
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 March 2021
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.633314
This article is part of the Research Topic Microbial Communities of Coastal Eutrophic Systems View all 10 articles
 Paraskevi Mara1
Paraskevi Mara1 Virginia P. Edgcomb1
Virginia P. Edgcomb1 Taylor R. Sehein2
Taylor R. Sehein2 David Beaudoin1
David Beaudoin1 Chuck Martinsen3
Chuck Martinsen3 Christina Lovely3
Christina Lovely3 Bridget Belcher4
Bridget Belcher4 Rebecca Cox1
Rebecca Cox1 Meghan Curran4
Meghan Curran4 Claire Farnan4
Claire Farnan4 Peter Giannini4
Peter Giannini4 Sarah Lott1
Sarah Lott1 Kyle Paquette4
Kyle Paquette4 Anna Pinckney4
Anna Pinckney4 Natalie Schafer4
Natalie Schafer4 Tonna-Marie Surgeon-Rogers5
Tonna-Marie Surgeon-Rogers5 Daniel R. Rogers4*
Daniel R. Rogers4*Coastal ecosystems are impacted by excessive nutrient inputs that cause degradation of water quality and impairments of ecosystem functioning. Regulatory and management efforts to enhance nutrient export from coastal ecosystems include sustainable oyster aquaculture that removes nitrogen in the form of oyster biomass and increases particulate export to underlying sediments where increased organic material may enhance microbial denitrification. To better understand the impacts of oyster aquaculture on nitrogen removal, we examined bacterial processes in sediments underlying three of the most common aquaculture methods that vary in the proximity of oysters to the sediments. Sediment samples underlying sites managed with these different aquaculture methods were examined using the 16S rRNA gene to assess microbial community structure, gene expression analyses to examine nitrogen and sulfur cycling genes, and nitrogen gas flux measurements. All sites were located in the same hydrodynamic setting within Waquoit Bay, MA during 2018 and 2019. Although sediments under the different oyster farming practices showed similar communities, ordination analysis revealed discrete community groups formed along the sampling season. Measured N2 fluxes and expression of key genes involved in denitrification, anaerobic ammonium oxidation (anammox), and dissimilatory nitrate reduction to ammonium (DNRA) increased during mid-summer and into fall in both years primarily under bottom cages. While all three oyster growing methods enhanced nitrogen removal relative to the control site, gene expression data indicate that the nitrogen retaining process of DNRA is particularly enhanced after end of July under bottom cages, and to a lesser extent, under suspended and floating bags. The choice of gear can also potentially increase processes that induce nitrogen retention in the form of ammonia in the underlying sediments over time, thus causing deviations from predicted nitrogen removal. If nitrogen removal is a primary objective, monitoring for these shifts is essential for making decisions about siting and size of aquaculture sites from year to year.
Coastal bays and estuaries receive excessive nutrient inputs from anthropogenic activities that have profound negative effects upon water quality (Smith, 2003). Over the past decades the use of chemical fertilizers has expanded, and population densities have increased along coastal zones. These have accelerated nutrient pollution and eutrophication has become a broad-scale concern for nearshore coastal and estuarine ecosystems (Howarth et al., 2011; Glibert et al., 2018; and discussed in Clements and Comeau, 2019). Coastal communities are now challenged to meet goals to decrease nitrogen inputs to coastal waters. Nitrogen controls primary production in coastal ecosystems and nitrogen in the form of nitrate (a common fertilizer component) can serve as a nutrient for many aquatic microorganisms that can utilize it as an energy source under both aerobic and anaerobic conditions (Howarth, 1988). Excessive inputs of nitrate can result in reduced oxygen levels (hypoxia) (Rabalais et al., 2014), enhanced algal growth, and increased duration and severity of harmful algal blooms (HABs) (Anderson et al., 2002; Glibert et al., 2005). The altered balance between nitrogen inputs and exports has potentially severe ecological and economic impacts and requires appropriate management to reduce eutrophication in coastal ecosystems (Humphries et al., 2016; Clements and Comeau, 2019).
Coastal areas in New England are impacted by nitrogen loads carried by sewage outfalls and river discharges, but the primary source of excess nitrogen is groundwater contaminated with septic fluids, fertilizers, and other contaminants that contribute to eutrophication (Correll et al., 1992; Valiela et al., 1997). While sewering can address significant fractions of these inputs, additional mitigation tools are needed to extract nutrients from coastal ecosystems. One potential tool for removing nitrogen from coastal estuaries and bays is shellfish aquaculture, and specifically oyster farming. Oysters are filter-feeders and their harvesting can remove nutrients from the water column because oysters filter blooms of primary producers and convert those nutrients into oyster biomass. As such, the harvesting of oysters can be beneficial for nitrogen removal, so-called “bio-extraction,” in New England’s estuaries and bays (Feinman et al., 2018; Clements and Comeau, 2019). The efficiency of oysters to assimilate and digest various sources of organic matter varies over the season which can lead to significant amounts of undigested particulate organic nitrogen (PON) being transported via their feces and pseudofeces to the sediment surface (Newell and Jordan, 1983). This biodeposition may enhance the microbial process of denitrification in underlying sediments (Kellogg et al., 2013). Denitrification releases nitrogen from the ecosystem in the form of nitrogen gas (N2) and this process naturally occurs in marine sediments. Oyster biodeposits can alter sediment chemistry since they contain twice as much the concentrations of carbon, nitrogen, and phosphorus compared to particles settling-out naturally from the water column (Jordan, 1987) and thus enhance denitrification rates of sediment bacterial communities (Newell et al., 2002).
Denitrification is a four-step metabolic process that converts nitrate to N2. The denitrification genes are assembled in clusters and encode functions for nitrate (narG/napA), nitrite (nirS/nirK), nitric oxide (norB/norC), and nitrous oxide (nosZ) reduction largely unique to denitrifying bacteria (Zumft, 1997). The process occurs in the membranes and the periplasm of the microbes and constitutes one of the basic energy flow pathways under anaerobic conditions. The energy for microbial denitrification in marine sediments derives from organic matter oxidation under suboxic (near-zero O2 values) or anoxic conditions by facultative heterotrophic anaerobes that use nitrate, nitrite, and nitrous oxide as terminal electron acceptors (Tiedje, 1988; Devol, 2015). Nitrate availability is important for denitrification, although denitrifiers are equipped with nitrate reduction enzymes (NapA and NarG) that share the same function but are induced at low (napA at <0.5 mM nitrate) and higher nitrate (narG at >1 mM of nitrate) concentrations based on experimental studies (Wang et al., 1999).
The importance of sediment denitrification underneath oyster aquaculture sites has been extensively debated, with studies suggesting different levels of significance of the process (Ray et al., 2020 and references therein). Rates measured at both bottom-planted and off-bottom oyster aquaculture sites in New England showed increased denitrification (Humphries et al., 2016; Vieillard, 2017; Feinman et al., 2018) that occasionally was comparable to the denitrification rates of restored oyster reefs (Humphries et al., 2016). This suggests that oyster farm biodeposits (e.g., faces, pseudofaces, particles) could enhance bacterial denitrification in marine sediments (Feinman et al., 2018), perhaps at rates equal to that of wild oyster beds.
Oyster biodeposits might also have the potential to promote alternative anaerobic metabolic processes, including dissimilatory nitrate reduction to ammonium (DNRA) or anaerobic ammonium oxidation (anammox). The process of DNRA reduces nitrate to nitrite (narG) and then nitrite to ammonium via the ammonia-forming nitrite reductase NrfA. DNRA is an active nitrate reduction process that can dominate over dentification in sediments below farmed bivalves, and it generates ammonium that can be partially regenerated into the water column (Nizzoli et al., 2006; Carlsson et al., 2012; Murphy et al., 2016). Enhanced fluxes of benthically produced ammonium into the water column has been documented below oyster aquaculture sites (Mazouni et al., 1996; Chapelle et al., 2000; Higgins et al., 2013; Lunstrum et al., 2018) may lead to retention rather than removal of reactive nitrogen (Lunstrum et al., 2018). However, to date, DNRA rates have been measured in association with three oyster aquaculture studies that show DNRA can either dominate over benthic denitrification (Erler et al., 2017; Lunstrum et al., 2018) or can have an insignificant (if any) role (Vieillard, 2017). The factors that control the competition between DNRA and denitrification and the impact oyster aquaculture can have on the balance of these processes needs further study (Burgin and Hamilton, 2007). Both processes are found across many different microbial lineages (Burgin and Hamilton, 2007; Braker et al., 2010) and there has been some suggestion that increased hydrogen sulfide concentrations in the environment may favor DNRA over denitrification (An and Gardner, 2002).
The impact of oyster biodeposits on sediment anaerobic ammonia oxidation is also not extensively characterized. Under anaerobic conditions anammox converts nitrite and ammonia to N2 that is removed from the marine sediments (Devol, 2015). Anammox usually involves the reduction of nitrite to nitric oxide (nirS/nirK) which is used for the oxidation of ammonia to hydrazine (Kartal and Keltjens, 2016). The last step of anammox involves hydrazine oxidoreductase (hzo) that oxidizes hydrazine to nitrogen gas (Hu et al., 2019). Both DNRA and anammox are strictly anaerobic processes that can be inhibited at oxygen concentrations as low as ∼1–2 μM (Mohan and Cole, 2007; Dalsgaard et al., 2014). Direct measurements of denitrification and anammox rates in sediments underneath bivalve farming have estimated that the contribution of anammox to nitrogen removal is minor (Carlsson et al., 2012; Erler et al., 2017; Lunstrum et al., 2018).
Microbial denitrification and other nitrogen cycling processes can be influenced by redox changes and sediment oxygen demand (SOD), temperature and salinity, availability of organic matter, and local hydrodynamics (Hoellein and Zarnoch, 2014). Also, the oyster stocking density and the available space for oyster aquaculturing can affect the quantity and quality of particles and thus impact nitrogen removal via “bio-extraction” and denitrification (Carmichael et al., 2012). Redox changes induced via hydrogen sulfide concentrations can alter rates of denitrification, DNRA, and anammox. The sulfur cycle via the oxidation and/or reduction of sulfides, sulfur intermediates, as well as via disproportionation influences the nitrogen cycle in marine sediments (e.g., Caffrey et al., 2019 and references therein; Jørgensen et al., 2019). Diverse aquaculture settings can potentially influence the above parameters and thus enhance or inhibit different nitrogen cycling processes in the underlying sediments. The type of gear used for aquaculture can also have an impact. Oyster aquaculture systems utilized in areas of New England include surface or floating bags, suspended oysters (oyster are suspended under a buoy), and bottom cage oysters (5–6 cm above the sediments). In this study, we examined net N2 release from sediments under different oyster aquaculture systems, whether the presence of oyster aquaculture altered sediment microbial community composition, and we investigated evidence for expression of genes involved in bacterial nitrogen metabolisms that impact N removal, including bacterial denitrification, anammox, and potentially, chemolithotrophic DNRA. We hypothesized that overall nitrogen production and individual nitrogen cycling processes might be differentially affected by different oyster culturing systems that hold oysters at different distances from the sediment, resulting in differences in net N2 release from underlying sediments. To test this hypothesis, we installed three different farming practices that included floating and suspended bags, and bottom cages within the same hydrodynamic setting in Waquoit Bay, MA, United States during the aquaculture season in two consecutive years. We collected sediment cores for 16S rRNA gene analyses, quantitative reverse transcription PCR (RT-qPCR), of denitrification, anammox, dissimilatory nitrate reduction to ammonium and sulfur oxidation genes, and metatranscriptome analyses addressing dissimilatory sulfate reduction (dsr genes). We also monitored nitrogen production fluxes under the different aquaculture sites. Waquoit Bay is typical of the setting of many oyster aquaculture sites in the region.
Our study site was established in Waquoit Bay, Falmouth Massachusetts (41°34′49″N, 70°31′27″W; Waquoit, MA, United States), within the boundaries of the Waquoit Bay National Estuarine Research Reserve in an area relatively undisturbed by human impacts. Waquoit Bay is a shallow bay encompassing approximately 825 acres. The bay has an average depth of 0.79 m (with a maximum depth of 2.7 m and tidal height of ∼0.5 m) and is tidally flushed through a narrow opening (∼115 m) to Vineyard Sound to the south of the study site (∼3.2 km) with salinity ranging from 0 (in the tributaries) to 32 PSU, with the highest salinities occurring in the center of the bay. Nitrogen loading is a major concern in the bay and is likely the driver behind the collapse of the Waquoit Bay eelgrass population over recent decades. Nitrogen enters the bay through atmospheric deposition, rivers (e.g., Childs River and Great River) and groundwater (accounting for 50%–80% of the N-load) (Talbot et al., 2003; Kroeger et al., 2006; Bowen et al., 2007). Our study site was carefully selected to best allow us to isolate the effects of using different aquaculture gear by co-locating sites for each of the three types of aquaculture gear as well as our control site at a location where all would be in the same hydrodynamic setting (exposure to winds, waves, and currents), have the same sediment characteristics, be the same distance from shore, and in the same depth of water in an area free of seagrass beds.
We used three commercially available oyster growth methods (surface, suspended, and bottom oyster aquaculture methods) that result in different proximity of oysters to the sediment. The different oyster methods were first installed in early March 2018 and 2019, with each treatment site covering an area of 80 m2 (Floating Bags), 108 m2 (Oyster Gro’), or 108 m2 (Bottom Cages) (Figure 1). Year old oysters were added in late April of each year. Oysters and all gear were removed in October 2018 and 2019 for over wintering.

Figure 1. Aerial image of oyster deployments and the sediment sampling sites in Waquoit Bay, MA (41°34′49″N, 70°31′27″W). Within the yellow box from left to right are the bottom cages, the floating bags, and the floating Oyster Gro cages (Image from Google Earth).
Between each gear type deployment there was a 5 m gap. For the Bottom Cage (BC) method we deployed 1.2 × 0.9 × 0.4 m3 cages that rested on the bottom and held the oysters 5–6 cm above the sediment. The BC setups allowed oysters to grow on the ocean bottom similar to wild oysters. The Floating Bag (FB) method utilized 0.5 × 0.8 m2 rigid floating bags with 6 mm2 mesh, attached to foam floats and aligned into a floating system that ensured adequate water circulation. The suspended Oyster Gro’ method (OG) utilized 0.46 × 0.89 m2 rigid grow-out bags with 6 mm2 mesh held within rigid metal wire frames suspended below buoys that allowed the oysters to be held about 30 cm below the sea surface and 0.5–2.0 m above the seafloor, depending on the tide. The BC oysters were constantly on the seafloor, while the FB and OG gear were exposed to intertidal changes but always in the water (Figure 2). For each growth method initial stocking weights of 1-year old oysters were 2.28 kg of oysters, targeting approximately 225 oysters per bag. This resulted with ∼200 kg of oysters initially in the FB, ∼205 kg in the OG and BC. Bottom cages and Oyster Gro’ systems hold six bags per unit. Our experimental design also included a control site (Figure 1). As mentioned, all sites (including the control site) were separated by approximately 5 m to limit effects of lateral transport of biodeposits between sites.
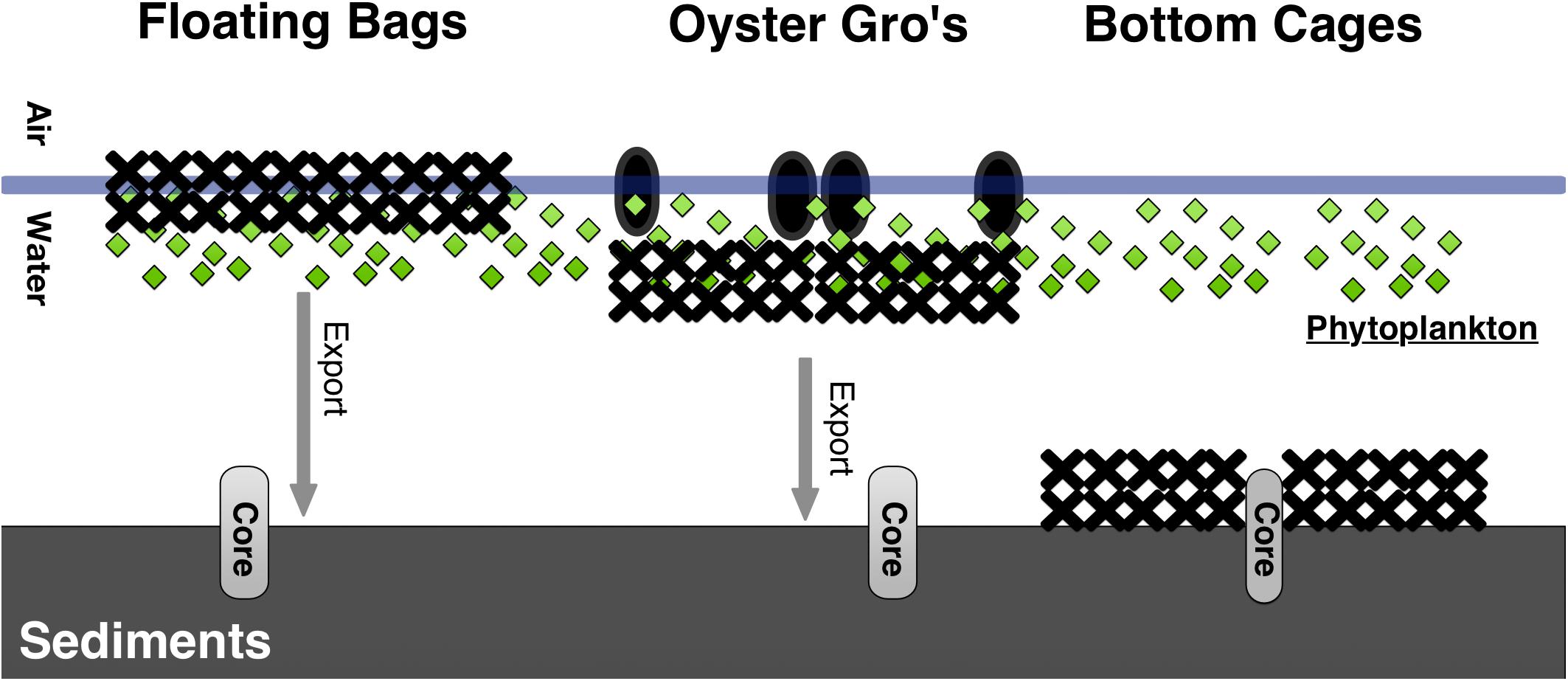
Figure 2. Schematic for the three different oyster aquaculture gear tested and their proximity to the sediment in Waquoit Bay. The Floating Bags and Oyster Gro’s were approximately 1–3 m and 0.5–2.0 m above the seafloor, depending on the tide, respectively.
Samples were collected bi-weekly during 2018 (May–September) and 2019 (April–October). Water column profiles for dissolved oxygen, temperature, and salinity were obtained in 15 cm intervals along the water column (max depth 60 cm) using a YSI Pro2030 handheld meter (YSI, Inc., yellow Springs, OH, United States) (Supplementary Table S1). We collected three sediment cores as biological replicates under each oyster method site and at the control site using a pole coring device (Aquatic Research Instruments, Hope, ID, United States) and polycarbonate core liners (internal diameter, 9.5 cm; total length, 0.5 m). All triplicate cores were collected near the center of each site (avoiding edge effects), directly under FB’s, OG’s, or BC’s, 0.5–3 m apart to limit spatial dependence between replicate cores (Figure 1). The top 3 cm of each core were sectioned immediately (within 5 min) in the field and homogenized for RNA, DNA, and porewater nutrient analyses. An extra set of two sediment cores per study site were collected for the measurements of N2 fluxes using flux chambers (see below). The same number of sediment cores were collected from our control site.
Replicate bottom water samples (∼50 ml) were collected from each core, filtered through a 0.45 μm sterivex filter (Fisher Scientific, United States) and frozen until analysis for nitrate, nitrite, and ammonium. Porewaters were collected from the upper 3-cm of sediments into 50 ml Falcon tubes. The Falcon tubes were stored on ice in the field and then at 10°C in the laboratory. For nitrate and nitrite measurements, 10 g of sediment were placed in Macrosep centrifuge tubes with a 0.45 μm membrane (Fisher Scientific, United States) and centrifuged for 5 min at 3000 rpm. The resulting filtrate was analyzed colorimetrically for nitrate + nitrite (NOx) using a salt compensated resorcinol method (Zhang and Fischer, 2006). Nitrite was measured colorimetrically according to Pai et al. (1990). Extractable ammonium was measured by incubating 10 g of sediment with 10 ml of a 1 M KCl and separating sediments from the aqueous solution using the 0.45 μm Macrosep tubes. The resultant ammonium was measured using the phenolhypochlorite method (Solorzano, 1969). Standards were salt matched and analyzed along with samples. The remaining sediments were oven-dried (80°C), ground with a mortar and pestle (<250 μm), weighed and encapsulated in Al cups, and analyzed for total carbon and total nitrogen using a Micro-Dumas (flash) combustion method at the Stable Isotope Ecology Laboratory at the University of Georgia.
The replicate sediment cores were collected to measure N2 fluxes. Collected cores from each site were submerged in a cooler containing site water and allowed to equilibrate for at least 1 h. After equilibration, cores were sealed with a cap that contained a magnetic stir bar and an oxygen optode (PSt3 sensor) attached to an Oxy10 SMA meter (PreSens Precision Sensing GmbH, Germany), that allowed real-time monitoring of oxygen concentrations using the PreSens Measurement Studio 2 software. The temperature of the incubation water was monitored continually. The sediment height and water height (no headspace) were recorded. Initial N2/Ar samples (t0) were collected with no headspace into 12-ml exetainers (739 W, Labco Ltd, Austin, TX, United States) containing 300 μl of saturated ZnCl2 (for preservation) from the incubation waters immediately after capping all the cores. Cores were incubated in the dark until 20%–25% of the oxygen in the overlying waters had been consumed. Then the overlying water from the core was collected into exetainers with no headspace as before and 50 ml of water was filtered through a 0.45 μm sterivex filter and prepared for NOx and ammonium analysis as described above. Exetainers were refrigerated (below in situ temperatures) until analysis. N2/Ar was measured on a membrane inlet mass spectrometer equipped with a water bath on the inlet, set to match sample collection temperature, a liquid nitrogen trap for CO2, a Cu-reduction column heated to 600°C to remove O2, and a quadrupole mass analyzer (RGA100, Stanford Research Systems, Sunnyvale, CA, United States). N2/Ar standards were made by equilibrating salinity matched waters at 4°C, room temperature (as measured, typically 21°C) and 30°C. Standards were analyzed every 10–15 samples to correct for any instrument drift. Excess N2 production was calculated as production above background (t0 samples as described above), scaled to water volume and normalized to incubation time (each core had the same surface area). All results for experimental site cores were compared to N2 production from the control cores.
We collected and homogenized the top 3 cm of each sediment core in ethanol-sterilized weigh boats. Homogenized sediments were aliquoted to sterile 15-ml Falcon tubes and 5-ml cryovials for subsequent extraction of bacterial RNA and DNA back in the laboratory. The RNA and DNA samples were flash frozen immediately with liquid nitrogen in the field and were stored to −80°C at the lab. All the RNA and DNA extractions occurred the day following the sampling.
We extracted 2 g of sediment for each sample in a UV-sterilized clean hood using the RNeasy PowerSoil® Total RNA Isolation Kit (Qiagen, United States) following the manufacturer’s protocol. To remove traces of contaminating DNA we treated our RNA extracts twice (2 × 30 min) with TURBO DNA-freeTM (Invitrogen, United States) as suggested by the manufacturer. To confirm absence of DNA from the RNA solutions we ran a PCR reaction using the bacterial primers BACT 1369F and PROK 1541F targeting the 16S rRNA. Each 25 μl reaction contained 0.5 U μl–1 Taq DNA Polymerase (Thermo Fisher Scientific), 1 × Taq DNA Polymerase reaction buffer (Thermo Fisher Scientific), 0.4 mM dNTPs (Thermo Scientific dNTP Mix), and 4 μM of each primer (final concentrations). These reactions were performed at 94°C for 5 min, followed by 35 cycles of 94°C (30 s), 55°C (30 s), and 72°C (45 s).
We quantified our total RNA extracts using a QubitTM RNA HS Assay Kit (Thermo Fisher Scientific) and prepared metatranscriptome libraries on selected samples using the KAPA RNA HyperPrepKit for Illumina Sequencing Platforms (Kapa Biosystems) according to manufacturer instructions. The metatranscriptome libraries were prepared only for selected samples collected from both the control site and the different oyster treatments in May (May_23), July (July_17), and September (Sep_25) 2018 and were stored at −80°C until sequencing using 150 bp paired-end Illumina NextSeq 550 (Georgia Genomics and Bioinformatics Core; University of Georgia).
Trimmomatic (v.0.32) was used to trim adapter sequences (leading = 20, trailing = 20, sliding window = 04:24, minlen = 50). Paired reads were further quality checked and trimmed using FastQC (v.0.11.7) and the FASTX-toolkit (v. 0.0.14). Downstream analyses utilized paired reads. After co-assembling reads with Trinity (v.2.6.6) (min length 150 bp) we used Bowtie2 (v.2.3.4.1) to align all sample reads and RSEM (v.1.3.0) to estimate the expression level of these reads. TMM was used to perform cross-sample normalization and to generate a TMM-normalized expression matrix. Within the Trinotate suite, TransDecoder (v.5.3.0) was used to identify coding regions within contigs and functional and taxonomic annotations were made by BLASTx and BLASTp against UniProt, Swissprot, and RefSeq non-redundant protein sequence (nr) databases (e-value threshold of 1e – 40).
Transcript abundances of denitrification, anammox, DNRA, and sulfur oxidation genes were determined with two-step quantitative RT-qPCR. The first step of RT-qPCR involved the reverse transcription of the extracted RNA into a single-stranded copy of cDNA using the QuantiTect Reverse Transcription Kit (Qiagen, United States) following the manufacturer’s protocol. The cDNAs were quantified using the QubitTM dsDNA HS Assay Kit and normalized to 10 ng reaction–1 to avoid concentration-related PCR variations. We performed the quantitative PCR amplification steps of RT-qPCR in triplicate to all genes of interest on a StepOnePlus Real Time PCR (ThermoFisher, United States) using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, United States). PCR protocols included 95°C (10 min) followed by 40 cycles of 95°C (30 s), annealing temperatures indicated in Supplementary Table S2 for 60 s, 72°C (30 s), and 85°C for 15 s. A melt curve was produced after each analysis. We generated the RT-qPCR standard curves with 101–107 dilutions of linearized plasmids containing known sequences of the targeted genes. The RT-qPCR efficiency ranged from 95% to 105% for all reactions, resulting in linear standard curves (r2 = 0.99%).
Genes involved in denitrification (narG, nirS/nirK, and nosZ), sulfur oxidation (soxA and soxB), DNRA (nrfA), and anammox (hzo) were amplified using the primers in Supplementary Table S2.
We extracted 0.5 g of sediment for each sample collected in 2018 in a UV-sterilized clean hood using the PowerSoil® DNA Isolation Kit (MOBIO) following manufacturer’s instructions. Bacterial and archaeal SSU rRNA gene fragments were PCR amplified and sequenced at Georgia Genomics and Bioinformatics Core (University of Georgia). The primers used were 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) targeting the hypervariable region 4-5 (V4-V5) of the SSU rRNA gene (Parada et al., 2016). Paired reads were quality checked and trimmed using FastQC (v. 0.11.7). We analyzed the sediment microbiome data using the QIIME2 platform (Bolyen et al., 2019), and used the DADA2 plugin provided in the QIIME2 pipeline to denoise and optimize the merging of the forward and reverse reads. Taxonomy was assigned to amplicon sequence variants (ASVs) using the scikit-learn multinomial naive Bayes classifier (q2-feature-classifier plugin; Bokulich et al., 2018) with the SILVA v132 database as the reference (Quast et al., 2013). Sequences were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRR13101827-SRR13101797, and SAMN16838263-SAMN16838385) database under project BioProject ID PRJNA679576.
The relative abundances of 16S rRNA data were determined by dividing the number of sequences in each taxon by the total number of sequences for each sample. These normalized relative abundances were used to estimate the average abundances of the different bacterial taxa described in Figures 4 and 5. We also calculated the Bray–Curtis dissimilarity index using the normalized relative abundances and performed a hierarchical clustering (complete linkage) to assess potential differences between the biological triplicates collected underneath the oyster treatments and the control site. Bray-Curtis dissimilarity index and the hierarchical clustering analyses were performed using the Vegan v2.5-6 packages in R (Dixon, 2003). We used the unweighted UniFrac distance to compare the phylogenetic similarity of the sediment bacterial communities between the different oyster culture methods. We generated the unweighted UniFrac principal component ordination plot (PCoA), in R using the package ggplot2 (Wickham, 2016). Our input data for the PCoA plot were the unweighted UniFrac PCoA results and the Shannon index feature-table generated by the QIIME2 pipeline.
We tested for the effects of the individual oyster treatments on the expression of key genes associated with nitrogen cycling, and on measurements of nitrogen flux. Seasonality may have impacted measurements, in addition to oyster treatment; therefore, two-way ANOVAs were performed to compare the measurements collected at all sites and to evaluate the effect of time. Given that marine sediments are heterogeneous by nature (e.g., random deposition of large organic particles, influences of Metazoa, and other disturbances) due to processes that occur on sub-millimeter scales, homogenized core horizons from 9.5 cm ID push core capture variation that occurs at this scale and triplicate cores collected 0.5–3 m apart at each site are reasonably independent to justify their use in ANOVAs. All gene expression datasets were normalized by log transformation and calculations were performed in R (version 3.6.1), with ANOVAs evaluated using the car package (version 3.0-10). All residuals were checked for normality using the Shapiro-Wilk test using the shapiro.test function in the stats package (version 3.6.2), with outliers removed from datasets when necessary to maintain the assumption of normality. Sliced least squares means (LS means) were calculated via Dunnett’s method using the lsmeans function in the lsmeans package, comparing each oyster treatment to the control site at each time point.
The average porewater nitrate (Supplementary Figure S1A) and ammonia concentrations in 2018 ranged from 37 to 103 mM, and 720 to 1387 μM respectively (Supplementary Figure S1B). Nitrate concentrations between the different oyster treatments were comparable, while nitrite concentrations were below detection in all samples. Spikes of high nitrate in samples from the OG and the control site were detected in May, June and early July, while FB showed elevated nitrate concentrations in June and early July. Porewater nitrate showed a generally negative correlation with N2 flux but this trend was only statistically significant in 2018 under the FB and OG treatments (Pearson’s correlation r −0.84 to −0.91, p < 0.5).
The average porewater nutrient concentrations in 2019 were generally steady over the season at all sites. The average nitrate ranged from 23.0 to 33.2 μM, presenting the highest concentrations in late June (Supplementary Figure S2A). Nitrite was below detection in all samples. The average ammonia concentrations were also comparable between the different oyster treatments and ranged from 355 to 493 μM (Supplementary Figure S2B). We detected positive correlations between ammonia and N2 fluxes in both 2018 and 2019, however these were not significant aside from under the FB site in 2018 (Pearson correlation r 0.88, p < 0.5). Bottom water nutrients were consistent between sites.
Total carbon and nitrogen were measured monthly at each site during both 2018 and 2019 sampling periods (Supplementary Table S3). The impact of the oyster aquaculture resulted in an increase on carbon content of the underlying sediment over the course of the growing season. BC sediments showed the largest impact with %C increasing ∼14%, followed by OG (∼8%), FB (∼4%), and the C (∼3%). Literature reports average sediment grain size in the area is ∼200 μm in the upper few cm (Maio et al., 2016).
In 2018 the water column temperature at all sites ranged from ∼18°C to 28°C over the sampling period June through September (Supplementary Table S1 and Figure S3A). Salinity ranged from ∼25 to 30 PSU, and dissolved oxygen ranged from ∼3.5 to 10 mg/L over the same period (Supplementary Table S1). In 2019 the water column temperature ranged from ∼12.5°C to 28°C over the sampling period from April to October (Supplementary Table S1). Salinity ranged from ∼18.5 to 26.5 PSU, and dissolved oxygen ranged from ∼0.5 to 12.5 mg/L (Supplementary Table S1 and Figure S3B). The water column temperature in 2019 was positively correlated with the N2 fluxes at all different treatment sites and the control site (Pearson correlations r 0.62–0.75, p < 0.5).
Flux chambers were used to estimate N2 production from the sediments across the sites. In all flux chamber experiments nitrate and ammonium flux to or from the sediments was negligible compared to concentration changes over the short incubation periods. The flux chambers showed production of N2 gas in sediments at the control as well as the different oyster aquaculture method sampling sites in 2018 and 2019 (Figures 3A,B). The N2 fluxes were comparable between the control and the different oyster treatments from late May until late June in 2018 (∼0.3 mmol N m–2 day–1). After June 2018 we observed a sudden twofold increase in N2 production at the aquaculture sites that reached up to a fivefold increase (in the case of BCs) when compared to the control. To test for significant differences between treatments over time we used a two-way ANOVA followed by the post hoc LS means analysis. In 2018, the overall comparison of the treatments and time point, as well as the interaction between the two variables, was found to be statistically significant (p < 0.05) (Figure 3A and Supplementary Table S4).
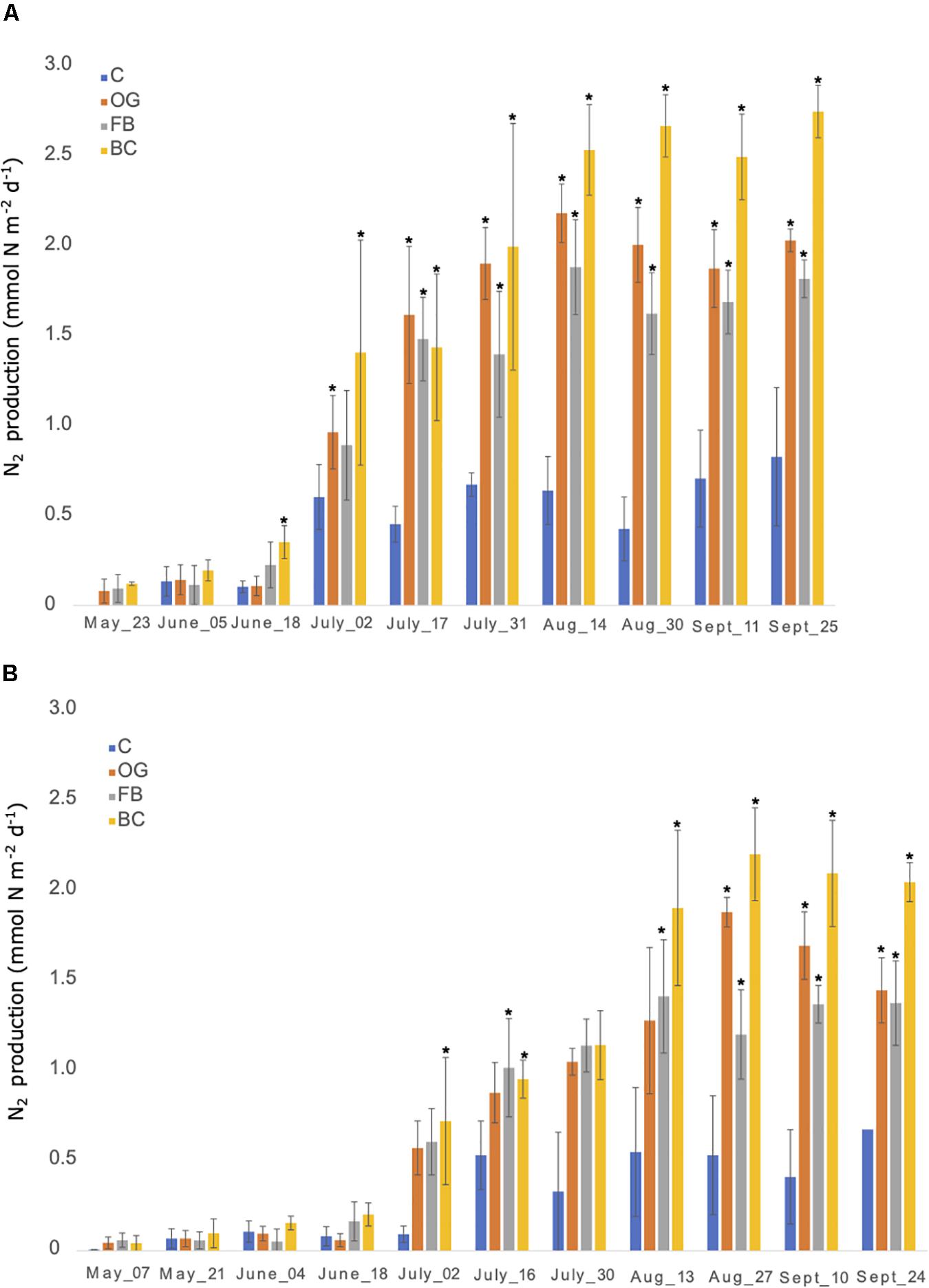
Figure 3. Nitrogen production rates measured from late May until end of September 2018 (A) and 2019 (B) using in situ flux chambers. C, Control; OG, Oyster Gro’; FB, Floating Bags; BC, Bottom Cages. Statistically significant differences are marked with an asterisk (t-test, p < 0.05).
In 2019 N2 production rates were comparable between the control site and the different treatment sites from late April until June and showed a sharp and statistically significant increase in N2 production (up to a fivefold increase) especially after July that continued until the end of the 2019 sampling period (Figure 3B and Supplementary Table S4).
The phylogenetic analysis of the bacterial communities underneath the oyster cultures and the control site was performed for the 2018 sampling period. The number of sequences per sample ranged from 1954 to 34,716, with a mean of 16,790 ± 6387 sequences. A total of 17,176 bacterial ASVs were recovered from the Waquoit sediment libraries. Hierarchical clustering of the biological triplicates from all sampling sites using the Bray–Curtis dissimilarity index revealed consistency of the microbial community structure amongst replicates for most samples (Supplementary Figures S4, S5).
Sediment community compositions underneath the different oyster treatments were generally similar to those at the control site. The dominant phyla at all sites were Proteobacteria, Cyanobacteria, and Bacteroidetes, while Planctomycetes was among the phyla with the lowest relative abundance at all sampling sites (Figure 4).
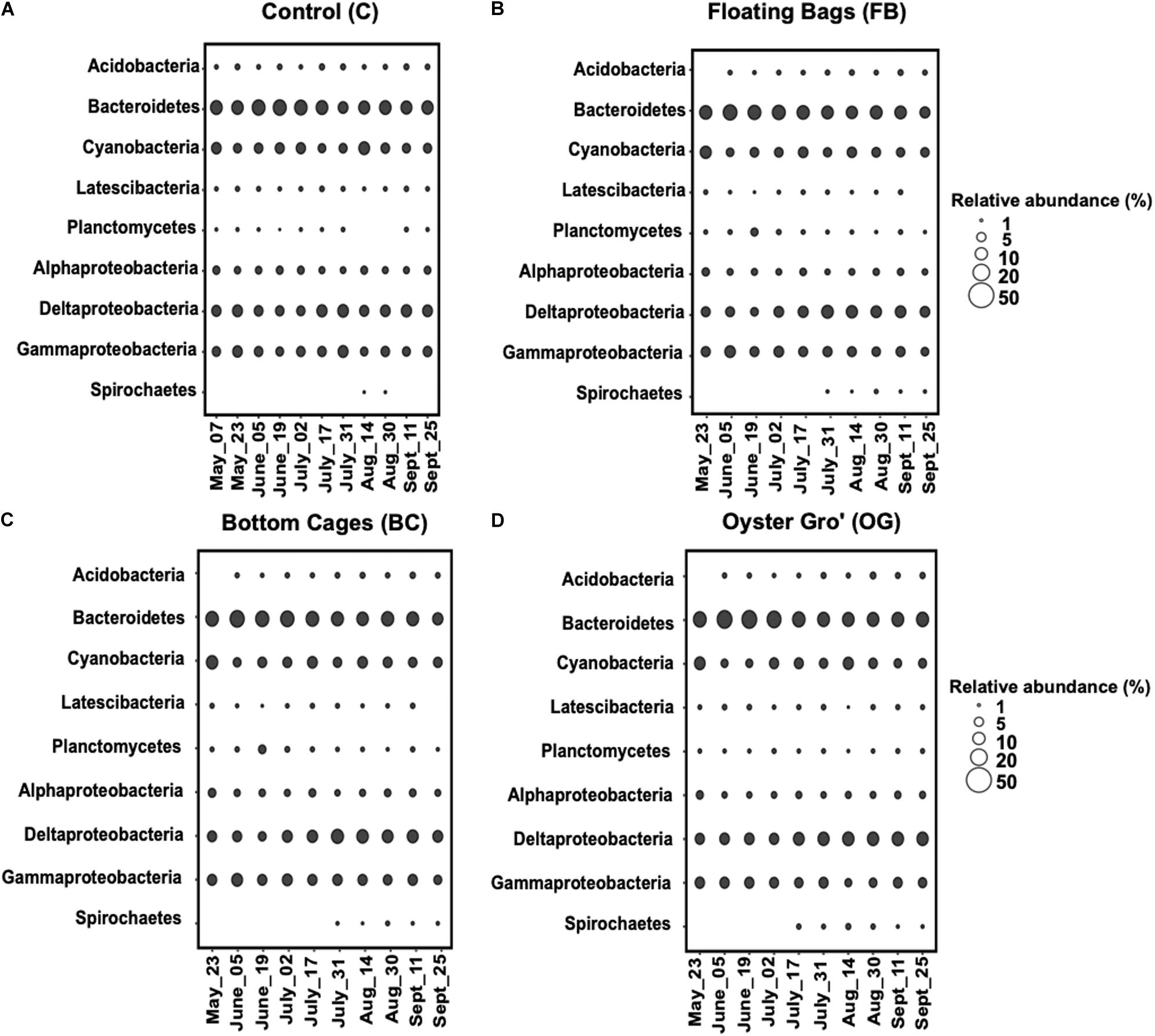
Figure 4. Average relative abundance of the bacterial communities comprising over 1% at phylum level (class level for Proteobacteria) based on 16S rRNA genesequencing per sampling date during May-September 2018. Control (A), Floating Bags (B), Bottom Cage (C), and Oyster Gro’ (D). Sampling dates are presented along the x-axis.
The relative abundance of Proteobacteria was similar, percentage-wise, between the control site and the oyster treatments throughout the sampling period (C: 41% ± 4.7%, BC: 39% ± 4.5%, OG: 38% ± 2.3%, FB: 40^ ± 3.9%). The relative abundance of Bacteroidetes was comparable between the control and different oyster aquaculture methods (C: 29% ± 5.2%, BC: 28% ± 5.4%, OG: 31.4% ± 6.8%, FB: 32.5% ± 11.5%). Planctomycetes had relative abundance of 1%–3% with higher abundances in the BC oyster treatment (C: 1.5% ± 0.5%, BC: 2.8% ± 2%, OG: 1.9% ± 0.3%, FB: 2.5% ± 0.6%). Regarding other less abundant phyla, Latescibacteria and Acidobacteria had relative abundances of 1.9%–2.5% and 2.5%–3%, respectively, and were comparable between the control and the oyster aquaculture sites. Spirochaetes were detected in the different oyster aquaculture treatments at relative abundances higher than 1% after the middle of July, while at the control site we detected Spirochetes only in August.
Within the Proteobacteria, the Deltaproteobacteria and Gammaproteobacteria classes were the most abundant, followed by the Alphaproteobacteria in all our samples (Figure 5). The ASVs affiliated with Alphaproteobacteria were primarily (∼90% of the relative abundance) within the family Mitochondria (order Rickettsiales), annotated to mitochondrial 16S rRNA gene from unicellular algae. The Gammaproteobacteria were mostly assigned to genera in Chromatiales (C: 29.3% ± 4.3%, BC: 22% ± 5.9%, OG: 29.7% ± 4.1%, FB: 22.3% ± 8.1%) and to Gammaproteobacteria incertae sedis. The most abundant classes in Deltaproteobacteria were Myxococcales (C: 9.6% ± 4%, BC: 13.6% ± 7.5%, OG: 8% ± 2.2%, FB: 15.2% ± 13.6%) and Desulfobacterales. Within Desulfobacterales, highly abundant taxa were in the Desulfobacteraceae family (C: 67.9% ± 6.5%, BC: 64% ± 11%, OG: 71.5% ± 7.2%, FB: 60.4% ± 14.2%). Flavobacteriales (C: 49.4% ± 12%, BC: 45.4% ± 12.7%, OG: 37.7% ± 15%, FB: 47.8% ± 13.4%) were the most abundant order within Bacteroidetes and specifically the Flavobacteriaceae family. The FB oyster site presented high (up to 68%) relative abundance of Flavobacteriales from May to June when compared to the control site and the rest of the oyster aquaculture treatments. The most abundant orders within ASVs assigned to Planctomycetes were the Phycisphaerales, Pirellulales, and Planctomycetales.
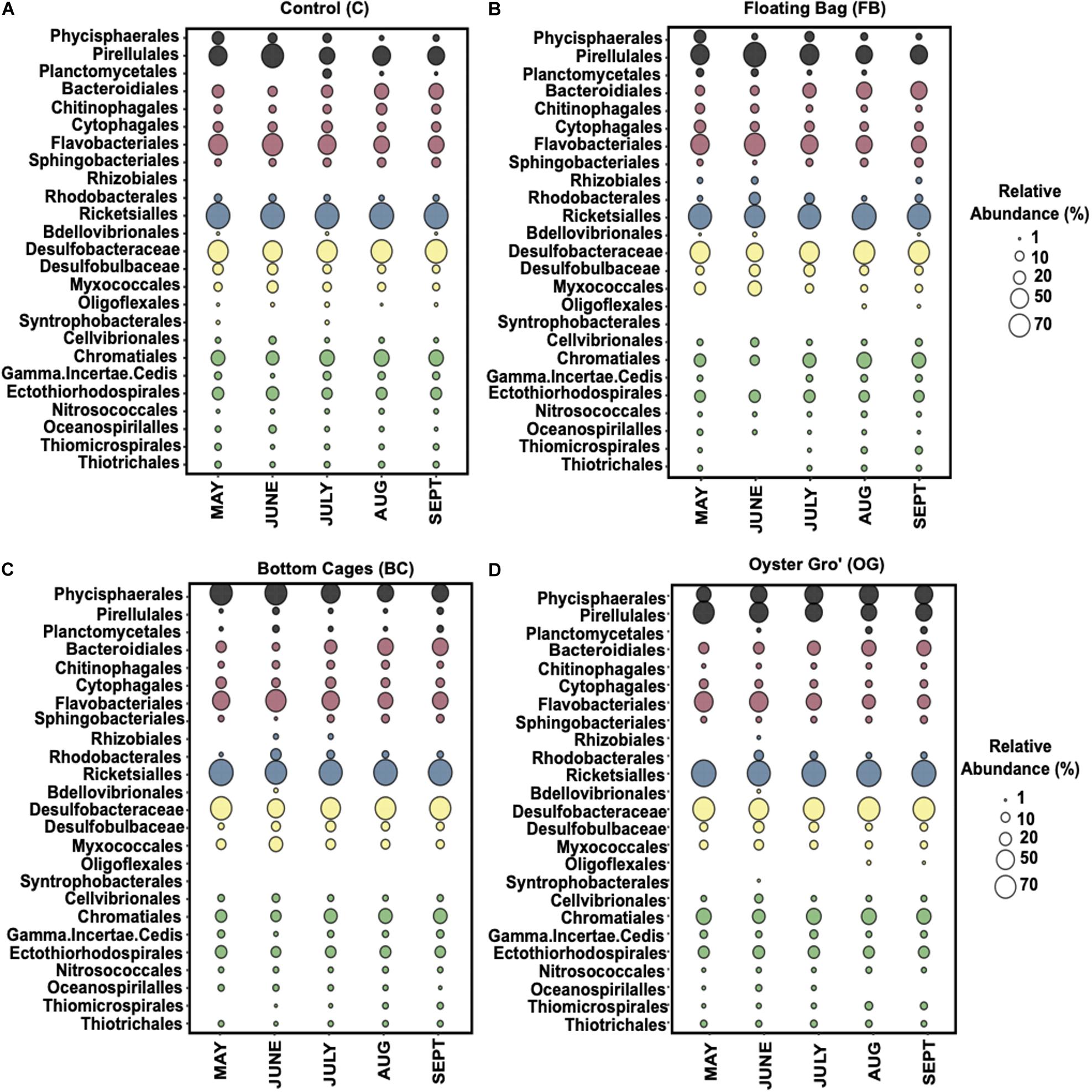
Figure 5. Average relative abundance of the bacterial communities comprising over 1% at order level (family level for Deltaproteobacteria) from May until September 2018 for Control (A), Floating Bags (B), Bottom Cages (C), and Oyster Gro’ (D) samples based on 16S rRNA gene sequencing. Planctomycetes: black, Bacteroidetes: red, Alphaproteobacteria: blue, Deltaproteobacteria: yellow, Gammaproteobacteria: green.
We used ordination analyses to identify possible (dis)similarities in community composition in sediments under the different oyster aquaculture treatments. The unweighted UniFRac principal coordinate analysis (PCoA) showed taxonomic overlap of the ASVs under the different oyster treatments as well as between the treatments and the control site (Figure 6). PCoA showed discrete phylogenetic groups that were formed at different times across the sampling season (Figure 6). A similar result was obtained when we generated a weighted UniFrac ordination plot using relative abundances.
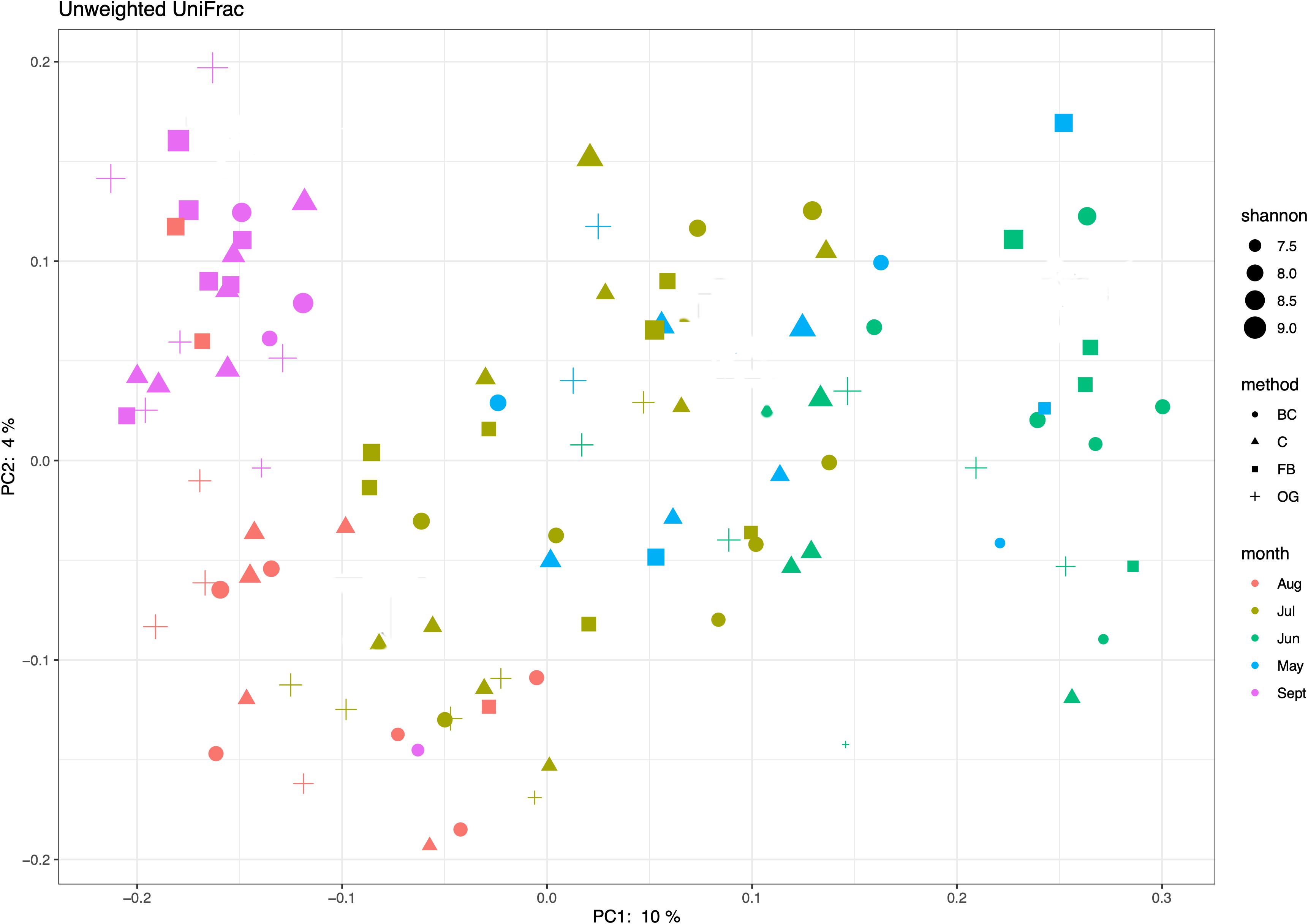
Figure 6. Principal coordinates analysis (PCoA) of bacterial phylogenetic similarity based on the unweighted UniFrac results and the Shannon index.
Analyses of gene expression by microbial communities can provide significant insights into microbial activities and interactions. Our RT-qPCR results for 2018 showed expression of the denitrification genes nirS, nirK, nosZ, and narG in sediments from all oyster treatments and the control site (Figure 7A). To estimate statistically significant effects of the oyster treatments on the expression of denitrification genes, we performed a two-way ANOVA for each denitrification gene comparing the three oyster treatments and control site measurements across the time series (Supplementary Table S5). Findings from the least squares means (LS means) showed that narG expression was similar between the control and the sediment samples collected below the Bottom Cage (BC), Oyster Gro’ (OG), and Floating Bag (FB) sites from May until middle of July 2018. However, at the end of July, we detected a sharp increase of narG expression relative to the control site under all aquaculture sites that continued until the end of September (Figure 7A). These increases were statistically significant when compared to the control based on the results of least squares means comparisons (p < 0.05). The number of narG transcripts detected under the different oyster treatments after the end of July was higher when compared to the number of copies detected earlier in the sampling period (May until mid of July 2018; Supplementary Figure S6). Elevated expression of narG after July was also detected in sediments at the control site, but was lower when compared to samples from under the aquaculture sites (Supplementary Figure S6). Differences in the narG expression were significant after July in both 2018 and 2019; however, t-ratios were greater in 2018, suggesting a larger deviation from the gene expression baseline detected in the control site. Elevated gene expression relative to the control was also detected for nirS, nirK, and nosZ (where measurable differences occurred) under the three oyster treatments (Figure 7A and Supplementary Figure S6). Expression of nosZ in sediments under the bottom cages was significantly lower than nosZ expression measured at the control site from late May to mid-July of 2018, with similar patterns observed in 2019.
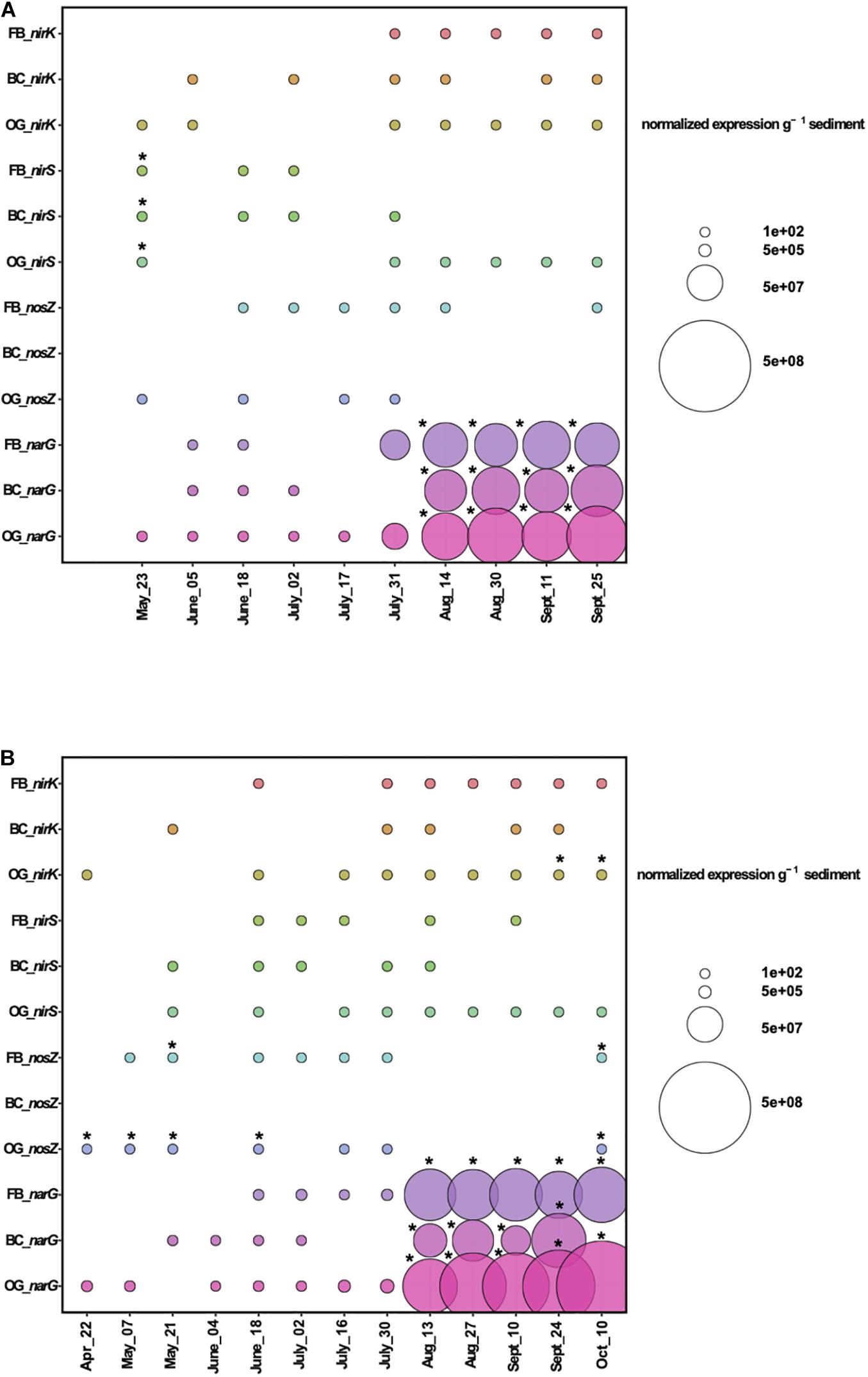
Figure 7. Expression of denitrification genes above control levels (gene expression values from the control were subtracted from the values from the treatments) during 2018 (A) and 2019 (B). FB, Floating Bags; BC, Bottom Cages and OG, Oyster Gro’. Statistically significant differences are shown with an asterisk (t-test, p < 0.05).
Similar patterns of expression of denitrification genes were also observed in 2019 (Figure 7B). Sliced LS means showed that narG expression was significantly higher under all treatments when comparing treatments to the control site at individual time points from mid-August to late September and overall comparisons of the treatments across the time series were found to be significant (Figure 7B and Supplementary Table S6). We detected elevated gene expression of nosZ under FB and OG oyster treatments that was statistically significant (primarily under the OG treatment) when compared to the control site on individual sampling days (p < 0.05). The two-way ANOVA results showed no statistically significant differences in nirS across all treatments. In 2019, nosZ also had significantly lower overall rates of expression in sediment samples collected under the BC.
We estimated expression of nrfA, the key gene in DNRA, both in 2018 and 2019. Our results showed that differences in expression levels of nrfA across treatments were statistically significant when compared to the control for both sampling seasons (Figure 8 and Supplementary Tables S5, S6). Closer examination of sampling dates reveals different nrfA expression trends between the three oyster treatments. In 2018, expression of nrfA was consistently elevated relative to the control site in sediments collected under the BC treatment, starting at the end of June. nrfA expression was also relatively elevated in samples collected under the FB site beginning in July, with statistically significant differences observed in late July through the end of sampling period. The expression levels of nrfA in the OG samples were significantly reduced relative to the control site (p < 0.05) from late July onward in 2018.
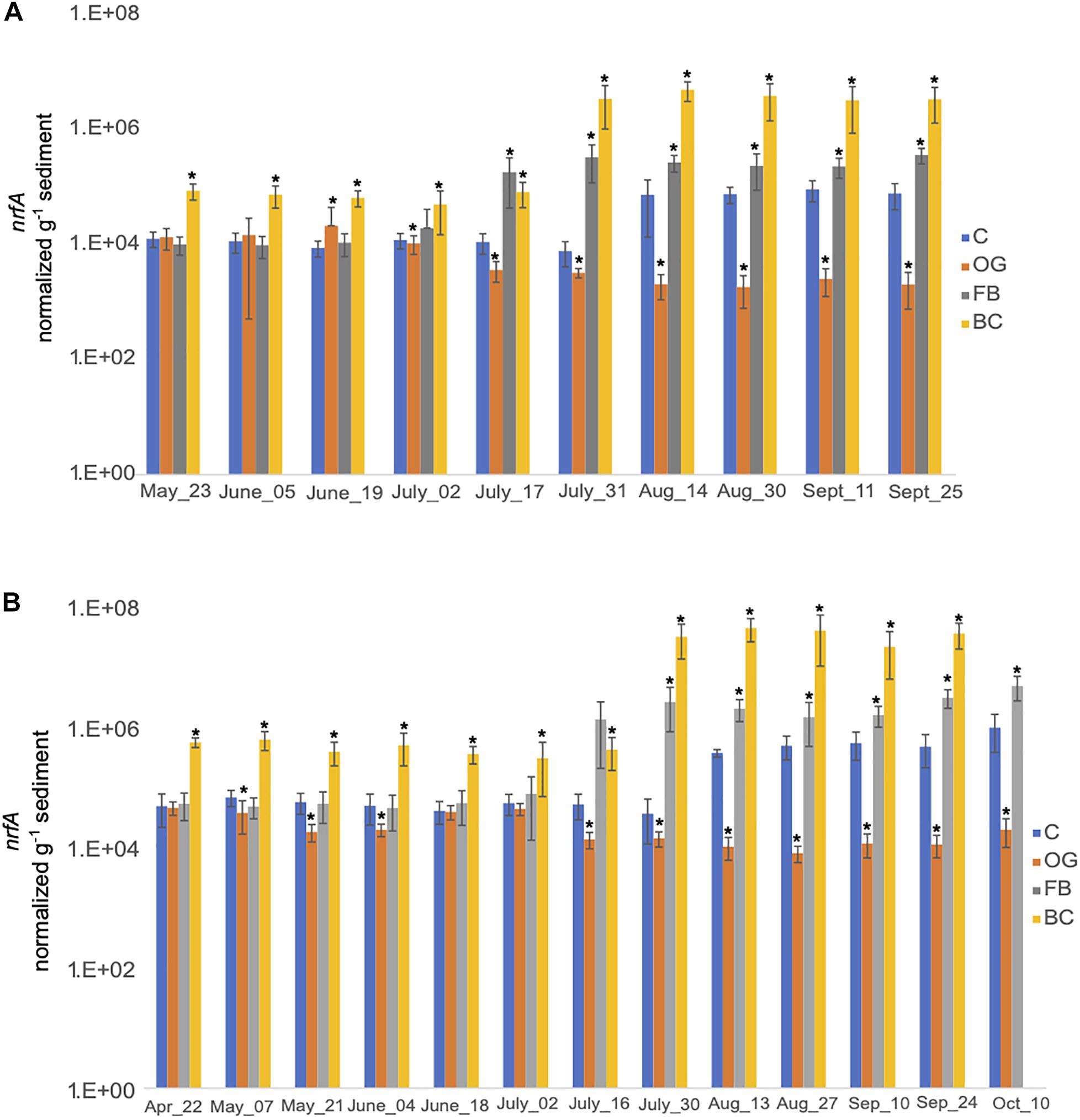
Figure 8. Average copy number of nrfA for the three cores collected under the three oyster treatments and at the control site over the course of the sampling period in 2018 (A) and 2019 (B). C, Control; OG, Oyster Gro’; FB, Floating Bags; BC, Bottom Cages. Statistically significant differences are marked with an asterisk (t-test, p < 0.05).
Trends in nrfA expression by sampling day were similar in 2019 (Figure 8B). We detected elevated expression of nrfA in BC and FB oyster treatment samples relative to the control site. OG sediment samples exhibited decreased expression relative to the control site across all sampling dates in 2019. The decreased nrfA expression levels were statistically significant across the time series (Figure 8B).
We estimated the expression of hzo genes, responsible for the final step of anammox, across all sites (Figure 9 and Supplementary Tables S5, S6). hzo expression across the three oyster treatment sites was found to be statistically significant for both 2018 and 2019 sampling seasons (Supplementary Tables S5, S6). In 2018, expression of hzo was relatively similar between the control site and the BC and FB treatments from May to late June (Figure 9A). Elevated expression of hzo was detected from mid-July through the end of the 2018 sampling period in BC sediments which was statistically significant when compared to the control. The hzo expression in FB sediments remained similar to the control site even after July, with the greatest change in expression levels observed in early September (Figure 9A). Patterns of hzo expression were the opposite in OG sediments relative to the other treatments and the control site. hzo expression in OG was slightly elevated primarily in May and early June when compared to the control and was followed by a sharp and statistically significant decrease from mid-July through the end of the 2018 sampling.
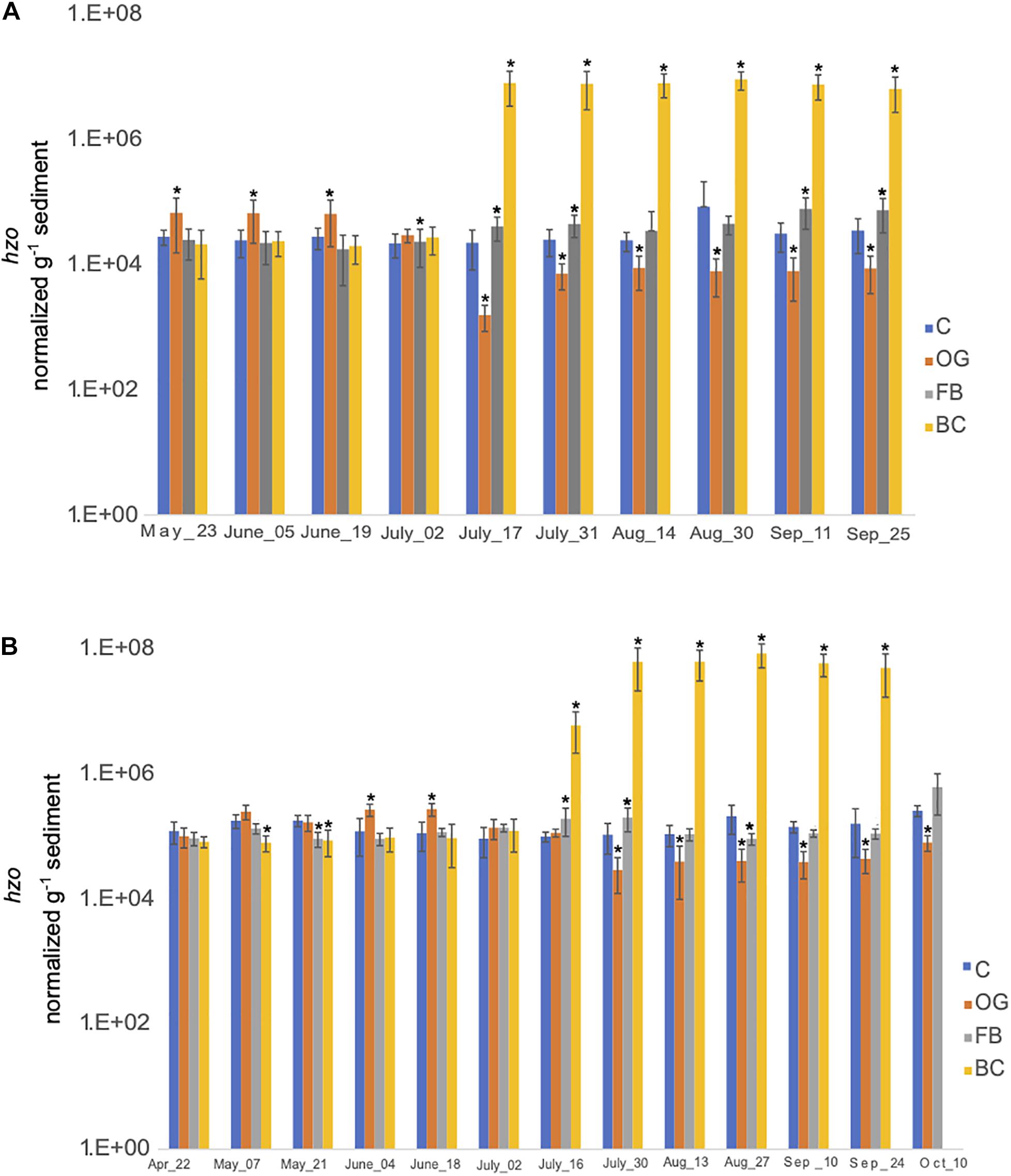
Figure 9. Average copy number of hzo for the three cores collected under the three oyster treatments and at the control site over the course of the sampling period in 2018 (A) and 2019 (B). C, Control; OG, Oyster Gro’; FB, Floating Bags; BC, Bottom Cages. Statistically significant differences are marked with an asterisk (t-test, p < 0.05).
Samples collected in 2019 demonstrated similar trends with 2018. t-ratios indicated similar hzo expression between the BC, FB, and the control site from April until June, with exception in May where we detected slightly reduced expression of hzo in BC and FB relative to the control. A sudden and statistically significant increase in hzo expression levels was detected in the BC sediments from late July and through the end of the 2019 sampling period. FB sediments had similar or significantly reduced hzo expression across the time series. Similar to 2018, expression of hzo in OG oyster site was slightly elevated early in the time series (primarily in June) when compared to the control, followed by a sharp and statistically significant decrease in expression levels from the end of July through the end of 2019 sampling period.
We detected similar expression levels of sulfur oxidizing genes, soxA (Figure 10A) and soxB (Figure 10B) in samples collected underneath the different oyster aquaculture methods and the control sites. Expression of both sox genes showed a gradual increase in the middle of July that continued until the end of September. Slightly enhanced expression levels of soxB were detected in samples collected from BC and FB sediments, when compared to the controls.
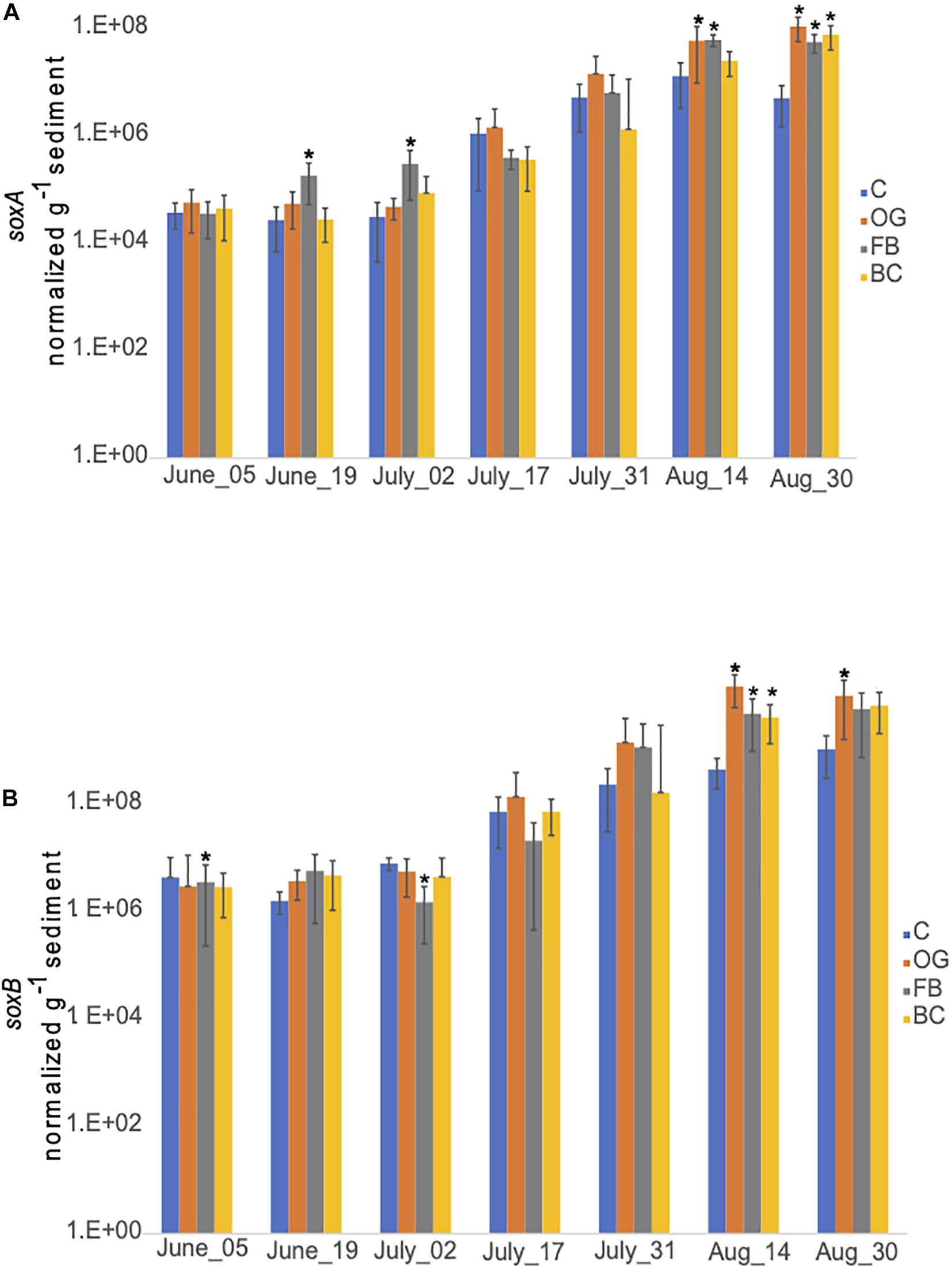
Figure 10. Average copy number of soxA (A) and soxB (B) for the three cores collected under the three oyster treatments and at the control site over the course of the sampling period in 2018. C, Control; OG, Oyster Gro’; FB, Floating Bags; BC, Bottom Cages. Statistically significant differences are marked with an asterisk (t-test, p < 0.05).
Expression of sox genes expression in 2019 was comparable between the samples collected at the different oyster treatments and the control site. Increase in expression was detected in all samples after the middle of July (data not shown).
Excessive nitrogen loading in coastal waters is a growing topic of concern to coastal communities. Many communities see aquaculture as a potential method of removing nitrogen from the coastal waters, by growing and removing biomass. However, nitrogen removed by biomass harvesting is only part of the equation. Farming activities can also impact the sediment chemistry and microbial communities. A recent study suggests that in coastal systems with excess nitrogen loading from anthropogenic activity, oyster aquaculture may have a positive long-term effect on sediment denitrification that persists over the years (Ray et al., 2020). In most coastal sediment-hosted microbial communities nitrogen removal is a natural result of the net activities of denitrification, DNRA, and anammox processes in coastal sediments. Here we investigate how the activity of these process, as reflected by net nitrogen removal and changes in gene expression for key nitrogen cycling processes may change as function of three popular oyster aquaculture methodologies; Bottom Cages (BC), Oyster Gro’ (OG), and Floating Bags (FB).
Our analyses of expression of genes for denitrification, DNRA, and anammox showed that genes associated with nitrogen cycling respond to changes that occur in the sediments underneath the different oyster treatments. BC, OG, and FB presented distinct expression patterns of key nitrogen-cycling genes over the season. We detected a statistically significant increase in the expression of narG under all sites relative to the control site after the end of July in both years (comparisons of LS means for each sampling event). This enhanced expression of narG could potentially be linked to enhanced N2 production rates and is consistent with greater and statistically significant N2 production detected in the sediments underneath the oyster aquaculture treatments relative to the control site. Although the presence of aquaculture increased expression of narG and resulted in greater production of N2 relative to background, the increase in N2 production at all sites over the installation period indicates that environmental factors (e.g., temperature), could also contribute to the enhanced N2 removal in Waquoit Bay. The expression of nirS, nirK, and nosZ were relatively consistent over the course of the growing season at all sites. Overall, our data do not show a clear link between N2 production and the expression of nirS, nirK, and nosZ, although the gene expression of nosZ presented statistically significant differences primarily between the OG treatment and the control site. This can be partially explained by the modular nature of denitrification since it is a nitrogen cycling process that can be performed in sediments in a cooperative way by a variety of phylogenetically unrelated microorganisms that participate in different steps of the process, and to differing degrees (Dalsgaard et al., 2014; Graf et al., 2014). Bacterial denitrifiers can host in their genome the full suite of denitrification genes or they can perform truncated denitrification that lacks the final step of the pathway (reduction of N2O to N2) (Braker et al., 2010; Graf et al., 2014). The expression of denitrification genes is controlled by parameters not yet fully understood that can vary among individual strains (Zumft, 1997; Wallenstein et al., 2006). Additionally, the sensitivity of denitrification gene transcripts (nirS, nosZ) to oxygen is known (Dalsgaard et al., 2014). Although our handling of sediments from the time cores were brought to shore and subsamples frozen was short (∼5–10 min), the process of subsampling may introduce enough oxygen to suppress nirS and nosZ transcription, as has been previously described (Dalsgaard et al., 2014). Collectively, these complexities illustrate the challenges of directly linking rates of nitrogen biogeochemical processes with gene expression data which can follow non-linear dynamics (Bowen et al., 2014), especially in natural ecosystems that usually host diverse bacterial communities. Finally, we need to point out that the commonly used gene-specific primer sets for RT-qPCR studies related to denitrification genes are usually designed based on sequence alignments found in publicly available database and for cultured microbes (Wallenstein et al., 2006). This can limit the RT-qPCR ability to target all variants of key denitrification genes (nirS, nirK, nosZ, norB/C), especially those deriving from uncultured taxa. Our Illumina-sequenced marker gene data (iTAGs) data showed that almost 60% of our ASVs were affiliated with uncultured taxa, which makes it difficult to assess to what degree our RT-qPCR primers were able to represent all variants of the key denitrification genes.
Our RT-qPCR data showed that the narG expression levels at the aquaculture sites were different when compared to those at the control site especially after July (Figure 7 and Supplementary Figure S6). This indicates that the different oyster treatments were able to trigger and maintain elevated narG expression in the underlying sediments after July. This might have occurred due to increasing carbon and nitrogen content in the underlying sediment, via the release of oyster biodeposits which are known to deliver organic nitrogen and carbon to the sediments (Newell et al., 2002; Smyth et al., 2013). Organic matter is a prerequisite for heterotrophic denitrification since denitrifiers oxidize organic carbon inputs using nitrate as electron acceptors to gain energy. Moderate organic additions to the sediments can increase activity of benthic denitrification while high inputs of organic matter can decrease denitrification rates (Caffrey et al., 1993). The biodeposits from bivalve cultures contain 2–3 times more carbon and nitrogen than unaggregated particles (Feinman et al., 2018 and references therein) and similarly to what observed in experimental studies (Caffrey et al., 1993), their moderate or excessive release to the sediments underneath bivalve cultures can enhance or diminish denitrification (Carlsson et al., 2012). Carlsson et al. (2012) suggested that the decline of denitrification under excessive organic matter inputs from bivalve farming is due to the production of hydrogen sulfide that can inhibit denitrification (Jensen et al., 2008), nitrification (Joye and Anderson, 2008), and anaerobic ammonia oxidation (anammox) (Porubsky et al., 2009), but also can favor dissimilatory nitrate reduction to ammonium (DNRA) (Caffrey et al., 2019).
While we did not directly measure rates of DNRA, our gene expression data for nrfA showed that DNRA is a process that likely occurs naturally in the Waquoit Bay. However, the expression of nrfA increased primarily under the BC site over the season whereas nrfA in sediments under the OG site significantly declined in both 2018 and 2019 sampling periods (Figures 8A,B). The expression of nrfA under the FB treatments was lower than under the BC treatment and comparable to what we observed in control sediments especially before the end of July. This may indicate that DNRA is an active process in the sediments underlying oyster treatments (especially for BCs), but that the suspended (primarily), and to a lesser extent, the floating oyster culturing methods limit or inhibit DNRA. The role of DNRA has been extensively debated with regard to whether or not oyster aquaculture can promote nitrogen retention over nitrogen release (denitrification). DNRA was identified as the dominant nitrate reduction process (∼80%–96%) under suspended oyster systems, and it was suggested that limited denitrification was due to competition for nitrate between denitrification and DNRA, and the labile nature of organic matter that favors DNRA (Erler et al., 2017; Lunstrum et al., 2018). Mortazavi et al. (2015) also detected low rates of net denitrification and high ammonium fluxes in sediments underneath cages and indicated DNRA as a potential process due to the high presence of hydrogen sulfide. Vieillard (2017) found low DNRA rates (< 130 μmol m–2 h–1) in the sediments underlying suspended nets and bottom bags/cages, with insignificant differences between oyster aquaculture sites and controls. Confounding interpretation of the role of DNRA in these systems is the observation of DNRA bacteria isolated from soils that contain “atypical” (due to their presence in non-denitrifying phyla) but functional nitrous oxide reductases genes (nosZ) that can contribute to nitrogen gas production (Sanford et al., 2012; Orellana et al., 2014). We do not know if these “atypical” nosZ genes are also expressed in marine sediment DNRA bacteria, but we argue that if they exist, then nitrogen cycling underneath bivalve cultures can be more complicated than previously anticipated.
The enhanced expression of nrfA detected after July, primarily in the BC oyster aquaculture, suggests that the biodeposits released from oyster treatments in close proximity to the sediment can enhance ammonium regeneration in the sediment. We hypothesize that this enhanced expression of nrfA especially in the BC sediments could be attributed to chemolithotrophic or fermentative DNRA, with the latter being less prominent. Fermentative DNRA couples nitrate reduction with organic matter fermentation and occurs under high carbon, non-sulfidic, and nitrate-limited conditions (Burgin and Hamilton, 2007; van den Berg et al., 2015). Oyster biodeposits, and/or active nitrification across oxic/anoxic interfaces can provide significant amounts of nitrate in the underlying sediments (Jensen et al., 1994; Newell et al., 2005 and references therein); thus, we consider that fermentative DNRA may not be the primary form of DNRA occurring underneath BC in our study. This is supported by our iTAG data that show low relative abundances (<0.5%) of fermentative bacteria that can perform DNRA and influence the competition between denitrification and DNRA in favor of DNRA (e.g., taxa affiliated to Clostridia, Vibrio, Desulfovibrio, Bacillus, and Pseudomonas sp.; Burgin and Hamilton, 2007; van den Berg et al., 2017).
Chemolithotrophic DNRA can dominate under sulfidic conditions in coastal sediments since it can couple nitrate reduction to the oxidation of reduced sulfur forms, including free sulfide (H2S, HS–, and S2–) and elemental sulfur (S0) (Burgin and Hamilton, 2007 and references therein). While seasonal sulfide concentration dynamics are not available because we did not measure sulfide concentrations directly during both 2018 and 2019 field seasons, sediments under the BC site had visibly greater accumulations of organic material including algae, and produced a noticeable sulfidic smell from mid-summer onward. This suggests that organic matter in biodeposits released from oysters accumulates at higher concentrations under BCs, possibly enhancing sulfate reduction, and increasing production of hydrogen sulfide and other sulfur species (e.g., S0, thiosulfates, and sulfites) below the BC site. Available metatranscriptome data was searched for expression of dsr genes. Expression of dsr genes supports the notion of active dissimilatory sulfate reduction under all treatments and also at the control site (Supplementary Table S7). The RT-qPCR results showed similar patterns of expression of the sulfur oxidizing genes soxA and soxB, under the different aquaculture sites and the control site (Figure 10). Both our metatranscriptome and RT-qPCR results indicate that sulfide production and oxidation occur naturally in Waquoit Bay sediments and also under the different oyster treatments.
Redox changes due to hydrogen sulfide accumulation can alter the rates of denitrification, anammox, and DNRA (Caffrey et al., 2019). Background hydrogen sulfide (H2S) measured in October (2020) at our study site when the aquaculture sites were not present was 53.2 ± 2.7 μM. Caffrey et al. (2019) estimated that exposure of hypoxic sediments to low sulfide (S–2) concentrations (∼52 μM) can promote DNRA in the expense of denitrification, with the latter remaining still active but with lower nitrogen production rates. We believe that the possible sulfide accumulation (if occurs) in the BC oyster aquaculture method via enhanced biodeposition was not critical to inhibit oyster-mediated N2 production via denitrification as has been previously described (Carlsson et al., 2012). A possible explanation for this can be the in situ biogeochemistry of the sediments in Waquoit Bay that can allow the oxidation of the sulfides that accumulate in the sediment with Mn and Fe oxides under anoxic conditions. Mn, and especially Fe, oxidants form metal-bound sulfides (e.g., iron sulfide; FeS) that do not inhibit denitrification (Brunet and Garcia-Gil, 1996). Downcore analyses of Waquoit sediments have shown black FeS underneath the oxic sediment layer (Charette et al., 2005). Mn and Fe, oxides are the dominant oxidants of sulfides in the coastal sediments with an active role on sulfur sediment dynamics (Jørgensen et al., 2019). Another possible explanation for the continuation of high N2 production rates underneath the BC site despite the potential sulfide accumulation can be the tolerance of the in situ microbial community to suboptimal sulfide conditions. Microbial communities from hypoxic sediment or the oxic/anoxic interfaces of estuarine and coastal sediments seem to tolerate periodic redox fluctuations that would occur with disturbances (weather events, animal burrowing, human activities, etc.) and are able to recover from sporadic exposure to oxic or highly sulfidic conditions (Marchant et al., 2017; Caffrey et al., 2019). This can be an adaptation of the in situ communities to overcome tidal cycles or periodic events that affect the redox regime of the sediments. We suggest that the proximity of the oyster gear to the seafloor can influence the fate of the nitrogen transformations since it can affect oxygen circulation or promote redox changes. Our data show that DNRA appears depressed under the OG site possibly due to the increased aeration caused by that gear, as discussed below.
The same explanations can apply to the enhanced hzo levels that we detected underneath the BC site. hzo is the key gene of anaerobic ammonium oxidation (anammox), and sulfide, at micromolar concentrations, has an inhibitory effect on anammox activity (Jensen et al., 2008). However, under our BC treatments we detected enhanced expression of hzo coincident with seasonal expression of narG, indicating possibly active anammox over the sampling season despite potential sulfide accumulation. Similar hzo expression levels between FBs treatments and at the control site were observed for most of the 2018 and 2019 sampling periods, while statically significant depressed expression levels of both hzo and nrfA were detected almost throughout the 2018 and 2019 sampling periods under the OG treatment (Figures 8 and 9). This suggests that the OG equipment may introduce oxygen to the underlying sediments by creating a “piston-pumping” activity in response to wind and waves that aerate the underlying sediments. While this action has not been quantified it could limit and/or inhibit the strictly anaerobic anammox and DNRA processes. Floating bags also appear to permit enough bottom water circulation to aerate surficial sediments enough to suppress these two processes relative to BCs, although not to the extent of the OG gear. Figure 5 shows that the control and FB and OG sites share high relative abundance of the order Pirellulales (Planctomycetes), compared to sediments below BCs. Bacterial representatives affiliated to this order can be aerobes (Dedysh et al., 2020), which can support our hypothesis of suppressed anammox activity under the OG and FB systems due to sufficient oxygen circulation underneath these gear.
Anammox is reported to have less influence than denitrification on nitrogen release from coastal ecosystems (Dalsgaard et al., 2005; Trimmer and Engström, 2011). This is consistent with observations at bivalve farming sites (Carlsson et al., 2012; Erler et al., 2017; Lunstrum et al., 2018). Our iTAGs indicate the relative abundance of Planctomycetes in our sediments is low (1%–3%); thus, we can infer that anammox makes a limited contribution to nitrogen removal in Waquoit Bay, however this requires further investigation. The potentially low anammox activity in Waquoit Bay may be insignificant when compared to the overall benefit of promoting nitrogen removal and possibly limiting or inhibiting ammonia regeneration using either the OG or FB oyster aquaculture methods. However, depending on in situ sediment biogeochemistry anammox activity can vary, so loss of anammox under oyster aquaculture activities might have a significant impact on overall nitrogen loss in other settings.
The possible “piston-pumping” caused by the OG gear (and to a lesser extent the FB) and consequent O2 circulation in surficial sediments did not appear to have a negative effect on the N2 production rates possibly induced via bacterial denitrification underneath OG and FB treatments at our study site. Denitrifiers are primarily facultative anaerobes in surficial sediments, with strict anaerobes appearing at increasing sediment depths where anoxia is more consistent (Tiquia et al., 2006). This idea is consistent with observations that denitrification in coastal sediments can be stimulated by frequent switches between oxic and anoxic conditions (influenced perhaps by tidal cycles and storm events), with denitrification occurring even under O2 exposure (Marchant et al., 2017 and references therein). Finally, as already discussed, denitrification is a modular process where distinct groups of microorganisms can mediate different steps of the process (Dalsgaard et al., 2014; Graf et al., 2014). Thus, a metabolic strategy that can be more resistant to sporadic changes in oxygen at coastal sediments. Finally, we note that sample handling (subsampling of cores in the field and dispensing to sterile containers prior to freezing) exposes sediment microbiota to oxygen, and while handling was consistent for all samples in this study, expression of genes for some oxygen-sensitive processes may be negatively affected to an unknown degree, and thus, be higher than we report.
The structure and composition of sediment bacterial communities were similar underneath the different aquaculture practices and the control site, and were dominated by three highly abundant phyla; Bacteroidetes, Cyanobacteria, and Proteobacteria (Figure 4). Bacteroidetes is a cosmopolitan phylum colonizing marine and freshwater sediments (Klier et al., 2018) with high abundances especially in coastal areas (Fernández-Gómez et al., 2013). Generally, Bacteroidetes are known for their ability to grow in association with particles and for their capacity to degrade polymers (Fernández-Gómez et al., 2013). Bacteroidetes may also be able to colonize biodeposits released from the oysters, however, this requires further investigation. Within Bacteroidetes there were five dominant orders including Bacteroidales, Flavobacteriales, Cytophagales, Chitinophagales, and Sphingobacteriales at both control and oyster aquaculture sites. Bacteroidales was the only taxon that showed a seasonal trend with elevated relative abundances after the end of June under all the examined aquaculture treatments (Figure 5). The seasonal increase of Bacteroidales can be due to increased presence of particles (biodeposits) and/or related to nitrogen cycling processes. For the latter we can only speculate that it is denitrification, since we know that Bacteroidetes have the nirK/nirS and nosZ genes in their genomes and thus, can perform specific steps of denitrification (Graf et al., 2014). These possibilities, however, require further investigation.
Proteobacteria were dominated by the classes Deltaproteobacteria, Gammaproteobacteria, and Alphaproteobacteria (Figure 4). Genera that belong to Proteobacteria can do denitrification sensus stricto (complete denitrification; they host nir/nor/nosZ genes in their genomes) while the majority of Proteobacteria have mainly the nir/nor genes (Graf et al., 2014). In Alphaproteobacteria, besides the family Mitochondria (order Rickettsiales) that dominated at all sites, the other less abundant taxa (<10% relative abundance) were Rhodobacterales (Rhodobacteraceae) and Rhizobiales. Members of the Rhodobacteraceae family are abundant in coastal sites with high nutrient availability and anoxic, or anoxic/sulfidic (euxinic) regimes (Pohlner et al., 2019). Genera that belong to Rhizobiales have nirS and nirK genes so they are also potential denitrifiers (Rasigraf et al., 2017).
Within Gammaproteobacteria the dominant order at all sites including our control site was Chromatiales. Genera and species affiliated to Chromatiales are sulfur oxidizers known to utilize the accumulated sulfides, or thiosulfates to produce S0 (Imhoff, 2005). Chromatiales can proliferate when sediments are nitrate-replete, possibly performing chemolithotrophic denitrification (Aoyagi et al., 2015). Chemolithotrophic denitrification might be a naturally occurring process at the Waquoit Bay sediments, and we suggest that it can take place underneath the different oyster treatments especially after July. Other dominant orders within Gammaproteobacteria were aerobic ammonia oxidizers (Nitrosococcales; Ward and O’Mullan, 2002), heterotrophic denitrifiers (Oceanospiralles; Llorens-Marès et al., 2015; Marchant et al., 2017), and other taxa related with sulfur/hydrogen sulfide oxidation (Thiotrichales, Thiomicrospirales, and Ectothiorhodospiraceae; Robinson et al., 2016). Finally, our iTAG data showed dominant sulfate reducing microorganisms (SRM) within the order Desulfobacterales (Desulfobacteraceae) under the different oyster aquaculture treatments and at the control site.
The taxonomic composition of our bacterial communities is consistent with previous studies of Waquoit Bay (Coskun et al., 2019), and with studies of bacterial communities underneath oyster aquaculture sites (Azandégbé et al., 2012). The principal component ordination analysis (PCoA) based on bacterial phylogenetic similarity of communities collected from all sites (control and oyster aquaculture) over the season indicated discrete groups formed along the sampling season, that cannot be explained solely by observed minor differences at the class/order level. A significant portion of our ASVs (∼60%) below the order/family taxonomic levels is affiliated with “uncultured” taxa, about which little is known. The observed discrete groups could be be driven by changes in these uncultured taxa. These taxa may also contribute (together with our assigned denitrifying and anammox phyla) to elevated nitrogen production rates that occurred after the end of June in both 2018 and 2019. Some of these uncultured taxa in Waquoit Bay have been described as “microbial dark matter” with micro-aerophilic and anaerobic lifestyles, and show metabolic flexibility under anoxic and euxinic conditions (Coskun et al., 2019). This supports further the idea that bacterial communities in coastal sediments might be more tolerant than previously assumed of fluctuations in oxygen and sulfide (Marchant et al., 2017; Caffrey et al., 2019), and with possible consequences for nitrogen cycling processes that occur underneath aquaculture sites.
Bacterial communities underlying our aquaculture systems enhanced nitrogen removal when compared to control sediments. Observable accumulations of organic material below each aquaculture site were evident, particularly under the BC’s (Supplementary Table S3). Future investigations would benefit from photo-documentation of the time-course evolution of increasing organic material below study sites due to oyster biodeposits and biofouling of gear. Additionally, from this study we have learned that sulfide concentration data prior to site installation and then gathered throughout the season in 2 or more years at the same site would be tremendously valuable for understanding biogeochemical shifts in sediments underlying oyster aquaculture, whether biodeposits drive sediments toward sulfide concentrations that start to depress the sulfide-sensitive processes of denitrification and anammox, and start to favor DNRA. While N2 production rates also increase over the course of the summer at the control site, this pattern is magnified at each of the treatment sites. The last two months of the growing season during both 2018 and 2019, had significant increases in N2 flux from the sediments under each of the aquaculture treatments compared to the control sediments with the daily flux rates increasing ∼two- to four-fold over the referenced control site (Figure 3). The variability of flux measurements within a particular site likely represents the inherent heterogeneity of the sediments (i.e., presence/absence of bioturbators, disturbances, local concentrations of organic matter, etc.). Additionally, we observe an increase in the carbon content at each site over the growing season (Supplementary Table S3). The addition of carbon to the sediments is constituent with the measured N2 production fluxes. Moreover, the overall patterns are consistent year to year. Flux rates, and carbon and nitrogen content of the sediments at our control site were on par with rates under each of our treatment sites at the beginning of the second aquaculture season (2019) when oysters were returned to the site, indicating that sediments under aquaculture sites in Waquoit Bay returned to control conditions during the winter. Taken together with the molecular data we view this system as primed for nitrogen removal, that is enhanced through the activity of the overlying oyster aquaculture. The activity of the oysters does not change the structure of the underlying microbial but rather increases the activity of the natural community, with individual gear types influencing the balance between different nitrogen cycling processes.
This study examines the effect of different oyster aquaculture methods to stimulate nitrogen removal from the underlying sediments. We show that all three aquaculture methods stimulated nitrogen removal (i.e., greater N2 flux relative to a control site). An increase in the abundance of genes associated with nitrogen cycling was observed under our aquaculture sites which suggests that underlying microbial communities actively respond to changes caused by the deployments. A smaller magnitude seasonal increase in the expression of nitrogen cycling genes at our control site was observed. We acknowledge that our control site could be influenced by our nearby oyster deployments via the tidal cycles that can transport suspended material. However, the shallow depths and east-west fetch of the bay would minimize these influences at our control site location. Additionally, N2 production rates were significantly different between the control site and the different treatment sites, which indicates that nitrogen removal processes are affected by released organic material from the oysters (and possibly accumulation of additional organic material under the BCs). Floating bags and the suspended oyster deployments (OG) can distribute biodeposits over larger areas than BCs, diluting the impact of denitrification and thus nitrogen removal (Lunstrum et al., 2018), but we argue OGs and FBs (to a lesser extent) can also limit DNRA because they increase oxygenation of surface sediments. The consequence of this is that OG and FB (to a lesser extent) methods can limit nitrogen retention in the overlaying sediment in the form of ammonium.
The hydrodynamic setting (water depth, bottom characteristics, exposure to wind and waves), the method of oyster cultivation, and the stocking density of the oysters, can all affect nitrogen cycling via influences on availability of organic matter, nitrate, and O2 (Lunstrum et al., 2018). More work is needed to understand the effect of these factors on net nitrogen removal. Sulfur and sulfur-speciation can affect nitrogen cycling and therefore should be considered prior to the deployment of oysters at potential aquaculture sites where nitrogen removal is a priority. Here, we show that nitrogen removal underneath the different oyster treatments is active and that DNRA, which can be counter-productive to nitrogen removal because it leads to retention of nitrogen in sediments in the form of ammonium, can be influenced by the oyster farming method. The dynamic complexity of these many factors can be the reason behind the diverse results researchers have obtained regarding impact of oyster culturing on nitrogen removal from coastal settings. Monitoring of sulfide and organic content of sediments at aquaculture sites can help stakeholders to predict whether anticipated nitrogen removal rates at their site will follow patterns observed in Waquoit Bay, or whether nitrogen-retaining processes may be favored over time. One way to combat nitrogen retention that should be the topic of further study is the relocation of the aquaculture site in subsequent years to optimize microbially mediated nitrogen removal.
Amplicon and RNA-Seq sequences were deposited into NCBI SRA BioProject # PRJNA579576 under accessions SRR13101311–SRR13101212 and SRR13101827–SRR13101797 respectively.
PM wrote the first version of the draft with contributions from DR and VE. PM, DR, VE, and TS incorporated changes after reviewers suggestions. VE, CM, and DR conceived and planned the experiments. CM and CL installed the aquaculture systems. PM, VE, DB, RC, MC, CF, PG, SL, KP, AP, NS, and DR conducted field and laboratory analyses. TS contributed to statistical analyses. TR was the collaboration lead. All authors contributed to the article and approved the submitted version.
This work was supported by the National Oceanic and Atmospheric Administration and National Estuarine Research Reserve System Science Collaborative, award NAI4NOS4190145 (subaward 3004686666) to DR and VE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Dr. Andrew Solow at WHOI for invaluable advice on applications of statistical analyses to our data. We would also like to acknowledge Dr. Maria G. Pachiadaki (WHOI) for her assistance with metatranscriptome and iTAG analyses and graphics. Dr. Lindsay Hinkle, Adam Ziegler, Fatima Seuffert (Stonehill College), and Nan Trowbridge (NOSAMS, WHOI) for their field assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.633314/full#supplementary-material
Supplementary Figure 1 | Average porewater nitrate (A) and ammonia (B) concentrations in 2018.
Supplementary Figure 2 | Average porewater nitrate (A) and ammonia (B) concentrations in 2019.
Supplementary Figure 3 | Water temperature, salinity, and oxygen measurements at 30 cm depth in 2018 (A) and 2019 (B). Uniform/consistent values were recorded at other depths in both years (Supplementary Table S1).
Supplementary Figure 4 | Hierarchical clustering of the three biological replicates from Control (A) and Floating Bags (B) oyster treatments using the Bray–Curtis dissimilarity index.
Supplementary Figure 5 | Hierarchical clustering of the three biological replicates from the Bottom Cages (A) and the Oyster Gro’ (B) oyster treatments using the Bray–Curtis dissimilarity index.
Supplementary Figure 6 | Average normalized abundance of denitrification genes expressed in the underlying sediments of the three oyster deployments during the biweekly sampling over the 2018 (A–D) and 2019 (A–D) sampling periods.
An, S., and Gardner, W. S. (2002). Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary. Mar. Ecol. Prog. Ser. 237, 41–50. doi: 10.3354/meps237041
Anderson, D. M., Glibert, P. M., and Burkholder, J. M. (2002). Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25, 704–726. doi: 10.1007/bf02804901
Aoyagi, T., Kimura, M., Yamada, N., Navarro, R. R., Itoh, H., Ogata, A., et al. (2015). Dynamic transition of chemolithotrophic sulfur-oxidizing bacteria in response to amendment with nitrate in deposited marine sediments. Front. Microbiol. 6:426. doi: 10.3389/fmicb.2015.00426
Azandégbé, A., Poly, F., Andrieux-Loyer, F., Kérouel, R., Philippon, X., and Nicolas, J. L. (2012). Influence of oyster culture on biogeochemistry and bacterial community structure at the sediment-water interface. FEMS Microbiol. Ecol. 82, 102–117. doi: 10.1111/j.1574-6941.2012.01410.x
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857.
Bowen, J. L., Babbin, A. R., Kearns, P. J., and Ward, B. B. (2014). Connecting the dots: linking nitrogen cycle gene expression to nitrogen fluxes in marine sediment mesocosms. Front. Microbiol. 5:429. doi: 10.3389/fmicb.2014.00429
Bowen, J. L., Kroeger, K. D., Tomasky, G., Pabich, W. J., Cole, M. L., Carmichael, R. H., et al. (2007). A review of land-sea coupling by groundwater discharge of nitrogen to New England estuaries: mechanisms and effects. J. Appl. Geochem. 22, 175–191. doi: 10.1016/j.apgeochem.2006.09.002
Braker, G., Fesefeldt, A., and Witzel, K. P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64, 3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998
Braker, G., Schwarz, J., and Conrad, R. (2010). Influence of temperature on the composition and activity of denitrifying soil communities. FEMS Microbiol Ecol. 73, 134–148.
Bru, D., Sarr, A., and Philippot, L. (2007). Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 73, 5971–5974. doi: 10.1128/aem.00643-07
Brunet, R. C., and Garcia-Gil, L. J. (1996). Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol. Ecol. 21, 131–138. doi: 10.1111/j.1574-6941.1996.tb00340.x
Burgin, A. J., and Hamilton, S. K. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 5, 89–96. doi: 10.1890/1540-9295(2007)5[89:hwotro]2.0.co;2
Caffrey, J., Sloth, N., Kaspar, H., and Blackburn, T. (1993). Effect of organic loading on nitrification and denitrification in a marine sediment microcosm. FEMS Microb. Ecol. 12, 159–167. doi: 10.1111/j.1574-6941.1993.tb00028.x
Caffrey, J. M., Bonaglia, S., and Conley, D. J. (2019). Short exposure to oxygen and sulfide alters nitrification, denitrification, and DNRA activity in seasonally hypoxic estuarine sediments. FEMS Microbiol. Lett. 366:fny288.
Cannon, J., Sanford, R. A., Connor, L., Yang, W. H., and Chee-Sanford, J. (2019). Optimization of PCR primers to detect phylogenetically diverse nrfA genes associated with nitrite ammonification. J. Microb. Meth. 160, 49–59. doi: 10.1016/j.mimet.2019.03.020
Carlsson, M., Engström, P., Lindahl, O., Ljungqvist, L., Petersen, J., Svanberg, L., et al. (2012). Effects of mussel farms on the benthic nitrogen cycle on the Swedish west coast. Aquac. Int. 2, 177–191. doi: 10.3354/aei00039
Carmichael, R. H., Walton, W., and Clark, H. (2012). Bivalve-enhanced nitrogen removal from coastal estuaries. Can. J. Fish. Aquat. Sci. 69, 1131–1149. doi: 10.1139/f2012-057
Chapelle, A., Menesguen, A., Deslous-Paoli, J.-M., Souchu, P., Mazouni, N., Vaquer, A., et al. (2000). Modelling nitrogen, primary production and oxygen in a Mediterranean lagoon: impact of oysters farming and inputs from the watershed. Ecol. Model. 127, 161–181. doi: 10.1016/s0304-3800(99)00206-9
Charette, M. A., Sholkovitz, E. R., and Hansel, C. M. (2005). Trace element cycling in a subterranean estuary: part 1. Geochemistry of the permeable sediments. Geochim. Cosmochim. Acta 69, 2095–2109. doi: 10.1016/j.gca.2004.10.024
Clements, J. C., and Comeau, L. A. (2019). Nitrogen removal potential of shellfish aquaculture harvests in eastern Canada: a comparison of culture methods. Aquac. Rep. 13:100183. doi: 10.1016/j.aqrep.2019.100183
Correll, D. L., Jordan, T. E., and Weller, D. E. (1992). Nutrient flux in a landscape: effects of coastal land use and terrestrial community mosaic on nutrient transport to coastal waters. Estuaries 15, 431–442. doi: 10.2307/1352388
Coskun, ÖK., Özen, V., Wankel, S. D., and Orsi, W. D. (2019). Quantifying population-specific growth in benthic bacterial communities under low oxygen using H218O. ISME J. 13, 1546–1559. doi: 10.1038/s41396-019-0373-4
Dalsgaard, T., Stewart, F. J., Thamdrup, B., De Brabandere, L., Revsbech, N. P., Ulloa, O., et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off Northern Chile. mBio 5:e01966-14.
Dalsgaard, T., Thamdrup, B., and Canfield, D. E. (2005). Anaerobic ammonium oxidation (anammox) in the marine environment. Res. Microbiol. 156, 457–464. doi: 10.1016/j.resmic.2005.01.011
Dedysh, S. N., Kulichevskaya, I. R., Beletsky, A. V., Ivanova, A. A., Rijpstra, I. C., Damsté, S. J. S., et al. (2020). Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst. Appl. Microbiol. 43:126050. doi: 10.1016/j.syapm.2019.126050
Devol, A. H. (2015). Denitrification, anammox, and N2 production in marine sediments. Ann. Rev. Mar. Sci. 7, 403–423. doi: 10.1146/annurev-marine-010213-135040
Dixon, R. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Erler, D. V., Welsh, D. T., Bennet, W. W., Meziane, T., Hubas, C., Nizzoli, D., et al. (2017). The impact of suspended oyster farming on nitrogen cycling and nitrous oxide production in a sub-tropical Australian estuary. Estuar. Coast. Shelf Sci. 192, 117–127. doi: 10.1016/j.ecss.2017.05.007
Feinman, S. G., Farah, Y. R., Bauer, J. M., and Bowen, J. L. (2018). The influence of oyster farming on sediment bacterial communities. Estuar. Coast. 41, 800–814. doi: 10.1007/s12237-017-0301-7
Fernández-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., González, J. M., et al. (2013). Ecology of marine bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Glibert, P. M., Anderson, D. A., Gentien, P., Granéli, E., and Sellner, K. G. (2005). The global, complex phenomena of harmful algal blooms. J. Oceanogr. 18, 136–147. doi: 10.5670/oceanog.2005.49
Glibert, P. M., Heil, C. A., Wilkerson, F. P., and Dugdale, R. C. (2018). “Nutrients and harmful algal blooms: dynamic kinetics and flexible nutrition,” in Global Ecology and Oceanography of Harmful Algal Blooms. Ecological Studies (Analysis and Synthesis), eds P. Glibert, E. Berdalet, M. Burford, G. Pitcher, and M. Zhou (Cham: Springer Nature). doi: 10.1007/978-3-319-70069-4_6
Graf, D. R. H., Jones, C. M., and Hallin, S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9:e114118. doi: 10.1371/journal.pone.0114118
Henry, S., Baudouin, E., Lopez-Gutierrez, J. C., Martin-Laurent, F., Brauman, A., and Philippot, L. (2004). Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59, 327–335. doi: 10.1016/j.mimet.2004.07.002
Higgins, C., Tobias, C., Piehler, M., Smyth, A., Dame, R., Stephenson, K., et al. (2013). Effect of aquacultured oyster biodeposition on sediment N2 production in Chesapeake Bay. Mar. Ecol. Progr. Ser. 473, 7–27. doi: 10.3354/meps10062
Hoellein, T. J., and Zarnoch, C. B. (2014). Oysters in an Eastern Estuary. Bull. Ecol. Soc. Am. 95, 82–84. doi: 10.1890/0012-9623-95.1.82
Howarth, R., Chan, F., Conley, D. J., Garnier, J., Doney, S. C., Marino, R., et al. (2011). Coupled biogeochemical cycles: eutrophication and hypoxia in temperate estuaries and coastal marine ecosystems. Front. Ecol. Environ. 9, 18–26. doi: 10.1890/100008
Howarth, R. W. (1988). Nutrient limitation of net primary production in marine ecosystems. Annu. Rev. Ecol. Evol. Syst. 19, 89–110. doi: 10.1146/annurev.es.19.110188.000513
Hu, Z., Wessels, H. J. C. T., van Alen, T., Jetten, M. S. M., and Kartal, B. (2019). Nitric oxide-dependent anaerobic ammonium oxidation. Nat. Commun. 10:1244.
Humphries, A. T., Ayvazian, S. G., Carey, J. C., Hancock, B. T., Grabbert, S., Cobb, D., et al. (2016). Directly measured denitrification reveals oyster aquaculture and restored oyster reefs remove nitrogen at comparable high rates. Front. Mar. Sci. 3:74. doi: 10.3389/fmars.2016.00074
Imhoff, J. F. (2005). “Order I. Chromatiales ord. nov,” in Bergey’s Manual of Systematic Bacteriology, (The Proteobacteria), Part B (The Gamma proteobacteria), 2nd Edn, Vol. 2, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. T. Garrity (New York: Springer), 1–3. doi: 10.1002/9781118960608.obm00096
Jensen, K., Sloth, N. P., Rixaard-Petersen, N., Rysgaard, S., and Revsbech, N. P. (1994). Regulationof the coupling between nitrification and denitrification in sediment as studied with a microsensor for nitrate. Appl. Environ. Microbial. 60, 2094–2100.
Jensen, M. M., Kuypers, M. M., Gaute, L., and Thamdrup, B. (2008). Rates and regulation of anaerobic ammonium oxidation and denitrification in the Black Sea. Limnol. Oceanogr. 53, 23–36. doi: 10.4319/lo.2008.53.1.0023
Jordan, S. J. (1987). Sedimentation and Remineralization Associated with Biodeposition by the American Oyster Crassostrea virginica (Gmelin). Doctoral thesis, University of Maryland, College Park, MD.
Jørgensen, B. B., Findlay, A. J., and Pellerin, A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10:849. doi: 10.3389/fmicb.2019.00849
Joye, S. B., and Anderson, I. C. (2008). “Nitrogen cycling in coastal sediments,” in Nitrogen in the Marine Environment, 2nd Edn, eds D. G. Capone, D. A. Bronk, M. R. Mulholland, and E. J. Carpenter (Amsterdam: Academic Press), 868–915.
Kartal, B., and Keltjens, J. T. (2016). Anammox biochemistry: a tale of Heme c Proteins. Trends Biochem Sci. 41, 998–1011. doi: 10.1016/j.tibs.2016.08.015
Kellogg, L. M., Cornwell, J. C., Owens, M. S., and Paynter, K. T. (2013). Denitrification and nutrient assimilation on a restored oyster reef. Mar. Ecol. Prog. Ser. 480, 1–19. doi: 10.3354/meps10331
Klier, J., Dellwig, O., Leipe, T., Jürgens, K., and Herlemann, D. (2018). Benthic bacterial community composition in the oligohaline-marine transition of surface sediments in the Baltic Sea based on rRNA analysis. Front. Microbiol. 9:236. doi: 10.3389/fmicb.2018.00236
Krishnani, K. K., Gopikrishna, G., Pillai, S. M., and Gupta, B. P. (2010). Abundance of sulphur-oxidizing bacteria in coastal aquaculture using soxB gene analyses. Aquac. Res. 41, 1290–1301. doi: 10.1111/j.1365-2109.2009.02415.x
Kroeger, K. D., Cole, M. L., York, J. K., and Valiela, I. (2006). Nitrogen loads to estuaries from waste water plumes: modeling and isotopic approaches. Ground Water 44, 188–200. doi: 10.1111/j.1745-6584.2005.00130.x
Li, M., and Gu, J.-D. (2011). Advances in methods for detection of anaerobic ammonium oxidizing (anammox) bacteria. Appl. Microbiol. Biotechnol. 90, 1241–1252. doi: 10.1007/s00253-011-3230-6
Llorens-Marès, T., Yooseph, S., Goll, J., Hoffman, J., Vila-Costa, M., Borrego, C. M., et al. (2015). Connecting biodiversity and potential functional role in modern euxinic environments by microbial metagenomics. ISME J. 9, 1648–1661. doi: 10.1038/ismej.2014.254
Lunstrum, A., McGlathery, K., and Smyth, A. (2018). Oyster (Crassostrea virginica) aquaculture shifts sediment nitrogen processes toward mineralization over denitrification. Estuar. Coast. 41, 1130–1146. doi: 10.1007/s12237-017-0327-x
Maio, C. V., Donnelly, J. P., Sullivan, R., Madsen, S. M., Weidman, C. R., Gontz, A. M., et al. (2016). Sediment dynamics and hydrographic conditions during storm passage, Waquoit Bay, Massachusetts. Mar. Geol. 381, 67–86. doi: 10.1016/j.margeo.2016.07.004
Marchant, H. K., Ahmerkamp, S., Lavik, G., Tegetmeyer, H. E., Graf, J., Klatt, J. M., et al. (2017). Denitrifying community in coastal sediments performs aerobic and anaerobic respiration simultaneously. ISME J. 11, 1799–1812. doi: 10.1038/ismej.2017.51
Mazouni, N., Gaertner, J.-C., Deslous-Paoli, J.-M., Landrein, S., and Geringer d’Oedenberg, M. (1996). Nutrient and oxygen exchanges at the water-sediment interface in a shellfish farming lagoon (Thau, France). J. Exp. Mar. Biol. Ecol. 205, 91–113. doi: 10.1016/s0022-0981(96)02594-4
Mohan, S. B., and Cole, J. A. (2007). “The dissimilatory reduction of nitrate to ammonia by anaerobic bacteria,” in Biology of the Nitrogen Cycle, eds H. Bohe, S. Ferguson, and W. E. Newton (Amsterdam: Elsevier), 93. doi: 10.1016/b978-044452857-5.50008-4
Mortazavi, B., Ortmann, A. C., Wang, L., and Bernard, R. (2015). Evaluating the impact of oyster (Crassostrea virginica) gardening on sediment nitrogen cycling in a subtropical estuary. Bull. Mar. Sci. 91, 323–341. doi: 10.5343/bms.2014.1060
Mosier, A. C., and Francis, C. A. (2010). Denitrifier abundance and activity across the San Francisco Bay estuary. Environ. Microb. Rep. 2, 667–676. doi: 10.1111/j.1758-2229.2010.00156.x
Mukhopadhyaya, P. N., Deb, C., Lahiri, C., and Roy, P. (2000). A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium. J. Bacteriol. 182, 4278–4287. doi: 10.1128/jb.182.15.4278-4287.2000
Murphy, A. E., Anderson, I. C., Smyth, A. R., Song, B., and Luckenbach, M. W. (2016). Microbial nitrogen processing in hard clam (Mercenaria mercenaria) aquaculture sediments: the relative importance of denitrification and dissimilatory nitrate reduction to ammonium (DNRA). Limn. Ocean. 61, 1589–1604. doi: 10.1002/lno.10305
Newell, R. I. E., Cornwell, J. C., and Owens, M. S. (2002). Influence of simulated bivalve biodeposition and microphytobenthos on sediment nitrogen dynamics: a laboratory study. Limn. Ocean. 47, 1367–1379. doi: 10.4319/lo.2002.47.5.1367
Newell, R. I. E., Fisher, T. R., Holyoke, R. R., and Cornwell, J. C. (2005). “Influence of eastern oysters on nitrogen and phosphorus regeneration in Chesapeake Bay, USA,” in The Comparative Roles of Suspension-Feeders in Ecosystems, NATO Science Series IV: Earth and Environmental Series, Vol. 47, eds R. F. Dame and S. Olenin (Dordrecht: Springer).
Newell, R. I. E., and Jordan, S. J. (1983). Preferential ingestion of organic material by the American Oyster Crassostrea virginica. Mar. Ecol. Prog. Ser. 13, 47–53. doi: 10.3354/meps013047
Nizzoli, D., Welsh, D. T., Fano, E. A., and Viaroli, P. (2006). Impact of clam and mussel farming on benthic metabolism and nitrogen cycling, with emphasis on nitrate reduction pathways. Mar. Ecol. Progr. Ser. 315, 151–165. doi: 10.3354/meps315151
Orellana, L. H., Rodriguez, R. L. M., Higgins, S., Chee-Sanford, J. C., Sanford, R. A., Ritalahti, K. M., et al. (2014). Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e001193-14.
Pai, S.-C., Yang, C.-C., and Riley, J. P. (1990). Formation kinetics of the pink azo dye in the determination of nitrite in natural waters. Anal. Chim. Acta 232, 345–349. doi: 10.1016/s0003-2670(00)81252-0
Parada, A., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pohlner, M., Dlugosch, L., Wemheuer, B., Mills, H., Engelen, B., and Reese, B. K. (2019). The majority of active Rhodobacteraceae in marine sediments belong to uncultured genera: a molecular approach to link their distribution to environmental conditions. Front. Microbiol. 10:659. doi: 10.3389/fmicb.2019.00659
Porubsky, W. P., Weston, N. B., and Joye, S. B. (2009). Benthic metabolism and the fate of dissolved inorganic nitrogen in intertidal sediments. Estuar. Coast. Shelf. Sci. 83, 392–402. doi: 10.1016/j.ecss.2009.04.012
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596.
Rabalais, N. N., Cai, W.-J., Carstensen, J., Conley, D. J., Fry, B., Hu, X., et al. (2014). Eutrophication-driven deoxygenation in the coastal ocean. J. Oceanogr. 27, 172–183. doi: 10.5670/oceanog.2014.21
Rasigraf, O., Schmitt, J., Jetten, M., and Lüke, C. (2017). Metagenomic potential for and diversity of N-cycle driving microorganisms in the Bothnian Sea sediment. Microbiol. Open 6:e00475. doi: 10.1002/mbo3.475
Ray, N. E., Al-Haj, A. N., and Fulweiler, R. W. (2020). Sediment biogeochemistry along an oyster aquaculture chronosequence. Mar. Ecol. Prog. Ser. 646, 13–27. doi: 10.3354/meps13377
Robinson, G., Caldwell, G. S., Wade, M. J., Free, A., Jones, C., and Stead, S. M. (2016). Profiling bacterial communities associated with sediment-based aquaculture bioremediation systems under contrasting redox regimes. Sci. Rep. 6:38850.
Sanford, R. A., Wagner, D. D., Wu, Q., Chee-Sanford, J. C., Thomas, S. H., Cruz-García, C., et al. (2012). Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 109, 19709–19714. doi: 10.1073/pnas.1211238109
Smith, V. H. (2003). Eutrophication of freshwater and coastal marine ecosystems a global problem. Environ. Sci. Pollut. Res. 10, 126–139. doi: 10.1065/espr2002.12.142
Smyth, A. R., Geraldi, N. R., and Piehler, M. F. (2013). Oyster-mediated benthic-pelagic coupling modifies nitrogen pools and processes. Mar. Ecol. Prog. Ser. 493, 23–30. doi: 10.3354/meps10516
Solorzano, L. (1969). Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14, 799–801. doi: 10.4319/lo.1969.14.5.0799
Talbot, J. M., Kroeger, K. D., Rago, A., Allen, M. C., Charette, M. A., and Abraham, D. M. (2003). Nitrogen flux and speciation through the subterranean estuary of Waquoit Bay, Massachusetts. Biol. Bull. Mar. Biol. Lab. Woods Hole 205:246.
Tiedje, J. M. (1988). “Ecology of denitrification and dissimilatory nitrate reduction to ammonium,” in Biology of Anaerobic Microorganisms, ed. A. J. B. Zehnder (New York, NY: John Wiley & Sons), 179–244.
Tiquia, S. M., Masson, S. A., and Devol, A. (2006). Vertical distribution of nitrite reductase genes (nirS) in continental margin sediments of the Gulf of Mexico. FEMS Microbiol. Ecol. 58, 464–475. doi: 10.1111/j.1574-6941.2006.00173.x
Tourna, M., Maclean, P., Condron, L., O’Callaghan, M., and Wakelin, S. A. (2014). Links between sulphur oxidation and sulphur-oxidising bacteria abundance and diversity in soil microcosms based on soxB functional gene analysis. FEMS Microbiol. Ecol. 3, 538–549. doi: 10.1111/1574-6941.12323
Trimmer, M., and Engström, P. (2011). “Distribution, activity, and ecology of anammox bacteria in aquatic environments,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (New York, NY: ASM Press), 201–236. doi: 10.1128/9781555817145.ch9
Valiela, I., McClelland, J., Hauxwell, J., Behr, P. J., Hersh, D., and Foreman, K. (1997). Macroalgal blooms in shallow estuaries: controls and ecophysiological and ecosystem consequences. Limnol. Oceanogr. 42, 1105–1118. doi: 10.4319/lo.1997.42.5_part_2.1105
van den Berg, E., van Dongen, U., Abbas, B., and van Loosdrecht, M. M. C. (2015). Enrichment of DNRA bacteria in a continuous culture. ISME J. 9, 2153–2161. doi: 10.1038/ismej.2015.26
van den Berg, E. M., Elisário, M. P., Kuenen, J. G., Kleerebezem, R., and van Loosdrecht, M. C. M. (2017). Fermentative bacteria influence the competition between denitrifiers and DNRA bacteria. Front. Microbiol. 8:1684. doi: 10.3389/fmicb.2017.01684
Vieillard, A. (2017). Impacts of New England Oyster Aquaculture on Sediment Nitrogen Cycling: Implications for Nitrogen Removal and Retention. Master thesis, University of Connecticut, Storrs CT.
Wallenstein, M. D., Myrold, D. D., Firestone, M., and Voytek, M. (2006). Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16, 2143–2152. doi: 10.1890/1051-0761(2006)016[2143:ecodca]2.0.co;2
Wang, H., Tseng, C. P., and Gunsalus, R. P. (1999). The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181, 5303–5308. doi: 10.1128/jb.181.17.5303-5308.1999
Ward, B. B., and O’Mullan, G. D. (2002). Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing gamma-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl. Environ. Microbiol. 68, 4153–4157. doi: 10.1128/aem.68.8.4153-4157.2002
Zhang, J.-Z., and Fischer, C. J. (2006). A simplified resorcinol method for direct spectrophotometric determination of nitrate in seawater. Mar. Chem. 99, 220–226. doi: 10.1016/j.marchem.2005.09.008
Keywords: nitrogen removal, oyster cultures, denitrification, anammox, DNRA
Citation: Mara P, Edgcomb VP, Sehein TR, Beaudoin D, Martinsen C, Lovely C, Belcher B, Cox R, Curran M, Farnan C, Giannini P, Lott S, Paquette K, Pinckney A, Schafer N, Surgeon-Rogers T-M and Rogers DR (2021) Comparison of Oyster Aquaculture Methods and Their Potential to Enhance Microbial Nitrogen Removal From Coastal Ecosystems. Front. Mar. Sci. 8:633314. doi: 10.3389/fmars.2021.633314
Received: 25 November 2020; Accepted: 11 February 2021;
Published: 24 March 2021.
Edited by:
Konstantinos Ar. Kormas, University of Thessaly, GreeceReviewed by:
Élise Lacoste, University of French Polynesia, French PolynesiaCopyright © 2021 Mara, Edgcomb, Sehein, Beaudoin, Martinsen, Lovely, Belcher, Cox, Curran, Farnan, Giannini, Lott, Paquette, Pinckney, Schafer, Surgeon-Rogers and Rogers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel R. Rogers, ZHJvZ2VyczJAc3RvbmVoaWxsLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.