- 1Marine College, Shandong University, Weihai, China
- 2State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
Although Verrucomicrobia is widely distributed in the marine environment, their physiological or cellular properties are poorly characterized because of the lack of cultured representatives. Under the selective pressure of two antibiotics, ofloxacin and norfloxacin, a Gram-stain-negative, aerobic coccus with exopolysaccharide (EPS) production ability was isolated from the coastal sediment of Xiaoshi Island, Weihai, China. These antibiotics inhibited bacterial growth, giving rise to the relatively slow-growing Verrucomicrobia that formed colonies on the isolation plates. It may be an effective method for the isolation of Puniceicoccaceae. From the taxonomic data obtained in this study, the new marine isolate NFK12T (=KCTC 72940T = MCCC 1H00424T) is proposed to be placed into a novel species within the genus Pelagicoccus for which the name Pelagicoccus enzymogenes sp. nov. is proposed. The EPS production of the strain NFK12T and the related strains were investigated and the effect of EPS produced by the strain NFK12T on the growth of other strains was examined. Besides, the effect of EPS on tolerance to ofloxacin and norfloxacin of the strain NFK12T was studied by measuring the biomass of the strain NFK12T. It was deduced that those strains that produced EPS tentatively protected themselves against the inhibitory effects of ofloxacin and norfloxacin.
Introduction
The bacterial phylum Verrucomicrobia is globally distributed in soil, marine, and freshwater environments, including the human gut (Bergmann et al., 2011; Freitas et al., 2012; Gómez-Gallego et al., 2016; Chiang et al., 2018). They have evolutionary and biological significance (Santarella-Mellwig et al., 2010; Patrick et al., 2017). Moreover, they are involved in the global cycling of methane and degradation of fucoidans and other complex polysaccharides, which are of immense significance to the understanding of ecology (Camp et al., 2009; Manuel et al., 2012; Sichert et al., 2020) and play key roles in the microbial communities of animal guts (Gómez-Gallego et al., 2016). Verrucomicrobia is nearly ubiquitous in the marine environment across a range of environmental conditions (Freitas et al., 2012). However, little is known about the important roles of this phylum in the marine environment.
Culture-independent sequencing studies of metagenomes from natural microbial communities in soil indicated that only a limited number of Verrucomicrobia species were cultured (Janssen, 2006). To date, few representatives have been cultivated; most of them are yet to be cultured, and only 60 species are (validly) described based on LPSN (LPSN, List of Prokaryotic Names with Standing in Nomenclature) (Parte et al., 2020). With the advent of novel genomics/bioinformatics techniques, the investigation of uncultured bacteria from sparsely studied phyla has become easier. However, axenic cultures are required for detailed studies of invisible physiological or cellular biological properties and unknown biochemical features (Wiegand et al., 2020). Besides, under the selective pressure of antibiotics, the members of PVC superphylum (Wagner and Horn, 2006) are more easily cultured, such as Planctomyces spp. and Pirellula spp. (Schlesner, 1994). Besides, Rhodopirellula spp. (Lage et al., 2012), Haloferula sargassicola MN1-1037T (Yoon et al., 2008b) and Luteolibacter pohnpeiensis A4T-83T (Yoon et al., 2008a) were successfully cultivated with the help of certain antibiotics. The inhibition of bacterial growth by the action of these antibiotics confers colony-forming abilities to the relatively slow-growing Planctomycetes and Verrucomicrobia.
Many marine bacteria produce exopolysaccharides (EPS), which assist the microbial communities in enduring extremes of temperature, salinity, and antimicrobial agents (Nichols et al., 2005; Liu et al., 2013). The extracellular polymeric substances are composed mainly of EPS, in which confer bacterial resistance to antibiotics (Hanna et al., 2018). Many studies have shown the association between EPS production and drug resistance of common pathogenic bacteria, especially Pseudomonas aeruginosa. Biofilm-forming bacteria are highly resistant to antibiotic therapy because the EPS acts as a barrier that provides anchorage and support and prevents the penetration of therapeutic agents (Venkatesan et al., 2015). Members of the phylum Kiritimatiellaeota and a member of the phylum Lentisphaerae, originally viewed as members of Verrucomicrobia, were reported to produce EPS (Cho et al., 2004; Vliet et al., 2020). However, this feature has not yet been described for other bacterial pure cultures of Verrucomicrobia. The relationship between EPS and antibiotic resistance in Verrucomicrobia is currently unknown
In this study, the EPS-producing strain NFK12T was isolated from coastal sediment. The EPS produced by strain the NFK12T was purified, and the effects of the EPS on the antibiotic tolerance and growth of NFK12T were studied. We hypothesized the possible roles of the EPS in the adaptation of the type strains of the family Puniceicoccaceae to the antibiotics. Based on the genomic analysis of the class Opitutae, we speculate that almost all bacteria belonging to this class produce EPS. The strain NFK12T (=KCTC 72940T = MCCC 1H00424T) is identified and classified into a novel species within the genus Pelagicoccus for which the name Pelagicoccus enzymogenes sp. nov. is proposed.
Materials and Methods
Sample Collection and Microbial Community Analysis
The sediment sample was collected from approximately 10 cm depth of the intertidal zone (37°31′ 36′′ N, 122°00′ 58′′ E) during September 2019. To obtain DNA for microbial community analysis, 5 mL of samples were centrifuged at 12,000 × g for 10 min, the supernatant was discarded, and the pellet was resuspended in 250 μL of sterile Milli-Q water. Then, DNA was extracted using the FastDNASpin Kit for Soil (MP Biomedicals, OH). The primer set used in the study was 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Mori et al., 2013), which was designed to amplify the V3–V4 region of the 16S rRNA gene and was demonstrated in silico to be universal for nearly all bacterial taxa. Sequencing was performed on a MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Antimicrobial Susceptibility Testing
The strains Oceanipulchritudo coccoides CK1056T (from our lab), Coraliomargarita sinensis WN38T (from our lab), Coraliomargarita akajimensis KCTC 12865T, Pelagicoccus mobilis KCTC 13126T, and Cerasicoccus arenae KCTC 12870T that were obtained from the Korean Collection for Type Cultures (KCTC) and Puniceicoccus vermicola JCM 14086T and Ruficoccus amylovorans JCM 31066T that were obtained from the Japan Collection of Microorganisms (JCM) were tested for antimicrobial susceptibility. The standard disk diffusion technique was used against 20 different antimicrobial disks, according to the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2018). Suspensions of the isolates equivalent to 0.5 McFarland standard turbidity were prepared, and MA plates were incubated at the optimal temperature before aseptic placement of antibiotic disks onto the surface. The antimicrobial activity was determined by measuring the diameters of the zones of inhibition formed around each paper disk. The tested antibiotics were categorized as sensitive, intermediate sensitive, or resistant, according to CLSI.
Bacterial Isolation and Culture Under Selective Pressure of Antibiotics
According to the results of antimicrobial susceptibility testing of strains of Puniceicoccaceae, all the studied strains (the related strains) were resistant to ofloxacin and norfloxacin. Therefore, the strains of phylum Verrucomicrobia were cultured under the selective pressure of ofloxacin and norfloxacin. Isolation was performed using marine agar 2216 (MA; BD) supplemented with ofloxacin (100 μg l–1) and norfloxacin (200 μg l–1). The sample was diluted with sterilized artificial seawater. Serial dilutions were spread onto MA plates and incubated under aerobic conditions at 30°C for 7 days. After incubation, a white colony was selected from the plate, and the NFK12T strain was obtained. The discrete colonies formed on the plate were selected and subjected to the identification procedure based on the analysis of the 16S rRNA gene sequence. Following purification with repeated streaking, the strain was routinely cultivated on MA at 30°C for 3 days. The strain was stored in sterile 15% (v/v) glycerol supplemented with 1% (w/v) NaCl suspensions at −80°C. The type strain, Pelagicoccus albus JCM 23202T, which was obtained from the Japan Collection of Microorganisms (JCM), was used as a related strain for comparative purposes.
16S rDNA Analysis
DNA was prepared using a bacterial DNA isolation kit (Takara). The 16S rRNA gene was amplified using primers 27F and 1492R (Lane, 1991). Sequencing was performed by the BGI Co. Ltd. (Qingdao, China), which resulted in a 1,406-bp almost complete 16S rRNA gene sequence. The cloned 16S rRNA gene sequence of the strain NFK12T was preliminarily identified by searching for matches in the EzBioCloud database1 and NCBI databases2. Comparative sequence analyses of the 16S rRNA gene sequences were used to determine the phylogenetic position of the NFK12T strain and related bacteria.
To definite its exact taxonomic status, we did a further phylogenetic and taxonomic investigation. The 16S rRNA gene sequences of the novel strain NFK12T and cultured type strains were aligned by using MAFFT version 7 (Katoh and Standley, 2013). The automated removal of spurious sequences and poorly aligned regions from the multiple sequence alignment was performed using trimAl (Capella-Gutierrez et al., 2009). The phylogenetic analysis was constructed by FastTree (Price et al., 2010) using GTR + CAT parameters and IQ-TREE (Jana et al., 2016) using the GTR + F + I + G4 model. Besides, these sequences were aligned by using MUSCLE (Edgar, 2004). A phylogenetic tree was reconstructed by using the neighbor-joining (Saitou and Nei, 1987) algorithm implemented in the software package MEGA (version 7.0) (Kumar et al., 2016). Phylogenetic trees were also generated with the maximum-likelihood (Felsenstein, 1981) and maximum-parsimony (Fitch, 1971) algorithms. Evolutionary distances were calculated using the Kimura 2-parameter model Bootstrap analysis was performed with 1,000 replications to provide confidence estimates for tree topologies.
In addition, 16S rRNA gene sequence of Puniceicoccaceae analysis was performed using the sequences available at the NCBI and EzBioCloud databases with completeness >90%. The 16S rRNA gene sequences of all of the strains of Puniceicoccaceae, including uncultured strains, were aligned using MAFFT version 7 (Katoh and Standley, 2013). The automated removal of spurious sequences and poorly aligned regions from the multiple sequence alignment was performed using trimAl (Capella-Gutierrez et al., 2009). The phylogenetic trees were constructed using IQ-TREE (Jana et al., 2016) by employing the GTR + F + I + G4 parameter and 1,000 bootstrap replications based on the 16S rRNA gene sequences from taxa with validly published names and the uncultured group containing almost 102 sequences. Bootstrap values were evaluated based on 1,000 replicates. Phylogenetic analysis was performed using MEGA version 7.0. The tree was rooted in Desulfocarbo indianensis SCBMT (KF032904), which was then pruned from the tree. The tree was collapsed and formatted using iTOL v4 (Letunic and Bork, 2019).
Genomic Analysis
The draft genome sequences were determined using the Shanghai OE Biotech Co., Ltd. (Shanghai, China). Sequencing was performed on an Illumina HiSeq Xten platform (Illumina Inc., San Diego, CA, United States). An Illumina shotgun library using the Illumina TruSeq Nano DNA LT Sample Prep Kit was reconstructed and sequenced in paired ends using the Illumina HiSeq platform. Raw sequencing data were generated using the Illumina base-calling software CASAVA v1.8.23, according to the manufacturer’s protocol. The sequenced reads were assembled using SOAPdenovo software (Li et al., 2009).
To determine its exact taxonomic status, further phylogenetic and taxonomic investigations were performed. Multilocus sequence analysis was performed for all the genomes reported in this study. All genomes were available from the NCBI and EzBioCloud databases. The genomes of the members of Puniceicoccaceae were identified using the Rapid Annotations using Subsystems Technology (RAST) server (Aziz et al., 2008), with a focus on EPS synthesis. Besides, gene content was annotated using the NCBI Prokaryotic Genome Annotation Pipeline. Gene genes involved in metabolic pathways were analyzed in detail by using information from the KEGG database (Kanehisa et al., 2016). The genomes were aligned using MUSCLE v.3.8.31. An up-to-date bacterial core gene set (UBCG) and a phylogenomics pipeline were utilized for phylogenetic tree construction. The phylogenetic analysis was constructed by FastTree (Price et al., 2010) using GTR + CAT parameters and IQ-TREE (Jana et al., 2016) using the GTR + F + I + G4 model. Phylogenetic analysis was performed with 1,000 bootstrap replicates based on 36 genomes. The genome of D. indianensis SCBMT served as the outgroup. The protein sequences (2,133 bp) of all the genomes reported in this study were subjected to phylogenetic analysis.
As measures of relatedness between the isolate and closely related taxa, the average amino acid identity (AAI) values, percentage of conserved proteins (POCP) values, the average nucleotide identity (ANI) (Yoon et al., 2017) values and the digital DNA-DNA hybridization (dDDH) values were determined. The average amino acid identity (AAI) between genomes were calculated by CompareM4 (Rodriguez-R and Konstantinidis, 2014). The POCP was calculated as described by Qin et al. (2014). The average nucleotide identity (ANI) values were calculated using JSpeciesWS5. The genome-to-genome distance calculator (GGDC 2.16) was used to calculate all dDDH (Goris et al., 2007) values.
Growth Conditions, Biochemical Characterization, and Chemotaxonomy
Phenotypic characterizations, biochemical characterization, and chemotaxonomy were performed using cells grown on MA at 33°C for 2 days. Cell morphology and size were examined by light microscopy (E600, Nikon), transmission electron microscopy (JEM-1200, Jeol), and scanning electron microscopy (Nova NanoSEM450, FEI). Physiological and biochemical experimental details have been described in previous studies (Feng et al., 2020). Motility was determined by the hanging-drop method and tested by inoculating the bacteria on the 0.5% agar. The Gram reaction was determined as described by Tindall et al. (2007). The temperature range for growth was determined on MA at 4, 10, 15, 20, 25, 28, 30, 33, 37, and 40. The NaCl concentrations for growth were determined by incubating the bacteria on modified marine broth 2216 made with 0.5% peptone, 0.1% yeast extract, artificial seawater (consisting 0.32% MgSO4, 0.22% MgCl2, 0.12% CaCl2, 0.07% KCl, 0.02% NaHCO3, w/v) and in the presence of 0.0–10.0% (w/v) NaCl at intervals of 0.5%. The effects of pH were determined by adding the appropriate buffers (Sangon), including MES (pH 5.5 and 6.0), PIPES (pH 6.5 and 7.0), HEPES (pH 7.5 and 8.0), Tricine (pH 8.5) and CAPSO (pH 9.0, and 9.5) to MB with a concentration of 20 mM and the pH of control groups were checked after autoclaved. OD600 values of the cultures were measured after incubation for 2 days at 33°C. Unless otherwise mentioned, all experiments were performed with three replicates. Catalase activity was determined by the observation of bubble formation in a 3% H2O2 solution. Oxidase activity was examined using an oxidase reagent kit (bioMérieux) according to the manufacturer’s instructions. Tests for the hydrolysis of starch, casein, alginate, carboxymethylcellulose, Tweens 20, 40, 60, and 80 were determined as described by Cowan (Cowan et al., 1966). The API 20E (bioMérieux) kits were performed according to the manufacturer’s instructions, using the biomass of strain NFK12 grown on MA at 33°C for 2 days. Production of other enzymes was assessed using API ZYM (bioMérieux) kits. Carbon source oxidation was checked in BIOLOG GEN III microplates7. All the API tests were performed according to the manufacturer’s instructions. All of the API and the BIOLOG tests were performed with two replicates. All tests were carried out simultaneously with the related type strain.
Polar lipids were determined using 2D thin-layer chromatography (TLC) (Minnikin et al., 1984). Isoprenoid quinones of the strain NFK12T were analyzed using reverse-phase HPLC (Kroppenstedt, 1982). The preparation and extraction of fatty acid methyl esters from biomass and their subsequent separation and identification by gas chromatography were performed as described elsewhere (Athalye et al., 2010). The fatty acids were extracted according to the standard protocol of MIDI (Sherlock Microbial Identification System, version 6.1). The fatty acids were methylated and analyzed by an Agilent 6890N gas chromatograph. Cellular fatty acids were identified using the TSBA40 database of the microbial identification system.
Identification of EPS-Producing Strains
The strains were subjected to Congo red staining to detect the presence of EPS. The strains NFK12T and P. albus JCM 23202T were cultured at 33°C for 3 days on Congo red agar plates (supplemented with 0.8 g/L Congo red) with MA (Freeman et al., 1989) or in MB with Congo red solution (0.3 g/L). Based on the results of genomic and phylogenomic analyses of Puniceicoccaceae, other strains of the family were also suspected to produce EPS. The type strains, O. coccoides CK1056T, C. akajimensis KCTC 12865T, P. mobilis KCTC 13126T, C. arenae KCTC 12870T, P. vermicola JCM 14086T, and R. amylovorans JCM 31066T, were inoculated on the Congo red agar plates. The cells were harvested, washed three times, and then resuspended in sterile artificial seawater (OD600 = 3.5). Congo red (150 μg/mL) and Trypan blue (20 μg/mL) were mixed evenly in a total volume of 125 μL with the suspensions of the cell strains. The samples were incubated at room temperature (RT) in the dark for 30 min and centrifuged at 12,000 × g for 10 min. Then, 200 μL of the supernatant was added to 96-well plates, and the optical density was measured at 490 and 585 nm for Congo red and Trypan blue, respectively.
EPS Purification
The NFK12T strain was cultured at 33°C with shaking at 200 rpm for 3 days to induce EPS production. The crude EPS from the bacterial isolates was obtained as previously described (Liu et al., 2013). The culture medium was heated in boiling water for 10 min to inactivate the enzymes and then cooled down to 25°C. Then, the medium was centrifuged (20 min, 10,000 g, and 4°C) to remove cells and coagulated proteins, and the supernatant was collected. Next, 80% (w/v) trichloroacetic acid was added to the supernatant to a final concentration of 4% (w/v) with gentle stirring and kept at 4°C for 10 h. Protein precipitates were removed by centrifugation, and then EPS were precipitated from the supernatant with three volumes of cold ethanol followed by overnight incubation at 4°C. After centrifugation, the precipitate was resuspended in deionized water and dialyzed (molecular weight cut-off: 14 kDa) for 3 days. The retentate was centrifuged to remove the insoluble material. The supernatant was lyophilized, and white crude polysaccharides (LCP) were obtained (Ai et al., 2008). Total EPS (expressed as mg l–1) was estimated by the phenol-sulfuric acid method using glucose as a standard (Dubois et al., 1956), and the amount of EPS produced by each isolate was calculated. The type species of the family Puniceicoccaceae were also cultured for EPS production, using the same method as that for the NFK12T strain.
Scanning electron microscopy-energy dispersive X-ray (SEM-EDX) (model Nova NanoSEM450; FEI) was employed to analyze the film cross-sections of the crude EPS from the bacterial isolates. Films were placed on a metallic stub and were covered with gold under vacuum in an argon atmosphere.
Effect of EPS on Bacterial Growth
The NFK12T strain was cultured at 33°C for 2 days on MA with or without EPS produced by P. enzymogenes (300 μg/mL). Since the process of extracting EPS from the strain NFK12T was complicated and chitosan as the only cationic marine polysaccharide is a random copolymer obtained from the alkaline deacetylation of chitin (Muxika et al., 2017), we consider whether it can be replaced by commercial chitosan for other related experiments. Therefore, the strain NFK12T was cultured at 33°C for 2 days on MA with chitosan (Sangon Biotech, 300 μg/mL). Samples were then removed and serially diluted to 10–6 in sterilized seawater and spotted on the modified MA agar plates for culturing at 33°C. Colony formation unit (CFU) counting was performed at 48 h. R. amylovorans JCM 31066T and Escherichia coli DH5α were cultured on modified NB agar plates at 25°C and on modified LB agar plates at 37°C, respectively. For the strain NFK12T, CFU counting was performed at 16 and 109 h using the same method. All experiments were performed in triplicate.
Measurement of Minimum Inhibitory Concentration (MIC)
Antimicrobial susceptibility testing revealed that the strain NFK12T was also resistant to ofloxacin (5 μg) and norfloxacin (10 μg). To investigate the MICs of ofloxacin and norfloxacin, the strain NFK12T was inoculated into a 250-mL Erlenmeyer flask containing 100 mL of MB and incubated for 37 h at 33°C and 200 rpm until reaching the late logarithmic phase. The cells were harvested, washed three times, and then resuspended in sterile artificial seawater (OD600 = 1.0). The sample was inoculated into a 96-well culture plate filled with MB medium. An equal volume of ofloxacin and norfloxacin at different concentrations (ofloxacin: 0–150 μg/mL at intervals of 10.0 μg/mL; norfloxacin: 0–300 μg/mL at intervals of 20.0 μg/mL) was mixed with the cell suspensions of the strain NFK12T in 96-well culture plates. Samples were incubated at 33°C in the dark for 2 days. Then, 200 μL of bacterial cultures were added to 96-well plates, and growth was measured by recording the optical density (OD) at 600 nm. For R. amylovorans JCM 31066T, the MIC was determined using the same method.
Effect of the EPS on Antibiotics Tolerance
The effect of EPS on the antibiotic tolerance of the strain NFK12T was studied by comparing biomass of EPS producing strains against non-EPS producing strains. Samples were then removed and serially diluted to 10–7 in sterilized seawater and spotted on MA agar plates supplemented with 180 μg/mL norfloxacin or 50 μg/mL ofloxacin with or without 300 μg/mL EPS or 300 μg/mL chitosan at 33°C. Colony formation unit counting was performed at 48 h. In addition, the strain NFK12T was diluted by 1.0% in MB containing 50 μg/mL with or without 300 μg/mL EPS or 300 μg/mL chitosan and incubated in a shaker (200 rpm) for 0, 6, 12, 24, 36, and 48 h at 30°C. Samples were then removed and serially diluted to 10–7 in sterilized seawater and spotted on MA agar plates for culturing at 30°C. CFU counting was performed at 48 h. R. amylovorans JCM 31066T, which had a very low yield of EPS production, were also cultured to verify the effect of EPS on antibiotic tolerance. The method was the same as that of the strain NFK12T. All experiments were performed in triplicate.
Results
Isolation and Identification of Strains
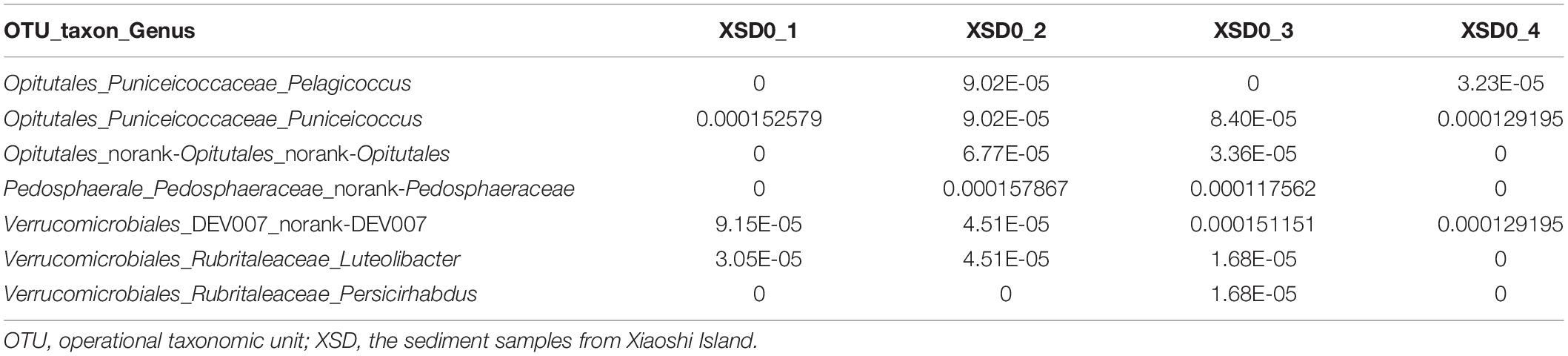
A novel strain exhibiting the defining features of the members of the genus Pelagicoccus was obtained based on its resistance to ofloxacin and norfloxacin. To culture marine Verrucomicrobia strains, sediment samples were collected from Xiaoshi Island (37°31 36′′ N, 122°00 58′′ E), Weihai, China, which is a national marine nature reserve. The samples were subjected to 16S high-throughput analysis, which revealed that the sequences of verrucomicrobia belonged to three families, Puniceicoccaceae, Pedosphaeraceae, and Rubritaleaceae, and two unclassified groups (Supplementary Appendix Table A). Among these, the species of Puniceicoccaceae were the most abundant and the genus Pelagicoccus was detected (Table 1). Furthermore, the antimicrobial susceptibility testing of the type strains of the family Puniceicoccaceae showed that they were resistant to ofloxacin (5 μg) and norfloxacin (10 μg) (Supplementary Appendix Table B).
Due to the results of the resistance to fluoroquinolones based on genomic analysis and antimicrobial susceptibility testing of the type strains of the family Puniceicoccaceae, we attempted to culture the strains of Puniceicoccaceae under the selective pressure of ofloxacin and norfloxacin. The strain NFK12T was isolated from marine agar plates containing ofloxacin (100 μg l–1) and norfloxacin (200 μg l–1). Based on the pairwise comparison of 16S rRNA gene sequences of species available at GenBank databases, the strain NFK12T exhibited the highest similarity to the most closely validly published species P. albus JCM 23202T with 98.5% similarity. Based on the 16S rRNA gene sequence similarity threshold value (98.65%) suggested for species delineation (Kim et al., 2014), the strain NFK12T represents a novel species within the Pelagicoccus clade. An evolutionary tree was constructed using the neighbor-joining method based on the 16S rRNA gene sequences. According to the results, the strain NFK12T formed an independent linage within the genus Pelagicoccus (Figure 1 and Supplementary Figure 1). The distinct phylogenomic lineage of strain NFK12T was also supported by the POCP values and average amino acid identity (AAI) values (Table 2). AAI was higher than the threshold proposed to include an organism in a given genus (60.0%). The POCP values were higher than 50% which the 50.0–60.0% POCP value proposed as a genus boundary. The ANI value of the strain NFK12T with the related strains were lower than 74.6%. In addition, the dDDH value (identities/HSP length) of the strain NFK12T with its related strains were lower than 20.3%. According to the proposed and generally accepted species boundary for dDDH values (i.e., <70% and ANI < 95%), the strain NFK12T was identified as a new species distinguishable from the related strain.
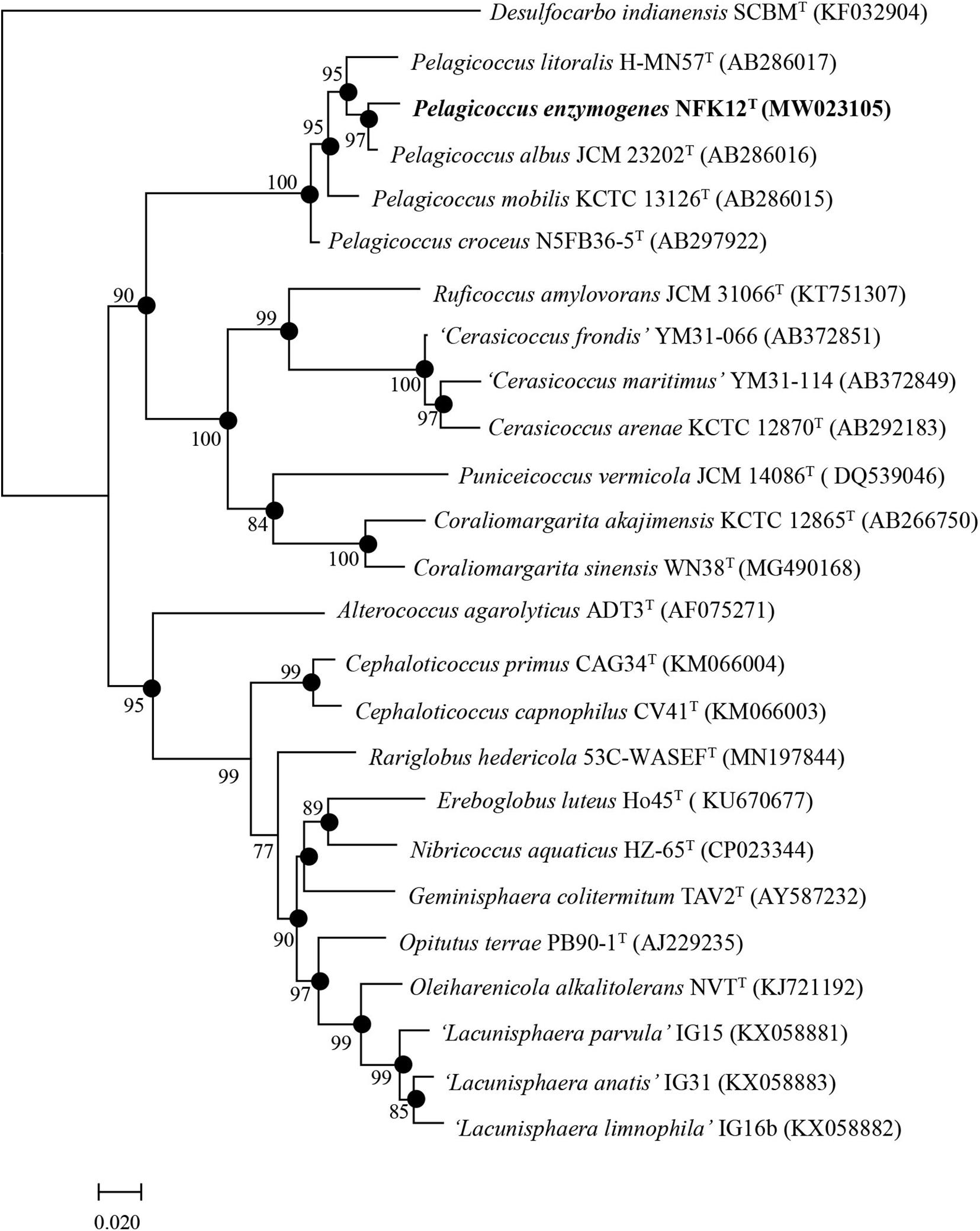
Figure 1. Phylogenetic trees based on the complete 16S rRNA gene sequences from bacterial taxa showing the taxonomic position of strain NFK12T. Filled circles indicate nodes overlapping on trees reconstructed using FastTree and IQ-TREE algorithms. Numbers on nodes represent bootstrap values (FastTree/IQ-TREE) based on 1,000 replications, and bootstrap values (>70%) are shown at branch nodes. Desulfocarbo indianensis SCBMT (KF032904) was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
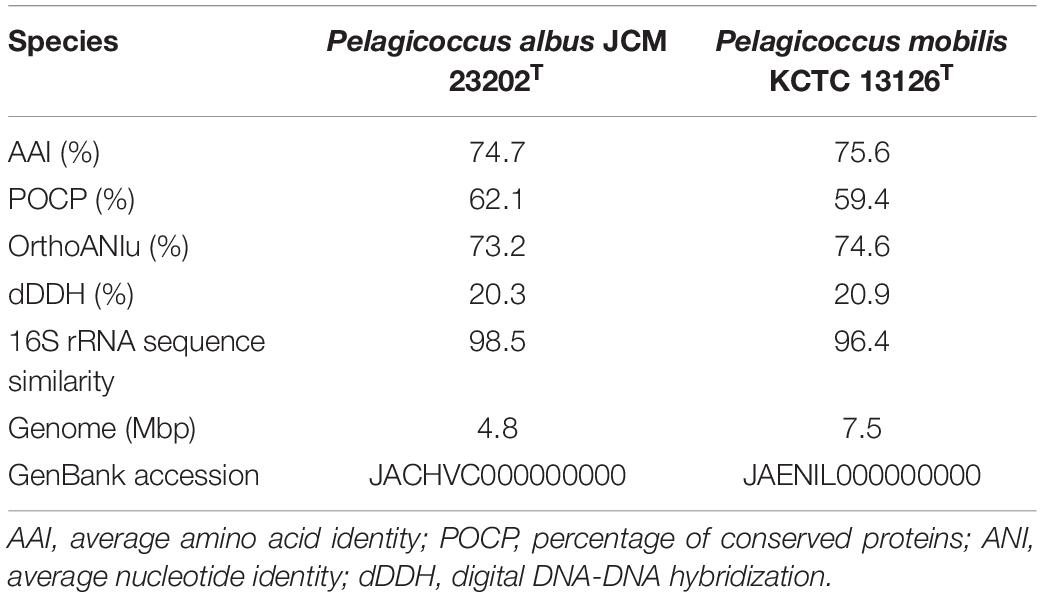
Table 2. The genome-to-genome distance between strain NFK12T and the phylogenetically related strains.
Morphological Characterization of Novel Strain
After 2 days, cells of the strain NFK12T exhibited a coccus shape with a diameter of 0.5–1.5 μm (Figures 2A–C). Through transmission electron microscopy (TEM), flagella were observed, but the number of flagella per cell of strain NFK12T is unclear (Figure 2A). A faint layer of extracellular slime was seen around the cells (Figure 2A), and cells of the strain NFK12T had randomly distributed finger-like extensions from which the fimbriae appendages projected, measuring more than 500 nm in diameter, as viewed under a scanning electron microscope (SEM) (Figures 2B,C). The electron micrographs indicate that the strain NFK12T produces peripheral EPS, which has a three-dimensional web-like morphology (Figure 2C), as described in Lentisphaera araneosa (Cho et al., 2004).
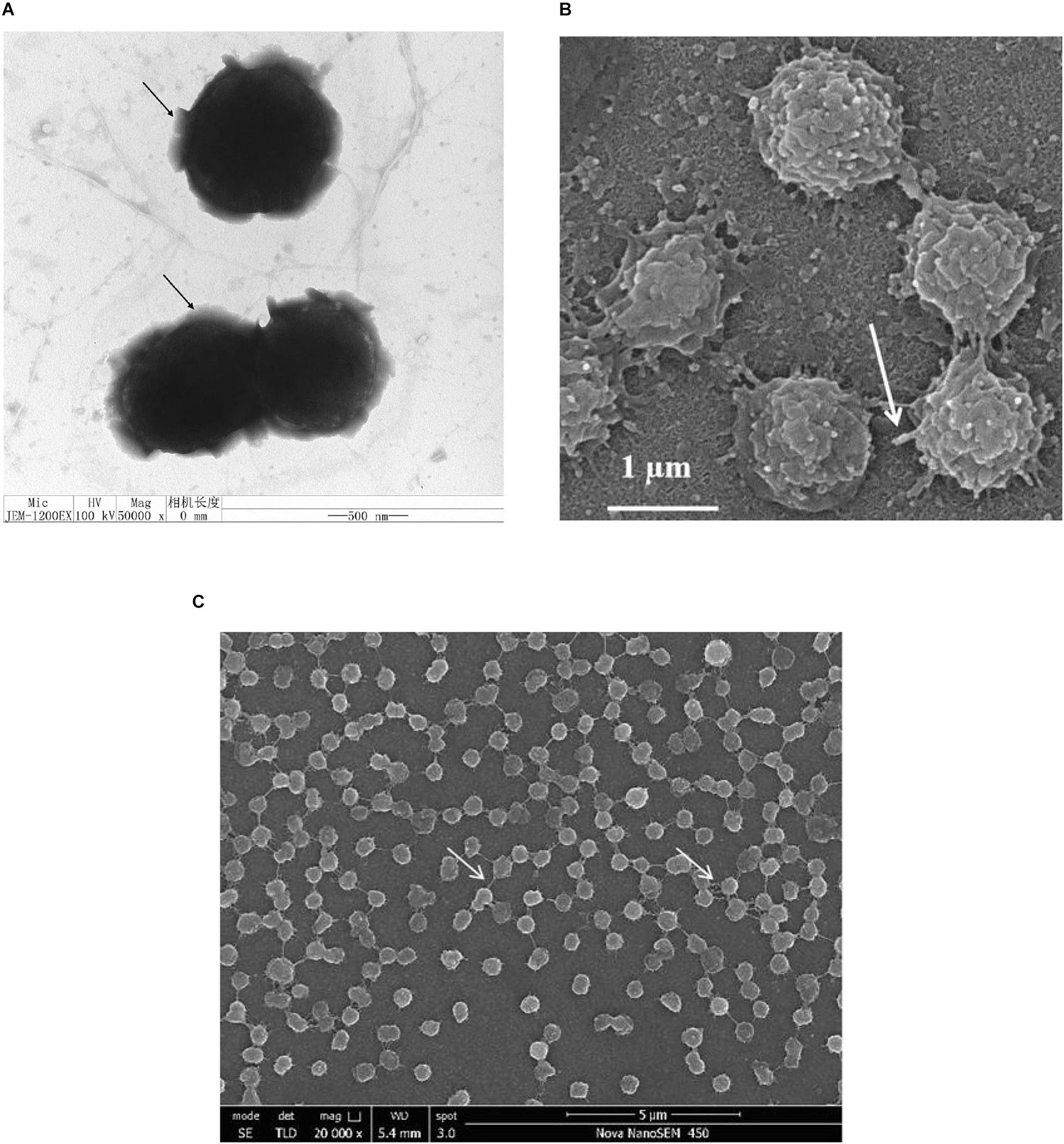
Figure 2. Shape and ultrastructure of cells of strain NFK12T. Cells were grown in MB at 33°C for 2 days. (A) Transmission electron microscopy. Arrows in panel a indicate ‘capsular’ EPS around the cells. Scale bar, 0.5 μm. (B) Scanning electron microscopy. Arrows indicate randomly distributed finger-like extensions from which fimbriae appendages. Scale bar, 1.0 μm. (C) Scanning electron microscopy. Arrows indicate ‘bridge’ extracellular polysaccharides (EPS). Scale bar, 5.0 μm.
EPS-Producing Ability of Puniceicoccaceae Species
The strains were subjected to Congo red staining to detect the presence of EPS. A positive result was indicated by red-stained colonies when the bacterial communities reached the late exponential stage of growth. The color of the strain NFK12T colonies was checked after 3 days, where the red-stained colonies indicated EPS positivity. The positive colonies were identified by the appearance of a red halo around the colonies (Supplementary Figure 2A). In addition, the red-stained cells gathered at the bottom, while the samples cultured in MB were centrifuged at 3,700 rpm for 10 min (10 min, 3,700 g, RT) (Supplementary Figure 2B). The results indicated that the strain NFK12T produced EPS, and we speculated that EPS production was one of the common features of Puniceicoccaceae. On the other hand, the results of Congo red and Trypan blue staining showed that the NFK12T strain and its related strains were EPS-producing isolates (Supplementary Figure 3). The EPS yield of the strain NFK12T under our experimental conditions was higher than that of its related strains. Among them, the yield of EPS produced by R. amylovorans JCM 31066T was meager (Figures 3A,B).
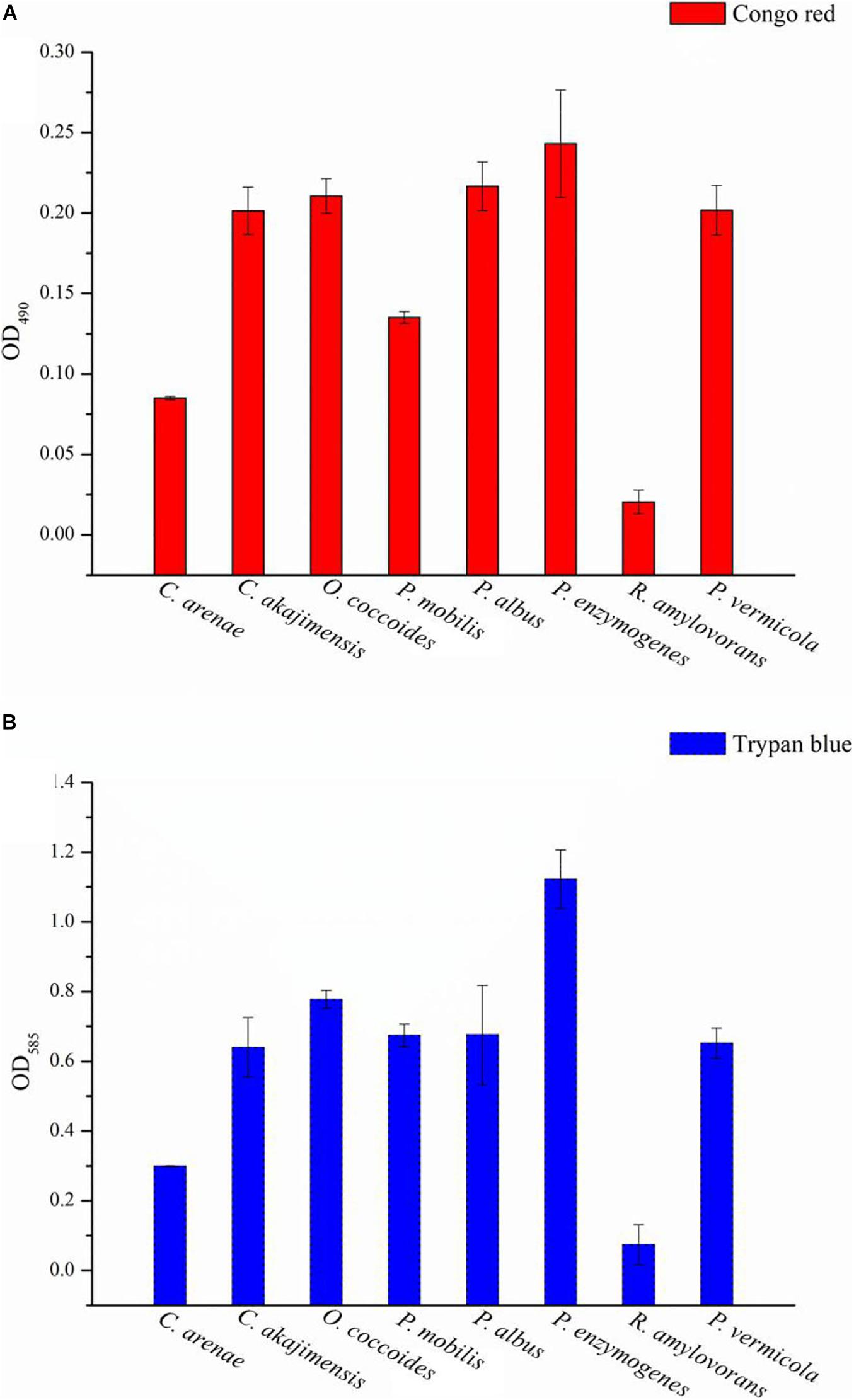
Figure 3. The EPS content of the type species strains of Puniceicoccaceae was assayed with Congo red and Trypan blue staining. Y-axis: all group data minus the optical densities (OD) of the control. The higher the value, the higher the EPS content of the strains. X-axis: the Latin name of strains. Congo red and Trypan blue are the name of the dye used.
Effect of EPS Derived From Strain NFK12T on Bacterial Growth
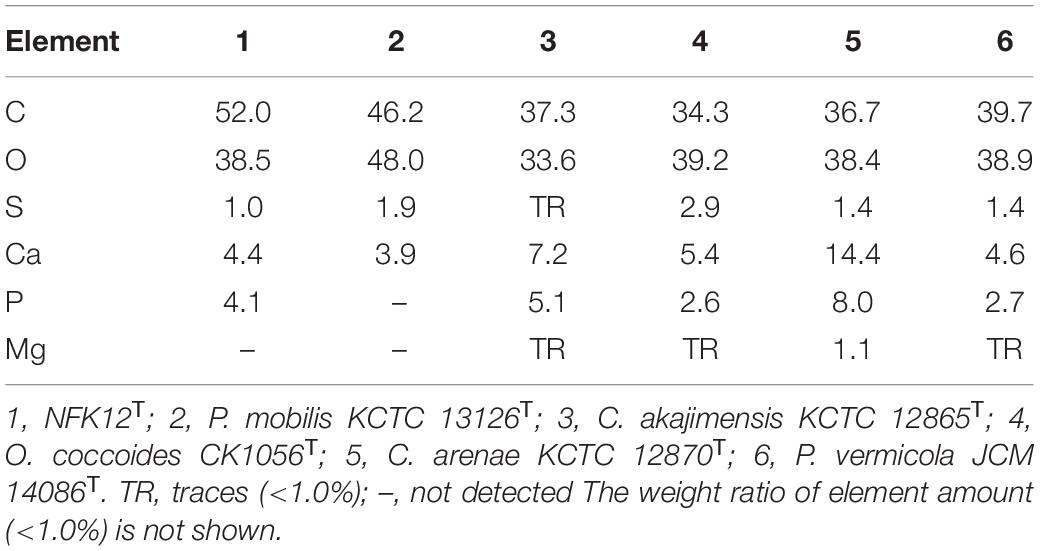
The yield of EPS production was characterized by gravimetric analysis of the lyophilized crude EPS. The strain NFK12T could produce EPS at a yield of 65.5 mg/L by the phenol-sulfuric acid method in MA at an optimal temperature of 33°C. Regarding SEM analysis, film cross-sections of the strain NFK12T are shown in Supplementary Figure 4. A compact structure was observed in all films, but the surface was not flat. Sulfated polysaccharides are prevalent in marine environments (William, 2017), and the EPS of NFK12T are polyanionic for the presence of phosphate and sulfate based on the EDX analysis. The yields of EPS synthesized by all strains were the same. The detailed differences in the EPS profiles between the strain NFK12T and its related strain are shown in Table 3. Among them, EPS production by R. amylovorans JCM 31066T was not detected by the phenol-sulfuric acid method.
The strain NFK12T, which produced the highest EPS, was selected for further analysis. For E. coli DH5α, only the colonies on the modified LB agar plates with chitosan were visible after 8 h, and more colonies on all plates gradually emerged over subsequent hours. However, the number of colonies was not significantly different on the plates with or without EPS/chitosan (Figure 4A). For R. amylovorans JCM 31066T and the strain NFK12T, the number of CFUs on the modified agar plates with chitosan or EPS was higher than that of the control. In particular, EPS and chitosan significantly promoted the growth of the NFK12T strain. Therefore, the uncultured strains of the genus Pelagicoccus and other rare bacteria could be supplemented by chitosan or EPS for optimum growth.
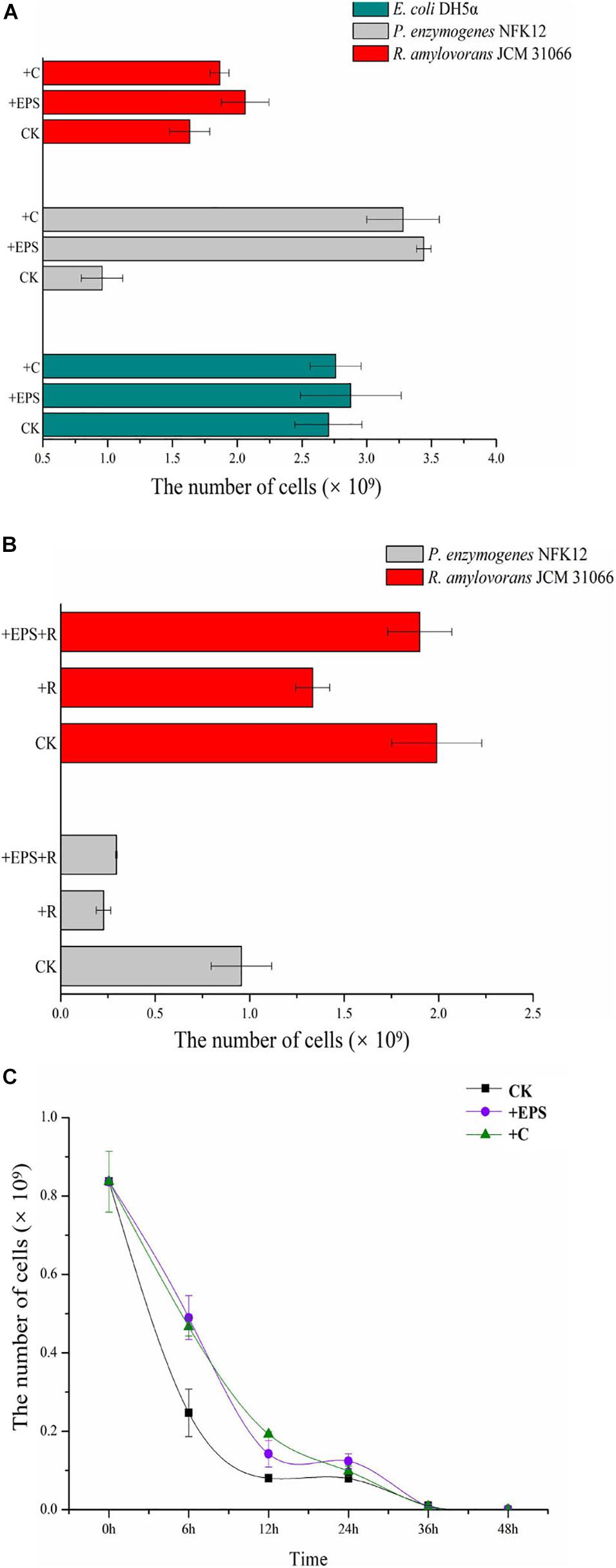
Figure 4. The CFU of the control and experimental groups and the number of CFU cells. (A) Effect of EPS and chitosan on the bacterial growth by comparing biomass of strains supplemented EPS against non-EPS supplementing strains. (B,C) The effect of EPS on the antibiotic tolerance of strain NFK12T was studied by measuring the biomass of strain NFK12T. EPS, exopolysaccharides; C, chitosan; R, norfloxacin; CK, control; +, supplemented.
Analysis of Antimicrobial Roles of the EPS
In agreement with previous findings (Venkatesan et al., 2015), the growth of the strain NFK12T with EPS in the presence of ofloxacin or norfloxacin was higher than that of the control. Because of antibiotic treatment, the strain NFK12T could be observed at sublethal concentrations of 180 μg/mL norfloxacin or 50 μg/mL ofloxacin. After treatment with norfloxacin, the number of CFUs on the plates with chitosan or EPS increased, but it was less than that on the plates without norfloxacin. Since the EPS yield of R. amylovorans JCM 31066T was meager, we speculated that the effect of chitosan or EPS on the strain might be different after antibiotic treatment. Therefore, we performed the same experiment after antibiotic treatment. The strain R. amylovorans JCM 31066T could be observed at sublethal concentrations of 260 μg/mL norfloxacin or 110 μg/mL ofloxacin. The result of R. amylovorans JCM 31066T was the same as that of the strain NFK12T (Figure 4B). It was concluded that EPS could promote bacterial resistance to norfloxacin. However, we observed that cells did not survive on MA with 50 μg/mL ofloxacin when the CFUs were counted at 48 h. Moreover, some cells resumed growth (Tregrowth = 132 h) after ofloxacin did not work. Therefore, the strain NFK12T was cultured in MB. After a bacterial population was treated with 50 μg/mL ofloxacin for 0, 6, 12, 24, 36, and 48 h, the surviving cells were cultured on fresh MA plates. With chitosan or EPS in MB, the colonies of the strain NFK12T were visible after 24 h on MA, while the colonies of the strain NFK12T were visible after 48 h in the case of control. More colonies gradually emerged over subsequent hours (Figure 4C). After the addition of chitosan or EPS, the number of CFUs of the NFK12T strain was higher than that of the control. After incubation in MB with ofloxacin for a different time, the number of CFUs of the NFK12T strain was significantly less than that of the control without ofloxacin treatment. This indicated that some cells were killed by ofloxacin. When chitosan or EPS was added, the number of CFUs increased. However, after incubation with ofloxacin for 36 and 48 h, no effect was observed with the addition of EPS. The number of CFUs was almost the same with or without EPS at 36 and 48 h. In other words, the EPS could tentatively improve the ofloxacin tolerance of the strain NFK12T, but ofloxacin still worked.
Phylogenetic Analysis of Puniceicoccaceae
The order Puniceicoccales, containing Puniceicoccaceae, and the order Opitutales, containing the family Opitutaceae, were proposed for the classification of species belonging to subdivision 4 (Cho et al., 2004). The genus Puniceicoccus (Choo et al., 2007) is the type genus of the family Puniceicoccaceae. Besides Puniceicoccus, the genera Cerasicoccus (Yoon et al., 2007a), Ruficoccus (Lin et al., 2017), Pelagicoccus (Yoon et al., 2007b), Coraliomargarita (Yoon et al., 2007c), and Oceanipulchritudo (Feng et al., 2020) have been described as members of the present family. In 2007, the genus Pelagicoccus was proposed by Yoon et al. (2007b) as a new member of the family Puniceicoccaceae. The strains of the genus Pelagicoccus have been isolated from the sea environment. As of December 2020, this genus contains four species: P. mobilis, P. albus, Pelagicoccus litoralis (Yoon et al., 2007b), and Pelagicoccus croceus (Yoon et al., 2007b). A few representatives of Puniceicoccaceae have been cultivated, most of which are yet to be cultured.
With the golden era of the sequencing technology revolution, the abundance, diversity, and ecology of microorganisms gained another dimension. A total of 102 16S rRNA gene sequences of Puniceicoccaceae were available from the NCBI and EzBioCloud databases with completeness >90%, and 80.4% of them belonged to uncultured strains. Cluster analysis of the 16S rRNA gene sequences, including the complete 16S rRNA gene sequence of the strain NFK12T, was performed, and the phylogenetic trees were constructed using the IQ-TREE server (Price et al., 2010). Of the gene sequences, 67.6% belonged to strains isolated from the marine environment, which could be divided into six groups (groups I–VI) (Supplementary Figure 5). This result was consistent with the previous study that subdivisions 1 and 4 generally dominated marine bacterial communities (Freitas et al., 2012). Most of the cultured species were grouped into groups V and VI. The strains of each group were from different sampling sites and environmental conditions. For group III, there were no cultured type species. Members of Puniceicoccaceae appear especially significant, but isolates do not represent the known phylogenetic breadth from culture-independent studies. Therefore, there seems to be a vast wealth of verrucomicrobial species remaining to be cultured and described.
Genomic Analysis of Puniceicoccaceae
The draft genome size of the strain NFK12T was determined to be 6,276,818 bp, arranged into 200 contigs. The G + C content was calculated to be 56.8 mol%, which was in the range of that of the genus Pelagicoccus (51.0–57.0 mol%). An average of 235× coverage depth was accomplished. According to the KEGG and RAST analyses, the membrane fusion protein of RND family multidrug efflux pump, multiple antibiotic resistance protein (marC), and multidrug resistance protein (MATE family) were found in the draft genome of strain the NFK12T.
In this study, we found that the majority of the new strains brought into the pure culture are affiliated with subdivision 1. In particular, for Puniceicoccaceae, thus far, only two genomes from validly described species (C. sinensis and C. akajimensis) are available. Therefore, we obtained the type species of Puniceicoccaceae and sequenced the draft genome sequences. Nineteen genomes of the uncultured members of Puniceicoccaceae were available at the NCBI and EzBioCloud databases, and the 15 genomes were identified as close neighbors after calculation of AAI with the published genome sequences of the members of Puniceicoccaceae (AAI > 55.0%) (Rodriguez-R and Konstantinidis, 2014). The 15 genomes were utilized for phylogenetic tree construction and were consistently clustered together within the same clade. When the whole genome sequences of these strains were analyzed, nearly all the branches (including the deeper branches) in the protein sequence tree were well supported by bootstrap replicates (Figure 5). In addition, the genomic analysis revealed EPS biosynthesis in those strains, which was almost consistent with the phenotype. Exopolysaccharide biosynthesis protein was predicted in 27 genomes of the class Opitutae after annotation using the RAST server and the NCBI Prokaryotic Genome Annotation Pipeline (Supplementary Appendix Table D). Interestingly, the results indicated that P. albus JCM 23202T consisted of EPS-producing isolates, but EPS biosynthesis protein was not predicted in the genome. However, it was less clear for us to arrive at that conclusion based on the draft genome. According to the phylogenetic trees of EPS biosynthesis protein sequences of the class Opitutae, the strains of Puniceicoccaceae and Opitutaceae clustered, respectively (Supplementary Figure 6).
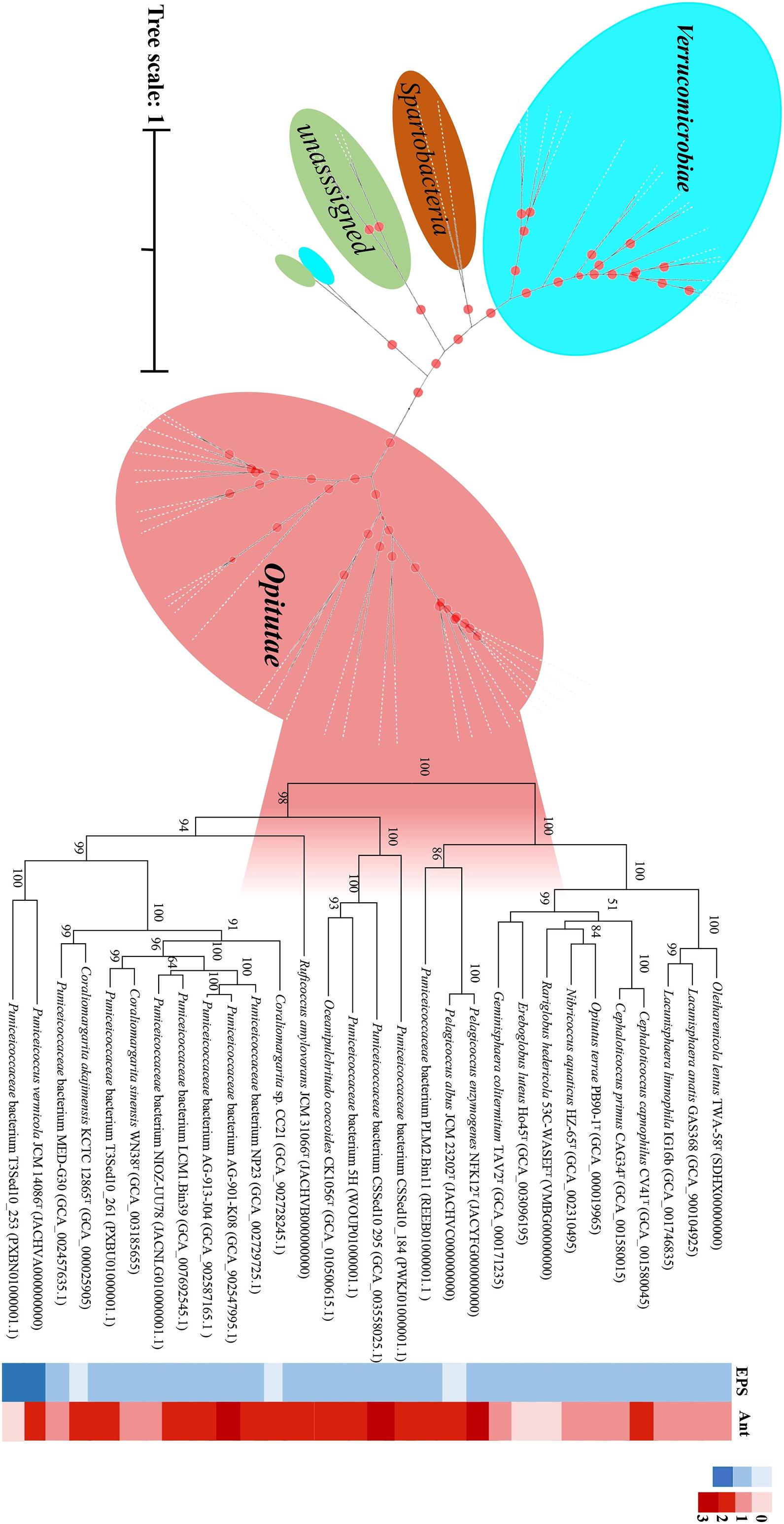
Figure 5. Phylogenetic analysis of the protein sequence computed using all the verrucomicrobial genomes reported in this study by IQ-TREE/FastTree. Desulfocarbo indianensis SCBMT was used as an outgroup to root the tree. The genomes of the exopolysaccharide (EPS)-producing strains are shown in the solid blue grid, and those of the fluoroquinolone-resistant strains are shown in the solid red grid.
Based on the results of genomic analysis, we suspect that almost all bacteria of this class produce EPS. The EPS-producing strains were screened out quickly at the first stage by adding Congo red to the medium. Therefore, besides supplementing with ofloxacin (100 μg l–1) and norfloxacin (200 μg l–1), Congo red solution (0.3 g/L) was added to the culture medium. By optimizing the isolation methodology, a large number of Puniceicoccaceae species could be obtained. In this study, we obtained two strains of the genus Coraliomargarita (R2-22 MW255631 and E2-5 MW255632). In future studies, we will use this method to isolate more species of this family and try to verify this method.
Taxonomy
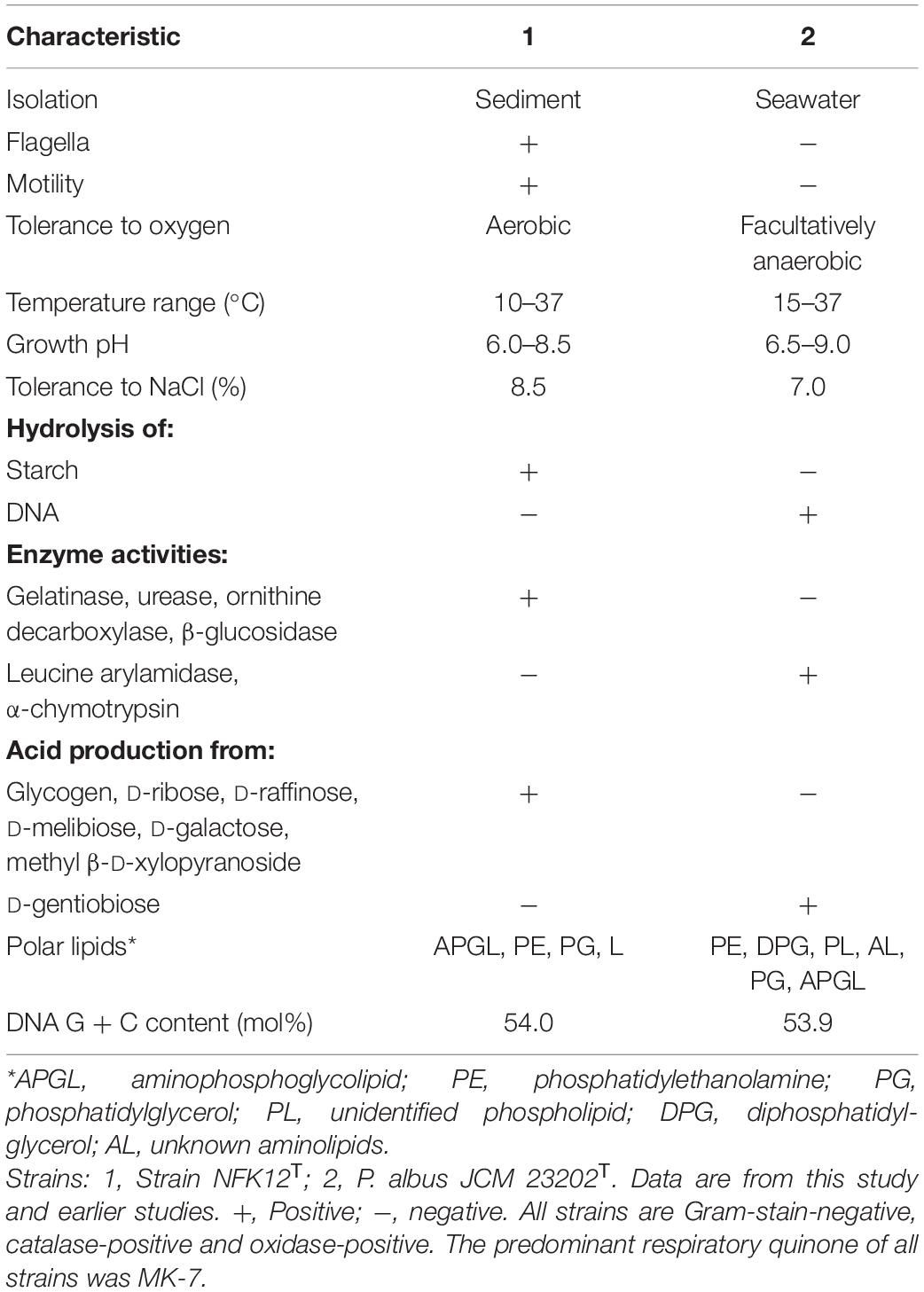
Based on 16S rRNA gene sequences analysis, phenotypic, genomic and chemotaxonomic, we suggest that strain NFK12T could be affiliated to genus Pelagicoccus currently. Consequently, we concluded that strain NFK12T could be placed in a novel species of the genus Pelagicoccus, for which the name P. enzymogenes sp. nov. is proposed. Other phenotypic characteristics from classical experiments are listed in Table 4 and the species description, while the comparative analyses with members of phylogenetically related strain are given in the species description. The detailed differences in the polar lipid profiles between strain NFK12T and the related strain are shown in Supplementary Figure 7. The fatty acid compositions of strain NFK12T and the related strains were similar, but there were differences with strain NFK12T in the proportions of some fatty acids, particularly summed feature 6 (C19:1 ω11c and/or C19:1 ω9c) (Supplementary Table 1).
Description of Pelagicoccus enzymogenes sp. nov.
Pelagicoccus enzymogenes (en.zy.mo’ge.nes. N.L. n. enzyma, enzyme; from Gr. mac. n. zumê, leaven; N.L. suff. -genes, producing; from Gr. v. gennaô, to produce; N.L.mac. adj. enzymogenes, enzyme-producing).
Cells are Gram-stain-negative, aerobic, cocci-shaped. Cells are motile by means of flagella, the mean cell size is 1.0 μm in diameter. Colonies on MA are round, white and glossy with entire margins, about 2.0 mm in diameter after incubation for 3 days at 33°C. Growth occurs at 10–37° (optimum, 33°), at pH 6.0–8.5 (optimum, 7.0–7.5) and in the presence of 1.0–8.5% (w/v) NaCl (optimum, 1.0–2.0%). Growth is not observed under anaerobic conditions on MA with or without 0.1% (w/v) KNO3. Nitrate is not reduced to nitrite. Catalase-positive and oxidase-positive. The strain was positive for hydrolyzes of Tweens 20, 40, and 60, alginate, starch, casein and cellulose but negative for DNase. Positive for ornithine decarboxylase, urease, gelatinase and citrate utilization. The following carbon sources are oxidized: dextrin, D-mannose, α-D-lactose, D-fucose, β-methyl-D-glucoside, L-rhamnose, glucuronamide, acetic acid and L-aspartic acid. Acid is produced from L-arabinose, D-ribose, methyl β-D-xylopyranoside, D-galactose, D-xylose, glycogen, D-raffinose, D-melibiose, D-lactose, D-maltose, D-cellobiose, D-tagatose and 5-ketogluconate. Positive for esterase (C4), gelatinase, tryptophan deaminase, α-galactosidase, β-galactosidase, β-glucuronidase, alkaline phosphatase, N-acetyl-β-glucosaminidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase. The predominant menaquinone is MK-7 and the major cellular polar lipids are aminophosphoglycolipid, phosphatidylethanolamine and one unidentified lipid. Main cellular fatty acids are anteiso-C15: 0, C16:0 and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c).
The type strain is NFK12T (=KCTC 72940T = MCCC 1H00424T), isolated from sediment of Xiaoshi Island, Weihai, China. The genomic DNA G + C content of the type strain is 56.8 mol%. The GenBank accession number for the 16S rRNA gene sequence of P. enzymogenes NFK12T is MW023105 and the draft genome has been deposited in GenBank under the accession number JACYFG000000000.
Disscusion and Conclusion
The ubiquitous and metabolically versatile environmental bacterium Verrucomicrobia plays important roles in natural ecosystems, such as soil, marine, and freshwater ecosystems, including the human gut. Bacteria belonging to the phylum Verrucomicrobia are commonly studied in the soil and gut environments. It has been shown that Verrucomicrobia is nearly ubiquitous in the marine environment (Freitas et al., 2012; Thomas et al., 2019; Sizikov et al., 2020), but little is known about the function and cellular features of this phylum in the marine environment. Besides, detailed studies on cellular and biochemical features of the phylum Verrucomicrobia necessitate axenic cultures. To address this issue, we focused on subdivision 4, which is nearly ubiquitous in the marine environment, but a limited number of species were cultured.
Except for R. amylovorans JCM 31066T, other strains have been isolated from the marine environment, which is consistent with a previous study (Freitas et al., 2012). The representatives of Puniceicoccaceae are Gram-negative, aerobic, or facultatively anaerobic cocci with nitrate reduction abilities. The major respiratory quinone was menaquinone-7 (MK-7). The DNA G + C content ranged from 52.3 to 60.3 mol% as determined during genome sequencing. The sequenced genomes were between 3.6 and 7.5 Mb (median 4.5 Mb) in size, with P. mobilis KCTC 13126T having the largest genome. Flagella were observed in the cells of strain NFK12T via TEM and SEM analyses. The cells of strain NFK12T had randomly distributed finger-like extensions from which fimbriae appendages project, as described in Lentisphaera araneosa and Kiritimatiella glycovorans, which were originally considered as the members of Verrucomicrobia (Cho et al., 2004; Vliet et al., 2019).
The existing studies included relatively few antibiotics and Verrucomicrobia organisms. To study the antibiotic susceptibility of the Verrucomicrobia phylum, we determined the susceptibility of the representatives of Puniceicoccaceae to 20 antibiotic families using the antibiotic disk diffusion method. Knowledge of antibiotic resistance in Puniceicoccaceae may therefore elucidate that in PVC organisms at large. It is generally believed that PVC bacteria are susceptible to antibiotics that target protein synthesis, such as chloramphenicol, tetracycline, and erythromycin, or those that target DNA replication, such as fluoroquinolone (Cayrou et al., 2010; Godinho et al., 2019). However, the results are different from those previously reported, we showed that the members of Puniceicoccaceae could be more easily cultured under the selective pressure of ofloxacin and norfloxacin. Ofloxacin and norfloxacin belong to the third-generation fluoroquinolones commonly used in the clinic, and quinolones target bacterial DNA and interfere with DNA cyclase. In order to produce their cytotoxic effect, quinolones must enter the cell through the bacterial membrane to reach their target, DNA-gyrase or topoisomerase IV, and induce cell death. The mechanisms of resistance to fluoroquinolones include: those mediated by gene mutations codifying for DNA gyrase and topoisomerase IV and leading to QRDR (Quinolone resistance determination zone); those characterized by changes in the permeability of the outer membrane which decrease intracellular penetration of the drug; and those caused by active endogenous carriers responsible for drug efflux (Smith, 1986). One of the bacterial resistance mechanisms to the quinolones is that mutations with chromosomal genes of DNA gyrase or DNA topoisomerase (Kojima, 1997). Thus, the resistance to fluoroquinolones was analyzed using genomic information. The protein that confers resistance to fluoroquinolones was predicted in 28 genomes of the class Opitutae after being annotated (Supplementary Appendix Table C). We suspect that almost all bacteria belonging to the family Puniceicoccaceae are resistant to fluoroquinolones. Then, we tried to isolate of Puniceicoccaceae in the presence of quinolones and successfully cultivated three species of Puniceicoccaceae. This study expands the current understanding of the mechanisms involved in EPS and antibiotic resistance of marine Puniceicoccaceae lineages. Besides, our findings focused on the common features among the representatives of Verrucomicrobia subdivision 4 while at the same time extending the scarce pool of cultivated species in this environmentally important phylum.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XF performed most of the experiments, wrote the main part of the manuscript, and functions as the first author. YG helped with the isolation of the novel strains and with cultivation measurements. YG helped with light microscopy and cell size measurements. M-QY contributed to the literature research and analyzed the sequencing data. XF, Z-JD, and YG designed the study and helped with experimental setups and design. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32070002 and 31770002) and National Science and Technology Fundamental Resources Investigation Program of China (2019FY100700).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work of scanning electron microscope was supported by the Physical-Chemical Materials Analytical & Testing Center of Shandong University at Weihai. Besides, we would like to thank Editage (www.editage.com) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.655735/full#supplementary-material
Abbreviations
MCCC, Marine Culture Collection of China; BD, Becton Dickinson; MEGA, molecular evolutionary genetics analysis; MA, marine agar 2216; MB, marine broth 2216; EDX, Energy Dispersive X-Ray Spectroscopy.
Footnotes
- ^ https://www.ezbiocloud.net
- ^ https://www.ncbi.nlm.nih.gov
- ^ http://support.illumina.com/
- ^ https://github.com/dparks1134/CompareM
- ^ http://jspecies.ribohost.com/jspeciesws/
- ^ http://ggdc.dsmz.de
- ^ http://www.biolog.com/
References
Ai, L., Zhang, H., Guo, B., Chen, W., Wu, Z., and Wu, Y. (2008). Preparation, partial characterization and bioactivity of exopolysaccharides from Lactobacillus casei LC2W. Carb. Poly. 74, 353–357. doi: 10.1016/j.carbpol.2008.03.004
Athalye, M., Noble, W. C., and Minnikin, D. E. (2010). Analysis of cellular fatty acids by gas chromatography as a tool in the identification of medically important coryneform bacteria. J. Appl. Bacteriol. 58, 507–512. doi: 10.1111/j.1365-2672.1985.tb01491.x
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 8:75. doi: 10.1186/1471-2164-9-75
Bergmann, G. T., Bates, S. T., Eilers, K. G., Lauber, C. L., Caporaso, J. G., Walters, W. A., et al. (2011). The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil. Biol. Biochem. 43, 1450–1455. doi: 10.1016/j.soilbio.2011.03.012
Camp, H. J. M. O. D., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cayrou, C., Raoult, D., and Drancourt, M. (2010). Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J. Antimicrob. Chemother. 65, 2119–2122. doi: 10.1093/jac/dkq290
Chiang, E., Schmidt, M. L., Berry, M. A., Biddanda, B. A., Burtner, A., Johengen, T. H., et al. (2018). Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS One 13:e0195112. doi: 10.1371/journal.pone.0195112
Cho, J. C., Vergin, K. L., Morris, R. M., and Giovannoni, S. J. (2004). Lentisphaera araneosa gen. nov., sp nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 6, 611–621. doi: 10.1111/j.1462-2920.2004.00614.x
Choo, Y. J., Lee, K., Song, J., and Cho, J.-C. (2007). Puniceicoccus vermicola gen. nov., sp. nov., a novel marine bacterium, and description of Puniceicoccaceae fam. nov., Puniceicoccales ord. nov., Opitutaceae fam. nov., Opitutales ord. nov. and Opitutae classis nov. in the phylum “Verrucomicrobia”. Int. J. Syst. Evol. Microbiol. 57, 532–537. doi: 10.1099/ijs.0.64616-0
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cowan, S. T., Steel, K. J., and Mccoy, E. (1966). Manual for the identification of medical bacteria. Proc. R. Soc. Med. 59:468.
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17, 368–376. doi: 10.1007/bf01734359
Feng, X., Zou, Q. H., Zhang, X. Y., Ye, M. Q., and Du, Z. J. (2020). Oceanipulchritudo coccoides gen. nov., sp. nov., isolated from marine sediment within the family Puniceicoccaceae. Int. J. Syst. Evol. Microbiol. 70, 5654–5664. doi: 10.1099/ijsem.0.004458
Fitch, W. M. (1971). Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Zool. 20, 406–416. doi: 10.2307/2412116
Freeman, D. J., Falkiner, F. R., and Keane, C. T. (1989). New method for detecting slime producing by coagulase negative Staphylococci. J. Clin. Pathol. 42, 872–874. doi: 10.1136/jcp.42.8.872
Freitas, S., Hatosy, S., Fuhrman, J. A., Huse, S. M., Mark Welch, D. B., Sogin, M. L., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6, 1499–1505. doi: 10.1038/ismej.2012.3
Godinho, O., Calisto, R., Øvreås, L., Quinteira, S., and Lage, O. M. (2019). Antibiotic susceptibility of marine Planctomycetes. Antonie Van Leeuwenhoek 112, 1273–1280. doi: 10.1007/s10482-019-01259-7
Gómez-Gallego, C., Pohl, S., Salminen, S., Vos, W. M. D., and Kneifel, W. (2016). Akkermansia muciniphila: A novel functional microbe with probiotic properties. Benef Microb. 7, 1–14. doi: 10.1007/978-1-4614-6418-1_793-1
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57(Pt 1), 81–91. doi: 10.1099/ijs.0.64483-0
Hanna, O., Donald, L. P., and Sheppard, D. C. (2018). Deacetylated microbial biofilm exopolysaccharides: It pays to be positive. PLoS Pathog. 14:e1007411. doi: 10.1371/journal.ppat.1007411
Jana, T., Lam-Tung, N., Arndt, V. H., and Quang, M. B. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 8, W232–W235.
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA Genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/aem.72.3.1719-1728.2006
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kojima, M. (1997). Mechanisms for the development of quinolone resistance. Nihon Rinsho Japanese J. Clin. Med. 55:1252.
Kroppenstedt, R. M. (1982). Separation of Bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J. Liq. Chromatogr. 5, 2359–2367. doi: 10.1080/01483918208067640
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 1870–1874. doi: 10.1093/molbev/msw054
Lage, O. M., Bondoso, J., and Viana, F. (2012). Isolation and characterization of Planctomycetes from the sediments of a fish farm wastewater treatment tank. Arch. Microbiol. 194, 879–885. doi: 10.1007/s00203-012-0821-2
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic acid techniques in bacterial systematics, eds E. Stackebrandt and M. Goodfellow, (New York, NY: Wiley), 115–175.
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2, W256–W259.
Lin, S. Y., Hameed, A., Liu, Y.-C., Hsu, Y.-H., Hung, M.-H., Lai, W. A., et al. (2017). Ruficoccus amylovorans gen. nov., sp. nov., an amylolytic and nitrate-reducing diazotroph of the family Puniceicoccaceae. Int. J. Syst. Evol. Microbiol. 67, 956–962. doi: 10.1099/ijsem.0.001723
Liu, S. B., Chen, X. L., He, H. L., Zhang, X. Y., Xie, B. B., Yu, Y., et al. (2013). Structure and ecological roles of a novel exopolysaccharide from the arctic sea ice bacterium Pseudoalteromonas sp. Strain SM20310. Appl. Environ. Microbiol. 79, 224–230. doi: 10.1128/aem.01801-12
Li, R., Yu, C., Li, Y., and Lam, T.-W. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Manuel, M. G., David, M. B., Brandon, K. S., Carol, A., Patrick, S. G. C., Krista, G. R., et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 7:e35314. doi: 10.1371/journal.pone.0035314
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., and Ohtsubo, Y. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16s rrna genes with a low possibility to amplify eukaryotic rrna genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Muxika, A., Etxabide, A., Uranga, J., Guerrero, P., and Caba, K. (2017). Chitosan as a bioactive polymer: Processing, properties and applications. Int. J. Biol. Macromol. 105, 1358–1368. doi: 10.1016/j.ijbiomac.2017.07.087
Nichols, C. A. M., Guezennec, J., and Bowman, J. P. (2005). Bacterial exopolysaccharides from extreme marine environments with special consideration of the Southern Ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar. Biotechnol. 7, 253–271. doi: 10.1007/s10126-004-5118-2
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Gker, M. (2020). List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Patrick, R., Ines, G., Christian, B., Olga, J., Sandra, W., Richard, R., et al. (2017). Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front. Microbiol. 13:202. doi: 10.3389/fmicb.2017.00202
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/jb.01688-14
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe 9, 111–118. doi: 10.1128/microbe.9.111.1
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406.
Santarella-Mellwig, R., Franke, J., Jaedicke, A., Gorjanacz, M., Bauer, U., Budd, A., et al. (2010). The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8:e1000281. doi: 10.1371/journal.pbio.1000281
Schlesner, H. (1994). The Development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17, 135–145. doi: 10.1016/s0723-2020(11)80042-1
Sichert, A., Corzett, C. H., Schechter, M. S., Unfried, F., and Hehemann, J. H. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5, 1026–1039. doi: 10.1038/s41564-020-0720-2
Smith, J. T. (1986). The mode of action of 4-quinolones and possible mechanisms of resistance. J. Antimicrob. Chemother. 1986, 21–29. doi: 10.1093/jac/18.supplement_d.21
Sizikov, S., Burgsdorf, I., Handley, K. M., Lahyani, M., Haber, M., and Steindler, L. (2020). Characterization of sponge-associated Verrucomicrobia: microcompartment-based sugar utilization and enhanced toxin-antitoxin modules as features of host-associated Opitutales. Environ. Microbiol. 22, 4669–4688. doi: 10.1111/1462-2920.15210
Tindall, B. J., Sikorski, J., Smibert, R. A., and Krieg, N. R. (2007). “Phenotypic characterization and the principles of comparative systematic,” in Methods for General and Molecular Microbiology, 3rd Edn, eds C. Reddy, T. Beveridge, J. Breznak, G. Marzluf, T. Schmidt, and L. Snyder, (Washington, DC: American Society of Microbiology), 330–393. doi: 10.1128/9781555817497.ch15
Thomas, F. A., Sinha, R. K., and Krishnan, K. P. (2019). Bacterial community structure of a glacio-marine system in the Arctic (Ny-lesund. Svalbard). Sci. Total Environ. 718:135264. doi: 10.1016/j.scitotenv.2019.135264
Venkatesan, N., Perumal, G., and Doble, M. (2015). Bacterial resistance in biofilm-associated bacteria. Future Microbiol. 2019, 1743–1750. doi: 10.2217/fmb.15.69
Vliet, D. M. V., Ayudthaya, S. P. N., Diop, S., Villanueva, L., Stams, A. J. M., and Sánchez-Andrea, I. (2019). Anaerobic degradation of sulfated polysaccharides by two novel Kiritimatiellales strains isolated from Black Sea sediment. Front. Microbiol. 18:253. doi: 10.3389/fmicb.2019.00253
Vliet, D. M. V., Lin, Y., Bale, N. J., Koenen, M., Villanueva, L., Stams, A. J. M., et al. (2020). Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., two marine anaerobes of the Pontiellaceae fam. nov. producing sulfated glycosaminoglycan-like exopolymers. Microorganisms 8:920. doi: 10.3390/microorganisms8060920
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249. doi: 10.1016/j.copbio.2006.05.005
Wiegand, S., Jogler, M., Boedeker, C., Pinto, D., Vollmers, J., Rivas-Marín, E., et al. (2020). Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 5, 126–140.
William, H. (2017). Marine Polysaccharide Sulfatases. Front. Mar. Sci. 4:6. doi: 10.3389/fmars.2017.00006
Yoon, J., Yasumoto-Hirose, M., Matsuo, Y., Nozawa, M., Matsuda, S., Kasai, H., et al. (2007). Pelagicoccus mobilis gen. nov., sp. nov., Pelagicoccus albus sp. nov. and Pelagicoccus litoralis sp. nov., three novel members of subdivision 4 within the phylum “Verrucomicrobia”, isolated from seawater by in situ cultivation. Int. J. Syst. Evol. Microbiol. 57, 1377–1385. doi: 10.1099/ijs.0.64970-0
Yoon, J., Matsuo, Y., Adachi, K., Nozawa, M., Matsuda, S., Kasai, H., et al. (2008a). Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum “Verrucomicrobia”, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 58, 998–1007. doi: 10.1099/ijs.0.65520-0
Yoon, J., Matsuo, Y., Katsuta, A., Jang, J. H., Matsuda, S., Adachi, K., et al. (2008b). Haloferula rosea gen. nov., sp. nov., Haloferula harenae sp. nov., Haloferula phyci sp. nov., Haloferula helveola sp. nov. and Haloferula sargassicola sp. nov., five marine representatives of the family Verrucomicrobiaceae within the phylum “Verrucomicrobia”. Int. J. Syst. Evol. Microbiol. 58, 2491–2500. doi: 10.1099/ijs.0.2008/000711-0
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H., and Yokota, A. (2007a). Cerasicoccus arenae gen. nov., sp. nov., a carotenoid-producing marine representative of the family Puniceicoccaceae within the phylum ‘Verrucomicrobia’, isolated from marine sand. Int. J. Syst. Evol. Microbiol. 57, 2067–2072. doi: 10.1099/ijs.0.65102-0
Yoon, J., Oku, N., Matsuda, S., Kasai, H., and Yokota, A. (2007b). Pelagicoccus croceus sp. nov., a novel marine member of the family Puniceicoccaceae within the phylum “Verrucomicrobia” isolated from seagrass. Int. J. Syst. Evol. Microbiol. 57, 2874–2880. doi: 10.1099/ijs.0.65286-0
Yoon, J., Yasumoto-Hirose, M., Katsuta, A., Sekiguchi, H., Matsuda, S., Kasai, H., et al. (2007c). Coraliomargarita akajimensis gen. nov., sp. nov., a novel member of the phylum “Verrucomicrobia” isolated from seawater in Japan. Int. J. Syst. Evol. Microbiol. 57, 959–963. doi: 10.1099/ijs.0.64755-0
Keywords: genome, antibiotic, exopolysaccharide, Verrucomicrobia, Puniceicoccaceae
Citation: Feng X, Gong Y, Ye M-Q and Du Z-J (2021) Antibiotic Modulation of Capsular Exopolysaccharide in Pelagicoccus enzymogenes sp. nov. Isolated From Marine Sediment. Front. Mar. Sci. 8:655735. doi: 10.3389/fmars.2021.655735
Received: 19 January 2021; Accepted: 08 March 2021;
Published: 23 March 2021.
Edited by:
Daniel P. R. Herlemann, Estonian University of Life Sciences, EstoniaReviewed by:
Akira Yokota, Tohoku University, JapanDhiraj Kumar Chaudhary, Korea University, South Korea
Copyright © 2021 Feng, Gong, Ye and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong-Jun Du, ZHV6b25nanVuQHNkdS5lZHUuY24=
 Xi Feng
Xi Feng Ya Gong1
Ya Gong1 Zong-Jun Du
Zong-Jun Du