- Key Laboratory of Applied Marine Biotechnology of Ministry of Education, School of Marine Sciences, Ningbo University, Ningbo, China
The P38 mitogen-activated protein kinase (MAPK) signal transduction pathway is widespread in organisms and plays important roles in immune activities. The infection mechanism of environmental gram-negative bacteria on crustaceans is an important scientific problem. In this study, the cDNA full-length sequence of Portunus trituberculatus P38 (PtP38) was cloned and its structure was analyzed by bioinformatics methods. To study the function of the PtP38 gene after a Gram-negative bacterial infection, we injected P. trituberculatus with LPS to activate the immune response instead of directly infecting with Gram-negative bacteria. With LPS stimulation, the expression of the PtP38 gene in different tissues increased significantly. At the same time, the expression of immune-related genes (ALF and crustin) in the hepatopancreas, activities of antioxidant enzymes [superoxide dismutase (SOD), catalase (CAT), and inducible nitric oxide synthase (iNOS) enzymes], and expression of apoptosis-related genes (caspase2 and caspase3) were increased significantly. To further conform the function of PtP38 in the immune response, we injected P. trituberculatus with P38 inhibitor and subsequently injected with LPS. The results showed that the expression of immune-related genes was inhibited, the activity of antioxidant enzymes was decreased, and the expression of apoptosis-related genes were inhibited. Thus, we speculated that PtP38 may increase the immune ability by improving the expression of antimicrobial peptides, increasing the activity of oxidative stress-related enzymes, and promoting cell apoptosis in infected P. trituberculatus. This study also laid the foundation for further study of the P38 MAPK signaling pathway and immune mechanism of P. trituberculatus.
Introduction
P38 mitogen-activated protein kinase (MAPK) is one of the important signaling pathways in the body (Seger and Krebs, 1995). It is widely present in various eukaryotes from yeast to humans (Lake et al., 2016) and can participate in inflammatory reactions, cell growth, cell differentiation, cell cycle, and cell death (Keshet and Seger, 2010). The P38 kinase activation site contains a conserved TGY (Thr-Gly-Tyr) motif (Ono and Han, 2000). Various extracellular stressors, such as UV, osmotic pressure, radiation, inflammatory cytokines, growth factors, and shock, as well as double phosphorylation of serine and threonine in the TGY motif, through the highly conserved “MAPKKK-MAPKK-MAPK” pathway, activate P38 MAPK (Cuenda and Rousseau, 2007; Yokota and Wang, 2016). Activated P38 MAPK directly or indirectly affects the activity of various transcription factors, specifically regulating the expression of multiple genes and, in turn, the cellular response.
In mammals, an activated P38 pathway promotes the production of inflammatory factors, such as TNF-a, IL-1, IL-4, IL-6, IL-8, and IL-12 (Guan et al., 1998), increases COX-2, iNOS, VCAM-1, and other proteins related to inflammation (Badger et al., 1998), promotes the proliferation and differentiation of immune cells (e.g., GM-CSF, EPO, CSF, and CD40) (Da Silva et al., 1997), and plays an important role in the immune mechanism of the organism. Intrathecal injection of the P38 MAPK antagonist SB203580 activated glucocorticoid receptors and decreased NF-κB, resulting in pain relief 3 days post-operation in spared nerve injury rats (Shao et al., 2018). Activation of the P38 MAPK stress response pathway can regulate insulin signaling and clear mitochondrial-produced ROS (Allahham et al., 2016). Relying on the P38 MAPK signal transduction, placental growth factor induces apoptosis and reduces gastric cancer cell migration (Akrami et al., 2016). The levels of P38 MAPK and P-P38 (Phosphorylated-P38) MAPK in the peripheral blood mononuclear cells of patients with pancreatitis were closely associated with the severity of illness and duration of disease (Wang and Li, 2016). P38 MAPK–hyaluronan-dependent reprogramming of the tumor microenvironment plays a critical role in driving lung tumorigenesis, while blocking this process could have far-reaching therapeutic implications (Brichkina et al., 2016).
Invertebrates rely mainly on the innate immune system to fight disease (Ren et al., 2017). The immune function of P38 in invertebrates is still in the preliminary research stage. LPS stimulation induces P38 protein phosphorylation in Drosophila, regulating the expression of various immune genes (Han et al., 1998). P38 mutant Drosophila is more susceptible to pathogenic bacteria and exhibits immunodeficiency (Chen et al., 2010; Chakrabarti et al., 2014; Cara et al., 2018). Pacific oyster Crassostrea gigas P38 is involved in the immune response by regulating the expression of inflammatory cytokines (Sun et al., 2019). After the in vivo infection with Vibrio splendidus, P38 mRNA expression in sea cucumber Apostichopus japonicus was significantly up-regulated (Zhan et al., 2018). After stimulation with Staphylococcus aureus, Vibrio harveyi, and White Spot Syndrome virus, the expression levels of Scylla paramamosain and Fenneropenaeus chinensis P38 MAPK were up-regulated (Yu et al., 2017). Additionally, knockdown of P38 by RNA interference (RNAi) resulted in a reduced expression of MKK3 (one of the P38 MAPKK) and ATF-2 (one of the P38 MAPK effector proteins) in Fenneropenaeus chinensis (He et al., 2018). However, no relevant study has been conducted on Portunus trituberculatus.
To study the roles of PtP38 in immune ability, PtP38 MAPK gene was cloned and characterized from P. trituberculatus. We detected the expression of antimicrobial peptides, activity of oxidative stress-related enzymes, and expressions of apoptosis related genes at the groups of LPS stimulation and P38 inhibitor and LPS stimulation. The results will lay the foundation for the further study of the P38 MAPK signaling pathway and immune mechanism in P. trituberculatus.
Materials and Methods
Laboratory Animals, Stimulation, and Sampling
The healthy P. trituberculatus used in this experiment was purchased from the Choupijiang Aquaculture Farm in Fenghua District, Ningbo City, Zhejiang Province, China, and weighed 250 (± 100) g. One hundred twenty crabs were randomly divided into three groups of 40 each, and they were housed in three indoor cement ponds. The natural seawater was continuously oxygenated. The water was regularly changed by replacing 1/3 of the original water. Feed bait was added regularly and dead crabs were removed. The crabs were acclimated for 1 week before the experiments.
LPS (Biyuntian Company) and the P38 inhibitor SB203580 (Biyuntian Company) were prepared as 2 and 1 mg/mL solutions in sterile PBS, respectively. The first group of crabs was weighed and then injected with 0.2 mL of LPS per 100 g of crab body weight from the soft membrane of the fourth step base (LPS group). The second group was injected with 0.2 mL of PBS per 100 g of crab body weight (control group). The third group was injected with 0.1 mL of SB203580 per 100 g of crab body weight 1 h ahead and then with 0.2 mL of LPS per 100 g of crab body weight as the SB203580 + LPS group (Yu et al., 2017). The culture conditions remained the same. Each group of crabs was sampled at 0, 3, 6, 12, 24, 48, 72, and 96 h after injection. Three crabs were taken at each time point. The heart, hepatopancreas, gills, muscles, and other tissues were collected and stored in liquid nitrogen.
Gene Cloning
The TRIzol method was used to extract the total RNA of P. trituberculatus. Agarose gel electrophoresis was used to detect the RNA integrity. The RNA concentration was measured using a nucleic acid quantifier. The extracted RNA was stored in a refrigerator at −80°C. First-strand cDNA was synthesized by reverse transcription, referring to the instructions of the Cwbio HiFiScript cDNA Synthesis Kit, and the product was stored at −20°C for future use.
Degenerate primers (Table 1) were designed and used in PCR of the first-strand cDNA. The PCR products were separated by gel electrophoresis and recovered by tapping. The purified product was ligated with the PMD18-T vector and transformed into E. coli DH5α competent cells. Flat, positive clones were picked by blue and white spot screening. After identification by M13F/M13R PCR, the bacterial solution was sent to a sequencing company to obtain the intermediate sequence of the gene.
After obtaining the intermediate fragment of the target gene, the full length of the target gene was obtained by RACE using 5′ RACE and 3′ RACE kits. Specific primers were designed based on the intermediate fragments (Table 1), and PCR was performed after reverse transcription. The 5′ and 3′ PCR products were separated by gel electrophoresis and were recovered by tapping. The purified product was ligated with the PMD18-T vector and transformed into E. coli DH5α competent cells. Flat, positive clones were picked by blue and white spot screening. After identification by M13F/M13R PCR, the bacterial solution was sent to a sequencing company to obtain the 5′ and 3′ ends of the target gene, thereby obtaining the full length of the P38 gene.
Bioinformatics Analysis
The ORF Finder1 website was used to predict the open reading frame of the P. trituberculatus P38 gene. The open reading frame was translated into amino acid sequences using the EMBL2 website. The SMART3 website was used to predict the P. trituberculatus P38 protein domain. The amino acid sequences of the P38 proteins from other species were obtained from the NCBI4 website, and multiple sequence alignments were performed using the Vector NTI software. A phylogenetic tree of the P38 protein from P. trituberculatus and other species was constructed using the MEGA5.1 software. The I-TASSER5 website was used to predict the secondary and tertiary structure of the P. trituberculatus P38 protein.
Quantitative PCR (qPCR) Analysis
RNA from the heart, hepatopancreas, gills, muscles, and other tissues of the control group at 0 h was extracted and used to analyze the tissue distribution of the PtP38 mRNA. RNA from the hepatopancreas, gills, and muscle tissues at each sampling time point of each experimental group was extracted for LPS stimulation experiments. Each group of samples was repeated in triplicate, and the quality and concentration of RNA were detected by agarose gel electrophoresis and nucleic acid quantifier. Referring to the Takara PrimeScriptTM RT reagent kit instruction manual, reverse transcription was performed to synthesize cDNA.
Based on the cDNA sequence of the PtP38 gene, fluorescent-specific primers were designed and synthesized (Table 1). Using GAPDH as an internal reference, PtP38 gene expression in each tissue was analyzed by qPCR using cDNA from the heart, hepatopancreas, gill, muscle, and other tissues of the control group at 0 h as the template.
Using the cDNA from the hepatopancreas, gills, and muscle tissues of each experimental group at each sampling time point as the template, qPCR was performed using GAPDH as an internal reference to detect the PtP38 gene expression after LPS stimulation.
The cDNA sequences of ALF, crustin, caspase2, and caspase3 were searched from the NCBI, and specific primers were designed and synthesized (Table 1). cDNA from the hepatopancreas was used as the template, and GAPDH was used as an internal reference. qPCR was used to detect the expression of ALF, crustin, caspase2, and caspase3 after LPS stimulation.
The qPCR reaction system comprised 5 μL of cDNA template, 1 μL each of forward and reverse primers, 10 μL of a 2X mastermix, and 3 μL of RNase-free water. The reaction procedure was as follows: 95°C for 2 min, followed by for 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Each sample was run in triplicate. The relative mRNA expression level was calculated using the 2–Δ Δ Ct method.
Enzyme Activity Test
The hepatopancreas tissue of each experimental group was accurately weighed at each sampling time point, and then, nine times the volume of normal saline was added according to the mass (g):volume (mL) ratio of 1:9. Under ice-water bath conditions, a 10% tissue homogenate was prepared. The homogenate was then subjected to centrifugation at 2,500 rpm for 10 min, and the supernatant was collected. The protein concentration was measured using the Cwbio BCA Protein Assay Kit.
SOD, CAT, and iNOS enzyme activity test kits (Nanjing Jiancheng Company) were used to detect the enzyme activities after LPS stimulation. Each sample was run in triplicate, and the corresponding enzyme activities were calculated according to the manufacturer’s instructions.
Data Analysis
Data analysis was performed using the SPSS 20.0 software. Independent sample t-test and Duncan’s multiple comparison test was used to analyze the significance of the data. P < 0.05 was considered to be statistically significant. All the data were expressed as the means ± standard deviations.
Experimental Results
P38 Gene Cloning
The full-length cDNA of PtP38 comprises a 5′ untranslated region of 119 bp, a 3′ untranslated region of 849 bp, and an open reading frame of 1,116 bp. Its coding region encodes 372 amino acids. The PtP38 amino acid sequence contains a S_TKc domain with a conserved TGY phosphorylation site and an ATRW substrate binding site (Figures 1A,B).

Figure 1. Sequence and domain architecture of PtP38. (A) Full-length cDNA of PtP38. The corresponding amino acid sequence is shown above the nucleotide. The figure shows that the full-length cDNA of PtP38 consists of a 119-bp 5′ untranslated region, an 849-bp 3′ untranslated region, and a 1,116-bp open reading frame. The coding region encodes 372 amino acids. The gray-highlighted sequence represents the S_TKc domain, and the underlined sequences denote the conserved TGY motif and ATRW sites. (B) The domain architecture of PtP38 is predicted by SMART (http://smart.embl-heidelberg.de/). The PtP38 amino acid sequence contains an S_TKc domain, with a conserved TGY phosphorylation site and an ATRW substrate binding site.
Bioinformatics Analysis
Multiple sequence alignment showed that PtP38 shares the highest identity with S. paramamosain (94%), followed by E. sinensis (93%) (Figure 2). The phylogenetic tree of the P38 protein was constructed by the neighbor-joining method using the MEGA5.1 software. The results showed that PtP38 has the closest relationship to Scylla paramamosain, followed by Eriocheir sinensis (Figure 3). Using the I-TASSER website, the secondary and tertiary structures of the PtP38 protein were predicted and analyzed. PtP38 protein was composed of two halves, a N-terminal domain and a C-terminal domain. The N-terminal domain was composed largely of β-sheet, whereas the C-terminal domain was largely helical (Figure 4).
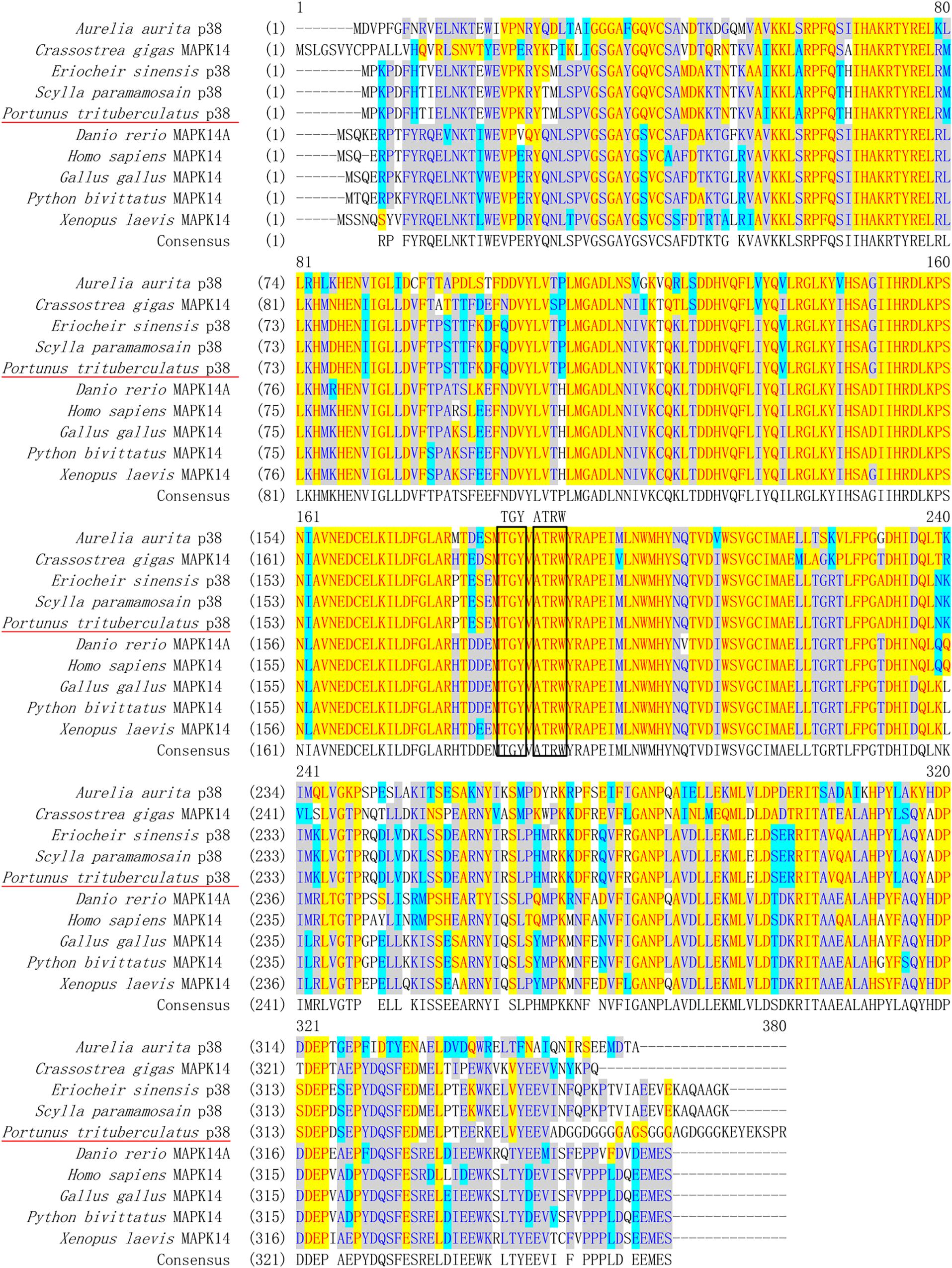
Figure 2. Multiple alignment of PtP38 with P38s from other species. The conserved amino acid residues in these sequences are shown in yellow. The conserved phosphorylated TGY motifs and ATRW sites are indicated by a black box. PtP38 shares the highest identity with Scylla paramamosain (94%), followed by Eriocheir sinensis (93%). The homology to Gallus, Python bivittatus, and Pacific oyster Crassostrea gigas is 73%, to Homo sapiens, Xenopus laevis, and Danio rerio is 72%, and to Aurelia aurita is 63%.
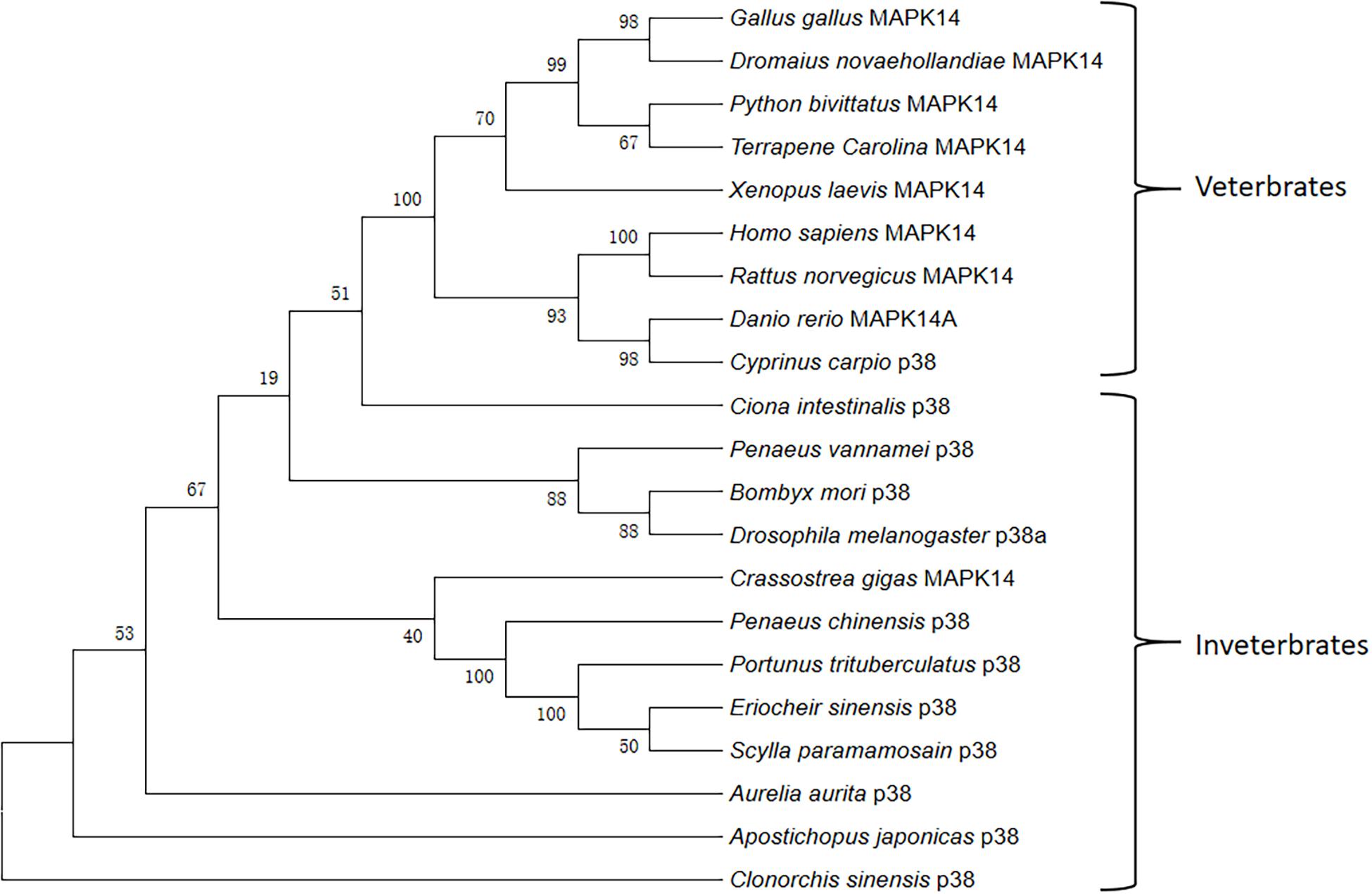
Figure 3. Phylogenetic relationships of PtP38 and representative P38s from other species. The phylogenetic tree of P38 protein was constructed by the neighbor-joining method using the MEGA5.1 software. PtP38 has the closest relationship to Scylla Paramamosain, followed by Eriocheir sinensis. The GenBank login number is as follows: Homo sapiens MAPK14 (NP_001306.1); Gallus MAPK14 (XP_001232616.1); Python bivittatus MAPK14 (XP_007426614.1); Xenopus laevis MAPK14 (NP_001080300.1); Danio rerio MAPK14A (NP_571797.1); Eriocheir sinensis P38 (AII32447.1); Scylla paramamosain P38 (AHH29322.1); Crassostrea gigas MAPK14 (EKC29510.1); Aurelia aurita P38 (ARJ54259.1); Dromaius novaehollandiae MAPK14 (XP_025967659.1); Terrapenecarolina triunguis MAPK14 (XP_024066297.1); Ciona intestinalis P38 (NP_001071958.1); Clonorchis sinensis P38 (GAA55102.1); Apostichopus japonicas P38 (ASU91381.1); Penaeus chinensis P38 (AIY23112.1); Penaeus vannamei P38 (AGG82488.1); Bombyx mori P38 (NP_001036996.1); Drosophila melanogaster P38a (NP_477163.1); Cyprinus carpio P38 (BAA96415.1); Rattus norvegicus MAPK14 (NP_112282.2).
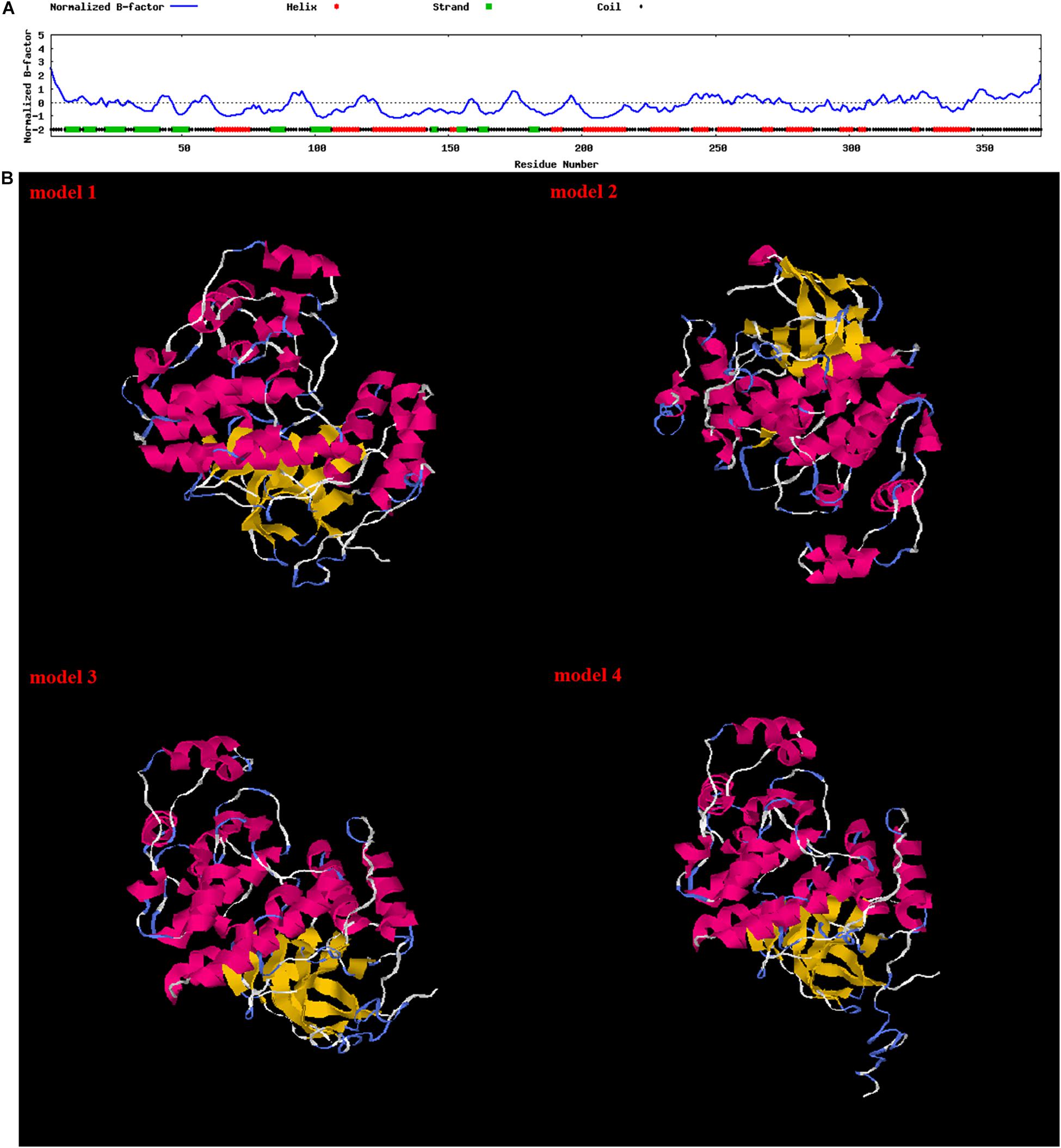
Figure 4. Secondary structure and tertiary structure of the PtP38 protein. (A) Secondary structure of the PtP38 protein predicted by the I-TASSER website. (B) Tertiary structure of the PtP38 protein predicted by the I-TASSER website. PtP38 protein was composed of two halves, a N-terminal domain and a C-terminal domain. The N-terminal domain was composed largely of β-sheet, whereas the C-terminal domain was largely helical.
Tissue Distribution of P38 mRNA
The expression of the PtP38 gene in the different tissues was analyzed by qPCR. The results showed that the P38 gene was expressed in the heart, muscle, hepatopancreas, gills, stomach, eyestalk, mucosa, testis, and ovaries. The highest transcription was observed in the gills, followed by the eyestalk and hepatopancreas. The lowest transcription was observed in the muscle (Figure 5).
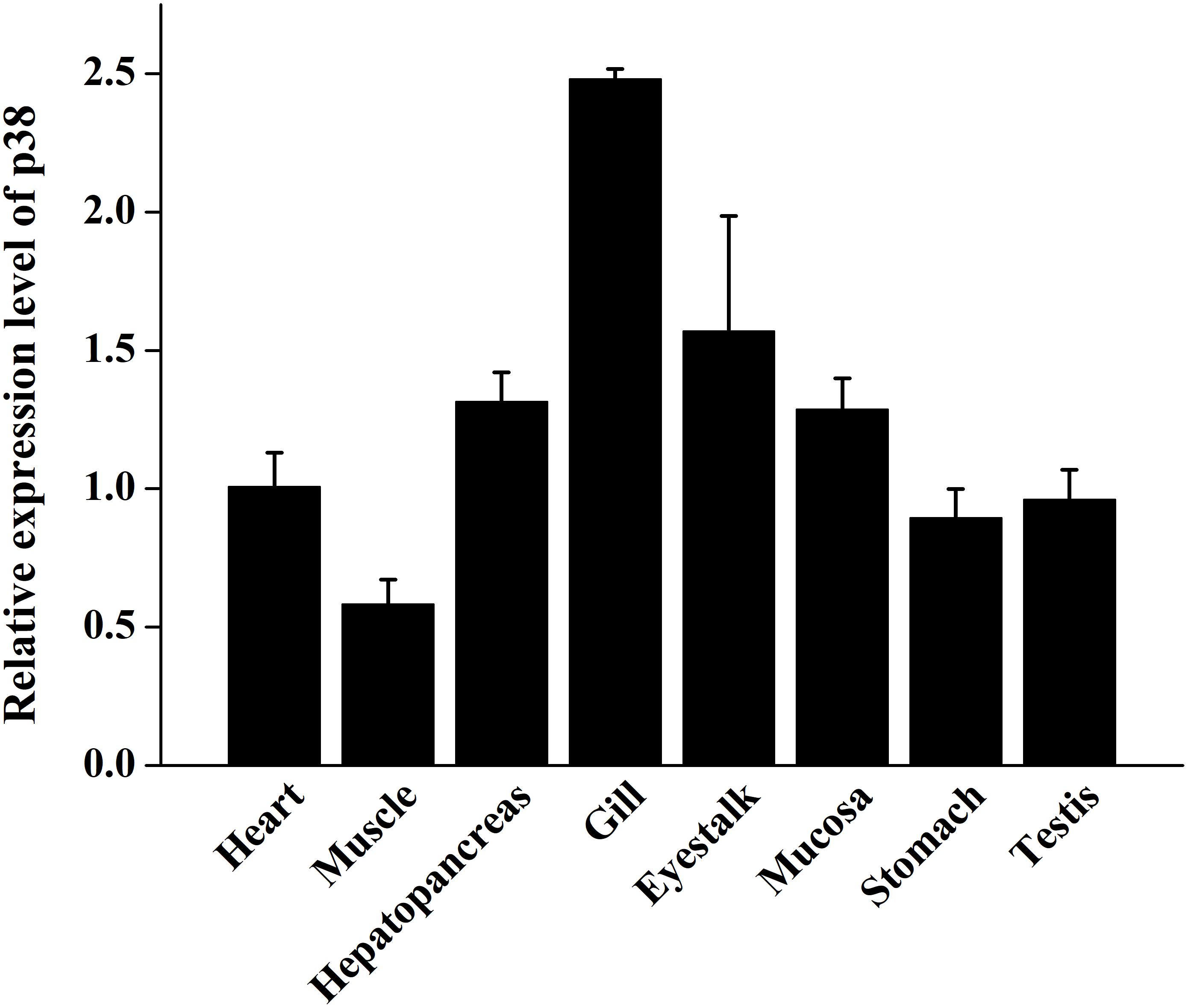
Figure 5. qRT-PCR analysis of the PtP38 transcription in different tissues of the untreated crabs. Using GAPDH as an internal reference, the relative expression level of PtP38 in the heart was set to 1. Three repeated experiments were performed for each sample. The results were expressed as means ± standard deviation.
LPS Stimulation
Expression Profile of P38 in Response to LPS Stimulation
Fluorescence quantitative PCR was used to analyze the relative expression of the P38 gene under LPS stimulation. The results showed that, after LPS stimulation, the relative expression of the P38 gene in the hepatopancreas, gills, and muscle increased significantly (Figure 6).
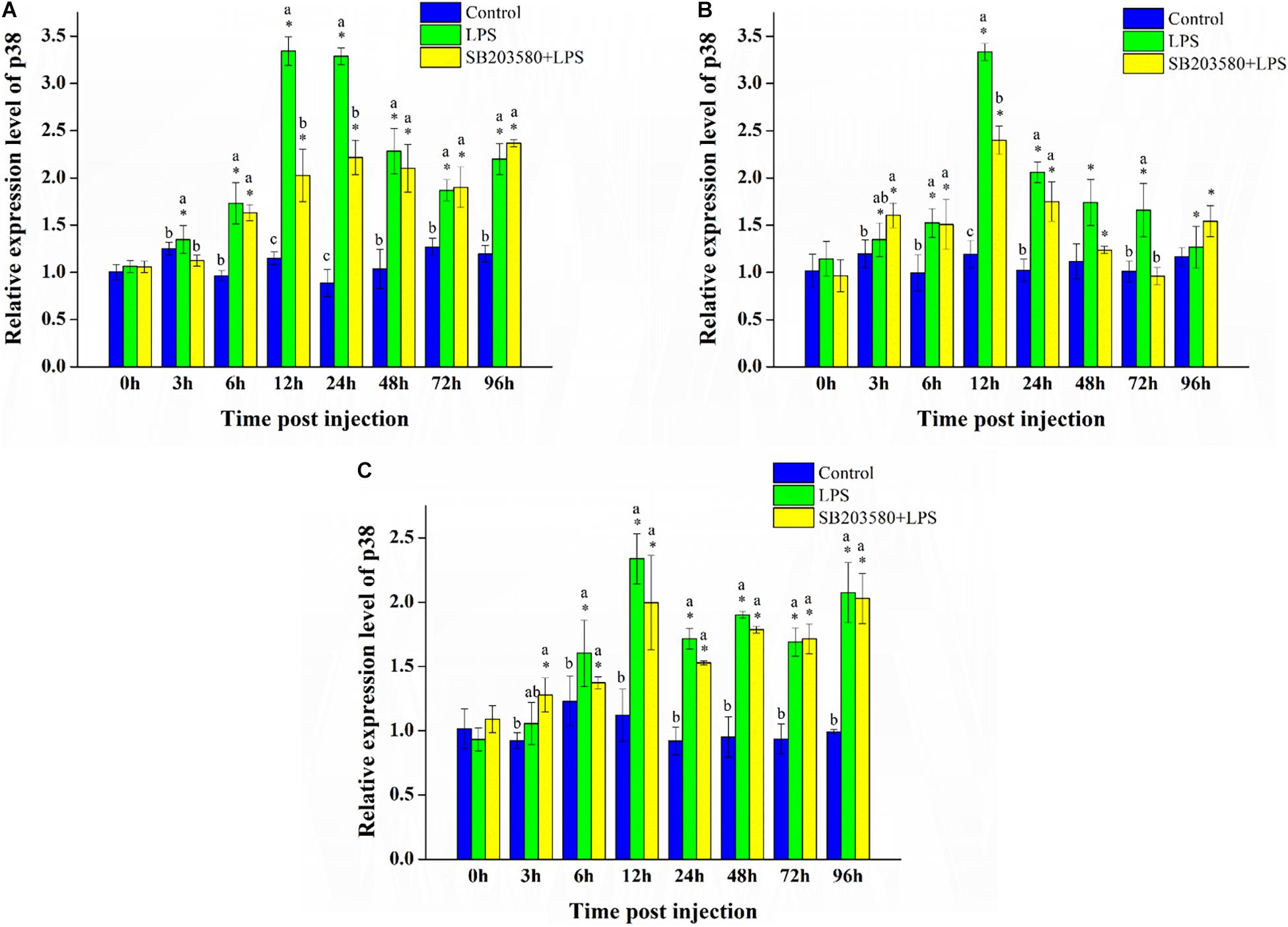
Figure 6. Analysis of the PtP38 gene expression under LPS stimulation. (A) Analysis of the PtP38 gene expression in the hepatopancreas under LPS stimulation. (B) Analysis of the PtP38 gene expression in the gills under LPS stimulation. (C) Analysis of the PtP38 gene expression in the muscle under LPS stimulation. Using GAPDH as an internal reference, the relative expression level of PtP38 in the control group at 0 h was set to 1. Error bars represent the SD of three independent repeats, and the results are expressed as means ± standard deviation. Independent sample t-test was used to analyze the significant changes at different time points in the same treatment group. *Indicates a significant difference between this time point with 0 h. Duncan’s multiple comparison test analyzed the significant changes between different treatments at the same time point. a, b, and c indicate significant differences between treatment groups. P < 0.05 was considered statistically significant.
In the hepatopancreas, independent sample t-test was used to analyze the significance of the changes at different time points of the same treatment. The results showed no significant change in the control group. Compared with 0 h, the relative expression of the P38 gene all changed significantly in the LPS group (P < 0.05). In the SB203580 + LPS group, the relative expression of the P38 gene changed significantly except at the 72-h LPS stimulation (P < 0.05). Duncan’s multiple comparison test was used to analyze the significance of the changes between the different treatment groups at the same time point. Except for the 0 and 3 h time points, the LPS and SB203580 + LPS groups showed a significantly increased P38 expression compared with the control group (P < 0.05). After LPS stimulation, the relative expression of the PtP38 gene in the hepatopancreas increased significantly, showing a tendency to increase first and then decrease (Figure 6A).
In the gills, independent sample t-test results showed no significant change in the control group. Compared with 0 h, the relative expression of the P38 gene changed significantly at each sampling time point in the LPS group (P < 0.05). In the SB203580 + LPS group, the relative expression of the P38 gene changed significantly except at the 72-h LPS stimulation (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the relative expression of the P38 gene in the LPS group increased significantly at 6, 12, 24, and 72 h (P < 0.05). Compared with the control group, the relative expression of the P38 gene in the SB203580 + LPS group increased significantly at 3, 12, 24, and 96 h (P < 0.05). Under LPS stimulation, the relative expression of the P38 gene in the gills increased significantly (Figure 6B).
In the muscle, independent sample t-test results showed no significant change in the control group. Compared with 0 h, the relative expression of the P38 gene changed significantly in the LPS group except at 3 h (P < 0.05). In the SB203580 + LPS group, the relative expression of the P38 gene changed significantly at each sampling time point (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the P38 expression in the LPS and SB203580 + LPS groups was significantly increased at 12, 24, 48, 72, and 96 h (P < 0.05). The relative expression of the P38 gene in the muscle increased significantly in response to LPS stimulation (Figure 6C).
Expression Profile of ALF and Crustin in Response to LPS Stimulation
Fluorescence quantitative PCR was used to analyze the relative expression of the ALF gene in the hepatopancreas under LPS stimulation. Independent sample t-test results showed no significant change in the control group. Compared with 0 h, the relative expression of the ALF gene was significantly changed in the LPS group (P < 0.05). In the SB203580 + LPS group, the relative expression of the ALF gene changed significantly except at the 3 h time point (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the relative expression of the ALF gene in the LPS and SB203580 + LPS groups increased significantly from 24 to 96 h (P < 0.05). Additionally, the relative expression of the ALF gene in the LPS and SB203580 + LPS groups was significantly different at the 3 and 24 h time points (P < 0.05). After LPS stimulation, the relative expression of the ALF gene in the hepatopancreas increased significantly. P38 protein functions by double phosphorylating the serine and threonine of the tripeptide domain TGY. Pretreatment with SB203580 specifically inhibited the phosphorylation of the P38 protein and abrogated its function, leading to an increased relative expression of the ALF gene in the hepatopancreas (Figure 7A).
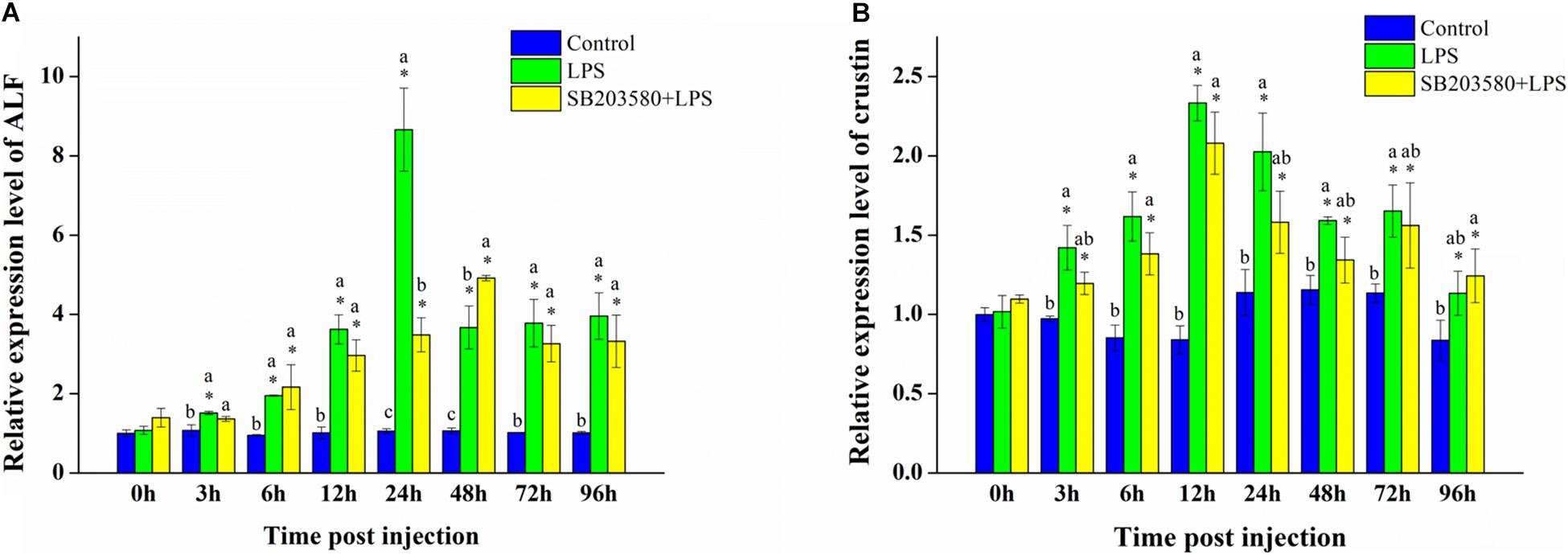
Figure 7. Analysis of the ALF and crustin gene expression in the hepatopancreas under LPS stimulation. (A) Analysis of the ALF gene expression in the hepatopancreas under LPS stimulation. (B) Analysis of the crustin gene expression in the hepatopancreas under LPS stimulation. Using GAPDH as an internal reference, the relative expression level of ALF in the control group at 0 h was set to 1. Error bars represent the SD of three independent repeats, and the results are expressed as means ± standard deviation. Independent sample t-test analyzed the significant changes at different time points in the same treatment group. *Indicates a significant difference between this time point with 0 h. Duncan’s multiple comparison test analyzed the significant changes between different treatments at the same time point. a, b, and c indicate significant differences between treatment groups. P < 0.05 was considered statistically significant.
Fluorescence quantitative PCR was used to analyze the relative expression of the crustin gene in the hepatopancreas under LPS stimulation. Independent sample t-test showed no significant change in the control group. Compared with 0 h, the relative expression of the crustin gene was significantly different in the LPS and SB203580 + LPS groups (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the LPS and SB203580 + LPS groups changed significantly at the 6 and 12 h time points (P < 0.05). The relative expression of the crustin gene in the hepatopancreas increased significantly after LPS stimulation, showing a trend of increasing first and then decreasing. Compared with the LPS group, the early injection of SB203580 abrogated the function of the P38 protein, inhibiting the increase in the relative expression of the crustin gene (Figure 7B).
SOD, CAT, and iNOS Enzyme Activity Analysis in Response to LPS Stimulation
The enzyme activity kit was used to calculate the SOD enzyme activity in the hepatopancreas under LPS stimulation. Independent sample t-test results showed no significant change in the control group. However, the SOD enzyme activity in the LPS and SB203580 + LPS groups both changed significantly (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the SOD enzyme activity in the LPS and SB203580 + LPS groups showed significant changes at the 12, 24, 48, and 96 h time points (P < 0.05). From 12 to 96 h, the LPS and SB203580 + LPS groups were significantly different (P < 0.05). The SOD enzyme activity in the hepatopancreas increased significantly after LPS stimulation, while the early injection of SB203580 inhibited the increase (Figure 8A).
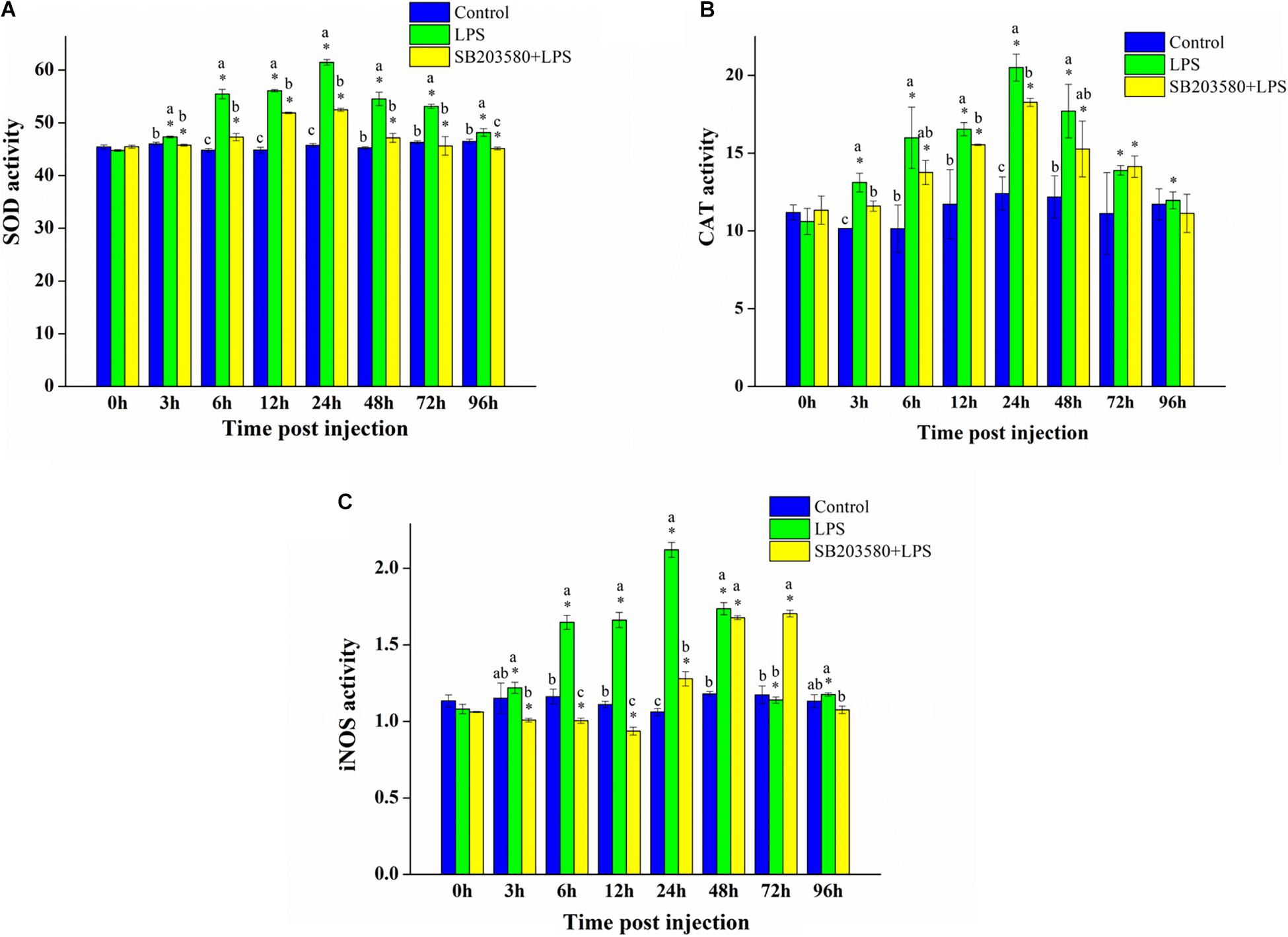
Figure 8. Analysis of the SOD, CAT, and iNOS enzyme activity analysis under LPS stimulation. (A) Analysis of the SOD enzyme activity analysis under LPS stimulation. (B) Analysis of the CAT enzyme activity analysis under LPS stimulation. (C) Analysis of the iNOS enzyme activity analysis under LPS stimulation. Error bars represent the SD of three independent repeats, and the results are expressed as means ± standard deviation. Independent sample t-test analyzed the significant changes at different time points in the same treatment group. *Indicates a significant difference between this time point with 0 h. Duncan’s multiple comparison test analyzed the significant changes between different treatments at the same time point. a, b, and c indicate significant differences between treatment groups. P < 0.05 was considered statistically significant.
The CAT enzyme activity in the hepatopancreas under LPS stimulation was measured using an enzyme activity kit. Independent sample t-test results showed no significant change in the control group. In the LPS group, the CAT enzyme activity changed significantly compared with that at 0 h (P < 0.05). In the SB203580 + LPS group, the activity of the CAT enzyme changed significantly from 6 to 72 h (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the LPS and SB203580 + LPS groups changed significantly at the 3, 6, 12, and 24 h time points (P < 0.05). The difference between the LPS and SB203580 + LPS groups was significant at the 3, 12, and 24 h time points (P < 0.05). After LPS stimulation, the activity of the CAT enzyme in the hepatopancreas increased significantly. After injection with SB203580 in advance to inhibit the phosphorylation of P38 protein, CAT enzyme activity was significantly reduced (Figure 8B).
The activity of the iNOS enzyme in the hepatopancreas tissues of P. trituberculatus stimulated by LPS was measured using an enzyme activity kit. Independent sample t-test results showed no significant change in the control group (P < 0.05). Compared with 0 h, the iNOS enzyme activity changed significantly in the LPS group (P < 0.05). In the SB203580 + LPS group, the activity of the iNOS enzyme changed significantly except at 96 h (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, the activity of the iNOS enzyme in the LPS group changed significantly from 6 to 48 h (P < 0.05). Except for the 0 and 48 h time points, the activity of the iNOS enzyme in the LPS group was significantly different from that in the SB203580 + LPS group (P < 0.05). After LPS stimulation, the iNOS enzyme activity in the hepatopancreas increased significantly and was significantly suppressed after the P38 protein lost its function (Figure 8C).
Expression Profile of Apoptosis-Related Genes Caspase2 and Caspase3 in Response to LPS Stimulation
Fluorescence quantitative PCR was used to analyze the relative expression of the caspase2 gene in the hepatopancreas under LPS stimulation. Independent sample t-test showed no significant change in the control group. In the LPS group, the relative expression of the caspase2 gene changed significantly except at the 3 h time point (P < 0.05). In the SB203580 + LPS group, the activity of the iNOS enzyme changed significantly (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control and SB203580 + LPS groups, the LPS group showed significant changes from 12 to 96 h (P < 0.05). After LPS stimulation, the relative expression of the caspase2 gene in the hepatopancreas showed an increasing trend. However, early injection of SB203580 significantly decreased the trend (P < 0.05) (Figure 9A).
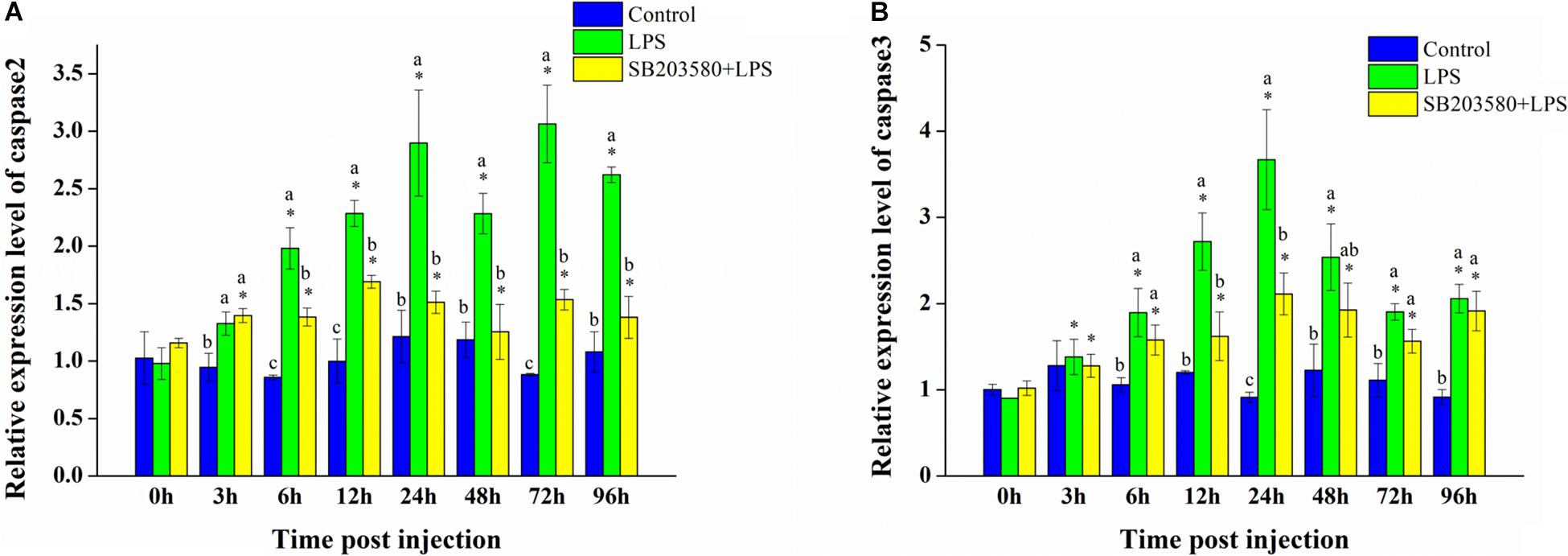
Figure 9. Analysis of caspase2 and caspase3 gene expression in the hepatopancreas under LPS stimulation. (A) Analysis of caspase2 gene expression in the hepatopancreas under LPS stimulation. (B) Analysis of caspase3 gene expression in the hepatopancreas under LPS stimulation. Using GAPDH as an internal reference, the relative expression level of caspase2 in the control group at 0 h was set to 1. Error bars represent the SD of three independent repeats, and the results are expressed as means ± standard deviation. Independent sample t-test analyzed the significant changes at different time points in the same treatment group. *Indicates a significant difference between this time point with 0 h. Duncan’s multiple comparison test analyzed the significant changes between different treatments at the same time point. a, b, and c indicate significant differences between treatment groups. P < 0.05 was considered statistically significant.
Fluorescence quantitative PCR was used to analyze the relative expression of the caspase3 gene in the hepatopancreas under LPS stimulation. Independent sample t-test showed no significant change in the control group. In the LPS and SB203580 + LPS groups, the relative expression of the caspase3 gene changed significantly (P < 0.05). Duncan’s multiple comparison test showed that, compared with the control group, caspase3 gene expression in the LPS and SB203580 + LPS groups changed significantly at the 6, 12, 24, and 96 h time points (P < 0.05). The difference in expression between the LPS and SB203580 + LPS groups was significant at the 12, 24, and 72 h time points (P < 0.05). After LPS stimulation, the relative expression of the caspase3 gene in the hepatopancreas increased significantly (P < 0.05). When SB203580 inhibited the function of the P38 protein, the relative expression of the caspase3 gene was significantly suppressed (P < 0.05) (Figure 9B).
Discussion
In this study, we cloned the full-length cDNA of P. trituberculatus PtP38. Multiple sequence alignment showed that PtP38 shares the highest identity with Scylla Paramamosain (Yu et al., 2017), followed by Eriocheir sinensis (Zhu et al., 2015). The PtP38 gene is expressed in the heart, muscle, hepatopancreas, gills, stomach, eyestalk, mucosa, testis, and ovaries. The expression is higher in the gills, eyestalk, and hepatopancreas, and lowest in the muscle. The gills, eyestalk, and hepatopancreas are important immune organs of crustaceans (Rõszer, 2014; Ikerd et al., 2015). High PtP38 expression indicates that PtP38 may be related to the immune function of P. trituberculatus.
LPS is the main component of the cell wall of Gram-negative bacteria, which can activate P38 MAPK, promote the expression of P38, and induce its phosphorylation (Mercau et al., 2014). In this study, we found that, after LPS stimulation, the expression of the PtP38 gene in the hepatopancreas, gills, and muscle significantly increased. The P38 protein functions by double phosphorylating the serine and threonine of the tripeptide domain TGY. SB203580 is a specific inhibitor of the P38 protein phosphorylation with extremely low immunotoxicity to the body and is widely used in P38 functional studies (Abhilasha et al., 2019). In this study, no significant change was found in the P38 gene expression between the SB203580 + LPS and LPS groups. SB203580 could significantly inhibit the phosphorylation level of the P38 protein in Scylla paramamosain, with no significant effect on the P38 gene expression (Yu et al., 2017).
ALF and crustin are important antimicrobial peptides that play important roles in the crustacean immune response (Yue et al., 2010). Antimicrobial peptides have a broad killing effect on bacteria, fungi, viruses, parasites, and even cancer cells (Liu et al., 2012). In this study, the expression of ALF and crustin in the hepatopancreas increased significantly after LPS stimulation, indicating that LPS stimulation mimicked Gram-negative bacterial infection and activated the body’s immune response. LPS stimulation promoted the expression of NF-κB (Ko et al., 2017), while the production of antimicrobial peptides was regulated by NF-κB (Li et al., 2018). In HeLa cells, shrimp ALF inhibited the inflammatory response caused by LPS stimulation through the NF-κB pathway (Lin et al., 2013). Inhibiting the function of the P38 protein suppressed the increase in the ALF and crustin expression in the hepatopancreas, indicating that P38 may regulate the expression of these two antimicrobial peptides. Similar results were obtained in Penaeus vannamei and Scylla Paramamosain (Yan et al., 2013; Yu et al., 2017). The P38 pathway could directly regulate the expression of NF-κB (Ji et al., 2009), while the production of antimicrobial peptides is regulated by NF-κB. We conclude that PtP38 may regulate the expression of NF-κB and expression of antimicrobial peptides such as ALF and crustin, participating in the immune response stimulated by LPS.
iNOS, SOD, and CAT are key enzymes to maintain ROS homeostasis and are closely related to the immune activity of the organism (Li et al., 2016). When the body develops bacterial infection, iNOS produces NO, which catalyzes the oxidation of NADPH and produces O2– to defend against disease (Kumar et al., 2018). SOD catalyzes O2– to produce H2O2, and CAT catalyzes H2O2 to produce H2O and O2, thereby eliminating the excess ROS to reduce oxidative damage to cells (Ighodaro and Akinloye, 2017). In this study, we found that, after LPS stimulation, the enzyme activities of iNOS, SOD, and CAT in the hepatopancreas increased significantly. Previous studies have shown that LPS stimulation could increase the iNOS enzyme activity and promote ROS production (Wu et al., 2018). After LPS stimulation, SOD and CAT enzyme activities increased to clear the excess ROS (Ighodaro and Akinloye, 2017). Early injection of P38 inhibitors reduced the activity of iNOS, SOD, and CAT in the hepatopancreas, indicating that P38 may regulate the activity of iNOS, SOD, and CAT. Studies have shown that P38 could directly regulate iNOS (Chang et al., 2004) and affect ROS (Jian et al., 2017). PtP38 may participate in the immune response stimulated by LPS by regulating the activities of the iNOS, SOD, and CAT enzymes.
Caspase2 is an upstream promoter required to activate apoptosis, and caspase3 cleaves most substrates in cells undergoing apoptosis (Fan et al., 2010); thus, they can be used as indicators of apoptosis (Ren et al., 2017). LPS stimulation induced apoptosis (Zhou et al., 2016). In this study, we also found that the expression of caspase2 and caspase3 in the hepatopancreas increased significantly after LPS stimulation. Apoptosis can remove abnormal cellular components, reducing inflammatory responses and maintaining cellular homeostasis (Ren et al., 2017). With the P38 protein’s loss of function, the increase in caspase2 and caspase3 expression in the hepatopancreas was inhibited, indicating that P38 may mediate apoptosis. Previous studies have shown that P38 could enhance the expression of c-myc (Mei et al., 2010), phosphorylate p53 (Roy et al., 2018), participate in Fas/FasL-mediated apoptosis (Qi et al., 2014), activate c-jun and c-fos (Xue et al., 2015), induce bax translocation (Owens et al., 2009), and enhance TNF-a expression (Yeh et al., 2008) to regulate cell apoptosis. Additionally, apoptosis is closely related to the NF-κB and ROS levels (Xia et al., 2017). It can be inferred that PtP38 may regulate apoptosis through the NF-κB, ROS, and other pathways, participating in the immune response stimulated by LPS.
Conclusion
In conclusion, we cloned the P38 gene from P. trituberculatus and studied its expression characteristics under LPS stimulation. We detected the expression of antimicrobial peptides, activity of oxidative stress-related enzymes, and expressions of apoptosis related genes at the groups of LPS stimulation and P38 inhibitor and LPS stimulation. According to the results, PtP38 likely plays an important immunoregulatory function under a Gram-negative bacteria infection, laying the foundation for the further study of the PtP38 MAPK signaling pathway and immune mechanism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
C-PL, C-CH, and J-QZ formulated and designed the experiments. C-PL, C-GW, and D-JT performed the experiments. C-PL and C-CH analyzed the data and wrote the manuscript. J-QZ and C-LW revised and proofread the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the Natural Science Foundation of China (No. 31602140), Natural Science Foundation of Zhejiang Province (No. LY20C190003), Major Agriculture Program of Ningbo (No. 2017C110007), Natural Science Foundation of Ningbo (No. 2018A610228), the K. C. Wong Magna Fund in Ningbo University, and the Collaborative Innovation Center for Zhejiang Marine High-efficiency and Healthy Aquaculture.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/orffinder/
- ^ http://web.expasy.org/translate/
- ^ http://smart.embl-heidelberg.de/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://zhanglab.ccmb.med.umich.edu/I-TASSER/
References
Abhilasha, K. V., Sumanth, M. S., Chaithra, V. H., Jacob, S. P., and Marathe, G. K. (2019). P38 MAP-kinase inhibitor protects against platelet-activating factor-induced death in mice. Free Radic. Biol. Med. 143, 275–287. doi: 10.1016/j.freeradbiomed.2019.08.019
Akrami, H., Mahmoodi, F., Havasi, S., and Sharifi, A. (2016). PlGF knockdown inhibited tumor survival and migration in gastric cancer cell via PI3K/Akt and P38MAPK pathways. Cell Biochem. Funct. 34, 173–180. doi: 10.1002/cbf.3176
Allahham, R., Deford, J. H., and Papaconstantinou, J. (2016). Mitochondrial-generated ROS down regulates insulin signaling via activation of the p38MAPK stress response pathway. Mol. Cell. Endocrinol. 419, 1–11. doi: 10.1016/j.mce.2015.09.013
Badger, A. M., Cook, M. N., Lark, M. W., Newman-Tarr, T. M., Swift, B. A., Barone, F. C., et al. (1998). SB 203580 Inhibits p38 mitogen-activated protein kinase, nitric oxide production, and inducible nitric oxide synthase in bovine cartilage-derived chondrocytes. J. Immunol. 161, 467–473.
Brichkina, A., Bertero, T., Loh, H. M., Nguyen, N. T. M., Emelyanov, A., and Rigade, S. (2016). p38MAPK builds a hyaluronan cancer niche to drive lung tumorigenesis. Genes Dev. 30, 2623–2636. doi: 10.1101/gad.290346.116
Cara, W., Neal, S., and Sara, C. (2018). P38b and JAK-STAT signaling protect against Invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog. 14:e1007020. doi: 10.1371/journal.ppat.1007020
Chakrabarti, S., Poidevin, M., Lemaitre, B., and Garsin, D. A. (2014). The Drosophila MAPK p38c regulates oxidative stress and lipid homeostasis in the intestine. PLoS Genet. 10:e1004659. doi: 10.1371/journal.pgen.1004659
Chang, P. C., Chen, T. H., Chang, C. J., Hou, C. C., Chan, P., and Lee, M. O. (2004). Advanced glycosylation end products induce inducible nitric oxide synthase (iNOS) expression via a p38 MAPK-dependent pathway. Kidney Int. 65, 1664–1675. doi: 10.1111/j.1523-1755.2004.00602.x
Chen, J., Xie, C., Tian, L., Hong, L., Wu, X., and Han, J. (2010). Participation of the P38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc. Natl. Acad. Sci. U.S.A. 107, 20774–20779. doi: 10.1073/pnas.1009223107
Cuenda, A., and Rousseau, S. (2007). p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773, 1358–1375. doi: 10.1016/j.bbamcr.2007.03.010
Da Silva, J., Pierrat, B., Mary, J. L., and Lesslauer, W. (1997). Blockade of p38 mitogen-activated protein kinase pathway inhibits inducible nitric-oxide synthase expression in mouse astrocytes. J. Biol. Chem. 272:28373. doi: 10.1074/jbc.272.45.28373
Fan, T. J., Han, L. H., Cong, R. S., and Liang, J. (2010). Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 37, 719–727. doi: 10.1111/j.1745-7270.2005.00108.x
Guan, Z., Buckman, S. Y., Pentland, A. P., Templeton, D. J., and Morrison, A. R. (1998). Induction of cyclooxygenase-2 by the activated MEKK1 → SEK1/MKK4 → p38 mitogen-activated protein kinase pathway. J. Biol. Chem. 273:12901. doi: 10.1074/jbc.273.21.12901
Han, Z. S., Enslen, H., Hu, X., Meng, X., Wu, I. H., and Barrett, T. (1998). A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression, immunity gene expression. Mol. Cell. Biol. 18, 3527–3539. doi: 10.1128/MCB.18.6.3527
He, Y., Yao, W., Liu, P., and Wang, Q. Y. (2018). Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp Fenneropenaeus chinensis in response to viral and bacterial infections. Gene 642, 381–388. doi: 10.1016/j.gene.2017.11.050
Ighodaro, O. M., and Akinloye, O. A. (2017). First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): their fundamental role in the entire antioxidant defence grid. Alexandria J. Med. 54, 287–293. doi: 10.1016/j.ajme.2017.09.001
Ikerd, J. L., Burnett, K. G., and Burnett, L. E. (2015). Effects of salinity on the accumulation of hemocyte aggregates and bacteria in the gills of Callinectes sapidus, the Atlantic blue crab, injected with Vibrio campbellii. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 183, 97–106. doi: 10.1016/j.cbpa.2014.12.030
Ji, G., Liu, D., Liu, J., Gao, H., and Shen, G. (2009). p38 mitogen-activated protein kinase up-regulates NF-κB transcriptional activation through RelA phosphorylation during stretch-induced myogenesis. Biochem. Biophys. Res. Commun. 391, 547–551. doi: 10.1016/j.bbrc.2009.11.095
Jian, K. L., Zhang, C., Shang, Z. C., Yang, L., and Kong, L. Y. (2017). Eucalrobusone C suppresses cell proliferation and induces ROS-dependent mitochondrial apoptosis via the p38 MAPK pathway in hepatocellular carcinoma cells. Phytomedicine 25, 71–82. doi: 10.1016/j.phymed.2016.12.014
Keshet, Y., and Seger, R. (2010). The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38. doi: 10.1007/978-1-60761-795-2_1
Ko, E. Y., Cho, S. H., Kwon, S. H., Eom, C. Y., Jeong, M. S., and Lee, W. W. (2017). The roles of NF-κB and ROS in regulation of pro-inflammatory mediators of inflammation induction in LPS-stimulated zebrafish embryos. Fish Shellfish Immunol. 68, 525–529. doi: 10.1016/j.fsi.2017.07.041
Kumar, A., Singh, K. P., Bali, P., Anwar, S., Kaul, A., and Singh, O. P. (2018). iNOS polymorphism modulates iNOS/NO expression via impaired antioxidant and ROS content in P. vivax and P. falciparum infection. Redox Biol. 15, 192–206. doi: 10.1016/j.redox.2017.12.005
Lake, D., Corrêa, S. A., and Müller, J. (2016). Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci. CMLS 73, 4397–4413. doi: 10.1007/s00018-016-2297-8
Li, M., Ma, C., Li, H., Peng, J., Zeng, D., and Chen, X. (2018). Molecular cloning, expression, promoter analysis and functional characterization of a new Crustin from Litopenaeus vannamei. Fish Shellfish Immunol. 73, 42–49. doi: 10.1016/j.fsi.2017.12.002
Li, M., Wang, J., Song, S., and Li, C. (2016). Molecular characterization of a novel nitric oxide synthase gene from P. trituberculatus and the roles of NO/O2 –-generating and antioxidant systems in host immune responses to Hematodinium. Fish Shellfish Immunol. 52, 263–277. doi: 10.1016/j.fsi.2016.03.042
Lin, M. C., Pan, C. Y., Hui, C. F., Chen, J. Y., and Wu, J. L. (2013). Shrimp anti-lipopolysaccharide factor (SALF),an antimicrobial peptide, inhibits proinflammatory cytokine expressions through the MAPK and NF-κB pathways in LPS-induced HeLa cells. Peptides 40, 42–48. doi: 10.1016/j.peptides.2012.11.010
Liu, Y., Cui, Z., Li, X., Song, C., Li, Q., and Wang, S. (2012). A new anti-lipopolysaccharide factor isoform (PtALF4) from the swimming crab P. trituberculatus exhibited structural and functional diversity of ALFs. Fish Shellfish Immunol. 32, 724–731. doi: 10.1016/j.fsi.2012.01.021
Mei, Y., Blomstrand, E., Chibalin, A. V., Krook, A., and Zierath, J. R. (2010). Marathon running increases ERK1/2 and p38 MAP kinase signalling to downstream targets in human skeletal muscle. J. Physiol. 536, 273–282. doi: 10.1111/j.1469-7793.2001.00273.x
Mercau, M. E., Astort, F., Giordanino, E. F., Martinez Calejman, C., Sanchez, R., and Caldareri, L. (2014). Involvement of PI3K/Akt and P38 MAPK in the induction of COX-2 expression by bacterial lipopolysaccharide in murine adrenocortical cells. Mol. Cell. Endocrinol. 384, 43–51. doi: 10.1016/j.mce.2014.01.007
Ono, K., and Han, J. (2000). The p38 signal transduction pathway: activation and function. Cell. Signal. 12, 1–13. doi: 10.1016/S0898-6568(99)00071-6
Owens, T. W., Valentijn, A. J., Upton, J. P., Keeble, J., Zhang, L., and Lindsay, J. (2009). Apoptosis commitment and activation of mitochondrial Bax during anoikis is regulated by p38MAPK. Cell Death Diff. 1’551–1562. doi: 10.1038/cdd.2009.102
Qi, S. Q., Fu, W. J., Wang, C. M., Liu, C. J., Quan, C., Kourouma, A., et al. (2014). BPA-induced apoptosis of rat Sertoli cells through Fas/FasL and JNKs/p38 MAPK pathways. Reprod. Toxicol. 50, 108–116. doi: 10.1016/j.reprotox.2014.10.013
Ren, X. Y., Yu, X., Gao, B., Liu, P., and Li, J. (2017). Characterization of three caspases and their pathogen-induced expression pattern in P. trituberculatus. Fish Shellfish Immunol. 66, 189–197. doi: 10.1016/j.fsi.2017.05.006
Rõszer, T. (2014). The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 358, 685–695. doi: 10.1007/s00441-014-1985-7
Roy, S., Roy, S., Rana, A., Akhter, Y., Hande, M. P., and Banerjee, B. (2018). The role of p38 MAPK pathway in p53 compromised state and telomere mediated DNA damage response. Mutation Res. Genet. Toxicol. Environ. Mutagenesis 836, 89–97. doi: 10.1016/j.mrgentox.2018.05.018
Seger, R., and Krebs, E. G. (1995). The MAPK signaling cascade. FASEB J. 9, 726–735. doi: 10.1096/fasebj.9.9.7601337
Shao, J. P., Rui, X., Ming, L., Zhao, Q. Z., Ren, X. H., Li, Z. H., et al. (2018). Glucocorticoid receptor inhibit the activity of NF-κB through p38 signaling pathway in spinal cord in the spared nerve injury rats. Life Sci. 208, 268–275. doi: 10.1016/j.lfs.2018.07.026
Sun, J. J., Wang, L. L., Wu, Z. J., Han, S., Wang, L. Y., Li, M. J., et al. (2019). P38 is involved in immune response by regulating inflammatory cytokine expressions in the Pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 91, 108–114. doi: 10.1016/j.dci.2018.10.011
Wang, B., and Li, L. H. (2016). Changes of p38 MAPK and P-p38 MAPK levels in peripheral blood mononuclear cells during inflammation reaction of acute and chronic pancreatitis. Chinese J. Clin. Res. 29, 5–20.
Wu, X. X., Gao, H. W., Hou, Y., Yu, J., Sun, W., Wang, Y., et al. (2018). Dihydronortanshinone, a natural product, alleviates LPS-induced inflammatory response through NF-κB,mitochondrial ROS, and MAPK pathways. Toxicol. Appl. Pharmacol. 335, 1–8. doi: 10.1016/j.taap.2018.06.007
Xia, G., Wang, X., Sun, H., Qin, Y., and Fu, M. (2017). Carnosic acid (CA) attenuates collagen-induced arthritis in db/db mice via inflammation suppression by regulating ROS-dependent p38 pathway. Free Radic. Biol. Med. 108, 418–432. doi: 10.1016/j.freeradbiomed.2017.03.023
Xue, Z. X., Gao, Y. S., Shen, L. X., and Xue, G. P. (2015). Effect of chrysophanol on down-regulated expression of c-fos and c-jun mRNA in mouse astrocytes exposed to ammonia and related mechanisms. Chinese J. Pharmacol. Toxicol. 29, 912–916. doi: 10.3867/j.issn.1000-3002.2015.06.006
Yan, H., Zhang, S., Li, C. Z., Chen, Y. H., Chen, Y. G., and Weng, S. P. (2013). Molecular characterization and function of a p38 MAPK gene from Litopenaeus vannamei. Fish Shellfish Immunol. 34, 1421–1431. doi: 10.1016/j.fsi.2013.02.030
Yeh, C. J., Lin, P. Y., Liao, M. H., Liu, H. J., Lee, J. W., Chiu, S. J., et al. (2008). TNF-α mediates pseudorabies virus-induced apoptosis via the activation of p38 MAPK and JNK/SAPK signaling. Virology 381, 55–66. doi: 10.1016/j.virol.2008.08.023
Yokota, T., and Wang, Y. (2016). p38 MAP kinases in the heart. Gene 575, 369–376. doi: 10.1016/j.gene.2015.09.030
Yu, Z., Geng, Y., Huang, A., Wang, K. Y., Huang, X. L., Chen, D. F., et al. (2017). Molecular characterization of a p38 mitogen-activated protein kinase gene from Scylla paramamosain, and its expression profiles during pathogenic challenge. J. Invertebr. Pathol. 144, 32–36. doi: 10.1016/j.jip.2017.01.001
Yue, F., Pan, L., Miao, J., Zhang, L., and Li, J. (2010). Molecular cloning, characterization and mRNA expression of two antibacterial peptides: crustin and anti-lipopolysaccharide factor in swimming crab P. trituberculatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 156, 77–85. doi: 10.1016/j.cbpb.2010.02.003
Zhan, Y. Y., Wang, Y., Li, K. Q., and Chang, Y. P. (2018). A novel p38 MAPK gene in the sea cucumber Apostichopus japonicus (Ajp38) is associated with the immune response to pathogenic challenge. Fish Shellfish Immunol. 81, 250–259. doi: 10.1016/j.fsi.2018.07.001
Zhou, Y., Wu, Z., Cao, X., Ding, L., Wen, Z. S., and Bian, J. S. (2016). HNO suppresses LPS-induced inflammation in BV-2 microglial cells via inhibition of NF-κB and p38 MAPK pathways. Pharmacol. Res. 111, 885–895. doi: 10.1016/j.phrs.2016.08.007
Keywords: Portunus trituberculatus, P38, MAPK, LPS, immunity
Citation: Lu C-P, Wei C-G, Zhu J-Q, Tang D-J, Wang C-L and Hou C-C (2021) PtP38 May Increase the Immune Ability of Portunus trituberculatus Stimulated by LPS Imitating a Gram-Negative Bacterial Infection. Front. Mar. Sci. 8:658733. doi: 10.3389/fmars.2021.658733
Received: 26 January 2021; Accepted: 22 March 2021;
Published: 03 May 2021.
Edited by:
Shengming Sun, Shanghai Ocean University, ChinaReviewed by:
Jitao Li, Yellow Sea Fisheries Research Institute (CAFS), ChinaHongyu Ma, Shantou University, China
Copyright © 2021 Lu, Wei, Zhu, Tang, Wang and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Lin Wang, d2FuZ2NodW5saW5AbmJ1LmVkdS5jbg==; Cong-Cong Hou, aG91Y29uZ2NvbmdAbmJ1LmVkdS5jbg==
 Cheng-Peng Lu
Cheng-Peng Lu Chao-Guang Wei
Chao-Guang Wei Chun-Lin Wang
Chun-Lin Wang Cong-Cong Hou
Cong-Cong Hou