- 1Fisheries College, Jimei University, Xiamen, China
- 2Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 3Qingdao Key Laboratory of Mariculture Epidemiology and Biosecurity, Qingdao, China
- 4Key Laboratory of Maricultural Organism Disease Control, Ministry of Agriculture, Qingdao, China
- 5Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Covert mortality nodavirus (CMNV) is an alphanodavirus mainly infecting marine shrimp and co-inhabiting organisms in aquaculture ponds. To evaluate the possibility of CMNV prevalence in the wild fish species, epidemiological survey of CMNV infection in Larimichthys polyactis, the dominant species in the Yellow Sea and East China Sea, were conducted in the present study. We performed CMNV RT-LAMP assay in samples of L. polyactis collected in August 2018 and 2019 and found that CMNV prevalence was 18% and 7%, respectively. The nucleotide sequences of both RdRp and capsid protein genes of CMNV from L. polyactis were 99% similar to those of CMNV isolated from shrimp. CMNV-positive L. polyactis exhibited necrosis of cardiac muscle, oocytes loosely arranging, severe cytoplasmic vacuolation of hepatocytes, moderate pyknosis of brain pyramidal cells, degenerate renal tubular cells with ill-defined margins, and declined spleen cells in the histological examination. Moreover, CMNV-positive signals were further observed in pyramidal cells of the brain, the cortical area of the kidney, oocyte growth rings, and in necrotic tissues of cardiac muscle, liver, and spleen in the in situ hybridization assay. The results revealed that CMNV had colonized in the wild populations of L. polyactis and the ecological risk of CMNV spread and epidemic in wild fish in the coastal water was non-negligible.
Introduction
Viral covert mortality disease (VCMD), also being named as running mortality syndrome (RMS), impacted greatly on the shrimp farming industry in the Southern East Asia and China in the past decade (Pooljun et al., 2016; Thitamadee et al., 2016; Zhang et al., 2017; Varela, 2018). Covert mortality nodavirus (CMNV), an alphanodavirus, was determined to be the causative agent for VCMD (Zhang et al., 2014; Zhang, 2019). CMNV could infect the most economic shrimp species including Penaeus vannamei, P. monodon, P. chinensis, P. japonicus, as well as a variety of co-inhabiting crustaceans in the shrimp ponds (Zhang et al., 2017; Liu et al., 2019). Recently, CMNV has been reported to switch its hosts to fish by cross-species transmission and could infect teleosteans, such as the gold fish Carassius auratus, olive flounder Paralichthys olivaceus, and Abe’s mangrove goby Mugilogobius abei (Zhang et al., 2018; Wang et al., 2019). However, the CMNV prevalence in the wild teleost species has not been well investigated and evaluated yet.
The small yellow croaker (Larimichthys polyactis), an important commodity species in China, Japan, and South Korea, is widely distributed in the Bohai sea, Yellow sea, and East China Sea (Guo et al., 2006; Li et al., 2006; Tian, 2007; Shan et al., 2017). L. polyactis was known as one of the “four major fishery products” in the East China Sea. However, the volumes of L. polyactis landings continuously declined since the 1970s, due to human activities and environmental degradation (Jin, 1996). Although the resource of L. polyactis had recovered significantly since the 1980s under the protection measures of the Chinese government and the scientific community, the landings were still decreasing (Cheng et al., 2004; Lin, 2004; Guo et al., 2006). Recently, its population showed a tendency to shrink, and adult individuals tended to show characters of reduced size and early maturation (Jin, 1996; Qi et al., 2020). In view of the key role of its playing in the marine ecology, the L. polyactis has been the focus of research on fish species in the Yellow Sea and northern East China Sea. Biological characteristics, ecological habits, and stock assessment of L. polyactis had been well reported (Liu, 1990; Jin, 1996; Xue et al., 2004; Guo et al., 2006; Wang and Sun, 2006; Zhang et al., 2008; Shan et al., 2017). A few studies have focused on its diseases, let alone how the disease affects the wild L. polyactis population.
In the present study, we first conducted a systematic investigation of CMNV presence and infection in L. polyactis from the Yellow Sea and northern East China Sea through molecular detection, histopathology, and in situ hybridization (ISH). The results revealed that CMNV, an aquaculture animal virus, had colonized in the wild populations of L. polyactis in the investigated seas.
Materials and Methods
Sample Collection
In August 2018 and August 2019, bottom trawl samplings were conducted at 120 designated sites (south of 38°20′N, west of 120°30′E, Figure 1) in the Yellow Sea and northern East China Sea. Three to five individuals of L. polyactis (Figure 2A) were sampled at every designated sampling site. From each individual, the tissues of liver, kidney, spleen, brain, heart, and ovary (if available) were dissected, and half of the tissues were preserved in 4% paraformaldehyde (4% PFA) solution (Sinopharm, Beijing, China) for histopathological and in situ hybridization analysis. Another half was further divided into two parts, one part was preserved in RNAstore solution (Tiangen, Beijing, China) for RT-PCR analyses, the other part was smeared on Whatman FTA® Elute card (GE Healthcare Life Sciences, Marlborough, MA, United States) for RT-LAMP analyses. The tissue-smeared FTA® cards were air dried for 30 min at room temperature, and then stored at –20°C.
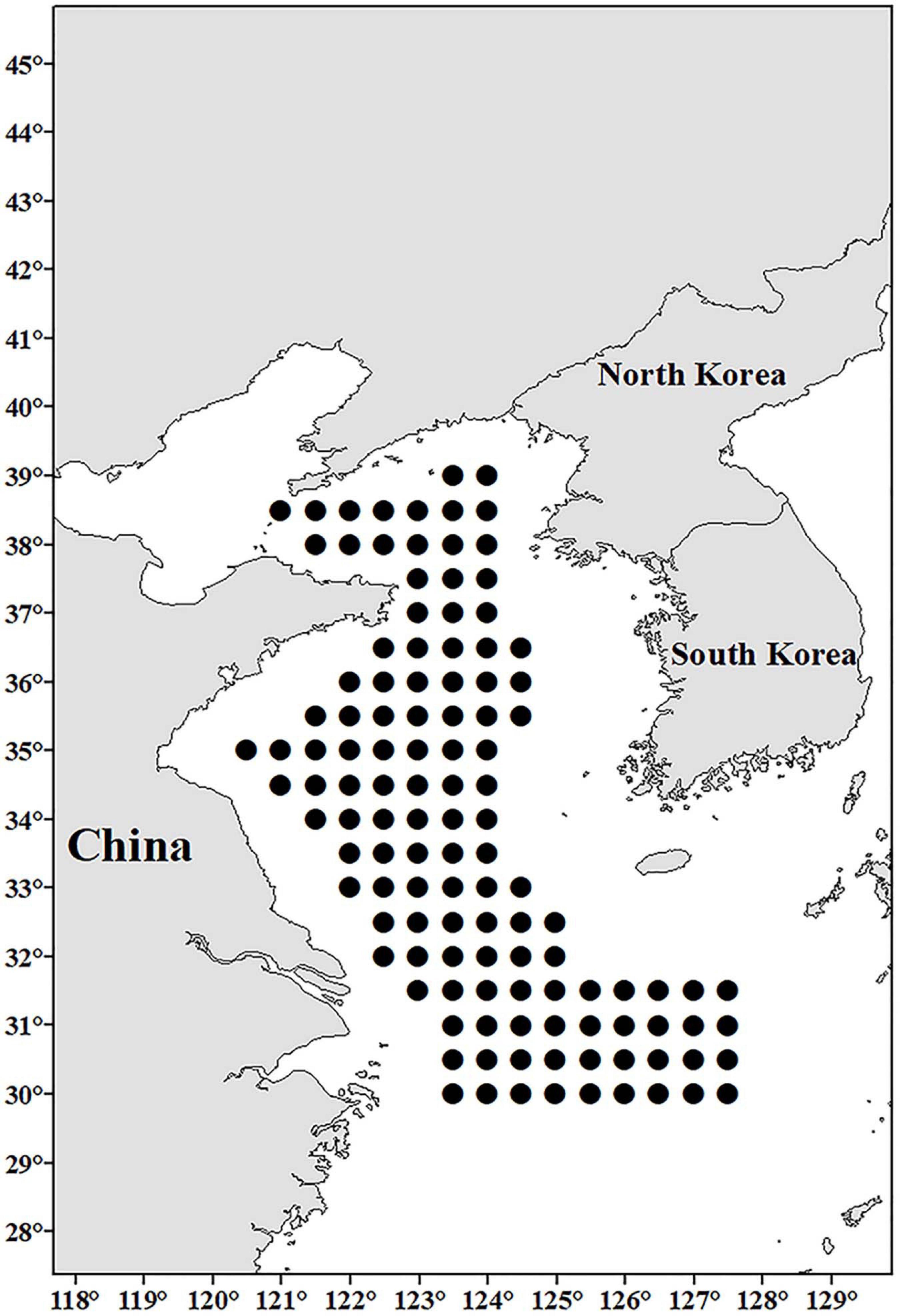
Figure 1. The designated sampling sites (black dots) in the Yellow Sea and northern East China Sea in the investigation.
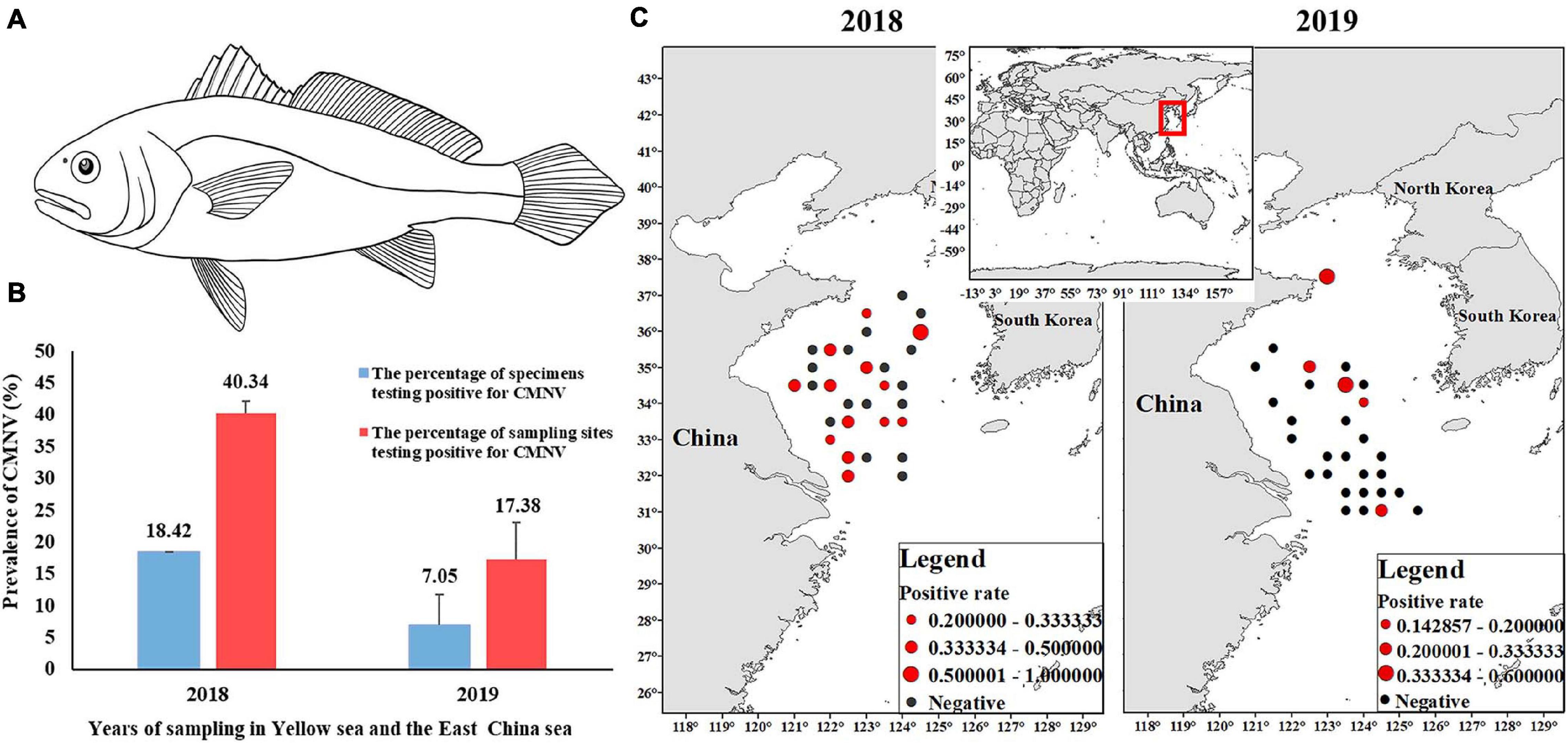
Figure 2. Prevalence of covert mortality nodavirus (CMNV) in Larimichthys polyactis of the Yellow Sea and the East China Sea in 2018 and 2019. (A) Pattern line drawing of L. polyactis; (B) CMNV-positive rates of samples and sampling sites; (C) prevalence rate and prevalence scope of CMNV. The black solid circles indicated the sites that L. polyactis were collected. The red solid circles indicate the sites the CMNV-positive L. polyactis were collected. The sizes of red solid circles represent the CMNV-positive rates.
Detection of Covert Mortality Nodavirus by RT-LAMP
From each of the tissue-smeared FTA® card, an area (about 2 mm2) was cut off and immersed into TE buffer to elute RNA. The eluted RNA was denatured at 70°C for 5 min, immediately placed on ice, and used as the template for CMNV RT-LAMP assay with the protocol described by Zhang et al. (2017).
RNA Extraction
The RNA was extracted from the CMNV RT-LAMP-positive RNA samples by using the Trizol RNA Extraction kit (Tiangen, Beijing, China), and then the amount and purity of the purified RNA was measured using the Nanodrop 2000 (Thermo Scientific, Waltham, MA, United States).
Primer Design, cDNA Synthesis, and PCR Amplification
To determine the targeted gene sequence of CMNV from the RT-LAMP-positive L. polyactis, six pairs of primers (Table 1) were designed based on the CMNV RNA1 and RNA2 genome segments (GenBank accession numbers MT270123 and MT270124) using the software Primer Premier 5 (Premier Biosoft, San Francisco, CA, United States).
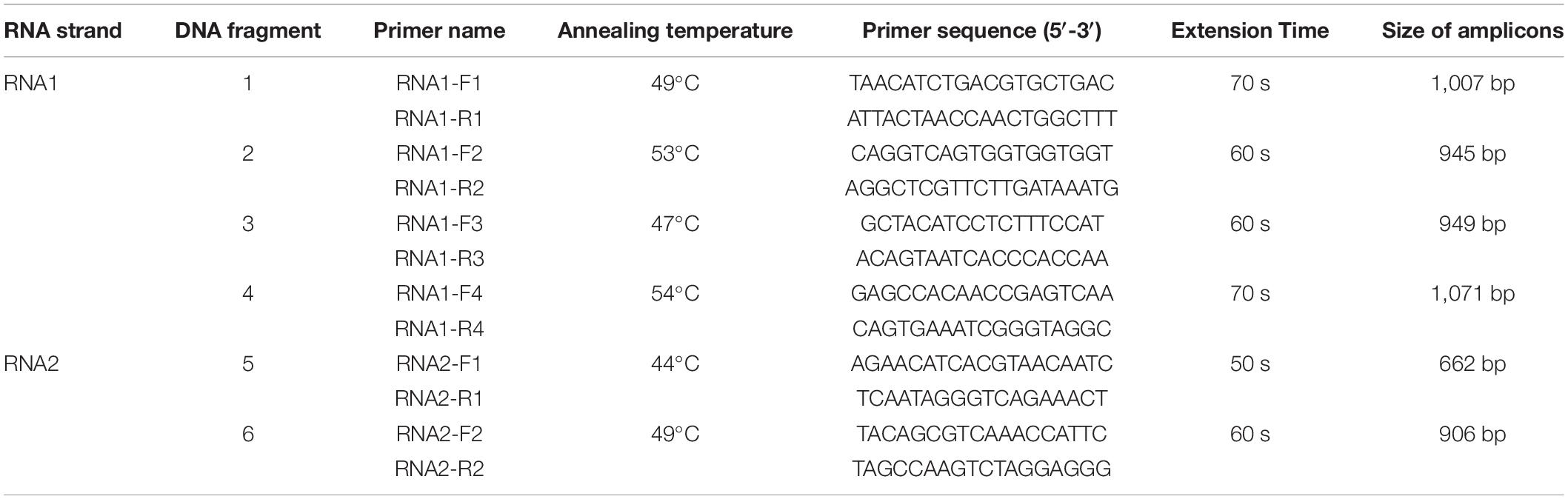
Table 1. PCR primer’s sequence, annealing temperature, and extension duration used to amplify RNA1 and RNA2 of covert mortality nodavirus (CMNV).
cDNA was synthesized from the extracted RNA with SMART® MLV-reverse transcriptase (TaKaRa) according to the procedure described by Xu et al. (2020).
PCR was carried out in a 25-μl reaction mixture containing 10 mM Tris/HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 1.5 mM dNTP, 0.2 μM primers, 2 μl of cDNA, and 2.5 U of TaKaRa EX Taq DNA polymerase (TaKaRa). The PCR was conducted according to a previous report (Xu et al., 2020) with modifications of annealing temperatures and extension time described in Table 1. DNA amplifications were performed for 35 cycles in all the reactions.
Sequence Analysis
A total of 20 viral genomic sequences (Supplementary Table 1) from the National Center for Biotechnology Information1 were downloaded and aligned with that of CMNV from L. polyactis by ClustalW as implemented (Tamura et al., 2011) using default settings. The phylogenetic tree was generated by the neighbor-joining method with bootstrap analysis (1,000 replicates) using MEGA 5.0.
Histopathological Section
The fish tissues were first preserved in 4% PFA for 12–24 h, and then transferred and stored into 70% ethanol. Paraffin sections were prepared and stained with hematoxylin and eosin-phloxine (H&E) according to the standard procedures (Lightner, 1996).
In situ RNA Hybridization
After examinng the H&E stained sections and determining the location of the lesions, the corresponding unstained sections were subjected to ISH with digoxigenin (DIG)-labeled CMNV RNA probe. In situ RNA hybridization was conducted according to the procedures reported (Piette et al., 2008; Chen et al., 2014). Final detection was performed with anti-digoxigenin antibody conjugated to alkaline phosphatase that was visualized using 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NTB). Nuclear Fast Red solution (Sigma-Aldrich, St. Louis, United States) was dropwise added onto the surface of the sections and stained the nucleus for 2 min. The sections were then visualized and photographed with a Nikon Eclipse E80i microscope (Nikon Co., Tokyo, Japan).
Results
Prevalence of Covert Mortality Nodavirus in Larimichthys polyactis
In August 2018, a total of 76 L. polyactis samples were collected from 30 sites out of the 120 designated sampling sites. The results of RT-LAMP analysis showed that 14 samples (18%) were detected to be CMNV positive (Figure 2B). The CMNV-positive samples were collected from 13 different sites, and the positive rate of sampling sites was determined to be 40% (Figures 2B,C).
In August 2019, a total of 102 L. polyactis samples were collected from 29 out of the 120 designated sampling sites. These 29 sites were different from those of 2018 (Figure 2C). RT-LAMP analysis results showed that seven samples (7%) from five sites were positive for CMNV (Figure 2B), and the positive rate of sampling sites was determined as 17% (Figures 2B,C).
In general, whether in 2018 or 2019, L. polyactis with positive CMNV were smaller than those with negative CMNV.
Covert Mortality Nodavirus (CMNV) RT-PCR, and Sequence Analysis
Larimichthys polyactis samples that were CMNV positive in the RT-LAMP assay were further subjected to RT-PCR analysis. Four fragments of the CMNV genomic RNA1, with the sizes of 1,007, 945, 949, and 1,071 bp, were amplified using the primer sets of RNA1-F1/R1, RNA1-F2/R2, RNA1-F3/R3, and RNA1-F4/R4, respectively (Figure 3A, lanes 1–4). Two fragments of the genomic RNA2, with the sizes of 662 and 906 bp, were generated with the primers RNA2-F1/R1 and RNA2-F2/R2, respectively (Figure 3A, lanes 5–6). All the overlapping amplicons were sequenced and then assembled according to the strategy shown in Figure 3B. The length of CMNVgenomic RNA1 and RNA2 fragments isolated from L. polyactis were determined as 3,228 and 1,448 bp, respectively. The genome sequence of L. polyactis-CMNV showed 99% similarity with the original CMNV isolate from shrimp P. vannamei (MT270123 and MT270124) in the BLASTn analysis. The deduced amino acid sequences of L. polyactis-CMNV RdRp and capsid protein were clustered with those from P. vannamei, respectively (Figure 4).
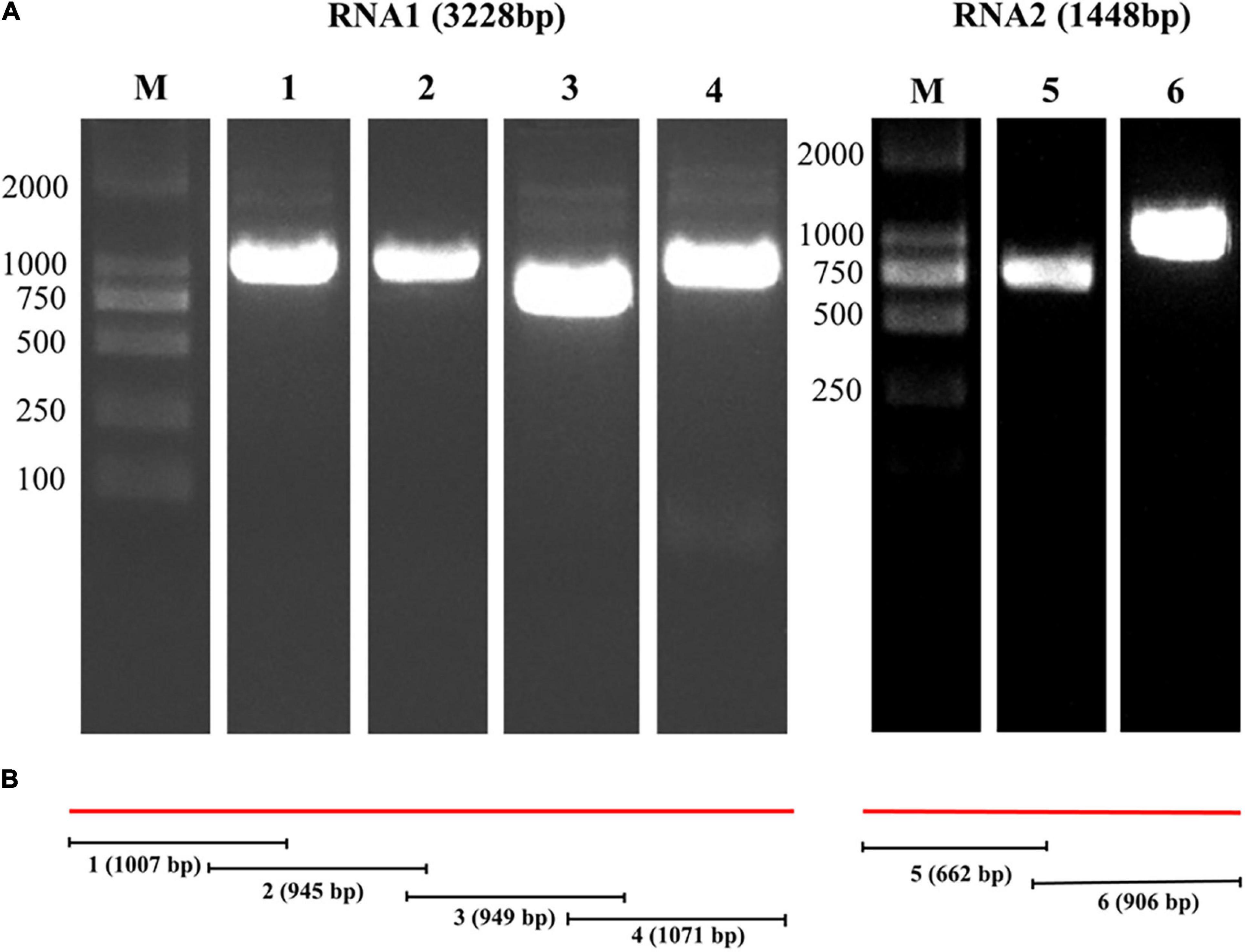
Figure 3. Assembly strategy and agarose gel electrophoresis of RT-PCR products of CMNV RNA1 and RNA2 segments from Larimichthys polyactis. (A) Agarose gel electrophoresis of RT-PCR products for cloning of the CMNV RNA1 and RNA2 strands. The images are cropped from the original image (Supplementary Figure 1). Lane M: 2,000 bp marker. Lanes 1–4: amplicons generated from the CMNV RNA1 with primers RNA1-F1/R1, RNA1-F2/R2, RNA1-F3/R3, and RNA1-F4/R4, respectively. Lanes 5–6: amplicons produced from RNA2 using primers RNA2-F1/R1 and RNA2-F2/R2. (B) Strategy of cloning and assembling the genomic RNA1 and RNA2 strands of CMNV by RT-PCR. The numbers 1–6 represent the size of the RT-PCR amplification products in lanes 1–6 in panel (A) and the position in the CMNV genome.
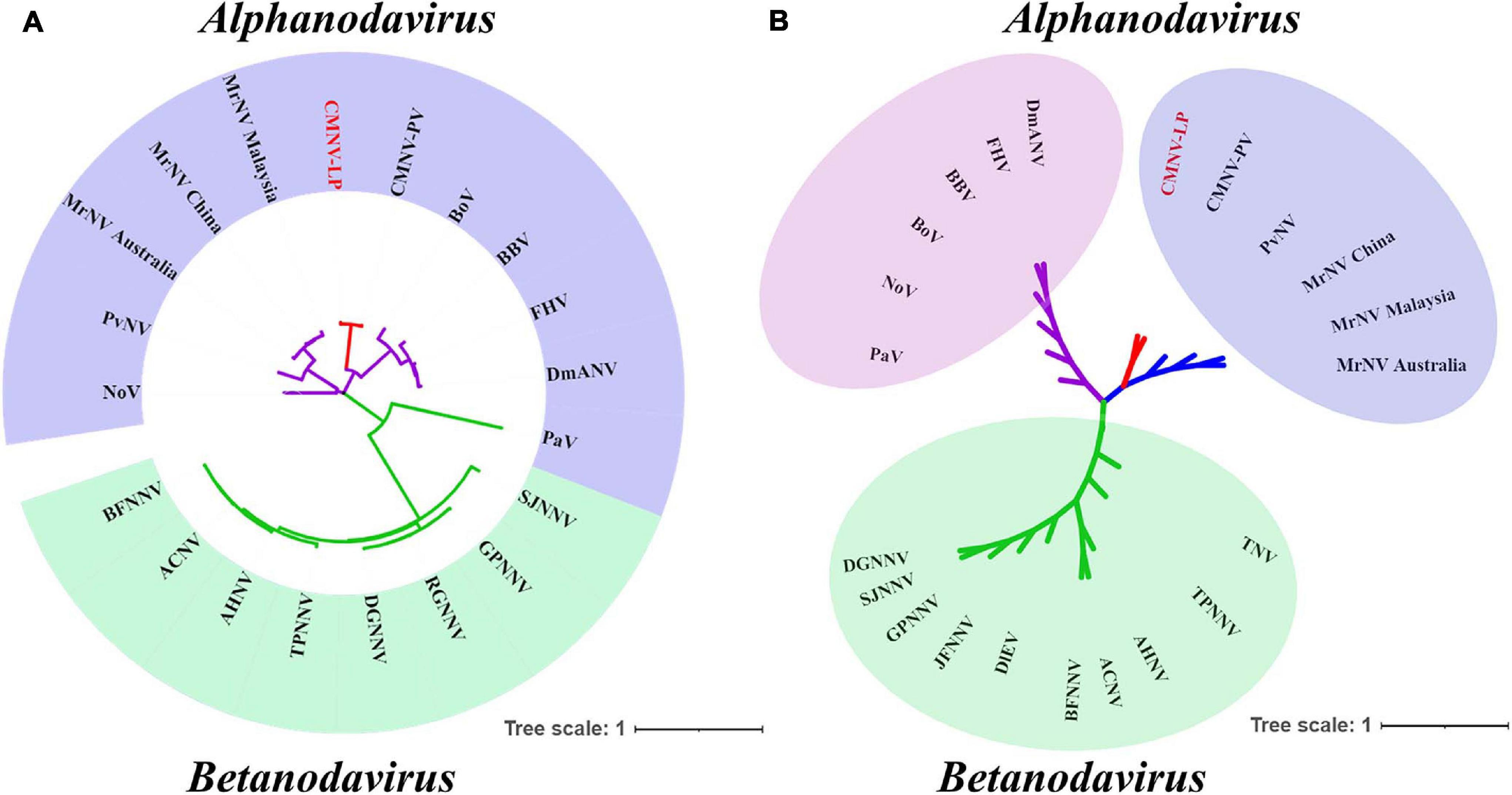
Figure 4. Phylogenetic tree based on the deduced amino acid sequences of RNA-dependent RNA polymerase (RdRp) and capsid protein from different CMNV isolates and other nodaviruses (for virus abbreviations see Supplementary Table 1). (A) Phylogenetic tree based on the RdRp. (B) Phylogenetic tree based on the capsid protein. CMNV-LP, CMNV in Larimichthys polyactis. Bar represents branch length of one substitution per site.
Histopathological Observation of Tissues From Covert Mortality Nodavirus (CMNV)-Positive Larimichthys polyactis
The tissues of L. polyactis that were determined to be positive for CMNV in RT-LAMP and RT-PCR assay were processed for histological evaluation. The histopathological observation revealed obvious histopathological changes and lesions in the tissues of the brain, kidney ovary cardiac muscle, liver, and spleen. Cerebral cortical tissue was severely vacuolated, and moderate karyopyknosis was noticed in pyramidal cells (Figure 5A). In the kidney, tubular degeneration and unclear boundaries, and vacuolar degeneration of the epithelial cells of the tubular wall were found. Meanwhile, the renal parenchyma showed diffuse hemorrhage, erythrocytes increased, and inflammatory cells exuded (Figure 5B). The oocytes in the ovary were loosely arranged (Figure 5C). Mild muscle fragmentation tending toward muscular lysis was noticed in cardiac muscle (Figure 5D). In the liver, the shape of the hepatocytes was deformed, and severe cytoplasmic vacuolation occurred in almost all hepatocytes (Figure 5E). Cells in the spleen were degenerated, and hemosiderin was obviously precipitated, forming “nodules” (Figure 5F). However, no typical lesions were observed in the above tissues of CMNV-free L. polyactis (Supplementary Figures 2A–F).
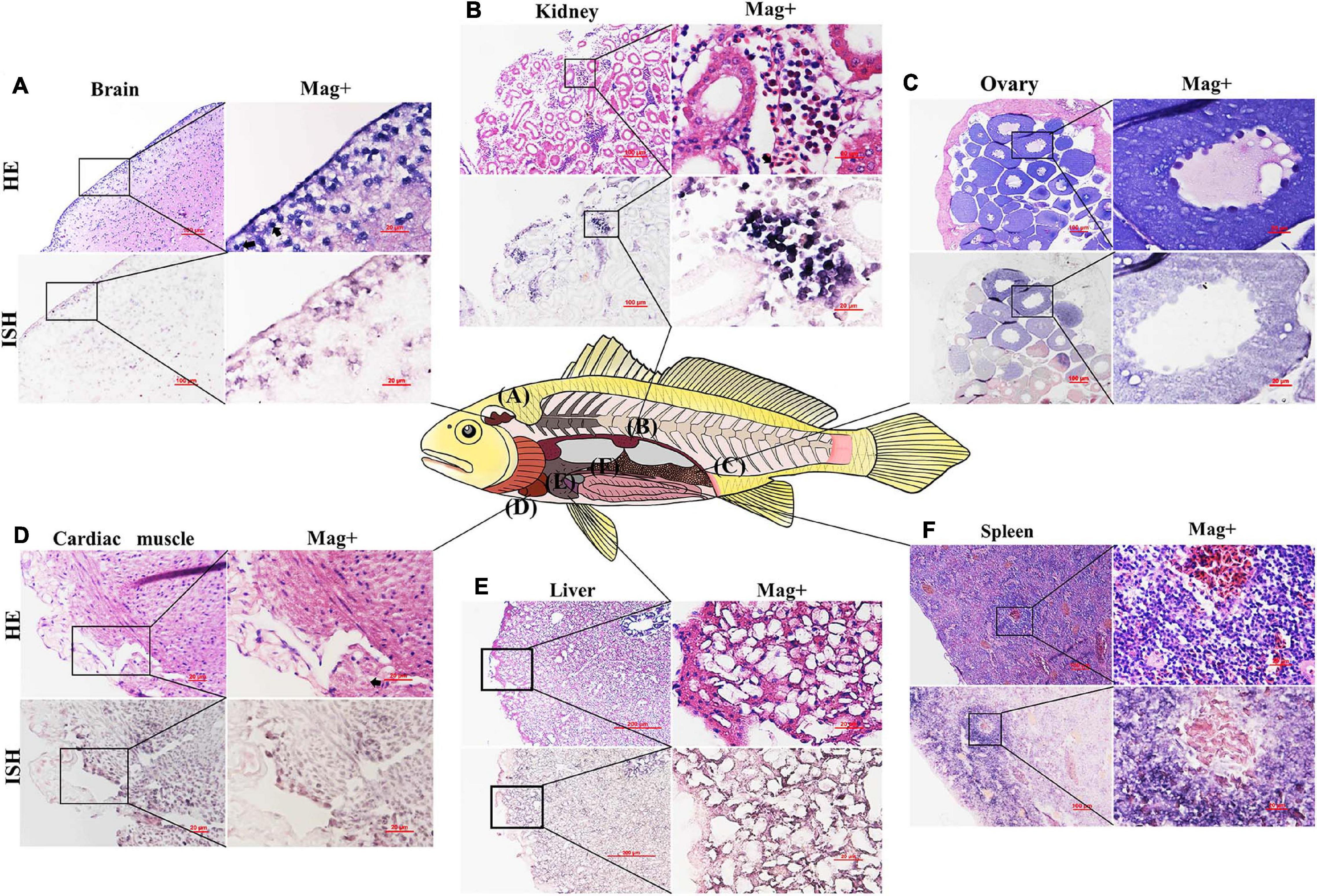
Figure 5. Micrographs of H&E staining and in situ hybridization for the brain, kidney, ovary, cardiac muscle, liver, and spleen of the Larimichthys polyactis naturally infected with CMNV. (A) brain, (B) kidney, (C) ovary, (D) cardiac muscle, (E) liver, and (F) spleen. Scale bar = 100 and 20 μm for low and high magnifications, respectively. HE, H&E stained; ISH, in situ hybridization; Mag+, high magnification.
In situ Hybridization Analysis of Tissues From Covert Mortality Nodavirus-Positive Larimichthys polyactis
The ISH assay results showed that purple hybridization signals of the CMNV probe could be observed in the pyramidal cells of the brain (Figure 5A), cortical tissue in the kidneys (Figure 5B), growth ring of the oocytes (Figure 5C), necrotic cardiac muscle cells (Figure 5D), necrotic hepatocytes (Figure 5E), and cells in the spleen (Figure 5F). However, no positive hybridization signals were detected on the sections without the addition of a CMNV DIG-labeled RNA probe (Supplementary Figures 3A–F).
Discussion
Up to now, CMNV had been prevalent in farming shrimp for 10 years and heavily attacked the production of farming shrimp. High incidence of CMNV infection in coastal ponds was causing increasing concerns of its spread to wild populations in the near seas. To evaluate the possibility of CMNV infection in the wild fish species, the infection and prevalence of CMNV in populations of wild L. polyactis, the dominant species in the Yellow Sea and the East China Sea, were investigated in the present study.
The investigation found that 18% (2018) and 7% (2019) of the L. polyactis samples collected from the Yellow sea and East China sea were CMNV positive. The rates of CMNV-positive sampling sites were determined to be 43.33 and 13.33% in 2018 and 2019, respectively. The results indicated that CMNV had colonized wildly in the Yellow Sea and East China Sea, but its prevalence in L. polyactis has declined in 2019 compared with that in 2018. Also, its large-scale spread might be related to the annual migration of L. polyactis populations in the Yellow Sea and East China Sea. The multiple sequence alignment and phylogenetic tree analysis showed that CMNV isolated from L. polyactis was highly homologous to the CMNV isolate from cultured Penaeus vannamei. It could be deduced that L. polyactis-CMNV isolate might originate from the aquacultured shrimp.
Wang et al. (2019) reported that CMNV infection in goldfish Carassius auratus could cause nervous tissue vacuolation and cardiac muscle necrosis. Zhang et al. (2018) found that CMNV infection in M. abei could induce extensive skeletal muscle necrosis, nervous tissue vacuolation in the retina of the eyes and the cerebellum of the brain. In this study, CMNV infection was revealed in multiple tissues of the L. polyactis individuals, including the brain, cardic muscle, liver, kidneys, spleen, and ovaries, by ISH analysis. The liver, the largest digestive gland of fish, has the functions of secretion, detoxification, storage, and excretion. Kidney and spleen are the main immune organs of fish. Hence, CMNV infection could affect the digestion and immune function of L. polyactis. Vertical transmission of CMNV has been reported in redgetail prawn Exopalaemon carinicauda (Liu et al., 2017). A high percentage of oocytes were found to be infected by CMNV, which had raised our concerns about the possibility of vertical transmission of CMNV in L. polyactis populations. However, L. polyactis is a benthic fish species, which will die due to the rupture of the swim bladder when caught during fishing (Liao, 1995), making it very difficult to obtain a live fish for the challenge test. Moreover, the technology of artificial cultivation and breeding of L. polyactis has not yet been achieved. Therefore, it is hard to perform challenge experiments to prove the vertical transmission ability of CMNV in L. polyactis at present.
The results of the present study raise concerns about the potential effects of CNNV on fish populations in coastal waters. A similar beta-nodavirus-like agent was suspected to be responsible for the mortality of mullets Liza saliens in the Caspian Sea (Mohammad et al., 2014; Maryam et al., 2015). Whether the decline in landings of L. polyactis was associated with CMNV infection remains to be determined. Lately, it was found that L. polyactis had gradually replaced anchovy (Engraulis japonicus) in the Bohai sea ecosystem and became an important food source for many larger predatory fish (Zhang, 2018). Although there was no direct evidence to prove the horizontal transmission ability of the CMNV from L. polyactis to other co-inhabiting fish or shrimp, the risk of CMNV propagation and spread in the wild fish species in the coastal waters could not be negligible considering that L. polyactis is a key prey in the Bohai sea ecosystem, especially preyed on by predators, and it might enhance the possibility of CMNV propagation and spread in the wild fish species in the coastal waters.
In conclusion, the molecular, histopathological, and epidemiological evidences in the present study indicated that CMNV had colonized in the populations of L. polyactis, one of the dominant species in the Yellow Sea and East China Sea. In addition, the ecological risk of CMNV spread and epidemic in wild fish in the coastal waters is non-negligible.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of the Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author Contributions
QZ, CY, and XS designed the experiments. TX, YL, and JH executed the experiments. TX and QZ analyzed the data. QW contributed to the sampling. TX wrote the manuscript. QZ, KT, and CY revised the manuscript. All authors interpreted the data, critically revised the manuscript for important intellectual contents, and approved the final version.
Funding
This work was supported by the National R&D Program of China (2017YFC1404503), the National Natural Science Foundation of China (32073016), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (NO. 2020TD39), the Projects of Marine S&T Fund of Shandong Province for Pilot, the National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0303-1), the Project of Species Conservation from the MARA-Marine Fisheries Resources Collection and Preservation, and the Central Public-Interest Scientific Institution Basal Research Fund, YSFRI, CAFS (NO. 20603022021022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the staff of the BEIDOU fishery research vessel from YSFRI, as well as the colleagues from the Division of Fishery Resources and Ecosystem, Yellow Sea Fisheries Research Institute, and Chinese Academy of Fishery Sciences, for sampling during the long-term surveys.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.670831/full#supplementary-material
Footnotes
References
Chen, S. L., Zhang, G. J., Shao, C. W., Huang, Q. F., and Liu, G. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260.
Cheng, J. H., Lin, L. S., Ling, J. Z., Li, J. S., and Ding, F. Y. (2004). Effects of summer close season and rational utilization on redlip croaker (Larimichthys polyactis Bleeker) resource in the East China Sea Region. J. Fish. Sci. China 11, 554–560. doi: 10.1016/S0960-0779(03)00420-X
Guo, X. P., Jin, X. S., and Dai, F. Q. (2006). Growth variations of small yellow croaker (Larimichthys polyactis) in the Bohai Sea. J. Fish. Sci. China 13, 243–249.
Jin, X. S. (1996). Studies on the ecology and population dynamics of yellow croakers in the Yellow Sea [J]. J. Fish. Sci. China 3, 32–46.
Li, J., Feng, F., and Yue, H. (2006). Twelve novel polymorphic microsatellites in a marine fish species, yellow croaker Larimichthys polyactis. Mol. Ecol. Resour. 6, 188–190. doi: 10.1111/j.1471-8286.2005.01188.x
Liao, X. D. (1995). The impact of water environment pressure on fish specific gravity and swim bladder. Bull. Biol. 030, 10–12.
Lightner, D. V. (1996). A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. Lodon: World Aquaculture Society Press.
Lin, L. S. (2004). Analysis of Existing Resources of Small Yellow Croakers in the East China Sea [J]. Mar. Fish. 26, 18–23.
Liu, S., Li, J. T., Tian, Y., Wang, C., Li, X. P., Xu, T. T., et al. (2017). Experimental vertical transmission of Covert mortality nodavirus (CMNV) in Exopalaemon carinicauda. J. Gen. Virol. 98, 652–661. doi: 10.1099/jgv.0.000731
Liu, S., Wang, X. H., Xu, T. T., Li, X. P., Du, L. C., and Zhang, Q. L. (2019). Vectors and reservoir hosts of covert mortality nodavirus (CMNV). J. Invertebr. Pathol. 154, 29–36. doi: 10.1016/j.jip.2018.03.011
Liu, X. S. (1990). Small Yellow Croaker Fishery Bureau of Ministry of Agriculture, Animal Husbandry and Fishery, Fishery Command of Yellow Sea District, Ministry of Agriculture. Survey and Regionalization of Fishery Resources in the Yellow and Bohai Sea Areas. Beijing: Ocean Press, 191–200.
Maryam, G., Mohammad, B., Mohades, G., Hasam, F., and Mohamad, J. Z. (2015). Haemato-biochemical disorders associated with nodavirus like-agent in adult leaping mullet Liza saliens (Risso, 1810) in the Caspian Sea[J]. Virus Dis. 27, 12–18. doi: 10.1007/s13337-015-0289-1
Mohammad, J. Z. R., Alireza, N., Mohaddes, G., Maryam, G., Somayeh, H. K., Giuseppe, B., et al. (2014). Vacuolating encephalopathy and retinopathy associated with a nodavirus-like agent: a probable cause of mass mortality of wild Golden grey mullet (Liza aurata) and Sharpnose grey mullet (Liza saliens) in Iranian waters of the Caspian Sea[J]. Virus Dis. 25, 430–436.
Piette, D., Hendrickx, M., Willems, E., Kemp, C. R., and Leyns, L. (2008). An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 3, 1194–1201. doi: 10.1038/nprot.2008.103
Pooljun, C., Direkbusarakom, S., Chotipuntu, P., Hirono, I., and Wuthisuthimethaveea, S. (2016). Development of a TaqMan real-time RT-PCR assay for detection of covert mortality nodavirus (CMNV) in penaeid shrimp. Aquaculture 464, 445–450. doi: 10.1016/j.aquaculture.2016.06.044
Qi, L., Alice, L., Zun, L. L., and Cody, S. S. (2020). Life history changes and fisheries assessment performance: a case study for small yellow croaker[J]. ICES J. Mar. Sci. 77, 645–654. doi: 10.1093/icesjms/fsz232
Shan, X. J., Li, X. S., Yang, T., Sharifuzzaman, S. M., Zhang, G. Z., and Jin, X. S. (2017). Biological responses of small yellow croaker (Larimichthys polyactis) to multiple stressors: a case study in the Yellow Sea, China[J]. Acta Oceanol. Sin. 36, 39–47. doi: 10.1007/s13131-017-1091-2
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thitamadee, S., Prachumwat, A., Srisala, J., Jaroenlak, P., Salachan, P. V., Sritunyalucksana, K., et al. (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69–87. doi: 10.1016/j.aquaculture.2015.10.028
Tian, Q. (2007). Health food from the ocean-yellow croaker [J]. Sea World, 1994. Biological food intake [J]. J. Ocean Univ. China 37, 75–82.
Varela, A. (2018). Pathologies of the hepatopancreas in marine shrimp cultivated in America and its differential diagnosis by histopathology. AquaTIC Magazine 50, 13–30.
Wang, C., Wang, X. H., Liu, S., Sang, S. W., and Zhang, Q. L. (2019). Preliminary study on the natural infection of Carassius auratus with covert mortality nodavirus (CMNV). Prog. Fish. Sci. 40, 25–32. doi: 10.19663/j.issn2095-9869
Wang, R. J., and Sun, S. (2006). The category composition and abundance of ichthyoplankton in the ecosystem of the Yellow Sea and the East China Sea. Acta Zool. Sin. 52, 28–44. doi: 10.1016/S1004-4132(06)60023-6
Xu, T. T., Liu, S., Li, X. P., and Zhang, Q. L. (2020). Genomic characterization of covert mortality nodavirus from farming shrimp: evidence for a new species within the family Nodaviridae. Virus Res. 286:198092. doi: 10.1016/j.virusres.2020.198092
Xue, Y., Jin, X. S., Zhang, B., and Liang, Z. L. (2004). Diet composition and seasonal variation in feeding habits of small yellow croaker Larimichthys polyactis in the central Yellow Sea. J. Fish. Sci. China 3, 237–243. doi: 10.3321/j.issn:1005-8737.2004.03.011
Zhang, B. (2018). The food relationship of Bohai Sea fish[J]. Prog. Fish. Sci. 039, 11–22. doi: 10.19663/j.issn2095-9869.20171103001
Zhang, B., Jin, X. S., and Dai, F. Q. (2008). Feeding habits of the two sciaenid fishes near the Changjiang estuary. Acta Zool. Sin. 54, 209–217.
Zhang, Q. L. (2019). Evidences for cross-species infection in fish of covert mortality nodavirus (CMNV). 12th Asian Fisheries and Aquaculture Forum, Iloilo Convention Center, Iloilo City, Pilipinas, 8-12 April 2019.Region. J. Fish. Sci. China 11, 554–560.
Zhang, Q. L., Liu, Q., Liu, S., Yang, H. L., Liu, S., Zhu, L. L., et al. (2014). A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 95, 2700–2709. doi: 10.1099/vir.0.070078-0
Zhang, Q. L., Shuang, L., Jun, L., Xu, T. T., Wang, X. H., Fu, G. M., et al. (2018). Evidence for cross-species transmission of covert mortality nodavirus to new host of Mugilogobius abei[J]. Front. Microbiol. 9:1447.
Keywords: covert mortality nodavirus, Larimichthys polyactis, coastal water, susceptible host, ecological risk
Citation: Xu TT, Li YX, Shan XJ, Hao JW, Wu Q, Tang KFJ, Zhang QL and Yao CL (2021) Natural Infection of Covert Mortality Nodavirus in Small Yellow Croaker in Coastal Water. Front. Mar. Sci. 8:670831. doi: 10.3389/fmars.2021.670831
Received: 23 February 2021; Accepted: 14 April 2021;
Published: 02 June 2021.
Edited by:
Jingqun Ao, Third Institute of Oceanography, State Oceanic Administration, ChinaReviewed by:
Changjun Guo, Sun Yat-sen University, ChinaXiaohong Huang, South China Agricultural University, China
Copyright © 2021 Xu, Li, Shan, Hao, Wu, Tang, Zhang and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingli Zhang, zhangql@ysfri.ac.cn; Cuiluan Yao, clyao@jmu.edu.cn; cnclyao@jmu.edu.cn
 Tingting Xu1,2,3,4,5
Tingting Xu1,2,3,4,5