- 1Roger Webb Forensic Science Institute, University of Central Oklahoma, Edmond, OK, United States
- 2Center for Wildlife Forensic Science and Conservation Studies, University of Central Oklahoma, Edmond, OK, United States
- 3Department of Biology, University of Central Oklahoma, Edmond, OK, United States
- 4Shoals Marine Laboratory, Cornell University and University of New Hampshire, Durham, NH, United States
- 5Department of Commerce, NOAA Greater Atlantic Regional Fisheries Office, National Marine Fisheries Service, Gloucester, MA, United States
Because Kemp’s ridley sea turtles (Lepidochelys kempii) are critically endangered and closely related to the vulnerable olive ridleys (L. olivacea), it is essential for forensic investigations and conservation efforts to distinguish these species when only skeletal elements remain. DNA extraction and analysis by DNA sequencing of genetic markers is the only method to determine species identity reliably, yet these methods are significantly compromised when DNA becomes degraded. To evaluate the role that time and environment play in obtaining high-quality DNA sequencing data, we placed skeletal elements of a terrestrial mammal (Bos taurus) and L. kempii in a supratidal and subtidal environment for 3 years. Bi-annual sampling revealed that after 3 years, mitochondrial DNA (mtDNA) consistently identified each species from each environment. Our results show that mtDNA recovery from bone and identification for Kemp’s ridley sea turtles was possible up to 3 years in both environments. All sequencing data obtained was accurate and robust, but DNA sequencing results were not consistent after 664-days of exposure. Our findings led us to conclude that if sufficient DNA is extracted from bone samples, then high-quality sequence data can be obtained, and the resulting sequence data accurately reflects the reference sequence for the given gene marker. This study provides evidence that DNA can be extracted and analyzed from challenging biological substrates, like bone, when these substrates are exposed to seasonally dynamic maritime environmental conditions for up to 3-years.
Introduction
Modern forensic and conservation investigations depend heavily on DNA samples for the identification of individuals and/or populations of species. However, these investigations often are hindered by DNA degradation and insufficient quantities of genetic material, which is influenced by time, type of tissue, and the environment. The importance of environmental exposure has been well documented in soft tissues (e.g., blood, saliva); for example, UV light (Hall et al., 2014), high levels of humidity and temperature (Dissing et al., 2010; Al-Kandari et al., 2016), and submergence in fresh or saltwater (Borde et al., 2008; Frippiat et al., 2017; Helmus et al., 2018; Meixner et al., 2020). However, these studies investigated a controlled set of variables in a laboratory setting, rather than field exposures, and often for only days or weeks.
The effects of environmental exposure on DNA degradation in hard tissues (bones, teeth) has also been examined, including UV exposure, high levels of humidity and temperature, burial, and submergence in fresh or saltwater (Perry et al., 1988; Schwartz et al., 1991; Alaeddini et al., 2010). Again, interpretations were limited by either brief time intervals or utilization of samples from a variety of locations to extend the exposure interval. Other complicating issues with DNA degradation studies in both soft and ossified tissue include the use of terrestrial mammals as a test organism, even in aqueous environments, and for studies of aquatic wildlife, the use of environmental DNA (eDNA) rather than decomposing tissue samples (Eichmiller et al., 2016; Sassoubre et al., 2016; Collins et al., 2018). Few, if any, studies have examined the feasibility of obtaining high-quality DNA sequencing results for species identification and conservation in marine vertebrate skeletal remains following environmental exposure. In addition to wildlife forensic and investigative applications, recent studies have demonstrated the usefulness of osteo-molecular identification techniques to emergent, novel studies of skeletochronology, dietary growth analyses, foraging ecology, geospacial satellite tracking, and migration ecology (Gredzens and Shaver, 2020; Ramirez et al., 2020). A more comprehensive knowledge of DNA longevity in maritime skeletal remains, particularly in endangered and threatened species, informs conservation, protection, and research initiatives.
All sea turtles are protected by the U.S. Endangered Species Act (ESA). Kemp’s ridleys (Lepidochelys kempii) are critically endangered and found only in the Gulf of Mexico and along the U.S. Atlantic coast (Wibbels and Bevan, 2019). They are closely related to olive ridleys (L. olivacea), which has only one subpopulation protected by the ESA and are commercially harvested worldwide (Abreu-Grobois and Plotkin, 2008). Both species migrate internationally, and conservation biologists have been able to identify and track subpopulations through sequence differences in mitochondrial DNA (mtDNA). Due to female philopatry, in which pregnant females usually return to the beaches where they hatched, and because mtDNA is maternally inherited, individual haplotypes can be associated with specific beaches of origin (Allard et al., 1994; Duchene et al., 2012; Patricio et al., 2017; Frandsen et al., 2020). Combining the genetic data with GPS and satellite tracking reveals migratory pathways for sea turtles (Dutton et al., 2019; Gredzens and Shaver, 2020). Mixed stock analyses, the analysis of a group of subjects from several populations, have been used to indicate where individuals originated (Bolker et al., 2007; Jones et al., 2018). As more haplotypes are identified and more mixed stocks are connected to their natal beaches, foraging grounds can be linked to specific natal beaches and foraging habitats can be monitored and protected for populations in decline.
Kemp’s ridley populations continue to suffer negative impacts associated with environmental perturbation, climate change, entanglement, marine debris, chemical pollution, and incidental by-catch (National Marine Fisheries Service [NMFS], and U.S. Fish and Wildlife Service [FWS], 2015). Additionally, mass morbidity/mortality events occur secondary to rapid unanticipated declines in near-coastal ocean water temperatures. Significantly impacted are juveniles who undergo seasonal migrations to shallow water feeding grounds toward the margins of their distributional range, often as far north as Nova Scotia, Canada (Liu et al., 2019). Inherent small size, increased surface-to-volume ratio, and estuarine foraging behaviors contribute to the victimization of these animals in periodic, hypothermic mass mortality events (National Marine Fisheries Service [NMFS], and U.S. Fish and Wildlife Service [FWS], 2015).
Integral to the process of monitoring sea turtles in this manner is the ability of an investigator to obtain high-quality DNA sequencing results from a wide variety of biological substrates. Most conservation studies obtain DNA from blood and tissues from living sea turtles, but only after a complicated and redundant permitting process (Paul and Sikes, 2013). While the analysis of living sea turtles is necessary to establish a reference dataset for sea turtle haplotypes, evidence collected during the investigation of environmental crimes, incidences of poaching, accidental deaths, and other unexplained mortality events often produces more challenging samples. Taphonomic changes in marine vertebrate remains are often rapidly progressive in oceanic and maritime-terrestrial environments (Lord and Burger, 1984a,b; Anderson and Bell, 2014). Frequently, evidentiary samples mainly consist of skeletal remains. This is particularly pronounced in marine vertebrates of small size. In many sea turtles, evolutionary adaptations associated with underwater locomotion have resulted in bones characterized by a more significant predominance of lighter cancellous bone (spongy bone) than seen in their terrestrial counterparts (Rhodin, 1985; Houssaye, 2012; Nakajima et al., 2014). Although the overall number of cells within a cancellous bone is increased, the amount of structural protection and support provided to the cells within a cancellous bone is decreased compared to the bone of terrestrial species.
Consequently, there is less cortical bone, the usual target for skeletal DNA extraction, in a sea turtle’s skeleton. It is unknown whether DNA extracted from sea turtle remains recovered from supratidal (terrestrial) and subtidal (ocean) environments can produce high-quality DNA sequences that can be used for species identification. Therefore, the intent of this study was to investigate the long-term viability of mtDNA for DNA sequencing analysis from environmental conditions that are representative of forensic casework. To investigate this phenomenon, mtDNA was extracted from Kemp’s ridley skeletal remains following prolonged exposure to supratidal and subtidal environments.
Materials and Methods
Sample Collection
Bone samples from two small juvenile (1–2 years old) male Kemp’s ridleys that expired due to cold shock and one domestic cow were used for this study. We used domestic cattle (Bos taurus) bones as a terrestrial mammal control specimen to evaluate the length of time amplifiable DNA can be obtained after environmental exposure and the effectiveness of Proteinase K (Pro K) vs. Collagenase Type II (CTII) in bone demineralization during DNA extraction. Our approach investigated if DNA sequencing data from the Kemp’s ridley remains were linked to the animal’s bone structure or the environmental factors alone. The Kemp’s ridley remains were recovered by a NOAA stranding/rehabilitation organization. Bone samples were obtained post-necropsy (NOAA Greater Atlantic Region ESA Permit TE01150C), disarticulated, and prepared according to McElreath (2018). On July 16, 2016, samples were placed in mesh bags and secured within lobster traps to prevent scavenging. One set of samples was submerged (= subtidal) to a depth of 3–9 m in a small cove at the Shoals Marine Laboratory (Appledore Island, ME) to protect the samples/traps from storms and to ensure submergence regardless of the tide. The other set was located 200 m inland (= supratidal). The locations were chosen to represent realistic environments where marine animal remains would typically be found. A terrestrial mammal comparison control was selected to facilitate the contemporaneous investigation of comparative early phase sea turtle and marine mammal maritime diagenesis.
The study sites were located on Appledore Island (42° 59′ 14″′ N, 70° 36′ 49″′ W) 9.5 km from the nearest mainland point at Rye, NH, and the largest/highest (39 ha, 0.8 km east-west, 1.0 km north-south; 20 m highest altitude) of nine islands forming the Isles of Shoals archipelago (McElreath, 2018). The Isles of Shoals falls within the Northern Coastal Shelf of the Gulf of Maine and is characterized by a variety of habitat types and complex bathymetry (Sowers et al., 2020). The Shoals are typified by ecologically diverse and climatically dynamic oceanic environs, in part due to proximity to the Gulf of Maine Coastal Current. The northern and eastern shores are exposed, subjected to the ever-present oceanic swells of varying amplitude and frequency and the violent Nor’easters (34–63 km), particularly in winter (Nichols and Nichols, 2008). The west and south shores are protected. Because of the small size of the island virtually all terrestrial vegetation is subjected to the damaging salt spray, although taxa more common along the eastern shore display increased tolerance to maritime exposure than those on the protected shore (reviewed in Nichols and Nichols, 2008). Marine algae also reflect the exposed/protected dichotomy, with some species more abundant and more adapted to one area over the other. Terrestrial temperatures peak from June through September (ca. 15–20°C) with lows in January and February (−1.0 to −0.5°C)1. Nearshore water temperatures at the Isles vary according to tides and protected/exposed coast (colder on exposed) but range from 2 to 21°C during the year. The study location is representative of seasonally dynamic, near-shore maritime environs of the Western North Atlantic.
The subtidal site was located in a small (40 × 80 m during low tide) extension of Babb’s Cove on the western shore, draining into the Cove via a 1–2 m channel during low tides but continuous with the Cove during high tides. On hot summer days during low tide, temperatures in the isolated pool can be considerably warmer than Babb’s Cove. The site was chosen because of the high degree of protection and concomitantly, the only beach on the island (a proxy for areas most likely to accumulate sea turtle remains), although intense westerly storms in the winter coinciding with the high tide can create water motion sufficient to move the submerged cages a few meters. The substratum is composed of small cobble and shell hash, continuously covered with diatom and microalgal films and seasonally abundant species of filamentous algae (all indicative of substratum stability). Crabs, small lobsters, snails, and microcrustaceans are abundant. Cages were placed in the deepest part of the pool (∼5 m), ensuring submergence throughout the tidal cycle (−0.5 to +3.4 m). Supralittoral cages were placed in short to moderate-height shrub thicket ∼95 m due east of the subtidal cages, an area never subjected to seawater submergence (although fine sea spray undoubtedly can reach the area during violent westerly storms). Grasses enveloped the cages during summer but died back in winter. In addition to logistical and research facilities considerations, the study location was selected to reflect the approximate northerly bounds of the Kemp’s ridley distributional range and maximize the potential negative impacts of environmental conditions and seasonal exposure on skeletal DNA longevity.
Twelve sets of Kemp’s ridley and cattle bone fragments, from all parts of the skeleton, were collected over 3 years: from the supratidal and subtidal environments in September and May/June until 2019, when they were collected only in September. Samples were double-sealed in plastic bags and shipped overnight in insulated packaging to the University of Central Oklahoma, where they were stored at −20°C upon arrival. The bone samples collected represented a variety of samples that could be encountered in forensic science and conservation casework. While the variation in bone sizes and structures evaluated is a strength of this study, we must also acknowledge that our sampling method could not resolve the role that the size and structure of a given bone sample had on obtaining a high-quality DNA sequence. In total, 26 bone samples representing 13 timepoints (including initial baseline samples) and two exposure conditions were analyzed. The number of time series analytical replicates was limited by regulatory restrictions on Kemp’s ridley carcass availability and the morphological juvenescence of the acquired specimens.
Bone Pulverization and Demineralization
A 0.5 g of each bone sample at each time point was excised using a Dremel tool and documented photographically. Each portion was cleaned according to the DNA Solutions “Bone Demineralization and Isolation Protocol” (DNA Solutions, 2016), then submerged individually into liquid nitrogen until brittle: ∼20 min for the L. kempii samples and ∼40 min for the B. taurus samples. Each portion of the bone samples was then pulverized with a Bone Morselizer (DDP Medical Supply, St. Petersburg, FL), returned to room temperature, and divided into two < ∼0.25 g subsamples. Each ∼0.25 g subsample was then treated with either Pro K or CTII. In accordance with Barrett (2015) protocol, 675 μL of Tissue and Cell Lysis Buffer (TCL Buffer; Epicentre/Lucigen, WI) was added to 750 μL of 0.5 M EDTA, 75 μL of Pro K or 50 mg/mL CTII, and 60 μL of 1M DTT. The samples were vortexed for 30 s. The Pro K treated samples were placed in a tube agitator at 56°C for 24 h, whereas the CTII treated samples were placed into a tube agitator at 37°C for 5 h and then at 56°C for an additional 19 h. Four reagent blanks (RB-CTII LK, RB-CTII BT, RB-ProK LK, and RB-ProK BT) were generated and processed with each turtle and cow sample set for the duration of the study. Finally, the resulting solutions were divided into three 300 μL replicates and transferred to 2 mL tubes; the remaining demineralization solution was stored at −20°C.
Extraction, Quantification, and Amplification
All replicate samples and reagent blanks were purified using the MasterPureTM Complete DNA and RNA Purification Kit (Epicentre/Lucigen) according to the manufacturer’s protocol (Epicentre, 2012), then quantified using the Qubit 4 Fluorometer (Thermo Fisher Scientific) and the Quant-iTTM PicoGreenTM dsDNA Assay Kit (Invitrogen). The working buffer for extraction contained: 10 μL of 20X TE buffer with 190 μL of ultra-pure water and 1 μL of Quant-iTTM PicoGreen® dsDNA reagent. Two microliters of each replicate DNA extraction were added to 198 μL of the working buffer and measured with the Quant-iTTM dsDNA high-sensitivity assay.
The mtDNA primer set for L. kempii was developed to achieve two specific outcomes: (1) provide sufficient genetic differentiation to distinguish between L. kempii and L. olivacea species and (2) maintain minimal sequence variation within the L. kempii population. The mitochondrial NADH:ubiquinone oxidoreductase core subunit 4 (MT-NAD4) genetic marker was selected for this study because it satisfied both conditions. As mentioned above, the intent of this study was to investigate the long-term viability of mtDNA for DNA sequencing analysis following prolonged environmental exposure. The selection of a genetic marker or markers that possessed high genetic polymorphisms would introduce an error that could not be accounted for or controlled otherwise. In short, our analysis would not be able to distinguish between a sequence polymorphism or DNA damage resulting from environmental exposure. Both mtDNA primers for L. kempii were generated using sequence data obtained from GenBank for the MT-NAD4 gene (accession number MN136058.1) and the B. taurus primer set was generated to amplify the mitochondrial cytochrome b (MT-CYB) gene (accession number KT260196), both using Geneious Prime software v2019.2.3 (Kearse et al., 2012). The MT-NAD4 amplicon was 371 bp long and was generated using the following primers: F–5′AAGCTCATGTAGAAGCCCCA-3′, R–5′TGTTCGGCTGTGAGTTCGTT-3′. The MT-CYB amplicon was 284 bp long and was generated using the following primers: F–5′ACCAGCCTGCTCTTCATCAC-3′, R-5′CGAGAGGTGCAGGAAGAAGG-3′. The PCR master mix for both the Pro K treated and CTII treated mtDNA contained: 17.1 μL DNA-free water, 1.2 μL of 10 μM forward mtDNA primer, 1.2 μL of 10 μM reverse mtDNA primer, 1 μL of GoTaq® Hot Start Polymerase (Promega), and 3.5 μL of Failsafe buffer K (Epicentre/Lucigen) for the L. kempii samples or buffer I (Epicentre/Lucigen) for the B. taurus samples. If the concentration of either Pro K or CTII purified DNA was > 10 ng/μL, 1 μL was added to the master mix; if <5 ng/μL, 3 μL of purified DNA was added. In both cases, the amount of water in the PCR master mix was adjusted to a reaction volume of 25 μL. The thermocycler (Applied Biosystems, GeneAmp PCR System 9700) was set to run under the following conditions: 2 min at 94°C; 15 s at 95°C, 1 min at 55°C, and 20 s at 72°C for 30 cycles; 10 min at 72°C, and hold at 4°C.
Cycle Sequencing
PCR amplicons were purified with ExoSAP-IT PCR Product Cleanup Reagent (Applied Biosystems) by combining 10 μL of the PCR amplicons and 4 μL of ExoSAP-IT, incubating in a thermocycler for 15 min at 37°C, and 15 min at 80°C. PCR amplicons generated for both L. kempii and B. taurus were prepared for sequencing as detailed in the BigDyeTM Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, 2016). The sequencing reaction master mix contained: 4 μL DNA-free water, 8 μL of BigDye Ready Reaction Mix (RR-100), and 4 μL 3 μM forward (or reverse) mtDNA per sample and was added to 4 μL of PCR amplified template DNA. The thermocycler was set to run under the following conditions: 1 min at 96°C; 10 s at 96°C, 5 s at 55°C, and 4 min at 60°C for 30 cycles; hold at 4°C. All samples were purified with Performa® Spin Columns (EdgeBio) according to the manufacturer’s protocol. Following the cycle sequencing reaction, all samples were placed onto the Applied Biosystems 3500 Genetic Analyzer with 10 μL of DNA standard from the BigDye 3.1v kit added into two wells for redundancy. Sequencing protocol was set for Short Sequencing (200–300 bp), running module FastSeq36_POP7 with dye set ZI, and the following settings modified from default: 2,480 s run time, 8.5 kV run voltage, 1.2 kV injection voltage, and 520 s data delay. The primary analysis protocol was set for BDTv3.1PA_Protocol-POP7 and was unmodified. No secondary analysis protocol was selected.
Sequence Data Analysis
Sequence data obtained were aligned with Geneious Prime software v2019.2.3 (Kearse et al., 2012), utilizing the Geneious Alignment tool with default settings. The resulting sequence alignments were manually trimmed to exclude the primer regions before extracting consensus sequences. A selection of reference gene regions were obtained from GenBank (accession numbers: L. kempii- MN136058.1, MN136059.1, MN136060.1, and MN136061.1; B. taurus- KT260195.1 and KT260195.1), aligned with the samples, and trimmed to the appropriate length for further comparison. These reference sequences were chosen primarily based on the availability of reference genomes at NCBI at the time of analysis. Maximum likelihood trees were estimated with the Hasegawa-Kishino-Yano (HKY) model and were generated with bootstrap resampling 1000 times using MEGA-X software (Kumar et al., 2018). L. kempii maximum likelihood trees were constructed using MT-NAD4 gene reference sequence data for L. olivacea (DQ486893.1, NC028634.1, and JX454991.1) and C. mydas (JX454976.1, JX454990.1, and NC000886.1) obtained from NCBI. B. taurus maximum likelihood trees were constructed using MT-CYB gene reference sequence data for B. javanicus (D34636.1 and D82889.1) and B. grunniens (NC006380.3). The quality scores of each sample’s untrimmed consensus sequence were opened using FASTQC2 and then compiled using MulitQC software (Ewels et al., 2016) to generate phred score graphs for each sequence at every nucleotide. Average phred scores and the average quantities were log10 normalized for repeated ANOVA analyses (SPSS Statistics software3). Phred score and DNA quantity values for both species were pooled for inter-species comparisons to determine if there were significant differences between the average values for both data sets. The analysis performed in this study sought to evaluate the persistence and quantity of skeletal mtDNA within sea turtle remains after prolonged exposure to subtidal and supratidal environments and determine how long a high-quality DNA sequencing result could be identified from the mtDNA within the remains.
Results
DNA Quantity
Overall, the L. kempii samples yielded a higher DNA concentration than the B. taurus samples at each time point and condition tested (Table 1). Compared to B. taurus samples, L. kempii samples prepared using Pro K yielded 9.3 times more DNA, while CTII samples yielded 6.6 times more DNA. Using the log10 of the average DNA quantities acquired from the triplicate samples, significantly (P < 0.05, one-way ANOVA) more DNA was recovered from the L. kempii (Pro K mean: 0.985 and CTII mean: 0.935) samples than from B. taurus (Pro K mean: 0.252 and CTII mean: 0.263) samples. For both L. kempii and B. taurus, the terrestrial samples resulted in substantially higher DNA yields than the marine samples; 9.5 times more for L. kempii and 37 times more for B. taurus samples. However, less variability was observed between exposure dates for the marine samples, but this could result from having less DNA coming from marine samples. Barrett (2015) demonstrated the effectiveness of CTII compared to Pro K for hard tissue DNA extractions in human bone samples that had not been exposed to damaging environments. However, in this experiment, the DNA yields between CTII and Pro K treated samples were not statistically significant. The averaged quantity of L. kempii and B. taurus DNA declined dramatically throughout the 3-year experiment. Interestingly, the DNA concentration for the terrestrial L. kempii samples increased at both the 323- and 664-day time point before declining (Figure 1A). While the increased yield is unclear, the variability of the bone type between time point may have played a role.
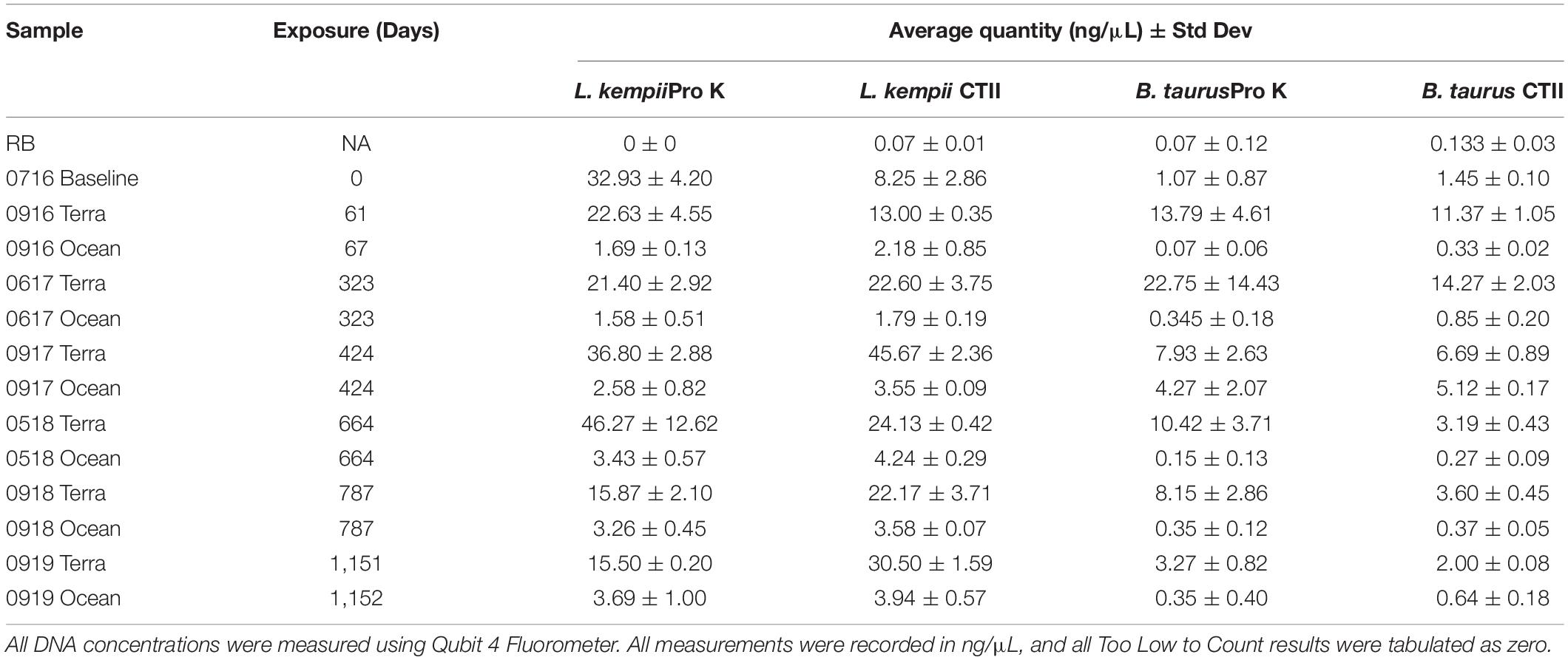
Table 1. Average and standard deviation values for DNA concentrations following DNA extraction from bone for Pro K and CTII treatment.
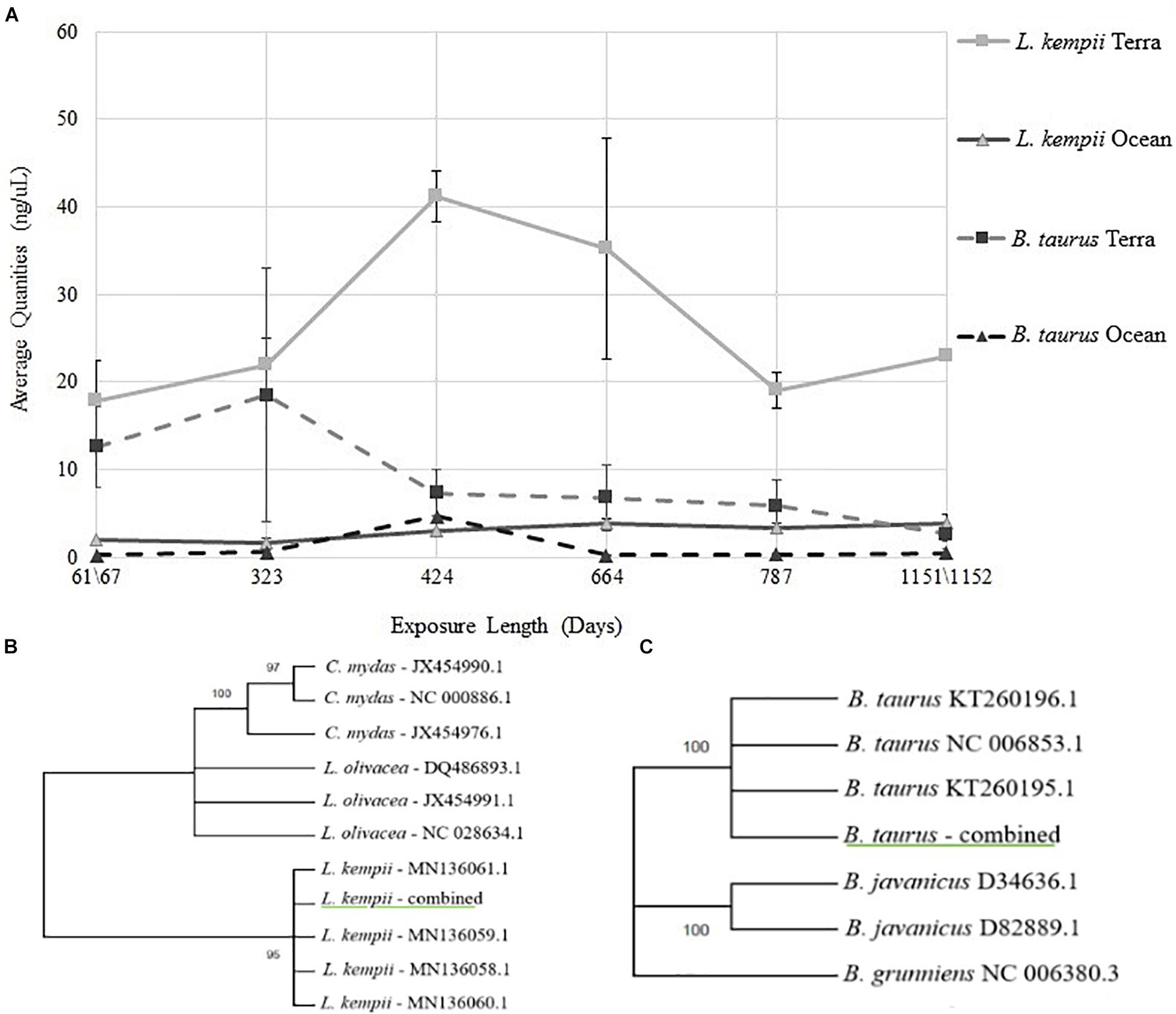
Figure 1. (A) Average DNA quantities recovered from L. kempii and B. taurus bone samples from terrestrial and marine environments over 3-years. DNA extracted from L. kempii and B. taurus bone samples were plotted as a function of exposure length in days ranging from 61/67- to 1151/1152-days. The four experimental conditions represented are L. kempii terrestrial exposure (L. kempii Terra), L. kempii marine exposure (L. kempii Ocean), B. taurus terrestrial exposure (B. taurus Terra), and B. taurus marine exposure (B. taurus Ocean). Because Pro K and CTII treatment groups demonstrated no statistical differences, all data represented in this figure is the aggregate of both the Pro K and CTII treatment groups. (B,C) Experimental DNA sequences generated from DNA extracted from L. kempii and B. taurus bone are underlined in green. All L. kempii and B. taurus sequence data were consistent no matter the experimental condition or time of exposure; therefore, sequence data for each species was simplified to “L. kempii—combined” and “B. Taurus—combined.” Reference sequences used in the construction of all tress are by list genus/species name and then by their GenBank accession number. All trees were constructed using the Hasegawa-Kishino-Yano model; bootstrap values are located at each node. (B) Maximum Likelihood tree for the L. kempii MT-NAD4 genetic marker. (C) Maximum Likelihood tree for the B. taurus MT-CYB genetic marker.
DNA Sequencing and Species Identifications
Throughout the 3-year study, 47 of the 52 experimental conditions and time points tested produced high-quality DNA sequencing results for both L. kempii MT-NAD4 and B. taurus MT-CYB genetic markers (Table 2). Under five experimental conditions, DNA extracted from L. kempii bone failed to produce viable DNA sequences, but the most surprising outcome was that high-quality DNA sequence data was obtained under the most extreme environmental condition tested, submerged for 1,152 days. In general, sequence quality, as measured by phred score, decreased as a function of time for both L. kempii MT-NAD4 and B. taurus MT-CYB gene markers no matter the extraction treatment (Pro K or CTII). At every experimental condition tested, the sequence quality scores for B. taurus MT-CYB varied considerably compared to L. kempii MT-NAD4 results, yet the sequencing quality variation did not affect the production of consensus sequences for these data. Examining the averaged phred scores across all data, one can observe that 41 of the 52 experimental conditions tested produced phred quality scores of 29 or greater (base call accuracy of ∼99.87%). Overall, there was no difference (P = 0.424, one-way ANOVA) in DNA quality between the L. kempii (Pro K mean: 1.4051 and CTII mean: 1.294) and B. taurus (Pro K mean: 1.5351 and CTII mean: 1.4704) samples (log10 of the averages of triplicate samples). All sequencing data produced across the entirety of the experiment, both for L. kempii and B. taurus samples, was consistent with the gene marker reference sequences obtained for NCBI. As such, the abundance of sequence data was simplified to “L. kempii—combined” and “B. taurus—combined” for maximum likelihood analysis. Maximum likelihood trees estimated using the HKY model showed the MT-NAD4 gene marker sequenced from DNA extracted from L. kempii bone samples grouping exclusively (bootstrap = 95) with the L. kempii reference sequences (Figure 1B). Similarly, the B. taurus MT-CYTB gene marker sequences grouped exclusively with B. taurus reference sequences (bootstrap = 100) and not with domestic yak (Bos grunniens) or banteng (Bos javanicus) (Figure 1C). When considered together, one can conclude that if sufficient DNA is extracted from bone samples, then high-quality sequence data can be obtained, and the resulting sequence data accurately reflects the reference sequence for the given gene marker.
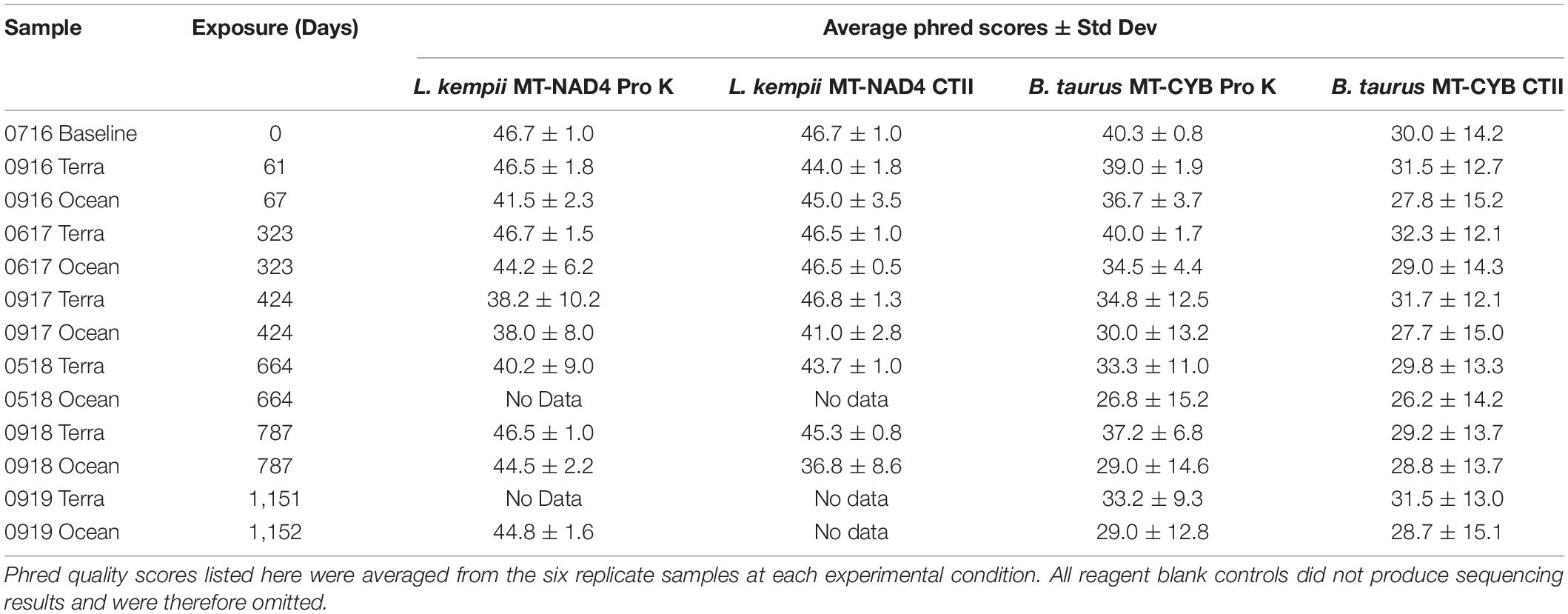
Table 2. Average and standard deviation values for phred scores following DNA sequencing for Pro K and CTII treatment.
Discussion
Mitochondrial and nuclear DNA has been successfully extracted from cetacean and shark remains, even after stranding incidents or many years on display as trophies (Morin et al., 2007; Ahonen and Stow, 2008). A pilot study conducted a controlled experiment using freshwater tubs exposed to the environment to determine individual kits’ success rate at extracting nuclear DNA (nuDNA) from waterlogged pig bones over 6 months. Cartozzo et al. (2018) observed a decrease in DNA quality from each porcine sample location over the duration, which they attributed to the hydrolysis of the DNA while submersed. Additionally, successful extractions of mtDNA that were of a high enough quality for species identification have been achieved from avian skeletal remains exposed to warm, shallow, hypersaline lagoon environments for up to 3 months (Koczur et al., 2017). Recently, a shipment of ivory was recovered after being shipwrecked for almost 500 years. de Flamingh et al. (2020) found that 71% of the tusks recovered had enough nuclear and mitochondrial DNA for species and population identifications. There have even been skeletal nuDNA identifications from human remains found fully submerged in freshwater environments after 3 years (Crainic et al., 2002; Cavalcanti et al., 2017) and partial success after 4 years for nuDNA identifications in saltwater environments (Fredericks et al., 2013). However, few studies of skeletal DNA extractions, human or marine animal, have been conducted with an extended time series or with consistent environmental exposure between samples to evaluate environmental effects on DNA quantity and sequence quality.
Previous work has demonstrated the detrimental effect of saltwater on human DNA recovery (Borde et al., 2008; Frippiat et al., 2017; Helmus et al., 2018; Meixner et al., 2020). Our results support the tenant that terrestrial samples generally yield higher DNA quantities than samples experiencing prolonged exposure to marine environments. Additionally, we found that the use of mtDNA gene markers to identify skeletal remains of Kemp’s ridley sea turtles is possible and accurate up to 3 years in maritime-terrestrial and submerged marine environments, however, the consistency of obtaining a sequencing result declined dramatically at and after 664 days of environmental exposure. In general, when comparing DNA quantity and sequencing quality, a correlation was not observed; however, obtaining a high-quality DNA sequencing result that can resolve species identity remains a possible and even probable outcome following 3 years of terrestrial and marine environmental exposure.
The increase in the amount of DNA extracted after 1 year was an unexpected result but may best be explained by variation in cortical thickness, non-source-sample contamination, and/or increased demineralization efficiency on more degraded samples. A positive relationship between cortical thickness and DNA extraction has been well documented in human skeletal remains (Latham and Miller, 2019) and might explain the variation observed in our study. Our study did not account for the contribution that bacterial genomes might have had to the overall DNA yields. Even though our bone DNA extraction protocol cleaned the bone’s external surface, it is unknown how pervasive bacteria are in 3-year-old bone samples. The observation that more DNA was extracted from terrestrial samples than their marine counterparts indicates that the coextraction of bacterial genomes is a real possibility. Surprisingly, the L. kempii samples in both environments yielded more DNA than the B. taurus samples, despite having thinner bones. The difference in bone morphology is the logical cause of these differences, and additional studies are needed to determine the role that bone physiology plays on DNA extraction processes.
Despite previous studies demonstrating the detrimental effects of saltwater on skeletal DNA (Perry et al., 1988; Schwartz et al., 1991; Alaeddini et al., 2010), our study shows that even with a lower concentration of extracted DNA, the marine L. kempii DNA sequences had comparable or better phred quality scores than terrestrial samples with higher DNA yields. Our DNA sequencing results revealed two important aspects related to DNA sequencing quality and accuracy following environment exposure: (1) the sequencing quality of the L. kempii MT-NAD4 or B. taurus MT-CYB gene markers is not affected by prolonged exposure, and (2) marine environments do not artificially alter the DNA sequence that is obtained. It is logical to conclude that the marine environment is not as detrimental to DNA as once thought. Additional research is needed, but consistent colder temperatures and lower UV radiation might begin to explain the persistence of DNA in marine environments. While our study cannot predict which experimental conditions will yield a DNA sequence, we can state that if sufficient DNA is extracted from bone samples, then high-quality sequence data can be obtained, and the resulting sequence data accurately reflects the reference sequence for the given gene marker.
Conclusion
In conclusion, mtDNA can be used to distinguish skeletal remains of the critically endangered Kemp’s ridley (L. kempii) from the closely related and vulnerable olive ridley (L. olivacea) after 3 years of continuous subtidal or supratidal exposure. For L. kempii samples, the quantity of DNA extracted from bone diminished as a function of time in both the marine and terrestrial environments, but DNA quantity was not predictive of a high-quality DNA sequencing result. Overall, when comparing the effects of prolonged subtidal or supratidal exposure to a marine reptile and a terrestrial mammal, marine exposure negatively impacts the amount of DNA extracted from both organisms. This study’s significance resides in the accurate identification of a challenging biological substrate (bone) under challenging conditions (subtidal or supratidal exposure) using a mitochondrial gene marker after 3 years. These findings further inform forensic and conservation communities that current DNA analysis techniques are potentially viable on ossified tissues following prolonged exposure to seasonally dynamic maritime environmental challenges; therefore, the breadth of biological substrates on which DNA analysis can be performed should be significantly expanded. Utilizing the information in this study, discovered sea turtle skeletal remains can now be more successfully analyzed, identified, and linked back to migration routes or poaching hotspots, thereby furthering our understanding of both natural and illegal mortality events adversely impacting Kemp’s ridley individuals and populations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW403879, MW403880, MW403881, MW403882, MW403883, MW403884, MW403885, MW403886, MW403887, MW403888, MW403889, MW403890, MW403891, MW403892, MW403893, MW403894, MW403895, MW403896, MW403897, MW403898, MW403899, MW403900, MW403901, MW403902.
Ethics Statement
Ethical review and approval was not required for the animal study because two sea turtles that had expired naturally were utilized by this study. Bone samples were obtained post-necropsy (NOAA Greater Atlantic Region ESA Permit TE01150C). The domestic cow sample was collected locally.
Author Contributions
These findings constitute an ongoing research and educational collaboration of the University of Central Oklahoma Center for Wildlife Forensic Science and Conservation Studies, the Shoals Marine Laboratory, and the Kirkpatrick Foundation. The study comprised the foundation for the W. Roger Webb Forensic Science Institute graduate studies of EK. Study conceptualization, implementation, and graduate program co-directorship was the purview of WL and JPC. Field logistics and specimen recovery were facilitated by JAC. Molecular analyses and data interpretation were assisted by MH. Sea turtle procurement and regulatory liaison was provided by KS. All authors contributed to data elucidation and manuscript preparation.
Funding
We would like to acknowledge significant research funding from the Kirkpatrick Foundation. The Kirkpatrick Foundation’s dedication to the conservation, protection and humane treatment of animals is unmatched, and we thank them for their support of our project as we work to protect the world’s endangered wildlife. Additionally, we would like to acknowledge the funding support from the University of Central Oklahoma’s Student Transformative Learning Record (STLR), a Department of Education Title III grant program. Field logistical and facilities support was provided by the Shoals Marine Laboratory. This study constitutes Shoals Marine Laboratory research contribution number 192.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research represents a collaborative effort of the Kirkpatrick Foundation; University of Central Oklahoma Center for Wildlife Forensic Science and Conservation Studies (C-FACS); NOAA, National Marine Fisheries Service Greater Atlantic Regional Fisheries Office (GARFO); New England Aquarium (NEQ); US Fish and Wildlife Service (USFWS); National Fish and Wildlife Forensic Laboratory (NFWFL); Shoals Marine Laboratory (SML) and University of New Hampshire Veterinary Diagnostics Laboratory. We would like to specifically thank, Dr. Brandt Cassidy (C-FACS), Dr. Amanda Waters (UCO), Dr. Sarah Morris (UCO). Dr. Inga Sidor (UNH), Robert D. Kenney (URI-GSO), Dr. Ed Espinoza (NFWFL), Abby Gleb (USFWS), Nicolle Hunter (NEA), Dr. Jennifer Seavey (SML), and Michael Rosen (SML) for their valued assistance at various stages in the research process. All data presented here was originally collect in conjunction with Field Wildlife Forensic Science, a C-FACS/SML collaborative field course and submitted as a master’s thesis with the University of Central Oklahoma (Krestoff, 2020).
Footnotes
- ^ https://www.ndbc.noaa.gov/view_climplot.php?station=iosn3&meas=at
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://www.ibm.com/products/spss-statistics
References
Abreu-Grobois, A., and Plotkin, P. (2008). Lepidochelys olivacea. The IUCN Red List of Threatened Species 2008 [Online]. Gland: IUCN.
Ahonen, H., and Stow, A. J. (2008). Shark jaws and teeth: an unexploited resource for population genetic studies. J. Fish Biol. 73, 450–455. doi: 10.1111/j.1095-8649.2008.01896.x
Alaeddini, R., Walsh, S. J., and Abbas, A. (2010). Molecular studies of time- and environment-dependent effects on bone DNA survival. Aust. J. Forensic Sci. 42, 211–220. doi: 10.1080/00450611003758264
Al-Kandari, N. M., Singh, J., and Sangar, V. C. (2016). Time-dependent effects of temperature and humidity on quantity of DNA in samples of human saliva, blood and semen in Kuwait. International J. Pharm. Sci. Res. 7, 2852–2873. doi: 10.13040/ijpsr.0975-8232.7(7).2852-73
Allard, M. W., Miyamoto, M. M., Bjorndal, K. A., Bolten, A. B., and Bowen, B. W. (1994). Support for natal homing in green turtles from mitochondrial DNA sequences. Copeia 1994, 34–41. doi: 10.2307/1446668
Anderson, G. S., and Bell, L. S. (2014). Deep coastal marine taphonomy: investigation into carcass decomposition in the saanich inlet, British Columbia using a baited camera. PLoS One 9:e110710. doi: 10.1371/journal.pone.0110710
Barrett, L. C. (2015). Effect of Collagenase Type 2 and Proteinase K Digestion on DNA Yield from Bone Samples Purified on the EZ1 Advanced XL. Doctoral dissertation.
Bolker, B. M., Okuyama, T., Bjorndal, K. A., and Bolten, A. B. (2007). Incorporating multiple mixed stocks in mixed stock analysis: ‘Many-to-Many’ analyses. Mol. Ecol. 16, 685–695. doi: 10.1111/j.1365-294X.2006.03161.x
Borde, Y., Tonnany, M., and Champod, C. (2008). A study on the effects of immersion in river water and seawater on blood, saliva, and sperm placed on objects mimicking crime scene exhibits. Can. Soc. Forensic Sci. J. 41, 149–163.
Cartozzo, C., Singh, B., Boone, E., and Simmons, T. (2018). Evaluation of DNA extraction methods from waterlogged bones: a pilot study. J. Forensic Sci. 63, 1830–1835. doi: 10.1111/1556-4029.13792
Cavalcanti, P., Carvalho, F., Carvalho, E. F., and Silva, D. A. (2017). A mini-primer set in a multiplex PCR fashion covering the MTDNA control region from submerged skeletal remains. Forensic Sci. Int. Genet. Suppl. Ser. 6, E469–E470. doi: 10.1016/j.fsigss.2017.09.194
Collins, R. A., Wangensteen, O. S., O’Gorman, E. J., Mariani, S., Sims, D. W., and Genner, M. J. (2018). Persistence of environmental DNA in marine systems. Commun. Biol. 1:185. doi: 10.1038/s42003-018-0192-6
Crainic, K., Paraire, F., Leterreux, M., Durigon, M., and de Mazancourt, P. (2002). Skeletal remains presumed submerged in water for three years identified using PCR-STR analysis. J. Forensic Sci. 47, 1025–1027.
de Flamingh, A., Coutu, A., Sealy, J., Chirikure, S., Bastos, A. D. S., Libanda-Mubusisi, N. M., et al. (2020). Sourcing elephant ivory from a sixteenth-century portuguese shipwreck. Curr. Biol. 31, 621–628.e4. doi: 10.1016/j.cub.2020.10.086
Dissing, J., Sondervang, A., and Lund, S. (2010). Exploring the limits for the survival of DNA in blood stains. J. Forensic Leg. Med. 17, 392–396. doi: 10.1016/j.jflm.2010.08.001
DNA Solutions (2016). Bone Demineralization and Isolation Protocol (B. Cassidy, Personal Communication, July 1, 2019).
Duchene, S., Frey, A., Alfaro-Núñez, A., Dutton, P. H., Thomas, P., Gilbert, M., et al. (2012). Marine turtle mitogenome phylogenetics and evolution. Mol. Phylogenet. Evol. 65, 241–250. doi: 10.1016/j.ympev.2012.06.010
Dutton, P. H., LeRoux, R. A., LaCasella, E. L., Seminoff, J. A., Eguchi, T., and Dutton, D. L. (2019). Genetic analysis and satellite tracking reveal origin of the green turtles in San Diego Bay. Mar. Biol. 166, 1–13. doi: 10.1007/s00227-018-3446-4
Eichmiller, J. J., Best, S. A. E., and Sorensen, P. W. (2016). Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 50, 1859–1867.
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048.
Frandsen, H. R., Figueroa, D. F., and George, J. A. (2020). Mitochondrial genomes and genetic structure of the kemp’s ridley sea turtle (Lepidochelys kempii). Ecol. Evol. 10, 249–262. doi: 10.1002/ece3.5891
Fredericks, J., Brown, K., Williams, A., and Bennett, P. (2013). DNA analysis of skeletal tissue recovered from the english channel. J. Forensic Leg. Med. 20, 757–759. doi: 10.1016/j.jflm.2013.04.013
Frippiat, C., Gastaldi, A., and Van Grunderbeeck, S. (2017). Persistence of immersed blood and hair DNA: a preliminary study based on casework. J. Forensic Leg. Med. 51, 1–8. doi: 10.1016/j.jflm.2017.07.009
Gredzens, C., and Shaver, D. J. (2020). Sattelite tracking can inform population-level dispersal to foraging grounds of post-nesting kemp’s ridley sea turtles. Front. Mar. Sci. 7:559. doi: 10.3389/fmars.2020.00559
Hall, A., Sims, L. M., and Ballantyne, J. (2014). Assessment of DNA damage induced by terrestrial UV irradiation of dried bloodstains: forensic implications. Forensic Sci. Int. Genet. 8, 24–32. doi: 10.1016/j.fsigen.2013.06.010
Helmus, J., Zorell, S., Bajanowski, T., and Poetsch, M. (2018). Persistence of DNA on clothes after exposure to water for different time periods-a study on bathtub, pond, and river. Int. J. Leg. Med. 132, 99–106. doi: 10.1007/s00414-017-1695-2
Houssaye, A. (2012). Bone histology of aquatic reptiles: what does it tell us about secondary adaptation to an aquatic life? Biol. J. Linn. Soc. 108, 3–21. doi: 10.1111/j.1095-8312.2012.02002.x
Jones, K., Jensen, M., Burgess, G., Leonhardt, J., van Herwerden, L., Hazel, J., et al. (2018). Closing the gap: mixed stock analysis of three foraging populations of green turtles (Chelonia mydas) on the Great Barrier Reef. Peerj 6:e5651. doi: 10.7717/peerj.5651
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Koczur, L. M., Williford, D., DeYoung, R. W., and Ballard, B. M. (2017). Back the dead: genetic data from avian carcasses. Wildl. Soc. Bull. 41, 796–803. doi: 10.1002/wsb.823
Krestoff, E. S. (2020). Nuclear and Mitochondrial DNA Quantity and Quality Evaluation of Kemp’s Ridley Sea Turtle (Lepidochelys Kempii) Bones After Oceanic and Terrestrial Environmental Exposure. Edmond, OK: M.Sc. University of Central Oklahoma.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Latham, K. E., and Miller, J. J. (2019). DNA recovery and analysis from skeletal material in modern forensic contexts. Forensic Sci. Res. 4, 51–59.
Liu, X., Manning, J., Prescott, R., Page, F., Zou, H., and Faherty, M. (2019). On simulating cold-stunned sea turtle strandings on Cape Cod, Massachusetts. PLoS ONE 14:e0204717. doi: 10.1371/journal.pone.0204717
Lord, W. D., and Burger, J. F. (1984b). Arthropods associated with herring gull (Larus argentatus) and Great Black-backed Gull Carrion on Islands in the Gulf of Maine. Environ. Entomol. 13, 1261–1268. doi: 10.1093/ee/13.5.1261
Lord, W. D., and Burger, J. F. (1984a). Arthropods associated with harbor seal (Phoca vitulina) carcasses stranded on islands along the New England Coast. Int. J. Entomol. 26, 282–285.
McElreath, C. B. (2018). Comparative Subtidal and Supratidal Taphonomic Changes in the Elemental Components of Marine Vertebrate Bones Using Energy Dispersive x-Ray Spectroscopy and Principle Component Analysis: Ecological and Forensic Application. Master’s thesis.
Meixner, E., Kallupurackal, V., Kratzer, A., Voegeli, P., Thali, M. J., and Bolliger, S. A. (2020). Persistence and detection of touch DNA and blood stain DNA on pig skin exposed to water. Forensic Sci. Med. Pathol. 16, 243–251. doi: 10.1007/s12024-020-00234-3
Morin, P. A., Hedrick, N. M., Robertson, K. M., and Leduc, C. A. (2007). Comparative mitochondrial and nuclear quantitative PCR of historical marine mammal tissue, bone, baleen, and tooth samples. Mol. Ecol. Notes 7, 404–411. doi: 10.1111/j.1471-8286.2007.01699.x
Nakajima, Y., Hirayama, R., and Endo, H. (2014). Turtle humeral microanatomy and its relationship to lifestyle. Biol. J. Linn. Soc. 112, 719–734. doi: 10.1111/bij.12336
National Marine Fisheries Service [NMFS], and U.S. Fish and Wildlife Service [FWS] (2015). Kemp’s Ridley Sea Turtle (Lepidochelys kempii)5-year Review: Summary and Evaluation. Silver spring, MD: National Marine Fisheries Service, 64.
Nichols, W. F., and Nichols, V. C. (2008). Nichols, W. F., and Nichols, V. C. (2008). The land use history, flora, and natural communities of the isles of shoals, rye, new hampshire and kittery, maine. Rhodora 110, 245–295.
Patricio, A. R., Formia, A., Barbosa, C., Broderick, A. C., Bruford, M., Carreras, C., et al. (2017). Dispersal of green turtles from africa’s largest rookery assessed through genetic markers. Mar. Ecol. Prog. Ser. 569, 215–225. doi: 10.3354/meps12078
Paul, E., and Sikes, R. S. (2013). Wildlife researchers running the permit maze. ILAR J. 54, 14–23. doi: 10.1093/ilar/ilt013
Perry, W. L., Bass, W. M., Riggsby, W. S., and Sirotkin, K. (1988). The autodegradation of deoxyribonucleic acid (DNA) in human rib bone and its relationship to the time interval since death. J. Forensic Sci. 33, 144–153.
Ramirez, M. D., Avens, L., Goshe, L. R., Snover, M. L., Cook, M., and Heppell, S. S. (2020). Regional variation in Kemp’s ridley sea turtle diet composition and its potential relationship with somatic growth. Front. Mar. Sci. 7:253. doi: 10.3389/fmars02020.00253
Rhodin, A. G. J. (1985). Comparative chondro-osseous development and growth of marine turtles. Copeia 3, 752–771. doi: 10.2307/1444768
Sassoubre, L. M., Yamahara, K. M., Gardner, L. D., Block, B. A., and Boehm, A. B. (2016). Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ. Sci. Technol. 50, 10456–10464.
Schwartz, T. R., Schwartz, E. A., Mieszerski, L., McNally, L., and Kobilinsky, L. (1991). Characterization of deoxyribonucleic acid (DNA) obtained from teeth subjected to various environmental-conditions. J. Forensic Sci. 36, 979–990.
Sowers, D., Dijkstra, J. A., Masetti, G., Mayer, L. A., Mello, K., and Malik, M. A. (2020). “Application of the coastal and marineecological classification standard to Gosnold Seamount, North Atlantic Ocean,” in Seafloor Geomorphology As Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitat, 2nd Edn, eds P. T. Harris and E. Baker (Amsterdam: Elsevier), 903–916.
Keywords: mitochondrial DNA, species identification, Lepidochelys kempii, environmental exposure, skeletal DNA extraction
Citation: Krestoff ES, Creecy JP, Lord WD, Haynie ML, Coyer JA and Sampson K (2021) Mitochondrial DNA Evaluation and Species Identification of Kemp’s Ridley Sea Turtle (Lepidochelys kempii) Bones After a 3-Year Exposure to Submerged Marine and Terrestrial Environments. Front. Mar. Sci. 8:646455. doi: 10.3389/fmars.2021.646455
Received: 26 December 2020; Accepted: 26 May 2021;
Published: 18 June 2021.
Edited by:
Fengping Wang, Shanghai Jiao Tong University, ChinaReviewed by:
Dong Dong, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaXupeng Cao, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, China
Copyright © 2021 Krestoff, Creecy, Lord, Haynie, Coyer and Sampson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James P. Creecy, amNyZWVjeUB1Y28uZWR1
 Elizabeth S. Krestoff
Elizabeth S. Krestoff James P. Creecy
James P. Creecy Wayne D. Lord1,2,3,4
Wayne D. Lord1,2,3,4 Kate Sampson
Kate Sampson