
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci. , 19 February 2021
Sec. Marine Biotechnology and Bioproducts
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.624624
 Donlaporn Sripan1†
Donlaporn Sripan1† Alisa Wilantho2†
Alisa Wilantho2† Khunnalack Khitmoh1
Khunnalack Khitmoh1 Doonyapong Wongsawaeng3
Doonyapong Wongsawaeng3 Jamal Ouazzani4
Jamal Ouazzani4 Suchana Chavanich5,6
Suchana Chavanich5,6 Sissades Tongsima2
Sissades Tongsima2 Naraporn Somboonna1,7*
Naraporn Somboonna1,7*The southeast Andaman Sea 52-m off the west coast of Phang Nga province, Thailand, is located in the Indian Ocean, representing a hotspot for marine biodiversity of the world. This study utilized metagenomics combined 16S rRNA gene (V3–V4) sequencing, and firstly revealed the microbiota and their metabolism potentials of this site at an epipelagic depth (150-m depth, TC150M), including comparison with its pelagic depth (30-m depth, TC30M) as well as other nearby Thailand and global ocean sites. Between TC150M and TC30M, the TC150M microbial metagenomics was an eight-fold higher, and the microbiota comprised, for examples more abundant Bacteroidetes while fewer Proteobacteria, than the TC30M. The microbial metabolic potentials of the TC150M were statistically higher in replication repair and metabolisms of amino acids, lipids, nucleotides, and xenobiotics biodegradation, etc. Following comparative microbiota analyses between three Andaman Sea sites and two Gulf of Thailand sites, the relatively great proportions of Bacteroidetes, Nitrospirae, Gemmatimonadetes, and Chlorobi characterized the southeast Andaman Sea. Nevertheless, the microbiota representing Thailand marine sites remained distinguished from the global ocean sites where beta diversities were close. Thai maritime sites showed proportionally higher Proteobacteria, Bacteroides, Nitrospirae, Gemmatimonadetes, and Chlorobi. Thus, the Thai marine microbiota database helps better understand our global ocean microbiota and microbial metabolic potentials. Here, the microbial metabolism potentials between Thailand and the global ocean sites of relatively close microbiota databases encompose the similar functions yet in statistically different frequencies. Our research provided the first preliminary marine microbiome comparison between the epipelagic and pelagic sea levels of the southeast Andaman Sea, Thailand.
An advance in next generation sequencing allows metagenomics to identify marine bacterial community (or microbiota), in which their biodiversity could have impact on nutrient and energy cyclings (Somboonna et al., 2012; Webster and Reusch, 2017; Kelly et al., 2018), and further an association to abiotic factors, such as Earth plate tectonics (Costello et al., 2017; Khitmoh et al., 2017; Zaffos et al., 2017) and global warming (Webster et al., 2016). Marine bacteria represent a sensitive biomarker of an environment, and play roles in ecosystems (Zaffos et al., 2017; Walter et al., 2019).
The Andaman Sea, located in the Indian Ocean, carries one of the highest aquatic life abundances on Earth. This region is considered one of the 25 endangered ecoregions. The 2004 Indian Ocean tsunami affected the southeast Andaman Sea off the west coast of Thailand. The Southeast Andaman Sea comprises a unique hydrography, of stratified inflow freshwater from the Irrawaddy River (India) and nutrient-rich water runoff from the mangrove forests of Thailand (Gupta et al., 1981; Varkey et al., 1996). Recently, the submarine landslide via the oceanic India Plate and the Burma Plate movement caused 9.1 magnitude Sumatra-Andaman earthquake (tsunami) that affect the coasts of many countries of South and Southeast Asia on 26 December 2004. This hit included the southeast islands of Phang Nga province of Thailand, Tachai (TC) and Phra Thong (PT) islands. The PT (N8.9334 E98.0976) and TC (N8.8516 E97.5201) locate approximately 15 and 52 km offshore of the southeast Andaman Sea coast of Thailand, respectively. Indeed, four tsunami events had been geographically revealed previously in this region, with the two most recent ones were in 2004 and the other approximately 600 years ago (Jankaew et al., 2008; Somboonna et al., 2014). These plate tectonics were reported to drive tropical reef bacteria and animal diversity dynamics, and might explain the hotspot species diversification in some tropical reef region (Leprieur et al., 2016; Zaffos et al., 2017). Nonetheless, only an open ocean level microbiota of this area has been explored.
Supportively, previous study of deeper marine ecosystems was found a group of bacteria involved in organic degradation than the surface of the water. Including the lack of sunlight and the greater pressure in the deeper sea level than that of the surface, these all could differentiate the bacterial diversity in the TC150 (Wannicke et al., 2015). Therefore, this research aimed to utilize a culture-independent, metagenomics derived 16S rRNA gene pyrosequencing to analyze the epipelagic-level bacterial community in this area (TC150M), and compare with nearby marine sites including TC at 30-m depth (TC30M), PT at 30-m depth (PT30M) (Khitmoh et al., 2017), surface marine site in the Gulf of Thailand (TW and TP) (Somboonna et al., 2012), and global ocean sites which bacterial communities were previously reported close to Thailand marine microbiota (GS001a, GS012, GS028, GS049, GS108b, GS110b, and GS122b) (Somboonna et al., 2017). Due to the difficulty of sample collection, the database of below an open ocean level is rare in Thailand. This database represents the first report of the epipelagic microbiota from the southeast Andaman Sea of Thailand, and supports the marine microbiota knowledge of the Indian Ocean.
The marine sample site TC150M (N8.8516 E97.5201) located 52 km offshore of the southeast Andaman Sea coast, Thailand (Supplementary Figure 1). This research area did not require any ethical approval. Water sample was collected at a depth of ∼150 m below the sea surface on 7 January 2013, 14:00–17:00 h. Three independent samples of 20 L of seawater were collected using Niskin Water Sampler (KC Denmark A/S, Silkeborg, Denmark). On-site water temperature, conductivity, salinity, dissolved oxygen, and pH were measured. Samples were transported immediately to laboratory.
Water sample was filtered through a four-layered sterile cheesecloth to remove organisms and debris of ≥30 μm in diameter sizes, and through a 0.22 μm filter (Merck Millipore, Burlington, MA, United States) to trap microorganisms of ≤0.22 μm. The total DNA was extracted using the Metagenomic DNA Isolation Kit for Water (Epicentre, Madison, WI, United States) (Somboonna et al., 2012). The isolated metagenomes had an apparent size of ∼40 kb as recommended by the manufacturer. The DNA concentration and quality were analyzed by agarose gel electrophoresis and absorbance at 260 and 280 nm, respectively.
The universal (conserved across genera) prokaryotic primers 338F and 786R, which target the V3 and V4 regions of the 16S rRNA gene (Lilliana et al., 2006; Mao et al., 2012; Somboonna et al., 2012) with appended 5′-AGACGACG-3′ tagged barcode (Parameswaran et al., 2007), were used to construct the amplicon library of the TC-150. For each of the individual triplicate sample, three PCR reactions were performed and each 25 μl PCR reaction comprised 1 × EmeraldAmp® GT PCR Master Mix (TaKaRa, Shiga, Japan), 0.3 μM of each primer and 100 ng of metagenomic DNA. The thermal cycling condition was 4 min at 95°C followed by 30 cycles of 94°C for 45 s, 50°C for 50 s, and 72°C for 90 s, and then a final extension at 72°C for 10 min. The PCR products from all replicates were equally pooled, purified by PureLink® Quick Gel Extraction Kit (Invitrogen, New York, United States) and pyrosequenced on an eight-lane Roche picotiter plate with the 454 GS FLX system (Roche, Branford, CT, United States) according to the manufacturer’s protocols.
The nucleotide sequences were deposited in NCBI sequence read archive1 under an accession number SRP072863.
The 16S rRNA gene sequences of less than 100 nucleotides in length (excluding the primers, barcode and sequencing adapter sequences) and those that did not pass the quality threshold cutoff by the sequencing machine (e.g., minimum sequencing quality score of <35 over a 50-bp window, >10 homopolymers, and chimera sequences) were discarded. Annotation to operational taxonomic unit (OTU) at phylum and genus levels were performed following Mothur’s standard operating procedures (SOP) (Schloss et al., 2009; Bulan et al., 2018), using Naïve Bayesian method. Possible contaminated sequences from mitochondria and chloroplasts were discarded. Alpha diversity (Good’s coverage index to estimate percent sequencing coverage, and rarefaction curve) and beta diversity analyses were performed using Mothur. The beta diversity coefficients (Thetayc) of the TC150M profile compared with TC30M and PT30M (Khitmoh et al., 2017), TW and TP (Somboonna et al., 2012), and global ocean sites GS001a, GS012, GS028, GS049, GS108b, GS110b, and GS122b profiles2 (Rusch et al., 2007; Zinger et al., 2011), were used to construct the neighbor-joining tree and PCoA in Mothur (Schloss et al., 2009). Potential metabolisms of the microbiota were predicted from the 16S rDNA sequences using PICRUSt in Mothur (Langille et al., 2013). The metabolic functions were categorized by KEGG (Kyoto Encyclopedia of genes and genomes) pathways, given level 1 representing pathways and level 2 representing categories of genes (COGs). For statistical analysis, analysis of molecular variance (AMOVA) were used to test for significant difference between groups (P < 0.05).
The physical properties of TC150M seawater were compared with those of TC30M (Khitmoh et al., 2017). The deeper sea TC150M was found 37.41% reduced dissolved oxygen (DO) (Table 1A, TC150M 4.3 mg/mL vs. TC30M 6.87 mg/m) and 29.61% reduced temperature (TC150M 19.99°C vs. TC30M 28.40°C).
The microbial metagenomic DNA concentration was 111.99 ng/L of seawater, with a purity (A260/A280) of 1.61 ± 0.05. This accounted an eight-fold higher abundance of the microbial metagenomic DNA compared with that of TC30M (Khitmoh et al., 2017), suggesting the more abundant microorganisms at the 150-m than the 30-m depth. The diversity in bacterial community and functional networks across sea depths had been reported elsewhere (Cram et al., 2015). The 16S rRNA gene V3-V4 library was amplified to the appropriate size [466 base pairs (bp), including primer, barcode, and adapter sequences] (Meyer et al., 2008; Somboonna et al., 2019), and pyrosequencing resulted in a respectable yield as observed by plateau rarefaction curve at 425 OTUs at genus level (Supplementary Figure 2). Of 39,094 raw reads, 99.98% were quality reads, and of the quality reads 99.17% could be annotated (Table 1B). Average read length after trimming of primer, barcode and adapter sequences were approximately 262 bp, as expected (Khitmoh et al., 2017).
The marine microbiota of TC150M comprised mainly bacterial phyla Proteobacteria (68.045%), followed by Bacteroidetes (11.301%), a group of unclassified bacterial phylum (7.672%), Nitrospirae, Gemmatimonadetes, Chlorobi, Acidobacteria, Spirochetes, Caldithrix, etc (Figure 1A). The common Proteobacteria genera were found a genus in family Piscirickettsiaceae (12.33%), a genus in class Gammaproteobacteria (11.93%) and genus Desulfococcus (5.69%), for examples; and 40.819% of Proteobacteria belonged to minor genera each <2% abundance (Figure 1B). Comparing with TC30M, the TC150M showed the close microbiota profiles at phylum and genus levels (Figures 2A,B). However, the microbiota between TC30M and PT30M were closer (Thetayc dissimilarity indices between TC30M-PT30M was 0.030643, and TC30M-TC150M was 0.034899) and thus the neighbor joining tree clustered these two together in prior to the clustering with the TC150M, and TP and TW, in order (Figure 2B). The prior pointed the differences of microbiota along surface versus bottom sea levels (in details, the relatively more abundant Bacteroidetes and the fewer Proteobacteria in the TC150M), and the latter suggested the relationship of marine microbiota in Thailand albeit the Andaman Sea and the Gulf of Thailand (GoT) (AMOVA, P = 0.579 between TC150M vs. Thailand sites and P = 0.046 between TC150M vs. global ocean sites). Comparative genome sequences of marine Bacteroidetes were reported a preference bacterial functions for growth and degradation of high molecular weight particles (e.g., polymers and glycosyl hydrolases) (Fernández-Gómez et al., 2013), which supported living in sea bottom marine. The Thetayc of marine microbiota of the TC150M with others of Thailand ranged 0.030643–0.037504, meanwhile of the TC150M with global ocean sites ranged 0.8937–0.9086. The higher the Thetayc dissimilarity indices infer the larger the community diversity. Additionally, a large number of unclassified genera in marine ecosystems, in ours as well as other global ocean sites (Figure 2A), using conventional 16S rRNA gene marker were consistent with previous reports, and that the tropical Indian Ocean remained unexplored and databases are limited to cultured bacteria (Royo-Llonch et al., 2017; Tully, 2019). Bacterial phyla that dominantly distinguished the group of TC150M, TC30M, and PT30M (sites in the southeast Andaman Sea), from the group of TP and TW (site in GoT), were found Bacteroidetes, Nitrospirae, Gemmatimonadetes, and Chlorobi (higher in the southeast Andaman Sea sites) and Actinobacteria (significantly higher in GoT sites) (Figure 2A; Khitmoh et al., 2017). These genus differences resulted in the principal coordinate analysis (PCoA) that separated the groups of the southeast Andaman Sea and the GoT (Figure 2C: right pane belonged Thailand sites, and left pane belonged GOS sites).
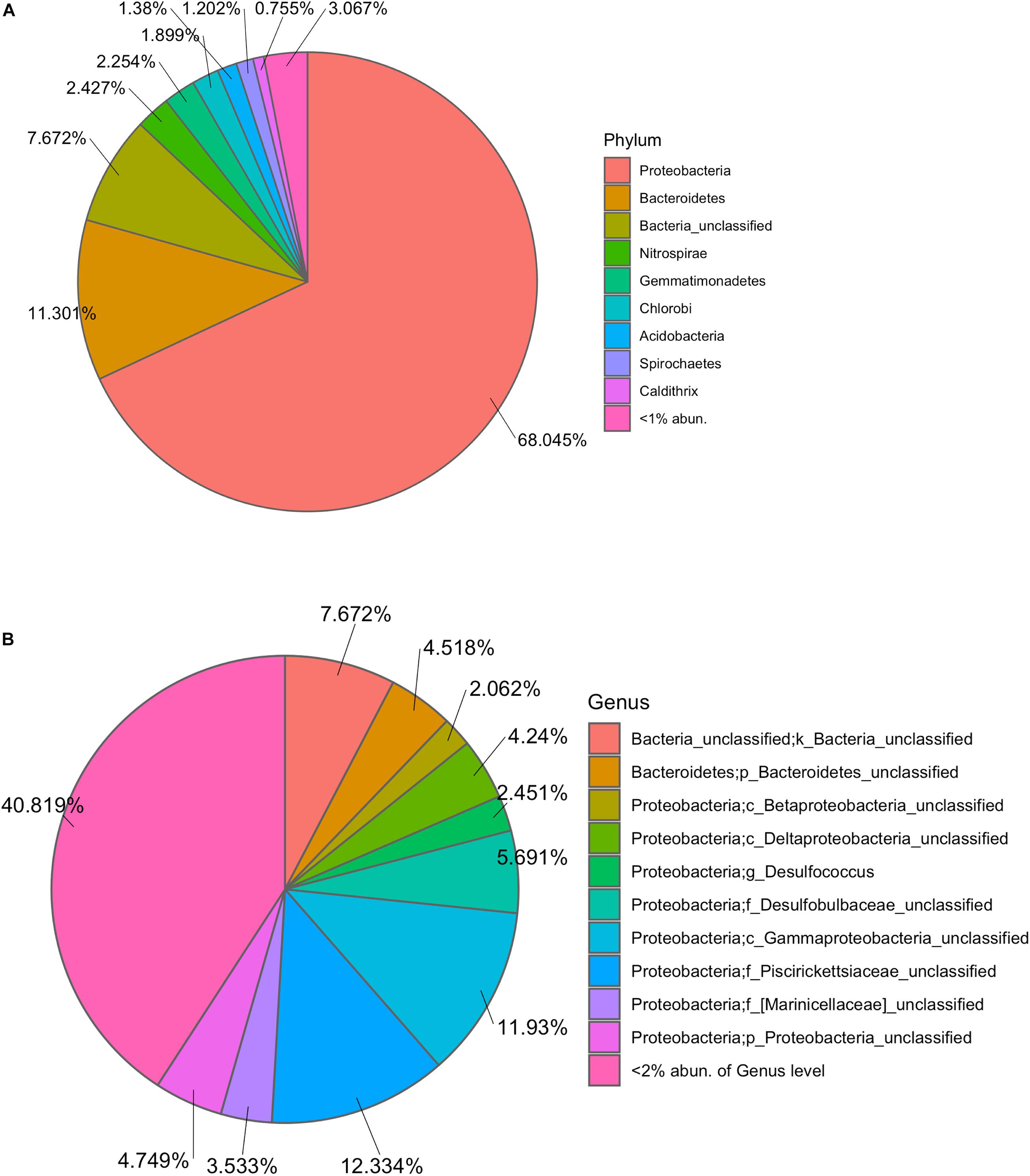
Figure 1. Taxonomic composition at phylum (A) and genus (B) levels. In (A), phyla of <2% relative abundance were not displayed names and percentages. In (B), species of unclassified genera were annotated f, family; o, order; c, class; or p, phylum, based on their deepest taxonomic ranks that can be identified.
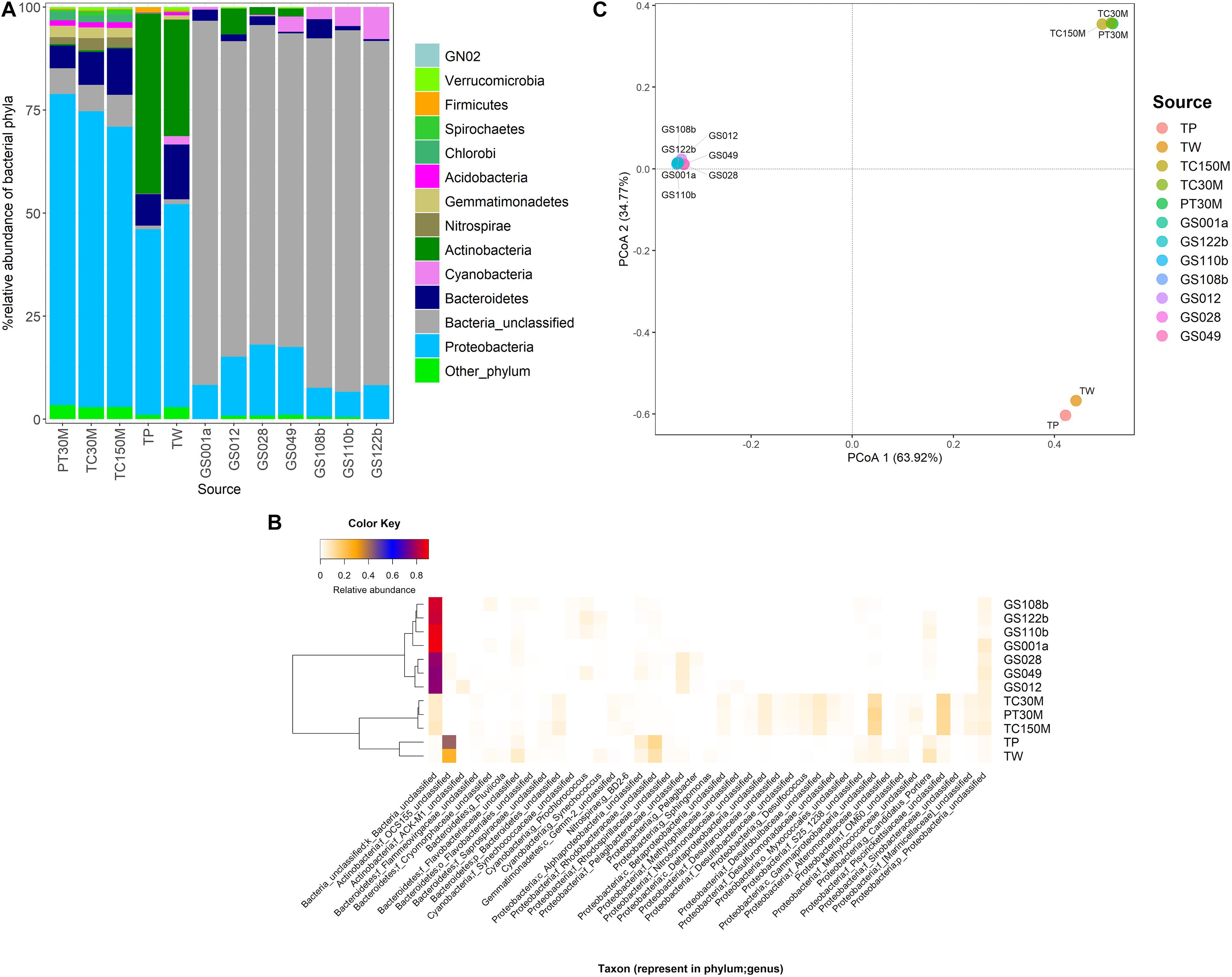
Figure 2. Prokaryotic communities at TC150M, compared with PT30M and TC30M (almost identical GPS coordinates), TP and TW (close GPS coordinates), and global ocean sites that were close to Thailand marine microbiota including GS001a, GS012, GS028, GS049, GS108b, GS110b, and GS122b (Khitmoh et al., 2017; Somboonna et al., 2017), at phylum (A) and genus (B) levels; and principal coordinates analysis (C) displaying beta diversity distances among sites. In (B), on the left represented a neighbor joining tree constructed from microbiota distances (Thetayc) among sites.
For examples, researches of deep seas of different locations, such as the north and south Atlantis, class Gammaproteobacteria were found more dominant at the relatively deeper sea levels (Agogué et al., 2011; Kai et al., 2017). Our results are consistent, as TC150 comprised the more relative abundance of Gammaproteobacteria than TC30 (TC150 35.29% and TC30 33.39%). Moreover, the marine sediment was reported relatively lower biodiversity than the sea surface, and Gammaproteobacteria could perform sulfate depletion function under anaerobic conditions (Wang et al., 2012). For phylum Bacteroidetes, which we found the relatively more abundant at the deeper sea level, the finding was consistent with Yin et al. (2013) that saprophytic bacteria in Bacteroidetes can degrade organic matters, which present relatively high in the deeper water level where sediment accumulates.
The microbiota derived potential metabolisms between the TC150M and TC30M were also compared. For instances, TC150M had significantly greater ability for amino acid metabolism (10.3%), replication repair (7.25%), lipid metabolism (3.834%), nucleotide metabolism (3.220%), and xenobiotics biodegradation and metabolism (2.923%), while significantly lower ability for membrane transport, energy metabolism and cell motility (Figure 3A). The results supported the dominance of Bacteroidetes in the TC150M. Together, our TC150M data provides a preliminary insight into a relationship of site and sea depth, which the degree of sunlight, water temperature and different macro-organism environment affected the bacterial diversity (Milici et al., 2016).
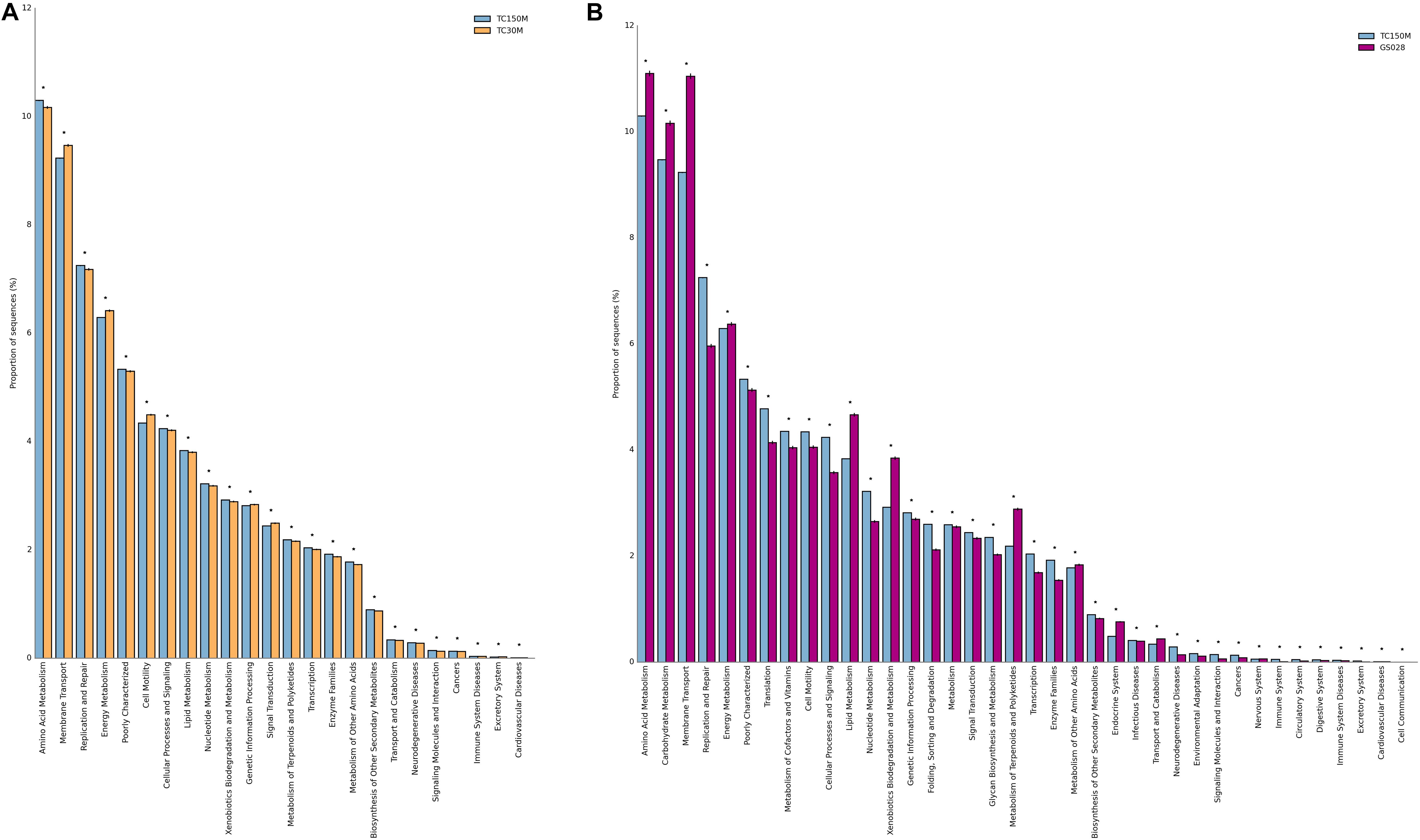
Figure 3. Metabolic potentials of TC150M, compared with TC30M (A) and GS028 (B). Only KEGG level 2 functions that were statistically different between the comparing sites were displayed (*P < 0.05).
Comparing with the global marine sites where beta diversity of microbiota profiles were close (Somboonna et al., 2017), the differences in either bacterial phylum and genus compositions and their relative frequencies could be distinguished (Figure 2). For instances, the proportionally higher Proteobacteria, Bacteroides, Nitrospirae, Gemmatimonadetes, and Chlorobi were found for Thailand marine sites. Figure 2C thereby separated the global ocean sites to the other direction (left) of Thailand marine sites. Between the TC150M and GS028 (a representative from global ocean sites) microbiota, the microbiota differences resulted in significantly high microbial metabolic potentials of replication repair, translation, cellular processes and signaling, and nucleotide metabolism in the TC150M, but the significantly high microbial metabolic potentials of amino acid, carbohydrate and energy metabolisms, and membrane transport in the GS028, for examples (Figure 3B). Nevertheless, relatively all microbial metabolic pathways (39–41 total KEGG level 2 functions) were present for both sites, in similar trends of frequencies, to suggest the common marine microbial functions that involve nutrient recyclings to facilitate sustainable phototroph-heterotroph marine species interactions (Christie-Oleza et al., 2017).
This study provided the first deep-sea (150 m) marine microbiota database from the southeast Andaman Sea of Thailand. Our comparative microbiota and potential microbial metabolisms analyses highlighted some unique bacterial diversity along the corresponding metabolic potentials in the TC150M, and supported the database and knowledge of the ecosystem of the epipelagic-level ocean microbiome. This research found the microbiota relationship between sites and mild difference at sea depth at epipelagic zone (150 m). Moreover, because of the limitation of sampling in this study, we are currently sampling ∼30–100 m depth of additional sites in order to more explicitly elucidate this epipelagic-level ocean microbiome.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP072863.
DS and AW analyzed the data. KK did molecular biology experiments and supported data analysis. DW collected the samples. JO, SC, and ST helped conceive of the study. NS conceived of the study, coordinated the experiments and data analysis, and wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the Thailand Research Fund (RSA6180046), 90th Anniversary of Chulalongkorn University Fund, EU-Horizon 2020 Project TASCMAR (634674), NRCT-JSPS Core-to-Core Program, and Microbiome Research Unit for Probiotics in Food and Cosmetics, Chulalongkorn University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.624624/full#supplementary-material
Supplementary Figure 1 | Map of TC150M site.
Supplementary Figure 2 | Estimated genus richness (rarefaction curve) of TC150M.
Agogué, H., Lamy, D., Neal, P. R., Sogin, M. L., and Herndl, G. J. (2011). Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol. Ecol. 20, 258–274. doi: 10.1111/j.1365-294x.2010.04932
Bulan, D. E., Wilantho, A., Krainara, P., Viyakarn, V., Chavanich, S., and Somboonna, N. (2018). Spatial and seasonal variability of reef bacterial communities in the upper Gulf of Thailand. Front. Mar. Sci. 5:441. doi: 10.3389/fmars.2018.00441
Christie-Oleza, J. A., Sousoni, D., Lloyd, M., Armengaud, J., and Scanlan, D. J. (2017). Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat. Microbiol. 2:17100. doi: 10.1038/nmicrobiol.2017.100
Costello, M. J., Tsai, P., Wong, P. S., Cheung, A. W. L., Basher, Z., and Chaudhary, C. (2017). Marine biogeographic realms and species endemicity. Nat. Commun. 8:1057. doi: 10.1038/s41467-017-01121-2
Cram, J. A., Xia, L. C., Needham, D. M., Sachdeva, R., Sun, F., and Fuhrman, J. A. (2015). Cross-depth analysis of marine bacterial networks suggests downward propagation of temporal changes. ISME J. 9, 2573–2586.
Fernández-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., and González, J. M. (2013). Ecology of marine Bacteroidetes: a comparative genome approach. ISME J. 7, 1026–1037.
Gupta, R. S., Moraes, C., George, M. D., Kureishy, T. W., Noronha, R. J., and Foudekar, S. P. (1981). Chemistry and hydrography of the Andaman Sea. Indian J. Mar. Sci. 10, 220–233.
Jankaew, K., Atwater, B. F., Sawai, Y., Choowong, M., Charoentitirat, T., Martin, M. E., et al. (2008). Medieval forewarning of the 2004 Indian ocean tsunami in Thailand. Nature 455, 1228–1231. doi: 10.1038/nature07373
Kai, W., Peisheng, Y., Rui, M., Wenwen, J., and Zongze, S. (2017). Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 8, 572–584. doi: 10.1080/21655979.2017.1284711
Kelly, L. E., Haas, A. F., and Nelson, C. E. (2018). Ecosystem microbiology of coral reefs: linking genomic, metabolomics and biogeochemical dynamics from animal symbioses to reefscape processes. mSystems 3:e00162-17. doi: 10.1128/mSystems.00162-17
Khitmoh, K., Wilantho, A., Wongsawaeng, D., Tongsima, S., and Somboonna, N. (2017). 16S rRNA gene sequencing assessment of the prokaryotic communities in the southeast Andaman Sea, Thailand and potential environmental alerts. Chiang Mai J. Sci. 44, 15–28.
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821.
Leprieur, F., Descombes, P., Gaboriau, T., Cowman, P. F., Parravicini, V., Kulbicki, M., et al. (2016). Plate tectonics drive tropical reef biodiversity. Nat. Commun. 7:11461. doi: 10.1038/ncomms11461
Lilliana, I. M., Deetta, K. M., James, E., Robert, T. S., and Kalai, M. (2006). Microbial metagenome profiling using amplicon length heterogeneity-polymerase chain reaction proves more effective than elemental analysis in discriminating soil specimens. J. Forensic Sci. 51, 1315–1322. doi: 10.1111/j.1556-4029.2006.00264.X
Mao, D. P., Zhou, Q., Chen, C. Y., and Quan, Z. X. (2012). Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol. 12:66. doi: 10.1186/1471-2180-12-66
Meyer, F., Paarmann, D., D’Souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386
Milici, M., Tomasch, J., Wos-Oxley, M. L., Wang, H., Jáuregui, R., Camarinha-Silva, A., et al. (2016). Low diversity of planktonic bacteria in the tropical ocean. Sci. Rep. 6:19054. doi: 10.1038/srep19054
Parameswaran, P., Jalili, R., Tao, L., Shokralla, S., Gharizadeh, B., Ronaghi, M., et al. (2007). A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35:e130. doi: 10.1093/nar/gkm760
Royo-Llonch, M., Ferrera, I., Cornejo-Castillo, F. M., Sánchez, P., Salazar, G., Stepanauskas, R., et al. (2017). Exploring microdiversity in novel Kordia sp. (Bacteroidetes) with proteorhodopsin from the tropical Indian Ocean via single amplified genomes. Front. Microbiol. 8:1317. doi: 10.3389/fmicb.2017.01317
Rusch, D. B., Halpern, A. L., Sutton, G., Heidelberg, K. B., Williamson, S., Yooseph, S., et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. doi: 10.1371/journal.pbio.0050077
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Somboonna, N., Assawamakin, A., Wilantho, A., Tangphatsornruang, S., and Tongsima, S. (2012). Metagenomic profiles of free-living archaea, bacteria and small eukaryotes in coastal areas of Sichang island, Thailand. BMC Genomics 13:S29. doi: 10.1186/1471-2164-13-S7-S29
Somboonna, N., Wilantho, A., Jankaew, K., Assawamakin, A., Sangsrakru, D., Tangphatsornruang, S., et al. (2014). Microbial ecology of Thailand tsunami and non-tsunami affected terrestrials. PLoS One 9:e94236. doi: 10.1371/journal.pone.0094236
Somboonna, N., Wilantho, A., Monanunsap, S., Chavanich, S., Tangphatsornruang, S., and Tongsima, S. (2017). Microbial communities in the reef water at Kham Island, lower Gulf of Thailand. PeerJ 5:e3625. doi: 10.7717/peerj.3625
Somboonna, N., Wilantho, A., Rerngsamran, P., Tongsima, S., and Chavanich, S. (2019). Marine bacterial diversity in coastal Sichang island, the upper Gulf of Thailand, in 2011 wet season. Front. Mar. Sci. 6:308. doi: 10.3389/fmars.2019.00308
Tully, B. J. (2019). Metabolic diversity within the globally abundant marine group II euryarchaea offers insight into ecological patterns. Nat. Commun. 10:271. doi: 10.1038/s41467-018-07840-4
Varkey, M. J., Murty, V. S. N., and Suryanarayanan, A. (1996). Physical oceanography of the Bay of Bengal and Andaman Sea. Oceanogr. Mar. Biol. 34, 1–70.
Walter, J. M., Bagi, A., and Pampanin, D. M. (2019). Insights into the potential of the Atlantic cod gut microbiome as biomarker of oil contamination in the marine environment. Microorganisms 7:209. doi: 10.3390/microorganisms7070209
Wang, Y., Sheng, H.-F., He, Y., Wu, J.-Y., Jiang, Y.-X., Tam, N. F.-Y., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Appl. Environ. Microbiol. 78, 8264–8271. doi: 10.1128/aem.01821-12
Wannicke, N., Frindte, K., Gust, G., Liskow, I., Wacker, A., Meyer, A., et al. (2015). Measuring bacterial activity and community composition at high hydrostatic pressure using a novel experimental approach: a pilot study. FEMS Microbiol. Ecol. 91:fiv036. doi: 10.1093/femsec/fiv036
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Webster, N. S., and Reusch, T. B. H. (2017). Microbial contributions to the persistence of coral reefs. ISME J. 11, 2167–2174. doi: 10.1038/ismej.2017.66
Yin, Q., Fu, B., Li, B., Shi, X., Inagaki, F., and Zhang, X.-H. (2013). Spatial variations in microbial community composition in surface seawater from the ultra-oligotrophic center to rim of the South Pacific Gyre. PLoS One 8:e55148. doi: 10.1371/journal.pone.0055148
Zaffos, A., Finnegan, S., and Peters, S. E. (2017). Plate tectonic regulation of global marine animal diversity. PNAS 114, 5653–5658. doi: 10.1073/pnas.1702297114
Keywords: metagenomics, 16S rRNA, diversity, pyrosequencing, southeast Andaman Sea, tsunami
Citation: Sripan D, Wilantho A, Khitmoh K, Wongsawaeng D, Ouazzani J, Chavanich S, Tongsima S and Somboonna N (2021) Marine Bacteria Community in a 150-m Depth Tachai Island, the Southeast Andaman Sea of Thailand. Front. Mar. Sci. 8:624624. doi: 10.3389/fmars.2021.624624
Received: 31 October 2020; Accepted: 01 February 2021;
Published: 19 February 2021.
Edited by:
Sachin Kumar, Sardar Swaran Singh National Institute of Renewable Energy, IndiaReviewed by:
Surajit De Mandal, South China Agricultural University, ChinaCopyright © 2021 Sripan, Wilantho, Khitmoh, Wongsawaeng, Ouazzani, Chavanich, Tongsima and Somboonna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naraporn Somboonna, bmFyYXBvcm4uc0BjaHVsYS5hYy50aA==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.