- 1College of Marine Sciences, Ningbo University, Ningbo, China
- 2Zhejiang Key Laboratory of Aquatic Germplasm Resources, Zhejiang Wanli University, Ningbo, China
- 3Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ningbo, China
The razor clam, Sinonovacula constricta is a commercially important bivalve in the western Pacific Ocean, yet little is known about the mechanisms of sex determination/differentiation and gametogenesis. In the present study, the comparative transcriptome analysis of adult gonads (female gonads and male gonads) was conducted to identify potential sex-related genes in S. constricta. The number of reads generated for each target library (three females and three males) ranged from 31,853,422 to 37,750,848, and 20,489,472 to 26,152,448 could be mapped to the reference genome of S. constricta (the map percentage ranging from 63.71 to 71.48%). A total of 8,497 genes were identified to be differentially expressed between the female and male gonads, of which 4,253 were female-biased (upregulated in females), and 4,244 were male-biased. Forty-five genes were identified as potential sex-related genes, including DmrtA2, Sox9, Fem-1b, and Fem-1c involved in sex determination/differentiation and Vg, CYP17A1, SOHLH2, and TSSK involved in gametogenesis. The expression profiles of 12 genes were validated by qRT-PCR, which further confirmed the reliability and accuracy of the RNA-Seq results. Our results provide basic information about the genes involved in sex determination/differentiation and gametogenesis, and pave the way for further studies on reproduction and breeding in S. constricta and other marine bivalves.
Introduction
The razor clam, Sinonovacula constricta is a marine bivalve, living in the lower to mid intertidal zones along the western Pacific Ocean coast (Morton, 1984; Wang and Xu, 1997). Razor clam farming began almost 500 years ago in southeastern China, particularly in the coastal areas of Fujian and Zhejiang Province (Wang et al., 1993). In China, S. constricta has become one of the most commercially important cultured clam species and has huge market potential. Therefore, it is urgent to further improve the technique in the field of artificial propagation and culture of this species. For the S. constricta selection program, the aim is to construct inbred lines with a variety of traits, such as rapid growth, that underlie successful reproduction.
Compared with other economically important molluscan species, S. constricta is diecious (i.e., with fixed gender; individuals do not change sex), and the gonad maturation period is from September to October (Wu and Xu, 2000). Furthermore, it is difficult to differentiate between female and male clams with external morphological characteristics, especially before sex maturity (Wang et al., 1993). Therefore, gonadal maturity and synchronicity cannot be estimated by appearance, yet they are important factors for successful artificial reproduction. In the artificial reproduction conditions of bivalves, the successful hatching of selected spat relies on production of gametes and embryos from synchronous breeders. Therefore, it is essential to achieve genetic improvement for controlled reproduction, which depends on understanding molecular mechanisms and factors that control reproduction (Vahirua-Lechat et al., 2008; Moullac et al., 2013). However, the mechanisms that underlie gonad development and sex determination/differentiation in S. constricta are poorly understood, and there is no gene yet identified as an actor involved in gonad development or sex determination.
Gender is the product of sex determination/differentiation. Genetic or environmental processes that establish the gender of an organism, lead to specific molecular cascades, which can change an undifferentiated gonad into an ovary or a testis (Penman and Piferrer, 2008; Piferrer and Guiguen, 2008). Previous genetic studies have shown that some genes expressed in a sexually dimorphic manner in the progress of activation of the ovarian or testis pathway or repression of the alternative pathway, and this process determines the resulting sex or gonad development in model organisms (Piferrer and Guiguen, 2008). In bivalves, studies have identified some genes related to sex determination/differentiation or gametogenesis, for example, Dmrt (Doublesex- and mab-3-related transcription factor) and SoxE involved in male determination in Crassostrea gigas (Naimi et al., 2009a; Santerre et al., 2014), and Foxl2 and Beta-catenin involved in female determination in Chlamys farreri (Liu et al., 2012; Li et al., 2014). In recent years, transcriptome sequencing analysis has been widely used to screen for potentially sex-related genes in bivalves. In C. gigas, genes involved in sex-determining pathways, such as SoxH and Foxl2 were identified by male and female gonad transcriptome analysis (Zhang et al., 2014). In Pinctada margaritifera, genes such as Dmrt, Fem-1, Foxl2, and Vitellogenin (Vg) were identified as potential sex differentiation/determining genes based on transcriptional change in male and female gonads at different development stages (Teaniniuraitemoana et al., 2014). In Patinopecten yessoensis, transcriptome sequencing revealed that the Foxl2 gene was ovary-biased and Dmrt1, SoxH, Fem-1, and Sox9 were testis-biased (Yang et al., 2016; Li et al., 2016; Zhou et al., 2019). In Tegillarca granosa, genes such as Sox, Foxl2, and Beta-catenin were identified as sex-related genes from the gonad transcriptome data (Chen et al., 2017). The Vg gene was identified to be expressed especially in female gonads in C. gigas (Matsumoto et al., 2003), C. farreri (Qin et al., 2012), and P. yessoensis (Yang et al., 2016). Clearly, studies on sex-related genes not only contribute to improving our knowledge about molecular mechanisms of sex determination/differentiation and gonad development, but also may be applicable to reproductive control of cultured mollusks. However, little is known about the sex-related genes in S. constricta.
Recently, the genomic sequence and a large number of transcriptomic data of S. constricta have been available (Niu et al., 2013; Ran et al., 2019; Dong et al., 2020), however, the genes related with sex determination/differentiation or gametogenesis were still not identified. In this study, we analyzed the female and male gonadal transcriptomes of S. constricta using Illumina sequencing technology, compared expression patterns of differentially expressed genes between female and male gonads, and identified potential sex-related genes. This study aims to identify genes that potentially involved in sex determination/differentiation or gametogenesis. Our results will be useful for further studies on reproduction control and artificial propagation practice in S. constricta.
Materials and Methods
Animal Sampling and RNA Extraction
Two-year-old mature clams were collected from the Technology Innovation Base of Marine and Fisheries of Ningbo, Zhejiang province, China and used for gonadal transcriptomic sequencing. During the reproduction season, the mature gonads of S. constricta are full of sperms or eggs. The gender of clams was confirmed by observing the presence of sperms or eggs under a microscope. Three female and three male clams were selected, and their gonad tissues were sampled and frozen in liquid nitrogen and then stored at −80°C until RNA extraction. The three females were designed as ScF1, ScF2, and ScF3 and three males were designed as ScM1, ScM2, and ScM3. Total RNA from each gonad tissue was isolated with Trizol reagent (Invitrogen, United States) according to manufacturer’s instructions. DNase I was used to remove DNA contamination from the isolated RNA, and the RNA quantity and quality were evaluated by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, United States).
cDNA Library Construction, Sequencing, and Reference-Based Assembly
Six cDNA libraries were constructed according to the standard protocol provided by Biomarker Technologies Co. (Beijing, China). Briefly, mRNA was purified from total RNA with oligo (dT) magnetic beads and fragmented with divalent cations buffer under elevated temperature. First-strand cDNA was generated using random hexamer-primed reverse transcription followed by the synthesis of second-strand cDNA using RNase H and DNA polymerase I. Then the double-strand cDNA was purified and washed with elution buffer followed by end reparation, dA-tailing and ligated to sequencing adapters. Afterward, adaptor-ligated fragments were purified, and suitable fragments were chosen as templates for PCR amplification. The prepared cDNA libraries were sequenced on the Illumina HiSeq 2500 platform (San Diego, United States) with 150 bp paired-end model.
The raw reads from the six samples were pre-processed by trimming the adaptor sequences and filtering the ambiguous or low-quality short reads with software (Grabherr et al., 2011). Then the resulting high-quality reads from all samples were mapped to the reference genome of S. constricta (WSYO00000000.1) using HISAT2 (Kim et al., 2015). String Tie software (Shen et al., 2014) were used to assemble mapped reads and compared with reference genome annotation to discover novel genes. All novel genes were subjected to annotation with different databases, including Swiss-Prot,1 NR (NCBI non-redundant protein database),2 GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes).3 BLASTX searches were conducted in the NR database and Swiss-Prot database with an e-value threshold of 1E–5.
Sample Relationship Analysis
The correlation coefficient between biological replicates was used to evaluate sample repeatability, and low correlation sample was removed for further analysis. In order to study the global transcriptomic differences and correlation among samples from the two sexes, Principal component analysis (PCA) was conducted and a heat map was generated. PCA was performed on the basic of the average fragments per kilobase of transcript per million mapped reads (FPKM) values for all expressed genes in each sample, and sample repeatability analysis was presented via heat map using R scripts.
Analysis of DEGs (Differentially Expressed Genes) and Function Enrichment
Gene expression level was generally estimated by mapping clean reads to the trinity transcript assembly by RSEM for each sample (Li and Dewey, 2011). Meanwhile, the gene expression level was presented in a normalized expressed value as FPKM. Afterward, reconstruction of transcript assemblies was performed by the reference genome annotation-based transcripts assembly program within the Cufflinks software package to obtain a comprehensive set of transcripts for further differential analysis. The DESeq2 R package was used to identify differentially expressed genes (DEGs) between the two sexes (Anders and Huber, 2010). DEGs were defined as |Log2FoldChange| > 1 and a false-discovery rate < 0.01. The transcriptional pattern variations between the two sexes were assessed by DEG union, and R scripts were used to generate a heat map of the DEGs.
For enrichment analysis, all DEGs were mapped to terms in the KEGG and GO databases using BLAST algorithm with an E-value of ≤ 1e–5, and the significantly (P < 0.05) enriched KEGG and GO terms in DEGs were identified compared with the transcriptome background.
Gene Expression Analysis by Quantitative Real-Time PCR (qRT-PCR)
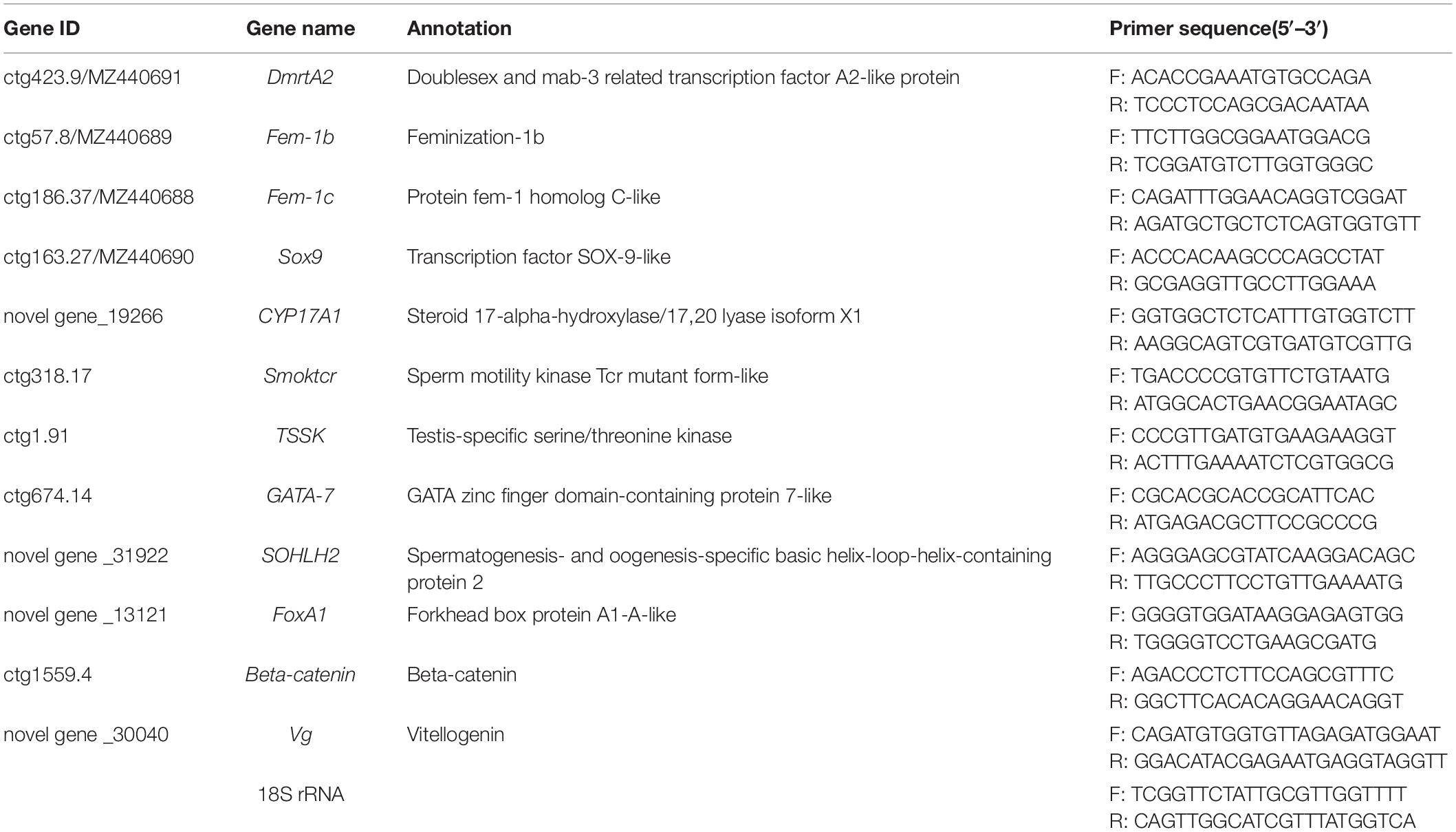
To validate the transcriptome sequencing data and the genes that might be potential sex-related genes obtained from DESeq analysis, 12 DEGs were chosen and quantified using quantitative real-time PCR (qRT-PCR). Total RNA was first extracted from each sample by using Trizol Reagent (Invitrogen), and cDNA was then synthesized using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Japan). Finally, qRT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, United States) according to manufacturer’s instructions on an ABI 7500 Fast Real-time PCR System (Applied Biosystems, United States). The 18S rRNA gene was chosen as the reference gene to serve as an internal control. The 12 pairs of prime sequences used for qRT-PCR analysis were listed in Table 1. Each 20 μL reaction system contained the following components: 10 μL iTaq Universal SYBR Green Supermix, 1 μL each primer, 0.8 μL cDNA, and 7.2 μL deionized water. The procedure of qRT-PCR was set as following: 94°C for 20 s and 40 cycles of 94°C for 3 s, 60°C for 15 s, and 72°C for 10 s. The gene relative expression level was calculated with the 2–△△ct methods, and the results were compared using one-way analysis of variance in SPSS 20.0 (SPSS, United States). P < 0.05 was considered to be statistically significant. The fold changes of 12 genes in female samples vs. male samples were calculated via FPKM, and the log2FoldChange values of these genes obtained by RNA-Seq and qRT-PCR were used for graphical presentation.
To detect tissue expression pattern of some DEGs, eight tissues, including hepatopancreas, gill, mantle, siphon, adductor muscle, foot, female gonad and male gonad, were collected from three female and three male individuals, respectively, at mature stage. And gene expressions levels were analyzed by qRT-PCR followed the aforementioned procedure.
Sequence Analysis of Sex-Related Genes
The amino acid sequences used for sequence analysis were all retrieved from GenBank. Protein domain structure analysis was performed with SMART software. Multiple sequence alignments were performed by the ClustalW2 program and phylogenetic trees with bootstrap values were constructed with MEGA 7.0 software using the neighbor-joining method. The numbers in the branches of phylogenetic trees represent the bootstrap values from 1,000 replicates, and all the sequence GenBank accession numbers were listed in Supplementary Table 1.
Results
Sequence Analysis and Gonad Transcriptome Assembly
To better understand the mechanism of sex determination/differentiation and gametogenesis in S. constricta, a comparative transcriptomic analysis was conducted. Six cDNA libraries (Female: ScF1, ScF2, and ScF3; Male: ScM1, ScM2, and ScM3) were sequenced and assembled. The number of reads generated for each library ranged from 31,853,422 to 37,750,848, with Q30 values ranging from 94.34 to 94.79% (Table 2). Among the clean reads, the number of reads that could be mapped to the reference genome ranged from 20,489,472 to 26,152,448, and the percentage of clean reads ranged from 63.71 to 71.48% in the different libraries (Table 2). After alignment with the reference genome, 30,651 genes (including 10,092 novel genes) were obtained from global gonad transcriptomes, and 24,198 genes (including 5,279 novel genes) were function-annotated in the GO, NR, KEGG and Swiss-Prot databases. We deposited the transcriptome data into the NCBI Sequence Read Archive (SRA) under accession numbers SRR9937008–SRR9937013.
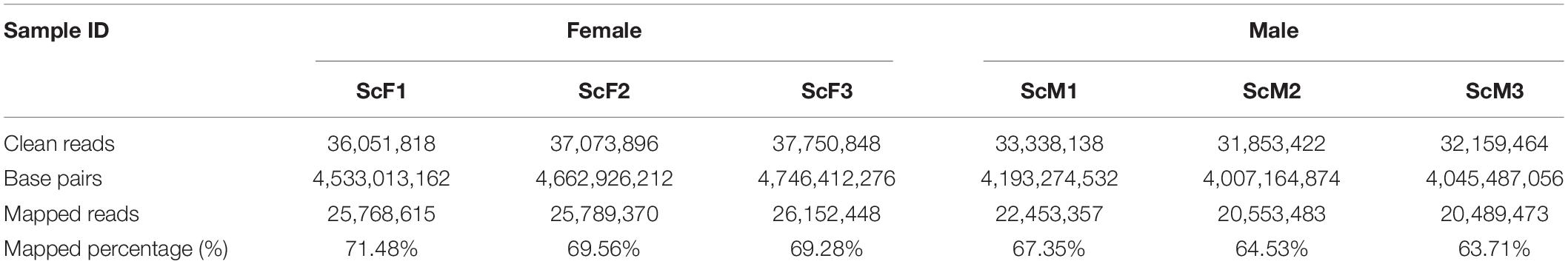
Table 2. Summary statistics of gonad transcriptome sequencing for S. constricta from ovary and testis.
DEGs Identification and Function Enrichment Analysis
PCA revealed strong clustering associated with sex, and there were obvious differences in transcript expression between ovaries and testis excluding the ScM1 sample (Figure 1A). Meanwhile, a heat map plot for sample relationship analysis also showed that sex identification of samples was exact, except for the ScM1 sample (Figure 1B). The Pearson’s correlation coefficient (r2) were 0.102 and 0.264 between ScM1 and ScM2, ScM1, and ScM3, respectively, with low coefficient, so the RNA-Seq data from the ScM1 sample were excluded from further analysis.
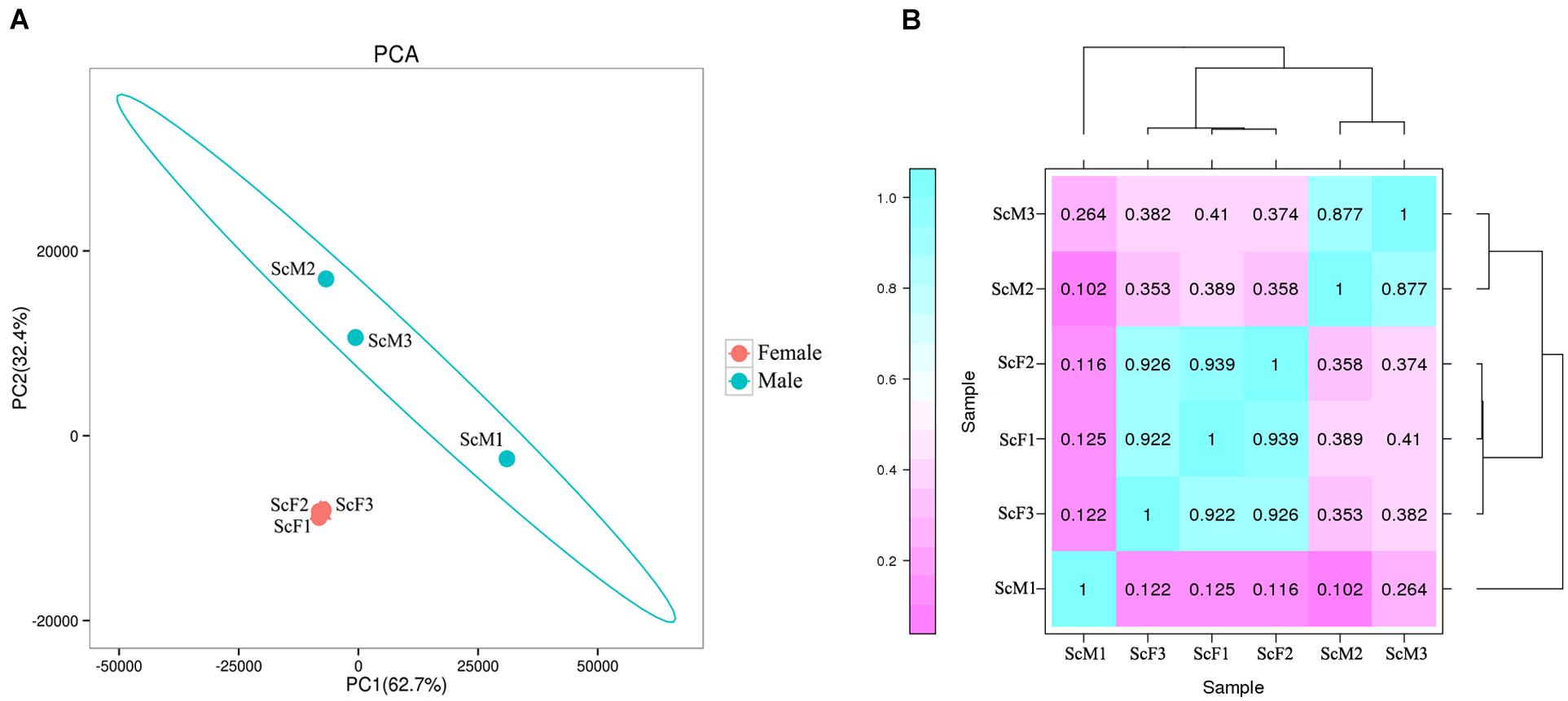
Figure 1. Expression pattern analysis of gonad samples in S. constricta. (A) Principal components analysis (PCA) showed the strong clustering associated with sex except for ScM1. (B) Sample correlation heatmap plot revealed exact sex identification except for ScM1. Dark green represents strong correlation and dark pink represents weak correlation, and the number in each column and row correspond to the correlation coefficient of one sample with the other five samples including itself.
For the differential expression analysis, 8,497 DEGs were identified between female gonad group and male gonad group. Compared to female gonad group, 4,244 genes were significantly up-regulated in male gonad group, accounting for 49.95% of all significant DEGs, and 4,253 genes were significantly down-regulated in male gonad group, accounting for 50.05% of all significant DEGs (Figure 2). Of all the significant DEGs, 6,823 genes were function-annotated in the GO, Swiss-Prot, KEGG and NR databases by BLASTX with a cut-off E-value of 1 × 10–5.
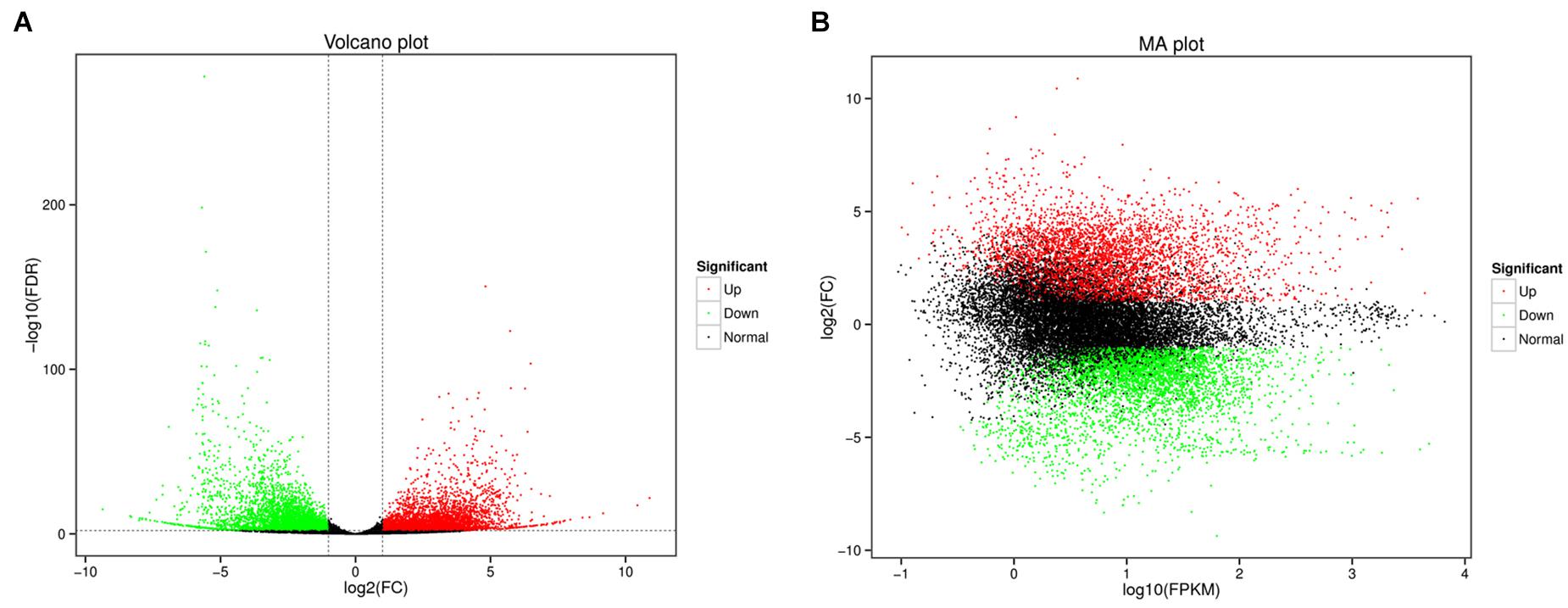
Figure 2. Volcano plot (A) and MA plot (B) of DEGs (Green dots represented downregulated genes, the red dots represented upregulated genes).
To explore the potential functions of these DEGs in sex determination/differentiation and gametogenesis, GO and KEGG pathway enrichment analyses were performed. GO functional enrichment analysis showed that 2,746 DEGs had a GO ID and could be categorized into 1,965 functional groups in three main categories: biological process, cellular component, and molecular function. Additionally, 151 GO terms were significantly enriched (P < 0.05). Biological process GO terms related to DNA repair (GO:0006281), DNA replication (GO:0006260), and nucleic acid phosphodiester bond hydrolysis (GO:0090305) were significantly enriched (P < 0.05), as were molecular function GO terms related to ATP binding (GO:0005524) and nucleic acid binding (GO:0003676) and cell cellular component GO terms related to nucleus (GO:0005634), catalytic step 2 spliceosome (GO:0071013), and Cdc73/Paf1 complex (GO:0016593) (Figure 3).
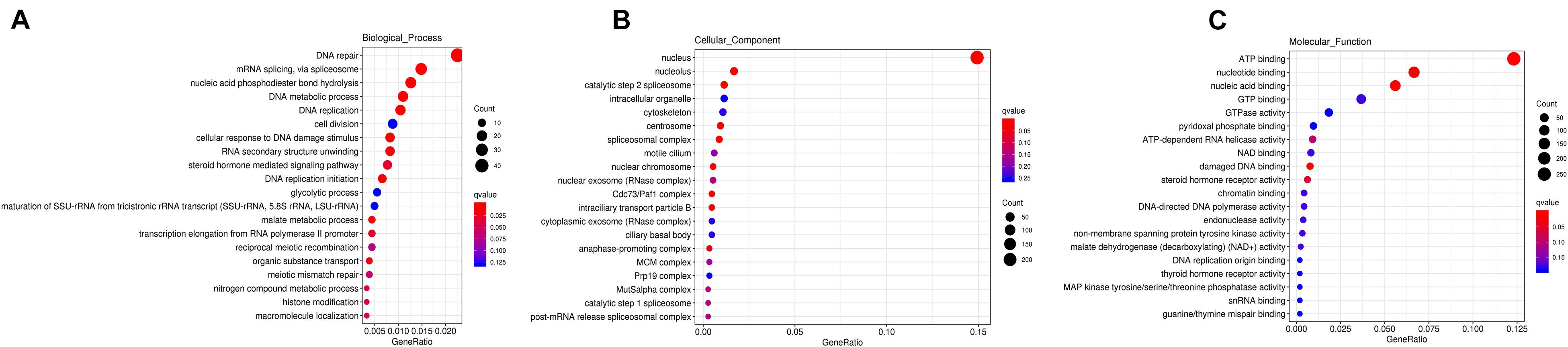
Figure 3. GO functional enrichment of DEGs in biological process (A), cellular component (B), and molecular function (C), respectively. The bigger the Rich factor or the smaller the q-value both indicates the more significant GO terms, and the bigger circle size indicates the higher number of genes.
In the KEGG analysis, 1,364 DEGs had KEGG assignations and were associated with 221 pathways. Figure 4 shows the top 20 enriched pathways. DNA replication, nucleotide excision repair, spliceosome, Fanconi anemia pathway, lysosome, RNA transport, and carbon metabolism were significantly enriched in KEGG pathways (P < 0.05), which showed patterns similar to those of the GO terms. Specifically, lysosome, endocytosis, phagosome, sphingolipid metabolism, carbon metabolism, and glycerophospholipid metabolism were significantly enriched in male gonad group (P < 0.05) (Figure 4A), whereas RNA transport, spliceosome, pyrimidine metabolism, ubiquitin mediated proteolysis, Fanconi anemia pathway, DNA replication, and nucleotide excision repair were significantly enriched in female gonad group (P < 0.05) (Figure 4B). Some of the DEGs were enriched in the progesterone-mediated oocyte maturation, estrogen signaling pathway, oocyte meiosis, oxytocin signaling pathway, ovarian steroidogenesis, mTOR signaling pathway, FoxO signaling pathway, Wnt signaling pathway, GnRH signaling pathway, ubiquitin-mediated proteolysis, and insulin secretion, which are known to be involved in male and female germ cell development. Some of these GO term and KEGG pathway enrichments were closely related to sex determination/differentiation or gametogenesis. At least 45 genes were identified as sex-related genes that might be involved in sex determination/differentiation or gametogenesis, and most of them were enriched in the GO function terms and KEGG pathways mentioned above (Supplementary Table 2).
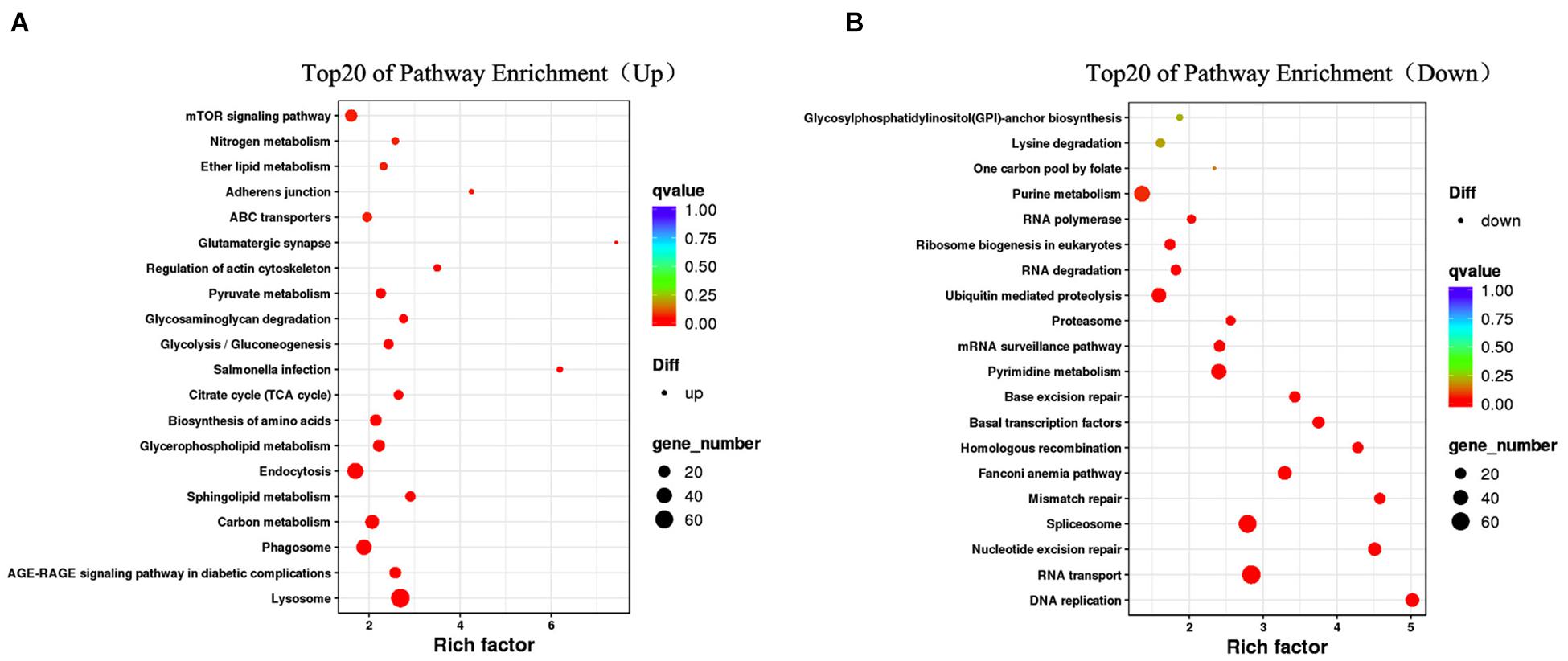
Figure 4. Top20 KEGG pathway enrichment of upregulated DEGs (A) and downregulated DEGs (B). The bigger the Rich factor or the smaller the q-value both indicates the more significant GO terms, and the bigger circle size indicates the higher number of genes.
Identification of DEGs Related to Sex Determination/Differentiation and Gonad Development
A heat map illustrating the hierarchical clustering of the genes differentially expressed between female and male gonads was conducted to visualize the overall gene expression pattern, and four clusters with similar gene expression patterns were generated (Figure 5). From the clusters, a catalog of 45 genes potentially involved in sex-determination/differentiation or gametogenesis was generated (Supplementary Table 1). The roles of these genes are unknown in S. constricta, but most of them have been identified as playing important roles in sex-determination/differentiation or gametogenesis in other organisms.
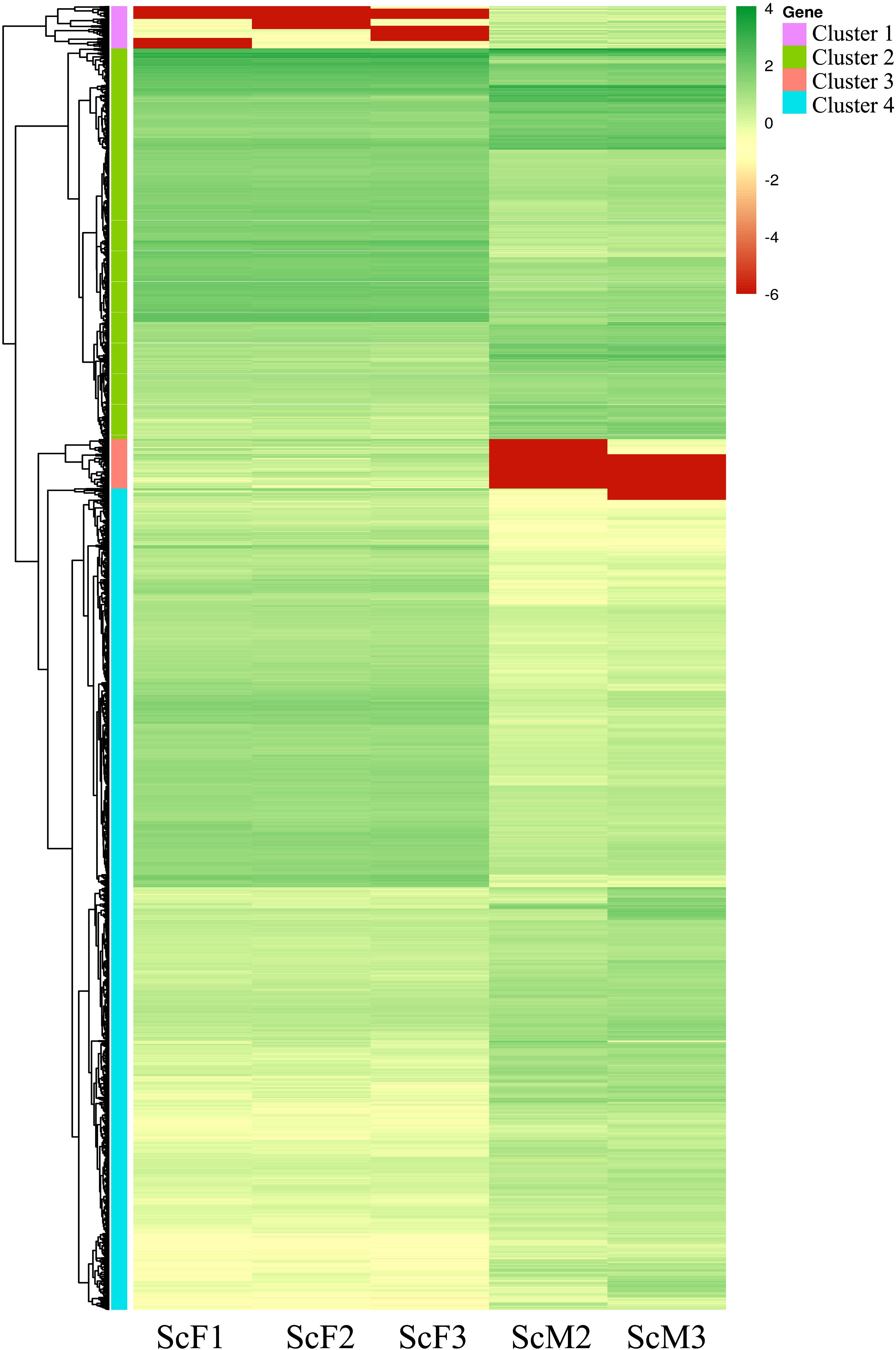
Figure 5. Heatmap of DEGs in female and male gonads. Different columns represent different samples, different rows represent different genes, and the colors represent the gene expression levels [log10(FPKM + 0.000001)] in the samples. Green represents highly expressed genes, and red represents weakly expressed genes.
Cluster 1 included 277 genes, of which 41 genes showed significant similarities to known proteins, and 64 genes possessed GO assignations. And several genes known to be involved in male sex determination/differentiation were identified, including Fem-1b (Fem-1b, ctg57.8) and Fem-1c (Fem-1c, ctg186.37). Genes such as E3 ubiquitin-protein ligase R2-like (R2, novel gene_12556) and sperm motility kinase Tcr mutant form-like (Smoktcr, ctg318.17 and ctg318.18) were also found, and they have been described as playing roles in spermatogenesis.
In cluster 2(including 2,545 genes), Beta-catenin (Beta-catenin, ctg1559.4, and ctg1559.5) and 3-oxo-5-alpha-steroid 4-dehydrogenase 1 (SRD5A1, ctg350.13) were detected, and they are known to be involved in sex determination/differentiation. Genes involved in oogenesis and oocyte maturation were also found, including mitotic apparatus protein p62-like isoform X2 (MP62, ctg197.11) and Lis1 (Lis1, ctg955.10). Some genes involved in catalyzing steroid biosynthesis and metabolism were also found, such as 17beta-HSD14 gene, which is of great importance to gametogenesis.
In cluster 3 (including 323 genes), we identified genes encoding proteins involved in spermatogenesis and oogenesis, such as spermatogenesis- and oogenesis-specific basic helix-loop-helix-containing protein 2 (SOHLH2, novel gene _31922) and spermatogenesis-associated protein 24-like (SPATA24, ctg9357.1). A few genes implicated in glycoprotein biosynthesis and metabolism were also found, such as vitellogenin (Vg, novel gene _30040) and vitellogenin-6 (Vg-6, novel gene _30041), which are important for female gonad development.
Of the 5352 genes in cluster 4, a large number of genes were found to be differentially expressed between females and males and involved in oogenesis or spermatogenesis. Genes potentially involved in male sex determination included double sex and mab-3 related transcription factor A2-like protein (DmrtA2, ctg423.9) and transcription factor Sox-9-like (Sox9, ctg163.27). Genes implicated in spermatid differentiation and development, including meiotic recombination protein SPO11-like (SPO11, ctg915.3) and testis-specific serine/threonine-protein (TSSK, ctg1.91; TSSK1, ctg1.92; TSSK4, ctg207.28), were found, as were genes associated with oogenesis, including cytochrome P450 26A1(CYP26A1, ctg146.9) and steroid 17-alpha-hydroxylase/17,20 lyase-like (CYP17A1, ctg67.144). Serine-protein kinase ATM-like isoform X2 (ATM, ctg637.23), which is involved in female gamete generation and oocyte development, was also identified.
Gene Expression and Sequence Analysis of Sex-Related Genes
Twelve genes were chosen for qRT-PCR analysis to validate the differential expression results identified by RNA-Seq. qRT-PCR results showed that the expression levels of DmrtA2, Sox9, Fem-1b, Fem-1c, CYP17A1, Smoktcr, TSSK, and GATA-7 in male gonads were significantly higher than those in female gonads (P < 0.05, Figure 6A), whereas SOHLH2, FoxA1, Beta-catenin, and Vg were significantly highly expressed in female gonads than in male gonads (P < 0.05, Figure 6B). The expression patterns of these DEGs validated by qRT-PCR were generally consistent with the RNA-Seq results (Figure 7), which further confirmed the reliability and accuracy of the RNA-Seq data.
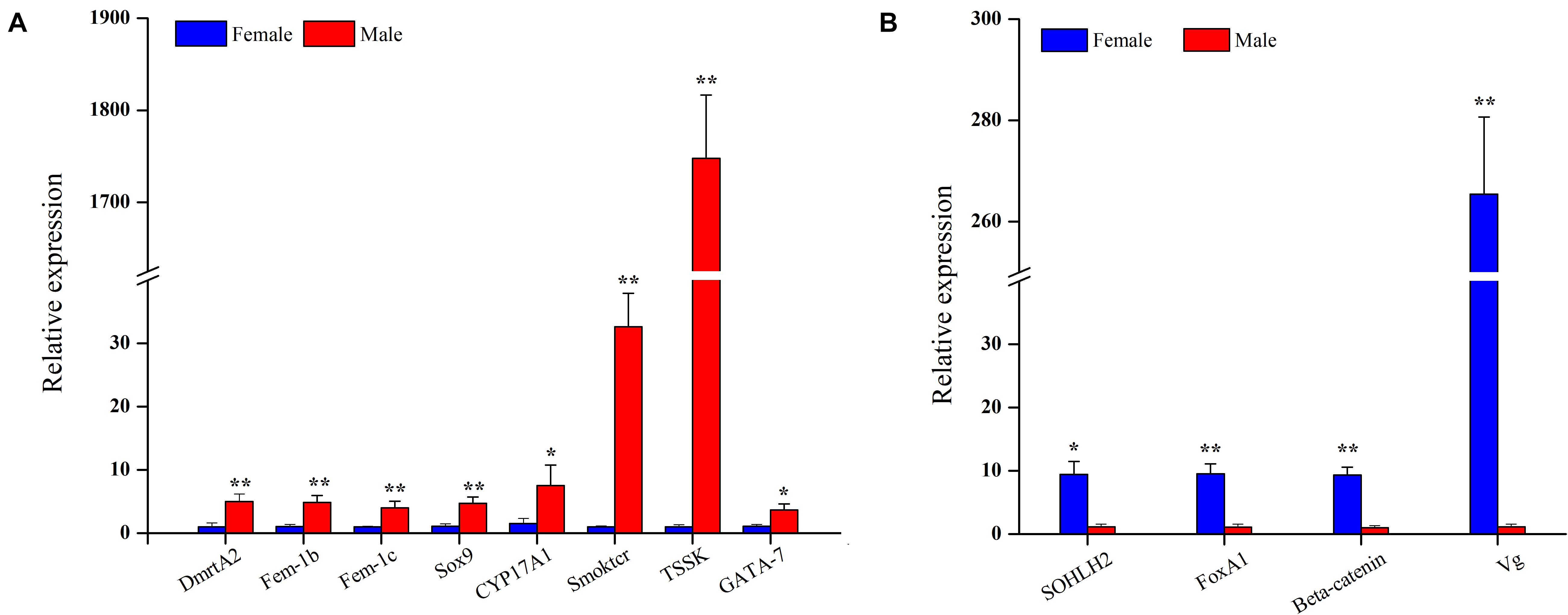
Figure 6. Relative expression level of male-biased genes (A) and female-biased genes (B) between female and male gonads in S. constricta (n = 4). Significant differences between female and male gonads are indicated with an asterisk for P < 0.05, and with two asterisks for P < 0.01.
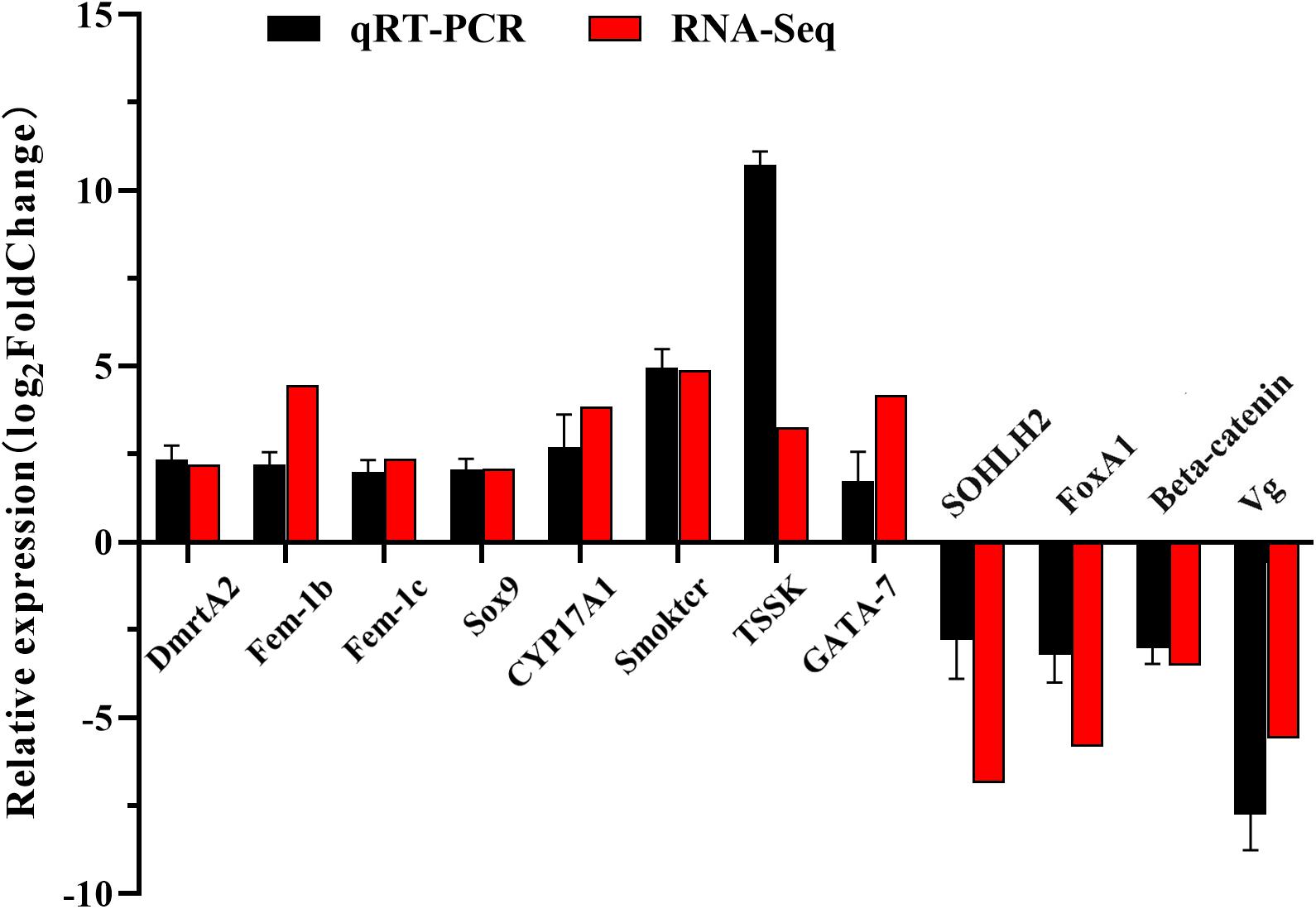
Figure 7. qRT-PCR and RNA-seq validation results. Expression comparisons of 12 sex-biased gene in female and male gonads in S. constricta. The Y-axis represented log2FoldChange determined by qRT-PCR and RNA-seq. The mean values and error bars were obtained from three biological and three technical replicates for qRT-PCR results.
To investigate potential sex-related genes of S. constricta, four crucial genes DmrtA2, Sox9, Fem-1b, and Fem-1c were further examined and analyzed, and named ScDmrtA2, ScSox9, ScFem-1b, and ScFem-1c in S. constricta, respectively.
ScDmrtA2
A full-length transcript of ScDmrtA2 (GenBank no.: MZ440691) was identified with an opening reading frame (ORF) of 1,170 bp encoding 389 amino acids. The deduced amino acids contain the DM domain (34–87 aa) consensus sequences (Figure 8A1). The phylogenetic tree showed that ScDmrtA2 was first clustered with DmrtA2 of Hyriopsis cumingii, and then grouped with the clade including scallops (Pecten maximus and Mizuhopecten yessoensis) and oysters (C. gigas and Crassostrea virginica). Finally, the molluscan DmrtA2 clade was clustered with the vertebrate DmrtA2 clade (Figure 8A2). Expression profile of ScDmrtA2 in different adult tissues revealed that ScDmrtA2 expressed significantly higher in mantle and gill (P < 0.05) than other tissues, and the expression level in male gonads is also significantly higher than that in female gonads (P < 0.05) (Figure 8A3).
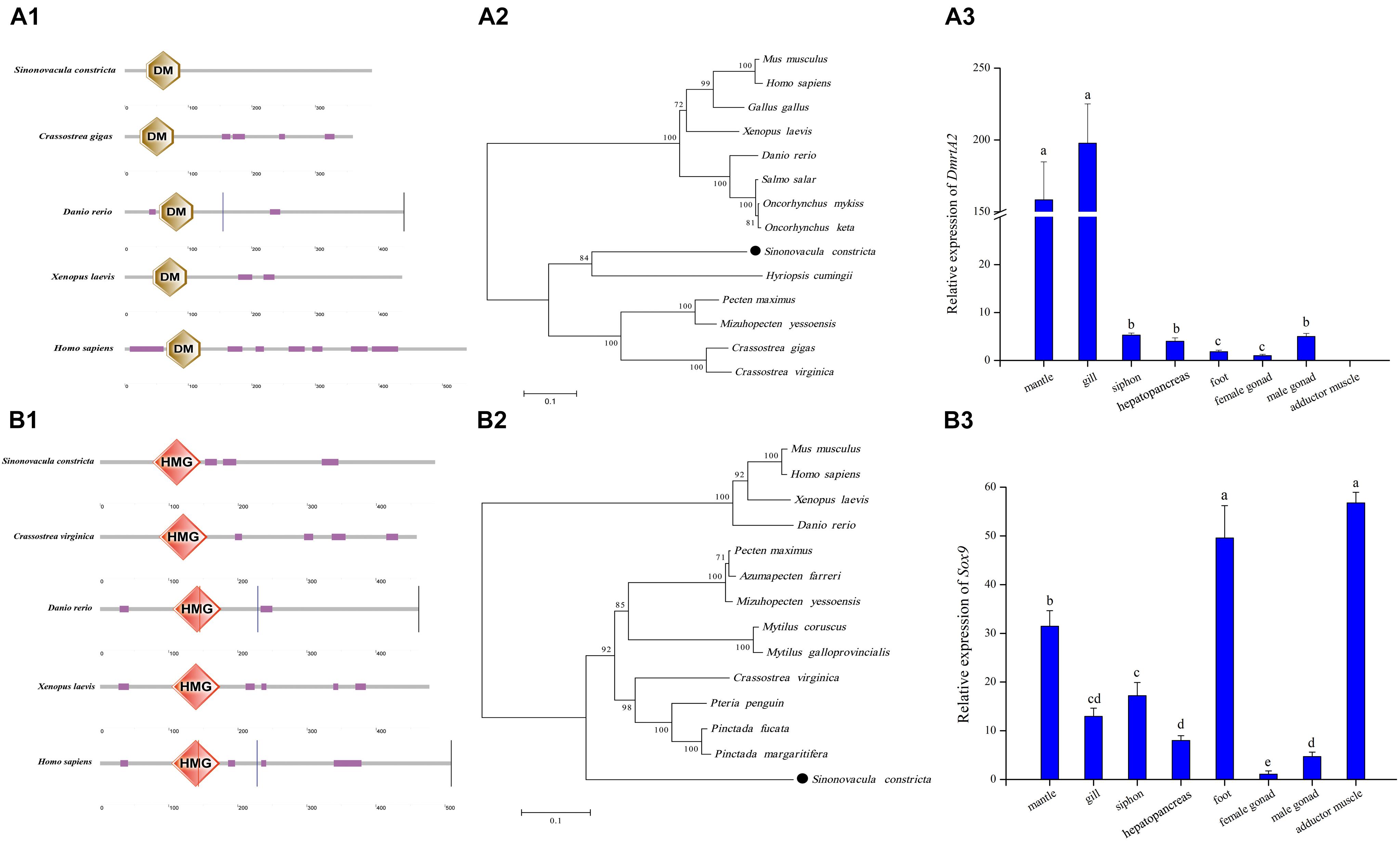
Figure 8. Analysis of ScDmrtA2 and ScSox9 protein domains and their expression profile in adult tissues with standard deviation as error bars (n = 3) in S. constricta. (A1) DmrtA2 domain structures of razor clam and selected model species. (A2) The phylogenetic tree generated with DmrtA2 proteins. (A3) Relative expression level of ScDmrtA2 in adult tissues. (B1) Sox9 domain analysis of razor clam and other model species. (B2) The phylogenetic tree generated using Sox9 proteins. (B3) Expression profile of ScSox9 in adult tissues.
ScSox9
ScSox9 (GenBank no.: MZ440690) was identified with ORF of 1,458 bp encoding 485 aa. An HMG domain consensus sequences were found from 76 to 146 aa in the deduced amino acids (Figure 8B1). The phylogenetic tree showed that ScSox9 was clustered with Sox9 of mollusks with the high bootstrap support (92), and then clustered with Sox9 of vertebrate species (bootstrap support 100) (Figure 8B2). Tissue expression analysis showed that ScSox9 was expressed in all nine examined tissues, and expressed significantly higher in foot and adductor muscle than in other tissues (P < 0.05) (Figure 8B3). Meanwhile, the relative expression level of ScSox9 in male gonads is about 4.25 folds higher than in female gonads.
ScFem-1b and ScFem-1c
Two Fem-1 family genes, Fem-1b (GenBank no.: MZ440689) and Fem-1c (GenBank no.: MZ440688), were identified with complete ORF sequences of 1,911 and 1,869 bp, respectively. The deduced amino acid sequences of Fem-1b and Fem-1c were 636 and 622 aa, respectively, both of which contain seven ankyrin (ANK) repeats at N-terminus and two ankyrin repeats at C-terminus. The ANK repeats are characteristics of the Fem family proteins (Figures 9A1,A2). The phylogenetic tree showed that ScFem-1b and ScFem-1c had closer relationships to other mollusks, and were both grouped with mollusks with high bootstrap support (Fem1b 92 and Fem-1c 100), respectively (Figure 9B). Both ScFem-1b and ScFem-1c were expressed in all tissues, and both of them expressed at highest level in male gonads among all detected tissues (P < 0.05) (Figures 9C1,C2).
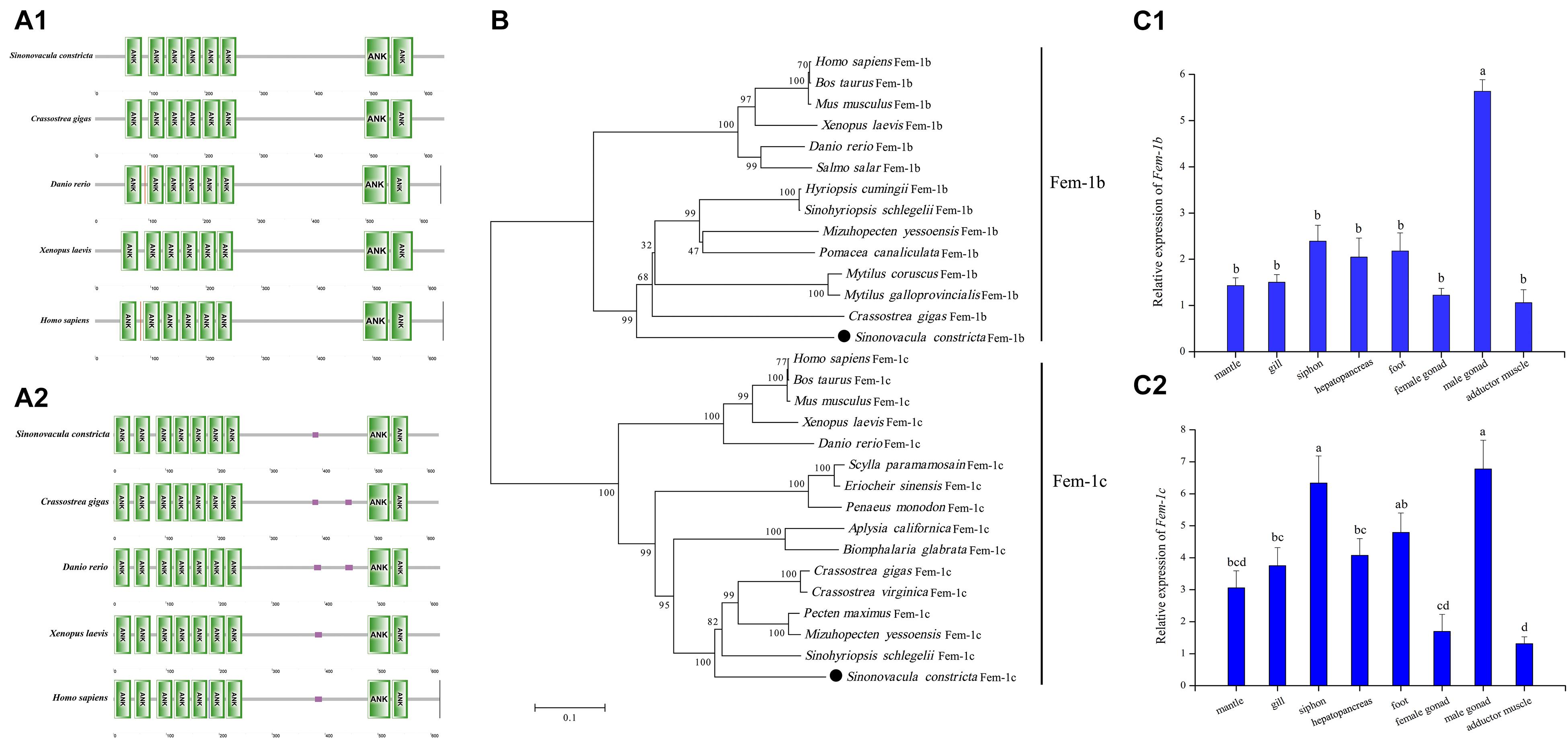
Figure 9. Sequence analysis of ScFem-1b and ScFem-1c and their expression profile in adult tissues with standard deviation as error bars (n = 3). Domain analysis of ScFem-1b (A1) and ScFem-1c (A2) with that of other model species. The phylogenetic tree of Fem-1b and Fem-1c proteins (B). Expression profile of ScFem-1b (C1) and ScFem-1c (C2) in adult tissues.
Discussion
By analyzing the female and male gonad transcriptome of S. constricta, we identified at least 45 sex-related genes, including DmrtA2, Sox9, Fem-1b, Fem-1c, and Beta-catenin potentially involved in sex determination or differentiation, and Vg, CYP17A1, TSSK, and SOHLH2 potentially involved in gametogenesis.
Potential Sex Determination/Differentiation Genes in S. constricta
Four potential male sex determining genes (DmrtA2, Sox9, Fem-1b, and Fem-1c) were all expressed significantly higher in male gonads than in female gonads (P < 0.05). In vertebrates, Dmrt is an important gene involved in sex determination/differentiation, which is a transcription factor and contains a characteristic zinc finger DM domain (Guo et al., 2005; Kopp, 2012). Mammals have at least seven DM domain genes, most of them showed obvious gonadal expression (Xu et al., 2013). DmrtA2 belongs to Dmrt family, and was expressed specially in testis in human, which suggested DmrtA2 might be involved in sexual development (Ottolenghi et al., 2002). In adult zebrafish, DmrtA2 was specially expressed in testis, ovary and brain, and the expression level is higher in testis than in ovary (Guo et al., 2004). Furthermore, zebrafish DmrtA2 expression was restricted in developing germ cells and brain, especially in spermatogonia, spermatocytes, spermatids, and sperm cells, which suggested that DmrtA2 was potentially involved in gonadal development and spermatogenesis (Guo et al., 2004; Xu et al., 2013). In this study, compared to female gonads, we detected significant elevation of DmrtA2 gene expression in the testis of S. constricta, which might be involved in male gonad development and spermatogenesis, and this would be consistent with the patterns in zebrafish. ScDmrtA2 were also detected to express significantly higher in mantle and gill than in other tissues (P < 0.05), and Yu et al. (2009) also found high expression of DmrtA2 in mantle, gill, digestive diverticulum and adductor muscle besides male and female gonads in Pinctada matensii, suggesting that DmrtA2 might exert other functions in mollusks.
We also found that the Sox9 gene, a homolog of the SoxE gene, was male-biased and therefore might be a potential sex determination gene in S. constricta. The transcription factor Sox9 containing an HMG domain, is a key gene in the mammalian testis determining pathway, and plays important roles in male gonadic differentiation and maintenance (Wagner et al., 1994; Knower et al., 2011). In P. margaritifera, the Sox9 gene was also identified as a potential sex determination gene (Teaniniuraitemoana et al., 2014). The expression profiles of SoxE in C. gigas suggested its important roles in early sex differentiation (Santerre et al., 2014).
Fem-1 gene, as a sex-determining gene for male body development and spermatogenesis, was first discovered in Caenorhabditis elegans (Ventura-Holman et al., 1998). In vertebrates, the Fem-1 family contains three conservative members: Fem-1a, Fem-1b, and Fem-1c, which are able to feminize nematode sex differentiation (Ventura-Holman and Maher, 2000; Krakow et al., 2001; Ventura-Holman et al., 2003). The Fem-1 gene identified in P. margaritifera was thought to be related to sex determination (Teaniniuraitemoana et al., 2014), and the Fem-1c gene in P. yessoensis showed upregulated expression in females (Yang et al., 2016). Using both qRT-PCR and RNA-Seq, we observed significant increases in mRNA levels of the Fem-1b and Fem-1c genes in male gonads of S. constricta, which suggested their functions in sex determination/differentiation.
FoxA1, FoxD2, and Beta-catenin were also identified to be involved in the female sex determination/differentiation in S. constricta. Fox gene family members are transcription factors and some of which play important roles in ovarian development (Shimeld et al., 2010). For example, the Foxl2 gene has been found to be involved in ovary differentiation and maintenance in vertebrates (Ottolenghi, 2005; Uhlenhaut et al., 2009). In mollusks, the Foxl2 gene has been identified in C. farreri (Liu et al., 2012), C. gigas (Naimi et al., 2009b; Zhang et al., 2014), P. yessoensis (Li et al., 2016; Yang et al., 2016), and P. margaritifera (Teaniniuraitemoana et al., 2014), and it is thought to be a master regulator of female sex determination or ovarian maintenance. However, we did not find the Foxl2 gene in our RNA-Seq results of S. constricta, although FoxA1 and FoxD2 were present and ovary-biased. In the canonical Wnt signaling pathway, Beta-catenin is a key transcriptional regulator and is essential for ovarian development and maintenance in mammals (Liu et al., 2009). Beta-catenin was found to be localized in the oocytes of the ovary, and its expression was significantly high in the ovaries in C. farreri (Li et al., 2014). Here, Beta-catenin was expressed in both male and female gonads, however, its expression level was significantly higher in the ovaries than in the testis, which suggested Beta-catenin might play important roles in ovarian differentiation and maintenance in S. constricta.
Identification of Genes Involved in Gametogenesis
In our study, some genes putatively involved in oogenesis and spermatogenesis were also identified. For example, Vg, Vg-6, and SOHLH2 were female-biased, and CYP17A1, TSSK, TSSK1, and TSSK4 were male-biased in S. constricta. Vg or Vg-6 as the storage protein and non-polar molecular carrier has been speculated to be involved in combining and transferring proteins, lipids, carotenoids and vitamin to oocytes during oogenesis (Wallace, 1985). In C. farreri and C. gigas, Vg was found to be specifically expressed in ovaries (Matsumoto et al., 2003; Qin et al., 2012). In S. constricta, the expression levels of Vg and Vg-6 were significantly higher in ovaries than in testis, suggesting their important roles in oocyte development. SOHLH2 is a universal transcription regulator of both male and female germline differentiation, with distinct and sex-specific downstream pathways that induce genes important to spermatogonia differentiation and coordinate oocyte differentiation without affecting meiosis I (Suzuki et al., 2011; Shin et al., 2017). In S. constricta, SOHLH2 was expressed in both male and female gonads, with significantly higher expression in female gonads.
CYP17A1, a cytochrome P450 monooxygenase, is involved in the biosynthesis of steroid hormones, such as corticoid and androgen, which occurred in adrenal glands and gonads. Therefore, it indicates that CYP17A1 is involved in gonad development (Yoshimoto et al., 2016). TSSK, TSSK1, and TSSK4 exhibited high expression levels in male gonads of S. constricta. TSSK family members play important roles in cytoplasm reconstruction at the late stage of spermatogenesis. Knockout of TSSK1 would result in spermatogonial or spermatocytic apoptosis deficiencies in mouse (Shang et al., 2010; Jha et al., 2013). Therefore, TSSK, TSSK1, and TSSK4 were characterized as male-specific genes in adult mature gonads, indicating their important roles in spermatogenesis.
Conclusion
In conclusion, we identified 45 sex-related genes that might be potentially involved in sex determination/differentiation or gametogenesis in S. constricta. These genes included DmrtA2, Sox9, Fem-1b, and Fem-1c for sex determination/differentiation and Vg, CYP17A1, SOHLH2, and TSSK for gametogenesis. Identification of sex-related genes not only contributes to our understanding of reproduction in S. constricta, but also may be applicable to developing methods to control reproduction to support sustainable development of the clam farming.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The Illumina transcriptome raw data are available in the Sequence Read Archive (SRA) database at NCBI under accession number SRR9937008-SRR9937010 for female gonad and SRR9937011-SRR9937013 for male gonad of S. constricta. The names of the repository/repositories and accession number(s) can be found below: NCBI Genbank, accession numbers: MZ440688, MZ440689, MZ440690, and MZ440691.
Author Contributions
YD and ZL conceived the project. XK collected the samples. HY, YD, and LH conducted the gonad transcriptome assembly, annotation, and transcriptome analysis. HY wrote the manuscript. LX revised the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2018YFD0901405), the National Natural Science Foundation of China (31902393), the National Marine Genetic Resource Center Program, the Ningbo Major Project of Science and Technology (2019B10005), and the China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank Hongqiang Xu for the sample collection, Dr. Wenfang Dai for her suggestions on data analysis, and we thank Dr. Jianfeng Ren for editing our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.725430/full#supplementary-material
Supplementary Table 1 | Forty-five sex-related genes identified in S. constricta.
Supplementary Table 2 | The information sequences used in this study.
Footnotes
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1038/npre.2010.4282.1
Chen, C., Xiao, G. Q., Chai, X. L., Lin, X. G., Fang, J., and Teng, S. S. (2017). Transcriptome analysis of sex-related genes in the blood clam Tegillarca granosa. PLoS One 12:e0184584. doi: 10.1371/journal.pone.0184584
Dong, Y. H., Zeng, Q. F., Ren, J. F., Yao, H. H., Lv, L. Y., He, L., et al. (2020). The chromosome-level genome assembly and comprehensive transcriptomes of the razor clam (Sinonovacula constricta). Front. Genet. 11:664. doi: 10.3389/fgene.2020.00664
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data 481 without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, Y., Cheng, H., Huang, X., Gao, S., Yu, H., and Zhou, R. (2005). Gene structure, multiple alternative splicing, and expression in gonads of zebrafish Dmrt1. Biochem. Biophys. Res. Commun. 330, 950–957. doi: 10.1016/j.bbrc.2005.03.066
Guo, Y., Qin, L., Shang, G., Xiang, Z., and Zhou, R. (2004). Molecular cloning, characterization, and expression in brain and gonad of Dmrt5 of zebrafish. Biochem. Biophys. Res. Commun. 324, 569–575. doi: 10.1016/j.bbrc.2004.09.085
Jha, K. N., Coleman, A. R., Wong, L., Salicioni, A. M., Howcroft, E., and Johnson, G. R. (2013). Heat shock protein 90 functions to stabilize and activate the testis-specific serine/threonine kinases, a family of kinases essential for male fertility. J. Biol. Chem. 288, 16308–16320. doi: 10.1074/jbc.M112.400978
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/NMETH.3317
Knower, K. C., Kelly, S., Ludbrook, L. M., Bagheri-Fam, S., Sim, H., Bernard, P., et al. (2011). Failure of SOX9 regulation in 46XY disorders of sex development with SRY, SOX9 and SF1 mutations. PLoS One 6:e17751. doi: 10.1371/journal.pone.0017751
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175–184. doi: 10.1016/j.tig.2012.02.002
Krakow, D., Sebald, E., King, L. M., and Cohn, D. H. (2001). Identification of human FEM1A, the ortholog of a C. elegans sex-differentiation gene. Gene 279, 213–219. doi: 10.1016/S0378-1119(01)00756-9
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Zhang, Z., Bi, Y., Yang, D., Zhang, L., and Liu, J. (2014). Expression characteristics of beta-catenin in scallop Chlamys farreri gonads and its role as a potential upstream gene of dax1 through canonical wnt signalling pathway regulating the spermatogenesis. PLoS One 9:e115917. doi: 10.1371/journal.pone.0115917
Li, Y., Zhang, L., Sun, Y., Ma, X. L., Wang, J., Li, R. J., et al. (2016). Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop Patinopecten yessoensis. Mar. Biotechnol. 18, 453–465. doi: 10.1007/s10126-016-9706-8
Liu, C. F., Bingham, N., Parker, K., and Yao, H. H. (2009). Sex-specific roles of beta-catenin in 794 mouse gonadal development. Hum. Mol. Genet. 18, 405–417.
Liu, X. L., Zhang, Z. F., Shao, M. Y., Liu, J. G., and Muhammad, F. (2012). Sexually dimorphic expression of foxl2 during gametogenesis in scallop Chlamys farreri, conserved with vertebrates. Dev. Genes Evol. 222, 279–286. doi: 10.1007/s00427-012-0410-z
Matsumoto, T., Nakamura, A. M., Mori, K., and Kayano, T. (2003). Molecular characterization of a cDNA encoding putative vitellogenin from the Pacific oyster Crassostrea gigas. Zool. Sci. 20, 37–42. doi: 10.2108/zsj.20.37
Morton, B. (1984). The functional morphology of Sinonovacula constricta with a discussion on the taxonomic status of the Novaculininae (Bivalvia). J. Zool. Lond. 202, 299–325. doi: 10.1111/j.1469-7998.1984.tb05085.x
Moullac, L. G., Soyez, C., Sham-Koua, M., Levy, P., Moriceau, J., Vonau, V., et al. (2013). Feeding the pearl oyster Pinctada margaritifera during reproductive conditioning. Aquac. Res. 44, 404–411. doi: 10.1111/j.1365-2109.2011.03045.x
Naimi, A., Martinez, A. S., Specq, M. L., Diss, B., Mathieu, M., and Sourdaine, P. (2009a). Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 134–142. doi: 10.1016/j.cbpb.2009.05.011
Naimi, A., Martinez, A. S., Specq, M. L., Mrac, A., Diss, B., Mathieu, M., et al. (2009b). Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp. Biochem. Phys. A 152, 189–196. doi: 10.1016/j.cbpa.2008.09.019
Niu, D., Wang, L., Sun, F., Liu, Z., and Li, J. (2013). Development of molecular resources for an intertidal clam, Sinonovacula constricta, using 454 transcriptome sequencing. PLoS One 8:e67456. doi: 10.1371/journal.pone.0067456
Ottolenghi, C. (2005). Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet. 14, 2053–2062. doi: 10.1093/hmg/ddi210
Ottolenghi, C., Fellous, M., Barbieri, M., and McElreavey, K. (2002). Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79, 333–343. doi: 10.1006/geno.2002.6711
Penman, D. J., and Piferrer, F. (2008). Fish gonadogenesis. Part I: genetic and environmental mechanisms of sex determination. Rev. Fish. Sci. 16, 16–34. doi: 10.1080/10641260802324610
Piferrer, F., and Guiguen, Y. (2008). Fish gonadogenesis. Part II: molecular biology and genomics of sex differentiation. Rev. Fish. Sci. 16, 35–55. doi: 10.1080/10641260802324644
Qin, Z., Li, Y., Sun, D., Shao, M., and Zhang, Z. (2012). Cloning and expression analysis of the vitellogenin gene in the scallop Chlamys farreri and the effects of estradiol-17b on its synthesis. Invertebr. Biol. 131, 312–321. doi: 10.1111/ivb.12006
Ran, Z., Li, Z., Yan, X., Liao, K., Kong, F., Zhang, L., et al. (2019). Chromosome-level genome assembly of the razor clam Sinonovacula constricta (Lamarck, 1818). Mol. Ecol. Resour. 19, 1647–1658. doi: 10.1111/1755-0998.13086
Santerre, C., Sourdaine, P., Adeline, B., and Martinez, A. S. (2014). Cg-SoxE and Cg-β-catenin, two new potential actors of the sex-determining pathway in a hermaphrodite lophotrochozoan, the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 167, 68–76. doi: 10.1016/j.cbpa.2013.09.018
Shang, P., Baarends, W. M., Hoogerbrugge, J., Ooms, M., Cappellen, W. A., Antonius, A. W., et al. (2010). Functional transformation of the chromatoid body in mouse spermatids requires testis-specific serine/threonine kinases. J. Cell. Sci. 123, 331–339. doi: 10.1242/jcs.059949
Shen, S., Park, J. W., Lu, Z. X., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601. doi: 10.1073/pnas.1419161111
Shimeld, S. M., Boyle, M. J., Brunet, T., Luke, G. N., and Seaver, E. C. (2010). Clustered Fox genes in lophotrochozoans and the evolution of the bilaterian Fox gene cluster. Dev. Biol. 340, 234–248. doi: 10.1016/j.ydbio.2010.01.015
Shin, Y. H., Ren, Y., Suzuki, H., Golnoski, K. J., Ahn, H. W., Mico, V., et al. (2017). Transcription factors SOHLH1 and SOHLH2 coordinate oocyte differentiation without affecting meiosis I. J. Clin. Invest. 127, 2106–2117. doi: 10.1172/JCI90281
Suzuki, H., Ahn, H. W., Chu, T., Bowden, W., Gassei, K., Orwig, K., et al. (2011). SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 361, 301–312. doi: 10.1016/j.ydbio.2011.10.027
Teaniniuraitemoana, V., Huvet, A., Levy, P., Klopp, C., Lhuillier, E., Gaertner-Mazouni, N., et al. (2014). Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics 15:491. doi: 10.1186/1471-2164-15-491
Uhlenhaut, N. H., Jakob, S., Anlag, K., Eisenberger, T., Sekido, R., Kress, J., et al. (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142. doi: 10.1016/j.cell.2009.11.021
Vahirua-Lechat, I., Laure, F., LeCoz, J. R., Bianchini, J. P., Bellais, M., and Le Moullac, G. (2008). Changes in fatty acid and sterol composition during oogenesis in the pearl oyster Pinctada margaritifera. Aquac. Res. 39, 1739–1746. doi: 10.1111/j.1365-2109.2008.02050.x
Ventura-Holman, T., Lu, D., Si, X., Izevbigie, E. B., and Maher, J. F. (2003). The Fem1c genes: conserved members of the Fem1 gene family in vertebrates. Gene 314, 133–139. doi: 10.1016/S0378-1119(03)00712-1
Ventura-Holman, T., and Maher, J. F. (2000). Sequence, organization, and expression of the human FEM1B gene. Biochem. Biophys. Res. Commun. 267, 317–320. doi: 10.1006/bbrc.1999.1942
Ventura-Holman, T., Seldin, M. F., Li, W., and Maher, J. F. (1998). The murine fem1 gene family: homologs of the Caenorhabditis elegans sex-determination protein FEM-1. Genomics 54, 221–230. doi: 10.1111/j.1365-2109.2008.02050.x
Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., et al. (1994). Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120. doi: 10.1016/0092-8674(94)90041-8
Wallace, R. A. (1985). “Vitellogenesis and oocyte growth in nonmammalian vertebrates,” in Oogenesis Developmental Biology, Vol. 1, ed. L. W. Browder (Boston MA: Springer), 127–177. doi: 10.1007/978-1-4615-6814-8-3
Wang, R. C., Wang, Z. P., and Zhang, J. Z. (1993). Marine Shellfish Aquaculture. Qingdao: Qingdao Ocean University Press, 322–324.
Wang, W. X., and Xu, Z. Z. (1997). Larval swimming and postlarval drifting behavior in the infaunal bivalve Sinonovacula constricta. Mar. Ecol. Prog. Ser. 148, 71–81. doi: 10.3354/meps148071
Wu, H. X., and Xu, A. G. (2000). A test on artificial cultivation of Sinonovacula constricta (Lamark). Mar. Sci. 24, 15–17.
Xu, S., Xia, W., Zohar, Y., and Gui, J. F. (2013). Zebrafish dmrta2 regulates the expression of cdkn2c in spermatogenesis in the adult testis. Biol. Reprod. 88:14. doi: 10.1095/biolreprod.112.105130
Yang, D., Yin, C., Chang, Y. Q., Dou, Y., Hao, Z. L., and Ding, J. (2016). Transcriptome analysis of male and female mature gonads of Japanese scallop Patinopecten yessonsis. Genes Genomics 38, 1041–1052. doi: 10.1007/s13258-016-0449-8
Yoshimoto, F. K., Gonzalez, E., Auchus, R. J., and Guengerich, F. P. (2016). Mechanism of 17α,20-lyase and new hydroxylation reactions of human cytochrome P450 17A1. J. Biol. Chem. 291, 17143–17164. doi: 10.1074/jbc.M116.732966
Yu, F. F., Gui, J. F., Zhou, L., Wang, M. F., and Yu, X. Y. (2009). Cloning and expression characterization of Dmrt5 in Pinctada Martensii. Acta. Hydrobiol. Sin. 33, 844–850. doi: 10.3724/SP.J.0000.2009.50844
Zhang, N., Xu, F., and Guo, X. (2014). Genomic analysis of the Pacific oyster (Crassostrea gigas) reveals possible conservation of vertebrate sex determination in a mollusc. G3 (Bethesda) 4, 2207–2217. doi: 10.1534/g3.114.013904
Keywords: Sinonovacula constricta, gonad, transcriptome analysis, sex determination/differentiation, gametogenesis
Citation: Yao H, Lin Z, Dong Y, Kong X, He L and Xue L (2021) Gonad Transcriptome Analysis of the Razor Clam (Sinonovacula constricta) Revealed Potential Sex-Related Genes. Front. Mar. Sci. 8:725430. doi: 10.3389/fmars.2021.725430
Received: 15 June 2021; Accepted: 11 August 2021;
Published: 30 August 2021.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, Chinese Academy of Sciences, ChinaReviewed by:
Xiaoting Huang, Ocean University of China, ChinaZhiguo Dong, Jiangsu Ocean University, China
Copyright © 2021 Yao, Lin, Dong, Kong, He and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghui Dong, ZG9uZ3lpbmdodWkxMThAMTI2LmNvbQ==; Liangyi Xue, eHVlbGlhbmd5aUBuYnUuZWR1LmNu
 Hanhan Yao1,2
Hanhan Yao1,2 Yinghui Dong
Yinghui Dong