- 1Reef Biology Research Group, Department of Marine Science, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 2Department of Microbiology, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 3Microbiome Research Unit for Probiotics in Food and Cosmetics, Chulalongkorn University, Bangkok, Thailand
- 4Centre National de la Recherche Scientifique, Institut de Chimie des Substances Naturelles ICSN, Gif-sur-Yvette, France
- 5Department of Marine Science, Faculty of Science, Center of Excellence for Marine Biotechnology, Chulalongkorn University, Bangkok, Thailand
Global warming has caused elevated seawater temperature and coral bleaching, including events on shallow reefs in the upper Gulf of Thailand (uGoT). Previous studies have reported an association between loss of zooxanthellae and coral bleaching. However, studies on the microbial diversity of prokaryotes and eukaryotes (microbiome) as coral holobionts are also important and this information is still limited in the uGoT. To address this shortcoming, this report provided baseline information on the prokaryotic (bacteria and archaea) and eukaryotic microbes of healthy and bleached colonies of four prevalent corals Acropora humilis, Acropora millepora, Platygyra sinensis, and Porites lutea and surrounding seawater and sediments, using 16S and 18S rRNA gene next-generation sequencing. Both prokaryotic and eukaryotic microbes showed isolated community profiles among sample types (corals, sediment, and seawater) (ANOSIM: P < 0.001, R = 0.51 for prokaryotic profiles and P < 0.001, R = 0.985 for eukaryotic microbe profiles). Among coral species, P. sinensis showed the most diverse prokaryotic community compared with the others (ANOSIM: P < 0.001, R = 0.636), and P. lutea showed the most diverse eukaryotic microbes (P = 0.014, R = 0.346). Healthy and bleached corals had some different microbiomes in species and their prevalences. For instance, the significant increase of Alphaproteobacteria in P. sinensis resulted in reduced prokaryotic community evenness and altered potential metabolic profiles (i.e., increased amino acid metabolism and genetic information processing and transcription, but decreased prokaryotic functions in cell motility, signaling, and transduction). For eukaryotic microbes, the loss of the algal Symbiodinium (colloquially known as zooxanthellae) in bleached corals such as P. lutea resulted in increased Chromista and Protista and, hence, clearly distinct eukaryotic microbe (including fungi) communities in healthy vs. bleached colonies of corals. Bleached corals were enriched in bacterial pathogens (e.g., Acinetobacter, Helicobacter, Malassesia, and Aspergillus) and decreased coral-beneficial prokaryotic and eukaryotic microbes (e.g., Rhizobiales and Symbiodinium). Additionally, this study identified microbiome species in bleached P. lutea that might help bleaching recovery (e.g., high abundance of Rhizobiales, Oceanospirillales, Flavobacteriales, and Alteromonadales). Overall, our coral-associated microbiome analyses identified altered diversity patterns of bacteria, archaea, fungi, and eukaryotic microbes between healthy and bleached coral species that are prevalent in the uGoT. This knowledge supports our ongoing efforts to manipulate microbial diversity as a means of reducing the negative impacts of thermal bleaching events in corals inhabiting the uGoT.
Introduction
Coral reefs represent one of the most productive and biodiverse ecosystems on earth, when normalized per unit area, than any other marine environments (Hatcher, 1990). Coral reefs provide many ecosystem services including being the major marine nutrient resources (carbon and nitrogen supplies) and habitat to many diverse marine organisms. Additionally, coral reefs are a source of bioactive compounds and provide coastal protection against waves and floods (Rosenberg et al., 2007; Burke et al., 2012; Bourne et al., 2013). However, during the past few decades, coral reefs have been extensively endangered by anthropogenic activities, directly (e.g., overfishing and coral collection) and indirectly (primarily by greenhouse gas pollution) (Hughes et al., 2003; Harvey et al., 2018). Accumulated greenhouse gas pollution affects UV radiation and global climate change (elevated atmosphere and seawater temperatures) and subsequently causes thermal coral bleaching events (Lesser et al., 2004; Burke et al., 2012; Bourne et al., 2013). When seawater temperature is too warm, corals expel their algal holobionts, of which one of the most predominant is Symbiodinium. Symbiodinium, alternatively known as zooxanthellae, are single-cell photosynthetic dinoflagellates that live in symbiosis with corals and several marine invertebrates (Hoegh-Guldberg and Smith, 1989). Symbiodinium spp. provide photosynthetic food to corals and protect corals from pathogens by competing against pathogens for food and space on coral bodies (Lesser et al., 2013). When Symbiodinium is expelled, corals become discolored (white) due to the absence of colored photosynthetic pigments from Symbiodinium; this is referred to as “coral bleaching” and importantly results in loss of food that is normally produced by Symbiodinium to feed the coral (Baker et al., 2008). Consequently, coral growth, reproduction, resistance to disease and stress, and survivability decline (Baker et al., 2008). These thermal bleaching events are considered the most problematic coral situation worldwide, including in the upper Gulf of Thailand (uGoT) that first recorded a thermal bleaching event in 2006 in Sattahip District, Samae San Island, Chon Buri Province (Chavanich et al., 2009).
Thermal events are the most well-known events that cause coral bleaching, but other factors may be involved, such as seawater acidification [which could indirectly be caused from greenhouse gas (CO2) pollution] (Anthony et al., 2011), increased levels of sediment that cover corals, smothering them and/or blocking sunlight (Peters, 1984), and dysbiosis of the coral-associated microbiome (Ritchie, 2006; Bourne et al., 2008a). In addition to Symbiodinium, corals live symbiotically with prokaryotic (bacteria and archaea) and eukaryotic microbes (Rohwer et al., 2002; Rosenberg et al., 2007). Scientists reported that coral prokaryotes, in particular, bacteria, are diverse and include species that are able to either provide food via photosynthesis, acquire and decompose organic and inorganic nutrients, and/or produce antibiotics and antioxidants to boost immunity of corals and promote resistance against pathogens and environmental stress (including coral bleaching). Coral-symbiotic bacteria also compete with coral pathogens for space and nutrients (Lesser et al., 2004; Rosenberg et al., 2007; Lema et al., 2012; Webster et al., 2016, Webster and Reusch, 2017).
For coral-associated fungi, some reports suggested their symbiotic roles in coral skeletal biomineralization (Le Campion-Alsumard et al., 1995), nitrogen fixation (Wegley et al., 2007), and UV protection (Dunlap and Shick, 1998), while others act as pathogens such as Aspergillus sydowii (Geiser et al., 1998; Smith and Weil, 2004), Rhytisma acernium, and Stephanocoenia intersepta (Sweet et al., 2013; Meyer et al., 2016). Other eukaryotic microbes, besides Symbiodinium, have been less documented, but they likely play major roles as photosynthesis and food providers, and/or coral resilience support to environmental stresses (Kramarsky-Winter et al., 2006; Harel et al., 2008) and disease (Bourne et al., 2008b; Sweet et al., 2013) is possible.
Profiling coral-associated prokaryotic and eukaryotic microbes (microbiome) is an important step required to understand coral holobionts and how, or if, these communities regulate coral health (e.g., against thermal bleaching). In Thailand, coral-associated bacteria and fungi microbiome studies have been limited. Thailand coral reefs are fringe type with three dominant coral genera: Acropora, Platygyra, and Porites (Phongsuwan et al., 2013). During the past few decades, corals have been reported to be continuously reduced in abundance, aerial coverage, and general health. For example, in Mun Island (Rayong Province) and Chang Island (Trat Province), the reported decreases ranged from approximately 37.4% in 1995 to 33.3% in 2006 and 22.2% in 2011, with more severe declines noted in 2011 related to thermal bleaching events (Phongsuwan et al., 2013; Pengsakun et al., 2019). Because the declines in corals have been continuous and are growing worse with time in the uGoT, we undertook studies aimed to reveal the healthy and bleached, coral-associated microbiome profiles to support restoration of Thailand coral reefs.
This report utilized 16S and 18S rRNA gene next-generation sequencing (NGS), to firstly identify both prokaryotic and eukaryotic microbe communities associated with healthy and bleached corals that are dominant in the uGoT (i.e., Acropora humilis, Acropora millepora, Platygyra sinensis, and Porites lutea), along with the microbiomes of the surrounding seawater and sediments, during a thermal bleaching event in 2016. Our studies included comparative alpha and beta community diversity analyses, correlation analyses, and comparisons of metabolic potentials of prokaryotic communities at three study sites in the uGoT [Tao Mo Island (T), Khao Ma Cho (M), and Samae San Island (S), in Sattahip District, Chon Buri Province]. Overall, we consider the microbiome knowledge gained from this study to be a crucial part of our understanding of coral reef health in the uGoT, which over time, will help us devise microbe-mediated strategies to protect corals from thermal bleaching events.
Materials and Methods
Sample Collections
Samples including coral species A. humilis (AH), A. millepora (AM), P. sinensis (PS), and P. lutea (PL), as well as sediment (S) and seawater (W), were collected from Tao Mo Island (T) (12°38′35.2″N, 100°51′43.3″E), Khao Ma Cho (M) (12°35′50.4″N, 100°56′52.5″E), and Samae San Island (S) (12°34′30.33″N, 100°57′29.55″E), in Sattahip District of Chon Buri Province, Thailand, during a midday of the great global thermal bleaching event in June–July 2016 (Figure 1). During the period of sample collections, the seawater temperature was approximately 32°C (minimum 30.66°C and maximum 33.74°C) at all three sites. For each coral species, healthy (H) and bleached (B) colonies were collected. Healthy and bleached coral colonies were determined via their appearance (i.e., white color for bleached coral) by on-site marine scientist divers (Chavanich, Jandang, and Viyakarn) ([opetwcitep]B9,B10[clotwcitep]Bulan et al., 2018a, b). At least three independent colonies of each species were sampled in each location. For each sample, a fragment approximately 5 cm in length and 5 cm in diameter was collected. The distance between each sampled colony was approximately 5 m. After a sample was collected underwater, it was placed in a plastic bag individually. For sediment samples, at least three samples (approximately 50 g of each) were collected just below each of the sampled coral colonies. Similar to the sediment samples, at least 3 L of seawater samples were collected directly above each of the sampled coral colonies. All samples were transported immediately to the laboratory and stored at −20°C. The abbreviations of the samples are as follows: sample (AH, AM, PS, PL, S, or W) followed by coral condition (H or B) (for coral), site (S, M, or T), and independent replicate number (1, 2, or 3). For instance, PLHS1 and PLBS3 represent coral P. lutea at Samae San Island of healthy replicate 1 and bleached replicate 3, respectively.
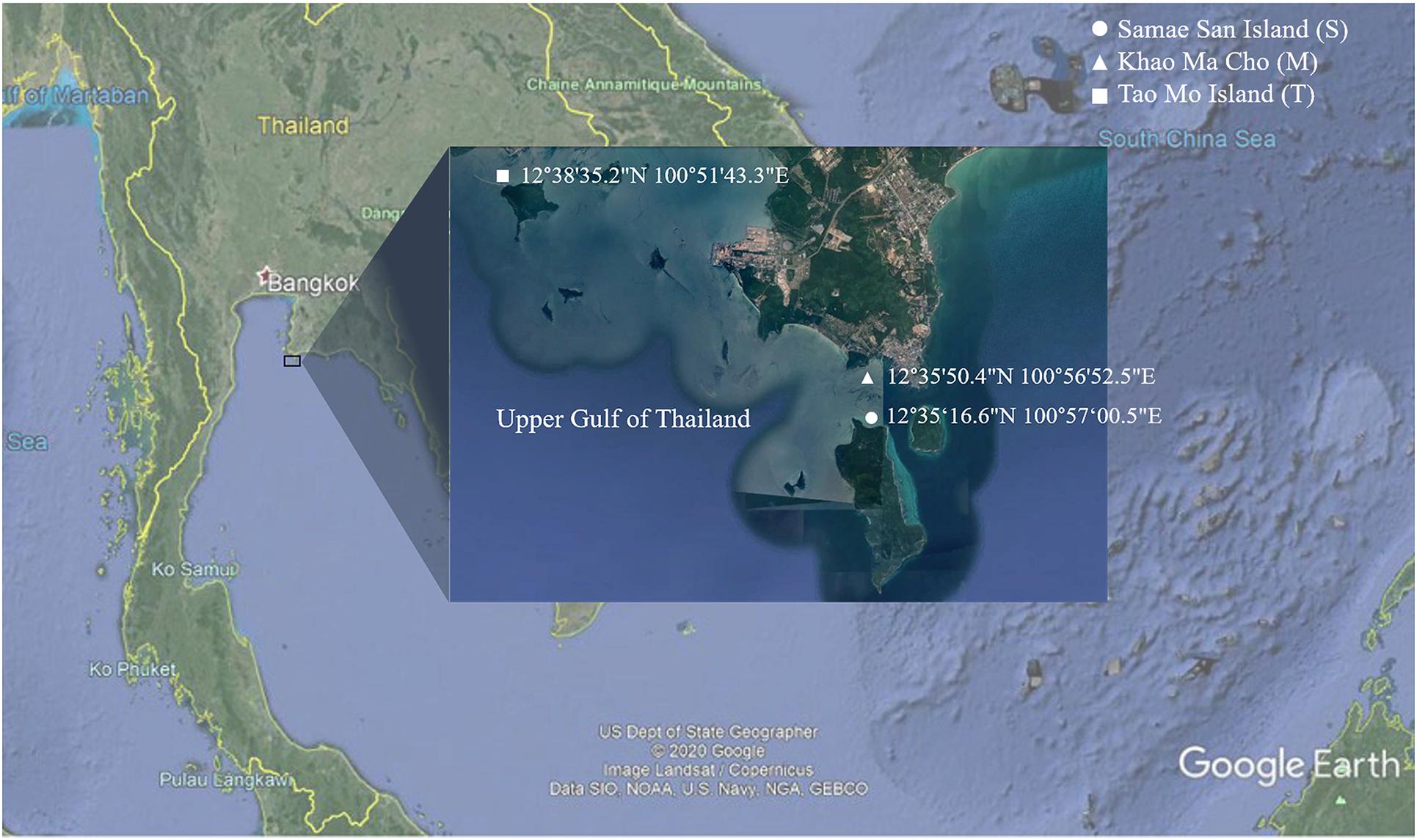
Figure 1. Map representing sampling sites of Samae San Island, Khao Ma Cho, and Tao Mo Island, in the upper Gulf of Thailand.
DNA Extraction
Metagenomes were extracted using Power Soil DNA Isolation Kit (for coral and sediment samples) and Power Water DNA Isolation Kit (seawater samples) (MoBio, Carlsbad, CA, United States), following the manufacturer’s instructions and previous literature ([opetwcitep]B9,B10[clotwcitep]Bulan et al., 2018a, b). The coral sample was ground using sterile mortar and pestle, and 1 g of coral and 1 g of sediment were used. For seawater, 2.5 L was filtered using a sterile 0.22-μm filter membrane (Merck Millipore, Burlington, MA, United States) and the filtered membrane was used. The quality and quantity of the extracted metagenomic DNA were checked by agarose gel electrophoresis (0.55% agarose gel w/v) and NanoDrop spectrophotometry, respectively.
16S and 18S rRNA Gene Library Preparation and Next-Generation Sequencing
Libraries of the V4 region of the 16S rRNA gene (for bacteria and archaea) and the V9 region of the 18S rRNA gene (for fungi and other small eukaryotes) were prepared by polymerase chain reaction (PCR) according to Caporaso et al. (2012). The universal prokaryotic primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) and the universal eukaryotic primers Illumina_Euk_1391F (5′-GTACACACCGCCCGTC-3′) and lllumina_EukBr (5′-TGATCCTTCTGCAGGTTCACCTAC-3′), with appended 5′ Illumina adapter and 3′ Golay barcode sequences, were used, respectively. Each 25-μl PCR reaction comprised 1 × EmeraldAmp® GT PCR Master Mix (TaKaRa, Shiga, Japan), 0.3 μM of each primer, and 75 ng of the metagenomic DNA. For the 16S rRNA gene, the PCR conditions were 94°C for 3 min and 30 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 1 min 30 s, followed by 72°C for 10 min. For the 18S rRNA gene, 10 μM of mammal-blocking primer (5′-GCCCGTCGCTACTACCGATTGGIIIIITTAGTGAGGCCCT3S pC3-3′) (Caporaso et al., 2012) was also included in the PCR recipe, and the PCR conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 65°C for 15 s (for mammal blocking primer), 57°C for 30 s (for universal eukaryotic primers), 72°C for 90 s, followed by 72°C for 10 min. Triplicate PCRs were performed and pooled for each sample to prevent stochastic bias. Amplicons of ∼381 bp (16S rDNA) and ∼260 bp (18S rDNA) in length were excised from agarose gels. The amplicons were purified using GF-1 Gel Extraction Kit (Vivantis Technologies Sdn Bhd, Selangor, Malaysia) and quantified with Picogreen using Qubit dsDNA HS assay kit (Invitrogen, Eugene, OR, United States) and Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States). Each barcoded sample (200 ng) was pooled and sequenced on MiSeq 300 NGS platform (Illumina, San Diego, CA, United States) at OMICS Science and Bioinformatics Center, Faculty of Science, Chulalongkorn University (Bangkok, Thailand).
Bioinformatic and Statistical Analyses
Sequences were processed according to Mothur’s standard operating procedures (SOP) (Schloss et al., 2009). For data cleaning, reads containing (i) ambiguous bases, (ii) >1 mismatch base in the primer region, (iii) >10 homopolymer, (iv) sequence length < 100 bp, and (v) chimera sequence were removed. Silva databases (version 1.32) were used to align the sequences and remove contaminated sequences (i.e., mitochondria and chloroplast sequences). Sequences that belong to corals were also removed for 18S rDNA sequences. For taxonomic classification, 16S and 18S rDNA sequences were classified prokaryotic and eukaryotic operational taxonomic units (OTUs) using Silva databases (version 1.32). Alpha diversity (Good’s coverage to estimate a sequencing coverage, Chao1 richness, and Shannon diversity indices) and beta diversity [thetayc dissimilarity index and non-metric multidimensional scaling (NMDS)] at the genus level were determined using Mothur (Schloss et al., 2009). Correlation analysis was performed by RStudio using VEGAN package (Oksanen et al., 2019). Potential metabolic function of the community was predicted using PICRUSt (version 1.1.4) following established protocols (Langille et al., 2013). The metabolic functions were categorized by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. For correlations between coral-associated prokaryotic and eukaryotic genera, the analyses were based on positive (or negative, or no) correlation between the corresponding genera relative frequency percentages.
Mean and standard deviation (mean ± SD) were computed. For statistical analysis, Student’s t-test and analysis of similarities (ANOSIM) were used to test for significant differences between and among groups (P < 0.05) for alpha and beta diversities, respectively. The significant differences for potential metabolic function were tested by White’s non-parametric t-test. Data visualization and statistical analyses were conducted using Microsoft Excel, metastats (Mothur), and RStudio version 1.3.10931.
Results
16S and 18S rRNA Gene Sequencing and Their Alpha Diversities
Quality scores (Q30) of 88.3% for 16S and 89.8% for 18S rRNA gene sequences were retrieved from the NGS runs. Note that Q30 represents an average sequence error rate of 1 in 1,000 or a corresponding base call accuracy of 99.9%; a higher Q30 percentage thereby infers a higher base call accuracy, and Illumina NGS runs should have Q30 score above 70% (Kastanis et al., 2018). After the Mothur’s SOP for quality read process, a total of 1,468,626 quality reads for prokaryotic sequences and 8,005,841 quality reads for eukaryotic microbe sequences were retrieved. The average quality reads per sample were 34,967 and 190,615 for prokaryotic and eukaryotic microbes, respectively. These numbers of quality reads per sample were considered sufficient sequencing depth, because they resulted in the computed Good’s coverage indices at genus-level OTUs to be all above 99%, except AHBT2 (97.36%) and PSHM1 (97.32%) prokaryotic communities (Supplementary Tables 1A,B). To prevent sequencing depth bias, every community profile was normalized to the same sequencing depth (6,846 quality sequences per 16S rRNA gene sample and 18,503 quality sequences per 18S rRNA gene sample) for analyses.
The alpha diversity indices of the samples across different sample types (corals, sediment, and seawater) were compared using Chao and Shannon indices (Supplementary Table 1 and Figure 2). Overall, the diversity of prokaryotes was found to be much greater than that of eukaryotic microbes for coral samples (Supplementary Table 1: prokaryotes avg. 437.25 ± 186 OTUs, eukaryotic microbes avg. 58.63 ± 27.46 OTUs) and sediments (prokaryotes avg. 650.33 ± 30.25 OTUs, eukaryotic microbes avg. 178.11 ± 18.42 OTUs). For seawater, the diversity of prokaryotes remained higher but was closer to that of eukaryotic microbes (prokaryotes avg. 329.56 ± 24.33 OTUs, eukaryotic microbes avg. 217.33 ± 24.99 OTUs).
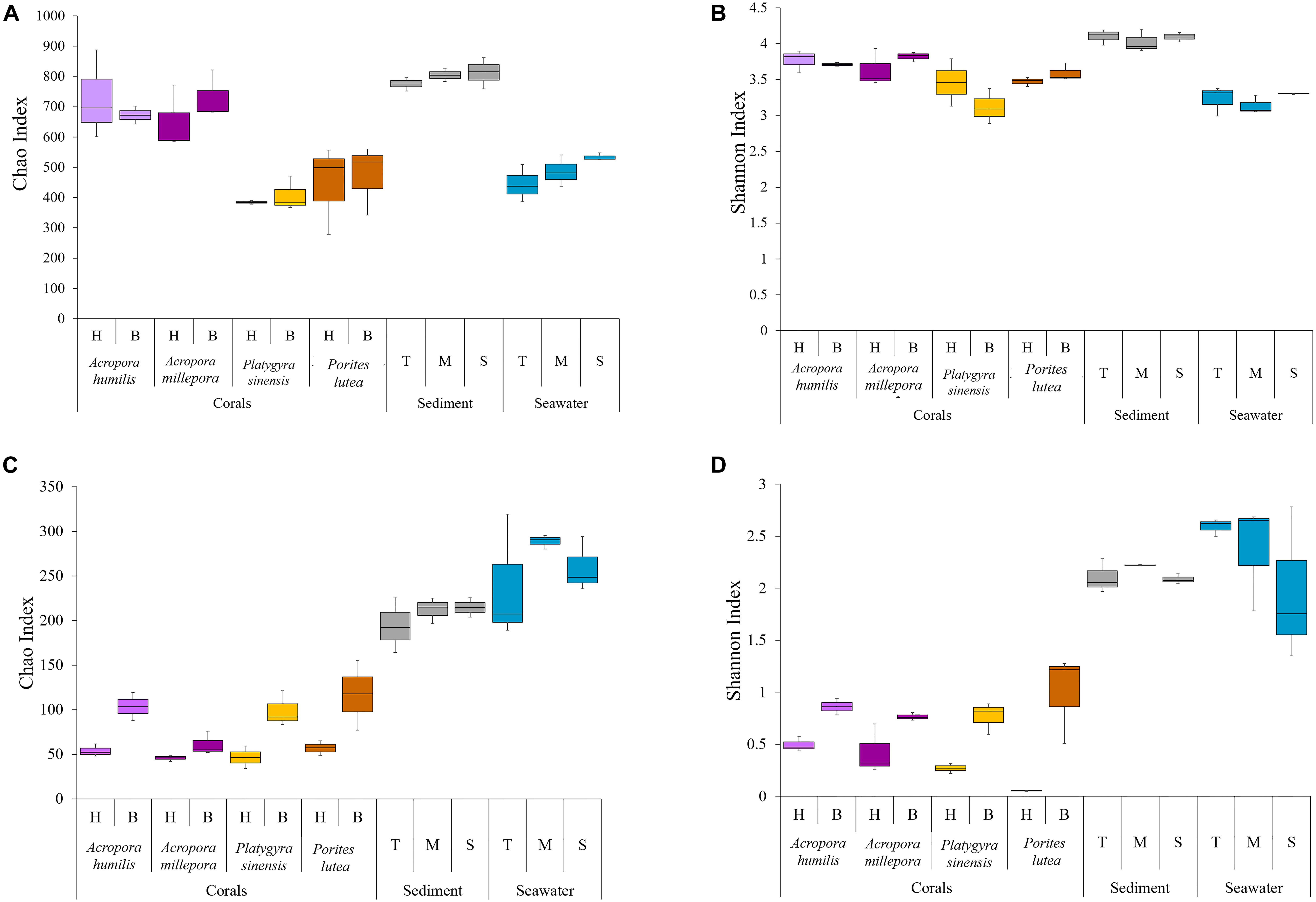
Figure 2. Alpha diversity indices of (A,B) prokaryotic Chao and Shannon and (C,D) small eukaryotic Chao and Shannon.
The greatest relative prokaryotic OTU richness (Chao index) was observed in sediment samples, followed by corals, and seawater represented the least OTU richness. Statistical tests demonstrated significant differences in the prokaryotic OTU diversity between the sediments and the other sample types (ANOSIM: P < 0.01). OTU richness varied among coral species: two Acropora had relatively greater prokaryotic diversity than Porites and Platygyra (Figure 2A: avg. Chao index of A. humilis was 705.63 ± 109.85, A. millepora 689.19 ± 94.85, P. sinensis 397.72 ± 42.0, and P. lutea 458.96 ± 119.44). Significant differences were determined between A. humilis vs. P. sinensis (t-test: P = 0.0002), A. humilis vs. P. lutea (P = 0.003), A. millepora vs. P. sinensis (P < 0.001), and A. millepora vs. P. lutea (P = 0.0021). Nonetheless, OTU evenness (Shannon indices) was found to be relatively similar among samples. No significant differences in Shannon indices were observed between the sediments (avg. 4.08 ± 0.11) and the other samples (3.45 ± 0.33) (P > 0.01), highlighting an evenness of individual distributions of the prokaryotic OTUs in each sample (Figure 2B: avg. Shannon index 3.28).
For eukaryotic microbes, seawater (Figures 2C,D: avg. Chao index 262.22 ± 44.44, avg. Shannon index 2.31 ± 0.53) and sediment (avg. Chao index 207.06 ± 20.41, avg. Shannon index 2.14 ± 0.11) samples had relatively greater alpha diversities than corals (avg. Chao index 74.19 ± 33.27, avg. Shannon index 0.57 ± 0.35). The statistically significant differences were observed when comparing the seawater and sediment samples against the coral samples, in both OTU richness (ANOSIM: P < 0.01) and evenness (P < 0.01). Furthermore, healthy coral samples showed lower alpha diversity than bleached coral samples, for instance, avg. Shannon index of healthy corals was 0.30 ± 0.21 and bleached corals was 0.84 ± 0.23 (Supplementary Table 1 and Figure 2). Statistically significant increases in both OTU richness and evenness between healthy and bleach conditions were found for three coral species: Chao1 indices for A. humillis (t-test: P = 0.027, given mean ± SD was 53.89 ± 6.91 for healthy and 114.28 ± 24.19 for bleached), P. sinensis (P = 0.009, given mean ± SD was 46.06 ± 12.62 for healthy and 98.76 ± 32.48 for bleached), and P. lutea (P = 0.03, given mean ± SD was 56.94 ± 8.42 for healthy and 116.78 ± 39.19 for bleached) and Shannon indices for A. humillis (P = 0.014, given mean ± SD was 0.49 ± 0.07 for healthy and 0.81 ± 0.12 for bleached), P. sinensis (P = 0.006, given mean ± SD was 0.24 ± 0.07 for healthy and 0.77 ± 0.15 for bleached), and P. lutea (P = 0.009, given mean ± SD was 0.05 ± 0.003 for healthy and 1.00 ± 0.429 for bleached).
Community Compositions and Beta Diversity Analyses of Prokaryotic Communities
The percent relative abundance of prokaryotic phylum compositions was found to be relatively close within the same sample types and (for corals) the same coral genus. The three corals (A. humilis, A. millepora, and P. sinensis) and sediment samples shared dominant prokaryotic phyla: Proteobacteria (Figure 3A: avg. 62.33 ± 15.93%) followed by Bacteroidetes (avg. 12.81 ± 7.02%). Seawater samples displayed different compositions: Cyanobacteria (avg. 48.62 ± 7.04%) followed by Proteobacteria (avg. 22.61 ± 2.77%). It is worth noting that the prokaryotic compositions were relatively consistent among sites for sediment and seawater samples (Figure 3A). This finding may support the close sediments and seawater environments among T, M, and S sites, yet a more diverse prokaryotic (in particular bacterial) compositions for corals may partly be owning to the genera of corals.
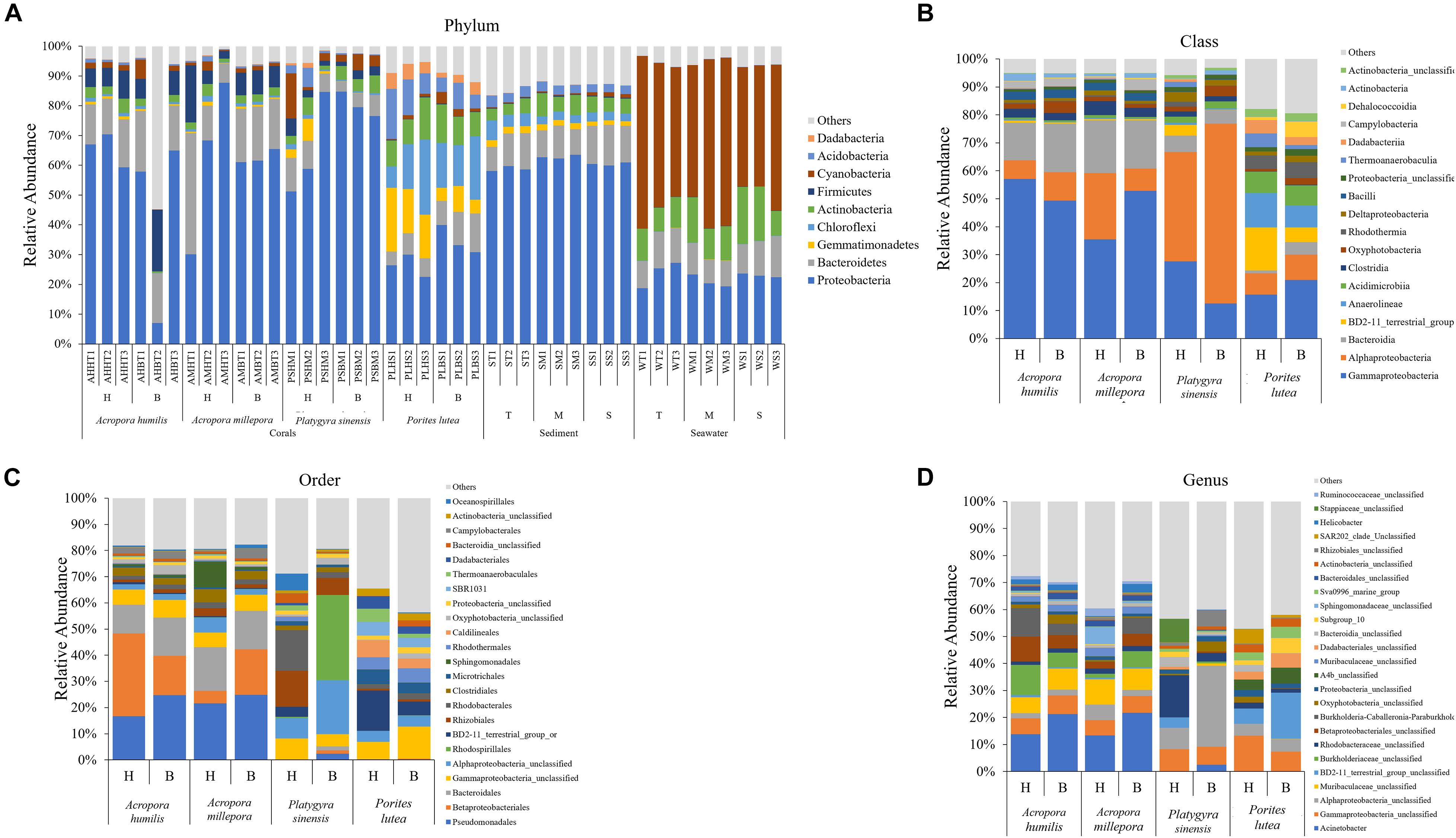
Figure 3. Percent relative abundance of prokaryotic communities analyzed at the (A) phylum, (B) class, (C) order, and (D) genus levels, respectively. Others represent genera with <1% abundance. For operational taxonomic units (out) classification where the genus could not be identified, the deepest classification level was given (g, genus; f, family; c, class; o, order, respectively).
Prokaryotic diversity was analyzed in detail by class, order, and species compositions, between bleached and healthy conditions, and across coral species (Figures 3B–D). Bacterial orders Rhizobiales and Rhodobacterales were found shared across all coral species and were among the top 20 most abundant in all cases, suggesting their important functions in corals. The prokaryotic compositions showed more variation between coral genera than the variations observed between healthy vs. bleached conditions. For example, relatively high percentages of class Alphaproteobacteria were found in P. sinensis (Figure 3B: healthy 39.01% and bleached 64.26%). Differences in diversity between healthy and bleached corals were observed, but these differences were non-statistically significant when analysis was conducted at the genus level (Figure 3D): A. humillis (ANOSIM: P = 0.21, R = 0.26), A. millepora (P = 0.20, R = 0.11), P. sinensis (P = 0.10, R = 0.96), and P. lutea (P = 0.10, R = 0.89). In addition, some classes that were reported in both healthy and bleached corals were found to be populated by different genera: for example, predominant Alphaproteobacteria in P. sinensis contained different genera between healthy and bleached conditions. Bleached Acropora species exhibited heightened proportions of bacterial genera Acinetobacter and Helicobacter. For bleached P. sinensis, Actinobacter and unclassified genera of class Alphaproteobacteria, class Oxyphotobacteria, and order Rhizobiales were instead found to increase, whereas unclassified genera of family Stappiaceae, family Rhodobacteriaceae, and class Gammaproteobacteria decreased. For bleached P. lutea, unclassified genera of BD2-11 terrestrial group, A4b, order Dadabacteriales, and bacteria of subgroup10 were found to increase, whereas unclassified genera of class Gammaproteobacteria, phylum Proteobacteria, class Oxyphotobacteria, class Bacteroidia, and SAR202 clade decreased.
Non-metric multidimensional scaling demonstrated clearly the community profile differences across coral genera and that A. humilis and A. millepora positions overlapped. Bleached P. sinensis demonstrated the most separation relative to their profiles in healthy samples (Figure 4A); this separation was largely due to the increased proportions of unclassified Alphaproteobacteria and Desulfobacterales along with decreases of the orders Rhizobiales, Rhodobacterales, Vibrionales, and Oceanospirillales (Figure 3). Correlation analysis further revealed the prokaryotic taxa contributing to each direction of the community; for example, P. sinensis was enriched with Alphaproteobacteria and Rhizobiales (consistent with Figure 3 results), while Proteobacteria was abundant in healthy, but not in stressed, P. sinensis. For Acropora and Porites genera, other specific bacteria taxa were associated with the changes in healthy vs. bleached corals (Figure 4A).
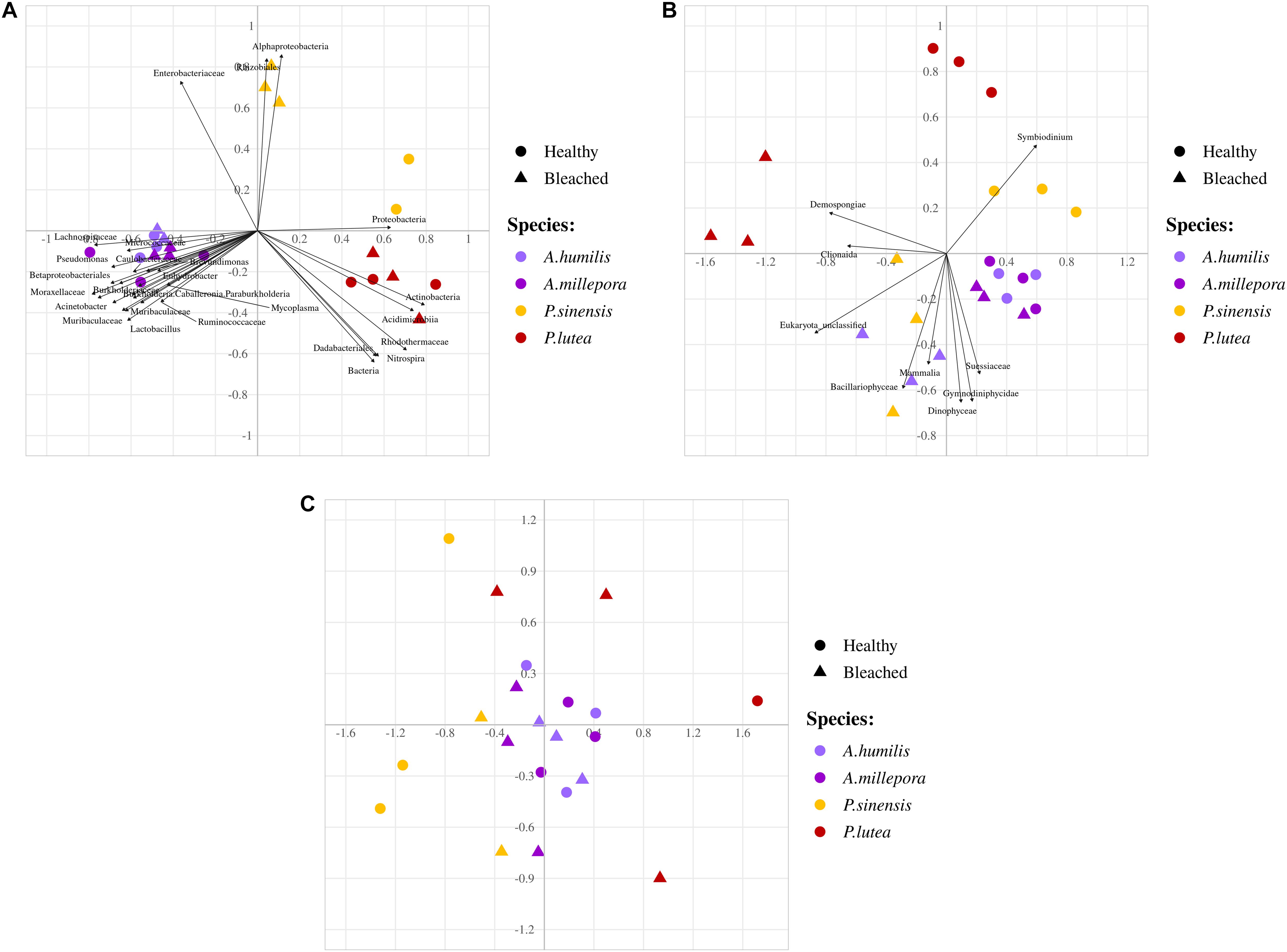
Figure 4. Non-metric multidimensional scaling analyses of (A) prokaryotes, (B) fungi and small eukaryotes, and (C) fungi communities at genus level OTUs. Arrows indicate significant correlation analysis of prokaryotic taxa to samples (P < 0.05).
Beta Diversity Analyses of Eukaryotic Microbe (Fungi and Small Eukaryote) Communities
The microbial 18S rRNA gene NGS yielded eukaryotic microbe profiles of fungi and small eukaryotes in the kingdoms of Chromista (avg. 61.58 ± 9.44%), Animalia (26.59 ± 3.41%), Plantae (0.41 ± 0.16%), and Protista (0.33 ± 0.09%) (Figure 5A). While sediments showed similar eukaryotic microbe compositions at all sites, seawater and corals showed community variations among sites especially at the S site. Corals A. humilis, A. millepora, and P. lutea demonstrated clearly different eukaryotic microbe compositions at the phylum level between healthy and bleached conditions compared with P. sinensis. A single-cell eukaryotic Dinoflagellata (algae) represented the most predominant phylum among corals and sediments. Of note was that the sediment eukaryotic microbe profiles contained many unclassified phyla. Next, we analyzed the relative abundances of important coral organisms of the genus Symbiodinium (phylum Dinoflagellata) (Figure 5B), simple eukaryotic Chromista (which contain photosynthetic organelles) and Protista (most of which contain photosynthetic organelles) (Figure 5C), and fungi (Figure 6). Comparing healthy and bleached corals, Symbiodinium was significantly reduced in most bleached corals, especially in bleached P. lutea: A. millepora (t-test: P = 0.04, given mean ± SD was 90.59 ± 3.85% for healthy and 82.06 ± 0.42% for bleached), P. sinensis (P = 0.004, given mean ± SD was 96.89 ± 0.38% for healthy and 88.41 ± 1.59% for bleached), and P. lutea (P = 0.016, given mean ± SD was 99.64 ± 0.02% for healthy and 65.94 ± 11.91% for bleached) (Figure 5B). The 10 topmost abundances of Chromista–Protista genera in corals were analyzed, and interestingly, the bleached corals had the higher number of unclassified genera of Dinophyceae and Alveolata and the genus Navicula (diatom) than those in their healthy corresponding coral species. Different Chromista–Protista OTUs showed fluctuation patterns across coral conditions, coral genera, and species. For instance, the unclassified genera of Suessiaceae, Phaeophyceae, Stramenopiles, and Embryophyta tended to be high in coral Acropora spp. The abundance of unclassified Gymnodiniphycidae genus was reduced in bleached A. humilis and A. millipora but increased in bleached P. sinensis (Figure 5C).
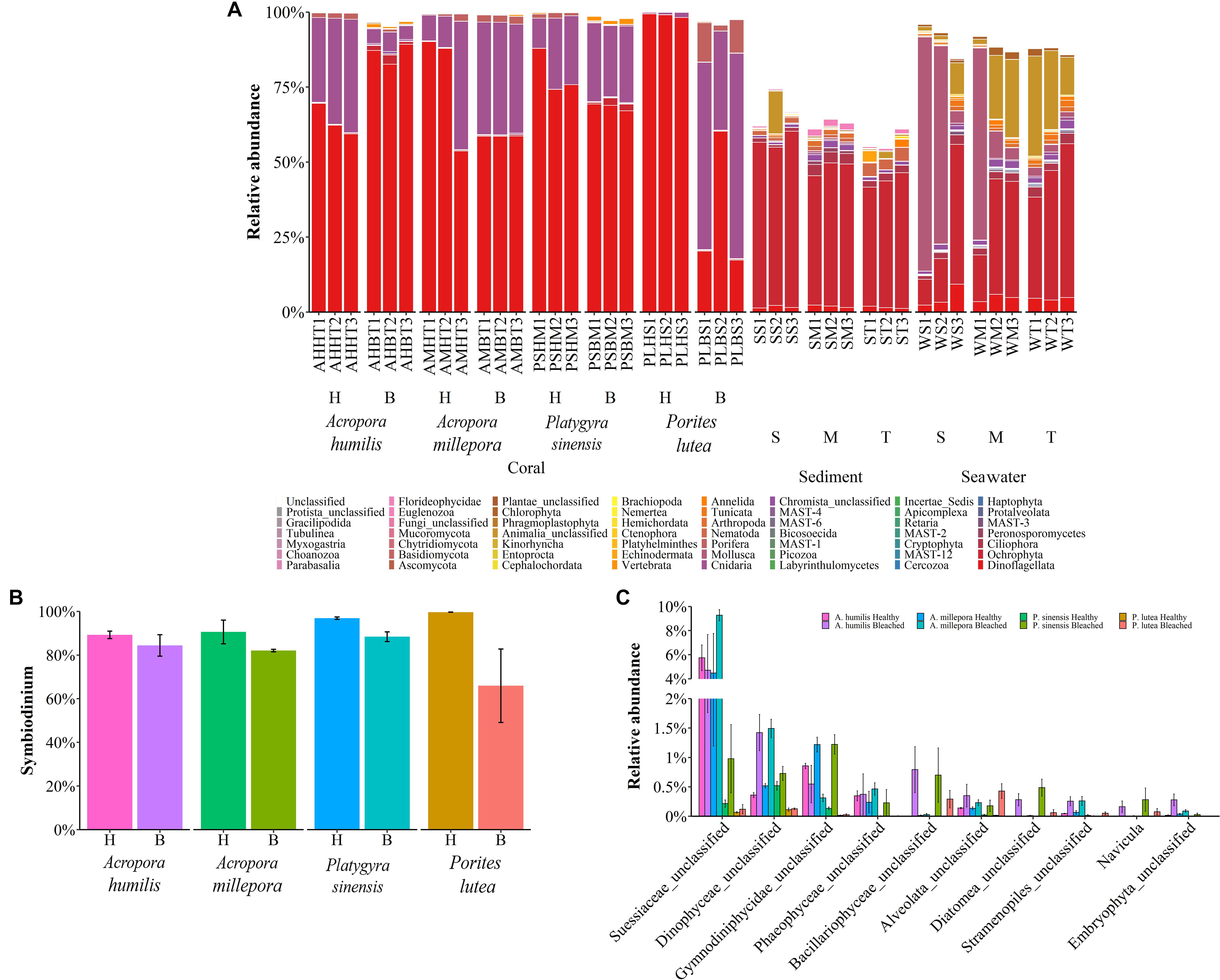
Figure 5. Percent relative abundance of (A) eukaryotic phyla across corals, sediments, and seawater; (B) Symbiodinium; and (C) simple eukaryotic Chromista and Protista across coral species.
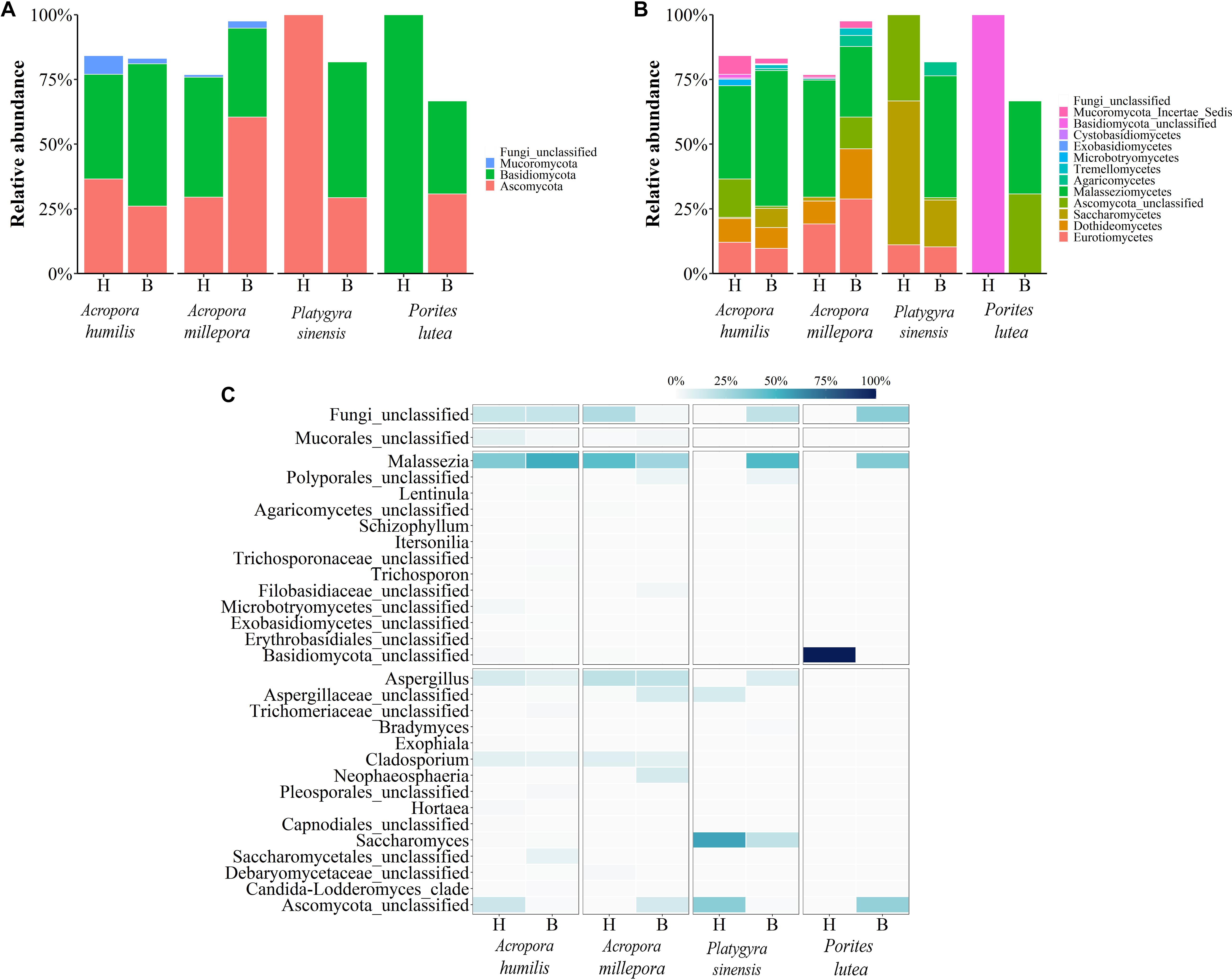
Figure 6. Relative abundance of fungal communities analyzed at the (A) phylum, (B) class, and (C) genus levels, respectively.
Analyzing the microbial community of bleached coral clusters showed that the bleached eukaryotic microbe cluster had rather community structure disparity from the healthy eukaryotic microbe cluster, especially for bleached P. lutea (Figure 4). Relatively higher frequencies of Symbiodinium showed positive correlation with healthy coral eukaryotic communities (Figure 5B). The correlation analysis also revealed that Demospongiae and Clionaida were statistically responsible for the separation of bleached from healthy corals, in particular, P. lutea (Figure 5B). Bacillariophyceae, Suessiaceae, Dinophyceae, and Gymnodiniphycidae were responsible for the correlation in the other bleached coral samples (Figures 4A, 5C).
For fungi communities, phyla Ascomycota and Basidiomycota were generally predominant in all samples. For healthy P. lutea, the single dominant Basidiomycota were unique (Figure 6A). P. lutea demonstrated the most fungal community dissimilarity between healthy and bleached conditions, followed by P. sinensis, A. millipora, and A. humilis, in order (Figure 6). For fungi, the same genus of corals did not always show consistent changes. Some findings of the differing fungi between healthy vs. bleached A. humilis were opposed to those in A. millepora: for instance, genera Malassezia (class Malasseziomycetes), unclassified Mucorales, and unclassified Ascomycota. The changes in bleached A. humilis shared commonality with those in bleached P. sinensis and P. lutea (Figure 6C). Correlation analysis of merely the fungi communities demonstrated no distinct community separations between healthy and bleached conditions, somewhat because of relatively high abundances of Saccharomyces and Malassezia in P. sinensis and the unclassified genus of Basidiomycota in P. lutea (Figures 4C, 6C).
Functional Potentials of Healthy and Bleached Coral-Associated Prokaryotic Communities
Functional potentials estimated from prokaryotic communities demonstrated bacterial functions in various categories, including membrane transport, amino acid and carbohydrate metabolisms, replication and repair, energy metabolism, translation, metabolism of cofactors and vitamins, xenobiotic biodegradation and metabolism, cellular processes and signaling, and lipid metabolism (Figure 7A; Hewson and Fuhrman, 2004; Tout et al., 2014). Nevertheless, the comparing pairs between healthy and bleached coral genera showed statistically significant differences of function frequencies involving immune system diseases for bleached Acropora. There were also statistical differences of function frequencies in cell motility, amino acid and nucleotide metabolism, cellular processes and signaling, genetic information processing, signal transduction, transcription, biosynthesis of secondary metabolites, transport and catabolism, and various body systems for Platygyra and xenobiotics biodegradation, nucleotide metabolism, transcription, and biosynthesis of secondary metabolites for Porites (t-test: P < 0.05) (Figure 7B). These differences in functional attributes highlighted key differences in bacterial community structure in healthy vs. bleached conditions of each coral genus.
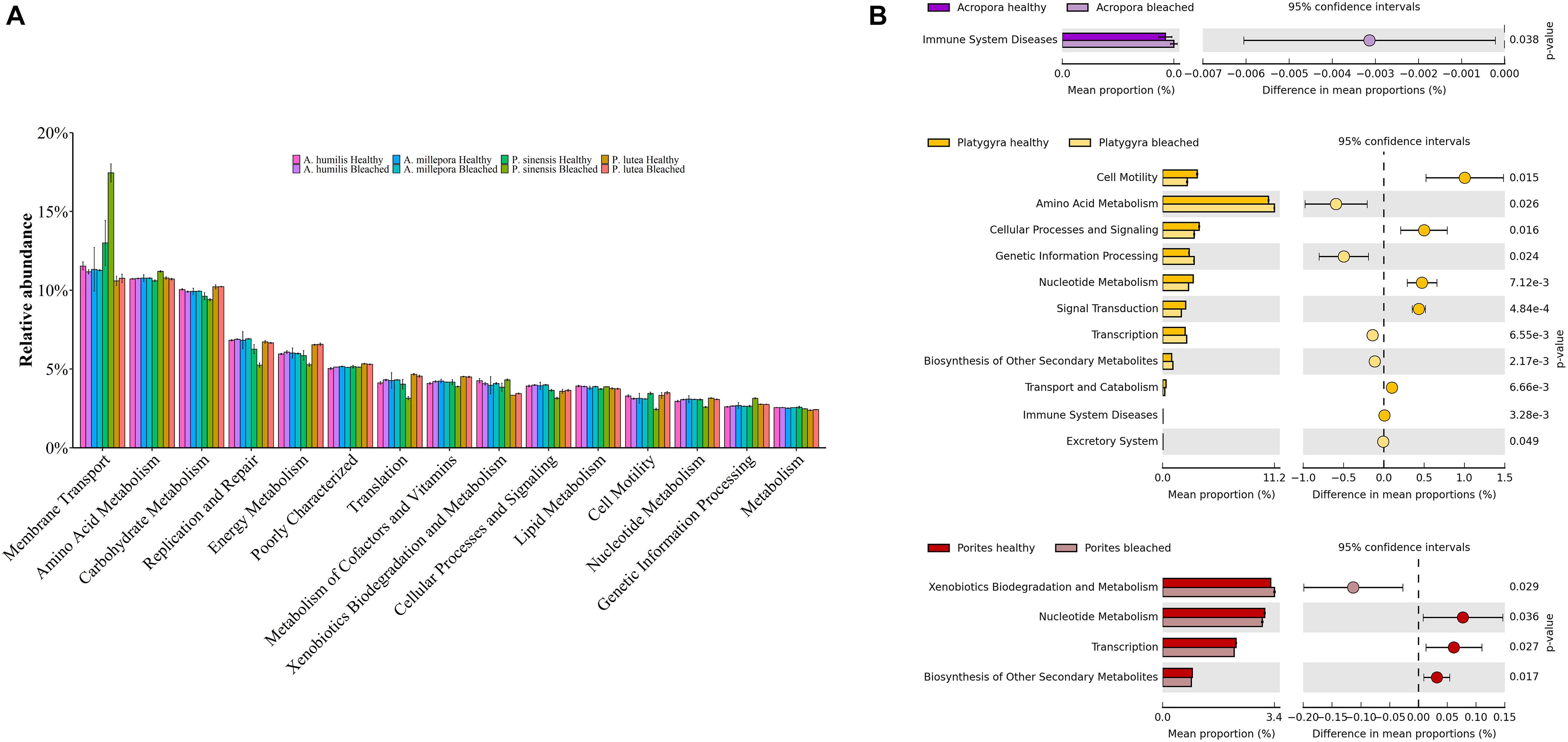
Figure 7. Functional potentials of (A) healthy and bleached coral-associated prokaryotic communities and (B) corresponding prokaryotic community pairs between healthy and bleached coral genera.
Correlations Between Coral-Associated Prokaryotic and Eukaryotic Genera
As prokaryotes and eukaryotes were reported to interact in a coral holobiont system ([opetwcitep]B4,B5[clotwcitep]Bernasconi et al., 2019a, b; Matthews et al., 2020), the coral-associated prokaryotic and eukaryotic genera were analyzed for correlation of prevalence. Although the statistics for the overall correlation between prokaryotic and eukaryotic communities were found not significant, many genera between prokaryotes and eukaryotes were found to be statistically significantly correlated (Supplementary Figure 1: green color). An unclassified eukaryotic genus of Suessiaceae showed relatively positive correlation to most number of bacterial genera, and the greatest positive correlation was determined between the unclassified eukaryotic genus of Suessiaceae and an unclassified prokaryotic genus of Lachnospiraceae (r = 0.719, P = 0.000033). This unclassified genus of Suessiaceae showed a strong positive correlation with other bacteria genera, such as Acinetobacter (r = 0.675, P = 0.00017), Helicobacter (r = 0.672, P = 0.00019), an unclassified genus of Muribaculaceae (r = 0.683, P = 0.00013), and an unclassified genus of Bacteroidales (r = 0.667, P = 0.00023). The pattern of these bacterial genera correlations with the unclassified genus of Suessiaceae was also observed with some other eukaryotes, including an unclassified genus of Stramenopiles, an unclassified genus of Embryophyta, and an unclassified genus of Dinophyceae. On the other hand, certain genera of bacteria (e.g., an unclassified genus of BD2-11 terrestrial group, Candidatus nitrosopumilus, and unclassified genera of Dadabacteriales and Nitrosopumilaceae) showed negative correlation to most eukaryotic genera except Symbiodinium (Supplementary Figure 1). The strongest negative correlation was found between an unclassified genus of Dinophyceae and Candidatus nitrosopumilus (r = −0.458, P = 0.023).
Discussion
Coral holobionts (i.e., microbiome) are known to be specific to sites and coral species and to be altered in bleached vs. healthy corals (Rohwer et al., 2002; [opetwcitep]B59,B58[clotwcitep]Pootakham et al., 2017, 2018). Previously, Roder et al. (2014) reported a higher abundance of coral pathogenic bacteria in bleached corals, and Littman et al. (2011) reported a shift in bacterial metabolisms (e.g., virulence genes, metabolisms of fatty acids, proteins, phosphorus, and sulfur) in bleached corals relative to healthy corals (Littman et al., 2011). The prokaryotic and eukaryotic microbe microbiomes associated with healthy and bleached coral species in the uGoT have not been extensively studied, yet microbial and small organisms play important roles in coral symbiosis in the uGoT, with important implications for coral health. Coral microbiomes play roles in providing nutrients for coral growth and reproduction and synthesize bioactive compounds that promote coral resistance to pathogens, as previously reported (Rosenberg et al., 2007; Wegley et al., 2007; Lesser et al., 2013; Webster and Reusch, 2017). We also know that greenhouse gas pollution has caused seawater temperature to rise; this shift in temperature has affected the microbiome of corals worldwide. This study firstly revealed both the prokaryotic and eukaryotic microbe microbiomes representing healthy and bleached samples of the four prevalent coral species (A. humilis, A. millipora, P. sinensis, and P. lutea) in the uGoT and also the microbiomes of underlying sediments and surrounding seawater over the corals. Samples were taken during the global thermal bleaching event in 2016 when significant bleaching occurred in the UGoT. Results reported here lay a firm foundation for understanding the coral microbiome in the UGoT, understanding how thermal bleaching events result in alterations in the microbiome and, with additional research, how it may be possible to promote coral recovery and health in future thermal bleaching events in the uGoT through manipulation of the coral microbiome.
In this study, different prokaryotic and eukaryotic microbiomes were found across different coral genera in healthy vs. bleached conditions. The greatest alpha diversity of prokaryotic OTUs was observed in sediments beneath the corals, which previous research reported the sediment may affect coral rates of growth, photosynthesis, larval settlement and survival, coral bleaching, and mortality (Tuttle et al., 2020). Sediment bacteria could play a role providing and/or recycling nutrient resources for coral growth/reproduction. In contrast, fungi and small eukaryotes had higher OTU diversity in seawater, which might be because these organisms are more independent on a requirement of surface area (i.e., coral surface) for colonization; also, some seawater small eukaryotes can harvest food from the water column like bacteria (Nielsen and Risgaard-Petersen, 2015; Mincer et al., 2016).
Gardner et al. (2019) previously reported that Acropora muricata and Acropora gemmifera are more bleaching sensitive than P. lutea, and raised a hypothesis that the high bacterial diversity found in Acropora may negatively affect bleaching resistance. In general, our results support this hypothesis, as we found that bleached corals exhibited a higher degree of both prokaryotic and eukaryotic microbe Shannon indices (which include both OTU richness and evenness) than healthy corals, consistent with previous reports that claimed an association between increased community diversity and corals undergoing a bleaching event (McDevitt-Irwin et al., 2017; Gardner et al., 2019; Sun et al., 2020).
Analyzing the beta diversity of the compositions of prokaryotes (and also fungi and other small eukaryotes) and the corresponding statistical analyses supported the general trends seen previously, namely, that microbiomes were specific to sample types (corals vs. sediment vs. seawater) ([opetwcitep]B9,B10[clotwcitep]Bulan et al., 2018a, b). The NMDS analyses further suggested that the compositions of prokaryotes (and also fungi and other small eukaryotes) were clustered separately by coral genera. For some coral genus microbiomes (i.e., Figures 4A,B, prokaryotic and eukaryotic microbe communities in healthy vs. bleached P. sinensis), bleached and healthy conditions showed diversified microbiomes.
The percent relative abundances and NMDS correlation analyses revealed differences in healthy and bleached samples, of each coral species. For instance, Rhizobiales, bacteria with high functions in nitrogen fixation (Lema et al., 2014), were high in bleached P. sinensis. Nonetheless, healthy and bleached Acropora demonstrated very close prokaryotic community clustering, a surprising finding since some bacteria, such as Acinetobacter, were found in diseased corals including dark spot syndrome disease (Sweet et al., 2013; Meyer et al., 2016). Other shared common altered species in bleached Acropora and P. sinensis included a decrease of beneficial bacteria, such as Rhizobiales, which Gardner et al. (2019) suggested was associated coral bleaching resistance. Interestingly, bleached P. lutea and P. sinensis shared higher prevalence for three of four beneficial bacteria taxa: Oceanospirillales (Kirkwood et al., 2010; Raina et al., 2016), Flavobacteriales (Kelly et al., 2014), Alteromonadales (Ceh et al., 2013), and Desulfobacterales (Gobet et al., 2012). Bleached Acropora species had higher prevalence for just two of four of these beneficial bacteria taxa. Note that Oceanospirillales could play a role in dimethylsulfoniopropionate (DMSP) degradation and antimicrobial compound production, which may be important during bleaching events (Kirkwood et al., 2010; Raina et al., 2016); we found these bacteria statistically higher in bleached A. millepora (t-test: P = 0.021) and P. lutea (P = 0.039) lending strength to the hypothesis that Oceanospirillales could play a role to support recovery from coral bleaching. Other orders of bacteria have diverse functional attributes; Flavobacteriales are energy scavengers from organic debris (Kelly et al., 2014), Alteromonadales are denoted as nitrogen fixers (Ceh et al., 2013), and Desulfobacterales are denoted as organic sulfate recyclers (Gobet et al., 2012). In our study of the uGoT, the communities of bleached corals tended to have higher percentages of Flavobacteriales (t-test: significant statistic for A. humilis, P = 0.025), Alteromonadales (significant statistics for A. millepora and P. sinensis, P = 0.015), and Desulfobacterales (significant statistics for P. sinensis, P = 0.018). Overall, our findings suggested that there are alterations of coral microbiome during bleached events, with the specific pattern of changes related to each coral examined. Increases of Vibrionales, Alteromonadales, and Rhizobiales were previously reported in stressed corals (Bulan et al., 2018b; Tout et al., 2015; McDevitt-Irwin et al., 2017). Vibrionales were previously reported to increase following a rise in seawater temperature, and some species of this bacterial order may act as coral pathogens (Kushmaro et al., 1998; Bulan et al., 2018a; Tout et al., 2015). The bacteria compositions in bleached P. lutea of our study were consistent with the bleached P. lutea collected at the Andaman Sea ([opetwcitep]B59,B58[clotwcitep]Pootakham et al., 2017, 2018); both uGoT and Anadaman Sea P. lutea had high percentages of Rhizobiales, Oceanospririllales, and Rhodobacterales in their microbiome.
An analysis of prokaryotic function potentials, estimated from the prokaryotic community composition, has shown that a variety of essential prokaryotic metabolic functions (such as membrane transport, amino acid and carbohydrate metabolisms, replication and repair, and energy metabolism) remained conserved following bleaching events (Badhai et al., 2016; Bulan et al., 2018a); however, the relative frequencies of the functions were sometimes found different in bleached vs. healthy corals. In our study, there were a significant increase of human immune system disease functions of bacteria in bleached Acropora species and the increase of xenobiotics biodegradation and metabolism in bleached P. lutea. Of note, there was a significant increase of metabolic function in bleached Acropora species, suggestive of the inflammatory disease state, which might play a role in supporting resistance to bleaching events. Although the immune system disease functional category of prokaryotes that we found belongs in human disease, corals are complex mutualisms with multiply associated microbiota and small eukaryotes, and our finding may support the modern concepts of learning immune responses in invertebrates and a coral holobiont immunity homeostasis ([opetwcitep]B52,B53[clotwcitep]Palmer, 2018a, b; Takagi et al., 2020). Nevertheless, the interpretation on this finding remains to be elucidated.
Moreover, fungi, Symbiodinium, and Chromista–Protista were analyzed in this study, as these microbes have been shown to have important relationships in coral symbiosis (Falkowski et al., 1984; Mieog et al., 2009). There is a well-known relationship for corals and Symbiodinium, e.g., the loss of Symbiodinium is associated with increased seawater temperatures and, thus, coral bleaching (Salih et al., 1997; Gardner et al., 2019). We too observed a reduction of Symbiodinium in all bleached coral species, with the largest reduction in P. lutea. Additionally, we have identified differing Symbiodinium clades in P. lutea (clade C15 in Porites and clade C3 in Acropora and Platygyra) following the established clade naming of Fisher et al. (2012). These differing Symbiodinium clades might have different heat resistance. Perhaps, P. lutea had a less heat-resistant Symbiodinium clade, so this coral species have adapted to heat stress through its prokaryotic (in particular bacteria) and eukaryotic microbe diversity. Following the loss of Symbiodinium, scientists have also observed that other fungi and small eukaryotes (Chromista–Protista) replace Symbiodinium in terms of providing photosynthesis and coral covering functions (Fine et al., 2005; del Campo et al., 2016; [opetwcitep]B4,B5[clotwcitep]Bernasconi et al., 2019a, b). Consistent with this “replacement” hypothesis, our analysis also found increased abundances of unclassified genera in Dinophyceae and Alveolata and genus Navicula in all bleached coral species. We suggest that Dinophyceae and Alveolata could provide photosynthesis (Gómez, 2012; Kim et al., 2013) and the diatom Navicula could replace nitrogen and phosphate recyclings (Kwon et al., 2013), for nutrients in corals that have lost Symbiodinium during a bleaching event. Moreover, the correlation analysis between coral-associated prokaryotic and eukaryotic genera highlighted a uniqueness of Symbiodinium that conferred a positive correlation to the otherwise prokaryotic genera that had negative correlations to other eukaryotic genera. Also, the correlation analysis revealed other eukaryotic genera that may be of importance to coral health and are actually phylogenetically associated with Symbiodinium, such as Suessiaceae, Stramenopiles, Dinophyceae, and Dinoflagellata (Pochon et al., 2014; Liu et al., 2018; Yorifuji et al., 2021).
In summary, this study firstly revealed both prokaryotic and eukaryotic microbiomes of four prevalent coral species in the uGoT and their surrounding niches (sediment and seawater) and compared healthy and bleached colonies of corals. Independently, triplicate sequencings per sample demonstrated that bleached Acropora, Platygyra, and Porites microbiomes were diverse. Overall, our findings were generally consistent with earlier work, but there were some key differences in our samples from the uGoT relative to reports for the same coral species in other geographic locations. We suggest that these geospecific differences in microbiomes in healthy or bleached conditions involve differences in marine biogeography, consistent with other results from corals in the uGoT (Somboonna et al., 2017). The results presented here will help lay a foundation to help minimize coral bleaching and/or maximize coral restoration following bleaching events; as long-term goals, we are actively working on techniques and strategies to improve beneficial microbiome members that would help protect or restore functions in heat-stressed corals. Finally, we note that this study was based on a single time point and a single bleaching event; we are gathering other coral microbiome bleaching event data to help us better understand which core microorganisms support bleaching resistance and coral reef restoration in the uGoT.
Data Availability Statement
Nucleic acid sequences in this study were deposited in an NCBI open access Sequence Read Archive database, accession number SRP291375 for 16S and 18S rDNA sequences.
Author Contributions
HK, CK, and MP did molecular biology experiments and data analysis. CK helped draft the manuscript. SJ and SC collected samples. KP, JO, SC, and VV conceived of the study. SC and VV helped revise the manuscript. NS conceived of the study, coordinated the experiments and data analysis, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by Thailand Research Fund (RSA6180046), EU-Horizon 2020 Project TASCMAR (634674), NRCT-JSPS Core-to-Core Program, Mubadala Petroleum (Thailand), Microbiome Research Unit for Probiotics in Food and Cosmetics, and the 90th Anniversary of Chulalongkorn University Fund. The authors declare that this study received funding from Mubadala Petroleum (Thailand). Mubadala Petroleum (Thailand) was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledged Gerry Plumley for English language editing. The authors acknowledged the Plant Genetic Conservation Project under the Royal Initiative of Her Royal Highness Princess Maha Chakri Sirindhorn and the Naval Special Warfare Command, Royal Thai Navy for their assistance in the field.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.643962/full#supplementary-material
Supplementary Figure 1 | Pearson’s correlations between coral associated prokaryotic and eukaryotic genera. Green color indicates positive correlation between species, red color indicates negative correlation, and white color indicates no correlation.
Supplementary Table 1 | Number of sequencing reads, Good’s coverage and alpha diversity indices of coral, sediment, and seawater samples of (A) 16S rDNA and (B) 18S rDNA profiles, at genus level OTUs.
Supplementary Table 2 | Taxonomy names along the number of OTUs and sequences in coral, sediment, and seawater samples of (A) 16S rDNA and (B) 18S rDNA profiles.
Footnotes
References
Anthony, K. R., Maynard, J. A., Diaz-Pulido, G., Mumby, P. J., Marshall, P. A., Cao, L., et al. (2011). Ocean acidification and warming will lower coral reef resilience. Glob. Change Biol. 17, 1798–1808. doi: 10.1111/j.1365-2486.2010.02364.x
Badhai, J., Ghosh, T. S., and Das, S. K. (2016). Composition and functional characterization of microbiome associated with mucus of the coral Fungia echinata collected from Andaman Sea. Front. Microbiol. 7:936. doi: 10.3389/fmicb.2016.00936
Baker, A. C., Glynn, P. W., and Riegl, B. (2008). Climate change and coral reef bleaching: an ecological assessment of long-term impacts, recovery trends and future outlook. Estuar. Coast. Shelf Sci. 80, 435–471. doi: 10.1016/j.ecss.2008.09.003
Bernasconi, R., Stat, M., Koenders, A., and Huggett, M. J. (2019a). Global networks of Symbiodinium-bacteria within the coral holobiont. Microb. Ecol. 77, 794–807. doi: 10.1007/s00248-018-1255-4
Bernasconi, R., Stat, M., Koenders, A., Paparini, A., Bunce, M., and Huggett, M. J. (2019b). Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 10:1529. doi: 10.3389/fmicb.2019.01529
Bourne, D. G., Boyett, H. V., Henderson, M. E., Muirhead, A., and Willis, B. L. (2008a). Identification of a ciliate (Oligohymenophorea: Scuticociliatia) associated with brown band disease on corals of the Great Barrier Reef. Appl. Environ. Microbiol. 74, 883–888. doi: 10.1128/aem.01124-07
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008b). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D., Webster, N., Rosenberg, E., Rosenberg, E., Stackebrandt, E., and DeLong, E. (2013). Coral Reef Microbial Communities. The Prokaryotes. New York: Springer.
Bulan, D. E., Wilantho, A., Krainara, P., Viyakarn, V., Chavanich, S., and Somboonna, N. (2018a). Spatial and seasonal variability of reef bacterial communities in the upper Gulf of Thailand. Front. Mar. Sci. 5:441. doi: 10.3389/fmars.2018.00441
Bulan, D. E., Wilantho, A., Tongsima, S., Viyakarn, V., Chavanich, S., and Somboonna, N. (2018b). Microbial and small eukaryotes associated with reefs in the upper Gulf of Thailand. Front. Mar. Sci. 5:436. doi: 10.3389/fmars.2018.00436
Burke, L., Reytar, K., Spalding, K., and Perry, A. (2012). Reefs at Risk Revisited in the Coral Triangle. Washington, DC: World Resources Institute, In The Nature Conservancy.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Ceh, J., Kilburn, M. R., Cliff, J. B., Raina, J. B., van Keulen, M., and Bourne, D. G. (2013). Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol. Evol. 3, 2393–2400. doi: 10.1002/ece3.642
Chavanich, S., Viyakarn, V., Loyjiw, T., Pattaratamrong, P., and Chankong, A. (2009). Mass bleaching of soft coral, Sarcophyton spp. in Thailand and the role of temperature and salinity stress. ICES J. Mar. Sci. 66, 1515–1519. doi: 10.1093/icesjms/fsp048
del Campo, J., Guillou, L., Hehenberger, E., Logares, R., López-García, P., and Massana, R. (2016). Ecological and evolutionary significance of novel protist lineages. Eur. J. Protistol. 55, 4–11. doi: 10.1016/j.ejop.2016.02.002
Dunlap, W. C., and Shick, J. M. (1998). Ultraviolet radiation-absorbing mycosporine-like amino acids in coral reef organisms: a biochemical and environmental perspective. J. Phycol. 34, 418–430. doi: 10.1046/j.1529-8817.1998.340418.x
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and Porter, J. W. (1984). Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709. doi: 10.2307/1309663
Fine, M., Meroz-Fine, E., and Hoegh-Guldberg, O. (2005). Tolerance of endolithic algae to elevated temperature and light in the coral Montipora monasteriata from the southern Great Barrier Reef. J. Exp. Biol. 208, 75–81. doi: 10.1242/jeb.01381
Fisher, P., Malme, M., and Dove, S. (2012). The effect of temperature stress on coral–Symbiodinium associations containing distinct symbiont types. Coral Reefs 31, 473–485. doi: 10.1007/s00338-011-0853-0
Gardner, S. G., Camp, E. F., Smith, D. J., Kahlke, T., Osman, E. O., Gendron, G., et al. (2019). Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol. Evol. 9, 938–956.
Geiser, D. M., Taylor, J. W., Ritchie, K. B., and Smith, G. W. (1998). Cause of sea fan death in the West Indies. Nature 394, 137–138. doi: 10.1038/28079
Gobet, A., Böer, S. I., Huse, S. M., Van Beusekom, J. E., Quince, C., Sogin, M. L., et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 6, 542–553. doi: 10.1038/ismej.2011.132
Gómez, F. (2012). A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Syst. Biodivers. 10, 267–275. doi: 10.1080/14772000.2012.721021
Harel, M., Ben-Dov, E., Rasoulouniriana, D., Siboni, N., Kramarsky-Winter, E., Loya, Y., et al. (2008). A new Thraustochytrid, strain Fng1, isolated from the surface mucus of the hermatypic coral Fungia granulosa. FEMS Microbiol. Ecol. 64, 378–387. doi: 10.1111/j.1574-6941.2008.00464.x
Harvey, B. J., Nash, K. L., Blanchard, J. L., and Edwards, D. P. (2018). Ecosystem-based management of coral reefs under climate change. Ecol. Evol. 8, 6354–6368. doi: 10.1002/ece3.4146
Hatcher, B. G. (1990). Coral reef primary productivity. A hierarchy of pattern and process. Trends Ecol. Evol. 5, 149–155. doi: 10.1016/0169-5347(90)90221-x
Hewson, I., and Fuhrman, J. (2004). Bacterioplankton species richness and diversity along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70, 3425–3433. doi: 10.1128/aem.70.6.3425-3433.2004
Hoegh-Guldberg, O., and Smith, G. J. (1989). The effect of sudden changes in temperature, light and salinity on the population density and export of zooxanthellae from the reef corals Stylophora pistillata Esper and Seriatopora hystrix Dana. J. Exp. Mar. Biol. Ecol. 129, 279–303. doi: 10.1016/0022-0981(89)90109-3
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Kastanis, G. J., Santana-Quintero, L. V., Sanchez-Leon, M., Lomonaco, S., Brown, E. W., and Allard, M. W. (2018). In-depth comparative analysis of Illumina® MiSeq run metrics: development of a wet-lab quality assessment tool. Mol. Ecol. Resour. 19, 377–387. doi: 10.1111/1755-0998.12973
Kelly, L. W., Williams, G. J., Barott, K. L., Carlson, C. A., Dinsdale, E. A., Edwards, R. A., et al. (2014). Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc. Natl. Acad. Sci. U.S.A. 111, 10227–10232. doi: 10.1073/pnas.1403319111
Kim, G. H., Jeong, H. J., Du Yoo, Y., Kim, S., Han, J. H., and Han, J. W. (2013). Still acting green: continued expression of photosynthetic genes in the heterotrophic Dinoflagellate Pfiesteria piscicida (Peridiniales, Alveolata). PLoS One 8:e68232. doi: 10.1371/journal.pone.0068232
Kirkwood, M., Todd, J. D., Rypien, K. L., and Johnston, A. W. (2010). The opportunistic coral pathogen Aspergillus sydowii contains dddP and makes dimethyl sulfide from dimethylsulfoniopropionate. ISME J. 4, 147–150. doi: 10.1038/ismej.2009.102
Kramarsky-Winter, E., Harel, M., Siboni, N., Dov, E. B., Brickner, I., Loya, Y., et al. (2006). Identification of a protist-coral association and its possible ecological role. Mar. Scol. Prog. Ser. 317, 67–73. doi: 10.3354/meps317067
Kushmaro, A., Rosenberg, E., Fine, M., Haim, Y. B., and Loya, Y. (1998). Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Scol. Prog. Ser. 171, 131–137. doi: 10.3354/meps171131
Kwon, H. K., Oh, S. J., and Yang, H.-S. (2013). Growth and uptake kinetics of nitrate and phosphate by benthic microalgae for phytoremediation of eutrophic coastal sediments. Bioresour. Technol. 129, 387–395. doi: 10.1016/j.biortech.2012.11.078
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Le Campion-Alsumard, T., Golubic, S., and Priess, K. (1995). Fungi in corals: symbiosis or disease? Interaction between polyps and fungi causes pearl-like skeleton biomineralization. Mar. Ecol. Prog. Ser. 117, 137–147. doi: 10.3354/meps117137
Lema, K. A., Bourne, D. G., and Willis, B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695. doi: 10.1111/mec.12899
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/aem.07800-11
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Lesser, M., Stat, M., and Gates, R. (2013). The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs 32, 603–611. doi: 10.1007/s00338-013-1051-z
Littman, R., Willis, B. L., and Bourne, D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660. doi: 10.1111/j.1758-2229.2010.00234.x
Liu, H., Stephens, T. G., González-Pech, R. A., Beltran, V. H., Lapeyre, B., Bongaerts, P., et al. (2018). Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1:95. doi: 10.1038/s42003-018-0098-3
Matthews, J. L., Raina, J.-B., Kahlke, T., Seymour, J. R., van Oppen, M. J. H., and Suggett, D. J. (2020). Symbiodiniaceae-bacteria interactions: rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 22, 1675–1687. doi: 10.1111/1462-2920.14918
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
Meyer, J. L., Rodgers, J. M., Dillard, B. A., Paul, V. J., and Teplitski, M. (2016). Epimicrobiota associated with the decay and recovery of Orbicella corals exhibiting dark spot syndrome. Front. Microbiol. 7:893. doi: 10.3389/fmicn.2016.00893
Mieog, J. C., Olsen, J. L., Berkelmans, R., Bleuler-Martinez, S. A., Willis, B. L., and van Oppen, M. J. (2009). The roles and interactions of symbiont, host and environment in defining coral fitness. PLoS One 4:e6364. doi: 10.1371/journal.pone.0006364
Mincer, T. J., Zettler, E. R., and Amaral-Zettler, L. A. (2016). “Biofilms on plastic debris and their influence on marine nutrient cycling, productivity, and hazardous chemical mobility,” in Hazardous Chemicals Associated with Plastics in the Marine Environment, eds H. Takada and H. Karapanagioti (Cham: Springer), 221–233. doi: 10.1007/698_2016_12
Nielsen, L. P., and Risgaard-Petersen, N. (2015). Rethinking sediment biogeochemistry after the discovery of electric currents. Ann. Rev. Mar. Sci. 7, 425–442. doi: 10.1146/annurev-marine-010814-015708
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). The Vegan Package. Vegan: Community Ecology Package. R Package Version 2.5-5.
Palmer, C. V. (2018a). Immunity and the coral crisis. Commun. Biol. 1:91. doi: 10.1038/s42003-018-0097-4
Palmer, C. V. (2018b). Warmer water affects immunity of a tolerant reef coral. Front. Mar. Sci. 5:253. doi: 10.3389/fmars.2018.00253
Pengsakun, S., Yeemin, T., Sutthacheep, M., Sansuvan, W., Klinthong, W., and Chamchoy, C. (2019). Monitoring of coral communities in the inner Gulf of Thailand influenced by the elevated seawater temperature and flooding. Acta Oceanol. Sin. 38, 102–111. doi: 10.1007/s13131-019-1376-8
Peters, E. C. (1984). A survey of cellular reactions to environmental stress and disease in Caribbean scleractinian corals. Helgol. Meeresunters. 37, 113–137. doi: 10.1007/bf01989298
Phongsuwan, N., Chankong, A., Yamarunpatthana, C., Chansang, H., Boonprakob, R., Petchkumnerd, P., et al. (2013). Status and changing patterns on coral reefs in Thailand during the last two decades. Deep Sea Res. 2 Top. Stud. Oceanogr. 96, 19–24. doi: 10.1016/j.dsr2.2013.02.015
Pochon, X., Putnam, H. M., and Gates, R. D. (2014). Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2:e394. doi: 10.7717/peerj.394
Pootakham, W., Mhuantong, W., Putchim, L., Yoocha, T., Sonthirod, C., Kongkachana, W., et al. (2018). Dynamics of coral-associated microbiomes during a thermal bleaching event. MicrobiologyOpen 7:e00604. doi: 10.1002/mbo3.604
Pootakham, W., Mhuantong, W., Yoocha, T., Putchim, L., Sonthirod, C., Naktang, C., et al. (2017). High resolution profiling of coral-associated bacterial communities using full-length 16S rRNA sequence data from PacBio SMRT sequencing system. Sci. Rep. 7:2774. doi: 10.1038/s41598-017-03139-4
Raina, J.-B., Tapiolas, D., Motti, C. A., Foret, S., Seemann, T., Tebben, J., et al. (2016). Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 4:e2275. doi: 10.7717/peerj.2275
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Roder, C., Arif, C., Bayer, T., Aranda, M., Daniels, C., Shibl, A., et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8, 31–39. doi: 10.1038/ismej.2013.127
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Salih, A., Hoegh-Guldberg, O., and Cox, G. (1997). “Bleaching responses of symbiotic dinoflagellates in corals: the effects of light and elevated temperature on their morphology and physiology,” in Proceedings of the Australian Coral Reef Society, Heron Island 50 year commemorative meeting (Heron Island).
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Mircobiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Smith, G. W., and Weil, E. (2004). “Aspergillosis of gorgonians,” in Coral Health and Disease, eds E. Rosenberg and Y. Loya (Berlin: Springer), 279–287. doi: 10.1007/978-3-662-06414-6_15
Somboonna, N., Wilantho, A., Monanunsap, S., Chavanich, S., Tangphatsornruang, S., and Tongsima, S. (2017). Microbial communities in the reef water at Kham Island, lower Gulf of Thailand. PeerJ 5:e3625. doi: 10.7717/peerj.3625
Sun, F., Yang, H., Wang, G., and Shi, Q. (2020). Combination analysis of metatranscriptome and metagenome reveal the composition and functional response of coral symbionts to bleaching during an El Niño Event. Front. Microbiol. 11:448. doi: 10.3389/fmicb.2020.00448
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Takagi, T., Yoshioka, Y., Zayasu, Y., Satoh, N., and Shinzato, C. (2020). Transcriptome analyses of immune system behaviors in primary polyp of coral Acropora digitifera exposed to the bacterial pathogen Vibrio coralliilyticus under thermal loading. Mar. Biotechnol. 22, 748–759. doi: 10.1007/s10126-020-09984-1
Tout, J., Jeffries, T. C., Webster, N. S., Stocker, R., Ralph, P. J., and Seymour, J. R. (2014). Variability in microbial community composition and function between different niches within a coral reef. Microb. Ecol. 67, 540–552.
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Tuttle, L. J., Johnson, C., Kolinski, S., Minton, D., and Donahue, M. J. (2020). How does sediment exposure affect corals? A systematic review protocol. Environ. Evid. 9:17. doi: 10.1186/s13750-020-00200-0
Webster, N. S., and Reusch, T. B. H. (2017). Microbial contributions to the persistence of coral reefs. ISME J. 11, 2167–2174. doi: 10.1038/ismej.2017.66
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Keywords: coral bleaching, coral reefs, microbiome, bacteria, fungi, small eukaryotes, next generation sequencing
Citation: Kusdianto H, Kullapanich C, Palasuk M, Jandang S, Pattaragulwanit K, Ouazzani J, Chavanich S, Viyakarn V and Somboonna N (2021) Microbiomes of Healthy and Bleached Corals During a 2016 Thermal Bleaching Event in the Upper Gulf of Thailand. Front. Mar. Sci. 8:643962. doi: 10.3389/fmars.2021.643962
Received: 19 December 2020; Accepted: 27 May 2021;
Published: 28 June 2021.
Edited by:
Anthony Lee Dellinger, Kepley BioSystems, Inc., United StatesReviewed by:
Patrick Cabaitan, University of the Philippines Diliman, PhilippinesLindsey Deignan, Nanyang Technological University, Singapore
Copyright © 2021 Kusdianto, Kullapanich, Palasuk, Jandang, Pattaragulwanit, Ouazzani, Chavanich, Viyakarn and Somboonna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naraporn Somboonna, TmFyYXBvcm4uU0BjaHVsYS5hYy50aA==; Voranop Viyakarn, Vm9yYW5vcC5WQGNodWxhLmFjLnRo
 Heru Kusdianto
Heru Kusdianto Chitrasak Kullapanich
Chitrasak Kullapanich Matanee Palasuk
Matanee Palasuk Suppakarn Jandang
Suppakarn Jandang Kobchai Pattaragulwanit2
Kobchai Pattaragulwanit2 Jamal Ouazzani
Jamal Ouazzani Suchana Chavanich
Suchana Chavanich Voranop Viyakarn
Voranop Viyakarn Naraporn Somboonna
Naraporn Somboonna