- 1Algal and Microbial Biotechnology Division, Faculty of Biosciences and Aquaculture, Nord University, Bodø, Norway
- 2School of Biological Sciences, Oceans Institute, The University of Western Australia, Crawley, WA, Australia
- 3New South Wales Fisheries, National Marine Science Centre, Coffs Harbour, NSW, Australia
- 4UMR 8227, CNRS, Algal Genetics Group, Integrative Biology of Marine Models, Sorbonne Université, UPMC University Paris 06, Station Biologique de Roscoff, Roscoff, France
- 5National Marine Science Centre, Southern Cross University, Coffs Harbour, NSW, Australia
- 6Marine Botany, University of Bremen, Bremen, Germany
- 7Division of Food Production and Society, Norwegian Institute of Bioeconomy Research (NIBIO), Bodø, Norway
- 8Key Laboratory of Experimental Marine Biology, Center for Ocean Mega-Science, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 9Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 10Ocean School, Yantai University, Yantai, China
Marine macrophytes, including seagrasses and macroalgae, form the basis of diverse and productive coastal ecosystems that deliver important ecosystem services. Moreover, western countries increasingly recognize macroalgae, traditionally cultivated in Asia, as targets for a new bio-economy that can be both economically profitable and environmentally sustainable. However, seagrass meadows and macroalgal forests are threatened by a variety of anthropogenic stressors. Most notably, rising temperatures and marine heatwaves are already devastating these ecosystems around the globe, and are likely to compromise profitability and production security of macroalgal farming in the near future. Recent studies show that seagrass and macroalgae can become less susceptible to heat events once they have been primed with heat stress. Priming is a common technique in crop agriculture in which plants acquire a stress memory that enhances performance under a second stress exposure. Molecular mechanisms underlying thermal priming are likely to include epigenetic mechanisms that switch state and permanently trigger stress-preventive genes after the first stress exposure. Priming may have considerable potential for both ecosystem restoration and macroalgae farming to immediately improve performance and stress resistance and, thus, to enhance restoration success and production security under environmental challenges. However, priming methodology cannot be simply transferred from terrestrial crops to marine macrophytes. We present first insights into the formation of stress memories in both seagrasses and macroalgae, and research gaps that need to be filled before priming can be established as new bio-engineering technique in these ecologically and economically important marine primary producers.
Introduction
Marine macrophytes, including seagrasses and macroalgae, form the foundational basis of some of the most productive and diverse coastal marine ecosystems on the planet (Larkum et al., 2006; Costanza et al., 2014; Klinger, 2015; Teagle et al., 2017) that provide ecosystem services worth US$ 28.9 ha–1 year–1 (Costanza et al., 2014). Moreover, macroalgae, traditionally cultivated in Asia (Chopin, 2017; Hu et al., 2021) at an annual value of US$ 13.3 billion (FAO, 2020), are increasingly recognized in Europe and America as a target for a new, highly profitable, and environmentally sustainable bioeconomy (Skjermo et al., 2014; Stévant et al., 2017; Grebe et al., 2019; Araújo et al., 2021).
Marine macrophytes are increasingly threatened by a variety of anthropogenic stressors, including coastal development, invasive species, agricultural run-offs, dredging, aquaculture, and rising sea levels (Orth et al., 2006; Krumhansl et al., 2016; Chefaoui et al., 2018; Filbee-Dexter and Wernberg, 2018). Nearly one-third of global seagrass areas have disappeared over the last 100 years (Waycott et al., 2009) and 60% of macroalgal forests have been in decline over the past 2–5 decades (Wernberg et al., 2019).
Above all, temperature is the most important range-limiting factor for marine macrophytes (Jueterbock et al., 2013; Repolho et al., 2017; Assis et al., 2018; Duarte et al., 2018; Martínez et al., 2018). Rising ocean temperatures, interfering with reproduction, development, and growth (Breeman, 1990; Short and Neckles, 1999), are fundamentally altering genetic diversity and adaptability (Coleman et al., 2020; Gurgel et al., 2020), and devastating macroalgal forests and seagrass meadows around the globe (Arias-Ortiz et al., 2018; Filbee-Dexter et al., 2020; Smale, 2020). In response, large-scale restoration efforts aim to avert severe ecological and economic consequences (Eger et al., 2020; Fredriksen et al., 2020; Layton et al., 2020; Tan et al., 2020; Vergés et al., 2020). Modeling studies, based on projected carbon emission scenarios, predict that poleward range shifts will intensify (Jueterbock et al., 2013; Valle et al., 2014; Assis et al., 2016, 2017; Chefaoui et al., 2018; Wilson and Lotze, 2019). Even if rising sea temperatures remain below lethal limits, they reduce macroalgal growth and performance (Nepper-Davidsen et al., 2019; Hereward et al., 2020; Smale et al., 2020), increase disease outbreaks and biofouling (Harley et al., 2012; Nepper-Davidsen et al., 2019; Qiu et al., 2019; Smale et al., 2020), and radically alter ecological interactions that determine persistence (Provost et al., 2017; Vergés et al., 2019)–thus compromising future sustainability of natural habitats, and production security of associated industries.
Priming Potential in Marine Macrophytes
Priming, a Common Technique for Crop Enhancement
In agriculture, priming (Box 1) is a commonly employed technique to enhance crop resistance to environmental challenges, including pathogen infections, hot, cold, dry, or saline conditions (Ibrahim, 2016; Pawar and Laware, 2018; Wojtyla et al., 2020); in some cases even across generations (transgenerational priming, Box 1) (Herman and Sultan, 2011; Lämke and Bäurle, 2017; Benson et al., 2020). For example, reproductive output of F3 Arabidopsis progeny increased five-fold under heat stress (30°C) if the F0 and F1 generations had previously experienced the same stress (Whittle et al., 2009). Seed priming also synchronizes germination and improves vigor, leading to improved crop establishment and yield (Pawar and Laware, 2018). Priming is now considered a promising strategy for crop production in response to future climate (Wang et al., 2017; Mercé et al., 2020), and may have large potential to alleviate negative climate change impacts on marine macrophytes as well as to enhance yield in macroalgae production.
BOX 1. Glossary of priming-related terms.
Priming–A plant’s ability to acquire a stress memory, enhancing its performance when exposed to a second stress by allowing it to respond faster, stronger, or in response to a lower threshold compared to a naïve plant (Figure 1A). Priming is often used synonymously with hardening, conditioning, or acclimation.
Stress memory–A stress-induced alteration in epigenetic state that may last under mitotic cell divisions and results in priming.
Transgenerational priming–Stability of a stress memory under meiotic cell divisions across at least one generation that benefits the progeny of primed parental plants.
Mechanisms Underlying Priming
Priming relies on the formation of a molecular stress memory (Box 1), a process that can include epigenetic mechanisms such as microRNAs (miRNAs), histone modifications, and DNA methylation (Iwasaki and Paszkowski, 2014; Balmer et al., 2015; Crisp et al., 2016; Hilker et al., 2016; Wojtyla et al., 2016; Gallusci et al., 2017; Lämke and Bäurle, 2017; Bäurle, 2018; Figure 2). Epigenetic mechanisms do not alter the DNA sequence but have the potential to change gene expression (Bossdorf et al., 2008). Stress memory based on non-coding RNA and histone modifications generally lasts no longer than several hours or days (Mathieu et al., 2007; Cedar and Bergman, 2009; Lämke and Bäurle, 2017; Kumar, 2018), with some exceptions (Huang et al., 2013; Bilichak et al., 2015; Morgado et al., 2017). In contrast, DNA methylation is more stable, and can even be heritable across generations (Boyko et al., 2010; Ou et al., 2012; Verhoeven and van Gurp, 2012; Bilichak and Kovalchuk, 2016; González et al., 2017). For example, mediation of transgenerational priming via inherited DNA methylation has been demonstrated in the plant Polygonum persicaria, in which demethylation of offspring with zebularine removed the adaptive effect of parental drought exposure in the form of longer root systems and greater biomass (Herman and Sultan, 2016).
Indications of Priming in Macrophytes
Recent studies show that seagrass can become less susceptible to heat events if it has been primed to stressful temperatures (Figure 1). For example, primed individuals (6 days at 29°C, 4°C above natural conditions) of the seagrass species Zostera muelleri and Posidonia australis showed significantly enhanced photosynthetic capacity, leaf growth, and chlorophyll a content after exposure to heat stress (32°C for 9 days) compared with naïve plants (Nguyen et al., 2020). Moreover, previous exposure of Zostera marina to simulated warming (15°C for 45 days, 2°C above control temperature) resulted in an increase in clonal shoot production and shoot length, as well as a decrease in leaf growth rates and in the ratio of below to above ground biomass (DuBois et al., 2020). Vegetatively grown shoots of primed parental plants could maintain biomass production under a second warming event (ca. 16°C for 40 days) but not shoots of naïve parental plants. The changes, which lasted for several years across multiple clonal generations after the stress was removed, would likely be adaptive in a warmer environment by reducing the respiratory burden of non-photosynthetic tissues. As discussed in Nguyen et al. (2020), heat priming may explain why the Mediterranean seagrass Posidonia oceanica did not suffer high mortality rates after intense and long-lasting heat-waves in 2012, 2015, and 2017 (Darmaraki et al., 2019), in contrast to an extensive die-off after the 2006 heatwave (Marbà and Duarte, 2010).
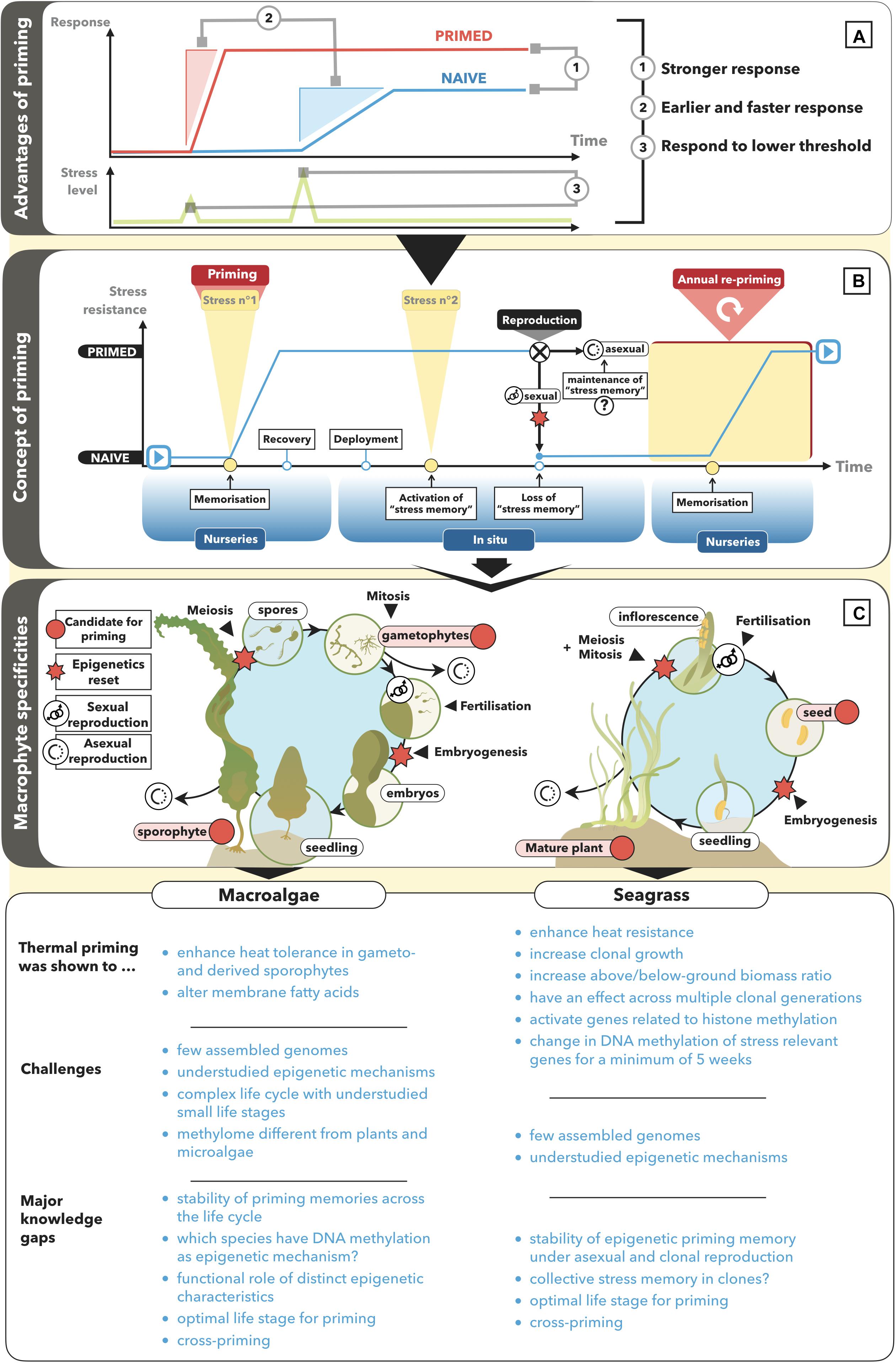
Figure 1. Concept of priming and potential for its application in macroalgae and seagrass. (A) A primed organism responds faster, earlier, stronger or to a lower threshold of a stressful triggering stimulus as compared with a naïve organism. (B) A naïve organism is primed by building up a memory of a certain stress stimulus. The stress memory is more likely to be heritable across asexually produced generations than across sexually produced generations because of epigenetic reprogramming under gametogenesis (meiosis) and embryogenesis. (C) At which stage priming is best applied depends on the stability/transfer of a priming memory across the life cycle stages of kelp and seagrass. The state of the art, challenges and knowledge gaps to establish priming as a novel bio-engineering technique in marine macrophytes are listed for macroalgae and seagrass, respectively.
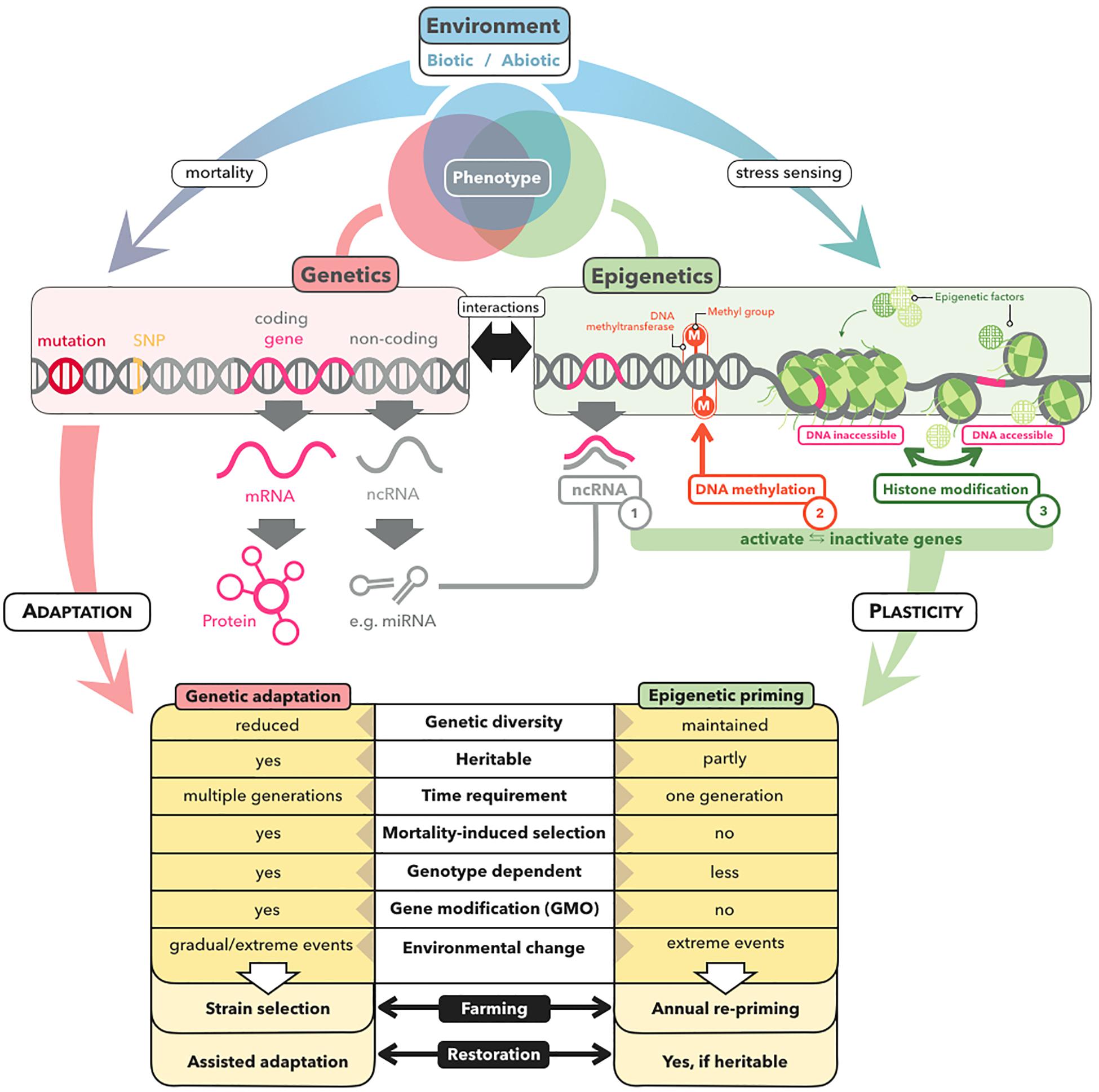
Figure 2. Genetic versus epigenetic mechanisms underlying stress adaptation and their relevance for restoration and farming of marine macrophytes. Environmental stress can alter the phenotype of marine macrophytes by positive selection of pre-adapted genotypes or of new beneficial mutations, resulting in genetic adaptation of the population within several generations. In contrast, epigenetic mechanisms, comprising ncRNAs, DNA methylation, and histone modifications, contribute to phenotypic plasticity by altering the expression patterns of genes within a single generation. The key characteristics with respect to the application potential of genetic and epigenetic mechanisms for farming and restoration of marine macrophytes, are listed at the bottom.
Evidence that epigenetic modifications contribute to form a thermal stress memory in seagrass is suggested by significant stress-induced regulation of methylation-related genes, in particular histone methyltransferases (Nguyen et al., 2020), and a change in DNA-methylation patterns that lasted for at least 5 weeks following exposure to heat stress (Jueterbock et al., 2020). A 5-week heat-stress memory is potentially long enough to heat-harden the same generation of previously exposed shoots. This methylation memory involved CG hyper-methylation and, thus, potentially constitutive upregulation (Zhang et al., 2006; Yang et al., 2014; Dubin et al., 2015; Niederhuth and Schmitz, 2017) of genes involved in the breakdown of heat-denatured proteins (Feder and Hofmann, 2002); which would be expected to provide a faster or stronger protective response upon exposure to a second heat stress.
In fucoid macroalgae, priming was shown to enhance resistance to dry and cold conditions (Schonbeck and Norton, 1979; Collén and Davison, 2001). Moreover, in the kelp Laminaria digitata, gametophyte exposure to low temperatures (5°C versus 15°C) significantly enhanced growth of the derived sporophytes under benign conditions (5 and 15°C) (Liesner et al., 2020). A small number of studies suggest that macroalgal performance under heat stress may be bio-engineered by thermal priming (Figure 1). First, priming the gametophyte generation of the kelp Alaria esculenta for 3 days at 22°C (compared with 12°C) enhanced their survival under increased temperatures, and the growth of the derived sporophyte generation (Quigley et al., 2018). Second, cultivation of Saccharina japonica gametophytes at 22–24°C increased the heat-tolerance of the derived sporophytes by 2°C (Wu and Pang, 1998) in Bricknell et al. (2021). Third, in the fucoid brown alga Fucus vesiculosus, storage of parental tissue at a higher temperature (14°C versus 4°C), or acclimation of embryos to 29°C significantly increased their survival by 30–50% under 33°C (Li and Brawley, 2004). Fourth, individuals of the red alga Bangia fuscopurpurea primed for 3 days at 28°C could survive 1 week at 32°C significantly better than naïve individuals (Kishimoto et al., 2019). The priming stress caused an increase in the saturation level of membrane fatty acids, suggesting that altered membrane fluidity is part of the species’ heat stress memory. However, this memory lasted for only 5 days after the primed individuals were returned to benign conditions (15°C).
Discussion – Prospects and Challenges of Priming in Marine Macrophytes
Distinguishing Priming From Selection
Just as thermal stress in natural settings can cause mortality and selection (Coleman and Wernberg, 2020; Coleman et al., 2020; Gurgel et al., 2020), priming induced mortality could inadvertently result in selection of pre-adapted genotypes–which may explain the observed transfer of positive effects from the primed gametophyte to the derived sporophyte generation of kelp (Quigley et al., 2018; Liesner et al., 2020). In order not to falsely ascribe improved stress tolerance to the formation of a molecular stress memory, it is critical to distinguish between priming and selection. This could be achieved through establishing correlations between positive priming effects and priming-induced epigenetic shifts that are independent from priming-induced genetic shifts by using partial mantel tests and multivariate redundancy analysis (Foust et al., 2016; Gugger et al., 2016; Herrera et al., 2016; Oksanen et al., 2016; Jueterbock et al., 2020). Moreover, tests for outlier loci that have become dominant allelic variants under positive selection (Narum and Hess, 2011; Günther and Coop, 2013; Ahrens et al., 2018) should be carried out in order to prove that positive priming effects cannot be explained by the survival of adapted genotypes.
Specificities of the Brown Algal Methylome
While the presence of cytosine methylation has been reported for green algae, red algae, dinoflagellates, and diatoms (Maumus et al., 2011; Tirichine and Bowler, 2011; Veluchamy et al., 2013; Bräutigam and Cronk, 2018; Lee J. M. et al., 2018), it is still not clear which brown algae share a lack of DNA methylation with the filamentous brown alga Ectocarpus sp. (Cock et al., 2010), in which epigenetic variation may be instead mediated at the chromatin level by histone modifications (Bourdareau et al., 2020) or via stress responsive miRNAs (Cock et al., 2017). Recently, DNA methylation was detected in the kelp S. japonica (Fan et al., 2019). The kelp methylome has been shown to change between life-cycle stages, to correlate with gene expression, and to differ from that of plants and microalgae. For example, methylation occurs predominately in CHH sequence contexts, which transfer methylation less reliably across mitotic cell divisions than CG sites (Law and Jacobsen, 2010). Moreover, DNA methylation appears to rely on a DNA methyltransferase (DNMT2) that is of low efficiency compared with other DNMTs, belonging to a class that mainly catalyzes tRNA methylation in plants and animals (Fan et al., 2019). How these specificities affect the functional role of the kelp methylome with respect to molecular stress memory remains unexplored.
Integrative Analyses
Parallel recording of epigenetic and transcriptomic priming responses can allow the identification of priming-induced epialleles that correlate with gene expression patterns and therefore potentially explain enhanced stress resistance (e.g., heat shock proteins). For example, that heat-induced methylation changes could be involved in stress acclimation of the red alga Pyropia haitanensis, was suggested by their correlation with the expression of stress-responsive genes (Yu et al., 2018). Penalized regression methods present promising integrative multi-locus models to test for statistical relationships between different “omics” data sets as they can overcome the challenge of having a small number of individuals (n) relative to the number of parameters (p) (Pineda et al., 2015; Lien et al., 2018; Zhong et al., 2019). Causal effects of DNA methylation on improved phenotypes may be possible to model with structural equation modeling (SEM), an established multivariate method that is relatively new to the field of molecular biology (Igolkina and Samsonova, 2018; Fatima et al., 2020). To demonstrate a causative relationship between priming memories and adaptive phenotypic changes requires experimental removal of DNA methylation, e.g., using Zebularine or 5-Azacytidine (Griffin et al., 2016), or targeted modifications of epigenetic marks, e.g., using the CRISPR-Cas system (Xu et al., 2016).
Inferences about the functional effect of molecular priming memories rely on the availability of annotated genomes, which are still scarce for marine macrophytes. Genomes have been published for six brown macroalgae: Ectocarpus sp. (Cock et al., 2010), S. japonica (Ye et al., 2015), Undaria pinnatifida (Shan et al., 2020), Cladosiphon okamuranus (Nishitsuji et al., 2016), Nemacystus decipiens (Nishitsuji et al., 2019), and Sargassum fusiforme (Wang et al., 2020). Published seagrass genomes include Z. marina (Olsen et al., 2016), Z. muelleri (Lee et al., 2016), and Halophila ovalis (Lee H. et al., 2018). Thus, the assembly and annotation of genomes, particularly of species of high ecological or commercial relevance, is a key priority in priming-related research.
Stability and Transfer of the Priming Memory
Multi-generational stability of the priming memory (transgenerational priming) is more important for the application of priming in restoration than in cultivation. Macroalgal cultivation naturally allows for annual re-priming during the few weeks the macroalgae are cultivated under controlled laboratory conditions. For example, for kelps, priming could be annually applied to either the haploid gametophyte cultures or to the young diploid sporophytes before being deployed at sea until harvest. While priming of the gametophytes would require the least resources (i.e., space and water), it is not clear to what extent epigenetic reprogramming during fertilization would affect transmission of a priming memory to the sporophyte generation. To characterize the transfer of priming memories via small life-cycle stages such as meiospores, gametes, and zygotes may become possible with new single-cell ‘omics technologies (Wang and Bodovitz, 2010; Zhu et al., 2020) that allow to sequence at DNA quantities which are too low for more traditional high-throughput sequencing technologies.
For restoration of kelp forests and macroalgae beds, thermal priming could be applied to the newly developed restoration tool “Green gravel,” where macroalgae are seeded on rocks and reared in the laboratory until reaching a size of 2–3 cm (Fredriksen et al., 2020). Specifically, priming could be used to enhance initial survival of gametophytes and juvenile sporophytes to the generally harsher conditions in degraded areas where an adult canopy is lacking. However, if not transferred across generations, any positive priming effect will last, at most, until the primed individuals have died, and will not provide long-term protection against recurrent stress. Some macroalgae grow vegetatively (e.g., Ecklonia brevipes; Coleman and Wernberg, 2018), allowing to compare the longevity of priming effects under different modes of reproduction in macroalgae being applied in a restoration context.
In seagrass meadows, priming memories are likely to be more stable across vegetatively/mitotically grown generations than across sexually produced generations because epigenetic marks are often reset during meiosis and embryogenesis (Figure 1C; Hirsch et al., 2012; Douhovnikoff and Dodd, 2014; Dodd and Douhovnikoff, 2016; González et al., 2017). While direct tests for predicted sexual-asexual differences in the transgenerational stability of epigenetic marks are virtually lacking (Verhoeven and Preite, 2014), a unique system to test these differences is provided by the dramatic range in clonal diversity and life history strategies of seagrasses, ranging from predominantly vegetative to predominantly sexual reproduction (Kilminster et al., 2015).
Clonal seagrass meadows further provide a unique potential to study whether communication of epigenetic information across physically connected shoots allows the acquisition of a collective stress memory to prepare interconnected ramets for a range of future environmental challenges (Latzel et al., 2016). The transport of epigenetic information from somatic tissue to the germline via miRNAs (small ncRNAs that can cross cell barriers) has been demonstrated in humans and the nematode Caenorhabditis elegans (Creemers et al., 2012; Devanapally et al., 2015; Sharma, 2015; Szyf, 2015). Whether such communication may extend across interconnected ramets of the same clone has never been tested.
Prospects to Explore Priming as Biotechnological Tool in Marine Macrophytes
Priming has a large potential to enhance restoration success of macroalgal forests and seagrass meadows, and to ensure production security of macroalgal biomass under environmental challenges. Because primed organisms are not considered genetically modified, they can be grown in countries where GMO restrictions apply. Moreover, priming would likely be a less controversial and more socially acceptable way to boost resilience in macrophytes relative to the proposed gene editing approaches (Coleman and Goold, 2019). However, priming cannot be simply transferred from terrestrial plants to marine macrophytes. In particular, brown and red macroalgae are distantly related to terrestrial plants, and kelps have complex heteromorphic life cycles with free-living gametophyte generations. Thus, in order to identify whether priming can be established as a novel bio-engineering technique for marine macrophytes, we need ambitious fundamental research that uses complex experimental setups combined with multivariate analyses that can integrate multiple high-throughput sequencing datasets to test at which intensity, duration, and life-cycle stage priming has a positive and long-term effect without inducing selection or high mortality. For priming to be of commercial value to the macroalgae farming industry, we must further assess whether the cost factor added to the cultivation process pays off by enhancing yield even in years where the macroalgae are not exposed to stress or by providing cross-protection to other relevant stressors (Hilker et al., 2016). Despite these knowledge gaps, priming should be explored as a tool to boost resilience of both seagrass and macroalgae to secure their ecological and economic values in future oceans.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
AJ wrote the first draft of the manuscript. AM created the figures. JC, MC, TW, LS, RR, JZ, and Z-MH contributed to the article by commenting and re-writing sections, and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a personal research talents grant from Nord University to AJ and Norwegian Research Council (project 243916). JC was supported by the Agence Nationale de la Recherche project Epicycle (ANR-19-CE20-0028-01). MC and TW were supported by Australian Research Council Grants DP190100058 and DP200100201. Z-MH was supported by the National Natural Science Foundation of China (31971395). Open access publication fees were covered by Nord University’s Open Access Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for their contribution to improve this manuscript.
References
Ahrens, C. W., Rymer, P. D., Stow, A., Bragg, J., Dillon, S., Umbers, K. D. L., et al. (2018). The search for loci under selection: trends, biases and progress. Mol. Ecol. 27, 1342–1356. doi: 10.1111/mec.14549
Araújo, R., Vázquez Calderón, F., Sánchez López, J., Azevedo, I. C., Bruhn, A., Fluch, S., et al. (2021). Current status of the algae production industry in Europe: an emerging sector of the blue bioeconomy. Front. Mar. Sci. 7:626389. doi: 10.3389/fmars.2020.626389
Arias-Ortiz, A., Serrano, O., Masqué, P., Lavery, P. S., Mueller, U., Kendrick, G. A., et al. (2018). A marine heatwave drives massive losses from the world’s largest seagrass carbon stocks. Nat. Clim. Chang. 8, 338–344. doi: 10.1038/s41558-018-0096-y
Assis, J., Araújo, M. B., and Serrão, E. A. (2018). Projected climate changes threaten ancient refugia of kelp forests in the North Atlantic. Glob. Chang. Biol. 24, e55–e66. doi: 10.1111/gcb.13818
Assis, J., Berecibar, E., Claro, B., Alberto, F., Reed, D., Raimondi, P., et al. (2017). Major shifts at the range edge of marine forests: the combined effects of climate changes and limited dispersal. Sci. Rep. 7:44348. doi: 10.1038/srep44348
Assis, J., Lucas, A. V., Bárbara, I., and Serrão, E. A. Á (2016). Future climate change is predicted to shift long-term persistence zones in the cold-temperate kelp Laminaria hyperborea. Mar. Environ. Res. 113, 174–182. doi: 10.1016/j.marenvres.2015.11.005
Balmer, A., Pastor, V., Gamir, J., Flors, V., and Mauch-Mani, B. (2015). The “prime-ome”: towards a holistic approach to priming. Trends Plant Sci. 20, 443–452. doi: 10.1016/j.tplants.2015.04.002
Bäurle, I. (2018). Can’t remember to forget you: chromatin-based priming of somatic stress responses. Semin. Cell Dev. Biol. 83, 133–139. doi: 10.1016/j.semcdb.2017.09.032
Benson, C. W., Mao, Q., and Huff, D. R. (2020). Global DNA methylation predicts epigenetic reprogramming and transgenerational plasticity in Poa annua L. Crop Sci. 1–12. doi: 10.1002/csc2.20337
Bilichak, A., Ilnytskyy, Y., Wóycicki, R., Kepeshchuk, N., Fogen, D., and Kovalchuk, I. (2015). The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 6:5. doi: 10.3389/fpls.2015.00005
Bilichak, A., and Kovalchuk, I. (2016). Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 67, 2081–2092. doi: 10.1093/jxb/erw066
Bossdorf, O., Richards, C. L., and Pigliucci, M. (2008). Epigenetics for ecologists. Ecol. Lett. 11, 106–115. doi: 10.1111/j.1461-0248.2007.01130.x
Bourdareau, S., Tirichine, L., Lombard, B., Loew, D., Scornet, D., Coelho, S., et al. (2020). Histone modifications during the life cycle of the brown alga Ectocarpus. Genome Biol. 22:12. doi: 10.1101/2020.03.09.980763
Boyko, A., Blevins, T., Yao, Y., Golubov, A., Bilichak, A., Ilnytskyy, Y., et al. (2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLoS One 5:e9514. doi: 10.1371/journal.pone.0009514
Bräutigam, K., and Cronk, Q. (2018). DNA methylation and the evolution of developmental complexity in plants. Front. Plant Sci. 9:1447. doi: 10.3389/fpls.2018.01447
Breeman, A. M. (1990). Expected effects of changing seawater temperatures on the geographic distribution of seaweed species. Expect. Eff. Clim. Chang. Mar. Coast. Ecosyst. 57, 69–76. doi: 10.1007/978-94-009-2003-3_9
Bricknell, I. R., Birkel, S. D., Brawley, S. H., Van Kirk, T., Hamlin, H., Capistrant-Fossa, K., et al. (2021). Resilience of cold water aquaculture: a review of likely scenarios as climate changes in the Gulf of Maine. Rev. Aquac. 13, 460–503. doi: 10.1111/raq.12483
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304. doi: 10.1038/nrg2540
Chefaoui, R. M., Duarte, C. M., and Serrão, E. A. (2018). Dramatic loss of seagrass habitat under projected climate change in the Mediterranean Sea. Glob. Chang. Biol. 24, 4919–4928. doi: 10.1111/gcb.14401
Chopin, T. (2017). Seaweed aquaculture – from the global, mostly Asian, picture to the opportunities and constraints of the Canadian scene. Bull. Aquacul. Assoc. Canada 1, 1–8.
Cock, J., Liu, F., Duan, D., Bourdareau, S., Lipinska, A. P., Coelho, S. M., et al. (2017). Rapid evolution of microRNA loci in the brown algae. Genome Biol. Evol. 9, 740–749. doi: 10.1093/gbe/evx038
Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621. doi: 10.1038/nature09016
Coleman, M. A., and Goold, H. D. (2019). Harnessing synthetic biology for kelp forest conservation1. J. Phycol. 751, 745–751. doi: 10.1111/jpy.12888
Coleman, M. A., Minne, A. J. P., Vranken, S., and Wernberg, T. (2020). Genetic tropicalisation following a marine heatwave. Sci. Rep. 10:12726. doi: 10.1038/s41598-020-69665-w
Coleman, M. A., and Wernberg, T. (2018). Genetic and morphological diversity in sympatric kelps with contrasting reproductive strategies. Aquat. Biol. 27, 65–73. doi: 10.3354/ab00698
Coleman, M. A., and Wernberg, T. (2020). Forum the silver lining of extreme events trends in ecology & evolution. Trends Ecol. Evol. 35, 1065–1067. doi: 10.1016/j.tree.2020.08.013
Collén, J., and Davison, I. R. (2001). Seasonality and thermal acclimation of reactive oxygen metabolism in Fucus vesiculosus (Phaeophyceae). J. Phycol. 37, 474–481. doi: 10.1046/j.1529-8817.2001.037004474.x
Costanza, R., de Groot, R., Sutton, P., van der Ploeg, S., Anderson, S. J., Kubiszewski, I., et al. (2014). Changes in the global value of ecosystem services. Glob. Environ. Chang. 26, 152–158. doi: 10.1016/j.gloenvcha.2014.04.002
Creemers, E. E., Tijsen, A. J., and Pinto, Y. M. (2012). Circulating MicroRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 110, 483–495. doi: 10.1161/CIRCRESAHA.111.247452
Crisp, P. A., Ganguly, D., Eichten, S. R., Borevitz, J. O., and Pogson, B. J. (2016). Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2:e1501340. doi: 10.1126/sciadv.1501340
Darmaraki, S., Somot, S., Sevault, F., and Nabat, P. (2019). Past variability of Mediterranean Sea marine heatwaves. Geophys. Res. Lett. 46, 9813–9823. doi: 10.1029/2019GL082933
Devanapally, S., Ravikumar, S., and Jose, A. M. (2015). Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc. Natl. Acad. Sci. U. S. A. 112, 2133–2138. doi: 10.1073/pnas.1423333112
Dodd, R. S., and Douhovnikoff, V. (2016). Adjusting to global change through clonal growth and epigenetic variation. Front. Ecol. Evol. 4:86. doi: 10.3389/fevo.2016.00086
Douhovnikoff, V., and Dodd, R. S. (2014). Epigenetics: a potential mechanism for clonal plant success. Plant Ecol. 216, 227–233. doi: 10.1007/s11258-014-0430-z
Duarte, B., Martins, I., Rosa, R., Matos, A. R., Roleda, M. Y., Reusch, T. B. H., et al. (2018). Climate change impacts on seagrass meadows and macroalgal forests: an integrative perspective on acclimation and adaptation potential. Front. Mar. Sci. 5:190. doi: 10.3389/FMARS.2018.00190
Dubin, M. J., Zhang, P., Meng, D., Remigereau, M. S., Osborne, E. J., Casale, F. P., et al. (2015). DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 4:e05255. doi: 10.7554/eLife.05255
DuBois, K., Williams, S. L., and Stachowicz, J. J. (2020). Previous exposure mediates the response of eelgrass to future warming via clonal transgenerational plasticity. Ecology 101:e03169. doi: 10.1002/ecy.3169
Eger, A. M., Vergés, A., Choi, C. G., Christie, H., Coleman, M. A., Fagerli, C. W., et al. (2020). Financial and institutional support are important for large-scale kelp forest restoration. Front. Mar. Sci. 7:535277. doi: 10.3389/fmars.2020.535277
Fan, X., Han, W., Teng, L., Jiang, P., Zhang, X., Xu, D., et al. (2019). Single-base methylome profiling of the giant kelp Saccharina japonica reveals significant differences in DNA methylation to microalgae and plants. New Phytol. 225, 234–249. doi: 10.1111/nph.16125
FAO (2020). The State of World Fisheries and Aquaculture 2020. Rome: Food and Agriculture Organization of the United Nations
Fatima, N., Ahmed, S. H., Chauhan, S. S., Mohammad, O., and Rehman, S. M. F. (2020). Structural equation modelling analysis determining causal role among methyltransferases, methylation, and apoptosis during human pregnancy and abortion. Sci. Rep. 10:12408. doi: 10.1038/s41598-020-68270-1
Feder, M. E., and Hofmann, G. E. (2002). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Filbee-Dexter, K., and Wernberg, T. (2018). Rise of turfs: a new battlefront for globally declining kelp forests. Bioscience 68, 64–76. doi: 10.1093/biosci/bix147
Filbee-Dexter, K., Wernberg, T., Grace, S. P., Thormar, J., Fredriksen, S., Narvaez, C. N., et al. (2020). Marine heatwaves and the collapse of North Atlantic kelp forests. Proc. R. Soc. B Biol. Sci. 10:13388. doi: 10.1038/s41598-020-70273-x
Foust, C. M., Preite, V., Schrey, A. W., Alvarez, M., Robertson, M. H., Verhoeven, K. J. F. F., et al. (2016). Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol. Ecol. 25, 1639–1652. doi: 10.1111/mec.13522
Fredriksen, S., Filbee-Dexter, K., Norderhaug, K. M., Steen, H., Bodvin, T., Coleman, M. A., et al. (2020). Green gravel: a novel restoration tool to combat kelp forest decline. Sci. Rep. 10:3983. doi: 10.1038/s41598-020-60553-x
Gallusci, P., Dai, Z., Génard, M., Gauffretau, A., Leblanc-Fournier, N., Richard-Molard, C., et al. (2017). Epigenetics for plant improvement: current knowledge and modeling avenues. Trends Plant Sci. 22, 610–623. doi: 10.1016/j.tplants.2017.04.009
González, A. P. R., Dumalasová, V., Rosenthal, J., Skuhrovec, J., and Latzel, V. (2017). The role of transgenerational effects in adaptation of clonal offspring of white clover (Trifolium repens) to drought and herbivory. Evol. Ecol. 31, 345–361. doi: 10.1007/s10682-016-9844-5
Grebe, G. S., Byron, C. J., Gelais, A. S., Kotowicz, D. M., and Olson, T. K. (2019). An ecosystem approach to kelp aquaculture in the Americas and Europe. Aquac. Rep. 15:100215. doi: 10.1016/j.aqrep.2019.100215
Griffin, P. T., Niederhuth, C. E., and Schmitz, R. J. (2016). A comparative analysis of 5-azacytidine-and zebularine-induced DNA demethylation. G3 (Bethesda) 6, 2773–2780. doi: 10.1534/g3.116.030262
Gugger, P. F., Fitz-Gibbon, S., PellEgrini, M., and Sork, V. L. (2016). Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol. Ecol. 25, 1665–1680. doi: 10.1111/mec.13563
Günther, T., and Coop, G. (2013). Robust identification of local adaptation from allele frequencies. Genetics 195, 205–220. doi: 10.1534/genetics.113.152462
Gurgel, C. F. D., Camacho, O., Minne, A. J. P., Wernberg, T., and Coleman, M. A. (2020). Marine heatwave drives cryptic loss of genetic diversity in underwater forests. Curr. Biol. 30, 1199–1206.e2. doi: 10.1016/j.cub.2020.01.051
Harley, C. D. G., Anderson, K. M., Demes, K. W., Jorve, J. P., Kordas, R. L., Coyle, T. A., et al. (2012). Effects of climate change on global seaweed communities. J. Phycol. 48, 1064–1078. doi: 10.1111/j.1529-8817.2012.01224.x
Hereward, H. F. R., King, N. G., and Smale, D. A. (2020). Intra-annual variability in responses of a canopy forming kelp to cumulative low tide heat stress: implications for populations at the trailing range edge. J. Phycol. 56, 146–158. doi: 10.1111/jpy.12927
Herman, J. J., and Sultan, S. E. (2011). Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front. Plant Sci. 2:102. doi: 10.3389/fpls.2011.00102
Herman, J. J., and Sultan, S. E. (2016). DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. R. Soc. B Biol. Sci. 283:20160988. doi: 10.1098/rspb.2016.0988
Herrera, C. M., Medrano, M., and Bazaga, P. (2016). Comparative spatial genetics and epigenetics of plant populations: heuristic value and a proof of concept. Mol. Ecol. 25, 1653–1664. doi: 10.1111/mec.13576
Hilker, M., Schwachtje, J., Baier, M., Balazadeh, S., Bäurle, I., Geiselhardt, S., et al. (2016). Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 91, 1118–1133. doi: 10.1111/brv.12215
Hirsch, S., Baumberger, R., and Grossniklaus, U. (2012). Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb. Symp. Quant. Biol. 77, 97–104. doi: 10.1101/sqb.2013.77.014605
Hu, Z. M., Shan, T. F., Zhang, J., Zhang, Q. S., Critchley, A. T., Choi, H. G., et al. (2021). Kelp aquaculture in China: a retrospective and future prospects. Rev. Aquac. 13, doi: 10.1111/raq.12524 [Epub ahead of print].
Huang, C., Xu, M., and Zhu, B. (2013). Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? Philos. Trans. R. Soc. B Biol. Sci. 368:20110332. doi: 10.1098/rstb.2011.0332
Ibrahim, E. A. (2016). Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 192, 38–46. doi: 10.1016/j.jplph.2015.12.011
Igolkina, A. A., and Samsonova, M. G. (2018). SEM: structural equation modeling in molecular biology. Biophysics 63, 139–148. doi: 10.1134/S0006350918020100
Iwasaki, M., and Paszkowski, J. (2014). Epigenetic memory in plants. EMBO J. 33, 1987–1998. doi: 10.15252/embj.201488883
Jueterbock, A., Boström, C., Coyer, J. A., Olsen, J. L., Kopp, M., Dhanasiri, A. K. S., et al. (2020). The seagrass methylome is associated with variation in photosynthetic performance among clonal shoots. Front. Plant Sci. 11:571646. doi: 10.3389/fpls.2020.571646
Jueterbock, A., Tyberghein, L., Verbruggen, H., Coyer, J. A., Olsen, J. L., and Hoarau, G. (2013). Climate change impact on seaweed meadow distribution in the North Atlantic rocky intertidal. Ecol. Evol. 3, 1356–1373. doi: 10.1002/ece3.541
Kilminster, K., McMahon, K., Waycott, M., Kendrick, G. A., Scanes, P., McKenzie, L., et al. (2015). Unravelling complexity in seagrass systems for management: Australia as a microcosm. Sci. Total Environ. 534, 97–109. doi: 10.1016/j.scitotenv.2015.04.061
Kishimoto, I., Ariga, I., Itabashi, Y., and Mikami, K. (2019). Heat-stress memory is responsible for acquired thermotolerance in Bangia fuscopurpurea. J. Phycol. 55, 971–975. doi: 10.1111/jpy.12895
Klinger, T. (2015). The role of seaweeds in the modern ocean. Perspect. Phycol. 2, 31–39. doi: 10.1127/pip/2015/0024
Krumhansl, K. A., Okamoto, D. K., Rassweiler, A., Novak, M., Bolton, J. J., Cavanaugh, K. C., et al. (2016). Global patterns of kelp forest change over the past half-century. Proc. Natl. Acad. Sci. U. S. A. 113, 13785–13790. doi: 10.1073/pnas.1606102113
Kumar, S. (2018). Epigenomics of plant responses to environmental stress. Epigenomes 2:6. doi: 10.3390/epigenomes2010006
Lämke, J., and Bäurle, I. (2017). Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18:124. doi: 10.1186/s13059-017-1263-6
Larkum, A. W. D. D., Orth, R. J., and Duarte, C. M. (2006). Seagrasses: biology, ecology, and conservation. Mar. Ecol. 27, 431–432. doi: 10.1111/j.1439-0485.2006.00138.x.
Latzel, V., Rendina González, A. P., and Rosenthal, J. (2016). Epigenetic memory as a basis for intelligent behavior in clonal plants. Front. Plant Sci. 7:1354. doi: 10.3389/fpls.2016.01354
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Layton, C., Coleman, M. A., Marzinelli, E. M., Steinberg, P. D., Swearer, S. E., Vergés, A., et al. (2020). Kelp forest restoration in Australia. Front. Mar. Sci. 7:74. doi: 10.3389/fmars.2020.00074
Lee, H., Golicz, A. A., Bayer, P. E., Severn-Ellis, A. A., Chan, C. K. K., Batley, J., et al. (2018). Genomic comparison of two independent seagrass lineages reveals habitat-driven convergent evolution. J. Exp. Bot. 69, 3689–3702. doi: 10.1093/jxb/ery147
Lee, H. T., Golicz, A. A., Bayer, P. E., Jiao, Y., Tang, H., Paterson, A. H., et al. (2016). The genome of a southern hemisphere seagrass species (Zostera muelleri). Plant Physiol. 172, 272–283. doi: 10.1104/pp.16.00868
Lee, J. M., Yang, E. C., Graf, L., Yang, J. H., Qiu, H., Zelzion, U., et al. (2018). Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in rhodophyta. Mol. Biol. Evol. 35, 1869–1886. doi: 10.1093/molbev/msy081
Li, R., and Brawley, S. H. (2004). Improved survival under heat stress in intertidal embryos (Fucus spp.) simultaneously exposed to hypersalinity and the effect of parental thermal history. Mar. Biol. 144, 205–213. doi: 10.1007/s00227-003-1190-9
Lien, T. G., Borgan, Ø, Reppe, S., Gautvik, K., and Glad, I. K. (2018). Integrated analysis of DNA-methylation and gene expression using high-dimensional penalized regression: a cohort study on bone mineral density in postmenopausal women. BMC Med. Genomics 11:24. doi: 10.1186/s12920-018-0341-2
Liesner, D., Shama, L. N. S., Diehl, N., Valentin, K., and Bartsch, I. (2020). Thermal plasticity of the kelp Laminaria digitata (Phaeophyceae) across life cycle stages reveals the importance of cold seasons for marine forests. Front. Mar. Sci. 7:456. doi: 10.3389/fmars.2020.00456
Marbà, N., and Duarte, C. M. (2010). Mediterranean warming triggers seagrass (Posidonia oceanica) shoot mortality. Glob. Chang. Biol. 16, 2366–2375. doi: 10.1111/j.1365-2486.2009.02130.x
Martínez, B., Radford, B., Thomsen, M. S., Connell, S. D., Carreño, F., Bradshaw, C. J. A., et al. (2018). Distribution models predict large contractions of habitat-forming seaweeds in response to ocean warming. Divers. Distrib. 24, 1350–1366. doi: 10.1111/ddi.12767
Mathieu, O., Reinders, J., Čaikovski, M., Smathajitt, C., and Paszkowski, J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130, 851–862. doi: 10.1016/j.cell.2007.07.007
Maumus, F., Rabinowicz, P., Bowler, C., and Rivarola, M. (2011). Stemming epigenetics in marine Stramenopiles. Curr. Genomics 12, 357–370. doi: 10.2174/138920211796429727
Mercé, C., Bayer, P. E., Fernandez, C. T., Batley, J., and Edwards, D. (2020). Induced methylation in plants as a crop improvement tool: progress and perspectives. Agronomy 10:1484. doi: 10.3390/agronomy10101484
Morgado, L., Preite, V., Oplaat, C., Anava, S., Ferreira de Carvalho, J., Rechavi, O., et al. (2017). Small RNAs reflect grandparental environments in apomictic dandelion. bioRxiv [Preprint] 099572. doi: 10.1101/099572
Narum, S. R., and Hess, J. E. (2011). Comparison of FST outlier tests for SNP loci under selection. Mol. Ecol. Resour. 11, 184–194. doi: 10.1111/j.1755-0998.2011.02987.x
Nepper-Davidsen, J., Andersen, D. T., and Pedersen, M. F. (2019). Exposure to simulated heatwave scenarios causes long-term reductions in performance in Saccharina latissima. Mar. Ecol. Prog. Ser. 630, 25–39. doi: 10.3354/meps13133
Nguyen, H. M., Kim, M., Ralph, P. J., Marín-Guirao, L., Pernice, M., and Procaccini, G. (2020). Stress memory in seagrasses: first insight into the effects of thermal priming and the role of epigenetic modifications. Front. Plant Sci. 11:494. doi: 10.3389/FPLS.2020.00494
Niederhuth, C. E., and Schmitz, R. J. (2017). Putting DNA methylation in context: from genomes to gene expression in plants. Biochim. Biophys. Acta Gene. Regul. Mech. 1860, 149–156. doi: 10.1016/j.bbagrm.2016.08.009
Nishitsuji, K., Arimoto, A., Higa, Y., Mekaru, M., Kawamitsu, M., Satoh, N., et al. (2019). Draft genome of the brown alga, Nemacystus decipiens, Onna-1 strain: fusion of genes involved in the sulfated fucan biosynthesis pathway. Sci. Rep. 9:4607. doi: 10.1038/s41598-019-40955-2
Nishitsuji, K., Arimoto, A., Iwai, K., Sudo, Y., Hisata, K., Fujie, M., et al. (2016). A draft genome of the brown alga, Cladosiphon okamuranus, S-strain: a platform for future studies of ‘mozuku’ biology. DNA Res. 23, 561–570. doi: 10.1093/dnares/dsw039
Oksanen, J. F., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2016). vegan: Community Ecology Package. R Packag. “vegan.”, doi: 10.4135/9781412971874.n145
Olsen, J. L., Rouzé, P., Verhelst, B., Lin, Y., Bayer, T., Collen, J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335. doi: 10.1038/nature16548
Orth, R. J., Carruthers, T. J. B., Dennison, W. C., Duarte, C. M., Fourqurean, J. W., Heck, K. L., et al. (2006). A global crisis for seagrass ecosystems. Bioscience 56, 987–996.
Ou, X., Zhang, Y., Xu, C., Lin, X., Zang, Q., Zhuang, T., et al. (2012). Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS One 7:e41143. doi: 10.1371/journal.pone.0041143
Pawar, V. A., and Laware, S. L. (2018). Seed priming a critical review. Int. J. Sci. Res. Biol. Sci. 5, 94–101. doi: 10.26438/ijsrbs/v5i5.94101
Pineda, S., Real, F. X., Kogevinas, M., Carrato, A., Chanock, S. J., Malats, N., et al. (2015). Integration analysis of three omics data using penalized regression methods: an application to bladder cancer. PLoS Genet. 11:e1005689. doi: 10.1371/journal.pgen.1005689
Provost, E. J., Kelaher, B. P., Dworjanyn, S. A., Russell, B. D., Connell, S. D., Ghedini, G., et al. (2017). Climate-driven disparities among ecological interactions threaten kelp forest persistence. Glob. Chang. Biol. 23, 353–361. doi: 10.1111/gcb.13414
Qiu, Z., Coleman, M. A., Provost, E., Campbell, A. H., Kelaher, B. P., Dalton, S. J., et al. (2019). Future climate change is predicted to affect the microbiome and condition of habitat-forming kelp. Proc. R. Soc. B Biol. Sci. 286, 1–10. doi: 10.1098/rspb.2018.1887
Quigley, C. T. C., Terry, C., and Quigley, C. (2018). Thermal and Microbial Effects On Brown Macroalgae: Heat Acclimation and the Biodiversity of the Microbiome. Ph. D. Thesis. The University of Maine, Orono, ME
Repolho, T., Duarte, B., Dionísio, G., Paula, J. R., Lopes, A. R., Rosa, I. C., et al. (2017). Seagrass ecophysiological performance under ocean warming and acidification. Sci. Rep. 7:41443. doi: 10.1038/srep41443
Schonbeck, M. W., and Norton, T. A. (1979). Drought-hardening in the upper-shore seaweeds Fucus spiralis and Pelvetia canaliculata. J. Ecol. 67:687–696. doi: 10.2307/2259120
Shan, T., Yuan, J., Su, L., Li, J., Leng, X., Zhang, Y., et al. (2020). First genome of the brown alga Undaria pinnatifida: chromosome-level assembly using PacBio and Hi-C technologies. Front. Genet. 11:140. doi: 10.3389/fgene.2020.00140
Sharma, A. (2015). Transgenerational epigenetic inheritance requires a much deeper analysis. Trends Mol. Med. 21, 269–270. doi: 10.1016/j.molmed.2015.02.010
Short, F. T., and Neckles, H. A. (1999). The effects of global climate change on seagrasses. Aquat. Bot. 63, 169–196. doi: 10.1016/S0304-3770(98)00117-X
Skjermo, J., Aasen, I. M., Arff, J., Broch, O. J., Carvajal, A., Forbord, S., et al. (2014). Report a New Norwegian Bioeconomy Based on Cultivation and Processing of Seaweeds: Opportunities and R&D Needs. Trondheim: SINTEF Fisheries and Aquaculture, 46.
Smale, D. A. (2020). Impacts of ocean warming on kelp forest ecosystems. New Phytol. 225, 1447–1454. doi: 10.1111/nph.16107
Smale, D. A., Pessarrodona, A., King, N., Burrows, M. T., Yunnie, A., Vance, T., et al. (2020). Environmental factors influencing primary productivity of the forest forming kelp Laminaria hyperborea in the northeast Atlantic. Sci. Rep. 10:12161. doi: 10.1038/s41598-020-69238-x
Stévant, P., Rebours, C., and Chapman, A. (2017). Seaweed aquaculture in Norway: recent industrial developments and future perspectives. Aquac. Int. 25, 1373–1390. doi: 10.1007/s10499-017-0120-7
Szyf, M. (2015). Nongenetic inheritance and transgenerational epigenetics. Trends Mol. Med. 21, 134–144. doi: 10.1016/j.molmed.2014.12.004
Tan, Y. M., Dalby, O., Kendrick, G. A., Statton, J., Sinclair, E. A., Fraser, M. W., et al. (2020). Seagrass restoration is possible: insights and lessons from Australia and New Zealand. Front. Mar. Sci. 7:617. doi: 10.3389/fmars.2020.00617
Teagle, H., Hawkins, S. J., Moore, P. J., and Smale, D. A. (2017). The role of kelp species as biogenic habitat formers in coastal marine ecosystems. J. Exp. Mar. Bio. Ecol. 492, 81–98. doi: 10.1016/j.jembe.2017.01.017
Tirichine, L., and Bowler, C. (2011). Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant J. 66, 45–57. doi: 10.1111/j.1365-313X.2011.04540.x
Valle, M., Chust, G., del Campo, A., Wisz, M. S., Olsen, S. M., Garmendia, J. M., et al. (2014). Projecting future distribution of the seagrass Zostera noltii under global warming and sea level rise. Biol. Conserv. 170, 74–85. doi: 10.1016/j.biocon.2013.12.017
Veluchamy, A., Lin, X., Maumus, F., Rivarola, M., Bhavsar, J., Creasy, T., et al. (2013). Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 4:2091. doi: 10.1038/ncomms3091
Vergés, A., Campbell, A. H., Wood, G., Kajlich, L., Eger, A. M., Cruz, D., et al. (2020). Operation crayweed: ecological and sociocultural aspects of restoring Sydney’s underwater forests. Ecol. Manag. Restor. 21, 74–85. doi: 10.1111/emr.12413
Vergés, A., McCosker, E., Mayer-Pinto, M., Coleman, M. A., Wernberg, T., Ainsworth, T., et al. (2019). Tropicalisation of temperate reefs: implications for ecosystem functions and management actions. Funct. Ecol. 33, 1000–1013. doi: 10.1111/1365-2435.13310
Verhoeven, K. J. F., and Preite, V. (2014). Epigenetic variation in asexually reproducing organisms. Evolution 68, 644–655. doi: 10.1111/evo.12320
Verhoeven, K. J. F., and van Gurp, T. P. (2012). Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS One 7:e38605. doi: 10.1371/journal.pone.0038605
Wang, D., and Bodovitz, S. (2010). Single cell analysis: the new frontier in “omics.”. Trends Biotechnol. 28, 281–290. doi: 10.1016/j.tibtech.2010.03.002
Wang, S., Lin, L., Shi, Y., Qian, W., Li, N., Yan, X., et al. (2020). First draft genome assembly of the seaweed Sargassum fusiforme. Front. Genet. 11:590065. doi: 10.3389/fgene.2020.590065
Wang, X., Liu, F. l., and Jiang, D. (2017). Priming: a promising strategy for crop production in response to future climate. J. Integr. Agric. 16, 2709–2716. doi: 10.1016/S2095-3119(17)61786-6
Waycott, M., Duarte, C. M., Carruthers, T. J. B., Orth, R. J., Dennison, W. C., Olyarnik, S., et al. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. U. S. A. 106, 12377–12381. doi: 10.1073/pnas.0905620106
Wernberg, T., Krumhansl, K., Filbee-Dexter, K., and Pedersen, M. F. (2019). Status and Trends for the World’s Kelp Forests. 57–78. Available online at: https://forskning.ruc.dk/en/publications/status-and-trends-for-the-worlds-kelp-forests (accessed November 13, 2020).
Whittle, C. A., Otto, S. P., Johnston, M. O., and Krochko, J. E. (2009). Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana this paper is one of a selection of papers published in a special issue from the National Research Council of Canada – Plant Biotechnology Institute. Botany 87, 650–657. doi: 10.1139/b09-030
Wilson, K. L., and Lotze, H. K. (2019). Climate change projections reveal range shifts of eelgrass Zostera marina in the Northwest Atlantic. Mar. Ecol. Prog. Ser. 620, 47–62. doi: 10.3354/meps12973
Wojtyla, Ł, Lechowska, K., Kubala, S., and Garnczarska, M. (2016). Molecular processes induced in primed seeds—increasing the potential to stabilize crop yields under drought conditions. J. Plant Physiol. 203, 116–126. doi: 10.1016/j.jplph.2016.04.008
Wojtyla, Ł, Paluch-Lubawa, E., Sobieszczuk-Nowicka, E., and Garnczarska, M. (2020). “Drought stress memory and subsequent drought stress tolerance in plants,” in Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants, eds M. A. Hossain, F. Liu, D. Burritt, M. Fujita, and B. Huang (Amsterdam: Elsevier), 115–131. doi: 10.1016/B978-0-12-817892-8.00007-6
Wu, C., and Pang, S. (1998). The Seaweed Resources of China. Seaweed Resource World, 26–47. Available online at: http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:THE+SEAWEED+RESOURCES+OF+CHINA#0 (accessed August, 2020).
Xu, X., Tao, Y., Gao, X., Zhang, L., Li, X., Zou, W., et al. (2016). A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2:16009. doi: 10.1038/celldisc.2016.9
Yang, X., Han, H., DeCarvalho, D. D., Lay, F. D., Jones, P. A., and Liang, G. (2014). Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590. doi: 10.1016/j.ccr.2014.07.028
Ye, N., Zhang, X., Miao, M., Fan, X., Zheng, Y., Xu, D., et al. (2015). Saccharina genomes provide novel insight into kelp biology. Nat. Commun. 6:6986. doi: 10.1038/ncomms7986
Yu, C., Xu, K., Wang, W., Xu, Y., Ji, D., Chen, C., et al. (2018). Detection of changes in DNA methylation patterns in Pyropia haitanensis under high-temperature stress using a methylation-sensitive amplified polymorphism assay. J. Appl. Phycol. 30, 2091–2100. doi: 10.1007/s10811-017-1363-4
Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S. W. L., Chen, H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201. doi: 10.1016/j.cell.2006.08.003
Zhong, H., Kim, S., Zhi, D., and Cui, X. (2019). Predicting gene expression using DNA methylation in three human populations. PeerJ 1:e6757. doi: 10.7717/peerj.6757
Keywords: DNA methylation, plasticity, stress memory, bio-engineering, seagrass, macroalgae farming, kelp restoration, heat hardening
Citation: Jueterbock A, Minne AJP, Cock JM, Coleman MA, Wernberg T, Scheschonk L, Rautenberger R, Zhang J and Hu Z-M (2021) Priming of Marine Macrophytes for Enhanced Restoration Success and Food Security in Future Oceans. Front. Mar. Sci. 8:658485. doi: 10.3389/fmars.2021.658485
Received: 25 January 2021; Accepted: 04 March 2021;
Published: 23 March 2021.
Edited by:
Ricardo A. Melo, University of Lisbon, PortugalReviewed by:
Daniel Robledo, Centro de Investigación y de Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), MexicoGareth Anthony Pearson, University of Algarve, Portugal
Copyright © 2021 Jueterbock, Minne, Cock, Coleman, Wernberg, Scheschonk, Rautenberger, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Jueterbock, YWxleGFuZGVyLWp1ZXRlcmJvY2tAd2ViLmRl
 Alexander Jueterbock
Alexander Jueterbock Antoine J. P. Minne
Antoine J. P. Minne J. Mark Cock
J. Mark Cock Melinda A. Coleman
Melinda A. Coleman Thomas Wernberg
Thomas Wernberg Lydia Scheschonk
Lydia Scheschonk Ralf Rautenberger
Ralf Rautenberger Jie Zhang8,9
Jie Zhang8,9 Zi-Min Hu
Zi-Min Hu