
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 23 July 2021
Sec. Marine Megafauna
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.676725
This article is part of the Research Topic Pathologic Findings in Stranded Marine Mammals: A Global Perspective View all 29 articles
 Ole Nielsen1*†
Ole Nielsen1*† Thaís C. S. Rodrigues2*†
Thaís C. S. Rodrigues2*† Vsevolod L. Popov3
Vsevolod L. Popov3 Kuttichantran Subramaniam2
Kuttichantran Subramaniam2 Thomas B. Waltzek2
Thomas B. Waltzek2 Dyanna M. Lambourn4
Dyanna M. Lambourn4 Stephen Raverty5
Stephen Raverty5As part of an ongoing investigation of harbor seal (Phoca vitulina) mortalities within Puget Sound, Washington State, United States, between October 2007 and July 2008, 25 seal cases were submitted for histopathology and ancillary diagnostic testing, including additional attempted virus isolation. In vitro granular and refractile cytopathic effects (CPE) were consistently observed in Vero.DogSLAMtag cells inoculated with tissue homogenates from three seals. Transmission electron microscopy of infected Vero.DogSLAMtag cells revealed cytoplasmic clusters of icosahedral viral particles morphologically consistent with members of the family Reoviridae. The complete genome of a novel species within the genus Orthoreovirus, tentatively named phocid orthoreovirus 1 (PhRV1), was determined by next-generation sequencing and confirmed by rt-PCR in isolates from the three harbor seals. This is the first report of an orthoreovirus infection associated with dead stranded harbor seals. Aside from the CPE and ultrastructural findings, no consistent signalment, gross pathology, histopathology, or ancillary diagnostic findings were identified with PhRV1 infection. Further research is needed to determine the prevalence, tissue tropism, transmission, pathogenicity, zoonotic potential, and host range of orthoreoviruses in pinnipeds. This study demonstrates the value of thorough necropsy investigations and a multidisciplinary team approach to advance our understanding of marine mammal health.
Harbor seals (Phoca vitulina) are long-lived phocids with natal philopatry and inhabit near shore waters of Europe, Asia, and North America. There are approximately 500,000 harbor seals worldwide with estimates up to 29,000 seals in Washington State in-shore waters, which is considered the Optimal Sustainable Population (OSP) for this stock (Jeffries et al., 2003). Harbor seals haul out, congregate and pup on land and forage in tidal and subtidal areas. These animals consume similar fish and invertebrate species as humans and share coastal zone habitats with people. Mortalities from anthropogenic activities such as fishing, illegal hunting, environmental degradation, industrial, agricultural and urban discharge and vessel strikes are major concerns for resource managers, biologists and the lay public. Harbor seals worldwide are subject to similar pressures and there are indications that some European populations are declining (Hanson et al., 2013); whereas, others have stabilized and are now increasing (Brasseur et al., 2018). Since enactment of the Marine Mammal Protection Act in 1972 by the United States government, harbor seal populations in the northeast Pacific Ocean have steadily increased to historic high levels (Brasseur et al., 2018).
Ongoing investigations of mortality, contaminant loads and health status in Washington State harbor seals have been conducted since the early 1980’s. Infectious disease and malnutrition were the leading causes of pup mortality between 2004 and 2009 in Washington State (Leahy, 2010; Huggins et al., 2013); whereas, in 1984, the leading causes of pup mortality were predation, malnutrition, and premature parturition (Steiger et al., 1989). With the advent of long-term post-mortem examination of pinnipeds, expanded diagnostic modalities and access to archived marine mammal case material, the causes of death of harbor seals has been expanded, refined and new pathogens identified including: helminth (Stroud and Dailey, 1978) and coccidian (Gibson et al., 2011) parasites, bacteria (Taurisano et al., 2018), and various DNA and RNA viruses.
Several families of single-stranded (Parvoviridae and Anelloviridae) and double-stranded (Herpesvirdae, Poxviridae, and Adenoviridae) DNA viruses have been characterized from harbor seals. Single-stranded DNA viruses include a Pacific harbor seal anellovirus (Ng et al., 2011) and a harbor seal B19-like parvovirus (Bodewes et al., 2013). Double-stranded DNA viruses characterized from harbor seals include at least one alphaherpesvirus and multiple gammaherpesviruses. Phocid alphaherpesvirus 1 (PhHV-1) was first isolated and characterized from juvenile harbor seals that died from liver necrosis and pneumonia during rehabilitation in the Netherlands in 1984 (Osterhaus et al., 1985; Borst et al., 1986). PhHV-1 has also been associated with adrenal and/or hepatic necrosis in harbor seal pups that died during rehabilitation after stranding along the northeastern Pacific coastlines of the United States and Canada (Gulland et al., 1997; Himsworth et al., 2010). Phocid gammaherpesvirus 2 (PhHV-2) was isolated and characterized from the mononuclear blood cells of an adult harbor seal that presented with severe respiratory disease along the German coastline of the Wadden Sea (Lebich et al., 1994; Harder et al., 1996). Other herpesviruses (PhHV-5, PhHV-6, and PhHV-7) were reported in harbor seals associated with ocular lesions (PhHV-6; Wright et al., 2015), gingivitis, glossitis and other health issues (PhHV-7; Bodewes et al., 2015). An endemic outbreak of seal parapoxvirus, proposed as a separate species within the genus Parapoxvirus, was described in association with ulcerative to proliferative cutaneous, and mucosal lesions in harbor seals (Becher et al., 2002; Müller et al., 2003). Phocid adenovirus 2 (PhAdV-2) has been identified in ocular lesions and in unaffected eyes of harbor seals (Wright et al., 2015).
Several families of single-stranded (Paramyxoviridae, Orthomyxoviridae, Picornaviridae, Coronaviridae, and Flaviviridae) and double-stranded (Reoviridae) RNA viruses have been characterized from harbor seals. Since the 1980’s, major epizootics have been documented in European harbor seals, including phocine distemper virus (PDV) which resulted in the loss of tens of thousands of harbor seals in 1988 and 2002 (Osterhaus and Vedder, 1988; Jensen et al., 2002). Influenza A virus has also been responsible for significant harbor seal morbidity and mortality events in European waters, as well as in the United States (Callan et al., 1995; Anthony et al., 2012; Krog et al., 2015; Bodewes et al., 2016). Seal coronavirus was reported in captive harbor seals associated with acute necrotizing enteritis and pulmonary edema (Bossart and Schwartz, 1990). West Nile flavivirus infection was reported in a harbor seal presenting with progressive signs of neurologic dysfunction and polioencephalomyelitis (Del Piero et al., 2006). Two distantly related picornaviruses have been isolated from stranded harbor seals along the Pacific and Atlantic coastlines of the United States (Anthony et al., 2015; Rodrigues et al., 2020) and a syncytium forming orthoreovirus was isolated from an aborted Steller sea lion (Eumetopias jubatus) fetus from British Columbia, Canada (Palacios et al., 2011).
Reoviruses possess segmented, double-stranded RNA genomes packaged within a double-shelled nucleocapsid of approximately 70–85 nm in diameter (Fields, 1996). The family Reoviridae comprises fifteen genera that infect a diverse range of hosts including: mammals, birds, fish, invertebrates, and even fungi (Attoui et al., 2012). Some reoviruses have been associated with disease in both birds and fish, resulting in significant economic losses to the poultry and aquaculture industries (Lu et al., 2015; Siah et al., 2015). Viruses within the genus Orthoreovirus are subdivided into those which induce syncytia in vitro (fusogenic) and those that do not (non-fusogenic). The species Mammalian orthoreovirus includes all the non-fusogenic mammalian orthoreoviruses. Mammalian orthoreoviruses (MRVs) are ubiquitous and can infect a range of hosts, with significant disease reported in highly susceptible species (Zhang et al., 2011). In this investigation, we characterized the signalment, clinical signs, gross pathology, and histopathology, as well as ultrastructural and molecular characteristics of a novel MRV associated with dead stranded harbor seal pups in Puget Sound, Washington State, United States.
From October 2007 to June 2008, a limited mortality event occurred among harbor seals along the coast of Puget Sound in Washington State, United States (Figure 1). In total, 25 harbor seals were included in this study which includes 15 weaned pups (WP) (eight males; seven females) and 10 adults (AD) (nine males; one female) (Table 1). Age classes were determined as described in Lambourn et al. (2013). Two seals had suspected serological titres to morbillivirus which prompted an increased effort in viral testing from October 2007 through July 2008 on cases that presented with evidence of respiratory disease.
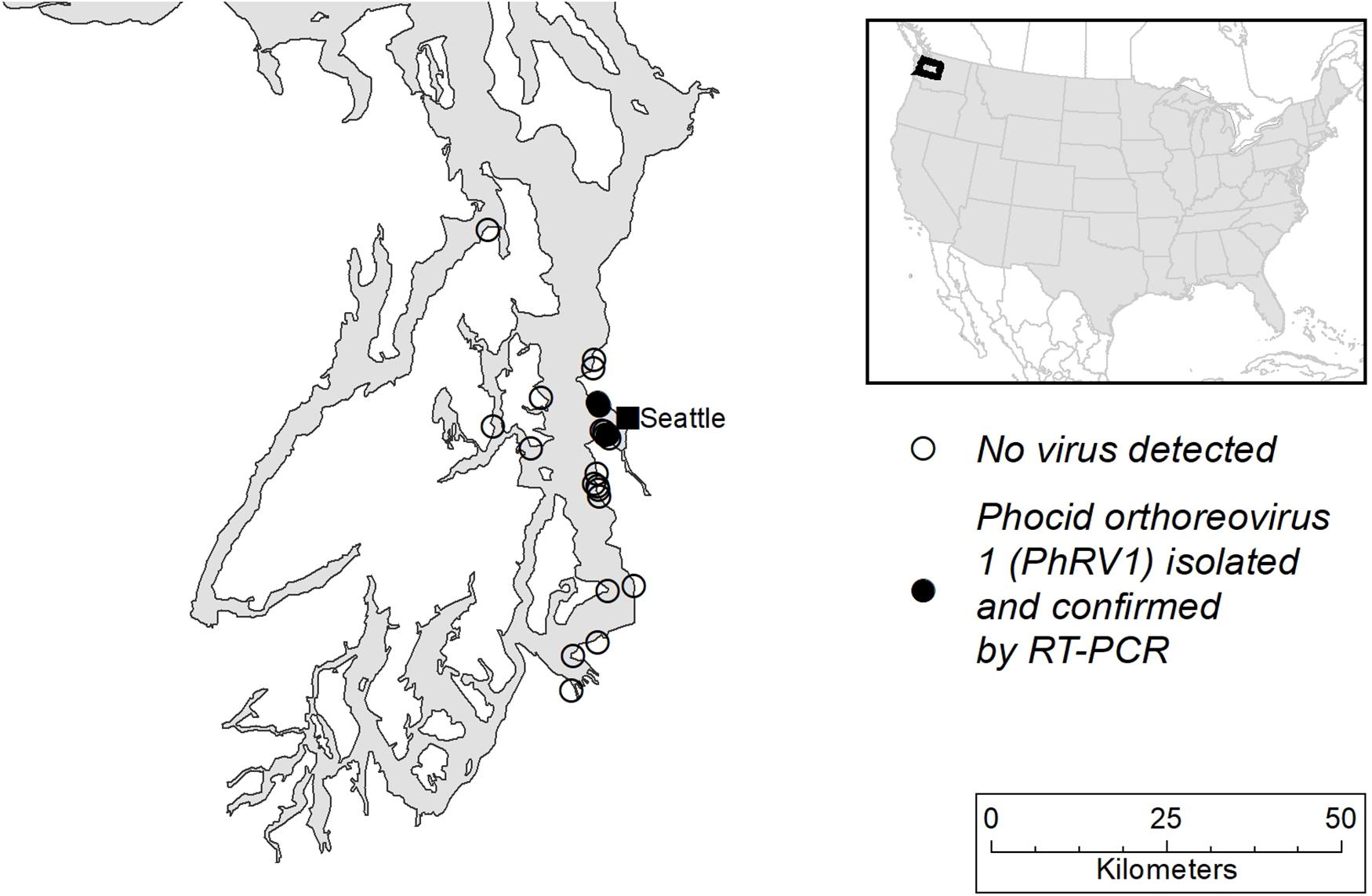
Figure 1. Map of Puget Sound, Washington State, United States showing the sites where seal carcasses were recovered.
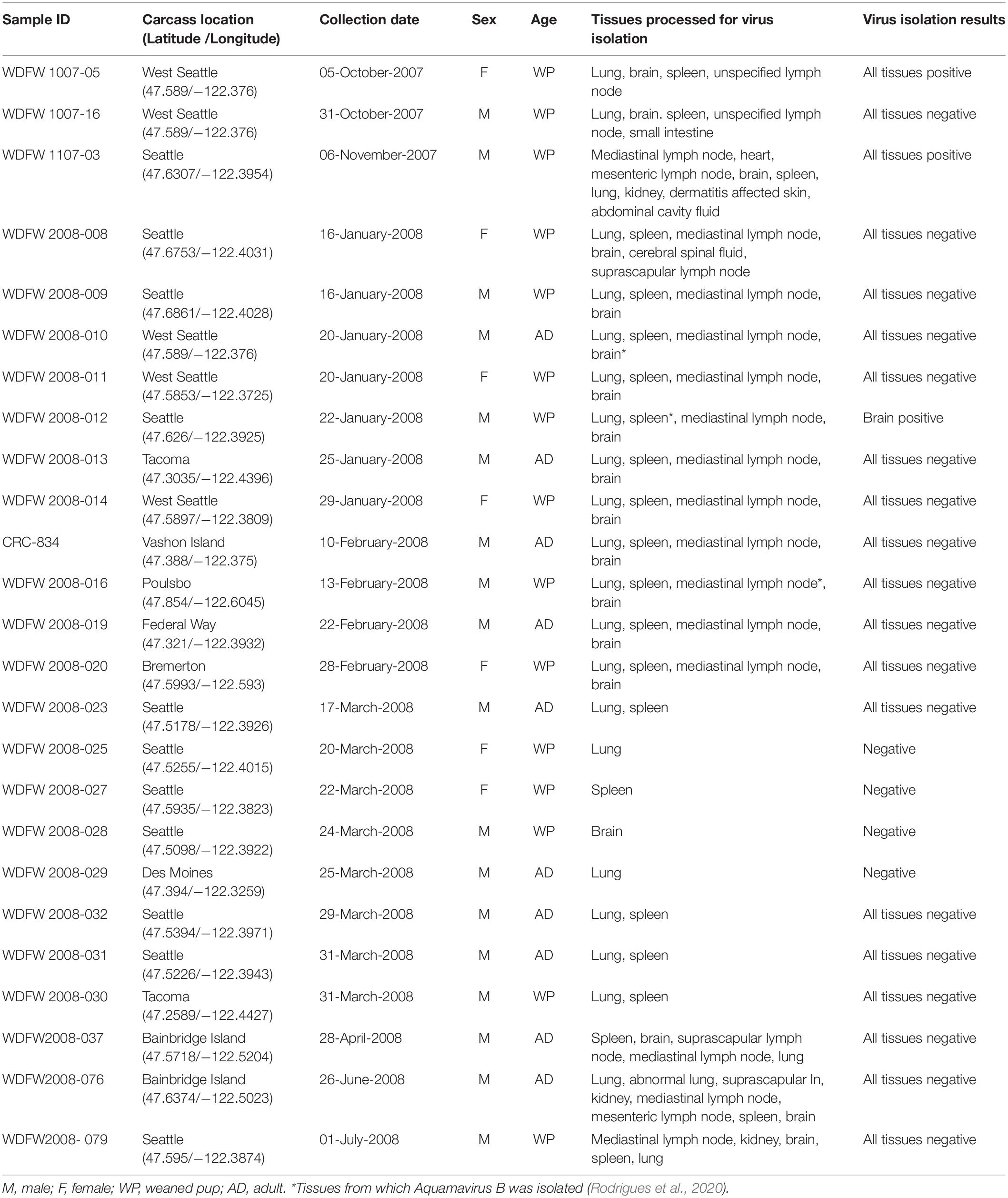
Table 1. Sample identification number, carcass collection location and date, sex, age class, tissues processed for virus isolation and results from 25 dead harbor seals from Puget Sound, Washington State between October 2007 and July 2008.
As part of a health assessment and investigation, systematic gross necropsies were conducted on code 2 and 3 harbor seal carcasses according to established protocols by biologists at the Washington Department of Fish and Wildlife and Cascadia Research Collective, Lakewood, WA, United States. Collected tissue samples were submitted for diagnostic testing to the Animal Health Center (AHC) in Abbotsford, BC, Canada. Frozen tissue samples were submitted for routine aerobic culture, Salmonella spp. enrichment culture of the small intestine, and when available, feces were submitted for routine fecal floatation (Blazevic and Ederer, 1975; Egwang and Slocombe, 1982). As part of routine surveillance for seal pathogens, pooled tissue samples from all 25 seals were screened by PCR for Brucella spp. (Lambourn et al., 2013; Wu et al., 2014) and canine distemper virus (Barrett et al., 1985). Tissue samples were preserved in 10% neutral buffered formalin and processed by an automated tissue processor, stained with hematoxylin and eosin and evaluated for histopathology. Additional samples were frozen at −80°C for virus isolation (Table 1).
Since morbillivirus infection was initially suspected as the cause of death in some of the seals, routine virus isolation (Chan and Hsuing, 1994) was performed using the Vero.DogSLAMtag cell line expressing a morbillivirus cell receptor that allows efficient morbillivirus isolation (Seki et al., 2003). Thawed tissue samples were homogenized to a 10% w/v cell free suspension in Hank’s Balanced Salts Solution (HBSS) containing antibiotics (penicillin 200 International Units/ml, streptomycin 200 μg/ml, gentamycin 50 μg/ml, and 0.25 μg/ml Fungizone) then clarified by low-speed centrifugation for 10 min (2,060 × g). A 0.5 ml aliquot of each sample was inoculated into each 70% confluent 25 cm2 tissue culture flask of Vero.DogSLAMtag cells. The inoculum was allowed to adsorb for 1 h at 37°C, then decanted, and a mixture of 5.0 ml of cell culture medium (DMEM/F-12), 4% fetal calf serum, and antibiotics was added. Flasks were incubated at 37°C and observed daily for cytopathic effects (CPE). Flasks were sub-cultured weekly for 6 weeks before discarding as negative. When CPE could be successfully passaged, presumptive virus isolates were harvested at passage two and frozen at −80°C for further study.
A 70% confluent culture of Vero.DogSLAMtag cells in a 75 cm2 flask was infected with a 1:100 dilution of a sample with extensive CPE (WDFW1107-03, mediastinal lymph node sample). After incubation and subsequent development of CPE, the infected cells were fixed in 15.0 ml of Karnovsky’s fixative (2% formaldehyde + 2% glutaraldehyde in 0.1 M cacodylate buffer pH 7.4) for 2 h at room temperature. The cells were harvested in 0.1 M cacodylate buffer, pelleted, and post-fixed in 1% OsO4 in 0.1 M cacodylate buffer pH 7.4. The sample was stained en bloc with 2% aqueous uranyl acetate (20 min at 60°C), dehydrated through a graded ethanol series, infiltrated and embedded in epoxy resin PolyBed 812 (Polysciences, Inc., Warrington, PA, United States). Ultrathin sections (60–90 nm) were cut on Leica EM UC7 ultramicrotome and stained with lead citrate prior to examination in a JEOL JEM 1400 electron microscope at 80 kV.
The cell suspensions of the WDFW1107-03 mediastinal lymph node sample viral isolate (presumptive) was frozen-thawed three times to release cell-associated virus and cell debris were removed by centrifugation at 5,509 × g at 4°C for 20 min using a Beckman JA-14 fixed angle rotor. Virus particles were pelleted from the clarified supernatant by ultracentrifugation at 100,000 × g at 4°C for 60 min in a Beckman Type 50.2 Ti rotor. RNA was extracted from the resuspended pellet using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s protocol. A cDNA library was generated using a NEBNext Ultra RNA Library Prep Kit (Illumina, San Diego, CA, United States) and sequenced on an Illumina MiSeq sequencer. De novo assembly of 2,099,690 paired-end reads was performed using SPAdes 3.5.0 (Bankevich et al., 2012). The assembled contigs were then subjected to BLASTX searches against the National Center for Biotechnology Information (NCBI) non-redundant protein database using OmicsBox 1.1. Putative open reading frames (ORFs) of all ten segments of the harbor seal virus were predicted using GeneMarkS1 (Besemer et al., 2001) and gene functions were predicted using BLASTP searches against the NCBI GenBank non-redundant protein sequence database.
The amino acid (aa) sequences of all ten proteins of orthoreoviruses (Supplementary Table 1), including the harbor seal virus discovered in this study, were separately aligned using the Multiple Alignment using Fast Fourier Transform 7.0 (MAFFT) with default parameters. Maximum Likelihood phylogenetic trees were generated in IQ-TREE 1.4.42 using 1,000 non-parametric bootstraps to test the robustness of the clades (Nguyen et al., 2015). The same aa alignments were used to perform genetic analyses within the Sequence Demarcation Tool v.1.2 (Muhire et al., 2014) with the MAFFT option implemented.
A specific endpoint reverse transcription PCR (RT-PCR) assay was designed to amplify a 390 bp region (including the primer sequences) within the segment 4 (that encodes the sigma 3 protein) of the harbor seal virus discovered in this study for screening RNA extracts from cell cultures displaying CPE (potential viral isolates). RNA from the tissue samples and cell culture supernatants were extracted using a QIAamp Viral RNA Mini Kit according to the manufacturer’s protocols. The assay was performed using a Qiagen OneStep RT-PCR Kit with 30 μL cocktails consisting of 1.2 μL Enzyme Mix, 6 μL 5X RT-PCR buffer, 6 μL 5x Q-Solution, 1.2 μL 10 mM dNTP Mix, 1.2 μL 20 μM forward (5′-TTGAAGAGGGTGGTTCTTGG-3′) and reverse (5′-CAGAGAAGACCGCAGGAATC-3′) primers, 8.4 μL RNase-free water and 4.8 μL extracted sample RNA. The RNA extracted from the WDFW1107-03 mediastinal lymph node sample cell culture supernatant was used as positive control and RNase-free water was used as the negative control. The reverse transcription step was conducted at 50°C for 30 min, followed by an initial cycle of 15 min at 95°C, 40 cycles of denaturation at 95°C, annealing at 56°C, and extension at 72°C for 30 s each, and a final elongation step at 72°C for 10 min. PCR products were subjected to electrophoresis on a 1% agarose gel stained with ethidium bromide. Bands of the expected size were purified using a QIAquick PCR Purification Kit and submitted to Functional Biosciences, Inc., Madison, WI, United States for Sanger sequencing.
Of the 25 harbor seals in this study, 11 were observed alive with clinical signs of respiratory distress, some noted with nasal discharge; nine seals (eight WP; one AD) were picked up and either euthanized immediately or died within 48 h of being in rehabilitation. The most significant gross findings were suboptimal nutritional condition in 21 seals, lungworms (n = 13; all WP) and variable puncture wounds on the flippers and torso (n = 4). Histopathology revealed pneumonia (n = 21 seals), enteritis (n = 20 seals), reactive lymphoid hyperplasia (n = 22 seals), hepatitis (n = 14 seals), hepatocellular hemosiderosis (n = 10 seals), and lymphoid depletion (n = 3 seals involving the thymus and lymph nodes) (Table 2). The changes were attributed to verminous pneumonia, enterocolonic helminthiasis and sepsis, presumably sequelae to malnutrition and other factors. The reactive lymphoid hyperplasia, lymphoid depletion, and iron sequestration within the liver were consistent with antigenemia. There were no pathognomonic lesions identified among the three weaned pups (WDFW1007-05, WDFW1107-03, and WDFW2008-012) in which RT-PCR confirmed the presence of the novel harbor seal virus discovered in this study. The seals presented with emaciation, pneumonia, parasitic enteritis, reactive lymphoid hyperplasia, and multifocal lacerations and punctures, that were also observed in unaffected conspecifics.
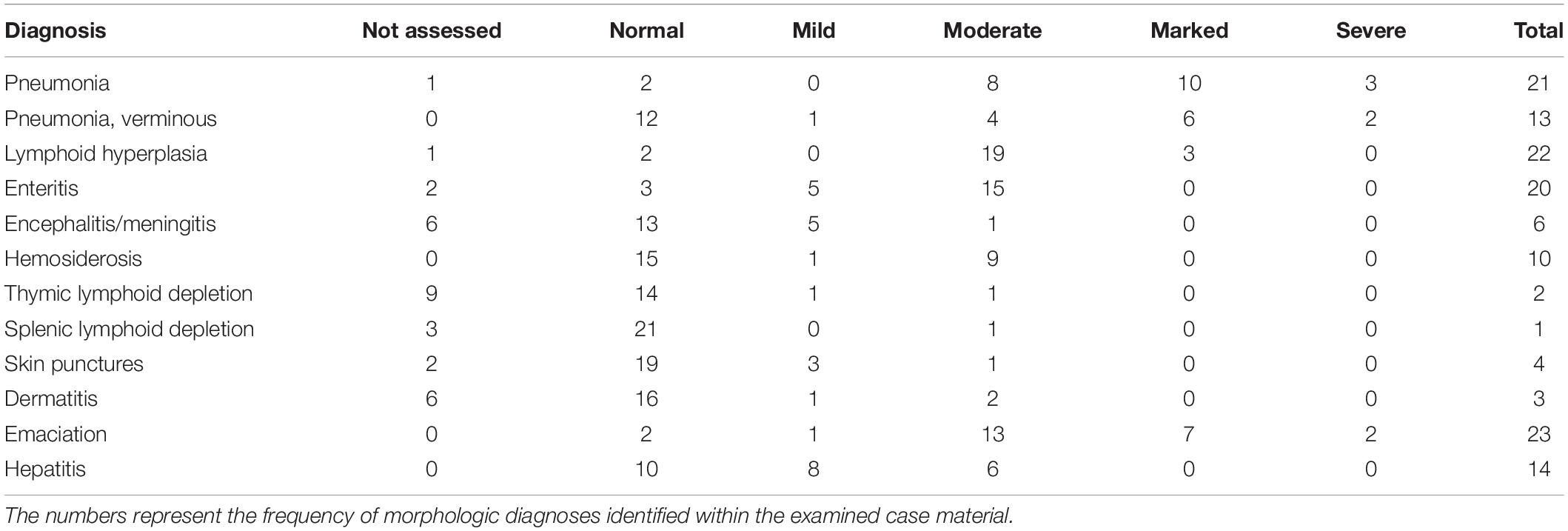
Table 2. Summary of the most significant and consistently observed gross and histologic findings for 25 harbor seals that stranded in Washington State between October 2007 and July 2008.
Aerobic culture yielded a plethora of individual or mixed bacterial flora from harvested tissues which likely represented opportunists, post mortem invaders, and occasional contaminants. The most frequently isolated bacteria in the cohort were Escherichia coli (n = 16 seals) and Streptococcus phocae (n = 11 seals), with occasional isolates of Psychrobacter spp. (n = 4 seals), Gemella haemolysans (n = 3 seals), Actinomycetes spp. (n = 3 seals), Enterococcus spp. (n = 2 seals), and individual cases of Salmonella enteritis, Pseudomonas aeruginosa, and other bacteria. Based on the degree of autolysis and associated histopathology, many isolates were attributed to post mortem overgrowth or contaminants.
Tissue samples from 25 animals were pooled (brain, lung, and lymph node) and PCR was conducted for Brucella spp. (n = 2 positive) and canine distemper virus (CDV) (n = 1 positive). Fecal parasitology identified light (n = 3 seals), moderate (n = 2 seals), or heavy (n = 1 seal) ascarid loads. Capillaria spp. and filariae were noted in the intestinal contents of two seals. No parasites were identified in three animals.
Following inoculation and incubation, cells infected with samples collected from three harbor seals showed identical CPE (Table 1). The infected cells became granular and developed a refractile appearance, began to round up, detach from the plastic substrate, and coalesced into clumps, which progressed over a few days until the entire cell sheet was involved (Figure 2). CPE developed as early as 18 days post infection (DPI) with some flasks developing CPE as late as 33 DPI. There was no evidence of syncytium formation, a hallmark of morbillivirus infection seen in the progression of the CPE therefore morbillivirus infection was no longer suspected. The presumptive viral isolates from the harbor seal WDFW1107-03 were chosen for expansion, TEM, and next-generation sequencing. Additional virus isolation attempts were also made from these seals and upon further characterization all additional isolates (from animals WDFW 2008-010, WDFW 2008-012, and WDFW-016) were aquamavirus B, a proposed new member of the family Picornaviridae (Rodrigues et al., 2020).

Figure 2. Cell culture (A,B) and transmission electron micrographs (C,D) of phocid orthoreovirus 1 (PhRV1) infecting Vero.DogSLAMtag cells. (A) Uninfected Vero.DogSLAMtag cells. Scale bar = 100 μm. (B) Passage two PhRV1 (WDFW2008-12 brain isolate) 11 DPI showing rounding up of infected cells with detachment of cells and the early stages of the disintigration of the cell monolayer. Scale bar = 100 μm. (C) Intracytoplasmic virus inclusion. N-fragment of cell nucleus with depleted chromatin in the nucleoplasm and chromatin condensation at the periphery of the nucleus. Bar = 1 μm. (D) Fragment of virus inclusion with particles ∼75 nm in diameter in different stages of maturation. Bar = 200 nm.
Transmission electron microscopy of ultrathin sections of the selected infected cells showed cytoplasmic inclusions consisting of fine fibrils and granules and contained virus particles measuring approximately 75 nm in diameter in different stages of maturity and some with empty shells. The nuclei of infected cells were depleted of most of the chromatin, the rest of it being condensed at the inner periphery of the perinuclear membrane (Figure 2). The CPE and virion morphology observed were typical of viruses within the family Reoviridae.
De novo assembly of the paired-end reads followed by BLASTX analyses recovered all 10 segments of the genome of a novel virus within the genus Orthoreovirus (Table 3). Phylogenetic analyses based on the aa sequences of 10 proteins showed the harbor seal virus formed an unique and a well-supported basal branch to the members of the species Mammalian orthoreovirus (Figure 3 and Supplementary Figures 1A–9A). Comparisons of the core proteins (segments L1–L3, M1, and S2) and outer capsid proteins (segments M2, S1, and S4) of the harbor seal virus to other orthoreoviruses showed the aa identities ranged between 25.9–77.2% and 18.8–60.0%, respectively (Table 4 and Supplementary Figures 1B–5B, 7B, 8B). The aa identities of the harbor seal virus non-structural proteins (segments M3 and S3) ranged between 24.0–53.1% as compared to other orthoreoviruses (Supplementary Figures 6B, 9B).
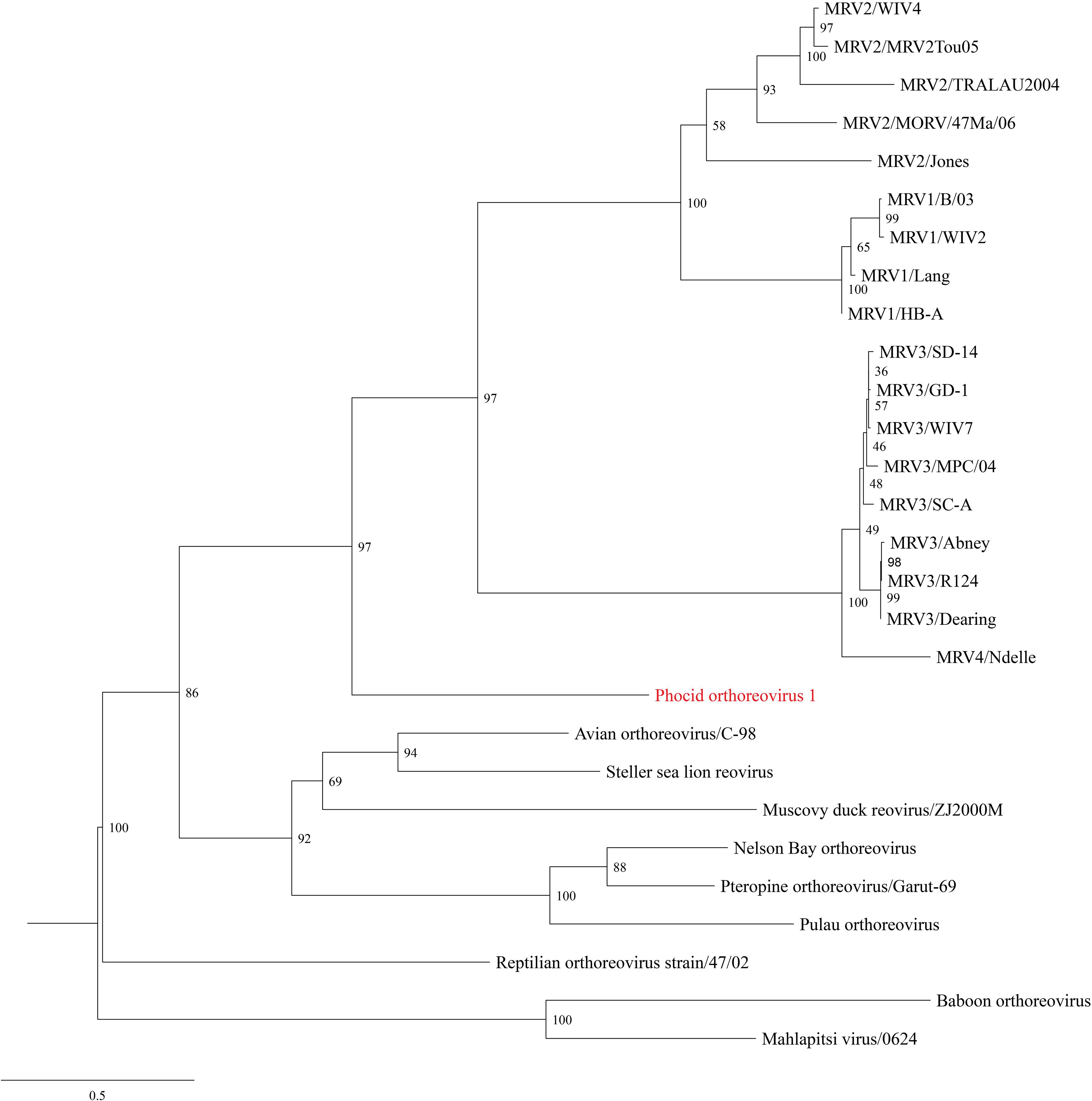
Figure 3. Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 (PhRV1) to representatives of the Orthoreovirus genus based on the aligned amino acid sequences of the sigma 1 protein (segment S1). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. Viral names are listed, followed by isolate/strain identification when available. Viral abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3; MRV4, mammalian orthoreovirus 4.

Table 4. Sequence identity matrix showing the amino acid (aa) percent identity of the phocid orthoreovirus 1 to 27 other orthoreoviruses based on the sigma 1 protein (segment S1).
The RNA extracted from cell cultures displaying identical CPE (Table 1) were tested by the RT-PCR designed in this study to confirm the presence of harbor seal virus RNA. The isolates from samples collected from the three harbor seals were positive by RT-PCR including: WDFW1007-05 (unspecified lymph node), WDFW1107-03 (mediastinal and mesenteric lymph nodes, heart, brain, spleen, lung, kidney, skin, and abdominal cavity fluid), and WDFW2008-012 (brain).
Based on the cytopathic, ultrastructural and sequencing studies, the harbor seal virus described in the present study is believed to be the first orthoreovirus isolated, in vitro propagated and genetically characterized from dead stranded harbor seals. Although an Avian orthoreovirus species designated the Steller sea lion reovirus (SSRV) had previously been reported from a Steller sea lion in the northeastern Pacific (Palacios et al., 2011), the harbor seal virus is believed to be the first Mammalian orthoreovirus discovered to infect marine mammals.
The harbor seal virus infected Vero.DogSLAMtag cells displayed characteristic in vitro cytopathology and virus particles with morphology and maturation stages consistent with non-fusogenic orthoreoviruses replicating in the cytoplasm of infected cells, as confirmed by TEM (Attoui et al., 2009; Quin et al., 2009). The fusogenic Avian orthoreovirus, SSRV, was isolated from multiple tissues of an aborted Steller sea lion fetus with syncytia also observed in infected Vero.DogSLAMtag cultures. CPE was reported in only 3–5 DPI with SSRV (Palacios et al., 2011); whereas, the harbor seal virus cell inoculations resulted in a more granular CPE after a much longer incubation period (18–33 DPI) and repeated blind passages.
The harbor seal virus genome sequence reported herein permitted phylogenetic and genetic analyses that support its inclusion as a novel species within the genus Orthoreovirus. Phylogenetic analyses based on aa sequences of the 10 proteins revealed the harbor seal virus formed a unique and well-supported branch as the sister species to all other mammalian orthoreoviruses. One criterion for the species demarcation of orthoreoviruses is the pairwise aa identity of the core proteins >85% represent different strains within a orthoreoviruses species and <65% represent different species. Additionally, the pairwise aa identity of the outer capsid proteins >55% represent different strains within a orthoreoviruses species and <35% represent different species (Attoui et al., 2012). The pairwise aa identity of the core proteins and outer capsid proteins of the harbor seal virus compared to other orthoreoviruses ranged between 25.9–77.2% and 18.8–60.0%, respectively. Given the unique phylogenetic position and sequence divergence of the harbor seal virus, we propose the formal species designation of phocid orthoreovirus 1 (PhRV1) to be considered for approval by the International Committee on Taxonomy of Viruses.
Phocid orthoreovirus 1 is most closely related to viruses within the Mammalian orthoreovirus species (MRV), that includes all non-fusogenic orthoreoviruses, with four recognized strains (MRV-1, MRV-2, MRV-3, and MRV-4; Attoui et al., 2012). MRV includes orthoreoviruses recovered from humans and terrestrial mammals, responsible for either symptomatic or asymptomatic infections in a range of hosts (Tyler, 2001). In humans, MRVs are primarily associated with respiratory and enteric infections. Similarly, in bats, three MRV strains have been described and infections typically present with hemorrhagic enteritis and interstitial pneumonia (Kohl et al., 2012). In mice and in non-human primates, orthoreovirus infections affect the respiratory, gastrointestinal, and nervous systems (Tyler, 2001; Attoui et al., 2009). A MRV-3 was described from the cerebrospinal fluid in a case of meningitis in a child in Colorado. When inoculated in newborn mice, this virus was capable of systemic spread and induced a lethal encephalitis (Tyler et al., 2004). Multiple MRV strains have been recovered from dogs with pneumonia or enteritis, sometimes in association with other pathogens, such as canine distemper virus (CDV) and canine parvovirus (Lou et al., 1963; Appel, 1987; Decaro et al., 2005). It has been suggested that MRV infection may act synergistically with other infectious agents, exacerbating concomitant or supervening infections (Appel, 1987; Ahasan et al., 2020).
In this study, PhRV1 was detected in harbor seals with non-specific pneumonia, enteritis, and non-suppurative encephalitis or meningitis; however, similar processes were identified in seals with no demonstrable virus. Morbillivirus infection was originally suspected as a leading cause of these mortalities but no histopathologic evidence of morbillivirus could be found in any of the examined tissues. Harbor seal WDFW 2008-012 was found to be coinfected with Aquamavirus B (isolated from spleen tissue; Rodrigues et al., 2020) and PhRV1 (isolated from the brain). Among the three weaned pups in which PhRV1 was confirmed by RT-PCR, no pathognomonic lesions were identified. The three animals presented with emaciation and multifocal skin lacerations and punctures, that were also observed in unaffected conspecifics. Due to the relatively small number of infected harbor seals, intercurrent disease [such as bacterial, viral (e.g., PhRV1), and verminous infections] and post mortem change in this case series, no specific gross or histopathology could be attributed to PhRV1. Efforts to enhance surveillance and detection of viral infection by PCR with systematic histopathology and possible development of in situ hybridization or immunohistochemistry assay may provide insights into the pathogenesis and incidence of pinniped infections. PhRV1 was recovered from multiple tissues from sampled animals, which may reflect a broad tissue tropism secondary to viremia, a trait common to other members of the family Reoviridae (Nibert et al., 1996; Tyler et al., 2004). For example, the SSRV was reported to be pancytotropic in a Steller sea lion fetus (Palacios et al., 2011).
Orthoreovirus infections have been reported worldwide in a broad range of host species and transmission is believed to be predominantly parental or via inhalation. These viruses typically feature a diverse host range and significant potential for extending across species barriers (Attoui et al., 2009; Wellehan et al., 2009). As segmented viruses, reoviruses are capable of gene segment reassortment, which increases their ability to generate unique strains (Wellehan et al., 2009). A study scored viruses infecting mammals for their biological properties considered advantageous to host switching, and concluded that reoviruses scored highest (Pulliam, 2008). Decaro et al. (2005) also suggested that because of the apparent lack of species barriers, MRVs may potentially spread from animals to humans and vice versa. A possible zoonotic transmission of Melaka virus, a bat orthoreovirus that is associated to respiratory disease, is assumed (Kohl et al., 2012). In the present study, PhRV1 was isolated in a cell line of African green monkey (Cercopithecus aethiops) origin. These cells are known to be deficient in the production of interferon and therefore more susceptible to infection by a number of different viruses (Emeny and Morgan, 1979). PhRV1 was isolated from multiple organs from three seals with CPE being detected after at least two and as many as four blind passages. Islam et al. (2003) also found that blind passaging an avian reovirus strain would allow adaptation in Vero cell cultures. This underscores the value of blind passaging for virus discovery.
In the present study, we isolated and characterized the genome and ultrastructural features of a novel orthoreovirus from wild harbor seals in Washington State. Although the virus was recovered from multiple tissues its pathogenicity has not yet been resolved. The Vero.DogSLAMtag cell line was permissive and facilitated primary isolation of PhRV1 in harbor seal tissues and may be effective on the isolation of similar viruses from other marine mammal species. Our study underscores the relevance of conventional virology and pathology techniques (such as virus isolation, histopathology, and TEM), coupled with high-throughput sequencing technologies for the discovery and characterization of pathogens in wild marine mammals. Further research is needed to unravel the transmission, host range, prevalence, and pathogenicity of PhRV1 in pinniped populations.
The datasets presented in this study can be found in online repositories. The repositories and accession numbers can be found below: MN820529 to MN820538, and SRR14601803.
Ethical review and approval was not required for the animal study because the animals were dead when investigated.
ON and TR: writing of the original draft manuscript, investigation, formal analysis, and data curation. VP, SR, and DL: review and editing of the manuscript, investigation, data curation, and funding acquisition. KS: conceptualization, investigation, formal analysis, software, and review and editing of the manuscript. TW: conceptualization, supervision, funding acquisition, and review and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding and support for this work was provided by the Washington Department of Fish and Wildlife and the John H. Prescott Marine Mammal Rescue Assistance Grant Program. The harbor seal samples were collected under Washington Department of Fish and Wildlife 109h authority and as part of the NW Marine Mammal Stranding Network. Parts were transferred for analysis to SR using NOAA Marine Mammal Stranding Agreement, SR Co-investigator under Permit 932-1905/MA-009526 issued under Section 104 (16 USC 1375).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor is currently organizing a Research Topic with one of the author SR.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Department of Fisheries and Oceans Canada (DFO) for supporting this study, Mark Ouellette (DFO) for providing the map, Jason Allen (DFO) for assisting with the photo microscopy, The University of Texas Medical Branch at Galveston for the transmission electron microscopy, and the Wildlife and Aquatic Veterinary Disease Laboratory (WAVDL, College of Veterinary Medicine, University of Florida) for the molecular diagnostics. The harbor seals and samples used in this study were collected under the NOAA Marine Mammal Stranding Agreement held by the West Seattle Seal Sitters Stranding Network and Cascadia Research Collective, the 109h Authority held by Washington Department of Fish and Wildlife. We would also like to thank volunteers and staff in the West Coast Marine Mammal Stranding Network that assisted with stranding response especially Robin Lindsey of West Seattle Seal Sitters, Paws Wildlife staff and Jessie Huggins from Cascadia Research Collective and Yasuke Yanagi, Department of Virology, Kyushu University, Fukuoka, Japan for providing the Vero.DogSLAMtag cell line. The expertise and efforts of the technical staff at the Animal Health Centre, Abbotsford, BC, Canada is gratefully acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.676725/full#supplementary-material
Supplementary Figure 1 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the lambda 1 protein (segment L3). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the lambda 1 protein (segment L3). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3.
Supplementary Figure 2 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the lambda 2 protein (segment L2). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the lambda 2 protein (segment L2). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3.
Supplementary Figure 3 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the lambda 3 protein (segment L1). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the lambda 3 protein (segment L1). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3; MRV4, mammalian orthoreovirus 4.
Supplementary Figure 4 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the Mu 1 protein (segment M2). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the Mu 1 protein (segment M2). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3; MRV4, mammalian orthoreovirus 4.
Supplementary Figure 5 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the Mu 2 protein (segment M1). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the Mu 2 protein (segment M1). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3.
Supplementary Figure 6 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the Mu NS protein (segment M3). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the Mu NS protein (segment M3). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3.
Supplementary Figure 7 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the sigma 2 protein (segment S2). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the sigma 2 protein (segment S2). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3; MRV4, mammalian orthoreovirus 4.
Supplementary Figure 8 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the sigma 3 protein (segment S4). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the sigma 3 protein (segment S4). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3; MRV4, mammalian orthoreovirus 4.
Supplementary Figure 9 | (A) Maximum Likelihood phylogram depicting the relationship of the phocid orthoreovirus 1 to representatives of the Orthoreovirus genus based on the amino acid (aa) sequences of the sigma NS protein (segment S3). Bootstrap values are given at each node and the branch lengths represent the number of inferred substitutions as indicated by the scale. (B) Sequence identity matrix showing the aa percent identity of the novel harbor seal virus to 27 other orthoreoviruses based on the sigma NS protein (segment S3). Virus name are listed, followed by isolate/strain identification when available. Virus abbreviations: MRV1, mammalian orthoreovirus 1; MRV2, mammalian orthoreovirus 2; MRV3, mammalian orthoreovirus 3.
Supplementary Table 1 | Orthoreoviruses used in the phylogenetic and genetic analyses.
Ahasan, M. S., Subramaniam, K., Campos Krauer, J. M., Sayler, K. A., Loeb, J. C., Goodfriend, O. F., et al. (2020). Three new Orbivirus species isolated from farmed white-tailed deer (Odocoileus virginianus) in the United States. Viruses 12:13. doi: 10.3390/v12010013
Anthony, S. J., Leger, J. S., Liang, E., Hicks, A. L., Sanchez-Leon, M. D., Jain, K., et al. (2015). Discovery of a novel hepatovirus (phopivirus of seals) related to human hepatitis A virus. mBio 6:e01180-15. doi: 10.1128/mBio.01180-15
Anthony, S. J., St. Leger, J. A., Pugliares, K., Ip, H. S., Chan, J. M., Carpenter, Z. W., et al. (2012). Emergence of fatal avian influenza in New England harbor seals. mBio 3:e00166-12. doi: 10.1128/mBio.00166-12
Appel, M. P. (1987). “Reovirus,” in Virus Infections in Carnivores, ed. M. P. Appel (New York, NY: Elsevier), 95–96.
Attoui, B. M., Brussaard, C. P., Chappell, J. D., Ciarlet, M., del Vas, M., Dermody, T. S., et al. (2012). “Family Reoviridae,” in Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, eds A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (Amsterdam: Elsevier Academic), 541–637.
Attoui, H., Mendez-Lopez, M. R., Rao, S., Hurtado-Alendes, A., Lizaraso-Caparo, F., Jaafar, F. M., et al. (2009). Peruvian horse sickness virus and Yunnan orbivirus, isolated from vertebrates and mosquitoes in Peru and Australia. Virology 394, 298–310. doi: 10.1016/j.virol.2009.08.032
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barrett, T., Shrimpton, S. B., and Russell, S. E. H. (1985). Nucleotide sequence of the entire protein coding region of canine distemper virus polymerase-associated (P) protein MRNA. Virus Res. 3, 367–372. doi: 10.1016/0168-1702(85)90436-8
Becher, P., König, M., Müller, G., Siebert, U., and Thiel, H. J. (2002). Characterization of sealpox virus, a separate member of the parapoxviruses. Arch. Virol. 147, 1133–1140. doi: 10.1007/s00705-002-0804-8
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Blazevic, D., and Ederer, G. (1975). Principles of Biochemical Tests in Diagnostic Microbiology. New York: John Wiley & Sons, 15–18.
Bodewes, R., Contreras, G. J. S., García, A. R., Hapsari, R., van de Bildt, M. W., Kuiken, T., et al. (2015). Identification of DNA sequences that imply a novel gammaherpesvirus in seals. J. Gen. Virol. 96, 1109–1114. doi: 10.1099/vir.0.000029
Bodewes, R., García, A. R., Wiersma, L. C. M., Getu, S., Beukers, M., Schapendonk, C. M. E., et al. (2013). Novel B19-like parvovirus in the brain of a harbor seal. PLoS One 8:e79259. doi: 10.1371/journal.pone.0079259
Bodewes, R., Zohari, S., Krog, J. S., Hall, M. D., Harder, T. C., Bestebroer, T. M., et al. (2016). Spatiotemporal analysis of the genetic diversity of seal influenza A(H10N7) virus, Northwestern Europe. J. Virol. 90, 4269–4277. doi: 10.1128/jvi.03046-15
Borst, G. H. A., Walvoort, H. C., Reijnders, P. J. H., Van Der Kamp, J. S., and Osterhaus, A. D. M. E. (1986). An outbreak of a herpesvirus infection in harbor seals (Phoca vitulina). J. Wildl. Dis. 22, 1–6. doi: 10.7589/0090-3558-22.1.1
Bossart, G. D., and Schwartz, J. C. (1990). Acute necrotizing enteritis associated with suspected coronavirus infection in three harbor seals (Phoca vitulina). J. Zoo Wildl. Med. 21, 84–87.
Brasseur, S., Peter, M. J. M., Reijnders, J. H., Cremer, J., Meesters, E., Kirkwood, R., et al. (2018). Echoes the from past: regional variations in recovery within a harbour seal population. PLoS One 13:e0189674. doi: 10.1371/journal.pone.0189674
Callan, R. J., Early, G., Kida, H., and Hinshaw, V. S. (1995). The appearance of H3 influenza viruses in seals. J. Gen. Virol. 76, 199–203. doi: 10.1099/0022-1317-76-1-199
Chan, V. F., and Hsuing, G. D. (1994). “Cell culture preparation,” in Hsuing’s Diagnostic Virology as Illustrated by Light and Electron Microscopy, 4th Edn, eds G. D. Hsiung, C. K. Y. Fong, and M. L. Landry (New Haven, CT: Yale University Press), 337–374.
Decaro, N., Campolo, M., Desario, C., Ricci, D., Camero, M., Lorusso, E., et al. (2005). Virological and molecular characterization of a mammalian orthoreovirus type 3 strain isolated from a dog in Italy. Vet. Microbiol. 109, 19–27. doi: 10.1016/j.vetmic.2005.05.014
Del Piero, F., Stremme, D. W., Habecker, P. L., and Cantile, C. (2006). West Nile flavivirus polioencephalomyelitis in a harbor seal (Phoca vitulina). Vet. Pathol. 43, 58–61. doi: 10.1354/vp.43-1-58
Egwang, T., and Slocombe, J. (1982). Evaluation of the Cornell-Wisconsin centrifugal floatation technique for recovering trichostrongylid eggs from bovine feces. Can. J. Comp. Med. 46, 133–137.
Emeny, J. M., and Morgan, M. J. (1979). Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43, 247–252. doi: 10.1099/0022-1317-43-1-247
Fields, B. N. (1996). “Reoviridae,” in Fields Virology, eds B. N. Fields, D. M. Knipe, and P. M. Howley (Philadelphia, PA: Raven Press), 1553–1555.
Gibson, A. K., Raverty, S., Lambourn, D. M., Huggins, J., Magargal, S. L., and Grigg, M. E. (2011). Polyparasitism is associated with increased disease severity in Toxoplasma gondii-infected marine sentinel species. PLoS Negl. Trop. Dis. 5:e1142. doi: 10.1371/journal.pntd.0001142
Gulland, F. M. D., Lowenstine, L. J., Lapointe, J. M., Spraker, T., and King, D. P. (1997). Herpesvirus infection in stranded pacific harbor seals of coastal California. J. Wildl. Dis. 33, 450–458. doi: 10.7589/0090-3558-33.3
Hanson, N., Thompson, D., Duck, C., Moss, S., and Lonergan, M. (2013). Pup mortality in a rapidly declining harbour seal (Phoca vitulina) population. PLoS One 8:e80727. doi: 10.1371/journal.pone.0080727
Harder, T. C., Harder, M., Vos, H., Kulonen, K., Kennedy-Stoskopf, S., Liess, B., et al. (1996). Characterization of phocid herpesvirus-1 and -2 as putative Alpha- and Gammaherpesviruses of North American and European pinnipeds. J. Gen. Virol. 77, 27–35. doi: 10.1099/0022-1317-77-1-27
Himsworth, C. G., Haulena, M., Lambourn, D. M., Gaydos, J. K., Huggins, J., Calambokidis, J., et al. (2010). Pathology and epidemiology of phocid herpesvirus-1 in wild and rehabilitating harbor seals (Phoca vitulina richardsi) in the northeastern pacific. J. Wildl. Dis. 46, 1046–1051. doi: 10.7589/0090-3558-46.3.1046
Huggins, J. L., Leahy, C. L., Calambokidis, J., Lambourn, D., Jeffries, S. J., Norman, S. A., et al. (2013). Causes and patterns of harbor seal (Phoca vitulina) pup mortality at Smith island, Washington, 2004–2010. Northwest. Nat. 94, 198–208. doi: 10.1898/12-14.1
Islam, K. M. D., Ahmed, T., Ahasan, M. M., Billah, M. M., Islam, M. E., and Hossain, K. M. (2003). Adaptation of reovirus on Vero cell line. J. Anim. Vet. Adv. 2, 425–429.
Jeffries, S., Huber, H., Calambokidis, J., and Laake, J. (2003). Trends and status of harbor seals in Washington State: 1978-1999. J. Wildl. Manage. 67, 207–218. doi: 10.2307/03076
Jensen, T., van de Bilt, M., Dietz, H. H., Andersen, T. H., Hammer, A. S., Kuiken, T., et al. (2002). Another phocine distemper outbreak in Europe. Science 297:209. doi: 10.1126/science.1075343
Kohl, C., Lesnik, R., Brinkmann, A., Ebinger, A., Radonić, A., Nitsche, A., et al. (2012). Isolation and characterization of three mammalian orthoreoviruses from European bats. PLoS One 7:e43106. doi: 10.1371/journal.pone.0043106
Krog, J. S., Hansen, M. S., Holm, E., Hjulsager, C. K., Chriél, M., Pedersen, K., et al. (2015). Influenza A(H10N7) virus in dead harbor seals, Denmark. Emerg. Infect. Dis. 21, 684–687. doi: 10.3201/eid2104.141484
Lambourn, D. M., Garner, M., Ewalt, D., Raverty, S., Sidor, I., Jeffries, S. J., et al. (2013). Brucella pinnipedialis infections in pacific harbor seals (Phoca vitulina richardsi) from Washington State, USA. J. Wildl. Dis. 49, 802–815. doi: 10.7589/2012-05-137
Leahy, C. L. (2010). Causes and Patterns of Harbor Seal (Phoca vitulina) Pup Mortality at Smith Island, Washington, 2004-2009. Master of Environmental Studies Thesis. Olympia, WAS: The Evergreen State College.
Lebich, M., Harder, T. C., Frey, H.-R., Visser, I. K. G., Osterhaus, A. D. M. E., and Liess, B. (1994). Comparative immunological characterization of type-specific and conserved B-cell epitopes of pinniped, felid and canid herpesviruses. Arch. Virol. 136, 335–347. doi: 10.1007/BF01321062
Lou, T. Y., Wenner, H. A., and McMillan, J. (1963). Natural and experimental infection of dogs with reovirus, type 1: pathogenicity of the strain for other animals. Am. J. Epidemiol. 77, 293–304. doi: 10.1093/oxfordjournals.aje.a120320
Lu, H., Tang, Y., Dunn, P. A., Wallner-Pendleton, E. A., Lin, L., and Knoll, E. A. (2015). Isolation and molecular characterization of newly emerging avian reovirus variants and novel strains in Pennsylvania, USA, 2011–2014. Sci. Rep. 5:14727. doi: 10.1038/srep14727
Muhire, B. M., Varsani, A., and Martin, D. P. (2014). SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277
Müller, G., Gröters, S., Siebert, U., Rosenberger, T., Driver, J., König, M., et al. (2003). Parapoxvirus infection in harbor seals (Phoca vitulina) from the German North Sea. Vet. Pathol. 40, 445–454. doi: 10.1354/vp.40-4-445
Ng, T. F., Wheeler, E., Greig, D., Waltzek, T. B., Gulland, F., and Breitbart, M. (2011). Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J. Gen. Virol. 92, 1318–1323. doi: 10.1099/vir.0.029678-0
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nibert, M. L., Margraf, R. L., and Coombs, K. M. (1996). Nonrandom segregation of parental alleles in reovirus reassortants. J. Virol. 70, 7295–7300.
Osterhaus, A. D. M. E., and Vedder, E. J. (1988). Identification of virus causing recent seal deaths. Nature 335:20. doi: 10.1038/335020a0
Osterhaus, A. D., Yang, H., Spijkers, H. E., Groen, J., Teppema, J. S., and van Steenis, G. (1985). The isolation and partial characterization of a highly pathogenic herpesvirus from the harbor seal (Phoca vitulina). Arch. Virol. 86, 239–251. doi: 10.1007/BF01309828
Palacios, G., Wellehan, J. F. X., Raverty, S., Bussetti, A. V., Hui, J., Savji, N., et al. (2011). Discovery of an orthoreovirus in the aborted fetus of a Steller sea lion (Eumetopias Jubatus). J. Gen. Virol. 92, 2558–2565. doi: 10.1099/vir.0.032649-0
Pulliam, J. R. C. (2008). Viral host jumps: moving toward a predictive framework. EcoHealth 5, 80–91. doi: 10.1007/s10393-007-0149-6
Quin, Q., Hastings, C., and Miller, C. L. (2009). Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 83, 11090–11101. doi: 10.1128/jvi.01239-09
Rodrigues, T. C., Nielsen, O., Burek-Huntington, K. A., Popov, V. L., Raverty, S., Lambourn, D. M., et al. (2020). Genomic characterization of picornaviruses isolated from ribbon (Histriophoca fasciata) and harbor (Phoca vitulina) seals. Front. Vet. Sci. 7:554716. doi: 10.3389/fvets.2020.554716
Seki, F., Ono, N., Yamaguchi, R., and Yanagi, Y. (2003). Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine slam (CD150) and their adaptability to marmoset b95a cells. J. Virol. 77, 9943–9950. doi: 10.1128/jvi.77.18.9943-9950.2003
Siah, A., Morrison, D. B., Fringuelli, E., Savage, P., Richmond, Z., Johns, R., et al. (2015). Piscine reovirus: genomic and molecular phylogenetic analysis from farmed and wild salmonids collected on the Canada/US Pacific Coast. PLoS One 10:e0141475. doi: 10.1371/journal.pone.0141475
Steiger, G. H., Calambokidis, J., Cubbage, J. C., Skilling, D. E., Smith, A. W., and Gribble, D. H. (1989). Mortality of harbor seal pups at different sites in the inland waters of Washington. J. Wildl. Dis. 25, 319–328. doi: 10.7589/0090-3558-25.3.319
Stroud, R. K., and Dailey, M. D. (1978). Parasites and associated pathology observed in pinnipeds stranded along the Oregon coast. J. Wildl. Dis. 14, 292–298. doi: 10.7589/0090-3558-14.3.292
Taurisano, N. D., Butler, B. P., Stone, D., Hariharan, H., Fields, P. J., Ferguson, H. W., et al. (2018). Streptococcus phocae in marine mammals of northeastern pacific and arctic Canada: a retrospective analysis of 85 postmortem investigations. J. Wildl. Dis. 54, 101. doi: 10.7589/2016-09-208
Tyler, K. L. (2001). “Mammalian reoviruses,” in Fields Virology, 4th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Lippincott Williams & Wilkins), 1729–1745.
Tyler, K. L., Barton, E. S., Ibach, M. L., Robinson, C., Campbell, J. A., O’Donnell, S. M., et al. (2004). Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. J. Infect. Dis. 189, 1664–1675. doi: 10.1086/383129
Wellehan, J. F. X., Childress, A. L., Marschang, R. E., Johnson, A. J., Lamirande, E. W., Roberts, J. F., et al. (2009). Consensus nested pcr amplification and sequencing of diverse reptilian, avian, and mammalian orthoreoviruses. Vet. Microbiol. 133, 34–42. doi: 10.1016/j.vetmic.2008.06.011
Wright, E. P., Waugh, L. F., and Goldstein, T. (2015). Evaluation of viruses and their association with ocular lesions in pinnipeds in rehabilitation. Vet. Ophthalmol. 18(Suppl. 1), 148–159. doi: 10.1111/vop.12235
Wu, Q., Mcfee, W. E., Goldstein, T., Tiller, R. V., and Schwacke, L. (2014). Real-time pcr assays for detection of Brucella spp. and the identification of genotype st27 in bottlenose dolphins (Tursiops truncatus). J. Microbiol. Methods 100, 99–104. doi: 10.1016/j.mimet.2014.03.001
Keywords: genome sequencing, marine mammal, mortality event investigation, pinniped, Reoviridae, transmission electron microscopy, virus isolation
Citation: Nielsen O, Rodrigues TCS, Popov VL, Subramaniam K, Waltzek TB, Lambourn DM and Raverty S (2021) A Novel Orthoreovirus Isolated From Dead Stranded Harbor Seals From Puget Sound, Washington State, United States. Front. Mar. Sci. 8:676725. doi: 10.3389/fmars.2021.676725
Received: 05 March 2021; Accepted: 10 June 2021;
Published: 23 July 2021.
Edited by:
Debra Lee Miller, The University of Tennessee, United StatesReviewed by:
Eric Delwart, Blood Systems Research Institute, United StatesCopyright © 2021 Nielsen, Rodrigues, Popov, Subramaniam, Waltzek, Lambourn and Raverty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ole Nielsen, b2xlLm5pZWxzZW5AZGZvLW1wby5nYy5jYQ==; Thaís C. S. Rodrigues, dGhhaXNjYXJuZWlyb18yNUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.