
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 31 May 2021
Sec. Marine Molecular Biology and Ecology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.656506
 Matthew Fuirst1,2*
Matthew Fuirst1,2* Christopher S. Ward3,4
Christopher S. Ward3,4 Caroline Schwaner2,4
Caroline Schwaner2,4 Zoie Diana4
Zoie Diana4 Thomas F. Schultz4
Thomas F. Schultz4 Daniel Rittschof4
Daniel Rittschof4The decorator worm Diopatra cuprea, a tube-forming marine polychaete common to intertidal and shallow subtidal waters, modifies habitats it occupies through microreef construction and algal gardening. While several studies have demonstrated that decorator worm tubes are hotspots of biogeochemical activity (i.e., nitrogen and sulfur cycling), it is still largely unclear whether the tube microbiome differs compositionally from the surrounding sediment and what distinct functional processes tube microbiomes may have. To address these unknowns, this study analyzed the bacterial communities of D. cuprea tubes and surrounding sediments using high-throughput 16S rRNA gene amplicon sequencing. Tubes and sediments were sampled at three sites along an anthropogenic stress gradient within the Newport River Estuary to also assess geographic variation of tube microbiomes and the possible influence of human disturbance. We found a clear distinction in the microbial community composition and diversity between tubes and surrounding sediment. Tube microbiomes were significantly enriched for the phyla Bacteriodetes, Actinobacteria, Verrucomicrobia, Deferribacteres, Latescibacteria, and Lentisphaerae. Chloroplast sequences of macroalgae and grass species were consistently abundant in tubes and nearly absent in surrounding sediment. Functional annotation of prokaryotic taxa (FAPROTAX)-based functional predictions suggested that tube microbiomes have higher potentials for aerobic chemoheterotrophy, sulfur compound respiration, nitrate reduction, methylotrophy, and hydrocarbon degradation than surrounding sediments. Tube microbiomes vary across sites, though dissimilarity is comparatively low compared to tube-to-sediment differences. Contrary to our hypothesis, the tubes at the most highly impacted site had the highest microbial diversity [i.e., amplicon sequence variant (ASV) richness and Shannon’s diversity], yet tubes from the medium impacted site actually had the lowest microbial diversity. Our findings show that D. cuprea tubes support a microbiome that is significantly distinct in composition and function from the surrounding sediment. Diopatra cuprea tubes appear to create unique microhabitats that facilitate numerous microbially-mediated biogeochemical processes in the marine benthic environment.
Advances in sequencing and bioinformatics approaches over the past several decades have fueled the study of the microbiome (Nelson et al., 2015), in the process revolutionizing our collective understanding of microbial influence on host physiology and ecosystem functions. One central concept that has emerged is that of the “holobiont,” the integrated collective of a host organism and all of its associated microbial and multicellular symbionts (Rohwer et al., 2002; Medina-Silva et al., 2018), for the contributions of the microbiome can considerably alter the physiology and ecology of the larger organism. Studies of marine polychaete microbiomes have revealed important dimensions imbued by the microbiomes to their hosts and in their environment, including metal resistance, contaminant degradation, and nutrient cycling (Dubilier et al., 2008; Li et al., 2009; Medina-Silva et al., 2018). But given their sheer diversity, ubiquity, and importance in shallow and pelagic food webs, marine polychaetes remain greatly understudied.
Many marine benthic organisms are ecosystem engineers, creating, maintaining, and modifying the physical and biological structure and heterogeneity of habitats (Gutiérrez et al., 2003; Callaway, 2006; Commito et al., 2008; Santos and Aviz, 2019). Mollusk beds, tubeworms, and reef-building organisms illustrate this role by providing substrata for epibiont attachment (Gutiérrez et al., 2003), solute transport (Hedman et al., 2011), and shelter for other organisms (O’Beirn et al., 2000; Gutiérrez et al., 2003; Kerry and Bellwood, 2012). Tube-building polychaetes create micro-reef structures that enhance the physical dynamics of benthic ecosystems and increase local biodiversity (Bailey-Brock, 1984; Thomsen et al., 2011; Berke, 2012; Santos and Aviz, 2019). They construct tubes by binding particles with biological glues, with some species adding bits of shell, algae, and other debris to build ornamented or decorated tubes (Berke et al., 2006; Santos and Aviz, 2019). Tubes are thought to constitute distinct microenvironments from the surrounding sediments, and they support microbiomes that contribute to microbial processes contributing to functions critical to the host and the marine benthic ecosystem (Medina-Silva et al., 2018; Rincón-Tomás et al., 2020). Characterizing the microbiome of tube-building worms can provide insights into biogeochemical processes between the interface of benthic organisms and surrounding sediments.
Burrowing marine animals, including intertidal polychaetes, are well-recognized contributors to the biogeochemistry of the benthic environment through excavation (mixing old and new sediments), grazing and eating, and excretion (Konhauser et al., 2020). More recently, studies have shown that intertidal polychaetes species host microbial symbionts capable of transformation, degradation, and detoxification. In separate studies, Ophelina sp. were found to have an abundance of metal-resistant bacterial groups (Neave et al., 2012), and the sediment-dwelling Capitella teleta were dominated by genera containing polyaromatic hydrocarbon (PAH)-degrading members (Hochstein et al., 2019), which possibly allow the polychaete hosts to inhabit polluted environments or serve as indicator of pollution. Similarly, the acorn worm Saccoglossus bromophenolosus contains microbial genera involved in sulfur oxidation (King, 2018). While recent studies have begun to shed light on the microbial composition and functions of marine polychaetes, most research to date has primarily focused on the internal symbionts of tube-forming polychaetes. There is substantially more limited information describing the microbiome of the external portion of the worm and how the microbiome of polychaetes differs from the surrounding sediment (Matsui et al., 2004; Medina-Silva et al., 2018). The interface between individuals and the sediment may promote a unique environment for microbial activity in intertidal ecosystems, as seen in cold seeps and coral reefs (Duperron et al., 2009; Röthig et al., 2017; Medina-Silva et al., 2018). Thus, determining the microbiota associated with tube-building worms relative to the surrounding sediment, and exploring how geographic location are linked to microbiome variation, may provide insights into the functional roles polychaetes have in the marine benthic environment.
The onuphid polychaete, Diopatra cuprea (hereafter D. cuprea) is a tube-building worm native and common to the intertidal and shallow subtidal waters of the western Atlantic (Paxton, 1998). The range of D. cuprea extends from Cape Cod, Massachusetts south to the coast of Brazil (Magnum et al., 1968). Diopatra cuprea constructs decorated vertical tubes, which are comprised of several glues. The tubes contain a hook-shaped “tube cap” at the top of the microreef that begins at the sediment surface (Magnum et al., 1968; Myers, 1972). Similar to other tube-forming polychaetes, D. cuprea tubes can influence the biological and physical aspects of benthic habitats and microhabitats by stabilizing sediments (Bailey-Brock, 1984) and promoting bacterial (Phillips and Lovell, 1999; Matsui et al., 2004), meiofaunal (Bell, 1985; Thomsen et al., 2011), and macrofaunal growth (Berke, 2012; Santos and Aviz, 2019). While limited in scope, previous studies found that D. cuprea worm tubes have sulfate reduction (Matsui et al., 2004) and nitrification potential (Mayer et al., 1995). While aspects of the ecological role of D. cuprea tubes are understood, the microbiome composition, diversity, and many of the microbial functions of the tubes have not been studied using modern microbiome approaches.
In this study, we used 16S rRNA gene amplicon sequencing to characterize the microbiome of D. cuprea across three different intertidal sites in the Newport River Estuary of North Carolina, United States. With this dataset, we compared the microbial communities between tubes and the surrounding marine sediment, as well as between sites. We investigated microbial taxa putatively involved in biogeochemical processes within D. cuprea tube tops. Our goals were to (i) determine how the microbial communities of D. cuprea differ from the surrounding sediment; (ii) evaluate how host-associated microbiome communities may vary across a landscape with various levels of anthropogenic influence; and (iii) identify whether host-associated microbial taxa are relevant to previously hypothesized D. cuprea biogeochemical functions.
We collected 24 D. cuprea tube samples (Bogue Sound = 8, Duke University Marine Lab = 8, North Bridge = 8; 24 total) and surrounding sediment samples (1 per site; 3 total) from three sites within the Newport River Estuary, North Carolina (NC), United States (Figure 1). Each of the sampling sites has historically been exposed to varying levels of shoreline development and water quality. We ranked the three sites as low (L), medium (M), and high (H) disturbance based on levels of fecal coliform bacterial contamination (a standard water quality parameter for the Department of Environmental Quality; North Carolina Environmental Quality, 2021) and surrounding land development according to Romano (2007). The least disturbed site, Bogue Sound (BG, 34°43'13" N 76°43'49" W), is a suburban shoreline. BG was classified as “L” due to previous studies showing the normal embryonic development of the sea urchin Lytechinus variegatus, indicating low pollution levels in this bay (Wessel et al., 1998; Gross et al., 2003; Romano, 2007). The fecal coliform monthly geometric mean for BG was 3.04 (North Carolina Environmental Quality, 2021). Additionally, BG has a substantially lower density of urban development surrounding the estuary (Romano, 2007). The “M” disturbance site was North Bridge (NB, 34°43'29" N 76°41'19" W), which crosses a 500-m channel from the Port of Morehead City and Calico Creek, a 4 km stretch of oyster habitat. The Morehead City sewage outfall is 4 km to the west of NB (fecal coliform monthly geometric mean = 3.27) and has been associated with human enteric viruses in 40% of eastern oyster (Crassostrea virginica) samples in Calico Creek, which is located near NB (Fong and Lipp, 2005; North Carolina Environmental Quality, 2021). Lastly, the “H” site was Duke University Marine Lab (DU), located on Pivers Island. DU is a university protected system (DU, 34°43'5" N 76°40'16" W) and is about 200 m across the Gallants Channel from Taylor Creek and about 2 km from the Beaufort Sewage Treatment Plant at the east end of Downtown Beaufort (Figure 1). DU also constant boat traffic entering the Beaufort Inlet making it more disturbed than the other two sites with a fecal coliform monthly geometric mean of 3.49 (North Carolina Environmental Quality, 2021). Both DU and NB are permanently closed to shellfish fisheries (North Carolina Environmental Quality, 2021).
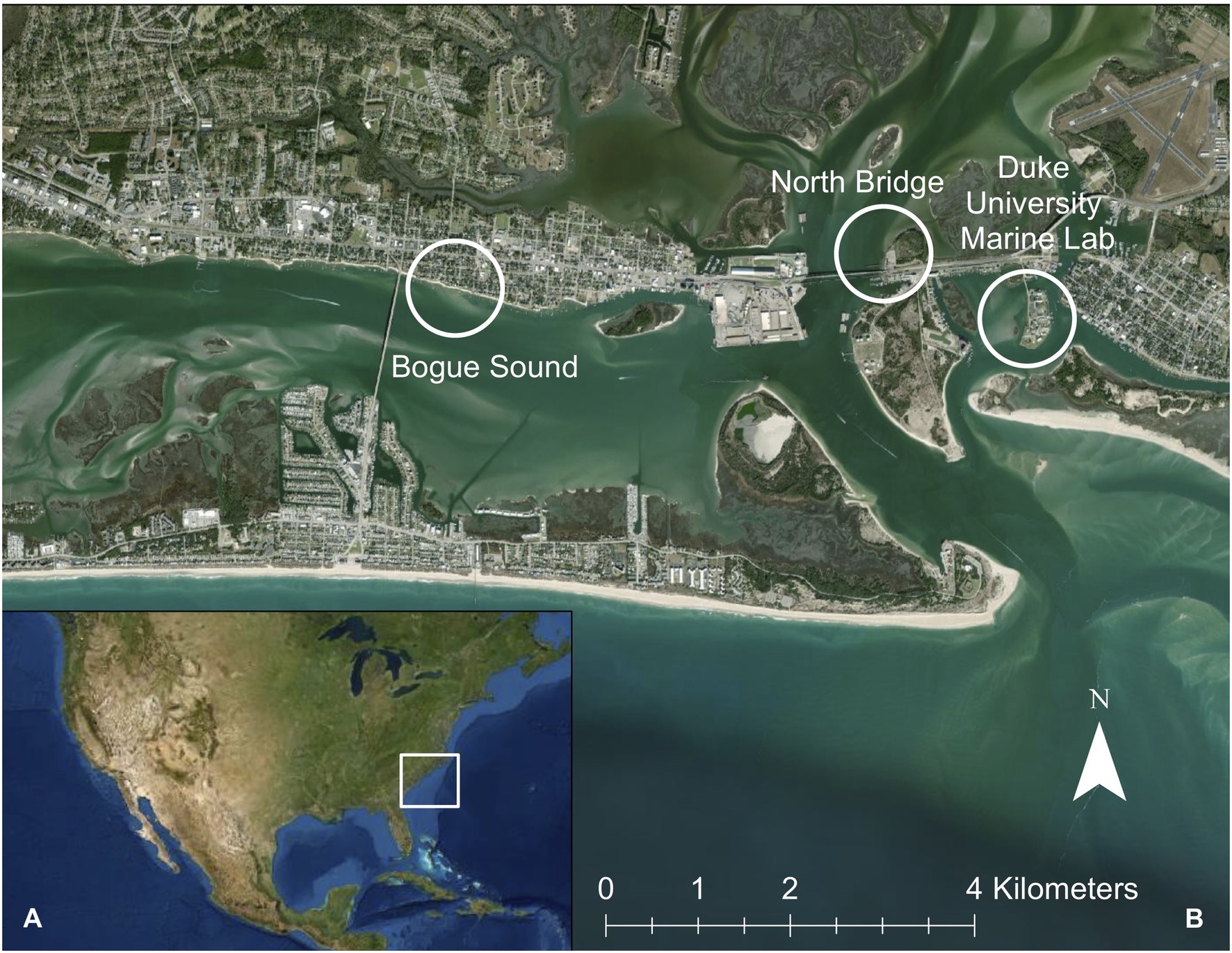
Figure 1. (A) Location of study site in Beaufort, NC, United States. (B) Distribution of three sample collection sites with inset of Newport River Estuary, Bogue Sound, and Beaufort Inlet. White circles indicate each of the three sites (Duke University Marine Lab, Bogue Sound, and North Bridge).
To ensure that the samples collected were from inhabited D. cuprea tubes, we collected tube tops from newly rebuilt tubes. Tube tops were clipped with sterilized scissors where the tube met the sediment surface and marked using small metal stake flags. We returned after two subsequent low tides to harvest the newly rebuilt tube tops (n = 8, BG; n = 8, DU; n = 8, NB) by clipping with scissors where the tubes met the sediment surface. Each tube top was cut into five pieces and placed in 2-ml tubes. We collected corresponding sediment samples (one from each site) in separate 2-ml tubes at each location. Immediately after collection, we stored all samples on dry ice in the field and transferred to −80°C upon returning to the lab.
We removed the seawater in each sample by centrifuging all samples and pipetting out the excess seawater from each test tube. All sediment and D. cuprea tube top samples prior to the DNA extractions. Microbial DNA was extracted from eight tube tops and one sediment sample from each of the three sites (27 total samples; n = 24 tube tops, n = 3 sediment) using PowerSoil DNA Isolation kit (MoBio Laboratories, Carlsbad, CA, United States) according to manufacturer’s instructions with slight modifications. Before extraction, we placed one of the five pieces of the tube top (instead of 0.25 mg of sample as recommended) in bead beater solution and agitated for 9-min at room temperature. For each sample, we obtained 150 μl of final DNA solution in Tris-EDTA buffer and stored at −20°C. DNA concentrations were quantified using a Qubit 2000 UV fluorometer and a dsDNA HS Assay kit (Thermo Fisher Scientific v 3.0). DNA concentrations obtained from the samples ranged from 5.2–29.9 ng per μl.
Samples were sent to the Center for Genomic and Computational Biology at Duke University, Durham, NC, United States where the V3–V4 region of 16S rRNA genes was amplified by PCR using 341f and 785r primers described in Klindworth et al. (2013). Libraries were individually barcoded, pooled in equal proportions, and sequenced using paired-end MiSeq platforms (2 × 300 bp; Illumina Inc., San Diego, CA, United States).
Sequences were demultiplexed using CASAVA (Illumina). All downstream sequence processing was performed using the DADA2 version 1.8.0 workflow (Callahan et al., 2016) in R (R Core Team, 2014, version 3.3.3). We chose ribosomal amplicon sequence variants (ASVs) to avoid the errors and limitations associated with operational taxonomic units (OTUs; Callahan et al., 2016). Reads were trimmed on the 5' to remove primer sequences and the 3' to remove low-quality nucleotides. Read pairs were then denoised using a DADA2-generated error model, merged, and de novo chimeras were removed. We obtained between 58,320 and 223,360 paired-end reads per tube sample, with an average of 154,796 paired end reads. We obtained between 62,514 and 168,797 paired-end reads per sediment sample, with an average of 119,499 paired end reads. The final output consisted of the final ASV sequences of 16S rRNA gene reads and an ASV table of the number of reads per ASV per sample (functionally analogous to an “OTU” table). ASVs were assigned using RDPtools and RDP release 11.5 classifiers (Wang et al., 2007) and taxonomy selected against the Greengenes database (version 13.8). The resulting ASV table was used for community analyses. One NB tube sample library failed to yield an adequate number of high-quality reads, and thus was excluded from further analyses (hereafter only seven NB tube samples were used). All sequencing data are available on the NCBI database and are under BioProject PRJNA18430 (Accessions SAMN18529213-SAMN1852239).
Alpha and beta diversity analyses were conducted in program R statistical software (R Core Team, 2014, version 3.3.3) using the phyloseq (v 1.24.2; McMurdie and Holmes, 2013) and vegan (v 2.5.2; Oksanen et al., 2015) packages, and community composition analyses were run using the DESeq2 package (v 1.14.1; Love et al., 2014). For alpha and beta diversity analyses, samples were rarified to a depth of 58,000 sequences per sample to avoid biases due to variation in library size across samples and capture a representative portion of the ASV diversity present. Using the rarified dataset, we estimated alpha diversity using ASV richness and the Shannon diversity index (Shannon, 1948). Kruskal-Wallis tests were used to compare Shannon values relative to site for all tube samples. When results were significant (p < 0.05), post hoc comparisons were made using Dunn’s test and Bonferroni correction. To examine community-wide differences in taxonomy and abundance, we used a non-metric multidimensional scaling (NMDS) analysis based on a Bray-Curtis dissimilarity index. Ordinations were performed on rarefied data and analyses of variance using distance matrices using the ADONIS function were run to test for differences between site and sample type (Anderson, 2001). We used DESeq2 on unrarefied data at several different taxonomic levels to test for differential abundances of bacterial taxa and to classify bacterial functions between sample groups.
To further investigate the potential functionality of microbial taxa, we used the functional annotation of prokaryotic taxa database (FAPROTAX) to classify taxa that may be involved in nutrient cycling processes (Louca et al., 2016). FAPROTAX is conservative in assigning function, only doing so if all members of a given taxonomic group have the trait in common.
In order to derive better taxonomic assignments for abundant chloroplast amplicons, ASV sequences were queried for highly similar sequences within NCBI’s database Nucleotide collection (nr/nt) using the megablast algorithm in the BLASTn suite and excluding uncultured sample sequences. All of the sequencing data analyzed in this study can be downloaded from the NCBI’s Sequence Read Archive (SRP #8914202).
The microbial communities of D. cuprea tube tops and surrounding sediment samples were determined through high-throughput amplicon sequencing of the 16S rRNA gene V3 and V4 hypervariable regions. After rarefaction, we detected a total of 28,971 unique ASVs in the dataset. The majority of ASVs had low prevalence across all sediment and tube samples, with 99.6% of ASVs having relative abundances below 0.1%.
Beta-diversity analyses revealed that tube communities had significantly different compositions from sediment samples. Plotting of NMDS analysis performed on Bray-Curtis community dissimilarities visualizes the distinct clustering of tube and sediment sample types (Figure 2; Supplementary Table S1) and is further supported by ADONIS model, with strong effects of sample type on Bray-Curtis dissimilarities (Sample Type: F24,1 = 4.0, R2 = 0.14, p = 0.001). Additionally, NMDS analysis indicates clustering by site. Site significantly predicted strong effects on Bray-Curtis distances in multivariate ADONIS models (Site: F23,2 = 2.8, R2 = 0.19, p = 0.001, Figure 2).
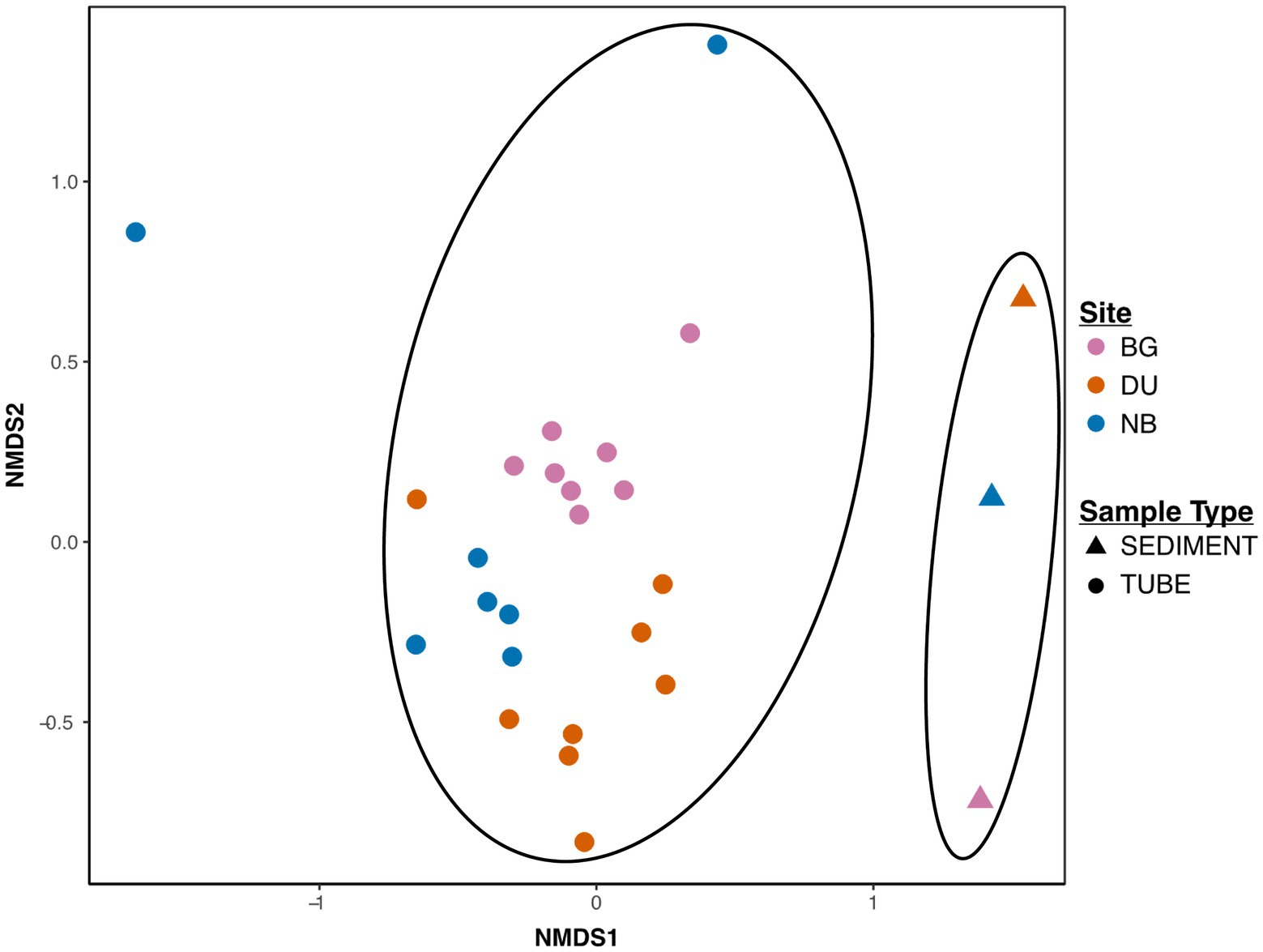
Figure 2. Beta diversity of microbiomes of tubes and sediment samples relative to each sample site. Data presented as a non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis dissimilarity values. Multivariate ADONIS models showed strong effects of sample type on Bray-Curtis dissimilarities (Sample Type: F24,1 = 4.0, R2 = 0.14, p = 0.001) and site (Site: F23,2 = 2.8, R2 = 0.19, p = 0.001) on distinct clustering of Bray-Curtis distances. Ellipses indicate distinct beta diversity of the microbiome between tubes and sediment samples.
The communities from newly constructed tube tops were highly diverse, with richness values ranging from 1,094 to 4,639 ASVs and an average Shannon diversity index (H') of 6.75 (Figure 3; Supplementary Table S2). These diversity values were similar to those of the surrounding sediment communities (1,450–3,800 ASVs; H' = 6.7). We also observed differences in tube microbiome diversity between sites. There was a significant difference in Shannon diversity of tubes between sites (Kruskal-Wallis p = 0.05), while ASV richness was not significant (Kruskal-Wallis p = 0.10). Tested site-to-site, ASV richness, and Shannon diversity were consistently highest for tubes from the DU site. Shannon diversity of DU tubes was significantly higher than NB (post hoc Dunn’s test p = 0.03), and BG tubes were marginally higher than NB (post hoc Dunn’s test p = 0.08).
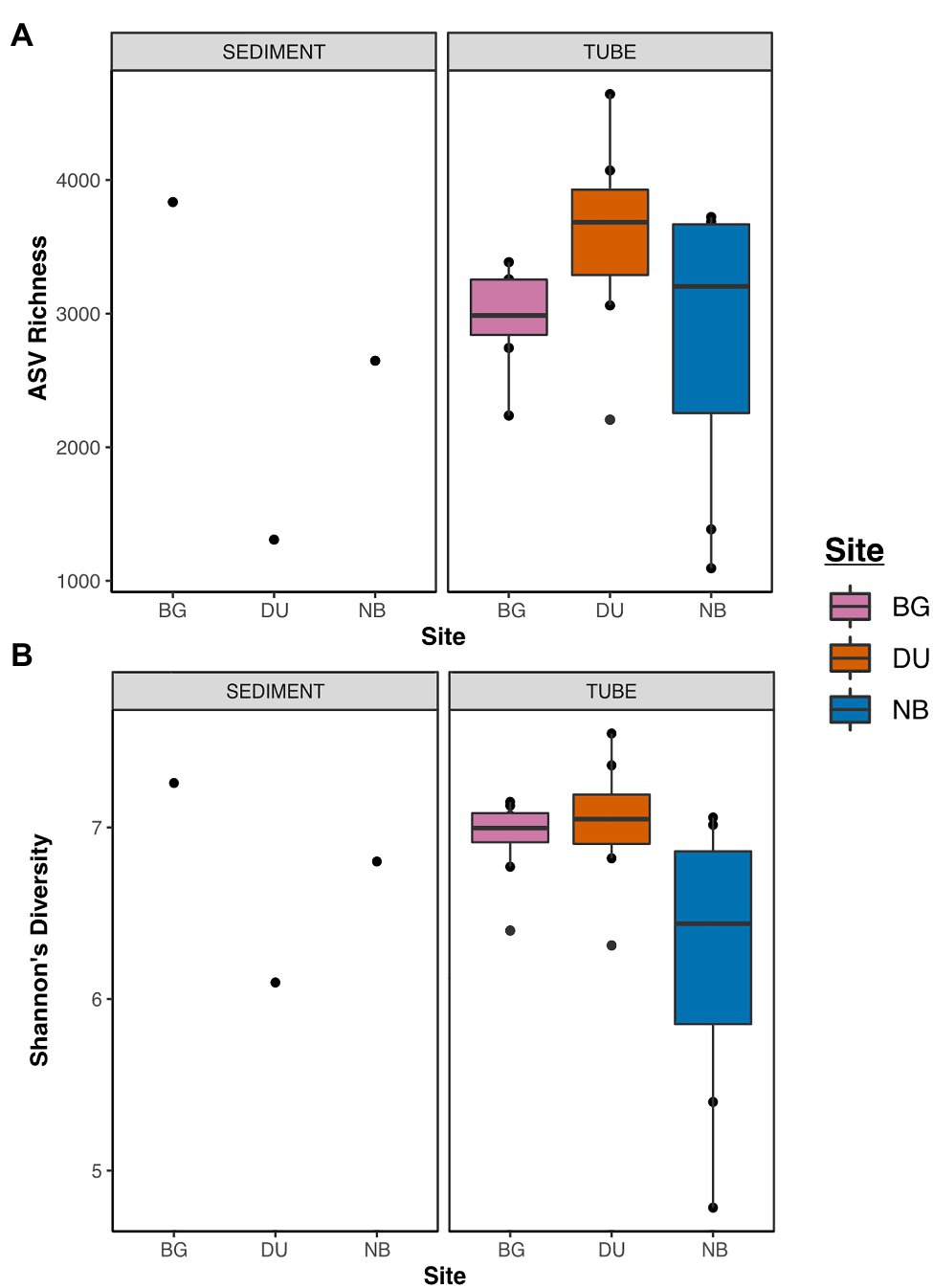
Figure 3. Dot and box-whisker plots of alpha diversities of sediment and tube samples across all three sites [Duke University Marine Lab (DU), Bogue Sound (BG), and North Bridge (NB)] relative to amplicon sequence variant (ASV) richness (A) and Shannon diversity (B).
Given previous findings that worm tubes have sulfate reduction and nitrification potential (Mayer et al., 1995; Matsui et al., 2004), we aimed to examine the broader range of biogeochemical processes to which the tube microbiome might contribute. Using the curated microbial function database FAPROTAX, we inferred microbial functions of the microbial taxa present in worm tubes. The most abundant functions present at >1% across the dataset were chloroplasts, aerobic chemoheterotrophy, sulfur compound respiration, fermentation, and anoxygenic photoautotrophy (Figure 4; Supplementary Table S3). Less abundant functions (<1 and >0.1%) fell into simple and complex carbon (methylotrophy/methanol oxidation and cellulolysis/hydrocarbon degradation, respectively), sulfur (oxidation/respiration), nitrogen (nitrate/urea), and iron (oxidation) biogeochemical processes.
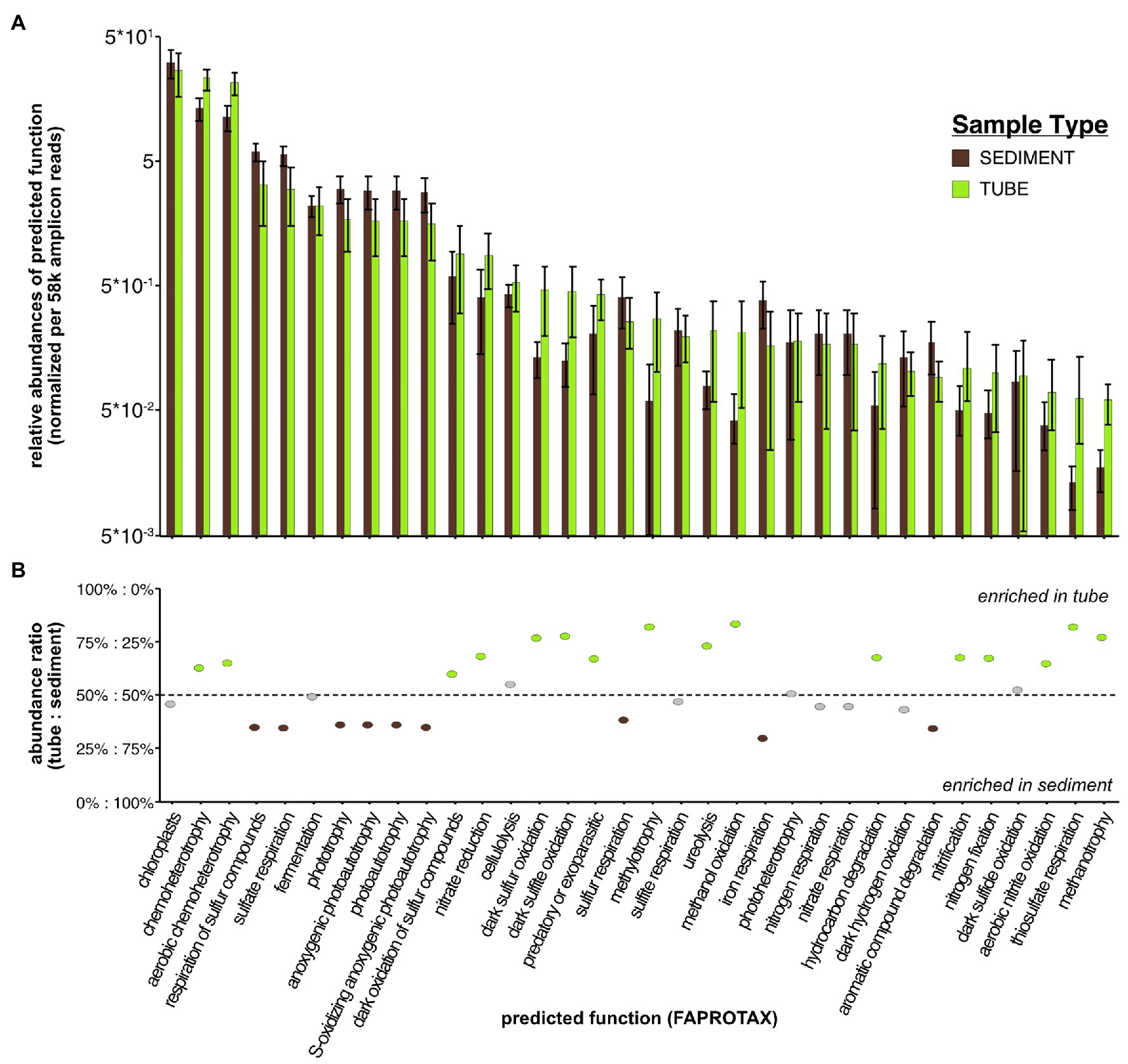
Figure 4. (A) Distribution of normalized abundances of predicted functions (via FAPROTAX) for sediment and tube microbial samples. (B) Ratio of abundance between tubes and sediments for each inferred functional group determined by FAPROTAX classification.
Considering the similarity in abundant taxonomic groups between tubes and sediments and between sites, distinct community compositions were likely the result of differences in relative abundances of most abundant taxa, differences at lower taxonomic classifications (e.g., genus-level), and/or presence/absence of rare taxa. ASVs from the worm tube communities primarily belonged to six dominant phyla: Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, Verrucomicrobia, and Planctomycetes, which accounted for over 80% of sequence reads (Figures 5, 6; Supplementary Table S4). Bacteroidetes (20% + 4.5), Actinobacteria (3% + 1.2), Verrucomicrobia (3% + 1.1), Deferribacteres (0.06% + 0.03), Latescibacteria (0.05% + 0.03), and Lentisphaerae (0.003% + 0.02) were all significantly more abundant in tubes compared to sediment communities (Supplementary Table S5). Only four phyla were differentially abundant between sample sites. The mean relative abundance of Deinococcus-Thermus was higher in BG (0.09% + 0.04) compared to DU (0.01% + 0.01), Firmicutes was more abundant in NB (0.7% + 0.3) compared to DU (0.2% + 0.1), Chloroplasts was more abundant in NB (33% + 12.0) compared to BG (21% + 4.0), and Fusobacteria was higher in BG (0.1% + 0.1) compared to NB (0.04% + 0.05). Between tubes and sediment samples, 48 families were differentially abundant. Within the top 20 most abundant families, Streptophya (1% + 4.0 tube vs. 0.003% + 0.005 sediment), Rhodobacteraceae (11% + 3.0 tube vs. 5% + 1.0 sediment), Granulosicoccaceae (0.8% + 0.6 tube vs. 0.2% + 0.08 sediment), Marinicella (0.4% + 0.7 tube vs. 0.01% + 0.01 sediment), and Rubritaleaceae (1% + 0.4 tube vs. 0.2% + 0.01 sediment) were all significantly more enriched in tubes (Supplementary Table S6). There were 140 ASVs that were differentially abundant between tubes and sediments and 385 ASVs that were differentially abundant between sites (Supplementary Table S7). Most notably, variants belonging to the genera Pseudoruegeria (2% + 1.0), Loktanella (5% + 2.0), Maribacter (3% + 1.0), Granulosicoccus (1% + 1.0), Rubritalea (2% + 1.0), Ilumatobacter (2% + 0.6), Tenacibaculum (1% + 1.0), Winogradskyella (1% + 0.7), and Erythrobacter (1% + 0.6) were all significantly more enriched in tubes compared to sediments by at least 1%.
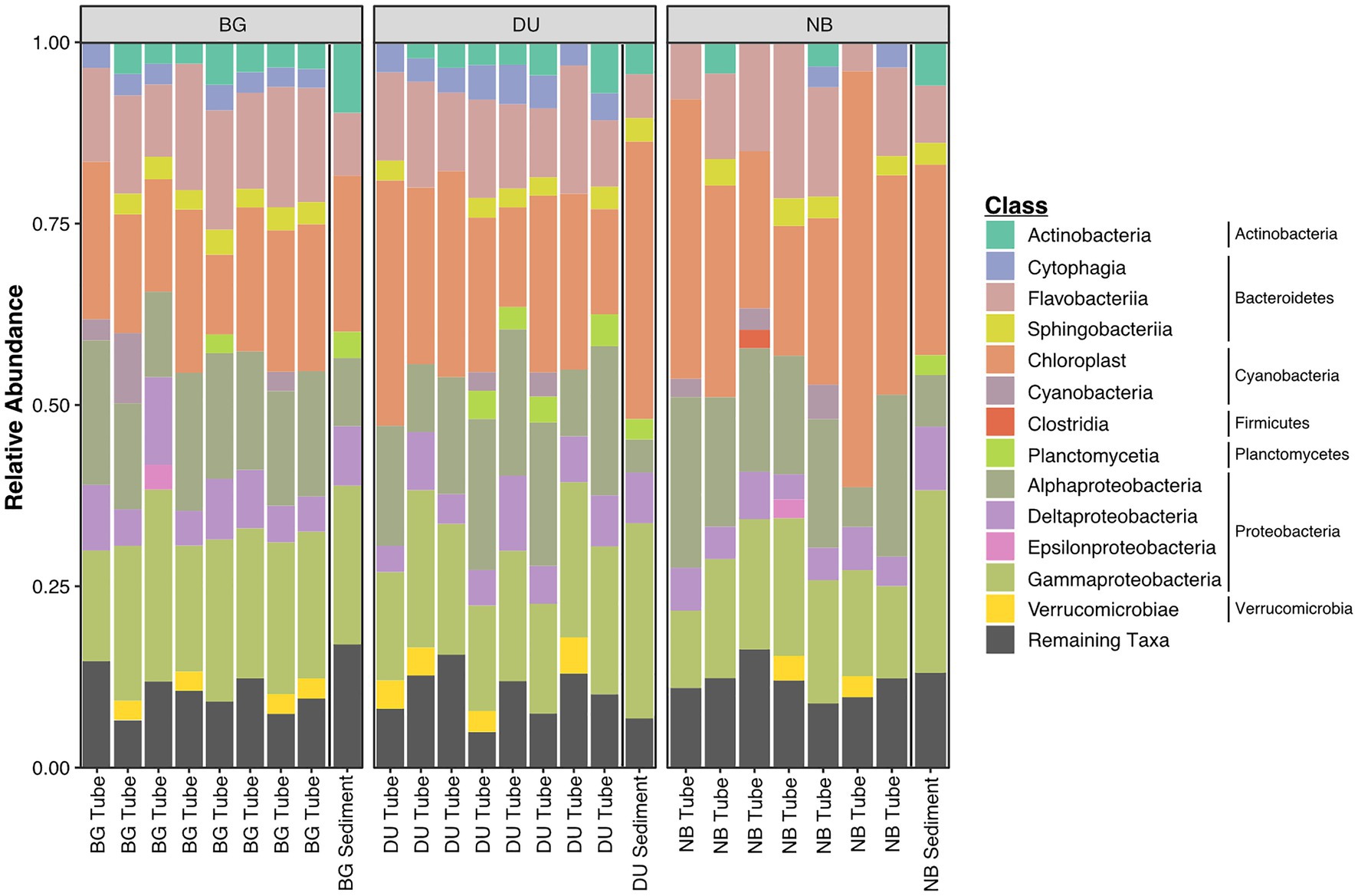
Figure 5. Relative abundance of microbial classes and phyla for 16S-rRNA amplicon sequence reads from tubes and sediments samples from all three sites (DU; Duke University Marine Lab, BG; Bogue Sound, and NB; North Bridge). Phylum that were >0.1% of the entire dataset were classified into appropriate class or phyla levels, whereas ASVs <0.1% were classified as “remaining taxa.”
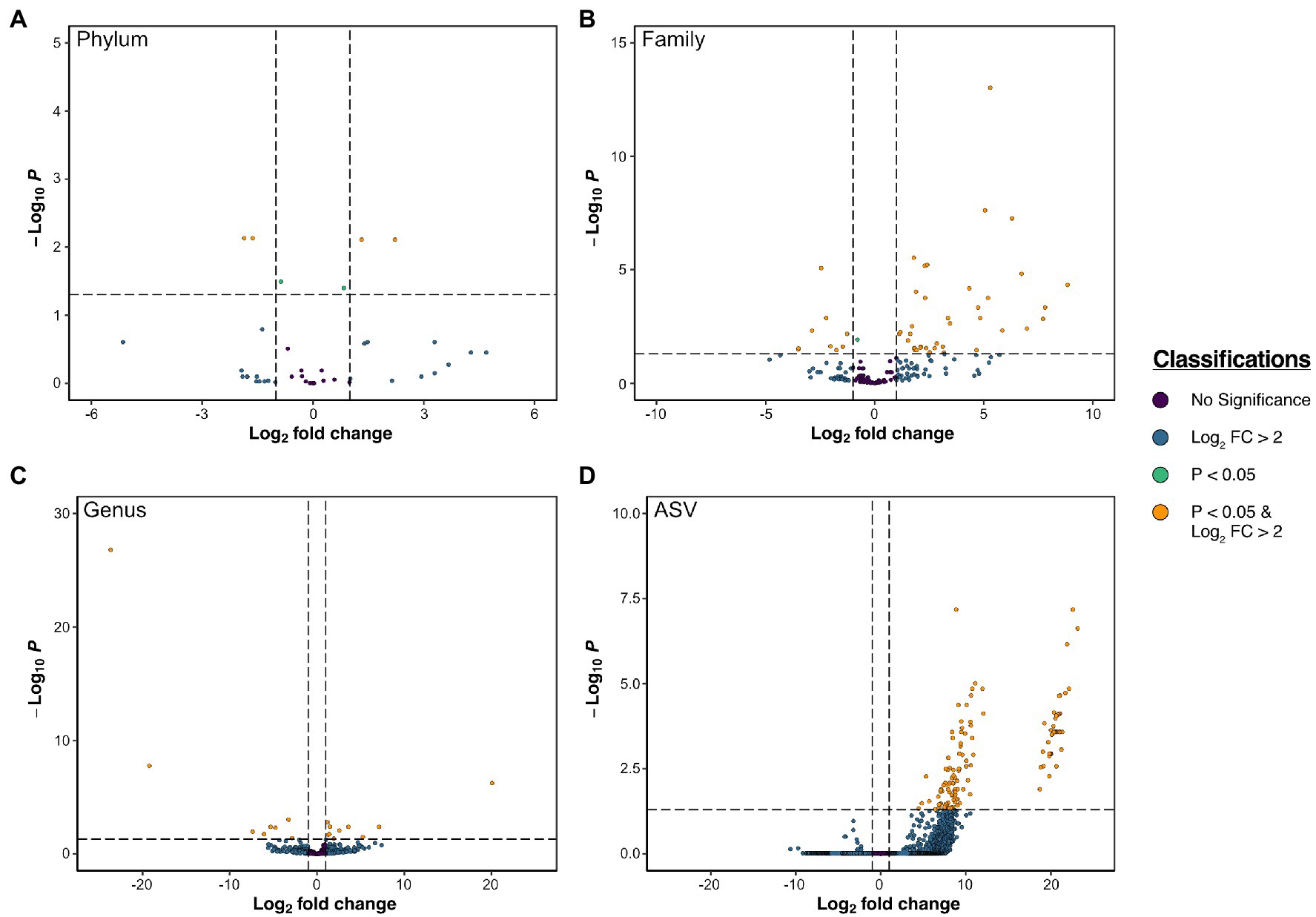
Figure 6. Volcano plots of differentially abundant taxa according to DESeq2 results between tubes and sediment at the phylum (A), family (B), genus (C), and ASV (D) level.
Using the same DESeq2 analysis approach used to compare bacterial relative abundances, we examined differential abundances of microbial function in tubes and sediments. We found unique microbial functions that were enriched in tubes compared to surrounding sediments: aerobic chemoheterotrophy (23% + 4.0), dark oxidation of sulfur compounds including dark sulfur/sulfate oxidation (0.8% + 0.6), methylotrophy (0.3% + 0.2), methanol oxidation (0.3% + 0.1), and ureolysis (0.2% + 0.1), in order of overall abundances of >0.1% (Supplementary Table S8). Interestingly, there were several depletions of functions in tubes, particularly for respiration of sulfate/sulfur compounds (6% + 1.0), anoxygenic photoautotrophy/oxidizing of sulfur (2% + 0.8), and phototrophy and photoautotrophy (both 1% + 0.6) – all anaerobic processes.
Chloroplast sequences were abundant in both tubes and sediments. Several of the most abundant ASVs enriched in tubes were assigned to Cyanobacteria/Chloroplast. Greengenes assignments were primarily Bacillariophyta, with Chlorophyta and Streptophyta in several tube communities. These ASVs were often present, in very high abundance, in the tubes and nearly completely absent in the surrounding sediment. We were interested in identifying particular chloroplast-containing organisms so we queried the ASV sequences against the NCBI nucleotide database since Greengenes classification of chloroplast sequences has limited taxonomic resolution. Many of the abundant chloroplast ASVs previously classified as microalgal by Greengenes were assigned to macroalgae and plants by BLAST – specifically Ectocarpus, Ulva, and a grass species, potentially Spartina spp. (saltmarsh cordgrass is commonly found in the area), or bamboo (Bambusoideae). Ectocarpus is present and abundant in tubes from all sites (up to 18% of the library). Ulva had high abundance in NB tubes (1 NB sample with 31% Ulva), and were present thought inconsistently abundant in tubes at other sites as well. The putative Spartina sp. was only in high abundance in NB tubes, within minimal detection in BG and DU.
Prior studies have demonstrated that D. cuprea tubes support a high abundance of varied microbial communities (Phillips and Lovell, 1999) and potential biogeochemical processes involved in decorator worm tubes (Mayer et al., 1995; Phillips and Lovell, 1999). However, the D. cuprea tube microbiome has not been revisited since the development of next generation sequencing. Using amplicon sequencing data, our results provide evidence that D. cuprea tubes host a distinct microbiome compared to the surrounding sediment and potentially contribute to multiple biogeochemical processes in the benthic environment. Furthermore, our results suggest that tube worm microbiome composition and diversity can vary even over small geographical scales within a connected waterbody, potentially due to land use effects, such as point source and non-point source pollution, tidal flows, and coastal development. Since coastal benthic communities are governed by a combination of physical, chemical, and biological processes, understanding the ecophysiology of marine tube-building worms is important in understanding coastal ecosystems.
We found clear evidence that tube and sediment communities are significantly distinct from each other at all taxonomic levels examined. Differentially abundant genera between tubes and sediment (enriched in tubes) included Maribacter, Tenacibaculum, Winogradskyella, Rubritalea, Ilumatobacter, Granulosicoccus, Pseudoruegeria, Erythrobacter, and Loktanella. These bacterial genera found enriched from the tubes indicate that the tubes might be acting as hotspots of biogeochemical activity.
Maribacter, Tenacibaculum, and Winogradskyella are all in the phylum Bacteriodetes. Bacteroidetes are able to degrade particulate matter, such as polymers, peptides, and biological adhesive proteins (Fernandez-Gomez et al., 2013). Thus, Bacteroidetes may be degrading and consuming the biological glues that bind the particles of the tubes or the plant and algal matter incorporated into the tubes. Bacteroidetes have many adaptations to grow attached to organic particles (Fernandez-Gomez et al., 2013), so could be incorporated into tubes through these associations. Maribacter is often associated with the surface of macroalgae and function in the degradation of algal polysaccharides (Bakunina et al., 2012). Maribacter might be enriched since decorator worms utilize algae from the intertidal sediment surface for construction of their tubes.
Maribacter species have the KEGG pathway “nitrogen metabolism,” and the other two genera of Bacteriodetes might also be involved in nitrogen cycling. Tenacibaculum is capable of heterotrophic nitrification-aerobic denitrification and specifically converts nitrite into gaseous Nitrogen (Huang et al., 2020). Interestingly, Winogradskyella contains dentification genes so might be involved in some steps of the dentification pathway (Villemur et al., 2019). Rubritalea, another enriched genus, is known to reduce nitrate to nitrite (Hedlund et al., 2015). Rubritalea is a member of the phylum Verrucomicrobia; marine Verrucomicrobia can also metabolize polysaccharides (Cardman et al., 2014), methane (Lee et al., 2009), and degrade phytodetritus (Gihring et al., 2009). While little information can be found on the genus Ilumatobacter, it belongs to the family Acidmicorbiaceae and Acidmicorbiaceae sp. are known to play a role in oxidation of ammonium in iron reducing conditions (Huang and Jaffe, 2018), further supporting the idea that tube tops are hot spots of biogeochemical activity, specifically nitrogen cycling.
Granulosicoccus, Pseudoruegeria, Erythrobacter, and Loktanella belong to the phylum Proteobacteria. Broadly, Proteobacteria are important in carbon cycling as they are able to metabolize glycolate, one of the most abundant sources of organic carbon in the ocean (Von Borzyskowski et al., 2019). Granulosicoccus have been isolated from seagrass (Kurilenko et al., 2010) and algae (Park et al., 2014). Similarly, Pseudoruegeria is associated with algae. Algae produces dissolved organic matter, oxygen, and sulfur, and the bacteria supplies the algae with the vitamins thiamin, biotin, and cobalamin (Cho et al., 2020). As mentioned before, tube tops contain plant material and algae, so it is not surprising genera associated with algae and sea grasses are enriched in tubes. Erythrobacter has been isolated from coral and might play a role in health of the animal by converting hydrogen sulfide to sulfate (Setiyono et al., 2019). While Erythrobacter metabolizes sulfated carotenoids, Loktanella is known to metabolize dimethylsulfoniopropionate, which can contribute to carbon and sulfur cycling (Curson et al., 2008). Due to these initial findings of enriched genera in tubes, we set out to further explore if the bacterial groups enriched in tubes could more confidently be attributed biogeochemical functions.
Core microbiota are important for maintaining diversity and composition of the microbial community and play roles in organic and inorganic processes (Mayer et al., 1995; Matsui et al., 2004; Berke et al., 2006). The FAPROTAX analysis revealed significant functional differences between tubes and surrounding sediment. Many of these functions indicate biogeochemical processes involved with the tube structure. The significant enrichment of functions involved with dark oxidation of sulfur/sulfate compounds is in concert with previous studies that showed elevated sulfate-reducing bacteria in D. cuprea tubes (Matsui et al., 2004). As mentioned above, Erythrobacter metabolizes sulfated carotenoids and Loktanella is known to metabolize dimethylsulfoniopropionate for carbon and sulfur cycling (Curson et al., 2008). Other functions included nitrogen and carbon biogeochemical processes. Many of the genera enriched in tubes are known to play a part in nitrogen cycling, such as converting nitrite into gaseous Nitrogen (Huang et al., 2020), dentification (Villemur et al., 2019), reducing nitrate to nitrite (Hedlund et al., 2015), and oxidation of ammonium (Huang and Jaffe, 2018).
Mayer et al. (1995) also found that particular macrofaunal tubes of D. cuprea had higher nitrification potential than sediment. Since sediments are subjected to high levels of disturbance, macrofaunal tubes can provide chemocline niches for microbial growth and increased microbial processes that marine sediments may not provide. D. cuprea tubes have been suggested to disrupt solenoidal eddies and decrease sediment scour, thus enhancing biogeochemical processes (Berke, 2012). The differences observed between the tubes and surrounding sediments likely reflect the differences in physicochemical conditions and composition of the material that the worms incorporate into the constructed tubes as ecosystem engineers (Berke, 2012), as well as the existing microbiome on those materials and the dramatic difference in physical conditions as tube tops are elevated in the tidal flow compared to sediment. The depletion of anaerobic processes in the tube suggests higher oxygen levels outside of the sediment, while enrichment of chemoheterotrophy may align with incorporated seaweed and plant material (Berke et al., 2006) and bioadhesive glue on the tube structure (Myers, 1972). The tube structure is hypothesized to bring oxygen, other dissolved gasses, and water into the surrounding sediment that may be hypoxic/anoxic (Matsui et al., 2004). Furthermore, analysis of the undecorated portion of the D. cuprea tube below the sediment surface by Konhauser et al. (2020) suggests that this area of the tube serves primarily as a physical barrier (potentially from predation) due to the low levels of trace metal reactivity, as compared to other burrowing worms (i.e., Terebellid crispus by Lalonde (2011)).
Decorator worms are known to incorporate shells, algae, and detrital plant material from the intertidal sediment surface (Berke et al., 2006). Instead of examining the composition of incorporated material through morphological analysis via microscopy, we used bioinformatics to compare chloroplast sequences abundant in both tubes and sediments. We found that particular sequences differed between two sample types. Classification using Greengenes database/RDP Classifier assigned most chloroplast sequences to Bacillariophyta (diatom), Chlorophyta (green alga), and Streptophyta (green alga), while the nucleotide BLAST to a larger reference database indicated that many of these chloroplast sequences belonged to macro-organisms, including seaweeds Ectocarpus and Ulva, grasses, and forams. Living in and using detrital material brings its own assemblage of bacteria, which can contribute to the tube microbiome signature. Bacteroidetes and Verrucomicrobia were associated with diatoms (Bruckner et al., 2008), and Latescibacteria are also associated with algae cells (Youssef et al., 2015). More specifically, Granulosicoccus have been isolated from seagrass (Kurilenko et al., 2010) and algae (Park et al., 2014), while Pseudoruegeria supplies algae with thiamin, biotin, and cobalamin (Cho et al., 2020), and Maribacter is found on the surface of macroalgae (Bakunina et al., 2012). Worms have been shown to collect algae and use it as building material to add structure to the tube tops (Berke et al., 2006; Berke, 2012) and may serve as a direct food supply for omnivorous worms (Magnum et al., 1968) or as an indirect food supply when the worm eats the algae-associated meiofuana (Bell and Coen, 1982). Thus, D. cuprea nitrogen excretion is hypothesized to stimulate algae growth (Giannotti and McGlathery, 2001), further strengthening the tube top’s role as a habitat modifier via algal “gardening” (Magnum et al., 1968; Brenchley and Tidball, 1980).
Human-induced disturbance can play a major role in the chemistry and hydrology of coastal environments (De Carlo et al., 2007), which can influence the microbiome communities of marine organisms. Previous studies in marine environments have supported the notion that microbial diversity decreases with increased human-induced disturbance (Cury et al., 2011; Drury et al., 2013; Wegner et al., 2013; Fuirst et al., 2018). The exposure to extrinsic disturbance can break apart certain phylotypes, thus reducing overall microbial diversity. For example, in situ studies on Pacific oysters (Crassotrea gigas) showed that increased stress from disturbance reduced the diversity of naturally occurring microbial assemblages (Wegner et al., 2013). Additionally, herring gulls (Larus argentatus) breeding in highly urbanized environments have shown lower microbial diversity than isolated populations (Fuirst et al., 2018).
While anthropogenic disturbances may lead to decreased microbial diversity in some systems, our results suggest that high-levels of nonpoint source pollutants may increase local bacterial diversity of D. cuprea tubes in benthic ecosystems. Tubes from DU, the site ranked as H, had the highest alpha diversity potentially due to the nearby sewage discharge (Coastal Resources Commission, 2007) and other pollutants present at the site, which can cause spikes in some microbial taxa in marine benthic environments (Shibata et al., 2004). However, it was surprising that NB (ranked M) has a lower Shannon’s diversity and ASV richness than both the DU and BG sites, given the proximity and high impact of nonpoint-source pollutants. We recommend that further research should incorporate more samples and more sites within the Newport River Estuary to disentangle how site-related environmental factors, such as water flow, pH, and nutrient input may cause changes to benthic microbial communities.
While our findings are informative, our limited sample size may cause biased estimates between sample groups that could be improved in further studies. This study provides an informative initial characterization of the D. cuprea tube and sediment microbiome and associated functions; however, due to our limited sample size and scope of the study, we were unable to investigate the assembly mechanisms of tubes and sediment. Future research could use niche/null models to investigate the relative contributions of deterministic and stochastic processes in community assembly, which may provide further insights into the ecology of host-associated microbiomes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI database, accession numbers SAMN18529213-SAMN18529239 and BioProject ID PRJNA718340.
CS, ZD, TS, and DR conceived the study, and planned and carried out the experiments. MF and CW processed the sequencing data, performed the analyses, and designed the figures. MF took the lead in writing the manuscript. All authors discussed the results and commented on the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by a grant from the Duke Microbiome Center to DR. Funding organizations played no role in the collection, analysis, and interpretation of data; writing the report; or the decision to submit the article for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Brian Taylor and Jason Dinh for assistance with field work. The authors would also like to acknowledge Beatriz Orihuela for assistance in the laboratory. We thank the Duke University School of Medicine for the use of the Microbiome Core Facility, which provided 16S sequencing services. We would also like to thank the Integrative Bioinformatics for Investigating and Engineering Microbiomes Graduate Traineeship at Duke University for their training and support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.656506/full#supplementary-material
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp
Bailey-Brock, J. H. (1984). Ecology of the tube-building polychaete Diopatra leuckarti Kinberg, 1865 (Onuphidae) in Hawaii: community structure, and sediment stabilizing properties. Zool. J. Linnean Soc. 80, 191–199.
Bakunina, I. Y., Nedashkovskaya, P. I., Kim, S. B., Zvyaginstseva, T. N., and Mikhailov, V. V. (2012). Diversity of glycosidase activities in the bacteria of the phylum bacteroidetes isolated from marine algae. Microbiology 81, 688–695. doi: 10.1134/S0026261712060033
Bell, S. S. (1985). Habitat complexity of polychaete tube-caps: influence of architecture on dynamics of a meioepibenthic assemblage. J. Mar. Res. 43, 647–671. doi: 10.1357/002224085788440358
Bell, S. S., and Coen, L. D. (1982). Investigations on epibenthic meiofauna I. abundances on and repopulation of the tube-caps of Diopatra cuprea (Polychaeta: Onuphidae) in a subtropical system. Mar. Biol. 67, 303–309. doi: 10.1007/BF00397671
Berke, S. K. (2012). Biogeographic variability in ecosystem engineering: patterns in the abundance and behavior of the tube-building polychaete Diopatra cuprea. Mar. Ecol. Prog. Ser. 447, 1–13. doi: 10.3354/meps09568
Berke, S. K., Miller, M., and Woodin, S. A. (2006). Modeling the energy-mortality trade-offs of invertebrate decorating behaviour. Evol. Ecol. Res. 8, 1409–1425.
Brenchley, G. A., and Tidball, J. G. (1980). Tube-cap orientations of Diopatra cuprea (Bosc) (Polychaeta): the compromise between physiology and foraging. Mar. Behav. Physiol. 7, 1–13.
Bruckner, C. G., Bahulikar, R., Rahalkar, M., Schink, B., and Kroth, P. G. (2008). Bacteria associated with benthic diatoms from lake constance: phylogeny and influences on diatom growth and secretion of extracellular polymeric substances. Appl. Environ. Microbiol. 74, 7740–7749. doi: 10.1128/AEM.01399-08
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callaway, R. (2006). Tube worms promote community change. Mar. Ecol. Prog. Ser. 308, 49–60. doi: 10.3354/meps308049
Cardman, Z., Arnosti, C., Durbin, A., Ziervogel, K., Cox, C., Steen, A. D., et al. (2014). Verrucomicrobia are candidates for polysaccharide-degrading Bacterioplankton in an Arctic Fjord of Svalbard. Appl. Environ. Microbiol. 80, 3749–3756. doi: 10.1128/AEM.00899-14
Cho, S. H., Lee, E., Ko, S., Jin, S., Song, Y., Ahn, C., et al. (2020). Elucidation of the biosynthetic pathway of vitamin B groups and potential secondary metabolite gene clusters via genome analysis of a marine bacterium Pseduoruegeria sp M32A2M. J. Microbiol. Biotechnol. 30, 505–514. doi: 10.4014/jmb.1911.11006
Coastal Resources Commission (2007). Core Land Use Plan. Town of Morehead City, North Carolina: Coastal Resources Commission, The Wooten Company Available at: https://moreheadcitync.org/DocumentCenter/View/230/Land-Use-Plan-PDF (Accessed May 5, 2021).
Commito, J. A., Como, S., Grupe, B. M., and Dow, W. E. (2008). Species diversity in the soft-bottom intertidal zone: biogenic structure, sediment, and macrofauna across mussel bed spatial scales. J. Exp. Mar. Biol. Ecol. 366, 70–81. doi: 10.1016/j.jembe.2008.07.010
Curson, A. R. J., Rogers, R., Todd, J. D., Brearley, C. A., and Johnston, A. W. B. (2008). Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ. Micrbiol. 10, 757–767. doi: 10.1111/j.1462-2920.2007.01499.x
Cury, J. C., Araujo, F. V., Coelho-Souza, S. A., Peixoto, R. S., Oliveira, J. A., Santos, H. F., et al. (2011). Microbial diversity of a Brazilian coastal region influenced by an upwelling system and anthropogenic activity. PLoS One 6:e16553. doi: 10.1371/journal.pone.0016553
De Carlo, E. H., Hoover, D. J., Young, C. W., Hoover, R. S., and Mackenzie, F. T. (2007). Impact of storm runoff from tropical watersheds on coastal water quality and productivity. J. Appl. Geochem. 22, 1777–1797. doi: 10.1016/j.apgeochem.2007.03.034
Drury, B., Rosi-Marshall, E., and Kelly, J. J. (2013). Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl. Environ. Microbiol. 79, 1897–1905. doi: 10.1128/AEM.03527-12
Dubilier, N., Bergin, C., and Lott, C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. doi: 10.1038/nrmicro1992
Duperron, S., De Beer, D., Zbinden, M., Boetius, A., Schipani, V., Kahil, N., et al. (2009). Molecular characterization of bacteria associated with the trophosome and the tube of Lamellibrachia sp., a siboglinid annelid from cold seeps in the eastern Mediterranean. FEMS Microbiol. Ecol. 69, 395–409. doi: 10.1111/j.1574-6941.2009.00724.x
Fernandez-Gomez, B., Richter, M., Schuler, M., Pinhassi, J., Acinas, S. G., Gonzalez, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Fong, T. T., and Lipp, E. K. (2005). Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. 69, 357–371. doi: 10.1128/MMBR.69.2.357-371.2005
Fuirst, M., Veit, R. R., Hahn, M., Dheilly, N., and Thorne, L. H. (2018). Effects of urbanization on the foraging ecology and microbiota of the generalist seabird Larus argentatus. PLoS One 13:e0209200. doi: 10.1371/journal.pone.0209200
Giannotti, A. L., and McGlathery, K. J. (2001). Consumption of Ulva lactuca (Chlorophyta) by the omnivorous mud snail Ilyanassa obsoleta. J. Phycol. 37, 1–7. doi: 10.1046/j.1529-8817.2001.037002209.x
Gihring, T. M., Humphrys, M., Mills, H. J., Huette, M., and Kostka, J. E. (2009). Identification of phytodetritus-degrading microbial communities in sublittoral Gulf of Mexico sands. Limnol. Oceanogr. 54, 1073–1083. doi: 10.4319/lo.2009.54.4.1073
Gross, J. M., Peterson, R. E., Wu, S. Y., and McClay, D. R. (2003). LvTbx2/3: a T-box family transcription factor involved in the formation of the oral/aboral axis of the sea urchin embryo. Development 130, 1989–1999. doi: 10.1242/dev.00409
Gutiérrez, J. L., Jones, C. G., Strayer, D. L., and Iribarne, O. O. (2003). Mollusks as ecosystem engineers: the role of shell production in aquatic habitats. Oikos 101, 79–90. doi: 10.1034/j.1600-0706.2003.12322.x
Hedlund, B. P., Yoon, J., and Kasai, H. (2015). Rubritalea. Bergey’s Manual of Systematics of Archaea and Bacteria.
Hedman, J. E., Gunnarsson, J. S., Samuelsson, G., and Gilbert, F. (2011). Particle reworking and solute transport by the sediment-living polychaetes Marenzelleria neglecta and Hediste diversicolor. J. Exp. Mar. Biol. Ecol. 407, 294–301. doi: 10.1016/j.jembe.2011.06.026
Hochstein, R., Zhang, Q., Sadowsky, M. J., and Forbes, V. E. (2019). The deposit feeder Capitella teleta has a unique and relatively complex microbiomes likely supporting its ability to degrade pollutants. Sci. Total Environ. 670, 547–554. doi: 10.1016/j.scitotenv.2019.03.255
Huang, S., and Jaffe, P. R. (2018). Isolation and characterization of an ammonium-oxidizing iron reducer: Acidimicrobiaceae sp. A6. PLoS One 13:e0194007. doi: 10.1371/journal.pone.0194007
Huang, F., Pan, L., He, Z., Zhang, M., and Zhang, M. (2020). Identification, interactions, nitrogen removal pathways and performances of culturable heterotrophic nitrification-aerobic denitrification bacteria from mariculture water by using cell culture metagenomics. Sci. Total Environ. 732:139268. doi: 10.1016/j.scitotenv.2020.139268
Kerry, J. T., and Bellwood, D. R. (2012). The effect of coral morphology on shelter selection by coral reef fishes. Coral Reefs 31, 415–424. doi: 10.1007/s00338-011-0859-7
King, G. M. (2018). Microbiomes of the Enteropneust, Saccoglossus bromophenolosus, and association marine intertidal sediments of Cod Cove, Maine. Front. Microbiol. 9:3066. doi: 10.3389/fmicb.2018.03066
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Konhauser, K. O., Hao, W., Li, U., Gunten, K., Bishop, B. A., Alessi, D. S., et al. (2020). Diopatra cuprea worm burrow parchment: a cautionary tale of infaunal surface reactivity. Lethaia 53, 47–61. doi: 10.1111/let.12335
Kurilenko, V. V., Christen, R., Zhukova, N. V., Kalinovaskaya, N. I., Mikhailov, V. V., Crawford, R. J., et al. (2010). Granulosicoccus cocccoides sp. Nov., isolated from leaves of seagrass (Zostera marina). Int. J. Syst. Evol. 60, 972–976. doi: 10.1099/ijs.0.013516-0
Lalonde, S. V., Dafoe, L. T., Pemberton, M. K., and Konhauser, K. O. (2011). Investigating the geochemical impact of burrowing animals: proton and cadmium adsorption on the mucus lining of Terebellied polychaete worms. Chem. Geol. 277, 44–51. doi: 10.1016/JCHEMGEO.2009.12.010
Lee, K. C., Webb, R. I., Janssen, P. H., Sangwan, P., Romeo, T., Staley, J. T., et al. (2009). Phylum Verrucomicrobia representative share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 9:5. doi: 10.1186/1471-2180-9-5
Li, M., Yang, H., and Gu, J. D. (2009). Phylogenetic diversity and axial distribution of microbes in the intestinal tract of the polychaete Neanthes glandicinta. Microb. Ecol. 58, 892–902. doi: 10.1007/s00248-009-9550-8
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Magnum, C. P., Santos, S. L., and Rhodes, W. R. (1968). Distribution and feeding in the onuphid polychaete, Diopatra cuprea (BOSC). Mar. Biol. 2, 33–40.
Matsui, G. Y., Ringelberg, D. B., and Lovell, C. R. (2004). Sulfate-reducing bacteria in tubes constructed by the marine infaunal polychaete Diopatra cuprea. Appl. Environ. Microbiol. 70, 7053–7065. doi: 10.1128/AEM.70.12.7053-7065.2004
Mayer, M., Schaffner, L., and Kemp, W. (1995). Nitrification potentials of benthic macrofaunal tubes and burrow walls: effects of sediment NH4+ and animal irrigation behavior. Mar. Ecol. Prog. Ser. 121, 157–169. doi: 10.3354/meps121157
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Medina-Silva, R., Oliveira, R. R., Trindade, F. J., Borges, L. G. A., Simao, T. L., Augustin, A. H., et al. (2018). Microbiota associated with tubes of Escarpia sp. from cold seeps in the southwestern Atlantic Ocean constitutes a community distinct from that of surrounding marine sediment and water. Atonie van Leeuwenhoek 111, 533–550. doi: 10.1007/s10482-017-0975-7
Myers, A. C. (1972). Tube-worm-sediment relationships of Diopatra cuprea (Polychaeta: Onuphidae). Mar. Biol. 17, 350–356. doi: 10.1007/BF00366746
Neave, M. J., Streten-Joyce, C., Glasby, C. J., McGuinness, K. A., Parry, D. L., and Gibb, K. S. (2012). The bacterial community associated with the marine polychaete Ophelina sp.1 (Annelida: Opheliidae) is altered by copper and zinc contamination in sediments. Microb. Ecol. 63, 639–650. doi: 10.1007/s00248-011-9966-9
Nelson, T. M., Apprill, A., Mann, J., Rogers, T. L., and Brown, M. V. (2015). The marine mammal microbiome: current knowledge and future directions. Microbiol. Aust. 13, 8–13. doi: 10.1071/ma15004
North Carolina Environmental Quality (2021). Polluted Area Proclamations. Available at: https://deq.nc.gov/polluted-area-proclamations/ (Accessed April 12, 2021).
O’Beirn, F. X., Luckenbach, M., Nestlerode, J. A., and Coates, G. M. (2000). Toward design criteria in constructed oyster reefs: oyster recruitment as a function of substrate type and tidal height. J. Shellfish Res. 19, 387–395.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2015). Vegan: community ecology package. Rpackage version 2.0–7.
Park, S., Jung, Y. T., Won, S. M., Park, J. M., and Yoon, J. H. (2014). Granulosicoccus undariae sp. nov., a member of the family Granulosicoccaceae isolated from a brown algae reservoir and emended description of the genus Granulosicoccus. Antonie van Leeuwenhoek 106, 845–852. doi: 10.1007/s10482-014-0254-9
Paxton, H. (1998). The Diopatra chiliensis confusion--redescription of D. chiliensis (Polychaeta, Onuphidae) and implicated species. Zool. Scr. 27, 31–48. doi: 10.1111/j.1463-6409.1998.tb00427.x
Phillips, T. M., and Lovell, C. R. (1999). Distributions of total and active bacteria in biofilms lining tubes of the onuphid polychaete Diopatra cuprea. Mar. Ecol. Prog. Ser. 183, 169–178. doi: 10.3354/meps183169
R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Rincón-Tomás, B., González, F. J., Somoza, L., Sauter, K., Madureira, P., Medialdea, T., et al. (2020). Siboglinidae tubes as an additional niche for microbial communities in the Gulf of Cádiz—A microscopical appraisal. Microorganisms 8:367. doi: 10.3390/microorganisms8030367
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Romano, J. A. (2007). Acute toxicity and sub-lethal effects of non-point source pollutants on invertebrates. [Dissertation]. [Beaufort, NC]: Duke University.
Röthig, T., Roik, A., Yum, L. K., and Voolstra, C. R. (2017). Distinct bacterial microbiomes associate with the deep-sea coral Eguchipsammia fistula from the Red Sea and from aquaria settings. Front. Mar. Sci. 4:259. doi: 10.3389/fmars.2017.00259
Santos, T. M. T., and Aviz, D. (2019). Macrobenthic fauna associated with Diopatra cuprea (Onuphidae: Polychaeta) tubes on a macrotidal sandy beach of the Brazilian Amazon coast. J. Mar. Biolog. 99, 751–759. doi: 10.1017/S0025315418000711
Setiyono, E., Pringgenies, D., Shioi, Y., Kanesaki, Y., Awai, K., and Brotosudarmo, T. H. P. (2019). Sulfur-containing carotenoids from a marine coral symbiont Erythrobacter flavus strain KJ5. Mar. Drugs 17:349. doi: 10.3390/md17060349
Shibata, T., Solo-Gabriele, H. M., Fleming, L. E., and Elmir, S. (2004). Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Res. 38, 3119–3131. doi: 10.1016/j.watres.2004.04.044
Thomsen, M. S., Muth, M. F., and McGlathery, K. J. (2011). Tube-forming polychaetes enhance invertebrate diversity and abundance in sandy sediments of Mozambique. Africa. Afr. J. Mar. 33, 327–332. doi: 10.2989/1814232X.2011.600433
Villemur, R., Payette, G., Geoffroy, V., Mauffrey, F., and Martineau, C. (2019). Dynamics of a methanol-fed marine denitrifying biofilm: 2-impact of environmental changes on the microbial community. PeerJ 7:e7467. doi: 10.7717/peerj.7467
Von Borzyskowski, L. S., Severi, F., Kruger, K., Hermann, L., Gilardet, A., Sippel, F., et al. (2019). Marine proteobacteria metabolize glycolate via the B-hydrozyaspartate cycle. Nature 575, 500–504. doi: 10.1038/s41586-019-1748-4
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wegner, K. M., Volkenborn, N., Peter, H., and Eiler, A. (2013). Disturbance induced decoupling between host genetics and composition of the associated microbiome. BMC Microbiol. 13, 1–12. doi: 10.1186/1471-2180-13-252
Wessel, G. M., Berg, L., Adelson, D. L., Cannon, G., and McClay, D. R. (1998). A molecular analysis of Hyalin--A substrate for cell adhesion in the hyaline layer of the sea urchin embryo. Dev. Biol. 192, 115–126. doi: 10.1006/dbio.1997.8793
Keywords: Diopatra cuprea, polychaete, microbiome, tube worm, biogeochemistry, microbial function
Citation: Fuirst M, Ward CS, Schwaner C, Diana Z, Schultz TF and Rittschof D (2021) Compositional and Functional Microbiome Variation Between Tubes of an Intertidal Polychaete and Surrounding Marine Sediment. Front. Mar. Sci. 8:656506. doi: 10.3389/fmars.2021.656506
Received: 20 January 2021; Accepted: 05 May 2021;
Published: 31 May 2021.
Edited by:
Irene Wagner-Doebler, Helmholtz Association of German Research Centers (HZ), GermanyReviewed by:
Vengatesen Thiyagarajan (Rajan), The University of Hong Kong, ChinaCopyright © 2021 Fuirst, Ward, Schwaner, Diana, Schultz and Rittschof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Fuirst, bWZ1aXJzdEB1b2d1ZWxwaC5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.