- 1Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 2STU-UMT Joint Shellfish Research Laboratory, Shantou University, Shantou, China
- 3Institute of Tropical Aquaculture and Fisheries, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
Identification and exploring the role of novel sex-related genes is a crucial step for understanding the regulation mechanism of sexual development. In the present study, we identified a novel sex-related gene (designated as Sp-Pol) upstream of a sex-specific single nucleotide polymorphism (SNP). SNP1888 together with Sp-Pol were mapped on LG32 (which is a sex-related linkage group) of a high-density genetic map. The full-length cDNA of Sp-Pol consists of 1703 bp with an open reading frame (ORF) encoding 204 amino acids, a 344 bp 5′-UTR and a 744 bp 3′-UTR. Phylogenetic analysis showed that Sp-Pol may need to be classified as a new gene family due to the very low sequence identity with other known genes (less than 25% identity). The sex-biased expression pattern of Sp-Pol starts from crablet stage V (C5) with approximately three- to four-fold higher in males than in females. Sp-Pol was expressed at a higher level in gonads compared to other tissues, with the highest expression level in the testis. In testis, a downward trend was observed in the expression level of Sp-Pol from the testis stage I (T1) to testis stage III (T3). After unilateral eyestalk ablation, the expression level of Sp-Pol significantly increased in testis and hepatopancreas in males, while it was downregulated in the hepatopancreas of females. Fluorescence in situ hybridization (FISH) assay revealed that Sp-Pol transcripts were strongly localized in the epithelia of seminiferous tubules of the testis, and in the ovary, it was detected in the oogonium cells. These findings showed that Sp-Pol may play crucial roles in the gonad development of S. paramamosain.
Introduction
Understanding the sexual differentiation mechanism is a key point for sexual manipulation and improving the reproductive efficiency of crustaceans (Niksirat et al., 2014, 2016; Cui et al., 2021a). Female and male crustaceans display different growth rate (Shi X. et al., 2019). For instance, male crayfish and prawns grow larger and faster than females, and all-male individual rearing is desired. In contrast, female crab and shrimp have a better growth performance than male individuals (Ventura et al., 2012). Female crustaceans are considered more valuable in terms of egg production because they can mate with more males with no negative impact on the percentage of berried females (Niksirat et al., 2015a; Harlıoğlu and Farhadi, 2017). In addition, males and females have different nutritional values and flesh quality (Niksirat et al., 2015b; Wu et al., 2019).
Crustacean female sex hormone (CFSH) and insulin-like androgenic gland hormone (IAG) are the two major hormones that regulate the sexual differentiation of female and male crustaceans (Ventura and Sagi, 2012; Zmora and Chung, 2014). Androgenic gland (AG) ablation and IAG silencing resulted in full sex reversal from male to female (feminization) in Macrobrachium rosenbergii (Rosen et al., 2010; Ventura and Sagi, 2012). However, the mechanisms underlying the regulation of sexual differentiation and most of the key factors upstream and downstream of the IAG in the “eyestalk-AG-testis” endocrine axis are still poorly understood (Guo et al., 2019). So far, the only functional sex reversal has been reported in M. rosenbergii by AG manipulation and IAG silencing (Aflalo et al., 2006; Ventura and Sagi, 2012). The IAG in M. rosenbergii is exclusively expressed only in AG rather than in the other tissues (Ventura et al., 2012; Ma et al., 2013). While in other crustaceans including shrimps, crayfish, and crabs (Mareddy et al., 2011; Li et al., 2012; Zhang et al., 2014; Shi L. et al., 2019; Jiang et al., 2020c) the IAG is specifically expressed in the AG, it also expressed in other tissues. Therefore, IAG may is not the only key regulator of sexual differentiation (Farhadi et al., 2021a).
Recently, several sex-related genes have been identified in crustaceans (Ma et al., 2012; Jin et al., 2013), suggesting that sexual differentiation in crustaceans is more complicated than previously expected. These sex-related genes include different types, namely Sxl, Fru, Fem-1, Tra, Tra-2, Dsx, Mab-3, Sry, Sox9, Foxl2, Wnt4, and Dmrt1 (Chandler et al., 2018; Yang et al., 2018; Zhang et al., 2018; Farhadi et al., 2021b). For example, it has been documented that CFSH, Sox, Dsx, Slx, Dmrt, GEM, and Fem-1 regulate the expression of IAG. IAG expression is negatively regulated by GEM, Fem-1, and CFSH (Jin et al., 2019; Shi L. et al., 2019; Zheng et al., 2019; Jiang et al., 2020b). In contrast, Slx, Dsx, and Dmrt silencing significantly decrease the expression of IAG indicating the existence of positive regulation (Abayed et al., 2019; Tan et al., 2020). Although most of these sex-related genes have been detected in both sexes, different expression patterns have been observed between the two sexes. IAG, Sox, Dmrt, Dsx, Vasa, and Slx exhibit male-biased expression patterns and play crucial roles in testis development and spermatogenesis. CFSH, Fem-1, Wnt4, FAMeT, Slo, Tra-2, BMPs, UCHLs, Erk2, Cdc2, EGFR, Vg, VgR, and VIH have higher expression levels in females and play important roles during ovarian development (Xu et al., 2018; Abayed et al., 2019; Jin et al., 2019; Shi L. et al., 2019; Zheng et al., 2019; Jiang et al., 2020a; Zhou et al., 2020; Farhadi et al., 2021b). Therefore, identification of sex-related genes and characterization of their functions are crucial steps for understanding the sex determination and sexual differentiation mechanism in crustaceans.
Mud crab (Scylla paramamosain) is an important economic crustacean species and mainly distributed in the south-eastern coastal area of China (Wang et al., 2005; Wu et al., 2019). There is a great interest in the culture of S. paramamosain due to their high price, large size, and fast growth in captivity (Shi X. et al., 2019; Cui et al., 2021b). The annual yield of S. paramamosain reached 236,769 tons in 2019, including 160,616 tons for aquaculture production and 79,153 tons for capture (Bureau of Fisheries Management, 2020).
In the present study, we verified a female-specific locus named SNP1888 and a novel sex-related gene (Sp-Pol). Then we mapped both SNP1888 and the gene on LG32 of a previously constructed high-density genetic map. The expression pattern of Sp-Pol in different tissues and different sexual development stages was analyzed. Unilateral eyestalk ablation was carried out to find out the regulation mechanism of Sp-Pol. Fluorescence in situ hybridization (FISH) localized the Sp-Pol mRNA in the testis and ovary. The present study provides novel insights into the gonad development of S. paramamosain.
Materials and Methods
Animals and Sampling
Samples including embryos, zoeae, megalopa, and crablets were obtained from a crab farm (named Raoping West Coast Biotechnology Co., Ltd.) and aquatic product market in Chaozhou City, Guangdong Province of China. After being transported to the laboratory, they were kept for adaption at room temperature. Then tissues including testis, ovary, heart, hepatopancreas, muscle, gill, thoracic ganglion, intestine, and stomach were quickly dissected and stored in RNA keeper solution (Vazyme Biotech Co., Ltd., Nanjing, China) at −80°C for RNA and DNA extraction. The total RNA was extracted using Trizol Reagent (Invitrogen, CA, United States) following the manufacturer’s protocol. First-strand cDNA was synthesized using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan). Genomic DNA was extracted using TIANamp Marine Animals DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) and treated with RNase A to remove residual RNA.
Verification of Sex-Specific SNP1888 and Discovery of Sp-Pol Gene
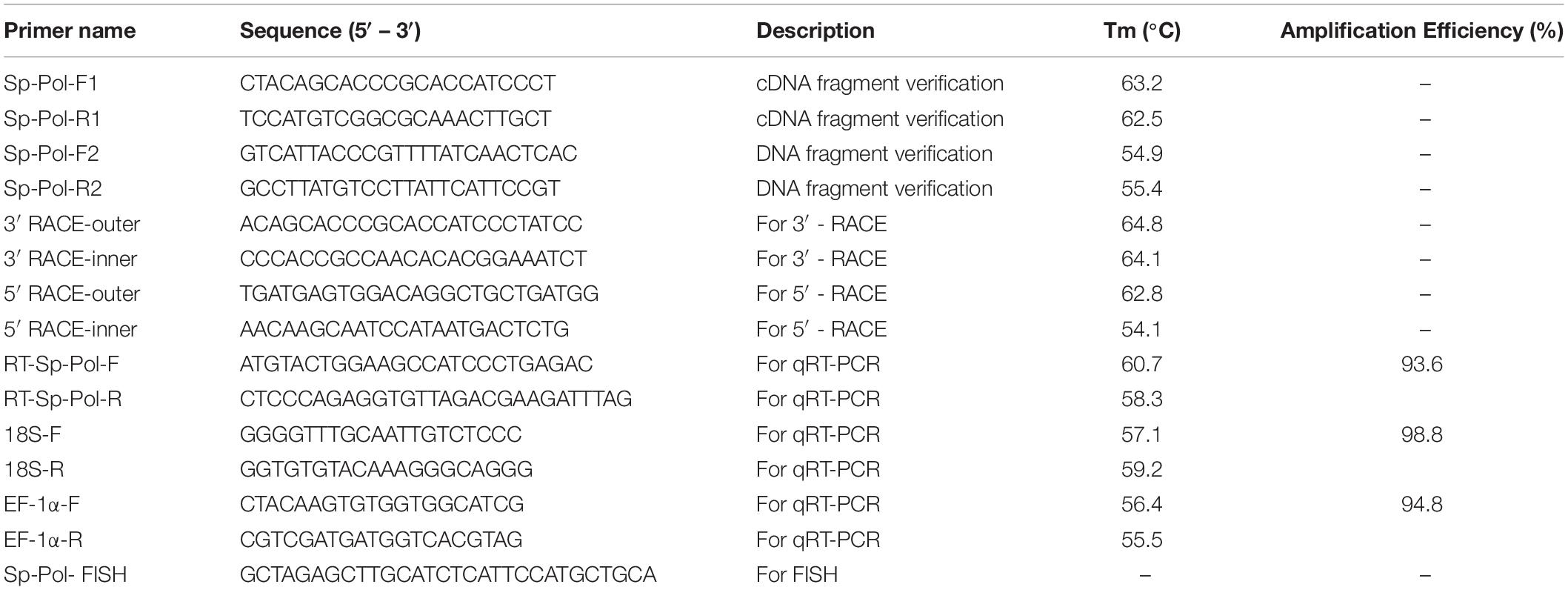
A potential female-specific SNP locus named SNP1888 was identified from Cluster_39896 by RAD-seq in our previous study (Shi et al., 2018). In order to verify SNP1888 is a real sex-specific locus, we first designed a pair of primers for PCR amplification, and then the PCR products of a total of 195 crabs (106 females and 89 males) were sequenced by bidirectional sequencing. The genotypes of SNP1888 were defined by the peak types according to the sequencing chromatograms: double peaks indicate heterozygous and single peak indicates homozygous. Based on gene clone technology and bioinformatics analysis, we identified a novel gene (designated as Sp-Pol) at the upstream of SNP1888 (SNP1888 is located at the 3′-UTR of Sp-Pol).
Cloning and Sequence Analysis of Sp-Pol
The Sp-Pol cDNA sequence was achieved by the rapid amplification of cDNA ends (RACE) technique using SMARTer RACE cDNA Amplification Kit (Clontech, United States). Nested PCR amplifications were carried out using a universal primer mix (UPM outer primer) and a nested universal primer (NUP inner primer), in conjunction with two gene-specific primers (GSPs) designed based on the cDNA sequence. PCR was performed on an Eppendorf Mastercycler® ep realplex (Eppendorf, Germany) with specific primers (Table 1) using the following conditions: denaturation at 94°C for 5 min; 35 cycles of amplification at 94°C for 30 s, 58°C for 30 s, 72°C for 30 s and final elongation at 72°C for 5 min. The PCR product was ligated into Simple T1 vector (Transgen, Beijing). The recombination was then transformed into Escherichia coli strain K12, and positive clones were sequenced. Then, positive clones were sequenced using an automatic DNA sequencer in Sangon Biotech Co., Ltd. Later, the gene sequence was blasted with the NCBI database to find its homologs.
Localization of SNP1888 and Sp-Pol
The sex-specific SNP1888 and Sp-Pol were localized in the high-density genetic linkage map of S. paramamosain. The obtained genotypes of SNP1888 in the mapping family with a total of 129 individuals by PCR amplification and Sanger sequencing (Shi et al., 2018) were integrated with all SNP genotypes obtained in our previous study (Waiho et al., 2019) to reconstruct the genetic map by Joinmap 5.0 software.
Bioinformatic Analysis
The sequences of Sp-Pol were analyzed based on the sequences in GenBank using BLASTN and BLASTX (available at1). The ORF was identified using Editseq program (Lasergene, United States). Compute pI/Mw tool2 was used to predict the molecular weight and isoelectric point of the Sp-Pol protein. NetPhos 3.1 Server3, NetOGlyc 4.0 Server4, and NetNGlyc 1.0 Server4 were used to predict the potential phosphorylation, O-glycosylation, and N-glycosylation sites, respectively. The three-dimensional (3D) protein structure and secondary structure of a protein were predicted using SWISS-MODE5 and self-optimized prediction method with alignment (SOPMA), respectively. The sequences were aligned using Bioedit software version 7.26. The phylogenetic diagram was generated by the neighbor-joining method (bootstrap analysis of 1,000 replicates) using Mega X software (Kumar et al., 2018).
Quantitative Real-Time PCR (qRT-PCR) and Expression Models Analysis
The qRT-PCR was used to investigate the Sp-Pol mRNA expression levels at different developmental stages and different tissues. The testis and ovary developmental stages were identified based on the morphology of gonads (Waiho et al., 2017; Wu et al., 2020). A LightCycler® 480 Instrument II (Roche, Switzerland) was used for the qRT-PCR analysis. The qRT-PCR using SYBR Green method (TIANGEN Biotech, Beijing, China) was performed under the following conditions: denaturation at 95°C for 3 min; 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 15 s. Specific primers for Sp-Pol and housekeeping genes (EF1α and 18S rRNA) are listed in Table 1. Gene expression data were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). In addition, the expression models analysis in mRNA of ovaries and testis were determined by Sanger sequencing through the peaks and their colors in the chromatogram.
Unilateral Eyestalk Ablation
Males (221.2 ± 23.3 g; N = 20) and females (211.4 ± 19.8 g; N = 30) S. paramamosain were randomly divided into two groups. Before the ablation of the eyestalk, animals were adapted for 7 days. Hot tweezers were used to remove the eyestalks. No eyestalk was removed in the crabs from the control group. Seven days later, gill, muscle, hepatopancreas, and gonads were dissected to detect the mRNA level of Sp-Pol.
Fluorescence in situ Hybridization (FISH)
After dissection, gonads samples (mean weight of male crabs: 107.4 ± 23.1 g; mean weight of female crabs: 124.8 ± 21.9 g) were used for the preparation of frozen sections. The OCT-embedding tissue was cut into 3 μm sections by a frozen section machine (Thermo). The fluorescein-labeled DNA probe (5′-FAM-GCT AGAGCTTGCATCTCATTCCATGCTGCA-3′) was synthesized based on the cDNA sequence of Sp-Pol by the Sangon Biotech Company (Shanghai, China) (Table 1). Before hybridization, slides were digested using proteinase K for 5 min. Then, sections were immersed in 75, 85, and 100% ethanol for 1 min, respectively. Hybridization was carried out at 65°C overnight with FAM-labeled Sp-Pol probe (10 μl for each slide). The sections were subjected to nuclear staining by DAPI and observed using Axio Observer 5 fluorescence microscopy (ZEISS, Germany).
Statistical Analysis
The experimental data were analyzed using SPSS version 21.0 (Chicago, IL, United States). The normality and homoscedasticity of the data were investigated using Kolmogorov-Smirnov and Levene’s test. The proportional data were normalized using square root arcsin transformation. To compare the expression level of Sp-Pol in different tissues and different developmental stages a one-way ANOVA was utilized. Significant differences between groups were determined using the Tukey’s HSD post hoc test in each case. The mRNA level of Sp-Pol between the eyestalk ablated and the control group was analyzed using an independent samples t-test. Significances were accepted at P < 0.05.
Results
Verification of Sex-Specific SNP1888
The position of SNP1888 in Cluster_39896 is shown in Figure 1A. With a pair of primers (F: CTACGGCA CCCGCACCATCCTT, R: AGCAAGTTTGCGCCGACATGGA), a specific product was obtained by PCR. According to the sequencing chromatograms, SNP1888 showed heterozygous (T/C) in all 106 females and homozygous (C/C) in all 89 males (Figures 1B,C).
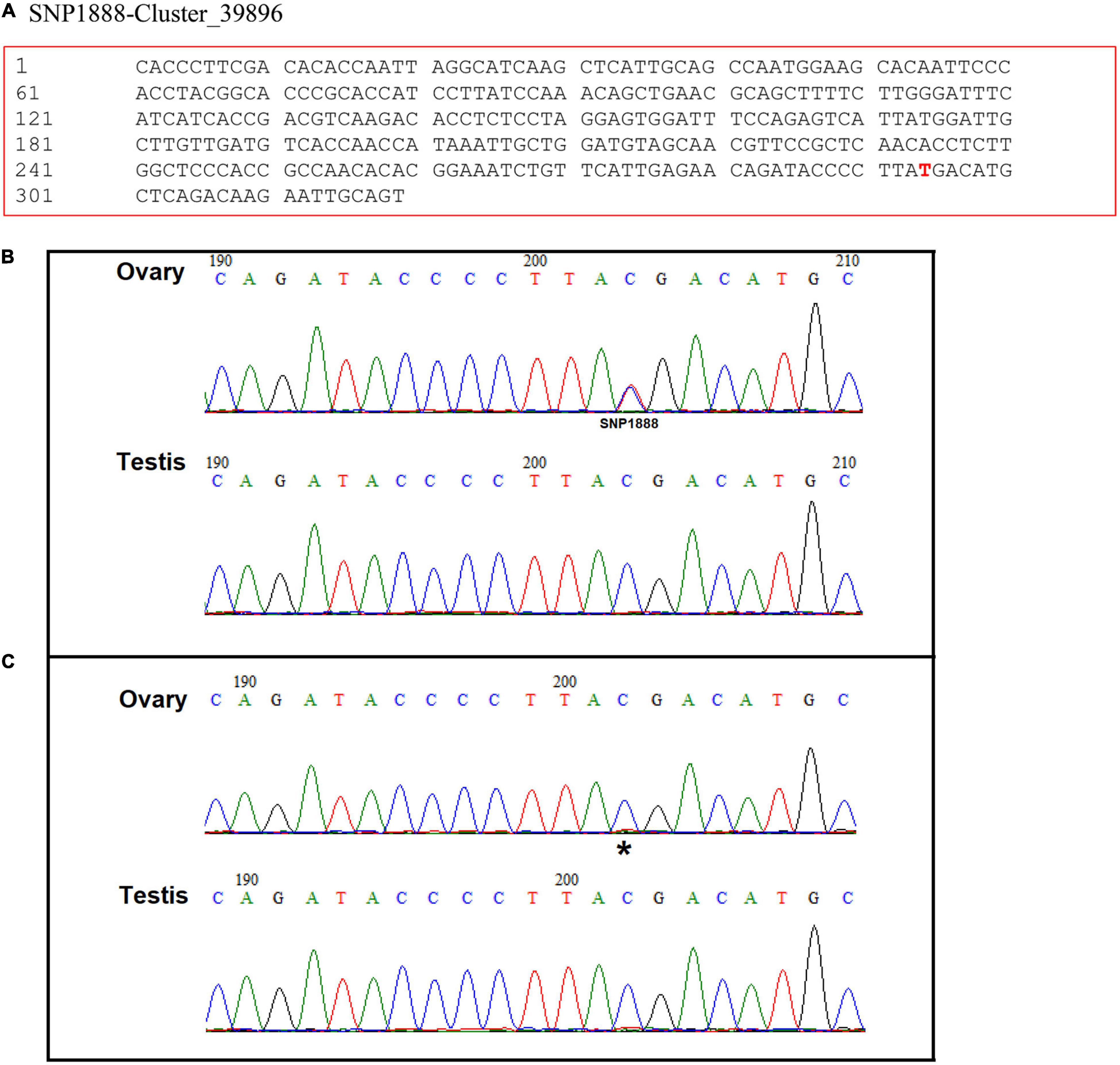
Figure 1. (A) The position of Seq-1888 and its sex-specific SNP marker on the sequence (SNP 1888:T/C). Sequencing chromatograms of the ovary and testis DNA (B) and cDNA (C) in Scylla paramamosain. Double peaks indicate heterozygous and single peak indicates homozygous. Sequencing data (i.e., sequencing data for SNP discovery) are available at NCBI Sequence Read Archive (accession number SRP135178). * shows the position of SNP 1888.
Characterization of Sp-Pol
The full-length cDNA of Sp-Pol was 1,703 bp and contains a 5′-UTR of 344 bp, a 3′-UTR of 744 bp, and an ORF of 615 bp encoding a total of 204 amino acid residues (aa) (Figure 2A). The deduced protein has a molecular mass of 22.74 kDa, with an estimated isoelectric point (PI) being 8.50. Analysis by SignalP 5.0 software revealed that the deduced peptide does not contain a putative signal peptide. Twenty-eight potential phosphorylation sites were identified at 14 serine residues (aa 6, 14, 25, 26, 29, 30, 31, 33, 40, 79, 100, 101, 131, and 173) and 14 threonine residues (aa 19, 20, 22, 27, 28, 54, 59, 73, 80, 81, 127, 136, 164, and 195). A potential N-glycosylation site (162NQTC165) and six potential O-glycosylation sites (Ser6, Ser14, Thr19, Ser26, Thr-27, Thr148) were also identified. The SOPMA program showed that the Sp-Pol protein contained 33.82% α helix, 3.43% β turn, 55.39% random coil, and 7.35% extended chain (Figure 2B).
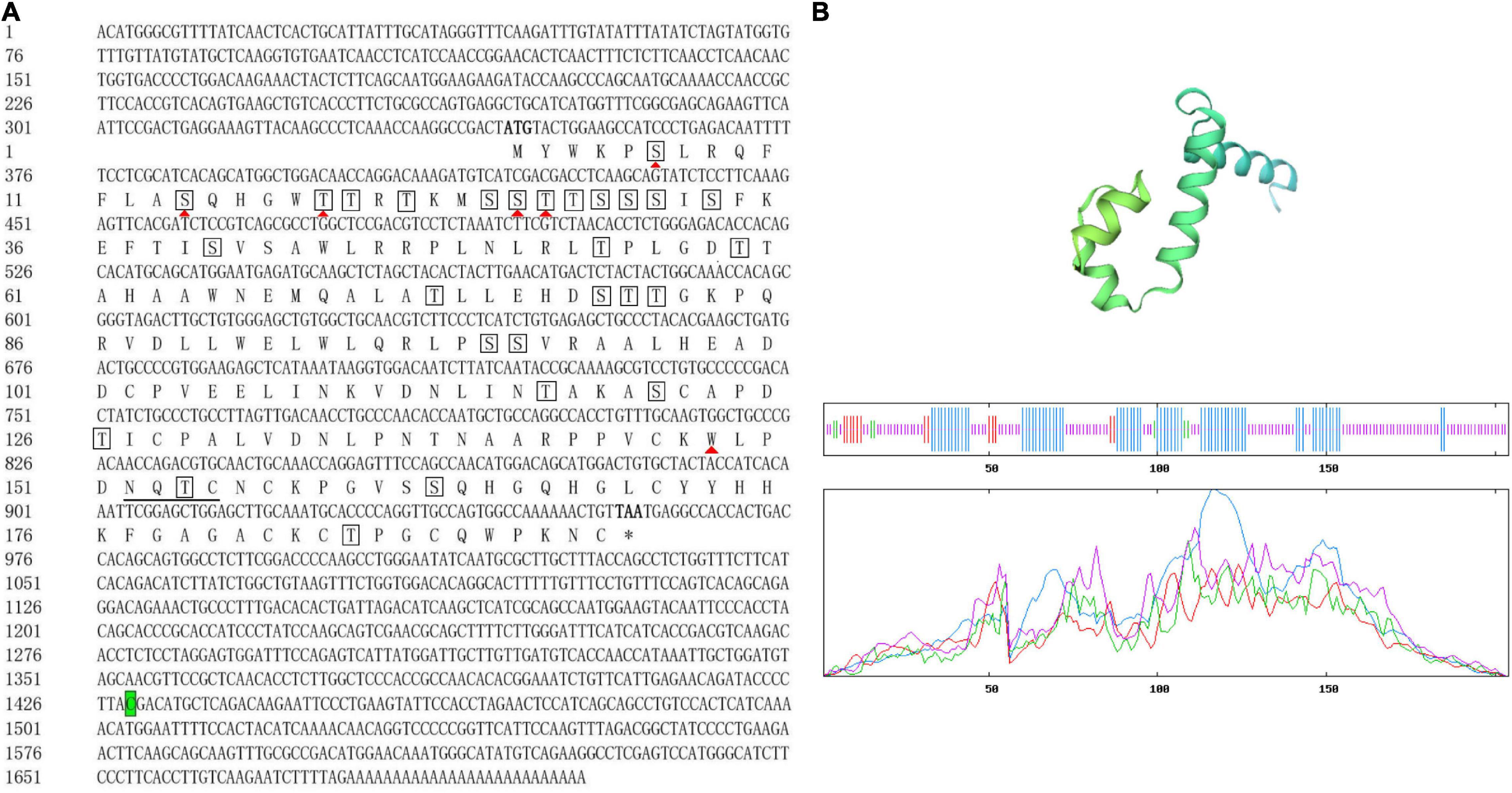
Figure 2. Sequences of full-length complementary DNA and deduced amino acids (aa) of Sp-Pol. The number of nucleotide and amino acid are listed on the left side. Start (ATG) and stop codon (TAA) of the open reading frame are both indicated in bold. Termination of aa is denoted by asterisks. Predicted O-glycosylation sites are indicated by red solid triangles (▲). The potential N-glycosylation site (162NQTC165) is underlined. Potential phosphorylation sites are in boxes. Letter with green background indicates the location of sex-specific SNP (SNP1888: C/T) (A). The three-dimensional (3D) protein structure of Sp-Pol and analysis of the secondary structure. The a-helix (blue), extended strand (red), random coil (purple), and b-turn (green) (B).
Mapping of Sex-Specific SNP1888 and Sp-Pol
The mapping results showed that the sex-specific SNP1888 with the Sp-Pol gene were mapped on LG32 of the sex-averaged high-density genetic linkage map (Figure 3). Hence, LG32 is suggested as a sex-related linkage group due to it already has 13 sex-specific SNPs (SNP1888, SNP1, SNP2, SNP3, SNP4, SNP5, SNP6, SNP7, SNP8, SNP9, SNP10, SNP11, SNP12) and exclusive sex QTLs of S. paramamosain (Waiho et al., 2019).
Multiple Alignments and Phylogenetic Analysis of Sp-Pol Protein
Bioinformatics analysis of the deduced amino acid sequence of Sp-Pol protein indicated its structural similarity to the Gag-Pol polyprotein family of peptides. Sp-Pol protein shared the highest identity with the Retrovirus-related Pol polyprotein from transposon opus (Penaeus vannamei) (ROT64510.1, 60.47% identity, bit score: 209, expectation value: 3.73e–61) (Figure 4). A phylogenetic tree was constructed by the neighbor-joining method using multiple alignments of 19 homologous proteins from Decapoda, Insecta, Echinodermata, and fish species (Figure 5). In this tree, the homologous proteins are divided into four separate clades: Decapoda, Insecta, Echinodermata, and fishes.
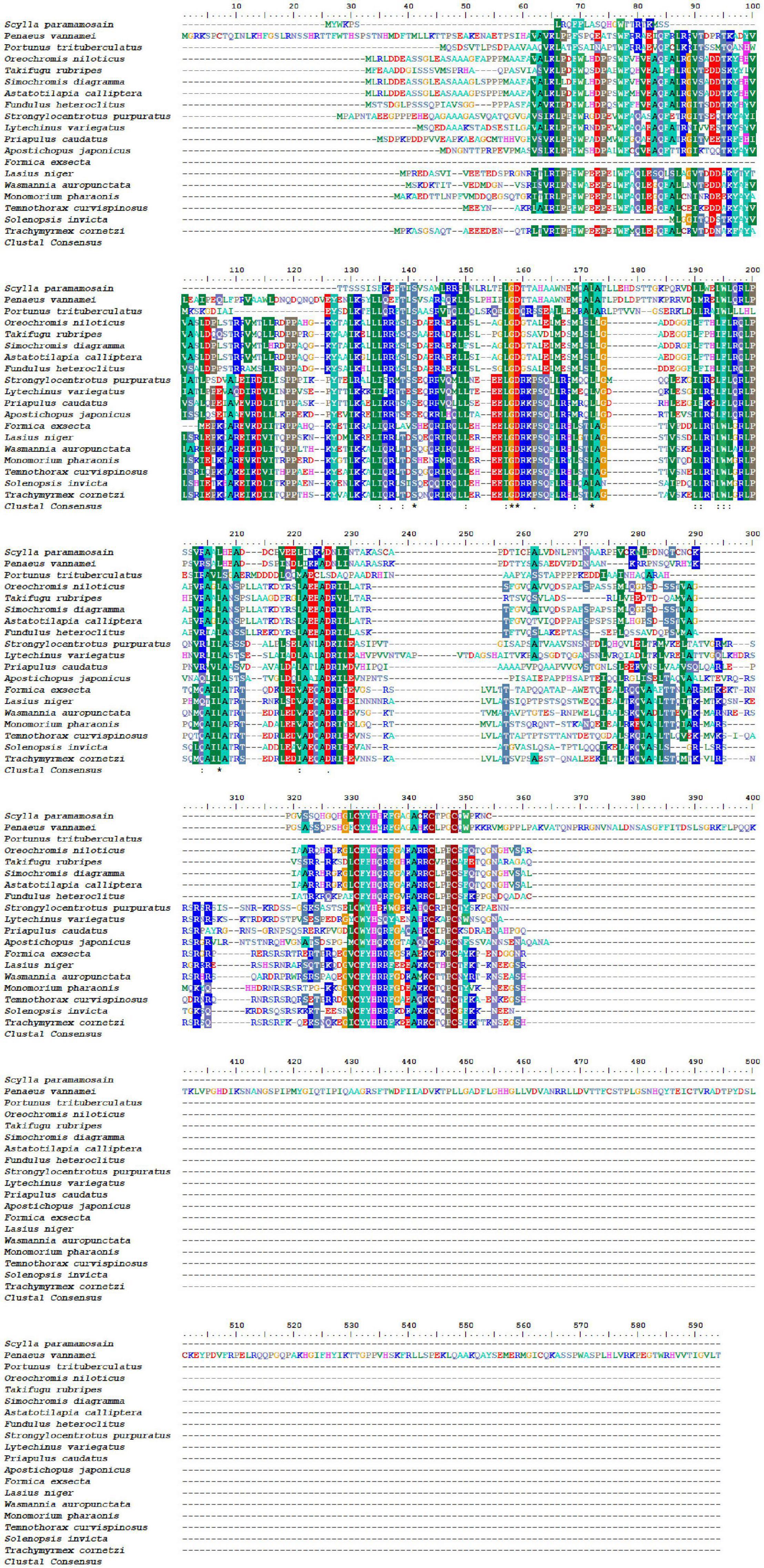
Figure 4. Multiple alignments of deduced amino acids of Sp-Pol protein and its homologs. GenBank accession numbers are as follows: Penaeus vannamei (ROT64510.1); Portunustri tuberculatus (MPC10902.1).
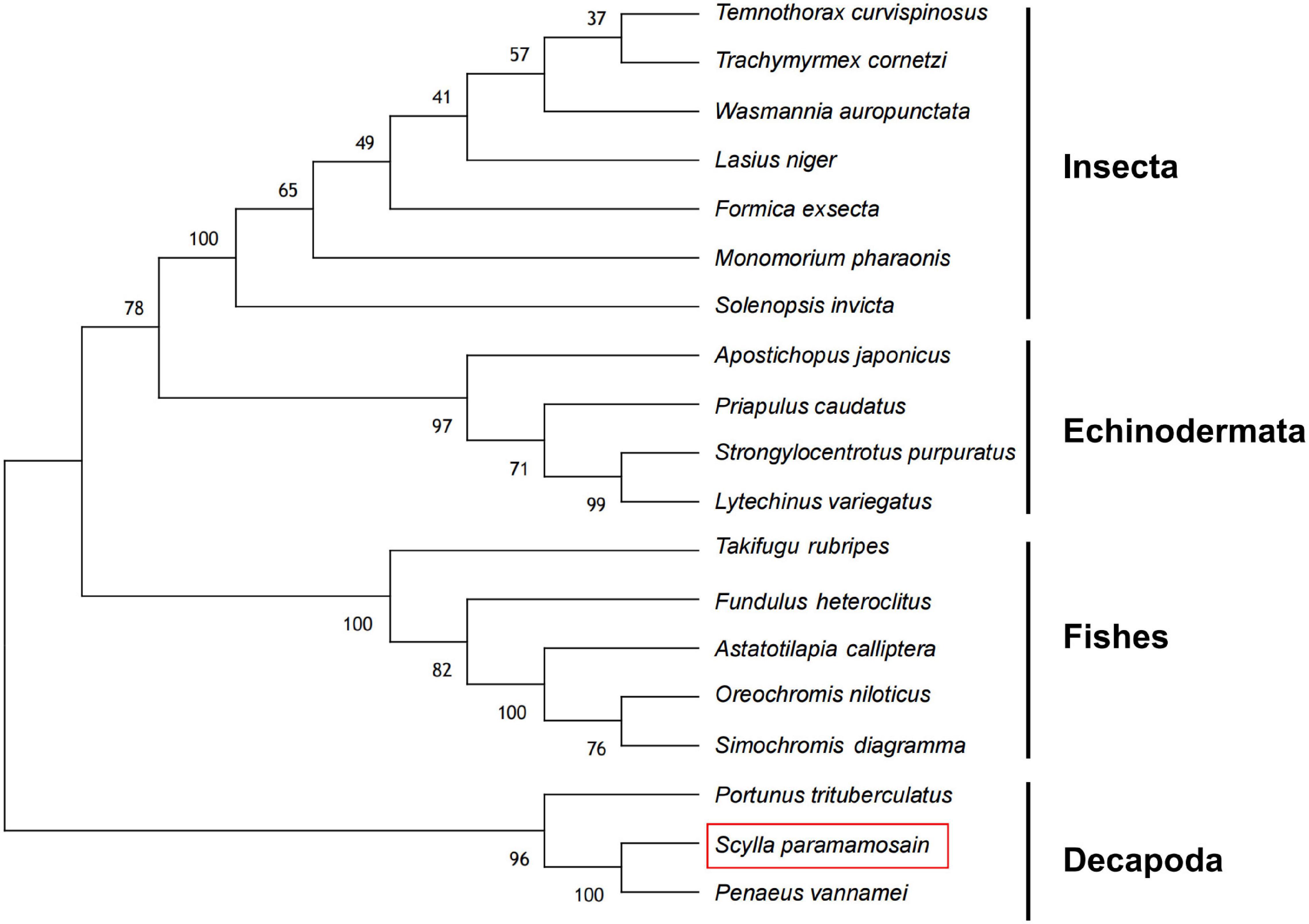
Figure 5. Phylogenetic analysis of Sp-Pol protein (red rectangle) from S. paramamosain and homologous proteins from other species. The amino acid sequence and GenBank accession number of the proteins are provided in the Supplementary Material.
Expression of Sp-Pol in Different Tissues and Different Developmental Stages of Gonads
Sp-Pol was expressed at a higher level in gonads compared to other tissues (P < 0.05). The highest expression of Sp-Pol was detected in testis with approximately four-fold higher than in the ovary (P < 0.05) (Figure 6A). Meanwhile, we found that Sp-Pol had mono-allelic expression in the ovary, whereas in the testis both alleles were expressed. In the ovary, the mono-allele which owing nucleotide acid C at the SNP1888 locus was expressed, and the other allele which owing T was not expressed (Figures 1B,C). The expression level of Sp-Pol in different development stages of the ovary and testis is presented in Figure 6B. The highest expression level of Sp-Pol was observed at the first stage of testis development (T-1), and then the expression level decreased significantly from T-1 to T-2, and T-3 (P < 0.05). In the ovary, the expression of Sp-Pol was low during all five stages of ovary development (O-1 to O-5). In addition, the expression profile of Sp-Pol during embryo (E), zoeae (Z-1 to Z-5), megalopa (M), and crablets (C-1 to C-6) was assessed (Figures 6C,D). An upward trend was observed for males at the crablet stage V (C-5) and the crablet stage VI (C-6) (P < 0.05) (Figure 6C). Among zoea stages, Sp-Pol had the highest expression at the zoea I stage (Figure 6D).
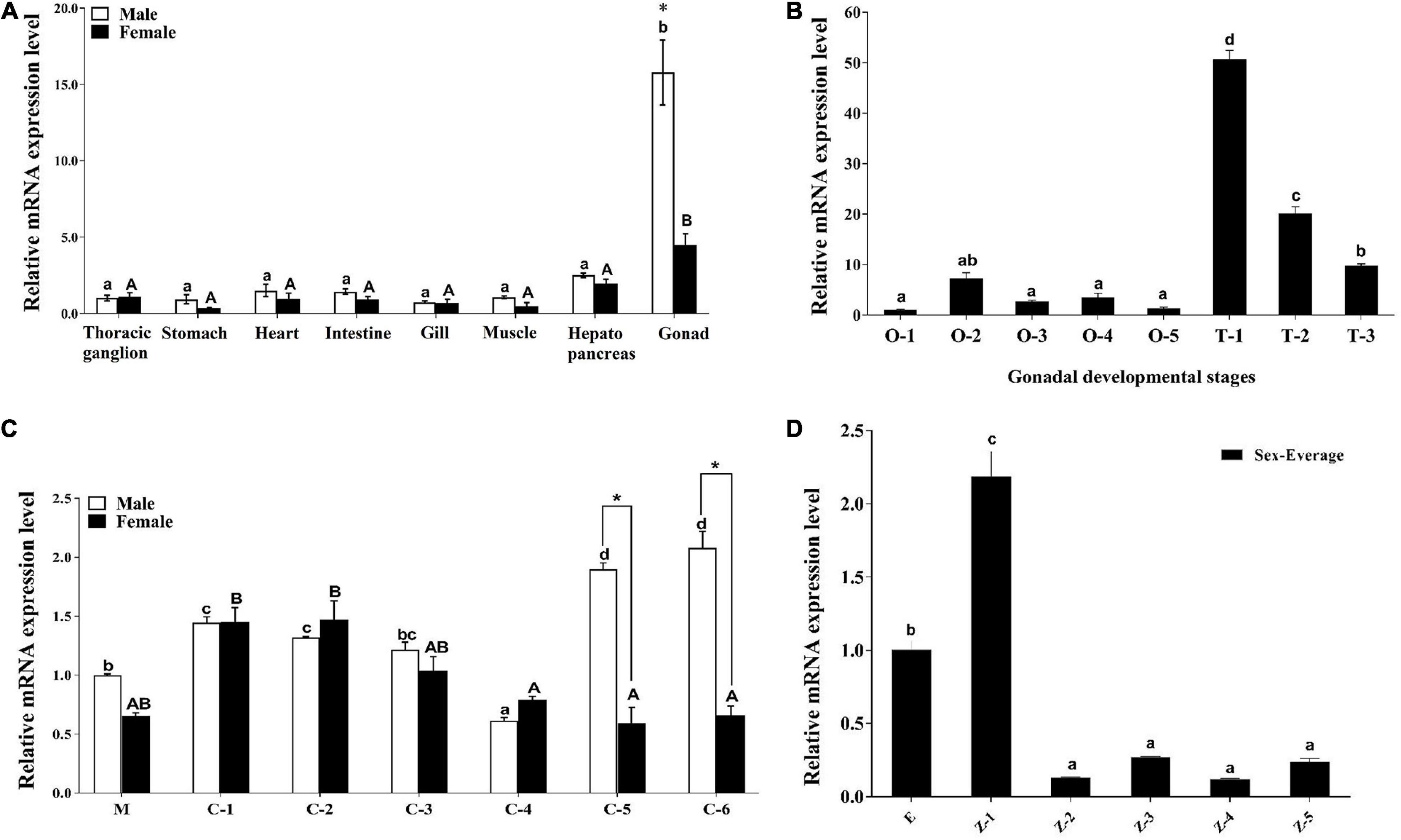
Figure 6. The expression profiles of Sp-Pol in different tissues and different sexual developmental stages. (A) The expression of Sp-Pol mRNA in different tissues. (B) The expression of Sp-Pol mRNA at different gonadal development stages (O, ovary; T, testis). (C) The expression of Sp-Pol mRNA during different juvenile stages (M, megalopa larva stage; C, crablet stage). (D) The expression of Sp-Pol mRNA during larvae development (E, eggs hatch; Z, zoea stage). Significant differences are indicated with different letters and * mark (P < 0.05). Values are mean (±SE).
The Effects of Unilateral Eyestalk Ablation on the mRNA Level of Sp-Pol
The eyestalk ablation significantly increased the expression level of Sp-Pol in the testis (4.7-fold) and hepatopancreas (2.1-fold) of males (P < 0.05) (Figure 7A). While in females, a downward trend was recorded in the Sp-Pol expression in hepatopancreas (2.2-fold) (P < 0.05) (Figure 7B). However, unilateral eyestalk ablation slightly increased the expression level of Sp-Pol in the ovary (1.5-fold), however, it was not statically significant (P = 0.23). Based on these results, Sp-Pol might be negatively regulated by eyestalk in testis and male hepatopancreas. Down-regulation of Sp-Pol in the female hepatopancreas showed the possible existence of positive regulation.
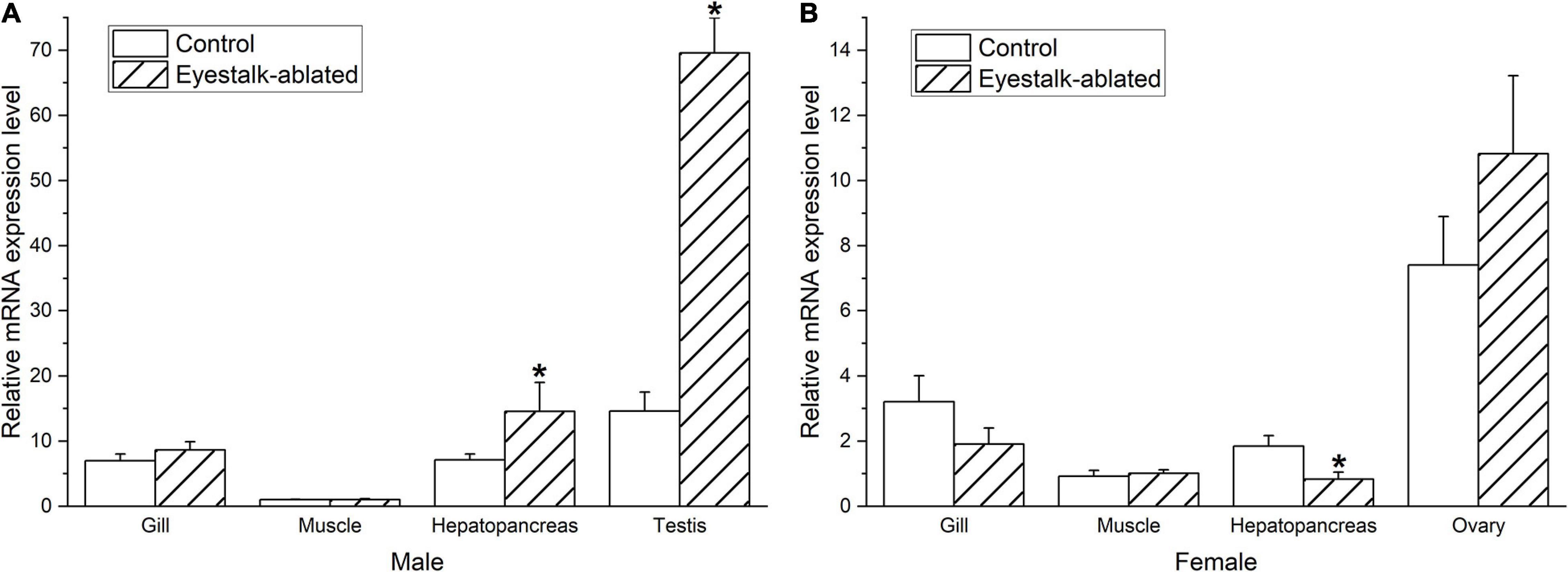
Figure 7. Effect of unilateral eyestalk ablation on Sp-Pol expression in different tissues of male (A) and female (B) S. paramamosain. *Indicates significant expression difference (P < 0.05). Values are mean (±SE).
The Localization of Sp-Pol
The FISH assay showed that Sp-Pol transcripts were detected in both ovary and testis (Figure 8). In the ovary, Sp-Pol was mainly expressed in oogonia cells (Figure 8A). In testis, Sp-Pol transcripts were strongly localized in epithelial cells of seminiferous tubules (Figure 8B).
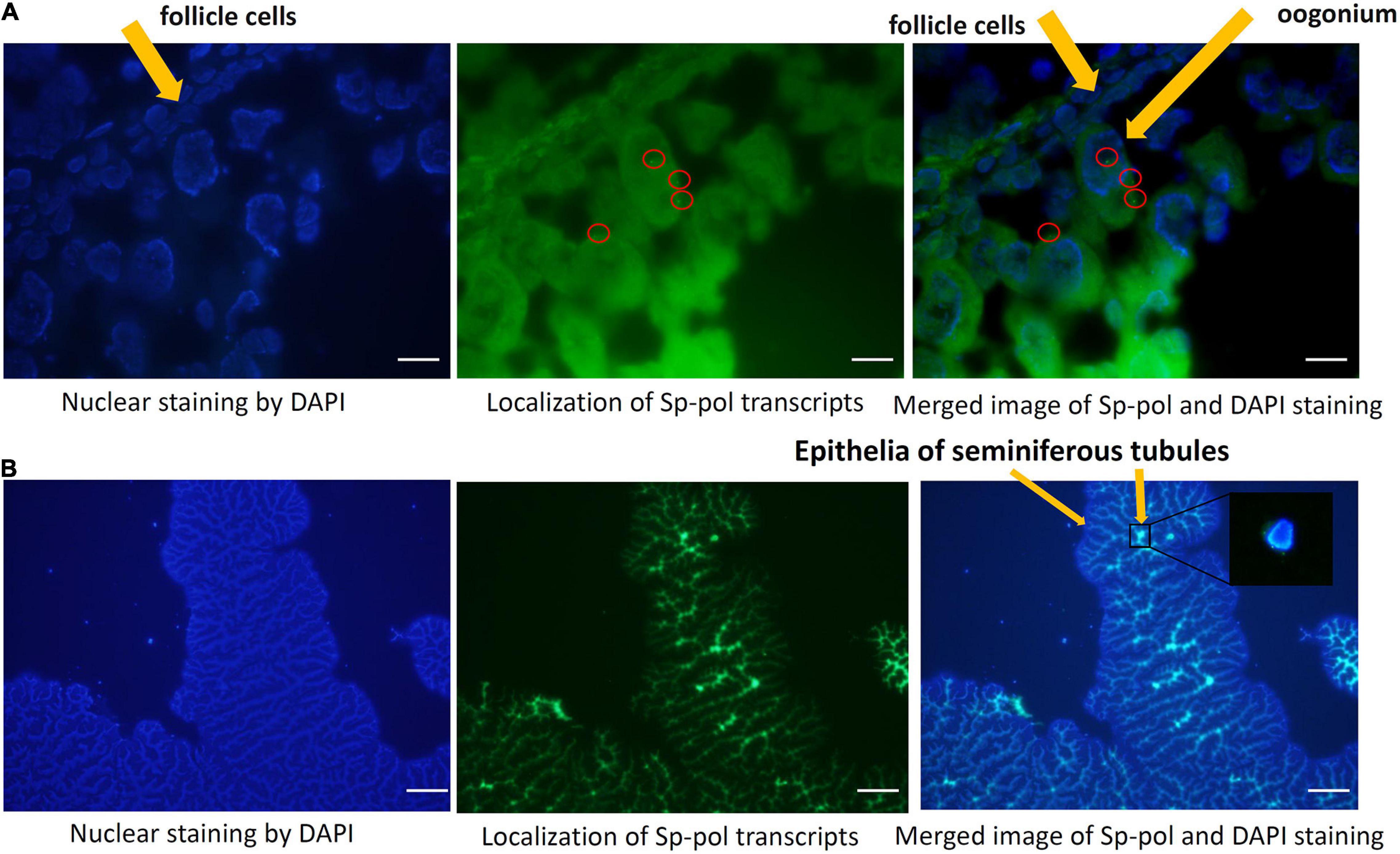
Figure 8. Localization of Sp-Pol transcripts in the ovary (A) and testis (B) of Scylla paramamosain by FISH. (A,B) were captured at × 100 and × 10 magnification, respectively. Sp-Pol mRNA signals (green dots) are presented inside the red circles.
Discussion
Limited information is available on the sex-determination system of brachyuran crabs. For decades, scientists believed that the XY/XX sex-determination system is the only sex-determination system for crabs (Legrand et al., 1987). Recently, besides the XY/XX (Fang et al., 2020), the WZ/ZZ sex-determination system has been found in some crab species such as Eriocheir sinensis (Cui et al., 2015), S. paramamosain, S. tranquebarica, and S. serrata (Shi et al., 2018). Sex-specific SNP marker is considered a powerful tool for understanding the sex-determination system (Pan et al., 2015). Detection of female-specific SNP markers provides insights into a WZ/ZZ sex determination system in S. paramamosain (Shi et al., 2018). In the present study, SNP1888 was mapped on the LG32 of the genetic map, which is a sex-related linkage group, and was located very close to the positions of other sex-specific SNPs (SNP1, SNP2, SNP3, SNP4, SNP5, SNP6, SNP7, SNP8, SNP9, SNP10, SNP11, SNP12) (Shi et al., 2018; Waiho et al., 2019). The presence of female-specific SNP markers and their locations on the female linkage map supports the existence of a WZ/ZZ sex determination system in S. paramamosain.
Phosphorylation plays important roles in metabolism, proliferation, apoptosis, differentiation, and cell division. Protein phosphorylation is catalyzed by a group of enzymes called kinases, which add a phosphate group to serine, threonine, tyrosine, or, to a lesser degree, histidine residues (Song et al., 2017). Twenty-eight putative phosphorylation sites (14 sites on serine and 14 sites on threonine residues) in Sp-Pol protein showed that Sp-Pol protein may be activated by some kinases enzymes. The prediction of six potential O-glycosylation and one N-glycosylation site indicates that glycosylation may also affect the function of Sp-Pol protein. Glycosylation, like phosphorylation, is important due to the various roles in protein folding, protein trafficking and localization, cell-cell interactions, and epitope recognition (Caragea et al., 2007). Glycosylation can be classified into four types: N-linked, O-linked, Glypiation, and C-linked. In N-linked glycosylation, the oligosaccharide chain is attached to the amide nitrogen of asparagine. In O-linked glycosylation, the glycan is attached to the hydroxyl oxygen of serine or threonine (Julenius et al., 2005).
Sp-Pol protein had the highest similarity to a Retrovirus-related Pol polyprotein gene from P. vannamei (60.47% identity) and was also slightly homologous (20–35% identity, bit score: 50–55, expectation value: 10–3–10–6) to a group of uncharacterized and hypothetical proteins from crustaceans, insects, echinoderms and fish species. Although Sp-Pol protein was closely related to a Retrovirus-related Pol polyprotein from P. vannamei and a hypothetical protein from swimming crab (Portunustri tuberculatus), it was distinct from other identified proteins. The very low similarity of Sp-Pol protein to other known homologous proteins and failure to predict functional domains indicates that this gene may not belong to the Gag-Pol polyprotein family and can be reclassified as a new family. Notably, the protein encoded by the Sp-Pol gene (204 aa) was quite smaller than Retrovirus-related Pol polyprotein from P. vannamei (541 aa) and other homologous proteins.
Spatio-temporal expression analysis showed that Sp-Pol was expressed in all the examined tissues and during all life stages, suggesting that it has a wide range of functions in different tissues and developmental stages. Expression of Sp-Pol in gonads was higher than other tissues including heart, hepatopancreas, muscle, gill, thoracic ganglion, intestine, and stomach. Comparing the expression of Sp-Pol between females and males, male crabs displayed a higher level of Sp-Pol expression compared to females. The higher expression of Sp-Pol in testis revealed that it is likely to play more important roles in the sexual development of males compared to females. The lower expression of Sp-Pol in the ovary compared to testis may be due to the mono-allelically expression pattern of this gene in the ovary. In the ovary, the mono-allele which owing nucleotide acid C at the SNP1888 locus was expressed, and the other allele which owing T was not expressed. The mono-allele expression could result in several different outcomes at the transcriptional level. There is a general trend for mono-allelically expressing cells to have fewer transcript levels than biallelically expressing cells (Eckersley Maslin and Spector, 2014).
It has been revealed that several sex-related genes such as CFSH, Sox, Dsx, Slx, Dmrt, GEM, and Fem-1 are involved in the “eyestalk-AG-testis” endocrine axis (Guo et al., 2019). These sex-related genes are mainly regulated by the neurohormones from eyestalk and play important roles in the IAG regulation (Shen et al., 2014; Ma et al., 2016; Shi L. et al., 2019; Zheng et al., 2019; Zhong et al., 2019).
Therefore, we hypothesized that Sp-Pol may be regulated by the eyestalk. Thus, we investigated the expression level of Sp-Pol after unilateral eyestalk ablation. Eyestalk ablation significantly up-regulated the expression level of Sp-Pol in the testis and male hepatopancreas and downregulated in female hepatopancreas. A similar expression pattern has been reported for IAG in the male blue crab, Callinectes sapidus. The expression of IAG was found to be up-regulated after eyestalk ablation in the testis and hepatopancreas (Chung et al., 2011). The significant changes of Sp-Pol mRNA in the hepatopancreas suggest Sp-Pol may be related to nutrient metabolism in the hepatopancreas. In decapods, the hepatopancreas is the major source for providing energy to gonad development (Sokolowicz et al., 2006). The increase of hepatopancreas weight during the reproductive season has been documented in several studies (i.e., material and energy storage for oogenesis) (Farhadi and Harlıoğlu, 2019). Vitellin (Vn) produced in the hepatopancreas is the main component of yolk proteins, which is stored in oocytes and is a nutritional source for animal embryo growth (Tseng et al., 2001). Vn is processed from its precursor, vitellogenin (Vg). In S. paramamosain, both the hepatopancreas and ovary are sites of Vg synthesis, but the hepatopancreas is the primary synthesis site (Huang et al., 2017). Therefore, further studies are required to evaluate the role of Sp-Pol in vitellogenesis and hepatic metabolism. Eyestalk ablation revealed that Sp-Pol expression may be regulated by inhibitory factors present in the eyestalk ganglia.
To investigate the potential role of Sp-Pol in gametogenesis, we performed FISH to locate Sp-Pol mRNA in the ovary and testis. In the ovary, Sp-Pol transcripts were detected in the oogonium cells but no signal was found in the follicle cells. In the testis, strong Sp-Pol signals were detected in the epithelia of seminiferous tubules. Signals decreased toward the center of the tubules of the testis, which means the expression level of Sp-Pol may progressively decrease from spermatogonia to spermatids. Spermatogenesis starts with the mitotic division of spermatogonia cells that locate on one side of the seminiferous tubules. Then, spermatogonia cells differentiate into primary spermatocytes. Spermatocytes divide into two equal haploid spermatids by Meiosis II. Later, spermatids differentiate into spermatozoa in the central region of the tubule (Zhang and Qiu, 2010). The role of the epithelium seminiferous tubules is not well studied in crustaceans. In fish, the cellular interactions between Sertoli cells and germ cells in the seminiferous epithelium play an important role during the spermatogenesis process (Batlouni et al., 2009). The structure of seminiferous epithelium changes from a Sertoli cell monolayer (containing spermatogonia cells during the non-reproductive season) to an active spermatogenic arrangement composed of spermatocytes and spermatozoa that fill the tubular lumen (reproductive season) (Batlouni et al., 2006). Further studies are required to investigate the roles (i.e., long-term and short-term RNA interference) and regulation mechanism of Sp-Pol in S. paramamosain. It would be interesting to study the relationship of Sp-Pol with other sexual genes such as IAG, CFSH, Sox gene family, Dmrt gene family, etc. Based on the observation of mono-allelic expression of Sp-Pol in the ovary, further studies are required to find whether Sp-Pol is an imprinted gene or regulate by DNA methylation mechanism.
Conclusion
In the present study, we first verified SNP1888 being a female-specific SNP marker and then we identified a novel sex-related gene (Sp-Pol) upstream of SNP1888. Furthermore, we mapped SNP1888 and Sp-Pol on a high-density genetic map. Despite the fact that Sp-Pol protein has low homology to other homologous proteins in other species, Sp-Pol protein may not belong to the Gag-Pol polyprotein family and can be classified as a new family. Sp-Pol exhibited different expression patterns in different tissues, different early development stages. Sp-Pol was expressed much higher in the testis than in the ovary, which might be related to the mono-allelically expression pattern of this gene in the ovary. The changes of Sp-Pol expression in testis and hepatopancreas after eyestalk ablation showed that Sp-Pol is regulated by eyestalk neurohormones. These results demonstrated that Sp-Pol may play important roles in the sexual development of S. paramamosain.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Ethics Committee of Shantou University.
Author Contributions
AF: conceptualization, data curation, formal analysis, investigation, methodology, visualization, and writing – original draft. XS: conceptualization, data curation, formal analysis, investigation, and methodology. YiZ: formal analysis and investigation. YuZ and HZ: methodology and editing. SL: methodology and review. MI: review and editing. HM: conceptualization, methodology, funding acquisition, project administration, resources, supervision, validation, review, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31772837), the Leading Talent Project of Special Support Plan of Guangdong Province (2019TX05N067), the Science and Technology Project of Guangdong Province (2018A050506080), and the “Sail Plan” Program for the Introduction of Outstanding Talents of Guangdong Province, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.727607/full#supplementary-material
Footnotes
- ^ www.ncbi.nlm.nih.gov/blast
- ^ https://web.expasy.org/compute_pi/
- ^ http://www.cbs.dtu.dk/services/NetPhos/
- ^ http://www.cbs.dtu.dk/services/NetOGlyc/
- ^ https://www.swissmodel.expasy.org/
- ^ http://bioedit.software.informer.com/7.2
References
Abayed, F. A. A., Manor, R., Aflalo, E. D., and Sagi, A. (2019). Screening for Dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. Gen. Comp. Endocrinol. 282:113205. doi: 10.1016/j.ygcen.2019.06.009
Aflalo, E. D., Hoang, T. T. T., Nguyen, V. H., Lam, Q., Nguyen, D. M., Trinh, Q. S., et al. (2006). A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 256, 468–478. doi: 10.1016/j.aquaculture.2006.01.035
Batlouni, S. R., Nóbrega, R. H., and França, L. R. (2009). Cell junctions in fish seminiferous epithelium. Fish Physiol. Biochem. 35, 207–217. doi: 10.1007/s10695-008-9253-y
Batlouni, S. R., Romagosa, E., and Borella, M. I. (2006). The reproductive cycle of male catfish Pseudoplatystoma fasciatum (Teleostei, Pimelodidae) revealed by changes of the germinal epithelium: an approach addressed to aquaculture. Anim. Reprod. Sci. 96, 116–132. doi: 10.1016/j.anireprosci.2005.11.012
Bureau of Fisheries Management (2020). China fishery statistics yearbook. Beijing: China Agriculture Press.
Caragea, C., Sinapov, J., Silvescu, A., Dobbs, D., and Honavar, V. (2007). Glycosylation site prediction using ensembles of support vector machine classifiers. BMC Bioinformatics 8:438. doi: 10.1186/1471-2105-8-438
Chandler, J. C., Elizur, A., and Ventura, T. (2018). The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825, 61–80. doi: 10.1007/s10750-017-3452-4
Chung, J. S., Manor, R., and Sagi, A. (2011). Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: implications for eyestalk regulation of IAG expression. Gen. Comp. Endocrinol. 173, 4–10. doi: 10.1016/j.ygcen.2011.04.017
Cui, W., Fang, S., Lv, L., Huang, Z., Lin, F., Wu, Q., et al. (2021a). Evidence of sex differentiation based on morphological traits during the early development stage of mud crab Scylla paramamosain. Front. Vet. Sci. 8:794. doi: 10.3389/fvets.2021.712942
Cui, W., Guan, M., Abu, S., Wu, F., Wu, Q., Tan, Q., et al. (2021b). Construction of a genetic linkage map and QTL mapping for sex indicate the putative genetic pattern of the F1 hybrid Scylla (Scylla serrata ♀× Scylla paramamosain ♂). Aquaculture. [Preprint].
Cui, Z., Hui, M., Liu, Y., Song, C., Li, X., Li, Y., et al. (2015). High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115, 206–215. doi: 10.1038/hdy.2015.26
Eckersley Maslin, M. A., and Spector, D. L. (2014). Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 30, 237–244. doi: 10.1016/j.tig.2014.03.003
Fang, S., Zhang, Y., Shi, X., Zheng, H., Li, S., Zhang, Y., et al. (2020). Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus) with implication of an XX/XY sex determination system. Genomics 112, 404–411. doi: 10.1016/j.ygeno.2019.03.003
Farhadi, A., and Harlıoğlu, M. M. (2019). The annual cycle of spermatozoa content in the vas deferens and some reproductive parameters in the narrow-clawed crayfish Pontastacus leptodactylus (Eschscholtz, 1823). Aquac. Rep. 13:100185. doi: 10.1016/j.aqrep.2019.100185
Farhadi, A., Cui, W., Zheng, H., Li, S., Zhang, Y., Ikhwanuddin, M., et al. (2021a). The Regulatory Mechanism of Sexual Development in Decapod Crustaceans. Front. Mar. Sci. 8:477. doi: 10.3389/fmars.2021.679687
Farhadi, A., Fang, S., Zhang, Y., Cui, W., Fang, H., Ikhwanuddin, M., et al. (2021b). The significant sex-biased expression pattern of Sp-Wnt4 provides novel insights into the ovarian development of mud crab (Scylla Paramamosain). Int. J. Biol. Macromol. 183, 490–501. doi: 10.1016/j.ijbiomac.2021.04.186
Guo, Q., Li, S., Lv, X., Xiang, J., Manor, R., Sagi, A., et al. (2019). Sex-Biased CHHs and Their Putative Receptor Regulate the Expression of IAG Gene in the Shrimp Litopenaeus vannamei. Front. Physiol. 10:1525. doi: 10.3389/fphys.2019.01525
Harlıoğlu, M. M., and Farhadi, A. (2017). Feminization strategies in crustacean aquaculture. Aquac. Int. 25, 1453–1468. doi: 10.1007/s10499-017-0128-z
Huang, X., Feng, B., Huang, H., and Ye, H. (2017). In vitro stimulation of vitellogenin expression by insulin in the mud crab, Scylla paramamosain, mediated through PI3K/Akt/TOR pathway. Gen. Comp. Endocrinol. 250, 175–180. doi: 10.1016/j.ygcen.2017.06.013
Jiang, Q., Lu, B., Lin, D., Huang, H., Chen, X., and Ye, H. (2020a). Role of crustacean female sex hormone (CFSH) in sex differentiation in early juvenile mud crabs, Scylla paramamosain. Gen. Comp. Endocrinol. 2020:113383. doi: 10.1016/j.ygcen.2019.113383
Jiang, Q., Lu, B., Wang, G., and Ye, H. (2020b). Transcriptional inhibition of Sp-IAG by crustacean female sex hormone in the mud crab, Scylla paramamosain. Int. J. Mol. Sci. 21:5300. doi: 10.3390/ijms21155300
Jiang, Q., Zheng, H., Zheng, L., Wang, Y., Wang, M., Xie, X., et al. (2020c). Molecular characterization of the insulin-like androgenic gland hormone in the swimming crab, Portunus trituberculatus, and its involvement in the insulin signaling system. Front. Endocrinol. 11:585. doi: 10.3389/fendo.2020.00585
Jin, S., Fu, H., Zhou, Q., Sun, S., Jiang, S., Xiong, Y., et al. (2013). Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, using illumina hiseq 2000. PLoS One 8:e76840. doi: 10.1371/journal.pone.0076840
Jin, S., Hu, Y., Fu, H., Jiang, S., Xiong, Y., Qiao, H., et al. (2019). Potential functions of Gem-associated protein 2-like isoform X1 in the oriental river prawn Macrobrachium nipponense: cloning, qPCR, in situ hybridization, and RNAi analysis. Int. J. Mol. Sci. 20:3995. doi: 10.3390/ijms20163995
Julenius, K., Mølgaard, A., Gupta, R., and Brunak, S. (2005). Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15, 153–164. doi: 10.1093/glycob/cwh151
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Legrand, J. J., Legrand−Hamelin, E., and Juchault, P. (1987). Sex determination in Crustacea. Biol. Rev. 62, 439–470. doi: 10.1111/j.1469-185x.1987.tb01637.x
Li, S., Li, F., Sun, Z., and Xiang, J. (2012). Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis. Gen. Comp. Endocrinol. 177, 246–255. doi: 10.1016/j.ygcen.2012.04.010
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, K. Y., Lin, J. Y., Guo, S. Z., Chen, Y., Li, J., and Le, et al. (2013). Molecular characterization and expression analysis of an insulin-like gene from the androgenic gland of the oriental river prawn, Macrobrachium nipponense. Gen. Comp. Endocrinol. 185, 90–96. doi: 10.1016/j.ygcen.2013.01.018
Ma, K. Y., Liu, Z. Q., Lin, J. Y., Li, J., and Le, and Qiu, G. F. (2016). Molecular characterization of a novel ovary-specific gene fem-1 homolog from the oriental river prawn, Macrobrachium nipponense. Gene 575, 244–252. doi: 10.1016/j.gene.2015.08.070
Ma, K., Qiu, G., Feng, J., and Li, J. (2012). Transcriptome analysis of the oriental river prawn, Macrobrachium nipponense using 454 pyrosequencing for discovery of genes and markers. PLoS One 7:e39727. doi: 10.1371/journal.pone.003972
Mareddy, V. R., Rosen, O., Thaggard, H. B., Manor, R., Kuballa, A. V., Aflalo, E. D., et al. (2011). Isolation and characterization of the complete cDNA sequence encoding a putative insulin-like peptide from the androgenic gland of Penaeus monodon. Aquaculture 318, 364–370. doi: 10.1016/j.aquaculture.2011.05.027
Niksirat, H., James, P., Andersson, L., Kouba, A., and Kozák, P. (2015a). Label-free protein quantification in freshly ejaculated versus post-mating spermatophores of the noble crayfish Astacus astacus. J. Proteomics 123, 70–77. doi: 10.1016/j.jprot.2015.04.004
Niksirat, H., Kouba, A., and Kozák, P. (2014). Post-mating morphological changes in the spermatozoon and spermatophore wall of the crayfish Astacus leptodactylus: insight into a non-motile spermatozoon. Anim. Reprod. Sci. 149, 325–334. doi: 10.1016/j.anireprosci.2014.07.017
Niksirat, H., Kouba, A., and Kozák, P. (2015b). Ultrastructure of egg activation and cortical reaction in the noble crayfish Astacus astacus. Micron 68, 115–121. doi: 10.1016/j.micron.2014.09.010
Niksirat, H., Vancová, M., Andersson, L., James, P., Kouba, A., and Kozák, P. (2016). Protein modification in the post-mating spermatophore of the signal crayfish Pacifastacus leniusculus: insight into the tyrosine phosphorylation in a non-motile spermatozoon. Anim. Reprod. Sci. 172, 123–130. doi: 10.1016/j.anireprosci.2016.07.009
Pan, Z. J., Li, X. Y., Zhou, F. J., Qiang, X. G., and Gui, J. F. (2015). Identification of sex-specific markers reveals male heterogametic sex determination in Pseudobagrus ussuriensis. Mar. Biotechnol. 17, 441–451. doi: 10.1007/s10126-015-9631-2
Rosen, O., Manor, R., Weil, S., Gafni, O., Linial, A., Aflalo, E. D., et al. (2010). A sexual shift induced by silencing of a single insulin-like gene in crayfish: ovarian upregulation and testicular degeneration. PLoS One 5:e15281. doi: 10.1371/journal.pone.0015281
Shen, H., Hu, Y., and Zhou, X. (2014). Sex-lethal gene of the Chinese mitten crab Eriocheir sinensis: cDNA cloning, induction by eyestalk ablation, and expression of two splice variants in males and females. Dev. Genes Evol. 224, 97–105. doi: 10.1007/s00427-014-0467-y
Shi, L., Han, S., Fei, J., Zhang, L., Ray, J. W., Wang, W., et al. (2019). Molecular characterization and functional study of insulin-like androgenic gland hormone gene in the red swamp crayfish, Procambarus clarkii. Genes 10:645. doi: 10.3390/genes10090645
Shi, X., Lu, J., Wu, Q., Waiho, K., Aweya, J. J., Fazhan, H., et al. (2019). Comparative analysis of growth performance between female and male mud crab Scylla paramamosain crablets: Evidences from a four-month successive growth experiment. Aquaculture 505, 351–362. doi: 10.1016/j.aquaculture.2019.02.062
Shi, X., Waiho, K., Li, X., Ikhwanuddin, M., Miao, G., Lin, F., et al. (2018). Female-specific SNP markers provide insights into a WZ/ZZ sex determination system for mud crabs Scylla paramamosain, S. tranquebarica and S. serrata with a rapid method for genetic sex identification. BMC Genomics 19, 981–993. doi: 10.1186/s12864-018-5380-8
Sokolowicz, C. C., Bond-Buckup, G., and Buckup, L. (2006). Dynamics of gonadal development of Aegla platensis Schmitt (Decapoda, Anomura, Aeglidae). Rev. Bras. Zool. 23, 1153–1158. doi: 10.1590/s0101-81752006000400023
Song, J., Wang, H., Wang, J., Leier, A., Marquez-Lago, T., Yang, B., et al. (2017). PhosphoPredict: A bioinformatics tool for prediction of human kinase-specific phosphorylation substrates and sites by integrating heterogeneous feature selection. Sci. Rep. 7, 1–19.
Tan, K., Li, Y., Zhou, M., and Wang, W. (2020). siRNA knockdown of MrIR induces sex reversal in Macrobrachium rosenbergii. Aquaculture 2020:735172. doi: 10.1016/j.aquaculture.2020.735172
Tseng, D. Y., Chen, Y. N., Kou, G. H., Lo, C.-F., and Kuo, C. M. (2001). Hepatopancreas is the extraovarian site of vitellogenin synthesis in black tiger shrimp, Penaeus monodon. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129, 909–917. doi: 10.1016/s1095-6433(01)00355-5
Ventura, T., and Sagi, A. (2012). The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 30, 1543–1550. doi: 10.1016/j.biotechadv.2012.04.008
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Rosen, O., and Sagi, A. (2012). Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 86:90.
Waiho, K., Fazhan, H., Jasmani, S., and Ikhwanuddin, M. (2017). Gonadal development in males of the orange mud crab, Scylla olivacea (Herbst, 1796)(Decapoda, Brachyura, Portunidae). Crustaceana 90, 1–19. doi: 10.1163/15685403-00003622
Waiho, K., Shi, X., Fazhan, H., Li, S., Zhang, Y., Zheng, H., et al. (2019). High-density genetic linkage maps provide novel insights into ZW/ZZ sex determination system and growth performance in mud crab (Scylla paramamosain). Front. Genet. 10, 298–314. doi: 10.3389/fgene.2019.00298
Wang, G. Z., Li, S. J., Zeng, C. S., Lin, S. J., Kong, X. H., Ai, C. X., et al. (2005). Status of biological studies and aquaculture development of the mud crab, Scylla serrata, in China: an experimental ecological studies. Aquac. Int. 13, 459–468. doi: 10.1007/s10499-005-1887-5
Wu, Q., Shi, X., Fang, S., Xie, Z., Guan, M., Li, S., et al. (2019). Different biochemical composition and nutritional value attribute to salinity and rearing period in male and female mud crab Scylla paramamosain. Aquaculture 513:734417. doi: 10.1016/j.aquaculture.2019.734417
Wu, Q., Waiho, K., Huang, Z., Li, S., Zheng, H., Zhang, Y., et al. (2020). Growth performance and biochemical composition dynamics of ovary, hepatopancreas and muscle tissues at different ovarian maturation stages of female mud crab, Scylla paramamosain. Aquaculture 515:734560. doi: 10.1016/j.aquaculture.2019.734560
Xu, Y. R., Wang, G. Y., Zhou, Y. C., and Yang, W. X. (2018). The characterization and potential roles of bone morphogenetic protein 7 during spermatogenesis in Chinese mitten crab Eriocheir sinensis. Gene 673, 119–129. doi: 10.1016/j.gene.2018.06.020
Yang, X., Ikhwanuddin, M., Li, X., Lin, F., Wu, Q., Zhang, Y., et al. (2018). Comparative transcriptome analysis provides insights into differentially expressed genes and long non-coding RNAs between ovary and testis of the mud crab (Scylla paramamosain). Mar. Biotechnol. 20, 20–34. doi: 10.1007/s10126-017-9784-2
Zhang, E. F., and Qiu, G. F. (2010). A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev. Genes Evol. 220, 151–159. doi: 10.1007/s00427-010-0336-2
Zhang, Y., Miao, G., Wu, Q., Lin, F., You, C., Wang, S., et al. (2018). Transcriptome sequencing and molecular markers discovery in the gonads of Portunus sanguinolentus. Sci. Data 5:180131.
Zhang, Y., Qiao, K., Wang, S., Peng, H., Shan, Z., and Wang, K. (2014). Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab, Scylla paramamosain. Aquaculture 433, 325–334. doi: 10.1016/j.aquaculture.2014.06.033
Zheng, J., Cheng, S., Jia, Y., Gu, Z., Li, F., Chi, M., et al. (2019). Molecular identification and expression profiles of four splice variants of Sex-lethal gene in Cherax quadricarinatus. Comp. Biochem. Physiol. B 234, 26–33. doi: 10.1016/j.cbpb.2019.05.002
Zhong, P., Zhou, T., Zhang, Y., Chen, Y., Yi, J., Lin, W., et al. (2019). Potential involvement of a DMRT family member (Mr−Dsx) in the regulation of sexual differentiation and moulting in the giant river prawn Macrobrachium rosenbergii. Aquac. Res. 50, 3037–3049. doi: 10.1111/are.14262
Zhou, L. X., Liu, X., Ye, B. Q., Liu, Y., Tan, S. P., Ma, K. Y., et al. (2020). Molecular characterization of ovary-specific gene Mrfem-1 and siRNA-mediated regulation on targeting Mrfem-1 in the giant freshwater prawn, Macrobrachium rosenbergii. Gene 754:144891. doi: 10.1016/j.gene.2020.144891
Keywords: eyestalk ablation, gonad development, sex-related gene, SNP, mud crab
Citation: Farhadi A, Shi X, Zhang Y, Zhang Y, Li S, Zheng H, Ikhwanuddin M and Ma H (2021) A Novel Imprinted Gene (Sp-Pol) With Sex-Specific SNP Locus and Sex-Biased Expression Pattern Provides Insights Into the Gonad Development of Mud Crab (Scylla paramamosain). Front. Mar. Sci. 8:727607. doi: 10.3389/fmars.2021.727607
Received: 19 June 2021; Accepted: 22 July 2021;
Published: 19 August 2021.
Edited by:
Shengming Sun, Shanghai Ocean University, ChinaCopyright © 2021 Farhadi, Shi, Zhang, Zhang, Li, Zheng, Ikhwanuddin and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Ma, bWFoeUBzdHUuZWR1LmNu
 Ardavan Farhadi
Ardavan Farhadi Xi Shi1,2
Xi Shi1,2 Mhd Ikhwanuddin
Mhd Ikhwanuddin Hongyu Ma
Hongyu Ma