- 1Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Guangzhou Scientific Observing and Experimental Station of National Fisheries Resources and Environment, Guangzhou, China
- 3Scientific Observing and Experimental Station of Fishery Resources and Environment in the Middle and Lower Reaches of Pearl River, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 4Key Laboratory of Aquatic Animal Immune Technology of Guangdong Province, Guangzhou, China
Grass carp (Ctenopharyngodon idellus) is one of the most economically important aquaculture species and is widely cultured in China. However, its wild populations in many rivers are increasingly declining, and seawater intrusion is one of the most important threats to their survival. However, the mechanisms underlying the decline due to salinity pressure are still unknown. Here, we performed a comparative transcriptome analysis of C. idellus larvae in response to salinity exposures; a total of 481 differentially expressed genes (DEGs) were identified. These DEGs were significantly enriched in eight Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, among which steroid biosynthesis was the most important one, with the highest enrichment score. The pathway plays an important role in the development of the testes and ovary. Interestingly, all DEGs in steroid biosynthesis showed a down regulation, indicating that salinity exposure may pose damage to the fertility of C. idellus. Furthermore, three immunity-associated pathways (cytokine–cytokine receptor interaction, Toll-like receptor signaling pathway, and NOD-like receptor signaling pathway) were also significantly enriched, suggesting impaired immunity and a high risk of disease infection under salinity exposure. Overall, damage to both fertility and immunity would decrease the number of offspring and increase the risk of death due to disease infection. Our results provide a potential molecular mechanism underlying the decline of wild C. idellus populations in the Pearl River.
Introduction
Seawater intrusion is a natural phenomenon along shores. However, seawater intrusion has intensified and frequently occurs downstream of rivers in recent years mainly due to global climate warming, which is accelerating the rise of sea levels (Melloul and Collin, 2006; Werner and Simmons, 2010; Werner et al., 2013). Seawater intrusion to inland river regions can increase the salinity concentrations and thus pose a threat to freshwater species (Nguyen et al., 2009). High salinity can affect physiological and biochemical reactions, such as fish growth and survival, by decreasing the digestive enzyme activity, immunity, and metabolic activity (Tong et al., 2007; Kang et al., 2013). Hence, it is an urgent need to understand the mechanisms underlying the negative influence of seawater intrusion on freshwater species.
Grass carp (Ctenopharyngodon idellus) is one of the most economically important aquaculture species due to its high value as food and is widely cultured in China. In 2019, grass carp yields account for 21.72% of the freshwater aquaculture products in China (Fishery Statistical Yearbook of China, 2020), highlighting its importance in freshwater aquaculture. Grass carp is naturally distributed in lakes, streams, rivers, and other water systems in the wild. However, its wild resource in many systems, such as the Pearl River, has declined rapidly in the past years (Fishery Statistical Yearbook of China, 2017, 2018, 2019, 2020). The decline not only poses a threat to the protection of biodiversity in freshwater realms but also has potential negative effects on the aquaculture industry of grass carp, given that wild populations can provide excellent germplasm resources to avoid inbreeding depression (Berthelot et al., 2014; Lu and Luo, 2020). Seawater intrusion is considered as one of the major environmental factors responsible for the decline of its wild populations (Melloul and Collin, 2006; Nguyen et al., 2009; Werner and Simmons, 2010; Kang et al., 2013). However, the mechanisms underlying the decline due to salinity pressure are still unknown.
RNA sequencing (RNA-seq) is a powerful technology and has been widely used for the identification of crucial genes targeted by external pressures in many aquaculture species, such as catfish (Ictalurus spp.) (Li et al., 2012), yellow catfish (Pelteobagrus fulvidraco) (Zou et al., 2015), Mandarin fish (Siniperca chuatsi) (Hu et al., 2015), top mouth culter (Culter alburnus) (Zhao et al., 2016), Gymnocypris namensis (Luo et al., 2020), and rainbow trout (Zhou et al., 2021). For example, RNA-seq was used to identify the differentially expressed genes (DEGs) of grass carp liver imposed by ammonia exposure. The results showed that the DEGs were significantly enriched in the MAPK, FOXO, AMPK, and NF-kB signaling pathways, cytokine–cytokine receptor interaction, and the apoptosis signaling pathway. These pathways are mainly involved in apoptosis, indicating that grass carp liver apoptosis was induced by ammonia exposure through apoptosis pathways (Jin et al., 2017). Moreover, the draft genome of grass carp (Hu and Chen, 2015) can provide a reference for read mapping, promoting accuracy of subsequent analysis.
In this study, we performed comparative transcriptome expression analyses using RNA-seq technology in grass carp after salinity exposure for 48 h to identify DEGs and characterize the function categories of these DEGs, aiming to understand the mechanisms underlying death due to salinity pressure in the wild. The study would allow us to comprehensively understand the molecular mechanisms behind the population decline of grass carp in the freshwater realm.
Materials and Methods
The procedures used in this study were approved by the Pearl River Fisheries Research Institute Animal Care Advisory Committee.
Sample Collection and Acclimation
Grass carp (2.0 ± 0.4 g) were purchased from an aquaculture farm (Huashan Yueqiangfeng Aquaculture Farm, Huadu District, Guangzhou, China) in November 2019. Fish were transported to our laboratory using plastic bags specially designed for fish transportation, and they were acclimatized in plastic tanks (4 × 200 L) with freshwater for 1 week. During the period of acclimation, grass carps were fed twice a day with commercial diet (Tongwei, Chengdu, China). Feeding was stopped 24 h before the experiment. After acclimation, a total of 90 fish were randomly selected and evenly divided into three groups: the first group was used for tolerance test to confirm the salinity concentration used in this experiment; the second and third groups were used as the experimental group and the control group, respectively.
Acute Salinity Stress
In order to confirm the salinity concentration used in the experimental group, we carried out a tolerance test of grass carp in different salinity gradients, which were prepared with sea crystal (25 kg/bag, purchased from Shanghai Yuepeng, Shanghai, China). The results showed that the 50% lethal concentration (LC50) for grass carp was 0.5 psu at 72 h. Hence, a salinity concentration of 0.5 psu was used for salinity exposure of grass carp. For the control group, there was only fresh water without sea crystal.
The experiment and control groups were kept at 26°C, pH 7.9–8.3. After 48 h, five individuals from each group were randomly chosen and dissected; the gills were collected and transferred into 2-ml tubes with RNA-EZ Reagents D RNA-Be-Locker A. The collected gill samples were kept at 4°C overnight and then transferred into –80°C until the extraction of total RNA.
RNA Extraction, Library Construction, and Transcriptome Sequencing
The total RNA of each sample was extracted according to the instruction manual of the TRlzol Reagent (Life Technologies, Carlsbad, CA, United States). RNA concentration and purity were measured using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, United States). Library construction was performed following the manufacturer’s recommendation in the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States). For the library fragments, purification was performed with the AMPure XP system (Beckman Coulter, Beverly, MA, United States) and size selection was carried out with USER Enzyme (NEB, Ipswich, MA, United States). Adaptor ligation cDNA was yielded at 37°C for 15 min followed by 5 min at 95°C. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. After purification of the PCR products with the AMPure XP system, library quality was assessed on the Agilent Bioanalyzer 2100 system. As a last step, clustering of the indexed samples was performed according to the instruction manual in the cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS (Illumina). The libraries were then sequenced on an Illumina platform and paired-end reads were generated.
Quality Filtration and Read Mapping
Raw data were firstly processed through in-house perl scripts. In this step, clean reads were obtained by removing the reads containing an adapter, ploy-N, and low-quality sequences (low-quality base Q < 10, but up to 50% of a whole read). At the same time, the Q20 (Ewing et al., 1998), Q30, GC content, and the sequence duplication level of the clean data were calculated. Hisat2 (Kim et al., 2015) was used to map the clean reads to the C. idellus genome (Wang et al., 2015). Only reads with a perfect match or with one mismatch were further analyzed and annotated based on the reference genome.
Identification of DEGs
Gene reads were normalized using FPKM (fragments per kilobase of exon per million fragments mapped) values by Cufflinks (Trapnell et al., 2010). DESeq2 (Anders and Huber, 2010) was used to identify the DEGs. DESeq2 provides statistical routines using a model based on a negative binomial distribution. The resulting p-values were adjusted using the Benjamini and Hochberg approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.01 and an absolute value of log2 ratio ≥ 2 were identified as DEGs.
Gene Ontology and KEGG Pathway Enrichment Analysis of DEGs
To achieve standardized gene functional classification, Gene Ontology (GO) analysis based on DEGs was performed by the GOseq R packages with Wallenius’ non-central hypergeometric distribution (Young et al., 2010), which can adjust for gene length bias in DEGs. KOBAS (Mao et al., 2005) was used to test the statistical enrichment of the DEGs in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Real-Time PCR Validation of Genes
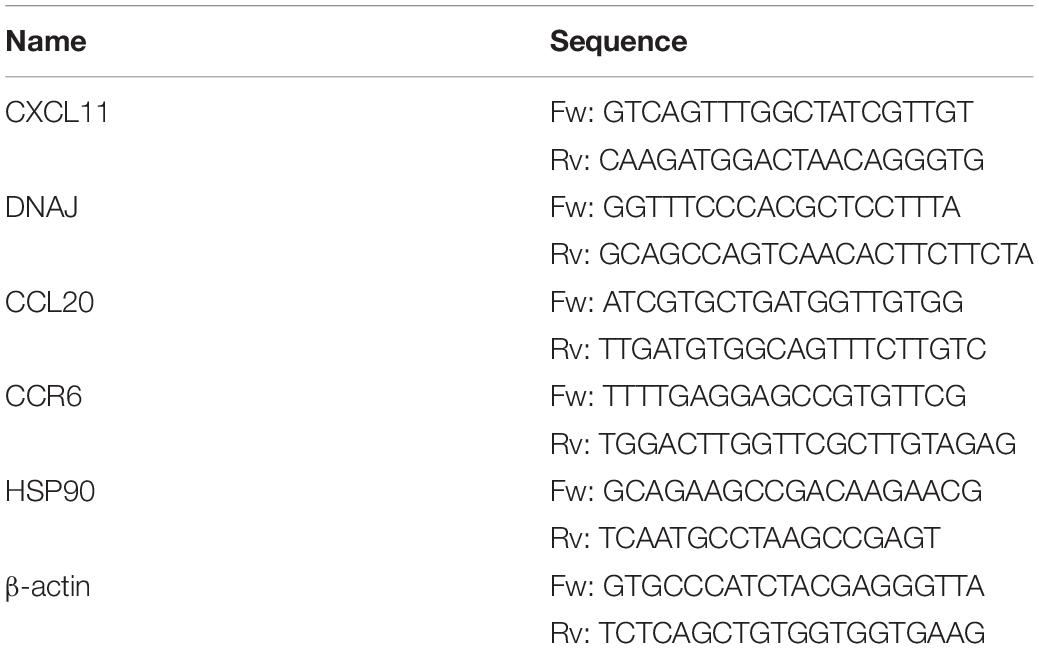
A total of five key genes including three immune-associated DEGs (CXCL11, CCL20, and CCR6) from cytokine–cytokine receptor interaction and two chaperones (HSP90 and DNAJB1) from the NOD-like receptor signaling pathway and protein processing in endoplasmic reticulum were selected for validation using quantitative real-time PCR (qPCR), with the same samples as those for transcriptome analysis. qPCR was performed on a Roche LightCycler 96 real-time PCR system (SN:10328) according to the manufacturer’s instruction. Each PCR reaction was carried out in a total volume of 12.5 μl containing 6.25 μl of the Biomarker 2X SYBR Green Fast qPCR Mix (Biomarker Technologies, Beijing, China), 2.5 μM of each primer, 2 μl of cDNA template, and 3.75 μl of RNase-free ddH2O. The PCR program was composed of a denaturation step of 3 min at 95°C, followed by 45 cycles of 95°C for 5 s, 60°C for 34 s, and 72°C for 10 s. To rule out potential contamination, the negative controls in each run were set up with ddH2O as templates. Melting curve analysis of the PCR products was performed at the end of each PCR reaction to confirm the specificity of the primers. β-actin (Yang et al., 2016) was used as an internal control to normalize the relative expression of mRNA with the 2–ΔΔCt method (Pfaffl, 2001). The forward and reverse primers of the genes for qPCR are shown in Table 1. Statistical analysis was performed using GraphPad Prism (version 8.0.0 for Windows, GraphPad Software, San Diego, CA, United States)1.
Results
Transcriptome Sequencing and DEGs
Transcriptome sequencing produced a total of 61.16 Gb clean data with an average of 6.12 Gb clean data for each sample (Table 2). The clean reads were firstly mapped to the reference genome of C. idellus (Wang et al., 2015). About 87.20% (± 0.62%) reads have only one mapping position, while 2.76% (± 0.13%) reads were mapped multiply (Table 2). The transcriptome sequencing data from this study were deposited in the NCBI Sequence Read Archive (SRA) with the accession number PRJNA725283. To understand the molecular mechanisms underlying the damage to grass carp caused by sea intrusion, we performed comparative transcriptome expression analyses to identify the DEGs between the salinity stress group and the control group. Compared to the control group, we identified a total of 481 DEGs, among which 273 DEGs showed down regulation and 208 DEGs were upregulated.
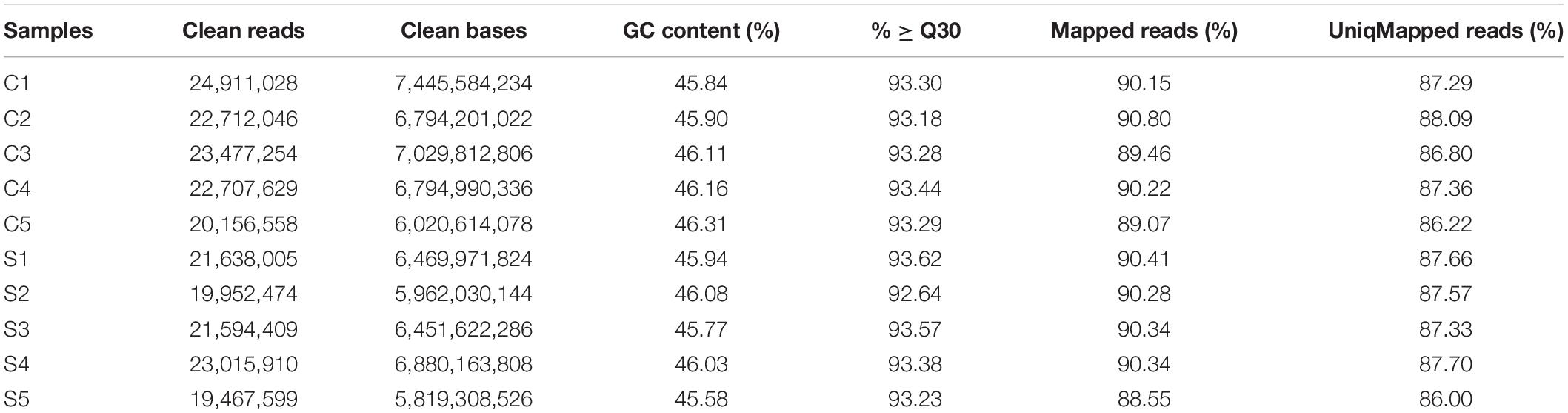
Table 2. Statistics of the sequencing data and sample sequence alignment with the selected reference genomes.
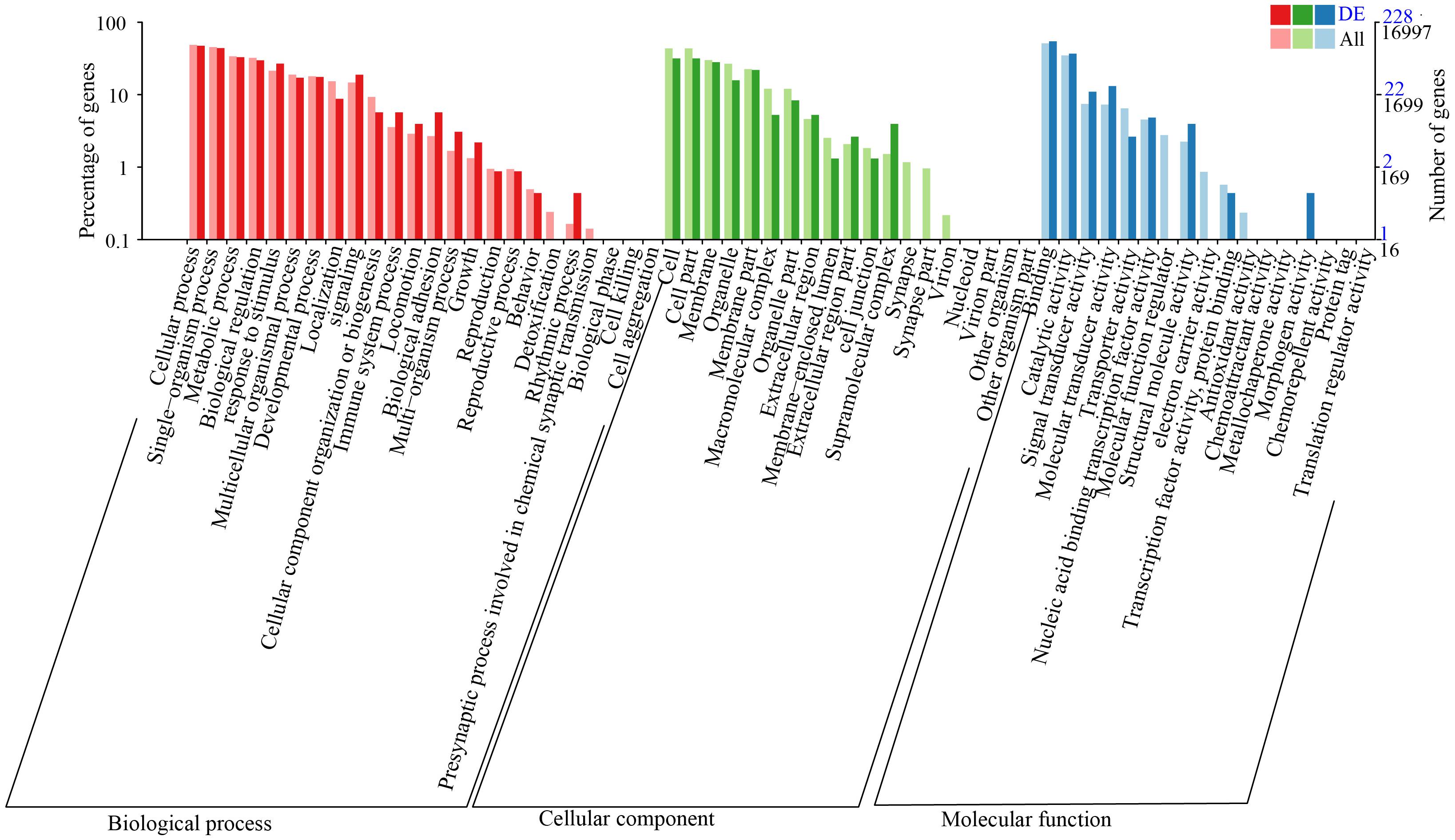
GO Analysis of the DEGs
In order to understand the functions of the DEGs, we firstly performed GO analysis (Figure 1). The results showed that these DEGs were divided into three categories: biological process, cellular component, and molecular function. The category enrichment bar plot based on the DEGs showed that there were significant GO terms (adjusted p < 0.05) in both biological process and molecular function, while no significant GO terms were detected in cellular component, highlighting the importance of both biological process and molecular function in response to salinity exposure (Figure 2). In the biological process category, response to lipopolysaccharide, inflammatory response, immune response, and receptor-mediated endocytosis were the most represented. Additionally, most molecular function-related DEGs were associated with phosphoenolpyruvate carboxykinase activity and squalene monooxygenase activity.
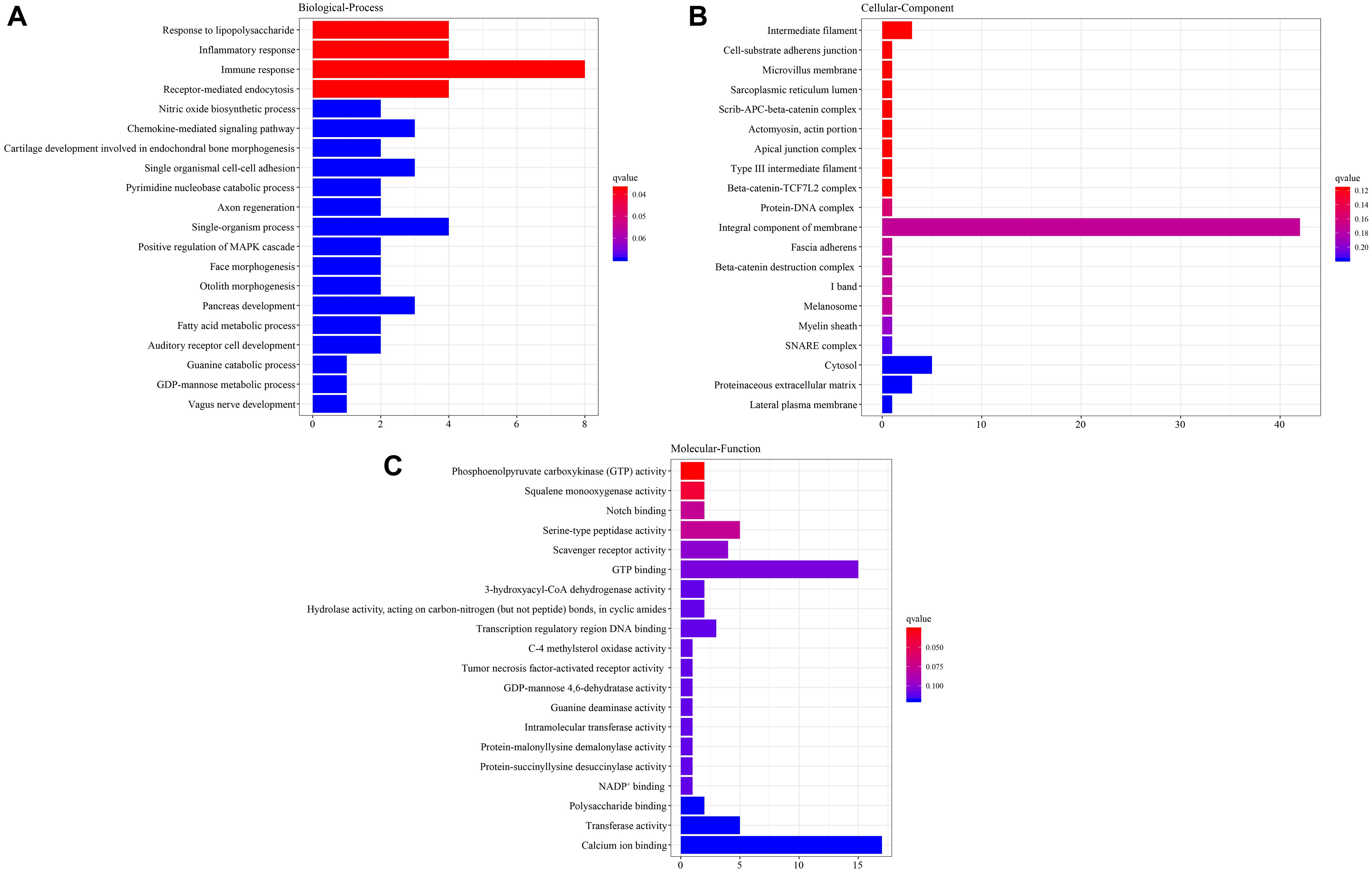
Figure 2. Bar plot of the Gene Ontology (GO) categories of the differentially expressed genes (DEGs). (A) Biological process. (B) Cellular component. (C) Molecular function.
KEGG Analysis of the DEGs
To identify the biological pathways that play an important role in response to salinity exposure, we performed KEGG enrichment analysis against the KEGG database. A total of eight pathways were significantly enriched (Figure 3), including one fertility-related pathway (steroid biosynthesis) with the highest enrichment score, three immunity-related pathways (cytokine–cytokine receptor interaction, Toll-like receptor signaling pathway, and NOD-like receptor signaling pathway), three metabolism-related pathways (fructose and mannose metabolism, purine metabolism, and pyruvate metabolism), and one ion exchange or signal transduction-related pathway (gap junction). Specifically, all DEGs of steroid biosynthesis, cytokine–cytokine receptor interaction, and fructose and mannose metabolism had downregulated expressions (Supplementary Table 1).
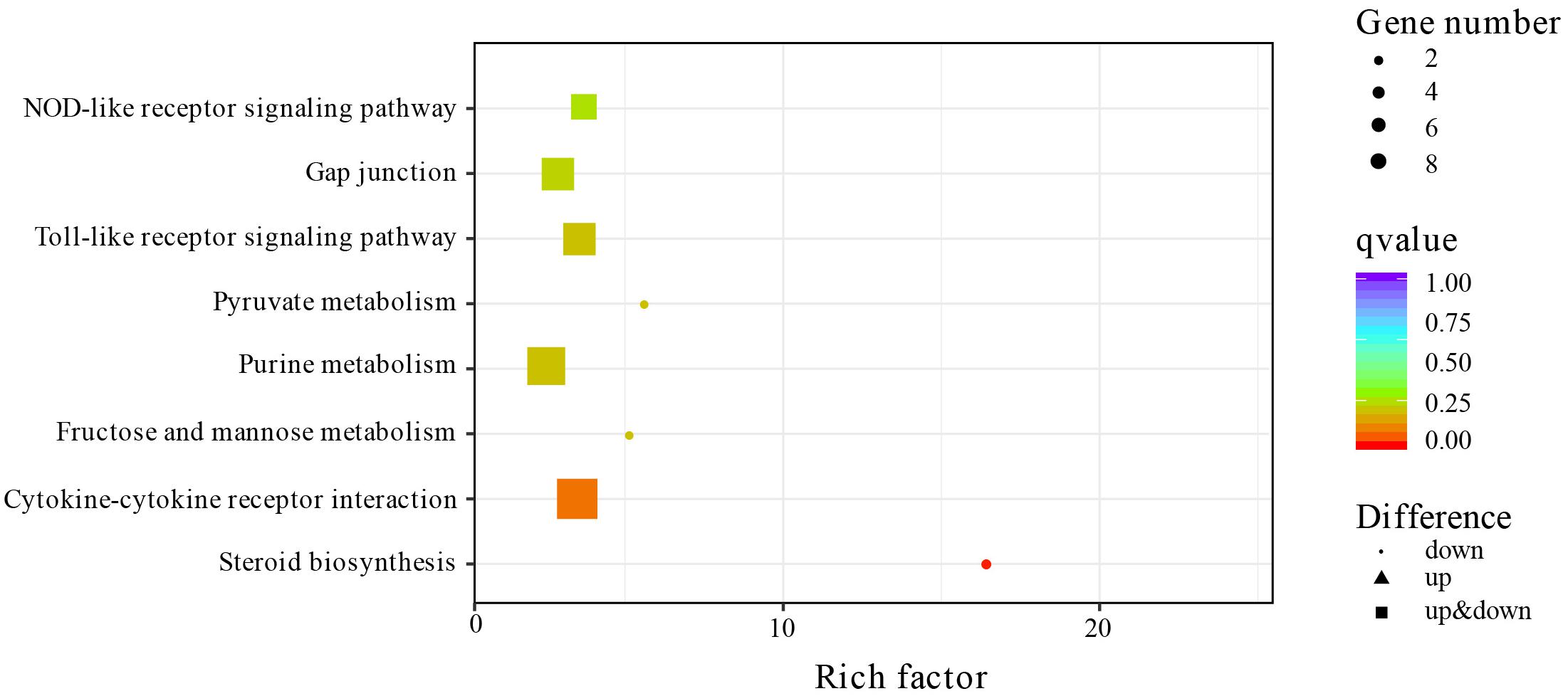
Figure 3. Scatter plot of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the differentially expressed genes (DEGs).
Validation by Real-Time PCR
In order to validate the expression levels of the DEGs in transcriptome sequencing, we performed qPCR to measure the expression levels of five DEGs: heat shock protein 90 alpha (HSP90), C–X–C motif chemokine 11 (CXCL11), chemokine ligand 20 (CCL20), DnaJ homolog subfamily B member 1 (DNAJB1), and C–C chemokine receptor type 6 (CCR6). For all genes, the expression tendencies between transcriptome sequencing and qPCR showed good consistency (Figure 4).
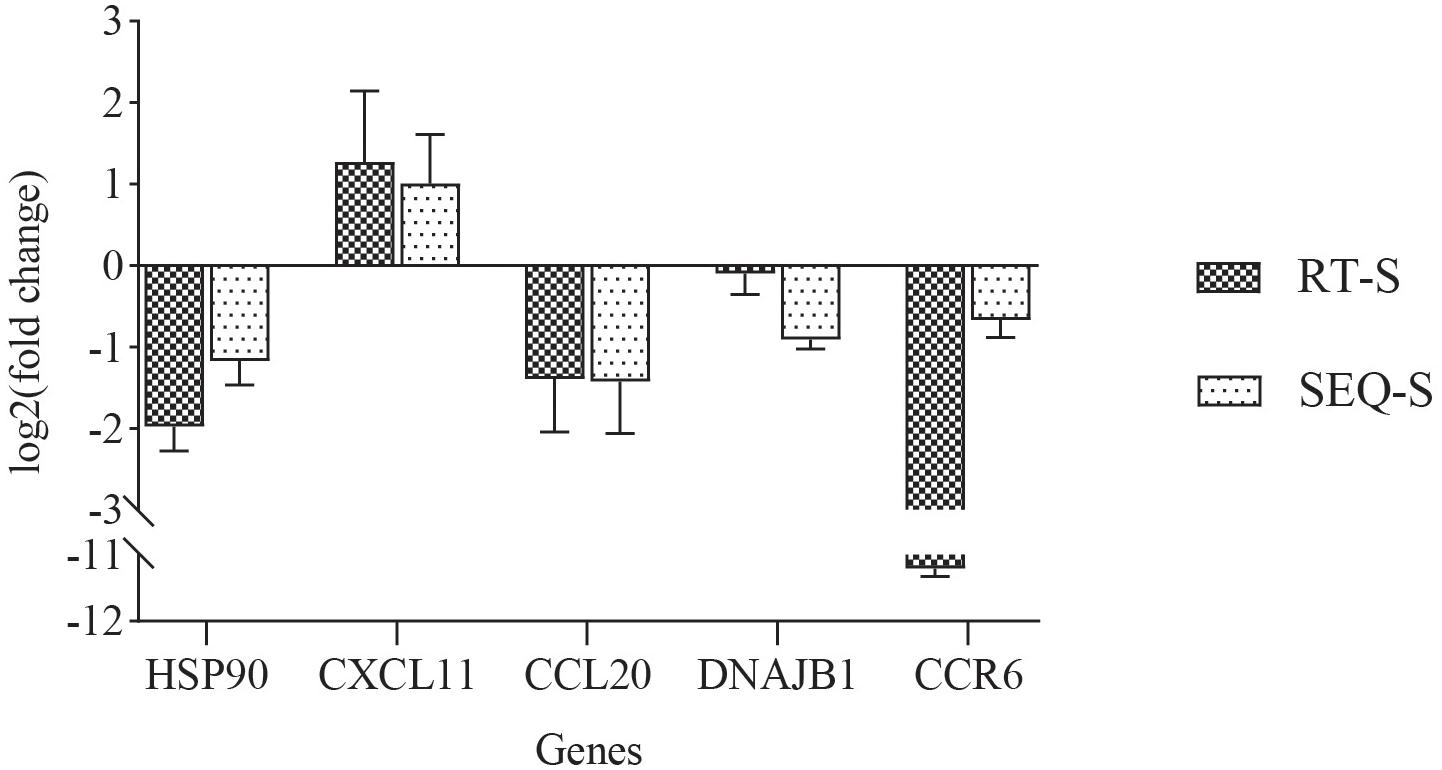
Figure 4. Comparison of the expression profiles of five differentially expressed genes (DEGs) determined by RNA-seq and qPCR in the gill. Each bar indicates the log2(fold change) of a gene’s expression compared to that in the control group.
Discussion
Seawater intrusion is one of the most important threats to freshwater ecosystems. High salinity due to seawater intrusion can induce a change of osmotic pressure, affecting physiology processes and even leading to large-scale death of freshwater species. Here, we identified a total of eight KEGG pathways with high enrichment, among which the steroid biosynthesis pathway had the highest enrichment score. In addition, GO analysis based on the DEGs in this study highlighted the importance of squalene monooxygenase, which participates in synthesis of sterol (Yamamoto and Bloch, 1970; Han et al., 2010). These results suggested that the synthesis of steroid was definitely influenced by salinity exposure. There were four DEGs in this pathway that were annotated to three proteins: lanosterol synthase, methylsterol monooxygenase, and squalene monooxygenase. The first one is widely detected in various organisms (Suzuki et al., 2006) and converts (S)-2,3-epoxysqualene to lanosterol in the biosynthesis of cholesterol (Wada et al., 2020), which is a precursor of steroid hormones and plays a role in maintaining Na+ balance in salt-sensitive hypertension (Cuka et al., 2019). The second one is localized on the endoplasmic reticulum membrane and is believed to function in cholesterol biosynthesis (Li and Kaplan, 1996). The last one is a sub-pathway of the lanosterol biosynthesis pathway (Yalinkaya and Lai, 2000). Steroid had been shown to play an important role in the early development of germ cells in Drosophila (Morris and Spradling, 2012). The downregulated expressions of all these DEGs in this pathway indicated that steroid biosynthesis in larval grass carp was inhibited by salinity exposure, further prohibiting the normal development of spermatocytogenesis and oogenesis and ultimately posing damage to the reproductive capacity of grass carp. The decreased fertility would reduce the number of offspring and may become one of the important mechanisms underlying the population decline of wild grass carp in rivers, such as the Pearl River.
Interestingly, three immunity-related pathways—cytokine–cytokine receptor interaction, Toll-like receptor signaling pathway, and NOD-like receptor signaling pathway—were also significantly enriched based on the DEGs between the salinity exposure group and the control group. Furthermore, four GO terms (response to lipopolysaccharide, inflammatory response, immune response, and receptor-mediated endocytosis) represented in biological process were also immunity-related processes (Raetz and Whitfield, 2002; Dauphinee and Karsan, 2006), providing further evidence of the affected immunity caused by salinity exposure. Cytokine–cytokine receptor interaction is critical in determining the effects of inflammation in the development of diseases (Reeth, 2000) and associates with innate and adaptive inflammatory host defenses (Chen et al., 2000). There are eight DEGs with annotation of seven proteins in the cytokine–cytokine pathway (Supplementary Table 1). Among which, interleukin-8 (IL-8) is a type of neutrophil-activating cytokine that plays a role in inflammation and host defense (Baggiolini and Clark-Lewis, 1992). CXCL11 is associated with inflammatory and angiogenic processes (Liu et al., 2011). CCL20 is the unique chemokine interacting with CCR6 and is essential in promoting the growth of immature dendritic cells and lymphocytes (Schutyser et al., 2003). Another crucial protein, Cytokine receptor family member b8 (preferred name: IL20Rα), is part of the IL20R complex and is associated with both pro- and anti-inflammatory functions (Ha et al., 2020). In this study, two crucial DEGs (IL-8 and CXCL11) were upregulated and three DEGs (CCL20, CCR6, and IL20Rα) were downregulated in the cytokine–cytokine receptor interaction pathway, suggesting that high salinity may change the activation of the DEGs, further damaging the function of cytokine–cytokine receptor interaction. Toll-like receptors (TLRs), the main components of the second pathway, are transmembrane molecules with a leucine-rich repeat motif in the external domain. The structure of Toll-like receptors is traditionally conserved, attributing to their important role in host defense (O’Neill, 2000; Kimbrell and Beutler, 2001; Kaisho and Akira, 2002; Janeway and Medzhitov, 2002). The Toll-like receptor signal pathway could induce inflammatory cytokines in macrophages and dendritic cells, triggering innate and adaptive immune responses (Krieg, 2002; Ozato et al., 2002; Tsujimura et al., 2002). TLR25 is a major member of the teleost TLR1 subfamily, belonging to the group “non-mammalian TLRs” (Temperley et al., 2008; Wang et al., 2016), and activates the transcriptomes of some pro-inflammatory cytokines and contributes to immune response (Lee et al., 2020). The downregulation of TLR25 highlights the dysfunction of the Toll-like receptor signaling pathway. As a main component of the third immunity-related pathway, NOD-like receptors are evolutionarily conserved intracellular proteins with at least 57 members and play key roles in host innate immune response (Franchi et al., 2010). They can be induced by both biotic factors such as bacteria (Swain et al., 2012a, b) and abiotic factors such as thermal stress (Basu et al., 2015). The observation suggested that the immunity of larval grass carp was largely affected by salinity exposure. Similar findings were also reported in other aquaculture species, such as Mauremys mutica (Gao et al., 2021), Apostichopus japonicus (Xu et al., 2016), Catla catla (Basu et al., 2015), and Labeo rohita (Swain et al., 2012a, b). It is widely recognized that impaired immunity may decrease the ability to deal with exogenous pathogens, making species more susceptible to illness or even mass mortality when populations suffer from adverse environmental challenges (Bowden et al., 2007). Taken together, impaired immunity may be another important mechanism responsible for the population decline of wild grass carp in rivers.
The maintenance of metabolism balance is a fundamental demand for the normal performance of physiological activities such as dealing with adverse environmental stress. Here, we identified one metabolism-related GO term (phosphoenolpyruvate carboxykinase activity) and three metabolism-related pathways (fructose and mannose metabolism, purine metabolism, and pyruvate metabolism). Pyruvate in the human body is produced by the degradation of glucose and glycogen, or the degradation of amino acids, and is involved in energy metabolism. In this study, three DEGs were detected in the pyruvate pathway and annotated to two proteins (phosphoenolpyruvate carboxykinase and mitochondrial trifunctional enzyme subunit beta). The former acts as the rate-limiting enzyme in gluconeogenesis, which is a process that converts various non-sugar substances into glucose or glycogen (Latorre et al., 2016). The latter is a protein of mitochondrial trifunctional enzyme that catalyzes the last part reactions of the mitochondrial beta-oxidation pathway, a major energy-producing process in tissues (Kamijo et al., 1994; Liang et al., 2018). Interestingly, the three genes in the pyruvate pathway were all downregulated. In addition, the expressions of the DEGs in both fructose and mannose metabolism showed similar patterns, suggesting that salinity exposure may inhibit the energy metabolism; thus, not enough energy was available for resisting environmental stress. Moreover, purines are essential components in energy-requiring enzymatic reactions, such as the metabolism of the nervous system (Jinnah et al., 2013). In this pathway, seven DEGs were detected and annotated to seven proteins, including guanylyl cyclases, 3′-phosphoadenosine 5′-prime-phosphosulfate, TWISTNB protein, ectonucleoside triphosphate diphosphohydrolase, NTPDase 3, NTPDase 8, cGMP-specific-3′,5′-cyclic phosphodiesterase, and guanine deaminase. They are mainly involved in the regulation of the nervous system (Snyder et al., 2000; Xu et al., 2000; Belcher et al., 2006). In this study, most of the DEGs in this pathway were downregulated, suggesting that the function of the nervous system is damaged, which may lead to uncoordinated movements in animals. This may be the reason for the unusual performance of the up or down swimming movements observed in the present study. Hence, the imbalance of metabolism may be another reason for the mortality and population decline of grass carp in the wild.
It is traditionally thought that salinity exposure can result in the imbalance of osmotic pressure and loss of integrant water in organisms. Gap junctions, dynamic hexameric protein channels, play important roles in ion exchange and balance of osmotic pressure. Inorganic ions or small molecules such as Na+ are necessary components in maintaining osmotic balance through gap junctions (Kumar and Gilula, 1996). In this study, there were four DEGs showing significant enrichment in the gap junction pathway. Among which, tubulin is one major component of microtubules that plays important roles in maintaining cell shape and transcription of cell signaling (Jordan and Wilson, 2004). The down regulation of tubulin 5 under salinity exposure indicated that the function of the gap junctions may be impaired, thus has potential to break the balance of osmotic pressure. The imbalance of osmotic pressure may cause tissue edema. Expectedly, intestinal edemas in the experiment group were observed, while no similar phenomenon was detected in the control group.
In conclusion, our results revealed that salinity exposure has a significant influence on gene expression in larval grass carp, and many DEGs were identified. The DEGs were mainly involved in fertility, immunity, metabolism, and osmotic pressure-related categories, which indicated that salinity exposure posed a significant influence on fertility, immunity, metabolism, and osmotic pressure. These changes may be the major mechanisms underlying the population decline of grass carp in river systems. Our study provided molecular evidence in support of the population decline of wild aquaculture species, which are conducive to the management of wild aquaculture species and recovery of germplasm resource.
Data Availability Statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, accession number is PRJNA725283.
Ethics Statement
The animal study was reviewed and approved by the Pearl River Fisheries Research Institute Animal Care Advisory Committee.
Author Contributions
JZ, JL, and XL contributed to the conception of the study. JZ, ZW, and YH performed the experiment. JZ, ZW, and JL contributed significantly to the analysis and manuscript preparation. JZ performed the data analyses and wrote the manuscript. JL and XL helped perform the analysis with constructive discussions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0900902) and the Project of Innovation Team of Survey and Assessment of the Pearl River Fishery Resources (2020TD-10).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Yangchun Gao, Ph.D, from the Institute of Zoology, Guangdong Academy of Sciences, for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.697813/full#supplementary-material
Footnotes
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Nat. Precedings 1:R106.
Baggiolini, M., and Clark-Lewis, I. (1992). Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 307, 97–101. doi: 10.1016/0014-5793(92)80909-z
Basu, M., Paichha, M., Swain, B., Lenka, S. S., Singh, S., Chakrabarti, R., et al. (2015). Modulation of TLR2, TLR4, TLR5, NOD1 and NOD2 receptor gene expressions and their downstream signaling molecules following thermal stress in the indian major carp catla (Catla catla). 3 Biotech 5, 1021–1030. doi: 10.1007/s13205-015-0306-5
Belcher, S. M., Zsarnovszky, A., Crawford, P. A., Hemani, H., Spurling, L., and Kirley, T. L. (2006). Immunolocalization of ecto-nucleoside triphosphate diphosphohydrolase 3 in rat brain: implications for modulation of multiple homeostatic systems including feeding and sleep-wake behaviors. Neuroscience 137, 1331–1346. doi: 10.1016/j.neuroscience.2005.08.086
Berthelot, C., Brunet, F., Chalopin, D., Juanchich, A., Bernard, M., Noël, B., et al. (2014). The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5:3657.
Bowden, T. J., Thompson, K. D., Morgan, A. L., Gratacap, R. M. L., and Nikoskelainen, S. (2007). Seasonal variation and the immune response: a fish perspective. Fish Shellfsh Immunol. 22, 695–706. doi: 10.1016/j.fsi.2006.08.016
Chen, Q., Ghilardi, N., Wang, H., Baker, T., Xie, M. H., Gurney, A., et al. (2000). Development of th1-type immune responses requires the type I cytokine receptor TCCR. Nature 407, 916–920. doi: 10.1038/35038103
Cuka, E., Lanzani, C., Simonini, M., Zagato, L., Citterio, L., Brioni, E., et al. (2019). Precision medicine: lanosterol synthase and CYP11B2 gene polymorphisms affect body Na+ in salt-sensitive hypertension. Am. J. Hypertens. 37:e74. doi: 10.1097/01.hjh.0000571104.75084.9e
Dauphinee, S. M., and Karsan, A. (2006). Lipopolysaccharide signaling in endothelial cells. Lab. Invest. 86, 9–22. doi: 10.1038/labinvest.3700366
Ewing, B., Hillier, L. D., Wendl, M. C., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. accuracy assessment. Genome Res. 8, 186–194. doi: 10.1101/gr.8.3.186
Fishery Statistical Yearbook of China (2017). Available online at: https://www.yearbookchina.com/navibooklist-n3018062509-1.html
Fishery Statistical Yearbook of China (2018). Available online at: https://www.yearbookchina.com/navibooklist-n3018112801-1.html
Fishery Statistical Yearbook of China (2019). Available online at: https://www.yearbookchina.com/navibooklist-n3019102806-1.html
Fishery Statistical Yearbook of China (2020). Available online at: https://www.yearbookchina.com/navibooklist-n3020013079-1.html
Franchi, L., Warner, N., Viani, K., and Nuñez, G. (2010). Function of nod-like receptors in microbial recognition and host defense. Immunol. Rev. 227, 106–128. doi: 10.1111/j.1600-065x.2008.00734.x
Gao, Y. C., Wei, Y. F., Cao, D. N., Ge, Y., and Gong, S. P. (2021). Transcriptome analysis reveals decreased immunity under heat stress in Mauremys mutica. Aquaculture 531:735894. doi: 10.1016/j.aquaculture.2020.735894
Ha, H. L., Wang, H. S., Claudio, E., Tang, W. H., and Siebenlist, U. (2020). IL-20-receptor signaling delimits IL-17 production in psoriatic inflammation. J. Invest. Dermatol. 140, 143–151. doi: 10.1016/j.jid.2019.06.127
Han, J. Y., In, J. G., Kwon, Y. S., and Choi, Y. E. (2010). Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry 71, 36–46. doi: 10.1016/j.phytochem.2009.09.031
Hu, W., and Chen, J. (2015). Whole-genome sequencing opens a new era for molecular breeding of grass carp (Ctenopharyngodon idellus). Sci. China Life Sci. 58, 619–620. doi: 10.1007/s11427-015-4864-x
Hu, X. Q., Fu, X. Z., Li, N. Q., Dong, X. X., and Zhao, L. J. (2015). Transcriptomic analysis of mandarin fish brain cells infected with infectious spleen and kidney necrosis virus with an emphasis on retinoic acid-inducible gene 1-like receptors and apoptosis pathways. Fish Shellfish Immun. 45, 619–629. doi: 10.1016/j.fsi.2015.05.007
Janeway, C. A., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216.
Jin, J. L., Wang, Y., Wu, Z. X., Hergazy, A., Lan, J. F., Zhao, L. J., et al. (2017). Transcriptomic analysis of liver from grass carp (Ctenopharyngodon idellus) exposed to high environmental ammonia reveals the activation of antioxidant and apoptosis pathways. Fish Shellfish Immun. 63, 444–451. doi: 10.1016/j.fsi.2017.02.037
Jinnah, H. A., Sabina, R. I., and Berghe, G. V. D. (2013). Metabolic disorders of purine metabolism affecting the nervous system. Handb. Clin. Neurol. 113, 1827–1836. doi: 10.1016/b978-0-444-59565-2.00052-6
Jordan, M. A., and Wilson, L. (2004). Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265. doi: 10.1038/nrc1317
Kaisho, T., and Akira, S. (2002). Toll-like receptors as adjuvant receptors. Biochim. Biophys. Acta 1589, 1–13.
Kamijo, T., Aoyama, T., Komiyama, A., and Hashimoto, T. (1994). Structural analysis of cDNAs for subunits of human mitochondrial fatty acid beta-oxidation trifunctional protein. Biochem. Biophys. Res. Commun. 199, 818–825. doi: 10.1006/bbrc.1994.1302
Kang, Z. Q., Deng, C. Z., Yu, H. J., Lin, X. R., and Huang, Y. C. (2013). Study advances in the effect of salinity change on fish. J. Fujian Fish. 35, 395–401.
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kimbrell, D. A., and Beutler, B. (2001). The evolution and genetics of innate immunity. Nat. Rev. Genet. 2, 256–267. doi: 10.1038/35066006
Krieg, A. M. (2002). CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20, 709–760.
Kumar, N. M., and Gilula, N. B. (1996). The gap junction review communication channel. Cell Vol. 84, 381–388. doi: 10.1016/s0092-8674(00)81282-9
Latorre, P., Burgos, C., Hidalgo, J., Varona, L., Carrodeguas, J. A., and López-Buesa, P. (2016). c.A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci. Rep. 6:19617.
Lee, P. T., Ho, T. H., Bao, T. N., Lin, Y. L., and Chiu, P. Y. (2020). Expression profile, subcellular localization and signaling pathway analysis of fish-specific TLR25 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 104, 141–154. doi: 10.1016/j.fsi.2020.05.028
Li, C., Zhang, Y., Wang, R. J., Lu, J. G., Nandi, S., Mohanty, S., et al. (2012). RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immun. 32, 816–827. doi: 10.1016/j.fsi.2012.02.004
Li, L. T., and Kaplan, J. (1996). Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J. Biol. Chem. 271, 16927–16933. doi: 10.1074/jbc.271.28.16927
Liang, K., Li, N. N., Wang, X., Dai, J. Y., Liu, P. L., Wang, C., et al. (2018). Cryo-EM structure of human mitochondrial trifunctional protein. Proc. Natl. Acad. Sci. U. S. A. 115, 7039–7044. doi: 10.1073/pnas.1801252115
Liu, Z., Chen, X., Wang, X., Chen, X., Song, C. H., Du, Y., et al. (2011). Chemokine CXCL11 links microbial stimuli to intestinal inflammation. Clin. Exp. Immunol. 164, 396–406. doi: 10.1111/j.1365-2249.2011.04382.x
Lu, G. Q., and Luo, M. K. (2020). Genomes of major fishes in world fisheries and aquaculture: status, application and perspective. Aquac. Fish. 5, 163–173. doi: 10.1016/j.aaf.2020.05.004
Luo, H., Liu, H. P., Zhang, J., Hu, B. J., Zhou, C. W., Xiang, M. B., et al. (2020). Full-length transcript sequencing accelerates the transcriptome research of Gymnocypris Namensis, an iconic fish of the Tibetan Plateau. Sci. Rep. 10:9668.
Mao, X. Z., Cai, T., Olyarchuk, J. G., and Wei, L. P. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Melloul, A., and Collin, M. (2006). Hydrogeological changes in coastal aquifers due to sea level rise. Ocean Coast. Manage. 49, 281–297. doi: 10.1016/j.ocecoaman.2006.03.009
Morris, L. X., and Spradling, A. C. (2012). Steroid signaling within drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS One 7:e46109. doi: 10.1371/journal.pone.0046109
Nguyen, F., Kemna, A., Antonsson, A., Engesgaard, P., Kuras, O., Ogilvy, R., et al. (2009). Characterization of seawater intrusion using 2d electrical imaging. Near Surf. Geophys. 7, 377–390. doi: 10.3997/1873-0604.2009025
Ozato, K., Tsujimura, H., and Tamura, T. (2002). Toll-like receptor signaling and regulation of cytokine gene expression in the immune system. Biotechniques 33, S66–S75.
O’Neill, L. A. J. (2000). The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. Signal. 2000, 1–11.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700.
Reeth, K. V. (2000). Cytokines in the pathogenesis of influenza. Vet. Microbiol. 74, 109–116. doi: 10.1016/s0378-1135(00)00171-1
Schutyser, E., Struyf, S., and Damme, J. V. (2003). The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14, 409–426. doi: 10.1016/s1359-6101(03)00049-2
Snyder, F. F., Yuan, R. G., Bin, J. C., Carter, K. L., and Mckay, D. J. (2000). Human guanine deaminase: cloning, expression and characterisation. Adv. Exp. Med. Biol. 486, 111–114. doi: 10.1007/0-306-46843-3_22
Suzuki, M., Xiang, T., Ohyama, K., Seki, H., Saito, K., Muranaka, T., et al. (2006). Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol. 47, 565–571. doi: 10.1093/pcp/pcj031
Swain, B., Basu, M., Sahoo, B. R., Maiti, N. K., Routray, P., Eknath, A. E., et al. (2012a). Molecular characterization of nucleotide binding and oligomerization domain (n.d.)-2, analysis of its inductive expression and down-stream signaling following ligands exposure and bacterial infection in rohu (Labeo rohita). Dev. Comp. Immunol. 36, 93–103. doi: 10.1016/j.dci.2011.06.018
Swain, B., Basu, M., and Samanta, M. (2012b). Molecular cloning and characterization of nucleotide binding and oligomerization domain-1 (n.d.) receptor in the indian major carp, rohu (Labeo rohita), and analysis of its inductive expression and down-stream signalling molecules following ligands exposure. Fish Shellfish Immunol. 32, 899–908. doi: 10.1016/j.fsi.2012.02.018
Temperley, N. D., Berlin, S., Paton, I. R., Griffin, D. K., and Burt, D. W. (2008). Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics 9:62. doi: 10.1186/1471-2164-9-62
Tong, Y., Chen, L. Q., Zhuang, P., Zhang, L. Z., Peng, S. M., Song, C., et al. (2007). Cortisol, metabolism response and osmoregulation of juvenile Acipenser schrenckii to ambient salinity stress. J. Fish. China 31, 38–44.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Baren, M. J. V., et al. (2010). Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsujimura, H., Nagamura-Inoue, T., Tamura, T., and Ozato, K. (2002). IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J. Immunol. 169, 1261–1269. doi: 10.4049/jimmunol.169.3.1261
Wada, Y., Kikuchi, A., Kaga, A., Shimizu, N., Ito, J., Onuma, R., et al. (2020). Metabolic and pathologic profiles of human LSS deficiency recapitulated in mice. PLoS Genet. 16:e1008628. doi: 10.1371/journal.pgen.1008628
Wang, Y. J., Han, J. R., Shu, J. J., and Chang, T. J. (2016). Identification and characteristic analysis of TLR28: a novel member of the TLR1 family in teleost. Dev. Comp. Immunol. 62, 102–107. doi: 10.1016/j.dci.2016.05.001
Wang, Y. P., Lu, Y., Zhang, Y., Ning, Z. M., Li, Y., Zhao, Q., et al. (2015). The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 47, 625–631.
Werner, A. D., Bakker, M., Post, V. E. A., Vandenbohede, A., Lu, C. H., Ataie-Ashtiani, B., et al. (2013). Seawater intrusion processes, investigation and management: recent advances and future challenges. Adv. Water Resour. 51, 3–26. doi: 10.1016/j.advwatres.2012.03.004
Werner, A. D., and Simmons, C. T. (2010). Impact of sea-level rise on sea water intrusion in coastal aquifers. Groundwater 47, 197–204. doi: 10.1111/j.1745-6584.2008.00535.x
Xu, D. X., Sun, L. N., Liu, S. L., Zhang, L. B., and Yang, H. S. (2016). Understanding the heat shock response in the sea cucumber Apostichopus japonicus, using iTRAQ-based proteomics. Int. J. Mol. Sci. 17:150.
Xu, Z. H., Otterness, D. M., Freimuth, R. R., Carlini, E. J., Wood, T. C., Mitchell, S., et al. (2000). Human 3‘-Phosphoadenosine 5‘-Phosphosulfate synthetase 1 (PAPSS1) and PAPSS2: gene cloning, characterization and chromosomal localization. Biochem. Biophys. Res. Commun. 268, 437–444. doi: 10.1006/bbrc.2000.2123
Yalinkaya, T., and Lai, Y. C. (2000). Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch. Biochem. Biophys. 374, 381–388. doi: 10.1006/abbi.1999.1629
Yamamoto, S., and Bloch, K. (1970). Studies of squalene epoxidase of rat liver. J. Biol. Chem. 245, 1670–1674. doi: 10.1016/s0021-9258(19)77144-0
Yang, Y., Yu, H., Li, H., Wang, A., and Yu, H. Y. (2016). Effect of high temperature on immune response of grass carp (Ctenopharyngodon idellus) by transcriptome analysis. Fish Shellfish Immun. 58, 89–95. doi: 10.1016/j.fsi.2016.09.014
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNAseq: accounting for selection bias. Genome Biol. 11:R14.
Zhao, L. J., Tu, J. G., Zhang, Y. L., Wang, J. F., and Yang, L. (2016). Transcriptomic analysis of the head kidney of topmouth culter (Culter alburnus) infected with flavobacterium columnare with an emphasis on phagosome pathway. Fish Shellfish Immun. 57, 413–418. doi: 10.1016/j.fsi.2016.09.001
Zhou, C. Q., Ka, W., Yuan, W. K., and Wang, J. L. (2021). The effect of acute heat stress on the innate immune function of rainbow trout based on the transcriptome. J. Therm. Biol. 96:102834. doi: 10.1016/j.jtherbio.2021.102834
Keywords: larval grass carp, RNA-seq, fertility, immunity, seawater intrusion
Citation: Zhang J, Wu Z, He Y, Li X and Li J (2021) Transcriptome Analysis Reveals Impaired Fertility and Immunity Under Salinity Exposure in Juvenile Grass Carp. Front. Mar. Sci. 8:697813. doi: 10.3389/fmars.2021.697813
Received: 20 April 2021; Accepted: 07 June 2021;
Published: 13 August 2021.
Edited by:
Zhi-Hua Li, Shandong University, ChinaReviewed by:
Shun Maekawa, National Pingtung University of Science and Technology, TaiwanBanikalyan Swain, University of Florida, United States
Copyright © 2021 Zhang, Wu, He, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Li, bGlqaWUxNTYxQDE2My5jb20=
 Jingjing Zhang
Jingjing Zhang Zhi Wu1,2,3,4
Zhi Wu1,2,3,4