- 1Department of Natural Sciences, Baruch College, City University of New York, New York, NY, United States
- 2The Graduate Center, Ph.D. Program in Biology, City University of New York, New York, NY, United States
- 3Division of Vertebrate Zoology, Department of Ichthyology, American Museum of Natural History, New York, NY, United States
- 4American Museum of Natural History, Sackler Institute for Comparative Genomics, New York, NY, United States
Since the initial discovery of Aqueoria victoria’s green fluorescence off the coast of Washington’s Puget Sound, biofluorescent marine organisms have been found across the globe. The variety of colors of biofluorescence as well as the variability in the organisms that exhibit this fluorescence is astounding. The mechanisms of biofluorescence in marine organisms are also variable. To fluoresce, some organisms use fluorescent proteins, while others use small molecules. In eels, green biofluorescence was first identified in Anguilla japonica. The green fluorescence in A. japonica was discovered to be caused by a fatty acid binding protein (UnaG) whose fluorescence is induced by the addition of bilirubin. Members of this class of proteins were later discovered in Kaupichthys eels (Chlopsid FP I and Chlopsid FP II). Here, we report the discovery and characterization of the first member of this class of green fluorescent fatty acid binding proteins from the moray eel Gymnothorax zonipectis. This protein, GymFP, is 15.6 kDa with a fluorescence excitation at 496 nm and an emission maximum at 532 nm upon addition of bilirubin. GymFP is 61% homologous to UnaG and 47% homologous to Chlopsid FP I. Here, we report de novo transcriptome assembly, protein expression, and fluorescence spectroscopic characterization of GymFP. These findings extend the fluorescent fatty acid binding proteins into a third family of true eels (Anguilliformes).
Introduction
Biofluorescence is a phenomenon that is widespread in the marine environment, being found in eels (Hayashi and Toda, 2009; Kumagai et al., 2013; Gruber et al., 2015; Krivoshik et al., 2020), sharks, and many other fish species (Sparks et al., 2014). Marine biofluorescence was first discovered in the 1960s by Osamu Shimomura in the jellyfish Aqueoria victoria (Shimomura et al., 1962). The structure of green fluorescent protein (GFP) is a beta barrel with the fluorophore forming from a spontaneous cyclization reaction of three amino acids: Serine-Tyrosine-Glycine (Barondeau et al., 2005). Derivatives of GFP were later identified in corals (Matz et al., 1999; Salih et al., 2000; Field et al., 2006; Leutenegger et al., 2007).
In 2009, a green fluorescent protein (UnaG) was discovered in an eel, Anguilla japonica (Hayashi and Toda, 2009; Kumagai et al., 2013). UnaG was found to be a member of the fatty acid binding protein family (Kumagai et al., 2013). Structural characterization of UnaG revealed a beta barrel structure (Kumagai et al., 2013). UnaG did not fluoresce spontaneously as GFP, instead it required the addition of bilirubin, a breakdown product of heme metabolism (Kumagai et al., 2013). Other Anguilla species were later discovered to have bilirubin inducible fluorescent fatty acid binding proteins in their transcriptome sequences (Funahashi et al., 2017). Our work led to the discovery of members of this class of proteins in two chlopsid eels: Kaupichthys hyoproroides and Kaupichthys n. sp. These proteins are called Chlopsid FP I and Chlopsid FP II, respectively (Gruber et al., 2015).
All fluorescent fatty acid binding proteins discovered to date have a Glycine-Proline-Proline (GPP) motif that resides on a loop that may act to protect bilirubin from solvent. This may be a reason for fluorescence in the eel fatty acid binding proteins (Gruber et al., 2015; Krivoshik et al., 2020). Work with Chlopsid FP I showed that mutation of the GPP motif to GGG significantly reduces quantum yield (Gruber et al., 2015), while deletion fully quenches fluorescence (Krivoshik et al., 2020).
Here, we report a new fluorescent fatty acid binding protein (GymFP) isolated from the widespread Indo-Pacific moray eel, Gymnothorax zonipectis. This discovery represents the third family of Anguilliformes (true eels), Muraenidae, from which FPs have been isolated and characterized. This is the first report of a fluorescent fatty acid binding protein from Muraenidae. Herein we report de novo transcriptome assembly of G. zonipectis, fluorescence characterization, and recombinant expression of GymFP.
Materials and Methods
Collection and Identification of Gymnothorax zonipectis
Specimens of G. zonipectis were collected during daylight SCUBA dives via the application of rotenone to a targeted shallow reef habitat (8–12 m) in Peava Lagoon, Western Province of the Solomon Islands (−8.784222 degrees S, 158.231345 degrees E). Immediately after collection, the G. zonipectis specimen (AMNH 277097) was placed in a narrow photographic tank and held flat against a thin plate glass (Figures 1A,B). Fluorescent macro images were produced in a dark room by covering the flash with interference bandpass excitation filters (Omega Optical, Inc., Brattleboro, VT; Semrock, Inc., Rochester, NY) to elicit fluorescence. Longpass (LP) and bandpass (BP) emission filters (Semrock) were attached to the front of the camera lens to block the excitation light and record emitted fluorescence. The eel was stored in a liquid nitrogen dry shipper and transported back to the American Museum of Natural History, New York, where it was immediately stored at −80°C. The specimen was then delivered to Baruch College.
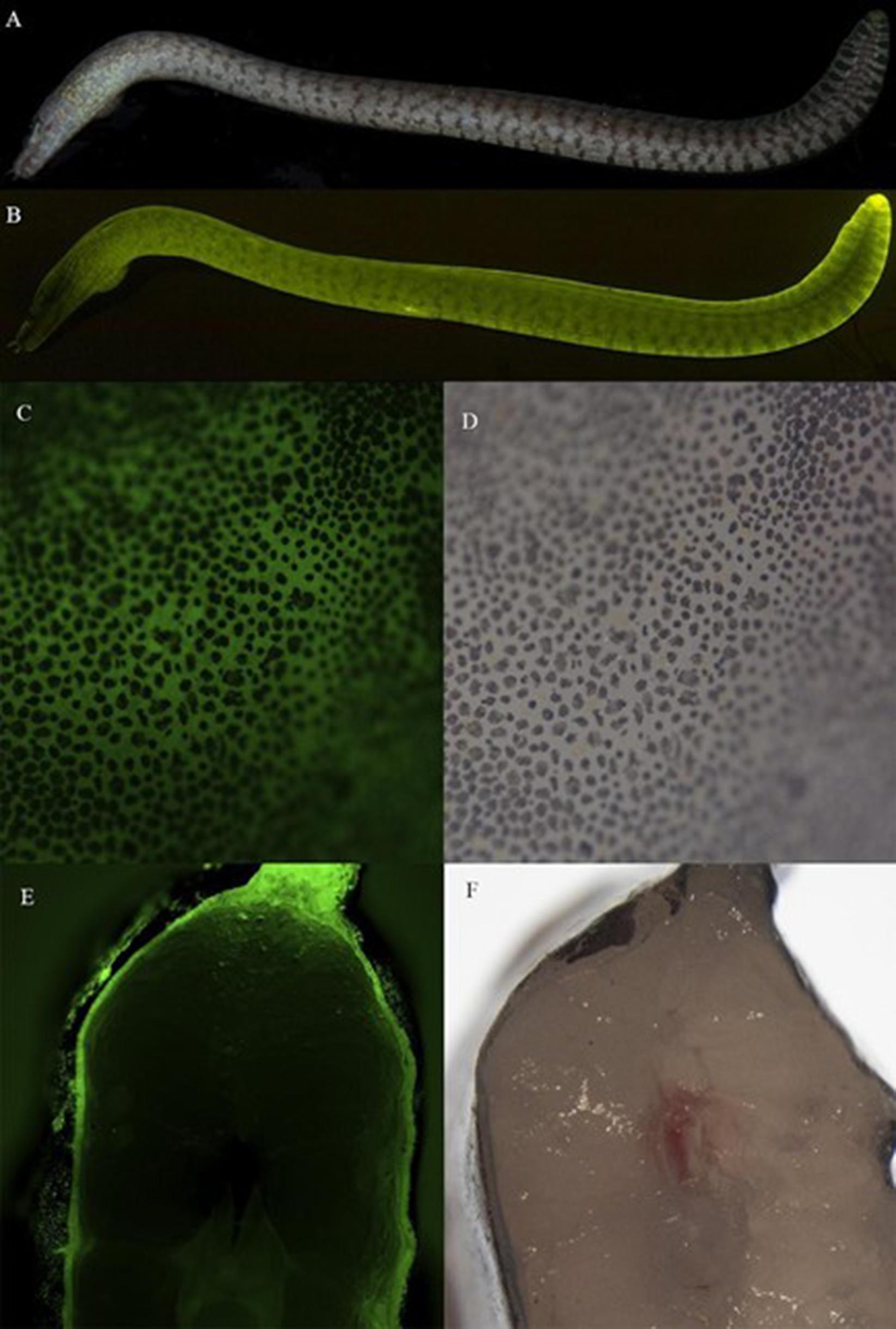
Figure 1. Details of green fluorescence of Gymnothorax zonipectis (AMNH 277097, 151 mm SL). (A) White light and (B) fluorescent emission photo of Gymnothorax zonipectis. (C,D) Close up of epidermis revealing dark pigmented regions (chromatophores) interspersed with green fluorescence arising from below the skin, and (E,F) cross-section through the mid body demonstrating fluorescence below the skin. Muraenids (morays), like most anguilliform eels, lack scales.
Research and collection permits were obtained from the Ministry of Fisheries and Marine Resources (MFMR), and the Ministry of Environment, Climate Change, Disaster Management and Meteorology (MECDM), Honiara, Solomon Islands. Fishes were collected and handled in accordance with AMNH Institutional Animal Care and Use Committee (IACUC) and American Fisheries Society (AFS) guidelines, as established for the safe and humane care and handling of vertebrate animals. Fieldwork was carried out in collaboration with and permitted by the Solomon Islands MFMR and MECDM, and facilitated in the Solomon Islands by the Wildlife Conservation Society (WCS, New York; Munda, Western Province, Solomon Islands).
Gymnothorax zonipectis was identified using the following criteria: Overall body coloration brown. The posterior 2–3 laterosensory pores on the upper and lower jaws are enclosed in vertically oriented white bars that are continuous across the lower jaw. Distinct irregular dark brown marking with pale borders posterior to orbit. Body with oblique pattern of darkly pigmented broken/discontinuous vertical bars (in four longitudinal series) that become more pronounced caudally. Bars become more darkly pigmented/larger with bright white borders on posterior fins (Supplementary Figure 1).
Determination of Fluorescent Properties From Tissue Samples
G. zonipectis was dissected using a Lightools Research LT-9900 Illumatool Tunable Lighting System (Lightools Research) to ensure that the samples taken contained fluorescent tissue. Cross-sectional images of specimens were generated using a Zeiss-Axio Zoom V16 stereo fluorescent microscope affixed with a Nikon D4 camera. These samples were homogenized in 1X PBS using a BeadBug homogenizer (Benchmark Scientific) and centrifuged at 15,000 rcf for 10 min.
RNA Extraction and de novo Transcriptome Assembly
RNA extraction of the fluorescent tissue from G. zonipectis was performed using the RNeasy Fibrous Tissue Mini Kit from Qiagen. Fluorescent tissue in G. zonipectis was found below the skin of the eel. The location of the fluorescence is shown in a photo of the cross section of the eel in Figure 1E. A 30 mg piece of fluorescent tissue was cut using a scalpel for RNA extraction. The extracted RNA sample (28 μL of 75 ng/μL) was then sent to GENEWIZ, LLC (South Plainfield, NJ) for de novo transcriptome assembly and bioinformatic analysis including a BLAST search was used to search for protein candidates.
Library Preparation With PolyA Selection and HiSeq Sequencing
RNA Library preparation was performed by Genewiz, LLC. RNA was quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). RNA integrity measurement was completed using TapeStation (Agilent Technologies, Palo Alto, CA, United States).
NEBNext Ultra RNA Library Prep Kit for Illumina was used in library preparation using manufacturer’s instructions (NEB, Ipswich, MA, United States). mRNAs were enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 15 min at 94°C. First and second strand cDNA were synthesized. cDNA fragments were end repaired and adenylated at 3′ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by PCR with limited cycles.
The Agilent TapeStation was used for library validation (Agilent Technologies, Palo Alto, CA, United States), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, United States).
After clustering the library on flowcell, the flowcell was loaded on the Illumina HiSeq instrument (4,000 or equivalent) according to manufacturer’s instructions. The sample was sequenced using a 2 × 150 bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
Data Analysis
Data analysis was performed by GENEWIZ, United States. Sequence reads were trimmed to remove possible adapter and nucleotides with poor quality (error rate < 0.01) using Trimmomatic v.0.36 (Bolger et al., 2014). Then Trinity v2.5, de novo assembler (Grabherr et al., 2011), was used on combined samples per species. One de novo assembled transcriptome was created with a minimum contig length of 200 bp. Transrate v1.0.3 (Smith-Unna et al., 2016) was used to generate statistics for the de novo assembled transcriptome. EMBOSS tool getorf (Rice et al., 2000) was then used to find the open reading frames within the de novo assembled transcriptome. The de novo transcriptome assembly was then annotated using Diamond BLASTx against the NCBI NR database.
Phylogetic Tree Generation
The phylogenetic tree is presented in Supplementary Figure 2A. The alignments are presented in Supplementary Figure 2B. Alignments were done using MAFFT v7.487 with the L-INS-i Iterative refinement method and default parameters (Figure 2A and Supplementary Figure 2B; Hofacker et al., 2002; Katoh et al., 2002, 2005; Katoh and Toh, 2008; Tabei et al., 2008; Katoh and Frith, 2012; Kuraku et al., 2013; Katoh and Standley, 2016; Yamada et al., 2016; Rozewicki et al., 2019). The best model of substitution was selected using ModelFinder (Kalyaanamoorthy et al., 2017). For generation of the phylogenetic tree (Supplementary Figure 2A), the following input data statistics were generated: 14 sequences with 146 amino-acid sites. Number of constant sites: 24 (= 16.4384% of all sites). Number of invariant (constant or ambiguous constant) sites: 24 (= 16.4384% of all sites). Number of parsimony informative sites: 91. Number of distinct site patterns: 143. The tree was constructed from 1,000 ultrafast bootstrap trees (Hoang et al., 2018).

Figure 2. (A) Alignment of GymFP, Chlopsid FP I, and UnaG generated by MAFFT (Hofacker et al., 2002; Katoh et al., 2002, 2005; Katoh and Toh, 2008; Tabei et al., 2008; Katoh and Frith, 2012; Kuraku et al., 2013; Katoh and Standley, 2016; Yamada et al., 2016; Rozewicki et al., 2019). (B) Predicted structure of GymFP generated by SWISS Model (Guex et al., 2009; Bienert et al., 2017; Bertoni et al., 2017; Waterhouse et al., 2018; Studer et al., 2020). In both, the GPP insert is shown in green and residues identical in all three sequences are shown in purple.
Bacterial Expression of GymFP
Plasmids were ordered from Genscript United States in a pET−24b(+) vector with kanamycin resistance for expression in BL21(DE3) E. coli with an N-terminal 6x histidine tag. Following a chemical transformation, BL21(DE3) were plated on agar containing a 1:1,000 dilution of kanamycin (50 mg/mL). Single colonies were selected and grown overnight in 5 mL LB media containing 5 μL kanamycin at 37°C in a shaking incubator. The resulting culture was transferred to 100 mL LB with 100 μL kanamycin and left to grow at 37°C until reaching an OD600 of 0.4. A 1:1,000 dilution of IPTG (0.1 M) was then added, and the culture was left to grow for another 3 h after which the culture was then centrifuged at 3,000 rcf for 30 min.
The pellets were then combined and resuspended into 5 mL of Tris-HCl (50 mM), NaCl (150 mM). Lysozyme (100 μL of 10 mg/mL) was added, and the sample was left at room temperature for 1 h. The sample was then centrifuged at 8,000 rcf for 30 min. The supernatant was purified using Nickel affinity chromatography. The elution buffer contained Tris-HCl (50 mM), NaCl (150 mM), and Imidazole (300 mM), pH 7.4. Protein A280 was measured using a Cary60 UV-Vis spectrophotometer. The extinction coefficient was calculated using the ExPasy ProtParam tool (Artimo et al., 2012) to be 21,430 M–1 cm–1. Bilirubin was dissolved in NaOH and diluted to 10 μM in 1X PBS (1.19 mM phosphates, 13.7 mM sodium chloride, 270 μM potassium chloride), pH 7.3, for further use in fluorescence assays.
Generation of Swiss Model
Swiss Model was used to construct a model of GymFP (Guex et al., 2009; Bienert et al., 2017; Bertoni et al., 2017; Waterhouse et al., 2018; Studer et al., 2020). The tool chose WT UnaG as the template (PDB code 4I3B, Kumagai et al., 2013). The model was rendered in PyMol (Schrödinger, 2015).
Fluorescence Analysis of GymFP
A Hitachi F-7,000 Fluorimeter was used to collect fluorescence spectra. Spectra were recorded of a 1:1 complex of bilirubin and purified GymFP (2.1 μM of each).
Results
Properties of Endogenous Gymnothorax zonipectis Fluorescence
Fluorescence is visible in the tissue of Gymnothorax zonipectis along the entire organism and is localized below the skin (Figure 1). Solubilization of fluorescent tissue from G. zonipectis was completed in 1X PBS (1.19 mM phosphates, 13.7 mM sodium chloride, 270 μM potassium chloride, pH 7.3). Following centrifugation, the fluorescent protein remained soluble in the supernatant. Boiling the homogenized samples fully quenched fluorescence.
Transcriptome
We searched the transcriptome for fatty acid-binding proteins. From there, we further narrowed the hits based on the presence of a GPP sequence at residues 58–60 (Gruber et al., 2015; Krivoshik et al., 2020). One sequence was found in G. zonipectis.
Alignments and Phylogenetic Tree
Sequences of GymFP, Chlopsid FP I, and UnaG were aligned to identify homologous residues (Figure 2A). Residues in purple are identical between the respective sequences. The GPP sequence motif is shown in green. GymFP is 61% identical to UnaG and 47% identical to Chlopsid FP I, according to the SIM Alignment tool on ExPasy (Huang and Miller, 1991). Alignments and phylogenetic analysis of fluorescent and non-fluorescent FABPs are included in Supplementary Figure 2. The tree illustrates not only the relation of the FPs to brain FABPs, but also the distinction of GymFP as it is phylogenetically distinct from the Anguilla and Kaupichthys FPs.
Properties of GymFP
GymFP is 139 amino acids in length and has a calculated molecular weight of 15.6 kDa, similar to UnaG (Hayashi and Toda, 2009; Kumagai et al., 2013) and Chlopsid FP (Gruber et al., 2015; Krivoshik et al., 2020). Bilirubin-bound GymFP has an excitation maximum at 496 nm and a peak emission at 532 nm (Figure 3). The apo protein does not fluoresce. Chopsid FP I has EX/EM of 488/525 nm and GymFP has 496/532 nm, which is similar to that of UnaG (497/532 nm) (Figure 4).
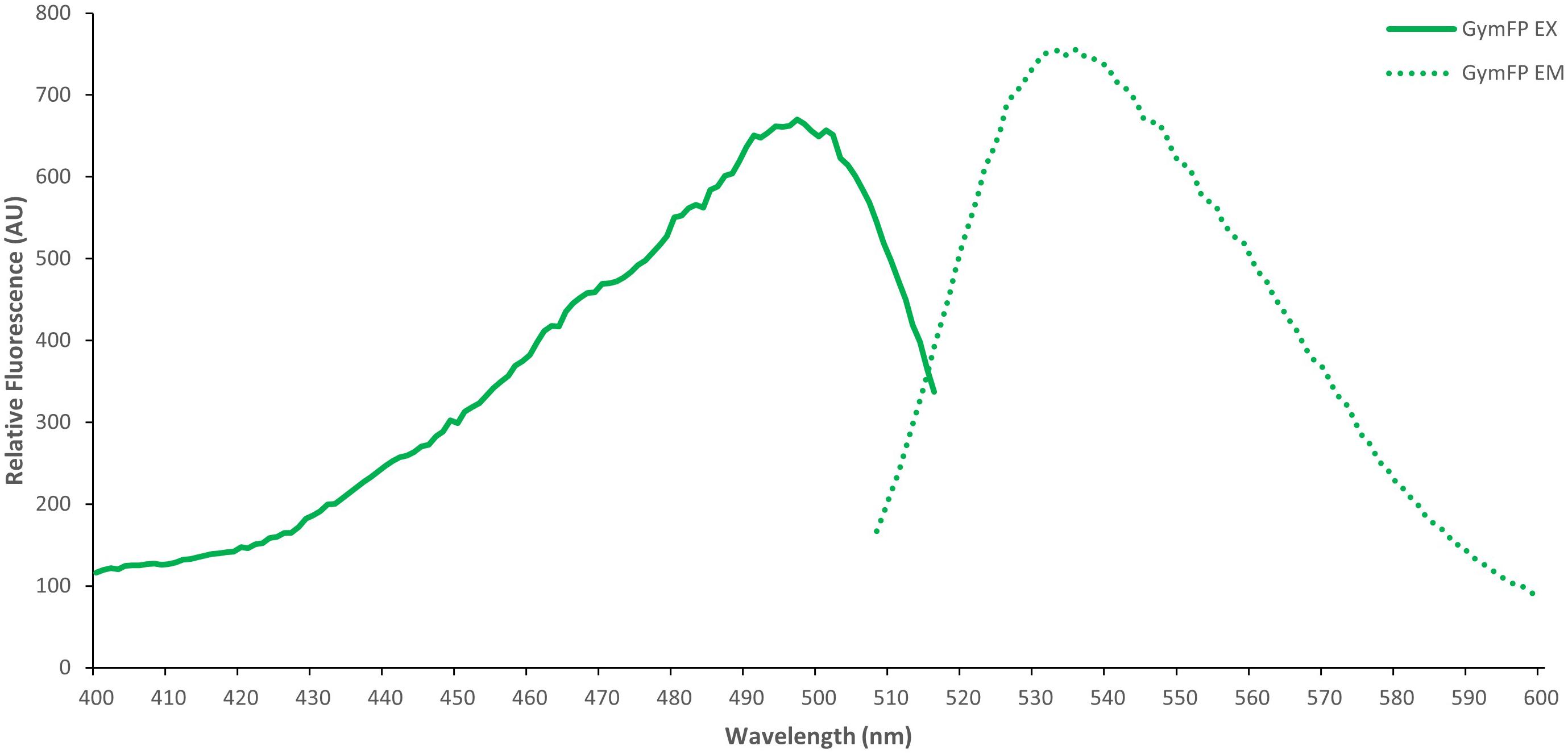
Figure 3. Excitation (EX) and emission spectra (EM) of GymFP with bilirubin (1:1 complex, 2.1 μM). GymFP was held in 50 mM Tris HCl, 150 mM NaCl, 300 mM Imidazole, pH 7.4. Bilirubin was held in 1X PBS (1.19 mM phosphates, 13.7 mM sodium chloride, 270 μM potassium chloride), pH 7.3.
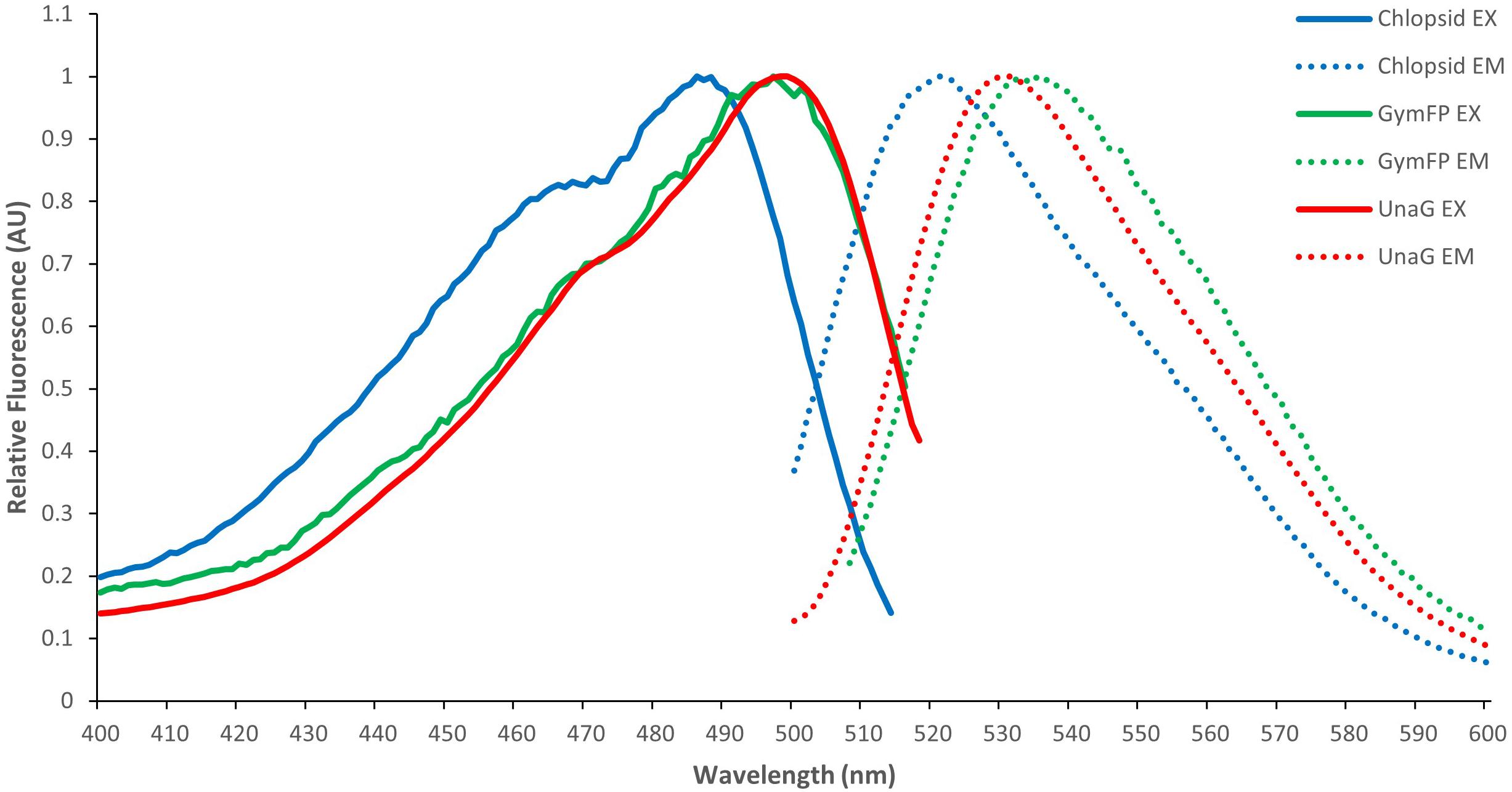
Figure 4. Normalized excitation (EX) and emission (EM) spectra of GymFP (green) compared to Chlopsid FP I (blue) and UnaG (red).
Discussion
Fluorescent proteins have now been reported and characterized in three anguilliform (true eels) families and genera, Anguillidae (Anguilla; Hayashi and Toda, 2009; Kumagai et al., 2013), Chlopsidae (false morays: Kaupichthys; Gruber et al., 2015; Krivoshik et al., 2020), and in this report, Muraenidae (morays: Gymnothorax). Interestingly, we observed that G. zonipectis has a different distribution of fluorescence in the tissue. In Anguilla and Kaupichthys fluorescence was observed throughout the musculature of the organism (Hayashi and Toda, 2009; Kumagai et al., 2013; Gruber et al., 2015). However, in G. zonipectis it was only found in a thin layer below the outer layer of the skin. The implications of this difference are not yet known.
Members of the genus Anguilla are catadromous and are known to undergo vast migrations (Tsukamoto, 2006) between habitats in freshwater and tropical and subtropical open ocean water. In Anguillidae, it has been hypothesized that fluorescence may have antioxidant properties in the muscle tissue, resulting from bilirubin binding (Kumagai et al., 2013). All members of Chlopsidae are marine, as are nearly all species of Muraenidae. In Chlopsidae, fluorescence has been proposed to potentially serve a visual function during full moon spawning. Previous studies have shown that these eels reproduce and spawn synchronized according to a lunar cycle (Lee et al., 2008). In the two species of chlopsid eels studied to date, Kaupichthys hyoproroides and Kaupichthys n. sp., two FPs were discovered (Chlopsid FP I and II) that are 94% homologous to each other (Gruber et al., 2015).
Previous work on muraenids has demonstrated that the undulated moray G. undulatus possesses rod visual pigments that allow them to detect wavelengths of 495 nm (Munz and McFarland, 1973), which suggests that Gymnothorax eels are capable of green-light detection. Additional studies of other moray species reveal that Gymnothorax favagineus has a rod at 487 ± 5.4 nm and single cone 501 ± 7.7 nm and Gymnothorax reticularis has a rod at 486 ± 4.0 nm and a single cone at 494 ± 5.8 nm (Wang et al., 2011). There is relatively sparse behavioral data associated with G. zonipectis and hence, at the moment, there is no clear indication if fluorescence plays a role in the eels’ visual ecology. Nocturnal feeding habits have been observed for individuals of this species (Böhlke and Randall, 2000), and while a Bronsonian knot has not been observed in G. zonipectis, it has been observed in other members of Gymnothorax (Kondo, 1955; Barley et al., 2016). Additional species of Gymnothorax have been shown to hunt collaboratively with groupers, including Plectropomus pessuliferus masrubri and the coral trout Plectropomus leopardus (Vail et al., 2013).
A transcriptome of G. zonipectis was completed and is now available under Bioproject PRJNA718586. A total of 273,073,027 reads were acquired and run through a BLAST search by GENEWIZ, LLC (South Plainfield, NJ) to generate a list of sequences. To search the hits from the transcriptome assembly, we looked for fatty acid binding proteins that contained a GPP sequence insert. We found one transcript matching this criteria and synthesized the corresponding gene for protein expression. A number of brain fatty acid-binding proteins were also found that did not contain this sequence. This further supports this sequence insert’s importance in eel fluorescence. None of these sequences contained GPP in residues 58–60 and were therefore not analyzed any further. In previous work with Chlopsid FP we had synthesized and expressed the homologous fatty acid binding protein analoges that did not contain GPP and they were found to be non-fluorescent.
The protein sequence discovered in G. zonipectis, which we refer to as GymFP, shares 61.0% identity with UnaG, isolated from Anguilla japonica, and 46.7% identity to Chlopsid FP I, isolated from Kaupichthys hyoproroides. Further characterization of this FP has uncovered many similarities with both UnaG and Chlopsid FP I, including the emission of green light and the use of bilirubin as a ligand. It is significant that this third eel FP also possesses a GPP motif. The consistency of this motif in all fluorescent fatty acid binding proteins is also observed in the grouping in the phylogenetic tree. We generated a phylogenetic tree based on 14 fatty acid binding proteins from fluorescent eels, non-fluorescent fish, and humans. The fluorescent sequences formed their own clade (Supplementary Figure 2A). This is consistent with other trees that have been previously published (Gruber et al., 2015; Funahashi et al., 2017). We also note that the Anguilla and Gymnothorax fluorescent fatty acid binding protein groups are sister, although this is not highly supported (36%).
We used Swiss Model (Guex et al., 2009; Bienert et al., 2017; Bertoni et al., 2017; Waterhouse et al., 2018; Studer et al., 2020) to generate a model of GymFP (Figure 2B). The template for the model was pdb code 4I3B, which is the WT structure of UnaG (Kumagai et al., 2013). The model shows the expected beta barrel structure. The GPP motif is highlighted in green and is in the same position found in the other fluorescent eel fatty acid binding proteins. In purple, we highlight those areas that are the same in UnaG, ChlopsidFP I and GymFP. It should be noted that we have not been able to determine sequence motifs responsible for shifting fluorescence excitation or emission to date (Krivoshik et al., 2020).
The lipocalin family of proteins, which includes fatty acid binding proteins and apolipoprotein D, contains other proteins that are able to use heme breakdown products to produce fluorescence. Sandercyanin, found in Stizostedion vitreum (previously Sander vitreus), is homologous to apolipoprotein D and is able to use biliverdin to produce red fluorescence (Ghosh et al., 2016).
In closing, this report provides the characterization of another member of the eel fluorescent fatty acid binding protein family. This is the first report of fluorescent protein characterization from an eel from the family Muraenidae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA718586.
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guidelines for the Use of Fishes in Research of the American Fisheries Society and the American Museum of Natural History’s Institutional Animal Care and Use Committee (IACUC).
Author Contributions
AG, DG, and JG conceptualized the project and wrote the manuscript. DG and JS were involved in the collection of specimens. JG obtained funding for this research. AG, SK, and JG performed the experiments and calculations. All authors reviewed and revised the manuscript.
Funding
Funding was provided by the NSF CAREER Award # 1652731 to JG. Field research in the Solomon Islands was funded by a National Science Foundation award to JS (DEB-1257555), and the American Museum of Natural History (AMNH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Alec Hughes (WCS) for logistical support and help obtaining permits, and to the crew of the A.R. Ford (Honiara) for considerable support in the field. We thank Corey Howell and The Wilderness Lodge for logistical and SCUBA diving assistance around Gatokae Island, and Robert Schelly and Tate Sparks for assistance with field collections and fluorescent photography. We would also like to thank Prof. Mercer Brugler for his help with phylogenetic tree preparation. We thank Genewiz, LLC for the RNA library preparation, transcriptome assembly and bioinformatics work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.678571/full#supplementary-material
Supplementary Video 1 | Biofluorescence in Gymnothorax funebris.
References
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Barley, S. C., Mehta, R. S., Meeuwig, J. J., and Meekan, M. G. (2016). To knot or not? Novel feeding behaviours in moray eels. Mar. Biodivers. 46, 703–705. doi: 10.1007/s12526-015-0404-y
Barondeau, D. P., Kassmann, C. J., Tainer, J. A., and Getzoff, E. D. (2005). Understanding GFP Chromophore Biosynthesis: controlling Backbone Cyclization and Modifying Post-translational Chemistry †, ‡. Biochemistry 44, 1960–1970. doi: 10.1021/bi0479205
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., and Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7:10480. doi: 10.1038/s41598-017-09654-8
Bienert, S., Waterhouse, A., de Beer, T. A. P., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 45, D313–D319. doi: 10.1093/nar/gkw1132
Böhlke, E. B., and Randall, J. E. (2000). A Review of the Moray eels (Angulliformes: Muraenidae) of the Hawaiian Islands, with Descriptions of Two New Species. Proc. Acad. Nat. Sci. Phila. 150, 203–278.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Field, S. F., Bulina, M. Y., Kelmanson, I. V., Bielawski, J. P., and Matz, M. V. (2006). Adaptive Evolution of Multicolored Fluorescent Proteins in Reef-Building Corals. J. Mol. Evol. 62, 332–339. doi: 10.1007/s00239-005-0129-9
Funahashi, A., Itakura, T., Hassanin, A. A. I., Komatsu, M., Hayashi, S., and Kaminishi, Y. (2017). Ubiquitous distribution of fluorescent protein in muscles of four species and two subspecies of eel (genus Anguilla). J. Genet. 96, 127–133. doi: 10.1007/s12041-017-0751-5
Ghosh, S., Yu, C.-L., Ferraro, D. J., Sudha, S., Pal, S. K., Schaefer, W. F., et al. (2016). Blue protein with red fluorescence. Proc. Natl. Acad. Sci. U. S. A. 113, 11513–11518. doi: 10.1073/pnas.1525622113
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gruber, D. F., Gaffney, J. P., Mehr, S., DeSalle, R., Sparks, J. S., Platisa, J., et al. (2015). Adaptive Evolution of Eel Fluorescent Proteins from Fatty Acid Binding Proteins Produces Bright Fluorescence in the Marine Environment. PLoS One 10:e0140972. doi: 10.1371/journal.pone.0140972
Guex, N., Peitsch, M. C., and Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, S162–S173. doi: 10.1002/elps.200900140
Hayashi, S., and Toda, Y. (2009). A novel fluorescent protein purified from eel muscle. Fish. Sci. 75, 1461–1469. doi: 10.1007/s12562-009-0176-z
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hofacker, I. L., Fekete, M., and Stadler, P. F. (2002). Secondary Structure Prediction for Aligned RNA Sequences. J. Mol. Biol. 319, 1059–1066. doi: 10.1016/S0022-2836(02)00308-X
Huang, X., and Miller, W. (1991). A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12, 337–357. doi: 10.1016/0196-8858(91)90017-D
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Frith, M. C. (2012). Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28, 3144–3146. doi: 10.1093/bioinformatics/bts578
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh, K., and Standley, D. M. (2016). A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32, 1933–1942. doi: 10.1093/bioinformatics/btw108
Katoh, K., and Toh, H. (2008). Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9:212. doi: 10.1186/1471-2105-9-212
Krivoshik, S. R., Guarnaccia, A. M., Fried, D. B., Gruber, D. F., and Gaffney, J. P. (2020). Disrupting Fluorescence by Mutagenesis in a Green Fluorescent Fatty Acid Binding Protein from a Marine Eel. Protein J. 39, 145–151. doi: 10.1007/s10930-020-09883-3
Kumagai, A., Ando, R., Miyatake, H., Greimel, P., Kobayashi, T., Hirabayashi, Y., et al. (2013). A Bilirubin-Inducible Fluorescent Protein from Eel Muscle. Cell 153, 1602–1611. doi: 10.1016/j.cell.2013.05.038
Kuraku, S., Zmasek, C. M., Nishimura, O., and Katoh, K. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22–W28. doi: 10.1093/nar/gkt389
Lee, T. W., Miller, M. J., Hwang, H. B., Wouthuyzen, S., and Tsukamoto, K. (2008). Distribution and early life history of Kaupichthys leptocephali (family Chlopsidae) in the central Indonesian Seas. Mar. Biol. 153, 285–295. doi: 10.1007/s00227-007-0804-z
Leutenegger, A., D’Angelo, C., Matz, M. V., Denzel, A., Oswald, F., Salih, A., et al. (2007). It’s cheap to be colorful. FEBS J. 274, 2496–2505. doi: 10.1111/j.1742-4658.2007.05785.x
Matz, M. V., Fradkov, A. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L., et al. (1999). Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973. doi: 10.1038/13657
Munz, F. W., and McFarland, W. N. (1973). The significance of spectral position in the rhodopsins of tropical marine fishes. Vision Res. 13, 1829–1874. doi: 10.1016/0042-6989(73)90060-6
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M., and Katoh, K. (2019). MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47, W5–W10. doi: 10.1093/nar/gkz342
Salih, A., Larkum, A., Cox, G., Kühl, M., and Hoegh-Guldberg, O. (2000). Fluorescent pigments in corals are photoprotective. Nature 408, 850–853. doi: 10.1038/35048564
Shimomura, O., Johnson, F. H., and Saiga, Y. (1962). Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59, 223–239. doi: 10.1002/jcp.1030590302
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Sparks, J. S., Schelly, R. C., Smith, W. L., Davis, M. P., Tchernov, D., Pieribone, V. A., et al. (2014). The Covert World of Fish Biofluorescence: a Phylogenetically Widespread and Phenotypically Variable Phenomenon. PLoS One 9:e83259. doi: 10.1371/journal.pone.0083259
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, R., Haas, J., and Schwede, T. (2020). QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. doi: 10.1093/bioinformatics/btz828
Tabei, Y., Kiryu, H., Kin, T., and Asai, K. (2008). A fast structural multiple alignment method for long RNA sequences. BMC Bioinformatics 9:33. doi: 10.1186/1471-2105-9-33
Vail, A. L., Manica, A., and Bshary, R. (2013). Referential gestures in fish collaborative hunting. Nat. Commun. 4:1765. doi: 10.1038/ncomms2781
Wang, F. Y., Tang, M. Y., and Yan, H. Y. (2011). A comparative study on the visual adaptations of four species of moray eel. Vision Res. 51, 1099–1108. doi: 10.1016/j.visres.2011.02.025
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Keywords: eels, fluorescence, fatty acid binding proteins, bilirubin, green fluorescence, marine fish, fluorescent proteins
Citation: Guarnaccia AM, Krivoshik SR, Sparks JS, Gruber DF and Gaffney JP (2021) Discovery and Characterization of a Bilirubin Inducible Green Fluorescent Protein From the Moray Eel Gymnothorax zonipectis. Front. Mar. Sci. 8:678571. doi: 10.3389/fmars.2021.678571
Received: 09 March 2021; Accepted: 16 August 2021;
Published: 24 September 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesCopyright © 2021 Guarnaccia, Krivoshik, Sparks, Gruber and Gaffney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean P. Gaffney, amVhbi5nYWZmbmV5QGJhcnVjaC5jdW55LmVkdQ==
 Andrew M. Guarnaccia
Andrew M. Guarnaccia Sara Rose Krivoshik
Sara Rose Krivoshik John S. Sparks
John S. Sparks David F. Gruber
David F. Gruber Jean P. Gaffney
Jean P. Gaffney