- 1Department of Biology, College of Charleston, Charleston, SC, United States
- 2Department of Biology, Middle Tennessee State University, Murfreesboro, TN, United States
- 3Department of Biology, Appalachian State University, Boone, NC, United States
- 4Department of Ecology and Evolution, Stony Brook University, Stony Brook, NY, United States
- 5Smithsonian Tropical Research Institute, Panama City, Panama
Marine sponges have been successful in their expansion across diverse ecological niches around the globe. Pioneering work attributed this success to both a well-developed aquiferous system that allowed for efficient filter feeding on suspended organic matter and the presence of microbial symbionts that can supplement host heterotrophic feeding with photosynthate or dissolved organic carbon. We now know that sponge-microbe interactions are host-specific, highly nuanced, and provide diverse nutritional benefits to the host sponge. Despite these advances in the field, many current hypotheses pertaining to the evolution of these interactions are overly generalized; these over-simplifications limit our understanding of the evolutionary processes shaping these symbioses and how they contribute to the ecological success of sponges on modern coral reefs. To highlight the current state of knowledge in this field, we start with seminal papers and review how contemporary work using higher resolution techniques has both complemented and challenged their early hypotheses. We outline different schools of thought by discussing evidence of symbiont contribution to both host ecological divergence and convergence, nutritional specificity and plasticity, and allopatric and sympatric speciation. Based on this synthesis, we conclude that the evolutionary pressures shaping these interactions are complex, with influences from both external (nutrient limitation and competition) and internal (fitness trade-offs and evolutionary constraints) factors. We outline recent controversies pertaining to these evolutionary pressures and place our current understanding of these interactions into a broader ecological and evolutionary framework. Finally, we propose areas for future research that we believe will lead to important new developments in the field.
Introduction
Sponges (Phylum Porifera) form one of the earliest branching lineages in the metazoan tree of life (Feuda et al., 2017; Nielsen, 2019). Despite their relatively simple body plan, they have expanded across diverse ecological niches within the marine environment on a global scale (Reiswig, 1973; Diaz and Rützler, 2001; McClintock et al., 2005; Wulff, 2006). With a well-developed aquiferous system, sponges are able to efficiently retain living particulate organic matter (phytoplankton and bacteria), detritus, and dissolved organic matter from the surrounding water (Maldonado et al., 2012; McMurray et al., 2018; Rix et al., 2020). Their ecological success on oligotrophic coral reefs (de Goeij et al., 2013), however, is likely also tied to an evolutionary investment in microbial symbionts that expand their metabolic capabilities into new niche dimensions (Easson and Thacker, 2014; Webster and Thomas, 2016; Bart et al., 2020; Freeman et al., 2020).
High diversity systems like coral reefs often display strong niche partitioning, with natural selection favoring adaptations that reduce competition by allowing coexisting organisms to fill unique ecological niches (Finke and Snyder, 2008; Northfield et al., 2010). Indeed, dominant functional groups (e.g., scleractinian corals, soft corals, sponges) on coral reefs have divergent heterotrophic feeding strategies (Porter, 1976; Reiswig, 1981) and variable (in both overall abundance and community composition) interactions with microbial symbionts (Vacelet and Donadey, 1977; Muscatine and Porter, 1977). In addition, the presence of slight nuances in host physiology, morphology, and symbiont productivity or identity across individual species can facilitate niche diversification within these groups and can contribute to high biodiversity within these crowded ecosystems (Porter, 1976; Baker et al., 2015; Freeman et al., 2020).
Pioneering work on sponges identified unique evolutionary solutions to the environmental challenges of coral reefs across different sponge species. Sponges were reported to vary in characteristics such as the complexity of their aquiferous system, tissue density, pumping rate, abundance of microbial symbionts, morphology, and exploitation of different nutrient sources (Reiswig, 1971, 1974; Vacelet and Donadey, 1977; Reiswig, 1981; Wilkinson, 1983). More contemporary work has expanded this story by identifying substantial interspecific variation in growth rates, reproductive effort, community composition and diversity of microbial symbionts, reliance on different heterotrophic nutrient pools, microbial symbiont nutrient transformations, metabolic plasticity, and chemical defense production (Loh and Pawlik, 2014; Wulff, 2017; McMurray et al., 2018; Pawlik et al., 2018; Pita et al., 2018; Zhang et al., 2019; Bell et al., 2020). Despite this wealth of research, both classic and contemporary papers frequently group species based on one characteristic (i.e., sponges either host productive symbionts or do not, have either high or low microbial abundance, or are either chemically defended or undefended); this trend toward binary categorizations has led to broad generalizations about sponge ecology and evolution (Wilkinson and Cheshire, 1990; Loh and Pawlik, 2014; McMurray et al., 2018). In reality, these binary categories frequently represent the extreme ends of a continuum. Thus, we hypothesize that nuanced differences in characteristics across individual species have allowed sponges to cope with diverse ecological and evolutionary pressures on coral reefs (Easson and Thacker, 2014; Freeman et al., 2020; Wulff, 2020).
Sponge microbial ecology is a rapidly advancing and multidimensional field that requires an integrated view of the complex interplay between hosts, symbionts, and their environments (Webster and Thomas, 2016). To date, however, there has not been a comprehensive synthesis that unites historical work with contemporary studies employing high-resolution techniques to elucidate the species-level variation in these interactions (Paul et al., 2019). This gap in our understanding is significant and has yielded an increasing frequency of over-generalizations that limit our understanding of the complex links between microbial symbionts and the evolutionary trajectory of individual host species (Freeman et al., 2015; Webster and Thomas, 2016). In this review, we therefore aim to synthesize data from both seminal and recent studies, discuss emerging schools of thought, highlight knowledge gaps, and propose testable hypotheses that will facilitate an improved understanding of sponge ecology and evolution on coral reefs.
We will broadly review nascent studies on sponge ecology and evolution, but will focus mostly in the Caribbean Sea, where sponges are a dominant structural and functional group that has been well studied over the past 30 years (Wilkinson, 1987; Webster and Thomas, 2016). Sponge cover in the Caribbean now exceeds that of reef building corals, with an average percent cover of 15.9% (range from ∼2 to 75%), and a species richness of more than 500 species (Diaz and Rützler, 2001; Miloslavich et al., 2010; Loh and Pawlik, 2014). Recent reviews have expounded on the diverse metabolic pathways and biogeochemical cycling present in marine sponges and their symbionts (Pita et al., 2018; Pawlik and McMurray, 2019; Zhang et al., 2019), so we will limit our discussion and review of these broad topics and instead focus on evidence of species-level differences in holobiont [both sponge hosts and their microbial symbionts (Zilber-Rosenberg and Rosenberg, 2008)] characteristics. We will focus mostly on shallow-water (<30 m deep) sponges and will address four main topics: (1) early work on sponge ecology and carbon metabolism; (2) microbial symbiont diversity and expansion of host resource use into niche dimensions beyond carbon metabolism; (3) ecological variation across host species; and (4) selective pressures that shape these associations and impact the evolution of life history strategies and speciation.
Pioneering Work in Sponge Ecology and Carbon Metabolism
Sponges have advanced, well-developed aquiferous systems that initiate water flow and remove particulate organic matter (POM) such as phytoplankton, bacteria, detritus, and even viruses from the water column (Reiswig, 1971, 1975a; Maldonado et al., 2012). The ability to efficiently exploit suspended sources of organic matter has contributed to the ecological success of sponges and fueled their expansion into diverse aquatic niches on a global scale (Reiswig, 1975b; Diaz and Rützler, 2001). Early work on this group, however, began to recognize that some sponge species might be better adapted to heterotrophic feeding on POM than others. In what remain some of the most in-depth studies on sponge physiology to date, Reiswig (1971; 1974; 1981) assessed the rates of respiration and water transport and the energy budgets of the Caribbean sponges Mycale laxissima, Tectitethya crypta, Verongula reiswigi, and Aplysina fistularis (see Reiswig, 1974, 1981; Pawlik et al., 2015 for original and current species names). Of these species, M. laxissima and T. crypta had low respiration rates, high and moderate water transport rates, respectively, and high pumping efficiency, indicating they dedicated a substantial portion of their energy budget [O2 consumed] to pumping water (Reiswig, 1974). In contrast, V. reiswigi, and A. fistularis extracted up to five times as much dissolved oxygen from the water as M. laxissima and T. crypta, had lower pumping rates, and expended less energy on pumping water. Particulate food resources in the surrounding water were sufficient to satisfy the energy requirements of M. laxissima and T. crypta, but V. reiswigi, and A. fistularis were only able to obtain 25 and 14% of their carbon budget from these particulate sources. This major deficit in their carbon budget was reconciled by the proposed assimilation of dissolved organic carbon (DOC) from the water column; abundant microbial symbiont communities within V. reiswigi, and A. fistularis (termed bacteriosponges; sensu Reiswig, 1981) were hypothesized to mediate this DOC assimilation. The use of DOC by V. reiswigi, and A. fistularis was not tested in his study, but Reiswig (1981) estimated that bacteriosponges might remove on average 22% of the available DOC from the surrounding water. This work had important conclusions that have shaped the trajectory of sponge research for the last 40 years: (1) microbial symbionts expand the metabolic capabilities of their host by allowing for the assimilation of dissolved sources of carbon, (2) there are fitness trade-offs (both metabolic costs and potential nutritional benefits) associated with hosting microbial symbionts, and (3) these interactions vary across host species due to differences in aquiferous system complexity, host physiology, and the abundance of microbial symbionts (Reiswig, 1971, 1974, 1981).
Additional work also found variable associations between microbes and their host sponges. Vacelet and Donadey (1977) used electron microscopy (Table 1) to survey the microbial communities within 13 sponge species and reported two general groups of sponges: (1) those with high bacterial abundance, dense tissue, and small choanocyte chambers and (2) species with low bacterial abundance, well-irrigated and low-density tissue, and well-developed aquiferous systems. Similar patterns were also observed by Wilkinson (1978a; 1978c), with considerable variation in microbial abundance and aquiferous system complexity across four tropical sponge species on the Great Barrier Reef (GBR). Although sponges were hypothesized to obtain a greater benefit from the abundant bacterial communities, the consistency of bacterial morphotypes among individuals of the same species led Vacelet and Donadey (1977) to propose that both groups formed true associations with bacterial symbionts that were distinct from surrounding bacteria in the water column. In addition, although sponges could be delineated into two groups based on their overall bacterial density, unique bacterial morphotypes were also found within host species from each group (Vacelet and Donadey, 1977; Wilkinson, 1978a,b,c). These observations, along with evidence that cultured bacteria (Table 1) from the four sponge species from the GBR could metabolize a wide range of compounds (Wilkinson, 1978b), provided initial evidence of functional (host physiology and symbiont metabolism) divergence across sponge species that is shaped at least in part by associations with microbial symbionts (Vacelet and Donadey, 1977; Wilkinson, 1978a,b,c; Reiswig, 1981).
Forty years later, the “bacteriosponges” of Reiswig (1981) and the sponges hosting dense bacterial communities observed by Vacelet and Donadey (1977) are now termed High Microbial Abundance (HMA; sensu Hentschel et al., 2003; Gloeckner et al., 2014) sponges and include the presence of archaea. HMA sponges generally have microbial densities exceeding 108–1010 cells per gram of sponge tissue. In contrast, Low Microbial Abundance (LMA) sponges have microbial numbers around 105–106 cells per gram of sponge tissue, a density of microbes similar to that of seawater (Gloeckner et al., 2014). Broad-scale trends in holobiont metabolism and host resource use are increasingly attributed to overall microbial abundance as HMA sponges generally have denser, less well irrigated tissue, lower pumping rates, and reduced aquiferous system complexity compared to LMA sponges. Moreover, high densities of microbes in HMA species are hypothesized to confer access to microbially mediated metabolic pathways (Hentschel et al., 2006; Weisz et al., 2007; Schläppy et al., 2010; Webster and Taylor, 2012; Poppell et al., 2013).
Heterotrophic Symbionts and Expansion of Host Carbon Metabolism
Sponges have long been hypothesized to assimilate DOC (Stephens and Schinske, 1961; Wilkinson and Garrone, 1980; Reiswig, 1981); this hypothesis has been supported by recent work (Jaeckle, 1995; Ribes et al., 1999; de Goeij et al., 2008; Ribes et al., 2012; Mueller et al., 2014; Hoer et al., 2018; Rix et al., 2020) demonstrating that some sponge species derive a substantial portion of their total carbon budget from dissolved compounds (Yahel et al., 2003; McMurray et al., 2016, 2018). Uptake of DOC by the sponge holobiont may have important implications for reef nutrient cycling as it is a central part of the ‘sponge-loop,’ where assimilated DOC is ultimately transferred to the benthic food web through the production and release of cellular detritus by the sponge (de Goeij et al., 2013). Dissolved organic matter (DOM) assimilation has been demonstrated by both sponge cells and microbial symbionts through stable isotope probing (SIP; Tables 2, 3) by feeding sponges 13C- and 15N-labeled compounds (diatom-, algae-, or coral-derived DOM and glucose) and tracing the incorporation of these isotopic labels into host and symbiont biomass (de Goeij et al., 2008; van Duyl et al., 2011; Rix et al., 2018) or using nanoscale Secondary Ion Mass Spectrometry (nanoSIMS) to visualize (at the cellular level) the assimilation of DOM by microbial symbionts and sponge filter feeding cells via pinocytosis (Achlatis et al., 2019; Rix et al., 2020).
The assimilation of ambient DOM by sponge and/or symbiont cells provides a rich and reliable nutritional resource not available to many other animals within reef ecosystems. Most studies (but see Fiore et al. (2017) and Letourneau et al. (2020) for higher-resolution studies) that have examined the removal of ambient DOM by sponges in situ have focused exclusively on dissolved organic carbon (DOC; Ribes et al., 1999; McMurray et al., 2016, 2018), the largest elemental component of DOM, and measured DOC removal by the sponge holobiont (unit including both host and symbiont cells) by sampling water prior to and following its passing through the sponge. However, although these studies have demonstrated that DOC can make up to 90% of the organic matter in seawater, many sponge species rely more on living particulate organic carbon (LPOC like phytoplankton and heterotrophic bacteria) and/or detritus than DOC to meet their energy requirements or rely on a combination of two or more of these sources of heterotrophic carbon (Figures 1, 2; McMurray et al., 2016, 2018; Morganti et al., 2017; Hoer et al., 2018; Wooster et al., 2019). This, along with variation in POC and DOC fluxes that is uncoupled to microbiome community structure across sponge species (Gantt et al., 2019), suggests that sponge species vary in their exploitation of different heterotrophic carbon pools and that these interspecific differences are shaped by characteristics of both the host and symbionts (Rix et al., 2020).
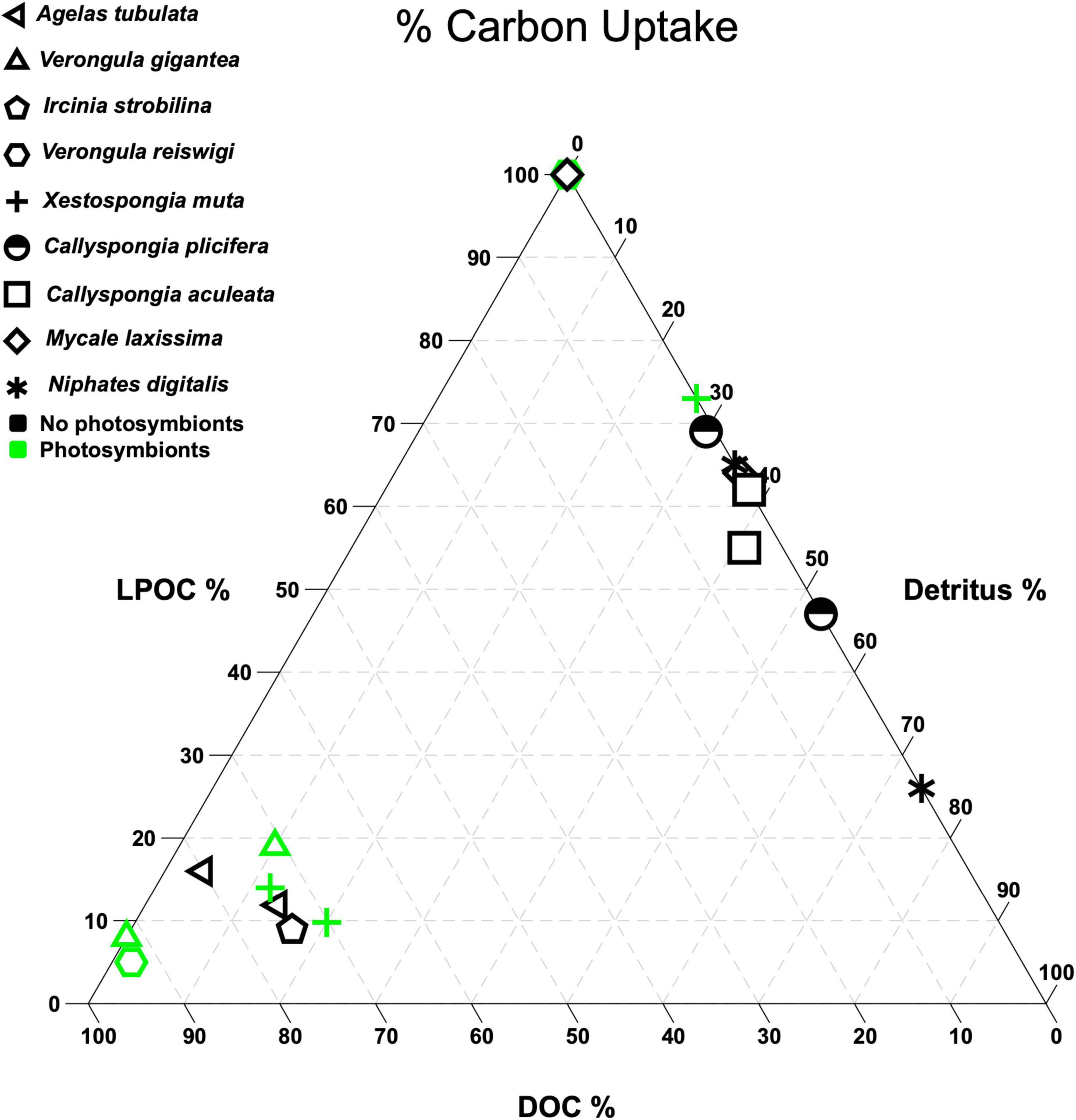
Figure 1. Ternary plot showing the % total carbon uptake (the percent contribution of each food type to the total organic carbon retained by each species). Food types are: dead organic carbon (detritus), living particulate organic carbon (LPOC), and dissolved organic carbon (DOC) (McMurray et al., 2016, 2018). Green symbols denote species or congeners that host communities of photosynthetic symbionts (Erwin and Thacker, 2007; Freeman et al., 2020). Multiple symbols for a species represent variation due to sample collection from different locations or times.
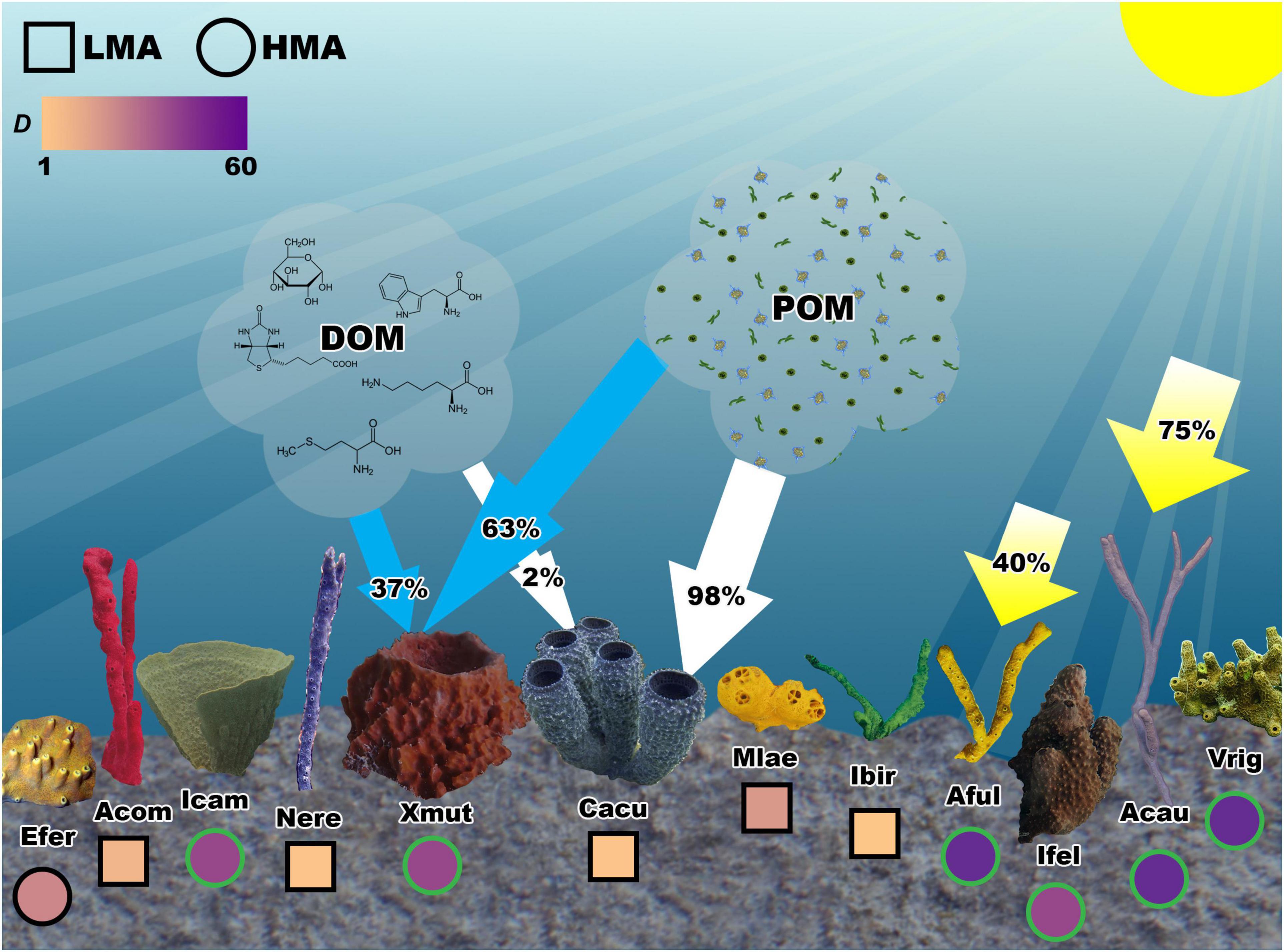
Figure 2. Conceptual model of 12 common sympatric sponge species in the Caribbean Sea. Caribbean sponges possess a diversity of morphologies and microbial symbioses that have helped them adopt novel strategies for obtaining nutrition from a variety of sources including particulate organic matter (POM), dissolved organic matter (DOM), and solar irradiance (yellow arrows). Relative average reliance on DOM and POM carbon sources is denoted with arrows from these clouds to two species Xestospongia muta (Xmut) and Callyspongia aculeata (Cacu) derived from McMurray et al. (2018). Relative reliance on carbon from photosymbionts is derived from Freeman and Thacker (2011). Continuous diversity values for the inverse Simpson’s Index (D) are represented by a color gradient in circles (HMA sponges) and squares (LMA sponges) below each sponge. Sponges that host abundant photosymbionts are denoted by a green outline of the circles or squares representing D.
Photosymbionts and the Expansion of Host Carbon Metabolism
Early work in sponge microbiology reported the presence of cyanobacterial cells (Vacelet and Donadey, 1977; Wilkinson, 1978a,b,c) that were hypothesized to supplement heterotrophic feeding with inputs of fixed carbon. To survey host sponge reliance on cyanobacterial symbionts, Wilkinson (1983) measured gross photosynthesis to respiration (P:R) ratios and 14CO2 fixation (Table 2) across ten of the most dominant sponge species present on the reefs of the Great Barrier Reef (GBR). These ten species varied in the abundance of photosynthetic symbionts in their tissue (measured by chlorophyll a [chl a] analysis), their shape, ratio of surface area to weight, and thickness. Seven of these species had instantaneous P:R values exceeding compensation (P:R of 1.0) at an irradiance of 200 μE m–2 s–1 (equivalent to 200 μmol photons m–2 s–1; future units in this review reported only in μmol photons m–2 s–1) and six of these species had P:R values exceeding 3.0 at an irradiance level of 400 μmol photons m–2 s–1 (the highest irradiance at 20 m depth, where these sponges are abundant). Productive sponge species generally had a thin, encrusting or dish/fan morphology, high surface area, an abundant photosymbiont community, and high rates of carbon fixation (counts of 14C in their tissue; Table 2; Wilkinson, 1983). Species lacking photosymbionts had lower P:R values and were unable to access this source of inorganic carbon.
Early work on the GBR eventually resulted in the delineation of three groups of sponges that vary in their reliance on photosymbiont-derived nutrition: phototrophic: small, thin, flattened species with P:R ratios of more than 1.5 suggesting that photosymbionts within these sponges provide more carbon than is needed to satisfy the respiration requirements of the host; mixotrophic: photosynthetic symbionts are present, but hosts have more diverse growth forms and have P:R < 1.5 suggestive of a reliance on a combination of photosymbiont-derived nutrition and heterotrophic filter feeding to meet their energy demands; and heterotrophic: massive and variable growth forms with no Cyanobacteria and P:R less than compensation point of 1.0 (Wilkinson and Trott, 1985).
Other methods for assessing photosymbiont metabolism (Table 2) have also revealed a continuum of host sponge reliance on photosymbiont-derived carbon. For instance, shading experiments have identified both obligate mutualisms and commensalism as some sponges held under reduced irradiance undergo substantial reductions in growth rate or experience mortality, while others experience a reduction in growth, but are able to maintain positive growth rates (Wilkinson and Vacelet, 1979; Roberts et al., 2006; Erwin and Thacker, 2008; Freeman and Thacker, 2011). In addition, although productivity (via P:R measurements) and photosymbiont-derived benefit to the host (via 13C enrichment of host cells) (Table 2) was minimal in a sponge species that did not host photosymbionts (Niphates erecta in the Caribbean) (Freeman et al., 2013), P:R values and host cell 13C enrichment was significantly elevated in five other species that host photosymbionts at an average concentration of ∼40x that found in N. erecta (Freeman et al., 2013). These five host species varied in their reliance on photosymbiont metabolism, with a potential link to the productivity and community composition of their photosymbionts (Aful and Acau in Figure 2; Erwin and Thacker, 2007, 2008; Thacker et al., 2007; Freeman and Thacker, 2011; Freeman et al., 2013). These recent results underscore that delineating sponge species into discrete categories based on P:R values alone has likely oversimplified the complexities of host reliance on photosymbiont metabolism.
Photosymbionts and Sponge Distribution
Wilkinson (1987) hypothesized that if photosymbionts reduce host sponge requirement for carbon derived from POM, then these symbionts may allow for the expansion of phototrophic sponges into oligotrophic waters where POM is limiting. To test this, extensive surveys were carried out across a transect from inshore to oceanic reefs on the Great Barrier Reef (GBR) (Wilkinson, 1987; Wilkinson and Cheshire, 1990). Sponge biomass was high on inner-shelf reefs (∼17 km from shore) but was up to 12x lower on mid-shelf (70 km from shore), outer-shelf (120 km from shore), and oceanic (220 km from shore) reefs (Wilkinson, 1987; Wilkinson and Cheshire, 1989, 1990). Small phototrophic sponges with flattened, foliose growth forms and high P:R values (Wilkinson, 1983) were absent from inshore reefs where sediment and nutrient loads were elevated (Done, 1982; Wilkinson and Trott, 1985; Wilkinson and Cheshire, 1989). Instead, there was a dominance of heterotrophic (63% of the biomass at 15 m) and mixotrophic species (37% of the biomass at 15 m) on these inner-shelf reefs. Phototrophic sponges represented more of the community by biomass on mid-shelf (40% phototrophic, 50% heterotrophic, and 10% mixotrophic), and at 20 m on outer-shelf (40% phototrophic, 44% heterotrophic, and 16% mixotrophic) and oceanic (68% phototrophic, 26% heterotrophic, and 6% mixotrophic) reefs (Figure 3A; Wilkinson, 1987; Wilkinson and Cheshire, 1990).
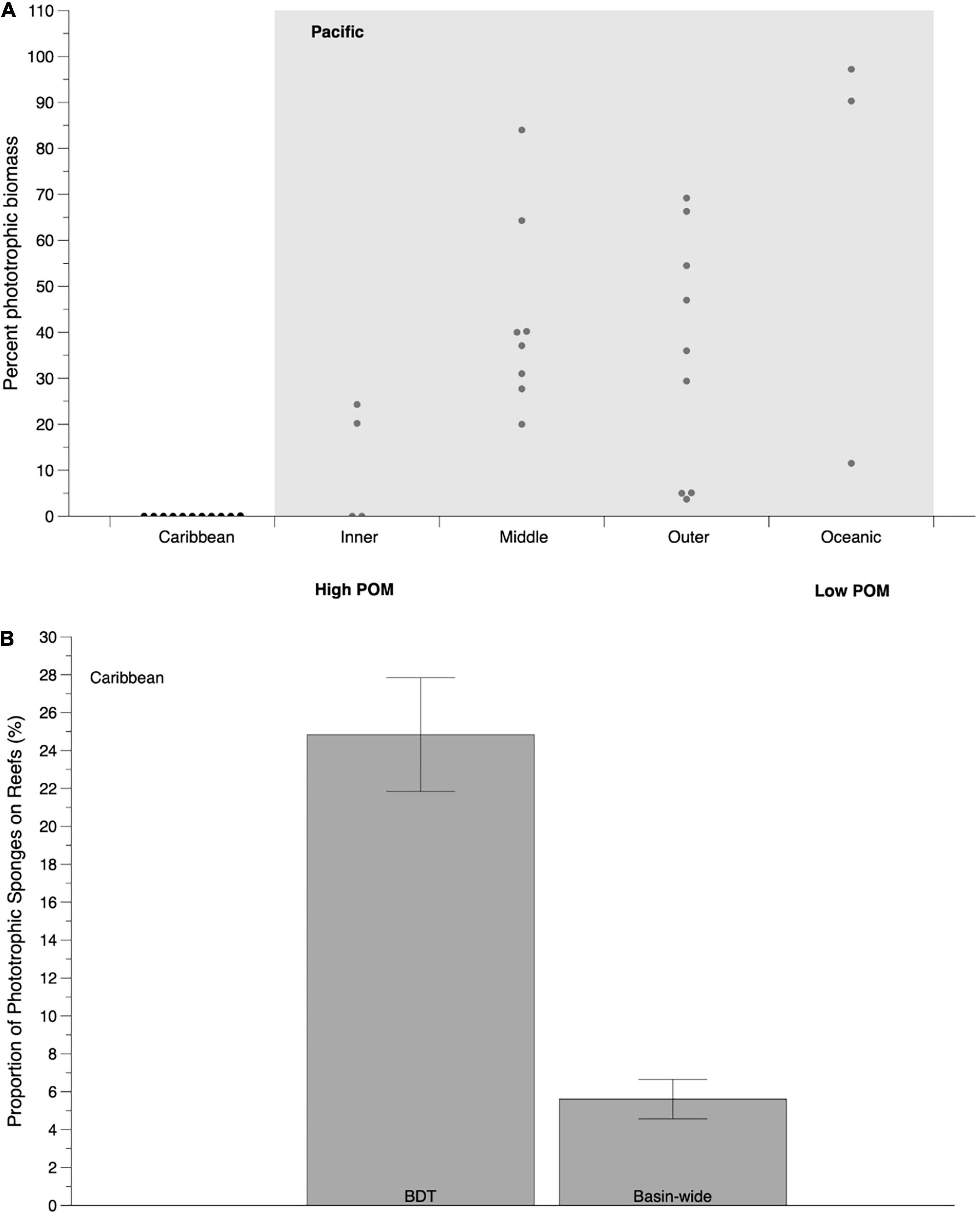
Figure 3. (A) Abundance patterns of phototropic sponges across an inshore to offshore gradient in the Pacific (including Great Barrier Reef and Fiji; light gray box) and Caribbean using data from Wilkinson (1987) and Wilkinson and Cheshire (1990). Abundance is represented as the proportion of overall sponge biomass composed of phototrophic (P:R > 1.5) sponges and each point represents data from a single site within each reef category or ocean basin. Low and high particulate organic matter (POM) predictions are based on Wilkinson and Cheshire (1990). (B) Prevalence of phototrophic sponges (proportion of overall sponge cover from phototrophic species) in the Caribbean (both within Bocas del Toro, Panama [BDT] and Caribbean-wide) based on contemporary methods (Thacker et al., 2007; Erwin and Thacker, 2008; Freeman et al., 2013) and distribution data from Loh and Pawlik (2014) and Easson et al. (2015).
These sponge distribution data highlight an important point that is often missed when referencing these studies. Although phototrophic sponges may generally increase in abundance along this inshore-offshore gradient, there is considerable variation in their abundance across individual sites and also the continued presence of heterotrophic and mixotrophic sponges on mid-, outer-self, and oceanic reefs that were assumed to largely be oligotrophic in terms of the abundance of particulate carbon (Figure 3B; Wilkinson, 1987; Wilkinson and Cheshire, 1989, 1990; Cheshire and Wilkinson, 1991; but see Wilkinson and Cheshire, 1988). This pattern suggests that sponges on the outer reefs of the GBR are either not carbon limited, there is unexplored interspecific variation in filter feeding efficiency within “heterotrophic” sponges, or there is reliance on microbial symbionts for other sources of carbon on these reefs (e.g., DOC); these hypotheses remain untested.
Similar cross-shelf surveys were carried out in the Caribbean (47 km wide survey in Belize compared to 220 km wide on the GBR) and 20 Caribbean sponge species were screened for photosynthetic activity by measuring oxygen consumption under dark and light conditions at a variety of irradiances (Wilkinson, 1987; Wilkinson and Cheshire, 1990; Table 2). As on the GBR, there was more sponge biomass on the inner-shelf reefs, and there was a higher incidence of species with photosynthetic symbionts from inner to outer reefs in Belize (31, 49, and 76% of the total sponge biomass on reefs from inner-shelf to outer-shelf and oceanic reefs) (Wilkinson and Cheshire, 1989, 1990). Unlike the GBR, however, sponge biomass in Belize and other sites across the Caribbean was between 2 and 12x greater than on comparable reefs on the GBR (Wilkinson and Cheshire, 1990). In addition, there was a lack of the thin, flattened foliose growth forms found on the GBR (Caribbean sponges adopt more massive, rope, or encrusting morphologies) and almost all (except for phototrophic boring sponges that host dinoflagellate symbionts) of the Caribbean sponges surveyed were considered to be heterotrophic (Figure 3A), with no “significant increase” above dark respiration rate when exposed to the highest irradiance level of 600 μmol photons m–2 s–1 (Wilkinson and Cheshire, 1990). These authors did not measure productivity in the GBR or Caribbean (see Figure 1 map in Koblentz-Mishke et al., 1970), but came to the general conclusion that phototrophic sponges were common in oligotrophic systems and absent from more productive reefs in the Caribbean (Wilkinson, 1987; Wilkinson and Cheshire, 1988; Wilkinson and Cheshire, 1990; Pawlik et al., 2015; Figure 3A).
These early papers have formed the basis for our understanding of sponge nutritional patterns at the level of entire ocean basins, but there are limitations to this work that should be discussed. First, the irradiance levels of 600 μmol photons m–2 s–1 used in Wilkinson and Cheshire (1990) on sponges in the Caribbean are not ecologically relevant. Caribbean sponges are regularly exposed to irradiances up to 1000 μmol photons m–2 s–1 even at a depth of 10 m (Vermeij and Bak, 2002). Thus, photosynthesis-irradiance (P-I) curves of Caribbean sponges do show P:R values above 1.5 when exposed to these higher irradiances and four “phototrophic” (based on P:R > 1.5) sponge species have now been identified in the Caribbean (Xestospongia bocatorensis, Haliclona walentinae, Aplysina fulva, and Neopetrosia subtriangularis; Thacker et al., 2007; Erwin and Thacker, 2008; Freeman et al., 2013). These species represent 25% of the sponge community on reefs in the Bocas del Toro (BDT) archipelago of Panama (a region highly influenced by allochthonous inputs from land; Aronson et al., 2014; Easson et al., 2015) and 5.6% of the entire sponge community in this ocean basin (Figure 3B; Loh and Pawlik, 2014). In addition, with over 30% of dominant Caribbean sponges hosting abundant photosymbiont communities (Wilkinson, 1987; Rützler, 1990; Erwin and Thacker, 2007, 2008; Thacker et al., 2007; Bell et al., 2020) and evidence that many of these symbionts fix inorganic sources of carbon and translocate fixed carbon to their host (Freeman et al., 2013), it is likely that more Caribbean sponges are heavily reliant on their photosymbionts. Finally, as the GBR and Caribbean Sea are now recognized to have comparable levels of POM, DOM, and inorganic nutrients (de Goeij et al., 2017), interocean differences in sponge biomass, morphologies, and reliance on photosymbionts are likely shaped by factors beyond just perceived nutrient limitation.
Although our understanding of sponge carbon metabolism has become more multidimensional in recent years (Freeman et al., 2013; Gantt et al., 2019; Rix et al., 2020), no studies to date have adopted a comprehensive analysis of the contribution of fixed carbon (Wilkinson, 1983; Freeman et al., 2013), and diverse pools of dissolved and particulate organic matter and detritus (Figures 1–3; Reiswig, 1981; Morganti et al., 2017; McMurray et al., 2018; Rix et al., 2020) to sponge holobiont carbon budgets. Without these data and standardized analyses across ocean basins, it is impossible to draw conclusions as to whether “heterotrophic” sponges are truly nutrient-limited or whether these species can meet their energy demands and expand into oligotrophic systems by assimilating DOC or hosting photosynthetic symbionts (Wilkinson et al., 1988; Wilkinson and Cheshire, 1990; Rix et al., 2020). For example, when considering only heterotrophic carbon sources, the sponge Xestospongia testudinaria appears to be nutrient limited on offshore sites of the Red Sea where DOC levels are low, but X. testudinaria and many other sponge species that assimilate DOC also host photosymbionts that likely contribute to the sponge carbon budget and may prevent nutrient limitation (Figure 1; McMurray et al., 2018; Wooster et al., 2019).
Specific Knowledge Gaps in Sponge Phototrophy and Carbon-Focused Nutrition in Sponges
Although studies on nutrient limitation in sponges have classically focused largely on carbon metabolism, it is increasingly apparent that there are diverse sources of carbon (dissolved, particulate, organic, and inorganic) that can be exploited by sponges on coral reefs (Rix et al., 2020). More holistic assessments of sponge carbon budgets are therefore needed along with a better understanding of how sponges supplement heterotrophic feeding with other metabolites from their symbionts or even phagotrophy of symbionts (Fiore et al., 2015a; Leys et al., 2017). This work will also improve our ability to elucidate potential fitness trade-offs associated with host reliance on photosynthetic and heterotrophic symbionts. In addition, the major heterotrophic resources described above can be further subdivided, so individual sponge species may exploit unique portions of these pools (Maldonado et al., 2012). Lastly, while photosymbionts and, in particular, Cyanobacteria have received considerable attention, there are many other symbiont groups that contribute to sponge metabolism and influence the exploitation and cycling of other elements. These points underscore the likely scenario that multiple selective pressures have shaped the evolution of sponge-microbe interactions and sponge species have diverged across multiple niche dimensions due to interactions with their microbial symbionts.
Sponge–Microbe Interactions and Host Expansion Into New Niche Dimensions
Coral reefs are generally considered to lack abundant organic matter needed to fuel heterotrophic metabolism, but these are complex and dynamic ecosystems with multidimensional nutritional niches. Hosting microbial symbionts thus greatly expands the ability of sponges to exploit additional resource pools and elements beyond just carbon. In this section, we highlight the complexities and nuances of both sponge microbial symbiont communities and resource pools on coral reefs.
Diversity in Sponge Microbial Communities
Sponges host a richness and diversity of microorganisms that is unparalleled in other marine organisms, with 52 phyla and candidate phyla across Bacteria, Archaea, and Eukarya (Thomas et al., 2016; Freeman et al., 2020). A few dominant groups within sponges include Gammaproteobacteria (relative abundance mean ± standard deviation; 31% ± 19%), Alphaproteobacteria (11% ± 17%), Chloroflexi (9% ± 11%), Thaumarchaeota (5% ± 7%), and Cyanobacteria (4% ± 7%; data from: Thomas et al., 2016; Table 4). Within each of these phyla are numerous individual taxa (often defined as Operational Taxonomic Units [OTUs] or more recently Amplicon Sequence Variants [ASVs]) that often form distinct clades (based on 16S rRNA gene sequencing) or “sponge-specific clusters” only found in sponges (Hentschel et al., 2006) or in low abundance within the environment (Taylor et al., 2013). Although sponges vary in the absolute abundance of their symbionts, species within both the HMA and LMA groups can differ substantially in the diversity, richness, and composition of their microbial communities and there is increasing evidence of high host specificity across individual species (Figure 2; Easson and Thacker, 2014; Thomas et al., 2016).
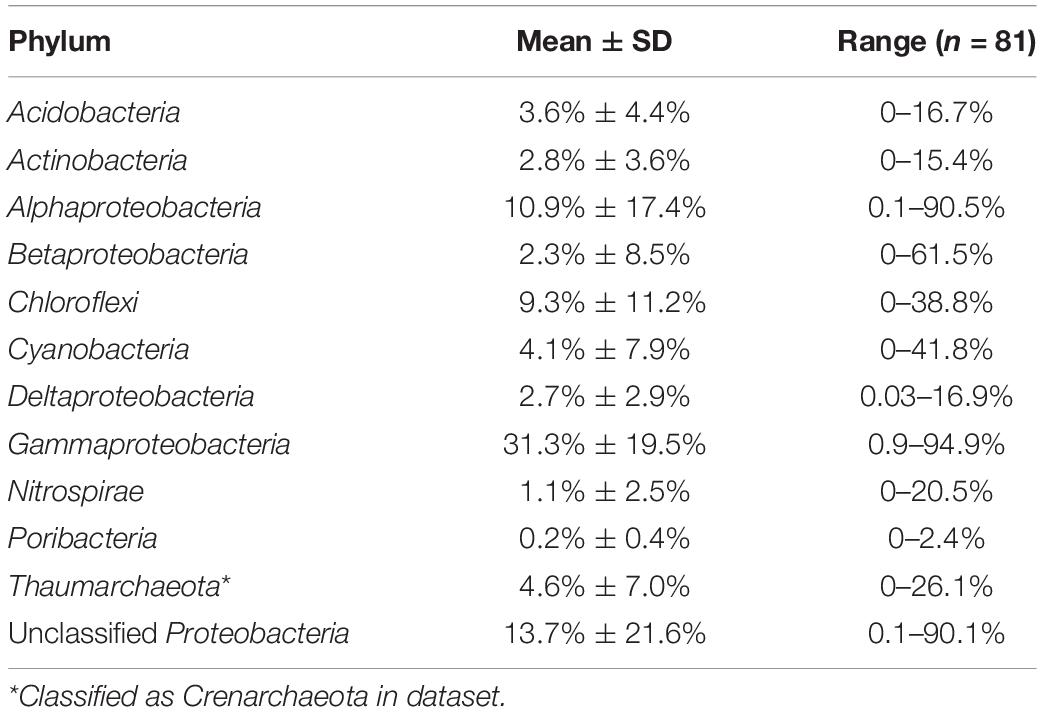
Table 4. Top Phyla present in sponges (data source: Thomas et al., 2016; 81 sponge species). Proteobacteria is separated into classes and unclassified (only classified to phylum).
Diversity of Resources and Symbiont Metabolism
Dissolved organic matter is largely comprised of refractory material derived from abiotic reactions and potentially some microbial activity (Ogawa et al., 2001; but see Kujawinski et al., 2016), while a small portion of the pool consists of labile biomolecules (i.e., metabolites) (<1% of total DOC, Nagata, 2000). The labile fraction is considered to be largely derived from phytoplankton exudates and cell lysates, and with a short turnover time (<hours to days, Cherrier et al., 1999; Figure 2), is critical to microbial metabolism and biological production (Nagata, 2000). The composition of this labile DOC is not fully characterized, but contains dissolved free amino acids (DFAA), vitamins, nucleosides/nucleotides, sugars, among other structurally diverse compounds (e.g., Suttle et al., 1991; Amon and Benner, 1996; Fiore et al., 2015b; Kujawinski et al., 2016). Sponges remove specific components of the labile pool of DOC (i.e., glycerol-3 phosphate, pantothenic acid, 5′-methylthioadenosine; Fiore et al., 2017) and release DOM in the exhalent seawater leaving their bodies (Fiore et al., 2017; Letourneau et al., 2020). In general, sponge microbes are well suited to assimilate and/or respire diverse labile metabolites (e.g., amino acids) and might even consume semi-labile to recalcitrant compounds (i.e., cellulose; Pawlik et al., 2016). For example, genes and transcripts involved in methylotrophy and other C1 metabolism were observed in several high throughput-sequencing based studies of sponge symbionts (Thomas et al., 2010; Fan et al., 2012; Radax et al., 2012; Li et al., 2014; Moitinho-Silva et al., 2014; Fiore et al., 2015a). Additionally, recent metagenomic analysis of six sponge microbiomes of the class Demospongiae supports the presence of functional guilds that target DOM like sialic acids derived from sponge tissue and carbohydrates derived from coral and algal DOM (Robbins et al., 2021). Symbiont-specific examples include the ability to consume diverse carbon sources including chitin, cellulose, and/or N-acetylglucosamine by Entotheonella (Liu et al., 2016) and Poribacteria (Siegl et al., 2011), and steroids by Actinobacteria, Alphaproteobacteria, and Gammaproteobacteria in sponges (Holert et al., 2018). Lastly, the ability to utilize carnitine and sulfated polysaccharides were among the noted metabolic traits of sponge symbiont genome analysis for the sponge Aplysina aerophoba (Slaby et al., 2017). These examples are not an exhaustive list of microbial symbiont DOM metabolism but demonstrate the diversity of metabolic substrates available for sponge hosts and their symbionts.
Although many sponge species rely heavily on dissolved organic matter (McMurray et al., 2018; Rix et al., 2020), the C:N ratio of DOM can vary depending on how labile or refractory it is (de Kluijver et al., 2021) and can be greater than 10 on oligotrophic reefs (Lesser et al., 2019). Thus, sponge species relying on DOM may need to supplement their diet with POM (Hadas et al., 2009) or form interactions with microbial symbionts that provide access to novel sources of nitrogen in order to prevent nutrient limitation (Southwell et al., 2008; Freeman and Thacker, 2011; de Goeij et al., 2017; Morganti et al., 2017; Pita et al., 2018; Paul et al., 2019; Zhang et al., 2019). The ability to fix nitrogen (N2) or assimilate exogenous nitrate (NO3) or ammonia (NH3) and recycle host-derived waste NH3 could therefore provide an adaptive advantage to some host species (Southwell et al., 2008; Liu et al., 2012; Zhang et al., 2014; Rädecker et al., 2015). Although nitrogen cycling is likely a core function of these communities (e.g., Fan et al., 2012), sponge species vary in their ability to utilize different nitrogen resource pools (PON/DON; e.g., Morganti et al., 2017) or carry out microbial transformations like nitrification, nitrogen fixation, nitrate assimilation, and transfer of organic nitrogen to the host (e.g., Mohamed et al., 2008; Southwell et al., 2008; Hoffmann et al., 2009; Freeman et al., 2013; Webster and Thomas, 2016; Steinert et al., 2020). Thus, nitrogen metabolism may be a critical component of niche partitioning among sponge species (e.g., Morganti et al., 2017; Freeman et al., 2020).
Sulfur metabolism, whether through oxidation and reduction of sulfur compounds for energy and the subsequent fixation of carbon or through assimilation of organic sulfur compounds (e.g., APS, DMSP), appears to be prevalent in sponge–microbe systems (Hoffmann et al., 2005a,b; Fiore et al., 2015a; Zhang D. et al., 2015; Li et al., 2016; Jensen et al., 2017; Lavy et al., 2018; Podell et al., 2019; Zhang et al., 2019; Engelberts et al., 2020). Sulfur-based symbioses have been documented in other invertebrates (e.g., Cavanaugh et al., 1981; Southward et al., 1981; Fisher and Childress, 1984; Dubilier et al., 2001) and have potential to be a common trait in marine invertebrate systems (Fiore et al., 2020). Work characterizing phosphorus cycling in sponges is at a nascent stage (see Zhang et al., 2019), but multiple studies have identified phosphorus cycling and metabolism of organic phosphorus-containing compounds as an important component in sponge–microbe systems (Zhang F. et al., 2015; Podell et al., 2019; Engelberts et al., 2020; Fiore et al., 2020) and further work is needed on this topic.
Microbial symbionts may also facilitate survival on coral reefs by supplying organic compounds like amino acids and vitamins that host sponges cannot synthesize or obtain from their diet. This form of metabolic interaction has been demonstrated in other systems (e.g., insects, Bennett and Moran, 2013; Salem et al., 2014; vertebrates, review by Neish, 2009; Degnan et al., 2014), and the potential for transfer of amino acids (e.g., lysine), vitamins (e.g., riboflavin, biotin), and other metabolites has been suggested by studies based on ‘omics’ analysis of sponge holobionts or sponge symbiont genomes (Hallam et al., 2006; Thomas et al., 2010; Liu et al., 2011, 2012; Siegl et al., 2011; Fan et al., 2012; Radax et al., 2012; Kamke et al., 2014; Fiore et al., 2015a; Moitinho-Silva et al., 2017; Bayer et al., 2020; Engelberts et al., 2020). This has been highlighted in recent reviews on the sponge holobiont (Taylor et al., 2007; Webster and Taylor, 2012; Webster and Thomas, 2016), but no studies that we are aware of have demonstrated transfer of specific nutrients to the host experimentally. Targeted experimental work is needed to uncover complex metabolic interactions between hosts and symbionts and how this varies across host species; this represents a significant gap in our understanding of the role of microbial symbionts in the survival of their hosts.
Specific Knowledge Gaps in Sponge Symbiont Diversity and Metabolism
Here, we have highlighted the taxonomic and functional diversity of microbial symbionts within sponges, but there is a need for additional work to understand the complex nutritional relationships between microbial symbionts within a sponge host. This includes the evolutionary origins of these microbe-microbe interactions and how they vary among diverse sponge species and over space and time. This will help to elucidate the ecological forces structuring microbial community composition and function across different species. Additionally, more work is needed to resolve microbially mediated benefits to the host sponge. For instance, DOM consumption by microbial symbionts could benefit the host in three ways: (1) excretion of essential metabolites for the host (e.g., vitamins, amino acids), (2) nutritional support for microbes that consume host waste (e.g., ammonium), and (3) nutritional support for microbes that are consumed by the host (i.e., ‘farming’ symbionts; Ilan and Abelson, 1995; Vacelet and Boury-Esnault, 1995; Hudspith et al., 2021). Lastly, while we have a basic understanding of overall biogeochemical cycling in sponges, we are still limited in our understanding of nitrogen, sulfur, and phosphorus metabolism by sponge symbionts, the potential connections between cycles of different elements (but see de Kluijver et al., 2021), and the potential intersections of these microbes and cycles with host metabolism and ecology.
Ecological Variation Among Host Species
As emergent techniques better resolve the nuances of sponge-microbe associations and sponge holobiont metabolism, it is increasingly clear that individual sponge species have unique solutions to the ecological challenges on coral reefs (e.g., obtaining food). Sponge species vary in the abundance, diversity, and community composition of their microbial communities (Figure 2; Easson and Thacker, 2014; Thomas et al., 2016; Gantt et al., 2019; Freeman et al., 2020) and host physiology [tissue density, pumping rates, and width of choanocyte chambers (Weisz et al., 2008; Poppell et al., 2013; Morganti et al., 2021)], exploit nutrient pools that are unique from sympatric sponge species (Freeman et al., 2020), converge on some specific metabolic pathways (Fan et al., 2012; Ribes et al., 2012; McMurray et al., 2018), and have varying levels of plasticity in both their microbial community structure and holobiont metabolism (McMurray et al., 2018). In this section, we highlight this variation and provide evidence that sponge species are filling unique functional or nutritional “niches” on coral reefs.
Host Specificity in Microbial Communities and Holobiont Metabolism
Host specificity in microbial community composition is increasingly reported in sponges across a variety of spatial (Anderson et al., 2010; Erwin et al., 2011, 2012a; Schöttner et al., 2013; Thomas et al., 2016; Dat et al., 2018; Easson et al., 2020; Freeman et al., 2020) and temporal (Erwin et al., 2012b; Cárdenas et al., 2019) scales. In fact, several recent studies have observed that up to 80% of the variation in microbial community structure is shaped by the identity of the host sponge (Easson and Thacker, 2014; Thomas et al., 2016; Freeman et al., 2020). In contrast, only about 19% of this variation can be attributed to the overall microbial abundance (HMA or LMA) within a host species (Figure 2; Freeman et al., 2020). With host specificity in microbial community composition and a strong phylogenetic signal between microbiome diversity and host phylogeny at local, regional, and global scales (Easson and Thacker, 2014; Thomas et al., 2016; Gantt et al., 2019; Freeman et al., 2020), this observed variation among host species might have important implications for sponge ecology and evolution. However, host specificity and microbiome diversity could also be driven by unique host characteristics that influence microbial habitats and differ among host species; therefore, experimental work is needed to better characterize the mechanisms by which microbiomes influence host sponge ecology and evolution. The fact that intraspecific variation in host sponge population genetics is linked to microbial community structure (Swierts et al., 2018; Griffiths et al., 2019; Easson et al., 2020) is also intriguing and implies a strong influence of the host sponge on its microbial community (see below).
Early work using stable isotopes (Table 3) by Weisz et al. (2007, 2008) demonstrated that species vary in their δ15N and δ13C values, with HMA species having depleted (lower) δ15N values than LMA species. This divergence in nitrogen metabolism was ascribed to microbial transformations of nitrogen in the HMA species and comparatively higher levels of heterotrophic feeding on organic matter in the water column in the LMA sponges (Weisz et al., 2008). In addition, evidence of variation in physiological traits (e.g., pumping rates and tissue density) among HMA and LMA species suggested that total microbial abundance might drive niche divergence across these sponge species (Weisz et al., 2007, 2008). Freeman and Thacker (2011) analyzed the stable isotope ratios of isolated sponge and microbial fractions and found that three HMA species each had a unique interaction with their microbial symbionts. For instance, transfer of nitrogen from symbionts to the host was documented in Aplysina fulva and A. cauliformis but not in N. subtriangularis or the LMA sponge Niphates erecta (Freeman and Thacker, 2011). Furthermore, experiments using inorganic compounds enriched in 13C and 15N found that photosymbiont carbon and nitrogen metabolism was highly variable across different HMA species (Freeman et al., 2013) and interspecific differences in host reliance on photosymbiont metabolism was also supported by shading experiments (Erwin and Thacker, 2008; Freeman and Thacker, 2011; Freeman et al., 2015).
More recently, Morganti et al. (2017) demonstrated trophic niche separation through a combination of variation in heterotrophic efficiency (bacterial retention) and uptake of DON/DOC across five sympatric Mediterranean species. Stable isotope surveys of 15 of the most common sponge species in the Caribbean have also indicated strong host specificity in broad scale trends of holobiont carbon and nitrogen metabolism, with 70–90% of the variation in the δ15N and δ13C values of sponge tissue driven by host identity (Freeman et al., 2014, 2020). Interestingly, although δ15N values of sponge tissue displayed a strong host-phylogenetic signal that is conserved across the Caribbean (Freeman et al., 2020), these patterns were not mirrored in δ13C values. This, along with Gantt et al. (2019) reporting that microbiome structure was uncoupled from C (POC/DOC fluxes) cycling and PO4 and NOx fluxes but was correlated with NH4 flux, suggests that it may benefit sponges to be flexible in carbon metabolism, but more constrained in their exploitation of nitrogen (see below for more discussion of this). Together, recent studies suggest that fine-scale trends in resource use are shaped at least in part by microbial community composition, but more work is needed to understand the functional importance of microbiome specificity in these interactions, the potential tradeoffs between host physiology and symbiont metabolism, and the role of both symbionts and the host in this divergence.
Convergence on Common Metabolic Pathways
Some metabolic pathways may be so critical to survival on coral reefs that different sponge species have evolved unique mechanisms to gain access to a specific resource or pathway. For instance, although two HMA species hosted unique communities of microbial symbionts, both species had high levels of nitrification, as well as DOC and NH4 uptake (Ribes et al., 2012). In addition, Fan et al. (2012) reported striking functional convergence in the microbiomes of six sponge species, implying the presence of core functions (e.g., nitrogen metabolism) of microbial symbionts across different sponge species. Finally, although HMA sponges are generally considered to assimilate DOM more efficiently than LMA sponges (McMurray et al., 2018) due to assimilation by their microbial symbionts (Rix et al., 2020), recent work has found that LMA species in the Red Sea (Dysidea avara) and the Indo Pacific (Cliona orientalis) are also able to exploit DOM directly via pinocytosis in specialized cells (choanocytes) within their aquiferous systems (Achlatis et al., 2019; Rix et al., 2020). Thus, sponges with different physiologies and interactions with microbial symbionts have unique adaptations for the exploitation of DOM.
Plasticity in Metabolism and Microbiomes
Flexibility in physiological traits that mediate resource use can be adaptive for organisms on coral reefs if it allows for the exploitation of temporally or spatially variable resource pools. Plasticity in pumping rates, the filtration rates of POC and DOC, and fluxes of dissolved nutrients (e.g., Carballo et al., 2006; Morley et al., 2016; Morganti et al., 2017; Wooster et al., 2019) have been demonstrated in some sponge species. In addition, intraspecific plasticity in sponge symbiotic microbial communities has been found (e.g., Burgsdorf et al., 2014; Sacristán-Soriano et al., 2020; and see review by Kiran et al., 2018) and may allow sponges to rapidly adapt to environmental conditions at a new site or over time. To expand on this, we highlight four examples of phenotypic and microbiome plasticity: (1) multiple studies on Xestospongia muta, (2) Ircinia felix, (3) Halisarca caerulea, and (4) a Caribbean-wide profile of 11 sponge species.
Arguably, one of the most well-studied sponges in the Caribbean is the giant barrel sponge, X. muta. Xestospongia muta harbors a diverse microbial community, including a high proportion of Cyanobacteria (Steindler et al., 2005; Montalvo and Hill, 2011; Fiore et al., 2013b). The symbiotic community of X. muta and nutrient fluxes from X. muta varied across multiple locations within the Caribbean (Fiore et al., 2013a,b; McMurray et al., 2016). Specifically, X. muta in the Florida Keys were a net sink for NOx, while those sampled in the Bahamas and Little Cayman Island were a net source of NOx (Fiore et al., 2013b). Relatively high individual variability was observed at each of these locations (Fiore et al., 2013b) and similar observations were made about X. muta pumping activity within one location (McMurray et al., 2014). Similarly, fluxes of DOC were net negative from X. muta in the Florida Keys and net positive from conspecifics at Carrie Bow Cay in Belize (McMurray et al., 2018). The ambient concentration of DOC was also higher in the Florida Keys compared to Belize (McMurray et al., 2018) and other studies have observed similar patterns between nutrient concentration and nutrient uptake by sponges (Morganti et al., 2017; Wooster et al., 2019). In some of the initial work on X. muta that included both nutrient fluxes and microbial community analysis, it was difficult to relate specific environmental variables, physiological traits, and the microbiome community composition to each other (e.g., Fiore et al., 2013a,b); however, later work uncovered several correlative relationships. For example, ammonium concentrations in the ambient seawater were higher in the Florida Keys than in the Bahamas and archaeal ammonia oxidation gene expression was higher in the X. muta samples from the Florida Keys than in the Bahamas (Fiore et al., 2015a). Additionally, while the microbiome of X. muta showed a consistent ‘core’ microbial community profile with depth, there was a shift in the relative proportion of several taxa over depth at two locations, notably Cyanobacteria and Chloroflexi (Olson and Gao, 2013; Morrow et al., 2016). Abiotic factors, including light and temperature, were likely driving the relative decrease and increase in Cyanobacteria and Chloroflexi, respectively, with depth at Little Cayman Island (Morrow et al., 2016). In contrast, inorganic nutrients were higher in the ambient seawater and there was a less observable difference in the symbiont community of X. muta over depth in the Bahamas (Morrow et al., 2016). These studies point to an inherent flexibility that X. muta maintains in its symbiotic community composition and function as well as in physiological traits when presented with different environmental variables.
Shifts in microbial composition and function were also demonstrated for the sponge I. felix at three locations in the Caribbean (Archer et al., 2017). Fluxes of DOC and inorganic nitrogen and phosphate generally correlated with the concentrations in ambient seawater, where sponges acted as a sink with higher ambient concentrations (Archer et al., 2017). Additionally, predicted functional genes of the symbiotic community differed by location; for genes associated with nitrogen metabolism, increases in predicted gene abundance correlated with higher ambient seawater concentrations of DIN. Similar to the work by Morrow et al. (2016) with X. muta, Archer et al. (2017) observed that abiotic factors were significantly correlated with changes in nutrient fluxes from I. felix and illustrate the context-dependent manner in which the functional profile of the microbial community and host-microbe interactions can change.
The encrusting sponge, H. caerulea, also exhibited plasticity in physiological traits and microbiome community composition (Lesser et al., 2019). Lesser et al. (2019) transplanted sponges between shallow (∼10 m) and deep (∼50 m) sites and observed correlative changes in detritus production, nutrient fluxes, nitrogen stable isotope signal, and microbiome composition in the transplanted sponges. Natural abundance stable isotope analysis revealed nutritional differences between shallow and deep sponges. This result, combined with shifts in detritus production, microbiome composition, and ambient POM and DOM concentrations over the depth profile indicate a plastic response by the sponge and microbial community to efficiently utilize available nutrients (Lesser et al., 2019). More work is needed, however, to assess how host reliance on symbionts and external sources of nutrients varies across these depth gradients.
Finally, to further visualize plasticity in sponge metabolism across different species, we leveraged data from 11 sponge species in Freeman et al. (2020) for which both stable isotope (δ13C and δ15N) and microbiomes were measured (Supplementary Figure 1) to provide proxies for broad-scale trends in metabolism and microbial community composition. Sponges were sampled from two reefs in Panama and one in the Florida Keys, United States that vary in their environmental conditions (Easson et al., 2015; Freeman et al., 2020). Variation in δ13C and δ15N values and microbiome community composition were calculated at two spatial scales (within site and Caribbean-wide) and visualized as a comparison of the mean distance to a species centroid (MDC) at individual sites compared to MDC across the Caribbean (Supplementary Figure 1).
The MDC for all three metrics was greater at the level of the Caribbean than within a site for all species, but the magnitude of this variation at different spatial scales varied across individual species (Supplementary Figure 1). For example, at the level of the Caribbean the species A. cauliformis showed low variability in δ13C, but moderate to high variability in δ15N and microbiome composition compared to co-occurring species. Aplysina cauliformis receives approximately 75% of its carbon from its photosynthetic symbionts (Freeman and Thacker, 2011), so a tightly conserved relationship with symbionts may buffer against variation in δ13C across large spatial scales. In contrast, Iotrochota birotulata exhibits low variability in δ15N and is dominated by a single symbiont group across all sites; this symbiont taxon may play an important role in nitrogen exploitation or cycling in this species. Although there are other interesting patterns in these figures, in short, these results clearly demonstrate that sympatric sponge species each have unique responses to the same environmental gradient.
Specific Knowledge Gaps in Ecological Variation Across Host Species
As evidence of intraspecific genetic variation of host sponges increases, it is unclear whether the plasticity or divergence documented above is actually caused by cryptic speciation. For instance, although three species of barrel sponges within the genus Xestospongia are recognized across the globe, there is new evidence that there are actually 17 different genotypes (Swierts et al., 2017). On a smaller scale, Deignan et al. (2018) found two unique genetic clusters of X. muta on Conch Reef in the Florida Keys. Thus, the ecological plasticity in X. muta outlined above may actually reflect ongoing ecological speciation. As a preliminary test of theories of niche differentiation under sympatric speciation, Kelly et al. (2021) examined the microbiomes associated with Panamanian species of Ircinia. Four newly described species of Ircinia were clearly differentiated from two known species using SNPs obtained from a 2bRAD protocol (Wang et al., 2012) and all six species displayed significant divergence in microbiome community structure, with an average of 38% within group Bray-Curtis dissimilarity (BCD) and an average of 46% between group BCD. Additional preliminary evidence from metagenomic sequencing of these Ircinia spp. suggests that each of these species hosts distinct microbial pathways for carbon fixation, nitrogen metabolism, and phosphorus metabolism, supporting predictions from theories of niche differentiation that may drive cryptic speciation. Finally, microbial community composition varied across unique populations of the sponges Cliona delitrix and Ircinia campana in the Caribbean (Chaves-Fonnegra et al., 2015; Griffiths et al., 2019; Easson et al., 2020), suggesting that divergence in microbial communities may occur as hosts are undergoing speciation over large spatial scales and adapting to new environmental conditions (Griffiths et al., 2019; Easson et al., 2020). More work is needed to test for cryptic speciation in other sponge species and to broaden our understanding of the selective pressures driving this speciation.
New Insights and Ecological and Evolutionary Implications
Numerous selective pressures shape trait evolution in organisms on coral reefs and different sponge species appear to have unique evolutionary solutions to these ecological challenges. In this section, we will place some of the patterns we have highlighted above into a broader evolutionary framework and provide novel insights into the ecological importance of microbial symbionts to sponge hosts.
Photosymbionts and Competition for Space With Corals
Although pioneering work by Wilkinson (1987) and Wilkinson and Cheshire (1990) attributed patterns of sponge biomass and nutrition to inshore-offshore resource gradients, other factors may certainly contribute to these patterns. For instance, competitive interactions between sponges and anthozoans are common (Bell et al., 2020) and may be influencing sponge distribution and evolution. On the GBR, sponges are common on inner sites where corals are rare (Done, 1982; Wilkinson and Cheshire, 1988), but sponges are a minor component (6.8% of bottom cover) of the benthic community on offshore sites compared to hard and soft corals (31 and 12.4% cover, respectively) (Reichelt et al., 1986; Wismer et al., 2009). In addition, there is an inverse relationship between coral and sponge cover within sites as the percent cover of sponges increases from 0.7 to 6.8% down the fore-reef slope to 20 m while the opposite is the case for hard corals (decreasing from 58.5 to 31.1% at 20 m, Wilkinson, 1981; Reichelt et al., 1986). On central and outer-shelf sites of the GBR, this shift with depth generally reflected an increase in the abundance of thin, foliose Dictyoceratid sponges that are predominantly phototrophic (Wilkinson, 1981; Wilkinson and Trott, 1985) down to 30 m (their 24-h compensation point) (Cheshire and Wilkinson, 1991). With irradiance levels on the GBR that can approach 600 μmol photons m–2 s–1, 430 μmol photons m–2 s–1, and 300 μmol photons m–2 s–1 at noon at 10, 20, and 18 m depths, respectively (Wilkinson, 1981; Cheshire et al., 1997), these obligate phototrophs must be well adapted for efficient photosynthesis at low irradiances (Cheshire and Wilkinson, 1991). Indeed, the irradiance at which there is photosynthetic compensation (where PGmax [gross oxygen production] equals R [respiration rate]) varies from 235 to 335 μmol photons m–2 s–1 and the irradiance at which 95% photosynthetic saturation occurs (the photokinetic parameter Ik) varied from 85 to 423 μmol photons m–2 s–1 depending on the sponge species and location on the GBR (Wilkinson, 1987; Cheshire and Wilkinson, 1991; Cheshire et al., 1997; Bannister et al., 2011). This variability, along with evidence that high hard coral cover on these reefs leaves only small (0 to <1 cm) substrate gaps between corals or between corals and sponges implies that competition between sponges and productive calcifying organisms is high and may be shaping sponge distribution and nutrition on the GBR (Wilkinson, 1981; Done, 1982; Reichelt et al., 1986).
Interocean variation in sponge biomass, morphology, and nutrition may be shaped by the fact that sponges in the Caribbean and GBR have evolved under unique selective pressures due to lower coral cover in the Caribbean than on the mid-shelf and outer reefs on the GBR (Done, 1982; Roff and Mumby, 2012). Although less is known about the photophysiology of Caribbean sponges, phototrophic sponges in this ocean do not reach photosynthetic saturation at low irradiances, are frequently found in shallow water (less than 15 m), and adopt a variety of growth forms (predominantly rope and encrusting and not the thin, foliose growth form of phototrophic sponges on the GBR) (Thacker et al., 2007; Erwin and Thacker, 2008; Pawlik et al., 2015). Interestingly, some of the most abundant phototrophic sponges on offshore and oceanic portions of the GBR (Phyllospongia lamellosa [now Phyllospongia foliascens] and Dysidea herbacea [now Lamellodysidea herbacea]) host the filamentous cyanobacteria Hormoscilla spongeliae, but this symbiont is comparatively rare across common sponges in the Caribbean. H. spongeliae has, however, been found in phototrophic sponges within the Caribbean (Diaz et al., 2007; Erwin and Thacker, 2007; Thacker et al., 2007). Thus, reduced niche space and high competition with corals may have selected for obligate interactions with photosymbionts in some sponge species on outer reefs of the GBR. In contrast, although Caribbean sponges obtain a nutritional benefit from their photosymbionts (Thacker et al., 2007; Erwin and Thacker, 2008; Freeman et al., 2013) they appear to be under less selective pressure to adopt growth forms that allow for efficient light capture at low irradiances and at the cost of reduced heterotrophic capacity. Lower coral cover in the Caribbean may also provide ideal conditions for adaptive radiation that fuels niche expansion, diversification, and growth (potentially leading to higher biomass) in groups like sponges (Gavrilets and Losos, 2009).
Microbial Symbionts and Resource Competition
Coral reefs represent a paradox of incredible biodiversity and productivity in low-nutrient ecosystems (de Goeij et al., 2013); an enduring challenge in coral reef ecology is to identify the ecological and evolutionary processes that support this biodiversity and facilitate species coexistence. Ecological theory posits that competition for limiting resources can lead to the exclusion of a species by a more efficient competitor (Gause, 1934). It is thus parsimonious to attribute the successful coexistence of sponges in the Caribbean, with all their different morphologies, physiologies, microbial symbiont communities, and nutrition to a lack of nutrient limitation (Wilkinson and Cheshire, 1990; Pawlik et al., 2015, 2018). However, competition also drives trait evolution, ecological divergence, and ultimately niche partitioning that can favor coexistence (Schoener, 1974). The sponge communities we see on present-day coral reefs (and all of their traits however similar or different) have been shaped by past competition and some of the species involved in past competitive interactions were likely driven to extinction. These “ghosts of competition past” may have helped to drive ecological divergence that contributes to the coexistence and low apparent interspecific competition for resources on modern reefs. Divergence in microbiome composition, along with strong host specificity and the exploitation of distinct nutritional niches by coexisting sponge species, suggests that microbial symbionts play an important role in host niche expansion and differentiation (Joy, 2013) and that ecological divergence across sponge species is maintained by strong selective pressure from competition (Easson and Thacker, 2014; Freeman et al., 2014, 2020).
Similar work on corals in the Caribbean demonstrates that this group also adopts diverse morphologies (Santavy et al., 2013) that influence their productivity and heterotrophic capacity. For instance, hard corals with low SA:V (surface area: volume) have larger polyps and are better adapted to heterotrophic feeding, while individuals with small polyps have high SA:V and are better adapted to capturing light and obtaining benefits from photosynthetic symbionts (Porter, 1976). Substantial differences in polyp size, productivity (P:R values), and host coral enrichment in 13C from SIP experiments (Table 2) were also demonstrated by Baker et al. (2015) across 11 Caribbean gorgonian species. These results imply that niche diversification is occurring in other dominant groups within this ocean basin.
Recent work suggests that competition can also drive clustering of species by traits that allow them to exploit a particularly plentiful or high-quality resource (D’Andrea et al., 2019). Although species within each cluster are competing with each other, they also experience competitive release from species within other clusters that are exploiting different resource pools or niches (D’Andrea et al., 2020). This hypothesis for coexistence is particularly interesting considering evidence of ecological convergence among marine sponges. For instance, species form clusters in broad scale trends of microbial abundance (HMA vs. LMA), reliance on specific nitrogen transformations, and DOC/LPOC/detritus assimilation (Fan et al., 2012; Ribes et al., 2012; McMurray et al., 2018). In particular, sponge species appear to cluster by their relative exploitation of three heterotrophic carbon pools (Figure 1), providing evidence of niche partitioning via heterotrophic feeding on different carbon sources.
Of course, species coexistence on coral reefs will be shaped by differentiation across multiple niche dimensions, so sponges that share the same heterotrophic carbon niche may differ in other traits. For instance, several species may form a cluster that exploits the DOC pool on Caribbean reefs, but species within this cluster also have unique microbiomes and/or physiologies that allow them to vary in their production, retention, or recycling of other elements (such as nitrogen) that might be more limiting on coral reefs (Easson and Thacker, 2014; Thomas et al., 2016; McMurray et al., 2018). More work is needed to test this hypothesis with traits like photosymbiont abundance and productivity, nitrogen metabolism, or chemical defense production, but the clustering of species along multidimensional resource axes for one element (Figure 1) and divergence when multiple elements are considered (Freeman et al., 2020) provides support for multidimensional niche partitioning in sponges.
The dissimilarity in sponge species traits that we see on modern reefs may also not be shaped solely by external evolutionary pressures like competition. Instead, because energy is finite, species are under intense selective pressure to optimize their life history strategy by balancing fitness tradeoffs associated with characteristics that influence their growth, survival, and reproduction (Pianka, 1970; Grime, 1977; Darling et al., 2012). Reiswig (1974) was the first to report variation in sponge life history strategies and relate these patterns to differential ecological success or habitat specialization. For instance, Mycale laxissima was well adapted for rapid colonization of new habitat due to its ability to efficiently acquire food, rapid growth, and allocation of substantial energy to reproductive effort at the expense of body size (Reiswig, 1974). In contrast, Tectitethya crypta and Verongula reiswigi, had lower reproductive rates and instead allocated energy to production and the maintenance of large, energy rich, and long-lived individuals (Reiswig, 1974). These results led Reiswig (1974) to assess that niche partitioning in sponges was “by means of energy channeling and reproduction.” Fitness trade-offs have since been reported across different sponge species, with perhaps the most well-known example pertaining to the production of chemical defenses in Caribbean sponges. Although the production of chemical defenses may come with an ecological benefit on reefs with high levels of predators (Loh and Pawlik, 2014), their production also comes with a cost that leads these species to have slower growth and reproductive rates (Pawlik et al., 2008; Leong and Pawlik, 2010). In contrast, chemically undefended sponges allocate more energy to reproduction and growth and are therefore able to rapidly recruit to new substrates (Pawlik et al., 2008).
There are surprising similarities between the conclusions of Reiswig (1974) and Pawlik et al. (2008) and classic ecological theory pertaining to the evolution of life history strategies. For instance, sponge species appear to broadly fit the r-K selection dichotomy, with some species allocating energy to fast growth and colonization (r-selected species) and other species adopting a strategy that optimizes longevity and survival in crowded ecosystems (K-selected species) (Pianka, 1970; Stearns, 1977). In addition, with a more in-depth consideration of traits, sponge species may fit within the classic C-S-R model proposed by Grime (1977). In this example, fast growing, fecund, and/or chemically undefended species that rapidly recruit to new habitat could be considered ruderal (R) or “weedy” species that favor disturbance, whereas slower growing, chemically defended sponges may be better suited for environments favoring competitive (C) or stress tolerant (S) life history strategies (Grime, 1977). Observations of primary succession on a shipwreck in the Florida Keys (Pawlik et al., 2008) have also provided preliminary support for classic ecological theory by documenting initial colonization by chemically undefended sponges followed 18 months later by the colonization by chemically defended sponges. These observations match predictions that pioneer species will be replaced over time by slower growing species that are better adapted for competitive environments (Connell, 1978; Grime, 1977). These differences in overall life history strategies represent another mechanism for niche divergence that could facilitate coexistence on Caribbean coral reefs (Schoener, 1974) and highlight the fact that community composition on Caribbean coral reefs is impacted by diverse ecological processes including competition, predation, and symbiosis (see recent debate on this outlined by Pawlik et al., 2018).
Although recent work has used trait-based classification to identify distinct life history strategies in corals (Darling et al., 2012), similar trait data is either unavailable or incomplete across sponge species. For example, the proximate biological composition of sponge taxa is expected to be highly correlated with feeding mode. However, although we found a set of 57 taxa with published information on carbohydrate, lipid, and protein composition, only 8–12 of these taxa have been examined in studies of feeding on detritus, living particulate organic carbon (LPOC) or DOC (see Supplementary Table 2). Despite these limitations, we can find evidence of multiple interesting correlations among sponge life-history traits (Figure 4). For example, energy content increases with increasing microbiome diversity (left panel Figure 4), while palatability is negatively correlated with energy (right panel Figure 4). Notably, when comparing palatability (a proxy for chemical defense) and microbiome diversity, we found no sponges with high palatability and high microbiome diversity, suggesting that metabolites inherent to high diversity microbiomes could substantially impact palatability (center panel Figure 4). It is also worth noting here that there are clear trends between overall microbial abundance (HMA vs. LMA) and palatability as many, but not all, sponges with low palatability were HMA species and some HMA species were also highly palatable (center panel Figure 4). These figures provide preliminary evidence of links and potential tradeoffs between individual life history characteristics of sponges, but more work is needed to both accumulate additional data on these characteristics and test for tradeoffs.
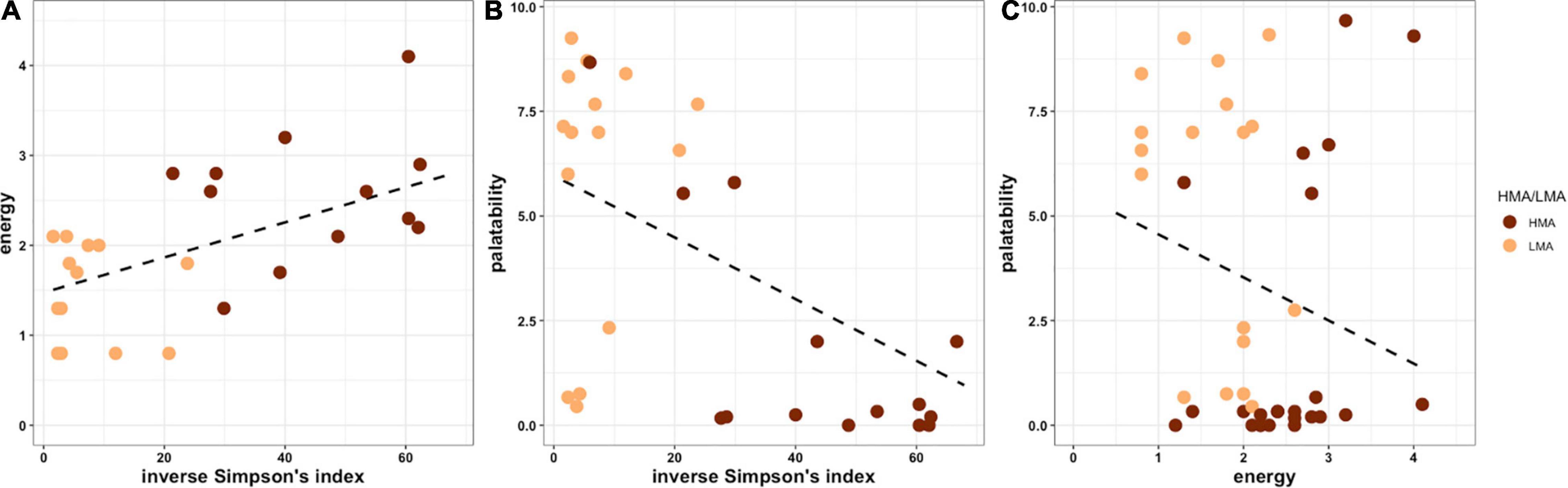
Figure 4. Relationships between multiple life-history traits of sponges. (A) Tissue energy content ([KJ ml– 1] from Chanas and Pawlik, 1995) vs. microbiome diversity (inverse Simpson’s index; Easson and Thacker, 2014; Thomas et al., 2016; Gantt et al., 2019; Freeman et al., 2020); (B) Palatability (mean number of pellets containing sponge chemical extracts that were consumed in trials with the bluehead wrasse Thalassoma bifasciatum; Loh and Pawlik, 2014) vs. microbiome diversity; (C) Palatability vs. energy.
Importantly, many major life-history traits of sponges display strong phylogenetic signal, implying that traits like palatability (a proxy for chemical defense), tissue energy content, microbiome diversity, and HMA/LMA status are linked to the evolutionary history of the host sponge (Figure 5 and Supplementary Tables 1, 2). However, this compilation of trait data also clearly shows the differential investigation of traits by sponge researchers. While many studies have examined palatability and proximate biochemical composition across multiple species, relatively few have quantified feeding on dissolved carbon vs. living particulate organic carbon or compared pumping rates with NOx production (Figure 5 and Supplementary Tables 1, 2). This data gap provides a clear priority for future work to better document life-history variation across a broader array of sponge diversity. For example, although multiple datasets measure microbiome diversity (Moitinho-Silva et al., 2017; Gantt et al., 2019; Freeman et al., 2020), few of these studies measure additional life history traits, limiting the integration of microbiome datasets into a broader understanding of sponge biology.
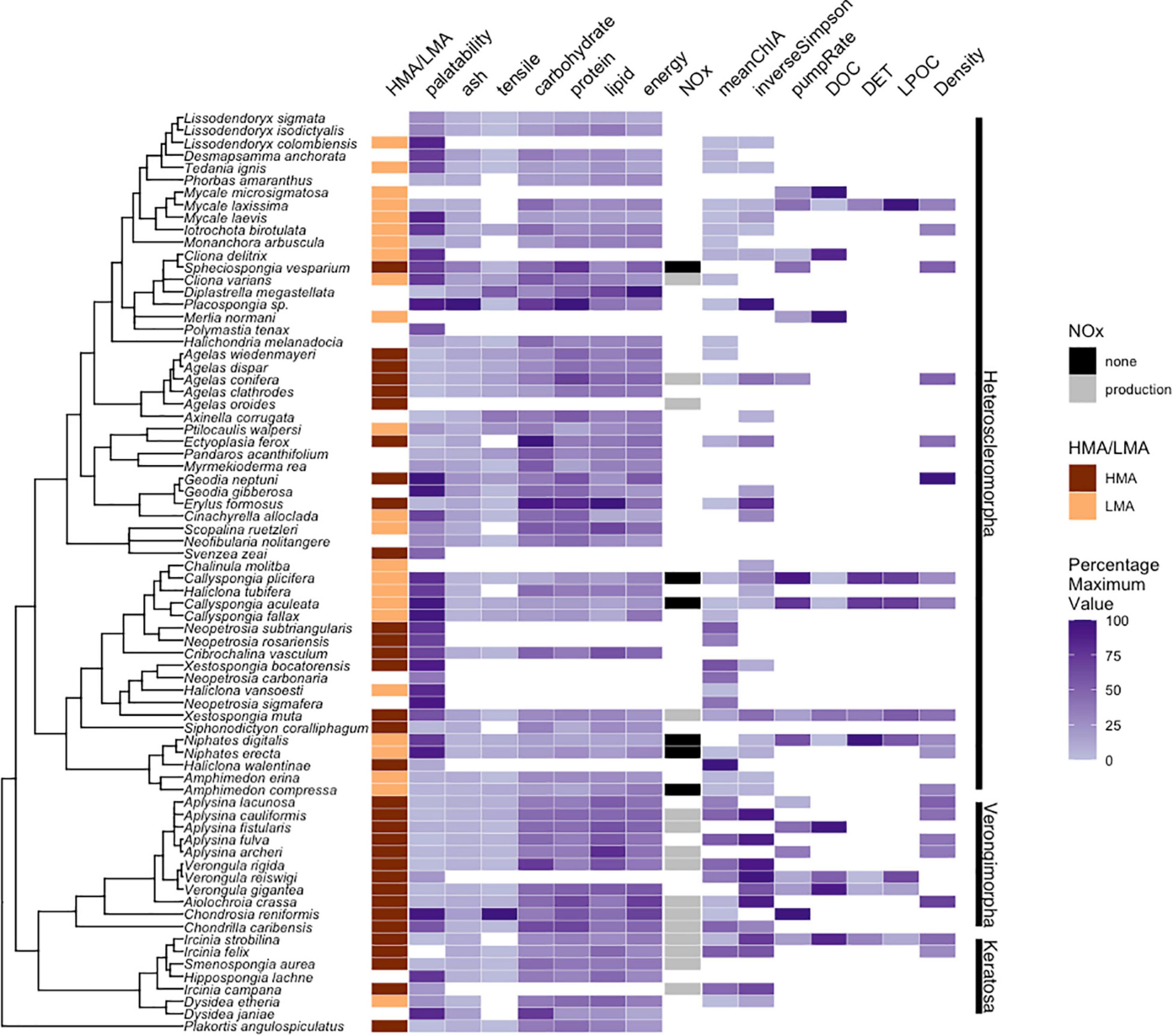
Figure 5. Major life history traits of sponges plotted against a phylogeny of common Caribbean species. Discrete traits include HMA/LMA status and NOx production. Continuous traits are displayed as a percentage of the maximum value of each trait. Vertical bars to the right indicate subclasses of class Demospongiae; Plakortis is a member of class Homoscleromorpha; palatability (mean number of pellets containing sponge chemical extracts that were consumed in trials with the bluehead wrasse Thalassoma bifasciatum; Loh and Pawlik, 2014), physical characteristics (ash, carbohydrate, protein, and lipid [mg ml– 1], tensile strength [N m– 2 × 105], and energy [KJ ml– 1] from Chanas and Pawlik, 1995), Chla, mean chlorophyll a concentrations (μg chl a g sponge tissue– 1); inverseSimpson, value of the inverse Simpson’s index for microbiome diversity; pumpRate, pumping rate of water through a sponge (l s– 1 l sponge– 1); DOC, DET, and LPOC, percent dissolved organic carbon, detritus, or living particulate organic carbon in diet, and Density = mean tissue density (g ml– 1). Analyses adapted from those in Freeman et al. (2020) using data and related references in tables within Supplementary Information.
Speciation
If microbiome metabolism is essential for the expansion and establishment of a host’s ecological niche (Phillips et al., 2017; Freeman et al., 2020), then the formation of sibling species at a single location (termed sympatric speciation) might be facilitated by changes in microbiome composition that yield different functional traits (Schluter, 2009), as predicted by theories of niche differentiation (D’Andrea and Ostling, 2016) that examine within-community, or alpha, niche differences (Ackerly et al., 2006). Alternatively, if speciation occurs subsequent to geographic isolation (termed allopatric speciation), differences in environmental conditions and selective pressures might select for larger differences in microbiome composition, particularly those associated with the new habitat (Loo et al., 2019). These between-community, or beta, niche differences are often associated with large-scale changes in environmental conditions (Ackerly et al., 2006). In instances where multiple speciation events occur in parallel, we might expect that these environmental forces act similarly across multiple host species.
The term “phylosymbiosis” has been proposed to describe the congruence of host phylogeny and dissimilarity (beta diversity) of microbiome community composition, without presuming that this pattern arises from coevolution or cospeciation (Brooks et al., 2016). For example, a recent survey of coral microbiomes found support for phylosymbiosis, but only limited support for codiversification of host and symbiont phylogenies, suggesting that changes in host habitat and recruitment of environmental microbes have a stronger impact on microbiome composition (Pollock et al., 2018). For sponges, a review of globally sampled hosts and microbiomes found strong support for phylosymbiosis (Thomas et al., 2016), but few studies have examined co-diversification among sponge and symbiont lineages, and have generally found weak support (Thacker et al., 2007; Reveillaud et al., 2014).
In addition, few studies have provided evidence of speciation being associated with changes in microbiome composition and functional traits. Kelly et al. (2021) examined multiple Ircinia spp. in the Caribbean, finding unique microbiomes associated with 12 distinct host lineages and suggested that these metagenomes code for distinct nutritional strategies. This study also found evidence for on-going hybridization among host sponge lineages, suggesting that the forces selecting for distinct microbiomes are stronger than neutral processes such as dispersal and hybridization. Likewise, Díez-Vives et al. (2020) found microbiome divergence correlated with population genetic differentiation within Petrosia ficiformis, while Easson et al. (2020) documented a similar pattern among populations of Cliona delitrix.
Specific Knowledge Gaps in Evolutionary Pressures and Life History Strategies
Many reported trends in sponge life-history evolution are driven by the relatively small number of species for which these traits have been quantified (Pawlik et al., 2018; Wulff, 2020). Indeed, surprisingly few of the more than 9,000 described sponge species can be included in our current comparisons. These preliminary results suggest that there are multiple dimensions of the ecological niches of sponges that are not related to nutrition and that sponge life history strategies are under pressure from diverse evolutionary forces including predation, competition, and mutualism (Pawlik, 2011; Pawlik et al., 2015; Wulff, 2020).
Our review of recent literature found few studies that explicitly compare genetic variation within species to variability in life-history traits, including microbiome composition and palatability. A clear priority for future research is to not only broaden the taxonomic coverage of surveys of life-history traits, but to examine how these traits vary within species. Identifying trade-offs within species would greatly facilitate studies connecting genotype to phenotype and the discovery of gene regulatory networks (Nuzhdin et al., 2012). When comparing among species, although several previous studies document phylosymbiosis, few studies have examined individual symbiont lineages in attempts to measure co-diversification or host switching, suggesting additional avenues for future research. Finally, as humans continue to impact reef habitats around the globe, additional work is also needed to determine how environmental disturbances shape host evolution and the role of microbial symbionts in host response or resilience to these stressors (Glasl et al., 2018; O’Brien et al., 2019; Ramsby et al., 2020).
Conclusion and Future Research
In our discussion above, we have outlined evidence that supports the notion that microbial symbiont communities have an important role in shaping the ecology and the evolutionary trajectory of host sponge species. We also highlight the multiple factors involved in contextualizing sponge evolution and the role of sponges in coral reef ecology. Here, we delineate five overarching research needs and themes derived from our synthesis. Two of these address necessary changes to specific experimental factors. First, it is critical to use appropriate irradiance values in experimental conditions to accurately assess photophysiology of photosymbionts and functional roles of these symbionts in hospite. If experiments cannot be carried out in situ and at depth, then irradiances used should be ecologically relevant and based on measured values at depth. Second, standardized combinations of modern analytical tools (Tables 1–3) are needed to assess how both hosts and symbionts contribute to the ecological success of sponges in different ocean basins and untangle the relationship between sponge community assemblages and nutrient composition and concentration. These methods should aim to provide detailed carbon and nitrogen budgets for (at least initially) the most ecologically dominant sponge species on reefs across regional environmental gradients and in different ocean basins. These studies should also assess the nutritional environments where these sponge species are found by measuring the concentrations of organic (both particulate and dissolved) and inorganic nutrients. In addition to these two experimental research needs, it is critical to broaden nutrient-focused geographic comparisons beyond carbon, as nitrogen in particular, but also sulfur, phosphorus, iron, and other trace nutrients likely have biologically meaningful consequences in sponge and microbial symbiont physiologies. More specifically, the decades-long focus on carbon has hindered our understanding of and constrained the conversation around sponges and reef nutrient dynamics. In addition, as we move beyond analyses of single traits toward variation in multiple nutritional niche dimensions, there is a clear need to address evolutionary questions pertaining to the selective pressures that help to form and maintain host–symbiont and symbiont–symbiont interactions, as well as variation in host specificity or plasticity in these interactions. Finally, more experimental studies are needed to test hypotheses being developed from the many descriptive-based studies, particularly involving high-throughput sequencing and large-scale ‘omics work (Mohanty et al., 2021). This will likely include the development and application of new tools that can facilitate such experimental studies (e.g., novel microscopy tools). Some of these research themes have been similarly highlighted in previous reviews; here we have built on these conversations with a historical perspective.
In summary, sponges have unique evolutionary solutions to the diverse ecological challenges on coral reefs. There is still a need for a variety of experimental approaches and scope including those targeting a particular factor of interest. However, we argue that the corresponding results should be placed in the context of the broader and more complicated picture of sponge holobiont ecology rather than viewed in isolation as a single factor (e.g., top–down vs. bottom–up processes). Furthermore, the field of sponge ecology should weave together the individually discovered narratives on molecular and genomic functional attributes of sponges and/or microbial symbionts (e.g., Letourneau et al., 2020; Robbins et al., 2021), underlying evolutionary patterns (e.g., Freeman et al., 2020; Kelly et al., 2021), and ecological interactions (e.g., Loh and Pawlik, 2014; Pawlik et al., 2018; Wulff, 2020), to better develop a framework for understanding ecological and molecular mechanisms relevant to the success of extant poriferans. This work is likely to have implications beyond the field of sponge biology.
Author Contributions
All the authors contributed to the development of this manuscript, conducted the literature reviews, and added to the writing and analyses of metadata. CJF led the writing of the manuscript.
Funding
This work was supported by the U.S. National Science Foundation (NSF) awards no. 1929293 to CJF, no. 1915949 to CGE, no. 1756171 to CLF, and no. 1756249 to RWT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff at the Smithsonian Tropical Research Institute in Bocas del Toro, Panama for their logistical support. We also thank J. Korn for her assistance creating and optimizing Figure 2.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.705053/full#supplementary-material
References
Achlatis, M., Pernice, M., Green, K., de Goeij, J. M., Guagliardo, P., Kilburn, M. R., et al. (2019). Single-cell visualization indicates direct role of sponge host in uptake of dissolved organic matter. Proc. R. Soc. B Biol. Sci. 286:20192153. doi: 10.1098/rspb.2019.2153
Ackerly, D. D., Schwilk, D. W., and Webb, C. O. (2006). Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology 87, S50–S61.
Amon, R. M. W., and Benner, R. (1996). Photochemical and microbial consumption of dissolved organic carbon and dissolved oxygen in the Amazon River system. Geochim. Cosmochim. Acta 60, 1783–1792. doi: 10.1016/0016-7037(96)00055-5
Anderson, S. A., Northcote, P. T., and Page, M. J. (2010). Spatial and temporal variability of the bacterial community in different chemotypes of the New Zealand marine sponge Mycale hentscheli. FEMS Microbiol. Ecol. 72, 328–342. doi: 10.1111/j.1574-6941.2010.00869.x
Archer, S. K., Stevens, J. L., Rossi, R. E., Matterson, K. O., and Layman, C. A. (2017). Abiotic conditions drive significant variability in nutrient processing by a common Caribbean sponge. Ircinia felix. Limnol. Oceanogr. 62, 1783–1793. doi: 10.1002/lno.10533
Aronson, R. B., Hilbun, N. L., Bianchi, T. S., Filley, T. R., and McKee, B. A. (2014). Land use, water quality, and the history of coral assemblages at Bocas del Toro. Panamá. Mar. Ecol. Prog. Ser. 504, 159–170. doi: 10.3354/meps10765
Baker, D. M., Freeman, C. J., Knowlton, N., Thacker, R. W., Kim, K., and Fogel, M. L. (2015). Productivity links morphology, symbiont specificity and bleaching in the evolution of Caribbean octocoral symbioses. ISME J. 9, 2620–2629. doi: 10.1038/ismej.2015.71
Bannister, R., Hoogenboom, M., Anthony, K., Battershill, C., Whalan, S., Webster, N., et al. (2011). Incongruence between the distribution of a common coral reef sponge and photosynthesis. Mar. Ecol. Prog. Ser. 423, 95–100. doi: 10.3354/meps08886
Bart, M. C., de Kluijver, A., Hoetjes, S., Absalah, S., Mueller, B., Kenchington, E., et al. (2020). Differential processing of dissolved and particulate organic matter by deep-sea sponges and their microbial symbionts. Sci. Rep. 10:17515. doi: 10.1038/s41598-020-74670-0
Bayer, K., Busch, K., Kenchington, E., Beazley, L., Franzenburg, S., Michels, J., et al. (2020). Microbial strategies for survival in the glass sponge Vazella pourtalesii. mSystems 5:e00473-20. doi: 10.1128/mSystems.00473-20
Bell, J. J., McGrath, E., Kandler, N. M., Marlow, J., Beepat, S. S., Bachtiar, R., et al. (2020). Interocean patterns in shallow water sponge assemblage structure and function. Biol. Rev. 95, 1720–1758. doi: 10.1111/brv.12637
Bennett, G. M., and Moran, N. A. (2013). Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol. Evol. 5, 1675–1688. doi: 10.1093/gbe/evt118
Botté, E. S., Nielsen, S., Abdul Wahab, M. A., Webster, J., Robbins, S., Thomas, T., et al. (2019). Changes in the metabolic potential of the sponge microbiome under ocean acidification. Nat. Commun. 10:4134. doi: 10.1038/s41467-019-12156-y
Brooks, A. W., Kohl, K. D., Brucker, R. M., van Opstal, E. J., and Bordenstein, S. R. (2016). Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14:e2000225. doi: 10.1371/journal.pbio.2000225
Bryson, S., Li, Z., Pett-Ridge, J., Hettich, R. L., Mayali, X., Pan, C., et al. (2016). Proteomic stable isotope probing reveals taxonomically distinct patterns in amino acid assimilation by coastal marine bacterioplankton. mSystems 1:e00027-15. doi: 10.1128/mSystems.00027-15
Burgsdorf, I., Erwin, P. M., López-Legentil, S., Cerrano, C., Haber, M., Frenk, S., et al. (2014). Biogeography rather than association with cyanobacteria structures symbiotic microbial communities in the marine sponge Petrosia ficiformis. Front. Microbiol. 5:529. doi: 10.3389/fmicb.2014.00529
Carballo, J. L., Ávila, E., Enríquez, S., and Camacho, L. (2006). Phenotypic plasticity in a mutualistic association between the sponge Haliclona caerulea and the calcareous macroalga Jania adherens induced by transplanting experiments. I: morphological responses of the sponge. Mar. Biol. 148, 467–478. doi: 10.1007/s00227-005-0104-4
Cárdenas, C. A., Font, A., Steinert, G., Rondon, R., and González-Aravena, M. (2019). Temporal stability of bacterial communities in Antarctic sponges. Front. Microbiol. 10:2699. doi: 10.3389/fmicb.2019.02699
Cavanaugh, C. M., Gardiner, S. L., Jones, M. L., Jannasch, H. W., and Waterbury, J. B. (1981). Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213, 340–342. doi: 10.1126/science.213.4505.340
Chanas, B., and Pawlik, J. R. (1995). Defenses of Caribbean sponges against predatory reef fish. II. Spicules, tissue toughness, and nutritional quality. Mar. Ecol. Prog. Ser. 127, 195–211. doi: 10.3354/meps127195
Chaves-Fonnegra, A., Feldheim, K. A., Secord, J., and Lopez, J. V. (2015). Population structure and dispersal of the coral-excavating sponge Cliona delitrix. Mol. Ecol. 24, 1447–1466. doi: 10.1111/mec.13134
Cherrier, J., Bauer, J. E., Druffel, E. R. M., Coffin, R. B., and Chanton, J. P. (1999). Radiocarbon in marine bacteria: Evidence for the ages of assimilated carbon. Limnol. Oceanogr. 44, 730–736. doi: 10.4319/lo.1999.44.3.0730
Cheshire, A. C., and Wilkinson, C. R. (1991). Modelling the photosynthetic production by sponges on Davies Reef, Great Barrier Reef. Mar. Biol. 109, 13–18. doi: 10.1007/BF01320226
Cheshire, A. C., Wilkinson, C. R., Seddon, S., and Westphalen, G. (1997). Bathymetric and seasonal changes in photosynthesis and respiration of the phototrophic sponge Phyllospongia lamellosa in comparison with respiration by the heterotrophic sponge Ianthella basta on Davies Reef. Great Barrier Reef. Mar. Freshw. Res. 48, 589–599. doi: 10.1071/mf96070
Connell, J. H. (1978). Diversity in tropical rain forests and coral reefs. Science 199, 1302–1310. doi: 10.1126/science.199.4335.1302
D’Andrea, R., Guittar, J., O’Dwyer, J. P., Figueroa, H., Wright, S. J., Condit, R., et al. (2020). Counting niches: Abundance-by-trait patterns reveal niche partitioning in a Neotropical forest. Ecology 101:e03019. doi: 10.1002/ecy.3019
D’Andrea, R., and Ostling, A. (2016). Challenges in linking trait patterns to niche differentiation. Oikos 125, 1369–1385. doi: 10.1111/oik.02979
D’Andrea, R., Riolo, M., and Ostling, A. M. (2019). Generalizing clusters of similar species as a signature of coexistence under competition. PLoS Comput. Biol. 15:e1006688. doi: 10.1371/journal.pcbi.1006688
Darling, E. S., Alvarez-Filip, L., Oliver, T. A., McClanahan, T. R., and C\̂text{o}té, I. M. (2012). Evaluating life-history strategies of reef corals from species traits. Ecol. Lett. 15, 1378–1386. doi: 10.1111/j.1461-0248.2012.01861.x
Dat, T. T. H., Steinert, G., Thi Kim, Cuc, N., Smidt, H., and Sipkema, D. (2018). Archaeal and bacterial diversity and community composition from 18 phylogenetically divergent sponge species in Vietnam. PeerJ 6:e4970. doi: 10.7717/peerj.4970
de Goeij, J., Lesser, M. P., and Pawlik, J. R. (2017). “Nutrient fluxes and ecological functions of coral reef sponges in a changing ocean,” in Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization, eds J. Carballo and J. Bell (Berlin: Springer), 373–410. doi: 10.1007/978-3-319-59008-0_8
de Goeij, J., van den Berg, H., van Oostveen, M., Epping, E., and van Duyl, F. (2008). Major bulk dissolved organic carbon (DOC) removal by encrusting coral reef cavity sponges. Mar. Ecol. Prog. Ser. 357, 139–151. doi: 10.3354/meps07403
de Goeij, J. M., Van Oevelen, D., Vermeij, M. J. A., Osinga, R., Middelburg, J. J., De Goeij, A. F. P. M., et al. (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342, 108–110. doi: 10.1126/science.1241981
de Kluijver, A., Bart, M. C., van Oevelen, D., de Goeij, J. M., Leys, S. P., Maier, S. R., et al. (2021). An integrative model of carbon and nitrogen metabolism in a common deep-sea sponge (Geodia barretti). Front. Mar. Sci. 7:1131. doi: 10.3389/fmars.2020.596251
Degnan, P. H., Taga, M. E., and Goodman, A. L. (2014). Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778. doi: 10.1016/j.cmet.2014.10.002
Deignan, L. K., Pawlik, J. R., and López-Legentil, S. (2018). Evidence for shifting genetic structure among Caribbean giant barrel sponges in the Florida Keys. Mar. Biol. 165:106. doi: 10.1007/s00227-018-3355-6
Diaz, C., and Rützler, K. (2001). Sponges: an essential component of Caribbean coral reefs. Bull. Mar. Sci. 69, 535–546. doi: 10.1016/s0013-4686(97)10172-4
Diaz, M. C., Thacker, R. W., Rützler, K., and Piantoni, C. (2007). “Two new haplosclerid sponges from Caribbean Panama with symbiotic filamentous cyanobacteria, and an overview of sponge-cyanobacteria associations,” in Porifera Research: Biodiversity, Innovation, and Sustainability, eds M. R. Custódio, G. Lobo-Hajdu, E. Hajdu, and G. Muricy (Rio de Janeiro: Museu Nacional Rio de Janeiro), 31–39.
Díez-Vives, C., Taboada, S., Leiva, C., Busch, K., Hentschel, U., and Riesgo, A. (2020). On the way to specificity - Microbiome reflects sponge genetic cluster primarily in highly structured populations. Mol. Ecol. 29, 4412–4427. doi: 10.1111/mec.15635
Done, T. J. (1982). Patterns in the distribution of coral communities across the central Great Barrier Reef. Coral Reefs 1, 95–107. doi: 10.1007/BF00301691
Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., et al. (2001). Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302. doi: 10.1038/35077067
Easson, C. G., Chaves-Fonnegra, A., Thacker, R. W., and Lopez, J. V. (2020). Host population genetics and biogeography structure the microbiome of the sponge Cliona delitrix. Ecol. Evol. 10, 2007–2020. doi: 10.1002/ece3.6033
Easson, C. G., Matterson, K. O., Freeman, C. J., Archer, S. K., and Thacker, R. W. (2015). Variation in species diversity and functional traits of sponge communities near human populations in Bocas del Toro. Panama. PeerJ 3:e1385. doi: 10.7717/peerj.1385
Easson, C. G., and Thacker, R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5:532. doi: 10.3389/fmicb.2014.00532
Engelberts, J. P., Robbins, S. J., de Goeij, J. M., Aranda, M., Bell, S. C., and Webster, N. S. (2020). Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 14, 1100–1110. doi: 10.1038/s41396-020-0591-9
Erwin, P. M., López-Legentil, S., González-Pech, R., and Turon, X. (2012a). A specific mix of generalists: bacterial symbionts in Mediterranean Ircinia spp. FEMS Microbiol. Ecol. 79, 619–637. doi: 10.1111/j.1574-6941.2011.01243.x
Erwin, P. M., Pita, L., López-Legentil, S., and Turon, X. (2012b). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368. doi: 10.1128/AEM.02035-12
Erwin, P. M., Olson, J. B., and Thacker, R. W. (2011). Phylogenetic diversity, host-specificity and community profiling of sponge-associated bacteria in the Northern Gulf of Mexico. PLoS One 6:e26806. doi: 10.1371/journal.pone.0026806
Erwin, P. M., and Thacker, R. W. (2007). Incidence and identity of photosynthetic symbionts in Caribbean coral reef sponge assemblages. J. Mar. Biol. Assoc. U. K. 87, 1683–1692. doi: 10.1017/S0025315407058213
Erwin, P. M., and Thacker, R. W. (2008). Phototrophic nutrition and symbiont diversity of two Caribbean sponge-cyanobacteria symbioses. Mar. Ecol. Prog. Ser. 362, 139–147. doi: 10.3354/meps07464
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 109, E1878–E1887. doi: 10.1073/pnas.1203287109
Ferris, M. J., Muyzer, G., and Ward, D. M. (1996). Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62, 340–346. doi: 10.1128/AEM.62.2.340-346.1996
Feuda, R., Dohrmann, M., Pett, W., Philippe, H., Rota-Stabelli, O., Lartillot, N., et al. (2017). Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr. Biol. 27, 3864.e4–3870.e4. doi: 10.1016/j.cub.2017.11.008
Finke, D. L., and Snyder, W. E. (2008). Niche partitioning increases resource exploitation by diverse communities. Science 321, 1488–1490. doi: 10.1126/science.1160854
Fiore, C. L., Baker, D. M., and Lesser, M. P. (2013a). Nitrogen biogeochemistry in the Caribbean cponge, Xestospongia muta: a source or sink of dissolved inorganic nitrogen? PLoS One 8:e72961. doi: 10.1371/journal.pone.0072961
Fiore, C. L., Jarett, J. K., and Lesser, M. P. (2013b). Symbiotic prokaryotic communities from different populations of the giant barrel sponge. Xestospongia muta. Microbiologyopen 2, 938–952. doi: 10.1002/mbo3.135
Fiore, C. L., Freeman, C. J., and Kujawinski, E. B. (2017). Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ 5:e2870. doi: 10.7717/peerj.2870
Fiore, C. L., Jarett, J. K., Steinert, G., and Lesser, M. P. (2020). Trait-based comparison of coral and sponge microbiomes. Sci. Rep. 10:2340. doi: 10.1038/s41598-020-59320-9
Fiore, C. L., Labrie, M., Jarett, J. K., and Lesser, M. P. (2015a). Transcriptional activity of the giant barrel sponge, Xestospongia muta Holobiont: molecular evidence for metabolic interchange. Front. Microbiol. 6:364. doi: 10.3389/fmicb.2015.00364
Fiore, C. L., Longnecker, K., Kido Soule, M. C., and Kujawinski, E. B. (2015b). Release of ecologically relevant metabolites by the cyanobacterium Synechococcus elongatus CCMP 1631. Environ. Microbiol. 17, 3949–3963. doi: 10.1111/1462-2920.12899
Fisher, C. R., and Childress, J. J. (1984). Substrate oxidation by trophosome tissue from Riftia pachyptila Jones (Phylum pogonophora). Mar. Biol. Lett. 5, 171–183.
Freeman, C. J., Baker, D. M., Easson, C. G., and Thacker, R. W. (2015). Shifts in sponge-microbe mutualisms across an experimental irradiance gradient. Mar. Ecol. Prog. Ser. 526, 41–53. doi: 10.3354/meps11249
Freeman, C. J., Easson, C. G., and Baker, D. M. (2014). Metabolic diversity and niche structure in sponges from the Miskito Cays, Honduras. PeerJ. 2:e695. doi: 10.7717/peerj.695
Freeman, C. J., Easson, C. G., Matterson, K. O., Thacker, R. W., Baker, D. M., and Paul, V. J. (2020). Microbial symbionts and ecological divergence of Caribbean sponges: a new perspective on an ancient association. ISME J. 14, 1571–1583. doi: 10.1038/s41396-020-0625-3
Freeman, C. J., and Thacker, R. W. (2011). Complex interactions between marine sponges and their symbiotic microbial communities. Limnol. Oceanogr. 56, 1577–1586. doi: 10.4319/lo.2011.56.5.1577
Freeman, C. J., Thacker, R. W., Baker, D. M., and Fogel, M. L. (2013). Quality or quantity: Is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J. 7, 1116–1125. doi: 10.1038/ismej.2013.7
Gantt, S. E., McMurray, S. E., Stubler, A. D., Finelli, C. M., Pawlik, J. R., and Erwin, P. M. (2019). Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 7:124. doi: 10.1186/s40168-019-0739-x
Gause, G. F. (1934). Experimental analysis of Vito Volterra’s mathematical theory of the struggle for existence. Science 79, 16–17. doi: 10.1126/science.79.2036.16-a
Gavrilets, S., and Losos, J. B. (2009). Adaptive radiation: contrasting theory with data. Science 323, 732–737. doi: 10.1126/science.1157966
Glasl, B., Smith, C. E., Bourne, D. G., and Webster, N. S. (2018). Exploring the diversity-stability paradigm using sponge microbial communities. Sci. Rep. 8:8425. doi: 10.1038/s41598-018-26641-9
Gloeckner, V., Wehrl, M., Moitinho-Silva, L., Gernert, C., Schupp, P., Pawlik, J. R., et al. (2014). The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol. Bull. 227, 78–88. doi: 10.1086/bblv227n1p78
Griffiths, S. M., Antwis, R. E., Lenzi, L., Lucaci, A., Behringer, D. C., and Butler, M. J. IV, et al. (2019). Host genetics and geography influence microbiome composition in the sponge Ircinia campana. J. Anim. Ecol. 88, 1684–1695. doi: 10.1111/1365-2656.13065
Grime, J. P. (1977). Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194. doi: 10.1086/283244
Hadas, E., Shpigel, M., and Ilan, M. (2009). Particulate organic matter as a food source for a coral reef sponge. J. Exp. Biol 212, 3643–3650. doi: 10.1242/jeb.027953
Hallam, S. J., Konstantinidis, K. T., Putnam, N., Schleper, C., Watanabe, Y., Sugahara, J., et al. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U.S.A. 103, 18296–18301. doi: 10.1073/pnas.0608549103
Hentschel, U., Fieseler, L., Wehrl, M., Gernert, C., Steinert, M., Hacker, J., et al. (2003). “Microbial diversity of marine sponges,” in Sponges (Porifera). Progress in Molecular and Subcellular Biology, ed. W. E. G. Müller (Berlin: Springer Berlin Heidelberg), 59–88. doi: 10.1007/978-3-642-55519-0_3
Hentschel, U., Usher, K. M., and Taylor, M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Hoer, D. R., Gibson, P. J., Tommerdahl, J. P., Lindquist, N. L., and Martens, C. S. (2018). Consumption of dissolved organic carbon by Caribbean reef sponges. Limnol. Oceanogr. 63, 337–351. doi: 10.1002/lno.10634
Hoffmann, F., Larsen, O., Thiel, V., Rapp, H. T., Pape, T., Michaelis, W., et al. (2005a). An anaerobic world in sponges. Geomicrobiol. J. 22, 1–10. doi: 10.1080/01490450590922505
Hoffmann, F., Larsen, O., Tore Rapp, H., and Osinga, R. (2005b). Oxygen dynamics in choanosomal sponge explants. Mar. Biol. Res. 1, 160–163. doi: 10.1080/17451000510019006
Hoffmann, F., Radax, R., Woebken, D., Holtappels, M., Lavik, G., Rapp, H. T., et al. (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ. Microbiol. 11, 2228–2243. doi: 10.1111/j.1462-2920.2009.01944.x
Holert, J., Cardenas, E., Bergstrand, L. H., Zaikova, E., Hahn, A. S., Hallam, S. J., et al. (2018). Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio 9:e02345-17. doi: 10.1128/mbio.02345-17
Hudspith, M., Rix, L., Achlatis, M., Bougoure, J., Guagliardo, P., Clode, P. L., et al. (2021). Subcellular view of host–microbiome nutrient exchange in sponges: insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 9:44. doi: 10.1186/s40168-020-00984-w
Ilan, M., and Abelson, A. (1995). The life of a sponge in a sandy lagoon. Biol. Bull. 189, 363–369. doi: 10.2307/1542154
Jaeckle, W. B. (1995). Transport and metabolism of alanine and palmitic acid by field-collected larvae of Tedania ignis (Porifera, Demospongiae): estimated consequences of limited label translocation. Biol. Bull. 189, 159–167. doi: 10.2307/1542466
Jensen, S., Fortunato, S. A. V., Hoffmann, F., Hoem, S., Rapp, H. T., Øvreås, L., et al. (2017). The relative abundance and transcriptional activity of marine sponge-associated microorganisms emphasizing groups involved in sulfur cycle. Microb. Ecol. 73, 668–676. doi: 10.1007/s00248-016-0836-3
Joy, J. B. (2013). Symbiosis catalyses niche expansion and diversification. Proc. R. Soc. B Biol. Sci. 280:20122820. doi: 10.1098/rspb.2012.2820
Kamke, J., Rinke, C., Schwientek, P., Mavromatis, K., Ivanova, N., Sczyrba, A., et al. (2014). The candidate phylum Poribacteria by single-cell genomics: new insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS One 9:e87353. doi: 10.1371/journal.pone.0087353
Kelly, J. B., Carlson, D. E., Low, J. S., Rice, T., and Thacker, R. W. (2021). The relationship between microbiomes and selective regimes in the sponge genus Ircinia. Front. Microbiol. 12:489. doi: 10.3389/fmicb.2021.607289
Kiran, G. S., Sekar, S., Ramasamy, P., Thinesh, T., Hassan, S., Lipton, A. N., et al. (2018). Marine sponge microbial association: towards disclosing unique symbiotic interactions. Mar. Environ. Res. 140, 169–179. doi: 10.1016/j.marenvres.2018.04.017
Kleiner, M., Dong, X., Hinzke, T., Wippler, J., Thorson, E., Mayer, B., et al. (2018). Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 115, E5576–E5584. doi: 10.1073/pnas.1722325115
Koblentz-Mishke, O. J., Volkovinski, V. V., and Kabanova, J. G. (1970). “Plankton primary productivity of the world ocean,” in Scientific Exploration of the South Pacific, ed. S. Wooster (Washington, DC: National Academy of Sciences), 183–193.
Kujawinski, E. B., Longnecker, K., Barott, K. L., Weber, R. J. M., and Kido Soule, M. C. (2016). Microbial community structure affects marine dissolved organic matter composition. Front. Mar. Sci. 3:45. doi: 10.3389/fmars.2016.00045
Lavy, A., Keren, R., Yu, K., Thomas, B. C., Alvarez-Cohen, L., Banfield, J. F., et al. (2018). A novel Chromatiales bacterium is a potential sulfide oxidizer in multiple orders of marine sponges. Environ. Microbiol. 20, 800–814. doi: 10.1111/1462-2920.14013
Leong, W., and Pawlik, J. R. (2010). Evidence of a resource trade-off between growth and chemical defenses among Caribbean coral reef sponges. Mar. Ecol. Prog. Ser. 406, 71–78. doi: 10.3354/meps08541
Lesser, M. P., Mueller, B., Pankey, M. S., Macartney, K. J., Slattery, M., and de Goeij, J. M. (2019). Depth-dependent detritus production in the sponge, Halisarca caerulea. Limnol. Oceanogr. 65, 1200–1216. doi: 10.1002/lno.11384
Letourneau, M. L., Hopkinson, B. M., Fitt, W. K., and Medeiros, P. M. (2020). Molecular composition and biodegradation of loggerhead sponge Spheciospongia vesparium exhalent dissolved organic matter. Mar. Environ. Res. 162:105130. doi: 10.1016/j.marenvres.2020.105130
Leys, S. P., Kahn, A. S., Fang, J. K. H., Kutti, T., and Bannister, R. J. (2017). Phagocytosis of microbial symbionts balances the carbon and nitrogen budget for the deep-water boreal sponge Geodia barretti. Limnol. Oceanogr. 63, 187–202. doi: 10.1002/lno.10623
Li, Z., Wang, Y., Li, J., Liu, F., He, L., He, Y., et al. (2016). Metagenomic analysis of genes encoding nutrient cycling pathways in the microbiota of deep-sea and shallow-water sponges. Mar. Biotechnol. 18, 659–671. doi: 10.1007/s10126-016-9725-5
Li, Z.-Y., Wang, Y.-Z., He, L.-M., and Zheng, H.-J. (2014). Metabolic profiles of prokaryotic and eukaryotic communities in deep-sea sponge Neamphius huxleyi indicated by metagenomics. Sci. Rep. 4:3895. doi: 10.1038/srep03895
Liu, F., Li, J., Feng, G., and Li, Z. (2016). New genomic insights into “Entotheonella” symbionts in Theonella swinhoei: mixotrophy, anaerobic adaptation, resilience, and interaction. Front. Microbiol. 7:1333. doi: 10.3389/fmicb.2016.01333
Liu, M., Fan, L., Zhong, L., Kjelleberg, S., and Thomas, T. (2012). Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 6, 1515–1525. doi: 10.1038/ismej.2012.1
Liu, M. Y., Kjelleberg, S., and Thomas, T. (2011). Functional genomic analysis of an uncultured δ-proteobacterium in the sponge Cymbastela concentrica. ISME J. 5, 427–435. doi: 10.1038/ismej.2010.139
Loh, T.-L., and Pawlik, J. R. (2014). Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc. Natl. Acad. Sci. U.S.A. 111, 4151–4156. doi: 10.1073/pnas.1321626111
Loo, W. T., Dudaniec, R. Y., Kleindorfer, S., and Cavanaugh, C. M. (2019). An inter-island comparison of Darwin’s finches reveals the impact of habitat, host phylogeny, and island on the gut microbiome. PLoS One 14:e0226432. doi: 10.1371/journal.pone.0226432
Maldonado, M., Ribes, M., and van Duyl, F. C. (2012). Nutrient fluxes through sponges: Biology, budgets, and ecological implications. Adv. Mar. Biol. 62, 113–182. doi: 10.1016/B978-0-12-394283-8.00003-5
McClintock, J. B., Amsler, C. D., Baker, B. J., and van Soest, R. W. M. (2005). Ecology of Antarctic marine sponges: an overview. Integr. Comp. Biol. 45, 359–368. doi: 10.1093/icb/45.2.359
McMurray, S., Pawlik, J., and Finelli, C. (2014). Trait-mediated ecosystem impacts: how morphology and size affect pumping rates of the Caribbean giant barrel sponge. Aquat. Biol. 23, 1–13. doi: 10.3354/ab00612
McMurray, S. E., Johnson, Z. I., Hunt, D. E., Pawlik, J. R., and Finelli, C. M. (2016). Selective feeding by the giant barrel sponge enhances foraging efficiency. Limnol. Oceanogr. 61, 1271–1286. doi: 10.1002/lno.10287
McMurray, S. E., Stubler, A. D., Erwin, P. M., Finelli, C. M., and Pawlik, J. R. (2018). A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Ser. 588, 1–14. doi: 10.3354/meps12466
Miloslavich, P., Díaz, J. M., Klein, E., Alvarado, J. J., Díaz, C., Gobin, J., et al. (2010). Marine biodiversity in the caribbean: regional estimates and distribution patterns. PLoS One 5:e11916. doi: 10.1371/journal.pone.0011916
Mohamed, N. M., Colman, A. S., Tal, Y., and Hill, R. T. (2008). Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10, 2910–2921. doi: 10.1111/j.1462-2920.2008.01704.x
Mohanty, I., Tapadar, S., Moore, S. G., Biggs, J. S., Freeman, C. J., and Gaul, D. A. (2021). Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures. mSystems 6, 1–17.
Moitinho-Silva, L., Nielsen, S., Amir, A., Gonzalez, A., Ackermann, G. L., Cerrano, C., et al. (2017). The sponge microbiome project. Gigascience 6, 1–7. doi: 10.1093/gigascience/gix077
Moitinho-Silva, L., Seridi, L., Ryu, T., Voolstra, C. R., Ravasi, T., and Hentschel, U. (2014). Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ. Microbiol. 16, 3683–3698. doi: 10.1111/1462-2920.12533
Montalvo, N. F., and Hill, R. T. (2011). Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl. Environ. Microbiol 77, 7207–7216. doi: 10.1128/AEM.05285-11
Morganti, T., Coma, R., Yahel, G., and Ribes, M. (2017). Trophic niche separation that facilitates co-existence of high and low microbial abundance sponges is revealed by in situ study of carbon and nitrogen fluxes. Limnol. Oceanogr. 62, 1963–1983. doi: 10.1002/lno.10546
Morganti, T. M., Ribes, M., Moskovich, R., Weisz, J. B., Yahel, G., and Coma, R. (2021). In situ pumping rate of 20 marine demosponges is a function of osculum area. Front. Mar. Sci. 8:19.
Morley, S. A., Berman, J., Barnes, D. K. A., de Juan Carbonell, C., Downey, R. V., and Peck, L. S. (2016). Extreme phenotypic plasticity in metabolic physiology of Antarctic Demosponges. Front. Ecol. Evol. 3:157. doi: 10.3389/fevo.2015.00157
Morrow, K. M., Fiore, C. L., and Lesser, M. P. (2016). Environmental drivers of microbial community shifts in the giant barrel sponge, Xestospongia muta, over a shallow to mesophotic depth gradient. Environ. Microbiol. 18, 2025–2038. doi: 10.1111/1462-2920.13226
Mueller, B., de Goeij, J. M., Vermeij, M. J. A., Mulders, Y., van der Ent, E., Ribes, M., et al. (2014). Natural diet of coral-excavating sponges consists mainly of dissolved organic carbon (DOC). PLoS One 9:e90152. doi: 10.1371/journal.pone.0090152
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Nagata, T. (2000). “Production mechanisms of dissolved organic matter,” in Microbial Ecology of the Oceans, ed. D. L. Kirchman (Hoboken, NJ: Wiley Series in Ecological and Applied Microbiology), 121–152.
Neish, A. S. (2009). Microbes in gastrointestinal health and disease. Gastroenterology 136, 65–80. doi: 10.1053/j.gastro.2008.10.080
Nielsen, C. (2019). Early animal evolution: a morphologist’s view. R. Soc. Open Sci. 6:190638. doi: 10.1098/rsos.190638
Northfield, T. D., Snyder, G. B., Ives, A. R., and Snyder, W. E. (2010). Niche saturation reveals resource partitioning among consumers. Ecol. Lett. 13, 338–348. doi: 10.1111/j.1461-0248.2009.01428.x
Nuzhdin, S. V., Friesen, M. L., and McIntyre, L. M. (2012). Genotype-phenotype mapping in a post-GWAS world. Trends Genet. 28, 421–426. doi: 10.1016/j.tig.2012.06.003
O’Brien, P. A., Webster, N. S., Miller, D. J., and Bourne, D. G. (2019). Host-microbe coevolution: Applying evidence from model systems to complex marine invertebrate holobionts. mBio 10:e02241-18. doi: 10.1128/mBio.02241-18
Ogawa, H., Amagai, Y., Koike, I., Kaiser, K., and Benner, R. (2001). Production of refractory dissolved organic matter by bacteria. Science 292, 917–920. doi: 10.1126/science.1057627
Olson, J. B., and Gao, X. (2013). Characterizing the bacterial associates of three Caribbean sponges along a gradient from shallow to mesophotic depths. FEMS Microbiol. Ecol. 85, 74–84. doi: 10.1111/1574-6941.12099
Paul, V. J., Freeman, C. J., and Agarwal, V. (2019). Chemical ecology of marine sponges: new opportunities through “-Omics.”. Integr. Comp. Biol. 59, 765–776. doi: 10.1093/icb/icz014
Pawlik, J. R. (2011). The chemical ecology of sponges on caribbean reefs: natural products shape natural systems. Bioscience 61, 888–898. doi: 10.1525/bio.2011.61.11.8
Pawlik, J. R., Burkepile, D. E., and Thurber, R. V. (2016). A vicious circle? Altered carbon and nutrient cycling may explain the low resilience of Caribbean coral reefs. Bioscience 66, 470–476. doi: 10.1093/biosci/biw047
Pawlik, J. R., Henkel, T. P., McMurray, S. E., López-Legentil, S., Loh, T. L., and Rohde, S. (2008). Patterns of sponge recruitment and growth on a shipwreck corroborate chemical defense resource trade-off. Mar. Ecol. Prog. Ser. 368, 137–143. doi: 10.3354/meps07615
Pawlik, J. R., Loh, T.-L., and McMurray, S. E. (2018). A review of bottom-up vs. top-down control of sponges on Caribbean fore-reefs: what’s old, what’s new, and future directions. PeerJ 6:e4343. doi: 10.7717/peerj.4343
Pawlik, J. R., and McMurray, S. E. (2019). The emerging ecological and biogeochemical importance of sponges on coral reefs. Ann. Rev. Mar. Sci. 12, 315–337. doi: 10.1146/annurev-marine-010419-010807
Pawlik, J. R., McMurray, S. E., Erwin, P., and Zea, S. (2015). A review of evidence for food limitation of sponges on Caribbean reefs. Mar. Ecol. Prog. Ser. 519, 265–283. doi: 10.3354/meps11093
Phillips, C. D., Hanson, J., Wilkinson, J. E., Koenig, L., Rees, E., Webala, P., et al. (2017). Microbiome structural and functional interactions across host dietary niche space. Integr. Comp. Biol. 57, 743–755. doi: 10.1093/icb/icx011
Pita, L., Rix, L., Slaby, B. M., Franke, A., and Hentschel, U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46. doi: 10.1186/s40168-018-0428-1
Podell, S., Blanton, J. M., Neu, A., Agarwal, V., Biggs, J. S., Moore, B. S., et al. (2019). Pangenomic comparison of globally distributed Poribacteria associated with sponge hosts and marine particles. ISME J. 13, 468–481. doi: 10.1038/s41396-018-0292-9
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9:4921. doi: 10.1038/s41467-018-07275-x
Poppell, E., Weisz, J., Spicer, L., Massaro, A., Hill, A., and Hill, M. (2013). Sponge heterotrophic capacity and bacterial community structure in high- and low-microbial abundance sponges. Mar. Ecol. 35, 414–424. doi: 10.1111/maec.12098
Porter, J. W. (1976). Autotrophy, heterotrophy, and resource partitioning in Caribbean reef-building corals. Am. Nat. 110, 731–742. doi: 10.1086/283100
Radax, R., Rattei, T., Lanzen, A., Bayer, C., Rapp, H. T., Urich, T., et al. (2012). Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. 14, 1308–1324. doi: 10.1111/j.1462-2920.2012.02714.x
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Ramsby, B., Heishman, J., Hoogenboom, M., Whalan, S., and Webster, N. S. (2020). Dissolved inorganic nutrient enrichment does not affect sponge growth or condition. Mar. Ecol. Prog. Ser. 634, 77–88. doi: 10.3354/meps13184
Reichelt, R. E., Loya, Y., and Bradbury, R. H. (1986). Patterns in the use of space by benthic communities on two coral reefs of the Great Barrier Reef. Coral Reefs 5, 73–79. doi: 10.1007/BF00270355
Reiswig, H. M. (1971). Particle feeding in natural populations of three marine Demosponges. Biol. Bull. 141, 568–591. doi: 10.2307/1540270
Reiswig, H. M. (1973). Population dynamics of three Jamaican Demospongiae. Univ. Miami - Rosentiel Sch. Mar. Atmos. Sci. 23, 191–226.
Reiswig, H. M. (1974). Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Bio. Ecol. 14, 231–249. doi: 10.1016/0022-0981(74)90005-7
Reiswig, H. M. (1975a). Bacteria as food for temperate-water marine sponges. Can. J. Zool. 53, 582–589. doi: 10.1139/z75-072
Reiswig, H. M. (1975b). The aquiferous systems of three marine Demospongiae. J. Morphol. 145, 493–502. doi: 10.1002/jmor.1051450407
Reiswig, H. M. (1981). Partial carbon and energy budgets of the bacteriosponge Verongula fistularis (Porifera: Demospongiae) in Barbados. Mar. Ecol. 2, 273–293. doi: 10.1111/j.1439-0485.1981.tb00271.x
Reveillaud, J., Maignien, L., Eren, A. M., Huber, J. A., Apprill, A., Sogin, M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209. doi: 10.1038/ismej.2013.227
Ribes, M., Coma, R., and Gili, J. (1999). Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 176, 179–190. doi: 10.3354/meps176179
Ribes, M., Jiménez, E., Yahel, G., López-Sendino, P., Diez, B., Massana, R., et al. (2012). Functional convergence of microbes associated with temperate marine sponges. Environ. Microbiol. 14, 1224–1239. doi: 10.1111/j.1462-2920.2012.02701.x
Rix, L., De Goeij, J. M., Van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2018). Reef sponges facilitate the transfer of coral-derived organic matter to their associated fauna via the sponge loop. Mar. Ecol. Prog. Ser. 589, 85–96. doi: 10.3354/meps12443
Rix, L., Ribes, M., Coma, R., Jahn, M. T., de Goeij, J. M., van Oevelen, D., et al. (2020). Heterotrophy in the earliest gut: a single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 14, 2554–2567. doi: 10.1038/s41396-020-0706-3
Robbins, S. J., Song, W., Engelberts, J. P., Glasl, B., Slaby, B. M., Boyd, J., et al. (2021). A genomic view of the microbiome of coral reef demosponges. ISME J. 15, 1641–1654. doi: 10.1038/s41396-020-00876-9
Roberts, D. E., Davis, A. R., and Cummings, S. P. (2006). Experimental manipulation of shade, silt, nutrients and salinity on the temperate reef sponge Cymbastela concentrica. Mar. Ecol. Prog. Ser. 307, 143–154. doi: 10.3354/meps307143
Roff, G., and Mumby, P. J. (2012). Global disparity in the resilience of coral reefs. Trends Ecol. Evol. 27, 404–413. doi: 10.1016/j.tree.2012.04.007
Rützler, K. (1990). “Associations between Caribbean sponges and photosynthetic organisms,” in New Perspectives in Sponge Biology, ed. K. Rützler (Washington, DC: Smithsonian Institution Press), 455–466.
Sacristán-Soriano, O., Turon, X., and Hill, M. (2020). Microbiome structure of ecologically important bioeroding sponges (family Clionaidae): the role of host phylogeny and environmental plasticity. Coral Reefs 39, 1285–1298. doi: 10.1007/s00338-020-01962-2
Salem, H., Bauer, E., Strauss, A. S., Vogel, H., Marz, M., and Kaltenpoth, M. (2014). Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. R. Soc. B Biol. Sci. 281:20141838. doi: 10.1098/rspb.2014.1838
Santavy, D. L., Courtney, L. A., Fisher, W. S., Quarles, R. L., and Jordan, S. J. (2013). Estimating surface area of sponges and gorgonians as indicators of habitat availability on Caribbean coral reefs. Hydrobiologia 707, 1–16. doi: 10.1007/s10750-012-1359-7
Schläppy, M. L., Schöttner, S. I., Lavik, G., Kuypers, M. M. M., de Beer, D., and Hoffmann, F. (2010). Evidence of nitrification and denitrification in high and low microbial abundance sponges. Mar. Biol. 157, 593–602. doi: 10.1007/s00227-009-1344-5
Schoener, T. W. (1974). Resource partitioning in ecological communities. Science 185, 27–39. doi: 10.1126/science.185.4145.27
Schöttner, S., Hoffmann, F., Cárdenas, P., Rapp, H. T., Boetius, A., and Ramette, A. (2013). Relationships between host phylogeny, host type and bacterial community diversity in cold-water coral reef sponges. PLoS One 8:e55505. doi: 10.1371/journal.pone.0055505
Schluter, D. (2009). Evidence for ecological speciation and its alternative. Science 323, 737–742. doi: 10.1126/science.1160006
Sharp, K. H., Eam, B., Faulkner, D. J., and Haygood, M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629. doi: 10.1128/AEM.01493-06
Siegl, A., Kamke, J., Hochmuth, T., Piel, J., Richter, M., Liang, C., et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70. doi: 10.1038/ismej.2010.95
Slaby, B. M., Hackl, T., Horn, H., Bayer, K., and Hentschel, U. (2017). Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 11, 2465–2478. doi: 10.1038/ismej.2017.101
Southward, A. J., Southward, E. C., Dando, P. R., Rau, G. H., Felbeck, H., and Flügel, H. (1981). Bacterial symbionts and low 13C/12C ratios in tissues of Pogonophora indicate unusual nutrition and metabolism. Nature 293, 616–619. doi: 10.1038/293616a0
Southwell, M. W., Popp, B. N., and Martens, C. S. (2008). Nitrification controls on fluxes and isotopic composition of nitrate from Florida Keys sponges. Mar. Chem. 108, 96–108. doi: 10.1016/j.marchem.2007.10.005
Stearns, S. C. (1977). The evolution of life history traits: a critique of the theory and a review of the data. Annu. Rev. Ecol. Syst. 8, 145–171. doi: 10.1146/annurev.es.08.110177.001045
Steindler, L., Huchon, D., Avni, A., and Ilan, M. (2005). 16S rRNA phylogeny of sponge-associated Cyanobacteria. Appl. Environ. Microbiol. 71, 4127–4131. doi: 10.1128/AEM.71.7.4127-4131.2005
Steinert, G., Busch, K., Bayer, K., Kodami, S., Arbizu, P. M., Kelly, M., et al. (2020). Compositional and quantitative insights into bacterial and archaeal communities of South Pacific deep-sea sponges (Demospongiae and Hexactinellida). Front. Microbiol. 11:716. doi: 10.3389/fmicb.2020.00716
Stephens, G. C., and Schinske, R. A. (1961). Uptake of amino acids by marine invertebrates. Limnol. Oceanogr. 6, 175–181. doi: 10.4319/lo.1961.6.2.0175
Suttle, C. A., Chan, A. M., and Fuhrman, J. A. (1991). Dissolved free amino acids in the Sargasso Sea: uptake and respiration rates, turnover times, and concentrations. Mar. Ecol. Prog. Ser. 70, 189–199. doi: 10.3354/meps070189
Swierts, T., Cleary, D. F. R., and de Voogd, N. J. (2018). Prokaryotic communities of Indo-Pacific giant barrel sponges are more strongly influenced by geography than host phylogeny. FEMS Microbiol. Ecol. 94:fiy194. doi: 10.1093/femsec/fiy194
Swierts, T., Peijnenburg, K. T. C. A., de Leeuw, C. A., Breeuwer, J. A. J., Cleary, D. F. R., and de Voogd, N. J. (2017). Globally intertwined evolutionary history of giant barrel sponges. Coral Reefs 36, 933–945. doi: 10.1007/s00338-017-1585-6
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/MMBR.00040-06
Taylor, M. W., Tsai, P., Simister, R. L., Deines, P., Botte, E., Ericson, G., et al. (2013). ‘Sponge-specific’ bacteria are widespread (but rare) in diverse marine environments. ISME J. 7, 438–443. doi: 10.1038/ismej.2012.111
Thacker, R. W. (2005). Impacts of shading on sponge-Cyanobacteria symbioses: a comparison between host-specific and generalist associations. Integr. Comp. Biol. 45, 369–376. doi: 10.1093/icb/45.2.369
Thacker, R. W., Diaz, M. C., Rützler, K., Erwin, P. M., Kimble, S. J. A., Pierce, M. J., et al. (2007). “Phylogenetic relationships among the filamentous cyanobacterial symbionts of Caribbean sponges and a comparison of photosynthetic production between sponges hosting filamentous and unicellular cyanobacteria,” in Porifera Research: Biodiversity, Innovation, and Sustainability, eds M. R. Custódio, G. Lobo-Hajdu, E. Hajdu, and G. Muricy (Rio de Janeiro: Museu Nacional Rio de Janeiro), 621–626.
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 1–12. doi: 10.1038/ncomms11870
Thomas, T., Rusch, D., DeMaere, M. Z., Yung, P. Y., Lewis, M., Halpern, A., et al. (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4, 1557–1567. doi: 10.1038/ismej.2010.74
Vacelet, J., and Boury-Esnault, N. (1995). Carnivorous sponges. Nature 373, 333–335. doi: 10.1038/373333a0
Vacelet, J., and Donadey, C. (1977). Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Bio. Ecol. 30, 301–314. doi: 10.1016/0022-0981(77)90038-7
van Duyl, F. C., Moodley, L., Nieuwland, G., van Ijzerloo, L., van Soest, R. W. M., Houtekamer, M., et al. (2011). Coral cavity sponges depend on reef-derived food resources: Stable isotope and fatty acid constraints. Mar. Biol. 158, 1653–1666. doi: 10.1007/s00227-011-1681-z
Vermeij, M. J. A., and Bak, R. P. M. (2002). How are coral populations structured by light? Marine light regimes and the distribution of Madracis. Mar. Ecol. Prog. Ser. 233, 105–116. doi: 10.3354/meps233105
Wang, S., Meyer, E., McKay, J. K., and Matz, M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810. doi: 10.1038/nmeth.2023
Webster, N. S., Cobb, R. E., and Negri, A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2, 830–842. doi: 10.1038/ismej.2008.42
Webster, N. S., and Taylor, M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Webster, N. S., and Thomas, T. (2016). The sponge hologenome. mBio 7:e00135-16. doi: 10.1128/mbio.00135-16
Weisz, J. B., Hentschel, U., Lindquist, N., and Martens, C. S. (2007). Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar. Biol. 152, 475–483. doi: 10.1007/s00227-007-0708-y
Weisz, J. B., Lindquist, N., and Martens, C. S. (2008). Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155, 367–376. doi: 10.1007/s00442-007-0910-0
Wilkinson, C., and Garrone, R. (1980). “Nutrition of marine sponges. Involvement of symbiotic bacteria in the uptake of dissolved organic carbon,” in Nutrition in the Lower Metazoa, eds D. C. Smith and Y. Tiffon (Oxford: Pergamon Press), 157–161. doi: 10.1016/B978-0-08-025904-8.50016-X
Wilkinson, C. R. (1978a). Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49, 161–167. doi: 10.1007/BF00387115
Wilkinson, C. R. (1978b). Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Mar. Biol. 49, 169–176. doi: 10.1007/BF00387116
Wilkinson, C. R. (1978c). Microbial associations in sponges. III. Ultrastructure of the in situ associations in coral reef sponges. Mar. Biol. 49, 177–185. doi: 10.1007/BF00387117
Wilkinson, C. R. (1981). “Significance of sponges with cyanobacteria symbionts on Davies Reef, Great Barrier Reef,” in Proceedings of the 64h International Coral Reef Symposium, Manila, 705–712.
Wilkinson, C. R. (1983). Net primary productivity in coral reef sponges. Science 219, 410–412. doi: 10.1126/science.219.4583.410
Wilkinson, C. R. (1987). Interocean differences in size and nutrition of coral reef sponge populations. Science 236, 1654–1657. doi: 10.1126/science.236.4809.1654
Wilkinson, C. R., and Cheshire, A. C. (1988). Cross-shelf variations in coral reef structure and function-influences of land and ocean. Proc. 6th Int. Coral Reef Symp. 1, 227–233.
Wilkinson, C. R., and Cheshire, A. C. (1989). Patterns in the distribution of sponge populations across the central Great Barrier Reef. Coral Reefs 8, 127–134. doi: 10.1007/BF00338268
Wilkinson, C. R., and Cheshire, A. C. (1990). Comparisons of sponge populations across the Barrier Reefs of Australia and Belize: evidence for higher productivity in the Caribbean. Mar. Ecol. Prog. Ser. 67, 285–294. doi: 10.3354/meps067285
Wilkinson, C. R., and Trott, L. A. (1985). Light as a factor determining the distribution of sponges across the central Great Barrier Reef. Proc. 5th Int. Coral Reef Symp. 5, 125–130.
Wilkinson, C. R., and Vacelet, J. (1979). Transplantation of marine sponges to different conditions of light and current. J. Exp. Mar. Bio. Ecol. 37, 91–104. doi: 10.1016/0022-0981(79)90028-5
Wilkinson, C. R., Cheshire, A. C., Klumpp, D. W., and McKinnon, A. D. (1988). “Nutritional spectrum of animals with photosynthetic symbionts-corals and sponges,” in Proceedings of the 6th International Coral Reef Symposium, Townsville: Coral Reef Symposium Executive Committee, 27–30.
Wismer, S., Hoey, A. S., and Bellwood, D. R. (2009). Cross-shelf benthic community structure on the Great Barrier Reef: relationships between macroalgal cover and herbivore biomass. Mar. Ecol. Prog. Ser. 376, 45–54. doi: 10.3354/meps07790
Wooster, M. K., McMurray, S. E., Pawlik, J. R., Morán, X. A. G., and Berumen, M. L. (2019). Feeding and respiration by giant barrel sponges across a gradient of food abundance in the Red Sea. Limnol. Oceanogr. 64, 1790–1801. doi: 10.1002/lno.11151
Wulff, J. (2017). Bottom-up and top-down controls on coral reef sponges: disentangling within-habitat and between-habitat processes. Ecology 98, 1130–1139. doi: 10.1002/ecy.1754
Wulff, J. L. (2006). Resistance vs recovery: morphological strategies of coral reef sponges. Funct. Ecol. 20, 699–708. doi: 10.1111/j.1365-2435.2006.01143.x
Wulff, J. L. (2020). Targeted predator defenses of sponges shape community organization and tropical marine ecosystem function. Ecol. Monogr. 91:e01438. doi: 10.1002/ecm.1438
Yahel, G., Sharp, J. H., Marie, D., Häse, C., and Genin, A. (2003). In situ feeding and element removal in the symbiont-bearing sponge Theonella swinhoei: Bulk DOC is the major source for carbon. Limnol. Oceanogr. 48, 141–149. doi: 10.4319/lo.2003.48.1.0141
Zhang, D., Sun, W., Feng, G., Zhang, F., Anbuchezhian, R., Li, Z., et al. (2015). Phylogenetic diversity of sulphate-reducing Desulfovibrio associated with three South China Sea sponges. Lett. Appl. Microbiol. 60, 504–512. doi: 10.1111/lam.12400
Zhang, F., Blasiak, L. C., Karolin, J. O., Powell, R. J., Geddes, C. D., and Hill, R. T. (2015). Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc. Natl. Acad. Sci. U.S.A. 112, 4381–4386. doi: 10.1073/pnas.1423768112
Zhang, F., Jonas, L., Lin, H., and Hill, R. T. (2019). Microbially mediated nutrient cycles in marine sponges. FEMS Microbiol. Ecol. 95:fiz155. doi: 10.1093/femsec/fiz155
Zhang, F., Vicente, J., and Hill, R. T. (2014). Temporal changes in the diazotrophic bacterial communities associated with Caribbean sponges Ircinia stroblina and Mycale laxissima. Front. Microbiol. 5:561. doi: 10.3389/fmicb.2014.00561
Keywords: Porifera, microbial symbionts, niche partitioning, competition, Caribbean
Citation: Freeman CJ, Easson CG, Fiore CL and Thacker RW (2021) Sponge–Microbe Interactions on Coral Reefs: Multiple Evolutionary Solutions to a Complex Environment. Front. Mar. Sci. 8:705053. doi: 10.3389/fmars.2021.705053
Received: 04 May 2021; Accepted: 22 June 2021;
Published: 20 July 2021.
Edited by:
Aldo Cróquer, The Nature Conservancy, Dominican RepublicReviewed by:
Ryan McMinds, University of South Florida, United StatesAlejandra Hernandez-Agreda, California Academy of Sciences, United States
Torsten Thomas, University of New South Wales, Australia
Copyright © 2021 Freeman, Easson, Fiore and Thacker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher J. Freeman, ZnJlZW1hbmNqQGNvZmMuZWR1
 Christopher J. Freeman
Christopher J. Freeman Cole G. Easson
Cole G. Easson Cara L. Fiore
Cara L. Fiore Robert W. Thacker
Robert W. Thacker