- 1Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, Ministry of Agriculture Rural Affairs, Guangzhou, China
- 2South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, Guangzhou, China
Development of effective conservation and management strategies requires assessments of ecosystem biodiversity status, especially in understudied hotspots of global fish diversity. Coral reefs are important habitats for fishes, with biodiversity hotspots known globally. We present the first data on molecular diversity of fishes of Mischief Reef, the largest atoll in the Nansha Islands. Partial sequences (650 bp) of mitochondrial COI gene (Cytochrome c oxidase subunit I) are used to identify 209 individuals, representing 101 species, referable to 62 genera, 27 families, 8 orders, and 1 class. The most abundant orders are the Perciformes (176 specimens, 84.21%), Tetraodontiformes (13 specimens, 6.22%), and Beryciformes (13 specimens, 6.22%). Mean Kimura 2-Parameter genetic distances within genera, families, and orders are 4.51, 13.90, and 17.63%, respectively. We record Monotaxis heterodon from this region for the first time—a species that may previously have been misidentified as M. grandoculis. In addition, we recognized possible cryptic species of Lethrinus olivaceus based on significantly diverging barcode sequences. Barcode data provide new insights into fish diversity of Mischief Reef, important for developing further researches on this fauna, and for its conservation.
Introduction
Coral reefs represent some of the most diverse of marine habitats and have been identified as biodiversity hotspots around the globe (Wilson et al., 2008; Hubert et al., 2012). Of species associated with them, fish are among the most conspicuous and fascinating. Unfortunately, some coral reef fishes have become critically endangered, threatened by a variety of activities, such as over-exploitation, habitat destruction, and pollution (Hixon, 2011; Friedlander et al., 2018).
Assessing the biodiversity of reef fishes is of critical importance in guiding conservation policy (Dawson et al., 2011). However, reliance on morphological characters to identify species can prove problematic because reef fishes are dominated by about 30 families, mostly perciform labroids, acanthuroids, chaetodontoids, and gobioids, many of which differ sexually, ontogenetically, or in general phenotypic plasticity (Radulovici et al., 2010). DNA barcoding—a molecular technique using mitochondrial cytochrome c oxidase I gene (COI) as a genetic marker (Hebert et al., 2003)—is now widely applied to identify adult and larval stages of fishes (Pegg et al., 2006; Lara et al., 2010; Weigt et al., 2012).
The South China Sea, in the western Pacific, can be viewed as a distinct ecosystem because of its archipelago and peninsula boundaries. Coral reefs in this area cover approximately 8,000 km2 (Yu and Zhao, 2009), with the largest concentration around the relatively remote Nansha Islands. Due to the vast sea area, perennial high temperature, and complex hydrology, the sea around the Nansha Islands has a diverse fish fauna (Li et al., 2016; Feng et al., 2020). Mischief Island, the largest atoll located in the eastern Nansha Islands, has a large and almost complete lagoon. Its tropical monsoon climate and warm waters render Mischief Reef an excellent location to develop marine fisheries. Major studies of the biodiversity of Nansha Islands have focused on more easily accessible islands, including Subi and Fiery Cross reefs (Yin et al., 2003; Shen et al., 2010; Wang et al., 2015), leaving the fish diversity of Mischief Reef poorly known, although several recent studies have explored environment pollution, ocean physics, and aquaculture around it (Lin et al., 2016; Chen et al., 2018; Sun et al., 2019).
In the present study, we investigate reef fish communities of Mischief Reef using morphology and molecular tools to provide insights into the diversity of fishes in this region. In addition, information generated in the present study will provide an adequate baseline that assist researchers, biodiversity managers, and policy makers to develop effective conservation measures for this ecosystem.
Materials and Methods
Ethics Statement
All experimental procedures were approved by the ethics committee of the Laboratory of Animal Welfare and Ethics of South China Sea Fisheries Research Institute. Methods involving animals were conducted in accordance with the Laboratory Animal Management Principles of China.
Sample Collection
Between May 23 and June 19, 2019, 258 fishes were sampled from Mischief Reef, mostly using gill or cast nets, or hand lines in the lagoon (Figure 1). For Acanthuridae and Chaetodontidae species, samples were caught on SCUBA (Self-Contained Underwater Breathing Apparatus) by hand net after light anesthetic with clove oil (50 ml of clove oil, 40 ml of ethanol, and 400 ml of seawater).
Specimens were identified to species based on morphology using appropriate taxonomic guides, then photographed and labeled, after which a muscle tissue sample was cut from it and stored in 95% ethanol, then frozen at −20°C before DNA extraction. Voucher specimens and tissue samples were deposited at the Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, Ministry of Agriculture Rural Affairs, China.
DNA Data Collection
Total genomic DNA was extracted from tissue samples using a DNeasy Blood and Tissue kit (Qiagen, The Netherlands) following manufacturer protocols. Fragments of DNA barcode regions were amplified using FishF1 (5′-TCA ACC AAC CAC AAA GAC ATT GGC AC-3′), FishF2 (5′-TCG ACT AAT CAT AAA GAT ATC GGC AC-3′), FishR1 (5-′TAG ACT TCT GGG TGG CCA AAG AAT CA-3′), and FishR2 (5′-ACT TCA GGG TGA CCG AAG AAT CAG AA-3′) primers (Ward et al., 2005). PCRs were run in a final volume of 25 μL, containing 12.5 μL of PCR Mix (Vazyme Biotech Co., Ltd), 1–2 μL of genomic DNA, and distilled water. PCR was carried out in an Eppendorf thermal cycler with 5 min initial denaturation at 94°C, 35 cycles of 45 s at 94°C for denaturation, 45 s at an annealing temperature, 45 s at 72°C for extension, and a final extension at 72°C for 10 min.
Data Analysis
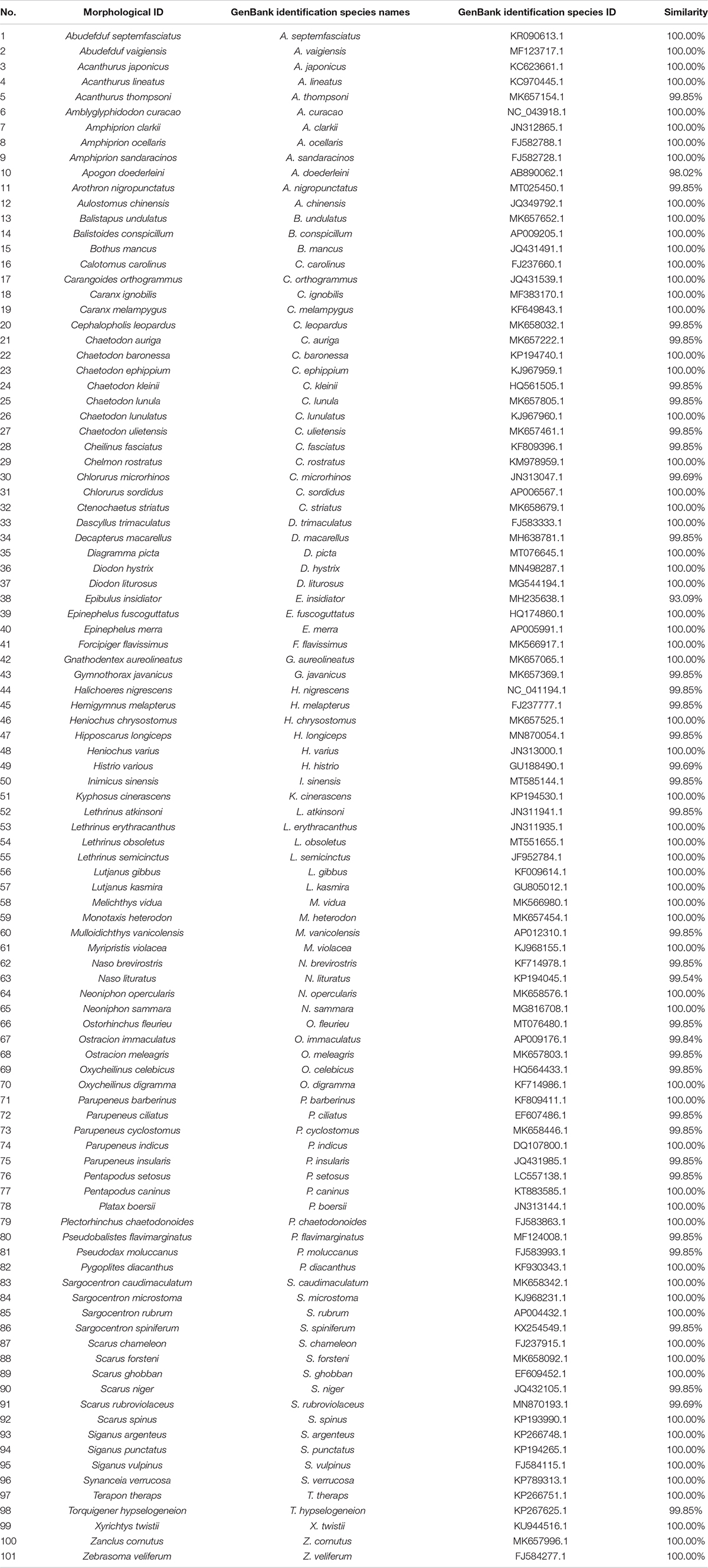
DNA barcode sequences were edited to remove ambiguous bases and primer reads, then aligned with DNASTAR (DNASTAR, Inc.) and MEGA ver. 7.0.14 softwares (Kumar et al., 2016). We also translated sequences into amino acids to check for premature stop codons or indels in the reading frame. For many reef fishes (Labridae, Scaridae, and Chaetodontidae) significant morphological differences exist between their different growth stages. To avoid misidentification using morphology, we compared our sequences to reference sequences from recently published taxonomic studies in the GenBank database (Nr/Nt database). We used a similarity threshold of 98% to assign specimens to species (Ward, 2009). Samples were reexamined in instances of conflict between molecular and morphological identification. Final identifications were compared with FishBase to determine new distribution records.
Genetic distances at different taxonomic levels (species, genus, family, and order) were calculated based on the Kimura 2-parameter (K2P) model performed in MEGA ver. 7.0.14 software (Kumar et al., 2016). For intra-generic comparisons, monotypic genera were excluded, as were families containing a single genus only; this criterion was applied for higher levels in genetic distance analysis. Then, we used the seaborn library of Python1 to draw heatmap of average K2P divergences between COI barcodes of families. MEGA ver. 7.0.14 software was also used to build a Maximum likelihood (ML) tree of all analyzed DNA barcode sequences based on the K2P model, with 5,000 bootstrap replications (Kumar et al., 2016).
Results
Species Identification and Fish Diversity
Based on morphology, the 258 collected fishes were attributed to 113 species. Despite repeat attempts, quality sequence reads could not be obtained from 43 specimens, so we excluded them from further analyses. The remaining 215 (87.76%) specimens (102 species based on morphology) were identified by amplification and nucleotide sequencing of a partial region of the COI mitochondrial gene, with sequences representing 103 species. Six specimens identified as Lethrinus olivaceus based on morphology were attributed to two species, with one sequence significantly different from five others (with 48 diverse sites, and a diverse ratio of 7.32%) (Figure 2). We could not differentiate these two species based on morphology. We therefore based fish diversity analyses on 209 specimens (Table 1) represented by 101 species in 62 genera, 27 families, 8 orders, and 1 class.

Figure 2. Sequences of six L. olivaceus specimens and accession numbers of sequences matched in Nt database.
The most abundantly fishes were in the orders Perciformes (176 specimens, 84.21%), Tetraodontiformes (13 specimens, 6.22%), and Beryciformes (13 specimens, 6.22%); other orders contributed 3.35% (7 specimens) to total abundance (Table 1). Four orders contained only one species: Bothus mancus (Pleuronectiformes), Histrio histrio (Lophiiformes), Gymnothorax javanicus (Anguilliformes), and Aulostomus chinensis (Gasterosteiformes). The most diverse family was Chaetodontidae, with 25 specimens attributed to 11 species, followed by Scaridae with 9 species, and Pomacentridae, Labridae, and Acanthuridae, with 8 species in each. The 11 specimens of Chaetodon comprised 7 species, followed by Scarus (7 specimens, 6 species). Of the 101 species, 56 were represented by single specimen.
Genetic Divergence
All amplified sequences were of 655 bp without deletions, insertions, or stop codons, indicating they represented functional mitochondrial COI sequences. Among the 655 sites, 290 were polymorphic and 281 were parsimony informative. Nucleotide diversity of the entire dataset was 0.1875, with 148 haplotypes and a diversity of 0.9949. Overall nucleotide composition and contents at each codon position were detailed in Table 2. The G content was 18.50%, indicating an obvious anti-guanine bias.
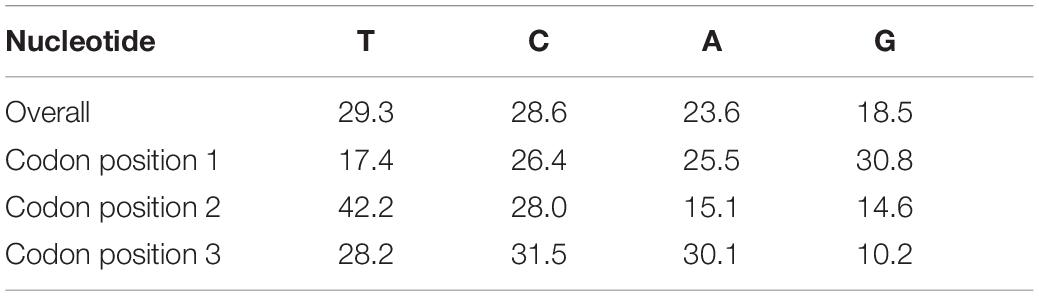
Table 2. Nucleotide composition, overall, and order-wise GC content and GC at each codon position 1, 2, and 3.
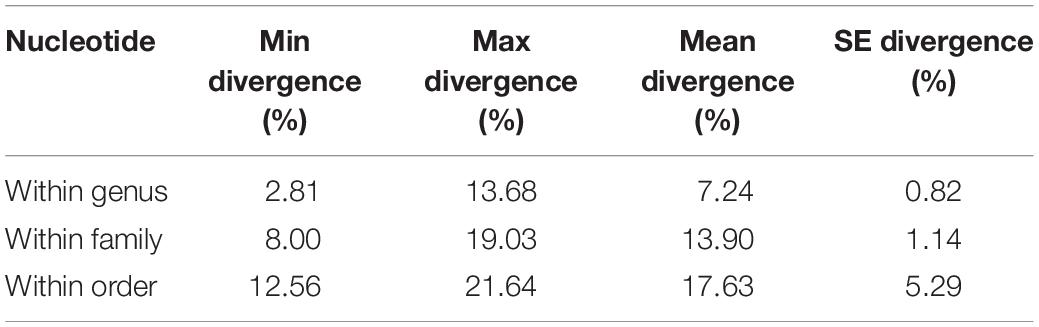
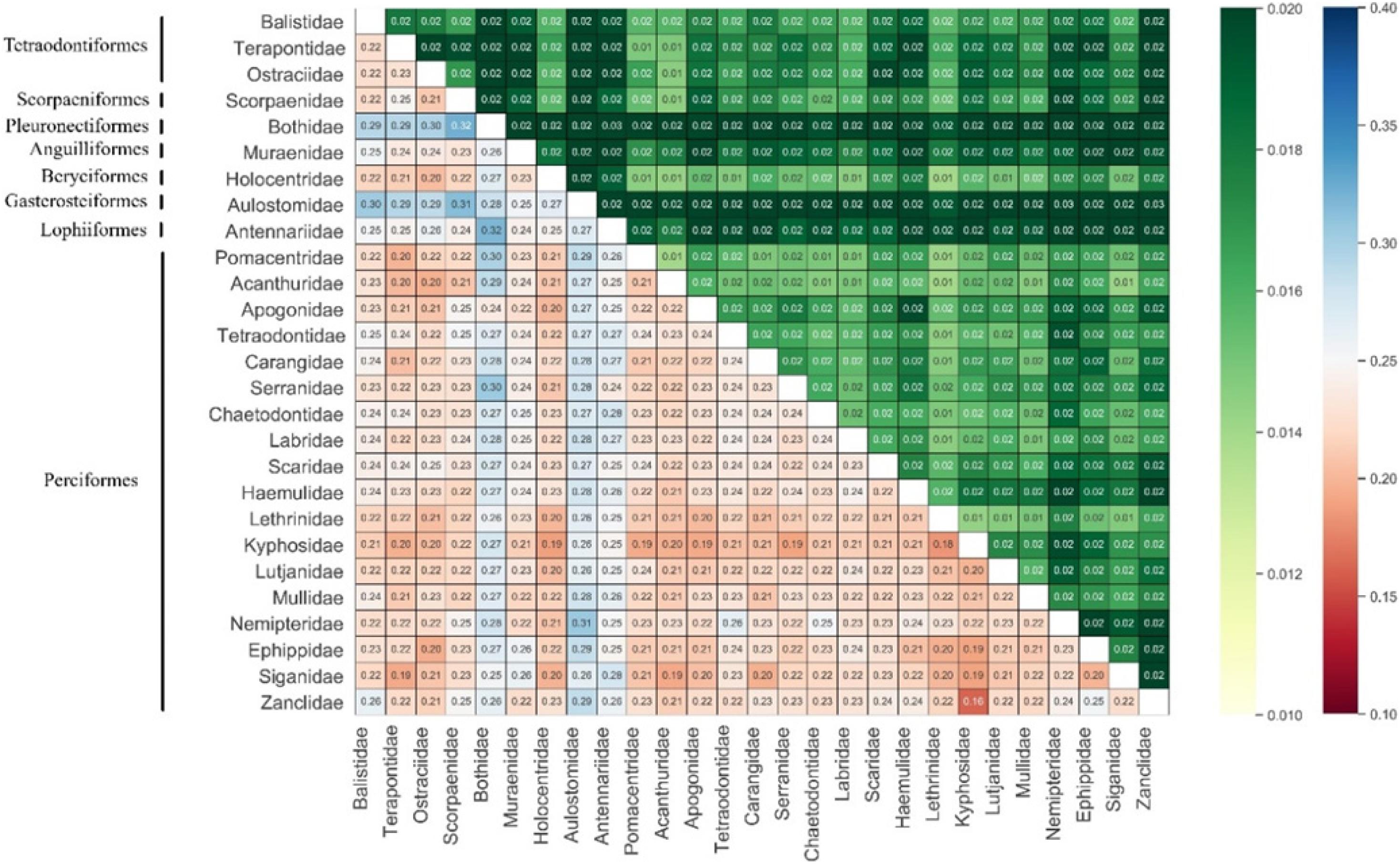
Most species identified using morphology were similarly identified by COI sequences, except for L. olivaceus, for which reason the six sequences were excluded from analyses. As expected, a hierarchical increase in the mean K2P genetic divergence with increasing taxonomic levels (from 7.24 to 17.63%) was observed (Table 3). We also calculated genetic divergence among genera and families; at the family level, the lowest divergence was observed between Zanclidae and Kyphosidae (16.28%), the highest between Scorpaenidae and Bothidae (32.19%) (Figure 3), and at the genus level, the lowest divergence was observed between Plectorhinchus and Diagramma (9.59%), and the highest was observed between Pygoplites and Bothus (33.82%).
The ML tree included 101 species (Figure 4). Unexpectedly, species of Tetraodontiformes did not cluster. The four species Balistapus undulatus, Balistoides conspicillum, Melichthys vidua, and Pseudobalistes flavimarginatus did cluster (Figure 4B). Diodon hystrix and D. liturosus clustered with Aulostomus chinensis in the Gasterosteiformes. Torquigener hypselogeneion clustered with Bothus mancus in Pleuronectiformes and then with Arothron nigropunctatus (Figure 4C). In addition, a single species of Anguilliformes (Gymnothorax javanicus) and Lophiiformes (Histrio histrio) clustered.
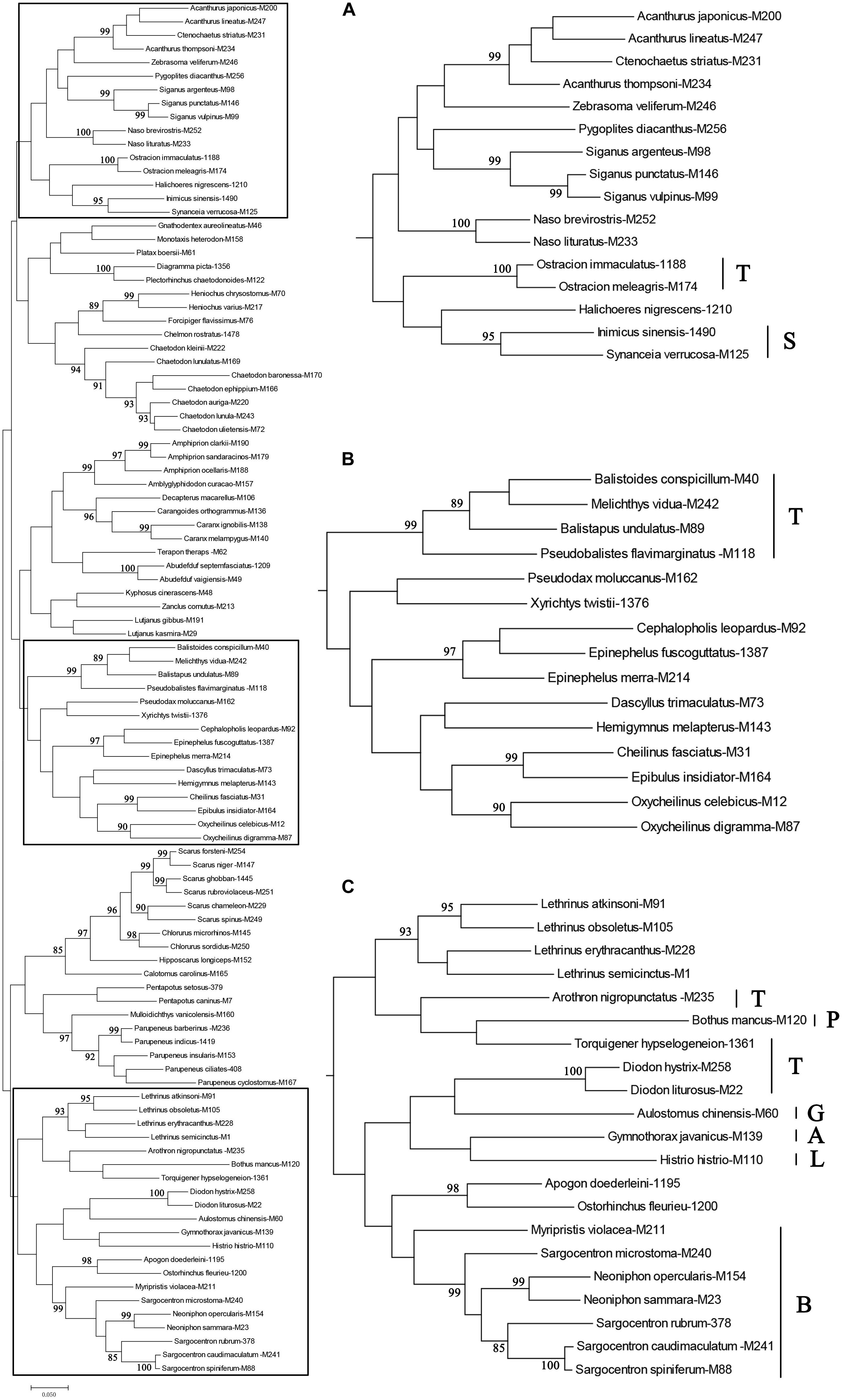
Figure 4. Maximum likelihood tree based on COI barcodes obtained from 101 species: A, Anguilliformes; B, Beryciformes; G, Gasterosteiformes; L, Lophiiformes; P, Pleuronectiformes; S, Scorpaeniformes; T, Tetraodontiformes. Numbers on branches are ML bootstrap values, those below 85% are hidden; (A–C) are larger versions of parts of the Maximum likelihood tree.
Discussion
As a core area of coral reefs in China, the Nansha Islands have a diverse array of species and rich mineral deposits and are well known for their tropical marine fisheries. However, numerous anthropogenic activities, such as increased marine transportation, over-exploitation of mineral resources, and a rapid increase in tourism, have contributed to deterioration in the marine ecosystem (Sun et al., 2019; Tan et al., 2020). While the fish diversity of Nansha Islands and nearby waters was reported by Chen et al. (2010) and Liu et al. (2012), knowledge of reef fish diversity in the Mischief Reef was limited. Because species represent basic units of biodiversity and are the foundation of ecosystem services to which the well-being of humans is closely linked (Barman et al., 2018), precise appraisals of biodiversity are needed to devise effective conservation measures.
Species Identification
Of 215 specimens examined, 209 were finally identified to species using morphological and molecular techniques. Six specimens referred to L. olivaceus based on morphology were referred to two species using DNA. Additionally, the six sequences were all referred to L. olivaceus by searching in database (Figure 2). Borsa et al. (2013) found two cranial morphotypes in L. olivaceus, and indicated one distributed from the Indian Ocean to the Coral Triangle and the other one distributed from the Coral Triangle to the western Central Pacific. The two morphotypes are concordant with reciprocally monophyletic mitochondrial lineages separated by a significant genetic difference, and their distributions range meet or overlap in the eastern part of the Coral Triangle, in Taiwan and in West Papua (Borsa et al., 2013). Deng et al. (2019) examined L. olivaceus from the Xisha, Zhongsha, and Nansha archipelagos in the South China Sea based on mitochondrial DNA control region, and identified two distinct lineages, one around Xisha and Zhongsha archipelagos and the other around Nansha archipelago. These researches illustrated a deep split between L. olivaceus, suggesting the possible occurrence of a cryptic species. We sequenced the homologous sequences (cytochrome b gene and control region) and compared the sequences of our samples and two monophyletic mitochondrial lineages of Borsa et al. (2013) and Deng et al. (2019). The results showed our L. olivaceus samples divided into two lineages, which is consistent with the previous study (Supplementary Figure 1). Furthermore, our result also showed that the distribution ranges meet or overlap in the Nansha Islands of South China Sea. For the further taxonomy studies of L. olivaceus, we suggest sequencing DNA barcodes of congeneric taxa, including specimens from type localities of two taxa currently considered junior synonyms (L. rostratus and L. waigiensis) to clarify the status of this species.
In the present study, Monotaxis heterodon was a new record species in South China Sea. Previous record showed that M. grandoculis was the single species of Monotaxis in South China Sea (Sun and Chen, 2013). So far, few studies have investigated the M. heterodon. Former researches considered that the genus Monotaxis was monotypic, and indicated M. heterodon was a junior synonym of M. grandoculis (Carpenter and Johnson, 2002). In contrast to earlier findings, other researchers found that both morphological characteristic and DNA barcodes of the two species were significantly different (Randall, 2005; Chen and Borsa, 2020; Limmon et al., 2020). Consistent with these literatures, the M. heterodon here was confirmed as a valid species based on morphologic characteristics and DNA barcodes.
Genetic Divergence
The mean K2P genetic distances hierarchy increased with increased taxonomic level, consistent with data from coral reef fishes of the Indo-Malay-Philippines Archipelago, Todos os Santos Bay, and marine fish of other areas (Lakra et al., 2011; Hubert et al., 2012; Duarte et al., 2017). Similar results were also found for freshwater fishes (Hubert et al., 2008; Barman et al., 2018). Previous studies have attempted to delineate species boundaries based on DNA barcode data (Meier et al., 2008; Bhattacharjee et al., 2012), with Hebert proposing a COI sequence threshold for conspecific and congeneric divergence—the 10 × rule—where a 10-fold difference in mean intraspecific variation was adequate to draw boundaries between species (Hebert et al., 2004). Our findings do not support this because we report much lower intergeneric genetic distance (9.59%) between Diagramma picta and Plectorhinchus chaetodonoides, but higher intrageneric genetic distances between taxa such as Epinephelus (9.81%), Parupeneus (9.88%), Lethrinus (10.33%), Acanthurus (10.41%), and Chaetodon (13.68%), consistent with Barman et al. (2018) and Guimarães-Costa et al. (2019). Because frequent overlap between intra- and interspecific divergence was also reported in earlier studies, it is difficult to generalize a threshold for genus- or higher-level resolution.
ML tree topology structure reveals convergence of congeneric taxa, although some species appear to be more closely related to those in other genera than within a genus. Species of Sargocentron appear to be more closely related to those of Neoniphon than to S. microstoma (Figure 1)—a finding broadly supporting other phylogenetic studies on the Holocentridae (Hubert et al., 2010; Dornburg et al., 2012). Dornburg et al. (2012) inferred the species-level phylogeny of the Holocentridae based on nuclear and mitochondrial genes and demonstrated that taxonomically diagnostic characters for Neoniphon and Sargocentron likely represent character states with a complex evolutionary history that do not reflect shared common ancestry (Dornburg et al., 2012). A similar result was found for the Acanthuridae, a clade containing Acanthurus and Ctenochaetus, which show a paraphyletic relationship, supporting Clements et al. (2003) and Sorenson et al. (2013). The ML tree for higher taxonomic levels (family and above) was also inconsistent with conventionally accepted phylogenetic relationships, with genera in the Tetraodontiformes scattered throughout it, and orders represented by single species or genera (e.g., Lophiiformes, Pleuronectiformes, Anguilliformes) not showing single branches. This inconsistency may be due to increased variability in the COI gene sequence at the level of family and higher. Since base substitutions among higher taxonomic levels tend to be saturated, this reduces resolution at high phylogenetic levels. In general, the COI gene may be unsuitable for phylogenetic studies above the level of family. The result reflects that of Xing et al. (2020) who reported that the COI gene sequence was unsuitable as a molecular marker for phylogenetic analysis of ophichthid fishes above the level of species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of the Laboratory of Animal Welfare and Ethics of South China Sea Fisheries Research Institute.
Author Contributions
BS: conceptualization, data curation, and formal analysis. QW, BS, and DS: funding acquisition, project administration, and resources. BS, YZ, GZ, and DS: investigation and methodology. BS, YZ, and GZ: software. YL, BS, and CY: supervision. BS and YL: validation and visualization. BS, DS, and YL: writing—original draft preparation. BS, CY, and QW: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Special Funds for the Construction of Modern Agricultural Industry Technology System (No. CARS-49), Ministry of Agriculture of China Special Financial Funding for Nansha Fisheries Management (South China Sea Fisheries Centre), the Central Public-interest Scientific Institution Basal Research Fund, the South China Sea Fisheries Research Institute (CAFS Nos. 2021SD14 and 2019CY0404), and the China–ASEAN Maritime Cooperation Fund (China–Vietnam Fishery Stock Enhancement and Conservation in Beibu Gulf).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Steve O’Shea, Ph.D., from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.618954/full#supplementary-material
Footnotes
References
Barman, A. S., Singh, M., Singh, S. K., Saha, H., Singh, Y. J., Laishram, M., et al. (2018). DNA barcoding of freshwater fishes of indo-myanmar biodiversity hotspot. Sci. Rep. 8:8579.
Bhattacharjee, M. J., Laskar, B. A., Dhar, B., and Ghosh, S. K. (2012). Identification and re-evaluation of freshwater catfishes through DNA barcoding. PLoS One 7:e49950. doi: 10.1371/journal.pone.0049950
Borsa, P., Hsiao, D. R., Carpenter, K. E., and Chen, W. J. (2013). Cranial morphometrics and mitochondrial DNA sequences distinguish cryptic species of the longface emperor (Lethrinus olivaceus), an emblematic fish of Indo-West Pacific coral reefs. CR. Biol. 336, 505–514. doi: 10.1016/j.crvi.2013.09.004
Carpenter, K. E., and Johnson, G. D. (2002). A phylogeny of sparoid fishes (Perciformes, Percoidei) based on morphology. Ichthyol. Res. 49, 114–127. doi: 10.1007/s102280200015
Chen, M. Q., Chen, X., Li, Y. N., Wu, Q. E., Yu, G., Ma, Z. H., et al. (2018). Cage culture of pearl oyster Pinctada martensii in Meiji Reef. Fish. Sci. 37, 379–383. (In Chinese).
Chen, W. H., Liu, X. D., and Lu, H. S. (2010). Research on seasonal changes of the fish structure community in Nansha Islands waters. J. Trop. Oceanogr. 29, 118–124.
Chen, W. J., and Borsa, P. (2020). Diversity, phylogeny, and historical biogeography of large-eye seabreams (Teleostei: Lethrinidae). Mol. Phylogenet. Evol. 151:106902. doi: 10.1016/j.ympev.2020.106902
Clements, K. D., Gray, R. D., and Choat, J. H. (2003). Rapid evolutionary divergences in reef fishes of the family Acanthuridae (Perciformes: Teleostei). Mol. Phylogenet. Evol. 26, 190–201. doi: 10.1016/s1055-7903(02)00325-1
Dawson, T. P., Jackson, S. T., House, J. I., Prentice, I. C., and Mace, G. M. (2011). Beyond predictions: biodiversity conservation in a changing climate. Science 332, 53–58. doi: 10.1126/science.1200303
Deng, Z., Song, N., Li, Y., Gao, T., and Han, Z. (2019). Population genetic structure and genetic diversity of coral reef species Lethrinus olivaceus in the south china sea. Pak. J. Zool. 51, 1289–1997.
Dornburg, A., Moore, J. A., Webster, R., Warren, D. L., Brandley, M. C., Iglesias, T. L., et al. (2012). Molecular phylogenetics of squirrelfishes and soldierfishes (Teleostei: Beryciformes: Holocentridae): reconciling more than 100 years of taxonomic confusion. Mol. Phylogenet. Evol. 65, 727–738. doi: 10.1016/j.ympev.2012.07.020
Duarte, L. A. G., Oliveira, E. J. F., Amorim, A., Silva, D. A., Carvalho, E. F., Mazzoni, R., et al. (2017). DNA Barcoding and Atlantic reef fishes: the molecular identification of a reef fish community from the Todos os Santos Bay, Bahia, Brazil. Forensic Sci. Int. Gen. Suppl. Ser. 6, e284–e285.
Feng, J., Li, Y., Lin, L. S., Lin, L. S., Wang, L. M., Zhang, R., et al. (2020). Preliminary study on taxonomic diversity of fish species based on the fishing surveys in southwestern sea area of nansha islands. Periodical Ocean Univ. China 50, 76–84. (In Chinese).
Friedlander, A. M., Donovan, M. K., Stamoulis, K. A., Williams, I. D., Brown, E. K., Conklin, E. J., et al. (2018). Human-induced gradients of reef fish declines in the Hawaiian Archipelago viewed through the lens of traditional management boundaries. Aquat. Conserv. Mar. Freshw. Ecosyst. 28, 146–157. doi: 10.1002/aqc.2832
Guimarães-Costa, A. J., Machado, F. S., Oliveira, R. R., Silva-Costa, V., Andrade, M. C., Giarrizzo, T., et al. (2019). Fish diversity of the largest deltaic formation in the Americas-a description of the fish fauna of the Parnaíba Delta using DNA Barcoding. Sci. Rep. 9:7530.
Hebert, P. D., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Trans. R. Soc. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D., Stoeckle, M. Y., Zemlak, T. S., and Francis, C. M. (2004). Identification of birds through DNA barcodes. PLoS Biol. 2:e312. doi: 10.1371/journal.pbio.0020312
Hixon, M. A. (2011). 60 years of coral reef fish ecology: past, present, future. Bull. Mar. Sci. 87, 727–765. doi: 10.5343/bms.2010.1055
Hubert, N., Delrieu-Trottin, E., Irisson, J. O., Meyer, C., and Planes, S. (2010). Identifying coral reef fish larvae through DNA barcoding: a test case with the families Acanthuridae and Holocentridae. Mol. Phylogenet. Evol. 55, 1195–1203. doi: 10.1016/j.ympev.2010.02.023
Hubert, N., Hanner, R., Holm, E., Mandrak, N. E., Taylor, E., Burridge, M., et al. (2008). Identifying canadian freshwater fishes through DNA barcodes. PLoS One 3:e2490. doi: 10.1371/journal.pone.0002490
Hubert, N., Meyer, C. P., Bruggemann, H. J., Guerin, F., Komeno, R. J., Espiau, B., et al. (2012). Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the centre-of-overlap hypothesis. PLoS One 7:e28987. doi: 10.1371/journal.pone.0028987
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lakra, W. S., Verma, M. S., Goswami, M., Lal, K. K., Mohindra, V., Punia, P., et al. (2011). DNA barcoding Indian marine fishes. Mol. Ecol. Resour. 11, 60–71. doi: 10.1111/j.1755-0998.2010.02894.x
Lara, A., León, J. L. P. D., Rodriguez, R., Casane, D., Cote, G., Bernatchez, L., et al. (2010). DNA barcoding of Cuban freshwater fishes: evidence for cryptic species and taxonomic conflicts. Mol. Ecol. Resour. 10, 421–430. doi: 10.1111/j.1755-0998.2009.02785.x
Li, Y., Zhang, J., Zhang, R., Song, P. Q., Zhong, Z. H., Wang, Y. P., et al. (2016). Fish diversity in southwestern seas of NANSHA Islands and the mouth of Beibu Bay. Biodivers. Sci. 24, 166–174. (In Chinese). doi: 10.17520/biods.2015203
Limmon, G., Delrieu-Trottin, E., Patikawa, J., Rijoly, F., Dahruddin, H., Busson, F., et al. (2020). Assessing species diversity of coral triangle artisanal fisheries: A DNA barcode reference library for the shore fishes retailed at Ambon harbor (Indonesia). Ecol. Evol. 10, 3356–3366. doi: 10.1002/ece3.6128
Lin, Z., Yu, K., Shi, Q., Chen, T., and Tao, S. (2016). Sea surface temperature variations during the last 100 years recorded in a porites coral from the mischief reef of Sansha City. Trop. Geogr. 36, 27–33. (In Chinese).
Liu, W., Zhang, Y. X., Chen, J. M., Mai, R. L., Zhu, H., Fu, Y. F., et al. (2012). Preliminary analysis of fish resources in the Nansha islands waters survey using light-purse seiner in spring season. J. Shanghai Ocean Univ. 21, 105–109. (In Chinese).
Meier, R., Zhang, G., and Ali, F. (2008). The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst. Biol. 57, 809–813. doi: 10.1080/10635150802406343
Pegg, G. G., Sinclair, B., Briskey, L., and Aspden, W. J. (2006). MtDNA barcode identification of fish larvae in the southern great barrier reef–Australia. Sci. Mar. 70, 7–12. doi: 10.3989/scimar.2006.70s27
Radulovici, A. E., Archambault, P., and Dufresne, F. (2010). DNA barcodes for marine biodiversity: moving fast forward? Diversity 2, 450–472. doi: 10.3390/d2040450
Randall, J. E. (2005). Reef and Shore Fishes of the South Pacific. New Caledonia to Tahiti and the Pitcairn Islands. Honolulu: University of Hawai’i Press.
Shen, P. P., Tan, Y. H., Huang, L. M., Zhang, J. L., and Yin, J. Q. (2010). Occurrence of brackish water phytoplankton species at a closed coral reef in Nansha Islands, South China Sea. Mar. Pollut. Bull. 60, 1718–1725. doi: 10.1016/j.marpolbul.2010.06.028
Sorenson, L., Santini, F., Carnevale, G., and Alfaro, M. E. (2013). A multi-locus timetree of surgeonfishes (Acanthuridae, Percomorpha), with revised family taxonomy. Mol. Phylogenet. Evol. 68, 150–160. doi: 10.1016/j.ympev.2013.03.014
Sun, D. R., and Chen, Z. (2013). South Sea Fish Retrieval Volume One. Beijing: China Ocean press. (In Chinese).
Sun, R. X., Yang, X., Li, Q. X., Wu, Y. T., Shao, H. Y., Wu, M. H., et al. (2019). Polycyclic aromatic hydrocarbons in marine organisms from Mischief Reef in the South China sea: implications for sources and human exposure. Mar. Pollut. Bull. 149:110623. doi: 10.1016/j.marpolbul.2019.110623
Tan, F., Yang, H., Xu, X., Fang, Z., Xu, H., Shi, Q., et al. (2020). Microplastic pollution around remote uninhabited coral reefs of Nansha Islands, South China Sea. Sci. Total Environ. 725:138383. doi: 10.1016/j.scitotenv.2020.138383
Wang, L., Zhang, N., Ma, Z., Guo, H., Zhu, C., and Zhang, D. (2015). Length-weight relationship and morphological studies of the Polydactylus sextarius, Nemipterus japonicus and Pampus argenteus from the fiery cross reef, South China sea. Int. J. Fish. Aquacult. 5:7.
Ward, R. D. (2009). DNA barcode divergence among species and genera of birds and fishes. Mol. Ecol. Resour. 9, 1077–1085. doi: 10.1111/j.1755-0998.2009.02541.x
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., and Hebert, P. D. N. (2005). DNA barcodig Australia’s fish species. Philosoph. Trans. R. Soc. B Biol. Sci. 360, 1847–1857.
Weigt, L. A., Baldwin, C. C., Driskell, A., Smith, D. G., Ormos, A., and Reyier, E. A. (2012). Using DNA barcoding to assess Caribbean reef fish biodiversity: expanding taxonomic and geographic coverage. PLoS One 7:e41059. doi: 10.1371/journal.pone.0041059
Wilson, S. K., Fisher, R., Pratchett, M. S., Graham, N. A. J., Dulvy, N. K., Turner, R. A., et al. (2008). Exploitation and habitat degradation as agents of change within coral reef fish communities. Glob. Change Biol. 14, 2796–2809. doi: 10.1111/j.1365-2486.2008.01696.x
Xing, B., Chen, X., Zhang, Z., Sun, R., Xiang, P., Lin, M., et al. (2020). Genetic identification of ophichthid fishes through DNA barcoding. Mitochondrial DNA Part B 5, 1970–1974. doi: 10.1080/23802359.2020.1756940
Yin, J. Q., Chen, Q. C., Tan, Y. H., and Zhang, J. L. (2003). Zooplanktonic community characteristics in waters of Zhubi coral reef (Nansha Islands) in spring. J. Trop. Oceanogr. 22, 1–8. (In Chinese).
Keywords: Mischief reef, Fish diversity, DNA barcoding, COI gene, Nansha Islands
Citation: Shan B, Liu Y, Yang C, Zhao Y, Zhang G, Wu Q and Sun D (2021) DNA Barcoding of Fish in Mischief Reef—Fish Diversity of a Reef Fish Community From Nansha Islands. Front. Mar. Sci. 7:618954. doi: 10.3389/fmars.2020.618954
Received: 19 October 2020; Accepted: 07 December 2020;
Published: 14 January 2021.
Edited by:
Libin Zhang, Institute of Oceanology (CAS), ChinaCopyright © 2021 Shan, Liu, Yang, Zhao, Zhang, Wu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dianrong Sun, c3VuZGlhbnJvbmdAeWVhaC5uZXQ=
 Binbin Shan
Binbin Shan Yan Liu1,2,3
Yan Liu1,2,3