- 1Marine Ecosystem Research Center, Korea Institute of Ocean Science and Technology, Busan, South Korea
- 2Department of Aquatic Life Medicine, Pukyong National University, Busan, South Korea
- 3Department of Biology, Gangneung-Wonju National University, Gangneung, South Korea
- 4Smithsonian Environmental Research Center, Edgewater, MD, United States
A tintinnid species, Helicostomella longa, infected by the parasitic dinoflagellate Euduboscquella triangula n. sp. was discovered from the southern coast of Korea in August of 2015 and 2016. Parasite morphology and development were analyzed by observation of live cells and protargol-stained specimens. The parasite was determined to be a new species in the genus Euduboscquella based on morphological and molecular data. A representative sequence of the novel species clustered in Euduboscquella group I. The morphological and developmental features of E. triangula were distinguished from those of its congeners by: (1) numerous shallow and intertwining grooves on an inconspicuous shield; (2) sporocytes initially forming a short chain, but separating after the second or third division regardless of spore type; (3) production of motile mushroom-shaped dinospores, non-motile spherical spores, and non-motile triangular spores. Dinospores were formed by ca. 28% of infections, while both non-motile spherical and triangular spores occurred at a frequency of ca. 36%. All spore types showed completely identical 18S rDNA sequences. Parasite prevalence was 15.5 and 8.3% on 17 and 24 August of 2015, respectively, with infection intensity on both dates being 1.3.
Introduction
The genus Euduboscquella (Coats et al., 2012) was erected for syndinian dinoflagellates previously included in the genus Duboscquella (Chatton, 1920). Reassignment of the syndinian species left the latter genus with a single dinokaryote species, Duboscquella (Gymnodinium) tintinnicola (Lohmann, 1908; Chatton, 1920), believed to act as an extracellular parasite of the tintinnid Cyttarocylis helix (basionym: Tintinnus helix; Claparède and Lachmann, 1858). By contrast, the nine current species of Euduboscquella are intracellular parasites of protists, with six described from tintinnid hosts, one from aloricate ciliates, and two from dinoflagellates (Chatton, 1952; Cachon, 1964; Coats, 1988; Coats et al., 2012; Jung et al., 2016). Of the species infecting tintinnids, four (E. anisospora, E. cnemata, E. costata, and E. crenulata) are known from only a single host taxon, while two (E. aspida and E. cachoni) have been reported from multiple host species (Cachon, 1964; Bachvaroff et al., 2012). In addition, a single tintinnid species can be parasitized by more than one species of Euduboscquella (Coats and Bachvaroff, 2013).
Environmental rDNA sequences for marine microbial eukaryotes have revealed pan-global distribution, high abundance, and striking diversity of Marine Alveolate (MALV) Group I, which encompasses the syndinean genera Euduboscquella and Ichthyodinium (Harada et al., 2007; Guillou et al., 2008; Bachy et al., 2011; Bachvaroff et al., 2012; de Vargas et al., 2015). Diversity of sequences within the Euduboscquella subclade of MALV Group I includes numerous cryptic species. Similarly, comparison of host and parasite rDNA phylogenies based on single cell isolates indicates that multiple tintinnid species are parasitized by undescribed Euduboscquella morphospecies (Bachvaroff et al., 2012).
Parasites infecting tintinnids have been reported worldwide, except for Australia and Antarctica (Coats and Bachvaroff, 2013; Jung et al., 2016), with most studies of Euduboscquella species conducted in Atlantic or Mediterranean environments (Chatton, 1952; Cachon, 1964; Coats, 1988; Bachvaroff et al., 2012; Coats et al., 2012). Horiguchi et al. (2006) were the first to clearly document the presence of Euduboscquella species in Asian waters by publishing a photograph of Euduboscquella aspida infecting Favella ehrenbergii from Rumoi, Hokkaido, Japan. The following year, Harada et al. (2007) provided brief morphological description and 18S rDNA sequences of Euduboscquella species parasitizing F. ehrenbergii in Japanese waters. Subsequently, Jung et al. (2016) described E. costata infecting Schmidingerella arcuata in Korean coastal waters and reported the 18S rDNA sequence of their new species to be identical with that of Euduboscquella sp. infecting S. arcuata collected from Assawoman Bay, United States (Bachvaroff et al., 2012). Bachvaroff et al. (2012) also, reported genetic analysis of Euduboscquella sp. infecting Favella panamensis from Masan Bay, Korea, without providing morphological description. Lastly, Amphorellopsis acuta, Helicostomella sp., Tintinnopsis sp., and T. kofoidi infected with parasites were reported from coastal waters of Russia and Japan adjacent to Korea (Hada, 1932; Konovalova, 2007); however, descriptions provided were insufficient for positive identification of the parasites as species of Euduboscquella. Therefore, many cryptic parasites may exist in coastal waters surrounding of Korea.
In the present study, we advance understanding of parasite diversity in tintinnids by describing a new species of Euduboscquella infecting Helicostomella longa from the southern coast of Korea. To describe the new species, we analyzed the morphology and development of life-history stages through observation of live cells and protargol-stained specimens. We also construct an 18S rDNA phylogeny and analyzed internal transcribed spacer 2 (ITS2) secondary structures to assess relationships between the new species and previously reported Euduboscquella species. Additionally, we estimated the potential impact of parasitism on host populations using data for parasite development time, parasitic prevalence, and infection intensity.
Materials and Methods
Sample Collection
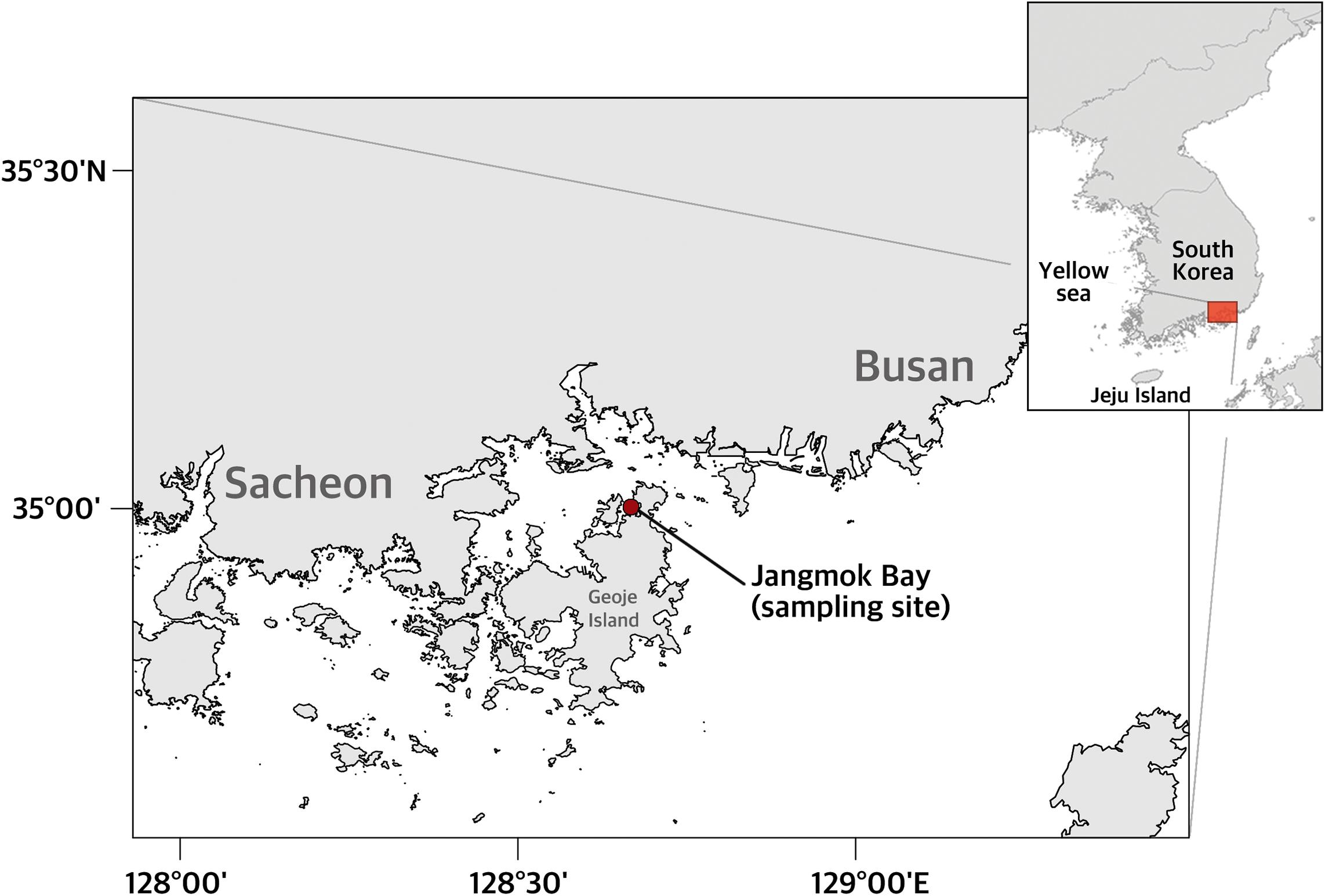
Surface water and net-tow samples were collected from Jangmok Bay, South Korea (34°59′38″N, 128°40′28″E) in August 2015 and 2016 (Figure 1). Surface water was obtained from a depth of 0.5 m using a 4-L Niskin sampler and distributed to each of two 2-L bottles. Vertical net-tow samples were collected from the upper 5 m using a 20-μm mesh plankton net and distributed to each of two 500-ml bottles. One sample for each set of bottles was immediately fixed with neutral Lugol’s iodine solution (final concentration of 0.04% iodine + 0.06% potassium iodide, w/v) and the other sample fixed with Bouin’s fluid (final concentration of 10%, w/v). The two types of preserved samples were stored at 4°C and room temperature, respectively. On selected dates when the tintinnid H. longa was abundant and infected by Euduboscquella triangula n. sp., additional, unfixed net-tow samples were collected, transported to the laboratory within 10 min, and maintained at near ambient seawater temperature for live observation. Water temperature and salinity were measured using a YSI (YSI Professional Plus Water Quality Instrument, YSI Incorporated, Yellow Springs, OH, United States) or a CTD (SBE 19plus, SeaBird Electronics, Washington, DC, United States).
In vivo and Cytological Observations
For live observation of parasite morphology and development, unfixed net-tow samples collected on August 2 and 3, 2016 and August 16, 2019 were transferred to a six-well plate and scanned at 100–320× using a Zeiss Axiovert 10 (Carl Zeiss Inc., Oberkochen, Germany) equipped with an AxioCam ICc1 rev. 4 to locate and photograph infected H. longa cells. Infected H. longa were isolated by fine glass microcapillaries, washed at least three times using 0.20-μm filtered site water (Advantec DISMIC 25CS020AS, Toyo Co., Tokyo, Japan), and transferred individually to wells of 96-well plates (SPL Life Sciences, Yeoju-gun, South Korea). To document developmental processes and life-history stages, specimens were periodically observed, photographed, and video recorded. For more detailed observation of morphology, the specimens were transferred to microscope slides and examined at 1000× using a Zeiss Axio Imager A2 and AxioCam ICc5 rev. 1 (Carl Zeiss Inc., Oberkochen, Germany).
To avoid observation of abnormal morphology caused by culture conditions, we confirmed the major characteristics (i.e., spore shape and shield grooves) using Lugol and Bouin’s preserved samples. Lugol-fixed specimens were de-stained for at least 15 min using 2% (w/v) sodium thiosulfate and then washed with distilled water. The Bouin’s preserved specimens were washed three times using distilled water only. Morphology of de-stained and washed specimens were examined as above.
For cytological observations, Bouin’s preserved samples collected on August 17 and 24, 2015 were processed using a silver impregnation technique, the Quantitative Protargol Staining of Montagnes and Lynn (1993) and examined using a Zeiss Axio Imager A2 as above. All morphological measurements were obtained using AxioVision SE64 4.9.1 software (Carl Zeiss Inc., Oberkochen, Germany), with mean values ± standard error (SE) of the mean provided in the text along with sample size (n). For assessment of relationships between nuclear attributes and trophont size, 20 suitably oriented and stained specimens were randomly selected and measured for each of five trophont size-categories (0–5, >5–10, >10–15, >15–20, >20–25 μm), with care taken to note whether specimens were from host cells infected by one or more parasites. Similar observations were made for seven specimens >25 μm in size. Other morphological attributes for E. triangula n. sp., parasite prevalence (percent of cells infected), and infection intensity (number parasites per infected cell) were determined by examining all specimens in protargol preparations (a total of 20 slides). Graphics and statistical comparisons were performed using OriginPro 2019b (OriginLab Corp., Northampton, MA, United States).
Molecular Analyses and Phylogeny
A Lugol-fixed sporocyte of E. triangula n. sp. isolated from the August 24, 2015 sample and living spores derived from specimens isolated and observed as described above were washed several times with sterile, filtered sea water and transferred to microcentrifuge tubes to produce 11 samples, 10 of which contained multiple spores, with each sample representing a different specimen of E. triangula n. sp. Genomic DNA was extracted from each sample using a RED-Extract-N-Amp Tissue PCR Kit (Sigma, St. Louis, MO, United States) and amplified by PCR. To amplify SSU to LSU rDNA regions, The PCR primers used were a modified EukA (5′-CTG GTT GAT YCT GCC AGT-3′) forward primer (Jung et al., 2012) and LSU rev. 4 (5′-GTT AGA CTY CTT GGT CCG TG-3′) reverse primer (Sonnenberg et al., 2007). Optimized PCR conditions were as follows: denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s with annealing at 58°C for 30 s and extension at 72°C for 3 min, and then a final extension step at 72°C for 7 min. No nested PCR was conducted. Three internal primers Dino18SF780 (5′-ACT TTG AGR AAA WBR GAG TG-3′), Dino18SF1470 (5′-CAG GTC TGT GAT GCC CTT MG-3′), and Dino18SR380 (5′-CTT CCT TAG ATG TGG TAG CC-3′) were used for DNA sequencing (Jung et al., 2016), with sequencing performed using an ABI 3700 sequencer (Applied Biosystems, Foster City, CA, United States). Geneious R10.2.5 (Kearse et al., 2012) and its internal plugin ClustalW (Thompson et al., 1994) were used to assemble and align sequence fragments with Euduboscquella spp. and other relevant sequences available from GenBank. p-Distances were calculated using MEGA v7.0.26 (Kumar et al., 2016). To determine an appropriate DNA substitution model for Bayesian inference (BI) and maximum likelihood (ML) analyses, we used the Akaike information criterion to identify the best-fit model using jModelTest 2.1.10 (Darriba et al., 2012). The model selected was GTR + I (0.3370) + G (0.5700). We evaluated phylogenetic relationships by using BI and ML analyses. BI assessment was performed using MrBayes 3.2.6, simulating a Markov chain Monte Carlo (MCMC) for 1,000,000 generations, 300,000 of which were discarded as burn-in (Ronquist et al., 2012). For ML analysis, PhyML version 3.1 was used (Guindon et al., 2010). Confidence in the resulting relationship was assessed using bootstrap procedure with 1000 replications for ML. The novel sequences were deposited in NCBI GenBank under the accession numbers MN388913 to MN388923.
Prediction of ITS2 Secondary Structure
We constructed ITS2 secondary structure with reference to Amoebophrya (HM483395), the closest taxon belonging to the syndinean group which was retrieved from the ITS2 Ribosomal RNA database (Ankenbrand et al., 2015). Secondary structure of the ITS2 sequences of E. triangula n. sp. (MN388913 to MN388923) and three congenic species (JN606065, JN934984, and JN934990) were built by energy minimization and constraint folding using the Mfold v2.3 program (Zuker, 2003). The predicted structures were edited and visualized using VARNA (Visualization Applet for RNA v3.93, Darty et al., 2009). We performed comparative analyses based on the E. triangula structure and classified the base changes into four types as follows: (1) compensatory base changes (CBCs; substitutions in both positions that retain canonical pair); (2) hemi-CBCs (canonical base pair to wobble base pair); (3) non-CBCs (canonical pair or wobble pair to any non-canonical pair); and (4) single nucleotide polymorphisms (SNPs).
Taxonomic Considerations
The International Code of Zoological Nomenclature (International Commission on Zoological Nomenclature, 1999) was followed for describing and naming E. triangula n. sp. The ZooBank LSID (Life Science Identifiers) of the new species has been registered in ZooBank.1
Results
In vivo Morphology and Development of Euduboscquella triangula n. sp.
Well-developed trophonts of E. triangula (39 μm maximum dimension) appeared as ovoid bodies in the host. Under differential interference contrast microscopy, the parasite had a translucent quality, with relatively homogeneous cytoplasm containing a few small granules and a large, ovoid nucleus (maximum size 14 by 12 μm) (Figure 2A). The shield (= episome) of the trophont was inconspicuous until very late in the infection cycle, and the margin of the shield was always difficult to discern. Numerous shallow, closely spaced intertwining grooves were visible on the surface of the shield for only a short time before the trophont began to emerge from its host cell (Figure 2B). The grooves were also evident in Lugol-fixed cells de-stained with sodium thiosulfate (Figure 2K). The tomont emerged latero-posteriorly from the infected zooid of H. longa and simultaneously began to ingest their host cell (Figure 2C). Emergence and ingestion of host material required 10–20 min, during which time the host continuously rotated about its longitudinal axis. Early in the emergence process, the parasite took on an elongate sac-like form, opening toward the oral end of the host lorica (see Supplementary Video 1). The sac remained open for a few minutes (5–10 min) during which time the host and parasite rotated in unison as host material was slowly drawn into the developing parasite food vacuole. The sac eventually closed, constricting and separating the remaining host cell. The parasite then stopped moving, but the host continued to rotate, resulting in separation of the two cells. Typically, the anterior portion of the host cell, including the oral ciliature, remained intact after the ingestion process, but usually persisted for only a short time. Excluding the oral ciliature, size of host cells averaged 51.0 ± 1.7 by 18.2 ± 0.7 μm (n = 3; range: 48.0–54.0 by 17.2–19.5 μm) immediately before parasite emergence and 11.6 ± 1.5 by 15.1 ± 0.4 μm (n = 3; range: 9.1–14.3 by 14.6–15.8 μm) immediately afterwards. Comparison of host cell volume assuming cylindrical geometry indicated that 84.1 ± 2.7% of host biomass (n = 3; range 79.7–88.9%) was converted to parasite biomass.
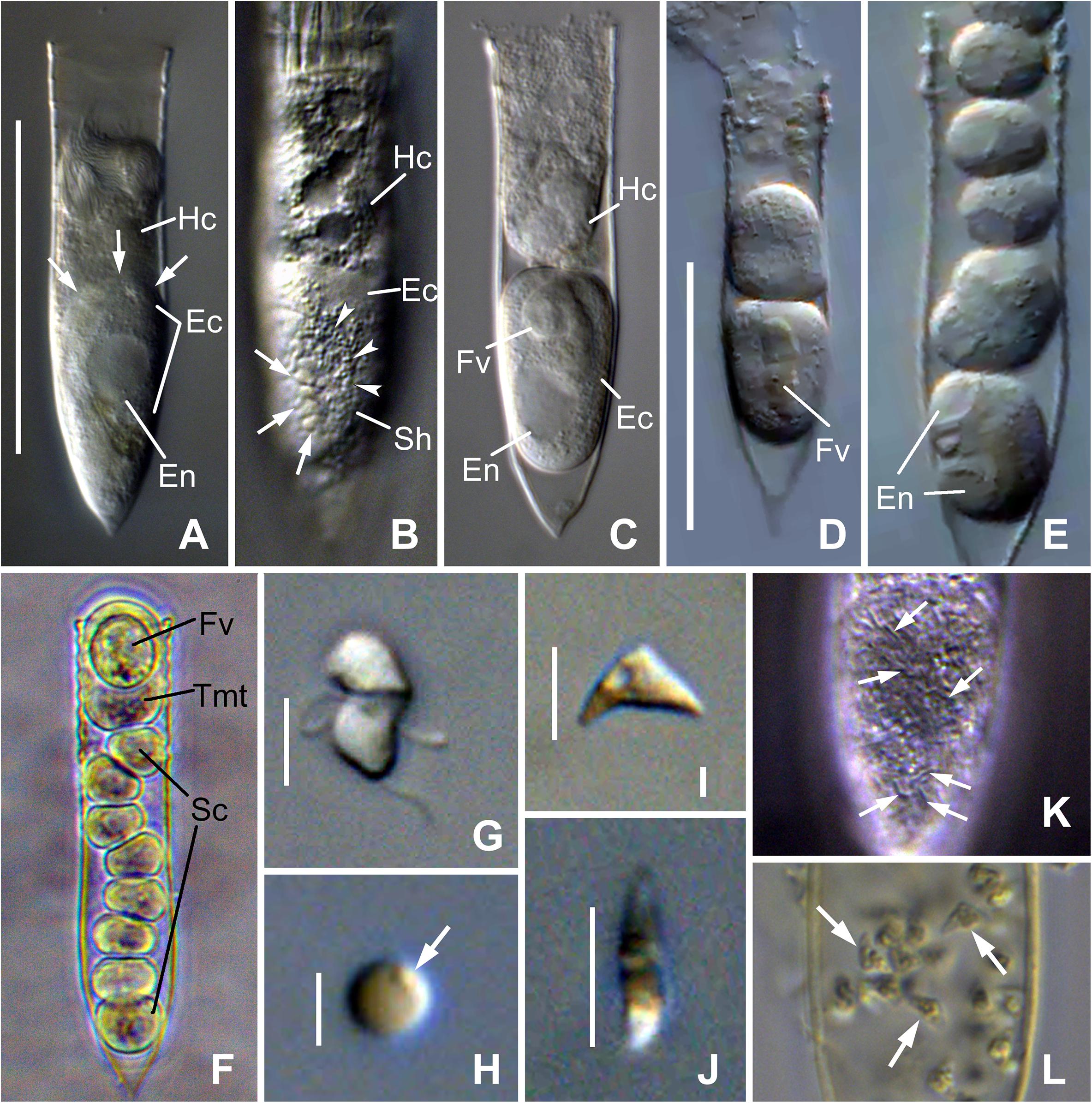
Figure 2. Photomicrograph of Euduboscquella triangula n. sp. from live (A–J), Lugol-fixed (K), and Bouin-fixed (L) specimens. (A) Lorica containing infected host cell with a late trophont of E. triangula (arrows indicate the margins of parasite cytoplasm). (B) Trophont showing shield margins (arrowheads) with intertwining grooves (arrows). (C) Tomont emerged from host with food vacuole. (D,E) Sporogenic division. (F) One tomont and eight sporocytes resulting from different trophonts of a double infected host. (G) Mushroom-shaped dinospore. (H) Non-motile spherical spore with granule on cell surface (arrow). (I,J) Lateral (I) and apical (J) view of non-motile triangular-shaped spores. (K) Shield with intertwining grooves (arrows) in Lugol-fixed specimen. (L) Triangular-shaped spores (arrows) in host lorica from field sample fixed with Bouin immediately after collection. Ec, Euduboscquella cytoplasm; En, Euduboscquella nucleus; Fv, food vacuole; Hc, Helicostomella longa cytoplasm; Sc, sporocytes; Sh, shield; Tmt, tomont. Scale bar: (A,D), 50 μm; (G,I,J), 5 μm; (H), 2 μm.
Tomonts, recently emerged parasite cells, were ovoid in shape (40.6 ± 3.6 by 17.7 ± 0.4 μm; n = 5; range: 30.5–51.4 by 16.4–18.7 μm), had a rigid surface, and appeared lodged in the host lorica. H. longa loricae were never found to contain two or more tomonts, but the lorica of one specimen contained a tomont and a chain of eight sporocytes (Figure 2F), indicating that the host cell had been infected by two trophonts which emerged sequentially over a short period of time. The parasite nucleus was initially surrounded by ingested host material that was not obviously enclosed by a membrane. Over time, however, the ingested material condensed into a spherical to elongate food vacuole and gradually decreased in size, sometimes developing a strong yellow color (Figures 2D, 3A).
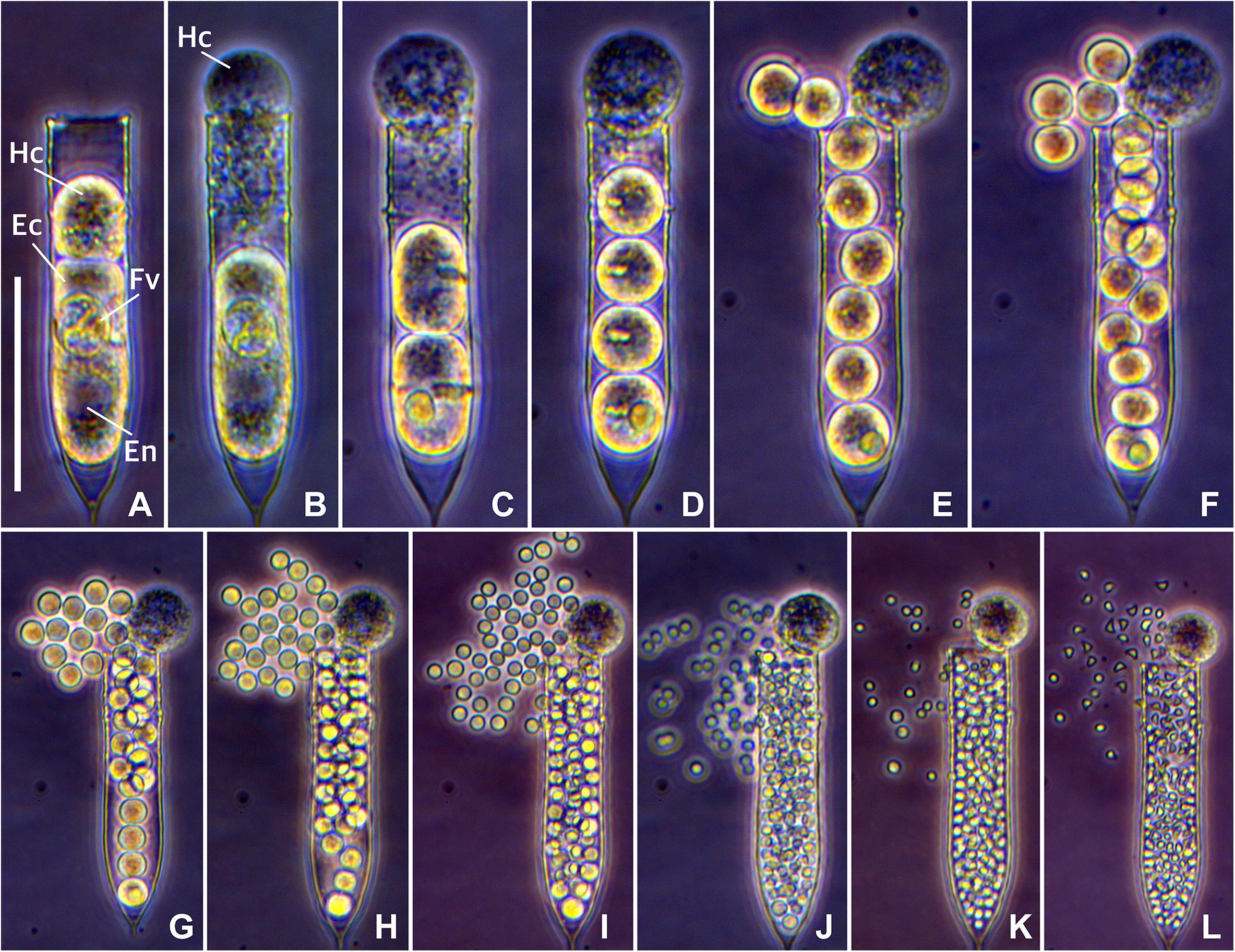
Figure 3. Time-lapse photomicrograph of Euduboscquella triangula sporogenesis resulting in the development of triangular-shaped spores. (A) Mature trophont emerging from the host (required about 10 min). (B) Tomont with food vacuole in lorica (persisted for about 30 min). (C–K) Sporogenic divisions occurring at 15–20 min intervals. (L) Spherical spores transformed into triangular-shaped spores (required about 2 hours). Fv, food vacuole; Hc, host cytoplasm; Ec, Euduboscquella cytoplasm; En, Euduboscquella nucleus. Scale bar = 50 μm.
The first two to three sporogenic divisions were transverse to the longitudinal axis of the tomont and produced closely set sporocytes aligned in a short string (Figures 2D, 3C–E). By completion of the fourth division, however, sporocytes began to drift apart and orientation of the division plane was subsequently difficult to assess. During division, the food vacuole passed to the posterior most sporocyte (relative to the host lorica), which was larger and divided more slowly than other sporocytes (Figures 2E, 3C–G). This pattern in early divisions was consistent, regardless of the type of spore produced. Each tomont of E. triangula formed one of three spore types (dinospores, non-motile spherical spores, and non-motile triangular spores; Figures 2G–J, 3L) that occurred with frequency as follows (n = 14): dinospores (28.6%); non-motile spherical spores and non-motile triangular spores (both 35.7%). The time from tomont formation to initiation of the first sporogenic division was 15–30 min, while time from the start of the first sporogenic division to completion of the final division was about 2.5 h for dinospores, about 3.5 h for non-motile spherical spores, and about 8 h for non-motile triangular spores (6 h division + 2 h for transformation from a spherical to a triangular shape) (Figure 3). Non-motile spherical spores were observed over several days, but never transformed into a triangular shape. Estimates for the duration of the extracellular developmental process, calculated by summing mean duration of the pre-division period with observations for time required for division and maturation of spores, averaged 5.1 ± 1.7 h (n = 3; range 2.9–8.6 h).
Tomonts produced 68–92 dinospores (mean = 81 ± 6; n = 4) that measured 8.2 ± 0.1 μm long by 5.5 ± 0.1 μm wide (n = 30; range: 7.0–9.5 by 4.5–6.6 μm). Dinospores had a mushroom-like shape, with the broad, slightly cone-shape episome offset to the left over the narrower, roughly cylindrical hyposome that curved dorso-posteriorly to a slightly pointed or rounded apex. Dinospores exhibited short rapid bursts of swimming alternating with longer periods when the flagella beat slowly and the cells were more or less stationary. Non-motile spherical spores numbered 450 ± 29 (n = 3; range: 400–500), measured 2.6 ± 0.03 μm in diameter (n = 30; range: 2.3–3.0 μm), and had a small granule on their surface (Figure 2H). Non-motile triangular spores developed from spherical sporocytes without granules on the surface and acquired their characteristic shape following the final sporogenic division. Triangular spores numbered 363 ± 19 (n = 4; range: 314–400) and were laterally compressed, with their triangular lateral surface being 6.2 ± 0.1 μm at the base (n = 30; range: 4.9–7.7 μm) and 2.9 ± 0.1 μm in height (n = 30; range: 2.3–3.7 μm). These unusually shaped spores were also observed in net-tow sample preserved with Bouin’s solution immediately after collection (Figure 2L). Dinospores lost motility and degenerated within 48 h, while spherical and triangular spores persisted for 2–3 week before degrading.
Cytology Based on Protargol Silver Impregnation
The smallest trophonts of E. triangula, presumably the earliest infections, were roughly spherical cells with a distended outer membrane surrounding a thin layer of lightly staining, homogeneous cytoplasm (Figure 4A, arrow). The cytoplasm contained a centrally positioned nucleus with darker staining nucleoplasm and a conspicuous primary (= initial) nucleolus pressed against the nuclear envelope (Figure 4B). As trophonts increased in size so did their nucleus and primary nucleolus, and additional smaller nucleoli developed just beneath the nuclear envelope (Figure 4D). In some instances, trophonts appeared enclosed in a vacuole, presumptively a parasitophorous vacuole, the origin of which was unclear (Figure 4E, arrows). Trophonts became more elongate as they grew, taking on an ovoid shape, and their cytoplasm acquired a vacuolated appearance (Figure 4F, arrow). With continued growth of the trophont, the enlarged primary nucleolus resorbed or fragmented, with the other nucleoli becoming larger (Figure 4G, arrows). Eventually, all the nucleoli resorbed leaving a granular nucleoplasm (Figure 4I). For convenience, cells with 1–10 nucleoli are hereafter consider early infections, cells with >10 nucleoli mid infections, and cells with resorbing nucleoli late infections.
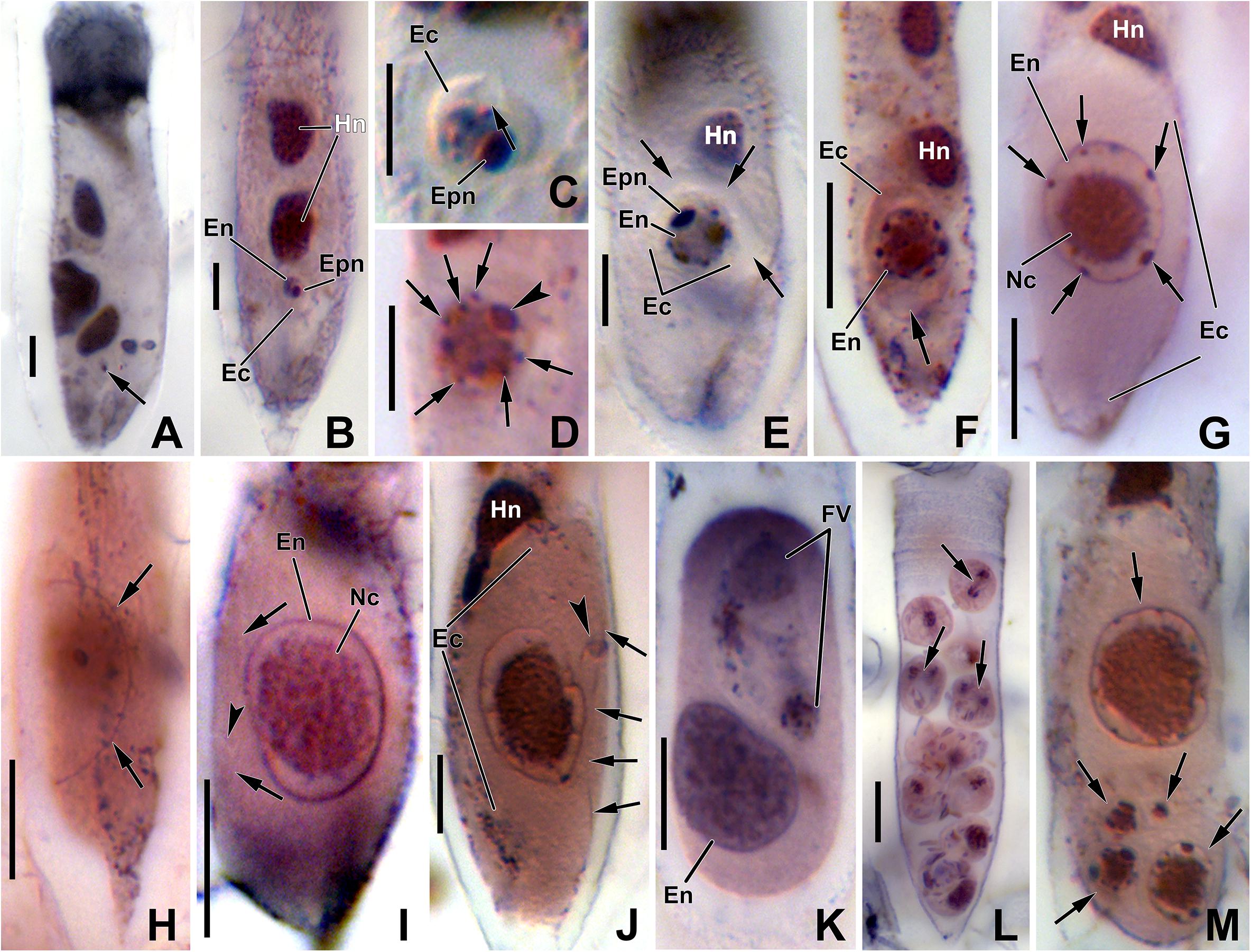
Figure 4. Photomicrograph of Euduboscquella triangula after protargol impregnation. (A) Very early trophont (arrow) with conspicuous nucleolus. (B) Growth of early trophont. (C) Trophont showing opened vacuole toward the external environment (arrow). (D) Enlarged primary nucleolus (arrowhead) and small nucleoli beneath the nuclear envelope (arrows). (E) Parasitophorous vacuole between parasite and host cytoplasm (arrows). (F) Elongated trophont showing vacuolated appearance (arrow). (G) Nucleoli (arrows) being reabsorbed during late stage. (H) The shield with encircling perinema (arrows). (I) Very late trophont showing granular nucleoplasm without nucleolus and with shield (arrows) having an inconspicuous shallow indentation (arrowhead). (J) Lamina pharyngea (arrowhead) adjacent to the perimena (arrows). (K) Tomont with food vacuole. (L) Sporocytes showing mitosis (arrows). (M) Five infections (arrows) in single host. Ec, Euduboscquella cytoplasm; En, Euduboscquella nucleus; Epn, Euduboscquella primary nucleolus; Fv, food vacuole; Hn, host nucleus; Nc, nucleoplasm. Scale bars: (A–E), 5 μm; (F–L), 10 μm.
Comparison of nuclear morphology with cell size indicated that changes in nuclear size (Figure 5A), primary nucleolar size (Figure 5B), and nucleolar number (Figure 5C) were positively related with trophont size. In addition, relationships between nuclear morphology and cell size appeared independent of infection intensity, as indicated by comparable plots for trophonts from host with single and multiple parasites. Cell length averaged 5.0 ± 0.3 μm (n = 27; range = 2.9–10.8) for early infections, 20.5 ± 1.0 μm (n = 72; range = 6.8–39.1) for mid infection, and 35.3 ± 0.7 μm (n = 8; range = 31.8–37.5 μm) for late infections.
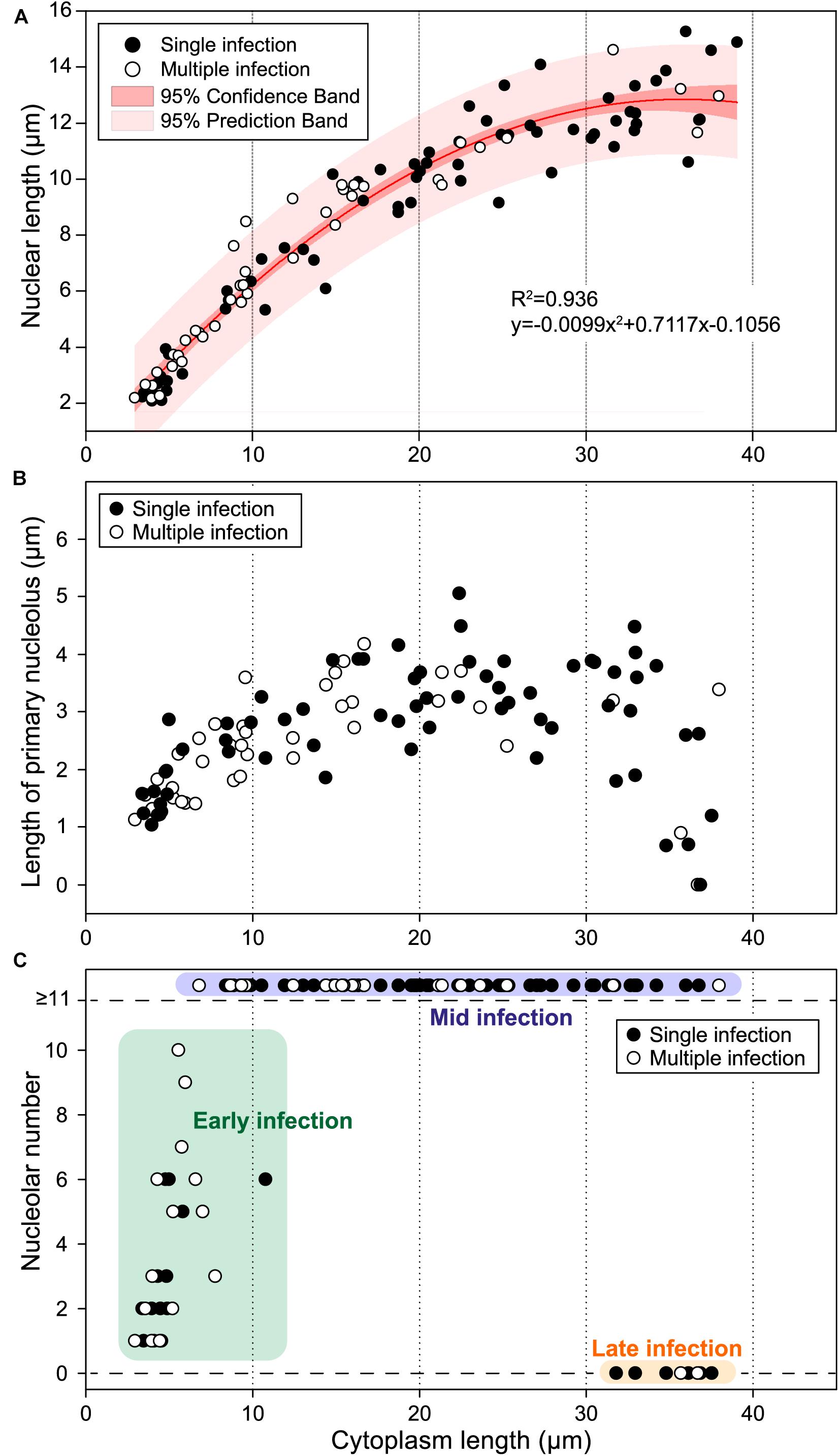
Figure 5. Nuclear size, primary nucleolar length, and nucleolar number relative to cytoplasm size for protargol-stained trophonts of Euduboscquella triangula. (A) Nuclear size positively correlated to cytoplasm size (R2 = 0.936). Single infection (filled circle), multiple infection (open circle), 95% confidence interval (dark pink band), 95% prediction interval (light pink band). (B) Primary nucleolus gradually increased in size during early to mid-stage, but began to decrease during the late stage. (C) Infections classified into three stages depending on the number and developmental state of nucleoli: early infection (green box), mid infection (purple box), and late infection (orange box).
Early infections sometimes had a large vacuole bridging the space between the outer surface of the parasite and the nucleus. The vacuole appeared fused with the cell membrane, thus opening to the exterior environment of host cytoplasm, while the surface adjacent to the nucleus was flattened and somewhat convex (Figure 4C, arrow). The flattened, convex surface may represent the developing shield of the trophont, but that was not confirmed by our preparations. The shield with encircling perinema (= perinematic ring) was, however, evident in some mid to late infections (Figure 4H, arrows), and ranged from 6.4 to 20.9 μm in maximum dimension. The surface of the shield lacked noticeable structure in protargol preparations, except in one mature trophont where a inconspicuous single short, shallow indentation was visible (Figure 4I, arrowhead). The lamina pharyngea was present in mature trophonts as a narrow funnel (Figure 4J, arrowhead), measuring ∼2 μm wide at its opening (Figure 4J, arrows), and extending below the shield to overlie the nucleus. No mature spores and only a few tomonts (Figure 4K) and sporocytes (Figure 4L) were encountered in our protargol preparations, making cytological characterization of those stages problematic.
Molecular Characterization of Euduboscquella triangula
A total of 11 novel rDNA sequences (SSU–ITS1–5.8S–ITS2–LSU regions; 2918–3013 bp) representing (1) a sporocyte (MN388913), (2) non-motile spherical spores (MN388914–MN388918), (3) non-motile triangular-shaped spores (MN388919–MN388921), and (4) dinospores (MN388922–MN388923) were obtained for E. triangula. Alignment of the 11 sequences identified a 2906 bp complementary region. The SSU rDNA (1731 bp) and 5.8S (153 bp) regions each had identical sequence; however, base polymorphism occurred at four loci in the ITS1 region (144 bp), eight loci in the ITS2 region (202 bp), and one locus in the LSU D1–D2 region (634 bp).
Genetic Dissimilarity
Pairwise analysis of genetic dissimilarity among 13 Euduboscquella rDNA sequences, including a representative sequence for E. triangula (MN388915), four sequences available in GenBank for E. crenulata, E. costata, and E. cachoni, and eight GenBank sequences from non-identified Euduboscquella species, showed the SSU and ITS1–LSU regions of E. triangula to, respectively, have a distance of 0.7 and 6.6% from E. crenulata, 5.5 and 26.6% from E. costata, and 4.5 and 23.2% from E. cachoni (Table 1). Compared with non-identified Euduboscquella spp., genetic dissimilarity of E. triangula ranged from 0.2 to 7.4% in the SSU and 1.6 to 34.8% in ITS1–LSU regions. The sequence for E. triangula showed the greatest similarity with Euduboscquella sp. ex Tintinnopsis sp. (JN934992), with SSU–LSU rDNA regions (2951 bp) having 98.9% identity.
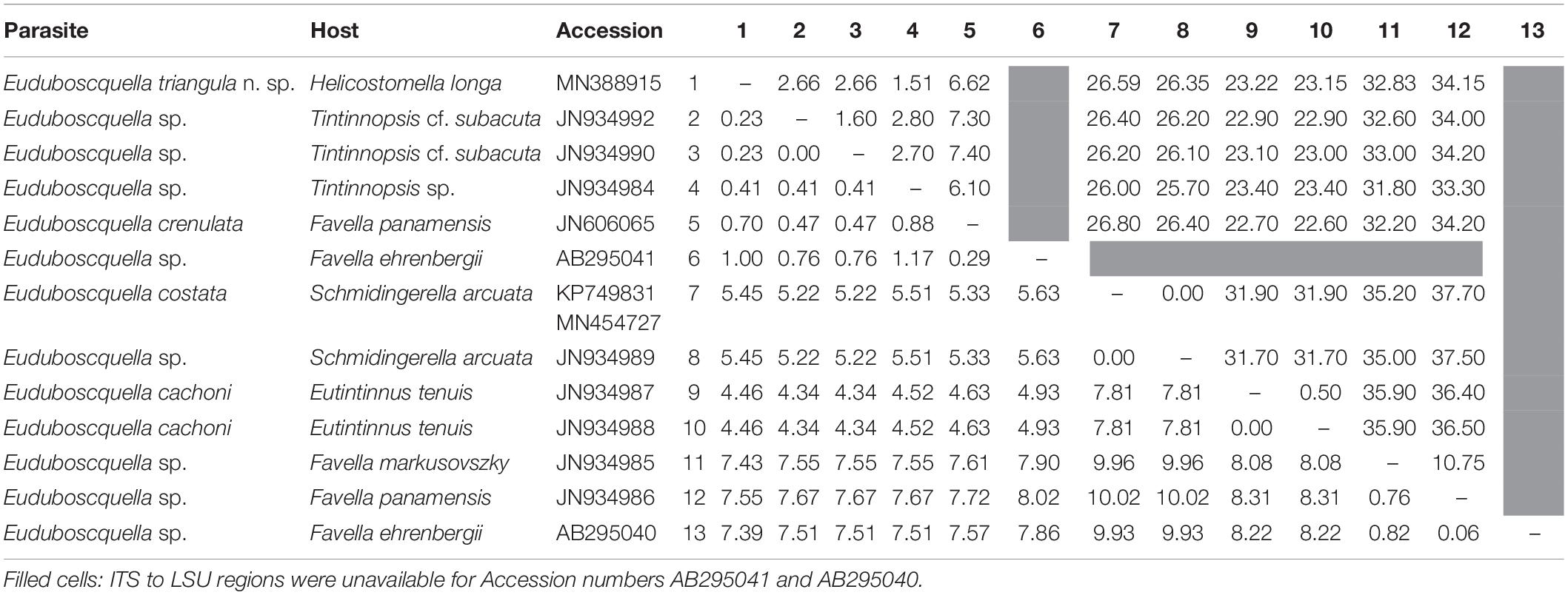
Table 1. Pairwise p-distance values (%) among SSU (1760 bp; bottom diagonal) and ITS1–5.8S–ITS2–LSU (1341 bp; upper diagonal) sequences for Euduboscquella.
Phylogeny
Phylogenetic trees inferred from SSU rDNA sequences using BI and ML analyses clustered the 13 Euduboscquella sequences together in a fully supported clade (BI/ML = 1.00/100) containing four groups (Figure 6). Groups I–III formed a highly supported subclade, with sequences distributed as follows: (1) non-identified Euduboscquella sp., E. crenulata ex F. panamensis, and E. triangula ex H. longa (Group I: AB295041, JN606065, JN934984, JN934990, JN934992, and MN388915; (2) non-identified Euduboscquella (Group II: AB295040 and JN934985–6); (3) E. cachoni ex E. tenuis (Group III: JN934987–8). Within Group I, E. triangula clustered with Euduboscquella sp. ex Tintinnopsis sp. (BI/ML = 0.95/69). Group IV containing sequences for E. costata ex S. arcuata and E. sp. ex S. arcuata (KP749831 and JN934989) formed a fully supported branch sister to the subclade represented by Groups I–III. Sequences for syndinean parasites belonging to the genera Amoebophrya, Syndinium, and Hematodinium sorted as a strongly supported clade basal to the Dinophyceae, while the sequence for Ichthyodinium chabelardi branched basal to the Euduboscquella clade.
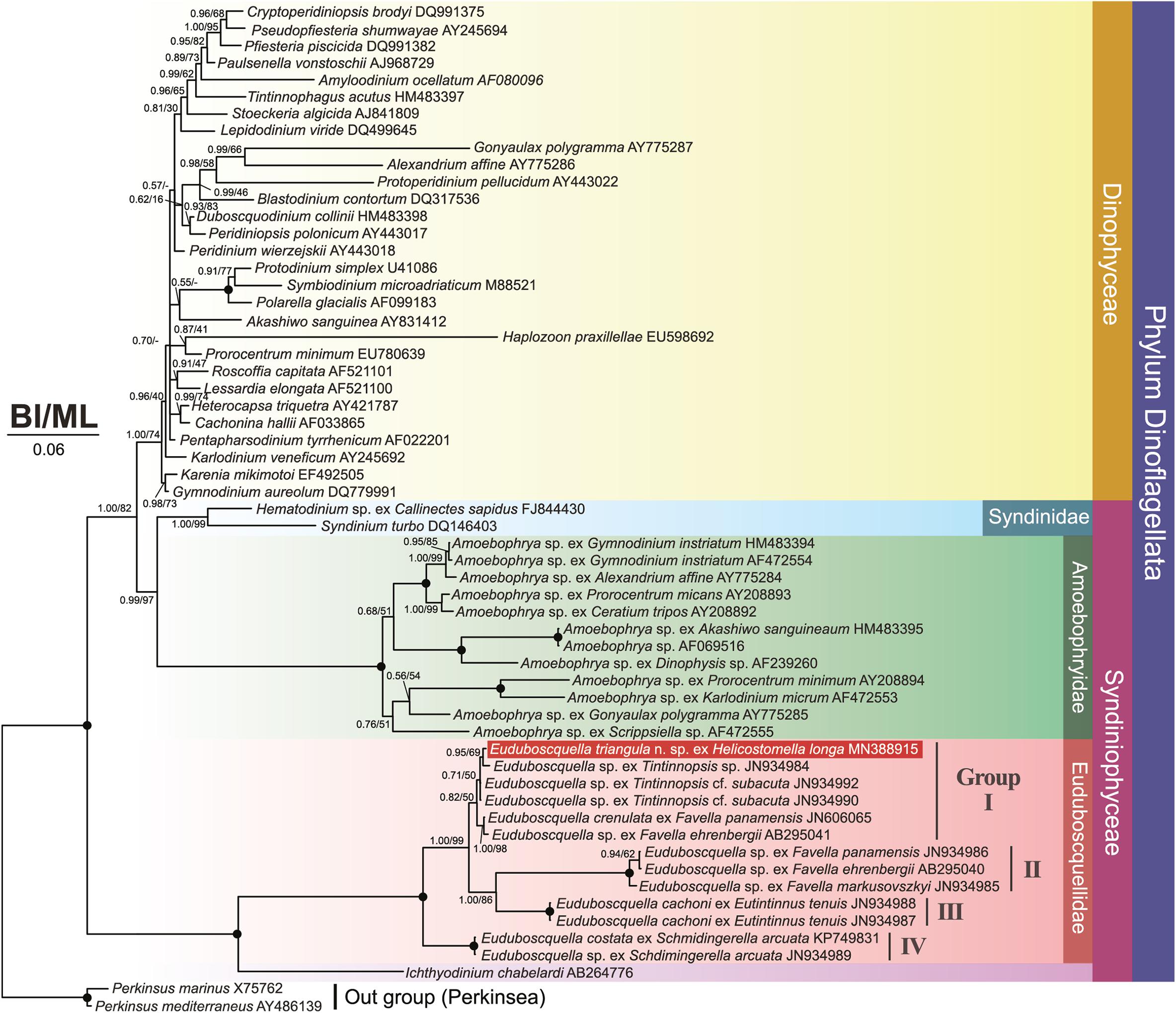
Figure 6. Bayesian interference (BI)/maximum likelihood (ML) phylogenetic trees inferred from SSU rDNA sequences (1606 bp). The tree contains 59 sequences of which 13 represent Euduboscquella species from known host taxa. The novel sequence for Euduboscquella triangula is indicated in white font. Posterior probabilities/bootstrap values for interior branches are shown for BI and ML trees, respectively. Fully supported branches (1.00/100%) are indicated by solid circles at branch nodes. Disagreements in BI and ML tree topology are indicated by a dash (–). Scale bar indicates six base substitutions per 100 nt.
ITS2 Secondary Structure
Internal transcribed spacer 2 secondary structures were evaluated for E. triangula n. sp. and three related species (two non-identified Euduboscquella spp. and E. crenulata) (Figure 7). The predicted secondary structures showed four helices (I–IV). Three uncertain base changes were presented in helices I–III due to ambiguous bases (see red letters in Figure 7; i.e., 44, 78, and 157th ambiguous base positioned in helix I–III) in retrieved sequences JN934984 and JN606065. Helices I and II showed little differences among the species compared. However, E. crenulata (JN606065) exhibited significantly different folding patterns from the other three species due to indels in helix III (one insertion, one deletion) and IV (one deletion), which resulted in variation in the lengths of those helices (Figure 7). Except for E. crenulata (JN606065) having very different structure in helices III and IV, the base changes for each helix were as follows: (1) one non-CBC and uncertain non-CBC were identified in helix I; (2) one h-CBC, three non-CBC, and one uncertain SNP were identified in helix II; (3) one CBC, three h-CBC, one non-CBC, one uncertain non-CBC, and one SNP were identified among three species in the longest helix, helix III; (4) one h-CBC present in the shortest helix, helix IV. In helix III, the SNP on a 130th base change was unique between E. triangula and each of the other compared species. Intraspecific variation occurred at one site in helix II, five sites in helix III, and one site helix IV.
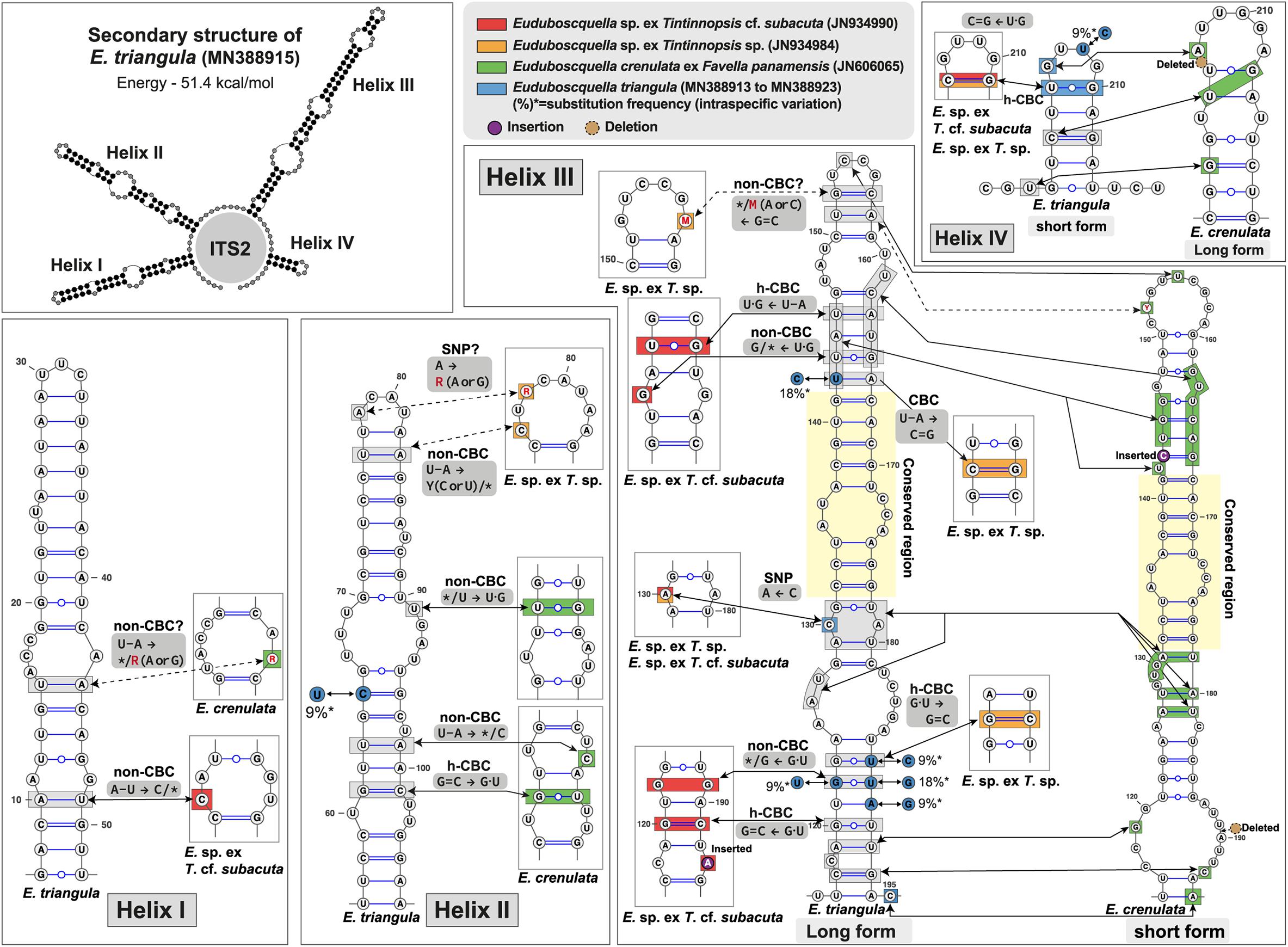
Figure 7. Predicted secondary structures of the internal transcribed spacer 2 (ITS2) of Euduboscquella species. The ITS2 helices are numbered I–IV. Base changes between four Euduboscquella sequences are indicated: the red box (E. sp. ex T. cf. subacuta; JN934990), the orange box (E. sp. ex T. sp.; JN934984), the green box (E. crenulata ex F. panamensis; JN606065), and the blue box (E. triangula n. sp.; MN388913–MN388923). Red letters indicate ambiguous bases. Blue circles are accompanied by substitution frequency of intraspecific variation. The insertion and deletion of bases are indicated by purple and brown circles, respectively. *Means missing (unpaired) base.
Parasite Prevalence, Infection Intensity, and Trophont Development Time
Protargol preparations for samples collected on August 17 and 24, 2015 enabled examination of 569 and 672 H. longa cells on the two dates, respectively. Parasite prevalence and infection intensity were 15.5% (95% confidence interval 12.9–18.0) with 1.3 ± 0.1 parasites/infected host (range: 1–5) on 17 August and 8.3% (95% confidence interval 6.3–10.3) with 1.3 ± 0.2 parasites/infected host (range: 1–3) on 24 August. When data for the two dates were combined, 131 H. longa cells were infected by one or more E. triangula trophonts, while 13 H. longa loricae contained extracellular parasite stages (i.e., tomonts, sporocytes, or spores) derived from one or more E. triangula trophonts. Based on extracellular development time of E. triangula (5.1 ± 1.7 h as above), the proportional representation of intracellular and extracellular parasite life-history stages, and the assumption of steady-state parasite population dynamics, trophont development time would be 51.8 ± 17.5 h (29.1–86.2 h); i.e. (parasite extracellular development time) (percent H. longa cells infected by trophonts)/(percent H. longa loricae containing extracellular parasite stages).
Taxonomical Information of Euduboscquella triangula
Phylum Dinoflagellata Bütschli, 1885
Subphylum Syndinea Corliss, 1984
Class Syndiniophyceae Loeblich III, 1976
Order Syndiniales Loeblich III, 1976
Family Euduboscquellidae (Coats et al., 2012)
Genus Euduboscquella (Coats et al., 2012)
Species E. triangula n. sp.
ZooBank LSID
urn:lsid:zoobank.org:act:F2BCD3C1-C602-4408-93A8-0C9CDC64FBB9.
Diagnosis
Shield inconspicuous until very late in infection cycle, with surface of shield covered by numerous shallow, intertwining grooves. Lamina pharyngea ∼2 μm wide at perinema. Three types of spores: motile mushroom-shaped dinospores measuring 7.0–9.5 by 4.5–6.6 μm in vivo with broad, slightly cone-shape episome offset to the left over narrower, roughly cylindrical hyposome curving dorso-posteriorly to a slightly pointed or rounded apex; non-motile spherical spores measuring 2.3–3.0 μm in vivo; non-motile triangular spores measuring 4.9–7.7 μm at base, 2.3–3.7 μm high in vivo. Sporogenesis producing chain of 4–8 cells before separation of sporocytes, regardless of spore type.
Type Host
Helicostomella longa (Brandt, 1906; Kofoid and Campbell, 1929).
Type Habitat
Marine coastal water of Geoje Island, South Korea.
Type Locality
Jangmok Bay, South Korea (34°59′38″N, 128°40′28″E).
Type Material
The name-bearing hapantotype is a slide of protargol-impregnated H. longa infected by E. triangula has been deposited in National Institute of Biological Resources (NIBRDN0000001794).
Etymology
The specific name is Latin triangula (adj., f. nom.) meaning triangular or three-cornered and is used in reference to the distinctive triangular-shaped spores of the species. The species epithet agrees with the feminine gender of the generic name Euduboscquella.
Discussion
Comparison of Euduboscquella triangula With Related Species Infecting Tintinnids
Of the morphological and morphogenetic attributes exhibited by Euduboscquella species, sporogenetic pattern, spore morphology, and shield (= episome) structure have proven most useful in distinguishing taxa. Architecture of the lamina pharyngea and host species are less useful, as the former shows limited variation, while host range for Euduboscquella species is not well understood. Coats and Bachvaroff (2013) distinguished three patterns of sporogenesis among species of Euduboscquella infecting tintinnids: (1) production of a tightly packed cluster of sporocytes that separate following the final division to produce dinokont spores (= dinospores) or non-dinokont spores (= spores with one or no flagella); (2) production of a very long string of sporocytes that mature into dinokont spores; and (3) production of a short string of sporocytes that separate, continue to divide, and produce non-dinokont spores. Subsequently, Jung et al. (2016) reported that sporocytes of E. costata separate after the second or third division, continue to divide, and produce either dinospores or non-dinokont spores. Sporogenesis in E. triangula matches that of E. costata, although spore morphology (see below) differs between the two species. Clearly, production of non-dinokont spores in both E. costata and E. triangula follows pattern 3 sporogenesis; however, their process of generating dinospores is not consistent with dinospore formation represented by pattern 1 or pattern 2 sporogenesis. Consequently, we consider dinospore formation in E. costata and E. triangula to be a fourth type of sporogenesis (pattern 4: formation of dinospores from sporocytes that separate early during sporogenesis). Thus, E. costata and E. triangula (pattern 3 and 4 sporogenesis) are distinguished by sporogenic pattern from E. cachoni (pattern 1), E. aspida and E. crenulata (patterns 2 and 3), and possibly from E. cnemata which from the description of Cachon (1964) appears to follow pattern 2. Coats and Bachvaroff (2013) interpreted the drawing of E. anisospora sporocytes provided in Chatton (1952) as representing pattern 2 sporogenesis (although incorrectly listed in Table 6.1 as pattern 3). Given observations reported here and in Jung et al. (2016), we believe that sporogenesis in E. anisospora is most appropriately classified as pattern 4, thus partially aligning E. anisospora with E. costata and E. triangula.
Reniform dinospores are produced by E. aspida (4–7 μm long) and E. cnemata (3–4 μm long), sigmoid dinospores by E. crenulata (7–10 μm long), and mushroom-shaped dinospores (bulbous episome offset over narrower hyposome) by E. anisospora (12.5 and 20 μm long), E. cachoni (5–8 μm long), E. costata (12–14 μm long), and E. triangula (7.0–9.5 μm long). Non-dinokont spores vary greatly in size and shape across Euduboscquella species, with E. aspida forming small bi-partite spores (2–3 μm long), E. cachoni small spherical spores (2–3 μm diameter) and spindle-shaped spores (4–8 μm long), E. costata oval spores (4.7–5.3 μm by 3.6–4.6 μm) and spherical to rectangular spores housed in a hyaline tubule (6.1–10.8 μm by 3.7–5.3 μm, excluding tubule), E. crenulata large spherical spores (10–14 μm diameter) and oviform spores (10–16 μm long), and E. triangula small spherical spores (2.3–3.0 μm in diameter) and triangular spores (4.9–7.7 μm at base by 2.3–3.7 μm in height). Thus E. triangula differs from E. aspida, E. cnemata, and E. crenulata in dinospore shape and from E. anisospora, E. cnemata, and E. costata in dinospore size, while differing from all species by its triangular, non-dinokont spore. That the unusually shaped triangular spores of E. triangula were observed in Bouin-fixed sample preserved immediately after collection, strongly supports their being are an integral part of the parasite’s life cycle, rather than an abnormal spore morphology caused by incubation conditions. Lastly, Gómez et al. (2011) provided, photographs of what appear to be triangular spores of an unidentified Euduboscquella species within the lorica of a Helicostomella species. Comparison of those spores with the triangular spores of E. triangula, however, is problematic as the authors did not provide relevant morphological data.
Grooves or furrows on the surface of the shield in mature trophonts are an important discriminating feature in species of Euduboscquella. The shields of E. aspida and E. cachoni have a single sagittal groove, while that of E. anisospore has 6–7 parallel grooves, those of E. cnemata and E. costata, an outwardly spiraling set of grooves numbering 20–30 and 25–40, respectively, that of E. crenulata, numerous deep, closely set intertwining grooves, and that of E. triangula, numerous shallow, closely set intertwining grooves. Shield structure in E. triangula is remarkably similar to that of E. crenulata, but in the former, the grooves are much shallower.
In SSU rDNA phylogeny, the Euduboscquella lineage split into four groups, with E. triangula ex H. longa (MN388915) located in the group I (Figure 6). Group I was subdivided into three clades showing very short branches: (1) E. triangula ex H. longa and Euduboscquella sp. ex Tintinnopsis sp. (JN934984); (2) Euduboscquella sp. ex Tintinnopsis cf. subacuta (JN934990); (3) E. crenulata ex F. panamensis (JN606065) and Euduboscquella sp. ex F. ehrenbergii (AB295041). Also, p-distance of SSU rDNA sequences within Group I showed values ≤ 1.0% (Table 1). Given the low genetic variation within Group I, we further analyzed sequences available for the highly variable ITS2 region of Euduboscquella species (note that the ITS2 sequence of E. sp. ex F. ehrenbergii is unavailable in GenBank). For use as a template in homology modeling of ITS2 secondary structure, Euduboscquella was first searched in the ITS2 database (Ankenbrand et al., 2015), but did not return a match. Alternatively, we constructed ITS2 secondary structure with reference to the closely related syndinean taxon Amoebophrya, which was available in Ankenbrand et al. (2015). Our estimated ITS2 secondary structure was quite different from that previously reported by Bachvaroff et al. (2012) when comparing the ITS2 sequence of E. crenulata with four other Euduboscquella sp. sequences, including two from parasites of Tintinnopsis spp., one from T. cf. subacuta, and one from T. major. Stem length reported by Bachvaroff et al. (2012) was shorter than that of E. triangula (MN388915): helix II (was 28 vs. 52 bp), helix III (35 vs. 95 bp), and helix IV (36 vs. 25 bp). Also, sequences positioned in stems II to IV by Bachvaroff et al. (2012) mostly overlapped the helix III region in our study. In our ITS2 secondary structures excluding uncertain base changes (Figure 7), Euduboscquella sp. ex T. cf. subacuta (JN934990) differed from E. triangula ex H. longa (MN388915) in helix I (one non-CBC located at the 2nd internal loop that formed a larger symmetrical loop), in helix III (two h-CBCs, two non-CBCs, and a new symmetrical internal loop induced by one insertion), and in helix IV (one h-CBC located at the hairpin loop). E. sp. ex T. sp. (JN934984) differed from E. triangula in helix II (one non-CBC induced formation of a larger hairpin loop), in helix III (one CBC occurred above the conserved region and one h-CBC below the 4th asymmetrical internal loop), and in helix IV (one h-CBC occurred at the hairpin loop). Although the SSU rDNA-based phylogenetic tree did not show a significant difference among the members of Group I, ITS–LSU p-distance values and ITS2 secondary structures exhibited distinct difference between E. triangula and the other species.
Potential Impact of Euduboscquella triangula on Helicostomella longa
Helicostomella longa infected by E. triangula typically dies shortly after emergence of the parasite. Since H. longa was infected by one to five E. triangula trophonts (Figure 4M), but extracellular stages derived from more than one trophont were rarely encountered in host loricae, one is left to ponder the fate of the superfluous parasite trophonts. Perhaps they simply perish along with the uneaten portion of the host cell, or as Cachon (1964) reported for other Euduboscquella species, they are ingested along with host cytoplasm as the most advanced trophont emerges from the host cell. Regardless of which is correct, it seems reasonable to estimate parasite induced mortality in H. longa following calculations of Coats and Heisler (1989); i.e., daily parasite induced mortality of host populations = % infected host cells/intracellular development time in days. Using data for samples collected on August 17 and 24, 2015, estimated parasite induced mortality for the two dates is 8.8 ± 2.5% (4.3–12.8%) and 4.7 ± 1.3% (2.3–6.8%), respectively. However, biovolumes of E. triangula tomonts and H. longa cells remaining immediately after parasite emergence indicates that only 84.1% of host biomass is transformed into parasite biomass. Thus, daily utilization of H. longa biomass by E. triangula (= daily parasite induced mortality of host populations multiplied by 84.1% cell biomass transformed to parasite biomass) on the dates above would represent 7.4 ± 2.1% (3.6–10.8%) and 4.0 ± 1.1% (1.9–5.8%) of host standing stock, respectively.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN388913–MN388923.
Author Contributions
JC, KK, DC, and YK: conceived and designed the study. JC and DC: morphological examinations. JC and JJ: molecular analyses. JC and KK: analyzed the data. All authors revised and contributed to the final manuscript.
Funding
This research was supported by Korean Institute of Ocean Science and Technology (KIOST) projects “Effect on ciliate plankton population of micro-parasite infection” (PE99962).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.720424/full#supplementary-material
Footnotes
References
Ankenbrand, M. J., Keller, A., Wolf, M., Schultz, J., and Förster, F. (2015). ITS2 database v: twice as much. Mol. Biol. Evol. 32, 3030–3032. doi: 10.1093/molbev/msv174
Bachvaroff, T. R., Kim, S., Guillou, L., Delwiche, C. F., and Coats, D. W. (2012). Molecular diversity of the syndinean genus Euduboscquella based on single-cell PCR analysis. Appl. Environ. Microbiol. 78, 334–345. doi: 10.1128/AEM.06678-11
Bachy, C., López-García, P., Vereshchaka, A., and Moreira, D. (2011). Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North pole at the end of the polar night. Front. Microbiol. 2:106. doi: 10.3389/fmicb.2011.00106
Brandt, K. (1906). Die tintinnodeen der plankton-expedition. tafelerklärungen nebst kurzer diagnose der neuen arten. Ergebn. Plankton Exped. Humboldt Stiftung 3, 1–33.
Cachon, J. (1964). Contribution a l’étude des péridiniens parasites. cytologie, cycles évolutifs. Ann. Sci. Nat. Zool. 6, 1–158.
Chatton, E. (1920). Les Péridiniens parasites: morphologie, reproduction, ethologie. Arch. Zool. Exp. Gen. 59, 1–475.
Chatton, E. (1952). “Classe des dinoflagellés ou péridiniens,” in Traité de Zoologie, ed P.-P. Grassé (Paris: Masson & Cie), 309–390.
Claparède, É, and Lachmann, J. (1858). Études sur les infusoires et les rhizopodes. Mém. Inst. Natn. Génev 5, 1–260.
Coats, D. W. (1988). Duboscquella cachoni N. Sp., a parasitic dinoflagellate lethal to its tintinnine host Eutintinnus pectinis. J. Protozool. 35, 607–617. doi: 10.1111/j.1550-7408.1988.tb04159.x
Coats, D. W., and Bachvaroff, T. R. (2013). “Parasites of tintinnids,” in The Biology and Ecology of Tintinnid Ciliates: Models for Marine Plankton, eds J. R. Dolan, D. J. S. Montagnes, and D. K. Stoecker (Chichester: Wiley-Blackwell), 145–170. doi: 10.1002/9781118358092.ch6
Coats, D. W., Bachvaroff, T. R., and Delwiche, C. F. (2012). Revision of the family duboscquellidae with description of Euduboscquella crenulata n. gen., n. sp. (Dinoflagellata, Syndinea), an intracellular parasite of the ciliate Favella panamensis Kofoid & Campbell, 1929. J. Eukaryot. Microbiol. 59, 1–11. doi: 10.1111/j.1550-7408.2011.00588.x
Coats, D. W., and Heisler, J. J. (1989). Spatial and temporal occurrence of the parasitic dinoflagellate Duboscquella cachoni and its tintinnine host Eutintinnus pectinis in Chesapeake Bay. Mar. Biol. 101, 401–409. doi: 10.1007/bf00428137
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). JMODELTEST 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Darty, K., Denise, A., and Ponty, Y. (2009). VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. doi: 10.1093/bioinformatics/btp250
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi: 10.1126/science.1261605
Gómez, F., Moreira, D., and López-García, P. (2011). Advances on the study of dinoflagellates (Dinophyceae) with the molecular phylogeny. Hidrobiológica 21, 343–364.
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R. M., Kirkham, A. R., Massana, R., et al. (2008). Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10, 3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hada, Y. (1932). Description of two new neritic Tintinnoinea, Tintinnopsis japonica and Tps. kofoidi with a brief note on an unicellular organisms parasitic on the latter. Proc. Imp. Acad. Jpn. 8, 209–212. doi: 10.2183/pjab1912.8.209
Harada, A., Ohtsuka, S., and Horiguchi, T. (2007). Species of the parasitic genus Duboscquella are members of the enigmatic marine alveolate group I. Protist 158, 337–347. doi: 10.1016/j.protis.2007.03.005
Horiguchi, T., Harada, A., and Ohtsuka, S. (2006). Taxonomic studies on parasitic dinoflagellates in Japan. Bull. Plankton Soc. Jpn. 53, 21–29.
International Commission on Zoological Nomenclature (1999). International Code of Zoological Nomenclature, 4 Edn. London: The International Trust for Zoological Nomenclature.
Jung, J.-H., Choi, J. M., Coats, D. W., and Kim, Y.-O. (2016). Euduboscquella costata n. sp. (Dinoflagellata, Syndinea), an intracellular parasite of the ciliate Schmidingerella arcuata: morphology, molecular phylogeny, life cycle, prevalence, and infection intensity. J. Eukaryot. Microbiol. 63, 3–15. doi: 10.1111/jeu.12231
Jung, J.-H., Park, K.-M., and Min, G.-S. (2012). Morphology, morphogenesis, and molecular phylogeny of a new brackish water ciliate, Pseudourostyla cristatoides n. sp., from songjiho lagoon on the coast of East Sea, South Korea. Zootaxa 3334, 42–54. doi: 10.11646/zootaxa.3334.1.3
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kofoid, C. A., and Campbell, A. S. (1929). A conspectus of the marine and fresh-water ciliata belonging to the suborder Tintinnoinea, with descriptions of new species principally from the Agassiz expedition to the eastern tropical Pacific 1904-1905. Univ. Calif. Publ. Zool. 34, 1–403.
Konovalova, G. V. (2007). Parasitic peridineae (dinoflagellates) and ellobiopsidae of coastal waters of the Sea of Japan. Biol. Morya 33, 167–175.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lohmann, H. (1908). Untersuchungen zur feststellung des vollständigen gehaltes des meeres an plankton. Wiss. Meeresunters. Kiel 10, 129–307.
Montagnes, D. J. S., and Lynn, D. H. (1993). “A quantitative protargol stain (QPS) for ciliates and other protists,” in Handbook of Methods in Aquatic Microbial Ecology, eds P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (Boca Raton, MIA: Lewis Publishers), 229–240. doi: 10.1201/9780203752746-28
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sonnenberg, R., Nolte, A. W., and Tautz, D. (2007). An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front. Zool. 4:6. doi: 10.1186/1742-9994-4-6
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Keywords: Euduboscquella, Helicostomella, marine parasite, syndinean dinoflagellate, tintinnid
Citation: Choi JM, Jung JH, Kim KH, Coats DW and Kim YO (2021) A Novel Parasitic, Syndinean Dinoflagellate Euduboscquella triangula Infecting the Tintinnid Helicostomella longa. Front. Mar. Sci. 8:720424. doi: 10.3389/fmars.2021.720424
Received: 04 June 2021; Accepted: 21 July 2021;
Published: 12 August 2021.
Edited by:
Hongbin Liu, Hong Kong University of Science and Technology, Hong Kong, SAR ChinaReviewed by:
Daniel Garcia-Souto, University of Vigo, SpainSung Joon Song, Seoul National University, South Korea
Copyright © 2021 Choi, Jung, Kim, Coats and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young Ok Kim, eW9raW1Aa2lvc3QuYWMua3I=
 Jung Min Choi
Jung Min Choi Jae Ho Jung
Jae Ho Jung Ki Hong Kim2
Ki Hong Kim2 D. Wayne Coats
D. Wayne Coats Young Ok Kim
Young Ok Kim