- 1Cooperative Institute for Marine and Atmospheric Studies, University of Miami, Miami, FL, United States
- 2Atlantic Oceanographic and Meteorological Laboratory, Ocean Chemistry and Ecosystem Division, National Oceanographic and Atmospheric Administration (NOAA), Miami, FL, United States
- 3Coral Morphologic, Miami, FL, United States
- 4Center for Satellite Applications and Research, Satellite Oceanography and Climate Division, National Oceanographic and Atmospheric Administration (NOAA), College Park, MD, United States
Healthy coral communities can be found on artificial structures (concrete walls and riprap) within the Port of Miami (PoM), Florida. These communities feature an unusually high abundance of brain corals, which have almost entirely vanished from nearby offshore reefs. These corals appear to be thriving in very low-quality waters influenced by dense ship and boat traffic, dredging, and numerous residential and industrial developments. The PoM basin is part of Biscayne Bay, an estuarine environment that experiences frequent freshwater input, high nutrient loading, hypoxia, and acidification. To investigate if there is a molecular basis behind the ability of these corals to persist within these highly “urbanized” waters, we compared whole transcriptome expression profiles from 25 PoM Pseudodiploria strigosa colonies against six conspecifics from a nearby offshore reef. We found that the urban corals exhibited higher expression of (1) transcripts encoding pattern-recognition receptors which may allow these corals to better sense and detect food particles and pathogenic invaders; (2) digestive and degradation-associated enzymes, which may suggest an elevated capacity for heterotrophy and pathogen digestion; and (3) transcripts related to innate immunity, defense, and cellular detoxification, which may collectively protect against pathogenic organisms and water pollution impacts. Large ribosomal subunit rRNA gene mapping revealed that P. strigosa colonies from the PoM sites predominantly hosted heat-tolerant endosymbionts from the genus Durusdinium while offshore conspecifics’ communities were dominated by symbionts in the genus Breviolum. These findings reveal transcriptomic plasticity and molecular mechanisms contributing to the persistence of these corals within a highly urbanized habitat.
Introduction
Reefs worldwide have suffered widespread coral mortality caused by anthropogenic stressors. This trend is especially apparent in the Florida Reef Tract, which has experienced multiple bleaching events (Manzello, 2015; van Woesik and McCaffrey, 2017; Manzello et al., 2018), and deadly outbreaks of infectious diseases (Precht et al., 2016; Walton et al., 2018). Contrary to surrounding reef areas, coral populations have persisted on artificial structures (concrete walls, riprap) in the Port of Miami (PoM) basin. These urban locations are home to high numbers of brain corals, while their Florida Reef Tract conspecifics have been almost entirely eradicated. The PoM waters are influenced by multiple stressors, including dense ship and boat traffic, river and stormwater runoff, eutrophication, and dredging activities. Further, these shallow sites experience variable temperatures and high turbidity, while eutrophication leads to occasional hypoxia and localized acidification (Caccia and Boyer, 2005; Briceño et al., 2011; Enochs et al., 2019; Millette et al., 2019). Despite these stressful conditions, the urban corals appear healthy, as we have not observed bleaching or disease during qualitative surveys since 2017. Thus, these urban corals might provide insight into coral survival with many environmental stressors. These coral populations might also provide a repository of stress-tolerant coral genotypes that could be used in coral conservation and restoration activities.
The study of coral communities exhibiting unique tolerance to abiotic and ecological stressors can contribute to understanding coral’s ability to withstand conditions influenced by future climate challenges. Specifically, corals living in marginal environments may provide hope for coral survival with climate change and environmental disturbances (review by Burt et al., 2020). Corals in marginal environments like highly urbanized waters, reefs near CO2 seeps, and shallow waters with higher-than-average water temperatures are of interest because corals surviving in those stressful environments provide information on coral acclimation and adaption potential. For example, corals near CO2 vents possess transcriptomic plasticity allowing them to live in highly acidified waters (Kenkel et al., 2018). In addition, corals in shallow water environments exhibit better adaptations to higher temperature extremes (Brener-Raffalli et al., 2019; Dziedzic et al., 2019). The investigations of coral populations in these marginal areas, such as those under anthropogenic disturbance and pollution, are increasing (Burt, 2014; Browne et al., 2015; Guest et al., 2016; Pizarro et al., 2017; Heery et al., 2018; Kenkel et al., 2018), but the cellular and molecular mechanisms of coral resilience to these stressful environments are not well investigated.
Coral transcriptomic studies can successfully compare variable responses of corals to environmental factors, including temperature (e.g., DeSalvo et al., 2010), pH (Lin et al., 2018), heavy metals (Schwarz et al., 2013), disease (Libro et al., 2013), and immune stimuli (Ocampo et al., 2015; Fuess et al., 2016). Those studies show how one factor or two environmental parameters influence coral transcriptomic response in a controlled experimental setting. These studies lead to the identification of molecular markers which respond to a specific stressor or a parameter. They provide a baseline for transcriptomic field-derived investigations where all environmental parameters are combined and acting synergistically or antagonistically.
Only a few studies have successfully used transcriptomics profiling to investigate coral performance in the field at different reef locations varying in environmental parameters (Bay and Palumbi, 2017; Brener-Raffalli et al., 2019; Helmkampf et al., 2019) or before and after bleaching events (Pinzon et al., 2015). The advantage of investigating coral transcriptomic performance in their natural location is the ability to measure real-world responses to complex combinations of environmental factors. Another strength of the in situ transcriptomic approach is that it requires only small pieces of coral tissue. In contrast, a full multifactorial experimental design requires numerous and larger coral fragments. The selection of a more coral-tissue conservative approach is becoming more important in South Florida areas, where the extensive coral decline makes experimental studies more logistically and morally challenging.
In this study, we applied whole transcriptome expression profiling (RNA-seq) and differential gene expression analysis to investigate the possible molecular mechanisms of coral persistence in the highly urbanized water of the PoM. Differential expression analysis between urban corals and offshore corals revealed a suite of molecular mechanisms that may play a role in the survival of these urban corals. Our results have important implications for leading the research into finding molecular markers of coral resistance to stressors. In addition, the PoM corals need to be frequently relocated to allow for new coastal development and modernization of existing infrastructures. Thus, the knowledge of corals’ acclimation to the existing conditions will assist in making decisions on the type of habitat suitable for their relocations.
Materials and Methods
Site Selection and Sample Collection
Four study sites (Supplementary Figure 1) were selected, with three within the PoM: one each along the north and south side of MacArthur Causeway (MACN and MACS, respectively) and one abutting the eastern seawall of Star Island (STAR). The control site, Emerald Reef (EMLR), was located 5.6 km offshore. Small tissue samples (2–4 cm3) were collected with hammer and chisel from 10, 10, 9, and 6 colonies of Pseudodiploria strigosa from MACN (∼1–2 m), MACS (∼1 m) STAR (∼1–2 m), EMLR (depth = 6–7 m), respectively, on February 14, 2020. Pieces of corals were kept in perforated plastic containers with seawater until surfacing, after which they were immediately frozen in liquid nitrogen.
RNA Extraction
RNA extractions were carried out using the Nucleospin TriPrep kit (Macherey Nagel) following the manufacturer’s protocol with a slight modification to the lysis and homogenization steps. Briefly, coral pieces were ground in liquid nitrogen using a mortar and pestle, with pulverized coral then transferred (without thawing) to 2-ml of lysis buffer (RP1) containing TCEP reducing agent and bashing beads (MN bead tubes type E, Macherey Nagel). Additional homogenization was achieved using a FastPrep bead-beater from MP Biomedicals at 6.0 m/s for 60 s. After extraction, total RNA was quantified using the Qubit BR RNA kit (Invitrogen) and shipped on dry ice to the Duke University Genomic Center. cDNA libraries were prepared using a Kapa Stranded mRNA kit (a poly-A enrichment method from Roche) and sequenced on two lanes of the SP-PE-150-bp flow cell and the Nova6000 sequencing machine (Illumina).
Transcriptome Assembly and Annotation
Raw fastq reads were subjected to quality control using FastQC v.0.11.5 (Brown et al., 2017). Illumina adapters were removed using Cutadapt v.1.14 (Martin, 2011). The reads were trimmed using Trimmomatic v.0.36 (Bolger et al., 2014) with the following settings: (1) a sliding window of 3 nucleotides averaging to a Phred score of 25, (2) leading and trailing sequence thresholds of Phred score 25, and (3) a minimum of 100 bp in length. The meta-transcriptome assembly was carried out using the Megahit assembler (Li et al., 2015). The quality of the transcriptome assembly was investigated using the TransRate tool (Smith-Unna et al., 2016), and protein-coding sequences (cds) with a minimum length of 99 amino acids were predicted using TransDecoder (Haas et al., 2013). The translated protein-coding sequences were queried against NCBI’s non-redundant protein database (downloaded in February 2020) using the Diamond Tool (Buchfink et al., 2015). The top diamond-matches were used for taxonomic classification with TaxonKit (Shen and Xiong, 2019). Protein sequences were functionally and taxonomically annotated using the Ghost Koala web server combined (Kanehisa et al., 2016). The refinement and verification of taxonomic assignments were conducted by BLASTn searches (Altschul et al., 1990) against a coral genomics database1 and dinoflagellate endosymbiont transcriptomes from NCBI’s Transcriptome Shotgun Assembly database and http://sampgr.org.cn/index.php website (for a complete list see Supplementary Table 5).
All protein-coding sequences were subjected to multiple similarity searches against several databases as described above. The final taxonomic assignments were based on the highest bit-score value from all matches. Functional annotation of predicted protein-coding sequences was completed using InterProScan (Zdobnov and Apweiler, 2001). The cellular localization of differentially expressed transcripts (DETs; discussed below) was predicted using the Wolf-Psort II webserver (Horton et al., 2007).
Differential Gene Expression Analysis
The quantification of transcripts was carried out using the Salmon tool under the “validate mapping” setting (Patro et al., 2017). We were interested in identifying coral transcripts whose expression differed significantly between each urban site and the control reef, thus three differential expression comparisons were made as follows: MACN vs. EMLR, STAR vs. EMLR, and MACS vs. EMLR. In addition, a fourth DE comparison was made between Durusdinium-dominated and Breviolum dominated corals dominated. The analysis was carried out only on the host transcriptome because all six samples from EMLR site and ten samples from MACN site showed zero expression values for about 50% of symbiont contigs, and the differential gene expression analysis is not suitable for presence (expressed) and absence (not expressed) comparison. Statistically significant DETs (p < 0.05 after Benjamini Hochberg false discover rate [FDR] correction and minimum twofold difference) were determined using the edgeR package after the TMM (trimmed mean of M-values) normalization method (Robinson et al., 2010; Dillies et al., 2013). This statistical analysis was carried out in the edgeR package for the R v.3.6.0 software using generalized linear models and F-likelihood tests. Principal components analysis (PCA) was conducted in R v.3.6.1 using the prcomp command in the Stats v.3.6.1. package and the TMM-normalized expression values converted to fragments per kilobase per million (FPKM). The PCA was calculated using a singular value decomposition (SVD) of the data matrix. For the heat map, both samples and transcripts were clustered using the k-means hierarchical clustering method and a Pearson correlation dissimilarity matrix, one of the best approaches for sample clustering based on gene expression data (Reeb et al., 2015). The heat map was generated using the Morpheus online tool available from https://software.broadinstitute.org/morpheus/.
Endosymbiont Type Determination
Twenty-two unique sequences of the large ribosomal subunit (28S) representing four genera in the family Symbiodiniaceae (formerly clades A–D; LaJeunesse et al., 2018) were selected from the GeneBank (NCBI) database (see Supplementary Table 4 for NCBI accession numbers and sequence information). These sequences were re-aligned with the reference tree from LaJeunesse et al. (2018) to verify their phylogenetic clustering with the correct genus. The re-alignments and phylogenetic tree reconstruction were accomplished using the Geneious aligner within the Geneious software version R10.2.62. After alignment, the 28S reference sequences were trimmed to a similar length (581–586 bp) and used as a template for read mapping with Bowtie2 (Langmead and Salzberg, 2012). Quality trimmed and filtered reads (Phred 25 see above) were mapped to the LSU reference sequences using Bowtie 2 settings were modified to include only 100% matches. The read quantification was retrieved using Samotools (Li et al., 2009). One sequence per genus with the highest number of mapped reads was used to calculate relative genus abundance. Percent abundance was calculated by dividing the number of reads mapped to each genus by the total number of mapped reads to all genera.
Results
Transcriptome Assembly, Taxonomic, and Functional Annotations
After quality (Phred 25) filtration, over 1 billion paired-end reads (between 20 and 48 million paired-end reads per sample) were obtained for RNA samples extracted from the 31 P. strigosa colonies. The fastq files containing the trimmed and quality filtered reads for the corresponding cDNA libraries were deposited at NCBI’s Short Read Archive under BioProject PRJNA693301 and BioSample IDs SAMN17387671- SAMN17387701 (Supplementary Table 1). Reads from all samples were used to assemble the metatranscriptome consisting of 383,263 contigs with a minimum length of 500 bp. The mean contig-length was 1,145 bp, and the N50 value was 1,314 bp (i.e., 50% of reads featured in contigs equal to or longer than 1,314 bp). After protein-coding prediction, 166,591 contigs containing 185,428 coding sequences (cds; some contigs contained more than one cds.) were maintained for downstream analyses; the 216,672 non-coding (ncRNA) contigs were excluded from further analysis. From the 185,428 cds, 38,931 (21%) were filtered out, given that they were found in less than 10 samples. Forty-eight thousand two hundred and eighty-three cds (33%) were assumed to be of host coral origin based on their top diamond-BLAST matches being with published cnidarian sequences in the reference databases. A total of 91,688 cds (63%) aligned most closely to published Symbiodiniaceae mRNA sequences. The remaining 6,526 cds (4%) aligned significantly to non-Symbiodiniaceae coral microbial constituents (e.g., archaea, bacteria, encrusting sponges, fungi, protists, etc.) and were removed since our focus was on the coral-dinoflagellate endosymbiosis exclusively.
Symbiont Composition
The relative abundance of symbiont types was determined based on the mapping of quality-filtered reads to a curated large ribosomal subunit rRNA gene database representing four major symbiont genera (Symbiodinium, Breviolum, Cladocopium, and Durusdinium). No reads mapped to Symbiodinium or Cladocopium LSU rRNA sequences, and the reads mapped to Breviolum and Durusdinium sequences were used to estimate percent symbiont abundance. The symbiont communities of all corals except one colony were either dominated by Breviolum or Durusdinium (i.e., >90% of symbionts). All offshore P. strigosa colonies predominantly hosted Breviolum (≥97% of total) (Figure 1). In contrast, all ten colonies of the urban site MACN contained exclusively (100% abundance) endosymbionts from the genus Durusdinium (Figure 1). The other urban sites had almost an equal mix of corals dominated by either Breviolum or Durusdinium (Figure 1). Only one colony harbored a mixed assemblage of symbionts and was not dominated by a single genus. This colony was located at MACS and contained 30% Durusdinium and 70% Breviolum (Figure 1). This colony was considered a Breviolum-dominated sample in differential expression analyses as the host transcription profile was more similar to the other Breviolum-dominated (>90% symbiont abundance) corals (Figure 2). Multiple species of Breviolum symbionts exist in Florida and the Caribbean. However, based on a subset of studies in which the symbiont species composition has been determined, it could be speculated that the P. strigosa hosts Breviolum minutum (B1 symbiont, e.g., Bongaerts et al., 2015; Cunning et al., 2017). Since the rDNA copy number of copies per cell for a few coral symbiont species (including B. minutum and D. trenchii) have been recently determined (Saad et al., 2020), we could calculate the relative abundance of the two symbiont species in P. strigosa colonies (Supplementary Figure 3 and Supplementary Table 6). Supplementary Figure 3 shows the possible symbiont species composition if we assume that the P. strigosa hosts only two symbiont species (B. minutum and D. trenchii); however, the true diversity of symbiont species could only be determined by the ITS2 sequence profiling (e.g., Hume et al., 2019).
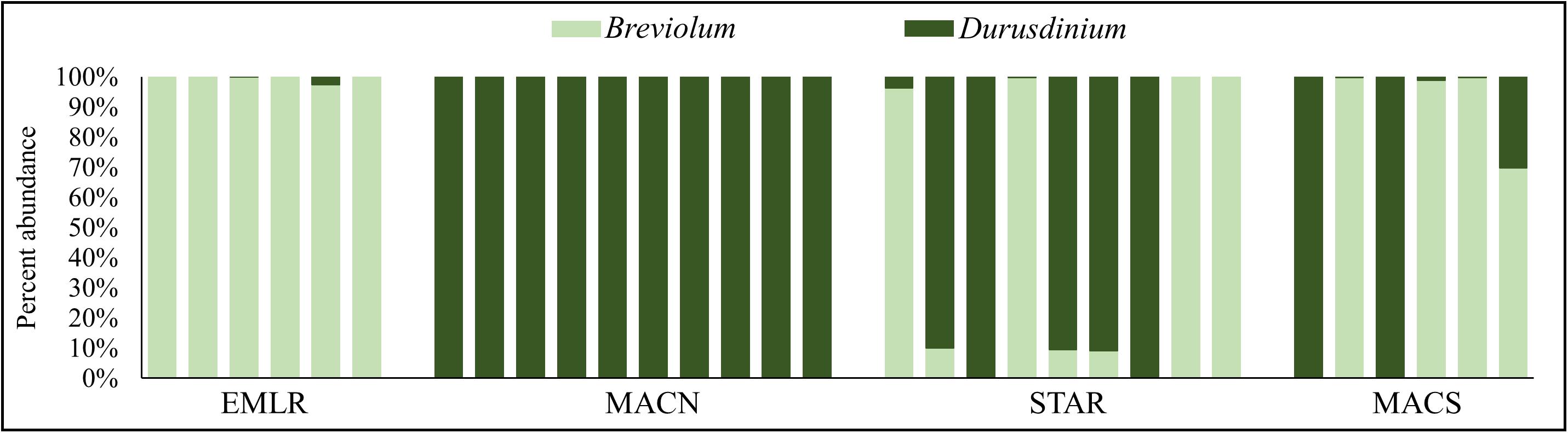
Figure 1. Genus-level endosymbiont composition of 31 individuals of P. strigosa collect from (A) an offshore reef (Emerald Reef, EMLR) and urban coral sites (B) MacArthur North (MACN), (C) MacArthur South (MACS), and (D) Star Island (STAR) near Port of Miami, FL.
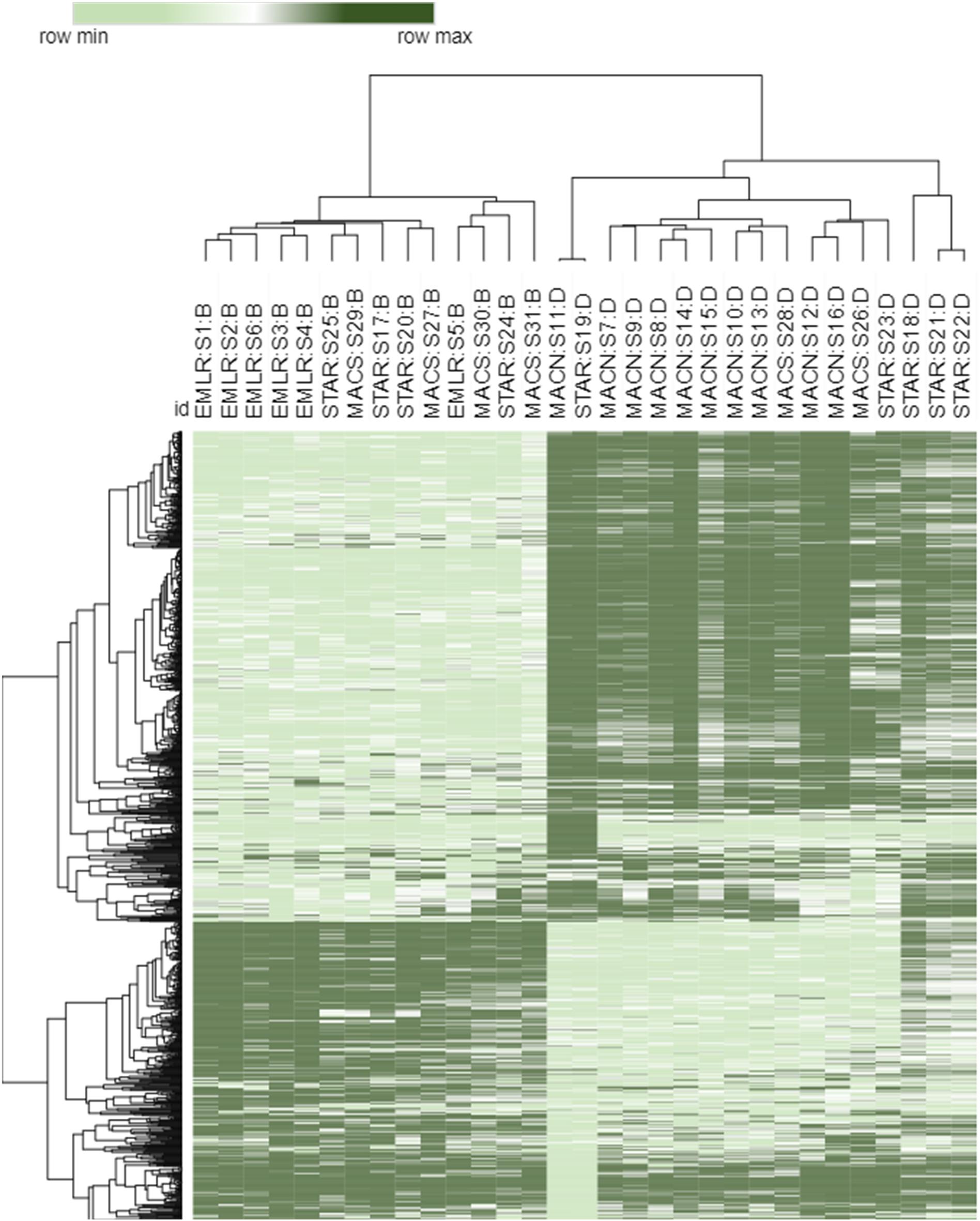
Figure 2. Heat map of top 1,000 transcripts contributing the highest weight to the PC1 axis of the PCA in Figure 3. Both samples and transcripts were clustered using the k-means complete hierarchical clustering method and a Euclidean distance dissimilarity matrix. The samples are labeled with a study site (MACN, MACS, STAR, EMLR), sample number (S1–S31) and dominant symbiont type (B-Breviolum and D-Durusdinium).
Principal Component Analysis
Sample similarity based on the P. strigosa transcriptome expression profiles were investigated using PCA, and samples were partitioned into three identifiable clusters within the PCA ordination (Figure 3A). The PC1 and PC2 together explained 26% of the total variability in gene expression. Samples from MACN and EMLR were most distinct from one another, with samples within each site demonstrating the strongest clustering. In contrast, samples from STAR and MACS showed greater variability and overlapped one another, as well as spanned the PC1 (15%) data space (Figure 3A). When dominant symbiont types were superimposed on the PCA, clear differentiation was observed between transcriptomes of Durusdinium-dominated and Breviolum-dominated corals (Figure 3B). In addition, endosymbiont type was the main factor contributing to the PCA structure based on hierarchical clustering analysis of the 1,000 transcripts contributing the most weight to the PC1 (Figure 2).
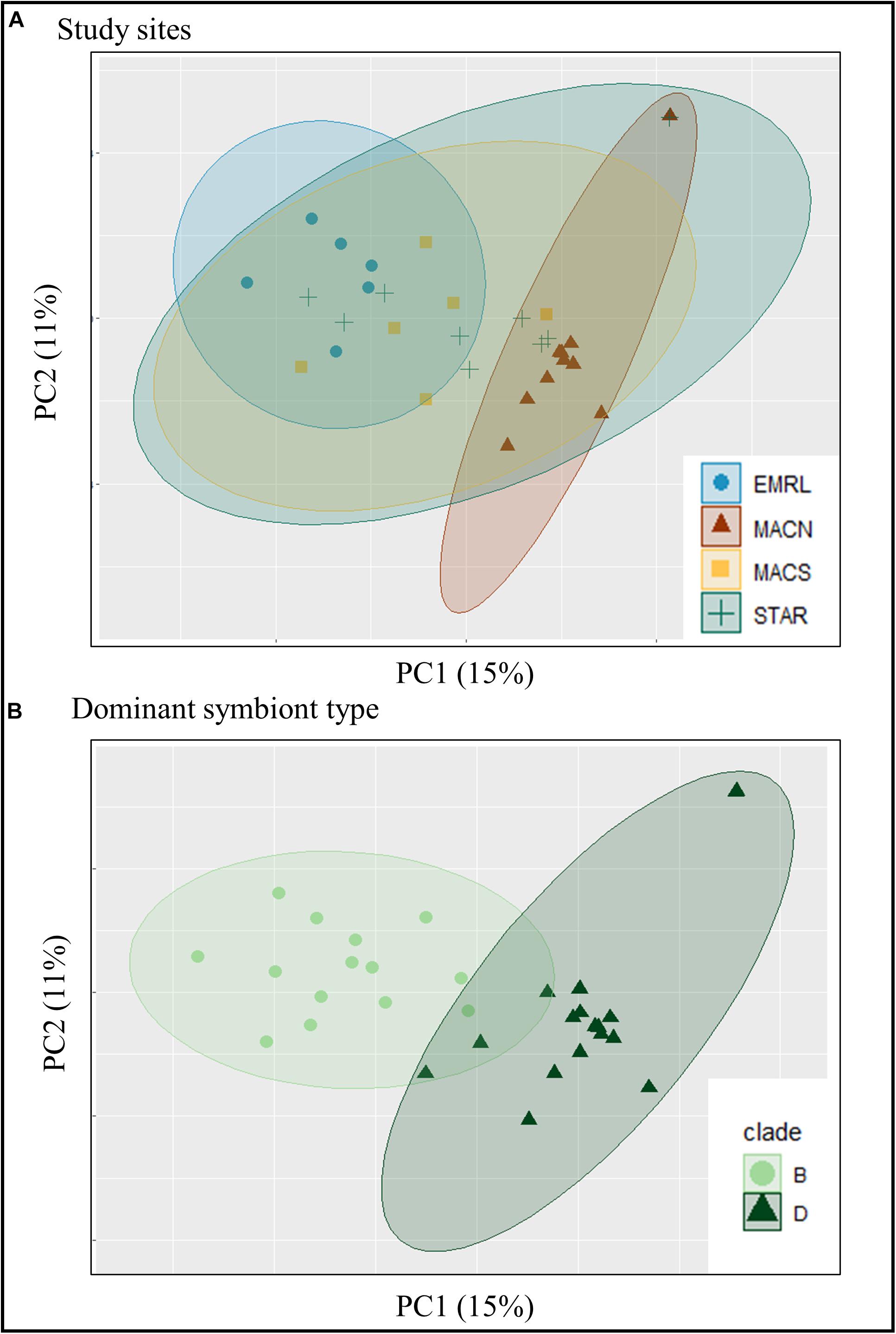
Figure 3. The similarity of 31 individuals of P. strigosa based on the transcriptome expression profiles (expression of 39,321 protein-coding sequences). Samples are color and symbol coded by the (A) study location and (B) dominant symbiont type. The ellipses represent the 95% clustering confidence level.
Statistically Significant Differentially Expressed Transcripts (DET)
Separate differential gene expression analyses were carried out to compare the transcriptomes of corals from each of the three urban sites to the transcriptomes of corals from the offshore reef. In addition, another differential expression analysis was conducted between corals hosting predominantly symbionts from the genus Breviolum (n = 14) vs. corals hosting mostly symbionts from the genus Durusdinium (n = 17). Only coral transcripts (39,321) that were expressed in a minimum of 27 samples were used for the analysis to avoid gene expression changes based on the presence (expressed) and absence (not expressed).
A total of 2,673 coral transcripts were statistically differentially expressed in at least one of the four differential analyses. Supplementary Table 3 lists all DETs and their taxonomic and functional annotations, fold change expression value, and FDR- adjusted p-values. The highest number of DETs (1,920 coral transcripts only) was found between MACN and EMLR, the two most distinct sites in the PCA (Figure 3A). Compared to the corals of EMLR, only 1,231 and 1,171 genes were differentially expressed between corals of the mixed-endosymbiont community urban sites, MACS, and STAR, respectively. This lower number of DETs might be due to the higher within-site variability and lower DET found between these two sites and the offshore site. In addition, 1,038 DETs were uncovered between corals hosting predominantly Durusdinium symbionts and those with Breviolum-dominated communities (regardless of study location).
The four most represented protein families for urban vs. offshore DETs were (1) pattern recognition receptors (n = 215 transcripts, Figures 4A,E), (2) digestive and degrading enzymes (n = 86, Figures 4B,F), (3) immune and defense-related proteins (n = 50, Figures 4C,G), and (4) transmembrane transporters (n = 83; Figures 4D,H). The DET distribution of the remaining functional categories (e.g., metabolism and genetic information processing) can be found in Supplementary Figure 3. The former 215 transcripts were expressed at higher levels in urban corals. They featured pattern recognition receptors (PRRs), a variety of lectins, scavenger receptors (SRCR), fibrinogens, and immunoglobulin domain-containing proteins (Figures 4A,E). The urban corals also exhibited higher expression of the 50 immune and defense-response-related transcripts, including seven membrane-attack-complex/perforin (MACPF) proteins and nine tumor necrosis factor-associated receptors (Figures 4B,F). Another pronounced difference between the urban and offshore corals was evident in the 86 digestive and degrading enzymes, including metallopeptidases, chymotrypsins, chitinases, lipases, and cathepsins (Figures 4C,G). The DETs also included 83 transcripts that encoded transmembrane transporters such as glucose and ammonium transporters and some lipid-transfer proteins that commonly occur in coral oocytes (Figures 4D,H).
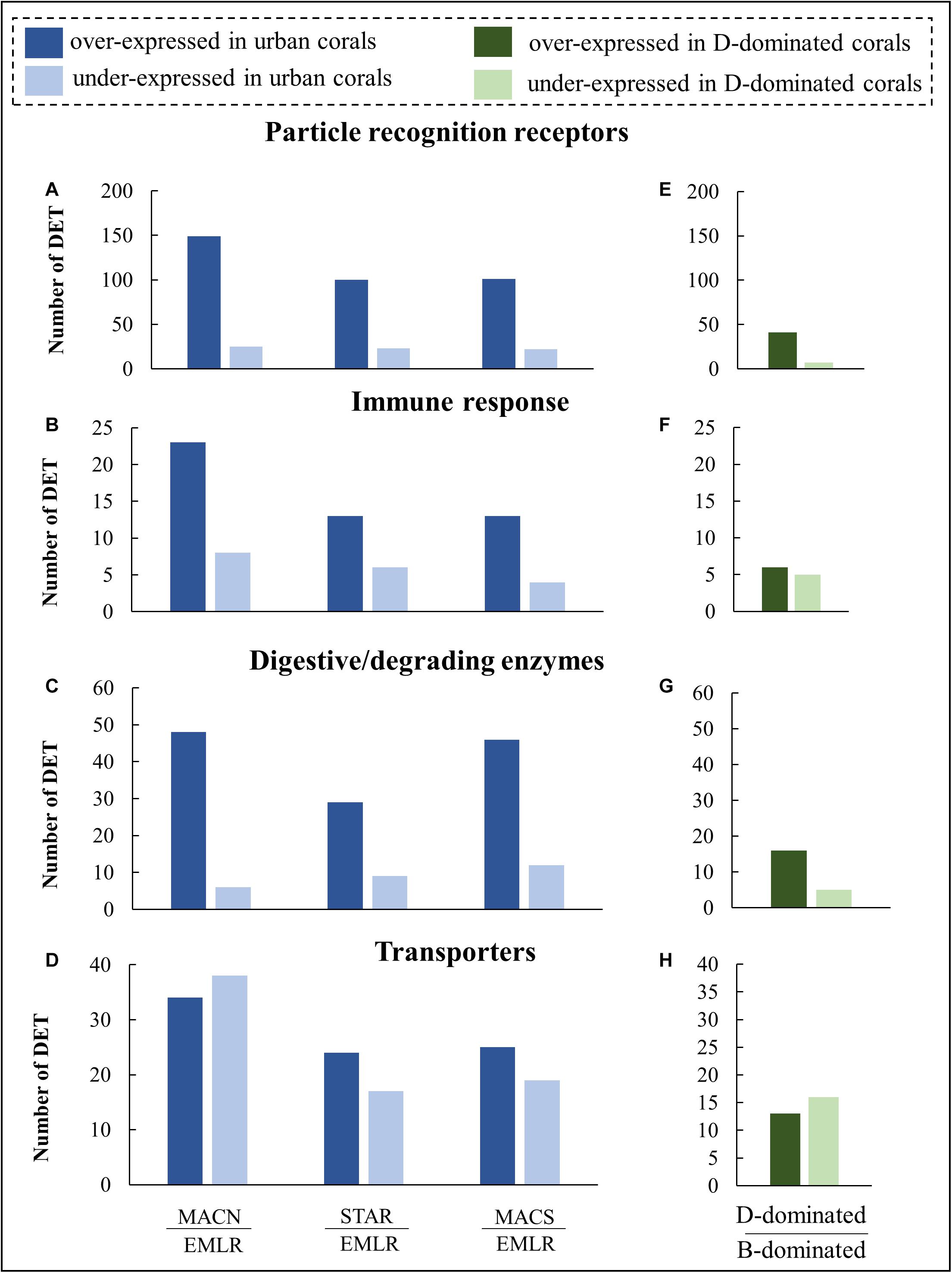
Figure 4. Distribution of DETs in P. strigosa colonies grouped into four molecular-function categories. Differential expression analyses were made between colonies from each urban site (MACN, MACS, STAR) and colonies from an offshore reef (EMLR) which was used as a control (A–D), and additional analysis was made between colonies (n = 17) dominated by Durusdinium symbionts (D-dominated) in comparison to colonies (n = 14) dominated by Breviolum (B-dominated) symbionts (E–H).
Discussion
Role of Symbiont Type in Coral Health and Resilience
Symbionts in the genus Durusdinium (formerly clade D) are more tolerant to environmental stressors, specifically high temperature. Durusdinium symbionts are associated with corals living in warmer waters and higher turbidity (Baker, 2003; Baker et al., 2004; Stat and Gates, 2011). For example, Durusdinium was the dominant symbiont type found in colonies of the coral Orbicella faveolata that were both bleaching-resistant and bleaching-resilient during the 2015 heat stress event in the Florida Keys (Manzello et al., 2019). In the present study, most (but not all) corals living in the PoM hosted Durusdinium symbionts predominantly. Similar to our results, a recent study also showed that Durusdinium symbionts contribute to the increased survival of Pocillopora acuta living in highly urbanized waters in Singapore (Poquita-Du et al., 2020). Interestingly, most P. strigosa symbiont assemblage studies have not found this coral species to partner with Durusdinium spp. in the Caribbean (Bongaerts et al., 2015); instead, P. strigosa typically harbors Breviolum spp. (Bongaerts et al., 2015) and, to a lesser extent, Cladocopium (Fabina et al., 2012). Indeed, Fabina et al. (2012) found no evidence for P. strigosa-Durusdinium associations in their meta-analysis of articles published between 1991 and 2010. Since only one Durusdinium species, D. trenchii, occurs in Atlantic waters and exhibits high clonality, it is believed to be an invasive species in the Caribbean (Pettay et al., 2015). The data from urbanized reefs (this study and Poquita-Du et al., 2020) demonstrate that Durusdinium endosymbionts are associated with this species in marginal environments. However, hosting Durusdinium may lead to compromised growth and lower calcification rates caused by the lower metabolic compatibility between the host and endosymbionts (Jones and Berkelmans, 2010; Pettay et al., 2015).
Effect of Symbiont Type on Host Transcriptome
The present study shows that the symbiont type has a strong influence on the host transcriptome profiles. PCA revealed that transcriptome profiles of corals that harbor predominantly Breviolum symbionts were more similar to each other and different from transcriptome profiles of corals that harbor Durusdinium symbionts (Figure 1). Previously, DeSalvo et al. (2010) reported that the symbiont type had a more significant impact on the coral transcriptome than the thermal treatment in O. faveolata. Similarly, Helmkampf et al. (2019) showed high modulation of the Montipora capitata transcriptomes by the presence of different symbiont types. In that study, the site location had a more significant impact on coral expression profiles than the symbiont type (Helmkampf et al., 2019). The current research shows the transcriptome profiles of P. strigosa were influenced by both the type of symbionts and the environmental factors at the study locations. The PC1and PC2 (site and symbiont type) accounted for only 26% of the variability in the transcriptome; thus, most of the changes remain unexplained. One possible hypothesis is that the urban and offshore populations vary genetically, and a population genetics study could explain the remaining differences. Many environmental parameters at the urban sites could have contributed to the differential expression, including the depth, which can influence light availability, distribution of oxygen, and turbidity. Determining the contribution of the depth alone as a single factor changing the differential expression was not an aim of the study. Still, it is known that the symbiont composition within coral hosts changes in response to depth relocation (Baker, 2003). Thus, the depth combined with symbiont type in the current study contributed significantly to the alteration of the host transcriptome.
Coral Response to Particles in the Water Column
Previous studies of water quality in Biscayne Bay, connected to the PoM, show poor water quality, with frequent bursts of nutrients and chlorophyll concentrations (Caccia and Boyer, 2005; Briceño et al., 2011; Enochs et al., 2019; Millette et al., 2019). It is also known that increased phytoplankton concentrations are usually coupled with high bacterial and microzooplankton abundances (Dubinsky and Berman-Frank, 2001). We hypothesize that these high levels of suspended particles explain why the urban corals exhibit an increased number of upregulated particle recognition receptors (PRRs), such as lectins, scavenger receptors, G-coupled receptors, and olfactomedin-like proteins, among others (for full list Supplementary Table 2). In general, lectins are carbohydrate-binding proteins found across phyla and implicated in many recognition processes, including food particle selection and defense (Arason, 1996; Lis and Sharon, 1998). Similarly, immunoglobulin domains and leucine-rich repeat (LRR) domains are major protein components of toll-like receptors (TLR) and nod-like receptors (NOD) involved in recognition of non-self-particles, including potentially pathogenic organisms (Miller et al., 2007). The cysteine-rich scavenger receptors (SRCR-domain), which were also upregulated in the urban corals, are evolutionarily conserved innate immune receptors known to bind bacteria (Yap et al., 2015). These PRRs are likely involved in recognizing food particles and pathogenic organisms and are used for symbiont recognition and acquisition. For example, Kvennefors et al. (2008) demonstrated the mannose-binding lectin from acroporid corals binds to both symbionts and pathogens, while tachylectin is an LPS-binding receptor (Parisi et al., 2020). This large set of PRRs likely enables urban corals to sense and recognize food particles, symbionts, and pathogenic organisms.
Coral Response to High Bacterial Loads and Potential Pathogens
Coastal waters, like Biscayne Bay, occasionally contain increased loads of terrestrial bacteria from surface runoff and sewage system leakage. For example, human-associated bacteria (Enterococci bacteria, a.k.a. fecal bacterial indicator) in Biscayne Bay are frequently reported to exceed the threshold of public safety standards (Dias et al., 2019). Thus, the PoM corals are exposed to a high level of coastal marine bacteria and occasionally to human and animal intestinal bacteria. Many of the PRRs showing increased expression in urban corals have been documented to play a role in this coral’s defense mechanisms, as they were found to be upregulated in corals after live-bacteria exposure (Ocampo et al., 2015). In addition, the C3-complement system is thought to play an essential role in cnidarian immune defense, specifically in an activation pathway that triggers a cascade that ultimately leads to the formation of membrane attack complex-perforins (MACPF), which carry out bacterial cell lysis (Hemmrich et al., 2007; Miller et al., 2007; Parisi et al., 2020). Seven of the twelve host MACPF proteins were upregulated in urban corals, suggesting a potential role of these proteins in coral defense against bacterial invasion. Alternatively, there is speculation that involvement of the coral C3-complement system in the regulation of symbiosis also exists (Davy et al., 2012). In the current study, the C3-complement protein was not differentially expressed between corals hosting different symbionts (Supplementary Table 2), but this does not exclude the possibility of its involvement in the onset of a symbiotic relationship. It has also been speculated that since a variety of lectins occur in all anthozoans, the C3-complement system in these animals might be activated by lectin-like receptors (Hemmrich et al., 2007; Dunn, 2009; Parisi et al., 2020). Thus, the upregulation of over 40 lectins and a C3-complement protein might suggest an intensiveactivation of the immune response in the surviving PoM corals, allowing them to tolerate high bacterial loads.
Other transcripts encoding putative immune-related proteins were also upregulated in urban corals, perhaps due to high bacterial loads in the water. For example, four transcripts annotated with the tumor necrosis factor-like domains (TNF) and nine transcripts annotated as tumor necrosis factor-associated receptor (TRAF) were differentially expressed between the urban and offshore corals (Supplementary Table 3). These TNF and TRAFS are well documented to participate in coral innate immunity (Miller et al., 2007; Mansfield and Gilmore, 2019). Similarly, nitric oxide is an essential antimicrobial molecule and a major component of the innate immune system (Parisi et al., 2020), while the NF-kappa-B-activating protein is a transcriptional factor that controls and regulates innate immune response (Mansfield and Gilmore, 2019). Even though the molecular function of pentraxins has not been investigated in cnidarians, they are components of the immune response in many other vertebrates and invertebrates (Garlanda et al., 2005; Armstrong, 2015).
Additional DETs between the urban and offshore corals were proteins involved in cell-to-cell adhesion, thus possibly playing a role in pathogen capture. These included cadherin-like proteins and dermatopontin-like proteins (Supplementary Table 2). The functional genomic studies of these proteins in invertebrates are limited but suggest that these are extracellular matrix proteins that act as adhesion/agglutination molecules (Okamoto and Fujiwara, 2006). Dermatopontin-related proteins are also one of the cytotoxins present in nematocysts of the fire coral Millepora dichotoma (Iguchi et al., 2008), which suggests that the dermatopontin-like proteins in other cnidarias might function as defense molecules.
Pseudodiploria strigosa individuals from the urban location showed higher expression of a variety of digestive enzymes carrying out degradation of proteins (peptidases), lipids (lipases, phospholipases), and carbohydrates (chitinases, glycoside hydrolases, mannosidases, pectin lyases, Supplementary Table 2). Many of these enzymes could be involved in heterotrophic feeding by extracellular digestion inside the gastric cavity (see section below), but it is also possible that some of these enzymes are involved in the intracellular digestion of bacteria and other invasive microorganisms after they are engulfed by the epithelial cells (Augustin et al., 2010). For example, the cysteine peptidases in the C1A family (a.k.a. cathepsins) are major endosomal/lysosomal enzymes needed to degrade pathogenic organisms (Colbert et al., 2009; Turk et al., 2012). Five cathepsins were differentially expressed between the urban and offshore corals, with two cathepsins being upregulated in urban corals. Immune-related phagocytosis has not been documented in stony scleractinian corals. However, in the model cnidarians, the epithelial cells of both the ecto- and endodermis mediate phagocytosis of bacterial cells (Augustin et al., 2010). Therefore, the potential existence of epithelial phagocytosis in stony corals could explain the upregulation of intracellular digestive enzymes in urban corals exposed to high bacterial concentrations.
Role of Coral Heterotrophy in Coral Health and Stress Tolerance
The benefits of heterotrophic feeding to coral health have been well investigated (see review by Houlbreque and Ferrier-Pages, 2009). Heterotrophic feeding is thought to increase coral resistance to bleaching and aid in recovery from bleaching (Fabricius et al., 2013; Wooldridge, 2014; Tremblay et al., 2016) as well as mitigate acidification stress (Towle et al., 2015). The availability of phytoplankton and zooplankton in Biscayne Bay and connected waterways may therefore provide urban corals with additional nutrition, which may, in turn, confer resilience. Multiple secreted protein-degrading enzymes (peptidases, Supplementary Table 2) were found to be at a higher expression in urban corals than the offshore corals. These include 12 different chymotrypsin-like peptidases (serine peptidases in S1 family, Supplementary Table 2) that are abundant in the coral digestive system, including mesenterial filaments (Raz-Bahat et al., 2017). In addition, there is some evidence that metallopeptidases in families M10, M12, and M14, members of which were upregulated in urban corals (Supplementary Table 2), participate in the coral digestive process inside the gastric cavity. For example, peptidases in the M12A family (astacins) are digestive enzymes in crayfish and sea urchins, but in other organisms might also be involved in other processes such as development, tissue remodeling, and extracellular matrix turnover (Cerda-Costa and Gomis-Ruth, 2014). The seven upregulated lipid degrading enzymes in urban corals include lipases and phospholipases (Supplementary Table 2), which were predicted to be secreted enzymes based on the localization prediction algorithm (see “Materials and Methods” for details). Thus, these enzymes are most likely secreted into the polyps’ gastric cavity for extracellular digestion. However, further studies on the enzymatic activity within the coral gastric cavity are required to confirm this hypothesis. Further, several carbohydrate-degrading enzymes, including glycoside hydrolases, mannosidases, alpha-L-arabinofuranosidase, chitinases, and others (Supplementary Table 2), were upregulated in the urban corals. Since corals preferentially feed on crustaceans such as copepods and crab larvae (Houlbreque and Ferrier-Pages, 2009), this could explain the upregulation of five different chitinases in the urban corals found in this study. Alternatively, the higher expression of chitinases might also protect corals from pathogenic fungi.
The data from this study also suggest that the benefit of heterotrophic feeding in urban corals could have reproductive implications. The urban corals had a higher expression of eight lipid transfer proteins, including four vitellogenins and four liptovitellins, which are known components of coral oocytes (Imagawa et al., 2004; Hayakawa et al., 2005; Shikina et al., 2013). This suggests that the urban corals at the time of collection (February) might have been undergoing oogenesis, which is not unusual for this coral species as it undergoes 1-year-long gametogenesis (Weil and Vargas, 2009). The higher expression of these reproductive genes found in this study may indicate that heterotrophic feeding provides extra nutritional resources that could be allocated to reproduction. The hypothesis of heterotrophic feeding and reproduction success in urban corals would need to be confirmed experimentally. Still, the transcriptomic data from this study suggest the possibility of heterotrophic feeding contributing to their healthy appearance and lack of external signs of stress.
Effect of a Coastal Eutrophication on Coral Transcriptome Response
Cellular and transmembrane transport carriers were differentially expressed between the urban and offshore corals, suggesting readjustment to the available nutrients in the water column. This is most likely due to different levels of inorganic nutrients at the two locations. Contrary to other transcript groups, a higher number of DET encoding transporters were upregulated in offshore corals. A more detailed analysis showed inorganic substance transporters were upregulated in urban corals while organic substance within cell transport was upregulated in offshore coral. The urban corals located in the PoM waters experience a much higher concentration of inorganic and organic nutrients such as nitrate, ammonia, and phosphate than the corals in the offshore reef waters (Caccia and Boyer, 2005; Briceño et al., 2011; Millette et al., 2019). Some evidence exists that the nutrients and photosynthates transported between the coral host and symbionts can be altered by the nutrient concentrations in the water column. More organic carbon is translocated to the host in low-nutrient waters (Dubinsky and Berman-Frank, 2001). The higher expression of the organic substance transporters (e.g., glucose, amino acid, and monocarboxylic acid, Supplementary Table 2) in the offshore corals indicated a pattern consistent with the above study. Specifically, the glucose transporter, which was annotated as solute carriers in family type 2 and is thought to participate in the glucose transport from symbiosome to host cytoplasm (Sproles et al., 2018), had higher expression in the offshore corals suggesting that they might be receiving more fixed carbon from their symbionts.
Signs of Possible Oxidative Stress in Response to Coastal Pollutants
Like the one in the current study, coastal waters near large metropolitan areas are known to contain many different pollutants, including heavy metals, pesticides, and other toxic products (Cantillo and Lauenstein, 2004; Briceño et al., 2011; Castro et al., 2013). Twelve ABC-transporters were DE between the offshore and urban corals (eight had higher expression in urban corals, with most of these belonging to the subfamilies B and C, specifically, ABCC4-type, Supplementary Table 2), which control substance efflux out of the cells (Goldstone, 2008). The ABC-B and ABC-C transporters protect organisms from various foreign substances, including pharmaceuticals and toxic chemicals (Goldstone, 2008). Since many of these showed higher expression in the urban corals, these may participate in detoxification and foreign substance removal from corals occupying contaminated waters. Signs of oxidative stress and detoxification in urban corals included the upregulation of 12 transcripts encoding thioredoxin-like proteins and five cytochrome P450-like proteins (Supplementary Table 2), all of which are predicted to be part of cnidarian defense against environmental pollution (Goldstone, 2008). In addition, oxidative stress-related genes (including peroxiredoxins, thioredoxins) were upregulated in corals exposed to toxic doses of heavy metals such as copper and cadmium (Schwarz et al., 2013; Zhou et al., 2018), while cytochrome P450 encoding transcripts were upregulated in corals exposed to a common insecticide (Wecker et al., 2018). Because a high concentration of copper is present in anti-biofouling paint, its concentration increases in areas near ports and shipping lanes and areas of high boat traffic like the one in PoM and the location of urban corals from this study. This may explain the increased expression of pathways related to oxidative stress and detoxification.
Conclusion
Corals in urbanized areas exhibit a low prevalence of bleaching and disease. Thus, we hypothesize that they are resistant to many environmental stressors such as eutrophication, coastal acidification, high turbidity, and increased bacterial loads and pollution. Here, we investigated possible molecular mechanisms of coral resistance to these environmental stressors using transcriptomic profiling and differential gene expression between urban and offshore corals. Our data demonstrate a large suite of molecules for which the mRNA expression plasticity may allow corals to withstand poor water quality and stressful conditions. Urban corals host symbionts that are better adapted to live and photosynthesize in shallow, turbid, and high-nutrient waters. Urban corals exhibited higher mRNA expression of pattern-recognition receptors, which may allow these corals to differentiate between food particles and potentially pathogenic organisms. In addition, numerous upregulated digestive and degrading enzymes suggested that the urban corals likely depend on heterotrophic feeding for a supplementary source of energy. Alternatively, the same enzymes might be used to protect the coral from potential pathogens. Increased expression of defense and innate immunity proteins in urban corals contributes to their resistance against foreign microorganisms, while the increased regulation of detoxification proteins protects these corals from toxic substances and water pollutants. The current description of the defense responses to stressful conditions may lead to the development of molecular markers of coral resilience, which could be used in coral monitoring, conservation, and restoration activities.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/, PRJNA693301.
Author Contributions
IE and DM were awarded the funding and provided the overview vision for the study with a crucial collaboration from CF, who provided the initial motivation, site selection, and background information for the study. AM and IE conceived and designed the study. GK, IB, and ER carried out the sample collection. ER completed the transcriptomic analysis and wrote the manuscript with editing contributions from co-authors. All authors gave final approval of this manuscript for publication.
Funding
This study was funded by the Omics Initiative of the National Oceanic Atmospheric Administration.
Conflict of Interest
CF was employed by Coral Morphologic.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank other member of the NOAA-AOML’s coral research lab including Ana Palacio, Nate Formel, Nicole Besemer, Michael S. Studivian, Michael Jankulak, Albert Boyd, Partic Kiel, and Nash Soderberg for their contributions to data interpretation and suggestions on ecological concepts included in the manuscripts. The scientific results and conclusions, as well as any views or opinions expressed herein, are those of the author(s) and do not necessarily reflect the views of NOAA or the Department of Commerce.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.695236/full#supplementary-material
Supplementary Figure 1 | Location of the study sites in Florida, near Port of Miami (PoM) including: an offshore reef (Emerald Reef, EMLR) and urban coral sites: MacArthur North (MACN), MacArthur South (MACS), and Star Island (STAR).
Supplementary Figure 2 | Number of P. strigosa’s DET grouped into eight molecular-function categories (A–H). Three differential expression analyses were made between colonies from each urban site (MACN, MACS, STAR) and colonies from an offshore reef (EMLR) which used as a control, and forth analysis was made between colonies (n = 17) hosting mostly Durusdinium (Duru.) symbionts in comparison to colonies (n = 14) hosting mostly Breviolum (Brev.) symbionts.
Supplementary Figure 3 | Species- level endosymbiont composition of 31 individuals of P. strigosa collect from (A) an offshore reef (Emerald Reef, EMLR) and urban coral sites (B) MacArthur North (MACN), (C) MacArthur South (MACS), and (D) Star Island (STAR) near Port of Miami, FL.
Supplementary Table 1 | The NCBIs Short Read Archive (SRA) accession numbers for cDNA libraries sequenced from 31 individuals of P. strigosa from three urban sites located in the Port of Miami (MACN, MACS, STAR) and one offshore reef site (EMLR). The quality-filtered reads (Phred score 25) were submitted under the NCBI SRA BioProject ID: PRJNA693301.
Supplementary Table 2 | Number of differentially expressed transcripts (DET) found between urban and offshore P. strigosa colonies, and between Durusdinium-dominated and Breviolum-dominated P. strigosa colonies. DET were considered statistically significantly different at a minimum of twofold mRNA change and FDR adjusted p-value < 0.05. Total is the number of DET annotated with the same protein domain, while values under each column are the number of DET that were significantly greater in the pair-wise comparison (urban vs. offshore and Durusdinium vs. Breviolum).
Supplementary Table 3 | List of 2673 DET found in the study, their associated fold change (logFC) in mRNA levels, FDR adjusted p-values, p-values, functional and taxonomic annotations and the cellular or molecular function used to categorize the DET into functional groups.
Supplementary Table 4 | NCBI accession number and metadata for 22 large-ribosomal-subunit gene sequences (LSU, 28S gene) used as references for symbiont composition estimate in this current study.
Supplementary Table 5 | List and source of coral endosymbiont reference transcriptomes used for taxonomic assignment of protein-coding sequences generated in this study.
Supplementary Table 6 | Comparison of symbiont composition based on reads mapped to the LSU gene-fragment and either corrected or uncorrected for the rDNA copy number per cell for the symbiont two species Breviolum minutum and Durusdinium trenchii hosted by Pseudodiploria strigosa colonies.
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arason, G. J. (1996). Lectins as defense molecules in vertebrates and invertebrates. Fish Shellf. Immunol. 6, 277–289. doi: 10.1006/fsim.1996.0029
Armstrong, P. B. (2015). Comparative biology of the pentraxin protein family: evolutionarily conserved component of innate immune system. Int. Rev. Cell Mol. Biol. 316, 1–47. doi: 10.1016/bs.ircmb.2015.01.002
Augustin, R., Fraune, S., and Bosch, T. C. (2010). How Hydra senses and destroys microbes. Semin. Immunol. 22, 54–58. doi: 10.1016/j.smim.2009.11.002
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Baker, A. C., Starger, C. J., McClanahan, T. R., and Glynn, P. W. (2004). Coral reefs: corals’ adaptive response to climate change. Nature 430:741. doi: 10.1038/430741a
Bay, R. A., and Palumbi, S. R. (2017). Transcriptome predictors of coral survival and growth in a highly variable environment. Ecol. Evol. 7, 4794–4803. doi: 10.1002/ece3.2685
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bongaerts, P., Carmichael, M., Hay, K. B., Tonk, L., Frade, P. R., and Hoegh-Guldberg, O. (2015). Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species. R. Soc. Open Sci. 2:140297. doi: 10.1098/rsos.140297
Brener-Raffalli, K., Vidal-Dupiol, J., Adjeroud, M., Rey, O., Romans, P., Bonhomme, F., et al. (2019). Gene expression plasticity and frontloading promote thermotolerance in Pocillopora corals. bioRxiv [Preprint]. doi: 10.1101/398602
Briceño, H. O., Boyer, J. N., and Harlem, P. W. (2011). Ecological Impacts on Biscayne Bay and Biscayne National Park from Proposed South Miami-Dade County Development, and Derivation of Numeric Nutrient Criteria for South Florida Estuaries and Coastal Waters. NPS TA# J5297-08-0085, Florida International University, Southeast Environmental Research Center Contribution # T530, 145. Available online at: http://serc.fiu.edu/wqmnetwork/BNP/Final%20Report%20BNP.pdf
Brown, J., Pirrung, M., and Mccue, L. A. (2017). FQC Dashboard integrates FastQC results into a web based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373
Browne, N. K., Tay, J. K., Low, J., Larson, O., and Todd, P. A. (2015). Fluctuations in coral health of four common inshore reef corals in response to seasonal and anthropogenic changes in water quality. Mar. Environ. Res. 105, 39–52. doi: 10.1016/j.marenvres.2015.02.002
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–63. doi: 10.1038/nmeth.3176
Burt, J. A. (2014). The environmental costs of coastal urbanization in the Arabian Gulf. City 18, 760–770. doi: 10.1080/13604813.2014.962889
Burt, J. A., Camp, E. F., Enochs, I. C., Johansen, J. L., Morgan, K. M., Riegl, B., et al. (2020). Insights from extreme coral reefs in a changing world. Coral Reefs 39, 495–507.
Caccia, V. G., and Boyer, J. N. (2005). Spatial patterning of water quality in Biscayne Bay, Florida as a function of land use and water management. Mar. Pollut. Bull. 50, 1416–1429. doi: 10.1016/j.marpolbul.2005.08.002
Cantillo, A. Y., and Lauenstein, G. G. (2004). Extent and Toxicity of Contaminated Marine Sediments in Southeastern Florida. NOAA Technical Memorandum NOS NCCOS 4. Silver Spring, MD: NOAA/NOS/NCCOS, 120.
Castro, J. E., Fernandez, A. M., Gonzalez-Caccia, V., and Gardinali, P. R. (2013). Concentration of trace metals in sediments and soils from protected lands in south Florida: background levels and risk evaluation. Environ. Monit. Assess. 185, 6311–6332. doi: 10.1007/s10661-012-3027-9
Cerda-Costa, N., and Gomis-Ruth, F. X. (2014). Architecture and function of metallopeptidase catalytic domains. Protein Sci. 23, 123–144. doi: 10.1002/pro.2400
Colbert, J. D., Matthews, S. P., Miller, G., and Watts, C. (2009). Diverse regulatory roles for lysosomal proteases in the immune response. Eur. J. Immunol. 39, 2955–2965. doi: 10.1002/eji.200939650
Cunning, R., Gates, R. D., and Edmunds, P. J. (2017). Using high-throughput sequencing of ITS2 to describe Symbiodinium metacommunities in St. John, US Virgin Islands. PeerJ 5:e3472. doi: 10.7717/peerj.3472
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. doi: 10.1128/MMBR.05014-11
DeSalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., and Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol. Ecol. 19, 1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x
Dias, M., Henn, C., and Day, K. (2019). Surfrider’s Clean Water Report. Available online at: https://bwtf.surfrider.org/resources (accessed January 13, 2021).
Dillies, M. A., Rau, A., Aubert, J., Hennequet-Antier, C., Jeanmougin, M., Servant, N., et al. (2013). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 14, 671–683. doi: 10.1093/bib/bbs046
Dubinsky, Z., and Berman-Frank, I. (2001). Uncoupling primary production from population growth in photosynthesizing organisms in aquatic ecosystems. Aquat. Sci. 63, 4–17. doi: 10.1007/PL00001343
Dunn, S. R. (2009). Immunorecognition and immunoreceptors in the Cnidaria. Invertebr. Surviv. J. 6, 71–74.
Dziedzic, K. E., Elder, H., Tavalire, H., and Meyer, E. (2019). Heritable variation in bleaching responses and its functional genomic basis in reef-building corals (Orbicella faveolata). Mol. Ecol. 28, 2238–2253. doi: 10.1111/mec.15081
Enochs, I. C., Manzello, D. P., Jones, P. R., Stamates, S. J., and Carsey, T. P. (2019). Seasonal carbonate chemistry dynamics on southeast florida coral reefs: localized acidification hotspots from navigational inlets. Front. Mar. Sci. 6:160. doi: 10.3389/fmars.2019.00160
Fabina, N. S., Putnam, H. M., Franklin, E. C., Stat, M., and Gates, R. D. (2012). Transmission mode predicts specificity and interaction patterns in coral-Symbiodinium networks. PLoS One 7:e44970. doi: 10.1371/journal.pone.0044970
Fabricius, K. E., Cseke, S., Humphrey, C., and De’ath, G. (2013). Does trophic status enhance or reduce the thermal tolerance of scleractinian corals? A review, experiment and conceptual framework. PLoS One 8:e54399. doi: 10.1371/journal.pone.0054399
Fuess, L. E., Pinzon, C. J., Weil, E., and Mydlarz, L. D. (2016). Associations between transcriptional changes and protein phenotypes provide insights into immune regulation in corals. Dev. Comp. Immunol. 62, 17–28. doi: 10.1016/j.dci.2016.04.017
Garlanda, C., Bottazzi, B., Bastone, A., and Mantovani, A. (2005). Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23, 337–366. doi: 10.1146/annurev.immunol.23.021704.115756
Goldstone, J. V. (2008). Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis. Cell Biol. Toxicol. 24, 483–502. doi: 10.1007/s10565-008-9107-5
Guest, J. R., Tun, K., Low, J., Verges, A., Marzinelli, E. M., Campbell, A. H., et al. (2016). 27 years of benthic and coral community dynamics on turbid, highly urbanized reefs off Singapore. Sci. Rep. 6:36260. doi: 10.1038/srep36260
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hayakawa, H., Nakano, Y., Andoh, T., and Watanabe, T. (2005). Sex-dependent expression of mRNA encoding a major egg protein in the gonochoric coral Galaxea fascicularis. Coral Reefs 24, 488–494. doi: 10.1007/s00338-005-0485-3
Heery, E. C., Hoeksema, B. W., Browne, N. K., Reimer, J. D., Ang, P. O., Huang, D., et al. (2018). Urban coral reefs: degradation and resilience of hard coral assemblages in coastal cities of East and Southeast Asia. Mar. Pollut. Bull. 135, 654–681. doi: 10.1016/j.marpolbul.2018.07.041
Helmkampf, M., Bellinger, M. R., Frazier, M., and Takabayashi, M. (2019). Symbiont type and environmental factors affect transcriptome-wide gene expression in the coral Montipora capitata. Ecol. Evol. 9, 378–392. doi: 10.1002/ece3.4756
Hemmrich, G., Miller, D. J., and Bosch, T. C. (2007). The evolution of immunity: a low-life perspective. Trends Immunol. 28, 449–454. doi: 10.1016/j.it.2007.08.003
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Houlbreque, F., and Ferrier-Pages, C. (2009). Heterotrophy in tropical scleractinian corals. Biol. Rev. Camb. Philos. Soc. 84, 1–17. doi: 10.1111/j.1469-185X.2008.00058.x
Hume, B. C. C., Smith, E. G., Ziegler, M., Warrington, H. J. M., Burt, J. A., Lajeunesse, T. C., et al. (2019). SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080. doi: 10.1111/1755-0998.13004
Iguchi, A., Iwanaga, S., and Nagai, H. (2008). Isolation and characterization of a novel protein toxin from fire coral. Biochem. Biophys. Res. Commun. 365, 107–112. doi: 10.1016/j.bbrc.2007.10.153
Imagawa, S., Nakano, Y., and Watanabe, T. (2004). Molecular analysis of a major soluble egg protein in the scleractinian coral Favites chinensis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 137, 11–19. doi: 10.1016/j.cbpc.2003.09.011
Jones, A., and Berkelmans, R. (2010). Potential costs of acclimatization to a warmer climate: growth of a reef coral with heat tolerant vs. sensitive symbiont types. PLoS One 5:e10437. doi: 10.1371/journal.pone.0010437
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kenkel, C. D., Moya, A., Strahl, J., Humphrey, C., and Bay, L. K. (2018). Functional genomic analysis of corals from natural CO2 -seeps reveals core molecular responses involved in acclimatization to ocean acidification. Glob. Chang. Biol. 24, 158–171. doi: 10.1111/gcb.13833
Kvennefors, E. C., Leggat, W., Hoegh-Guldberg, O., Degnan, B. M., and Barnes, A. C. (2008). An ancient an variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev. Comp. Immunol. 32, 1582–1592. doi: 10.1016/j.dci.2008.05.010
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e2576. doi: 10.1016/j.cub.2018.07.008
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Libro, S., Kaluziak, S. T., and Vollmer, S. V. (2013). RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. PLoS One 8:e81821. doi: 10.1371/journal.pone.0081821
Lin, Z., Wang, L., Chen, M., and Chen, J. (2018). The acute transcriptomic response of coral-algae interactions to pH fluctuation. Mar. Genom. 42, 32–40. doi: 10.1016/j.margen.2018.08.006
Lis, H., and Sharon, N. (1998). Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 98, 637–674. doi: 10.1021/cr940413g
Mansfield, K. M., and Gilmore, T. D. (2019). Innate immunity and cnidarian-Symbiodiniaceae mutualism. Dev. Comp. Immunol. 90, 199–209. doi: 10.1016/j.dci.2018.09.020
Manzello, D. P. (2015). Rapid recent warming of coral reefs in the Florida Keys. Sci. Rep. 5:16762. doi: 10.1038/srep16762
Manzello, D. P., Enochs, I. C., Kolodziej, G., Carlton, R., and Valentino, L. (2018). Resilience in carbonate production despite three coral bleaching events in 5 years on an inshore patch reef in the Florida Keys. Mar. Biol. 165:99. doi: 10.1007/s00227-018-3354-7
Manzello, D. P., Matz, M. V., Enochs, I. C., Valentino, L., Carlton, R. D., Kolodziej, G., et al. (2019). Role of host genetics and heat-tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Glob. Chang. Biol. 25, 1016–1031. doi: 10.1111/gcb.14545
Martin, M. (2011). Cutadapt removed adapter sequences from high throughput sequenecing reads. EMBnet J. 17:200. doi: 10.14806/ej.17.1.200
Miller, D. J., Hemmrich, G., Ball, E. E., Hayward, D. C., Khalturin, K., Funayama, N., et al. (2007). The innate immune repertoire in cnidaria–ancestral complexity and stochastic gene loss. Genome Biol. 8:R59. doi: 10.1186/gb-2007-8-4-r59
Millette, N. C., Kelble, C., Linhoss, A., Ashby, S., and Visser, L. (2019). Using spatial variability in the rate of change of chlorophyll a to improve water quality management in a subtropical oligotrophic estuary. Estuar. Coast. 42, 1792–1803. doi: 10.1007/s12237-019-00610-5
Ocampo, I. D., Zarate-Potes, A., Pizarro, V., Rojas, C. A., Vera, N. E., and Cadavid, L. F. (2015). The immunotranscriptome of the Caribbean reef-building coral Pseudodiploria strigosa. Immunogenetics 67, 515–530. doi: 10.1007/s00251-015-0854-1
Okamoto, O., and Fujiwara, S. (2006). Dermatopontin, a novel player in the biology of the extracellular matrix. Connect. Tissue Res. 47, 177–189. doi: 10.1080/03008200600846564
Parisi, M. G., Parrinello, D., Stabili, L., and Cammarata, M. (2020). Cnidarian Immunity and the repertoire of defense mechanisms in anthozoans. Biology 9:283. doi: 10.3390/biology9090283
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pettay, D. T., Wham, D. C., Smith, R. T., Iglesias-Prieto, R., and Lajeunesse, T. C. (2015). Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc. Natl. Acad. Sci. U.S.A. 112, 7513–7518. doi: 10.1073/pnas.1502283112
Pinzon, J. H., Kamel, B., Burge, C. A., Harvell, C. D., Medina, M., Weil, E., et al. (2015). Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 2:140214. doi: 10.1098/rsos.140214
Pizarro, V., Rodriguez, S. C., Lopez-Victoria, M., Zapata, F. A., Zea, S., Galindo-Martinez, C. T., et al. (2017). Unraveling the structure and composition of Varadero Reef, an improbable and imperiled coral reef in the Colombian Caribbean. PeerJ 5:e4119. doi: 10.7717/peerj.4119
Poquita-Du, R. C., Huang, D., Chou, L. M., and Todd, P. A. (2020). The contribution of stress-tolerant endosymbiotic dinoflagellate Durusdinium to Pocillopora acuta survival in a highly urbanized reef system. Coral Reefs 39, 745–755. doi: 10.1007/s00338-020-01902-0
Precht, W. F., Gintert, B. E., Robbart, M. L., Fura, R., and Van Woesik, R. (2016). Unprecedented disease related coral mortality in Southeastern Florida. Sci. Rep. 6:31374. doi: 10.1038/srep31374
Raz-Bahat, M., Douek, J., Moiseeva, E., Peters, E. C., and Rinkevich, B. (2017). The digestive system of the stony coral Stylophora pistillata. Cell Tissue Res. 368, 311–323. doi: 10.1007/s00441-016-2555-y
Reeb, P. D., Bramardi, S. J., and Steibel, J. P. (2015). Assessing dissimilarity measures for sample-based hierarchical clustering of RNA sequencing data using plasmode datasets. PLoS One 10:e0132310. doi: 10.1371/journal.pone.0132310
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saad, O. S., Lin, X., Ng, T. Y., Li, L., Ang, P., and Lin, S. (2020). Genome size, rDNA copy, and qPCR assays for Symbiodiniaceae. Front. Microbiol. 11:847. doi: 10.3389/fmicb.2020.00847
Schwarz, J. A., Mitchelmore, C. L., Jones, R., O’dea, A., and Seymour, S. (2013). Exposure to copper induces oxidative and stress responses and DNA damage in the coral Montastraea franksi. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 157, 272–279. doi: 10.1016/j.cbpc.2012.12.003
Shen, W., and Xiong, J. (2019). TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. bioRxiv [Preprint]. doi: 10.1101/513523
Shikina, S., Chen, C. J., Chung, Y. J., Shao, Z. F., Liou, J. Y., Tseng, H. P., et al. (2013). Yolk formation in a stony coral Euphyllia ancora (Cnidaria, Anthozoa): insight into the evolution of vitellogenesis in nonbilaterian animals. Endocrinology 154, 3447–3459. doi: 10.1210/en.2013-1086
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Sproles, A. E., Kirk, N. L., Kitchen, S. A., Oakley, C. A., Grossman, A. R., Weis, V. M., et al. (2018). Phylogenetic characterization of transporter proteins in the cnidarian-dinoflagellate symbiosis. Mol. Phylogenet. Evol. 120, 307–320. doi: 10.1016/j.ympev.2017.12.007
Stat, M., and Gates, R. D. (2011). Clade D Symbiodiniumin from scleractinian corals: a “nugget” of hope, selfish opportunist, an ominous sign, or all the above? J. Mar. Biol. 2011, 1–9. doi: 10.1155/2011/730715
Towle, E. K., Enochs, I. C., and Langdon, C. (2015). Threatened Caribbean coral is able to mitigate the adverse effects of ocean acidification on calcification by increasing feeding rate. PLoS One 10:e0123394. doi: 10.1371/journal.pone.0123394
Tremblay, P., Gori, A., Maguer, J. F., Hoogenboom, M., and Ferrier-Pages, C. (2016). Heterotrophy promotes the re-establishment of photosynthate translocation in a symbiotic coral after heat stress. Sci. Rep. 6:38112. doi: 10.1038/srep38112
Turk, V., Stoka, V., Vasiljeva, O., Renko, M., Sun, T., Turk, B., et al. (2012). Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88. doi: 10.1016/j.bbapap.2011.10.002
van Woesik, R., and McCaffrey, K. R. (2017). Repeated thermal stress, shading, and directional selection in the Florida reef tract. Front. Mar. Sci. 4:182. doi: 10.3389/fmars.2017.00182
Walton, C. J., Hayes, N. K., and Gilliam, D. S. (2018). Impacts of a regional, multi-year, multi-species cora disease outbreak in Southeast Florida. Front. Mar. Sci. 5:323. doi: 10.3389/fmars.2018.00323
Wecker, P., Lecellier, G., Guibert, I., Zhou, Y., Bonnard, I., and Berteaux-Lecellier, V. (2018). Exposure to the environmentally persistent insecticide chlordecone induces detoxification genes and causes polyp bail-out in the coral P. damicornis. Chemosphere 195, 190–200. doi: 10.1016/j.chemosphere.2017.12.048
Weil, E., and Vargas, W. L. (2009). Comparative aspects of sexual reproduction in the Caribbean coral genus Diploria (Scleractinia: Faviidae). Mar. Biol. 157, 413–426. doi: 10.1007/s00227-009-1328-5
Wooldridge, S. A. (2014). Formalising a mechanistic linkage between heterotrophic feeding and thermal bleaching resistance. Coral Reefs 33, 1131–1136. doi: 10.1007/s00338-014-1193-7
Yap, N. V., Whelan, F. J., Bowdish, D. M., and Golding, G. B. (2015). The evolution of the scavenger receptor cysteine-rich domain of the class A of scavenger receptors. Front. Immunol. 6:342. doi: 10.3389/fimmu.2015.00342
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan – an integration platform for the signature recognition methods in InterPro. Bioinfomatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Keywords: symbiodiniaceae, Pseudodiploria strigosa, transcriptome, differential gene expression analysis, urban corals
Citation: Rubin ET, Enochs IC, Foord C, Mayfield AB, Kolodziej G, Basden I and Manzello DP (2021) Molecular Mechanisms of Coral Persistence Within Highly Urbanized Locations in the Port of Miami, Florida. Front. Mar. Sci. 8:695236. doi: 10.3389/fmars.2021.695236
Received: 14 April 2021; Accepted: 24 June 2021;
Published: 20 July 2021.
Edited by:
Cliff Ross, University of North Florida, United StatesReviewed by:
John Everett Parkinson, University of South Florida, United StatesColleen Bove, University of North Carolina at Chapel Hill, United States
Copyright © 2021 Rubin, Enochs, Foord, Mayfield, Kolodziej, Basden and Manzello. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ewelina T. Rubin, ZXdlbGluYS5ydWJpbkBub2FhLmdvdg==
 Ewelina T. Rubin
Ewelina T. Rubin Ian C. Enochs
Ian C. Enochs Colin Foord
Colin Foord Anderson B. Mayfield
Anderson B. Mayfield Graham Kolodziej
Graham Kolodziej Isabelle Basden1
Isabelle Basden1 Derek P. Manzello
Derek P. Manzello