- Department of Life Sciences, Texas A&M University-Corpus Christi, Corpus Christi, TX, United States
Uncertainties from sampling biases present challenges to ecologists and evolutionary biologists in understanding species sensitivity to anthropogenic climate change. Here, we synthesize possible impediments that can constrain research to assess present and future seagrass response from climate change. First, our knowledge of seagrass occurrence information is prevalent with biases, gaps and uncertainties that can influence inferences on species response to global change. Second, research on seagrass diversity has been focused on species-level metrics that can be measured with data from the present – but rarely accounting for the shared phylogenetic relationships and evolutionary distinctiveness of species despite species evolved and diversified from shared ancestors. Third, compared to the mass production of species occurrence records, computational tools that can analyze these datasets in a reasonable amount of time are almost non-existent or do not scale well in terms of computer time and memory. These impediments mean that scientists must work with incomplete information and often unrepresentative data to predict how seagrass diversity might change in the future. We discuss these shortfalls and provide a framework for overcoming the impediments and diminishing the knowledge gaps they generate.
Introduction
Human activities, through fossil fuel emissions and widespread deforestation, have contributed to increased global temperature above pre-industrial levels (IPCC, 2018). As a consequence, global increases in temperature and atmospheric carbon dioxide can influence species by altering their growth rates, physiological functions, sexual reproduction, distribution, community composition, and primary productivity (Short and Neckles, 1999; Campbell et al., 2006). Such changes in environmental climate outside species’ tolerable thresholds will cause some species to relocate in order to stay within their tolerance zones (Bradshaw and Holzapfel, 2001; Parmesan, 2006; Miller-Rushing and Primack, 2008; Anderson et al., 2012; MacLean et al., 2018). For instance, species on land generally ascend to higher elevations or latitudes as temperatures warm, but may run out of room, which can lead to local extirpation (Parmesan et al., 1999; Freeman et al., 2018). The sensitivity and responsivity of seagrasses or other marine species, whose distributional ranges lie at the land-sea margin and with very different evolutionary histories may show different responses to climate change. Seagrasses are a major vascular plant clade of about 70 species belonging to the Alismatales, an order that includes ∼4,000 other non-marine species (Berry, 2019). They are widely distributed across marine coastlines or estuarine environments, often growing submerged in marine water (Hemminga and Duarte, 2000). Seagrasses display a wide variety of morphological diversity including turtlegrass (Thalassia testudinum) which forms long and jointed rhizomes, rhizome matts in Posidonia, ribbonlike leaves in eelgrass (Zostera marina), and paddle-shaped leaves in paddle grass (Halophila decipiens) (Figure 1). They play key ecosystem roles including primary productivity, nutrient cycling, and carbon sequestration (Hemminga and Duarte, 2000; Duarte, 2002; Les et al., 2002; Orth et al., 2006a,b; McGlathery et al., 2007; Nordlund et al., 2018). Seagrass meadows are an important nursery ground for many invertebrates and fishes (Beck et al., 2001), and directly provide food for marine herbivores including manatees, dugongs, and green sea turtles (Green and Short, 2003; Larkum et al., 2006). As threats from global climate change intensify, the impacts across seagrass communities are mixed. Some studies have found a decline in seagrass habitats especially in Australasia with decline rates of about 110 km2 per year (Waycott et al., 2009). This pattern is not true in North America and Europe where seagrass communities are no longer in decline, but in fact show positive trajectories in some cases (de los Santos et al., 2019), perhaps as a result of the proliferation of seagrass monitoring and conservation programs such as Seagrass-Watch1 and SeagrassSpotter.2 Indeed, the vulnerability to the impacts of climate change on seagrass communities may be scale or context dependent (Day et al., 2008).

Figure 1. Morphological diversity of selected species of seagrasses. (A) Thalassia testudinum (turtle grass) bed with view of jointed rhizomes, San Salvador Island, Bahamas. (B) Posidonia oceanica (Neptune grass) meadow with view of rhizome matts, Portofino, Italy. (C) Zostera marina (eelgrass) with ribbon-like blades. (D) Halophila decipiens (paddle grass) with paddle-shaped blades. (https://commons.wikimedia.org and https://calphotos.berkeley.edu/).
A number of studies indicate that global climate change can impact seagrass communities in a variety of ways. Short and Neckles (1999) reviewed the potential effects of climate change on seagrass growth rates, reproduction and spatial distributions; Duarte et al. (2018) explored relationships between climate change and phenotypic variation in seagrasses (including physiological variation, propagation success, and herbivore resistance); whereas Erry et al. (2019) used a mesocosm experiment to assess response of a multi-trophic seagrass ecosystem to several global change factors. The findings overwhelmingly demonstrated that these factors in unison could lead to deleterious effects on seagrass ecosystems if they are unable to rapidly adapt to changes in climate. Similar trends have been observed for specific seagrass locations e.g., Great Barrier Reef (Waycott et al., 2007), Mediterranean (Pergent et al., 2014), tropical Pacific Ocean (Waycott et al., 2011), and Western Australia (Arias-Ortiz et al., 2018; Strydom et al., 2020); or in selected species (e.g., Chefaoui et al., 2018). Other threats to seagrass populations can be attributed to overexploitation, physical modification, nutrient and sediment pollution, and introduction and spread of invasive species (Zieman, 1976; Ralph et al., 2006; Moksnes et al., 2008; Bryars et al., 2011; Dewsbury et al., 2016). By contrast, research to elucidate effects of global climate change on seagrass meadows and how to improve the prediction of future risks under varying scenarios of climate change have received less attention (Pernetta et al., 1994; Bijlsma et al., 1995; Short and Neckles, 1999).
Here, we argue that the extension of research agenda to assess seagrasses’ response to climate change may be constrained by at least three factors. First, our knowledge of seagrass occurrence information is widespread with biases, gaps and uncertainties that can influence downstream inferences. Second, most of the research on seagrass diversity has been focused on species-level metrics (e.g., species richness, endemism or threat) that can be measured with data from the present – but rarely accounting for the shared phylogenetic relationships and evolutionary distinctiveness of species. Species are not independent units but are lineages that evolve and diversify from shared ancestors (Diniz-Filho et al., 2013). Third, compared to the mass production of species occurrence records, computational tools that can analyze these datasets in a reasonable amount of time are almost non-existent or do not scale well in terms of computer time and memory. These impediments mean that scientists must work with incomplete information and often unrepresentative data to predict how seagrass diversity might change in the future. These shortfalls need be carefully recognized and remedied. The objectives of this review are therefore to first identify the knowledge gaps to understanding seagrasses’ response to climate change, and secondly propose strategies and tools to overcome these impediments.
Knowledge Gaps in Seagrass Sampling Practices
Global change has become a central focus of modern ecology. Yet, our knowledge of how anthropogenic drivers affect seagrass evolutionary diversity is limited by a lack of biological data spanning the Anthropocene that equally represents all seagrass species. We define the Anthropocene as a period of profound human impact on biodiversity, characterized by widespread migration by humans as initiated by the Columbian Exchange circa 1492 (Nunn and Qian, 2010). The vast amounts of specimens of seagrasses deposited in herbaria can serve as a historical lens into the ecological processes by which present-day seagrass diversity arose, are maintained, and may evolve in the future. However, occurrence records archived in herbaria and museums are non-randomly collected over space and time, and thus present biases and uncertainties that can complicate ecological inferences (e.g., Boakes et al., 2010; Meyer et al., 2016; Daru et al., 2018; Dias Tarli et al., 2018). As a consequence, the use of occurrence records has not fully permeated the field of global change biology. The gap between specimen availability and use is widening as hundreds of thousands of specimens are being mobilized through massive digitization efforts worldwide. We argue that sampling biases in seagrass occurrence records can manifest in at least three ways: geographic, taxonomic, and temporal biases (Figure 2). We distinguish between the biases and describe how these limitations can inhibit progress in understanding seagrass response to global change.
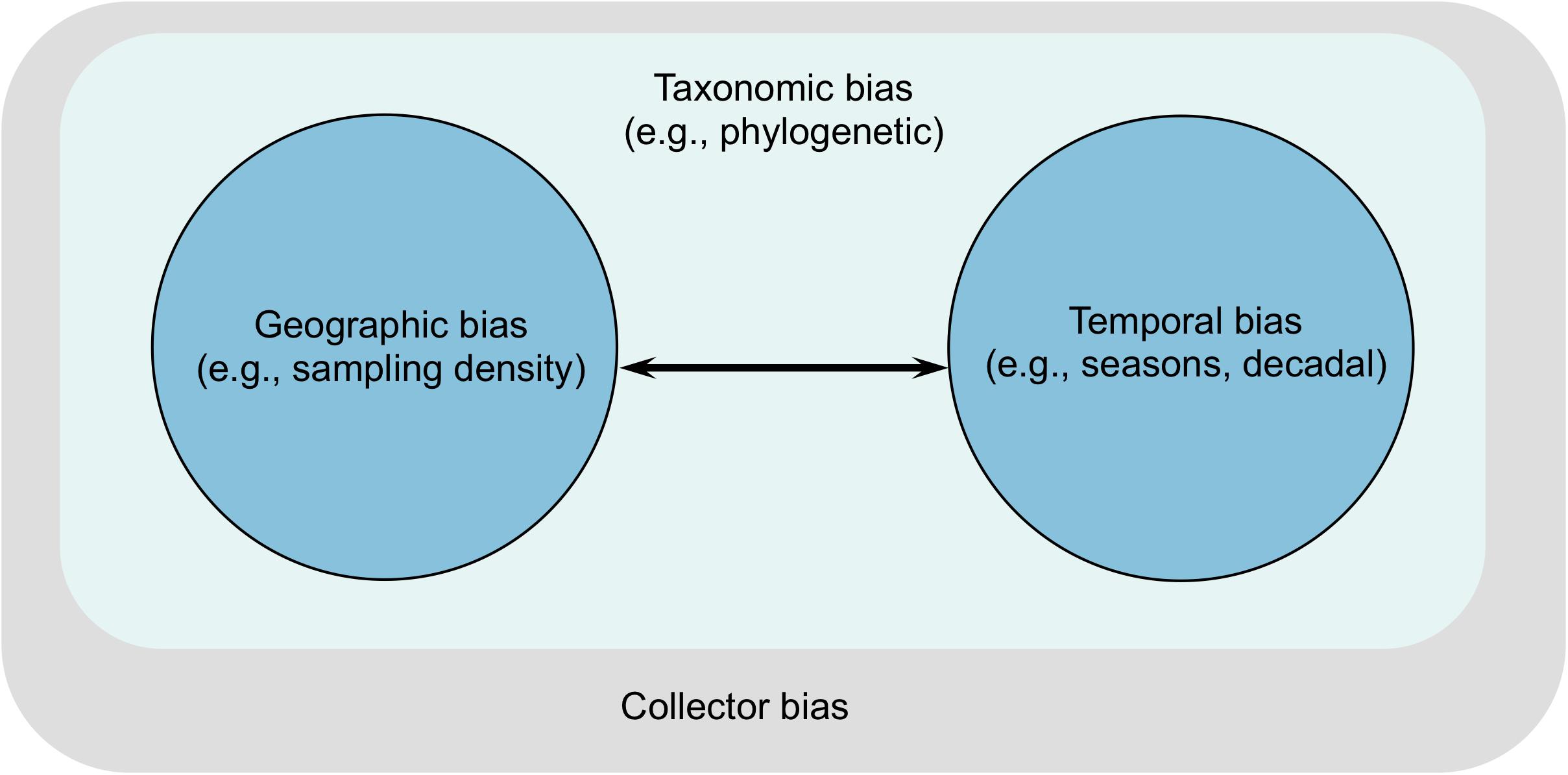
Figure 2. Interactions between sampling biases indicating the extent of influence of each bias on the others. Taxonomic bias affects all other biases whereas arrows indicate direction of influence between the other two. However, all three types of biases ultimately reflect the personal preferences, biases, and proclivities of collectors.
Biases in Geographic Sampling
Geographic bias is the disproportionate sampling of a species in some regions of its range relative to others (Meyer et al., 2016; Stropp et al., 2016; Daru et al., 2018; Menegotto et al., 2019). Seagrass geographic data is commonly available as point records or polygons. Point records are commonly derived from major data hubs such as the Global Biodiversity Information Facility (Edwards et al., 2000; GBIF.org, 2020), United Nations Environment World Conservation Monitoring Centre (UNEP-WCMC, and Short, 2020) or Ocean Biodiversity Information Facility (OBIS) whereas polygons are derived from the International Union for the Conservation of Nature’s (IUCN) spatial database and United Nations Environment World Conservation Monitoring Centre (Green and Short, 2003; UNEP-WCMC, and Short, 2020). Despite the fundamental importance of occurrence data for species distribution modeling, the sampling of seagrasses across most of their ranges are underrepresented in collections (Green and Short, 2003). For instance, extensive spatial gaps exist across regions that harbor high concentrations of seagrass diversity, especially in Western and Central Indo-Pacific, whereas Europe and North America are well sampled (Figure 3) (see Methods and Source Data file in Supplementary Material for details). This pattern is consistent with previous studies. For example, Waycott et al. (2009) found wide sampling gaps in West Africa, northeast South America, and the northwest Pacific area of the United States, most of which correspond to seagrass areas of endemism. Moreover, since biogeographic patterns are scale dependent, varying along spatial grains, geographic extents and taxonomic treatments (Jarzyna and Jetz, 2018; Daru et al., 2020a; GBIF.org, 2020), the extent to which geographic biases in seagrass sampling vary with spatial extent, grain size and taxonomic treatment remains poorly explored. However, it has been predicted that as grain size decreases, the knowledge gap in geographic sampling correspondingly increases (Hortal et al., 2015).
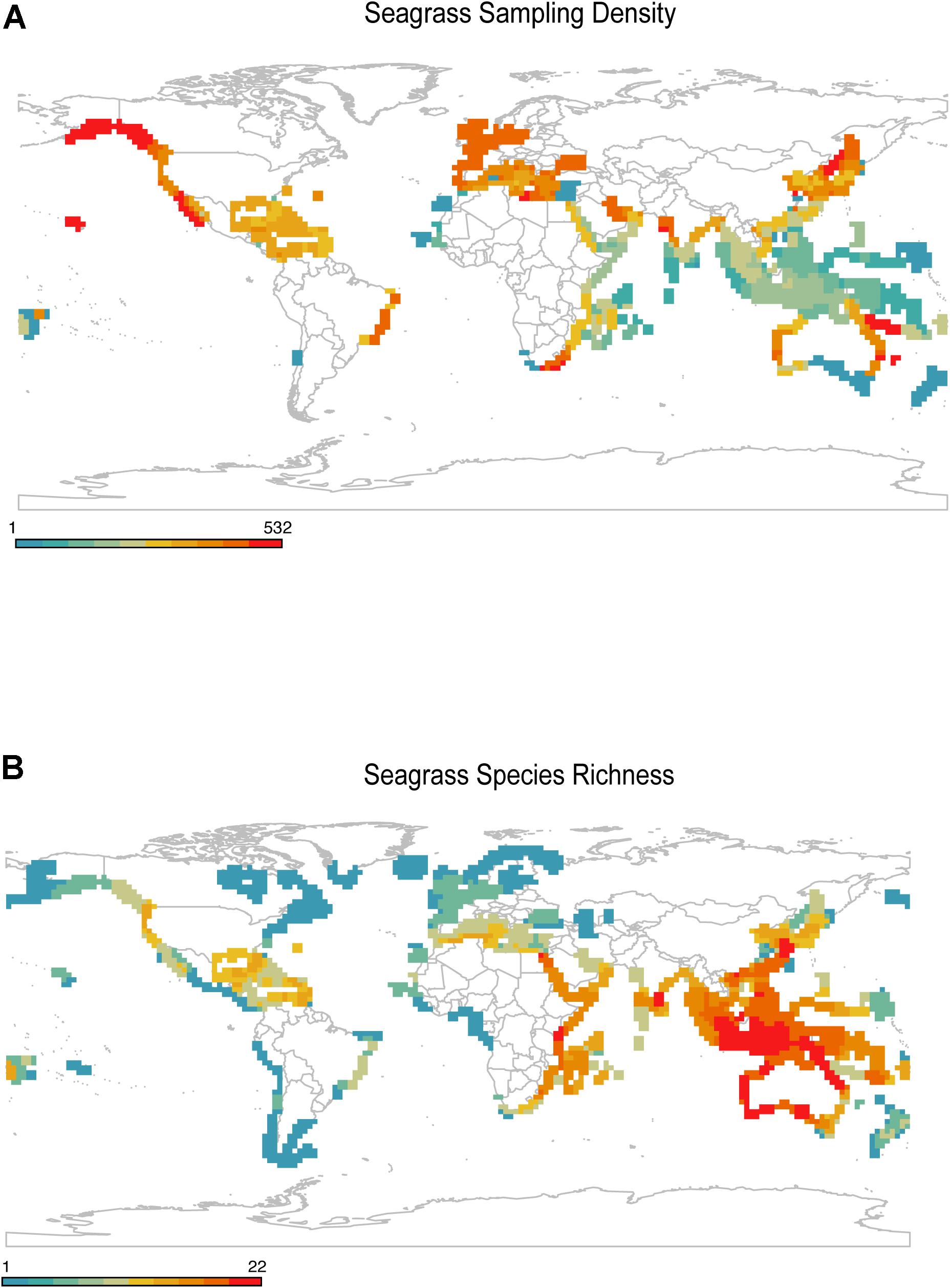
Figure 3. Gaps in geographic sampling of seagrasses. (A) Seagrass occurrence records showed strong density of sampling in temperate regions, while sampling within the tropics was generally low. (B) Geographic distribution of seagrass known species richness based on expert delineated polygons. Source data are provided as a Source Data file.
The mismatch between observed seagrass diversity and maps of survey efforts can be attributed to several factors: (1) knowing data exists in the first place and where it is, (2) harvesting data collected in native languages not common to science, (3) getting permission to access data collected under commercial license or from uncooperative governments, (4) validating data both spatially and taxonomically, (5) the difficulty in sampling specimens especially species in remote and inaccessible waters e.g., Halophila decipiens occurring >70 m deep in the Central Indo-Pacific (Short et al., 2007) or large parts of Northern Australia that are only accessible by helicopter, (6) lack of reliable research infrastructure e.g., West Papua and Papua New Guinea, (7) un-inhabited reef lagoons in large parts of the tropics and Western Pacific, (8) the cost of gathering long-term data (Wolfe et al., 1987), (9) perhaps a reversing trend of seagrass loss in Europe, North America, and subtropical Atlantic, e.g., increasing population trends in Cymodocea nodosa (Schäfer et al., 2021), Zostera marina and Zostera noltei (de los Santos et al., 2019; Guerrero-Meseguer et al., 2021), and (10) budget constraints for seagrass research. If seagrass species observations are made near accessible areas e.g., seaports, harbors or marine research stations, their application in analysis of species distribution modeling can compromise model performance (Kadmon et al., 2004; Lobo and Tognelli, 2011; Bystriakova et al., 2012; Kramer-Schadt et al., 2013; Varela et al., 2014). In practice, this means that most observations only reflect the climate space of accessible areas (e.g., Daru, 2021), and correspondingly areas of human activities where surface temperatures are higher than in surrounding natural areas (Kalnay and Cai, 2003). Additionally, regions known to contain seagrass meadows (e.g., Canada, Indonesia, and Russia) have inadequately mapped distributions, while other currently mapped regions most likely only represent a small portion of seagrass diversity (McKenzie et al., 2020). Targeting the places that are underrepresented in future collecting expeditions could remedy these limitations and aid in evaluating how species are responding to recent and future environmental change across biomes.
Biases in Temporal Sampling
The sampling of seagrasses can manifest as temporal bias – the unbalanced collecting of specimens in some years or parts of a given year. This can influence conclusions drawn from analyses of such non-randomly sampled collections records (Syfert et al., 2013). Temporal data is increasingly used in a wide range of applications in ecology and evolutionary studies including tracking changes in phenology – the timing of seasonal events such as flowering, leafing, and fruiting – and monitoring the spread of invasive species (Iler et al., 2013; Veeneklaas et al., 2013; Daru et al., 2019; Meerdink et al., 2019). Yet, while there is general agreement that climate change can influence phenological patterns by disrupting the timing of life cycle events and consequently drive changes in fitness and population demography (Ovaskainen et al., 2013; CaraDonna et al., 2014; Thackeray et al., 2016; Kharouba and Wolkovich, 2020), most have been observed in terrestrial species and to a lesser extent in marine flowering plants. In a meta-analysis of GBIF occurrence records over the course of 250 years (1770–2020) to understand the nature and evolution of seagrass sampling (GBIF.org, 2020), sparser records were observed in earlier years and high collection densities between the 1900s and present-day (Figure 4). Although over the 250-year time span, occurrence data was absent for a total of 131 years. Seasonally, seagrass specimens were overwhelmingly biased toward spring and summer months (regardless of hemisphere location) for most marine ecoregions including Temperate Southern Africa, Temperate Australasia, Temperate Northern Pacific, and Temperate Northern Atlantic (Figure 5; see Methods and Source Data file in Supplementary Material). Interestingly, these periods are spanned by comprehensive time series data of ocean climate including sea temperature and salinity (Benway et al., 2019). This means that the time series of changes in seagrass communities across years or seasons are fewer than the available climate records (cf. Duarte, 1992). As a consequence, the non-random sampling of seagrasses in some years or parts of a year could mean that occurrence records are not reliable sources of phenological change driven by climate or population demography. If seagrasses are collected only when it is climatically convenient coupled with lack of reproductive structures on most specimens (Pearson et al., 2020), botanists may miss important phenological events such as winter bud formation, which protects the embryonic shoot of species during development and elongation (van der Schoot et al., 2013). Similarly, climate change can influence population demography through range change (Hunter et al., 2010; Dalgleish et al., 2011; Hugo, 2011; Gaillard et al., 2013; Selwood et al., 2015) or facilitate the spread of invasive species (Hellmann et al., 2008; Clements and Ditommaso, 2011; Vicente et al., 2013; Hou et al., 2014; Thapa et al., 2018). However, the skewed sampling of seagrass occurrence data suggests that the data is insufficient to track demographic changes or monitor spread of invasive species. We recognize that several aspects can influence seagrass sampling across years or seasons. For instance, some seagrass species are annuals, completing their life cycle within one growing season (e.g., Halophila decipiens). Other reasons include inaccessibility to most sites in the West Indo-Pacific during monsoon times, resulting in overrepresentation of specimens during maximum growing season/flowering season.
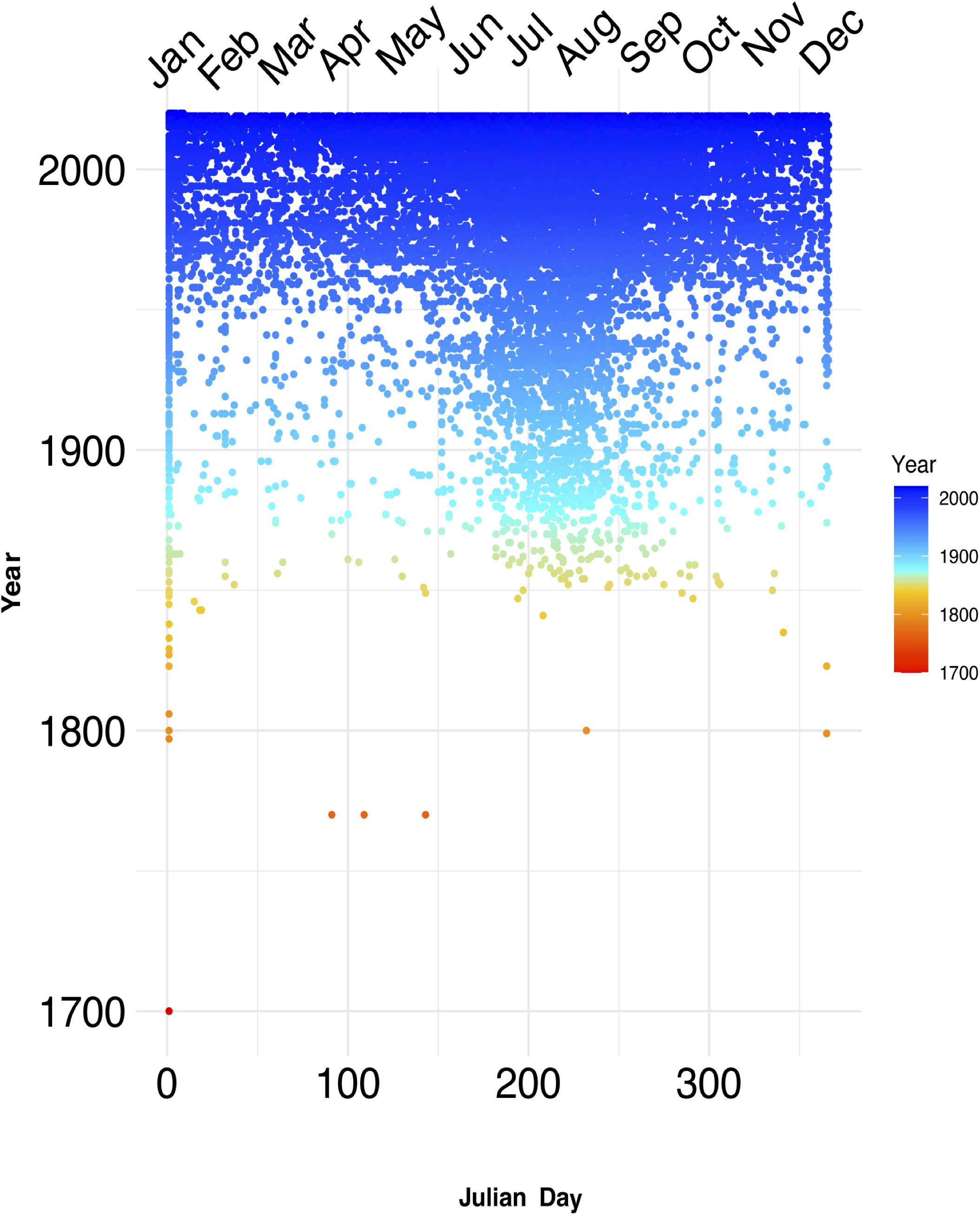
Figure 4. Temporal sampling of seagrasses reveal drastic increases midway throughout the 18th century. Temporal data from seagrass records over the course of three centuries (roughly 1700–2000) display dense amount of sampling records accumulating after 1850. Each dot represents an occurrence record of a seagrass in Julian day of year format, with the color gradient representing recent years with colder color tones, and older years represented by warmer color tones. These data also support the previously identified global trend of increased sampling occurring predominantly within the summer months (early June through early October). Source data are provided as a Source Data file.
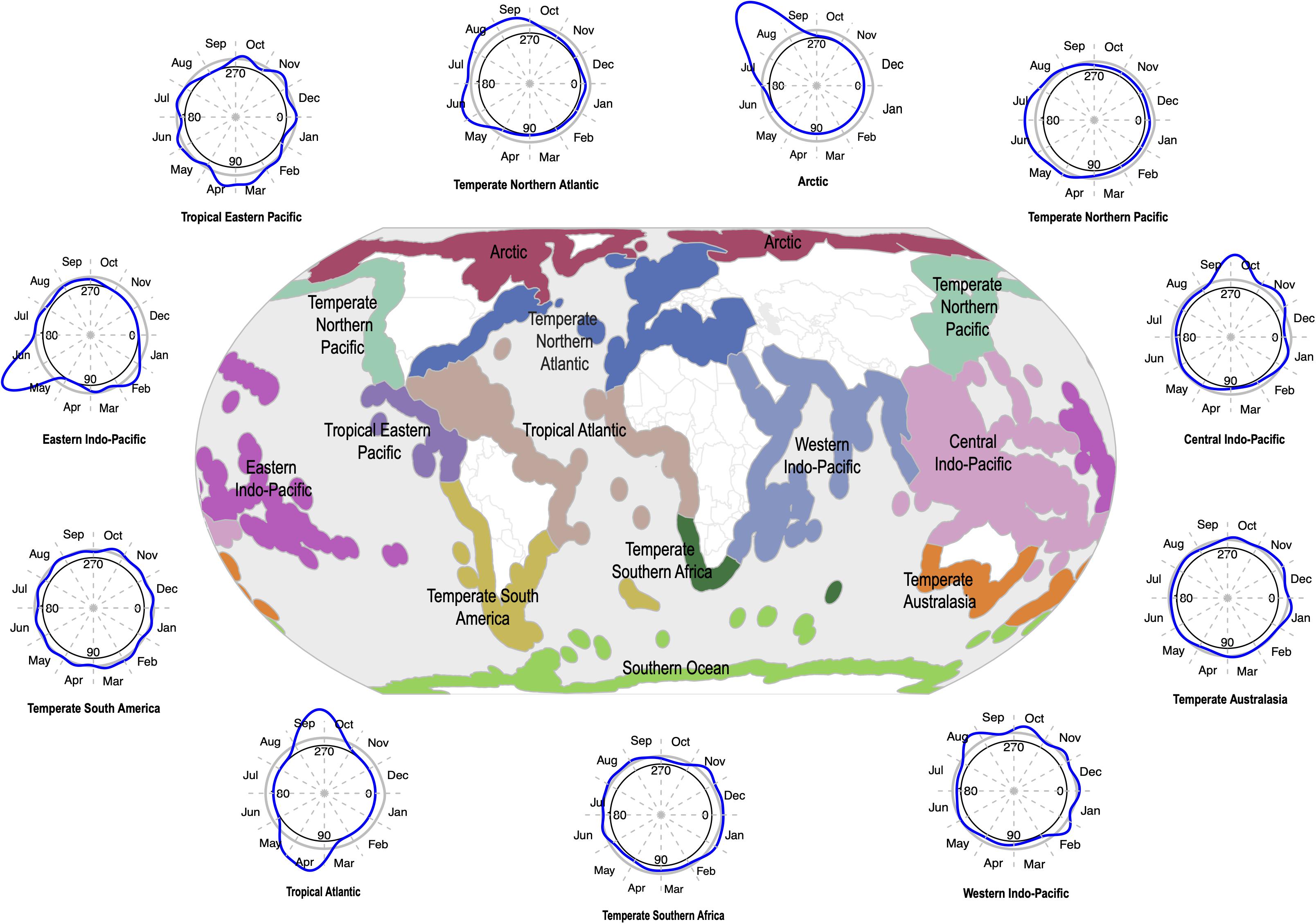
Figure 5. Temporal trends in seagrass sampling are not consistent across seasons within marine ecoregions of the world (MEOWs). Temporal data from seagrass occurrence records were converted into Julian day of year format in order to analyze trends in the monthly sampling of seagrasses for all MEOWs. The blue line around each temporal sampling plot represents seagrass sampling density in monthly intervals over an extensive time period (1770–2019), with corresponding temporal sampling plot for each MEOW. Seagrass sampling rates increase during summer seasons associated with northern and southern hemispheres. The central plot provides a reference for the geographic location of each MEOW included in the analysis. Source data are provided as a Source Data file.
Biases in Taxonomic Sampling
The sampling and collection of seagrass data may be disproportionately higher in some taxa over others (Hortal, 2008). Taxonomic bias can manifest as phylogenetic bias and be assessed by testing for phylogenetic signal in collection frequency. A strong phylogenetic signal – closely related species share similar collecting frequency – would suggest phylogenetic bias in collections (Daru et al., 2018). Phylogenetic bias can hamper prospects of identifying species that are climate change indicators and those most likely to be affected by future climate change, especially given that species’ response to climate change tends to be phylogenetically non-random (Willis et al., 2008; Davis et al., 2010; Davies et al., 2013). A phylogenetic analysis of long-term monitoring data in Concord Massachusetts, for instance, revealed a strong association between change in abundance with flowering time response such that the response traits are shared among closely related plant species (Willis et al., 2008). However, taxonomically non-random collection may mask such patterns and therefore bias conclusions of seagrass response to climate change. These data limitations may result from a research focus on specific seagrass lineages over other groups or simply lack of data on some species. For example, Coyer et al. (2013) estimated divergence times in 20 species in the family Zosteraceae at 14.4 Ma, whereas Dilipan et al. (2018) assessed phylogenetic relationships by focusing on only family Hydrocharitaceae. Not only do these clade-based approaches point to different divergence times, but the phylogenetic reconstructions also used different gene regions with likely different rates of evolution. Seagrass occurrence data on GBIF tends to display a weak phylogenetic signal in the tendency of closely related species to be sampled similarly; with an average of ∼9 specimens per species representing most Halophila, and ∼6–9 specimens per species representing most Zostera, whereas Halodule and Posidonia had far fewer records (Figure 6; see Methods and Source Data file in Supplementary Material for details).
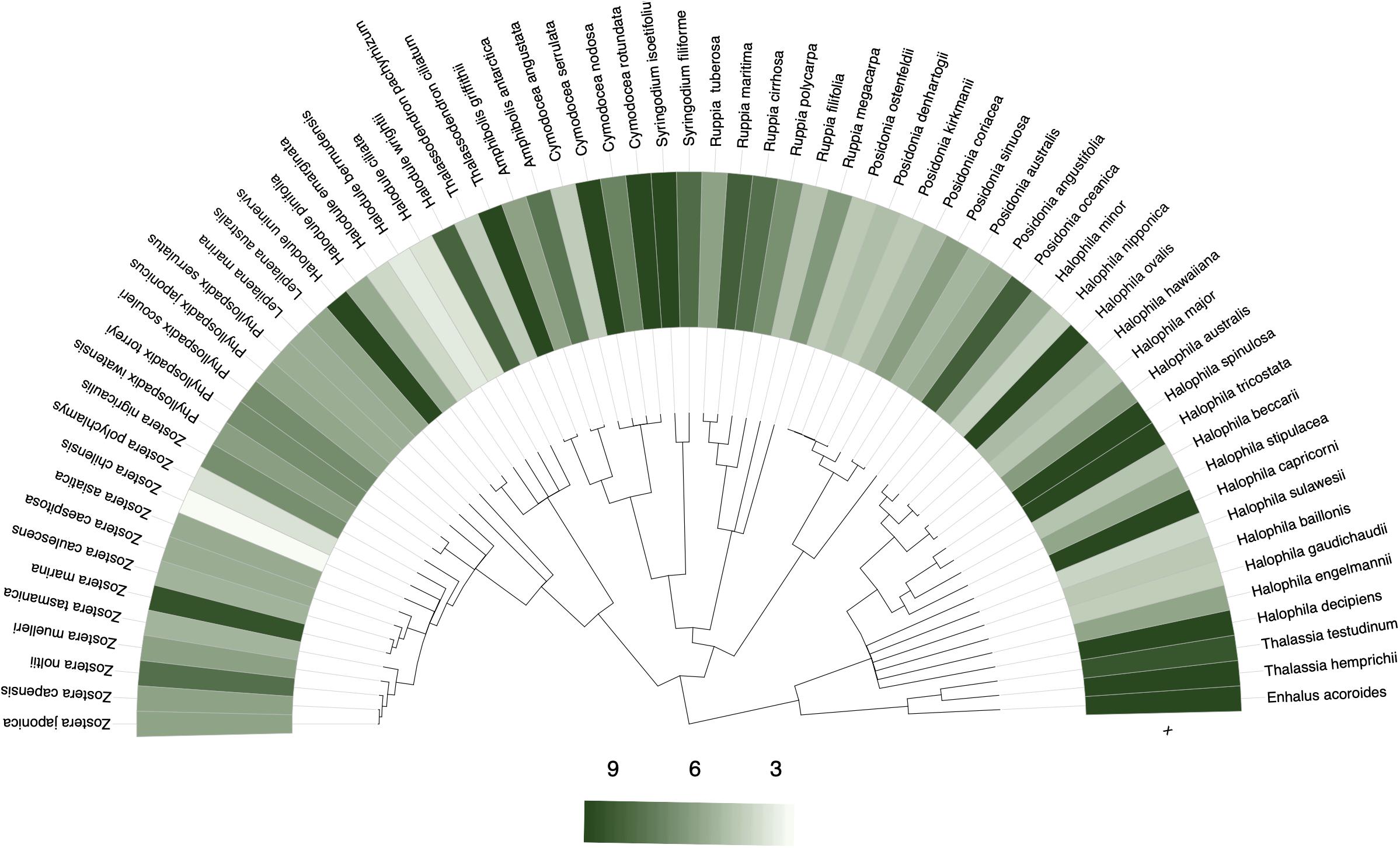
Figure 6. Phylogenetic bias in seagrass sampling. Phylogenetic distribution of the number of specimens sampled per seagrass species to assess the tendency of closely related species to be similarly collected. No statistically significant phylogenetic signals were detected, although there was slight favoring for sampling of the Thalassia, Enhalus, and Halophila genera over other seagrass genera. Source data are provided as a Source Data file.
Another factor that can induce taxonomic bias is the lack of comprehensive phylogeny for seagrass species. Inferring evolutionary patterns based only on phylogeny of the taxa within the community of interest without fully accounting for the overall phylogenetic diversity of the entire lineage can potentially lead to spurious results (Park et al., 2018). The available DNA sequences of seagrasses in GenBank/EBI are sufficient to construct a molecular phylogenetic tree for only 55 (of 72) species (Daru and le Roux, 2016). The 17 species without available DNA sequences are often manually grafted to the molecular tree in a multichotomy to the node of their close relatives using a Bayesian framework (Thomas et al., 2013). Such incomplete sampling or misplaced taxa on the phylogeny can influence the final tree topology and compromise rates of evolution (Nee et al., 1994; FitzJohn et al., 2009), especially when biases are also geographically non-random (Daru et al., 2018). Even with complete DNA sequences for all seagrass species, there are large uncertainties in the estimation of divergence times, and unknown evolutionary models linking phylogenies to underlying ecological traits and life history variation (Diniz-Filho et al., 2013). Moreover, the polyphyletic nature of seagrasses, drawing from several lineages within the Alismatales, might also compound our understanding of phylogenetic sampling biases.
The aforementioned sampling biases can combine with each other in several ways (Figure 2). Taxonomic bias can influence all other biases because it reflects knowledge gaps on the fundamental unit of ecology and evolutionary biology. Geographic bias is strongly influenced by temporal bias as limited accumulation of data over time can alter accurate estimations of species’ range size or population demographic history (Pybus et al., 2000; Drummond et al., 2005). Similarly, geographic bias can compromise estimates of species’ phenological response to climate change or demographic change, owing to lack of geographical coverage in many regions (Poelen et al., 2014). Ultimately, these sampling biases are human artifacts such that any personal preferences, biases, and proclivities of collectors can greatly skew our understanding of seagrass diversity.
Gaps in Knowledge of Seagrass Evolutionary Diversity
Understanding what drives variation in the distribution of biodiversity can provide insights into the ecological and historical processes underlying community assembly (Cavender-Bares et al., 2009) and for prioritizing conservation (Kreft and Jetz, 2010; Holt et al., 2013; Daru and le Roux, 2016). However, data gaps in the sampling of seagrasses (as outlined above) can influence estimates of broad-scale patterns and underlying processes (e.g., extinction, speciation and niche conservatism). Traditionally, identifying broad-scale patterns in seagrasses has been based on species-level metrics (e.g., species richness, and endemism) (Short et al., 2007; Mtwana Nordlund et al., 2016; Duffy et al., 2019). Although indispensable in providing baseline biodiversity knowledge, these metrics alone fail to detect the substantial evolutionary and conservation implications captured by the shared phylogenetic relationships and evolutionary distinctiveness of species (Mace et al., 2003; Redding and Mooers, 2006; Cadotte, 2013). Recent approaches harmonized metrics that consider evolutionary components, for example, phylogenetic diversity (Faith, 1992), evolutionary distinctiveness (Redding and Mooers, 2006), phylogenetic endemism (Rosauer et al., 2009), or a combination of these metrics. As pressures from climate change induced by anthropogenic activity mount, we will eventually observe range shifts and losses that can erase unique evolutionary history (Waycott et al., 2009). There is some evidence that evolutionarily distinct temperate seagrass assemblages might be disproportionately at risk of extinction (Daru et al., 2017a), which could elevate losses of phylogenetic diversity (Redding et al., 2008). However, the associated directionality of species’ responses to climate change and impact on phylogenetic diversity under a scenario of non-random extinction is unclear (Purvis et al., 2000). This means that as global temperatures increase, tropical seagrass species might be capable of expanding their distributions (Beca-Carretero et al., 2020) into regions traditionally utilized only by temperate seagrass species. This can induce selection pressures on temperate species that can result in the loss of distinct evolutionary diversity of seagrasses as the available climate space for temperate species is reduced by warming temperatures. Such pressures would inhibit our ability to understand the evolutionary history of seagrasses, as evolutionarily distinct species are lost or greatly reduced.
The global decline of seagrasses along a latitudinal gradient is imbalanced, with greater declines documented in temperate than tropical regions, requiring urgent conservation action (Hauxwell et al., 2001; Orth et al., 2006a,b; Moksnes et al., 2008; Bryars et al., 2011; Erry et al., 2019). The recent finding that temperate seagrass assemblages tend to be those that are most evolutionarily unique also warrants concern given that their extinction would result in a greater loss of phylogenetic diversity (Daru et al., 2017a). In this regard, the familial membership of threatened seagrass species across marine ecoregions (see Methods and Source Data file in Supplementary Material) showed a tendency of threatened species in the Temperate Northern Pacific and Tropical Eastern Pacific clustering within similar families (Figure 7). This phylogenetic and taxonomic structuring suggests that evolutionary history is an important predictor of species decline, possibly reflecting a non-random pattern of extinction risk (Purvis et al., 2000). Van Allen et al. (2012) demonstrated the importance of life-history traits for predicting how natural assemblages are likely to be impacted by anthropogenic and climatic disturbances using modeled declines in population growth rates under simulated stochastic disturbance. With regard to species extinctions and extinction risk, an important link has been identified between the loss of species and the loss of unique evolutionary history (National Research Council [NRC-US], 2008). Furthermore, the extinction of evolutionarily distinct or paleoendemic species can elevate losses of evolutionary history (Veron et al., 2015; Daru et al., 2017b). These patterns might be indicative that seagrasses are characterized by species that subtends longer phylogenetic branches perhaps representing once diverse clades that have been lost through historical extinctions.
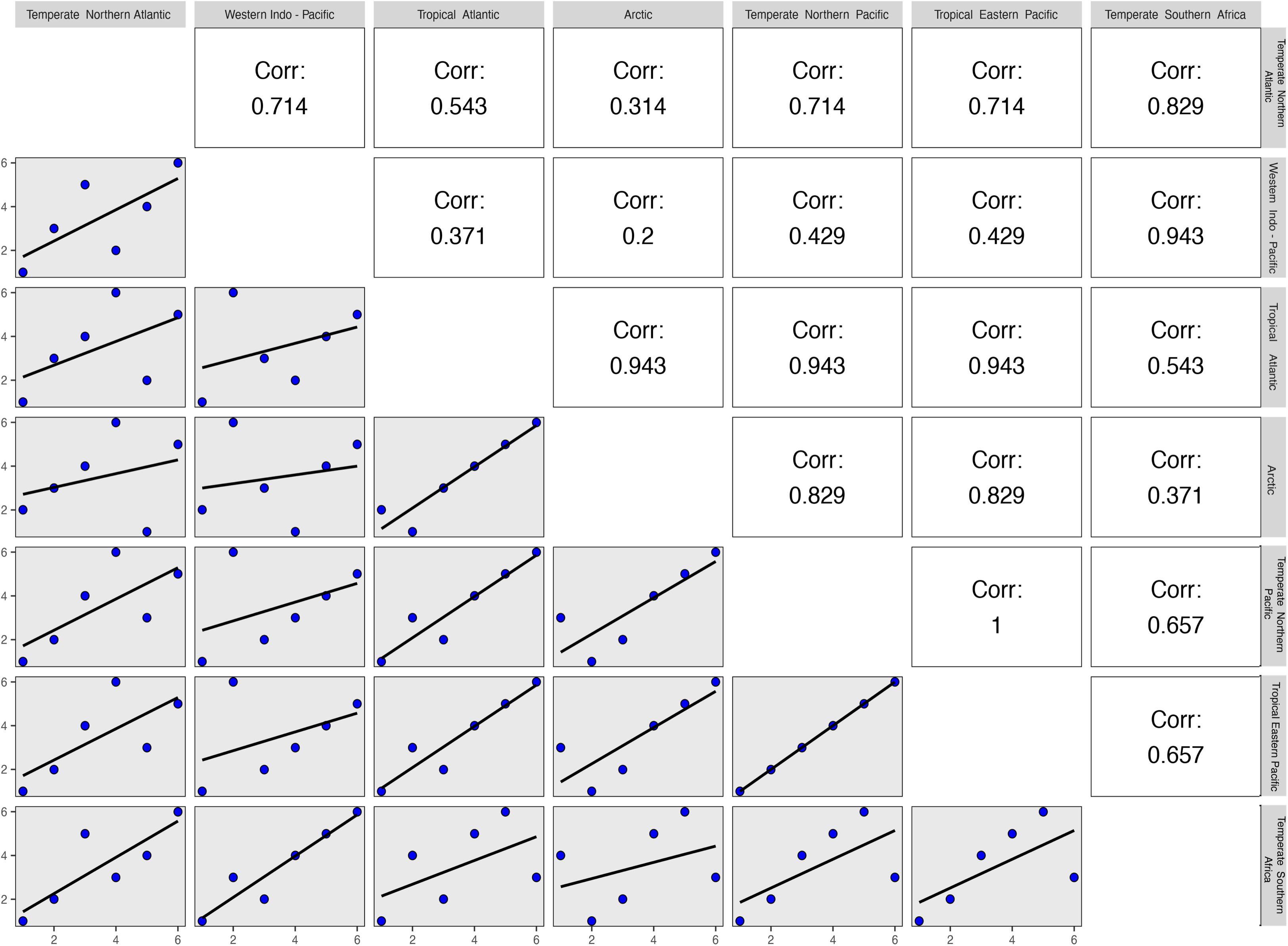
Figure 7. Correlations of family ranks possessing threatened seagrass species across marine ecoregions of the world (MEOWs). The pairwise correlational analysis assigned values based on the level of overlap of seagrass families across MEOWs that possessed seagrass species classified as threatened by the International Union for Conservation of Nature. Low correlation values were generally reported between temperate and tropical MEOWs, indicating that the threatened seagrass species in these regions are unique to those areas. Source data are provided as a Source Data file.
As seagrasses are increasingly threatened along their taxonomic structure spanning several marine ecoregions, we argue that seagrass extinctions are unlikely to be random. Previously, Short et al. (2011) determined that roughly 14% of seagrass species were at an elevated risk of extinction based on the IUCN’s Red List of Threatened Species criteria. Currently, the IUCN indicates that 31% (22 out of 72) of seagrass species are in global decline, and 22% lack information for proper assessment of conservation status (IUCN, 2020). Therefore, the question of why some species persist while others decline across regions will require an understanding of the shared evolutionary history underlying changes in species richness and composition (Waycott, 1999; Arnaud-Haond et al., 2010; Massa et al., 2013). With many species’ ranges greatly reduced or unknown, it is even more challenging to track patterns in seagrass population successes or failures that could be indicative of their resilience to climate change. In the absence of these key insights for the adaptive potential of seagrass species, we are unable to fully predict how individual species of seagrasses will respond to drastic, widespread environmental changes.
In order to facilitate effective conservation action, it is important to accurately determine which species are currently at the greatest risk for extinction, and which species will be at risk in the future. One successful approach has been to collect expert opinion data to prioritize seagrass management actions at regional scales (Grech et al., 2012) for species that may be unequally impacted. To this end, phylogenetic information can be very useful for predicting vulnerabilities at individual or familial levels (Gallagher et al., 2015). For example, families with a high proportion of species in global decline include Zosteraceae, Hydrocharitaceae, Posidoniaceae, and Cymodoceaceae; with Zosteraceae contributing about half of the total number of species in decline (Figure 8). Therefore, Zosteraceae and other evolutionarily similar families may possess a phylogenetic signal for extinction pressures. Families with seagrasses having unknown population trends include Hydrocharitaceae, Cymodoceaceae, Ruppiaceae, Posidoniaceae, and Zosteraceae according to the IUCN (see Methods and Source Data file in Supplementary Material for details). These groups are of high conservation concern given that species associated with these families may be currently threatened or already in decline without notice. Such population trends, or lack thereof, imply that certain species of seagrasses may be too heavily impacted in the future to prevent complete losses or extinctions given the rapid pace of climatic change.
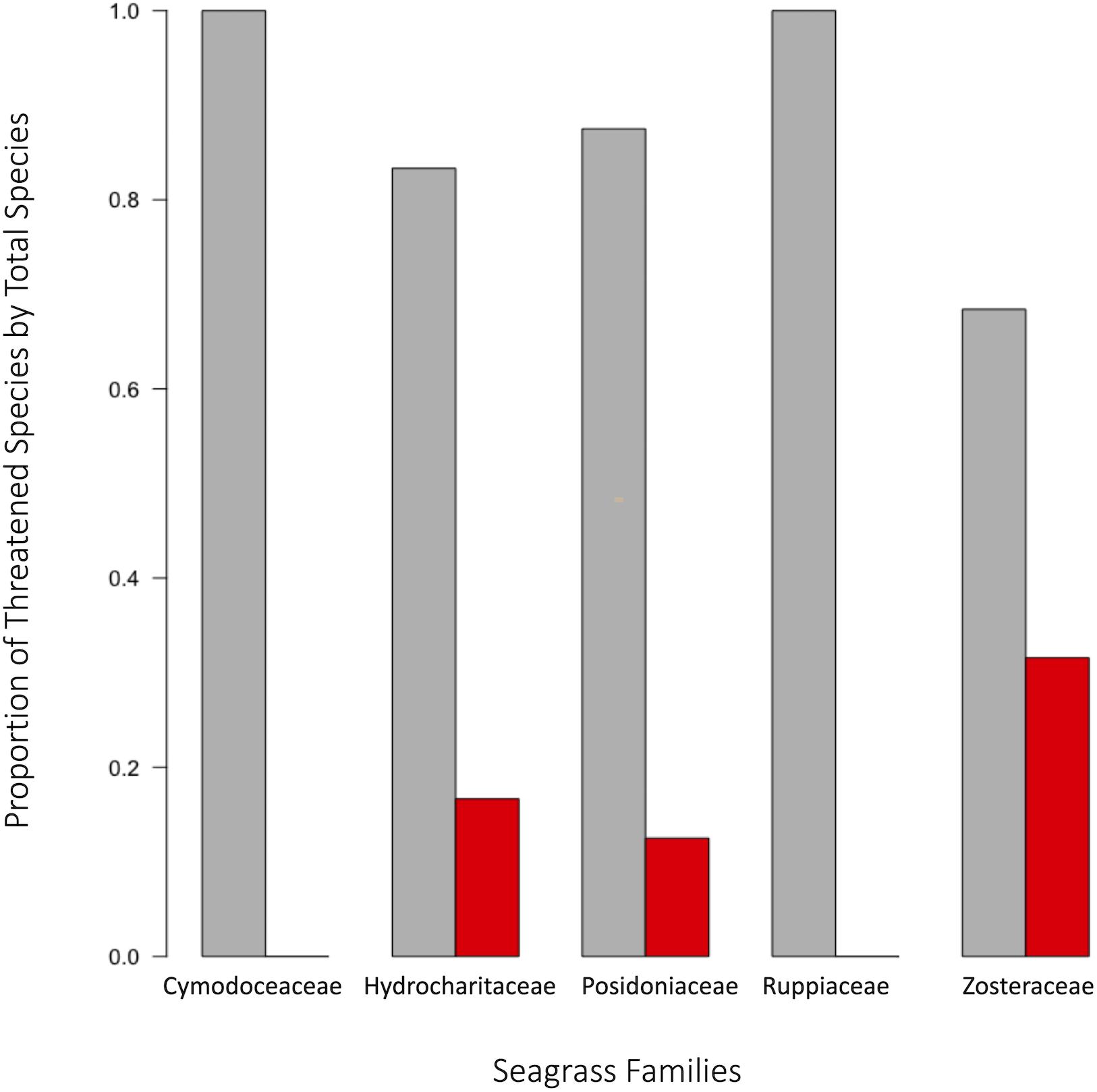
Figure 8. Taxonomic distribution of extinction risk in seagrass. Population status of seagrasses were assessed using the classifications set forth by the International Union for Conservation of Nature. Proportion of threatened species was assessed as number of threatened species in a family divided by the total number of species assessed within that family. When comparing the proportions of threatened species per family to the calculated 95% confidence interval, three families were significant: Zosteraceae, Posidoniaceae, and Hydrocharitaceae. Source data are provided as a Source Data file.
Shortfalls in Computational Tools for Assessing Species Response to Climate Change
It is possible that the aforementioned impediments can be solved by increasing biological knowledge and computational capacity. However, compared to the mass production of occurrence records and climate data, tools that can analyze these datasets in a reasonable amount of time are almost non-existent or do not scale well in terms of computer time, memory, or other resources. This is particularly true for seagrasses that have wide geographic ranges, colonizing every coastline. As a consequence, ecologists and conservationists wishing to address questions related to seagrass response to climate change may be deterred by lack of analytical tools.
The occurrence data typically used for species distribution modeling is generated from massive digitization of museum records and citizen science campaigns (e.g., Seagrass-Watch, see text footnote 1) and are often available as point records; whereas global oceanographic variables are measured by instruments on satellites daily (NOAA Climate.gov, 2020), which increase the size of the dataset many-folds. This exponential increase in species occurrences and oceanographic information inflate the size of running time for modeling algorithms (Farley et al., 2018; Allen et al., 2019), and consequently increases the challenges for visualizing downstream patterns. In Figure 9, the number of seagrass occurrence records in GBIF has increased over time. Where there used to be access to only a few dozen records, the rapid expansion of biodiversity occurrence data has now made it common for there to be a few thousand records per species (see Source Data file in Supplementary Material). This poses computational challenges for researchers. For analysis of species distribution modeling under different representative concentration pathway scenarios, for instance, researchers rapidly run into a spatial scale exponentiation problem. At a spatial resolution of 0.5 degrees (equivalent to ∼50 km at the equator) covering the geographic ranges of seagrasses, there are 201,600 possible pixels for the algorithm to evaluate from. Computing probabilities across a 201,600-possibility data frame is a challenge. Such large-scale analysis can easily reach thousands of bytes and analysis using current tools would be prohibitively expensive computationally.
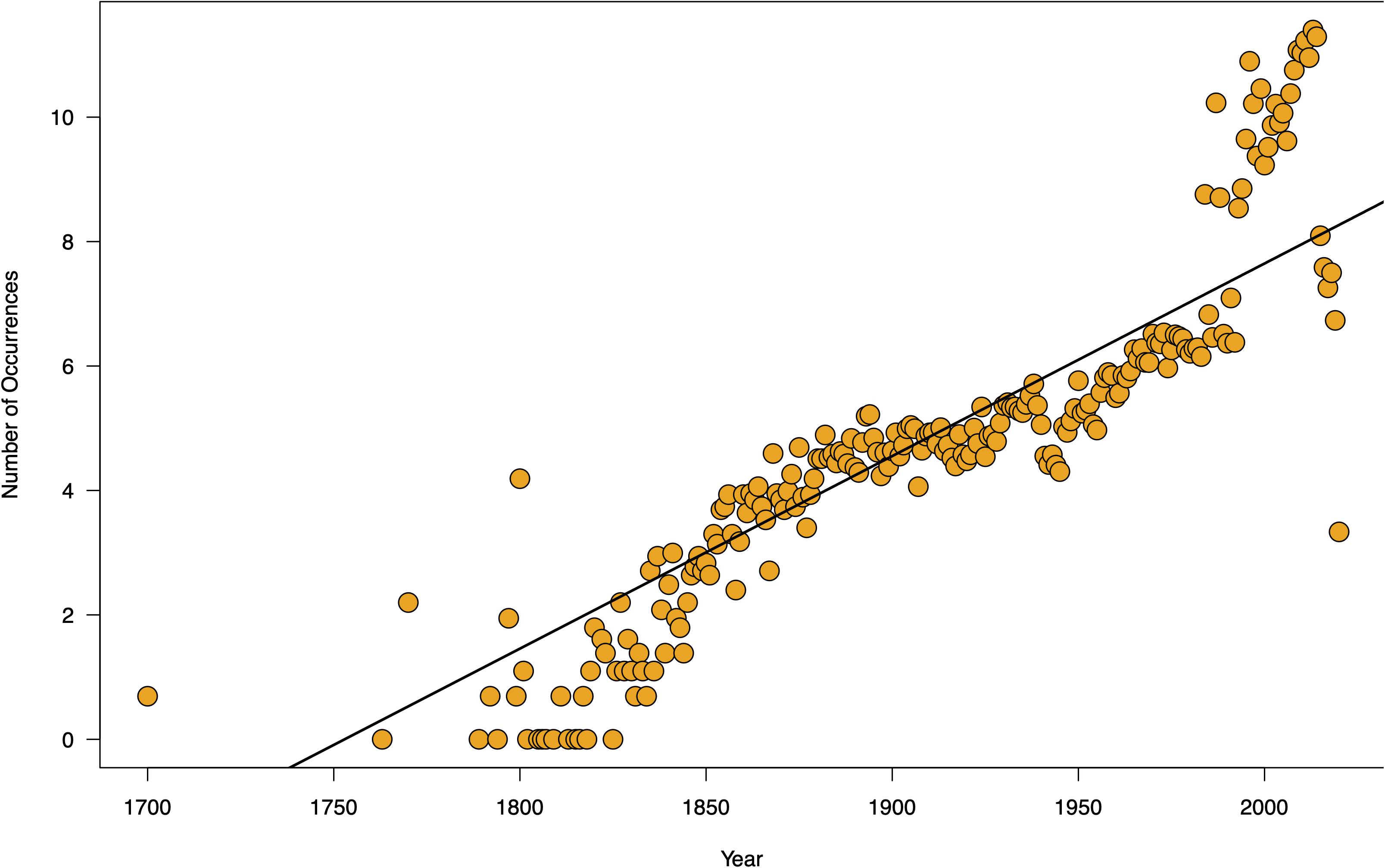
Figure 9. Temporal change in the amount of seagrass occurrence records over time. Seagrass point records downloaded from GBIF were mapped over time based on the chronological date listed in the occurrence data for each record to demonstrate that seagrass occurrences have greatly increased within recent decades. This indicates that analyses with these data will be computationally expensive. Source data are provided as a Source Data file.
Presently, the software that can facilitate analysis of species distribution modeling of seagrasses includes maxent (Phillips et al., 2017), dismo (Hijmans et al., 2011), biomod2 (Thuiller et al., 2014), esdm (Woodman et al., 2019), ModEco (Guo and Liu, 2010), SDMtoolbox 2.0 (Brown et al., 2017), ArcGIS, and ARCMap. Several of these packages contain some statistical capabilities by integrating occurrence information and climate data. For instance, biomod2 facilitates species distribution modeling by averaging across different methods including generalized additive models, generalized linear models, generalized boosting trees, maximum entropy, and random forest (Thuiller et al., 2014). However, these packages differ in their inferences, and analytical and computational capacity to process the massively mobilized occurrence records spanning tens of thousands of pixels across the globe (depending on the measurement scale). Some of these packages are developed for use in command-line while others are graphical user-interface (GUI). Most packages are developed to address a specific biological question and may have restricted analytical options that can limit computational flexibility. Ultimately, scientists wishing to address more complex hypotheses will have to use a compilation of multiple computational workflows.
More recent approaches to scale existing software to handle the exponential growth of biodiversity datasets include developing parallel algorithms (McCallum and Weston, 2011) and using modern computational architectures, such as multicore systems, graphics processing units, and supercomputers (Maruyama et al., 2011). The advantages of these methods are that they provide reproducible source codes. However, they might require the user to have a good background in high performance computing. These limitations should not detract from exploring other outstanding questions that remained to be addressed with the available tools: (1) What are the effects of reduced area and increased isolation of marine habitats? (2) Where will seagrass species disperse to under alternative scenarios of climate change? and (3) How have anthropogenic activities e.g., marine pollution, sedimentation, and coastal urbanization changed the geography of seagrasses?
Overcoming the Impediments
Seagrass occurrence records are increasingly being utilized in biogeographical investigations and prioritizing conservation (Valle et al., 2014; Chefaoui et al., 2018; Jayathilake and Costello, 2018; Beca-Carretero et al., 2020; Heck et al., 2021). As possible solutions for the geographic uncertainty, we suggest enhanced funding for local, regional, and global inventories such as SeagrassNet for seagrass habitats in the Western Pacific (Short et al., 2006), Seagrass-Watch in Australasia (McKenzie et al., 2000, 2009), ResilienSEA3 in West Africa, Texas Seagrass Monitoring program,4 SeagrassSpotter (see text footnote 2) a global tool for locating seagrasses, or Zostera Experimental Network5 for eelgrass (Zostera marina). Overcoming gaps in geographic sampling can also include collectors using best practices for collecting and vouchering specimens such as capturing accurate geolocations. It could also require the digitization and mobilization of vouchered seagrass specimens stored in herbaria and museums across the world. The iNaturalist project is a platform for sharing species observations along with geographic coordinates for terrestrial organisms and can be leveraged for filling in the data gaps in seagrass sampling. Kew’s Plants of the World Online portal (POWO) provides distribution information on the seed-bearing plants of the world based on level 3 of the Taxonomic Diversity Working Group distribution scheme which corresponds to country borders (POWO, 2019) and can be extended to cover seagrasses as well. High resolution cameras attached to unmanned aerial vehicles can be deployed to survey seagrasses in remote and inaccessible waters; however, special permits can often be required to access some sites (Johnston, 2019).
Species distribution models – the statistical estimation of species geographic distributions based on only some known occurrences and environmental conditions (Peterson et al., 2011) – can also provide an unbiased and easily interpretable estimate of improving representativeness and coverage of seagrass distributions. For example, a recent species distribution model predicts more than two-fold increase in the potential global distribution of seagrasses (Jayathilake and Costello, 2018). However, the accuracy of this prediction has attracted particular scrutiny because of inconsistent measures and widespread sampling gaps in seagrass occurrence records (McKenzie et al., 2020). Additionally, modeling approaches can contribute other useful measurements of seagrass meadows such as assessing ecosystem services as well as estimating broad-scale seagrass resources as was exemplified by Collier et al. (2021) who used historical data to accurately predict the below-ground biomass of five seagrass species. Because geographic scale is an important consideration in ecological analyses (Jarzyna and Jetz, 2018; Daru et al., 2020a), a multi-scale approach varying along spatial extents (local, regional and global) and grain resolutions should be considered in assessing seagrass response to global change and model testing. Temporal bias can be diminished by carrying out new field surveys that are more consistent and evenly distributed across seasons and years. Collectors should use best practices such as capturing and documenting accurate dates of collection. For the taxonomic bias: increased support for marine plant taxonomy and advances in taxonomic publications could minimize biases. Next-generation DNA sequencing combined with bioinformatics (Taberlet et al., 2012) will help diminish taxonomic bias such as sequencing old herbarium specimens of very rare species such as Halodule bermudensis. The rapid growth of large databases such as GenBank,6 SeagrassDB (Sablok et al., 2018), and Treebase,7 allows researchers to download available phylogenies or DNA sequences to build their own (Morell, 1996; Piel et al., 2000; Page, 2007). Taxonomic bias can also be reduced by targeting future collecting in poorly sampled clades.
Improvement of analytical and computational tools is an important priority for handling the analyses for large-scale comparative analyses of seagrass species. For instance, the US National Science Foundation-funded software BiotaPhy facilitates integration, data collection and analysis by connecting to existing data repositories such as the Open Tree of Life, iDigBio, and Lifemapper (BiotaPhy, 2020), whereas the open-source package sampbias allows quantification of geographic sampling biases in species distribution data (Zizka et al., 2020). The R software package phyloregion – designed for biogeographic regionalization and macroecology – can overcome some computational challenges (Daru et al., 2020b). It contains tools for biogeographical regionalization, macroecology, conservation, and visualizing biodiversity patterns, and has potential application in diverse fields including evolution, microbial diversity, systematics, ecology, phylogenetics, and many others (Daru et al., 2020b). We expect that the proliferation of more open-source analytical tools to greatly facilitate comprehensive understanding of seagrass sensitivity to ecological change driven by anthropogenic causes.
Concluding Remarks
Here, we outlined impediments that limit progress in understanding seagrass sensitivity to global change induced by human activities. These knowledge gaps are interconnected and represent only few of the possible issues related to research in seagrass diversity and evolution. Taxonomic bias can influence all other types of biases as it reflects knowledge gaps on the fundamental unit of ecology and evolutionary biology. The geographic and temporal biases are strongly related and capture knowledge gaps about species distributions in space and time, respectively. Even when the aforementioned impediments are resolved, many of the critical questions about seagrass sensitivity to global change, can be out of reach for scientists without the right analytical tools. The recent development of efficient and replicable computational tools, massive mobilization of natural history collections, and increased funding for seagrass research could remedy these shortcomings. Most of the management tools designed for use in developed countries can be extended to remote areas in developing countries where most seagrass diversity resides e.g., the Central Indo-Pacific. Although research on a single taxon or selected taxa is useful to a certain extent, species are lineages that evolve and diversify from shared ancestors, suggesting an integrative approach that accounts for their shared phylogenetic relationships.
Author Contributions
BHD conceived and designed the study. BMR ran the analyses with help from BHD. BMR wrote the manuscript with substantial contributions from BHD. Both authors approved the submitted version.
Funding
This study was supported by a US National Science Foundation grant no. 2031928.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Texas A&M University-Corpus Christi for logistic and financial support, and Kristen Ruggles for comments on style and language.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.608867/full#supplementary-material
Footnotes
- ^ https://www.seagrasswatch.org/
- ^ https://seagrassspotter.org/
- ^ http://resiliensea.org/3
- ^ http://www.texasseagrass.org/
- ^ http://zenscience.org
- ^ http://www.ncbi.nlm.nih.gov/genbank
- ^ http://www.treebase.org
References
Allen, J. M., Folk, R. A., Soltis, P. S., Soltis, D. E., and Guralnick, R. P. (2019). Biodiversity synthesis across the green branches of the tree of life. Nat. Plants 5, 11–13. doi: 10.1038/s41477-018-0322-7
Anderson, J. T., Panetta, A. M., and Mitchell-Olds, T. (2012). Evolutionary and ecological responses to anthropogenic climate change. Plant Physiol. 160, 1728–1740. doi: 10.1104/pp.112.206219
Arias-Ortiz, A., Serrano, O., Masqué, P., Lavery, P. S., Mueller, U., Kendrick, G. A., et al. (2018). A marine heatwave drives massive losses from the world’s largest seagrass carbon stocks. Nat. Clim. Chang. 8, 338–344. doi: 10.1038/s41558-018-0096-y
Arnaud-Haond, S., Marbà, N., Diaz-Almela, E., Serrão, E. A., and Duarte, C. M. (2010). Comparative analysis of stability—genetic diversity in seagrass (Posidonia oceanica) meadows yields unexpected results. Estuaries Coasts 33, 878–889. doi: 10.1007/s12237-009-9238-9
Beca-Carretero, P., Teichberg, M., Winters, G., Procaccini, G., and Reuter, H. (2020). Projected rapid habitat expansion of tropical seagrass species in the mediterranean sea as climate change progresses. Front. Plant Sci. 11:1762. doi: 10.3389/fpls.2020.555376
Beck, M., Heck, K. Jr., Able, K., Childers, D., Eggleston, D., Gillanders, B., et al. (2001). The identification, conservation, and management of estuarine and marine nurseries for fish and invertebrates. BioScience 51, 633–641. doi: 10.1641/0006-3568(2001)051[0633:ticamo]2.0.co;2
Benway, H. M., Lorenzoni, L., White, A. E., Fiedler, B., Levine, N. M., Nicholson, D. P., et al. (2019). Ocean time series observations of changing marine ecosystems: an era of integration, synthesis, and societal applications. Front. Mar. Sci. 6:393. doi: 10.3389/fmars.2019.00393
Berry, P. E. (2019). Alismatales. Available online at: https://www.britannica.com/plant/Alismatales (accessed November 27, 2020).
Bijlsma, L., Ehler, C., Klein, R., Kulshrestha, S., Mclean, R., Mimura, N., et al. (1995). “Coastal zones and small islands,” in Climate Change 1995—Impacts, Adaptations and Mitigation of Climate Change: Scientific-Technical Analyses, eds R. T. Watson, M. C. Zinyowera, and R. H. Moss (Cambridge: Cambridge University Press), 289–324.
BiotaPhy (2020). BiotaPhy Project. Available online at: https://biotaphy.github.io/ (accessed October 14, 2020).
Boakes, E. H., McGowan, P. J. K., Fuller, R. A., Chang-qing, D., Clark, N. E., O’Connor, K., et al. (2010). Distorted views of biodiversity: spatial and temporal bias in species occurrence data. PLoS Biol. 8:e1000385. doi: 10.1371/journal.pbio.1000385
Bradshaw, W. E., and Holzapfel, C. M. (2001). Genetic shift in photoperiodic response correlated with global warming. Proc. Natl. Acad. Sci. U.S.A. 98, 14509–14511. doi: 10.1073/pnas.241391498
Brown, J., Bennett, J., and French, C. (2017). SDMtoolbox 2.0: the next generation python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ. 5:e4095. doi: 10.7717/peerj.4095
Bryars, S., Collings, G., and Miller, D. (2011). Nutrient exposure causes epiphytic changes and coincident declines in two temperate Australian seagrasses. Mar. Ecol. Prog. Ser. 441, 89–103. doi: 10.3354/meps09384
Bystriakova, N., Peregrym, M., Erkens, R., Bezsmertna, O., and Schneider, H. (2012). Sampling bias in geographic and environmental space and its effect on the predictive power of species distribution models. Syst. Biodivers. 10, 305–315. doi: 10.1080/14772000.2012.705357
Cadotte, M. W. (2013). Experimental evidence that evolutionarily diverse assemblages result in higher productivity. Proc. Natl. Acad. Sci. U.S.A. 110, 8996–9000. doi: 10.1073/pnas.1301685110
Campbell, S. J., McKenzie, L. J., and Kerville, S. P. (2006). Photosynthetic responses of seven tropical seagrasses to elevated seawater temperature. J. Exp. Mar. Biol. Ecol. 330, 455–468. doi: 10.1016/j.jembe.2005.09.017
CaraDonna, P. J., Iler, A. M., and Inouye, D. W. (2014). Shifts in flowering phenology reshape a subalpine plant community. Proc. Natl. Acad. Sci. U.S.A. 111, 4916–4921. doi: 10.1073/pnas.1323073111
Cavender-Bares, J., Kozak, K., Fine, P., and Kembel, S. (2009). The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. doi: 10.1111/j.1461-0248.2009.01314.x
Chefaoui, R. M., Duarte, C. M., and Serrão, E. A. (2018). Dramatic loss of seagrass habitat under projected climate change in the Mediterranean Sea. Glob. Change Biol. 24, 4919–4928. doi: 10.1111/gcb.14401
Clements, D. R., and Ditommaso, A. (2011). Climate change and weed adaptation: can evolution of invasive plants lead to greater range expansion than forecasted? Weed Res. 51, 227–240. doi: 10.1111/j.1365-3180.2011.00850.x
Collier, C. J., Langlois, L. M., McMahon, K. M., Udy, J., Rasheed, M., Lawrence, E., et al. (2021). What lies beneath: predicting seagrass below-ground biomass from above-ground biomass, environmental conditions and seagrass community composition. Ecol. Indic. 121:107156. doi: 10.1016/j.ecolind.2020.107156
Coyer, J. A., Hoarau, G., Kuo, J., Tronholm, A., Veldsink, J., and Olsen, J. L. (2013). Phylogeny and temporal divergence of the seagrass family Zosteraceae using one nuclear and three chloroplast loci. Syst. Biodivers. 11, 271–284. doi: 10.1080/14772000.2013.821187
Dalgleish, H. J., Koons, D. N., Hooten, M. B., Moffet, C. A., and Adler, P. B. (2011). Climate influences the demography of three dominant sagebrush steppe plants. Ecology 92, 75–85. doi: 10.1890/10-0780.1
Daru, B. H. (2021). Exploring a new way to think about climate regions. eLife 10:e67422. doi: 10.7554/eLife.67422
Daru, B. H., Farooq, H., Antonelli, A., and Faurby, S. (2020a). Endemism patterns are scale dependent. Nat. Commun. 11:2115. doi: 10.1038/s41467-020-15921-6
Daru, B. H., Karunarathne, P., and Schliep, K. (2020b). phyloregion: R package for biogeographic regionalization and macroecology. Methods Ecol. Evol. 11, 1483–1491. doi: 10.1111/2041-210X.13478
Daru, B. H., Holt, B. G., Lessard, J.-P., Yessoufou, K., and Davies, T. J. (2017a). Phylogenetic regionalization of marine plants reveals close evolutionary affinities among disjunct temperate assemblages. Biol. Conserv. 213, 351–356. doi: 10.1016/j.biocon.2016.08.022
Daru, B. H., Elliott, T. L., Park, D. S., and Davies, T. J. (2017b). Understanding the processes underpinning patterns of phylogenetic regionalization. Trends Ecol. Evol. 32, 845–860. doi: 10.1016/j.tree.2017.08.013
Daru, B. H., and le Roux, P. C. (2016). Marine protected areas are insufficient to conserve global marine plant diversity. Glob. Ecol. Biogeogr. 25, 324–334. doi: 10.1111/geb.12412
Daru, B. H., Park, D. S., Primack, R. B., Willis, C. G., Barrington, D. S., Whitfeld, T. J. S., et al. (2018). Widespread sampling biases in herbaria revealed from large-scale digitization. New Phytol. 217, 939–955. doi: 10.1111/nph.14855
Daru, B. H., Kling, M. M., Meineke, E. K., and van Wyk, A. E. (2019). Temperature controls phenology in continuously flowering Protea species of subtropical Africa. Appl. Plant Sci. 7:e1232. doi: 10.1002/aps3.1232
Davies, T. J., Wolkovich, E. M., Kraft, N. J. B., Salamin, N., Allen, J. M., Ault, T. R., et al. (2013). Phylogenetic conservatism in plant phenology. J. Ecol. 101, 1520–1530. doi: 10.1111/1365-2745.12154
Davis, C. C., Willis, C. G., Primack, R. B., and Miller-Rushing, A. J. (2010). The importance of phylogeny to the study of phenological response to global climate change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3201–3213. doi: 10.1098/rstb.2010.0130
Day, J. W., Christian, R. R., Boesch, D. M., Yáñez-Arancibia, A., Morris, J., Twilley, R. R., et al. (2008). Consequences of climate change on the ecogeomorphology of coastal wetlands. Estuaries Coasts 31, 477–491. doi: 10.1007/s12237-008-9047-6
de los Santos, C. B., Krause-Jensen, D., Alcoverro, T., Marbà, N., Duarte, C. M., van Katwijk, M. M., et al. (2019). Recent trend reversal for declining European seagrass meadows. Nat. Commun. 10:3356. doi: 10.1038/s41467-019-11340-4
Dewsbury, B. M., Bhat, M., and Fourqurean, J. W. (2016). A review of seagrass economic valuations: gaps and progress in valuation approaches. Ecosyst. Serv. 18, 68–77. doi: 10.1016/j.ecoser.2016.02.010
Dias Tarli, V., Grandcolas, P., and Pellens, R. (2018). The informative value of museum collections for ecology and conservation: a comparison with target sampling in the Brazilian Atlantic forest. PLoS One 13:e0205710. doi: 10.1371/journal.pone.0205710
Dilipan, E., Lucas, C., Papenbrock, J., and Thangaradjou, T. (2018). Tracking the phylogeny of seagrasses: inferred from 18S rRNA gene and ancestral state reconstruction of morphological data. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 88, 497–504. doi: 10.1007/s40011-016-0780-5
Diniz-Filho, J. A. F., Loyola, R. D., Raia, P., Mooers, A. O., and Bini, L. M. (2013). Darwinian shortfalls in biodiversity conservation. Trends Ecol. Evol. 28, 689–695. doi: 10.1016/j.tree.2013.09.003
Drummond, A. J., Rambaut, A., Shapiro, B., and Pybus, O. G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192. doi: 10.1093/molbev/msi103
Duarte, B., Martins, I., Rosa, R., Matos, A. R., Roleda, M. Y., Reusch, T. B. H., et al. (2018). Climate change impacts on seagrass meadows and macroalgal forests: an integrative perspective on acclimation and adaptation potential. Front. Mar. Sci. 5:190. doi: 10.3389/fmars.2018.00190
Duarte, C. (2002). The future of seagrass meadows. Environ. Conserv. 29, 192–206. doi: 10.1017/S0376892902000127
Duarte, C. M. (1992). Nutrient concentration of aquatic plants: patterns across species. Limnol. Oceanogr. 37, 882–889. doi: 10.4319/lo.1992.37.4.0882
Duffy, J. E., Benedetti-Cecchi, L., Trinanes, J., Muller-Karger, F. E., Ambo-Rappe, R., Boström, C., et al. (2019). Toward a coordinated global observing system for seagrasses and marine macroalgae. Front. Mar. Sci. 6:317. doi: 10.3389/fmars.2019.00317
Edwards, J., Lane, M. A., and Nielsen, E. (2000). Interoperability of biodiversity databases: biodiversity information on every desktop. Science 289, 2312–2314. doi: 10.1126/science.289.5488.2312
Erry, D. I. P., Taveley, T. H. S., Eyanova, D. I. D., Aden, S. U. B., Upont, S. A. M. D., and Al, P. E. T. (2019). Global environmental changes negatively impact temperate seagrass ecosystems. Ecosphere 10:e02986. doi: 10.1002/ecs2.2986
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Farley, S. S., Dawson, A., Goring, S. J., and Williams, J. W. (2018). Situating ecology as a big-data science: current advances, challenges, and solutions. BioScience 68, 563–576. doi: 10.1093/biosci/biy068
FitzJohn, R. G., Maddison, W. P., and Otto, S. P. (2009). Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. doi: 10.1093/sysbio/syp067
Freeman, B. G., Scholer, M. N., Ruiz-Gutierrez, V., and Fitzpatrick, J. W. (2018). Climate change causes upslope shifts and mountaintop extirpations in a tropical bird community. Proc. Natl. Acad. Sci. U.S.A. 115, 11982–11987. doi: 10.1073/pnas.1804224115
Gaillard, J.-M., Mark Hewison, A. J., Klein, F., Plard, F., Douhard, M., Davison, R., et al. (2013). How does climate change influence demographic processes of widespread species? Lessons from the comparative analysis of contrasted populations of roe deer. Ecol. Lett. 16, 48–57. doi: 10.1111/ele.12059
Gallagher, A. J., Hammerschlag, N., Cooke, S. J., Costa, D. P., and Irschick, D. J. (2015). Evolutionary theory as a tool for predicting extinction risk. Trends Ecol. Evol. 30, 61–65. doi: 10.1016/j.tree.2014.12.001
GBIF.org (2020). GBIF Occurrence Download. Available online at: https://doi.org/10.15468/dl.t7xgct (accessed January 17, 2020).
Grech, A., Chartrand, K., Erftemeijer, P., Fonseca, M., McKenzie, L., Rasheed, M., et al. (2012). A comparison of threats, vulnerabilities and management approaches in global seagrass bioregions. Environ. Res. Lett. 7:024006. doi: 10.1088/1748-9326/7/2/024006
Green, E. P., and Short, F. T. (2003). World Atlas of Seagrasses. Prepared by UNEP World Conservation Monitoring Centre. Berkeley, CA: University of California, 332.
Guerrero-Meseguer, L., Veiga, P., Guerrero-Meseguer, L., and Sampaio, L. (2021). Resurgence of Zostera marina in the Ria de Aveiro lagoon, Portugal. Aquat. Bot. 169:103338. doi: 10.1016/j.aquabot.2020.103338
Guo, Q., and Liu, Y. (2010). ModEco: an integrated software package for ecological niche modeling. Ecography 33, 637–642. doi: 10.1111/j.1600-0587.2010.06416.x
Hauxwell, J., Cebrián, J., Furlong, C., and Valiela, I. (2001). Macroalgal canopies contribute to eelgrass (Zostera marina) decline in temperate estuarine ecosystems. Ecology 82, 1007–1022. doi: 10.2307/2679899
Heck, K. L., Samsonova, M., Poore, A. G. B., and Hyndes, G. A. (2021). Global patterns in seagrass herbivory: why, despite existing evidence, there are solid arguments in favor of latitudinal gradients in seagrass herbivory. Estuaries Coasts 44, 481–490. doi: 10.1007/s12237-020-00833-x
Hellmann, J. J., Byers, J. E., Bierwagen, B. G., and Dukes, J. S. (2008). Five potential consequences of climate change for invasive species. Conserv. Biol. 22, 534–543. doi: 10.1111/j.1523-1739.2008.00951.x
Hemminga, M., and Duarte, C. (2000). Seagrass Ecology. Cambridge: Cambridge University Press, doi: 10.1017/CBO9780511525551
Hijmans, R. J., Phillips, S., Leathwick, J., and Elith, J. (2011). Package ‘Dismo’. Available online at: http://cran.r-project.org/web/packages/dismo/index.html (accessed November 17, 2020).
Holt, B., Lessard, J.-P., Borregaard, M., Fritz, S., Araújo, M., Dimitrov, D., et al. (2013). An update of wallace’s zoogeographic regions of the world. Science 339, 74–78. doi: 10.1126/science.1228282
Hortal, J. (2008). Uncertainty and the measurement of terrestrial biodiversity gradients. J. Biogeogr. 35, 1335–1336. doi: 10.1111/j.1365-2699.2008.01955.x
Hortal, J., de Bello, F., Diniz-Filho, J. A. F., Lewinsohn, T. M., Lobo, J. M., and Ladle, R. J. (2015). Seven shortfalls that beset large-scale knowledge of biodiversity. Annu. Rev. Ecol. Evol. Syst. 46, 523–549. doi: 10.1146/annurev-ecolsys-112414-054400
Hou, Q. Q., Chen, B. M., Peng, S. L., and Chen, L.-Y. (2014). Effects of extreme temperature on seedling establishment of nonnative invasive plants. Biol. Invasions 16, 2049–2061. doi: 10.1007/s10530-014-0647-8
Hugo, G. (2011). Future demographic change and its interactions with migration and climate change. Glob. Environ. Change 21, S21–S33. doi: 10.1016/j.gloenvcha.2011.09.008
Hunter, C. M., Caswell, H., Runge, M. C., Regehr, E. V., Amstrup, S. C., and Stirling, I. (2010). Climate change threatens polar bear populations: a stochastic demographic analysis. Ecology 91, 2883–2897. doi: 10.1890/09-1641.1
Iler, A. M., Høye, T. T., Inouye, D. W., and Schmidt, N. M. (2013). Long-term trends mask variation in the direction and magnitude of short-term phenological shifts. Am. J. Bot. 100, 1398–1406. doi: 10.3732/ajb.1200490
IPCC (2018). Summary for Policymakers of IPCC Special Report on Global Warming of 1.5°C Approved by Governments. (2018, October 8). Available online at: https://www.ipcc.ch/2018/10/08/summary-for-policymakers-of-ipcc-special-report-on-global-warming-of-1-5c-approved-by-governments/ [December 03, 2020]
Jarzyna, M. A., and Jetz, W. (2018). Taxonomic and functional diversity change is scale dependent. Nat. Commun. 9:2565. doi: 10.1038/s41467-018-04889-z
Jayathilake, D. R. M., and Costello, M. J. (2018). A modelled global distribution of the seagrass biome. Biol. Conserv. 226, 120–126. doi: 10.1016/j.biocon.2018.07.009
Johnston, D. W. (2019). Unoccupied aircraft systems in marine science and conservation. Annu. Rev. Mar. Sci. 11, 439–463. doi: 10.1146/annurev-marine-010318-095323
Kadmon, R., Farber, O., and Danin, A. (2004). Effect of roadside bias on the accuracy of predictive maps produced by bioclimatic models. Ecol. Appl. 14, 401–413. doi: 10.1890/02-5364
Kalnay, E., and Cai, M. (2003). Impact of urbanization and land-use change on climate. Nature 423, 528–531. doi: 10.1038/nature01675
Kharouba, H. M., and Wolkovich, E. M. (2020). Disconnects between ecological theory and data in phenological mismatch research. Nat. Clim. Change 10, 406–415. doi: 10.1038/s41558-020-0752-x
Kramer-Schadt, S., Niedballa, J., Pilgrim, J. D., Schröder, B., Lindenborn, J., Reinfelder, V., et al. (2013). The importance of correcting for sampling bias in MaxEnt species distribution models. Divers. Distrib. 19, 1366–1379. doi: 10.1111/ddi.12096
Kreft, H., and Jetz, W. (2010). A framework for delineating biogeographical regions based on species distributions. J. Biogeogr. 37, 2029–2053. doi: 10.1111/j.1365-2699.2010.02375.x
Larkum, W. D., Orth, R. J., and Duarte, C. M. (eds) (2006). Seagrasses: Biology, Ecology and Conservation. Dordrecht: Springer.
Les, D. H., Moody, M. L., Jacobs, S. W. L., and Bayer, R. J. (2002). Systematics of seagrasses (Zosteraceae) in Australia and New Zealand. Syst. Bot. 27, 468–484.
Lobo, J., and Tognelli, M. (2011). Exploring the effects of quantity and location of pseudo-absences and sampling biases on the performance of distribution models with limited point occurrence data. J. Nat. Conserv. 19, 1–7. doi: 10.1016/j.jnc.2010.03.002
Mace, G. M., Gittleman, J. L., and Purvis, A. (2003). Preserving the tree of life. Science 300:1707. doi: 10.1126/science.1085510
MacLean, S. A., Rios Dominguez, A. F., de Valpine, P., and Beissinger, S. R. (2018). A century of climate and land-use change cause species turnover without loss of beta diversity in California’s Central Valley. Glob. Change Biol. 24, 5882–5894. doi: 10.1111/gcb.14458
Maruyama, N., Nomura, T., Sato, K., and Matsuoka, S. (2011). “Physis: an implicitly parallel programming model for stencil computations on large-scale GPU-accelerated supercomputers,” in Proceedings of the 2011 International Conference for High Performance Computing, Networking, Storage and Analysis, (New York, NY: Association for Computing Machinery), doi: 10.1145/2063384.2063398
Massa, S. I., Paulino, C. M., Serrão, E. A., Duarte, C. M., and Arnaud-Haond, S. (2013). Entangled effects of allelic and clonal (genotypic) richness in the resistance and resilience of experimental populations of the seagrass Zostera noltii to diatom invasion. BMC Ecol. 13:39. doi: 10.1186/1472-6785-13-39
McGlathery, K. J., Sundbäck, K., and Anderson, I. (2007). Eutrophication in shallow coastal bays and lagoons: the role of plants in the coastal filter. Mar. Ecol. Prog. Ser. 348, 1–18. doi: 10.3354/meps07132
McKenzie, L., Nordlund, L., Jones, B., Cullen-Unsworth, L., Roelfsema, C., and Unsworth, R. (2020). The global distribution of seagrass meadows. Environ. Res. Lett. 15:074041. doi: 10.1088/1748-9326/ab7d06
McKenzie, L. J., Long, L., Coles, R. G., and Roder, C. A. (2000). Seagrass-watch: community based monitoring of seagrass resources. Biol. Mar. Mediterr. 7, 393–396.
McKenzie, L. J., Yoshida, R. L., Mellors, J. E., and Coles, R. G. (2009). “Seagrass-watch,” In Proceedings of a Workshop for Monitoring Seagrass Habitats in Indonesia. Sanur: Coral Triangle Center.
Meerdink, S. K., Roberts, D. A., Roth, K. L., King, J. Y., Gader, P. D., and Koltunov, A. (2019). Classifying california plant species temporally using airborne hyperspectral imagery. Remote Sens. Environ. 232:111308. doi: 10.1016/j.rse.2019.111308
Menegotto, A., Rangel, T., Schrader, J., Weigelt, P., and Kreft, H. (2019). A global test of the subsidized island biogeography hypothesis. Glob. Ecol. Biogeogr. 29, 320–330. doi: 10.1111/geb.13032
Meyer, C., Weigelt, P., and Kreft, H. (2016). Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19, 992–1006. doi: 10.1111/ele.12624
Miller-Rushing, A. J., and Primack, R. B. (2008). Global warming and flowering times in thoreau’s concord: a community perspective. Ecology 89, 332–341. doi: 10.1890/07-0068.1
Moksnes, P., Gullstro, M., Tryman, K., and Baden, S. (2008). Trophic cascades in a temperate seagrass community. Oikos 117, 763–777. doi: 10.1111/j.2008.0030-1299.16521.x
Morell, V. (1996). TreeBASE: the roots of phylogeny. Science 273:569. doi: 10.1126/science.273.5275.569
Mtwana Nordlund, L., Koch, E. W., Barbier, E. B., and Creed, J. C. (2016). Seagrass ecosystem services and their variability across genera and geographical regions. PLoS One 11:e0163091. doi: 10.1371/journal.pone.0163091
National Research Council [NRC-US] (2008). “In the light of evolution: volume ii: biodiversity and extinction,” in 9 Extinction as the Loss of Evolutionary History, eds J. C. Avise, S. P. Hubbell, and F. J. Ayala (Washington, DC: National Academies Press).
Nee, S., May, R. M., and Harvey, P. H. (1994). The reconstructed evolutionary process. Philos. Trans. R. Soc. Lond. B Biol. Sci. 344, 305–311. doi: 10.1098/rstb.1994.0068
NOAA Climate.gov (2020). NOAA Climate.gov: Science & Information for a Climate-Smart Nation. Available online at: https://www.climate.gov/ [December 03, 2020]
Nordlund, L. M., Jackson, E. L., Nakaoka, M., Samper-Villarreal, J., Beca-Carretero, P., and Creed, J. C. (2018). Seagrass ecosystem services – What’s next? Mar. Pollut. Bull. 134, 145–151. doi: 10.1016/j.marpolbul.2017.09.014
Nunn, N., and Qian, N. (2010). The columbian exchange: a history of disease. Food, and Ideas. J. Econ. Perspect. 24, 163–188. doi: 10.1257/jep.24.2.163
Orth, R. J., Harwell, M. C., and Inglis, G. J. (2006a). “Ecology of seagrass seeds and seagrass dispersal processes,” In Seagrasses: Biology, Ecology and Conservation, eds A. W. D. Larkum, R. J. Orth, and C. M. Duarte (Dordrecht: Springer), 111–133. doi: 10.1007/1-4020-2983-7_5
Orth, R. J., Carruthers, T. J. B., Dennison, W. C., Duarte, C. M., Fourqurean, J. W., Heck, K. L., et al. (2006b). A global crisis for seagrass ecosystems. BioScience 56, 987–996.
Ovaskainen, O., Skorokhodova, S., Yakovleva, M., Sukhov, A., Kutenkov, A., Kutenkova, N., et al. (2013). Community-level phenological response to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 13434–13439. doi: 10.1073/pnas.1305533110
Page, R. D. (2007). TBMap: a taxonomic perspective on the phylogenetic database TreeBASE. BMC Bioinformatics 8:158. doi: 10.1186/1471-2105-8-158
Park, D. S., Worthington, S., and Xi, Z. (2018). Taxon sampling effects on the quantification and comparison of community phylogenetic diversity. Mol. Ecol. 27, 1296–1308. doi: 10.1111/mec.14520
Parmesan, C. (2006). Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669. doi: 10.1146/annurev.ecolsys.37.091305.110100
Parmesan, C., Ryrholm, N., Stefanescu, C., Hill, J. K., Thomas, C. D., Descimon, H., et al. (1999). Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature 399, 579–583. doi: 10.1038/21181
Pearson, K. D., Nelson, G., Aronson, M. F. J., Bonnet, P., Brenskelle, L., Davis, C. C., et al. (2020). Machine learning using digitized herbarium specimens to advance phenological research. BioScience 70, 610–620. doi: 10.1093/biosci/biaa044
Pergent, G., Bazairi, H., Bianchi, C., Boudouresque, C., Buia, M., Calvo, S., et al. (2014). Climate change and mediterranean seagrass meadows: a synopsis for environmental managers. Mediterr. Mar. Sci. 15, 462–473. doi: 10.12681/mms.621
Pernetta, J. C., Leemans, R., Elder, D., Humphrey, S., and Brouns, J. J. (1994). Impacts of climate change on ecosystems and species: marine and coastal ecosystems, eds J. C. Pernetta, R. Leemans, D. Elder, and S. Humphrey (International Union for Conservation of Nature: Gland), 2, 59–72.
Peterson, A. T., Soberón, J., Pearson, R. G., Anderson, R. P., Martínez-Meyer, E., Nakamura, M., et al. (2011). Monographs in Population Biology, Vol. 49. Princeton, NJ: Princeton University Press.
Phillips, S. J., Dudík, M., and Schapire, R. E. (2017). MaxEnt Software for Modeling Species Niches and Distributions (Version 3.4.1).
Piel, W. H., Donoghue, M. J., Sanderson, M. J., and Netherlands, L. (2000). “TreeBASE: a database of phylogenetic information,” in Proceedings of the 2nd International Workshop of Species 2000, Tsukuba.
Poelen, J. H., Simons, J. D., and Mungall, C. J. (2014). Global biotic interactions: an open infrastructure to share and analyze species-interaction datasets. Ecol. Inform. 24, 148–159. doi: 10.1016/j.ecoinf.2014.08.005
POWO (2019). Plants of the World Online. Available online at: http://www.plantsoftheworldonline.org/ [Accessed December 7, 2020]
Purvis, A., Agapow, P.-M., Gittleman, J. L., and Mace, G. M. (2000). Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330. doi: 10.1126/science.288.5464.328
Pybus, O. G., Rambaut, A., and Harvey, P. H. (2000). An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155, 1429–1437.
Ralph, P., Tomasko, D., Moore, K., Seddon, S., Macinnis-Ng, C., Larkum, A., et al. (2006). “Human impacts on seagrasses: eutrophication, sedimentation, and contamination,” in In Seagrasses: Biology, Ecology and Conservation, eds A. W. D. Larkum, R. J. Orth, and C. Duarte (Netherlands: Springer), 567–593. doi: 10.1007/1-4020-2983-7_24
Redding, D., Hartmann, K., Mimoto, A., Bokal, D., Devos, M., and Mooers, A. (2008). Evolutionarily distinct species capture more phylogenetic diversity than expected. J. Theor. Biol. 251, 606–615. doi: 10.1016/j.jtbi.2007.12.006
Redding, D. W., and Mooers, A. Ø (2006). Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678. doi: 10.1111/j.1523-1739.2006.00555.x
Rosauer, D. A. N., Laffan, S. W., Crisp, M. D., Donnellan, S. C., and Cook, L. G. (2009). Phylogenetic endemism: a new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 18, 4061–4072. doi: 10.1111/j.1365-294X.2009.04311.x
Sablok, G., Hayward, R. J., Davey, P. A., Santos, R. P., Schliep, M., Larkum, A., et al. (2018). SeagrassDB: an open-source transcriptomics landscape for phylogenetically profiled seagrasses and aquatic plants. Sci. Rep. 8:2749. doi: 10.1038/s41598-017-18782-0
Schäfer, S., Monteiro, J., Castro, N., Gizzi, F., Henriques, F., Ramalhosa, P., et al. (2021). Lost and found: a new hope for the seagrass Cymodocea nodosa in the marine ecosystem of a subtropical Atlantic Island. Reg. Stud. Mar. Sci. 41:101575. doi: 10.1016/j.rsma.2020.101575
Selwood, K. E., McGeoch, M. A., and Mac Nally, R. (2015). The effects of climate change and land-use change on demographic rates and population viability. Biol. Rev. 90, 837–853. doi: 10.1111/brv.12136
Short, F., Carruthers, T., Dennison, W., and Waycott, M. (2007). Global seagrass distribution and diversity: a bioregional model. J. Exp. Mar. Biol. Ecol. 350, 3–20. doi: 10.1016/j.jembe.2007.06.012
Short, F., Koch, E., Creed, J., Magalhães, K., Fernandez, E., and Gaeckle, J. (2006). SeagrassNet monitoring across the americas: case studies of seagrass decline. Mar. Ecol. 27, 277–289. doi: 10.1111/j.1439-0485.2006.00095.x
Short, F. T., and Neckles, H. A. (1999). The effects of global climate change on seagrasses. Aquat. Bot. 63, 169–196. doi: 10.1016/S0304-3770(98)00117-X
Short, F. T., Polidoro, B., Livingstone, S. R., Carpenter, K. E., Bandeira, S., Bujang, J. S., et al. (2011). Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 144, 1961–1971. doi: 10.1016/j.biocon.2011.04.010
Stropp, C. J., Ladle, R., Malhado, A., Hortal, J., Gaffuri, J., Temperley, W. H., et al. (2016). Mapping ignorance: 300 years of collecting flowering plants in Africa. Glob. Ecol. Biogeogr. 25, 1085–1096. doi: 10.1111/geb.12468
Strydom, S., Murray, K., Wilson, S., Huntley, B., Rule, M., Heithaus, M., et al. (2020). Too hot to handle: unprecedented seagrass death driven by marine heatwave in a World Heritage Area. Glob. Change Biol. 26, 3525–3538. doi: 10.1111/gcb.15065
Syfert, M. M., Smith, M. J., and Coomes, D. A. (2013). The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS One 8:e55158. doi: 10.1371/journal.pone.0055158
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x
Thackeray, S. J., Henrys, P. A., Hemming, D., Bell, J. R., Botham, M. S., Burthe, S., et al. (2016). Phenological sensitivity to climate across taxa and trophic levels. Nature 535, 241–245. doi: 10.1038/nature18608
Thapa, S., Chitale, V., Rijal, S. J., Bisht, N., and Shrestha, B. B. (2018). Understanding the dynamics in distribution of invasive alien plant species under predicted climate change in Western Himalaya. PLoS One 13:e0195752. doi: 10.1371/journal.pone.0195752
Thomas, J., Lonsdale, J., Salvatore, M., Phillips, R., Lo, E., Shad, S., et al. (2013). The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585. doi: 10.1038/ng.2653
Thuiller, W., Georges, D., and Engler, R. (2014). biomod2: Ensemble Platform for Species Distribution Modelling. Available online at: https://www.researchgate.net/publication/309762991_biomod2_Ensemble_Platform_for_Species_Distribution_Modeling (accessed February 26, 2020).
UNEP-WCMC, and Short, F. T. (2020). Global Distribution of Seagrasses (Version 7.0). Seventh Update to the Data Layer Used in Green and Short (2003). Cambridge: UN Environment World Conservation Monitoring Centre.
Valle, M., Chust, G., del Campo, A., Wisz, M. S., Olsen, S. M., Garmendia, J. M., et al. (2014). Projecting future distribution of the seagrass Zostera noltii under global warming and sea level rise. Biol. Conserv. 170, 74–85. doi: 10.1016/j.biocon.2013.12.017
Van Allen, B. G., Dunham, A. E., Asquith, C. M., and Rudolf, V. H. (2012). Life history predicts risk of species decline in a stochastic world. Proc. Biol. Sci. 279, 2691–2697. doi: 10.1098/rspb.2012.0185
van der Schoot, C., Paul, L., and Rinne, P. (2013). The embryonic shoot: a lifeline through winter. J. Exp. Bot. 65, 1699–1712. doi: 10.1093/jxb/ert413
Varela, S., Anderson, R. P., García-Valdés, R., and Fernández-González, F. (2014). Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography 37, 1084–1091. doi: 10.1111/j.1600-0587.2013.00441.x
Veeneklaas, R. M., Dijkema, K. S., Hecker, N., and Bakker, J. P. (2013). Spatio-temporal dynamics of the invasive plant species Elytrigia atherica on natural salt marshes. Appl. Veg. Sci. 16, 205–216. doi: 10.1111/j.1654-109X.2012.01228.x
Veron, S., Davies, T., Cadotte, M., Clergeau, P., and Pavoine, S. (2015). Predicting loss of evolutionary history: where are we? Biol. Rev. Camb. Philos. Soc. 92, 271–291. doi: 10.1111/brv.12228
Vicente, J. R., Fernandes, R. F., Randin, C. F., Broennimann, O., Gonçalves, J., Marcos, B., et al. (2013). Will climate change drive alien invasive plants into areas of high protection value? An improved model-based regional assessment to prioritise the management of invasions. J. Environ. Manage. 131, 185–195. doi: 10.1016/j.jenvman.2013.09.032
Waycott, M. (1999). Genetic factors in the conservation of seagrasses. Pac. Conserv. Biol. 5, 269–276. doi: 10.1071/pc000269
Waycott, M., Collier, C., McMahon, K., Ralph, P., McKenzie, L., Udy, J., et al. (2007). “Vulnerability of seagrasses in the great barrier reef to climate change,” in Climate Change and the Great Barrier Reef: A Vulnerability Assessment, eds J. E. Johnson and P. A. Marshall (Townsville: Great Barrier Reef Marine Park Authority), 193–236.
Waycott, M., Duarte, C. M., Carruthers, T. J. B., Orth, R. J., Dennison, W. C., Olyarnik, S., et al. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. U.S.A. 106, 12377–12381. doi: 10.1073/pnas.0905620106
Waycott, M., McKenzie, L. J., Mellors, J. E., Ellison, J. C., Sheaves, M. T., Collier, C., et al. (2011). Vulnerability of Mangroves, Seagrasses and Intertidal Flats in the Tropical Pacific to Climate Change. Available online at: https://hdl.handle.net/20.500.12348/1069 (accessed May 26, 2020).
Willis, C. G., Ruhfel, B., Primack, R. B., Miller-Rushing, A. J., and Davis, C. C. (2008). Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc. Natl. Acad. Sci. U.S.A. 105, 17029–17033. doi: 10.1073/pnas.0806446105
Wolfe, K. H., Li, W. H., and Sharp, P. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNA. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Woodman, S. M., Forney, K. A., Becker, E. A., Deangelis, M., Lee Hazen, E., Palacios, D. M., et al. (2019). esdm: A tool for creating and exploring ensembles of predictions from species distribution and abundance models. Methods Ecol. Evol. 10, 1923–1933. doi: 10.1111/2041-210X.13283
Zieman, J. C. (1976). The ecological effects of physical damage from motorboats on turtle grass beds in Southern Florida. Aquat. Bot. 2, 127–139. doi: 10.1016/0304-3770(76)90015-2
Keywords: marine macrophytes, sampling biases, computational infrastructure, Alismatales, biodiversity shortfalls, climate change
Citation: Rock BM and Daru BH (2021) Impediments to Understanding Seagrasses’ Response to Global Change. Front. Mar. Sci. 8:608867. doi: 10.3389/fmars.2021.608867
Received: 21 September 2020; Accepted: 08 February 2021;
Published: 24 March 2021.
Edited by:
Inés G. Viana, Instituto Español de Oceanografía, SpainReviewed by:
Rob Coles, James Cook University, AustraliaBenjamin L. Jones, Project Seagrass, United Kingdom
Copyright © 2021 Rock and Daru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brianna M. Rock, YnJpcm9jazM1QGdtYWlsLmNvbQ==; Barnabas H. Daru, YmFybmFiYXMuZGFydUB0YW11Y2MuZWR1
 Brianna M. Rock
Brianna M. Rock Barnabas H. Daru
Barnabas H. Daru