- 1Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China, Qingdao, China
- 2College of Life Sciences, Yantai University, Yantai, China
Heat shock proteins (hsps) are cellular chaperones that are involved in developmental stages and stress responses. Hsp40 is the major subfamily of hsps, but has not been fully characterized in Japanese flounder (Paralichthys olivaceus), especially their roles in immune response. In this study, a comprehensive identification and analysis of hsp40 in flounder is presented, including gene structures, evolutionary relationships, conserved domains, molecular evolution analysis, and expression patterns. Sequence features and phylogenetic analysis revealed that hsp40 genes could be grouped into 40 distinct subfamilies and most of them (96%) in Japanese flounder possessed no less than two introns. Molecular evolution analysis indicated that the hsp40 genes were conservative during evolution and were functional-constrained. Meanwhile, hsp40 genes were found to express in different embryonic and larval stages and might play the role of sentinel in healthy organisms. Furthermore, hsp40 genes’ expression profiles after Edwardsiella tarda injection were determined in Japanese flounder without precedent, and 88% (44/50) of hsp40 genes showed differential expression patterns after bacterial challenge. Our findings provide basic and useful resources for understanding the immune responsibilities of hsp40 genes in flatfish.
Introduction
In fisheries, fish are often affected by a variety of stresses during growth and development, including abiotic and biotic stresses such as poor water quality, thermal stress, environmental pollution, osmotic pressure, as well as bacterial and viral infections, which may influence the balance between fish and the environment and cause the stress responses of fish (Chen et al., 2010; Roberts et al., 2010; Zhang et al., 2011; Eissa, 2014; Eissa et al., 2018; Lee et al., 2018; Wen et al., 2019; Xu Z. N. et al., 2019). Over the course of long-term evolution, fish proceed changes in reaction to a variety of stressors, which are named the “general adaptation syndrome” (GAS) (Pickering, 1998). “Cellular stress response” is one feature of GAS that has been little studied.
Heat shock proteins (hsps), first reported in Drosophila under heat stress, are a superfamily of stress proteins expressed ubiquitously in most species from bacteria to human beings (Ritossa, 1962; Whitley et al., 1999). Although first discovered in response to thermal stress, hsps were also found to participate in a great number of stress conditions including radiation (UV), heavy metals, pesticides, hypoxia, oxygen radicals, anti-inflammatory drugs, malignant transformation, bacterial, and viral infection (Fuller et al., 1994; Sørensen et al., 2003; Gehrmann et al., 2004; Multhoff, 2006; Akira et al., 2008). Based on the molecular weight, sequence homology, and domain structures, the hsp superfamily can be classified into several subfamilies, including Hsp90, Hsp70/Hsp110, Hsp10/Hsp60, Hsp40, and Hsp20 (sHsp) families (Gething, 1997).
Hsp40 proteins (also referred to DnaJ proteins) constitute one of the largest subfamilies among the hsp superfamily. Each member of hsp40 proteins contains the J domain (JD), a 70-amino-acid domain with similarity to the initial 73 amino acids of the Escherichia coli hsp40 (Georgopoulos et al., 1980; Zylicz et al., 1985). The conserved JD is necessary for hsp40 to bind to hsp70 and regulate the ATPase activity of hsp70 proteins (Ohtsuka and Hata, 2000; Qiu et al., 2006; Li et al., 2009). Except for JD, members of E. coli hsp40 proteins typically have three other distinct regions: glycine/phenylalanine-rich region (G/F domain), cysteine-rich region (CRR domain), and variable C-terminal domain (CTD) (Bork et al., 1992; Hennessy et al., 2005; Qiu et al., 2006). Based on the homology of hsp40 proteins of E. coli, DnaJ proteins are divided into three categories: Type I DnaJ proteins (DnaJA) possess all four regions of DnaJ protein in E. coli; Type II hsp40 proteins (DnaJB) lack the CRR domain; and Type III hsp40 proteins (DnaJC) only possess the JD, which is not necessarily located at N-terminus of the protein (Cheetham and Caplan, 1998; Kampinga et al., 2009). The concept of type IV DnaJ protein family was raised, which owns a “J-like” domain (Walsh et al., 2004; Botha et al., 2007; Morahan et al., 2011) containing a wide range of mutations in a highly conserved histidine, proline, and aspartic acid–HPD motif located between helices II and III in the DnaJ domain (Tsai and Douglas, 1996; Mayer et al., 1999; Hennessy et al., 2000). Cooperating with hsp70, another molecular chaperone that couples the cycles of ATP binding, hydrolysis, and ADP release, the hsp40 proteins are involved in numerous cellular functions, including regulation of protein folding, translocation, and assembly (Cheetham and Caplan, 1998; Ohtsuka and Hata, 2000).
Except for the traditional functions, previous studies suggested that hsps may actually play important roles in immune reactions (Srivastava, 2002; Roberts et al., 2010). Hsps took part in humoral and cellular responses in innate immunity (Sung and MacRae, 2011) and also played a role as the danger signal in vitro to communicate innate immune responses by activating various cells (Chen et al., 1999; Kol et al., 2000; Singh-Jasuja et al., 2000). Besides, hsps can induce a variety of cytokines including interleukin-12, nitric oxide, tumor necrosis-a, interleukin-1β, and several chemokines (Basu et al., 2000; Lehner et al., 2000; Moré et al., 2001; Panjwani et al., 2002). Additionally, in adaptive immunity, hsps can also function as powerful danger signals and antigen carriers. For example, Hsp60, Hsp70, and Hsp90 act as ligands for a number of clusters of differentiation and cell-surface receptors (Ohashi et al., 2000; Basu et al., 2001; Vabulas et al., 2001; Habich et al., 2002). Furthermore, the increase of hsp gene expressions induced by stressors influences the immune resistance of aquatic animals (Wilhelm et al., 2005; Sung and MacRae, 2011). However, little is known about the participation of hsp40 genes in the immune reactions of flatfish except the differential expression of three hsp40 genes in the embryonic cells of Japanese flounder after virus infection (Dong et al., 2006).
Because of various advantages such as rapid growth rate and delicious taste, Japanese flounder is a high-value flatfish in Asian countries including China, Japan, and Korea (Fuji et al., 2006). Nevertheless, the development of industrial farming has caused the Japanese flounder’s susceptibility to various pathogens like bacteria, parasites, and viruses, resulting in numerous diseases that are infectious and severe losses in aquaculture recently (Isshiki et al., 2001; Moustafa et al., 2010). Edwardsiellosis is a serious illness caused by Edwardsiella tarda, which has led to sizable economic losses all over the world in aquaculture (Hoshina, 1962; Meyer and Bullock, 1973; Yasunaga, 1982; Bang et al., 1992; Nougayrede et al., 1994). Japanese flounder that suffers from Edwardsiellosis often shows various symptoms including a swollen abdomen, pigmentation loss, dermal damages, and spiral movement; as a result, Edwardsiellosis has affected the flounder breeding industry tempestuously (Bang et al., 1992; Moon et al., 2014). Recently, immune responses in different organs of Japanese flounder after E. tarda affection have been reported (Takano et al., 2006; Taechavasonyoo et al., 2013; Li et al., 2014; Liu et al., 2017; Thanasaksiri et al., 2017).
Up to now, the systematic efforts to reveal the roles of hsps in economically important fish’ immune reactions have not been completed. In order to avoid commercial losses caused by E. tarda in aquaculture, more in-depth comprehension about the involvement of hsps in the resistance to E. tarda is of great importance. Since the first discovery of hsp40 in bacteria, thousands of hsp40 genes have been identified in prokaryotes and eukaryotes (Yochem et al., 1978; Song et al., 2014; Chen T. et al., 2018; Huang et al., 2018; Xu Y. et al., 2019). Previous researches illustrated that the expression of several hsp subfamilies from Japanese flounder could be affected after the infection of several pathogens, e.g., Streptococcus parauberis (Dong et al., 2006; Chen et al., 2010; Sung and MacRae, 2011; Cha et al., 2013; Wei et al., 2013). Nevertheless, with respect to the roles of hsp40 genes in the disease resistance in Japanese flounder, extensive understanding is lacking. In this study, we conducted the genome-wide characterization of 50 hsp40 genes, including sequence information, selective pressures, and phylogenies. We also determined their expression profiles in embryonic and larval stages in E. tarda-infected and healthy tissues, to give early reference about the characteristics of hsp40 genes in the immune reaction in Japanese flounder.
Materials and Methods
Identification of hsp40 Family Members in Flounder Genome
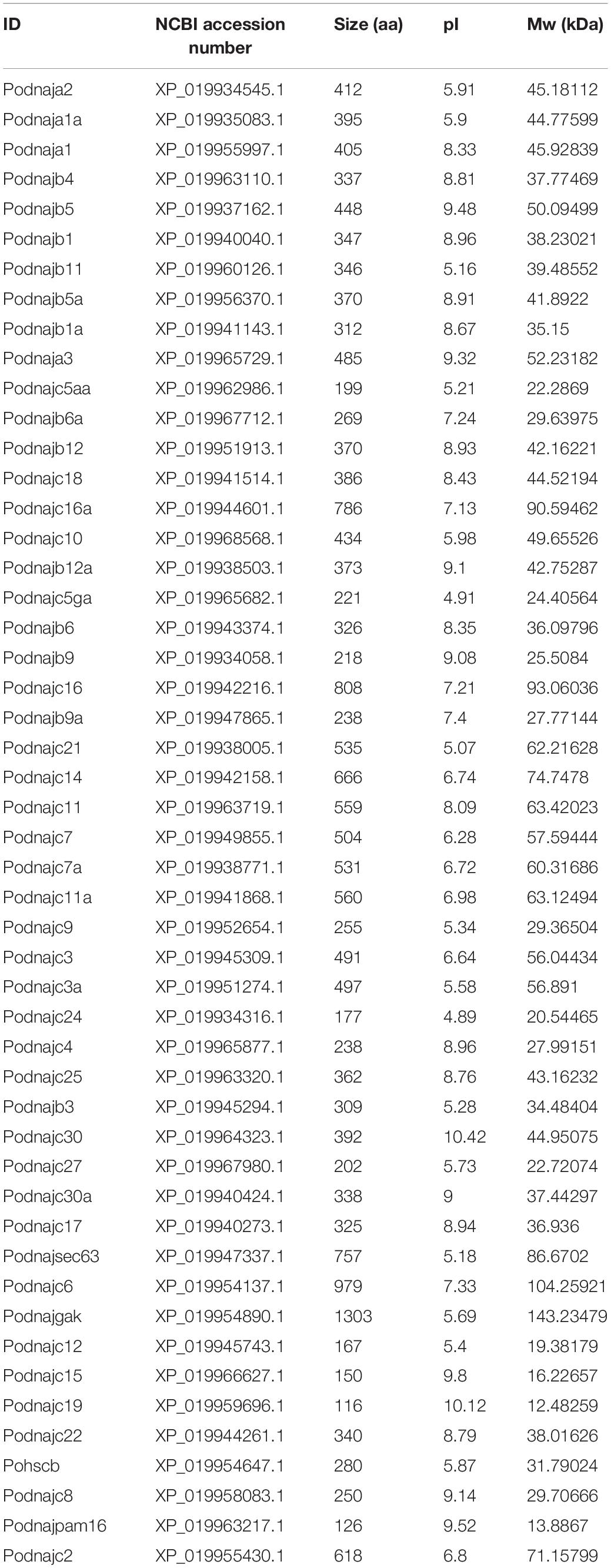
To identify the hsp40 genes, the whole genome database of Japanese flounder (NCBI accession number MPLB00000000.1) was searched using available hsp40 sequences from teleosts [zebrafish (Danio rerio), medaka (Oryzias latipes), tilapia (Oreochromis niloticus), channel catfish (Ictalurus punetaus), yellow catfish (Tachysurus fulvidraco), spotted gar (Lepisosteus oculatus), and fugu (Takifugu rubripes)] from the Heat Shock Protein Database Information Resource (Sinha et al., 2012). The E-value was set at 1e-10 to acquire as many candidate hsp40 genes as possible. Redundant sequences were removed. Afterward, Pfam (El-Gebali et al., 2018) and SMART (Letunic and Bork, 2017) databases were applied to confirm the conserved hsp40 domain, namely, DnaJ_CXXCXGXG (PF00684), DnaJ_C (PF01556), DnaJ (PF00226), and Pam16 (PF03656), and sequences without any hsp40 domain were excluded. Newly identified hsp40 genes of flounder were named following Zebrafish Nomenclature Guidelines and the Guidelines for the nomenclature of the human hsps (Kampinga et al., 2009), and the genes were named after the zebrafish orthologs whenever possible (Table 1).
Phylogenetic Tree Conduction of hsp40 Gene Families
The amino acid sequences of hsps of seven teleosts above and Japanese flounder were used for phylogenetic analysis. The MEGA7 (Kumar et al., 2016) software was used to construct a phylogenetic tree, using parameters of the WAG model and the maximum-likelihood method. Further, we applied the Evolview (Zhang et al., 2012) to visualize the tree.
Sequence Structure and Motif Prediction
We utilized the MEME program (Bailey et al., 2009) to evaluate the motifs in the hsp40 sequences; the parameters were as follows: any number of repetitions, maximum of eight motifs, and an optimum motif width of 6–200 amino acid residues. Then, we used the Gene Structure Display Server (GSDS1) to identify exon–intron organizations of the hsp40 genes from Japanese flounder. Then, TBtools software was applied to construct a diagrammatic sketch (Chen C. et al., 2018). Finally, the ProtParam Tool (Walker, 2005) predicted the biophysical properties of each hsp40 protein.
Molecular Evolution Analysis
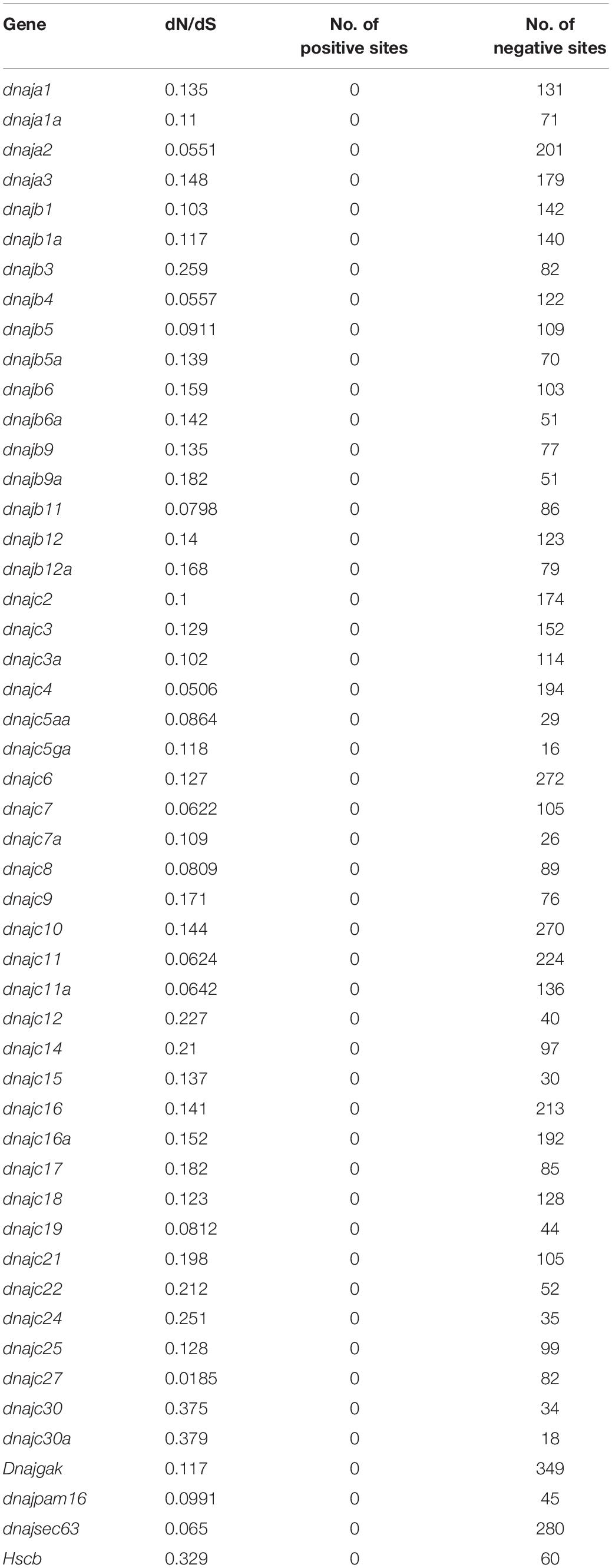
To investigate the selective pressure of hsp40 genes, the relative rates of non-synonymous substitutions (dN) and synonymous substitutions (dS) were used to represent the natural selective pressure of eight different teleosts above (Kryazhimskiy and Plotkin, 2008). According to previous research, a dN/dS ratio of greater than 1 forecasts positive selection and less than 1 forecasts negative selective pressure, whereas a ratio equal to 1 hints neutral selection (Nei and Gojobori, 1986). We applied the ClustalW for codon-based alignment and removed the terminator. Then, the single likelihood ancestor counting (SLAC) method (Kosakovsky Pond and Frost, 2005) in Datamonkey (Delport et al., 2010) was utilized to predict selective pressure on individual codons (sites) within the CDS of the hsp40 genes. SLAC is good at exploring non-neutral evolution in over 50 sequences, using likelihood-based branch lengths, nucleotide and codon substitution parameters, and ancestral sequence reconstructions (Pond and Frost, 2005).
Subcellular Localization and Secondary Structure Analysis of hsp40 Proteins
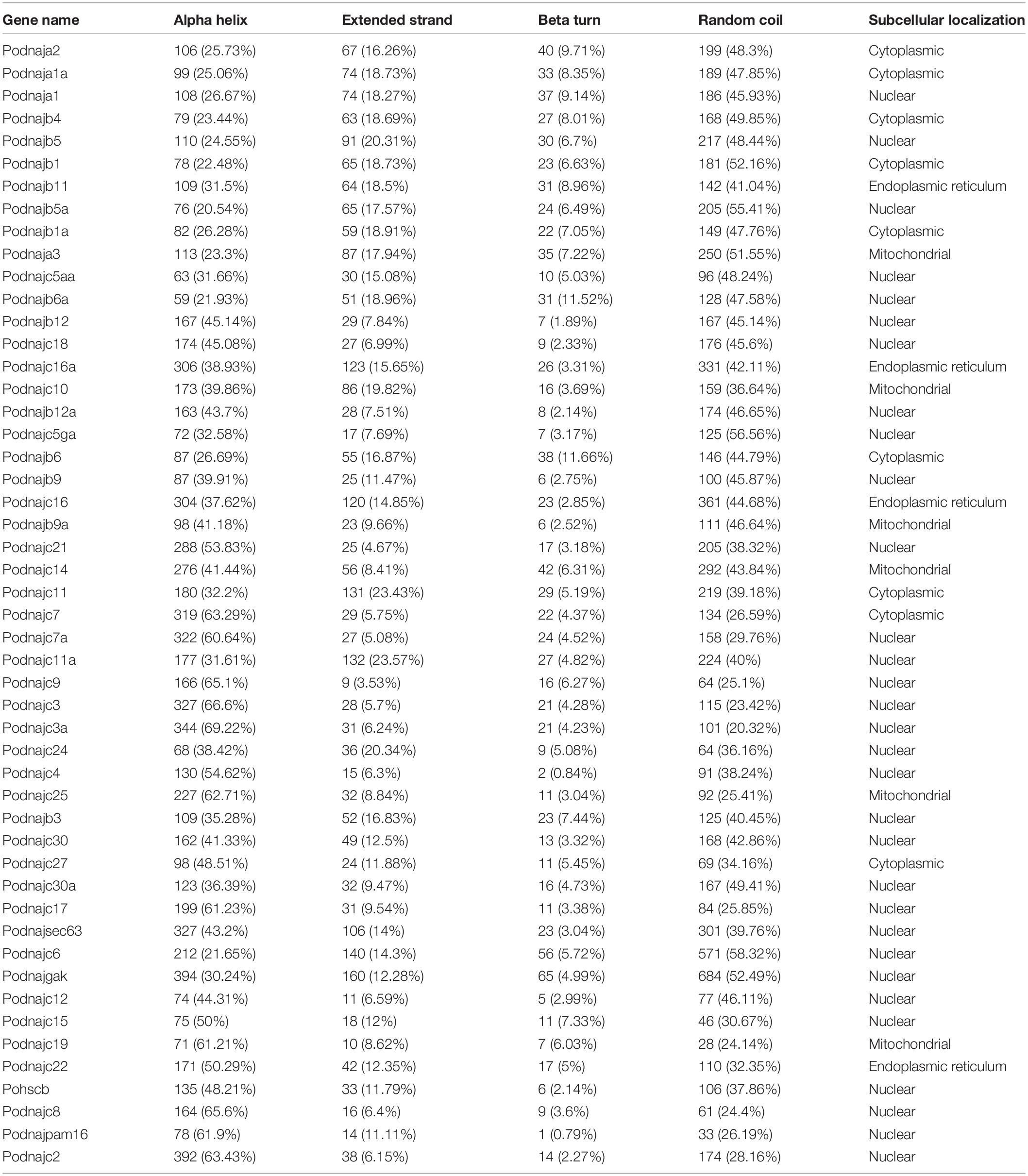
The secondary structure of hsp40 genes in Japanese flounder was predicted by SOPMA (Geourjon and Deleage, 1995) with the following parameters: output width, 70; number of conformational states, four (helix, sheet, turn, and coil); similarity threshold, 8; and window width, 17. The subcellular localization was performed using the amino acid sequences of hsp40 proteins by PSORT (protein subcellular localization prediction tool) (Nakai, 1999).
Expression Profiles in Developmental Stages, Challenged and Unchallenged Tissues
We analyzed the expression profiles of hsp40 genes in unchallenged Japanese flounder tissues and E. tarda-infected Japanese flounder, namely, 11 tissues (heart, spleen, liver, kidney, intestines, muscle, brain, gill, stomach, testis, and ovary) and six embryonic and larval stages (stages 1–6) (NCBI accession number: SRX500343, PRJNA319595).
To analyze the expression levels of hsp40 mRNA in Japanese flounder blood, gill, and kidney samples during E. tarda infection, we applied the RNA-seq data from previous research of our lab (Liu et al., 2017; Li et al., 2018a, b). The individuals for infection provided by the Yellow Sea Aquatic Product Co. Ltd. in China were approximately 1 year old with an average body length of 16.3 ± 1.5 cm (mean ± SD) and an average weight of 70.5 ± 7.9 g (mean ± SD). They were acclimatized in aerated seawater at 19°C for 7 days before injection. The E. tarda strain EIB202 was acquired from the Key Laboratory of Microbial Oceanography, Ocean University of China. It was a chloramphenicol, tetracycline, rifampicin, and streptomycin strain isolated from an outbreak in farmed turbot in Shandong province of China (Xiao et al., 2008; Wang et al., 2009). We did not detect pathogenic E. tarda from the Japanese flounder before the experiment utilizing primers based on the specific esaV gene of pathogenic pathogen (Tan et al., 2005). We incubated the E. tarda strain in Luria–Bertani (LB) medium to mid-logarithmic stage at 28°C and then harvested it by centrifugation and re-suspended it to a final concentration of 2 × 107 colony-forming units (CFU) ml–1 in Ringer’s solution.
Before the formal injection, we performed two pre-experiments to confirm that the injections did make healthy individuals sick and even die. After we validated that E. tarda infections were virulent, the formal injections were performed. The Japanese flounder was randomly spilt into three groups: 60 individuals as bacteria-challenge experiment group (BCEG), 60 in the Ringer’s solution control group (RSCG), and 10 as the blank control group (BCG). Japanese flounder in BCEG was injected intraperitoneally with 1 ml of the abovementioned pathogen suspension. Additionally, RSCG individuals were injected with the same dosage of Ringer’s solution, and individuals in BCG have not been injected. Then, their kidney, gill, and blood samples were extracted and stored in liquid nitrogen until RNA extraction. Three time points (0, 8, and 48 h) were chosen to sample collection at each group: BCG for 0 h (Bl-BC), BCEG for 8 h (Bl-8hE), RSCG for 8 h (Bl-8hC), BCEG for 48 h (Bl-48hE), and RSCG for 48 h (Bl-48hC). We have set the necessary biological duplication, and equal molar ratios of two individuals’ RNA were pooled as one replicate for further study. Sequencing libraries were constructed using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (#E7530L, NEB, United States) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Raw reads were cleaned by removing adaptor sequences, low-quality sequences (Sanger base quality < 20), and reads with unknown nucleotides larger than 10%. The TopHat–Cufflinks–Cuffmerge–Cuffdiff pipeline was used to analyze the clean data by using default parameters, and then the parameter fragments per kilobase of transcript per million mapped reads (FPKM) was used to quantify the abundance of assembled transcripts (Trapnell et al., 2012; Ghosh and Chan, 2016; Li et al., 2018b). Thirty sequencing libraries from four groups were constructed totally. The raw sequencing reads were submitted to Sequence Read Archive (SRA) in NCBI with accession numbers PRJNA359626, PRJNA359627, SRR5713071, SRR5713072, SRR5713073, SRR5713074, SRR5713075, SRR5713076, SRR5713077, SRR5713078, SRR5713079, and SRR5713080. Last, the expression levels in unchallenged (RSCG) and challenged groups (BCEG) were examined to find out further information about hsp40 genes’ differential expression in reaction to E. tarda injection (Liu et al., 2017; Li et al., 2018a, b). Then, we applied R package pheatmap to visualize the profiles in different tissues (Kolde, 2018).
Results
Sequence Extraction From Japanese Flounder Genome
We applied 57 zebrafish hsp40 sequences extracted from the Heat Shock Protein Database Information Resource as a query to search against the Japanese flounder genome. Afterward, SMART and Pfam databases were applied to validate the putative hsp40 genes by the standard of hsp40 domains (PF00226, PF00684, PF01556, and PF03656). Sequences that were repetitive or lacked any hsp40 domains were deleted. As a result, 50 hsp40 sequences (dnaja1, dnaja1a, dnaja2, dnaja3, dnajb1, dnajb1a, dnajb3, dnajb4, dnajb5, dnajb5a, dnajb6, dnajb6a, dnajb9, dnajb9a, dnajb11, dnajb12, dnajb12a, dnajc2, dnajc3, dnajc3a, dnajc4, dnajc5aa, dnajc5ga, dnajc6, dnajc7, dnajc7a, dnajc8, dnajc9, dnajc10, dnajc11, dnajc11a, dnajc12, dnajc14, dnajc15, dnajc16, dnajc16a, dnajc17, dnajc18, dnajc19, dnajc21, dnajc22, dnajc24, dnajc25, dnajc27, dnajc30, dnajc30a, dnajgak, dnajpam16, dnajsec63, and hscb) identified from the Japanese flounder genome were named based on the Guidelines for the nomenclature of the human hsps as well as the Zebrafish Nomenclature Guidelines (Table 1).
Phylogenetic Analysis
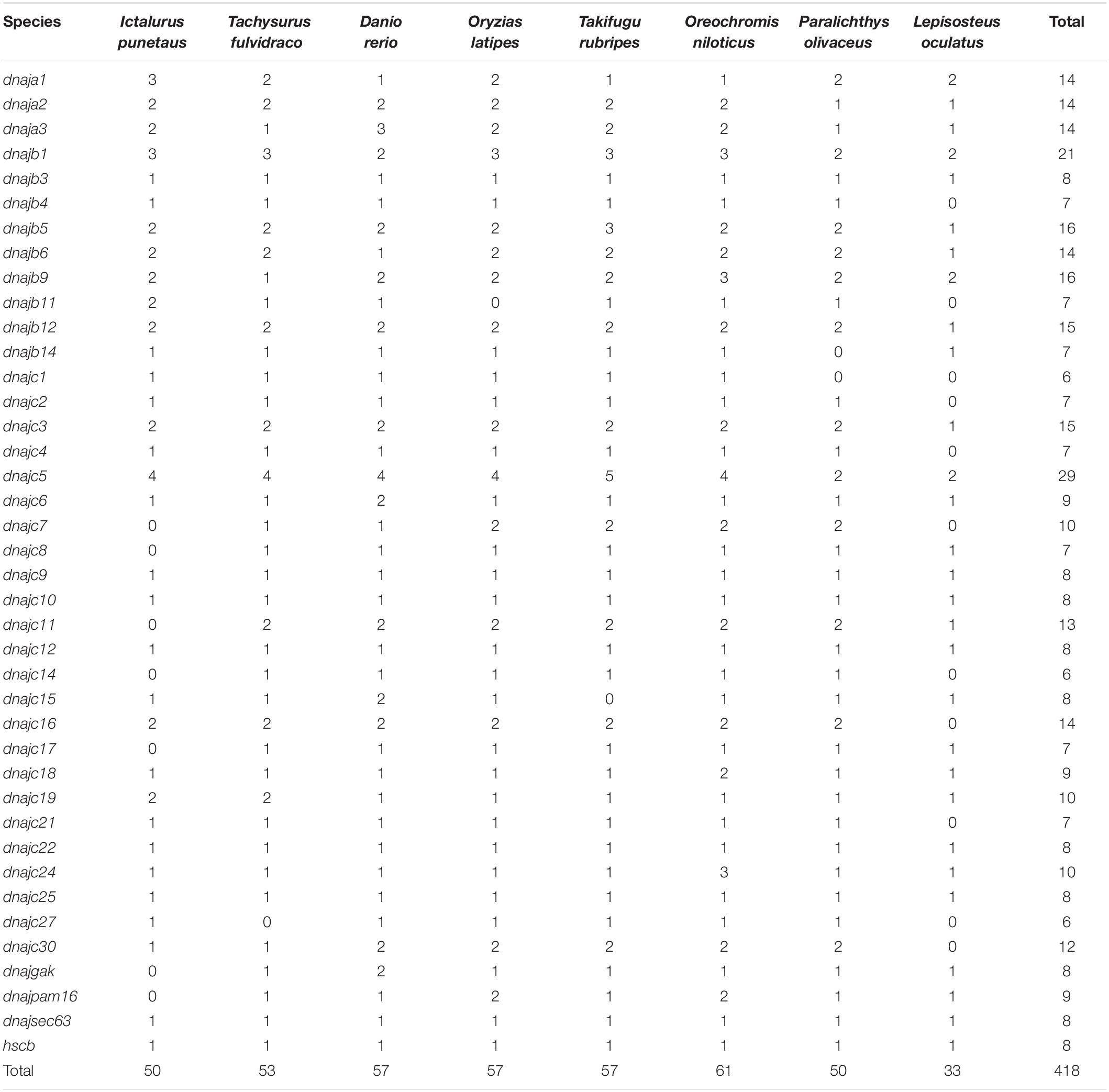
To evaluate the evolutionary history of the hsp40 genes collected from eight teleosts above, a phylogenetic tree (maximum-likelihood method, WAG model) was constructed by MEGA7 (Figure 1). The 418 hsp40 genes fell into 40 specific subfamilies: 14 dnaja1, 14 dnaja2, 14 dnaja3, 21 dnajb1, eight dnajb3, seven dnajb4, 16 dnajb5, 14 dnajb6, 16 dnajb9, seven dnajb11, 15 dnajb12, seven dnajb14, six dnajc1, seven dnajc2, 15 dnajc3, seven dnajc4, 29 dnajc5, nine dnajc6, 10 dnajc7, seven dnajc8, eight dnajc9, eight dnajc10, 13 dnajc11, eight dnajc12, six dnajc14, eight dnajc15, 14 dnajc16, seven dnajc17, nine dnajc18, 10 dnajc19, seven dnajc21, eight dnajc22, 10 dnajc24, eight dnajc25, six dnajc27, 12 dnajc30, eight dnajgak, nine dnajpam16, eight dnajsec63, and eight hscb. Each of the homologous hsp40 genes from eight teleosts was divided into the same cluster. Furthermore, hsp40 genes whose protein shows high similarity in structure share a close range in the phylogenetic tree.
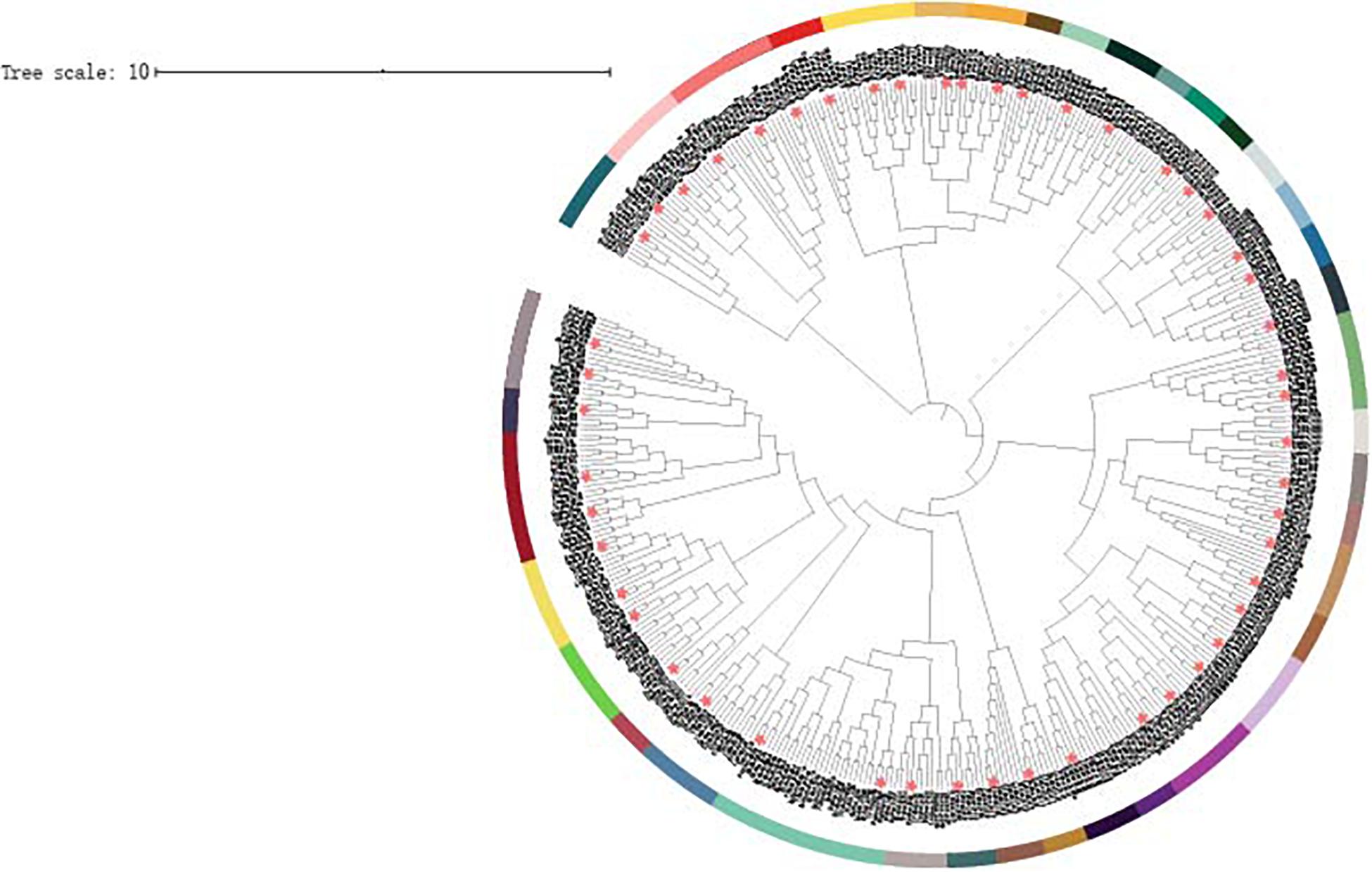
Figure 1. Phylogenetic analysis of 418 hsp40 genes constructed using ML method and WAG models. Different subfamilies were distinguished in different colors, and the Japanese flounder hsp40 genes were asterisk labeled. Abbreviations: On, Oreochromis niloticus; Tr, Takifugu rubripes; Dr, Danio rerio Ol, Oryzias latipes; Ip, Ictalurus punetaus; Tf, Tachysurus fulvidraco; Lo, Lepisosteus oculatus.
Exon–Intron Organizations and Motif Patterns
Gene structure and motif patterns of hsp40 genes were analyzed to illustrate their similarities and differences and provide deeper comprehension about their evolutionary relationship (Figure 2). As for the 50 hsp40 genes of Japanese flounder, two (dnajc30 and dnajc30a) had no introns, belonging to the “no intron” group, whereas other genes had no less than two introns and were divided into the “multiple introns” group. We identified eight evolutionary-conserved motifs from flounder hsp40 genes. Results indicated that hsp40 genes had diverse motif patterns, and genes with close phylogenetic relationship had similar motif patterns. Additionally, the biophysical properties are listed in Table 2. The amino acid numbers of hsp40 proteins were with the scope of 116 (dnajc19) to 1303 (dnajgak); the molecular masses were between 12.48259 kDa (dnajc19) and 143.23479 kDa (dnajgak), and the pI values are confined from 4.89 (dnajc24) to 10.42 (dnajc30).

Figure 2. Phylogenetic analysis, exon/intron organization, and motif analysis of hsp40 genes. (A) Phylogenetic tree of 50 hsp40 proteins. The maximum-likelihood phylogenetic tree was constructed using amino acid sequences of 50 hsp40 proteins with MEGA7 software. (B) Distributions of conserved motifs in hsp40 genes. Eight motifs were distinguished in different colored boxes. (C) Gene structure of hsp40 genes. Yellow lines represent CDSs and green boxes with the same length represent UTRs. The length of exons can be inferred by the scale.
Molecular Evolution Analysis of the hsp40 Genes
In order to get more information about the evolutionary history of hsp40 gene interspecies, we used the synonymous rate to infer whether fixation of non-synonymous mutations is strengthened or weakened by selection pressure, and the coding sequences of the 50 hsp40 genes were applied for calculating the dN/dS ratio (Table 3). In this study, the dN/dS ratio of all the 50 hsp40 genes was less than 1, indicating pronounced negative selective pressure. We cannot find any positive selection sites of all the hsp40 genes from Japanese flounder.
Subcellular Localization and Secondary Structure Analysis of hsp40 Proteins
Subcellular localization analysis showed that the hsp40 genes of Japanese flounder were mainly expressed in the cytoplasm, nucleus, mitochondria, and endoplasmic reticulum (ER) (Table 4), and less expressed in peroxisome, cytoskeleton, and Golgi apparatus. In detail, most hsp40 genes were expressed mainly in the nucleus, nine genes (dnaja1a, dnaja2, dnajb1, dnajb1a, dnajb4, dnajb6, dnajc7, dnajc11, and dnajc27) were mainly expressed in the cytoplasm, six genes (dnaja3, dnajb9a, dnajc10, dnajc14, dnajc19, and dnajc25) were expressed highly in mitochondria, and four genes (dnajb11, dnajc16, dnajc16a, and dnajc22) were mainly expressed in ER. The secondary structure of hsp40 proteins consisted of alpha helix, extended strand, beta turn, and random coil (Table 4). Among the 50 hsp40 proteins in Japanese flounder, danjb12 was alpha helix = random coil > extended strand > beta turn, dnajc9 was alpha helix > random coil > beta turn > extended strand, 19 proteins (dnajc2, dnajc3, dnajc3a, dnajc4, dnajc7, dnajc7a, dnajc8, dnajc10, dnajc15, dnajc17, dnajc19, dnajc21, dnajc22, dnajc24, dnajc25, dnajc27, dnajsec63, hscb, and dnajpam16) were alpha helix > random coil > extended strand > beta turn, and the other 29 proteins were random coil > alpha helix > extended strand > beta turn. In summary, the alpha helix and random coil were the main components of the hsp40 secondary structure.
Expression Profiles Analysis
The expression profiles of hsp40 genes at embryo and larval developmental stages and tissues extracted from healthy individuals and after E. tarda injection were illustrated using our previous data. In order to visualize the expression profiles in detail, heat maps with the phylogenetic tree were constructed (Figure 3). Results showed that most hsp40 genes participated in six embryonic and larval stages of Japanese flounder, while three of them (dnajb1a, dnajb5, and dnajb12a) were expressed only at a certain part of developmental stages. Furthermore, there were six genes (dnaja2, dnajb3, dnajc3a, dnajc4, dnajc5aa, and dnajc19) that showed high expression levels at all stages and 12 genes (dnaja1, dnaja1a, dnaja3, dnajb6a, dnajb9, dnajb11, dnajb12, dnajc8, dnajc9, dnajc10, dnaj16, and dnajc21) had relatively high expression levels at one or several developmental stages. Under normal conditions, all hsp40 genes were expressed to maintain homeostasis, as a reserve in case of sudden pathogen invasion. The expression of hsp40 genes in different tissues had various patterns, and some genes showed preferential expression in certain tissues. For instance, several hsp40 genes were highly expressed in one specific tissue, but had low expression levels or not expressed in other tissues, namely, dnajc22 in the heart, dnajc3a in the gill, dnaja3 and dnajc21 in the muscle, dnajb11 in the stomach, dnajb6 and dnajc7 in the intestines, and dnajb5, dnajb12a, dnajc5aa, dnajc6, and dnajc11a in the brain. Additionally, we also found that dnajb1a had weak expression levels in 11 tissues, which required deeper studies.
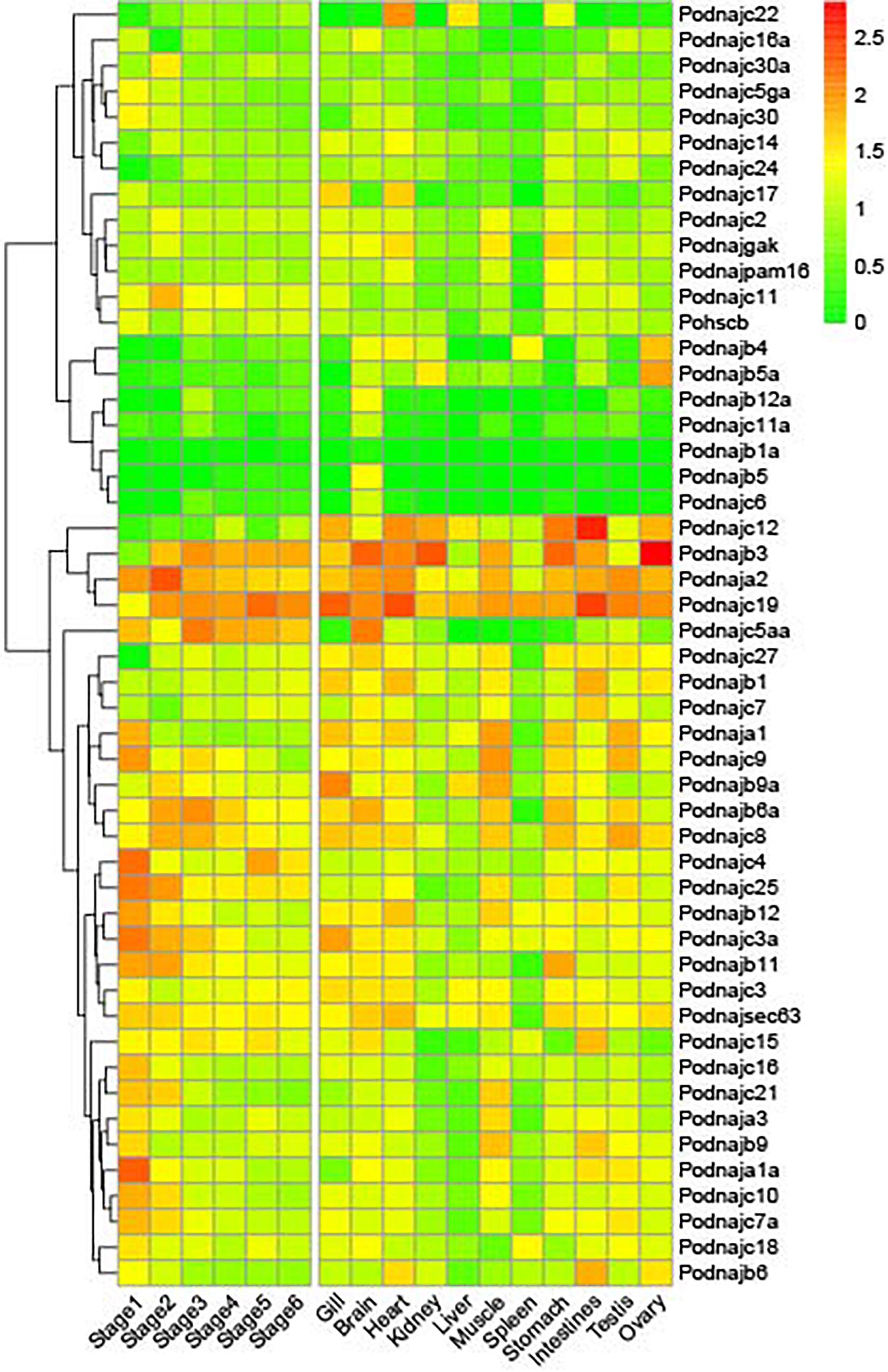
Figure 3. Expression patterns of hsp40 genes in 11 tissues and six embryonic and larval stages in healthy individuals. Each cell in the heat map corresponds to an expression level and the numbers in cell are FPKM values. Six stages of embryonic and larval development: stage 1 (from two cells to morula), stage 2 (from early gastrula to late somites), stage 3 (from hatching stage to 2 days after hatching), stage 4 (before metamorphosis), stage 5 (metamorphosis stages 1–2), and stage 6 (metamorphosis stages 3–5).
To investigate the role of hsp40 family members in response to E. tarda, we analyzed the expression levels of the 50 hsp40 genes from three tissues and three time points after E. tarda infection from the RNA-seq data of previous research (Figure 4); the summary of the comparison of Ringer’s solution and bacterial-challenge group, namely, log2(fold_change) and p-value, is shown in Supplementary Table 1. In total, 41 hsp40 genes except dnajb1a, dnajb4, dnajb5, dnajb6a, dnajb12a, dnajc5aa, dnajc6, dnajc22, and dnajc30 were significantly influenced by Ringer’s solution and the differential expression showed a tissue- and gene-specific pattern. After the injection with Ringer’s solution in the blood, the expression of eight genes (dnaja1, dnajb5a, dnajc3, dnajc16a, dnajc18, dnajc24, hscb, and dnajsec63) was significantly influenced at 8 h and the amount of 10 genes’ transcripts (dnaja3, dnajb9, dnajb9a, dnajc4, dnajc7, dnajc11a, dnajc12, dnajc15, dnajc18, and dnajc25) was not influenced until 48 h, whereas that of eight genes (dnaja1a, dnajb1, dnajb3, dnajb12, dnajc5ga, dnajc9, dnajc10, and dnajgak) was influenced from 8 to 48 h. After Ringer’s injection in the gill, gene expression of 11 genes (dnaja2, dnajb9a, dnajc10, dnajc11, dnajc14, dnajc15, dnajc16, dnajc17, dnajc18, dnajc21, and dnajc30a) was prominently influenced at 8 h whereas the expression of eight genes (dnaja1a, dnajb1, dnajb9, dnajc2, dnajc12, dnajc19, dnajgak, and hscb) was influenced at 48 h. In addition, gene expression of 13 genes (dnaja3, dnajb3, dnajb6, dnajb11, dnajc3, dnajc3a, dnajc5ga, dnajc7a, dnajc8, dnajc25, dnajc27, dnajsec63, and dnajpam16) was influenced by Ringer’s solution throughout all the time points investigated. After the injection with Ringer’s solution in the kidney, the expression of 11 genes (dnaja2, dnajb9a, dnajc10, dnajc11, dnajc14, dnajc15, dnajc16, dnajc17, dnajc18, dnajc21, and dnajc30a) was up- or downregulated at 8 h, whereas that of eight genes (dnaja1a, dnajb1, dnajb9, dnajc2, dnajc12, dnajc19, dnajgak, and hscb) was influenced at 48 h. The expression of 13 genes (dnaja3, dnajb3, dnajb6, dnajb11, dnajc3, dnajc3a, dnajc5ga, dnajc7a, dnajc8, dnajc25, dnajc27, dnajsec63, and dnajpam16) was influenced from 8 to 48 h.
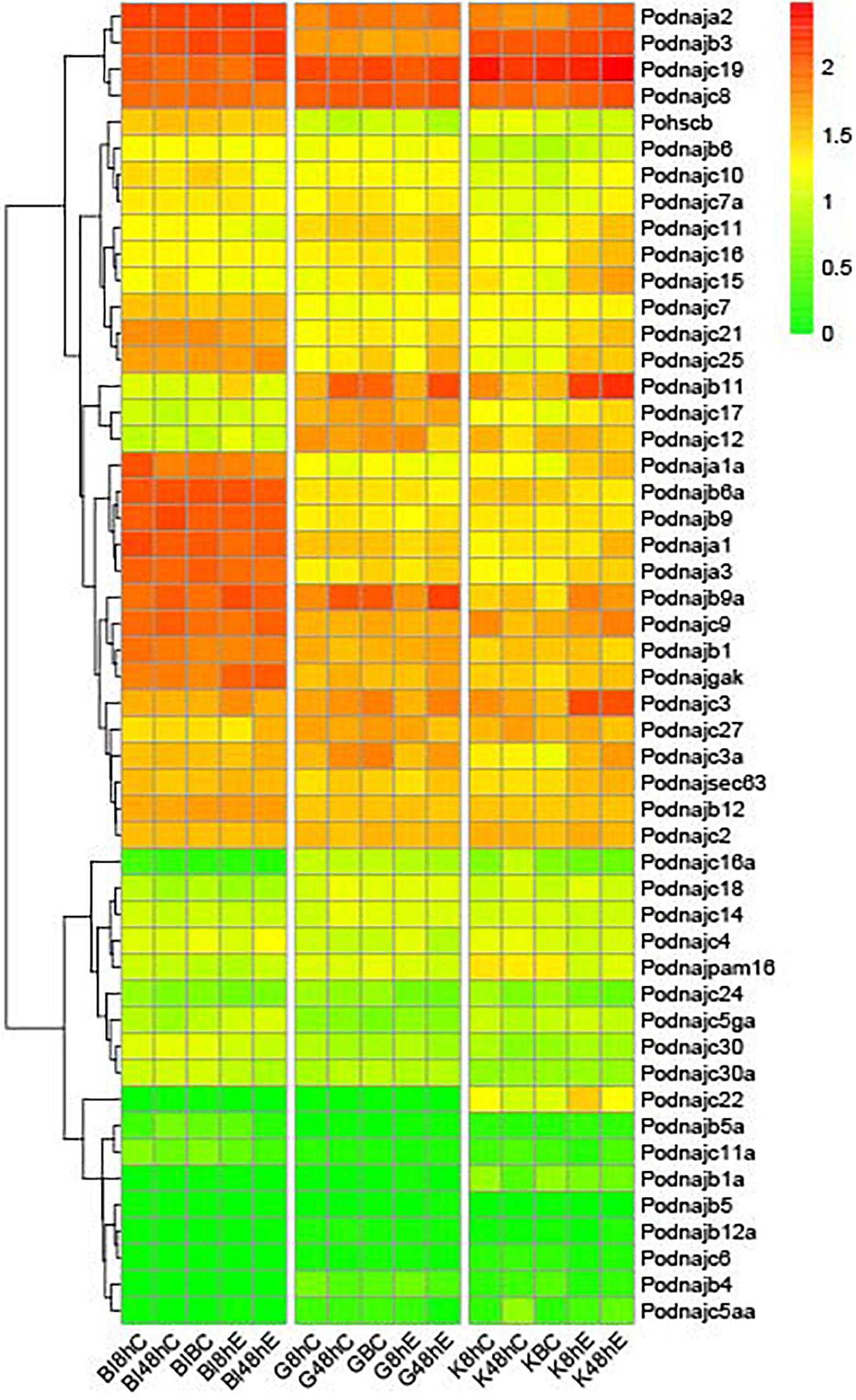
Figure 4. Expression profiles of hsp40 genes after E. tarda injection. Each row represents a gene, and each column represents a time point. The relative expression level is indicated by the color bar on the top right. G represents gill, K represents kidney, and Bl represents blood. 0 h represents the blank control group at the beginning of the experiment, C 8 h and C 48 h indicate Ringer’s solution control group, whereas E 8 h and E 48 h indicate E. tarda-challenged experimental group, and BC represents blank control.
Discussion
The industrial farming of Japanese flounder was affected by E. tarda infection seriously (Egusa, 1976; Nakatsugawa, 1983; Miyazaki and Kaige, 1985; Isshiki et al., 2001; Moustafa et al., 2010; Park et al., 2012). Previous studies indicated that hsps participated in disease defense and resistance of teleosts, e.g., channel catfish (Xie et al., 2015; Song et al., 2016). However, despite these observations, a deeper understanding of the molecular mechanism of Japanese flounder immune responses is required. In this study, we identified 61 hsp40 genes from tilapia, 57 from medaka, 57 from fugu, 57 from zebrafish, 53 from yellow catfish, 50 from channel catfish, 50 from Japanese flounder, and 33 from spotted gar, indicating a relatively similar hsp gene number. In order to elucidate the evolution history of hsp40 proteins, we conducted a maximum-likelihood phylogenetic gene tree of eight teleosts above. As shown in Figure 1, each of the hsp40 genes was divided into 40 subfamilies (dnaja1, dnaja2, dnaja3, dnajb1, dnajb3, dnajb4, dnajb5, dnajb6, dnajb9, dnajb11, dnajb12, dnajb14, dnajc1, dnajc2, dnajc3, dnajc4, dnajc5, dnajc6, dnajc7, dnajc8, dnajc9, dnajc10, dnajc11, dnajc12, dnajc14, dnajc15, dnajc16, dnajc17, dnajc18, dnajc19, dnajc21, dnajc22, dnajc24, dnajc25, dnajc27, dnajc30, dnajgak, dnajpam16, dnajsec63, and hscb) with no obvious extension between species, and Japanese flounder harbored most genes, indicating a high evolutionary conservativeness. Although we conducted complete searches with all Japanese flounder genomic resources available, the dnajb14 and dnajc1 have not been found in the Japanese flounder genome. Together with selective pressure analysis, dN/dS analysis of all hsp40 genes experienced pronounced negative selection, indicating that there were no non-synonymous nucleotide changes at that codon. There existed three genes (dnajc30, dnajc30a, and hscb) whose dN/dS ratio was more than 0.3, which is greater than that of others, hinting that they may experience a higher evolutionary dynamic. Previous studies have found that the evolution of new genes was usually accompanied by changes that occurred in both their sequence and structure, while mutation is the original condition in the evolution of genes. Furthermore, positive Darwinian selection may be another important power forcing new genes’ evolution. In addition, natural selection pressure might affect particular sites but not the whole gene, namely, site-specific selection (Koester et al., 2012). These results revealed that the hsp40 genes between teleosts above were evolutionary-constrained and they were function-conserved; therefore, negative selection might account for the loss of genes in the Japanese flounder genome. In addition, according to the subcellular localization analysis, we assume that most hsp40 genes function in the nucleus of cells because of their expression preference. As for the structural characteristics of hsp40 proteins, the results above could predict their potential roles and ultimately provide their sequence–structure–function relationships by binding and acting with other proteins.
During embryogenesis, there exist vigorous cell proliferation and differentiation as well as gene expression and protein synthesis, the intra- and extracellular environments experience steady changes, and cells are extremely sensitive to external stimuli (Haanen and Vermes, 1996). Thus, the change and function of hsps may be more vital (Walsh et al., 1997; Neuer et al., 1999). Hsp genes are regarded as chaperones in morphologic development of cells and organisms and are believed to be related to normal and abnormal development of embryo (Neuer et al., 1999; Brown et al., 2007). Nevertheless, among the hsp superfamily, compared to the abundant researches about hsp20 and hsp10/60 genes with respect to embryonic and larval development, there is a lack of systemic studies about the role of hsp40 genes in normal development of embryo and larva (Mao and Shelden, 2006; Elicker and Hutson, 2007; Xu et al., 2011; Middleton and Shelden, 2013; Wang et al., 2017). As for the model species, hsp40 gene was reported to participate in the embryonic development of mouse forelimbs (Zhu et al., 2010). With regard to marine animals, we also found reports about the roles of hsp40 genes in sea urchin cilia regeneration during embryogenesis (Casano et al., 2003). Furthermore, previous research also indicated that hsp40 genes were upregulated after pathogen injection in Japanese flounder embryonic cells (FECs) (Dong et al., 2006). Herein, results indicated that hsp40 genes had different expression profiles during different embryonic and larval stages and six of them (dnaja2, dnajb3, dnajc3a, dnajc4, dnajc5aa, and dnajc19) had high expression levels at all developmental stages, which hints that hsp40 genes may be involved in the development of Japanese flounder embryo and larva.
Previous studies have shown the roles of hsp40 genes in the reactions of a large number of stressors of aquatic animals; however, they emphasized on the abiotic stresses such as thermal, acidity/alkalinity, and salinity challenges (Chen T. et al., 2018; Huang et al., 2018; Xu Y. et al., 2019). As for the immune response, hsp40 genes were found to be significantly regulated after Edwardsiella ictaluri and Flavobacterium columnare challenges in channel catfish (Song et al., 2014). Interestingly, in this study, we also found that a large percentage of hsp40 family members, with a number of 44 genes (88%), were significantly influenced after pathogen injection. Though the mechanisms behind are ambiguous, hsp40 genes were dramatically regulated after E. tarda challenge. These results indicate that hsp40 genes participated in pathogen reactions and disease resistance against pathogens. In detail, the expression patterns showed a tissue-dependent feature after E. tarda infection: 35 genes were up- or downregulated in kidney, whereas 34 genes were regulated in blood while 31 were regulated in gill. In summary, 44 out of 50 hsp40 genes participated in E. tarda defense reactions. After E. tarda infection in the blood, expression of three genes (dnaja1a, dnajc11a, and dnajc21) decreased from 8 to 48 h, whereas that of seven genes (dnaja2, dnajb3, dnajb12, dnajc3, dnajc5ga, dnajsec63, and dnajgak) was upregulated significantly. Gene expression of dnajc19 and dnajpam16 decreased at 8 h and then rose at 48 h, and gene expression of dnajb5a and dnajc7a was upregulated at 8 h but downregulated at 48 h. Gene expression of dnajb9a, dnajb11, and dnajc11 was upregulated at 8 h but returned to the original level at 48 h, whereas that of nine genes (dnaja1, dnaja3, dnajb1, dnajb6, dnajc9, dnajc10, dnajc16a, dnajc18, and dnajc24) was downregulated at 8 h and kept in a normal standard at 48 h. In addition, of the 50 hsp40 genes, nine did not show significant differential regulation until 48 h, among them, five (dnajc3a, dnajc4, dnajc17, dnajc25, and dnajc27) were upregulated at 48 h, whereas three genes (dnajc15, dnajc30, and dnajc30a) were downregulated at 48 h. After E. tarda injection in the gill, expression of dnaja2 was dramatically increased from 8 to 48 h, whereas that of dnajc24 decreased significantly after injection. The expression of dnajc14 increased at 8 h and followed by a drop at 48 h. Gene expression of dnajc19 dropped at 8 h and rose to a high level at 48 h. Besides, the expression of 19 hsp40 genes (dnaja1a, dnajb1a, dnajb3, dnajb4, dnajb5, dnajb5a, dnajb12a, dnajc4, dnajc5aa, dnajc5ga, dnajc6, dnajc9, dnajc10, dnajc11a, dnajc17, dnajc22, dnajc30, dnajc30a, and hscb) was stable throughout all time period. Interestingly, dnajb12 and dnajc18 showed a different response pattern, which was upregulated at 8 h but returned to the original level at 48 h, while that of dnaja1 and dnajb9 was downregulated at 8 h but restored to a normal level at 48 h. Additionally, gene expression of 16 genes (dnaja3, dnajb1, dnajb6, dnajb9a, dnajb11, dnajc2, dnajc3, dnajc7, dnajc8, dnajc11, dnajc15, dnajc16, dnajc21, dnajc25, dnajsec63, and dnajgak) was kept in a normal standard and showed a significant increase at 48 h, and the gene expression of seven genes (dnajb6a, dnajc3a, dnajc7a, dnajc12, dnajc16a, dnajc27, and dnajpam16) was kept normal and decreased prominently at 48 h. Besides, after E. tarda injection in the kidney, the gene expression of 20 genes (dnaja1, dnaja1a, dnaja2, dnaja3, dnajb3, dnajb9a, dnajb11, dnajc3, dnajc3a, dnajc5ga, dnajc8, dnajc10, dnajc11, dnajc15, dnajc16, dnajc21, dnajc22, dnajc25, dnajcsec63, and dnajgak) was prominently elevated after administration, and the expression of dnajb6a, dnajpam16, and hscb was downregulated from 8 to 48 h. The expression of dnajb1 and dnajc27 was elevated at 8 h and followed by a decrease at 48 h. Gene expression of six genes (dnajb6, dnajc12, dnajc17, dnajc19, and dnajc30) was kept in a normal standard and showed a significant increase at 48 h, whereas that of dnajc16a was kept normal but decreased at 48 h. Moreover, the expression of dnajb9, dnajc7, and dnajc18 was upregulated at 8 h, whereas the original level was restored at 48 h, and gene expression of dnajc24 was downregulated but restored to the normal standard at 48 h. Notably, 15 genes (dnajb1a, dnajb4, dnajb5, dnajb5a, dnajb12, dnajb12a, dnajc2, dnajc4, dnajc5aa, dnajc6, dnajc7a, dnajc9, dnajc11a, dnajc14, and dnajc30a) did not exhibit up- or downregulation from 8 to 48 h. In a word, results showed that a large percentage of hsp40 gene in gill, blood, and kidney samples were likely to be involved in reaction to E. tarda injection, with the exception of dnajb1a, dnajb4, dnajb5, dnajb12a, dnajc5aa, and dnajc6. Interestingly, there existed a tissue-specific response pattern in these 44 regulated hsp40 genes; namely, most hsp40 genes had different reaction profiles in three tissues examined except dnaja2, which was upregulated in gill, kidney, and blood tissues from 8 to 48 h. In addition, there were still 14 hsp40 genes (dnaja1, dnajb3, dnajb6, dnajc3, dnajc5ga, dnajc16a, dnajc17, dnajc18, dnajc19, dnajc24, dnajc25, dnajc27, dnajgak, and dnajsec63) that showed a similar response pattern in two of three particular tissues, among them, dnaja1, dnajc19, and dnajc25 had the same patterns in blood and gill, while dnajb3, dnajc3, dnajc5ga, dnajc17, dnajsec63, and dnajgak had the same profiles in blood and kidney, whereas the remaining five genes had the same expression patterns in gill and kidney. Furthermore, the 44 regulated hsp40 genes had different response efficiency: dnajc2, dnajc4, dnajc17, dnajc30, and dnajc30a did not show an up- or downregulation until 48 h after E. tarda injection, while the other 39 genes respond rapidly at 8 h after injection. Results above hint that five genes (dnajc2, dnajc4, dnajc17, dnajc30, and dnajc30a) showed a lower speed in regulating E. tarda challenge in flounder.
In total, 44 out of 50 hsp40 genes seem to show a response in reaction to E. tarda administrations. The regulatory mechanism behind has not yet been fully validated nevertheless. Therefore, more in-depth research is needed to elucidate the mechanisms of differential expression and to verify the characteristics of hsp40 genes in immune defenses. In summary, a full set of 50 hsp40 genes derived from the Japanese flounder genome were identified and characterized in this study. As the largest subfamily of hsp superfamily that is involved in many vital physiological processes, hsp40 genes were divided into 40 subfamilies in phylogenetic analysis, and selective pressure analysis indicated that hsp40 genes experienced pronounced purifying selection. Additionally, we investigated the expression levels of the hsp40 genes in E. tarda-injected and unchallenged organisms. In healthy individuals, hsp40 genes played the sentinel role. In challenged individuals, 44 hsp40 genes were up- or downregulated after E. tarda injection, hinting that they might play a role as a portion of the disease response while some of differential expression genes may participate in disease defense against E. tarda. These findings give elementary reference for in-depth validation of the characteristics of hsp40 genes in the process of immune reactions and molecular evolutionary history in Japanese flounder.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the Ocean University of China and the China Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training.
Author Contributions
WY contributed to conceptualization, methodology, software, writing—original draft, writing—review and editing, and visualization. YQ contributed to software and resources. JQ contributed to software. XL contributed to formal analysis. QZ contributed to funding acquisition. XW contributed to conceptualization, methodology, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD0900601).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XS declared a shared affiliation with several of the authors, WY, YQ, JQ, QZ, and XW to the handling editor at the time of review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.596534/full#supplementary-material
Footnotes
References
Akira, S., Bauer, S., and Hartmann, G. (2008). Toll Like Receptors (TLRs) and Innate Immunity. Berlin: Springer.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208.
Bang, J.-D., Chun, S.-K., Park, S.-I., and Choi, Y.-J. (1992). Studies on the biochemical and serological characteristics of Edwardsiella tarda isolated from cultured flounder, Paralichthys olivaceus. J. Fish Pathol. 5, 29–35.
Basu, S., Binder, R. J., Ramalingam, T., and Srivastava, P. K. (2001). CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 14, 303–313. doi: 10.1016/s1074-7613(01)00111-x
Basu, S., Binder, R. J., Suto, R., Anderson, K. M., and Srivastava, P. K. (2000). Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int. Immunol. 12, 1539–1546. doi: 10.1093/intimm/12.11.1539
Bork, P., Sander, C., Valencia, A., and Bukau, B. (1992). A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem. Sci. 17:129. doi: 10.1016/0968-0004(92)90319-5
Botha, M., Pesce, E., and Blatch, G. L. (2007). The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Intern. J. Biochem. Cell Biol. 39, 1781–1803. doi: 10.1016/j.biocel.2007.02.011
Brown, D. D., Christine, K. S., Showell, C., and Conlon, F. L. (2007). Small heat shock protein Hsp27 is required for proper heart tube formation. Genesis 45, 667–678. doi: 10.1002/dvg.20340
Casano, C., Gianguzza, F., Roccheri, M. C., Giorgi, R. D., Maenza, L., and Ragusa, M. A. (2003). Hsp40 is involved in cilia regeneration in sea urchin embryos. J. Histochem. Cytochem. 51, 1581–1587. doi: 10.1177/002215540305101202
Cha, I. S., Kwon, J., Park, S. B., Jang, H. B., Nho, S. W., Kim, Y. K., et al. (2013). Heat shock protein profiles on the protein and gene expression levels in olive flounder kidney infected with Streptococcus parauberis. Fish Shellf. Immunol. 34, 1455–1462. doi: 10.1016/j.fsi.2013.03.355
Cheetham, M. E., and Caplan, A. J. (1998). Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperon. 3:28. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv [Preprint], doi: 10.1101/289660
Chen, T., Lin, T., Li, H., Lu, T., Li, J., Huang, W., et al. (2018). Heat shock protein 40 (HSP40) in Pacific white shrimp (Litopenaeus vannamei): molecular cloning, tissue distribution and ontogeny, response to temperature, acidity/alkalinity and salinity stresses, and potential role in ovarian development. Front. Physiol. 9:1784. doi: 10.3389/fphys.2018.01784
Chen, W., Syldath, U., Bellmann, K., Burkart, V., and Kolb, H. (1999). Human 60-kDa heat-shock protein: a danger signal to the innate immune system. J. Immunol. 162, 3212–3219.
Chen, Y.-M., Kuo, C.-E., Wang, T.-Y., Shie, P.-S., Wang, W.-C., Huang, S.-L., et al. (2010). Cloning of an orange-spotted grouper Epinephelus coioides heat shock protein 90AB (HSP90AB) and characterization of its expression in response to nodavirus. Fish Shellf. Immunol. 28, 895–904. doi: 10.1016/j.fsi.2010.02.004
Delport, W., Poon, A. F., Frost, S. D., and Kosakovsky Pond, S. L. (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. doi: 10.1093/bioinformatics/btq429
Dong, C.-W., Zhang, Y.-B., Zhang, Q.-Y., and Gui, J.-F. (2006). Differential expression of three Paralichthys olivaceus Hsp40 genes in responses to virus infection and heat shock. Fish Shellf. Immunol. 21, 146–158. doi: 10.1016/j.fsi.2005.11.002
Egusa, S. (1976). Some bacterial diseases of freshwater fishes in Japan. Fish Pathol. 10, 103–114. doi: 10.3147/jsfp.10.103
Eissa, N. (2014). Probiotic effect on molecular antioxidant profiles in yellow perch, Perca flavescens. Glob. Jo. Fish. Aquac. Res. 1, 16–29.
Eissa, N., Wang, H.-P., Yao, H., and Abou-ElGheit, E. (2018). Mixed Bacillus species enhance the innate immune response and stress tolerance in yellow perch subjected to hypoxia and air-exposure stress. Sci. Rep. 8, 1–10.
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2018). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432.
Elicker, K. S., and Hutson, L. D. (2007). Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene 403, 60–69. doi: 10.1016/j.gene.2007.08.003
Fuji, K., Kobayashi, K., Hasegawa, O., Coimbra, M. R. M., Sakamoto, T., and Okamoto, N. (2006). Identification of a single major genetic locus controlling the resistance to lymphocystis disease in Japanese flounder (Paralichthys olivaceus). Aquaculture 254, 203–210. doi: 10.1016/j.aquaculture.2005.11.024
Fuller, K., Issels, R., Slosman, D., Guillet, J.-G., Soussi, T., and Polla, B. (1994). Cancer and the heat shock response. Eur. J. Cancer 30, 1884–1891.
Gehrmann, M., Brunner, M., Pfister, K., Reichle, A., Kremmer, E., and Multhoff, G. (2004). Differential up-regulation of cytosolic and membrane-bound heat shock protein 70 in tumor cells by anti-inflammatory drugs. Clin. Cancer Res. 10, 3354–3364. doi: 10.1158/1078-0432.ccr-03-0382
Georgopoulos, C. P., Lundquist-Heil, A., Yochem, J., and Feiss, M. (1980). Identification of the E. coli dnaJ gene product. Mol. Gen. Genet. MGG 178, 583–588. doi: 10.1007/bf00337864
Geourjon, C., and Deleage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Gething, M.-J. (1997). Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Oxford: OUP Oxford.
Ghosh, S., and Chan, C.-K. K. (2016). “Analysis of RNA-Seq data using TopHat and Cufflinks, Plant Bioinformatics,” in Plant Bioinformatics. Methods in Molecular Biology, Vol. 1374, ed. D. Edwards (New York, NY: Humana Press), 339–361. doi: 10.1007/978-1-4939-3167-5_18
Haanen, C., and Vermes, I. (1996). Apoptosis: programmed cell death in fetal development. Eur. J. Obstetr. Gynecol. Reprod. Biol. 64, 129–133. doi: 10.1016/0301-2115(95)02261-9
Habich, C., Baumgart, K., Kolb, H., and Burkart, V. (2002). The receptor for heat shock protein 60 on macrophages is saturable, specific, and distinct from receptors for other heat shock proteins. J. Immunol. 168, 569–576. doi: 10.4049/jimmunol.168.2.569
Hennessy, F., Cheetham, M. E., Dirr, H. W., and Blatch, G. L. (2000). Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperon. 5, 347–358. doi: 10.1379/1466-1268(2000)005<0347:aotloc>2.0.co;2
Hennessy, F., Nicoll, W. S., Zimmermann, R., Cheetham, M. E., and Blatch, G. L. (2005). Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14, 1697–1709. doi: 10.1110/ps.051406805
Hoshina, T. (1962). On a new bacterium, Paracolobactrum anguillimortiferum n. sp. Bull. Jpn. Soc. Sci. Fish. 28, 162–164. doi: 10.2331/suisan.28.162
Huang, X., Li, S., Gao, Y., and Zhan, A. (2018). Genome-wide identification, characterization and expression analyses of heat shock protein-related genes in a highly invasive ascidian Ciona savignyi. Front. Physiol. 9:1043. doi: 10.3389/fphys.2018.01043
Isshiki, T., Nishizawa, T., Kobayashi, T., Nagano, T., and Miyazaki, T. (2001). An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed Japanese flounder Paralichthys olivaceus in Japan. Dis. Aquat. Organ. 47, 87–99. doi: 10.3354/dao047087
Kampinga, H. H., Hageman, J., Vos, M. J., Kubota, H., Tanguay, R. M., Bruford, E. A., et al. (2009). Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperon. 14, 105–111.
Koester, J. A., Swanson, W. J., and Armbrust, E. V. (2012). Positive selection within a diatom species acts on putative protein interactions and transcriptional regulation. Mol. Biol. Evol. 30, 422–434. doi: 10.1093/molbev/mss242
Kol, A., Lichtman, A. H., Finberg, R. W., Libby, P., and Kurt-Jones, E. A. (2000). Cutting edge: heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. J. Immunol. 164, 13–17. doi: 10.4049/jimmunol.164.1.13
Kosakovsky Pond, S. L., and Frost, S. D. (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222. doi: 10.1093/molbev/msi105
Kryazhimskiy, S., and Plotkin, J. B. (2008). The population genetics of dN/dS. PLoS Genet. 4:e1000304. doi: 10.1371/journal.pgen.1000304
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, H. B., Kim, D. S., Gil, H. W., and Park, I.-S. (2018). Pysiological responses of diploid and triploid far eastern catfish, Silurus asotus to water temperature stress. Dev. Reprod. 22:165. doi: 10.12717/dr.2018.22.2.165
Lehner, T., Bergmeier, L. A., Wang, Y., Tao, L., Sing, M., Spallek, R., et al. (2000). Heat shock proteins generate β-chemokines which function as innate adjuvants enhancing adaptive immunity. Eur. J. Immunol. 30, 594–603. doi: 10.1002/1521-4141(200002)30:2<594::aid-immu594>3.0.co;2-1
Letunic, I., and Bork, P. (2017). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496.
Li, J., Qian, X., and Sha, B. (2009). Heat shock protein 40: structural studies and their functional implications. Protein Pept. Lett. 16, 606–612. doi: 10.2174/092986609788490159
Li, S., Li, X., Gen, X., Chen, Y., Wei, J., and Sun, J. (2014). Identification and characterization of lipopolysaccharide-induced TNF-alpha factor gene from Japanese flounder Paralichthys olivaceus. Vet. Immunol. Immunopathol. 157, 182–189. doi: 10.1016/j.vetimm.2013.11.006
Li, Z., Liu, X., Cheng, J., He, Y., Wang, X., Wang, Z., et al. (2018a). Transcriptome profiling provides gene resources for understanding gill immune responses in Japanese flounder (Paralichthys olivaceus) challenged with Edwardsiella tarda. Fish Shellf. Immunol. 72, 593–603. doi: 10.1016/j.fsi.2017.11.041
Li, Z., Liu, X., Liu, J., Zhang, K., Yu, H., He, Y., et al. (2018b). Transcriptome profiling based on protein-protein interaction networks provides a core set of genes for understanding blood immune response mechanisms against Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev. Compar. Immunol. 78, 100–113. doi: 10.1016/j.dci.2017.09.013
Liu, X., Li, Z., Wu, W., Liu, Y., Liu, J., He, Y., et al. (2017). Sequencing-based network analysis provides a core set of gene resource for understanding kidney immune response against Edwardsiella tarda infection in Japanese flounder. Fish Shellf. Immunol. 67, 643–654. doi: 10.1016/j.fsi.2017.06.051
Mao, L., and Shelden, E. (2006). Developmentally regulated gene expression of the small heat shock protein Hsp27 in zebrafish embryos. Gene Expr. Patter. 6, 127–133. doi: 10.1016/j.modgep.2005.07.002
Mayer, M. P., Laufen, T., Paal, K., Mccarty, J. S., and Bukau, B. (1999). Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289, 1131–1144. doi: 10.1006/jmbi.1999.2844
Meyer, F., and Bullock, G. (1973). Edwardsiella tarda, a new pathogen of channel catfish (Ictalurus punctatus). Appl. Microbiol. 25:155. doi: 10.1128/aem.25.1.155-156.1973
Middleton, R. C., and Shelden, E. A. (2013). Small heat shock protein HSPB1 regulates growth of embryonic zebrafish craniofacial muscles. Exp. Cell Res. 319, 860–874. doi: 10.1016/j.yexcr.2013.01.002
Miyazaki, T., and Kaige, N. (1985). Comparative histopathology of edwardsiellosis in fishes. Fish Pathol. 20, 219–227. doi: 10.3147/jsfp.20.219
Moon, J. Y., Hong, Y.-K., Kong, H. J., Kim, D.-G., Kim, Y.-O., Kim, W.-J., et al. (2014). A cDNA microarray analysis to identify genes involved in the acute-phase response pathway of the olive flounder after infection with Edwardsiella tarda. Vet. Immunol. Immunopathol. 161, 49–56. doi: 10.1016/j.vetimm.2014.07.002
Morahan, B. J., Strobel, C., Hasan, U., Czesny, B., Mantel, P., Marti, M., et al. (2011). functional analysis of the exported Type IV HSP40 protein PfGECO in Plasmodium falciparum gametocytes. Eukaryot. Cell 10, 1492–1503. doi: 10.1128/ec.05155-11
Moré, S. H., Breloer, M., and von Bonin, A. (2001). Eukaryotic heat shock proteins as molecular links in innate and adaptive immune responses: Hsp60-mediated activation of cytotoxic T cells. Int. Immunol. 13, 1121–1127. doi: 10.1093/intimm/13.9.1121
Moustafa, E. M. M., Naota, M., Morita, T., Tange, N., and Shimada, A. (2010). Pathological study on the scuticociliatosis affecting farmed Japanese flounder (Paralichthys olivaceus) in Japan. J. Vet. Med. Sci. 72, 1359–1362. doi: 10.1292/jvms.10-0026
Multhoff, G. (2006). “Heat shock proteins in immunity,” in Molecular Chaperones in Health and Disease. Handbook of Experimental Pharmacology, Vol. 172, eds K. Starke and M. Gaestel (Berlin: Springer), 279–304.
Nakai, K. (1999). PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35. doi: 10.1016/s0968-0004(98)01336-x
Nakatsugawa, T. (1983). Edwardsiella tarda isolated from cultured young flounder. Fish. Pathol. 18, 99–101.
Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426.
Neuer, A., Spandorfer, S., Giraldo, P., Jeremias, J., Dieterle, S., Korneeva, I., et al. (1999). Heat shock protein expression during gametogenesis and embryogenesis. Infect. Dis. Obstet. Gynecol. 7, 10–16. doi: 10.1155/s1064744999000034
Nougayrede, P., Vuillaume, A., Vigneulle, M., Faivre, B., Luengo, S., and Delprat, J. (1994). First isolation of Edwardsiella tarda from diseased turbot (Scophthalmus maximus) reared in a sea farm in the Bay of Biscay. Bull. Eur. Assoc. Fish Pathol. 14, 128–129.
Ohashi, K., Burkart, V., Flohé, S., and Kolb, H. (2000). Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J. Immunol. 164, 558–561. doi: 10.4049/jimmunol.164.2.558
Ohtsuka, K., and Hata, M. (2000). Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperon. 5:98. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2
Panjwani, N. N., Popova, L., and Srivastava, P. K. (2002). Heat shock proteins gp96 and hsp70 activate the release of nitric oxide by APCs. J. Immunol. 168, 2997–3003. doi: 10.4049/jimmunol.168.6.2997
Park, S. B., Aoki, T., and Jung, T. S. (2012). Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 43:67. doi: 10.1186/1297-9716-43-67
Pickering, A. D. (1998). “Stress responses of farmed fish,” in Biology of Farmed Fish, eds K. D. Black and A. D. Pickering (Sheffield: Sheffield Academic Press), 222–255.
Pond, S. L. K., and Frost, S. D. (2005). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. doi: 10.1093/bioinformatics/bti320
Qiu, X.-B., Shao, Y.-M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. CMLS 63, 2560–2570. doi: 10.1007/s00018-006-6192-6
Ritossa, F. M. (1962). A new puffing pattern induced by heat shock and DNP in Drosophila. Experientia 18, 515–523.
Roberts, R., Agius, C., Saliba, C., Bossier, P., and Sung, Y. (2010). Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J. Fish. Dis. 33, 789–801. doi: 10.1111/j.1365-2761.2010.01183.x
Singh-Jasuja, H., Hilf, N., Scherer, H. U., Arnold-Schild, D., Rammensee, H.-G., Toes, R. E., et al. (2000). The heat shock protein gp96: a receptor-targeted cross-priming carrier and activator of dendritic cells. Cell Stress Chaperon. 5:462. doi: 10.1379/1466-1268(2000)005<0462:thspga>2.0.co;2
Sinha, D., Veedin Rajan, V. B., Esthaki, V. K., and D’Silva, P. (2012). HSPIR: a manually annotated heat shock protein information resource. Bioinformatics 28, 2853–2855. doi: 10.1093/bioinformatics/bts520
Song, L., Li, C., Xie, Y., Liu, S., Zhang, J., Yao, J., et al. (2016). Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellf. Immunol. 49, 154–162. doi: 10.1016/j.fsi.2015.12.009
Song, L., Zhang, J., Li, C., Yao, J., Jiang, C., Li, Y., et al. (2014). Genome-wide identification of Hsp40 genes in channel catfish and their regulated expression after bacterial infection. PLoS One 9:e115752. doi: 10.1371/journal.pone.0115752
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Srivastava, P. (2002). Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2, 185–194. doi: 10.1038/nri749
Sung, Y., and MacRae, T. (2011). Heat shock proteins and disease control in aquatic organisms. J. Aquac. Res. Dev. S2:006.
Taechavasonyoo, A., Kondo, H., Nozaki, R., Suzuki, Y., and Hirono, I. (2013). Identification of novel interleukin 1 beta family genes in Japanese flounder Paralichthys olivaceus. Fish Shellf. Immunol. 34, 393–396. doi: 10.1016/j.fsi.2012.10.001
Takano, T., Kondo, H., Hirono, I., Saito-Taki, T., Endo, M., and Aoki, T. (2006). Identification and characterization of a myeloid differentiation factor 88 (MyD88) cDNA and gene in Japanese flounder, Paralichthys olivaceus. Dev. Comparat. Immunol. 30, 807–816. doi: 10.1016/j.dci.2005.11.003
Tan, Y., Zheng, J., Tung, S., Rosenshine, I., and Leung, K. (2005). Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151, 2301–2313. doi: 10.1099/mic.0.28005-0
Thanasaksiri, K., Hirono, I., and Kondo, H. (2017). Molecular cloning and expression analysis of NOD-like receptor 5 in Japanese flounder (Paralichthys olivaceus) after injection with two different formalin-killed pathogenic bacteria and poly (I: C). Dev. Comparat. Immunol. 67, 481–484. doi: 10.1016/j.dci.2016.08.017
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tsai, J., and Douglas, M. G. (1996). A Conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271, 9347–9354. doi: 10.1074/jbc.271.16.9347
Vabulas, R. M., Ahmad-Nejad, P., da Costa, C., Miethke, T., Kirschning, C. J., Häcker, H., et al. (2001). Endocytosed HSP60s use toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. J. Biol. Chem. 276, 31332–31339. doi: 10.1074/jbc.m103217200
Walsh, D., Li, Z., Wu, Y., and Nagata, K. (1997). Heat shock and the role of the HSPs during neural plate induction in early mammalian CNS and brain development. Cell. Mol. Life Sci. CMLS 53, 198–211. doi: 10.1007/pl00000592
Walsh, P., Bursać, D., Law, Y. C., Cyr, D., and Lithgow, T. (2004). The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571. doi: 10.1038/sj.embor.7400172
Wang, P., Xuac, P., Zhouac, L., Zengac, S., and Liac, G. (2017). Molecular cloning, characterization, and expression analysis of HSP60 in mandarin fish Siniperca chuatsi. Isra. J. Aquac. Bamidgeh 69:13.
Wang, Q., Yang, M., Xiao, J., Wu, H., Wang, X., Lv, Y., et al. (2009). Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS One 4:e7646. doi: 10.1371/journal.pone.0007646
Wei, T., Gao, Y., Wang, R., and Xu, T. (2013). A heat shock protein 90 β isoform involved in immune response to bacteria challenge and heat shock from Miichthys miiuy. Fish Shellf. Immunol. 35, 429–437. doi: 10.1016/j.fsi.2013.04.045
Wen, X., Hu, Y., Zhang, X., Wei, X., Wang, T., and Yin, S. (2019). Integrated application of multi-omics provides insights into cold stress responses in pufferfish Takifugu fasciatus. BMC Genom. 20:563. doi: 10.1186/s12864-019-5915-7
Whitley, D., Goldberg, S. P., and Jordan, W. D. (1999). Heat shock proteins: a review of the molecular chaperones. J. Vasc. Surg. 29, 748–751.
Wilhelm, V., Soza, C., Martínez, R., Rosemblatt, M., Burzio, L. O., and Valenzuela, P. D. (2005). Production and immune response of recombinant Hsp60 and Hsp70 from the salmon pathogen Piscirickettsia salmonis. Biol. Res. 38, 69–82.
Xiao, J., Wang, Q., Liu, Q., Wang, X., Liu, H., and Zhang, Y. (2008). Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquac. Res. 40, 13–17.
Xie, Y., Song, L., Weng, Z., Liu, S., and Liu, Z. (2015). Hsp90, Hsp60 and sHsp families of heat shock protein genes in channel catfish and their expression after bacterial infections. Fish Shellf. Immunol. 44, 642–651. doi: 10.1016/j.fsi.2015.03.027
Xu, X.-Y., Shen, Y.-B., Fu, J.-J., Liu, F., Guo, S.-Z., Yang, X.-M., et al. (2011). Molecular cloning, characterization and expression patterns of HSP60 in the grass carp (Ctenopharyngodon idella). Fish Shellf. Immunol. 31, 864–870. doi: 10.1016/j.fsi.2011.07.028
Xu, Y., Zheng, G., Liu, G., Yang, Q., and Yu, X. (2019). Molecular cloning, characterization of Pomacea canaliculata HSP40 and its expression analysis under temperature change. J. Therm. Biol. 81, 59–65. doi: 10.1016/j.jtherbio.2019.02.006
Xu, Z.-N., Zheng, G.-D., Wu, C.-B., Jiang, X.-Y., and Zou, S.-M. (2019). Identification of proteins differentially expressed in the gills of grass carp (Ctenopharyngodon idella) after hypoxic stress by two-dimensional gel electrophoresis analysis. Fish Physiol. Biochem. 45, 743–752. doi: 10.1007/s10695-018-0599-5
Yasunaga, N. (1982). Characteristics of fish pathogen Edwardsiella isolated from several species of cultured marine fishes. Bull. Nagasaki Pref. Instit. Fish. 8, 57–65.
Yochem, J., Uchida, H., Sunshine, M., Saito, H., Georgopoulos, C., and Feiss, M. (1978). Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol. Gen. Genet. MGG 164, 9–14. doi: 10.1007/bf00267593
Zhang, H., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572.
Zhang, X., Dai, L., Wu, Z., Jian, J., and Lu, Y. (2011). Expression pattern of heat shock protein 90 gene of humphead snapper Lutjanus sanguineus during pathogenic Vibrio harveyi stress. J. Fish Biol. 79, 178–193. doi: 10.1111/j.1095-8649.2011.03012.x
Zhu, Y., Zhu, J., Wan, X., Zhu, Y., and Zhang, T. (2010). Gene expression of sHsps, Hsp40 and Hsp60 families in normal and abnormal embryonic development of mouse forelimbs. Toxicol. Lett. 193, 242–251. doi: 10.1016/j.toxlet.2010.01.016
Keywords: heat shock protein, Hsp40, Japanese flounder, Edwardsiella tarda, immune response
Citation: Yan W, Qiao Y, Qu J, Liu X, Zhang Q and Wang X (2021) The hsp40 Gene Family in Japanese Flounder: Identification, Phylogenetic Relationships, Molecular Evolution Analysis, and Expression Patterns. Front. Mar. Sci. 7:596534. doi: 10.3389/fmars.2020.596534
Received: 19 August 2020; Accepted: 01 December 2020;
Published: 14 January 2021.
Edited by:
Anthony Lee Dellinger, Kepley BioSystems, Inc., United StatesReviewed by:
Xiuzhen Sheng, Ocean University of China, ChinaSeongbin Park, Mississippi State University, United States
Fu Yuanshuai, Shanghai Ocean University, China
Copyright © 2021 Yan, Qiao, Qu, Liu, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xubo Wang, d2FuZ3h1Ym9Ab3VjLmVkdS5jbg==
 Weijie Yan1
Weijie Yan1 Quanqi Zhang
Quanqi Zhang Xubo Wang
Xubo Wang