
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci. , 14 May 2021
Sec. Coral Reef Research
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.668043
This article is part of the Research Topic Coral Reef Research Methods View all 22 articles
 Supisara Pipithkul1
Supisara Pipithkul1 Sota Ishizu2
Sota Ishizu2 Akifumi Shimura2
Akifumi Shimura2 Hiroyuki Yokochi3
Hiroyuki Yokochi3 Satoshi Nagai4
Satoshi Nagai4 Hironobu Fukami1*
Hironobu Fukami1* Nina Yasuda1*
Nina Yasuda1*Acropora pruinosa is a threatened zooxanthellate scleractinian coral that is distributed in the temperate areas along the coastline of Japan and the northern area of the South China Sea. Since A. pruinosa propagates both asexually and sexually, assessing clonal diversity and genetic connectivity among populations is important for conservation. In addition, high morphological variations in the field create confusion during species identification. To examine the existence of hidden genetic lineages, clonality, and genetic connectivity of A. pruinosa for conservation, we applied microsatellite analysis. Clustering analysis indicated two distinct geographically separated genetic lineages: one is distributed in the west, and the other is distributed in the east. The two lineages co-existed in Nishidomari, Kochi. There was no obvious difference in morphological characteristics between the two lineages. Although the factors influencing the observed distribution patterns remain unknown, there is a possibility that the two lineages might have diverged somewhere in the north-western Kyushu and north-eastern Pacific coast habitats in the past, and then periodically colonized the current habitats. A low clonal diversity was observed in most of the populations, indicating a high rate of asexual reproduction associated with their branching morphologies. In addition, there are strong genetic structuring in this species, indicating weak connectivity among populations. These results indicated a low larval dispersal potential among populations and that populations are basically sustained by a high rate of clone propagation and self-seeding. The existence of cryptic lineages and genetically isolated populations with high clonality emphasized the importance of conservation of A. pruinosa.
Coral reefs provide habitat, shelter, and food for many marine organisms, helping with nutrient recycling in the ocean and other ecosystem services (Principe et al., 2011). In Japan, the distribution of coral communities stretches from the south-western areas toward the eastern part of the Japan mainland (Nishihira, 2004). However, coral communities have been threatened by ocean acidification, climate change, and human activities (Goldberg and Wilkinson, 2004), and reef-building coral species are now listed to be threatened or vulnerable species (IUCN, 2020). Thus, basic knowledge for the proper conservation planning for coral species is needed. Most coral species can reproduce both asexually and sexually and disperse from their natal populations to cause genetic connectivity among different populations during pelagic larval period. Genetic diversity and clonality assessment may also play a vital role in predicting the adaption of populations to climate and environment changes (Gorospe et al., 2015). Thus, information about clonality, genetic diversity, and genetic connectivity between populations is required for the protection of species. In this context, the molecular population genetic approach, which is one of the promising methods for examining the clonality and genetic structure of species, can provide useful information for conservation purposes (O’Brien, 1994).
Acropora pruinosa is a hermatypic coral with an arborescent growth form (underwater photos in Figure 1). Veron (1992) suggested that this species is presumably endemic to the Japan mainland and Hong Kong. This unusual and limited distribution as well as sudden decrease of A. pruinosa population in some areas (Nanto et al., 2009; Nomura, 2009) led the species to be listed in the Japanese Red list1 (Ministry of the Environment, 2017). Acropora pruinosa is a hermaphrodite and broadcast-spawning species. This coral species releases its gametes into the water column once a year in summer, approximately 106 ± 10 min after sunset (Suzuki and Fukami, 2012). After fertilization of gametes in the water column, larvae of A. pruinosa move to the surface and float in the ocean for 3-10 days before settling on the ocean floor (Iwase et al., 2009). Larvae of A. pruinosa have a high settlement rate, and usually settle on places slightly exposed to sunlight (Iwase et al., 2009). The overall early life ecology is quite similar to that of other Acropora spp. (Iwase et al., 2009) and it exhibits genetic connectivity among different populations (e.g., Nakabayashi et al., 2019).
Similar to other coral species, A. pruinosa has a high morphological diversity, especially in terms of the thickness and length of branches, causing confusion in species delimitation. This species is often confused with Acropora tumida in Japan; arborescent or caespitose colonies with thicker and robust branches have been identified incorrectly as A. tumida (Nishihara and Veron, 1995). Sugihara et al. (2015) showed that real A. tumida exhibits a corymbose colony form but not an arborescent or caespitose colony form like A. pruinosa. Examining possible hidden lineages using molecular markers and comparisons between morphologies and genetically different lineages are of great importance for determining conservation units.
The aims of this study were to provide useful information to conserve the threatened reef-building species, A. pruinosa; assess the clonality, genetic diversity, and genetic structure of A. pruinosa to examine the existence of possible hidden lineages; assess the concordance between genetically distinct lineages and morphological characteristics; and to examine connectivity among different populations of A. pruinosa. To this end, we applied codominant, highly polymorphic microsatellite markers by collecting samples from most of the Japanese habitats of A. pruinosa.
In total, 331 small fragment colonies (about 5-10 cm long) of A. pruinosa were collected by SCUBA diving from 17 temperate sites along the Kuroshio and Tsushima currents covering most of the species’ distribution in Japan from 2009 to 2015 (Table 1 and Figure 1). A small fragment (about 5 mm) of each coral sample was collected and preserved in CHAOS solution for DNA extraction (Fukami et al., 2004). Coral samples were collected from independent colonies that are mutually distantly apart whenever possible. However, the distance between the sampled colonies varied depending on the size of the population, and the shortest distance between colonies was about 1 m in a small population, such as the Enashi population (E). The rest of the samples were bleached for morphological examination.
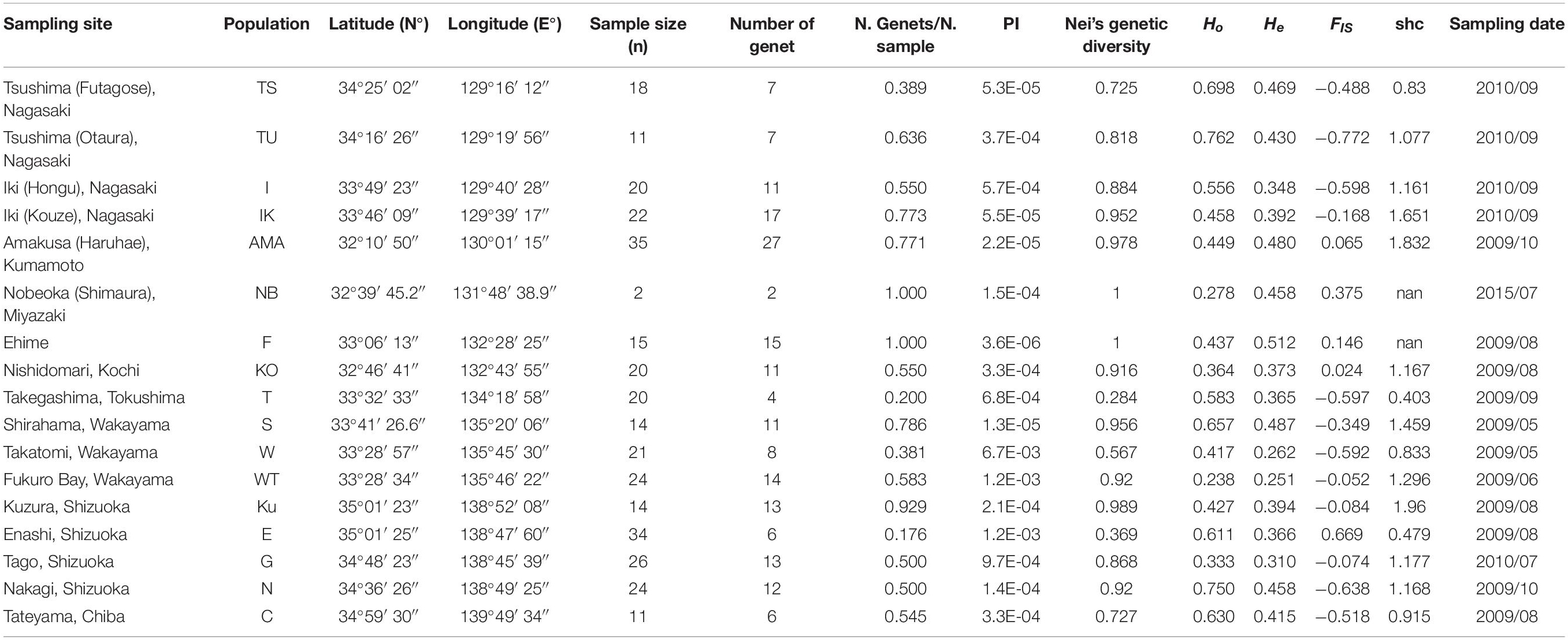
Table 1. List of the sample site, population name, latitude (in the northern hemisphere), longitude (in the eastern hemisphere), sample size (n), number of genet in each population, number of genet divided by sample size (N. Genets/n), cumulative probability of identity of the 9 microsatellite markers (PI), Nei’s genetic diversity, mean observed heterozygosity (Ho), mean expected heterozygosity (He), fixation index (FIS), and Shannon index, which was corrected for sample size (shc), and the sampling date for all populations (yyyy/mm) of Acropora pruinosa.
Genotypes of the corals were determined using 9 microsatellite loci (Shinzato et al., 2014; van Oppen et al., 2016). Nine loci were amplified independently and then mixed into 3 PCR product sets using different dyes and non-overlapping loci size: Plex 1 includes 8346m3, 8499m4, and 10366m5; Plex 2 includes 441m6 and 7961m4, and Amil 2-2; and Plex 3 includes Amil 2-10, Amil 2-12, and Amil 2-23. The total PCR reaction volume was 8.14 μl, including 1 μl of template DNA, 0.07 μl of each 50 mM primer, 4 μl of master mix (GoTaq® Promega), and 3 μl of water. The PCR program was 95°C for 5 min, followed by 40 cycles at 95°C for 30 s, 40°C for 30 s, 72°C for 30 s, and a final extension step at 72°C for 5 min.
The PCR products were diluted by 20, 50, 100, 200, 400, and 800 times depending on the quality of the samples after electrophoresing on agarose gel. For each sample, 1 μl of the diluted PCR product was added to 14 μl of highly deionized formamide containing 0.2 μl of the GeneScan-600 ZX (LIZ) size standard (Applied Biosystems, Foster City, CA, United States) for genotyping on an Applied Biosystems 3730xl DNA analyzer (GeneMapper ver. 6, Applied Biosystems) into determine the allele sizes of all samples. All the genotyping loci were manually checked and modified. Wherever there was ambiguously, we re-extracted genomic DNA and re-amplified it for accurate genotyping.
We analyzed the clonal structure of each population, Nei’s genetic diversity, and Shannon index based on 9 microsatellite loci by Genodive and then calculated the probability of identity using GenAlex ver. 6.51b2 (Peakall and Smouse, 2006, 2012). This software was used to identify possible clones and clonal diversity in each population (asexually reproduced by fragmentation) and all the clonal samples except for one genet, which was excluded from the subsequent population genetic analyses. We then measured observed heterozygosity and expected heterozygosity to estimate genetic diversity in each population.
We used STRUCTURE software (Pritchard et al., 2000; Falush et al., 2003, 2007; Hubisz et al., 2009), using the admixture model without any prior information. We used the following settings to assign each individual to a pre-defined number of hypothetical clusters (K): a burn-in of 100,000 chains followed by 100,000 Markov chain Monte Carlo replications. We performed ten independent runs for each K = 1 to 10 to confirm convergence. We used STRUCTURE HARVESTER (Earl and VonHoldt, 2012) to estimate delta K and applied CLUMPAK (Kopelman et al., 2015) to calculate and visualize the coefficient of ancestry for each sample across 10 runs for the most likely values of K (Delta K).
We conducted principal coordinates analysis (PCoA) of 13 populations (N > 7) including Futagose and Otaura of Tsushima, Hongu and Kouze of Iki island, Amakusa, Ehime, Kochi, Shirahama, Takatomi, Fukuro Bay, Kuzura, Tago, and Nakagi using GenAlex 6.51b2 (Peakall and Smouse, 2012) to visualize the genetic relationship among different populations and to confirm the result of STRUCTURE analysis.
We measured the following morphological characteristics: thickness of branch under 1 cm from the tip, and outer and inner diameters of the axial corallite (Supplementary Figure 1). We also counted the septal number of axial corallites and observed the radial corallites and structure of the coenosteum (Supplementary Table 2). Vernier caliper was used to measure the branch thickness, and a digital microscope VHX-1000 (Keyence) was used for all others. We randomly chose 44 samples and collected 2 or 3 sets of data per 1 sample, according to the number of their branches. We calculated the average, standard deviation, and maximum and minimum values over all samples and within each cryptic lineage. We also analyzed the data with a non-parametric statistical method, Mann-Whitney-Wilcoxon Rank Sum, to test if there is a significant difference between two lineages.
To assess genetic connectivity of the samples from different populations, we calculated global FST as well as pairwise FST of 13 populations (N > 7) including Futagose and Otaura of Tsushima, Hongu and Kouze of Iki island, Amakusa, Ehime, Kochi, Shirahama, Takatomi, Fukuro Bay, Kuzura, Tago, and Nakagi using GenAlex ver. 6.51b2 (Table 2). We also calculated global FST for eastern and western cryptic lineages.
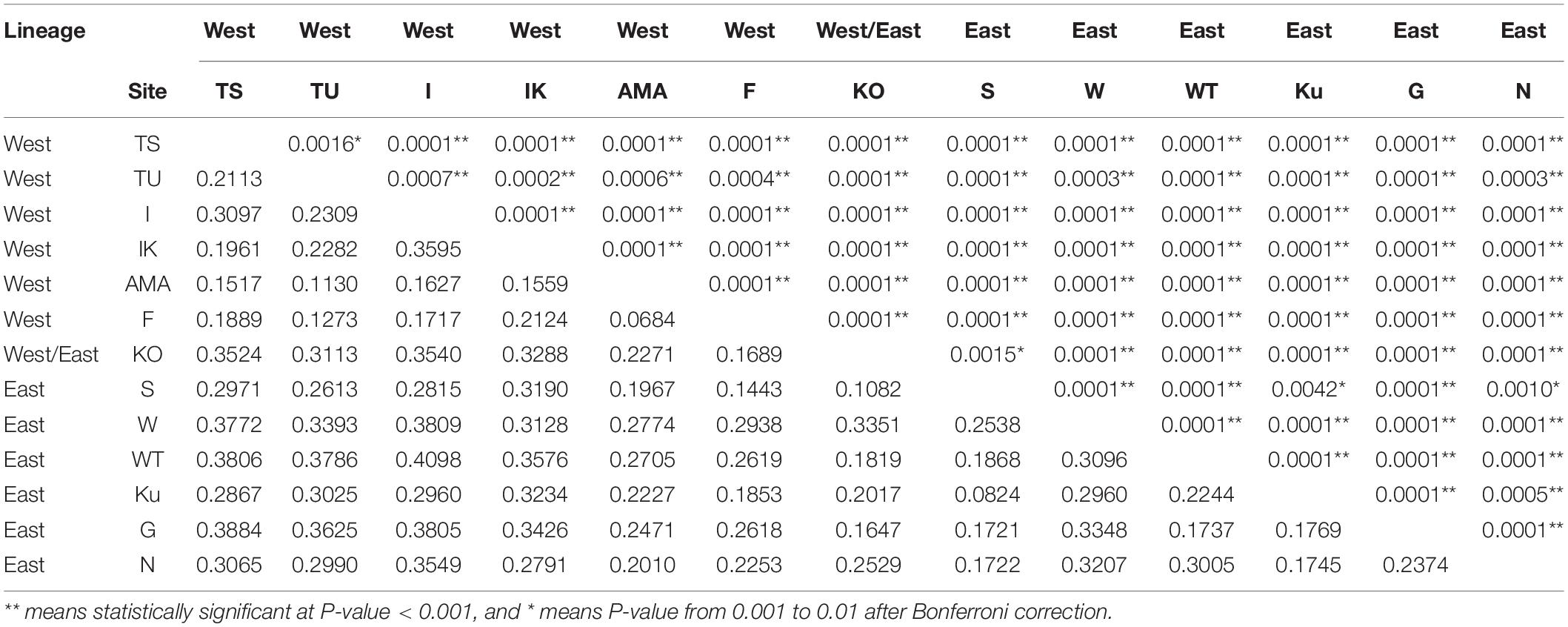
Table 2. Pairwise FST values (below) and P value (above) of the 13 Japanese populations of Acropora pruinosa.
We found a relatively high proportion of asexual reproduction in each A. pruinosa population; clonal diversity using the 9 resemblance microsatellites loci ranged from 84 to 337 (Genotyped raw data available in Supplementary Table 1). The probability of identity averaged 0.00076 and ranged from 0.0067 (Takatomi, Wakayama) to 0.0000036 (Ehime) (Table 1). Thus, we assumed that the 9 microsatellite markers cannot be used to completely identify clones, but the possible error rate is still ignorable and thus is powerful enough to estimate clonality and genetic diversity in each population in this study. High frequencies of asexual reproduction with a small number of genets compared to the number of individuals were especially found in Tsushima, Iki, Amakusa, Kochi, Takegashima, Takatomi, Fukuro Bay, Enashi, Tago, Nakagi, and Tateyama. Nei’s genetic diversity of each population showed the lowest value (0.284) at Takegashima and then Enashi, which indicated that each genet within a population contains quite similar genes, when comparing to other populations. Observed and expected heterozygosity ranged from 0.238 to 0.762 and 0.251 to 0.512 (Table 1).
The STRUCTURE analysis revealed delta K = 2, and there were two distinct cryptic lineages within Japanese populations of A. pruinosa (Figure 2A). The distribution of the two lineages clearly corresponded to the geographic regions: western (Tsushima, Iki, Amakusa, Nobeoka, Ehime) and eastern (Takegashima, Shirahama, Takatomi, Fukuro Bay, Kuzura, Enashi, Tago, Nakagi, Tateyama) lineages, and the two co-existed in Kochi. The PCoA also clearly suggested a distinction between western and eastern populations, and the Kochi population showed more resemblance to the population of the eastern lineage than to that of the western lineage (Figure 2B).
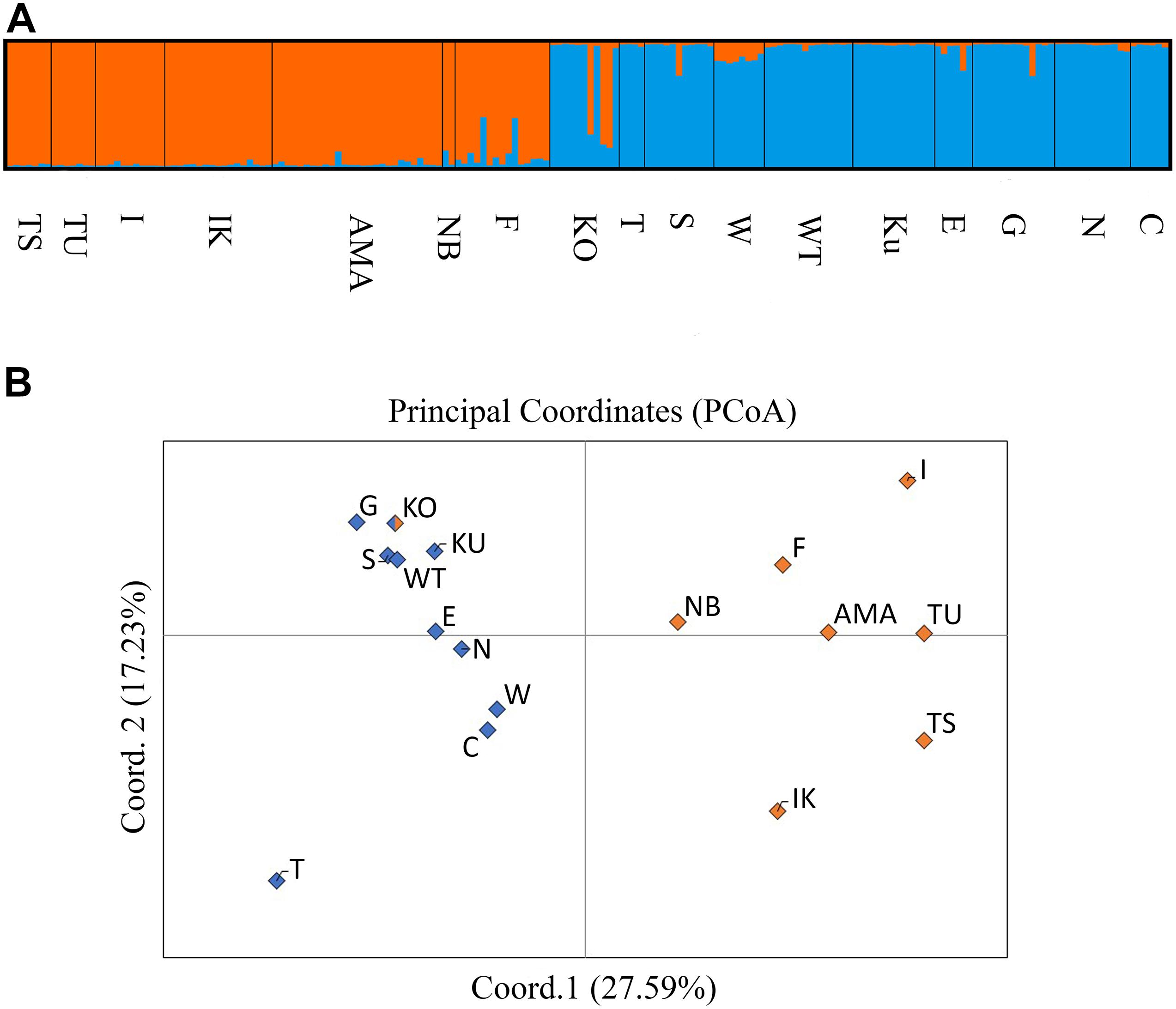
Figure 2. (A) Bar plot data from STRUCTURE analysis at K = 2 that analyzed all A. pruinosa samples across Japanese populations indicated the clearly distinct western and eastern lineages, which co-existed in Kochi. (B) PCoA indicates the clearly separated cryptic lineages from Japanese populations revealed by STRUCTURE at K = 2; the first two coordinates explain 27.59% (x-axis) and 17.23% (y-axis) of the data.
We measured the inner and outer diameters of axial corallites and the thickness of branches under 1 cm from the tip. The average lengths of all samples were 998 (S.D ± 149.3), 2,076 (S.D ± 243.0), and 3,076 (S.D ± 673.8) μm for inner diameter of axial corallites, outer diameter of corallite, and the thickness of branches under 1 cm from the tip, respectively (Supplementary Table 2). The lengths of the inner diameter ranged from 730 to 1,678 μm, and those of the outer diameter ranged from 1,630 to 3,011 μm; the thickness of branches under 1 cm from the tip ranged from 2,047 to 5,780 μm. The averages of the inner and outer diameters of axial corallites, and the thickness of branches under 1 cm from the tip for the western lineage were 1021.2 (S.D ± 156.7), 2071.4 (S.D ± 219.6), and 3628.6 (S.D ± 721.3), respectively, while those for the eastern lineage were 962.3 (S.D ± 130.5), 2082.5 (S.D ± 277.1), and 3823.7 (S.D ± 579.8), respectively (Supplementary Table 2).
The septal number of axial corallites was 11.7 (S.D ± 1.2) for all samples, and 12.0 (S.D ± 1.3) for the western lineage, and 11.5 (S.D ± 1.1) for the eastern lineage (Supplementary Table 2). In all samples, six primary septa were distinct with nearly 2/3 to 1 radius of inner diameter. In all samples, radial corallites usually had 6 primary septa with nearly 2/3 to 1 radius of inner diameter, and the coenosteum was reticulate.
After Mann-Whitney-Wilcoxon Rank Sum test to compare the two lineages, we found the P-values for the inner and outer diameters of axial corallites, the thickness of branches under 1 cm from the tip, and septal number of axial corallites to be 0.980, 0.584, 0.011, 0.926, respectively, suggesting a significant difference for only the thickness of branches under 1 cm from the tip. However, the range of the thickness of branches under 1 cm from the tip overlapped between the eastern and western lineages, indicating that the thickness of branches under 1 cm from the tip alone could not distinguish the two species (Supplementary Table 3).
The global FST among all populations based on 9 microsatellite loci was large (FST = 0.245, P < 0.001). Pairwise FST values among 13 populations were also high, even within the same lineage; in the western lineage, pairwise FST values ranged from 0.0684 (P < 0.001 between F and AMA) to 0.3595 (P < 0.001 between I and IK), and the in eastern lineage, pairwise FST values ranged from 0.0824 (P = 0.0042 between S and Ku) to 0.3096 (P < 0.001 between W and WT). All the FST values for all populations were statistically significant (P < 0.01) after Bonferroni correction (Table 2).
This study provided basic important information for conservation of the threatened reef-building coral species A. pruinosa; a relatively high proportion of asexual reproduction and low genetic diversity in each population, weak genetic connectivity among populations, presence of eastern and western cryptic genetic lineages were observed in Japan. All these facts emphasize the threatened status of A. pruinosa, highlighting the importance of effective conservation management of the species.
Principal coordinates analysis and STRUCTURE results revealed the existence of two genetically different lineages distributed in the east and west and co-existed in Kochi. While there was a significant difference in the thickness of branches under 1 cm from the tip between the eastern and western lineages, it was not possible to distinguish the lineages using this morphological trait alone because there was a substantial overlap in the thickness of branches under 1 cm from the tip between the two lineages. Recently, Taguchi et al. (2020) reported that A. pruinosa in Nishidomari, Kochi, where is the same sampling site in our study, had two different chromosome number, 28 and 29, in the same embryo. This difference might reflect the two genetically different lineages, but further detailed microscopic examination of morphology, ecological and physiological experiments will be required to determine the species status of the eastern and western lineages of A. pruinosa.
The existence of genetically separated cryptic lineages and their distribution are enigmatic. One possible explanation is that two distinct refugia, such as the north-eastern (somewhere in the eastern Pacific coast) and north-western (somewhere in the western Kyushu area) refugia, resulted in the distinct genetic lineages during the past climate change. A. pruinosa is a temperate coral that has a relatively higher tolerance to cold temperature than other coral species, including the widely distributed (tropical to temperate) species Acropora hyacinthus and Acropora solitaryensis (Higuchi et al., 2015). A. pruinosa is not distributed in subtropical areas in Japan. This trait of adapting well to cold temperature suggests that A. pruinosa might survive in colder habitats, such as the refugia in the north-eastern (Pacific) and north-western (western Kyushu) habitats, during the past warming period when corals suffered from coral bleaching in the warmer areas due to elevated water temperature. After such a warming period, the distribution of the species might expand to the southwest and southeast again to the current locations with time. Population genetic analysis on Japanese turban shell Turbo cornutus also showed a similar distribution of two distinct mitochondrial haplotype clusters, one of which is distributed along the Tsushima current to the west and the other is distributed along the Kuroshio current to the east. The two types also co-existed in the Seto Inland Sea (Kojima et al., 1997). The authors suggested that the two distinct lineages might have formed by two geographically separated refugia in the Japan Sea (western Kyushu) and on the Pacific coasts during the past climate change. Gene flow through a narrow strait called the Kanmon Strait resulted in the co-existence of the two types in the Seto Inland Sea (Kojima et al., 1997). Similarly, in A. pruinosa, the co-existence of the western and eastern lineages in Kochi and western lineage in Nobeoka and Ehime might be attributed to gene flow through the Kanmon Strait from the north-western refugia.
A high ratio of asexual reproduction was found in A. pruinosa. Despite the similar habitats and sexual reproductive biology with A. hyacinthus and A. solitaryensis (Iwase et al., 2009), A. pruinosa showed a higher ratio of asexual reproduction than A. hyacinthus (Nakabayashi et al., 2019) and A. solitaryensis (Noreen et al., 2013). It is likely that coral growth form is associated with the ratio of asexual reproduction; the arborescent growth form is more fragile to environmental factors than the corymbose or tabular form (Wallace, 1999).
In addition to the high clonality, we found a low genetic diversity of A. pruinosa in each population, indicating that this species will be vulnerable to environmental changes. Furthermore, the limited genetic connectivity of A. pruinosa among populations suggests that each population is highly depends on local reproduction (self-recruitment), including asexual reproduction, implying a higher risk of local extinction in the future in the face of climate change.
Acropora pruinosa has a more limited genetic connectivity than other Acropora species, such as Acropora digitifera (Nakajima et al., 2010), A. solitaryensis (Noreen et al., 2013), and A. hyacinthus (Nakabayashi et al., 2019), despite having a similar early life ecology and the same larval dispersal duration (Iwase et al., 2009) to these species. It is possible that the arborescent morphological character, associated with a high frequency of asexual reproduction (fragmentations) of A. pruinosa, might be responsible for the limited genetic connectivity among populations. Laboratory experiments examining the fecundity of fragmented arborescent form A. formosa (= Acropora muricata) indicated that there is a trade-off of energy between reproduction and survival (Okubo et al., 2007). The high frequency of asexual reproduction in the arborescent A. pruinosa leads to a lower number of sexually reproduced larvae that disperse among different populations, resulting in the limited genetic connectivity of A. pruinosa.
This study revealed that the threatened status of A. pruinosa is much more serious than expected due to the overall low genetic diversity and the existence of cryptic lineages (possibly different species). Whether the two cryptic lineages are different species or not, genetically different lineages with different evolutional histories should be conserved separately to sustain genetic and biodiversity as they might have adapted to different environments. Thus, the existence of two separated cryptic lineages emphasized the importance of more precise conservation because cryptic lineages would have different heat tolerance capacities and might respond differently to climate change. Although we need more detailed morphological, ecological, and physiological analyses of the two lineages to examine species boundaries, our results cast a doubt on the “unusual distribution” of a single species, A. pruinosa. Given that we found distinct cryptic lineages and limited larval dispersal of A. pruinosa, even within Japan, it will be worth examining if A. pruinosa in geographically separated Hong Kong is really the same species. If A. pruinosa in Hong Kong is a genetically distinct endemic species, it requires more attention for conservation.
The overall low genetic diversity and high rate of asexual reproduction and self-seeding across the Japanese populations suggested that A. pruinosa is potentially vulnerable to environmental changes in the future because a low genetic diversity limits the ability of populations to adapt and evolve in response to climate changes and there is a low chance of replenishment by recruitment from other neighboring populations (Nei et al., 1975; Spielman et al., 2004). Alternatively, the high rate of clonal growth might be an important reproductive strategy for the preservation of the genetic diversity of populations or for the maintenance of population persistence during poor sexual recruitment periods (Baums et al., 2006).
This study unveiled evolutionary distinct cryptic lineages of the threatened coral species A. pruinosa that are distributed in the east and west of the temperate Japanese coast. These two lineages might have resulted from the eastern and western refugia during the past climate change. This study also demonstrated a much higher ratio of asexual reproduction and more limited connectivity of A. pruinosa among different populations than other Acropora spp. Even though A. pruinosa (arborescent form) has almost the same early life ecology (spawning period, pelagic larval duration, and larval behavior) as other Acropora spp., a much higher rate of asexual reproduction and limited connectivity were found for this species than for the corymbose or tabular form Acropora spp., highlighting the importance of coral growth form on meta-population structure. All these information emphasize the threatened status of A. pruinosa, requiring urgent and effective conservation to avoid extinction in the face of climate change.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
HY, HF, and NY conceived the study. HY and HF collected the samples. SI and HF conducted morphological analysis. SP, SI, AS, SN, and NY conducted molecular experiment. SP and NY analyzed the data and drafted the manuscript. All authors checked and edited the final manuscript.
This study was supported by a research grant from the Kuroshio Biological Research Foundation, Grant-in-Aid for Young Scientists (A) (17H04996), the Program to Disseminate Tenure Track System in University of Miyazaki, and the Environment Research and Technology Development Fund (4RF-1501), Ministry of the Environment, Japan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful for Ms. Ritsuko Kubota for helping with sequencing. The following helped with sampling in the field: Eriko Hatakeyama, Midori Kogi, Masanobu Sanpei, Muneyoshi Okabe, Ryuji Yamaguchi, Keiichi Nomura, Takuma Mezaki, and Kaoru Sugihara.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.668043/full#supplementary-material
Baums, I. B., Miller, M. W., and Hellberg, M. E. (2006). Geographic variation in clonal structure in a reef-building Caribbean coral, Acropora palmata. Ecol. Monogr. 76, 503–519.
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Falush, D., Stephens, M., and Prtichard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Falush, D., Stephens, M., and Prtichard, J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7:4. doi: 10.1111/j.1471-8286.2007.01758.x
Fukami, H., Budd, A. F., Levitan, D. R., Jara, J., Kersanach, R., and Knowlton, N. (2004). Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution 58, 324–337. doi: 10.1111/j.0014-3820.2004.tb01648.x
Goldberg, J., and Wilkinson, C. (2004). “Global threats to coral reefs: coral bleaching, global climate change, disease, predator plagues, and invasive species,” in Status of Coral Reefs of the World: 2004, ed. C. Wilkinson (Townsville, AU: Australian Institute of Marine Science), 67–92.
Gorospe, K. D., Donahue, M. J., and Karl, S. A. (2015). The importance of sampling design: spatial patterns and clonality in estimating the genetic diversity of coral reefs. Mar. Biol. 162, 917–928. doi: 10.1007/s00227-015-2634-8
Higuchi, T., Agostini, S., Casareto, B. E., Suzuki, Y., and Yuyama, I. (2015). The northern limit of corals of the genus Acropora in temperate zones is determined by their resilience to cold bleaching. Sci. Rep. 5:18467. doi: 10.1038/srep18467
Hubisz, M. J., Falush, D., Stephens, M., and Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 5, 1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x
IUCN (2020). The IUCN Red List of Threatened Species. Version 2020-3. Available online at: https://www.iucnredlist.org (accessed Febuary 07, 2021).
Iwase, F., Nakano, S., Aki, H., Okada, N., and Shimizu, R. (2009). Field test on breeding technique of the zoogamy for Acropora tumida. J. Jpn. Soc. Civil Eng. 65, 1216–1220. doi: 10.2208/kaigan.65.1216
Kojima, S., Segawa, R., and Hayashi, I. (1997). Genetic differentiation among populations of the Japanese truban shell Turbo (Batillus) cornutus corresponding to warm currents. Mar. Ecol. Prog. Ser. 150, 149–155.
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., and Mayrose, I. (2015). CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191.
Ministry of the Environment (2017). The Japanese Red list of Coral Species (in Japanese). Available online at: https://www.env.go.jp/nature/kisho/sango_tokusei.html (accessed December 18, 2020).
Nakabayashi, A., Yamakita, T., Nakamura, T., Aizawa, H., Kitano, Y. F., Iguchi, A., et al. (2019). The potential role of temperate Japanese regions as refugia for the coral Acropora hyacinthus in the face of climate change. Sci. Rep. 9:1892. doi: 10.1038/s41598-018-38333-5
Nakajima, Y., Nishikawa, A., Iguchi, A., and Sakai, K. (2010). Gene flow and genetic diversity of a broadcast-spawning coral in northern peripheral populations. PLoS One 5:e11149. doi: 10.1371/journal.pone.0011149
Nanto, C., Kishimoto, T., and Ueno, S. (2009). Changes in Acropora tumida community in Kuzura, Sugura Bay, Central Japan and effects of fences for protecting the coral. Bull. Inst. Oceanic Res. Develop. Tokai Univ. 30, 13–20.
Nei, M., Maruyama, T., and Chakraborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution 29, 1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x
Nishihira, M. (2004). “Hermatypic coral of Japan,” in Coral Reefs of Japan, eds Japan Coral Reef Society and The Ministry of Environment of Japan (Japan: Ministry of Environment), 10–13.
Nomura, K. (2009). Recent changes in coral communities in Kushimoto, the southernmost part of Honshu, Japan. J. Jpn. Coral Reef Soc. 11, 39–49. doi: 10.3755/jcrs.11.39
Noreen, A. M. E., Van Oppen, M. J. H., and Harrison, P. L. (2013). Genetic diversity and differentiation among high-latitude broadcast-spawning coral populations disjunct from the core range. Mar. Ecol. Prog. Ser. 491, 101–109. doi: 10.3354/meps10480
O’Brien, S. J. (1994). A role for molecular genetics in biological conservation. Proc. Natl. Acad. Sci. U.S.A. 91, 5748–5755. doi: 10.1073/pnas.91.13.5748
Okubo, N., Motokawa, T., and Omori, M. (2007). When fragmented coral spawn? Effect on size and timing on survivorship and fecundity of fragmentation in Acropora formosa. Mar. Biol. 151, 353–363. doi: 10.1007/s00227-006-0490-2
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts46
Principe, P. P., Bradley, P., Yee, S. H., Allen, P. E., and Campbell, D. E. (2011). Quantifying Coral Reef Ecosystem Services. North Carolina: U.S. Environmental Protection Agency, Office of Research and Development, Research Triangle Park.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Shinzato, C., Yasuoka, Y., Mungpakdee, S., Arakaki, N., Fujie, M., Nakajima, Y., et al. (2014). Development of novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology. Front. Mar. Sci. 1:11. doi: 10.3389/fmars.2014.00011
Spielman, D., Brook, B. W., Briscoe, D. A., and Frankham, R. (2004). Does inbreeding and loss of genetic diversity decrease disease resistance? Conserve. Genet. 5, 439–448.
Sugihara, K., Nomura, K., Yokochi, H., Shimoike, K., Kajiwara, K., Suzuki, G., et al. (2015). Zooxanthellate Scleractinian Corals of Tanegashima Island, Japan. Tsukuba: Center for Environmental Biology and Ecosystem Studies, National Institute for Environmental Studies.
Suzuki, G., and Fukami, H. (2012). Evidence of genetic and reproductive isolation between two morphs of subtropical-dominant coral Acropora solitaryensis in the non-reef region of Japan. Zool. Sci. 29, 134–140.
Taguchi, T., Tagami, E., Mezaki, T., Vacarizas, J. M., Canon, K. L., Avila, T. N., et al. (2020). Karyotypic mosaicism and molecular cytogenetic markers in the scleractinian coral Acropora pruinosa Brook, 1982 (Hexacorallia, Anthozoa, Cnidaria). Coral Reefs 39, 1415–1425. doi: 10.1007/s00338-020-01975-x
van Oppen, M. J. H., Underwood, J. N., Muirhead, A. N., and Peplow, L. (2016). Ten microsatellite loci for the reef-building coral Acropora millepora (Cnidaria, Scleractinia) from the Great Barrier Reef, Australia. Mol. Ecol. Notes 7:3. doi: 10.1111/j.1471-8286.2006.01610.x
Veron, J. E. N. (1992). Conservation of biodiversity: a critical time for the hermatypic coral of Japan. Coral Reefs 11, 13–21. doi: 10.1007/BF00291930
Keywords: scleractinia, temperate coral, biodiversity, cryptic lineages, clonality, asexual reproduction, morphological variation
Citation: Pipithkul S, Ishizu S, Shimura A, Yokochi H, Nagai S, Fukami H and Yasuda N (2021) High Clonality and Geographically Separated Cryptic Lineages in the Threatened Temperate Coral, Acropora pruinosa. Front. Mar. Sci. 8:668043. doi: 10.3389/fmars.2021.668043
Received: 15 February 2021; Accepted: 16 April 2021;
Published: 14 May 2021.
Edited by:
Tamotsu Oomori, University of the Ryukyus, JapanReviewed by:
David A. Paz-García, Centro de Investigaciónes Biológicas del Noroeste (CIBNOR), MexicoCopyright © 2021 Pipithkul, Ishizu, Shimura, Yokochi, Nagai, Fukami and Yasuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hironobu Fukami, aGlyb2Z1a2FtaUBjYy5taXlhemFraS11LmFjLmpw; Nina Yasuda, TmluYTI3QGNjLm1peWF6YWtpLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.