- 1Marine Scotland Science, Marine Laboratory, Aberdeen, United Kingdom
- 2Marine Biological Association of the UK, Citadel Hill Laboratory, Plymouth, United Kingdom
- 3Orkney Islands Council Marine Services, Orkney, United Kingdom
- 4Coastal and Freshwater, Cawthron Institute, Nelson, New Zealand
- 5School of Biological Sciences, University of Aberdeen, Aberdeen, United Kingdom
The presence and diversity of marine non-native species, the number of new invasions, and the impact on native communities and habitats are important metrics used to assess the health of marine ecosystems. Monitoring for marine non-native species, using traditional approaches such as rapid assessment surveys (RASs), requires taxonomic expertise and may still fail to detect rare or inconspicuous species. This study reports a validation process for a quantitative PCR (qPCR) assay based on the cytochrome oxidase 1 gene, designed to detect highly invasive tunicate Didemnum vexillum by targeting environmental DNA (eDNA) present in water samples. The D. vexillum qPCR assay showed high sensitivity, with the threshold limit of detection (LOD) and modeled LOD3 (based on triplicate qPCR reactions) estimated as 9.187 and 1.117 copies reaction–1, respectively and the limit of quantification (LOQ) was calculated as 18 copies reaction–1. Analyses of water samples collected from selected Pacific oyster farms and recreational marinas in Scotland showed 100% concordance between the historical data on presence of D. vexillum from RASs and detection of D. vexillum eDNA. Consistency of detection of D. vexillum eDNA among different sampling points within each infected sampling site varied, ranging between 100% positive throughout the site to some sampling points testing “negative” or only as “suspected” for D. vexillum. Sites with lower within-site detection consistency included sites with a low density of D. vexillum as reported by RASs or were sites undergoing D. vexillum management. The present pilot monitoring program demonstrates the potential to generate important data on presence of D. vexillum. This program will be scaled up across large geographic regions and used in the first instance to focus and target the traditional RASs to D. vexillum eDNA-positive sites in a cost-effective way, with an aim to verify the species presence by visual observation and direct Sanger sequencing of positive qPCR products.
Introduction
The carpet sea squirt Didemnum vexillum Kott, 2002 is a colonial tunicate native to the Northwest Pacific Ocean including Japan (Stefaniak et al., 2012). Analyses based on molecular data have revealed two distinct D. vexillum clades, one clade (clade B) appears to be restricted to the native region, while the other clade (clade A) has been introduced to temperate coastal waters worldwide, and is often highly invasive in these new habitats (Smith et al., 2012; McKenzie et al., 2017). Biological characteristics of D. vexillum such as rapid colony growth via asexual reproduction, early maturation, tolerance to a wide range of water temperature and depth and ability to spread and reproduce from small, detached pieces of colonies, all contribute to the success of D. vexillum as an invader (Valentine et al., 2007a, 2009; Bullard and Whitlatch, 2009; Morris and Carman, 2012; Forrest et al., 2013; Stefaniak and Whitlatch, 2014; Ordóñez et al., 2015; Fletcher et al., 2018). In addition, a wide variety of substrates, including both natural substrates [e.g., gravel, boulders, subtidal rocks (Bullard et al., 2007; Valentine et al., 2007b), bryozoans, eelgrass, macroalgae, sponges, tubeworms (Valentine et al., 2007b; Hitchin, 2012; Carman et al., 2014; Vercaemer et al., 2015), and bivalve species (Fletcher et al., 2013a; Vercaemer et al., 2015; Forrest and Atalah, 2017)] and artificial man-made structures [e.g., harbor, marina, and aquaculture installations (Pederson et al., 2005; Coutts and Forrest, 2007; Hitchin, 2012; Bishop et al., 2015b; Cottier-Cook et al., 2019)] have been reported as suitable for the attachment and successful establishment of D. vexillum colonies.
Numerous studies have reported the ability of D. vexillum to alter marine benthic habitats by forming dense mats and reducing areas suitable for settlement and larval recruitment (Bullard et al., 2007; Valentine et al., 2007b; Lengyel et al., 2009; Lengyel, 2013; Kaplan et al., 2017). Experimental work using suspended settlement panels has also shown how D. vexillum can rapidly colonize available space. However, the ability of D. vexillum to establish long-term on natural substrates, occupied by highly biodiverse communities, seems to be site specific and dependent on the composition of native organisms, the abundance of natural predators, and the availability of free space (Forrest et al., 2013; Janiak et al., 2013; Fletcher et al., 2018). International shipping, regional recreational boating and aquaculture are considered as the major introductory pathways for D. vexillum (e.g., Tidbury et al., 2016). Artificial surfaces in ports, harbors and recreational marinas often support fouling communities that are distinct from those occurring on neighboring natural seabeds, with a greater preponderance of non-native species (e.g., Glasby et al., 2007; Dafforn et al., 2012; Simkanin et al., 2012; Lopez-Legentil et al., 2015; Rogers et al., 2016). D. vexillum can also form large biofouling colonies in aquaculture settings, resulting in large economic losses due to direct impact on biomass of farmed species and trade restrictions (Dijkstra and Nolan, 2011; Fitridge et al., 2012; Fletcher et al., 2013a; Forrest and Atalah, 2017). In the United Kingdom, a comprehensive assessment classified the risk of impact of D. vexillum on the local economy and environment as high, with the species’ establishment very likely and eradication difficult once well-established (GBNNSS, 2011). The report also highlighted the difficulty of eradication and the numerous lessons learnt by unsuccessful attempts in New Zealand and Wales (Coutts and Forrest, 2007; Forrest and Hopkins, 2013; Sambrook et al., 2014).
Accurate information on non-native species diversity, number of new invasions, and impact on native communities and habitats are important metrics necessary to assess the health of marine ecosystems and inform management decisions (EU-COM, 2008). Risk-based and regular monitoring for non-native species, specifically targeting their introductory hot spots, is the most cost-effective strategy to ensure rapid detection of invaders to protect the marine environment and economy. Long-term monitoring programs often apply rapid assessment survey (RAS) protocols (Cohen et al., 2005; Arenas et al., 2006; Bishop et al., 2015a; Nall et al., 2015; Kakkonen et al., 2019) where a large proportion of non-native species identification is conducted in the field, although for some taxa laboratory examination is required. Field identification enables rapid reporting of findings, with potential for early management interventions. However, the accuracy of morphological species identification relies on the knowledge and experience of the scientists conducting the RAS. The identification of D. vexillum, directly in the field is often difficult as the colonies can vary substantially in color and appearance, and full morphological identification involves laboratory processing of mature brooding colonies to enable scoring of several internal characters (Lambert, 2009). Difficulties in D. vexillum identification in the field has been highlighted recently by the discovery of a new species, Didemnum pseudovexillum, which can only be distinguished morphologically from D. vexillum by detailed microscopic examination of larval and spicule morphology or by DNA sequencing (Turon et al., 2020).
In the last decade, the continuous advancement of DNA-based techniques in aquatic monitoring has managed to overcome some of the uncertainty associated with morphology-based identification by utilizing well-characterized markers such as the cytochrome oxidase I (COI) gene and the small subunit ribosomal RNA region coupled with molecular tools based on the detection of environmental DNA (eDNA), i.e., DNA released externally by organisms into the environment (e.g., Rees et al., 2014; Barnes and Turner, 2016; Thomsen et al., 2016). Primarily, two approaches have been applied for the detection of eDNA. The first, targeted single-species detection uses conventional PCR, quantitative real-time (qPCR), or digital PCR and utilizes specific primers and/or probe. The second, multi-species detection approach by metabarcoding uses generic primer pairs and high throughput sequencing (see review by Zaiko et al., 2018). Recent studies have demonstrated that the detection of invasive non-native species by targeting their eDNA can be incorporated into routine marine biosecurity surveillance either at species (Simpson et al., 2017; Wood et al., 2019; LeBlanc et al., 2020) or community level (Holman et al., 2019; Rey et al., 2019). This study presents the results of a D. vexillum monitoring program carried out between 2017 and 2019 in selected recreational marinas and shellfish aquaculture farms in Scotland based on detection of target species eDNA by qPCR and describes the assay validation process for the use of this assay in the Northern Hemisphere.
Materials and Methods
Sampling Sites, eDNA Sampling and DNA Extraction
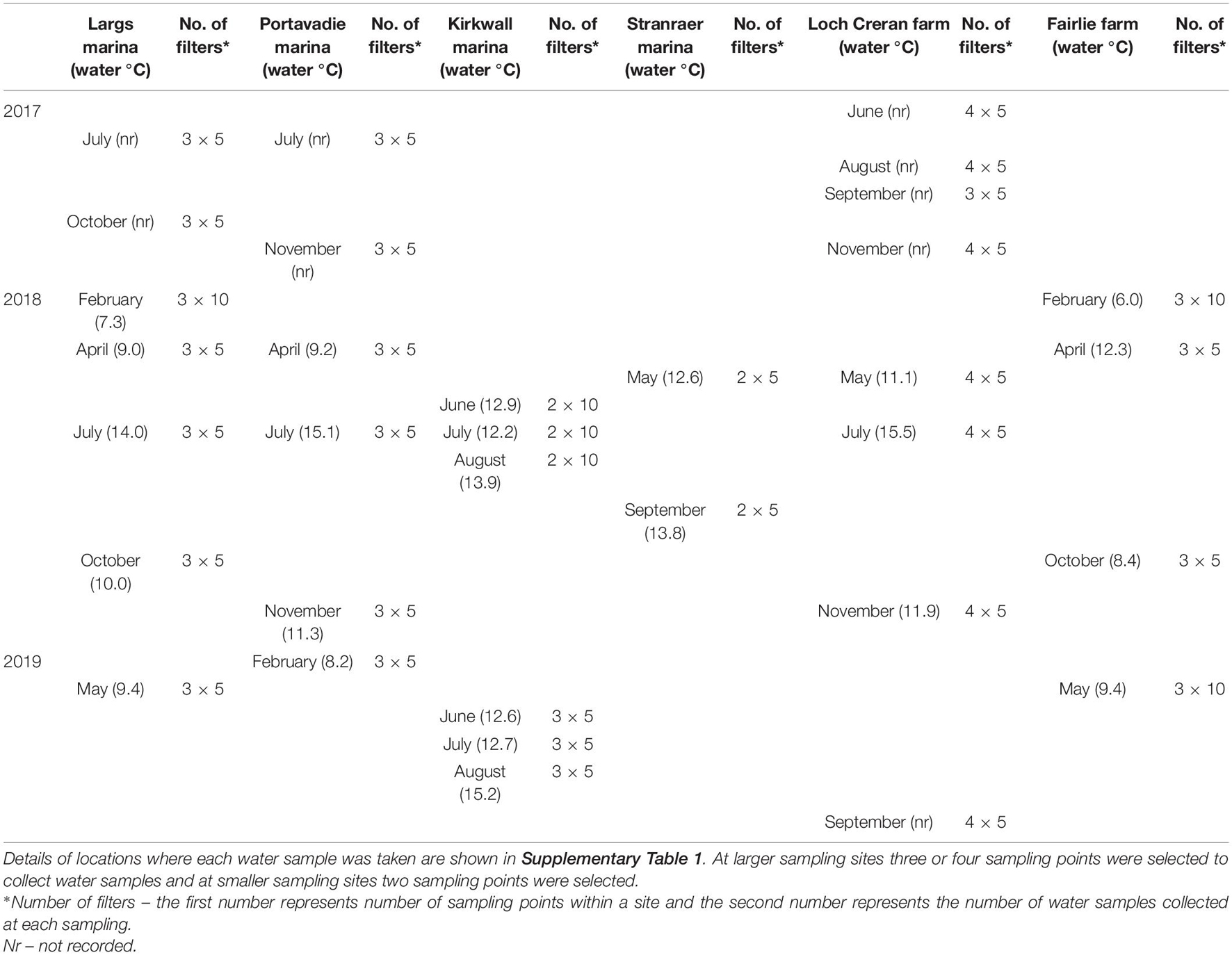
Two Pacific oyster, (Crassostrea gigas, Thunberg, 1793), aquaculture sites and four recreational marina sites were included in the present study (Table 1). The selection of sampling sites was based on known presence or absence of D. vexillum resulting from previous RAS. Individual 0.5 L water samples (within the top 50 cm of the water column) were collected using sterile Gosselin HDPE plastic bottles (Fisher Scientific). At the aquaculture sites, water samples were collected from the shoreline during low tide and at the recreational marinas water samples were collected from the pontoons. The number of different sampling points (between 2 and 4) was decided based on the size of each sampling site. The number of water samples collected at each sampling point varied between sites and years as summarized in Table 1. The detailed GPS locations of each sampling point are recorded in Supplementary Table 1.
Water samples were filtered in the field within 1 h of collection, using sterile 50 mL syringes (Fisher Scientific) and sterile 0.22 μm Sterivex filters (VWR Merck). During each sampling visit, 0.5 L sterile seawater (seawater from an offshore location, autoclaved in the laboratory and transported to the field in a sterile disposable bottle) was processed as an “in field” negative sampling control following the same procedures as for the field samples. Filters were kept on ice in an insulated polystyrene box, transported into the laboratory, within 12–24 h of collection, and stored at −20°C prior to processing. Total DNA was extracted from the filters using DNeasy Blood and Tissue extraction kits (Qiagen) using the protocol described by Spens et al. (2017). DNA was eluted in 100 μL of AE buffer and stored at −80°C until sample analysis.
Didemnum vexillum Quantitative PCR (qPCR) Design–in vivo and in vitro Specificity
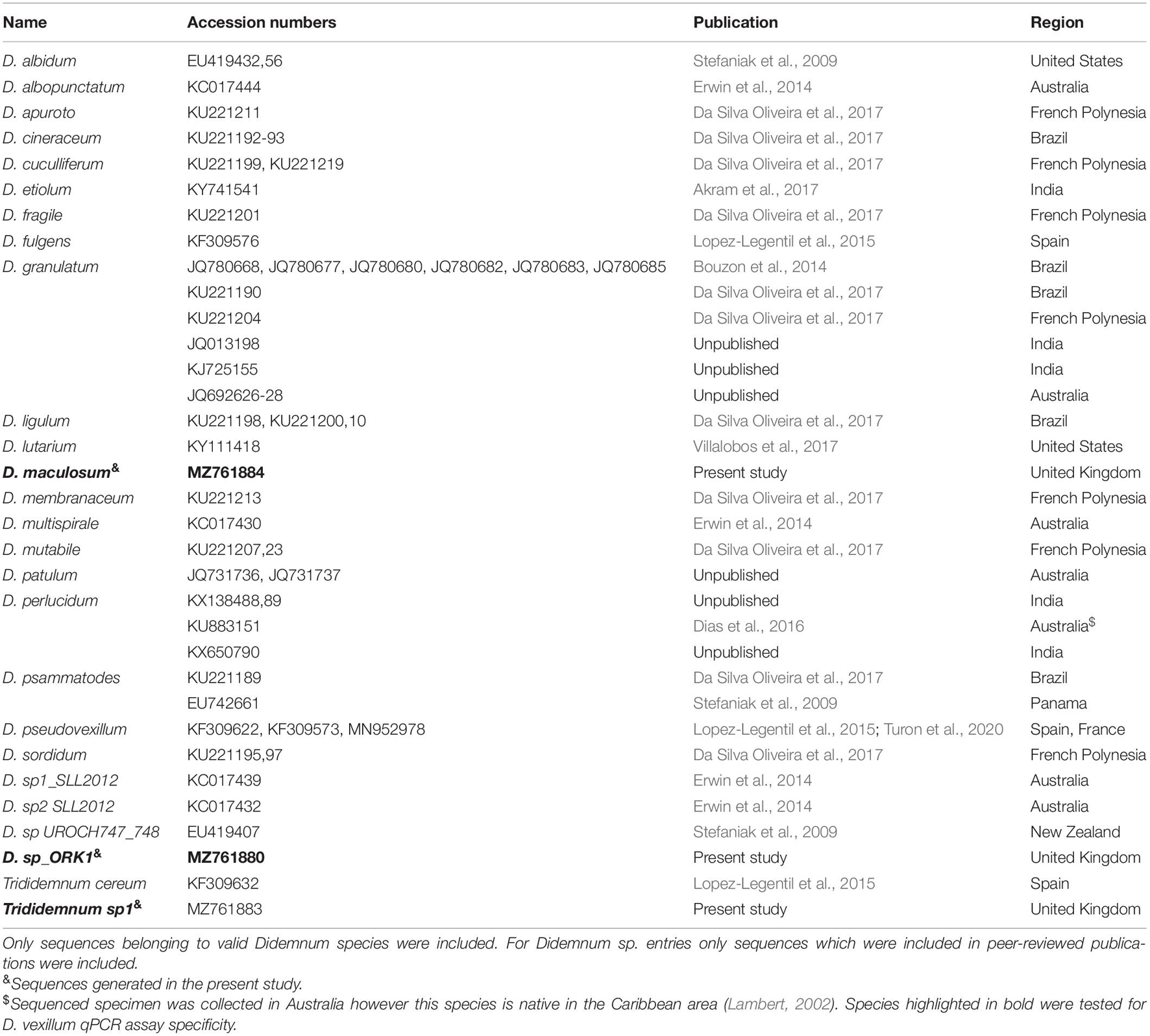
A TaqManTM qPCR assay, consisting of forward and reverse primers and a probe, was designed from a multisequence alignment of mitochondrial COI gene sequences from D. vexillum, closely related didemnid species (Table 2) and other tunicate species commonly found in the United Kingdom (Supplementary Table 2). To increase probe melting temperature (Tm) and ensure a higher specificity of the assay, considering the low GC content of the D. vexillum COI gene, a locked nucleic acid (LNA) probe-based qPCR assay was designed at the 5′ end of the COI gene. Where possible, the unique nucleotides within the D. vexillum probe region, relative to the majority species included in the alignment, were selected as LNA bases (Figure 1). When selecting the D. vexillum-specific probe region, a particular focus was given to closely related species that are reported from similar geographic regions to the present study, such as Didemnum albidum, Didemnum fulgens, Didemnum maculosum, D. pseudovexillum and Didemnum sp. ORK1 plus Trididemnum cereum and Trididemnum sp.1. These taxa were discriminated from D. vexillum by a minimum of three LNAs within the probe region (Figure 1). Sequences were aligned using Muscle version 3.8.425 software implemented in Geneious version 2020.1.1.
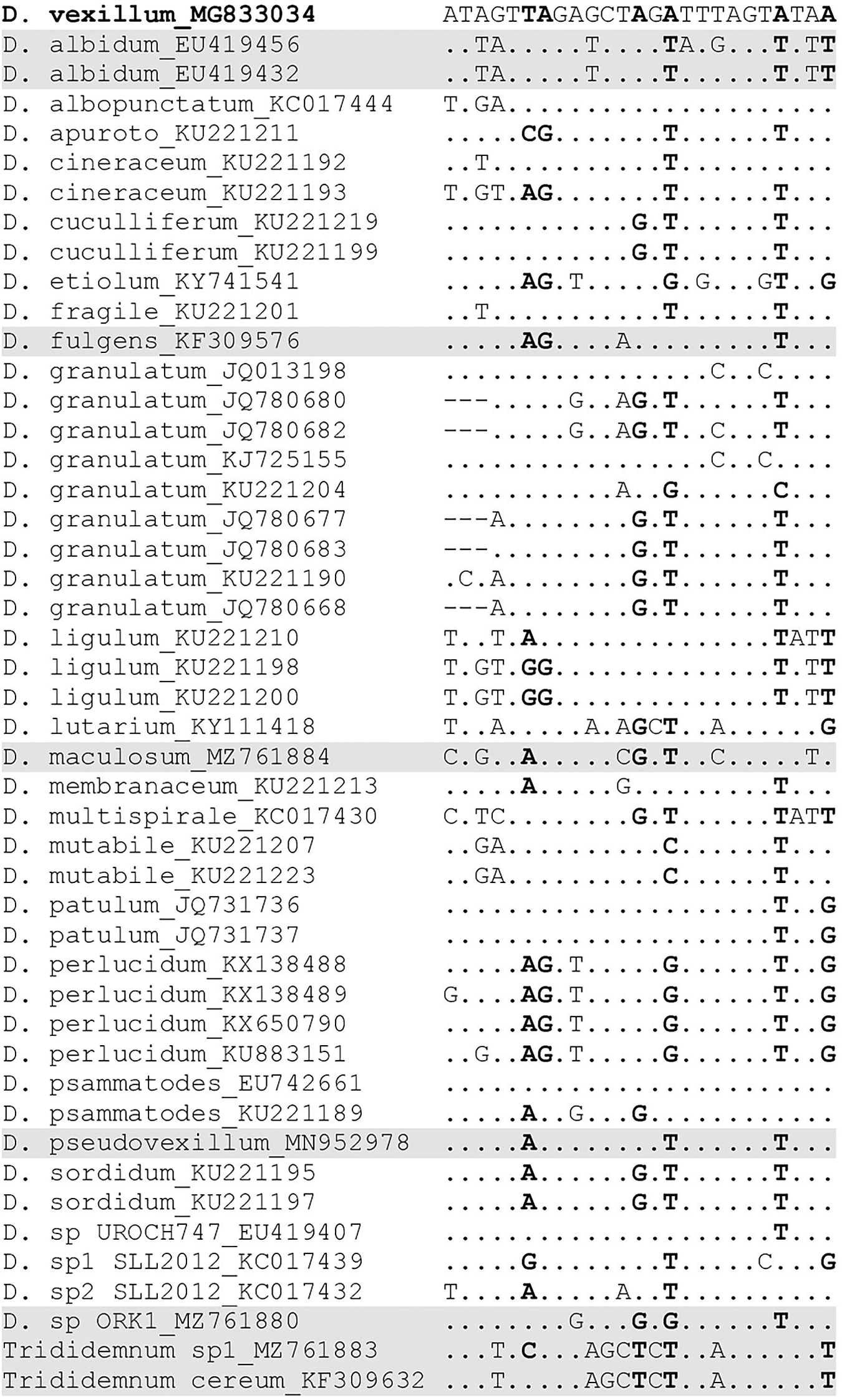
Figure 1. Multisequence alignment of D. vexillum and 26 species of tunicates from genera Didemnum and Trididemnum demonstrating nucleotide diversity in the D. vexillum qPCR assay probe region. Consensus with D. vexillum probe is shown as dots, with mismatched nucleotides shown. The locked nucleic acid bases (LNA) are highlighted in bold. The taxa highlighted in gray are closely related species co-occurring in the same geographic location as D. vexillum.
In silico assay specificity analysis was carried out using primer-BLAST (Ye et al., 2012). Assay specificity was further tested in vitro using genomic DNA (approximately 30 ng μL–1) extracted from D. vexillum colonies, belonging to eight different haplotypes [haplotypes 1, 2, 3, 5, 9, and 10 from D. vexillum clade A and haplotypes 14 and 17 from D. vexillum clade B (Smith et al., 2015)] (Supplementary Figure 1) and 21 tunicate species commonly co-existing with D. vexillum and/or common in the studied geographic region (Supplementary Table 2). Tissue samples stored in 100% ethanol (Sigma) were processed according to Graham et al. (2015) and the partial COI gene fragment was sequenced using the universal tunicate primers published in Stefaniak et al. (2009) to confirm species identity.
Different combinations of the TaqManTM assay primers (final concentration: 50, 300, and 900 nM) and fluorogenic-labeled probe specific for D. vexillum (final concentration: 50, 100, and 250 nM) were tested as recommended by the manufacturer1. The best-performing combination was 900 nM for both primers and 50 nM for the probe and these conditions were used throughout the whole study. Reaction plates (Micro-Amp Optical 96-well plate, Applied Biosystems) were prepared in a UV-cabinet located in a dedicated clean, positive-pressure room (no target DNA present) and template DNA was added in a UV-cabinet located in a separate dedicated pre-PCR room. DNA derived from preserved tissue or water samples was handled in separate UV-cabinets. UV-cabinets and all pipettes were cleaned before and after each use by 10% microsol and 70% ethanol and weekly with 1% sodium hypochloride solution followed by 10% microsol and 70% ethanol. In addition to primers and probe, the qPCR reaction contained 2× PerfeCTa qPCR Toughmix Low ROX Mastermix (QuantaBio, VWR) and sterile water (Sigma) up to a total volume of 20 μl. Reactions were run in triplicate, including three non-target controls (NTC) on the QuantStudio 5 Real time PCR System (Applied Biosystems) under the following cycling conditions: 1 cycle of 50°C for 2 min and 95°C for 10 min to activate/deactivate uracil-N-glycosylase (UNG) to degrade potential contaminant from previous PCR runs, and 45 cycles of 95°C for 15 s and 55°C for 1 min.
Limit of Detection (LOD) and Limit of Quantification (LOQ) of Didemnum vexillum qPCR Assay
A synthetic double-stranded DNA construct, containing the D. vexillum qPCR amplicon sequence, was ordered from www.amsbio.com. The construct also contained an artificial universal control sequence (VIC 5′-ACCGTCTAGCATCCAGT-3′ MGB) targeted by a second specific probe, to distinguish between “true” D. vexillum positive detection and a potential contamination with the positive control construct (Snow et al., 2009). An eleven-point 10-fold serial dilution of the construct (each in technical triplicates) was set up in a dedicated UV cabinet located in the post-PCR room and 10–5 (0.1 pg reaction–1) dilution was established as a working solution for the D. vexillum positive control, resulting in Cq (quantification cycle) values of around 20.
To calculate the D. vexillum qPCR assay limit of detection (LOD) and limit of quantification (LOQ), a seven-point 10-fold serial dilution of the synthetic construct working solution was performed to generate a standard curve, ranging between 5.548 × 105 and 0.555 copies reaction–1 (using a copy-number calculator in www.idtdna.com). Ten technical replicates for each dilution were run on the QuantStudio 5 Real time PCR System, using the cycling conditions described above. LOD was estimated from the standard seven-point curve using two methods. The first method used a discrete threshold approach and LOD was defined as the lowest standard concentration of DNA template that produced at least 95% positive replicates. The second method used a more flexible curve fitting approach as described in Klymus et al. (2020) to estimate an effective LOD for three replicates (LOD3). LOQ was defined as the lowest standard concentration that could be quantified with a coefficient of variation (CV) below 35% according to Klymus et al. (2020). All LOD and LOQ values were estimated using the R package ‘‘ednar’’2.
Detection and Quantification of Didemnum vexillum eDNA in Mesocosm and Field Water Samples
To test the sensitivity of the D. vexillum assay under experimental conditions, a mesocosm containing thirteen shucked Pacific oyster shells each harboring small patches of D. vexillum (approximately 2 cm2 of colony per shell) was created in a 320 L plastic tank (12°C seawater) with a flow-through of 150 L/h. The tank was housed in a bio-secure aquarium in Marine Scotland Science, Aberdeen, United Kingdom. The water was filtered through fluid sand towers to achieve filtration to approximately 10 μm and passed through UV prior to entering the tank room. Prior to pumping into experimental tanks water was passed again through UV and filtered through series of 5 and 1 μm filter socks. After 24 h exposure, 500 mL of water was collected using a sterile bottle and filtered immediately through a sterile 0.22 μm Sterivex filter. DNA was extracted from the filter as described above. A five-point serial dilution of the extracted DNA from the mesocosm was set up and eight replicates for each dilution were run on the QuantStudio 5 Real time PCR System, using the cycling conditions described above.
DNA extracted from all water samples (mesocosm, field study) was used as a template in qPCR reactions. Preparation and set up of qPCR reaction plates and cleaning of UV-cabinets was carried out as stated above. DNA was added to reaction plates in the pre-PCR room in a separate UV-cabinet to where DNA extracted from target organism tissue was handled. Each water sample was run in triplicate, alongside the seven-point standard curve (also in triplicates) which was used both for target copy number quantification and as a positive amplification control. All appropriate negative controls, in “field” negative, extraction negative and three non-target PCR controls (NTC) were included on each plate. A mean copy number of the target D. vexillum DNA in a reaction was calculated for all water samples which showed a positive detection of the target (Cq ≤ 41) in at least two out of three technical replicates (Wilcox et al., 2013; Agersnap et al., 2017). D. vexillum status at the studied sites was classified as: (1) detected (if at least one water sample showed a positive detection in all three technical replicates with a mean copy number of target reaction–1 > LOD3, or if at least 2 samples returned two out of three technical replicates with a mean copy number of target/reaction−1 > LOD3 (Goldberg et al., 2016), (2) suspected (if only one water sample showed a positive detection in two out of three technical replicates with a mean copy number of target reaction–1 > LOD or if multiple samples showed two out of three technical replicates with a mean copy number of target reaction–1 < LOD3), (3) not detected.
Results
Didemnum vexillum qPCR Assay Specificity
Didemnum vexillum species-specific LNA (nucleotides in brackets) probe DvexProbe (FAM 5′-ATAGT{T}{A}GAGCT{A}G{A}TTTAGT{A}TA{A}-3′ BHQ1) was designed to complement forward DvexFP (5′-CGACTAATCATAAAGATATTAGAACA-3′) and reverse primer DVexRP (5′-TTCTTGTAGAACTTAATTCTATTCG-3′), resulting in a 111 bp amplicon of the COI gene (positioned at bases 26–136 of the complete D. vexillum COI gene, GenBank accession number KM259616). The assay specificity was tested on eight different haplotypes of D. vexillum from both clades A and B (Supplementary Figure 1), including all three haplotypes from clade A that have been detected in the United Kingdom (Graham et al., 2015). Tissues from all tested haplotypes returned Cq values of approximately 20 in all triplicates.
Nucleotide differences in the probe region of the D. vexillum qPCR assay and mismatches to 46 sequences from 26 species (genera Didemnum and Trididemnum) are visualized in Figure 1. Variability across the whole amplicon is shown in Supplementary Figure 2. Sequence ambiguities were observed for Didemnum psammatodes COI, with the sequence entry published by Stefaniak et al. (2009) (Accession number EU472661) for this species from Panama being identical to D. vexillum in the assay probe region, whereas the entry published by Da Silva Oliveira et al. (2017) (Accession number KU221189) from Brazil showed three nucleotide differences (two of these bases were LNA) (Figure 1). Considering D. psammatodes has not been reported from the Northern Europe to date, we did not consider this to be an issue for the present study.
Due to the design of the D. vexillum qPCR assay at the 5′ end of COI gene, there are significant gaps in the NCBI nucleotide database for many closely related tunicates in the region of the DvexFP primer. Only two species of terrestrial insects, Bombus lucorum and Athalia proxima, were returned by the primer-BLAST, as possible non-target species (both with four mismatches in each primer). To further demonstrate specificity of the D. vexillum assay, DNA extracted from 21 tunicate species (Table 2 and Supplementary Table 2), was run in triplicate and no amplification was observed. In particular, no amplification was shown for closely related species found commonly co-existing with D. vexillum at the study sampling sites (Table 1), such as D. maculosum, Didemnum sp. ORK1 and Trididemnum sp.1 (Figure 1).
Didemnum vexillum qPCR Assay Sensitivity and Detection of eDNA in Mesocosm Experiment
Detection of D. vexillum target was recorded in all seven serial dilution points generated from the positive control construct, ranging between 5.548 × 105 and 0.555 copies/reaction. The lowest concentration at which all ten technical replicates detected the target was 55.48 copies/reaction (average Cq values = 34.53), 90% of replicates showed detection at the concentration of 5.548 copies/reaction (average Cq values = 38.25) and only five replicates gave a Cq value at the concentration of 0.5548 copies reaction–1 (average = 39.42). No amplification was observed in NTCs. The assay efficiency calculated from the straight-line equation (y = −3.370x + 40.063; R2 = 0.993) generated from the positive control DNA-based standard curve was 98.02%. The threshold LOD and LOQ were calculated at 9.187 and 18 copies reaction–1 and the modeled LOD3 was estimated at 1.117 copies reaction–1 (Figure 2).
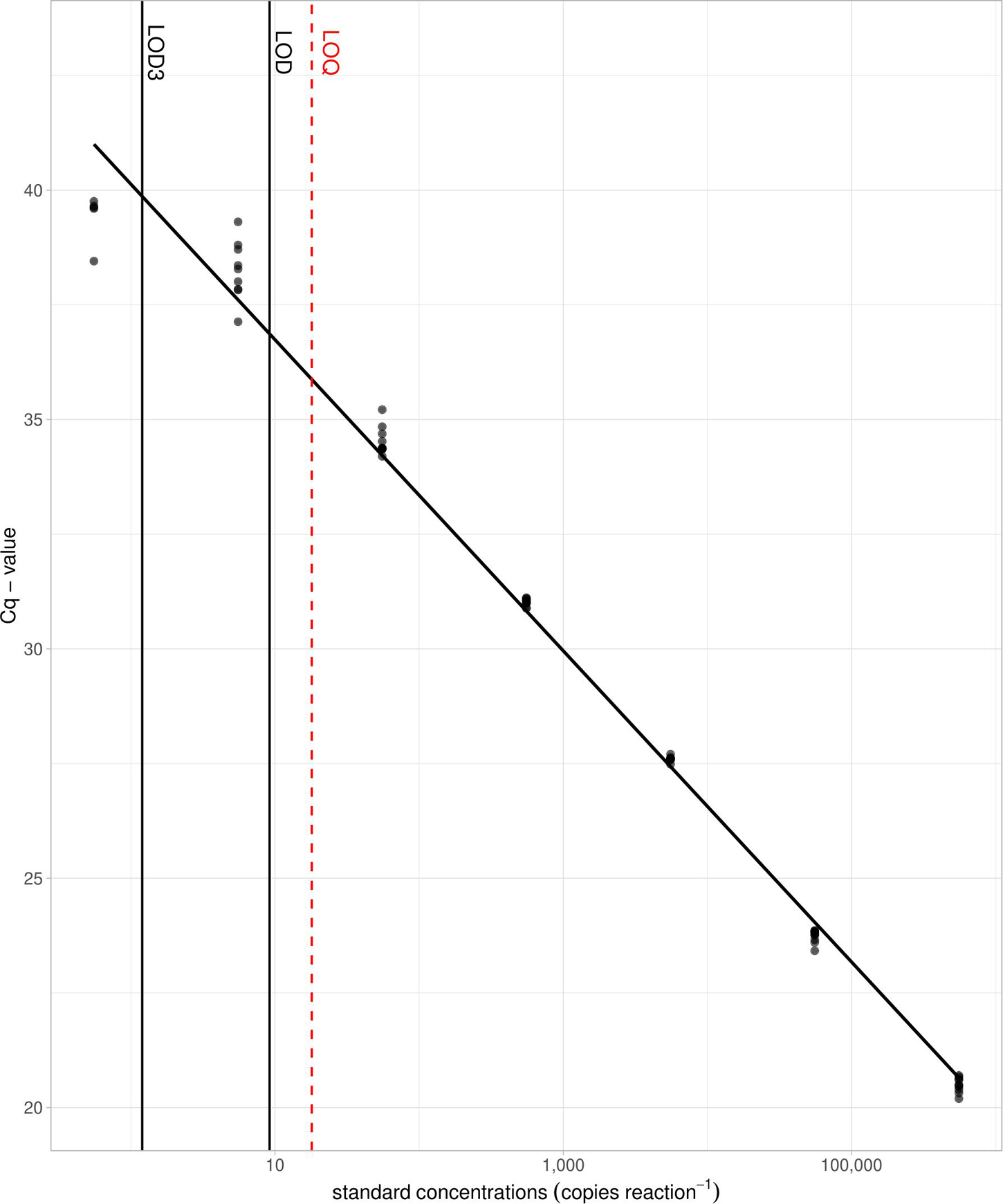
Figure 2. Limit of detection (LOD) and limit of quantification (LOQ) derived from a 10-fold serial dilution of the synthetic construct containing the D. vexillum target sequence. Threshold LOD (9.187 copies reaction– 1) and modeled LOD3 (1.117 copies reaction– 1) for the D. vexillum qPCR assay are shown as solid lines and LOQ (18 copies reaction– 1) is shown as dashed line. Ten technical replicates per dilution were run, with a reaction efficiency of 98.02%.
Sensitivity of the D. vexillum qPCR assay in detecting DNA released by living colonies was assessed in the aquarium mesocosm, where D. vexillum colonies attached to oyster shells were held for 24 h prior to water collection. The target was detected in a sample of 500 mL of water (average Cq value of 25.70), with estimated concentration of 3416.69 copies reaction–1 (average value from 8 replicates). Positive detection of D. vexillum (in all eight replicates) was recorded in the next four serial dilutions of extracted DNA (the lowest average concentration of 3.72 copies reaction–1 in 1 × 10–4 dilution of DNA). In the fifth dilution point of extracted DNA, D. vexillum was detected in only one out of eight technical replicates (Cq = 39.54), with estimated concentration of 1.44 copies reaction–1 (Figure 3).
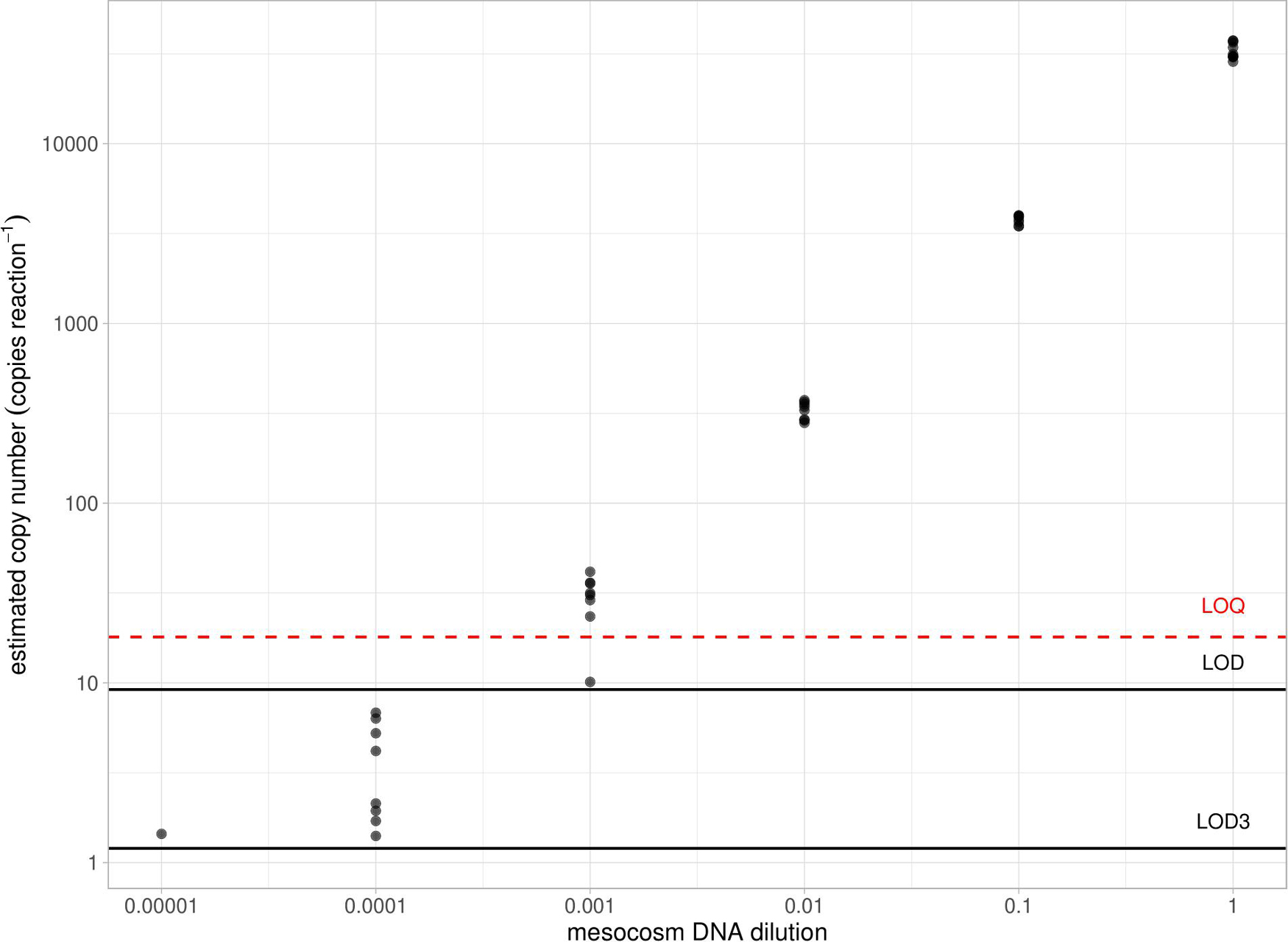
Figure 3. Sensitivity of the D. vexillum qPCR assay in the in situ mesocosm experiment. Threshold LOD (9.187 copies reaction– 1) and modeled LOD3 (1.117 copies reaction– 1) for D. vexillum qPCR assay are shown in solid lines and LOQ (18 copies reaction– 1) is shown in dashed line.
Detection of Didemnum vexillum eDNA in Field Samples Collected at Pacific Oyster Farms and Recreational Marinas
Efficiency of qPCR runs on field samples varied between 92.34 and 109.31% (average 99.27%) with R2 ranging from 0.9830 to 0.9974 (Supplementary Table 3). No amplification was shown in any field blank controls, negative extraction controls or NTCs. No detection of D. vexillum eDNA was shown at Kirkwall and Stranraer marinas throughout the study period. Positive eDNA detection of D. vexillum was found in samples from the Pacific oyster farms in Loch Creran and Fairlie and in Largs and Portavadie marinas throughout the study period, based on the modeled LOD3 (Figure 4). eDNA signal identity from all D. vexillum positive sites was verified by sequencing of the qPCR product and the sequences showed 100% identity to D. vexillum (Accession number MG833034, Cottier-Cook et al., 2019). There was 100% concordance between detection of D. vexillum eDNA and physical observation of the colonies by both site operators and authors of this study.
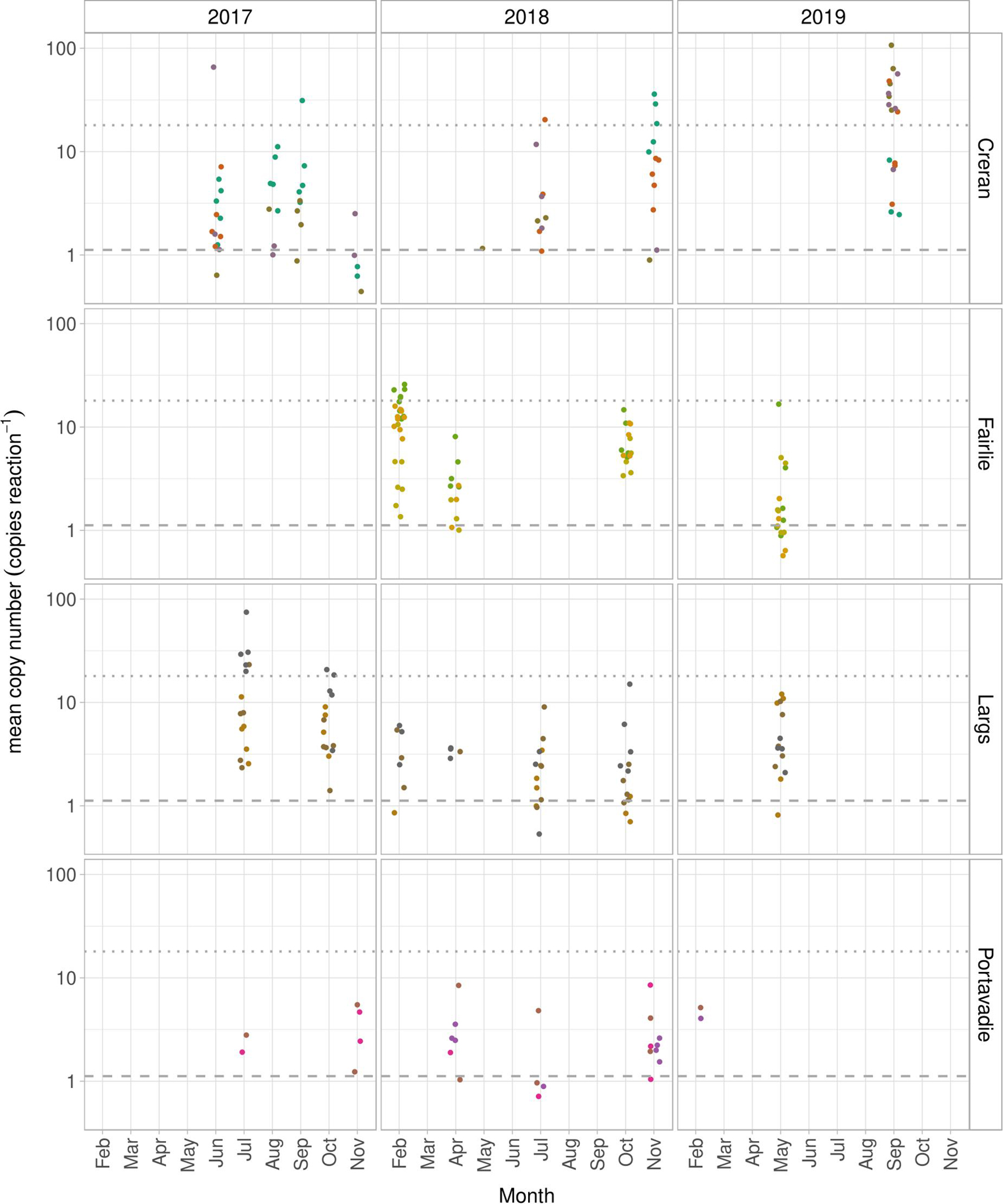
Figure 4. Positive detection of D. vexillum eDNA expressed as mean copy number per reaction at Loch Creran and Fairlie farms and Largs and Portavadie marinas. The modeled LOD3 (1.117 copies reaction– 1) for the D. vexillum qPCR assay is show in dashed line and LOQ (18 copies reaction– 1) is shown in dotted line. Sampling points within the sampling sites are visualized in different colors.
Proportions of water samples collected at the four D. vexillum-positive sites between 2017 and 2019, showing D. vexillum eDNA concentrations > modeled LOD3 (1.117 copies reaction–1) (Table 3), are presented in Table 3. D. vexillum eDNA concentrations only occasionally reached the threshold set for LOQ (18 copies reaction–1) (Table 3 and Figure 4) and therefore it was not feasible to infer any accurate quantitative trends in concentration of D. vexillum eDNA between the sampling dates and/or sites.
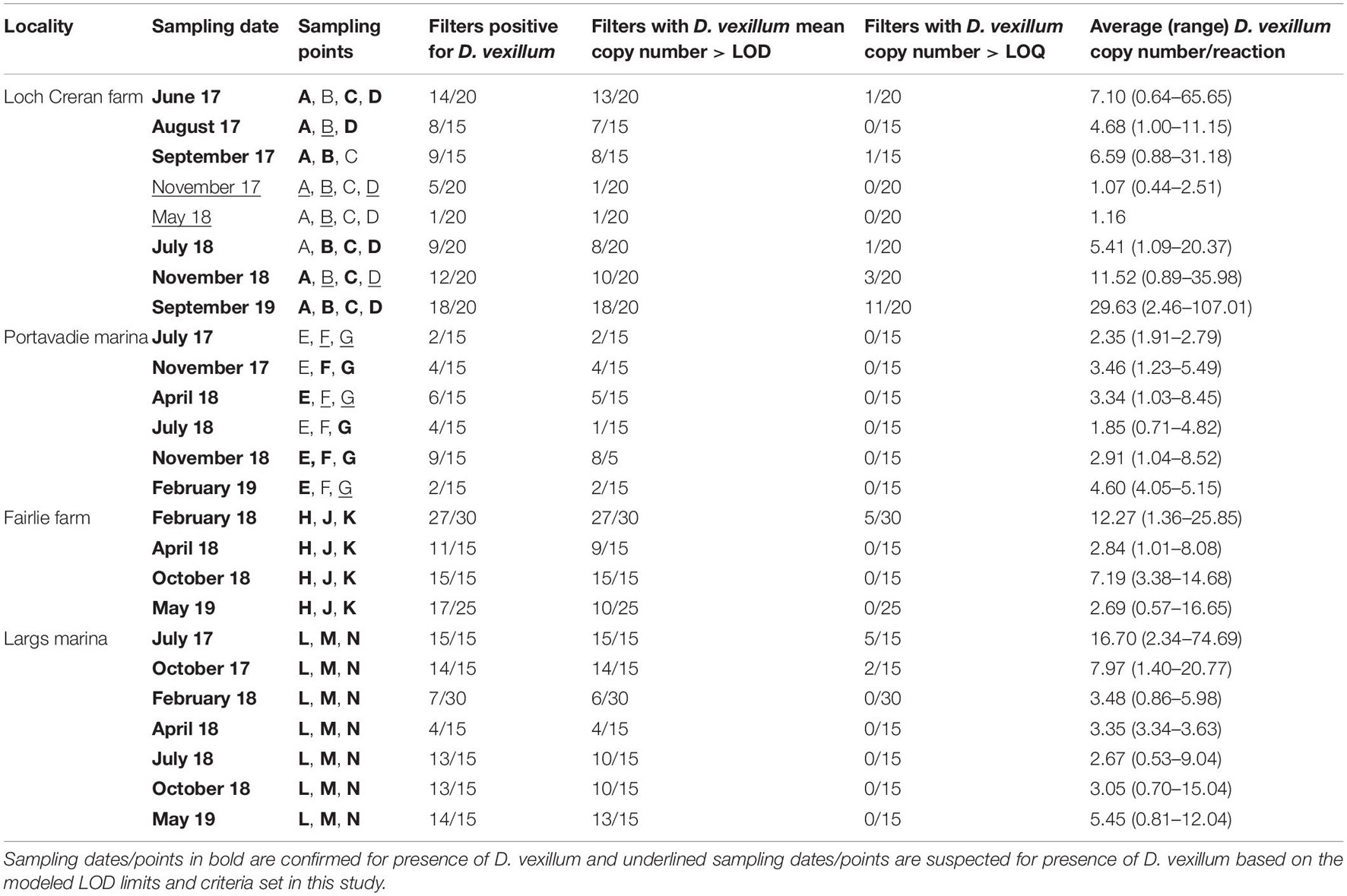
Table 3. Summary of water samples showing positive detection of D. vexillum at each of the studied sites, including average and range of target species copy number per reaction.
At the Fairlie farm and Largs marinas, a high proportion of water samples showed D. vexillum eDNA concentration/reaction > modeled LOD3 and the eDNA was detected consistently in all sampling points throughout the studied period. At Portavadie marina, D. vexillum eDNA concentration/reaction appeared to be lower (always <10 copies reaction–1) compared to the Fairlie farm and Largs marina and occasionally one out of three sampling points tested “negative” or only as “suspected” for D. vexillum (Table 3 and Figure 4). Overall, D. vexillum status was confirmed as “detected” at all sampling dates at the Fairlie farm and Largs and Portavadie marinas.
In the Loch Creran farm the proportion of water samples testing positive for D. vexillum, with eDNA concentration > modeled LOD3, varied (Table 3). There was an apparent reduction in D. vexillum-positive water samples in November 2017 and a further reduction was recorded in May 2018. During these particular sampling dates, only 1/20 water samples detected D. vexillum eDNA at concentration > modeled LOD3 (in 2/3 technical replicates) (Table 3) and according to the stringent eDNA detection criteria set up in this study, the status was reported as D. vexillum “suspected.” In all other sampling dates, the status of D. vexillum was determined as “detected.” Inconsistency in D. vexillum eDNA detection among the four sampling points within the Loch Creran farm was also higher compared to the other sites (Table 3).
Discussion
Rapid, sensitive and cost-effective monitoring tools are required to obtain accurate baseline information on presence and distribution of marine non-native species from high-risk sites. Monitoring and management strategies are essential mechanisms by which the EU Member States can ensure the Good Environmental Status (GES) of their seas required by the Marine Strategy Framework Directive (MSFD) and United Kingdom Marine Strategy. Under the MSFD Descriptor 2 marine non-native species baselines should be measured using indicators such as number of new introductions, total number of established non-native species and their distribution within the member state, plus their impact on the ecosystem if established (EU-COM, 2008).
While traditional RAS are known to be an effective approach to detect new introductions and monitor known non-native species populations (Cohen et al., 2005; Arenas et al., 2006; Bishop et al., 2015a; Nall et al., 2015; Kakkonen et al., 2019), these require the field deployment of taxonomists or trained parataxonomists and may still fail to detect inconspicuous species or the early stages of an incursion. Furthermore, RAS are not a suitable tool to measure the potential impact of invasive non-native species on native communities and habitats. In contrast, DNA-based tools can performed well, with the ease of collecting water samples offering the potential for scaling up monitoring efforts. In some countries progress toward incorporating eDNA into routine surveillance programs has already been initiated, demonstrating how these tools can contribute to improved monitoring of marine non-native species (Roux et al., 2020; Wood et al., 2019; LeBlanc et al., 2020). Performance and ability of targeted tools such as qPCR to support marine biosecurity scored high in a review conducted by Zaiko et al. (2018). Based on a set of predefined parameters, including tool feasibility, precision, cost-effectiveness, quantitative ability and early warning applicability, qPCR tools could aid biosecurity, risk assessments, baseline surveys, early warning, pathway and impact managements of non-native species. In addition, recent reviews suggest that environmental genomics, either the more conventional taxonomy-based approaches or de novo taxonomy-independent bioindicators, may provide powerful tools to understand changes in marine communities in response to a variety of anthropogenic stressors and therefore qualitatively and quantitatively describe the impact of non-native species (Cordier et al., 2020).
Specificity of Didemnum vexillum qPCR Assay
The present study demonstrates both analytical and field validation for a D. vexillum qPCR assay, based on the COI gene, for use in the Northern Hemisphere. When using a qPCR approach to target non-native species, ensuring assay specificity is an essential component of the validation process (Goldberg et al., 2016; Thalinger et al., 2021). Specificity testing included closely related species, many of them occurring in the studied geographic area as well as co-occurring species distantly related to D. vexillum. The probe region was selected to maximize the number of mismatches to ensure assay specificity. Furthermore, the D. vexillum qPCR assay specificity was increased by mismatches in primer regions (Wilcox et al., 2013), including one mismatch at the 3′ end of the forward primer and a high overall variability in the reverse primer for the majority of closely and distantly related co-occurring species (Supplementary Figure 1). Giving the variability of the COI region within D. vexillum and, on the other hand, the similarity of the COI region of D. vexillum to other didemnids, it was difficult to design a robust and specific assay that could be used to detect the species world-wide. There was one base pair variation in the assay probe region for D. vexillum between haplotypes 9 (from clade A) and 13, 15–18 (from clade B) (Smith et al., 2015) but after in vitro testing of haplotypes 9 and 17 it appeared that this level of variation did not impact the ability of this assay to detect these haplotypes.
With regards to the assay’s specificity relative to other tunicate species, no amplification was observed in the in vitro specificity analysis, suggesting sufficient specificity of the D. vexillum assay for use in the studied region (see Supplementary Table 2). It is important to emphasize the geographic region for which the assay was validated. In the present study the D. vexillum assay was validated for use in the Northern Hemisphere and will not necessary be suitable for regions where D. psammatodes is expected to be present. Therefore, other published D. vexillum qPCR assays may thus be better suited for studies and monitoring programs in the Southern Hemisphere (Simpson et al., 2017). Furthermore, it is highly recommended to confirm the presence of a target species by direct Sanger sequencing of positive qPCR products, especially when eDNA detection in a given locality is a new record or unexpected finding. Repeating the specificity analysis with closely related non-target species relevant to the particular geographic region is also advised.
Since approximately 2016, the validation process for targeted single-species qPCR assays has begun to routinely incorporate in silico and in vitro analyses (Thalinger et al., 2021), providing a valuable first insight into qPCR assay specificity. However, species identification for many different groups of tunicates remains challenging due to cryptic diversity and issues with species misidentification leading to errors in entries in the public sequence repositories. For example, Viard et al. (2019) documented confusion between the invasive species Botrylloides diegensis and the native European species Botrylloides leachii, which highlights the need for well-curated reference material and sequence data. The reliance on poorly curated sequences can lead to the design of DNA-based tools which misinterpret the native versus non-natives status of species. The ability to prevent or at least recognize false positive detections is essential when using eDNA-based tools for monitoring of non-native species due to the high costs associated with follow-up surveys and management interventions.
Sensitivity of Didemnum vexillum qPCR Assay
In addition to the specificity of the eDNA assay, clear definitions of the limit of detection (LOD) and quantification (LOQ) are essential to generate robust and accurate data. Different templates are used to generate the LOD and LOQ calibration plots with either target species genomic DNA (Gillum et al., 2014; Miralles et al., 2016, 2019; Mauvisseau et al., 2019; LeBlanc et al., 2020), synthetic DNA constructs (Larson et al., 2017; Harper et al., 2018; Mirimin et al., 2019; Roux et al., 2020), or a PCR product of the target gene (Wood et al., 2019) being commonly utilized. The standards in calculating and reporting these parameters continue to vary widely in the published literature and consequently the LOD and LOQ are reported either as a copy number or concentration of a given target per reaction or microliter. Moreover, the criteria used to set LOD and LOQ limits vary significantly, making results of eDNA studies difficult to interpret and compare to each other. The present study adopted the latest proposed criteria for LOD and LOQ set out in Klymus et al. (2020), and the methods and reporting of results comply with the minimum reporting criteria for eDNA studies proposed by Goldberg et al. (2016). Using a synthetic DNA construct, the D. vexillum LOD was identified as 9 copies reaction–1 and LOD3 as 1.117 copies reaction–1. LOD3 value is comparable to other published eDNA assays (Agersnap et al., 2017; Harper et al., 2018; Roux et al., 2020) and hence was selected as the cut-off point for the field samples collected in this study. Some attempts to confirm detection limits using water samples collected from D. vexillum in a mesocosm were made in the present study. It appears that D. vexillum eDNA can be detected at very low levels, consistently at 3.72 copies reaction–1 and sporadically (one of eight replicates) at 1.44 copies reaction–1. However, this result was only for one water sample from one mesocosm tank (after 24 h exposure) and therefore additional experiments with larger numbers of samples are required to accurately predict the number of qPCR replicates needed to detect D. vexillum eDNA at low concentration. In this study, it was not financially possible to carry out more than three technical qPCR replicates for each filter sample collected, and it is important to view the D. vexillum-negative detections in this context. It is expected that more qPCR replicates will be needed if eDNA detection is to be used to support management interventions such as eradications.
Furthermore, the criteria upon which water samples and/or sites are classified as either positive, suspected or negative for a given target are often not clearly specified. This study takes into consideration remarks by Goldberg et al. (2016) on reproducibility among collected samples and inferring a positive detection from a result in a single well. The present study only accepted the detection of the target based on one positive sample if all three technical replicates gave a reproducible Ct value ≤ 41 (Wilcox et al., 2013; Agersnap et al., 2017) and mean copy number > modeled LOD3. There is urgent need to further develop the standards for analysis of eDNA data and agree a set of consistent parameters and criteria for estimating the LOD and LOQ. Without this standardization, the lack of detailed methodology and inconsistent reporting will continue to create uncertainty regarding the robustness of eDNA detection methods for use in non-native species management and decision making.
Consistency in Detection of Didemnum vexillum eDNA and Correlation With Traditional Surveys and Abundance
Concordance between the results of eDNA-based and traditional surveys has been demonstrated in numerous studies, with some studies reporting a higher detection success for some marine invasive species using eDNA, either detected by a single-species approach (for example Muñoz-Colmenero et al., 2018; LeBlanc et al., 2020) or metabarcoding (for example Holman et al., 2019; Rey et al., 2019). The present D. vexillum qPCR assay demonstrated 100% concordance between physical observation of this species and detection of eDNA by qPCR in both marina and aquaculture farm settings. The fact that no D. vexillum eDNA was detected at Kirkwall and Stranraer marinas is in keeping with results of traditional surveys carried out at these sites. At Kirkwall marina, a dedicated marine non-native species monitoring program has been on-going since 2013, including RASs and morphological identification of biofouling in scrapes from marina hard surfaces and deployed settlement panels. This monitoring program has resulted in the recording of nineteen non-native species but no D. vexillum was found since the monitoring began (Kakkonen et al., 2019). At Stranraer marina, RA surveys and settlement panels have been deployed between 2016 and 2018 (unpublished data, Solway Firth Partnership) and as in Kirkwall marina, no D. vexillum was recorded.
Presence of D. vexillum in Largs and Portavadie marinas and Loch Creran and Fairlie farms was confirmed by the authors of this study both visually and by partial COI sequencing of collected tissue material. For the majority samples/sites, the eDNA expressed as mean copy number per reaction was less than the assay LOQ. Therefore, a quantitative assessment and comparison of D. vexillum eDNA was not feasible in this study. For the Fairlie farm (where D. vexillum is present on Pacific oyster bags and exposed to air during the tidal cycle) and Largs marina (where D. vexillum is present on marine infrastructures and remains constantly submerged) there is also no estimate of D. vexillum abundance based on traditional surveys but its presence has been reported also by site operators (Beveridge et al., 2011). eDNA-based monitoring revealed a high proportion of water samples positive for D. vexillum and the detection was consistent in all sampling points throughout the studied period.
The abundance of D. vexillum at Portavadie marina (D. vexillum present on marine infrastructures and constantly submerged) is thought to be very low: visual observations from pontoons and two remotely operated underwater vehicle (ROV)-based surveys in August 2018 and February 2019 (unpublished data, Marine Scotland Science) detected only 10 small colonies. This low abundance was reflected in low concentration of D. vexillum eDNA and lower proportion of positive water samples at each sampling point compared to Largs and Fairlie sites. eDNA data collected in Portavadie marina shows an increase in false negatives when detection probabilities are low, and this may be due to low concentrations of the target species eDNA, inconsistent release of eDNA and/or imperfect mixing of eDNA in the water column (Ficetola et al., 2015; Furlan et al., 2016). Whilst the sampling effort in the present study was sufficient to confirm D. vexillum at Portavadie marina, a further study is being prepared to better understand the false negative rates at sites where D. vexillum is present at low abundance and to inform sampling and laboratory strategies.
The abundance of D. vexillum at the Loch Creran farm (D. vexillum present on Pacific oyster bags and exposed to air during the tidal cycle) was expected to be the highest of all studied sites, based on the results of annual farm surveys between 2016 and 2019 (unpublished data). The Loch Creran farm site is substantially larger than the other sites and is an active site in terms of movement of shellfish stock within and out of the farm. In terms of LOQ, the D. vexillum eDNA concentrations were only sporadically higher than the LOQ threshold, although in September 2019 a large proportion of samples crossing this threshold was observed (Table 3 and Figure 4). In November 2017, there was an apparent decrease in the proportion of water samples for which the mean copy number/reaction reached values higher modeled LOD3. This decrease fits well with operational activities ongoing on the site during this time, and may be linked to the removal of Pacific oyster bags colonized by D. vexillum. Reduced detection of D. vexillum eDNA in May 2018 could therefore be a reflection of the reduced abundance of D. vexillum on the site following the management interventions in the previous year. Studies utilizing eDNA to assess efficacy and success of aquatic invasive species eradication programs are beginning to emerge, especially for freshwater species in lentic habitats (Davison et al., 2017; Furlan et al., 2019). In the marine environment, subject to constant mixing of the water column by tidal currents and wind action, a more intensive sampling strategy will be needed to achieve high detection probabilities at low target densities (Wood et al., 2019) if eDNA-based methods to be used as a tool to support invasive species eradications. In addition, the number of biological and technical replicates will need to be carefully considered before applying these methods to inform marine invasive species eradication programs.
Seasonality in Detection of Didemnum vexillum eDNA
Fluctuations in eDNA concentrations associated with a target organism’s life strategies have been observed in freshwater (Buxton et al., 2018) and marine environments (Takahara et al., 2019), with higher concentration of eDNA being detected during the summer months, associated with increased growth and/or spawning seasons. The impact of seasonality and the D. vexillum life cycle on the detectability of eDNA was not the primary objective of the present study and therefore only limited inference can be made. In Loch Creran and Fairlie farms and Largs and Portavadie marinas the status of D. vexillum appears to be established and during the winter months when D. vexillum is not expected to release larvae and exhibits only limited growth (Valentine et al., 2009; Fletcher et al., 2013b), 90 and 20% of water samples tested positive for D. vexillum (>modeled LOD3) at Fairlie farm and Largs marina (February 2018) respectively. A smaller proportion of samples (13% of samples > modeled LOD3) tested positive for D. vexillum in February 2018 in Portavadie marina, and this could reflect a further decrease in concentration of D. vexillum eDNA during slow growth period at this site. As a result of our limited data further studies are required to fully explore the variability of eDNA detection over D. vexillum reproductive cycles and across seasons to optimize sampling strategies and inform management decisions.
Data Availability Statement
The data presented in the study are deposited in the https://github.com/alexd106/ednar repository. The sequence generated in this study are deposited in Genbank with accession numbers MZ761879-84.
Author Contributions
IM, JG, and AD conceptualized the idea and design for the study. IM designed the qPCR assay, analyzed the qPCR data, and wrote the first of the drafts. IM and JG developed the laboratory workflows. JG, FB, and IM conducted the laboratory work. AD carried out statistical analysis. JG, IM, JK, J-PL, GH, ED, FB, and LB conducted water collection and filtering. JB, JK, and KS provided essential reference tissue material. JB, JK, KS, and AD provided a critical feedback on the first draft. All authors contributed to the second draft and gave final approval for publication.
Funding
This work was funded by Marine Scotland and Marine Scotland Science, Scottish Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the site operators, owners and Solway Firth Partnership for allowing access and sample collection at studied sites visited during this study. Thanks also to Frank Armstrong, Katy Beaton, Maria Campbell, Pablo Dias, James Dooley, Judith Horrill, Nial McLeod, Warren Murray, Andrea Taylor, Joe Triscott, and Bill Turrell for contributing to field work and sample collection. The authors thank National Museums Scotland and particularly Fiona Ware for the loan of reference material (specimen register number NMS.Z.2015.82.8, 9 and 14 and NMS.Z.2018.2.2) which was used in the present study. KS thank the Japan Society for the Promotion of Science for post-doctoral fellowship funding.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.728456/full#supplementary-material
Supplementary Figure 1 | Variability within the D. vexillum qPCR assay forward primer and probe region for 23 D. vexillum COI haplotypes. Variable nucleotides are shown in green and haplotypes tested in the present study are shown in bold. Haplotypes 1–10 belong to clade A and haplotypes 11–23 belong to clade B (Smith et al., 2015).
Supplementary Figure 2 | Multisequence alignment of D. vexillum and 26 other species from the genera Didemnum and Trididemnum demonstrating nucleotide diversity in the complete D. vexillum qPCR assay amplicon. Consensus with the D. vexillum assay is shown as dots, with mismatched nucleotides identified.
Supplementary Table 1 | Location of each sampling point.
Supplementary Table 2 | List of tunicate species coexisting with D. vexillum in the United Kingdom included to assess D. vexillum qPCR assay specificity. All tissue material was confirmed to species by sequencing of the partial COI gene using primers published in Stefaniak et al. (2009) and showed 100% identity to the published sequences. &Novel sequences obtained in the present study.
Supplementary Table 3 | Quantitative PCR (qPCR) performance parameters.
Footnotes
- ^ https://www.eurogentec.com/assets/10a3bf2d-eedd-452b-b26a-c1e85568c87b/guide-en-rtqpcr-sample-test-guide.pdf
- ^ https://github.com/alexd106/ednar
References
Agersnap, S., Larsen, W. B., Knudsen, S. W., Strand, D., Thomsen, P. F., Hesselsøe, M., et al. (2017). Monitoring of noble, signal and narrow-clawed crayfish using environmental DNA from freshwater samples. PLoS One 12:e0179261. doi: 10.1371/journal.pone.0179261
Akram, S., Arshan, K. M. L., and Abdul, J. H. (2017). DNA barcoding and phylogenetic analysis of five ascidians (Phlebobranchia) distributed in Gulf of Mannar, India. Mitochondrial DNA A DNA Mapp. Seq. Anal. 29, 581–586. doi: 10.1080/24701394.2017.1325479
Arenas, F., Bishop, J. D. D., Carlton, J. T., Dyrynda, P. J., Farnham, W. F., Gonzalez, D. J., et al. (2006). Alien species and other notable records from a rapid assessment survey of marinas on the south coast of England. J. Mar. Biol. Assoc. UK 89, 1329–1337. doi: 10.1017/S0025315406014354
Barnes, M. A., and Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. doi: 10.1007/s10592-015-0775-4
Beveridge, C., Cook, E. J., Brunner, L., MacLeod, A., Black, K., Brown, C., et al. (2011). Initial Response to the Invasive Carpet Sea Squirt, Didemnum vexillum, in Scotland. Scottish Natural Heritage Commissioned Report No. 413. Inverness: Scottish Natural Heritage.
Bishop, J. D. D., Wood, C. A., Lévêque, L., Yunnie, A. L. E., and Viard, F. (2015a). Repeated rapid assessment surveys reveal contrasting trends in occupancy of marinas by non-indigenous species on opposite sides of the western English Channel. Mar. Pollut. Bull. 95, 699–706. doi: 10.1016/j.marpolbul.2014.11.043
Bishop, J. D. D., Wood, C. A., Yunnie, A. L. E., and Griffiths, C. A. (2015b). Unheralded arrivals: non-native sessile invertebrates in marinas on the English coast. Aquat. Invasions 10, 249–264. doi: 10.3391/ai.2015.10.3.01
Bouzon, J. L., Vargas, S. M., Oliveira Neto, J. F., Stoco, P. H., and Brandini, F. P. (2014). Cryptic species and genetic structure in Didemnum granulatum Tokioka, 1954 (Tunicata: Ascidiacea) from the southern Brazilian coast. Braz. J. Biol. 74, 923–932. doi: 10.1590/1519-6984.00213
Bullard, S. G., and Whitlatch, R. (2009). In situ growth of the colonial ascidian Didemnum vexillum under different environmental conditions. Aquat. Invasions 4, 275–278. doi: 10.3391/ai.2009.4.1.27
Bullard, S. G., Lambert, G., Carman, M. R., Byrnes, J., Whitlatch, R. B., Ruiz, G., et al. (2007). The colonial ascidian Didemnum sp. A: current distribution, basic biology and potential threat to marine communities of the northeast and west coasts of North America. J. Exp. Mar. Biol. Ecol. 342, 99–108. doi: 10.1016/j.jembe.2006.10.020
Buxton, A. S., Groombridge, J. J., and Griffiths, R. A. (2018). Seasonal variation in environmental DNA detection in sediment and water samples. PLoS One 13:e0191737. doi: 10.1371/journal.pone.0191737
Carman, M. R., Grunden, D. W., and Ewart, D. (2014). Coldwater reattachment of colonial tunicate Didemnum vexillum fragments to natural (eelgrass) and artificial (plastic) substrates in New England. Aquat. Invasions 9, 105–110. doi: 10.3391/ai.2014.9.1.09
Cohen, A. N., Harris, L. H., Bingham, B. L., Carlton, J. T., Chapman, J. W., Lambert, C. C., et al. (2005). Rapid assessment survey for exotic organisms in southern California bays and harbors, and abundance in port and non-port areas. Biol. Invasions 7, 995–1002. doi: 10.1007/s10530-004-3121-1
Cordier, T., Alonso-Sáez, L., Apothéloz-Perret-Gentil, L., Aylagas, E., Bohan, D. A., Bouchez, A., et al. (2020). Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol. Ecol. 30, 2937–2958. doi: 10.20944/preprints202001.0278.v1
Cottier-Cook, E. J., Minchin, D., Gielser, R., Graham, J., Mogg, A. O. M., Sayer, M. D. J., et al. (2019). Biosecurity implications of the highly invasive carpet sea-squirt Didemnum vexillum Kott, 2002 for a protected area of global significance. Manag. Biol. Invasions 10, 311–323. doi: 10.3391/mbi.2019.10.2.07
Coutts, A. D. M., and Forrest, B. M. (2007). Development and application of tools for incursion response: lessons learned from the management of the fouling pest Didemnum vexillum. J. Exp. Mar. Biol. Ecol. 342, 154–162. doi: 10.1016/j.jembe.2006.10.042
Da Silva Oliveira, F. A., Michonneau, F., and da Cruz Lotufo, T. M. (2017). Molecular phylogeny of Didemnidae (Ascidiacea: Tunicata). Zool. J. Linn. Soc. 180, 603–612. doi: 10.1093/zoolinnean/zlw002
Dafforn, K. A., Glasby, T. M., and Johnston, E. L. (2012). Comparing the invasibility of experimental “reefs” with field observations of natural reefs and artificial structures. PLoS One 7:e38124. doi: 10.1371/journal.pone.0038124
Davison, P. I., Copp, G. H., Créach, V., Vilizzi, L., and Britton, J. R. (2017). Application of environmental DNA analysis to inform invasive fish eradication operations. Sci. Nat. 104:35. doi: 10.1007/s00114-017-1453-9
Dias, P. J., Rocha, R., Godwin, S., Tovar-Hernandez, M. A., Delahoz, M. V., McKirdy, S., et al. (2016). Investigating the cryptogenic status of the sea squirt Didemnum perlucidum (Tunicata, Ascidiacea) in Australia based on a molecular study of its global distribution. Aquat. Invasions 11, 239–245. doi: 10.3391/ai.2016.11.3.02
Dijkstra, J. A., and Nolan, R. (2011). Potential of the invasive colonial ascidian, Didemnum vexillum, to limit escape response of the sea scallop, Placopecten magellanicus. Aquat. Invasions 6, 451–456. doi: 10.3391/ai.2011.6.4.10
Erwin, P. M., Pineda, M. C., Webster, N., Turon, X., and Lopez-Legentil, S. (2014). Down under the tunic: bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. Microb. Ecol. 8, 575–588. doi: 10.1038/ismej.2013.188
EU-COM (2008). Directive 2008/56/EC of the European Parliament and of the Council Establishing a Framework for Community Action in the Field of Marine Environmental Policy (Marine Strategy Framework Directive). Official Journal of the European Union L164, 19e40. Brussels: EU-COM.
Ficetola, G. F., Pansu, J., Bonin, A., Coissac, E., Giguet-Covex, C., De Barba, M., et al. (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Res. 15, 543–546. doi: 10.1111/1755-0998.12338
Fitridge, I., Dempster, T., Guenther, J., and de Nys, R. (2012). The impact and control of biofouling in marine aquaculture: a review. Biofouling 28, 649–669. doi: 10.1080/08927014.2012.700478
Fletcher, L. M., Atalah, J., and Forrest, B. M. (2018). Effect of substrate deployment timing and reproductive strategy on patterns in invasiveness of the colonial ascidian Didemnum vexillum. Mar. Environ. Res. 141, 109–118. doi: 10.1016/j.marenvres.2018.08.006
Fletcher, L. M., Forrest, B. M., and Bell, J. J. (2013a). Impacts of the invasive ascidian Didemnum vexillum on green-lipped mussel Perna canaliculus aquaculture in New Zealand. Aquac. Environ. Interact. 4, 17–30. doi: 10.3354/aei00069
Fletcher, L. M., Forrest, B. M., Atalah, J., and Bell, J. J. (2013b). Reproductive seasonality of the invasive ascidian Didemnum vexillum in New Zealand and implications for shellfish aquaculture. Aquac. Environ. Interact. 3, 197–211. doi: 10.3354/aei00063
Forrest, B. M., and Atalah, J. (2017). Significant impact from blue mussel Mytilus galloprovincialis biofouling on aquaculture production of green-lipped mussels in New Zealand. Aquac. Environ. Interact. 9, 115–126. doi: 10.3354/aei00220
Forrest, B. M., and Hopkins, G. A. (2013). Population control to mitigate the spread of marine pests: insights from management of the Asian kelp Undaria pinnatifida and colonial ascidian Didemnum vexillum. Manag. Biol. Invasions 4, 317–326. doi: 10.3391/mbi.2013.4.4.06
Forrest, B. M., Fletcher, L. M., Atalah, J., Piola, R. F., and Hopkins, G. A. (2013). Predation limits spread of Didemnum vexillum into natural habitats from refuges on anthropogenic structures. PLoS One 8:e82229. doi: 10.1371/journal.pone.0082229
Furlan, E. M., Gleeson, D., Hardy, C. M., and Duncan, R. P. (2016). A framework for estimating the sensitivity of eDNA surveys. Mol. Ecol. Res. 16, 641–654. doi: 10.1111/1755-0998.12483
Furlan, E. M., Gleeson, D., Wisniewski, C., Yick, J., and Duncan, R. P. (2019). eDNA surveys to detect species at very low densities: a case study of European carp eradication in Tasmania, Australia. J. Appl. Ecol. 56, 2505–2517. doi: 10.1111/1365-2664.13485
GBNNSS (2011). Didemnum vexillum: GB Non-Native Species Organism Risk Assessment. Available online at: https://secure.fera.defra.gov.uk/nonnativespecies/index.cfm?sectionid=51 (accessed March 22, 2011).
Gillum, J. E., Jimenez, L., White, D. J., Goldstien, S. J., and Gemmell, N. J. (2014). Development and application of a quantitative real-time PCR assay for the globally invasive tunicate Styela clava. Manag. Biol. Invasions 5, 133–142. doi: 10.3391/mbi.2014.5.2.06
Glasby, T. M., Connell, S. D., Holoway, M. G., and Hewitt, C. L. (2007). Nonindigenous biota on artificial structures: could habitat creation facilitate biological invasions? Mar. Biol. 151, 887–895. doi: 10.1007/s00227-006-0552-5
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Graham, J., Collins, C., Lacaze, J. P., Brown, L., and McCollin, T. (2015). Molecular identification of Didemnum vexillum Kott, 1982 from sites around the UK coastline. BioInvasions Rec. 4, 171–177. doi: 10.3391/bir.2015.4.3.03
Harper, L. R., Griffiths, N. P., Lason-Handley, L., Sayer, C. D., Read, D. S., Harper, K. J., et al. (2018). Development and application of environmental DNA surveillance for the threatened crucian carp (Carassius carassius). Freshw. Biol. 64, 93–107. doi: 10.1111/fwb.13197
Hitchin, B. (2012). New outbreak of Didemnum vexillum in North Kent: on stranger shores. Porcupine Mar. Nat. Hist. Soc. Newslett. 31, 43–48.
Holman, L. E., de Bruyn, M., Creer, S., Carvalho, G., Robidart, J., and Rius, M. (2019). Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci. Rep. 9:11559. doi: 10.1038/s41598-019-47899-7
Janiak, D. S., Osman, R. W., and Whitlatch, R. B. (2013). The role of species richness and spatial resources in the invasion success of the colonial ascidian Didemnum vexillum Kott, 2002 in eastern Long Island Sound. J. Exp. Mar. Biol. Ecol. 443, 12–20. doi: 10.1016/j.jembe.2013.02.030
Kakkonen, J. E., Worsfold, T. M., Ashelby, C. W., Taylor, A., and Beaton, K. (2019). The value of regular monitoring and diverse sampling techniques to assess aquatic non-native species: a case study from Orkney. Manag. Biol. Invasions 10:46. doi: 10.3391/mbi.2019.10.1.04
Kaplan, K. A., Hart, D. R., Hopkins, K., Gallager, S., York, A., Taylor, R., et al. (2017). Evaluating the interaction of the invasive tunicate Didemnum vexillum with the Atlantic sea scallop Placopecten magellanicus on open and closed fishing grounds of Georges Bank. ICES J. Mar. Sci. 74, 2470–2479.
Klymus, K. E., Merkes, C. M., Allison, M. J., Goldberg, C. S., Helbing, C. C., Hunter, M. E., et al. (2020). Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2, 271–282. doi: 10.1002/edn3.29
Lambert, G. (2002). Nonindigenous ascidians in tropical waters. Pac. Sci. 56, 291–298. doi: 10.1353/psc.2002.0026
Lambert, G. (2009). Adventures of a sea squirt sleuth: unraveling the identity of Didemnum vexillum, a global ascidian invader. Aquat. Invasions 4, 5–28. doi: 10.3391/ai.2009.4.1.2
Larson, E. R., Renshaw, M. A., Gantz, C. A., Umek, J., Chandra, S., Lodge, D. M., et al. (2017). Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia 800, 173–185. doi: 10.1007/s10750-017-3210-7
LeBlanc, F., Belliveau, V., Watson, E., Coomber, C., Simard, N., DiBacco, C., et al. (2020). Environmental DNA (eDNA) detection of marine aquatic invasive species (AIS) in Eastern Canada using a targeted species-specific qPCR approach. Manag. Biol. Invasions 11, 201–217. doi: 10.3391/mbi.2020.11.2.03
Lengyel, N. L. (2013). Characterizing the Spread and Impacts of the Invasive Colonial Tunicate Didemnum Vexillum on Georges Bank Paper 24. Open Access Master’s Thesis. South Kingstown, RI: University of Rhode Island.
Lengyel, N. L., Collie, J. S., and Valentine, P. C. (2009). The invasive colonial ascidian Didemnum vexillum on Georges Bank–ecological effects and genetic identification. Aquat. Invasions 4, 143–152. doi: 10.3391/ai.2009.4.1.15
Lopez-Legentil, S., Legentil, M. L., Erwin, P. M., and Turon, X. (2015). Harbor networks as introduction gateways: contrasting distribution patterns of native and introduced ascidians. Biol. Invasions 17, 1623–1638. doi: 10.1007/s10530-014-0821-z
Mauvisseau, Q., Troth, C., Young, E., Burian, A., and Sweet, M. (2019). The development of an eDNA based detection method for the invasive shrimp Dikerogammarus haemobaphes. Manag. Biol. Invasions 10, 449–460. doi: 10.3391/mbi.2019.10.3.03
McKenzie, C. H., Reid, V., Lambert, G., Matheson, K., Minchin, D., Pederson, J., et al. (2017). Alien species alert: Didemnum vexillum Kott, 2002: invasion, impact, and control. ICES Coop. Res. Rep. 335:33.
Miralles, L., Dopico, E., Devlo-Delva, F., and Garcia-Vazquez, E. (2016). Controlling populations of invasive pygmy mussel (Xenostrobus securis) through citizen science and environmental DNA. Mar. Pollut. Bull. 110, 127–132. doi: 10.1016/j.marpolbul.2016.06.072
Miralles, L., Parrondo, M., Hernándes de Rojas, A., Garcia-Vazquez, E., and Borrell, Y. J. (2019). Development and validation of eDNA markers for the detection of Crepidula fornicata in environmental samples. Mar. Pollut. Bull. 146, 827–830. doi: 10.1016/j.marpolbul.2019.07.050
Mirimin, L., Hickey, A., Barrett, D., DeFaoite, F., Boschetti, S., Ventatesch, S., et al. (2019). Environmental DNA detection of Arctic char (Salvelinus alpinus) in Irish lakes: development and application of a species-specific molecular assay. Environ. DNA 2, 221–233. doi: 10.1002/edn3.60
Morris, J. A. Jr., and Carman, M. R. (2012). Fragment reattachment, reproductive status, and health indicators of the invasive colonial tunicate Didemnum vexillum with implications for dispersal. Biol. Invasions 14, 2133–2140. doi: 10.1007/s10530-012-0219-8
Muñoz-Colmenero, M., Ardura, A., Clusa, L., Miralles, L., Gower, F., Zaiko, A., et al. (2018). New specific molecular marker detects Ficopomatus enigmaticus from water eDNA before positive results of conventional sampling. J. Nat. Conserv. 43, 173–178. doi: 10.1016/j.jnc.2017.12.004
Nall, C., Guerin, A., and Cook, E. (2015). Rapid assessment of marine non-native species in northern Scotland and a synthesis of existing Scottish records. Aquat. Invasions 10, 107–121. doi: 10.3391/ai.2015.10.1.11
Ordóñez, V., Pascual, M., Fernández-Tejedor, M., Pineda, M. C., Tagliapietra, D., and Turon, X. (2015). Ongoing expansion of the worldwide invader Didemnum vexillum (Ascidiacea) in the Mediterranean Sea: high plasticity of its biological cycle promotes establishment in warm waters. Biol. Invasions 17, 2075–2085. doi: 10.1007/s10530-015-0861-z
Pederson, J., Bullock, R., Carlton, J., Dijkstra, J., Dobroski, N., Dyrynda, P., et al. (2005). Marine Invaders in the Northeast: Rapid Assessment Survey of Non-Native and Native Marine Species of Floating Dock Communities, August 2003. Cambridge, MA: MIT Sea Grant College Program.
Rees, H., Madisson, B. C., Middleditch, D. J., Patmore, J. R. M., and Gough, K. C. (2014). The detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 51, 1450–1459. doi: 10.1111/1365-2664.12306
Rey, A., Basurko, O. C., and Rodriguez-Ezpeleta, N. (2019). Considerations for metabarcoding-based port biological baseline surveys aimed at marine nonindigenous species monitoring and risk assessments. Ecol. Evol. 10, 2452–2465. doi: 10.1002/ece3.6071
Rogers, T. L., Byrnes, J. E., and Stachowicz, J. J. (2016). Native predators limit invasion of benthic invertebrate communities in Bodega Harbor, California, USA. Mar. Ecol. Prog. Ser. 545, 161–173. doi: 10.3354/meps11611
Roux, M.-L. D., Giblot-Ducray, D., Bott, N. J., Wiltshire, K. H., Deveney, M. R., Westfall, K. M., et al. (2020). Analytical validation and field testing of a specific qPCR assay for environmental DNA detection of invasive European green crab (Carcinus maenas). Environ. DNA 2, 309–320. doi: 10.1002/edn3.65
Sambrook, K., Holt, R. H. F., Sharp, R., Griffith, K., Roche, R. C., Newstead, R. G., et al. (2014). Capacity, capability and cross-border challenges associated with marine eradication programmes in Europe: the attempted eradication of an invasive non-native ascidian, Didemnum vexillum in Wales, United Kingdom. Mar. Policy 48, 51–58. doi: 10.1016/j.marpol.2014.03.018
Simkanin, C., Davidson, I. C., Dower, J. F., Jamieson, G., and Therriault, T. W. (2012). Anthropogenic structures and the infiltration of natural benthos by invasive ascidians. Mar. Ecol. 33, 499–511. doi: 10.1111/j.1439-0485.2012.00516.x
Simpson, T. S., Dias, P. J., Snow, M., Muñoz, J., and Berry, T. (2017). Real-time PCR detection of Didemnum perlucidum (Monniot, 1983) and Didemnum vexillum (Kott, 2002) in an applied routine marine biosecurity context. Mol. Biol. Res. 17, 443–453. doi: 10.1111/1755-0998.12581
Smith, K. F., Abbott, C. L., Saito, Y., and Fidler, A. E. (2015). Comparison of whole mitochondrial genome sequences from two clades of the invasive ascidian, Didemnum vexillum. Mar. Genomics 19, 75–83. doi: 10.1016/j.margen.2014.11.007
Smith, K. F., Stefaniak, L., Saito, Y., Gemmill, C. E. C., Cary, S. C., and Fidler, A. E. (2012). Increased inter-colony fusion rates are associated with reduced COI haplotype diversity in an invasive colonial ascidian Didemnum vexillum. PLoS One 7:e30473. doi: 10.1371/journal.pone.0030473
Snow, M., McKay, P., and Matejusova, I. (2009). Development of a widely applicable positive control strategy to support detection of infectious salmon anaemia virus (ISAV) using Taqman real-time PCR. J. Fish Dis. 32, 151–156. doi: 10.1111/j.1365-2761.2008.00972.x
Spens, J., Evans, A. R., Halfmaerten, D., Knudsen, S. W., Sengupta, M. E., Mak, S. S. T., et al. (2017). Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol. Evol. 8, 635–645. doi: 10.1111/2041-210X.12683
Stefaniak, L. M., and Whitlatch, R. B. (2014). Life history attributes of a global invader: factors contributing to the invasion potential of Didemnum vexillum. Aquat. Biol. 21, 221–229. doi: 10.3354/ab00591
Stefaniak, L. M., Lambert, G., Gittenberger, A., Zhang, H., Lin, S., and Whitlatch, R. B. (2009). Genetic conspecificity of the worldwide populations of Didemnum vexillum Kott, 2002. Aquat. Invasions 4, 29–44. doi: 10.3391/ai.2009.4.1.3
Stefaniak, L. M., Zhang, H., Gittenberger, A., Smith, K., Holsinger, K., Lin, S., et al. (2012). Determining the native region of the putatively invasive ascidian Didemnum vexillum Kott, 2002. J. Exp. Mar. Biol. Ecol. 422–423, 64–71. doi: 10.1016/j.jembe.2012.04.012
Takahara, T., Ikebuchi, T., Doi, H., and Minamoto, T. (2019). Using environmental DNA to estimate the seasonal distribution and habitat preferences of a Japanese basket clam in Lake Shinji, Japan. Estuar. Coast. Shelf Sci. 31, 15–20. doi: 10.1016/j.ecss.2019.02.022
Thalinger, B., Deiner, K., Harper, L. R., Rees, H. C., Blackmann, R. C., Sint, D., et al. (2021). A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environ. DNA 3, 823–836. doi: 10.1002/edn3.189
Thomsen, P. F., Møller, P. R., Sigsgaard, E. E., Knudsen, S. W., Jørgensen, O. A., and Willerslev, E. (2016). Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS One 11:e0165252. doi: 10.1371/journal.pone.0165252
Tidbury, H. J., Taylor, N. G. H., Copp, G. H., Garnacho, E., and Stebbing, P. (2016). Predicting and mapping the risk of introduction of marine non-indigenous species into Great Britain and Ireland. Biol. Invasions 18, 3277–3292. doi: 10.1007/s10530-016-1219-x
Turon, X., Casso, M., Pascual, M., and Viard, F. (2020). Looks can be deceiving: Didemnum pseudovexillum sp. nov. (Ascidiacea) in European harbours. Mar. Biodivers. 50:48. doi: 10.1007/s12526-020-01083-7
Valentine, P. C., Carman, M. R., Blackwood, D. S., and Heffron, E. J. (2007a). Ecological observations on the colonial ascidian Didemnum sp. in a New England tide pool habitat. J. Exp. Mar. Biol. Ecol. 342, 109–121. doi: 10.1016/j.jembe.2006.10.021
Valentine, P. C., Carman, M. R., Dijkstra, J., and Blackwood, D. S. (2009). Larval recruitment of the invasive colonial ascidian Didemnum vexillum, seasonal water temperatures in New England coastal and offshore waters, and implications for spread of the species. Aquat. Invasions 4, 153–168. doi: 10.3391/ai.2009.4.1.16
Valentine, P. C., Collie, J. S., Reid, R. N., Asch, R. G., Guida, V. G., and Blackwood, D. S. (2007b). The occurrence of the colonial ascidian Didemnum sp. on Georges Bank gravel habitat — ecological observations and potential effects on groundfish and scallop fisheries. J. Exp. Mar. Biol. Ecol. 342, 179–181. doi: 10.1016/j.jembe.2006.10.038
Vercaemer, B., Sephton, D., Clément, P., Harman, A., Stewart-Clark, S., and DiBacco, C. (2015). Distribution of the non-indigenous colonial ascidian Didemnum vexillum (Kott, 2002) in the Bay of Fundy and on offshore banks, eastern Canada. Manag. Biol. Invasions 6, 385–394. doi: 10.3391/mbi.2015.6.4.07
Viard, F., Roby, C., Turon, X., Bouchemousse, S., and Bishop, J. (2019). Cryptic diversity and database errors challenge non-indigenous species surveys: an illustration with Botrylloides spp. in the English Channel and Mediterranean Sea. Front. Mar. Sci. 6:615. doi: 10.3389/fmars.2019.00615
Villalobos, S. M., Lambert, G., Shenkar, N., and López-Legentil, S. (2017). Distribution and population dynamics of key ascidians in North Carolina harbors and marinas. Aquat. Invasions 12, 447–458. doi: 10.3391/ai.2017.12.4.03
Wilcox, T. M., McKelvey, K. S., Young, M. K., Jane, S. F., Lowe, W. H., Whiteley, R. J., et al. (2013). Robust detection of rare species using environmental DNA: the importance of primer specificity. PLoS One 8:e59520. doi: 10.1371/journal.pone.0059520
Wood, S. A., Pochon, X., Ming, W., von Ammon, U., Woods, C., Carter, M., et al. (2019). Considerations for incorporating real-time PCR assays into routine marine biosecurity surveillance programmes: a case study targeting the Mediterranean fanworm (Sabella spallanzanii) and club tunicate (Styela clava). Genome 62, 137–146. doi: 10.1139/gen-2018-0021
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13:134. doi: 10.1186/1471-2105-13-134
Keywords: eDNA, invasive species, qPCR, aquaculture, monitoring, marina, Didemnum vexillum
Citation: Matejusova I, Graham J, Bland F, Lacaze J-P, Herman G, Brown L, Dalgarno E, Bishop JD, Kakkonen JE, Smith KF and Douglas A (2021) Environmental DNA Based Surveillance for the Highly Invasive Carpet Sea Squirt Didemnum vexillum: A Targeted Single-Species Approach. Front. Mar. Sci. 8:728456. doi: 10.3389/fmars.2021.728456
Received: 23 June 2021; Accepted: 27 July 2021;
Published: 09 September 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, United StatesXinqing Zheng, Third Institute of Oceanography, State Oceanic Administration, China
Copyright © 2021 Matejusova, Graham, Bland, Lacaze, Herman, Brown, Dalgarno, Bishop, Kakkonen, Smith and Douglas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iveta Matejusova, SXZldGEuTWF0ZWp1c292YUBnb3Yuc2NvdA==; Alex Douglas, YS5kb3VnbGFzQGFiZG4uYWMudWs=
 Iveta Matejusova
Iveta Matejusova Jennifer Graham1
Jennifer Graham1 Fiona Bland
Fiona Bland John D. Bishop
John D. Bishop Jenni E. Kakkonen
Jenni E. Kakkonen Kirsty F. Smith
Kirsty F. Smith Alex Douglas
Alex Douglas