
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 10 February 2021
Sec. Marine Conservation and Sustainability
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.628142
Giant clams are an important ecological component of coral reefs in the Red Sea, as they enhance the reef’s productivity and provide habitat that can increase diversity. Three species of giant clams, namely Tridacna maxima, T. squamosa, and T. squamosina have been described within the Red Sea. However, due to its scarcity, information about the distribution and ecology of T. squamosina in the Saudi Arabian Red Sea is still lacking. This study used DNA barcoding to confirm the identity of the rare T. squamosina in the Farasan Banks. Six mtCOI fragments (500 bp) of T. squamosina were successfully amplified using the SQUA-primers for the first time. We used our data along with 18 reference sequences (16S) from the online database to assess the genetic diversity and population structure of T. squamosina. Low genetic diversity among the T. squamosina populations inferred from the 16S sequences implies a recent bottleneck for this species, which is supported by their historically higher diversity based on the coalescent-based estimator. Given the small population abundance and limited genetic variation of T. squamosina, it may warrant immediate local protections such as biobanking and fertility preservation programs as well as effective integrated coastal zone management plans.
The recognition of giant clams (subfamily Tridacninae) as a contributor to net primary productivity and biomass of Red Sea coral reefs is driving a growing research interest in their abundance, ecological roles and population genetic structure (Roa-Quiaoit, 2005; Richter et al., 2008; Pappas et al., 2017; Rossbach et al., 2019a,b; Fauvelot et al., 2020). They are a common food source within the coral reef community, with 75 known predators, including fishes such as wrasses, triggerfishes, and emperor fishes (Neo et al., 2015). Among these predatory fishes, 11 species naturally occur in the Red Sea (Golani and Bogorodsky, 2010), suggesting that giant clams are an important food source for coral reef food webs in this region. Three species of giant clams, specifically Tridacna maxima, T. squamosa, and T. squamosina are found in the Red Sea. The first two species are listed in the International Union for Conservation of Nature (IUCN) Red List of Threatened Species while the latter is a relatively recent resurrected species that has not been assessed for IUCN Red Listing, which requires information on their ecology, habitat, and density (Neo, 2020).
To date, at least 31 reported sightings of T. squamosina have been documented from the Red Sea since their first description during the Pola Red Sea Expeditions 1895/96 and 1897/98 (Huber and Eschner, 2011). The observations from the literature include 16 individuals from Roa-Quiaoit (2005) and Richter et al. (2008), seven individuals from Huber and Eschner (2011), two individuals from Fauvelot et al. (2020) and six individuals from Rossbach et al. (2020). The past and present known distribution of T. squamosina are Egypt, Jordan, Yemen, Saudi Arabia, and possible sightings in Israel based on photographic anecdotes (Richter et al., 2008; Huber and Eschner, 2011; Neo et al., 2017; Fauvelot et al., 2020; Rossbach et al., 2020). Although a recently published analysis using genome skimming has provided highly credible phylogenetic relationship between T. squamosina and the other Tridacninae (Tan et al., 2021), information about their population genetic structure across the Red Sea is still lacking.
Cryptic diversity within giant clams was evident through the recent rediscovery of T. noae (Su et al., 2014; Borsa et al., 2015) and T. elongatissima (Fauvelot et al., 2020) with the aid of molecular tools. Tridacna squamosina is another example of a cryptic species, having previously been identified to be a local variant of T. squamosa in the Red Sea. This species has been recently resurrected by Richter et al. (2008) using mitochondrial gene marker (16S) with detailed descriptions on its morphological characteristics. Mitochondrial cytochrome c oxidase subunit I (mtCOI) gene sequencing is a rapid and powerful tool for accurate identification of giant clam species in the Indo-Pacific (Nuryanto et al., 2007). However, to our knowledge, there was no reference mtCOI sequences of T. squamosina available prior to the study conducted by Fauvelot et al. (2020). Although the authors have successfully amplified the mtCOI sequences of two T. squamosina individuals collected from the Red Sea, only short fragments (i.e., 234 base pairs) were obtained. Given the scarcity of reference sequences for this species, our study aims to amplify the mitochondrial sequences of the rare T. squamosina using the Tridacna specific primers, to improve barcode reference libraries and investigate their population structure. This paper highlights the limited genetic diversity of T. squamosina in the Red Sea, which indicates a need for local conservation programs to maintain and even enhance their low populations.
Six specimens of Tridacna squamosina from a reef in Farasan Banks, as part of the large-scale giant clam survey that covered 58 unique reefs (∼1,300 km) along the eastern Red Sea. For each T. squamosina, we recorded the water depth and shell length. In situ images (camera model: Canon G7x) with their mantles exposed were taken to catalog species (Figure 1). Biopsies of the mantle tissues of each clam were cut using surgical scissor and forceps, the samples were then stored in seawater at −80°C. We combined our results with those of Richter et al. (2008) and Fauvelot et al. (2020), to document the known distribution of T. squamosina in the Red Sea (Figure 2).
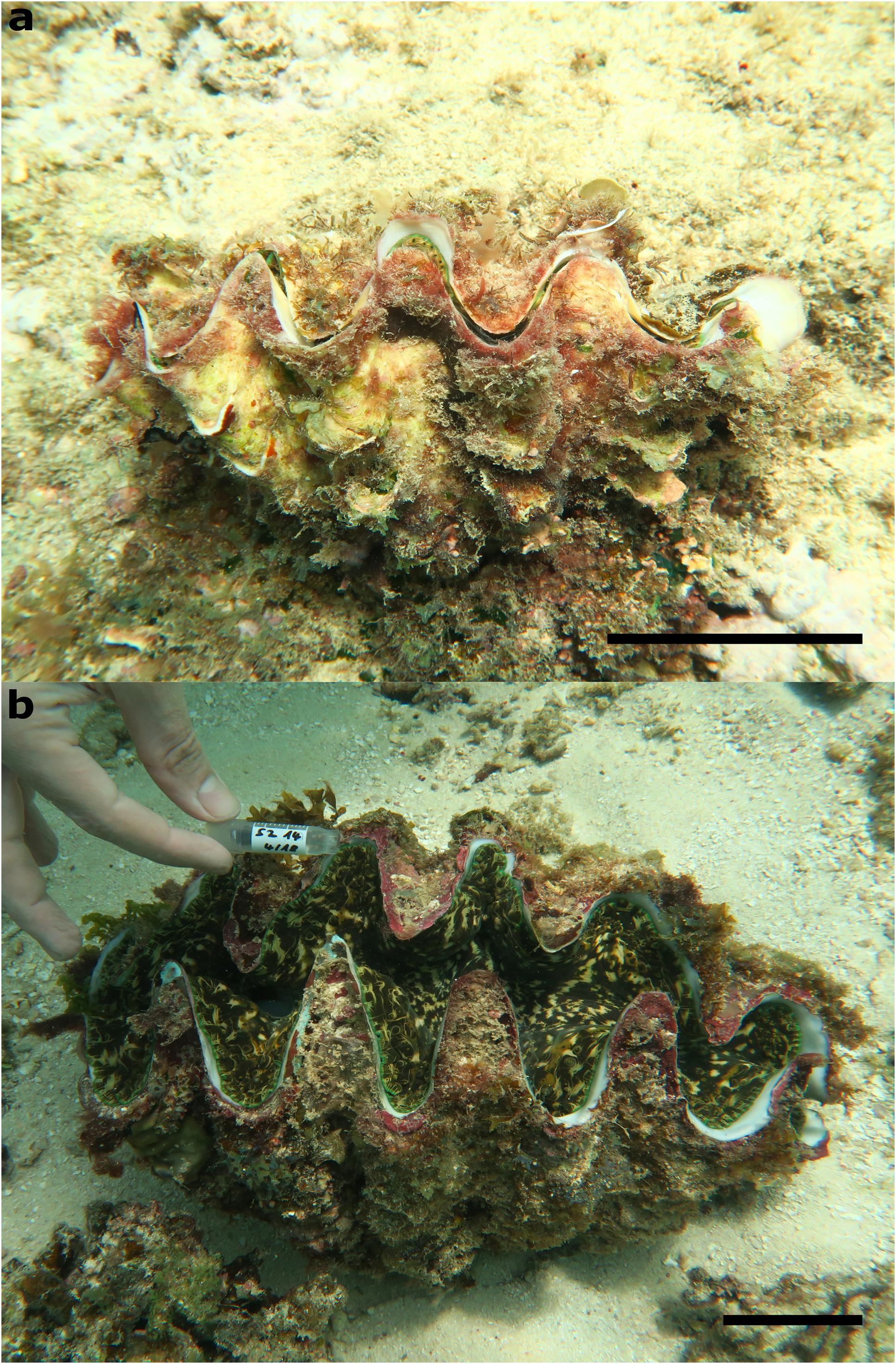
Figure 1. Specimens of the (a) smallest and (b) largest T. squasomina specimens that were found in Farasan Banks. Scale bar = 5 cm.
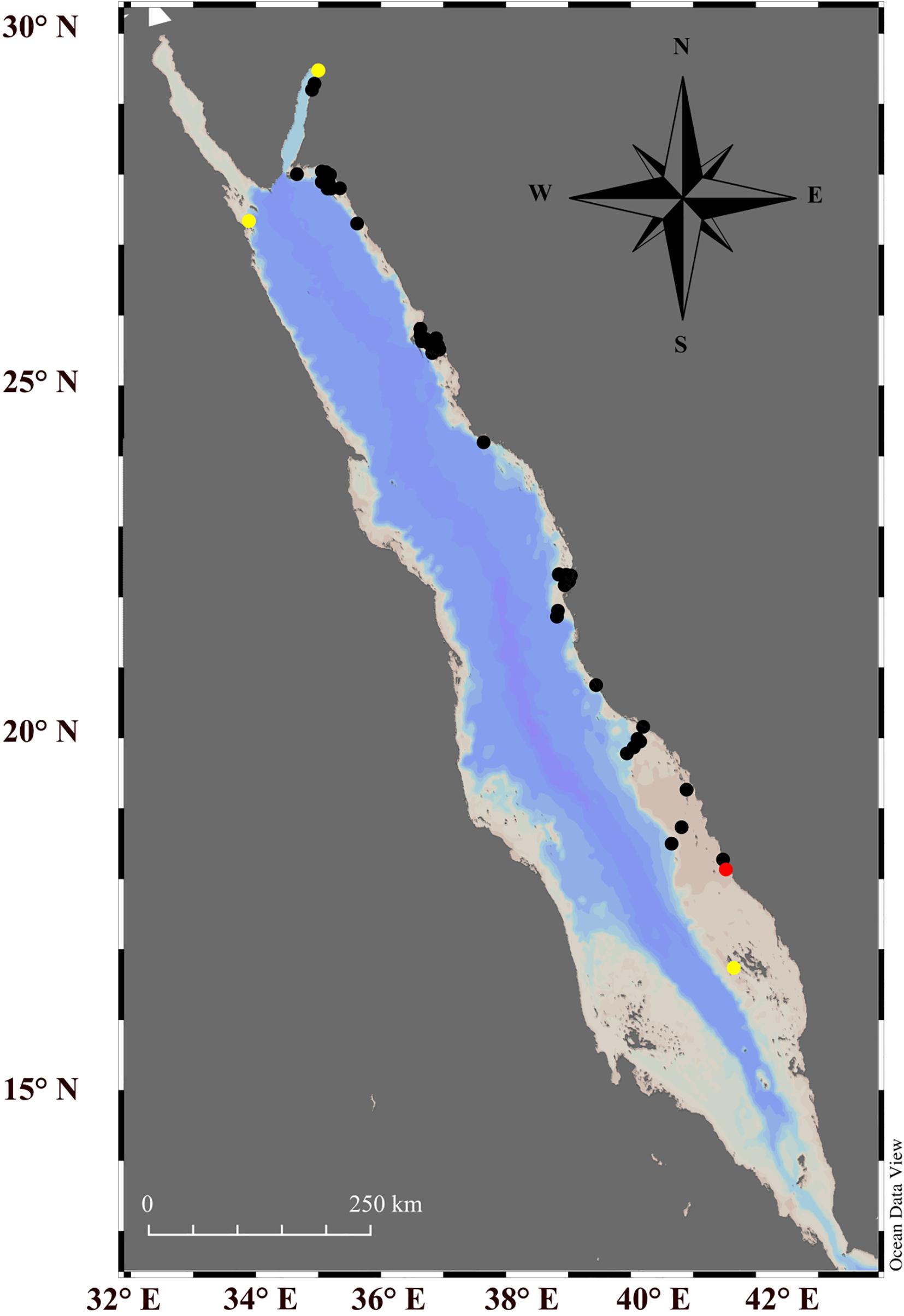
Figure 2. Fifty-eight reefs studied during a large-scale giant clam survey along the eastern Red Sea (black and red dots). The red dot indicates the sampling site where the specimens of Tridacna squamosina were found and sampled for this study while yellow dots indicate the other three locations where T. squamosina were reported in the previous studies.
DNA was extracted using the QIAGEN DNeasy® Blood and Tissue Kit (QIAGEN, Hilden, Germany). Two mitochondrial gene markers (mtCOI and 16S) were amplified and sequenced. First, a fragment of the mtCOI gene for the giant clam was amplified via polymerase chain reaction (PCR) with SQUA-R1 (5′–ATG TAT AAA CAA AAC AGG ATC–3′) and SQUA – F3 (5′–CAT CGT TTA GAG TAA TAA TTC G–3′) (DeBoer et al., 2008). PCR was performed using the QIAGEN® Multiplex PCR Kit (QIAGEN, Valencia, CA, United States) in a total volume of 25 μL containing 2.5 μL of genomic DNA, 12.5 μL of QIAGEN Multiplex PCR master mix, forward and reverse primers at a concentration of 10 mM (1.25 μL each) and RNase-Free water to adjust the volume. The protocol for isolating the mtCOI gene locus was modified from DeBoer et al. (2008) as follows: initial denaturation at 95°C for 15 min, 35 cycles of 94°C for 1 min to denature the DNA, 45°C for 1.5 min to anneal the DNA, 72°C for 1 min for elongation of the DNA, and a final elongation step at 72°C for 5 min. The second fragment used for barcoding was part of the large 16S ribosomal RNA gene which was amplified using the 16sar-L (5′–CGC CTG TTT ATC AAA AAC AT–3′) and 16sbr-H (5′–CCG GTC TGA ACT CAG ATC ACG T–3′) primers (Richter et al., 2008); PCR was conducted using the same conditions as the mtCOI gene except the annealing temperature and time were set to 43°C and 30 s respectively. The PCR products were checked using UV light after running in a 1% agarose gel (1 × TAE) pre-stained with SYBR Safe dye (Invitrogen Corp., Carlsbad, CA, United States) under 100 V for an hour. Amplicons were bead-cleaned using Agencourt AMPure XP (Beckman Coulter) and both forward and reverse strands were sequenced using Sanger sequencing at the KAUST Bioscience Core Lab.
Sequences were aligned and trimmed to a common length of 500 base pairs (bp) for mtCOI and 410 bp for 16S, using Geneious R9 (Biomatters Ltd., Auckland, New Zealand). To infer the phylogenetic relationship of Red Sea T. squamosina, six 16S sequences and 18 reference sequences (MN068557, MN068558, and AM909726–AM909741; Supplementary Table 1) were included in the analysis. The dataset was exported to DnaSP to estimate the genetic diversity. A 16S haplotype network was constructed in PopArt1 using TCS network approach. Using the same 16S dataset, we estimated the value of π and θ from the Tajima’s test of neutrality to gain insight into the species’ population history (see Templeton, 1993) using MEGA 7.0.14. We used Arlequin v.3.5 (Excoffier and Lischer, 2010) to test for genetic structure with Analysis of MOlecular Variance (AMOVA). As there is a genetic break around 19°N reported among various reef species due to the heterogeneous environmental conditions in the region (Froukh and Kochzius, 2007; Nanninga et al., 2014; Giles et al., 2015), we calculated the AMOVA by grouping the samples into two geographic clusters based on the proximity of their environmental variables. The northern population consists of populations from Jordan and Egypt (SST: ∼26°C, salinity: ∼40) and the southern population consists of Farasan Banks population (SST: ∼22°C, salinity: ∼37). As we had only one individual from the Farasan Island, this locality was omitted from population analyses. We used the same software to calculate Wright Fisher’s FST to infer the relative level of genetic difference between the northern and southern populations. As the low number of mtCOI sequences (i.e., less than 20 samples) obtained in current study can cause incomplete estimates of genetic diversity within species (Phillips et al., 2019), the mtCOI sequences from current study were combined with the short reference sequences (MN068808 and MN068809; Supplementary Table 1) to visualize the haplotype network and no further analysis is done.
All individuals were found shallower than 3 m water depth, with shell lengths ranged from 15 to 30 cm (mean = 22.5 ± 4.6 cm). Twelve sequences from six individuals were successfully amplified using the Tridacna specific primers (six for mtCOI and 16S, respectively), and they were deposited in GenBank with accession numbers MW411954–MW411959 (mtCOI), MW407155–MW407160 (16S). The genetic diversity of T. squamosina based on the 16S sequences from the pooled dataset was 0.163 ± 0.099. From the 16S haplotype network, three unique haplotypes, representing the 24 individuals of Red Sea T. squamosina were detected. Five out of six samples collected in the current study shared the same haplotype with other samples found in the northern Red Sea and Farasan Islands, in the southern Red Sea (H1) (Figure 3A). The Tajima estimator (π) and coalescent estimator of genetic diversity (θ) values were 0.001 and 0.003, respectively. AMOVA analysis showed that 99.81% of variations measured in the overall population are found within individuals of the same populations (Table 1). The fixation index (FST) among populations was 0.00189, which was not significant. One individual (G6) from current study shared the same point mutation with the Egypt specimen (MN068808) at the 51st position based on the mtCOI alignment (i.e., 234 bp) in Geneious R9 (Supplementary Figure 1), congruent with the haplotype network as depicted in Figure 3B.
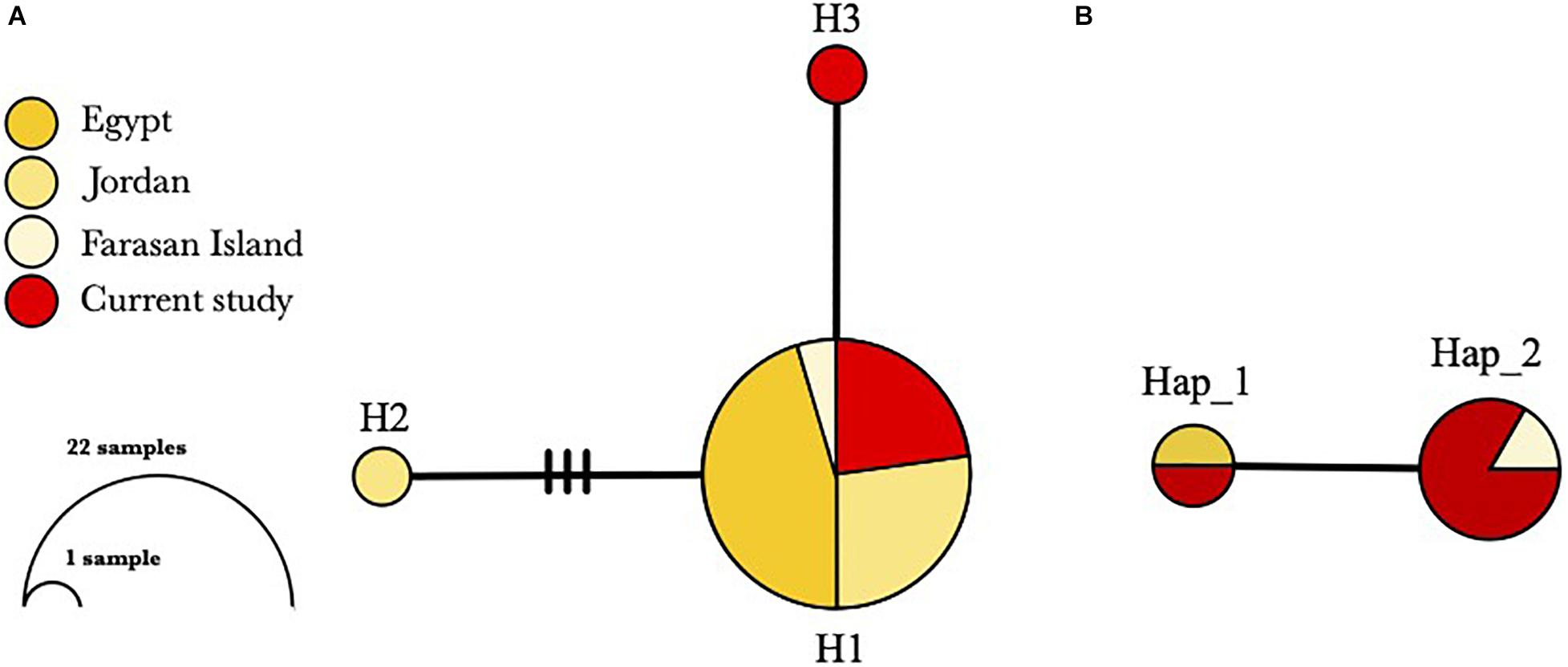
Figure 3. Haplotype networks based on (A) 24 16S sequences and (B) eight mtCOI sequences of T. squamosina in the Red Sea. Each circle corresponds to one haplotype (H for 16S; Hap for mtCOI) and the size of each circle is proportional to its frequency. Connecting lines between haplotypes represent one mutational step with additional mutational steps represented by the hatches.
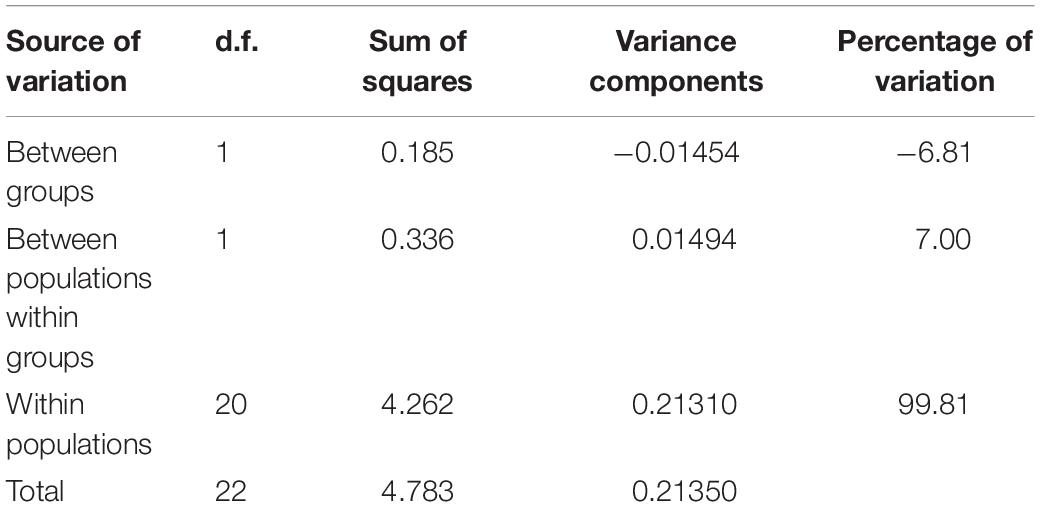
Table 1. Analysis of molecular variance of three populations in the Red Sea. Group refers to the pooling of Jordan and Egypt together (northern population) and Farasan Banks alone (southern population).
This study confirms the occurrence of Tridacna squamosina in the South-eastern Saudi Arabian Red Sea, based on DNA sequences. Roa-Quiaoit (2005) reported that T. squamosina is geographically restricted at the marginal tip of the Gulf of Aqaba, where the cooler water temperatures are assumed to favor this species. However, our results combined with the finding of a single individual from Farasan Island by Fauvelot et al. (2020), confirm that this species also occurs in the warmer, productive waters of the southern Red Sea. Apart from the environmental factors, their current distribution pattern could be affected by the settlement behavior of giant clam larvae and the geographical events. The disjointed distribution of T. squamosina under two extreme environmental conditions in the Red Sea is perplexing, given that the environmental conditions in the central Red Sea should also be suitable for this species. The most parsimonious explanation for this distribution pattern is a local extinction in the central Red Sea as suggested by Pappas et al. (2017) from their field survey data. The ecology and habitat of T. squamosina we describe here is similar to that of the bear paw clam (Hippopus hippopus), which has suffered substantial range contraction and local extinction in Pacific Islands as a result of human harvesting (Seeto et al., 2012) as well as glacio-eustatic sea level fluctuations (Paulay, 1996). Unlike most other Tridacninae species, T. squamosina is not permanently attached to a hard substrate, thus making it more susceptible to manual harvesting by hand [see Neo et al. (2017) and references therein]. This species may have been the main target for human collection in the Red Sea coast during the last interglacial period (>1,250,000 years ago) (Richter et al., 2008), but the ongoing exploitation of Tridacna stocks in the northern, central, and southern Red Sea was also evident in the past decades (Bodoy, 1984; Gladstone, 2000; Ashworth et al., 2004). The decline in wild Tridacna stocks in the Red Sea could also be linked with coastal development around the major cities. In early 2000s, reduced coral cover (i.e., 20%, at 5 m) was observed in central Red Sea (e.g. Jeddah) due to the urban tourism and city development, in comparison to less disturbed reefs in the North and South (Kotb et al., 2004). High sedimentation can hinder the settlement and recruitment of giant clam larvae (Guest et al., 2008), thus jeopardizing their recruitment success in the central Red Sea. Hence, we hypothesize that the disjoint distribution of T. squamosina is a result of extirpation of populations between the Gulf of Aqaba in the North and the Farasan regions in the South by the combined effects of historical overexploitation and coastal development.
The overall size of T. squamosina found in this study (15–30 cm; six individuals) was generally smaller than reported by Richter et al. (2008) (27–34 cm; five individuals), but meaningful conclusions from this remains limited. The sexual maturation of giant clams can be inferred from their gonad status (Richter et al., 2008) and hence, biopsies of the gonadal tissue can help to estimate their age. However, we did not perform gonad biopsies due to logistic limitations and further investigations are needed to verify clam age. Mohammed and Yassien (2008) reported that high coral cover plays a major role in the bivalve assemblages in the Red Sea, it is however, the opposite for T. squamosina. All individuals were found free-living on the sandy-rubble flats with extremely low coral cover (i.e., ∼5%, see Anton et al., 2020), supporting the hypothesis that this species might have overcome the competitive advantages of its sympatric congeners by inhabiting shallow areas with low coral cover. With respect to our genetic results, we obtained T. squamosina mtCOI sequences that were twice the length (i.e., 500 bp) of those from Fauvelot et al. (2020). The reasons for this large difference are unclear. Our results confirm that the use of SQUA-primers for DNA barcoding is successful for amplification, identification and discrimination of the rare species from its congeners in the Red Sea. Low mitochondrial genetic diversity inferred from the pooled 16S dataset suggests that this population is exposed to an increased risk of extinction (Markert et al., 2010), but the data should be treated with caution as low genetic diversity could also result from a small population size. Low levels of genetic diversity observed in marine organisms is often associated with overexploitation and habitat degradation in the past (Rodrigues et al., 2008), suggesting that T. squamosina has experienced a recent population bottleneck. The number of haplotypes (n = 3) and variable sites were extremely low (n = 5/410, 1%). This level of variation is comparable to the rare bear paw clams (Hippopus hippopus) in the Philippines using the same marker (n = 1/498, 0.2%; Lizano and Santos, 2014). As the surveyed population is too small to make a comparison, it is challenging to distinguish between a species having naturally low levels of genetic variation and a significant loss in variation as the result of a population bottleneck. To overcome this limitation, we looked into the species’ population history. The coalescent estimator of genetic diversity (θ = 0.003) (Watterson, 1975) was larger than Tajima estimator (π = 0.001) (Tajima, 1983), suggesting historically higher diversity for T. squamosina.
The genetic structure of T. squamosina based on 16S mtDNA marker revealed in this study is important for ecological conservation and restoration. The fact that major proportion of the variability (99.81%) is present within populations inferred from AMOVA analysis implies that outbreeding depression is not a concern, and sourcing donor clams for future restocking programs does not have to be restricted to certain sites or environmental conditions. The low, non-significant FST showing weak mitochondrial DNA (mtDNA) differentiation between northern and southern populations of T. squamosina in the Red Sea could be attributed to high gene flow. Such a weak population structure noticed in the present study is in accordance with Lim et al. (2020), where a panmictic population was identified for its sister species, T. maxima in the same region. However, the limited sample size of T. squamosina specimens collected in this study did not allow us to fully address the true population structure. Thus, the genetic differentiation could be overestimated in our scenario. While mtDNA is one of the popular choices of genetic markers in conservation genetics, it has a few drawbacks in interpreting population connectivity as discussed in Zhang and Hewitt (2003) and Wan et al. (2004). Firstly, the maternal inheritance of mtDNA restricts it to exploring events at the maternal angle, which may underrepresent the species’ true dispersal potential. Secondly, it has limited use in investigating recent loss of genetic variation and any individual-level events such as identity, individual dispersal, and mating systems. Future studies should include the use of microsatellite markers to study the demographic connectivity of Red Sea giant clams at these spatial scales, which was tested in other endangered marine bivalves based on the isolation by distance (IBD) hypothesis (Wesselmann et al., 2018).
In conclusion, our study suggests that T. squamosina is an endangered species in the Red Sea. Given their already low population abundance and genetic variability, this species deserves an enhanced monitoring effort in Saudi Arabia. Overharvesting of Red Sea giant clams was probably one of the main drivers for their historical population decline (Gladstone, 2000; Ashworth et al., 2004). Nowadays, fishing pressure seems to have a rather negligible impact on the present giant clams stocks in the region (Rossbach et al., 2020). The particularly sharp decline in T. squamosina densities, however, may be caused by its weak attachment to the substratum by its byssus threads, making it more vulnerable to collection by hand than the other Tridacninae species in the same region. Nonetheless, we call for actions to protect and conserve the remaining population of T. squamosina along with biobanking and fertility preservation programs to safeguard remnant populations in the Red Sea, paralleling successful efforts in cryopreserving the gametes and embryos of other marine bivalve molluscs elsewhere in the world (Marina et al., 2020). Lastly, integrated coastal zone management plans need to be implemented in the coastal cities to mitigate the impacts of coastal developments and wastewater discharges on nearby reefs. Hence, we urge the relevant authorities to work closely with the coastal communities toward conservation of giant clams, such has been proposed on listing Tridacna spp. as a key taxa of high conservation priority in Saudi Arabia (AbuZinada et al., 2004).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SR collected the samples. KL conducted the barcoding with assistance from NG and ES. KL took the lead in writing the manuscript. All authors discussed the results and commented on the manuscript.
This work was funded by King Abdullah University of Science and Technology (KAUST), through baseline funding to CD (BAS/1/1071-01-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the KAUST BioScience Core Lab for sequence data generation and Felix Ivo Rossbach for assistance with field work. KL acknowledges support from the IMBRSea MSs program. ES acknowledges fellowships from FCT under grant (SFRH/BSAB/150485/2019) and Pew Marine.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.628142/full#supplementary-material
Supplementary Figure 1 | Alignment of eight T. squamosina mtCOI sequences showing the two specimens sharing the same point mutation site on the 51st position.
Supplementary Table 1 | Summary information of Red Sea Tridacna squamosina sequences downloaded from GenBank used in the phylogenetic analysis and comparison purpose.
AbuZinada, A. H., Robinson, E. R., Nader, I. A., and Al Wetaid, Y. I. (2004). First Saudi Arabian National Report on the Convention on Biological Diversity. Available online at: https://www.cbd.int/doc/world/sa/sa-nr-01-en.pdf (accessed October 26, 2020).
Anton, A., Randle, J. L., Garcia, F. C., Rossbach, S., Ellis, J. I., Weinzierl, M., et al. (2020). Differential thermal tolerance between algae and corals may trigger the proliferation of algae in coral reefs. Global Change Biol. 26, 4316–4327. doi: 10.1111/gcb.15141
Ashworth, J. S., Ormond, R. F. G., and Sturrock, H. T. (2004). Effects of reef-top gathering and fishing on invertebrate abundance across take and no-take zones. J. Exp. Mar. Biol. Ecol. 303, 221–242. doi: 10.1016/j.jembe.2003.11.017
Bodoy, A. (1984). “Assessment of human impact on giant clams (Tridacna maxima) near Jeddah, Saudi Arabia,” in Proceedings of the Symposium on Coral Reef Envrionment of the Red Sea, Jeddah.
Borsa, P., Fauvelot, C., Andréfouet, S., Chai, T.-T., Kubo, H., and Liu, L.-L. (2015). On the validity of Noah’s giant clam Tridacna noae (Röding, 1798) and its synonymy with Ningaloo giant clam Tridacna ningaloo Penny & Willan, 2014. Raffles Bull. Zool. 63, 484–489.
DeBoer, T. S., Subia, M. D., Erdmann, M. V., Kovitvongsa, K., and Barber, P. H. (2008). Phylogeography and limited genetic connectivity in the endangered boring giant clam across the Coral Triangle. Conserv. Biol. 22, 1255–1266. doi: 10.1111/j.1523-1739.2008.00983.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fauvelot, C., Zuccon, D., Borsa, P., Grulois, D., Magalon, H., Riquet, F., et al. (2020). Phylogeographical patterns and a cryptic species provide new insights into Western Indian Ocean giant clams phylogenetic relationships and colonization history. J. Biogeogr. 47, 1086–1105. doi: 10.1111/jbi.13797
Froukh, T., and Kochzius, M. (2007). Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Mol. Ecol. 16, 1359–1367. doi: 10.1111/j.1365-294X.2007.03236.x
Giles, E. C., Saenz-Agudelo, P., Hussey, N. E., Ravasi, T., and Berumen, M. L. (2015). Exploring seascape genetics and kinship in the reef sponge Stylissa carteri in the Red Sea. Ecol. Evol. 5, 2487–2502. doi: 10.1002/ece3.1511
Gladstone, W. (2000). The ecological and social basis for management of a Red Sea marine-protected area. Ocean Coast. Manag. 43, 1015–1032. doi: 10.1016/s0964-5691(00)00070-3
Golani, D., and Bogorodsky, S. V. (2010). The fishes of the Red Sea- reappraisal and updated checklist. Zootaxa 2463, 1–100. doi: 10.11646/zootaxa.2463.1.1
Guest, J. R., Todd, P. A., Goh, E., Sivalonganathan, B. S., and Reddy, K. P. (2008). Can giant clam (Tridacna squamosa) populations be restored on Singapore’s heavily impacted coral reefs? Aquat. Conserv. 18, 570–579. doi: 10.1002/aqc.888
Huber, M., and Eschner, A. (2011). Tridacna (Chametrachea) costata Roa-Quiaoit, Kochzius, Jantzen, Al-Zibdah & Ritcher from the Red Sea, a junior synonym of Tridacna squamosina Sturany, 1899 (Bivalvia, Tridacnidae). Ann. Naturhist. Mus. Wien B 112, 153–162.
Kotb, M., Abdulaziz, M., Al-Agwan, Z., Al-Shaikh, K., Al-Yami, H., Banajah, A., et al. (2004). “Status of coral reefs in the red sea and gulf of Aden in 2004,” in Status of Coral Reefs of the World: 2004, Vol. 1, ed. C. Wilkinson (Townsville, QLD: Australian Institute of Marine Science), 137–154.
Lim, K. K., Rossbach, S., Geraldi, N. R., Schmidt-Roach, S., Serrao, E. A., and Duarte, C. M. (2020). The small giant clam, Tridacna maxima exhibits minimal population genetic structure in the Red Sea and genetic differentiation from the Gulf of Aden. Front. Mar. Sci. 7:570361. doi: 10.3389/fmars.2020.570361
Lizano, A. M. D., and Santos, M. D. (2014). Updates on the status of giant clams Tridacna spp. and Hippopus hippopus in the Philippines using mitochondrial CO1 and 16S rRNA genes. Philipp. Sci. Lett. 7, 187–200.
Marina, D.-S., Nelly, S., Marc, S., and Jacky, C. (2020). “State of the art in cryopreservation of bivalve spermatozoa,” in Cryopreservation of Fish Gametes, eds J. Betsy and S. Kumar (Singapore: Springer), 239–284. doi: 10.1007/978-981-15-4025-7_11
Markert, J. A., Champlin, D. M., Gutjahr-Gobell, R., Grear, J. S., Kuhn, A., McGreevy, T. J., et al. (2010). Population genetic diversity and fitness in multiple environments. BMC Evolution. Biol. 10:205. doi: 10.1186/1471-2148-10-205
Mohammed, T. A., and Yassien, M. H. (2008). Bivalve assemblages on living coral species in the Northern Red Sea, Egypt. J. Shellfish Res. 27, 1217–1223. doi: 10.2983/0730-8000-27.5.1217
Nanninga, G. B., Saenz-Agudelo, P., Manica, A., and Berumen, M. L. (2014). Environmental gradients predict the genetic population structure of a coral reef fish in the Red Sea. Mol. Ecol. 23, 591–602. doi: 10.1111/mec.12623
Neo, M. L. (2020). “Conservation of giant clams (Bivalvia: Cardiidae),” in Encyclopedia of the World’s Biomes, Vol. 4, eds M. I. Goldstein and D. A. DellaSala (Amsterdam: Elsevier), 527–538. doi: 10.1016/b978-0-12-409548-9.11780-4
Neo, M. L., Eckman, W., Vicentuan, K., Teo, S. L. M., and Todd, P. A. (2015). The ecological significance of giant clams in coral reef ecosystems. Biol. Conserv. 181, 111–123. doi: 10.1016/j.biocon.2014.11.004
Neo, M. L., Wabnitz, C. C. C., Braley, R. D., Heslinga, G. A., Fauvelot, C., Van Wynsberge, S., et al. (2017). Giant clams (Bivalvia: Cardiidae: Tridacninae): a comprehensive update of species and their distribution, current threats and conservation status. Oceanogr. Mar. Biol. 55, 87–387. doi: 10.1201/b21944-5
Nuryanto, A., Duryadi, D., Soedharma, D., and Blohm, D. (2007). Molecular phylogeny of giant clams based on mitochondrial DNA cytochrome c oxidase I gene. HAYATI J. Biosci. 14, 162–166. doi: 10.4308/hjb.14.4.162
Pappas, M. K., He, S., Hardenstine, R. S., Kanee, H., and Berumen, M. L. (2017). Genetic diversity of giant clams (Tridacna spp.) and their associated Symbiodinium in the central Red Sea. Mar. Biodivers. 47, 1209–1222. doi: 10.1007/s12526-017-0715-2
Paulay, G. (1996). Dynamic clams: changes in the bivalve fauna of Pacifc Islands as a result of sea-level fluctuations. Am. Malacol. Bull. 12, 45–57.
Phillips, J. D., Gillis, D. J., and Hanner, R. H. (2019). Incomplete estimates of genetic diversity within species: Implications for DNA barcoding. Ecol. Evol. 9, 2996–3010. doi: 10.1002/ece3.4757
Richter, C., Roa-Quiaoit, H., Jantzen, C., Al-Zibdah, M., and Kochzius, M. (2008). Collapse of a new living species of giant clam in the Red Sea. Curr. Biol. 18, 1349–1354. doi: 10.1016/j.cub.2008.07.060
Roa-Quiaoit, H. A. F. (2005). The Ecology and Culture of Giant Clams (Tridacnidae) in the Jordanian Sector of the Gulf of Aqaba, Red Sea. Ph.D. thesis, Universitat Bremen, Bremen.
Rodrigues, R., Schneider, H., Santos, S., Vallinoto, M., Sain-Paul, U., and Sampaio, I. (2008). Low levels of genetic diversity depicted from mitochondrial DNA sequences in a heavily exploited marine fish (Cynoscion acoupa, Sciaenidae) from the Northern coast of Brazil. Genet. Mol. Biol. 31, 487–492. doi: 10.1590/s1415-47572008000300015
Rossbach, S., Anton, A., and Duarte, C. M. (2020). Drivers of the abundance of Tridacna spp. giant clams in the Red Sea. Front. Mar. Sci. 7:592852. doi: 10.3389/fmars.2020.592852
Rossbach, S., Cardenas, A., Perna, G., Duarte, C. M., and Voolstra, C. R. (2019a). Tissue-specific microbiomes of the Red Sea giant clam Tridacna maxima highlight differential abundance of Endozoicomonadaceae. Front. Microbiol. 10:2661. doi: 10.3389/fmicb.2019.02661
Rossbach, S., Saderne, V., Anton, A., and Duarte, C. M. (2019b). Light-dependent calcification in Red Sea giant clam Tridacna maxima. Biogeosciences 16, 2635–2650. doi: 10.5194/bg-16-2635-2019
Seeto, J., Nunn, P. D., and Sanjana, S. (2012). Human-mediated prehistoric marine extinction in the tropical Pacific? Understanding the presence of Hippopus hippopus (Linn. 1758) in ancient shell middens on the Rove Peninsula, southwest Viti Levu Island, Fiji. Geoarchaeology 27, 2–17. doi: 10.1002/gea.21385
Su, Y., Hung, J.-H., Kubo, H., and Liu, L.-L. (2014). Tridacna noae (Röding, 1798) – a valid gaint clam species separated from T. maxima (Röding, 1798) by morphological and genetic data. Raffles Bull. Zool. 62, 124–135.
Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460. doi: 10.1093/genetics/105.2.437
Tan, E. Y. W., Quek, Z. B. R., Neo, M. L., Fauvelot, C., and Huang, D. (2021). Genome skimming resolves the giant clam (Bivalvia: Cardiidae: Tridacninae) tree of life. Coral Reefs doi: 10.1007/s00338-020-02039-w
Templeton, A. R. (1993). The “eve” hypotheses: a genetic critique and reanalysis. Am. Anthropol. 95, 51–72. doi: 10.1525/aa.1993.95.1.02a00030
Wan, Q. H., Wu, H., Fujihara, T., and Fang, S. G. (2004). Which genetic marker for which conservation genetics issue? Electrophoresis 25, 2165–2176. doi: 10.1002/elps.200305922
Watterson, G. A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276. doi: 10.1016/0040-5809(75)90020-9
Wesselmann, M., Gonzalez-Wanguemert, M., Serrao, E. A., Engelen, A. H., Renault, L., Garcia-March, J. R., et al. (2018). Genetic and oceanographic tools reveal high population connectivity and diversity in the endangered pen shell Pinna nobilis. Sci. Rep. 8:4770. doi: 10.1038/s41598-018-23004-2
Keywords: rare species, extinction, conservation biology, Western Indian Ocean, Tridacna
Citation: Lim KK, Rossbach S, Geraldi NR, Serrão EA and Duarte CM (2021) Phylogeographic Analysis Suggests a Recent Population Bottleneck in the Rare Red Sea Tridacna squamosina. Front. Mar. Sci. 8:628142. doi: 10.3389/fmars.2021.628142
Received: 11 November 2020; Accepted: 21 January 2021;
Published: 10 February 2021.
Edited by:
Rochelle Diane Seitz, College of William & Mary, United StatesReviewed by:
Mei Lin Neo, National University of Singapore, SingaporeCopyright © 2021 Lim, Rossbach, Geraldi, Serrão and Duarte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos M. Duarte, Y2FybG9zLmR1YXJ0ZUBrYXVzdC5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.