- 1MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
- 2Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
The Chinese soft-shelled turtle (Pelodiscus sinensis) is among the most primitive amphibians and reptiles in nature. On account of its environmental suitability and unique hibernation habit, the peculiar physiological phenomenon in P. sinensis attracted the attention of researchers in the field of marine science. The present study aimed to investigate the molecular mechanisms underlying the periodic variation of lipid droplet (LD) in the liver of P. sinensis. Histological results indicated that accumulated LD in the liver of P. sinensis during non-hibernation was gradually consumed during hibernation. RNA-Seq results revealed that genes responsible for carbohydrate catabolism were down-regulated during hibernation, while genes involved in lipid oxidation were up-regulated. These results suggest that energy metabolism in the liver of P. sinensis changes during hibernation, i.e., the energy generation mode shifted from carbohydrate catabolism to lipid oxidation. Further analysis of RNA-Seq results indicated that both lipolysis and autophagy could promote the degradation of hepatic LD during hibernation. To further determine the relationship between lipolysis and autophagy in the process of LD breakdown, we applied the inhibitors of lipolysis and autophagy (diethylumbelliferyl phosphate and 3-Methyladenine) in cultured primary hepatocytes of P. sinensis. The results indicated that lipolysis is the main way for LD degradation in the hepatocyte of P. sinensis. These data provide clear evidence about the seasonal changes in hepatocytes, corresponding with the different energy generation mode in the liver of P. sinensis.
Introduction
Marine biodiversity is not only the important sustainer of the marine ecosystem but also provides a broader space for human survival and development (Beaugrand et al., 2010). As the important component of marine organisms, sea turtles are reckoned to be threatened or endangered due to natural predators (Eckrich and Owens, 1995), harvest for consumption (Mancini and Koch, 2009), illegal sea turtle shell trade (Foran and Ray, 2016), artificial lighting (Lorne and Salmon, 2007), marine pollution (Bugoni et al., 2001), and climate change (Hays et al., 2003). In consequence, restoring or maintaining sea turtle habitats and developing an artificial culture of turtles (Lovich et al., 2018) are better ways to protect wild turtles. As the most extensive cultured turtles in the world, the abundant experience in breeding Chinese soft-shelled turtles (Pelodiscus sinensis) could provide us new inspirations in improving survival rates of marine life.
On account of the sea turtle’s marine and terrestrial attributes over the life cycle, the impacts of climate change would likely devastatingly affect the endangered sea turtles. Besides, as ectothermic animals, the physiological index, such as the body temperature, heart rate, and metabolism, will decline following the external environmental temperature (Melzner et al., 2009). Simultaneously, high temperatures under global warming scenarios could also decrease the resistance of sea turtles to emerging diseases (Dang et al., 2015). Once they get into hibernation, the extreme physiological challenges (repressed metabolism and the large accumulation of lactic acid) are caused by anoxic mud or water depleted of O2 where the turtle hibernating (Jackson, 2002), and then threaten the capacity for turtle’s long-term survival. In consequence, the physiological function adjustment during hibernation underlies the capacity for the long-term survival of sea turtles.
The liver, the maximal digestive gland in the body, is a vital metabolic organ. It promotes the decomposition and absorption of lipids through secreting bile and participates in the synthesis, storage, metabolism, transformation, and decomposition of various substances (Gebhardt, 1992). In the liver, energy substances are in the form of hepatic glycogen and LD. Along with the exhaustion of hepatic glycogen, TGs stored in LDs are decomposed to fatty acids (FAs) and glycerol, which could be transformed into ketone bodies and glucose to maintain the body’s energy balance (Rui, 2011). During hibernation, animals face not only hypothermia but also food scarcity. How animals acclimatize to the hostile environment is still unknown. Previous transcriptome sequencing studies in the liver and heart of black bear (Fedorov et al., 2011) and the liver of thirteen-lined ground squirrel (Nelson et al., 2009) and greater horseshoe bat (Xiao et al., 2015) revealed that carbohydrate catabolism-related genes were down-regulated, while lipid β-oxidation-related genes were up-regulated during hibernation compared to non-hibernation period. These results indicated that hibernating animals are replacing carbohydrates with lipid as the predominant energy source during hibernation. Although these previous studies have unmasked the change of hepatic metabolism among the non-hibernation and hibernation period, the molecular regulation mechanism underlying this shift remained unresolved.
Given the considerable environmental suitability and hibernation habit of order Testudines, we looked forward to seeking inspiration from Chinese soft-shelled turtle (P. sinensis) for improving survival rates of marine life. Our previous researches have clarified that the periodic variation of the energy metabolism mechanism throughout the year is of great importance in improving the survival rate of P. sinensis (Huang et al., 2019). Consequently, the present study aimed to ulteriorly investigate the molecular mechanism in the shift of metabolic substrates during hibernation.
Materials and Methods
Animals
In the present study, 40 matured Chinese soft-shelled turtles were captured in the wild ponds of Nanjing, Southeastern China (GPS coordinates N 32.07 E 118.78). Twenty of them were from hibernation periods and the other 20 turtles were arrested in non-hibernation periods. Intraperitoneal injection of sodium pentobarbital (20 mg/kg) (P3761, Sigma-Aldrich, Saint Louis, MO, United States) was applied in anesthesia of turtles and they were killed by cervical bleeding. A part of liver tissue was immersed in 2.5% glutaraldehyde (G5882, Sigma-Aldrich, Saint Louis, MO, United States) and 4% paraformaldehyde (P6148, Sigma-Aldrich, Saint Louis, MO, United States) for at least 24 h, respectively. Some fresh liver tissues would test for the content of triglycerides, total cholesterol, and non-esterified free FA. The remnant liver was kept in an ultra cold storage freezer (−80°C). All processes of handling turtles complied with the Animal Research Institute Committee guidelines of Nanjing Agriculture University, China. The Science and Technology Agency of Jiangsu Province and Nanjing Agricultural University Veterinary College approved the sampling procedures. The approval ID is SYXK (SU) 2010-0005. All efforts were made to minimize the animals’ suffering.
Immunofluorescence
A total of 5 μm paraffin slices of a different group (non-hibernation and hibernation period) were firstly deparaffinized and then hydrated. The internal peroxidase activity was wiped out using 3% H2O2. Then, 1% Triton X-100 was applied to penetrate the cytomembrane. Following this, 5% bovine albumin (BSA) was added to block the non-specific protein binding site for 30 min under 37°C. The lipid droplets (LDs) were stained with BODIPY 493/503 (D3922, Invitrogen, Waltham, MA, United States). The staining results were observed using the light microscope (DP73, Olympus, Tokyo, Japan).
Transmission Electron Microscopy
The liver of the Chinese soft-shelled turtle was directly trimmed into a 1-mm3 block and immersed in 2.5% glutaraldehyde for at least 24 h. Subsequently, the blocks were immersed in 1% osmium tetroxide for 1 h and dehydrated in ethyl alcohol. Acetone was used for the substitution of ethyl alcohol in the liver tissues. Then, the liver tissues were embedded using Epon812. The slides were sectioned employing ultramicrotome and observed by electron microscope (Hitachi H-7650).
Oil Red O Staining
Six livers from different turtles in the non-hibernation group and six livers from different turtles in the hibernation group were stained in this experiment. A total of 8 μm frozen slices of liver were immersed in 10% neutral formalin (c1040410015, Nanjing Reagent, Nanjing, China) for at least 10 min. The slices were stained with Oil Red O reagent (D027-1-1, Jiancheng, Nanjing, China) for 10 min after rinsing with distilled water. Next, the liver slices were rinsed with distilled water at 37°C for nearly 15 s. After dying with hematoxylin (H8070, Solarbio, Beijing, China) for 3 min, the frozen slices of liver were washed with running water for approximately 60 s. The slides were observed under an Olympus DP73 light microscope.
Periodic Acid-Schiff Staining
Six livers from different turtles in the non-hibernation group and six livers from different turtles in the hibernation group were stained in this experiment. A total of 5 μm paraffin sections of liver were deparaffinized and then hydrated. The slices were immersed in periodic acid for 5 min. After rinsing with distilled water, slices were stained using Schiff’s dye (G1281, Solarbio, Beijing, China) liquor for 15 min. The slices were then rinsed with running water for 7 min. After staining with hematoxylin for 1 min, the slides were washed in running water for 5 min. The slices were observed via a light microscope (DP73, Olympus, Tokyo, Japan).
RNA-Sequencing and Analysis
In this experiment, the total RNA from turtle livers from the non-hibernation (n = 6) and hibernation (n = 6) groups was isolated using the TRIzol reagent (Invitrogen, United States). The concentrations of RNA were determined by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, United States). The integrity of RNA samples was verified by measuring the ratio of ODs at 260 and 280 nm. Only the RNA samples that had a ratio >1.8 were used for the subsequent cDNA synthesis. The first-strand cDNA was synthesized from RNA using a cDNA synthesis kit (TaKaRa, China) according to the manufacturer’s instructions. The RNA and cDNA were used for RNA-Seq and quantitative real-time PCR procedures. The library was constructed using the Illumina HiSeq 2500 platform. Eukaryotic mRNAs with polyA tails were enriched using magnetic beads with Oligo (dT) Primer and interrupted via ultrasonic waves. Fragmented mRNA was used as a template and random oligonucleotides were used as a primer. The first strand of cDNA was synthesized utilizing the M-MuLV reverse transcriptase system. The second strand of cDNA was synthesized from dNTPs utilizing the DNA polymerase I system. After end reparation and polyA tail addition, cDNA with a length of around 200 bp was filtrated using AMPure XP beads. The Illumina HiSeq 2500 platform was used in building the two libraries. The sequence data were deposited in the NCBI Sequence Read Archive (SRA1) with accession numbers SRR11586153 (CK, non-hibernation group) and SRR11586154 (T, hibernation group). We judged the significant differences in gene expression using the threshold value of |log2 (fold-change)| ≥ 1 with a false discovery rate (FDR) ≤ 0.05. The fold change was calculated as follows: fold-change = RPKM in T/RPKM in CK. To ensure the quality of data, the original data should be filtered before information analysis as this reduces analysis interference caused by invalid data. First, quality control of raw reads was performed using fastp2 to filter low-quality data and thus obtain clean reads. The steps in read filtering were as follows: (1) remove reads containing adapter; (2) remove reads for which N ratio is greater than 10%; (3) remove reads which are all A base; (4) remove low-quality reads. Quality control of raw reads was performed using fastp (see text footnote 2) to filter low-quality data, and the “R” package was also used in data analysis. The reference genome was from the Chinese soft-shelled turtle, P. sinensis (doi: 10.1038/ng.2615). All differentially expressed genes (DEGs) were given Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations. GO terms with P < 0.05 were considered significantly enriched among the DEGs. The main biological functions of DEGs were determined according to GO function enrichment analysis. KEGG is a database of biological systems that links genomes to living organisms through the process of pathway mapping. After multiple inspections and corrections, pathways with P < 0.05 in KEGG3 were defined as significantly enriched pathways for the DEGs.
RT-PCR
Total RNA from turtle livers from the non-hibernation and hibernation groups was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol and was incubated with 20 U/ml DNaseI (Sigma-Aldrich, Saint Louis, MO, United States) for 15 min at 37°C to remove genomic DNA contamination. Next, the RNA quality and quantity were examined using a NanoPhotometer spectrophotometer (IMPLEN GmbH, Munich, Germany) and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States). The RNA integrity number (RIN) of all samples was greater than 8.0. HiScript III RT SuperMix for qPCR (R323-01, Vazyme, Nanjing, China) was used in the reverse transcription of extracted RNA. Real-time fluorescence quantitative PCR (Q-PCR) assays were performed with the LightCycler 480 Sequence Detection System (Roche, Switzerland) using an SYBR Green PCR Master Mix (Vazyme Biotech Ltd., China). The PCR reactions were conducted using the following program: 94°C for 5 min, followed by 40 cycles of amplification with denaturation at 95°C for 30 s and annealing at 60°C for 30 s. Melting curves were recorded from 60 to 95°C to verify a single-product generation at the end of the reaction. Specific primers for detective genes were designed by Primer3 Input (version 0.4.0) (Supplementary Table 1) and β-actin served as the internal control. The relative changes in gene expression between different groups were calculated using the 2-ΔΔCT method.
Isolating LD and Verifying the Quality of Isolated LDs
Lipid droplets in the liver of Chinese soft-shelled turtles were isolated according to an optimized approach of Ding et al. (2013). The purity of LDs was confirmed using Colloidal blue (P1305, Solarbio, Beijing, China) and Nile red staining (19123, Sigma-Aldrich, Saint Louis, MO, United States).
Western Blotting
The livers kept in ultra cold storage freezer were split using RIPA lysis buffer (R0278, Sigma-Aldrich, Saint Louis, MO, United States) and centrifuged for 10 min at 12,000g. The protein concentration of different groups was detected using the BCA protein assay kit (ab102536, Abcam, Cambridge, MA, United States). Samples (100 μg of protein per lane) were added into different concentrations of SDS-PAGE gel based on electrophoresis on the different molecular weights of protein. The next step is transferring proteins onto PVDF membranes (ISEQ00010, Millipore, MA, United States). After immersing in 5% non-fat milk for 30 min, the membranes were interacted with antibodies overnight at 4°C. After washing via TBST, the membranes were incubated with HRP conjucted goat anti-rabbit IgG (1:5000, TransGen Biotech, HS101) for 2 h at room temperature. After rinsing, the conjunct antibodies on PVDF membranes were proceeded to chemiluminescence using an automatic chemiluminescence analysis system (5200 Multi, Tannon, Shanghai, China). Protein Ladder was used to identify and test for each protein. Protein bands were quantitatively analyzed using Quantity One software (Bio-Rad, Hercules, CA, United States).
Cell Culture
Primary hepatocytes were isolated from the fresh liver of Chinese soft-shelled turtles. After disinfecting turtles with 0.01% (W/V) potassium permanganate and 75% ethyl alcohol successively, livers were sampled in a biosafety cabinet. After washing with Dulbecco’s phosphate-buffered saline (DPBS) (D1040, Solarbio, Beijing, China) containing penicillin-streptomycin (P1400, Solarbio, Beijing, China), the liver tissues were cut into 1 mm3 pieces. High concentrations of hepatocytes were obtained via trypsin (T1320, Solarbio, Beijing, China) digesting and centrifugation. Isolated hepatocytes were cultured using RPMI Medium 1640 in the incubator under 32°C. Six hours after culturing, 3MA (M9281, Sigma-Aldrich, Saint Louis, MO, United States) and diethylumbelliferyl phosphate (DEUP) (D7692, Sigma-Aldrich, Saint Louis, MO, United States) was added to cultured cells. After treatment for 24 h, cells were collected for corresponding experiments western blot and transmission electron microscope (TEM) experiments.
Statistical Analysis
All data were presented as mean ± standard error of the mean (SEM). The statistical data was obtained by importing data into SPSS for windows version 22.0 statistical package (SPAS Inc., Chicago, IL, United States). The normality and the equality of variances of data were assessed by Shapiro–Wilk test and Levene’s test, respectively. All data conformed to normal distribution. The significant difference between two groups was analyzed by either unpaired Student’s t-test or Mann–Whitney U test. The data were considered statistically significant when P < 0.05.
Results
Variation of LD (Main Components) in the Liver of Chinese Soft-Shelled Turtle
As the primary ingredient of LD, triglyceride and cholesterol levels decreased during hibernation (Figure 1A and Supplementary Figure 1). Under light microscope, BODIPY 493/503 and Oil Red O staining results unmasked decreased LD number during hibernation in the liver of P. sinensis (Figures 1B,C). By TEM, the decline in numbers of LDs per hepatocyte further confirmed LD variation during hibernation (Figure 1D). Additionally, glycogen content decreased during hibernation via Periodic Acid-Schiff staining (Supplementary Figure 2). The results mentioned above indicated that LD and its main components were decreased during hibernation in the liver of the Chinese soft-shelled turtle.
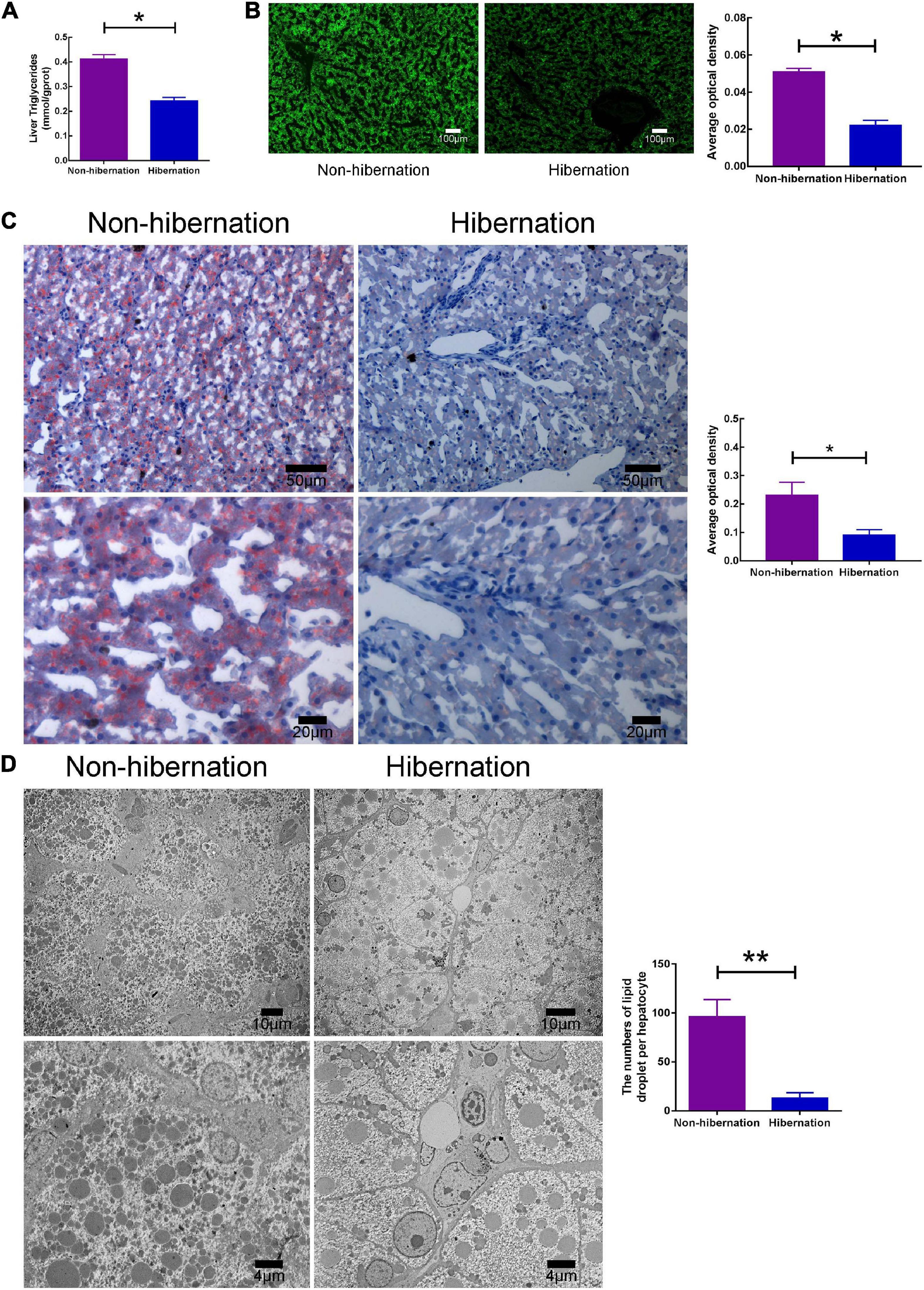
Figure 1. The content of lipid droplet (LD) was decreased during hibernation, compared to non-hibernation. (A) Triglyceride (TG) levels in the liver of hibernation and non-hibernation group. (B) Liver tissues stained with BODIPY 493/503. (C) Oil Red O staining of liver. (D) Transmission electron microscopy (TEM) observation of hepatocytes. *P < 0.05; **P < 0.01.
RNA-Seq and Identification of Differentially Expressed Genes
Two cDNA libraries in the present study were built from the turtle liver of non-hibernation (CK) and hibernation (T) using Illumina Hiseq 2500. The results of sequencing quality assessment were found to be highly reliable. After removing low-quality reads and adapter sequences, 66661832 and 46619602 clean reads were acquired from the above two libraries, among which 84.47 and 82.51% reads were mapped to reference genome of Chinese soft-shelled turtle (Supplementary Table 2). The reference genome of P. sinensis includes 24,494 known genes. In the libraries, 17,061 and 16,096 known genes were detected in non-hibernation (CK) and hibernation (T), wherein 69.65 and 65.71% matched to the reference genome. Simultaneously, 6116 and 6053 novel genes were separately sequenced in non-hibernation (CK) and hibernation (T) group.
To identify the expression of sequenced genes, the read counts in the CK and T libraries were analyzed using the FPKM method. The thresholds of |log2{FPKM(T)/FPKM(CK)| > 0 and FDR < 0.05 were adopted to differentiate DEGs. Based on it, total 8730 DEGs were successfully identified which include 3991 remarkably up-regulated and 4739 remarkably down-regulated genes in the hibernation (T) library compared to the non-hibernation (CK) library (Supplementary Figure 3).
GO and KEGG Enrichment Analysis
The threshold of P-value < 0.05 was applied in performing functional annotation of DEGs. GO includes three ontologies: molecular function (F), cellular component (C), and biological process (P) (Supplementary Figure 4). In all 110 notably enriched GO terms were acquired for up-regulated and down-regulated genes. Top 20 enriched GO terms were exhibited in Supplementary Figure 5. In the biological process, most up-regulated and down-regulated genes were involved in the cellular process (GO: 0009987), single-organism process (GO: 0044699), metabolic process (GO: 0008152), biological regulation (GO: 0065007), and regulation of biological process (GO: 0050789). In molecular function, most DEGs were enriched in binding (GO: 0005488) and catalytic activity (GO: 0003824). In cellular component, cell (GO: 0005623), cell part (GO: 0044464), and organelle (GO: 0043226) included the most DEGs (Supplementary Figure 5).
The threshold of Q-value < 0.05 was wielded in conducting pathway enrichment analysis of DEGs. Top 20 enriched KEGG pathways were exhibited in Supplementary Figure 6. Based on this, nine significantly enriched KEGG pathways were acquired regarding LD degradation. Among the DEGs, “Glycolysis/Gluconeogenesis” (ko00010) related to carbohydrate catabolism was enriched. The DEGs were also enriched in “Fatty acid biosynthesis” (ko00061), “Glycerolipid metabolism” (ko00561), “Fatty acid metabolism” (ko01212), “Fatty acid degradation” (ko00071), “Cholesterol metabolism” (ko04979), and “Oxidative phosphorylation” (ko00190) which involved in lipid and FA metabolism. As the two main decomposition ways of LD degradation, “Regulation of lipolysis in adipocyte” (ko04923) and “Autophagy-animal” (ko04140) were enriched.
Identification of Crucial DEGs Engaged in Hepatic LD Degradation During Hibernation
During hibernation, carbohydrates will firstly be consumed and then lipids (Noreika et al., 2016). Regarding carbohydrate metabolism, five up-regulated genes (e.g., G6PC, GYS2, etc.) and 13 (e.g., HK1, PFKFB3, etc.) down-regulated genes were identified in “Glycolysis/Gluconeogenesis” (ko00010) (Supplementary Table 3 and Figure 2A). A total of 31 up-regulated genes (e.g., GK, ALDH2, etc.) and 14 (e.g., GPAT4, DGAT2, etc.) down-regulated genes involved in lipid and FA metabolism were distinguished among the DEGs (Supplementary Table 4 and Figure 2B). Nine up-regulated genes (e.g., PNPLA2, PPARA, etc.) and three (e.g., ME1, DGAT2, and G0S2) down-regulated genes involved in lipolysis were distinguished among the DEGs (Supplementary Table 5 and Figure 2C). Eight up-regulated genes (e.g., MAP1LC3B, ATG16L1, etc.) and 10 (e.g., PIK3R1, NBR1, etc.) down-regulated genes involved in autophagy were distinguished among the DEGs (Supplementary Table 6 and Figure 2D).
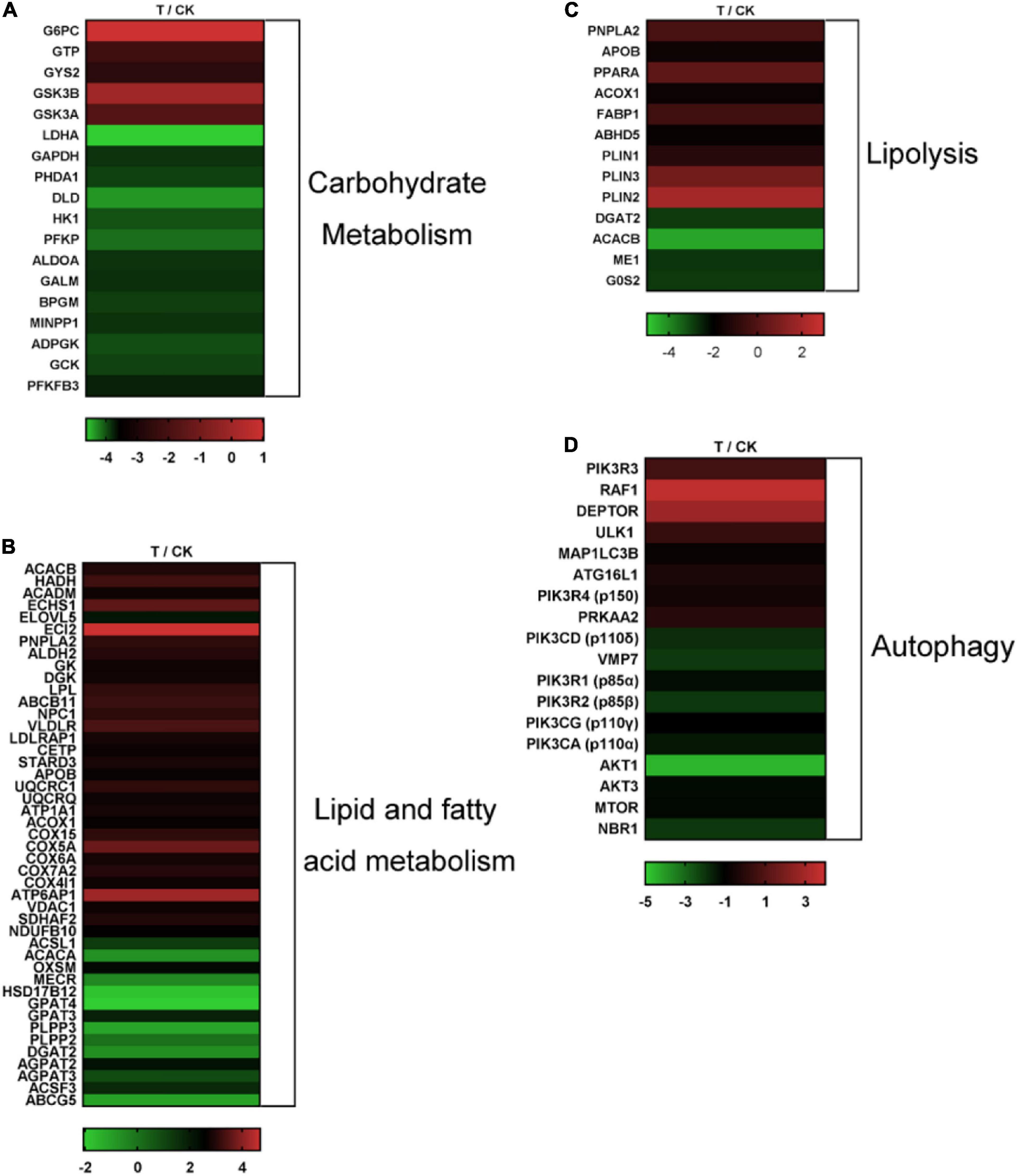
Figure 2. Heat map diagrams of expression patterns of critical DEGs among the two libraries. (A) Heat map diagrams of DEGs involved in carbohydrate metabolism. (B) Heat map diagrams of DEGs involved in lipid and FA metabolism. (C) Heat map diagrams of DEGs involved in lipolysis. (D) Heat map diagrams of DEGs involved in autophagy.
Verification of Sequencing Results via RT-qPCR and Western Blot
To substantiate these identified DEGs during hibernation in the liver of Chinese soft-shelled turtle, 19 genes were randomly chosen to conduct RT-qPCR analysis. These chosen genes comprised four carbohydrate metabolism genes (i.e., G6PC, GYS2, HK1, and PFKFB3), five lipid and FA metabolism genes (i.e., ACADM, ACSL1, ACACA, GPAT4, and AGPAT3), five lipolysis genes (i.e., PNPLA2, PPARA, DGAT2, ME1, and G0S2), and five autophagy genes (i.e., PIK3R3, MAP1LC3B, ATG16L1, PIK3R1, and NBR1). Additionally, the relative mRNA levels of these selected genes detected by RT-qPCR were compared with the read counts computed by the FPKM method. Results of comparison manifested that the dynamic changes of gene expression were closely fitted to the variations of the transcript abundance acquired via RNA-Seq (Supplementary Figure 6), implying the pinpoint accuracy and quality of transcriptome sequencing.
To further confirm whether the protein expression patterns coincide with the RNA-Seq results, we detected the chosen 19 proteins by western blot. These chosen proteins included four carbohydrate metabolism proteins (i.e., G6PC3, GYS2, HK1, and PFKFB3), five lipid and FA metabolism proteins (i.e., ACADM, ACSL1, ACACA, GPAM, and AGPAT3), five lipolysis proteins (i.e., ATGL, PPARα, DGAT2, ME1, and G0S2) and five autophagy proteins (i.e., PIK3R3, MAP1LC3B, ATG16L1, PIK3R1, and NBR1). Western blot results indicated that the dynamic variation of protein expression was tightly matched with the changes of transcript abundance attained through RNA-Seq (Supplementary Figures 7, 8). This further proved the pinpoint accuracy and quality of transcriptome sequencing.
Lipophagy Was Activated During Hibernation in the Liver of the Chinese Soft-Shelled Turtle
In our previous study (Huang et al., 2019), we have shown that lipolysis promoted LD breakdown during hibernation in the liver of Chinese soft-shelled turtles. In the present study, RNA-Seq results manifested that lipolysis and autophagy (two main degradation pathways for LD breakdown) related genes were significantly increased during hibernation. In consequence, we detected the change of autophagy level in the liver among non-hibernation and hibernation turtles. Western blot results showed that the ratio of LC3-II/LC3-I was increased and p62 protein expression was increased in the turtle’s liver during hibernation (Figures 3A,C). And this means that autophagy was activated in the liver of Chinese soft-shelled turtles during hibernation. To further verify how autophagy promotes hepatic LD breakdown, we isolated LD from the liver as previously described (Ding et al., 2013). The increase in the ratio of LC3-II/LC3-I and LAMP1 protein expression signified increased autophagy level in LDs (Figures 3B,D). Furthermore, under TEM, we observed that LD was enwrapping or entirely encased by isolation membranes (IMs) or autophagosomes (APs) in hepatocytes (Figure 4). The above results proved that the activation of lipophagy in hepatic LDs promoted LD breakdown during hibernation in the liver of Chinese soft-shelled turtles.
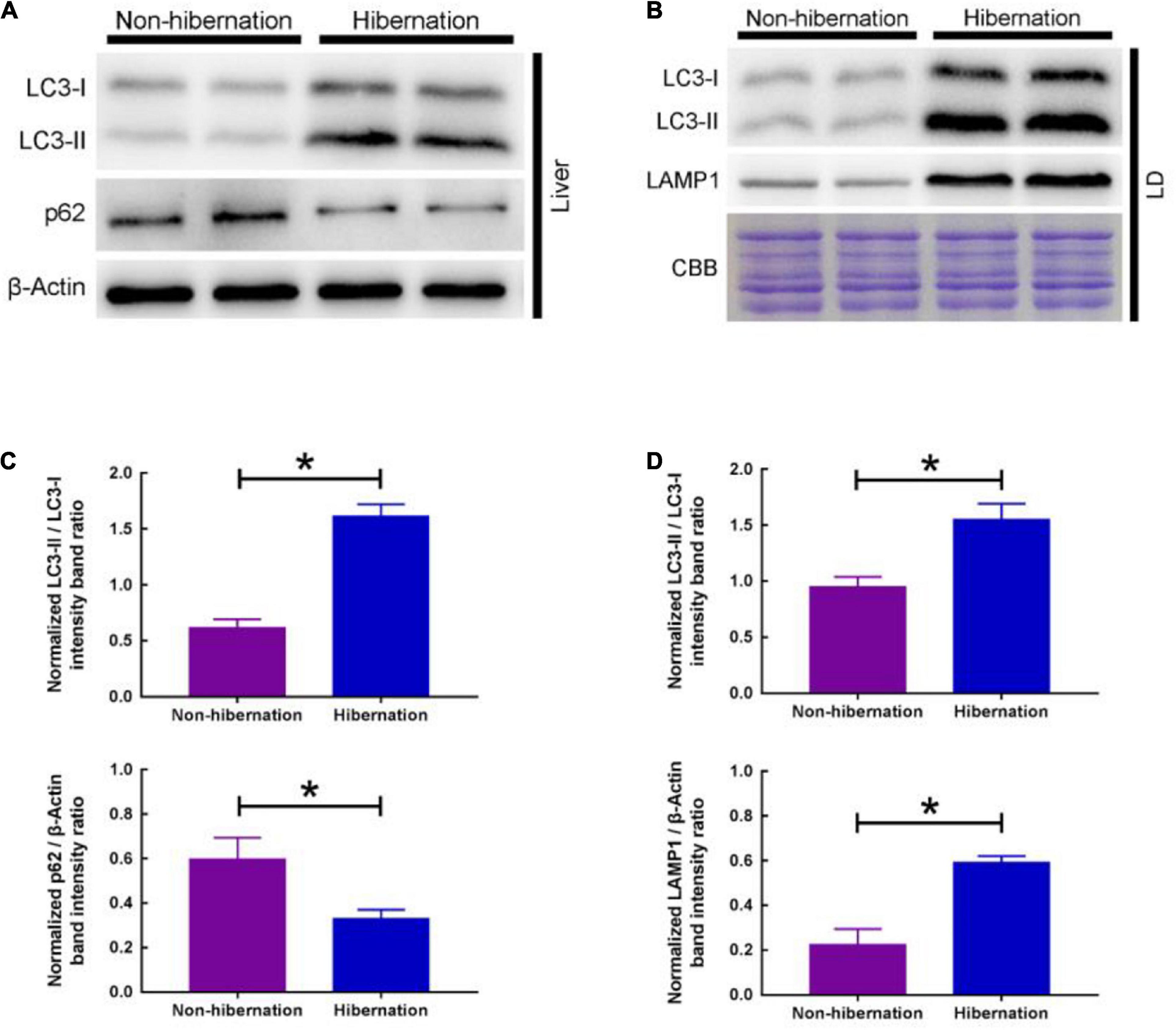
Figure 3. Change of autophagy level in the liver and LD of P. sinensis between non-hibernation and hibernation. (A) Expression of autophagy protein (LC3 and p62) in liver tissue by western blotting; (B) expression of autophagy protein (LC3 and LAMP1) in LD by western blotting; (C) statistics of the western blotting results in liver tissue; (D) statistics of the western blotting results in LD. *P < 0.05.
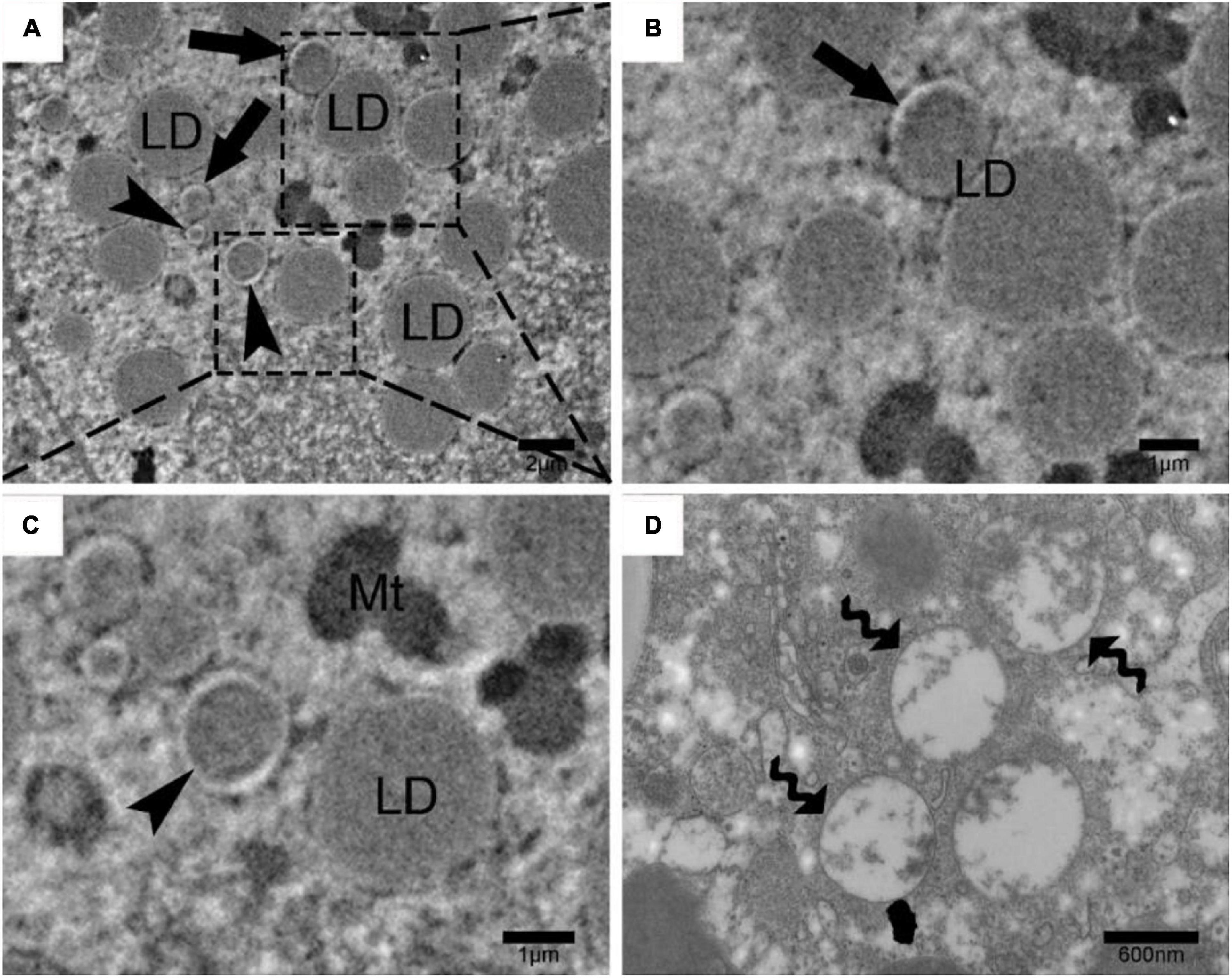
Figure 4. Lipophagy and autophagosome in Chinese soft-shelled turtle (Pelodiscus sinensis) hepatocytes during hibernation by transmission electron microscopy. (A) Ultrastructure of P. sinensis hepatocytes during hibernation; (B) lipid droplet that enwrapping by isolation membranes; (C) lipid droplet that entirely enwrapped by autophagosome; (D) autophagosome in P. sinensis hepatocytes during hibernation. Scale bar = 2 μm (A), 1 μm (B,C), 600 nm (D).
Transcriptome sequencing in the present study manifested that PI3K-AKT signaling pathway was significantly enriched (Supplementary Figure 9). Then, we verified the mRNA level of PI3K-AKT signaling pathway-related genes. Among them, the gene expression level of AKT, ULK1, Beclin-1, PIK3C3, and ATG8 was increased, whereas the gene expression level of PIK3R1 and p62 was decreased during hibernation (Figure 5A). Furthermore, western blot analysis manifested that the protein expression of PIK3R1 was decreased, while the phosphorylation of AKT was increased (Figure 5B). In the meantime, the phosphorylation of mTOR (the pivotal autophagy protein) was decreased and the phosphorylation of ULK1 was elevated. Other critical autophagy proteins expression, such as Beclin-1 and PIK3C3, were significantly increased during hibernation (Figure 5C). The above RT-qPCR and western blot analysis indicated that PI3K-AKT signaling pathway was activated during hibernation in the liver of Chinese soft-shelled turtles. These provide further evidence that hepatic LD breakdown during hibernation was likely to be promoted via PI3K-AKT-mTOR signaling pathway.
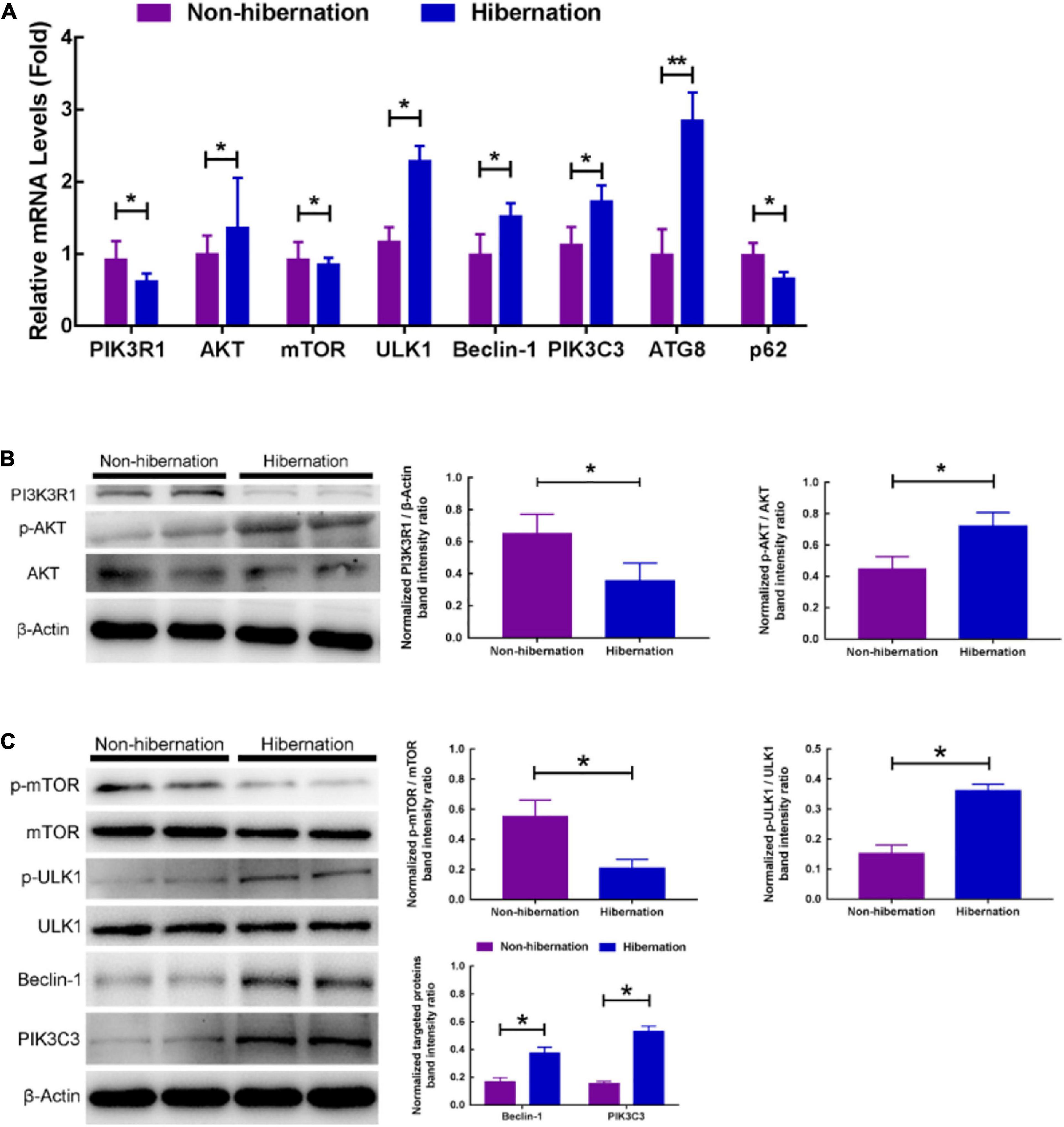
Figure 5. Hepatic LDs breakdown in the liver of hibernating turtles was likely to be promoted via PI3K-AKT-mTOR signaling pathway. (A) The relative mRNA levels of genes related to PI3K-AKT-mTOR pathway; (B) protein expression of PI3K3R1 and phosphorylation of AKT in the liver of different groups; (C) protein expression of phosphorylation of mTOR, phosphorylation of ULK1, Beclin-1, and PIK3C3 in the liver of different groups. *P < 0.05; **P < 0.01.
Reverse Validation in vitro (Inhibition of Lipolysis and Autophagy)
Our previous study (Huang et al., 2019) and the above results indicated that both lipolysis and autophagy could promote the degradation of LDs in the liver of P. sinensis during hibernation. To further determine the relationship between lipolysis and autophagy, we applied the inhibitors of lipolysis and autophagy (DEUP and 3MA) in cultured primary hepatocytes from the liver of Chinese soft-shelled turtle. After suppressing autophagy with 3MA (the inhibitor of PI3K), the ratio of LC3II/LC3I was significantly decreased, whereas the expression of p62 was increased (Supplementary Figure 10), implying that autophagic activity was decreased. Compared with the control group, the content of triglyceride (TG) in hepatocytes was significantly increased after 3MA treatment (Figure 6A), while the content of free fatty acids (FFA) was significantly decreased (Figure 6B), indicating that inhibition of autophagy would affect LD degradation. Additionally, after DEUP (the lipolysis inhibitor) inhibition, the TG content in cultured hepatocytes increased significantly (Figure 6A), while the FFA content decreased (Figure 6B), indicating that inhibition of lipolysis could also affect the degradation of LDs. At this point, the autophagy level in cultured hepatocytes was slightly increased (the ratio of LC3II/LC3I did not change, but the expression of p62 decreased) (Supplementary Figure 11), indicating that inhibition of lipolysis would arouse compensatory increase of autophagy. Furthermore, after treating with 3MA and DEUP simultaneously, TG content in hepatocytes increased significantly (Figure 6A), while the FFA content decreased (Figure 6B). However, the changes of TG and FFA content in the co-treatment group have little difference with DEUP treatment but have a significant difference with 3MA treatment (Figure 6). Similar changes in the number of LDs in cultured hepatocytes were observed under TEM (Figure 7). The above results indicated that lipolysis is the main way for LD degradation in the hepatocyte of Chinese soft-shelled turtles (P. sinensis).
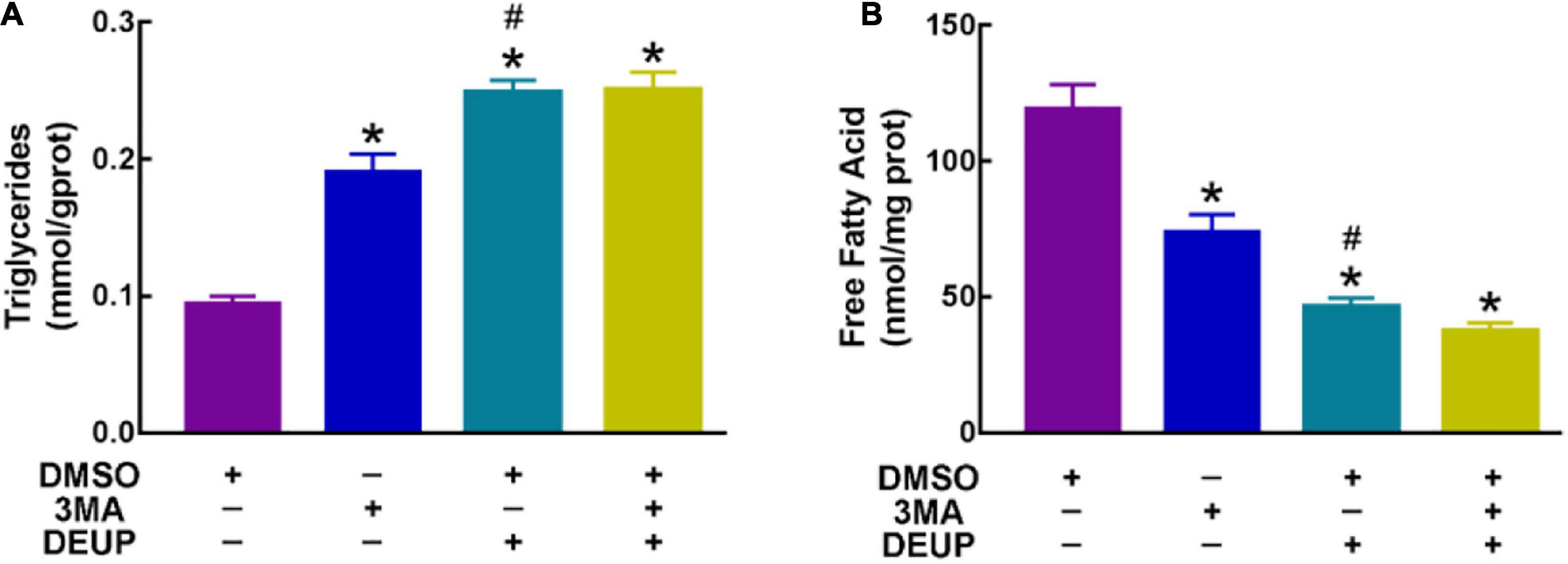
Figure 6. Content variation of triglyceride (TG) and free fatty acids (FFA) in primary hepatocyte from Chinese soft-shelled turtle (Pelodiscus sinensis) under different treatments. (A) Content variation of TG in primary hepatocyte from Chinese soft-shelled turtle (Pelodiscus sinensis); (B) content variation of FFA in primary hepatocyte from Chinese soft-shelled turtle (Pelodiscus sinensis). *P < 0.05; #P > 0.05.
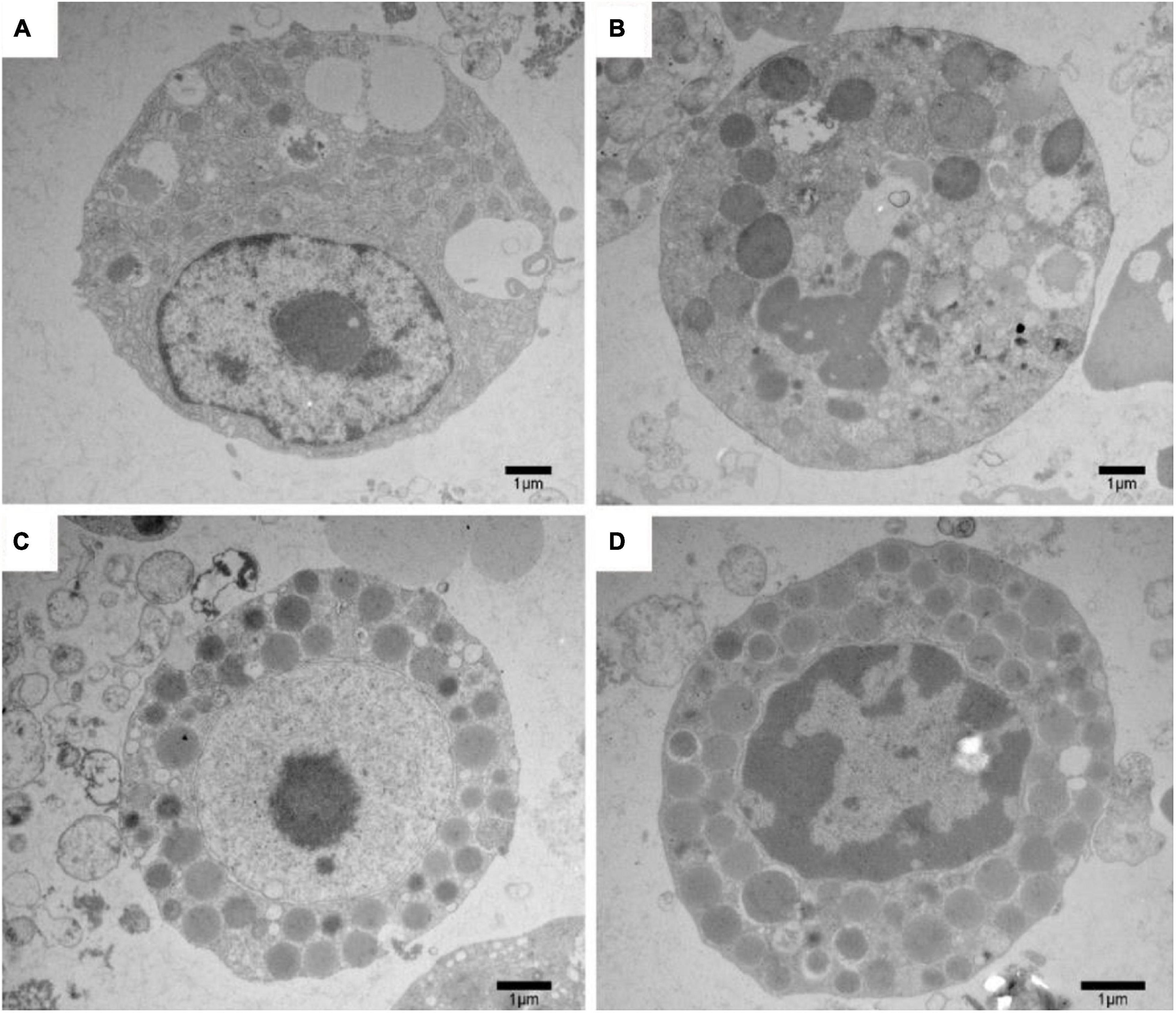
Figure 7. Ultrastructural change of P. sinensis hepatocytes after different treatments under transmission electron microscopy. (A) Ultrastructural change of P. sinensis hepatocytes in control group; (B) ultrastructural change of P. sinensis hepatocytes after 3MA treatments; (C) ultrastructural change of P. sinensis hepatocytes after DEUP treatments; (D) ultrastructure of P. sinensis hepatocytes after 3MA and DEUP treatments. Scale bar = 1 μm.
Discussion
Sea turtles are sensitive to the changes of ambient temperature based on their biological characteristics (Schofield et al., 2009). Under global warming, the rise in sea level took away their habitats and then affected their reproduction (Fish et al., 2005). The temperature of the sand determines the gender of the hatchling sea turtles in a nest because reptiles are temperature-dependent sex determination. Generally, the turtles’ eggs in a cooler nest tend to be males, while the eggs in the warmer nest are likely to become females (Ge et al., 2018). Accompanying the increasing temperatures of the nest, researchers predicted that more female turtles will be hatched than males, and then posed a great threat to the genetic diversity of marine life (Velo-Antón et al., 2011). Besides, the variation of ocean temperature could make a huge impact on food resources for sea turtles and even nearly all marine life. The important food source for sea turtles, Coral reefs, were also in great danger (Jensen et al., 2018). The extreme environmental conditions seriously destroyed the coral reef ecosystems all over the world. And parts of coral reefs were disappearing by bleaching due to rising temperature (Baker, 2001).
To determine the effective means for improving survival rates of sea turtles, we reviewed previous studies in the artificial culture of Chinese soft-shelled turtle (P. sinensis). As an important kind of special aquaculture animals, P. sinensis have been bred in China for decades. To improve the survival rates of Chinese soft-shelled turtles, researchers tried to change the diet formula. They found that Chinese soft-shelled turtles have higher utilization of lipid than other fish and grow better when the fat level in the feed is between 4 and 7% (Huang et al., 2005). Moreover, the supplementation of protein in fodder also significantly improved the growth performance and weight due to its higher demand than other animals (Jia et al., 2005). Researchers also realized supplementation of carbohydrates in fodder could effectively improve the utilization of animal feed and fat consumption in the turtles (Huanling et al., 2005). Different from protein and lipid, vitamins play an important role in the development of vision and the maintenance of the respiratory, nervous, and reproductive systems in animals (Clagett-Dame and Knutson, 2011). The supplementation of vitamins in fodder improved the resistibility and then raised the survival rates (Su and Huang, 2019). In addition to these, the development of bones and carapace of P. sinensis rely on mineral substance (Wang and Huang, 2015), and the supplementation of microelement in fodder dramatically improved the resistibility and then raised the survival rates (Chu et al., 2007). On this account, the lessons learned from the aquaculture of P. sinensis could enlighten the artificial culture of sea turtles.
Despite multiple possible trials (Chen et al., 2018) which have been verified in increasing the survival of turtles, researchers found that the physiological phenomenon in hibernating animals still needs further to explore. In previous studies, researchers found that hibernation might improve the survival rates by slowing down the shortening of telomeres (Turbill et al., 2013). Additionally, researches showed that Syrian hamsters could develop liver fibrosis during hibernation, which is rapidly resolved out of hibernation (Hendriks et al., 2020). This phenomenon indicates that mechanisms underlying this might provide new sight into preventing or reversing emerging diseases in the turtles. Furthermore, the maintenance of physiological function could also get inspiration from the ways grizzly bears deal with hibernation and their fluctuating weight (Rivet et al., 2017). These implied that hibernating animal is a good experimental animal model for improving the present situation of turtles.
In animals, carbohydrates and lipid both provide energy needs for maintenance, growth, and production. Understanding the dynamic switching of energy generation mode will undoubtedly improve the survival rates of sea turtles during hibernation. In the liver of Chinese soft-shelled turtle (P. sinensis), genes involved in carbohydrate catabolism were down-regulated, whereas genes relevant to lipid and FA catabolism were up-regulated during hibernation, and this transition could inspire the artificial culture of sea turtles. In the laboratory simulation environment, despite Kemp’s ridley (Lepidochelys kempi) sea turtles (Moon et al., 1997) were able to acclimate to cold temperatures behaviorally, the food consumption was significantly decreased. Based on the role of hepatic LDs breakdown in maintaining the Chinese soft-shelled turtle’s homeostasis during hibernation, the artificial culture of sea turtles could be promoted by a high-fat diet. In previous studies, the effects of fasting and increased feeding on energy metabolism were investigated in immature greens (Chelonia mydas) and Kemp’s ridley (L. kempi) sea turtles and found that lipids were important for sea turtles during fasting (Moon et al., 1999). However, they only described the connection between the metabolic adjustment of sea turtles and fasting periods and did not investigate the impact of hibernation. In consequence, the role of hepatic LDs breakdown in maintaining the body’s homeostasis of Chinese soft-shelled turtle (P. sinensis) during hibernation could provide researchers with new inspirations for improving the survival rates of raised sea turtles during hibernation.
Conclusion
In conclusion, the shifts of hepatic energy utilization and its adaptive mechanisms in Chinese soft-shelled turtle successfully ensured its wintering. Deviant hepatic lipid breakdown may contribute to the increased incidence of liver diseases in sea turtles. The periodic variation of hepatocellular ultrastructure and molecular mechanisms underlying this phenomenon in P. sinensis might provide us new inspirations in decreasing the resistance of sea turtles to emerging diseases. Furthermore, the Chinese soft-shelled turtle (P. sinensis) could be used as a better experimental animal model for the protection of marine biodiversity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR11586153 and SRR11586154.
Ethics Statement
The animal study was reviewed and approved by the Science and Technology Agency of Jiangsu Province, and Nanjing Agricultural University Veterinary College approved the sampling procedures. The approval ID is SYXK (SU) 2010-0005.
Author Contributions
QC and YH conceived and designed the experiments. YH, SY, XB, and YS performed the experiments. YH and SY analyzed the data. YH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Natural Science Foundation of China (Grant numbers: 31672505 and 31872433), Priority Academic Program Development of Jiangsu Higher Education Institutions, China, and Practice Innovation Program of Jiangsu Province (KYCX18-0713).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank YS for his excellent technical assistance in TEM experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.633425/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/sra/
- ^ https://github.com/OpenGene/fastp
- ^ http://www.genome.jp/kegg/
References
Baker, A. C. (2001). Reef corals bleach to survive change. Nature 411, 765–766. doi: 10.1038/35081151
Beaugrand, G., Edwards, M., and Legendre, L. (2010). Marine biodiversity, ecosystem functioning, and carbon cycles. Proc. Natl. Acad. Sci. USA 107, 10120–10124. doi: 10.1073/pnas.0913855107
Bugoni, L., Krause, L., and Petry, M. V. (2001). Marine debris and human impacts on sea turtles in southern Brazil. Mar. Pollut. Bull. 42, 1330–1334. doi: 10.1016/s0025-326x(01)00147-3
Chen, Y., Zhang, Y.-F., Qian, H.-C., Wang, J.-L., Chen, Z., Ordovas, J. M., et al. (2018). Supplementation with turmeric residue increased survival of the Chinese soft-shelled turtle (Pelodiscus sinensis) under high ambient temperatures. J. Zhejiang Univ. Sci. B 19, 245–252. doi: 10.1631/jzus.b1600451
Chu, J.-H., Chen, S.-M., and Huang, C.-H. (2007). Effect of dietary iron concentrations on growth, hematological parameters, and lipid peroxidation of soft-shelled turtles, Pelodiscus sinensis. Aquaculture 269, 532–537. doi: 10.1016/j.aquaculture.2007.03.004
Clagett-Dame, M., and Knutson, D. (2011). Vitamin A in reproduction and development. Nutrients 3, 385–428. doi: 10.3390/nu3040385
Dang, W., Lu, H., Gao, Y., Xu, N., Qu, T., and Liu, Y. (2015). Molecular analysis of inducible Heat shock protein 70 of Pelodiscus sinensis and its effects during pathogen (Aeromonas hydrophila) infection. Aquaculture 442, 93–99. doi: 10.1016/j.aquaculture.2015.02.030
Ding, Y., Zhang, S., Yang, L., Na, H., Zhang, P., Zhang, H., et al. (2013). Isolating lipid droplets from multiple species. Nat. Protoc. 8, 43–51. doi: 10.1038/nprot.2012.142
Eckrich, C. E., and Owens, D. W. (1995). Solitary versus arribada nesting in the olive ridley sea turtles (Lepidochelys olivacea): a test of the predator-satiation hypothesis. Herpetologica 51, 349–354.
Fedorov, V. B., Goropashnaya, A. V., Tøien, Ø, Stewart, N. C., Chang, C., Wang, H., et al. (2011). Modulation of gene expression in heart and liver of hibernating black bears (Ursus americanus). BMC Genomics 12:171.
Fish, M. R., Côté, I. M., Gill, J. A., Jones, A. P., Renshoff, S., and Watkinson, A. R. (2005). Predicting the impact of sea-level rise on Caribbean sea turtle nesting habitat. PLoS One 19, 482–491. doi: 10.1111/j.1523-1739.2005.00146.x
Foran, D. R., and Ray, R. L. (2016). Mitochondrial DNA profiling of illegal tortoiseshell products derived from hawksbill sea turtles. J. Froensic Sci. 61, 1062–1066. doi: 10.1111/1556-4029.13062
Ge, C., Ye, J., Weber, C., Sun, W., Zhang, H., Zhou, Y., et al. (2018). The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360, 645–648. doi: 10.1126/science.aap8328
Gebhardt, R. (1992). Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol. Ther. 53, 275–354. doi: 10.1016/0163-7258(92)90055-5
Hays, G. C., Broderick, A. C., Glen, F., and Godley, B. J. (2003). Climate change and sea turtles: a 150-year reconstruction of incubation temperatures at a major marine turtle rookery. Global Change Biol. 9, 642–646. doi: 10.1046/j.1365-2486.2003.00606.x
Hendriks, K. D., Joschko, C. P., Hoogstra-Berends, F., Heegsma, J., Faber, K.-N., and Henning, R. H. (2020). Hibernator-derived cells show superior protection and survival in hypothermia compared to non-hibernator cells. Int. J. Mol. Sci. 21:1864. doi: 10.3390/ijms21051864
Huang, C.-H., Lin, W.-Y., and Chu, J.-H. (2005). Dietary lipid level influences fatty acid profiles, tissue composition, and lipid peroxidation of soft-shelled turtle, Pelodiscus sinensis. Comp. Biochem. Phsyiol. A Mol. Integr. Physiol. 142, 383–388. doi: 10.1016/j.cbpa.2005.09.004
Huang, Y., Chen, H., Yang, P., Bai, X., Shi, Y., Vistro, W. A., et al. (2019). Hepatic lipid droplet breakdown through lipolysis during hibernation in Chinese soft-shelled turtle (Pelodiscus sinensis). Aging 11, 1990–2002. doi: 10.18632/aging.101887
Huanling, Y., Yong, L., Junbo, W., Liping, Z., and Weixing, Y. (2005). Chinese soft-shelled turtle egg powder lowers serum cholesterol, increases faecal neutral steroids and bile acid excretion, and up-regulates liver cytochrome P450 mRNA level in rats. Br. J. Nutr. 94, 315–320. doi: 10.1079/bjn20051485
Jackson, D. (2002). Hibernating without oxygen: physiological adaptations of the painted turtle. J. Physiol. 543, 731–737. doi: 10.1113/jphysiol.2002.024729
Jensen, M. P., Allen, C. D., Eguchi, T., Bell, I. P., LaCasella, E. L., Hilton, W. A., et al. (2018). Environmental warming and feminization of one of the largest sea turtle populations in the world. Curr. Biol. 28, 154–159 e154.
Jia, Y., Yang, Z., Hao, Y., and Gao, Y. (2005). Effects of animal–plant protein ratio in extruded and expanded diets on nitrogen and energy budgets of juvenile Chinese soft-shelled turtle (Pelodiscus sinensis Wiegmann). Aquac. Res. 36, 61–68. doi: 10.1111/j.1365-2109.2004.01184.x
Lorne, J. K., and Salmon, M. (2007). Effects of exposure to artificial lighting on orientation of hatchling sea turtles on the beach and in the ocean. Endanger. Species Res. 3, 23–30. doi: 10.3354/esr003023
Lovich, J. E., Ennen, J. R., Agha, M., and Gibbons, J. W. (2018). Where have all the turtles gone, and why does it matter? BioScience 68, 771–781. doi: 10.1093/biosci/biy095
Mancini, A., and Koch, V. (2009). Sea turtle consumption and black market trade in Baja California Sur, Mexico. Endanger. Species Res. 7, 1–10. doi: 10.3354/esr00165
Melzner, F., Gutowska, M., Langenbuch, M., Dupont, S., Lucassen, M., Thorndyke, M., et al. (2009). Physiological basis for high CO2 tolerance in marine ectothermic animals: pre-adaptation through lifestyle and ontogeny? Biogeosciences 6, 2313–2331. doi: 10.5194/bg-6-2313-2009
Moon, D.-Y., Owens, D. W., and MacKenzie, D. S. (1999). The effects of fasting and increased feeding on plasma thyroid hormones, glucose, and total protein in sea turtles. J. Zool. Sci. 16, 579–586. doi: 10.2108/zsj.16.579
Moon, D. Y., MacKenzie, D. S., and Owens, D. W. (1997). Simulated hibernation of sea turtles in the laboratory: i. feeding, breathing frequency, blood pH, and blood gases. J. Exp. Zool. 278, 372–380. doi: 10.1002/(sici)1097-010x(19970815)278:6<372::aid-jez4>3.0.co;2-l
Nelson, C. J., Otis, J. P., Martin, S. L., and Carey, H. V. (2009). Analysis of the hibernation cycle using LC-MS-based metabolomics in ground squirrel liver. Physiol. Genomics 37, 43–51. doi: 10.1152/physiolgenomics.90323.2008
Noreika, N., Madsen, N. E., Jensen, K., and Toft, S. (2016). Balancing of lipid, protein, and carbohydrate intake in a predatory beetle following hibernation, and consequences for lipid restoration. J. Insect Physiol. 88, 1–9. doi: 10.1016/j.jinsphys.2016.02.004
Rivet, D., Nelson, O. L., Vella, C., Jansen, H., and Robbins, C. (2017). Systemic effects of a high saturated fat diet in grizzly bears (Ursus arctos horribilis). Can. J. Zool. 95, 797–807. doi: 10.1139/cjz-2016-0271
Rui, L. (2011). Energy metabolism in the liver. Compr. Physiol. 4, 177–197. doi: 10.1002/cphy.c130024
Schofield, G., Bishop, C. M., Katselidis, K. A., Dimopoulos, P., Pantis, J. D., and Hays, G. (2009). Microhabitat selection by sea turtles in a dynamic thermal marine environment. J. Anim. Ecol. 78, 14–21. doi: 10.1111/j.1365-2656.2008.01454.x
Su, Y. T., and Huang, C. H. (2019). Estimation of dietary vitamin K requirement of juvenile Chinese soft-shelled turtle, Pelodiscus sinensis. Aquac. Nutr. 25, 1327–1333. doi: 10.1111/anu.12953
Turbill, C., Ruf, T., Smith, S., and Bieber, C. (2013). Seasonal variation in telomere length of a hibernating rodent. Biol. Lett. 9:20121095. doi: 10.1098/rsbl.2012.1095
Velo-Antón, G., Becker, C. G., and Cordero-Rivera, A. (2011). Turtle carapace anomalies: the roles of genetic diversity and environment. PLoS One 6:e18714. doi: 10.1371/journal.pone.0018714
Wang, C.-C., and Huang, C. (2015). Effects of dietary vitamin C on growth, lipid oxidation, and carapace strength of soft-shelled turtle, Pelodiscus sinensis. Aquaculture 445, 1–4. doi: 10.1016/j.aquaculture.2015.04.009
Keywords: the Chinese soft-shelled turtle (Pelodiscus sinensis), lipid droplet, marine life, hibernation, liver
Citation: Huang Y, Yang S, Bai X, Shi Y and Chen Q (2021) Molecular and Cellular Mechanisms of Lipid Droplet Breakdown in the Liver of Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Front. Mar. Sci. 8:633425. doi: 10.3389/fmars.2021.633425
Received: 25 November 2020; Accepted: 22 March 2021;
Published: 09 April 2021.
Edited by:
Samad Rahimnejad, University of South Bohemia in České Budějovice, CzechiaReviewed by:
Khor Waiho, University of Malaysia Terengganu, MalaysiaXiaowu Chen, Shanghai Ocean University, China
Youji Wang, Shanghai Ocean University, China
Copyright © 2021 Huang, Yang, Bai, Shi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiusheng Chen, Y2hlbnFzaDMwNUBuamF1LmVkdS5jbg==
 Yufei Huang
Yufei Huang Sheng Yang1
Sheng Yang1 Qiusheng Chen
Qiusheng Chen