- 1Laboratory of Protozoological Biodiversity and Evolution in Wetland, College of Life Sciences, Shanxi Normal University, Xi’an, China
- 2Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 3Department of Zoology, Comenius University in Bratislava, Bratislava, Slovakia
- 4College of Marine Life Sciences, Ocean University of China, Qingdao, China
Morphogenesis of ciliated protists attracts a lot of attention, because their huge morphological diversity is related to formation of ciliary structures during cell division. In the present work, the morphology and morphogenesis as well as the phylogenetic position of a new, marine hypotrich ciliate, Nothoholosticha luporinii n. sp., were investigated. The new species is characterized by having a combination of the following features: a bicorona whose anterior row contains four frontal cirri and posterior row includes only two cirri, a single buccal cirrus, midventral complex composed of about 30 cirral pairs, one pretransverse cirrus, 3–6 transverse cirri, one left and one right marginal cirral row; three bipolar dorsal kineties; contractile vacuole located in about 2/3 of the body length, two types of cortical granules, and many macronuclear nodules scattered throughout the cytoplasm. The morphogenesis of N. luporinii follows the ontogenetic mode of Pseudokeronopsis, a well-known and closely related genus except that the macronucleus fuses into a single mass in the middle fission stage. Phylogenetic analyses based on the rDNA operon classify Nothoholosticha in the family Pseudokeronopsidae and support the distinctness of the new taxon as well as the monophyletic origin of the subfamily Nothoholostichinae.
Introduction
Ciliates (phylum Ciliophora Doflein, 1901), a highly diverse and ubiquitously distributed group of unicellular microbial eukaryotes, play substantial roles in various ecosystems. A lot of attention has been therefore paid to their diversity, function and evolution (e.g., Bharti et al., 2019; Hu et al., 2019; Jung and Berger, 2019; Kaur et al., 2019; Luo et al., 2019; Wang et al., 2019; Yan et al., 2019; Gong et al., 2020; Shao et al., 2020; Sheng et al., 2020; Wang Y. R. et al., 2020; Zhang et al., 2020). Hypotrichs (subclass Hypotrichia Stein, 1859) are not only among the most differentiated ciliate groups, but also among the most confused ones in terms of their systematics and phylogeny (for reviews, see Berger, 1999, 2006, 2008, 2011; Luo et al., 2017; Song and Shao, 2017; Luo et al., 2018; Lyu et al., 2018; Kim and Min, 2019; Chen et al., 2020; Dong et al., 2020; Paiva, 2020; Park et al., 2020; Wang J. et al., 2020; Xu et al., 2020). In the present study, we focus on the hypotrich family Pseudokeronopsidae, which was established by Borror and Wicklow (1983). Hitherto, this family includes the following genera: Antiokeronopsis Fan et al., 2014b, Apoholosticha Fan et al., 2014a, Heterokeronopsis Pan et al., 2013, Nothoholosticha Li et al., 2009, Pseudokeronopsis Borror and Wicklow, 1983 (type genus), Tetrakeronopsis Paiva et al., 2014, and Uroleptopsis Kahl, 1932 (Li et al., 2009, 2016; Pan et al., 2013; Fan et al., 2014a, b; Paiva et al., 2014; Hu et al., 2015). Rather recently, the family Pseudokeronopsidae was divided into two subfamilies by Paiva et al. (2014): Nothoholostichinae Paiva et al., 2014, with an atypical bicorona whose anterior portion is formed by four frontal cirri, and Pseudokeronopsinae Borror and Wicklow, 1983, with a typical bicorona whose anterior portion is formed by more than four frontal cirri. As a result, Apoholosticha, Heterokeronopsis, Nothoholosticha, and Tetrakeronopsis were classified within the Nothoholostichinae, and only the three remaining genera, Antiokeronopsis, Pseudokeronopsis, and Uroleptopsis, were assigned to the Pseudokeronopsinae. The monophylies of both subfamilies are supported not only by the cirral pattern of the bicorona, but also by molecular analyses (Fan et al., 2014a; Huang et al., 2014; Paiva et al., 2014; Hu et al., 2015; Li et al., 2016).
So far, only two species have been assigned to the genus Nothoholosticha, namely, N. fasciola (Kahl, 1932) Li et al., 2009 (type species) and N. flava Li et al., 2016. In this study, a new species, Nothoholosticha luporinii n. sp., has been discovered in the intertidal sediment of Chizhou Island near the city of Shenzhen in the southern China. Its morphology, ontogenesis, and complete ribosomal operon (SSU rDNA, the ITS1-5.8S-ITS2 region, and LSU rDNA) have been studied to further extended our knowledge about the diversity and phylogeny of pseudokeronopsids.
Materials and Methods
Sampling and Cultivation
Samples including sea water and sediment were collected from the intertidal zone in the Daya Bay, Chizhou Island, near the city of Shenzhen, southern China (22°38′11″N, 114°38′32″E) on 1st April 2018, when the water temperature was 26°C and salinity was 32‰. The original sample was divided into aliquots that were used to establish raw cultures in Petri dishes. Single specimens of Nothoholosticha luporinii n. sp. were isolated from the raw cultures and used to set up clonal cultures in filtered in situ sea water at room temperature (25°C). Some rice grains were added to stimulate the growth of bacteria, which served as prey organisms for ciliates.
Taxonomic Methods and Terminology
Nothoholosticha luporinii n. sp. was investigated using a combination of detailed in vivo observation and protargol impregnation. Living cells were observed under a microscope Olympus BX 53 (Olympus Corporation, Tokyo, Japan) using bright field illumination and differential interference contrast optics at a magnification of 100–1,000×. Protargol impregnation followed the Wilbert’s method and served to reveal the nuclear apparatus and ciliary pattern of the new species (Wilbert, 1975). Also, the morphogenetic processes were rebuilt from the protargol-impregnated preparations. Stained cells were investigated mostly at high magnification (1,000×).
In vivo measurements were made from microphotographs of freely swimming specimens, while measurements on protargol-impregnated specimens were conducted using an ocular micrometer. Illustrations of living cells were based on free-hand sketches and photographs, while those of impregnated specimens were made at 1,000× magnification with the help of a drawing device. All illustrations were finally processed in Adobe PhotoShop CS5. To distinguish parental and daughter structures during the morphogenetic processes, new (daughter) structures are painted solid, while old ciliary structures are depicted by contour. General terminology and systematics mostly follow Berger (2006) and Lynn (2008).
DNA Extraction, PCR Amplification, and Sequencing
Single cells were picked, carefully washed five times in filtered in situ marine water, and lysed in 45 μl of Cell Lysis Buffer (DNeasy Blood and Tissue Kit, Qiagen, Hilden, Germany). To confirm the sequencing results, altogether three different types of samples were prepared: the first contained just one cell, the second comprised two cells, and finally the third included three cells. Genomic DNA was extracted with the DNeasy Blood and Tissue Kit, but only 1/4 of the suggested volume for all reagents was used as suggested by Lu et al. (2020). Amplification of SSU rDNA was achieved with the primers 82-F (5′-GAA ACT GCG AAT GGC TC-3′) (Jerome et al., 1996) and 18S-R (5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′) (Medlin et al., 1988). Fragments containing ITS-5.8S rDNA and LSU rDNA were amplified with the primers ITS-F (5′-GTA GGT GAA CCT GCG GAA GGA TCA TTA-3′) (Miao et al., 2008) and 28S-R2 (5′-AAC CTT GGA GAC CTG AT-3′) (Moreira et al., 2007), using the same thermo cycler program as described by Huang et al. (2014). PCR products were purified using the EasyPure® Quick Gel Extraction Kit (TransGen Biotech Co., Ltd., Beijing, China) and subsequently cloned using the pEASY® -Blunt Cloning Kit (TransGen Biotech Co., Ltd., Beijing, China). Recombinant plasmids were sequenced in both directions on an ABI-PRISM 3730 automatic sequencer (Applied Biosystems, Tsingke Biological Technology Company, Qingdao, China) with the PCR primers. To obtain high quality sequences, two internal sequencing primers were used for SSU rDNA: 900F (5′-CGA TCA GAT ACC GTC CTA GT-3′) and 900R (5′-ACT AGG ACG GTA TCT GAT CG-3′), and also two internal primers for LSU rDNA: F2 (5′-GGA GTG TGT AAC AAC TCA CCT GC-3′) and R3 (5′-CAT TCG GCA GGT GAG TTG TTA CAC-3′) (Zhao et al., 2014). Subsequently, the newly obtained sequences were carefully inspected, trimmed, and assembled into contigs using SeqMan Pro ver. 7.1.0 (Anson and Myers, 1997). Sequences obtained from all samples were identical and therefore only those derived from the single-cell sample were included into the subsequent phylogenetic analyses.
Molecular Phylogeny
The newly obtained sequences were blasted against the nucleotide NCBI database1. The BLASTn algorithm revealed that the new species belongs to the core urostylids (subclass Hypotrichia). Sequences of all related urostylids, except for those without associated publication information, were included into the phylogenetic analyses. The taxon sampling in the single-gene dataset (SSU rDNA) and in the concatenated, multi-gene dataset (SSU rDNA + ITS1-5.8S-ITS2 + LSU rDNA) mostly followed Huang et al. (2014) and Zhao et al. (2014). SeaView ver. 4 was used to prepare the concatenated dataset (Galtier et al., 1996; Gouy et al., 2010). Oxytricha granulifera (accession no. AF508762), Stylonychia lemnae (accession no. AF508773), Stylonychia mytilus (accession no. AF508774), Sterkiella nova (accession no. AF508771), and Sterkiella histriomuscorum (accession no. FJ545743) were used as outgroup taxa. GenBank accession numbers are provided in Supplementary Tables S1, S2. Sequences were aligned online using the MAFFT algorithm on the GUIDANCE2 server2 with the following parameters: the 6mer pairwise method, the maximum number of 100 iterations, and 100 bootstrap repeats (Landan and Graur, 2008; Sela et al., 2015). The 5′ and 3′ ends of the resulting alignments were trimmed manually in the program BioEdit ver. 7.0 (Hall, 1999). The number of unmatched nucleotides and the pairwise SSU rDNA sequence identities within the subfamily Nothoholostichinae were calculated in the program BioEdit, using the “sequence difference count matrix” and “sequence identity matrix” options, respectively. The single-gene alignment contained 1,530 nucleotide positions, while the multi-gene alignment comprised 3,078 positions.
Maximum likelihood (ML) analyses were performed in RAxML-HPC2 ver. 8.2.10 on XSEDE (Stamatakis, 2014) on the CIPRES Science Gateway,3 using the GTR + gamma evolutionary model and 1,000 bootstrap replicates. Bayesian inference (BI) was carried out in MrBayes ver. 3.2.6 on XSEDE (Ronquist et al., 2012) with the GTR + I + G evolutionary model selected by MrModeltest ver. 2.2 via the Akaike information criterion (Nylander, 2004). Markov chain Monte Carlo simulations were run for six million generations with a sampling frequency of 100 and a burn-in of 6,000 trees (10%). Remaining trees were used to calculate the 50%-majority rule consensus trees and their posterior probabilities. ML and BI trees were computed as unrooted and were rooted using the outgroup taxa in FigTree ver. 1.2.34.
Results
Systematics
Subclass Hypotrichia Stein, 1859
Family Pseudokeronopsidae Borror and Wicklow, 1983
Subfamily Nothoholostichinae Paiva et al., 2014
Genus Nothoholosticha Li et al., 2009
Nothoholosticha luporinii n. sp.
Zoobank registration number of work.
urn:lsid:zoobank.org:pub:8E7FD944-5497-4AF0-9FC4-0B8280B 07F41
Zoobank registration number of new species.
urn:lsid:zoobank.org:act:AB581D7E-CB84-42C0-8528-1836369 B3185
Diagnosis
Size in vivo 130–280 × 25–60 μm. Many macronuclear nodules. One contractile vacuole located near left body margin in about 2/3 of body length. Two types of cortical granules: big ones colourless, 1.5–2.0 μm in length, irregularly ellipsoid or slightly blood-cell shaped, and densely distributed throughout cortex; small ones bright brown-reddish in color, spherical, clustered in groups around dorsal bristles or sparsely arranged along cirral rows. Six frontal cirri arranged in two rows (four cirri in anterior row and two cirri in posterior row), 3–6 frontoterminal cirri, one buccal cirrus, one pretransverse cirrus, 3–6 transverse cirri, midventral complex composed of 17–43 cirral pairs, 38–84 left and 43–91 right marginal cirri. Three dorsal kineties. Adoral zone bipartite, composed of 7–13 crown and 23–39 lapel membranelles.
Type Locality
Sediment from the intertidal zone of Daya Bay, Chizhou Island, Shenzhen, southern China (22°38′11″N, 114°38′32″E).
Type Material
The protargol slide (no. ZTY2018040101_1) with the holotype specimen (Figures 1K,L, 2I) marked with an ink circle, and eight paratype slides (no. ZTY2018040101_2–9), are deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China.
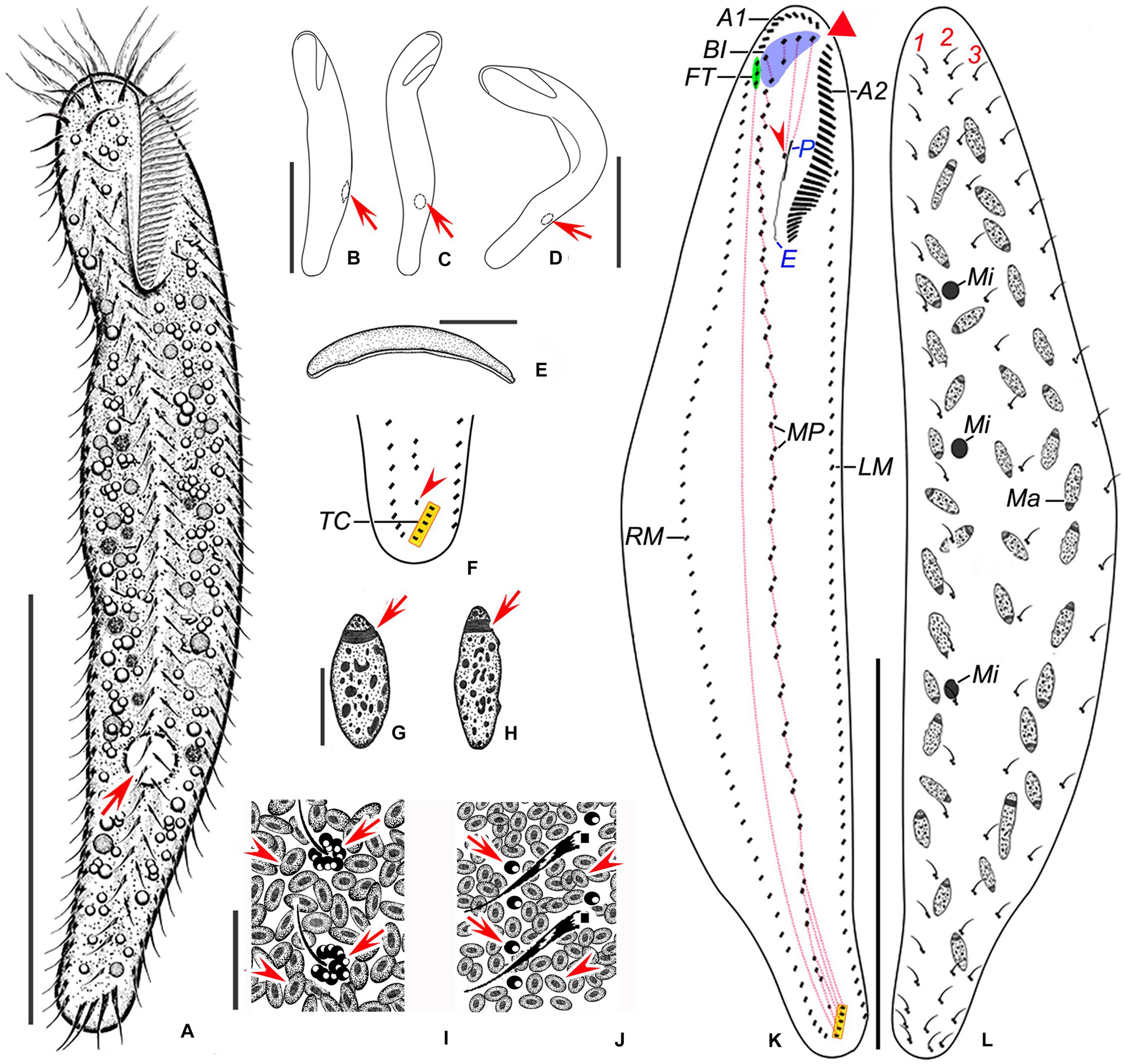
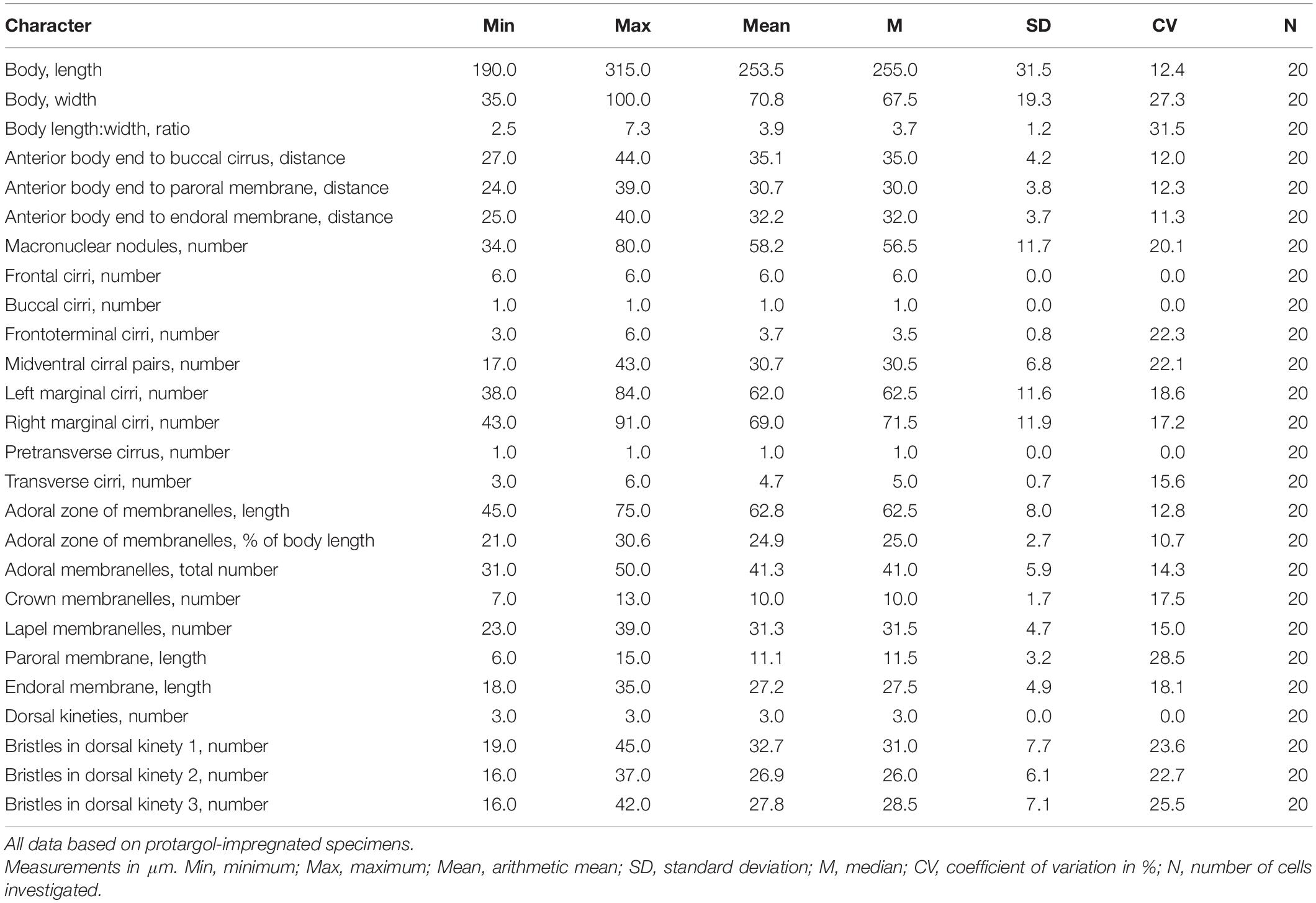
Figure 1. Nothoholosticha luporinii n. sp. from life (A–E,I,J) and after protargol impregnation (F–H,K,L). (A) Ventral view of a representative individual, arrow marks the contractile vacuole. (B–D) Overviews, showing the body flexibility, arrows mark contractile vacuoles. (E) Lateral overview. (F) Detail of the posterior body region, showing the single pretransverse cirrus (arrowhead) and transverse cirri (yellow rectangle). (G,H) Macronuclear nodules, arrows indicate replication bands. (I,J) Dorsal (I) and ventral (J) surface views, showing the arrangement of cortical granules, arrowheads denote the larger cortical granules, arrows point the smaller ones. (K,L) Ventral (K) and dorsal (L) views of the holotype specimen, showing the ciliary pattern and the nuclear apparatus. Red triangle in (K) marks the conspicuous gap between two parts of the adoral zone of membranelles, arrowhead marks the buccal cirrus, yellow rectangle delimits transverse cirri. Frontal-ventral-transverse cirri originating from the same anlagen are connected by dotted lines. A1 and A2, crown and lapel adoral membranelles; BI, bicorona; E, endoral membrane; FT, frontoterminal cirri; LM, left marginal cirral row; Ma, macronuclear nodules; Mi, micronuclei; MP, midventral cirral pairs; P, paroral membrane; RM, right marginal cirral row; TC, transverse cirri; 1–3, dorsal kineties. Scale bars = 5 μm (G–J) and 100 μm (A–E,K,L).
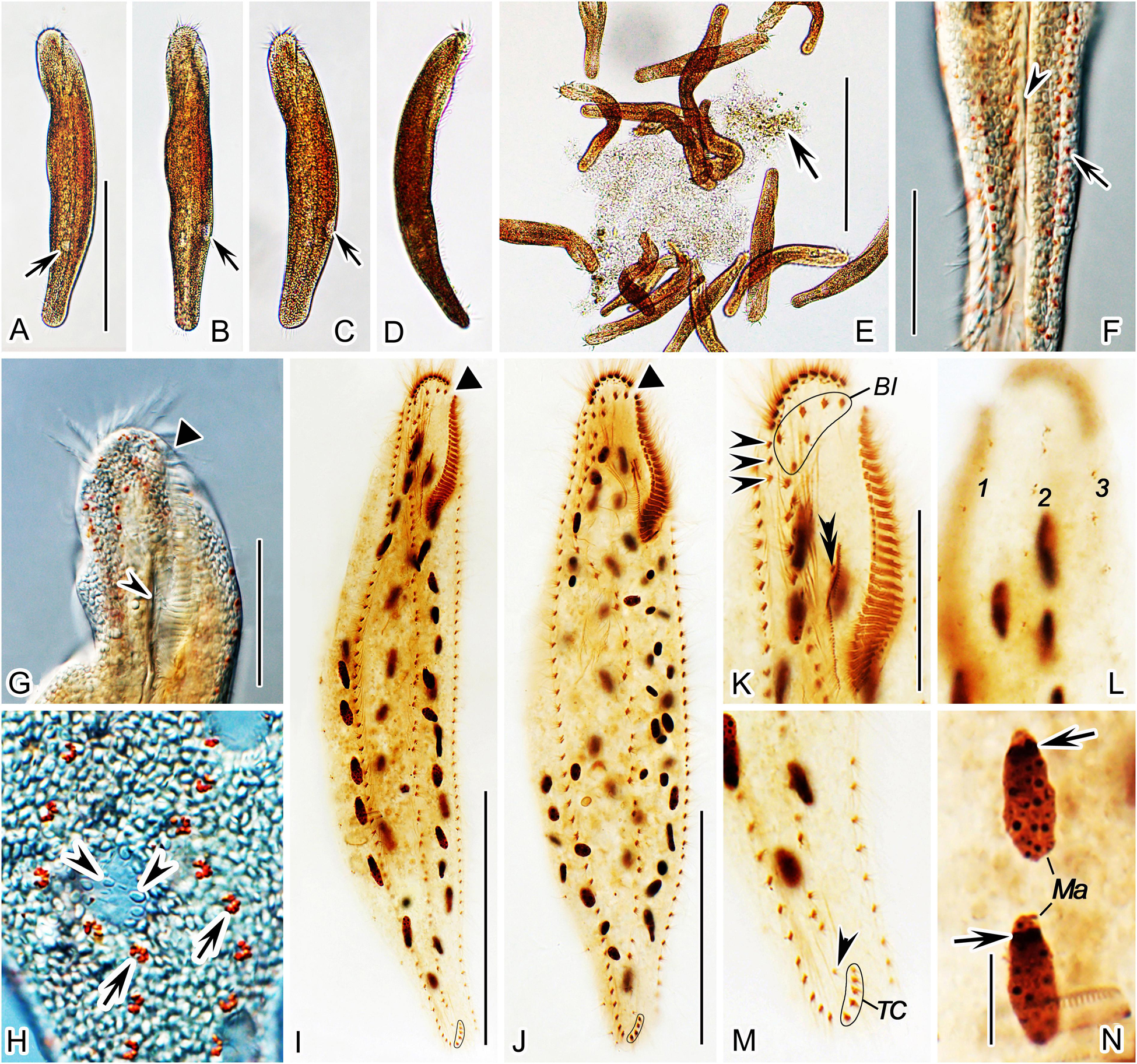
Figure 2. Nothoholosticha luporinii n. sp. from life (A–H) and after protargol impregnation (I–N). (A–D) Ventral (A–C) and lateral (D) overviews of different individuals, showing the body shape and the localization of the contractile vacuole (arrows in A–C). (E) Free-foraging individuals, arrow indicates a mass of microalgae. (F) Ventral view of the posterior body portion, showing the red cortical granules (arrow) and the longitudinal groove along the midventral cirral complex (arrowhead). (G) Ventral view of the anterior body portion, showing the gap between the two parts of the adoral zone (triangle) and the buccal cavity (arrowhead). (H) Dorsal surface view, showing the arrangement of colourless (arrowheads) and bright red (arrows) cortical granules. (I,J) Ventral view of the holotype (I) and the paratype (J) specimen, showing the ciliary pattern and the nuclear apparatus. Triangles mark the gap between the two parts of the adoral zone of membranelles. (K,L) Ventral (K) and dorsal (L) views of the anterior body portion of the same specimen. Arrowheads in (K) indicate the frontoterminal cirri, double-arrowhead shows the buccal cirrus. (M) Ventral view of the posterior body portion, arrowhead denotes the single pretransverse cirrus. (N) Details of the macronuclear nodules, arrows mark replication bands. BI, bicorona; Ma, macronuclear nodules; TC, transverse cirri; 1–3, dorsal kineties. Scale bars = 5 μm (N), 35 μm (G,K–M), 100 μm (A–D,I,J), and 200 μm (E).
Gene Sequences
The nuclear SSU rDNA, ITS1-5.8S-ITS2 and LSU rDNA sequences have been deposited in GenBank under the following accession nos. MW035040, MW035039, and MW035042.
Dedication
We dedicate this species to Prof. Dr. Pierangelo Luporini (University of Camerino, Italy) in recognition of his great contributions to ciliatology.
Morphological Description of Nothoholosticha luporinii n. sp.
Size of specimens from fresh raw cultures about 130–280 × 25–60 μm, usually 200 × 35 μm in vivo. Body elongate ellipsoidal with conspicuous longitudinal groove along midventral cirral complex (Figure 2F, arrowhead), anterior end broadly rounded and wider than posterior one; dorsoventrally flattened from about 2:1 to 3:2; rather flexible but not contractile (Figures 1A–E, 2A–E). About 34–80 ellipsoidal macronuclear nodules scattered throughout cytoplasm, individual nodules approximately 3–5 × 6–11 μm in size after protargol impregnation; 1–3 globular micronuclei but exact number difficult to determine because hardly distinguishable from similar-sized and impregnated cytoplasmic inclusions (Figures 1G,H,L, 2I,J,N). Contractile vacuole approximately 15 μm across during diastole, located in posterior two thirds of body length near left body margin (Figures 1A–D, 2A–C, arrows). Cytoplasm colourless, transparent at high magnifications, packed with macronuclear nodules, lipid droplets, and food vacuoles. Cortex flexible, contains two types of granules: type I bigger, i.e., 1.5–2.0 μm in diameter, irregularly ellipsoidal or slightly blood cell-shaped, colourless, narrowly arranged underneath cortex, possibly mitochondria (Figures 1I,J, 2F–H, arrowheads); type II smaller, i.e., about 0.5 μm in diameter, bright brown-reddish, clustered in a flower-like pattern around dorsal bristles or sparsely arranged along cirral rows (Figures 1I,J, 2F,H, arrows), provides cells with a reddish-brown appearance under low magnifications (40×, 100× and 200×; Figures 2A–E) and with a yellowish appearance under moderate magnification (400×) (Figures 2F,G). Crawls moderately slowly on debris particles, sometimes swims by rotation about main body axis.
Cirri about 10–15 μm long in vivo; number of frontal, buccal, and pretransverse cirri invariable, while number of frontoterminal cirri, midventral cirral pairs, transverse, and marginal cirri rather highly variable (CV = 15.6–22.3%) (Table 1). Frontal cirri approximately 13 μm long in vivo, arranged in an atypical bicorona, invariably four cirri in anterior coronal row and constantly two cirri in posterior coronal row (Figure 1K, blue area, Figure 2K). Buccal cirrus about 10 μm long in vivo, situated right of mid-portion of paroral membrane (Figure 1K, red arrowhead, Figure 2K, black double arrowhead). Three to six frontoterminal cirri, about 10 μm long in vivo, located posterior to distal end of crown adoral membranelles (Figure 1K, green area, Figure 2K, black arrowheads). Midventral complex consists of 17–43 cirral pairs arranged in a zigzag pattern, left cirrus of midventral pairs slightly longer than right one, i.e., about 10 vs. 9 μm (Figures 1K, 2I,J); posterior most cirrus of midventral complex, labeled as a pretransverse cirrus, distinctly shifted toward transverse cirri and hence more or less separated from midventral complex (Figures 1F, 2M, arrowhead). Three to six transverse cirri, about 13–15 μm long in vivo, arranged in an oblique row (Figures 1F,K, yellow rectangle, Figures 2I,J,M). One left and one right marginal cirral row, composed of 38–84 and 43–91 cirri, respectively, individual cirri about 11 μm long in vivo (Figures 1K, 2I,J).
Dorsal bristles about 4 μm long in vivo, arranged in three bipolar rows. All three dorsal kineties begin subapically and extend to posterior body end (Figures 1L, 2L). Dorsal kinety 1 composed of 19–45 dikinetids, kinety 2 of 16–37, and kinety 3 of 16–42 (Table 1). Caudal cirri absent.
Adoral zone of membranelles occupies 20–30% of body length; bipartite, i.e., divided into crown and lapel region separated by a conspicuous gap (Figures 1A,K, 2I–K and Table 1). Crown region composed of 7–13 membranelles arranged in an arch-shaped pattern along anterior cell pole, length of membranellar cilia 18–22 μm. Lapel region composed of 23–39 membranelles forming a Gonostomum-like pattern, i.e., extends along left body margin to one fifth or third of body length, where it bends rather abruptly rightwards to run almost in parallel with undulating membranes, length of membranellar cilia up to 17 μm. Undulating membranes arranged in a Pseudokeronopsis-like pattern, i.e., paroral and endoral almost straight, extend in parallel and only partially overlap. Endoral membrane commences posterior to buccal cirrus and runs to buccal vertex, 18–35 μm long after protargol impregnation. Paroral membrane begins anterior to endoral, remarkably shorter than endoral, i.e., only 6–15 μm long in protargol preparations (Figures 1K, 2I–K).
Morphogenesis of Nothoholosticha luporinii n. sp.
Oral Primordium and Cirral Streaks
Morphogenesis commences with de novo formation of small groups of basal bodies adjacent to left cirri of the midventral complex about in the mid-body (Figure 6A). Groups of proliferating basal bodies join to form a longitudinal field, i.e., the oral primordium of the opisthe. Simultaneously, the proter’s oral primordium develops as a single anarchic field of closely spaced basal bodies in the region of the buccal vertex (Figures 3A, 6B,C). New adoral membranelles differentiate within the oral primordium of both the proter and the opisthe in a posteriad direction (Figures 3B,C, 6D–F). The undulating membranes (UM) anlage (streak I) forms to the right of and possibly from the oral primordium both in the proter and the opisthe (Figures 3C, 4A, 6F, arrowheads). The anterior portion of the UM anlage splits a single cirrus that migrates anteriorly to become the leftmost frontal cirrus in the anterior row of the bicorona (Figures 4C, 6J,K, arrowheads). Then, the UM anlage divides longitudinally to give rise to the paroral membrane and the endoral membrane (Figures 4C, 6J–N, arrowheads). Meanwhile, multiple frontal-midventral-transverse (FVT) cirral anlagen develop as series of oblique streaks to the right of the oral primordia (Figures 3C, 4A,C, 6F,J,K, arrows). Streak II (FVT anlage I) generates the second frontal cirrus of the anterior row of the bicorona as well as the buccal cirrus, which migrates toward the newly formed paroral membrane in mid-dividers; streak III (FVT anlage II) splits the third frontal cirrus of the anterior row of the bicorona as well as the left frontal cirrus of the posterior row of the bicorona; streak IV (FVT anlage III) produces the rightmost frontal cirrus of the anterior row of the bicorona as well as the right frontal cirrus of the posterior row of the bicorona; the rearmost (rightmost) streak develops the rightmost transverse cirrus and the 3–6 frontoterminal cirri, which migrate anteriorly to their species-specific position during the late division stages; streak n-1 (n represents the last FVT cirral anlage) provides the rearmost midventral cirral pair, a single pretransverse cirrus and one transverse cirrus. Streaks n-2 to n-5 (deduced from morphometric data) contribute one midventral cirral pair and a single transverse cirrus each (Figures 4C, 5A,B, 6L,N–Q). The remaining streaks (FVT anlagen) provide one midventral cirral pair each (Figures 5A,B). When the formation of the new oral apparatus is almost completed in each daughter cell, new cirri migrate to their final positions.
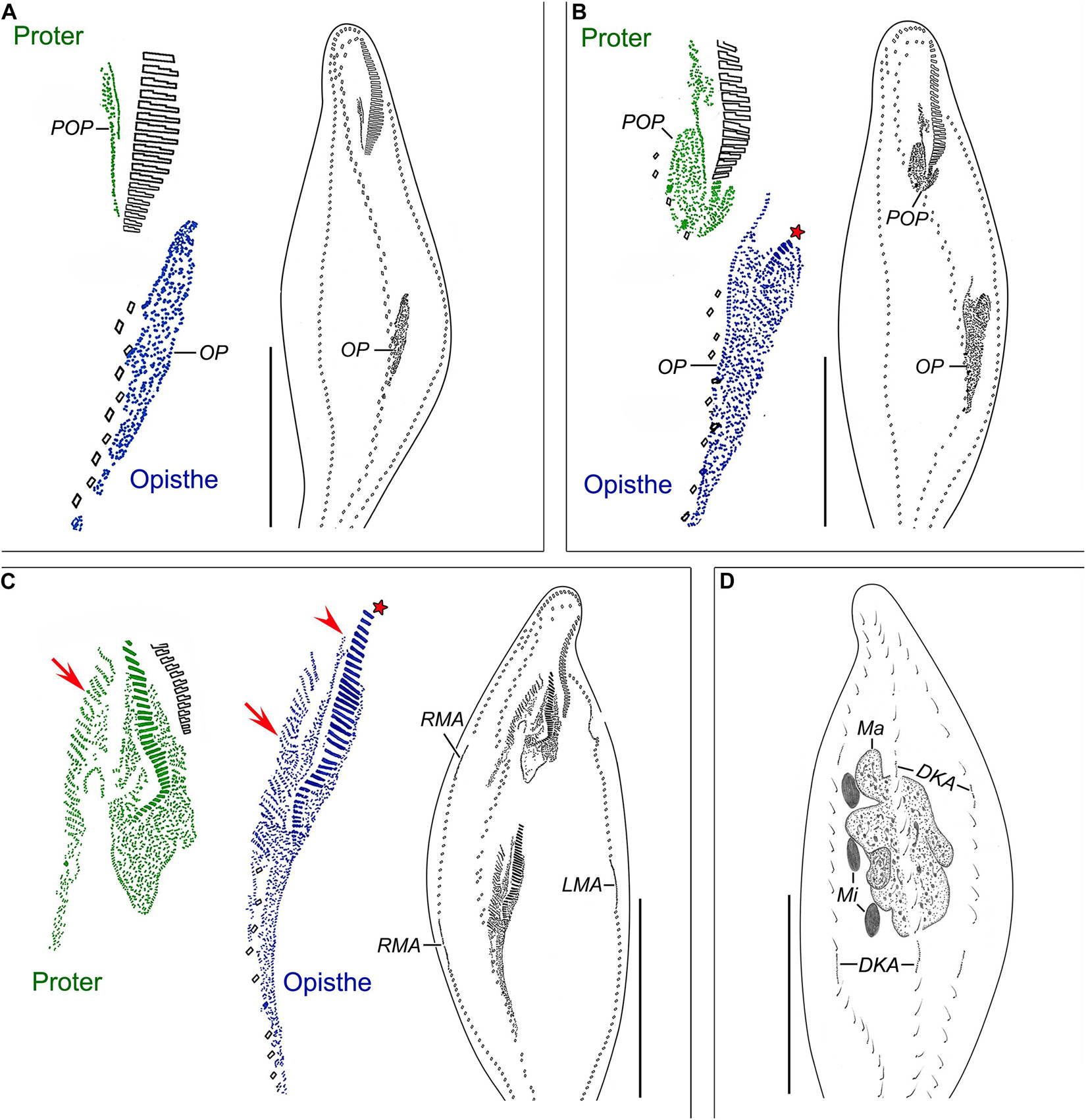
Figure 3. Nothoholosticha luporinii n. sp., early dividers (A,B) and a mid-divider (C,D) after protargol impregnation. Magnified details of the proter’s ciliature are in green colour, while those of the opisthe’s ciliature are in blue colour. (A) Ventral view of an early divider, showing the formation of oral primordia in the proter and the opisthe. (B) Ventral view of another early divider, whose opisthe’s oral primordium begins to differentiate into adoral membranelles (red asterisk). (C,D) Ventral (C) and dorsal (D) views of the same early mid-divider. Arrows indicate formation of frontal-ventral–transverse cirral anlagen, arrowhead marks the undulating membranes anlage, asterisk denotes the newly formed adoral membranelles from the opisthe’s oral primordium. DKA, dorsal kineties anlagen; LMA, left marginal cirral row anlagen; Ma, macronuclear mass; Mi, micronuclei; OP, opisthe’s oral primordium; POP, proter’s oral primordium; RMA, right marginal cirral row anlagen. Scale bars = 120 μm.
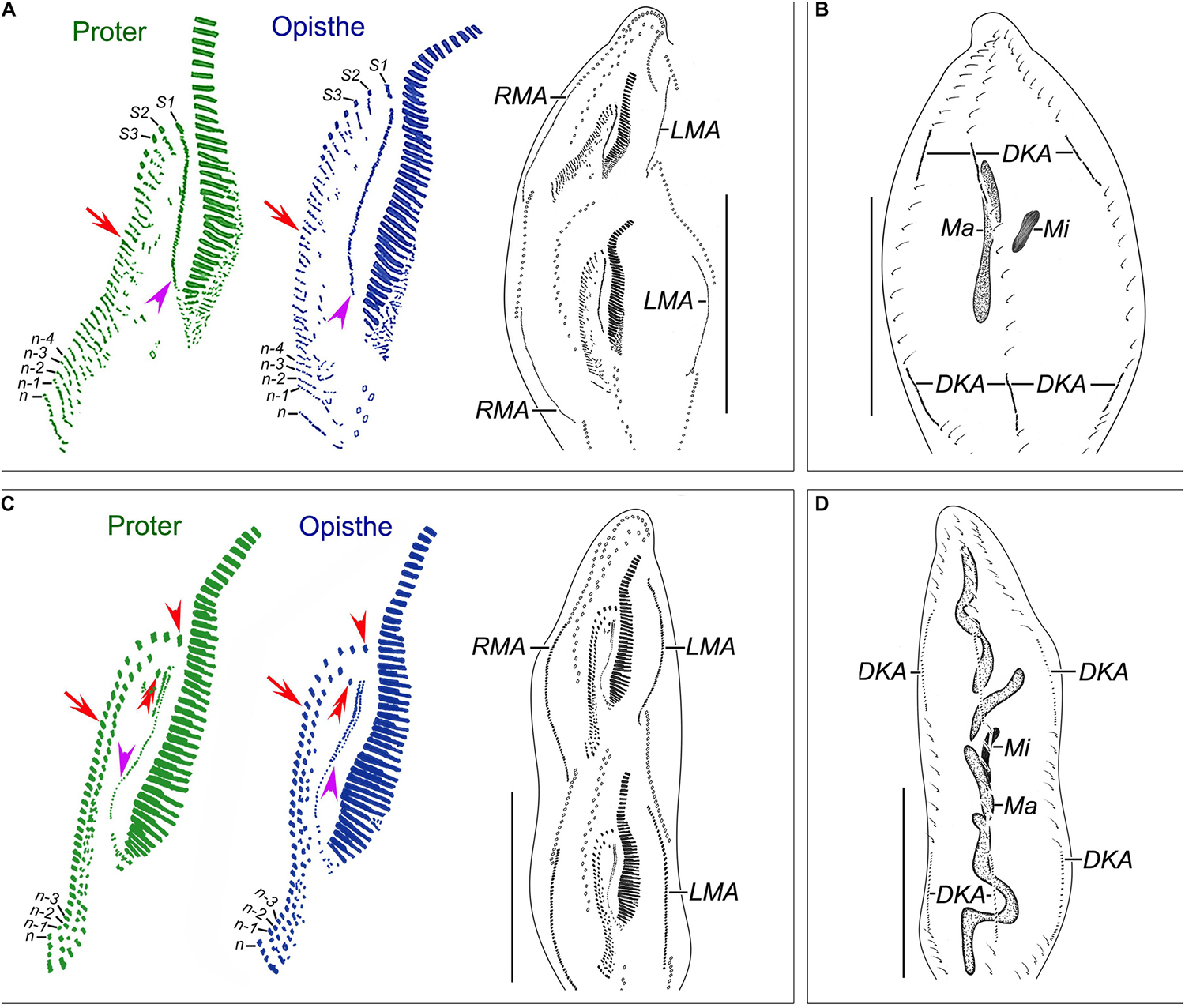
Figure 4. Nothoholosticha luporinii n. sp., a late mid-divider (A,B) and a late divider (C,D) after protargol impregnation. Magnified details of the proter’s ciliature are in green colour, while those of the opisthe’s ciliature are in blue colour. (A,B) Ventral (A) and dorsal (B) views of a late mid-divider, showing the undulating membrane anlagen (purple arrowheads), frontal-midventral-transverse cirral anlagen (red arrows), the anlagen for marginal cirral rows and dorsal kineties, and the dividing macronuclear mass. (C,D) Ventral (C) and dorsal (D) views of a late divider, showing the buccal cirri (red double-arrowheads), frontal cirri (red arrowheads), undulating membranes (purple arrowheads), frontal-midventral-transverse cirral anlagen differentiating into cirri (red arrows), and dividing macronucleus. DKA, dorsal kinety anlagen; LMA, left marginal cirral row anlagen; Ma, macronucleus; Mi, micronuclei; n, last frontal-midventral-transverse cirral anlage; n–1, n–2, n–3, n–4, second, third, fourth, and fifth rearmost frontal-midventral-transverse cirral anlagen; RMA, right marginal cirral row anlagen; S1–3, first, second, and third streaks. Scale bars = 140 μm.
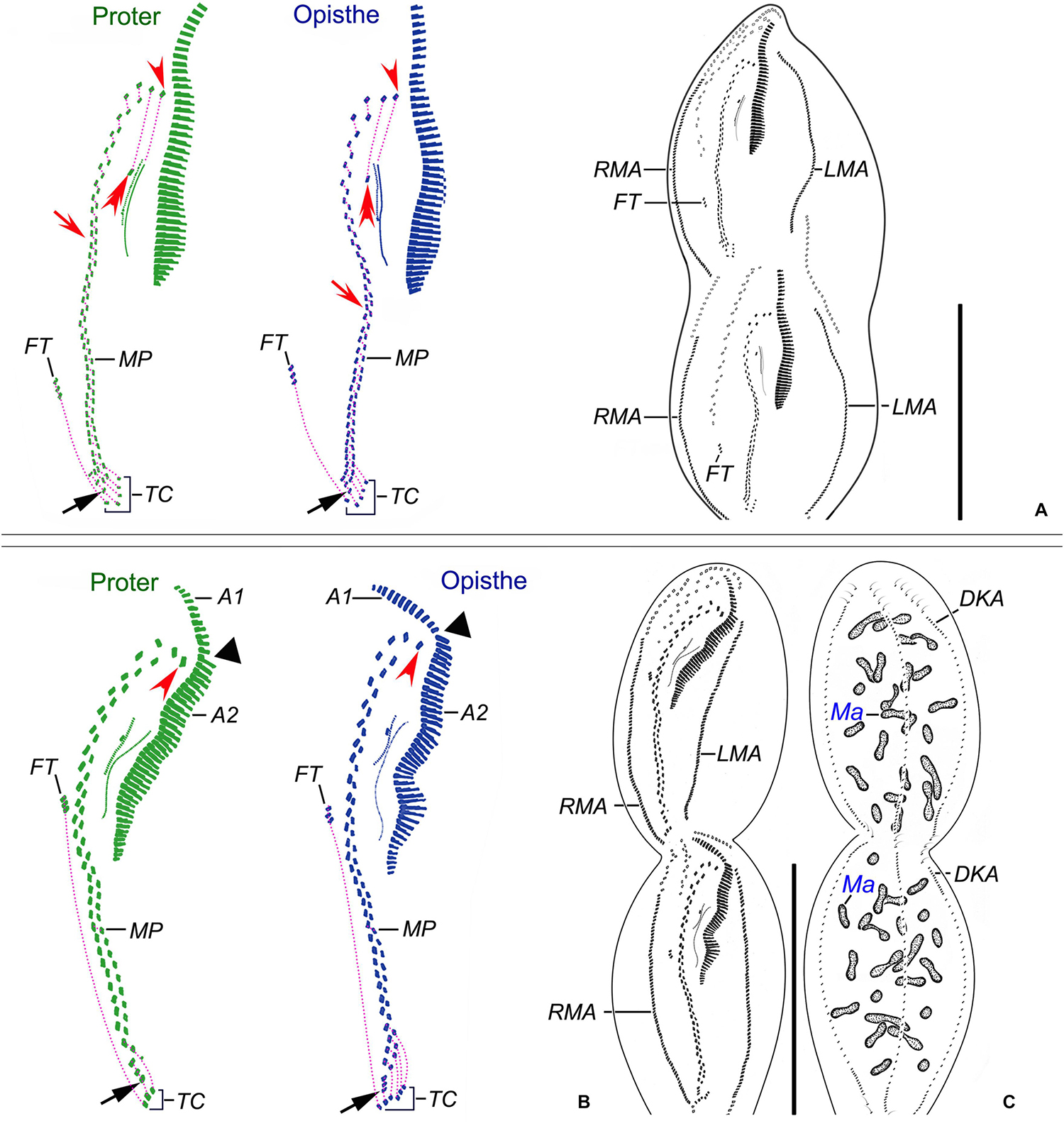
Figure 5. Nothoholosticha luporinii n. sp., a late (A) and a very late (B,C) divider after protargol impregnation. Magnified details of the proter’s ciliature are in green colour, while those of the opisthe’s ciliature are in blue colour. Frontal-ventral-transverse cirri originating from the same anlagen are connected by dotted lines. (A) Ventral view of a late divider, showing the buccal cirri (red double-arrowheads), the leftmost frontal cirri (red arrowheads), the single pretransverse cirri (black arrows), and the migrating frontoterminal cirri. (B,C) Ventral (B) and dorsal (C) views of a very late divider, showing the formation of a gap in the adoral zone of membranelles (black triangles) as well as the formation of the anlagen for marginal cirral rows and dorsal kineties. The leftmost frontal cirri are marked by red arrowheads, the pretransverse cirri by black arrows. A1 and A2, crown and lapel adoral membranelles; DKA, dorsal kinety anlagen; FT, frontoterminal cirri; LMA, left marginal cirral row anlagen; Ma, macronuclear nodules; MP, midventral cirral pairs; RMA, right marginal cirral row anlagen; TC, transverse cirri. Scale bars = 140 μm.
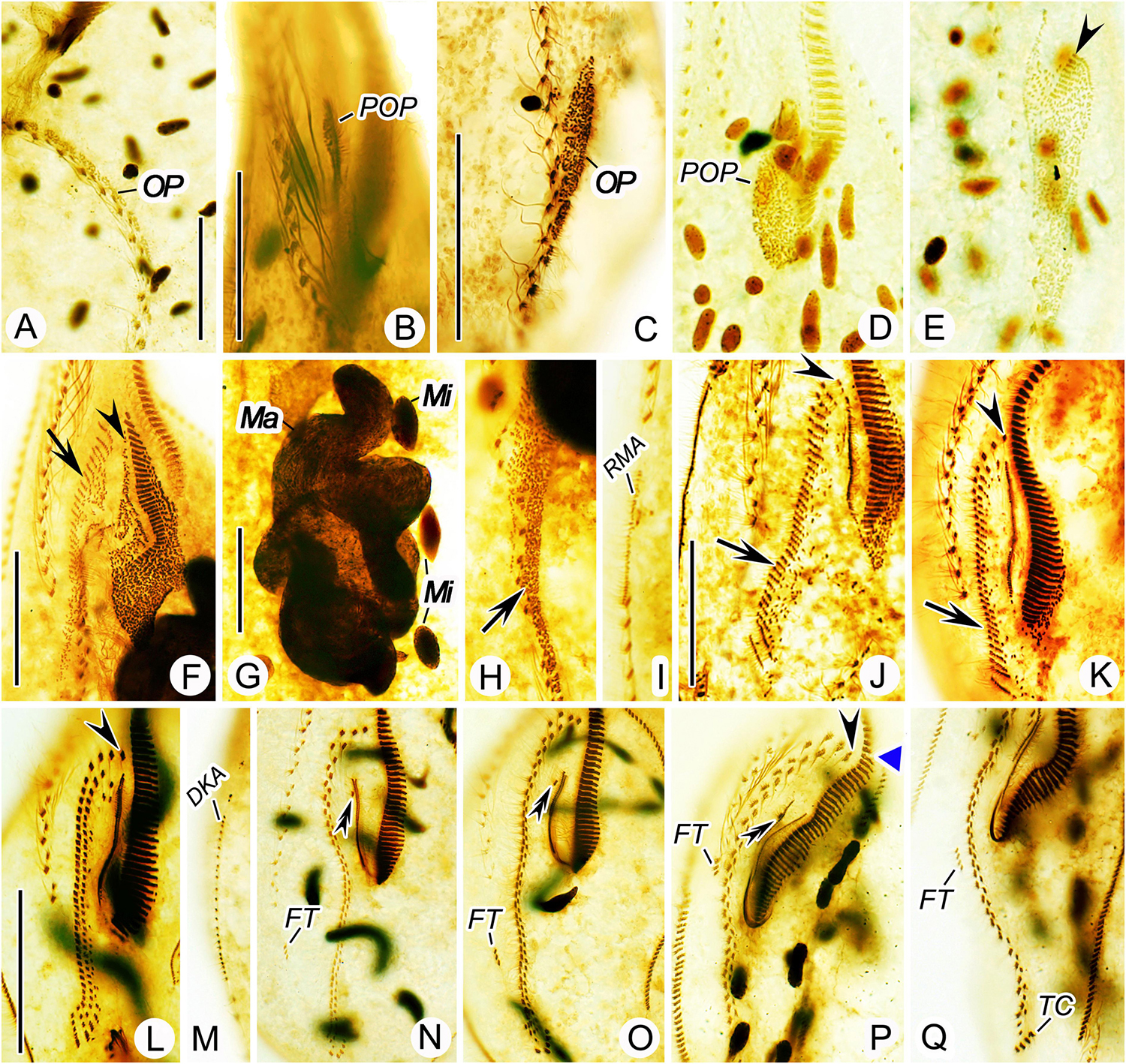
Figure 6. Nothoholosticha luporinii n. sp., various division stages after protargol impregnation. (A) Ventral view of an early divider, showing the opisthe’s oral primordium. (B,C) Ventral view of an early divider, showing the oral primordium in the proter (B) and the opisthe (C). (D,E) Ventral view of another early divider, arrow marks the proter’s oral primordium (D) and arrowhead denotes the differentiating adoral membranelles from the opisthe’s oral primordium (E). (F–I) Ventral view of an early mid-divider, showing the undulating membranes anlage (arrowheads) and the frontal-midventral-transverse cirral anlagen (arrows) in (F,H), the fusing macronuclear nodules and dividing fusiform micronuclei in (G), the right marginal cirral row anlage in (I). (J,K) Ventral view of a mid-divider, showing the leftmost frontal cirrus originating from the undulating membranes anlage (arrowheads) and the frontal-midventral-transverse cirral anlagen (arrows) in the proter (J) and the opisthe (K). (L–Q) Details of different late dividers, showing the leftmost frontal cirrus (arrowheads) in (L,P), the buccal cirrus (double-arrowheads) in (N–P), the forming gap in the adoral zone of membranelles (blue triangle) in (P), the migrating frontoterminal cirri in (N–Q), the transverse cirri in (Q), and the dorsal kinety anlage in (M). DKA, dorsal kinety anlage; FT, frontoterminal cirri; Ma, macronuclear nodules; Mi, micronuclei; OP, opisthe’s oral primordium; POP, proter’s oral primordium; RMA, right marginal cirral row anlage; TC, transverse cirri. Scale bars = 30 μm (F–K), 40 μm (A–E), and 60 μm (L–Q).
Marginal and Dorsal Anlagen
The proter’s left and right marginal cirral row anlagen form within the parental marginal cirral rows, very likely by dissociation of some cirri, about at level of the growing proter’s oral primordium. Similarly, the opisthe’s left and right marginal cirral row anlagen develop within the parental marginal cirral rows about at level of the opisthe’s oral primordium (Figures 3C, 6I). The marginal row anlagen extend posteriorly, gradually producing new cirri already in early mid-dividers and stretch toward both ends of the dividing cell to form new ones for each daughter cell (Figures 4A,C, 5A,B). The morphogenesis of the dorsal side ciliature begins in mid-dividers (Figure 3D). Specifically, within-row primordia appear in the parental dorsal kineties at two sites, viz., anterior and posterior to the prospective fission area (Figure 4B). The new dorsal kineties elongate and obtain their characteristic positions in late dividers (Figures 4D, 5C, 6M).
Nuclear Division
The macronuclear nodules fuse in mid-dividers to a branched mass (Figures 3D, 6G), becoming oblong before and during the cell fission (Figure 4B). The elongate macronucleus divides into two pieces in late mid-dividers (Figure 4D). Each piece develops into a tree-dimensional macronuclear reticulum that gradually fragments into individual ellipsoidal nodules in very late dividers (Figure 5C). Micronuclei divide only once during the middle stages of binary fission (Figures 3D, 4B,D, 6G). More specifically, the micronuclei become spindle-shaped when the macronuclear nodules are fused into a branched mass (Figures 3D, 6G). Then, the micronuclei assume a dumbbell-shaped morphology as the macronuclear mass elongates into an oblong structure constricted in the middle (Figure 4B). Finally, the daughter micronuclei are connected by an internal fiber bundle that conspicuously elongates in late dividers. During the post-divisional patterning, the micronuclei move to the scattered macronuclear nodules (Figure 5C).
Phylogenetic Analyses
Both maximum likelihood (ML) and Bayesian (BI) trees were constructed to determine the phylogenetic position of N. luporinii n. sp. (Figures 7, 8). Although the taxon sampling slightly differed between the single- and multi-gene datasets, N. luporinii was consistently assigned to the order Urostylida with very strong or full statistical support. All members of the family Pseudokeronopsidae, including N. luporinii, always clustered together with Anteholosticha pulchra with full statistical support. Monophylies of the Pseudokeronopsinae and Nothoholostichinae were strongly to fully statistically supported in phylogenetic analyses of both single- and multi-gene datasets. The Pseudokeronopsinae clustered with the Nothoholostichinae in the multi-gene trees, but with very weak support (50% ML, 0.79 BI). On the other hand, their sister-group relationship was not recognized in the single-gene trees, as the Pseudokeronopsinae grouped with A. pulchra, but with very weak support (63% ML, 0.62 BI).
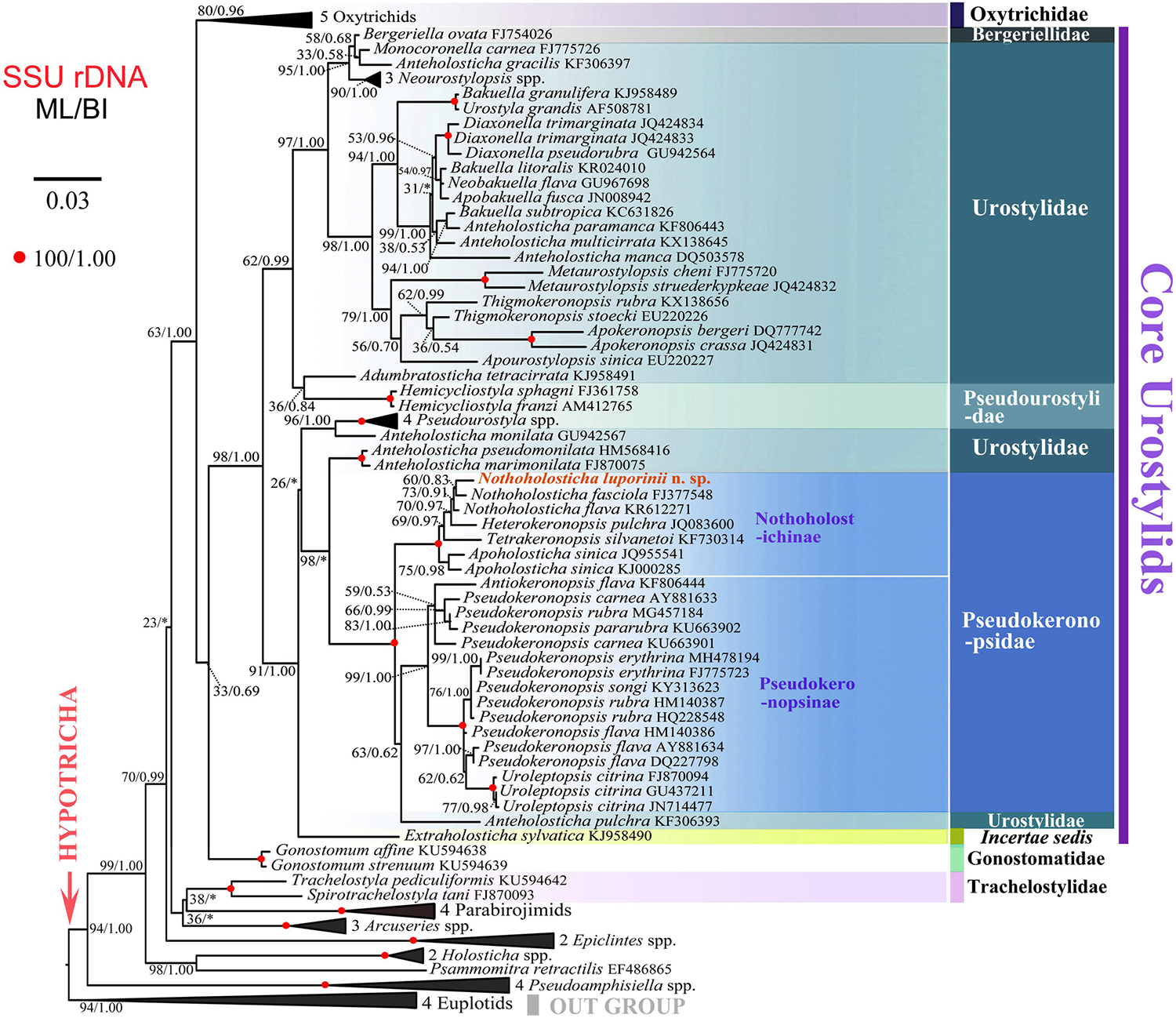
Figure 7. Maximum likelihood tree inferred from SSU rDNA sequences, showing the systematic position of Nothoholosticha luporinii n. sp. (indicated in red). Numbers near branches denote bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI). Asterisk indicates the disagreement between ML and BI trees. GenBank accession numbers are provided after species names. For taxon sampling and GenBank accession numbers in the collapsed clades, see Supplementary Table S1. Scale bar corresponds to three substitutions per 100 nucleotide positions.
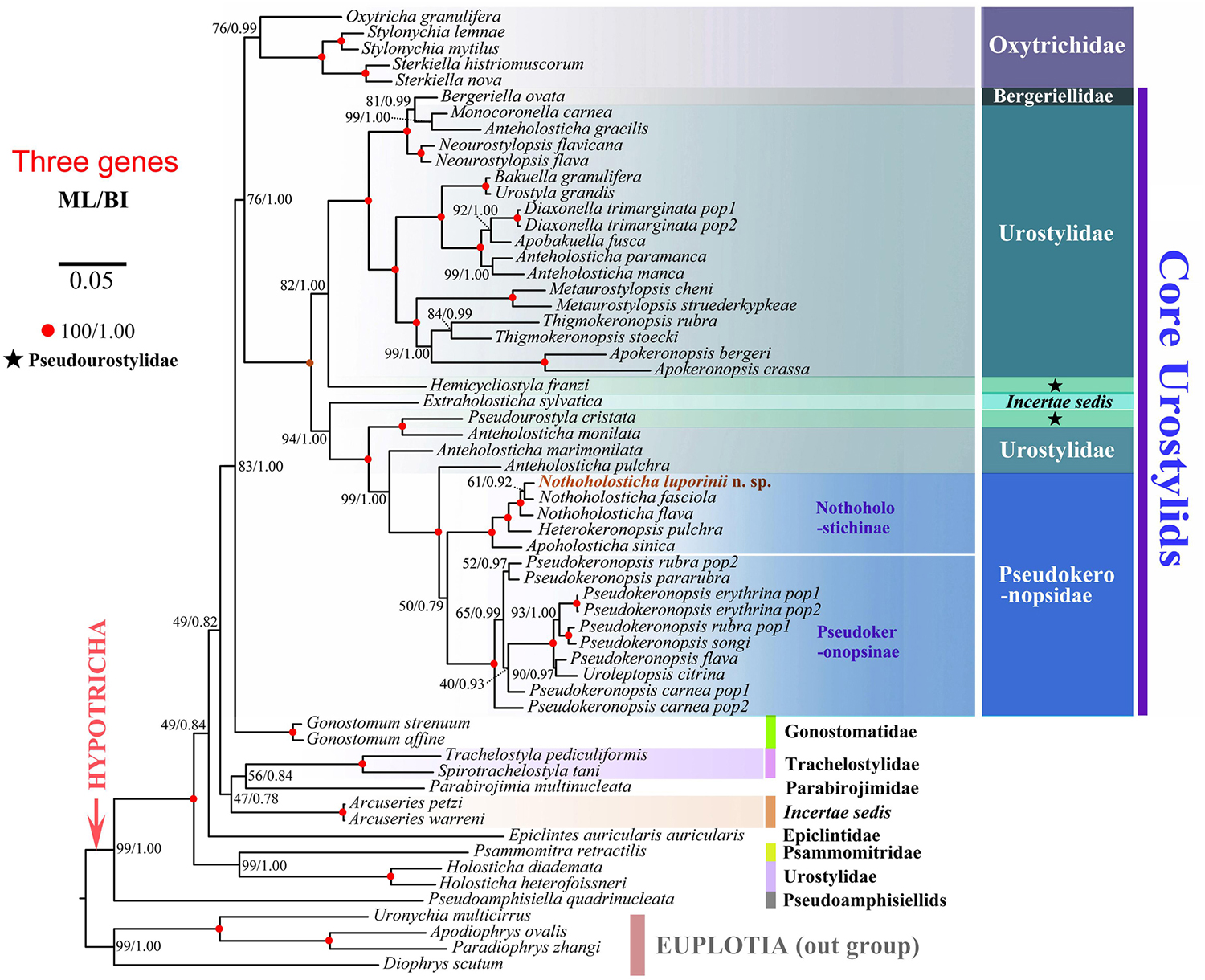
Figure 8. Maximum likelihood tree inferred from SSU rDNA, ITS region and LSU rDNA sequences, showing the systematic position of Nothoholosticha luporinii n. sp. (indicated in red). Numbers near branches denote bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI). GenBank accession numbers are provided in Supplementary Table S2. Scale bar corresponds to five substitutions per 100 nucleotide positions.
According to the single-gene and multi-gene trees, N. luporinii clustered with full statistical support in the monophyletic subfamily Nothoholostichinae, which encompasses N. fasciola, N. flava, Heterokeronopsis pulchra Pan et al., 2013, and Apoholosticha sinica Fan et al., 2014a. Within this subfamily, the genus Nothoholosticha was depicted as monophyletic with very weak support in the single-gene analyses (73% ML, 0.91 BI), but with full statistical support in the multi-gene analyses. Nothoholosticha luporinii was shown to be most closely related to N. fasciola, but this relationship was very poorly statistically supported in all analyses. According to the multi-gene analyses, Nothoholosticha was sister to the monotypic genus Heterokeronopsis. And, finally, Apoholosticha was placed in a sister position to the Nothoholosticha-Heterokeronopsis cluster in the multi-gene trees.
The number of unmatched nucleotide positions and the pairwise SSU rDNA sequence similarities among members of the subfamily Nothoholostichinae are summarized in Table 2. Nothoholosticha luporinii differs from N. flava by 13 nucleotide positions (99.1% sequence identity), from N. fasciola by 14 nucleotides (99.1% identity), from H. pulchra by 21 nucleotides (98.6% identity), from T. silvanetoi by 26 nucleotides (98.3% identity, the 121 Ns positions in the T. silvanetoi sequence were coded according to other related species as this region was fully conserved), from A. sinica population 1 by 26 nucleotides (98.3% identity), and from A. sinica population 2 by 25 nucleotides (98.4% identity).
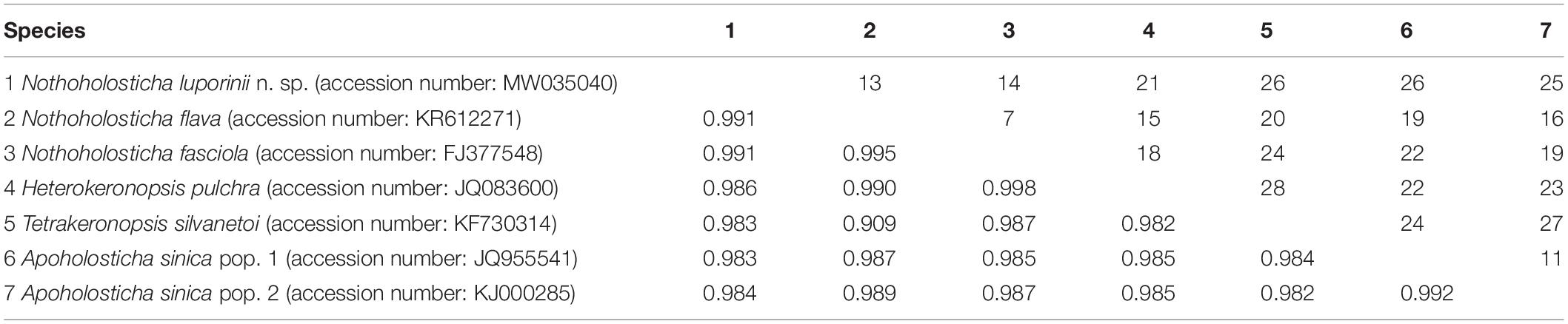
Table 2. Numbers of unmatched nucleotides (above diagonal) and pairwise similarities (below diagonal) of SSU rDNA sequences among members of the subfamily Nothoholostichinae.
Discussion
The New Species Nothoholosticha luporinii
Nothoholosticha luporinii n. sp. can be easily separated from members of the genera Anteholosticha, Antiokeronopsis, Pseudokeronopsis, and Uroleptopsis by having an atypical bicorona composed of four anterior and two posterior frontal cirri (vs. only three frontal cirri in Anteholosticha and more than four anterior coronal cirri in the three latter genera); from members of the genus Apoholosticha by possessing (vs. lacking) buccal cirri; and from Heterokeronopsis by exhibiting (vs. lacking) frontoterminal and transverse cirri. On the other hand, the new species highly resembles taxa assigned to the genera Nothoholosticha and Tetrakeronopsis (Figure 9). Hitherto, Nothoholosticha comprises two species, N. fasciola and N. flava, while Tetrakeronopsis is monotypic and includes only T. silvanetoi.
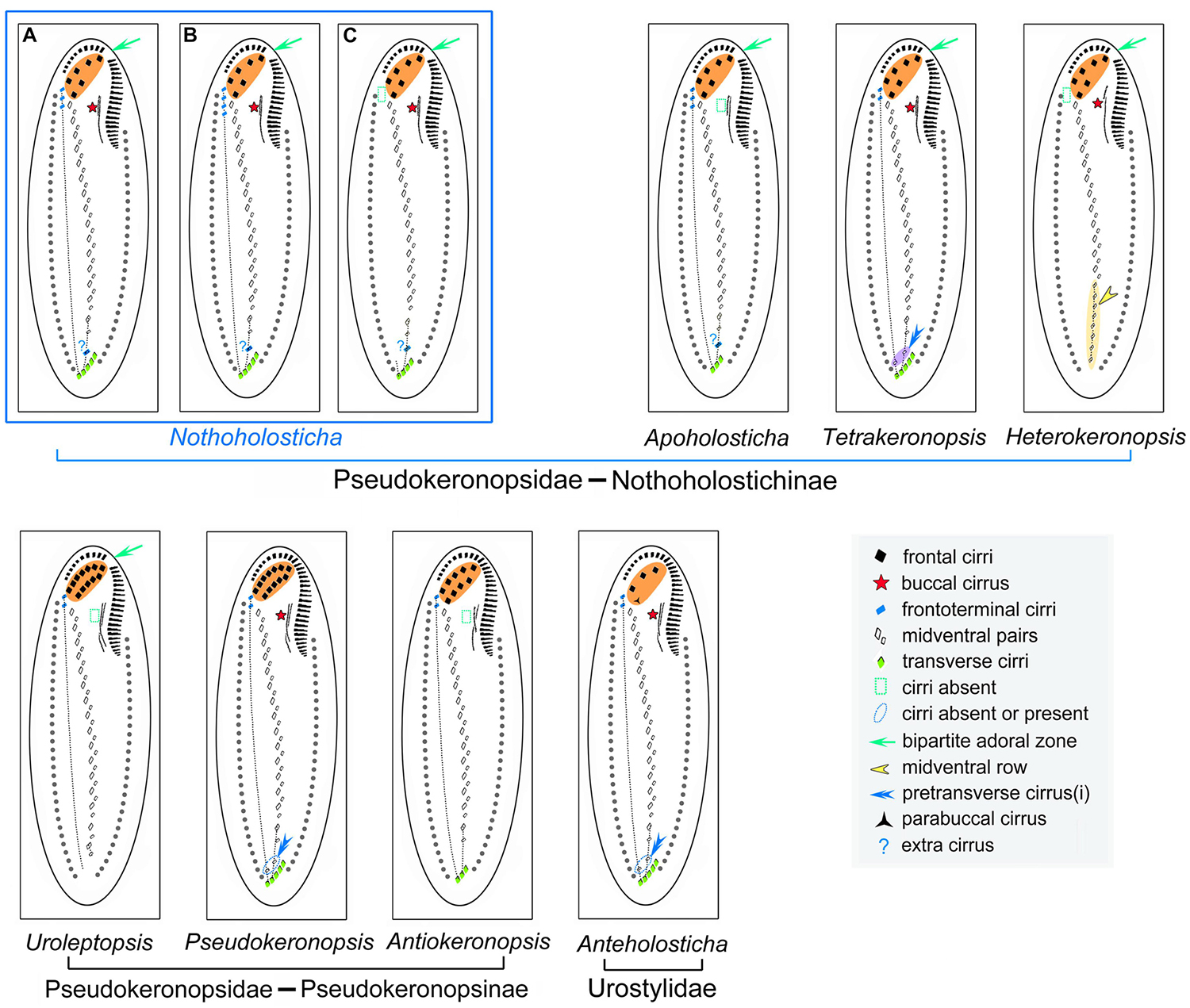
Figure 9. Comparison of cirral patterns of some related urostylid genera. Cirri originating from the two rightmost (rearmost) frontal-midventral-transverse anlagen are connected by a dotted line. (A) N. luporinii n. sp. (B) N. flava. (C) N. fasciola. Note that the “extra” cirrus is regarded as a pretransverse cirrus in N. luporinii (A), while as a midventral cirrus in the two remaining Nothoholosticha species (B,C).
Our newly discovered species cannot be confused with N. fasciola, the type species of Nothoholosticha, because it lacks frontoterminal cirri (Li et al., 2009). On the other hand, N. luporinii highly resembles N. flava in the body shape and size as well as in the number and arrangement of the macronuclear nodules and cirri (Li et al., 2016). Morphologically, N. luporinii can be distinguished from N. flava only by the lower number of crown (7–13 vs. 13–18) and lapel (23–39 vs. 36–47) adoral membranelles. However, the proper identification requires molecular data, because both differentiating characters partially overlap. The SSU rDNA sequences of N. luporinii and N. flava differ in 13 nucleotide positions (Table 2), which undoubtedly supports the distinctness of both taxa.
Because of the high morphological similarity, it might be speculated whether N. luporinii should not be classified as a subspecies of N. flava. However, the 13 different nucleotide positions in the SSU rDNA stand strongly against this suggestion. This is, indeed, a very pronounced genetic difference as the SSU rRNA gene is highly conservative with a rate of only 1.24–3.96 × 10–4 substitutions per site per one million years (Wright and Lynn, 1997; Vd’ačný, 2015; Vd’ačný et al., 2019). Kumar et al. (2017) showed that one base pair difference in SSU rDNA is sufficient to separate two closely related hypotrich taxa. Although Bistichella variabilis (HQ699895), Uroleptoides magnigranulosus (AM412774), and Orthoamphisiella breviseries (AY498654) are classified in different genera on the basis of morphological data, they differ only by 5–9 nucleotide positions (99.5–99.7% identity) in their SSU rDNA sequences (He and Xu, 2011). In this light, we find the species status of N. luporinii proposed in the present study to be justified.
Finally, N. luporinii can be distinguished from T. silvanetoi by the higher number of frontoterminal cirri (3–6 vs. invariably 2), the lower number of pretransverse cirri (1 vs. 2), the lower number of crown (7–13 vs. 12–17) and lapel (23–39 vs. 40–52) adoral membranelles as well as by the arrangement of the smaller type of bright brown–reddish cortical granules (clustered in a flower-like pattern around dorsal bristles vs. loosely scattered throughout the cortex). The morphological differences are supported also by the genetic differences in SSU rDNA sequences. Thus, there are 21 unmatched nucleotides between N. luporinii and T. silvanetoi (Table 2).
Generic Classification of Nothoholosticha luporinii
Generic classification of hypotrichs is traditionally based on the cirral and oral patterns as well as on the morphogenesis of the ventral and the dorsal ciliature (for reviews, see Berger, 1999, 2006, 2008, 2011). The generic classification of N. luporinii is a difficult matter, because its cirral pattern is a mixture of features found in the type species of Nothoholosticha (an “extra” cirrus between the midventral complex and transverse cirri) and the monotypic genus Tetrakeronopsis (multiple frontoterminal cirri). More specifically, N. fasciola, the type species of Notholosticha, lacks frontoterminal cirri (Li et al., 2009), which are present not only in two further Nothoholosticha species, N. flava and N. luporinii, but also in T. silvanetoi (Paiva et al., 2014; Li et al., 2016; present study). The extra cirrus, situated between the midventral complex and transverse cirri, is interpreted as the last cirrus of the midventral complex in N. fasciola and N. flava (Li et al., 2009, 2016), while as a pretransverse cirrus in N. luporinii. Regardless of the terminology, it is very likely the same cirrus, which is derived from the second rearmost frontal-midventral-transverse cirral streak n-1 (Figures 4C, 5A,B). Tetrakeronopsis exhibits even two cirri between the midventral complex and transverse cirri. Because these two extra cirri are situated ahead of the rightmost transverse cirri, they also might be designated as pretransverse cirri. The right pretransverse cirrus is derived from the rearmost streak n and the left pretransverse cirrus from the second rearmost streak n-1 (Paiva et al., 2014). In this light, all Nothoholosticha species have retained only the left pretransverse cirrus and N. fasciola further lost the frontoterminal cirri, which are derived from the rearmost streak n in N. luporinii and Tetrakeronopsis (Figure 9). Because N. luporinii is more closely related to N. fasciola than to T. silvanetoi in SSU DNA phylogenies (Figure 7), we classify our new species in the genus Nothoholosticha. However, the results of the present phylogenetic analyses question the generic diagnostic value of the loss of frontoterminal and pretransverse cirri in N. fasciola, which are derived from the two rearmost frontal-midventral-transverse cirral anlagen. Nevertheless, we prefer to await discovery of further species to improve the diagnosis of the genus Nothoholosticha.
Classification Framework of the Subfamily Nothoholostichinae
The structure of the bicorona represents an important subfamily-level character that was used to divide the family Pseudokeronopsidae into two subfamilies by Paiva et al. (2014): the Nothoholostichinae, with an atypical bicorona whose anterior portion is formed by four frontal cirri, and the Pseudokeronopsinae, with a typical bicorona whose anterior portion is formed by more than four frontal cirri. The present phylogenetical analyses corroborate this subdivision as well as the monophyletic origin of the Nothoholostichinae (Figures 7, 8). Interestingly, there are as many as three monotypic genera within the subfamily Nothoholostichinae. However, they can be easily distinguished from each other either by the loss or by the retention of some cirri, with respect to the type genus Nothoholosticha. Thus, Apoholosticha lost the buccal cirrus, Heterokeronopsis lost the frontoterminal and transverse cirri, while Tetrakeronopsis maintained both the left and the right pretransverse cirrus (Pan et al., 2013; Fan et al., 2014a; Paiva et al., 2014; Hu et al., 2015; Figure 9), which seems to be the plesiomorphic condition in the family Pseudokeronopsidae. Remarkably, frontoterminal cirri were lost in Heterokeronopsis and N. fasciola, but not in any other Nothoholosticha species. Homoplastic nature of this character thus questions its taxonomic significance at genus level. On the other hand, the loss of the buccal cirrus in Heterokeronopsis and of the right pretransverse cirrus in Nothoholosticha might serve as good generic characters at the present state of knowledge. Likewise, the long midventral row found in Heterokeronopsis, seems to be another good generic diagnostic feature for separation of genera within the subfamily Nothoholostichinae.
Multi-Gene Phylogenetic Analyses of the Family Pseudokeronopsidae
The SSU rRNA gene is very conservative and it is generally known that it bears mainly information for deeper nodes of phylogenetic trees. Therefore, manifold studies pursued to improve the knowledge about phylogenetic interrelationships within the subclass Hypotrichia using mostly sequences of that gene (e.g., Luo et al., 2017, 2018; Song and Shao, 2017; Lyu et al., 2018; Kim and Min, 2019; Chen et al., 2020; Dong et al., 2020; Paiva, 2020; Park et al., 2020; Wang J. et al., 2020; Xu et al., 2020). On the other hand, the ITS1-5.8S-ITS2 region as well as the LSU rRNA gene are much faster evolving parts of the rDNA operon and hence bear phylogenetic signal also for more recent divergences (Abraham et al., 2019). Therefore, the concatenation of SSU rDNA, ITS1-5.8S-ITS2 region and LSU rDNA might lead to better resolved phylogenetic trees, as also evidenced in the present study (Figures 7, 8). For instance, Anteholosticha pulchra, a typical urostylid, clustered in a sister position to the subfamily Pseudokeronopsinae in the SSU rDNA tree, causing non-monophyly of the family Pseudokeronopsidae (Figure 7), as defined by Paiva et al. (2014). On the other hand, A. pulchra was placed outside the Pseudokeronopsidae in the multi-gene trees although the statistical support remained poor (Figure 8). This position is, however, much more consistent with morphological classifications, because A. pulchra possesses three enlarged frontal cirri while pseudokeronopsids have a bicorona. The internal branching pattern within the subfamilies Pseudokeronopsinae and Nothoholostichinae as well as the placement of the new species within the genus Nothoholosticha were also much better statistically supported in multi-gene than in the single-gene analyses (Figures 7, 8). Likewise, the sister-group relationship of Uroleptopsis citrina and Pseudokeronopsis flava within the Pseudokeronopsinae obtained much better support in the multi-gene trees. Thus, the whole rDNA operon strongly suggests that the genus Pseudokeronopsis is non-monophyletic and might be split into multiple genera in future. Pseudokeronopsis species are, however, highly similar in terms of their nuclear apparatus and cirral patterns (for details, see Li et al., 2017). This was the main reason why multiple species were synonymized, misidentified, or their species status was questioned. Nevertheless, the distinctness of most Pseudokeronopsis species was corroborated by analyses of SSU rDNA sequences along with ITS-5.8S rDNA sequences (Li et al., 2017). To summarize, the combination of the traditionally used SSU rDNA sequences with ITS region and LSU rDNA sequences improves phylogenetic inferences and classifications of pseudokeronopsids in specific and of ciliates in general.
Data Availability Statement
GenBank accession numbers of sequences used in phylogenetic analyses can be found in the Supplementary Material.
Author Contributions
WS and CS conceptualized the project. TZ and TC performed the laboratory work. TZ, YW, and JM prepared the data sets and conducted analyses. TZ and PV wrote the first draft of the article. YW, CS, and WS revised the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Project numbers: 32070428 and 32030015) and by the Slovak Research and Development Agency (grant number APVV-19-0076) and by the Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and Slovak Academy of Sciences (grant number VEGA 1/0041/17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.610886/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://guidance.tau.ac.il/ver2/
- ^ http://www.phylo.org
- ^ http://tree.bio.ed.ac.uk/software/figtree/
References
Abraham, J. S., Sripoorna, S., Maurya, S., Makhija, S., Gupta, R., and Toteja, R. (2019). Techniques and tools for species identification in ciliates: a review. Int. J. Syst. Evol. Microbiol. 69, 877–894. doi: 10.1099/ijsem.0.003176
Anson, E. L., and Myers, E. W. (1997). ReAligner: a program for refining DNA sequence multi-alignments. J. Comput. Biol. 4, 369–383. doi: 10.1089/cmb.1997.4.369
Berger, H. (1999). Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Monogr. Biol. 78, 1–1080. doi: 10.1007/978-94-011-4637-1
Berger, H. (2006). Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monogr. Biol. 85, 1–1304. doi: 10.1007/1-4020-5273-1_1
Berger, H. (2008). Monograph of the Amphisiellidae and Trachelostylidae (Ciliophora, Hypotricha). Monogr. Biol. 88, 1–737. doi: 10.1007/978-1-4020-8917-6
Berger, H. (2011). Monograph of the Gonostomatidae and Kahliellidae (Ciliophora, Hypotricha). Monogr. Biol. 90, 1–741. doi: 10.1007/978-94-007-0455-8
Bharti, D., Kumar, S., Terza, A. L., and Chandra, K. (2019). Morphology and ontogeny of Tetmemena pustulata indica nov. subspec. (Ciliophora, Hypotricha), from the Thane Creek, Mumbai, India. Eur. J. Protistol. 71:125629. doi: 10.1016/j.ejop.2019.125629
Borror, A., and Wicklow, B. (1983). The suborder Urostylina Jankowski (Ciliophora, Hypotrichida): morphology, systematics and identification of species. Acta Protozool. 22, 97–126.
Chen, L., Dong, J., Xin, Y., Warren, A., Ning, Y., and Zhao, Y. (2020). Morphology and molecular phylogeny of a new hypotrich ciliate, Anteholosticha songi nov. spec., and an American population of Holosticha pullaster (Muller, 1773) Foissner et al., 1991 (Ciliophora, Hypotricha). Eur. J. Protisotol. 72:125646. doi: 10.1016/j.ejop.2019.125646
Dong, J., Li, L., Fan, X., Ma, H., and Warren, A. (2020). Two Urosoma species (Ciliophora, Hypotrichia): a multidisciplinary approach provides new insights into their ultrastructure and systematics. Eur. J. Protistol. 72:125661. doi: 10.1016/j.ejop.2019.125661
Fan, Y., Chen, X., Hu, X., Shao, C., Al-Rasheid, K. A. S., Al-Farraj, S. A., et al. (2014a). Morphology and morphogenesis of Apoholosticha sinica n.g., n. sp. (Ciliophora, Hypotrichia), with consideration of its systematic position among urostylids. Eur. J. Protistol. 50, 78–88. doi: 10.1016/j.ejop.2013.06.003
Fan, Y., Pan, Y., Huang, J., Lin, X., Hu, X., and Warren, A. (2014b). Molecular phylogeny and taxonomy of two novel brackish water hypotrich ciliates, with the establishment of a new genus, Antiokeronopsis gen. n. (Ciliophora, Hypotrichia). J. Eukaryot. Microbiol. 61, 449–462. doi: 10.1111/jeu.12125
Galtier, N., Gouy, M., and Gautier, C. (1996). SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548. doi: 10.1093/bioinformatics/12.6.543
Gong, R., Jiang, Y., Vallesi, A., Gao, Y., and Gao, F. (2020). Conjugation in Euplotes raikovi (Protista, Ciliophora): new insights into nuclear events and macronuclear development from micronucleate and amicronucleate cells. Microorganisms, 8:162. doi: 10.3390/microorganisms8020162
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
He, Y., and Xu, K. (2011). Morphology and small subunit rDNA phylogeny of a new soil ciliate, Bistichella variabilis n. sp. (Ciliophora, Stichotrichia). J. Eukaryot. Microbiol. 58, 332–338. doi: 10.1111/j.1550-7408.2011.00554.x
Hu, X., Fan, Y., and Warren, A. (2015). New record of Apoholosticha sinica (Ciliophora, Urostylida) from the UK: morphology, 18S rRNA gene phylogeny and notes on morphogenesis. Int. J. Syst. Evol. Microbiol. 40, 78–92. doi: 10.1016/j.ejop.2016.09.005
Hu, X., Lin, X., and Song, W. (2019). Ciliate atlas: Species Found in the South China Sea. Beijing: Science Press. doi: 10.1007/978-981-13-5901-9
Huang, J., Chen, Z., Song, W., and Berger, H. (2014). Three-gene based phylogeny of the Urostyloidea (Protista, Ciliophora, Hypotricha), with notes on classification of some core taxa. Mol. Phylogenet. Evol. 70, 337–347. doi: 10.1016/j.ympev.2013.10.005
Jerome, C. A., Lynn, D. H., and Simon, E. M. (1996). Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can. J. Zool. 74, 1898–1906. doi: 10.1139/z96-214
Jung, J. H., and Berger, H. (2019). Monographic treatment of Paraholosticha muscicola (Ciliophora, Keronopsidae), including morphological and molecular biological characterization of a brackish water population from Korea. Eur. J. Protistol. 68, 48–67. doi: 10.1016/j.ejop.2018.12.004
Kaur, H., Negi, S. R. K., and Kamra, K. (2019). Morphological and molecular characterization of Neogastrostyla aqua nov. gen., nov. spec. (Ciliophora, Hypotrichia) from River Yamuna, Delhi; comparison with Gastrostyla-like genera. Eur. J. Protistol. 68, 68–79. doi: 10.1016/j.ejop.2019.01.002
Kim, K. S., and Min, G. S. (2019). Morphology and molecular phylogeny of Oxytricha seokmoensis sp. nov. (Hypotrichia: Oxytrichidae), with notes on its morphogenesis. Eur. J. Protistol. 71:125641. doi: 10.1016/j.ejop.2019.125641
Kumar, S., Bharti, D., Shazib, S. U. A., and Shin, M. K. (2017). Discovery of a new hypotrich ciliate from petroleum contaminated soil. PLoS One 12:e0178657. doi: 10.1371/journal.pone.0178657
Landan, G., and Graur, D. (2008). Local reliability measures from sets of co-optimal multiple sequence alignments. Pacific Symp. Biocomput. 13, 15–24. doi: 10.1142/9789812776136_0003
Li, J., Chen, X., and Xu, K. (2016). Morphology and small subunit rDNA phylogeny of two new marine urostylid ciliates, Caudiholosticha marina sp. nov. and Nothoholosticha flava sp. nov. (Ciliophora, Hypotrichia). J. Eukaryot. Microbiol. 63, 460–470. doi: 10.1111/jeu.12290
Li, J., Zhan, Z., and Xu, K. (2017). Systematics and molecular phylogeny of the ciliate genus Pseudokeronopsis (Ciliophora, Hypotrichia). J. Eukaryot. Microbiol. 64, 850–872. doi: 10.1111/jeu.12420
Li, L., Zhang, Q., Hu, X., Warren, A., Al-Rasheid, K. A. S., Al-Khedheiry, A. A., et al. (2009). A redescription of the marine hypotrichous ciliate, Nothoholosticha fasciola (Kahl, 1932) nov. gen., nov. comb. (Ciliophora: Urostylida) with brief notes on its cellular reorganization and SSU rRNA gene sequence. Eur. J. Protistol. 45, 237–248. doi: 10.1016/j.ejop.2009.01.004
Lu, X., Wang, Y., Al-Farraj, S. A., El-Serehy, H., Huang, J., and Shao, C. (2020). The insights into the systematic relationship of Gastrostyla-affinitive genera, with report on a new saline soil ciliate genus and new species (Protozoa, Ciliophora). BMC Evol. Biol. 20:92. doi: 10.1186/s12862-020-01659-8
Luo, X., Gao, F., Yi, Z., Pan, Y., Al-Farraj, S. A., and Warren, A. (2017). Taxonomy and molecular phylogeny of two new brackish hypotrichous ciliates, with the establishment of a new genus (Protozoa, Ciliophora). Zool. J. Linn. Soc. 179, 475–491. doi: 10.1111/zoj.12451
Luo, X., Huang, J., Li, L., Song, W., and Bourland, W. A. (2019). Phylogeny of the ciliate family Psilotrichidae (Protista, Ciliophora), a curious and poorly-known taxon, with notes on two algae-bearing psilotrichids from Guam, USA. BMC Evol. Biol. 19:125. doi: 10.1186/s12862-019-1450-z
Luo, X., Yan, Y., Shao, C., Al-Farraj, S. A., Bourland, W. A., and Song, W. (2018). Morphological, ontogenetic and molecular data support strongylidiids as being closely related to Dorsomarginalia (Protozoa, Ciliophora) and reactivation of the family Strongylidiidae Fauré-Fremiet, 1961. Zool. J. Linn. Soc. 184, 237–254. doi: 10.1093/zoolinnean/zly001
Lynn, D. H. (2008). The ciliated Protozoa: Characterization, Classification, and Guide to the Literature. Dordrecht: Springer.
Lyu, Z., Wang, J., Huang, J., Warren, A., and Shao, C. (2018). Multigene-based phylogeny of Urostylida (Ciliophora, Hypotrichia), with establishment of a novel family. Zool. Scr. 47, 243–254. doi: 10.1111/zsc.12267
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Miao, M., Warren, A., Song, W., Wang, S., Shang, H., and Chen, Z. (2008). Analysis of the internal transcribed spacer 2 (ITS2) region of scuticociliates and related taxa (Ciliophora, Oligohymenophorea) to infer their evolution and phylogeny. Protist 159, 519–533. doi: 10.1016/j.protis.2008.05.002
Moreira, D., von der Heyden, S., Bass, D., López-García, P., Chao, E., et al. (2007). Global eukaryote phylogeny: combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol. Phylogenet. Evol. 44, 255–266. doi: 10.1016/j.ympev.2006.11.001
Nylander, J. (2004). MrModeltest, v.2. Uppsala: Evolutionary Biology Centre. Sweden: Uppsala University.
Paiva, T. D. (2020). Systematic redefinition of the Hypotricha (Alveolata, Ciliophora) based on combined analyses of morphological and molecular characters. Protist 171:125755. doi: 10.1016/j.protis.2020.125755
Paiva, T. D., de Albuquerque, A. F., Borges, B. N., and Harada, M. L. (2014). Description and phylogeny of Tetrakeronopsis silvanetoi gen. nov., sp. nov. (Hypotricha, Pseudokeronopsidae), a new benthic marine ciliate from Brazil. PLoS One 9:e88954. doi: 10.1371/journal.pone.0088954
Pan, Y., Li, J., Li, L., Hu, X., Al-Rasheid, K. A. S., and Warren, A. (2013). Ontogeny and molecular phylogeny of a new marine ciliate genus, Heterokeronopsis g. n. (Protozoa, Ciliophora, Hypotricha), with description of a new species. Eur. J. Protistol. 49, 298–311. doi: 10.1016/j.ejop.2012.08.008
Park, K. M., Jung, J. H., Kim, J. H., Min, G. S., and Kim, S. (2020). Morphology, morphogenesis, and molecular phylogeny of a new freshwater ciliate, Gonostomum jangbogoensis n. sp. (Ciliophora, Hypotricha), from Victoria Land, Antarctica. Eur. J. Protistol. 73:125669. doi: 10.1016/j.ejop.2019.125669
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Sheng, Y., Duan, L., Cheng, T., Qiao, Y., and Stover, N. A., and Gao, S. (2020). The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Sci. China Life Sci. 63, 1534–1542. doi: 10.1007/s11427-020-1689-4
Stamatakis, A. (2014). RAxML v.8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vd’ačný, P. (2015). Estimation of divergence times in litostomatean ciliates (Ciliophora: Intramacronucleata), with Bayesian relaxed clock and 18S rRNA gene. Eur. J. Protistol. 51, 321–334. doi: 10.1016/j.ejop.2015.06.008
Vd’ačný, P., Rajter, L’, Stoeck, T., and Foissner, W. (2019). A proposed timescale for the evolution of armophorean ciliates: clevelandellids diversify more rapidly than metopids. J. Eukaryot. Microbiol. 66, 167–181. doi: 10.1111/jeu.12641
Wang, J., Zhao, Y., Lu, X., Lyu, Z., Warren, A., and Shao, C. (2020). Does the Gonostomum-patterned oral apparatus in Hypotrichia carry a phylogenetic signal? Evidence from morphological and molecular data based on extended taxon sampling using three nuclear genes (Ciliophora, Spirotrichea). Sci. China Life Sci. 63. doi: 10.1007/s11427-020-1667-3
Wang, Y. R., Jiang, Y., Liu, Y., Li, Y., Katz, L. A., Gao, F., et al. (2020). Comparative studies on the polymorphism and copy number variation of mtSSU rDNA in ciliates (Protista, Ciliophora): implications for pylogenetic, environmental, and ecological research. Microorganisms 8:316. doi: 10.3390/microorganisms8030316
Wang, Y. Y., Sheng, Y., Liu, Y., Zhang, W., Cheng, T., Duan, L., et al. (2019). A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res. 47, 11771–11789. doi: 10.1093/nar/gkz1053
Wilbert, N. (1975). Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 64, 171–179.
Wright, A. D. G., and Lynn, D. H. (1997). Maximum ages of ciliate lineages estimated using a small subunit rRNA molecular clock: crown eukaryotes date back to the Paleoproterozoic. Arch. Protistenkd. 148, 329–341. doi: 10.1016/s0003-9365(97)80013-9
Xu, W., Wang, Y., Cheng, T., Yu, Y., El-Serehy, H., Al-Farraj, S. A., et al. (2020). Reevaluation of the ‘well-known’ Paraurostyla weissei complex, with notes on the ontogenesis of a new Paraurostyla species (Ciliophora, Hypotrichia). Eur. J. Protistol. 73:125672. doi: 10.1016/j.ejop.2020.125672
Yan, Y., Maurer-Alcalá, X. X., Knight, R., Pond, S. L. K., and Katz, L. A. (2019). Single-cell transcriptomics reveal a correlation between genome architecture and gene family evolution in ciliates. mBio 10, e2524–e2519. doi: 10.1128/mBio.02524-19
Zhang, T., Dong, J., Cheng, T., Duan, L., and Shao, C. (2020). Reconsideration on taxonomy of the marine ciliate Neobakuella aenigmatica Moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia). Mar. Life Sci. Technol. 2, 97–108. doi: 10.1007/s42995-020-00032-4
Keywords: ciliated protists, ontogenesis, phylogeny, rDNA operon, integrative taxonomy
Citation: Zhang T, Wang Y, Cheng T, Ma J, Vd’ačný P, Song W and Shao C (2020) Systematics and Multi-Gene Phylogeny of the Subfamily Nothoholostichinae (Ciliophora, Hypotrichia), With Integrative Description of a New Marine Species Nothoholosticha luporinii n. sp.. Front. Mar. Sci. 7:610886. doi: 10.3389/fmars.2020.610886
Received: 27 September 2020; Accepted: 11 November 2020;
Published: 08 December 2020.
Edited by:
Hongbo Pan, Shanghai Ocean University, ChinaReviewed by:
Santosh Kumar, Zoological Survey of India, IndiaBarbara Nascimento Borges, Federal University of Pará, Brazil
Copyright © 2020 Zhang, Wang, Cheng, Ma, Vd’ačný, Song and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Shao, c2hhb2NoZW5Ac25udS5lZHUuY24=
†These authors have contributed equally to this work
 Tengyue Zhang
Tengyue Zhang Yurui Wang
Yurui Wang Ting Cheng
Ting Cheng Jiyang Ma
Jiyang Ma Peter Vd’ačný
Peter Vd’ačný Weibo Song
Weibo Song Chen Shao
Chen Shao