- 1State Key Laboratory of Marine Environmental Science, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2Tianjin Key Laboratory of Aqua-Ecology and Aquaculture, College of Fisheries, Tianjin Agricultural University, Tianjin, China
- 3Fujian Key Laboratory of Genetics and Breeding of Marine Organisms, Xiamen University, Xiamen, China
Heterosis is a widely distributed phenomenon in mollusks. It is vital in aquaculture by bringing beneficial traits into hybrids. People have utilized the heterosis theory in aquaculture for years. However, the molecular basis of heterosis remains elusive. Evident growth and survival heterosis were shown in the hybrid (“Dongyou-1”) of two Haliotis diversicolor geographic genotypes (Japan and Taiwan). To explore the molecular basis underlying the hybrid abalone’s heterosis, we conducted comparative mRNA and miRNA transcriptional analysis in the hybrid and parental genotypes. Differentially expression analysis identified 5,562 differentially expressed genes (DEGs) and 102 differentially expressed miRNAs (DEMs) between the three genotypes. 1,789 DEGs and 71 DEMs were found to be non-additively expressed in the hybrid. Meanwhile, both the expression level dominance pattern (ELD) and expression level overdominance pattern (ELOD) were found in the DEGs and DEMs, showing the existence of dominance and overdominance models in the hybrid’s transcriptome and post-transcriptional regulation. Functional analysis showed the non-additively expressed genes, ELD genes, and ELOD genes were significantly enriched in growth, immunity, and stress response related pathways, while some of the pathways were regulated by the mRNA-miRNA interactions. The expression levels of FGF, C1Q, HC, CAT, SEGPX, and MGST were significantly up-regulated in the hybrid compared to the middle parent value. In conclusion, we identified the existence of non-additivity, dominance, and overdominance models in the transcriptome and miRNAome of the H. diversicolor hybrid; these models facilitate the advantageous parental alleles’ integration into the hybrid, contributing to the hybrid’s growth and survival heterosis.
Introduction
Heterosis, or hybrid vigor, refers to the phenomenon that hybrids often show advantages in growth, fitness, or resistance relative to the parents (Chen, 2013). Heterosis is widespread in plants and animals, such as Arabidopsis, maize, rice, cotton, mice, donkey, chicken, dog, oyster, abalone, and mussel (Chen, 2013; McKeown et al., 2013). Farmers and researchers have utilized the heterosis theory to improve agricultural species’ genotypes and phenotypes for years (Goff, 2011; Veitia and Vaiman, 2011; Baranwal et al., 2012; McKeown et al., 2013). Heterosis is also common in the interspecific hybrids of abalones. For example, H. discus hannai ♀ × H. discus ♂ and H. discus hannai ♀ × H. fulgens were superior in growth or viability than the parents (Inoue et al., 1986; You et al., 2015). The reciprocal hybrids of H. gigantea and H. discus hannai were more resistant to heat and bacteria than the parents (Luo et al., 2010; Liang et al., 2014, 2018). Besides, heterosis also exists in the intraspecific hybrids of abalones. For instance, the hybrid of the H. discus hannai China genotype and Japan genotype showed more considerable shell length, shell width, wet weight, and a higher survival rate (17.9, 22.1, 61.9, and 80%, respectively) compared to the China genotype (Zhang et al., 2004). Some abalone hybrids have been successfully applied in abalone aquaculture due to their superior traits (You et al., 2015; Cook, 2019).
Although abalone heterosis is important in aquaculture, its molecular mechanisms remain unclear. Traditional analyses of molluscan species’ heterosis mainly focused on the relationship between isozyme heterozygosity and heterosis, or heterosis-related genes and proteins. For example, a positive correlation between organisms’ isozyme heterozygosity and growth rate was identified in mussels and oysters (Gentili and Beaumont, 1988; Hedgecock et al., 1995, 1996). The hybrid H. discus hannai ♀ × H. gigantea ♂ had higher activities of immunity-related enzymes (superoxide dismutase, acid phosphatase, alkaline phosphatase, and myeloperoxidase) and higher mRNA level of heat shock protein 70 gene upon heat stress compared to the parents (Liang et al., 2014). Similarly, the hybrid Haliotis discus hannai ♀ × H. fulgens ♂ showed higher phenoloxidase activities, superoxide dismutase, and higher mRNA levels of several immunity-related genes compared to the maternal genotype (Shen et al., 2020). These studies give us a deeper understanding of molluscan heterosis by identifying heterosis-related molecules. However, to have a comprehensive understanding of molluscan heterosis, we need to conduct further genome-scale studies of heterosis using omic tools.
Transcriptome sequencing is the most frequently used omic tool in heterosis studies. It has been utilized in the heterosis studies of many species such as Arabidopsis, rice, wheat, corn, fish, and oyster (Hedgecock et al., 2007; Goff, 2011; Chen, 2013; Gao et al., 2013). Although different species have different parent-hybrid gene expression variations, some common trends could still be concluded. The parental alleles’ expression in the hybrid could be divided into several patterns: additivity, non-additivity, dominance, and overdominance (Baranwal et al., 2012). For the non-additivity, dominance, and overdominance patterns, the heterozygote’s gene expression value is unequal to the middle parental expression value (MPV) (Chen, 2010); in other words, a new allelic expression pattern is generated in the hybrid. Hence, the non-additivity, dominance, and overdominance patterns are vital to the heterosis generation (Springer and Stupar, 2007; Fujimoto et al., 2012). The differentially expressed genes (DEGs) among parents and hybrids are functionally enriched in various biological pathways such as “cell growing and aging”, “stress response”, and “signal transduction” (Chen, 2013). These pathways are closely related to organisms’ growth, fitness, and resistance. Besides, the role of microRNAs (miRNAs) in the heterosis generation obtains much attention in recent years. miRNAs can negatively regulate gene expression through epistasis (Sha et al., 2014). The miRNAs expression variation between hybrids and parents has been investigated in many species such as Arabidopsis, wheat, and corn (Greaves et al., 2012; Chen, 2013). A common finding is the number of non-additively expressed miRNAs (NEMs) in hybrids positively correlates with the parents’ genetic distance. The NEMs could further cause allele expression changes in hybrids (Groszmann et al., 2011, 2013; Ng et al., 2012). To conclude, the transcriptional analysis could help us understand how parental mRNAs and miRNAs are integrated and expressed in hybrids. Meanwhile, the joint analysis of transcriptional data and physiological data could help us identify essential genes or pathways related to the heterosis generation.
Haliotis diversicolor is an important economic abalone species cultured in China’s continental coasts. This species was initially introduced from Taiwan and then successively cultured for several generations. However, due to frequent inbreeding, the TW’s survival rate continuously declined. To solve this problem, we introduced several different geographical genotypes of H. diversicolor to hybridize with TW. The hybrid genotypes showed significantly different growth and survival traits compared to the parents. Within the hybrid genotypes, the hybrid of H. diversicolor Taiwan genotype and Japan genotype (JP) showed the most desirable traits; it was TW-like in growth and JP-like in survival (You et al., 2015). This hybrid (named DY) has been widely used in China’s abalone aquaculture. However, we know little about the molecular mechanisms underlying DY’s heterosis. In this study, we conducted comparative transcriptional analysis in DY and its parents (JP and TW) to compare the mRNAs and miRNAs differences between the three genotypes. We wish to identify the important biological molecules and functions determining DY’s heterosis and investigate the miRNA-mRNA interactions in the abalone’s heterosis generation.
Materials and Methods
Abalones and Phenotype Measurements
Abalones of the three H. diversicolor genotypes (JP, TW, and DY) were used. The abalone breeding and cultivation were performed at “Fuda” abalone company in Jinjiang City, Fujian Province, China. The artificial fertilization of JP ♀ × JP ♂, TW ♀ × JP ♂, and TW ♀ × TW ♂ was conducted in October 2015. The obtained abalone seeds were labeled and mixed cultured under the same conditions (reared in running seawater; fed with fresh alga Gracilaria confervoides; salinity 33‰; pH 8.0) for 2 years. The three genotypes’ survival rates were recorded every month from October 2016 to March 2017. The shell length and wet weight of 100 abalones from each genotype were measured in October 2017.
Acclimation
For each genotype, 30 abalones with a normal appearance and good vitality were used for the following experiments. The abalones were acclimated under the controlled laboratory conditions (reared in still seawater with aeration, the seawater was replaced every day; fed with fresh alga Gracilaria confervoides; temperature 24 ± 1°C; salinity 33‰; pH 8.0) for 14 days. No abalone was dead during the acclimation.
RNA Extraction
For each genotype, ten abalones were used for the transcriptome and miRNAome sequencing. Total RNA was extracted from each abalone’s muscle using the TRIpure reagents (Invitrogen, United States). The RNA quality was tested using 1% agarose gels and Agilent 2,100 Bioanalyzer (Agilent Technologies, United States). The ten RNA samples from each genotype were equally mixed together (400 ng RNA from each sample) and then equally divided into two RNA pools (2 μg RNA for each pool). The two RNA pools were then used to construct the cDNA library and small RNA library separately.
cDNA Library Construction and mRNAs Sequencing
For each genotype, one RNA pool, as described below, was used for the poly (A) + mRNA selection using oligo (dT) magnetic beads (Invitrogen, United States). The cDNA library was constructed using the ScriptSeq kit (Epicentre, United States). For the mRNAs sequencing, a total of three cDNA libraries were constructed. The libraries were amplified by 12–15 cycles of PCR and then sequenced in two lanes on the HiSeq 2,500 sequencer (Illumina, United States) at BGI-tech company (Shenzhen, China).
De novo Assembly and Functional Annotation
The generated raw reads were first filtered by removing the adaptors, ambiguous reads (those with ≥10% ambiguous nucleotides), and low-quality reads (those containing ≥20% bases whose quality scores ≤10) then assembled using Trinity (Grabherr et al., 2011), with min_kmer_cov set to 2 and other parameters set to defaults. The obtained contigs and unigenes were then aligned to the non-redundant (NR) protein database, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) using KAAS (Mao et al., 2005; Moriya et al., 2007), with an E-value cut-off of 10–5.
Differentially Expression Analysis
The read counts were normalized as FPKM (Fragments Per Kilobase of exon model per Million mapped reads) values. Then statistical analyses were applied on FPKM of genes in the parents and hybrid using DEseq1. DEGs between any two populations were defined to have a significant expression level against each other (fold-change ≥1.5, p-value ≤ 0.01). Non-additively expressed genes (NEG) and NEMs in the hybrid were defined to have a significantly different expression level against MPV (fold-change ≥1.3, p-value ≤ 0.01).
Small RNAs Sequencing
As described before, the three RNA pools (one from each genotype) were used to construct three small RNAs libraries for small RNAs sequencing according to the manufactures of TruSeq Small RNA Sample Prep Kits (Illumina, United States). For each library, 1 μg of total RNA was used. The three libraries were then sequenced on the HiSeq 2500 sequencer (Illumina, United States) at BGI-tech company (Shenzhen, China).
Bioinformatic Analysis of Small RNAs Sequencing Data
The 49 nt sequence tags from Hiseq sequencing went through the data cleaning analysis to get credible clean tags without adaptor and unknown nucleotides (>10%). Only 18–32 nt sequences were used for further study. Then the length distribution of the clean tags and common and specific sequences between samples were summarized. Then clean tags were aligned to the Rfam database2 using bowtie (Langmead et al., 2009) to identify miRNA sequences. The known miRNA sequences were obtained by aligning the clean tags to the miRNase database3 by BLAST4, and the novel miRNA sequences were predicted using miRDeep2 (Friedländer et al., 2012).
Differentially Expression Analysis and Target Genes Prediction
Differentially expressed miRNAs (DEMs) between the parents and hybrid were analyzed as the method described in “2.7 Differentially expression analysis”. The target genes of the DEMs were identified by targetscan (García et al., 2011), miRanda5, PITA6, and RNAhybrid (Rehmsmeier et al., 2004). The overlapping target genes predicted by the software were kept for further analysis.
Twelve Expression Patterns of DEGs and DEMs
To further dissect parental alleles’ expression in DY, the DEGs and DEMs between the parents and hybrid were binned into 12 categories according to the previous studies (Rapp et al., 2009; Zhang et al., 2019). To investigate the possible miRNA-mRNA interactions for abalone heterosis, we achieved the negative interaction between the DEMs and DEGs by integrating DEGs’ expression profiles and DEMs with the DEMs targeting genes information as described before (Zhang et al., 2019).
Functional Analysis of DEGs and DEMs
Gene Ontology analysis of the DEGs and DEMs (target genes of the DEMs) was conducted by Blast2GO7 with a significant threshold of FDR < 0.05. KEGG pathway analysis of the DEGs and DEMs was performed through the hypergeometric test using the corrected p-value < 0.05 by KOBAS (Mao et al., 2005).
Expression Profiles of Growth, Immune, and Stress Response Genes
Based on NR annotation data and previous studies (Song et al., 2015; Wang et al., 2018b; Bouallegui, 2019), 30 growth-related genes, 52 immunity-related genes, and 29 stress response genes were identified in the three genotypes. For comparison, each genotype’s gene expression level was normalized by comparing to the MPV. Besides, each gene group (genes with the same functions) was clustered between the three genotypes based on the Pearson correlation algorithm.
Data Visualization
Heat maps of gene expression were drawn using Morpheus8 while the genes were clustered based on the Pearson correlation algorithm. Venn diagrams were drawn by Venny9.
Real-Time PCR Validation
To validate the RNA-seq results, we assessed the expression levels of six DEGs (unigene 34268, unigene 47208, unigene 47561, unigene 32067, unigene 19086, and unigene 49863) and six DEMs (miR-4755-5p, miR-6317, miR-7255-5p, miR-4326, miR-2836, and miR-263b) by real-time PCR, according to the method described below (Liang et al., 2018). Briefly, the real-time PCR experiment was conducted in a 7500 fast qPCR system (ABI, United States) using specific qPCR reagents (Thermo, United States). The cycling parameters used were: 95°C for 7 min, 35 cycles at 95°C for 20 s, and 60°C for 1 min. The relative mRNA level of target genes was calculated based on the Ct values of this gene and β-actin normalized to that of the cDNA standard.
Results
Phenotypes of the Three H. diversicolor Genotypes
The three genotypes’ survival rates from October 2016 to March 2017 are shown in Supplementary Figure 1. During the 6 months, the JP or DY’s survival rate was significantly higher than that of TW. By March 2017, JP, DY, and TW’s survival rates were 93.56, 95.56, and 37.56%, separately. The DY’s shell length and wet weight were similar to that of TW, higher than that of JP (Supplementary Table 1). These results suggest DY has JP-like resistance and TW-like growth ability.
Summary of Transcriptomes and miRNAomes
After the sequencing quality control, an average of 54 million raw reads and 52 million clean reads were obtained for each transcriptome sample. The average Q20 percentage of the samples was 97.01% (Supplementary Table 2). After assembling, 85,979, 75,220, and 72,936 unigenes were generated (with an average length of 750, 667, and 672 nt) in JP, DY, and TW, respectively. The N50 of all unigenes was 2,166, and the mean unigene length was 1,075 nt (Supplementary Table 3).
An average of 11,455 thousand raw reads and 11,318 thousand clean reads were obtained for each miRNAome sample. Most of the reads were 20–30 bp in length (Supplementary Table 4). After alignment and target genes prediction, 3,186, 2,903, and 3,142 known miRNAs were identified in JP, DY, and TW, corresponding to 68,633, 68,623, and 68,632 target genes, respectively. 20, 14, and 9 novel miRNAs were identified in JP, DY and TW, corresponding to 34,363, 31,985, and 26,692 target genes, respectively (Supplementary Table 5).
DEGs and DEMs
Differentially expression analysis identified 3,661, 3,048, 1,778 DEGs and 65, 63, and 68 DEMs between JP and TW, DY and JP, and DY and TW, separately (Figure 1). The DEGs numbers suggest a distant gene expression pattern between the two parental genotypes, while DY’s gene expression pattern was more like TW other than JP. The number of DEMs was almost the same between any two genotypes. Besides, there were 99 overlapping DEGs and one overlapping DEM between the three genotypes.
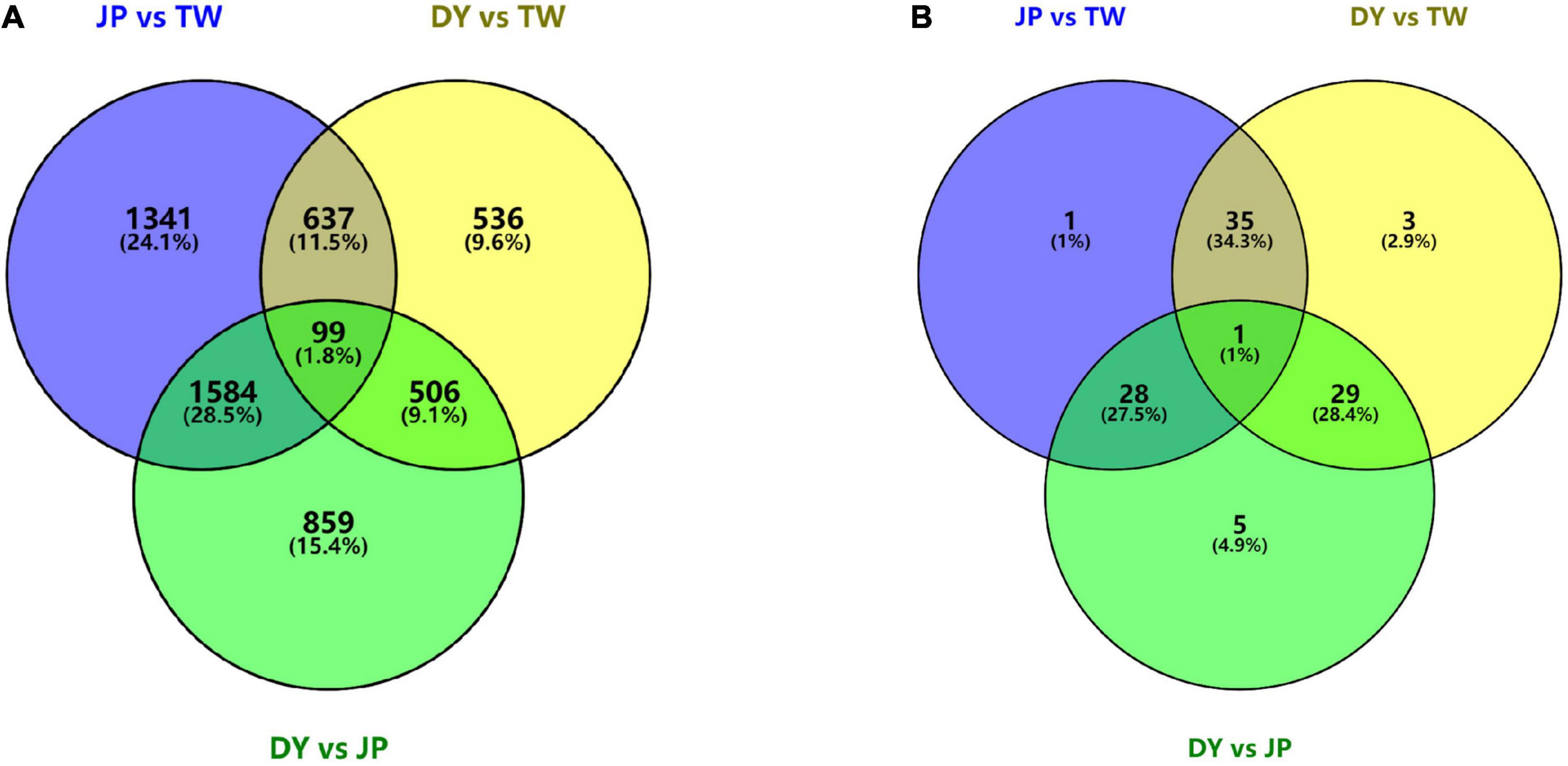
Figure 1. Number of (A) differentially expressed genes and (B) differentially expressed miRNAs between the three H. diversicolor genotypes.
A total of 1,789 NEGs and 71 NEMs were identified. Heatmap clustering shows most of the NEGs could be divided into two patterns (Figure 2): (1) Overdominance/Underdominance, including “DY > TW > JP”, “DY > JP > TW”, “DY > JP = TW”, “DY < JP < TW”, “DY < TW < JP”; and (2) Dominance, including “DY = JP < TW”, “DY = TW < JP”. Most of the NEMs were dominant patterned (“DY > JP = TW”, “DY < JP < TW”) or underdominant patterned (“DY = JP < TW”, “DY = TW < JP”). These findings indicate the coexist of non-additivity, dominance, overdominance, and underdominance models in the hybrid’s transcriptome and miRNAome.
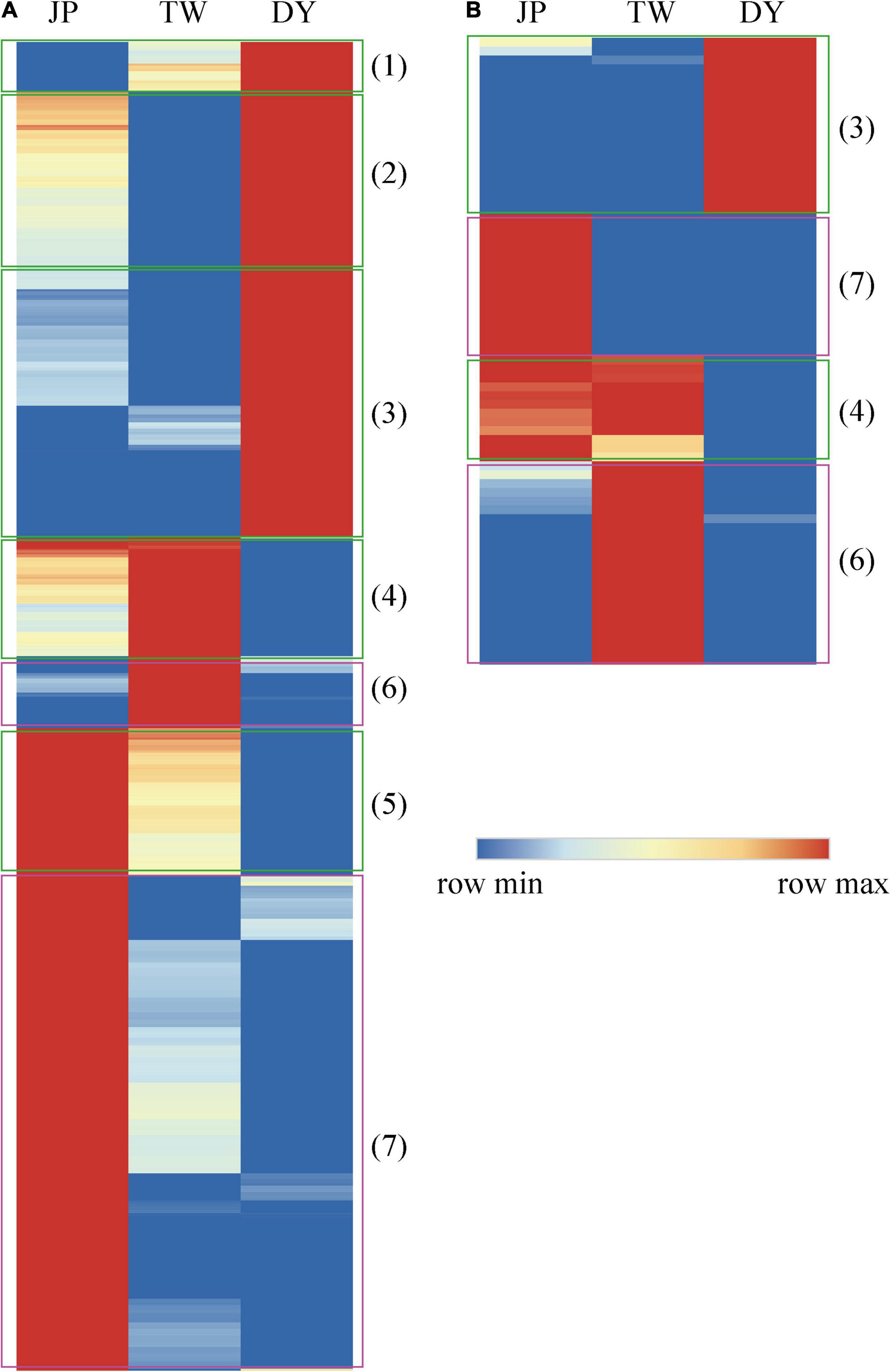
Figure 2. Heatmap clustering of the (A) non-additively expressed genes and (B) non-additively expressed miRNAs between the three H. diversicolor genotypes. Relative gene expression levels in the three genotypes were normalized to the middle parental value MPV. Red and blue bars represent up- and down-regulated expression levels of individual gene, respectively. Green boxes indicate genes/miRNAs with overdominant or underdominant expression patterns: (1) “DY > TW > JP;” (2) “DY > JP > TW;” (3) “DY > JP = TW;” (4) “DY < JP < TW;” or “DY < JP = TW;” and (5) “DY < TW < JP.” Pink boxes indicate genes/miRNAs with dominant expression patterns: (6) “DY = JP < TW” and (7) “DY = TW < JP.”
Functional Analysis of NEGs
The NEGs were mainly enriched in “biological process” category GO terms. The “development process”, “chromosome”, and “phosphoric ester hydrolase activity” were the most enriched GO terms in the biological process, cellular component, and molecular function categories, respectively (Supplementary Figure 2). KEGG pathway analysis of the NEGs identified 35 significantly enriched pathways (p < 0.05) (Figure 3). Most of the enriched pathways were related to “organismal systems (digestive, endocrine, excretory, immune, sensory, and circulatory systems)”, “diseases (immune, infectious, neurodegenerative, and cardiovascular diseases)”, and “metabolisms (amino acid, xenobiotics, and carbohydrate metabolisms)”. The enrichment results suggest the hybrid is different from the parents’ average performance (MPV) in growth, immunity, and stress-response-related biological functions; these differences may contribute to the DY’s superior phenotypic traits.
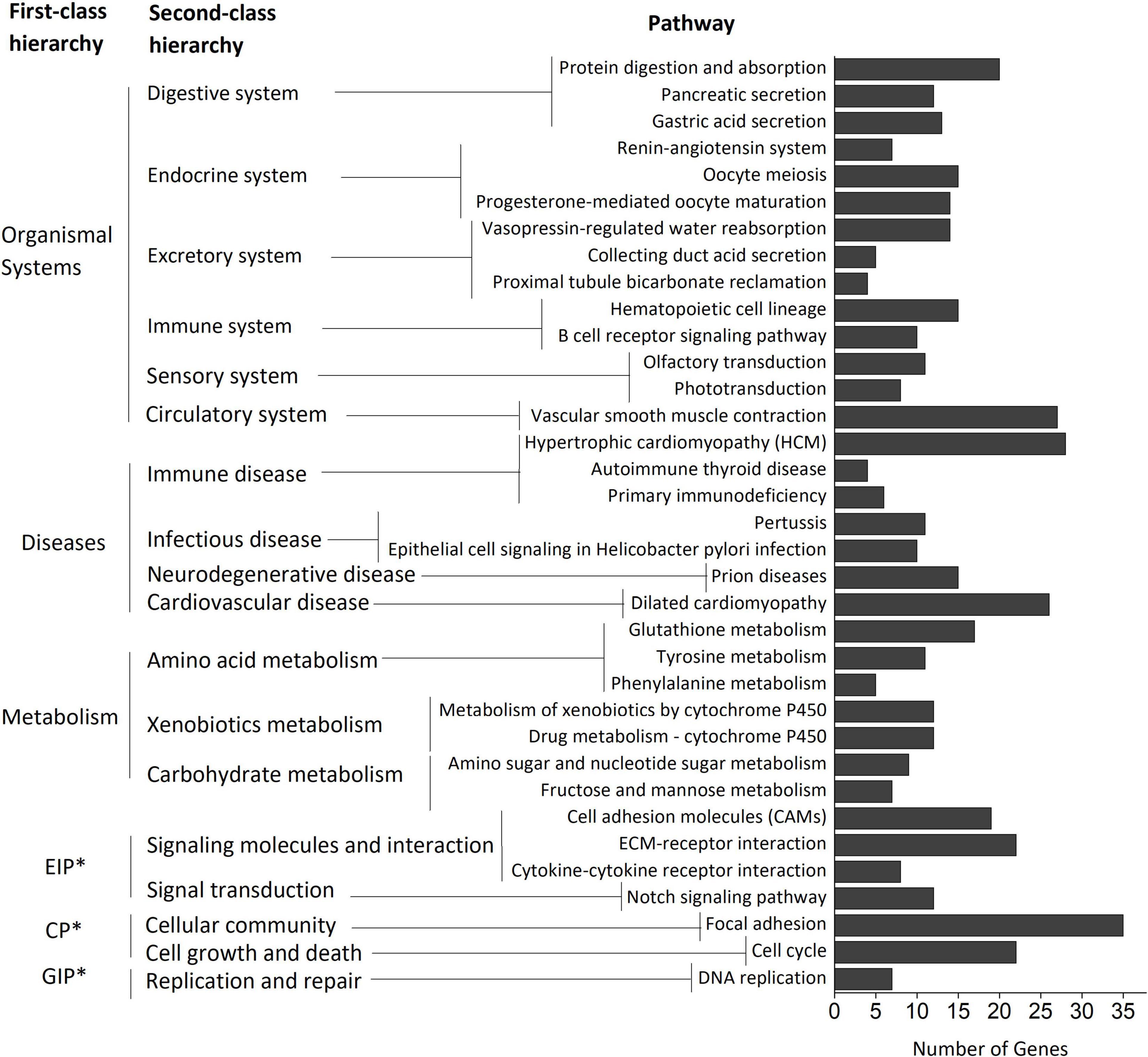
Figure 3. KEGG pathway enrichment of the NEGs. This figure consists of the pathways, the second-class hierarchy, and the first-class hierarchy. The first-class hierarchy and second-class hierarchy rank from top to bottom according to the number of pathways included in the hierarchy. The pathways rank from top to bottom according to the enrichment significance of each pathway. Each bar represents the enrichment count of transcripts in a KEGG pathway. *EIP, environmental information processing; CP, cellular processes; GIP, genetic information processing.
Twelve Expression Patterns of DEGs and Functional Analysis
The 12 categories are shown in Figure 4: (1) Additivity (I and II); (2) Expression level dominance by JP (ELD-J, III and IV); (3) Expression level dominance by TW (ELD-T, V and VI); (4) Expression level underdominance (ELUD, VII, VIII, and IX); and (5) Expression level overdominance (ELOD, X, XI, and XII). Notably, a large number of ELD patterns were identified, with the proportion of 52.28% (1,226/2,345) in DEGs and 61.64% (45/73) in DEMs. The second large category was the ELOD pattern, with the proportion of 8.70% (204/2,345) in DEGs and 24.66% (18/73) in DEMs. Besides, the number of ELD-T patterns was significantly higher than that of ELD-J patterns in DEGs, while in DEMs the two patterns had similar numbers. These findings indicate the widespread of ELD and ELOD patterns in the hybrid’s transcriptome and miRNAome.
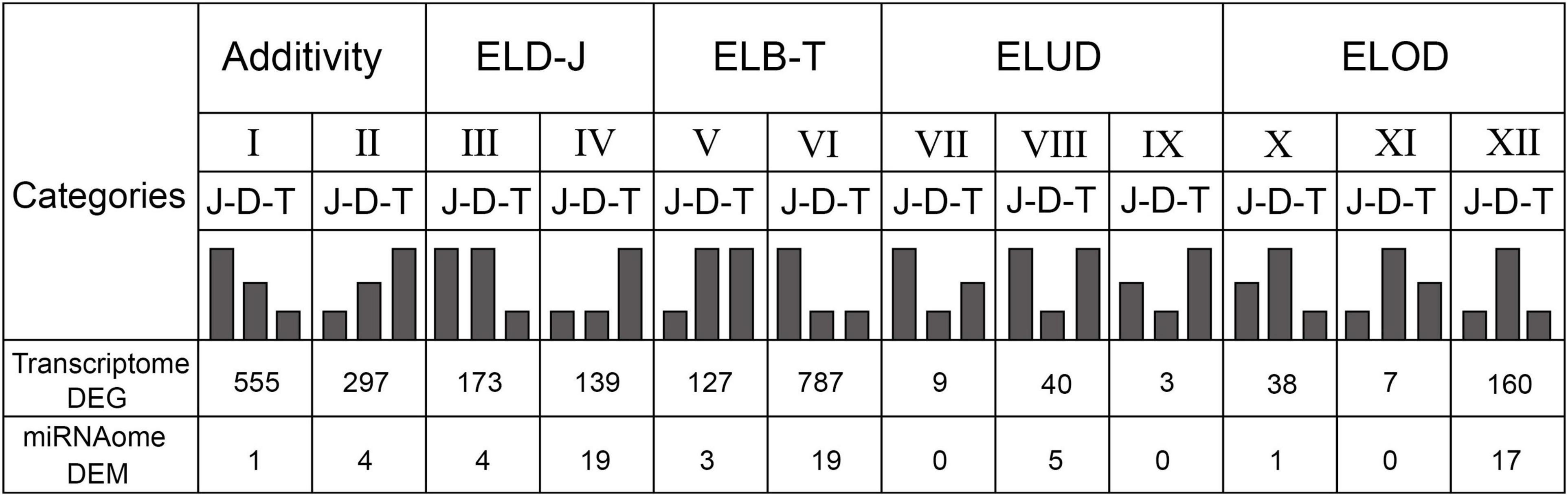
Figure 4. Twelve differential expression patterns in the DEGs and DEMs between the hybrid abalone and its parents. ELD-J: expression level dominance by JP; ELD-T: expression level dominance by TW; ELUD: expression level underdominance; ELOD: expression level overdominance; J: JP; D: DY; T: TW.
Gene Ontology analysis showed the ELD-J genes were mostly enriched in “biological adhesion” and “gland development” (biological processes); “extracellular region” and “late endosome” (cellular components); “isomerase activity” and “binding-related activities” (molecular functions). The ELD-T genes were most enriched in “development process” and “organ development” (biological processes), “non-membrane-bounded organelle” (cellular components), “binding-related activities” (molecular functions). The ELOD genes were mainly enriched in “transport-related processes” (biological processes), “membrane-related components” (cellular components), and “transmembrane-related functions” (molecular functions) (Supplementary Figures 3–6). These results suggest the ELD-J and ELOD genes are more related to cellular interaction and signaling transduction functions, while the ELD-T genes are more related to growth and development functions.
KEGG pathways analysis identified 11, 11, 7, and 18 significant (p < 0.05) pathways from the ELD-J, ELD-T, ELOD, and ELUD genes separately (Figure 5 and Supplementary Figure 7). Pathways enriched from the ELD-J genes were mainly related to “diseases (immune disease)”, “organismal systems (digestive, nervous, and circulatory systems)”, and “environment information processes”. Pathways enriched from ELD-T genes were mostly related to “diseases (cardiovascular, infectious, and neurodegenerative diseases)” and “metabolism (carbohydrate and lipid metabolisms)”. Pathways enriched from the ELOD genes were mainly related to “metabolisms (amino acid, cofactors, and vitamins metabolisms)”, “organismal systems (digestive, endocrine, immune, and excretory systems)”, and “environmental information processing (signal transduction, signaling molecules, and interaction)”. The above pathways’ functions are closely related to organisms’ growth, immune response, and organism’s reaction to external environments. Hence, the ELD and ELOD genes are crucial to the DY’s heterosis generation.
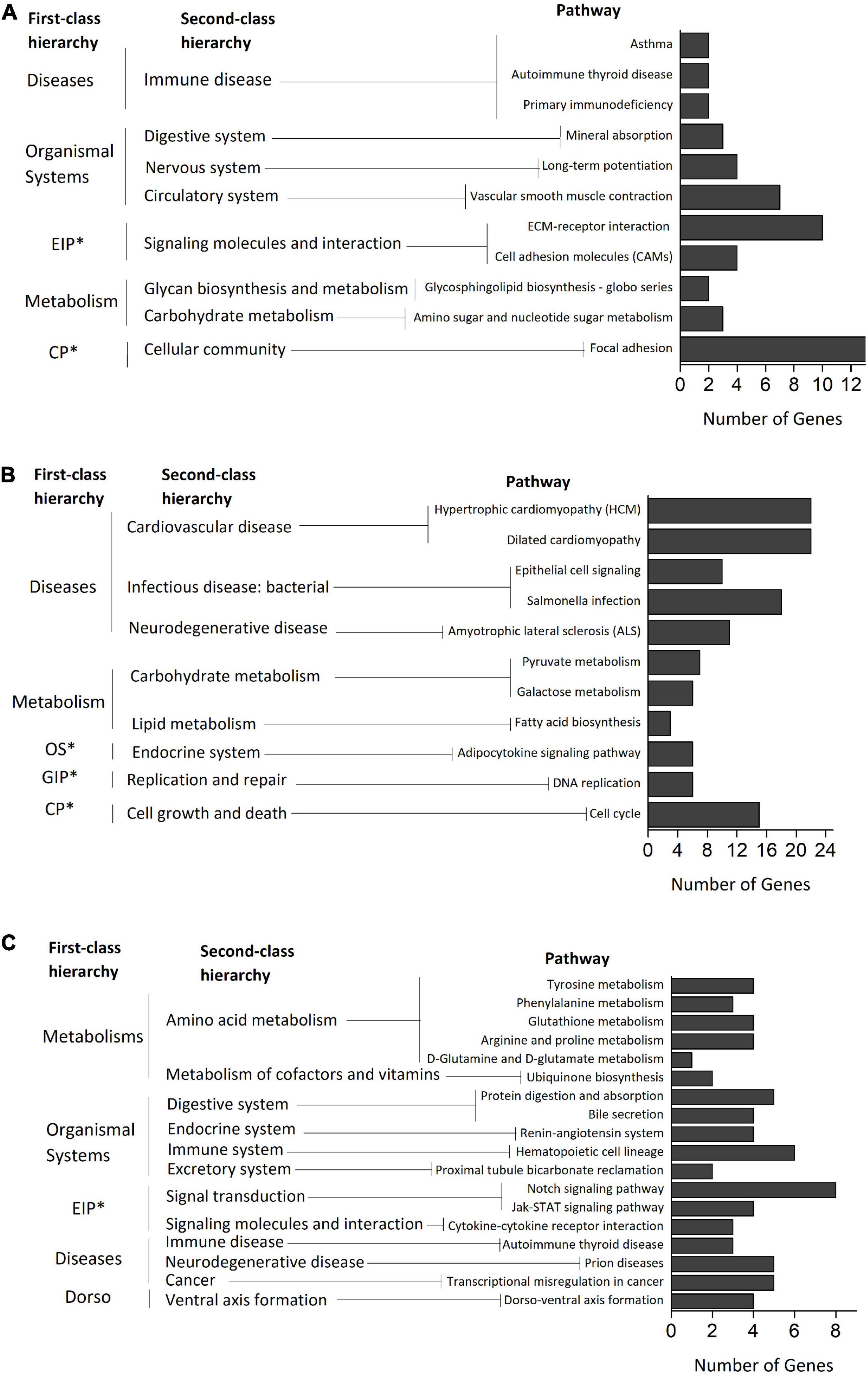
Figure 5. KEGG pathway enrichment of the (A) ELD-J genes, (B) ELD-T genes, and (C) ELOD genes. *EIP, environmental information processing; CP, cellular processes; OS, organismal systems; GIP, genetic information processing.
Interestingly, by comparing the pathways enriched from the ELD and ELOD genes with those enriched from the NEGs, we found 63.67% (7/11) of the ELD-J pathways, 45.46% (5/11) of the ELD-T pathways, and 50.00% (9/18) of the ELOD pathways were overlapped with the NEGs pathways (Supplementary Datasheet 1). The overlapped ELD pathways were mostly related to “immune disease”, “nervous and circulatory systems”, and “signaling molecules and interaction”, while the overlapped ELOD pathways were mainly related to “amino acid metabolism” and “organismal systems (digestive, endocrine, immune, and excretory systems)”. The above results suggest the ELD and ELOD patterns partially act together with the non-additivity pattern in altering the hybrid’s growth, immunity, and stress response related functions, which might be a vital mechanism for the emergency of abalone heterosis.
Interactions Between DEGs and DEMs
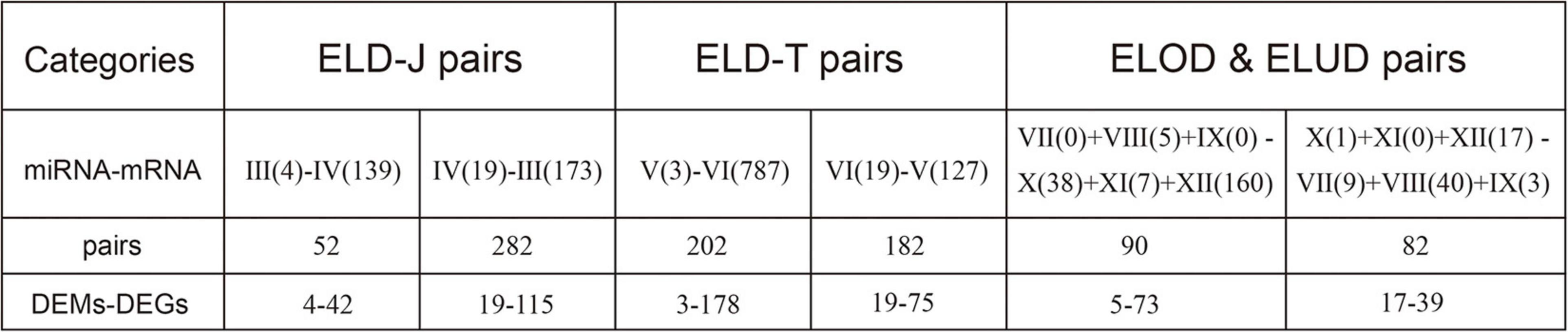
As Figure 6 shows, the most common class in the miRNA-mRNA interactions was ELD-J “IV(19)–III(173),” involving 19 DEMs and 115 DEGs. Besides, 50.32% of the ELD-J genes (157/312), 27.68% of the ELD-T genes (253/914), and 43.58% of the ELOD/ELOD genes (112/257) were involved in the miRNA-mRNA interactions, suggesting a significant role of miRNA regulation in the parent-hybrid DEGs in favoring of the paternal expression genes and transgressive expression genes.
Gene Ontology analysis showed the ELD-J pairs genes were mostly enriched in “cell adhesion” and “biological adhesion” (biological processes); “nucleolus” (cellular components); “DNA binding” and “transcription regulator activity” (molecular functions). The ELD-T pairs genes were most enriched in “development process” and “organ development” (biological processes), “organelle” (cellular components), “binding-related activities” (molecular functions). The ELOD/ELUD pairs genes were mainly enriched in “DNA integration” (biological processes), “respiratory-related complexes” (cellular components), and “oxidoreductase activity (NADPH)” (molecular functions) (Supplementary Figures 8–10). The enriched GO terms of ELD, ELOD, and ELUD pairs genes are quite similar to that of the ELD, ELOD, and ELUD genes.
KEGG pathway analysis identified 17, 6, and 10 significant pathways (p < 0.05) from the ELD-J pairs (52 + 282), ELD-T pairs (202 + 182), and transgressive regulation pairs genes (90 + 82) respectively (Figure 7). The pathways related to “organismal systems (nervous, circulatory, excretory, and digestive)” and “metabolisms (glycan, cofactors and vitamins, and amino acid)” were enriched from the ELD-J pairs genes, while pathways related to “diseases (cardiovascular and infectious)” and “environmental information processing” were enriched from the ELD-T and ELOD/ELOD pairs genes. By comparing the KEGG pathways enriched from the ELD and ELOD genes with those enriched from ELD pairs and ELOD pairs genes, we found 54.55% (6/11) of the ELD-J pathways, 45.45% (5/11) of the ELD-T pathways (45.45%), and 44.44% (8/18) of the ELOD pathways overlapped with the corresponding miRNA-mRNA pairs pathways (Supplementary Datasheet 1). These results suggest the significant miRNA regulations in the DEGs’ expression; these miRNA-mRNA interactions may partly contribute to DY’s heterosis through inheriting or transgressive regulation mechanisms.
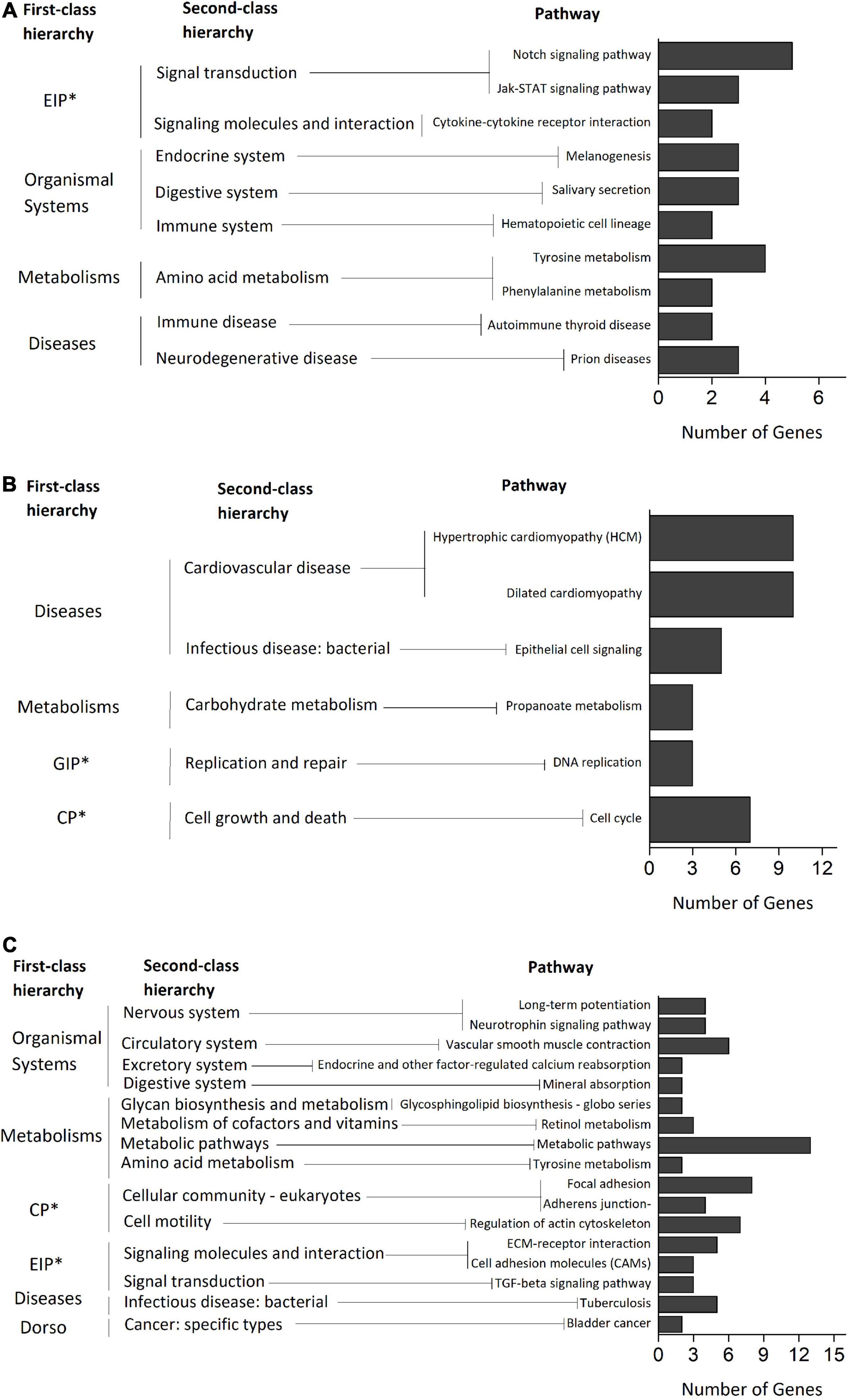
Figure 7. KEGG pathway enrichment of the (A) ELD-J pairs genes, (B) ELD-T pairs genes, and (C) ELOD/ELUD pairs genes. ∗EIP, environmental information processing; GIP, genetic information processing; CP, cellular processes.
Expression of Growth, Immunity, and Stress Response Related Genes
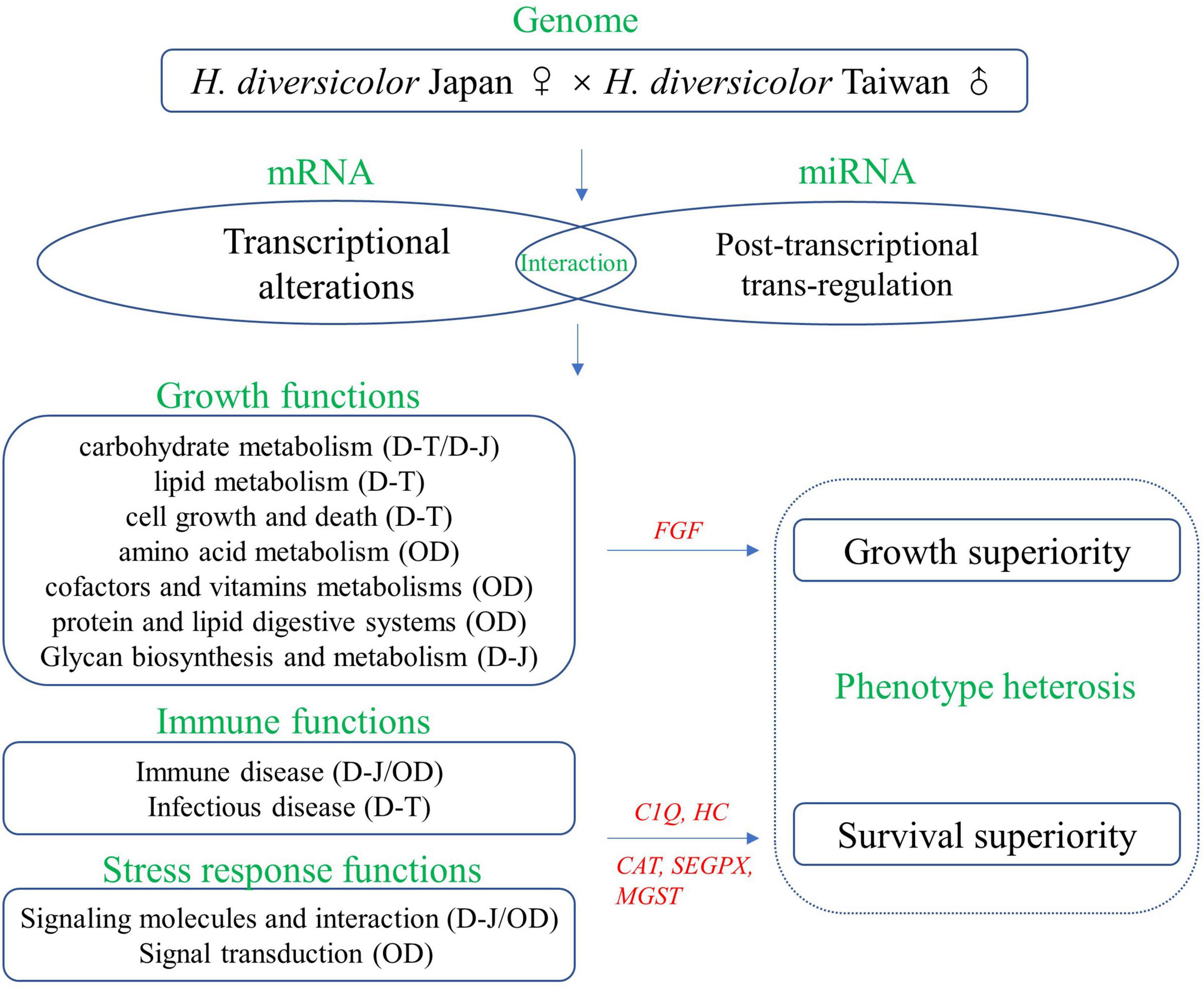
As is shown in Figures 8–10, most of the growth-related genes showed higher expression levels in DY and TW other than JP. Notably, the fibroblast growth factor receptor gene (FGFR) was overdominant expressed in DY. These results confirm the existence of dominance and overdominance models in DY’s growth-related genes expression.
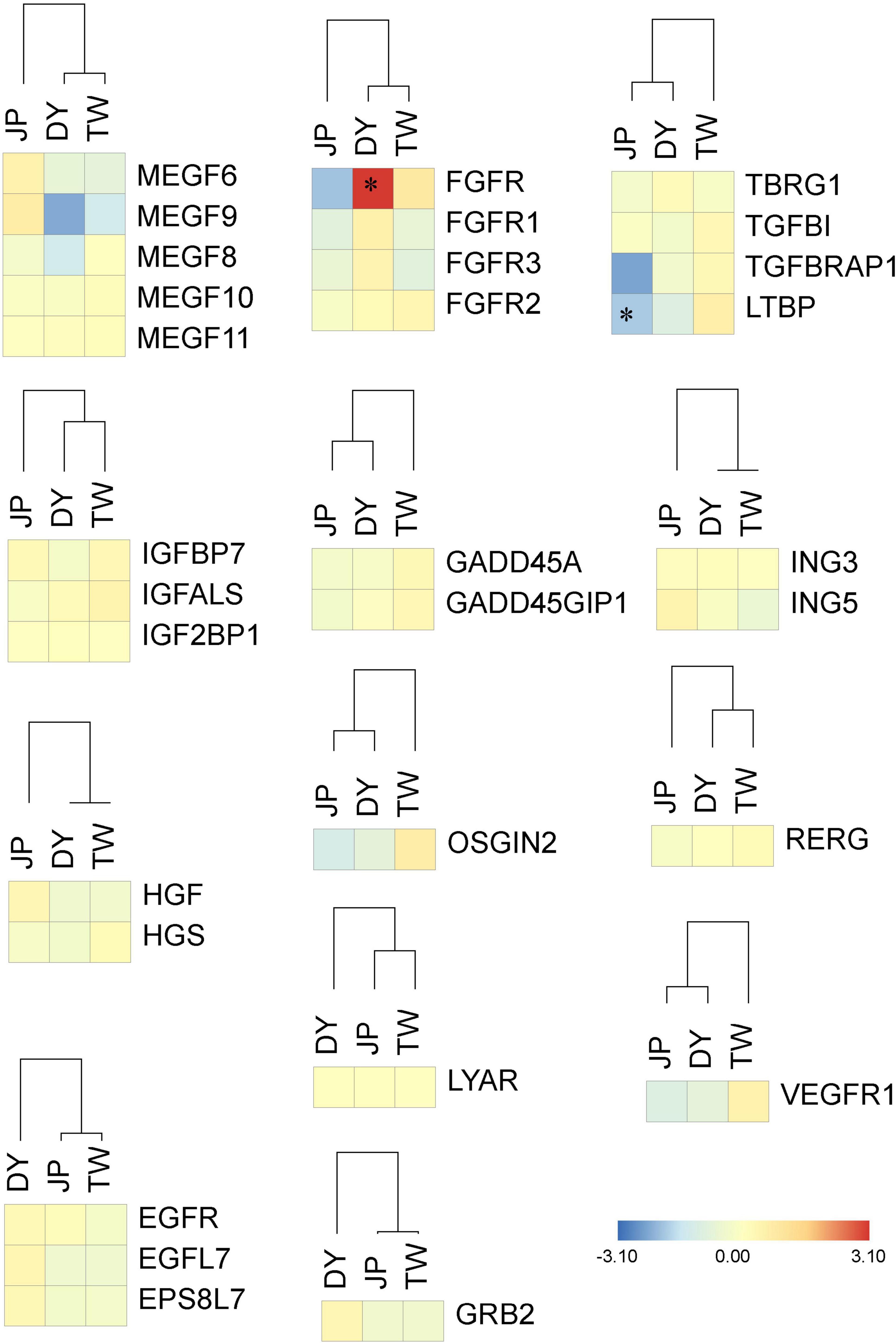
Figure 8. Relative expression of growth-related genes in the three H. diversicolor genotypes. Relative gene expression levels in the three genotypes were normalized to the MPV. Red and blue bars represent up- and down-regulated expression fold change against the MPV, respectively. ∗represents the significant difference of the gene expression level compared to the MPV.
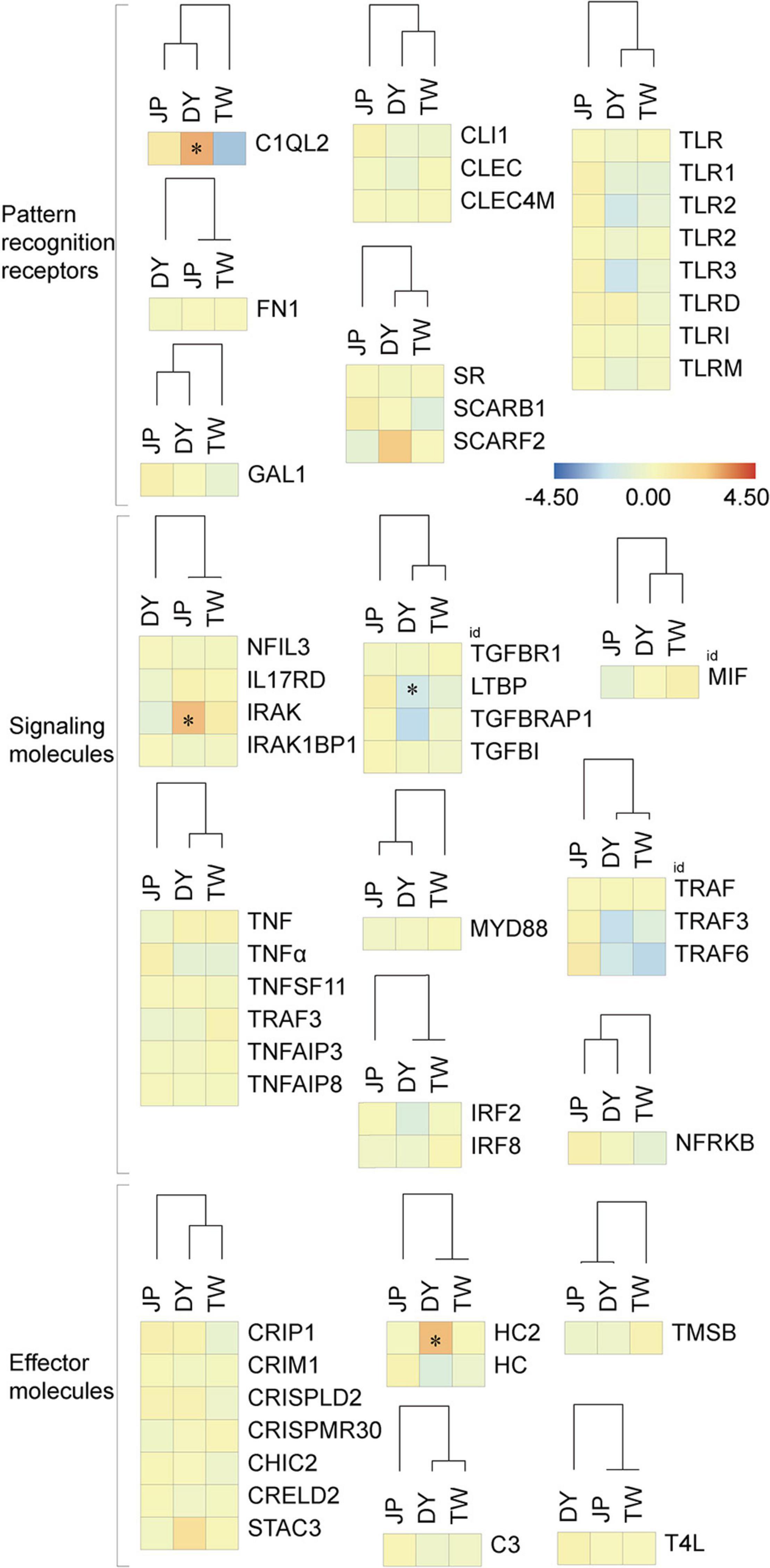
Figure 9. Relative expression of immunity-related genes in the three H. diversicolor genotypes. * represents the significant difference of the gene expression level compared to the MPV.
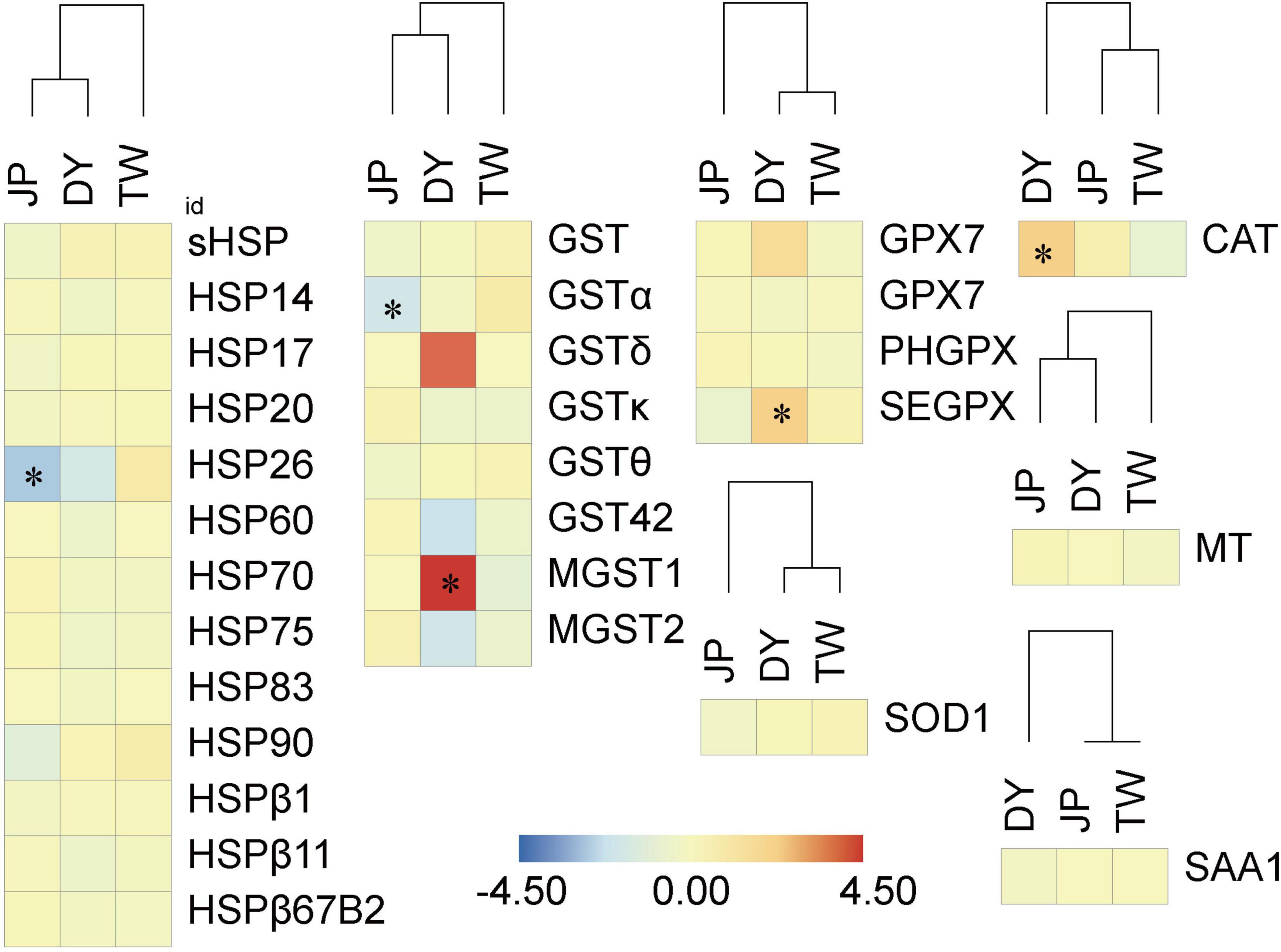
Figure 10. Relative expression of stress response genes in the three H. diversicolor genotypes. * represents the significant difference of the gene expression level compared to the MPV.
DY had a significant maternal expression of most immunity-related genes (13 out of the 20 gene groups) and a significant paternal expression of most stress response genes (four out of the seven gene groups). This indicates the similar survival phenotype of JP and DY (Supplementary Figure 1) might be due to their similar expression of stress response molecules, other than immunity-related molecules. Besides, we also found DY has significantly higher expression levels of complement C1 gene (C1QI2), hemocyanin gene (HC-2), catalase gene (CAT), glutathione peroxidase gene (SEGPX), and glutathione s-transferase gene (MGST1) than MPV. These genes might also be the important molecules related to DY’s heterosis generation.
Validation of the mRNA and miRNA Sequencing
The expression levels of six DEGs (ELD_J+, Unigene 34268; ELD_J−, Unigene 47208; ELD-T+, Unigene 47561; ELD-T−, Unigene 32067; ELUD, Unigene 19086; ELOD, Unigene 49863) and six DEMs (ELD_J+, miR-4755-5p; ELD_J−, miR-6317; ELD-T+, miR-7255-5p; ELD- T-, miR-4326; ELUD, miR-2836; and ELOD, miR-263b) were in accordance with their expression patterns revealed by the mRNA and miRNA sequencing (Supplementary Figures 11, 12), proving the reliability of the sequencing results.
Discussion
In recent years, transcriptional analysis has been conducted in various aquatic species to compare the allelic expression variations between the parents and hybrids, such as brook charr (Bougas et al., 2010, 2013), putterfish (Gao et al., 2013), grouper (Sun et al., 2016a, b), crucian (Ren et al., 2016), and bream (Zheng et al., 2019). However, the transcriptional analysis of molluscan heterosis was only reported in the oysters (Hedgecock et al., 2007; Yang et al., 2018) and the sea cucumber (Wang et al., 2018a). In this study, we compared the growth and survival phenotypes of two H. diversicolor genotypes and the hybrid, analyzed the transcriptomes and miRNAomes of the three genotypes, and predicted the miRNA-mRNA interactions. We wish to explore the molecular mechanisms of the H. diversicolor heterosis through this study and lay the foundation for molluscan heterosis analysis.
DY integrated profitable maternal growth and paternal advantageous survival phenotypes (Supplementary Figure 1 and Supplementary Table 1). In our previous study, DY was JP-like in low-temperature and air exposure resistance (You et al., 2019). The hybrids’ inheritance of parental advantages is not uncommon in mollusks. For example, the F1 hybrid of H. discus hannai and H. kamtschatkana was fast in growth like H. discus hannai and strong in resistance like H. kamtschatkana (Hoshikawa et al., 1998). The F1 hybrid of H. discus hannai and H. gigantea had maternal growth superiority and paternal resistance superiority (Luo et al., 2010; Liang et al., 2014, 2018). A similar phenomenon was also observed in the oyster (Guo et al., 2008), mussle (Beaumont et al., 2004), and scallop (Zheng et al., 2006). The hybrids’ vigorous phenotypes may result from the interactions between two distant parental subgenomes.
A total of 5,562 DEGs and 102 DEMs were identified between the three genotypes (Figure 1), indicating a significant fluctuation in the hybrid’s transcriptome and miRNAome during hybridization. Besides, 1,789 NEGs and 71 NEMs were identified (Figure 2). Non-additively expression (NE) is important in the heterosis formation as it represents a novel expression pattern in hybrids (Guo et al., 2006; Meyer et al., 2012). NE is thought to originate from the interactions between two parental genomes. These interactions result in the hybrid’s unequal gene expression values compared with the average parental expression values (Hochholdinger and Hoecker, 2007; Chen, 2013). NE is commonly seen in mollusks’ hybrids, such as oysters (Hedgecock et al., 2007; Yan et al., 2017) and pearl oysters (Yang et al., 2018). In our previous proteomic analysis of the three Haliotis diversicolor genotypes, 20 proteins were non-additively (overdominance) expressed in the hybrid, consisting 43.48% of the total identified proteins (Di et al., 2013). Functional analysis showed the mollusks’ NEGs were frequently enriched in protein metabolism, energy regulation, and stress response related pathways. In this study, the abalone’s NEGs were also enriched in biological processes, and their functions were closely related to organismal development, diseases, and metabolisms (Figure 3 and Supplementary Figure 2). These findings suggest the contribution of NEGs to DY’s phenotypic superiorities. Besides, we also identified 71 NEMs. NEMs have been proven crucial regulators in hybrids’ heterosis formation (Barber et al., 2012; Fang et al., 2013). Although research data of abalone miRNAs is quite limited, we could still classify the 71 NEMs into three functional categories according to the miRNA studies of other species: (1) Development, metabolism, and energy homeostasis: miR-196, miR-263, miR-9, miR-278, let-7i, and miR-33 (Reinhart et al., 2000; Hornstein et al., 2005; Aurelio and Stephen, 2006; Rayner et al., 2010; Nian et al., 2019; Zhou et al., 2020); (2) Immune response: miR-150, miR-1, miR-133, miR-4326, miR-152, miR-200, and miR-891 (Xiao et al., 2007; Xu et al., 2007, 2017; Yang et al., 2007; Park et al., 2008; Tsuruta et al., 2011; Liao et al., 2019); and (3) Stress response and oxidation resistance: miR-2898, miR-885, and miR-466 (Aliaksandr et al., 2012; Alural et al., 2014; Deb and Sengar, 2020). The NEMs’ functions also pattern with DY’s vigorous phenotypes, highlighting the role of miRNA regulation in the abalone heterosis formation.
The ELD and ELOD expression patterns have been identified in the hybrids of oyster (Hedgecock et al., 2007), pearl oyster (Yang et al., 2018), and various fish species (Bougas et al., 2013; Gao et al., 2013; Ren et al., 2016; Sun et al., 2016a; Zhang et al., 2019). In this study, large amounts of ELD and ELOD genes were also identified in the hybrid abalone (Figure 4). Interestingly, the ELD-J genes were mostly high-parent patterned (type III) rather than low-parent patterned (type IV), while the ELD-T genes were mostly low-parent patterned (type VI). In the yellow catfish and the Cyprinidae’s hybrids, both the maternal and paternal ELD genes were high-parent patterned (Zhou et al., 2015; Zhang et al., 2019). The authors of the two studies propose the hybrids may inherit advantageous molecules from both parents to obtain heterosis. In our case, the hybrid abalone may inherit more advantageous alleles from the paternal genotype other than the maternal genotype. Besides, the number of ELOD genes was much higher than ELUD genes, indicating the overdominance models are more common in the hybrid than the underdominance models. This phenomenon agrees with the yellow catfish and Cyprinidae studies. The overdominance model is an essential mechanism of transgressive heterosis by creating a more robust heterozygous gene expression pattern (Baranwal et al., 2012; Chen, 2013). Moreover, the ELD and ELOD patterns were also found in the DEMs (Figure 6), indicating the dominance and overdominance models exist in the transcriptome and post-transcriptional regulation of the hybrid abalone.
Most of the ELD genes, ELOD genes, ELD pairs genes, and ELOD pairs genes were functionally related to metabolisms, organismal developments, diseases, and environmental information processing (Figure 5). Considering the DY’s heterosis in growth and survival, we could discuss these genes by three functional categories: (1) Growth and development, (2) Immune defense, and (3) Stress response.
(1) Growth and development
The growth-related pathways were mostly enriched from ELD-T and ELOD genes, including metabolism, growth, digestion related pathways. ELD-J genes were also enriched in two metabolism pathways even though the enrichment significances were relatively small (Figure 5). These pathways are crucial for organisms to keep normal growth functions (Gao et al., 2013). In other aquatic species, ELD or ELOD genes’ functions are also frequently related to growth (Zhang et al., 2017; Wang et al., 2018b). Hence, hybrids’ growth heterosis could result from their similar growth-related functions with the more robust progeny, or their transgressive functions beyond both parents. In this study, DY was TW-dominant in the carbohydrate and lipid metabolisms; overdominant in the protein and lipid digestions, amino acids, and vitamins metabolisms. These dominant or overdominant biological functions may work together in forming DY’s growth phenotypes. Besides, the “carbohydrate metabolism,” “cell growth and death,” and “amino acid metabolism” pathways were also enriched from the ELD-T pairs and ELOD/ELUD pairs genes (Figure 7), suggesting the existence of miRNA regulations in these biological functions. The overlapping of ELD gene pathways and ELD pairs genes pathways was also observed in the hybrid yellow catfish (Zhang et al., 2019).
The 30 growth-related genes were mostly ELD-T, or ELOD expressed in DY (Figure 8). The dominant or overdominant expression of growth-related genes was identified in almost all the comparative transcriptional analyzes between hybrids and parents (Di et al., 2013, 2015; Sun et al., 2016a; Zhang et al., 2017; Wang et al., 2018a; Zheng et al., 2019). Functions of the ELD and ELOD genes differed within different species, showing the different heterosis generation mechanisms. Notably, the FGFR gene was overdominant expressed in DY. The FGFR family genes are tyrosine kinase receptors controlling the functions of fibroblast growth factor (FGF) genes by binding and activating processes (Dailey et al., 2005; Beenken and Mohammadi, 2009). Four FGFR transcripts were found to be highly expressed in DY, indicating DY’s superior FGF-related functions. The proteomic analysis of the three H. diversicolor genotypes also found several growth-related proteins to be overdominant expressed (ATP synthase subunit, fructose-bisphosphate aldolase, and triosephosphate isomerase) or partial dominant expressed (enolase, arginine kinase, and taurine dehydrogenase) in the hybrid (Di et al., 2013). These results suggest the ELD and ELOD expression patterns of growth-related molecules exist in the hybrid’s transcriptional and proteomic levels.
(2) Immune defense
In molluscan heterosis studies, the immunity-related molecules draw less attention than the growth-related molecules. However, the immune ability is also crucial to hybrids by improving the organisms’ resistance (Miller et al., 2015; Ma et al., 2018; Zhao et al., 2019). In our study, the diseases related pathways were mainly enriched from the ELD-J and ELD-T genes, suggesting the integration of parental immune characteristics into the hybrid. The ELOD genes were also enriched in several immunity-related pathways, indicating the DY’s transgressive performance in some immune functions. However, DY’s survival phenotype highly conformed with JP and differed from TW (Supplementary Figure 1). Hence, DY’s immunity-related molecules’ expression may not be enough to explain its survival heterosis toward TW. Besides, the “infectious disease” and “immune disease” pathways were also enriched from the ELD pairs and ELOD/ELUD pairs genes, suggesting the role of miRNA regulations in some immunity-related pathways during hybridization.
The mollusks’ immunity-related molecules could be divided into three functional categories: pattern recognition receptors, signaling molecules, and effector molecules (Wang et al., 2013a; Song et al., 2015; Bouallegui, 2019). Of the 52 immunity-related genes investigated, DY and TW had lower expression levels of most genes (11 out of the 19 gene families) than JP. However, DY showed an overdominant expression pattern in two genes, C1QI2 and HC-2 (Figure 9). The C1Q gene is a classic peptidoglycan recognition protein gene containing the versatile charge pattern recognition globular C1Q domain in the C-terminus, engaging a wide range of ligands and trigger a serial of immune responses (Wang et al., 2013a; Song et al., 2015). Hemocyanins are multifunctional proteins existing in invertebrate hemolymph, which contribute to mollusks’ innate immunity through phenoloxidase-like activities (Zhuang et al., 2015). Hence, DY may have transgressive performances in the complement system and hemocyanin-related functions. Interestingly, in aquatic species, unlike growth-related genes, which usually show overdominant expression patterns in the hybrids, the immunity-related genes seem more likely to be dominant or partial-dominant expressed in the hybrids (Yan et al., 2017; Zhang et al., 2017; Yang et al., 2018; Shen et al., 2020). This phenomenon might be due to aquatic hybrids’ immune system characteristic, which needs further investigation.
(3) Stress response
The stress response molecules, including antioxidant enzymes and acute-phase molecules, protect organisms’ cells from the damage of excess reactive oxygen species (ROS) or superoxide anion (O2–) generating upon environmental stressors, or help organisms maintain regular molecular and cellular homeostasis (Aguirre et al., 2005; Vaák and Meloni, 2011; Wang et al., 2013b). In our study, the “environmental information processing” pathways were enriched from the ELD-J and ELOD genes (Figure 5). Environmental information processing is usually the first step of organisms’ stress responses. These results suggest DY is JP-dominant or overdominant in stress response related functions. This hypothesis is further supported by the expression of the 29 stress response genes in the hybrid. DY showed JP-dominant or overdominant expression in most gene groups. Notably, the CAT, SEGPX, and MGST genes were highly expressed in DY compared with the MPV. These genes are antioxidant enzyme genes whose functions include removing excess ROS (Chelikani et al., 2004), reducing organic hydroperoxide and hydrogen peroxidase (Arthur, 2000), and catalyzing the nucleophilic attack of the glutathione’s sulfur atom in organisms (Blanchette et al., 2007). Oxidative stress is common for mollusks as they mostly live in high-velocity water environments. Hence, the ability to reduce oxidative stress is crucial for mollusks to maintain physiological equilibrium. Similarly, in the abalone hybrid of H. discus hannai and H. gigantea, three antioxidant enzymes were found to be overdominant expressed (Di et al., 2015). The positive relationship between the hybrid abalone’s stronger antioxidant functions and its superior survival rates could be concluded.
Conclusion
This study investigated the transcriptomes and miRNAomes of two H. diversicolor geographic genotypes and their hybrid (Figure 11). Evident non-additivity, ELD, and ELOD expression patterns were identified in the DEGs and DEMs, showing the widespread existence of dominant and overdominant models in the hybrid’s heterosis formation. Functional analysis of the ELD and ELOD genes illustrated DY had maternal growth-related functions and paternal stress-response functions. Meanwhile, DY was overdominant in several functional pathways. The existence of miRNA regulation was found in several important functional pathways. The role of FGF, C1Q, HC, CAT, SEGPX, and MGST genes in the abalone heterosis generation is noteworthy. Our study provides a case of molecular heterosis study in mollusk species, which may help disclose the mysteries underlying this complex phenomenon.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA database under BioProject accession PRJNA701659.
Author Contributions
SL conducted the experiments and wrote the manuscript. WY helped in the abalone culture, experiments design, and manuscript revision. XL helped in the abalone culture and experiments design. JK helped in the abalone culture. MH helped in the lab experiments. YG helped in the manuscript revision. CK sponsored the study and participated in the experiments design. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the China Agriculture Research System of MOF and MARA, The National Natural Science Foundation of China (Grant No. 31902369), Tianjin Natural Science Foundation (Grant No. 18JCQNJC84500), Tianjin Seed Industry Science and Technology Major Project (Grant No. 19ZXZYSN00080), College Student Innovation and Entrepreneurship Training Program (Grant No. 202010061010), and Tianjin Key Research and Development Program (Grant No. 19YFZCSN00070).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.667636/full#supplementary-material
Footnotes
- ^ http://www.bioconductor.org/packages/release/bioc/html/DESeq/
- ^ http://Rfam.sanger.ac.uk/
- ^ http://www.mirbase.org/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.miranda.org/
- ^ https://tools4mirs.org/software/target_prediction/pita/
- ^ https://www.blast2go.com/
- ^ https://software.broadinstitute.org/morpheus/
- ^ http://bioinformatics.psb.ugent.be/webtools/Venn/
References
Aguirre, J., Ríos-Momberg, M., Hewitt, D., and Hansberg, W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13, 111–118. doi: 10.1016/j.tim.2005.01.007
Aliaksandr, D., Michael, B., and Joseph, S. (2012). Glucose depletion activates mmu-miR-466h-5p expression through oxidative stress and inhibition of histone deacetylation. Nucleic. Acids. Res. 40, 7291–7302. doi: 10.1093/nar/gks452
Alural, B., Duran, G. A., Tufekci, K. U., Allmer, J., Onkal, Z., Tunali, D., et al. (2014). EPO mediates neurotrophic, neuroprotective, antioxidant, and anti-apoptotic effects via downregulation of miR-451 and miR-885-5p in SH-SY5Y neuron-like cells. Front. Immunol. 5:475. doi: 10.3389/fimmu.2014.00475
Arthur, J. R. (2000). The glutathione peroxidases. Cell. Mol. Life. Sci. 57, 1825–1835. doi: 10.1007/pl00000664
Aurelio, A., and Stephen, M. C. (2006). Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes. Dev. 20:913. doi: 10.1101/gad.374406
Baranwal, V. K., Mikkilineni, V., Zehr, U. B., Tyagi, A. K., and Kapoor, S. (2012). Heterosis: emerging ideas about hybrid vigour. J. Exp. Bot. 63, 6309–6314. doi: 10.1093/jxb/ers291
Barber, W. T., Zhang, W., Win, H., Varala, K. K., Dorweiler, J. E., Hudson, M. E., et al. (2012). Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. U. S. A. 109, 10444–10449. doi: 10.1073/pnas.1202073109
Beaumont, A. R., Turner, G., Wood, A. R., and Skibinski, D. O. F. (2004). Hybridisations between Mytilus edulis and Mytilus galloprovincialis and performance of pure species and hybrid veliger larvae at different temperatures. J. Exp. Mar. Biol. Ecol. 302, 177–188. doi: 10.1016/j.jembe.2003.10.009
Beenken, A., and Mohammadi, M. (2009). The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug. Discov. 8, 235–253. doi: 10.1038/nrd2792
Blanchette, B., Feng, X., and Singh, B. R. (2007). Marine glutathione S-transferases. Mar. Biotechnol. 9, 513–542. doi: 10.1007/s10126-007-9034-0
Bouallegui, Y. (2019). Immunity in mussels: an overview of molecular components and mechanisms with a focus on the functional defenses. Fish. Shellfish. Immun. 89, 158–169. doi: 10.1016/j.fsi.2019.03.057
Bougas, B., Audet, C., and Bernatchez, L. (2013). The influence of parental effects on transcriptomic landscape during early development in brook charr (Salvelinus fontinalis, Mitchill). Heredity 110, 484–491. doi: 10.1038/hdy.2012.113
Bougas, B., Granier, S., Audet, C., and Bernatchez, L. (2010). The transcriptional landscape of cross-specific hybrids and its possible link with growth in brook charr (Salvelinus fontinalis Mitchill). Genetics 186, 97–107. doi: 10.1534/genetics.110.118158
Chelikani, P., Fita, I., and Loewen, P. C. (2004). Diversity of structures and properties among catalases. Cell. Mol. Life. Sci. 61, 192–208. doi: 10.1007/s00018-003-3206-5
Chen, Z. J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant. Sci. 15, 57–71. doi: 10.1016/j.tplants.2009.12.003
Chen, Z. J. (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14, 471–482. doi: 10.1038/nrg3503
Cook, P. A. (2019). Worldwide abalone production statistics. J. Shellfish. Res. 38, 401–404. doi: 10.2983/035.038.0222
Dailey, L., Ambrosetti, D., Mansukhani, A., and Basilico, C. (2005). Mechanisms underlying differential responses to FGF signaling. Cytokine Growth. F. R. 16, 233–247. doi: 10.1016/j.cytogfr.2005.01.007
Deb, R., and Sengar, G. S. (2020). Expression pattern of bta-mir-2898 miRNA and their correlation with heat shock proteins during summer heat stress among native vs crossbred cattle. J. Therm. Biol. 94:102771. doi: 10.1016/j.jtherbio.2020.102771
Di, G., Luo, X., You, W., Zhao, J., Kong, X., and Ke, C. (2015). Proteomic analysis of muscle between hybrid abalone and parental lines Haliotis gigantea Reeve and Haliotis discus hannai Ino. Heredity 114, 564–574. doi: 10.1038/hdy.2014.124
Di, G., You, W., Yu, J., Wang, D., and Ke, C. (2013). Genetic changes in muscle protein following hybridization between Haliotis diversicolor reeve Japan and Taiwan populations revealed using a proteomic approach. Proteomics 13, 845–859. doi: 10.1002/pmic.201200351
Fang, R. Q., Li, L. Y., and Li, J. X. (2013). Spatial and temporal expression modes of MicroRNAs in an elite rice hybrid and its parental lines. Planta 238, 259–269. doi: 10.1007/s00425-013-1881-5
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids. Res. 40, 37–52. doi: 10.1093/nar/gkr688
Fujimoto, R., Taylor, J. M., Shirasawa, S., Peacock, W. J., and Dennis, E. S. (2012). Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. U. S. A. 109, 7109–7114. doi: 10.1073/pnas.1204464109
Gao, Y., Zhang, H., Gao, Q., Wang, L. L., Zhang, F. C., Siva, V. S., et al. (2013). Transcriptome analysis of artificial hybrid pufferfish jiyan-1and its parental species: implications for pufferfish heterosis. PLoS One 8:e58453. doi: 10.1371/journal.pone.0058453
García, D. M., Baek, D., Shin, C., Bell, G. W., Grimson, A., and Bartel, D. P. (2011). Weak seed-pairing stability and high target-site abundance decrease the proficiency of Lsy-6 and other miRNAs. Nat. Struct. Mol. Biol. 18, 1139–1146. doi: 10.1038/nsmb.2115
Gentili, M. R., and Beaumont, A. R. (1988). Environmental stress, heterozygosity, and growth rate in Mytilus edulis L. J. Exp. Mar. Biol. Ecol. 120, 145–153. doi: 10.1016/0022-0981(88)90085-8
Goff, S. A. (2011). A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. New. Phytol. 189, 923–937. doi: 10.1111/j.1469-8137.2010.03574.x
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without areference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greaves, I. K., Groszmann, M., Ying, H., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2012). Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. U. S. A. 109, 3570–3575. doi: 10.1073/pnas.1201043109
Groszmann, M., Greaves, I. K., Albertyn, Z. I., Scofield, G. N., Peacock, W. J., and Dennis, E. S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. U. S. A. 108, 2617–2622. doi: 10.1073/pnas.1019217108
Groszmann, M., Greaves, I. K., Fujimoto, R., Peacock, W. J., and Dennis, E. S. (2013). The role of epigenetics in hybrid vigour. Trends Genet. 29, 684–690. doi: 10.1016/j.tig.2013.07.004
Guo, M., Rupe, M. A., Yang, X. F., Crasta, O., Zinselmeier, C., Smith, O. S., et al. (2006). Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 113, 831–845. doi: 10.1086/498428
Guo, X., Wang, Y., Debrosse, G., Bushek, D., and Ford, S. E. (2008). Building a superior oyster for aquaculture. Jersey Shore 1, 7–9.
Hedgecock, D., Lin, J., DeCola, S., Haudenschild, C. D., Meyer, E., Manahan, D. T., et al. (2007). Transcriptomic analysis of growth heterosis in larval Pacific oysters (Crassostrea gigas). Proc. Natl. Acad. Sci. U. S. A. 7, 2313–2318. doi: 10.1073/pnas.0610880104
Hedgecock, D., McGoldrick, D. J., and Bayne, B. L. (1995). Hybrid vigor in Pacific oysters: an experimental approach using crosses among inbred lines. Aquaculture 137, 285–298. doi: 10.1016/0044-8486(95)01105-6
Hedgecock, D., McGoldrick, D. J., Manahan, D. T., Vavra, J., Appelmans, N., and Bayne, B. L. (1996). Quantitative and molecular genetic analyses of heterosis in bivalve molluscs. J. Exp. Mar. Biol. Ecol. 203, 49–59. doi: 10.1016/0022-0981(96)02569-5
Hochholdinger, F., and Hoecker, N. (2007). Towards the molecular basis of heterosis. Trends Plant Sci. 12, 427–432. doi: 10.1016/j.tplants.2007.08.005
Hornstein, E., Mansfield, J. H., Yekta, S., Hu, K. H., Harfe, B. D., Mcmanus, M. T., et al. (2005). The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature 438, 671–674. doi: 10.1038/nature04138
Hoshikawa, H., Sakai, Y., and Kijima, A. (1998). Growth characteristics of the hybrid between pinto abalone, Haliotis kamtschatkana Jonas, and ezo abalone, H. discus hannai Ino, under high and low temperature. J. Shellfish. Res. 17, 673–677.
Inoue, K., Kito, H., Uki, N., and Kikuchi, S. (1986). Influence of the high temperature on the growth and survival of three species of abalone. Bull. Tohoku. Reg. Fish. Res. Lab. 1, 73–78.
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Liang, S., Luo, X., You, W., and Ke, C. (2018). Hybridization improved bacteria resistance in abalone: evidence from physiological and molecular responses. Fish. Shellfish Immun. 72, 679–689. doi: 10.1016/j.fsi.2017.11.009
Liang, S., Luo, X., You, W., Luo, L., and Ke, C. (2014). The role of hybridization in improving the immune response and thermal tolerance of abalone. Fish. Shellfish Immun. 39, 69–77. doi: 10.1016/j.fsi.2014.04.014
Liao, Y. W., Ho, B. C., Chen, M. H., and Yu, S. L. (2019). Host relieves lnc-IRAK3-3-sequestered miR-891b to attenuate apoptosis in Enterovirus 71 infection. Cell. Microbiol. 21:e13043. doi: 10.1111/cmi.13043
Luo, X., Ke, C., You, W., Wang, D., and Chen, F. (2010). Molecular identification of interspecific hybrids between Haliotis discus hannai Ino and Haliotis gigantea Gmelin using amplified fragment-length polymorphism and microsatellite markers. Aquac. Res. 41, 1827–1834. doi: 10.1111/j.1365-2109.2010.02568.x
Ma, J., Zhang, D., Cao, Y., Wang, L., Li, J., Lubberstedt, T., et al. (2018). Heterosis-related genes under different planting densities in maize. J. Exp. Bot. 69, 5077–5087. doi: 10.1093/jxb/ery282
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
McKeown, P. C., Fort, A., Duszynska, D., Sulpice, R., and Spillane, C. (2013). Emerging molecular mechanisms for biotechnological harnessing of heterosis in crops. Trends Biotechnol. 31, 549–551. doi: 10.1016/j.tibtech.2013.06.008
Meyer, R. C., Witucka-Wall, H., Becher, M., Blacha, A., Boudichevskaia, A., Dormann, P., et al. (2012). Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 71, 669–683. doi: 10.1111/j.1365-313X.2012.05021.x
Miller, M., Song, Q., Shi, X., Juenger, T. E., and Chen, Z. J. (2015). Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 6:7453. doi: 10.1038/ncomms8453
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic. Acids. Res. 35, 182–185. doi: 10.1093/nar/gkm321
Ng, D., Lu, J., and Chen, Z. J. (2012). Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 15, 154–161. doi: 10.1016/j.pbi.2012.01.007
Nian, X. G., Chen, W. F., Bai, W. W., Zhao, Z. W., and Zhang, Y. (2019). miR-263b controls circadian behavior and the structural plasticity of pacemaker neurons by regulating the LIM-only protein beadex. Cells 8:923. doi: 10.3390/cells8080923
Park, S. M., Gaur, A. B., Lengyel, E., and Peter, M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Gene. Dev. 22:894. doi: 10.1101/gad.1640608
Rapp, R. A., Udall, J. A., and Wendel, J. F. (2009). Genomic expression dominance in allopolyploids. BMC. Biol. 7:18. doi: 10.1186/1741-7007-7-18
Rayner, K. J., Suárez, Y., Dávalos, A., Parathath, S., Fitzgerald, M. L., Tamehiro, N., et al. (2010). MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. doi: 10.1126/science.1189862
Rehmsmeier, M., Steffen, P., Hochsmann, M., and Giegerich, R. (2004). Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517. doi: 10.1261/rna.5248604
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Ren, L., Li, W., Tao, M., Qin, Q., Luo, J., Chai, J., et al. (2016). Homoeologue expression insights into the basis of growth heterosis at the intersection of ploidy and hybridity in Cyprinidae. Sci. Rep. 6:27040. doi: 10.1038/srep27040
Sha, Z., Gong, G., Wang, S., Lu, Y., Wang, L., Wang, Q., et al. (2014). Identification and characterization of Cynoglossus semilaevis microRNA response to Vibrio anguillarum infection through high-throughput sequencing. Dev. Comp. Immunol. 44, 59–69. doi: 10.1016/j.dci.2013.11.014
Shen, Y., He, T., Luo, X., Ke, C., and You, W. (2020). Comparative immune response during the juvenile and adult stages of two abalones under Vibrio harveyi challenge. Fish. Shellfish Immun. 98, 109–111. doi: 10.1016/j.fsi.2020.01.004
Song, L., Wang, L., Zhang, H., and Wang, M. (2015). The immune system and its modulation mechanism in scallop. Fish. Shellfish Immun. 46, 65–78. doi: 10.1016/j.fsi.2015.03.013
Springer, N. M., and Stupar, R. M. (2007). Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res. 17, 264–275. doi: 10.1016/j.aquatox.2007.07.004
Sun, Y., Guo, C., Wang, D., Li, X. F., Xiao, L., Zhang, X., et al. (2016a). Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus ♀×E. lanceolatus ♂). BMC. Genet. 17:24. doi: 10.1186/s12863-016-0328-y
Sun, Y., Huang, Y., Hu, G., Zhang, X., Ruan, Z., Zhao, X., et al. (2016b). Comparative transcriptomic study of muscle provides new insights into the growth superiority of a novel grouper hybrid. PLoS One 11:e0168802. doi: 10.1371/journal.pone.0168802
Tsuruta, T., Kozaki, K. I., Uesugi, A., Furuta, M., Hirasawa, A., Imoto, I., et al. (2011). miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 71, 6450–6462. doi: 10.1158/0008-5472.CAN-11-0364
Vaák, M., and Meloni, G. (2011). Chemistry and biology of mammalian metallothioneins. J. Biol. Inorg. Chem. 16:1067. doi: 10.1007/s00775-011-0799-2
Veitia, R. A., and Vaiman, D. (2011). Exploring the mechanistic bases of heterosis from the perspective of macromolecular complexes. Faseb J. 25, 476–482. doi: 10.1096/fj.10-170639
Wang, L., Qiu, L., Zhou, Z., and Song, L. (2013a). Research progress on the mollusc immunity in China. Dev. Comp. Immunol. 39, 2–10. doi: 10.1016/j.dci.2012.06.014
Wang, L., Yang, C., and Song, L. (2013b). The molluscan HSP70s and their expression in hemocytes. Invert. Surviv. J. 10, 77–83.
Wang, Z., Cui, J., Song, J., Gou, M., Wang, H., Gao, K., et al. (2018a). Integration of small RNAs and mRNAs by high-throughput sequencing reveals a complex regulatory network in Chinese sea cucumber, Russian sea cucumber and their hybrids. Comp. Biochem. Phys. D. 29, 1–13. doi: 10.1016/j.cbd.2018.10.003
Wang, Z., Cui, J., Song, J., Wang, H., Gao, K., Qiu, X., et al. (2018b). Comparative transcriptome analysis reveals growth-related genes in juvenile Chinese sea cucumber, Russian sea cucumber, and their hybrids. Mar. Biotechnol. 20, 193–205. doi: 10.1007/s10126-018-9796-6
Xiao, C., Calado, D. P., Galler, G., Thai, T. H., Patterson, H. C., Jing, W., et al. (2007). miR-150 controls b cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159. doi: 10.1016/j.cell.2016.04.056
Xu, C., Lu, Y., Pan, Z., Chu, W., Luo, X., Lin, H., et al. (2007). The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell. Sci. 120, 3045–3052. doi: 10.1242/jcs.098830
Xu, G., Zhang, Z., Zhang, L., Chen, Y., Li, N., Lv, Y., et al. (2017). miR-4326 promotes lung cancer cell proliferation through targeting tumor suppressor APC2. Mol. Cell. Biochem. 443:151–157. doi: 10.1007/s11010-017-3219-2
Yan, L., Su, J., Wang, Z., Yan, X., Yu, R., Ma, P., et al. (2017). Transcriptomic analysis of Crassostrea sikamea × Crassostrea angulata hybrids in response to low salinity stress. PLoS One 12:e0171483. doi: 10.1371/journal.pone.0171483
Yang, B., Lin, H., Xiao, J., Lu, Y., Luo, X., Li, B., et al. (2007). The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13, 486–491. doi: 10.1038/nm1569
Yang, J., Luo, S., Li, J., Zheng, Z., Du, X., and Deng, Y. (2018). Transcriptome analysis of growth heterosis in pearl oyster Pinctada fucata martensii. Febs. Open. Bio. 8, 1794–1803. doi: 10.1002/2211-5463.12502
You, W., Guo, Q., Fan, F., Ren, P., Luo, X., and Ke, C. (2015). Experimental hybridization and genetic identification of Pacific abalone Haliotis discus hannai and green abalone H. fulgens. Aquaculture 448, 243–249. doi: 10.1016/j.aquaculture.2015.05.043
You, W., Wang, B., Luo, X., and Ke, C. (2019). Environmental stress tolerance and immune response for the small abalone hybrids. Aquac. Int. 27, 105–123. doi: 10.1007/s10499-018-0310-y
Zhang, G., Li, J., Zhang, J., Liang, X., Zhang, X., Wang, T., et al. (2019). Integrated analysis of transcriptomic, miRNA and proteomic changes of a novel hybrid yellow catfish uncovers key roles for miRNAs in heterosis. Mol. Cell. Proteomics 18, 1437–1453. doi: 10.1074/mcp.RA118.001297
Zhang, G., Que, H., Liu, X., and Xu, H. (2004). Abalone mariculture in China. J. Shellfish Res. 23, 947–950.
Zhang, H., Xu, X., He, Z., Zheng, T., and Shao, J. (2017). De novo transcriptome analysis reveals insights into different mechanisms of growth and immunity in a Chinese soft-shelled turtle hybrid and the parental varieties. Gene 605, 54–62. doi: 10.1016/j.gene.2016.12.003
Zhao, Y., Hu, F., Zhang, X., Wei, Q., Dong, J., Bo, C., et al. (2019). Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC. Plant. Biol. 19:273. doi: 10.1186/s12870-019-1878-8
Zheng, G., Wu, C., Liu, J., Chen, J., and Zou, S. (2019). Transcriptome analysis provides new insights into the growth superiority of a novel backcross variety, Megalobrama amblycephala female × (M. amblycephala female × Culter alburnus male) male. Aquaculture 512:734317. doi: 10.1016/j.aquaculture.2019.734317
Zheng, H. P., Zhang, G. F., Guo, X., and Liu, X. (2006). Heterosis between two stocks of the bay scallop Argopecten irradians irradians Lamarck (1819). J. Shellfish Res. 25, 807–812.
Zhou, M., Jia, X., Wan, H., Wang, S., and Wang, Y. (2020). miR-9 and miR-263 regulate the key genes of the ERK pathway in the ovary of mud crab Scylla paramamosain. Mar. Biotechnol. 22, 594–606. doi: 10.1007/s10126-020-09981-4
Zhou, Y., Ren, L., Xiao, J., Zhong, H., Wang, J., Hu, J., et al. (2015). Global transcriptional and miRNA insights into bases of heterosis in hybridization of Cyprinidae. Sci. Rep. 5:13847. doi: 10.1038/srep13847
Keywords: heterosis, abalone, Haliotis diversicolor, transcriptome, miRNAome, growth, resistance
Citation: Liang S, You W, Luo X, Ke J, Huang M, Guo Y and Ke C (2021) Integrated Analysis of mRNA and miRNA Changes in Two Haliotis diversicolor Genotypes and Their Hybrid. Front. Mar. Sci. 8:667636. doi: 10.3389/fmars.2021.667636
Received: 14 February 2021; Accepted: 12 April 2021;
Published: 04 June 2021.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Chang-Ming Bai, Yellow Sea Fisheries Research Institute (CAFS), ChinaHongyu Ma, Shantou University, China
Copyright © 2021 Liang, You, Luo, Ke, Huang, Guo and Ke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caihuan Ke, Y2hrZUB4bXUuZWR1LmNu
†These authors have contributed equally to this work
 Shuang Liang
Shuang Liang Weiwei You
Weiwei You Xuan Luo
Xuan Luo Jinwei Ke1,3
Jinwei Ke1,3