- 1College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 3CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Delimiting ichthyoplankton is fundamental work for monitoring the recruitment process and identifying the spawning and nursing grounds of fishes. Nevertheless, it is extremely difficult to identify the fish during the early stages at the species level based on morphological characters because of the paucity of diagnostic features. In this study, we investigated the fish larval community through large-scale ecosystemic sampling in South China Sea (SCS) during 2013 and 2017 using DNA barcodes. To maintain the morphologies of fish larvae, we preserved the larvae in formalin and developed a technique to recover their DNA. Among the 3,500 chosen larvae, we successfully extracted DNA from 2,787 larval samples and obtained 1,006 high-quality sequences. Blast searches showed that 408 larvae (i.e., 40.5%) could be unambiguously identified to species, 413 larvae (i.e., 41.1%) were ambiguously species delimitation, and 185 larvae (i.e., 18.4%) showed a low match similarity with target sequences. A total of 101 species were identified, among which 38 and 33 species corresponded to demersal and reef-associated species, whereas the remaining 30 species corresponded to benthopelagic, pelagic-oceanic, bathypelagic, and pelagic-neritic species. High-quality larval photographs of the 101 diagnosed species showed intact morphological characters and thus provided a reference for identifying fish species during the early stages based on morphological characters. Our study highlighted the possibility of recovering and amplifying DNA from formalin-fixed samples and provided new insight into the fish larval community in the SCS.
Introduction
Perennial and continuous anthropogenic impacts, for example, global warming, water extraction, species invasions, overfishing, and habitat degradation, have been documented to severely decrease fish diversity in both the sea and inland water bodies (Olden et al., 2007; Rahel et al., 2008). South China Sea (SCS), an important biodiversity hot spot for marine ecosystems, has been reported to harbor more than 3,300 fish species (Alien et al., 2000; Morton and Blackmore, 2001; Shao et al., 2008; Huang et al., 2015). However, the illegal fishing, mangrove damage resulting from irrational usage and deforestation, uncontrolled wastewater emissions, and degeneration of coral reefs constantly occurring in the SCS over the last several decades have put the sustainability of fish populations at serious risk (Macusi et al., 2011; Huang et al., 2015); therefore, protecting the fish resources in the SCS is an urgent task.
Larvae, the early phase of fish without initiative migration potential, offer an ideal opportunity to obtain important information on reproductive biology, preferred reproduction sites and times, and population recruitment success rates (Bialetzki et al., 2005; Cao et al., 2007; Reynalte-Tataje et al., 2012). This information is also comparatively important for ecological monitoring and developing management and conservation plans. However, empirical studies on larvae are currently limited by the difficulty in accurately delimiting larvae at the species level based on morphological characters, even for expert taxonomists. The main limiting factor is plastic and labile diagnostic characters at various development stages (Choat and Doherty, 1993; Cao et al., 2007). Accordingly, a reliable and efficient approach that can identify fish larvae to species is urgently needed.
DNA barcoding, an identification method using molecular makers, has created new opportunities for the delimitation of fish larvae, regardless of their developmental stage and plastic morphological characters (Hubert et al., 2010, 2015; Valdez-Moreno et al., 2010; Ko et al., 2013; Frantine-Silva et al., 2015; Kimmerling et al., 2018). Many previous studies consistently demonstrated that DNA barcoding could be a reliable molecular method of identifying fish larvae (Hubert et al., 2010, 2015; Valdez-Moreno et al., 2010; Ko et al., 2013; Frantine-Silva et al., 2015). For example, Valdez-Moreno et al. (2010) used DNA barcodes to connect adults and early life stages of marine fishes in the Yucatan Peninsula, Mexico. Hubert et al. (2015) successfully identified 373 of 505 coral reef fish larvae in open waters in the vicinity of Moorea by applying DNA barcodes. In addition, an increasing number of sequences of fish taxa deposited in an extensive database (BOLD: The Barcode of Life Data System)1 will aid in delimiting both adult and larvae. By early 2020, more than 19,000 fish species including more than 260,000 specimens had been deposited in the BOLD database (Ratnasingham and Hebert, 2007).
Most earlier DNA-based studies that focused on fish larvae used high concentrations of alcohol to preserve samples (Hubert et al., 2010, 2015; Valdez-Moreno et al., 2010; Ko et al., 2013; Frantine-Silva et al., 2015; Kimmerling et al., 2018). However, alcohol will damage the morphological features of fish larvae to some degree, which influences the morphological studies of larvae. As a result, to maintain the morphological characters of fish larvae, we stored larvae in aqueous formalin and attempted to extract sufficient DNA for sequence amplification. This technique can overcome the major challenge of recovering DNA sequences from formalin-fixed samples. Generally known, formalin fixation of specimens will damage DNA via fragmentation, base modification, and cross-linkage within the DNA itself or between DNA and proteins (Quach et al., 2004; Wong et al., 2014). Randomly and fragmented formalin-fixed DNA poses a difficulty in Sanger sequencing.
To date, several DNA barcode studies have been conducted on the adult fish in the SCS and its adjacent areas (Wang et al., 2009, 2012; Zhang and Hanner, 2012; Chang et al., 2017; Hou et al., 2018), providing a large number of reference sequences for the fish fauna in this sea. Here, we sampled fish larvae from 230 sites in the SCS from 2013 to 2017 (Figure 1), preserved them in neutral formalin solution, and sequenced standardized molecular tags located in the mitochondrial cytochrome c oxidase I (COI) gene for larval delimitation. The aims of this study were (i) to assess the feasibility of recovering and amplifying DNA fragments from formalin-fixed samples, (ii) to determine how many samples can be successfully identified through DNA barcodes, and (iii) to evaluate the value of information on the reproduction and recruitment of fish populations and the distribution patterns of ichthyoplankton communities.
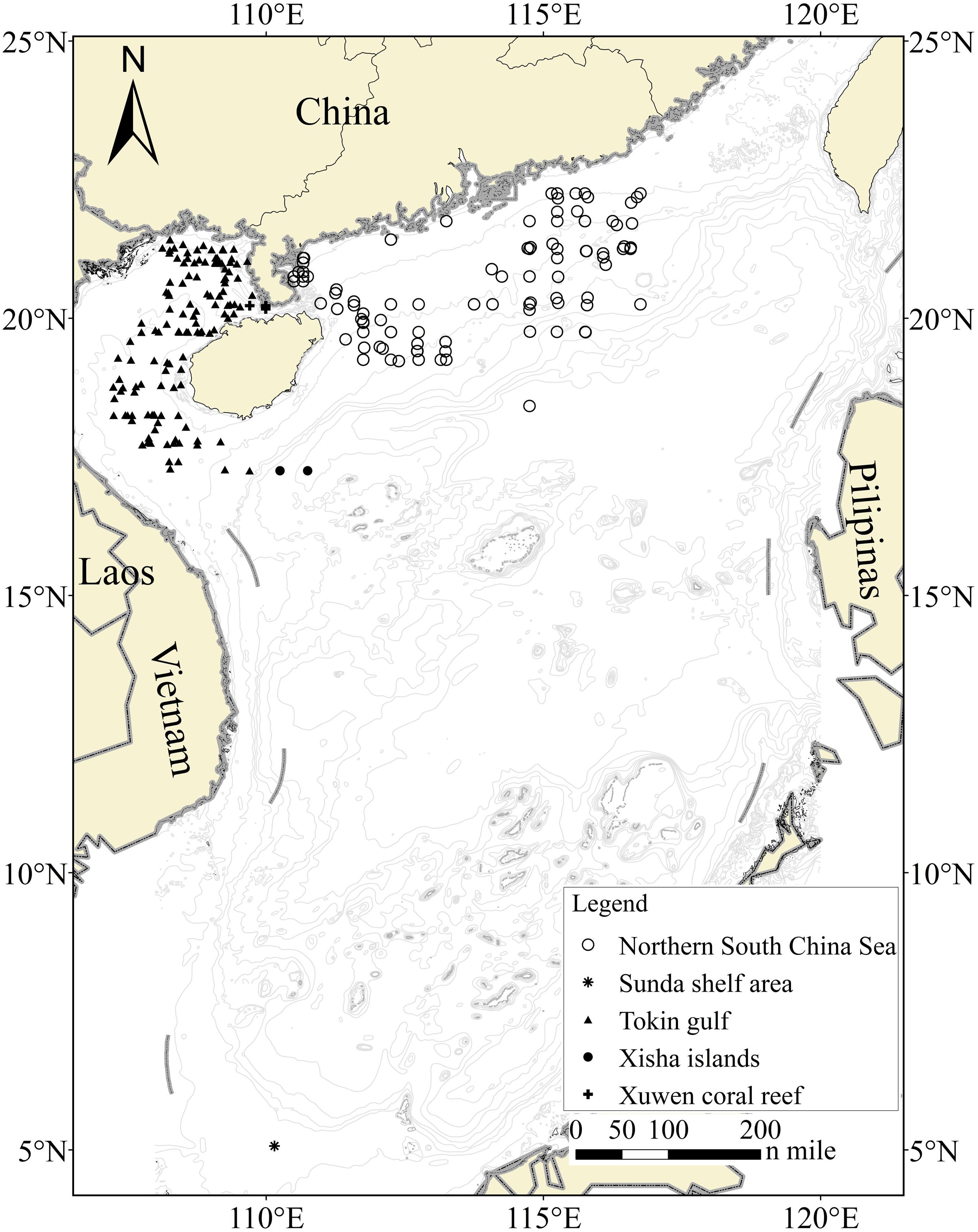
Figure 1. Sampling locations for fish larvae from five areas. Each station yielded one or more samples, and the stations that failed to obtain larval COI sequences were not shown.
Materials and Methods
Sampling
Extensive ichtyoplankton sampling was conducted during a 5-year fishery survey, which covered six regions in the SCS (i.e., Xuwen National Coral Reef Nature Reserve, the Gulf of Tokin, the Xisha Islands, the northern SCS, the southwestern shelf area, and the Sunda Shelf area; Figure 1) during 2013 and 2017. We collected fish larvae from a total of 230 sites with a seabed water depth ranging from 3 to 1,638 m. Two types of nets, i.e., set and zooplankton nets, were used for larval sampling. The set net was used to sample specimens on the edge of the Xuwen National Coral Reef Nature Reserve established for coral reef conservation. With respect to the remaining sites, larvae were collected using an 80-cm-diameter zooplankton net (length = 2.7 m, mesh size = 0.505 mm, and bore diameter = 400 μm) deployed in vertical and horizontal trawls. For vertical trawls, nets were lowered down to approximately the sea bottom if the seabed water depth was less than 200 m and dragged with a hauling speed of 1.5 m/s. Each site was sampled twice or thrice per survey voyage. For horizontal trawls, nets were dragged for 15 min at a speed of 1.5–2.2 knots/h. Given that the sampling boats were 35–45 m long, turbulent water created by the propeller could lead to serious disturbances of abundance and diversity, even if a sampling rope more than 30 m long were tired on the stern. Thus, we affixed a steel hanger of 5.5 m long and 9 cm in diameter to both sides of the midship, which began from the four outside-welded rings. Among the four rings, two rings were used to hang the net on the ship, and the remaining two rings were used to suspend the zooplankton net. An auxiliary line connected to the front plankton net could be used to recycle nets and complete operations multiple times (Supplementary Figure 1).
At the nature reserve of Xuwen Coral Reef national, fish larvae were sampled monthly between 5 PM and 6 AM from March 2013 to March 2015. We collected four, four, and two samples for the Gulf of Tokin, the northern SCS, and the Sunda Shelf area, respectively. In the mouth of Gulf of Tokin to northwestern Xisha Islands and Southwest shelf area, only one sampling was conducted. Fish larvae were mainly sampled between 7 PM and 6 AM from November 2013 to September 2017. A total of 205,000 fish larvae were obtained during our 5-year survey.
Preservation, Photography, DNA Extraction, and Polymerase Chain Reaction Amplification
For the purpose of obtaining undamaged morphological characteristics of fish larvae for photography and sequencing, the ichthyoplankton samples were instantly preserved in 4% neutral formalin solution. Given that formalin fixation can lead to DNA corrosion and damage and thus hamper amplification of the target COI gene, especially in fragile larvae, we developed a formalin-based preservation and fixation technique for ocean fish larvae. We first added nearly 0.175 g of NaOH powder to neutralize 500 mL of seawater and then used the neutralized seawater to prepare a 4% neutral formalin solution. The 4% formalin solution was preserved at 4°C for 24 h before being applied to preserve fish larvae. Samples preserved in the neutral formalin solution still required cold preservation (4°C) for less than 10 days.
In the laboratory, larvae with different morphological features were manually selected to produce silhouette images. A total of 3,500 fish larvae were chosen during our 5-year sampling. The selected larvae were initially photographed using a Zeiss microscope (Axioplan 2 imaging E) and then used for DNA extraction (Figure 2). After the photography was completed, each sample was preserved in GTE solution (100 mmol/L glycine, 10 mmol/L Tris, and 1 mmol/L EDTA) to eliminate the formalin utilizing a 2-mL centrifuge tube and given a serial number. The preservation fluid was replaced once per day for 7 days. The samples were moved into a 1.5-mL centrifuge tube, air dried for 3–5 h, and then boiled by adding 1 mL of GTE solution for 20 min. Samples were dried using absorbent paper; placed in a bioclean centrifuge tube; mixed with 30 μL sodium dodecyl sulfate, 10 μL DTT, 20 μL protease K, and 200 μL STE; and heated in a 55°C water bath for 4–5 h. When the solution was clear, the samples were centrifuged at 1,500 revolutions/min (rpm) for 30 s, mixed fully with 300 μL of NaCl solution (6 M) for 20 min, and centrifuged for 15 min at 13,000 rpm. Then, 400 μL of supernatant liquor was extracted into a new bioclean centrifuge tube. We then added 400 μL of isopropanol, mixed the solution slightly for 10 min, stored the mixture at −20°C for 2–3 h, and centrifuged the mixture for 15 min at 13,000 rpm. After that, we removed the supernatant liquor, added 800 μL of 70% ethanol, centrifuged the solution for 10 min at 13,000 rpm, and removed the supernatant liquor. Finally, the samples were air dried for 60 min and resolved in 30 μL of TE.
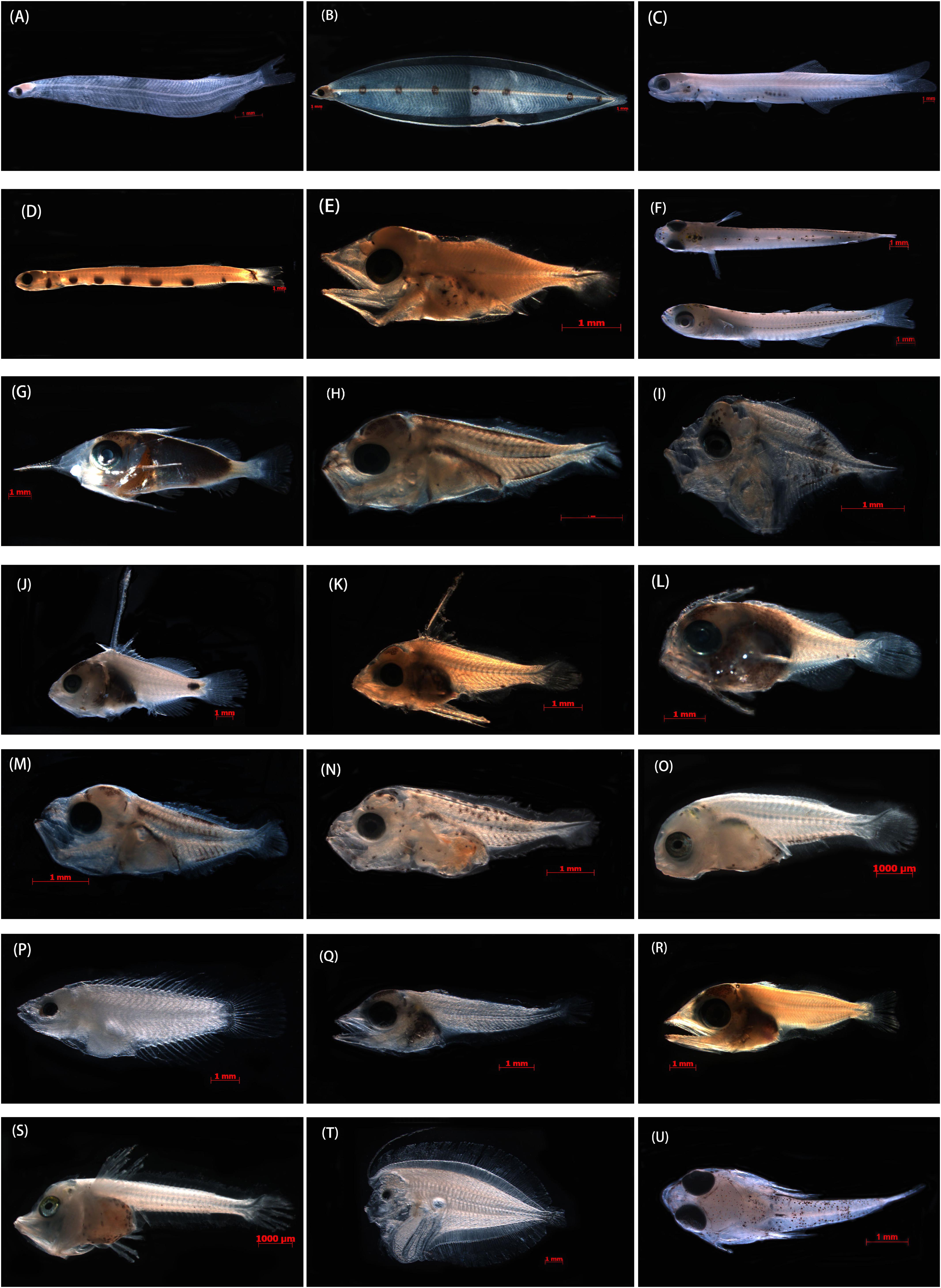
Figure 2. Photographs of the specimens that produced good-quality sequences from formalin-fixed samples and that were identified to species taxa in the SCS. (A) Megalops cyprinoides, XWZ163, 10.54 mm TL; (B) Gnathophis heterognathos, GDYH1, 35.87 mm TL; (C) Encrasicholina heteroloba, XWZ54, 27.10 mm TL; (D) Trachinocephalus myops, GDYH352 26.50 mm TL; (E) Lampanyctus tenuiformis, GDYH275, 5.12 mm TL; (F) Hypoatherina valenciennei, XWZ79, 13.75 mm TL; (G) Sargocentron punctatissimum, GDYH209, 12.41 mm TL; (H) Decapterus tabl, GDYH648, 4.91 mm TL; (I) Mene maculate, BBWZ702, 3.74 mm TL; (J) Triso dermopterus, DSZ48, 12.28 mm TL; (K) Lutjanus russellii, GDYH264, 6.01 mm TL; (L) Priacanthus macracanthus, GDYH36, 6.38 mm TL; (M) Selar crumenophthalmus, GDYH193, 4.52 mm TL; (N) Trachurus japonicus, BBWZ145, 5.81 mm TL; (O) Pagrus major, FCZ10, 7.31 mm TL; (P) Thalassoma amblycephalum, DSZ32, 9.43 mm TL; (Q) Thunnus albacares, DSZ15, 7.44 mm TL; (R) Katsuwonus pelamis, GDYH24, 8.99 mm TL; (S) Lepidotrigla alata, FCZ019, 8.32 mm TL; (T) Bothus pantherinus, DSZ43, 11.46 mm TL; and (U) Callionymus curvicornis, XWZ101, 6.29 mm TL.
C Oxidase I sequences (∼648 bp) were amplified and sequenced using the universal primers FishF1 and FishR1 (Ward et al., 2005). The polymerase chain reaction (PCR) contained approximately 100 ng of template DNA, 1 μL of each primer (10 pmol), 4 μL of 10 × reaction buffer, 1.2 μL of dNTPs (10 mM each), and 5.0 U of Taq DNA polymerase in a total volume of 40 μL. The PCR conditions and processes include initial denaturation at 95°C for 2 min, 35 cycles at 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were sequenced bidirectionally on an ABI 3730XL DNA system following the manufacturer’s protocol (Perkin-Elmer Applied Biosystems, Foster City, CA, United States).
Data Analyses
We initially checked the tracer files and assembled sequences using SEQMAN in Lasergene version 7.0 (DNASTAR Inc., Madison, WI, United States). High-quality sequences were aligned and manually edited using MEGA v6.0 (Tamura et al., 2013). Collection data, sequences, tracer files, and primer details were deposited in the BOLD database.
Larval identification was performed by blast searches in BOLD. We retained reference sequences of both the first and second best interspecific matches in BOLD and documented the percentages of sequence matches. Three criteria similar to those used in Hubert et al. (2015) were adopted in our study: (i) sequences with greater than 99% similarity and exhibiting greater than 1% divergence between sequences and the nearest neighbor species were unambiguously tagged with that species name (case I); (ii) if the genetic divergence was less than the 1% threshold, we considered identification to species uncertain (case II); and (iii) sequences with matches less than 99% were deemed unidentified samples (case III). Canonical 98% similarity and 2% genetic divergence criteria were utilized. Genetic distance was calculated in MEGA6 based on the Kimura 2-parameter model (K2P, Kimura, 1980). A neighbor-joining (NJ) tree based on the K2P model was reconstructed to illustrate lineage diversity via phylogenetic topology. The NJ tree was constructed with 1,000 bootstrap replicates using MEGA v6.0. To compare the results of NJ tree, maximum-likelihood (ML) tree was implemented in RAXML-VI-HPC (Stamatakis, 2006) with the GTR + I + G model and 1,000 non-parametric bootstrap replicates. The optimal nucleotide substitution model (GTR + I + G) was selected based on the Akaike information criterion in MRMODELTEST version 2.3 (Nylander, 2004). Furthermore, to group specimens into operational taxonomic units (OTUs), Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012) was performed online2 using the default value for relative gap width (X = 1.5) and K2P distance metrics.
Results
During our 5-year sampling period, we collected more than 205,000 larvae, among which 3,500 larvae were chosen for the molecular experiment on the basis of their morphological characters. A total of 2,787 larvae (i.e., 80%) were successfully amplified, and only 1,006 samples (36%) showed high-quality sequences with no evidence of insertions, deletions, or stop codons. The larvae collected from the southwestern shelf area failed to produce high-quality DNA or sequences as the preservation time in the aqueous formalin was too long (>30 days). The 1,006 sequences obtained had a length of 604 nucleotides after alignment and trimming of noisy sites, which were subjected to subsequent analyses. Sample information and sequences for the 1,006 larvae were deposited in BOLD (see text footnote 1, Project Codes LFSCS).
Using rigorous 99% similarity and 1% genetic divergence among species thresholds, we found that approximately 81.6% of the sequences (821/1,006) had a match of 99–100%. The match results for nearest neighbors ranged from 81.24 to 100%. Among the 821 sequences, only 408 sequences fit the criteria of case I (i.e., ∼40.5%). A total of 413 sequences showed percentages of similarity with both the best match and the nearest neighbor ranging from 99 to 100% and genetic divergence from the nearest neighbor of less than 1%, indicating that 41.1% of the samples were not ambiguously assigned species names (case II). Less than 99% similarity between the analyzed sequences and the best-matching sequences was shown for the remaining 185 sequences, suggesting that 18.4% of the samples could not be identified to species (case III; Supplementary Figure 2 and Supplementary Table 1). In contrast, using a canonical 98% similarity threshold and 2% divergence among species (Hebert et al., 2003; April et al., 2011), 287 (i.e., ∼28.5%), 567 (i.e., ∼56.4%), and 152 (i.e., ∼15.1%) sequences were assigned to cases I, II, and III, respectively, with the largest proportion of sequences classified as case II.
One hundred one species belonging to 48 families were identified from the 408 larval sequences (case I) according to the canonical 99% similarity and 1% genetic divergence criteria. Apogonidae (eight species, 26 individuals), Myctophidae (seven species, 25 individuals), Sciaenidae (five species, 21 individuals), Oxudercidae (five species, 11 individuals), and Bothidae (five species, 10 individuals) were the most abundant families.
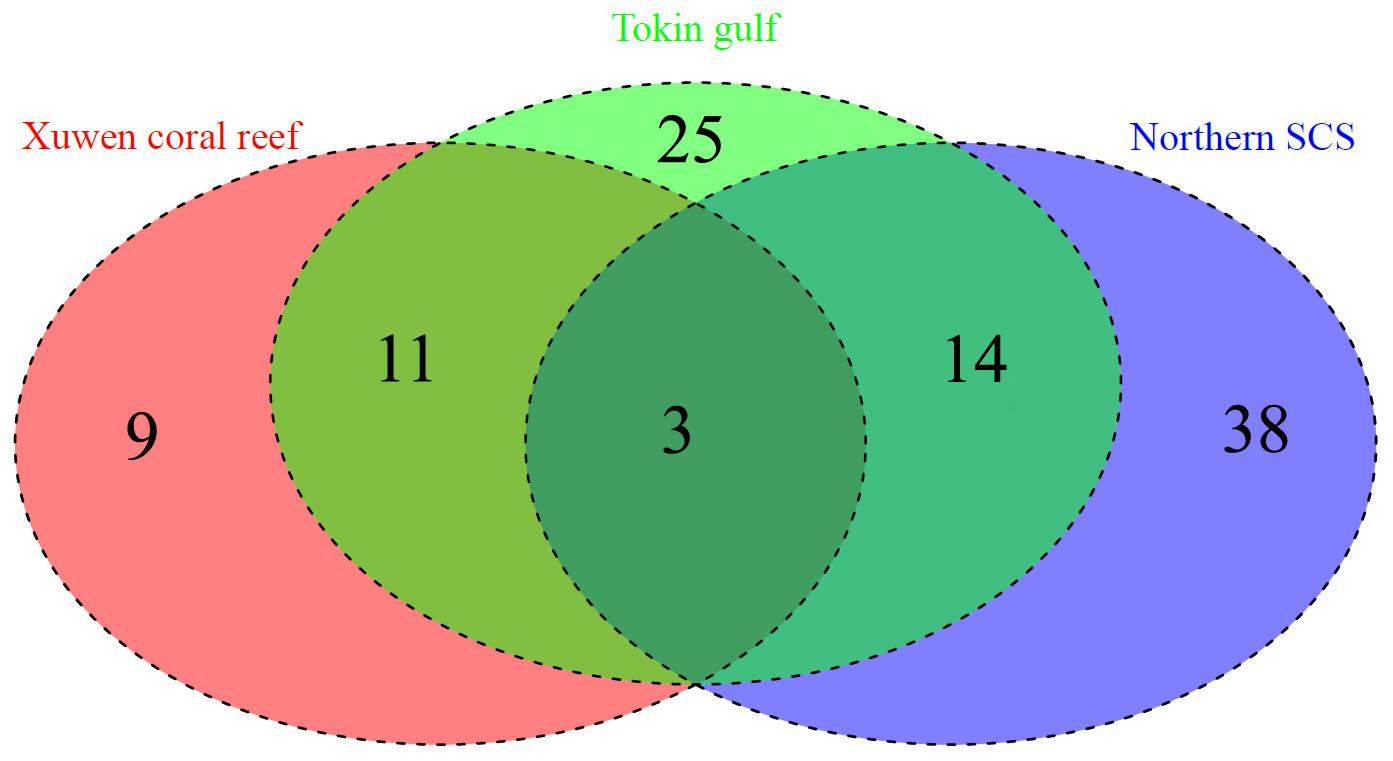
Among the 101 identified species, the Xuwen coral reef area harbored 23 species, the Gulf of Tonkin harbored 53 species, and the northern SCS harbored 55 species. Because of DNA extraction failure or low-quality sequences, only one species (two individuals from the Xisha Islands and one individual from the Sunda Shelf area) each was obtained from the Xisha Islands and Sunda Shelf areas. Venn diagram analysis showed that the Xuwen coral reef area, Gulf of Tonkin, and northern SCS shared only three species, i.e., Omobranchus punctatus (Valenciennes, 1836), Sillago asiatica (Mckay, 1982), and Terapon jarbua (Forsskål, 1775). In addition, the Xuwen coral reef area, Gulf of Tonkin, and northern SCS harbored 9, 25, and 38 exclusive species, respectively (Figure 3). Using habitat data for adult fish species downloaded from FishBase3, 38, 33, 12, 9, 5, and 4 species were demersal, reef associated, benthopelagic, pelagic–oceanic, bathypelagic, and pelagic–neritic, respectively (Figure 4).
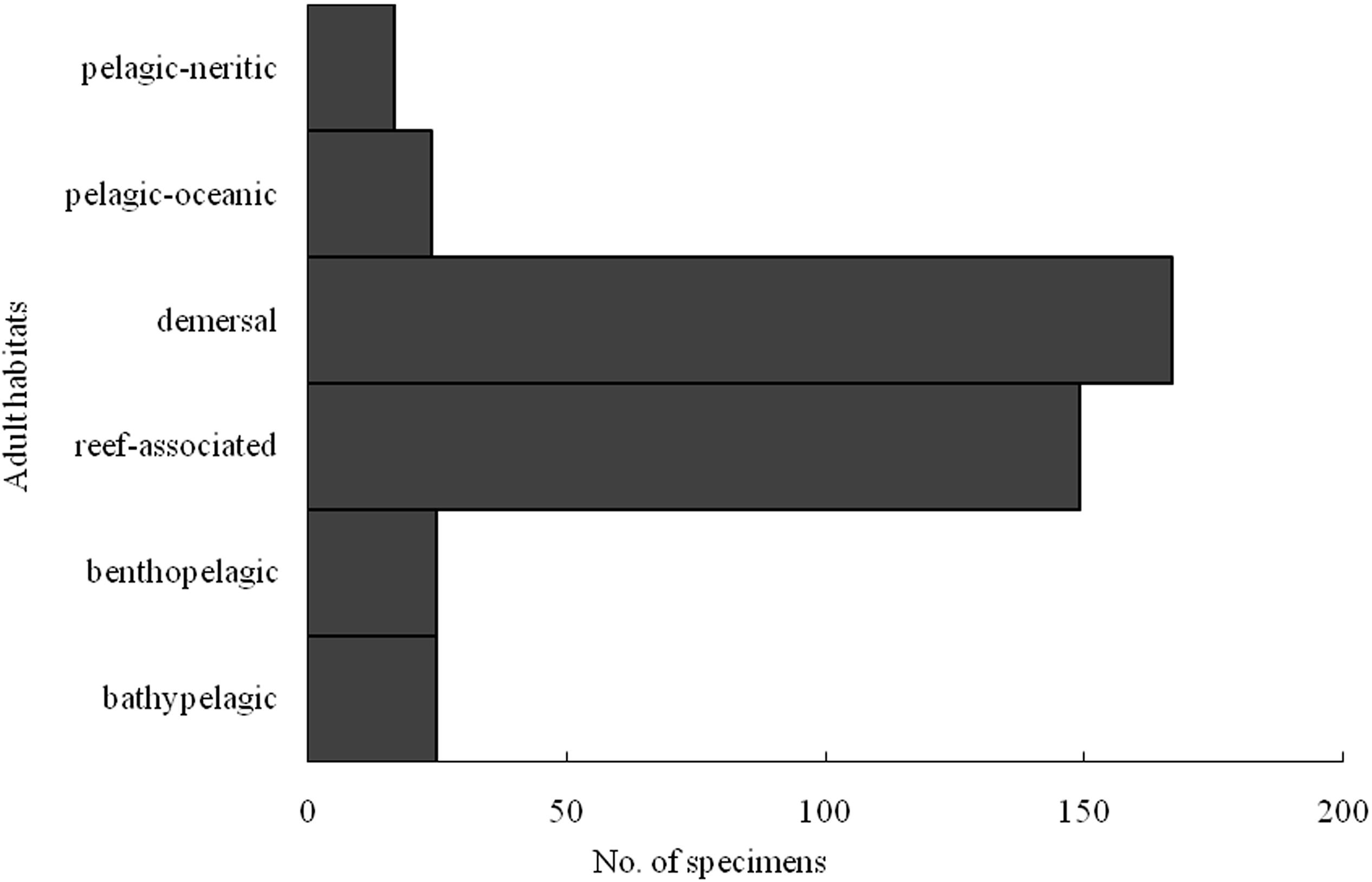
Figure 4. Number specimens of larval fish identified by COI sequences in case I according to adult habitat distribution.
According to morphological features, we assigned 1,006 larvae to 213 morphological forms. The NJ tree based on the K2P distance model and ML tree consistently yielded 241 lineages with high support values (Supplementary Figures 3, 4). The ABGD analyses conducted with the K2P models consistently produced initial partitions with 241 OTUs (P = 0.0215–0.0599).
Discussion
DNA Extraction and Sequence Amplification From Formalin-Fixed Larvae
To obtain high-quality morphological characteristics of fish larvae and to conduct further molecular analyses, we preserved the larvae in aqueous formalin rather than highly concentrated alcohol. A total of 3,500 larval photographs with corresponding serial numbers were obtained. Although formalin has many negative effects on DNA recovery and extraction from specimens (Quach et al., 2004; Wong et al., 2014), our study successfully extracted and amplified DNA from 73.3% of formalin-fixed larval samples and obtained a moderate number of high-quality sequences, highlighting the opportunities to obtain genomic data from formalin-fixed specimens. As a result, our modified approaches to preserve fresh specimens and to extract and amplify DNA from formalin-fixed samples can be effectively utilized in molecular analyses that require intact morphological data.
To date, many reports have attempted to extract and amplify DNA from formalin-fixed samples and have made much progress (Wirgin et al., 1997; Xia et al., 2007; Zhang, 2010; Hykin et al., 2015; Chambers and Hebert, 2016). However, the success rates in most previous studies were lower than the rate yielded in our study. For example, Chambers and Hebert (2016) yielded only five high-quality sequences from 208 formalin-fixed samples. The relatively high success rate of DNA extraction and sequence amplification observed in our study might be attributed to the short preservation time (<10 days). Many earlier studies demonstrated that the quantities of extracted DNA and the success rates of sequencing decreased with the duration of formalin fixation (Legrand et al., 2002; Diaz-Viloria et al., 2005; Palero et al., 2010; Zhang, 2010). Failure to obtain high-quality DNA and sequences from the Xisha Islands, southwestern shelf area, and Sunda Shelf area in our study (formalin fixation > 10 days) provided a direct line of evidence. Furthermore, our study yielded high-quality sequences that exceeded 600 bp in length, whereas most past studies obtained fragmented sequences that were less than 500 bp in length (Palero et al., 2010; Zhang, 2010; Paireder et al., 2013). Previous documents showed that samples preserved in formalin for longer durations tended to yield short sequences (Palero et al., 2010; Zhang, 2010; Paireder et al., 2013). Given that such short DNA sequences provide limited information for genetic studies, researchers should reduce the duration of preservation in formalin as much as possible to ensure adequate sequence length in future genetic analyses.
DNA Barcodes of the Larvae in the SCS
Using a 99% similarity threshold and a 1% threshold for species boundaries, we successfully assigned 40.5% of sequences to species and delineated 101 unambiguous species. Furthermore, we provided a high-quality photographic library of fish larvae assigned to species. Because the fish larvae were fixed in formalin, their morphological features were better maintained than those of fish larvae preserved in ethanol. In past investigations with larval and/or planktonic delimitation based on molecular techniques, the samples were preserved in ethanol, which damaged their morphological characters (Frantine-Silva et al., 2015; Hubert et al., 2015; Ardura et al., 2016; Collet et al., 2018; Liu et al., 2018). Hence, our study is one of the first reports in which DNA-based species identification was applied to a large number of fish larvae fixed in formalin. Therefore, it provided a unique opportunity to examine their morphology. This ability to examine their morphology is particularly important, as the larval morphology of most SCS fishes remains unknown.
Our analyses revealed O. punctatus (Valenciennes, 1836), S. asiatica (McKay, 1982), and T. jarbua (Forsskål, 1775) in three sea areas. This is an expected phenomenon as the three species are widespread species that live in coastal oceans. Furthermore, many endemic species were found in the three sea regions (Figure 3). The main reason for this finding is the different ichthyofaunas of the three regions. Previous reports argued that the fish of the Xuwen National Coral Reef Nature Reserve, the Gulf of Tonkin, and the northern SCS belonged to different ichthyofaunas (Qiu, 1988; Qiu, 1990; Yang et al., 2008; Wang et al., 2011a,b). Reef fishes, demersal fishes, and pelagic-neritic fishes represented the vast majority of species, which largely agreed with the results of previous fishery surveys in the SCS (Qiu, 1988, 1990, 1996; Ma et al., 2006).
The rate of successful identification in our study was similar to that in studies on fish larvae in the eastern Atlantic Ocean and Taiwanese waters (Ko et al., 2013; Ardura et al., 2016) but much lower than the rates obtained in many previous DNA barcode studies on fish larvae (Frantine-Silva et al., 2015; Hubert et al., 2015; Steinke et al., 2016; Liu et al., 2018), which may be mainly explained by the incompleteness of the DNA barcode reference libraries used for the SCS fishes. Despite the more than 3,300 fish species documented in the SCS, barcode sequences for fewer than 1,000 species have been published (Wang et al., 2009, 2012; Zhang and Hanner, 2012; Chang et al., 2017; Hou et al., 2018). Considering that our analyses uncovered 213 forms based on morphological features, 241 lineages through the NJ tree and 241 OTUs from ABGD, indicating that many specimens could not be assigned to species, we missed a large amount of the diversity in the larval fish community. As a result, further extensive studies on this unidentified diversity should be conducted in a timely manner.
A total of 41.1% of the sequences were more than 99% similar to sequences attributed to two different species (i.e., case II), suggesting that misidentification of these samples occurred. These cases may have resulted for the following reasons: hybridization, recent divergence, inadequate taxonomy, and erroneous identification (Meyer and Paulay, 2005; Ivanova et al., 2007). Hybridization between species is a major problem for DNA barcode identification because a hybrid will be identified as its maternal species. When closely related species only recently diverged, they can exhibit close genetic relationships or even share identical haplotypes in two isolated gene pools. Some examples of incomplete lineage sorting due to recent divergence have been recently described in the SCS and its adjacent areas (Chang et al., 2017; Hou et al., 2018), such as two species of Uranoscopus (U. oligolepis Bleeker, 1878 and U. kaianus Günther, 1880) observed by Hou et al. (2018) and multiple species of Thunnus found by Chang et al. (2017) and Hou et al. (2018). Inadequate taxonomy can lead to two different species sharing barcodes if the two nominal species derive from the same gene pool, in which case they should be treated as the same species. Erroneous identifications within the reference library could produce an apparent case of haplotype sharing. A large number of closely related species living in the SCS are difficult to delimit (Zhang and Hanner, 2012; Hou et al., 2018), and thus, specimens of the same species may be incorrectly given two different names by different identifiers. In addition, many sequences were identified based on specimens collected in different oceans and likely resulted from misidentification due to different retrieval systems and multiple independent identifications. Given this information, we argue that more geographically distinct populations of closely related species and species-specific markers should be included in studies to delimit these species with due consideration of spatial variation. In addition, explicit morphological characters among these species should be measured, and synonyms need to be revised through the efforts of researchers from different countries.
Approximately 18.4% of the sequences (case III) obtained poor matches (<99% similarity). Moreover, the match similarity of some sequences was even less than 90%. These results reflected the taxonomic incompleteness of the DNA barcode reference libraries in BOLD for the SCS fishes. Accordingly, these cases highlight that DNA barcodes for a large number of fish taxa in the SCS are still lacking, a gap that deserves more attention in future research. Establishing DNA barcode reference libraries for SCS fishes is an enormous task that will require efforts from many researchers from different countries over a long period of time. Additionally, cryptic diversity has a non-negligible effect on the failure to assign sequences to species. If cryptic speciation occurs, sequences of the same species will show deep genetic divergence and thus will not be matched. Cryptic diversity seems to be a common phenomenon within SCS fishes (Zhang and Hanner, 2012; Chang et al., 2017; Hou et al., 2018), e.g., Apogon ellioti (Day, 1875), Naucrates ductor (Linnaeus, 1758), and Psenopsis anomala (Temminck and Schlegel, 1758) (Hou et al., 2018).
Here, we successfully obtained high-quality DNA and corresponding barcode sequences from fish larvae fixed in formalin and linked the morphologies of fish species during the early stage with their molecular information, which facilitated the diagnosis of fish larvae based on morphological features. The approach used in this study may be extended to genetic studies that require both morphological and genomic data. Among the 1,006 analyzed sequences, 40.5% were unambiguously assigned to 101 species. The delimitation of fish larvae at the species level was ambiguous for approximately 41% of the sequences as misidentification occurred in the DNA barcode reference libraries. Furthermore, approximately 18.4% of the sequences were poorly matched with DNA barcode reference libraries because of the incompleteness of the libraries. To overcome this limitation, substantial efforts will be required to improve the completeness of DNA barcode reference libraries for SCS fishes. Although exact species names could not be obtained for many larvae, the identification of 101 species through the DNA barcode-based method provided new insight into the fish larval communities in the SCS.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank MW379416 – MW379823.
Ethics Statement
The animal study was reviewed and approved by the Guangdong Ocean University.
Author Contributions
GH and HZ analyzed the data and completed the first draft. GH, YC, SW, and JW performed the field survey. GH and WC conducted the experiments. WC and HZ provided the guidance on data analysis and structure. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 31702347 and 42090044), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB42000000), the Special Fund for Economic Development of the Marine Economy of Guangdong Province (No. GDME-2018E004), undergraduate innovation and Entrepreneurship training program of Guangdong province (S202010566005), the start-up project of Guangdong Ocean University (R19006), and Youth Innovation Promotion Association CAS and S&T Program of Fangchenggang (Fangke AB20014024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the editor and reviewers for their constructive comments on our manuscript. We thank numerous colleagues for their generous support of this study, the 14 crew members of Beiyu60011 and all the survey ships for their help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.634575/full#supplementary-material
Footnotes
References
Alien, G. R., Amaoka, K., Anderson, W. D. Jr., Bellwood, D. R., Bohlke, E. B., Bradbury, M. G., et al. (2000). A checklist of the fishes of the South China Sea. Raffles. B. Zool. 8, 569–667.
April, J., Mayden, R. L., Hanner, R. H., and Bernatchez, L. (2011). Genetic calibration of species diversity among North America’s freshwater fishes. Proc. Natl. Acad. Sci. U.S.A. 108, 10602–10607. doi: 10.1073/pnas.1016437108
Ardura, A., Morote, E., Kochzius, M., and Garcia-Vazquez, E. (2016). Diversity of planktonic fish larvae along a latitudinal gradient in the Eastern Atlantic Ocean estimated through DNA barcodes. PeerJ 4:e2438. doi: 10.7717/peerj.2438
Bialetzki, A., Nakatani, K., Sanches, P. V., Baumgartner, G., and Gomes, L. C. (2005). Larval fish assemblage in the Baía River (Mato Grosso do Sul State, Brazil): temporal and spatial patterns. Environ. Biol. Fish. 73, 37–47. doi: 10.1007/s10641-004-3795-3
Cao, W., Chang, J., Qiao, Y., and Duan, Z. (2007). Fish Resources of Early Life History Stages in Yangtze River. Beijing: China Water power Press.
Chambers, E. A., and Hebert, P. D. (2016). Assessing DNA barcodes for species identification in north american reptiles and amphibians in natural history collections. PLoS One 11:e0154363. doi: 10.1371/journal.pone.0154363
Chang, C. H., Shao, K. T., Lin, H. Y., Chiu, Y. C., Lee, M. Y., Liu, S. H., et al. (2017). DNA barcodes of the native ray-finned fishes in Taiwan. Mol. Ecol. Resour. 17, 796–805. doi: 10.1111/1755-0998.12601
Choat, J., and Doherty, P. J. (1993). A comparison of towed nets, purse seine, and light-aggregation devices for sampling larvae and pelagic juveniles of coral reef fishes. Fish. Bull. 91, 195–209. doi: 10.1002/elan.201300220
Collet, A., Durand, J.-D., Desmarais, E., Cerqueira, F., Cantinelli, T., Valade, P., et al. (2018). DNA barcoding post-larvae can improve the knowledge about fish biodiversity: an example from La Reunion, SW Indian Ocean. Mitochond. DNA Part A 29, 905–918. doi: 10.1080/24701394.2017.1383406
Diaz-Viloria, N., Sanchez-Velasco, L., and Perez-Enriquez, R. (2005). Inhibition of DNA amplification in marine fish larvae preserved in formalin. J. Plankton Res. 27, 787–792. doi: 10.1093/plankt/fbi052
Frantine-Silva, W., Sofia, S., Orsi, M., and Almeida, F. (2015). DNA barcoding of freshwater ichthyoplankton in the Neotropics as a tool for ecological monitoring. Mol. Ecol. Resour. 15, 1226–1237. doi: 10.1111/1755-0998.12385
Hebert, P. D., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hou, G., Chen, W. T., Lu, H. S., Cheng, F., and Xie, S. G. (2018). Developing a DNA barcode library for perciform fishes in the South China Sea: species identification, accuracy and cryptic diversity. Mol. Ecol. Resour. 18, 137–146. doi: 10.1111/1755-0998.12718
Huang, D., Licuanan, W. Y., Hoeksema, B. W., Chen, C. A., Ang, P. O., Huang, H., et al. (2015). Extraordinary diversity of reef corals in the South China Sea. Mar. Biodivers. 45, 157–168. doi: 10.1007/s12526-014-0236-1
Hubert, N., Delrieu-Trottin, E., Irisson, J. O., Meyer, C., and Planes, S. (2010). Identifying coral reef fish larvae through DNA barcoding: a test case with the families Acanthuridae and Holocentridae. Mol. Phylogenet. Evol. 55, 1195–1203. doi: 10.1016/j.ympev.2010.02.023
Hubert, N., Espiau, B., Meyer, C., and Planes, S. (2015). Identifying the ichthyoplankton of a coral reef using DNA barcodes. Mol. Ecol. Resour. 15, 57–67. doi: 10.1111/1755-0998.12293
Hykin, S. M., Bi, K., and Mcguire, J. A. (2015). Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS One 10:e0141579. doi: 10.1371/journal.pone.0141579
Ivanova, N. V., Zemlak, T. S., Hanner, R. H., and Hebert, P. D. N. (2007). Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Resour. 7, 544–548. doi: 10.1111/j.1471-8286.2007.01748.x
Kimmerling, N., Zuqert, O., Amitai, G., Gurevich, T., Armoza-Zvuloni, R., Kolesnikov, I., et al. (2018). Quantitative species-level ecology of reef fish larvae via metabarcoding. Nat. Ecol. Evol. 2:306. doi: 10.1038/s41559-017-0413-2
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Ko, H. L., Wang, Y. T., Chiu, T. S., Lee, M. A., Leu, M. Y., Chang, K. Z., et al. (2013). Evaluating the accuracy of morphological identification of larval fishes by applying DNA barcoding. PLoS One 8:e53451. doi: 10.1371/journal.pone.0053451
Legrand, B., de Mazancourt, P., Durigon, M., Khalifat, V., and Crainic, K. (2002). DNA genotyping of unbuffered formalin fixed paraffin embedded tissues. Forensic Sci. Int. 125, 205–211. doi: 10.1016/S0379-0738(01)00641-7
Liu, M., Wang, D., Gao, L., Tian, H., Liu, S., Chen, D., et al. (2018). Species diversity of drifting fish eggs in the Yangtze River using molecular identification. PeerJ 6:e5807. doi: 10.7717/peerj.5807
Ma, C. H., You, K., Li, F. Q., and Zhang, M. C. (2006). A study on the relationship of the fish biodiversity and the faunal distribution in the South China Sea. Perio. Ocean Univ. China 36, 665–670. doi: 10.1360/aps050066
Macusi, E. D., Katikiro, R. E., Deepananda, K. A., Jimenez, L. A., Conte, A. R., and Fadli, N. (2011). Human induced degradation of coastal resources in Asia Pacific and implications on management and food security. J. Nat. Stud. 9, 13–28.
Meyer, C. P., and Paulay, G. (2005). DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 3:e422. doi: 10.1371/journal.pbio.0030422
Morton, B., and Blackmore, G. (2001). South China Sea. Mar. Pollut. Bull. 42, 1236–1263. doi: 10.1016/S0025-326X(01)00240-5
Nylander, J. (2004). MrModeltest v2. Program Distributed by the Author, ed. J. A. A. Nylander (Uppsala: Evolutionary Biology Centre, Uppsala University).
Olden, J. D., Hogan, Z. S., and Zanden, M. J. V. (2007). Small fish, big fish, red fish, blue fish: size-biased extinction risk of the world’s freshwater and marine fishes. Glob. Ecol. Biogeogr. 16, 694–701. doi: 10.1111/j.1466-8238.2007.00337.x
Paireder, S., Werner, B., Bailer, J., Werther, W., Schmid, E., Patzak, B., et al. (2013). Comparison of protocols for DNA extraction from long-term preserved formalin fixed tissues. Anal. Biochem. 439, 152–160. doi: 10.1016/j.ab.2013.04.006
Palero, F., Hall, S., Clark, P. F., Johnston, D., Mackenzie-Dodds, J., and Thatje, S. (2010). DNA extraction from formalin-fixed tissue: new light from the deep sea. Sci. Mar. 74, 465–470. doi: 10.3989/scimar.2010.74n3465
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294x.2011.05239.x
Qiu, Y. S. (1988). The regional changes of fish community on the northern continental shelf of South China Sea. J. Fish. China 12, 303–313.
Qiu, Y. S. (1990). A preliminary analysis of fish species groups of the northern continental shelf of South China Sea. J. Fish. China 14, 267–276.
Qiu, Y. S. (1996). Ordinations of fish communities in the inshore waters of Guangdong province. Acta Ecol. Sin. 16, 576–583.
Quach, N., Goodman, M. F., and Shibata, D. (2004). In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 4:1. doi: 10.1186/1472-6890-4-1
Rahel, F. J., Bierwagen, B., and Taniguchi, Y. (2008). Managing aquatic species of conservation concern in the face of climate change and invasive species. Conserv. Biol. 22, 551–561. doi: 10.1111/j.1523-1739.2008.00953.x
Ratnasingham, S., and Hebert, P. D. (2007). Bold: the barcode of life data system. Mol. Ecol. Resour. 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Reynalte-Tataje, D. A., Nuñer, A. P., Nunes, M. C., Garcia, V., Lopes, C. A., and Zaniboni-Filho, E. (2012). Spawning of migratory fish species between two reservoirs of the upper Uruguay River, Brazil. Neotrop. Ichthyol. 10, 829–835. doi: 10.1590/S1679-62252012000400016
Shao, K. T., Ho, H. C., Lin, P. L., Lee, P. F., Lee, M. Y., Tsai, C. Y., et al. (2008). A checklist of the fishes of southern Taiwan, northern South China Sea. Raffles. B. Zool. 19, 233–271.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Steinke, D., Connell, A. D., and Hebert, P. D. (2016). Linking adults and immatures of South African marine fishes. Genome 59, 959–967. doi: 10.1139/gen-2015-0212
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Valdez-Moreno, M., Vásquez-Yeomans, L., Elías-Gutiérrez, M., Ivanova, N. V., and Hebert, P. D. (2010). Using DNA barcodes to connect adults and early life stages of marine fishes from the Yucatan Peninsula, Mexico: potential in fisheries management. Mar. Freshw. Res. 61, 655–671. doi: 10.1071/MF09222
Wang, X. H., Qiu, Y. S., Du, F. Y., Lin, Z. J., Sun, D. R., and Huang, S. L. (2011a). Fish community pattern and its relations to environmental factors in the Beibu Gulf. J. Fish. China 34, 1579–1586. doi: 10.3724/SP.J.1231.2010.06827
Wang, X. H., Qiu, Y. S., Du, F. Y., Lin, Z. J., Sun, D. R., and Huang, S. L. (2011b). Spatio-temporal variability of fish diversity and dominant species in the Beibu Gulf. J. Fish. Sci. China 18, 427–436. doi: 10.3724/SP.J.1011.2011.00415
Wang, Z. D., Guo, Y. S., Chen, R. L., Xiao-Ying, H. E., Liu, C. W., and Liu, Y. (2009). COI Barcoding sequences of teleosts in the South China Sea. Oceanol. Limnol. Sin. 40, 608–614. doi: 10.1016/S1874-8651(10)60059-2
Wang, Z. D., Guo, Y. S., Liu, X. M., Fan, Y. B., and Liu, C. W. (2012). DNA barcoding South China Sea fishes. Mitochondrial DNA 23, 405–410. doi: 10.3109/19401736.2012.710204
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., and Hebert, P. D. (2005). DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Wirgin, I., Maceda, L., Stabile, J., and Mesing, C. (1997). An evaluation of introgression of Atlantic coast striped bass mitochondrial DNA in a Gulf of Mexico population using formalin-preserved museum collections. Mol. Ecol. 6, 907–916. doi: 10.1046/j.1365-294X.1997.00271.x
Wong, S. Q., Li, J., Tan, A. Y., Vedururu, R., Pang, J. M., Do, H., et al. (2014). Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med. Genom. 7:23. doi: 10.1186/1755-8794-7-23
Xia, Y. Z., Zhang, C. G., Chen, Y. Y., Yan, S., and Sun, Y. H. (2007). Discussing on the feasibility of large fragments DNA extraction from formalin-fixed fish specimens. Acta Hydro. Sin. 31, 485–491.
Yang, G. H., Hou, X. Q., Chen, C. L., and Sun, X. L. (2008). Fish species composition in the coast reef of coast Xuwen county. Fish. Sci. 27, 533–538. doi: 10.3969/j.issn.1003-1111.2008.10.009
Zhang, J. (2010). Exploiting formalin-preserved fish specimens for resources of DNA barcoding. Mol. Ecol. Resour. 10, 935–941. doi: 10.1111/j.1755-0998.2010.2838.x
Keywords: fish larvae, DNA barcode, South China Sea, spawning ground, fish conservation
Citation: Hou G, Chen Y, Wang S, Wang J, Chen W and Zhang H (2021) Formalin-Fixed Fish Larvae Could Be Effectively Identified by DNA Barcodes: A Case Study on Thousands of Specimens in South China Sea. Front. Mar. Sci. 8:634575. doi: 10.3389/fmars.2021.634575
Received: 28 November 2020; Accepted: 17 February 2021;
Published: 25 March 2021.
Edited by:
Jinghui Fang, Yellow Sea Fisheries Research Institute (CAFS), ChinaReviewed by:
Zhiqiang Han, Zhejiang Ocean University, ChinaMing Liu, Morgan State University, United States
Copyright © 2021 Hou, Chen, Wang, Wang, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weitao Chen, bmN1c2tjaGVud2VpdGFvQDE2My5jb20=; Hui Zhang, emhhbmdodWlAcWRpby5hYy5jbg==
†Lead contact
 Gang Hou
Gang Hou Yanying Chen
Yanying Chen Sijin Wang1
Sijin Wang1 Jinrun Wang
Jinrun Wang Hui Zhang
Hui Zhang