- 1Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 2The Shraga Segal Department of Microbiology, Immunology, and Genetics, Faculty of Health Sciences, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 3Regenerative Medicine and Stem Cell Research Center, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 4Dead Sea and Arava Science Center, Masada National Park, Jerusalem, Israel
- 5National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 6Department of Life Sciences, Achva Academic College, Arugot, Israel
Novel monosex biotechnologies in crustacean aquaculture involve the induction of sex reversal through manipulations of the androgenic gland (AG), and its most prominent factor, the insulin-like androgenic gland (IAG) hormone, during early developmental stages. In the giant freshwater prawn Macrobrachium rosenbergii, all-female populations are desirable for aquaculture, since the females can be stocked at higher densities and exhibit more uniform growth than males. Recently, a novel biotechnology for all-female aquaculture was developed based on injection into post-larvae of suspended primary AG cells from mature M. rosenbergii males. However, this biotechnology depends on the availability of appropriate male donors and it requires delicate surgical procedures on the small endocrine AG to produce appropriate quantities of cells for the sex manipulation. We therefore established a new platform for the production of M. rosenbergii IAG (Mr-IAG) in hematopoietic cells. The method rests on the induction of ectopic Mr-IAG expression under the white spot syndrome virus (WSSV) immediate-early 1 (IE1) promoter via a novel lentiviral vector. Our results demonstrate that M. rosenbergii primary cells infected with the Mr-IAG lentiviruses are capable of transcription, translation, and secretion of Mr-IAG in culture. Our new platform, which produces easy-to-harvest cells in abundance, could replace the AG cells used in the first step of the above-mentioned biotechnology for all-female aquaculture and, importantly, pave the way for producing monosex populations in other edible crustacean aquaculture species. In addition, a lentiviral system for crustacean cells provides a useful tool for basic and applied research in crustacean species.
Introduction
The increasing demand for monosex crustacean aquaculture populations requires the development of biotechnological tools to overcome labor-intensive manual sorting efforts and hence to offer the more profitable approach of all-male or all-female populations. Currently available biotechnological tools for monosex strategies are based on fundamental research, which has proceeded apace to uncover the mechanisms of crustacean sex determination and differentiation processes. For example, it is known that the androgenic gland (AG) is a key endocrine factor in crustacean sexual differentiation, since its presence induces the development of the male reproductive system (Nagamine et al., 1980; Malecha et al., 1992), while its ablation enhances the feminization of males (Nagamine et al., 1980; Sagi et al., 1990, 1997). The most prominent AG factor responsible for masculine development is the insulin-like androgenic gland (IAG) hormone, believed to be the universal sexual differentiation switch (IAG-switch) in decapod species (Levy and Sagi, 2020). Since its first discovery in the Australian red claw crayfish Cherax quadricarinatus (Manor et al., 2007), the IAG peptide has been found in most decapod species studied thus far, playing a conserved role through IAG-switch-mediated sexual differentiation (Levy and Sagi, 2020).
In the study reported here, we focused on a biotechnology for monosex production of an important aquaculture species (FAO, 2020), the giant freshwater prawn, Macrobrachium rosenbergii. This species exhibits male superiority and differential growth, with a dominance hierarchy being evident for the three adult male morphotypes—small males at the bottom, followed by orange-claw males, and then by the dominant blue-claw males (characterized by two fully developed AGs), which constitute only a small portion of M. rosenbergii populations. The females are smaller than the males and exhibit a more uniform growth pattern, but they can be stocked at higher densities than the males, whose territorial and aggressive behavior precludes the use of high stocking densities (Sagi et al., 1986; Kuris et al., 1987; Levy et al., 2017). Therefore, all-female aquaculture, giving higher total yields under higher stocking densities, would appear to be the preferable mode of monosex aquaculture (Malecha, 2012; Levy et al., 2017). Since it was originally believed that, even under low stocking densities, all-male M. rosenbergii populations would be the most profitable, research on monosex biotechnologies has focused on all-male populations, with that on all-female aquaculture lagging behind. Nonetheless, a start has been made on developing a novel approach for high-stocking-density all-female M. rosenbergii aquaculture that is based on sex reversal during an early developmental stage (Ventura et al., 2012; Levy et al., 2016). The first step in this three-step approach to produce all-female populations is sex reversal by the administration of a single injection of suspended primary AG cells from mature M. rosenbergii blue-claw males into post-larvae (Levy et al., 2016). Thus, this biotechnology depends on the availability of appropriate male donors and requires complex surgical procedures of the small endocrine AG to supply appropriate quantities of cells required for sex manipulation. The main goal of the present study was therefore to seek a biotechnologically more efficient source of IAG-expressing cells. To this end, we sought to develop a primary crustacean cell culture that would ectopically express M. rosenbergii IAG (Mr-IAG) and would thereby provide an abundant, easy-to-harvest source of the hormone. In the context of this study, the development of the cell-culture technology provides a source of Mr-IAG for the novel biotechnology that relies on a sex reversal procedure, but in the wider context of crustacean aquaculture it will contribute a universal tool for cell transplantation for the facile generation of monosex populations.
In parallel with the biotechnological developments in crustacean aquaculture over the past few decades, numerous studies on marine invertebrate cell cultures have focused on developing scientific tools to study gene expression, regulation, and molecular functions during cellular events. These tools are usually achieved by overexpression or knockdown of particular target genes, all requiring an efficient way to deliver DNA into crustacean cell cultures. A variety of techniques have been exploited for this purpose, including lipofection (Tapay et al., 1995; Claydon and Owens, 2008), electroporation (Lin and Söderhäll, 2011; Han et al., 2015; Shi et al., 2018), retroviral transduction (Shike et al., 2000; Hu et al., 2008, 2010; Han et al., 2015; Puthumana et al., 2015; Pu et al., 2017), and baculovirus transduction (Lu et al., 2005; Puthumana et al., 2016). However, all have yielded low efficiencies of gene transfer and expression, and there remains a need for a robust system that can deliver transgenes into crustacean cells.
We therefore aimed to develop a novel DNA delivery platform based on lentiviruses, which are commonly used for gene insertion into mammalian cells, since these vectors have been shown to be superior to the retroviral systems in earlier use (Vigna and Naldini, 2000; Durand and Cimarelli, 2011). Importantly, lentiviruses can efficiently infect terminally differentiated cells, thereby driving the stable expression of genes of interest due to their integration into the host genome (Vigna and Naldini, 2000; Durand and Cimarelli, 2011; Lundstrom, 2019). The lentiviral replication cycle consists of several steps: attaching to and entering the host cell; reverse transcription of viral RNA in the cytoplasm of the infected host cell; and integration of the viral cDNA into the host genomic DNA, a crucial process for replication that generates a provirus that persists as long as the host cell survives (Acheampong et al., 2003). The integration stage is catalyzed by viral integrase, and various host cellular proteins function as cofactors for the integration of lentiviral genes, for example, the p75 isoform of lens epithelium-derived growth factor (LEDGF/p75). LEDGF interacts with and promotes the activity of viral integrase, and its expression is thus required for the association of integrase with mitotic chromosomes (Busschots et al., 2005; Hombrouck et al., 2007).
The particular advantage offered by lentiviral vectors in a study such as ours is their ability to infect quiescent non-dividing primary cells since cultures of primary crustacean cells exhibit a reduced ability to proliferate, and usually remain in a stationary state for several days (Rinkevich, 2005, 2011). However, this advantage is offset by the disadvantage that commercially constructed lentiviruses are raised to infect mammalian cells, and they therefore contain a constitutive mammalian promoter that is designed to regulate gene expression in mammalian cells. It was therefore necessary to find an alternative promoter that would be suitable for affecting gene expression in crustacean primary cells. To date, only one study has demonstrated the use of lentiviral technique in a decapod species (a penaeid shrimp) but with a relatively low efficiency of gene expression (Chen et al., 2019). We therefore reviewed a number of possible candidate promoters from the white spot syndrome virus (WSSV). This virus is one of the major pathogens infecting aquacultured crustaceans, resulting in high rates of mortality, and significant economic losses to the shrimp industry worldwide (Sánchez-Martínez et al., 2007; Verbruggen et al., 2016). WSSV is a double-stranded circular DNA virus whose genes can be classified as immediate early (IE), delayed early, late, and very late. IE gene products are expressed prior to the replication cycle, encoding transcriptional factors under strong promoter regulation (Sánchez-Paz, 2010; Lin et al., 2013). Among these promoters is IE1, the promoter of the immediate early 1 gene (IE1, or WSV069), which is activated by several host crustacean transcription factors (Liu et al., 2007; Huang et al., 2010; Wang et al., 2011; Qiu et al., 2014). Therefore, the IE1 promoter was chosen in the present study to replace the mammalian promoter in the commercially constructed lentiviral vector.
Among the studies aimed at identifying functional crustacean tissues and culturing primary cells with the potential to proliferate (Rinkevich, 2005, 2011; Jayesh et al., 2012), a number have focused on crustacean hematopoietic tissue (Johansson et al., 2000; Soderhall et al., 2003; Lin and Söderhäll, 2011). A previous study in our laboratory identified hematopoietic tissue in M. rosenbergii and verified transglutaminase as a marker (data not shown), as has been reported for Pacifastacus leniusculus (Lin et al., 2008; Lin and Söderhäll, 2011) and recently for M. rosenbergii (Sirikharin et al., 2019). Hence, our current study was conducted on an M. rosenbergii primary hematopoietic cell culture. We thus first established a novel lentiviral platform for efficient transduction of a hematopoietic M. rosenbergii primary cell culture, resulting in enhanced expression of the reporter fluorescent proteins ZsGreen and DsRed under the WSSV IE1 promoter. Thereafter, we generated a new lentiviral plasmid encoding Mr-IAG and ZsGreen proteins under the WSSV IE1 promoter and successfully expressed the Mr-IAG hormone in transduced M. rosenbergii hematopoietic cells. This is, to the best of our knowledge, the first demonstration of the ectopic expression of Mr-IAG in cell types other than naturally expressing AG cells. Further studies will focus on using the Mr-IAG lentiviral infected M. rosenbergii hematopoietic cells to replace AG cells in the biotechnology for all-female M. rosenbergii aquaculture and will also pave the way for producing monosex populations in other crustacean species and for other applications.
Materials and Methods
Lentiviral Plasmid Construction
A pHAGE2 lentiviral plasmid that includes the red fluorescent protein DsRed and the green fluorescent protein ZsGreen, under human elongation factor-1 alpha (EF1α), was first modified to include a constitutive promoter for crustacean cells and later to include Mr-IAG, as follows. The WSSV IE1 promoter sequence (AF332093.3; WSV069 promoter 36228–36732 bp) (Li et al., 2009) was synthesized in a pUC57 plasmid (by Hylabs, Israel) and ligated into the pHAGE2 plasmid to replace the original EF1α by using SpeI and NotI restriction enzymes and T4-ligase (New England Biolabs), thereby generating a plasmid encoding for DsRed and ZsGreen proteins under the WSSV IE1 promoter (Figure 1). Thereafter, the full sequence of Mr-IAG mRNA was amplified from an Mr-IAG-pGEM plasmid previously prepared in our laboratory (Ventura et al., 2009) by using specifically designed primers that also added a restriction site at each end of the Mr-IAG sequence (NotI and BamHI sites were added, using 5′-ataGCGGCCGCATGGGATACTGGAATGCCGAG and 5′-ataGGATCCCTACCTGGAACTGCAGGTGTT, respectively). The DsRed sequence was then replaced with the Mr-IAG sequence by restriction and ligation, as above, to produce the WSSV IE1-Mr-IAG-IRES-ZsGreen plasmid, designated here as the Mr-IAG lentiviral plasmid (Figure 2).
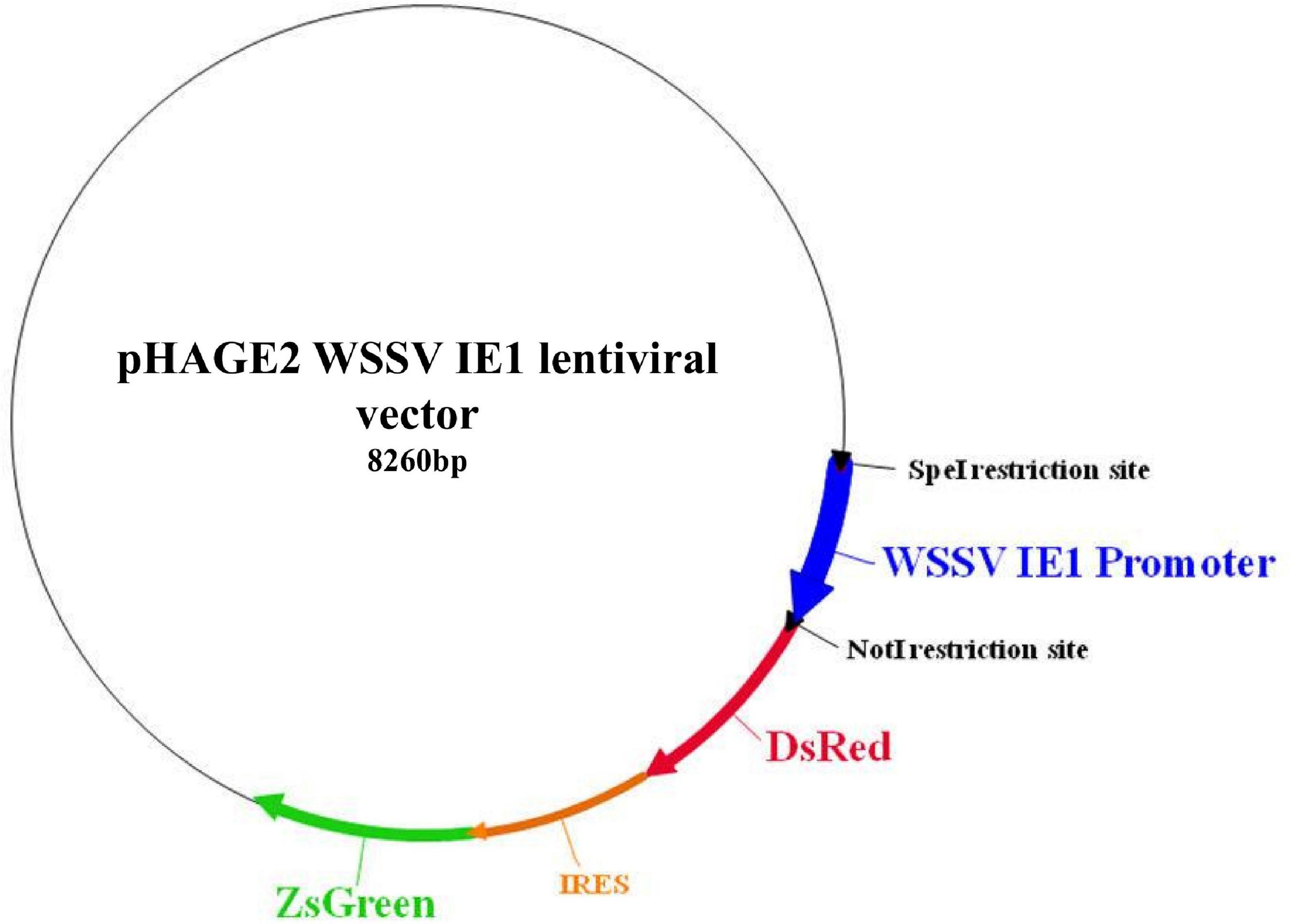
Figure 1. Plasmid map of the novel WSSV IE1 lentiviral plasmid after insertion of the WSSV IE1 promoter region into a pHAGE2 lentiviral plasmid. The sequence of the human EF1α promoter was restricted out by using SpeI and NotI restriction enzymes, followed by ligation of the WSSV IE1 sequence into the lentiviral plasmid (dark blue arrow). Functional elements are shown, including the relevant restriction sites and the red fluorescent (DsRed) and green fluorescent (ZsGreen) coding genes under the regulation of the WSSV IE1 promoter. The novel lentiviral plasmid was sequenced, aligned, and verified.
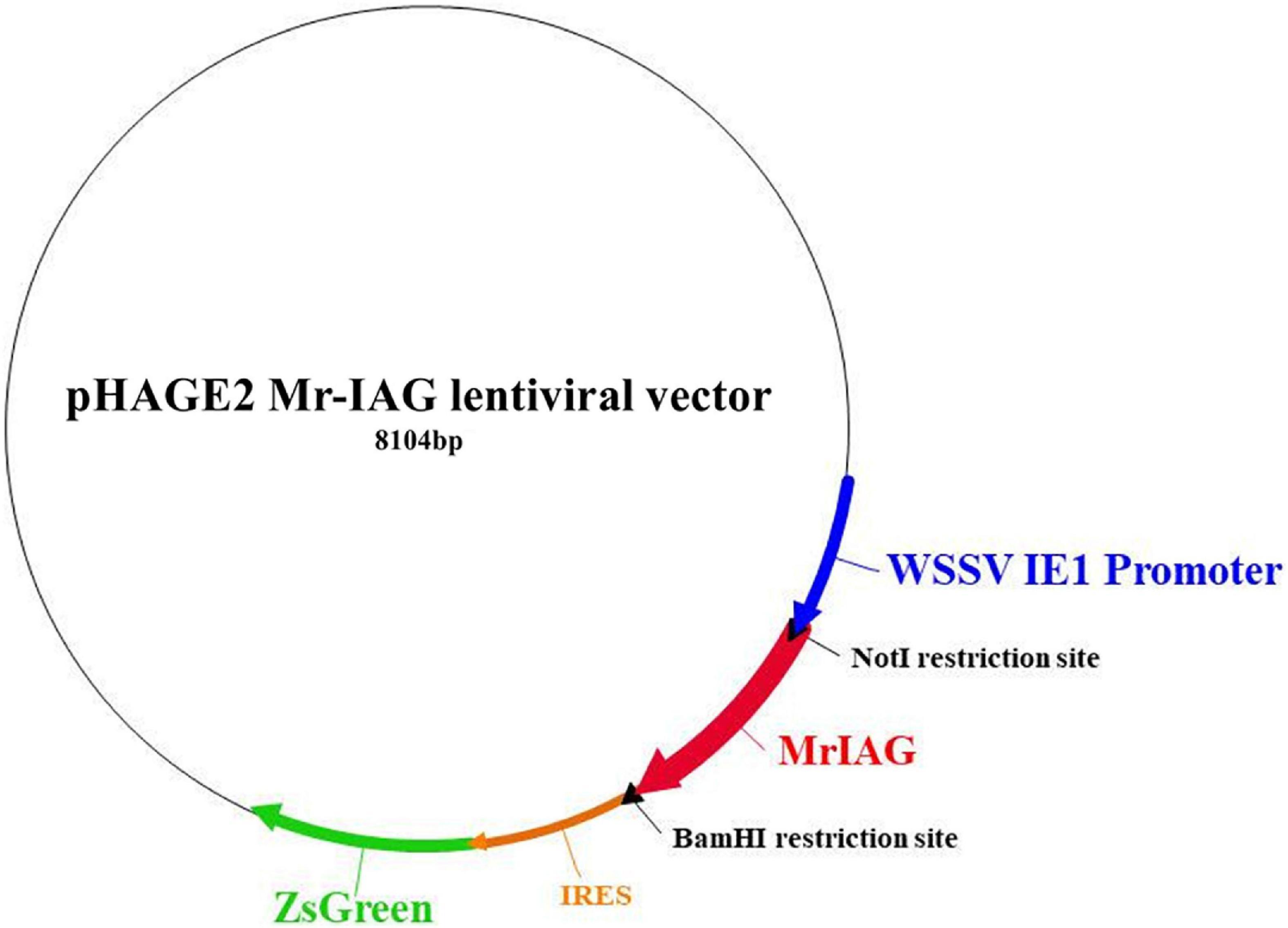
Figure 2. Insertion of the Mr-IAG sequence into the WSSV IE1 pHAGE2 lentiviral plasmid. The sequence of Mr-IAG mRNA was inserted and ligated into the WSSV IE1 lentiviral plasmid (bold red arrow) instead of DsRed, which was restricted out by using NotI and BamHI restriction enzymes. The novel Mr-IAG lentiviral plasmid was sequenced, aligned, and verified.
Lentivirus Production in the HEK293T Cell Line
The HEK293T cell line was used to produce the lentivirus (Benskey and Manfredsson, 2016; Sena-Esteves and Gao, 2018). Co-transfection of the pHAG2 vector (12 μg) together with a fourth-generation packaging lentiviral mix, containing Tat, Rev, Hgpm2, and VSVG helper genes (3 μg) in a final ratio of 12:1:1:2:1 (Mostoslavsky et al., 2005; Olender et al., 2020) was performed with polyethylenimine (Sigma-Aldrich) according to the manufacturer protocol. Four hours following the transfection, the media was replaced. The cell culture medium was collected on days 2–4 and then centrifuged at 1,000 g for 2 min to pellet cell debris and 0.22 μm filtered (Millipore). The supernatant was then concentrated by using an ultracentrifuge (Beckman Coulter) at 17,000 g for 90 min at 4°C. The lentiviral fraction (the pellet) was aliquoted and stored at −80°C.
Animals
Macrobrachium rosenbergii blue claw males were obtained from artificial ponds at Ben-Gurion University of the Negev, Beersheba, Israel, as described previously (Levy et al., 2016).
Primary Androgenic Gland Cell Culture
Hypertrophy and hyperplasia of the AGs of eight M. rosenbergii blue claw males were achieved by surgical removal of the neuroendocrine X organ-sinus gland complex, located in the eyestalk, as previously described (Khalaila et al., 2002). Eight days post-endocrine manipulation, the M. rosenbergii blue-claw males were anesthetized for 5 min in ice-cold water followed by 2 min in ice-cold water supplemented with 0.2% hypochlorite for disinfection purposes. Thereafter, the animals were dissected, and their hypertrophied androgenic glands (hAGs) were isolated under a dissecting microscope. The glands from two animals were immediately transferred for RNA purification, serving as a positive control for qPCR analysis (as described below). The other glands were pooled into an Eppendorf tube and were further treated as stated below for the hematopoietic primary cell culture.
Primary Hematopoietic Cell Culture
For each experiment, two to four M. rosenbergii blue-claw males were anesthetized for 5 min in ice-cold water, followed by 2 min in ice-cold water supplemented with 0.2% hypochlorite for disinfection purposes. The animals were dissected, and the sheet-like thin hematopoietic tissue covering the dorsal and dorsolateral sides of the stomach was isolated. Tissue samples were pooled into an Eppendorf tube containing Leibovitz L-15 medium (Sigma-Aldrich) supplemented with GlutaMAXTM (Gibco) and a penicillin-streptomycin-amphotericin B solution (PSA, Biological Industries); centrifuged at 800 g for 5 min at room temperature (RT); and washed twice. Thereafter, cells were resuspended in 500–1000 μl of a specific enzyme mix [0.1% (w/v) collagenase type I and 0.1% (w/v) collagenase type IV (both from Sigma-Aldrich) in L-15 medium supplemented with GlutaMax and PSA], chopped into 1 mm tissue pieces using surgical microscissors, and spun at 15 rpm for 45 min at RT. The enzymatically digested hematopoietic tissues were then triturated using a 1 ml pipette tip and filtered through a 40-μm mesh, followed by centrifugation at 800 g for 5 min at RT. Cells were washed twice with 0.5 ml of Leibovitz L-15 medium with GlutaMAX, PSA, and 10% (v/v) fetal bovine serum (FBS) (Biological Industries). Finally, cells were suspended in 150 μl of the growth medium, counted, and examined for viability by using a trypan blue exclusion assay (Biological Industries).
Hematopoietic cells were seeded at 2 × 105 cells per well in a 96-well plate and grown for all the experiments in NutriStem® hPSC XF Medium (Biological Industries), a specific medium for human stem cell cultures, based on the stem-like nature of hematopoietic cells. Osmolality was adjusted to 420 mOsm with crustacean physiological saline (Sirikharin et al., 2019). Plates were incubated in a CO2 incubator at 28°C with 5% CO2.
Transduction of Lentiviruses in Primary Cell Culture
The day following the seeding of primary M. rosenbergii hematopoietic cell cultures, the cultures were transduced with WSSV IE1 lentiviruses or Mr-IAG lentiviruses (20 μl per well at a seeding density of 2 × 105 cells in 96-well). Two days after transduction, a fresh medium was added. Cultures were then analyzed to monitor the expression of fluorescent proteins (using confocal microscopy or flow cytometry) or the presence of lentiviral sequences (PCR analyses).
Genomic DNA and RNA Preparation and PCR Analyses
Four days following transduction, genomic DNA (gDNA) was extracted from M. rosenbergii primary cell cultures using DNeasy Blood and Tissue kit (Qiagen), yielding 200–400 ng/μl of gDNA from each well and then examined by PCR for the presence of lentiviral sequences in the genomic fraction using specific primers (for ZsGreen, IE1-DsRed, Mr-IAG, and Mr-Actin sequences) as shown in Table 1. RNA extraction from tissue and cells was performed using EZ-RNA Total RNA Isolation Kit (Biological Industries).
Confocal Microscopy
Four days following transduction of M. rosenbergii primary cell cultures by WSSV IE1 lentiviruses or Mr-IAG lentiviruses, cells were transferred onto glass adhesive slides, fixed with paraformaldehyde, and stained with the nucleic acid stain DAPI (4′,6-diamidino-2-phenylindole) to visualize nuclear DNA. Infected cells were monitored under confocal microscopy on an Olympus FV1000 laser scanning confocal microscope for fluorescent DsRed and ZsGreen signals and compared to the signal for uninfected control cells as a reference for autofluorescent signaling.
FACS Analysis
Four days following transduction of M. rosenbergii primary cell cultures by WSSV IE1 lentiviruses or Mr-IAG lentiviruses, cells were collected into an Eppendorf tube and washed twice in crustacean physiological saline (420 mOsm). Cells were stained with DAPI (1 μg/ml) for the fluorescent detection of dead cells. The cells were analyzed using Beckman Coulter® GalliosTM flow cytometer. The flow cytometry was analyzed using FlowJo V10 software and Kaluza Beckman Coulter® software. Due to substantial autofluorescence of the M. rosenbergii primary cells, in order to determine specific signal the analysis of the ZsGreen protein was performed using two dimensional gates with DsRed and ZsGreen. The two-dimensional gating strategy is enabling gating below the autofluorescence diagonal of the specific signal.
Immunohistochemistry
Cells were seeded on an eight-well cell culture slide (SPL, Korea) at a density of 3 × 105 cells per well, and each well was infected with 20 μl of Mr-IAG lentivirus solution. Four days post-transduction, the medium was discarded, and cells were washed twice with crustacean physiological saline, and fixed with 4% paraformaldehyde for 15 min at RT. Cells were blocked and permeabilized with blocking solution containing 2% normal goat serum, 0.1% Triton X-100, 0.05% Tween-20 in PBS for 1 h at RT. Thereafter, cells were incubated overnight at 4°C, in a humidity chamber, with rabbit anti-Mr-IAG antibody, as previously described and validated (Ventura et al., 2011). The primary antibody was diluted to 1:200 in blocking solution. After three washes with PBS, the cells were incubated for 1 h at RT with donkey anti-rabbit IgG Alexa Fluor-546 (Thermo Fisher Scientific, United States) diluted to 1:500 in PBS containing 0.2% fish skin gelatin (Sigma, Germany). Non-specific staining was performed with blocking solution without primary antibody. Following three washes with PBS, slides were mounted with Fluoromount-G® containing DAPI (Southern Biotech, United States) and examined under an Olympus Fluoview FV1000 laser scanning confocal microscope.
Sandwich ELISA
Four days following transduction of M. rosenbergii primary cell cultures by Mr-IAG lentiviruses, the growth medium (supernatants) was collected from each well and preserved for quantitative measurement of Mr-IAG protein secreted from infected and control cultures. On the day before the sandwich ELISA was performed, the plates were coated with 150 μl of capture antibody, namely, mouse polyclonal specific anti-Mr-IAG, which had previously been generated and validated in our laboratory (Sharabi et al., 2016). The capture antibody was diluted (1:1000) in 50 mM carbonate buffer (CB), pH 9.6, and the coated plates were covered with adhesive film and kept at 4°C. On the day of the assay, each plate was washed three times with 200 μl of PBS (all the solutions and washes were discarded, and the remaining drops were removed by patting the plates dry with a paper towel). Thereafter, blocking solution (1% BSA and 0.05% Tween-20 in PBS) was added for 1 h at RT. After washing, the above-mentioned supernatants were added at different dilutions (50, 75, and 100 μl of collected growth medium samples; n = 6 for each dilution), and the plates were covered and incubated for 2 h at 37°C. After washing, the detection antibody, namely, rabbit anti-Mr-IAG (1:200 in CB), was added in 150 μl of blocking solution for 2 h at 37°C (Sharabi et al., 2016). After three washes, secondary anti-rabbit alkaline phosphatase-conjugated antibody (1:10,000, Sigma-Aldrich) was added, followed by incubation for 1 h. For detection of alkaline phosphatase enzymatic activity, its substrate, p-nitrophenyl phosphate (Sigma-Aldrich), was added at 1 μg/ml in 0.1 M glycine buffer, pH 10.4, and incubated for 1 h. The enzymatic activity was detected at 405 nm using an ELISA plate reader (Tecan).
Bioinformatic Analyses of the Macrobrachium rosenbergii Lens Epithelium-Derived Growth Factor Gene
The sequence of the M. rosenbergii lens epithelium-derived growth factor (MrLEDGF) gene was searched using BLAST of a known Litopenaeus vannamei LEDGF (XP_027235252.1) against a previously published transcriptomic library of M. rosenbergii (Sharabi et al., 2016). The filtered candidate transcript was then computationally translated into a protein by using the translate tool from the ExPASy Proteomics Server (Gasteiger et al., 2005)1, and the longest open reading frame was selected as the putative protein sequence, similar to the ORF of the known L. vannamei LEDGF. The identification and annotation of the MrLEDGF protein in the filtered transcript were done using SMART (Schultz et al., 1998).
Relative Quantification of Gene Expression Levels
For qPCR, total RNA was isolated from cultures infected with WSSV IE1 lentiviruses, from cultures infected with Mr-IAG lentiviruses, or from uninfected cultures (n = 7 for each). Total RNA isolation was performed using EZ-RNA Total RNA Isolation Kit (Biological Industries, Beit Haemek, Israel), and the first-strand cDNA was synthesized by reverse transcription using the qScript cDNA Kit (Quanta BioSciences, Gaithersburg, MD, United States) with 1 μg of total RNA. Relative quantification of transcript levels was performed using Roche Diagnostics FastStart Universal Probe Master Mix (Basel, Switzerland) and Roche Universal Probe Library probes. The following primers and probes were used: Probe #68 for MrLEDGF, qMrLEDGFF, and qMrLEDGFR, and Probe #25 for Mr-IAG, qMr-IAGF, and qMr-IAGR. M. rosenbergii 18S, which served as a normalizing gene, was also quantified by means of real-time RT PCR using the primers, qMr18SF and qMr18SR, Probe #106. All primer sequences are shown in Table 2. Reactions were performed with the ABI Prism7300 Sequence Detection System, Applied Biosystems (Foster City, CA, United States). Statistical analysis for relative transcript levels between the treatment groups was performed using the non-parametric Kruskal-Wallis rank sum test, followed by multiple pair-wise comparisons using the Wilcoxon rank-sum test; p < 0.05 was considered statistically significant.
Results
Lentiviral Particles Are Expressed in Macrobrachium rosenbergii Cells
To obtain a culture that was as long-lasting as possible, the first step was to confirm that our technique for modifying the commercially produced plasmid was indeed successful. To this end, we examined the exogenous expression of two fluorescent proteins inserted into cultured M. rosenbergii primary hematopoietic cells transduced with the novel lentiviral WSSV IE plasmid that had been constructed and generated in our laboratory (as described in section “Materials and Methods”; Figure 1). The expression of lentiviral sequences in samples of gDNA extracted from WSSV IE1 lentiviral infected cells was thus examined. As shown in Figure 3, the infected cells exhibited high levels of lentiviral sequences: ZsGreen and IE1-DsRed sequences were detected in three separate samples of infected cells, whereas in control uninfected cells, no amplification products of ZsGreen and IE1-DsRed sequences were detected (while the positive control Mr-Actin was detected in all genomic DNA extractions) (Figure 3). The expression of the reporter genes was further confirmed by confocal microscopy through examination of the expression of the two fluorescent proteins, ZsGreen and DsRed, encoded by the WSSV IE1 lentivirus. These proteins were detected (with the relevant lasers, 488 and 546 nm, respectively) in infected cells but not in control uninfected cells (Figure 4). The detection of the green and red fluorescent signals correlated with the nucleic-acid stained (DAPI) and brightfield images, implying that these fluorescent signals originated inside the cells. It is noteworthy that fluorescent signals were not detected in cells infected with EF1α lentiviruses, used as a negative control (data not shown).
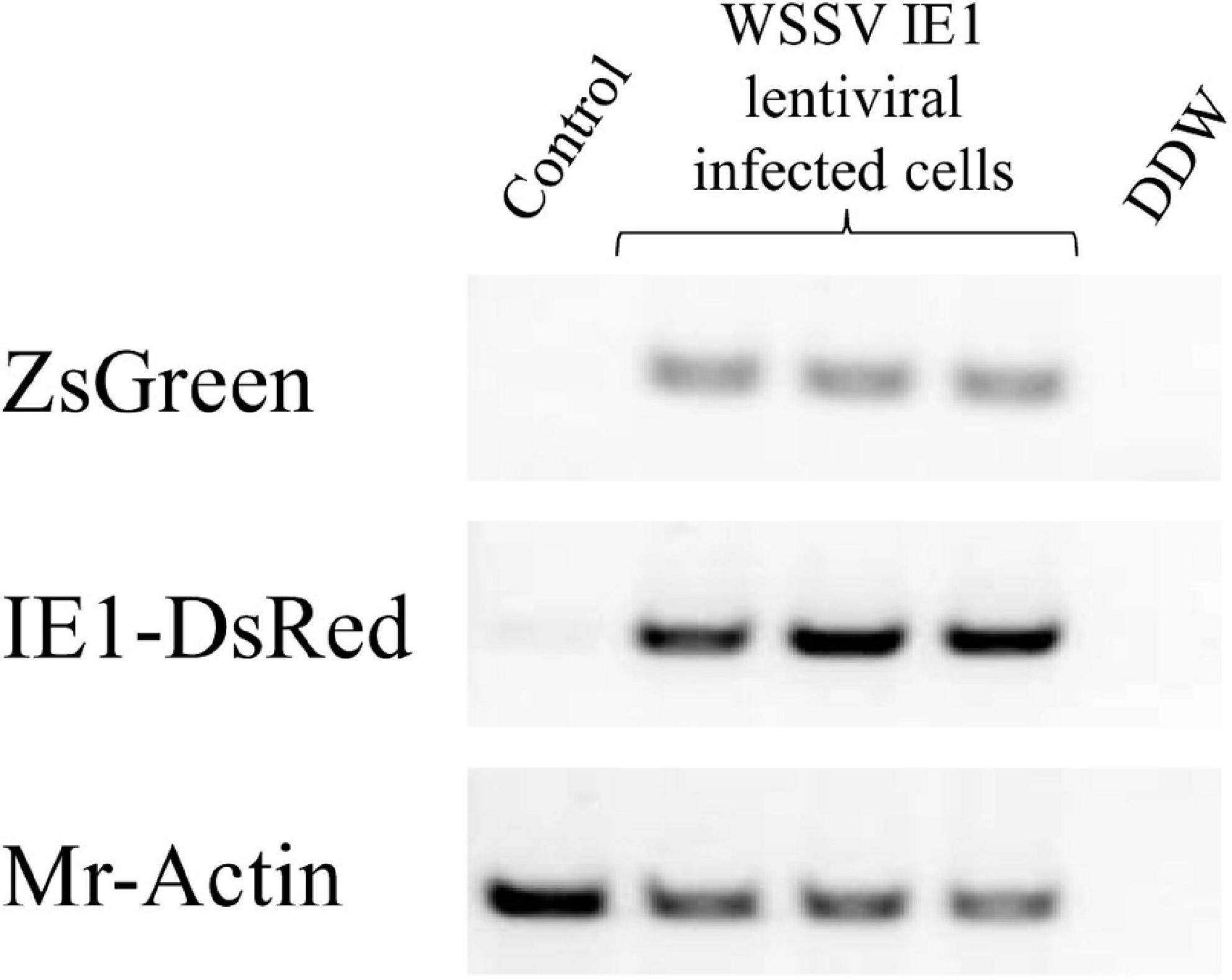
Figure 3. PCR analysis (using specific primers) of lentiviral sequences (ZsGreen and IE1-DsRed) in three different genomic DNA extractions from M. rosenbergii primary cells following transduction with WSSV IE1 lentiviruses. Mr-actin served as the positive control for M. rosenbergii cell cultures. Control, uninfected M. rosenbergii cells; DDW, double distilled water.
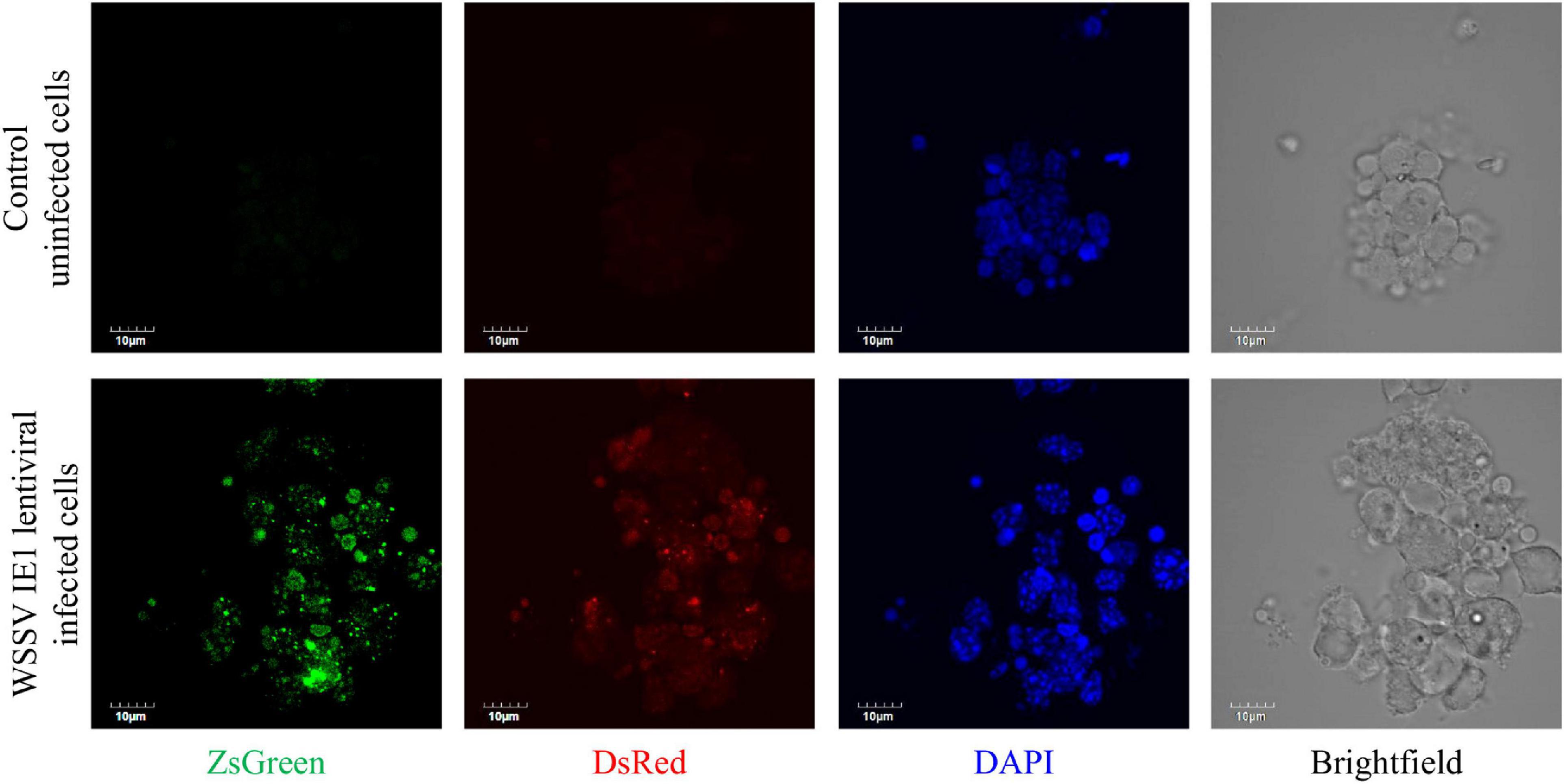
Figure 4. Expression of lentiviral particles in an M. rosenbergii primary cell culture. Representative images of WSSV IE1-infected M. rosenbergii primary cell culture (lower panel) or control uninfected cells (upper panel) grown only in culture medium. Cultures were examined by confocal microscopy (using the appropriate lasers, 488 nm for ZsGreen and 546 nm for DsRed). Nucleic acid staining was performed with DAPI (and visualized with a 405 nm laser). Scale bar = 10 μm.
Ectopic Macrobrachium rosenbergii Insulin-Like Androgenic Gland Expression in Macrobrachium rosenbergii Primary Cell Culture
To further evaluate the ability of our lentiviral system to insert novel genes into our cultured cells with the aim to achieve ectopic protein expression, we then constructed a new lentiviral plasmid encoding Mr-IAG and ZsGreen proteins under the WSSV IE1 promoter (as described in section “Materials and Methods”; Figure 2). The expression of Mr-IAG in M. rosenbergii is known to be specific to AG cells (Ventura et al., 2009), and thus any detection of Mr-IAG protein in cell culture would indeed be the result of the insertion of the gene by our lentiviral system, under the WSSV IE1 promoter. Following the transduction of the cell culture with Mr-IAG lentiviruses, the expression of the ZsGreen fluorescent protein was examined by confocal microscopy. As shown in Figure 5A, cells infected with WSSV IE1 or Mr-IAG lentiviruses expressed ZsGreen (middle panel and lower panel, respectively), whereas control uninfected cells did not (upper panel). Expression of the DsRed fluorescent protein was detected only in cell cultures infected with WSSV IE1 lentiviruses, as was to be expected from our constructed lentiviral plasmids, and control uninfected cells were visible only in the DAPI-stained and brightfield panels, also as was to be expected. To evaluate the efficiency of the lentiviral transduction in cell culture, we determined the percentage of infected cells by using FACS analysis. Populations of live cells that were also ZsGreen-positive, reaching 30–40% of the tested cells, were detected in both the WSSV IE1- and Mr-IAG-lentiviral infected cells, suggesting high lentiviral transduction efficiency and resulting in lentiviral protein expression in M. rosenbergii primary cell cultures (Figures 5B,C). In the cells infected with WSSV IE1 lentiviruses, ZsGreen-positive cells also expressed DsRed (Figures 5B,C). This co-expression of the two fluorescent proteins in the infected cells seen under the confocal microscope was confirmed by the elevated Y-axis in the two-dimensional FACS analysis (Figures 5A–C).
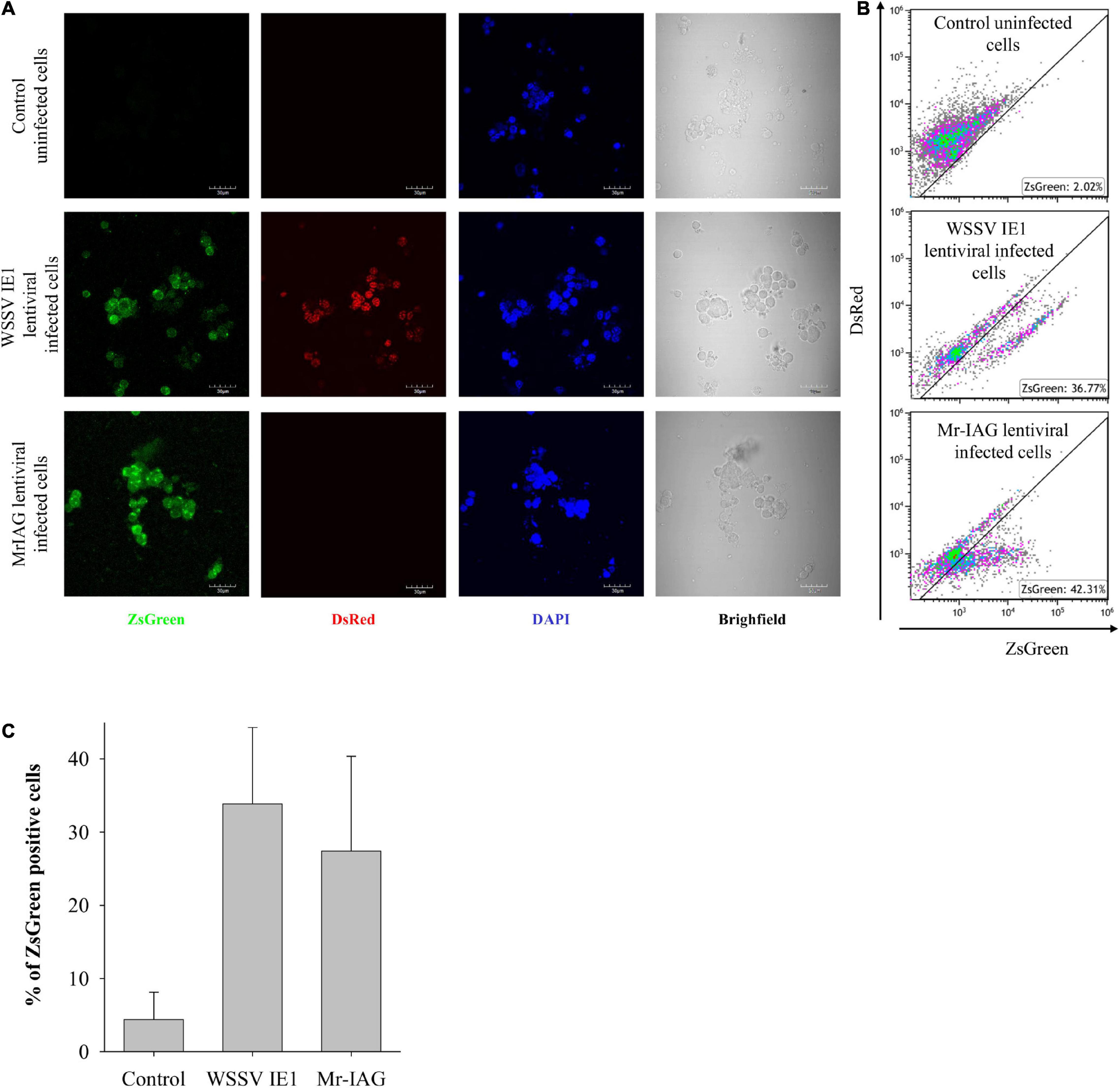
Figure 5. Efficient infection of M. rosenbergii primary cell cultures with WSSV IE1 and Mr-IAG lentiviruses. M. rosenbergii primary cell cultures were infected as indicated. (A) Representative confocal microscopy images of uninfected cells, WSSV IE1-infected cells, and Mr-IAG-infected cells. Scale bar = 10 μm. (B,C) FACS analysis of cells washed once with 1.4 × PBS (for molarity correction). Data are single discriminated and compensated. (B) Due to substantial autofluorescence of the M. rosenbergii primary cells, in order to determine specific signal, the analysis of the ZsGreen protein was performed using two-dimensional gates with DsRed and ZsGreen. The two-dimensional gating strategy is enabling gating below the autofluorescence diagonal of the specific signal. (C) Bar graph of three experiments analyzed by Flow Cytometry of ZsGreen positive populations; SD is indicated.
Furthermore, the insertion of Mr-IAG lentiviruses into the cells was supported by the detection of the Mr-IAG sequence in genomic DNA samples prepared from M. rosenbergii cells infected with Mr-IAG lentiviruses compared to WSSV IE1-infected and uninfected cells (Figure 6). In the Mr-IAG- infected or WSSV IE1-infected cells, enhanced expression of ZsGreen was detected, as expected, whereas DsRed was detected only in the WSSV IE1-lentiviral infected cells, and Mr-Actin was detected only in genomic DNA extractions from M. rosenbergii cells.
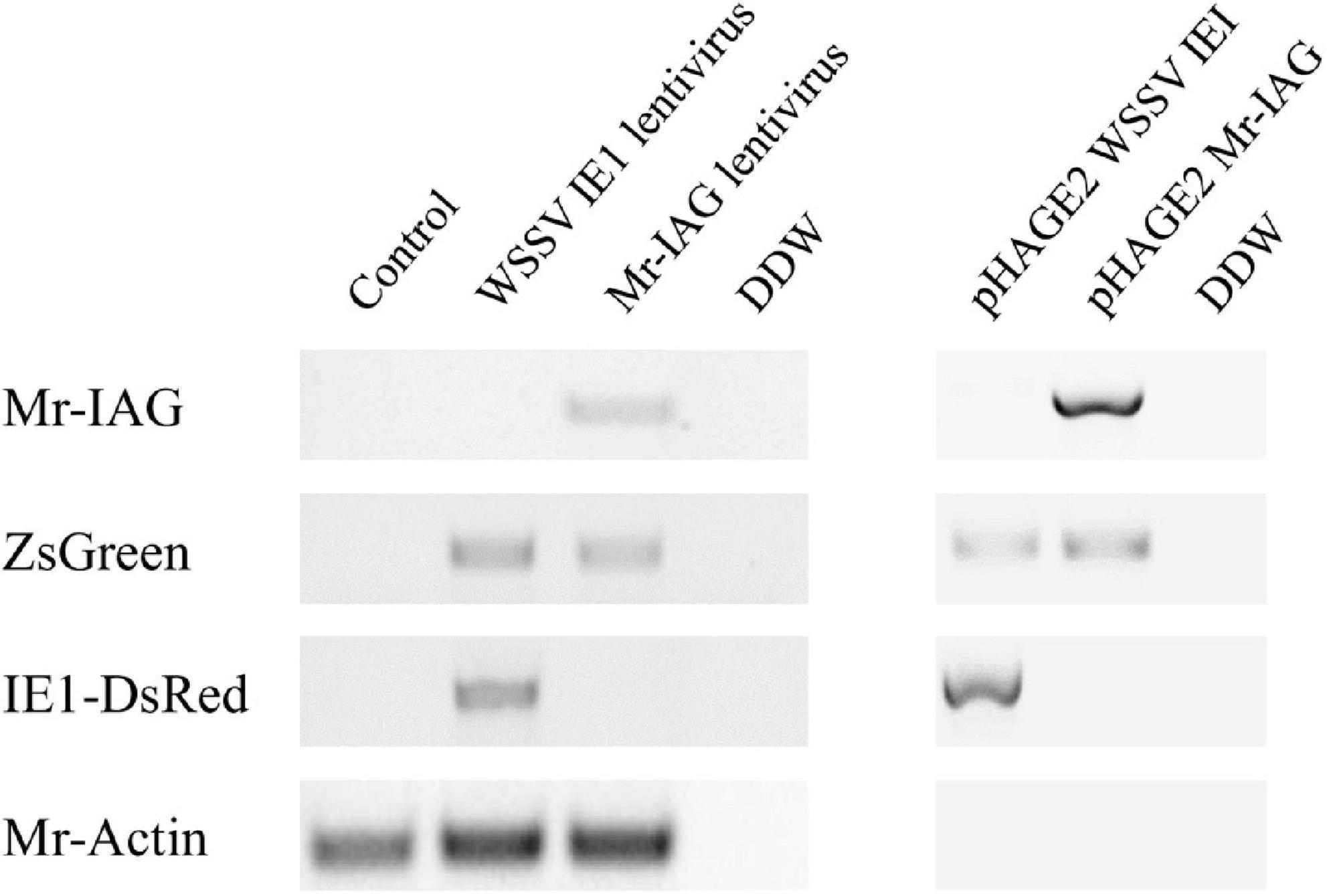
Figure 6. Detection of Mr-IAG sequence in gDNA of M. rosenbergii primary cells. Representation of lentiviral elements (Mr-IAG, ZsGreen, and IE1-DsRed) detected in an M. rosenbergii transduced cell culture (representative figure of three replicates). Extractions of genomic DNA were performed 96 h post-transduction (left panel). pHAGE2 WSSV IE1 and pHAGE2 Mr-IAG vectors served as the positive control for lentiviral sequences (right panel). Mr-Actin served as the positive control for M. rosenbergii cell cultures.
In addition, the expression profile of Mr-IAG in the infected cells was monitored by qPCR. M. rosenbergii cells infected with Mr-IAG lentiviruses exhibited significantly higher levels of Mr-IAG, compared to WSSV IE1-infected cells and control cells (Figure 7). Importantly, the levels of Mr-IAG in Mr-IAG-infected cells were similar to those in hAG RNA prepared directly from AG tissue.
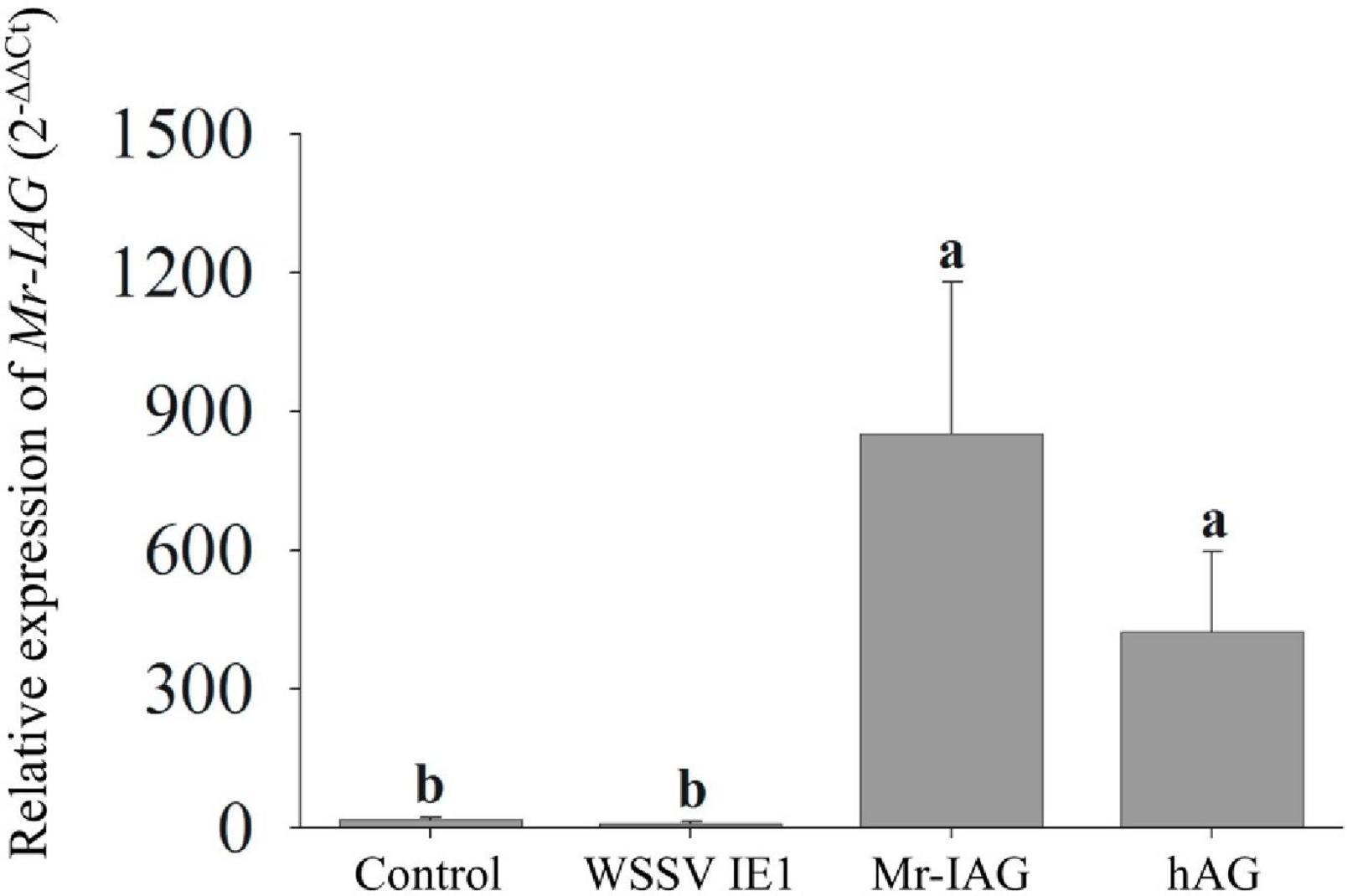
Figure 7. qPCR analysis of Mr-IAG expression levels in Mr-IAG transduced M. rosenbergii primary cell cultures. The relative levels of Mr-IAG expression in uninfected cells, WSSV IE1-infected cells, and Mr-IAG-infected cells were determined by qPCR. hAG served as the positive control. For all samples n = 5. Different letters represent groups that are significantly different (p < 0.05); error bars represent standard error.
The ectopic expression and secretion of Mr-IAG hormone in the cultured cells were determined by immunostaining and ELISA immunoassay, respectively, with specific Mr-IAG antibodies that had previously been generated and validated for Mr-IAG specificity in our laboratory (Ventura et al., 2011; Sharabi et al., 2016). As shown in Figure 8A, the expression of Mr-IAG was evident in the Mr-IAG lentivirus-infected cells and in AG primary culture cells but not in uninfected control cells (red color in Figure 8A). Since the synthesis of the Mr-IAG hormone occurs naturally in AG cells, these cells served as the positive control for Mr-IAG staining. As Mr-IAG lentiviruses carry the ZsGreen reporter gene, transduction also resulted in green fluorescent signals, exhibited only in Mr-IAG infected cells (middle panel). This finding was supported by the fact that co-staining of Mr-IAG and ZsGreen was found in Mr-IAG lentiviral infected cells but not in AG cells, which immunostained only for the Mr-IAG protein.
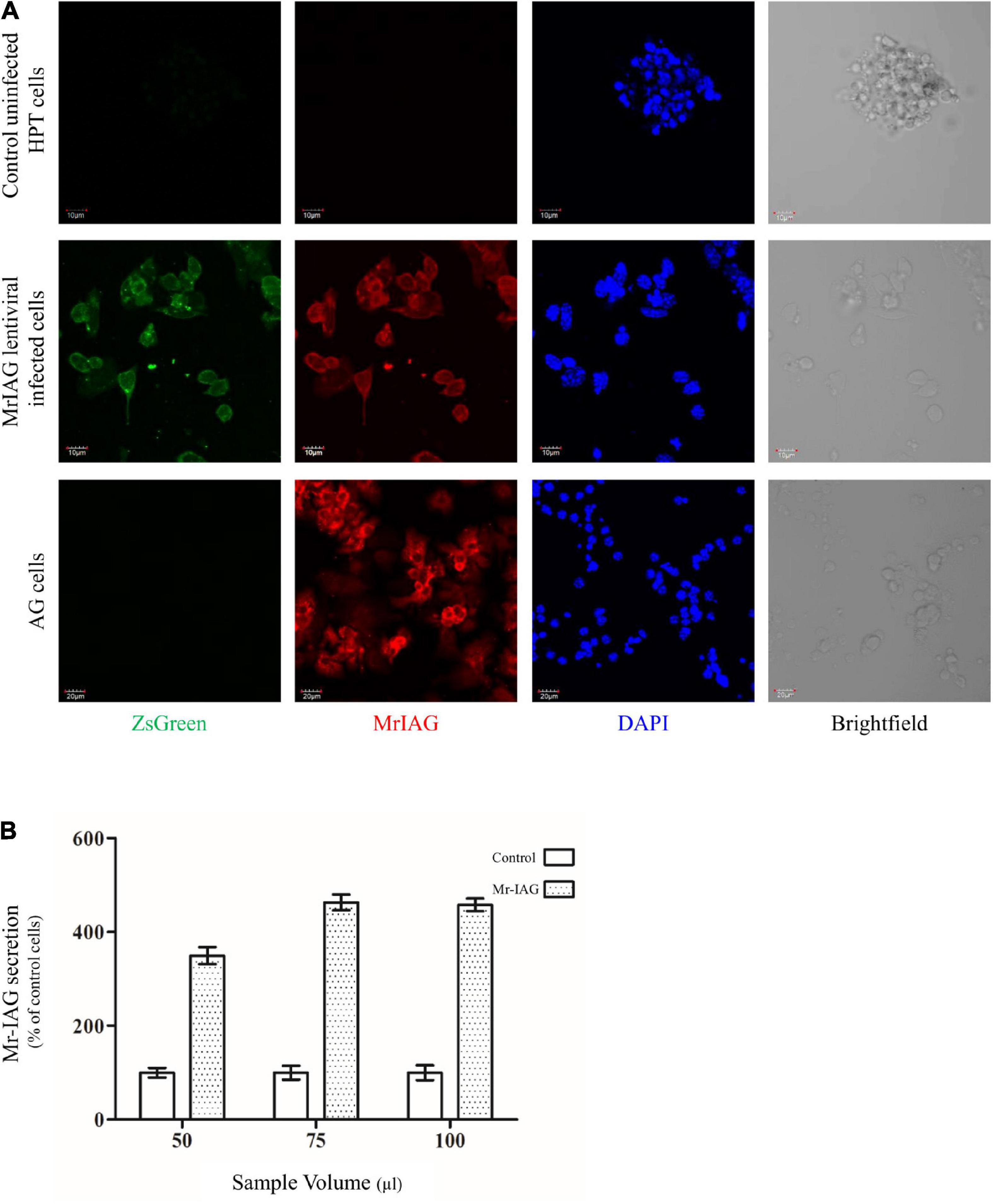
Figure 8. Mr-IAG hormone synthesis and secretion by an M. rosenbergii hematopoietic primary cell culture following transduction with the Mr-IAG lentivirus. (A) Representative images of control uninfected M. rosenbergii primary cells, Mr-IAG-infected cells, and AG primary cells, which served as the positive control. Cells were immunostained with a specific antibody against Mr-IAG and examined using confocal microscopy with the appropriate lasers (488 nm for ZsGreen and red signal at 546 nm for secondary donkey anti-rabbit IgG Alexa Fluor) for Mr-IAG detection. Nucleic acid staining was performed with DAPI. Scale bar = 10 or 20 μm, as indicated. (B) Relative quantitative secretion of Mr-IAG into the culture medium by Mr-IAG lentivirus-infected M. rosenbergii cells. Four days following transduction of M. rosenbergii cells with Mr-IAG lentiviruses, Mr-IAG protein was detected using sandwich ELISA in different volumes of cell culture medium (50, 75, and 100 μl) from Mr-IAG infected cells (spotted bars) or uninfected cells (clear bars). Y-axis represents the percentage of Mr-IAG hormone secreted relative to control in each volume.
Next, since the Mr-IAG hormone has a signal peptide serving for excretion from the cells, the levels of secreted Mr-IAG in the culture medium were quantified by Mr-IAG sandwich ELISA. Mr-IAG-infected cells demonstrated relatively higher – by 4.5-fold – levels of Mr-IAG vs control uninfected cells (Figure 8B). Taken together, our results indicate that Mr-IAG-infected cells are able to transcribe (Figure 7), translate, and finally ectopically express and secrete (Figure 8) the Mr-IAG protein as a result of Mr-IAG lentiviral transduction.
Macrobrachium rosenbergii Lens Epithelium-Derived Growth Factor Indicates Lentiviral Integration Into the Genome of Macrobrachium rosenbergii Cells
The sequence of MrLEDGF was found in the M. rosenbergii transcriptomic library, having a typical domain organization of PWWP in its N-terminal and an LEDGF domain in the C-terminal (Busschots et al., 2005), as shown in Figure 9. Expression levels of MrLEDGF were significantly higher in the WSSV IE1- and Mr-IAG-infected cells than in the control unifected cells, as shown in Figure 10. Since integrase-mediated integration of lentiviral sequences into genomic host DNA takes place through the host LEDGF cofactor, the elevated level of MrLEDGF in primary infected cells implies DNA integration of the lentiviral particles.
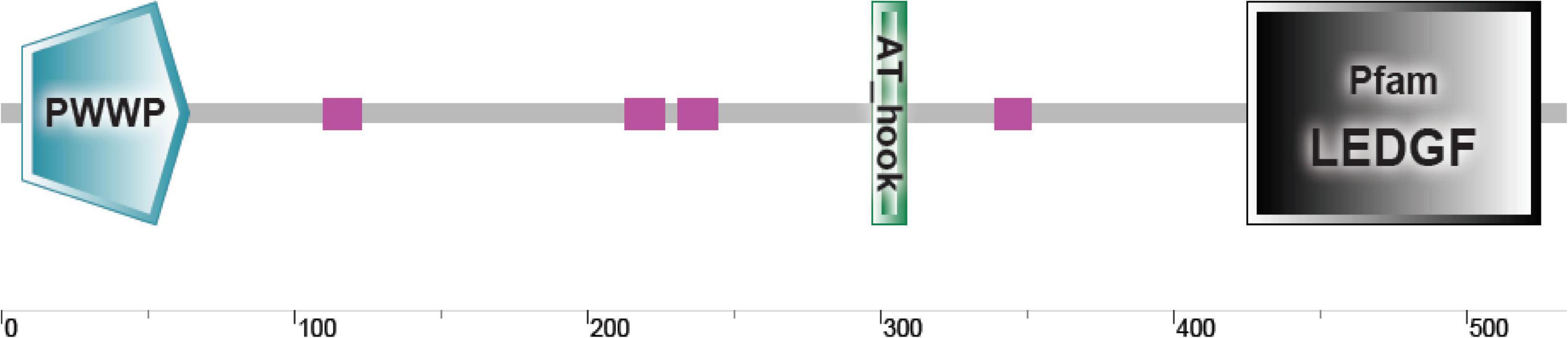
Figure 9. MrLEDGF sequence domain organization. Schematic representation of newly discovered MrLEDGF and its domain organization as predicted by SMART.
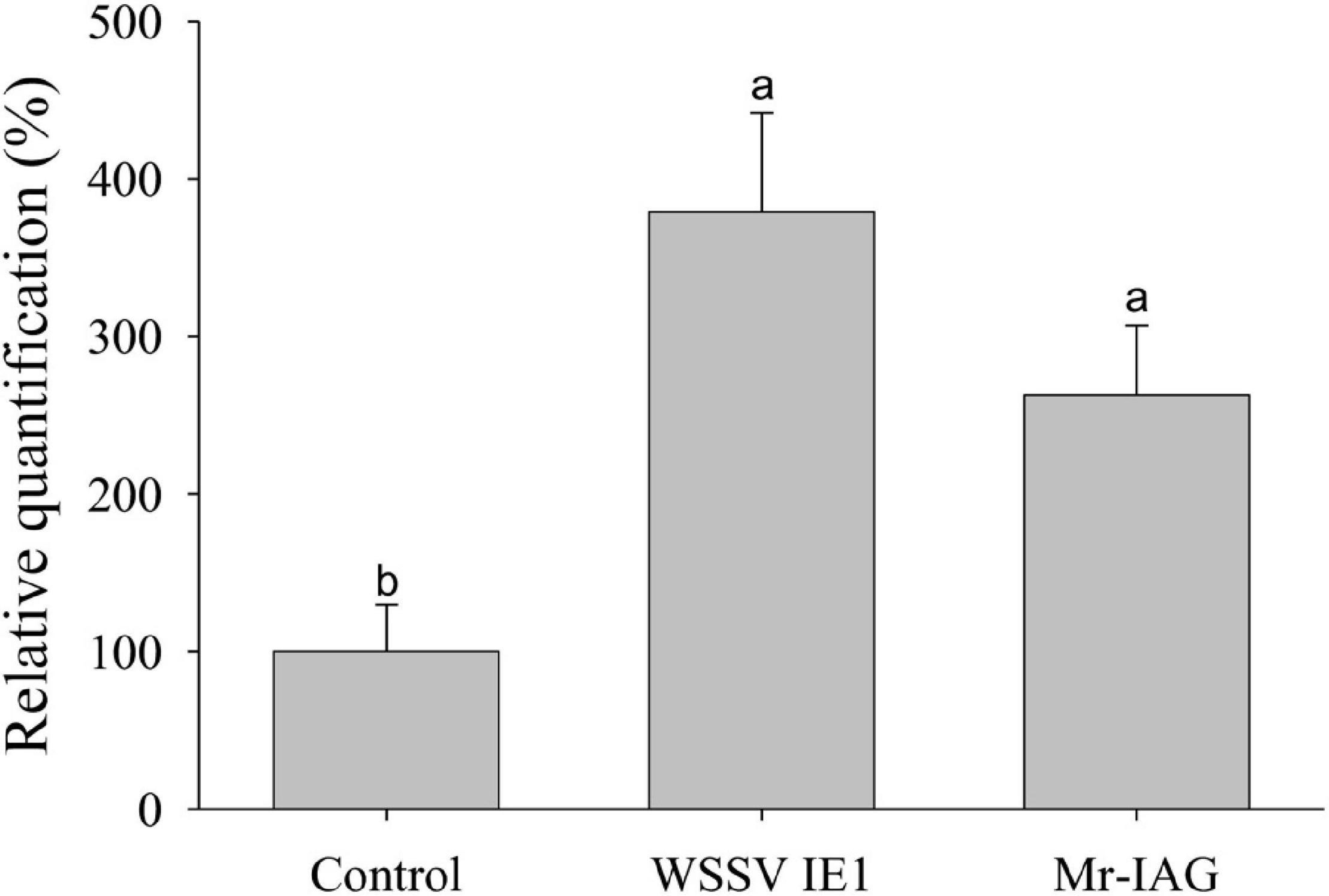
Figure 10. qPCR analysis of MrLEDGF expression level in Mr-IAG transduced M. rosenbergii primary cell cultures. The relative quantification of the levels of MrLEDGF expression in uninfected cells, WSSV IE1-infected cells, and Mr-IAG-infected cells were determined by qPCR and normalized to control group and presented as relative quantification (%). For all samples n = 7. Different letters represent groups that are significantly different (p < 0.05); error bars represent standard error.
Discussion
In view of the dimorphic growth pattern of many crustacean aquaculture species, biotechnological tools – based on sex determination and differentiation mechanisms – have been developed to increase yields through monosex culture (Aflalo et al., 2006; Malecha, 2012; Ventura et al., 2012; Levy et al., 2016). Our goal was to establish a method that could improve the first step of a recently established biotechnology for all-female M. rosenbergii aquaculture in which sex reversal is achieved by an injection into post-larvae of primary AG cells from mature M. rosenbergii males (Levy et al., 2016). In other words, we sought to replace the AG-based cell source of Mr-IAG with Mr-IAG expressed in primary cells in culture. In the present study, we demonstrate, for the first time, the successful ectopic expression and secretion of Mr-IAG following lentiviral transduction of an M. rosenbergii primary hematopoietic cell culture.
The difficulties associated with the delivery of genes into crustacean cells continue to be an obstacle in the study of cellular and molecular mechanisms of crustacean species. Although considerable research using a variety of techniques has focused on the establishment of an efficient platform to insert and express a foreign gene in primary crustacean cell cultures, this research has met only with limited success, due to the difficulty of delivering foreign DNA into primary crustacean cell cultures, which enter a mitosis-arrest phase a short time after harvesting (Tapay et al., 1995; Shike et al., 2000; Lu et al., 2005; Claydon and Owens, 2008; Hu et al., 2008, 2010; Li et al., 2011; Han et al., 2015; Puthumana et al., 2015, 2016; Pu et al., 2017; Shi et al., 2018). We thus sought to leverage the ability of lentiviruses to infect both dividing and non-dividing cells by constructing a lentiviral vector that would be efficiently expressed in primary crustacean cell cultures. The lentiviral vector used here is designed in such a way that it can contain a target gene under the WSSV IE1 promoter, which is known to be very active in crustacean cells. Recently, the use of lentiviral transduction was demonstrated for the first time in penaeid shrimp cells (Chen et al., 2019). However, the drawback associated with the system used in that study was that the expression of the reporter gene gave a relatively low intensity of fluorescent signals, as a result of the use of a lentiviral vector pLVX-PEF1α -Pie1 that contains a human promoter and an additional WSSV promoter, unlike the present study. The lentiviral system was composed of second-generation structural and envelope components and additional envelope proteins of WSSV to produce the lentiviruses in the cell lines (Chen et al., 2019). In contrast, we constructed a lentiviral platform containing only one promoter, the WSSV IE1 promoter, and together with a fourth-generation packaging mix easily produce the desired lentiviruses. Moreover, our efficient lentiviral platform for the transduction of primary M. rosenbergii cells gained constitutive expression ex vivo, implying that the host cells had transcribed and translated the lentiviral sequences. Notably, although any large gene of interest could be inserted into the lentiviral vector, there remains a packging limit of a few thousands of base pairs (Kumar et al., 2001).
The replacement of the human constitutive EF1α promoter region on the lentiviral plasmid with the WSSV IE1 promoter produced an efficient lentiviral gene delivery platform for primary cultures of M. rosenbergii hematopoietic cells. Previous studies have attempted to improve gene delivery systems by using WSSV promoters to induce the expression of reporter genes, but only with limited success, possibly due to their use of an inapproriate electroporation technique for gene delivery into primary cell cultures, yielding low levels of expression of the reporter gene (Li et al., 2011; Shi et al., 2018). In the not so distant past, Pu and his colleagues attempted to overcome the tropism of pseudotyped retroviruses by the inclusion of VP19 and VP28 into the envelope of the packaged retrovirus (Pu et al., 2017), but our use of a lentivirus vector instead of a retrovirus has circumvented this need, since the successful expression of the lentiviral proteins in M. rosenbergii hematopoietic primary cell culture demonstrates high tropism of the lentiviruses to these cells. Our study suggests that the commonly used packaging lentiviral vector is capable of enhancing the expression of the desired target gene.
Next, we aimed to ectopically express – in M. rosenbergii hematopoietic cells – the Mr-IAG hormone, which is a key regulator within the IAG-switch controlling sexual differentiation (Ventura et al., 2012; Levy and Sagi, 2020). In M. rosenbergii, MrIAG is uniquely expressed in AG cells (Ventura et al., 2009), and thus detection of the MrIAG mRNA or the Mr-IAG protein in the hematopoietic cells may be regarded as proof of lentiviral insertion. Indeed, we constructed a novel lentiviral vector that includes the MrIAG full mRNA sequence under WSSV IE1 promoter, which yielded a 30–40% population of positive cells compared to uninfected cells. In contrast to previous studies that evaluated fluorescent signals only microscopically (Li et al., 2011; Puthumana et al., 2015, 2016; Pu et al., 2017; Shi et al., 2018; Chen et al., 2019), the quantitative data from flow cytometry analysis in our study has the advantage of offering efficient fluorescent detection and the validation of cell viability. Importantly, Mr-IAG levels in the primary hematopoietic cells were similar to those in the hypertrophied AG cells, suggesting that the MrIAG-transduced hematopoietic cells could indeed replace these cells in the first step of the biotechnology for the generation of all-female aquaculture. To summarize, M. rosenbergii primary cells infected with Mr-IAG lentiviruses acquire the ability to transcribe, translate, and secrete Mr-IAG in culture, namely, a new trait of ectopic expression of a target protein that results from the lentiviral transduction. In the longer term, the replacement of AG cells with the novel Mr-IAG expressing cells in the biotechnology for the production of all-female M. rosenbergii aquaculture, followed by a series of in vivo optimization experiments, will improve and facilitate the widespread implementation of the biotechnology.
Let us now take a brief look at the broader implications of this study. Since lentiviruses cause stable transfection by integration of their sequences into the host genomic DNA (Vigna and Naldini, 2000; Durand and Cimarelli, 2011), cells in primary culture could acquire novel protein expression, which will remain as long as the culture is maintained. Our results suggest that lentiviral integration did indeed occur, since the levels of MrLEDGF, which plays a role in promoting viral cDNA integration into host genomic DNA through its interaction with the viral integrase (Busschots et al., 2005; Hombrouck et al., 2007), increased following lentiviral transduction of cells. Thus, our novel lentiviral platform could provide a useful tool for the ectopic expression of any gene of interest.
The platform is likely to find broad application in the production of monosex aquaculture populations. The strategy for growing monosex crustacean populations evolved from fish-based aquaculture, for example, the production of monosex male populations of tilapia (Oreochromis spp.) via the administration of the synthetic hormone 17α-methyltestosterone (17α-MT), which acts similarly to naturally produced testosterone (Green and Teichert-Coddington, 2000; Wahby and Shalaby, 2010). Previous studies in M. rosenbergii failed to demonstrate sex reversal through feeding the animals with 17α-MT (Baghel et al., 2004; Ohs et al., 2006), but there are other biotechnological methods for sex reversal that are based on microsurgical and cell transplantation methods (Aflalo et al., 2006; Levy et al., 2016) that are more sustainable and that do not include hormonal treatments that might affect other organisms in co-culture. The induction of ectopic hormone expression in a primary cell culture through lentiviral transduction would provide a technology for sex reversal – by injection of the hormone-expressing cells into a live animal – without causing any gene modifications or damaging the genome of the animal. As in monosex fish cultures in which sex reversal is generated by administration of a single factor (such as a synthetic hormone), the lentiviral method introduces a single factor causing sex reversal via manipulation of the IAG-switch (Levy and Sagi, 2020). Since the latter switch is conserved in all decapod crustaceans studied thus far (Levy and Sagi, 2020), our platform could also be harnessed for monosex population biotechnologies in other aquaculturally important crustacean species.
Conclusion
This study shows for the first time the establishment of an efficient and effective lentiviral transduction platform for primary cells, which will enable the manipulation of gene expression in primary crustacean cell culture. Induction of the expression of any target gene can be achieved through lentivirus transduction, and the current study serves as an example for a future commercial use. Ultimately, the exploitation of lentiviral gene delivery in the manipulation of other signaling pathways, such as cell proliferation, could reveal the factors that affect and elicit cell proliferation, eventually promoting the development of continuous crustacean cell lines for molecular studies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
NRD and SW conceived the manuscript, conducted the experiments, data analysis, and figure production, and wrote the manuscript. YG and BR conducted flow cytometry analysis. SA conducted real-time PCR analysis and bioinformatic analyses. All authors contributed to the final manuscript preparation.
Funding
This work was supported by the Israel Ministry of Science, Technology and Space (Grant No. 13610-3 to NRD). The work of BR was supported by the Israel Science Foundation (ISF, Grants Nos. 1416/19 and 2841/19). Further partial support was provided by Enzootic Genetics and Innovation, the Israel Science Foundation (ISF) within the ISF-UGC joint research program framework (Grant No. 2728/2016), and the ISF-NSFC joint research program (Grant No. 2368/2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Acheampong, E., Rosario-Otero, M., Dornburg, R., and Pomerantz, R. J. (2003). Replication of Lentiviruses. Front. Biosci. 8:156–174. doi: 10.2741/935
Aflalo, E. D., Hoang, T. T. T., Nguyen, V. H., Lam, Q., Nguyen, D. M., Trinh, Q. S., et al. (2006). A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 256, 468–478. doi: 10.1016/j.aquaculture.2006.01.035
Baghel, D. S., Lakra, W. S., and Satyanarayana Rao, G. P. (2004). Altered sex ratio in giant fresh water prawn, Macrobrachium rosenbergii (de Man) using hormone bioencapsulated live Artemia feed. Aquac. Res. 35, 943–947. doi: 10.1111/j.1365-2109.2004.01104.x
Benskey, M. J., and Manfredsson, F. P. (2016). “Lentivirus production and purification,” in Gene Therapy for Neurological Disorders: Methods and Protocols, ed. F. P. Manfredsson (New York, NY: Springer), 107–114. doi: 10.1007/978-1-4939-3271-9_8
Busschots, K., Vercammen, J., Emiliani, S., Benarous, R., Engelborghs, Y., Christ, F., et al. (2005). The interaction of LEDGF/p75 with integrase is Lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280, 17841–17847. doi: 10.1074/jbc.m411681200
Chen, X., Chen, Y., Shen, X., Zuo, J., and Guo, H. (2019). The improvement and application of Lentivirus-mediated gene transfer and expression system in penaeid shrimp cells. Mar. Biotechnol. 21, 9–18. doi: 10.1007/s10126-018-9862-0
Claydon, K., and Owens, L. (2008). Attempts at immortalization of crustacean primary cell cultures using human cancer genes. In Vitro Cell. Dev. Biol. Anim. 44, 451–457. doi: 10.1007/s11626-008-9141-x
Durand, S., and Cimarelli, A. (2011). The inside out of lentiviral vectors. Viruses 3, 132–159. doi: 10.3390/v3020132
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S.e, Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607. doi: 10.1385/1-59259-890-0:571
Green, B. W., and Teichert-Coddington, D. R. (2000). Human food safety and environmental assessment of the use of 17α-methyltestosterone to produce male tilapia in the United States. J. World Aquac. Soc. 31, 337–357. doi: 10.1111/j.1749-7345.2000.tb00885.x
Han, Q., Dong, D., Zhang, X., Liang, C., Lu, Q., and Guo, H. (2015). Problems with the use of liposome-and retrovirus-mediated gene transfer methods in the primary lymphoid cells of the Oka organs of the greasyback shrimp, Metapenaeus ensis (De Haan, 1844). Crustaceana 88, 1351–1365. doi: 10.1163/15685403-00003498
Hombrouck, A., De Rijck, J., Hendrix, J., Vandekerckhove, L., Voet, A., Maeyer, M. D., et al. (2007). Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathogens 3:e47. doi: 10.1371/journal.ppat.0030047
Hu, G.-b, Wang, D., Wang, C.-h, and Yang, K.-f (2008). A novel immortalization vector for the establishment of penaeid shrimp cell lines. In Vitro Cell. Dev. Biol. Anim. 44, 51–56. doi: 10.1007/s11626-007-9076-7
Hu, G.-B., Wang, D., and Chen, J.-X. (2010). Retroviral delivery of simian virus 40 large t antigen gene into primary cultured ovary cells of the penaeid shrimp, penaeus chinensis : indirect evidence of retroviral integration. J. World Aquac. Soc. 41, 137–143. doi: 10.1111/j.1749-7345.2009.00321.x
Huang, X.-D., Zhao, L., Zhang, H.-Q., Xu, X.-P., Jia, X.-T., Chen, Y.-H., et al. (2010). Shrimp NF-κB binds to the immediate-early gene ie1 promoter of white spot syndrome virus and upregulates its activity. Virology 406, 176–180. doi: 10.1016/j.virol.2010.06.046
Jayesh, P., Seena, J., and Singh, I. S. (2012). Establishment of shrimp cell lines: perception and orientation. Indian J. Virol. 23, 244–251. doi: 10.1007/s13337-012-0089-9
Johansson, M. W., Keyser, P., Sritunyalucksana, K., and Söderhäll, K. (2000). Crustacean haemocytes and haematopoiesis. Aquaculture 191, 45–52. doi: 10.1016/S0044-8486(00)00418-X
Khalaila, I., Manor, R., Weil, S., Granot, Y., Keller, R., and Sagi, A. (2002). The eyestalk-androgenic gland-testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 127, 147–156. doi: 10.1016/S0016-6480(02)00031-X
Kumar, M., Keller, B., Makalou, N., and Sutton, R. E. (2001). Systematic determination of the packaging limit of Lentiviral vectors. Hum. Gene Ther. 12, 1893–1905. doi: 10.1089/104303401753153947
Kuris, A. M., Ra’anan, Z., Sagi, A., and Cohen, D. (1987). Morphotypic differentiation of male malaysian giant prawns, Macrobrachium Rosenbergii. J. Crustac. Biol. 7, 219–237. doi: 10.2307/1548603
Levy, T., and Sagi, A. (2020). The “IAG-Switch”-a key controlling element in decapod crustacean sex differentiation. Front. Endocrinol. 11:651. doi: 10.3389/fendo.2020.00651
Levy, T., Rosen, O., Eilam, B., Azulay, D., Aflalo, E. D., Manor, R., et al. (2016). A single injection of hypertrophied androgenic gland cells produces all-female aquaculture. Mar. Biotechnol. 18, 554–563. doi: 10.1007/s10126-016-9717-5
Levy, T., Rosen, O., Eilam, B., Azulay, D., Zohar, I., Aflalo, E. D., et al. (2017). All-female monosex culture in the freshwater prawn Macrobrachium Rosenbergii-a comparative large-scale field study. Aquaculture 479, 857–862. doi: 10.1016/j.aquaculture.2017.07.039
Li, F., Ke, W., Yan, X., and Xu, L. (2011). Gene transfection and expression in the primary culture of crayfish hemocytes. Fish Shellfish Immunol. 31, 161–163. doi: 10.1016/j.fsi.2011.04.002
Li, F., Li, M., Ke, W., Ji, Y., Bian, X., and Yan, X. (2009). Identification of the immediate-early genes of white spot syndrome virus. Virology 385, 267–274. doi: 10.1016/j.virol.2008.12.007
Lin, F., Huang, H., Ke, W., Hou, L., Li, F., and Yang, F. (2013). Characterization of white spot syndrome virus immediate-early gene promoters. J. Gen. Virol. 94(Pt 2), 387–392. doi: 10.1099/vir.0.047274-0
Lin, X., and Söderhäll, I. (2011). Crustacean hematopoiesis and the astakine cytokines. Blood 117, 6417–6424. doi: 10.1182/blood-2010-11-320614
Lin, X., Söderhäll, K., and Söderhäll, I. (2008). Transglutaminase activity in the hematopoietic tissue of a crustacean. Pacifastacus leniusculus, importance in hemocyte homeostasis. BMC Immunol. 9, 1–11. doi: 10.1186/1471-2172-9-58
Liu, W. J., Chang, Y. S., Wang, A. H., Kou, G. H., and Lo, C. F. (2007). White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 81, 1461–1471. doi: 10.1128/jvi.01880-06
Lu, L., Wang, H., Manopo, I., Yu, L., and Kwang, J. (2005). Baculovirus-mediated promoter assay and transcriptional analysis of white spot syndrome virus orf427 gene. Virol. J. 2:71. doi: 10.1186/1743-422x-2-71
Lundstrom, K. (2019). RNA viruses as tools in gene therapy and vaccine development. Genes 10:189. doi: 10.3390/genes10030189
Malecha, S. (2012). The case for all-female freshwater prawn, Macrobrachium rosenbergii (De Man), culture. Aquac. Res. 43, 1038–1048. doi: 10.1111/j.1365-2109.2011.03007.x
Malecha, S. R., Nevin, P. A., Ha, P., Barck, L. E., Lamadrid-Rose, Y., Masuno, S., et al. (1992). Sex-ratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii. Aquaculture 105, 201–218. doi: 10.1016/0044-8486(92)90087-2
Manor, R., Weil, S., Oren, S., Glazer, L., Aflalo, E. D., Ventura, T., et al. (2007). Insulin and gender: an insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 150, 326–336. doi: 10.1016/j.ygcen.2006.09.006
Mostoslavsky, G., Kotton, D., Fabian, A., Gray, J., Lee, J.-S., and Mulligan, R. (2005). Efficiency of transduction of highly purified murine hematopoietic stem cells by Lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 11, 932–940. doi: 10.1016/j.ymthe.2005.01.005
Nagamine, C., Knight, A. W., Maggenti, A., and Paxman, G. (1980). Effects of androgenic gland ablation on male primary and secondary sexual characteristics in the Malaysian prawn, Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae), with first evidence of induced feminization in a nonhermaphroditic decapod. Gen. Comp. Endocrinol. 41, 423–441. doi: 10.1016/0016-6480(80)90048-9
Ohs, C., D’Abramo, L., and Kelly, A. (2006). Effect of Dietary Administration of 17α-methyltestosterone on the sex ratio of postlarval freshwater prawn, Macrobrachium rosenbergii, during the nursery stage of culture. J. World Aquac. Soc. 37, 328–333. doi: 10.1111/j.1749-7345.2006.00045.x
Olender, L., Bujanover, N., Sharabi, O., Goldstein, O., and Gazit, R. (2020). Cyclosporine H improves the multi-vector Lentiviral Transduction of murine haematopoietic progenitors and Stem Cells. Sci. Rep. 10:1812. doi: 10.1038/s41598-020-58724-x
Pu, L., Wang, J., Zhang, X., and Guo, H. (2017). Development of pseudotyped retroviral system for effective gene transfer and expression in penaeid shrimp cells. Aquaculture 467, 198–210. doi: 10.1016/j.aquaculture.2016.04.014
Puthumana, J., Philip, R., and Bright Singh, I. S. (2016). Transgene expression in Penaeus monodon cells: evaluation of recombinant baculoviral vectors with shrimp specific hybrid promoters. Cytotechnology 68, 1147–1159. doi: 10.1007/s10616-015-9872-y
Puthumana, J., Prabhakaran, P., Philip, R., and Singh, I. S. (2015). Attempts on producing lymphoid cell line from Penaeus monodon by induction with SV40-T and 12S EIA oncogenes. Fish Shellfish Immunol. 47, 655–663. doi: 10.1016/j.fsi.2015.08.010
Qiu, W., Zhang, S., Chen, Y. G., Wang, P. H., Xu, X. P., Li, C. Z., et al. (2014). Litopenaeus vannamei NF-κB is required for WSSV replication. Dev. Comp. Immunol. 45, 156–162. doi: 10.1016/j.dci.2014.02.016
Rinkevich, B. (2005). Marine invertebrate cell cultures: new millennium trends. Mar. Biotechnol. 7, 429–439. doi: 10.1007/s10126-004-0108-y
Rinkevich, B. (2011). Cell cultures from marine invertebrates: new insights for capturing endless stemness. Mar. Biotechnol. (N. Y.) 13, 345–354. doi: 10.1007/s10126-010-9354-3
Sagi, A., Cohen, D., and Milner, Y. (1990). Effect of androgenic gland ablation on morphotypic differentiation and sexual characteristics of male freshwater prawns, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 77, 15–22. doi: 10.1016/0016-6480(90)90201-v
Sagi, A., Ra’anan, Z., Cohen, D., and Wax, Y. (1986). Production of Macrobrachium rosenbergii in monosex populations: yield characteristics under intensive monoculture conditions in cages. Aquaculture 51, 265–275. doi: 10.1016/0044-8486(86)90318-2
Sagi, A., Snir, E., and Khalaila, I. (1997). Sexual differentiation in decapod crustaceans: role of the androgenic gland. Invertebr. Reprod. Dev. 31, 55–61. doi: 10.1080/07924259.1997.9672563
Sánchez-Martínez, J. G., Aguirre-Guzmán, G., and Mejía-Ruíz, H. (2007). White spot syndrome virus in cultured shrimp: a review. Aquac. Res. 38, 1339–1354. doi: 10.1111/j.1365-2109.2007.01827.x
Sánchez-Paz, A. (2010). White spot syndrome virus: an overview on an emergent concern. Vet. Res. 41:26.
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Sena-Esteves, M., and Gao, G. (2018). Production of high-titer retrovirus and Lentivirus Vectors. Cold Spring Harb. Protoc. 2018, 273–280. doi: 10.1101/pdb.prot095687
Sharabi, O., Manor, R., Weil, S., Aflalo, E. D., Lezer, Y., Levy, T., et al. (2016). Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 157, 928–941. doi: 10.1210/en.2015-1391
Shi, H., Ruan, L., Söderhäll, I., Söderhäll, K., and Xu, X. (2018). Transfection of crayfish hematopoietic tissue cells. Dev. Comp. Immunol. 88, 70–76. doi: 10.1016/j.dci.2018.07.005
Shike, H., Shimizu, C., Klimpel, K. S., and Burns, J. C. (2000). Expression of foreign genes in primary cultured cells of the blue shrimp Penaeus stylirostris. Mar. Biol. 137, 605–611. doi: 10.1007/s002270000381
Sirikharin, R., Utairungsee, T., Srisala, J., Roytrakul, S., Thitamadee, S., and Sritunyalucksana, K. (2019). Cell surface transglutaminase required for nodavirus entry into freshwater prawn hemocytes. Fish Shellfish Immunol. 89, 108–116. doi: 10.1016/j.fsi.2019.03.052
Soderhall, I., Bangyeekhun, E., Mayo, S., and Soderhall, K. (2003). Hemocyte production and maturation in an invertebrate animal; proliferation and gene expression in hematopoietic stem cells of Pacifastacus leniusculus. Dev. Comp. Immunol. 27, 661–672. doi: 10.1016/s0145-305x(03)00039-9
Tapay, L. M., Lu, Y., Brock, J. A., Nadala, E. C. Jr., and Loh, P. C. (1995). Transformation of primary cultures of shrimp (Penaeus stylirostris) lymphoid (Oka) organ with Simian virus-40 (T) antigen. Proc. Soc. Exp. Biol. Med. 209, 73–78. doi: 10.3181/00379727-209-43880
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Khalaila, I., Rosen, O., et al. (2011). Expression of an androgenic gland-specific insulin-like peptide during the course of prawn sexual and morphotypic differentiation. ISRN Endocrinol. 476283:11.
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Raviv, S., Glazer, L., et al. (2009). Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 150, 1278–1286. doi: 10.1210/en.2008-0906
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Rosen, O., and Sagi, A. (2012). Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, macrobrachium rosenbergii1. Biol. Reprod. 86:90. doi: 10.1095/biolreprod.111.097261
Verbruggen, B., Bickley, L. K., van Aerle, R., Bateman, K. S., Stentiford, G. D., Santos, E. M., et al. (2016). Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses 8:23. doi: 10.3390/v8010023
Vigna, E., and Naldini, L. (2000). Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2, 308–316. doi: 10.1002/1521-2254(200009/10)2:5<308::aid-jgm131>3.0.co;2-3
Wahby, O. M., and Shalaby, S. H. (2010). Oral administration of testosterone in fish diet affect sex differentiation and testis development in tilapia. Res. J. Agric. Biol. Sci. 6, 946–952.
Keywords: androgenic gland (AG), hematopoietic cells, insulin-like androgenic gland switch (IAG-switch), lentivirus, Macrobrachium rosenbergii, monosex aquaculture, white spot syndrome virus
Citation: Rotem-Dai N, Weil S, Greenshpan Y, Abehsera S, Manor R, Ofir R, Gazit R, Rosental B, Aflalo ED and Sagi A (2021) Lentiviral-Transduced Ectopic Expression of Androgenic Hormone in a Crustacean Hematopoietic Primary Cell Culture. Front. Mar. Sci. 8:677679. doi: 10.3389/fmars.2021.677679
Received: 08 March 2021; Accepted: 22 September 2021;
Published: 14 October 2021.
Edited by:
Bin Wu, Zhejiang University, ChinaReviewed by:
Chuner Cai, Friedrich Schiller University Jena, GermanyHaihui Ye, Jimei University, China
Copyright © 2021 Rotem-Dai, Weil, Greenshpan, Abehsera, Manor, Ofir, Gazit, Rosental, Aflalo and Sagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amir Sagi, c2FnaWFAYmd1LmFjLmls; orcid.org/0000-0002-4229-1059
 Noa Rotem-Dai
Noa Rotem-Dai Simy Weil
Simy Weil Yariv Greenshpan
Yariv Greenshpan Shai Abehsera1
Shai Abehsera1 Roi Gazit
Roi Gazit Benyamin Rosental
Benyamin Rosental Eliahu D. Aflalo
Eliahu D. Aflalo Amir Sagi
Amir Sagi
