- 1Department of Microbiology and Department of Earth and Planetary Sciences, The University of Tennessee, Knoxville, Knoxville, TN, United States
- 2CologneAMS, University of Cologne, Köln, Germany
- 3Department of Physical and Environmental Sciences, Texas A&M University-Corpus Christi, Corpus Christi, TX, United States
- 4Institute for Chemistry and Biology of the Marine Environment (ICBM), Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
- 5MARUM – Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany
- 6Helmholtz Institute for Functional Marine Biodiversity (HIFMB), University of Oldenburg, Oldenburg, Germany
- 7Department of Geosciences, Baylor University, Waco, TX, United States
- 8Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA, United States
- 9School of Oceanography, University of Washington, Seattle, WA, United States
- 10Alfred-Wegener-Institut Helmholtz-Zentrum für Polar- und Meeresforschung, Bremerhaven, Germany
- 11Marine Science Institute, University of Texas at Austin, Port Aransas, TX, United States
- 12Department of Marine Chemistry, Leibniz Institute for Baltic Sea Research, Rostock, Germany
- 13Department of Marine Microbiology and Biogeochemistry, NIOZ Royal Netherlands Institute for Sea Research and Utrecht University, Den Burg, Netherlands
- 14Department of Geological Sciences and Institute of Arctic and Alpine Research (INSTAAR), University of Colorado Boulder, Boulder, CO, United States
- 15Skidaway Institute of Oceanography, University of Georgia, Savannah, GA, United States
Advances in sampling tools, analytical methods, and data handling capabilities have been fundamental to the growth of marine organic biogeochemistry over the past four decades. There has always been a strong feedback between analytical advances and scientific advances. However, whereas advances in analytical technology were often the driving force that made possible progress in elucidating the sources and fate of organic matter in the ocean in the first decades of marine organic biogeochemistry, today process-based scientific questions should drive analytical developments. Several paradigm shifts and challenges for the future are related to the intersection between analytical progress and scientific evolution. Untargeted “molecular headhunting” for its own sake is now being subsumed into process-driven targeted investigations that ask new questions and thus require new analytical capabilities. However, there are still major gaps in characterizing the chemical composition and biochemical behavior of macromolecules, as well as in generating reference standards for relevant types of organic matter. Field-based measurements are now routinely complemented by controlled laboratory experiments and in situ rate measurements of key biogeochemical processes. And finally, the multidisciplinary investigations that are becoming more common generate large and diverse datasets, requiring innovative computational tools to integrate often disparate data sets, including better global coverage and mapping. Here, we compile examples of developments in analytical methods that have enabled transformative scientific advances since 2004, and we project some challenges and opportunities in the near future. We believe that addressing these challenges and capitalizing on these opportunities will ensure continued progress in understanding the cycling of organic carbon in the ocean.
Introduction
The growth of marine organic biogeochemistry over the past several decades has been largely driven by advances in sampling tools, analytical methods and data handling capabilities. Max Blumer, one of the founding fathers of marine organic biogeochemistry, suggested in 1975 that limitations of analytical techniques are the major roadblock to understanding the chemical complexities of nature (Blumer, 1975). He asked whether we can conduct “realistic studies of nature that acknowledge the limitations of our present analytical powers and gaps in our understanding.” It is therefore not surprising that every symposium, workshop or review on marine organic biogeochemistry since (reviewed by Wakeham and Lee, 2019) has repeated one theme – continuing development of analytical capabilities is necessary to affect progress in the future.
There has always been a strong feedback between analytical advances and scientific advances, but analytical technology should not be the driving force. Rather, process-based scientific questions should drive analytical developments. Several of the challenges identified at the 2004 symposium honoring the late John Hedges (Lee et al., 2004) remain. At the same time, the impressive growth of ‘omics technologies since 2004 have changed the context in which marine organic biogeochemistry operates. For our purposes, we define ‘omics as the set of analytical technologies and associated algorithms and databases that shine light on the internal workings of ecosystems via the sequences of biological molecules such as DNA, RNA, and protein, and the concentrations and structures of lipids and small organic molecules that are essential to life. Accordingly, we used the following themes to guide the discussions of this working group on analytical methodologies:
(i) Multidisciplinary investigations generating diverse data. Integration of disparate data sets, including better global coverage and mapping, is critical to understanding processes in the ocean.
(ii) “Molecular headhunting,” or attempts to characterize the structures present in marine organic matter for their own sake, are being subsumed into process-driven targeted investigations requiring new analytical capabilities.
(iii) There are still major gaps in characterizing the composition and behavior of macromolecules – the molecularly uncharacterized component of marine organic matter after Hedges et al. (2000).
(iv) Improvements are needed in protocols for making in situ rate measurements of key biogeochemical processes.
Here, we present a perspective review on advances in analytical methods since 2004 derived from a 2019 Hanse-Wissenschaftskolleg Workshop on Marine Organic Geochemistry, and on some pressing analytical needs for the next decade or two. We do not attempt to create a totally comprehensive review of advances and needs; such an effort would require a book. Rather, we highlight advances and needs that workshop participants found particularly important, exciting, or underappreciated, as summarized in Figure 1. We hope that this review will serve as a guide and inspiration to those attempting to expand the limits of analytical chemistry to better understand the cycling of organic matter in the ocean.
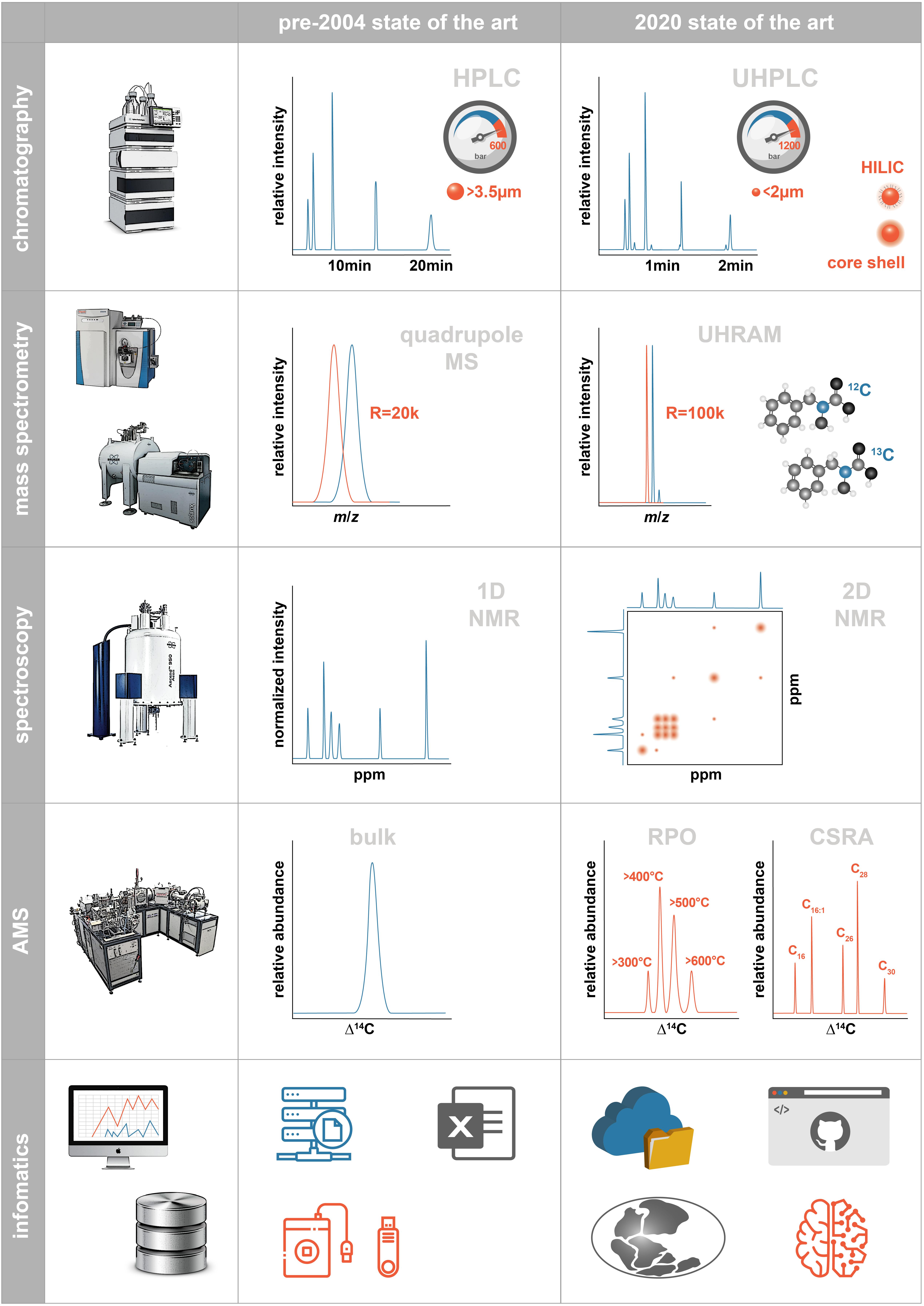
Figure 1. Schematic illustrating the state of analytical methodologies available for marine organic biogeochemistry prior to 2004 and at present. Abbreviations are described in the text.
Advances in Analytical Instrumentation
Nano-Elemental Analysis
Advances in continuous flow elemental analysis-isotope ratio mass spectrometry (EA-IRMS) have facilitated the tandem isotopic analysis of carbon and nitrogen in suspended, sinking, and sedimentary organic matter for describing C and N cycling processes in the ocean. Modifications to EA-IRMS systems allow for small samples of ca. 10–100 nmol, termed “nano-EA” (Polissar et al., 2009; Ogawa et al., 2010; Langel and Dyckmans, 2014), and have been applied to the study of isolated biomarker compounds (Junium et al., 2015; Fulton et al., 2018; Isaji et al., 2020) and samples with low content of organic matter (Junium et al., 2018; Cui et al., 2019; Murray et al., 2019).
Other techniques to minimize sample size include the spooling wire microcombustion (SWiM) method. SwiM has been applied to the C isotope analysis of sorted microbial cells (Eek et al., 2007), biomarkers (Pearson et al., 2016), and diatoms from sediments (Hansman and Sessions, 2016). The “denitrifier method” (Sigman et al., 2001) has been applied to the N isotopic analysis of biomarker compounds, phytoplankton, and dissolved organic matter (Robinson et al., 2004; Knapp et al., 2005; Higgins et al., 2009).
Chromatography
Gas chromatography (GC) and high-performance liquid chromatography (HPLC) are common in the analysis of molecular biomarkers in marine biogeochemistry (Wakeham et al., 2007, 2009). GC and HPLC systems are coupled to a range of detectors, but most compound identifications are made by mass spectrometry (MS). Chromatographic methods generally target specific compound classes useful in biogeochemical proxy development, and recent developments have increased the analytical capacity to resolve previously intractable components. Comprehensive two-dimensional GC (GC × GC) has been applied to biomarker studies and unresolved complex mixtures in petroleomics (reviewed by Eiserbeck et al., 2014) and non-targeted marine contaminant research (Hoh et al., 2012). It also has great potential in the study of metabolomics (Higgins Keppler et al., 2018).
Relatively large (m/z 500-2000) polar molecules (pigments and intact polar lipids) are most effectively separated by HPLC, with some of the most recent analytical advances summarized by Wörmer et al. (2015). Garrido et al. (2012) reviewed methods for pigment analysis, and Higgins et al. (2012) summarized pigment-based phytoplankton chemotaxonomy. Intact polar lipid analysis by HPLC is used in environmental proxy development (e.g., Rütters et al., 2002; Sturt et al., 2004; Van Mooy et al., 2009; Hopmans et al., 2016; Schubotz et al., 2018), and the range of compounds that can be detected during an individual analysis has expanded (Collins et al., 2016). Underivatized small polar metabolites are also analyzed by HPLC (Genta-Jouve et al., 2014; Johnson et al., 2017). Most of the recent technical improvements include Ultra-HPLC (UHPLC or UPLC) systems operating at higher pressure than conventional HPLC systems to rapidly resolve molecular peaks, in addition to chromatographic columns containing sub-2-μm solid-core particles that enable large increases in efficiency. UHPLC has been applied to the analysis of pigments (Zhang et al., 2016), intact polar lipids (Besseling et al., 2020), lipid biomarkers in temperature proxy calculations (de Bar et al., 2017), and organic ligands (Wichard, 2016).
The introduction of hydrophilic interaction liquid chromatography (HILIC) columns resulted in enhanced peak resolution and reduced analysis time of IPLs using normal-phase chromatography, which separates molecules according to their polar headgroups (Wörmer et al., 2013). On the other hand, intact polar lipid (IPL) analysis using reverse-phase chromatography provides compound separation by alkyl chain hydrophobicity (Wörmer et al., 2013), which improves the separation of compounds with the same headgroup and slight differences in the core lipid structure, and also allows for the simultaneous analysis of less polar compounds such as glycerol dialkyl glycerol tetraethers (GDGTs), pigments, alkenones, bacteriohopanepolyols (BHPs), and quinones. Core and intact archaeal isoprenoidal GDGTs can be now analyzed in a single run using reversed-phase chromatography (Zhu et al., 2013), while the analysis of core bacterial and archaeal GDGTs has been improved by the use of ethylene bridged hybrid (BEH) HILIC columns (Hopmans et al., 2016). The detection of BHPs has been improved by the analysis of non-derivatized compounds using either atmospheric pressure chemical ionization (APCI) (Talbot et al., 2016) or electrospray ionization (ESI) (Rush et al., 2019).
Fourier-Transform Ion Cyclotron Resonance Mass Spectrometry (FT-ICR-MS)
Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS), with its unsurpassed mass resolution and mass accuracy (UHRMS – ultrahigh-resolution MS), provides the primary tool to study the composition of complex mixtures of organic compounds not accessible via other methods (Comisarow and Marshall, 1974; Kujawinski et al., 2004; Koch et al., 2005; Marshall and Chen, 2015). FT-ICR-MS provides elemental formulae, involving primarily C, H, and O and lesser amounts of N, S, and P. Elemental formulae may be converted into molecular formulae, and elemental compositions are visualized in van Krevelen plots of H/C and O/C ratios where they may be compared with the same ratios in potential biochemical precursors (lipids, proteins, carbohydrates), between samples, and across various spatial scales. H/C and O/C ratios that diverge from precursors indicate organic matter (OM) that has been degraded or transformed. FT-ICR-MS as a semi-quantitative fingerprinting method can be combined with targeted assays quantifying specific fractions of the natural organic matter (NOM) pool such as amino acids, carbohydrates or lipids. The unconventional type of information derived from FT-ICR-MS analyses – relative intensities of up to tens of thousands of peaks – warrants the use and development of new tools for data evaluation and interpretation. We discuss these needs in more detail below, in the section labeled “Untargeted high-resolution analysis: FT-ICR-MS and Orbitrap.”
FT-ICR-MS has enabled detailed analysis of dissolved organic matter (DOM) composition across gradients from marine to freshwater biomes (Medeiros et al., 2015; Osterholz et al., 2016) and ocean basins (Lechtenfeld et al., 2014; Hansman et al., 2015; Martínez-Pérez et al., 2017) to sediment porewaters (Schmidt et al., 2011; Seidel et al., 2014) to understand its biotic and abiotic turnover. The detection limit below femto-or even sub-attomolar concentrations enabled the description of bacterial metabolites (Kujawinski, 2011; Schwedt et al., 2015; Noriega-Ortega et al., 2019; see further description below), trace metal-DOM complexes (Lechtenfeld et al., 2011; Waska et al., 2015), or anthropogenic contaminants (Wagner et al., 2015b; Powers and Gonsior, 2019).
A critical caveat for FT-ICR-MS analysis is that a single chemical formula could represent structurally distinct compounds, which may potentially exhibit divergent chemical properties. The extent to which this is a problem in practice is a subject of substantial controversy. Coupling FT-ICR-MS to trapped ion mobility mass spectrometry (TIMS-FT-ICR-MS) provides new insights into the presumed high structural diversity of DOM. Lower limits of isomeric diversity could be established (Tose et al., 2018; Leyva et al., 2019), while upper limit estimates still range widely and are dependent on detection limit and resolving power.
Orbitrap Mass Spectrometers
Orbitraps, a new type of Fourier-transform MS (Hu et al., 2005), use an electrostatic field rather than the superconducting magnets of FT-ICR-MS. These are available to a much wider range of labs than FT-ICR-MS because purchase prices as well as maintenance expenses are much lower than for FT-ICR-MS. Although maximum mass accuracy is slightly lower for Orbitraps than for FT-ICR-MS, acquisition rates are faster (15 Hz vs. 1 Hz), compatible with LC resolution allowing detection of multiple mass spectra across a chromatographic peak. This provides (ultra-) high-resolution accurate mass (HRAM) determination with resolution of up to 1,000,000 at m/z 200 and mass accuracies of <1 ppm and can be used for targeted and untargeted analyses. Extended mass range Orbitrap systems now have analytical windows of up to m/z 20,000 allowing, for example, for intact protein analysis. Orbitrap also offers superior resolving power at the low m/z end of the analytical window in comparison to other HRAM instruments such as quadrupole time-of-flight (Q-TOF), important for structural elucidation in metabolomics and lipidomics. Thus, Orbitrap mass spectrometers provide the potential to greatly expand the number of labs that can use high-resolution mass spectrometry to characterize marine OM. Coupled with UHPLC and ESI, Orbitrap technology has proven to be a very powerful tool for proteomics, metabolomics, and lipidomics in marine organic geochemical research (Poulson-Ellestad et al., 2014; Saito et al., 2014; Hunter et al., 2015; Cantarero et al., in press) and allows the deconvolution of isotopic patterns of all common stable isotopes (compound-specific isotope analysis; CSIA) and position-specific isotope analysis (PSIA) of molecular fragment ions including multiply substituted isotopologues (Eiler et al., 2017). PSIA holds promise to intensively increase our understanding of biogeochemical pathways, cell physiology and metabolic state, dietary patterns, and degradation effects among other potential applications.
Analysis of Polar, Biologically Labile Organic Molecules
A central goal of organic biogeochemistry has been to track the composition of biologically relevant molecules as they are produced, consumed or transformed by small and large marine organisms (Moran et al., 2016). These molecules can be categorized according to the major biochemicals within cells (i.e., lipids, proteins, carbohydrates, and nucleic acids), their monomeric counterparts (i.e., amino acids, sugars, and nucleobases) and other metabolic intermediates. Small polar intermediates in this last biochemical class have received attention in the last 5 years as advances in sample acquisition, liquid chromatography, and mass spectrometry have expanded the analytical window of marine organic compounds. These compounds can be detected and quantified in both dissolved (Johnson et al., 2017) and particulate (Durham et al., 2019; Johnson et al., 2020) pools, and include molecules such as growth substrates, metabolic cofactors, and signaling molecules (the collective is often referred to as “metabolites,” inferring a biological origin and sink).
Methods for metabolite analysis couple liquid chromatography to high-resolution mass spectrometry (LC/MS), thus targeting molecules that are polar and soluble in aqueous solution. The two most popular LC-column materials are reversed-phase C18 (or C8) and hydrophilic-interaction liquid chromatography (HILIC) and can be used in parallel to detect the largest suite of compounds within a single sample (Boysen et al., 2018). Detection limits with triple-quadrupole or Orbitrap mass spectrometry fall within the femtogram to picogram range (per 5–10 μL injection). Thus, reasonable signals are achievable with a few hundred mL to a few L of seawater for both dissolved and particulate metabolites. Smaller volumes can be used for metabolite analysis by gas chromatography (GC) MS (Sogin et al., 2019) but fewer molecules are accessible to the derivatization required for GC analysis.
Targeted and untargeted analyses of polar metabolites generate large, complex data sets that require novel tools to analyze. Many of these tools cross disciplinary boundaries from biomedicine to marine science. Of interest to the marine science community will be advances in programs for chromatographic data analysis (MS-DIAL, Lai et al., 2018; MZmine, Pluskal et al., 2010; XCMS, Smith et al., 2006), programs to make existing tools easier to use (Li and Li, 2019; McLean and Kujawinski, 2019), and improvements in websites used to search and store mass spectrometry data (METLIN, Guijas et al., 2018; MetaboLights, Haug et al., 2020; the Metabolomics Workbench, Sud et al., 2016; MassIVE-KB, Wang et al., 2018). Several computational tools increase our ability to identify unknown compounds through searches and in silico calculations (CSI:FingerID, Dührkop et al., 2019; ClassyFire, Feunang et al., 2016; MetFrag, Ruttkies et al., 2019). The structural similarities in MS2 spectra have proven especially useful in identifying compounds (MolNetEnhancer, Ernst et al., 2019; MetDNA, Shen et al., 2019; MS2LDA, van der Hooft et al., 2020), which is evident in the expanded list of tools now available at Global Natural Products Social Networking (Wang et al., 2016; Aron et al., 2020). For example, users can search mass spectrometry data using MASST (Wang et al., 2020) and reuse publicly available data via ReDU (Jarmusch et al., 2019). Collectively, these tools allow marine scientists to expand our understanding of OM in seawater (Kharbush et al., 2016; Hartmann et al., 2017; Longnecker and Kujawinski, 2017; Petras et al., 2017).
Metabolite analyses offer a strong connection to genome-based assessments of community metabolism, both potential (diversity analysis or metagenomics) and realized (metatranscriptomics or metaproteomics). Metabolic capabilities can be inferred from diversity analysis while metabolic pathways can be identified through metagenomics. Pathway activity can be monitored through metatranscriptomics and metaproteomics and their end result within elemental cycles can be tracked through metabolomics. Early studies have used diversity analysis to connect culture-based metabolic inferences with metabolite analyses (Allen et al., 2008; Johnson et al., 2016; Heal et al., 2017). Field studies are connecting community composition (and inferred metabolic potential) with metabolite concentrations to postulate microbe-microbe interactions (Durham et al., 2019) and OM-related processes (Johnson et al., 2020). This field is rapidly expanding with new method developments presented at major conferences; significant advances in our understanding of microbe–microbe interactions through chemical exchange are likely in the near future.
Nuclear Magnetic Resonance (NMR)
Nuclear magnetic resonance spectroscopy (NMR) is a group of analytical techniques that rely on measuring how NMR-active nuclei (i.e., 13C, 1H, 15N, 31P) resonate in the presence of a strong magnetic field. Within the same nucleus (e.g., 13C), the resonance frequency differs slightly based on shielding of local electrons, deshielding from adjacent chemical functional groups, and coupling with other nuclei. These differences in resonance allow identification of the chemical functional groups in which nuclei reside, and in some cases larger-scale structural information.
In the 1980s and 1990s, cross-polarization (CP) and magic angle spinning (MAS) techniques were coupled to allow characterization of functional group abundance in marine DOM isolate fractions such as XAD-resin-extracted DOM, ultrafiltered DOM (Benner et al., 1992; Abdulla et al., 2010a,b), and DOM concentrated by reverse osmosis–electrodialysis (Koprivnjak et al., 2009), as well as ultrafiltered particulate organic matter (POM) (particles 0.1–60 μm; Sannigrahi et al., 2005) and sinking particles (Liu et al., 2009). One of the major drawbacks of CP-13C NMR is underestimating de-protonated carbons (e.g., carbonyl and substituted aromatic carbon), especially in the presence of paramagnetic iron, which makes it a semi-quantitative technique (Pfeffer et al., 1984; Mao et al., 2000; Abdulla et al., 2010a). Direct polarization (DP-MAS-13C NMR) is more quantitative, and has been used to measure changes in the composition of DOM isolated from coastal and open oceans sites (Helms et al., 2015; Cao et al., 2018). However, DP- 13C NMR requires a longer acquisition time and its spectrum has a lower signal to noise (S/N) ratio relative to CP-13C NMR.
Recently, many sophisticated solid-state spectra-editing techniques (e.g., 13C chemical shift anisotropy filter, CH selection, CH2 selection) have been applied to resolve overlapping peaks within marine DOM (Helms et al., 2015; Cao et al., 2018). Two-dimensional correlation spectroscopy analysis techniques on CP-MAS-13C NMR spectra have been used to track the changes of different DOM components under different perturbations and to correlate 13C NMR with other spectroscopy techniques such as Fourier-transform infrared spectroscopy (FTIR) and FT-ICR-MS (Abdulla et al., 2010b, 2013a,b).
DOM has also been characterized by 1H NMR, but this technique can be challenging due to interference from 1H in water. Therefore, 1H NMR has typically required extraction, drying, and redissolution in D2O (e.g., Repeta et al., 2002). However, recent advances in high magnetic field NMR (currently up to 900 MHz or 21 Tesla) and the development of water suppression techniques allow analysis of marine DOM using 1H NMR techniques without any isolation or pretreatment on open ocean and sediment pore water samples (Lam and Simpson, 2008; Zheng and Price, 2012; Fox et al., 2018).
Fewer studies have reported marine DOM and POM spectra for 31P and 15N (McCarthy et al., 1997; Clark et al., 1998; Kolowith et al., 2001; Aluwihare et al., 2005; Maie et al., 2006; Sannigrahi et al., 2006). Chemical shifts from multiple nuclei are measured in two-dimensional (2D) NMR spectroscopy techniques, which allows resolution of overlapping peaks, verification of the interpretation of the chemical shifts, and identification of specific structures. These 2D-NMR techniques include correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), heteronuclear single-quantum correlation spectroscopy (HSQC), heteronuclear multiple-bond correlation spectroscopy (HMBC) and 1H-13C heteronuclear correlation (HETCOR) (e.g., Repeta et al., 2002; Hertkorn et al., 2006, 2013; Cao et al., 2018).
Natural-Level Radiocarbon Analysis
Advancements in accelerator mass spectrometry (AMS) technology have dramatically reduced the cost and sample size required for 14C analysis. It is now possible to measure natural abundance 14C in samples containing as low as a few tens to hundred μg C rather than the routine 1 mg C (Pearson et al., 1998; Santos et al., 2007; Shah and Pearson, 2007). Compact low energy AMS systems (e.g., the Mini Carbon Dating System; Synal et al., 2007) represent a milestone toward (ultra-) small-scale 14C analyses (<10 μg C at a precision of ±2‰ for modern samples Wacker et al., 2010). At this point, 14C measurements are analytically no longer size-limited, but rather by the sample pre-treatment methods and the associated blank C contamination (Santos et al., 2010). These improvements in sample size requirements have also allowed for natural-level compound-specific radiocarbon analysis, which has improved our understanding of carbon metabolisms of marine organisms, sedimentary processes in the ocean, and continental carbon cycling (Wakeham and McNichol, 2014; Druffel et al., 2016; Van der Voort et al., 2018). Methods allowing for the isolation of individual compounds, mostly based on preparative gas chromatography or preparative liquid chromatography, continue to be developed for multiple compound classes. Compound classes for which compound-specific radiocarbon analytic protocols have been established include alkanes, alkanoic acids (Eglinton et al., 1996), benzene polycarboxylic acids (BPCAs) (Gierga et al., 2014), PAHs (Reddy et al., 2002) and phospholipid fatty acids (Druffel et al., 2010; Wakeham and McNichol, 2014), aliphatic alcohols, sterols and hopanols (Pearson et al., 2001), lignin phenols (Hou et al., 2010; Ingalls et al., 2010), GDGTs (Ingalls et al., 2006; Birkholz et al., 2013), amino acids (Bour et al., 2016; Ishikawa et al., 2018), pigments (Kusch et al., 2010), and diatom bound organic compounds (Ingalls et al., 2004).
Several recent developments suggest further improvements to natural abundance 14C analysis. New CO2-accepting ion sources allow peripheral instruments, such as elemental analyzers, other oxidation or hydrolysis systems and potentially gas chromatography, to interface directly to AMS (Bronk Ramsey et al., 2004; Ruff et al., 2010; Haghipour et al., 2019). This eliminates the need for an offline graphitization step, which is labor-intensive and potentially introduces 14C contamination. Moreover, positive ion mass spectrometry (PIMS) uses an electron cyclotron resonance (ECR) plasma ion source generating high ion beam currents, which may allow further reductions in sample size. Finally, saturated absorption cavity ring-down spectroscopy (SCAR), a technique based on optical absorption of CO2 gas, is now capable of measuring 14C at natural abundance levels (Galli et al., 2016). The accuracy (±3%) and sample size required (6 mg) are not currently acceptable for most marine organic geochemical applications, but because this technique is inherently simpler than mass spectrometry, it has the potential to dramatically reduce the cost of natural abundance 14C analysis.
Improvements have also focused on peripheral instrumentation. Ramped pyrolysis/oxidation (RPO) involves progressively heating OM to 685°C under helium, then oxidizing the pyrolysis products to CO2 (Rosenheim et al., 2008). This procedure exploits differences in stability of organic molecules to quantify and characterize different pools of OM. Coupled with radiocarbon dating, RPO is used to independently measure the age and reactivity distribution of fresh autochthonous OC and diagenetically stabilized allochthonous OM in terrestrial and marine settings. Recent applications of RPO include studies of riverine organic carbon (OC) (Rosenheim and Galy, 2012), oil-spill impacted sediments (Pendergraft et al., 2013), permafrost OC cycling in the Arctic (Zhang et al., 2017), weathering and recycling of petrogenic OC (Hemingway et al., 2018), the fate of terrestrial OC in the marine environment (Bao et al., 2019), soil OC preservation (Grant et al., 2019) and a global assessment of the controls on OC persistence in the environment (Hemingway et al., 2019). Because it characterizes the reactivity and age distribution of the entire OC pool, RPO is complementary to compound-specific 14C, which is much more specific but characterizes only a small portion of the bulk OC. Serial UV oxidation protocols aim at the separation of distinct DOC subpools for 14C analysis (Beaupré et al., 2007; Beaupré and Druffel, 2012). In both RPO and UV-oxidation, sample size limits have been pushed in recent years and more attention has been put toward minimizing potential blank C contamination.
Enzyme Assays
Field measurements along spatial and temporal gradients have been the mainstay for characterizing OM cycling in the ocean. However, process-based measurements, such as on-deck or laboratory incubations, are essential to deconvolute environmental factors affecting OM decomposition, or to quantify rates of distinct processes, although these rates should be regarded as “potential.” An essential step along the pathway from POM to DOM is extracellular enzymatic hydrolysis, in which high molecular weight DOM is cleaved into fragments smaller than ∼600–1000 Da, allowing for microbial uptake (Arnosti, 2011). Various simple substrate proxies, often fluorogenic or with a fluorescent tag, are utilized to evaluate hydrolytic rates in different aquatic environments (e.g., Hoppe, 1983; Somville and Billen, 1983). Interpreting results requires caution since natural OM is far more complex than these simple proxies in terms of its structural diversity and three-dimensional conformation and organic matter uptake mechanisms are complex (Reintjes et al., 2017). Nevertheless, these simple substrates integrate the activities of diverse enzymes reasonably well (Steen et al., 2015). Substrate proxies better resembling NOM in terms of structural complexity, such as polysaccharides and peptides with fluorescent tags, provide further insight (Arnosti, 1995; Pantoja et al., 1997). Extracellular enzymatic hydrolysis is often instant and can outpace the uptake of hydrolysis products by microbes (Arnosti, 2004; Liu et al., 2013), and hydrolysis rate is highly dependent on substrate structure and environment (Liu et al., 2010; Arnosti et al., 2011). These methods collectively provide a bridge between genome/proteome-based methods to understand microbial metabolism and methods for the analysis of organic carbon composition.
Advances in Data Processing
The mandate for scientists to describe experiments in sufficient detail that they could be reproduced by others dates to at least the 17th century (Stodden, 2010). With respect to obtaining and physically processing samples, clearly written descriptions of analytical methods in manuscripts can suffice to make methods reproducible. However, many of the new analytical systems described above generate large quantities of data which must be extensively transformed and reduced prior to presentation, frequently requiring custom-written computer code. Accordingly, an increasing number of analytical chemists have learned to become programmers on the side. While this opens exciting new opportunities for discovery, it also places new burdens on analytical chemists to clearly describe the computational methods by which we obtain our results. Such descriptions are as essential to reproducibility as are descriptions of the chemical conditions by which a sample was analyzed.
Fortunately, several trends have facilitated reproducible data analysis. First, (relatively) user-friendly, free and open-source software languages such as R and python, and to some extent Julia, have been developed into sophisticated analytical platforms, complete with packages for almost any type of statistical analysis that is common in the literature and high-quality integrated development environments. These languages have rapidly gained popularity compared to closed-source platforms such as SAS and SPSS (Muenchen, 2019).
Second, tools to integrate code with text, such as the R Markdown document specification and JupyterLab allow seamless integration of text, code, and code results (Baumer and Udwin, 2015; Perkel, 2016, 2018). This makes it easy to relate code to the results that it produces, and to annotate code and results with a plain-language explanation of why the analysis was done in the way that it was. Such tools have long been available (Knuth, 1984; Ramsey, 1994; Leisch, 2002) but used the cumbersome TeX/LaTeX document preparation system (Knauff and Nejasmic, 2014) and did not support the languages most often used for data analysis.
Third, the availability of version control systems such as git and mercurial encourage integration of version control into the process of writing data analysis scripts and packages. Version control encourages a systematic approach to updating documents and tracking contributions by multiple authors, a key aspect of project management that scientists often manage informally (Cham, 2012). Associated hosting services such as GitHub and GitLab, each of which are commercial services that offer a free tier, make it easy to share and collaborate on code. These tools collectively take some time to learn (and can be frustrating even for experienced users; Munroe, 2015) but once adopted, they improve research productivity (Wilson, 2006; Ram, 2013).
Fourth, systems to facilitate the preservation and sharing of raw data have developed considerably. Repositories such as PANGAEA, National Centers for Environmental Information (NCEI), and Biological-Chemical Oceanography Data Management Office (BCO-DMO) have grown rapidly over the past decade. As of early 2020, PANGAEA has over 392,000 unique, publicly available data sets (Grobe et al., 2006). Concurrently, academics have developed best-practices and principles for data sharing and preservation, although there is disagreement in the literature as to how best to reconcile ideals of how best to share and preserve data with the practicalities of doing so with limited time and resources (Wilkinson et al., 2016; Tierney and Ram, 2020).
Finally, a broad ecosystem of support services for the above three developments has emerged. Evidence-based short courses such as Software Carpentry and Data Carpentry offer low-cost, evidence-based lessons in computational skills aimed specifically at scientific researchers (Wilson, 2014). Websites such as stackoverflow.com host questions about programming, and provide incentives for users to provide clear, accurate answers. Integrated Development Environments such as R Studio and JupyterLab, which are explicitly designed for data analysists, make the process of code development and debugging much easier (Gandrud, 2013). These trends have led to the development of a range of open-source tools that are valuable for marine organic geochemistry, particularly instruments that generate large quantities of data such as ultra-high resolution mass spectrometers, for instance XCMS (Smith et al., 2006) and LOBSTAHS (Collins et al., 2016), and many of the tools listed in the “Analysis of polar, biologically labile organic molecules” section.
All of these trends have deep roots in computer science. For instance, Donald Knuth coined the term “literate programming” to describe the practice of mixing explanatory text with computer code in 1984 (Knuth, 1984) and the idea of reproducible data analysis has a long history in the computer science literature (as reviewed in Stodden, 2010). The main advances since 2004 have stemmed from the fact that the tools for reproducible data analysis have become much more widespread and accessible to researchers, such as marine organic geochemists, whose primary expertise lies outside of computer science. The new accessibility of these tools, combined with an increased need for them to be used by marine organic geochemists, suggests a bright future for reproducible data analysis in our community. In any case, increased transparency in data analysis is far preferable to what Rossini et al. (2003) identified as the alternative: “blind faith in our colleagues’ programming skills.”
Drawing From Other Fields
Progress in adjacent fields has also led to improved understanding of marine organic geochemistry. Most notably, the explosive progress in DNA and RNA sequencing technology, coupled with advances in bioinformatic tools to process those data, have led to a phase shift in our ability to draw inferences from biological sequence data. Hedges et al. (2000) predicted that “fast-developing capabilities [to identify marine microbes by taxonomy] should help compensate for our inability to culture most microorganisms.” Microbial taxonomy does offer some insight into environmental function, but this insight is severely limited (Royalty and Steen, 2019). However, it is now routine for individual labs to assemble incomplete or even complete, closed genomes of the most abundant microbes in a marine environment, in the context of a single project. Given microbial genomes, researchers can make reasonable predictions about broad nature of microbial interactions with organic matter (Rinke et al., 2013; Bird et al., 2019). It remains the case that bacterial and archaeal cells in seawater and sediments are only distantly related to microbes that have been grown in culture (Lloyd et al., 2018), and that the most common method of determining microbial abundance, surveys of primer-amplified 16S gene abundance, are biased toward taxa represented in culture collections (Steen et al., 2019). However, new culturing techniques and a renewed focus on culturing previously uncultured taxa have led to the culturing of quite a few abundant, environmentally important marine microbes (Könneke et al., 2005; Katayama et al., 2019; Imachi et al., 2020). Studies of microbes in pure culture act as “ground truthing” for inferences based on culture-independent techniques.
Other fields of ocean science have also impacted marine organic geochemistry. Improved understanding of the importance of mesoscale eddies to ocean physics have led biogeochemists to look for, and discover, differences in OM cycling in mesoscale eddies, for instance differences in D:L-amino acid transformation rates apparently due to different microbial communities within vs. outside of eddies (e.g., Zhang et al., 2009; Sarma et al., 2019). Remote sensing techniques have advanced substantially since 2001, due to improvements in sensors, the recent availability of inexpensive research drones, and improvements in the algorithms used to remove the overprint of the atmosphere on light reaching satellites from the ocean (e.g., Werdell et al., 2013). Coupled with improved understanding of DOM optical characteristics (Coble, 2007; Helms et al., 2008), this permits more accurate and robust measurements of parameters such as DOC concentration, chromophoric dissolved organic matter (CDOM) content, and even indirect estimations of parameters such as methylmercury concentrations (Fichot et al., 2016; Slonecker et al., 2016).
Challenges and Opportunities for the Next Decade(s)
The technical advances described create new challenges and opportunities for the next decade-plus of marine organic geochemistry research (Table 1). While a few of these challenges and opportunities are specific to individual technologies, for the most part they rely on integrating advances from different analytical techniques and fields of knowledge. Below, we describe some directions for future research that our working group found most exciting.
New Lipid Biomarkers and New Uses of Old Lipid Biomarkers
Biomarker lipids show the presence of particular source organisms or biosynthetic pathways and can be proxies for past environmental parameters, including temperature, pH, and input of soil organic matter to marine systems. Lipid chemical stability enables their preservation in the geological record and thus, their extensive use as paleo-indicators. Novel HPLC-MS techniques have greatly widened the range of compounds amenable for analysis, both in terms of size and polarity (Schouten et al., 2000; Sturt et al., 2004), revealing a large diversity of microbial lipids in their intact (polar) forms in marine environments (e.g., Van Mooy and Fredricks, 2010; Schubotz et al., 2018) and make possible environmental proxies (e.g., the GDGT-based TEX86 sea surface temperature proxy; see Schouten et al., 2013b). These techniques facilitate quantification of microbial intact polar lipids, which provide high taxonomic resolution (Schubotz et al., 2009), and which can yield insight into microbial processes such as ammonia oxidation rates (Hurley et al., 2016). Adding compound-specific isotope analysis in the context of natural isotope abundances (e.g., Schubotz et al., 2009; Pearson et al., 2016) or tracer experiments (e.g., Kellermann et al., 2016) can give further insight into elemental flow within ancient or currently active communities.
However, analytical refinements, along with extensive large-scale environmental and laboratory-based surveys, have highlighted the complexity in interpreting the biomarker record solely from a single influencing parameter point of view (Hurley et al., 2016; Sollai et al., 2019). Recent advances have focused on increasing the resolution and sensitivity of lipid analysis. Wörmer et al. (2014) introduced, for example, the use of direct laser-ablation of lipids from sediment core surfaces drastically improving spatial sampling resolution. High resolution mass spectrometry technology, such as orbitraps, has decreased detection limits and shown value as a tool to characterize and identify novel lipid structures (Moore et al., 2016; see also below). The identification of carboxyl-rich alicyclic material (CRAM; Hertkorn et al., 2006; see also below), carotenoid degradation products (CDP; Arakawa et al., 2017) and material derived from linear terpenoids (MDLT; Woods et al., 2011; Arakawa and Aluwihare, 2015) have shone a light on some of the most abundant classes of recalcitrant NOM in seawater. Although these structures have been inferred from NMR and ultrahigh-resolution mass spectrometry, and they appear related to biologically produced source molecules, there is not yet direct proof of how those molecules are produced, whether biotically or abiotically. Fractionation of DOM through hydrophilic interaction chromatography (HILIC) and multidimensional separation (2-D HPLC) coupled with ultrahigh-resolution mass spectrometry or conjunction with NMR experiments (Woods et al., 2011; Spranger et al., 2019) represent promising tools to determine if these materials might be useful as biomarkers or indicators of biogeochemical processes. At the same time, the data produced by these high-resolution instruments have highlighted the lack of pipelines for ‘omics’ data that are available for environmental lipidomic work.
To fully exploit the potential of these techniques, high-throughput methods need to be developed. For example, LOBSTAHS is an open-source lipidomics workflow, written in R, for high-throughput annotation and putative identification of lipids in high-mass-accuracy HPLC-MS data that can identify thousands of compounds (>14,000 unique entries) in a sample (Collins et al., 2016). In our view, the improved accessibility to high-resolution mass spectrometers and lipidomics workflow for high-throughput annotation and putative identification of lipids, not only enhance the number of structures that scientists can identify for targeted lipidomics, but they also allow us to explore the patterns of unknown compounds in sample sets with contrasting conditions. The latter has the potential of enhancing the amount of biologically and biogeochemically relevant information obtained from non-targeted, exploratory lipidomic analysis.
Future challenges require better comprehension of the biomarker producers, through understanding the environmental constraints and the biosynthetic origin of lipid biomarkers. For example, targeting the diversity of lipid biosynthetic genes has already helped us identify limitations of using long-established hopane biomarkers (Welander et al., 2010). Lipid isotope probing assays, recently adapted to use radiotracer applications (Evans et al., 2018), hold high promise in revealing lipid biosynthetic pathways in pure cultures, but also in environmental settings. By working together with microbiologists in the future, we will fulfill the need for a better grasp of the effects of microbial physiology on biomarker synthesis.
More and Improved Data Comparisons
State-of-the-art analytical techniques allow rapid chemical measurements of small samples. Some of these techniques, such as FT-ICR-MS, allow rapid measurements from small samples that can generate large amounts of data, with greater efficiency. Consequently, the amount of data available is expanding, presenting new challenges for data analysis, integration and intercalibration. Additionally, small variations in sample collection, physicochemical experiments, extraction protocols, fractionations, mass spectrometry analysis and data processing can lead to challenges in intercomparing data from multiple studies.
Differences in data processing algorithms can also impact acquired data files. As described above, computational methods to transform raw data should be understood as part of the analytical method, not as something downstream of the analysis. For example, comparison of three software tools and their parameters for processing mass spectral data showed large differences in the results depending on the tools and parameters chosen, specifically for the degree of false positive or negative peak detections (Cajka and Fiehn, 2016). This is especially important in the area of peak picking and identification in untargeted mass spectrometry analysis. There is a need to build on existing, successful data intercomparison (Schouten et al., 2013a) and sharing of information. Strengthened international cooperation and connections between diverse research fields could play a pivotal role in standardizing protocols and advancing understanding in the field of marine organic biogeochemistry.
Untargeted High-Resolution Analysis: Ultrahigh-Resolution Mass Spectrometry
Advances in sample preparation, analysis and interpretation for FT-ICR-MS of NOM are ongoing and will aid in further deciphering the structural diversity of NOM. Molecular analysis of DOM from marine samples is hindered by the salt matrix in which the compounds are present. Extraction with commercially available PPL columns (Dittmar et al., 2008) and analysis of the obtained methanol extract in ESI-negative mode represent the most widely applied combination. Different isolation techniques with variable efficiency have been proposed and compared (e.g., Sleighter and Hatcher, 2008; Green et al., 2014; Schmidt et al., 2014; Li et al., 2017; Stücheli et al., 2018). Because ESI primarily ionizes polar to semi-polar compounds and in itself represents a complex technique affecting compounds differently based on polarity, molecular size, acid/base character and concentration, no single ionization method captures the whole suite of compounds present in NOM (Ohno and Bro, 2006; Hertkorn et al., 2008; Reemtsma, 2009). Accordingly, extraction and ionization methods need to be tailored to the scientific question (Kido Soule et al., 2010; Sleighter et al., 2012).
FT-ICR-MS analyzers provide the highest accuracy and resolving power (>1,000,000 at m/z 400, Junot et al., 2014) for exact molecular formula determination. Direct infusion techniques are able to produce detailed information on sample composition in relatively short time, allowing fingerprinting of complex mixtures in high throughput studies. As one molecular formula can represent an unknown number of isomers (Hertkorn et al., 2008; Zark et al., 2017) and co-suppression of signals may occur when analyzing highly complex mixtures of organics such as (marine) DOM (Patriarca et al., 2018). Chemical separation techniques, such as liquid chromatography or electrophoresis, are applied prior to sample injection to overcome these issues and to provide more detailed structural information behind molecular formula assignments (Hawkes et al., 2018). While these techniques are commonly used in conjunction with tandem mass spectrometers including Orbitrap, we expect one-dimensional as well as multi-dimensional chromatographic separations prior to FT-ICR-MS analysis to become more prevalent in the future (Ghaste et al., 2016). However, the relatively slow acquisition rates of FT-ICR-MS instruments limit their applicability to hyphenated approaches often used in metabolomics and lipidomics research (Junot et al., 2010). Coupling FT-ICR-MS to trapped ion mobility mass spectrometry (TIMS-FT-ICR-MS) provided new insights into the presumed high structural diversity of DOM. Lower limits of isomeric diversity have been established (Tose et al., 2018; Leyva et al., 2019), while upper limit estimates still range widely and are dependent on detection limit and resolving power. Additionally, fragmentation approaches have been applied to confirm molecular formula attributions and to elucidate the structural diversity of DOM (Wagner et al., 2015a; Kujawinski et al., 2016; Zark et al., 2017).
The complex, high-dimensional information provided by ultrahigh-resolution MS requires development of new mathematical tools. The first, fundamental step is the assignment of unique molecular formulas. All tools in one way or another use mathematical transformations to improve resolving power, mass accuracy and sensitivity (e.g., Kilgour et al., 2013), or apply chemical and stochastic rules (Dittmar and Koch, 2006; Kind and Fiehn, 2007). After formula assignment, elemental ratios, molecular mass or mass defect analyses (Hughey et al., 2001) can provide first insights into NOM composition. Due to the high number of molecular formulae and increasing sample throughput, the application of multivariate statistics has been established in the field (Kujawinski et al., 2009; Sleighter et al., 2010; Longnecker and Kujawinski, 2016).
Untargeted High-Resolution Analysis: Coupled NMR and Ultrahigh-Resolution Mass Spectrometry
The alliance of NMR and ultrahigh-resolution mass spectrometry will continue to provide valuable insights into the sources of a refractory DOM components (Hertkorn et al., 2006; Aluwihare and Meador, 2008; Abdulla et al., 2013b; DiDonato and Hatcher, 2017). For example, combined NMR and FT-ICR-MS analyses show that the major components of the polysaccharide fraction of HMW-DOM are acylated polysaccharides (APS; N-acyl amino acids bound with neutral and amino sugars and non-acylated heteropolysaccharides) and CRAM (Hertkorn et al., 2006) with smaller amounts of aromatics and aromatic N-heterocyclics. CRAM is the most abundant identified component of DOM. Increasing depth in the water column conserves increasingly branched CRAM and heteroatom-containing components at the expense of carbohydrates, contributing to the resistance of DOM to biodegradation (Hertkorn et al., 2013).
Additional information may be gained by utilizing different NMR techniques. Higher magnetic field NMR instruments will likely not advance solid-state NMR techniques. This is because higher magnetic fields require a higher spinning speed for the sample rotor to remove the spinning sideband, which is not currently feasible (see Mopper et al., 2007 for a more complete explanation). However, higher magnetic field NMR and NMR Cryoprobes may allow the use of water suppression 1H NMR techniques to analyze the entire marine DOM composition even in the deep ocean (Lam and Simpson, 2008; Fox et al., 2018). Diffusion ordered spectroscopy (DOSY) and 2D-NMR techniques (1H-1H-TOCSY and 1H-1H-COSY) estimate the molecular weight of DOM components, resolve overlapping peaks, confirm the interpretation of the chemical shifts, and help to identify the structures of specific DOM components. Coupling 1H NMR techniques with offline chromatography and high-resolution mass spectrometry techniques will advance our understanding of the entire marine DOM transformation and cycling characterization of marine DOM (Simpson et al., 2004).
Chemometric statistical methods allow combining NMR data with mass spectrometry data using multivariate statistics, for example to identify structural components and pathways of metabolic perturbations or to determine the biotransformation of metabolites on short timescales (Jaeger and Aspers, 2014). These methods use the intrinsic covariance between signal intensities in the same and related molecules measured by different techniques across great numbers of samples (Crockford et al., 2006). Another approach identifies single metabolites by generating theoretical NMR and MS/MS spectra for possible chemical structures and by comparing them directly against experimental NMR and ultrahigh-resolution MS2 spectra of the unknowns (Boiteau et al., 2018). This approach does not require identified compounds from experimental metabolomics databases, providing means for the identification of unknown and uncatalogued metabolites in natural DOM. Further development of chemometric tools will be needed in the future.
Coupling Untargeted Chemical and Microbiological Assays
Information on microbial metabolite transformations will enhance our understanding of carbon cycling in the oceans. Parallel trends between the complex, but closely connected non-living and living worlds are established by combining chemical with, e.g., untargeted microbiological assessments (Kujawinski, 2011; Osterholz et al., 2016). The application of stable isotope tracers will enable the delineation of causal links (e.g., Biddle et al., 2006; Orsi et al., 2018; Seyler et al., 2018). Understanding structural diversity as a meaningful property of DOM adds a new facet to the interpretation of UHRMS data. Borrowing on traditional ecological concepts, this approach may increase the understanding of the long-term stability of carbon compounds dissolved in the ocean, sustaining highly diverse microbial communities (Dittmar, 2015; Osterholz et al., 2016). Likewise, bioassay experiments employing metabolic profiling via NMR and FT-ICR-MS show that bacterial DOM has a chemical composition and structural diversity similar to refractory natural DOM in seawater (Lechtenfeld et al., 2015). In the future, including DOM fingerprint information into numeric models such as ocean circulation models can provide mechanistic explanations to observations and may enable us to better place our findings within the bigger picture of global carbon cycling.
Consensus Reference Materials
A broad range of chemical reference materials for ocean sciences remains elusive, despite the recognition that accuracy of data depends on calibration and intercomparison over time and among laboratories. To that end, the US National Research Council prepared a report identifying the most critically needed reference materials and recommending the most appropriate approaches for their development, with an emphasis on organic materials (Committee on Reference Materials for Ocean Science, 2002). Some of the recommendations of this committee remain unfulfilled.
The marine DOC community has adopted the consensus reference material available from D. Hansell’s lab (University of Miami) to determine reproducibility and accuracy of oceanic DOC analyses (Sharp et al., 2002). When several researchers promoted the need for a collection of humic and fulvic acids to be made available to the scientific community, the International Humic Substances Society (IHSS) was organized in 1981. Since then, NOM research has benefited significantly from sharing common standard and reference materials with known elemental and stable isotopic ratios. Intercomparison of UHRMS data is challenging, as the “true” result is not known. A first interlaboratory comparison including 17 different high-resolution mass spectrometers and NOM reference material from the IHSS showed that while differences exist, common trends in elemental ratios are reproduced (Hawkes et al., 2020). Efforts such as this one will lead to the development of consensus values for quality control of the complex analyses. However, a marine reference material similar to the freshwater-sourced NOM from IHSS suitable also for fingerprinting techniques such as FT-ICR-MS or Orbitrap is missing to date, calling upon the community to establish such a material.
Reference materials for biological, particulate, and sedimentary OM are also conspicuously absent, in part because of logistical difficulties in preparing and certifying them, and perhaps more complex, a lack of consensus as to what would best constitute an appropriate reference material and what organic parameters should be targeted. Intercomparison exercises have been conducted for organic contaminants in biological tissues (mussel) and sediments (New York/New Jersey estuary) in the marine environment (Schantz et al., 2008), but comparable exercises for marine biogeochemical materials have generally not been carried out. These difficulties are partially alleviated by targeting specific biomarkers or ratios of biomarkers. Two successful small-scale round-robin laboratory intercomparison exercises, in which a few kilograms of sediment or extracts/isolates were distributed to a limited number of laboratories, are the alkenone intercomparison (Rosell-Melé et al., 2001) and the GDGT intercomparison (Schouten et al., 2013a). It should be noted that in both of these round robin exercises, the targeted parameters were biomarker ratios (UK’37 and TEX86 and BIT, respectively) rather than a rigorous assessment of absolute concentrations. Nonetheless, the marine organic biogeochemical community needs to address this shortfall.
Concluding Remarks
Marine organic biogeochemistry has made significant contributions toward a better understanding of the fundamental processes that affect element cycles in the ocean, especially for carbon and nitrogen. As new questions arise and new paradigms are developed, there is a never-ending need for novel analytical methodologies, ranging from advanced instrumentation, cross-disciplinary field observations and experimentation, to new thinking about how the massive amounts of data generated by biogeochemists is handled and interpreted (Figure 1). This review has summarized the state of the art with respect to analytical instrumentation in use by marine organic biogeochemists (e.g., chromatography, mass spectrometry, NMR spectroscopy, radiocarbon analysis, enzyme assays) and chemometrics for data handling. Key challenges for the future are also highlighted. These include identifying new biomarkers, employing advanced high-resolution mass spectrometry and NMR spectroscopy for characterizing dissolved organic matter and its behavior, and coupling chemical analyses with microbiological assays. The review concludes with a plea to move forward in the long-standing but as yet unrealized effort to develop and distribute reference materials – whether certified standard reference materials (SRMs) for intercalibration exercises or consensus reference materials for intercomparisons – to the biogeochemical community. Identifying and preparing reference materials, while not an easy task, is critical to guarantee that data generated by laboratories worldwide will truly be applicable and useable worldwide.
Author Contributions
SW conceived the workshop and convened the group of authors. AS, SK, and SW organized the manuscript and lead the writing. SK created the figure. All co-authors contributed text and ideas. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The Hanse-Wissenschaftskolleg Delmenhorst, Germany, sponsored the “Marine Organic Biogeochemistry” workshop in April 2019, of which this working group report was a part. The workshop was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project number: 422798570. The Geochemical Society provided additional funding for the conference. AS was supported by DOE grant DE-SC0020369.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdulla, H. A. N., Minor, E. C., Dias, R. F., and Hatcher, P. G. (2010a). Changes in the compound classes of dissolved organic matter along an estuarine transect: a study using FTIR and 13C NMR. Geochim. Cosmochim. Acta 74, 3815–3838. doi: 10.1016/j.gca.2010.04.006
Abdulla, H. A. N., Minor, E. C., Dias, R. F., and Hatcher, P. G. (2013a). Transformations of the chemical compositions of high molecular weight DOM along a salinity transect: using two dimensional correlation spectroscopy and principal component analysis approaches. Geochim. Cosmochim. Acta 118, 231–246. doi: 10.1016/j.gca.2013.03.036
Abdulla, H. A. N., Minor, E. C., and Hatcher, P. G. (2010b). Using two-dimensional correlations of 13C NMR and FTIR to investigate changes in the chemical composition of dissolved organic matter along an estuarine transect. Environ. Sci. Technol. 44, 8044–8049. doi: 10.1021/es100898x
Abdulla, H. A. N., Sleighter, R. L., and Hatcher, P. G. (2013b). Two dimensional correlation analysis of Fourier transform ion cyclotron resonance mass spectra of dissolved organic matter: a new graphical analysis of trends. Anal. Chem. 85, 3895–3902. doi: 10.1021/ac303221j
Allen, A. E., LaRoche, J., Maheswari, U., Lommer, M., Schauer, N., Lopez, P. J., et al. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443. doi: 10.1073/pnas.0711370105
Aluwihare, L. I., and Meador, T. (2008). Chemical composition of marine dissolved organic nitrogen. Nitrogen Mar. Environ. 95–140. doi: 10.1016/b978-0-12-372522-6.00003-7
Aluwihare, L. I., Repeta, D. J., Pantoja, S., and Johnson, C. G. (2005). Two chemically distinct pools of organic nitrogen accumulate in the ocean. Science 308, 1007–1010. doi: 10.1126/science.1108925
Arakawa, N., and Aluwihare, L. (2015). Direct identification of diverse alicyclic terpenoids in suwannee river fulvic acid. Environ. Sci. Technol. 49, 4097–4105. doi: 10.1021/es5055176
Arakawa, N., Aluwihare, L. I., Simpson, A. J., Soong, R., Stephens, B. M., and Lane-Coplen, D. (2017). Carotenoids are the likely precursor of a significant fraction of marine dissolved organic matter. Sci. Adv. 3:e1602976. doi: 10.1126/sciadv.1602976
Arnosti, C. (1995). Measurement of depth- and site-related differences in polysaccharide hydrolysis rates in marine sediments. Geochim. Cosmochim. Acta 59, 4247–4257. doi: 10.1016/0016-7037(95)00247-W
Arnosti, C. (2004). Speed bumps and barricades in the carbon cycle: substrate structural effects on carbon cycling. Mar. Chem. 92, 263–273. doi: 10.1016/j.marchem.2004.06.030
Arnosti, C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Ann. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Arnosti, C., Steen, A. D., Ziervogel, K., Ghobrial, S., Jeffrey, W. H. W. H., and Wanunu, M. (2011). Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS One 6:e28900. doi: 10.1371/journal.pone.0028900
Aron, A. T., Gentry, E. C., McPhail, K. L., Nothias, L.-F., Nothias-Esposito, M., Bouslimani, A., et al. (2020). Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 15, 1954–1991.
Bao, R., Zhao, M., McNichol, A., Wu, Y., Guo, X., Haghipour, N., et al. (2019). On the origin of aged sedimentary organic matter along a river-shelf-deep ocean transect. J. Geophys. Res. Biogeosci. 124, 2582–2594. doi: 10.1029/2019JG005107
Baumer, B., and Udwin, D. (2015). R Markdown. Wiley Interdiscip. Rev. Comput. Stat. 7, 167–177. doi: 10.1002/wics.1348
Beaupré, S. R., and Druffel, E. R. M. (2012). Photochemical reactivity of ancient marine dissolved organic carbon. Geophys. Res. Lett. 39:L18602. doi: 10.1029/2012GL052974
Beaupré, S. R., Druffel, E. R. M., and Griffin, S. (2007). A low-blank photochemical extraction system for concentration and isotopic analyses of marine dissolved organic carbon. Limnol. Oceanogr. Methods 5, 174–184. doi: 10.4319/lom.2007.5.174
Benner, R., Pakulski, J. D., McCarthy, M., Hedges, J. I., and Hatcher, P. G. (1992). Bulk chemical characteristics of dissolved organic matter in the ocean. Science 255, 1561–1564. doi: 10.1126/science.255.5051.1561
Besseling, M. A., Hopmans, E. C., Bale, N. J., Schouten, S., Damsté, J. S. S., and Villanueva, L. (2020). The absence of intact polar lipid-derived GDGTs in marine waters dominated by Marine Group II: implications for lipid biosynthesis in Archaea. Sci. Rep. 10:294.
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Bird, J. T., Tague, E. D., Zinke, L., Schmidt, J. M., Steen, A. D., Reese, B. K., et al. (2019). Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in Baltic Sea sediments. mBio 10:e02376-18.
Birkholz, A., Smittenberg, R. H., Hajdas, I., Wacker, L., and Bernasconi, S. M. (2013). Isolation and compound specific radiocarbon dating of terrigenous branched glycerol dialkyl glycerol tetraethers (brGDGTs). Org. Geochem. 60, 9–19. doi: 10.1016/j.orggeochem.2013.04.008
Blumer, M. (1975). Organic compounds in nature: limits of our knowledge. Angew. Chemie Int. Ed. Engl. 14, 507–514. doi: 10.1002/anie.197505071
Boiteau, R. M., Hoyt, D. W., Nicora, C. D., Kinmonth-Schultz, H. A., Ward, J. K., and Bingol, K. (2018). Structure elucidation of unknown metabolites in metabolomics by combined NMR and MS/MS prediction. Metabolites 8:8. doi: 10.3390/metabo8010008
Bour, A. L., Walker, B. D., Broek, T. A. B., and McCarthy, M. D. (2016). Radiocarbon analysis of individual amino acids: carbon blank quantification for a small-sample high-pressure liquid chromatography purification method. Anal. Chem. 88, 3521–3528. doi: 10.1021/acs.analchem.5b03619
Boysen, A. K., Heal, K. R., Carlson, L. T., and Ingalls, A. E. (2018). Best-matched internal standard normalization in liquid chromatography-mass spectrometry metabolomics applied to environmental samples. Anal. Chem. 90, 1363–1369. doi: 10.1021/acs.analchem.7b04400
Bronk Ramsey, C., Ditchfield, P., and Humm, M. (2004). Using a gas ion source for radiocarbon AMS and GC-AMS. Radiocarbon 46, 25–32. doi: 10.1017/s003382220003931x
Cajka, T., and Fiehn, O. (2016). Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 88, 524–545. doi: 10.1021/acs.analchem.5b04491
Cantarero, S., Henríquez-Castillo, C., Dildar, N., Vargas, C., von Dassow, P., Cornejo-D’Ottone, M., et al. (in press). Size-fractionated contribution of microbial biomass to suspended organic matter in the Eastern Tropical South Pacific oxygen minimum zone. Front. Mar. Sci. doi: 10.3389/fmars.2020.540643
Cao, X., Aiken, G. R., Butler, K. D., Huntington, T. G., Balch, W. M., Mao, J., et al. (2018). Evidence for major input of riverine organic matter into the ocean. Org. Geochem. 116, 62–76. doi: 10.1016/j.orggeochem.2017.11.001
Cham, J. (2012). Piled Higher and Deeper Comics. Available online at: http://phdcomics.com/comics/archive.php?comicid=1531 (accessed July 1, 2020).
Clark, L. L., Ingall, E. D., and Benner, R. (1998). Marine phosphorus is selectively remineralized. Nature 393:426. doi: 10.1038/30881
Coble, P. G. (2007). Marine optical biogeochemistry: the chemistry of ocean color. Chem. Rev. 107, 402–418. doi: 10.1021/cr050350
Collins, J. R., Edwards, B. R., Fredricks, H. F., and Van Mooy, B. A. S. S. (2016). LOBSTAHS: an adduct-based lipidomics strategy for discovery and identification of oxidative stress biomarkers. Anal. Chem. 88, 7154–7162. doi: 10.1021/acs.analchem.6b01260
Comisarow, M. B., and Marshall, A. G. (1974). Fourier transform ion cyclotron resonance spectroscopy. Chem. Phys. Lett. 25, 282–283.
Committee on Reference Materials for Ocean Science (2002). Chemical Reference Materials: Setting the Standards for Ocean Science. Washington, DC: The National Academies Press.
Crockford, D. J., Holmes, E., Lindon, J. C., Plumb, R. S., Zirah, S., Bruce, S. J., et al. (2006). Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies. Anal. Chem. 78, 363–371. doi: 10.1021/ac051444m
Cui, L., Wang, X., and Feng, L. (2019). Determination of nitrogen isotopes on samples with tens of nmol of N using the combination of an elemental analyzer, a GasBench interface and an isotope ratio mass spectrometer: an evaluation of blank N contributions and blank-correction. Rapid Commun. Mass Spectrom. 33, 74–80. doi: 10.1002/rcm.8309
de Bar, M. W., Hopmans, E. C., Verweij, M., Dorhout, D. J. C., Damsté, J. S. S., and Schouten, S. (2017). Development and comparison of chromatographic methods for the analysis of long chain diols and alkenones in biological materials and sediment. J. Chromatogr. A 1521, 150–160. doi: 10.1016/j.chroma.2017.09.037
DiDonato, N., and Hatcher, P. G. (2017). Alicyclic carboxylic acids in soil humic acid as detected with ultrahigh resolution mass spectrometry and multi-dimensional NMR. Org. Geochem. 112, 33–46. doi: 10.1016/j.orggeochem.2017.06.010
Dittmar, T. (2015). “Reasons behind the long-term stability of dissolved organic matter,” in Biogeochemistry of Marine Dissolved Organic Matter, eds D. A. Hansell and C. A. Carlson (New York, NY: Elsevier Inc), 369–385. doi: 10.1016/B978-0-12-405940-5.00007-8
Dittmar, T., Koch, B., Hertkorn, N., and Kattner, G. (2008). A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 6, 230–235. doi: 10.4319/lom.2008.6.230
Dittmar, T., and Koch, B. P. (2006). Thermogenic organic matter dissolved in the abyssal ocean. Mar. Chem. 102, 208–217. doi: 10.1016/j.marchem.2006.04.003
Druffel, E. R. M., Beaupré, S. R., and Ziolkowski, L. A. (2016). “Radiocarbon in the oceans,” in Radiocarbon and Climate Change: Mechanisms, Applications and Laboratory Techniques, eds E. Schuur, E. Druffel, and S. Trumbore (Cham: Springer), 139–166. doi: 10.1007/978-3-319-25643-6_5
Druffel, E. R. M., Zhang, D., Xu, X., Ziolkowski, L. A., Southon, J. R., Dos Santos, G. M., et al. (2010). Compound-specific radiocarbon analyses of phospholipid fatty acids and n-alkanes in Ocean sediments. Radiocarbon 52, 1215–1223. doi: 10.1017/S0033822200046294
Dührkop, K., Fleischauer, M., Ludwig, M., Aksenov, A. A., Melnik, A. V., Meusel, M., et al. (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302. doi: 10.1038/s41592-019-0344-8
Durham, B. P., Boysen, A. K., Carlson, L. T., Groussman, R. D., Heal, K. R., Cain, K. R., et al. (2019). Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat. Microbiol. 4, 1706–1715. doi: 10.1038/s41564-019-0507-5
Eek, K. M., Sessions, A. L., and Lies, D. P. (2007). Carbon-isotopic analysis of microbial cells sorted by flow cytometry. Geobiology 5, 85–95. doi: 10.1111/j.1472-4669.2006.00094.x
Eglinton, T. I., Aluwihare, L. I., Bauer, J. E., Druffel, E. R. M., and McNichol, A. P. (1996). Gas chromatographic isolation of individual compounds from complex matrices for radiocarbon dating. Anal. Chem. 68, 904–912. doi: 10.1021/ac9508513
Eiler, J., Cesar, J., Chimiak, L., Dallas, B., Grice, K., Griep-Raming, J., et al. (2017). Analysis of molecular isotopic structures at high precision and accuracy by Orbitrap mass spectrometry. Int. J. Mass Spectrom. 422, 126–142. doi: 10.1016/j.ijms.2017.10.002
Eiserbeck, C., Nelson, R. K., Reddy, C. M., and Grice, K. (2014). Advances in Comprehensive Two-Dimensional Gas Chromatography (GC× GC). London: Royal Society of Chemistry.
Ernst, M., Kang, K. B., Caraballo-Rodríguez, A. M., Nothias, L.-F., Wandy, J., et al. (2019). MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9:144. doi: 10.3390/metabo9070144
Evans, T. W., Könneke, M., Lipp, J. S., Adhikari, R. R., Taubner, H., Elvert, M., et al. (2018). Lipid biosynthesis of Nitrosopumilus maritimus dissected by lipid specific radioisotope probing (lipid-RIP) under contrasting ammonium supply. Geochim. Cosmochim. Acta 242, 51–63. doi: 10.1016/j.gca.2018.09.001
Feunang, Y. D., Eisner, R., Knox, C., Chepelev, L., Hastings, J., Owen, G., et al. (2016). ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8:61.
Fichot, C. G., Downing, B. D., Bergamaschi, B. A., Windham-Myers, L., Marvin-Dipasquale, M., Thompson, D. R., et al. (2016). High-resolution remote sensing of water quality in the san francisco bay-delta estuary. Environ. Sci. Technol. 50, 573–583. doi: 10.1021/acs.est.5b03518
Fox, C. A., Abdulla, H. A., Burdige, D. J., Lewicki, J. P., and Komada, T. (2018). Composition of dissolved organic matter in pore waters of anoxic marine sediments analyzed by 1H nuclear magnetic resonance spectroscopy. Front. Mar. Sci. 5:172. doi: 10.3389/fmars.2018.00172
Fulton, J. M., Arthur, M. A., Thomas, B., and Freeman, K. H. (2018). Pigment carbon and nitrogen isotopic signatures in euxinic basins. Geobiology 16, 429–445. doi: 10.1111/gbi.12285
Galli, I., Bartalini, S., Ballerini, R., Barucci, M., Cancio, P., De Pas, M., et al. (2016). Spectroscopic detection of radiocarbon dioxide at parts-per-quadrillion sensitivity. Optica 3, 385–388. doi: 10.1364/OPTICA.3.000385
Garrido, J. L., Airs, R. L., Rodríguez, F., van Heukelem, L., and Zapata, M. (2012). “New HPLC separation techniques,” in Phytoplankton Pigments, eds S. Roy and C. A. Llewellyn (Cambridge: Cambridge University Press), 165–194. doi: 10.1017/cbo9780511732263.008
Genta-Jouve, G., Croué, J., Weinberg, L., Cocandeau, V., Holderith, S., Bontemps, N., et al. (2014). Two-dimensional ultra high pressure liquid chromatography quadrupole/time-of-flight mass spectrometry for semi-targeted natural compounds identification. Phytochem. Lett. 10, 318–323. doi: 10.1016/j.phytol.2014.10.029
Ghaste, M., Mistrik, R., and Shulaev, V. (2016). Applications of fourier transform ion cyclotron resonance (FT-ICR) and orbitrap based high resolution mass spectrometry in metabolomics and lipidomics. Int. J. Mol. Sci. 17:816. doi: 10.3390/ijms17060816
Gierga, M., Schneider, M. P. W., Wiedemeier, D. B., Lang, S. Q., Smittenberg, R. H., Hajdas, I., et al. (2014). Purification of fire derived markers for μg scale isotope analysis (δ13C, Δ14C) using high performance liquid chromatography (HPLC). Org. Geochem. 70, 1–9. doi: 10.1016/j.orggeochem.2014.02.008
Grant, K. E., Galy, V. V., Chadwick, O. A., and Derry, L. A. (2019). Thermal oxidation of carbon in organic matter rich volcanic soils: insights into SOC age differentiation and mineral stabilization. Biogeochemistry 144, 291–304. doi: 10.1007/s10533-019-00586-1
Green, N. W., Perdue, E. M., Aiken, G. R., Butler, K. D., Chen, H., Dittmar, T., et al. (2014). An intercomparison of three methods for the large-scale isolation of oceanic dissolved organic matter. Mar. Chem. 161, 14–19. doi: 10.1016/j.marchem.2014.01.012
Grobe, H., Diepenbroek, M., Dittert, N., Reinke, M., and Sieger, R. (2006). “Archiving and distributing earth-science data with the PANGAEA information system,” in Antarctica, eds D. K. Fütterer, D. Damaske, G. Kleinschmidt, H. Miller, and F. Tessensohn (Berlin: Springer-Verlag), 403–406. doi: 10.1007/3-540-32934-x_51
Guijas, C., Montenegro-Burke, J. R., Domingo-Almenara, X., Palermo, A., Warth, B., Hermann, G., et al. (2018). METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 90, 3156–3164. doi: 10.1021/acs.analchem.7b04424
Haghipour, N., Ausin, B., Usman, M. O., Ishikawa, N., Wacker, L., Welte, C., et al. (2019). Compound-specific radiocarbon analysis by elemental analyzer-accelerator mass spectrometry: precision and limitations. Anal. Chem. 91, 2042–2049. doi: 10.1021/acs.analchem.8b04491
Hansman, R. L., Dittmar, T., and Herndl, G. J. (2015). Conservation of dissolved organic matter molecular composition during mixing of the deep water masses of the northeast Atlantic Ocean. Mar. Chem. 177, 288–297. doi: 10.1016/j.marchem.2015.06.001
Hansman, R. L., and Sessions, A. L. (2016). Measuring the in situ carbon isotopic composition of distinct marine plankton populations sorted by flow cytometry. Limnol. Oceanogr. Methods 14, 87–99. doi: 10.1002/lom3.10073
Hartmann, A. C., Petras, D., Quinn, R. A., Protsyuk, I., Archer, F. I., Ransome, E., et al. (2017). Meta-mass shift chemical profiling of metabolomes from coral reefs. Proc. Natl. Acad. Sci. U.S.A. 114, 11685–11690. doi: 10.1073/pnas.1710248114
Haug, K., Cochrane, K., Nainala, V. C., Williams, M., Chang, J., Jayaseelan, K. V., et al. (2020). MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48, D440–D444.
Hawkes, J. A., D’Andrilli, J., Agar, J. N., Barrow, M. P., Berg, S. M., Catalán, N., et al. (2020). An international laboratory comparison of dissolved organic matter composition by high resolution mass spectrometry: are we getting the same answer? Limnol. Oceanogr. Methods 18, 235–258. doi: 10.1002/lom3.10364
Hawkes, J. A., Patriarca, C., Sjöberg, P. J. R., Tranvik, L. J., and Bergquist, J. (2018). Extreme isomeric complexity of dissolved organic matter found across aquatic environments. Limnol. Oceanogr. Lett. 3, 21–30. doi: 10.1002/lol2.10064
Heal, K. R., Qin, W., Ribalet, F., Bertagnolli, A. D., Coyote-Maestas, W., Hmelo, L. R., et al. (2017). Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc. Natl. Acad. Sci. U.S.A. 114, 364–369. doi: 10.1073/pnas.1608462114
Hedges, J., Eglinton, G., Hatcher, P., Kirchman, D., Arnosti, C., Derenne, S., et al. (2000). The molecularly-uncharacterized component of nonliving organic matter in natural environments. Org. Geochem. 31, 945–958. doi: 10.1016/s0146-6380(00)00096-6
Helms, J. R., Mao, J., Chen, H., Perdue, E. M., Green, N. W., Hatcher, P. G., et al. (2015). Spectroscopic characterization of oceanic dissolved organic matter isolated by reverse osmosis coupled with electrodialysis. Mar. Chem. 177, 278–287. doi: 10.1016/j.marchem.2015.07.007
Helms, J. R., Stubbins, A., Ritchie, J. D., Minor, E. C., Kieber, D. J., and Mopper, K. (2008). Absorption spectral slopes and slope rations as indicators of molecular weight, source, and photoleaching of chromophoric dissolved organic matter. Limnol. Oceanogr. 53, 955–969. doi: 10.4319/lo.2008.53.3.0955
Hemingway, J. D., Hilton, R. G., Hovius, N., Eglinton, T. I., Haghipour, N., Wacker, L., et al. (2018). Microbial oxidation of lithospheric organic carbon in rapidly eroding tropical mountain soils. Science 360, 209–212. doi: 10.1126/science.aao6463
Hemingway, J. D., Rothman, D. H., Grant, K. E., Rosengard, S. Z., Eglinton, T. I., Derry, L. A., et al. (2019). Mineral protection regulates long-term global preservation of natural organic carbon. Nature 570, 228–231. doi: 10.1038/s41586-019-1280-6
Hertkorn, N., Benner, R., Frommberger, M., Schmitt-Kopplin, P., Witt, M., Kaiser, K., et al. (2006). Characterization of a major refractory component of marine dissolved organic matter. Geochim. Cosmochim. Acta 70, 2990–3010. doi: 10.1016/j.gca.2006.03.021
Hertkorn, N., Frommberger, M., Witt, M., Koch, B. P., Schmitt-Kopplin, P., and Perdue, E. M. (2008). Natural organic matter and the event horizon of mass spectrometry. Anal. Chem. 80, 8908–8919. doi: 10.1021/ac800464g
Hertkorn, N., Harir, M., Koch, B. P., Michalke, B., and Schmitt-Kopplin, P. (2013). High-field NMR spectroscopy and FTICR mass spectrometry: powerful discovery tools for the molecular level characterization of marine dissolved organic matter. Biogeosciences 10, 1583–1624. doi: 10.5194/bg-10-1583-2013
Higgins, H. W., Wright, S. W., and Schlüter, L. (2012). “Quantitative interpretation of chemotaxonomic pigment data,” in Phytoplankton Pigments, eds S. Roy and C. A. Llewellyn (Cambridge: Cambridge University Press), 257–313. doi: 10.1017/cbo9780511732263.010
Higgins, M. B., Robinson, R. S., Casciotti, K. L., McIlvin, M. R., and Pearson, A. (2009). A method for determining the nitrogen isotopic composition of porphyrins. Anal. Chem. 81, 184–192. doi: 10.1021/ac8017185
Higgins Keppler, E. A., Jenkins, C. L., Davis, T. J., and Bean, H. D. (2018). Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC Trends Anal. Chem. 109, 275–286. doi: 10.1016/j.trac.2018.10.015
Hoh, E., Dodder, N. G., Lehotay, S. J., Pangallo, K. C., Reddy, C. M., and Maruya, K. A. (2012). Nontargeted comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry method and software for inventorying persistent and bioaccumulative contaminants in marine environments. Environ. Sci. Technol. 46, 8001–8008. doi: 10.1021/es301139q
Hopmans, E. C., Schouten, S., and Sinninghe Damsté, J. S. (2016). The effect of improved chromatography on GDGT-based palaeoproxies. Org. Geochem. 93, 1–6. doi: 10.1016/j.orggeochem.2015.12.006
Hoppe, H.-G. (1983). Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl-substrates. Mar. Ecol. Prog. Ser. 11, 299–308. doi: 10.3354/meps011299
Hou, J., Huang, Y., Brodsky, C., Alexandre, M. R., McNichol, A. P., King, J. W., et al. (2010). Radiocarbon dating of individual lignin phenols: a new approach for establishing chronology of late quaternary lake sediments. Anal. Chem. 82, 7119–7126. doi: 10.1021/ac100494m
Hu, Q., Noll, R. J., Li, H., Makarov, A., Hardman, M., and Graham Cooks, R. (2005). The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443. doi: 10.1002/jms.856
Hughey, C. A., Hendrickson, C. L., Rodgers, R. P., Marshall, A. G., and Qian, K. (2001). Kendrick mass defect spectrum: a compact visual analysis for ultrahigh-resolution broadband mass spectra. Anal. Chem. 73, 4676–4681. doi: 10.1021/ac010560w
Hunter, J. E., Frada, M. J., Fredricks, H. F., Vardi, A., and Van Mooy, B. A. S. (2015). Targeted and untargeted lipidomics of Emiliania huxleyi viral infection and life cycle phases highlights molecular biomarkers of infection, susceptibility, and ploidy. Front. Mar. Sci. 2:81. doi: 10.3389/fmars.2015.00081
Hurley, S. J., Elling, F. J., Könneke, M., Buchwald, C., Wankel, S. D., Santoro, A. E., et al. (2016). Influence of ammonia oxidation rate on thaumarchaeal lipid composition and the TEX86 temperature proxy. Proc. Natl. Acad. Sci. U.S.A. 113, 7762–7767. doi: 10.1073/pnas.1518534113
Imachi, H., Nobu, M. K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., et al. (2020). Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577, 519–525.
Ingalls, A. E., Anderson, R. F., and Pearson, A. (2004). Radiocarbon dating of diatom-bound organic compounds. Mar. Chem. 92, 91–105. doi: 10.1016/j.marchem.2004.06.019
Ingalls, A. E., Ellis, E. E., Santos, G. M., McDuffee, K. E., Truxal, L., Keil, R. G., et al. (2010). HPLC purification of higher plant-dervied lignin phenols for compound specific radiocarbon analysis. Anal. Chem. 82, 8931–8938. doi: 10.1021/ac1016584
Ingalls, A. E., Shah, S. R., Hansman, R. L., Aluwihare, L. I., Santos, G. M., Druffel, E. R. M., et al. (2006). Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. U.S.A. 103, 6442–6447. doi: 10.1073/pnas.0510157103
Isaji, Y., Ogawa, N. O., Boreham, C. J., Kashiyama, Y., and Ohkouchi, N. (2020). Evaluation of δ13C and δ15N uncertainties associated with the compound-specific isotope analysis of geoporphyrins. Anal. Chem. 92, 3152–3160. doi: 10.1021/acs.analchem.9b04843
Ishikawa, N. F., Itahashi, Y., Blattmann, T. M., Takano, Y., Ogawa, N. O., Yamane, M., et al. (2018). Improved method for isolation and purification of underivatized amino acids for radiocarbon analysis. Anal. Chem. 90, 12035–12041. doi: 10.1021/acs.analchem.8b02693
Jaeger, M., and Aspers, R. (2014). Chapter Five - Covariance NMR and small molecule applications. Annu. Rep. NMR Spectrosc. 83, 271–349. doi: 10.1016/b978-0-12-800183-7.00005-8
Jarmusch, A. K., Wang, M., Aceves, C. M., Advani, R. S., Aguire, S., Aksenov, A. A., et al. (2019). Repository-scale co-and re-analysis of tandem mass spectrometry data. bioRxiv [Preprint]. doi: 10.1101/750471
Johnson, W. M., Kido Soule, M. C., and Kujawinski, E. B. (2016). Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 10, 2304–2316. doi: 10.1038/ismej.2016.6
Johnson, W. M., Kido Soule, M. C., and Kujawinski, E. B. (2017). Extraction efficiency and quantification of dissolved metabolites in targeted marine metabolomics: matrix effects in marine metabolomics. Limnol. Oceanogr. Methods 15, 417–428. doi: 10.1002/lom3.10181
Johnson, W. M., Longnecker, K., Kido Soule, M. C., Arnold, W. A., Bhatia, M. P., Hallam, S. J., et al. (2020). Metabolite composition of sinking particles differs from surface suspended particles across a latitudinal transect in the South Atlantic. Limnol. Oceanogr. 65, 111–127. doi: 10.1002/lno.11255
Junium, C. K., Arthur, M. A., and Freeman, K. H. (2015). Compound-specific δ15N and chlorin preservation in surface sediments of the Peru Margin with implications for ancient bulk δ15N records. Geochim. Cosmochim. Acta 160, 306–318. doi: 10.1016/j.gca.2014.12.018
Junium, C. K., Dickson, A. J., and Uveges, B. T. (2018). Perturbation to the nitrogen cycle during rapid Early Eocene global warming. Nat. Commun. 9:3186. doi: 10.1038/s41467-018-05486-w
Junot, C., Fenaille, F., Colsch, B., and Bécher, F. (2014). High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom. Rev. 33, 471–500. doi: 10.1002/mas.21401
Junot, C., Madalinski, G., Tabet, J. C., and Ezan, E. (2010). Fourier transform mass spectrometry for metabolome analysis. Analyst 135, 2203–2219. doi: 10.1039/c0an00021c
Katayama, T., Nobu, M. K., Kusada, H., Meng, X.-Y., Yoshioka, H., Kamagata, Y., et al. (2019). Membrane-bounded nucleoid discovered in a cultivated bacterium of the candidate phylum ‘Atribacteria.’. bioRxiv [Preprint]. doi: 10.1101/728279
Kellermann, M. Y., Yoshinaga, M. Y., Wegener, G., Krukenberg, V., and Hinrichs, K. U. (2016). Tracing the production and fate of individual archaeal intact polar lipids using stable isotope probing. Org. Geochem. 95, 13–20. doi: 10.1016/j.orggeochem.2016.02.004
Kharbush, J. J., Allen, A. E., Moustafa, A., Dorrestein, P. C., and Aluwihare, L. I. (2016). Intact polar diacylglycerol biomarker lipids isolated from suspended particulate organic matter accumulating in an ultraoligotrophic water column. Org. Geochem. 100, 29–41. doi: 10.1016/j.orggeochem.2016.07.008
Kido Soule, M. C., Longnecker, K., Giovannoni, S. J., Kujawinski, E. B., Soule, M. C. K., Longnecker, K., et al. (2010). Impact of instrument and experiment parameters on reproducibility of ultrahigh resolution ESI FT-ICR mass spectra of natural organic matter. Org. Geochem. 41, 725–733. doi: 10.1016/j.orggeochem.2010.05.017
Kilgour, D. P. A. A., Wills, R., Qi, Y., O’Connor, P. B., and O’Connor, P. B. (2013). Autophaser: an algorithm for automated generation of absorption mode spectra for FT-ICR MS. Anal. Chem. 85, 3903–3911. doi: 10.1021/ac303289c
Kind, T., and Fiehn, O. (2007). Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 8:105. doi: 10.1186/1471-2105-8-105
Knapp, A. N., Sigman, D. M., and Lipschultz, F. (2005). N isotopic composition of dissolved organic nitrogen and nitrate at the Bermuda Atlantic Time-series study site. Global Biogeochem. Cycles 19, 1–15. doi: 10.1029/2004GB002320
Knauff, M., and Nejasmic, J. (2014). An efficiency comparison of document preparation systems used in academic research and development. PLoS One 9:e115069. doi: 10.1371/journal.pone.0115069
Koch, B., Witt, M., Engbrodt, R., Dittmar, T., and Kattner, G. (2005). Molecular formulae of marine and terrigenous dissolved organic matter detected by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Geochim. Cosmochim. Acta 69, 3299–3308. doi: 10.1016/j.gca.2005.02.027
Kolowith, L. C., Ingall, E. D., and Benner, R. (2001). Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46, 309–320. doi: 10.4319/lo.2001.46.2.0309
Könneke, M., Bernhard, A. E., De La Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Koprivnjak, J.-F., Pfromm, P. H., Ingall, E., Vetter, T. A., Schmitt-Kopplin, P., Hertkorn, N., et al. (2009). Chemical and spectroscopic characterization of marine dissolved organic matter isolated using coupled reverse osmosis–electrodialysis. Geochim. Cosmochim. Acta 73, 4215–4231. doi: 10.1016/j.gca.2009.04.010
Kujawinski, E. B. (2011). The impact of microbial metabolism on marine dissolved organic matter. Ann. Rev. Mar. Sci. 3, 567–599. doi: 10.1146/annurev-marine-120308-081003
Kujawinski, E. B., Del Vecchio, R., Blough, N. V., Klein, G. C., Marshall, A. G., Delvecchio, R., et al. (2004). Probing molecular-level transformations of dissolved organic matter: insights on photochemical degradation and protozoan modification of DOM from electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 92, 23–37. doi: 10.1016/j.marchem.2004.06.038
Kujawinski, E. B., Longnecker, K., Barott, K. L., Weber, R. J. M., and Kido Soule, M. C. (2016). Microbial community structure affects marine dissolved organic matter composition. Front. Mar. Sci. 3:45. doi: 10.3389/fmars.2016.00045
Kujawinski, E. B., Longnecker, K., Blough, N. V., Del Vecchio, R., Finlay, L., Kitner, J. B., et al. (2009). Identification of possible source markers in marine dissolved organic matter using ultrahigh resolution mass spectrometry. Geochim. Cosmochim. Acta 73, 4384–4399. doi: 10.1016/j.gca.2009.04.033
Kusch, S., Kashiyama, Y., Ogawa, N. O., Altabet, M., Butzin, M., Friedrich, J., et al. (2010). Implications for chloro-and pheopigment synthesis and preservation from combined compound-specific δ13C, δ15N, and Δ14C analysis. Biogeosci. Discuss. 7, 6265–6294.
Lai, Z., Tsugawa, H., Wohlgemuth, G., Mehta, S., Mueller, M., Zheng, Y., et al. (2018). Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53–56. doi: 10.1038/nmeth.4512
Lam, B., and Simpson, A. J. (2008). Direct 1H NMR spectroscopy of dissolved organic matter in natural waters. Analyst 133, 263–269. doi: 10.1039/b713457f
Langel, R., and Dyckmans, J. (2014). Combined 13C and 15N isotope analysis on small samples using a near-conventional elemental analyzer/isotope ratio mass spectrometer setup. Rapid Commun. Mass Spectrom. 28, 1019–1022. doi: 10.1002/rcm.6878
Lechtenfeld, O. J., Hertkorn, N., Shen, Y., Witt, M., and Benner, R. (2015). Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 6:6711. doi: 10.1038/ncomms7711
Lechtenfeld, O. J., Kattner, G., Flerus, R., McCallister, S. L., Schmitt-Kopplin, P., and Koch, B. P. (2014). Molecular transformation and degradation of refractory dissolved organic matter in the Atlantic and Southern Ocean. Geochim. Cosmochim. Acta 126, 321–337. doi: 10.1016/j.gca.2013.11.009
Lechtenfeld, O. J., Koch, B. P., Geibert, W., Ludwichowski, K. U., and Kattner, G. (2011). Inorganics in organics: quantification of organic phosphorus and sulfur and trace element speciation in natural organic matter using HPLC-ICPMS. Anal. Chem. 83, 8968–8974. doi: 10.1021/ac201765a
Lee, C., Wakeham, S. G., and Benner, R. H. (2004). Symposium on new approaches in marine organic biogeochemistry: a tribute to the life and science of John I. Hedges. Mar. Chem. 92, 1–3. doi: 10.1016/j.marchem.2004.06.013
Leisch, F. (2002). “Sweave: dynamic generation of statistical reports using literate data analysis,” in Compstat, eds W. Härdle and B. Rönz (Heidelberg: Physica-Verlag HD), 575–580. doi: 10.1007/978-3-642-57489-4_89
Leyva, D., Tose, L. V., Porter, J., Wolff, J., Jaffé, R., and Fernandez-Lima, F. (2019). Understanding the structural complexity of dissolved organic matter: Isomeric diversity. Faraday Discuss. 218, 431–440. doi: 10.1039/c8fd00221e
Li, Y., Harir, M., Uhl, J., Kanawati, B., Lucio, M., Smirnov, K. S., et al. (2017). How representative are dissolved organic matter (DOM) extracts? A comprehensive study of sorbent selectivity for DOM isolation. Water Res. 116, 316–323. doi: 10.1016/j.watres.2017.03.038
Li, Y., and Li, L. (2019). Mass accuracy check using common background peaks for improving metabolome data quality in chemical isotope labeling LC-MS. J. Am. Soc. Mass Spectrom. 30, 1733–1741. doi: 10.1007/s13361-019-02248-w
Liu, Z., Kobiela, M. E., McKee, G. A., Tang, T., Lee, C., Mulholland, M. R., et al. (2010). The effect of chemical structure on the hydrolysis of tetrapeptides along a river-to-ocean transect: AVFA and SWGA. Mar. Chem. 119, 108–120. doi: 10.1016/j.marchem.2010.01.005
Liu, Z., Liu, S., Liu, J., and Gardner, W. S. (2013). Differences in peptide decomposition rates and pathways between hypoxic and oxic coastal environments. Mar. Chem. 157, 67–77. doi: 10.1016/j.marchem.2013.08.003
Liu, Z., Mao, J., Peterson, M. L., Lee, C., Wakeham, S. G., and Hatcher, P. G. (2009). Characterization of sinking particles from the northwest Mediterranean Sea using advanced solid-state NMR. Geochim. Cosmochim. Acta 73, 1014–1026. doi: 10.1016/j.gca.2008.11.019
Lloyd, K. G., Steen, A. D., Ladau, J., Yin, J., and Crosby, L. (2018). Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3:e00055-18. doi: 10.1128/mSystems.00055-18
Longnecker, K., and Kujawinski, E. B. (2016). Using network analysis to discern compositional patterns in ultrahigh-resolution mass spectrometry data of dissolved organic matter. Rapid Commun. Mass Spectrom. 30, 2388–2394. doi: 10.1002/rcm.7719
Longnecker, K., and Kujawinski, E. B. (2017). Mining mass spectrometry data: using new computational tools to find novel organic compounds in complex environmental mixtures. Org. Geochem. 110, 92–99. doi: 10.1016/j.orggeochem.2017.05.008
Maie, N., Parish, K. J., Watanabe, A., Knicker, H., Benner, R., Abe, T., et al. (2006). Chemical characteristics of dissolved organic nitrogen in an oligotrophic subtropical coastal ecosystem. Geochim. Cosmochim. Acta 70, 4491–4506. doi: 10.1016/j.gca.2006.06.1554
Mao, J. D., Hu, W. G., Schmidt-Rohr, K., Davies, G., Ghabbour, E. A., and Xing, B. (2000). Quantitative characterization of humic substances by solid-state carbon-13 nuclear magnetic resonance. Soil Sci. Soc. Am. J. 64, 873–884. doi: 10.2136/sssaj2000.643873x
Marshall, A. G., and Chen, T. (2015). 40 years of Fourier transform ion cyclotron resonance mass spectrometry. Int. J. Mass Spectrom. 377, 410–420. doi: 10.1016/j.ijms.2014.06.034
Martínez-Pérez, A. M., Osterholz, H., Nieto-Cid, M., Álvarez, M., Dittmar, T., and Álvarez-Salgado, X. A. (2017). Molecular composition of dissolved organic matter in the Mediterranean Sea. Limnol. Oceanogr. 62, 2699–2712. doi: 10.1002/lno.10600
McCarthy, M., Pratum, T., Hedges, J., and Benner, R. (1997). Chemical composition of dissolved organic nitrogen in the ocean. 390, 150–154. doi: 10.1038/36535
McLean, C., and Kujawinski, E. B. (2019). AutoTuner: high fidelity, robust, and rapid parameter selection for metabolomics data processing. bioRxiv [Preprint]. doi: 10.1101/812370
Medeiros, P. M., Seidel, M., Ward, N. D., Carpenter, E. J., Gomes, H. R., Niggemann, J., et al. (2015). Fate of the Amazon River dissolved organic matter in the tropical Atlantic Ocean. Glob. Biogeochem. Cycles 29, 677–690. doi: 10.1002/2015GB005115
Moore, E. K., Hopmans, E. C., Rijpstra, W. I. C., Villanueva, L., and Damsté, J. S. S. (2016). Elucidation and identification of amino acid containing membrane lipids using liquid chromatography/high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 30, 739–750. doi: 10.1002/rcm.7503
Mopper, K., Stubbins, A., Ritchie, J. D., Bialk, H. M., and Hatcher, P. G. (2007). Advanced instrumental approaches for characterization of marine dissolved organic matter: extraction techniques, mass spectrometry, and nuclear magnetic resonance spectroscopy. Chem. Rev. 107, 419–442. doi: 10.1021/cr050359b
Moran, M. A., Kujawinski, E. B., Stubbins, A., Fatland, R., Aluwihare, L. I., Buchan, A., et al. (2016). Deciphering Ocean Carbon in a Changing World. National Academy of Sciences. Available online at: http://www.pnas.org/content/113/12/3143.abstract (accessed March 13, 2017).
Muenchen, R. (2019). The Popularity of Data Science Software. Available online at: http://r4stats.com/articles/popularity/ (accessed July 5, 2020).
Munroe, R. (2015). xkcd: Git. Available online at: https://xkcd.com/1597/ (Accessed July 1, 2020).
Murray, J., Prouty, N. G., Peek, S., and Paytan, A. (2019). Coral Skeleton δ 15 N as a tracer of historic nutrient loading to a coral reef in Maui, Hawaii. Sci. Rep. 9:5579.
Noriega-Ortega, B. E., Wienhausen, G., Mentges, A., Dittmar, T., Simon, M., and Niggemann, J. (2019). Does the chemodiversity of bacterial exometabolomes sustain the chemodiversity of marine dissolved organic matter? Front. Microbiol. 10:215. doi: 10.3389/fmicb.2019.00215
Ogawa, N. O., Nagata, T., Kitazato, H., and Ohkouchi, N. (2010). “Ultra-sensitive elemental analyzer/isotope ratio mass spectrometer for stable nitrogen and carbon isotope analyses,” in Earth Life Isotopes, eds N. Ohkouchi, I. Tayasu, and K. Koba (Kyoto: Kyoto University Press), 339–353.
Ohno, T., and Bro, R. (2006). Dissolved organic matter characterization using multiway spectral decomposition of fluorescence landscapes. Soil Sci. Soc. Am. J. 70, 2028–2037. doi: 10.2136/sssaj2006.0005
Orsi, W. D., Wilken, S., del Campo, J., Heger, T., James, E., Richards, T. A., et al. (2018). Identifying protist consumers of photosynthetic picoeukaryotes in the surface ocean using stable isotope probing. Environ. Microbiol. 20, 815–827. doi: 10.1111/1462-2920.14018
Osterholz, H., Kirchman, D. L., Niggemann, J., and Dittmar, T. (2016). Environmental drivers of dissolved organic matter molecular composition in the Delaware Estuary. Front. Earth Sci. 4:95. doi: 10.3389/feart.2016.00095
Pantoja, S., Lee, C., and Marecek, J. F. (1997). Hydrolysis of peptides in seawater and sediment. Mar. Chem. 57, 25–40. doi: 10.1016/s0304-4203(97)00003-0
Patriarca, C., Bergquist, J., Sjöberg, P. J. R., Tranvik, L., and Hawkes, J. A. (2018). Online HPLC-ESI-HRMS method for the analysis and comparison of different dissolved organic matter samples. Environ. Sci. Technol. 52, 2091–2099. doi: 10.1021/acs.est.7b04508
Pearson, A., Hurley, S. J., Walter, S. R. S., Kusch, S., Lichtin, S., and Zhang, Y. G. (2016). Stable carbon isotope ratios of intact GDGTs indicate heterogeneous sources to marine sediments. Geochim. Cosmochim. Acta 181, 18–35. doi: 10.1016/j.gca.2016.02.034
Pearson, A., McNichol, A. P., Benitez-Nelson, B. C., Hayes, J. M., and Eglinton, T. I. (2001). Origins of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound-specific Δ14C analysis. Geochim. Cosmochim. Acta 65, 3123–3137. doi: 10.1016/s0016-7037(01)00657-3
Pearson, A., McNichol, A. P., Schneider, R. J., von Reden, K. F., and Zheng, Y. (1998). Microscale AMS 14C measurement at NOSAMS. Radiocarbon 40, 61–75. doi: 10.1017/s0033822200017902
Pendergraft, M. A., Dincer, Z., Sericano, J. L., Wade, T. L., Kolasinski, J., and Rosenheim, B. E. (2013). Linking ramped pyrolysis isotope data to oil content through PAH analysis. Environ. Res. Lett. 8:44038.
Perkel, J. (2016). Democratic databases: science on GitHub. Nature 538, 127–128. doi: 10.1038/538127a
Perkel, J. M. (2018). Why Jupyter is data scientists’ computational notebook of choice. Nature 563, 145–147.
Petras, D., Koester, I., Da Silva, R., Stephens, B. M., Haas, A. F., Nelson, C. E., et al. (2017). High-resolution liquid chromatography tandem mass spectrometry enables large scale molecular characterization of dissolved organic matter. Front. Mar. Sci. 4:405.
Pfeffer, P. E., Gerasimowicz, W. V., and Piotrowski, E. G. (1984). Effect of paramagnetic iron on quantitation in carbon-13 cross polarization magic angle spinning nuclear magnetic resonance spectrometry of heterogeneous environmental matrixes. Anal. Chem. 56, 734–741. doi: 10.1021/ac00268a032
Pluskal, T., Castillo, S., Villar-Briones, A., and Orešiè, M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. doi: 10.1186/1471-2105-11-395
Polissar, P. J., Fulton, J. M., Junium, C. K., Turich, C. C., and Freeman, K. H. (2009). Measurement of 13C and 15N isotopic composition on nanomolar quantities of C and N. Anal. Chem. 81, 755–763. doi: 10.1021/ac801370c
Poulson-Ellestad, K. L., Jones, C. M., Roy, J., Viant, M. R., Fernández, F. M., Kubanek, J., et al. (2014). Metabolomics and proteomics reveal impacts of chemically mediated competition on marine plankton. Proc. Natl. Acad. Sci. U.S.A. 111, 9009–9014. doi: 10.1073/pnas.1402130111
Powers, L., and Gonsior, M. (2019). Non-targeted screening of disinfection by-products in desalination plants using mass spectrometry: a review. Curr. Opin. Environ. Sci. Health 7, 52–60. doi: 10.1016/j.coesh.2018.11.001
Ram, K. (2013). Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 8:7. doi: 10.1186/1751-0473-8-7
Reddy, C. M., Pearson, A., Xu, L., McNichol, A. P., Benner, B. A., Wise, S. A., et al. (2002). Radiocarbon as a tool to apportion the sources of polycyclic aromatic hydrocarbons and black carbon in environmental samples. Environ. Sci. Technol. 36, 1774–1782. doi: 10.1021/es011343f
Reemtsma, T. (2009). Determination of molecular formulas of natural organic matter molecules by (ultra-) high-resolution mass spectrometry, Status and needs. J. Chromatogr. A 1216, 3687–3701. doi: 10.1016/j.chroma.2009.02.033
Reintjes, G., Arnosti, C., Fuchs, B. M., and Amann, R. (2017). An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 11, 1640–1650. doi: 10.1038/ismej.2017.26
Repeta, D. J., Quan, T. M., Aluwihare, L. I., and Accardi, A. M. (2002). Chemical characterization of high molecular weight dissolved organic matter in fresh and marine waters. Geochim. Cosmochim. Acta 66, 955–962. doi: 10.1016/s0016-7037(01)00830-4
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Robinson, R. S., Brunelle, B. G., and Sigman, D. M. (2004). Revisiting nutrient utilization in the glacial Antarctic: evidence from a new method for diatom-bound N isotopic analysis. Paleoceanography 19, A3001. doi: 10.1029/2003PA000996
Rosell-Melé, A., Bard, E., Emeis, K. C., Grimalt, J. O., Müller, P., Schneider, R., et al. (2001). Precision of the current methods to measure the alkenone proxy U37K’ and absolute alkenone abundance in sediments: results of an interlaboratory comparison study. Geochem. Geophys. Geosyst. 2. doi: 10.1029/2000GC000141
Rosenheim, B. E., Day, M. B., Domack, E., Schrum, H., Benthien, A., and Hayes, J. M. (2008). Antarctic sediment chronology by programmed-temperature pyrolysis: Methodology and data treatment: PYROLYSIS OF ANTARCTIC SEDIMENTS. Geochem. Geophys. Geosyst. 9:Q04005. doi: 10.1029/2007GC001816
Rosenheim, B. E., and Galy, V. (2012). Direct measurement of riverine particulate organic carbon age structure. Geophys. Res. Lett. 39:L19703. doi: 10.1029/2012GL052883
Rossini, A. J., Lumley, T., and Leisch, F. (2003). On the edge: Statistics & computing: Reproducible statistical research. Chance 16, 41–45.
Royalty, T., and Steen, A. D. (2019). Partitioning of microbial function among taxonomic ranks across the tree of life. bioRxiv [Preprint]. doi: 10.1101/520973
Ruff, M., Fahrni, S., Gäggeler, H. W., Hajdas, I., Suter, M., Synal, H. A., et al. (2010). On-line radiocarbon measurements of small samples using elemental analyzer and MICADAS gas ion source. Radiocarbon 52, 1645–1656. doi: 10.1017/S003382220005637X
Rush, D., Talbot, H. M., van der Meer, M. T. J., Hopmans, E. C., Douglas, B., and Sinninghe Damsté, J. S. (2019). Biomarker evidence for the occurrence of anaerobic ammonium oxidation in the eastern Mediterranean Sea during Quaternary and Pliocene sapropel formation. Biogeosciences 16, 2467–2479. doi: 10.5194/bg-16-2467-2019
Rütters, H., Sass, H., Cypionka, H., and Rullkötter, J. (2002). Phospholipid analysis as a tool to study complex microbial communities in marine sediments. J. Microbiol. Methods 48, 149–160. doi: 10.1016/s0167-7012(01)00319-0
Ruttkies, C., Neumann, S., and Posch, S. (2019). Improving MetFrag with statistical learning of fragment annotations. BMC Bioinformatics 20:376. doi: 10.1186/s12859-019-2954-7
Saito, M. A., McIlvin, M. R., Moran, D. M., Goepfert, T. J., DiTullio, G. R., Post, A. F., et al. (2014). Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science 345, 1173–1177. doi: 10.1126/science.1256450
Sannigrahi, P., Ingall, E. D., and Benner, R. (2005). Cycling of dissolved and particulate organic matter at station Aloha: insights from 13C NMR spectroscopy coupled with elemental, isotopic and molecular analyses. Deep Sea Res. Part I Oceanogr. Res. Pap. 52, 1429–1444. doi: 10.1016/j.dsr.2005.04.001
Sannigrahi, P., Ingall, E. D., and Benner, R. (2006). Nature and dynamics of phosphorus-containing components of marine dissolved and particulate organic matter. Geochim. Cosmochim. Acta 70, 5868–5882. doi: 10.1016/j.gca.2006.08.037
Santos, G. M., Southon, J. R., Drenzek, N. J., Ziolkowski, L. A., Druffel, E., Xu, X., et al. (2010). Blank assessment for ultra-small radiocarbon samples: chemical extraction and separation versus AMS. Radiocarbon 52, 1322–1335. doi: 10.1017/S0033822200046415
Santos, G. M., Southon, J. R., Griffin, S., Beaupre, S. R., and Druffel, E. R. M. (2007). Ultra small-mass AMS 14C sample preparation and analyses at KCCAMS/UCI Facility. Nucl. Inst. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 259, 293–302. doi: 10.1016/j.nimb.2007.01.172
Sarma, V. V. S. S., Yadav, K., and Behera, S. (2019). Role of eddies on organic matter production and f-ratios in the Bay of Bengal. Mar. Chem. 210, 13–23. doi: 10.1016/j.marchem.2019.01.006
Schantz, M. M., Parris, R. M., and Wise, S. A. (2008). NIST Intercomparison Exercise Program for Organic Contaminants in the Marine Environment: Description and Results of the 2007 Organic Intercomparison Exercises. Gaithersburg, MD: National Institute of Standards and Technology, 1–279.
Schmidt, F., Koch, B. P., Elvert, M., Schmidt, G., Witt, M., and Hinrichs, K. U. (2011). Diagenetic transformation of dissolved organic nitrogen compounds under contrasting sedimentary redox conditions in the black sea. Environ. Sci. Technol. 45, 5223–5229. doi: 10.1021/es2003414
Schmidt, F., Koch, B. P., Witt, M., and Hinrichs, K. U. (2014). Extending the analytical window for water-soluble organic matter in sediments by aqueous Soxhlet extraction. Geochim. Cosmochim. Acta 141, 83–96. doi: 10.1016/j.gca.2014.06.009
Schouten, S., Hopmans, E. C., Pancost, R. D., and Damste, J. S. (2000). Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc. Natl. Acad. Sci. U.S.A. 97, 14421–14426. doi: 10.1073/pnas.97.26.14421
Schouten, S., Hopmans, E. C., Rosell-Melé, A., Pearson, A., Adam, P., Bauersachs, T., et al. (2013a). An interlaboratory study of TEX 86 and BIT analysis of sediments, extracts, and standard mixtures. Geochem. Geophys. Geosystems 14, 5263–5285. doi: 10.1002/2013GC004904
Schouten, S., Hopmans, E. C., and Sinninghe Damsté, J. S. (2013b). The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org. Geochem. 54, 19–61. doi: 10.1016/j.orggeochem.2012.09.006
Schubotz, F., Wakeham, S. G., Lipp, J. S., Fredricks, H. F., and Hinrichs, K. U. (2009). Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ. Microbiol. 11, 2720–2734. doi: 10.1111/j.1462-2920.2009.01999.x
Schubotz, F., Xie, S., Lipp, J. S., Hinrichs, K.-U., and Wakeham, S. G. (2018). Intact polar lipids in the water column of the eastern tropical North Pacific: abundance and structural variety of non-phosphorus lipids. Biogeosciences 15, 6481–6501. doi: 10.5194/bg-15-6481-2018
Schwedt, A., Seidel, M., Dittmar, T., Simon, M., Bondarev, V., Romano, S., et al. (2015). Substrate use of Pseudovibrio sp. growing in ultra-oligotrophic seawater. PLoS One 10:e0121675. doi: 10.1371/journal.pone.0121675
Seidel, M., Beck, M., Riedel, T., Waska, H., Suryaputra, I. G. N. A., Schnetger, B., et al. (2014). Biogeochemistry of dissolved organic matter in an anoxic intertidal creek bank. Geochim. Cosmochim. Acta 140, 418–434. doi: 10.1016/j.gca.2014.05.038
Seyler, L. M., McGuinness, L. R., Gilbert, J. A., Biddle, J. F., Gong, D., and Kerkhof, L. J. (2018). Discerning autotrophy, mixotrophy and heterotrophy in marine TACK archaea from the North Atlantic. FEMS Microbiol. Ecol. 94:fiy014.
Shah, S. R., and Pearson, A. (2007). Ultra-Microscale (5–25 μg C) Analysis of Individual Lipids by 14 C AMS: assessment and correction for sample processing blanks. Radiocarbon 49, 69–82. doi: 10.1017/s0033822200041904
Sharp, J. H., Carlson, C. A., Peltzer, E. T., Castle-Ward, D. M., Savidge, K. B., and Rinker, K. R. (2002). Final dissolved organic carbon broad community intercalibration and preliminary use of DOC reference materials. Mar. Chem. 77, 239–253. doi: 10.1016/s0304-4203(02)00002-6
Shen, X., Wang, R., Xiong, X., Yin, Y., Cai, Y., Ma, Z., et al. (2019). Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 10:1516.
Sigman, D. M., Casciotti, K. L., Andreani, M., Barford, C., Galanter, M., and Böhlke, J. K. (2001). A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal. Chem. 73, 4145–4153. doi: 10.1021/ac010088e
Simpson, A. J., Tseng, L. H., Simpson, M. J., Spraul, M., Braumann, U., Kingery, W. L., et al. (2004). The application of LC-NMR and LC-SPE-NMR to compositional studies of natural organic matter. Analyst 129, 1216–1222. doi: 10.1039/b408064e
Sleighter, R. L., Chen, H., Wozniak, A. S., Willoughby, A. S., Caricasole, P., and Hatcher, P. G. (2012). Establishing a measure of reproducibility of ultrahigh-resolution mass spectra for complex mixtures of natural organic matter. Anal. Chem. 84, 9184–9191. doi: 10.1021/ac3018026
Sleighter, R. L., and Hatcher, P. G. (2008). Molecular characterization of dissolved organic matter (DOM) along a river to ocean transect of the lower Chesapeake Bay by ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 110, 140–152. doi: 10.1016/j.marchem.2008.04.008
Sleighter, R. L., Liu, Z., Xue, J., and Hatcher, P. G. (2010). Multivariate statistical approaches for the characterization of dissolved organic matter analyzed by ultrahigh resolution mass spectrometry. Environ. Sci. Technol. 44, 7576–7582. doi: 10.1021/es1002204
Slonecker, E. T., Jones, D. K., and Pellerin, B. A. (2016). The new Landsat 8 potential for remote sensing of colored dissolved organic matter (CDOM). Mar. Pollut. Bull. 107, 518–527. doi: 10.1016/j.marpolbul.2016.02.076
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R., and Siuzdak, G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. doi: 10.1021/ac051437y
Sogin, E. M., Puskás, E., Dubilier, N., and Liebeke, M. (2019). Marine metabolomics: a method for nontargeted measurement of metabolites in seawater by gas chromatography–mass spectrometry. mSystems 4:e00638-19.
Sollai, M., Villanueva, L., Hopmans, E. C., Reichart, G. J., and Sinninghe Damsté, J. S. (2019). A combined lipidomic and 16S rRNA gene amplicon sequencing approach reveals archaeal sources of intact polar lipids in the stratified Black Sea water column. Geobiology 17, 91–109. doi: 10.1111/gbi.12316
Somville, M., and Billen, G. (1983). A method for determining exoproteolytic activity in natural waters. Limnol. Oceanogr. 28, 190–193. doi: 10.4319/lo.1983.28.1.0190
Spranger, T., Van Pinxteren, D., Reemtsma, T., Lechtenfeld, O. J., and Herrmann, H. (2019). 2D liquid chromatographic fractionation with ultra-high resolution MS analysis resolves a vast molecular diversity of tropospheric particle organics. Environ. Sci. Technol. 53, 11353–11363. doi: 10.1021/acs.est.9b03839
Steen, A. D., Crits-Christoph, A., Carini, P., DeAngelis, K. M., Fierer, N., Lloyd, K. G., et al. (2019). High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 13, 3126–3130. doi: 10.1038/s41396-019-0484-y
Steen, A. D., Vazin, J. P. J. P., Hagen, S. M. S. M., Mulligan, K. H. K. H., and Wilhelm, S. W. S. W. (2015). Substrate specificity of aquatic extracellular peptidases assessed by competitive inhibition assays using synthetic substrates. Aquat. Microb. Ecol. 75, 271–281. doi: 10.3354/ame01755
Stodden, V. (2010). The Scientific Method in Practice: Reproducibility in the Computational Sciences. MIT Sloan Research Paper No. 4773-10. Available online at: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=1550193 (accessed February 9, 2010).
Stücheli, P. E., Niggemann, J., and Schubert, C. J. (2018). Comparison of different solid phase extraction sorbents for the qualitative assessment of dissolved organic nitrogen in freshwater samples using FT-ICR-MS. J. Limnol. 77, 400–411.
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M., and Hinrichs, K.-U. (2004). Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18, 617–628. doi: 10.1002/rcm.1378
Sud, M., Fahy, E., Cotter, D., Azam, K., Vadivelu, I., Burant, C., et al. (2016). Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470.
Synal, H. A., Stocker, M., and Suter, M. (2007). MICADAS: a new compact radiocarbon AMS system. Nucl. Inst. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 259, 7–13. doi: 10.1016/j.nimb.2007.01.138
Talbot, H. M., Sidgwick, F. R., Bischoff, J., Osborne, K. A., Rush, D., Sherry, A., et al. (2016). Analysis of non-derivatised bacteriohopanepolyols by ultrahigh-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 30, 2087–2098. doi: 10.1002/rcm.7696
Tierney, N. J., and Ram, K. (2020). A Realistic Guide to Making Data Available Alongside Code to Improve Reproducibility. Available online at: http://arxiv.org/abs/2002.11626 (accessed July 1, 2020).
Tose, L. V., Benigni, P., Leyva, D., Sundberg, A., Ramírez, C. E., Ridgeway, M. E., et al. (2018). Coupling trapped ion mobility spectrometry to mass spectrometry: trapped ion mobility spectrometry-time-of-flight mass spectrometry versus trapped ion mobility spectrometry-Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 32, 1287–1295. doi: 10.1002/rcm.8165
van der Hooft, J. J. J., Mohimani, H., Bauermeister, A., Dorrestein, P. C., Duncan, K. R., and Medema, M. H. (2020). Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 49, 3297–3314. doi: 10.1039/D0CS00162G
Van der Voort, T. S., Mannu, U., Blattmann, T. M., Bao, R., Zhao, M., and Eglinton, T. I. (2018). Deconvolving the fate of carbon in coastal sediments. Geophys. Res. Lett. 45, 4134–4142. doi: 10.1029/2018GL077009
Van Mooy, B. A. S., and Fredricks, H. F. (2010). Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: water-column distribution, planktonic sources, and fatty acid composition. Geochim. Cosmochim. Acta 74, 6499–6516. doi: 10.1016/j.gca.2010.08.026
Van Mooy, B. A. S., Fredricks, H. F., Pedler, B. E., Dyhrman, S. T., Karl, D. M., Koblížek, M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. doi: 10.1038/nature07659
Wacker, L., Bonani, G., Friedrich, M., Hajdas, I., Kromer, B., Nìmec, M., et al. (2010). MICADAS: routine and high-precision radiocarbon dating. Radiocarbon 52, 252–262. doi: 10.1017/S0033822200045288
Wagner, S., Dittmar, T., and Jaffé, R. (2015a). Molecular characterization of dissolved black nitrogen via electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Org. Geochem. 79, 21–30. doi: 10.1016/j.orggeochem.2014.12.002
Wagner, S., Riedel, T., Niggemann, J., Vähätalo, A. V., Dittmar, T., and Jaffé, R. (2015b). Linking the molecular signature of heteroatomic dissolved organic matter to watershed characteristics in world rivers. Environ. Sci. Technol. 49, 13798–13806. doi: 10.1021/acs.est.5b00525
Wakeham, S. G., Amann, R., Freeman, K. H., Hopmans, E. C., Jørgensen, B. B., Putnam, I. F., et al. (2007). Microbial ecology of the stratified water column of the Black Sea as revealed by a comprehensive biomarker study. Org. Geochem. 38, 2070–2097. doi: 10.1016/j.orggeochem.2007.08.003
Wakeham, S. G., and Lee, C. (2019). Limits of our knowledge, part 2: selected frontiers in marine organic biogeochemistry. Mar. Chem. 212, 16–46. doi: 10.1016/j.marchem.2019.02.005
Wakeham, S. G., Lee, C., Peterson, M. L., Liu, Z., Szlosek, J., Putnam, I. F., et al. (2009). Organic biomarkers in the twilight zone-Time series and settling velocity sediment traps during MedFlux. Deep. Res. Part II Top. Stud. Oceanogr. 56, 1437–1453. doi: 10.1016/j.dsr2.2008.11.030
Wakeham, S. G., and McNichol, A. P. (2014). Transfer of organic carbon through marine water columns to sediments-insights from stable and radiocarbon isotopes of lipid biomarkers. Biogeosciences 11, 6895–6914. doi: 10.5194/bg-11-6895-2014
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837.
Wang, M., Jarmusch, A. K., Vargas, F., Aksenov, A. A., Gauglitz, J. M., Weldon, K., et al. (2020). Mass spectrometry searches using MASST. Nat. Biotechnol. 38, 23–26.
Wang, M., Wang, J., Carver, J., Pullman, B. S., Cha, S. W., and Bandeira, N. (2018). Assembling the community-scale discoverable human proteome. Cell Syst. 7, 412–421.
Waska, H., Koschinsky, A., Ruiz Chancho, M. J., and Dittmar, T. (2015). Investigating the potential of solid-phase extraction and Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) for the isolation and identification of dissolved metal-organic complexes from natural waters. Mar. Chem. 173, 78–92. doi: 10.1016/j.marchem.2014.10.001
Welander, P. V., Coleman, M. L., Sessions, A. L., Summons, R. E., and Newman, D. K. (2010). Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes. Proc. Natl. Acad. Sci. U.S.A. 107, 8537–8542. doi: 10.1073/pnas.0912949107
Werdell, P. J., Franz, B. A., Bailey, S. W., Feldman, G. C., Boss, E., Brando, V. E., et al. (2013). Generalized ocean color inversion model for retrieving marine inherent optical properties. Appl. Opt. 52, 2019–2037. doi: 10.1364/AO.52.002019
Wichard, T. (2016). Identification of metallophores and organic ligands in the chemosphere of the marine Macroalga Ulva (Chlorophyta) and at Land-Sea Interfaces. Front. Mar. Sci. 3:131. doi: 10.3389/fmars.2016.00131
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). Comment: the FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
Wilson, G. (2006). Software carpentry: getting scientists to write better code by making them more productive. Comput. Sci. Eng. 8, 66–69. doi: 10.1109/MCSE.2006.122
Wilson, G. (2014). Software Carpentry: lessons learned. F1000Res. 3:62. doi: 10.12688/f1000research.3-62.v2
Woods, G. C., Simpson, M. J., Koerner, P. J., Napoli, A., and Simpson, A. J. (2011). HILIC-NMR: toward the identification of individual molecular components in dissolved organic matter. Environ. Sci. Technol. 45, 3880–3886. doi: 10.1021/es103425s
Wörmer, L., Elvert, M., Fuchser, J., Lipp, J. S., Buttigieg, P. L., Zabel, M., et al. (2014). Ultra-high-resolution paleoenvironmental records via direct laser-based analysis of lipid biomarkers in sediment core samples. Proc. Natl. Acad. Sci. U.S.A. 111, 15669–15674. doi: 10.1073/pnas.1405237111
Wörmer, L., Lipp, J. S., and Hinrichs, K.-U. (2015). “Comprehensive analysis of microbial lipids in environmental samples through HPLC-MS protocols,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. McGenity, K. Timmis, and B. Nogales (Berlin: Springer), 289–317. doi: 10.1007/8623_2015_183
Wörmer, L., Lipp, J. S., Schröder, J. M., and Hinrichs, K.-U. (2013). Application of two new LC–ESI–MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Org. Geochem. 59, 10–21. doi: 10.1016/j.orggeochem.2013.03.004
Zark, M., Christoffers, J., and Dittmar, T. (2017). Molecular properties of deep-sea dissolved organic matter are predictable by the central limit theorem: evidence from tandem FT-ICR-MS. Mar. Chem. 191, 9–15. doi: 10.1016/j.marchem.2017.02.005
Zhang, J., Liu, D., Cao, P., Wang, Y., Keesing, J. K., Li, J., et al. (2016). A highly sensitive method for analyzing marker phytoplankton pigments: ultra-high-performance liquid chromatography-tandem triple quadrupole mass spectrometry: analyzing marker phytoplankton pigments: UHPLC-MS/MS. Limnol. Oceanogr. Methods 14, 623–636. doi: 10.1002/lom3.10117
Zhang, X., Bianchi, T. S., Cui, X., Rosenheim, B. E., Ping, C., Hanna, A. J. M., et al. (2017). Permafrost organic carbon mobilization from the watershed to the colville river delta: evidence from 14 C ramped pyrolysis and lignin biomarkers. Geophys. Res. Lett. 44, 11,491–11,500. doi: 10.1002/2017GL075543
Zhang, Y., Sintes, E., Chen, J., Zhang, Y., Dai, M., Jiao, N., et al. (2009). Role of mesoscale cyclonic eddies in the distribution and activity of Archaea and Bacteria in the South China Sea. Aquat. Microb. Ecol. 56, 65–79. doi: 10.3354/ame01324
Zheng, G., and Price, W. S. (2012). Direct hydrodynamic radius measurement on dissolved organic matter in natural waters using diffusion NMR. Environ. Sci. Technol. 46, 1675–1680. doi: 10.1021/es202809e
Keywords: chemometrics, natural marine organic matter, FT-ICR-MS, analytical challenges, HR-NMR, marine organic biogeochemistry
Citation: Steen AD, Kusch S, Abdulla HA, Cakić N, Coffinet S, Dittmar T, Fulton JM, Galy V, Hinrichs K-U, Ingalls AE, Koch BP, Kujawinski E, Liu Z, Osterholz H, Rush D, Seidel M, Sepúlveda J and Wakeham SG (2020) Analytical and Computational Advances, Opportunities, and Challenges in Marine Organic Biogeochemistry in an Era of “Omics”. Front. Mar. Sci. 7:718. doi: 10.3389/fmars.2020.00718
Received: 21 March 2020; Accepted: 06 August 2020;
Published: 02 September 2020.
Edited by:
Christian Lønborg, Aarhus University, DenmarkReviewed by:
Elise Sabina Morrison, University of Florida, United StatesDaniel Petras, University of California, San Diego, United States
Copyright © 2020 Steen, Kusch, Abdulla, Cakić, Coffinet, Dittmar, Fulton, Galy, Hinrichs, Ingalls, Koch, Kujawinski, Liu, Osterholz, Rush, Seidel, Sepúlveda and Wakeham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stuart G. Wakeham, c3R1YXJ0Lmcud2FrZWhhbUBnbWFpbC5jb20=
 Andrew D. Steen
Andrew D. Steen Stephanie Kusch
Stephanie Kusch Hussain A. Abdulla
Hussain A. Abdulla Nevenka Cakić
Nevenka Cakić Sarah Coffinet
Sarah Coffinet Thorsten Dittmar
Thorsten Dittmar James M. Fulton
James M. Fulton Valier Galy
Valier Galy Kai-Uwe Hinrichs
Kai-Uwe Hinrichs Anitra E. Ingalls
Anitra E. Ingalls Boris P. Koch
Boris P. Koch Elizabeth Kujawinski
Elizabeth Kujawinski Zhanfei Liu
Zhanfei Liu Helena Osterholz
Helena Osterholz Darci Rush
Darci Rush Michael Seidel
Michael Seidel Julio Sepúlveda
Julio Sepúlveda Stuart G. Wakeham
Stuart G. Wakeham