
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci., 05 August 2021
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.697269
This article is part of the Research TopicMicrobial Ecology of the Arabian/Persian GulfView all 9 articles
The Arabian Gulf ranks among the world’s most arid and warm regions; the land has high salinity levels with many Sabkhas and receives little precipitation. This region holds about one-third of the world’s oil supply. Qatar is the leading gas producer worldwide, which raises many concerns about the pollution of the sea, groundwater, and soil. Thus, the Arabian Gulf area has paid particular attention to environmental studies since the environmental status of this region imposed unique biological diversity, and microbial ecology has gained special importance following the identification of promising roles of microorganisms. This review article discusses the microbial ecology at the main habitats of the State of Qatar. We discuss important principles for successful ecological restoration and future perspectives of using biological approaches to solve many problems related to health, the economy, and agriculture. There are at least five microbial communities that have been recognized at the Qatari habitats: marine environment, salt marshes and mangrove forests, the arid lands (including dune communities), wetlands (including pond communities), and Rawdahs (including the Ghaf tree communities). Although, the environmental conditions of this region are almost the same, these habitats are compared with those at other countries of the Arabian Gulf whenever necessary, as each habitat has its own peculiar characteristics. Some case studies are presented to describe the biochemical characterizations of bacterial isolates from soils and leaf surface of native plants, including halophytes and xerophytes at these habitats. These studies rarely went beyond the general identification at species levels. There is a discussion about the possible roles of microorganisms at the rhizosphere, non-rhizosphere, and phyllosphere, and using plant exudates to control microbial activity. However, modern approach (culture-independent methods) addressing these topics has opened the door for deeper investigations, and to explore the roles played by microorganisms at these habitats. These methods have already begun during the last decade as serious step to solve many environmental issues. In the future, it is very likely that microorganisms will be used to tackle many pollution issues, as well as health, agricultural, and economic problems.
The Arabian Gulf (Supplementary Figure 1) ranks among the world’s most arid, warm, and humid (air) areas; moreover, the land has high salinity, with many scattered Sabkhas and saline patches, with scarce rainfall (Abdel-Bari et al., 2007b; Yasseen and Al-Thani, 2013). The State of Qatar is relatively small among the countries of this region; it is located in the middle of the western coast of the Arabian Gulf, its land extends as an outcrop from the eastern part of the Arabia Peninsula toward the Gulf (Figure 1). The Arabian Gulf region holds about one-third of the world’s oil supply, and Qatar is among the leading gas and oil producers worldwide, which put a great pressure on the biodiversity of this country and raising many concerns about the pollution at various habitats.
Figure 1. Qatar as extended land from the Arabia peninsula. Qatar is largely surrounded by seawater of the Arabian Gulf.
Obviously, the main source of salts at the coastal areas, including the mangrove forests and salt marshes, is attributed to Arabian Gulf water, and the intrusion of seawater into the underground water might have contributed in the creation of Sabkhas and other salt patches around the country. Moreover, the high evaporation during most months of the year could increase the salinity problem at these areas of the country. Thus, the biodiversity in the State of Qatar is affected in a way or another by the marine environment of Arabian Gulf (Abulfatih et al., 2001). Moreover, in such desert and saline habitats under harsh environmental conditions, many flora, fauna, and microorganisms (Batanouny, 1981; Abulfatih et al., 2001; Abdel-Bari, 2012) play various biological roles. Those roles related to microbes have been attracting much interest (Al-Thani and Yasseen, 2018a; Al-Thani and Yasseen, 2020), and recent works suggested many possible activities that these microorganisms might play, such as contributing to energy equilibrium (Abulfatih et al., 2001; Al-Thani and Yasseen, 2018b; Yasseen et al., 2018). Recent publications have described the activities of microorganisms in the phytoremediation of polluted soils and waters (Al-Thani and Yasseen, 2020, 2021). On the other hand, successful ecological restoration and maintaining a healthy environment are based on the following main principles: (1) Information, (2) Problems, (3) Plans, (4) Solutions, and (5) Monitoring (Yasseen and Al-Thani, 2013). Therefore, knowledge of microbial ecology is the information needed to solve the problems related to agriculture, human life and health, and wildlife. Such problems need plans to setup solutions and active monitoring systems to follow the success of any restoration program. Good example can be given here, a pioneer project was done for restoration program of the coastal vegetation habitat during pipeline installation at Ras Laffan area. The details of this project can be found in the report of Al-Ansi et al. (2004). In fact, gas companies have pledged to the Supreme Council for Environmental Nature Reserve (SCENR) to implement Environmental Impact Assessment (EIA) for any plan to change the natural habitat (Yasseen and Al-Thani, 2007; Yasseen and Abu-Al-Basal, 2008; Yasseen and Al-Thani, 2013). The lessons learnt from such projects can be utilized in any future project regarding the restoration programs to tackle any environmental issues.
In Qatar, some groups of microorganisms have been largely ignored in recent decades. The availability of specialists and technical facilities might be the main reasons behind this; for example, plant biodiversity and ecology and fungi received considerable importance in the 1980s. Some published articles and books (Batanouny, 1981; Moubasher and Al-Subai, 1987) have documented native plants and fungi, giving some details about the distribution and habitat and their roles in the ecosystem. This article addresses mainly the ecology of bacteria in Qatar and other parts of the Arabian Gulf, and whenever possible, other microorganisms are covered. The possible biological roles at various habitats in Qatar are discussed, as well as the modern biotechnological methods used for their study. Until relatively recently, there were no serious scientific reports about the bacteria species in the Qatari waters and soils. Recent efforts by scientific institutions with the State of Qatar and research centers at Qatar University and other universities will be discussed below. However, more is needed to identify and describe novel bacteria species in the Gulf waters and soils around the country.
Various habitats at the Arabian Gulf region, coastal and inland, support a range of ecosystems; to play multiple functions and offer environmental benefits that contribute to the productivity (Naser, 2014). In Qatar, there are at least five habitats that encompass various bacterial communities: (1) marine environment, (2) salt marshes and mangrove forests, (3) the arid lands (including dune communities), (4) wetlands (including pond communities), and (5) Rawdahs (including Ghaf tree communities). Some case studies are addressed in detail; however, a general discussion is given about other locations in the Arabian Gulf region or elsewhere. Moreover, concise clarifications about the microbial communities at wastewater ponds are given (Yasseen, 2014a; Al-Thani and Yasseen, 2020, 2021).
The water of the Arabian Gulf surrounding the Qatari peninsula has peculiar chemical and physical characteristics, as salinity levels ranged between 39 and 57 psu (Practical Salinity Unit: it is equivalent g/kg), and the temperature is fluctuated between 15 and 39°C (Fotedar et al., 2015). More data about the climate conditions of the Gulf region can be found in many reports and articles (Yasseen and Al-Thani, 2013), however, the recent work of Rakib et al. (2021) listed more data about the distinct characteristics of the Gulf at the Qatari coasts. Fotedar et al. (2016) have discussed the changes in the global climate as a serious challenge facing governments in the development of the promising programs to sustain the wellbeing of people. One important habitat at the Arabian Gulf is the marine environment; it is unique with harsh environmental conditions, which influences sea salinity and temperature, thereby having great impact on the water density and currents. Marine habitats under such environmental conditions have been under increasing pressure from the anthropogenic and industrial activities due to expansion at oil and gas sectors and as result of great development at the coastal habitats that led these habitats to become much vulnerable especially coral reefs, seagrass beds, and commercial fisheries (Burt et al., 2017).
Till 2013, no serious microbial studies had been performed in Qatari seawaters to isolate and identify the microbial flora at the marine environment. Since then, many studies have been conducted to identify the bacteria communities surrounding the Qatari peninsula. By adopting modern biotechnology (culture independent methods) and phylogenetic analysis, Fotedar (2013) has isolated and cultured 50 bacteria species that inhabited the marine environment in Qatar. These included: 88% of the strains belonged to Proteobacteria: Cobetia marina, Halobacillus profundi, Pseudoalteromonas agarivorans, Pseudoalteromonas piscicida, Pseudoalteromonas rubra, Pseudoalteromonas prydzensis, Ruegeria mobilis, Shewanella loihica, Virgibacillus dokdonensis, Vibrio harveyi, Vibrio nereis, Vibrio nigripulchritudo, Vibrio parahaemolyticus, followed by 10% of CFB group bacteria: Tenacibaculum mesophilum, and 2% of GC, Firmicutes: Bacillus boroniphilus. The Vibrio species have been very interesting since most of these bacteria are infectious. Thus, Anand et al. (2016) conducted follow up study on the genus Vibrio; isolated 52 isolates around the Qatari peninsula using modern techniques. They identified 13 different luminous and non-luminous Vibrio species: Vibrio alginolyticus (50%) was the predominant species followed by V. harveyi (11%), Vibrio owensii (7%), Vibrio mediterranei (6%), V. nigripulchritudo (6%), Vibrio rotiferianus (6%), Vibrio coralliilyticus (2%), Vibrio hepatarius (2%), Vibrio maritimus (2%), V. parahaemolyticus (2%), Vibrio proteolyticus (2%), Vibrio shilonii (2%), and Vibrio tubiashii (2%). The study has concluded that some pathogenic species were found at the fishing ports giving a warning alert of the potential sanitary risk and suggesting further investigations to prevent infections. Other microbial species (such as Halomonas aquamarina, H. elongata, Pseudomonas fragi, P. stutzeri, V. alginolyticus, and Vibrio fluvialis) were identified and recognized to have negative impact on some facilities of desalination plants. These bacteria proved active in the biodegradation of antiscalants (called scale inhibitors) commonly used in SWRO (Sea Water Reverse Osmosis); used in desalination technique to reduce membrane scaling caused by the accumulation of salts (Ashfaq et al., 2019). They come to conclusion that new polymers/nanomaterials are needed to resist the biological degradation caused by these microorganisms and others to reduce both membrane scaling and biofouling in SWRO.
On the other hand, marine sediments and their contents of biotic and abiotic components participate substantially to the global mass, as 70% of the Earth surface covered by sea water. Thus, microbial communities of the marine sediments contribute considerably to the biotic part of the sediments and play key role in the degradation of organic compounds accumulated due to the anthropogenic and industrial activities (Catania et al., 2018; Hoshino et al., 2020). Unfortunately, not much work has been done yet about the microbiology of the sediments at the marine habitat surrounding the Qatari peninsula. This topic might attract scientists at research centers to look at the microbial ecology of the marine sediment habitat. Research centers at Qatar university were engaged in investigating the content of heavy metals at the sediments of the sea water surrounding the State of Qatar. Some interesting results worth to be mentioned here, Al-Naimi et al. (2015) found that the content of heavy metals was within the international standards criteria, and the data were comparable to previous studies conducted around Qatar, and a recent study was carried out on the THg of the sediments of Arabian Gulf around Qatar (Hassan et al., 2019) showed that mercury levels at all the locations of the study were relatively low.
These include the mangrove forests and coastal and inland salt marshes (Sabkhas), and these habitats might cover all the extremophiles including halo-thermophiles and cyanobacteria.
Mangrove forests play significant functions at the marine ecosystem, and the multiple ecological benefits of having mangrove forests at the coastal line can be listed as follows: (1) basis for marine food chain, (2) create breeding habitat, (3) restricted harbors for protection of offspring, (4) filtering and cleaning pollutants and improving water quality, (5) stabilization of bottom sediments, (6) protecting the shorelines from erosion, cyclones, and tidal waves, (7) source of fuel wood and food for living organisms at the coastal line, and (8) other biological roles in mitigation of CO2 and cooling the area due to photosynthesis process (Nagelkerken et al., 2008). More details can be found at the two links: Ecological Importance - mangrove.org; MANGROVE - FOREST OF THE TIDE — Tancred Production (wordpress.com). Therefore, the awareness of many scientists to the above facts has led Burt and Batholomew (2019) to set up some important measures to maintain the coastal line including artificial coastal defense structures by promoting mangrove forests. Studies on mangrove forests (Figure 2) started seriously in the early 2000s. Two study locations were described by Mahasneh (2000); Alkhor and Al-Dhakhira and that encompass most mangrove forests. These coastal locations are situated at the north-eastern coast of the country (25° 45″ N, 51° 30′ E), and the environmental parameters and physical and chemical characteristics were described by Mahasneh (2002) and Abdel-Bari et al. (2007b) (Supplementary Table 1). These studies attempted to complete some projects in other countries in the Arabian Gulf region and other parts of the Asia continent. Early studies showed that many bacterial isolates of Gram-positive were salt-tolerant (up to 11% salinity) and accumulated toxic ions from the seawater. Moreover, these isolates showed resistance to the high concentration of streptomycin and possibly other antibiotics, such as chloramphenicol and tetracycline (Amir et al., 1993). Studies across the Arabian Gulf region started earlier than those in Qatar. For example, some reports (Mahasneh and Al-Sayed, 1997) have indicated that after the war of 1991 at the Arabian Gulf, some changes in the bacterial communities (coliforms, Vibrio cholerae, V. parahaemolyticus and Fecal streptococci) have taken place in the Bahraini pelagic and nearshore waters and pearl oyster (Pinctada radiata). High counts of bacteria at these sites were observed during the warmer months, and pearl oysters had the highest counts, especially the coliforms, all through the study period of March 1991 to March 1992; after the Gulf war of 1991. Such results might be a direct or indirect effects of oil spills during the military activities. In support of such findings, a study was conducted at the mangrove forests in United Arab Emirates (UAE) in 2002 (El-Tarabily, 2002) which showed that total aerobic and anaerobic hydrocarbon-utilizing bacteria were significantly higher in the polluted than in the nonpolluted sediments.
Al-Sayed et al. (2005) studied the Bahraini coasts and found that mangrove forests harbor at least five bacterial communities: anoxygenic phototrophs (AP), oxygenic phototrophs (OP), organotrophs (OT), total coliforms (TC), fecal coliforms (FC), and haloalkaliphiles (HA). All these microorganisms require Na+ concentration of at least 0.5 mol⋅l–1 and an alkaline pH of 9 for optimal growth. Their unique features enable them to make significant contributions to a wide array of biotechnological applications. A recent Vietnamese study (Tam, 2017) found that rhizospheric bacterial isolates from mangrove forest soil samples (at Ca Mau Peninsula, Vietnam) had ammonium synthesis and phosphate solubilization abilities. These isolates belonged to Bacilli, and six strains were Gamma-Proteobacteria.
In Qatar, Mahasneh (2000, 2001a,b, 2002) has performed several studies on mangrove forests, focusing on the heterotrophic aerobic marine bacteria. Great numbers of bacteria were found in the sediments of mangrove forests (about 105–109 cfu/g dry soil or 1 ml of seawater). Such an amount is necessary to keep this system in equilibrium. All these bacteria help degrade the leaf litter and possibly other nutrients from natural sources of various biological origins (Twilley, 1985; Mahasneh, 2000). Almost all degraded components are useful for various activities to maintain the natural life in mangrove forests (Mahasneh, 2001a). However, such degradation might be affected by seasonal atmospheric temperatures, which show wide fluctuations during the summer and winter; on the other hand, salinity and pH show a narrow annual range of fluctuations. Comparing the sediment and water of mangrove forests with locations lack of mangrove trees, the highest activity of bacteria was found at mangrove forests, especially during the hot months of the year (June–September). The high bacterial counts were accompanied by those of highest proteolytic, cellulolytic, amylolytic, and lipolytic activities (Mahasneh, 2001a,b). It has been reported that cellulase-producing bacteria were dominant over others. Such variation in the activities of key enzymes might be related to many antifouling substances. Future studies should investigate the bacteria’s ability to produce antibiotics and microbial competition, and microbial antagonistic interactions (Mahasneh, 2002). Many microorganisms have been identified to play significant roles in decomposing leaf litters of mangrove plants, including Rhizophora mucronata and Avicennia marina (Kathiresan et al., 2011). These microbes include two genera of total heterotrophic bacteria (THB), four species of Lactobacillus, two species of Azotobacter, two species of Actinobacteria, three isolates of fungus, four species of yeasts, two species of Thraustochytrids, and four species of Trichoderma. Recent works (Singh et al., 2020) have suggested that microflora at mangrove habitats play significant biological and biotechnological roles; these roles include: (1) depositing hydrocarbons, (2) recycling nutrients, (3) consuming gasses that affect global warming, (4) destroying pollutants, (5) treating anthropogenic waste, (6) bioremediation, (7) carbon fixation, and (8) producing biosurfactants. Other products produced by microflora in the mangrove ecosystem include drugs, enzymes, vaccines, antimicrobial agents, immune modulators, anticancer treatments, insecticides, and vitamins. Moreover, some bacteria found in mangrove forests, such as Zobellella denitrificans ZD1, can consume sludge and wastewater to produce polymers that can be used instead of petroleum-based plastics1.
Habitats of coastal line, inland sabkhas, and inland dry lands in Qatar (Supplementary Figure 2) and other parts of the Arabian Gulf region are rich in plants called halophytes and xerophytes (Batanouny, 1981; Abdel-Bari et al., 2007b; Norton et al., 2009; Abdel-Bari, 2012). These plants have developed various structures, physiological characteristics, and biochemical pathways associated with their abilities to resist and adapt to salinity and drought (Yasseen and Al-Thani, 2007; Yasseen and Abu-Al-Basal, 2008, 2010). The details of mechanisms adopted by these plants have been discussed in many articles and research monographs (Abdel-Bari et al., 2007b; Yasseen and Al-Thani, 2013; Yasseen, 2014b, 2016). It is interesting to report that many of these plants excrete materials, inorganic and organic. Such substances might attract various microorganisms by creating preferred mini habitats on the plant organs (roots, stems, leaves, and reproductive organs) (Godfrey, 1976; Fahmy and Al-Thani, 2006); a system as such might encourage many microorganisms to thrive, playing various roles in the biological life of halophytes.
Early reports (Mercier and Lindow, 2000; Lindow and Brandl, 2003) revealed that the aerial surfaces of higher plants growing under natural conditions are normally covered with considerable and varied populations of microorganisms2 (Gougoulias et al., 2014; Hanin et al., 2016; Al-Thani and Yasseen, 2017; Al-Thani and Yasseen, 2018b). However, a limited number of microbial species can grow extensively on healthy leaves unless the plant tissues start to senesce or are damaged (Fahmy and Ouf, 1999; Fahmy and Al-Thani, 2006). For example, microorganisms (e.g., fungal species) inhabit the surface of senescent leaves of Tetraena spp., a native xero-halophyte, and possibly others. These microbes appeared to be adapted to stressful conditions of such microhabitats (dry conditions, high salt contents of their leachates, and high convective heat). Microorganisms show different abilities to adapt to such extreme micro-habitats; Ouf (1993) showed that the presence of salt crusts excreted by the epidermal salt glands and/or the occurrence of leachates on the plant surfaces might affect the colonization and growth of microorganisms.
On the other hand, some differences between the phyllosphere and rhizosphere have been observed; the presence of root exudates at the rhizosphere might encourage or inhibit the soil microorganisms, as suggested by some early findings (Quesada et al., 1982). During the last two decades, evidence has been presented that root exudates regulate plant’s relationships with their microbial communities at the rhizosphere. More investigations have already confirmed the role of exudates of various types at the rhizosphere to attract and repel microorganisms at the narrow zone of soil surrounding the root system (Walker et al., 2003). Negative and positive interactions have been reported by various authors (Narula et al., 2009), including: mycorrhiza, nitrogen-fixing symbionts; plant growth-promoting rhizobacteria endophytes and biological control organisms; and allelopathy and antimicrobial effects on nematodes. Moreover, despite the huge number of microbes living in the soil, few examples were found of plant-associated microbes acting as pathogens. Instead, most root exudates are of antimicrobial actions and might boost many activities related to growth under extreme environmental conditions (Huck et al., 1991; Walker et al., 2003; Srividya et al., 2008; Ahmed et al., 2013; Bizuye et al., 2013). Researchers should now conduct comprehensive investigations to explore the reality of the role of microbes at the rhizosphere, including contributions to resisting extreme biotic and abiotic factors (Glick, 2014; Hanin et al., 2016; Al-Thani and Yasseen, 2018a,b, 2021; Yasseen et al., 2018).
The details of the soil analysis at these locations were given in a research book (Fahmy and Al-Thani, 2006). Supplementary Table 2 shows the results of mechanical analysis, and the chemical and physical properties of soil. During the last two decades, investigations were carried out at Sabkhas and mangrove forests at Al-Dhakhira-Qatar (Figure 3). Among the objectives of these studies were isolation and biochemical characterizations of bacteria, including actinomycetes from plant surfaces (green and senescent parts) and those at rhizosphere and non-rhizosphere soils that support halophyte plants.

Figure 3. Mangrove forests and halophytes at Al-Dhakhira. (A) Heavy traces of salts at the phyllosphere parts of Avicennia marina plants at low tides. (B) Dense growth of halophytic plants in a Sabkha seen at high tide.
Also, the significance of the different biochemical activities of these microorganisms in their adaptation to salt stress of the soil and the surface of those plant species at such extreme habitat were discussed. The details of this project’s experimental works and results can be found in a research book by Fahmy and Al-Thani (2006).
Counts of bacteria were determined using standard methods described elsewhere (Bagwell et al., 1998; Barakate et al., 2002). Looking at the total bacterial counts in the soil supporting halophytes growing in different locations at the coastal salt marsh of Al-Dhakhira showed clear differences between rhizosphere and non-rhizosphere (Table 1), and the results indicated no consistency. Some species, such as A. marina, Halopeplis perfoliata, Cressa cretica, Aeluropus lagopoides, and Tetraena qatarensis, showed higher bacteria counts at rhizosphere than non-rhizosphere, while Arthrocnemum macrostachyum and Suaeda vermiculata showed the opposite trend. Such outcomes need further investigation focused on the root exudates, the type of bacteria, and their activities (González-López and Ruano-Rosa, 2020). The bacteria count at the phyllosphere was determined using standard methods (Richardson, 1985; Yang et al., 2001; Hashidoko et al., 2002). As shown in Table 2, the results revealed no consistency in the bacteria counts between green and senescent parts. Moreover, some plant species, such as S. vermiculata and C. cretica, showed no differences between those parts, and again this needs further investigation (Lindow and Brandl, 2003; Kembel et al., 2014).
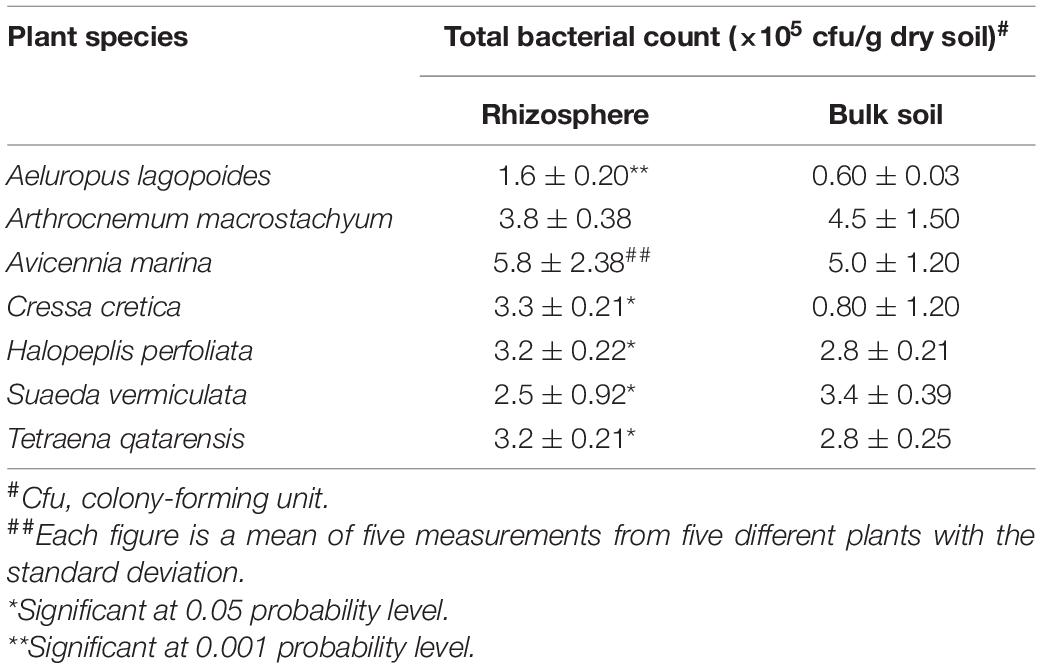
Table 1. Total bacteria count in soil supporting halophytes growing in different coastal salt marshes at Al-Dhakhira region, Qatar (Fahmy and Al-Thani, 2006).
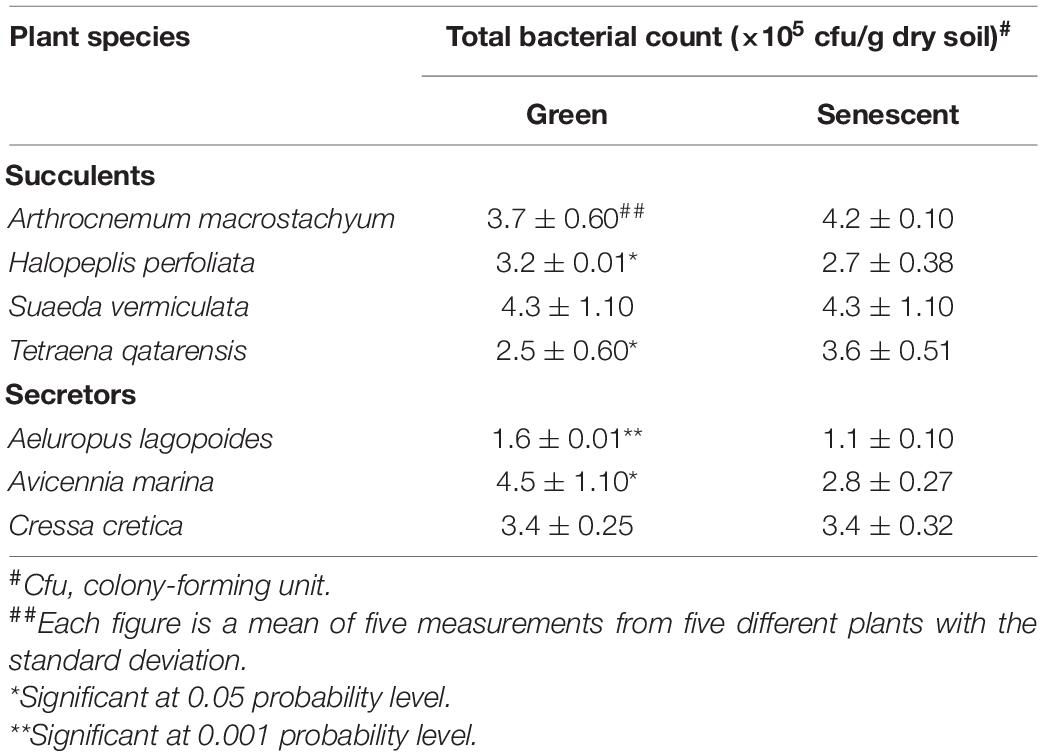
Table 2. Total bacteria count on the phyllosphere of green and senescent parts of some halophyte plants in the coastal salt marshes at Al-Dhakhira region-Qatar (Fahmy and Al-Thani, 2006).
The above results reveal differences in the bacteria counts in this area compared to other studies in the Arabian Gulf region and even with previous investigations at the same sites of the current study. For example, in the salt marshes at Shuaiba lagoon in Saudi Arabia, Zaki et al. (1980) found that the total counts ranged from 81 × 106 to 1484 × 106 cfu/g dry soil, while a study conducted by Al-Thani and Mahasneh (2002) at Al-Dhakhira region-Qatar showed lower bacteria counts than those given in Table 1.
Such contradictory results are expected as many factors might have affected bacteria counts at salt marshes. These include the physicochemical properties of the soil, salinity levels, and dilution of seawater due to flooding conditions which affect aeration and nutrient availability; some reports concluded that these factors might have a significant impact on the bacteria counts through the decomposition rates of the organic matter of the soil (Zaki et al., 1980; Fahmy and Al-Thani, 2006). On the other hand, the nature of root exudates and the number of detritivores (such as meiofauna which feed mainly on bacteria) have a significant role in the bacteria counts as found by many authors (Alexander, 1977; Wahbeh and Mahasneh, 1984; Bardgett and Griffiths, 1997; Zahran, 1999).
The results of Table 2 show inconsistency regarding the relationships between the main resistance mechanisms adopted by halophytes and the activity of the plant parts with total bacteria counts. For example, A. macrostachyum and T. qatarensis showed higher bacteria counts at senescent parts, while H. perfoliata, A. lagopoides, and A. marina showed the opposite trend; others showed no change in the total bacteria counts (such as: S. vermiculata and C. cretica). However, Fahmy and Al-Thani (2006) have concluded that the bacteria counts showed non-significant differences between the phyllosphere of the green and the senescent parts of the studied halophytes. Looking at the individual plants in both groups, no clear conclusion can be made about the factors affecting the total bacteria counts on the green and senescent parts of the phyllosphere. Some of these factors have been discussed in the published articles. These factors are either organic and inorganic materials deposited on the surface of the green parts (mainly leaves and branches) and senescent parts of the plants. However, the origin of these materials varies when comparing the outside plant body and the internal parts of the plant.
There is some basic information that needs to be presented here: (1) organic and inorganic materials are secreted on the outer surface of the plant body from the internal parts of the plants; and (2) these materials might have originated from the atmosphere (Fahmy and Ouf, 1992, 1999; Fahmy et al., 2004; Fahmy and Al-Thani, 2006). Earlier works have indicated that leachates can directly affect microorganisms; such an effect might stimulate or inhibit the growth (Godfrey, 1976). Unfortunately, no further investigation or follow-up has been done to look at the nature of such promoting or delaying of the growth of microorganisms under such mini-habitats.
The colonial status of each isolate was assured by microscopic inspection, and Gram-staining characteristics and cell morphologies (cell form and colony color) were determined by standard methods (Gerhardt et al., 1981; Hensyl, 1994). The cocci forms were dominant in the isolates from the rhizospheric and non-rhizospheric soils, followed by bacilli; Spirilla and Streptomyces were less abundant. On the other hand, the bacilli forms were dominant at the phyllosphere of the studied plants, followed by cocci, with a few spirilla forms. The previously published data on cell forms at the rhizosphere and phyllosphere at saline habitats in the Arabia region (Egypt and Arabian Gulf regions) confirmed the results of this case study: that the Gram-positive bacilli and cocci forms are predominant (Zahran et al., 1992, 1995; Al-Thani and Mahasneh, 2002; Fahmy and Al-Thani, 2006). Saudi Arabia Sabkhas showed that the most commonly isolated bacterial species were Bacillus subtilis and Lactobacillus murinus (Alotaibi et al., 2020). Bacillus spp., on the other hand, were activated at soil polluted with chlorophenol compounds at the industrial zone at Umm-Saied city (Qatar) (Al-Thani et al., 2007; Personal observation). Moreover, most isolates were Gram-positive at rhizosphere soils; few isolates were found Gram-negative, such as the rhizosphere of A. macrostachyum and both rhizosphere and non-rhizosphere of Limonium axillare (Supplementary Figure 3). However, the situation was not the same at the phyllosphere of the studied plants since most of the bacteria at the green and senescent parts of the phyllosphere were Gram-negative. On the other hand, the color of colonies revealed that the white and/or off-white colors were predominated in the rhizosphere and at the phyllosphere; other colors were found at the non-rhizosphere, such as pink colonies. Fahmy and Al-Thani’s (2006) study of the coastal salt marshes concluded that the pigmented bacteria represented 40% of the bacterial flora isolated from the study area’s rhizosphere soils at the Al-Dhakhira-Qatar. Such an outcome is similar to the conclusions obtained from previous studies (Lindow and Brandl, 2003): pigmentation of the bacteria confers protection of the epiphytic bacteria against UV radiation. Many applications of bacterial pigments were reported in many articles, including aspects covering the pathogenesis, industrial, and taxonomic significance. More details about the roles of colorful bacteria can be found3. The most prominent listed functions were: resistance to heavy metals and antibiotics and protection at phagocytosis and radiation. Further study is needed to reach a clear conclusion about the reasons for the predominance of certain colored colonies at such mini-habitats at Al-Dhakhira, Qatar. As a family of bacteria having some pathogens and saprophytes, the Enterobacteriaceae count did not show any presence among these halophyte species. However, Salsola soda showed 300 cfu/g dry soil of non-rhizosphere, while the rhizosphere of Salicornia europaea contains 1,000 cfu/g dry soil.
Many biochemical tests were done for bacteria, including actinomycetes from the rhizosphere and non-rhizosphere soils and phyllosphere (green and senescent parts) supporting the halophyte plants at Al-Dhakhira-Qatar. Nine tests were used to differentiate between groups and even to identify some genera bacteria. To summarize these results: a low percentage of bacteria isolates was positive at the urease test (URE), while more positivity reactions were found with the starch (STA) test at both rhizosphere and phyllosphere. Regarding the reduction of nitrate (NO3-R), bacteria at both rhizosphere soils and phyllosphere showed non-consistence and almost similar reactions and demonstrated as high as 72.7% in both rhizosphere and non-rhizosphere. The hydrolysis of lactose (LAC) tests showed the same reactions of NO3-R tests and infrequent capacity to hydrolyze both lactose (LAC) and casein (CAS). Two more tests were done for the parameters PHE/PA (Phenylalanine decarboxylation), and IND (Indole) reduction, all isolates of the rhizosphere and non-rhizosphere, and phyllosphere (green and senescent) showed a negative reaction. Another interesting observation was that all bacteria isolates showed positive tests for gelatin liquefaction (GEL) and catalase (CAT) activities.
A common observation regarding the capability of bacteria isolates is that most of the isolated bacteria can utilize various substrates, as confirmed by the study of Fahmy and Al-Thani (2006). These authors concluded that isolates from the soil, rhizosphere, and phyllosphere could attack at least 50% of the substrates. Such findings indicate that the soil and shoot surface of the halophytic species have sufficient metabolites necessary to support bacteria which have diverse biochemical activities that enable them to deal with various nutrients of the salt marshes. The biological meanings of the reaction tests can give more features of bacteria isolates at the rhizosphere soil and the phyllosphere. We elaborate below.
Regarding URE, the negative test result of urease activity in the phyllosphere means that the urease-producing bacteria (specifically: Proteus spp.) are absent, while such isolates of bacteria are present mainly in non-rhizospheric soil (Hogarth, 1999; Zahran, 1999). On the contrary, the low percentage of urease-producing bacteria in the soil reflects two possibilities: (1) the saline conditions inactivate the urease action or (2) the soil bacteria depend on other sources to obtain their nitrogen requirements. The second option is valid in this investigation since the rhizosphere isolates and soil showed positive gelatin liquefaction activity. The NO3-R test is useful for differentiating bacteria that able to reduce NO3 (such as: Enterobacteriaceae) from those that cannot. This test showed that 72.7% of the total bacteria isolates exhibited positive tests in the soil and the rhizosphere. The bacteria recovered from the green parts of plants showed a lower percentage of activity than the senescent parts. Looking at the total nitrogen levels in salt marsh soils, the amount of biologically active available forms of nitrogen is low (Adam, 1990). Moreover, the isolates of bacteria give positive tests with LAC and STA from one side and with GEL from the other side, meaning that these bacteria metabolize various substances as carbon sources and nitrogen sources, respectively. The LAC test is used to identify Gram-negative enteric bacteria, the STA test to differentiate species from the genera Clostridium and Bacillus, and the GEL test to differentiate between species in the genera Staphylococcus and Clostridium. Indole production is a key test for identifying enteric bacteria. In fact, an IND negative test is a good indication of the absence of some infectious genera and species [e.g., Salmonella, Escherichia coli (E. coli), Campylobacter, Shigella]. The catalase production by all the bacterial isolates indicates that these are aerobic bacteria (Fahmy and Al-Thani, 2006). In this case study, the CAT test showed that most bacteria are catalase-positive, but anaerobic genera bacteria, such as Streptococcus, Enterococcus, Lactobacillus, and Clostridium, are catalase-negative. The results of the CAS test showed that limited bacteria isolates could hydrolyze proteins (such as milk protein) at the rhizosphere soils. In contrast, more isolates can hydrolyze such proteins at the phyllosphere. Finally, all bacteria isolates showed negative results of the PHE/PA test. This was a good indication of the absence of Enterobacteriaceae from others having Gram-negative and rods. Therefore, more modern techniques are needed to further investigate and identify the bacteria inhabiting salt marshes.
Many locations around the Arabian Gulf region can be considered arid lands. Studies in Qatar have not covered the identification of bacteria species at the desert land in detail. However, an investigation on the Streptomyces genus was done that included the arid lands. Moreover, unpublished data revealed the identification of some microbial species, including bacteria and yeast.
A comprehensive survey was done at 123 locations around Qatar to investigate the ecology and biodiversity of Streptomycets. The main physical and chemical characteristics of the soil at these locations showed a narrow range of moisture content and pH; for example, the soil moisture content varied between 0.2 and 8.8%, and such soil at these locations was alkaline as pH values varied within a narrow range between 7.6 and 9.5, which is quite favorable for the growth and proliferation of Streptomycets. EC of soil extracts, on the other hand, showed wide range of fluctuations between 1.2 and 40.1 dSm–1, and high EC values were recorded in some sites, while most of them showed low values. More details can be found in a research report of Al-Thani and Mahasneh (2002). The genus Streptomyces is common in the Qatari habitats, especially those of drylands and Rawdahs. These are aerobic, Gram-positive, spore-forming, filamentous bacteria that produce extensive branching aerial hyphae and pigmented colonies (Figure 4). It is interesting to notice the higher Streptomyces counts at the non-rhizospheric soil compared to the canopy of the trees.
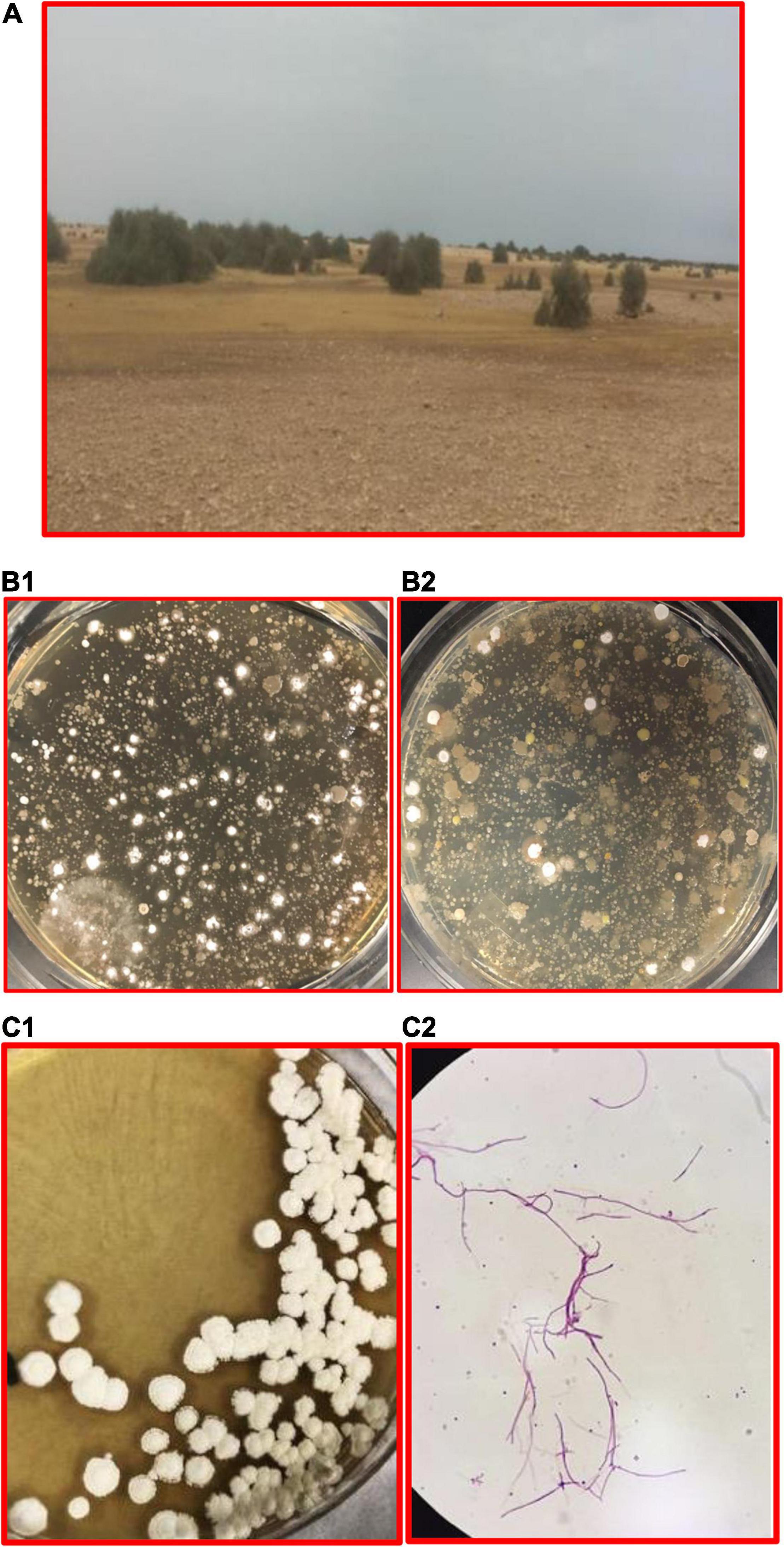
Figure 4. Streptomyces is found at various Qatari lands. (A) The site of study at a Rawdah, (B1) control soil sample from a Rawdah (non-rhizospheric), the bacteria count = 2.75 × 103 cfu/g dry soil, (B2) soil sample collected from under the canopy of the tree at the same location, the bacteria count = 7.5 × 102 cfu/g dry soil. (C1) Pure cultures of Streptomyces, (C2) filaments of Streptomyces, Gram-positive, magnification of 100×. From figures (B1,B2): the white colonies represent the Streptomyces colonies; more Streptomyces bacteria are found at (B1) than (B2). However, in general, more colonies are found in the soil under the canopy of trees compared to those at the non-rhizospheric soil.
There are multiple reasons that can account for this finding: (a) competition between Streptomyces and other living organisms might limit this bacteria to thrive; (b) Rawdahs are good places for various social trip activities, thereby some traces of foodstuffs left by some visitors could limit the growth of Streptomyces; (c) some protozoans feed on bacteria at the canopy; (d) the environmental factors (e.g., UV sunlight, moisture, a litter of the trees, presence of organic substances) at the non-rhizospheric soil might affect the presence of microorganisms; and more bacterial colonies are found at the canopy as compared to the non-rhizospheric soil. Further studies are needed to determine and evaluate the factors affecting the presence of Streptomyces at the Qatari lands. Al-Thani and Mahasneh (2002) gave a preliminary assessment of Streptomyces distributions in Qatar. The authors indicate a wide range of microbial counts (1.2 × 104–12 × 104 cfu/g dry soil). Still, the counts distributed evenly around the country, even though some differences were observed between the studied sites. Moreover, the total count of Streptomyces spp. has accounted for about 40% of the total microbial counts. Early results from Kuwait showed higher microbial counts in many locations, for example, maximum counts may reach 1156 × 104/g soil at the rhizosphere of some native plants such as Rhanterium epapposum; a plant is common in some Arabian Gulf countries like Kingdom of Saudi Arabia (KSA) and Qatar (Elwan et al., 1985).
These results suggest that species of this genus are predominated in the Qatari soil and sites of decaying vegetation; such outcomes confirmed similar findings and are comparable to many international reports (Huddleston et al., 1997; Gupte and Naik, 1999). This report described the morphology of colonies of 34 isolates recognized in this investigation. These colonies were mainly spherical, smooth, elevated with a diameter ranging between 1 and 5 mm with some exceptional cases; they might reach 10 mm. All these strains could utilize various carbohydrate sources (D-glucose, D-xylose, D-galactose, D-lactose, D-sucrose, D-sorbitol, D-mannitol, and starch). Some unique patterns of distribution were detected between the studied locations. For example, all white and orange strains were found only in the northern part of Qatar; the transparent strains were predominated in the southern part of the country, while strains of other colors such as gray, black, and brown appeared restricted in the middle part of the country, some ecological factors might be behind such pattern of distribution of these strains such as moisture, pH, or possibly other factors. Most (42.4%) of the colonies were white, while 18.2% were gray, and the rest were of various other colors. Comparing these results with those found elsewhere at the Arabian Gulf countries, the grey strains were dominant in the desert and saline habitats of Kuwait (72%), followed by red strains (14%) and then yellow, blue, violet and green. It was interesting to report that Elwan et al. (1985) reported the frequency of the presence of individual species of Streptomyces, for example, S. olivaceus was comparatively frequent (12%) followed by others such as S. albogriseolus, S. resistomycificus, S. lydicus, S. echinatus, S. plicatus and S. fumanus, other species represented by 0.4–3.1%. One common characteristic compatible with the results obtained from Qatar, that each particular species found in one community not found in others, such observation was found even with the rhizosphere of plant species; each one has its own Streptomyces sp. Another study from Saudi Arabia seemed consistent with above investigation that grey strain was dominant over the total Streptomyces spp. (Al-Zahrani and Al-Harbi, 2004).
This genus of interest to the biotechnology industry, as species belonging to this genus have been used to produce over two-thirds of the clinically useful antibiotics of natural origin, as well as agents for biocontrol against the pathogenic fungi of both roots and seeds (Williams and Vickers, 1986; Okami and Hotta, 1988; Kortemaa et al., 1994; Tahoven et al., 1995; Al-Zahrani and Al-Harbi, 2004). Al-husnan and Alkahtani (2016) recognized some isolates of Streptomyces with anti-bacterial and anti-fungal activities. They concluded that the soil of the Arabia Peninsula in the Arabian Gulf region is an interesting source of new antibiotics and possibly more bioactive compounds and might be utilized for a more promising future in biotechnology. Recently, plant growth-promoting activities introduced by this genus have been recognized to protect crops against pests (Amaresan et al., 2018). Promoting growth includes improving nutrition acquisition such as nitrogen, phosphorus, potassium, and other essential elements and production of phytohormones, such as indole acetic acid, cytokinins, gibberellins, or suppression of plant disease. This study has concluded that the application of Streptomyces species could improve the productivity of various plant species, including crops, under various stressful environmental conditions (Dias et al., 2017). Many roles of Streptomyces isolates were listed as plant growth-promoting rhizobacteria (PGPR) to improve plant growth and yield and act as agents of biocontrol. More details of the roles of microorganisms, including PGPR growth-promoting rhizobacteria under harsh environmental conditions in the Arabian Gulf region, were recently discussed (Al-Thani and Yasseen, 2018a,b). These roles cover a wide range of biological activities. Moreover, Streptomyces spp. might include saprophytes of beneficial action to plants. Simultaneously, a few might be pathogenic, Vurukonda et al. (2018) reported that some species producing various degrading enzymes such as lytic enzymes, which can degrade insoluble organic polymers chitin and cellulose, breaking them to substituent sugars for binding and uptake by multiple ABC transporters. In a recent report by Manteca and Yagüe (2019), Streptomyces was confirmed as an important natural source with many biotechnological roles, including antibiotics and harbors many cryptic secondary metabolite pathways. Moreover, other reports have introduced these bacteria to provide new anti-fungal and anti-parasite drugs for people and livestock and compounds to prevent cancer after transplant operations4. More innovative new medicines should be developed by gathering financial support from the industrial activities of oil and gas and the goodwill of scientific research centers and decision-makers of the Arabian Gulf countries.
Sand dunes are the main part of the Qatari inland, and the soil characteristics of such habitats were reported in all environmental, ecological, and biodiversity studies. Therefore, we refer the interested readers to many monographs, research reports and articles to look at the environmental and ecological parameters of these sites (Batanouny, 1981; Abulfatih et al., 2001; Abdel-Bari, 2012; Yasseen and Al-Thani, 2013; Al-Thani and Yasseen, 2018a,b). The land of the desert at the Arabia Peninsula is a repository of gas and oil, but there are also other precious treasures worth investing in. Desert plants and their associated microorganisms have the potential to make important contributions to various sectors of biotechnology (Al-Thani and Yasseen, 2018a,b). Figure 5 shows the dunes at the Qatari desert and the bacterial communities at the rhizosphere, and non-rhizosphere soils of the desert plants, as high counts are found at the former relative to the latter. It was interesting to report that the non-rhizospheric soil of dune contains various types of bacteria, about five to seven types of colonies with total bacterial counts: 8 × 103 cfu/g dry soil, while the rhizospheric soil contains many types of colonies and the total bacterial counts were 14 × 103 cfu/g dry soil.
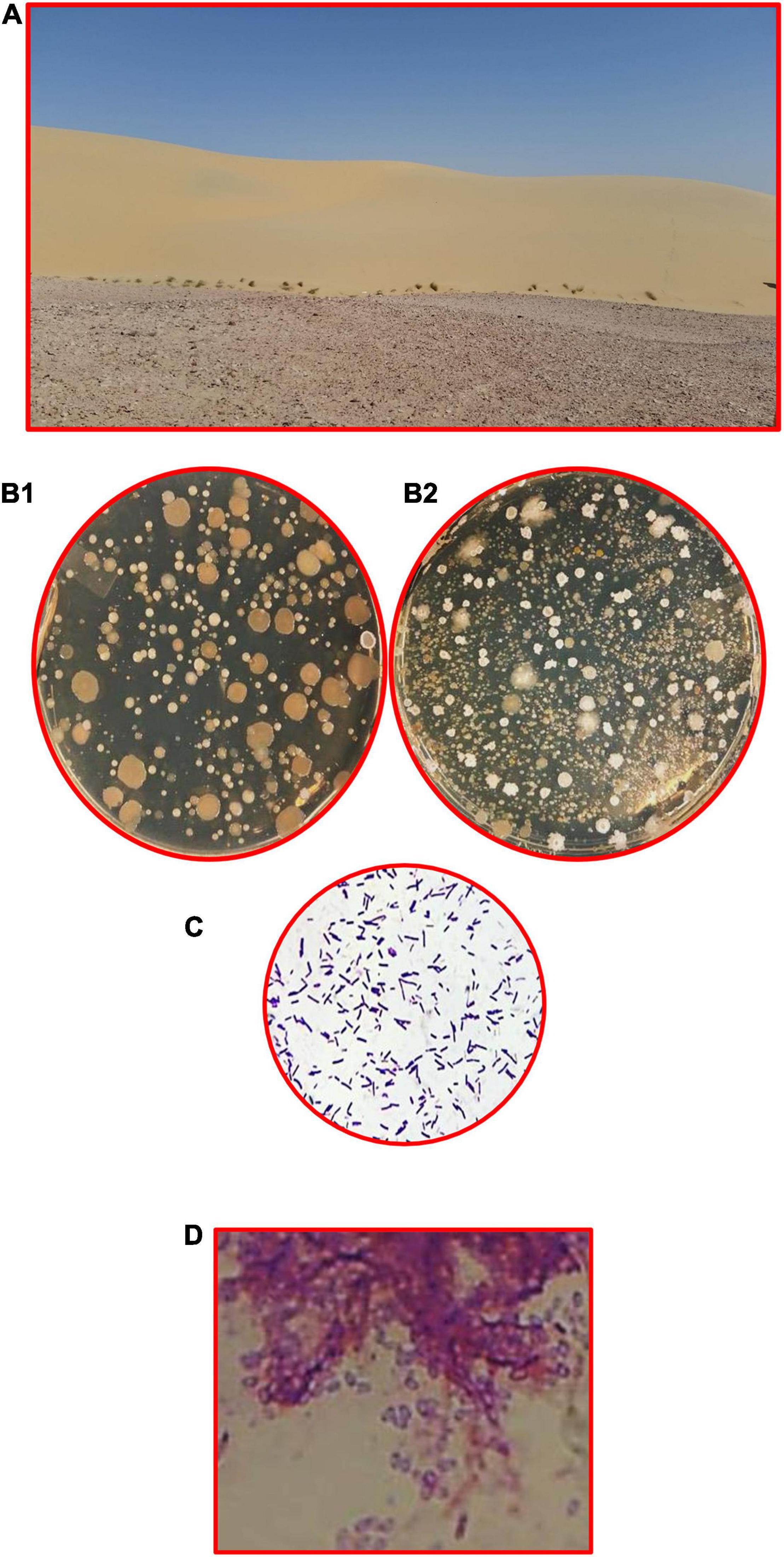
Figure 5. Dunes are rich in BSC, including bacteria. (A) Qatari desert rich of xerophytes, (B1) non-rhizosphere soil of dune contains various types of bacteria (five to seven types of colonies, total bacterial counts: 8 × 103 cfu/g dry soil), (B2) rhizosphere soil contains high microbial activities (many types of colonies, total bacterial counts: 14 × 103 cfu/g dry soil), the most common bacteria are Gram-positive bacilli, spore-forming and non-spore-forming. (C) Non-spore forming and (D) spore-forming (endospores).
Desert plants and microorganisms can be considered as databases and/or banks of information for future biotechnology research in many sectors, such as agriculture, health, and the economy. Little attention had been paid to the microbial ecology of the desert habitats. However, with the new development of the modern approach, as well as growing awareness among scientists of the role of microorganisms in solving many problems facing mankind, during the last decade, great efforts have been paid to the microbiology of desert soil. Some data (personal observations) indicate that the following bacteria species are present in the raw Qatari soils: (1) Micrococcus luteus, Kocuria kristinae, and Streptomyces (Al-Thani and Mahasneh, 2002) (Actinobacteria); (2) Staphylococcus spp. (mainly: Staphylococcus sciuri and Staphylococcus lentus), Lactococcus lactis, Bacillus spp. (mainly: Bacillus megaterium) (Firmicutes); and (3) Pseudomonas spp. (mainly: Pseudomonas aeruginosa, Pseudomonas stutzeri, and Pseudomonas putida), Stenotrophomonas maltophilia, Sphingomonas paucimobilis, Burkholderia spp. (mainly: Burkholderia cepacia and Burkholderia pseudomallei), Enterobacter cloacae (Proteobacteria). Other species can also be found, especially when the soil is exposed to various abiotic and biotic factors or treated with organic and inorganic agents from oil and gas activities (such as Bacillus spp., Pseudomonas geniculata and Achromobacter xylosoxidans) (Al-Thani et al., 2007, 2009), and some of the above species might be activated and/or thrived under those conditions.
During the last decade, great efforts have been made to draw attention to potential contributions of microorganisms to solving the many problems facing humans (Glick, 2012, 2014; Hanin et al., 2016; Al-Thani and Yasseen, 2018b, 2020, 2021; Yasseen et al., 2018). Moreover, some recent works [e.g., Egamberdievea et al. (2017) and Yan et al. (2020)] have suggested that root-associated microbes offer some support to the growth of plants by producing phytohormones against abiotic stresses. Richer et al. (2012) investigated biological soil crusts (BSC), mainly cyanobacteria, of Qatari land. These authors concluded that assessing the coverage of BSC across the State of Qatar will be a useful baseline to monitor environmental disturbance and protect the loss of valuable soils in this arid environment. Al-Thani and Yasseen (2018a) have listed the main useful roles played by BSC in the arid habitats: (1) increasing the fertilization of soil; (2) stabilization of soil surface; (3) monitoring the pollution caused by oil and gas activities; and (4) various roles related to the industry, medicine, food preparation, and cleaning the environment (Müller, 2001; Nimis et al., 2002; Boustie and Grube, 2005; Nascimbene et al., 2007; Melgarejo et al., 2008). The role of cyanobacteria was confirmed by Richer et al. (2015): that BSC of cyanobacteria stabilizes the desert soils. These authors explain the mechanism of such a role that sand of the desert is bound with some polysaccharides extruded by cyanobacteria. The rainy season might trigger the physiological activities of these living organisms, such as photosynthesis, to achieve their actions. Moreover, such activities might end in the production of some toxins that are persistent in the environment. For many years, toxins can accumulate in the desert soils, causing human health problems, especially when desert soils are disturbed by storms, constructions, and military activities.
The environmental parameters around the wasteland ponds are almost the same as recorded by the meteorological station in Doha city and reported by Abulfatih et al. (2002) in their research book: Ecology of wastewater ponds in Qatar. Climate of this area shows two extremes during the year, it is very hot, muggy between June and August, and pleasant between November and February. The average annual rain fall is 81 mm, average maximum temperature is 31°C, average minimum temperature is 22°C, absolute maximum temperature is 47°C, absolute minimum temperature is 1°C, average morning relative humidity is 71% and average afternoon relative humidity is 43%. Moreover, this country is occasionally affected by sandstorms that originate in United Arab Emirates and the Kingdom of Saudi Arabia in the summer time (Abulfatih et al., 2001). A team studied the ecology of wastewater ponds in Qatar from Qatar University (Supplementary Figure 4) (Abulfatih et al., 2002). The implications of the physical and chemical characteristics of wastewaters at these ponds were discussed recently (Al-Thani and Yasseen, 2020, 2021).
One part of this investigation was concentrating on bacterial ecology, and Al-Thani (2002, 2003) has listed the main bacteria genera at these ponds. These genera and species include: (A) Gram-negative bacteria: Aeromonas hydrophila, P. aeruginosa, Klebsiella pneumoniae, Chromobacterium violaceum, and E. coli; and (B) Gram-positive bacteria: Streptomyces spp., Bacillus spp., and Macrococcus spp. The degree of the hazardous effect of these ponds was discussed in recent articles, and the decision-makers are fully aware of the future risk of these ponds continuing to receive wastewater (Elobaid et al., 2018; Al-Thani and Yasseen, 2020, 2021). Therefore, these ponds are being drained and replaced by new lagoons remote of the residential areas; however, lessons should be learned so that such pollution risks should be avoided in the future. The microbial ecology of the new lagoon at Al-Karaana (southwest of Doha city), for example, has not been studied yet. Therefore, any plan to investigate the biological roles of microorganisms at such anthropogenic lagoons and other industrial wastewater ponds to find out their environmental impact, lessons should be learned from the old ponds.
Recently, more details were reported regarding the ecology of microorganisms (algae and bacteria) and their possible biological roles in cleaning wastewater from organic and inorganic pollutants. There is concern about the future water sources in Qatar and at all the Arabian Gulf States. Thus, recent works have suggested using industrial wastewater (IWW) as a promising future water source after water is passed through a series of treatments and processes. These activities involve modern approaches and the contribution of native plants, algae, and bacteria through environmental, genetic, and biological approaches (Al-Thani and Yasseen, 2021). We refer the reader to these recent works to look at some details of the microbial ecology at those ponds and the perspectives of promising roles of microorganisms.
Rawdahs are deep depressions or low leveled areas with good soil and water; the soils of Rawdahs in Qatar and all other Gulf States are the most fertile (Supplementary Figure 5) and have freshwater sources from wells. These depressions can reach a maximum depth of 1.5 m and are filled with fine soil enriched with clay, silt, and minerals washed down from the surrounding higher areas during the rainy seasons. Thus, many of them were transformed into farmlands and are considered good sites as rangelands for domestic animals, such as camels and cattle (Abulfatih et al., 2001; Abdel-Bari, 2012). The soil is rich in biological soil crusts (BSC) with various communities of bacteria, fungi, algae, lichens, cyanobacteria, and mosses (Figure 6), which play significant biological roles in improving the nutritional status of the soil and inhibiting its deterioration (Richer et al., 2012).

Figure 6. The soil of the Rawdahs is rich in biological soil crusts (BSC), which have various biological roles.
Many investigations have measured the soil salinity (ECe) and pH at Rawdahs; however, many Rawdahs have low soil salinity levels (4–6 dSm–1) and acceptable pH ranges (6–8), and such figures might give a good indication of the quality of the soil at these locations (Abulfatih et al., 2001; Al-Thani and Yasseen, 2018a). At Rawdahs, many mesophytes, xerophytes, and facultative halophytes can be found, giving the impression of the adaptability of many plants to such habitats. More details about the microbial ecology of Rawdahs, including BSC, can be found in some recent literature (Al-Thani and Yasseen, 2018a). Case studies of the microbial communities at these locations will be discussed below, and especially for the private farms of Ghaf trees.
The microbiological community around the Ghaf trees (Prosopis cineraria) habitat does not exclusively represent the arid land and dry saline lands at the Arabian Gulf region (Supplementary Figure 6) but also includes Rawdhas of low saline soils (Figure 7). This tree is long-living, multi-purpose (fuel, wood, food and medicine, silviculture), drought-resistant, well adapted to high temperatures, and tolerant of infestation. The Qatar University and Environmental Studies Center (ESC) has conducted studies of the bacteria adjacent and associated with these trees (Abdel-Bari et al., 2007a). Supplementary Table 3 shows the soil characteristics at the subcanopy of P. cineraria trees at a private farm.

Figure 7. Many trees can be found at many private farms around Qatar. The second author was sitting near Ghaf trees during one of his research trips to Rawdahs.
The rhizosphere is the soil section that extends about 5 mm outward from the root surface and has a series of gradients of organic components (pH, O2, CO2, and H2O). The rhizosphere can be categorized into two main regions: (1) rhizosphere soil, the area around the root system; (2) the rhizoplane, the soil in direct contact with plant root. Some microorganisms, called endophytes, also inhabit the internal root tissue. The plant root releases organic and inorganic substances that are classified into five types: (a) exudates, (2) secretions, (3) lysates, (4) plant mucilage, (5) mucigel (Rovira et al., 1979). Exudates are low molecular weight compounds leaked non-metabolically from intact cells. Secretions are compounds leaked metabolically from the active plant. Lysates are compounds released by the autolysis of older cells. Plant mucilage compounds are carbohydrates released from, e.g., the root cap cells and primary cell wall. Mucigel compounds are gelatinous material of the plant and/or microbial origin; they lubricate the root tips to ease the growth of roots deep into the soil. These substances might play active roles related to the availability of nutrients and phytoremediation of various compounds (organic and inorganic origin). It has been reported that the rhizospheres of some arid plants (e.g., Prosopis juliflora) offer niches for bacteria that exert beneficial effects, including nutrient transformation, plant growth promotion, disease suppression, resistance to drought and salinity, and might remediate pollutants of various origin (Jothibasu et al., 2012; Yasseen and Al-Thani, 2013; Yasseen, 2014a). Al-Thani (2007) showed that soil populations of bacteria at the rhizosphere were quite different from the non-rhizosphere sites. Table 3 shows that the rhizoplane had highest populations that decrease steadily (toward the center and intermediate subcanopy) and then showed lowest counts at the outer canopy soil. The high content of organic matter, soil nutrients, and clay moisture at the rhizoplane and the sub-canopy locations might be why there is a significant difference in the bacterial counts between these locations. The study’s outcomes confirmed those of previous studies reporting that non-rhizosphere soils showed little bacterial activity (Bachman and Kinzel, 1992; Whitford, 2002).

Table 3. The number of bacterial colonies recorded from soil samples obtained from various locations at the rhizosphere and outer canopy soil of Ghaf (Prosopis cineraria) trees (Al-Thani, 2007).
Unfortunately, these studies did not identify the bacterial species at the rhizosphere and outer canopy soil of the Ghaf trees. The traditional methods for microbial identification are based on phenotypic characteristics, which are not accurate compared to genotypic methods (Matsumoto and Sugano, 2013). Moreover, in many clinical situations (Mignard and Flandrois, 2006), these methods are time-consuming and fail to identify some bacteria groups (for example, Gram-positive rods). Other studies have found the rhizosphere of P. juliflora harbors more diversified bacteria communities from various phyla than the non-rhizosphere areas, which means that such bacteria communities might boost the nutritional status for crops and wild plants (Jothibasu et al., 2012). In this study, the molecular approach to investigate the bacterial activity at the rhizosphere of P. juliflora revealed that the culturable microbes were higher than those remote of the rhizosphere region, and the root exudates of this plant might influence such activity. The bacterial communities at the rhizosphere are mainly affected by soil environmental factors, such as soil water content, soil nutrients, electrical conductivity, and salinity (Jia et al., 2020), which give rhizosphere microorganisms peculiarities compared to those found at the above-ground parts of plants; the phyllosphere. Both the rhizosphere and phyllosphere accommodate most of the bacterial communities found on the plant body, and these groups mainly include phyla bacteria: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (Bulgarelli et al., 2013).
The green parts of terrestrial plants represent the major biological active surface that provides essential elements of life that contribute substantially to CO2 fixation and biomass production, the liberation of O2 by photosynthesis, and secondary metabolites production. The term phyllosphere refers to the above-ground part of the plant, which is normally colonized by various types of microbes, such as bacteria, fungi, yeasts, and possibly others like algae, and such interactions might be detrimental or beneficial for either microbe of the host plants, which have been classified as neutralism, commensalism, and synergism, among others (Montesinos, 2003; Yasseen and Al-Thani, 2013). The rhizosphere and phyllosphere are both affected by different physical and chemical factors, and the nitrogen content of a plant affects the phyllosphere. Other factors related to soil environment and soil solution might have major impacts on the rhizosphere bacterial communities (Jia et al., 2020). Such factors might account for the peculiarities of these bacterial habitats; for example, pigmented bacteria, which are rarely found in the rhizosphere, dominate the leaf surfaces of green plants (Fokkema and Schippers, 1986; Zeng et al., 2014; Jia et al., 2020). The living organisms that inhabit the aerial parts of the plants are called epiphytes. Most of the studies on the microbiology of the phyllosphere have focused on leaves, the dominant part of the plant. However, bacteria are the most abundant inhabitants of the phyllosphere, and the average number of bacterial cells was 106–108 cells/g of a leaf (Hirano and Upper, 2000). Epiphytic bacterial populations show significant variation in size among and within plants of the same species over any period during the growing season (Thompson et al., 1993; Jacques et al., 1995; Al-Thani, 2007). The presence of such microbial communities on the aerial parts of plants might boost the positive interactions, thereby providing many features to the metabolic activities, such as resistance against biotic and abiotic factors, stress tolerance, growth promotion, nutrient acquisition, and disease resistance (Stone et al., 2018). The mechanisms of many activities offered by microorganisms to the host plants (adjacent or associated) were recently discussed in relation to resistance to abiotic resistance, including pollutants of oil and gas activities (Al-Thani and Yasseen, 2018a,b, 2020, 2021). Moreover, plants with large total leaf area, such as cucumber, beans, and other native plants, like Amaranthus spp., might accommodate more microorganisms than those of small and narrow leaves of crops (e.g., barley and wheat) or native plants (Juncus rigidus, Sporobolus spp.) and even those of waxy texture leaves (Kinkel et al., 2000; Al-Thani, 2007).
In conclusion, the functional roles of microbiota at aerial parts of plants are of interest; bacteria inhabiting the phyllosphere are linked to the biology of their host, as these bacteria might offer protection against a variety of bacteria, including pathogens. They might also influence plant reproduction (Massoni et al., 2020). Moreover, environmental factors (such as light) exert indirect effects on plant aerial tissues, such as modification of moisture on leaf surfaces, change of microbial lifestyle, alteration of phyllosphere composition, and diversity, among others (Carvalho and Castillo, 2018).
New approaches have been suggested in many aspects, including identification, biotechnology, and utilizing the desert lands for future wellbeing and prosperity. Regarding the identification of bacteria, the traditional approach based on phenotypic characteristics is time-consuming and might fail to identify and cover all microorganisms. This was replaced by more accurate genotypic methods (Embley and Stackebrandt, 1996), and using the 16S rRNA sequence has emerged as the most preferred modern method (Mignard and Flandrois, 2006; Matsumoto and Sugano, 2013). These methods have been referred to as: culture-independent methods; used widely and successfully since the ninetieths of the last century. Using such methods, numerous studies have identified a range of bacteria, cyanobacteria, and yeast for various uses under routine conditions (Redfield et al., 2002), these methods were better approach than the traditional ones in many aspects such as: environmental microbiology, environmental pollution, food processing, etc. (Su et al., 2012; Cocolin et al., 2013; Stefani et al., 2015). In fact, the cost is not a limiting factor as this technique provides great benefits and accuracy compared to traditional methods. In Qatar, works adopted this modern approach; 16S rRNA gene amplicons and metagenomic sequence analysis, described the most common microbial groups at the desert and marine habitats. Thus, serious breakthrough started a decade ago when a sequence of 16S rRNA gene of a naturally occurring wild type strain from the Qatari soil of Streptomyces coelicolor QU66C has been determined using modern technology. This strain has been deposited in the International Nucleotide Sequence Database (INSD), and assigned an accession number of AB588124, and it is known as: Streptomyces coelicolor (AB588124) (QU66c-2002) (Mahasneh et al., 2011). Also, Abdul Majid et al. (2016) have recognized three main bacteria phyla typically found at Qatari Barchan dunes using this technique: Actinobacteria (58%), Firmicutes (27%), and Proteobacteria (15%). More investigations are needed to determine the microbial communities at all the above habitats in Qatar and the Arabian Gulf region.
In recent years, by adopting modern technology, more studies have been identifying marine microorganisms at the Qatari coastal water of the Arabian Gulf. Identification of these new species was based on sequence analysis of the internal transcribed spacer (ITS) regions and D1/D2 domains of the large subunit (LSU) rRNA gene. It is interesting to report that some novel fungi species, including yeast, were isolated from the hypersaline marine environment using that technology; these new strains proved efficient in metabolizing some organic compounds (Fotedar et al., 2018, 2019b). These studies were followed by further investigations to explore their characterizations (Fotedar et al., 2019a).
Regarding the roles of microorganisms in the Qatari ecosystem, many desert and marine inhabitants have been involved in many aspects of life, including modern biotechnology, health and agriculture, and wildlife. Gathering international records, Tables 4, 5 show the reported roles played by these microorganisms.
Also, it is interesting to report that some data (personal observations) revealed the roles of yeast species at the Qatari soils during bioremediation of polluted soils with IWW; these species were identified using API procedure5 include: (1) Candida spp. as (Candida tropicalis, Candida format, and Candida guilliermondii); (2) Cryptococcus spp. as (Cryptococcus humicola, Cryptococcus albidus, and Cryptococcus laurentii); (3) Kloeckera spp.; (4) Pichia angusta spp.; and (5) Trichosporon mucoides, and possibly others. These yeast species show significant activation and growth at soil polluted with IWW, giving clear signs of their bioremediation processes. Further comprehensive studies are needed to clarify their roles in remediating polluted soils due to the oil and gas activities, at least at the Qatari soil. On the other hand, various fungi including yeast species from Ascomycota and Basidiomycota at the marine habitat of Qatar were identified. One interesting study of Fotedar et al. (2015) which had extended for 3 years using modern technology [sequence analyses of the internal transcribed spacers (ITS1/ITS2) and the D1/D2 domains of the large subunit (LSU) of the ribosomal DNA (rDNA)], showed that 37 genera were identified from these main divisions. Significant number of yeast species were found at sites influenced by anthropogenic activities, such as fishing harbors along the Eastern coast of Qatar. Candida spp. (Ascomycota) are common at land and sea; they are the most common cause of fungal infections worldwide. Thus, the study has concluded the following species: Candida spp. (24%) (Ascomycota), Rhodotorula spp. (16%) (Basidiomycota), Kondoa aeria (8%) (Basidiomycota), Aureobasidium spp. (7%) (Ascomycota), and Hortaea spp. (7%) (Ascomycota), were among the most frequently identified yeast species. However, these yeast species were varied during the main seasons of the year. For example, the dominant ones during the summer season were Kondoa spp. (Basidiomycota) (34%), Knufia spp. (24%) (Ascomycota), and Hypocrea spp. (13%) (Ascomycota), whereas Debaryomyces spp. (37%) (Ascomycota), Clavispora spp. (23%) (Ascomycota), Issatchenkia spp. (17%) (Ascomycota) were dominant species during the winter season. It very interesting to report a novel yeast species isolated from marine water in Qatar; it was characterized and identified and given the name Kondoa qatarensis (Fotedar et al., 2019b). Further investigation is needed to look at the roles might have at Qatari marine habitats.
Some recent reports are of particular interest. For example, that symbiotic and/or associated bacteria can boost plant growth, control of pathogens, or even alleviate abiotic factors like drought, salinity, and pollution of organic and inorganic components (Egamberdievea et al., 2017; Al-Thani and Yasseen, 2018b; Arif et al., 2020). Kaushal (2019) listed the many useful roles of rhizosphere bacteria in (1) phytoremediation and bioremediation; (2) promoting growth by production of siderophore (high-affinity iron-chelating compounds that are secreted by microorganisms such as bacteria and fungi and serve primarily to transport iron across cell membranes) and phytohormones (including auxins, gibberellins, cytokinins, ethylene, and abscisic acid), mobilize mineral phosphate, nitrogen fixation, and inhibition of plant ethylene synthesis; (3) increasing resistance to adverse conditions (drought, salinity, compost toxicity); (4) disease interactions; (5) improving survival of seedlings; (6) increasing biomass; and (7) immobilizing of heavy metals through bioaccumulation, biotransformation to less soluble forms, chemisorption and biomineralization of heavy metals. Yan et al. (2020) have concluded that microbes associated with plants at the rhizosphere confer plants with many traits to support plant growth under various stresses, including heavy metal tolerance. Moreover, plant exudates might attract some microorganisms to activate biological functions (Al-Thani and Yasseen, 2021). Also, Basu et al. (2021) have suggested that microorganisms are key players in many agricultural and ecological processes, including increasing fertility by promoting the availability of major elements such as nitrogen, phosphorus, and sulfur.
During the last decade, great attention has been paid to the promising roles of microorganisms in the Qatari ecosystem. For example, Al-Thani and Yasseen (2017) studied the halo-thermophilic bacteria and cyanobacteria adjacent to some halophytes: S. vermiculata, L. axillare, and T. qatarensis at the salt marshes and Sabkhas. These authors found that the most common bacteria groups are Gram-positive rods (such as Bacillus spp.) of various forms (with pointed ends occurred singly, pairs and short chains), and some cyanobacteria genera such as Anabaena and Nostoc, and suggested significant roles including nutrition, pollution, production of antibiotics. This study shows that the populations of thermo-halophilic bacteria were highest in soils adjacent to L. axillare compared to S. vermiculata and T. qatarensis (7.2, 0.4, and 0.65 cfu/g dry soil × 102, respectively). Further investigation is needed to examine the reasons behind such significant differences between the bacteria counts at the plant’s rhizosphere. Investigations also need to address the origin of metabolic pathways in these microorganisms and native plants (Al-Thani and Yasseen, 2017).
Later, discussions have shed light on the roles of BSC in such harsh environments of the Arabian Gulf region and the presence of extremophiles in salt glands and the plant body in providing some important resistance traits to the native plants and other biological needs. Some mechanisms were introduced and suggested that microorganisms support native plants against various types of environmental stresses at the Arabian Gulf region: biofilm formation, polymers production, chemotaxis, phytohormone production, nitrogen fixation, phosphate solubilization, production of phytohormone-degrading enzymes, and osmolytes biosynthesis (Al-Thani and Yasseen, 2018a,b). Moreover, microorganisms (algae and bacteria) and native plants at Qatari ponds are promising for bioremediation, phytoremediation, and phytoremediation of polluted soils and water. IWW pollution has become a significant issue because of increasing investments in the oil and gas sectors and increasing demand for quality water for various uses in agriculture and domestic uses (Al-Thani and Yasseen, 2020, 2021).
Microorganisms are also likely to contribute to the development of alternative energy sources to replace petroleum diesel. Simultaneously, many governments have begun to turn toward renewable energy (e.g., wind and solar). Recently, the idea of biodiesel has emerged as a cost-effective and environmentally friendly biological approach. The Arabian Gulf region is well-suited to biodiesel projects. Wilson et al. (2012) have reported that microalgae (e.g., Dunaliella and Chlorella, and possibly others) are available in seawater and that seawater is the best growth medium for such living organisms. Simultaneously, the deserts of this region, including the land of Qatar, are sufficiently large to accommodate such projects, assuming that such lands are not used for agricultural activities.
Finally, microorganisms such as bacteria and algae can be considered as bio-indicators for many environmental issues. For example, the presence of some algae and cyanobacteria in waters and/or soils can be considered as a sign of pollution, while some other algae species are found only in fresh water and a sign of water quality (Yasseen et al., 2001). Also, the presence of some bacteria species in water might cause some illnesses and sign of pollution (Karbasdehi et al., 2017). More details can be found in some published works (Al-Thani and Yasseen, 2021). Therefore, considering the international standards and guidelines for water used in drinking water and irrigation as obligation in any work related to water and soil used by humans.
It seems likely that microbes are poised to make significant contributions to future technological endeavors and the overcoming of the many problems facing humanity (Makhalanyane et al., 2015). Microorganisms, including bacteria and yeasts, have emerged as biological tools (Glick, 2014; Hanin et al., 2016). The countries of the Arabian Gulf have the ecological, scientific, and financial resources to make significant contributions to such progress (Bhatnagar and Bhatnagar, 2005). The microorganisms found within Qatari habitats have shown promise for use in bioremediation and phytoremediation (Al-Thani and Yasseen, 2021), and various aspects of human life including health, agriculture, and wildlife.
Written informed consent was obtained from the relevant individual for the publication of any potentially identifiable images or data included in this article.
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank the Environmental Studies Center (ESC), Qatar University, for the technical and financial support of the research projects reported in this article. The authors acknowledge that the editing service was done by American Manuscript Editing; AME, Kevin.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.697269/full#supplementary-material
Abdel-Bari, E. M., Fahmy, G. M., Al-Thani, N. J., Al-Thani, R. F., and Abdel-Dayem, M. S. (2007a). The Ghaf Tree, Prosopis cineraria in Qatar. Doha: Environmental Studies Centers, Qatar University.
Abdel-Bari, E. M., Yasseen, B. T., and Al-Thani, R. F. (2007b). Halophytes in the State of Qatar. Doha: Environmental Studies Center, Qatar University.
Abdel-Bari, E. M. M. (2012). The Flora of Qatar, The Dicotyledons, The Monocotyledons, Vol. 1. Doha: Environmental Studies Centre, Qatar University, 2.
Abdul Majid, S., Graw, M. F., Chatziefthimiou, A. D., Nguyen, H., Richer, R., Louge, M., et al. (2016). Microbial characterization of Qatari Barchan sand dunes. PLoS One 11:e0161836. doi: 10.1371/journal.pone.0161836
Abouelkheir, S. S., Abdelghany, E. A., Ghozlan, H. A., and Sabry, S. A. (2020). Characterization of biofilm forming marine Pseudoalteromonas spp. J. Mar. Sci. 2, 31–37. doi: 10.30564/jms.v2i1.1412 (accessed May 28, 2021).
Abulfatih, H. A., Abdel Bari, E. M., Alsubaey, A., and Ibrahim, Y. M. (2001). Vegetation of Qatar. Doha: Scientific and Applied Research Centre (SARC), Qatar University.
Abulfatih, H. A., Al-Thani, R. F., Al-Naimi, I. S., Swelleh, J. A., Elhag, E. A., and Kardousha, M. M. (2002). Ecology of Wastewater Ponds in Qatar. Doha: Scientific and Applied Research Centre (SARC), University of Qatar.
Adam, P. (1990). Saltmarsh Ecology. Cambridge: Cambridge University Press, doi: 10.1017/CBO9780511565328
Ahmed, I., Yokota, A., and Fujiwara, T. (2007). A novel highly boron tolerant bacterium, Bacillus boroniphilus sp. nov., isolated from soil, that requires boron for its growth. Extremophiles 11, 217–224. doi: 10.1007/s00792-006-0027-0
Ahmed, R. N., Sani, A., Ajijolakewu, A. K., and Alamu, F. B. (2013). Soil screening for antibiotic–producing microorganisms. Adv. Environ. Biol. 7, 7–11.
Al-Ansi, M. A., Abdel-Bari, E. M., Yasseen, B. T., and Al-Khayat, J. A. (2004). Coastal Restoration: Restoration of a Coastal Vegetation Habitat at Ras Raffan Industrial City. SARC, University of Qatar.
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genom. 15:482. doi: 10.1186/1471-2164-15-482
Alexander, A., Singh, V. K., Mishra, A., and Jha, B. (2019). Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS One 14:e0222405. doi: 10.1371/journal.pone.0222405
Al-husnan, L. A., and Alkahtani, M. D. F. (2016). Molecular identification of Streptomyces producing antibiotics and their antimicrobial activities. Ann. Agric. Sci. 61, 251–255. doi: 10.1016/j.aoas.2016.06.002
Al-Naimi, H. A., Al-Ghouti, M. A., Al-Shaikh, I., Al-Yafe, M., and Al-Meer, S. (2015). Metal distribution in marine sediment along the Doha Bay, Qatar. Environ. Monit. Assess. 187:130. doi: 10.1007/s10661-015-4352-6
Alotaibi, M. O., Sonbol, H. S., Alwakeel, S. S., Suliman, R. S., Fodah, R. A., Abu Jaffal, A. S., et al. (2020). Microbial diversity of some sabkha and desert sites in Saudi Arabia. Saudi J. Biol. Sci. 27, 2778–2789. doi: 10.1016/j.sjbs.2020.06.038
Al-Sayed, H. A., Ghanem, E. H., and Saleh, K. M. (2005). Bacterial community and some physico-chemical characteristics in a subtropical mangrove environment in Bahrain. Mar. Pollut. Bull. 50, 147–155. doi: 10.1016/j.marpolbul.2004.10.002
Al-Thani, R. F. (2002). “Coliform bacteria of wastewater ponds in Qatar, part 2,” in Ecology of Wastewater Ponds in Qatar, eds H. A. Abulfatih, R. F. Al-Thani, I. S. Al-Naimi, J. A. Swelleh, E. A. Elhag, and M. M. Kardousha (Doha: Scientific and Applied Research Centre (SARC), University of Qatar), 129–145.
Al-Thani, R. F. (2003). The coliform bacteria in the wastewater ponds of Doha city, Qatar. Qatar Univ. Sci. J. 23, 99–107.
Al-Thani, R. F. (2007). “Microbiological analysis of Prosopis cineraria (L.) Druce in the State of Qatar,” in The Ghaf Tree Prosopis Cineraria in Qatar, eds E. M. M. Abdel-Bari, G. M. Fahmy, N. J. Al-Thani, R. F. Al-Thani, and M. S. Abdel-Dayem (Doha: National Council for Culture, Arts and Heritage, Environmental Studies Center, QU).
Al-Thani, R. F., Abd-El-Haleem, D., and Al- Shammri, M. (2007). Isolation, biochemical and molecular characterization of 2-chlorophenol degrading Bacillus isolates. Afric. J. Biotechnol. 6, 2675–2681. doi: 10.5897/ajb2007.000-2428
Al-Thani, R. F., Abd-El-Haleem, D. A., and Al-Shammri, M. (2009). Isolation and characterization of polyaromatic hydrocarbons-degrading bacteria from different Qatar soils. Afric. J. Micro. Res. 3, 761–766.
Al-Thani, R. F., and Mahasneh, I. A. K. (2002). Ecology and Distribution of Streptomyces in the Soil of Qatar. Legal deposit No.- at qul 212/2004. Doha: Environmental Studies Centre (ESC), Qatar University, 69.
Al-Thani, R. F., and Yasseen, B. T. (2017). Halo-Thermophilic bacteria and heterocyst cyanobacteria found adjacent to halophytes at Sabkhas–Qatar: preliminary study and possible roles. Afric. J. Micro. Res. 11, 1346–1354. doi: 10.5897/ajmr2017.8648
Al-Thani, R. F., and Yasseen, B. T. (2018a). Biological soil crusts and extremophiles adjacent to native plants at Sabkhas and Rawdahs, Qatar: the possible roles. Front. Environ. Microbiol. 4:55–70. doi: 10.11648/j.fem.20180402.13
Al-Thani, R. F., and Yasseen, B. T. (2018b). Solutes in native plants in the Arabian Gulf region and the role of microorganisms: future research. J. Plant Ecol. 11, 671–684. doi: 10.1093/jpe/rtx066
Al-Thani, R. F., and Yasseen, B. T. (2020). Phytoremediation of Polluted soils and waters by native Qatari plants: future perspectives. Environ. Pollut. 259:113694. doi: 10.1016/j.envpol.2019.113694
Al-Thani, R. F., and Yasseen, B. T. (2021). Perspectives of future water sources in Qatar by phytoremediation: biodiversity at ponds and modern approach. Int. J. Phytoremediation 4, 1–24. doi: 10.1080/15226514.2020.1859986
Al-Zahrani, S H., and Al-Harbi, A. A. (2004). Studies on soil Streptomyces from Saudia Arabia. Egypt. J. Microbiol. 39, 59–65.
Amaresan, N., Kumar, K., Naik, J. H., Bapatla, K. G., and Mishra, R. K. (2018). “Streptomyces in plant growth promotion: mechanisms and role, Chapter 8,” in New and Future Developments in Microbial Biotechnology and Bioengineering, Actinobacteria: Diversity and Biotechnological Applications, eds B. P. Singh, V. K. Gupta, and A. K. Passari (Amsterdam: Elsevier), 125–135. doi: 10.1016/B978-0-444-63994-3.00008-4
Amir, R., Ahmed, N., and Talat, M. (1993). Salt-tolerance in mangrove bacteria. Pak. J. Mar. Sci. 2, 129–135.
Anand, A., Zeyara, A., Al Malaki, A., Al-Ghanim, M. M., Hitha, P. K., and Taj, et al. (2016). “Isolation and identification of potentially pathogenic vibrio species from qatari coastal seawaters,” in Proceedings of the Qatar Foundation Annual Research Conference Proceedings Volume 2016 EESP2323 (Ar Rayyan: Hamad bin Khalifa University Press (HBKU Press)).
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Ashfaq, M. Y., Al-Ghouti, M. A., Qiblawey, H., Rodrigues, D. F., and Hu, Y., and Zouari, C. (2019). Isolation, identification and biodiversity of antiscalant degrading seawater bacteria using MALDI-TOF-MS and multivariate analysis. Sci. Total Environ. 656, 910–920. doi: 10.1016/j.scitotenv.2018.11.477
Bachman, G., and Kinzel, H. (1992). Physiological and ecological aspects of the interactions between plant roots and rhizosphere soil. Soil. Biol. Biochem. 24, 5543–5552.
Bagwell, C. E., Piceno, Y. M., Lucas, A. A., and Lovell, C. R. (1998). Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl. Environ. Microbiol. 64, 4276–4282. doi: 10.1128/AEM.64.11.4276-4282.1998
Baker-Austin, C., JOliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio spp. Infections. Nat. Rev. Dis. Primers 4, 1–19. doi: 10.1038/s41572-018-0005-8
Barac, T., Toghavi, S., Borremans, B., Provoost, A., Oeyen, L., Colpaert, J. V., et al. (2004). Engineered endophytic bacteria improved phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 2, 583–588. doi: 10.1038/nbt960
Barakate, M., Ouhdouch, Y., and Oufdou, Kh, and Beaulien, C. (2002). Characterization of rhizospheric soil streptomycetes from Moroccan habitats and their antimicrobial activities. World J. Microbiol. Biotechnol. 18, 49–54. doi: 10.1023/A:1013966407890
Bardgett, R. D., and Griffiths, B. (1997). “Ecology and biology of soil protozoa, nematodes and microarthropods,” in Modern Soil Microbiology, eds J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (New York, NY: Marcel Dekker, Inc), 129–163.
Basu, S., Kumar, G., Chhabra, S., and Prasad, R. (2021). “Role of soil microbes in biogeochemical cycle for enhancing soil fertility, Chapter 13,” in New and Future Developments in Microbial Biotechnology and Bioengineering, ed. V. K. Gupta (Amsterdam: Elsevier), 149–157. doi: 10.1016/B978-0-444-64325-4.00013-4
Benaiges-Fernandez, R., Palau, J., Offeddu, F. G., Cama, J., Urmeneta, J., Soler, J. M., et al. (2019). Dissimilatory bioreduction of iron (III) oxides by Shewanella loihica under marine sediment conditions. Mar. Environ. Res. 151:104782. doi: 10.1016/j.marenvres.2019.104782
Bhatnagar, A., and Bhatnagar, M. (2005). Microbial diversity in desert ecosystems. Curr. Sci. 89, 91–100.
Bizuye, A., Moges, F., and Andualem, B. (2013). Isolation and screening of antibiotic producing actinomycetes from soils in Gondar town, North West Ethiopia. Asian Pac. J. Trop. Dis. 3, 375–381. doi: 10.1016/s2222-1808(13)60087-0
Boustie, J., and Grube, M. (2005). Lichens-a promising source of bioactive secondary metabolites. Plant Genet. Resour. 3, 273–278. doi: 10.1079/pgr200572
Bowman, J. P. (1998). Pseudoalteromonas prydzensis sp. nov., a psychrotrophic, halotolerant bacterium form Antarctic sea ice. Appl. Environ. Microbiol. 83:e00175-17. doi: 10.1128/AEM.00175-17
Bulgarelli, D., Schlaeppi, K., Spaepen, S., van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Burt, J. A., and Batholomew, A. (2019). Towards more sustainable coastal development in the Arabian Gulf: opportunities for ecological engineering in an urbanized seascape. Mar. Pollut. Bull. 142, 93–102. doi: 10.1016/j.marpolbul.2019.03.024
Burt, J. A., Ben-Hamadou, R., Abdel-Moati, M. A. R., Fanning, L., Kaitibie, S., Al-Jamali, F., et al. (2017). Improving management of future coastal development in Qatar through ecosystem-based management approaches. Ocean Coast. Manag. 148, 171–181. doi: 10.1016/j.ocecoaman.2017.08.006
Carvalho, S. D., and Castillo, J. A. (2018). Influence of light on plant–phyllosphere interaction. Front Plant Sci. 9:1482. doi: 10.3389/fpls.2018.01482
Catania, V., Cappello, S., Di Giorgi, V., Santisi, S., Di Maria, R., Mazzola, A., et al. (2018). Microbial communities of polluted sub-surface marine sediments. Mar. Pollut. Bull. 131, 396–406.
Chen, H. M., Chi, H., Chiu, N. C., and Huang, F. Y. (2015). Kocuria kristinae: a true pathogen in pediatric patients. J. Microbiol. Immunol. Infect. 48, 80–84. doi: 10.1016/j.jmii.2013.07.001
Choudhury, J. D., Pramanik, A., Webster, N. S., Llewellyn, L. E., Gachhui, R., and Mukherjee, J. (2015). The pathogen of the great barrier reef sponge Rhopaloeides odorabile is a New Strain of Pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar. Biotechnol. (NY) 17, 463–478. doi: 10.1007/s10126-015-9627-y
Cocolin, L., Alessandria, V., Dolci, P., Gorra, R., and Rantsiou, K. (2013). Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int. J. Food Microbiol. 167, 29–43. doi: 10.1016/j.ijfoodmicro.2013.05.008
Dias, M. P., Bastos, M. S., Xavier, V. B., Cassel, E., Astarita, L. V., and Santarém, E. R. (2017). Plant growth and resistance promoted by Streptomyces spp. in tomato. Plant Physiol. Biochem. 118, 479–493. doi: 10.1016/j.plaphy.2017.07.017
Egamberdievea, D., Wirth, S. J., Alqarawi, A. Z., Abd-Allah, E. F., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8:2104. doi: 10.3389/fmicb.2017.02104
Elobaid, E. A., Sadooni, F. N., Lebbe, L., Claus, J., Boeckaert, C., Muylaert, K., et al. (2018). “Environmental Evaluation Risk Assessment and Management Options of Abu Nakhla Treated Sewage Effluent (TSE) Unlined Pond Doha Qatar,” in Proceedings of the Qatar Foundation Annual Research Conference Proceedings 2018, EEPD470 (Ar Rayyan: Hamad bin Khalifa University Press (HBKU Press)), doi: 10.5339/qfarc.2018.EEPD470
El-Tarabily, K. A. (2002). Total microbial activity and microbial composition of a mangrove sediment are reduced by oil pollution at a site in the Arabian Gulf. Can. J. Microbiol. 48, 176–182. doi: 10.1139/w01-140
Elwan, S. H., Diab, A., and Al-Gounaim, M. Y. (1985). Ecology of the streptomycetes flora in the Desert Soil of Kuwait. Syst. Appl. Microbiol. 6, 99–104. doi: 10.1016/S0723-2020(85)80020-5
Embley, T. M., and Stackebrandt, E. (1996). “The use of 16S ribosomal RNA sequences in microbial ecology,” in Molecular Approaches to Environmental Microbiology, eds R. W. Pickup and J. R. Saunders (London: Horwood), 39–62.
Fahmy, G. M., and Al-Thani, R. F. (2006). Ecology of Halophytes and their Bacterial Inhabitants in the Coastal Salt Marsh of Al-Dhakhira, Qatar. Doha: Environmental Studies Centre (ESC), University of Qatar.
Fahmy, G. M., and Ouf, S. A. (1992). The mucilage glands of Limonium delicatulum (Plumbaginaceae): Secretion and fungal colonization. Zagazig J. Agric. Res. 19, 773–788.
Fahmy, G. M., and Ouf, S. A. (1999). Significance of microclimate on phylloplane mycoflora of green and senescing leaves of Zygophyllum album L. J. Arid Environ. 41, 257–276. doi: 10.1006/jare.1998.0474
Fahmy, G. M., Zeid, I. M., Hassan, L. M., and Farahat, E. A. M. M. (2004). “Impacts of Fuel Oil (mazot) combustion products of brick-klins on air quality and on two economic plants,” in Proceedings of 3th International Conference on Biological Sciences, Tanta University, Vol. 3, Tanta, 463–481.
Feto, N. A. (2016). “Bacillus spp. and their biotechnological roles in green industry,” in Bacilli and Agrobiotechnology, eds M. Islam, M. Rahman, P. Pandey, C. Jha, and A. Aeron (Cham: Springer), doi: 10.1007/978-3-319-44409-3_7
Fokkema, N. J., and Schippers, B. (1986). “Phyllosphere vs rhizosphere as environments for saprophyte colonization,” in Microbiology of the Phyllosphere, eds N. J. Fokkema and J. van den Heuvel (London: Cambridge University Press), 137–159.
Fotedar, R. (2013). “Identification of bacteria from the marine environment surrounding Qatar,” in Proceedings of the Qatar Foundation Annual Research Forum Proceedings Volume 2013 EEP-074 (Ar Rayyan: Hamad bin Khalifa University Press (HBKU Press)). doi: 10.5339/qfarf.2013.EEP-074
Fotedar, R., Fell, J. W., Boekhout, T., Kolecka, A., Zeyara, A., Kaul, R., et al. (2019a). Cystobasidium halotolerans sp. nov., a novel basidiomycetous yeast species isolated from the Arabian Gulf. Int J Syst Evol Microbiol. 69, 839–845. doi: 10.1099/ijsem.0.003250
Fotedar, R., Kolecka, A., Boekhout, T., Fell, J. W., Anand, A., Al Malaki, A., et al. (2018). Naganishia qatarensis sp. nov., a novel basidiomycetous yeast species from a hypersaline marine environment in Qatar. Int. J. Syst. Evol. Microbiol. 68, 2924–2929. doi: 10.1099/ijsem.0.002920
Fotedar, R., Kolecka, A., Boekhout, T., Fell, J. W., Zeyara, A., Al-Malki, A., et al. (2019b). Kondoa qatarensis f. a., sp. nov., a novel yeast species isolated from marine water in Qatar. Int. J. Syst. Evol. Microbiol. 69, 486–492. doi: 10.1099/ijsem.0.003182
Fotedar, R., Zeyara, A., Al Malaki, A., Fell, J. W., Stoeck, T., Filker, S., et al. (2016). “Biodiversity, Sustainability and Climate Change, with Perspectives from Qatar,” in Proceedings of the QScience Proceedings, Qatar University Life Science Symposium 2016: Volume 2016 (Ar Rayyan: Hamad bin Khalifa University Press (HBKU Press)), 32. doi: 10.5339/qproc.2016.qulss.32
Fotedar, R., Zeyara, A., Kolecka, A., Al Malaki, A., Fell, J. W., Bukhari, S. J., et al. (2015). “Distribution of yeasts in the Arabian Gulf surrounding Qatar,” in Proceedings of the QScience Proceedings, Qatar University Life Science Symposium-QULSS 2015 Global Changes: The Arabian Gulf Ecosystem, Volume 2015 (Ar Rayyan: Hamad bin Khalifa University Press (HBKU Press)), 10. doi: 10.5339/qproc.2015.qulss2015.10
Fujita, M. J., Nakano, K., and Sakai, R. (2013). Bisucaberin B, a linear hydroxamate class siderophore from the marine bacterium Tenacibaculum mesophilum. Molecules 18, 3917–3926. doi: 10.3390/molecules18043917
Gerhardt, P., Murray, R. G. E., Costilow, R. N., Nester, K. W., Wood, W. A., Krieg, N. R., et al. (1981). Manual of Methods for General Bacteriology. Washington DC: American Society for Microbiology.
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Godfrey, B. E. S. (1976). “Leachates from aerial parts of plants and their relation to plant surface microbial populations,” in Microbiology of Aerial Plant Surfaces, eds C. H. Dickinson and T. F. Preece (London: Academic Press), 433–439. doi: 10.1016/b978-0-12-215050-0.50023-0
González-López, O., and Ruano-Rosa, D. (2020). “Root exudates, a key factor in the plant-bacteria interaction mechanisms, Chapter 8,” in Molecular Aspects of Plant Beneficial Microbes in Agriculture, eds V. Sharma, R. Salwan, and L. Tawfeeq (London: Academic Press)111–121. doi: 10.1016/B978-0-12-818469-1.00009-2
Gougoulias, C., Clark, J. M., and Shaw, L. J. (2014). The role of soil microbes in the global carbon cycle: tracking the belowground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 94, 2362–2371. doi: 10.1002/jsfa.6577
Grothusen, H., Castillo, A., Henríquez, P., Navas, E., Bohle, H., Araya, C., et al. (2016). First complete genome sequence of Tenacibaculum dicentrarchi, an emerging bacterial pathogen of salmonids. Genome Announc. 4:e01756-15. doi: 10.1128/genomeA.01756-15
Gupte, T. E., and Naik, S. R. (1999). Isolation, taxonomic and fermentation studies on a new strain of Streptomyces arenae var ukrainiana producing a tetraene antibiotic. World J. Microbiol. Biotechnol. 15, 545–552.
Hanin, M., Ebel, C., Ngom, M., Laplaze, L., and Masmoudi, K. (2016). New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 7:1787.
Hashem, A., Tabassum, B., and Abd-Allah, E. F. (2019). Bacillus subtilis: a plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 26, 1291–1297. doi: 10.1016/j.sjbs.2019.05.004
Hashidoko, Y., Itoh, E., Yokota, K., Yoshida, T., and Tahara, S. (2002). Characterization of five phyllosphere bacteria isolated from Rosa rugosa leaves, and their phenotypic and metabolic properties. Biosci. Biotechnol. Biochem. 66, 2474–2478. doi: 10.1271/bbb.66.2474
Hassan, H., Abou-Elezz, A., Abuasali, M., and Al-Saadi, H. (2019). Baseline concentrations of mercury species within sediments from Qatar’s coastal marine zone. Mar. Pollut. Bull. 142, 595–602. doi: 10.1016/j
Hensyl, W. R. (1994). Bergey’s Manual of Determinative Bacteriology, 9th Edn. London: Williams and Wilkins Co.
Hirano, S. S., and Upper, C. D. (2000). Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae- a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64, 624–653. doi: 10.1128/mmbr.64.3.624-653.2000
Holmström, C., and Kjelleberg, S. (1999). Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30, 285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x
Hoshino, T., Doi, H., Uramoto, G.-I., Wörmer, L., Adhikari, R. R., Xiao, N., et al. (2020). Global diversity of microbial communities in marine sediment. PNAS 117, 27587–27597. doi: 10.1073/pnas.1919139117
Hua, N.-P., Kanekiyo, A., Fujikura, K., Yasuda, H., and Naganuma, T. (2007). Halobacillus profundi sp. nov. and Halobacillus kuroshimensis sp. nov., moderately halophilic bacteria isolated from a deep-sea methane cold seep. Int. J. Syst. Evol. Microbiol. 57(Pt 6), 1243–1249. doi: 10.1099/ijs.0.64817-0
Huck, T. A., Porter, N., and Bushell, M. E. (1991). Positive selection of antibiotic-producing soil isolates. J. Gen. Microbiol. 137, 2321–2329. doi: 10.1099/00221287-137-10-2321
Huddleston, A. S., Cresswell, N., Neves, M. C., Beringer, J. E., Baumbers, S., Thomas, D. I., et al. (1997). Molecular detection of streptomycin-producing Streptomyces in Brazilian soils. Appl. Environ. Microbiol. 63, 1288–1297. doi: 10.1128/aem.63.4.1288-1297.1997
Jaafar, R. S. (2019). The potential role of Sphingomonas paucimobilis in bioremediation of soils contaminated with hydrocarbon and heavy metal. Malays. J. Sci. (MJS) 38, 48–58. doi: 10.22452/mjs.vol38no3.5
Jacques, M., Kinkel, L. L., and Morris, C. E. (1995). Population sizes, immigration, and growth of epiphytic bacteria on leaves of different ages and positions of field-grown endive (Cichorium endivia var. latifolia). Appl. Environ. Microbiol. 61, 899–906. doi: 10.1128/aem.61.3.899-906.1995
Jia, T., Yao, Y., Wang, R., Wu, T., and Chai, B. (2020). Dynamics relationship of phyllosphere and rhizosphere bacterial communities during the development of Bothriochloa ischaemum in copper tailings. Front. Microbiol. 11:869. doi: 10.3389/fmicb.2020.00869
Jothibasu, K., Chinnadurai, C., Sundaram, S. P., Kumar, K., and Balachandar, D. (2012). Molecular profiling of rhizosphere bacterial communities associated with Prosopis juliflora and Parthenium hysterophorus. J. Microbiol. Biotechnol. 22, 301–310. doi: 10.4014/jmb.1109.09025
Karbasdehi, V. N., Dobaradaran, S., Nabipour, I., Ostovar, A., Arfaeinia, H., Vazirizadeh, A., et al. (2017). Indicator bacteria community in seawater and coastal sediment: the Persian Gulf as a case. J. Environ. Health Sci. Eng. 15:6 doi: 10.1186/s40201-017-0266-2
Kathiresan, K., Saravanakumar, K., Anburaj, R., Gomathi, V., Abirami, G., Sahu, S. K., et al. (2011). Microbial enzyme activity in decomposing leaves of mangroves. Int. J. Adv. Biotechnol. Res. 2, 382–389.
Kaushal, M. (2019). “Chapter 12 - Climatic Resilient Agriculture for Root, Tuber, and Banana Crops using Plant Growth-Promoting Microbes,” in Climate Change and Agricultural Ecosystems, eds K. K. Choudhary, A. Kumar, and A. K. Singh (Sawston: Woodhead Publishing), 307–329. doi: 10.1016/B978-0-12-816483-9.00012-8
Kembel, S. W., O’Connor, T. K., Arnold, H. K., Hubbell, S. P., Wright, S. J., and Green, J. L. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U.S.A. 111, 13715–13720. doi: 10.1073/pnas.1216057111
Kinkel, L., Wilson, M., and Lindow, S. (2000). Plant species and plant incubation conditions influence variability in epiphytic bacterial population size. Microb. Ecol. 39, 1–11. doi: 10.1007/s002489900182
Kortemaa, H., Rita, H., Haahtela, K., and Smolander, A. (1994). Root-colonization ability of antagonistic Streptomyces grisieviridis. Plant Soil. 163, 77–83. doi: 10.1007/bf00033943
Lei, X., Chen, Y., Shao, Z., Chen, Z., Li, Y., Zhu, H., et al. (2015). Effective harvesting of the microalgae Chlorella vulgaris via flocculation-flotation with bioflocculant. Bioresour. Technol. 198, 922–925. doi: 10.1016/j.biortech.2015.08.095
Lelchat, F., Cérantola, S., Brandily, C., Colliec-Jouault, S., Baudoux, A.-C., Ojima, T., et al. (2015). The marine bacteria Cobetia marina DSMZ 4741 synthesizes an unexpected K-antigen-like exopolysaccharide. Carbohydr. Polym. 124, 347–356. doi: 10.1016/j.carbpol.2015.02.038
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere, minireview. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/aem.69.4.1875-1883.2003
Löfblom, J., Rosenstein, R., Nguyen, M. T., Ståhl, S., and Götz, F. (2017). Staphylococcus carnosus: from starter culture to protein engineering platform. Appl. Microbiol. Biotechnol. 101, 8293–8307. doi: 10.1007/s00253-017-8528-6
Lv, Q., Zhang, B., Xing, X., Zhao, Y., Cai, R., Wang, W., et al. (2018). Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: novel approach and mechanisms investigation. J. Hazard. Mater. 347, 141–149. doi: 10.1016/j.jhazmat.2017.12.070
Ma, E. S., Wong, C. L., Lai, K. T., Chan, E. C., Yam, W. C., and Chan, A. C. (2005). Kocuria kristinae infection associated with acute cholecystitis. BMC Infect. Dis. 5:60. doi: 10.1186/1471-2334-5-60
Mahasneh, A. M. (2000). Distribution, Abundance and Seasonal Variation of Aerobic Heterotrophic Marine Bacteria Attached to A. Marina Leaves and Surrounding Water at the Qatari Coast. A report prepared for SARC. Doha: University of Qatar, 22–52.
Mahasneh, A. M. (2001a). Seasonal incidence of some heterotrophic aerobic marine bacteria in an Avicennia marina habitat along a Qatari coast. Online J. Biol. Sci. 1, 666–670. doi: 10.3923/jbs.2001.666.670
Mahasneh, A. M. (2001b). Bacterial decomposition of Avicennia marina leaf litter from Al-Khor (Qatar-Arabian Gulf). Online J. Biol. Sci. 1, 717–719. doi: 10.3923/jbs.2001.717.719
Mahasneh, A. M. (2002). Heterotrophic marine bacteria attached to leaves of Avicennia marina L. along the Qatari coast (Arabian Gulf). Online J. Biol. Sci. 2, 740–743. doi: 10.3923/jbs.2002.740.743
Mahasneh, A. M., and Al-Sayed, H. A. (1997). Seasonal incidence of some heterotrophic aerobic bacteria in Bahrain pelagic and near shore waters and Oysters. Int. J. Environ. Studies 51, 301–312. doi: 10.1080/00207239708711088
Mahasneh, I., Al-Thani, R. F., Abuannadi, M. A., and Qandil, M. A. (2011). A complete sequence of the 16S rRNA gene of a novel Streptomyces coelicolor (AB588124) (QU66c-2002) isolated from the soil of Qatar. Biotechnology (Faisalabad) 10, 167–174. doi: 10.3923/biotech.2011.167.174
Makhalanyane, T. P., Valverde, A., Gunnigle, E., Frossard, A., Ramond, J.-B., and Cowan, D. A. (2015). Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 39, 203–221. doi: 10.1093/femsre/fuu011
Manteca, A., and Yagüe, P. (2019). “Streptomyces as a source of antimicrobials: novel approaches to activate cryptic secondary metabolite pathways,” in Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods, ed. R. Croatia. (London, UK: Intech Open Limited) doi: 10.5772/intechopen.81812
Masai, E., Katayama, Y., and Fukuda, M. (2007). Genetic and biochemical investigations on bacterial catabolic pathway for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71, 1–15. doi: 10.1271/bbb.60437
Masák, J., Čejková, A., Schreiberová, O., and Řezanka, T. (2014). Pseudomonas biofilms: possibilities of their control. FEMS Microbiol. Ecol. 89, 1–14. doi: 10.1111/1574-6941.12344
Massoni, J., Bortfeld-Miller, M., Jardillier, L., Salazar, G., Sunagawa, S., and Vorhol, J. A. (2020). Consistent host and organ occupancy of phyllosphere bacteria in a community of wild herbaceous plant species. ISME J. 14, 245–258. doi: 10.1038/s41396-019-0531-8
Matejkova, O. N. J. (2010). Stenotrophomonas maltophilia: significant contemporary hospital pathogen–review. Folia Microbiol. 55, 286–294. doi: 10.1007/s12223-010-0043-4
Matsumoto, T., and Sugano, M. (2013). 16S rRNA gene sequence analysis for bacterial identification in the clinical laboratory. Rinsho Byori 61, 1107–1115.
Melgarejo, M., Sterner, O., Vila-Castro, J., and Mollinedo, P. (2008). More investigations in potent activity and relationship structure of the lichen antibiotic (+)- using acid its derivate dibenzoylusnic acid. Rev. Boliviana Quim. 25, 24–29.
Mercier, J., and Lindow, S. E. (2000). Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66, 369–374. doi: 10.1128/aem.66.1.369-374.2000
Mignard, S., and Flandrois, J. P. (2006). 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J. Microbiol. Methods 67, 574–581. doi: 10.1016/j.mimet.2006.05.009
Miura, N., Motone, K., Takagi, T., Aburaya, S., Watanabe, S., Aoki, W., et al. (2019). Ruegeria sp. Strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Mar. Biotechnol. 21, 1–8. doi: 10.1007/s10126-018-9853-1
Montesinos, E. (2003). Plant-associated microorganisms: a view from the scope of microbiology. Int. Microbiol. 6, 221–223. doi: 10.1007/s10123-003-0141-0
Morya, R., Salvachúa, D., and Thakur, I. S. (2020). Burkholderia: an untapped but promising bacterial genus for the conversion of aromatic compounds. Trends Biotechnol. 38, 963–975. doi: 10.1016/j.tibtech.2020.02.008
Moubasher, A. H., and Al-Subai, A. A. T. (1987). Soil Fungi in the State of Qatar. Doha: The Scientific and Applied Research Council, University of Qatar, 108.
Müller, K. (2001). Pharmaceutically relevant metabolites from lichens. Appl. Microbiol. Biotechnol. 56, 9–16. doi: 10.1007/s002530100684
Nagelkerken, I., Blaber, S. J. M., Bouillon, S., Green, P., Haywood, M., Kirton, L. G., et al. (2008). The habitat function of mangroves for terrestrial and marine fauna: a review. Aquat. Bot. 89, 155–185. doi: 10.1016/j.aquabot.2007.12.007
Narula, N., Kothe, E., and Behl, R. (2009). Role of root exudates in plant-microbe interactions. J. Appl. Bot. Food Qual. 82, 122–130.
Nascimbene, J., Nimis, P. L., and Marini, L. (2007). Testing indicators of epiphytic lichen diversity: a case study in N Italy. Biodivers. Conserv. 16, 3377–3383. doi: 10.1007/s10531-006-9084-z
Naser, H. A. (2014). “Marine ecosystem diversity in the Arabian Gulf: threats and conservation, chapter 12,” in Biodiversity - The Dynamic Balance of the Planet, Marine Ecosystem Diversity in the Arabian Gulf: Threats and Conservation. INTECH. doi: 10.5772/57425
Nimis, P. L., Scheidegger, C., and Wolseley, P. A. (2002). Monitoring with Lichens–onitoring Lichens. Bryologist 105:408.
Norton, J., Abdul Majid, S., Allan, D., Al Safran, M., Böer, B., and Richer, R. (2009). An Illustrated Checklist of the Flora of Qatar. United Nations Educational, Scientific and Cultural Organization, UNESCO Office in Doha. Qatar Foundation. Doha: MAERSK OIL QATAR AS.
Odu, N. N., and Akujobi, C. O. (2012). Protease production capabilities of Micrococcus luteus and Bacillus species isolated from abattoir environment. J. Microbiol. Res. 2, 127–132. doi: 10.5923/j.microbiology.20120205.03
Okami, Y., and Hotta, K. (1988). “Search and discovery of new antibiotics,” in Actinomycetes in Biotechnology, eds M. Goodfellow, S. T. Williams, and M. Mordarski (London: Academic Press), 33–67. doi: 10.1016/b978-0-12-289673-6.50007-5
Ouf, S. A. (1993). Mycological studies on the angiosperm root parasite Cynomorium coccineum L. and two of its halophytic hosts. Biol. Plant. 35, 591–602. doi: 10.1007/BF02928037
Patel, P., Patel, K., Dhandhukia, P., and Thakker, J. N. (2021). Plant growth promoting traits of marine Micrococcus sp. with bio-control ability against Fusarium in chickpea plant. Vegetos 34, 94–101. doi: 10.1007/s42535-021-00191-4
Quesada, E., Ventosa, A., Rodriguez-Valera, F., and Ramos-Cormenzana, A. (1982). Types and properties of some bacteria isolated from hypersaline soils. J. Appl. Bacteriol. 53, 155–161. doi: 10.1111/j.1365-2672.1982.tb04671.x
Rakib, F., Al-Ansari, E. M. A. S., Husrevoglu, Y. S., Yigiterhan, O., Al-Maslamani, I. A., Aboobacker, V. M., et al. (2021). Observed variability in physical and biogeochemical parameters in the central Arabian Gulf. Oceanologia 63, 227–237. doi: 10.1016/j.oceano.2020.12.003
Rasheed, M. N., Al-Saadi, B. Q. H., Hasan, O. M., Salman, N. Y., and Faisal, S. (2021). Study the role of Ph in curli biogenesis gene expression in Enterobacter Cloacae local isolates. Indian J. Forensic Med. Toxicol. 15, 1260–1264.
Redfield, E., Barns, S. M., Belnap, J., Daane, L. L., and Kuske, C. R. (2002). Comparative diversity and composition of cyanobacteria in three predominant soil crusts of the Colorado Plateau. FEMS Microbiol. Ecol. 40, 55–63. doi: 10.1111/j.1574-6941.2002.tb00936.x
Richards, G. P., Watson, M. A., Needleman, D. S., Uknalis, J., Boyd, E. F., and Fay, J. P. (2017). Mechanisms for Pseudoalteromonas piscicida-induced killing of vibrios and other bacterial pathogens. Appl. Environ. Microbiol. 83:e00175-17. doi: 10.1128/aem.00175-17
Richer, R., Anchassi, D., El-Assaad, I., El-Matbouly, M., Ali, F., Makki, I., et al. (2012). Variation in the coverage of biological soil crusts in the State of Qatar. J. Arid Environ. 78, 187–190. doi: 10.1016/j.jaridenv.2011.10.009
Richer, R., Banack, S. A., Metcalf, J. S., and Cox, P. A. (2015). The persistence of cyanobacterial toxins in desert soils. J. Arid Environ. 112, 134–139. doi: 10.1016/j.jaridenv.2014.01.023
Roberts, D. P., Derby, P. D., Yucel, I., Buyer, J., Holtman, M. A., and Kobayashi, D. Y. (1999). Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl. Environ. Microbiol. 65, 2513–2519. doi: 10.1128/aem.65.6.2513-2519.1999
Rovira, A. D., Foster, R. C., and Martin, J. K. (1979). “Note on terminology: origin, nature, and nomenclature of organic materials in the rhizosphere,” in The Soil-Root Interface, eds J. C. Harley and R. S. Russell (London: Academic Press), 1–14. doi: 10.1016/b978-0-12-325550-1.50007-0
Sanders, J. W., Venema, G., and Kok, J. (1999). Environmental stress responses in Lactococcus lactis. FEMS Microbiol. Rev. 23, 483–501.
Sansinenea, E. (2019). “Bacillus spp.: As Plant Growth-Promoting Bacteria,” in Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms, eds H. Singh, C. Keswani, M. Reddy, E. Sansinenea, and C. García-Estrada (Singapore: Springer), doi: 10.1007/978-981-13-5862-3_11
Santos, P. E.-D. L., Bustillos-Cristales, R., and Caballero-Mellado, J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. App. Environ. Microbiol. 67, 2790–2798. doi: 10.1128/aem.67.6.2790-2798.2001
Saxena, A. K., Kumar, M., Chakdar, H., Anuroopa, N., and Bagyaraj, D. J. (2019). Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 128, 1583–1594. doi: 10.1111/jam.14506
Setiyono, E., Adhiwibawa, M. A. S., Indrawati, R., Prihastyanti, M. N. U., Shioi, Y., and Brotosudarmo, T. H. P. (2020). An indonesian marine bacterium, Pseudoalteromonas rubra, produces antimicrobial prodiginine pigments. ACS Omega 5, 4626–4635. doi: 10.1021/acsomega.9b04322
Singh, Y. D., Singh, M. C., and Panda, M. K. (2020). “Biotechnological aspects of mangrove microorganisms, Chapter 18,” in Biotechnological Utilization of mangrove resources, 1st ed, eds J. K. Patra, R. R. Mishra, and H. Thatoi (Cambridge, MA: Academic Press), 381–398. doi: 10.1016/b978-0-12-819532-1.00018-4
Sitaraman, R. (2015). Pseudomonas spp. as models for plant-microbe interactions. Front. Plant Sci. 6:787. doi: 10.3389/fpls.2015.00787
Sousa, J. A. d. J., and Olivares, F. L. (2016). Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications. Chem. Biol. Technol. Agric. 3, 24. doi: 10.1186/s40538-016-0073-5
Srividya, A. R., Saitha, G. S., and Suresh, B. (2008). Study of the soil isolates for antimicrobial activity. Indian J. Pharm. Sci. 70, 812–815. doi: 10.4103/0250-474x.49132
Stefani, F. O. P., Bell, T. H., Marchand, C., de la Providencia, I. E., El Yassimi, A., et al. (2015). Culture-dependent and -independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLOS One 10:e0128272. doi: 10.1371/journal.pone.0128272
Stone, B. W. G., Weingarten, E. A., and Jackson, C. R. (2018). The role of the phyllosphere microbiome in plant health and function. Ann. Plant Rev. 1. doi: 10.1002/9781119312994.apr0614 (accessed August, 2018).
Su, C., Lei, L., Duan, Y., Zhang, K.-Q., and Yang, J. (2012). Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl. Microbiol. Biotechnol. 93, 993–1003. doi: 10.1007/s00253-011-3800-7
Su, Y., Tang, K., Liu, J., Wang, Y., Zheng, Y., and Zhang, X.-H. (2019). Quorum sensing system of ruegeria mobilis Rm01 controls lipase and biofilm formation. Front. Microbiol. 9:3304. doi: 10.3389/fmicb.2018.03304
Tahoven, R., Hannukkala, A., and Avikainen, H. (1995). Effect of seed dressing treatment of Streptomyces grisieviridis on barley and spring wheat in field experiments. Agric. Sci. Finl. 4, 419–427. doi: 10.23986/afsci.72619
Tam, H. T. (2017). Isolation and characterization of bacteria of mangrove rhizosphere in the Mekong Delta, Vietnam. Int. J. Innov. Engin. Technol. (IJIET). 9, 68–79. doi: 10.21172/ijiet.91.11
Thompson, I. P., Baily, M. J., Fenlon, J. S., Fermor, T. R., Lilley, A. K., Lynch, J. M., et al. (1993). Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil. 150, 177–191. doi: 10.1007/bf00013015
Trentin, D. S., Gorziza, D. F., Abraham, W. R., Antunes, A. L. S., Lerner, C., Mothes, B., et al. (2011). Antibiofilm activity of Cobetia marina filtrate upon Staphylococcus epidermidis catheter-related isolates. Braz. J. Microbiol. 40, 1329–1333. doi: 10.1590/S1517-838220110004000013
Twilley, R. R. (1985). The exchange of organic carbon in mangrove forest in a south west Florida estuary. Est. Coast. Shelf Sci. 20, 543–557. doi: 10.1016/0272-7714(85)90106-4
Vaish, M., Price-Whelan, A., Reyes-Robles, T., Liu, J., Jereen, A., Christie, S., et al. (2018). Roles of Staphylococcus aureus Mnh1 and Mnh2 antiporters in salt tolerance, alkali tolerance, and pathogenesis. J. Bacteriol. 200:e00611-17. doi: 10.1128/JB.00611-17
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19, 1–26. doi: 10.3390/ijms19040952
Wahbeh, M. I., and Mahasneh, A. M. (1984). Heterotrophic bacteria attached to leaves, rhizomes and root of three seagrass species from Aqaba (Jordan). Aquat. Bot. 20, 87–96. doi: 10.1016/0304-3770(84)90029-9
Walker, T. S., Bais, H. P., Grotewold, E., Jorge, M., and Vivanco, J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Williams, S. T., and Vickers, J. C. (1986). The ecology of antibiotic production. Microb. Ecol. 12, 43–52. doi: 10.1007/BF02153221
Wilson, R. J., Salama, G., and Farag, I. H. (2012). Microalgae growth in Qatar for CO2 capture and biodiesel feedstock production. Glob. J. Eng. Res. Chem. Eng. 12, 2249–4596. Available online at: https://globaljournals.org/GJRE_Volume12/1-Microalgae-Growth-in-Qatar-for.pdf
Yan, A., Wang, Y., Tan, S. N., Yusof, M. L. M., Ghosh, S., and Chen, Z. (2020). Phytoremediation: a promising approach for revegetation of heavy metal-polluted Land. Front. Plant Sci. 11:359. doi: 10.3389/fpls.2020.00359
Yang, C. H., Crowley, D. E., Borneman, J., and Keen, N. T. (2001). Microbial phyllosphere populations are more complex than previously realized. Proc. Nat. Acad. Sci. U.S.A. 98, 3889–3894. doi: 10.1073/pnas.051633898
Yang, S., Zhang, X., Cao, Z., Zhao, K., Wang, S., Chen, M., et al. (2014). Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 7, 611–620. doi: 10.1111/1751-7915.12148
Yasseen, B. T. (2014a). Phytoremediation of industrial wastewater from oil and gas fields using native plants: the research perspectives in the state of Qatar. Sch. Res. Lib. Cent. Eur. J. Exp. Biol. 3, 6–23.
Yasseen, B. T. (2014b). Adaptation of Plants in Qatar (Booklet). Legal Deposit No. 2015/10. Doha: Environmental Studies Centre, Qatar University.
Yasseen, B. T. (2016). Traits of wild plants in qatar peninsula and research perspectives. J. Biol. Nat. 5, 52–66.
Yasseen, B. T., and Abu-Al-Basal, M. A. (2008). Ecophysiology of Limonium axillare and Avicennia marina from the coastline of Arabian Gulf-Qatar. J. Coast. Conserv. Plan. Manag. 12, 35–42. doi: 10.1007/s11852-008-0021-z
Yasseen, B. T., and Abu-Al-Basal, M. A. (2010). Ecophysiology of chenopodiaceae at the coastline of arabian gulf-qatar: possible destruction and prespective conservation. Eur. J. Sci. Res. 39, 90–104.
Yasseen, B. T., Abulfatih, H. A., Nasher, A. K., Abid, K. Y., and Al-Mofti, M. B. (2001). Preliminary assessment of pollution due to faulty sewage system in north of Sana, a, Republic of Yemen. Dirasat Eng. Sci. 28, 89–96.
Yasseen, B. T., and Al-Thani, R. F. (2007). Halophytes and associated properties of natural soils in the Doha area, Qatar. Aquat. Ecosyst. Health Manag. 10, 320–326. doi: 10.1080/14634980701519462
Yasseen, B. T., and Al-Thani, R. F. (2013). “Ecophysiology of wild plants and conservation perspectives in the State of Qatar,” in Agricultural Chemistry, eds M. Stoytcheva and R. Zlatev (Rijeka: InTech), 37–70. doi: 10.5772/55305
Yasseen, B. T., Al-Thani, R. F., Alhadi, F. A., and Abbas, R. A. A. (2018). Soluble sugars in plants under stress at the arabian gulf region: possible roles of microorganisms. J. Plant Biochem. Physiol. 6:224. doi: 10.4172/2329-9029.1000224
Yoon, J.-H., Kang, S.-J., Lee, S.-Y., Lee, M.-H., and Tae Oh, K. (2005). Virgibacillus dokdonensis sp. nov., isolated from a Korean island, Dokdo, located at the edge of the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 55 (pt 5), 1833–1837. doi: 10.1099/ijs.0.63613-0
Zahran, H. H. (1999). Diversity, adaptation and activity of the bacterial flora in saline environments. Biol. Fertil. Soils 25, 211–223. doi: 10.1007/s003740050306
Zahran, H. H., Ahmad, M. S., and Afkar, E. A. (1995). Isolation and characterization of nitrogen -fixing moderate halophilic bacteria from saline soils of Egypt. J. Basic Microbiol. 35, 269–275. doi: 10.1002/jobm.3620350412
Zahran, H. H., Moharram, A. M., and Mohammad, H. A. (1992). Some ecological and physiological studies on bacteria isolated from salt-affected soils of Egypt. J. Basic Microbiol. 32, 405–413. doi: 10.1002/jobm.3620320612
Zaki, M. M., Hamed, A. S., Sejiny, M. J., Baeshin, N. A., and Younes, H. A. (1980). Halophilic bacteria in soil and rhizosphere of some littoral salt marsh plants at Shuaiba lagoon, Saudi Arabia. Bull. Fac. Sci. 4, 91–100.
Zeaiter, Z., Marasco, R., Booth, J. M., Prosdocimi, E. M., Mapelli, F., Callegari, M., et al. (2019). Phenomics and genomics reveal adaptation of Virgibacillus dokdonensis strain 21D to its origin of isolation, the seawater-brine interface of the Mediterranean Sea deep hypersaline anoxic basin discovery. Front. Microbiol. 10:1304. doi: 10.3389/fmicb.2019.01304
Zeng, Y., Feng, F., Medová, H., Dean, J., and Koblížek, M. (2014). Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc. Natl. Acad. Sci. U.S.A. 111, 7795–7800. doi: 10.1073/pnas.1400295111
Zhou, L., Li, H., Zhang, Y., Han, S., and Xu, H. (2016). Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Braz. J. Microbiol. 47, 271–278. doi: 10.1016/j.bjm.2016.01.001
Keywords: microorganisms, marine environment, salt marshes, arid lands, Rawdahs, wetland ponds, bioremediation, biotechnology
Citation: Al-Thani RF and Yasseen BT (2021) Microbial Ecology of Qatar, the Arabian Gulf: Possible Roles of Microorganisms. Front. Mar. Sci. 8:697269. doi: 10.3389/fmars.2021.697269
Received: 19 April 2021; Accepted: 08 July 2021;
Published: 05 August 2021.
Edited by:
Shady A. Amin, New York University Abu Dhabi, United Arab EmiratesReviewed by:
Hojeong Kang, Yonsei University, South KoreaCopyright © 2021 Al-Thani and Yasseen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bassam T. Yasseen, YmFzc2FtX3RhaGFhQHlhaG9vLmNvLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.