
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 03 May 2021
Sec. Marine Pollution
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.640425
This article is part of the Research TopicCombined Effects of Climate Change Factors with Marine PollutantsView all 4 articles
 Kexin Xu1†
Kexin Xu1† Yinghui Wang1†
Yinghui Wang1† Shanshan Lian1,2*
Shanshan Lian1,2* Naina Hu1
Naina Hu1 Xiaomei Chen1
Xiaomei Chen1 Xiaoting Dai1
Xiaoting Dai1 Lingling Zhang1,2
Lingling Zhang1,2 Shi Wang1,2,3
Shi Wang1,2,3 Jingjie Hu1,3
Jingjie Hu1,3 Xiaoli Hu1,4
Xiaoli Hu1,4 Zhenmin Bao1,3,4
Zhenmin Bao1,3,4C1Q (Complement 1Q) is an important recognition molecule in the immunological complement system, which could also be putatively involved in the stress responses induced by endotoxins or exotoxins, potentially through detoxification processes. Marine bivalves are well adapted to highly complex aquatic environments with various stressors. As filter feeders, they have to cope with highly potent microalgae-derived neurotoxins, such as paralytic shellfish toxin (PSTs). Zhikong scallops, Chlamys farreri, are commercially important bivalve with the remarkable ability to accumulate PSTs. Exploring the C1Qs related to PST accumulation in C. farreri could benefit our understanding of the adaptations of bivalve species. In the present study, we systematically analyzed C1Q genes in C. farreri. In total, 97 CfC1Q genes mainly from the expanded C1Q-B subfamily were identified, from which the C1QL, C1QTNF, and C1QDC1 members in C. farreri were revealed to be under positive selection. Spatiotemporal expression analysis revealed that most CfC1QLs and CfC1QDC1s were highly expressed during the post-umbo stage and in hepatopancreas, while most CfC1QTNF members were highly expressed after the creeping larva stage and in mantle. The hepatopancreas and kidney in C. farreri are two toxin-rich organs with the highest concentrations of PSTs, acting as major “centers” for toxin accumulation and transformation, respectively. Therefore, after feeding the scallops with PST-producing dinoflagellates Alexandrium minutum and Alexandrium catenella, we determined the expression patterns of CfC1Qs in these two organs. In kidney, more than 85% of CfC1QLs and CfC1QDC1s showed drastic up-regulation with both diets. However, among these members with significant induction, a different response manner was detected after feeding with A. minutum and A. catenella, respectively as acute and chronic response patterns. In comparison, far fewer CfC1Qs showing significant up-regulation in hepatopancreas with both toxic diets and only mild regulation pattern could be found. This organ-, toxin-, and time-dependent genetic regulation of CfC1Qs may contribute to the virulence difference on account of the tissue-specific or dinoflagellate-specific different toxin analogs composition, implying the possible involvement of CfC1Qs in PST transport and homeostasis. Our findings imply the functional diversity of scallop C1Q genes in coping with PST accumulation, which might be developed as potential molecular indicators for monitoring toxin accumulation in edible mollusks or provide insight into the lineage-specific adaptation of scallops for dealing with microalgal toxin challenges.
The C1Q proteins, also known as C1Q-domain-containing (C1QDC) proteins, widely exist in various species, from bacteria to mammals. Besides the conserved globular C1Q domain, they usually possess a leading signal peptide and a collagen-like region (CLR) (Liu et al., 2013). Previously, the vertebrate C1Q proteins were revealed to play a key role during immune and inflammation regulation (Kishore and Reid, 2000; Tom Tang et al., 2005; Huang et al., 2008; Yuzaki, 2008). As the first subcomponent of the complement C1 complex, C1Q is the target recognition protein of the classical complement pathway (Gestal et al., 2010). It plays a key role in initiating the complement activation through binding to antibody–antigen complexes, pathogen surfaces, apoptotic cells, or polyanionic structures (Gestal et al., 2010). At present, based on sequence homology, structure similarity, and functional relatedness, in-depth studies of C1Q proteins have been carried out in human (Homo sapiens) and zebrafish (Danio rerio). C1Q proteins have been classified into three major subfamilies: C1Q-A, -B, and -C (Tom Tang et al., 2005; Mei and Gui, 2008). In addition to the immunological functions, several C1Q proteins were also found to act as important neuronal transduction factors, mediating CNS (Central Nervous system) synapse development and in response to neurotoxicity as well as a variety of neuronal injuries (Stevens et al., 2007; Ghebrehiwet et al., 2012; Benoit et al., 2013). For example, C1Q was found to protect immature and mature primary neurons against fibrillar amyloid-β toxicity in mammals (Li et al., 2008; Benoit et al., 2013) and the precerebellin (CBLN) proteins (C1Q-B subfamily) were found to act as a new class of transneuronal regulatory factors, regulating synaptic development and synaptic plasticity in many regions of the brain (Yuzaki, 2008; Boulanger, 2009).
Marine bivalves are filter-feeding species that feed mainly on microalgae, and they can bioaccumulate various microalgae derived toxins, posing a health hazard to humans and other animals through food chains (Anderson et al., 2012; Borcier et al., 2017). Among these toxins, paralytic shellfish toxins (PSTs) are one of the most potent neurotoxins. PSTs attack the sodium channels (Nav) on nerve cell membranes through cross-pore binding at multi-polar active sites, inducing Na+-flux disruption and interfering transmission of an action potential (Bricelj et al., 2005; Borcier et al., 2017). The accumulation of PSTs can not only affect the biological function of bivalves, such as filtration/feeding activities, immune response, antioxidant response, and even reproduction, but also become a major concern in terms of their sanitary quality (Li et al., 2002; Fabioux et al., 2015). In invertebrate mollusks, the conservative functions of activating the complement system have been found in bivalve C1Q proteins. In Bay scallop (Argopecten irradians), AiC1QDC-1 and AiC1QDC-2 molecules exhibited significant yeast agglutination activity, and they could function as pattern-recognition receptors (PRRs) to recognize different pathogen-associated molecular patterns (PAMPs), triggering immune defense (Kong et al., 2010; Wang et al., 2012). In Zhikong scallop (Chlamys farreri), the CfC1Q (C1Q in C. farreri) proteins were found to combine multiple PAMPs to assist with pathogenic bacteria elimination (Zhang et al., 2008). They were also found to serve as pattern recognition receptors and bind with human heat-aggregated IgG in a dose-dependent manner (Zhang et al., 2008; Wang et al., 2012). Further, several lines of evidence have revealed that C1Q genes could be involved in toxin metabolism in bivalves, not only for PSTs, but also for amnesic shellfish poisoning (ASP) and environmental toxins (heavy metals, for instance). For example, several C1Qs have been identified as responsive genes during transcriptional research on mussel (Mytilus galloprovincialis) when exposed to PST-producing Alexandrium (Gerdol et al., 2014). ASP is caused by a species of the diatom genus Pseudo-nitzschia, which produces the toxin domoic acid. According to the differential gene expression analysis in mussels (M. galloprovincialis) naturally exposed to Pseudo-nitzschia, C1Qs were found to be significantly enriched and researchers pointed out that C1Qs could be involved in detoxification processes, in response to oxidative stress and/or in immunological processes (Pazos et al., 2017). In mussel (M. coruscus), time-dependent responsive pattern of a novel C1Q was revealed after copper and cadmium exposure, which might be developed as a potential indicator for monitoring heavy metals’ pollution (Liu et al., 2014). In particular, the number of C1Q genes was found to be enormously expanded in the genome of bivalves, such as Pinctada fucata and Crassostrea gigas (Takeuchi et al., 2016), which might be related to their adaptive functions. All these findings suggest that, besides the microbial recognition capability, the C1Q proteins could function in a toxin-responsive manner in bivalves, which may contribute to the adaptation of scallops for dealing with microalgal toxin challenge and merits further study.
Zhikong scallops (Hu et al., 2010) are commercially important shellfish, which thrive in environments from intertidal zones to deep oceans as semi-sessile filter feeders, with the outstanding ability to accumulate PSTs (Li et al., 2017). C. farreri are naturally distributed along the coasts of Northern China, Korea, Japan, and Eastern Russia. Most previous studies on bivalve C1Q proteins usually focus on a certain member that participated in the classical complement activation. In this study, we genome-widely identified the C1Q genes in C. farreri. The C1QL, C1QTNF, and C1QDC1 members from the expanded subfamily C1Q-B were revealed to be under positive selection. To explore these C1Qs’ involvement during PSTs accumulation, we chose two major strains of the PST-producer Alexandrium (A. catenella and A. minutum) to feed the scallops. In C. farreri, hepatopancreas and kidney are the main repository of PSTs during uptake and depuration, and dynamic PST metabolites were detected, contributing to PSTs detoxification (Hu et al., 2019; Lou et al., 2020). We systematically assessed the CfC1Q responses in hepatopancreas and kidney after feeding with toxic A. catenella and A. minutum, revealing the variable regulation of scallop C1Qs between organs and between different toxic microalgae species. Our findings imply the functional diversity of scallop C1Q genes in coping with microalgae-derived toxins, providing a better understanding on their adaptive evolution.
Based on our previous work (Li et al., 2017), the whole genome of a 2-years-old C. farreri was sequenced using the Illumina HiSeq 2000 platform through the construction and sequencing of both short-insert (180, 300, and 500 bp) and long-insert (2, 5, 10, 20, and 30 kb) DNA libraries. Thirteen adult tissues/organs of the scallop were chosen for transcriptome sequencing, including striated muscle, smooth muscle, foot, hepatopancreas, kidney, female gonad, male gonad, gill, eyes, mantle, cerebral ganglion, and visceral ganglion. All these data have been deposited on NCBI database. The available C1Q protein sequences of representative species, including humans (Homo sapiens), chickens (Gallus gallus), frogs (Xenopus laevis), zebrafish (Danio rerio), and mouse (Mus musculus), were downloaded from NCBI1 and UNIPROT2 databases. The downloaded orthologous C1Q sequences were used as query sequences for transcriptome and whole-genome blast searching in C. farreri, with the E value threshold as 1E-05. To confirm the completeness of C1Q gene family and to identify all possible C1Q gene loci in the genome, TBLASTN was used to verify the cDNA sequences by comparing the transcriptome sequences to the whole genome sequences. Further, sequence analysis was performed through C1Q HMM searching method to ensure the completeness of C1Q proteins, and the position of conserved C1Q Pfam domain (PF00386) was illustrated for C. farreri in Supplementary Table 1.
To confirm the predicted amino acid sequences, the candidate C1Q genes sequences of C. farreri were submitted to the ORF Finder program3 to predict the open reading frame (ORF), then the ORFs were translated into amino acid sequences. The translated sequences were submitted to the SMART program4 to identify conserved domains. Compute pl/Mw tool5 was used to predict the information of isoelectric point (PI) and molecular weight. The protein structures of all the identified C1Q proteins were drawn with IBS1.0.3 software (Adema et al., 2017) and Geneious 7.1.76 was used to predict protein secondary structure. A web server GSDS (Gene Structure Display Server)7 was used to draw gene structure schematic diagrams. Based on the whole genome sequence, we analyzed the distribution of C1Q genes in genome and their gene linkage relationship.
To determine which subfamily the CfC1Q genes belong to, the phylogenetic analysis was performed using MEGA7 (Kumar et al., 2016). All CfC1Q proteins as well as C1Q proteins from human (Homo sapiens), chicken (Gallus gallus), frog (Xenopus laevis), zebrafish (Danio rerio), and mouse (Mus musculus) were employed to construct a phylogenetic tree. The C1Q protein sequences from these reference species were obtained from public databases such as NCBI (see footnote) and UNIPROT (see footnote). Multi-sequence alignment was performed using ClustalW (Thompson et al., 2003), and then was edited by Genedoc software (Nicholas, 1997). Poisson model and uniform rates were selected as the best fit model from MEGA 7.0. The tree was constructed based on neighbor-joining (NJ) method with a bootstrap of 1,000 replicates. Under the least stringent parameters, the poorly aligned amino acid positions were excluded using Gblocks, as implemented in the online version of the program (Castresana, 2000). In total, this analysis involved 235 amino acids across six animals.
In order to test for positive selection at individual amino acid codons, the specific site model was implemented using the program of the PAML (Yang, 2007; Xun et al., 2020) under maximum-likelihood (ML) and Nei and Gojobori (NG) approach. Site model supposes that different branches of the phylogenetic tree are subject to the same selection pressure, but different amino acid positions experience different selection pressures (Gao et al., 2019). To measure the selective pressure at the protein level, this model allows the ratio to vary among sites of the expanded C1Qs to estimate Non-synonymous (dN) and Synonymous (dS) substitution ratios (dN/dS or ω). Different positions of the same sequence have different ω values. In the situation of a ω = 1, <1, or >1 indicates neutral evolution, purifying selection/negative selection, or positive selection, respectively. In total, eight different hypothetical models were tested. Model M0 (single ratio) assumes that all sites have the same value of ω, whereas model M3 (discrete) assumes that all sites have a simple discrete distribution trend. Model M1a (neutral) assumes that there are two classes of sites in proteins with 0 < ω < 1 (conservative sites) and ω = 1 (neutral sites), and the ratio of these two types of sites is p0 and p1, respectively. M2a (selection) adds a proportion (p2) to account for a class of sites where ω 2 is estimated from the data and can be > 1. Model M7 (beta) assumes that ω at all sites belongs to the matrix (0, 1) and is in beta distribution, whereas model M8 (beta and ω) additionally contains ω value (ω > 1). If the data fitted by M8 (M2a) is significantly better than M7 (M1a), we will apply the Bayesian Empirical Bayes (BEB) estimation method to determine the site category estimated to be positively selected.
The expression profiles of Zhikong scallop across all developmental stages and in adult organs/tissues were analyzed based on the RNA-seq data from our previous research (Li et al., 2017). Heat map of expression patterns of CfC1Q genes was drawn using custom R scripts8 (Robinson et al., 2010).
Three hundred 2-years-old adult C. farreri were collected from the hatchery of Xunshan Group Co. Ltd. (Shandong, China). The scallops without shell damage were transported on ice to the laboratory and acclimated for 1 week in a recirculated aquarium system equipped with a filtering system. For the next 3 weeks, the scallops were fed with non-toxic microalgae Isochrysis galbana for depuration. After depuration, scallops were separated into six ventilated tanks independently as six different experimental group (50 scallops/each group). Three groups were fed with toxic A. minutum (AM-1 strain) and the other three groups were fed with toxic A. catenella (ACDH strain). Each scallop was fed once a day with 2,500 cells/mL for microalgae in a 3L volume and it could filter nearly all the microalgal cells within the same day (Navarro et al., 2006; García-Lagunas et al., 2013). Before feeding with toxic microalgae, samples were taken as control (0 day). After feeding with A. minutum or A. catenella, samples were taken at different test time points (1, 3, 5, 10, and 15 days) as treatment group. In detail, the specimens that were sampled at 1 and 3 days were taken as the acute response group and the specimens that were sampled at 5, 10, and 15 days were taken as chronic response group. At each test time point, three random scallops were selected and the hepatopancreas and kidney sample from each scallop were dissected and stored in liquid nitrogen directly for further RNA-seq analysis.
According to the phenol–chloroform method of RNA extraction (Shafer et al., 1997; Fanson et al., 2000), total RNA was extracted from 1mg sampled tissue specimen obtained from section “Feeding the Scallops With Two Different Toxic Dinoflagellates: A. minutum and A. catenella.” RNA-seq library was constructed using NEB Next mRNA Library Prep Kit, following the manufacturer’s instructions, and was subjected to PE125 sequencing on the Illumina HiSeq 2000 platform (Park et al., 2019). After the RNA-seq data were pre-processed, quality filtering was carried out to screen out the sequences. The raw sequencing sequence was first processed with low quality pass rate, and then the sequence errors with NN were removed after the adapter sequences were removed. More than 80% of the sequences with a site quality value greater than 30 points were regarded as high-quality sequences and reserved for subsequent analysis. The RNA-seq sequences were aligned to the genome with Tophat (ver2.0.9). Expression of all C1Q genes was normalized and represented in the form of RPKM (Reads Per Kilobase of exon model per Million mapped reads) based on R software, following the formula total exon reads/mapped reads (millions)/exon length (Kb) (Hu et al., 2019). RPKM of the C1Q genes obtained from RNA-seq data were used to analyze their expression profile in kidney and hepatopancreas with different toxic dinoflagellates diets. Using the R package edgeR, the significant differences between the test group and the control group and the differentially expressed genes (DEGs) were detected with | log 2 FC | > 2 and p < 0.05. Using MeV 4.90 software9, a heat map was drawn.
In this study, a total of 97 C1Q genes were identified based on the genome and transcriptome of C. farreri. There were 77 members of C1QL, 7 members of C1QTNF, and 12 members of C1QDC1. The basic information (gene name, genomic location, number of exons, protein length, number of C1Q domains, isoelectric point, and molecular weight) of these C1Q genes was summarized in Supplementary Table 1. The coding sequence (CDS) of the longest C1Q gene was 2,331 bp, while the CDS of the shortest C1Q gene was only 97 bp. The exon number of cfC1Q genes varies from 1 to 10 (Figure 1), with encoding protein lengths ranging from 49 to 758 amino acids (Figure 2). The predicted molecular weights of CfC1Q proteins ranged from 5.1–84.9 kDa, with PIs from 3.97 to 9.49. At the N-terminal of all 97 CfC1Q proteins, the conserved C1Q domain could be detected, from which 89 members contained one C1Q domain and eight members contained two C1Q domains. In addition, there were 21 CfC1Q proteins containing 1–4 coiled coil structures at the C-terminus. Further, we found most CfC1Qs contained one or more α-helix structures at the N-terminus or C-terminus, and there were multiple β-sheets, corners, and coiled coil structures in the middle (Supplementary Figure 1). According to the results of multiple sequence alignment of C1Q domain amino acids, most of the aromatic amino acids of C. farreri were relatively conservative, such as phenylalanine (F), tyrosine (Y), and tryptophan (W) (Supplementary Figure 2). It was consistent with previous findings that those aromatic amino acid residues participated in architecting the hydrophobic core of C1Q center (Liu et al., 2013). Genomic distribution analysis showed that C1Q gene linkage phenomenon was widespread in C. farreri (Supplementary Table 2). There were 66 C1Q genes found unequally distributed on 44 scaffolds, and among them, 35 genes were found located in terms of gene clusters. Of note, scaffold26237, scaffold64279, and scaffold56669 contained the biggest C1Q gene cluster, consisting of four C1QL members. Scaffold48207 and scaffold49013 were found to consist of three C1QL and C1QDC1 members, respectively. In addition, out of the 18 chromosomes that C1Q genes located, chromosome 5 contained the largest number of C1Q genes, ten of which were all members of C1QL, followed by chromosomes 18 and 19, both of which contained seven C1Q genes.
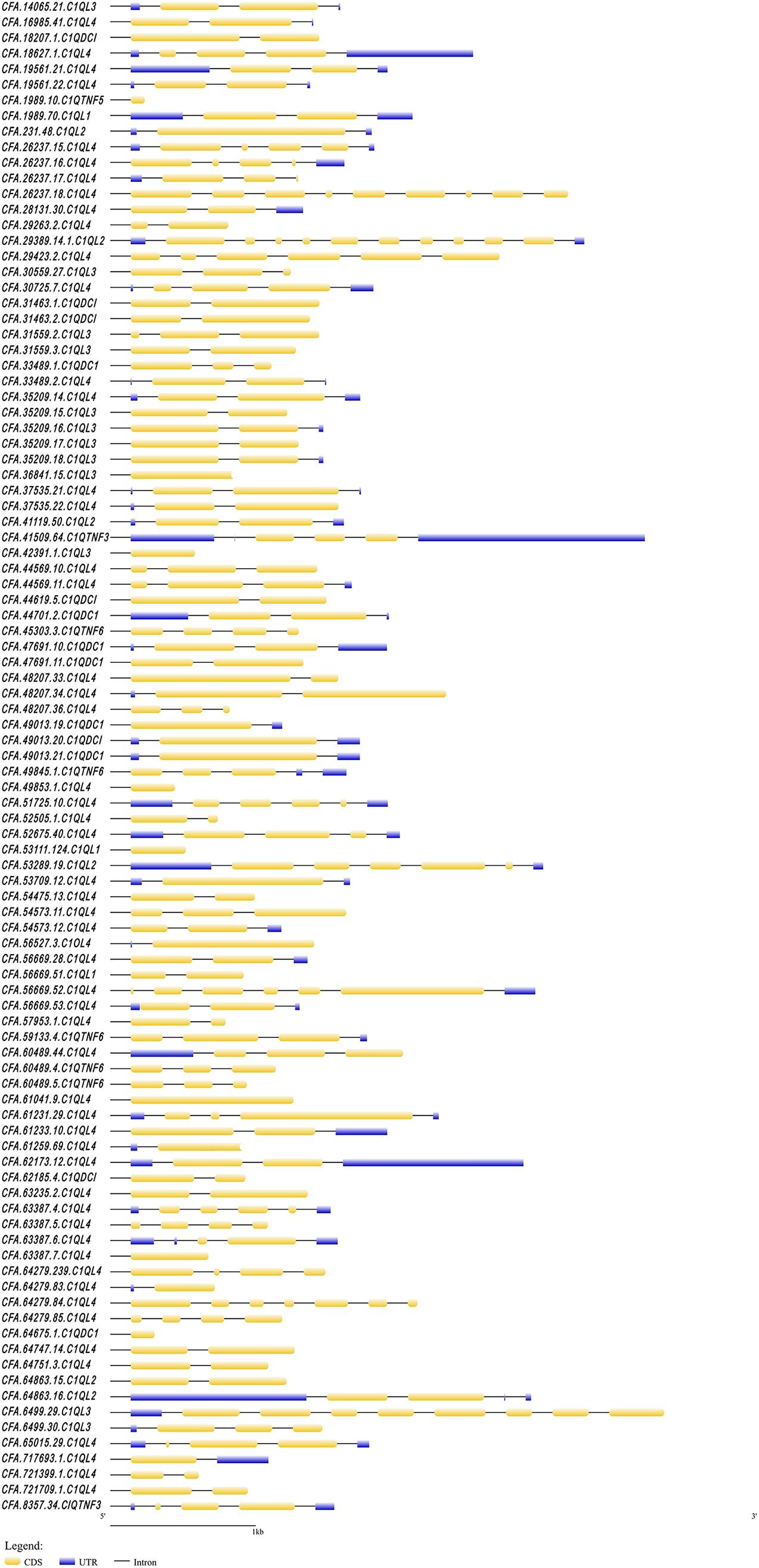
Figure 1. The structure of C1Q genes in C. farreri. The yellow boxes indicate the CDS regions. The blue boxes indicate the UTR regions.

Figure 2. The structure of C1Q proteins in C. farreri. The navy-blue boxes indicate the conserved C1Q domains. The orange boxes indicate coiled coil. The green boxes indicate low complexity regions (for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method (Figure 3). We noticed that subfamily C1Q-B (red) and C1Q-C (pink) were clustered together first. In comparison, members of C1Q-A subfamily from vertebrate were found clustered together as a branch (blue). All CfC1Qs except one protein, CFA.C1QTNF5.1989.10 (green), were found clustered into C1Q-B subfamily, revealing the significant expansion of this subfamily in C. farreri. To put it further, members of the C1Q-B subfamily were subdivided into four subcategories, namely C1QDC1, C1QL, CBLN, C1QTNF, and, respectively, represented as yellow, light blue, dark purple, and rose red. Of note, the CBLN members were found lost in C. farreri. The expansion of CfC1QLs was most obvious, in which 77 members were identified, accounting for 80% of CfC1Q members.
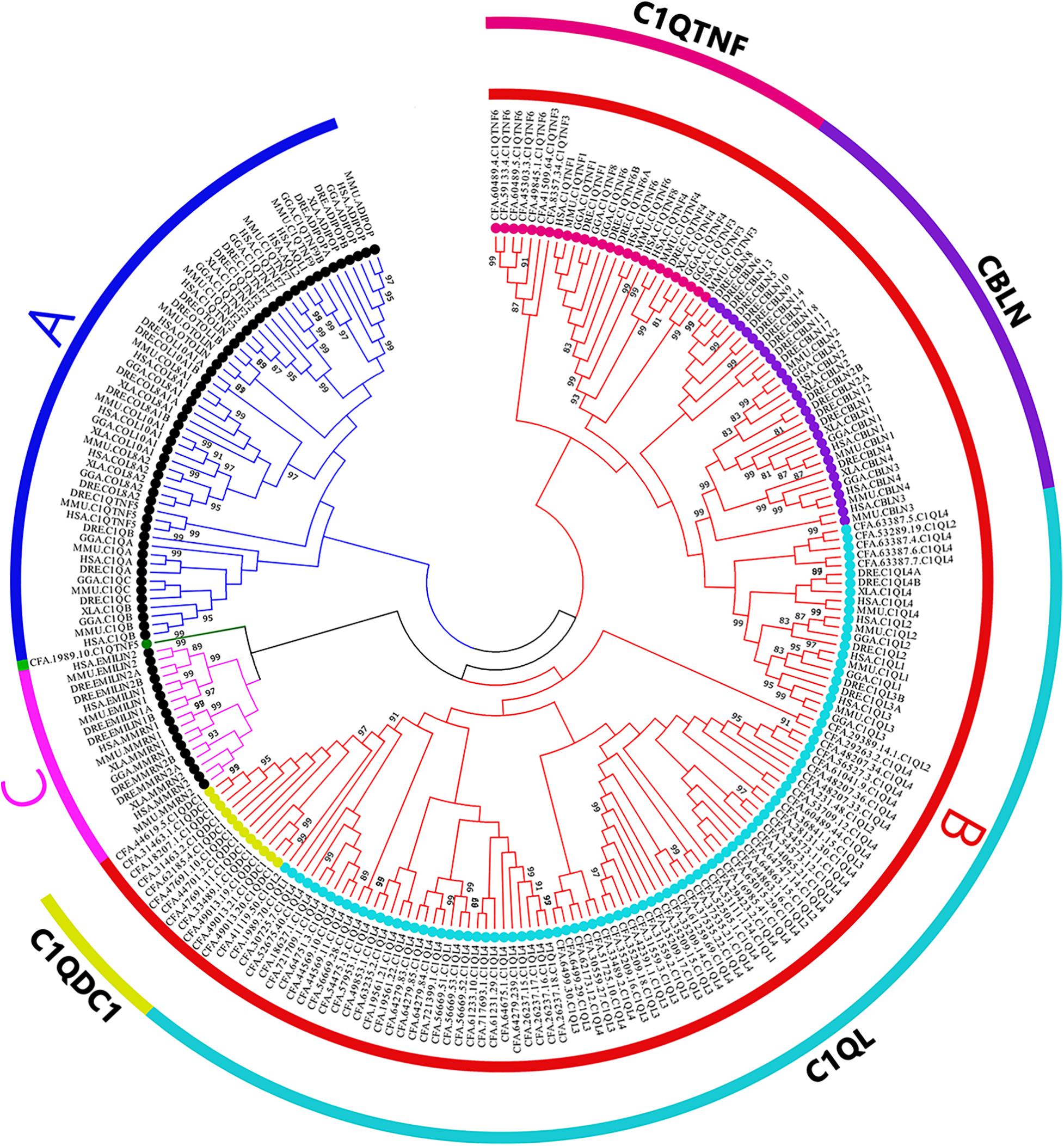
Figure 3. Phylogenetic trees of selected organisms’ C1Q amino acid sequences. The whole amino acid sequence of C1Q proteins were used to perform multiple alignment and phylogenetic analysis using NJ method computed using the poisson correction method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown above the branches. In the figure, different symbols were used to represent different subfamilies. Black solid dots represent A and C subfamily, colored solid dots represent B subfamily (HSA; MMU, Mus musculus; GGA, Gallus gallus; XLA, Xenopus laevis; DRE, Danio rerio; CFA).
In order to explore whether or not the expanded CfC1Q genes were impacted under positive selective pressure, we assess their non-synonymous and synonymous substitution divergences (dN/dS, ω) using the codon models implemented in PAML. As shown in Table 1, for all three major subcategories of expanded CfC1Q-B subfamily, M0 (one-ratio) was rejected in favor of the alternative model M3 (discrete) (M3 vs. M0, p < 0.05), revealing their different ω ratios. Meanwhile, in comparison between the M7 and M8 models, the p-value of the chi-square test for CfC1QLs, CfC1QTNFs, and CfC1QDC1s, respectively, reached 1e-09, 9e-04, and 0.027, demonstrating that they all allow for the positive selection. Furthermore, one positive selection site in the C1QL subfamily and seven positive selection sites in the C1QTNF subfamily was worth noting, which further supports the existence of positive selection for the lineage specific expanded subfamilies.
Expression patterns of CfC1Q genes during different developmental stages (Figure 4) and in different adult tissues (Figure 5) were analyzed. For CfC1QLs and CfC1QDC1s, most genes began to express on a large scale until umbo-early larva stage, which continue to elevate their expression level and reach peak expression either in umbo-post larva or after creeping larva stage (with a log2RPKM ranging from 0 to 524.48). For most CfC1QTNFs, their period of high expression shifted later, usually during the creeping larva stage. However, one C1QTNF3 (CFA.8357.34) showed elevated expression during zygote cleavage and one C1QTNF6 (CFA.49845.1) showed elevated expression in blastula. Similarly, the tissue expression pattern of CfC1QLs and CfC1QDC1s is consistent, most of which showed hepatopancreas dominant expression. Of note, three CfC1QLs (CFA.64863.15, CFA.60489.44, and CFA.64279.84) and two CfC1QDC1s (CFA.49013.20 and CFA. 62185.4) out of these 90 genes showed obvious different tissue-dominant expression, differentially in female gonad, gill, kidney, and smooth muscle. Meanwhile, diverse tissue expression could be detected for C1QTNF genes. Similar to the situation of CfC1QLs and CfC1QDC1s, three C1QTNF6 genes (CFA.60489.5, CFA.45303.3, and CFA.49845.1) exhibited high mRNA levels, respectively, in kidney, female gonad, and smooth muscle. Two other members of C1QTNF6 and both C1QTNF3 members showed highest expression in mantle, suggesting their differential biological function in comparison with hepatic major expressed CfC1Qs.
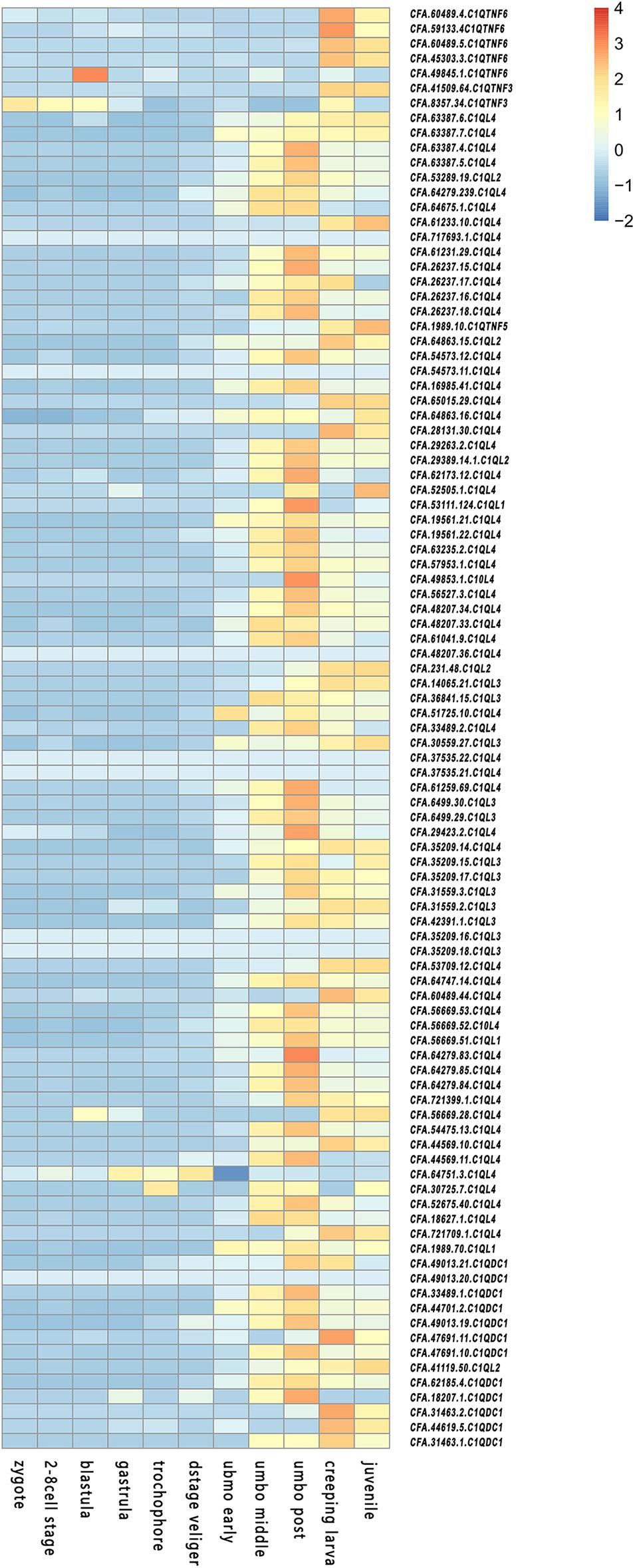
Figure 4. Temporal expression profiles of C1Q during embryonic and larval development in C. farreri.
In kidney, the organ that has been proven to transform incoming PSTs into derivatives with higher toxicity (Hu et al., 2019), more than 85% of CfC1QLs and CfC1QDC1s showed drastic up-regulation with both diets (Figure 6 and Supplementary Table 3). After feeding A. minutum, 26 C1QL members and 2 C1QDC1 members were significantly up regulated (| log2FC| > 2). After feeding A. catenella, 18 C1QL members and 1 C1QDC1 member were significantly up-regulated (| log2FC| > 2). However, among the members with significant induction, most of them showed different response manners after feeding with A. minutum or A. catenella. With A. minutum diets, 26 CfC1Qs showed acute responses, while with A. catenella diets, chronic responses could be detected in 13 CfC1Qs. After feeding with A. minutum or A. catenella, all C1QTNF6 genes showed down regulation tendencies on day 10 and day 15. With AM-1 diet, two members (CFA.59133.4 and CFA.49845.1) showed significantly reduced expression on day 5 and day 1, respectively. All C1QTNF6 showed significant downregulation with ACDH diet, most of which responded acutely on day 1 or day 3, while only one gene (CFA.59133.4) responded chronically on day 5.
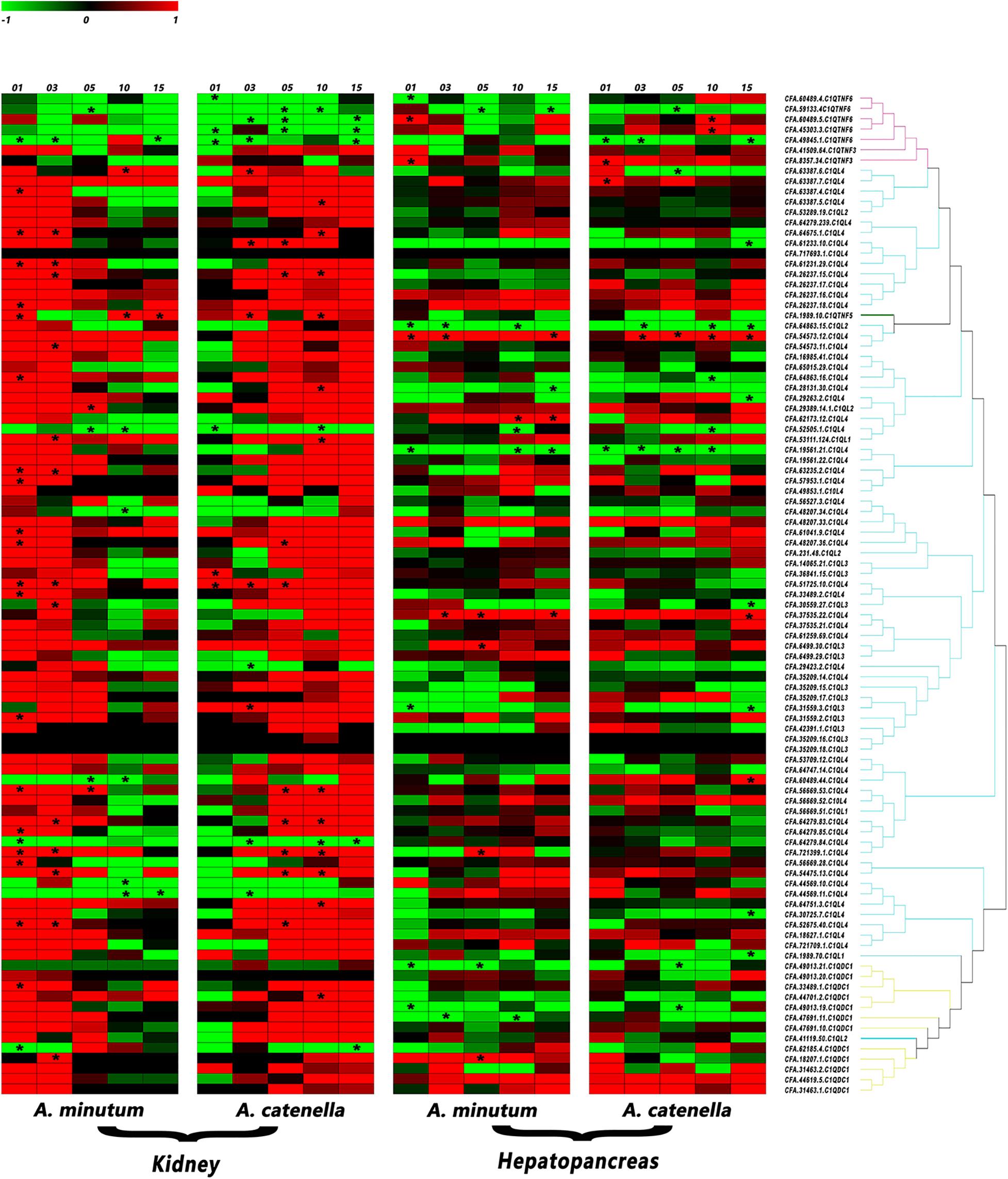
Figure 6. Heatmap of C1Q genes expression in C. farreri hepatopancreas and kidneys after feeding with AM-1 and ACDH. The red branch represents the C1QTNF member, the blue branch represents the C1QL member, and the yellow branch represents the C1QDC1 member. * indicated the significantly regulated C1Q genes with | log2 FC | > 2and p < 0.05 (for interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
In hepatopancreas, the main organ for accumulating the incoming microalgae-derived PSTs, most CfC1Qs only showed mild regulation pattern with toxic dinoflagellates diets (Figure 6 and Supplementary Table 3). Together, a total of 29 gene members showed significant responses with two toxic diets, among which 11 and 18 members, respectively, showed up- and down-regulation patterns. Different from the situation in kidney, for C1QTNFs, C1QLs, and C1QDC1s, no obvious or consistent regulation pattern could be found between or within these categories. However, we did find several members which showed consistent regulation with both toxic diets. For example, after feeding with AM-1 and ACDH diets, significantly declined expression of three C1QL members (CFA.64863.15, CFA.19561.21, CFA.52505.1) and one C1QTNF6 (CFA.59133.4) could be detected, in acute and chronic manners, respectively. Besides, expressions of one C1QTNF3 (CFA.8357.34) and one C1QL (CFA.54573.12) were both significantly and acutely enhanced after feeding with AM-1 and ACDH diets. Meanwhile, the dinoflagellate-type dependent response manner that was mentioned in kidney was also detected for several other genes. For example, with AM-1 and ACDH diet, C1QTNF6 (CFA.60489.5) and C1QL (CFA.37535.22) showed acute and chronic up-regulation, respectively, not to mention that four members (CFA.62173.12, CFA.6499.30, CFA.721399.1, and CFA.18207.1) only showed significant up-regulation with AM-1 diet.
Filter-feeding marine bivalves are able to cope with highly potent microalgae-derived neurotoxins, such as paralytic shellfish toxins (PSTs). Exploring the genes related to toxin resistance in bivalves could benefit our understanding of the adaptations of these species. C1Q proteins are active molecules with multiple biological functions (Mj and Mf, 2016). They can activate the classical pathway of complement, clear pathogens, phagocytose, and lyse bacteria, and regulate inflammation (Kishore and Reid, 2000). It was reported that C1Qs could be involved in the detoxification processes in bivalves, in response to the induced stresses and the affected physiological state (Pazos et al., 2017). Zhikong scallops possess the remarkable ability to accumulate PSTs. In this study, we systematically identified CfC1Q genes and analyzed their responsive pattern after feeding with PST-producing dinoflagellates. The candidate responsive CfC1Qs might be developed as potential molecular indicators for monitoring toxin accumulation in edible scallops.
The C1Q proteins that widely existed in many species are structurally conserved. The typical C1Q domain can form a jam roll shape composed of 10 β-strands. Eight conserved amino acid residues exist in the center of the C1Q domain, among which five aromatic amino acid residues constitute the central hydrophobic core, and these conserved amino acid residues play an important role in forming or stabilizing the hydrophobic core of the C1Q domain (Gaboriaud et al., 2012). As shown in Figure 4, most of the aromatic amino acids of C. farreri were relatively conservative, which may suggest a similar hydrophobic function (Liu et al., 2013). On a side note, the existence of the conserved C1Q domain also reflects its role as one kind of charge pattern recognition molecule in order to bind certain ligands (Gestal et al., 2010). In C. farreri, approximately 90% C1Q proteins contain 1-2 C1Q domains at the N-terminal, and 0–1 coiled coil structures at the C-terminal, consistent with the C1Q structural characteristics of D. rerio and H. sapiens. Of note, there are nine CfC1Q proteins that contain multiple curled helices, and one member possesses three C1Q domains, showing a distinguishable structure that might be specific in bivalve lineage.
Despite their wide existence, the retention or loss of C1Q genes occurred during the evolution of metazoans (Supplementary Table 4). Fungi and Plantae seem to be completely free of C1Q genes (Yuzaki, 2008). According to a previous study, a total of seven C1Q genes have been identified in sea urchin (Hibino et al., 2006), and only two C1Qs were found in sea squirt Ciona intestinalis (Azumi et al., 2003). Then, the number of C1Qs seems to increase during evolution, for instance, 50 C1Q genes were discovered in amphioxus Branchiostoma floridae, 52 genes in zebrafish D. rerio, and 31 in human (Tom Tang et al., 2005; Huang et al., 2008; Mei and Gui, 2008; Yuzaki, 2008). Further, we found that the C1Q genes had quite obvious gene expansion in mollusks. For example, in mussel Mytilus californianus and M. galloprovincialis, respectively, 52 and 168 genes could be found, and in oyster C. gigas, 321 C1Q genes have been found (Gerdol et al., 2011; Zhang et al., 2015). In the present study, 97 C1Qs were identified in scallop C. farreri, from which the C1Q-B subfamily was found being mainly expanded. Phylogenetic analysis showed that proteins of C1QDC1 and C1QL were first clustered together, suggesting they may share a common ancestor gene. Besides, the CBLN members were lost in C. farreri, and the CfC1QTNF members were isolated in a single cluster and separated from the C1QTNF members of vertebrates, indicating the independent gene replication events in the bivalve lineage. Tandem gene duplication and an unequal crossing over drove the expansion of the C1Q gene family in bivalves, which represented an important evolutionary advantage in these filter-feeding organisms (Gerdol et al., 2019). Under the circumstances of various stressors in the ocean, the expansion of bivalve C1Q genes may provide insights into lineage specific adaptations. Adaptive evolution is usually driven by positive selection, the acquisition of new genes combined with the modification of existing genes (Ellegren, 2014). Our results showed that, for all three major subcategories of expanded CfC1Q-B subfamily, they all allow for the positive selection, and one positive selection site in the CfC1QL subfamily and seven positive selection sites in the CfC1QTNF subfamily was revealed. These results also remind us that the C1Q gene family could be a good candidate gene family for lineage specific molecular evolution analyzation.
Expression of C1Q genes in C. gigas was found to be tightly regulated during early development, most of which were not expressed at all until the larva settling (Gerdol et al., 2015). Researchers pointed out that, during the planktonic/benthic life transition, the cumulative expression of C1Q genes in the frame of a radical reorganization of body parts may serve to suit a sedentary existence. Consistent expression patterns during the development of C. farreri were found. Staggered expression peaks for different CfC1Q subcategories could be detected, which may suggest their functional differentiation during development. Most CfC1QLs and CfC1QDC1s showed obvious elevated expression in umbo larva, which might be involved with the formation of eye points and motor organs. However, most CfC1QTNFs began to express in a large scale during the metamorphosis development and may contribute to stress tolerance to assist with body reorganization.
The specificity of C1Q genes’ expression in a certain tissue may be related to the tissue specific functions (Yang et al., 2013). From Figure 6, we can see that C1Q genes were highly specifically expressed in hepatopancreas, consistent with previous study results in Pacific oyster, giant freshwater prawn, and Nile tilapia (Fabioux et al., 2015; Ye et al., 2015). Hepatopancreas is an important organ for digestion and absorption, and it is equivalent to the fat body of insects or the liver of mammals, serving as the first defense barrier against pathogen invasion (Gross et al., 2001). Therefore, we speculate that the CfC1Q genes play important roles in resisting external aggressions in the organism and maintaining the steady state of the internal environment. Besides, most C1QTNF members showed relative high expression level in mantle, the tissue that possess numerous tentacles and eyes to perceive the surrounding environment (Land, 1966). The enhancement of sense skills expands the range of exercise against predators and is conducive for food intake, and mantle-specific high level of C1QTNF expression might contribute to future studies.
Zhikong scallops could tolerate and accumulate potent neurotoxic PSTs. PSTs comprise several structural variants, such as saxitoxin (STX), sulfated gonyautoxins (GTX), and N -sulfocarbamoyl toxins (B- and C-toxins), of which STX is the most neurotoxic (Thottumkara et al., 2014). Our previous study found that Zhikong scallop possess two sodium channel genes (Nav1 and Nav2), and Nav1 has a potentially toxin-resistant T mutation at position 1,425 (Li et al., 2017). This mutation has been proven to be able to yield a significant resistance to STX and TTX in rat (Jost et al., 2008). Alexandrium spp. is one of the major PSTs-producing dinoflagellates. It has been reported that A. minutum (AM-1 strain) mainly produces gonyautoxins (mainly GTX1-4), while A. catenella (ACDH strain) mainly produces N-sulfocarbamoyl toxins (C1/2) (Tian et al., 2010; Li et al., 2017; Blossom et al., 2019). Hepatopancreas and kidney are two major bivalve toxic organs (Tian et al., 2010; Li et al., 2017), and diverse regulation of CfC1Qs presented in hepatopancreas and kidney after challenge with A. minutum or A. catenella. Compared with hepatopancreas, it is obvious that the responses of most CfC1Qs were more intense in kidney, which may attribute to the more toxic analogs’ transformation in kidney (Joerink et al., 2006; Tellez-Bañuelos et al., 2009; Xu et al., 2018). Besides, dinoflagellate type dependent gene regulation was also detected in kidney. For CfC1QLs and CfC1QDC1s, acute and chronic responses could be, respectively, detected after feeding with A. minutum and A. catenella. This result reinforced our inference that different toxin composition and virulence could be the causal factors for CfC1Q genes’ responsive pattern. Meanwhile, although not significant, the induction of the CfC1QTNF3 (CFA.41509.64) in kidney could be detected just like the regulation of CfC1QLs and CfC1QDC1s. However, the responses of five CfC1QTNF6 members in kidney are obviously regulated in the opposite way, demonstrating the functional differentiation for CfC1Q genes. In both kidney and hepatopancreas, two C1Qs (CFA.721399.1 and CFA.18207.1) were significantly up-regulated with AM-1 diet, and two members (CFA.59133.4 and CFA.49845.1) were significantly down-regulated with ACDH diet. Only one member (CFA.52505.1) showed consistent significant downregulation with both AM-1 and ACDH diet and in both kidney and hepatopancreas, and these five genes could be good candidates as tissue responsive indicator genes with PSTs challenge.
The present study demonstrated a systematic genomic and transcriptomic survey of C1Q genes in C. farreri. Phylogenetic analysis indicated a bivalve-specific gene duplication event may occur for C1Q genes, and expansion of CfC1Q-B subfamily was revealed. Diverse spatiotemporal expression patterns of C1Q genes were observed, and their response manners to the stresses of PST-producing A. minutum and A. catenella varied between scallop tissues or between dinoflagellate strains. These findings may suggest the adaptive functional diversity of expanded C1Q genes in C. farreri, which could serve as a potential candidate gene family for lineage specific molecular evolution analysis and several members could be developed as potential indicator genes for PST challenges. We believe our study provides valuable information that would offer insights into the evolution and functional characteristics of C1Q genes in bivalves, and in the future, further in-depth research on the functional divergence for different members would be needed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The accession numbers of genomic data are SRX1305705, SRX2486272, SRX2486273, SRX2486281–SRX2486284, SRX2486300, and SRX2913253–SRX2913260. The accession numbers of transcriptomic data are SRX2444844–SRX2444876, SRX2508197–SRX2508199, SRX2444668–SRX2444682, SRX2445405–SRX2445440, and SRR11840040–SRR11840053.
SL, SW, and ZB conceived and designed the study. KX and YW performed the experiments. XC and XD participated in data analysis. KX and SL wrote the manuscript. LZ and JH participated in the revision of the article. All authors contributed to the article and approved the submitted version.
We acknowledge the grant support from the National Natural Science Foundation of China (32002373), the Fundamental Research Funds for the Central Universities (202064008), the Project of Sanya Yazhouwan Science and Technology City Management Foundation (SKJC-KJ-2019KY01), and the Taishan Scholar Project Fund of Shandong Province of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.640425/full#supplementary-material
Supplementary Figure 1 | The secondary structures of C1Q domain. The pink cylinder stands for alpha helixes; the orange arrows stand for beta strands. The curved arrows stand for turns and the wavy lines stand for coils.
Supplementary Figure 2 | Alignment of deduced C1Q amino acid sequences. The gray and black shadow, respectively, show the most shared amino acids and the conserved amino acid group (such as aromatic amino acids) in C. farreri.
Supplementary Table 1 | Summary of the C1Q gene family in Chlamys farreri.
Supplementary Table 2 | Distribution of C1Q genes on chromosome of Chlamys farreri genome.
Supplementary Table 3 | Summary of C1Q genes regulated expression in C. farreri kidney and hepatopancreas after feeding the scallops with PST-producing AM-1 and ACDH.
Supplementary Table 4 | Distribution of C1Q genes in different species.
Adema, C. M., Hillier, L. W., Jones, C. S., Loker, E. S., Knight, M., Minx, P., et al. (2017). Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 8:15451. doi: 10.1038/ncomms15451
Anderson, D. M., Cembella, A. D., and Hallegraeff, G. M. (2012). Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 4, 143–176. doi: 10.1146/annurev-marine-120308-081121
Azumi, K., De Santis, R., De Tomaso, A., Rigoutsos, I., Yoshizaki, F., Pinto, M. R., et al. (2003). Genomic analysis of immunity in a urochordate and the emergence of the vertebrate immune system: “waiting for Godot.” Immunogenetics 55, 570–581. doi: 10.1007/s00251-003-0606-5
Benoit, M. E., Hernandez, M. X., Dinh, M. L., Benavente, F., Vasquez, O., and Tenner, A. J. (2013). C1q-induced LRP1B and GPR6 proteins expressed early in Alzheimer Disease mouse models, are essential for the C1q-mediated protection against Amyloid-β neurotoxicity∗. J. Biol. Chem. 288, 654–665. doi: 10.1074/jbc.M112.400168
Blossom, H. E., Markussen, B., Daugbjerg, N., Krock, B., Norlin, A., and Hansen, P. J. (2019). The cost of toxicity in microalgae: direct evidence from the Dinoflagellate Alexandrium. Front. Microbiol. 10:1065. doi: 10.3389/fmicb.2019.01065
Borcier, E., Morvezen, R., Boudry, P., Miner, P., Charrier, G., Laroche, J., et al. (2017). Effects of bioactive extracellular compounds and paralytic shellfish toxins produced by Alexandrium minutum on growth and behaviour of juvenile great scallops Pecten maximus. Aquat. Toxicol. 184, 142–154. doi: 10.1016/j.aquatox.2017.01.009
Boulanger, L. M. (2009). Immune Proteins in Brain Development and Synaptic Plasticity. Neuron 64, 93–109. doi: 10.1016/j.neuron.2009.09.001
Bricelj, V. M., Connell, L., Konoki, K., MacQuarrie, S. P., Scheuer, T., Catterall, W. A., et al. (2005). Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 434, 763–767. doi: 10.1038/nature03415
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evolut. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Ellegren, H. (2014). Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evolut. 29, 51–63. doi: 10.1016/j.tree.2013.09.008
Fabioux, C., Sulistiyani, Y., Haberkorn, H., Hégaret, H., Amzil, Z., and Soudant, P. (2015). Exposure to toxic Alexandrium minutum activates the detoxifying and antioxidant systems in gills of the oyster Crassostrea gigas. Harmful Algae 48, 55–62. doi: 10.1016/j.hal.2015.07.003
Fanson, B. G., Osmack, P., and Di Bisceglie, A. M. (2000). A comparison between the phenol–chloroform method of RNA extraction and the QIAamp viral RNA kit in the extraction of hepatitis C and GB virus-C/hepatitis G viral RNA from serum. J. Virol. Methods 89, 23–27. doi: 10.1016/S0166-0934(00)00192-0
Gaboriaud, C., Frachet, P., Thielens, N. M., and Arlaud, G. J. (2012). The human C1q globular domain: structure and recognition of non-Immune selfl igands. Front. Immunol. 2:92. doi: 10.3389/fimmu.2011.00092
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., and Ho, S. Y. W. (2019). EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
García-Lagunas, N., Romero-Geraldo, R., and Hernández-Saavedra, N. Y. (2013). Genomics study of the exposure effect of gymnodinium catenatum, a paralyzing toxin producer, on Crassostrea gigas’ defense system and detoxification genes. PLoS One 8:e72323. doi: 10.1371/journal.pone.0072323
Gerdol, M., De Moro, G., Manfrin, C., Milandri, A., Riccardi, E., Beran, A., et al. (2014). RNA sequencing and de novo assembly of the digestive gland transcriptome in Mytilus galloprovincialis fed with toxinogenic and non-toxic strains of Alexandrium minutum. BMC Res. Notes 7:722. doi: 10.1186/1756-0500-7-722
Gerdol, M., Greco, S., and Pallavicini, A. (2019). Extensive tandem duplication events drive the expansion of the C1q-Domain-containing gene family in bivalves. Mar. Drugs 17:583. doi: 10.3390/md17100583
Gerdol, M., Manfrin, C., De Moro, G., Figueras, A., Novoa, B., Venier, P., et al. (2011). The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis: A widespread and diverse family of immune-related molecules. Dev. Comparat. Immunol. 35, 635–643. doi: 10.1016/j.dci.2011.01.018
Gerdol, M., Venier, P., and Pallavicini, A. (2015). The genome of the Pacific oyster Crassostrea gigas brings new insights on the massive expansion of the C1q gene family in Bivalvia. Dev. Comparat. Immunol. 49, 59–71. doi: 10.1016/j.dci.2014.11.007
Gestal, C., Pallavicini, A., Venier, P., Novoa, B., and Figueras, A. (2010). MgC1q, a novel C1q-domain-containing protein involved in the immune response of Mytilus galloprovincialis. Dev. Comparat. Immunol. 34, 926–934. doi: 10.1016/j.dci.2010.02.012
Ghebrehiwet, B., Hosszu, K., Valentino, A., and Peerschke, E. I. B. (2012). The C1q family of proteins: insights into the emerging non-traditional functions. Front. Immunol. 3:52. doi: 10.3389/fimmu.2012.00052
Gross, P. S., Bartlett, T. C., Browdy, C. L., Chapman, R. W., and Warr, G. W. (2001). Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific White Shrimp, Litopenaeus vannamei, and the Atlantic White Shrimp, L. setiferus. Dev. Comparat. Immunol. 25, 565–577. doi: 10.1016/S0145-305X(01)00018-0
Hibino, T., Loza-Coll, M., Messier, C., Majeske, A. J., Cohen, A. H., Terwilliger, D. P., et al. (2006). The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349–365. doi: 10.1016/j.ydbio.2006.08.065
Hu, X., Guo, H., He, Y., Wang, S., Zhang, L., Wang, S., et al. (2010). Molecular characterization of Myostatin gene from Zhikong scallop Chlamys farreri (Jones et Preston 1904). Genes Genet. Syst. 85, 207–218. doi: 10.1266/ggs.85.207
Hu, B., Li, M., Yu, X., Xun, X., Lu, W., Li, X., et al. (2019). Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 234, 62–69. doi: 10.1016/j.chemosphere.2019.06.034
Huang, S., Yuan, S., Guo, L., Yu, Y., Li, J., Wu, T., et al. (2008). Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 18, 1112–1126. doi: 10.1101/gr.069674.107
Joerink, M., Ribeiro, C. M. S., Stet, R. J. M., Hermsen, T., Savelkoul, H. F. J., and Wiegertjes, G. F. (2006). Head kidney-derived macrophages of Common Carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J. Immunol. 177, 61–69. doi: 10.4049/jimmunol.177.1.61
Jost, M. C., Hillis, D. M., Lu, Y., Kyle, J. W., Fozzard, H. A., and Zakon, H. H. (2008). Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evolut. 25, 1016–1024. doi: 10.1093/molbev/msn025
Kishore, U., and Reid, K. B. M. (2000). C1q: Structure, function, and receptors. Immunopharmacology 49, 159–170. doi: 10.1016/S0162-3109(00)80301-X
Kong, P., Zhang, H., Wang, L., Zhou, Z., Yang, J., Zhang, Y., et al. (2010). AiC1qDC-1, a novel gC1q-domain-containing protein from bay scallop Argopecten irradians with fungi agglutinating activity. Dev. Comparat. Immunol. 34, 837–846. doi: 10.1016/j.dci.2010.03.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evolut. 33, 1870–1874. doi: 10.1093/molbev/msw054
Land, M. F. (1966). Activity in the optic nerve of Pecten maximus in response to changes in light intensity, and to pattern and movement in the optical environment. J. Exp. Biol. 45, 83–99.
Li, M., Ager, R. R., Fraser, D. A., Tjokro, N. O., and Tenner, A. J. (2008). Development of a humanized C1q A chain knock-in mouse: assessment of antibody independent ß-amyloid induced complement activation. Mol. Immunol. 45, 3244–3252. doi: 10.1016/j.molimm.2008.02.022
Li, S.-C., Wang, W.-X., and Hsieh, D. P. H. (2002). Effects of toxic dinoflagellate Alexandrium tamarense on the energy budgets and growth of two marine bivalves. Mar. Environ. Res. 53, 145–160. doi: 10.1016/S0141-1136(01)00117-9
Li, Y., Sun, X., Hu, X., Xun, X., Zhang, J., Guo, X., et al. (2017). Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8:1721. doi: 10.1038/s41467-017-01927-0
Liu, G., Pang, Y., Liu, X., and Li, Q.-W. (2013). [Structure, distribution, classification, and function of C1q protein family: a review]. Yi Chuan 35, 1072–1080. doi: 10.3724/sp.j.1005.2013.01072
Liu, H.-H., Xiang, L.-X., and Shao, J.-Z. (2014). A novel C1q-domain-containing (C1qDC) protein from Mytilus coruscus with the transcriptional analysis against marine pathogens and heavy metals. Dev. Comparat. Immunol. 44, 70–75. doi: 10.1016/j.dci.2013.11.009
Lou, J., Cheng, J., Xun, X., Li, X., Li, M., Zhang, X., et al. (2020). Glutathione s-transferase genes in scallops and their diverse expression patterns after exposure to PST-producing dinoflagellates. Mar. Life Sci. Technol. 2, 252–261. doi: 10.1007/s42995-020-00050-2
Mei, J., and Gui, J. (2008). Bioinformatic identification of genes encoding C1q-domain-containing proteins in zebrafish. J. Genet. Genomics 35, 17–24. doi: 10.1016/S1673-8527(08)60003-X
Navarro, J. M., Muñoz, M. G., and Contreras, A. M. (2006). Temperature as a factor regulating growth and toxin content in the dinoflagellate Alexandrium catenella. Harmful Algae 5, 762–769. doi: 10.1016/j.hal.2006.04.001
Park, Y.-S., Kim, S., Park, D.-G., Kim, D. H., Yoon, K.-W., Shin, W., et al. (2019). Comparison of library construction kits for mRNA sequencing in the Illumina platform. Genes Genom. 41, 1233–1240. doi: 10.1007/s13258-019-00853-3
Pazos, A. J., Ventoso, P., Martínez-Escauriaza, R., Pérez-Parallé, M. L., Blanco, J., Triviño, J. C., et al. (2017). Transcriptional response after exposure to domoic acid-producing Pseudo-nitzschia in the digestive gland of the mussel Mytilus galloprovincialis. Toxicon 140, 60–71. doi: 10.1016/j.toxicon.2017.10.002
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Shafer, R. W., Levee, D. J., Winters, M. A., Richmond, K. L., Huang, D., and Merigan, T. C. (1997). Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J. Clin. Microbiol. 35, 520–522. doi: 10.1128/jcm.35.2.520-522.1997
Stevens, B., Allen, N. J., Vazquez, L. E., Howell, G. R., Christopherson, K. S., Nouri, N., et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. doi: 10.1016/j.cell.2007.10.036
Takeuchi, T., Koyanagi, R., Gyoja, F., Kanda, M., Hisata, K., Fujie, M., et al. (2016). Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool. Lett. 2:3. doi: 10.1186/s40851-016-0039-2
Tellez-Bañuelos, M. C., Santerre, A., Casas-Solis, J., Bravo-Cuellar, A., and Zaitseva, G. (2009). Oxidative stress in macrophages from spleen of Nile tilapia (Oreochromis niloticus) exposed to sublethal concentration of endosulfan. Fish Shellf. Immunol. 27, 105–111. doi: 10.1016/j.fsi.2008.11.002
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using clustalW and clustalX. Curr. Protoc. Bioinformat. 2003, 2.3.1–2.3.22.
Thottumkara, A. P., Parsons, W. H., and Du Bois, J. (2014). Saxitoxin. Angewandte Chemie Int. Edn. 53, 5760–5784.
Tian, H., Gao, C., Wang, Z., Sun, P., Fan, S., and Zhu, M. (2010). Comparative study on in vitro transformation of paralytic shellfish poisoning (PSP) toxins in different shellfish tissues. Acta Oceanol. Sin. 29, 120–126. doi: 10.1007/s13131-010-0015-1
Tom Tang, Y., Hu, T., Arterburn, M., Boyle, B., Bright, J. M., Palencia, S., et al. (2005). The complete complement of C1q-domain-containing proteins in Homo sapiens. Genomics 86, 100–111. doi: 10.1016/j.ygeno.2005.03.001
Wang, L., Wang, L., Zhang, H., Zhou, Z., Siva, V. S., and Song, L. (2012). A C1q domain containing protein from scallop Chlamys farreri serving as pattern recognition receptor with heat-aggregated IgG binding activity. PLoS One 7:e43289. doi: 10.1371/journal.pone.0043289
Xu, C., Li, E., Suo, Y., Su, Y., Lu, M., Zhao, Q., et al. (2018). Histological and transcriptomic responses of two immune organs, the spleen and head kidney, in Nile tilapia (Oreochromis niloticus) to long-term hypersaline stress. Fish Shellf. Immunol. 76, 48–57. doi: 10.1016/j.fsi.2018.02.041
Xun, X., Cheng, J., Wang, J., Li, Y., Li, X., Li, M., et al. (2020). Solute carriers in scallop genome: gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere 241:124968. doi: 10.1016/j.chemosphere.2019.124968
Yang, L., Liu, X., Liu, W., Li, X., Qiu, L., Huang, J., et al. (2013). Characterization of complement 1q binding protein of tiger shrimp, Penaeus monodon, and its C1q binding activity. Fish Shellf. Immunol. 34, 82–90. doi: 10.1016/j.fsi.2012.10.002
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evolut. 24, 1586–1591. doi: 10.1093/molbev/msm088
Ye, T., Huang, X., Wang, X.-W., Shi, Y.-R., Hui, K.-M., and Ren, Q. (2015). Characterization of a gC1qR from the giant freshwater prawn, Macrobrachium rosenbergii. Fish Shellf. Immunol. 43, 200–208. doi: 10.1016/j.fsi.2014.12.030
Yuzaki, M. (2008). Cbln and C1q family proteins – New transneuronal cytokines. Cell Mol. Life Sci. 65, 1698–1705. doi: 10.1007/s00018-008-7550-3
Zhang, H., Song, L., Li, C., Zhao, J., Wang, H., Qiu, L., et al. (2008). A novel C1q-domain-containing protein from Zhikong scallop Chlamys farreri with lipopolysaccharide binding activity. Fish Shellf. Immunol. 25, 281–289. doi: 10.1016/j.fsi.2008.06.003
Keywords: PSTs challenge, bivalve mollusk, expression profiling, complement 1Q, genomic analysis
Citation: Xu K, Wang Y, Lian S, Hu N, Chen X, Dai X, Zhang L, Wang S, Hu J, Hu X and Bao Z (2021) Expansion of C1Q Genes in Zhikong Scallop and Their Expression Profiling After Exposure to the Toxic Dinoflagellates. Front. Mar. Sci. 8:640425. doi: 10.3389/fmars.2021.640425
Received: 11 December 2020; Accepted: 06 April 2021;
Published: 03 May 2021.
Edited by:
Zhi-Hua Li, Shandong University, ChinaReviewed by:
Sebastian Boltana, University of Concepcion, ChileCopyright © 2021 Xu, Wang, Lian, Hu, Chen, Dai, Zhang, Wang, Hu, Hu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Lian, bGlhbnNoYW5zaGFuQG91Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.