiotics participate in the immune response, which can help organisms resist invasion, and increase their immunity. Recent studies have shown that antibiotics can treat some immune diseases and prevent immune system disorders (Stevens, 1996; Pomorska-Mól and Pejsak, 2012; Yang et al., 2017). In our study, biosynthesis of antibiotics signaling pathway was significantly enriched, suggesting that antibiotics act as an important part in the immune response of A. fangsiao larvae. Many key immune-related genes in this pathway were up-regulated with the growth of infected A. fangsiao larvae, indicating that antibiotics can guide a variety of immune responses to eliminate invading pathogens.
As a regulatory factor, TNF exists in various organisms. It can send signals to proteins in the immune system, promote the interaction between immune cells, and enhance the immune ability of organisms (Ferreira et al., 1993; Vielhauer and Mayadas, 2007; Croft, 2009; George et al., 2014; Li et al., 2021). In the immune system, TNF also acts as a signal protein to regulate the release of other cytokines. Moreover, TNF plays an important role in promoting T cell proliferation, interferon expression, and accelerating the efficiency of chemokines (Palanki et al., 2000; Vielhauer and Mayadas, 2007; Lougaris et al., 2017; Li et al., 2021). In our research, multiple immune-related genes including NFKB1, ATF4, and MAP2K4 were significantly enriched in this pathway. NFKB1 plays a major part in the regulation of immune response in organisms, which increases the resistance of organisms by regulating expression levels of proinflammatory factors, and inflammation-related proteins in immune cells. The other two genes are also useful in regulating the immune response of organisms (Sethi et al., 2007; Vielhauer and Mayadas, 2007; Huang et al., 2019). This result suggested that immune genes responded quickly after infection, which increasing the immune activity of A. fangsiao larvae, and effectively eliminated the invading pathogens.
Among all hub genes, CASP3, CREBBP, and PTK2 were enriched in more than two immune-related signaling pathways, and possessed higher numbers of protein–protein interactions. CASP3 is a regulator of cell apoptosis. It can maintain the dynamic balance of cell number and the healthy growth of organisms by regulating the apoptosis of pathological cells and aging cells. The up-regulation of CASP3 is helpful to clear the diseased cells, and improve the survival rate and vitality of organism larvae (Lowe and Lin, 2000; Suzuki et al., 2020). As an acetyltransferase, CREBBP exists widely in lymphocytes of various organisms. It is a key gene regulating immune response, resisting pathogen invasion, and promoting immune cell expression. The low expression or loss of function of CREBBP leads to the disorder of enhancer network and B cell signal, and causes the disorder of expression of other immune cells (Jiang et al., 2016; Zhang et al., 2017; Jia et al., 2018). PTK2 is a protein kinase widely existing in various tissues, and organs of organisms, which acts as an extremely important disease regulator. At the same time, PTK2 is involved in the regulation of cell adhesion and movement, which is conducive to substance transport, and the expression levels of immune genes (Zhang et al., 2017; Yi et al., 2021). In this study, CREBBP was up-regulated within the first 12 h of infection, and slightly down-regulated within the last 12 h, indicating an important role in the immune regulation of A. fangsiao larvae after infection. PTK2 was significantly up-regulated within 24 h after infection and down-regulated within 4 to 12 h. The causes of PTK2 down-regulation need to be explained by subsequent studies. The above two genes had obvious effects on larval immunity, which is beneficial for the larvae to resist the invasion of pathogens. However, with the prolongation of infection time, CASP3 was down-regulated. This result is not consistent with our study, which needs additional studies to explain.
In addition to the above hub genes and KEGG signaling pathways, our results identified other key DEGs and pathways involved in A. fangsiao larval immunity, including pathways in cancer signaling pathway, NF-kappa B signaling pathway, the PLCG1 gene, the ITGA4 gene, and the MTOR gene. The above two signaling pathways and three genes have been reported to be closely related to the immune response in various organisms (Wang et al., 2008; Thurnher et al., 2013; Darzi et al., 2017; Dieli-Crimi et al., 2018; Hong et al., 2019; Shiseki et al., 2019). The signaling pathways and genes details of the immune response mechanisms of A. fangsiao larvae infected with V. anguillarum require additional studies to further elucidate mechanistic details.
We performed transcriptome analyses of gene expression in primary incubation A. fangsiao larvae within 24 h of V. anguillarum infection, and established a protein–protein interaction network. Twenty hub DEGs enriched in multiple KEGG signaling pathways or with multiple protein–protein interaction relationships were identified. In our research, protein–protein interaction networks were first used to explore differences on A. fangsiao larval immunity. Our results provide valuable genetic resources for understanding the immunity of invertebrate larvae. Meanwhile, the data serve as a foundation for additional research into immune processes in invertebrates.
The original contributions presented in the study are publicly available in NCBI using accession numbers SRR15204591 – SRR15204602 at the following link: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SAMN20309867&o=acc_s%3Aa.
This research was conducted in accordance with the protocols of the Institutional Animal Care and Use Committee of the Ludong University (protocol number LDU-IRB20210308NXY) and the China Government Principles for the Utilization and Care of Invertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No. 2, October 31, 1988. http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
ZL and JY designed and supervised the study. XB, YL, JZ, XC, XX, and BL prepared the samples. XB, YL, YF, GS, WW, and XL analyzed all sequencing data. XB and ZL wrote the manuscript. All authors have read and approved the final manuscript.
This research was supported by China Agriculture Research System of MOF and MARA, the Natural Science Foundation of Shandong Province (No. ZR2019BC052), the National Natural Science Foundation of China (No. 42006077), and the “Entrepreneurship and Innovation Talents” programme of Jiangsu Province of China.
WW was employed by the company Jiangsu Baoyuan Biotechnology Co., Ltd. BL was employed by the company Yantai Haiyu Marine Science and Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.731517/full#supplementary-material
Aragona, F., D’Urso, L., and Marcolongo, R. (1998). Immunologic Aspects of Bovine Injectable Collagen in Humans. Eur. Urol. 33, 129–133. doi: 10.1159/000019544
Bao, P., Sun, X., Liu, Q., Zhang, Y., and Liu, X. (2019). Synergistic effect of a combined live Vibrio anguillarum and Edwardsiella piscicida vaccine in turbot. Fish Shellfish Immunol. 88, 84–90. doi: 10.1016/j.fsi.2019.02.014
Barcia, R., and Ramos-Martínez, J. I. (2011). Stress-based modulation of the immune response in molluscan hemocytes: a two-receptor model. Invertebr. Survival J. 8, 56–58.
Brezas, A., and Hardy, R. W. (2020). Improved performance of a rainbow trout selected strain is associated with protein digestion rates and synchronization of amino acid absorption. Sci. Rep. 10:4678.
Brondijk, T. H. C., Ruiter, T. D., Ballering, J., Wienk, H., Lebbink, R. J., Boelens, R., et al. (2010). Crystal structure and collagen-binding site of immune inhibitory receptor LAIR-1: unexpected implications for collagen binding by platelet receptor GPVI. Blood 115, 1364–1373. doi: 10.1182/blood-2009-10-246322
Bubici, C., Papa, S., Pham, C. G., Zazzeroni, F., and Franzoso, G. (2004). NF-κB and JNK: An Intricate Affair. Cell Cycle 3, 1524–1529. doi: 10.4161/cc.3.12.1321
Budelmann, B. U. (1994). Cephalopod sense organs, nerves and the brain: Adaptations for high performance and life style. Mar. Behav. Physiol. 25, 13–33. doi: 10.1080/10236249409378905
Bulfon, C., Prearo, M., Volpatti, D., Byadgi, O., Righetti, M., Maniaci, M. G., et al. (2020). Resistant and susceptible rainbow trout (Oncorhynchus mykiss) lines show distinctive immune response to Lactococcus garvieae. Fish Shellfish Immunol. 105, 457–468. doi: 10.1016/j.fsi.2020.06.040
Castellanos-Martínez, S., Arteta, D., Catarino, S., and Gestal, C. (2014). De Novo Transcriptome Sequencing of the Octopus vulgaris Hemocytes Using Illumina RNA-Seq Technology: Response to the Infection by the Gastrointestinal Parasite Aggregata octopiana. PLoS One 9:e107873. doi: 10.1371/journal.pone.0107873
Chae, W., and Bothwell, A. L. M. (2018). Canonical and Non-Canonical Wnt Signaling in Immune Cells. Trends Immunol. 39, 830–847. doi: 10.1016/j.it.2018.08.006
Cong, M., Song, L., Wang, L., Zhao, J., Qiu, L., Li, L., et al. (2008). The enhanced immune protection of Zhikong scallop Chlamys farreri on the secondary encounter with Listonella anguillarum. Compar. Biochem. Physiol. Part B 151, 191–196. doi: 10.1016/j.cbpb.2008.06.014
Croft, M. (2009). The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol. 9, 271–285. doi: 10.1038/nri2526
Curtis, K. J., Kim, Y. S., Perdomo, J. M., Silk, D. B. A., and Whitehead, J. S. (1978). Protein digestion and absorption in the rat. J. Physiol. 274, 409–419.
Damian, S., Gable, A. L., David, L., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2018). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613.
Darzi, L., Boshtam, M., Shariati, L., Kouhpayeh, S., Gheibi, A., Rahimmanesh, I., et al. (2017). The silencing effect of miR-30a on ITGA4 gene expression in vitro: an approach for gene therapy. Res. Pharm. Sci. 12, 456–464. doi: 10.4103/1735-5362.217426
Dieli-Crimi, R., Martínez-Gallo, M., Franco-Jarava, C., Antolin, M., Blasco, L., Paramonov, I., et al. (2018). Th1-skewed profile and excessive production of proinflammatory cytokines in a NFKB1-deficient patient with CVID and severe gastrointestinal manifestations. Clin. Immunol. 195, 49–58. doi: 10.1016/j.clim.2018.07.015
Ernst, J., and Bar-Joseph, Z. (2006). STEM: a tool for the analysis of short time series gene expression data. BMC Bioinform. 7:191.
Famili, F., Perez, L. G., Naber, B. A., Noordermeer, J. N., Fradkin, L. G., and Staal, F. J. (2016). The non-canonical Wnt receptor Ryk regulates hematopoietic stem cell repopulation in part by controlling proliferation and apoptosis. Cell Death Dis. 7:e2479. doi: 10.1038/cddis.2016.380
Ferreira, S. H., Lorenzetti, B. B., Cunha, F. Q., and Poole, S. (1993). Bradykinin release of TNF-α plays a key role in the development of inflammatory hyperalgesia. Inflam. Res. 38, 7–9.
George, P. D. C., Agoramoorthy, G., and Chakraborty, C. (2014). TNF/TNFR: drug target for autoimmune diseases and immune-mediated inflammatory diseases. Front. Biosci. 19:1028–1040. doi: 10.2741/4265
Ginger, K., Vera, C., Dineshram, R., Dennis, C., Adela, L., Yu, Z., et al. (2013). Larval and Post-Larval Stages of Pacific Oyster (Crassostrea gigas) Are Resistant to Elevated CO2. PLoS One 8:e64147. doi: 10.1371/journal.pone.0064147
Herman, R. A. (2020). Increasing allergy: are antibiotics the elephant in the room? Allergy Asthma Clin. Immunol. 16, 35–36.
Ho, X. D., Phung, P., Le, V. Q., Nguyen, V. H., Reimann, E., Prans, E., et al. (2017). Whole transcriptome analysis identifies differentially regulated networks between osteosarcoma and normal bone samples. Exp. Biol. Med. 242, 1802–1811. doi: 10.1177/1535370217736512
Hoel, K., Salonius, K., and Lillehaug, A. (1997). Vibrio antigens of polyvalent vaccines enhance the humoral immune response to Aeromonas salmonicida antigens in Atlantic salmon (Salmo salar L.) Fish Shellfish Immunol. 7, 71–80. doi: 10.1006/fsim.1996.0063
Hong, B. S., Ryu, H. S., Kim, N., Kim, J., Lee, S., Moon, H., et al. (2019). Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 79, 1520–1534.
Huang, W., Qiu, S., Tong, X., He, Y., Wang, Z., and Dong, Y. (2019). Activating transcription factor 4 is required for high glucose inhibits proliferation and differentiation of MC3T3-E1 cells. J. Recept. Signal Transduct. 39, 409–414.
Iehata, S., Inagaki, T., Okunishi, S., Nakano, M., Tanaka, R., and Maeda, H. (2009). Colonization and probiotic effects of lactic acid bacteria in the gut of the abalone Haliotis gigantea. Fisheries Sci. 75, 1285–1293. doi: 10.1007/s12562-009-0138-5
Jia, D., Augert, A., Kim, D., Eastwood, E., Wu, N., Kim, K., et al. (2018). Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Dis. 8, 1422–1437. doi: 10.1158/2159-8290.cd-18-0385
Jiang, Y., Ortega-Molina, A., Geng, H., Ying, H., Hatzi, K., Parsa, S., et al. (2016). CREBBP Inactivation Promotes the Development of HDAC3 Dependent Lymphomas. Cancer Dis. 7, 38–53. doi: 10.1158/2159-8290.cd-16-0975
Jürgensen, H. J., Putten, S. V., Nørregaard, K. S., Bugge, T. H., Engelholm, L. H., Behrendt, N., et al. (2020). Cellular uptake of collagens and implications for immune cell regulation in disease. Cell. Mole. Life Sci. 77, 3161–3176. doi: 10.1007/s00018-020-03481-3
Kaplan, M. B., Mooney, T. A., McCorkle, D. C., and Cohen, A. L. (2013). Adverse Effects of Ocean Acidification on Early Development of Squid (Doryteuthis pealeii). PLoS One 8:e63714. doi: 10.1371/journal.pone.0063714
Kresina, T. F., Rosner, I. A., Goldberg, V. M., and Moskowitz, R. W. (1984). Immunoglobulin G-induced experimental chronic immune synovitis: Cell-mediated immunity to native interstitial collagen molecules and their constituent polypeptide chains. Cell. Immunol. 87, 504–516. doi: 10.1016/0008-8749(84)90019-4
Kuppevelt, T. H. V., Veerkamp, J. H., and Timmermans, J. A. H. (1995). Immunoquantification of type I, III, IV and V collagen in small samples of human lung parenchyma. Internat. J. Biochem. Cell Biol. 27, 775–782. doi: 10.1016/1357-2725(95)00047-s
Lanna, A., Gomes, D. C. O., Muller-Durovic, B., McDonnell, T., Escors, D., Gilroy, D. W., et al. (2017). A sestrin-dependent Erk/Jnk/p38 MAPK activation complex inhibits immunity during aging. Nat. Immunol. 18, 354–363. doi: 10.1038/ni.3665
Lee, J., and Pilch, P. F. (1994). The insulin receptor: structure, function, and signaling. Am. Physiol. 266, 319–334.
Li, J., Gao, D., Wang, Q., Wang, J., and Wang, Q. (2015). Efficacy of Vibrio anguillarum antigen administered by intraperitoneal injection route in Japanese flounder, Paralichthys olivaceus (Temminck et Schlegel) Aquacult. Res. 36, 1104–1111. doi: 10.1111/j.1365-2109.2005.01327.x
Li, K., Qiu, H., Yan, J., Shen, X., Wei, X., Duan, M., et al. (2021). The involvement of TNF-α and TNF-β as proinflammatory cytokines in lymphocyte-mediated adaptive immunity of Nile tilapia by initiating apoptosis. Dev. Compar. Immunol. 115:103884. doi: 10.1016/j.dci.2020.103884
Li, Z., Fan, T., Liu, X., Liu, X., Wang, W., Wang, Q., et al. (2019). Characterization and Functional Study on Octopus ocellatus Interleukin-17. J. Ocean Univ. China 18, 1443–1450. doi: 10.1007/s11802-019-4116-y
Li, Z., Liu, X., Liu, J., Zhang, K., Yu, H., He, Y., et al. (2017). Transcriptome profiling based on protein–protein interaction networks provides a core set of genes for understanding blood immune response mechanisms against Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev. Compar. Immunol. 78, 100–113. doi: 10.1016/j.dci.2017.09.013
Lougaris, V., Patrizi, O., Baronio, M., Tabellini, G., Tampella, G., Damiati, E., et al. (2017). NFKB1 regulates human NK cell maturation and effector functions. Clin. Immunol. 175, 99–108. doi: 10.1016/j.clim.2016.11.012
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550.
Makino, H., Shikata, K., Wieslander, J., Wada, J., Kashihara, N., Yoshioka, K., et al. (2010). Localization of Fibril/Microfibril and Basement Membrane Collagens in Diabetic Glomerulosclerosis in Type 2 Diabetes. Diabetic Med. 11, 304–311. doi: 10.1111/j.1464-5491.1994.tb00276.x
Naskar, D., Maiti, G., Chakraborty, A., Roy, A., Chattopadhyay, D., and Sen, M. (2014). Wnt5a-Rac1-NF-kB homeostatic circuitry sustains innate immune functions in macrophages. J. Immunol. 192, 4386–4397. doi: 10.4049/jimmunol.1302817
Nie, L., Zhou, Q., Qiao, Y., and Chen, J. (2017). Interplay between the gut microbiota and immune responses of ayu (Plecoglossus altivelis) during Vibrio anguillarum infection. Fish Shellfish Immunol. 68, 479–487. doi: 10.1016/j.fsi.2017.07.054
Nikola, S., Taeg, O. G., Hyung-Hwan, K., Beaulieu, L. M., Falet, H., Kaminski, K., et al. (2017). Decreased Thromboembolic Stroke but not Atherosclerosis or Vascular Remodeling in Mice with ROCK2-deficient Platelets. Cardiovas. Res. 2017, 1307–1317. doi: 10.1093/cvr/cvx071
Pai, S. G., Carneiro, B. A., Mota, J. M., Costa, R., Leite, C. A., Barroso-Sousa, R., et al. (2017). Wnt/beta-catenin pathway: modulating anticancer immune response. J. Hematol. Oncol. 10, 101–112.
Palanki, M. S., Erdman, P. E., Gayo-Fung, L. M., Shevlin, G. I., Sullivan, R. W., Suto, M. J., et al. (2000). Inhibitors of NF-kB and AP-1 gene expression: SAR studies on the pyrimidine portion of 2-Chloro-4-trifluoromethylpyrimidine-5-[N-(3′,5′-bis(trifluoromethyl)phenyl)carboxamide]. J. Med. Chem. 43, 3995–4004. doi: 10.1021/jm0001626
Pomorska-Mól, M., and Pejsak, Z. (2012). Effects of antibiotics on acquired immunity in vivo – Current state of knowledge. Polish J. Vet. Aciences 15, 583–588. doi: 10.2478/v10181-012-0089-0
Puigvert, J. C., Stechow, L. V., Siddappa, R., Pines, A., Bahjat, M., Haazen, L., et al. (2013). Systems biology approach identifies the kinase Csnk1a1 as a regulator of the DNA damage response in embryonic stem cells. Sci. Signal. 6:ra5.
Rowley, A. F., and Powell, A. (2007). Invertebrate immune systems specific, quasi-specific, or nonspecific? J. Immunol. 179, 7209–7214. doi: 10.4049/jimmunol.179.11.7209
Ruan, Y., Ogana, H., Gang, E., Kim, H. N., and Kim, Y. M. (2021). Wnt Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 1270, 107–121. doi: 10.1007/978-3-030-47189-7_7
Salazar, K. A., Joffe, N. R., Dinguirard, N., Houde, P., and Castillo, M. G. (2015). Transcriptome Analysis of the White Body of the Squid Euprymna tasmanica with Emphasis on Immune and Hematopoietic Gene Discovery. PLoS One 10:e0119949. doi: 10.1371/journal.pone.0119949
Schepis, A., Sepich, D., and Nelson, W. J. (2012). αE-catenin regulates cell-cell adhesion and membrane blebbing during zebrafish epiboly. Dev. 139, 537–546. doi: 10.1242/dev.073932
Schultz, J. H., and Adema, C. M. (2017). Comparative immunogenomics of molluscs. Dev. Compar. Immunol. 75, 3–15. doi: 10.1016/j.dci.2017.03.013
Sethi, G., Ahn, K. S., Xia, D., Kurie, J. M., and Aggarwal, B. B. (2007). Targeted Deletion of MKK4 Gene Potentiates TNF-Induced Apoptosis through the Down-Regulation of NF-κB Activation and NF-κB-Regulated Antiapoptotic Gene Products. J. Immunol. 179, 1926–1933. doi: 10.4049/jimmunol.179.3.1926
Shiseki, M., Ishii, M., Miyazaki, M., Osanai, S., Wang, Y., Yoshinaga, K., et al. (2019). Reduced PLCG1 expression is associated with inferior survival for myelodysplastic syndromes. Cancer Med. 9, 460–468. doi: 10.1002/cam4.2717
Silva, T. M., Melo, E. S., Lopes, A. C. S., Veras, D. L., Duarte, C. R., Alves, L. C., et al. (2013). Characterization of the bacterial microbiota of Biomphalaria glabrata (Say, 1818) (Mollusca: Gastropoda) from Brazil. Lett. Appl. Microbiol. 57, 19–25. doi: 10.1111/lam.12068
Staal, F. J. T., Luis, T. C., and Tiemessen, M. M. (2008). WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 8, 581–593. doi: 10.1038/nri2360
Stevens, D. L. (1996). Immune modulatory effects of antibiotics. Curr. Opin. Infect. Dis. 9, 165–169. doi: 10.1097/00001432-199606000-00007
Sun, S. (2017). The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558. doi: 10.1038/nri.2017.52
Suzuki, T., Ichii, O., Nakamura, T., Horino, T., Elewa, Y. H. A., and Kon, Y. (2020). Immune-associated renal disease found in caspase 3-deficient mice. Cell Tissue Res. 379, 323–335. doi: 10.1007/s00441-019-03084-w
Takai, K. K., Hattori, S., and Irie, S. (2001). Type V collagen distribution in liver is reconstructed in coculture system of hepatocytes and stellate cells; the possible functions of type V collagen in liver under normal and pathological conditions. Cell Struct. Funct. 26, 289–302. doi: 10.1247/csf.26.289
Thurnher, M., Gruenbacher, G., and Nussbaumer, O. (2013). Regulation of mevalonate metabolism in cancer and immune cells. Biochim. et Biophys. Acta 1831, 1009–1015. doi: 10.1016/j.bbalip.2013.03.003
Vázquez, J. A., Docasal, S. F., Mirón, J., González, M. P., and Murado, M. A. (2006). Proteases production by two Vibrio species on residuals marine media. J. Industr. Microbiol. Biotechnol. 33, 661–668.
Venturutti, L., Teater, M., Zhai, A., Chadburn, A., Babiker, L., Kim, D., et al. (2020). TBL1XR1 Mutations Drive Extranodal Lymphoma by Inducing a Pro-tumorigenic Memory Fate. Cell 182, 1–20.
Vielhauer, V., and Mayadas, T. N. (2007). Functions of TNF and its receptors in renal disease: distinct roles in inflammatory tissue injury and immune regulation. Semin. Nephrol. 27, 286–308.
Wang, L., Qiu, L., Zhou, Z., and Song, L. (2013). Research progress on the mollusc immunity in China. Dev. Compar. Immunol. 39, 2–10. doi: 10.1016/j.dci.2012.06.014
Wang, L., Wang, W., Yang, C., Sun, J., and Song, L. (2019). The immune recognition mechanisms in mollucs. Fish Shellfish Immunol. 91, 461–462. doi: 10.1016/j.fsi.2019.04.263
Wang, X., Yue, P., Kim, Y. A., Fu, H., Khuri, F. R., and Sun, S. (2008). Enhancing Mammalian Target of Rapamycin (mTOR)–Targeted Cancer Therapy by Preventing mTOR/Raptor Inhibition-Initiated, mTOR/Rictor Independent Akt Activation. Cancer Res. 68, 7409–7418. doi: 10.1158/0008-5472.can-08-1522
Wei, X., Xu, J., Yang, J., Liu, X., Zhang, R., Wang, W., et al. (2015). Involvement of a Serpin serine protease inhibitor (OoSerpin) from mollusc Octopus ocellatus in antibacterial response. Fish Shellfish Immunol. 42, 79–87.
Wei, X., Zhao, T., Ai, K., Li, H., Jiang, X., Li, C., et al. (2018). Role of scavenger receptor from Octopus ocellatus as a co-receptor of Toll-like receptor in initiation of TLR-NF-kB signaling during anti-bacterial response. Dev. Compar. Immunol. 84, 14–27. doi: 10.1016/j.dci.2018.01.023
Yang, J. H., Bhargava, P., McCloskey, D., Mao, N., Palsson, B. O., Collins, J. J., et al. (2017). Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 22, 757–765. doi: 10.1016/j.chom.2017.10.020
Yi, L., Zhou, L., Luo, J., and Yang, Q. (2021). Circ-PTK2 promotes the proliferation and suppressed the apoptosis of acute myeloid leukemia cells through targeting miR-330-5p/FOXM1 axis. Blood Cells Mole. Dis. 86:102506. doi: 10.1016/j.bcmd.2020.102506
Yu, B., Chang, J., Liu, Y., Li, J., Kevork, K., Al-Hezaimi, K., et al. (2014). Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-κB. Nat. Med. 20, 1009–1017. doi: 10.1038/nm.3586
Zhang, J., Vlasevska, S., Wells, V. A., Nataraj, S., Holmes, A. B., Duval, R., et al. (2017). The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Dis. 7, 322–337. doi: 10.1158/2159-8290.cd-16-1417
Zhang, Q., Lu, Y., Zheng, H., Liu, H., and Li, S. (2016). Differential immune response of vitellogenin gene to Vibrio anguillarum in noble scallop Chlamys nobilis and its correlation with total carotenoid content. Fish Shellfish Immunol. 50, 11–15. doi: 10.1016/j.fsi.2016.01.001
Zhang, X., Shi, J., Sun, Y., Habib, Y. J., Yang, H., Zhang, Z., et al. (2018). Integrative transcriptome analysis and discovery of genes involving in immune response of hypoxia/thermal challenges in the small abalone Haliotis diversicolor. Fish Shellfish Immunol. 84, 609–624. doi: 10.1016/j.fsi.2018.10.044
Zhao, X., Wang, Q., Jiao, Y., Huang, R., Deng, Y., Wang, H., et al. (2012). Identification of Genes Potentially Related to Biomineralization and Immunity by Transcriptome Analysis of Pearl Sac in Pearl Oyster Pinctada martensii. Mar. Biotechnol. 14, 730–739. doi: 10.1007/s10126-012-9438-3
Keywords: Amphioctopus fangsiao, Vibrio anguillarum, larval growth, transcriptome, protein-protein interaction networks, immunity, infecting
Citation: Bao X, Li Y, Zhang J, Chen X, Xu X, Feng Y, Sun G, Liu X, Li B, Wang W, Li Z and Yang J (2021) Transcriptome Profiling Based on Different Time Points After Hatching Provides a Core Set of Gene Resource for Understanding Larval Immune Response Mechanisms Against Vibrio anguillarum Infection in Amphioctopus fangsiao. Front. Mar. Sci. 8:731517. doi: 10.3389/fmars.2021.731517
Received: 27 June 2021; Accepted: 06 September 2021;
Published: 29 September 2021.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jun Li, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Bao, Li, Zhang, Chen, Xu, Feng, Sun, Liu, Li, Wang, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zan Li, bGl6YW5seG1AMTYzLmNvbQ==; Jianmin Yang, bGFkZGVydXBAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.