- 1Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
- 2Department of Biology, San Diego State University, San Diego, CA, United States
- 3Department of Oceanography and Sea Grant College Program, Daniel K. Inouye Center for Microbial Oceanography: Research and Education, School of Ocean and Earth Science and Technology, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 4Department of Marine Microbiology and Biogeochemistry, Royal Netherlands Institute for Sea Research (NIOZ), Den Burg, Netherlands
- 5Collaborative Mass Spectrometry Innovation Center, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA, United States
The rapidly advancing field of metabolomics encompasses a diverse suite of powerful analytical and bioinformatic tools that can help to reveal the diversity and activity of chemical compounds in individual organisms, species interactions, and entire ecosystems. In this perspective we use examples from studies of coral reefs to illustrate ways in which metabolomics has been and can be applied to understand coastal ecosystems. Examples of new insights that can be provided by metabolomics include resolving metabolite exchange between plants, animals and their microbiota, identifying the relevant metabolite exchanges associated with the onset and maintenance of diverse, microbial mutualisms characterizing unknown molecules that act as cues in coral, reproduction, or defining the suites of compounds involved in coral-algal competition and microbialization of algal-dominated ecosystems. Here we outline sampling, analytical and informatic methods that marine biologists and ecologists can apply to understand the role of chemical processes in ecosystems, with a focus on open access data analysis workflows and democratized databases. Finally, we demonstrate how these metabolomics tools and bioinformatics approaches can provide scientists the opportunity to map detailed metabolic inventories and dynamics for a holistic view of the relationships among reef organisms, their symbionts and their surrounding marine environment.
Introduction
Coastal ecosystems where benthic and pelagic habitats intermingle and the complex ecological dynamics of each biome interact: benthic suspension feeders process the plankton while benthic primary producers exude photosynthate, each variously scouring light, carbon, nutrients and oxygen according to their metabolic needs. Diverse plants, algae, vertebrates, invertebrates, protists, bacteria and viruses bloom at this ecotone, thriving on the physical and chemical complexity born from the interface of land and water. So too does the chemical diversity and interactivity in this environment bloom. Understanding the ecology of coastal organisms demands a window into the cacophony of metabolic resources that perfuse and surround the denizens of the reef: the metabolome.
Over the last two decades, analytical tools to detect, distinguish, characterize, and quantify complex chemotypes from a single environmental sample have emerged. The most prominent of these are ultra-high resolution mass spectrometry (Dittmar and Paeng, 2009; Hawkes et al., 2016; Osterholz et al., 2016) and tandem mass spectrometry [MS/MS, the latter often coupled to various types of liquid and gas chromatographic separation methods (LC, GC)]. Improvements in sample throughput, data management and bioinformatics make it possible to track these compounds increasing and decreasing in abundance through time and space in an open market of chemical dynamics (Steen et al., 2020). These advances are not trivial; in fact, this decade marks the rapid and exponential arrival of cost efficient, interpretable, and deployable untargeted mass spectrometric experimentation (da Silva et al., 2015). It is the goal of this article to demonstrate to coastal ecologists how metabolomics can inform organismal physiology and ecology as well as reveal the biochemical roles of organisms in ecosystems. Herein we outline approaches that marine organismal ecologists, coastal ecosystem scientists and resource managers can employ to sample, query and interpret the metabolome of organisms and ecosystems at the interface of benthic and pelagic habitats, using coral reef systems as one concrete example.
Holobiont Metabolomics: Identifying Drivers of Host-Symbiont Physiology, Symbiosis, and Disease
In marine holobionts, environmental fluctuations and disturbances influence relationships between multicellular hosts and symbionts, with far reaching consequences ranging from physiological variations to dysbiosis and disease (Apprill, 2017). Coral growth and calcification, for example, are influenced by the performance of algal symbionts (Symbiodiniaceae) which are affected by light, temperature and nutrient availability. In addition to the zooxanthellae symbiosis, the coral colony is host to hundreds, if not thousands, of other viral, bacterial, archaeal, protist, and fungal symbionts that comprise the coral holobiont (Rohwer et al., 2002; Knowlton and Rohwer, 2003; Casas et al., 2004; Kellogg, 2004; Wegley et al., 2004, 2007; Marhaver et al., 2008; Bourne et al., 2016; Ainsworth et al., 2017; Thurber et al., 2017). The diversity and complexity of these host-microbe relationships has become increasingly clear, yet no shortage of questions have emerged regarding the establishment and coexistence of these vital partnerships (Ainsworth and Gates, 2016). The current knowledge about coral microbiomes can be combined with metabolomic studies to reveal how microbial symbionts protect host fitness by conferring resistance to stressors, recovery from bleaching, and prevention of diseases.
A research priority after two decades of genomic research on coral holobionts is understanding the extent to which metabolic complementation occurs between hosts and their microbial symbionts (Gerardo, 2013; Shoguchi et al., 2013). The genomes of host and symbionts frequently lack key genes in important metabolic pathways, indicating an interdependence on the mutualistic exchange of metabolic intermediates between holobiont members (Ferrier-Pagès et al., 2005; Shinzato et al., 2011; Matthews et al., 2018). Recent studies have used metabolomic approaches to explore how functionally divergent endosymbionts impact the nutritional resources and fitness of their hosts (Matthews et al., 2018, 2020a) and careful work at the scale of bacterial-epithelial interactions has demonstrated how even taxonomically homogenous endosymbionts may facilitate spatial heterogeneity in metabolite profiles (Geier et al., 2020). Hermatypic corals colonized by two different species of Symbiodiniaceae exhibit similar metabolite profiles in both host and symbiont, illustrating the potential for more thermally robust strains to colonize diverse hosts without negatively impacting resource allocation (Matthews et al., 2020a). A variety of tools employed to examine the metabolomes of thermally stressed corals have demonstrated significant differences between stressed and control corals (Sogin et al., 2016; Farag et al., 2018; Williams et al., 2021) and Symbiodiniaceae (Petrou et al., 2018; Lawson et al., 2019) while others have demonstrated changes in metabolite exchange between host and symbiont (Hillyer et al., 2017, 2018) or phenotypic differences in whole tissue metabolomes (Roach et al., 2021) or symbiocyte acid-base homeostasis (Innis et al., 2021). In a recent review, Matthews et al. (2020b) posit that Bacteria-Symbiodiniaceae metabolomic interactions that regulate stability between host and algal symbionts are crucial for holobiont resilience to environmental stress, extending areas of investigation into more complex aspects of intra-holobiont metabolomics.
Imbalances in nutrient availability can destabilize symbiotic partnerships with algal symbionts (Wiedenmann et al., 2013) as well as commensal bacteria (Klinges et al., 2019) causing lower bleaching thresholds and higher disease incidence, respectively. Diazotrophic Bacteria and ammonia-oxidizing Archaea associated with corals have been described widely (Beman et al., 2007; Lesser et al., 2007; Pernice et al., 2012; Rädecker et al., 2015) but metabolomics opens up the capability to investigate the exchange of organic nutrient sources within coral holobionts, providing means to examine myriad questions in the field related to the exchange of metabolomic products between symbionts (Figure 1A). The exchange of metabolites in competitive interactions between corals and macroalgae have been implicated in altering the growth of coral-associated microbiota, inducing dysbioses that potentially initiate proliferation of alternative communities that may diminish holobiont fitness, ultimately reducing competitive dominance at coral-algal interfaces (Roach et al., 2020).
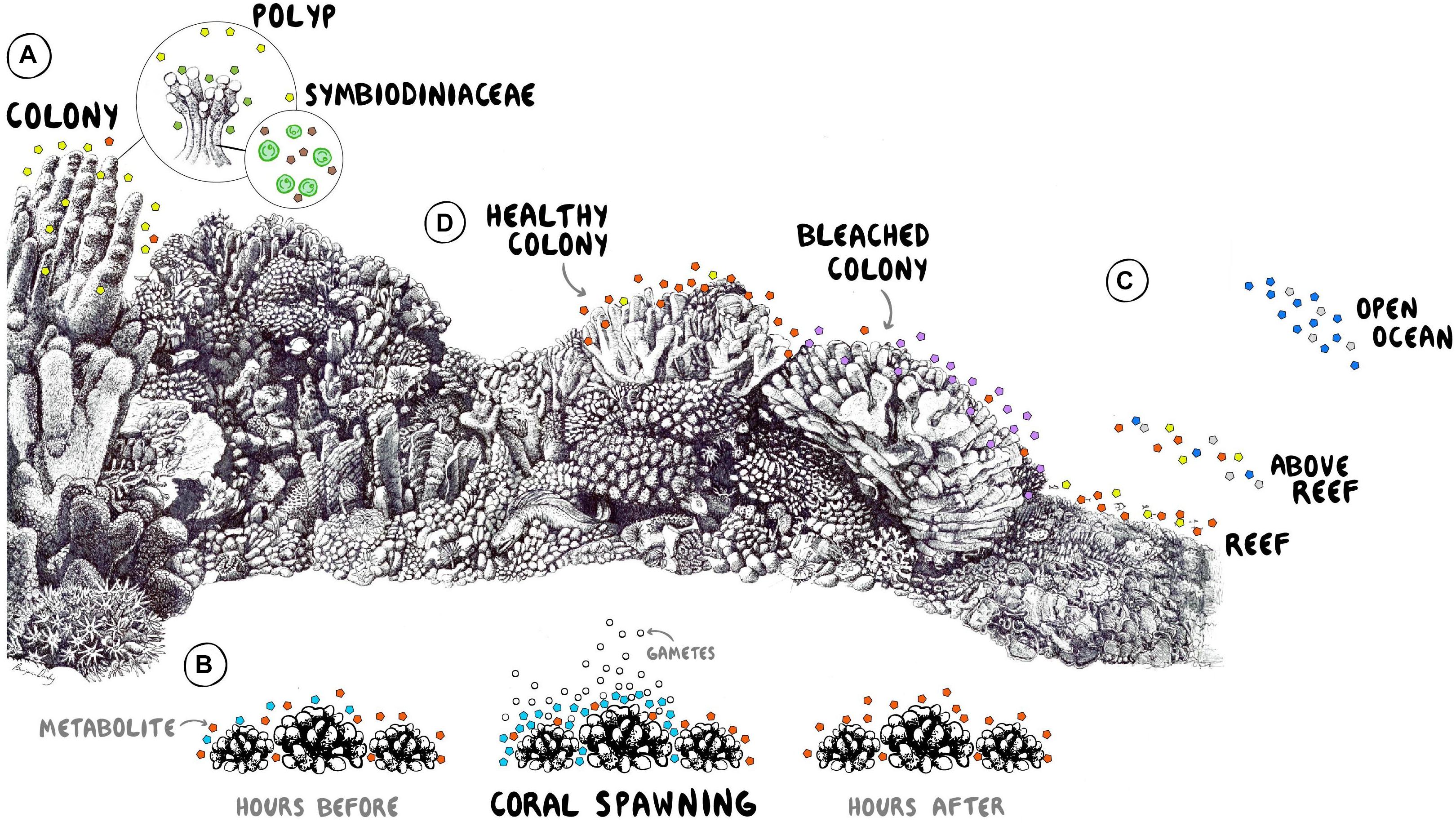
Figure 1. Conceptual illustration of metabolomic studies on coral reefs. (A) Determination of metabolites involved in the recruitment, establishment, and maintenance of symbionts. (B) Identification of chemical signals facilitating spawning synchrony and mating success. (C) Spatial comparisons of exometabolomes across coral reef habitats and the adjacent ocean waters. (D) Characterization of exometabolite stress biomarkers during a warming event. Coral reef drawing by Ben Darby. Graphic design by Shayle Matsuda.
Beyond the metabolomic diversity associated with the microbiome, unique host genotypes also exhibit distinct metabolite profiles that reflect physiological state and can reveal mechanisms of resilience (Lohr et al., 2019b) and photoacclimation (Lohr et al., 2019a). Different coral genera have been shown to exhibit not only unique metabolomes but also distinct profiles of metabolite modifications, hinting at a more nuanced aspect of metabolomic characterization with potential to identify both compounds and reactions that distinguish metabolomes (Hartmann et al., 2017). Comparative metabolomics of coral holobionts interacting with corals or algae demonstrated distinct metabolomes activated by non-self interactions (Quinn et al., 2016), emphasizing that host metabolomes alone may add to the suite of tools we can use for assessing organismal physiology and phenotype.
The Holobiont-Water Interface: Exometabolites Regulate Organismal and Host-Symbiont Interactions
Communication among marine organisms is primarily dictated by chemical exchange (Hay, 2009). While a diversity of molecules used for communication have been discovered across the tree of life, most signaling compounds that mediate reproduction, larval settlement, symbiont recruitment and pathogen defense remain wholly uncharacterized (Levitan et al., 2011). Organismal interactions ranging from mutualistic to commensal to antagonistic or parasitic are chemically mediated through the water by e.g., allelopathic chemicals, toxins, morphogens, and chemoattractants for symbionts and microbes (Barott and Rohwer, 2012; Sneed et al., 2014; Tout et al., 2015).
In marine organismal ecology, metabolomics is particularly powerful for revealing otherwise invisible signaling systems and uncovering candidate molecules involved therein. For example, the reproductive signaling compounds produced by economically important marine organisms such as corals are largely unknown. Corals worldwide suffer from decreased reproduction due to Allee effects and recruitment failure (Knowlton, 2001; Vermeij et al., 2011), and recent evidence shows that several coral species from the Red Sea have lost mating synchrony (Fogarty and Marhaver, 2019; Shlesinger and Loya, 2019). A better mechanistic understanding of the chemical cues that synchronize coral spawning could potentially open new avenues for intervention, whether in the field or in captive breeding programs. In hermaphroditic, mass-spawning corals, it has been posited that daylight and sunset times affect the coordination and initiation of the early stages of spawning (including the setting or “staging” of egg-sperm bundles in coral mouths) whereas the precise timing of the release of these gamete bundles appears to be driven by a local threshold concentration of pheromones within a localized patch of coral colonies (Levitan et al., 2011). Metabolomic tools can help to characterize these pheromone systems and determine which coral hormones induce conspecific neighbors to spawn. Figure 1B conceptually illustrates how specific compounds can be distinguished from spawning colonies of the grooved brain coral Diploria in Curacao (Arts and Haas, unpublished data), demonstrating the potential for untargeted metabolomic studies to identify putative cues involved in reproduction and synchronous spawning.
After spawning and fertilization occur, the behavior of coral larvae and settled juveniles is also influenced by local chemical cues which can either induce or suppress larval settlement, metamorphosis, polyp division and growth (Kuffner et al., 2006; Hadfield, 2011; Tebben et al., 2011; Sneed et al., 2014). Recent advances in gamete collection, in vitro fertilization, and larval propagation have created the potential for larger-scale restoration efforts to repopulate reefs with coral juveniles (e.g., Chamberland et al., 2017a,b; van Oppen et al., 2017) yet post-settlement survival remains a major bottleneck to scale and success (Boström-Einarsson et al., 2020; Randall et al., 2020). A mechanistic understanding of the biochemistry that underpins coral settlement and survival can be accomplished by identifying specific metabolites involved in these processes; this could help drive the development of novel settlement substrates and grow-out methods.
Other important metabolic dynamics at the holobiont-water interface include the biochemical strategies used by juvenile coral recruits to acquire and regulate their extra- and intracellular symbionts, which are obtained either vertically from parent colonies or horizontally from the environment; these symbionts may help corals resist pathogens and establish competitive dominance on the reef benthos. For example, the acquisition of Alphaproteobacterial symbionts in the family Rhodobacteraceae by coral offspring both in brooding (Sharp et al., 2012) and broadcast spawning corals (Apprill et al., 2009; Ceh et al., 2013) suggests a key role for these symbionts, but, thus far, we have only a limited understanding of the benefits that these symbionts may confer to their hosts (Sharp and Ritchie, 2012). There is evidence that some coral symbionts produce protective metabolites such as antimicrobial peptides (Ritchie, 2006) and quorum-quenching molecules (Teplitski and Ritchie, 2009; Ma et al., 2018) that could both serve to protect coral spawn and larvae against uncontrolled bacterial colonization. Other bacterial symbionts promote the coral-Symbiodiniaceae symbiosis by supplying vitamin B12, an exogenously acquired cofactor essential for de novo synthesis of methionine (Croft et al., 2005; Lawson et al., 2018). A number of metabolomic tools could help to illuminate the biochemical mechanisms that underly these species interactions. For example, untargeted characterization of metabolites from bacterial isolates collected during spawning or targeted examination of egg bundles and newly developed larvae could help to resolve the protective roles of the microbes associated with each stage of coral reproduction and survival (Thompson et al., 2015).
Experimental evidence has shown that chemical gradients play an important role in bacterial chemotaxis and the establishment of coral symbioses. Specific amino acids (e.g., tryptophan and aspartic acid), nitrogen sources, and dimethylsulfoniopropionate (DMSP) all elicit chemoattractant behavior by natural assemblages of coral-associated bacteria (Tout et al., 2015). These chemotactic cues are presumed to attract beneficial taxa to the coral microbiome, although these gradients may also serve as signals for pathogens to locate coral hosts or even serve as indicators of stress or susceptibility. Compounds such as DMSP are regulators of pathogenesis; for example, the infochemical DMSP stimulates the chemotaxis of the coral pathogen Vibrio coralliilyticus toward its coral host (Garren et al., 2014; Sogin et al., 2016; Farag et al., 2018). When corals are heat stressed, concentrations of DMSP are elevated, promoting even greater pathogen recruitment toward susceptible hosts; combined with the release of temperature-regulated virulence factors, this mechanism rapidly promotes coral disease states (Boroujerdi et al., 2009; Kimes et al., 2012; Garren et al., 2014).
The Water Column Exometabolome: Complex Mixtures of Biochemicals Influence Acquisition, Export, Distribution, and Sequestration of Carbon and Nutrients
Highly complex and productive coral reef biomes thrive in regions that are depleted in mineral nutrients. Tightly coupled exchanges and efficient recycling of available resources are therefore crucial. Organic matter provided by primary producers is one of the main vectors for the transfer of energy that fuels the entire reef system. Autotrophic holobionts like corals, sponges, or algae, and even microbes exude a significant proportion of carbon fixed through photosynthesis as dissolved organic matter (DOM) into the surrounding reef waters (Haas et al., 2011, 2013). These exudates are rich substrates for microbial growth and remineralization (Haas et al., 2011; Nelson et al., 2013) and the coupled processes of production and consumption potentially establish a significant flux of organic nutrient recycling. Holobionts have been shown to establish a sphere of microbial associates that extends beyond the narrow boundary layers, variously termed the aurabiome (Walsh et al., 2017) or ecosphere (Weber et al., 2019), that is likely driven by the organic milieu exuded into the surrounding water column by reef primary producers (Figure 1C). Seminal work has also shown distinct metabolomic gradients in this sphere of influence that differ between diseased and healthy corals and contain suites of compounds associated with animal-microbe interactions (Ochsenkühn et al., 2018). Establishing the exometabolome of reef ecosystems will unveil a new analytical window to understand their persistence, their ability to support a productive fishery and ultimately their resilience to both local and global anthropogenic threats such as nutrient pollution, deoxygenation and warming, acidification, respectively.
Extensive research has illustrated the multiple effects of DOM from different exometabolome sources in coral reef ecosystems (Haas et al., 2011; Rix et al., 2017; Kelly et al., 2018, 2019). For instance, exudates derived from corals promote recycling into the benthic community, reducing the loss of energy and nutrients from the reef ecosystem (Wild et al., 2004). Coral exudates potentially foster a distinct microbial community throughout the reef waters enriched in more oligotrophic taxa with higher growth efficiencies (Nelson et al., 2013) that can potentially increase the net productivity of the entire system (Haas et al., 2013) in part through suspension feeding on bacterioplankton (McNally et al., 2017). In contrast, the exometabolome of fleshy algae has been shown to foster less efficient and potentially pathogenic bacterioplankton communities (Nelson et al., 2013) and algal dominated reefs are commonly associated with increased microbial biomass (Dinsdale et al., 2008; Bruce et al., 2012; McDole et al., 2012), reduced trophic transfer to higher trophic levels (Silveira et al., 2015) and higher abundances of copiotrophic bacteria (Haas et al., 2016), leading to greater respiration of organic resources and contributing to the overall degradation of the coral reef ecosystem. The influence of emergent sponges on DOM dynamics in coral reefs has also been shown to be instrumental via the sponge loop (De Goeij et al., 2013, 2017; Silveira et al., 2015). While almost all these studies emphasize the importance of the source and composition of DOM in supporting the energetic budgets of reef ecosystems, very few have chemically characterized coral reef exometabolomes (e.g., Haas and Wild, 2010; Nelson et al., 2013; Fiore et al., 2017; Letourneau et al., 2020; Weber et al., 2020; Olinger et al., 2021). Further characterization of DOM exudates is required for elucidating the detailed mechanisms behind these selective processes and could provide insights into enhanced coral growth versus fleshy macroalgae competition and may even guide new conservation approaches. Because our working group and others have reported preliminary evidence of significant changes in microbial activity during warming and deoxygenation events (Johnson et al., in press; Quinlan, unpublished data) and specific coral-derived metabolites have been characterized during bleaching (Sogin et al., 2016; Farag et al., 2018; Williams et al., 2021), we anticipate that stress biomarkers can be measured from reef waters in response to environmental disturbances, providing a new avenue for early detection and monitoring (Figure 1D).
Another vital area where characterization of dissolved metabolites can rapidly advance coastal ecosystem ecology is in the realm of anthropogenic pollutants. Terrigenous sources of pesticides and pesticide breakdown products (DDT, DDE, glyphosates, benzamides, imidazolinones) are widespread, and little is known of the distribution or transformation of these compounds in the coastal marine environment (Mackintosh et al., 2016). Additionally, cleaning products, fluoropolymers, pharmaceuticals and a variety of compounds broadly referred to as persistent organic pollutants (POPs) have all been associated with a wide range of toxicities to coastal organisms (Trego et al., 2018). Metabolomic analyses of DOM can provide insight into the prevalence and distribution of POPs and other xenobiotics that are amenable to the extraction and analysis methods employed (Lara-Martín et al., 2020). Even natural organic inputs from streams, estuaries and groundwater sources have been shown to exhibit unique organic signatures (e.g., Nelson et al., 2015), and untargeted metabolomics of this DOM as it is modified in the marine environment can better define the connectivity of coastal systems and the potential for allochthonous organic matter to play a role in reef nutrient and food web dynamics.
Approaches for Applying Untargeted Metabolomics in Coastal Marine Ecology From Holobiont Physiology to Ecosystem Processes
Encouraging the application of a complex new methodology to an established area of study demands a pathway to facilitate early adoption. In addition to advances in analytical instrumentation, such as tandem mass spectrometers, the field of metabolomics has made great strides in recent years in the democratization of bioinformatic tools and chemical databases, both of which facilitate the adoption of these methods by disparate fields. Our goal here is to introduce one approach that the authors have used to acquire and interpret biochemical data from coral reef ecosystems by recommending bioinformatic methods bundled into the Global Natural Products Social Molecular Networking (GNPS; Wang et al., 2016) system; we have used these approaches for several years and have had success in elucidating unknown metabolites in diverse ecosystems.
Collecting samples for metabolomic analyses in the field or at remote experimental stations is relatively straightforward and can be done even without access to refrigerants, although use of ultra-low temperature freezers (−80°C or lower) is recommended where possible. Biomass samples should be frozen or solvent-extracted as quickly as possible as metabolites are typically dynamic especially in organismal tissues. In general, metabolomic samples are extracted in organic solvents such as methanol. Small quantity tissue biopsies (mg to g scales) can be directly extracted in small volumes (mL) and chilled or frozen until processing can be undertaken. The extraction of dissolved metabolites from seawater can be done by using the widely adopted approach of Dittmar et al. (2008) where filtered, acidified water is passed slowly (no more than 1 L per h) over a solid phase extraction resin to bind metabolites that are subsequently eluted in methanol: PPL, a hydrophobic non-polar styrene-divinylbenzene polymer (Agilent), is sold in straight barrel cartridges containing a packed bed of resin that has been demonstrated to isolate a wide range of non-polar and semi-polar DOM compounds (Dittmar et al., 2008; Li et al., 2016; Petras et al., 2017). Extraction techniques and data acquisition vary considerably, but for the purposes of this Perspective we assume the extracted metabolites are processed via liquid chromatographic separation and analyzed via electrospray ionization tandem mass spectrometry. This approach is widely applied for untargeted analysis of small to medium sized molecules (150–3000 Da), the mass spectrometers are widely available and the analytical methods are well documented (Petras et al., 2017).
Statistical, bioinformatic and visualization analyses of tandem mass spectrometry datasets derived from environmental samples and their application to questions in coastal marine ecology are outlined in Figure 2. Questions associated with chemodiversity (Figures 2A,B), including basic investigations of the number (richness) and relative abundances (evenness) of biochemicals found in different organisms or habitats are optimized by methods that seek to identify putative compounds and quantify their relative abundances (translate-able to absolute abundances when run with appropriate standards in “targeted” metabolomics). Programs that find and quantify ion features in mass spectrometry data (defined as detected ion signal for an eluting compound with a representative MS/MS spectrum) are diverse and here we apply MZmine 2 as a free, widely used and well-supported option (Pluskal et al., 2010). For example, analyses of metabolite profiles in holobiont tissues may seek to understand when and where chemodiversity increases and decreases as an indicator of organismal response to environmental stressors (Figure 2A). Changes in chemodiversity in the water column may be associated with pulses of metabolite exchange associated with signaling or allelopathic interactions (Figure 2B). Questions of comparative metabolomics (Figures 2C,D), where ion features must be tracked across multiple samples to examine changes in chemical composition, require informatic approaches that leverage large sample sets to better align (Pluskal et al., 2010) and resolve ion features, such as the feature alignment and annotation tools built into the Feature-based Molecular Networking workflow in the GNPS ecosystem (Wang et al., 2016; Nothias et al., 2020). When unknown ion features interpreted as metabolites can be tracked across samples then analyses of beta diversity, including compositional similarity and distance based multivariate approaches, become possible (Oksanen et al., 2019). Resolving holobiont metabolomic profiles associated with pathogen infection or water column metabolite profiles induced by thermal stress is a crucial area of research in coastal benthic ecology, and these comparative approaches may allow identification of both suites of covarying metabolites that illuminate a new aspect of physiological response (Figure 2C) and/or wholesale changes in multivariate metabolome structure (Figure 2D).
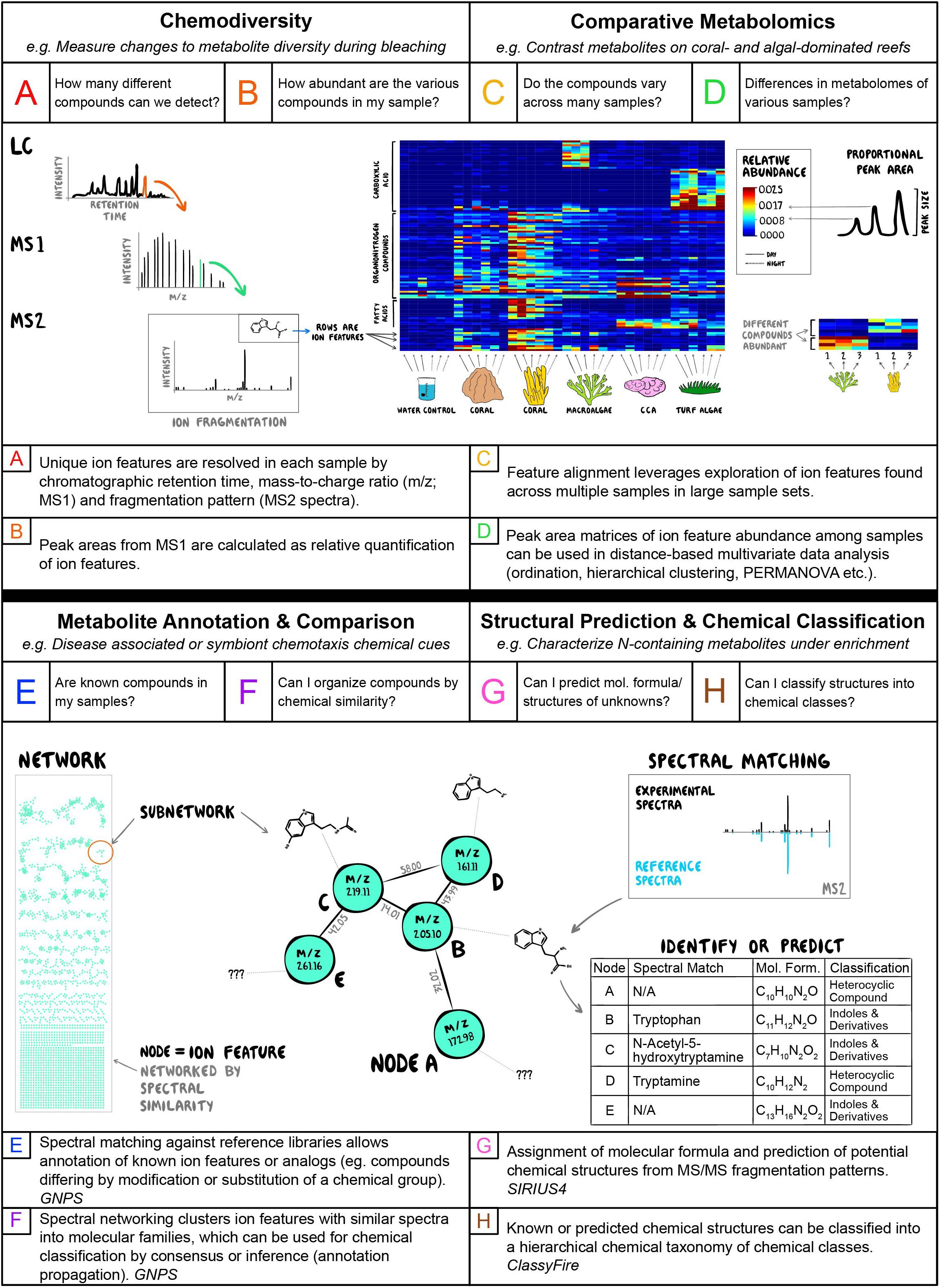
Figure 2. Summary of research areas and tools in coral reef science appropriate for metabolomics analysis, including specific questions asked with metabolomic data (A–H, top of panels), and example public access tools and informatic workflows recommended (A–H, bottom of panels).
Detecting and annotating unknown metabolites is the holy grail of untargeted metabolomics. While high resolution mass measurements may allow inference of ion feature molecular formulae, tandem mass spectrometry using ion fragmentation (MS2 spectra) patterns can, when coupled with sophisticated informatics, allow characterization of unknown metabolites (Figures 2E–H). Analyses of holobiont metabolomes, particularly those of vertebrates and plants, are likely to resolve features with MS2 spectra that are exact or near-exact matches to known compounds, allowing annotation via large public databases facilitated within GNPS (Wang et al., 2016) and unveiling key details of the chemical ecology of host organisms (Figure 2E). In more chemically diverse or poorly characterized habitats such as sediments, water or invertebrate holobionts, even unannotated MS2 spectra can be analyzed via Feature Based Molecular Networking (Wang et al., 2016; Nothias et al., 2020) to organize groups of ion features with similar spectral characteristics into networks (Shannon et al., 2003) that are likely to be structurally related (Figure 2F). In studies of chemical signaling in benthic interactions mediated through the water column (Figure 1), suites of unknown molecules enriched in areas of, for example, coral-algal interaction could be networked to identify new molecular families important in antagonistic host or microbial processes (e.g., Roach et al., 2020). Another advantage of acquiring MS2 spectra in untargeted metabolomics is the potential for higher confidence assignment of molecular formulas through fragmentation trees and network based algorithms (SIRIUS and ZODIAC; Dührkop et al., 2019; Ludwig et al., 2020) or in silico prediction of molecular structures (CSIFingerID, NAP, MS2LDA; Dührkop et al., 2015; van Der Hooft et al., 2016; da Silva et al., 2018). Even when exact structural predictions are limited by database breadth, emerging ontologies for hierarchical chemical classification (Feunang et al., 2016) allow organization of unknown ion features into meaningful chemical classes useful for understanding the chemical ecology of systems (Figures 2G,H). Networking and structural prediction can be combined with chemical classification to provide order to the chaos and help illuminate gradients in broad or narrow classes of chemicals that inform coastal ecology. Such approaches are particularly useful for tracking compounds in highly dynamic environments such as the water column, where anthropogenic pollutants, lipid-based signaling compounds, carbohydrate-based photosynthetic exudates and proteinaceous breakdown products intermingle and are metabolized by microbial processes into varied reactants and products.
Conclusion
Metabolomic approaches can be used to obtain new knowledge about these precious coastal ecosystems that comprise some of the most critically endangered habitats worldwide. New instruments that are becoming widely accessible combined with the development of open source informatic tools allow for greater capacity to use these biochemical approaches across disciplines where chemodiversity is less well understood. Integrating metabolomic data with experimental and field studies has the potential to:
(i) Answer specific questions about the establishment and maintenance of symbiotic partnerships between corals and their associated microbiota, including the determination of signaling compounds for the recruitment of symbionts, antimicrobial compounds produced by commensal bacteria, or compounds exchanged between symbionts and host that provide essential nutrients and support energetic budgets;
(ii) Understand trade-offs between directed establishment of thermal tolerant species of Symbiodiniaceae and host phenotypic responses;
(iii) Identify chemical signals instrumental in mating synchrony and larval settlement as well as those involved in the acquisition of microbial symbionts;
(iv) Identify microbial- or host-derived metabolites that serve to protect juveniles against uncontrolled bacterial colonization or pathogens;
(v) Compare the exometabolomes of coral reefs dominated by reef-builders or fleshy benthic constituents to identify DOM compounds that select for opportunistic pathogens or enrich for copiotrophic microbial guilds which alter ecosystem metabolism; and
(vi) Track the distribution of terrigenous pollutants and their incorporation into coastal food webs.
Our broad perspective is that knowledge gained from metabolomic approaches will improve mechanistic understanding of relationships between hosts and their microbial consortia, and the chemical exchanges between organisms and their environment.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
LWK, CN, and AH drafted the manuscript. SM designed the figures. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
Funding was provided by the United States National Science Foundation awards OCE-2118618, OCE-2023298, and OCE-2023509 to LWK, CN, LA, and PD, respectively. This is publication number 11369 of the University of Hawaii School of Ocean and Earth Science and Technology and publication number UNIHI-SEAGRANT-JC-21-04 of UH Sea Grant. This paper was funded in part by a grant/cooperative agreement from the National Oceanic and Atmospheric Administration, Project A/AS-1, which is sponsored by the University of Hawaii Sea Grant College Program, SOEST, under Institutional Grant No. NA18OAR4170076 from NOAA Office of Sea Grant, Department of Commerce.
Author Disclaimer
The views expressed in this article are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ainsworth, T. D., Fordyce, A. J., and Camp, E. F. (2017). The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 25, 980–991. doi: 10.1016/j.tim.2017.06.007
Ainsworth, T. D., and Gates, R. D. (2016). Corals’ microbial sentinels. Science 352, 1518–1519. doi: 10.1126/science.aad9957
Apprill, A. (2017). Marine animal microbiomes: toward understanding host-microbiome interactions in a changing ocean. Front. Mar. Sci. 4:222. doi: 10.3389/fmars.2017.00222
Apprill, A., Marlow, H. Q., Martindale, M. Q., and Rappé, M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Barott, K. L., and Rohwer, F. L. (2012). Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. doi: 10.1016/j.tim.2012.08.004
Beman, J. M., Roberts, K. J., Wegley, L., Rohwer, F., and Francis, C. A. (2007). Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73, 5642–5647. doi: 10.1128/aem.00461-07
Boroujerdi, A. F., Vizcaino, M. I., Meyers, A., Pollock, E. C., Huynh, S. L., Schock, T. B., et al. (2009). NMR-based microbial metabolomics and the temperature-dependent coral pathogen Vibrio coralliilyticus. Environ. Sci. Technol. 43, 7658–7664. doi: 10.1021/es901675w
Boström-Einarsson, L., Babcock, R. C., Bayraktarov, E., Ceccarelli, D., Cook, N., Ferse, S. C. A., et al. (2020). Coral restoration - A systematic review of current methods, successes, failures and future directions. PLoS One 15:e0226631. doi: 10.1371/journal.pone.0226631
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bruce, T., Meirelles, P. M., Garcia, G., Paranhos, R., Rezende, C. E., de Moura, R. L., et al. (2012). Abrolhos bank reef health evaluated by means of water quality, microbial diversity, benthic cover, and fish biomass data. PLoS One 7:e36687. doi: 10.1371/journal.pone.0036687
Casas, V., Kline, D. I., Wegley, L., Yu, Y., Breitbart, M., and Rohwer, F. (2004). Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6, 1137–1148. doi: 10.1111/j.1462-2920.2004.00647.x
Ceh, J., Kilburn, M. R., Cliff, J. B., Raina, J.-B., van Keulen, M., and Bourne, D. G. (2013). Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol. Evol. 3, 2393–2400. doi: 10.1002/ece3.642
Chamberland, V. F., Petersen, D., Guest, J. R., Petersen, U., Brittsan, M., and Vermeij, M. J. A. (2017a). New seeding approach reduces costs and time to outplant sexually propagated corals for reef restoration. Sci. Rep. 7:18076. doi: 10.1038/s41598-017-17555-z
Chamberland, V. F., Snowden, S., Marhaver, K. L., Petersen, D., and Vermeij, M. J. A. (2017b). The reproductive biology and early life ecology of a common Caribbean brain coral, Diploria labyrinthiformis (Scleractinia: Faviinae). Coral Reefs 36, 83–94. doi: 10.1007/s00338-016-1504-2
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B 12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
da Silva, R. R., Dorrestein, P. C., and Quinn, R. A. (2015). Illuminating the dark matter in metabolomics. PNAS 112, 12549–12550. doi: 10.1073/pnas.1516878112
da Silva, R. R., Wang, M., Nothias, L.-F., van der Hooft, J. J., Caraballo-Rodríguez, A. M., Fox, E., et al. (2018). Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 14:e1006089. doi: 10.1371/journal.pcbi.1006089
De Goeij, J. M., Lesser, M. P., and Pawlik, J. R. (2017). “Nutrient fluxes and ecological functions of coral reef sponges in a changing ocean,” in Climate Change, Ocean Acidification and Sponges, eds J. L. Carballo and J. J. Bell (Cham: Springer International Publishing), 373–410. doi: 10.1007/978-3-319-59008-0_8
De Goeij, J. M., Van Oevelen, D., Vermeij, M. J., Osinga, R., Middelburg, J. J., De Goeij, A. F., et al. (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342, 108–110. doi: 10.1126/science.1241981
Dinsdale, E. A., Pantos, O., Smriga, S., Edwards, R. A., Angly, F., Wegley, L., et al. (2008). Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3:e1584. doi: 10.1371/journal.pone.0001584
Dittmar, T., Koch, B., Hertkorn, N., and Kattner, G. (2008). A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 6, 230–235. doi: 10.4319/lom.2008.6.230
Dittmar, T., and Paeng, J. (2009). A heat-induced molecular signature in marine dissolved organic matter. Nat. Geosci. 2, 175–179. doi: 10.1038/ngeo440
Dührkop, K., Fleischauer, M., Ludwig, M., Aksenov, A. A., Melnik, A. V., Meusel, M., et al. (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302. doi: 10.1038/s41592-019-0344-8
Dührkop, K., Shen, H., Meusel, M., Rousu, J., and Böcker, S. (2015). Searching molecular structure databases with tandem mass spectra using CSI: fingerID. Proc. Natl. Acad. Sci. U.S.A. 112, 12580–12585. doi: 10.1073/pnas.1509788112
Farag, M. A., Meyer, A., Ali, S. E., Salem, M. A., Giavalisco, P., Westphal, H., et al. (2018). Comparative metabolomics approach detects stress-specific responses during coral bleaching in soft corals. J. Proteome Res. 17, 2060–2071. doi: 10.1021/acs.jproteome.7b00929
Ferrier-Pagès, C., Houlbrèque, F., Wyse, E., Richard, C., Allemand, D., and Boisson, F. (2005). Bioaccumulation of zinc in the scleractinian coral Stylophora pistillata. Coral Reefs 24, 636–645. doi: 10.1007/s00338-005-0045-x
Feunang, Y. D., Eisner, R., Knox, C., Chepelev, L., Hastings, J., Owen, G., et al. (2016). ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminformatics 8:61.
Fiore, C. L., Freeman, C. J., and Kujawinski, E. B. (2017). Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ 5:e2870. doi: 10.7717/peerj.2870
Fogarty, N. D., and Marhaver, K. L. (2019). Coral spawning, unsynchronized. Science 365, 987–988. doi: 10.1126/science.aay7457
Garren, M., Son, K., Raina, J.-B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Geier, B., Sogin, E. M., Michellod, D., Janda, M., Kompauer, M., Spengler, B., et al. (2020). Spatial metabolomics of in situ host-microbe interactions at the micrometre scale. Nat. Microbiol. 5, 498–510. doi: 10.1038/s41564-019-0664-6
Gerardo, N. M. (2013). The give and take of host-microbe symbioses. Cell Host Microbe 14, 1–3. doi: 10.1016/j.chom.2013.07.001
Haas, A. F., Fairoz, M. F., Kelly, L. W., Nelson, C. E., Dinsdale, E. A., Edwards, R. A., et al. (2016). Global microbialization of coral reefs. Nat. Microbiol. 1, 1–7.
Haas, A. F., Nelson, C. E., Rohwer, F., Wegley-Kelly, L., Quistad, S. D., Carlson, C. A., et al. (2013). Influence of coral and algal exudates on microbially mediated reef metabolism. PeerJ 1:e108. doi: 10.7717/peerj.108
Haas, A. F., Nelson, C. E., Wegley Kelly, L., Carlson, C. A., Rohwer, F., Leichter, J. J., et al. (2011). Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS One 6:e0027973. doi: 10.1371/journal.pone.0027973
Haas, A. F., and Wild, C. (2010). Composition analysis of organic matter released by cosmopolitan coral reef-associated green algae. Aquat. Biol. 10, 131–138. doi: 10.3354/ab00271
Hadfield, M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hartmann, A. C., Petras, D., Quinn, R. A., Protsyuk, I., Archer, F. I., Ransome, E., et al. (2017). Meta-mass shift chemical profiling of metabolomes from coral reefs. Proc. Natl. Acad. Sci. U.S.A. 114, 11685–11690. doi: 10.1073/pnas.1710248114
Hawkes, J. A., Dittmar, T., Patriarca, C., Tranvik, L., and Bergquist, J. (2016). Evaluation of the Orbitrap mass spectrometer for the molecular fingerprinting analysis of natural dissolved organic matter. Anal. Chem. 88, 7698–7704. doi: 10.1021/acs.analchem.6b01624
Hay, M. E. (2009). Marine chemical ecology: chemical signals and cues structure marine populations, communities, and ecosystems. Annu. Rev. Mar. Sci. 1, 193–212. doi: 10.1146/annurev.marine.010908.163708
Hillyer, K. E., Dias, D., Lutz, A., Roessner, U., and Davy, S. K. (2018). 13 C metabolomics reveals widespread change in carbon fate during coral bleaching. Metabolomics 14:12.
Hillyer, K. E., Dias, D. A., Lutz, A., Wilkinson, S. P., Roessner, U., and Davy, S. K. (2017). Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs 36, 105–118. doi: 10.1007/s00338-016-1508-y
Innis, T., Allen-Waller, L., Brown, K. T., Sparagon, W., Carlson, C., Kruse, E., et al. (2021). Marine heatwaves depress metabolic activity and impair cellular acid-base homeostasis in reef-building corals regardless of bleaching susceptibility. Glob. Change Biol. 27, 2728–2743. doi: 10.1111/gcb.15622
Johnson, M. D., Scott, J. S., Leray, M., Lucey, N., Rodriguez Bravo, L. M., Wied, W., et al. (in press). Rapid ecosystem-scale consequences of acute deoxygenation on a Caribbean coral reef. Nat. Commun. 12:24777.
Kellogg, C. A. (2004). Tropical Archaea: diversity associated with the surface microlayer of corals. Mar. Ecol. Prog. Ser. 273, 81–88. doi: 10.3354/meps273081
Kelly, L. W., Haas, A. F., and Nelson, C. E. (2018). Ecosystem microbiology of coral reefs: linking genomic, metabolomic, and biogeochemical dynamics from animal symbioses to reefscape processes. mSystems 3:e00162-17. doi: 10.1128/mSystems.00162-17
Kelly, L. W., Nelson, C. E., Haas, A. F., Naliboff, D. S., Calhoun, S., Carlson, C. A., et al. (2019). Diel population and functional synchrony of microbial communities on coral reefs. Nat. Commun. 10:1691. doi: 10.1038/s41467-019-09419-z
Kimes, N. E., Grim, C. J., Johnson, W. R., Hasan, N. A., Tall, B. D., Kothary, M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846. doi: 10.1038/ismej.2011.154
Klinges, J. G., Rosales, S. M., McMinds, R., Shaver, E. C., Shantz, A. A., Peters, E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938–2953. doi: 10.1038/s41396-019-0482-0
Knowlton, N., and Rohwer, F. (2003). Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 162, S51–S62.
Kuffner, I. B., Walters, L. J., Becerro, M. A., Paul, V. J., Ritson-Williams, R., and Beach, K. S. (2006). Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar. Ecol. Prog. Ser. 323, 107–117. doi: 10.3354/meps323107
Lara-Martín, P. A., Chiaia-Hernández, A. C., Biel-Maeso, M., Baena-Nogueras, R. M., and Hollender, J. (2020). Tracing urban wastewater contaminants into the Atlantic ocean by nontarget screening. Environ. Sci. Technol. 54, 3996–4005. doi: 10.1021/acs.est.9b06114
Lawson, C. A., Possell, M., Seymour, J. R., Raina, J.-B., and Suggett, D. J. (2019). Coral endosymbionts (Symbiodiniaceae) emit species-specific volatilomes that shift when exposed to thermal stress. Sci. Rep. 9, 1–11.
Lawson, C. A., Raina, J.-B., Kahlke, T., Seymour, J. R., and Suggett, D. J. (2018). Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ. Microbiol. Rep. 10, 7–11. doi: 10.1111/1758-2229.12599
Lesser, M. P., Falcón, L. I., Rodríguez-Román, A., Enríquez, S., Hoegh-Guldberg, O., and Iglesias-Prieto, R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 346, 143–152. doi: 10.3354/meps07008
Letourneau, M. L., Hopkinson, B. M., Fitt, W. K., and Medeiros, P. M. (2020). Molecular composition and biodegradation of loggerhead sponge Spheciospongia vesparium exhalent dissolved organic matter. Mar. Environ. Res. 162:105130. doi: 10.1016/j.marenvres.2020.105130
Levitan, D. R., Fogarty, N. D., Jara, J., Lotterhos, K. E., and Knowlton, N. (2011). Genetic, spatial, and temporal components of precise spawning synchrony in reef building corals of the Montastraea annularis species complex. Evol. Int. J. Organ. Evol. 65, 1254–1270. doi: 10.1111/j.1558-5646.2011.01235.x
Li, Y., Harir, M., Lucio, M., Kanawati, B., Smirnov, K., Flerus, R., et al. (2016). Proposed guidelines for solid phase extraction of Suwannee River dissolved organic matter. Anal. Chem. 88, 6680–6688. doi: 10.1021/acs.analchem.5b04501
Lohr, K. E., Camp, E. F., Kuzhiumparambil, U., Lutz, A., Leggat, W., Patterson, J. T., et al. (2019a). Resolving coral photoacclimation dynamics through coupled photophysiological and metabolomic profiling. J. Exp. Biol. 222:jeb195982.
Lohr, K. E., Khattri, R. B., Guingab-Cagmat, J., Camp, E. F., Merritt, M. E., Garrett, T. J., et al. (2019b). Metabolomic profiles differ among unique genotypes of a threatened Caribbean coral. Sci. Rep. 9, 1–11.
Ludwig, M., Nothias, L.-F., Dührkop, K., Koester, I., Fleischauer, M., Hoffmann, M. A., et al. (2020). Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nat. Mach. Intellig. 2, 629–641. doi: 10.1038/s42256-020-00234-6
Ma, Z.-P., Song, Y., Cai, Z.-H., Lin, Z.-J., Lin, G.-H., Wang, Y., et al. (2018). Anti-quorum sensing activities of selected coral symbiotic bacterial extracts from the South China Sea. Front. Cell. Infect. Microbiol. 8:144. doi: 10.3389/fcimb.2018.00144
Mackintosh, S. A., Dodder, N. G., Shaul, N. J., Aluwihare, L. I., Maruya, K. A., Chivers, S. J., et al. (2016). Newly identified DDT-related compounds accumulating in Southern California bottlenose dolphins. Environ. Sci. Technol. 50, 12129–12137. doi: 10.1021/acs.est.6b03150
Marhaver, K. L., Edwards, R. A., and Rohwer, F. (2008). Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x
Matthews, J. L., Cunning, R., Ritson-Williams, R., Oakley, C. A., Lutz, A., Roessner, U., et al. (2020a). Metabolite pools of the reef building coral Montipora capitata are unaffected by Symbiodiniaceae community composition. Coral Reefs 39, 1727–1737. doi: 10.1007/s00338-020-01999-3
Matthews, J. L., Oakley, C. A., Lutz, A., Hillyer, K. E., Roessner, U., Grossman, A. R., et al. (2018). Partner switching and metabolic flux in a model cnidarian-dinoflagellate symbiosis. Proc. R. Soc. B 285:20182336. doi: 10.1098/rspb.2018.2336
Matthews, J. L., Raina, J. B., Kahlke, T., and Seymour, J. R. (2020b). Symbiodiniaceae-bacteria interactions: rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 22, 1675–1687. doi: 10.1111/1462-2920.14918
McDole, T., Nulton, J., Barott, K. L., Felts, B., Hand, C., Hatay, M., et al. (2012). Assessing coral reefs on a Pacific-wide scale using the microbialization score. PLoS One 7:e43233. doi: 10.1371/journal.pone.0043233
McNally, S. P., Parsons, R. J., Santoro, A. E., and Apprill, A. (2017). Multifaceted impacts of the stony coral Porites astreoides on picoplankton abundance and community composition. Limnol. Oceanogr. 62, 217–234. doi: 10.1002/lno.10389
Nelson, C. E., Donahue, M. J., Dulaiova, H., Goldberg, S. J., La Valle, F. F., Lubarsky, K., et al. (2015). Fluorescent dissolved organic matter as a multivariate biogeochemical tracer of submarine groundwater discharge in coral reef ecosystems. Mar. Chem. 177, 232–243. doi: 10.1016/j.marchem.2015.06.026
Nelson, C. E., Goldberg, S. J., Kelly, L. W., Haas, A. F., Smith, J. E., Rohwer, F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7, 962–979. doi: 10.1038/ismej.2012.161
Nothias, L.-F., Petras, D., Schmid, R., Dührkop, K., Rainer, J., Sarvepalli, A., et al. (2020). Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 17, 905–908.
Ochsenkühn, M. A., Schmitt-Kopplin, P., Harir, M., and Amin, S. A. (2018). Coral metabolite gradients affect microbial community structures and act as a disease cue. Commun. Biol. 1, 1–10.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., et al. (2019). Vegan: Community Ecology Package, Version 2.4-4p.
Olinger, L. K., Strangman, W. K., McMurray, S. E., and Pawlik, J. R. (2021). Sponges with microbial symbionts transform dissolved organic matter and take up organohalides. Front. Mar. Sci. 8:665789. doi: 10.3389/fmars.2021.665789
Osterholz, H., Singer, G., Wemheuer, B., Daniel, R., Simon, M., Niggemann, J., et al. (2016). Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 10, 1717–1730. doi: 10.1038/ismej.2015.231
Pernice, M., Meibom, A., Van Den Heuvel, A., Kopp, C., Domart-Coulon, I., Hoegh-Guldberg, O., et al. (2012). A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324. doi: 10.1038/ismej.2011.196
Petras, D., Koester, I., da Silva, R., Stephens, B. M., Haas, A. F., Nelson, C. E., et al. (2017). High-resolution liquid chromatography tandem mass spectrometry enables large scale molecular characterization of dissolved organic matter. Front. Mar. Sci. 4:405. doi: 10.3389/fmars.2017.00405
Petrou, K., Nielsen, D. A., and Heraud, P. (2018). Single-cell biomolecular analysis of coral algal symbionts reveals opposing metabolic responses to heat stress and expulsion. Front. Mar. Sci. 5:110. doi: 10.3389/fmars.2018.00110
Pluskal, T., Castillo, S., Villar-Briones, A., and Orešič, M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. doi: 10.1186/1471-2105-11-395
Quinn, R. A., Vermeij, M. J., Hartmann, A. C., Galtier d’Auriac, I., Benler, S., Haas, A., et al. (2016). Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. R. Soc. B Biol. Sci. 283:20160469. doi: 10.1098/rspb.2016.0469
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Randall, C. J., Negri, A. P., Quigley, K. M., Foster, T., Ricardo, G. F., Webster, N. S., et al. (2020). Sexual production of corals for reef restoration in the Anthropocene. Mar. Ecol. Prog. Ser. 635, 203–232. doi: 10.3354/meps13206
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rix, L., de Goeij, J. M., van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2017). Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct. Ecol. 31, 778–789. doi: 10.1111/1365-2435.12758
Roach, T. N., Little, M., Arts, M. G., Huckeba, J., Haas, A. F., George, E. E., et al. (2020). A multiomic analysis of in situ coral-turf algal interactions. Proc. Natl. Acad. Sci. U.S.A. 117, 13588–13595. doi: 10.1073/pnas.1915455117
Roach, T. N. F., Dilworth, J., H, C. M., Jones, A. D., Quinn, R. A., and Drury, C. (2021). Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol. 5, 495–503. doi: 10.1038/s41559-020-01388-7
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharp, K. H., Distel, D., and Paul, V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. doi: 10.1038/ismej.2011.144
Sharp, K. H., and Ritchie, K. B. (2012). Multi-partner interactions in corals in the face of climate change. Biol. Bull. 223, 66–77. doi: 10.1086/bblv223n1p66
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Shlesinger, T., and Loya, Y. (2019). Breakdown in spawning synchrony: a silent threat to coral persistence. Science 365, 1002–1007. doi: 10.1126/science.aax0110
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Silveira, C. B., Silva-Lima, A. W., Francini-Filho, R. B., Marques, J. S. M., Almeida, M. G., Thompson, C. C., et al. (2015). Microbial and sponge loops modify fish production in phase-shifting coral reefs. Environ. Microbiol. 17, 3832–3846. doi: 10.1111/1462-2920.12851
Sneed, J. M., Sharp, K. H., Ritchie, K. B., and Paul, V. J. (2014). The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. R. Soc. B Biol. Sci. 281:20133086. doi: 10.1098/rspb.2013.3086
Sogin, E. M., Putnam, H. M., Anderson, P. E., and Gates, R. D. (2016). Metabolomic signatures of increases in temperature and ocean acidification from the reef-building coral, Pocillopora damicornis. Metabolomics 12:71.
Steen, A. D., Kusch, S., Abdulla, H. A., Cakić, N., Coffinet, S., Dittmar, T., et al. (2020). Analytical and computational advances, opportunities, and challenges in marine organic biogeochemistry in an Era of “Omics.”. Front. Mar. Sci. 7:718. doi: 10.3389/fmars.2020.00718
Tebben, J., Tapiolas, D. M., Motti, C. A., Abrego, D., Negri, A. P., Blackall, L. L., et al. (2011). Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One 6:e19082. doi: 10.1371/journal.pone.0019082
Teplitski, M., and Ritchie, K. (2009). How feasible is the biological control of coral diseases? Trends Ecol. Evol. 24, 378–385. doi: 10.1016/j.tree.2009.02.008
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2015). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4:176. doi: 10.3389/fcimb.2014.00176
Thurber, R. V., Payet, J. P., Thurber, A. R., and Correa, A. M. (2017). Virus-host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 15, 205–216. doi: 10.1038/nrmicro.2016.176
Tout, J., Jeffries, T. C., Petrou, K., Tyson, G. W., Webster, N. S., Garren, M., et al. (2015). Chemotaxis by natural populations of coral reef bacteria. ISME J. 9, 1764–1777. doi: 10.1038/ismej.2014.261
Trego, M. L., Hoh, E., Kellar, N. M., Meszaros, S., Robbins, M. N., Dodder, N. G., et al. (2018). Comprehensive screening links halogenated organic compounds with testosterone levels in male Delphinus delphis from the Southern California bight. Environ. Sci. Technol. 52, 3101–3109. doi: 10.1021/acs.est.7b04652
van Der Hooft, J. J. J., Wandy, J., Barrett, M. P., Burgess, K. E., and Rogers, S. (2016). Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. U.S.A. 113, 13738–13743. doi: 10.1073/pnas.1608041113
van Oppen, M. J. H., Gates, R. D., Blackall, L. L., Cantin, N., Chakravarti, L. J., Chan, W. Y., et al. (2017). Shifting paradigms in restoration of the world’s coral reefs. Glob. Change Biol. 23, 3437–3448. doi: 10.1111/gcb.13647
Vermeij, M. J. A., Bakker, J., van der Hal, N., and Bak, R. P. M. (2011). Juvenile coral abundance has decreased by more than 50% in only three decades on a small Caribbean Island. Diversity 3, 296–307. doi: 10.3390/d3030296
Walsh, K., Haggerty, J. M., Doane, M. P., Hansen, J. J., Morris, M. M., Moreira, A. P. B., et al. (2017). Aura-biomes are present in the water layer above coral reef benthic macro-organisms. PeerJ 5:e3666. doi: 10.7717/peerj.3666
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837.
Weber, L., Armenteros, M., Soule, M., Longnecker, K., Kujawinski, E. B., and Apprill, A. (2020). Extracellular reef metabolites across the protected Jardines de la Reina, Cuba reef system. Front. Mar. Sci. 7:582161. doi: 10.3389/fmars.2020.582161
Weber, L., Gonzalez-Díaz, P., Armenteros, M., and Apprill, A. (2019). The coral ecosphere: a unique coral reef habitat that fosters coral-microbial interactions. Limnol. Oceanogr. 64, 2373–2388. doi: 10.1002/lno.11190
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Wegley, L., Yu, Y., Breitbart, M., Casas, V., Kline, D. I., and Rohwer, F. (2004). Coral-associated archaea. Mar. Ecol. Prog. Ser. 273, 89–96. doi: 10.3354/meps273089
Wiedenmann, J., D’Angelo, C., Smith, E. G., Hunt, A. N., Legiret, F.-E., Postle, A. D., et al. (2013). Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 160–164. doi: 10.1038/nclimate1661
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y., and Jørgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Keywords: biogeochemistry, coral reefs, coral holobiont, dissolved organic matter, metabolomics, microbial ecology, nutrient cycling
Citation: Wegley Kelly L, Nelson CE, Aluwihare LI, Arts MGI, Dorrestein PC, Koester I, Matsuda SB, Petras D, Quinlan ZA and Haas AF (2021) Molecular Commerce on Coral Reefs: Using Metabolomics to Reveal Biochemical Exchanges Underlying Holobiont Biology and the Ecology of Coastal Ecosystems. Front. Mar. Sci. 8:630799. doi: 10.3389/fmars.2021.630799
Received: 18 November 2020; Accepted: 28 June 2021;
Published: 23 July 2021.
Edited by:
Simon K. Davy, Victoria University of Wellington, New ZealandReviewed by:
Daniel J. Thornhill, National Science Foundation (NSF), United StatesLuke Thompson, Atlantic Oceanographic and Meteorological Laboratory (NOAA), United States
Copyright © 2021 Wegley Kelly, Nelson, Aluwihare, Arts, Dorrestein, Koester, Matsuda, Petras, Quinlan and Haas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda Wegley Kelly, bHdlZ2xleUBnbWFpbC5jb20=; Craig E. Nelson, Y3JhaWcubmVsc29uQGhhd2FpaS5lZHU=
†These authors have contributed equally to this work
 Linda Wegley Kelly
Linda Wegley Kelly Craig E. Nelson
Craig E. Nelson Lihini I. Aluwihare
Lihini I. Aluwihare Milou G. I. Arts
Milou G. I. Arts Pieter C. Dorrestein5
Pieter C. Dorrestein5 Irina Koester
Irina Koester Shayle B. Matsuda
Shayle B. Matsuda Daniel Petras
Daniel Petras Zachary A. Quinlan
Zachary A. Quinlan Andreas F. Haas
Andreas F. Haas