- 1State Key Laboratory of Biocontrol, Institute of Aquatic Economic Animals and Guangdong Provincial Key Laboratory for Aquatic Economic Animals, Life Sciences School, Sun Yat-sen University, Guangzhou, China
- 2Southern Laboratory of Ocean Science and Engineering, Zhuhai, China
- 3College of Marine Sciences, South China Agricultural University, Guangzhou, China
- 4Yangjiang Polytech, Department Food & Environmental Engineering, Yangjiang, China
Asynchrony of sexual maturity is a huge limitation in the reproduction of grouper sperm. Cold storage of sperm is an effective method to solve the problem of asynchronization. However, sperms gradually lose their activity with the prolonged storage time in vitro. In order to explore causes, the effects of cold storage on transcriptome, proteome and oxidation resistance of giant grouper sperm were analyzed. Firstly, the absolute RNA quantity and consistent transcripts existed in each spermatozoon were estimated. With the prolonged storage, the RNA quantity gradually decreased both in the cytoplasm and in the mitochondria of the spermatozoon. The decreased transcripts were mainly enriched with energy metabolism and stress response. Similar to RNAs, the absolute protein quantity was also significantly decreased during the storage of sperm. Decreased proteins were mainly enriched with the oxidative phosphorylation pathway. Proteins involved in the oxidative phosphorylation showed a faster degradation rate compared to the average total protein. In addition, the oxidation resistance and adenosine triphosphate (ATP) contents showed a significant decrease in the sperm during storage in vitro. These results implied that damages of transcriptome, proteome, and oxidation resistance have negative effects on the normal functions of sperm, especially their energy metabolism. The present study provides essential foundation for improving the storage of sperm in vitro.
Introduction
Artificial insemination is one of the most important methods for reproduction in the grouper industry. The assisted reproductive technology has been widely applied in intraspecific and interspecific cross-breeding of the grouper. Asynchrony of sexual maturity always exists in cross-breeding, which limits the development of grouper industry. Thus, the conservation of sperm is pivotal to grouper reproduction, hybridization, and transportation of sperms. In vitro, the motility, and normal functions of the fish sperm only last for a short time, such as 2 days for the common carp, Cyprinuscarpio, at 4°C (Saad et al., 1988), 3 days for Atlantic salmon, Salmo salar, at 4°C (Parodi et al., 2017), and 6 days for the rainbow trout, Oncorhynchus mykiss, at 4°C (Aguilar-Juárez et al., 2014). Recently, several studies related to the physiological change of the sperm during storage in vitro have been reported, which suggested that sperm inactivation is mainly caused by the production of reactive oxygen species, increase of DNA fragments, and damages of the plasma membrane, mitochondria, and the structure of flagellum (Contreras et al., 2019). These studies provide valuable information for the understanding of sperm damage.
Published studies associated with the RNA damage of the sperm during storage are few. Traditional views suggested that spermatozoon is a highly differentiated cell with few cytoplasm and a condensed nucleus (Braun, 2001). Transcriptions and translations are inactive in sperms (Boerke et al., 2007). Thus, the changes of transcripts during sperm preservation received less attention. However, recent studies demonstrated that abundant messenger RNAs (mRNAs) exist in sperms and they are closely related to various biological processes, such as motility, fertilization, capacitation, and early embryo development (Krawetz, 2005; Hwang et al., 2013). Gur and Breitbart (2006) reported that inhibition of protein translation in sperms induce a significant decrease of sperm motility, capacitation, and fertilization capacity. Another study discovered that mRNAs existed in murine sperm could be delivered into the fertilized oocyte and play important roles in the zygotic development (Fang et al., 2014). Transcripts also exist abundantly in sperms of fish species; several reports have discovered that the changes of sperm transcriptome are done under the process of cryopreservation (Yang et al., 2019). Proteins are very important for the biological process of the sperms. Many studies have demonstrated that proteins have effects on the biological functions of sperm, such as motility, capacitation, acrosome reaction, and fertilization (Djourabchi Borojerdi et al., 2020; Wu et al., 2020). In fish species, there was no report about the effects of storage in vitro on transcriptome and proteome levels.
Epinephelus lanceolatus, a giant grouper, is the largest coral reef-dwelling bony fish species found all over the world; its length and weight can reach up to 2.7 m and 400 kg, respectively (Williams, 2009). This species is considered as the ideal male parent in interspecific crossing of grouper species due to its excellent growth traits and high yield of sperm. Up to now, at least five hybrids including kelp grouper (E. moara) ♀ × giant grouper ♂ (Chen et al., 2018), orange-spotted grouper (E. coioides) ♀ × giant grouper ♂ (Tang et al., 2017), brown-marbled grouper (E. fuscogutatus) ♀ × giant grouper ♂ (Sun et al., 2016), humpback grouper (Cromileptesaltivelis) ♀ × giant grouper ♂ (Chen et al., 2017), red-spotted grouper (E. akaara) ♀ × giant grouper ♂ (Guo et al., 2018) have been reported. Among them, brown-marbled grouper ♀ × giant grouper ♂ is the most successful hybrid; its annual yield accounts for over 70% of the total output of grouper in China (Wang et al., 2019). In addition, asynchrony of sexual maturity is common during the interspecific hybridization of grouper. Thus, prolonging the storage time of the giant grouper sperm is pivotal to grouper industry and breeding studies. Up to now, cryopreservation of giant grouper sperm has been reported (Tian et al., 2015; Fan et al., 2020). However, for short-term storage, cold preservation is more convenient.
In the present study, firstly, we analyzed the changes in the motility, velocity, fertilization, and hatchability of sperm during storage in vitro. Then, we discovered the profiles of transcripts and proteins in sperm and assessed the effects of storage in vitro on them. At last, the change of adenosine triphosphate (ATP) contents and oxidation resistance in sperm during storage in vitro were measured. The research provides important information for sperm conservation and artificial insemination.
Materials and Methods
Broodstock Management and Sperm Collection
The matured broodstocks used in the experiment were obtained from Morning Sea Aquatic Co., Ltd (Hainan, China). Male fish (46–68 kg) were bred in an outdoor rectangular cement pool (6 × 6 × 2 m) using a seawater circulating system. The water temperature and salinity were maintained at 29–32°C and 29–33%; the broodstocks were fed once every 3 days with frozen fish. Matured female brown-marbled groupers (E. fuscoguttatus) were applied to provide spawn; they were bred in another rectangular cement pool with the same condition. The female broodstocks were fed every 3 days with frozen fish. Feeding was stopped for 24 h before the experiment.
Five candidate matured male fish were selected for the experiment. After anesthetizing with eugenol (Sangon Biotech, China), the urogenital pore was dried using a clean towel and the initial semen was removed. Clean semen was collected by a syringe (10 ml), without needle, after stripping by an abdominal massage. At this process, the semen was protected from contamination of blood, excrement, and seawater. The quality of sperm was evaluated and the best three sperm samples with motility >90% were applied for further analysis. The semen samples were stored in several 50 ml tubes with crushed ice and were immediately used for further experiment.
Sperm Quality Evaluation
The methods of sperm quality evaluation were followed as per the protocols of our laboratory, with some modifications (Yang et al., 2017, 2020b). Sperm quality parameters including motility, curvilinear velocity (VCL), straight-line velocity (VSL), and sperm type were measured using Integrated Semen Analysis Systems (ISAS 2.0, Spain). The motility evaluation of sperm was carried out below 100 magnified negative-phase field with a microscope. Spermatozoa were activated using seawater filtered by syringe-driven filter with a final dilution ratio of 1:100. About 0.1% of bovine serum albumin (BSA) (w/v) was added to prevent sperm from being attached to the microscope slide. Approximately 500–1,000 spermatozoa in 3–5 random sights were evaluated for each sample. The VCL of spermatozoa with <20 μm/s was defined as motile. The spermatozoa of each sample were divided into four degrees i.e., Levels A, B, C, and D based on VCL ≥ 100 μm/s, 100 μm/s > VCL ≥ 60 μm/s, 60 μm/s > VCL ≥ 20 μm/s and VCL <2 0 μm/s, respectively.
Fertilization and Hatch
In this experiment, we employed matured, female brown-marbled grouper for fertilization and hatch analysis due to high hatchability of the brown-marbled grouper ♀ × giant grouper ♂. Fertilization and the hatch potential of FS and stored sperm were evaluated. In the sample, the well-developed eggs were used for artificial insemination. Approximately, 100 eggs were distributed into each plastic Perti dish, and excess sperm (1 μl) were added and gently stirred. After mixing, 10 ml filtered seawater was added slowly. Twenty minutes later, the seawater was removed using 80-mesh gauze, and the eggs were transferred into a beaker containing 50 ml seawater having the temperature of 28°C and 30% salinity. After 2 h of incubation, most of the eggs have been developed to the 64-cell stage; the developmental malformation and fertilization ratio were observed and assessed under 4 × microscope. The hatchability was measured after hatching larvae from the fertilized eggs.
Isolation and Quantification of Total RNA of Sperm
Fresh sperms (FS) and 2 day cold storage sperms (C2) were used to isolate total RNA. Total RNA was extracted from 100 μl sperm using TRIzol reagent (Thermo Fisher Scientific, MA, USA). In brief, 100 μl sperm of each sample was applied to RNA isolation and 1 ml of TRIzol reagent was added to the sample and homogenized using the tissue breaker JX-2010 (Jingxin, Shanghai, China). After 5 min of incubation, 200 μl of chloroform was added and homogenized, and incubated on ice for 5 min. The samples were centrifuged for 15 min at 12,000 × g at 4°C. About 500 μl of colorless, upper aqueous phase was transferred into a new tube. The remaining upper aqueous phase in the old tube was quantified using a pipettor. About 500 μl of isopropanol was added and mixed with the colorless upper aqueous phase. After 5 min of incubation at 4°C, the mixtures were centrifuged for 10 min at 12,000 × g at 4°C. The supernates were removed and added to 1 ml of 75% ethyl alcohol with 0.5 M lithium chloride to wash the sediments. After 1 h of incubation at −20°C, the samples were centrifuged at 12,000 × g for 15 min. After removing the supernatant, the washing processes were repeated for one more time. The sediments were dried for 10 min at room temperature. About 100 μl of RNAse-free water was added for dissolving the sediments. The quantity and quality of the RNA samples were determined using a microplate spectrophotometer (BioTek Istruments Inc., VT, USA) and electrophoresized on 1% agarose gel. In addition, the sperm numbers per milliliter was calculated using a hemocytometer.
The total average absolute RNA contents of each sperm were calculated using the following formula:
The concentration indicates the measured concentration of RNA solution. The actual volume indicates the total volume of the upper aqueous phase and isolated volume indicates the upper aqueous phase using the RNA isolation.
Transcriptome Library of Sperm Preparation and Sequencing
The FS and C2 with three repeats were applied to construct a transcriptome library. A total of six RNA samples with an equal amount of RNA contents were used for the preparation of the complementary DNA (cDNA) library. The RNA_sequencing (RNA-seq) libraries were prepared using NEBNextUltraTM RNA Library Prep Kit (New England Biolabs, MA, USA) following the protocols of the manufacturer. The quality and quantity of the library were measured using Agilent 2100 Bioanalyzer (Agilent Technologies Inc., IL, USA). Finally, the libraries were sequenced in Novogene Co. Ltd. using the Illumina-Hiseq 2500 platform and PE150 strategy.
Estimation of Commonly Expressed mRNA in Each Spermatozoon
For estimating the consistent transcripts expressed in each spermatozoon, we refer to Fang's method with some optimization (Fang et al., 2014). On an average, a transcript is less than one copy in one spermatozoon, which indicates that the transcript is randomly existed in a part of spermatozoa, and a transcript that has more than one copy in one spermatozoon indicates that the transcript is most likely existed in each spermatozoon. The mapped transcripts per kilobase million (TPM) of exon model represents the abundance of a transcript regardless of its length, which is a suitable parameter for estimating the commonly expressed transcripts in each spermatozoon. Based on the average counts of transcripts in each spermatozoon, the lowest numerical value of the TPM of transcript commonly expressed in each spermatozoon can be estimated. Firstly, the TPM of each expressed transcript were calculated using the following formula:
Exon mapped readsi indicates the counts of one transcript and exon lengthi indicates the length of this transcript. Secondly, the average length of all the expressed transcripts was calculated.
Lengthi indicates the length of transcripti and n indicates the number of expressed transcripts. According to RNA contents in each spermatozoon estimated above, the average molecular weight of ribonucleotide in RNA measures 340 Da. The number of total transcripts in a spermatozoon was calculated using the following formula:
The transcripts number indicates the number of total transcripts in a single spermatozoon and N indicates the Avogadro's constant. To ensure that transcripti exists in each spermatozoon, the average counts of the tanscriptishould be more than 1 in each spermatozoon. Based on that, the minimum TPM of transcripticommonly existed in the sperm can be calculated as follows:
The m value indicates the minimum TPM of transcripti existing in each spermatozoon.
The m value serves as the threshold value for each sample to screen the consistently expressing transcripts in each spermatozoon. We took the intersection of the transcripts with TPM ≥ m in each sample as the commonly expressed RNAs in all spermatozoa.
Differential Expression Analyses
Raw data were filtered to remove the adapters, poly-N, and low-quality reads using Trimmomatic version 0.38 (Bolger et al., 2014). Clean reads were mapped to giant grouper genome (unpublished) using TopHat2 Version 2.1.1 (Kim et al., 2013). The mapping ratio was counted using samtools flagstat (Li, 2011). The counts of transcripts were calculated using HTSeq version 0.12.4 (Anders et al., 2015). The counts were weighted by the RNA contents in each spermatozoon. Final DEGs were defined by FDR statistic method. The FDR with a cut-off value of 0.05 and log2 fold change with a cut-off absolute value of 1 were used to identify the significant differentially expressed genes (DEGs). For a better understanding of the effects of the biological processes of sperm, the transcripts were investigated by gene set enrichment analysis (GSEA) (Mootha et al., 2003). The transcripts with an average TPM of more than 200 in the fresh sperm were used for GSEA. The TPM of transcripts in the cold-stored sperm were weighted by their RNA contents per sperm. All GSEA analyses were carried out using the GSEA version 4.1.0 software (Mootha et al., 2003). The h.all.v7.2.symbols.gmt [Hallmarks] was served as the background for genes. The genes are ranked using the Signal2Noise method to produce a gene list, and other parameters were set as default.
Mitochondrial Gene Expression in Sperm
The mitochondrial genome of the giant grouper genome (NC_011715.1) was mapped onto the RNA-seq data using the BWA software (Li and Durbin, 2009); the mapped data were then sorted using samtools. The counts of genes were obtained using HTSeq version 0.12.4. The TPM were calculated using the above mentioned method. The quality of mitochondrial RNA in each sperm was estimated using the following formula:
The value of m indicates the mitochondrial RNA contents per sperm (in pg), m.mapping.ratio indicates the mapping ratio of clean data on the mitochondrial genome, and g.mapping.ratio indicates the mapping ratio of clean data on the genome.
The TPM of the mitochondrial RNA were weighted based on the mitochondrial RNA contents per sperm. The differential expression analysis was analyzed based on the false discovery rate (FDR) statistic methods.
Quantification of Protein in the Sperm
Three sperm samples were collected from each group (FS, C1, C2, C3, and C4) to quantify protein contents. In simple, 100 μl sperm was added into 900 μl of 0.9% saline solution and was homogenized. The lysate was centrifuged at 4,000 r/min for 10 min at 4°C and the supernatant was transferred to a clean tube. The protein quantity was measured using Total Protein Assay Kit (BCA method) (Jiancheng Bioengineering Institute, Nanjing) according to the instructions of the manufacturer.
Proteome Library Preparation and Sequencing
Fresh sperm and C2 were used for constructing proteome libraries. About 100 μl sperm from three individuals of each group were added into 600 μl of Lysis buffer (50 mM Tris buffer, 8M Urea, 1% SDS, Ph = 8), followed by 5 min of ultrasonication under 4°C. The lysate was centrifuged at 13,000 r/min for 20 min at 4°C and the supernatant was transferred to a clean tube. The supernatant was mixed with cold acetone with 10 mM DDT for 2 h. After centrifuging at 13,000 r/min for 20 min, the sediments were collected and washed two times with cold acetone. The protein precipitation was dissolved by 8 M Urea (50 mM Tris buffer, 8 M Urea, pH = 8). Protein quality was assessed using Coomassie (Bradford) Protein Assay Kit (Thermo Fisher Scientific, MA, USA) according to the instructions of the manufacturer.
A total of 120 μg protein for each sample was taken to add TMT label mixed with acetonitrile and incubated at room temperature for 2 h. The labeling reactions were terminated by ammonium hydroxide with a final concentration of 8%. Peptides tagged by tandem mass of each sample were mixed in equal proportion.
Peptide fraction separation was performed using a C18 column (Waters BEH C18 4.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC system. The lyophilized powder of peptides was dissolved in Solution A (2% acetonitrile, adjusted pH to 10.0 using ammonium hydroxide) and centrifuged at 12,000 × g for 10 min at room temperature. A column oven was set at 50°C. Solution B (98% acetonitrile) was used to develop a gradient elution. The flow rate was set at 1 ml/min, and fractions were collected in a tube per minute under a controller based on UV absorbance at 214 nm. All fractions were collected as ten fractions and dried under vacuum, and then, reconstituted in 0.1% of formic acid (FA) and water for LC-MS/MS analysis.
For transition library construction, shotgun proteomics analyses were performed on EASY-nLCTM 1200 UHPLC system (Thermo Fisher Scientific, MA, USA) coupled with a Q Exactive HF (X) mass spectrometer (Thermo Fisher Scientific, MA, USA). About 1 μg of sample was injected into a C18 Nano-Trap column (2 cm × 75 μm, 3 μm). Peptides were separated in an analytical column (15 cm × 1 50 μm, 1.9 μm) using a linear gradient elution. The separated peptides were analyzed by MS/MS using Q Exactive HF (X) mass spectrometer (Thermo Fisher Scientific, MA, USA), with ion source of Nanospray Flex™ (ESI). Spray voltage and ion transport capillary temperature were set at 2.3 kV and 320°C.
Proteomic Data Analysis
Peptide identification and quantitation were carried out using Proteome Discoverer 2.2 (PD 2.2, Thermo) based on the giant nuclear genome and mitochondrial genome. The parameters are set as follows: mass tolerance for the precursor ion was 10 ppm and mass tolerance for the product ion was 0.02 Da. Carbamidomethyl was specified as fixed modifications, Oxidation of methionine (M) and TMTplex was specified as dynamic modification, acetylation and the TMT plex were specified as N-Terminal modification in PD 2.2. A maximum of 2 miscleavage sites were allowed. Maximum FDRs were set to 1% for peptide and protein identification. The protein quantitation results were weighted by protein concentration in sperm and statistically analyzed by T-test. The proteins, whose quantitation were significantly different between experimental and control groups (p < 0.01 and |log2FC| >1.2) were defined as differentially expressed proteins (DEPs). The sperm proteins were enriched using GSEA, and the parameters were the same as mentioned above. In addition, the average degradation rate of proteins was estimated using following formula:
Average degradation rate of proteins involved in the pathways were standardized by average degradation rate of total proteins.
Enzymatic Determination in Sperm
Five sperm samples were collected from each group (FS, C1, C2, C3, and C4) with motility >90% for below experiments. The ATP contents were quantified using ATP assay kit (Jiancheng Bioengineering Institute, Nanjing, China). Sample preparation was described as follows: About 100 μl sperm was added into 900 μl at 95°C in double distilled water, fully ground and incubated at 95°C for 10 min. The lysate was centrifuged at 4,000 r/min for 10 min at 4°C and the supernatant was transferred to a clean tube. The ATP quantification details were prepared according to the instructions of the manufacturer (http://www.njjcbio.com/). Oxidation resistance in the sperm were analyzed, and the total antioxidant capacity (T-AOC) was analyzed using the Total Antioxidant Capacity Assay Kit (ABTS method) (Jiancheng Bioengineering Institute, Nanjing, China). Superoxide dismutase (SOD) was quantified using the SOD Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China). Total glutathione (T-GSH), oxidized glutathione disulfide (GSSG), and reduced glutathione (GSH) were quantified using Total Glutathione/Oxidized Glutathione Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China). The sample preparation was done in the following way: About 100 μl sperm was added into 0.9% saline solution and homogenized. The lysate was centrifuged at 4,000 r/min for 10 min at 4°C and the supernatant was transferred to a clean tube. The quantification details were prepared according to the instructions provided by Jiancheng Bioengineering Institute (http://www.njjcbio.com/).
Statistical Analysis
Difference between different groups was analyzed using SPSS 22.0 software (SPSS Inc., Chicago, IL, United States) and One-way ANOVA method. The significant difference was marked using different alphabets (p < 0.05).
Results
Sperm Changes During Storage in vitro
The initial sperm motility was 97.70 ± 1.05%, in prolonged cold storage; the sperm motility gradually decreased. Compared to FS, the motility of 1 day stored (95.70 ± 1.60%) and 2 day stored sperm (96.24 ± 2.02%) were slightly lower than that of FS. On the fourth day, the motility (74.53 ± 4.45%) was significantly lower than that of the FS (p < 0.05; Figure 1A). For VCL, the FS reached up to 108.21 ± 3.88 μm/s. Subsequently, the VCL consistently decreased with 84.85 ± 2.15 μm/s on the first day (p < 0.05), 74.07 ± 1.51 μm/s on the second day (p < 0.05), 63.62 ± 0.98 μm/s on the third day (p < 0.05), and 60.42 ± 1.73 μm/s on the fourth day (p < 0.05; Figure 1B). The changes in the VSL showed similar trend as in VCF during cold storage (Figure 1C). In FS, A-level spermatozoa reached up to 42.93 ± 2.84%, and then significantly decreased to 20.37 ± 2.15% on the first day (p < 0.05), 9.53 ± 2.70% on the second day (p < 0.05), 3.13 ± 0.35% on the third day (p < 0.05), and 1.24 ± 0.45% on the fourth day (p < 0.05; Figure 1D). The B-level spermatozoa accounted for 28.82 ± 0.63% in FS and significantly increased in the first 2 days, then decreased, and were significantly lower than FS on the fourth day (Figure 1D). The C-level spermatozoa gradually increased with prolonged cold storage and were significantly higher than FS after the second (p < 0.05), third (p < 0.05) and fourth days (p < 0.05; Figure 1D). The D-level spermatozoa gradually increased and showed significant difference after the fourth day (p < 0.05; Figure 1D). In zygotes fertilized by FS, the rate of normal, abnormal, and unfertilized eggs were found to be 93.15 ± 2.22, 5.19 ± 2.33, and 1.67 ± 1.20%, respectively. With the prolonged cold storage, the fertilization rate gradually decreased and showed significant difference after the second day (77.02 ± 2.20%; p < 0.05), while the abnormal and unfertilized eggs gradually increased and showed significant difference after the second (15.03 ± 2.54%; p < 0.05) and third days (7.94 ± 2.54%; p < 0.05), respectively (Figure 1E). The hatching rates of fertilized eggs showed a decreasing trend, from 89.33 ± 0.88% in FS to 20.67 ± 1.45% in 4 day stored sperm (p < 0.05) (Figure 1F). The rate of malformed larva gradually increased during storage and significantly increased after 2 days (p < 0.05; Figures 1G,H).
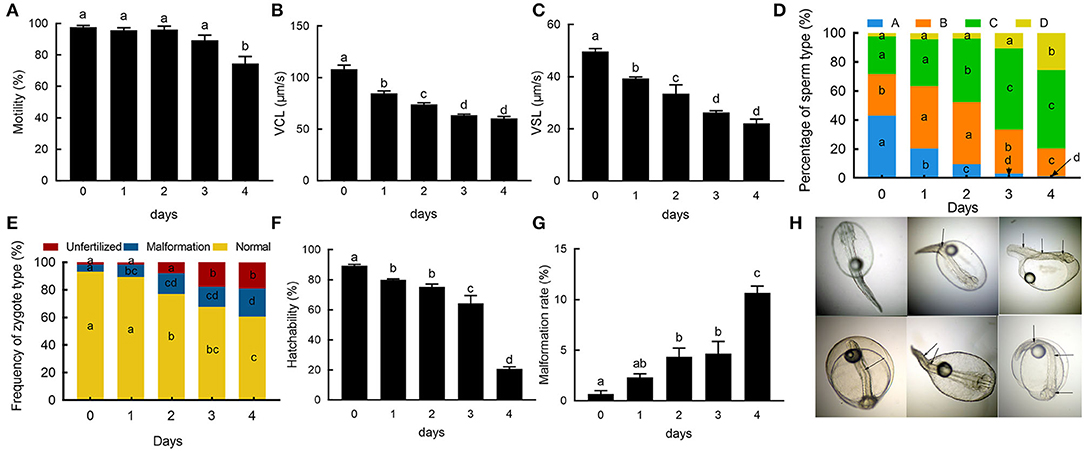
Figure 1. The sperm changes during cold storage. (A) The changes of sperm motility during cold storage. (B) The changes of curvilinear velocity (VCL) of sperm during cold storage. (C) The changes of straight-line velocity (VSL). (D) The rate of changes of the sperm type during cold storage. Blue, orange, green, and yellow indicate “A, B, C, and D” type spermatozoa, respectively. (E) The rate of changes of zygote types. Yellow, blue, and red indicate normal, malformed, and unfertilized zygote. (F) Changes of hatchability. (G) Malformation rate of fry using sperm on different days; (H) Normal and malformed fries. Different letters indicate the significant difference among groups; Arrows point to the deformity of fries.
RNA-Sequencing and Estimation of Commonly Expressed mRNA in Each Spermatozoon
In FS, a total of 21,286 transcripts were identified. Firstly, we obtained the top 30 transcripts with the highest expression level in sperm; the transcripts were mainly involved in motility, energy metabolism, cell division, peptide biosynthetic process, etc. (Figure 2A). MD52 is the most highly expressed gene in sperm. For estimating the commonly expressed mRNAs in each spermatozoon, the minimum TPM values of the transcripts commonly existed in each spermatozoon were calculated depending on the absolute RNA contents in each sperm and RNA-seq data; the TPM values in three samples were found to be 285.62, 218.39, and 222.76, respectively. Based on the threshold value, a total of 1,677 transcripts were collected (Figure 2B). These transcripts were involved in energy metabolism, ribosome, protein metabolism, RNA transport, and lysosome (Figure 2C). In sperm, most transcripts related to oxidative phosphorylation, glycolysis, and citrate cycle were highly expressed (Figures 2D–F and Supplementary Figure 2). Based on the database of gene ontology (GO), the commonly expressed transcripts were mainly enriched with translation, proton transport, and ATP metabolic process (Supplementary Figure 1).
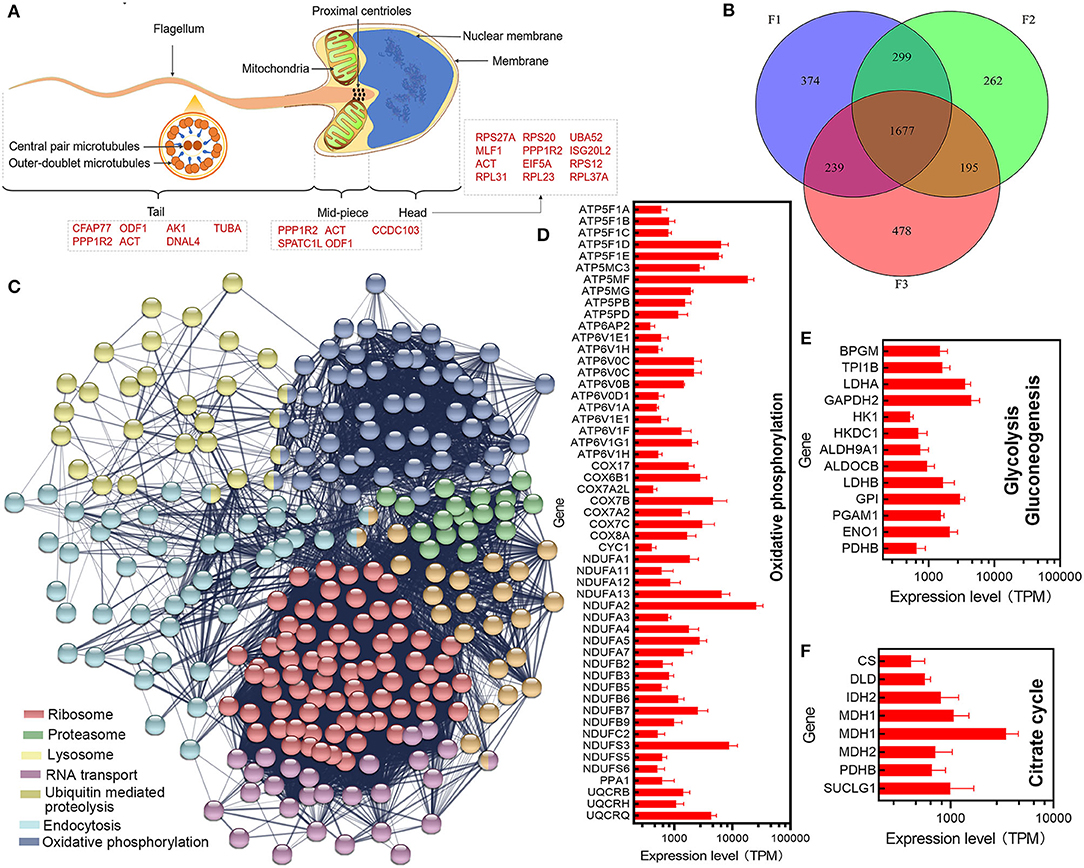
Figure 2. Estimation of commonly expressed transcripts in each spermatozoon. (A) Distribution of highly expressed transcripts. (B) Intersection of commonly expressed transcripts in each sample. (C) Significant enriched pathway of commonly expressed transcripts in each spermatozoon. (D–F) Expression levels of transcripts involved in the oxidative phosphorylation, glycolysis/gluconeogenesis, and citrate cycle pathway, respectively.
Comparative Transcriptome Analysis Between Fresh Sperm and Cold-Stored Sperm
The absolute RNA contents in sperm decreased gradually during cold storage in vitro. The RNA contents of FS were found to be 6.78 ± 0.67 μg/ml, while after 2 days of cold storage, the RNA contents significantly decreased to 3.87 ± 0.08 μg/ml (Figure 3A). The valid RNA content per spermatozoon was 0.045 ± 0.0041 pg in FS, and it significantly decreased to 0.025 ± 0.0025 pg in C2 (Figure 3B). The TPM values were weighted based on the valid RNA contents per spermatozoon (Figure 3C). Most transcripts were decreased during cold storage (Figure 3D). Among them, there were 76 DEGs (Figure 3E). Based on GSEA, decreased transcripts were significantly enriched into pathways related to immunity (complement, interferon response) and stress response (hypoxia) (Figure 3F). In addition, numerous transcripts involved with glycolysis were also decreased (Figure 3G).
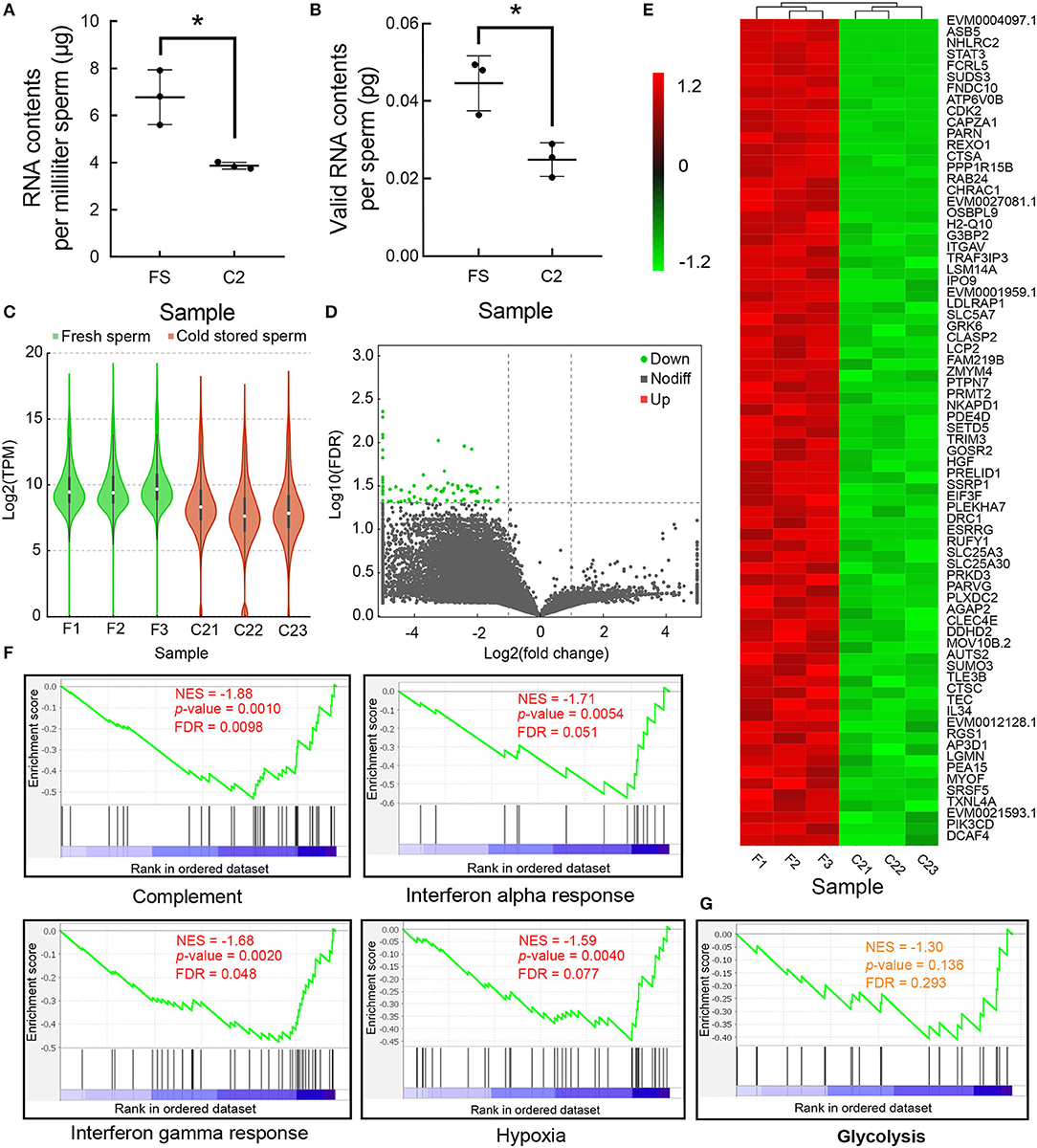
Figure 3. The changes of messenger RNAs (mRNAs) during cold storage. (A) The changes of RNA contents per milliliter fresh sperm (FS) and C2. (B) The changes of RNA contents per sperm at FS and C2. (C) The changes in the TPM transcripts during cold storage. (D) The volcano graph of all transcripts. Green indicates the downregulated differentially expressed genes (DEGs). (E) The heatmap of DEGs. (F) The four most-enriched gene set during cold storage based on GSEA. (G) The enrichment of transcripts involved in glycolysis. Asterisk indicated significant difference (p < 0.05).
Transcriptome Analyses of Mitochondria in Sperm
The transfer RNAs (tRNAs) in mitochondria were not detected in RNA-seq data (Figure 4A). In the mitochondria of sperm, the two highest expressed transcripts include 16s rRNA and 12s rRNA (Figure 4C), followed by COX1, COX3, and ATP6 transcripts. In addition, the expression profiles of the transcripts in mitochondria from multiple tissues were compared, except sperm; the expression levels of 12srRNA of other tissues are all very low (Figure 4B). The RNA contents in the mitochondria of each spermatozoon were estimated based on the mapping rate of the mitochondrial RNA. In FS, the mitochondrial RNA content in each spermatozoon was 0.0030 ± 0.00039 pg, in C2, the content was significantly decreased with the value of 0.0017 ± 0.00006 pg (Figure 4D). However, there was no significant difference in the proportion of mitochondrial RNA and total RNA in FS and C2 (Figure 4E). The TPM values of mitochondrial RNA were weighted based on the RNA contents in the mitochondria of spermatozoon. The results showed that all mitochondrial RNAs were decreased (Figure 4F), and nine transcripts of them were significantly decreased (Figure 4G).
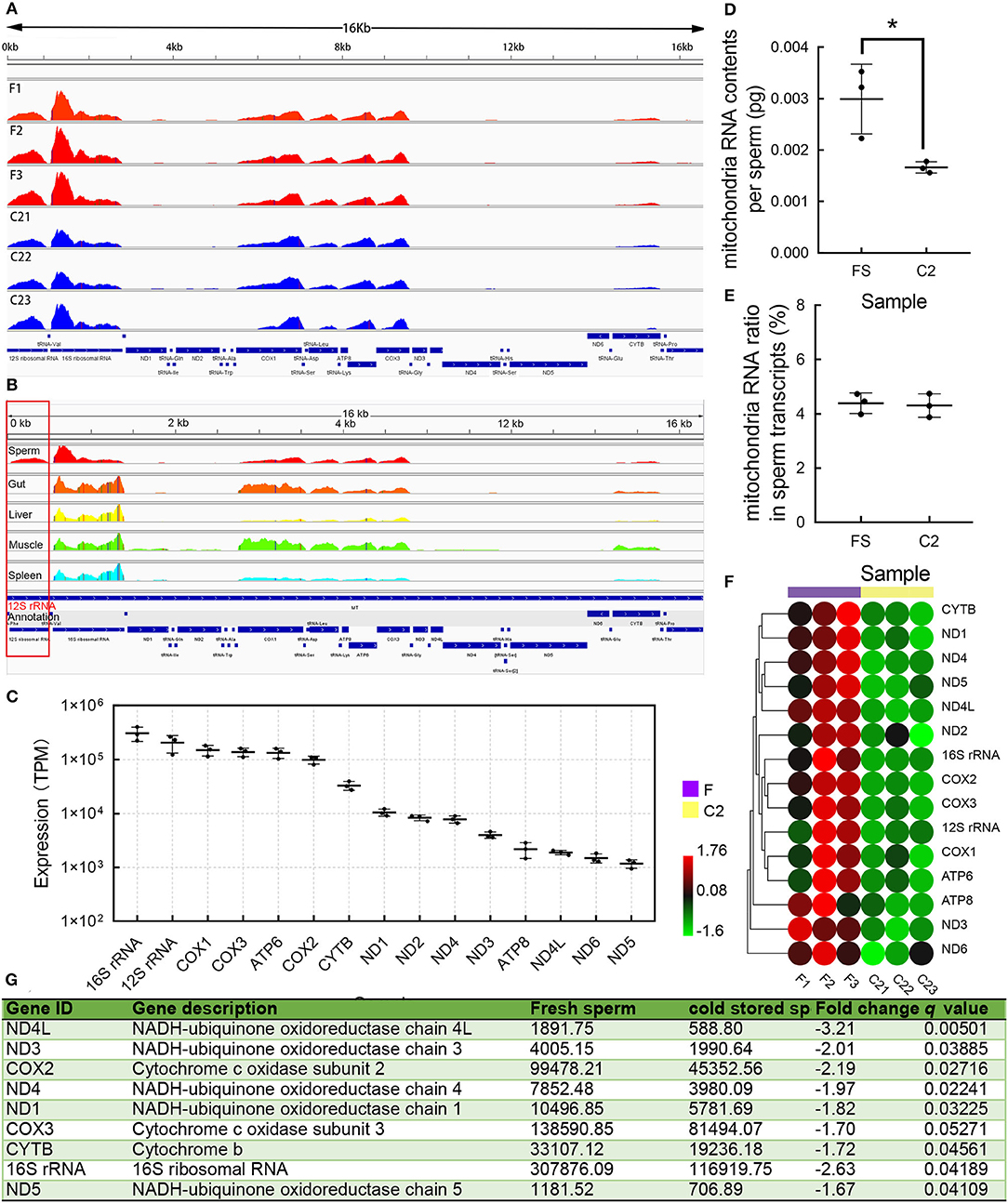
Figure 4. The components and changes of mitochondrial transcript. (A) The expression profiles of mitochondrial messenger RNA (mRNA). (B) The expression profiles of mitochondrial mRNA in sperm and other tissues in the giant grouper. The 12s rRNA showed a high expression level in sperm and low expression levels in other tissues. (C) The expression levels of mitochondrial mRNA. (D) The mitochondrial mRNA contents per sperm. (E) Mitochondrial mRNA ratio changes and whole mRNA ratio change. (F) The heatmap of mitochondrial mRNA during cold storage. (G) The differentially expressed transcripts during cold storage. Asterisk indicated significant difference (p < 0.05).
Proteome Analyses of Sperm
In FS, 3,195 proteins were detected and they were found to be widely distributed across 24 chromosomes (Figure 5A). We set a high threshold (4,096 or 212) of expression to collect the highest expressed protein in sperm. We obtained 19 proteins, which were closely associated with signal transduction, translation, stress response, immunity, metabolism etc. (Figure 5A and Supplementary Figure 3). In addition, 1,174 proteins were obtained after removing the lower expressed proteins with an expressed threshold (256 or 28) (Figure 5A). These proteins were aligned to GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway databases (Supplementary Figures 4, 5), which showed that these proteins were mainly associated with energy metabolism, complement system, and aminoacyl-tRNA biosynthesis. The expression profiles of mRNAs and proteins in sperm were compared, and they showed no significant consistency with a low Pearson coefficient of 0.11 (Figure 5F).
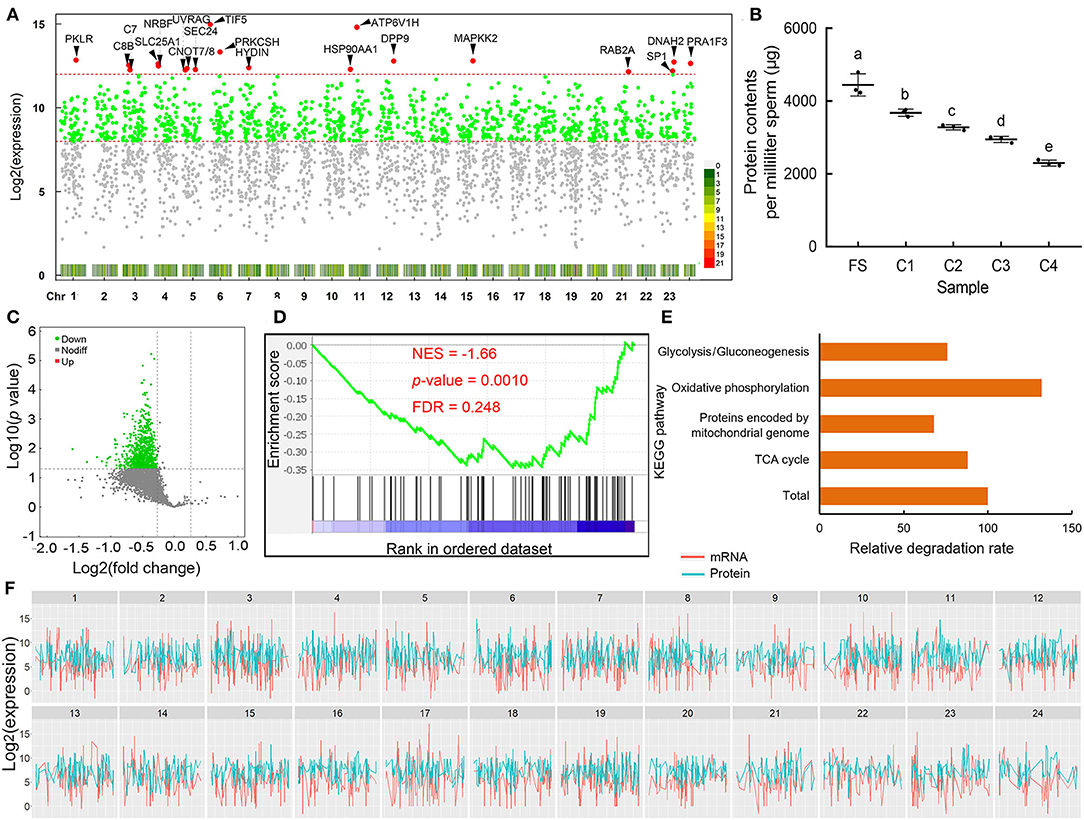
Figure 5. The components and change of protein in sperm. (A) The distribution and density of sperm proteins. The red, green, and gray points indicate the highest expressed, high expressed, and low expressed proteins. (B) The changes of total protein contents in sperm during cold storage. (C) The volcano plot of proteins during storage; green points indicate the decreased proteins. (D) The enrichment of proteins related to oxidative phosphorylation. (E) Relative degradation rate of genes associated with energy metabolism. (F) The expression profiles of transcripts and proteins in sperm; green and red indicate proteins and transcripts, respectively. Different letters indicated significant difference (p < 0.05).
The absolute protein concentrations were found to be 4444.30 ± 176.10, 3680.94 ± 57.50, 3279.85 ± 40.40, 2949.93 ± 48.84, and 2296.54 ± 46.65 μg in FS, 1, 2, 3, and 4 days cold-stored sperm, respectively. The protein concentrations gradually decreased during cold storage (Figure 5B). The expression levels of proteins were weighted based on the protein concentration and applied to differential expression analysis. The results showed that most of the proteins decreased (Figure 5C), and proteins involved in the oxidative phosphorylation pathway was significantly enriched using GSEA (Figure 5D). In addition, proteins involved in oxidative phosphorylation showed a faster degradation rate and proteins encoded by mitochondrial genome showed a lower degradation rate compared to the total average of proteins (Figure 5E).
Change of Oxidation Resistance in Sperm
The total antioxidant capacity) and SOD gradually decreased in the sperm during cold storage (Figures 6A,B). With prolonged cold storage, the total TGSH slowly decreased; at the 4 day, the TGSH was significantly lower than that of the FS (Figure 6C). The GSSG) significantly increased and the GSH significantly decreased during cold storage (Figures 6D,E). In addition, we observed significant decrease of ATP contents in sperm during prolonged cold storage (Figure 6F).
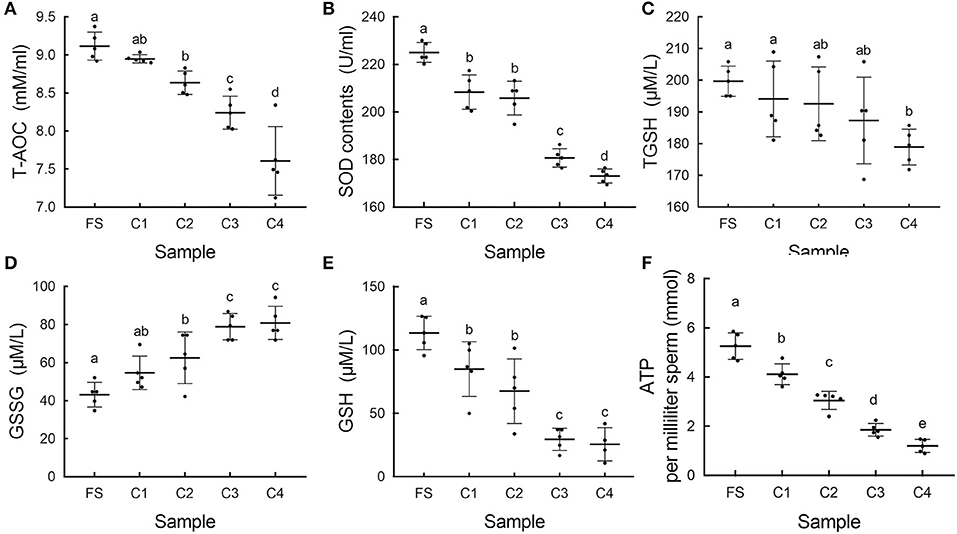
Figure 6. The change of oxidation resistance during cold storage. (A) The changes of total antioxidant capacity (T-AOC) in sperm during storage in vitro. (B) The changes of superoxide dismutase (SOD) in sperm. (C) The changes of total glutathione (TGSH) in sperm. (D) The changes of oxidized glutathione disulfide (GSSG) in sperm. (E) The changes of reduced glutathione (GSH) in sperm. (F) The changes of adenosine triphosphate (ATP) contents in sperm. Different letters indicated significant difference (p < 0.05).
Discussion
Giant grouper is the most valuable male parent in the cross-breeding of groupers. Sperm storage is an assurance for the reproduction of hybrid grouper. Understanding of the mechanism of sperm inactivation in vitro is essential for prolonging the storage time. Here, we focused on the effects of storage in vitro on functions, such as transcriptome, proteome, and oxidation resistance of sperm. First, we analyzed the changes in the motility, velocity, fertilization, and hatchability of sperm during storage in vitro. Then, the profiles of the transcripts and the proteins in sperm were discovered and their change during cold storage in vitro was displayed. In addition, we discovered the decrease of ATP and oxidation resistance of sperm during prolonged storage time in vitro. The studies provide important information for the improvement of sperm storage.
During storage time in vitro, the motility, velocity, Type A spermatozoa, fertility, and hatchability gradually decreased in sperm. On the fourth day, the hatchability of sperm severely decreased, which is perhaps due to the fact that the sperm cannot keep up the normal function owing to severe damage at multiple aspects. We suggest that the maximum time of cold storage for giant grouper sperm is 3 days. The maximum time for cold storage is similar to lots of other fish species. For example, the semen of Masusalmon (Oncorhynchus masou) preserved at 4°C for 4 days had relatively high sperm quality (Scott and Baynes, 1980); for patagonian blenny (Eleginops maclovinus), high sperm mobility was maintained for 3 days under 4°C (Ulloa-Rodriguez et al., 2018). The results provide valuable information for sperm conservation of giant grouper. Compared to cryopreservation of giant grouper sperm (Tian et al., 2015), the 2 day stored sperm at 4°C showed a similar fertility rate and hatchability. The fertility rate and hatchability of 3 day stored sperm are lower than those of the cryopreserved sperm, though their motility is similar.
Firstly, 30 highest expressed genes from FS were obtained. These genes were closely associated with sperm motility and functions, energy metabolism, and cell division. For example, outer dense fibers (ODF), spermatogenesis and centriole-associated 1 like (SPATC1L) protein, Tubulin and Coiled-coil domain-containing protein 103 (CCDC103) play important roles in cytoskeleton stability and in the motion of sperm tail (Panizzi et al., 2012; Yang et al., 2012; Parab et al., 2017; Kim et al., 2018). Adenylate kinase isoenzyme 1 is involved in the energy metabolism for sperm motility (Cao et al., 2006). Myeloid leukemia factor 1 (MLF1) and tumor protein D52 (TPD52) is involved in cell cycle (Winteringham et al., 2004; Zhao et al., 2017). These transcripts may play important roles both in sperm functions and in the early development of embryo.
Transcripts per kilobase million is a normalization method to remove the effect of exon length of transcripts (Zhao et al., 2020). Based on the computational formula of TPM, it is directly proportional to the number of transcripts. In addition, spermatozoa contained few or even no rRNA (Miller et al., 2005; Fang et al., 2014; El Fekih et al., 2017), which provides a favorable condition for the estimation of consistent transcripts. The average number of transcripts in each spermatozoon can be estimated based on TPM values and average RNA contents in each spermatozoon. The average number of transcripts is more than 1; they are probably considered to be existing in each spermatozoon. Based on above strategies, the threshold of TPM for screening consistent transcripts in each spermatozoon could be estimated.
A total of 1,677 transcripts were obtained as consistent transcripts in spermatozoa. The common existence of these transcripts implied their importance in sperm. These transcripts existed in each spermatozoon were mainly involved in energy metabolism, ribosome, protein metabolism, RNA transport, and acrosomal reaction (Shi et al., 2018). The other remaining transcripts in spermatozoon may be the residual of transcripts associated with sperm maturation and spermatogenesis. During the last phase of spermatogenesis, the spermatids are transformed into spermatozoon. The DNA condenses into tightly compact nucleus in spermatozoon head. Transcription is stopped, where almost all of the cytoplasm is discarded, and a mass of mRNA related to spermatogenesis were also degraded (Hecht, 1998).
The number of common genes in the giant grouper is less than that (4,885) in the mouse (Fang et al., 2014). The difference may be caused by two major reasons. One is the difference in the RNA contents between the two species, where the RNA content of the sperm was 0.045 ± 0.0041 pg per spermatozoon in the giant grouper, while the value was approximately 0.1 pg in murine sperm (Pessot et al., 1989; Concha et al., 1993). The other reason is that we applied a stricter threshold. We obtained these commonly existing transcripts by taking the intersection of three duplicates, which was more strictly done than that from the one mixed sample (Fang et al., 2014). Here, the total RNAs were isolated from the spermatozoa using a traditional phase separation and precipitation method, which is also known as TRIzol method. Here, we used 75% ethyl alcohol with 0.5 M lithium chloride to wash the precipitation to reduce the loss of RNA. Lithium chloride facilitates the precipitation of RNAs and keeps the integrity of RNAs to ensure the correction of RNA quantification (Cathala et al., 1983).
During cold storage, the amount of RNAs per spermatozoon was significantly decreased. Most of the mRNAs were closely related to stress response, immunity, and energy supply. These transcripts were transcribed before the transformation of spermatids and stored as ribonucleoprotein particles, which wait for translation in sperm at the right time (Steger, 1999). The significant decrease of RNA contents probably affects the normal functions of spermatozoon and in the early development of embryo. In addition, during storage in vitro, the mRNAs were damaged by multiple harmful factors. Timely degradation of impaired mRNAs is beneficial to the survival of sperm. We discovered several high-expressed mRNA deadenylases in the sperm using tandem mass tag (TMT)-labeled quantitative proteomic analysis. These RNA enzymes (CCR4-NOT transcription complex submits) possess 3'-5' poly(A) exoribonuclease activity and closely relate to bulk mRNA degradation and translational repression (Cooke et al., 2010). They may play important roles in the programmed degradation of sperm mRNAs. During cold storage in vitro, multiple genes involved in immunity, resistance to hypoxia, and energy metabolism were significantly decreased. Bacteria in semen induce serious injury to sperm. Multiple studies have reported that the presence of microorganisms negatively affect the motility and fertility of the sperm (Stoss and Refstie, 1983; Viveiros et al., 2010). With the reduction of genes associated with immunity, sperm is more susceptible to injury of bacteria. Respiration is a common physiological activity of sperm during storage, and oxygen contributes to the maintenance of the normal function of sperm (Contreras et al., 2019). Genes associated with hypoxia tolerance may play important roles in sperm survival during cold storage under hypoxia condition. Moreover, the energy metabolism is closely related to ATP production, which is the most important energy resource of sperm. The decrease of these genes would reduce the sperm quality during cold storage in vitro.
In sperm, mitochondria are energy-supplying organelles, which also provide space for RNA translation. Gur and Breitbart (2006) observed that the inhibition of protein translation in mitochondria significantly reduced sperm motility, capacitation, and fertilization rate. Cytoplasmic mRNAs can enter mitochondria and utilize the mitochondrial ribosome to be translated as proteins (Gur and Breitbart, 2006). In mitochondrial genome, there are 13 protein-coding genes, 22 tRNA, 16S rRNA, and 12S rRNA. The raw data were mapped against the mitochondrial genome. We obtained expression profiles of 13 protein-coding genes, 12S rRNA, and 16S rRNA. During RNA sequencing library construction, the mRNAs were purified using magnetic beads with Oligo dT sequence. The tRNAs cannot be detected due to lack of poly A tails. The 16S rRNA and 12S rRNA are not only detected by RNA-seq, but their expression levels are the highest in the mitochondria of sperms as well. The results suggested that there were poly A tails at the end of 16S rRNA and 12S rRNA in the mitochondria of sperm. Abundance of 16S rRNA is essential for intra-mitochondrial translation (Arroyo et al., 2016); its high expressed level guarantees the normal function of sperm. Moreover, compared to other tissues, the expression level of 12S rRNA is very high in the mitochondria of sperm. This phenomenon may be a specialization of mitochondria for specific translation system in sperm. The 12S rRNA is a component of the 28S which is the small subunit of 55S mitochondrial ribosome (Sharma et al., 2003). The 55S ribosomes are involved in protein translation in sperm (Gur and Breitbart, 2006). High reserve of 12S rRNA facilitates the normal translation of mRNA in sperm.
During cold storage, the absolute RNA contents gradually decreased in the mitochondria of sperm. There were similar degradation rates for mitochondrial RNA and whole RNA. All transcripts showed a declining trend, where nine of them significantly decreased, which include five NADH dehydrogenases subunits (ND1, ND3, ND4, ND4L, and ND5) and two cytochrome c oxidase subunits (COX2 and COX3). They were involved in the respiratory chain (Ohnishi et al., 2018; Hartley et al., 2019), which is a complicated enzyme complex that drives ATP synthesis by a series of hydrogen transfer reactions and electron transfer reactions (Ohnishi et al., 2018). The damage of mitochondrial transcripts may induce ATP reduction and in the dysfunction of sperm.
In the current study, the protein profiles of sperm were obtained using TMT-labeled quantitative proteomic analysis. A total of 3,195 proteins were detected; these proteins were widely distributed throughout the whole genome. The highest expressed proteins were associated with energy metabolism, motility, stress response, and protein degradation. In sperm, ATP is the direct energy source for sperm functions, including mobility, capacitation, hyperactivation, fertilization, etc. Two metabolic processes, glycolysis and oxidative phosphorylation, are the most important ways to produce ATP (Du Plessis et al., 2015). Several highly expressed proteins were involved in the two processes, such as pyruvate kinase (PKLR) (Israelsen and Vander Heiden, 2015) and tricarboxylate transport protein (SLC25A1) (Chaouch et al., 2014). Movement speed is an important parameter, and sperm movement mainly depends on the ciliary swing. Hydin and Dynein axonemal heavy chain 2 (DNAH2) are the components of cilia that are pivotal for sperm motility (Dawe et al., 2007; Ciolfi et al., 2016). Sperm has defense capability for external environment. For example, UV radiation resistance-associated gene protein (UVRAG) plays an important role in the maintenance of chromosomal stability and DNA double-strand break (DSB) repair (Zhao et al., 2012). The heat shock protein, HSP 90-alpha (HSP90AA1) as molecular chaperone, assists in the correct folding, stabilization, and activation of proteins. Complement component C7 (C7) and complement component C8 beta chain (C8B) play key roles in the innate and adaptive immune responses (Haas et al., 1983). In sperm, several proteins were related to protein degradation, such as dipeptidyl peptidase 9 (DPP9) (Gall et al., 2013) and E3 ubiquitin-protein ligase SP1 (Ling and Jarvis, 2015). These proteins may play important roles in protein attenuation during the cold storage of sperm. It is worth noting that the highest expressed protein is eukaryotic translation initiation factor 5 (TIF5), which is essential for polypeptide synthesis. High expression of TIF5 implies that the transcription is not completely silent in sperm.
After removing lower expressed proteins, 1,174 proteins were remained. Many of them were involved in energy metabolism, including carbon metabolism, pentose phosphate, citrate cycle, and oxidative phosphorylation. These processes provide enough energy for the functions of sperm. Proteins associated with complement system also were abundant in sperm. These proteins facilitate to protect sperms from pathogenic microorganisms. In addition, proteins involved in aminoacyl-tRNA biosynthesis were also significantly enriched in both KEGG and GO database; the existence of these proteins suggests that the translation is not silent in sperm.
Absolute protein quantity gradually decreased during cold storage in sperm. Almost all proteins were decreased. There were numerous proteins involved in the degradation of polypeptide and amino acids. Protein degradation helps sperm to remove damaged proteins, remobilize, and recycle amino acids and transform amino acids to the energy material (Goldberg, 2003). The protein degradation provides essential energy and reduces injured proteins, which contributes to the maintenance of normal sperm l functions. In addition, numerous proteins involved in oxidative phosphorylation were significantly decreased. Proteins involved in oxidative phosphorylation showed a faster degradation rate. The main source of reactive oxygen species (ROS) is due to the leakage of electrons from the electron transport chain during the process of oxidative phosphorylation. The ROS is one of the main reasons for reducing the protein activity (Trenzado et al., 2006; Aitken et al., 2012). Proteins involved in oxidative phosphorylation are the first to be affected by ROS. Interestingly, proteins encoded by mitochondrial genome showed a lower degradation rate compared to the total average of proteins. Only one protein, the ND4, showed a significant decrease. This is probably because of the direct utilization of mitochondrial ribosomes by mitochondrial genes to translate proteins as a supplement. In general, sperm proteins were damaged widely, which negatively affect the normal functions of sperm, especially their energy metabolism. In addition, we discovered a significant decline of ATP contents in sperm during cold storage, and the energy metabolism in sperm is unable to satisfy the power requirement of the normal functions of sperm. A recently published study shows that L-carnitine could contribute to the maintenance of antioxidant capacities and enhancing the ATP content and mitochondrial activity in ejaculated sperm (Yang et al., 2020a), which may be a feasible method to prolong the cold storage of giant grouper sperm. Recent studies show that cryopreservation induced significant decrease of several genes involved in the metabolism, oxidation resistance, and signal transduction in the sperm of common carp (Li et al., 2010), which showed similar changes in our research to some extent. However, after cryopreservation, two transferrins were upregulated in the sperm of common carp, which may protect spermatozoa against oxidative damage (Li et al., 2010). The result is different from that in our studies, probably because of the different strategies of storage.
In addition, the correlation of the expression level between transcripts and proteins in sperm were analyzed; there was no correlation between their expression levels with a low Pearson coefficient of 0.11 in sperm. Transcripts existed in the sperm are transcribed during spermatogenesis processes, then some of them were degraded during the transformation of spermatids, and the remaining transcripts were stored for further translation (Steger, 1999). During transformation of spermatids, almost all of the cytoplasm was discarded and the protein components had changed radically (Teves et al., 2020). Thus, a huge difference in the expression level between transcripts and proteins is quite possible.
During storage in vitro, oxidation resistance is significantly decreased in sperm, which suggests that the sperm were threatened by ROS. The main source of ROS is due to the leakage of electrons during electron transport chain in the mitochondria of sperm. The damage of mitochondria during storage speeds up the production of ROS. The ROS causes damage to organelles, membrane structure, DNA and RNA integrity, and protein activity (Trenzado et al., 2006; Aitken et al., 2012). In the present experiment, ROS is an important reason for the damage of transcripts and proteins. Recent studies also show that oxidative stress significantly interrupts oxidative phosphorylation, which causes the loss of motility and velocity of spermatozoa during short storage in vitro (Shaliutina et al., 2013). Addition of antioxidants in the storage medium may be an effective method to prevent sperm from peroxide damage and prolong the storage time in vitro.
Conclusion
In our study, we explored the effects of storage in vitro on functions, such as transcriptome, proteome, and oxidation resistance of the giant grouper sperm. We observed that the motility, velocity, fertilization, and hatchability gradually decreased during cold storage, where the maximum storage time of the sperm is for 3 days. Then, the profiles of the transcripts and proteins in the sperm were analyzed, and their functions and roles in sperm were predicted. During storage, the contents of transcripts and proteins gradually decreased. Expression levels of genes involved in immunity, hypoxia resistance, and energy metabolism were decreased remarkably. Proteins related to energy metabolism were heavily affected. During storage, the oxidation resistance of sperm gradually decreased. The oxidative stress is an important treat to spermatozoa structures and functions during storage in vitro.
Data Availability Statement
The raw data of transcriptome have been deposited in genome sequence archive (https://bigd.big.ac.cn/gsa/). The access numbers of project is PRJCA005270. The access number of raw data were CRA004258. Run numbers were CRX242448–CRX242453.
Ethics Statement
This animal study was reviewed and approved by the Animal Care and Use Committee in School of Life Sciences, Sun Yat-Sen University.
Author Contributions
YY, ZM, and XL designed the study and drafted the paper. YY, SY, BF, and WH collected the samples. YY and TW performed the laboratory work. YY and ZM performed analyses. JX and XW contributed technical assistance. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by China Agriculture Research System of MOF and MARA (CARS-47), National Natural Science Foundation of China (31872572), Yang Fan Innovative & Entrepreneurial Research Team Project (No.201312H10), Huizhou Swan Project (20170214023102296), and Science and Technology Planning of Guangzhou (201804020013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.716047/full#supplementary-material
References
Aguilar-Juárez, M., Ruiz-Campos, G., and Paniagua-Chávez, C. G. (2014). Cold storage of the sperm of the endemic trout Oncorhynchus mykiss nelsoni: a strategy for short-term germplasm conservation of endemic species. Rev. Mex. Biodivers. 85, 294–300. doi: 10.7550/rmb.36352
Aitken, R. J., Jones, K. T., and Robertson, S. A. (2012). Reactive oxygen species and sperm function-in sickness and in health. J. Androl. 33, 1096–1106. doi: 10.2164/jandrol.112.016535
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Arroyo, J. D., Jourdain, A. A., Calvo, S. E., Ballarano, C. A., Doench, J. G., Root, D. E., et al. (2016). A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 24, 875–885. doi: 10.1016/j.cmet.2016.08.017
Boerke, A., Dieleman, S. J., and Gadella, B. M. (2007). A possible role for sperm RNA in early embryo development. Theriogenology 68, S147–S155. doi: 10.1016/j.theriogenology.2007.05.058
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30,2114–2120. doi: 10.1093/bioinformatics/btu170
Braun, R. E. (2001). Packaging paternal chromosomes with protamine. Nat. Genet. 28, 10–12. doi: 10.1038/ng0501-10
Cao, W., Haig-Ladewig, L., Gerton, G. L., and Moss, S. B. (2006). Adenylate kinases 1 and 2 are part of the accessory structures in the mouse sperm flagellum. Biol. Reprod. 75, 492–500. doi: 10.1095/biolreprod.106.053512
Cathala, G., Savouret, J. F., Mendez, B., West, B. L., Karin, M., Martial, J. A., et al. (1983). A method for isolation of intact, translationally active ribonucleic acid. DNA 2, 329–335. doi: 10.1089/dna.1983.2.329
Chaouch, A., Porcelli, V., Cox, D., Edvardson, S., Scarcia, P., De Grassi, A., et al. (2014). Mutations in the mitochondrial citrate carrier SLC25A1 are associated with impaired neuromuscular transmission. J. Neuromusc. Dis. 1, 75–90. doi: 10.3233/JND-140021
Chen, J. X., Ye, Z. F., Yu, Z. S., Wang, J., Li, P. S., Chen, X., et al. (2017). The complete mitochondrial genome of the hybrid grouper (Cromileptes altivelis female x Epinephelus lanceolatus male) with phylogenetic consideration. Mitochondrial DNA Part B Resour. 2, 171–172. doi: 10.1080/23802359.2017.1303346
Chen, Z. F., Tian, Y. S., Wang, P. F., Tang, J., Liu, J. C., Ma, W. H., et al. (2018). Embryonic and larval development of a hybrid between kelp grouper Epinephelus moara female x giant grouper E.lanceolatus male using cryopreserved sperm. Aquac. Res. 49, 1407–1413. doi: 10.1111/are.13591
Ciolfi, S., Mencarelli, C., and Dallai, R. (2016). The evolution of sperm axoneme structure and the dynein heavy chain complement in cecidomid insects. Cytoskeleton 73, 209–218. doi: 10.1002/cm.21291
Concha, I.i, Urzua, U., Yañez, A., Schroeder, R., Pessot, C., and Burzio, L. O. (1993). U1 and U2 snRNA are localized in the sperm nucleus. Exp. Cell Res. 204, 378–381. doi: 10.1006/excr.1993.1046
Contreras, P., Dumorné, K., Ulloa-Rodríguez, P., Merino, O., Figueroa, E., Farías, J. G., et al. (2019). Effects of short-term storage on sperm function in fish semen: a review. Rev. Aquacult. 12, 1373–1389. doi: 10.1111/raq.12387
Cooke, A., Prigge, A., and Wickens, M. (2010). Translational repression by deadenylases. J. Biol. Chem. 285, 28506–28513. doi: 10.1074/jbc.M110.150763
Dawe, H. R., Shaw, M. K., Farr, H., and Gull, K. (2007). The hydrocephalus inducing gene product, Hydin, positions axonemal central pair microtubules. BMC Biol. 5:33. doi: 10.1186/1741-7007-5-33
Djourabchi Borojerdi, A. S., Welchowski, T., Peng, W., Buchen, A., Novak, N., Haidl, G., et al. (2020). Human spermatozoa of male patients with subfertility express the interleukin-6 receptor. Andrologia 52:e13511. doi: 10.1111/and.13511
Du Plessis, S. S., Agarwal, A., Mohanty, G., and Van Der Linde, M. (2015). Oxidative phosphorylation versus glycolysis: what fuel do spermatozoa use? Asian J. Androl. 17, 230–235. doi: 10.4103/1008-682X.135123
El Fekih, S., Nguyen, M. H., Perrin, A., Beauvillard, D., Morel, F., Saad, A., et al. (2017). Sperm RNA preparation for transcriptomic analysis: review of the techniques and personal experience. Andrologia 49:e12767. doi: 10.1111/and.12767
Fan, B., Yang, S., Wang, L., Chen, X., Liu, X., Zhang, Y., et al. (2020). Hybridization of tiger grouper (Epinephelus fuscoguttatus ♀) x giant grouper (Epinephelus lanceolatus ♂) using cryopreserved sperm. Cryobiology 95, 84–89. doi: 10.1016/j.cryobiol.2020.06.001
Fang, P., Zeng, P., Wang, Z. X., Liu, M., Xu, W. J., Dai, J. B., et al. (2014). Estimated diversity of messenger RNAs in each murine spermatozoa and their potential function during early zygotic development. Biol. Reprod. 90:11. doi: 10.1095/biolreprod.114.117788
Gall, M. G., Chen, Y., Vieira De Ribeiro, A. J., Zhang, H., Bailey, C. G., Spielman, D. S., et al. (2013). Targeted inactivation of dipeptidyl peptidase 9 enzymatic activity causes mouse neonate lethality. PLoS ONE 8:e78378. doi: 10.1371/journal.pone.0078378
Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. doi: 10.1038/nature02263
Guo, Y., Li, W. P., Liu, Y. L., Li, P. S., Wei, R. P., Liu, Y., et al. (2018). The complete mitochondrial genome of the hybrid grouper Epinephelus akaara female x Epinephelus lanceolatus male. Mitochondrial DNA Part B Resour. 3, 599–600. doi: 10.1080/23802359.2018.1473727
Gur, Y., and Breitbart, H. (2006). Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 20, 411–416. doi: 10.1101/gad.367606
Haas, G. G. Jr., Schreiber, A. D., and Blasco, L. (1983). The incidence of sperm-associated immunoglobulin and C3, the third component of complement, in infertile men. Fertil. Steril. 39, 542–547. doi: 10.1016/S0015-0282(16)46948-7
Hartley, A. M., Lukoyanova, N., Zhang, Y., Cabrera-Orefice, A., Arnold, S., Meunier, B., et al. (2019). Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc(1). Nat. Struct. Mol. Biol. 26, 78–83. doi: 10.1038/s41594-018-0172-z
Hwang, J. Y., Mulligan, B. P., Kim, H.-M., Yang, B.-C., and Lee, C.-K. (2013). Quantitative analysis of sperm mRNA in the pig: relationship with early embryo development and capacitation. Reprod. Fertil. Dev. 25, 807–817. doi: 10.1071/RD12160
Israelsen, W. J., and Vander Heiden, M. G. (2015). Pyruvate kinase: function, regulation and role in cancer. Semin. Cell Dev. Biol. 43, 43–51. doi: 10.1016/j.semcdb.2015.08.004
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, J., Kwon, J. T., Jeong, J., Kim, J., Hong, S. H., Kim, J., et al. (2018). SPATC1L maintains the integrity of the sperm head-tail junction. EMBO Rep. 19:e45991. doi: 10.15252/embr.201845991
Krawetz, S. A. (2005). Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 6, 633–642. doi: 10.1038/nrg1654
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, P., Hulak, M., Koubek, P., Sulc, M., Dzyuba, B., Boryshpolets, S., et al. (2010). Ice-age endurance: the effects of cryopreservation on proteins of sperm of common carp, Cyprinus carpio L. Theriogenology 74, 413–423. doi: 10.1016/j.theriogenology.2010.02.024
Ling, Q., and Jarvis, P. (2015). Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534. doi: 10.1016/j.cub.2015.08.015
Miller, D., Ostermeier, G. C., and Krawetz, S. A. (2005). The controversy, potential and roles of spermatozoal RNA. Trends Mol. Med. 11, 156–163. doi: 10.1016/j.molmed.2005.02.006
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. doi: 10.1038/ng1180
Ohnishi, T., Ohnishi, S. T., and Salerno, J. C. (2018). Five decades of research on mitochondrial NADH-quinone oxidoreductase (complex I). Biol. Chem. 399, 1249–1264. doi: 10.1515/hsz-2018-0164
Panizzi, J. R., Becker-Heck, A., Castleman, V. H., Al-Mutairi, D. A., Liu, Y., Loges, N. T., et al. (2012). CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 44, 714–719. doi: 10.1038/ng.2277
Parab, S., Dalvi, V., Mylavaram, S., Kishore, A., Idicula-Thomas, S., Sonawane, S., et al. (2017). Tubulin acetylation: a novel functional avenue for CDYL in sperm. Cytoskeleton 74, 331–342. doi: 10.1002/cm.21381
Parodi, J., Guerra, G., Cuevas, M., Ramírez-Reveco, A., and Romero, F. (2017). Effects of storage time on the motility, mortality and calcium levels of Atlantic salmon Salmo salar spermatozoa. J. Fish Biol. 90, 1506–1516. doi: 10.1111/jfb.13245
Pessot, C. A., Brito, M., Figueroa, J., Concha, I.i, Yañez, A., and Burzio, L. O. (1989). Presence of RNA in the sperm nucleus. Biochem. Biophys. Res. Commun. 158, 272–278. doi: 10.1016/S0006-291X(89)80208-6
Saad, A., Billard, R., Theron, M. C., and Hollebecq, M. G. (1988). Short-term preservation of carp (Cyprinus carpio) semen. Aquaculture 71, 133–150. doi: 10.1016/0044-8486(88)90280-3
Scott, A. P., and Baynes, S. M. (1980). A review of the biology, handling and storage of salmonid spermatozoa. J. Fish Biol. 17, 707–739. doi: 10.1111/j.1095-8649.1980.tb02804.x
Shaliutina, A., Hulak, M., Gazo, I., Linhartova, P., and Linhart, O. (2013). Effect of short-term storage on quality parameters, DNA integrity, and oxidative stress in Russian (Acipenser gueldenstaedtii) and Siberian (Acipenser baerii) sturgeon sperm. Anim. Reprod. Sci. 139, 127–135. doi: 10.1016/j.anireprosci.2013.03.006
Sharma, M. R., Koc, E. C., Datta, P. P., Booth, T. M., Spremulli, L. L., and Agrawal, R. K. (2003). Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108. doi: 10.1016/S0092-8674(03)00762-1
Shi, W. J., Jiang, Y. X., Huang, G. Y., Zhao, J. L., Zhang, J. N., Liu, Y. S., et al. (2018). Dydrogesterone causes male bias and accelerates sperm maturation in zebrafish (Danio rerio). Environ. Sci. Technol. 52, 8903–8911. doi: 10.1021/acs.est.8b02556
Steger, K. (1999). Transcriptional and translational regulation of gene expression in haploid spermatids. Anat. Embryol. 199, 471–487. doi: 10.1007/s004290050245
Stoss, J., and Refstie, T. (1983). Short-term storage and cryopreservation of milt from Atlantic salmon and sea trout. Aquaculture 30, 229–236. doi: 10.1016/0044-8486(83)90165-5
Sun, Y., Guo, C. Y., Wang, D. D., Li, X. F., Xiao, L., Zhang, X., et al. (2016). Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus female x E. lanceolatus male). BMC Genetics 17:10. doi: 10.1186/s12863-016-0328-y
Tang, Z. J., Chen, J. X., Tang, L., Chen, X., Li, S. S., Liu, Y., et al. (2017). The complete mitochondrial genome of the hybrid grouper Epinephelus coioides female x Epinephelus akaara male with phylogenetic consideration. Mitochondrial DNA Part B Resour. 2, 31–32. doi: 10.1080/23802359.2016.1241680
Teves, M. E., Roldan, E. R. S., Krapf, D., Strauss, J. F., Iii, B. V., and Sapao, P. (2020). Sperm differentiation: the role of trafficking of proteins. Int. J. Mol. Sci. 21:3702. doi: 10.3390/ijms21103702
Tian, Y., Jiang, J., Wang, N., Qi, W., Zhai, J., Li, B., et al. (2015). Sperm of the giant grouper: cryopreservation, physiological and morphological analysis and application in hybridizations with red-spotted grouper. J. Reprod. Dev. 61, 333–339. doi: 10.1262/jrd.2014-087
Trenzado, C., Hidalgo, M. C., García-Gallego, M., Morales, A. E., Furné, M., Domezain, A., et al. (2006). Antioxidant enzymes and lipid peroxidation in sturgeon Acipenser naccarii and trout Oncorhynchus mykiss. A comparative study. Aquaculture 254, 758–767. doi: 10.1016/j.aquaculture.2005.11.020
Ulloa-Rodriguez, P., Contreras, P., Dumorne, K., Lee-Estevez, M., Diaz, R., Figueroa, E., et al. (2018). Patagonian blenny (Eleginops maclovinus) spermatozoa quality after storage at 4 degrees C in Cortland medium. Anim. Reprod. Sci. 197, 117–125. doi: 10.1016/j.anireprosci.2018.08.019
Viveiros, A. T. M., Isaú, Z. A., Figueiredo, H. C. P., Leite, M. A. S., and Maria, A. N. (2010). Gentamycin controls bacterial growth during refrigerated storage of piracanjuba, Brycon orbignyanus, semen. J. World Aquac. Soc. 41, 57–65. doi: 10.1111/j.1749-7345.2009.00333.x
Wang, D., Chen, X., Zhang, X., Li, J., Yi, Y., Bian, C., et al. (2019). Whole genome sequencing of the giant grouper (Epinephelus lanceolatus) and high-throughput screening of putative antimicrobial peptide genes. Mar. Drugs 17:503. doi: 10.3390/md17090503
Williams, K. C. (2009). A review of feeding practices and nutritional requirements of postlarval groupers. Aquaculture 292, 141–152. doi: 10.1016/j.aquaculture.2009.04.026
Winteringham, L. N., Kobelke, S., Williams, J. H., Ingley, E., and Klinken, S. P. (2004). Myeloid Leukemia Factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27Kip1 accumulation. Oncogene 23, 5105–5109. doi: 10.1038/sj.onc.1207661
Wu, H., Gao, J., Wang, X., Leung, T. Y., Duan, Y. G., and Chiu, P. C. N. (2020). Platelet-activating factor induces acrosome reaction via the activation of extracellular signal-regulated kinase in human spermatozoa. Andrologia 52:e13565. doi: 10.1111/and.13565
Yang, K., Meinhardt, A., Zhang, B., Grzmil, P., Adham, I. M., and Hoyer-Fender, S. (2012). The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol. Cell. Biol. 32, 216–225. doi: 10.1128/MCB.06158-11
Yang, K., Wang, N., Guo, H. T., Wang, J. R., Sun, H. H., Sun, L. Z., et al. (2020a). Effect of L-carnitine on sperm quality during liquid storage of boar semen. Asian Australas. J. Anim. Sci. 33, 1763–1769. doi: 10.5713/ajas.19.0455
Yang, S., Han, L., Huang, R., Liufu, Y., Meng, Z., and Lin, H. (2017). Optimization of conditions for the cryopreservation of yellow catfish (Pelteobagrus fulvidraco) sperm. Cryobiology 76, 104–110. doi: 10.1016/j.cryobiol.2017.03.009
Yang, S., Huang, W., Guo, W., Chen, X., Fan, B., and Meng, Z. (2020b). Effects of gametes post-activation, egg storage and sperm-egg ratio on in vitro fertilization outcomes in the brown-marbled grouper (Epinephelus fuscoguttatus). Anim. Reprod. Sci. 219:106529. doi: 10.1016/j.anireprosci.2020.106529
Yang, Y., Liu, D., Wu, L., Huang, W., Yang, S., Xia, J., et al. (2019). Comparative transcriptome analyses reveal changes of gene expression in fresh and cryopreserved yellow catfish (Pelteobagrus fulvidraco) sperm and the effects of Cryoprotectant Me2SO. Int. J. Biol. Macromol. 133, 457–465. doi: 10.1016/j.ijbiomac.2019.04.050
Zhao, S., Ye, Z., and Stanton, R. (2020). Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 26, 903–909. doi: 10.1261/rna.074922.120
Zhao, Z., Liu, H., Hou, J., Li, T., Du, X., Zhao, X., et al. (2017). Tumor protein D52 (TPD52) inhibits growth and metastasis in renal cell carcinoma cells through the PI3K/Akt signaling pathway. Oncol. Res. 25, 773–779. doi: 10.3727/096504016X14774889687280
Keywords: grouper, sperm, proteome, transcriptome, storage
Citation: Yang Y, Wang T, Yang S, Wu X, Huang W, Fan B, Xia J, Meng Z and Liu X (2021) The Effects of Storage in vitro on Functions, Transcriptome, Proteome, and Oxidation Resistance of Giant Grouper Sperm. Front. Mar. Sci. 8:716047. doi: 10.3389/fmars.2021.716047
Received: 28 May 2021; Accepted: 23 June 2021;
Published: 26 July 2021.
Edited by:
Jorge G. Farias, University of La Frontera, ChileReviewed by:
John Quiñones Diaz, University of La Frontera, ChilePatricio Ulloa-Rodríguez, Catholic University of Maule, Chile
Copyright © 2021 Yang, Wang, Yang, Wu, Huang, Fan, Xia, Meng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zining Meng, bWVuZ3puQG1haWwuc3lzdS5lZHUuY24=; Xiaochun Liu, bHNzbHhjQG1haWwuc3lzdS5lZHUuY24=
 Yang Yang1,2
Yang Yang1,2 Junhong Xia
Junhong Xia Zining Meng
Zining Meng Xiaochun Liu
Xiaochun Liu