
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 31 March 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.655714
This article is part of the Research Topic Molecular Physiology in Molluscs, Volume II View all 26 articles
Giant clams display light-enhanced inorganic phosphate (Pi) absorption, but how the absorbed Pi is translocated to the symbiotic dinoflagellates living extracellularly in a tubular system is unknown. They can accumulate Pi in the kidney, but the transport mechanism remains enigmatic. This study aimed to elucidate the possible functions of sodium-dependent phosphate transporter protein 1-homolog (PiT1-like), which co-transport Na+ and H2PO4–, in these two processes. The complete cDNA coding sequence of PiT1-like, which comprised 1,665 bp and encoded 553 amino acids (59.3 kDa), was obtained from the fluted giant clam, Tridacna squamosa. In the kidney, PiT1-like was localized in the plasma membrane of nephrocytes, and could therefore absorb Pi from the hemolymph. As the gene and protein expression levels of PiT1-like were up-regulated in the kidney during illumination, PiT1-like could probably increase the removal of Pi from the hemolymph during light-enhanced Pi uptake. In the ctenidial epithelial cells, PiT1-like had a basolateral localization and its expression was also light-dependent. It might function in Pi sensing and the absorption of Pi from the hemolymph when Pi was limiting. In the outer mantle, PiT1-like was localized in the basolateral membrane of epithelial cells forming the tertiary tubules. It displayed light-enhanced expression levels, indicating that the host could increase the translocation of Pi from the hemolymph into the tubular epithelial cells and subsequently into the luminal fluid to support increased Pi metabolism in the photosynthesizing dinoflagellates. Taken together, the accumulation of Pi in the kidney of giant clams might be unrelated to limiting the availability of Pi to the symbionts to regulate their population.
Tropical waters are low in nutrients, but coral reefs, which are hubs of a wide variety of marine organisms including scleractinian corals and giant clams, can be found in shallow tropical waters with adequate irradiance. Specifically, giant clams (genus: Tridacna or Hippopus) are inhabitants of the Indo-Pacific reef ecosystems. They can grow rapidly despite the shortage of nutrients and are the largest of all bivalves, because they live in symbiosis with mainly three genera (Symbiodinium, Cladocopium, and Durusdinium; LaJeunesse et al., 2018) of phototrophic dinoflagellates belonging to the family Symbiodiniaceae (Hernawan, 2008; Weber, 2009; DeBoer et al., 2012; Ikeda et al., 2017; Lim et al., 2019). In giant clams, symbiotic dinoflagellates (zooxanthellae) reside extracellularly in a branched tubular system that originates from the digestive tract of the host (Norton and Jones, 1992). They are located mainly inside numerous tertiary tubules in the colorful and extensible outer mantle that contains pigments and iridocytes (Norton and Jones, 1992). During insolation, the outer mantle can be fully extended, and the host iridocytes can deflect light of appropriate wavelengths to the dinoflagellates to promote photosynthesis. Photosynthesizing dinoflagellates can donate as much as 95% of photosynthates to the host clam to fulfill its energy and nutrition requirements (Klumpp and Griffiths, 1994). In turn, the host must provide the symbionts with nutrients such as inorganic carbon (Ci), nitrogen (N), and phosphorous (P), because they have no access to the ambient seawater (Furla et al., 2005). As a consequence, the clam host displays many light-dependent physiological phenomena (Ip et al., 2006, 2015; see Ip and Chew, 2021 for a review), including light-enhanced uptake of Ci (Hiong et al., 2017a; Ip et al., 2018; Koh et al., 2018; Chew et al., 2019), N (Hiong et al., 2017b; Chan et al., 2018, 2019; Ip et al., 2020), and P (Chan et al., 2020). P is one of the essential elements for living systems, and inorganic phosphate (Pi) is needed for syntheses of ATP, phospholipids and genetic materials (Knowles, 1980), as well as for intracellular signaling (Olsen et al., 2006). Dissolved Pi comprises dihydrogen phosphate (H2PO4–), hydrogen phosphate (HPO42–), and phosphate (PO43–), with HPO42– and H2PO4– being the major components in water at pH 7.
Despite being deprived of access to planktonic/particulate matter, the fluted giant clam, T. squamosa, can survive and grow in Millipore-filtered seawater for more than 10 months with light as the sole energy source (Fitt and Trench, 1981). Without feeding, the clam host needs to receive adequate supplies of energy and nutrients from its phototrophic symbionts (Klumpp and Griffiths, 1994) in order to grow and conduct light-enhanced calcification. As symbionts are P-deficient, the host must supply them with Pi, which is absorbed from the ambient seawater, through the hemolymph and tubular fluid (Jackson et al., 1989). Although the concentration of Pi in seawater is low (0.08–0.50 μmol l–1; Rivkin and Swift, 1985; Godinot et al., 2009), T. squamosa can absorb exogenous Pi, and the rate of Pi absorption is higher in light than in darkness (Chan et al., 2020; Ip and Chew, 2021). T. squamosa expresses a homolog of sodium-dependent phosphate transport protein 2a (NPT2a-like) in the apical membranes of the epithelia of three organs (Chan et al., 2020). These include the upper epithelium of the colorful outer mantle, the seawater-facing epithelium of the whitish inner mantle, and the epithelium covering the ctenidial filaments. Hence, the apical NPT2a-like in these epithelia can participate in Pi absorption from the ambient seawater. In addition, the protein abundance of NPT2a-like in the outer mantle, but not those in the other two organs, is up-regulated by illumination, denoting the outer mantle as a crucial site of light-enhanced Pi absorption (Chan et al., 2020). It is probable that the host could deliver the Pi absorbed through the outer mantle epithelium directly to symbionts residing therein, and regulate the supply of Pi absorption partially through NPT2a-like. As such, the photosynthesizing symbionts are spared from competing with other host’s organs for the limiting Pi available in the hemolymph (Chan et al., 2020). In particular, the kidneys of giant clams are known to absorb and accumulate Pi, although the transporter involved remains unknown (Belda and Yellowlees, 1995).
It has been reported that the intracellular Pi concentration and the rate of division in cultured dinoflagellates isolated from Tridacna gigas increase after exposure to 10 μmol l–1 Pi. By contrast, the density of dinoflagellates and their N:P ratio remain unchanged in T. gigas exposed to 10 μmol l–1 Pi (Belda and Yellowlees, 1995). Therefore, it has been suggested that the host clam could absorb and store Pi in the kidney, thereby controlling the availability of Pi to the symbionts so as to regulate their population (Belda and Yellowlees, 1995). However, the kidney of T. squamosa has only weak expression of NPT2a-like, leading Chan et al. (2020) to postulate that other types of phosphate transporter could be present in the host’s kidney tissues and the epithelium of the host’s zooxanthellal tubules (Chan et al., 2020).
Phosphate transporters are members of the solute carrier family of proteins (SLC), and they consist of SLC17 (Type I), SLC34 (Type II), and SLC20 (Type III) (Virkki et al., 2007). Members of SLC20 and SLC34 are involved in the secondary active uptake of Pi in cells (Virkki et al., 2007; Forster et al., 2013). NPT2s are members of SLC34 that bind to HPO42– and transport it together with Na+ (Virkki et al., 2007). On the other hand, SLC20 consists of sodium-dependent phosphate transport proteins (PiTs; Forster et al., 2013) that co-transport Na+ and H2PO4– (Virkki et al., 2007). PiTs are regarded as house-keeping transporters involved in phosphate metabolism in mammals, and it consists of sodium-dependent phosphate transporter protein 1 (PiT1) (SLC20A1) and PiT2 (SLC20A2) (Forster et al., 2013). In mammals, PiT1 is vital in embryonic liver development and in bone metabolism, while PiT2 has a more specific role in phosphate homeostasis in the brain (Forster et al., 2013). Furthermore, PiT1 and PiT2 can function as Pi sensing and signaling mechanisms by binding with external Pi but without translocation (Bon et al., 2017).
The populations of giant clams have been decreasing due to combinations of overharvesting for food and aquarium trade, pollution, and anthropogenic activities (Watson, 2015). Particularly, a decrease in the internal population of symbiotic dinoflagellates in giant clams can occur due to increased ocean temperature resulting from global warming (Addessi, 2001). As the symbiont population could be controlled by the Pi concentration in the tubular fluid, it is essential to understand the molecular mechanisms of Pi transport in the host’s kidney and those tissues that are involved in the supply of Pi to the symbionts. Therefore, this study was undertaken to clone and sequence a homolog of PiT1 (PiT1-like) of host origin from the kidney of T. squamosa. Efforts were made to examine the gene expression of PiT1-like in various organs of T. squamosa to test the hypothesis that it was expressed predominantly in the kidney. Subsequently, we discovered that PiT1-like was also expressed strongly in the ctenidium and moderately in the outer mantle. Hence, in order to elucidate the cellular and subcellular localization of PiT1-like in the kidney, ctenidium and outer mantle, an anti-PiT1-like polyclonal antibody was custom-made to perform immunofluorescence microscopy. Furthermore, quantitative real-time PCR (qPCR) and western blotting were performed to determine the effects of light exposure on the transcript levels and protein abundances of PiT1-like/PiT1-like in these three organs of T. squamosa.
Individuals of T. squamosa specimens (520 ± 180 g with shells; n = 19) were purchased from Xanh Tuoi Tropical Fish Ltd., in Vietnam. They were acclimated in three glass tanks (L90 cm × W62 cm × H60 cm) under a 12 h light:12 h dark regime for one month before experimentation. Each tank contained 350 L of recirculating seawater and 6 or 7 clams. The water conditions were as follows: salinity, 30–32; temperature, ∼26°C; pH, 8.1–8.3; hardness, 143–179 ppm; calcium, 380–420 ppm; phosphate, <0.25 ppm; nitrate, 0 ppm; nitrite, 0 ppm; total ammonia, <0.25 ppm. The underwater light intensity (PPFD) at the level of the clam was ∼120 μmol m–2 s–1.
Four individuals (controls; n = 4) were sampled directly and randomly from all the three glass tanks, and killed for tissue sampling after 12 h of darkness in the normal 12 h light:12 h dark regime. Other individuals were then exposed to light for 3, 6, or 12 h. At each specific time points, four individuals (n = 4) were randomly selected and killed for tissue sampling. Individual clams were anesthetized with 0.2% phenoxyethanol before being forced open to cut the adductor muscles. Collected tissue samples were stored at −80°C until analyses.
After being anesthetized in 0.2% phenoxyethanol, three individuals exposed to darkness for 12 h (n = 3) were killed for the sampling of tissues for immunofluorescence microscopy. Samples were immersion-fixed in 3% paraformaldehyde in seawater at 4°C overnight and processed according to the method of Hiong et al. (2017a).
The total RNA from tissue samples was isolated using TRI ReagentTM (Sigma-Aldrich Co. St. Louis, MO, United States). Samples were homogenized with silica beads (425–600 μm; Sigma-Aldrich) in TRI Reagent® using a Mini-GTM tissue homogenizer (SPEX® Sample Prep, Metuchen, NJ, United States). Remnant of DNA was removed by PureLinkTM RNA Mini Kit (Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, United States), and the purified RNA was quantified by a Shimadzu BioSpec-nano spectrophotometer (Shimadzu Corporation, Tokyo, Japan). After verification of the integrity of RNA by electrophoresis, cDNA was synthesized from the purified total RNA using the RevertAidTM first-strand cDNA synthesis kit (Thermo Fisher Scientific Inc.).
A partial PiT1-like sequence was isolated using a pair of PCR primers (forward: 5′-TGATCGTTGGTTCCTGGTT-3′; reverse: 5′-TCGCCAATCGTATTGATGAC-3′) designed from the conserved regions of the sodium-dependent phosphate transporter 1 of Octopus vulgaris (XM_029794590.1), Crassostrea gigas (XM_020063507.2), Crassostrea virginica (XM_022469514.1), Mizuhopecten yessoensis (XM_021500463.1), and Priapulus caudatus (XM_014826122.1). The PCR was conducted using a 9902 Veriti 96-well thermal cycler (Applied Biosystems, Carlsbad, CA, United States) and DreamTaqTM polymerase (Thermo Fisher Scientific Inc.). The thermal cycling conditions included an initial denaturation at 95°C held for 3 min, followed by 40 cycles of denaturation, annealing and extension at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, respectively, and a final extension for 10 min at 72°C. Agarose gel electrophoresis was used to separate the PCR products, and the Wizard® SV Gel and PCR clean-up system (Promega Corporation, Madison, WI, United States) was used to extract the targeted bands.
Following the method of Chan et al. (2020), the pGEM-T Easy vector system (Promega Corporation) was used to clone and transform the extracted PCR product into JM109 Escherichia coli competent cells. Multiple clones of PiT1-like were sequenced bi-directionally. BlastN1 was used to confirm the identity of the sequences obtained. Analysis of multiple sequences revealed the absence of isoforms. The full coding sequence of PiT1-like was obtained by conducting 5′ and 3′ RACE-PCR (SMARTerTM RACE cDNA amplification kit; Clontech Laboratories, Mountain View, CA, United States) with specific RACE primers (forward: 5′-TTCCTGCAAATCCTTACTGCTGTGTT-3′, reverse: 5′-CAA TTCGGATCCTTTGTAGAAGACAG-3′).
BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific Inc.) was used to prepare samples for sequencing, and sequencing was performed using a 3130XL Genetic Analyzer (Thermo Fisher Scientific Inc.). Sequences obtained were assembled and analyzed using BioEdit version 7.2.5. The GenBank Accession number of the full coding sequence of PiT1-like obtained from T. squamosa is MN256447.
The nucleotide sequence was translated into PiT1-like amino acid sequence using the ExPASy Proteomic server2. The deduced amino acid sequence was aligned with selected PiT1-like sequences from various species of mollusks and vertebrates to confirm its identity. The TOPCONS server was used to predict the transmembrane regions (TMs).
The gene expression of PiT1-like in the outer mantle, inner mantle, ctenidium, kidney, heart, hepatopancreas, adductor muscle, byssal retractor muscle, and foot muscle of T. squamosa was examined quantitatively by qPCR with gene-specific qPCR primers (forward: 5′-CAATCCTGGTTCGTCTGCTG-3′; reverse: 5′-TCTCAACAAGAACTCCCTCCTC-3′). qPCR was carried out with a StepOnePlusTM Real-Time PCR System (Thermo Fisher Scientific) and each reaction contained various quantities of cDNA of sample, 0.3 μmol l–1 each of specific forward and reverse primers and 5 μl of qPCRBIO SyGreen Mix (PCR Biosystems Ltd., London, United Kingdom). The cycling conditions consisted of an initial 20 s denaturation and enzyme activation at 95°C, 40 cycles of 95°C for 3 s and 60°C for 30 s. Absolute quantification of transcript level of PiT1-like from the kidney, ctenidum, and outer mantle were performed in triplicates. The specific qPCR primers had a amplification efficiency of 93%. The transcript level in the sample was estimated using a standard curve constructed according to the method of Hiong et al. (2017a).
For immunofluorescence microscopy and western blotting, a commercial firm (GenScript, Piscataway, NJ, United States) was engaged to produce a custom-made rabbit polyclonal anti-PiT1-like antibody against the epitope TPSSGFSIEVGSAA (residue 468–481) of PiT1-like of T. squamosa. For western blotting, an anti-α-tubulin antibody, 12G10 (Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City, IA, United States), was used to detect the reference protein, α-tubulin.
Proteins were extracted from samples of kidney, ctenidium, and outer mantle following the method of Hiong et al. (2017a). SDS-PAGE (10%) electrophoresis was applied to separate proteins of kidney (20 μg), ctenidium (50 μg) or outer mantle (100 μg) samples. Separated proteins were blotted onto a nitrocellulose membrane, which was then incubated for 1 h at 25°C with anti-PiT1-like (2 μg ml–1) or anti-α-tubulin (12G10, 0.05 μg ml–1) antibody. The membrane was processed using the Pierce Fast Western Blot kit, SuperSignal® West Pico Substrate (Thermo Fisher Scientific Inc.). Visualization of protein bands was achieved using a ChemiDocTM MP Imaging System (Bio-Rad Laboratories Inc., Hercules, CA, Untied States). Optical density of the protein bands were determined using ImageJ (version 1.50i, NIH) and a 21-step Tiffen (The Tiffen Company, Rochester, NY, Untied States) transmission photographic step tablet (EK1523406T). Protein abundance of PiT1-like was expressed as the optical density of the PiT1-like band normalized with that of the α-tubulin band. As the protein expression level of PiT1-like was the highest in kidney, a peptide competition assay (PCA) was performed to validate the specificity of the custom-made anti PiT1-like antibody on kidney exposed to light for 12 h (Supplementary Figure 1). The anti-PiT1-like antibody was incubated with neutralizing peptide (Genscript) in a ratio of 1:5 for 1 h prior to western blotting.
Samples of the kidney, ctenidium, and outer mantle were excised and immersion-fixed overnight in 3% paraformaldehyde in seawater at 4°C, and processed according to the method of Hiong et al. (2017b). The paraffin-embedded samples were sectioned (3 μm) using a Leitz 1512 rotary microtome (Leica Biosystems, Germany) and collected on slides. Antigen retrieval was performed by treating deparaffinized sections with heated solution of 0.05% citraconic anhydride (Nacalai Tesque, Japan), pH 7.4, for 5 min with a further 10 min incubation in 1% sodium dodecyl sulfate (SDS) solution at room temperature. The section was blocked in 1% BSA in TPBS (0.05% Tween-20 in phosphate-buffered saline: 10 mmol l–1 Na2HPO4, 1.8 mmol l–1 KH2PO4, 137 mmol l–1 NaCl, 2.7 mmol l–1 KCl, pH 7.4) for 20 min. The blocked section was incubated with the anti-PiT1-like antibody, which had been diluted to 2.5 μg ml–1 in Signal Enhancer HIKARI Solution A (Nacalai Tesque), for 1 h in a humid chamber. It was then incubated for another 1 h at 37°C with the fluorochrome-coupled goat anti-rabbit gamma globulin (Alexa Fluor 488; Life Technologies Corporation) diluted to 2.5 μg ml–1 in Signal Enhancer HIKARI Solution B (Nacalai Tesque). The slide was washed again with TPBS, counter-stained with fluorescing DNA stain, 4′ 6-diamidino-2-phenylindole (DAPI) and mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific Inc.).
Digital photos were taken using an Olympus DP73 camera (Olympus Corporation, Tokyo, Japan) fitted to an Olympus BX60 epifluorescence microscope equipped with a differential interference contrast (DIC) slider (Olympus U-DICT), as well as corresponding filter sets (Olympus U-MWU, U-MWIG, and U-MNIBA) and various objectives. DIC images were taken to visualize tissue structure. For the ctenidium and the outer mantle, nuclei were stained with DAPI (in blue) and visualized using the U-MWU filter (emission at 420 nm). Autofluorescence (in red) of the plastid of symbiotic dinoflagellates in the outer mantle was captured using the U-MWIG band pass filter (emission at 580 nm). PiT1-like immunostaining (in green) was visualized using the U-MNIBA band pass filter (emission at 515–550 nm). All images were processed using the cellSens Standard v1.15 software (Olympus Corporation) and prepared using the Adobe Photoshop CS6 (Adobe Systems, CA, United States). The overlaid of images was performed using Adobe Photoshop CC.
Results were presented as mean + SEM. Statistical analysis was performed using SPSS Statistics version 19 software (IBM Corporation, Armonk, NY, United States). Levene’s test was used to examine the homogeneity of variance among the means. Depending on the homogeneity of variances, one-way analysis of variance (ANOVA), followed by Dunnett T3 post hoc test or Tukey post hoc test, was used to evaluate differences among means. Differences among means with a p-value less than 0.05 were regarded as statistically significant.
The complete cDNA coding sequence of PiT1-like obtained from the kidney of T. squamosa (GenBank accession number MN256447) comprised 1,665 bp, and the deduced amino acid sequence had 553 amino acids with an estimated molecular mass of 59.3 kDa.
An analysis of sequence similarities among PiT1-like of T. squamosa and PiT1 proteins of various species obtained from GenBank (Table 1) indicated that it had the highest sequence similarity with those of mollusks (50.8–54.4%), followed by those of cnidarians, brachiopods and echinoderms (38.4–48.6%), and it had the lowest sequence similarity with those of plants and chlorophytes (8.8–14.3%). The host origin of PiT1-like of T. squamosa was validated by phenogramic analysis (Figure 1). The low similarity of PiT1-like of T. squamosa with various NPT2s confirmed that it was indeed a PiT1 transport protein (Table 1). In addition, the N and C terminals of PiT1-like of T. squamosa contained the ProDom domain (Bottger and Pedersen, 2011) and the 12 amino acid-long signature sequence (Bottger and Pedersen, 2005) characteristic of the PiT family. Typical of PiT proteins, there were 11 predicted TMs in PiT1-like of T. squamosa. An alignment with other PiT1s (Homo sapiens, Xenopus laevis, and Exaiptasia diaphana) revealed the conservation of several important residues that are critical for Pi transport (D39, E66, H403, D407, E476, and S494, numbered according to PiT1-like of T. squamosa in Figure 2). Other conserved residues included those that constituted part of the substrate binding site (V77, I86, and V88) and those responsible for substrate selectivity (S78 and R82) of Pi binding (S125).
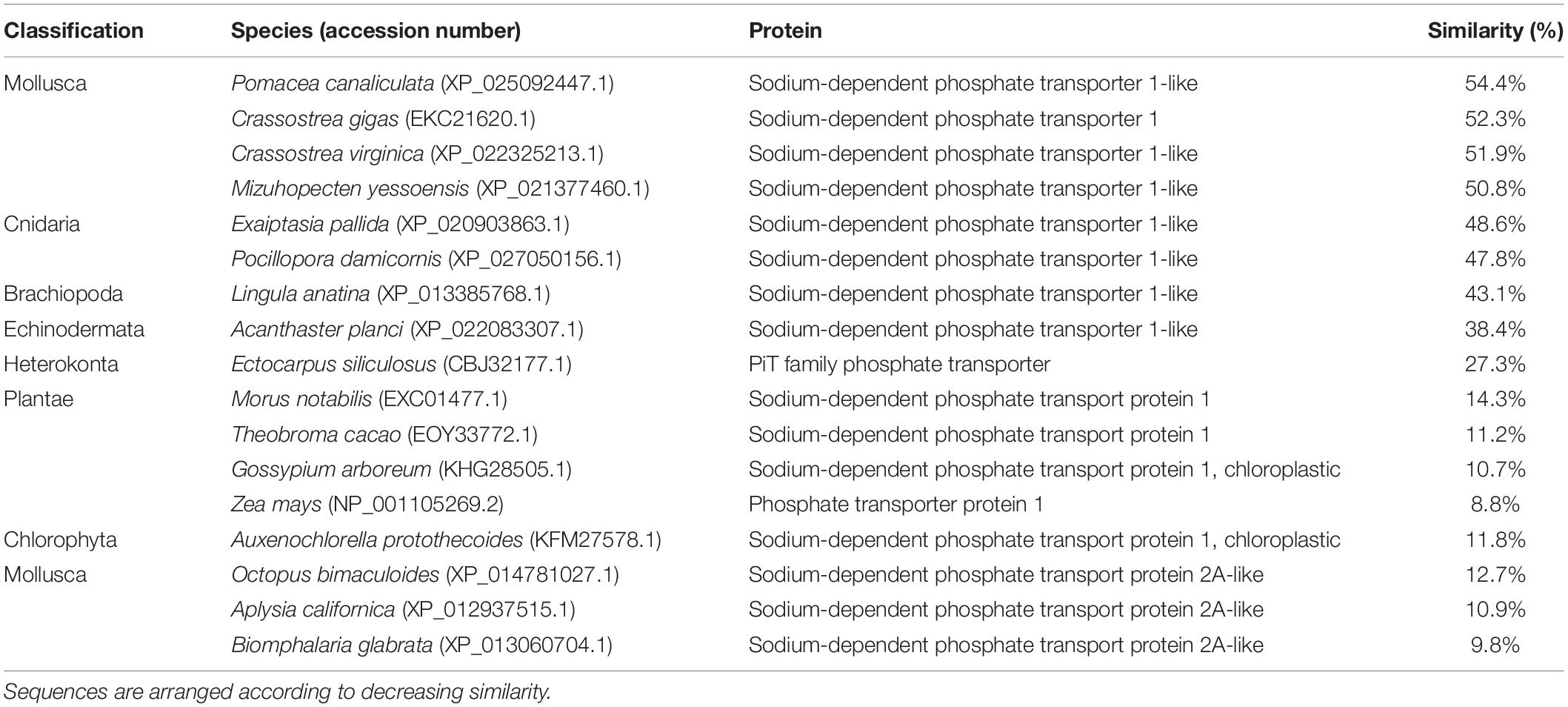
Table 1. Percentage similarity of the deduced amino acid sequence of sodium-dependent phosphate transporter protein 1 (PiT1-like) obtained from T. squamosa and sodium-dependent transport proteins from other species obtained from GenBank.
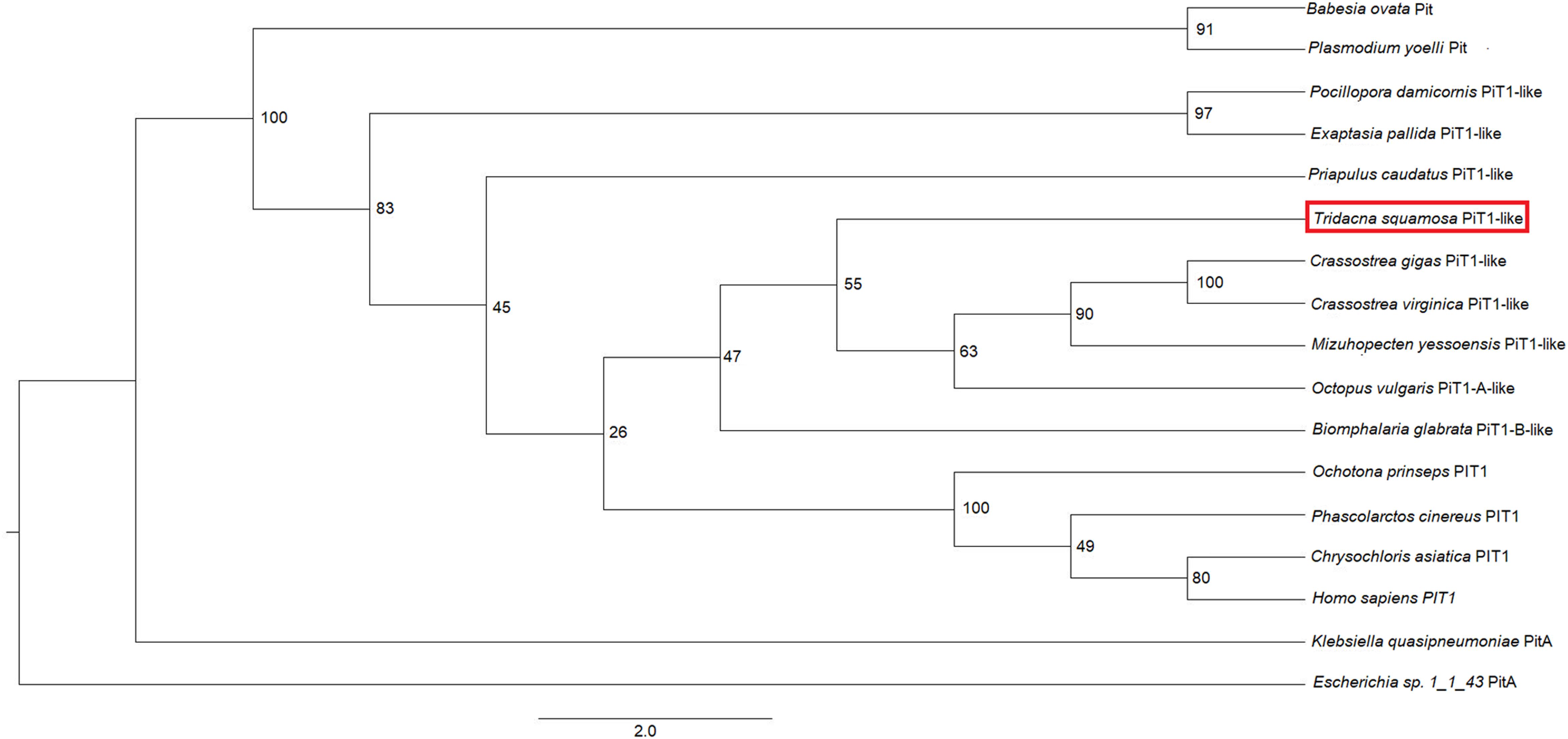
Figure 1. Phenogramic analysis of the homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) from Tridacna squamosa. Numbers shown at each branch point represent bootstrap values from 1,500 replicates. PitA from Escherichia sp. 1_1_43 PitA is used as the outgroup.

Figure 2. A multiple sequence alignment of the deduced amino acid sequence of sodium-dependent phosphate transporter protein 1-homolog (PiT1-like) obtained from Tridacna squamosa, Homo sapiens PiT1 (NP_005406.3), Xenopus laevis PiT1 (NP_001087494.1), Exaiptasia diaphana PiT1 (XP_020903863.1) and the PiT2 paralog from Homo sapiens (Q08357.1). The 11 transmembrane regions (TMs) predicted using the TOPCONS server have been delineated in black with the region number above. The two ProDom (PD001131) domains are underlined in red. The 12 amino acid-long signature sequence of the PiT family is marked with a black box. The red boxes denote the conserved P-X-S motif at the N and C terminal (where X is a hydrophobic amino acid). Residues identified as critical for Pi transport in human PiT1 protein are indicated with asterisks while residues that are important for Pi transport in human PiT2 paralog are marked with hash. Residues marked by open triangles constitute part of the substrate binding site(s) whereas the residue marked by a closed triangle binds Pi. Residues indicated with closed circles play a role in substrate selectivity and coordination.
In T. squamosa, PiT1-like was expressed strongly in the kidney and ctenidium, and moderately in the outer mantle, inner mantle, heart, and adductor muscle. It was only weakly expressed in the hepatopancreas, byssal retractor muscle, and foot muscle (Figure 3). Hence, efforts were focused subsequently on elucidating the possible functions of PiT1-like in the kidney, ctenidium, and outer mantle of T. squamosa.
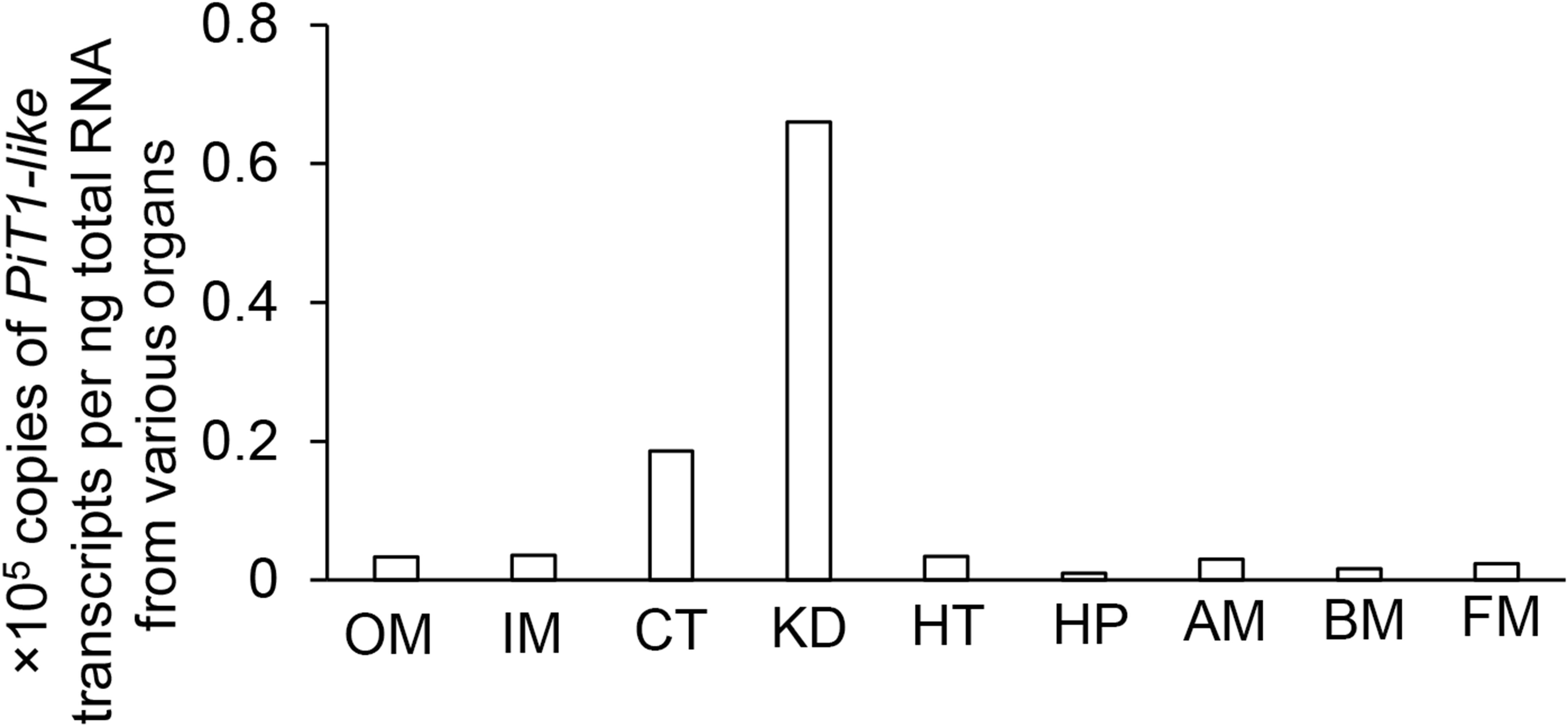
Figure 3. Gene expression of a homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) in the outer mantle (OM), inner mantle (IM), ctenidium (CT), kidney (KD), heart (HT), hepatopancreas (HP), adductor muscle (AM), byssal retractor muscle (BM), and foot muscle (FM) of Tridacna squamosa exposed to light for 12 h.
The kidneys of giant clams are known to consist of nephrocytes, which are large globular cells with indistinct boundaries and little visible cytoplasmic contents (Norton and Jones, 1992). In the kidney of T. squamosa, PiT1-like was localized in the membrane surrounding a type of nephrocytes (Figure 4). In the ctenidium of T. squamosa, PiT1-like had a basolateral localization in the filaments and tertiary water channels (Figure 5). In addition, PiT1-like was detected in the epithelial cells of the tertiary zooxanthellal tubules in the outer mantle (Figure 6). In order to identify the location of PiT1-like in the tubular epithelium, it was essential to capture, albeit with some difficulties, the nuclei of the tubular epithelial cells. Nonetheless, based on three separate locations of the nuclei of the tubular epithelial cells in the image presented in Figure 6 and many similar observations in other slide preparations, PiT1-like appeared to have a basolateral localization in the tubular epithelium.
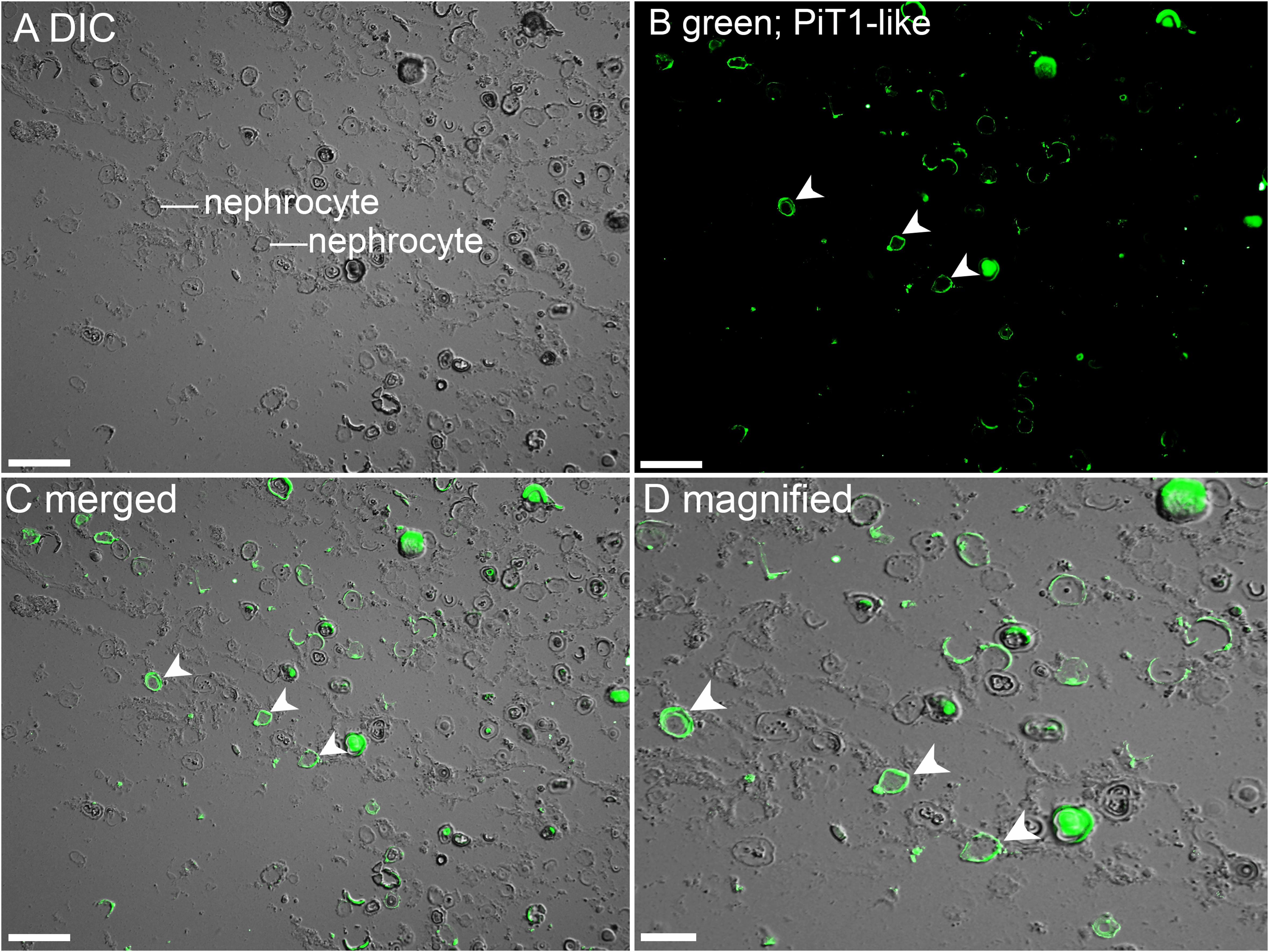
Figure 4. Immunofluorescence localization of a homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) in the kidney of Tridacna squamosa exposed to 12 h of light. (A) Differential interference contrast (DIC) image to demonstrate the histological structure of kidney nephrocytes. (B) Green immunofluorescence staining of the membrane of kidney nephrocytes (labeled with arrowheads) indicates presence of PiT1-like protein. (C) Merged images of DIC with PiT1-like labeling with (D) a magnified view. Scale bar: (A–C), 50 μm; (D), 20 μm.
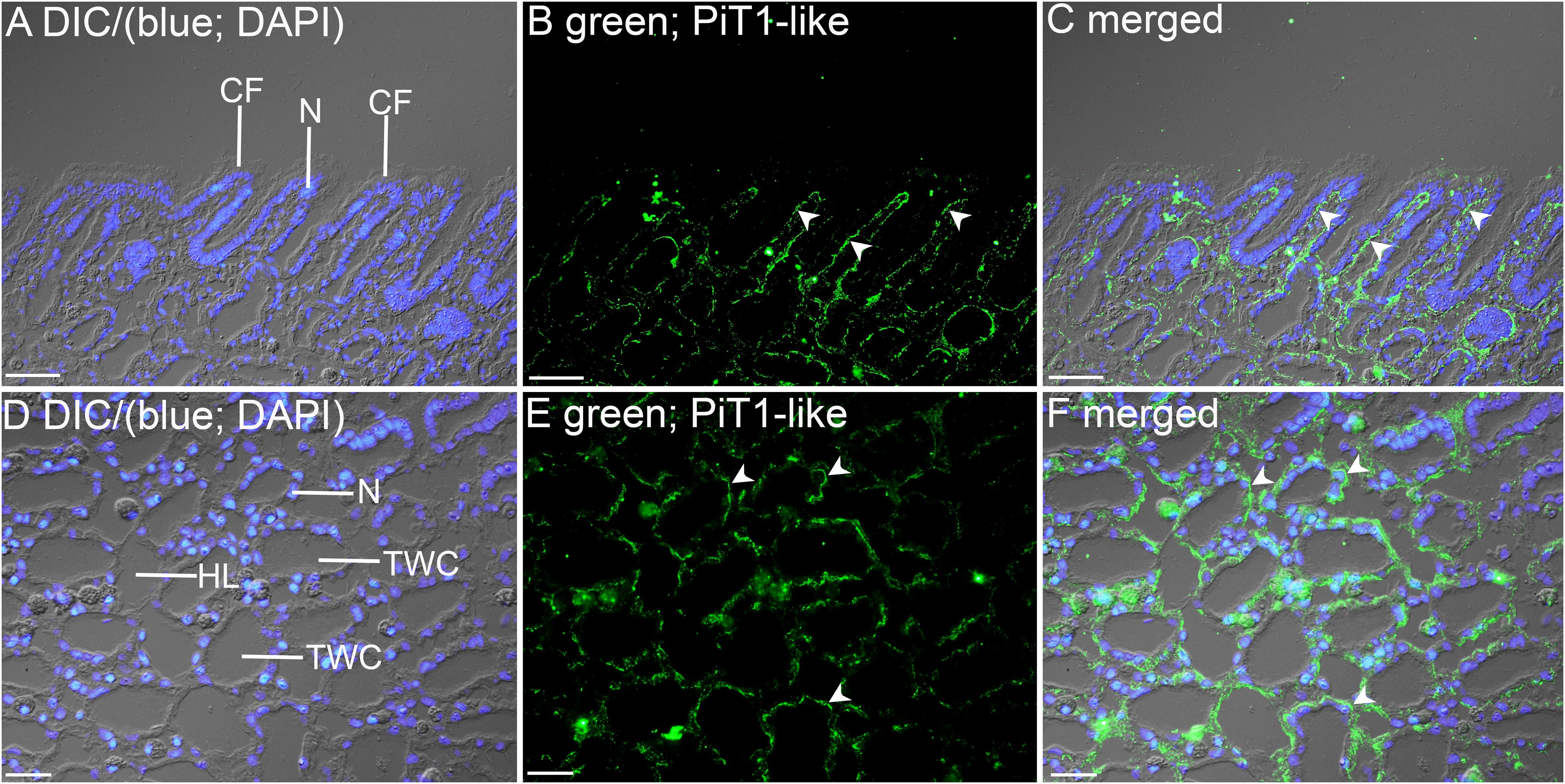
Figure 5. Immunofluorescence localization of a homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) in the filament (A–C) or the tertiary water channels (D–F) in the ctenidium of Tridacna squamosa exposed to 12 h of light. (A,D) Differential interference contrast (DIC) image is overlaid with DAPI nucleus (N) stain in blue to demonstrate the histological structure of ctenidium filament (CF) and tertiary water channel (TWC). (B,E) PiT1-like-immunofluorescence is displayed in green. (C,F) The green and blue channels are overlaid with DIC. Arrowheads in (B,C) show extensive basolateral staining of PiT1-like on the epithelium (EP) of CFs. (E,F) indicates the presence of PiT1-like on the basolateral membrane of the tertiary water channels. HL, hemolymph. Scale bar: 20 μm.
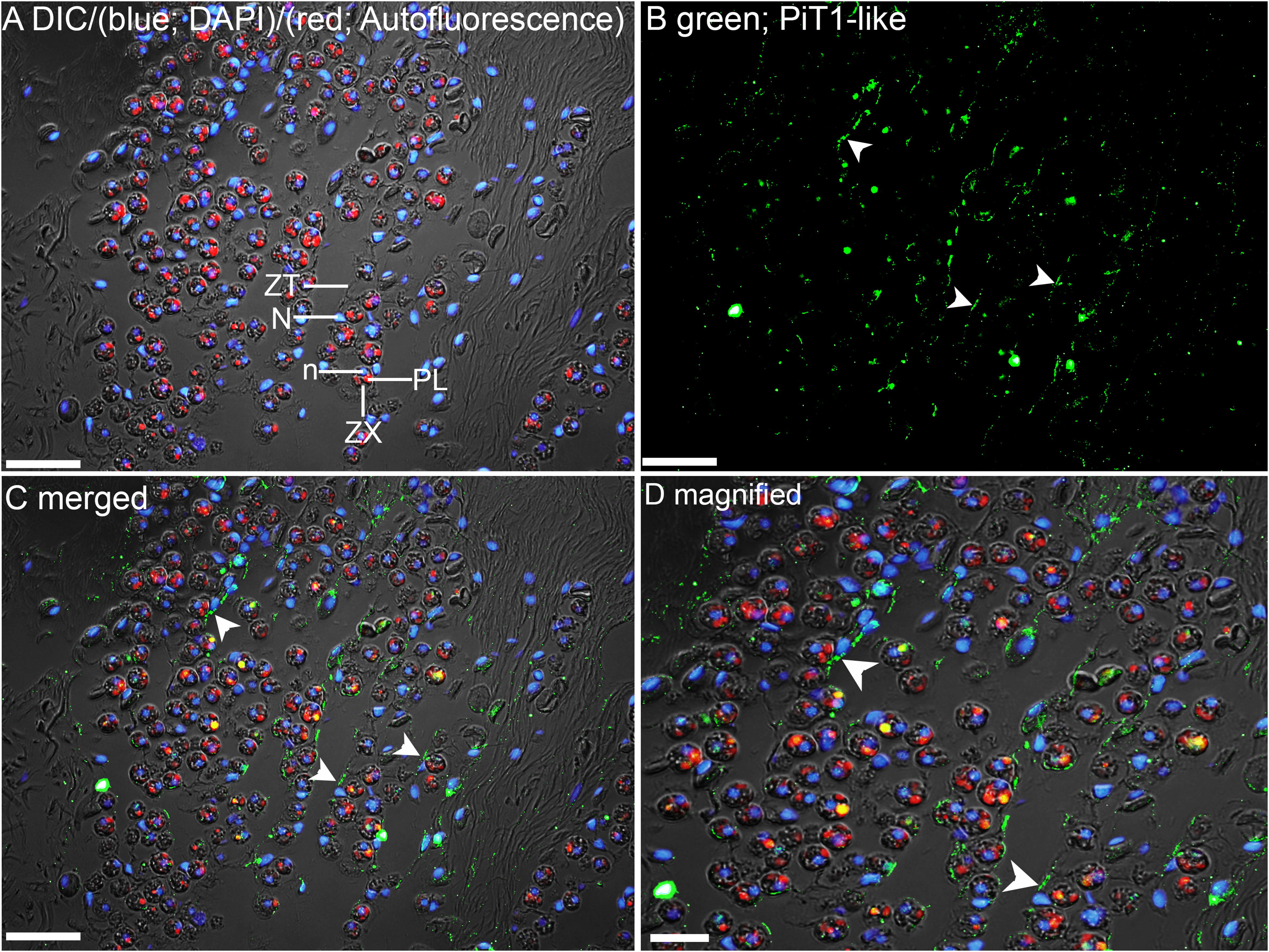
Figure 6. Immunofluorescence localization of a homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) in the outer mantle of Tridacna squamosa exposed to 12 h of light. (A) Overlaid image of differential interference contrast (DIC), auto-fluorescence of symbiotic dinoflagellates (zooxanthellae, ZX) in red, merged with that of DAPI (blue) staining of zooxanthellal nucleus (n) and nucleus (N) of host tubular epithelium (ZT). Red auto-fluorescence of ZX was due to plastid (PL). (B) Green immunofluorescence staining of the epithelial cell linings of the basolateral membrane of ZT (labeled with arrowheads) indicates presence of PiT1-like protein. (C) Merged images of DIC, DAPI, red, and PiT1-like immunolabeling. (D) Magnified view of (C). Scale bar: 20 μm.
In T. squamosa, the transcript level of PiT1-like in the kidney was approximately 10-fold higher than those in the ctenidium and outer mantle (Figure 7). There were significant increases in the transcript levels of PiT1-like in the kidney at hour 12 of light exposure (Figure 7A), in the ctenidium at hour 6 or 12 of light exposure (Figure 7B), and in the outer mantle at hour 3 of light exposure (Figure 7C), when compared with the corresponding values of the control clams kept in darkness for 12 h.
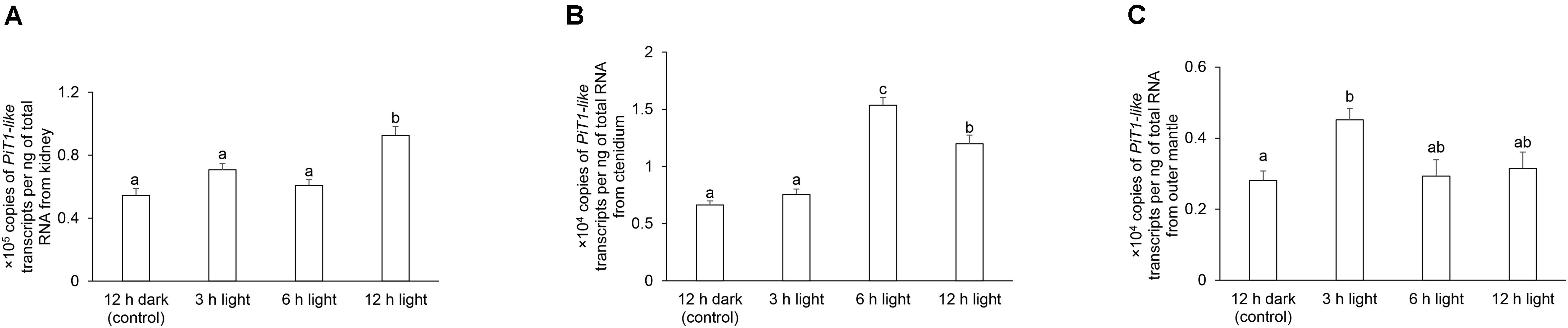
Figure 7. Transcript levels of a homolog of sodium-dependent phosphate transporter protein 1 (PiT1-like) from the (A) kidney, (B) ctenidium, and (C) outer mantle of Tridacna squamosa kept in darkness for 12 h (control), or exposed to light for 3, 6, or 12 h. Transcript levels of PiT1-like expressed as ×105 copies of transcript per ng total RNA for kidney, or ×104 copies of transcript per ng total RNA for ctenidium and outer mantle. Results represent means + SEM (n = 4). Means not sharing the same letter are significantly different (p-value < 0.05).
As the kidney and ctenidium had apparently higher protein abundance of PiT1-like than the outer mantle, the protein loads of samples of kidney, ctenidium, and outer mantle for electrophoresis were 20, 50, and 100 μg (Figure 8), respectively. The protein abundance of PiT1-like increased significantly in the kidney of T. squamosa after 6 or 12 h of exposure to light, as compared to the control (Figure 8A). In comparison, the protein abundance of PiT1-like increased significantly in the ctenidium and outer mantle only after 12 h of exposure to light as compared to the control (Figures 8B,C). The specificity of anti-PiT1-like antibody was validated by PCA, whereby the PiT1-like band of the kidney sample was abolished using the peptide-neutralized antibody (Supplementary Figure 1).
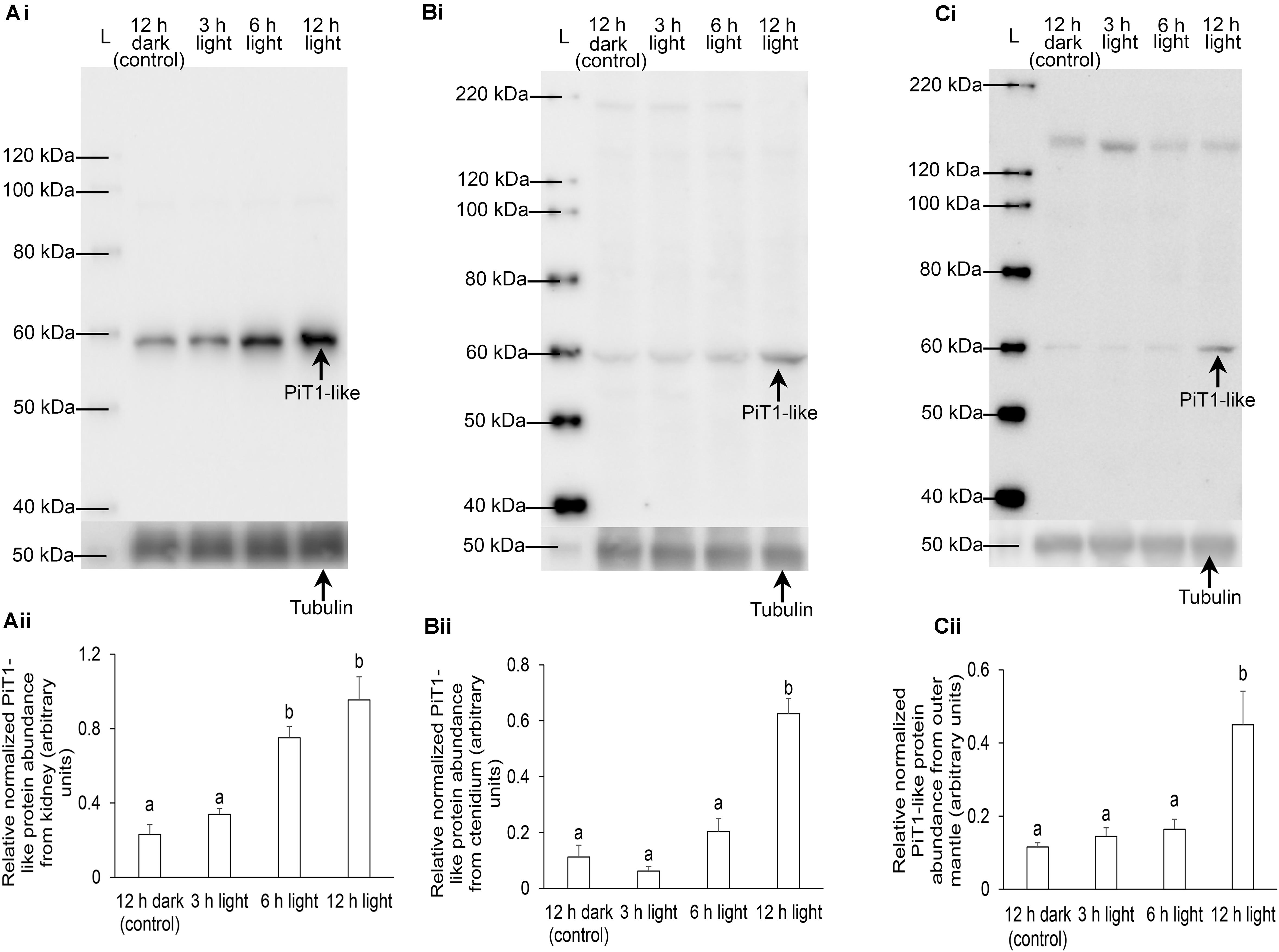
Figure 8. Protein abundance of a homolog of sodium-dependent phosphate transporter protein 1 from the (A) kidney, (B) ctenidium, and (C) outer mantle of Tridacna squamosa kept in darkness for 12 h (control) or exposed to light for 3, 6, or 12 h. (i) A representative blot. L represents ladder. (ii) The optical density of the PiT1-like band for 20, 50, and 100 μg of protein from kidney, ctenidium, and outer mantle, respectively, was expressed as arbitrary units and normalized with respect to that of tubulin from each tissue. Results represent means + SEM (n = 4). Means not sharing the same letter are significantly different (p-value < 0.05).
PiT1-like of T. squamosa consists of two homology ProDom domains (PD001131), one at the N terminal region and another at the C terminal region (Salaün et al., 2004; Bottger and Pedersen, 2011), which are characteristic of PiT proteins. Moreover, two related sequence motifs (of 12 amino acids long), which are designated as signature sequences of the PiT family and involved in cation translocation (Bottger and Pedersen, 2005), can be found in the PD001131 domains of PiT1-like of T. squamosa. All conserved amino acids known to be critical for Pi transport are located inside the two PD001131 domains of PiT1-like. Based on the characterization of human PiT2 (Bottger and Pedersen, 2011), residues D39, E66, H403, D407, E476, and S494 in PiT1-like of T. squamosa could be involved in the translocation of H2PO4– (Bottger and Pedersen, 2002, 2005, 2011), although the exact mechanism remains elusive (Beck et al., 2009). The two conserved glutamate residues situated in the TMs could act to couple Na+ transport with Pi uptake (Bottger et al., 2006). Similar to PiT1 of H. sapiens and X. laevis, PiT1-like of T. squamosa comprises several conserved residues, such as V77, S78, R82, I86, V88, and S125, which constitute the substrate binding site(s) (Ravera et al., 2013; Bon et al., 2017). In particular, S125 may be responsible for binding of H2PO4– (Bon et al., 2017) while S78 and R82 are responsible for substrate selectivity and coordination (Ravera et al., 2013). As V77, S78, R82, I86, and V88 are located in the extracellular loop region 2, they may be involved in the formation of a hydrophilic pore through which substrates might interact with the binding sites during the transport cycle (Ravera et al., 2013). With conserved substrate binding sites, PiT1-like of T. squamosa can co-transport Na+ and H2PO4– against the uphill H2PO4– electrochemical gradient at the expense of the downhill electrochemical gradient of Na+ through the plasma membrane into the cell.
PiT1-like expression was detected in many organs of T. squamosa, indicating a possible house-keeping role in Pi homeostasis. However, PiT1-like was expressed strongly in the kidney of T. squamosa, which is known to contain much more phosphate than other organs (Belda and Yellowlees, 1995), denoting a probable role in the uptake and accumulation of Pi. The strong expression of PiT1-like in the ctenidium denotes an important physiological function, but whether it could be involved in exogenous Pi absorption depends on its subcellular localization. Furthermore, the moderate gene expression level of PiT1-like in the colorful outer mantle suggests an important role in the supply of Pi to the dinoflagellates that are found in high density therein. In order to elucidate the functions of PiT1-like, it was essential to determine its cellular and subcellular localization in these three organs by immunofluorescence microscopy.
Extracellular Pi can act as a signaling molecule that directly alters gene expression and cellular physiology. Cells can detect changes in extracellular Pi levels through a Pi-sensing mechanism (Camalier et al., 2013; Michigami, 2013) that may involve the MAPK ERK1/2 pathway (Khoshniat et al., 2011). In human skeletal cells, PiT1 and PiT2 heterodimerization can mediate Pi signaling independent of Pi uptake (Bon et al., 2017). The deletion of PiT1 or PiT2 impedes phosphorylation mediated by the Pi-dependent MAPK ERK1/2 signaling (Bon et al., 2017). If PiT1-like has an apical localization in epithelial cells in T. squamosa, it could not be involved in sensing the concentration of Pi in the hemolymph. However, if it has a basolateral localization in epithelial cells and direct contact with the hemolymph, it could possibly function as a transporter to translocate H2PO4– from the hemolymph into the cell, as well as a sensing mechanism to detect hemolymph H2PO4– but without transporting it. Although it would be difficult to decipher these two functions of PiT1-like in T. squamosa, attempts were made to interpret our results based on the physiological functions of the three organs studied.
Symbiotic animals absorb Pi from the ambient seawater not only for their own metabolism, but also for the needs of the symbionts (Jackson and Yellowlees, 1990). Pi uptake in corals (Jackson and Yellowlees, 1990; Godinot et al., 2009) and giant clams (Chan et al., 2020) are light-dependent. For giant clams, it has been postulated that the host could maintain low concentrations of Pi (<0.1 μM) in the hemolymph to control the internal population of symbiotic dinoflagellates (Belda and Yellowlees, 1995). Without knowledge of the mechanism involved, Belda and Yellowlees (1995) proposed that the kidney of giant clams could rapidly take up Pi circulating in the hemolymph to limit the availability of Pi to the symbionts. Our results demonstrate for the first time that the plasma membrane of the globular nephrocytes in the kidney of T. squamosa displayed a strong immunolabeling of PiT1-like. Nephrocytes are located adjacent to the hemolymph sinuses. Hence, they can probably sense the extracellular Pi concentration and actively absorb H2PO4– from the hemolymph through PiT1-like, driven by the electrochemical potential gradient of Na+ (Virkki et al., 2007). Furthermore, the gene and protein expression levels of PiT1-like increased significantly in the kidney of T. squamosa during illumination, indicating its involvement in the increased removal of Pi from the hemolymph during light-enhanced Pi uptake to maintain a low hemolymph Pi concentration. As Pi is scarce in seawater, the host probably accumulates Pi in nephrocytes to cater for its long-term growth and development, but whether the accumulation of Pi in nephrocytes is meant to control the populations of symbionts (Belda and Yellowlees, 1995) is debatable. Results obtained from the outer mantle of T. squamosa indicate that the host clam could probably increase the supply of Pi to the symbionts through PiT1-like during illumination, as they would logically have a higher demand for Pi in light than in darkness.
The ctenidium of T. squamosa expresses NPT2a-like in the apical membrane of its epithelial cells (Chan et al., 2020). The inwardly directed Na+ electrochemical gradient would drive HPO42– from the ambient seawater into the epithelial cells through the apical NPT2a-like. Then, Pi can be translocated through some unknown basolateral transporter to the hemolymph. Our results indicate that the ctenidial epithelial cells of T. squamosa also expressed PiT1-like in the basolateral membrane. The basolateral localization prescribes that PiT1-like would transport H2PO4– from the hemolymph into the epithelial cells. Enterocytes of human intestine also express an apical NPT2b as well as basolateral PiT1 and PiT2 (Kiela and Ghishan, 2018), whereby NPT2b absorbs Pi from the intestinal lumen under normal circumstances, and PiT1/PiT2 function to import Pi from the blood for cellular Pi homeostasis during periods of dietary insufficiency (Kiela and Ghishan, 2018). Hence, the possibility of the basolateral PiT1-like in the ctenidial epithelial cells of T. squamosa absorbing Pi from the hemolymph during Pi-limiting situation cannot be ignored. However, the light-enhanced expression of PiT1-like in the ctenidial epithelial cells might suggest that the capacity for Pi-sensing in these cells is enhanced by light. As Pi binding rather than Pi uptake is the key factor in mediating Pi signaling through the PiT protein, it is possible that the basolateral PiT1-like in the ctenidial epithelial cells could act as part of a Pi-sensing mechanism, and that the physiological activities of these epithelial cells, including Pi uptake, can be regulated by the Pi concentration in the hemolymph, although future works are needed to verify these propositions.
The outer mantle of T. squamosa can be regarded as a crucial site of light-enhanced Pi absorption through NPT2a-like (Chan et al., 2020), and the Pi absorbed can be delivered directly to the dinoflagellates therein. As PiT1-like had a basolateral localization in the epithelial cells that formed the tertiary tubules in the outer mantle of T. squamosa, it could act as a mechanism for Pi sensing and signaling. It could also actively transport H2PO4– from the hemolymph into the tubular epithelial cells against its concentration gradient, and then Pi could be transported through some unknown apical transporter into the luminal fluid of the zooxanthellal tubules. It has been postulated previously that the growth and reproduction of symbionts are determined by the low concentration of Pi in the hemolymph (Belda and Yellowlees, 1995). However, symbiotic dinoflagellates are surrounded by the luminal fluid of the zooxanthellal tubules, and the ionic concentrations in the luminal fluid are most likely different from those in the hemolymph. In fact, PiT1-like could logically facilitate and regulate the translocation of Pi from the hemolymph into the luminal fluid surrounding the symbionts.
Importantly, the transcript level and protein abundance of PiT1-like/PiT1-like increased significantly in the outer mantle at hour 3 and hour 12 of light exposure, respectively. The increase in the protein abundance of PiT1-like in the outer mantle occurred between hour 6 and hour 12, which aligned well with the light–enhanced expression of RuBisCO protein therein (Poo et al., 2020), and with the light-enhanced host’s carbon concentration mechanism (Ip et al., 2017, 2018). In light, photosynthesizing dinoflagellates need Pi to increase the synthesis of ATP during the light-dependent process of photosynthesis. The localization and light-dependency of PiT1-like expression in the outer mantle provide novel insights into the interaction between the host clam and the symbionts.
Results obtained from this study have three implications. Firstly, the same host transporter (PiT1-like) could serve different physiological functions in different organs (kidney, ctenidium, and outer mantle). Secondly, the host could probably regulate the supply of Pi to the symbionts through PiT1-like of the tubular epithelial cells. Thirdly, the increase in the supply of Pi to the symbionts, together with the absorption of Pi by nephrocytes, led to the low concentration of Pi in the hemolymph. When taken together, these results denote that the accumulation of Pi in the kidney of giant clams is unlikely to be dedicated for the control of the internal dinoflagellate population, but to act as a Pi reserve for the host’s growth and development due to the scarcity of Pi in the environment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Institutional (National University of Singapore Institutional Animal Care and Use Committee) approval was not required for research on invertebrates including giant clams. Nonetheless, giant clams were anesthetized with 0.2% phenoxyethanol before tissue sample collection.
YI and SC: conceptualization, writing – original draft preparation, and supervision. YI: methodology and funding acquisition. YI, MB, JP, and SC: formal analysis and investigation and writing – review and editing. WW: resources. All authors contributed to the article and approved the submitted version.
This study was supported by the Singapore Ministry of Education through a grant to YI (R-154-000-B69-114).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.655714/full#supplementary-material
Supplementary Figure 1 | Peptide competition assay (PCA) on anti-PiT1-like antibody. The presence of PiT1-like band of a kidney sample exposed to light for 12 h (A) disappeared when anti-PiT1-like antibody was neutralized with the immunizing peptide (B). L, ladder.
Addessi, L. (2001). Giant clam bleaching in the lagoon of takapoto atoll (french polynesia). Coral Reefs 19:220.
Beck, L., Leroy, C., Salaun, C., Margall-Ducos, G., Desduoets, C., and Friedlander, G. (2009). Identification of a novel function of PiT1 critical for cell proliferation and independent of its phosphate transport activity. J. Biol. Chem. 284, 31363–31374. doi: 10.1074/jbc.M109.053132
Belda, C. A., and Yellowlees, D. (1995). Phosphate acquisition in the giant clam-zooxanthellae symbiosis. Mar. Biol. 124, 261–266.
Bon, N., Couasnay, G., Bourgine, A., Sourice, S., Beck-Cormier, S., Guicheux, J., et al. (2017). Phosphate (Pi)-regulated heterodimerization of the high-affinity sodium-dependent Pi transporters PiT1/Slc20a1 and PiT2/Slc20a2 underlies extracellular Pi sensing independently of Pi uptake. J. Biol. Chem. 293, 2102–2114. doi: 10.1074/jbc.M117.807339
Bottger, P., Hede, S. E., Grunnet, M., Høyer, B., Klærke, D. A., and Pedersen, L. (2006). Characterization of transport mechanisms and determinants critical for Na+-dependent Pi symport of the PiT family paralogs human PiT1 and PiT2. Am. J. Physiol. Cell. Physiol. 291, 1377–1387. doi: 10.1152/ajpcell.00015.2006
Bottger, P., and Pedersen, L. (2002). Two highly conserved glutamate residues critical for type III sodium-dependent phosphate transport revealed by uncoupling transport function from retroviral receptor function. J. Biol. Chem. 277, 42741–42747. doi: 10.1074/jbc.M207096200
Bottger, P., and Pedersen, L. (2011). Mapping of the minimal inorganic phosphate transporting unit of human PiT2 suggests a structure universal to PiT-related proteins from all kingdoms of life. BMC Biochem. 12:21. doi: 10.1186/1471-2091-12-21
Bottger, P., and Pedersen, L. (2005). Evolutionary and experimental analyses of inorganic phosphate transporter PiT family reveals two related signature sequences harboring highly conserved aspartic acids critical for sodium-dependent phosphate transport function of human PiT2. FEBS J. 272, 3060–3074. doi: 10.1111/j.1742-4658.2005.04720.x
Camalier, C. E., Yi, M., Yu, L. R., Hood, B. L., Conrads, K. A., Lee, Y. J., et al. (2013). An integrated understanding of the physiological response to elevated extracellular phosphate. J. Cell. Physiol. 228, 1536–1550.
Chan, C. Y. L., Hiong, K. C., Boo, M. V., Choo, C. Y. L., Wong, W. P., Chew, S. F., et al. (2018). Light exposure enhances urea absorption in the fluted giant clam Tridacna squamosa and up-regulates the protein abundance of a light-dependent urea active transporter DUR3-like in its ctenidium. J. Exp. Biol. 221:jeb176313. doi: 10.1242/jeb.176313
Chan, C. Y. L., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2019). Increased apical sodium-dependent glucose transporter abundance in the ctenidium of the giant clam Tridacna squamosa upon illumination. J. Exp. Biol. 222:jeb195644. doi: 10.1242/jeb.195644
Chan, C. Y. L., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2020). Light-enhanced phosphate absorption in the fluted giant clam, Tridacna squamosa, entails an increase in the expression of sodium-dependent phosphate transporter 2a in its colourful outer mantle. Coral Reefs 39, 1055–1070. doi: 10.1007/s00338-020-01930-w
Chew, S. F., Koh, C. Z. Y., Hiong, K. C., Choo, C. Y. L., Wong, W. P., Neo, M. L., et al. (2019). Light-enhanced expression of carbonic anhydrase 4-like supports shell formation in the fluted giant clam Tridacna squamosa. Gene 683, 101–112. doi: 10.1016/j.gene.2018.10.023
DeBoer, T. S., Baker, A. C., Erdmann, M. V., Ambariyanto, Jones, P. R., and Barber, P. H. (2012). Patterns of Symbiodinidium distribution in three giant clam species across the biodiverse bird’s head region of indonesia. Mar. Ecol. Prog. Ser. 444, 117–132. doi: 10.3354/meps09413
Fitt, W. K., and Trench, R. K. (1981). Spawning, development and acquisition of zooxanthellae by Tridacna squamosa (mollusca, bivalvia). Biol. Bull. 161, 213–235. doi: 10.2307/1540800
Forster, I. C., Hernando, N., Biber, J., and Murer, H. (2013). Phosphate transporters of the SLC20 and SLC34 families. Mol. Aspects Med. 34, 386–395. doi: 10.1016/j.mam.2012.07.007
Furla, P., Allemand, D., Shick, J. M., Ferrier-Pagès, C., Richier, S., Plantivaux, A., et al. (2005). The symbiotic anthozoan: a physiological chimera between alga and animal. Integ. Comp. Biol. 45, 595–604. doi: 10.1093/icb/45.4.595
Godinot, C., Ferrier-Pages, C., and Grover, R. (2009). Control of phosphate uptake by zooxanthellae and host cells in the scleractinian coral stylophora pistillata. Limnol. Oceanogr. 54, 1627–1633. doi: 10.4319/lo.2009.54.5.1627
Hernawan, U. E. (2008). Review: symbiosis between the giant clams (bivalvia: cardiidae) and zooxanthellae (dinophyceae). Jurusan Biol. 9, 53–58. doi: 10.13057/biodiv/d090113
Hiong, K. C., Cao-Pham, A. H., Choo, C. Y. L., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2017a). Light-dependent expression of a Na+/H+ exchanger 3-like transporter in the ctenidium of the giant clam, Tridacna squamosa, can be related to increased H+ excretion during light-enhanced calcification. Physiol. Rep. 5:e13209. doi: 10.14814/phy2.13209
Hiong, K. C., Choo, C. Y. L., Boo, M. V., Ching, B., Wong, W. P., Chew, S. F., et al. (2017b). A light-dependent ammonia-assimilating mechanism in the ctenidia of a giant clam. Coral Reefs 36, 311–323.
Ikeda, S., Yamashita, H., Kondo, S., Inoue, K., Morishima, S., and Koike, K. (2017). Zooxanthellal genetic varieties in giant clams are partially determined by species-intrinsic and growth-related characteristics. PLoS One 12:e0172285. doi: 10.1371/journal.pone.0172285
Ip, Y. K., and Chew, S. F. (2021). Light-dependent phenomena and related molecular mechanisms in giant clam-dinoflagellate associations: a review. Front. Mar. Sci. 8:627722. doi: 10.3389/fmars.2021.627722
Ip, Y. K., Ching, B., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., et al. (2015). Light induces changes in activities of Na+/K+-ATPase, H+/K+-ATPase and glutamine synthetase in tissues involved directly or indirectly in light-enhanced calcification in the giant clam, Tridacna squamosa. Front. Physiol. 6:68. doi: 10.3389/fphys.2015.00068
Ip, Y. K., Hiong, K. C., Lim, L. J. Y., Choo, C. Y. L., Boo, M. V., Wong, W. P., et al. (2018). Molecular characterization light-dependent expression and cellular localization of a host vacuolar-type H+-ATPase (VHA) subunit A in the giant clam Tridacna squamosa indicate the involvement of the host VHA in the uptake of inorganic carbon and its supply to the symbiotic zooxanthellae. Gene 659, 137–148. doi: 10.1016/j.gene.2018.03.054
Ip, Y. K., Hiong, K. C., Teng, J. H. Q., Boo, M. V., Choo, C. Y. L., Wong, W. P., et al. (2020). The fluted giant clam (Tridacna squamosa) increases nitrate absorption and upregulates the expression of a homolog of SIALIN (H+:2NO3– cotransporter) in the ctenidium during light exposure. Coral Reefs 39, 451–465. doi: 10.1007/s00338-020-01907-9
Ip, Y. K., Koh, C. Z. Y., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., et al. (2017). Carbonic anhydrase 2-like in the giant clam, Tridacna squamosa: characterization, localization, response to light, and possible role in the transport of inorganic carbon from the host to its symbiont. Physiol. Rep. 5:e13494. doi: 10.14814/phy2.13494
Ip, Y. K., Loong, A. M., Hiong, K. C., Wong, W. P., Chew, S. F., Reddy, K., et al. (2006). Light induces an increase in the pH of, and a decrease in the ammonia concentration in, the extrapallial fluid of the giant clam Tridacna squamosa. Physiol. Biochem. Zool. 79, 656–664.
Jackson, A. E., Miller, D. J., and Yellowlees, D. (1989). Phosphorus metabolism in the coral-zooxanthellae symbiosis: characterization and possible roles of two acid phosphatases in the algal symbiont Symbiodinium sp. Proc. R. Soc. Lond. B 238, 193–202. doi: 10.1098/rspb.1989.0076
Jackson, A. E., and Yellowlees, D. (1990). Phosphate uptake by zooxanthellae isolated from corals. Proc. R. Soc. Lond. B 242, 204–210. doi: 10.1098/rspb.1989.0076
Khoshniat, S., Bourgine, A., Julien, M., Petit, M., Pilet, P., Rouillon, T., et al. (2011). Phosphate-dependent stimulation of MGP and OPN expression in osteoblasts via the ERK1/2 pathway is modulated by calcium. Bone 48, 894–902.
Kiela, P. R., and Ghishan, F. K. (2018). Chapter 59 Molecular mechanisms of intestinal transport of calcium, phosphate and magnesium. In Physiology of Gastrointestinal Tract, Sixth Edn. Amsterdam: Elsevier Inc, 1405–1449.
Klumpp, D. W., and Griffiths, C. L. (1994). Contributions of phototrophic and heterotrophic nutrition to the metabolic and growth requirements of four species of giant clam (tridacnidae). Mar. Ecol. Progr. Ser. 115, 103–115. doi: 10.3354/meps115103
Knowles, J. R. (1980). Enzyme-catalyzed phosphoryl transfer reactions. Ann. Rev. Biochem. 49, 877–919. doi: 10.1146/annurev.bi.49.070180.004305
Koh, C. Z. Y., Hiong, K. C., Choo, C. Y. L., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2018). Molecular characterization of a dual domain carbonic anhydrase from the ctenidium of the giant clam, Tridacna squamosa, and its expression levels after light exposure, cellular localization, and possible role in the uptake of exogenous inorganic carbon. Front. Physiol. 9:281. doi: 10.3389/fphys.2018.00281
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Lim, S. S. Q., Huang, D., Soong, K., and Neo, M. L. (2019). Diversity of endosymbiotic symbiodiniaceae in giant clams at dongsha atoll, northern south china sea. Symbiosis 78, 251–262. doi: 10.1007/s13199-019-00615-5
Michigami, T. (2013). Extracellular phosphate as a signaling molecule. Contrib. Nephrol. 180, 14–24.
Norton, J. H., and Jones, G. W. (1992). The giant clam: an anatomical and histological atlas. Canberra Australia: Australian Centre for International Agricultural Research.
Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., et al. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648. doi: 10.1016/j.cell.2006.09.026
Poo, J. S. T., Choo, C. Y. L., Hiong, K. C., Boo, M. V., Wong, W. P., Chew, S. F., et al. (2020). Phototrophic potential and form II ribulose-1,5-bisphosphate carboxylase/oxygenase expression in five organs of the fluted giant clam, Tridacna squamosa. Coral Reefs 39, 361–374. doi: 10.1007/s00338-020-01898-7
Ravera, S., Murer, H., and Forster, I. C. (2013). An externally accessible linker region in the sodium-coupled phosphate transporter PiT-1 (SLC20A1) is important for transport function. Cell. Phys. Biochem. 32, 187–199. doi: 10.1159/000350135
Rivkin, R. B., and Swift, E. (1985). Phosphorus metabolism of oceanic dinoflagellates: phosphate uptake, chemical composition and growth of Pyrocystis noctiluca. Mar. Biol. 88, 189–198. doi: 10.1007/BF00397166
Salaün, C., Maréchal, V., and Heard, J. M. (2004). Transport-deficient Pit2 phosphate transporters still modify cell surface oligomers structure in response to inorganic phosphate. J. Mol. Biol. 340, 39–47. doi: 10.1016/j.jmb.2004.04.050
Virkki, L. V., Biber, J., Murer, H., and Forster, I. C. (2007). Phosphate transporters: a tale of two solute carrier families. Am. J. Physiol. Renal. Physiol. 293, 643–654. doi: 10.1152/ajprenal.00228.2007
Watson, S. A. (2015). Giant clams and rising CO2: Light may ameliorate effects of ocean acidification on a solar-powered animal. PLoS One 10:e0128405. doi: 10.1371/journal.pone.0128405
Keywords: photosynthesis, shell formation, Symbiodiniaceae, symbiosis, zooxanthellae
Citation: Ip YK, Boo MV, Poo JST, Wong WP and Chew SF (2021) Sodium-Dependent Phosphate Transporter Protein 1 Is Involved in the Active Uptake of Inorganic Phosphate in Nephrocytes of the Kidney and the Translocation of Pi Into the Tubular Epithelial Cells in the Outer Mantle of the Giant Clam, Tridacna squamosa. Front. Mar. Sci. 8:655714. doi: 10.3389/fmars.2021.655714
Received: 19 January 2021; Accepted: 12 March 2021;
Published: 31 March 2021.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Zhang Yuehuan, South China Sea Institute of Oceanology (CAS), ChinaCopyright © 2021 Ip, Boo, Poo, Wong and Chew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuen K. Ip, ZGJzaXB5a0BudXMuZWR1LnNn; orcid.org/0000-0001-9124-7911
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.