- 1School of Biological, Earth and Environmental Sciences, University of New South Wales, Sydney, NSW, Australia
- 2School of Environmental and Life Sciences, The University of Newcastle, Ourimbah, NSW, Australia
Coral bleaching events in the marine environment are now occurring globally, and the frequency and severity of these events are increasing. Critically, these events can cause the symbiosis between Symbiodiniaceae and their coral hosts to break down, but how the microbial community within the coral responds to bleaching is still equivocal. We investigated the impact of thermal stress exposure on the meta-organism responses of the generalist scleractinian coral species Pocillopora damicornis. Using mesocosms to recreate warming scenarios previously observed at Heron Island, we show that P. damicornis symbiont densities and photophysiological parameters declined at a similar rate under thermal stress regardless of the length of pre-bleaching thermal stress, defined here as temperatures above the monthly maximum mean (MMM) for Heron Island but below the local bleaching threshold (MMM + 2°C). However, we find that the P. damicornis microbiome remains stable over time regardless of the degree of thermal stress and the accumulation of pre-bleaching thermal stress. Our study therefore suggests that while P. damicornis is physiologically impacted by bleaching temperatures, the microbial community identified through 16S rRNA sequencing remains unchanged at the ASV level throughout bleaching. Understanding the capacity of a generalist species to withstand bleaching events is imperative to characterizing what coral species will exist on coral reefs following disturbances, as it has been suggested that the success of environmental generalist species may simplify community structure and lead to changes in biodiversity following environmental disturbance.
Introduction
In the marine environment, rising sea surface temperatures (SST) have been linked to the longest and most severe coral bleaching events on record (Hughes et al., 2017, 2018; Eakin et al., 2019; Sully et al., 2019), with reductions in the live coral cover of >98% following bleaching events (Vargas-Ángel et al., 2019). Bleaching affects the coral meta-organism or holobiont, considered to be the Symbiodiniaceae, bacteria, archaea, viruses, and other microscopic eukaryotes and prokaryotes living in the coral host in addition to the cnidarian animal (Rohwer et al., 2002; Kellogg, 2004; Rosenberg et al., 2007; Bosch and McFall-Ngai, 2011; Blackall et al., 2015; Ainsworth et al., 2016). While these micro-organisms are generally considered to be fundamentally important for the survival of the coral animal, with functional roles that may include nutrient provisioning, nitrogen fixation, and prevention of infection by pathogens (Rosenberg et al., 2007; Lema et al., 2012; Bourne et al., 2016; Peixoto et al., 2017), rising SST and the resultant bleaching events can cause the symbiosis between Symbiodiniaceae and their coral hosts to break down (Hoegh-Guldberg and Smith, 1989). This can result in a reduction of dinoflagellate cell densities, photosynthetic pigments, and carbon shared with coral hosts (Brown, 1997; Warner et al., 1999; Fitt et al., 2000). Nonetheless, the biological mechanisms of coral response to bleaching are equivocal and have been suggested to be driven by host plasticity (Bellantuono et al., 2012; Palumbi et al., 2014), host genotype (Dziedzic et al., 2019; Drury and Lirman, 2021), symbiont type (Berkelmans and van Oppen, 2006; Sampayo et al., 2008; Howells et al., 2012), bacterial community dynamics (Ziegler et al., 2017), or a combination of the above.
We therefore examined the coral physiology and microbiome responses to bleaching in Pocillopora damicornis. P. damicornis, one of the most widespread and generalist species of scleractinian coral in the pan-Pacific, has historically been characterized as a “loser” in terms of bleaching susceptibility (Harriott, 1985; Loya et al., 2001; van Woesik et al., 2011), with between 10 and 50% of Pocillopora spp. bleaching in the 2016 mass bleaching event on the Great Barrier Reef (Great Barrier Reef Marine Park Authority, 2017). However, bleaching susceptibility of P. damicornis and genetically distinct but morphometrically similar species within its species complex (Schmidt-Roach et al., 2014) can differ by latitude; for example, on the Great Barrier Reef, bleaching thresholds vary across the 6 degrees of latitude (Ulstrup et al., 2006). In addition, a recent study of P. damicornis colonies from Djibouti, French Polynesia, New Caledonia, and Taiwan found that host haplotype and thermal regime were stronger drivers of Symbiodinium and bacterial assemblages than geographic origin, further emphasizing the role of P. damicornis as an environmental generalist (Brener-Raffalli et al., 2018). In addition, corals in the genus Pocillopora have been found to make up 84% of the total recovery on the reef following disturbance, despite being reported as thermally susceptible species (Pérez-Rosales et al., 2021).
Although P. damicornis is well characterized as a thermally sensitive coral species, the aforementioned studies and the cosmopolitan distribution of P. damicornis suggest that changes in P. damicornis physiology in response to bleaching temperatures could be related to local environmental conditions. The well-studied reproductive biology, feeding ability, and widespread availability of P. damicornis (Harriott, 1983; Harii et al., 2002) have also made it a low-cost and successful target for coral restoration efforts on degraded reefs in the Tropical Eastern Pacific (Yeemin et al., 2006; Toh et al., 2013; Lizcano-Sandoval et al., 2018; Ishida-Castañeda et al., 2020). Nonetheless, evidence related to how coral microorganisms function and contribute to the success of P. damicornis corals under future ocean conditions, especially in response to the rising frequency and severity of coral bleaching, is still equivocal. Therefore, the widespread distribution of P. damicornis and potential use as a restoration target make it an ideal study species for disentangling the response of the coral meta-organism.
While P. damicornis has been extensively characterized as a bleaching-susceptible and thermally sensitive coral species, the role of the coral microbiome in bleaching is still unclear. It has been shown that the microbiome of reef corals differs over thermally variable habitats (Ziegler et al., 2017), and there is potentially selection for beneficial bacterial taxa that are important to sustaining coral health under warming thermal regimes (Osman et al., 2020). However, thermal stress has also been linked to shifts in the native microbial community that may have a negative impact on the coral host, e.g., dysbiosis (Petersen and Round, 2014). A dysbiotic state has been described as one where there is a change to bacterial community structure through an increase in opportunistic pathogens (Bourne et al., 2008; Mouchka et al., 2010), such as higher proportions of Vibrio and Acidobacteria in bleached corals than healthy corals (Mouchka et al., 2010). Higher proportions of opportunistic pathogens can shift the community toward an unhealthy composition like that found in association with diseased corals (Rosenberg et al., 2009; Vega Thurber et al., 2009; Li et al., 2020). Dysbiosis can also be defined by individuals that vary more in microbial community composition, e.g., increased beta diversity (McDevitt-Irwin et al., 2017; Zaneveld et al., 2017), with heat stress producing stochastic changes in the bacterial microbiome (Zaneveld et al., 2016). Accordingly, a bacterial community with a non-significant amount of change has been suggested to modulate the response of corals to thermal stress. Examples include corals from highly variable warm environments bleaching minimally and maintaining their original bacterial community composition throughout heat stress (Ziegler et al., 2017), Turbinaria reniformis maintaining a diverse and stable (e.g., not changing from original community composition prior to stress) microbiome throughout heat stress with minimal physiological decline (Grottoli et al., 2018), and low variation in bacterial diversity within individual colonies of Pocillopora acuta showing no visible signs of bleaching during the 2016 bleaching event on the GBR (Epstein et al., 2019). However, the relationship between changes, or a lack thereof, in the community structure of the bacterial microbiome and host physiology during bleaching events requires further research. Findings from these studies further highlight the complexity and importance of comprehensive responses of the coral host, algal symbionts, and associated bacteria to heat stress.
Given the widespread nature of P. damicornis and its potential for use in coral restoration efforts, it is important to determine if these generalist species have the capacity to acclimate to or overcome future environmental conditions. The present study investigated the impact of summertime sea surface temperature scenarios on P. damicornis physiological and microbial community response by exposing colonies of P. damicornis to the predominant summertime SST trajectory recorded on the GBR during bleaching events at Heron Island, where a sustained sub-bleaching temperature pulse (∼32°C) precedes bleaching conditions (∼34°C), to better understand how the community structure of the P. damicornis bacterial microbiome corresponds with changes in the coral physiology in response to thermal stress.
Materials and Methods
Coral Collection
A total of 18 colonies of the scleractinian coral P. damicornis were collected from the shallow fringing reef flat (1–2 m depth) on the southern side of Heron Island, Australia (23.4423°S, 151.9148°E). Colonies were collected from >3 m apart and separated by distinct sand patches to ensure that colonies were not clonal (Permits: G17/39039.1, G19/41974.1). A temperature logger was also deployed at the site of collection to record in situ reef temperatures throughout the experiment (HOBO MX-2201, Onset, United States). Immediately after collection, colonies were transported to Heron Island Research Station and divided equally between two outdoor flow-through raceway tanks (1000 L) exposed to ambient light and seawater temperatures. Colonies were tagged and left in the flow-through tanks to acclimate for 11 days prior to the start of the experiment.
Experimental Conditions
All four 1000-L flow-through tanks were filled with sand-filtered ambient seawater pumped in from 1 to 3 m depth on Heron Island reef flat. Following acclimation, six colonies were allocated randomly to two heating regimes replicating two durations of SST pre-stress periods prior to thermal stress events. Broadly, the temperature in the heat treatment tanks was increased daily by ∼ 0.5–1°C/day until reaching a target temperature and then maintained at that temperature, with an overnight relaxation in SST of a similar amplitude between the daily minimum and peak maximum temperatures to what was observed for summertime SST on the reef (∼ ± 3−4°C) and in ambient conditions (Figure 1). The sustained sub-bleaching stress period tested here re-creates a temperature stress trajectory that is characteristic of 75% of past thermal stress events on the GBR (n = 277, Ainsworth et al., 2016). Specifically, heat trajectories were experimentally created as follows:
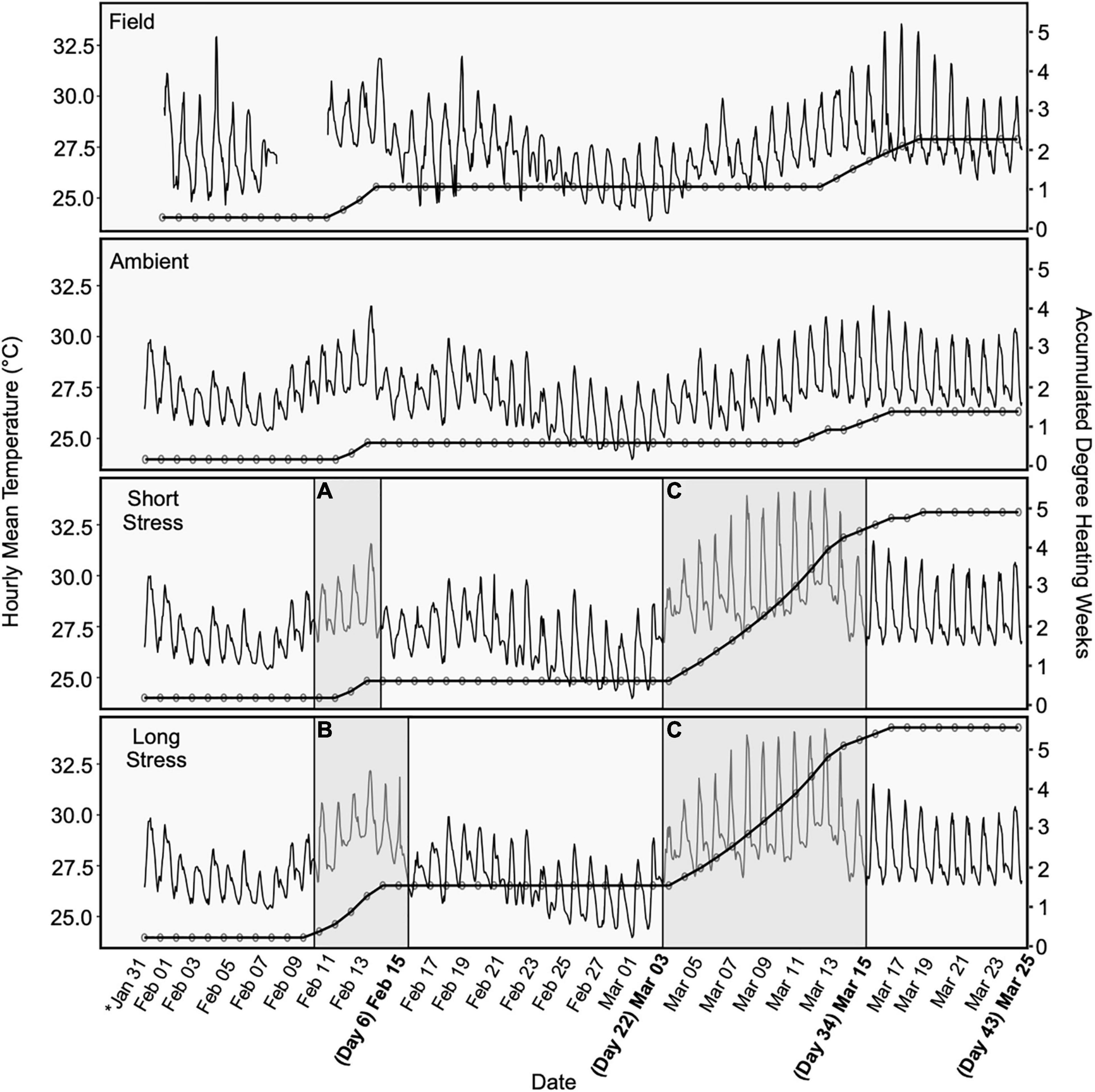
Figure 1. Mean temperature (averaged by hour) for each warming trajectory, overlain with accumulated degree heating weeks (DHWs). Periods of warming are shaded showing initial sub-lethal temperature increases for (A) the short stress scenario (4 days) and (B) the long stress scenario (6 days), and an increase to bleaching temperatures for (C) both scenarios. Bold dates represent sample timepoints. DHWs continued to accumulate following the end of the warming experiment, due to above-average ambient/in situ seawater temperatures. Field temperature data for 9–11 Feb are missing due to logger malfunction.
(1) Short stress: Colonies were exposed to a gradual stepwise increase in peak daytime seawater temperature to ∼2°C below the thermal bleaching threshold (32°C) over a 4 day period (Figure 1A), followed by a recovery period of 15 days at ambient seawater temperature (∼27°C), and then another gradual increase in seawater temperature to the thermal bleaching threshold (34°C) over a 12 day period (Figure 1C).
(2) Long stress: Colonies were exposed to a gradual stepwise increase in peak daytime seawater temperature to ∼2°C below the thermal bleaching threshold (32°C) over a 4 day period and then held at 32°C for 2 days (Figure 1B), followed by a recovery period of 15 days at ambient seawater temperature (∼27°C), and then another gradual increase in seawater temperature to the thermal bleaching threshold (34°C) over a 12 day period (Figure 1C).
The remaining six colonies were held in the ambient tanks for the duration of the experiment and exposed to the normal thermal variance from the reef lagoon the colonies were sourced from. Following the completion of heating trajectories, all corals were randomly distributed to ambient tanks for a 10-day recovery period.
Photosynthetically active radiation (PAR) was measured continuously ∼20 cm below the surface in each tank using a PAR logger (Odyssey, Dataflow Systems, NZ) calibrated to a 2π quantum sensor (LI-192 connected to an LI-1400 meter, LiCor, United States) and temperature in each tank (°C) was continuously recorded every 5 min using HOBO MX-2201 temperature loggers (Onset, United States). Because HOBO temperature loggers may record higher temperatures than surrounding seawater due to internal heating of the transparent plastic casing (Bahr et al., 2016), loggers were deployed beneath a small PVC cover that provided shade but did not limit water flow. To provide a numeric estimate of amount of heat stress accumulated, in situ temperatures were translated to the NOAA Coral Reef Watch Degree Heating Weeks (DHWs) (Liu et al., 2014), which are used to identify local temperature thresholds linked to coral bleaching on a 5 km scale. In brief, “experimental Degree Heating Weeks (eDHWs)” were adapted from the formula used to calculate DHWs from satellite-derived SST, which measures heat accumulation relative to increases greater than 1°C above the Monthly Maximum Mean (MMM) for a location (e.g., Heron Island: MMM + 1 = 28.3°C, so eDHWs accumulate when daily in situ temperatures average >28.3°C).
Sampling Procedure
Six coral fragments (“wild”) were sampled on the Heron Island reef flat 17 days prior to the start of the experiment to characterize a baseline for the P. damicornis microbiome at the study site (day 0). Following the acclimation period, corals were sampled on days 6 (following pre-stress), 22 (end of first recovery period), 34 (end of second stress), and 43 (end of second recovery period) (Figure 1). At each sampling timepoint, one branch was randomly sampled from the center of each colony between 1,200 and 1,300 h using bone cutters sterilized with 100% molecular-grade ethanol between each sample. The bottom 1 cm of each branch (n = 18 per timepoint) was immediately snap-frozen in Whirl-Pak© bags dipped in liquid nitrogen and stored at −80°C for later 16S rRNA gene sequencing. The remaining upper portion of each branch (3–4 cm) was used for determination of Symbiodiniaceae cell density (n = 18 per timepoint).
Symbiont Densities
Immediately following sampling, fragments at each sampling timepoint were washed with 50 mL of 0.45 μm-filtered seawater (FSW) and stripped of tissue using a -inch pistol grip blow gun (Ozito, Australia) fitted to a compressed air tank. The resulting algal slurry was centrifuged and washed to remove residual coral tissue, and the resulting pelleted Symbiodiniaceae cells were then resuspended in 5 mL of FSW. Symbiont cell densities in 5 mL aliquots were determined from an average of 6 replicate Neubauer hemocytometer counts, standardized to the liquid volume of the original slurry (50 mL), and normalized to coral surface area. The surface area of each fragment was determined using the wax dipping method (Stimson and Kinzie, 1991).
Photophysiology
Replicate coral fragments (n = 5 per tank) were used to measure photosynthetic efficiency of in hospite Symbiodiniaceae cells, using a Pulse-Amplitude Modulated (PAM) fluorometer (MAXI Imaging PAM, Waltz, Effeltrich, Germany). Imaging PAM analysis was done between 19:30 and 21:00 every 3 days during recovery periods and daily during heating periods (n = 3 technical replicates per fragment). The dark fluorescence yield (F0) of each fragment was measured with a weak pre-stress of light, followed by a saturating pulse of 2,700 μmol quanta m–2 s–1 of photosynthetically active radiation (PAR) for 800 ms to measure maximal fluorescence (Fm). The maximal dark-adapted photosystem II (PSII) quantum yield of chlorophyll fluorescence (Fv/Fm) was then calculated according to the following equation:
In addition, measurements on each fragment were taken every ∼2–3 days using the pre-programmed “induction curve + recovery” kinetic recording setting, used to quantify the dark-recovery ability of Symbiodiniaceae cells after a period of illumination (induction recovery, IR). In short, after an initial pulse of actinic light, 15 saturating pules of PAR were applied to each fragment at ∼ 20 s intervals for 5 min. A recovery phase followed this, where a further 10 saturation pulses were applied over a 5-min period without actinic illumination. A total of 25 measurements of effective quantum yield were taken over 10 min for each fragment at each timepoint to construct an IR curve.
Statistics
All statistical analyses were performed in R. A linear mixed-effects model with “Day” and “Treatment” as main effects, “Day × Treatment” as an interaction effect, and “Tank” nested within “Treatment” as a random effect was used for comparisons of symbiont cell density. For symbiont densities, data were log-transformed to meet assumptions of normality and homogeneity of variance. A similar linear mixed-effects model was used for analyses of photophysiology. The model used “Day” and “Treatment” as main effects, “Day × Treatment” as an interaction effect, and “Fragment” nested within “Tank” as a random effect to test (a) changes in dark-adapted yield over time by treatment, (b) changes in quantum yield at the end of the induction period, and (c) changes in quantum yield at the end of the recovery period. For the IR curve data, the last data points of the induction period (point 15, 00:05:05) and the recovery curves (point 25, 00:09:24) were selected for analyses to characterize the ability of the symbiont population to recover from light stress. Yield data were Box Cox transformed for (a), (b), and (c) to meet assumptions of homogeneity of variance. Significant differences for symbiont cell density and yield were determined between treatments or treatments by day using Tukey post hoc pairwise comparisons.
16S rRNA Gene Sequencing
A 1–2 cm fragments (1.26 ± 0.05 g, mean plus SE, n = 68) were homogenized by bead beating in 15 mL tubes using a mix of 4 × 6.35 mm zirconium oxide-coated ceramic grinding spheres (Matrix M, MP Biomedicals) and 1.08 g of 1.4 mm ceramic spheres (Matrix D, MP Biomedicals). DNA extraction followed manufacturer protocols using a QIAamp DNA Mini Kit, Qiagen with minor adjustments as briefly outlined here. Two mL of lysis buffer (Buffer ATL, Qiagen) was added to each tube, and a FastPrep-24 5G homogenizer fitted with a TeenPrep adapter head was programmed to run 3 rounds of 20 s each (6.0 m/s) to homogenize the sample. Following homogenization, 360 μL of paste from each sample was transferred to a 2 mL microcentrifuge tube and mixed with 40 μL Proteinase K. All samples in 2 mL tubes were incubated overnight at 56°C and then purified using a silica-membrane-based nucleic acid technique. Following the remainder of the manufacturer’s protocol, quantities of AL buffer and 100% molecular-grade ethanol were increased in proportion to the increased ATL buffer volume prior to vortexing and adding to the collection column (QIAamp DNA Mini Kit, Qiagen). Extracted DNA concentration and purity were quantified using a Qubit Fluorometer and Qubit dsDNA broad-spectrum assay kits (Life Technologies, Thornton, NSW, Australia). Extracted DNA was stored at −20°C prior to PCR amplification and sequencing. DNA extraction, amplification, and sequencing were performed on negative controls (no sample template) run as every 12th sample as well. Sequencing was performed at the Ramaciotti Centre for Genomics (UNSW, Sydney, NSW, Australia) on the Illumina MiSeq platform following manufacturer’s protocol. For bacterial communities, 16S rRNA gene libraries were generated using the 27F and 519R primers (V1-V3 region).
Sequence Analysis
Sequence data were analyzed using Quantitative Insights Into Microbial Ecology version 2 (QIIME2, Bolyen et al., 2019). After denoising and primer removal using the DADA2 pipeline (Callahan et al., 2016), taxonomy was assigned to amplicon sequence variants (ASVs) in QIIME2 using a Naive Bayes classifier trained on the Greengenes 13_8 database (McDonald et al., 2012), a reference database commonly used in microbiome analysis (Knight et al., 2018). Chloroplasts, mitochondria, and unassigned ASVs (classification absent at a phylum level) were excluded from the final ASV table. Quality control was done in R (version 3.6.1) using the package decontam at a threshold of 0.1, which implements a statistical classification procedure that identifies contaminants in sequencing data (Davis et al., 2018). In brief, decontam identifies and filters out contaminants that appear more frequently in negative controls than experimental samples (Davis et al., 2018). Further exploratory analyses were done in R using the package phyloseq (McMurdie and Holmes, 2013). The identified bacteria were annotated based upon three hypothetical categories (sensu Hernandez-Agreda et al., 2018):
(1) Community: The overall microbial community was characterized as all taxa present in P. damicornis samples after filtering (Hernandez-Agreda et al., 2018) and broadly represents the bacterial community of P. damicornis at Heron Island. Indicator species, defined as the taxa found significantly more often in one treatment group compared to another, were analyzed on community samples agglomerated to class level (De Caceres and Legendre, 2009).
(2) Core: The coral core microbiome was determined for species-specific community taxa at a prevalence threshold of ≥80%, e.g., present in equal to or greater than 80% of samples (Ainsworth et al., 2015; Hernandez-Agreda et al., 2018), shared among individuals of the species within and across distinct sampling timepoints (Shade and Handelsman, 2012).
(3) Environmentally responsive: The environmentally responsive community was composed of the remaining core taxa at a prevalence threshold of ≥80% that were distinct to each treatment and timepoint, e.g., not shared among all individuals and changing in response to environmental conditions (Hernandez-Agreda et al., 2018).
All downstream statistical analyses were performed in R; the package vegan was used for multivariate statistics, indicspecies for indicator species analysis, and ggplot2 for data visualization. Alpha diversity metrics were analyzed separately for each treatment using unrarefied data, as rarefaction can result in bias of alpha diversity metrics when taxa are unobserved due to rarefaction (McMurdie and Holmes, 2014; Willis, 2019). Kruskal–Wallis tests were used for non-parametric distributions to compare changes in alpha diversity metrics (Chao1, Shannon, and Simpson) over time, as residuals were not normally distributed (determined via Shapiro test). A one-factorial permutational analysis of variance (PERMANOVA) was run using in package vegan using 999 permutations to test for differences between timepoints for each treatment using a weighted UniFrac distance matrix on rarefied data. Weighted UniFrac analysis incorporates abundances of ASVs when calculating distance matrices, and is useful for examining differences in community structure.
Results
In situ reef temperatures averaged 27.2°C each day (n = 53 days), equivalent to a thermal exposure of ∼ 2.2 DHWs over the course of the experiment due to several warmer days where the average temperature for the days reached above the MMM + 1°C. However, no bleaching was evident in the Heron Island lagoon or reef slope during Jan – Mar 2019. Similarly, ambient experimental conditions accumulated ∼1.4 eDHWs due to the normal summertime water temperature variation on the reef flat (source water) (Figure 1) and no signs of bleaching were evident in the experimental ambient corals. Within the simulated heat stress conditions, the short stress conditions averaged a max temperature of 30.3 ± 0.4°C (mean ± SE, n = 4 days) during the pre-stress period and 32.9 ± 0.6°C during the second heating period (mean ± SE, n = 12 days), equivalent to the accumulation of ∼ 4.8 eDHWs. Long stress conditions averaged a max temperature of 31.2 ± 0.4°C (mean ± SE, n = 6 days) during the pre-stress period and 32.8 ± 0.6°C (mean ± SE, n = 12 days) during the second heating period, equivalent to the accumulation of ∼ 5.4 eDHWs. No mortality, identified as bleached coral skeleton with tissue loss and/or visible microbial biofilm growth (Leggat et al., 2019), was observed through the duration of the experiment within any tanks or on the reef flat. All coral colonies in the heat treatments paled visibly, and both bleached and pale colonies were observed to have polyps extended and feeding at night throughout the experiment.
Symbiont Densities
Symbiont densities were quantified on days 6, 22, 34, and 43 to determine the extent of loss of algal symbionts consistent with bleaching in response to variable scenarios of heat stress (Figure 2A). A significant (p < 0.0500) decline in symbiont cells was observed in corals exposed to the short or long stress periods on days 34 and 43 of the experiment, respectively, at the end of prolonged exposure to bleaching temperatures and after a post-heating relaxation period. However, there was no significant decline in symbiont densities in the pre-stress period at day 6 (following the initial sub-bleaching temperature pulse for both treatments) or during the relaxation period at day 22 (following initial sub-bleaching temperature pulse). On average, the symbiont densities of healthy P. damicornis throughout the experiment were 0.57 × 106 ± 0.06 cells cm–2 (± SE, n = 22). Symbiont densities of corals held in ambient conditions did not differ over time (p > 0.900 for all timepoint comparisons). By contrast, on days 34 and 43 combined, symbiont densities averaged 0.18 × 106 ± 0.02 cells cm–2 (± SE, n = 11) in corals exposed to short stress conditions and 0.13 × 106 ± 0.04 cells cm–2 (± SE, n = 12) in corals exposed to long stress conditions. On day 34, symbiont densities of corals in both short stress and long stress conditions declined significantly from ambient corals (short stress: t = 2.463, p = 0.0470; long stress: t = 4.252, p = 0.0004). On day 43, symbiont densities in corals exposed to long stress conditions declined significantly from ambient corals (t = 4.147, p = 0.0006) but did not differ from those corals exposed to short stress conditions (t = 2.425, p = 0.052, Figure 2A).
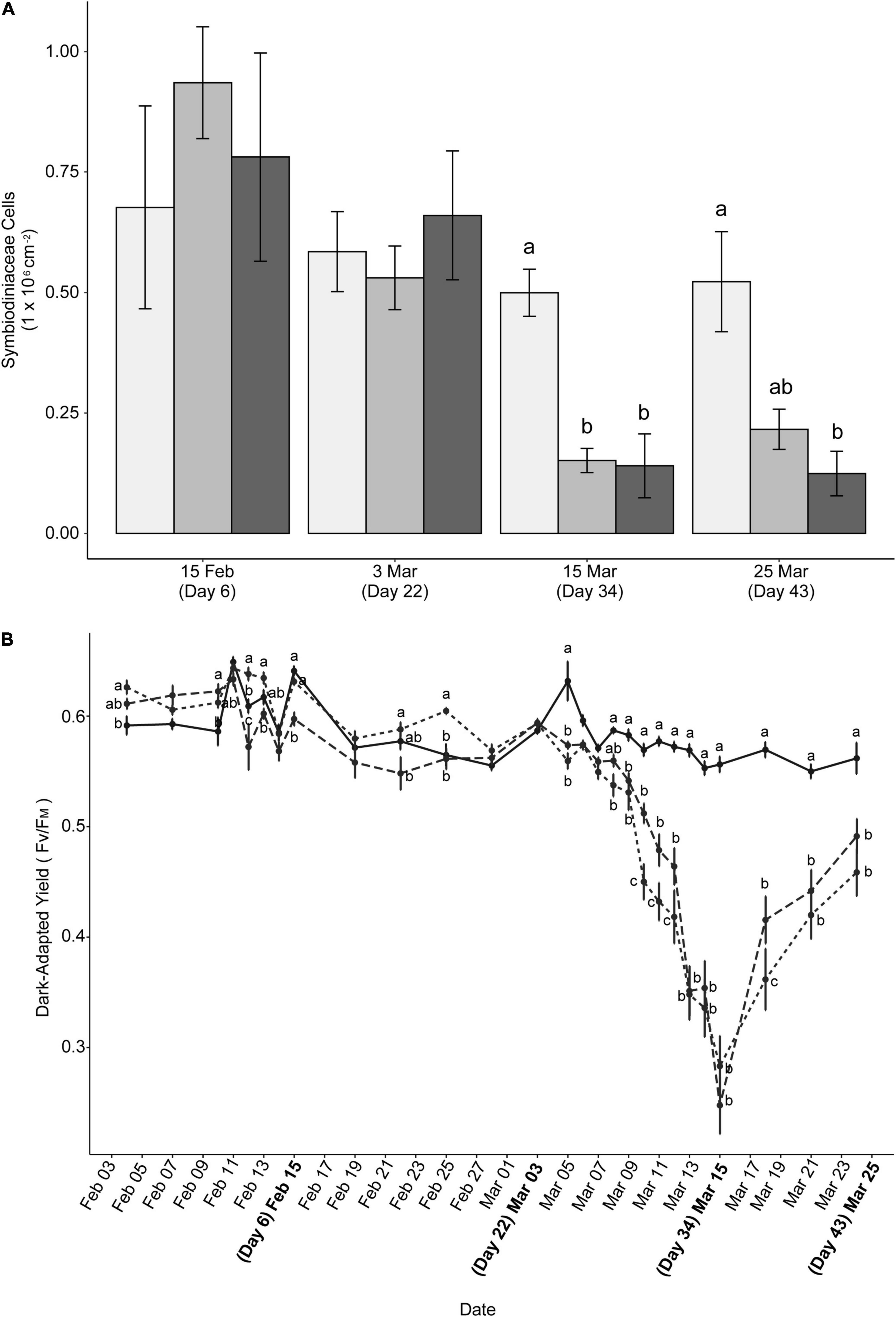
Figure 2. (A) Symbiodiniaceae cell densities in response to warming trajectories. Cell densities are displayed for ambient (light gray), short stress (medium gray), and long stress (dark gray) scenarios. Data are mean ± SE (n = 6). (B) Dark-adapted yield of colonies in ambient (solid line), short stress (short dashes), and long stress (long dashes) scenarios. Bolded dates represent sample timepoints. For both (A,B), different lowercase letters indicate significant differences between groups on the same day (Tukey post hoc test, p < 0.05). Timepoints without letters indicate no significant differences between treatments on that day.
Photophysiology
Measurements of PSII quantum yield were taken every 2–3 days throughout the experiment to determine if PSII function decreased in response to heat stress scenarios. Similar to symbiont densities, significant declines in photosynthetic response were found in the heat treatments (Figure 2B). Dark-adapted yield of corals exposed to long stress conditions declined significantly (t = 3.636, p = 0.0009) from ambient conditions following the initial warming period. On day 6, yield (± SE) for corals exposed to long stress conditions averaged 0.598 ± 0.006, contrasting with 0.641 ± 0.004 for ambient and 0.631 ± 0.003 for corals exposed to short stress conditions. Dark-adapted yield converged among treatment and ambient conditions for several days (including day 22) prior to the start of the second warming period, where corals exposed to both short and long stress conditions then began to decline in yield from ambient corals (Day 24; short stress: t = 5.993, p < 0.0001; long stress: t = 4.923, p < 0.0001). Corals exposed to long stress conditions began to increase PSII quantum yield sooner than those exposed to shorter stress conditions (Mar 18, t = −2.650, p = 0.0224) but the yield of both heat conditions at the final sampling date (day 43) did not significantly differ from each other (t = −1.979, p = 0.1182).
Induction recovery curves for effective quantum yield of PSII were used to estimate the ability of symbionts to respond to short-term light stress. Curves were analyzed at the three timepoints closest to sampling dates during the second heating and final recovery period: 7 Mar (day 22, Figure 3A), 15 Mar (day 34, Figure 3B), and 24 Mar (day 43, Figure 3C) at both points 15 and 25 on each curve (Figures 3D,E). On day 34, effective quantum yield declined significantly from ambient for all experimental conditions at point 15 (short stress: t = 15.778, p < 0.0001; long stress: t = 15.121, p < 0.0001) and point 25 (short stress: t = 11.790, p < 0.0001; long stress: t = 12.761, p < 0.0001). Significant differences in effective quantum yield in corals exposed to ambient, short stress, and long stress conditions were found on day 43 at point 15 (p < 0.030 for all); at point 25, ambient and short stress treatments differed (p = 0.002) but long stress did not differ from either.
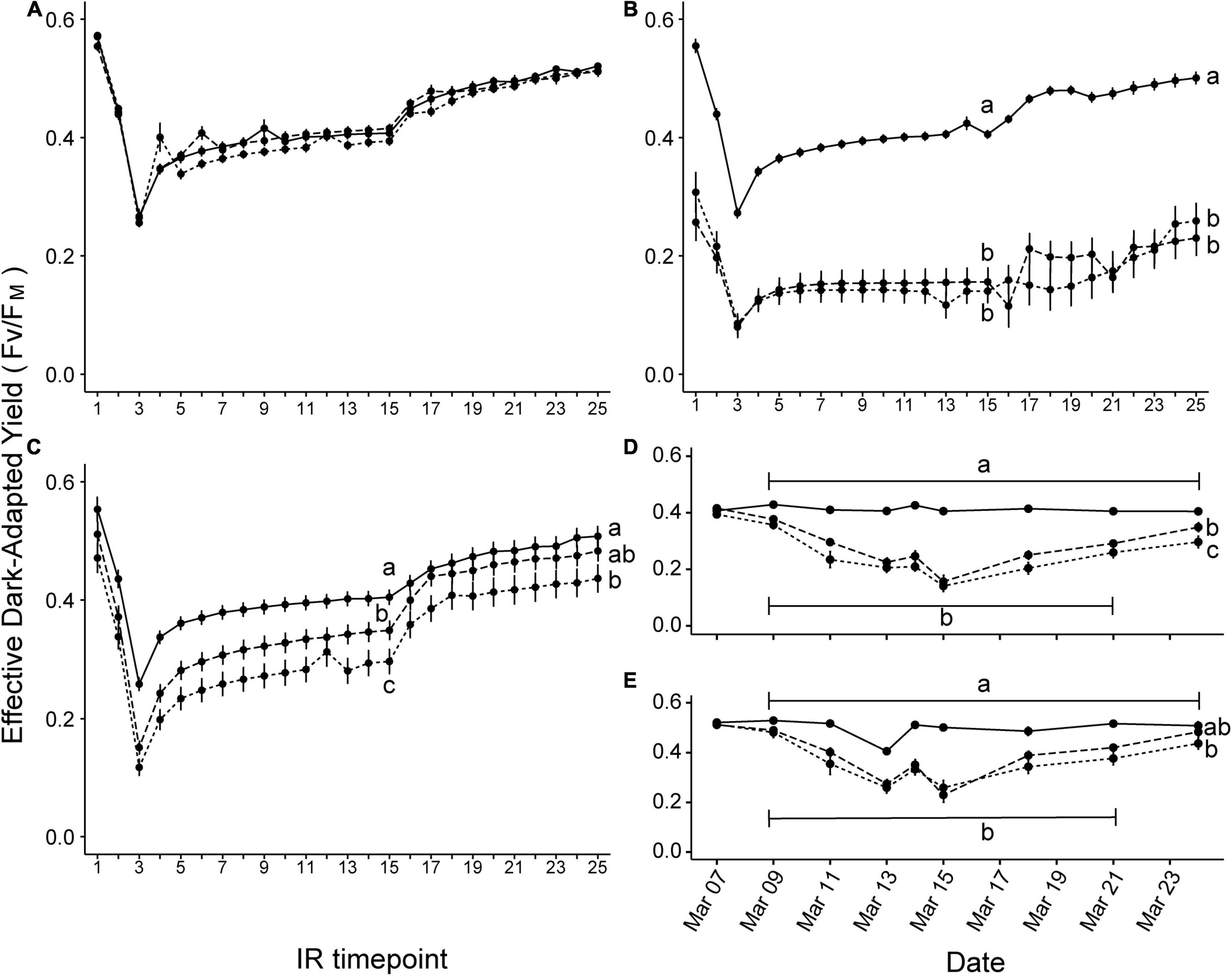
Figure 3. Symbiodiniaceae induction + recovery curves for effective dark-adapted yield of photosystem II in ambient (solid line), short stress (short dashes), and long stress (long dashes) scenarios. Plots show the effective dark-adapted yield for measurements taken nearest to each sampling timepoint during the final heating period, e.g., (A) Day 22, (B) Day 34, and (C) Day 43. Different lowercase letters indicate significant differences between treatments at the end of the induction recovery period (IR timepoint 15) and the dark recovery period (IR timepoint 25) (Tukey post hoc test, p < 0.05). Plots (D,E) show the change in effective quantum yield values of IR timepoint 15 and 25, respectively, across treatments over the entire warming period, indicating a trend of negative response to heat stress followed by gradual recovery.
Coral Microbiome
The Illumina MiSeq sequencing of bacterial 16S rRNA gene amplicons resulted in 6,875,709 sequences from 68 samples, seven negative controls, and one positive control. Quality control and removal of chloroplasts, mitochondria, unassigned ASVs (classification absent at a phylum level), singletons, and potential contaminants resulted in the retention of 6,537,144 sequences with a mean of 96,134 reads per sample. DADA2 analysis yielded 8,893 distinct ASVs for analysis of the P. damicornis bacterial community. As significant changes in symbiont densities and photophysiology were observed between treatment and ambient conditions, we grouped corals into two categories for microbial analyses: controls (ambient and wild samples, combined initial timepoint hereafter referred to as day 0) and heat stress (long and short pre-stress samples). Control samples contained an average of 104,961 reads (range 1,079–157,222) and heat stress samples an average of 91,319 reads (range 4,180–166,552). The mean number of ASVs per sample for control samples was 170 (range 27–1,140, n = 24 samples) and 138 for heat stress samples (n = 43 samples). A total of 4,078 ASVs were identified for the control community and 6,101 ASVs for the heat stress community; of these, 1,286 ASVs were present in both treatments. These unrarefied communities were used for all analyses excluding beta diversity. For beta diversity metrics, reads were rarefied to a depth of 22,900 reads per sample, resulting in 3,314 ASVs for controls (n = 23 samples) and 5,421 ASVs for heat stress (n = 43 samples).
Beta Diversity Was Found to Differ Between Bacterial Communities Over Timepoints in Both Treatments, but Not Alpha Diversity
Bacterial communities changed over time in both controls (PERMANOVA on weighted UniFrac distance, F = 1.764, p = 0.018) and in heat treatments (PERMANOVA on weighted UniFrac distance, F = 2.032, p = 0.001). There was significant variation between timepoints in both controls and in heat treatments for weighted UniFrac beta diversity metrics using reads rarefied to 22,900 reads (Figure 4C). However, further pairwise statistical tests revealed that the same timepoints differed for both controls and heat treatments (0/6 versus 22 – controls: p = 0.042, heat stress: p = 0.040; 22 versus 43: p < 0.001 for both heat stress and controls). Conversely, Kruskal-Wallis tests for all measures of alpha diversity showed that Shannon diversity in P. damicornis did not differ over time in controls (χ2 = 1.455, p = 0.6931) or heat treatments (χ2 = 2.480, p = 0.4789). ASV richness (Chao1) was also similar over time in controls (χ2 = 3.067, p = 0.3815) and heat treatments (χ2 = 5.920, p = 0.1156). Simpson diversity also did not differ between timepoints for controls (χ2 = 1.860, p = 0.6106) (Figure 4A) or heat treatments (χ2 = 1.602, p = 0.6589) (Figure 4B). There were no significant differences over time between treatments in Shannon diversity (χ2 = 4.5143, p = 0.7190), Chao1 (χ2 = 9.0204, p = 0.2512), or Simpson (χ2 = 3.8261, p = 0.7996).
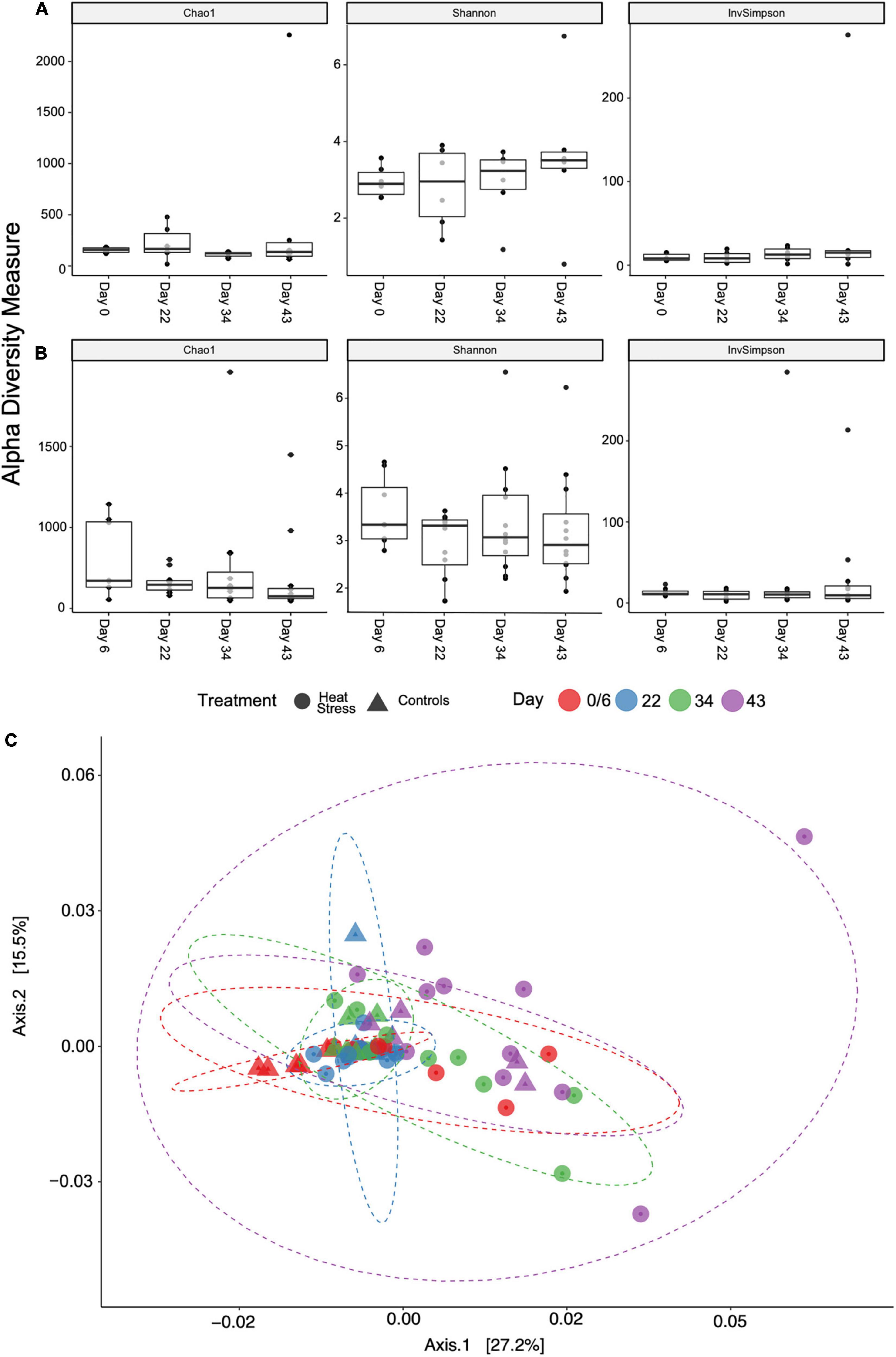
Figure 4. Alpha diversity metrics sorted by treatment over time for (A) control and (B) heat stress treatments. There were no statistical differences between timepoints. Samples were rarefied to a depth of 22,900 reads per sample for beta diversity (C). PCoA plot shows weighted UniFrac distance, sorted by treatment and sampling date. Ellipses are normally distributed data ellipses.
Coral Microbiome Composition Was Found to Be Similar Between Heat Treated and Ambient (Control) Coral Colonies
Phylum-level assignment of bacterial ASVs indicated the dominance of Proteobacteria in all samples, making up 53% of the observed number of bacterial taxa. The next most dominant phyla consisted of Bacteroidetes (26%), Cyanobacteria (6%), and Planctomycetes (5%). The top four most prevalent phyla in the controls were similar to the P. damicornis microbial community of all samples combined, composed of Proteobacteria (54%), Bacteroidetes (29%), Cyanobacteria (5%), and Planctomycetes (4%) (Figure 5). Within those phyla, the top four most prevalent classes were Gammaproteobacteria, Alphaproteobacteria, Flavobacteriia, and Saprospirae (Figure 5). The top four most prevalent phyla (and the percent they made up of the total observed number of bacterial taxa) in the heat stress treatments were similar to the control community, consisting of Proteobacteria (54%), Bacteroidetes (26%), Cyanobacteria (7%), and Planctomycetes (5%) (Figure 5). The top four most prevalent classes for the heat treatments were also Gammaproteobacteria, Alphaproteobacteria, Flavobacteriia, and Saprospirae. Indicator species analysis was conducted on the ambient and heat stress communities on days 34 and 43 combined, where declines in photophysiology and symbiont densities were indicative of severe bleaching, to determine which taxa were significantly different between bleached and healthy groups. Of 148 taxa observed (agglomerated to genus level), two taxa differed significantly between groups: a Xenococcus spp. and a Piscirickettsia spp.
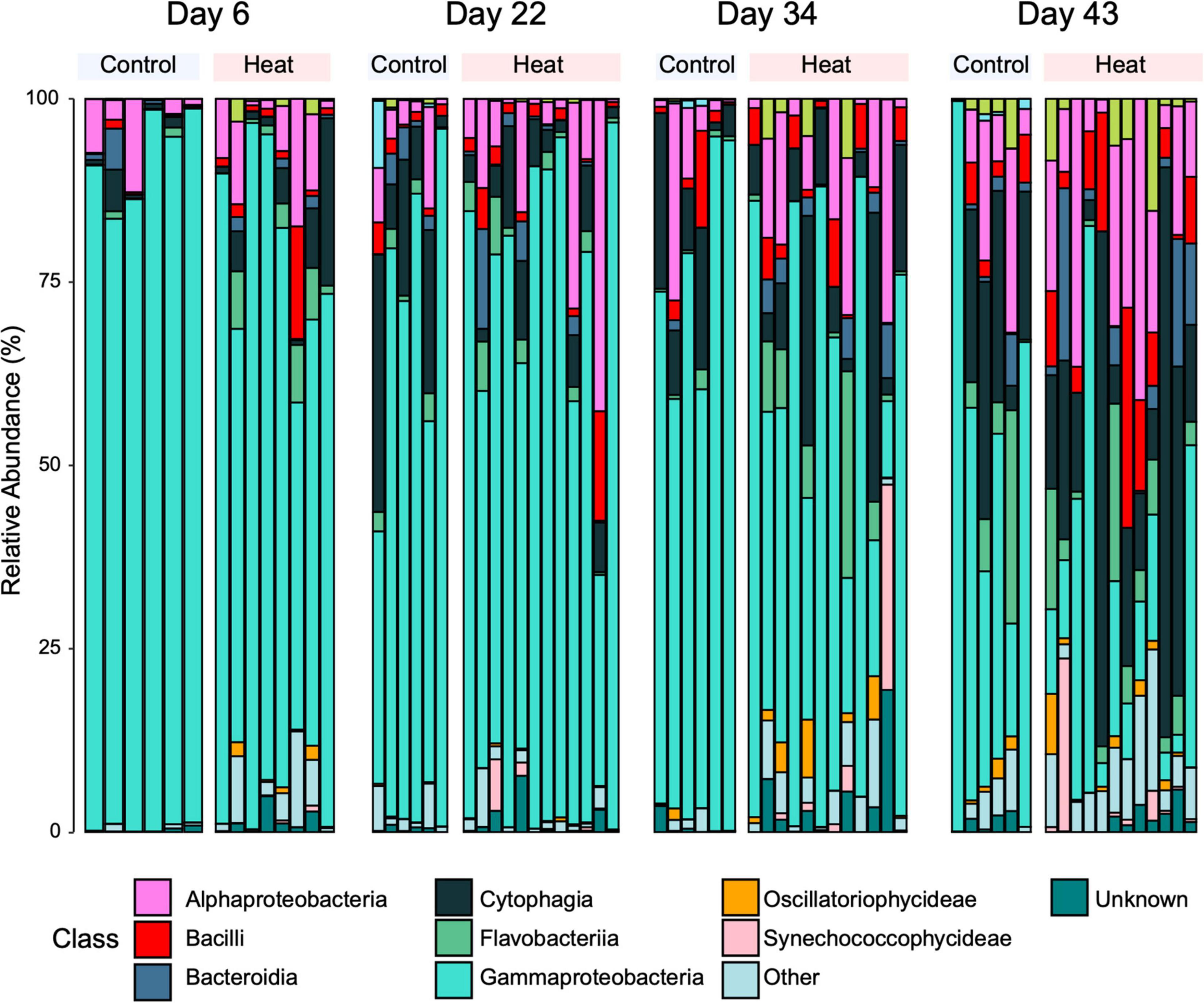
Figure 5. Relative percent abundance of ASVs in the community microbiome, sorted into top 10 bacterial classes. Samples are grouped into treatment by timepoint.
Bacterial Amplicon Sequence Variants of Interest Do Not Differ Between Treatments Over Time
We further analyzed several taxonomic groups of interest commonly studied in the coral microbiome: the order Vibrionales and the family Endozoicomonaceae. Vibrionales, a family containing known coral pathogens, occurred in only two samples in the controls (on day 0 and day 22) at less than 0.01% relative abundance (Figure 6A). The order Vibrionales was comprised of the genera Pseudoalteromonas (day 6) and an unclassified Vibrionales (day 22). In the heat treatments, the order Vibrionales appeared at all timepoints and was composed of the genera Pseudoalteromonas on days 6, 22, and 34 and an unclassified Vibrionales on day 43 (Figure 6B). One taxa could be identified to species level: Pseudoalteromonas ruthenica, which was present in two heat treatment samples on day 6. The mean relative abundance of Vibrionales was low (between 0.001 and 0.277%) and did not differ between treatments across timepoints (two-factor ANOVA, F = 0.065, p = 0.828). Since sequences assigned to Pseudoalteromonadaceae using Greengenes can often be erroneously classified as Vibrionales (instead of Alteramonadales; Edgar, 2018; Lydon and Lipp, 2018), it is likely that the mean relative abundance of Vibrionales was even lower than 0.277%, further supporting that Vibrionales did not have an impact between treatments across timepoints. Additionally, Vibrio spp. can often be improperly assigned to the Pseudoalteromonadaceae family instead of Vibrionales, however, an NCBI BLAST search of sequences of interest showed that all Pseudoalteromonadaceae in this study were classified as Pseudoalteromonas. The family Endozoicomonaceae was not identified in any samples. Taxonomic classification using Greengenes can also often place the type description of the genus Endozoicomonas within the family Hahellaceae (Kurahashi and Yokota, 2007). In the controls, Hahellaceae appeared in one sample on day 43 at 0.13% relative abundance. In the heat treatments, Hahellaceae appeared in two samples on day 43, at 0.01 and 0.02% relative abundances.
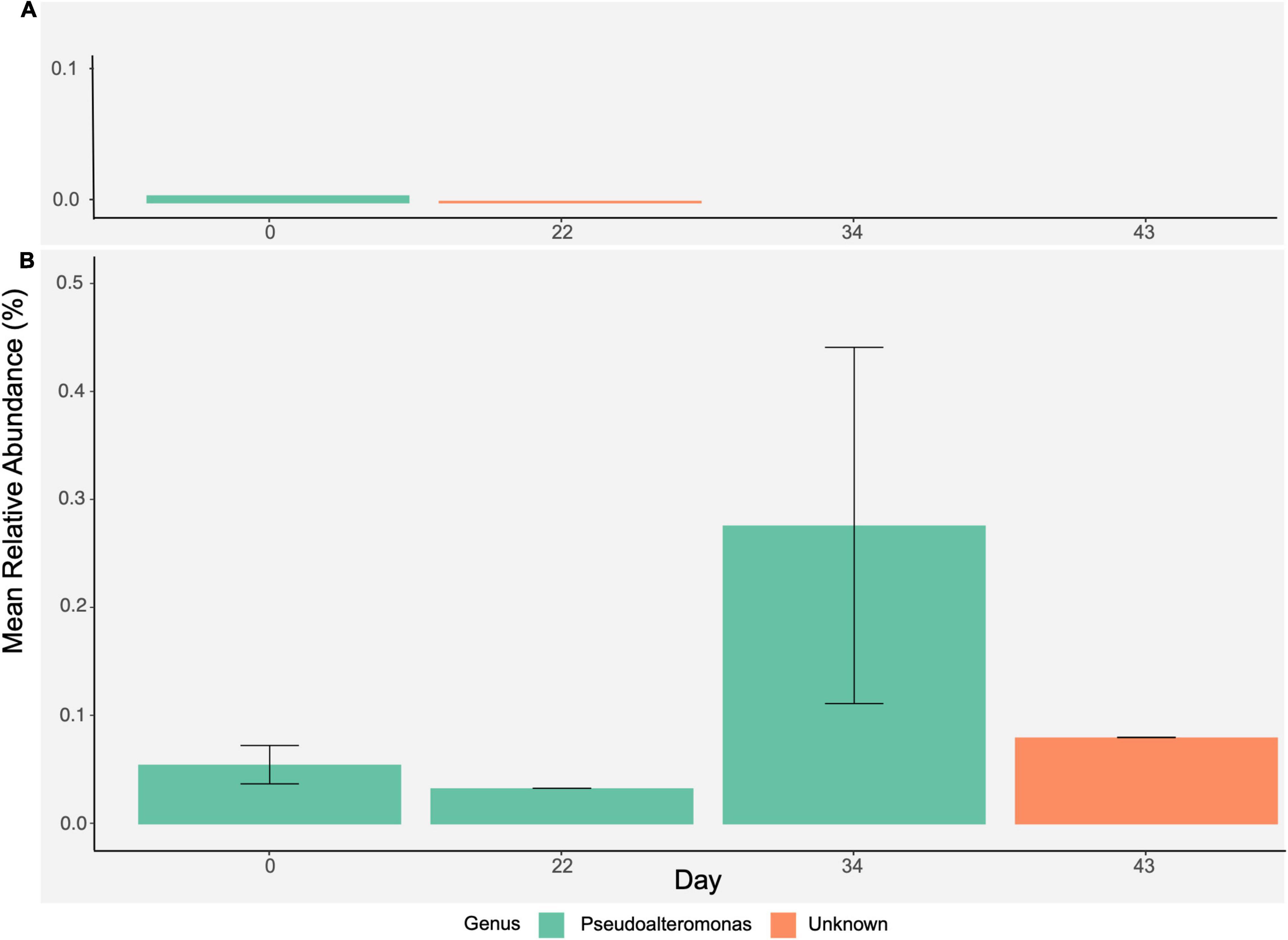
Figure 6. Further analysis of order Vibrionales. Bar plots represent mean relative abundance (± SE) of Vibrionales over time by (A) control and (B) heat stress treatments and show a breakdown of genera present within Vibrionales.
Coral Core Microbiome
The core microbiome was also determined at each treatment and timepoint at a >80% prevalence threshold, chosen as a conservative representation of the core microbiome based on our study design and small number of sample replicates at each timepoint (sensu Hernandez-Agreda et al., 2016). This led us to identify four ASVs present across all timepoints within corals held under ambient conditions (Figure 7A). For the heat stress treatment, three ASVs were identified as consistently associated taxa across all timepoints (Figure 7A). Of the ASVs identified, a Bacilli (Staphylococcus aureus) and an unclassified Gammaproteobacteria were found in both control and heat-stressed core microbiomes. Two unclassified Gammaproteobacteria were found at all four timepoints only in the control core microbiome and one Bacteroidetes (identified to genus as Candidatus Amoebophilus) was found at all four timepoints only in the heat-stressed core microbiome. The sum of the relative abundances of core ASVs per sample averaged between 40 and 60% per timepoint for controls and between 31 and 58% for heat treatments and did not change over time for either treatment (two-factor ANOVA, F = 1.082, p = 0.368) (Figures 7B,C). The most abundant core ASV, found between 5 and 32% mean relative abundance in both control and heat treatments, was an unclassified Gammaproteobacteria previously identified by Bayer et al. (2013) as an uncultured bacterium clone P6-H03 in Stylophora pistillata.
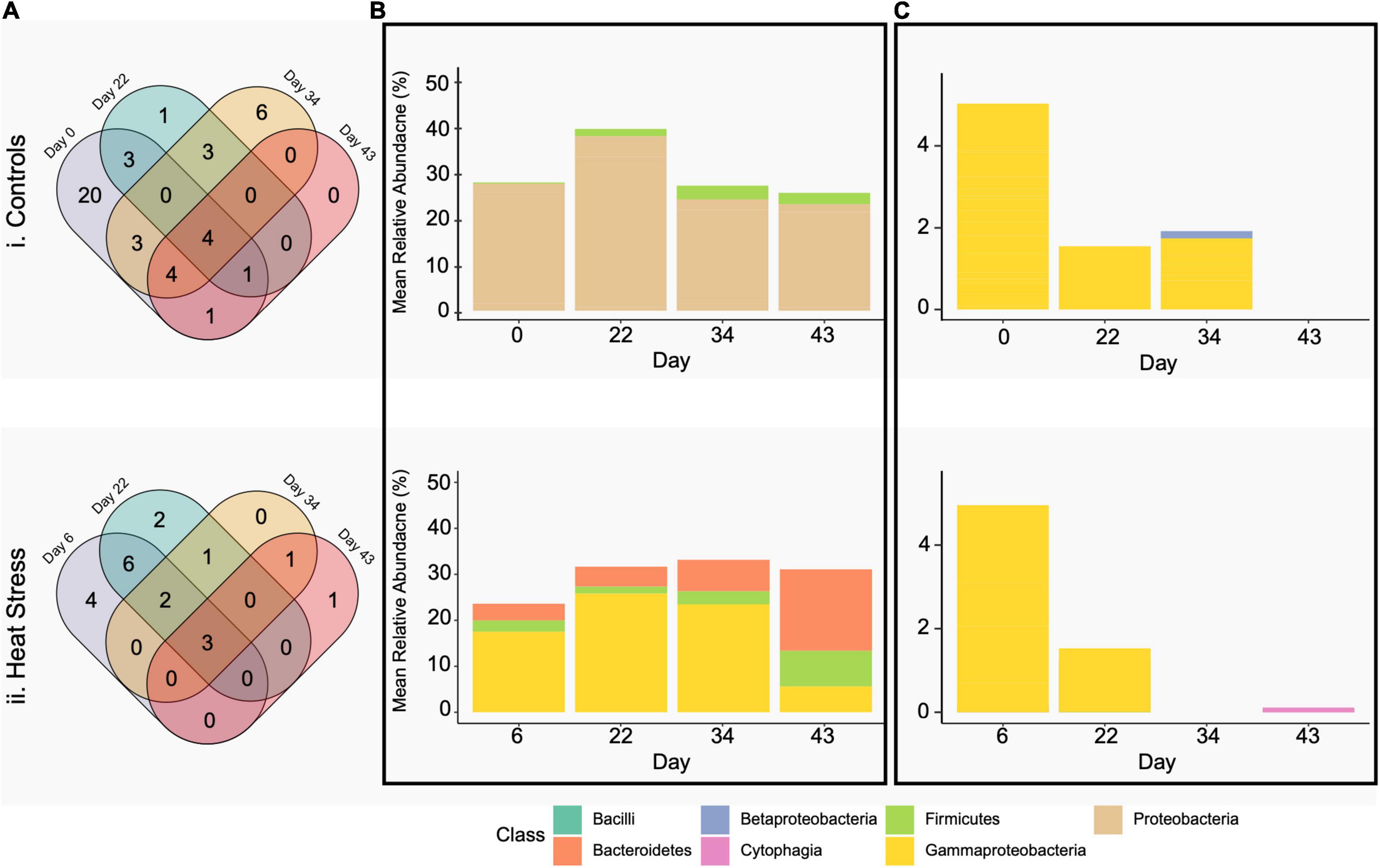
Figure 7. Venn diagrams (A) show common bacterial phylotypes shared between P. damicornis colonies by treatment crossed with timepoint at 80% prevalence, separated into controls (i) and heat stress (ii). Bar plots are average percent relative abundance by class of bacterial phylotypes present in the P. damicornis (B) core and (C) environmentally responsive microbiome.
In corals held under control conditions we find 20 core ASVs were present only on day 0, 1 on day 22, 6 on day 34, and 0 on day 43 (Figure 7A). In corals held under heat conditions, we find 4 ASVs were present only on day 6, 2 on day 22, 0 on day 34, and 1 on day 43 (Figure 7A). All core taxa were identified as unclassified Gammaproteobacteria, with the exception of one Betaproteobacteria (identified as Pelomonas puraquae) present on day 34 in controls and one Bacteroidetes (identified to genus as Candidatus Amoebophilus) present on day 43 in heat treatments. The portion of the core community prevalent in 80% of samples at each timepoint for each treatment made up fewer than 0.05% of the number of P. damicornis bacterial community ASVs.
Discussion
In this study, we investigated the impact of summertime sea surface temperature scenarios on the physiological and microbial community responses of the generalist coral species P. damicornis to determine how fine-scale changes in coral physiology due to different bleaching scenarios corresponded with changes in the coral microbiome. We found that symbiont densities, dark-adapted yield, and effective quantum yield all declined in response to heat stress regardless of the length of exposure to pre-stress warming. The 68–77% difference in symbiont densities between heat-stressed and control colonies in the present study, as well as declines in Fv/FM consistent with other studies of heat-stressed P. damicornis at Heron Island (max temperatures of 31°C, Ulstrup et al., 2008; Tout et al., 2015), indicate that P. damicornis in the heat treatments severely physiologically bleached relative to the control colonies.
Despite severe bleaching in the heat-stressed samples in the present study, we found that the P. damicornis bacterial community at Heron Island did not increase in diversity in heat-stressed colonies. Rather, diversity changed similarly over time in both controls and heat treatments. Similar to Epstein et al. (2019), no clear pattern emerged in which timepoints were driving changes in diversity. Both control and heat stress samples changed over time, suggesting that any bacterial instability was not driven by the heat stress event. Instead, it is possible that the changes observed could be a product of experimental conditions that both control and heat stress samples were subject to, such as fragmentation of the tissues, sample handling, and altered bacterial loads of corals in aquarium tanks versus those found on the reef flat (Kline et al., 2006; Kooperman et al., 2007; Ainsworth and Hoegh-Guldberg, 2009). The ordination plot (Figure 4C) also indicates that one or two outlier samples may have driven this outcome, suggesting overall bacterial stability in both the control and heat treatments. Community structure also resembled that of healthy P. damicornis and other coral species, with Gammaproteobacteria and Alphaproteobacteria as the most commonly identified taxa (Rohwer et al., 2002; Bourne and Munn, 2005). These trends differ from two commonly observed microbial community responses to heat stress: first, the “Anna Karenina Principle” (AKP) of dysbiosis, which suggests that microbial changes induced by perturbations are stochastic and lead to unstable community states instead of a specific microbiome configuration associated with stress (Zaneveld et al., 2017); second, the complementary hypothesis to the AKP in which a shift toward a pathogenic community similar to that found in diseased corals is observed in heat-stressed corals (Bourne et al., 2008; Vega Thurber et al., 2009; Tout et al., 2015). There was no observed shift toward an increase in the pathogenic Vibrio spp., which are likely either a consequence of bleaching or play an active role in bleaching (Morrow et al., 2018). Additionally, there were no pronounced changes in ASV relative abundance in heat treatments, such as the >70% reduction in relative abundance of Clostridium, Endozoicomonas, Nitrincolaceae, and Terasakiellaceae observed at temperatures >31°C by Li et al. (2020). There were also no significant changes in abundance in the core microbiome (e.g., Figure 7B) and in the environmentally responsive community (Figure 7C), where unique phylotypes were associated with ≥80% of control and heat-stressed samples over time but not correlated to the increase of heat stress. The higher number of core taxa (20) in the controls at day 0, which included wild samples, is likely due to inherent differences between corals in the natural reef environment and in tanks (Ainsworth and Hoegh-Guldberg, 2009). The two indicator taxa identified at the genus level (Xenococcus and Piscirickettsia) have both been found on coral reefs previously. Xenococcus is a unicellular cyanobacteria found associated with benthic cyanobacterial mats (Brocke et al., 2018). Piscirickettsia is a known pathogen in salmonid fishes (Fryer and Hedrick, 2003) that has been identified as one of 15 taxa potentially associated with diseased coral tissue in Acropora muricata (Sweet and Bythell, 2015). Changes in beta diversity that have been interpreted as dysbiosis in the coral microbiome are usually accompanied by other compelling evidence of a coral stress response to heat stress, with examples including increased Vibrionales and Oscillatoriales (Zaneveld et al., 2016), significant increases in pathogenic Geitlerinema, Leptolyngbya, Oscillatoria, and Sphingomonas (Casey et al., 2015), or changes in alpha diversity and 24 indicator bacterial species for heat-stressed corals (Pootakham et al., 2019). Therefore, while a significant difference in beta diversity was observed here, evidence for a destabilization of the coral microbiome was limited to two infrequently reported indicator species, further suggesting that the P. damicornis microbiome did not shift toward a dysbiotic state that could be associated with a diseased state leading to bleaching (Rosenberg et al., 2007; Vega Thurber et al., 2009; Zaneveld et al., 2016).
However, our results are consistent with Epstein et al.’s (2019) findings of stable microbiome composition in P. acuta, a species included in the P. damicornis species complex (Schmidt-Roach et al., 2014). Epstein et al. indicate that while the P. acuta microbiome showed some evidence of variation through time, overall stability (defined by Epstein et al., as no marked increase in diversity) was maintained during the months of the experiment that corresponded with two DHWs of heat stress and mass bleaching on the GBR (Epstein et al., 2019). The constrained core microbiome shown here is also similar to what has been shown for the environmental generalist species Pachyseris speciosa, where only 9 bacterial phylotypes out of 173,690 were present in over 90% of coral individuals regardless of abiotic environmental factors (Hernandez-Agreda et al., 2016). In addition, one of the core taxa identified here (Staphylococcus aureus) is prevalent on human skin and in mucus membranes (Krishna and Miller, 2012), and may have been present in all samples as a contaminant irrespective of heat treatment. A study exposing P. damicornis to heat treatments found the most abundant phyla to be similar to the present study, with relative abundances >5% in Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Halanaerobiaeota, Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, and Verrucomicrobia, and while bacterial communities destabilized at temperatures above 31°C, no increases or decreases of any ASV relative abundances were observed in corals up to 31°C (Li et al., 2020). The disease-associated bacterial taxa that were observed to occur in high abundance in corals at >31°C in Li et al., such as Sphingomonadaceae, Flavobacteriia, Rhodobacteraceae, did not increase in the present study. Similar stability has been observed in the core microbiome across depths (5 – 130 m) and geographic region (7,000 km apart) (7 core phylotypes, Ainsworth et al., 2015). Taken together, these results suggest that in the present study, phyla present at the community level as well as at a high prevalence (≥80%) in the P. damicornis microbiome are not associated differentially with healthy or bleached corals.
Here, we show that P. damicornis underwent a significant decrease in photosystem health and a loss of endosymbiotic Symbiodiniaceae densities indicative of bleaching irrespective of length of bleaching pre-stress, yet the data herein suggest that the community structure of the coral microbiome is unchanged. While rapid microbial community change can potentially aid in the adaptability of the coral host (Peixoto et al., 2017; Pootakham et al., 2018) or the destabilization of the coral meta-organism (Zaneveld et al., 2016; McDevitt-Irwin et al., 2017), an unchanged microbiome may also facilitate resistance to small fluctuations in environmental conditions for the environmental generalist P. damicornis. It has even been suggested that Pocillopora may be a “microbiome regulator,” maintaining a constant microbiome through microbial regulation (Ziegler et al., 2019). For example, the microbiome of the thermally tolerant Turbinaria reniformis remained unchanged when exposed to thermal stress, contrasting with microbiome response of the thermally sensitive Acropora millepora and suggesting that a stable microbiome may contribute to resilience against dynamic environmental conditions (Grottoli et al., 2018). These findings are supported by those of Epstein et al. (2019), who found that stability in the microbiome of the coral P. acuta corresponded with retained pigmentation in corals and no marked increase in pathogenic taxa during a mass bleaching event. In general, studies have shown that if heat stress is mild (e.g., does not result in bleaching or mortality, as in the case of Epstein et al., 2019), the coral microbiome remains stable. However, whether this stability is a result of mild heat stress or in response to mild heat stress remains equivocal, and it is possible that stability in the microbial community may not be reflective of functional changes in microbial taxa. While the present study analyzed prokaryotes, the other microeukaryotes of the coral microbiome (e.g., fungi, endolithic microalgae, and protists; Ainsworth et al., 2017) not included here may also be responding to heat stress; this suggests that there is an underlying strategy employed by P. damicornis to cope with heat stress that could not be identified through analysis of physiology and the microbial community alone. Conversely, an inflexible microbiome could also contribute to a breakdown of the meta-organism relationship between host, symbionts, and microbiome when exposed to severe stress, as observed in the present experiment through a bleaching response. For example, when Orbicella faveolata colonies in the Caribbean bleached during a natural in situ warming anomaly, no coordinated shift in the coral microbiome through time was observed (Tracy et al., 2015). These contrasting results highlight that the ability of a stable microbiome to potentially confer thermal tolerance is likely highly dependent on the severity and duration of the thermal stress event, as well as biological factors of the coral host and its symbioses.
The critical differences between studies observing alternative microbial states (synonymous with microbial dysbiosis) in the bleached coral microbiome, either through increased beta diversity or increased abundance of pathogenic taxa, and the present study are potentially reflective of specific differences in species biology, morphology, and the extent of mutualistic symbioses, commensalism, and pathogenesis of the different members of the coral’s highly diverse microbiome. For example, the microbial communities of Acropora spp. have been shown to change in response to differing levels of anthropogenic stress and to changes in the surrounding environment (Bourne et al., 2008; Grottoli et al., 2018; Ziegler et al., 2019), however, microbial stability in response to heat stress or across different environmental conditions has been observed in P. damicornis (Brener-Raffalli et al., 2018), and in closely related members of its species complex (Ziegler et al., 2016, 2019; Neave et al., 2017; Pogoreutz et al., 2018; Epstein et al., 2019). Our study highlights several possible roles of P. damicornis as an environmental generalist meta-organism occupying a variety of distinct environmental niches on the observed microbial response. First, P. damicornis, as a brooding environmental generalist, is native to both tropical and subtropical reefs in the Indo-Pacific and can be found equally on reef slopes, reef lagoons, mangroves, wharves, and other areas without strong water movement (Hoeksema and Cairns, 2021). In contrast, Acropora spp. are mass-spawning environmental specialists on tropical Indo-Pacific reef locations, favoring sites with a high oxygen content due to good circulation and strong movement of water as well as access to food from oceanic waters (Wallace, 2011). Given the high reproductive rates and widespread distribution of Pocilloporids across environmental conditions, the general stability of the coral microbiome may be reflective of a plastic mechanism by which P. damicornis is able to adjust to local environmental conditions and exist as a generalist. Since bleaching response in scleractinian corals is a combination of traits specific to the host (Fitt et al., 2009; Bellantuono et al., 2012), microbial community (Gilbert et al., 2012; Grottoli et al., 2018; Morrow et al., 2018), and algal symbionts (Fitt et al., 2001; Berkelmans and van Oppen, 2006; Howells et al., 2012), it is likely that the differences in ecological niches between generalist and specialist species may drive at least some portion of the observed divergent microbial responses to thermal stress between corals, such as those reported between Acropora and Pocillopora spp.
Secondly, the P. damicornis growth form and morphology is likely to have influenced microbial response. P. damicornis is well-characterized as plastic on a structural level, having many different morphologies and species within its complex (Todd, 2008; Schmidt-Roach et al., 2014; Johnston et al., 2017). For the species P. acuta, it has been suggested that the closer branching of the “chunky” morph (Smith et al., 2017) may contribute to a stable bacterial community by controlling intra-colonial light amplification during thermal stress and reducing the stress within the colony (Epstein et al., 2019). Therefore, it is a possibility that the tightly clustered morph of P. damicornis found at Heron Island influenced the stability of the bacterial community observed herein by reducing light amplification midway down colony branches (where samples were taken for microbial analyses), while showing symbiont loss at the colony tips (where samples were taken for symbiont counts), highlighting the importance of considering micro-habitats within the coral when sampling. Differential distribution of symbionts along a coral branch may account for the disconnect between symbiont loss and changes in the microbial community observed herein. Additionally, P. damicornis is an imperforate coral species, meaning that it does not have a porous skeletal matrix with intercalating tissues (Yost et al., 2013). This corresponds with a shallower tissue depth and more highly concentrated symbionts in P. damicornis host tissues than in perforate species, raising the possibility that symbionts in imperforate corals may encounter higher light levels due to thinner coral tissues (Terán et al., 2010). Future studies targeting the microbial communities of the tissue (e.g., through decalcification or airbrushing; Ainsworth et al., 2015) instead of homogenizing the entire coral fragment, including the microbial communities within the surface mucus layers, tissue, and skeleton (Rosenberg et al., 2007; Ainsworth et al., 2010), may also capture finer-scale changes in the host physiology and microbial community in response to heat stress. Therefore, drivers of symbiont performance through sub-surface habitat interactions that are not discernable from an exterior morphological perspective may account for why symbiont loss in the present study was observed, but not a change in the community structure of the microbiome.
In the present study, P. damicornis bleached under thermal stress yet the microbial community did not change over time, likely due to host biology and changes in host physiology occurring either prior to or uncoupled from changes in the microbiome. Neither an increase in diversity nor an increase in pathogenic taxa was observed, both of which have been used as indicators of thermal stress. It is still equivocal whether alternative states of the coral microbiome linked to bleaching are a consequence of bleaching (Ainsworth et al., 2008) or play an active role in the development of bleaching (Zhou et al., 2020), therefore it is possible that changes (or a lack thereof) occurring at the microbial community level are offset from the photosystem health and symbiont density indicators of bleaching used in the present study. As the present study analysis was a community-level approach to quantifying the coral microbiome, it is also possible that subtle changes in the microbiome remain undetected, as do functional changes of the in hospite bacteria. The appearance of stability in the gross coral microbiome may not always capture these fine-scale shifts in a small number of taxa (Tracy et al., 2015; van Oppen and Blackall, 2019). For example, metagenomic studies that have recorded no measurable increases in Vibrio bacteria in bleached corals, similar to the present study, have observed a significant increase in virulence genes from the Vibrio family (Vega Thurber et al., 2009). Therefore, future studies would benefit from employing multi-omics approaches to explore the roles and functions of the microbiota present in the coral meta-organism to better understand how the host and microbiome interact with eachx other in response to environmental change. Taken together, these findings highlight the importance of a multi-faceted approach when studying coral response to bleaching to more accurately capture the associations and interactions between the biology, physiology, and bleaching response of generalist coral species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA762381).
Author Contributions
JB, TA, and WL planned the experiments and contributed to the experiments. JB conducted the experiments. JB analyzed the data and wrote the manuscript with advice, contributions, review, and editing from TA and WL. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by an Australian Research Council Discovery Project Grant (DP180103199) to TA and WL and the Australian Coral Reef Society Student Award to JB. JB was also supported by the UNSW Scientia Ph.D. Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research was conducted at Heron Island Research Station. We thank M. Caley, A. Fordyce, C. Lantz, C. Page, M. Pappas, and the staff of the Heron Island Research Station for guidance and logistical assistance in both the field and the lab.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.664063/full#supplementary-material
References
Ainsworth, T. D., and Hoegh-Guldberg, O. (2009). Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 4, 289–296. doi: 10.3354/ab00102
Ainsworth, T. D., Fine, M., Roff, G., and Hoegh-Guldberg, O. (2008). Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J. 2, 67–73.
Ainsworth, T. D., Fordyce, A. J., and Camp, E. F. (2017). The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 25, 980–991. doi: 10.1016/j.tim.2017.06.007
Ainsworth, T. D., Heron, S. F., Carlos Ortiz, J., Mumby, P. J., Grech, A., Ogawa, D., et al. (2016). Climate change disables coral bleaching protection on the Great Barrier Reef. Science 352, 338–342. doi: 10.1126/science.aac7125
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J.-B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Ainsworth, T. D., Vega Thurber, R., and Gates, R. D. (2010). The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Bahr, K. D., Jokiel, P. L., and Rodgers, K. S. (2016). Influence of solar irradiance on underwater temperature recorded by temperature loggers on coral reefs. Limnol. Oceangr. Methods 14, 338–342.
Bayer, T., Neave, M. J., Areej Alsheikh-Hussain, M. A., Yum, L. K., Mincer, T., Hughen, K., et al. (2013). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Bellantuono, A. J., Hoegh-Guldberg, O., and Rodriguez-Lanetty, M. (2012). Resistance to thermal stress in corals without changes in symbiont composition. Proc. R. Soc. B Biol. Sci. 279, 1100–1107. doi: 10.1098/rspb.2011.1780
Berkelmans, R., and van Oppen, M. J. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Blackall, L. L., Wilson, B., and van Oppen, M. J. H. (2015). Coral - the world’s most diverse symbiotic ecosystem. Mol. Ecol. 24, 5330–5347. doi: 10.1111/mec.13400
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bosch, T. C. G., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology 114, 185–190. doi: 10.1016/J.ZOOL.2011.04.001
Bourne, D. G., and Munn, C. B. (2005). Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7, 1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–357. doi: 10.1146/annurev-micro-102215-095440
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Brener-Raffalli, K., Clerissi, C., Vidal-Dupiol, J., Adjeroud, M., Bonhomme, F., Pratlong, M., et al. (2018). Thermal regime and host clade, rather than geography, drive Symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato. Microbiome 6:39. doi: 10.1186/s40168-018-0423-6
Brocke, H. J., Piltz, B., Herz, N., Abed, R. M. M., Palinska, K. A., John, U., et al. (2018). Nitrogen fixation and diversity of benthic cyanobacterial mats on coral reefs in Curaçao. Coral Reefs 37, 861–874. doi: 10.1007/s00338-018-1713-y
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Casey, J. M., Connolly, S. R., and Ainsworth, T. D. (2015). Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5, 1–11. doi: 10.1038/srep11903
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. doi: 10.1186/s40168-018-0605-2
De Caceres, M., and Legendre, P. (2009). Associations Between Species And Groups Of Sites: Indices And Statistical Inference. Available online at: https://cran.r-project.org/web/packages/indicspecies/citation.html (accessed July 14, 2021).
Drury, C., and Lirman, D. (2021). Genotype by environment interactions in coral bleaching. Proc. R. Soc. B. 288:20210177. doi: 10.1098/rspb.2021.0177
Dziedzic, K. E., Elder, H., Tavalire, H., and Meyer, E. (2019). Heritable variation in bleaching responses and its functional genomic basis in reef-building corals (Orbicella faveolata). Mol. Ecol. 28, 2238–2253. doi: 10.1111/mec.15081
Eakin, C. M., Sweatman, H., and Brainard, R. E. (2019). The 2014-2017 global-scale coral bleaching event: insights and impacts. Coral Reefs 38, 539–545. doi: 10.1007/s00338-019-01844-2
Edgar, R. (2018). Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 6:e5030. doi: 10.7717/peerj.5030
Epstein, H. E., Torda, G., and van Oppen, M. J. (2019). Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 38, 373–386. doi: 10.1007/s00338-019-01783-y
Fitt, W. K., Brown, B. E., Warner, M. E., and Dunne, R. P. (2001). Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51–65. doi: 10.1007/s003380100146
Fitt, W. K., Gates, R. D., Hoegh-Guldberg, O., Bythell, J. C., Jatkar, A., Grottoli, A. G., et al. (2009). Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. J. Exp. Mar. Biol. Ecol. 373, 102–110. doi: 10.1016/J.JEMBE.2009.03.011
Fitt, W. K., McFarland, F. K., Warner, M. E., and Chilcoat, G. C. (2000). Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol. Oceanogr. 45, 677–685. doi: 10.4319/lo.2000.45.3.0677
Fryer, J. L., and Hedrick, R. (2003). Piscirickettsia salmonis: a Gram-negative intracellular bacterial pathogen of fish. J. Fish Dis. 26, 241–262. doi: 10.1046/j.1365-2761.2003.00460.x
Gilbert, J. A., Hill, R., Doblin, M. A., and Ralph, P. J. (2012). Microbial consortia increase thermal tolerance of corals. Mar. Biol. 159, 1763–1771. doi: 10.1007/s00227-012-1967-9
Great Barrier Reef Marine Park Authority (2017). 2016 Coral Bleaching Event On The Great Barrier Reef. Available online at: https://hdl.handle.net/11017/3206. [Accessed Feb 2, 2021].
Grottoli, A. G., Dalcin Martins, P., Wilkins, M. J., Johnston, M. D., Warner, M. E., Cai, W. J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13:e0191156. doi: 10.1371/journal.pone.0191156
Harii, S., Kayanne, H., Takigawa, H., Hayashibara, T., and Yamamoto, M. (2002). Larval survivorship, competency periods and settlement of two brooding corals, Heloipora coerulea and Pocillopora damicornis. Mar. Biol. 141, 39–46. doi: 10.1007/s00227-002-0812-y
Harriott, V. J. (1983). Reproductive seasonality, settlement, and post-settlement mortality of Pocillopora damicornis at Lizard Island, Great Barrier Reef. Coral Reefs 2, 151–157. doi: 10.1007/bf00336721
Harriott, V. J. (1985). Mortality rates of scleractinian corals before and during a mass bleaching event. Mar. Ecol. Prog. Ser. 21, 81–88. doi: 10.3354/meps021081
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., and Ainsworth, T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 7:e00560–16. doi: 10.1128/mBio.00560-16
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., Herrera, C., and Ainsworth, T. (2018). Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9, e812–e818. doi: 10.1128/mBio.00812-18
Hoegh-Guldberg, O., and Smith, G. J. (1989). The effect of sudden changes in temperature, light and salinity on the population density and export of zooxanthellae from the reef corals Stylophora pistillata and Seriatopora hystrix. J. Exp. Mar. Biol. Ecol. 129, 279–303. doi: 10.1016/0022-0981(89)90109-3
Hoeksema, B. W., and Cairns, S. (2021). World List Of Scleractinia: Pocillopora Damicornis (Linnaeus, 1758). Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=206953 on [accessed Jan 23, 2021].
Howells, E. J., Beltran, V. H., Larsen, N. W., Bay, L. K., Willis, B. L., and van Oppen, M. J. H. (2012). Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat. Clim. Chang. 2, 116–120. doi: 10.1038/nclimate1330
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Ishida-Castañeda, J., Pizarro, V., Lopez-Victoria, M., and Zapata, F. A. (2020). Coral reef restoration in the Eastern Tropical Pacific: feasibility of the coral nursery approach. Restor. Ecol. 28, 22–28. doi: 10.1111/rec.13047
Johnston, E. C., Forsman, Z. H., Flot, J. F., Schmidt-Roach, S., Pinzón, J. H., Knapp, I. S., et al. (2017). A genomic glance through a fog of plasticity and diversification in Pocillopora. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-06085-3
Kellogg, C. A. (2004). Tropical Archaea: diversity associated within the surface micro layer of corals. Mar. Ecol. Prog. Ser. 273, 81–88. doi: 10.3354/meps273081
Kline, D., Kuntz, N. M., Breitart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser 314, 119–125. doi: 10.3354/meps314119
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Kooperman, N., Ben-Dov, E., Kramasky-Winter, E., Braak, Z., and Kushmaro, A. (2007). Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276, 106–113. doi: 10.1111/j.1574-6968.2007.00921.x
Krishna, S., and Miller, L. S. (2012). Host-pathogen interactions between the skin and Staphylococcus aureaus. Curr. Opin. Microbiol. 15, 28–35. doi: 10.1016/j.mib.2011.11.003
Kurahashi, M., and Yokota, A. (2007). Endozoicomonas elysicola gen. nov., sp. nov., a γ-proteobacterium isolated from the sea slug Elysia ornate. Syst. Appl. Microbiol. 30, 202–206. doi: 10.1016/j.syapm.2006.07.003
Leggat, W. P., Camp, E. F., Sugget, D. J., Heron, S. F., Fordyce, A. J., Gardner, S., et al. (2019). Rapid coral decay is associated with marine heatwave mortality events on reefs. Curr. Biol. 29, 2723–2730.e4. doi: 10.1016/j.cub.2019.06.077
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microb. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Li, J., Long, L., Zou, Y., and Zhang, S. (2020). Microbial community and transcriptional responses to increased temperatures in coral Pocillopora damicornis holobiont. Environ. Microbiol. 23, 826–843. doi: 10.1111/1462-2920.15168
Liu, G., Heron, S. F., Eakin, C. M., Muller-Karger, F. E., Vega-Rodriguez, M., Guild, L. S., et al. (2014). Reef-scale thermal stress monitoring of coral ecosystems: new 5-km global products from NOAA Coral Reef Watch. Remote Sens. 6, 11579–11606. doi: 10.3390/rs61111579
Lizcano-Sandoval, L. D., Londoño-Cruz, E., and Zapata, F. A. (2018). Growth and survival of Pocillopora damicornis (Scleractinia: Pocilloporidae) coral fragments and their potential for coral reef restoration in the Tropical Eastern Pacific. Mar. Biol. Res. 14, 887–897. doi: 10.1080/17451000.2018.1528011
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Lydon, K. A., and Lipp, E. K. (2018). Taxonomic annotation errors incorrectly assign the family Pseudoalteromonadaceae to the order Vibrionales in Greengenes: implications for microbial community assessments. PeerJ 6:e5248. doi: 10.7717/peerj.5248
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comp. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Morrow, K. M., Muller, E., and Lesser, M. P. (2018). “How does the coral microbiome cause, respond to, or modulate the bleaching process?,” in Coral Bleaching, eds M. van Oppen and J. Lough (Cham: Springer), 153–188. doi: 10.1007/978-3-319-75393-5_7
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Appril, A., et al. (2017). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Osman, E. O., Suggett, D. J., Voolstra, C. R., Pettay, D. T., Clark, D. R., Pogoreutz, C., et al. (2020). Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 8, 1–16. doi: 10.1007/bf02539790
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., and Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898. doi: 10.1126/science.1251336
Peixoto, R. S., Rosado, P. M., Leite, D. C., de, A., Rosado, A. S., and Bourne, D. G. (2017). Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8:341. doi: 10.3389/fmicb.2017.00341
Pérez-Rosales, G., Brandl, S. J., Chancerelle, Y., Siu, G., Martinez, E., Parravicini, V., et al. (2021). Documenting decadal disturbance dynamics reveals archipelago-specific recovery and compositional change on Polynesian reefs. Mar. Pollut. Bull. 170:112659. doi: 10.1016/j.marpolbul.2021.112659
Petersen, C., and Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 16, 1024–1033. doi: 10.1111/cmi.12308
Pogoreutz, C., Rädecker, N., Cárdenas, A., Gärdes, A., Wild, C., and Voolstra, C. R. (2018). Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol. Evol. 8, 2240–2252. doi: 10.1002/ece3.3830
Pootakham, W., Mhuanton, W., Putchim, L., Yoocha, T., Sonthirod, C., Kongkachana, W., et al. (2018). Dynamics of coral-associated microbiomes during a thermal bleaching event. Microbiol. Open 7, 1–16. doi: 10.1002/mbo3.604
Pootakham, W., Mhuantong, W., Yoocha, T., Putchim, L., Jomchai, N., Sonthirod, C., et al. (2019). Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiol. Open. 8:e935. doi: 10.1002/mbo3.935
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10.
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rosenberg, E., Kushmaro, A., Kramarsky-Winter, E., Banin, E., and Yossi, L. (2009). The role of microorganisms in coral bleaching. ISME J. 3, 139–146. doi: 10.1038/ismej.2008.104
Sampayo, E. M., Ridgway, T., Bongaerts, P., and Hoegh-Guldberg, O. (2008). Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. U.S.A. 105, 10444–10449. doi: 10.1073/pnas.0708049105
Schmidt-Roach, S., Miller, K. J., Lundgren, P., and Andreakis, N. (2014). With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool. J. Lin. Soc. 170, 1–33. doi: 10.1111/zoj.12092
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Smith, H., Epstein, H., and Torda, G. (2017). The molecular basis of differential morphology and bleaching thresholds in two morphs of the coral Pocillopora acuta. Sci. Rep. 7:10066. doi: 10.1038/s41598-017-10560-2
Stimson, J., and Kinzie, R. A. (1991). The temporal pattern and rate of release of zooxanthellae from the reef coral Pocillopora damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J. Exp. Mar. Biol. Ecol. 153, 63–74. doi: 10.1016/S0022-0981(05)80006-1
Sully, S., Burkepile, D. E., Donovan, M. K., Hodgson, G., and van Woesik, R. (2019). A global analysis of coral bleaching over the past two decades. Nat. Commun. 10:1264. doi: 10.1038/s41467-019-09238-2
Sweet, M., and Bythell, J. (2015). White syndrome in Acropora muricata: nonspecific bacterial infection and ciliate histophagy. Mol. Ecol. 24, 1150–1159. doi: 10.1111/mec.13097
Terán, E., Méndez, E. R., Enríquez, S., and Iglesias-Prieto, R. (2010). Multiple light scattering and absorption in reef-building corals. Appl. Optics. 49, 5032–5042. doi: 10.1364/AO.49.005032
Todd, P. A. (2008). Morphological plasticity in scleractinian corals. Biol. Rev. 83, 315–337. doi: 10.1111/j.1469-185x.2008.00045.x
Toh, T. C., Ng, C. S. L., and Chou, L. M. (2013). “Enhancing coral reef resilience through ecological restoration: concepts and challenges,” in Proceedings of the Asian Conference on Sustainability, Energy and the Environment, Osaka, 528–545.
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Tracy, A. M., Koren, O., Douglas, N., Weil, E., and Harvell, C. D. (2015). Persistent shifts in Caribbean coral microbiota are linked to the 2010 thermal anomaly. Environ. Microbiol. Rep. 7, 471–479. doi: 10.1111/1758-2229.12274
Ulstrup, K. E., Berkelmans, R., Ralph, P. J., and van Oppen, M. J. (2006). Variation in bleaching sensitivity of two coral species across a latitudinal gradient on the Great Barrier Reef: the role of zooxanthellae. Mar. Ecol. Prog.Ser. 314, 136–148.
Ulstrup, K. E., Hill, R., Van Oppen, M. J. H., Larkum, A. W. D., and Ralph, P. J. (2008). Seasonal variation in the photo-physiology of homogeneous and heterogeneous Symbiodinium consortia in two scleractinian corals. Mar. Ecol. Prog. Ser. 361, 139–150. doi: 10.3354/meps07360
van Oppen, M. J. H., and Blackall, L. L. (2019). Coral microbiome dynamics, functions, and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
van Woesik, R., Sakai, K., Ganase, A., and Loya, Y. (2011). Revisiting the winners and the losers a decade after coral bleaching. Mar. Ecol. Prog. Ser. 434, 67–76. doi: 10.3354/meps09203
Vargas-Ángel, B., Huntington, B., Brainard, R. E., Venegas, R., Oliver, T., Barkley, H., et al. (2019). El Niño-associated catastrophic coral mortality at Jarvis Island, central Equatorial Pacific. Coral Reefs 38, 731–741.
Vega Thurber, R., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Wallace, C. (2011). “Acropora,” in Encyclopedia of Modern Coral Reefs, ed. D. Hopley (Dordrecht: Springer), 3–9.
Warner, M. E., Fitt, W. K., and Schmidt, G. W. (1999). Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc. Natl. Acad. Sci. U.S.A. 96, 8007–8012. doi: 10.1073/pnas.96.14.8007
Willis, A. D. (2019). Rarefaction, alpha diversity, and statistics. Front. Microbiol. 10:2407. doi: 10.3389/fmicb.2019.02407
Yeemin, T., Suttacheep, M., and Pettongma, R. (2006). Coral reef restoration projects in Thailand. Ocean Coast. Manag. 49, 562–575. doi: 10.1016/j.ocecoaman.2006.06.002
Yost, D. M., Wang, L. H., Fan, T. Y., Chen, C. S., Lee, R. W., Sogin, E., et al. (2013). Diversity in skeletal architecture influences biological heterogeneity and Symbiodinium habitat in corals. Zoology 116, 262–269. doi: 10.1016/j.zool.2013.06.001
Zaneveld, J. R., Burkepile, D. E., Shantz, A. A., Pritchard, C. E., McMinds, R., Payet, J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7, 1–12. doi: 10.1038/ncomms11833
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Zhou, J., Lin, Z. J., Cai, Z. H., Zeng, Y. H., Zhu, J. M., and Du, X. P. (2020). Opportunistic bacteria use quorum sensing to disturb coral symbiotic communities and mediate the occurrence of coral bleaching. Environ. Microbiol. 22, 1944–1962. doi: 10.1111/1462-2920.15009
Ziegler, M., Grupstra, C. G., Barreto, M. M., Eaton, M., BaOmar, J., Zubier, K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-10969-5
Ziegler, M., Roik, A., Porter, A., Zubier, K., Mudarris, M. S., Ormond, R., et al. (2016). Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar. Pollut. Bull. 2, 629–640. doi: 10.1016/j.marpolbul.2015.12.045
Keywords: generalist, coral microbiome, bleaching, photophysiology, climate change
Citation: Bergman JL, Leggat W and Ainsworth TD (2021) The Meta-Organism Response of the Environmental Generalist Pocillopora damicornis Exposed to Differential Accumulation of Heat Stress. Front. Mar. Sci. 8:664063. doi: 10.3389/fmars.2021.664063
Received: 04 February 2021; Accepted: 13 November 2021;
Published: 06 December 2021.
Edited by:
Marcelino T. Suzuki, Sorbonne Université, FranceReviewed by:
Hannah Epstein, Oregon State University, United StatesJie Li, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Bergman, Leggat and Ainsworth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica L. Bergman, ai5iZXJnbWFuQHVuc3cuZWR1LmF1
 Jessica L. Bergman
Jessica L. Bergman William Leggat
William Leggat Tracy D. Ainsworth
Tracy D. Ainsworth