
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 13 April 2021
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.636636
Endophytes are microorganisms that live inside the plant tissue without causing external symptoms. All plants in nature harbor endophytes. Some endophytes produce and secrete biologically active compounds, known as secondary metabolites, which can help the host plant cope with bacterial, fungal, and other pest pathogens. Endophytes are isolated from aquatic plants and algae. Diseases are detrimental in the aquaculture industry where chemical pesticides and antibiotics are widely used in an attempt to cope with fish pathogens. However, the ability to prevent disease outbreaks in aquaculture is currently extremely limited. Here, we isolated 173 bacterial and fungal endophytes from 16 Mediterranean seaweed and 4 algae from fresh or thermo-mineral water. We found 88 endophytes (51%) with biological activity against four common aquaculture pathogens. Fifty endophytes (29%) caused mortality of at least one of these four pathogens. We identified 23 bioactive endophytes, 18 of which are from the Bacilli class. Our findings suggest that macroalgae from different aquatic environments can serve as a good source of potential biocontrol agents against aquaculture diseases. To the best of our knowledge, there are no published studies demonstrating the use of algal endophytes to control aquaculture diseases. Our study may lead to finding new molecules for use as novel environmentally friendly products that will solve one of the most challenging problems for the growing aquaculture industry: pathogens and pests.
Endophytes are a group of endosymbiotic microorganisms, often bacteria or fungi, that colonize intercellular sites in plants in natural ecosystems (Liarzi and Ezra, 2014; Gouda et al., 2016). In these mutualistic relations, the plant provides a living niche and food for the endophyte, while the endophyte provides protection against pathogens and pests and induces tolerance to abiotic stressors such as drought and salinity (Bacon and White, 2016). Some endophytes produce and secrete biologically active compounds that prevent bacteria, fungi and plant pests from growing in the host plant. These compounds are called secondary metabolites. They are produced by the endophytes and have varying roles. In recent years, endophytes and their secondary metabolites are increasingly recognized for their medicinal and industrial uses, as well as for the production of new biological control agents for agriculture (Masand et al., 2015; Ismail et al., 2016). Endophytes are also found in aquatic plants, including algae. Several studies have described the isolation of fungal and bacterial endophytes (Schulz et al., 2002; Flewelling et al., 2015; Manomi et al., 2015; Mandelare et al., 2018) and the identification of novel compounds with antibiotic activity from seaweed (Ismail et al., 2016).
For the last 20 years, aquaculture has been considered the fastest growing agricultural food sector. From 1980 to 2000, aquaculture production grew from 7.8 to 43 million tons, with an annual growth rate of 9%. Yet, from 2000 to 2018, the growth rate was reduced to 5.5% (43–114.5 million tons, respectively), and still, it continues to grow faster than other major food-producing sectors (FAO, 2018). The need to find additional protein sources to feed a growing world population has led to this remarkable growth rate.
Pathogens are a major and harmful factor in the aquaculture industry, affecting aquatic vertebrates and invertebrates worldwide. They cause huge economic losses, in excess of 6 billion USD per year (The International Bank for Reconstruction and Development/The World Bank, 2014). In addition to well-known parasites and viral diseases in aquaculture, other diseases caused by bacteria, fungi and oomycetes (water molds) are common, considered to be responsible for about 30% of all losses in aquaculture. Today, the ability to control disease outbreaks in aquaculture is very limited, consisting mainly of pesticide and antibiotic use. Malachite green, a toxic chemical compound and suspected carcinogen, was extensively used as the major chemical control against fungal diseases and parasites. Yet, it has been globally banned since 2000, as both the compound and residues from its breakdown products were found in fish flesh long after marketing (van West, 2006). Other chemicals are used to control aquaculture diseases (e.g., formaldehyde, copper sulfate, chlorine, hydrogen peroxide, and others). However, growing concerns regarding eventual harm to the consumer and environmental pollution are prompting research into alternative sources and products to control aquaculture pathogens.
In this study, endophytes isolated from algae, originating from different water sources and locations in Israel were tested for their biological activity against four major aquaculture pathogens: (i) Photobacterium damselae subsp. damselae, a gram-negative marine bacterium causing pasteurellosis (photobacteriosis), a disease characterized by wound infections and hemorrhagic septicemia. It affects many economically important fish species cultured in inland aquaculture facilities and in offshore net cages, e.g., turbot (Psetta maxima), rainbow trout (Oncorhynchus mykiss), eel (Anguilla spp.), gilt-head bream (Sparus aurata), and European bass (Dicentrarchus labrax), among others (Romalde, 2002; Rivas et al., 2013; Zhang et al., 2019); (ii) Streptococcus iniae, a gram-positive bacterium accounting for high losses in farmed marine and freshwater finfish in warmer regions, causing reduced growth rates and increased mortality, as well as changed (non-marketable) appearance. Strep. iniae affects a wide range of cultured species, including red drum (Sciaenops ocellatus), tilapia (Oreochromis spp.), rainbow trout, European bass, barramundi (Lates calcarifer), common carp (Cyprinus carpio), and more (Abutbul et al., 2004; Agnew and Barnes, 2007; Francis et al., 2014); (iii) Aeromonas salmonicida, a gram-negative bacterium that is the causative agent of furunculosis and fish septicemia. It affects major aquaculture species, in particular salmonids, causing significant economic losses (Wiklundl, 1998; Menanteau-Ledouble et al., 2016; Duarte et al., 2018); (iv) Saprolegnia parasitica, an oomycete causing saprolegniosis, a disease characterized by visible white or gray patches of filamentous mycelium on the body or fins of freshwater fish. Saprolegnia infections were previously controlled with Malachite green but with its ban, this oomycete has become an important fish pathogen, mainly of catfish (Ictalurus spp.), salmon (Salmo spp.), and trout species (van West, 2006).
Attempts have been made to find alternatives for the chemicals used to control these pathogens, for example, plant extracts (rosemary and others) (Abutbul et al., 2004; District and Nadu, 2009; Genovese et al., 2012; Yonar et al., 2019). Additionally, microorganisms are commonly applied as beneficial agents (probiotics) in aquaculture (Hoseinifar et al., 2018; Wang et al., 2019). Nevertheless, little is known about metabolite-secreting endophytes or their use in controlling aquaculture diseases. The use of active microorganisms originating from the target itself or organisms living in the same environment, is an approach that has often been implemented for biological control in agriculture, but less so in aquatic environments. Therefore, this study aimed to examine the presence of biologically active culturable endophytes, in macroalgae collected from different water sources and locations in Israel and assess their bioactive, in vitro potential, as a novel source for metabolites and biological control agents in disease control of aquaculture pathogens.
Macroalgae were collected from various marine and freshwater sources of Israel (Table 1). Algae were collected randomly from easily accessible locations, close to the shore. The algae were brought to the laboratory for analysis in sealed plastic boxes containing the ambient water. During transport, the boxes were kept in a chilled environment (±25°C). Upon arrival at the laboratory, algae were kept in the transport boxes and air was bubbled through the ambient water until endophyte isolation. In each collection site, one sample of the host algae was used for culturable endophytes isolation as dilated in Table 1.
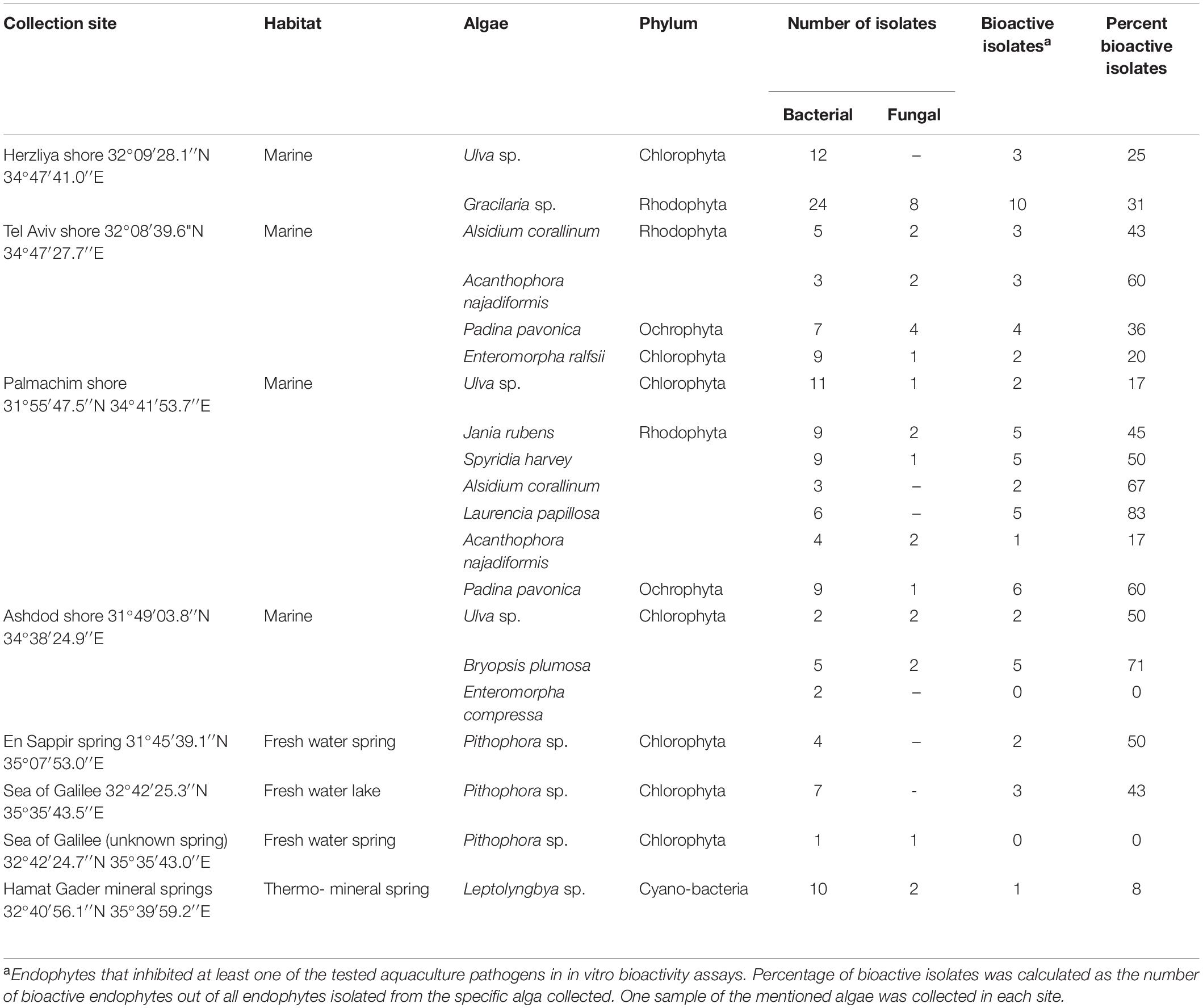
Table 1. Algae and isolated endophytes from different water sources in Israel, with percentage of bioactive isolates.
Culturable endophytes were isolated under sterile conditions in a laminar hood. The sampled algae were washed with freshwater and surface-sterilized by dipping algal pieces twice for 3 s each in 70% ethanol, followed by two 1-min washes in distilled water. To ensure algal surface sterilization, water from the algae’s last wash was tested for bacterial and fungal presence by inoculation on appropriate agar media. Lack of bacterial or fungal growth confirmed algal surface sterility. The treated algae were cut into small pieces (∼0.5 by 0.5 cm) and placed on growth media: nutrient agar (NA) for bacteria and potato dextrose agar (PDA) for fungi (Acumedia, Lansing, MI, United States), in 90-mm petri plates. At least two plates of the different media, with 10 pieces, for each algae were used for endophytes isolation and collection. The plates were incubated at 25°C for 8 days, allowing endophytes to grow out of the algal pieces (Figures 1A,B). The growing bacteria and fungi were collected and reinoculated on appropriate growth media. A single colony/spore culture was established for each of the endophytes, to ensure culture purity.
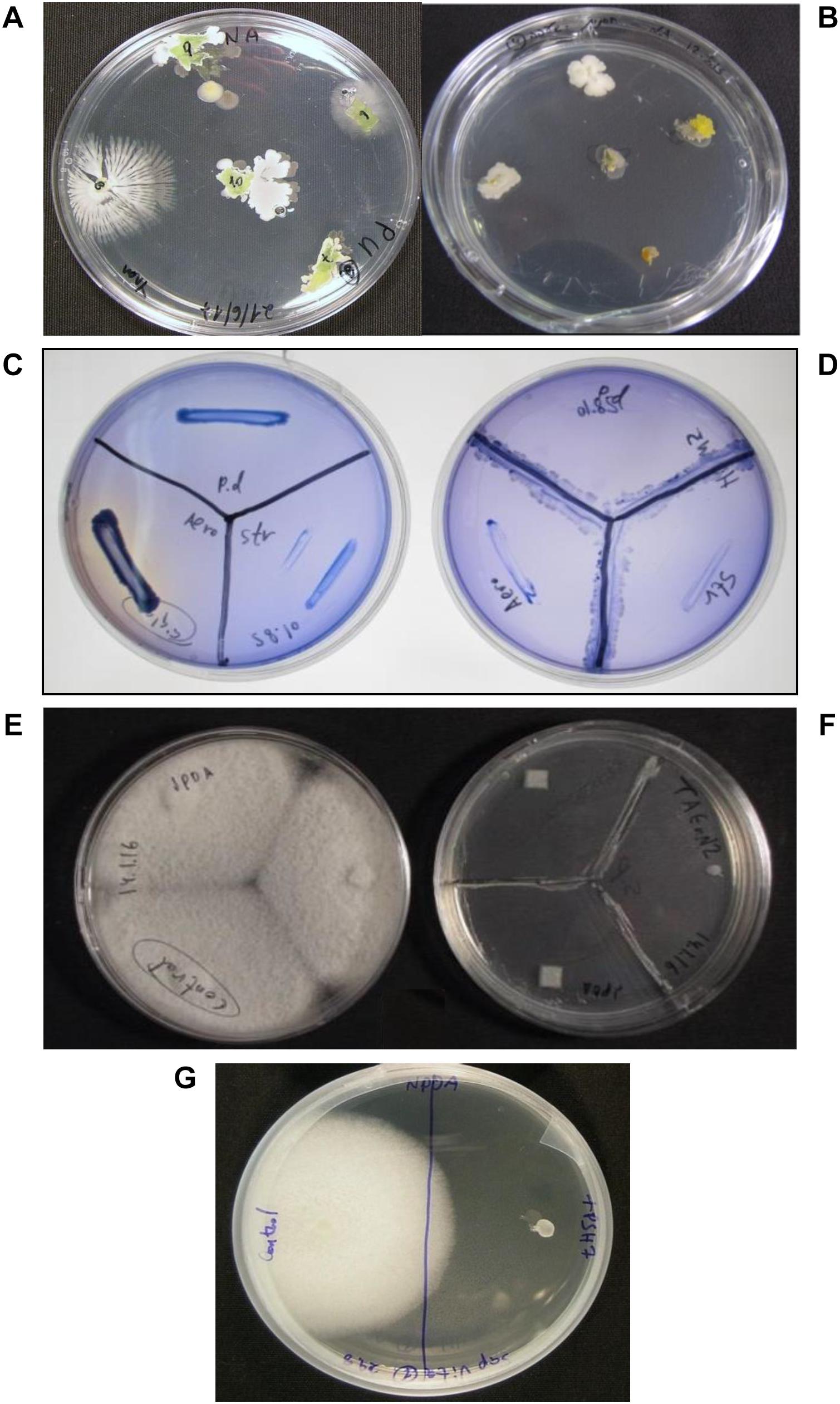
Figure 1. Isolation and bioactivity assessment of endophytes from algae. Endophytes were isolated by placing sterilized algal pieces on growth media in petri plates and incubating at 25°C until initiation of bacterial and fungal growth from the algal tissue (A,B). The endophytes were then tested in vitro in bioactivity assays (C–F) by growing them on supporting growth media in a “Y” shape prior to placing the pathogens as shown in (D,F). Inhibition was measured in comparison to controls (no endophytes) (C,E). Pathogen viability test (G) exhibited temporary inhibition or loss of viability of the pathogen. This test was performed by transferring the pathogen from the bioactivity test to a new growth plate in the absence of the endophyte. Regrowth of the pathogen after 3 days of incubation indicated inhibition by the endophyte, while no growth (G, right) indicated loss of viability. Endophytes were tested for activity against the oomycete pathogen: S. parasitica (F,G, right) and three bacterial pathogens: P. damselae, Strep. iniae and A. salmonicida (D). Control plates: pathogens in the absence of the endophytes (C,E,G, left).
Endophytic bacteria and fungi were tested for biological activity against three bacterial pathogens (P. damselae, Strep. iniae, A. salmonicida) and a pathogenic oomycete (S. parasitica). In vitro assays were performed on synthetic media supporting both the endophytes and the pathogens: tryptic soy agar (TSA, Acumedia) for bacteria, PDA for fungi, and NPDA (1/2 NA + 1/2 PDA), concocted in our laboratory, for both fungi and bacteria in the same plate. The assays were performed as follows: the tested endophytes were inoculated in a “Y shape” on the appropriate growth medium in a 90-mm petri plate. The plates were incubated at 25°C for 5–7 days. The pathogens were subsequently introduced by inoculation at the edges of the plate. The plates were then incubated for another week (25°C) and examined for pathogen growth during that period (Figures 1C–F). S. parasitica growth was examined quantitatively (percentage) by comparing the growth radius of the oomycete colony to the growth of a colony in the absence of endophyte (control). Challenged bacterial pathogen growth was estimated qualitatively using four levels of inhibition compared to the control: (1) very strong inhibition – no pathogen growth (+++), (2) moderate inhibition – low pathogen growth (++), (3) weak inhibition – slight pathogen growth (+), and (4) no inhibition – pathogen growth equal to control (–) (Table 2). At least two repetitions for each endophyte against each of the pathogens were performed.
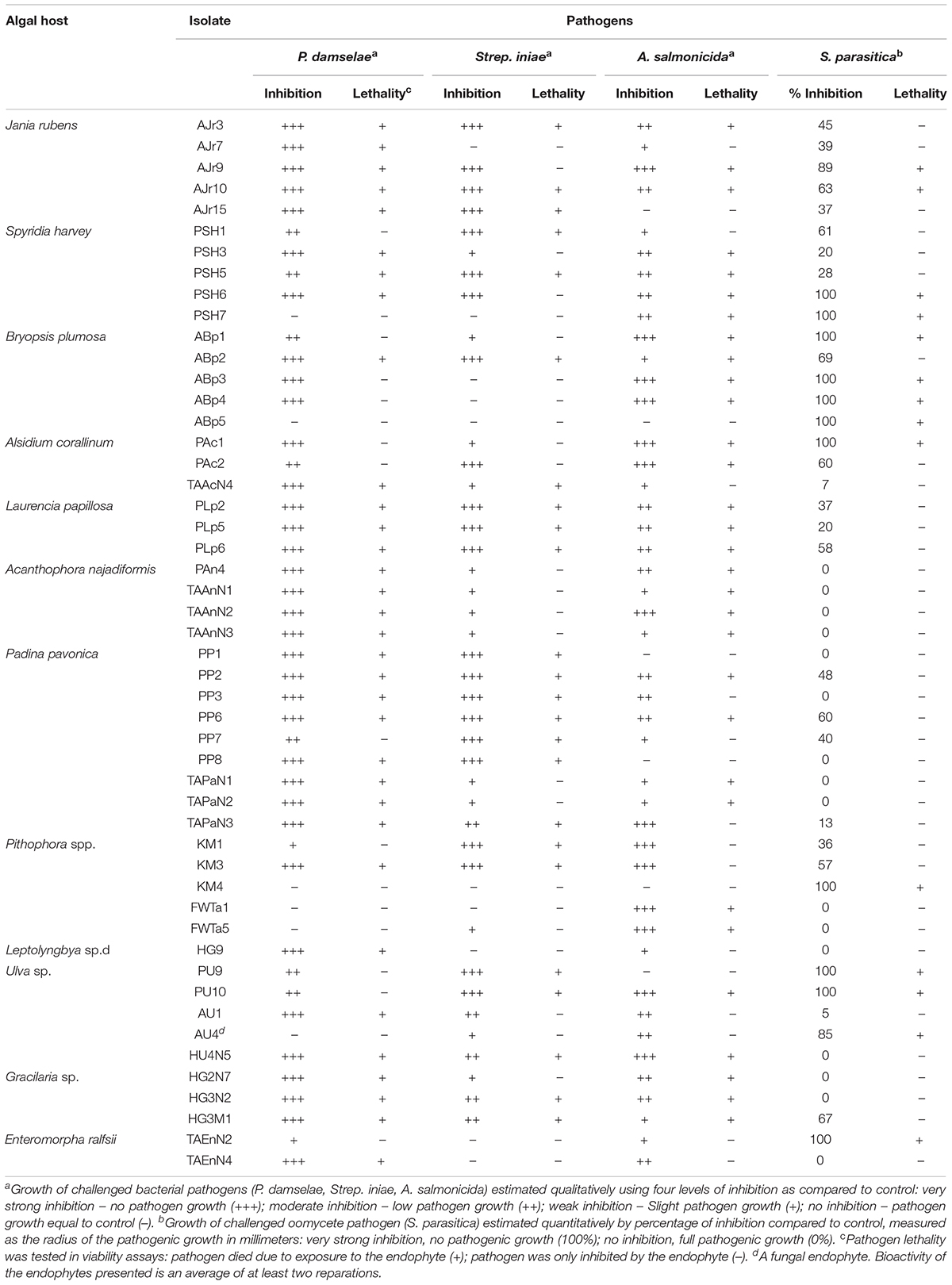
Table 2. Highly active endophytes from different algae, sorted by their ability to inhibit and/or kill the four major aquaculture pathogens.
Pathogens that were fully inhibited by the endophytes were further examined for viability. The inhibited pathogen from the bioassay plates was re-cultured on a new growth medium plate without endophyte and incubated under optimal growth conditions for a week. Unlike dead pathogens, inhibited pathogens generally grew on the new media a few days later (Figure 1G).
Endophyte identification was performed to the genus level by amplification and alignment of conserved rDNA sequences (internal transcribed spacer [ITS] for fungi and 16S for bacteria) and comparison with the National Center for Biotechnology Information (NCBI) GenBank database. Identification to the species level was done by sequencing at least three different conserved sequences.
Single-colony cultures of bacterial endophytes were incubated overnight at 25°C, 150 rpm, in Luria-Bertani (LB) broth medium (Acumedia). After 24 h, the cultures were centrifuged (6000 rpm, 10 min) and 100 mg of pellet (wet weight) was used for DNA extraction with a fungal/bacterial DNA extraction kit according to the manufacturer’s protocol (ZYMO Research, Irvine, CA, United States). Pure cultures of fungal endophytes were incubated on PDA plates for 4 days, 100 mg of fungal hypha (wet weight) were harvested, and DNA was extracted using the DNA extraction kit according to the manufacturer’s protocol (ZYMO Research).
PCR was performed using a labcycler (SensoQuest, Göttingen, Germany) in 25 μl PCR mixture containing 2 μl extracted DNA (approximately 50 ng), 12.5 μl DreamTaq green PCR master mix (Thermo Fisher Scientific, Waltham, MA, United States), 1 μl of each primer (10 μM) and 8.5 μl PCR-grade water. For bacterial isolates, the 16S region was amplified with the primers P3MOD and Pc5B (Kuske et al., 1998) and the following cycling program: 95°C for 5 min, 34 cycles at 95°C for 60 s, 50°C for 60 s and 72°C for 90 s, with a final extension at 72°C for 5 min. For fungal isolates, the ITS region was amplified with primers ITS1 and ITS4 (White et al., 1990) and the following cycling program: 96°C for 5 min, 39 cycles at 96°C for 45 s, 55°C for 45 s and 72°C for 60 s, with a final extension at 72°C for 5 min. For identification of Bacillus spp. to the species level, rpoB, recA, or gyrB sequences were amplified, respectively, with primers rpoB1206 and rpoBR3202 (Ki et al., 2009) and the program: 95°C for 3 min, 35 cycles at 95°C for 20 s, 55°C for 30 s and 72°C for 90 s, with a final extension at 72°C for 5 min; primers recA-F and recA-R (Mohkam et al., 2016) and the program: 95°C for 5 min, 30 cycles at 95°C for 30 s, 45°C for 30 s and 72°C for 60 s, with a final extension at 72°C for 5 min; and primers UP-1 and UP-2r (Yamamoto and Harayama, 1995) and the program: 95°C for 5 min, 30 cycles at 95°C for 60 s, 60°C for 60 s and 72°C for 120 s, with a final extension at 72°C for 8 min. For identification of endophytic fungi to the species level, the β-tubulin region was amplified with primers Bt1a and Bt1b (Glass and Donaldson, 1995) and the following cycling program: 95°C for 5 min, 39 cycles at 95°C for 45 s, 54°C for 60 s and 72°C for 90 s, with a final extension at 72°C for 5 min. In addition, the EF-1α region was amplified using primers EF1-728F and EF1-986R (Carbone and Kohn, 1999) and the program: 95°C for 5 min, 39 cycles at 95°C for 45 s, 51°C for 45 s and 72°C for 90 s, with a final extension at 72°C for 5 min.
PCR products (3 μl) were subjected to agarose gel electrophoresis (1.2%) and visualization under UV light, by soaking the gel in 0.5 μg/ml ethidium bromide solution for 3 min followed by washing in water for 5 min. PCR product (22 μl) was purified using the fragment DNA purification kit (iNtRON Biotechnology, South Korea). The purified PCR product (30 μl of ∼20 ng/μl) was sent for sequencing (Macrogen Europe, Netherlands). The sequencing results were compared to the NCBI GenBank database using the Basic Local Alignment Search Tool (BLAST)1 for the identification of bacterial/fungal species (Table 3).
Isolate ABp5 was sequenced (whole-genome sequencing, NovoGene, Beijing, China). Genome reads were compared to a known reference genome using gene annotations. Annotated fragments were compared to NCBI GenBank using BLAST.
Twenty macroalgae were collected from different locations in Israel. Twelve different genera were sampled from different marine and freshwater sources and locations. The algal genera and site locations are presented in Table 1.
We isolated 31 fungal and 142 bacterial endophytes from the 20 collected algae. Most of the macroalgae were collected from the Mediterranean shoreline (Herzliya, Tel Aviv, Palmachim, and Ashdod). The highest number of endophytes (148) were isolated from algae collected in this environment. Amongst the selected algal samples, Gracilaria sp. recorded the highest endophytic bacterial strains, followed by Ulva sp. and cyanobacteria (Table 1). Comparison of the number of bacterial endophytes isolated from the same genus of alga from the different locations revealed variations according to the sampling location. For example, the Ulva sp. from Herzliya and Palmachim areas produced a similar number of bacterial endophytes (12 and 11, respectively), compared to only two endophytes from the Ulva sp. collected from Ashdod. In general, the Ashdod location was poorer in endophytes than the other locations sampled along the Mediterranean shoreline. The same was found for the only alga collected in freshwater. We isolated more bacterial endophytes from the Pithophora sp. collected from the Sea of Galilee (freshwater lake) than from the Pithophora sp. collected from a spring running into the lake (Table 1). The number of fungal isolates was much lower than the number of bacteria in all algae at all locations (Table 1).
All 173 isolates were tested for their ability to inhibit the four aquaculture pathogens in in vitro assays (Tables 1, 2). Fifty endophytes (1 fungal and 49 bacterial) showed strong activity (caused loss of viability) against at least one of the four pathogens tested, and were classified as highly active endophytes (HAE; 50 isolates); active endophytes (AE; 38 isolates) had moderate inhibitory ability; non-active endophytes (NAE; 85 isolates) had no bioactivity at all (Figure 2). Only one endophyte (AJr10) was highly active against all four pathogens, while 11 were active against all bacterial pathogens. Four endophytes were lethal only to the oomycete (ABp5, KM4, AU4, and TAEnN2) (Figure 3 and Table 2).
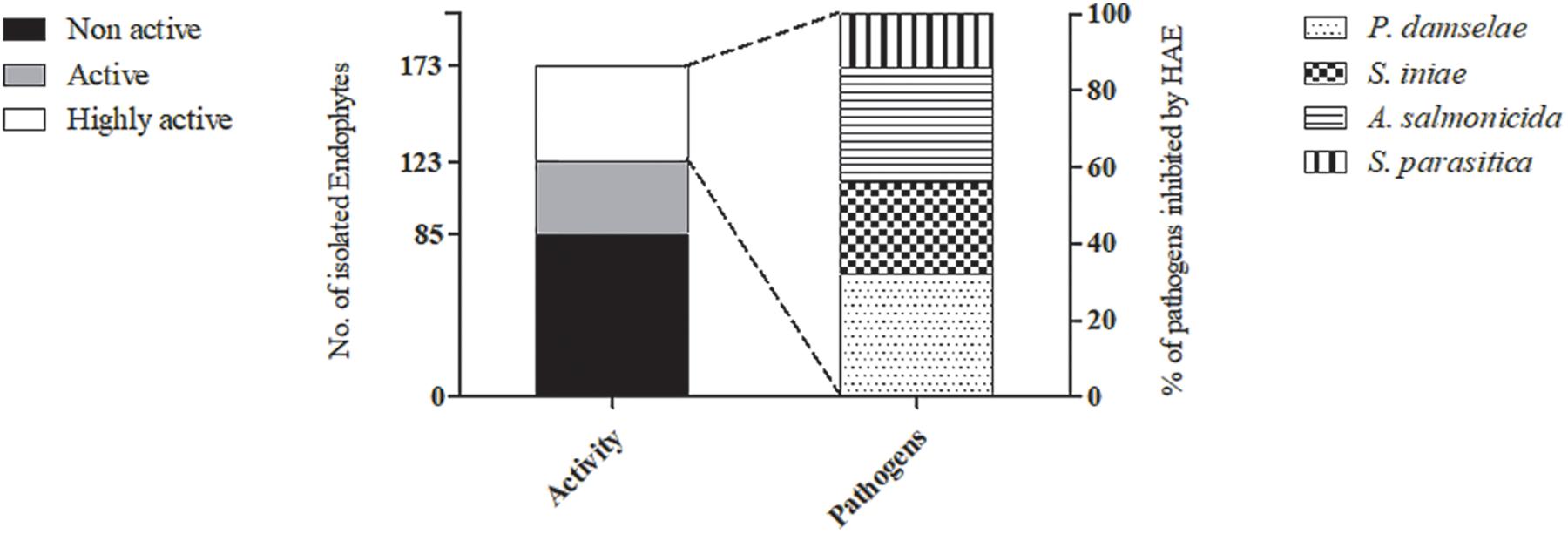
Figure 2. Bioactivity of the endophytes isolated from different algae. The number of endophytes isolated from the 20 different algae, classified by bioactivity level, is demonstrated (primary Y-axis). The highly active endophytes (HAE) are featured by their activity against each of the four pathogens (secondary Y-axis).
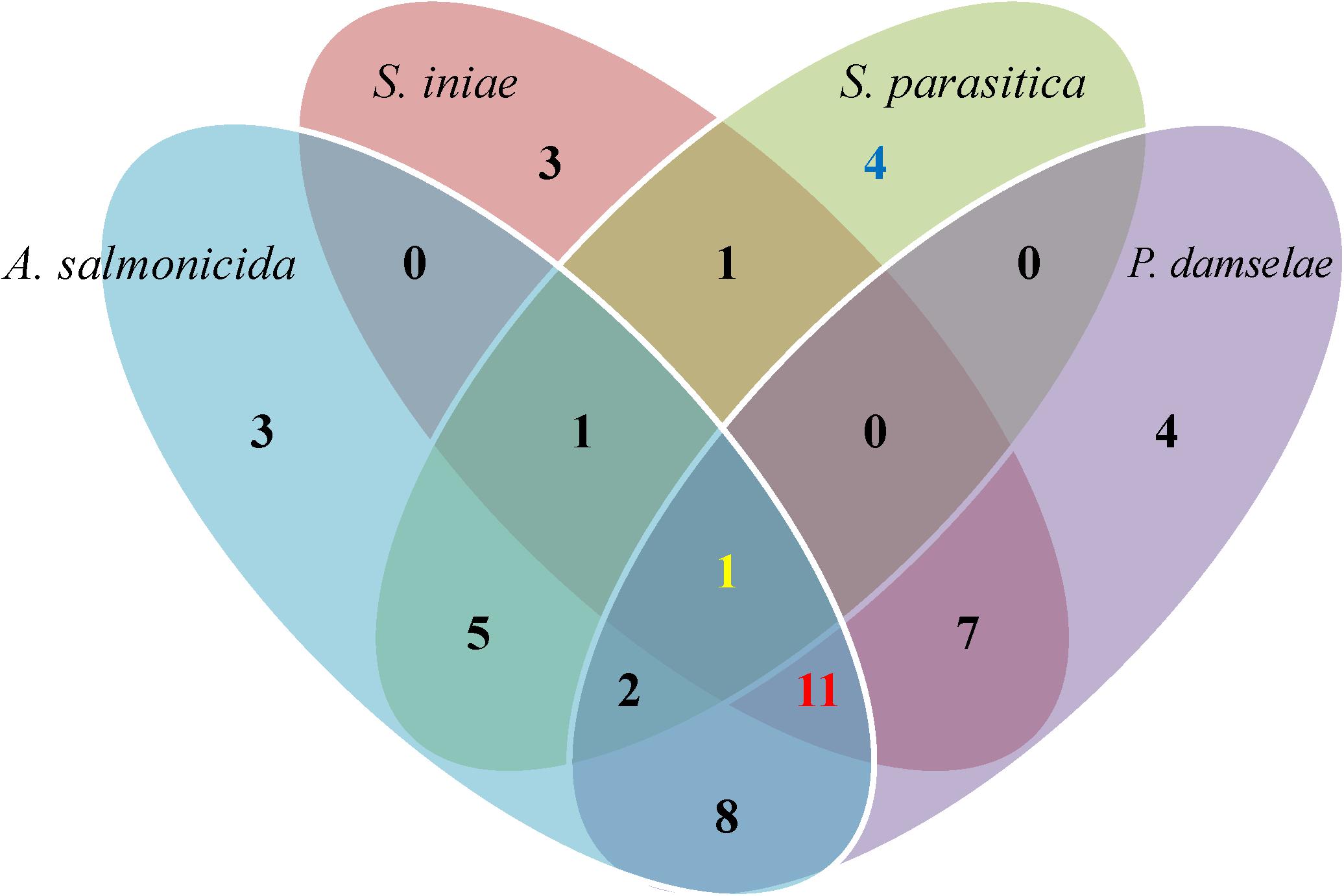
Figure 3. Venn diagram of the highly active endophytes’ distribution against the four tested aquaculture pathogens. Fifty highly active endophytes were isolated from the different algae and sorted according to their bioactivity against three bacterial pathogens (A. salmonicida, Strep. iniae and P. damselae) and one oomycete (S. parasitica). Each ellipse color represents a pathogen. Only 1 endophyte was active against all four pathogens (yellow number), 4 endophytes were active specifically against the oomycete S. parasitica (blue number), and 11 were specifically active against all three bacterial pathogens (red number).
We further analyzed the endophytes’ bioactivity with reference to the sampled alga, endophyte activity against pathogens, and sampling sites.
To analyze the distribution of active endophytes in the collected algae, we normalized the number of bioactive isolates from each algal genus to percentages (Figure 4 and Table 1). The average percentage of HAE for all sampled algae was 29, with the highest percentage isolated from the seawater alga Bryopsis plumosa and the lowest percentage from the cyanobacterium Leptolyngbya sp. The highest percentages of NAE (83) were isolated from Enteromorpha spp. (seawater), whereas B. plumosa had no NAE (Figure 4). At the phylum level, algae from the phylum Rhodophyta had the highest percentage of active isolates (eight sampled algae had 49.5% active isolates), while the phylum Chlorophyta had nine algae sampled with 30.6% active isolates. The alga Padina pavonica of the phylum Ochrophyta was sampled twice and had 48% active isolates. The only cyanobacteria sampled was Leptolyngbya sp., and it had the lowest percentage of bioactive isolates (Table 1).
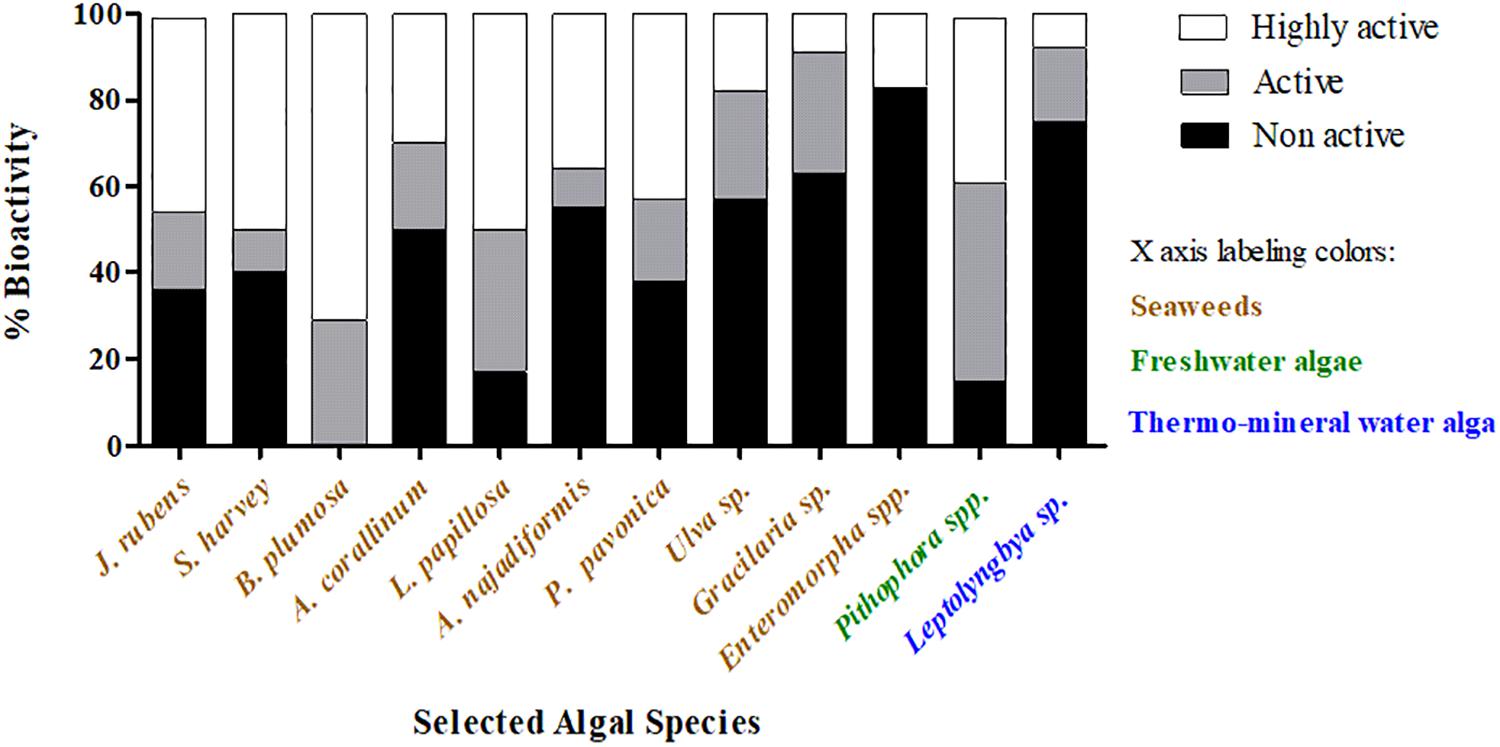
Figure 4. Distribution of the isolated endophytes by algal genera and bioactivity level. Endophytes were isolated from the indicated algae and tested for their bioactivity against aquaculture pathogens. The percentage of bioactivity in each alga was calculated by the number of active/non-active isolates out of the total isolated endophytes. Black bars represent the non-active endophytes, and gray and white bars represent active and highly active isolates, respectively.
The four pathogens used to test the bioactivity of the endophytes isolated from the different algae represent some of the most influential disease agents in aquaculture, and specifically in fish farming. P. damselae was the most susceptible pathogen to the endophytes tested in this study (Figure 2). Thirty-three endophytes from the different algae inhibited and caused mortality of P. damselae (Figure 3 and Table 2). Only two out of the six HAE (33%) from the freshwater algae Pithophora spp. and Leptolyngbya sp. inhibited P. damselae, whereas 32 of the 44 HAE (73%) from seaweed were active against this pathogen. The highest number of active endophytes against the bacterial pathogens were isolated from Padina pavonica (Figure 5 and Table 2). Although these endophytes inhibited all four pathogens, none was sufficiently active to cause mortality of the pathogenic oomycete S. parasitica (as observed in the viability tests) (Table 2). A. salmonicida was the second-most susceptible pathogen to the bioactive endophytes. The only alga from which no isolates were active against A. salmonicida was the cyanobacterium Leptolyngbya sp. (Figure 5). In the present study, the fungal endophyte AU4 isolated from the seaweed Ulva sp. had the ability to inhibit, but not kill Strep. iniae (weak inhibition) and A. salmonicida (moderate inhibition), but had strong mortality-inducing activity (85% inhibition) against the oomycete S. parasitica (Table 2 and Figure 5).
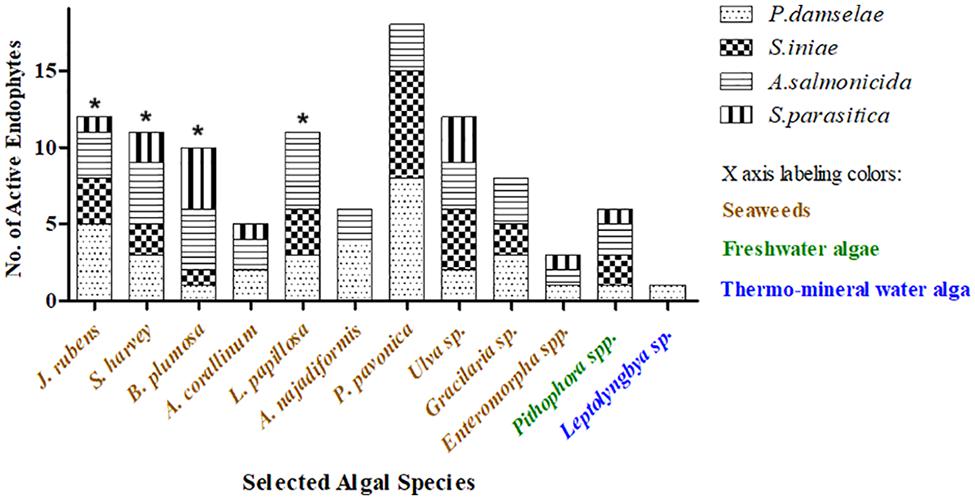
Figure 5. Distribution of the isolated endophytes by algal genus and their activity against aquaculture pathogens. Endophytes were isolated from the indicated algae and tested for their bioactivity against one oomycete and three bacterial pathogens. Endophyte activity is demonstrated as the sum of all inhibition events by the isolated endophytes of a certain alga. Each pathogen is marked by a different pattern in the bars. * The number of active endophytes from a particular alga can be greater than the sum of isolates from that same alga because the same isolate can be active against more than one pathogen.
Although the algal sampling sites along the Mediterranean shoreline of Israel were relatively close to one another (45 km at most from Herzliya to Ashdod), the diversity of algal genera, the number of endophytes isolated from the different algae, and the activity of those endophytes were high. This may be attributed to the differences in habitat in terms of abiotic parameters (nutrients, temperature, salinity, pollution, etc.) that has significantly elevated the diversity of endophytes in the algae. Nevertheless, we found the number of isolates from Ashdod shoreline to be somewhat low (13%), albeit with a high percentage of HAE (54%). Herzliya, Tel Aviv, and Palmachim shorelines produced higher numbers of endophytes with lower percentages of active isolates (29.5, 36, and 45%, respectively) (Table 1).
The alga collected from Hamat Gader hot springs had a high number of isolates but a low number of active endophytes (Table 1). Interestingly, isolates from the Sea of Galilee showed higher bioactivity than isolates from a spring that flows into the Sea of Galilee (NAE), even though they were isolated from the same algal genus (Table 1).
Altogether 23 active endophytes were identified at the genus level using rDNA conserved sequences of 16S for bacterial and ITS 5.8S for fungal isolates. Six endophytes were identified to the species level using the conserved sequences rpoB, recA, and gyrB for bacterial and EF-1α and β-tubulin for fungal endophytes in addition to the rDNA sequences. All sequences were deposited to the gene bank in the NCBI and are presented in Table 3. The molecular identification of the endophytes revealed that most of the active ones isolated from the different algae belong to the Bacillus spp. Isolate ABp5 of Bryopsis plumosa from Ashdod’s shoreline was identified as Kocuria flava, HG9 of Leptolyngbya sp. from the thermal springs in Hamat Gader was identified as a Lysinibacillus sp., TAEnN2 isolated from Enteromorpha ralfsii at the Tel Aviv shoreline was identified as Staphylococcus haemolyticus, KM4 of Pithophora sp. from the Sea of Galilee (freshwater) was identified as Pseudomonas alcaligenes and the fungal isolate AU4 from the Ulva sp. collected at Ashdod was identified as a Beauveria sp. (Table 3).
Nearly 200 fungal and bacterial culturable endophytes were isolated in this study, from 20 algae belonging to 12 different algal species sampled from varying water sources and locations in Israel. Collection sites and algal species were chosen randomly. All endophytes were tested for bioactivity against aquaculture pathogens (three bacteria and one oomycete). In general, a larger number of endophytes was isolated from the Mediterranean shoreline with a clear dominance of bacterial endophytes. Endophyte presence and variations in the different algae is most probably driven by multiple abiotic parameters including temperature, light, salinity, nutrient pollution, etc. Here we did not investigate these drivers. Rather, the objective of our study was to examine the differing activities of the endophytes isolated from the various hosts.
Several studies have shown variations in endophytic diversity according to algal host genus (Suryanarayanan et al., 2010; Flewelling et al.,2013a,b). Our algal sampling is not representative for a statistical based study of the diversity and abundance of host-endophyte prevalence. Nevertheless examining the number of culturable endophytes isolated from the different algae collected, did not find substantial differences in the number of isolates between the different algal genera. We did isolate different numbers of endophytes from the same algal genus at varying locations (Table 1). This supports the possibility that the inhabitation of algae by endophytic microorganisms is not only a factor of aptness; environmental factors may be involved as well.
As noted above, environmental drivers (e.g., changes in temperature, nutrients) will impact the diversity of endophytes inhabiting the algae. Thus, different endophytes were identified from the same host (Ulva sp.) that was collected at three locations: Ashdod (close to both an industrial port and a power plant), Herzliya (collected at a marina), and from Palmachim, an open site distanced from anthropogenic inputs (Table 1). Moreover, different isolates were identified from the freshwater alga Pithophora sp. sampled from the Sea of Galilee (three out of seven bacterial endophytes were active against the pathogens) and from a small nearby spring (where only one non-active isolate was characterized). This can be explained by the exposure of the algae to a potentially higher variety of microorganisms in the lake compared to the spring, possibly due to habitat differences in abiotic parameters in the spring vs. the lake.
To understand the significance and influence of the factors involved, on the presence and isolation of HAE in the different environments, we performed multiple principal component analysis (PCA) (Figure 6). In this analysis, we tested the influence of sampling location, aquatic habitat (type of water), and algal-host affiliation (phyla and genera levels) on the presence of HAE from the isolated endophytes. All together 20 possible combinations were tested (not shown) with only one combination displaying significant correlation. We found habitat to be the only factor significantly influencing the presence of HAE isolates from the sampled algae (circled in light blue). The PCA also demonstrated some degree of clustering by location (circled in red). The majority of the algae from Palmachim (five out of seven) and Ashdod locations provided higher abundance of HAE, while Herzliya’s algae clustered with lower presence of HAE. Samplings near Tel Aviv are scattered between these two clusters. The same is seen for the Sea of Galilee. Algae sampled from the Sea of Galilee and Ein Sapir Spring are clustered together showing high HAE while samples from a spring near the lake are positioned separately and represent poor HAE. As there was only one active isolate from the hot springs – its affiliation (and thus location on the PCA plot) to the other endophytes cannot be determined accurately (table 1).
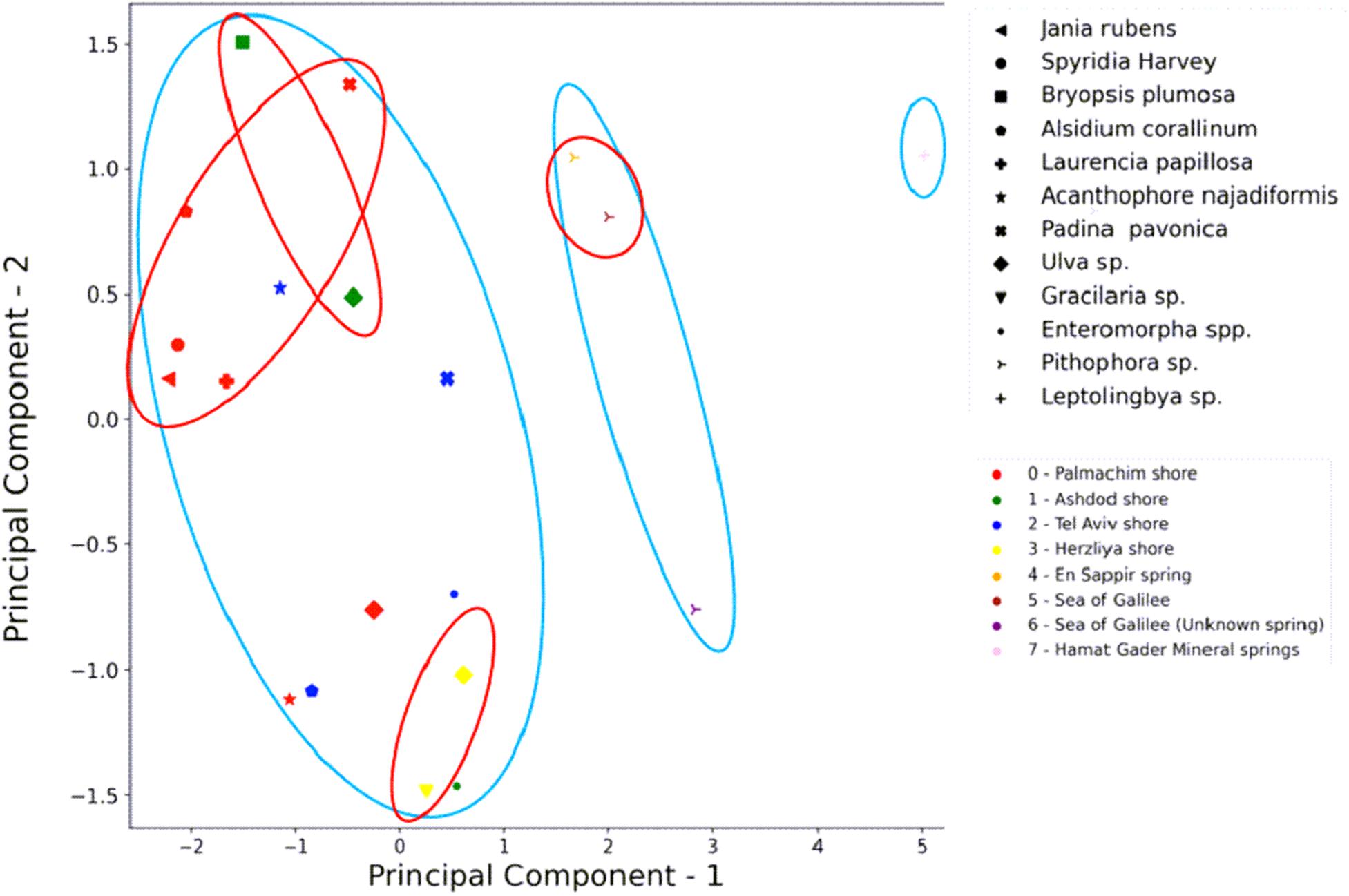
Figure 6. PCA analysis of highly active endophytes (HAE) isolation in regard to habitat, location, and algal genera. HAE isolation from the different algal genera is most influenced by the “habitat” (type of water: marine, freshwater, or thermo-mineral). An influence of the location is noticeable. Component 1 is the “habitat,” component 2 is the HAE isolate incidence. Different shapes represent algal genera. The colors represent the location of algal sampling. Light blue circles are the clustering by habitat (left marine, right thermo-mineral). Red circles are the clustering by location.
Suryanarayanan et al. (2010) and Flewelling et al. (2013b) explored the differences in endophytic numbers and diversity from the same algal phyla. While differences in geography, climate, and seasonality appeared to impact abundance and diversity of endophytes (Flewelling et al., 2013b), the authors suggested that a more statistically robust sampling of isolates is required to make any definite conclusions. We suggest this is the case in our study too.
Approximately 30% of the tested endophytes killed at least one of the four pathogens tested (HAE). Of these 50 HAE, 11 killed all three bacterial pathogens, while four isolates demonstrated effective bioactivity specifically against the oomycete pathogen (Figures 2, 3 and Table 2). Twenty-three HAE belonging mostly to the Bacilli (class) were identified by genera (Table 3). Bacilli are frequently used in agriculture for biological control of pathogens and are known for their bioactive secondary metabolite production (Jacobsen et al., 2004; Shafi et al., 2017). Therefore, it is not surprising to find them among the HAE bioactive isolates we tested. Of the pathogens tested, the marine pathogen P. damselae was the most susceptible. When examining the activity of endophytes isolated from the freshwater algae against P. damselae, only 20% of the isolates were active, compared to 75% of the active endophytes isolated from seaweed. Antimicrobial activity against such pathogens would be expected in the ambient environment as, throughout their evolutionary history, endophytes may have competed with the pathogens for different resources (i.e., pathogens spend part of their life cycle inside or on algae as saprophytes). Thus endophytes would have evolved strategies to combat these competitors, including the secretion of active metabolites, as has been demonstrated in many agricultural biocontrol studies (Liarzi and Ezra, 2014; Masand et al., 2015).
We found the alga Bryopsis plumosa to have the highest percentage of HAE (71%) of the 12 different algal genera collected. No NAE were isolated from this alga. Bryopsis spp. have extraordinary wound-repair and propagation mechanisms whereby they form protoplasts that can survive in the absence of a cell membrane for several minutes before regenerating into new individuals (Hollants et al., 2011). Protection at this sensitive stage is thus essential and resident microorganisms may help control pathogens. Moreover, to guarantee the involvement of specific microorganism, algae such as Bryopsis spp. have intrinsic mechanisms that can attract, select, and maintain endophytes of possible ecological importance within their cells (Hollants et al., 2011). Our observation of only HAE in Bryopsis plumosa would appear to support this notion. We further assume these endophytes may help protect this seaweed from marine pathogens during the wound-repair and susceptible protoplast stages. We isolated a relatively high number of endophytes (N = 12, Table 1) from the cyanobacterium Leptolyngbya sp., yet only one isolate (HG9) was highly active against one of the pathogens (P. damselae). As we do not have a statistically representative number of replicates, it is hard to make any conclusions as to the reasons. This might be due to the high water temperature at the site Leptolyngbya sp. was sampled from (37–42°C). The literature, as much as we could find, lacks data regarding bioactive bacteria or fungi in alga of thermo-mineral water environments, which are sometimes also rich in cyanobacteria. The thermophilic unicellular cyanobacterium Gloeocapsa sp. demonstrates antibacterial and antifungal activities (Gacheva et al., 2013). Endophytes are known to induce tolerance to high temperatures, such as the endophytic fungus Paecilomyces formosus that induces thermo-tolerance in cucumber (Khan et al., 2012). This algal host would be expected to have endophytes that help it cope with the high temperatures and the high mineral concentrations in which it grows. Yet, this remains unclear presently. It would be interesting to test the ability of the endophytes isolated from Leptolyngbya sp. to induce temperature tolerance in other algae and plants.
Kharwar and Strobel (2010) described endophyte diversity in a tropical rainforest, an ecosystem in which the evolutionary race to survive is a major force; competition is high for limited resources and selection pressure is at its peak. Accordingly, rainforests can serve as a source for novel biologically active compounds (Kharwar and Strobel, 2010). Endophytes, living within algal tissue avoid the harsher marine environment outside. Yet, as in the tropical forest, competition for space and nutrients within the algal tissue potentially stimulates the production of highly bioactive metabolites against competitor microorganisms. Moreover, if this actually occurs, then some of the highly bioactive metabolites from seaweed endophytes may serve as a new source of bioactive metabolites protecting aquaculture from pathogens.
The bioactive endophytes isolated in this study from macroalgae, secreted metabolites that were active against at least one of the four tested aquaculture pathogens (in 51% of the in vitro assays) (Figures 2, 3, and 5). This finding is consistent with research demonstrating that antifungal and antibacterial bioactivity from macroalgal endophytes, varies from 50 to 83% (Schulz et al., 2002; Flewelling et al.,2013a,b; Girão et al., 2019). However, we could find no reference in the mentioned studies of the viability of the pathogens after exposure to the endophytes. Here, using viability assays, we show that 57% of the active isolates (51% of total isolates) not only inhibited but also killed the pathogens to which they were exposed.
This study evaluated the potential of endophytes isolated from macroalgae as a source of biological control and new secondary metabolites for use in aquaculture.
Ismail et al. (2016) were working with macroalgae and their commensal bacteria (epiphytes) as a source for antimicrobials against several fish pathogens. Their work demonstrates the potential of algae associated microbes as biological control agents against one of the most influencing problems of aquaculture industry.
To the best of our knowledge, there are no published studies demonstrating the use of algal endophytes to control aquaculture diseases. Our study may lead to finding new molecules for use as novel, environmentally friendly, products that will solve one of the most challenging problems for the growing aquaculture industry: pathogens and pests.
Our findings show the potential of algal endophytes for future applications in the biological control of aquaculture diseases. For example, one possibility is to use the endophytes as a probiotic additive to the fish feed. It may be added directly as bacterial biomass mixed with the feed or inside the host algae after re introduction. Another option is to use the active metabolites as pretreatment to inland growth facilities or as pharmaceuticals to control diseases in those facilities. Further study is needed to reinforce our findings. The metabolites secreted by the active endophytes that we isolated will be identified using analytical chemistry methods. This first step is mandatory to eliminate unacceptable compounds, such as carcinogens, poisons, and other harmful metabolites, in the long procedure of developing a new control product for disease and pest management in aquaculture.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
DE, YD, and IB wrote the manuscript. All the authors discussed the results and contributed to the final manuscript. YD and LG collected the algae and isolated the endophytes. YD performed the all bioactivity experiments, identification of isolates, and data analysis. DE and IB supervised the project.
This work was financed by the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development (MOARD); grant number 20-02-0122 and Copia Agro Israel.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Mrs. Rita Smirnov and Mr. Tamir Ofek from The Central Laboratory for Fish Health, Nir David, Israel for providing the four-aquaculture pathogens, Dr. Gabi Banet from the Dead Sea-Arava Science Center, Israel for taxonomic identification of the cyanobacterium Leptolyngbya sp. and Ms. Bar Ezra for helping with the PCA analysis.
Abutbul, S., Golan-Goldhirsh, A., Barazani, O., and Zilberg, D. (2004). Use of Rosmarinus officinalis as a treatment against Streptococcus iniae in tilapia (Oreochromis sp.). Aquaculture 238, 97–105. doi: 10.1016/j.aquaculture.2004.05.016
Agnew, W., and Barnes, A. C. (2007). Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 122, 1–15. doi: 10.1016/j.vetmic.2007.03.002
Bacon, C. W., and White, J. F. (2016). Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 68, 87–98. doi: 10.1007/s13199-015-0350-2
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
District, K. K., and Nadu, T. (2009). Dietary medicinal plant extracts improve growth, immune activity and survival of tilapia Oreochromis mossambicus. J. Fish Biol. 74, 1462–1475. doi: 10.1111/j.1095-8649.2009.02212.x
Duarte, J., Pereira, C., Moreirinha, C., Salvio, R., Lopes, A., Wang, D., et al. (2018). New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: an in vitro preliminary study. Aquaculture 495, 970–982. doi: 10.1016/j.aquaculture.2018.07.002
FAO (2018). The State of World Fisheries and Aquaculture 2018 - Meeting the Sustainable Development Goals. Avaliable at: http://www.fao.org/3/i9540en/i9540en.pdf (accessed July 9, 2018).
Flewelling, A. J., Currie, J., Gray, C. A., and Johnson, J. A. (2015). Endophytes from marine macroalge: promising sources of novel natural products. Curr. Sci. 109, 88–111.
Flewelling, A. J., Ellsworth, K. T., Sanford, J., Forward, E., Johnson, J. A., and Gray, C. A. (2013a). Macroalgal endophytes from the atlantic coast of Canada: a potential source of antibiotic natural products? Microorganisms 1, 175–187. doi: 10.3390/microorganisms1010175
Flewelling, A. J., Johnson, J. A., and Gray, C. A. (2013b). Isolation and bioassay screening of fungal endophytes from North Atlantic marine macroalgae. Bot. Mar. 56, 287–297. doi: 10.1515/bot-2012-0224
Francis, P. M., Suming, Z., Jiting, Z., Xiaoye, Z., Shuwei, A., and Guoliang, W. (2014). Massive mortality associated with Streptococcus iniae infection in cage-cultured red drum (Sciaenops ocellatus) in Eastern China. Afr. J. Microbiol. Res. 8, 1722–1729. doi: 10.5897/ajmr2014.6659
Gacheva, G., Gigova, L., Ivanova, N., Iliev, I., Toshkova, R., Gardeva, E., et al. (2013). Suboptimal growth temperatures enhance the biological activity of cultured cyanobacterium Gloeocapsa sp. J. Appl. Phycol. 25, 183–194. doi: 10.1007/s10811-012-9852-y
Genovese, G., Faggio, C., Gugliandolo, C., Torre, A., Spanò, A., Morabito, M., et al. (2012). In vitro evaluation of antibacterial activity of Asparagopsis taxiformis from the Straits of Messina against pathogens relevant in aquaculture. Mar. Environ. Res. 73, 1–6. doi: 10.1016/j.marenvres.2011.10.002
Girão, M., Ribeiro, I., Ribeiro, T., Azevedo, I. C., Pereira, F., Urbatzka, R., et al. (2019). Actinobacteria isolated from Laminaria ochroleuca: a source of new bioactive compounds. Front. Microbiol. 10:683. doi: 10.3389/fmicb.2019.00683
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330.
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7:1538. doi: 10.3389/fmicb.2016.01538
Hollants, J., Decleyre, H., Leliaert, F., De Clerck, O., and Willems, A. (2011). Life without a cell membrane: challenging the specificity of bacterial endophytes within Bryopsis (Bryopsidales. Chlorophyta). BMC Microbiol. 11:255. doi: 10.1186/1471-2180-11-255
Hoseinifar, S. H., Sun, Y.-Z., Wang, A., and Zhou, Z. (2018). Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 9:2429. doi: 10.3389/fmicb.2018.02429
Ismail, A., Ktari, L., Ahmed, M., Bolhuis, H., and Boudabbous, A. (2016). Antimicrobial activities of bacteria associated with the brown alga Padina pavonica. Front Microbiol. 7:10723. doi: 10.3389/fmicb.2016.01072
Jacobsen, B. J., Zidack, N. K., and Larson, B. J. (2004). The role of Bacillus-based biological control agents in integrated pest management systems: plant diseases. Phytopathology 94, 1272–1275. doi: 10.1094/PHYTO.2004.94.11.1272
Khan, A. L., Hamayun, M., Radhakrishnan, R., Waqas, M., Kang, S. M., Kim, Y. H., et al. (2012). Mutualistic association of Paecilomyces formosus LHL10 offers thermotolerance to Cucumis sativus. Antonie Van Leeuwenhoek 101, 267–279. doi: 10.1007/s10482-011-9630-x
Kharwar, R. N., and Strobel, G. (2010). “Fungal endophytes: an alternative source of bioactive compounds for plant protection,” in Natural Products in Plant Pest Management, ed. N. K. Dubey (Wallingford: CABI), 218–241.
Ki, J. S., Zhang, W., and Qian, P. Y. (2009). Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J. Microbiol. Methods 77, 48–57. doi: 10.1016/j.mimet.2009.01.003
Kuske, C. R., Banton, K. L., Adorada, D. L., Stark, P. C., Hill, K. K., and Jackson, P. J. (1998). Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64, 2463–2472. doi: 10.1128/AEM.64.7.2463-2472.1998
Liarzi, O., and Ezra, D. (2014). “Endophyte-mediated biocontrol of herbaceous and non-herbaceous plants,” in Advances in Endophytic Research, eds V. Verma, C. Vijay, and C. Gange Alan (New Delhi: Springer), 335–369.
Mandelare, P. E., Adpressa, D. A., Kaweesa, E. N., Zakharov, L. N., and Loesgen, S. (2018). Coculture of two developmental stages of a marine-derived Aspergillus alliaceus results in the production of the cytotoxic bianthrone allianthrone A. J. Nat. Prod. 81, 1014–1022. doi: 10.1021/acs.jnatprod.8b00024
Manomi, S., Job, N., and Philip, R. (2015). Bioactive potential of endophytic fungi from macroalgae. Int. J. Res. Mar. Sci. 4, 27–32. doi: 10.1007/s11274-015-1950-y
Masand, M., Jose, P. A., Menghani, E., and Jebakumar, S. R. D. (2015). Continuing hunt for endophytic actinomycetes as a source of novel biologically active metabolites. World J. Microbiol. Biotechnol. 31, 1863–1875.
Menanteau-Ledouble, S., Kumar, G., Saleh, M., and El-Matbouli, M. (2016). Aeromonas salmonicida: updates on an old acquaintance. Dis. Aquat. Organ. 120, 49–68. doi: 10.3354/dao03006
Mohkam, M., Nezafat, N., Berenjian, A., Mobasher, M. A., and Ghasemi, Y. (2016). Identification of Bacillus probiotics isolated from soil rhizosphere using 16S rRNA, recA, rpoB gene sequencing and RAPD-PCR. Probiotics Antimicrob. Proteins 8, 8–18. doi: 10.1007/s12602-016-9208-z
Rivas, A. J., Lemos, M. L., and Osorio, C. R. (2013). Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front. Microbiol. 4:283. doi: 10.3389/fmicb.2013.00283
Romalde, J. L. (2002). Photobacterium damselae subsp. piscicida: an integrated view of a bacterial fish pathogen. Int. Microbiol. 5, 3–9. doi: 10.1007/s10123-002-0051-6
Schulz, B., Boyle, C., Draeger, S., Römmert, A.-K., and Krohn, K. (2002). Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol. Res. 106, 996–1004. doi: 10.1017/S0953756202006342
Shafi, J., Tian, H., and Ji, M. (2017). Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip. 31, 446–459. doi: 10.1080/13102818.2017.1286950
Suryanarayanan, T. S., Thirunavukkarasu, N., and Geetha, V. (2010). Internal mycobiota of marine macroalgae from the Tamilnadu coast: distribution, diversity and biotechnological potential. Bot. Mar. 53, 457–468. doi: 10.1515/BOT.2010.045
The International Bank for Reconstruction and Development/The World Bank (2014). Reducing Disease Risk in Aquaculture. Avaliable at: http://documents.worldbank.org/curated/en/110681468054563438/pdf/882570REPLACEM00NAME0Reantaso0Melba.pdf (accessed January 1, 2014).
van West, P. (2006). Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20, 99–104. doi: 10.1016/j.mycol.2006.06.004
Wang, C., Liu, Y., Sun, G., Li, X., and Liu, Z. (2019). Growth, immune response, antioxidant capability, and disease resistance of juvenile Atlantic salmon (Salmo salar L.) fed Bacillus velezensis V4 and Rhodotorula mucilaginosa compound. Aquaculture 500, 65–74. doi: 10.1016/j.aquaculture.2018.09.052
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Snisky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wiklundl, T. (1998). Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis. Aquat. Organ. 32, 49–69. doi: 10.3354/dao032049
Yamamoto, S., and Harayama, S. (1995). PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61, 1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995
Yonar, M. E., Mişe Yonar, S., İspir, Ü, and Ural, M. Ş (2019). Effects of curcumin on haematological values, immunity, antioxidant status and resistance of rainbow trout (Oncorhynchus mykiss) against Aeromonas salmonicida subsp. achromogenes. Fish Shellfish Immunol. 89, 83–90. doi: 10.1016/j.fsi.2019.03.038
Keywords: endophytes, bacteria and fungi, aquaculture, biocontrol, alga, environmentally friendly, sustainable, disease and pathogen
Citation: Deutsch Y, Gur L, Berman Frank I and Ezra D (2021) Endophytes From Algae, a Potential Source for New Biologically Active Metabolites for Disease Management in Aquaculture. Front. Mar. Sci. 8:636636. doi: 10.3389/fmars.2021.636636
Received: 01 December 2020; Accepted: 15 March 2021;
Published: 13 April 2021.
Edited by:
Chris Dupont, J. Craig Venter Institute, United StatesReviewed by:
Ramasamy Ramasubburayan, Manonmaniam Sundaranar University, IndiaCopyright © 2021 Deutsch, Gur, Berman Frank and Ezra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Ezra, ZGV6cmFAdm9sY2FuaS5hZ3JpLmdvdi5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.