- 1Graduate School of Science and Technology for Innovation, Yamaguchi University, Yamaguchi, Japan
- 2Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Japan
- 3Faculty of Science, The University of the Ryukyus, Nishihara, Japan
Numerous genes involved in calcification, algal endosymbiosis, and the stress response have been identified in corals by large-scale gene expression analysis, but functional analysis of those genes is lacking. There are few experimental examples of gene expression manipulation in corals, such as gene knockdown by RNA interference (RNAi). The purpose of this study is to establish an RNAi method for coral juveniles. As a first trial, the genes encoding green fluorescent protein (GFP, an endogenous fluorophore expressed by corals) and thioredoxin (TRX, a stress response gene) were selected for knockdown. Synthesized double-stranded RNAs (dsRNAs) corresponding to GFP and TRX were transformed into planula larvae by lipofection method to attempt RNAi. Real-time PCR analysis to verify knockdown showed that GFP and TRX expression levels tended to decrease with each dsRNA treatment (not significant). In addition, stress exposure experiments following RNAi treatment revealed that planulae with TRX knockdown exhibited increased mortality at elevated temperatures. In GFP-knockdown corals, decreased GFP fluorescence was observed. However, the effect of GFP-knockdown was confirmed only in the coral at the initial stages of larval metamorphosis into polyps, but not in planulae and 1 month-old budding polyps. This study showed that lipofection RNAi can be applied to coral planulae and polyps after settlement, and that this method provides a useful tool to modify expression of genes involved in stress tolerance and fluorescence emission of the corals.
Introduction
RNA interference (RNAi) is a sequence-specific gene silencing phenomenon in which double-stranded RNA (dsRNA) causes a reduction of the complementary RNA (Fjose et al., 2001). The RNAi method has been widely used in model organisms and cell cultures for functional genomic study (Buckingham et al., 2004; Perrimon and Mathey-Prevot, 2007). Recently, large-scale sequencing analyses and metabolome analyses have been increasingly used in coral–algae symbiosis, and studies of the identification of symbiosis-, stress-, and skeletogenesis-related genes are increasing (Hillyer et al., 2016; Thomas and Palumbi, 2017; Hou et al., 2018; Yuyama et al., 2018; Ying et al., 2019; Zhang et al., 2019; Shinzato et al., 2021). Following identification, the functions of these genes need to be determined by genetic manipulation and examination of their expression localization. However, there have been only a few attempts to manipulate the expression of coral genetic material by molecular approaches, such as RNAi (Yasuoka et al., 2016; Cleves et al., 2018). Meanwhile, there are some reports of RNAi in other members of the phylum Cnidarian, such as hydra and sea anemone. RNAi experiments in hydra have revealed the genes involved in cephalic and foot development, budding, and axis determination (Lohmann et al., 1999; Amimoto et al., 2006; Galliot et al., 2007; Jakob and Schierwater, 2007; Chera et al., 2009). These findings have helped clarify how body plans evolved in metazoan organisms. In addition, a recent gene expression silencing experiment using a marine hydrozoan model (Hydractinia symbiolongicarpus) has shown that knockdown of an exogenous green fluorescent protein (GFP) was successful by electroporation of shRNAs (Quiroga-Artigas et al., 2020). Knockdown of actin (ACT) and caspases in sea anemones (Aiptasia pallida) has been reported to reduce their gene expression levels (Dunn et al., 2007). Although sea anemones have been used as models of coral–algae symbiosis (Gabay et al., 2018; Jones et al., 2018; Li et al., 2018), the symbiotic relationship between coral-algae and sea anemones has different aspects: algal in corals contribute significantly to host skeletogenesis (Yuyama and Higuchi, 2014). Establishing RNAi technology for corals will enable further analysis of coral-specific phenomena, while morpholino oligo injection and CRISPR/Cas9-mediated genome editing are also an promising approach for gene knockdown in corals (Yasuoka et al., 2016; Cleves et al., 2018).
The objective of this study was to establish an RNAi method for use in coral. Although coral larvae and eggs are easy to manipulate genetically, their spawning period is limited to once or twice a year. A series of experiments using juvenile polyps and planula larvae of Acropora tenuis showed that the larvae could be used for fluorescence observation and stress experiments (Yuyama et al., 2005, 2012b, 2016). Given that they can be maintained in a small container, and the incubation conditions are similar to Aiptasia, it is possible that the RNAi method (lipofection method) performed in sea anemones could be performed in young A. tenuis (Dunn et al., 2007). Therefore, we performed RNAi by lipofection in A. tenuis, referring to the method performed in Aiptasia.
In this study, thioredoxin (TRX) and GFP were targeted by RNAi. TRX is a redox-regulating protein that functions as an indicator of oxidative stress in an organism (Arnér and Holmgren, 2000; Yoshida et al., 2003). TRX expression is up-regulated in corals in high temperature environments (Edge et al., 2005); we confirmed this in planula larvae by real-time PCR as well in preliminary experiments. GFP is a fluorescent protein naturally present in coral, and thus its expression in planula larvae and juvenile polyps is easily monitored using fluorescence stereomicroscopy (Strader et al., 2015; Yuyama et al., 2018). GFP is expressed throughout tissues in the planula, and is especially abundant in the endoderm (Haryanti and Hidaka, 2019). The GFP intensity of juvenile polyps was reportedly changed due to the algal symbiosis and stress exposure (Yuyama et al., 2012a). We synthesized dsRNA probes containing sequences complementary to TRX and GFP, transfected them into coral, and verified RNAi-mediated knockdown using real-time PCR. The effectiveness of TRX knockdown was assessed by calculating the survival rate of TRX-dsRNA-treated corals exposed to high temperature. GFP-dsRNA silencing was confirmed by fluorescence stereomicroscopy. In these experiments, ACT dsRNA transfection was used to confirm the specificity of each knockdown.
Materials and Methods
Coral Samples
Collection of A. tenuis larvae was performed as described previously (Iwao et al., 2002) at the Akajima Marine Science Laboratory (Okinawa, Japan). In 2013, the annual mean seawater temperature in the coastal area in Okinawa was 24.5°C, and the highest temperature of 31.2°C was recorded in early August (Higuchi et al., 2015). More than five parental colonies were used to produce larvae. A portion of the larvae were induced to undergo metamorphosis by exposure to 22 μM Hym 248 in filtered sea water (Iwao et al., 2002) in glass Petri dishes (55 mm in diameter) at 24°C under a 12-h light (20 μE/m2/s):12-h dark cycle. No significant change in the growth rate of larvae was observed within the same peptide stimulation batch. Seawater was filtered using a 0.22-μm filter (Millipore, Billerica, MA, United States).
Preparation of RNAi Target Genes
The coral TRX sequence used for RNAi was identified from A. tenuis as follows. Total RNA was isolated from A. tenuis aposymbiotic juvenile polyps using an Absolutely RNA reverse transcription (RT)-PCR Miniprep Kit (Stratagene, La Jolla, CA, United States) and TRX cDNA was prepared using a SMART rapid amplification of cDNA ends (RACE) cDNA amplification kit (Takara Bio, CA, United States). Using the cDNA, fragments of approximately 200 bp containing TRX coding sequences were amplified by RT-PCR with degenerate primers (5′-TTRCANGGNCCRCACCA-3′ and 5′-YTTRCANGGNCCRCACCAYTC-3′). Then, 5′ and 3′ RACE was carried out to identify cDNA sequences that were 5′ and 3′ of the fragments, using the cDNA described above. In 3′ RACE, RT was performed with gene-specific primers (5′-GGATCCGAGAAAGAATAATGAA-3′), whose sequences were designed from isolated fragments, and subsequent nested PCR was performed with specific primers (5′-CTATCCTGACAAGCTGTTGG-3′). For 5′ RACE, the primers 5′-CCAACAGCTTGTCAGGATAG-3′ were used as forward primers. The RACE PCR products were cloned into the pGEM-T Easy vector and sequenced on both strands by Macrogen1. A homology search in the ‘‘Protein-All’’ database was conducted on the DNA Data Bank of Japan (DDBJ) website2. A search of the Pfam database was performed using the “Motif Search” program on the Genome Net website (http://www.genome.jp/ja/; Bioinformatics Center, Institute for Chemical Research, Kyoto University, Japan). The TRX sequences identified here been registered in DDBJ (accession no. LC532156). The gene encoding GFP, identified by Yuyama et al. (2012a; accession no. AY646066; Yuyama et al., 2012a), was used for RNAi. In addition, the ACT gene of A. tenuis (accession no. BJ999688), identified by Kii et al. (2007), was used in these experiments.
Preparation of dsRNA for RNAi
An RNAi experiment was performed in accordance with the previously published method of Dunn et al., 2007. The dsRNAs were synthesized using the MEGAscript RNAi kit (Ambion, Austin, TX, United States). dsRNAs specific for TRX (311 bp), GFP (607 bp), and ACT (499 bp) were generated. The fragments were amplified from A. tenuis cDNA using specific primers flanked with the T7 RNA polymerase promoter sequence (Table 1). After amplifying the target sequence by PCR using each primer, sequences of PCR product were confirmed with sanger-sequencing by MACROGEN JAPAN (Tokyo, Japan). Fifty planula larvae or five polyps were placed in each well of a 24-well plate with 1 mL of filtered seawater (0.22-μm filter) with 0.2 μg/mL ampicillin and 0.5 μg/mL kanamycin. The liposomal compound (DMRIE-C, Invitrogen/Gibco, Co Dublin, Ireland) was used as an RNA transfection agent. Then, 1 mL of filtered seawater with 0.5 μg/mL or 1 μg/mL dsRNA and DMRIE-C was further added to the coral. For control treatments of TRX- or GFP-targeting RNAi, filtered seawater containing only DMRIE-C or ACT- dsRNA + DMRIE-C was prepared. After incubation for 3 days, corals were fixed in RNAlater for real-time PCR analysis, or the following assay (Verification of knockdown by stress exposure and fluorescence observation) was performed.
Real-Time PCR
RNA was extracted from 50 planula larvae in each well. Samples were fixed in RNAlater (Ambion) after 72 h of incubation with dsRNA. Since planulae had better RNA extraction efficiency than primary polyps, we performed real-time PCR using planulae. Total RNA was extracted using the PureLink RNA Mini Kit (Thermo Fisher Scientific, Tokyo, Japan). Then, first strand cDNAs were synthesized from 20 ng of total RNA using the Superscript VILO Master Mix (Invitrogen, Carlsbad, CA, United States) for the PCR template. For real-time PCR, the expression levels of TRX and GFP were determined in triplicate using SYBR PreMix ExTaq II (Sigma, Tokyo, Japan). Primers for real-time PCR analyses were designed using Primer Express 3.0 software (Applied Biosystems, Foster City, CA, United States; Table 2). It has been confirmed that PCR using each primer set and cDNA of A. tenuis yields the desired PCR product by electrophoresis. Genome analysis of A. tenuis has revealed that there are multiple isoforms of GFP (Satoh et al., 2021). Here, we used primers that detect only the GFP of accession no. AY646066, that showed high expression levels in planulae and polyps (Yuyama et al., 2012a). This is the only sequence isolated from the cDNA library of the similar origin A. tenuis. Each PCR reaction contained 10 μL of PCR master mix of SYBR PreMix ExTaq II (Sigma), 1.5 mM forward primer, 1.5 mM reverse primer, and 2.0 μL of cDNA template. The PCR conditions were as follows: denaturation at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C for 30 s and annealing at 58°C for 30 s, and extension at 72°C for 30 s. The mean values were calculated using the Thermal Cycler Dice Real-time System (TAKARA, Sigma). The data were normalized to the expression levels of tubulin and elongation factor of A. tenuis. As candidates for expression normalization, the tubulin, elongation factor, and ribosomal protein L5 genes, which have been used previously for normalization in real-time PCR (Yuyama et al., 2012b), were selected and their expression levels tested using NormFinder (Andersen et al., 2004). Elongation factor and tubulin were found to be relatively stable, and the average expression levels of these genes were used for normalization. As a control group, corals of ACT -targeting RNAi were used.
Verification of Knockdown by Stress Exposure and Fluorescence Observation
To investigate the effect of GFP-targeting RNAi, the GFP expression of planula larvae and juvenile polyps was observed by fluorescence microscope (Olympus SZX16, Olympus, Tokyo, Japan). In this experiment, three types of corals at different stages of growth were used: planula larvae, early-stage morphogenetic polyps 2 days after metamorphosis induction, and polyps 1 month after metamorphosis. At day 3 after dsRNA treatment, corals were tested to confirm the effect of each treatment on GFP expression. Epifluorescence photomicrographs of Juvenile polyps were taken under Olympus SZX16 (Olympus, Tokyo, Japan) using a digital camera (Tucsen Photonics, Fuzhou, China). Images were analyzed using imageJ software (Wayne Rasband, National Institutes of Health, Bethesda, MD, United States) to estimate fluorescence intensity in each polyp (n = 3). The amount of GFP in the polyp was estimated using the RawIntDen function of imageJ. Photographs of polyps before and after RNAi treatment were stacked and compared their fluorescent intensity and the rate of change in the amount of GFP (after-RNAi/pre-RNAi) was calculated. One image per polyp was used for imageJ analyses. To investigate the effect of TRX-targeting RNAi, planula larvae were incubated at high temperature (31.5°C); these experiments were performed in triplicate containing 90 planula larvae. 3 days after dsRNA (0.5 μg/mL) treatment in a 24-well plate as described above, thirty treated larvae were picked up and added to 50 mL of filtered seawater in a Falcon centrifuge tubes (Fisher scientific) and incubated at 31.5°C for 1 day. The survival rate after 1 day was confirmed. A coral exposed to DMRIE-C alone and corals treated with ACT-dsRNA for 3 days was used as a control. We prepared three 50 mL experimental sets for each treatment group.
Statistical Analyses
Statistical analyses were performed using Python (Python 3.8.5 version, Anaconda 3, Continuum Analytics, Inc., United States). The survival rates were analyzed for treatment using a Turkey-Kramer HSD (honestly significant difference) test. To confirm whether the response was specific to GFP or TRX dsRNA, the same assay was performed with corals treated with ACT dsRNA. Student’s t-test was used for comparing the relative gene expression levels and fluorescence levels after RNAi treatment.
Results and Discussion
As the initial knockdown trial in corals, TRX and GFP were used for RNAi targets. From A. tenuis planula larvae, cDNA clones that comprised a full-length open reading frame of GFP and TRX, respectively, were isolated, and dsRNA were synthesized from these clones. We introduced these TRX and GFP dsRNA into planula larvae at two concentrations (0.5 and 1 μg/mL) by lipofection (Dunn et al., 2007). Larvae incubated in 1 μg/mL each dsRNA, melted during the 3 days incubation period. Larvae treated with 0.5 μg/mL dsRNA showed no phenotypic changes, and all 50 planula larvae in the well of a multi-well plate survived. Based on these results, 0.5 μg/mL dsRNA was used in subsequent experiments.
The effect of RNAi on the expression levels of each gene was examined by real-time PCR (Figure 1). Gene expression levels of GFP and TRX tended to decrease in the GFP-ds RNA treated and TRX-ds RNA treated groups, respectively, compared to the control group (ACT dsRNA treated larvae) on day 3. GFP-RNAi had lower knockdown efficiency than TRX-RNAi. This may be due to the presence of complex gene regulatory networks that control GFP expression, such as the involvement of multiple isoforms in GFP expression. Since the expression levels of each gene varied greatly and the number of replicates was small (n = 3), there was no significant decrease in gene expression compared to the control treatment group.
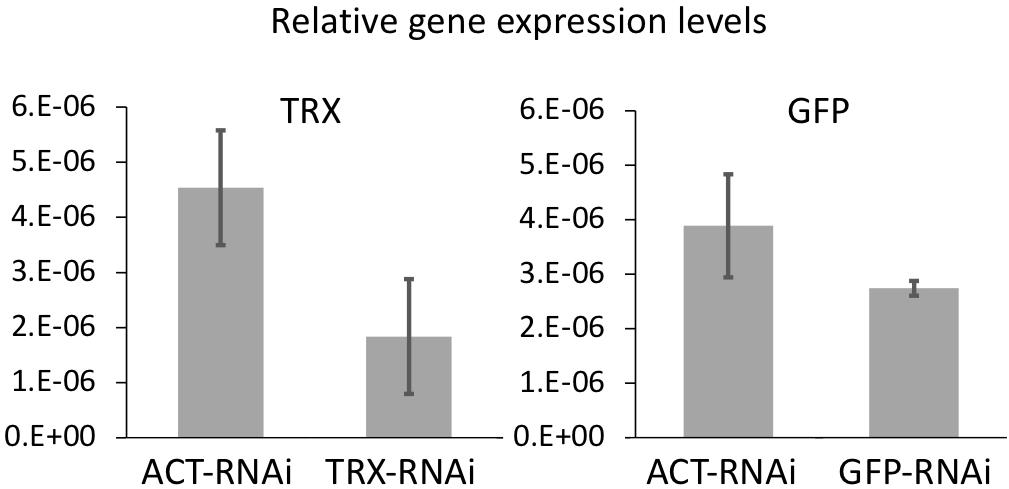
Figure 1. The mean value of GFP and TRX expression levels in control and each dsRNA-treated corals measured by quantitative real-time PCR. Coral treated with actin (ACT) ds-RNA was used as a control to confirm gene-specific RNAi. Corals incubated for 3 days after RNAi treatment were used. Expression levels were normalized to the expression levels of two reference genes (elongation factor and β-tubulin). Experiments were conducted in triplicate with 50 planula larvae each. Error bars are standard error.
Coral TRX expression was previously reported to respond to high temperature stress (Edge et al., 2005; Maor-Landaw and Levy, 2016); therefore, we confirmed the TRX knockdown using high-temperature incubation experiments. Coral planula larvae are vulnerable to environmental changes; the effects of stress exposure on planula larvae can be assessed by survival rates. To assess the TRX dsRNA-specific response to temperature stress, control larvae treated with ACT dsRNA were also assessed. As shown in Figure 2, the survival rate was 8.9% in TRX dsRNA-treated planula larvae, while the survival rates of ACT dsRNA-treated population and the control population (treated with DMRIE-C reagent) were 25.6 and 32.2%, respectively. The survival rate of TRX-treated larvae under non-stress conditions was 100%. TRX-knockdown corals exhibited a significantly reduced survival rate (p < 0.05, HSD test) compared with the other treatment groups under high temperature conditions. It is suggested that TRX knockdown reduces the ability of coral to relieve oxidative stress, making the coral vulnerable to temperature stress. However, the survival rate of the control group was low, suggesting that acute stress responses other than TRX knockdown are also significantly involved in the death of planula larvae treated with TRX-dsRNA. TRX has two thiol residues, and their redox reaction might be involved in defense against oxidative stress either directly removing active oxygen or regulating the activity of target enzymes as signal transducer of intracellular oxidative stress (Cunningham et al., 2015). Use of the redox state of TRX as indicator of intracellular oxidative stress has been suggested (Hidaka, 2016). Considering our results, further analyses of TRX functions via gene knockdown experiment are necessary to reveal the role of TRX in coral stress response.
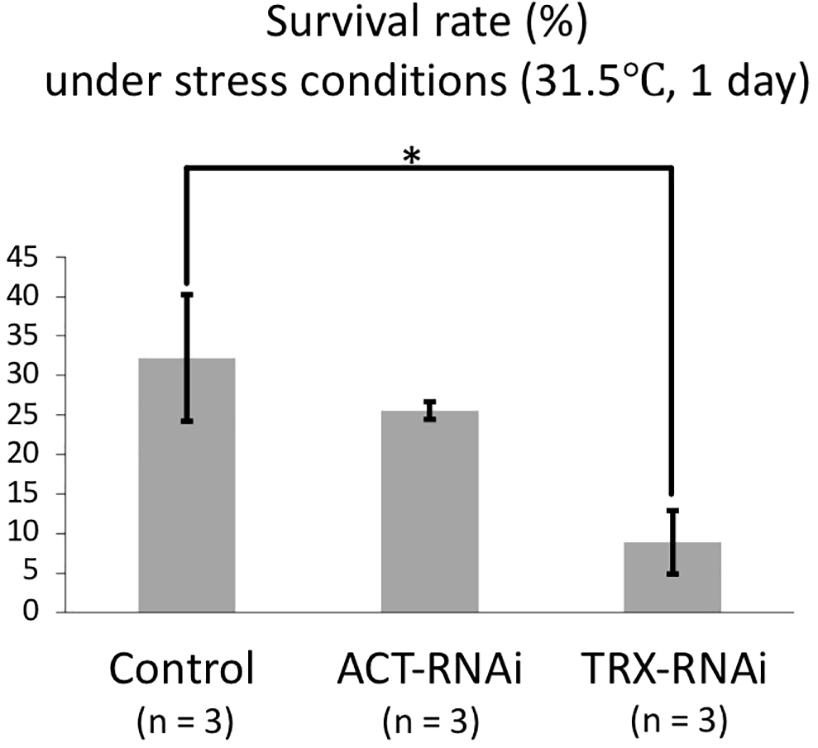
Figure 2. Survival rate of dsRNA-treated and control planula larvae at high temperature (31.5°C, 1 day). Three replicaties were prepared for each treatment (control, ACT-RNAi, and TRX-RNAi). Each replicate contains 30 larvae, and survival rates were calculated based on the numbers of individuals at the beginning and end of the experiment. The average and standard error of three replicates are shown. Error bars are standard error. Asterisk indicates a significant difference (p < 0.05).
To determine whether GFP dsRNA suppress GFP expression, coral polyps were observed under fluorescence microscope. The coral at different growth stages were used to observe the effect of RNAi treatment: juvenile polyps immediately after metamorphosis (day 3 after Hym treatment), polyps 1 month after metamorphosis, and planula larvae. A decrease in GFP fluorescence was observed only in polyps immediately after metamorphosis (Figure 3a). The quantification of the GFP fluorescence in the photo image showed that the GFP fluorescence decreased significantly after application of GFP-targeting RNAi (Figure 3b, P < 0.05, t-test). In planula larvae, GFP fluorescence was strong, and only one of four individuals showed a large difference before and after treatment (data not shown). Haryanti and Hidaka (2019) showed that acroporid planula larvae exhibited strong green fluorescence, and the fluorescence was decreased after metamorphosis. The inherently high expression level of GFP in planula larvae might be a reason for the lower RNAi effect on GFP intensity. It was possible that the larvae had a fast GFP turnover and that resulted in a weak knockdown efficiency. Polyps at 1 month after metamorphosis appeared to be minimally affected by RNAi targeting of GFP. At 1 month after metamorphosis, corals are covered with mesh-like calcareous skeleton; these corals might be less receptive to the application of dsRNA. Thus, polyps immediately after metamorphosis were optimal for observing the effects of GFP dsRNA. As mentioned above, there are multiple isoforms of GFP in A tenuis (Satoh et al., 2021). The GFP-dsRNA and its four isoforms have 253–579 bp homologous regions, and it is possible that multiple GFP isoforms were knocked down during RNAi processing, resulting in increased efficacy. Our results suggest that RNAi may not be successful in corals in which the body wall is covered with mesh-like calcareous skeleton, even when targeting multiple isoforms.
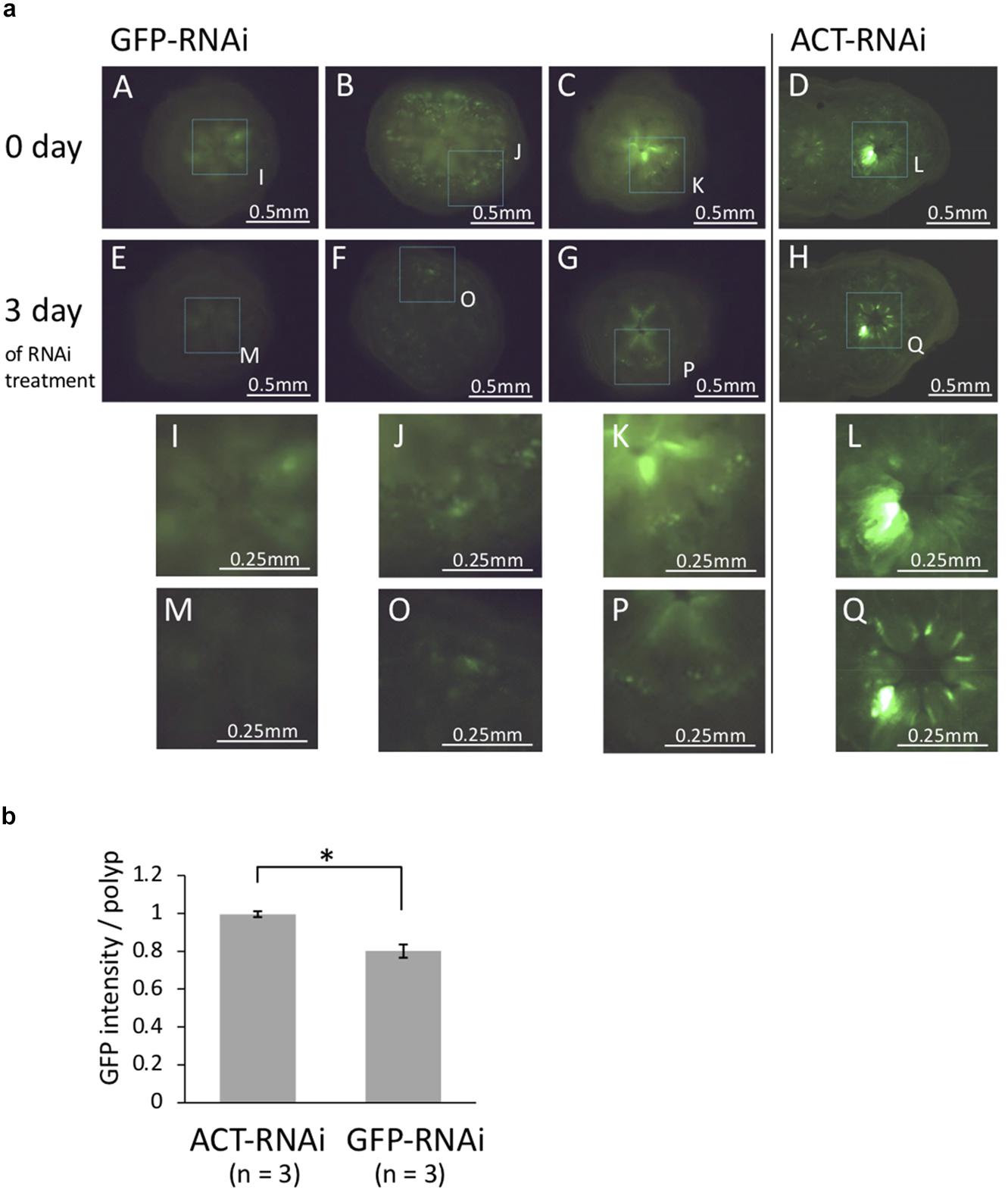
Figure 3. (a) Fluorescence microscopy images of juvenile polyps (day 6 after metamorphosis) treated with GFP-dsRNA and ACT-dsRNA. Corals treated with ACT-dsRNA were prepared as a control group to confirm the response was specific to GFP-dsRNA. GFP-dsRNA-treated corals showed a tendency toward decreased green auto-fluorescence. (A–E, B–F, C–G, and D–H) Show the same colonies before-after treatment with each dsRNA. (I–Q) show higher magnification of dashed box region in A–H. The same part in the photos before-after dsRNA treatment is surrounded by dotted lines. The same area of the two figures (before-after treatment) is surrounded by dotted lines and enlarged. (b) The rate of change in GFP content (after-RNAi/pre-RNAi). The GFP contents were estimated from juvenile polyps (n = 3) treated with GFP-dsRNA or ACT-dsRNA using the RawIntDen function of imageJ. The green fluorescence intensity was decreased in corals treated with GFP-dsRNA. Bars represent the mean GFP fluorescence intensity of corals treated with ds-GFP or ds-ACT. Error bars are standard error. Asterisk indicate significant differences (p < 0.05).
In summary, RNAi using juvenile corals resulted in a reduction in target gene expression although there was statistically no significant difference from the control group. Moreover, RNAi targeting TRX and GFP confirmed the involvement of each gene in coral stress tolerance in the planula stage or GFP intensity 3 days after settlement, significantly. However, the effect of RNAi on GFP was confirmed only in the corals at the initial stages after metamorphosis, and no effect on GFP color development was observed in planulae and 1 month-old budding juveniles. There are still improvements to be made in this method for corals. We used dsRNA based on previous studies, but the use of other interfering RNAs, such as morpholino antisense oligonucleotides and small interfering RNAs (siRNAs), could also be useful. The use of siRNAs and morpholino antisense oligonucleotides is likely to produce a more pronounced effect than RNAi (Layden et al., 2013; Guo et al., 2018). In future studies, it might be desirable to make sure that dsRNA is actually incorporated by all or certain cell types using fluorescent dye-conjugated dsRNA, and to use suitable positive and negative control dsRNAs in addition to the controls used in the present study. In addition, the results of this study suggest the effects of RNAi are reduced in skeletonized corals, and planula larvae and polyps are appropriate for genetic manipulation. However, as planula larvae can only be obtained once or twice a year, the number of RNAi experiments that can be performed is limited. Furthermore, the genetic diversity of planula larvae may have affected RNAi efficiency. In the future, coral cell clumps could be useful for advancement the RNAi method (Nesa and Hidaka, 2008). Alternatively, the use of sea anemones and hydra as models to elucidate common symbiotic relationships may be informative. Here, we showed that RNAi by lipofection can be applied to juvenile corals, and the effect on gene expression, coral stress tolerance and fluorescence intensity were tested. It is expected that such RNAi experiments will be utilized for functional identification of coral-specific genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author Contributions
IY: study conception and design, data collection, and draft manuscript preparation. TH: statistic analysis, drafting and writing. MH: drafting and writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a research grant by the Japan Society for the Promotion of Science (IY, #14J40135 and #19H03026), the Environment Research and Technology Development Fund (No. 4-1806) from the Ministry of the Environment in Japan, and the Kurita Water and Environment Foundation (18B083).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the members of Laboratory for DNA data analysis in NIG (National Institute of Genetics), especially Hisako Tashiro and Chie Iwamoto for supporting our incubation experiments, and also thank the member of Laboratory of Molecular Evolution of Microbes in University of Tsukuba.
Footnotes
References
Amimoto, Y., Kodama, R., and Kobayakawa, Y. (2006). Foot formation in Hydra: a novel gene, anklet, is involved in basal disk formation. Mech. Dev. 123, 352–361. doi: 10.1016/j.mod.2006.03.002
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Arnér, E. S., and Holmgren, A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x
Buckingham, S. D., Esmaeili, B., Wood, M., and Sattelle, D. B. (2004). RNA interference: from model organisms towards therapy for neural and neuromuscular disorders. Hum. Mol. Genet. 13, 275–288. doi: 10.1093/hmg/ddh224
Chera, S., Buzgariu, W., Ghila, L., and Galliot, B. (2009). Autophagy in Hydra: a response to starvation and stress in early animal evolution. Biochim. Biophys. Acta 1793, 1432–1443. doi: 10.1016/j.bbamcr.2009.03.010
Cleves, P. A., Strader, M. E., Bay, L. K., Pringle, J. R., and Matz, M. V. (2018). CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc. Natl. Acad. Sci. 115, 5235–5240. doi: 10.1073/pnas.1722151115
Cunningham, G. M., Roman, M. G., Flores, L. C., Hubbard, G. B., Salmon, A. B., Zhang, Y., et al. (2015). The paradoxical role of thioredoxin on oxidative stress and aging. Arch. Biochem. Biophys. 576, 32–38. doi: 10.1016/j.abb.2015.02.025
Dunn, S. R., Phillips, W. S., Green, D. R., and Weis, V. M. (2007). Knockdown of actin and caspase gene expression by RNA interference in the symbiotic anemone Aiptasia pallida. Biol Bull. 212, 250–258. doi: 10.2307/25066607
Edge, S. E., Morgan, M. B., Gleason, D. F., and Snell, T. W. (2005). Development of a coral cDNA array to examine gene expression profiles in Montastraea faveolata exposed to environmental stress. Mar. Pollut. Bull. 51, 507–523. doi: 10.1016/j.marpolbul.2005.07.007
Fjose, A., Ellingsen, S., Wargelius, A., and Seo, H. C. (2001). RNA interference: mechanisms and applications. Biotechnol. Annu. Rev. 7, 31–57. doi: 10.1016/s1387-2656(01)07032-6
Gabay, Y., Weis, V. M., and Davy, S. K. (2018). Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model dnidarian-dinoflagellate symbiosis. Biol. Bull. 234, 1–10. doi: 10.1086/696365
Galliot, B., Miljkovic-Licina, M., Ghila, L., and Chera, S. (2007). RNAi gene silencing affects cell and developmental plasticity in hydra. C. R. Biol. 330, 491–497. doi: 10.1016/j.crvi.2007.01.008
Guo, X., Wang, Y., Sinakevitch, I., Lei, H., and Smith, B. H. (2018). Comparison of RNAi knockdown effect of tyramine receptor 1 induced by dsRNA and siRNA in brains of the honey bee, Apis mellifera. J. Insect Physiol. 111, 47–52. doi: 10.1016/j.jinsphys.2018.10.005
Haryanti, D., and Hidaka, M. (2019). Developmental changes in the intensity and distribution pattern of green fluorescence in coral larvae and juveniles. Galaxea J. Coral Reef Stud. 21, 13–25. doi: 10.3755/galaxea.21.1_13
Hidaka, M. (2016). “Life History and Stress Response of Scleractinian Corals” in Coral Reef Science: Strategy for Ecosystem Symbiosis and Coexistence With Humans Under Multiple Stresses Coral Reefs of the World, ed. H. Kayanne (Tokyo: Springer). 1–24. doi: 10.1007/978-4-431-54364-0_1
Higuchi, T., Agostini, S., Casareto, B. E., Suzuki, Y., and Yuyama, I. (2015). Northern limit of corals of the genus Acropora in temperate zones is determined by their resilience to cold bleaching. Sci. Rep. 5:18467. doi: 10.1038/srep18467
Hillyer, K. E., Tumanov, S., Villas-Bôas, S., and Davy, S. K. (2016). Metabolite profiling of symbiont and host during thermal stress and bleaching in a model cnidarian–dinoflagellate symbiosis. J. Exp. Biol. 219, 516–527. doi: 10.1242/jeb.128660
Hou, J., Xu, T., Su, D., Wu, Y., Cheng, L., Wang, J., et al. (2018). RNA-Seq reveals extensive transcriptional response to heat stress in the stony coral Galaxea fascicularis. Front. Genet. 9:37. doi: 10.3389/fgene.2018.00037
Iwao, K., Fujisawa, T., and Hatta, M. (2002). A cnidarian neuropeptide of the GLWamide family induces metamorphosis of reef-building corals in the genus Acropora. Coral Reefs 21, 127–129. doi: 10.1007/s00338-002-0219-8
Jakob, W., and Schierwater, B. (2007). Changing hydrozoan bauplans by silencing Hox-like genes. PLoS One 2:e694. doi: 10.1371/journal.pone.0000694
Jones, V. A. S., Bucher, M., Hambleton, E. A., and Guse, A. (2018). Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci. Rep. 8:16437. doi: 10.1038/s41598-018-34773-1
Kii, S.-I., Tanaka, J., and Watanabe, T. (2007). Guanine-cytosine contents of the host and symbiont cDNA in a symbiotic coral. Fish. Sci. 73, 1362–1372. doi: 10.1111/j.1444-2906.2007.01479.x
Layden, M. J., Röttinger, E., Wolenski, F. S., Gilmore, T. D., and Martindale, M. Q. (2013). Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone. Nematostella vectensis. Nat. Protoc. 8, 924–934. doi: 10.1038/nprot.2013.009
Li, Y., Liew, Y. J., Cui, G., Cziesielski, M. J., Zahran, N., Michell, C. T., et al. (2018). DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci. Adv. 4:eaat2142. doi: 10.1126/sciadv.aat2142
Lohmann, J. U., Endl, I., and Bosch, T. C. G. (1999). Silencing of developmental genes in Hydra. Dev. Biol. 214, 211–214. doi: 10.1006/dbio.1999.9407
Maor-Landaw, K., and Levy, O. (2016). Gene expression profiles during short-term heat stress; branching vs. massive Scleractinian corals of the Red Sea. PeerJ. 4:e1814. doi: 10.7717/peerj.1814
Nesa, B., and Hidaka, M. (2008). “Thermal stress increases oxidative DNA damage in coral cell aggregates,” in Proceedings of the 11th International Coral Reef Symposium, Vol. 5, Fort Lauderdale, FL. 144–148.
Perrimon, N., and Mathey-Prevot, B. (2007). Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175, 7–16. doi: 10.1534/genetics.106.069963
Quiroga-Artigas, G., Duscher, A., Lundquist, K., Waletich, J., and Schnitzler, C. E. (2020). Gene knockdown via electroporation of short hairpin RNAs in embryos of the marine hydroid Hydractinia symbiolongicarpus. Sci. Rep. 30:12806. doi: 10.1038/s41598-020-69489-8
Satoh, N., Kinjo, K., Shintaku, K., Kezuka, D., Ishimori, H., Yokokura, A., et al. (2021). Color morphs of the coral, Acropora tenuis, show different responses to environmental stress and different expression profiles of fluorescent-protein genes. G3 11:jkab018. doi: 10.1093/g3journal/jkab018
Shinzato, C., Khalturin, K., Inoue, J., Zayasu, Y., Kanda, M., Kawamitsu, M., et al. (2021). Eighteen coral genomes reveal the evolutionary frigin of Acropora strategies to accommodate environmental changes. Mol. Biol. Evol. 38, 16–30. doi: 10.1093/molbev/msaa216
Strader, M. E., Davies, S. W., and Matz, M. V. (2015). Differential responses of coral larvae to the colour of ambient light guide them to suitable settlement microhabitat. R. Soc. Open Sci. 2:150358. doi: 10.1098/rsos.150358
Thomas, L., and Palumbi, S. R. (2017). The genomics of recovery from coral bleaching. Proc. R. Soc. B Biol. Sci. 284:20171790. doi: 10.1098/rspb.2017.1790
Yasuoka, Y., Shinzato, C., and Satoh, N. (2016). The mesoderm-rorming gene brachyury regulates ectoderm-endoderm demarcation in the coral Acropora digitifera. Curr. Biol. 26, 2885–2892. doi: 10.1016/j.cub.2016.08.011
Ying, H., Hayward, D. C., Cooke, I., Wang, W., Moya, A., Siemering, K. R., et al. (2019). The whole-genome sequence of the coral Acropora millepora. Genom. Biol. Evol. 11, 1374–1379. doi: 10.1093/gbe/evz077
Yoshida, T., Oka, S., Masutani, H., Nakamura, H., and Yodoi, J. (2003). The role of thioredoxin in the aging process: involvement of oxidative stress. Antioxid. Redox Signal. 5, 563–570. doi: 10.1089/152308603770310211
Yuyama, I., Harii, S., and Hidaka, M. (2012a). Algal symbiont type affects gene expression in juveniles of the coral Acropora tenuis exposed to thermal stress. Mar. Environ. Res. 76, 41–47. doi: 10.1016/j.marenvres.2011.09.004
Yuyama, I., Ito, Y., Watanabe, T., Hidaka, M., Suzuki, Y., and Nishida, M. (2012b). Differential gene expression in juvenile polyps of the coral Acropora tenuis exposed to thermal and chemical stresses. J. Exp. Mar. Biol. Ecol. 43, 17–24. doi: 10.1016/j.jembe.2012.06.020
Yuyama, I., Hayakawa, H., Endo, H., Iwao, K., Takeyama, H., Maruyama, T., et al. (2005). Identification of symbiotically expressed coral mRNAs using a model infection system. Biochem. Biophys. Res. Commun. 336, 793–798. doi: 10.1016/j.bbrc.2005.08.174
Yuyama, I., and Higuchi, T. (2014). Comparing the effects of symbiotic algae (Symbiodinium) clades C1 and D on early growth stages of Acropora tenuis. PLoS One 9:e98999. doi: 10.1371/journal.pone.0098999
Yuyama, I., Ishikawa, M., Nozawa, M., Yoshida, M., and Ikeo, K. (2018). Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci. Rep. 8:16802. doi: 10.1038/s41598-018-34575-5
Yuyama, I., Nakamura, T., Higuchi, T., and Hidaka, M. (2016). Different stress tolerances of juveniles of the coral Acropora tenuis associated with clades C1 and D Symbiodinium. Zool. Stud. 55:9.
Keywords: screractinian coral, RNAi, stress response, thioredoxin, GFP
Citation: Yuyama I, Higuchi T and Hidaka M (2021) Application of RNA Interference Technology to Acroporid Juvenile Corals. Front. Mar. Sci. 8:688876. doi: 10.3389/fmars.2021.688876
Received: 31 March 2021; Accepted: 21 June 2021;
Published: 09 August 2021.
Edited by:
James Davis Reimer, University of the Ryukyus, JapanReviewed by:
Isabelle Domart-Coulon, Muséum National d’Histoire Naturelle, FranceKátia Cristina Cruz Capel, University of São Paulo, Brazil
Copyright © 2021 Yuyama, Higuchi and Hidaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ikuko Yuyama, eXV5YW1haUBnbWFpbC5jb20=
 Ikuko Yuyama
Ikuko Yuyama Tomihiko Higuchi
Tomihiko Higuchi Michio Hidaka
Michio Hidaka
