
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 20 May 2021
Sec. Coral Reef Research
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.665789
Seawater dissolved organic matter (DOM) is a large reservoir of carbon composed of a complex and poorly characterized mixture of molecules. Sponges have long been known to consume dissolved organic carbon (DOC) from this mixture, but the role of microbial sponge symbionts in this process is complex, and the molecules involved remain largely unknown. In order to better understand how sponge processing changes seawater DOM, we used untargeted metabolomics to characterize DOM in samples of incurrent and excurrent seawater taken from sponges on the fore-reef off Carrie Bow Cay, Belize, over 2 years. We collected samples from three sponge species each with either high or low microbial abundance (HMA, LMA) to explore the relationship between symbiont abundance and DOM alterations. Analyses revealed that sponges took up metabolites and changed the composition of seawater DOM, but only for the three HMA species, and none of the LMA species, implicating microbial symbionts in this uptake. Using a new mass spectra classification tool, we found that putative compositions of features depleted in the excurrent samples of HMA sponges were similar in both years and were dominated by organic acids and derivatives (74%) and organic nitrogen compounds (19%). Interestingly, HMA sponges also took up halogenated compounds (containing chlorine or bromine), providing evidence of a previously unknown mechanism of halide cycling. The metabolites taken up by HMA sponges may be used as a food source or as building blocks of chemical defenses, selective advantages that may have guided the evolution of microbial symbioses in sponges.
Sponges are important members of most aquatic habitats, ranging from the tropics to the polar seas, and from the intertidal zone to the deep-sea (Wörheide et al., 2005). On Caribbean coral reefs, the success of these ancient animals is evident in the abundance and diversity of sponge species occupying the benthos (Loh and Pawlik, 2014). There is increasing evidence that current conditions on many Caribbean reefs are favoring the spread of sponges, as reefs that were once dominated by reef-building corals are now degraded by the ongoing effects of climate change, diseases, and overfishing (Gardner et al., 2003; Bell et al., 2013; Loh et al., 2015; Gochfeld et al., 2020). On the fore-reef of the Florida Keys reef tract, for example, populations of the giant barrel sponge Xestospongia muta more than doubled in size and increased in biomass by 39% between 2000 and 2012 (McMurray et al., 2015). Massive sponges such as X. muta are efficient filter feeders capable of overturning a 30 m water column in as little as 2.3 days (McMurray et al., 2014). As they pump seawater through their aquiferous system, sponges assimilate dietary carbon in forms ranging in size from particulate to dissolved. Some sponges acquire most of their dietary carbon from the dissolved fraction of organic carbon (DOC), including Xestospongia spp., for which DOC can comprise 60–96% of total organic carbon uptake (Yahel et al., 2003; McMurray et al., 2016; Hoer et al., 2018; Wooster et al., 2019). Efforts to quantify and isotopically trace the DOC taken up by sponges have revealed the biogeochemical importance of this group and have inspired novel hypotheses, such as the “sponge loop,” which proposes that sponges retain carbon on coral reefs by converting carbon from DOC to forms such as particulate detritus or biomass that are accessible to higher trophic levels (de Goeij et al., 2013; McMurray et al., 2018).
The DOC assimilated by sponges is part of the pool of dissolved organic matter (DOM), the largest reservoir of carbon in the ocean (~600 Gt C) that is made up of a complex mixture of molecules present at vanishingly small concentrations (Carlson and Hansell, 2015; Moran et al., 2016). An estimated 60–70% of DOM has been characterized at least to the level of functional groups (Repeta, 2015), but only about 1% of DOM can be matched to spectral libraries (Petras et al., 2017). Liquid chromatography-mass spectrometry (LC-MS)-based untargeted metabolomics analyses are commonly used for molecular characterization of DOM, and ongoing improvements in extraction methods, LC-MS instrument mass resolution, and data analysis workflows are advancing our understanding of the composition and dynamics of DOM (Dittmar et al., 2008; Kido Soule et al., 2015; Longnecker et al., 2015). There are limits to the isolation and characterization of DOM due to imperfect extraction efficiencies and chromatographic biases inherent to LC-MS, but this technique is uniquely able to resolve single metabolites across the breadth of DOM (Petras et al., 2017). One recent study used LC-MS to characterize DOM in seawater before and after processing by sponges, and it concluded that exhalant seawater had a unique composition of DOM which may act as a signature of sponge processing (Fiore et al., 2017). Studies using stable isotopes have shown that sponges prefer to take up locally-produced DOC over oceanic DOC (van Duyl et al., 2011), supporting the role of sponges as recyclers of carbon produced as DOC by benthic primary producers (Haas et al., 2011; Nelson et al., 2013; Brocke et al., 2015). Sponges may also prefer to take up algae-derived DOC over coral-derived DOC, where the former is produced in large quantities and enriched in neutral sugars, while the latter is produced mostly as colloidal mucus and enriched in proteins and lipids (Rix et al., 2017). Studies exploring sponge alteration of DOM on a molecular level (Fiore et al., 2017) and preferred DOC food sources (van Duyl et al., 2011; Rix et al., 2017) converge on the goal to know both the chemical identities and sources of DOM taken up by sponges, and these insights will be important for predicting how carbon cycles on coral reefs will respond to ongoing shifts in benthic community structure.
Sponge species are categorized as either high or low microbial abundance (HMA, LMA), depending on the density of symbiotic microbes inhabiting their mesohyl tissues (Gloeckner et al., 2014). HMA sponges contain more abundant (by 2–4 orders of magnitude) and more diverse microbial symbionts compared to LMA sponges, and sponge microbiomes are species-specific and stable across seasons and geographic regions (Hentschel et al., 2003; Taylor et al., 2007; Erwin et al., 2012; Pita et al., 2013; Moitinho-Silva et al., 2017). The putative metabolic functions of sponge symbionts include nitrogen cycling, CO2 fixation, and production of secondary metabolites (Taylor et al., 2007; Fan et al., 2012; de Goeij et al., 2013; Thomas et al., 2016). Sponge symbionts have also been implicated in DOC uptake since Reiswig's pioneering studies of sponge nutrition which linked DOC uptake to the presence of microbial symbionts in the HMA sponge host, in contrast to LMA sponges that primarily fed on particles (Reiswig, 1974). This early evidence that DOC uptake is exclusive to HMA species (and is therefore likely microbially-mediated) has since been contradicted by some reports of DOC uptake by LMA sponges (de Goeij et al., 2008; Mueller et al., 2014; Morganti et al., 2017; Rix et al., 2020), but corroborated by other reports of DOC uptake by HMA but not LMA species (Hoer et al., 2018; McMurray et al., 2018; Freeman et al., 2020), stimulating ongoing debate over which, between sponge host and symbiont, takes up DOC. Together, the sponge host and its associated microbes form a holobiont that functions like an ecosystem nested within and able to influence the surrounding reef system (Pita et al., 2018). This, plus the general grouping of sponge species as either HMA or LMA, makes the sponge holobiont a useful model for better understanding the relationship between microbial diversity, metabolic patterns, and biogeochemical cycles (DeLong, 2009); experiments spanning HMA and LMA species can decouple the effect of host and symbionts and reveal how microbiomes influence ecological and biogeochemical processes (e.g., DOC uptake and carbon recycling).
In this study, we used LC-MS to analyze seawater going into (incurrent, or In) and coming out of (excurrent, or Ex) individual sponges on the fore-reef off Carrie Bow Cay, Belize, to characterize and compare seawater DOM before and after processing by three HMA and three LMA sponge species. We sought to answer the following questions: Does the processing of seawater by sponges change the chemical composition of DOM? Are changes to DOM dependent on the abundance of microbial symbionts in the host sponge? Is uptake or production more important for driving changes to DOM? Of the compounds taken up by sponges, are there chemical or structural characteristics that may inform the origin of these compounds or how they are used by the sponge?
Seawater going into (incurrent) and coming out of (excurrent) individual tube- and vase-shaped sponges was collected and analyzed by liquid chromatography-mass spectrometry (LC-MS) to characterize changes in seawater dissolved organic matter (DOM). Samples were collected on the fore reef near the Carrie Bow Cay (CBC) field station, Belize, at 13–23 m depths, over 2 years (July 2018 and March 2019). In 2018, we sampled n = 3 individuals each of three high microbial abundance (HMA) species (Agelas tubulata, Xestospongia muta, and Verongula reiswigi) and three low microbial abundance (LMA) species (Callyspongia vaginalis, Niphates digitalis, and Mycale laxissima). In 2019, we sampled n = 7 individuals each of two HMA species (X. muta and A. tubulata; Supplementary Table 1); the choice to sample these two species with increased replication in 2019 was based on preliminary analysis of the 2018 data. All six species targeted herein are common in the Caribbean and were subjects of a recent study that calculated flux of bulk dissolved organic carbon (DOC) from incurrent and excurrent seawater samples (McMurray et al., 2018). All materials used to collect and store samples were made of glass or chemically inert plastic to minimize contamination. Glassware was combusted (500°C for 4 h) prior to each trip, and all materials were sample-rinsed between uses during each trip. Samples were prepared for LC-MS analysis using high purity (Fisher Optima) solvents and additives. Sections 2.2–2.6 summarize the workflow from sample collection to data analyses. We followed this workflow twice, analyzing 2018 and 2019 sample sets in parallel, before comparing general patterns across the years.
Samples were collected from sponge individuals >10 m apart and with an atrium (inner empty space) large enough that a diver could maneuver a sample syringe inside without touching the sponge. Before sampling, sponge pumping was confirmed by applying a small amount of fluorescein dye above the sponge atrium and checking for upwards movement indicative of excurrent flow. This dye was allowed to dissipate for at least 5 min before sampling began. Two divers then collected paired 1 L incurrent and excurrent (In/Ex) seawater samples from the sponge using 50 mL syringes (Norm-Ject Air-Tite) attached via Teflon tubing and a polycarbonate check valve to a 1 L foil sample bag (FlexFoil PLUS, SKC; Supplementary Figure 1A). The syringes contained no latex, plasticizers, PVC, rubber gaskets, or silicone oil, and the check valve allowed for continuous pumping of sample to gradually fill up the foil bag (Supplementary Figure 1B). The incurrent seawater sample was collected adjacent to the ostia lining the external sponge surface, and the excurrent sample was collected from ~5 cm below the osculum within the atrium of each sponge and at a rate lower than the excurrent water velocity to avoid mixing with ambient seawater. In/Ex samples were collected simultaneously. The In/Ex method is commonly used to measure the in situ metabolic processes of active suspension feeders, such as sponges that usually stop feeding upon being disturbed or removed from their natural habitat (Yahel et al., 2005). Samples were transported back to the lab at CBC immediately (~15 min) after collection for filtration and solid phase extraction (SPE) as described below in section 2.3. Experimental controls were also made by passing 500 mL water (Fisher Optima) through the sampling mechanism (syringe, foil bag, tubing), filtration apparatus, and SPE apparatus. These were analyzed alongside experimental samples for post-processing removal of contaminants.
Solid phase extraction (SPE) followed previously described methods (Dittmar et al., 2008). Briefly, samples were filtered (0.2 μm PTFE filter, Omnipore), acidified to pH 2 using HCl, and then passed at a rate <5 mL min−1 through SPE cartridges (1 g/6 cc PPL, Agilent BondElut) that had been previously conditioned with 6 mL methanol and washed with 18 mL water, 18 mL methanol, and 18 mL acidified water (pH 2). After passing the sample through the cartridges, cartridges were de-salted with 18 mL acidified water, and dried under vacuum pressure (<10 kPa) until no moisture was visible in the cartridge. Sample DOM was then eluted by passing 6 mL methanol through the cartridge and into scintillation vials (Traceclean, VWR). Extracts were stored at −20°C and transported on ice in coolers to University of North Carolina Wilmington (UNCW) for analysis.
Prior to LC-MS analysis, 2 mL aliquots of extracts from 1 L In/Ex samples and 4 mL aliquots of extracts from 500 mL experimental controls were filtered (0.2 μm PTFE syringe filter, VWR), dried using a vacuum centrifuge, and redissolved in 30 μL of 99% methanol and 1% formic acid. A solvent blank containing only the 99% methanol and 1% formic acid solution and pooled QC sample containing equal quantities of each extract were also prepared.
Triplicate injections of each sample were analyzed using an ACQUITY I-Class UPLC (Waters, USA) coupled to a Xevo G2-XS QToF mass spectrometer (Waters, UK) with an electrospray ionization (ESI) source. For chromatography, a CORTECS UPLC C18 column (2.1 × 100 mm, 1.6 μm particle size, Waters, UK) was used for the reversed phase separation. The column was maintained at 30°C and the sample chamber was maintained at 10°C. The mobile phase consisted of solvent A (water + 0.1% formic acid) and solvent B (acetonitrile + 0.01% formic acid) pumped at 0.5 mL min−1. A 15 min gradient was used with the following elution conditions: 0–0.5 min, 95% phase A, 5% phase B; 0.5–5 min, 95–50% phase A, 5–50% phase B; 5–10 min, 50–0% phase A, 50–100% phase B; 10–12 min, 0% phase A, 100% phase B; 12–12.5 min, 0–95% phase A, 100–5% phase B; 12.5–15 min, 95% phase A, 5% phase B. The injection volume was 3 μL. MS data were acquired in positive ESI mode, in profile format, and using MSE to collect information on both parent and fragment ions. Source conditions were as follows: capillary voltage, 2.5 kV; sample cone, 80 V; source temperature, 100°C; desolvation temperature, 550°C; cone gas flow rate, 100 L h−1; desolvation gas (N2) flow rate, 800 L h−1. Lockspray was injected at 10 μL min−1 for accurate mass acquisition, using lockmass Leucine–enkephalin producing a reference ion at m/z 556.2771. MSE data encompassed two scan functions spanning a mass range of 50–1,800 m/z, with 0.1 s scan time, but with varied collision energies: function 1 (low energy for parent ions) had a 6 eV collision energy, and function 2 (high energy for fragment ions) had a collision energy ramp of 15–25 eV. The raw LC-MS data from both years are available in the MetaboLights repository (https://www.ebi.ac.uk/metabolights), accession number MTBLS2199 and MTBLS2200 (Haug et al., 2020).
Waters. raw datasets (Figure 1A) were centroided and lockmass corrected using the automatic peak detection tool in Masslynx (Waters, US; Figure 1B), and converted to .mzXML in msconvert (Chambers et al., 2012), applying the scan event filter to isolate function 1 which contained the parent ions (Figure 1C). The resulting .mzXML files were imported into R for further processing and analysis using the package xcms (Smith et al., 2006; R Core Team, 2020). The centWave algorithm (Tautenhahn et al., 2008) was used for peak picking, and the Obiwarp method (Prince and Marcotte, 2006) was used for alignment of retention times. Peaks were then matched across samples to produce mass features (i.e., correspondence), and for samples in which any feature was not detected, peaks were filled by integrating the area under the curve within the region defined from other samples (Figure 1D).
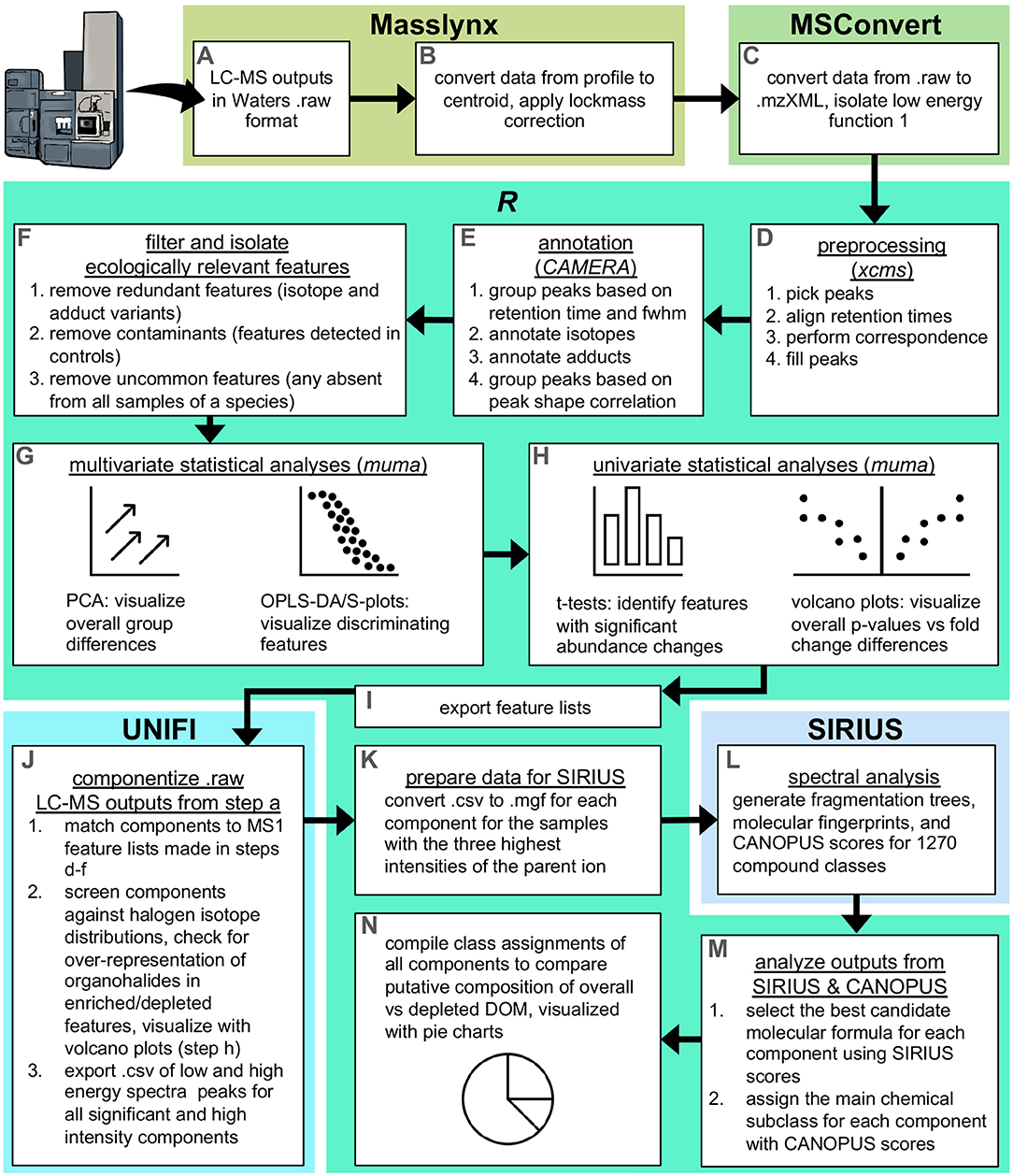
Figure 1. Summary of the data analysis workflow. The process started in Masslynx, where (A). Raw LC-MS outputs were (B) centroided and subsequently (C) converted to .mzXML in MSconvert (Chambers et al., 2012), and then (D–H) processed and statistically analyzed in R using XCMS, CAMERA, and muma (Smith et al., 2006; Kuhl et al., 2012; Gaude et al., 2013). Feature lists were then (I) exported to use as targets for (J) componentization in UNIFI. This produced fragmentation peaks that were (K) converted in R to be compatible for (L) spectral analysis in SIRIUS using the CANOPUS submodule (Böcker et al., 2009; Dührkop et al., 2015, 2019, 2020; Böcker and Dührkop, 2016; Djoumbou Feunang et al., 2016), followed by (M,N) analysis of SIRIUS/CANOPUS outputs in R.
The preprocessing steps converted raw spectra into tables of features with abundances for each sample. The R package CAMERA (Kuhl et al., 2012) was then used to annotate putative isotopes and adducts, and group features according to peak shape (Figure 1E). First, features were roughly grouped according to retention time and full width at half max (fwhm). Features were then screened to annotate isotopes ([M+1], [M+2], and [M+3]) and adducts (M+H, M+Na, and M+K). Lastly, features were finely grouped into pseudospectra according to peak shape using a minimum correlation threshold of 0.75. Annotating and grouping features from the same parent compound allowed removal of all but one feature representative of that parent compound, and this was useful for reducing data complexity and redundancy for subsequent data analyses. Within each pseudospectra, features were removed if they were isotopes or lower intensity variants of adduct pairs/triads. This semi-conservative approach eliminated features that were most likely to be redundant, while minimizing the loss of novel features.
Features were further filtered to remove all but the most ecologically relevant. First, any features detected in experimental controls or solvent blanks were removed, as these represented likely contaminants or impurities from solvents, glassware, or instruments. Also removed were uncommon features, defined here as those not detected in any samples (incurrent or excurrent) of one or more species. This left “common” features, or those that were detected in at least one sample (incurrent or excurrent) of at least one individual of each species (Figure 1F).
Raw data from 2018 and 2019 samples comprised a total of 12,801 and 11,878 features, respectively, reduced to 10,595 and 9,544 features after removal of adduct variants and isotopologues. Removal of contaminants and uncommon features left 2,188 and 1,875 features in 2018 and 2019 sample sets, respectively, and data analyses proceeded using these remaining features.
Multivariate analyses were conducted on half-minimum imputed, normalized, and range-scaled data using the R package muma (Gaude et al., 2013) (Figure 1G). First, principle component analysis (PCA) was used as an unsupervised check for In/Ex groupings within and among species and species groups (HMA/LMA). A PCA that shows grouping is an important prerequisite for supervised analyses, such as orthogonal partial least squares-discriminant analysis (OPLS-DA), where the model uses known experimental groupings to refit sample scores and maximize experimental variation along one PC (Worley and Powers, 2016). For the PCA biplots with clear In/Ex groupings, analyses proceeded with OPLS-DA in order to determine which features had the greatest influence on In/Ex differences. Univariate analyses were also conducted on half-minimum imputed, normalized, and log2 transformed data (Figure 1H). In/Ex differences in the abundance of single features were assessed for statistical significance (p < 0.05) using the univariate function in muma, which first tests for normality using a Shapiro-Wilk test, then computes either a Welch's t-test or non-parametric Wilcoxon-Mann Whitney U test, before correcting for multiple testing using the Benjamini-Hochberg method (Gaude et al., 2013). A feature list was then exported, and this list contained unique IDs, m/z, retention times, and significance (from univariate analyses) of all features (Figure 1I).
Fragmentation spectra were generated via componentization of MSE data in UNIFI Scientific Information System (Waters, US; Figure 1J). MSE data comprised full-scan data in low and high energy ranges, and the proprietary algorithm within UNIFI resolved individual components from these scans by matching low energy peaks (precursor ions) to putative high energy fragments at the same retention time and removing interfering ions (Goshawk et al., 2015). The feature lists containing m/z and retention times were used as targets for matching with candidate components in UNIFI. UNIFI was also used to screen matched components against theoretical isotope distributions corresponding to halogenated ions (chlorine or bromine); feature components were classified as halogenated at a confidence level of 90% or greater.
Components matching significant and high intensity (detector counts >5,000) features were exported from UNIFI as .csv files of m/z and intensity values of peaks from low and high energy fragmentation spectra, for analysis in SIRIUS 4 (Dührkop et al., 2019). Within SIRIUS is the submodule CANOPUS (Class Assignment aNd Ontology Prediction Using mass Spectrometry), which can predict compound classes from fragmentation spectra even in the absence of structural reference data or MS/MS training data (Dührkop et al., 2020). For each feature, .csvs were converted to .mgfs for the three samples with the most intense peak of the parent ion (Figure 1K), imported into the SIRIUS GUI, and run using the following sub-modules and settings: SIRIUS (10 ppm deviation, C H N O P Cl Br allowed elements, 10 candidates), CSI:FingerID (all included databases), CANOPUS selected (Figure 1L). For each feature, SIRIUS scored 10 molecular formula candidates based on similarities to isotope patterns and fragmentation trees (Böcker et al., 2009; Böcker and Dührkop, 2016). For each candidate, CANOPUS scores were assigned to 1,270 chemical classes spanning 11 levels of the hierarchical ClassyFire chemontology (Djoumbou Feunang et al., 2016). CANOPUS scores are provided by a deep neural network (DNN) trained on molecular fingerprints of over a million compound structures. The input of the DNN is the molecular fingerprint predicted by CSI:FingerID, which uses a support vector machine that is trained on MS/MS spectra of over 20,000 compounds and uses the predicted fragmentation tree as input (Dührkop et al., 2015, 2020; Ludwig et al., 2018). We analyzed SIRIUS outputs and CANOPUS scores in R, with a series of simple rules that first picked the best molecular formula candidate, and then the highest scoring chemical class of that candidate (Figure 1M). For each feature, the best candidate was defined as that with the highest score in two out of the three samples. Classification then proceeded for that candidate using the CANOPUS scores from the sample with the most intense peak of the parent ion. From this set of 1,270 scores, the top subclass classification was chosen based on which scored highest for subclass, class, and superclass, to resemble how “main classes” are assigned in the CANOPUS tab on the SIRIUS GUI, and ensuring scores >0.5. We compiled these putative classifications to investigate DOM composition, and compare overall DOM to the fraction of DOM that was significantly depleted by the sponge (Figure 1N).
Analyses of 2018 and 2019 samples comprised 2,188 and 1,875 mass features, respectively. Principle component analysis (PCA) biplots for 2018 samples revealed differences between HMA and LMA species (Figure 2A). Incurrent samples from all six species were relatively clustered together along the x-axis, while excurrent samples of the three HMA species showed a notable shift to the right along the x-axis. The arrows connecting incurrent and excurrent samples from each HMA individual also exhibited relatively uniform length and direction, reflecting similarly enriched or depleted abundances of the mass features underlying the detected DOM. However, there was no such separation or grouping of In/Ex samples from LMA species (Figure 2A). Results from 2018 samples were the basis for targeting two HMA species (Agelas tubulata, Xestospongia muta) for sampling in 2019.
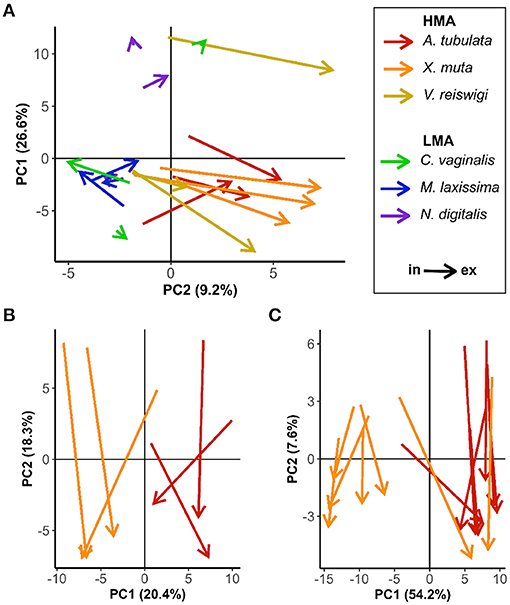
Figure 2. PCA biplots of In/Ex samples. Arrows connect incurrent and excurrent samples for an individual sponge. (A) 2018 all six species; (B) 2018 A. tubulata and X. muta samples; (C) 2019 A. tubulata and X. muta samples.
The biplots of A. tubulata and X. muta samples from 2018 and 2019 show some notable similarities in how samples were grouped along the x and y axes (Figures 2B,C). Separation between the two species along the x-axis in both 2018 and 2019 is likely due to external factors that introduced variation in the incurrent DOM. All 2018 samples were collected over 3 days (19–21 July 2018), and at least one HMA and one LMA species were sampled on each day. However, all X. muta (n = 3) were sampled on the first day, and all A. tubulata (n = 3) were sampled on the third day; sequential sampling of the two species likely introduced temporal variation to the incurrent DOM that may explain the horizontal separation in Figure 2B. All 2019 samples were also collected over 3 days (28 February–2 March 2019). Several individuals of both species (n = 4 X. muta, n = 3 A. tubulata) were sampled on the second day, but only X. muta (n = 3) were sampled on the first day, and only A. tubulata (n = 4) were sampled on the third day. Additionally, samples from all n = 7 X. muta and n = 3 A. tubulata were collected from individuals at depths of 13–16 m, while samples from n = 4 A. tubulata were collected from individuals at slightly deeper depths (18–23 m). The separation of the two species along the x-axis in Figure 2C may be explained by variability in the incurrent DOM caused by a combination of sequential sampling and sampling at different depths. Despite variable compositions of incurrent DOM due to these and other external factors, In/Ex separation is clearly defined along the y-axis for both species in both years (Figures 2B,C).
Statistical analyses were conducted on 2018 and 2019 samples collected from A. tubulata and X. muta, the two HMA species targeted for sampling in both years. Of the features whose abundances differed significantly between incurrent and excurrent samples, most were significantly depleted in excurrent samples (i.e., excurrent-depleted), reflecting uptake by the two HMA species. Excurrent-depleted features outnumbered excurrent-enriched features by an order of magnitude in 2018, and by a factor of three for each species in 2019 (Supplementary Table 2). Another difference between excurrent-depleted and -enriched features was their magnitude of change in raw abundance. Excurrent-depleted features had fold-change reductions of up to 73 and 57 in 2018 and 2019, respectively, while excurrent-enriched features never exceeded an 8-fold increase in abundance. Furthermore, S-plots of OPLS-DA results showed clustering of excurrent-depleted features in the regions of minimum p[1], meaning they were among the most important drivers of In/Ex differences (Supplementary Figure 2). The abundance, fold-change differences, and discriminating influence of excurrent-depleted features underscores their importance as key metabolites whose uptake transformed seawater DOM.
We noted 15 features that were significantly excurrent-depleted by A. tubulata and X. muta and putatively identified in both 2018 and 2019 samples, based on similar m/z and retention times (Supplementary Table 3). These features were also depleted by the third HMA sponge, V. reiswigi, but they were notably not depleted by any of the three LMA species, as shown by plots of In/Ex abundances of two of the 15 features in Figure 3 and plots of the remaining features in Supplementary Figure 3.
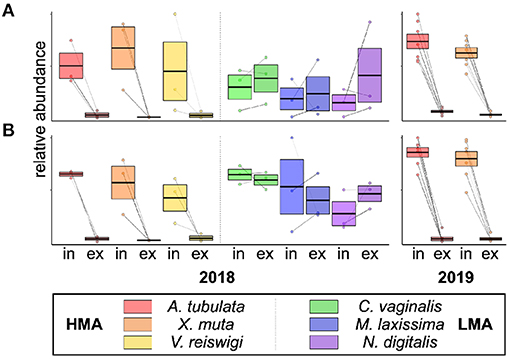
Figure 3. Relative abundances of two of the 15 features that were significantly excurrent-depleted by HMA species and present in samples from both 2018 (n = 3 species−1) and 2019 (n = 7 species−1). Box-plots show the median ± 95% CI for incurrent and excurrent samples of each species, with lines connecting the incurrent and excurrent samples from the same individual. Shown are abundances of feature IDs (A) 12 and (B) 5, and IDs correspond to the rows in Supplementary Table 2. For box-plots of all 15 features, see Supplementary Figure 3.
Feature classifications revealed interesting year-to-year similarities in the excurrent-depleted DOM, in contrast to year-to-year differences in overall DOM. A total of n = 293 features from 2018 samples and n = 359 features from 2019 samples were analyzed and classified using the CANOPUS tool in SIRIUS (Dührkop et al., 2019, 2020). The putative composition of this overall DOM showed high variability between 2018 and 2019. Classifications spanned 10 chemical classes, dominated by organic acids and derivatives (66 and 80% in 2018 and 2019, respectively), organic nitrogen compounds (17 and 7%, respectively), and lipids and lipid-like molecules (5 and 8%, respectively; Supplementary Figures 4, 5). Interestingly, compared to overall DOM, the subset of features that were depleted in the excurrent samples of HMA sponges showed much less variability between 2018 and 2019. The putative composition of this excurrent-depleted DOM (n = 108 features in 2018; n = 65 features in 2019) was dominated (85–90%) by organic acids and derivatives and organic nitrogen compounds, including an almost identical percentage of features putatively classified as amines, (18–19%) despite a 10% reduction of organic nitrogen compounds in the overall DOM from 2018 to 2019. Excurrent-depleted features were also represented by fewer chemical classes, with an absence of compounds putatively classified as hydrocarbons and lipids and lipid-like molecules, classes that comprised 5–8% of the overall DOM from either year (Figure 4, Supplementary Figures 4C, 5C).
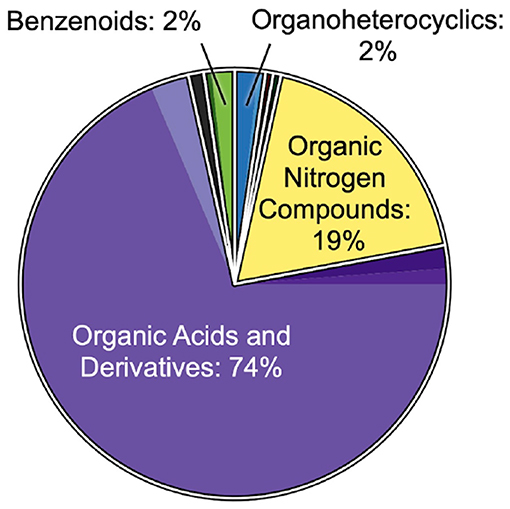
Figure 4. Putative compositions of combined 2018 and 2019 excurrent-depleted features (n = 108 and 65 features, respectively). Classifications are from CANOPUS (Dührkop et al., 2020) and based on the ClassyFire nomenclature (Djoumbou Feunang et al., 2016). The top-level groupings labeled and outlined in white represent superclasses, and the filled color of each superclass is subdivided into shades that correspond to the subclasses listed in Supplementary Figure 4A.
Halogenation was another similarity among excurrent-depleted features. Overall, 6% of mass features from both years were classified as halogenated in UNIFI (n = 93 chlorinated and 40 brominated in 2018; n = 100 chlorinated and 13 brominated in 2019). When visualized on volcano plots, halogenated features clustered disproportionately with excurrent-depleted features and approached a minimum among excurrent-enriched features (Figure 5). Halogenation was indicated for 10–20% of significantly excurrent-depleted features (n = 22 out of 115 in 2018; n = 7 out of 73 in 2019), including six of the 15 features that were excurrent-depleted by HMA sponges and present in samples from both years (Figure 3A, Supplementary Figure 3, Supplementary Table 3). Screening of excurrent-depleted and excurrent-enriched features in SIRIUS and UNIFI did not return any reliable matches to specific metabolites.
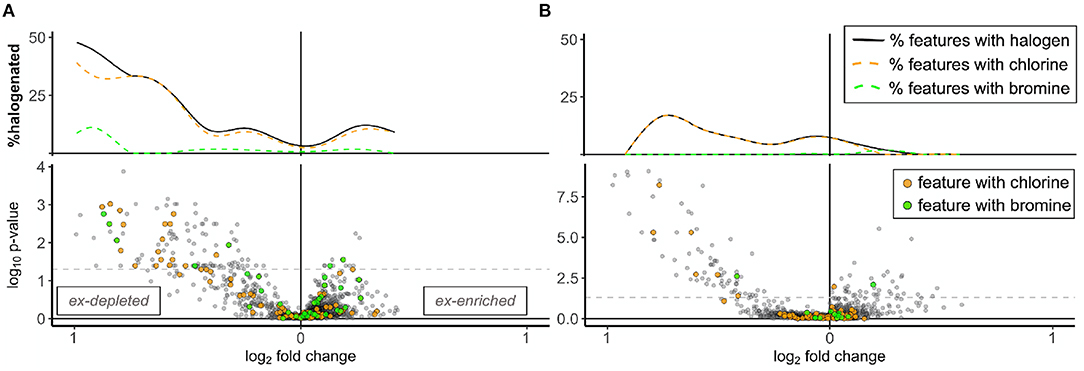
Figure 5. Histograms and volcano plots showing the dispersion of halogenated features in combined A. tubulata and X. muta samples for each year. Panels show samples from (A) 2018 and (B) 2019. Excurrent-depleted features are shown on the left (x <0) and excurrent-enriched features on the right (x >0) of each plot. Dashed gray lines on the volcano plot signify p = 0.05, above which all features were significantly more abundant in incurrent or excurrent samples. Colors denote halogenation, with orange representing chlorines and green representing bromines. Histograms above volcano plots show the percentage of total features determined to be chlorinated (orange dashed line), brominated (green dashed line) and total (solid black line).
Seawater dissolved organic matter (DOM) is a complex mixture of molecules that is constantly modified by biotic and abiotic processes (Carlson and Hansell, 2015; Moran et al., 2016). By pumping large volumes of seawater and taking up dissolved organic carbon-containing compounds (DOC), sponges can also change the composition of seawater DOM (Fiore et al., 2017; Pawlik and McMurray, 2020). The application herein of In/Ex sampling across six common Caribbean sponge species revealed distinct differences in how DOM was altered by sponges with high and low microbial abundances (HMA, LMA). While LMA sponges made negligible changes to seawater DOM, the DOM profile of In/Ex samples from HMA sponges revealed considerable and uniform In/Ex differences driven by mass features that were depleted by the sponge (i.e., excurrent-depleted features). Differences between HMA and LMA sponges and the observation of organohalide uptake by HMA sponges offers new insights into the possible evolutionary origin of complex sponge-microbe symbioses and consequences of sponge DOM uptake on Caribbean coral reefs.
The abundance of excurrent-depleted features suggests that changes to DOM were driven by its removal from seawater, and differences between HMA and LMA sponges implicate symbiotic microbes in this removal. Microbial involvement has been cited to explain differences in carbon and nitrogen metabolism between HMA and LMA sponges. For example, lower δ15N in the tissues of HMA species was attributed to HMA sponges deriving more nitrogen from microbial symbionts than LMA species that rely on external nitrogen (Weisz et al., 2007). Symbiotic microbes have also been linked to DOC uptake, including in similar In/Ex sampling experiments on the same six species as used herein that reported DOC uptake only by HMA and not LMA sponges (Hoer et al., 2018; McMurray et al., 2018). There are conflicting reports of DOC uptake by LMA sponges (de Goeij et al., 2008; Mueller et al., 2014; Morganti et al., 2017), and this could be possible if DOC can be taken up and utilized by sponge cells. Direct assimilation of DOC by host sponge cells has been observed in a number of recent studies that used innovative mass spectral imaging techniques to visualize DOC uptake on a cellular level (Achlatis et al., 2019; Rix et al., 2020; Hudspith et al., 2021). The traditional understanding favors prokaryotes as the main agents of DOC uptake; while it is a rare food source for eukaryotes, DOC processing by free-living prokaryotic microbes is globally ubiquitous (de Goeij et al., 2008; Kujawinski, 2011; Jiao et al., 2014). Like their free-living counterparts, prokaryotic sponge symbionts may be able to process DOC and pass nutritional products to their host. Transcriptomic analysis of the holobiont of X. muta demonstrated that amino acids and vitamins could be anabolized by the microbiome but only catabolized by the sponge host, suggesting that sponge symbionts biosynthesize materials needed by their host (Fiore et al., 2015). An abundance of membrane transporters found throughout the microbiome genome may help move resources between symbiont and host (Fan et al., 2012).
The HMA/LMA differences observed herein may be due to greater influence by HMA sponge symbionts, which are more abundant and diverse than LMA sponge symbionts (Hentschel et al., 2003; Moitinho-Silva et al., 2017). Using a combination of in situ sampling and manipulative experiments, Rix et al. (2020) quantified the contribution of microbial symbionts to sponge heterotrophy, and found that the microbial symbionts of HMA sponge Aplysina aerophoba were responsible for 65–89% of DOM uptake, while the microbial symbionts of LMA sponge Dysidea alvara contributed <5% to DOM uptake; this variability in microbial contribution paralleled the order of magnitude differences in microbial abundance between the HMA and LMA species. Another recent study found no relationship between DOC uptake and overall microbial community structure (Gantt et al., 2019). This relationship may not be apparent if sponges have flexible carbon metabolisms and opportunistically feed on DOC (Pawlik et al., 2018), or if certain symbiont subgroups contribute disproportionately to the carbon metabolism of their host. Some subgroups of sponge symbionts can have an outsized influence on host sponge carbon uptake. For example, on deep-sea methane seeps, methane oxidizing (MOX) bacteria found in two distantly-related sponges suggested convergent acquisition of these methanotrophs that gave their host access to an otherwise inaccessible carbon source (Rubin-Blum et al., 2019). Unlike methane, DOM is a complex mixture, but studies of free-living microbes have shown that processing of a diversity of compounds in DOM does not require diverse microbial assemblages, and even single strains can take up as much labile DOM as a natural assemblage (Pedler et al., 2014). Disproportionate contribution to uptake of DOM compounds by symbiont subgroups could explain the HMA/LMA differences observed herein and elsewhere (Hoer et al., 2018; McMurray et al., 2018), despite no apparent link to microbial community structure on the whole (Gantt et al., 2019).
The differences that we report between HMA and LMA sponges may be due to factors other than microbially-mediated uptake. In general, HMA sponges pump more slowly than LMA sponges, which may allow more of the DOM to be processed during the extra time spent passing through the sponge aquiferous system (McMurray et al., 2014; Hoer et al., 2018). However, if pumping rates were the main drivers of In/Ex differences, we would expect to see In/Ex differences and DOM uptake by LMA Mycale laxissima, which is an exceptionally slow-pumping species (Gantt et al., 2019), yet our data for M. laxissima samples resembled that of the other LMA species. Inconsistencies may also arise when DOC is defined by seawater filtration methods that use filters with different pore sizes. In the present study, DOM was operationally defined as what passed through the 0.2 μm filter, but apparent DOC uptake by LMA sponges may be observed when using filters with larger pore sizes (0.7 μm) that allow the passage of more filtrate that is not truly dissolved (e.g., picoplankton, colloids) and that may be amenable to uptake by particle-feeding sponges (de Goeij et al., 2008). For example, a recent study that reported direct DOM consumption by sponge cells cautioned that this organic matter may have been a part of the colloidal fraction of the 0.7 μm DOM (Rix et al., 2020). Future work is needed to determine what types of DOM are taken up by host sponge cells compared to their microbial symbionts; mass spectrometric based approaches offer much promise to this end, especially the spatial information provided by mass spectral imaging (e.g., NanoSIMS; Rix et al., 2020) and molecular-level resolution of untargeted metabolomics and data analysis techniques used herein.
The present study used a sampling technique that was similar to Fiore et al. (2017), which also concluded that sponges changed the composition of seawater DOM. Our findings that only HMA species changed seawater DOM is at odds with Fiore et al. (2017) which reported a similar signature of sponge processing in samples from both of the two species analyzed in their study: HMA Ircinia campana and LMA Spheciospongia vesparium. These conflicting results may be due in part to the complicated status of S. vesparium; this species has been recognized as an exception to the traditional HMA/LMA dichotomy owing to low pumping rates characteristic of HMA sponges (Weisz et al., 2008) and low symbiont abundance characteristic of LMA sponges (Poppell et al., 2014). Although not an ideal representative of the LMA group, the novelty of S. vesparium could be useful for future studies on interspecific differences across the HMA/LMA spectrum, for example in order to decouple the effect of symbiont abundance and pumping rates, or to narrow down the symbiont subgroups that may be responsible for DOM alterations observed by Fiore et al. (2017).
Halogenated organic compounds are abundant in marine ecosystems and derived from several sources: as natural products made by many types of organisms (e.g., bacteria, cyanobacteria, macroalgae, sponges, cnidarians, polychaetes), as environmental contaminants, and as products of abiotic reactions (e.g., volcanic activity, photolysis) (Ahn et al., 2009; Wiseman et al., 2011; Leri et al., 2015; Liu et al., 2020). The abundance of organohalides that were excurrent-depleted in this study was unexpected. We did not directly test whether DOM alterations, including depletion of organohalides, were due to direct metabolism either by sponge microbial symbionts or host cells. However, if organohalide depletion was due to direct metabolism and not some other process (e.g., adsorption), this further implicates sponge symbionts in DOM uptake because sponge cells lack the necessary dehalogenase and halogenase enzymes that have been described for sponge-associated microbes. For example, metagenomics-based analysis of the Dysidea spp. holobiont revealed gene clusters for biosynthesis of polybrominated diphenyl ethers (PBDEs) that were present in cyanobacterial symbionts but distinctly missing from the sponge host (Agarwal et al., 2017). Metaproteomics-based analysis of the Aplysina aerophoba holobiont also revealed transport and degradation of halogenated compounds to be common active processes of the microbiome of the sponge (Chaib De Mares et al., 2018). There are many strategies used by microbes to remove halogens from organic compounds for respiratory and metabolic purposes, including oxidation, substitution, hydration, and reduction. Reductive dehalogenation is the most prevalent and well-studied of these mechanisms, and it is used by many organohalide respiring bacteria (OHRB) to liberate a halogen and use it as an electron acceptor in respiration (Häggblom and Bossert, 2004; Atashgahi et al., 2018). Sponge-associated OHRB were first reported in A. aerophoba (Ahn et al., 2003, 2009), and have since been found in a number of sponge species, including from the Caribbean (Horna-Gray, 2015). The microbiome of six species of Aplysina from the Caribbean and Mediterranean were also recently shown to contain a variety of halogenases and dehalogenases, and the diversity of these enzymes was positively correlated with sponge microbial diversity (Gutleben et al., 2019).
The prevalence of OHRB and other sponge symbionts capable of halogenation and dehalogenation suggests some benefit to the sponge host. These symbionts are likely involved in the production of halogenated secondary metabolites that can be concentrated in the tissues of sponges (Turon et al., 2000). Sacristán-Soriano et al. (2011) reported that concentrations of brominated alkaloids increased with increasing abundance of Chloroflexi symbionts, and proposed that the sponge host and microbe may work together to produce the metabolites, with the former producing the inactive precursor, and the latter activating it through enzymatic halogenation. The sponge holobiont may also benefit from the product that remains after dehalogenation; removal of halogens can eliminate the toxicity of metabolites and make them available as a food source (Hug et al., 2013). In marine environments where free-living microbes rapidly consume the most labile of DOC compounds (Moran et al., 2016), dehalogenation could provide a reliable backup food source. This food source may either be used by the dehalogenating symbiont itself, or by a mutualistic associate of the dehalogenating symbiont. The former is possible if sponge symbionts perform catabolic reductive dehalogenation for use of the carbon byproduct and not the halogen (Chen et al., 2013). The latter is possible if the dehalogenating sponge symbiont is an OHRB that uses the halogen for respiration and exchanges the carbon byproduct for growth factors as part of a mutualism (Shelton and Tiedje, 1984; Atashgahi et al., 2018). Sponges may take up a range of natural and synthetic organohalides if uptake is mediated by OHRB, a group known for its ability to act on a diversity of compounds using a variety of dehalogenase enzymes (Atashgahi et al., 2018). Many industrial compounds containing halogens (e.g., insecticide DDT, industrial coolant PCB, and flame retardant PBDEs) are harmful to and persistent in the environment, prompting efforts to reduce or eliminate these organic pollutants (Häggblom and Bossert, 2004). OHRB have shown some promise for bioremediation (Wang et al., 2019), and sponges may also represent an all-in-one solution owing to their ability to pump seawater while removing organohalides from it.
A growing concern on Caribbean reefs is the rise of benthic cyanobacterial mats (BCM) made of a consortia of species including Moorea producens (formerly Lyngbya majuscula) and Oscillatoria spp., and known to produce large quantities of DOC (Brocke et al., 2015). Decadal surveys of coral reefs of Curaçao and Bonaire between 1973 and 2013 revealed BCM benthic cover had increased from 0.1 to 22% and corresponded to a smaller but significant increase in sponge benthic cover from 0.5 to 2.3% (de Bakker et al., 2017). Species, such as M. producens are known to synthesize several polychlorinated defensive metabolites, such as barbamide, nordysidenin, and dysidenamide, and these compounds have structures and uncommon trichloromethyl functionalities that closely resemble defensive peptides from the sponge Dysidea herbacea, including dysidenin and the diketopiperazine dihydrodysamide C (König et al., 2006). Herbamide B has even been isolated from both M. producens and D. herbacea (Flatt et al., 2005). Similarities between cyanobacterial and sponge defensive metabolites have been cited as evidence that cyanobacterial symbionts are involved in the biosynthesis of sponge chemical defenses (Flatt et al., 2005; König et al., 2006). Sponge chemical defenses may also resemble cyanobacterial chemical defenses if they come from the same source, for example if sponge symbionts take up metabolites released by nearby BCM and modify and store them in the tissues of their host as a defense against predation. This theory that sponges recycle cyanobacterial metabolites is consistent with our findings that sponges took up organohalides. Additionally, this theory offers a parsimonious explanation for the occurrence of such similar halogenated defensive metabolites in neighboring sponges and cyanobacteria. Exchanges of these halogenated defenses may also explain tandem increases in benthic cover of BCM and sponges (de Bakker et al., 2017), consistent with the hypothesized “vicious circle” in which sponges and benthic autotrophs benefit from the DOC and nitrogenous wastes produced by one another, to the detriment of reef building corals (Pawlik et al., 2016).
Sponge uptake of DOC has been the recent focus of several research groups and the basis for new hypotheses regarding coral reef ecosystem function, such as the sponge loop and the vicious circle (de Goeij et al., 2013; Pawlik et al., 2016). However, whether DOC uptake is common across HMA and LMA sponge species has been the subject of conflicting studies. By sampling 3 each of HMA and LMA species, we found that sponge processing of seawater and uptake of metabolites altered the composition of seawater DOM, but only for HMA, and not LMA, species, suggesting that sponge symbionts were involved in this uptake. LC-MS-based untargeted metabolomics analyses resolved single mass features across a range of polarities and sizes, and associated mass spectral isotope and fragmentation peaks enabled metabolite annotation in UNIFI and SIRIUS and a closer look into the types of DOM being taken up by sponges. Grouping of DOM metabolites based on CANOPUS classifications revealed that compositions of excurrent-depleted DOM were similar across the 2 years that samples were collected. Annotation of halogenated mass features in the DOM revealed that sponges took up organohalides. Moreover, organohalides were over-represented in excurrent-depleted DOM, and the extent of this uptake could be consequential to halide cycling on coral reefs. The uptake of compounds such as organohalides implicate microbial symbionts that have the enzymes needed to process them, and these compounds may eventually be transformed into food or chemical defenses that benefit the sponge host. The uptake and use of this complex chemical mixture may be evidence of the selective forces that led to complex sponge-microbe symbioses, for example if the divergence of HMA and LMA species was facilitated by acquisition of specialized microbial communities that helped early HMA sponges improve competitiveness or overcome food-limitation (Freeman et al., 2020). This specialized strategy may have consequences on present-day reefs, as reef-building corals are replaced by growing populations of sponges whose rapid processing of seawater and complex microbiomes allow them to alter seawater chemistry in ways that may further drive the demise of the corals that build the reefs.
The datasets presented in this study can be found in the MetaboLights repository (https://www.ebi.ac.uk/metabolights), accession numbers MTBLS2199 and MTBLS2200.
LO, WS, SM, and JP designed and performed the research. LO analyzed the data and wrote the paper. All authors edited the paper.
This research was funded by the National Science Foundation (NSF 1558580).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Many thanks to Laura Gaitan Daza, Melissa Dionesotes, and Michael Wooster for assistance in the field, to Ralph Mead for helpful guidance on sample preparation and analysis/interpretation of mass spectrometry data, and the staff of Smithsonian Institution's Carrie Bow Cay Field Station in Belize for logistical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.665789/full#supplementary-material
Achlatis, M., Pernice, M., Green, K., de Goeij, J. M., Guagliardo, P., Kilburn, M. R., et al. (2019). Single-cell visualization indicates direct role of sponge host in uptake of dissolved organic matter. Proc. R. Soc. B Biol. Sci. 286:20192153. doi: 10.1098/rspb.2019.2153
Agarwal, V., Blanton, J. M., Podell, S., Taton, A., Schorn, M. A., Busch, J., et al. (2017). Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 13:537. doi: 10.1038/nchembio.2330
Ahn, Y. B., Kerkhof, L. J., and Häggblom, M. M. (2009). Desulfoluna spongiiphila sp. nov., a dehalogenating bacterium in the Desulfobacteraceae from the marine sponge Aplysina aerophoba. Int. J. Syst. Evol. Microbiol. 59, 2133–2139. doi: 10.1099/ijs.0.005884-0
Ahn, Y. B., Rhee, S. K., Fennell, D. E., Kerkhof, L. J., Hentschel, U., and Häggblom, M. M. (2003). Reductive dehalogenation of brominated phenolic compounds by microorganisms associated with the marine sponge Aplysina aerophoba. Appl. Environ. Microbiol. 69, 4159–4166. doi: 10.1128/AEM.69.7.4159-4166.2003
Atashgahi, S., Haggblom, M. M., and Smidt, H. (2018). Organohalide respiration in pristine environments: implications for the natural halogen cycle. Environ. Microbiol. 20, 934–948. doi: 10.1111/1462-2920.14016
Bell, J. J., Davy, S. K., Jones, T., Taylor, M. W., and Webster, N. S. (2013). Could some coral reefs become sponge reefs as our climate changes? Glob. Change Biol. 19, 2613–2624. doi: 10.1111/gcb.12212
Böcker, S., and Dührkop, K. (2016). Fragmentation trees reloaded. J. Cheminform. 8:5. doi: 10.1186/s13321-016-0116-8
Böcker, S., Letzel, M. C., Lipták, Z., and Pervukhin, A. (2009). SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics 25, 218–224. doi: 10.1093/bioinformatics/btn603
Brocke, H. J., Wenzhoefer, F., de Beer, D., Mueller, B., van Duyl, F. C., and Nugues, M. M. (2015). High dissolved organic carbon release by benthic cyanobacterial mats in a Caribbean reef ecosystem. Sci. Rep. 5:8852. doi: 10.1038/srep08852
Carlson, C. A., and Hansell, D. A. (2015). “DOM sources, sinks, reactivity, and budgets,” in Biogeochemistry of Marine Dissolved Organic Matter, eds C. A. Carlson and D. A. Hansell (New York, NY: Elsevier Inc.), 65–126. doi: 10.1016/B978-0-12-405940-5.00003-0
Chaib De Mares, M., Jiménez, D. J., Palladino, G., Gutleben, J., Lebrun, L. A., Muller, E. E. L., et al. (2018). Expressed protein profile of a Tectomicrobium and other microbial symbionts in the marine sponge Aplysina aerophoba as evidenced by metaproteomics. Sci. Rep. 8:11795. doi: 10.1038/s41598-018-30134-0
Chambers, M. C., Maclean, B., Burke, R., Amodei, D., Ruderman, D. L., Neumann, S., et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920. doi: 10.1038/nbt.2377
Chen, K., Huang, L., Xu, C., Liu, X., He, J., Zinder, S. H., et al. (2013). Molecular characterization of the enzymes involved in the degradation of a brominated aromatic herbicide. Mol. Microbiol. 89, 1121–1139. doi: 10.1111/mmi.12332
de Bakker, D. M., van Duyl, F. C., Bak, R. P. M., Nugues, M. M., Nieuwland, G., and Meesters, E. H. (2017). 40 years of benthic community change on the Caribbean reefs of Curaçao and Bonaire: the rise of slimy cyanobacterial mats. Coral Reefs 36, 355–367. doi: 10.1007/s00338-016-1534-9
de Goeij, J. M., Moodley, L., Houtekamer, M., Carballeira, N. M., and van Duyl, F. C. (2008). Tracing 13C-enriched dissolved and particulate organic carbon in the bacteria-containing coral reef sponge Halisarca caerulea: evidence for DOM feeding. Limnol. Oceanogr. 53, 1376–1386. doi: 10.4319/lo.2008.53.4.1376
de Goeij, J. M., van Oevelen, D., Vermeij, M. J. A., Osinga, R., Middelburg, J. J., de Goeij, A. F. P. M., et al. (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342, 108–110. doi: 10.1126/science.1241981
DeLong, E. F. (2009). The microbial ocean from genomes to biomes. Nature 459, 200–206. doi: 10.1038/nature08059
Dittmar, T., Koch, B., Hertkorn, N., and Kattner, G. (2008). A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 6, 230–235. doi: 10.4319/lom.2008.6.230
Djoumbou Feunang, Y., Eisner, R., Knox, C., Chepelev, L., Hastings, J., Owen, G., et al. (2016). ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8:61. doi: 10.1186/s13321-016-0174-y
Dührkop, K., Fleischauer, M., Ludwig, M., Aksenov, A. A., Melnik, A. V., Meusel, M., et al. (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302. doi: 10.1038/s41592-019-0344-8
Dührkop, K., Nothias, L. F., Fleischauer, M., Reher, R., Ludwig, M., Hoffmann, M. A., et al. (2020). Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 39, 462–471. doi: 10.1038/s41587-020-0740-8
Dührkop, K., Shen, H., Meusel, M., Rousu, J., and Böcker, S. (2015). Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. U.S.A. 112, 12580–12585. doi: 10.1073/pnas.1509788112
Erwin, P. M., Pita, L., López-Legentil, S., and Turon, X. (2012). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368. doi: 10.1128/AEM.02035-12
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 109, E1878–E1887. doi: 10.1073/pnas.1203287109
Fiore, C. L., Freeman, C. J., and Kujawinski, E. B. (2017). Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ 5:e2870. doi: 10.7717/peerj.2870
Fiore, C. L., Labrie, M., Jarett, J. K., and Lesser, M. P. (2015). Transcriptional activity of the giant barrel sponge, Xestospongia muta holobiont: molecular evidence for metabolic interchange. Front. Microbiol. 6:364. doi: 10.3389/fmicb.2015.00364
Flatt, P. M., Gautschi, J. T., Thacker, R. W., Musafija-Girt, M., Crews, P., and Gerwick, W. H. (2005). Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar. Biol. 147, 761–774. doi: 10.1007/s00227-005-1614-9
Freeman, C. J., Easson, C. G., Matterson, K. O., Thacker, R. W., Baker, D. M., and Paul, V. J. (2020). Microbial symbionts and ecological divergence of Caribbean sponges: a new perspective on an ancient association. ISME J. 14, 1571–1583. doi: 10.1038/s41396-020-0625-3
Gantt, S. E., McMurray, S. E., Stubler, A. D., Finelli, C. M., Pawlik, J. R., and Erwin, P. M. (2019). Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 7:124. doi: 10.1186/s40168-019-0739-x
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A., and Watkinson, A. R. (2003). Long-term region-wide declines in Caribbean corals. Science 301, 958–960. doi: 10.1126/science.1086050
Gaude, E., Chignola, F., Spiliotopoulos, D., Spitaleri, A., Ghitti, M., Garcia-Manteiga, J. M., Musco, S. M., and Giovanna (2013). muma, an R package for metabolomics univariate and multivariate statistical analysis. Curr. Metabol. 1, 180–189. doi: 10.2174/2213235X11301020005
Gloeckner, V., Wehrl, M., Moitinho-Silva, L., Gernert, C., Schupp, P., Pawlik, J. R., et al. (2014). The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol. Bull. 227, 78–88. doi: 10.1086/BBLv227n1p78
Gochfeld, D. J., Olson, J. B., Chaves-Fonnegra, A., Smith, T. B., Ennis, R. S., and Brandt, M. E. (2020). Impacts of hurricanes Irma and Maria on coral reef sponge communities in St. Thomas, U.S. Virgin Islands. Estuar. Coasts 1, 1–13. doi: 10.1007/s12237-020-00694-4
Goshawk, J., Eatough, D., and Wood, M. (2015). Componentization Following 3D-Peak Detection in the UNIFI Scientific Information System. Milford, MA: Waters Corporation.
Gutleben, J., Koehorst, J. J., McPherson, K., Pomponi, S., Wijffels, R. H., Smidt, H., et al. (2019). Diversity of tryptophan halogenases in sponges of the genus Aplysina. FEMS Microbiol. Ecol. 95:fiz108. doi: 10.1093/femsec/fiz108
Haas, A. F., Nelson, C. E., Wegley Kelly, L., Carlson, C. A., Rohwer, F., Leichter, J. J., et al. (2011). Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS ONE 6:e27973. doi: 10.1371/journal.pone.0027973
Häggblom, M. M., and Bossert, I. D. (2004). “Halogenated organic compounds–a global perspective,” in Dehalogenation, eds M. M. Häggblom and I. D. Bossert (Norwell, MA: Kluwer Academic Publishers), 3–29. doi: 10.1007/0-306-48011-5_1
Haug, K., Cochrane, K., Nainala, V. C., Williams, M., Chang, J., Jayaseelan, K. V., et al. (2020). MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48, D440–D444. doi: 10.1093/nar/gkz1019
Hentschel, U., Fieseler, L., Wehrl, M., Gernert, C., Steinert, M., Hacker, J., et al. (2003). “Microbial diversity of marine sponges,” in Sponges (Porifera), W. E. G. Müller (Berlin, Heidelberg: Springer), 59–88. doi: 10.1007/978-3-642-55519-0_3
Hoer, D. R., Gibson, P. J., Tommerdahl, J. P., Lindquist, N. L., and Martens, C. S. (2018). Consumption of dissolved organic carbon by Caribbean reef sponges. Limnol. Oceanogr. 63, 337–351. doi: 10.1002/lno.10634
Horna-Gray, I. D. C. (2015). Sponge-associated dehalogenating microorganisms and isotope analysis of their dehalogenation of brominated phenols (Ph.D. thesis), Rutgers University, New Brunswick, NJ, United States.
Hudspith, M., Rix, L., Achlatis, M., Bougoure, J., Guagliardo, P., Clode, P. L., et al. (2021). Subcellular view of host–microbiome nutrient exchange in sponges: insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 9:44. doi: 10.1186/s40168-020-00984-w
Hug, L. A., Maphosa, F., Leys, D., Löffler, F. E., Smidt, H., Edwards, E. A., et al. (2013). Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. B Biol. Sci. 368:20120322. doi: 10.1098/rstb.2012.0322
Jiao, N., Robinson, C., Azam, F., Thomas, H., Baltar, F., Dang, H., et al. (2014). Mechanisms of microbial carbon sequestration in the ocean–future research directions. Biogeosci. Discuss. 11, 7931–7990. doi: 10.5194/bg-11-5285-2014
Kido Soule, M. C., Longnecker, K., Johnson, W. M., and Kujawinski, E. B. (2015). Environmental metabolomics: analytical strategies. Mar. Chem. 177, 374–387. doi: 10.1016/j.marchem.2015.06.029
König, G. M., Kehraus, S., Seibert, S. F., Abdel-Lateff, A., and Müller, D. (2006). Natural products from marine organisms and their associated microbes. ChemBioChem 7, 229–238. doi: 10.1002/cbic.200500087
Kuhl, C., Tautenhahn, R., Böttcher, C., Larson, T. R., and Neumann, S. (2012). CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 84, 283–289. doi: 10.1021/ac202450g
Kujawinski, E. B. (2011). The impact of microbial metabolism on marine dissolved organic matter. Annu. Rev. Mar. Sci. 3, 567–599. doi: 10.1146/annurev-marine-120308-081003
Leri, A. C., Mayer, L. M., Thornton, K. R., Northrup, P. A., Dunigan, M. R., Ness, K. J., et al. (2015). A marine sink for chlorine in natural organic matter. Nat. Geosci. 8, 620–624. doi: 10.1038/ngeo2481
Liu, J., Adrian, L., and Häggblom, M. M. (2020). Transcriptomic and proteomic responses of the organohalide-respiring bacterium Desulfoluna spongiiphila to growth with 2,6-dibromophenol as the electron acceptor. Appl. Environ. Microbiol. 86:e02146-19. doi: 10.1128/AEM.02146-19
Loh, T. L., McMurray, S. E., Henkel, T. P., Vicente, J., and Pawlik, J. R. (2015). Indirect effects of overfishing on Caribbean reefs: sponges overgrow reef-building corals. PeerJ 3:e901. doi: 10.7717/peerj.901
Loh, T. L., and Pawlik, J. R. (2014). Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc. Natl. Acad. Sci. U.S.A. 111, 4151–4156. doi: 10.1073/pnas.1321626111
Longnecker, K., Futrelle, J., Coburn, E., Kido Soule, M. C., and Kujawinski, E. B. (2015). Environmental metabolomics: databases and tools for data analysis. Mar. Chem. 177, 366–373. doi: 10.1016/j.marchem.2015.06.012
Ludwig, M., Dührkop, K., and Böcker, S. (2018). Bayesian networks for mass spectrometric metabolite identification via molecular fingerprints. Bioinformatics 34, i333–i340. doi: 10.1093/bioinformatics/bty245
McMurray, S., Stubler, A., Erwin, P., Finelli, C., and Pawlik, J. (2018). A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Series 588, 1–14. doi: 10.3354/meps12466
McMurray, S. E., Finelli, C. M., and Pawlik, J. R. (2015). Population dynamics of giant barrel sponges on Florida coral reefs. J. Exp. Mar. Biol. Ecol. 473, 73–80. doi: 10.1016/j.jembe.2015.08.007
McMurray, S. E., Johnson, Z. I., Hunt, D. E., Pawlik, J. R., and Finelli, C. M. (2016). Selective feeding by the giant barrel sponge enhances foraging efficiency. Limnol. Oceanogr. 61, 1271–1286. doi: 10.1002/lno.10287
McMurray, S. E., Pawlik, J. R., and Finelli, C. M. (2014). Trait-mediated ecosystem impacts: how morphology and size affect pumping rates of the Caribbean giant barrel sponge. Aquat. Biol. 23, 1–13. doi: 10.3354/ab00612
Moitinho-Silva, L., Steinert, G., Nielsen, S., Hardoim, C. C. P., Wu, Y. C., McCormack, G. P., et al. (2017). Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 8:752. doi: 10.3389/fmicb.2017.00752
Moran, M. A., Kujawinski, E. B., Stubbins, A., Fatland, R., Aluwihare, L. I., Buchan, A., et al. (2016). Deciphering ocean carbon in a changing world. Proc. Natl. Acad. Sci. U.S.A. 113, 3143–3151. doi: 10.1073/pnas.1514645113
Morganti, T., Coma, R., Yahel, G., and Ribes, M. (2017). Trophic niche separation that facilitates co-existence of high and low microbial abundance sponges is revealed by in situ study of carbon and nitrogen fluxes: trophic niche separation in sponges. Limnol. Oceanogr. 62, 1963–1983. doi: 10.1002/lno.10546
Mueller, B., Goeij, J. M. d., Vermeij, M. J. A., Mulders, Y., Ent, E. v. d., Ribes, M., et al. (2014). Natural diet of coral-excavating sponges consists mainly of dissolved organic carbon (DOC). PLoS ONE 9:e90152. doi: 10.1371/journal.pone.0090152
Nelson, C. E., Goldberg, S. J., Kelly, L. W., Haas, A. F., Smith, J. E., Rohwer, F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7, 962–979. doi: 10.1038/ismej.2012.161
Pawlik, J. R., Burkepile, D. E., and Thurber, R. V. (2016). A vicious circle? Altered carbon and nutrient cycling may explain the low resilience of Caribbean coral reefs. BioScience 66, 470–476. doi: 10.1093/biosci/biw047
Pawlik, J. R., Loh, T. L., and McMurray, S. E. (2018). A review of bottom-up vs. top-down control of sponges on Caribbean fore-reefs: what's old, what's new, and future directions. PeerJ 6:e4343. doi: 10.7717/peerj.4343
Pawlik, J. R., and McMurray, S. E. (2020). The emerging ecological and biogeochemical importance of sponges on coral reefs. Annu. Rev. Mar. Sci. 12, 315–337. doi: 10.1146/annurev-marine-010419-010807
Pedler, B. E., Aluwihare, L. I., and Azam, F. (2014). Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207. doi: 10.1073/pnas.1401887111
Petras, D., Koester, I., Da Silva, R., Stephens, B. M., Haas, A. F., Nelson, C. E., et al. (2017). High-resolution liquid chromatography tandem mass spectrometry enables large scale molecular characterization of dissolved organic matter. Front. Mar. Sci. 4:405. doi: 10.3389/fmars.2017.00405
Pita, L., López-Legentil, S., and Erwin, P. M. (2013). Biogeography and host fidelity of bacterial communities in Ircinia spp. from the Bahamas. Microb. Ecol. 66, 437–447. doi: 10.1007/s00248-013-0215-2
Pita, L., Rix, L., Slaby, B. M., Franke, A., and Hentschel, U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46. doi: 10.1186/s40168-018-0428-1
Poppell, E., Weisz, J., Spicer, L., Massaro, A., Hill, A., and Hill, M. (2014). Sponge heterotrophic capacity and bacterial community structure in high- and low-microbial abundance sponges. Mar. Ecol. 35, 414–424. doi: 10.1111/maec.12098
Prince, J. T., and Marcotte, E. M. (2006). Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal. Chem. 78, 6140–6152. doi: 10.1021/ac0605344
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Core Team.
Reiswig, H. (1974). Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14, 231–249. doi: 10.1016/0022-0981(74)90005-7
Repeta, D. J. (2015). “Chemical characterization and cycling of dissolved organic matter,” in Biogeochemistry of Marine Dissolved Organic Matter, eds C. A. Carlson and D. A. Hansell (New York, NY: Elsevier Inc), 21–63. doi: 10.1016/B978-0-12-405940-5.00002-9
Rix, L., de Goeij, J. M., van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2017). Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct. Ecol. 31, 778–789. doi: 10.1111/1365-2435.12758
Rix, L., Ribes, M., Coma, R., Jahn, M. T., de Goeij, J. M., van Oevelen, D., et al. (2020). Heterotrophy in the earliest gut: a single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 14, 2554–2567. doi: 10.1038/s41396-020-0706-3
Rubin-Blum, M., Antony, C. P., Sayavedra, L., Martínez-Pérez, C., Birgel, D., Peckmann, J., et al. (2019). Fueled by methane: deep-sea sponges from asphalt seeps gain their nutrition from methane-oxidizing symbionts. ISME J. 13, 1209–1225. doi: 10.1038/s41396-019-0346-7
Sacristán-Soriano, O., Banaigs, B., Casamayor, E. O., and Becerro, M. A. (2011). Exploring the links between natural products and bacterial assemblages in the sponge Aplysina aerophoba. Appl. Environ. Microbiol. 77, 862–870. doi: 10.1128/AEM.00100-10
Shelton, D., and Tiedje, J. (1984). Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl. Environ. Microbiol. 48, 840–848. doi: 10.1128/AEM.48.4.840-848.1984
Smith, C. A., Want, E. J., O'Maille, G., Abagyan, R., and Siuzdak, G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. doi: 10.1021/ac051437y
Tautenhahn, R., Böttcher, C., and Neumann, S. (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9:504. doi: 10.1186/1471-2105-9-504
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/MMBR.00040-06
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Turon, X., Becerro, M. A., and Uriz, M. J. (2000). Distribution of brominated compounds within the sponge Aplysina aerophoba: coupling of X-ray microanalysis with cryofixation techniques. Cell Tissue Res. 301, 311–322. doi: 10.1007/s004410000233
van Duyl, F. C., Moodley, L., Nieuwland, G., van Ijzerloo, L., van Soest, R. W., Houtekamer, M., et al. (2011). Coral cavity sponges depend on reef-derived food resources: stable isotope and fatty acid constraints. Mar. Biol. 158, 1653–1666. doi: 10.1007/s00227-011-1681-z
Wang, S., He, J., Shen, C., and Manefield, M. J. (2019). Editorial: Organohalide respiration: new findings in metabolic mechanisms and bioremediation applications. Front. Microbiol. 10:526. doi: 10.3389/fmicb.2019.00526
Weisz, J. B., Hentschel, U., Lindquist, N., and Martens, C. S. (2007). Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar. Biol. 152, 475–483. doi: 10.1007/s00227-007-0708-y
Weisz, J. B., Lindquist, N., and Martens, C. S. (2008). Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155, 367–376. doi: 10.1007/s00442-007-0910-0
Wiseman, S. B., Wan, Y., Chang, H., Zhang, X., Hecker, M., Jones, P. D., et al. (2011). Polybrominated diphenyl ethers and their hydroxylated/methoxylated analogs: environmental sources, metabolic relationships, and relative toxicities. Mar. Pollut. Bull. 63, 179–188. doi: 10.1016/j.marpolbul.2011.02.008
Wooster, M. K., McMurray, S. E., Pawlik, J. R., Morán, X. A. G., and Berumen, M. L. (2019). Feeding and respiration by giant barrel sponges across a gradient of food abundance in the Red Sea. Limnol. Oceanogr. 64, 1790–1801. doi: 10.1002/lno.11151
Wörheide, G., Solé-Cava, A. M., and Hooper, J. N. A. (2005). Biodiversity, molecular ecology and phylogeography of marine sponges: patterns, implications and outlooks. Integr. Compar. Biol. 45, 377–385. doi: 10.1093/icb/45.2.377
Worley, B., and Powers, R. (2016). PCA as a practical indicator of OPLS-DA model reliability. Curr. Metabol. 4, 97–103. doi: 10.2174/2213235X04666160613122429
Yahel, G., Marie, D., and Genin, A. (2005). InEx—a direct in situ method to measure filtration rates, nutrition, and metabolism of active suspension feeders. Limnol. Oceanogr. 3, 46–58. doi: 10.4319/lom.2005.3.46
Keywords: Porifera, dissolved organic matter, metabolomics, chemical defense, symbiosis, coral reef
Citation: Olinger LK, Strangman WK, McMurray SE and Pawlik JR (2021) Sponges With Microbial Symbionts Transform Dissolved Organic Matter and Take Up Organohalides. Front. Mar. Sci. 8:665789. doi: 10.3389/fmars.2021.665789
Received: 08 February 2021; Accepted: 21 April 2021;
Published: 20 May 2021.
Edited by:
Wei Jiang, Guangxi University, ChinaReviewed by:
Cristiane Cassiolato Pires Hardoim, São Paulo State University, BrazilCopyright © 2021 Olinger, Strangman, McMurray and Pawlik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren K. Olinger, bC5vbGluZ2VyMTI5OTNAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.