- 1Ocean and Earth Science, National Oceanography Centre Southampton, University of Southampton, Southampton, United Kingdom
- 2National Oceanography Centre, Southampton, United Kingdom
Abyssal microbes drive biogeochemical cycles, regulate fluxes of energy and contribute to organic carbon production and remineralization. Therefore, characterizing the spatial variability of benthic microbes is important for understanding their roles in benthic environments and for conducting baseline assessments of areas of the seabed that might be targeted by commercial mining activities. Yet, detailed assessments of the spatial distributions of benthic microbial communities in these regions are still incomplete, and these efforts have not yet considered the influence of seafloor topography and heterogeneity on microbial distributions across a range of scales. In this study, we investigated the composition and spatial variability of benthic microbial assemblages found in sediments and polymetallic nodules collected from the Clarion Clipperton Zone (CCZ) in the equatorial Pacific (4000–4300 m water depth). We used 16S rRNA gene sequences to characterize these communities. The upper 20 cm of abyssal sediments harbored diverse and distinctive microbial communities in both sediments and their associated polymetallic nodules, with high similarity across topographical areas of the seabed. Assemblage composition differed vertically through the sediment, by habitat and across small to mesoscales. Potential carbon-fixing microbes formed more than 25% relative abundance of sediment assemblages, which were dominated by ammonia-oxidizing Archaea Nitrosopumilus. Non-photosynthetic Cyanobacteria were more frequent in the deeper sediment layers and nodules. Sediment communities had a higher abundance of taxa involved in nitrogen cycling, such as Nitrosopumilus, Nitrospina, Nitrospira, AqS1 (Nitrosococcaceae), and methanogens wb1-A12 (NC10 phylum). In contrast, nodules were more enriched in Alphaproteobacteria, Gammaproteobacteria, Planctomycetes, Acidobacteria, Bacteroidetes, Nanoarchaeaeota, and Calditrichaeota. Microbes related to potential metal-cycling (Magnetospiraceae and Kiloniellaceae), organic carbon remineralization (Woeseia), and sulfur-oxidizing Thiohalorhabdaceae were also more enriched in nodules. Our results indicate that benthic microbial community composition is driven by sediment profile depth and seafloor heterogeneity at small and mesoscales. The most abundant microbial taxa within the sediments were nitrifying and putative carbon-fixing microbes, and may have key ecological roles in mediating biogeochemical cycles in this habitat.
Introduction
The high abundances of polymetallic nodules and the possibility of deep-sea mining has heightened scientific and commercial interest in the environment of the Clarion Clipperton Zone (CCZ) in the eastern Pacific (Wedding et al., 2015). In an area of seafloor covering ∼ 6 million km2 (Wedding et al., 2013), seventeen exploration licenses for nodule mining have been granted since 2001 by the International Seabed Authority (ISA) (Jones et al., 2020). In addition, the ISA have also designated nine Areas of Particular Environmental Interest (APEIs), protected from mining activities and these conservation areas surround the belt of mining contract areas (Lodge et al., 2014). The aim of these conservation areas is to form a network of sites that represent the range of habitats and communities of the CCZ. Ideally, these sites will help to preserve biodiversity in the region, should any mining exploration and activities take place (Wedding et al., 2013). The potential impacts of mining on the benthic assemblages are poorly understood (Jones et al., 2017). Recovery is likely to be very slow, from decades to millions of years (Jones et al., 2017), particularly as nodule formation and growth can range from thousands to millions of years (Kerr, 1984). The process of nodule removal would remove the top layers of sediment and generate plumes near the seabed from the action of the mining collector vehicle as well as in the water column from discharge of sediment-laden water from shipboard dewatering of nodules (Jones et al., 2018). This activity could lead to widespread ecological disturbance on the seafloor. These effects could result from direct physical disturbance of communities, the removal of nodule habitat by the collector, and/or to the resettling of plume sediments to the surrounding seabed over 100’s to 1000’s km (Rolinski et al., 2001; Smith et al., 2008a). Recovery of benthic assemblages is likely to be very slow in an area with naturally low sedimentation rates (approximately 0.35 cm/kyr; Mewes et al., 2014) and stable physical conditions (Levin et al., 2016). Additionally, the CCZ is a food limited environment, and the benthic ecosystems are structured by a very low flux of particulate organic carbon (POC; fluxes 1 mg Corg m–2 d–1; Volz et al., 2018) from oligotrophic overlying waters (Smith et al., 2008a; Levin et al., 2016). As a result of a low POC flux, the soft sediments of the CCZ are oxygenated down to 300 cm below seafloor (cmbsf: Mewes et al., 2014; Volz et al., 2018; Menendez et al., 2019).
Despite a low input of carbon, there are highly diverse benthic microbial communities in this region that inhabit the sediment and nodules (Durbin and Teske, 2011; Tully and Heidelberg, 2013; Wu et al., 2013; Blöthe et al., 2015; Shulse et al., 2016; Lindh et al., 2017; Molari et al., 2020). Benthic microbes form a key part of benthic ecosystems by mediating biogeochemical cycles, regulating fluxes of energy and contributing to organic carbon production and remineralization. As well as the input of energy into abyssal ecosystems from photosynthetically derived particulate organic carbon (Ruhl et al., 2008; Smith et al., 2008b, 2009), microbially mediated inorganic C-fixation by chemolithoautotrophic microbes, (Middelberg, 2011; Molari et al., 2013) appear to provide an important carbon subsidy to benthic ecosystems in the CCZ (Sweetman et al., 2019), and more generally (Brunnegård et al., 2004; Tully and Heidelberg, 2016).
Owing to our limited knowledge of microbial communities and their functions in these environments, our ability to predict the impact of commercial mining activities on benthic microbial communities inhabiting the sediment and nodules is also limited. Benthic impact experiments designed to mimic the consequences of mining have indicated that along with metazoans, microbial assemblage recovery is poor (Gjerde et al., 2016; Jones et al., 2017). Microbial abundance in sediments has been reduced by up to 50% following sediment disturbance from plowing at the abyssal DISCOL site (Peru Basin) and recovery times for organic matter (OM) remineralization rates and microbial activity to return to baseline are more than 50 years (De Jonge et al., 2020; Vonnahme et al., 2020).
The CCZ is heterogenous, with bathymetry varying over 2000 m (Washburn et al., 2021), and clear variability in visible seafloor habitats (Simon-Lledó et al., 2019a). The CCZ is characterized by landscape-scale topographical features, such as ridges, valleys and flat plains (Macdonald et al., 1996). This topography influences the diversity and distribution of metazoans (Cosson et al., 1997; Durden et al., 2015, 2020; Stefanoudis et al., 2016; Leitner et al., 2017; Simon-Lledó et al., 2019a), possibly owing to the influence of topography on bottom currents, sediment grain size and food supply (Durden et al., 2015; Morris et al., 2016). These factors would also be likely to influence benthic microbial communities, but have not been assessed at a landscape scale. Such information is an important component of baseline environmental assessment for commercial mining activities and subsequent monitoring (Ingels et al., 2020), particularly as benthic microbes represent an important energy and nutrient source for abyssal ecosystems. Previous work examining the regional-scale structure of CCZ benthic microbial communities using 16S rRNA gene sequencing has indicated that microbial communities may have a spatial variability over distances of less than 1000 km (Tully and Heidelberg, 2013; Lindh et al., 2017), but finer-scale patterns or their controls are unknown.
Here we characterize bacterial and archaeal assemblages via amplicon sequencing of the 16S rRNA genes, from sediment and nodule samples collected from one of the CCZ conservation areas, APEI-6, in order to determine environmental controls on microbial community structure and compare these with one of the proximal exploration areas (UK-1). Specifically, we tested the following hypotheses: (1) Benthic microbes vary in their spatial distribution from small scales (cm) to larger scales (100’s m) within sediments at APEI-6 analogous to that previously observed in metazoans and (2) the broad landscape-scale features of APEI-6 (ridges, valleys, and plains) will influence the diversity and distribution of benthic microbial assemblages.
Materials and Methods
Sample Collection
Sediment samples and polymetallic nodules were collected during the RRS James Cook cruise JC120, April-May 2015 to the eastern CCZ in the northern equatorial Pacific. Samples were obtained from 20 megacores (Bowers and Connelly type; Barnett et al., 1984) that were deployed in the south western part of APEI-6 (Sampling area centered: 17° 10 N, 122° 75 W) and 2 megacores deployed in the UK-1 Exploration Contract Area, ∼750 km away from APEI-6 (13° 28 N, 116° 35 W) (Table 1). Megacores were deployed in four distinct topographical landscape areas in APEI-6 that were each defined by bathymetric data: Deep-Plain, Flat, Ridge and Trough [Figure 1; definition details in Simon-Lledó et al. (2019a)]. Megacores of sediment were sliced at 1 cm intervals down to 10 cm, below which slices were taken every 2 cm. The following sediment depth layers were used for analysis of eDNA studies: 0–1 cm below seafloor (cmbsf), 1–2, 5–6, 10–12, and 20–22 cmbsf. Only sediment from the center of the core that had not been in contact with the sides of the megacore tube was sampled. Sediment was placed into sterile Whirl-pak bags and frozen at −80°C immediately on board. Surface nodules in the top 1 cm of the megacore were removed with forceps, rinsed with filtered seawater and frozen immediately at −80°C in sterile Whirl-pak bags for microbial analysis.
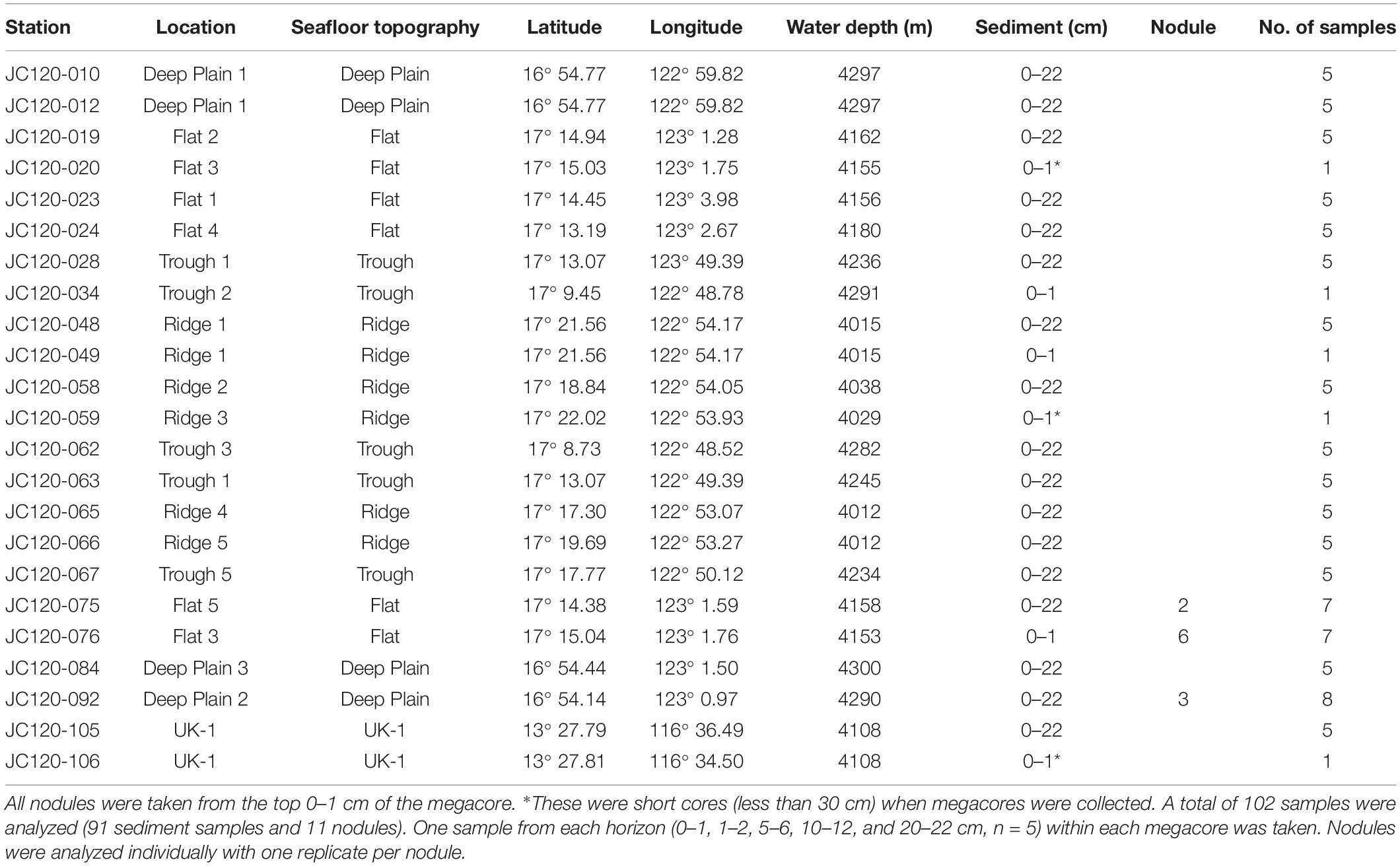
Table 1. Sampling site locations of megacore deployments in APEI-6 SW and UK-1 (UK Seabed Resources Limited eastern contract area) that were collected during cruise JC120 with location and depths shown for each sample taken.
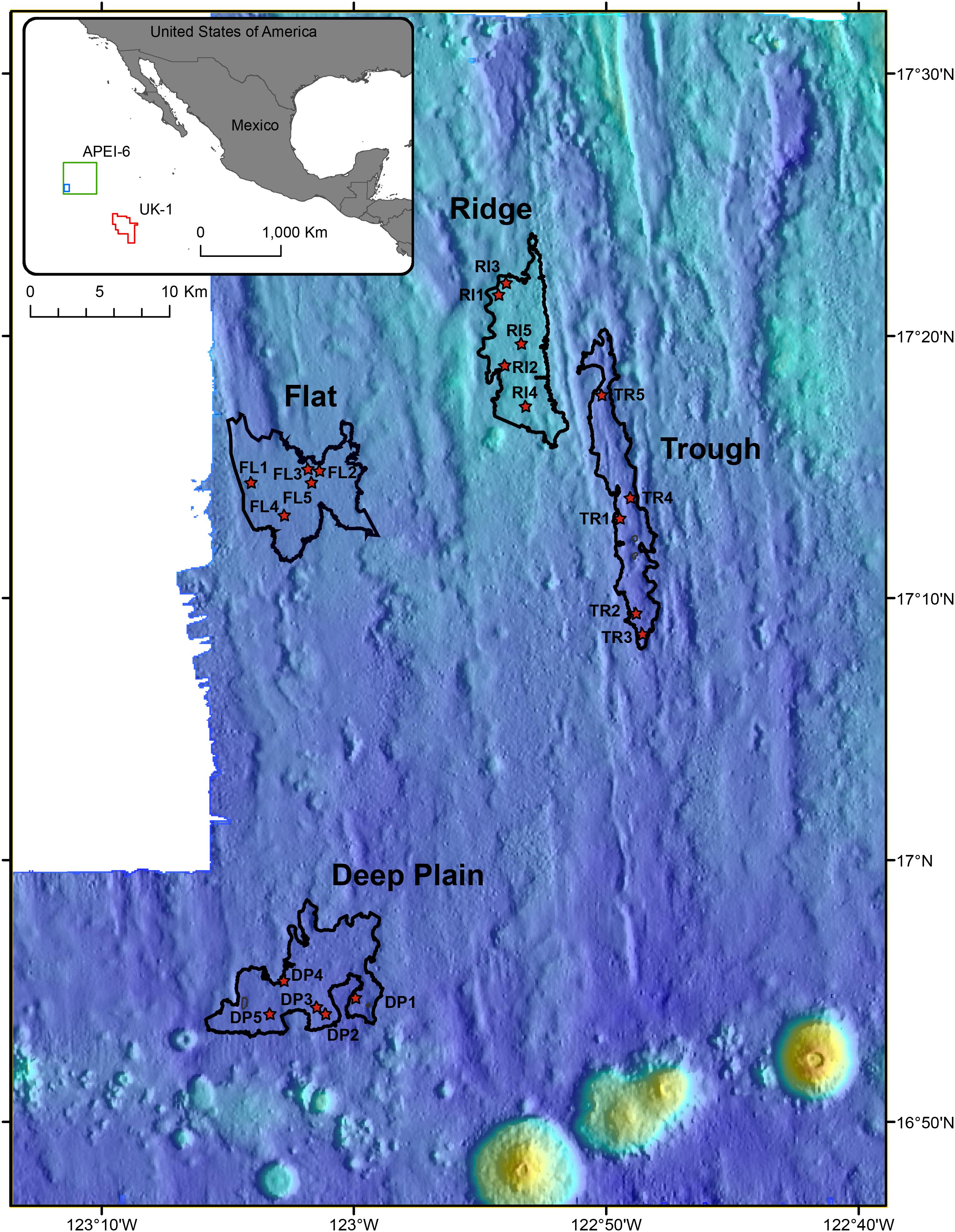
Figure 1. Location of the Area of Particular Environmental Interest (APEI-6) and UK-1 exploration contract area in the Clarion-Clipperton Zone (CCZ) of the Pacific Ocean, where sampling occurred (insert map). Coring locations within the four landscape regions of the APEI-6 are shown with a red star. Red polygon shows location of UK-1, the green box indicates the location of APEI-6, and the blue box delineates the location of the sampling sites within APEI-6.
Environmental data was collected during the JC120 cruise and included sediment grain size, nodule density (no. m–2), total carbon (TC,% sediment dry weight), total organic carbon (TOC, wt%), total nitrogen (TN, wt%), the ratio of total organic carbon to total nitrogen (TOC:TN), the ratio of total carbon to total nitrogen (TC:TN) and carbonate content (CaCO3, wt%). Manganese (Mn, wt%) values are from Menendez et al. (2019). Collection and calculation of these values is detailed in Simon-Lledó et al. (2019a) and Menendez et al. (2019).
DNA Extraction and Sequencing
Genomic DNA was extracted from 500 mg of wet sediment and 500 mg of crushed nodule samples using the FastDNA Spin Kit for Soil (MP Biomedicals, United States) following the manufacturer’s protocol. Nodules were first crushed with an autoclaved pestle and mortar. Additional extraction blanks containing only the FastDNA Spin Kit reagents were processed with the sediment samples. The concentrations of DNA from all samples was below 0.1 ng/μl and required further concentration. DNA was concentrated using the Zymo Clean & Concentrator-5 kits with a 2:1 DNA Binding Buffer ratio and eluted into 50 μl sterile, DNase-free water. The V4 region of the 16S bacterial and archaeal rRNA gene was amplified by the polymerase chain reaction (PCR), following the Earth Microbiome Project (EMP) protocol (Thompson et al., 2017), using the oligonucleotide primers Pro515f/Pro806r. The amplified 16S rRNA gene products and extraction blanks were prepared with the Nextera XT v2 Kit (Illumina, San Diego, CA, United States) and sequenced on an Illumina MiSeq platform at the Environmental Genomics Sequencing Facility (University of Southampton, National Oceanography Centre, Southampton).
Data Analysis
The demultiplexed sequences were analyzed with the microbiome analysis software package QIIME 2 (Quantitative Insights Into Microbial Ecology), version 2019.1 (Bolyen et al., 2019). The DADA2 pipeline (Callahan et al., 2016) within QIIME 2 was implemented for sequence quality control and chimera removal. Amplicon Sequence Variants (ASVs) or features were resolved using the DADA2 denoise-single method. Features that were observed in the PCR blanks were considered to be contaminants and were filtered from the samples. A Naïve Bayes classifier that was pre-trained on the V4 region of reference sequences from the Silva database (version 132; Quast et al., 2013) was used to classify representative sequences of ASVs in our dataset and clustered at 99% identity. A rooted phylogenetic tree was generated within QIIME2, using MAFFT and FastTree, which was used for calculating diversity metrics.
The generated feature table contained 48,198 ASVs and 4,383,878 reads, with an average of 36,839 reads per sample. The feature table was normalized to 4600 sequences per sample in QIIME 2 prior to abundance and diversity analysis, to account for uneven sampling depth and library sizes. Samples were categorized by substrate (sediment and nodules), topographical landscape and sediment horizon. Relative abundances were calculated on ASV feature tables with raw counts that had been normalized by library size to get the proportions of species within each library and then square-root transformed. Alpha diversity (observed features (q = 0), Shannon-Wiener index, evenness (Pielou’s evenness values, Chao-1 index) and beta diversity (Bray-Curtis distance) were calculated in QIIME 2 on the normalized samples and formatted along with the generated feature-table for further analysis in R. Community composition and statistical analyses were performed using the “vegan” package in R v 3.3.2 (Oksanen, 2007, 2017). To determine if there were significant differences between alpha diversity in sediments and nodules at APEI-6 (Flat and Deep Plain sites), a one-way ANOVA test was performed in “vegan.” Given the low numbers of replicates, the data from different topographies were combined for the testing. Tables of ASV counts were double square-root transformed and Bray-Curtis dissimilarity measures applied to determine variations in community composition between the four topographic areas and sediment layers in APEI-6. Non-metric multidimensional scaling (NMDS) ordination was used to visualize community variation between geographic location (APEI-6 vs. UK1), topographical landscape and sediment horizon, along with environmental parameters, with the “envfit” method in the “vegan” package (Oksanen, 2007).
Permutational multivariate analysis of variance (PERMANOVA; Anderson et al., 2001) and post hoc pair-wise tests were performed on Bray-Curtis dissimilarity matrices using PRIMER v.7 (Clarke and Gorley, 2015). A mixed effect model was used to partition variance among factors and to test whether microbial assemblage composition varied significantly between factors at APEI-6 (20 megacore samples, Table 1). In this model, topography and sediment horizon were treated as fixed factors. To account for potential covariance between depth strata sampled from individual megacores and to reflect the stratified random sampling within topographical regions, megacore was treated as a random factor nested within topographical region. This model is analogous to a classical repeated measures ANOVA design. Topography contained four levels: Deep Plain, Flat, Ridge, and Trough. Sediment Horizon contained five levels: 0–1, 1–2, 5–6, 10–12, and 20–22 cm. The main effects, topography and depth, the nested factor, core, and the interaction term, topography × depth, were included in the model. Methods of permutation used was “method (ii) permutation of residuals under a reduced model.” Nodules and the UK-1 samples were excluded from the PERMANOVA mixed-effect model due to lack of sufficient replication.
We were not able to carry out comparative statistics on UK-1 samples as there was only one full megacore taken from this region. The second megacore from UK-1 was a short core owing to a large nodule that was retrieved in the eDNA core, so only the top 1 cm of undisturbed sediment could be recovered. Nevertheless, samples from UK-1 and nodule samples were qualitatively evaluated using taxon abundance plots and NMDS. To aid in visualization, ASVs were grouped by taxonomy at phylum, class and genus levels, and the higher classification taxonomic groups that represented more than 1% of the total abundance in at least one substrate type, one depth horizon or one topographical landscape type were further analyzed and discussed. Graphical outputs from abundance and statistical analysis were displayed using the “ggplot2” package in R (Wickham, 2016). Raw sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under BioProject ID PRJNA694451.
Results
Microbial Diversity at APEI-6 and UK-1
After the data were normalized, 547,400 reads comprising of 36,815 ASVs were identified in 91 sediment and 11 nodule samples (Table 1). Of these ASVs from APEI-6, 19% were classified as Archaea and 81% as Bacteria. In UK-1, the proportion of Archaea was slightly higher, with 21% of ASVs classified as Archaea and 79% as Bacteria.
Sediment assemblages in the upper 2 cm of sediment were more diverse than polymetallic nodule-associated assemblages in APEI-6 (One-way ANOVA, P = 0.005, F1,46 = 8.99, R2 = 0.17). Alpha diversity (Figures 2A–C) in UK-1 sediments (Observed ASVs: mean = 755, SE ± 82; Shannon diversity 8.75 ± 0.15) is higher compared to alpha diversity in APEI-6 sediments (ASVs: 675 ± 14; Shannon: 8.72 ± 0.01) and nodules (ASVs: 288 ± 8; Shannon: 7.34 ± 0.18). Despite non-significant alpha diversity between sediment horizons or between topographical regions within APEI-6, mid to deeper layers (5–22 cm) of sediment at APEI-6 were more diverse than those at the sediment surface, except at Ridge, where the reverse was true. Sediments at Flat sites were the most diverse, with the 5–6 cm layer being the most diverse overall (Observed ASVs 2000 ± 447, Shannon; 9.35 ± 0.52) and least diverse in the 20–22 cm horizon at Ridge (Observed ASVs 534 ± 114, Shannon; 8.36 ± 0.38). At UK-1, greater alpha diversity was also observed in the surface layers, particularly in the 1–2 cm horizon (Observed ASVs 1089 ± 412; Shannon 9.15 ± 0.68). Estimated richness, (Chao1, Figure 2D) was also higher overall at UK-1 (978 ± 159), compared to APEI-6 sediments (805 ± 46).
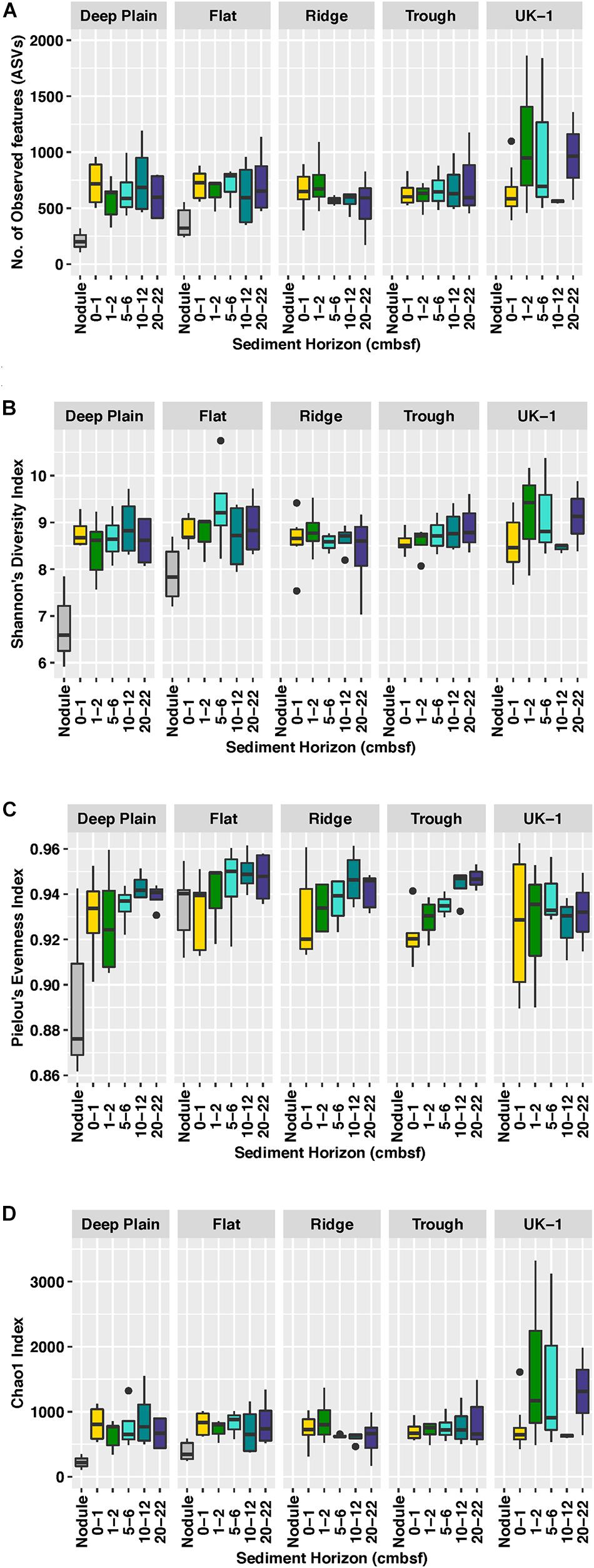
Figure 2. Comparisons of diversity indices between sediment and nodule-associated microbial communities, at APEI-6 and UK-1, based on (A) Number of observed features or amplicon sequence variants (ASVs), (B) Shannon’s Diversity Index, (C) Pielou’s Evenness Index, and (D) Chao1 richness Index. Boxplots show the median (horizontal line), interquartile ranges (box limits), range (whiskers), and outliers (black dots). Diversity indices were calculated on samples that had been rarefied to account for uneven sampling depth.
General Patterns in Microbial Community Composition
Non-metric multidimensional scaling plots were used to visualize differences in microbial community composition between the sediment layers at APEI-6, UK-1, and nodule samples (Figure 3). Microbial assemblages associated with nodules clustered separately from sediment samples within APEI-6 (Figure 3A). Benthic topography only explained 2.7% of the variance in beta diversity and was not statistically significant. We observed a significant influence of sediment horizon depth on assemblage composition within APEI-6, (PERMANOVA, p = 0.001, F4,83 = 6.977, Table 2) accounting for 23% of variance in community composition and structure of sediment assemblages overall. The interaction between topography and depth was not statistically significant, but accounted for 9.6% of the variance (Table 2). We observed a relatively large between core variance (20%), suggesting a degree of heterogeneity in microbial assemblage composition between megacores within topographical regions (PERMANOVA, p = 0.001, F18,83 = 1.3212, Table 2). We also noted that UK-1 sediment samples formed a distinct cluster from APEI-6 sediment and nodules (Figure 3B). It was not possible, however, to perform statistical analysis on UK-1 samples, owing to lack of replication.
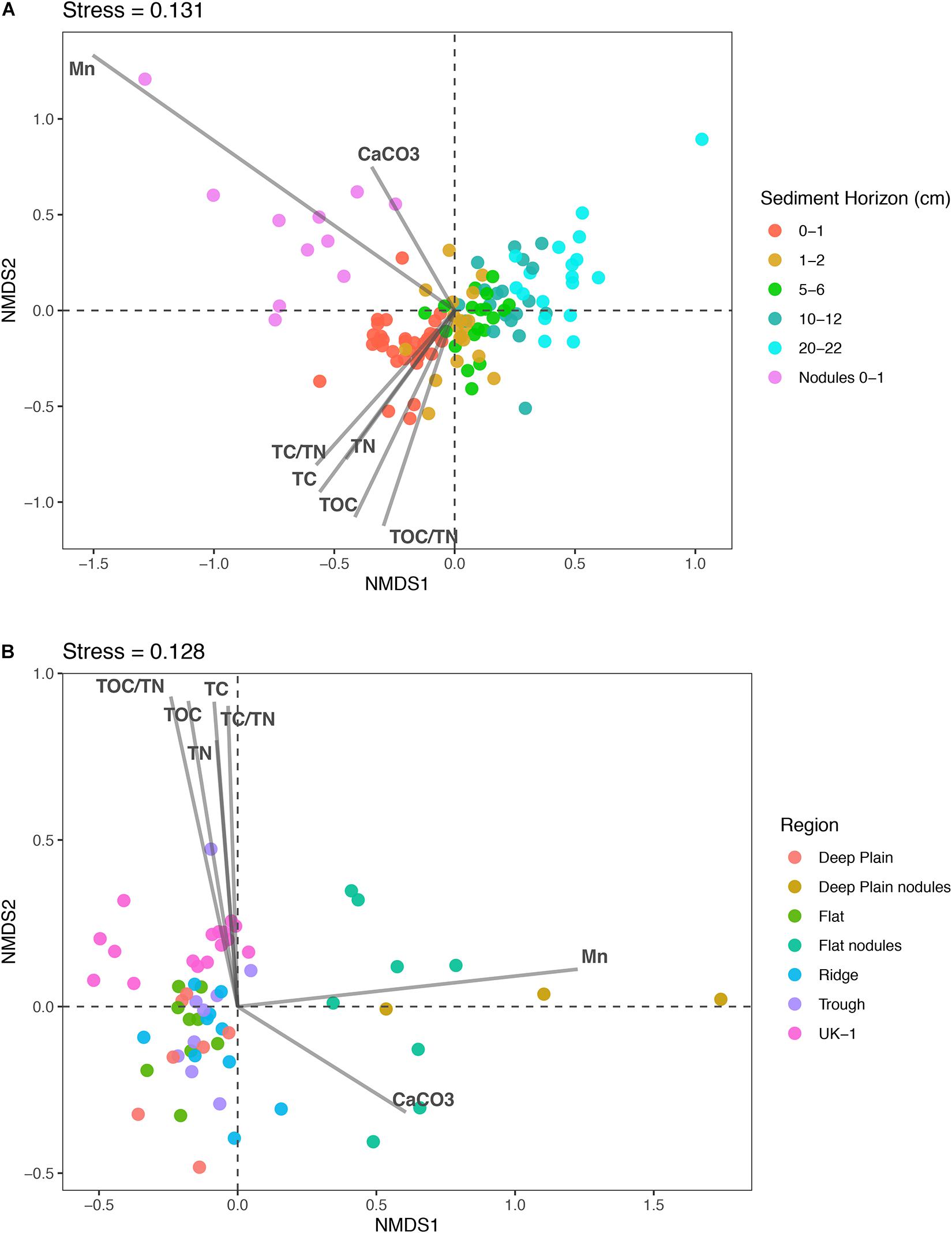
Figure 3. Non-metric multidimensional scaling (nMDS) plots of microbial community dissimilarity based on Bray-Curtis distances. The points represent individual samples and their proximity to one another indicates their similarity. The closer the points are to each other, the more similar the samples are to each other based on community structure and composition. (A) Nodule-associated and sediment communities through the sediment profile (0–22 cm) from APEI-6 and UK-1 in the CCZ. Nodules are from the surface sediments (0–1 cm) of the Deep Plain and Flat regions of APEI-6. (B) Sediment communities in the top 2 cm of sediment across the four regions in APEI-6 and sediment at UK-1. Nodules are from the surface sediments (0–1 cm) of the Deep Plain and Flat regions of APEI-6. The relationship of environmental variables to the samples are shown with arrows, where the greater the arrow length, the stronger the correlation. Environmental variables are from Jones et al., 2021, in prep.) and Menendez et al. (2019).
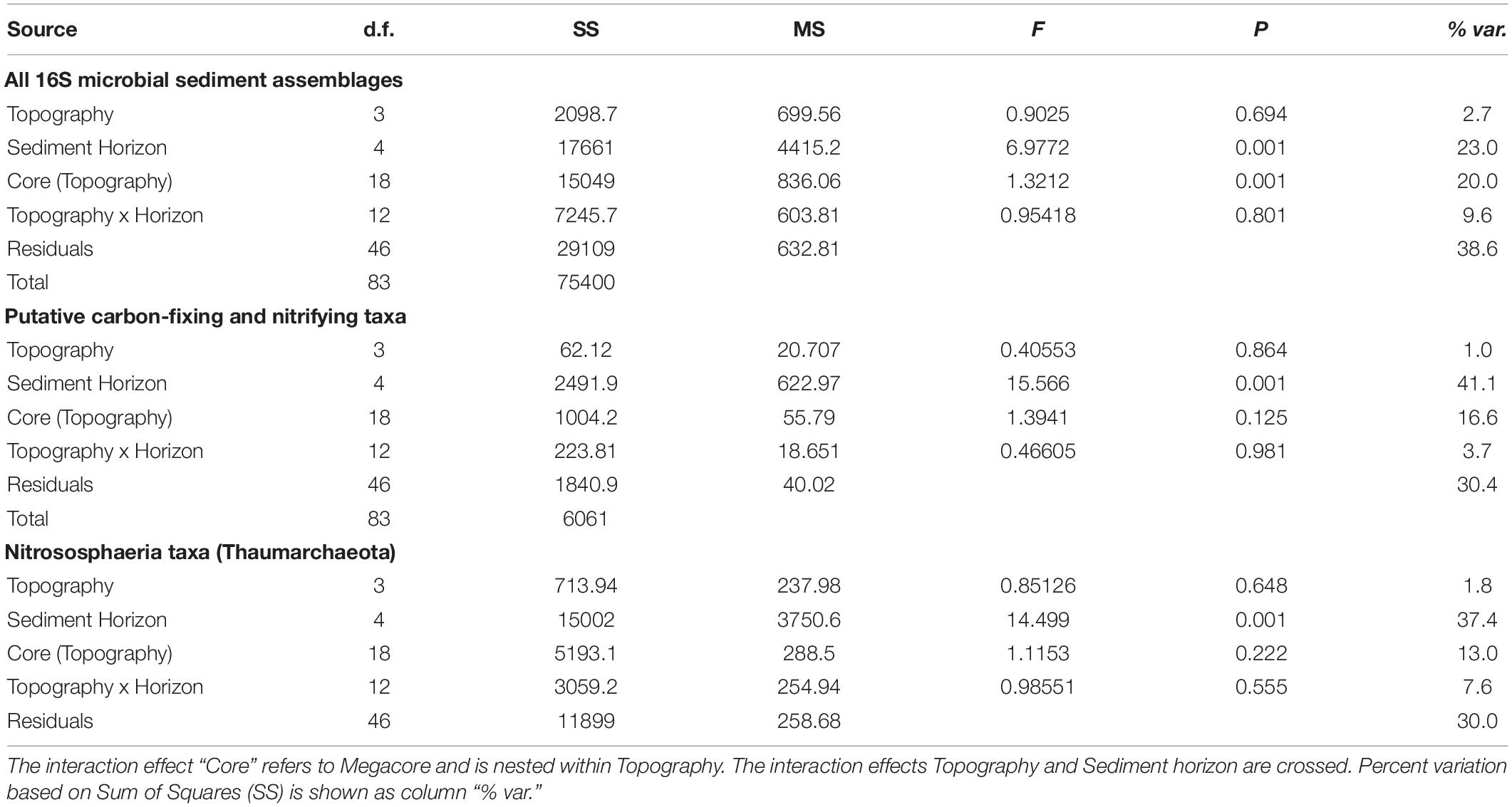
Table 2. Output permutational multivariate analysis of variance (PERMANOVA) on relative abundance counts that have been square-root transformed prior to creation of Bray-Curtis dissimilarity matrices for sediment microbial assemblages at APEI-6, showing the following: all 16S microbial assemblages; putative carbon-fixing and nitrifying taxa within sediment microbial assemblages; and taxa within the Nitrososphaeria class in the Thaumarchaeota phylum.
Environmental variables were largely homogenous within APEI-6. Relative proportions of TC, TOC, TN, TOC:TN, TC:TN, CaCO3, Mn, and mean grain size were very similar across the four sampled regions of APEI-6 with no significant differences detected. Nodule coverage was significantly different between the study areas (Simon-Lledó et al., 2019b), with greatest nodule density observed at Deep Plain (Table 3).
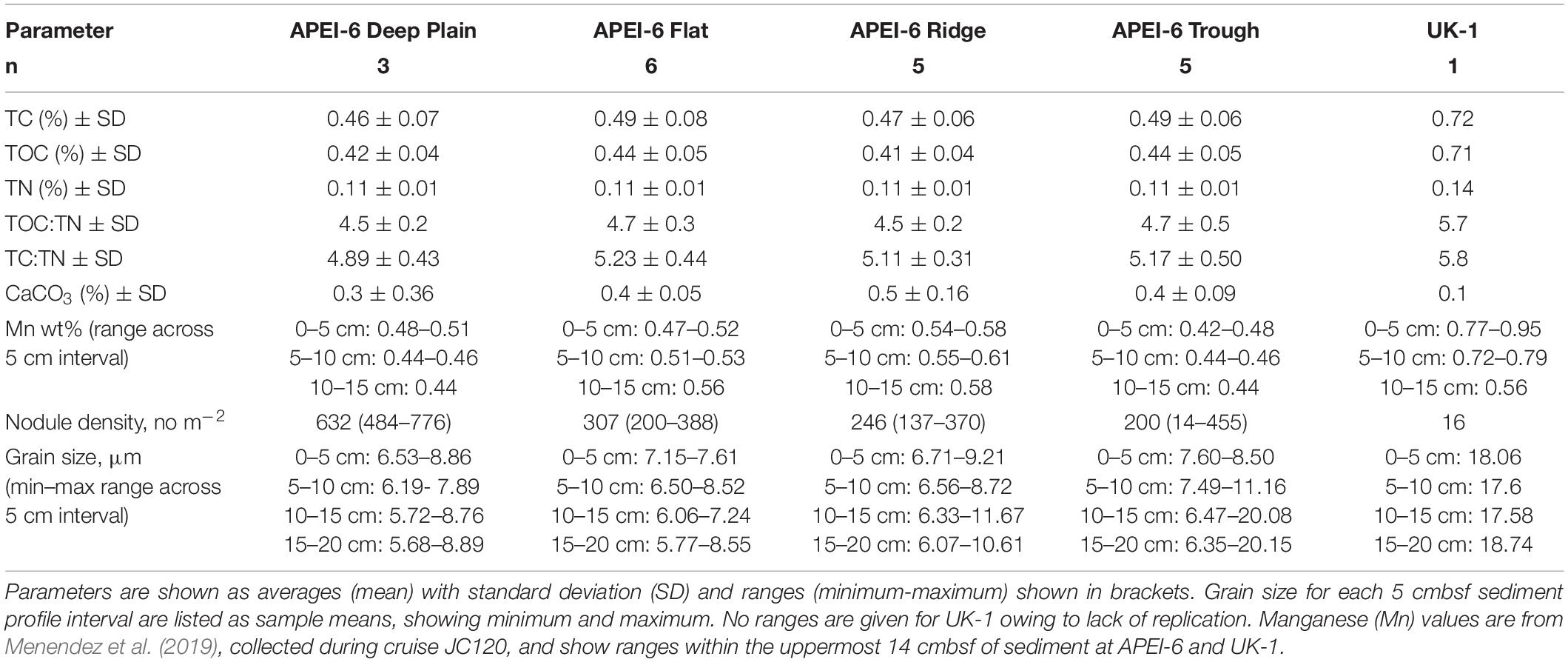
Table 3. Sediment biogeochemistry and environmental data for each APEI-6 SW landscape type and UK-1 (UK Seabed Resources Limited eastern contract area) which were collected during cruise JC120 (Jones et al., 2021, in prep.).
Microbial Assemblage Composition of Sediments at APEI-6 and UK-1
Overall, 32% of the community in the whole dataset consisted of Proteobacteria, including Gammaproteobacteria (13%), Alphaproteobacteria (12%), Deltaproteobacteria (5%), and Betaproteobacteria (1.5%) (Figure 4). Sequences assigned to the Thaumarchaeota phylum comprised a significant proportion of the sediment microbial assemblages overall (UK-1 = 20%, APEI-6 = 19%). This proportion was lower in the nodules at APEI-6 (11%). The dominant class within Thaumarchaeota was Nitrososphaeria, which composed 12–23% of sediment communities and 11% of the nodule-associated communities. Within both sediment and nodule communities, the most dominant genera within Nitrososphaeria was Candidatus Nitrosopumilus, which comprised on average 12% of the sediment communities and 7% of the nodule-associated assemblages (Figure 5). Within sediments, the highest abundance of Nitrosopumilus was in the surface sediments, ranging from 10–14.5% (±2.10) across all topographical landscapes in APEI-6 and within UK-1. Overall, Nitrosopumilus was more enriched in UK-1 sediments (10%) than those of APEI-6 (5%). The highest abundances of Nitrosopumilus were found at Trough in APEI-6 (14.5% ± 1.29) and UK-1 (14%) in the top cm of sediment.
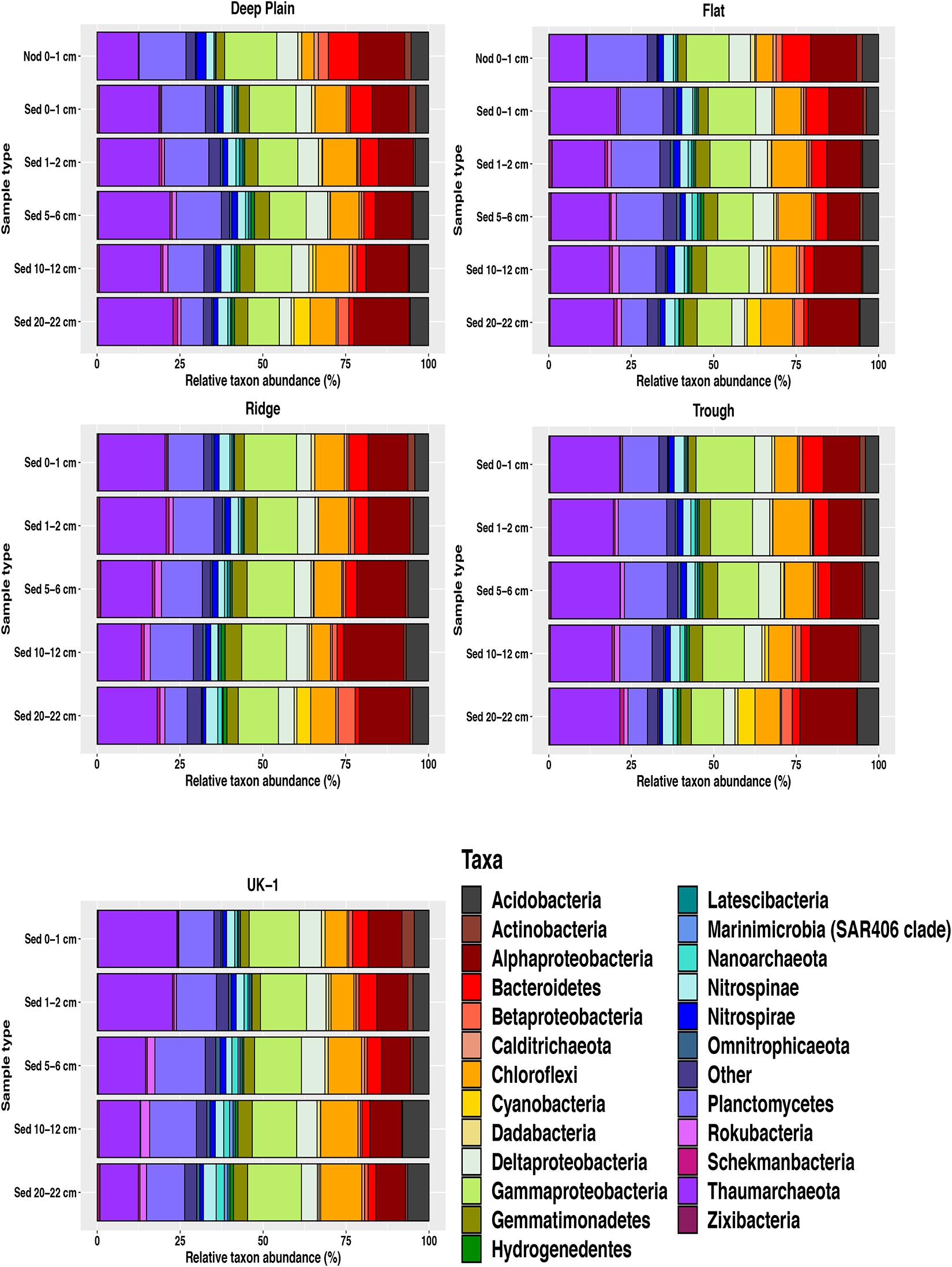
Figure 4. Relative abundances of amplicon sequence variants (ASVS) at phylum level nodule-associated and sediment (0–22 cm) from APEI-6 and UK-1 in the CCZ. Nodules are from the surface sediments (0–1 cm) of the Deep Plain and Flat regions of APEI-6. Only taxa ≥1% relative abundance are shown. Category “Other” represents taxa <1% relative abundance. Relative abundances were calculated on samples that had first been normalized to account for uneven sampling depth, by scaling sequence counts by library sizes.
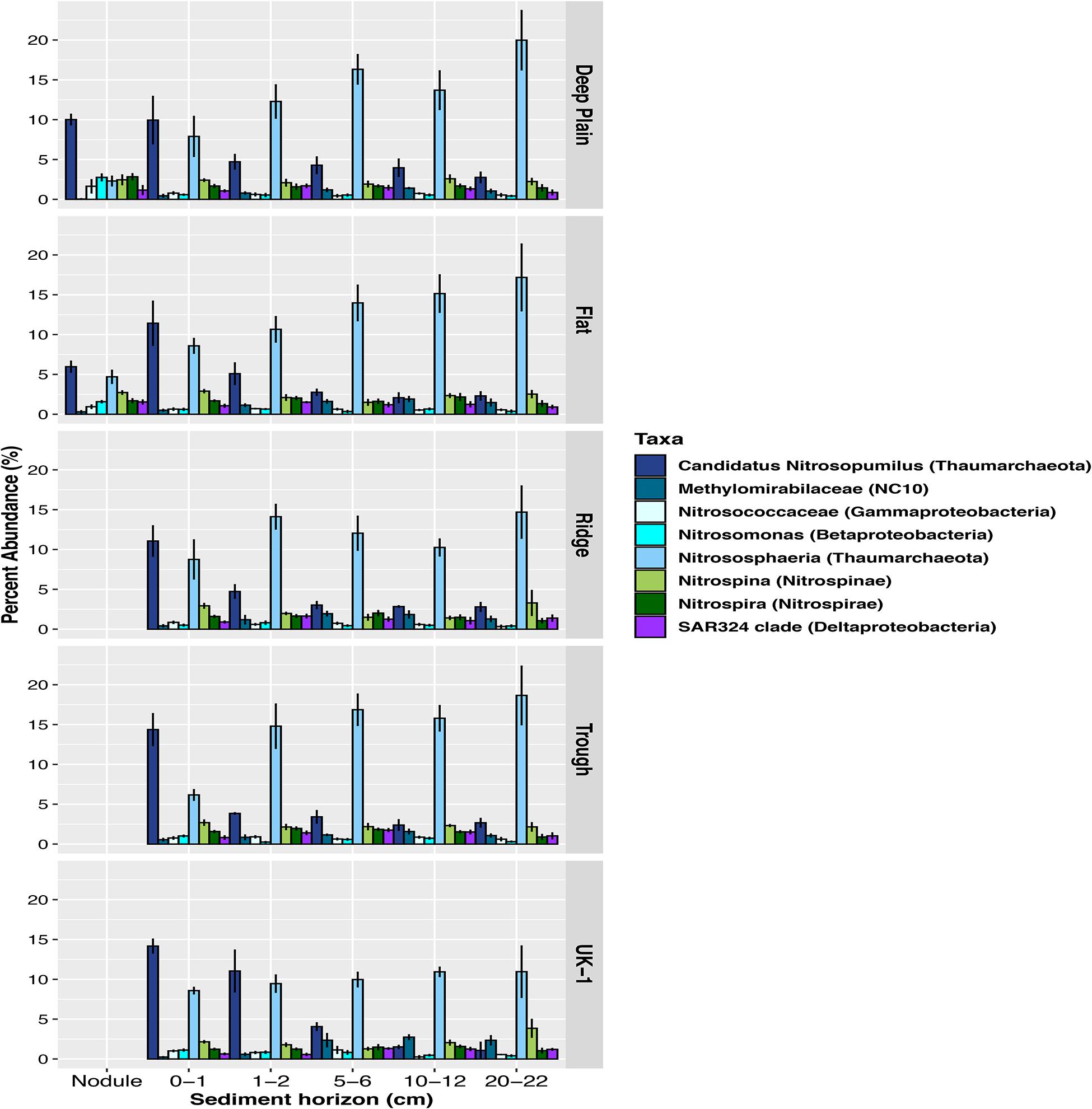
Figure 5. Relative abundances of amplicon sequence variants (ASVs) of the most abundant putative carbon-fixing and nitrifying microbes in sediments and nodules within each region at APEI-6 and at UK-1, defined at genera or family level. The class Nitrososphaeria includes all taxa within that class excluding the genera Candidatus Nitrosopumilus, which is shown in a separate category. Only taxa ≥1% relative abundance are shown. Relative abundances were calculated on samples that had first been normalized to account for uneven sampling depth, by scaling sequence counts by library sizes.
Other prevalent groups that were also well-represented in sediment assemblages at phylum level were Planctomycetes (11.5%), Chloroflexi (9%), Acidobacteria (5%), Bacteroidetes (4%), Gemmatimonadetes (4%), Nitrospinae (3%), Nitrospirae (2%), and Actinobacteria (1%). Abundant taxa at class level in sediment assemblages in APEI-6 and UK-1 included Dehalococcoidia (Chloroflexi, 7%), Phycisphaerae (Planctomycetes, 7%), Bacteroidia (Bacteroidetes, 3%), Nitrospinia (Nitrospinae, 2.5%), Gemmatimonadetes (Gemmatimonadetes, 2%), Nitrospira (Nitrospirae, 1.5%), Planctomycetacia (Planctomycetes, 1.5%), and OM190 (Planctomycetes, 1.5%). The most dominant genera in sediment assemblages were Ca. Nitrosopumilus (6.5–10%), the Urania-1B-19 marine sediment group (Planctomycetes, Phycisphaerae, 3%), uncultured bacterium in the Kiloniellaceae family (Alphaproteobacteria, 2.5–3%), Woeseia (Gammaproteobacteria (2.5–3%), uncultured Nitrosopumilales archaeon in the Nitrososphaeria class (Thaumarchaeota, 2–2.5%), uncultured bacterium in the Magnetospiraceae family (Alphaproteobacteria, 1.5–2%), Nitrospina (Nitrospinae, 1.5%), and Nitrospira (Nitrospirae, 1.5%).
Microbial community composition differed among the vertical layers of the sediment, and these differences were influenced by topography (Table 2). For example, the abundance of taxa within the Nitrososphaeria class (Thaumarchaeota) varied through the sediment profile within each topographical region, explaining 37.4% of the variance in beta diversity (PERMANOVA, p = 0.001, F3,83 = 14.50, Table 2; Figure 5). However, at Deep Plain, Thaumarchaeota increased with depth through the sediment, whereas at Ridge, the opposite was observed. In contrast, at Flat and Trough, the top one cm of sediment contained a high abundance of Thaumarchaeota (>20% of sequences), which declined with depth to the mid-layers, then increased in abundance again in the 20–22 cm layer. At UK-1, the top 2 cm had the highest observed abundances of Thaumarchaeota (24%). However, the abundance of this group rapidly declined through the sediment profile, to 11.5% in the deepest layer. Certain groups of microbial taxa increased in abundance with increasing depth through the sediment profile. The abundance of Acidobacteria and Rokubacteria followed this pattern at both APEI-6 and UK-1. Rokubacteria were most abundant at UK-1 and peaked at 10–12 cm before declining in abundance in deeper layers. Alphaproteobacteria only increased in deeper layers at the APEI, whereas the reverse was true in UK-1 sediments. Cyanobacteria were largely absent in sediments, except for the 20–22 cm layer in APEI-6. Other groups such as Actinobacteria and Bacteroidetes decreased in abundance with depth through APEI-6 and UK-1 sediments. Gammaproteobacteria and Nitrospina declined in abundance in mid-layers (5–6 cm) of sediment at both locations, before increasing in deeper layers again (10–22 cm). Other taxa followed the opposite pattern, such as Chloroflexi, Gemmatimonadetes, Nitrospira, and Planctomycetes, which reached a peak in relative abundance in the 5–6 cm layers before declining deeper into the sediment profile.
Microbial Assemblage Composition of Nodules at APEI-6
Nodule-associated communities were dominated by a higher abundance of Proteobacteria than sediments, forming 37% of total sequences, compared to 32% in sediments, driven by the classes Gammaproteobacteria (14%), Alphaproteobacteria (14%), Deltaproteobacteria (6.5%), and Betaproteobacteria (2%). The dominance of Gammaproteobacteria was driven by the families Thiohalorhabdaceae, Woeseiaceae (Woeseia genera), Arenicellaceae and the KI89A clade. Within Alphaproteobacteria, the most abundant families were Kiloniellaceae and Magnetospiraceae and the most represented groups within Deltaproteobacteria were the Orders Myxococcales and the SAR324 clade. Betaproteobacteria were largely dominated by ASVs affiliated with Nitrosomonas genera. Nodule communities also had a higher abundance of the Planctomycetes phylum, which formed 17% of assemblages associated with nodules, compared to 11.5% of sequences in sediments. This difference was driven by the enrichment of classes Phycisphaerae, OM190, Planctomycetacia and Pla4 lineage. The Bacteroidetes phylum was also more abundant in nodule communities, forming 9% of nodule communities compared to 4% in sediment communities and enriched by uncultured genera in the Rhodothermaceae family and the Cyclobacteriaceae family. Nodule-associated assemblages were also depleted in certain phyla compared to sediments, namely Thaumarchaeota (11% nodules, 19% sediments), Chloroflexi (5% nodules, 9% sediments), Gemmatimonadetes (2.5% nodules, 4% sediments), and Rokubacteria (0.2% nodules, 1% sediments).
We also observed differences in community composition and abundance of the main taxa found within the microbiomes of nodules retrieved from Deep Plain and Flat (Supplementary Tables 1, 2). The most dominant genera in nodule-associated communities, Ca. Nitrosopumilus, on average formed 10% relative abundance from Deep Plain nodules, and 6% from nodules from Flat (Figure 5). The family Thiohalorhabdaceae within the Gammaproteobacteria, formed 5% relative abundance in Deep Plain nodules, compared with 2% in Flat nodules. There was also a greater number of taxa that exceeded 1% relative abundance in nodule assemblages from Deep Plain than Flat. Groups such as Cohaesibacter (Alphaproteobacteria), AqS1 (Gammaproteobacteria), Pelagibius (Alphaproteobacteria), JdFR-76 (Calditrichaeota) and LS-NOB (Nitrospinae) exceeded 1% relative abundance in Deep Plain nodule assemblages but were below the 1% cut-off in nodules from Flat. Aquibacter (Bacteroidetes) and the Pir4 lineage (Planctomycetes) were present only in the most abundant taxa in nodule-associated assemblages from Flat but not Deep plain nodules.
Discussion
Spatial Differences in Benthic Assemblages Between Substrates
Microbial assemblages in sediments were more diverse than those associated with polymetallic nodules at both APEI-6 and UK-1, a finding that agrees with studies conducted in other areas of the Pacific (Tully and Heidelberg, 2013; Wu et al., 2013; Zhang et al., 2014; Shulse et al., 2016; Lindh et al., 2017; Molari et al., 2020). Microbes related to potential metal-cycling (Magnetospiraceae and Kiloniellaceae), organic carbon remineralization (Woeseia), and sulfur-oxidizing Thiohalorhabdaceae were also more enriched in nodules than sediments. Other ASVs that were more prevalent in nodules included AqS1, Cohaesibacter, JdFR-76, Pelagibius, Nitrospina, and Nitrospira. These ASVs have been previously reported in other nodule-associated communities (Wu et al., 2013; Blöthe et al., 2015; Molari et al., 2020).
Spatial Differences in Sediment Benthic Assemblages
Previously, abyssal plain habitats were considered to be homogenous landscapes, and the microbial constituents inhabiting the sediments were assumed to be cosmopolitan in their distribution. Yet, the distributions of metazoans, such as megafauna (Durden et al., 2015, 2020; Leitner et al., 2017; Simon-Lledó et al., 2019a), macrofauna (Cosson et al., 1997), and meiofauna (Stefanoudis et al., 2016), are influenced by seafloor heterogeneity.
Bathymetric variation of the seafloor can modify bottom current velocities, which in turn effect local sedimentation rates (Mewes et al., 2014) and POM deposition (Durden et al., 2015; Morris et al., 2016). In the CCZ, however, environmental features were generally homogenous between the landscape regions. Grain size distribution, relative proportions of TC, TOC, TN, TOC:TN, TC:TN, CaCO3, and Mn did not vary significantly between the landscape regions of this area (Table 3; Menendez et al., 2019; Simon-Lledó et al., 2019a), which has also been reported in nearby areas of the CCZ (Khripounoff et al., 2006; Volz et al., 2018). Sediments at the CCZ have been reported to be oxygenated (50 μmol L–1) down to 2–3 m depth (Haeckel et al., 2001; Mewes et al., 2014; Volz et al., 2018; Menendez et al., 2019), and across the four landscape types at APEI-6, the oxygen penetration depth (OPD) was similar, at >2.0 m depth (Menendez et al., 2019; Jones et al., 2021, in prep.). One factor that did vary between the landscape regions, however, was nodule density across the seafloor, which was twice as high at Deep Plain, compared to Flat, Ridge and Trough (Table 3; Simon-Lledó et al., 2019a).
At APEI-6, we observed spatial differences in the structure and composition of sediment assemblages, across small scales (cm), through the sediment depth profiles, to mesoscales (m to km) between cores within topographical regions. Across a range of 100’s meters to kilometers, the structure of assemblages varied between coring locations within each topographical region, a phenomenon previously observed in meiofauna (Stefanoudis et al., 2016).
Benthic assemblages also varied at larger scales of hundreds of km, between APEI-6 and UK-1. However, the lack of replicates at UK-1 prevented robust statistical analysis. We noted higher alpha diversity at UK-1 compared to APEI-6, a pattern also reported by Lindh et al. (2017). Putative chemolithoautotrophic microbes, such as Nitrosopumilus, were more prevalent in UK-1 sediments, particularly in surface layers. Another group, non-photosynthetic Cyanobacteria, Melainabacteria group bacterium S15B-MN24 CBMW_12, was found in the deeper sediment layers of APEI-6 but were largely absent from UK-1 sediments. Differences in assemblages between the two locations may be driven by varying biogeochemistry. The chemical composition of sediments at UK-1 differs from that at APEI-6, with higher proportions of total carbon (TC), total organic carbon (TOC), total nitrogen (TN), TOC:TN and TC:TN ratios, and CaCO3 (Table 3), as well as minerals Mn, Ni, and Cu in the sediments of UK-1 (Menendez et al., 2019). Differences in the biogeochemistry of sediments of the neighboring APEI-3 compared to contract areas, as well as lower faunal abundances have also been reported (Volz et al., 2018). The UK-1 area is also closer to the equatorial region of higher primary production in the overlying waters than APEI-6, giving rise to a higher flux of POC to the sediments (Menendez et al., 2019). As a result, the oxygen penetration depth (OPD) in UK-1 sediments is shallower (∼150 cmbsf) than it is at APEI-6 (>300 cmbsf) (Menendez et al., 2019), which is likely to influence benthic assemblage structure and composition.
High Abundance of Chemoautotrophic and Nitrogen Cycling Taxa
In the dark ocean, it is now recognized that chemolithoautotrophic microbes form a significant source of organic carbon through inorganic carbon fixation (Middelberg, 2011; Swan et al., 2011; Molari et al., 2013; Tully and Heidelberg, 2016; Sweetman et al., 2019), which could be equal to or slightly greater than phytoplankton export production in some regions (Reinthaler et al., 2010). Nitrification is an important process in benthic nitrogen cycling and accounts for a large proportion of the chemoautotrophic carbon fixation in the deep ocean (Herndl et al., 2005; Wuchter et al., 2006; Middelberg, 2011). Although not well constrained, using reduced compounds to drive energy for carbon fixation, alongside anabolic processes of inorganic carbon incorporation, may be potentially important strategies for supplementing recalcitrant carbon in oligotrophic deep-sea sediments (Brunnegård et al., 2004; Molari et al., 2013; Sweetman et al., 2019; Trembath-Reichert et al., 2021). Ammonia (NH3) released from organic matter decay is converted to nitrite (NO2) mediated by ammonia-oxidizing archaea (AOA) or ammonia-oxidizing bacteria (AOB), in the first step (Bock and Wagner, 2013). The second reaction, the oxidation of nitrite to nitrate (NO3) is catalyzed by nitrite-oxidizing bacteria (NOB), providing an important source of organic material to the benthic community (Middelberg, 2011). Indeed, benthic impact experiments have indicated that microbial nitrification was reduced following sediment disturbance and plowing (Vonnahme et al., 2020), hence likely impacting nutrient cycling and the availability of labile organic matter (Haeckel et al., 2001).
Across all four of the topographical landscape types of APEI-6 and within UK-1, both sediment and nodule-associated communities were dominated by putatively chemoautotrophic and nitrogen-cycling microbes, up to 29% in sediments and 21% from nodules (Figure 6). We observed a community of nitrifying microbes at CCZ, that was numerically dominated by AOA Nitrosopumilus (Nitrososphaeria class), which oxidize ammonia to nitrite (Könneke et al., 2005) and have previously been observed in deep-sea sediments in this region, despite the low input of phytodetritus (Nitahara et al., 2011, 2017; Shulse et al., 2016). There were also other nitrifiers present, including the AOA: Candidatus Nitrosopelagicus, Candidatus Nitrosopumilus, Candidatus Nitrosotenuis, taxa within the Nitrososphaeria class; ammonia-oxidizing bacteria (AOB) Nitrosomonas (Betaproteobacteria), and AqS1 (Gammaproteobacteria); and nitrite-oxidizing bacteria (NOB) Nitrospina (Nitrospinae) and Nitrospira (Nitrospirae).
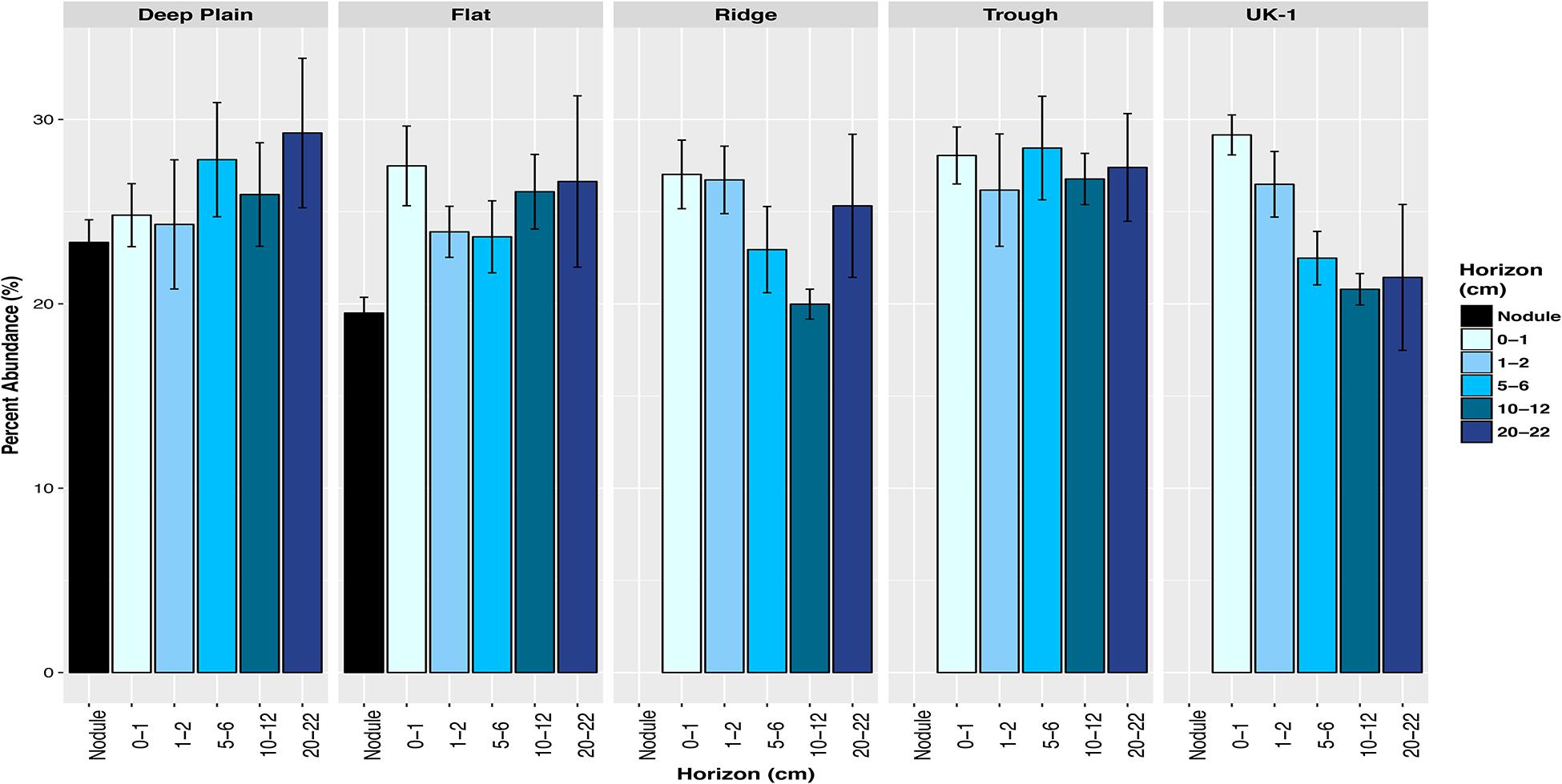
Figure 6. Total relative abundances of amplicon sequence variants (ASVs) of putative carbon-fixing and nitrifying microbes in sediments and nodule-associated communities at APEI-6 and UK-1. Error bars represent standard deviation of the 3 or more replicates for each category. Relative abundances were calculated on samples that had first been normalized to account for uneven sampling depth, by scaling sequence counts by library sizes.
The relatively high abundances of sequences corresponding to AOA Ca. Nitrosopumilus and NOB genera Nitrospina and Nitrospira have also been reported in other oligotrophic sediments with Fe-Mn deposits in the South Pacific Gyre (Tully and Heidelberg, 2016; Kato et al., 2019), in the CCZ (Nitahara et al., 2011, 2017: Shulse et al., 2016) and in the Peru Basin (Molari et al., 2020). The high proportions of these AOA and NOB groups at CCZ suggest a coupling between ammonia and nitrite oxidation. This co-occurrence has also been noted in marine environments, from water column (Mincer et al., 2007; Santoro et al., 2010), to shallow sediments (Herbert, 1999; Mills et al., 2008) and deep sediments (Shulse et al., 2016; Tully and Heidelberg, 2016; Molari et al., 2020) to freshwater environments such as oligotrophic lakes (Parro et al., 2019). For the AOA at least, greater numbers have been reported in oligotrophic sediments with low ammonium concentrations, and as AOA have a higher affinity for ammonia, they outcompete AOB in these environments (Martens-Habbena et al., 2009). The nitrate produced by the NOB may be used in other microbial metabolisms, such as oxidation of organic molecules and reduced sulfur compounds (Parro et al., 2019).
In addition to nitrification, carbon fixation in abyssal ecosystems might take place through other metabolic pathways and microbial lineages. Sediment assemblages at APEI-6 and UK-1 contained the methanotrophic taxon wb1-A12 (family Methylomirabilaceae, phylum NC10). This group are nitrite-dependent anaerobic methane oxidizing (N-damo) bacteria that utilize nitrate or nitrite as an electron acceptor to oxidize methane into CO2 (Ettwig et al., 2010). In addition, we observed the SAR324 clade (Deltaproteobacteria) in sediment assemblages, a metabolically flexible taxon capable of heterotrophy as well as autotrophy, that is capable of oxidizing reduced sulfur compounds to fix inorganic carbon (Swan et al., 2011; Sheik et al., 2014). Hence, the ubiquity of these microbial groups, in particular the nitrifiers, in the sediment might provide a source of organic carbon and nitrate for the benthic communities at CCZ. Indeed, microbial carbon incorporation mechanisms in deep-sea environments that lack high quantities of reduced compounds are variable (Trembath-Reichert et al., 2021) and warrant further study in these environments.
As many of the microbial taxa in the CCZ have neither been cultured or genomically sequenced, and with few in situ microbial activity or cultivation experiments available for this region (Gillard et al., 2019; Sweetman et al., 2019; Vonnahme et al., 2020), it is difficult to definitely infer metabolic capability from 16S sequences alone. For this reason, metagenomics should be conducted to confirm the metabolic functions that we hypothesize, here, and to link those back to taxonomic markers. Developing a comprehensive microbial ecology of the CCZ will contribute to our knowledge of microbially mediated biogeochemical cycling and will help to develop a mechanistic understanding of the contribution that benthic microbial communities provide to the overall resilience of nodule-associated communities.
Conclusion
Differences in microbial community composition at APEI-6 were predominantly driven by depth through the sediment profile and seafloor heterogeneity at small and mesoscales. We observed a diverse and varied microbial assemblage through the sediment, both within APEI-6 and at UK-1. Communities differed between nodules and sediments. The predominance of nitrifying and putative carbon-fixing microbes within sediment assemblages suggests a key ecological role of these groups of taxa in the CCZ, and our results emphasize the importance of microbial communities in mediating carbon, nitrogen and sulfur cycling in this habitat.
Understanding the diversity and functional roles of benthic microbes is vital for the sustainable use and preservation of ecosystems targeted for commercial-scale deep-sea mining. Therefore, ascertaining the composition and diversity of benthic microbial assemblages in both APEI-6 and mining exploration contract regions of the CCZ will be essential ahead of mining activities and will be a useful tool for monitoring in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject ID PRJNA694451.
Author Contributions
CY and DJ performed sampling. AH processed the samples, analyzed the data, and wrote the manuscript with the contribution from all authors. All authors conceived the study.
Funding
This work was part of the Managing Impacts of Deep-seA reSource exploitation (MIDAS) European Union Seventh Framework Programme (FP7/2007-2013; grant agreement no. 603418) and UK National Environmental Research Council (NERC) Seabed Mining And Resilience To EXperimental impact (SMARTEX) project (Grant Reference NE/T003537/1). Funding was jointly supported by a studentship from the NERC (Grant NE/L002531/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the captain, crew and scientists of the RRS James Cook for their assistance during the JC120 research cruise. We are grateful to P. Lam for helpful advice and suggestions with data analysis and interpretation. We are grateful to R. Jeffreys, University of Liverpool, for carrying out the sediment biogeochemical analysis. We would like to thank N. Pratt and A. Baylay for their technical support and assistance with Illumina sequencing (Environmental Sequencing Facility, National Oceanography Centre, Southampton). We also thank two reviewers for their constructive and helpful comments and suggestions, which significantly improved the quality of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.663420/full#supplementary-material
Supplementary Table 1 | Most abundant taxa associated with polymetallic nodules sampled from the top cm of sediment in the “Deep plain” region of APEI-6. Taxa forming ≥1% average relative abundances (% of total sequences) are given along with standard deviation (SD).
Supplementary Table 2 | Most abundant taxa associated with polymetallic nodules sampled from the top cm of sediment in the “Flat” region of APEI-6. Taxa forming ≥1% average relative abundances (% of total sequences) are given along with standard deviation (SD).
Supplementary Table 3 | Pair-wise post hoc tests for fixed term “Sediment Horizon” for sediment microbial assemblages at APEI-6, showing the following: all 16S microbial assemblages; putative carbon-fixing and nitrifying taxa within sediment microbial assemblages; and taxa within the Nitrososphaeria class in the Thaumarchaeota phylum.
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070
Barnett, P. R. O., Watson, J., and Connelly, D. (1984). The multiple corer for taking virtually 712 undisturbed samples from shelf, bathyal and abyssal sediments. Oceanologica Acta 7, 399–408.
Blöthe, M., Wegorzewski, A., Müller, C., Simon, F., Kuhn, T., and Schippers, A. (2015). Manganese-cycling microbial communities inside deep-sea manganese nodules. Environ. Sci. Technol. 49, 7692–7700. doi: 10.1021/es504930v
Bock, E., and Wagner, M. (2013). “Oxidation of Inorganic Nitrogen Compounds as an Energy Source,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), doi: 10.1007/978-3-642-30141-4_64
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brunnegård, J., Grandel, S., Ståhl, H., Tengberg, A., and Hall, P. O. J. (2004). Nitrogen cycling in deep-sea sediments of the Porcupine Abyssal Plain, NE Atlantic. Prog. Oceanogr. 63, 159–181. doi: 10.1016/j.pocean.2004.09.004
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cosson, N., Sibuet, M., and Galeron, J. (1997). Community structure and spatial heterogeneity of the deep-sea macrofauna at three contrasting stations in the northeast Atlantic. Deep Sea Res. Part I. 44, 247–269. doi: 10.1016/S0967-0637(96)00110-0
De Jonge, D. S. W., Stratmann, T., Lins, L., Vanreusel, A., Purser, A., Marcon, Y., et al. (2020). Abyssal food-web model indicates faunal carbon flow recovery and impaired microbial loop 26 years after a sediment disturbance experiment. Prog. Oceanogr. 2020:102446. doi: 10.1016/j.pocean.2020.102446
Durbin, A. M., and Teske, A. (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
Durden, J. M., Bett, B. J., and Ruhl, H. A. (2020). Subtle variation in abyssal terrain induces significant change in benthic megafaunal abundance, diversity, and community structure. Prog. Oceanogr. 186, 1–11. doi: 10.1016/j.pocean.2020.102395
Durden, J. M., Bett, B. J., Jones, D. O. B., Huvenne, V. A. I., and Ruhl, H. A. (2015). Abyssal hills – hidden source of increased habitat heterogeneity, benthic megafaunal biomass and diversity in the deep sea. Prog. Oceanogr. 137, 209–218. doi: 10.1016/j.pocean.2015.06.006
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–550. doi: 10.1038/nature08883
Gillard, B., Chatzievangelou, D., Thomsen, L., and Ullrich, M. S. (2019). Heavy-metal-resistant microorganisms in deep-sea sediments disturbed by mining activity: an application toward the development of experimental in vitro systems. Front. Mar. Sci. 6, 1–12. doi: 10.3389/fmars.2019.00462
Gjerde, K. M., Weaver, P., Billett, D., Paterson, G., Colaco, A., Dale, A., et al. (2016). Implications of MIDAS Results for Policy Makers: Recommendations for Future Regulations. Managing Impacts of Deep Sea Resource Exploitation Consortium. 16. Available online at: https://www.eu-midas.net/sites/default/files/downloads/MIDAS_recommendations_for_policy_lowres.pdf (accessed date 20th October 2020)
Haeckel, M., König, I., Reich, V., Weber, M. E., and Suess, E. (2001). Pore water profiles and numerical modelling of biogeochemical processes in Peru Basin deep-sea sediments. Deep-Sea Res. Pt. II. 48, 3713–3736. doi: 10.1016/S0967-0645(01)00064-9
Herbert, R. A. (1999). Nitrogen cycling in coastal marine ecosystems. FEMS 23, 563–590. doi: 10.1111/j.1574-6976.1999.tb00414.x
Herndl, G. J., Reithaler, T., Teira, E., van Aken, H., Veth, C., Pernthaler, A., et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71, 2303–2309. doi: 10.1128/aem.71.5.2303-2309.2005
Ingels, J., Vanreusel, A., Pape, E., Pasotti, F., Macheriotou, L., Arbizu, P. M., et al. (2020). Ecological variables for deep-ocean monitoring must include microbiota and meiofauna for effective conservation. Nat. Ecol. Evol. 2020:1335. doi: 10.1038/s41559-020-01335-6
Jones, D. O. B., Amon, D. J., and Chapman, A. S. A. (2018). Mining Deep-Ocean Mineral Deposits: What are the Ecological Risks? Elements 14, 325–330. doi: 10.2138/gselements.14.5.325
Jones, D. O. B., Amon, D. J., and Chapman, A. S. A. (2020). “Chapter 5: Deep-sea mining: processes and impacts,” in Natural Capital and Exploitation of the Deep Ocean, eds M. Baker, E. Ramirez-Llodra, and P. Tyler (Oxford: Oxford University Press).
Jones, D. O. B., Kaiser, S., Sweetman, A. K., Smith, C. R., Menot, L., Vink, A., et al. (2017). Biological responses to disturbance from simulated deep-sea polymetallic nodule mining. PLoS One 12:e0171750. doi: 10.1371/journal.pone.0171750
Jones, D. O. B., Simon-Lledó, E., Amon, D. J., Bett, B. J., Caulle, C., Clement, L., et al. (2021). Environment, ecology, and potential effectiveness of an area protected from deep-sea mining (Clarion Clipperton Zone, abyssal Pacific).
Kato, S., Hirai, M., Ohkuma, M., and Suzuki, K. (2019). Microbial metabolisms in an abyssal ferromanganese crust from the Takuyo-Daigo Seamount as revealed by metagenomics. PLoS One 2019:224988. doi: 10.1371/journal.pone.0224888
Kerr, R. A. (1984). Manganese nodules grow by rain from above. Science 223, 576–577. doi: 10.1126/science.223.4636.576
Khripounoff, A., Caprais, J. C., Crassous, P., and Etoubleau, J. (2006). Geochemical and biological recovery of the disturbed seafloor in polymetallic nodule fields of the Clarion-Clipperton Fracture Zone (CCFZ) at 5,000 m depth. Limnol. Oceanogr. 51, 2033–2041. doi: 10.4319/lo.2006.51.5.2033
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidising marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Leitner, A. B., Neuheimer, A. B., Donlon, E., Smith, C. R., and Drazen, J. C. (2017). Environmental and bathymetric influences on abyssal bait-attending communities of the Clarion Clipperton Zone. Deep Sea Res. Part I. 125, 65–80. doi: 10.1016/j.dsr.2017.04.017
Levin, L. A., Mengerink, K., Gjerde, K. M., Rowden, A. A., Van Dover, C. L., Clark, M. R., et al. (2016). Defining “serious harm” to the marine environment in the context of deep-seabed mining. Mar. Policy 74, 245–259. doi: 10.1016/j.marpol.2016.09.032
Lindh, M. V., Maillot, B. M., Shulse, C. N., Gooday, A. J., Amon, D. J., Smith, C. R., et al. (2017). From the surface to the deep-sea: bacterial distributions across polymetallic nodule fields in the Clarion-Clipperton Zone of the Pacific Ocean. Front. Microbiol. 8, 1–12. doi: 10.3389/fmicb.2017.01696
Lodge, M., Johnson, D., Le Gurun, G., Wengler, M., Weaver, P., and Gunn, V. (2014). Seabed mining: International Seabed Authority environmental management plan for the Clarion-Clipperton Zone. A partnership approach. Mar. Policy 49, 66–72. doi: 10.1016/j.marpol.2014.04.006
Macdonald, K. C., Fox, P. J., Alexander, R. T., Pockalny, R., and Gente, P. (1996). Volcanic growth faults and the origin of Pacific abyssal hills. Nature 380:125. doi: 10.1038/380125a0
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Menendez, A., James, R. H., Lichtschlag, A., Connelly, D., and Peel, K. (2019). Controls on the chemical composition of ferromanganese nodules in the Clarion-Clipperton Fracture Zone, eastern equatorial Pacific. Mar. Geo. 409, 1–14. doi: 10.1016/j.margeo.2018.12.004
Mewes, K., Mogollón, J. M., Picard, A., Rühlemann, C., Kuhn, T., Nöthen, K., et al. (2014). Impact of depositional and biogeochemical processes on small scale variations in nodule abundance in the Clarion-Clipperton Fracture Zone. Deep Sea Res. Part I. 91, 125–141. doi: 10.1016/j.dsr.2014.06.001
Middelberg, J. J. (2011). Chemoautotrophy in the ocean. Geophys. Res. Lett. 38, 1–4. doi: 10.1029/2011GL049725
Mills, H. J., Hunter, E., Humphrys, M., Kerkhof, L., McGuiness, L., Huettel, M., et al. (2008). Characterization of nitrifying, denitrifying and overall bacterial communities in permeable marine sediments of the Northeastern Gulf of Mexico. Appl. Environ. Microbiol. 74, 4440–4453. doi: 10.1128/aem.02692-07
Mincer, T. J., Church, M. J., Trent Taylor, L., Preston, C., Karl, D. M., and DeLong, E. F. (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x
Molari, M., Janssen, F., Vonnahme, T., Wenzhöfer, F., and Boetius, A. (2020). Microbial communities associated with sediment and polymetallic nodules of the Peru Basin. Biogeosciences 2020:11. doi: 10.5194/bg-2020-11
Molari, M., Manini, E., and Dell’Anno, A. (2013). Dark inorganic carbon fixation sustains the functioning of benthic deep-sea ecosystems. Global Biogeochem. Cycles. 27, 212–221. doi: 10.1002/gbc.20030
Morris, K. J., Bett, B. J., Durden, J. M., Benoist, N. M. A., Huvenne, V. A. I., Jones, D. O. B., et al. (2016). Landscape-scale spatial heterogeneity in phytodetrital cover and megafauna biomass in the abyss links to modest topographical variation. Sci. Rep. 6, 1–10. doi: 10.1038/srep34080
Nitahara, S., Kato, S., Urabe, T., Usui, A., and Yamagishi, A. (2011). Molecular characterization of the microbial community in hydrogenetic ferromanganese crusts of the Takuyo-Daigo Seamount, northwest Pacific. FEMS 321, 121–129. doi: 10.1111/j.1574-6968.2011.02323.x
Nitahara, S., Kato, S., Usui, A. S., Urabe, T., Suzuki, K., and Yamagishi, A. (2017). Archaeal and bacterial communities in deep-sea hydrogenetic ferromanganese crusts on old seamounts of the northwestern Pacific. PLoS One 12:e0173071. doi: 10.1371/journal.pone.0173071
Oksanen, J. (2007). Multivariate analyses of ecological communities in R: vegan tutorial. 39. Available online at: http://cc.oulu.fi/jarioksa/opetus/metodi/vegantutor.pdf (accessed on August 15th, 2020).
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). Vegan: Community ecology package. R Package version 2.4-2.
Parro, V., Puente-Sánchez, F., Cabrol, N. A., Gallardo-Carreño, I., Moreno-Paz, M., Blanco, Y., et al. (2019). Microbiology and nitrogen cycle in the benthic sediments of a glacial oligotrophic deep Andean lake as analog of ancient Martian lake-beds. Front. Microbiol. 10, 1–14. doi: 10.3389/fmicb.2019.00929
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Reinthaler, T., van Aken, H. M., and Herndl, G. J. (2010). Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep Sea Res. II. 57, 1572–1580. doi: 10.1016/j.dsr2.2010.02.023
Rolinski, S., Segschneider, J., and Sündermann, J. (2001). Long-term propagation of tailing from deep-sea mining under variable conditions by means of numerical simulations. Deep Sea Res. Part I. 48, 3469–3485. doi: 10.1016/S0967-0645(01)00053-4
Ruhl, H. A., Ellena, J. A., and Smith, K. L. Jr. (2008). Connections between climate, food limitation, and carbon cycling in abyssal sediment communities. PNAS 105, 17006–17011. doi: 10.1073/pnas.0803898105
Santoro, A. E., Casciotti, K. L., and Francis, C. A. (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12, 1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x
Sheik, C. S., Jain, S., and Dick, G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Shulse, C. N., Maillot, B., Smith, C. R., and Church, M. J. (2016). Polymetallic nodules, sediments and deep waters in the North Pacific exhibit highly diverse and distinct bacterial, archaeal and microeukaryotic communities. MicrobiologyOpen 6, 1–16. doi: 10.1002/mbo3.428
Simon-Lledó, E., Bett, B. J., Huvenne, V. A. I., Schoening, T., Benoist, N. M. A., Jeffreys, R. M., et al. (2019a). Megafaunal variation in the abyssal landscape of the Clarion Clipperton Zone. Progr. Oceanogr. 170, 119–133. doi: 10.1016/j.pocean.2018.11.003
Simon-Lledó, E., Bett, B. J., Huvenne, V. A. I., Schoening, T., Benoist, N. M. A., and Jones, D. O. B. (2019b). Ecology of a polymetallic nodule occurrence gradient: Implications for deep-sea mining. Limnol. Oceanogr. 64, 1883–1894. doi: 10.1002/lno.11157
Smith, C. R., De Leo, F. C., Bernardino, A. F., Sweetman, A. K., and Martinez Arbizu, P. (2008b). Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528. doi: 10.1016/j.tree.2008.05.002
Smith, C. R., Gaines, S., Friedlander, A., Morgan, C., Thurnherr, A., Mincks, S., et al. (2008a). Preservation reference areas for nodule mining in the Clarion-Clipperton Zone: rationale and recommendations to the International Seabed Authority. Manoa. 2008:2008.
Smith, K. L., Ruhl, H. A., Bett, B. J., Billett, D. S. M., Lampitt, R. S., and Kaufmann, R. S. (2009). Climate, carbon cycling and deep-ocean ecosystems. PNAS 106, 19211–19218. doi: 10.1073/pnas.0908322106
Stefanoudis, P. V., Schiebel, R., Mallet, R., Durden, J. M., Bett, B. J., and Gooday, A. J. (2016). Agglutination of benthic foraminifera in relation to mesoscale bathymetric features in the abyssal NE Atlantic (Porcupine Abyssal Plain). Mar. Micropaleontol. 123, 15–28. doi: 10.1016/j.marmicro.2015.12.005
Swan, B. K., Martinez-Garcia, M., Preston, C. M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacterial lineages in the dark ocean. Science 333, 1296–1300. doi: 10.1126/science.1203690
Sweetman, A. K., Smith, C. R., Shulse, C. N., Maillot, B., Lindh, M., Church, M. J., et al. (2019). Key role of bacteria in the short-term cycling of carbon at the abyssal seafloor in a low particulate organic carbon flux region of the eastern Pacific Ocean. Limnol. Oceanogr. 64, 694–713. doi: 10.1002/lno.11069
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463.
Trembath-Reichert, E., Shah Walter, S. R., Fontánez Ortis, M. A., Carter, P. D., Girguis, P. R., and Huber, J. A. (2021). Multiple carbon incorporation strategies support microbial survival in cold subseafloor crustal fluids. Sci. Adv. 7:18.
Tully, B. J., and Heidelberg, J. F. (2013). Microbial communities associated with ferromanganese nodules and the surrounding sediments. Front. Microbiol. 4, 1–10. doi: 10.3389/fmicb.2013.00161
Tully, B. J., and Heidelberg, J. F. (2016). Potential Mechanisms for Microbial Energy Acquisition in Oxic Deep-Sea Sediments. Appl. Environ. Microbiol. 82, 4232–4243. doi: 10.1128/aem.01023-16
Volz, J. B., Mogollón, J. M., Geibert, W., Martínez Arbizu, P., Koschinsky, A., and Kasten, S. (2018). Natural spatial variability of depositional conditions, biogeochemical processes and element fluxes in sediment of the eastern Clarion-Clipperton Zone, Pacific Ocean. Deep Sea Res. Part I. 140, 159–172. doi: 10.1016/j.dsr.2018.08.006
Vonnahme, T. R., Molari, M., Janssen, F., Wenzhöfer, F., Haeckel, M., Tischack, J., et al. (2020). Effects of a deep-sea mining experiment on seafloor microbial communities and functions after 26 years. Sci. Adv. 6, 1–14.
Washburn, T. W., Jones, D. O. B., Wei, C.-L., and Smith, C. R. (2021). Environmental Heterogeneity Throughout the Clarion-Clipperton Zone and the Potential Representativity of the APEI Network. Front. Mar. Sci. 8:661685. doi: 10.3389/fmars.2021.661685
Wedding, L. M., Friedlander, A. M., Kittinger, J. N., Watling, L., Gaines, S. D., Bennett, M., et al. (2013). From principles to practice: a spatial approach to systematic conservation planning in the deep sea. Proc. R. Soc. B. 280, 1–10. doi: 10.1098/rspb.2013.1684
Wedding, L. M., Reiter, S. M., Smith, C. R., Gjerde, K. M., Kittinger, J. N., Friedlander, A. M., et al. (2015). Managing mining of the deep seabed. Science 349, 144–145.
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis, 2nd Edn. New York, NY: Springer Publishing Company, Incorporated.
Wu, Y. H., Liao, L., Wang, C. S., Ma, W. L., Meng, F. X., Wu, M., et al. (2013). A comparison of microbial communities in deep-sea polymetallic nodules and the surrounding sediments in the Pacific Ocean. Deep Sea Res. Part I. 79, 40–49. doi: 10.1016/j.dsr.2013.05.004
Wuchter, C., Abbas, B., Coolen, M. J. L., Herfort, L., van Bleijswijk, J., Timmers, P., et al. (2006). Archaeal nitrification in the ocean. PNAS 104, 12317–12322. doi: 10.1073/pnas.0600756103
Keywords: 16S rRNA, microbial diversity, nitrifiers, deep-sea mining, polymetallic nodules, sediment, Clarion-Clipperton Zone
Citation: Hollingsworth AL, Jones DOB and Young CR (2021) Spatial Variability of Abyssal Nitrifying Microbes in the North-Eastern Clarion-Clipperton Zone. Front. Mar. Sci. 8:663420. doi: 10.3389/fmars.2021.663420
Received: 02 February 2021; Accepted: 06 July 2021;
Published: 26 July 2021.
Edited by:
Adrian Glover, Natural History Museum (United Kingdom), United KingdomReviewed by:
Massimiliano Molari, Max Planck Institute for Marine Microbiology (MPG), GermanyGustavo Fonseca, Federal University of São Paulo, Brazil
Copyright © 2021 Hollingsworth, Jones and Young. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anita L. Hollingsworth, YS5sLmhvbGxpbmdzd29ydGhAc291dGhhbXB0b24uYWMudWs=
 Anita L. Hollingsworth
Anita L. Hollingsworth Daniel O. B. Jones
Daniel O. B. Jones C. Robert Young
C. Robert Young