- 1Centre for Chemical Biology, Universiti Sains Malaysia, Bayan Lepas, Malaysia
- 2Lee Kong Chian Natural History Museum, Natural University of Singapore, Singapore, Singapore
- 3School of Biological Sciences, Universiti Sains Malaysia, Minden, Malaysia
- 4Faculty of Science and Marine Environment, Universiti Malaysia Terengganu, Kuala Terengganu, Malaysia
- 5Advanced Genomics and Bioinformatics Division, Malaysia Genome Institute, National Institutes of Biotechnology Malaysia, Selangor, Malaysia
Introduction
Phylogenetically, crabs (Brachyura) are among the most diverse crustaceans, with representatives in marine, freshwater, and terrestrial niches, encompassing over 7,000 species in 98 families (Ng et al., 2008; De Grave et al., 2009). The Gecarcinidae crab family, consist of six genera encompassing 20 species, showing distinct adaptations for a terrestrial lifestyle, with larval development taking place in the oceans (Hartnoll, 1988; Greenaway, 1999; Ng and Shih, 2015). Interestingly, while the conquest of terrestrial habitats is reported to encompass long-term adaptations, a much more rapid process has been reported to occur in terrestrial crabs (Schubart et al., 1998). During this sea to land transition, animals have developed a number of physiological and anatomical adaptations associated with gas exchange, salt and water balance, nitrogenous excretion, thermoregulation, reproduction, feeding, and diet (Bliss and Mantel, 1968; Powers and Bliss, 1983; Greenaway, 1999; Richardson and Araujo, 2015). The Gecarcoidea genus consists of Gecarcoidea natalis, which is endemic to Christmas Island and the Cocos (Keeling) Islands while Gecarcoidea lalandii is widely distributed in the Indo-West pacific region (Lai et al., 2017). Both these species release larvae from above the water, a behavior speculated to confer protection to the ovigerous females (Liu and Jeng, 2007). Land crabs are considered opportunistic omnivores, feeding on carrion, insects, mammalian feces, and plant material (Wolcott and Wolcott, 1984; Ortega-Rubio et al., 1997). Nevertheless, the nature of their habitat has driven some of the species to become more herbivorous, foraging mostly on leaf litter, vascular plants foliage, seeds, and fruits (Linton and Greenaway, 2007; Wolcott and O'connor, 2015). This shift was shown to be crucial in promoting the diversification of crustaceans (Poore et al., 2017).
A dichotomy between terrestrial and marine-based diets is the concentration of long-chain polyunsaturated fatty acids (LC-PUFA) such as eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and arachidonic acid (ARA) in the food chain (Hixson et al., 2015). The importance of this difference is magnified by the reliance of terrestrial organisms on the aquatic environment for supply of LC-PUFA (Gladyshev et al., 2009). Aquatic primary producers and invertebrates, especially marine species, possess all the necessary enzymes required to biosynthesize the LC-PUFA from shorter chain fatty acid substrates. Two classes of enzymes, the fatty acyl desaturase (Fads) and elongases of very long-chain fatty acid (Elovl), work together sequentially, inserting a double bond at defined locations of the fatty acyl backbone and elongating the fatty acyl chain through addition of two carbon units. Work on various vertebrate species, mainly on teleost, have elucidated the complete gamut of Fads and Elovls responsible for the biosynthesis of EPA/DHA and ARA from linolenic acid (LNA) and linoleic acid (LA), respectively. Similar corresponding works on various aquatic invertebrates such as molluscs and echinoderms have also revealed the presence of Fads and Elovl orthologs and LC-PUFA biosynthesis activities (Monroig and Kabeya, 2018). The presence of functional Elovl in crustaceans were only recently shown in two marine brachyuran crabs (Mah et al., 2019; Sun et al., 2020; Ting et al., 2020). Given the limited amount of LC-PUFA in the terrestrial food chain, very little is known about how terrestrial crustaceans adapt in terms of LC-PUFA biosynthesis capacity.
Transcriptome reconstruction is a rapid and economical approach useful for systematic gene models characterization in species without any genome reference. RNA sequencing via next-generation sequencing technology is widely used for this purpose (Grabherr et al., 2011; Mutz et al., 2013). However, short reads require large computational assembly and are often insufficient to capture entire end-to-end transcripts, limiting the accuracy of gene model prediction (Steijger et al., 2013). The long-read isoform (Iso-Seq) sequencing method (Pacific Biosciences, PacBio) analyse full-length transcripts as a single sequence read without the need for further assembly, making it an ideal for identifying and characterizing novel transcripts and transcript isoforms (Eid et al., 2009; Roberts et al., 2013; Rhoads and Au, 2015). This technique was used for transcriptome analysis in crustacean species such as Litopenaeus vannamei (Zeng et al., 2018; Wan et al., 2019), Penaeus monodon (Pootakham et al., 2020), and Scylla paramamosain (Wan et al., 2019). In consideration of the lack of any terrestrial crab reference genome, the aim of this study was to apply the PacBio Iso-Seq technique to profile the transcriptome of G. lalandii (Decapoda, Brachyura, Gecarcinidae) with the objective of identifying relevant transcripts for LC-PUFA biosynthesis Fads and Elovl enzymes. Our study provides the first gene catalogs for a terrestrial crab species, adding to currently available Brachyura transcriptome datasets.
Data Description
Sample Collection and Total RNA Extraction
G. lalandii sample was collected at Rawa Island, Johor, Malaysia (2.5204°N, 103.9760°E) in June 2018. Three adult male crabs weighing 120–150 g were sampled. We selected hepatopancreas tissue to obtain transcript library as hepatopancreas is responsible for the storage of organic matter and metabolism of nutrients (Vogt, 1994; Wen et al., 2001; Abol-Munafi et al., 2016). The tissue was sampled immediately after euthanization, snap-frozen in liquid nitrogen, and stored at −80 °C prior to RNA extraction. Total RNA was isolated from the tissues of three crabs using the Qiagen RNeasy mini kit (Qiagen, Germany) following manufacturer's recommendations and pooled together for library preparation. Potential DNA contamination was eliminated by applying on-column DNase digestion using RNase-Free DNase Set (Qiagen, Germany). RNA concentration was determined using a Qubit® 2.0 Fluorometer (Life Technologies, USA), and RNA quality was assessed using an Agilent 2,100 Bioanalyzer (Agilent Technologies, USA). RNA with RNA integrity number (RIN) value above 9 was used for library construction.
PacBio cDNA Library Preparation and Sequencing
Sequencing library was prepared according to the PacBio Iso-Seq protocol. The Clontech SMARTer PCR cDNA Synthesis Kit with Oligo(dT) primers was used to generate the first and second cDNA strand from polyA mRNA. Size fractionation and selection on ≤ 4 kb and ≥ 4 kb bin were performed using the BluePippinTM Size selection system (Saga Science, USA). SMRT bell library was constructed with the PacBio DNA Template Prep Kit 1.0, and sequencing run was performed on a PacBio Sequel platform.
Iso-Seq Data Analyses
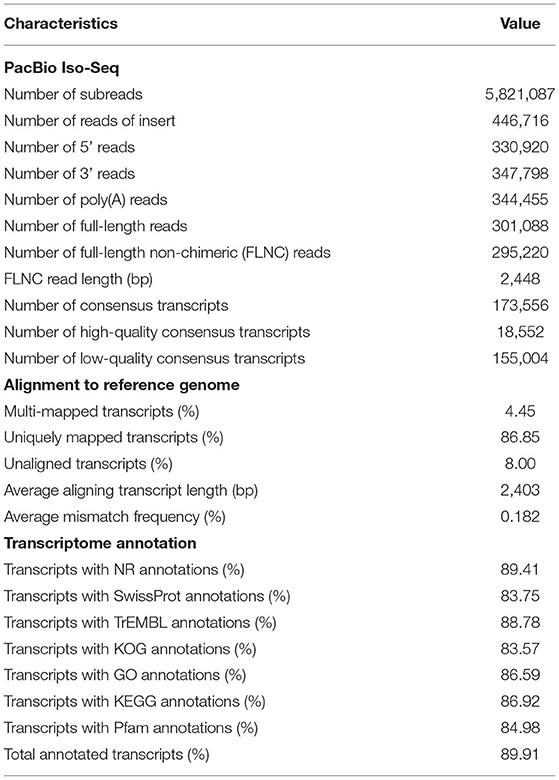
The sequence data was processed through the RS_IsoSeq (version 2) protocol. Reads of insert ROIs (previously known as circular consensus sequence) were generated from raw subreads using the SMRT Link (version 5.1) software (Gordon et al., 2015). A total of 11.43 Gb raw data was generated by 5,821,087 of subreads, which were classified into 446,716 of ROI reads (Table 1). The ROIs were classified based on the presence of 5′ and 3′ adapters as well as the poly(A) tail into full-length and non-full length reads. Sequences containing both the 5′ and 3′ primers and having a poly(A) tail signal preceding the 3′ primer were considered to be full-length (FL) ROIs. ROI reads comprised of 295,220 full-length non-chimeric transcripts with an average read length of 2,448 bp. FL ROIs sequences were then passed through the isoform-level clustering (ICE algorithm). ROI sequences were used to correct errors in the isoform sequences using the Quiver algorithm. The Quiver polishing process produced high-quality and low-quality isoform sequences corresponding to a predicted accuracy of ≥ 99% or below, respectively. The isoform-level clustering and final polishing steps yielded 18,552 and 2,76,668 of high-quality and low-quality consensus isoforms, respectively. The completeness of consensus transcripts was assessed by benchmarking universal single-copy orthologs (BUSCO) (version 3.1.0) (Simão et al., 2015). Our G. lalandii hepatopancrease transcriptome only captured 67% of the conserved arthropod genes, likely due to our sampling from a single tissue (Supplementary Figure 1). The isoform sequences were mapped to the Portunus trituberculus reference genome (Tang et al., 2020) (a chromosome-level genome assembly of Brachyura) using GMAP (version 2015-09-29) (Wu and Watanabe, 2005). More than 92% of consensus transcripts were mapped to the P. trituberculus genome and the average transcript length aligning to the genome was 2,403 bp.
Functional Annotation of Isoforms
The obtained high-quality isoforms were annotated by conducting a local BLASTx (version 2.7.1) search against the protein databases, namely the GenBank NCBI non-redundant protein sequences (NR), SwissProt, TrEMBL, euKaryotic Ortholog Groups (KOG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The BLAST hits to NR were processed by functional annotation with BLAST2GO (version 5.1) (Conesa et al., 2005). The coding region prediction of high-quality isoforms was performed using ANGLE (version 2.2) (Shimizu et al., 2006). Hmmscan (version 3.1b) was used to search sequence protein domains against the Pfam database.
In general, most of the transcripts (89.91%) exhibited homology with at least one protein database. Matches to the specific databases were as follows: 89.41% to NR, 88.78% to TrEMBL, 86.92% to KEGG, 84.98% to Pfam, 83.75% to SwissProt, and 83.57% transcripts aligned to the KOG databases. Further, 86.59 % of transcripts were assigned to multiple Gene Ontology (GO) classification terms. Among them, “metabolic process,” “cellular process,” and “response to stimulus” were the most represented terms in the biological process (Supplementary Figure 2). In the cellular component, the majority of the transcripts were represented by “cellular anatomical entit,” “intracellular,” and “protein-containing complex.” Within the molecular function category, “catalytic activity,” “binding,” and “transporter activity” had the highest number of transcripts. The top hit species distribution of matches with known sequences indicates that the G. lalandii transcripts had the highest number of hits to the Eriocheir sinensis, Penaeus vannamei and Metacarcinus magister (Supplementary Figure 3).
Analyses of Fads and Elovl Orthologs in G. lalandii
KEGG annotation revealed the presence of several genes implicated in the polyunsaturated fatty acid biosynthesis pathway of G. lalandii (Supplementary Figure 4). The transcriptomic databases for selected crustacean species used for comparative analyses were downloaded from the NCBI and JGI databases. The downloaded sequences were analyzed using BLAST for Fads and Elovl sequences. Fads and Elovl identified were aligned using MUSCLE in MEGAX (Kumar et al., 2018) (Supplementary Figures 5–8). Two Elovl elongases were found in the G. lalandii transcripts with highest BLAST hits matching Elovl 6 and a putative novel Elovl, respectively. The best fit model was predicted using ModelFinder (Kalyaanamoorthy et al., 2017), and phylogenetic tree was built using IQ-TREE (version 1.6.12) based on the predicted model (Nguyen et al., 2015). Maximum likelihood phylogenetic analysis of Elovl from various crustacean transcriptomes also placed the two Elovl sequences in their respective ortholog clusters (Figure 1A). Elovl6, Elovl7, ElovlA, Elovl B and the putative new Elovl all shared the common ancestors while the Elovl4 formed a separate clade. To date, the Elovl4 and Elovl6 have been shown to have in vitro PUFA elongation capacities in marine brachyuran species (Sun et al., 2020; Ting et al., 2020). Interestingly, while G. lalandii appear to have a Elovl6 ortholog, there was no Elovl4.
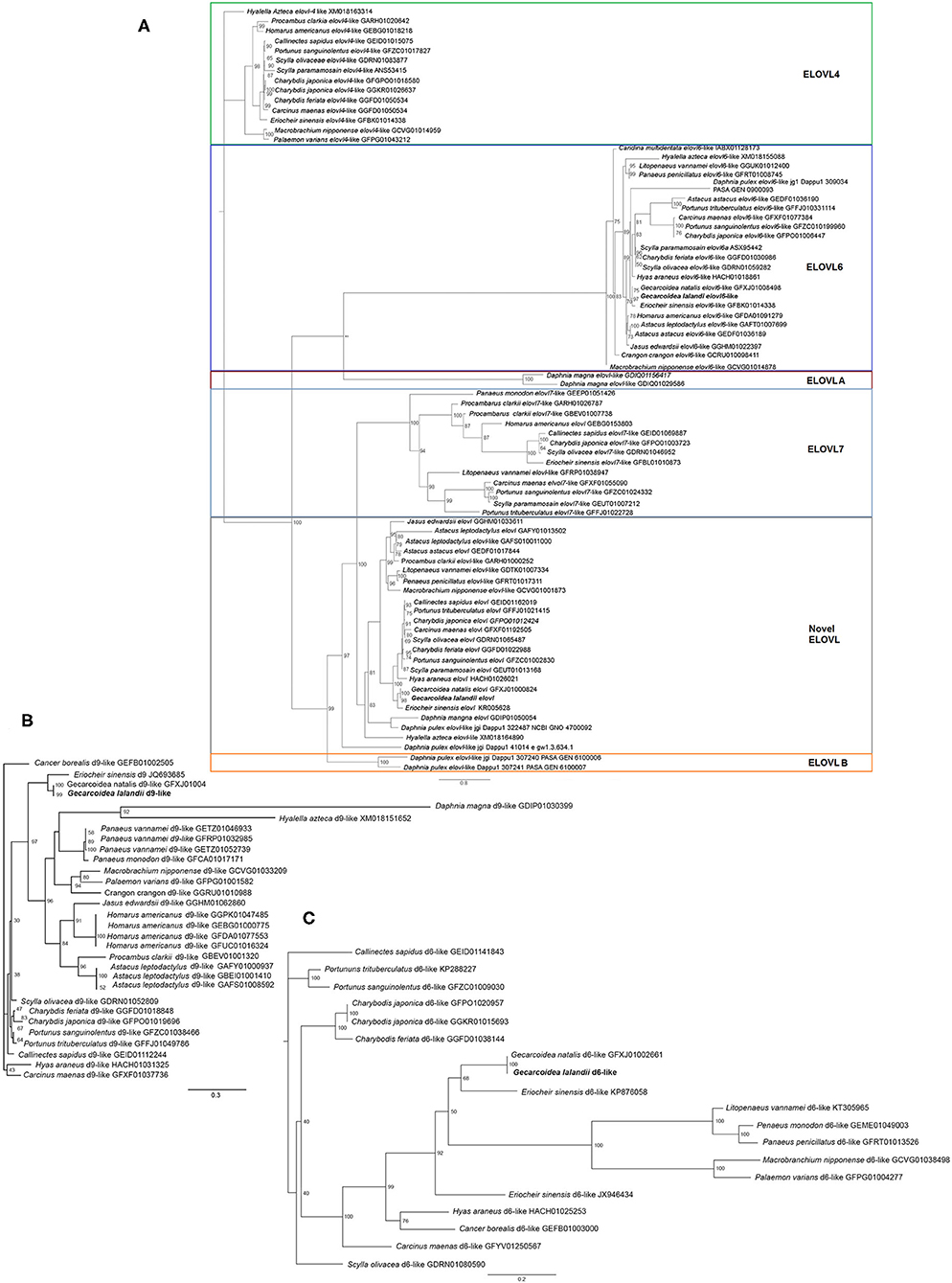
Figure 1. Phylogenetic analyses of crustacean (A) elongase (VT + I + G4, bootstraps value = 1,000), (B) acyl-CoA desaturase 9 (LG + G4, bootstraps value = 1000), and (C) delta-6 desaturase (JTTDCMut + I + G4, bootstraps value = 1000) sequences using IQ-TREE v1.6.12. The different colored boxes were drawn to facilitate viewing of the different elongase families.
Sequence alignment analysis was also performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the conserved motif was highlighted. Motif analysis was done using WebLogo (https://weblogo.berkeley.edu/examples.html), where the amino acids of histidine boxes were represented based on their frequency of occurrence. Alignment of the conserved histidine box in each respective Elovl revealed the conservation of the LHxxHH motif in all the elongases (Supplementary Figure 9). Functional characterization of these elongases for comparison with the marine crab species will yield insights on the LC-PUFA biosynthesis capacities of brachyuran living in habitats with different availability of LC-PUFA in the food chain.
Transcripts of two putative Fads orthologs were also obtained from G. lalandii (Figures 1B,C). The desaturase Δ6-like Fads have been reported in several crustacean species, although its capacity to desaturate PUFA substrates and participate in the LC-PUFA biosynthesis pathway remained vague (Wu et al., 2018; Mah et al., 2019). Another G. lalandii Fads was clustered alongside Δ9 Fads or stearyl-CoA desaturase, which has high affinity for saturated fatty acids (Monroig et al., 2017). Collectively these transcripts will facilitate the deciphering of LC-PUFA biosynthesis in regard to adaptation to terrestrial living.
Re-use potential
Here, we present a long-read RNA sequencing dataset of G. lalandii hepatopancreas that was generated by using the PacBio platform. Specifically, our analyses exemplify applicability of the dataset for mining two orthologs of two classes of enzymes known to work in concert in LC-PUFA biosynthesis, the elongase and fatty acyl desaturase. This transcript assembly serves as a resource for gene mining, which will aid in the functional characterization of various enzymes relevant to the biosynthetic pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR10903072; https://www.ncbi.nlm.nih.gov/, GIKV00000000.
Author Contributions
EQ and AA collected and identified the samples. ST and N-SL designed the experiments and analyzed the data. K-KS and M-NM-I participated in the data analysis and checking. AS-C conceived and supervised the study. ST, N-SL, and AS-C wrote the manuscript. Revision of data and manuscript after first round of peer-review were by all authors.
Funding
We thank the Malaysian Ministry of Higher Education for funding the project under the FRGS Grants Scheme (203/PCCB/6711650).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.713928/full#supplementary-material
References
Abol-Munafi, A. B., Mukrim, M. S., Amin, R. M., Azra, M. N., Azmie, G., and Ikhwanuddin, M. (2016). Histological profile and fatty acid composition in hepatopancreas of blue swimming crab, Portunus pelagicus (Linnaeus, 1758) at different ovarian maturation stages. Turk. J. Fish. Aquat. Sc. 16, 251–258. doi: 10.4194/1303-2712-v16_2_04
Bliss, D. E., and Mantel, L. H. (1968). Adaptations of crustaceans to land: a summary and analysis of new findings. Am. Zool. 8, 673–685. doi: 10.1093/icb/8.3.673
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
De Grave, S., Pentcheff, N. D., Ahyong, S. T., Chan, T. Y., Crandall, K. A., Dworschak, P. C., et al. (2009). A classification of living and fossil genera of decapod crustaceans. Raffles B. Zool. 1–109.
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Gladyshev, M. I., Arts, M. T., and Sushchik, N. N. (2009). “Preliminary estimates of the export of omega-3 highly unsaturated fatty acids (EPA+DHA) from aquatic to terrestrial ecosystems,” in Lipids in Aquatic Ecosystems, eds. M. Kainz, M.T. Brett & M.T. Arts (New York, NY: Springer New York), 179–210. doi: 10.1007/978-0-387-89366-2_8
Gordon, S. P., Tseng, E., Salamov, A., Zhang, J., Meng, X., Zhao, Z., et al. (2015). Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 10:e0132628. doi: 10.1371/journal.pone.0132628
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greenaway, P. (1999). “Physiological diversity and the colonisation of land,” in Proceedings of the Fourth International Crustacean Congress, eds. F. R. Schram, and J. C. Von Vaupel Klein (Leiden, Germany), 823–842.
Hartnoll, R. G. (1988). “Evolution, systematics, and geographical distribution,” in Biology of the Land Crabs, eds. W.W. Burggren, and B.R. Mcmahon (Cambridge: Cambridge University Press), 6–54. doi: 10.1017/CBO9780511753428.003
Hixson, S. M., Sharma, B., Kainz, M. J., Wacker, A., and Arts, M. T. (2015). Production, distribution, and abundance of long-chain omega-3 polyunsaturated fatty acids: a fundamental dichotomy between freshwater and terrestrial ecosystems. Environ. Rev. 23, 414–424. doi: 10.1139/er-2015-0029
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai, J. C. Y., Shih, H. T., and Ng, P. K. L. (2017). The systematics of land crabs of the genus Gecarcoidea and recognition of a pseudocryptic species, G. humei from the eastern Indian Ocean (Crustacea : Decapoda : Gecarcinidae). Invertebr. Syst. 31, 406–426. doi: 10.1071/IS16052
Linton, S. M., and Greenaway, P. (2007). A review of feeding and nutrition of herbivorous land crabs: adaptations to low quality plant diets. J. Comp. Physiol. B. 177, 269–286. doi: 10.1007/s00360-006-0138-z
Liu, H. C., and Jeng, M. S. (2007). Some reproductive aspects of Gecarcoidea lalandii (Brachyura: Gecarcinidae) in Taiwan. Zool. Stud. 46, 347–354.
Mah, M. Q., Kuah, M. K., Ting, S. Y., Merosha, P., Janaranjani, M., Goh, P. T., et al. (2019). Molecular cloning, phylogenetic analysis and functional characterisation of an Elovl7-like elongase from a marine crustacean, the orange mud crab (Scylla olivacea). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 232, 60–71. doi: 10.1016/j.cbpb.2019.01.011
Monroig, Ó., De Llanos, R., Varó, I., Hontoria, F., Tocher, R. D., Puig, S., et al. (2017). Biosynthesis of polyunsaturated fatty acids in Octopus vulgaris: molecular cloning and functional characterisation of a stearoyl-coA desaturase and an elongation of very long-chain fatty acid 4 protein. Mar. Drugs 15:82. doi: 10.3390/md15030082
Monroig, Ó., and Kabeya, N. (2018). Desaturases and elongases involved in polyunsaturated fatty acid biosynthesis in aquatic invertebrates: a comprehensive review. Fish. Sci. 84, 911–928. doi: 10.1007/s12562-018-1254-x
Mutz, K. O., Heilkenbrinker, A., Lönne, M., Walter, J. G., and Stahl, F. (2013). Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 24, 22–30. doi: 10.1016/j.copbio.2012.09.004
Ng, P. K. L., Guinot, D., and Davie, P. J. F. (2008). Systema Brachyurorum: part I. An annotated checklist of extant Brachyuran crabs of the world. Raffles B. Zool. 17, 1–286.
Ng, P. K. L., and Shih, H. T. (2015). The land crabs of the Discoplax longipes A. Milne-Edwards, 1867 species group, with description of a new species from Guam (Crustacea: Decapoda: Brachyura: Gecarcinidae). Zootaxa 3980:27. doi: 10.11646/zootaxa.3980.3.3
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ortega-Rubio, A., Jímenez, M. L., Llinas, J., and Arnaud, G. (1997). Some ecological aspects of the land crab, Gecarcinus planatus Stimpson, at Socorro Island, Colima, Mexico. J. Arizona-Nevada Acad. Sci. 30, 17–22.
Poore, A. G. B., Ahyong, S. T., Lowry, J. K., and Sotka, E. E. (2017). Plant feeding promotes diversification in the Crustacea. Proc. Natl. Acad. Sci. U. S. A. 114, 8829–8834. doi: 10.1073/pnas.1706399114
Pootakham, W., Uengwetwanit, T., Sonthirod, C., Sittikankaew, K., and Karoonuthaisiri, N. (2020). A novel full-length transcriptome resource for black tiger shrimp (Penaeus monodon) developed using isoform sequencing (Iso-Seq). Front. Mar. Sci. 7:172. doi: 10.3389/fmars.2020.00172
Powers, L. W., and Bliss, D. E. (1983). “Terrestrial adaptations,” in The Biology of Crustacea, eds. F. J. Vernberg, and W. B. Vernberg (New York, NY: Academic Press), 272–333.
Rhoads, A., and Au, K. F. (2015). PacBio sequencing and its applications. Genom. Proteom. Bioinf. 13, 278–289. doi: 10.1016/j.gpb.2015.08.002
Richardson, A., and Araujo, P. (2015). “Lifestyles of terrestrial crustaceans,” in The Natural History of the Crustacea, Lifestyles and Feeding Biology, ed. L. W. Martin Thiel (Oxford: Oxford University Press), 299–336.
Roberts, R. J., Carneiro, M. O., and Schatz, M. C. (2013). The advantages of SMRT sequencing. Genome Biol. 14:405. doi: 10.1186/gb-2013-14-6-405
Schubart, C. D., Diesel, R., and Hedges, S. B. (1998). Rapid evolution to terrestrial life in Jamaican crabs. Nature 393, 363–365. doi: 10.1038/30724
Shimizu, K., Adachi, J., and Muraoka, Y. (2006). ANGLE: a sequencing errors resistant program for predicting protein coding regions in unfinished cDNA. J. Bioinform. Comput.Biol. 4, 649–664. doi: 10.1142/S0219720006002260
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Steijger, T., Abril, J. F., Engström, P. G., Kokocinski, F., Abril, J. F., Akerman, M., et al. (2013). Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184. doi: 10.1038/nmeth.2714
Sun, P., Zhou, Q., Monroig, O., Navarro, J. C., Jin, M., Yuan, Y., et al. (2020). Cloning and functional characterization of an elovl4-like gene involved in the biosynthesis of long-chain polyunsaturated fatty acids in the swimming crab Portunus trituberculatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 242:110408. doi: 10.1016/j.cbpb.2020.110408
Tang, B., Zhang, D., Li, H., Jiang, S., Zhang, H., Xuan, F., et al. (2020). Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). GigaScience 9:giz161. doi: 10.1093/gigascience/giz161
Ting, S. Y., Janaranjani, M., Merosha, P., Sam, K.-K., Wong, S. C., Goh, P.-T., et al. (2020). Two elongases, Elovl4 and Elovl6, fulfill the elongation routes of the LC-PUFA biosynthesis pathway in the orange mud crab (Scylla olivacea). J. Agric. Food Chem. 68, 4116–4130. doi: 10.1021/acs.jafc.9b06692
Vogt, G. (1994). Life-cycle and functional cytology of the hepatopancreatic cells of Astacus astacus (Crustacea, Decapoda). Zoomorphology 114, 83–101. doi: 10.1007/BF00396642
Wan, H., Jia, X., Zou, P., Zhang, Z., and Wang, Y. (2019). The single-molecule long-read sequencing of Scylla paramamosain. Sci. Rep. 9:12401. doi: 10.1038/s41598-019-48824-8
Wen, X., Chen, L., Ai, C., Zhou, Z., and Jiang, H. (2001). Variation in lipid composition of Chinese mitten-handed crab, Eriocheir sinensis during ovarian maturation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130, 95–104. doi: 10.1016/S1096-4959(01)00411-0
Wolcott, D. L., and O'connor, N. J. (2015). Herbivory in crabs: adaptations and ecological considerations. Am. Zool. 32, 370–381. doi: 10.1093/icb/32.3.370
Wolcott, D. L., and Wolcott, T. G. (1984). Food quality and cannibalism in the red land crab, Gecarcinus lateralis. Physiol. Zool. 57, 318–324. doi: 10.1086/physzool.57.3.30163720
Wu, D. L., Huang, Y. H., Liu, Z. Q., Yu, P., Gu, P. H., Fan, B., et al. (2018). Molecular cloning, tissue expression and regulation of nutrition and temperature on Delta6 fatty acyl desaturase-like gene in the red claw crayfish (Cherax quadricarinatus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 225, 58–66. doi: 10.1016/j.cbpb.2018.07.003
Wu, T. D., and Watanabe, C. K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. doi: 10.1093/bioinformatics/bti310
Keywords: Andaman Islands purple crab, Gecarcoidea lalandii, Iso-Seq, transcriptome, long-chain polyunsaturated fatty acid
Citation: Ting SY, Lau N-S, Sam K-K, Quah ESH, Ahmad AB, Mat-Isa M-N and Shu-Chien AC (2021) Long-Read Sequencing Reveals the Repertoire of Long-Chain Polyunsaturated Fatty Acid Biosynthetic Genes in the Purple Land Crab, Gecarcoidea lalandii (H. Milne Edwards, 1837). Front. Mar. Sci. 8:713928. doi: 10.3389/fmars.2021.713928
Received: 24 May 2021; Accepted: 22 June 2021;
Published: 20 July 2021.
Edited by:
Ylenia Carotenuto, Stazione Zoologica Anton Dohrn, ItalyReviewed by:
Vittoria Roncalli, Stazione Zoologica Anton Dohrn Napoli, ItalyChristine Nicole Shulse, Los Medanos College, United States
Copyright © 2021 Ting, Lau, Sam, Quah, Ahmad, Mat-Isa and Shu-Chien. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Chong Shu-Chien, YWxleEB1c20ubXk=
 Seng Yeat Ting1
Seng Yeat Ting1 Nyok-Sean Lau
Nyok-Sean Lau Ka-Kei Sam
Ka-Kei Sam Evan S. H. Quah
Evan S. H. Quah Alexander Chong Shu-Chien
Alexander Chong Shu-Chien