- 1Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao, China
- 2College of Fisheries and Key Laboratory of Mariculture of the Education Ministry of China, Ocean University of China, Qingdao, China
- 3Ecology Group, Technische Universität Kaiserslautern, Kaiserslautern, Germany
- 4Department of Zoology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Department of Life Sciences, Natural History Museum, London, United Kingdom
The morphology and taxonomy of three scuticociliates found in China, viz. Citrithrix smalli sp. nov., Homalogastra binucleata sp. nov., and Uronema orientalis Pan et al., 2015, were investigated. The small subunit ribosomal RNA (SSU rRNA) gene of these species, and the cytochrome c oxidase subunit I (COI) gene of Uronema orientalis, were sequenced and compared with those of related taxa to determine their systematic positions. The new monotypic genus Citrithrix gen. nov. is characterized by its lemon-shaped body, posteriorly located cytostome, dominant oral groove, and the compact structure of its multi-rowed membranelles 1 and 2 (M1, M2). Based on both morphological and molecular data, this new genus cannot be assigned to any known family and thus, a new family, Citrithrixidae fam. nov., is proposed within the order Philasterida. Homalogastra binucleata sp. nov., a brackish water form (salinity 2‰), differs from all congeners in having two macronuclear nodules. Uronema orientalis closely resembles the type population in all respects other than having fewer somatic kineties.
This article is registered in ZooBank under: urn:lsid:zoobank.org:pub:5727F18E-5421-446D-B22C-774783539FE4.
Introduction
Ciliates in the subclass Scuticociliatia Small, 1967 are usually small in size, speciose and can be found in various environments such as aquatic as well as terrestrial habitats all over the world (Foissner et al., 1982, 1994; Lynn and Strüder-Kypke, 2005; Jankowski, 2007; Lynn, 2008). However, many scuticociliates lack a detailed description using modern methods resulting in problems of species identification, delineation and systematics (Borror, 1972; Carey, 1992; Jankowski, 2007; Song et al., 2009; Gao et al., 2016).
Recent studies on the ciliate fauna in coastal and freshwater habitats in China have revealed a much higher diversity of scuticocliates than was previously assumed (Song et al., 2009; Gao et al., 2012, 2013, 2014, 2017; Pan H. et al., 2016; Pan X. et al., 2016; Hu et al., 2019; Zhang et al., 2019; Liu et al., 2020; Pan et al., 2020). Among these, the philasterids are generally well-studied in terms of the application of modern silver staining and molecular methods to determine their taxonomy (Song et al., 2009; Liu et al., 2017). Six new genera and 22 new species have been added to the order Philasterida Small, 1967 since the beginning of the Twenty-first century (Aescht, 2001; Gong et al., 2007; Jankowski, 2007; Pan X. et al., 2016; Pan et al., 2020), which implies the diversity of this group may be underestimated.
During recent studies of marine and brackish water ciliates in China, three scuticociliates were investigated. Based on further morphological as well as molecular analyses, one of the species has been assigned to new genus and new family. The molecular phylogeny of all three species was investigated based on sequence data for the small subunit ribosomal RNA (SSU rRNA) gene and, for one species, the cytochrome c oxidase 1 (COI) gene.
Materials and Methods
Collection and Isolation
Citrithrix smalli sp. nov. and Uronema orientalis Pan et al., 2015 were both collected from a bathing beach near the Zhanqiao Pier, Qingdao, China (Figure 1), the former from marine water on reefs (36°03'43” N; 120°19'12” E) in September 2016, and the latter from the intertidal zone (36°03'42” N; 120°19'25” E) in March 2017. Homalogastra binucleata sp. nov. was collected from an inland brackish water sewage ditch in Lanzhou (36°05'37” N; 103°44'58” E), China, in April, 2017 (Figure 1). In all three cases the sample comprised water and sediment after gently stirring the water.
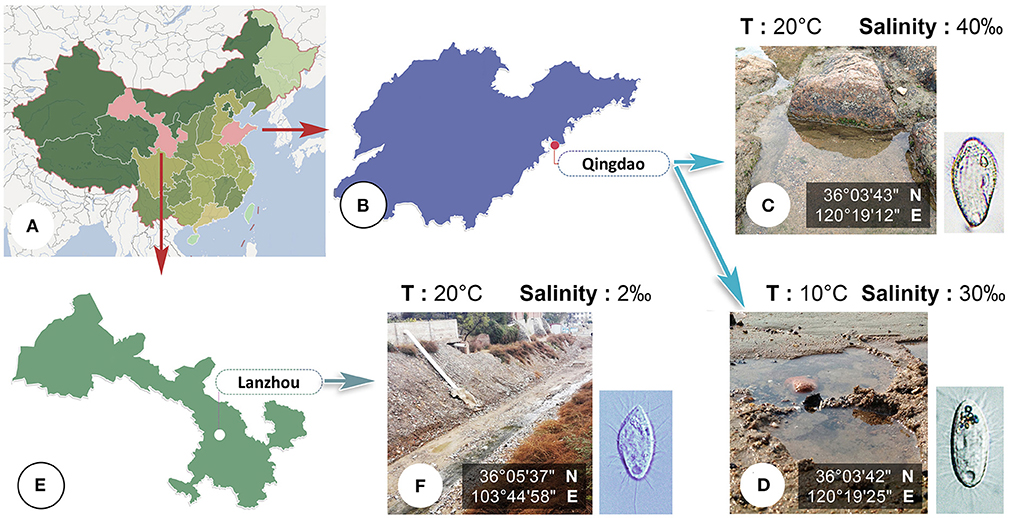
Figure 1. (A–F) Map, sampling sites and habitats. (A) Map of China. (B) Shandong Province, showing the location of Qingdao. (C,D) Photographs of sampling sites in Qingdao where Citrithrix smalli sp. nov. (C) and Uronema orientalis Pan et al., 2015 (D) were collected. (E) Gansu Province, showing the location of Lanzhou. (F) Photograph of sampling site in Lanzhou where Homalogastra binucleata sp. nov. was collected.
Morphological Studies
Observations of specimens in vivo and specimens stained with silver were performed to reveal the general morphology and ciliary pattern based on the methods described by Bai et al. (2020). The protargol silver proteinate reagent was made in-house according to Pan et al. (2013). Counts, measurements, and drawings of specimens were made according to Qu et al. (2020). Terminology and systematics followed Lynn (2008).
DNA Extraction, PCR Amplification and Sequencing
Genomic DNA extraction, polymerase chain reaction (PCR), and gene sequencing were conducted, mainly according to Chi et al. (2020). The SSU rRNA gene was amplified using the primers 82F (Jerome et al., 1996) and 18S-R (Medlin et al., 1988). The primers for the COI gene amplification were COI-NEW-17-F1 and COI-NEW-812-R2 (Zhang et al., 2019). To minimize the amplification errors, Q5 Hot Start High-Fidelity 2× Master Mix (New England BioLabs) was used (Wang et al., 2017). PCR for the SSU rRNA and COI genes were performed according to Chi et al. (2020) and Lynn and Strüder-Kypke (2006), respectively.
Phylogenetic Analyses
The SSU rRNA gene sequences of the three newly obtained isolates were aligned with 65 sequences from 50 related taxa, retrieved from GenBank. The accession numbers of these sequences are provided in the phylogenetic trees. Eight sequences of pleuronematids were selected as the outgroup. Alignments were conducted, edited, and trimmed according to Zhang et al. (2020). Comparisons of SSU rRNA gene sequences of Citrithrix smalli sp. nov., Homalogastra binucleata sp. nov., and Uronema orientalis with related sequences were performed, and the number of unmatched nucleotides and sequence identities were calculated, using the program BioEdit version 7.0.5.2 (Hall, 1999). The COI gene sequence of the newly isolated population of Uronema orientalis was compared with that of U. orientalis (MH605553).
Two maximum likelihood (ML) analyses, i.e., RAxML (Stamatakis, 2014) and IQTREE, and one Bayesian inference (BI) analysis, were carried out according to Wang et al. (2019). The IQ-trees were constructed on IQTREE 2.0.6 (Minh et al., 2020) with 104 ultrafast bootstrap replicates (Hoang et al., 2018). The best-fit model (GTR + F + R3) was selected based on the Bayesian information criterion (BIC) using the in-built ModelFinder program (Kalyaanamoorthy et al., 2017).
BI analysis was performed with the best-fit model GTR + I + G, selected by the Akaike Information Criterion using MrModeltest 2 (Nylander, 2004). Markov chain Monte Carlo (MCMC) simulations were run with two sets of four chains for 107 generations at a sampling frequency of 102 and a burn-in of 104 trees (10%).
Tree topologies were visualized using SeaView version 4 (Gouy et al., 2010).
Results
Morphological Study
Subclass: Scuticociliatia Small, 1967
Order: Philasterida Small, 1967
Citrithrixidae fam. nov.
ZooBank. urn:lsid:zoobank.org:act:F65C9B97-42EE-47A2-ABA8-DC8B581C289F
Diagnosis. Free-living philasterid with highly developed and compact membranelles 1 and 2; cytostome located in a long and conspicuous buccal grove; somatic ciliature of Uronematidae-type.
Type genus. Citrithrix gen. nov.
Citrithrix gen. nov.
ZooBank. urn:lsid:zoobank.org:act:9A7480F9-A487-40B5-9784-815368E5E09B
Diagnosis. Body lemon-shaped to cylindrical in outline; cytostome located in posterior half of cell within a conspicuous, concave oral groove; multi-rowed membranelles 1 and 2 extremely close-set and highly developed; membranelle 3 small; paroral membrane extends anteriorly to level of mid-region of membranelle 2; somatic kineties composed of dikinetids and monokinetids; single caudal cilium.
Etymology. The genus-group name Citrithrix is a composite of citri- (Latin noun; lemon) and thrix (Greek noun; hair~ciliate s.l.). It alludes to the typical lemon-shaped body. Feminine gender.
Type species. Citrithrix smalli sp. nov.
Citrithrix smalli sp. nov.
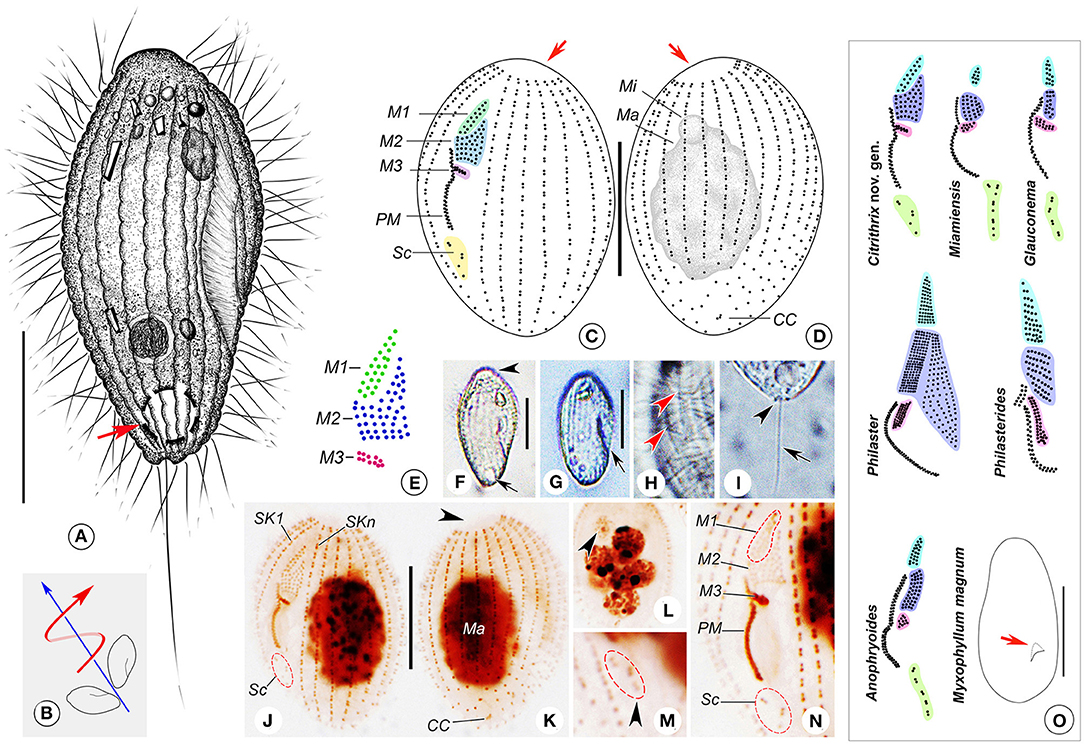
Figure 2. (A–O) Citrithrix smalli sp. nov. from life (A,B,F–I), after protargol staining (C–E,J–N), and a comparison with related genera (O). (A) Right ventrolateral view of a representative cell, arrow shows the contractile vacuole. (B) Swimming trace, red arrow indicates the body rotation, blue arrow shows the swimming direction. (C,D) Left ventrolateral (C) and right dorsolateral (D) views of the holotype specimen, arrows indicate the apical plate. (E) Oral structure showing three-rowed membranelle 1. (F,G) Right ventrolateral views, showing the variation in body shape, arrow and arrowhead in F point to the apical plate and contractile vacuole, respectively, arrow in G indicates the oral groove. (H) Detail of pellicle, arrowheads mark the grooves on the ciliary rows. (I) Lateral view of rear end of cell, arrowhead points to the small depression at basal position, arrow indicates the caudal cilium. (J,K) Left ventrolateral (J) and right dorsolateral (K) views of the holotype specimen, arrowhead shows the glabrous apical plate. (L) Nuclear apparatus of an individual with several macronuclear nodules, arrowhead indicates the micronucleus. (M) Arrowhead marks the scutica. (N) Detailed view of oral apparatus. (O) An oral structure comparison with related genera and a body size and shape comparison with Myxophyllum magnum, all redrawn based on previous studies except for the new genus (Cawthorn et al., 1996; Song, 2000; Song and Wilbert, 2000; Xu and Song, 2000; Ma et al., 2006; Pan et al., 2015b). Arrow in M. magnum points to the buccal field. CC, caudal cilium; M1–3, membranelle 1–3; Ma, macronucleus; Mi, micronucleus; PM, paroral membrane; Sc, scutica; SK1, somatic kinety 1; SKn, somatic kinety n. Scale bars: 15 μm (A,C,D,F,G,J,K); 100 μm (O).
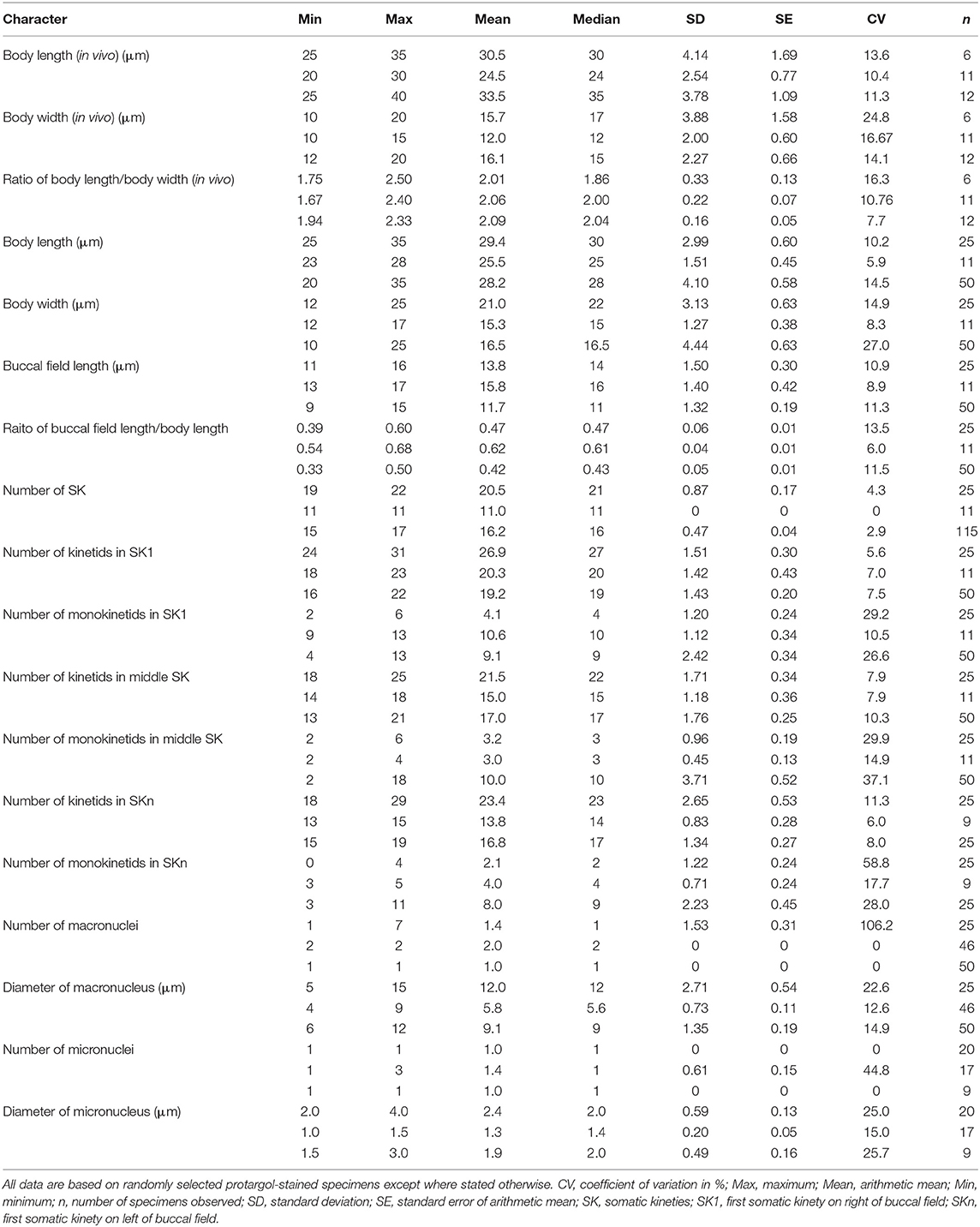
Table 1. Morphometric data for Citrithrix smalli sp. nov. (upper row), Homalogastra binucleata sp. nov. (middle row) and Uronema orientalis Pan et al., 2015 (lower row).
ZooBank. urn:lsid:zoobank.org:act:94C52D9F-DE13-4D33-B40A-4328D9D88FD3
Diagnosis. Cell size about 25–35 × 10–20 μm in vivo; 19–22 somatic kineties; single macronucleus; M1 composed of two or three rows; M2 irregular pentagon-shaped; contractile vacuole terminally positioned; marine biotope.
Dedication. We dedicate the new species to Prof. Eugene B. Small, University of Maryland, USA, in recognition of his significant contributions to the taxonomy and classification of ciliates in general and scuticociliates in particular.
Type locality and habitat. Seawater on reefs at a sandy beach in Qingdao (36°03'18” N; 120°20'22” E), northern China. Salinity 40‰, water temperature about 20°C.
Deposition of type slide. One protargol slide containing the holotype specimen and several paratype specimens (registration number: LMJ2016091901) was deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China.
SSU rRNA gene sequence. The length is 1640 bp, G + C content 45.00% and GenBank accession number MT982807.
Description. Body about 25–35 × 10–20 μm in vivo, lemon- or spindle-shaped, widest part about one-third down length of cell (Figures 2A,F,G). Anterior end prominently truncated with apical plate about one-quarter to one-third of maximum body width (Figures 2A,F). Buccal field deeply concaved, length one-third to half of cell length; cytostome located in posterior half of cell (Figures 2A,F,G). Pellicle conspicuously notched with longitudinal ridges between ciliary rows (Figures 2A,F–H). No extrusomes recognizable. Cytoplasm colorless to slightly grayish, usually containing a few bar-shaped crystals in anterior region of cell (Figures 2A,F,G). Most cells with single ellipsoidal macronucleus located in mid-body region, 10–15 μm in diameter (Figures 2D,J,K); two out of 25 specimens examined with several (six to seven) spherical macronuclear nodules, each about 5–6 μm across (Figure 2L). Micronucleus closely associated with macronucleus, ~3 μm across (Figures 2D,J,L). Single contractile vacuole caudally positioned, about 5 μm in diameter during diastole (Figures 2A,F), pulsating at intervals of 20–45 s. Somatic cilia about 7 μm long, densely arranged; single caudal cilium ~15 μm long, emerging from a small depression at posterior end of cell (Figures 2A,I).
Locomotion by swimming moderately fast in upper layer of water, with anterior end swinging from side to side and body rotating continuously about longitudinal axis (Figure 2B).
Nineteen to 22 somatic kineties (SK), each commencing anteriorly around apical plate and extending almost to posterior end of cell (Figures 2C,D,J,K). SK composed of closely arranged dikinetids in anterior four-fifths of kinety and loosely arranged monokinetids in posterior one-fifth (Figures 2C,D,J,K). Somatic kinety 1 (SK1, first kinety on right of buccal field) composed of 24–31 kinetids of which 2–6 are monokinetids; mid somatic kinety on dorsal side composed of 18–25 kinetids of which 2–6 are monokinetids; somatic kinety n (SKn, first kinety on left of buccal field) composed of 18–29 kinetids including 0–4 monokinetids. Caudal complex consisting of three argentophilic granules (Figures 2D,K).
Oral apparatus consisting of three membranelles (M1–3) and one paroral membrane (PM). Characteristically M1 and M2 closely apposed, gap between them difficult to observe (Figure 2E). M1 comprising two or three longitudinal kinety rows that are progressively shortened at anterior ends (five cells of 25 examined having three-rowed M1) (Figures 2C,E,J,N). Rightmost row in M1 composed of eight or nine basal bodies, commencing anteriorly about one-sixth down length of cell; second row with seven or eight basal bodies; third row containing five or six basal bodies (Figures 2C,E,J,N). M2 irregular pentagon-shaped, usually consisting of two parts: upper left part with basal bodies arranged in triangle shape, and lower part with five or six horizontal kinety rows basically arranged in rectangle shape (Figures 2C,E,J,N). M3 slightly distant from M2, horizontally oriented, with six to 10 basal bodies (Figures 2C,E,J,N). PM on right side of oral groove, with basal bodies arranged in zig-zag pattern, length about one-quarter to one-third of cell length, anterior end commencing at level of middle portion of lower (rectangular) part of M2, posterior end terminating in mid-region of cell (Figures 2C,E,J,N). Scutica (Sc) located below posterior end of PM, usually consisting of three pairs of basal bodies (Figure 2C) or two pairs with additional one or two posteriorly located basal bodies (Figures 2C,E,J,N).
Genus: Homalogastra Kahl, 1926
Homalogastra binucleata sp. nov.
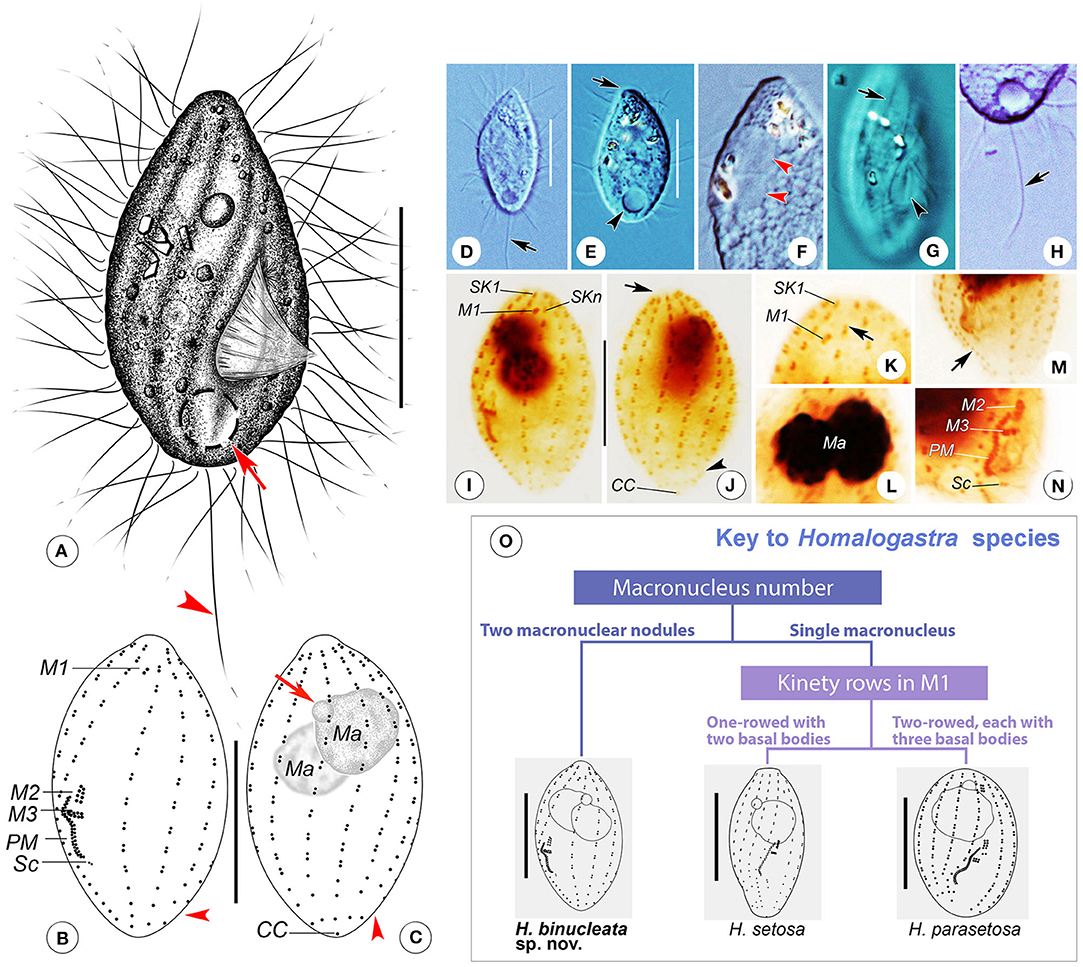
Figure 3. (A–O) Homalogastra binucleata sp. nov. from life (A,D–H), after protargol staining (B,C,I–N) and key to species of Homalogastra (O). (A) Right ventrolateral view of a representative individual, arrow shows the contractile vacuole, arrowhead points to the caudal cilium. (B,C) Left ventrolateral (B) and right dorsolateral (C) views of the holotype specimen, arrowheads in B and C indicate the gap between last two basal bodies in somatic kineties, arrow in C points to the micronucleus. (D,E) Cells showing the variations in body shape, arrow in D marks the caudal cilium, arrow in E indicates the apical plate, arrowhead shows the contractile vacuole. (F) Left lateral view of a compressed cell, arrowheads show the two macronuclear nodules. (G) Detail of pellicle, arrow marks the grooves on the ciliary rows, arrowhead points to the buccal field. (H) Detailed view, arrow indicates the caudal cilium. (I,J) Left ventrolateral (I) and right dorsolateral (J) views of the holotype specimen, arrow in J shows the apical plate and arrowhead points to the gap between last two basal bodies in somatic kineties. (K) Anterior ventral side of protargol-stained cell, arrow indicates the shortened anterior end of SKn. (L) Nuclear apparatus, showing the two macronuclear nodules. (M) Posterior half of stained cell, arrow points to the densely arranged monokinetids in SK1. (N) Oral apparatus except for membranelle 1. (O) A species key of Homalogastra. Two congeners are redrawn based on previous studies (Foissner et al., 1982; Pomp and Wilbert, 1988). CC, caudal cilium; M1–3, membranelle 1–3; Ma, macronucleus; PM, paroral membrane; Sc, scutica; SK1, somatic kinety 1; SKn, somatic kinety n. Scale bars: 12 μm.
ZooBank. urn:lsid:zoobank.org:act:F828E465-8119-4D59-9DD8-6B58C7870678
Diagnosis. Body spindle- or pear-shaped, about 20–30 × 10–15 μm in vivo; invariably two spherical macronuclear nodules; 11 somatic kineties; membranelle 1 highly reduced with only one pair of basal bodies, conspicuously separated from other membranelles; single caudal cilium; contractile vacuole caudally positioned; freshwater or brackish water habitat.
Etymology. The species-group name binucleata (having two nuclei) is a composite of the Latin numeral bi- (Latin numeral; two) and nucleatus, -a, -um [Latin adjective (m; f; n); kernel-like], and indicates the two macronuclear nodules, a diagnostic feature of the species.
Type locality and habitat. A brackish water sewage ditch in Lanzhou (36°05'37” N; 103°44'58” E), China. Salinity 2‰, water, temperature about 20°C.
Deposition of type slides. One protargol slide containing the holotype specimen (registration number: QZS2017043001-1) and one protargol slide with paratype specimens (registration number: QZS2017043001-2) were deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China.
SSU rRNA gene sequence. The length is 1643 bp, G + C content 43.64% and GenBank accession number MT982808.
Description. Body about 20–30 × 10–15 μm in vivo, spindle-shaped, with oral groove in mid- to posterior region, cytostome located about two-thirds down length of cell (Figures 3A,D,E,G). Anterior end conspicuously pointed, with a small apical plate (Figures 3A,D–F). Pellicle with twisted shallow grooves along cilia rows (Figures 3A,G). No extrusomes detectable. Cytoplasm colorless, usually containing several irregular-shaped crystals in anterior half of cell (Figures 3D–F). One contractile vacuole caudally located, about 4–5 μm in diameter during diastole, pulsating at intervals of 35–45 s (Figures 3A,D,E,H). Invariably two spherical macronuclear nodules (in 46 individuals examined), closely apposed, each nodule about 5 μm in diameter (Figures 3C,I,J,L). Usually one micronucleus, about 1.5 μm in diameter, in anterior half of body. Somatic cilia about 7 μm long; single caudal cilium about 15–20 μm in length (Figures 3A,H).
Locomotion by rapid swimming while rotating continuously about longitudinal body axis or by crawling on substrate.
Constantly 11 somatic kineties (Figures 3B,C,I,J). Anterior ends commencing below apical plate except for SKn, which shortened anteriorly and commencing slightly above level of M1 (Figures 3B,I). SK1 comprising dikinetids in anterior three-fifths of body and densely arranged monokinetids in posterior two-fifths (Figures 3B,I,M). Other kineties composed of dikinetids in anterior four-fifths of kinety and loosely arranged monokinetids in posterior one-fifth (Figures 3B,C,I,J). Conspicuous gap between last two basal bodies in each somatic kinety with exception of SK1 and SKn (Figures 3B,C,I,J). SK1 composed of 18–23 kinetids of which 9–13 are monokinetids; mid somatic kinety on dorsal side composed of 14–18 kinetids including 2–4 monokinetids; SKn composed of 13–15 kinetids including 3–5 monokinetids. Caudal cilium monokinetid.
Oral apparatus typical of genus. M1 with two basal bodies longitudinally arranged, located apically in buccal field and clearly separated from M2 and M3 (Figures 3B,I,K). M2 two-rowed, each row with four basal bodies, situated in mid-region of cell (Figures 3B,I,N). M3 also two-rowed, each row comprising about eight to 10 basal bodies, horizontally oriented and located close to M2 (Figures 3B,I,N). Paroral membrane on right side of oral groove, two-rowed with basal bodies arranged in a zig-zag pattern; anterior end commencing at level of middle portion of M2, terminating posteriorly about two-thirds down length of cell, occupying about one-fifth of body length (Figures 3B,I,N). Scutica located below posterior end of paroral membrane, consisting of one pair of basal bodies (Figures 3B,N).
Family: Uronematidae Thompson, 1964
Genus: Uronema Dujardin, 1841
Uronema orientalis Pan et al., 2015
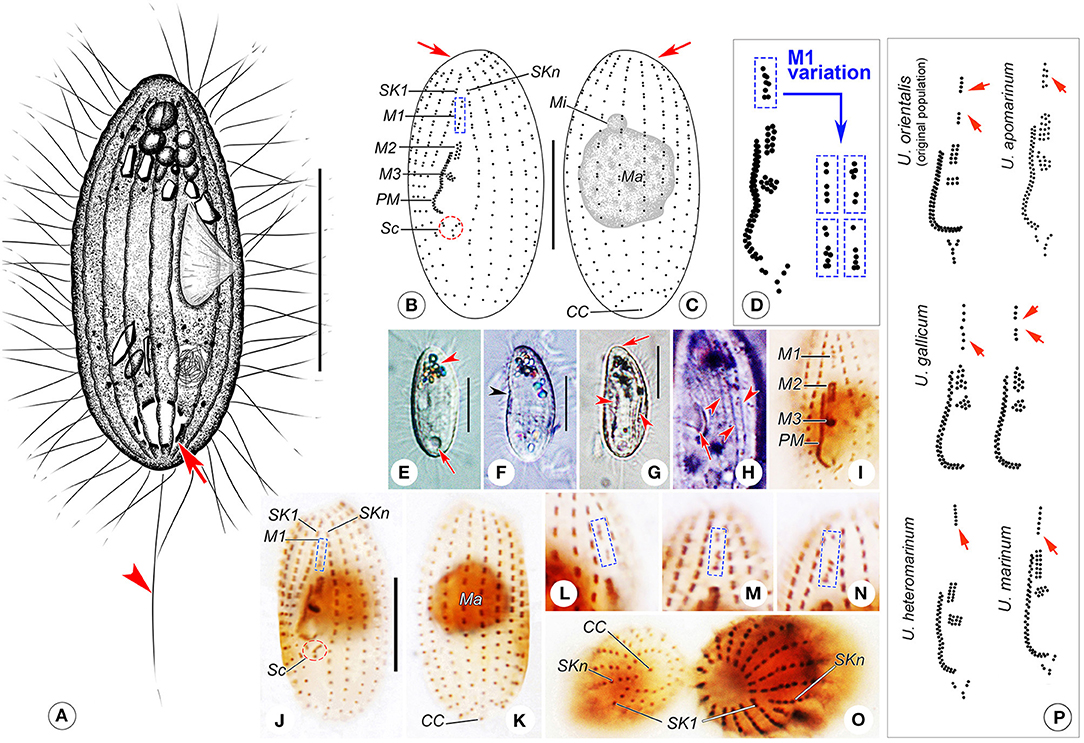
Figure 4. (A–P) Uronema orientalis Pan et al., 2015 from life (A,E–H), after protargol staining (B–D,I–O), and comparison of the oral apparatus with that in related population/species (P). (A) Right ventrolateral view of a representative cell, arrow shows the contractile vacuole and arrowhead points to the caudal cilium. (B,C) Ventral (B) and dorsal (C) views of a specimen with a gap in the mid-region of M1, arrows indicate the apical plate. (D) Oral apparatus of the Qingdao population of Uronema orientalis, and the variation of M1 structure. (E) Left ventrolateral view, arrow marks the contractile vacuole, arrowhead shows the clustered crystals at anterior of cell. (F) Left lateral view, arrowhead indicates the depressed buccal field on ventral side. (G) Individual with prominent depressions (arrowheads), arrow shows the apical plate. (H) Detail of pellicle, showing the buccal field (arrow) and ridges between ciliary rows (arrowheads). (I) Oral apparatus of a protargol-stained cell. (J,K) Ventral (J) and dorsal (K) views of a protargol-stained specimen, showing the ciliature and nuclear apparatus. (L–N) Variety of M1 structures that are bipartite. (O) Protargol-stained cells, revealing the structures of the posterior end (left cell) and the anterior end (right cell). (P) Comparison of oral apparatus with that in related population/species, all redrawn based on previous studies (Uronema orientalis, original population: Pan et al., 2015a; U. apomarinum: Liu et al., 2020; U. gallicum: Pérez-Uz and Song, 1995; U. heteromarinum: Pan et al., 2010; U. marinum: Song et al., 2009), arrows point to M1. CC, caudal cilium; M1–3, membranelle 1–3; Ma, macronucleus; Mi, micronucleus; PM, paroral membrane; Sc, scutica; SK1, somatic kinety 1; SKn, somatic kinety n. Scale bars: A, E–G: 15 μm; B, C, I–K: 10 μm.
Improved diagnosis. Marine Uronema with 15 to 20 somatic kineties, single macronucleus and single micronucleus; body size about 25–55 × 12–30 μm in vivo, with narrowed anterior end; cytostome constantly sub-equatorial; membranelle 1 one-rowed or partly two-rowed, occasionally divided into two parts; membranelle 2 two-rowed; contractile vacuole caudally located, contractile vacuole pore positioned at end of the second somatic kinety.
Deposition of voucher slides. Three voucher slides containing protargol-stained specimens (registration numbers: LMJ2017031004, LMJ2017031003-1, LMJ2017031003-2) were deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China.
SSU rRNA gene sequence. The length is 1633 bp, G + C content 42.38% and GenBank accession no. MT982806.
COI gene sequence. The length is 764 bp, G + C content 27.75% and GenBank accession no. MT981149.
Description of the Qingdao population. Body about 25–40 × 12–20 μm in vivo, basically cylindrical in shape, anterior end truncated, with a glabrous apical plate about one-third of maximum body width, posterior end rounded (Figures 4A,E,G). Buccal cavity about one-quarter of body length, cytostome located in mid-body region (Figures 4A,E,F). Pellicle thin with inconspicuous notches and, in some individuals, prominent depressions on ventral and dorsal sides (Figure 4G). Extrusomes not detected. Cytoplasm colorless to grayish. Several irregular-shaped crystals usually clustered in anterior third of cell, forming a “black spot” when viewed at low magnification (Figures 4A,E–G). One spherical macronucleus, 8–12 μm in diameter, located in mid-region of cell; single spherical micronucleus, about 2 μm in diameter, adjacent to macronucleus (Figures 4C,K). One caudally located contractile vacuole, about 5 μm in diameter, pulsating at intervals of 20–50 s (Figures 4A,E). Somatic cilia about 5–7 μm in length; single caudal cilium about 12–15 μm long (Figure 4A).
Locomotion by swimming moderately fast without fixed pattern, or by crawling on substrates; sometimes suspended in water with anterior end moving in circles.
Fifteen to 17 somatic kineties, usually 16 (only two cells with 15 somatic kineties out of 115 cells examined). Each kinety commencing around apical plate and extending almost to rear end of cell (Figures 4B,C,I–K,O). SKn extending posteriorly further than other somatic kineties (Figures 4B,O). Generally, kineties composed of closely arranged dikinetids in anterior half and loosely arranged monokinetids in posterior half (Figures 4B,C,I–K). First dikinetid in SKn closely apposed to that in SK1, leaving a conspicuous space between first two dikinetids in SKn (Figures 4B,J,O). SK1 composed of 16–22 kinetids of which 4–13 are monokinetids; middle somatic kinety on dorsal side composed of 13–21 kinetids including 2–18 monokinetids; SKn composed of 15–19 kinetids including 3–11 monokinetids. Caudal cilium monokinetid (Figures 4C,K,O).
Oral apparatus typical of genus. M1 located about one-fifth down length of cell, about equal to M2 in length; in most cases, M1 partly two-rowed, that is, about eight basal bodies arranged in five transverse rows of which first and fourth rows each has only one basal body (Figure 4D). Occasionally (in eight out of 79 individuals examined) with gap in anterior or mid-portion dividing M1 into two parts, five or six transverse rows in total, making M1 slightly longer than M2 (Figures 4B,D,J,L–N). M2 located about one-third down length of cell, consisting of two equal-length longitudinal kinety rows (Figures 4B,D,I,J). M3 comprising about eight to 10 basal bodies arranged in a small patch below M2 (Figures 4B,D,I,J). Paroral membrane on right side of buccal field, comprising two rows of basal bodies arranged in a zig-zag pattern, about one-fifth of body length, commencing anteriorly at level of mid-portion of M2 and terminating posteriorly about two-thirds down length of cell (Figures 4B,D,I,J). Scutica located below posterior end of PM, usually consisting of three pairs of basal bodies arranged in Y-shape (Figures 4B,D,I,J).
Molecular Phylogenies and Sequence Comparisons
The topologies of the SSU rRNA gene trees constructed by two ML methods (RAxML and IQTREE) and one BI method (MrBayes) are generally congruent. Therefore, only the RAxML tree is presented here with support values from all algorithms (Figure 5).
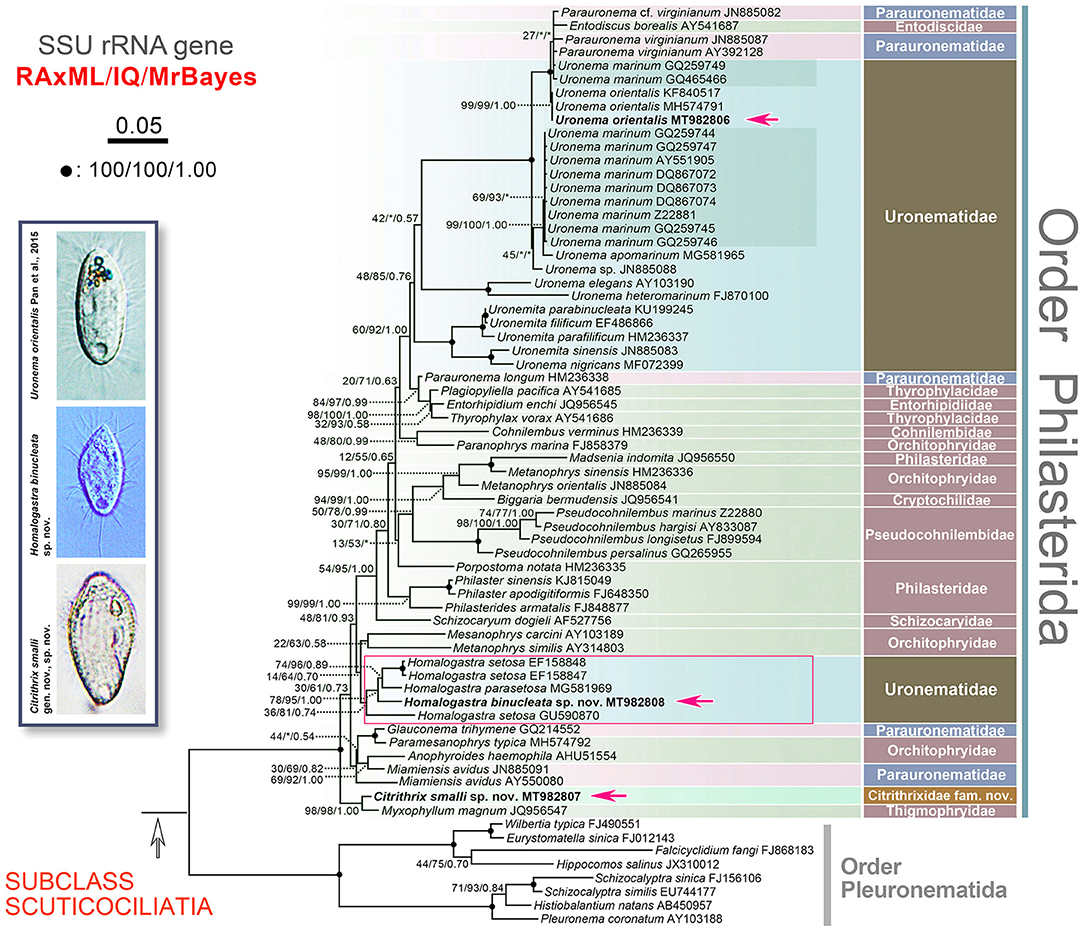
Figure 5. RAxML tree inferred from SSU rRNA gene sequence data, red arrows point out the three newly sequenced populations. Numbers at nodes denote RAxML bootstrap values/IQTREE ultrafast bootstrap values/Bayesian inferences posterior probabilities. Asterisks (*) indicate mismatches of the topologies between RAxML, IQTREE and/or Bayesian inferences conducted in MrBayes. Fully supported (100%/100%/1.00) nodes are marked with black dots. The scale bar corresponds to five substitutions per 100 nucleotide positions. All branches are drawn to scale. The systematic classification mainly follows Lynn (2008).
Citrithrix smalli sp. nov. MT982807 clusters with Myxophyllum magnum JQ956547 with strong to full support (98% RAxML, 98% IQ, 1.00 MrBayes) (Figure 5). The clade formed by C. smalli and M. magnum, a highly specialized parasitic form, then clusters with full support (100% RAxML, 100% IQ, 1.00 MrBayes) with the group containing all other representative sequences of the order Philasterida used in this study (Figure 5). Currently, few data are available to infer the evolutionary relationships and taxonomic rank of the C. smalli + M. magnum clade, but this basal branch very likely represents a lineage at about suborder level.
We compared the sequence identity of the SSU rRNA gene among C. smalli sp. nov., M. magnum, Anophyroides haemophila, Miamiensis avidus, Glauconema trihymene and Paramesanophrys typica, and found that C. smalli sp. nov. MT982807 differs in 28–71 nucleotides from the others with a sequence identity ranging from 95.6 to 98.2% (Figure 6A).
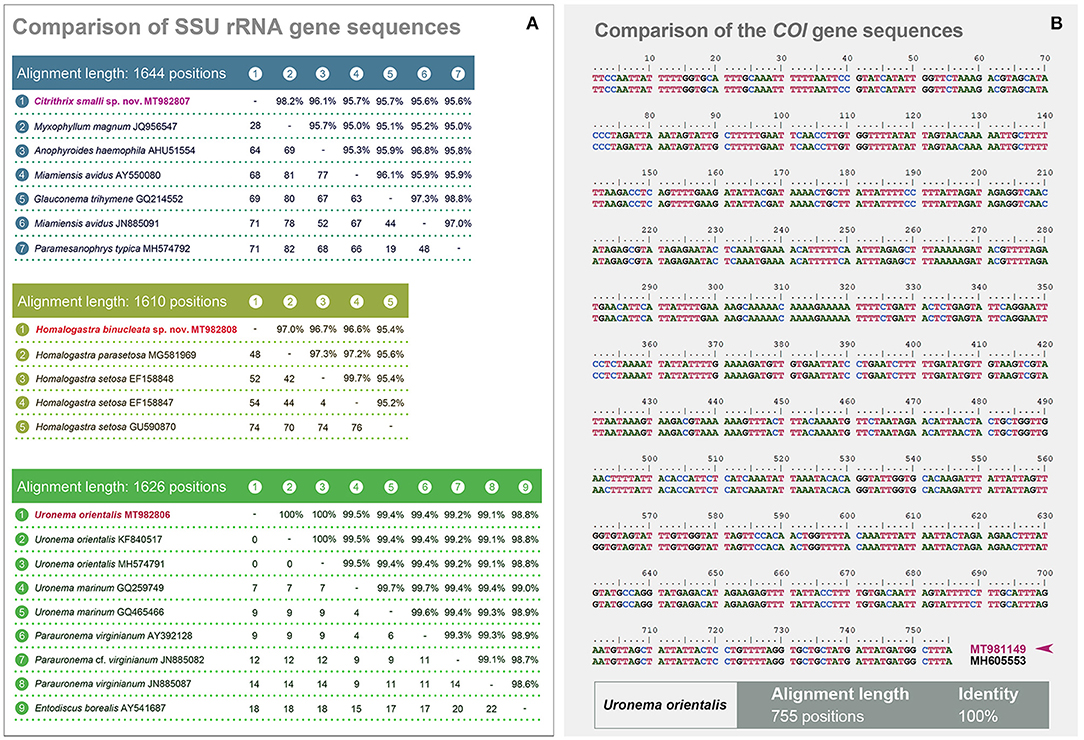
Figure 6. (A, B) Comparisons of SSU rRNA and the COI gene sequences. (A) Comparisons of the SSU rRNA gene sequences of Citrithrix smalli sp. nov. (upper matrix), Homalogastra binucleata sp. nov. (middle matrix), and Uronema orientalis Pan et al., 2015 (lower matrix), with their related sequences. The lengths of each alignment after trimming both ends are shown. The lower left values in each matrix are numbers of unmatched nucleotides, while the upper right numbers indicate the sequence identity. (B) Comparison of the COI gene sequences between Uronema orientalis MT981149 (from the present study, with arrowhead) and Uronema orientalis MH605553 (from the original population). The final alignment length is 755 positions, with a 100% sequence identity.
All five sequences of the genus Homalogastra group together forming a clade in the SSU rRNA gene tree (Figure 5). Homalogastra binucleata sp. nov. falls outside the group formed by H. setosa EF158847, H. setosa EF158848, and H. parasetosa MG581969, with medium to full support (78% RAxML, 95% IQ, 1.00 MrBayes). However, H. setosa GU590870 branches separately from these four Homalogastra sequences rather than clustering with the two strains of H. setosa. In the SSU rRNA gene sequence alignment of the five Homalogastra species/strains, H. binucleata sp. nov. MT982808 differs from the other Homalogastra sequences in 48 to 74 nucleotides and has a sequence identity ranging from 95.4 to 97.0% (Figure 6A).
Apart from Homalogastra, other species/strains of Uronematidae cluster with Parauronematidae and Entodiscidae (48% RAxML, 85% IQ, 0.76 MrBayes). The genus Uronema is not monophyletic (Figure 5). The newly sequenced Uronema orientalis MT982806 clusters with two U. orientalis sequences (KF840517, MH574791) from the same population (Pan et al., 2015a) with high or full statistical support (99% RAxML, 99% IQ, 1.00 MrBayes), and then groups with the Parauronematidae + Entodiscidae + Uronema marinum (GQ259749; GQ465466) clade (Figure 5). Sequence comparison of the SSU rRNA gene shows that there is no difference between U. orientalis MT982806 and two previous U. orientalis sequences (KF840517, MH574791) (Figure 6A). The COI gene sequence of the present population of U. orientalis (MT981149) is identical to that of the original population (MH605553) after trimming both ends of the alignment, giving 755 nucleotides in the final alignment (Figure 6B).
Discussion
Morphological Comparison and Phylogenetic Analyses of Citrithrix smalli gen. nov., sp. nov.
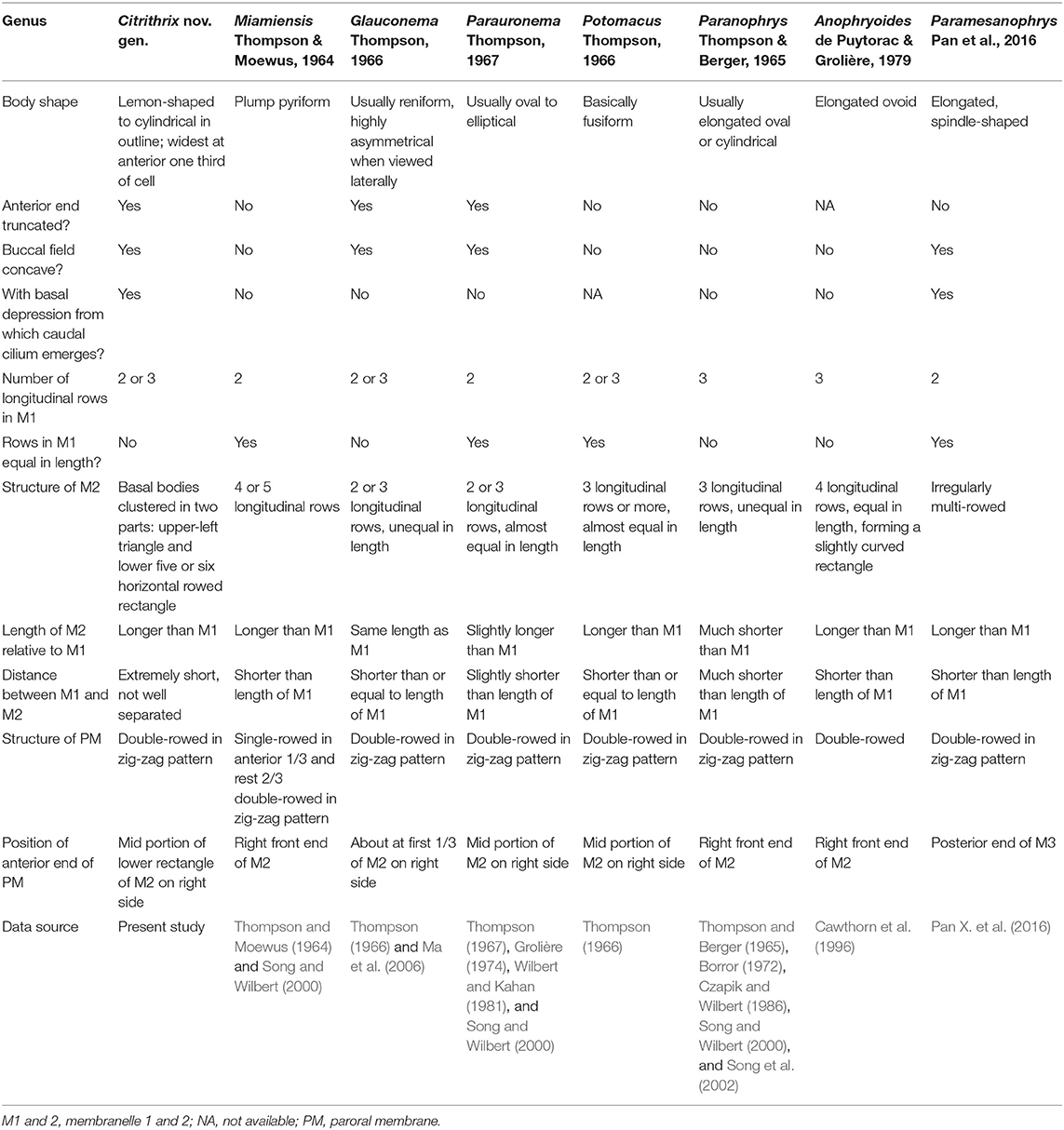
Citrithrix smalli gen. nov., sp. nov. has a distinct oral structure (extremely close-set M1 and M2 when compared with morphologically similar genera) which slightly resembles that of parauronematids (Figure 2O). However, the new taxon can be distinguished from all four known genera of parauronematids [i.e., Miamiensis, Glauconema, Parauronema, and Potomacus (Table 2)] by the long, conspicuously deep oral groove (vs. absent or inconspicuous in the latter four genera) and the closely apposed M1 and M2 (vs. M1 and M2 clearly separated in the latter four genera) (Thompson and Moewus, 1964; Thompson, 1966, 1967; Grolière, 1974; Wilbert and Kahan, 1981; Song and Wilbert, 2000; Ma et al., 2006). In addition, Citrithrix gen. nov. is not closely related to the parauronematids in the SSU rRNA gene tree (Figure 5).
Three genera in the family Orchitophryidae, namely Paranophrys, Anophryoides, and Paramesanophrys, should be compared with Citrithrix gen. nov. since the M1 and M2 of all these genera are multi-rowed (Table 2). Citrithrix gen. nov. differs from them mainly in the presence of an oral groove, the arrangement of M1 and M2, and the position of the anterior end of the PM (Thompson and Berger, 1965; Borror, 1972; Czapik and Wilbert, 1986; Cawthorn et al., 1996; Song and Wilbert, 2000; Song et al., 2002; Pan X. et al., 2016).
These morphological differences are reflected in the molecular analyses in the present study: Citrithrix smalli sp. nov. is well-separated from the families Parauronematidae, Orchitophryidae and Philasteridae, which belong to the core part of the order Philasterida, and occupies the basal position within the order suggesting that Citrithrix gen. nov. may represent the ancestral group of philasterids (Figure 5).
Although Citrithrix smalli sp. nov. clusters with Myxophyllum magnum JQ956547 and shows a sequence identity of 98.2% (Figures 5, 6A), the former differs significantly from the latter in its morphology and ecology, e.g., the number of somatic kineties, the oral structure, and the free-living (vs. parasitic) life-style (Xu and Song, 2000).
In conclusion, both the morphological and the molecular data indicate that our new taxon cannot be assigned to any extant family. A new family, Citrithrixidae fam. nov., is thus proposed within the order Philasterida.
Morphological Comparison and Phylogenetic Analyses of Homalogastra binucleata sp. nov.
Based on its body size and shape, conspicuous oral groove and oral structure, Homalogastra binucleata sp. nov. corresponds well with the genus diagnosis of Homalogastra (Liu et al., 2020). Hitherto there are only two known congeners, i.e., H. setosa (type species) and H. parasetosa (Table 3).
Homalogastra binucleata sp. nov. can easily be separated from both congeners by having two (vs. one) macronuclear nodules. In addition, it differs from H. parasetosa in the structure of its M1 which comprises a pair of basal bodies in H. binucleata sp. nov. (and H. setosa) vs. M1 two-rowed, each row with three basal bodies, in H. parasetosa (Figure 3O) (Kahl, 1931; Buitkamp, 1977; Foissner et al., 1982; Pomp and Wilbert, 1988; Alekperov, 2005; Liu et al., 2020).
In the SSU rRNA gene tree, H. binucleata sp. nov. nests within the Homalogastra clade (Figure 5). The SSU rRNA gene sequence of our new species has the highest sequence identity with H. parasetosa MG581969 (97%), although there are 48 unmatched nucleotides (Figure 6A). Therefore, clarification of the phylogenetic relationships among Homalogastra species awaits data from more gene markers and from additional populations and species. Such data might also be helpful to determine how their morphological characteristics reflect their evolutionary relationships.
Morphological Comparison and Phylogenetic Analyses of Qingdao Population of Uronema orientalis Pan et al., 2015
Uronema orientalis was established by Pan et al. (2015a) mainly according to its bipartite M1 and its high number of somatic kineties. The Qingdao population resembles the original population in cell size, shape and oral structure, but it differs from the latter by having: fewer somatic kineties, i.e., 15–17, usually 16 (vs. constantly 20 in the type population); each kinety with mono- and dikinetids (vs. each kinety usually comprises monokinetids only in the type population); M1 partly two-rowed that occasionally with a gap (vs. one-rowed and divided by a gap, but the stability of M1 structure is not available in the original report). Besides, the extrusomes in the current population are not detected (vs. bar-shaped extrusomes, 4 μm in length in the original population) (Pan et al., 2015a) (Figures 4D,J,L–N,P).
Based on the morphological differences, the current population can be a new subspecies of Uronema orientalis. However, the locations of the inhabitation of the current and the original populations are not geographically separate. In addition, the sequence comparisons of the SSU rRNA and COI genes of the two populations show that the sequences of each gene are identical (Figure 6), strongly supporting the identity of the current population as U. orientalis. Therefore, it is reasonable to treat the current population as U. orientalis Pan et al., 2015 with some morphological variations.
Morphological Comparison of Uronema orientalis Pan et al., 2015 With Closely Related Congeners
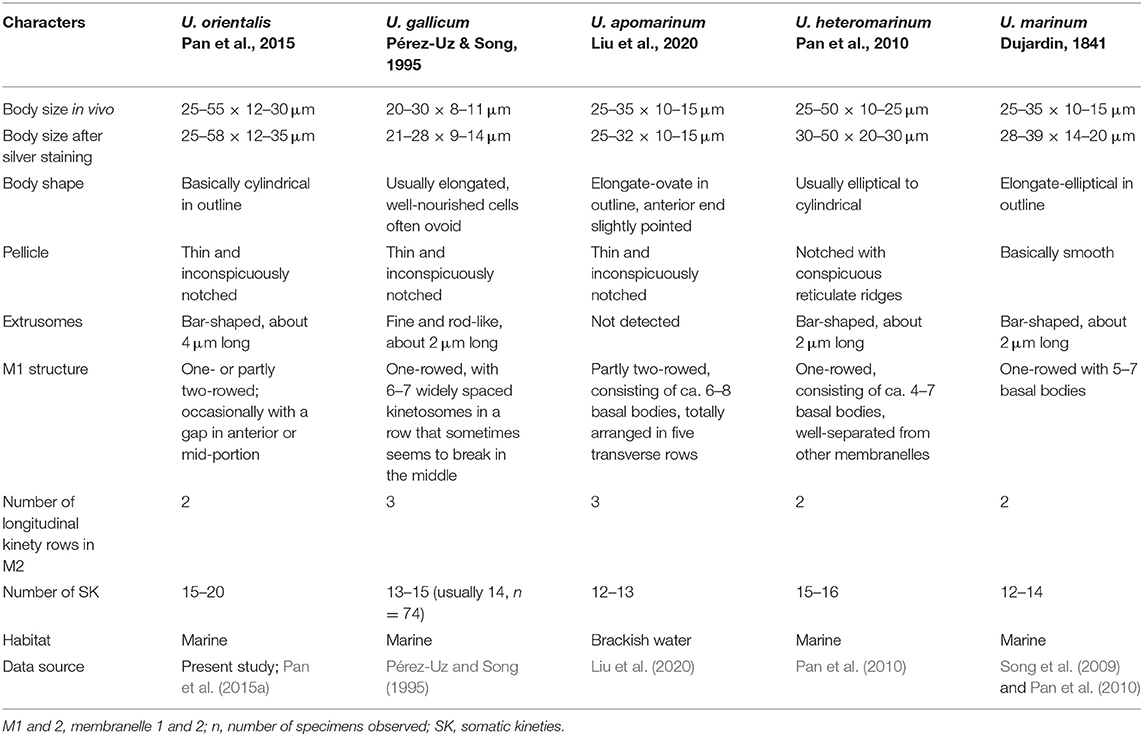
Considering the structure of the oral apparatus and the somatic ciliature, four Uronema species should be compared with Uronema orientalis Pan et al., 2015, namely U. gallicum Pérez-Uz and Song, 1995, U. apomarinum Liu et al., 2020, U. heteromarinum Pan et al., 2010, and U. marinum Dujardin, 1841 (Figure 4P; Table 4).
Uronema orientalis most closely resembles U. gallicum in the M1 structure, that is, occasionally bipartite with a gap in the anterior or mid-region of M1 dividing it into two parts (Figures 4D,J,L–N,P). However, the latter can be distinguished from the former by its slimmer body shape (body width in vivo 8–11 μm vs. 12–30 μm in U. orientalis), and in having fewer somatic kineties (13–15 vs. 15–20 in U. orientalis) (Pérez-Uz and Song, 1995; Pan et al., 2015a).
Uronema orientalis can be separated from the other three Uronema species mainly by the number of somatic kineties and by the structure of M1 and M2 (Song et al., 2009; Pan et al., 2010; Liu et al., 2020) (Figures 4B–D,P; Table 4).
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
XH and WS conceived and designed the paper. ML, ZQ, and LJ carried out the live observation and protargol staining. CW analyzed the data. ML, CW, XH, ZQ, LJ, SA-F, HE-S, AW, and WS wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Marine S and T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0406-1), the National Natural Science Foundation of China (project numbers: 41976086, 41706168), and the Researchers Supporting Project Number (RSP-2020/7), King Saud University, Riyadh, Saudi Arabia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many thanks are due to Ms. Jiyang Ma (OUC) for providing the samples, and to Dr. Zhe Wang for the advice on the IQTREE 2.0.6 program.
References
Aescht, E. (2001). Catalogue of the Generic Names of Ciliates (Protozoa, Ciliophora). Linz: Biologiezentrum des Oberösterreichischen Landesmuseums.
Alekperov, I. (2005). Atlas Svobodnozhivushchikh Infuzorii (Atlas of Free-living Ciliates). Baku: Institute of Zoology NAS of Azerbaijan.
Bai, Y., Wang, R., Song, W., Suzuki, T., and Hu, X. (2020). Redescription of five tintinnine ciliates (Alveolata: Ciliophora: Oligotrichea) from coastal waters of Qingdao, China. Mar. Life Sci. Technol. 2, 209–221. doi: 10.1007/s42995-020-00034-2
Borror, A. C. (1972). Tidal marsh ciliates (Protozoa): morphology, ecology, systematics. Acta Protozool. 10, 29–71.
Buitkamp, U. (1977). Die Ciliatenfauna der Savanne von Lamto (Elfenbeinküste). Acta Protozool. 16, 249–276.
Cawthorn, R. J., Lynn, D. H., Despres, B., MacMillan, R., Maloney, R., Loughlin, M., et al. (1996). Description of Anophryoides haemophila n. sp. (Scuticociliatida: Orchitophryidae), a pathogen of American lobsters Homarus americanus. Dis. Aquat. Organ. 24, 143–148. doi: 10.3354/dao024143
Chi, Y., Duan, L., Luo, X., Cheng, T., Warren, A., Huang, J., et al. (2020). A new contribution to the taxonomy and molecular phylogeny of three, well-known freshwater species of the ciliate genus Spirostomum (Protozoa: Ciliophora: Heterotrichea). Zool. J. Linn. Soc. 189, 158–177. doi: 10.1093/zoolinnean/zlz115
Czapik, A., and Wilbert, N. (1986). Sur une nouvelle espèce de cilié Paranophrys carnivora sp. n. (Scuticociliatida). Acta Protozool. 25, 427–432.
Foissner, W., Adam, H., and Foissner, I. (1982). Morphologie, infraciliatur und silberliniensystem einiger wenig bekannter Scuticociliatida (Protozoa: Ciliophora). Zool. Jb. Syst. 109, 443–468.
Foissner, W., Berger, H., and Kohmann, F. (1994). Taxonomische und ökologische revision der ciliaten des saprobiensystems - Band III: Hymenostomata, Prostomatida, Nassulida. Informationsber. Bayer. Landesamtes Wasserwirtschaft 1/94, 1–548.
Gao, F., Gao, S., Wang, P., Katz, L. A., and Song, W. (2014). Phylogenetic analyses of cyclidiids (Protista, Ciliophora, Scuticociliatia) based on multiple genes suggest their close relationship with thigmotrichids. Mol. Phylogenet. Evol. 75, 219–226. doi: 10.1016/j.ympev.2014.01.032
Gao, F., Huang, J., Zhao, Y., Li, L., Liu, W., Miao, M., et al. (2017). Systematic studies on ciliates (Alveolata, Ciliophora) in China: progress and achievements based on molecular information. Eur. J. Protistol. 61, 409–423. doi: 10.1016/j.ejop.2017.04.009
Gao, F., Katz, L. A., and Song, W. (2012). Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol. Phylogenet. Evol. 64, 308–317. doi: 10.1016/j.ympev.2012.04.008
Gao, F., Katz, L. A., and Song, W. (2013). Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 68, 55–63. doi: 10.1016/j.ympev.2013.03.018
Gao, F., Warren, A., Zhang, Q., Gong, J., Miao, M., Sun, P., et al. (2016). The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6:24874. doi: 10.1038/srep24874
Gong, J., Choi, J. K., Roberts, D. M. L., Kim, S. Y., and Min, G. S. (2007). Morphological descriptions of new and little-known benthic ciliates from Ganghwa tidal flat, Korea. J. Eukaryot. Microbiol. 54, 306–316. doi: 10.1111/j.1550-7408.2007.00268.x
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Grolière, C. A. (1974). Étude comparée de la stomatogenèse chez quelques cilies hymenostomes des genres Paralembus Kahl, 1933 Philaster Fabre-Domergue, 1885 Parauronema Thompson, 1967, Tetrahymena Furgasson, 1940. Protistologica 10, 319–331.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hu, X., Lin, X., and Song, W. (2019). Ciliate Atlas: Species Found in the South China Sea. Beijing: Science Press. doi: 10.1007/978-981-13-5901-9
Jankowski, A. V. (2007). “Phylum Ciliophora Doflein, 1901. Review of taxa,” in Protista: Handbook on Zoology, ed A. F. Alimov (St. Petersburg: Nauka), 415–993.
Jerome, C. A., Simon, E. M., and Lynn, D. H. (1996). Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can. J. Zool. 74, 1898–1906. doi: 10.1139/z96-214
Kahl, A. (1931). Urtiere oder Protozoa I: wimpertiere oder Ciliata (Infusoria) 2. Holotricha. Tierwelt Dtl. 21, 181–398.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Liu, M., Li, L., Zhang, T., Fan, X., Yi, Z., and Lin, X. (2020). Two new scuticociliates from southern China: Uronema apomarinum sp. nov. and Homalogastra parasetosa sp. nov., with improved diagnoses of the genus Homalogastra and its type species Homalogastra setosa (Ciliophora, Oligohymenophorea). Int. J. Syst. Evol. Microbiol. 70, 2405–2419. doi: 10.1099/ijsem.0.004046
Liu, W., Jiang, J., Xu, Y., Pan, X., Qu, Z., Luo, X., et al. (2017). Diversity of free-living marine ciliates (Alveolata, Ciliophora): faunal studies in coastal waters of China during the years 2011–2016. Eur. J. Protistol. 61, 424–438. doi: 10.1016/j.ejop.2017.04.007
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature. 3rd Edn. Dordrecht: Springer.
Lynn, D. H., and Strüder-Kypke, M. (2005). Scuticociliate endosymbionts of echinoids (phylum Echinodermata): phylogenetic relationships among species in the genera Entodiscus, Plagiopyliella, Thyrophylax, and Entorhipidium (phylum Ciliophora). J. Parasitol. 91, 1190–1199. doi: 10.1645/GE-445R.1
Lynn, D. H., and Strüder-Kypke, M. C. (2006). Species of Tetrahymena identical by small subunit rRNA gene sequences are discriminated by mitochondrial cytochrome c oxidase I gene sequences. J. Eukaryot. Microbiol. 53, 385–387. doi: 10.1111/j.1550-7408.2006.00116.x
Ma, H., Song, W., Warren, A., Roberts, D., Gong, J., and Al-Rasheid, K. A. S. (2006). Redescription of the marine scuticociliate Glauconema trihymene Thompson, 1966 (Protozoa: Ciliophora): life cycle and stomatogenesis. Zootaxa 1296, 1–17. doi: 10.11646/zootaxa.1296.1.1
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Pan, H., Hu, J., Jiang, J., Wang, L., and Hu, X. (2016). Morphology and phylogeny of three Pleuronema species (Ciliophora, Scuticociliatia) from Hangzhou Bay, China, with description of two new species, P. binucleatum n. sp. and P. parawiackowskii n. sp. J. Eukaryot. Microbiol. 63, 287–298. doi: 10.1111/jeu.12277
Pan, H., Huang, J., Hu, X., Fan, X., Al-Rasheid, K. A. S., and Song, W. (2010). Morphology and SSU rRNA gene sequences of three marine ciliates from Yellow Sea, China, including one new species, Uronema heteromarinum nov. spec. (Ciliophora, Scuticociliatida). Acta Protozool. 49, 45–59.
Pan, M., Chen, Y., Liang, C., and Pan, X. (2020). Taxonomy and molecular phylogeny of three freshwater scuticociliates, with descriptions of one new genus and two new species (Protista, Ciliophora, Oligohymenophorea). Eur. J. Protistol. 74:125644. doi: 10.1016/j.ejop.2019.125644
Pan, X., Bourland, W. A., and Song, W. (2013). Protargol synthesis: an in-house protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Pan, X., Fan, X., Al-Farraj, S. A., Gao, S., and Chen, Y. (2016). Taxonomy and morphology of four “ophrys-related” scuticociliates (Protista, Ciliophora, Scuticociliatia), with the description of a new genus, Paramesanophrys gen. nov. Eur. J. Taxon. 2016, 1–18. doi: 10.5852/ejt.2016.191
Pan, X., Huang, J., Fan, X., Ma, H., Al-Rasheid, K. A. S., Miao, M., et al. (2015a). Morphology and phylogeny of four marine scuticociliates (Protista, Ciliophora), with descriptions of two new species: Pleuronema elegans spec. nov. and Uronema orientalis spec. nov. Acta Protozool. 54, 31–43.
Pan, X., Yi, Z., Li, J., Ma, H., Al-Farraj, S. A., and Al-Rasheid, K. A. S. (2015b). Biodiversity of marine scuticociliates (Protozoa, Ciliophora) from China: description of seven morphotypes including a new species, Philaster sinensis spec. nov. Eur. J. Protistol. 51, 142–157. doi: 10.1016/j.ejop.2015.02.005
Pérez-Uz, B., and Song, W. (1995). Uronema gallicum sp. n. (Protozoa: Ciliophora) a new marine scuticociliate from the coastal area of Calais. Acta Protozool. 34, 143–149.
Pomp, R., and Wilbert, N. (1988). Taxonomic and ecological studies of ciliates from Australian saline soils: colpodids and hymenostomate ciliates. Mar. Freshw. Res. 39, 479–495. doi: 10.1071/MF9880479
Qu, Z., Weinisch, L., Fan, X., Katzenmeier, S., Stoeck, T., and Filker, S. (2020). Morphological, phylogenetic and ecophysiological characterization of a new ciliate, Platynematum rosellomorai (Oligohymenophorea, Scuticociliatia), detected in a hypersaline pond on Mallorca, Spain. Protist 171:125751. doi: 10.1016/j.protis.2020.125751
Song, W. (2000). Morphological and taxonomical studies on some marine scuticociliates from China Sea, with description of two new species, Philasterides armatalis sp. n. and Cyclidium varibonneti sp. n. (Protozoa: Ciliophora: Scuticociliatida). Acta Protozool. 39, 295–322.
Song, W., Ma, H., Wang, M., and Zhu, M. (2002). Comparative studies on two closely related species Uronemella filificum (Kahl, 1931) and Uronema elegans Maupas, 1883 with redescription of Paranophrys marina Thompson et Berger, 1965 (Ciliophora: Scuticociliatida) from China seas. Acta Protozool. 41, 263–278.
Song, W., Warren, A., and Hu, X. (eds.) (2009). Free-living Ciliates in the Bohai and Yellow Seas, China. Beijing: Science Press.
Song, W., and Wilbert, N. (2000). Redefinition and redescription of some marine scuticociliates from China, with report of a new species, Metanophrys sinensis nov. spec. (Ciliophora, Scuticociliatida). Zool. Anz. 239, 45–74.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Thompson, J. C. (1966). Glauconema trihymene n. g., n. sp., a hymenostome ciliate from the Virginia coast. J. Protozool. 13, 393–395. doi: 10.1111/j.1550-7408.1966.tb01927.x
Thompson, J. C. (1967). Parauronema virginianum n. g., n. sp., a marine hymenostome ciliate. J. Protozool. 14, 731–734. doi: 10.1111/j.1550-7408.1967.tb02069.x
Thompson, J. C., and Berger, J. (1965). Paranophrys marina n. g., n. sp., a new ciliate associated with a hydroid from the northeast Pacific (Ciliata: Hymenostomatida). J. Protozool. 12, 527–531. doi: 10.1111/j.1550-7408.1965.tb03252.x
Thompson, J. C., and Moewus, L. (1964). Miamiensis avidus n. g., n. sp., a marine facultative parasite in the ciliate order Hymenostomatida. J. Protozool. 11, 378–381. doi: 10.1111/j.1550-7408.1964.tb01766.x
Wang, C., Zhang, T., Wang, Y., Katz, L. A., Gao, F., and Song, W. (2017). Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impact of copy number variation and experimental error. Proc. R. Soc. B Biol. Sci. 284:20170425. doi: 10.1098/rspb.2017.0425
Wang, Y., Wang, C., Jiang, Y., Katz, L. A., Gao, F., and Yan, Y. (2019). Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci. China Life Sci. 62, 203–214. doi: 10.1007/s11427-018-9422-5
Wilbert, N., and Kahan, D. (1981). Ciliates of Solar Lake on the Red Sea shore. Arch. Protistenkd. 124, 70–95. doi: 10.1016/S0003-9365(81)80004-8
Xu, K., and Song, W. (2000). Studies on pathogenetic ciliates from marine molluscs II. Philasterine ciliates (Protozoa, Ciliophora, Scuticociliatida). J. Ocean Univ. Qingdao 30, 224–229.
Zhang, T., Dong, J., Cheng, T., Duan, L., and Shao, C. (2020). Reconsideration of the taxonomy of the marine ciliate Neobakuella aenigmatica Moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia). Mar. Life Sci. Technol. 2, 97–108. doi: 10.1007/s42995-020-00032-4
Keywords: biodiversity, ciliated protozoa, ciliature, molecular systematics, new taxa
Citation: Liu M, Wang C, Hu X, Qu Z, Jiang L, Al-Farraj SA, El-Serehy HA, Warren A and Song W (2020) Taxonomy and Molecular Phylogeny of Three Species of Scuticociliates From China: Citrithrix smalli gen. nov., sp. nov., Homalogastra binucleata sp. nov. and Uronema orientalis Pan et al., 2015 (Protozoa, Ciliophora, Oligohymenophorea), With the Proposal of a New Family, Citrithrixidae fam. nov. Front. Mar. Sci. 7:604704. doi: 10.3389/fmars.2020.604704
Received: 10 September 2020; Accepted: 05 November 2020;
Published: 01 December 2020.
Edited by:
Hongbo Pan, Shanghai Ocean University, ChinaReviewed by:
Xuming Pan, Harbin Normal University, ChinaMann Kyoon Shin, University of Ulsan, South Korea
Copyright © 2020 Liu, Wang, Hu, Qu, Jiang, Al-Farraj, El-Serehy, Warren and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaozhong Hu, eGlhb3pob25naHVAb3VjLmVkdS5jbg==; Weibo Song, d3NvbmdAb3VjLmVkdS5jbg==
†These authors have contributed equally to this work
 Mingjian Liu
Mingjian Liu Chundi Wang
Chundi Wang Xiaozhong Hu1,2*
Xiaozhong Hu1,2* Zhishuai Qu
Zhishuai Qu Limin Jiang
Limin Jiang Weibo Song
Weibo Song