- 1Simon F. S. Li Marine Science Laboratory, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China
- 2State Key Laboratory of Agrobiotechnology, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China
- 3Department of Chemistry, City University of Hong Kong, Hong Kong, China
- 4Department of Biology, Queen’s University, Kingston, ON, Canada
- 5Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
Horseshoe crabs are a group of marine chelicerates that contain only four extant species, some of which are endangered. Their hemolymph has been widely used in medical applications for endotoxin detection. Nevertheless, there is limited information on the profiles of their hemolymph proteins and their gut microbial diversity. In this study, we performed the first detailed investigation of the hemolymph proteomics and gut microbiota of two Asian horseshoe crabs Tachypleus tridentatus and Carcinoscorpius rotundicauda. Among the identified proteins being cataloged in the juvenile and adult hemolymph, unexpectedly, sesquiterpenoid signaling pathway proteins including Heat shock protein 83 (HSP83), Chd64, and a juvenile hormone binding protein (JHBP) were revealed. This provides evidence for the presence of functional sesquiterpenoid hormonal systems in these marine chelicerates. consumption of certain horseshoe crab species often leads to tetrodotoxin poisoning and the horseshoe crab is thought to possess a tetrodotoxin resistance mechanism. As such, sodium channels were analyzed and found to have critical amino acid residues that are similar to the toxin resistant pufferfish sodium channels. The source of the toxin is unknown so we investigated the gut microbiota, and found that Clostridium and Vibrio were the most dominant bacteria in T. tridentatus and C. rotundicauda, respectively. Together, this study provides a framework for further understanding of sesquiterpenoids and gut microbiota of these marine chelicerates.
Introduction
The Chelicerata is the second largest subphylum in the Arthropoda after the Insecta, comprising more than 77,000 described living species. Horseshoe crabs represent a group of marine chelicerates in the order Xiphosura and its earliest fossil records can be traced back to the Ordovician period (Rudkin et al., 2008; Van Roy et al., 2010). Currently, there are only four extant horseshoe crab species, including Limulus polyphemus found in the Americas, and Carcinoscorpius rotundicauda, Tachypleus gigas, and Tachypleus tridentatus distributed in Asia (Vestbo et al., 2018). These animals generally live in shallow water regions with soft sandy or muddy bottoms, mainly the estuaries. They are also a good indicator for tracking the health of the coastal regions given their reproduction is dependent on the quality of water and the nearby micro-ecosystem (Chen et al., 2004).
The hemolymph of horseshoe crabs is well known for its blue color, as well as for its great medical importance in detecting contaminants in vaccines and injectable medicines. Specifically, coagulation proteins released from crab amebocytes detect endotoxin, a type of pyrogen that presents on the surface of the outer membrane of gram-negative bacteria, that could be life-threatening when introduced intravenously (Maloney et al., 2018). The Limulus amebocyte lysate (LAL) test is an in vitro test that is commonly used for the detection of gram-negative bacteria since the 1970s based on the fact that a coagulative protein in the Limulus amebocyte was found to be highly sensitive to bacterial endotoxins (Levin and Bang, 1964, 1968). It had also been suggested that the LAL test could be more ethical and sustainable than the traditional rabbit pyrogen test (Mehmood, 2019).
Despite its medical importance to human society, consumption of its body tissues, including viscera and eggs of the mangrove horseshoe crab C. rotundicauda could result in tetrodotoxin poisoning (Fusetani et al., 1982; Kungsuwan et al., 1987; Tanu and Noguchi, 1999; Kanchanapongkul, 2008; Dao et al., 2009; Suleiman et al., 2017; Zheng et al., 2019). A previous study suggested that the horseshoe crabs could possess a tetrodotoxin neutralizing system in their hemolymph (Ho et al., 1994), and yet, how such toxin tolerance is achieved remains unclear. Another question comes from the origin and/or biosynthesis of endogenous tetrodotoxin in these animals. Some bacteria including Pseudoalteromonas, Pseudomonas, and Vibrio have been demonstrated to produce tetrodotoxin (Jal and Khora, 2015; Lorentz et al., 2016), and yet, the gut microbiota composition remains unknown in both C. rotundicauda and T. tridentatus.
The horseshoe crabs represent an understudied basal group of arthropods and have genuine utility in their own unique biology; they can also provide insights into arthropod comparative biology. In this study, we performed the proteomic profiling of hemolymph of the two Asian species T. tridentatus and C. rotundicauda to reveal their hemolymph composition. In addition, we comparatively analyzed the sodium channels and gut microbiota in T. tridentatus and C. rotundicauda, to provide insights on the possible tetrodotoxin tolerant mechanisms and diversity of gut microbes in the two species of Asian horseshoe crabs.
Materials and Methods
Experimental Animals
Two adult (one male and one female) horseshoe crabs of T. tridentatus were purchased from local fish markets in Sai Kung of Hong Kong. Juvenile horseshoe crabs of T. tridentatus (four individuals) and C. rotundicauda (four individuals) with carapace width about 4–5 cm were collected from Ha Pak Nai and Sha Tau Kok in Hong Kong respectively. Two juvenile individuals of each species were used in gut microbiota analysis, while the others were used in hemolymph proteomics study. All horseshoe crabs (except those being taken for gut microbiota analyses) were acclimated in a culture tank with 30 ppt of artificial seawater (ASW) at room temperature with a photoperiod of 12:12 h (Light:Dark) before carrying out experiments.
Hemolymph Proteomics (ESI-Nano-LC MS/MS Analysis)
Hemolymph of both species of horseshoe crabs were drawn by syringe from the junction between the prosoma and opisthosoma on the dorsal side, and was directly extracted for protein. A protein sample (10 mg) was mixed with 20 μL of ProteoMiner beads (BioRad) and incubated on a rotational shaker for 2 h at room temperature throughout the process. The unbound excess proteins were removed by bead washing with PBS thrice followed by deionized water twice. The captured proteins were eluted with elution buffer I (8 M urea, 2% CHAPS, and 5% acetic acid) once and elution buffer II (6 M urea, 2 M thiourea, and 5% acetic acid) twice and the elution was combined. The eluates were reduced by 25 mM DTT for 1 h at room temperature and alkylated with 5 mM iodoacetamide for 30 min in dark at room temperature. The urea concentration of the samples was diluted to 1 M with 25 mM ammonium bicarbonate buffer. Sequencing-grade trypsin (Promega) was added to each sample in 1:20 ratio and incubated overnight at 37°C. The lysates were then mixed with the same volume of 2% trifluoroacetic acid and loaded onto a high pH reverse-phase fractionation spin column (Thermo Fisher Scientific) and fractionated into 4 fractions with increasing ACN concentrations (10, 15, 20, and 50%). The fractionated peptides were dried by vacuum drying and resuspended in 2% ACN with 0.5% formic acid. The nano-LC separation was performed using a Dionex UltiMate 3000 RSLC nano system. Peptides (1 ug) were loaded onto a 25 cm-long, 75 μm-i.d. C18 column and eluted from the column at a constant flow rate of 0.3 μL/min with a linear gradient from 2 to 35% of ACN over 120 min. The eluted peptides were analyzed by an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). MS and MS/MS scans were acquired in the Orbitrap with a mass resolution of 60,000 and 15,000 respectively. MS scan range was from 375 to 1,500 m/z. The AGC target for MS and MS/MS were 4e5 and 5e4 respectively; and the maximum injection time for MS and MS/MS were 50 ms and 250 ms respectively. Precursor isolation windows were set to 1.6 m/z. Data were analyzed by Proteome Discoverer version 2.3 with SEQUEST as a search engine to search against the proteome database generated from T. tridentatus and C. rotundicauda genomes (Kenny et al., 2016). The searching parameters were as follows: oxidation of methionine (+15.9949 Da) and carbamidomethylation of cysteine (+57.0215 Da) was set as dynamic modification; precursor-ion mass tolerance, 10 ppm; fragments-ion mass tolerance, 0.02 Da, protein-level false discovery rate was calculated by Percolator at an experimental q-value (exp. q value) threshold of 0.05. Proteins were quantified utilizing the precursor ion quantification module of Proteome Discoverer. BLASTP was used to search the detected hemolymph proteins against Nr (non-redundant) database and further annotated the GO function via BLAST2GO package. Short peptides within 200 amino acids were used to predict antimicrobial peptides by dbAMP (Jhong et al., 2019). Multi-alignment of protein sequences with other known proteins obtained from NCBI database was carried out by MAFFT (Katoh et al., 2019), and phylogenetic tree was constructed with MEGAX (Kumar et al., 2018) using both Neighbor-Joining (NJ) and Maximum-Likelihood (ML) methods with 1,000 bootstrap.
Sodium Channel Gene Analysis
The amino acid sequences of known sodium channel proteins of pufferfish Tetraodon nigroviridis and Takifugu rubripes were used to search against the gene models of T. tridentatus and C. rotundicauda using the BLASTP algorithm with an E-value threshold of less than 1E–5. Reciprocal BLASTP was then performed on NCBI database. In addition, online NCBI Conserved Domain Search (CD Search) Tool was used to annotate the conserved functional domains of predicted sodium channel proteins. The alignment of the amino acid sequences was performed using MUSCLE (Edgar, 2004).
Gut Microbiota Analysis
The guts of the juvenile horseshoe crabs T. tridentatus and C. rotundicauda with carapace width about 4–5 cm collected from the field was immediately removed on return to the laboratory. About 10 mg of feces was removed from the gut and preserved in 95% ethanol at 4°C. The DNA in the fecal sample was extracted using Purelink Genomic Mini Kit (Invitrogen) according to the instructions provided by the manufacturer. The extracted DNA samples were then sent to Novagene (Hong Kong) Company Limited for bacterial amplicon sequencing (16S V3–V4 region with 100 k raw tags/sample) using the Novaseq PE250 platform (sequencing information are shown in Supplementary Table 1). After trimming and quality-filtering, the qualified reads were used as input of QIIME 2 pipeline (q2cli version 2019.10.0) (Bolyen et al., 2019). The classification results [operational taxonomic units (OTUs)] were generated using the qiime feature-classifier classify command with Greengenes database (13-8-99 version, DeSantis et al., 2006). Two replicates of each species were analyzed, and only assigned OTUs with read counts more than 10 were used for further analysis.
Results
Protein Content in Asian Horseshoe Crabs Hemolymph
To reveal the protein content of horseshoe crab hemolymph, proteomic analysis of the hemolymph proteins from both T. tridentatus and C. rotundicauda juveniles and T. tridentatus adults were carried out. A total of 1,056 and 1,107 proteins with high confidence values (exp. q value <0.05) were identified in C. rotundicauda and T. tridentatus juveniles respectively. In addition, a total of 1,254 proteins were detected in T. tridentatus adult hemolymph. The molecular weight distribution of C. rotundicauda and T. tridentatus hemolymph proteins was both observed with ∼80% within 80 kDa (Figure 1A). All these proteins were subjected to functional annotation with gene ontology (GO) (Figure 1B, Supplementary Tables 1, 2). In C. rotundicauda and T. tridentatus juveniles, ∼78% hemolymph proteins were associated with biological processes (GO:0008150) and molecular functions (GO:0003674) (Figures 1C,D). The majority of these proteins in two horseshoe crabs were associated in similar functional categories (Figures 1C,D) and KEGG pathway (Supplementary Figures 1,2), revealing conserved functions of horseshoe crab hemolymph proteins between different species.
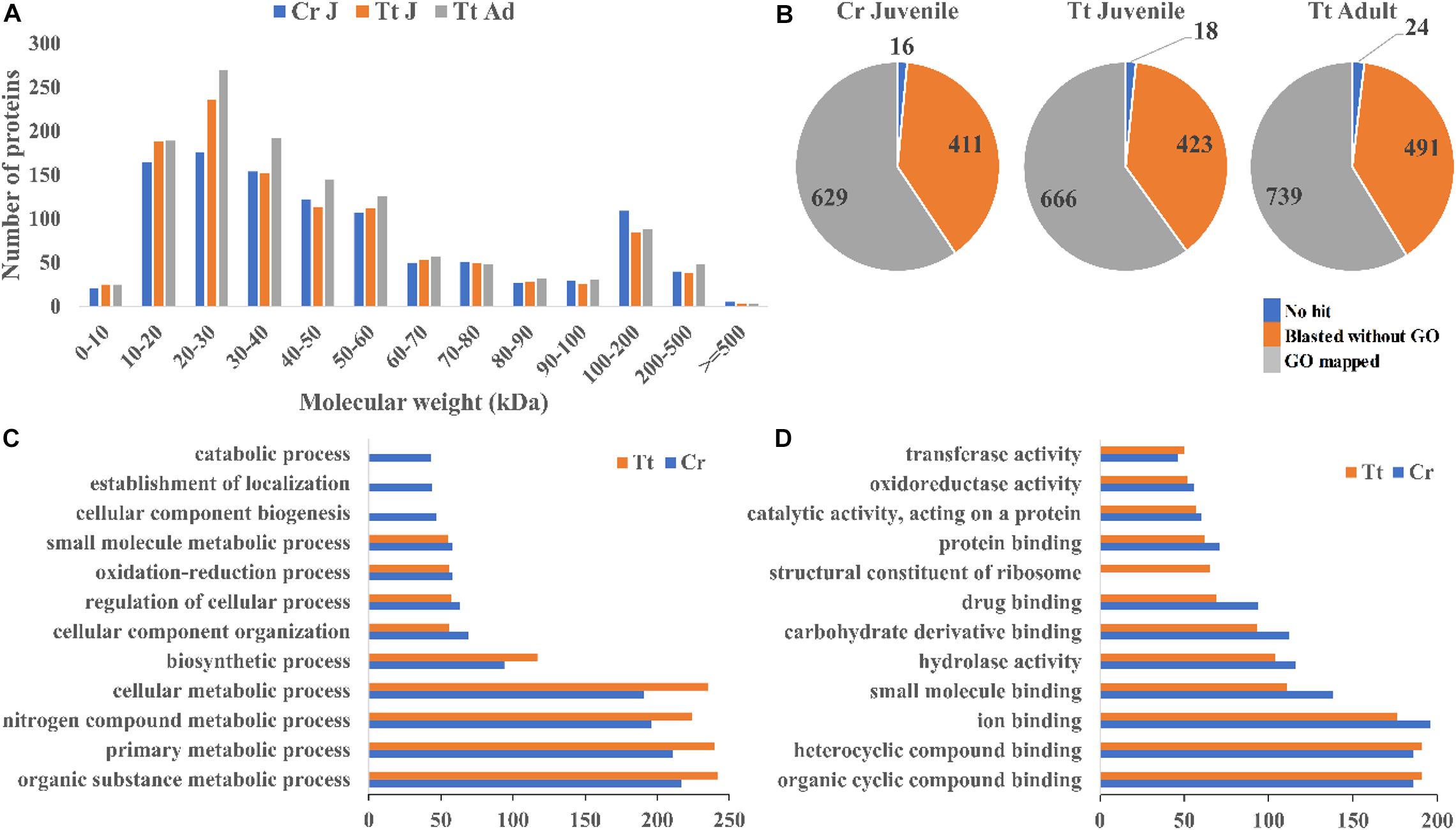
Figure 1. Landscape of the proteins detected in the hemolymph of C. rotundicauda (Cr) and T. tridentatus (Tt). (A) Size distribution of detected hemolymph proteins; (B) Protein numbers with the blast and functional annotation (GO) results in Cr juvenile (Cr J), Tt juvenile (Tt J), and adult (Tt Ad) hemolymph respectively; (C,D) Level 3 Gene ontology (GO) terms distribution based on biological processes (C) and molecular function (D) of juvenile C. rotundicauda and T. tridentatus hemolymph proteins.
Proteins involved in sesquiterpenoid hormone signaling pathways that affects the physiology and development of arthropods, including Hsp83, Chd64, and hemolymph juvenile hormone binding protein (hJHBP) were also identified in the hemolymph of these two species of horseshoe crabs for the first time. Phylogenetic analysis revealed a close relationship of Chd64 and hJHBP to similar proteins identified in crustaceans (Figure 2, Supplementary Tables 1, 2 and Supplementary Figures 3,4). Abundances of these proteins contributed 0.09, 0.06, and 0.02% to the total hemolymph proteins of adult T. tridentatus, juvenile T. tridentatus, and C. rotundicauda, respectively.

Figure 2. Phylogenetic relationship of juvenile hormone binding protein (JHBP) family. Maximum-likelihood phylogenetic tree showing potential hemolymph JHBP (hJHBP) revealed in horseshoe crab hemolymph. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.
In addition, Limulus clotting factor C, and other proteins involved in coagulation and related immune responses in spiders (Sanggaard et al., 2016) such as hemocyanins, von Willebrand factor related proteins, complement component, alpha-2-macroglobulin, histones, lectins, and coagulation factor were also identified in the hemolymph of two horseshoe crab species (Supplementary Tables 1, 2).
Further, 10 and 15 antimicrobial peptides (AMPs) were identified in the C. rotundicauda and T. tridentatus hemolymph samples, respectively (Supplementary Table 3), including the previously characterized tachyplesin and tachystatin (Miyata et al., 1989; Osaki et al., 1999).
Sodium Channels in Horseshoe Crabs
To reveal the possible relationships between sodium channels and tetrodotoxin resistance in horseshoe crabs, the sodium channel genes in the two Asian horseshoe crab species were examined and compared. A total of 15 and 7 sodium channel genes were annotated in C. rotundicauda and T. tridentatus respectively (Figures 3A,B and Supplementary Table 4). By comparing the amino acid sequences to the orthologs in pufferfish and other tetrodotoxin-tolerant species (Jost et al., 2008), amino acid replacements that lead to tetrodotoxin resistance in other animals were also identified in the two species (Figure 3C).
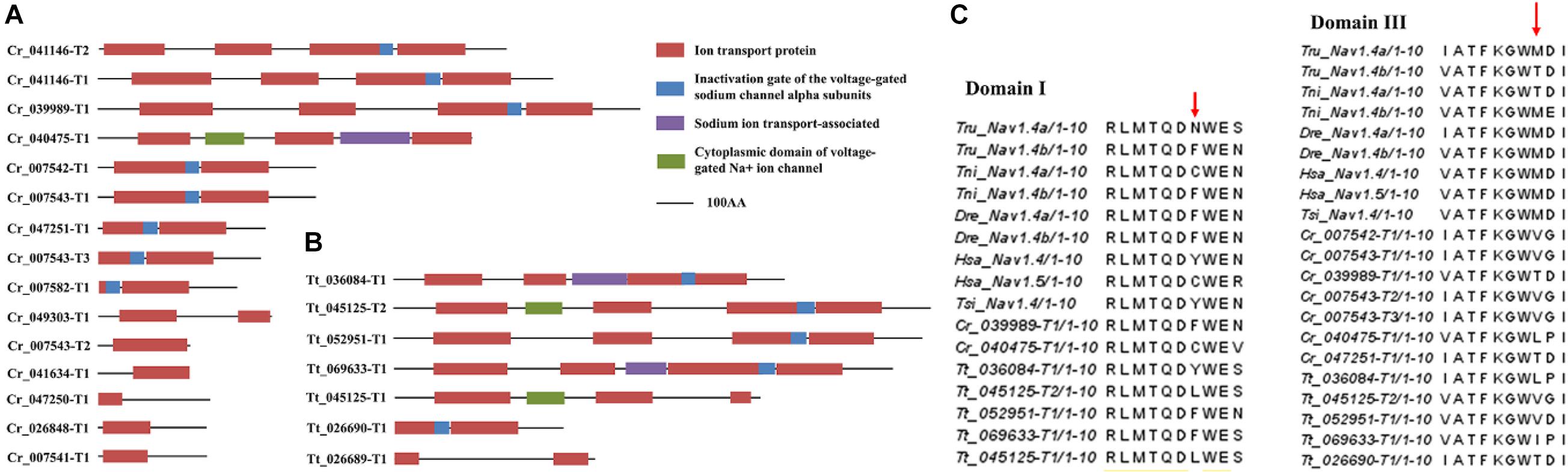
Figure 3. Protein domains of identified sodium channels in panels (A) C. rotundicauda, (B) T. tridentatus and (C) Tetrodotoxin-resistant related site replacements. Sequences used in the alignment: Takifugu rubripes Nav1.4a channel (ABB29441); Takifugu rubripes Nav1.4b channel (ABB29442); Tetraodon nigroviridis Nav1.4a channel (ABB29443); Tetraodon nigroviridis Nav1.4b channel (ABB29444); Danio rerio Nav1.4a channel (ABB29445); Danio rerio Nav1.4b channel (ABB29446); Homo sapiens skeletal muscle Nav1.4 channel (P35499); Homo sapiens cardiac Nav1.5 channel (Q14524) and Thamnophis sirtalis skeletal muscle Nav1.4 (AAW68224).
Variations in the Diversities of Gut Microbiome Between Two Horseshoe Crabs
To understand and compare the gut microbiome diversity of two horseshoe crab species, 16S amplicon sequencing of their gut fecal samples was conducted. 349 and 184 OTUs were obtained from T. tridentatus and C. rotundicauda gut, respectively (Figure 4A and Supplementary Tables 5, 6). Distribution of these OTUs at phylum level were examined and shown in Figure 4B. Top phyla found in T. tridentatus were Firmicutes (33.34%), Proteobacteria (30.85%), Tenericutes (14.56%), and Fusobacteria (12.99%), while Proteobacteria (50.40%), Tenericutes (26.22%), and Fusobacteria (18.05%) accounted for the top phyla found in C. rotundicauda. Among the top 5 most abundant genera found in the T. tridentatus and C. rotundicauda gut (Figure 4C), the most abundant were found to be Clostridium (22.61%) and Vibrio (36.20%) in T. tridentatus and C. rotundicauda, respectively, and the abundance of Propionigenium, Photobacterium, and Burkholderia were similar in the two horseshoe crab species. Further, the abundance of 66 common genera identified in both species was also compared (Figure 4D). This is the first result revealing the diversity and variations of gut microbiota in wild juveniles of the two Asian horseshoe crab species, and their significance remains to be determined.
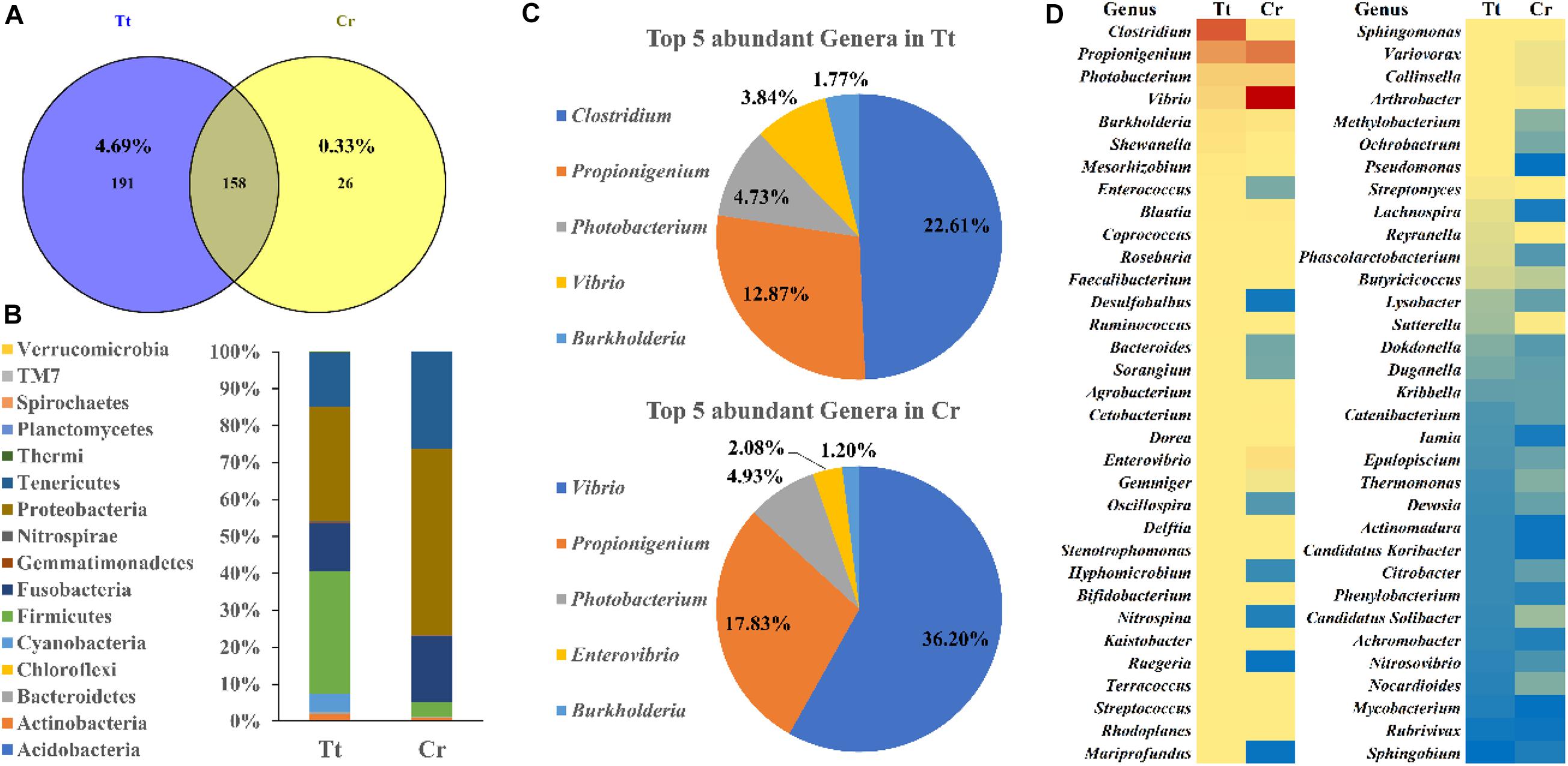
Figure 4. Gut microbiota diversity of two Asian horseshoe crabs C. rotundicauda (CR) and T. tridentatus (Tt). (A) Comparing OTUs identified in two horseshoe crabs; (B) Phylum abundance of identified OTUs in the gut of T. tridentatus and C. rotundicauda; (C) Top 5 most abundant genera identified in the two horseshoe crabs; (D) Comparison of the abundance of common genera in both species. The abundance is shown in blue to red (low to high).
Discussion
The hemolymph protein content of T. tridentatus and C. rotundicauda is, in general, comparable to other arthropods including termites and the Atlantic horseshoe crab, which have a majority of <80 kDa protein components (D’Amato et al., 2010; Zheng et al., 2019). Further, the identification of various conserved chelicerate peptides, including the Limulus clotting factor C, arachnid immunity proteins and coagulation factors, display a conserved hemolymph biology across lineages in the chelicerates. Unexpectedly, sesquiterpenoid pathway proteins were also identified in both species of horseshoe crabs.
The sesquiterpenoid hormones are well known to play important roles in arthropod development and physiology (Cheong et al., 2015; Qu et al., 2018). Although the majority of the functional studies on these hormones have been carried out on insects and crustaceans, recent studies showed that the sesquiterpenoid hormone system was established in the last common ancestor of Arthropoda (Qu et al., 2015, 2018). Here, we found that sesquiterpenoid signaling pathway components Hsp83, Chd64, and hJHBP are present in the hemolymph of both species of horseshoe crabs. Moreover, examination of the transcriptomes of different tissues of T. tridentatus and C. rotundicauda (Nong et al., 2020) also revealed the expression profiles of these hormonal pathway genes (Supplementary Figure 5). These data further supports the existence of sesquiterpenoid hormone system in marine chelicerates.
Hsp83 is a molecular chaperone that assists in the nuclear import of JH-Met complex (He et al., 2014). Chd64 is a protein in the JH signaling pathway that binds directly to the JH response element and forms in complexes that may link juvenile hormone production with ecdysone production (Li et al., 2007; Tarczewska et al., 2015). To date, the hJHBP has only been identified and functionally tested in some lepidopterans (Orth et al., 2003; Suzuki et al., 2011), which was used to deliver the juvenile hormone to target tissue and protect it from degradation (Hidayat and Goodman, 1994; Debski et al., 2004). The transportation of JH in hemolymph within other insects or non-insect arthropods is still not clear. Previously, only cytosolic JHBP (cJHBP) was characterized in non-insect arthropods (Qu et al., 2015). There are different types of sesquiterpenoid hormones in various arthropods [e.g., JH0, I, II, III, JHB3, and methyl farnesoate (MF)], Lepidoptera species exclusively possess JH0, JHI, and JHII in contrast to most of the other insects which use JH III, while the non-insect arthropods use MF as the major form (Goodman and Cusson, 2012; Qu et al., 2018; Bendena et al., 2020; Tsang et al., under review). The specific clade of Lepidoptera hJHBP (Figure 2) might be co-evolved with the special types of JH usage. Our finding of hJHBP in horseshoe crabs thus provides an opportunity to elucidate sesquiterpenoid transport in the chelicerates. Early in 1970s, Jegla et al. had demonstrated that injection of insect ecdysone and related analogs stimulated the molting of first stage American horseshoe crabs Limulus (Jegla and Costlow, 1970; Jegla et al., 1972). Although these studies have proven the functional ecdysone hormonal systems in horseshoe crabs, there is very little research about the sesquiterpenoid hormonal system in this group of arthropods. Our findings shed new lights on this aspect, providing useful information for future in vivo research (e.g., sesquiterpenoids treatment, hormonal measurement, genetic manipulation etc.).
Carcinoscorpius rotundicauda is one of the toxic marine invertebrates that has been known to cause tetrodotoxin poisoning, yet the cause of toxicity is still an unknown (Kungsuwan et al., 1987; Tanu and Noguchi, 1999; Kanchanapongkul, 2008; Suleiman et al., 2017). Given tetrodotoxin acts as a voltage-gated sodium channel blocker, animals that can accumulate high concentrations without adverse effects, such as pufferfish, have evolved with adaptive evolution in sodium channels to become toxin-resistant (Jost et al., 2008; Llewellyn, 2009). We thus analyzed the numbers of sodium channel genes in the genomes of C. rotundicauda and T. tridentatus (Figure 3). More sodium-channel genes could be identified in C. rotundicauda than in T. tridentatus. In addition, examination of expression profiles of these identified sodium-channel genes in transcriptomes of various tissues of both species revealed that C. rotundicauda exhibited higher expression levels of these genes (Supplementary Figure 6). Critical amino acid positions that potentially lead to toxin resistance in other animals were identified in these crabs. Further experiments such as pharmacologically treatment and toxin challenges are warranted to confirm these correlations.
This study also investigates the potential production of tetrodotoxin by its symbiotic microorganisms. In recent, two studies reported on gut microbiome in early stages (first and second instar) of cultivated T. tridentatus (Miao et al., 2020) and wild adults of T. tridentatus and C. rotundicauda (Wang et al., 2020), and this study provides the first result revealing the diversity and variations of gut microbiota in wild juveniles of the two Asian horseshoe crab species. We then analyzed if any species of bacteria known to produce tetrodotoxin (Magarlamov et al., 2017) was present in the gut of horseshoe crabs. In pufferfish Arothron hispidus and Vibrio harveyi strains isolated from this species were found to be able to produce tetrodotoxin (Campbell et al., 2009). However, only very low portion of Vibrio harveyi was identified in both C. rotundicauda (1.36%) and T. tridentatus (1.12%) (Supplementary Tables 5, 6), suggesting this strain might not be the major source of tetrodotoxin in horseshoe crabs. Moreover, other known tetrodotoxin-producing bacteria species were not found in these two horseshoe crab species. Intriguingly, a large proportion of Vibrio was identified in the gut of C. rotundicauda which existed in much lower proportion in T. tridentatus. Such finding could potentially mirror the correlations of Vibrio sp. with the accumulation of tetrodotoxin in various aquatic species (Magarlamov et al., 2017; Turner et al., 2017; Leao et al., 2018). On a different line, ingestion of toxic flatworm eggs had also been suggested as the source of tetrodotoxin in pufferfish Takifugu alboplumbeus (Okabe et al., 2019). Further, since tetrodotoxin-producing strains have been isolated from skin slime and kidneys of pufferfish (Campbell et al., 2009), toxin-producing microbes could be present in other body parts of horseshoe crab instead of gut. Thus, whether the toxic nature of C. rotundicauda is due to the endogenous symbiotic bacteria (contribution of other Vibrio sp. strains) and/or exogenous source via diet perhaps via determination of fatty acids profile (Kwan et al., 2019) remains to be further determined.
It is becoming clear that certain symbiotic microbiota can affect host reproduction and physiology. In arthropods, perhaps the better-known example is that of the endosymbiotic Wolbachia and that of the insects, which has been used to control mosquito populations (Flores and O’Neill, 2018). Indeed, the interactions between gut flora and the endocrine system of insects have also been documented (e.g., Shin et al., 2011; Storelli et al., 2011; Zheng et al., 2017). With this study, the possibility of determining the pathways of sesquiterpenoid hormone action and the effects of gut microbiota in horseshoe crab physiology, should permit the exploration of interactions between sesquiterpenoid regulation/metabolism and gut microbiota in this understudied group. This may provide a good system for examining the cross-kingdom regulation of sesquiterpenoid production and action in arthropods.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JHLH conceived and supervised the study. ZQ carried out the data mining. TCNL carried out the proteomics analyses. WN provided the gut microbiome and other bioinformatics analyses. HYY collected animals in the field and provided other samples for the experiments. IHTL and WLS analyzed the sodium channel genes. ZQ, TCNL, WLS, and JHLH drafted the first draft of manuscript. All authors commented on the manuscript, and approved its submission.
Funding
This research was supported by The Chinese University of Hong Kong Direct Grant (4053248), and Hong Kong Research Grant Council General Research Fund (14100919) (JHLH). WLS and IHTL were supported by Ph.D. studentships provided by The Chinese University of Hong Kong.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.579706/full#supplementary-material
References
Bendena, W. G., Hui, J. H. L., Chin-Sang, I., and Tobe, S. S. (2020). Neuropeptide and microRNA regulators of juvenile hormone production. Gen. Comp. Endocr. 295:113507. doi: 10.1016/j.ygcen.2020.113507
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. biotech. 37, 852–857.
Campbell, S., Harada, R. M., DeFelice, S. V., Bienfang, P. K., and Li, Q. X. (2009). Bacterial production of tetrodotoxin in the pufferfish Arothron hispidus. Nat. Prod. Res. 23, 1630–1640. doi: 10.1080/14786410903003780
Chen, C. P., Yeh, H. Y., and Lin, P. F. (2004). Conservation of the horseshoe crab at Kinmen, Taiwan: strategies and practices. Biodivers. Conserv. 13, 1889–1904. doi: 10.1023/b:bioc.0000035868.11083.84
Cheong, S. P. S., Huang, J., Bendena, W. G., Tobe, S. S., and Hui, J. H. L. (2015). Evolution of ecdysis and metamorphosis in arthropods: the rise of regulation of juvenile hormone. Integr. Comp. Biol. 55, 878–890. doi: 10.1093/icb/icv066
D’Amato, A., Cereda, A., Bachi, A., Pierce, J. C., and Righetti, P. G. (2010). In depth exploration of the hemolymph of Limulus polyphemus via Combinatorial Peptide Ligand Libraries. J. Proteome Res. 9, 3260–3269. doi: 10.1021/pr1002033
Dao, H. V., Takata, Y., Sato, S., Fukuyo, Y., and Kodama, M. (2009). Frequent occurrence of the tetrodotoxin-bearing horseshoe crab Carcinoscorpius rotundicauda in Vietnam. Fish. Sci. 75, 435–438. doi: 10.1007/s12562-008-0041-5
Debski, J., Wyslouch-Cieszynska, A., Dadlez, M., Grzelak, K., Kludkiewicz, B., Kolodziejczyk, R., et al. (2004). Positions of disulfide bonds and N-glycosylation site in juvenile hormone binding protein. Arch. Biochem. Biophys. 421, 260–266. doi: 10.1016/j.abb.2003.10.019
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/aem.03006-05
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Flores, H. A., and O’Neill, S. L. (2018). Controlling vector-borne diseases by releasing modified mosquitoes. Nat. Rev. Microbiol. 16, 508–518. doi: 10.1038/s41579-018-0025-0
Fusetani, N., Endo, H., Hashimoto, K., and Takahashi, K. (1982). Occurrence of potent toxins in the horseshoe crab Carcinoscorpius rotundicauda. Toxicon 20, 662–664. doi: 10.1016/0041-0101(82)90061-7
Goodman, W. G., and Cusson, M. (2012). “The juvenile hormones,” in Insect Endocrinology, ed. L. I. Gilbert (Amsterdam: Elsevier), 310–365.
He, Q., Wen, D., Jia, Q., Cui, C., Wang, J., Palli, S. R., et al. (2014). Heat shock protein 83 (Hsp83) facilitates methoprene-tolerant (Met) nuclear import to modulate juvenile hormone signaling. J. Biol. Chem. 289, 27874–27885. doi: 10.1074/jbc.m114.582825
Hidayat, P., and Goodman, W. G. (1994). Juvenile hormone and hemolymph juvenile hormone binding protein titers and their interaction in the hemolymph of fourth stadium Manduca sexta. Insect Biochem. Mol. Biol. 24, 709–715. doi: 10.1016/0965-1748(94)90058-2
Ho, B., Yeo, D. S., and Ding, J. L. (1994). A tetrodotoxin neutralizing system in the haemolymph of the horseshoe crab, Carcinoscorpius rotundicauda. Toxicon 32, 755–762. doi: 10.1016/0041-0101(94)90001-9
Jal, S., and Khora, S. S. (2015). An overview on the origin and production of tetrodotoxin, a potent neurotoxin. J. Appl. Microbiol. 119, 907–916. doi: 10.1111/jam.12896
Jegla, T. C., and Costlow, J. D. (1970). Induction of molting in horseshoe crab larvae by polyhydroxy steroids. Gen. Comp. Endocr. 14, 295–302. doi: 10.1016/0016-6480(70)90058-4
Jegla, T. C., Costlow, J. D., and Alspaugh, J. (1972). Effects of ecdysones and some synthetic analogs on horseshoe crab larvae. Gen. Comp. Endocr. 19, 159–166. doi: 10.1016/0016-6480(72)90016-0
Jhong, J. H., Chi, Y. H., Li, W. C., Lin, T. H., Huang, K. Y., and Lee, T. Y. (2019). dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 47, 285–297.
Jost, M. C., Hillis, D. M., Lu, Y., Kyle, J. W., Fozzard, H. A., and Zakon, H. H. (2008). Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 25, 1016–1024. doi: 10.1093/molbev/msn025
Kanchanapongkul, J. (2008). Tetrodotoxin poisoning following ingestion of the toxic eggs of the horseshoe crab Carcinoscorpius rotundicauda, a case series from 1994 through 2006. Southeast Asian J. Trop. Med. Public Health 39, 303–306.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kenny, N. J., Chan, K. W., Nong, W., Qu, Z., Maeso, I., Yip, H. Y., et al. (2016). Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity 116, 190–199. doi: 10.1038/hdy.2015.89
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 33, 1870–1874.
Kungsuwan, A., Nagashima, Y., Noguchi, T., Shida, Y., Suvapeepan, S., Suwansakornkul, P., et al. (1987). Tetrodotoxin in the horseshoe crab Carcinoscorpius rotundicauda Inhabiting Thailand. Nippon Suisan Gakk 53, 261–266. doi: 10.2331/suisan.53.261
Kwan, B. K. Y., Hu, M., Wang, Y., Cheung, S. G., and Shin, P. K. S. (2019). Fatty acids from controlled feeding as dietary markers of juvenile Chinese horseshoe crab, Tachypleus tridentatus. J. Mar. Biol. Assoc. U. K. 99, 421–428. doi: 10.1017/s0025315418000279
Leao, J. M., Lozano-Leon, A., Giraldez, J., Vilarino, O., and Gago-Martinez, A. (2018). Preliminary results on the evaluation of the occurrence of tetrodotoxin associated to Marine vibrio spp. in Bivalves from the Galician Rias (Northwest of Spain). Mar. Drugs 16:81. doi: 10.3390/md16030081
Levin, J., and Bang, F. B. (1964). A description of cellular coagulation in the Limulus. Bull. Johns Hopkins Hosp. 115, 337–345.
Levin, J., and Bang, F. B. (1968). Clottable protein in Limulus: its localization and kinetics of its coagulation by endotoxin. Thromb.Haemost. 19, 186–197. doi: 10.1055/s-0038-1651195
Li, Y., Zhang, Z., Robinson, G. E., and Palli, S. R. (2007). Identification and characterization of a juvenile hormone response element and its binding proteins. J. Biol. Chem. 282, 37605–37617. doi: 10.1074/jbc.m704595200
Llewellyn, L. E. (2009). “Sodium channel inhibiting marine toxins,” in Marine Toxins as Research Tools, eds N. Fusetani and W. Kem (Berlin: Springer), 67–97. doi: 10.1007/978-3-540-87895-7_3
Lorentz, M. N., Stokes, A. N., Rößler, D. C., and Lötters, S. (2016). Tetrodotoxin. Curr. Biol. 26, 870–872.
Magarlamov, T. Y., Melnikova, D. I., and Chernyshev, A. V. (2017). Tetrodotoxin-producing bacteria: detection, distribution and migration of the toxin in aquatic systems. Toxins 9:166. doi: 10.3390/toxins9050166
Maloney, T., Phelan, R., and Simmons, N. (2018). Saving the horseshoe crab: a synthetic alternative to horseshoe crab blood for endotoxin detection. PLoS Biol. 16:e2006607. doi: 10.1371/journal.pbio.2006607
Mehmood, Y. (2019). “What is Limulus amebocyte lysate (LAL) and its applicability in endotoxin quantification of pharma products,” in Growing and Handling of Bacterial Cultures, ed. M. Mishra (London: IntechOpen).
Miao, F., Zhao, Z., Li, Q., Song, J., Wang, Y., and Hu, M. (2020). Impact of initial feeding and molting on Tachypleus tridentatus gut microbiota. Curr. Microbiol. 77, 2847–2858. doi: 10.1007/s00284-020-02108-x
Miyata, T., Tokunaga, F., Yoneya, T., Yoshikawa, K., Iwanaga, S., Niwa, M., et al. (1989). Antimicrobial Peptides, Isolated from horseshoe crab hemocytes, Tachyplesin II, and Polyphemusins I and II: chemical structures and biological activity1. J. Biochem. 106, 663–668. doi: 10.1093/oxfordjournals.jbchem.a122913
Nong, W., Qu, Z., Li, Y., Barton-Owen, T., Wong, A. Y. P., Yip, H. Y., et al. (2020). Horseshoe crab genomes reveal the evolutionary fates of genes and microRNAs after three rounds (3R) of whole genome duplication. bioRxiv [Preprint]. doi: 10.1101/2020.04.16.045815
Okabe, T., Oyama, H., Kashitani, M., Ishimaru, Y., Suo, R., Sugita, H., et al. (2019). Toxic flatworm egg plates serve as a possible source of tetrodotoxin for Pufferfish. Toxins 11:402. doi: 10.3390/toxins11070402
Orth, A. P., Tauchman, S. J., Doll, S. C., and Goodman, W. G. (2003). Embryonic expression of juvenile hormone binding protein and its relationship to the toxic effects of juvenile hormone in Manduca sexta. Insect Biochem. Mol. Biol. 33, 1275–1284. doi: 10.1016/j.ibmb.2003.06.002
Osaki, T., Omotezako, M., Nagayama, R., Hirata, M., Iwanaga, S., Kasahara, J., et al. (1999). Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J. Biol. Chem. 274, 26172–26178. doi: 10.1074/jbc.274.37.26172
Qu, Z., Bendena, W. G., Tobe, S. S., and Hui, J. H. L. (2018). Juvenile hormone and sesquiterpenoids in arthropods: biosynthesis, signaling, and role of MicroRNA. J. Steroid Biochem. Mol. Biol. 184, 69–76. doi: 10.1016/j.jsbmb.2018.01.013
Qu, Z., Kenny, N. J., Lam, H. M., Chan, T. F., Chu, K. H., Bendena, W. G., et al. (2015). How did arthropod sesquiterpenoids and ecdysteroids arise? Comparison of hormonal pathway genes in non-insect arthropod genomes. Genome Biol. Evol. 7, 1951–1959.
Rudkin, D. M., Young, G. A., and Nowlan, G. S. (2008). The oldest horseshoe crab: a new xiphosurid from late ordovician konservat-lagerstätten deposits. Manitoba Canada. Palaeontology 51, 1–9. doi: 10.1111/j.1475-4983.2007.00746.x
Sanggaard, K. W., Dyrlund, T. F., Bechsgaard, J. S., Scavenius, C., Wang, T., Bilde, T., et al. (2016). The spider hemolymph clot proteome reveals high concentrations of hemocyanin and von Willebrand factor-like proteins. Biochim. Biophys. Acta. Proteins Proteomics 1864, 233–241. doi: 10.1016/j.bbapap.2015.11.004
Shin, S. C., Kim, S. H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell. Metab. 14, 403–414. doi: 10.1016/j.cmet.2011.07.012
Suleiman, M., Muhammad, J., Jelip, J., William, T., and Chua, T. H. (2017). An outbreak of tetrodotoxin poisoning from consuming horseshoe crabs in sabah. Southeast Asian J. Trop. Med. 48, 197–203.
Suzuki, R., Fujimoto, Z., Shiotsuki, T., Tsuchiya, W., Momma, M., Tase, A., et al. (2011). Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori. Sci. Rep. 1:133. doi: 10.1038/srep00133
Tanu, M. B., and Noguchi, T. (1999). Tetrodotoxin as a toxic principle in the horseshoe crab Carcinoscorpius rotundicauda collected from bangladesh. J. Food Hyg. Saf. 40, 426–430. doi: 10.3358/shokueishi.40.6_426
Tarczewska, A., Koztowska, M., Dobryszycki, P., Kaus-Drobek, M., Dadlez, M., and Ozyhar, A. (2015). Insight into the unfolding properties of Chd64, a small, single domain protein with a globular core and disordered tails. PLoS One 10:e0137074. doi: 10.1371/journal.pone.0137074
Turner, A. D., Dhanji-Rapkova, M., Coates, L., Bickerstaff, L., Milligan, S., O’Neill, A., et al. (2017). Detection of tetrodotoxin shellfish poisoning (TSP) toxins and causative factors in bivalve molluscs from the UK. Mar. Drugs 15, 277. doi: 10.3390/md15090277
Van Roy, P., Orr, P. J., Botting, J. P., Muir, L. A., Vinther, J., Lefebvre, B., et al. (2010). Ordovician faunas of burgess shale type. Nature 465, 215–218. doi: 10.1038/nature09038
Vestbo, S., Obst, M., Quevedo Fernandez, F. J., Intanai, I., and Funch, P. (2018). Present and potential future distributions of asian horseshoe crabs determine areas for conservation. Front. Mar. Sci. 5:164. doi: 10.3389/fmars.2018.00164
Wang, P., Ning, Y., Huang, J., Dai, Z., Wang, H., Wang, Y., et al. (2020). Intestinal tract microbe communities associated with horseshoe crabs from beibu gulf. China. Curr Microbiol. 77, 3330–3338. doi: 10.1007/s00284-020-02140-x
Zheng, H., Powell, J. E., Steeele, M. L., Dietrich, C., and Moran, N. A. (2017). Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 134, 4775–4780. doi: 10.1073/pnas.1701819114
Keywords: hemolymph, proteomics, sesquiterpenoids, gut microbiota, horseshoe crabs
Citation: Qu Z, Leung TCN, Nong W, Yip HY, Lee IHT, Cheung SG, Ming NS, So WL, Bendena WG, Tobe SS and Hui JHL (2020) Hemolymph Proteomics and Gut Microbiota of Horseshoe Crabs Tachypleus tridentatus and Carcinoscorpius rotundicauda. Front. Mar. Sci. 7:579706. doi: 10.3389/fmars.2020.579706
Received: 13 August 2020; Accepted: 27 October 2020;
Published: 02 December 2020.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Tara Essock-Burns, University of Hawaii, United StatesKimberly B. Ritchie, University of South Carolina Beaufort, United States
Copyright © 2020 Qu, Leung, Nong, Yip, Lee, Cheung, Ming, So, Bendena, Tobe and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jerome H. L. Hui, amVyb21laHVpQGN1aGsuZWR1Lmhr
†These authors have contributed equally to this work
 Zhe Qu
Zhe Qu Thomas C. N. Leung
Thomas C. N. Leung Wenyan Nong
Wenyan Nong Ho Yin Yip1,2†
Ho Yin Yip1,2† Siu Gin Cheung
Siu Gin Cheung Ngai Sai Ming
Ngai Sai Ming William G. Bendena
William G. Bendena Stephen S. Tobe
Stephen S. Tobe Jerome H. L. Hui
Jerome H. L. Hui