
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 14 July 2021
Sec. Coral Reef Research
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.706308
This article is part of the Research TopicCoral Reef Research MethodsView all 22 articles
A correction has been applied to this article in:
Corrigendum: In vitro Symbiosis of Reef-Building Coral Cells With Photosynthetic Dinoflagellates
 Kaz Kawamura1*†‡
Kaz Kawamura1*†‡ Satoko Sekida2‡
Satoko Sekida2‡ Koki Nishitsuji3†
Koki Nishitsuji3† Eiichi Shoguchi3†
Eiichi Shoguchi3† Kanako Hisata3
Kanako Hisata3 Shigeki Fujiwara1†
Shigeki Fujiwara1† Noriyuki Satoh3*†
Noriyuki Satoh3*†
Coral reefs are the biodiversity hot spots of the oceans, but they have suffered from increasing environmental stresses caused principally by anthropogenic global warming. The keystone species of coral reefs are scleractinian corals, which maintain obligatory symbiotic relationships with photosynthetic dinoflagellates. Understanding cellular and molecular mechanisms of symbiosis is therefore essential for future preservation of coral reefs. To date, however, almost no in vitro experimental systems have been devised to illuminate such mechanisms. To this end, our previous study established stable in vitro cell culture lines, including IVB5, originating from planula larvae of the scleractinian coral, Acropora tenuis. Here, we show that soon after mixture with the dinoflagellate, Breviolum minutum, flattened amorphous coral cells with endodermal properties exhibited elevated locomotor activity using filopodia and lamellipodia and interacted with dinoflagellates. Several minutes thereafter, coral cells began to incorporate B. minutum, and in vitro symbiosis appeared to have been accomplished within 30 min. Nearly a half of the coral cells had incorporated algal cells within 24 h in a reproducible manner. Coral cells that harbored algal cells gradually became round and less mobile, and the algal cells sometimes settled in vacuole-like structures in coral cell cytoplasm. This symbiosis state was maintained for at least a month. The IVB5 line of A. tenuis therefore provides an experimental system to explore cellular and molecular mechanisms involved in coral-dinoflagellate symbiosis at the single-cell level, results of which may be useful for future preservation of coral reefs.
During evolution of cnidarians, the lineage leading to scleractinian corals or stony, reef-building corals acquired the capacity to establish obligatory symbiosis with photosynthetic dinoflagellates. In this endosymbiosis, corals provide shelter for their algal symbionts, which supply most of their photosynthetic products to the host corals (Yellowlees et al., 2008). This alga-animal symbiosis resulted in extraordinal prosperity of scleractinian corals, which produce stony reefs by depositing calcium carbonate skeletons. Although coral reefs cover only 0.2∼0.3% of the marine surface, they harbor an estimated one-third of all described marine species (Reaka-Kudla, 2001; Wilkinson, 2008). Coral reefs therefore support the most biodiverse ecosystems in the oceans. These ecosystems also support human life in tropical and subtropical countries by virtue of fisheries, tourism, and culture (Spaldinga et al., 2017). Coral reefs, however, are in crisis due to environmental changes, including increased seawater temperatures, acidification, and pollution, mainly caused by human activities (Hughes et al., 2017; Sully et al., 2019). These stresses cause collapse of coral-dinoflagellate symbioses, resulting in white, dead reefs, a process known as coral bleaching, leading to loss of coral reefs and all the species they support (Hoegh-Guldberg et al., 2007; Hughes et al., 2017; Sully et al., 2019). Conservation of coral reefs is therefore one of the most urgent environmental crises facing humanity.
Most scleractinian corals incorporate photosynthetic dinoflagellates of the family Symbiodiniaceae through the digestive tract into the gastrodermis (Coffroth and Santos, 2005), although some species inherit symbionts vertically (Loh et al., 2001). Modulation of coral host inert immunity allows symbiosis of given species of algae in their cytoplasm (Davy et al., 2012). Although many studies has attempted to explore cellular and molecular mechanisms of coral-dinoflagellate endosymbiosis (Dove, 2004; Davy et al., 2012; Yuyama et al., 2018; Weis, 2019; Rosset et al., 2021; Yoshioka et al., 2021), many questions remain, especially regarding recognition mechanisms involved in the initial contact of animal and algal cells, cellular mechanisms allowing dinoflagellate endocytosis and maintenance of endosymbiosis, especially at the single-cell level (Davy et al., 2012). Recently, our knowledge of mechanisms involved in cnidaria-dinoflagellate symbiosis has been advanced by studies using the sea anemone, Exaiptasia diaphana (Weis, 2019), and the soft coral, Xenia sp. (Hu et al., 2020). Simultaneously, we need to understand symbiosis mechanisms of reef-building corals. Thus, many studies have attempted to culture in vitro lines of stony coral cells, by which mechanisms of coral-dinoflagellate symbiosis might be tackled. However, most attempts have failed to establish stable in vitro cultures, although primary cultures of cells or cell aggregates have been accomplished (Rinkevich, 2011; Domart-Coulon and Ostrander, 2016).
In a previous study, we developed in vitro coral cell lines originating from planula larvae of the scleractinian coral, Acropora tenuis. After several trials to improve culture media, we succeeded in producing various stable in vitro lines of coral cells, twenty of which have been cryo-preserved (Kawamura et al., 2021). Most cells of several lines are dark, flattened, amorphous cells with lamellipodia and locomotor activity. Judging from their morphology, behavior, and higher expression of endoderm-related genes, they likely originated from larval endoderm. Then we examined whether these cells could interact in vitro with dinoflagellates when mixed in culture media or seawater. We report here the occurrence of in vitro symbiosis between coral cells and dinoflagellates in culture dishes.
Detailed methods for production of stable in vitro culture lines of coral cells are described in Kawamura et al. (2021). Although the IVB5 line was not described in that report, the essence of establishment of this line was same as those reported there. Briefly, basic seawater medium consisted of natural seawater, one-fifth volume of H2O, 10 mM HEPES (final pH 6.8), and the antibiotics penicillin (100 U/mL), streptomycin (100 μg/mL), and amphotericin B (0.25 μg/mL). Immediately before use, the basic medium was mixed with Dulbecco’s modified Eagle’s medium (DMEM) containing 15% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL) at a ratio of 9:1. In the secondary cell culture, plasmin (166-24231, FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) was added to the growth medium at a final concentration of 2 μg/mL. Dissociated cells were centrifuged at 300 x g for 5 min and resuspended in growth medium at a density of 2–5 x 107 cells/mL. Aliquots (0.5 mL each) were dispensed to a 24-well multiplate and maintained at 20°C by adding 0.2 mL of fresh growth medium to the old medium twice a week. Proliferating cells were replated in new multiplates every month.
A culturable dinoflagellate, Breviolum minutum (LaJeunesse et al., 2018), was used in this study. B. minutum was previously named Symbiodinium minutum (LaJeunesse et al., 2012). The B. minutum strain used was originally harbored by the Caribbean coral, Orbicella faveolata (previously Montastraea faveolata), maintained in the laboratory of Dr. Mary Alice Coffroth, at State University at New York, Buffalo, United States and then in the laboratories of Okinawa Institute of Science and Technology Graduate University and Kochi University. The culture ID of this strain is Mf1.05b (McIlroy and Coffroth, 2017), which is currently provided through National Institute of Environmental Science, Tsukuba, Japan under https://mcc.nies.go.jp/strainList.do?strainId=3806 upon request. For symbiosis experiments with the A. tenuis IVB5 line, the B. minutum strain was precultured at 20°C with standard IMK medium using an incubator (SANYO MIR-554) under 12 h:12 h dark and light conditions for more than 10 months. A fluorescent lamp producing 20 μmol/m2/s was used for the algae culture.
A 200–250-μL drop of culture medium containing B. minutum was added to each well of a 24-well multiplate, which contained approximately 1 mL of medium for culturing cells of the IVB5 line. A 24-well multiplate that contained cultured coral cells and algae was put in a translucent, moist container and exposed to natural lighting at 20–22°C throughout culture and observation. Immediately after addition, interactions between the coral cells and the dinoflagellates were observed using an inverted microscope (Olympus CKX41) equipped with a color digital camera (WRAYMER SR300). Photos obtained by consecutive observation with time-lapse video (2–3-s interval) was converted to a time-lapse video using iPhoto. Pictures and videos were also taken with an ordinary microscope (Nikon Eclipse 80i) equipped with a differential interference contrast (DIC) apparatus.
In a previous study, we made two rabbit antibodies against synthetic oligopeptides, one corresponding to a part of A. tenuis Snail protein (a zinc finger transcriptional repressor) and the other to a part of A. tenuis Fat1 protein (a Fat-like cadherin-related tumor suppressor homolog) (for details, see Kawamura et al., 2021). To confirm that cells of the IVB5 line that engulfed algae are of A. tenuis, we carried out immunocytochemistry using the antibodies. Cultured cells suspended in the culture medium were centrifuged at 300 x g for 5 min and resuspended in phosphate-buffered salt solution (PBS). Cells were fixed with 4% paraformaldehyde in PBS for 15 min in an ice bath. After quenching with 200 mM glycine for 2 min and permeabilizing with 0.1% Triton X-100 for 10 min, cells were incubated in a mixture of 0.25% blocking reagent (Roche, Mannheim, Germany) and 5% skim milk in PBS for 30 min. Then, they were reacted with the rabbit primary antibody diluted 400-fold with PBS for 1 h and with goat anti-rabbit secondary antibody labeled with fluorescein isothiocyanate (FITC) (Vector Laboratory, Burlingame, CA, USA) diluted 200-fold with PBS for 30 min. After washing by centrifugation for 5 min twice with PBS, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). They were observed by means of a confocal microscopy system (ECLIPSE C1si, Nikon Co. LTD., Tokyo, Japan).
Seven hours after mixing coral cells and dinoflagellates, cells were fixed for semi-thin sectioning for light microscopy. Cells were fixed in 3% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) containing 0.2 M sucrose and 1% tannic acid for 2 h at 4°C. Samples were rinsed with the buffer containing 0.2 M sucrose and post-fixed in buffer containing 1% osmium tetroxide and 0.2 M sucrose for 2 h at 4°C. Post-fixed specimens were dehydrated in an acetone series and embedded in Spurr’s resin. Semi-thin sections (0.5–1.5 μm) were cut with a diamond knife using a Leica Ultracut UCT ultra-microtome (Leica Microsystems, Germany), stained with 0.5% toluidine blue O and observed through a light microscope (Nikon Eclipse 80i).
IVB5 is one of 20 cryo-reserved cell lines established from A. tenuis planula larvae in 2020 (Figure 1A). After replating the culture line several times, cells were frozen in 2-mL serum tubes in liquid nitrogen for permanent preservation. A few months later, cells in a tube were melted back into culture medium to proliferate as the original line of cells did (Figure 1A). Although the IVB5 line is polyclonal and contains several types of cells with different morphologies, the majority are dark, flattened, amorphous cells, 20∼30 μm in length (Figure 1A). The line also contains a few brilliant cells (Figure 1A) and small elongated cells (Figure 1A). A large vacuole (Figures 1B1,B3) and several small vesicles (Figures 1B1,B2) are found in the cytoplasm of flattened amorphous cells. They extend lamellipodia and sometimes filopodia as well (Figures 1B2,B3) and show moderate locomotor activity (Supplementary Movie 1).
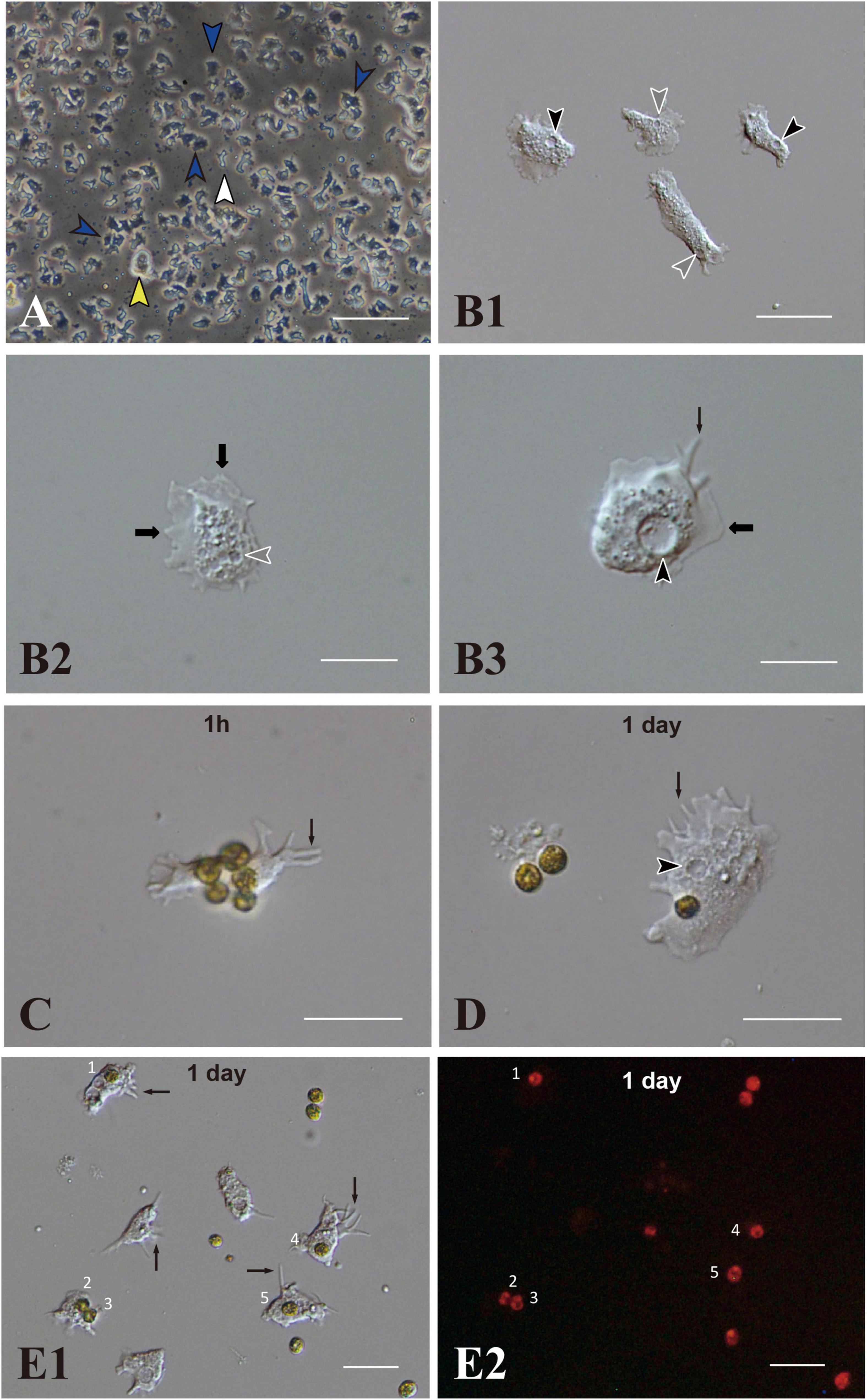
Figure 1. Acropora tenuis cells of the IVB5 line and symbiosis with the photosynthetic dinoflagellate, Breviolum minutum. (A) A vertical microscope image of IVB5 line. Most cells are dark, flattened, amorphous cells (blue arrowheads), while a few brilliant cells (yellow arrowhead) and small cells of different morphology (white arrowhead) are present as well. (B–D) Differential interference microscopy images showing morphological features of flattened, amorphous cells. (B1) Lower and (B2,B3) higher magnification. Flattened amorphous cells contain a large vacuole (black arrowheads) and several small vesicles (white arrowheads) in the cytoplasm. They extend lamellipodia (bold b black arrows) and/or filopodia (thin black arrows) and show locomotor activity. (C–E) Coral cell engulfment of dinoflagellates, (C) 1 h (1 h) and (D,E) 1 day after mixturing cells of the two taxa. (E2) is a dark-field image of (E1), showing red auto-fluorescence of dinoflagellates. Cells endocytosed dinoflagellates numbered 1–5. Filopodia are indicated by arrows and vacuoles by black arrowheads. Scale bars, 100 μm in (A), 50 μm in (B1), 20 μm in (B2,B3,C,D), and 50 μm in (E1,E2).
Using immunocytochemistry, we examined dark, flattened, amorphous cells that did not engulf symbiotic dinoflagellates (Supplementary Figure 1A) and cells that did (Supplementary Figures 1B,1C). Although the former maintained their flattened, amorphous morphology, the latter became spherical (described later). Both showed distinct fluorescent signals in response to A. tenuis-specific antibodies (Supplementary Figures 1A–1C). In the case of anti-AtSnail, signals were restricted to the nucleus (Supplementary Figure 1C), while in the case of anti-AtFat1, fluorescent signals appeared throughout the entire cell bodies (Supplementary Figures 1A,1B). In contrast, negative control cells that were stained with non-immunized rabbit serum did not exhibit FITC signals, but confirmed DAPI signals (Supplementary Figure 1D). These results indicate that cells of the IVB5 line that engulfed symbiotic algae are of Acropora tenuis.
Breviolum minutum “(Figures 1C,D) has been maintained in our laboratories for 8 years. During the proliferation stage, brown cells appear globular, approximately 8 μm in diameter, do not extend flagella, and show no locomotor activity. On the other hand, during steady state, some algal cells extend flagellae and swim in the culture medium. The B. minutum genome has been sequenced (Shoguchi et al., 2013). The identity of B. minutum used in this study was identified by partial genome sequence to be strain ITS2-type B1 (Supplementary Figure 2).”
A 200–250-μL drop of culture medium containing B. minutum was added to each well of a 24-well multiplate that contained subconfluent coral cells in approximately 1 mL of growth medium. Most immobilized B. minutum gradually settled at the bottom of the culture plate wells after 5 or 6 min. The first change detected after mixing animal and algal cells was increased locomotor activity of flattened amorphous cells. Immediately after mixing, coral cells developed filopodia and actively extended and retracted pseudopodia (Figures 1C,D,E1, arrows). They crept faster than those in plates that lacked dinoflagellates (Supplementary Movie 1). Interactions between cells of the two taxa occurred shortly after mixing (Figure 1C), followed by coral cell phagocytosis of dinoflagellates (Figure 1D). Coral cells incorporated coccoid cells, but not thecate motile cells. One day after mixing, one or two and sometimes three dinoflagellates were found in the cytoplasm of individual cultured coral cells (Figures 1E1,E2). We repeated experiments more than five times and obtained the same results, indicating that this in vitro symbiosis is reproducible.
We examined whether the culture medium affects the interaction between coral and dinoflagellate cells. Three media, the conditioned cell growth medium that had been used for cell culture for 2 weeks or more, newly prepared cell growth medium, and basic seawater medium, were examined. One day after inoculation in the conditioned medium, 50.2 ± 30.2% (number of observation fields, n = 12, which contained approximately 20 cells) of cultured host cells incorporated algae (Supplementary Figure 3A). In newly prepared growth medium, algal uptake was observed in 45.7 ± 28.8% (n = 14) of all cells (Supplementary Figure 3B), and in seawater medium, 45.5 ± 30.3% (n = 7) (Supplementary Figure 3C). These results indicate that 1 day after inoculation, coral cell symbiosis with algal cells occurred similarly in each of the three media in approximately 50% of the cultured coral cells. Several days after mixing, coral cells did not show further incorporation of dinoflagellates. Even when dinoflagellates were present near or attached to coral cells, they appeared to show no interest in dinoflagellates.
Two examples of in vitro symbiosis are described below. In the first case (Supplementary Movie 2), three coral cells and four dinoflagellates are present in the frames (Figure 2A). At 0:00:00 (starting time of observation, Figure 2A), coral cell b extended a filopodium further and further (Figure 2B), and after approximately 20 sec, the filopodium contacted dinoflagellate x (Figure 2C). Maintaining contact with dinoflagellate x, the filopodium of cell b became thicker and thicker (Figures 2C–E), and ∼2 min after contact, dinoflagellate x was phagocytosed by the thickened filopodium or a part of the cytoplasm of cell b (Figure 2F). Dinoflagellate x was moved to the center of cell b (from Figures 2F–J). Cell b still extended lamellipodia and showed active locomotion. The accomplishment of in vitro symbiosis between b and x, from contact of the two cells until the settlement of x in the cytoplasm of b, took only five min. In addition, during this time, cell c contacted dinoflagellate y with its cell membrane or a thin lamellipodium (Figure 2A). Cell c actively shifted the membrane around y (Figures 2A–C) and quickly incorporated y into the cytoplasm (Figure 2D). With symbiotic dinoflagellate y in the cytoplasm, cell c exhibited locomotion using pseudopodia (Figures 2E–J). Cell c also promptly engulfed dinoflagellate y within a few min after their first encounter.
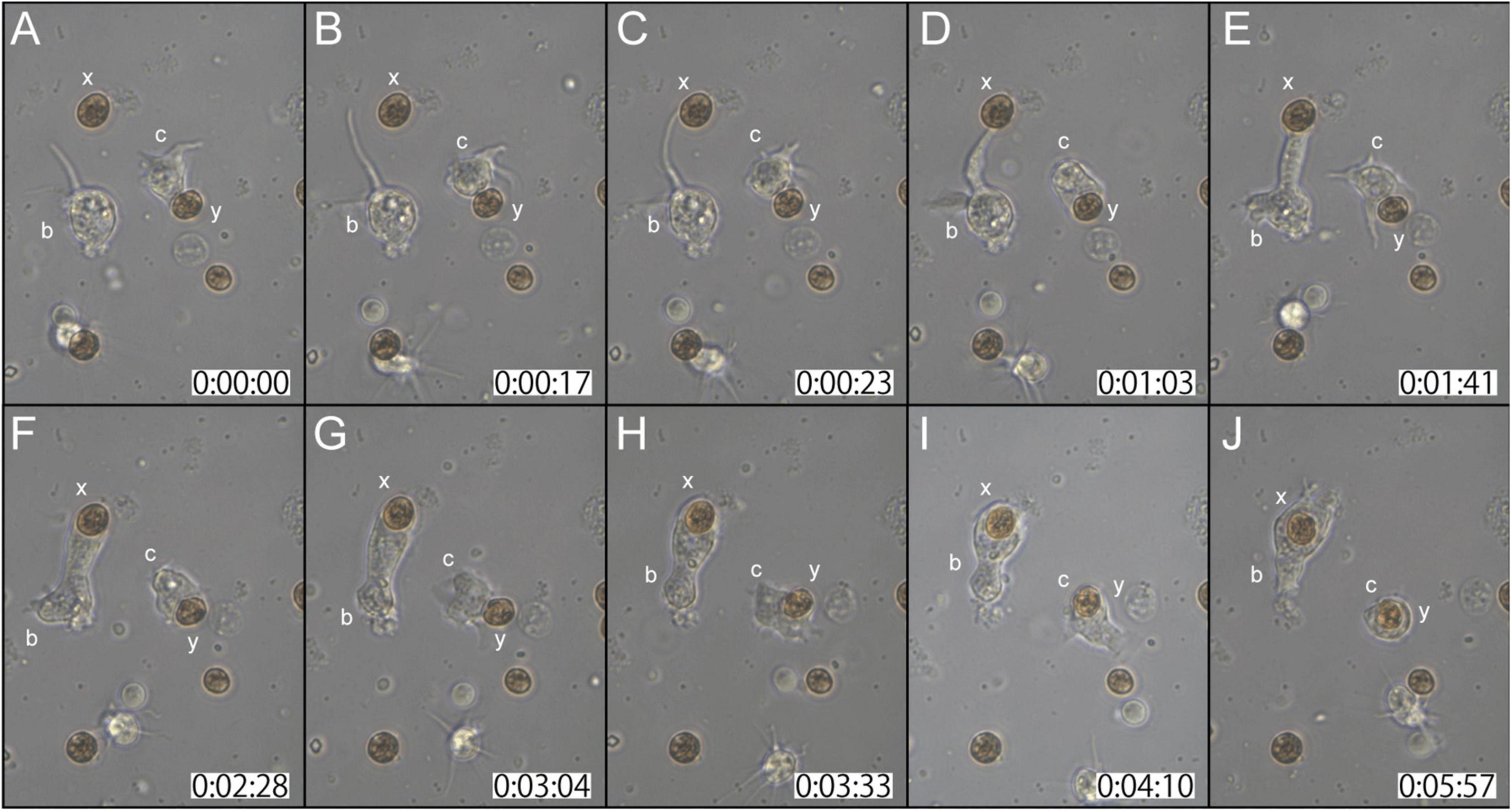
Figure 2. A time-lapse video of symbiotic interactions of coral cells (b and c) and dinoflagellates (x and y). The starting point of observations is shown at 0 h: 00 min: 00 s. (A–J) Serial pictures are taken at time intervals indicated at the bottom. Coral cell b engulfs symbiont x, whereas coral cell c endocytoses symbiont y. See the detailed description in the text.
In the second case (Supplementary Movie 3), at the start of observations, coral cell b contacted the cell wall of dinoflagellate x with its lamellipodium (Figure 3A). Although cell b showed extensive lamellipodial activity, the interaction between b and x appeared not to proceed further (Figures 3A–C). In the interim, neighboring cell c that had already engulfed dinoflagellate y, extended lamellipodia toward x (Figures 3B,C). Within 30 s thereafter, dinoflagellate x was engulfed by cell c (Figure 3D). While actively shuffling its membrane, an interaction between cells b and c continued for a while (Figures 3E–H). Finally, cell c departed from b, with two dinoflagellates, x and y, in its cytoplasm (Figures 3I,J). This in vitro symbiosis between coral cells and dinoflagellates was accomplished within ∼5 min (Figure 3).
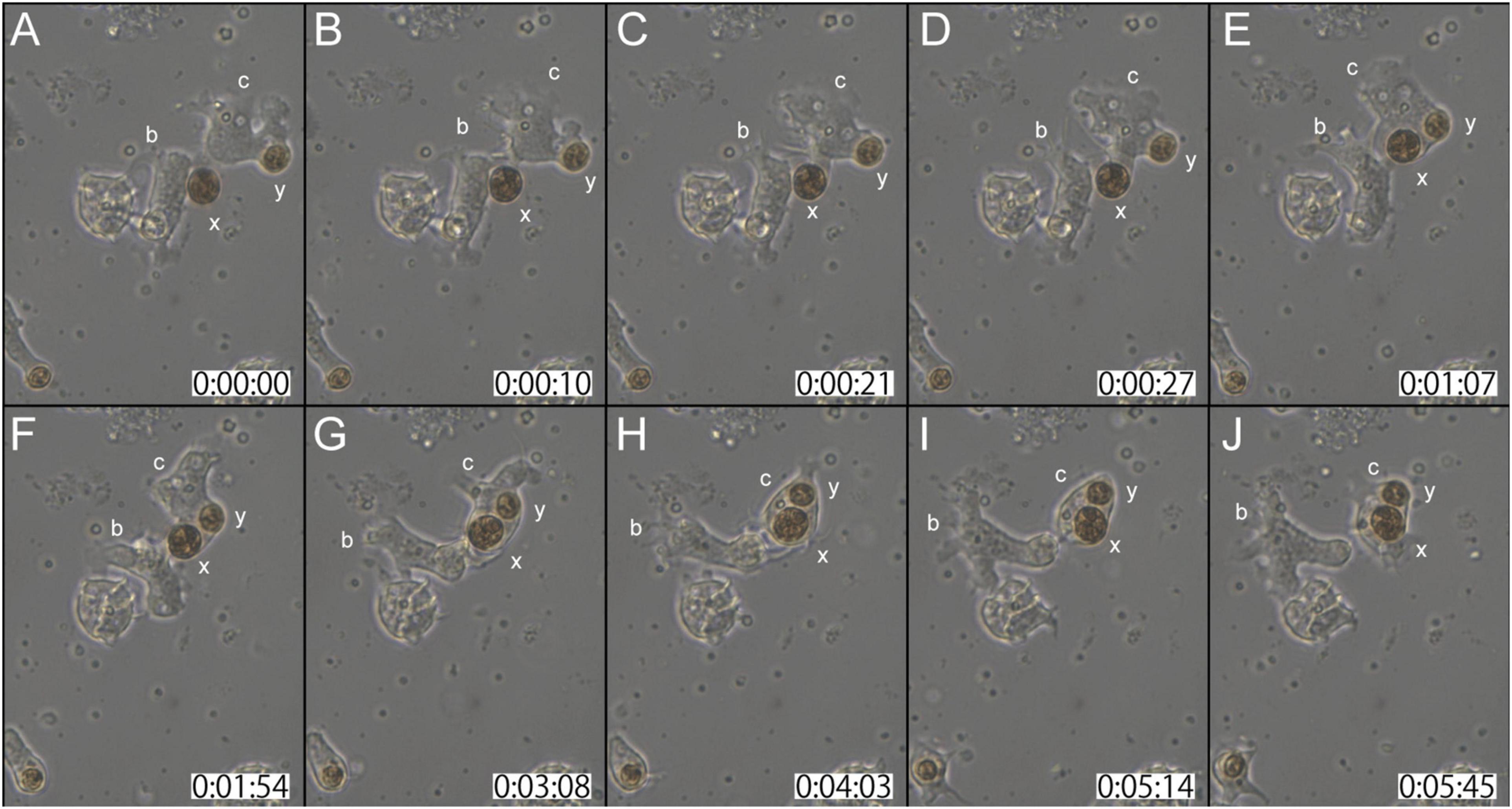
Figure 3. Another time-lapse video of symbiotic interactions of coral cells (b and c) and dinoflagellates (x and y). The starting point of observations is shown at 0 h: 00 min: 00 s. (A–J) Serial pictures are taken at time intervals indicated at the bottom. Coral cell b interacted with symbiont x, but did not incorporate it, whereas coral cell c endocytosed both x and y. See the detailed description in the text.
As described above, flattened amorphous coral cells repeatedly extended and retracted filopodia and lamellipodia around algal cells. Many coral cells had engulfed dinoflagellates within ∼5 min after mixing.
No cell types other than the flattened amorphous cells showed this behavior. Coral cell membranes around the region that attached to algal cells showed extensive shuffling activity, which may be necessary for the cells to endocytose algal cells. Sometimes coral cells endocytosed dinoflagellates and then exocytosed them (Supplementary Movie 4). Some cells repeated this process. In addition, single coral cells sometimes incorporated multiple symbionts (Figures 1E, 3). The number of coral cells that incorporated dinoflagellates increased throughout the day of mixing (see next section).
We examined and confirmed the interaction of coral cells with algal cells by observation of semi-thin sections of the cells, 7 h after mixture of the two types of cells (Figure 4). Figure 4A shows a micrograph in which a coral cell filopodium extended over an algal cell. In the next step, more than a half of the alga body was covered by coral cell membrane (Figure 4B). Then, another section revealed complete engulfment of an algal cell in host coral-cell cytoplasm (Figure 4C). Figures 4D,E show the relationship between engulfed algal cells and a large vacuole of host cell cytoplasm. In Figure 4D, an engulfed algal cell was close to the vacuole, but the two were separated by membranes. In Figure 4E, however, the cytoplasmic membrane enclosing an alga and the membrane of a vacuole appeared fused into a continuous structure. Figure 4F shows that an engulfed algal cell became fragmented in the host coral cell cytoplasm. The semi-thin section undoubtedly demonstrates coral cell engulfment of dinoflagellates.
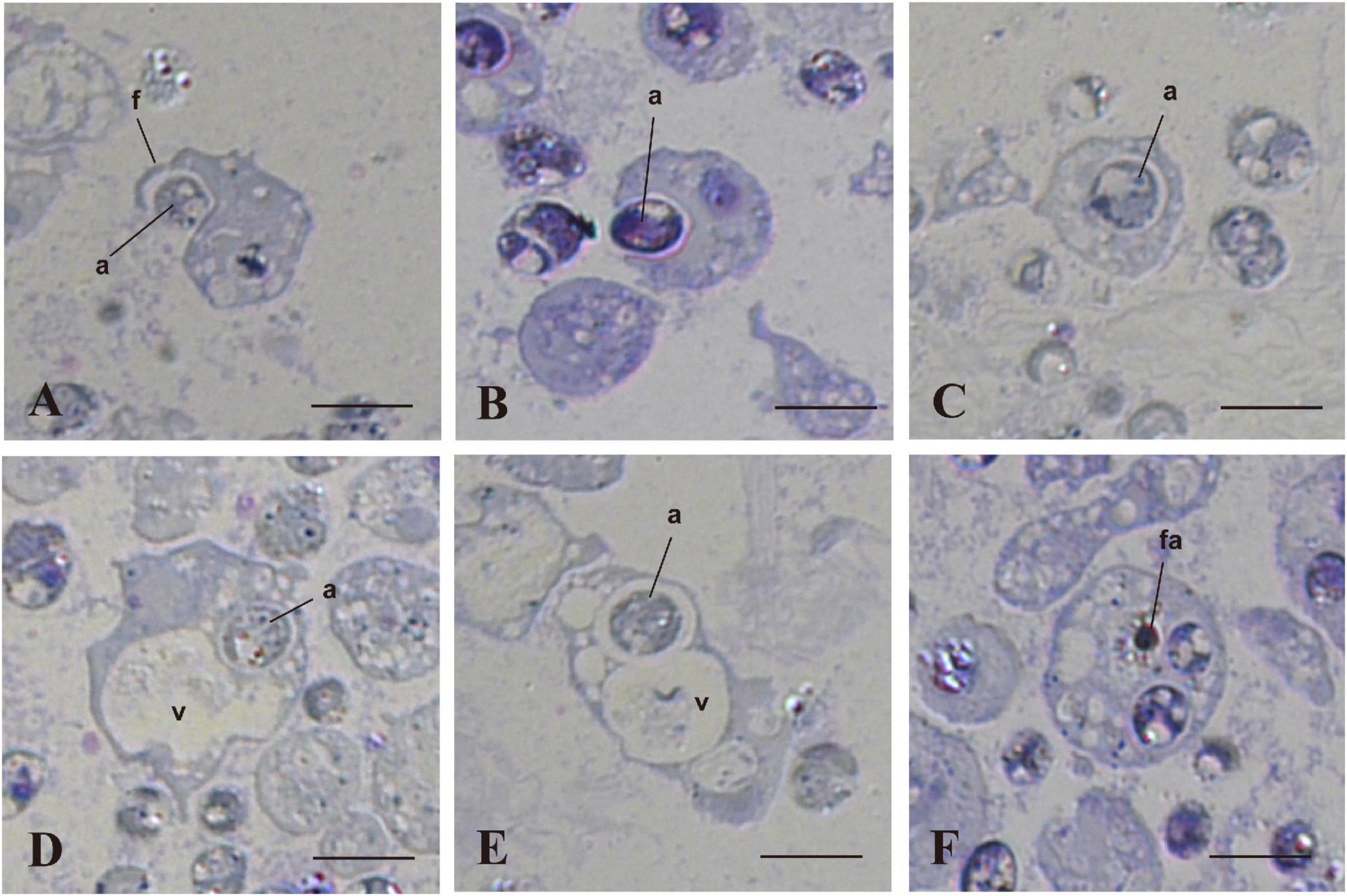
Figure 4. In vitro interactions between coral cells and algae, 7 h after coral cell-algae mixture. Semi-thin sections were stained with toluidine blue. (A) Coral cell extending filopodium over an alga. (B) Coral cell engulfing an alga. (C) Alga incorporated into coral cell cytoplasm. (D) Algal phagosome in association with a coral cell vacuole. (E) Algal phagosome fused with a host vacuole. (F) Fragmented alga in a coral cell. a, algae; f, filopodium; fa, fragmented alga; v, vacuole. Bars, 10 μm.
One half to 1 day after inoculation, some engulfed algal cells remained in the cytoplasm of host coral cells (Figure 5A). On the other hand, some other algal cells were present within large vacuoles in the cytoplasm (Figure 5B). Algal cells in the cytoplasm (not in vacuoles) were approximately 12 μm in diameter (Figure 5A), which was somewhat larger than those in vacuoles (Figure 5B) and non-engulfed algal cells, which were usually 8–10 μm in diameter (Figures 5C,D, arrows). This may indicate swelling of dinoflagellates in host coral cell cytoplasm. In addition, some algal cells began to exhibit fragmentation (Figure 5C), and debris remained in the cytoplasm (Figure 5D; Supplementary Movie 5). These changes were consistent with the observation of semi-thin sections (Figure 4).
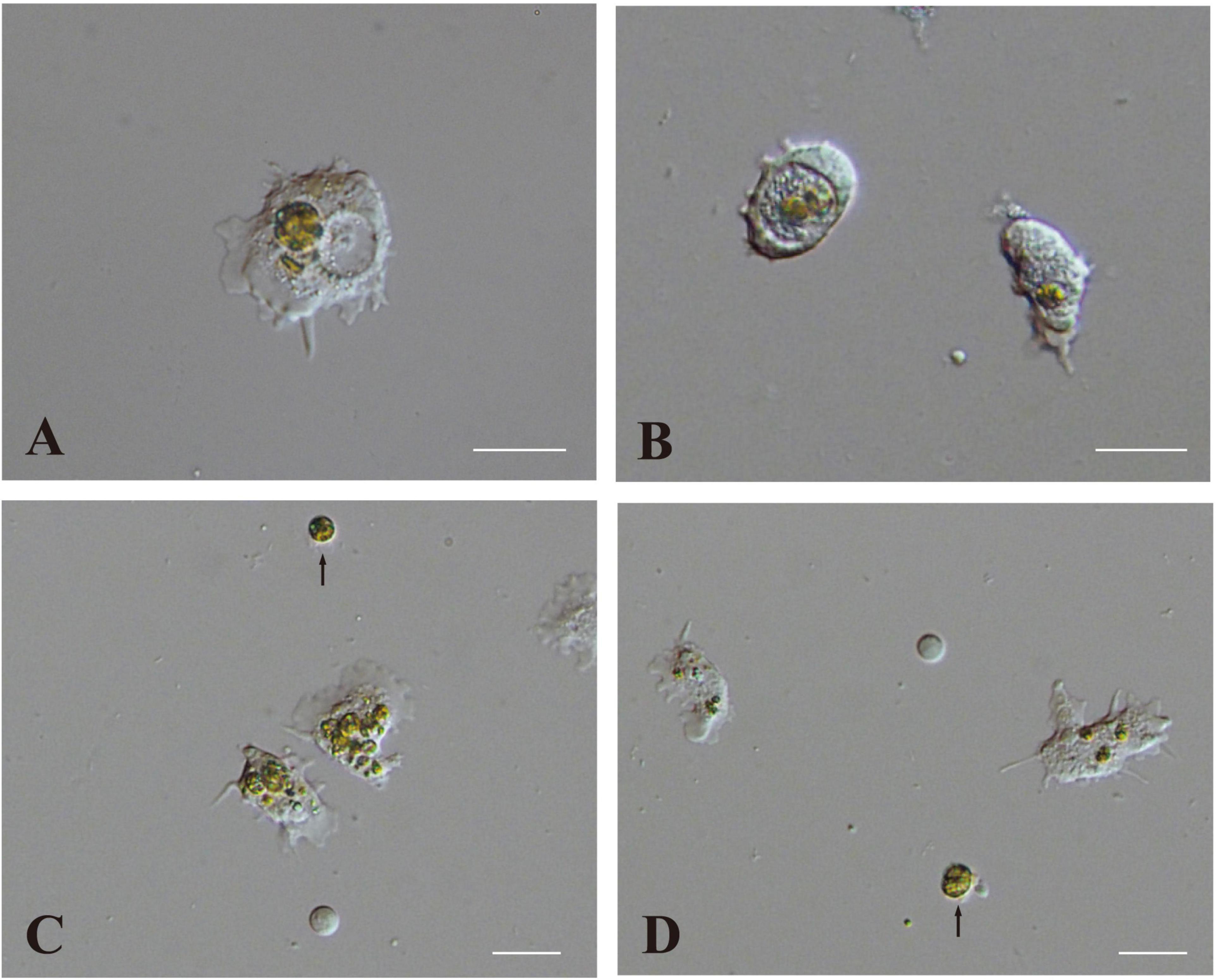
Figure 5. Two modes of interaction between coral cells and dinoflagellates. (A,B) Settlement of dinoflagellates in coral cell cytoplasm, (A) outside and (B) inside a vacuole. (C,D) Fragmentation followed by collapse of dinoflagellates in coral cell cytoplasm. Fragmentation appears more severe in (C) than (D). Dinoflagellates not engulfed by coral cells are indicated with arrows. Scale bar, 20 μm.
After 2 days of culture, cells with unfragmented algae survived in the culture dish (Figure 6A, broken circles). They withdrew pseudopods, stopped moving, and assumed a spherical form (Figure 5B). Spherical cells with algal cells increased in number until about a week of cell culture (Figures 6A,B,C1). In many cases of 7-day-old cell culture, algae were in the cytoplasm of host cells. Approximately 80% of spherical cells contained 1–3 unfragmented algae in their cell bodies (Figure 6C3). In some cases, algal cells were observed within endogenous large vacuoles in host cells (Figure 6C2), indicating translocation of engulfed algae from cytoplasmic vesicles to vacuoles. However, it is uncertain at present which phagosome-like vesicles or vacuolar organelles become genuine, long-sustainable symbiosomes, because it became more and more difficult to discriminate between vacuolar membranes and phagosomal membranes. In 22-day-old symbionts, algal cells were still found in cultured host cells, although host cells possessing algae had significantly decreased in number (Figures 6D1,D2). At present, we failed in subculturing coral cells with symbionts, because the symbionts die after replating.
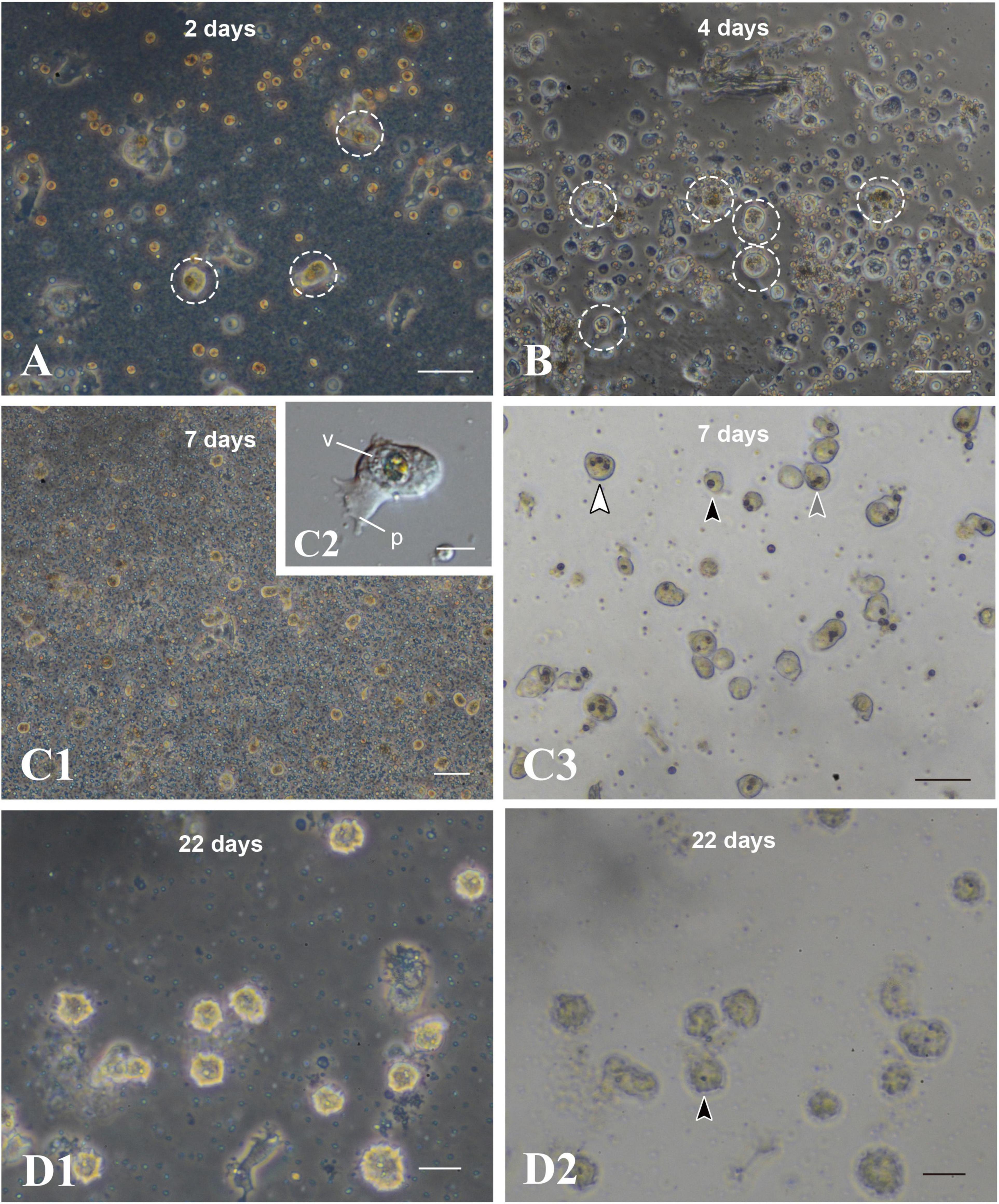
Figure 6. Maintenance of coral cells with symbiotic dinoflagellates. (A) 2 days, (B) 4 days, (C) 7 days, and (D) 22 days after mixture of the two types of cells. Coral cells with symbiotic dinoflagellates (broken circles) became spherical, without pseudopodial extensions, and exhibit less and less locomotor activity. Those cells became globular, bright cells. (C3,D2) are bright field images of (C1,D1), showing one (black arrowhead), two (gray arrowhead), and three algae (white arrowhead) in single cells. p, pseudopodium; v, vacuole. Scale bars, 50 μm in (A,B,C3), 100 μm in (C1), 10 μm in (C2), and 20 μm in (D1,D2).
We reported here the occurrence of in vitro symbiosis of cells of the reef-building coral, Acropora tenuis, with ITS2-type B1 dinoflagellates (Breviolum minutum). Dark flattened amorphous cells of the IVB5 line have properties of endoderm and engulfed algal cells by actively shuffling cell membranes and by extending filopodia and lamellipodia. Phagocytosis itself was completed within 5 min after contact of coral cells with algal cells. Approximately half of flattened amorphous coral cells achieved symbiosis in the first day of the mixture, irrespective of the type of culture medium. This in vitro symbiosis is highly reproducible; thus, this in vitro system will facilitate exploration of cellular and molecular mechanisms involved in coral-dinoflagellate symbiosis at the single-cell level. Knowledge obtained in future studies using this system may advance our understanding of coral biology associated with bleaching, thereby providing cues for methodological improvements of coral reef preservation.
The phagocytotic behavior of flattened amorphous coral cells is very dynamic, and phagocytosis occurs very quickly (Figures 2, 3). It is uncertain whether in vivo phagocytosis of planula larva endoderm cells is like that of this in vitro system. However, such dynamic and prompt symbiosis tempts us to speculate that the first evolutionary step of coral-dinoflagellate interactions was active coral cell phagocytosis of dinoflagellates, moving through the gastric cavity into coral gastroderm cells. It is likely that corals have maintained this high potential for phagocytosis throughout evolution so that when they are cultured in vitro, they show high ancestral phagocytotic activity.
One of the questions raised by this in vitro system is whether symbiosis is accomplished by flattened amorphous cells alone (without mixture or indirect help of other types of cells) or only in a mixture containing other types of cells (Figure 1A). The IVB5 line is polyclonal and contains other types of cells beside flattened amorphous cells. The cellular composition of the IVB5 line therefore resembles that of planula larvae, which consist of outer ectoderm and inner endoderm. Such a complex, harmonious mixture of cells may be required for flattened amorphous cells of endodermal origin to initiate symbiosis with algal cells. This should be examined in the future using monoclonal, flattened, amorphous cells. Nevertheless, the IVB5 line serves as an in vitro system for studying coral cell symbiosis with photosynthetic dinoflagellates at the single-cell level.
There are many questions regarding cellular and molecular mechanisms involved in coral-dinoflagellate symbiosis. Three major overlapping, interrelated processes involved in this symbiosis are (1) the onset of symbiosis and mechanisms of partner recognition, (2) maintenance and dynamic homeostasis, and (3) dysbiosis that leads to bleaching (reviewed by Davy et al., 2012; Weis, 2019).
In relation to the first process of coral-dinoflagellate symbiosis, classification of symbiotic species of the family Symbiodiniaceae has recently been revised and these taxa are classified into Symbiodinium (previously Clade A Symbiodinium), Breviolum (Clade B), Cladocopium (Clade C), Durusdinium (Clade D), Effrenium (Clade E), and others (LaJeunesse et al., 2018). Of these, Symbiodinium, Breviolum, Cladocopium, and Durusdinium comprise major coral symbionts. Extensive studies have been carried out to examine specificity, preferences, or exchange of dinoflagellates of different genera hosted by different species of corals (in case of A. tenuis, e.g., Yamashita et al., 2013, 2014, 2018). In our laboratory conditions, Acropora tenuis larvae harbor Symbiodinium, Breviolum, and Cladocopium, although ratios of symbiosis differ depending on the algal species (Shoguchi et al., unpublished data). Future studies should examine whether dinoflagellates other than Breviolum are engulfed by the IVB5 line. If the IVB5 line shows different acceptance ratios, we might address the question of symbiont recognition and specificity using the IVB5 line and different species of dinoflagellates.
As to the second issue of maintenance and dynamic homeostasis, algal cells need to be enclosed by an endomembrane to form “symbiosome,” which is a critical component of coral-dinoflagellate symbiosis (Wakefield and Kempf, 2001; Davy et al., 2012; Rosset et al., 2021). How does its composition and/or structures differ from the plasma membrane in function, and what is the role of membrane trafficking in its regulation and maintenance (Weis, 2019)? In the IVB5-line system, algae engulfed by cultured cells likely have two destinations: fragmentation or settlement in vacuoles (Figures 4, 5). Fragmentation of Breviolum minutum does not occur during the culture process, as far as our experience goes. On the other hand, fragmentation of engulfed algae occurred in cells of the IVB5 line. It is natural to assume that in the cytoplasm, a phagosomal membrane encircling the algal cell fuses with lysosomal membranes to make secondary lysosomes as part of the normal pathway for degradation of foreign material (Davy et al., 2012). Therefore, the escape of engulfed algae from the lysosomal digestion by host cells is essential to establish symbiosis. If algal cells are transferred from the phagosome to a vacuole, they may escape lysosomal digestion. However, at present, it is uncertain whether fragmentation always occurs in algae that were not engulfed by vacuoles. Future studies should explore the role of organelles as well as intracellular membranes in the establishment of symbiosis in vitro.
The IVB5 line also provides a system to explore molecular mechanisms involved in coral-dinoflagellate symbiosis. Studies should strive to duplicate results of previous transcriptomic studies (Yuyama et al., 2018; Yoshioka et al., 2021) and of single-cell RNA-seq analyses (Hu et al., 2020). If genes that play pivotal roles in the symbiosis are found in future studies, an introduction of CRISPR/Cas9-mediated genome editing method seems challenging (Cleves et al., 2018). Since genomes of both Acropora tenuis (Shinzato et al., 2021) and Breviolum minutum (Shoguchi et al., 2013) have been decoded, molecular biological studies may be facilitated using genomicinformation. Transcriptome analyses of gene expression changes in the IVB5-line cells after interaction and phagocytosis of algal cells are now in progress.
KK and NS conceived and designed the study and prepared the manuscript. ES and SS cultured dinoflagellates. KK, SS, KN, KH, SF, and NS carried out analyses. All authors commented on it.
This work was supported by an OIST fund for collaboration with Kochi University, and also funded in part by the Mrs. Sumiko Imano Memorial Foundation. Sachiko McAlinn and Miki McAlinn were acknowledged for their funding support.
The handling editor declared a shared committee with one of the authors, NS, at time of review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank all members of the Marine Genomics Unit at OIST for their support. Steven D. Aird (https://www.sda-technical-editor.org) is acknowledged for editing the manuscript. Professor Virginia Weis, one of the reviewers of this manuscript, kindly noted the mistake in our transcriptome presentation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.706308/full#supplementary-material
Supplementary Movie 1 | A time-lapse video showing locomotor activity of flattened amorphous cells. Soon after replating, the cells extend lamellipodia and filopodia and show moderate locomotor activity. Frames were taken 20 sec intervals.
Supplementary Movie 2 | A time-lapse video showing that coral cells engulf dinoflagellates into the cytoplasm using filopodia and lamellipodia. For more detailed, see Figure 2 and the main text. Frames were taken 2–3 sec intervals.
Supplementary Movie 3 | A time-lapse video showing another case in which coral cells engulf dinoflagellates into the cytoplasm using filopodia and lamellipodia. For more detailed, see Figure 3 and the main text. Frames were taken 2–3 sec intervals.
Supplementary Movie 4 | A time-lapse video showing extensive shuffling activity of coral cell membranes around the region that attached to algal cells, by which coral cells endocytose algal cells and then exocytose them. Some coral cells repeat this process. Frames were taken 2–3 sec intervals.
Supplementary Movie 5 | A time-lapse video showing coral cells that incorporated dinoflagellates in host coral-cell cytoplasm, and fragmentation of dinoflagellates in the coral-cell cytoplasm. Frames were taken 20 sec intervals.
Cleves, P. A., Strader, M. E., Bay, L. K., Pringle, J. R., and Matz, M. V. (2018). CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc. Natl. Acad. Sci. U.S.A. 115, 5235–5240. doi: 10.1073/pnas.1722151115
Coffroth, M. A., and Santos, S. R. (2005). Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156, 19–34. doi: 10.1016/j.protis.2005.02.004
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. doi: 10.1128/mmbr.05014-11
Domart-Coulon, I., and Ostrander, G. K. (2016). “Coral cell and tissue methods,” in Diseases of Coral, eds C. M. Woodley, C. A. Downs, A. W. Bruckner, J. W. Porter, and S. B. Galloway (Hoboken, NJ: Wiley), 489–505. doi: 10.1002/9781118828502.ch37
Dove, S. G. (2004). Scleractinian corals with photoprotective host pigments are hypersensitive to thermal bleaching. Mar. Ecol. Prog. Ser. 272, 99–116. doi: 10.3354/meps272099
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., Harvell, C. D., et al. (2007). Coral reefs under rapid climate changes and ocean acidification. Science 318, 1737–1742.
Hu, M., Zheng, X., Fan, C.-H., and Zheng, Y. (2020). Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 582, 534–538. doi: 10.1038/s41586-020-2385-7
Hughes, T. P., Kerry, J. T., Alvarez-Noriega, M., Alvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377.
Kawamura, K., Nishitsuji, K., Shoguchi, E., Fujiwara, S., and Satoh, N. (2021). Establishing sustainable cell lines of a coral, Acropora tenuis. Mar. Biotechnol. 23, 373–388. doi: 10.1007/s10126-021-10031-w
LaJeunesse, T. C., Parkinson, J. E., and Reimer, J. D. (2012). A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J. Phycol. 48, 1380–1391. doi: 10.1111/j.1529-8817.2012.01217.x
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., and Santos, S. R. (2018). Systematic revision of symbiodiniaceae high- lights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Loh, W. K. W., Loi, T., Carter, D., and Hoegh-Guldberg, O. (2001). Genetic variability of the symbiotic dinoflagellates from the wide ranging coral species Seriatopora hystrix and Acropora longicyathus in the Indo-West Pacific. Mar. Ecol. Prog. Ser. 222, 97–107. doi: 10.3354/meps222097
McIlroy, S. E., and Coffroth, M. A. (2017). Coral ontogeny affects early symbiont acquisition in laboratory-reared recruits. Coral Reefs 36, 927–932. doi: 10.1007/s00338-017-1584-7
Reaka-Kudla, M. L. (2001). “Coral reefs: biodiversity and conservation,” in Contemporary Approaches to the Study of Biodiversity, eds H. M. Hernandez, A. N. Garcia Aldrete, F. Alvarez, and M. Ulloa (Laramie, WY: Instituto de Biodiversity), 221–243.
Rinkevich, B. (2011). Cell cultures from marine invertebrates: insights for capturing endless stemness. Mar. Biotechnol. 13, 345–354. doi: 10.1007/s10126-010-9354-3
Rosset, S.L., Oakley, C. A., Ferrier-Page‘s, C., Suggett, D.J., Weis, V. M., and Davy, S. K. (2021). The molecular language of the cnidarian-dinoflagellate symbiosis. Trends Microbiol. 29, 320–333. doi: 10.1016/j.tim.2020.08.005
Sebe-Pedros, A., Saudemont, B., Chomsky, E., Plessier, F., Mailhé, M. P., Renno, J., Loe-Mie, Y., et al. (2018). Cnidarian cell types diversity and regulation revealed by whole-organism single-cell RNA-seq. Cell 173, 1520–1534.e20.
Shinzato, C., Khalturin, K., Inoue, J., Zayasu, Y., Kanda, K., Kawamitsu, M., Yoshioka, Y., Yamashita, H., Suzuki, G., and Satoh, N. (2021). Eighteen coral genomes reveal the evolutionary origin of Acropora strategies to accommodate environmental changes. Mol. Biol. Evol. 38, 16–30. doi: 10.1093/molbev/msaa216
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., Takeuchi, T., Hisata, K., Tanaka, M., Fujiwara, M., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Spaldinga, M., Burke, L., Wood, S. A., Ashpolee, J., Hutchisone, J., and zu Ermgassene, P. (2017). Mapping the global value and distribution of coral reef tourism. Mar. Policy 82, 104–113. doi: 10.1016/j.marpol.2017.05.014
Sully, S., Burkepile, D. E., Donovan, M. K., Hodgson, G., and van Woesik, R. (2019). A global analysis of coral bleaching over the past two decades. Nat. Commun. 10:1264.
Wakefield, T. S., and Kempf, S. C. (2001). Development of host- and symbiont-specific monoclonal antibodies and confirmation of the origin of the symbiosome membrane in a cnidarian–dinoflagellate symbiosis. Biol. Bull. 200, 127–143. doi: 10.2307/1543306
Weis, Y. M. (2019). Cell biology of coral symbiosis: foundational study can inform solution to the coral reef crisis. Integr. Comp. Biol. 59, 845–855. doi: 10.1093/icb/icz067
Wilkinson, C. (2008). Status of Coral Reefs of the World: 2008. Townsville, AU: Global Coral Reef Monitoring Network.
Yamashita, H., Suzuki, G., Hayashibara, T., and Koike, K. (2013). Acropora recruits harbor “rare” Symbiodinium in the environmental pool. Coral Reefs 32, 355–366. doi: 10.1007/s00338-012-0980-2
Yamashita, H., Suzuki, G., Kai, S., Hayashibara, T., and Koike, K. (2014). Establishment of coral–algal symbiosis requires attraction and selection. PLoS One 9:e97003. doi: 10.1371/journal.pone.0097003
Yamashita, H., Suzuki, G., Shinzato, C., Jimbo, M., and Koike, K. (2018). Symbiosis process between Acropora larvae and Symbiodinium differs even among closely related Symbiodinium types. Mar. Ecol. Prog. Ser. 592, 119–128. doi: 10.3354/meps12474
Yellowlees, D., Rees, T. A., and Leggat, W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694. doi: 10.1111/j.1365-3040.2008.01802.x
Yoshioka, Y., Yamashita, H., Suzuki, G., Zayaus, Y., Tada, I., Kanda, M., et al. (2021). Whole-genome transcriptome analyses of native symbionts reveal host coral genomic novelties for establishing coral–algae symbioses. Genome Biol. Evol. 13:evaa240. doi: 10.1093/gbe/evaa240
Yuyama, I., Ishikawa, M., Nozawa, M., Yoshida, M. A., and Ikeo, K. (2018). Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci. Rep. 8:16802.
Keywords: in vitro symbiosis, corals, dinoflagellates, Acropora, endoderm, phagocytosis
Citation: Kawamura K, Sekida S, Nishitsuji K, Shoguchi E, Hisata K, Fujiwara S and Satoh N (2021) In vitro Symbiosis of Reef-Building Coral Cells With Photosynthetic Dinoflagellates. Front. Mar. Sci. 8:706308. doi: 10.3389/fmars.2021.706308
Received: 07 May 2021; Accepted: 21 June 2021;
Published: 14 July 2021.
Edited by:
James Davis Reimer, University of the Ryukyus, JapanReviewed by:
Virginia M. Weis, Oregon State University, United StatesCopyright © 2021 Kawamura, Sekida, Nishitsuji, Shoguchi, Hisata, Fujiwara and Satoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaz Kawamura, a2F6dWtAa29jaGktdS5hYy5qcA==; Noriyuki Satoh, bm9yaXNreUBvaXN0Lmpw
†ORCID: Kaz Kawamura, orcid.org/0000-0003-2118-9511; Koki Nishitsuji, orcid.org/0000-0002-4015-7139; Eiichi Shoguchi, orcid.org/0000-0003-3136-5558; Shigeki Fujiwara, orcid.org/0000-0001-9464-180X; Noriyuki Satoh, orcid.org/0000-0002-4480-3572
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.