- 1Benthic Resources and Processes, Institute of Marine Research, Bergen, Norway
- 2Oceanography and Climate, Institute of Marine Research, Bergen, Norway
- 3Marine Ecology Research Group, NORCE Norwegian Research Centre AS, Bergen, Norway
- 4Department of Marine Sciences, University of Gothenburg, Gothenburg Global Biodiversity Centre, Gothenburg, Sweden
An effective management of vulnerable marine ecosystems is dependent on thorough knowledge of their location. Multibeam bathymetric mapping and targeted remotely operated vehicle (ROV) surveys are currently used to map areas impacted by industrial activities when vulnerable species are expected. However, multibeam bathymetric mapping is not always a possibility and surveying large areas using ROVs is expensive. Here, we developed a species-specific eDNA assay targeting a 178 bp fragment in the control region of the mitochondrial DNA of the cold-water coral (CWC) Lophelia pertusa. The aim was to test if concentrations of L. pertusa eDNA in seawater, determined using droplet digital PCR (ddPCR) technology, could be used to assess the broad scale distribution of CWCs in a region, to supplement multibeam mapping and direct targeted ROV surveys. Our assay successfully amplified L. pertusa DNA from seawater. In laboratory we documented an exponential decay rate of the targeted DNA fragment and a linear correlation between coral biomass and eDNA concentrations in flow through microcosms. The ability of the method to detect CWC reefs in situ was tested in the fjords south of Bergen, Norway, where such reefs are common. We tested five sites with, and five sites without, known reefs. Lophelia pertusa eDNA was detected in all 10 sites. However, concentrations were elevated by 5 to 10 times in water sampled off the two large reefs growing on vertical surfaces. Water sampled 10 m above CWC reefs growing on the flat seabed did not produce an equally clear eDNA signal, nor did single CWC colonies growing on vertical surfaces. Treating the eDNA as a passive particle with no active vertical or horizontal movement, we successfully modeled the dispersal of eDNA from the known CWC reefs in the region and achieved a good fit with measured eDNA concentrations. In all, our study demonstrated a great potential for eDNA measurements as a cost-efficient tool for a rapid screening of the broad scale distribution of CWC reefs growing on vertical surfaces (so called wall reefs) that cannot be imaged using traditional ship mounted downward looking multibeam echo-sounders and difficult to detect using ROVs alone.
Introduction
Cold-water coral (CWC) reefs compose one of the most diverse and productive ecosystems in the marine realm (Rogers, 1999; Roberts et al., 2009; Cathalot et al., 2015; Jensen et al., 2015), yet human impact on these reefs is significant and rising. The reefs face both local and global threats, including physical destruction (Heifetz et al., 2009), chemical pollution (Fisher et al., 2014) as well as warming and acidifying waters as a consequence of climate change (Hoegh-Guldberg et al., 2017). Knowledge of species distributions is critical to ecological management and conservation but mapping deep-sea environments is time consuming and expensive, and rarely carried out to full extent.
Norway’s long and jagged coastline provides excellent conditions for CWC growth. On the shelf and inside the deep fjords more than 6000 Lophelia pertusa reefs have been registered, representing roughly 25% of the known global occurrence of the species. Only a fraction of the Norwegian deep water has, however, been mapped and the 6000 reefs known today likely comprise only a small fraction of the true reef count. On the shelf, reefs can be mapped using a combination of multi-beam bathymetric surveys, identifying features on the seafloor resembling coral reefs, followed by surveys of interesting targets using either towed video rigs or remotely operated vehicles (ROVs). Furthermore, species distribution models (recently developed for the Norwegian shelf) can inform surveyors as to where targeted video investigations are needed (Burgos et al., 2020; Sundahl et al., 2020). In fjords, however, L. pertusa often grows on the vertical or overhanging side walls, forming so called wall reefs (Järnegren and Kutti, 2014). These reefs cannot be imaged using traditional ship mounted downward looking multibeam echo-sounders. Moreover, species distribution models have not yet been developed for fjord settings because the lack of input data for such models (e.g., observed species distributions and substrate maps). Wall reefs have been described from several fjords (Järnegren and Kutti, 2014) and are likely a big part of the Norwegian L. pertusa population but have largely been overlooked to date. Examining new methods (i.e., molecular, acoustic or mathematical) that can help in selecting areas for targeted video surveys, within the 90 000 km2 of the Norwegian coastal zone, is therefore desirable.
Lophelia pertusa reefs are ecologically and functionally highly important members of the benthic ecosystem. The reef’s framework is built solely by L. pertusa but the reefs are highly species rich as the framework provides habitat for a diversity of benthic invertebrates, microorganisms and fish (Rogers, 1999; Roberts et al., 2009). Furthermore, the reefs play a major role in the circulation of organic carbon (Cathalot et al., 2015). The reefs are fragile and slow growing (Mortensen and Rapp, 1998; Freiwald, 2002) and are vulnerable to bottom trawling (Fosså et al., 2002; Stone, 2006), oil and gas exploration and production (Rogers, 1999; Hall-Spencer et al., 2002; Larsson et al., 2013), as well as aquaculture production in open sea-cages (Kutti et al., 2015), all of which are large and influential industries on the Norwegian shelf and coastal zone. Localization of reef formations is imperative in order to prevent irreversible damage to the reefs by industrial activity.
All organisms shed or expel DNA (at different rates) into the surrounding environment via e.g., production of feces, urea and saliva and loss of skin cells and mucus (Taberlet et al., 2018). Using appropriate study designs (outlined e.g., in Goldberg et al., 2016) the extraction and measurements of this extracellular DNA in water samples can be employed to study biodiversity and/or to discover cryptic and introduced species, in shallow waters as well as the deep sea (e.g., Port et al., 2016; Everett and Park, 2018; Holman et al., 2019). Although the quantity of eDNA in a water sample at a given site will vary in time, dependent on factors such as hydrodynamic conditions and temperature (Harrison et al., 2019), there are many examples of strong links between the abundance of marine benthic organisms and the concentration of cognate eDNA in water samples (e.g., Doi et al., 2015; Uthicke et al., 2018). For tropical reefs, metabarcoding counts of coral eDNA correlate well with coral cover based on visual surveys (Nichols and Marko, 2019). Furthermore, droplet digital PCR (ddPCR) has lately been successfully applied to quantify eDNA and correlate it to the abundance of crown of thorn sea-stars on tropical coral reefs (Uthicke et al., 2018).
Here, we present an assessment of quantitative eDNA detection as a suitable tool for localization of L. pertusa CWC reefs in Norwegian fjords. First, we assessed the degradation and shedding rate of L. pertusa DNA in seawater microcosm experiments. Second, we tested the suitability of eDNA techniques to localize CWC reefs by quantifying L. pertusa eDNA in deep-water sampled in five locations known to host L. pertusa coral colonies of varying extent and form and five locations without corals. Thirdly, we tested if the observed levels of eDNA varied in accordance to the predicted passive dispersion of genetic material from known reefs using a hydrodynamic model. We conclude by outlining the strengths and weaknesses of quantifications of eDNA for the mapping of CWC reefs and discuss how eDNA analysis can assist in expediting reef mapping, thereby assisting in the mitigation of deleterious effects of industrial activity on CWC reefs.
Materials and Methods
Coral Collection
Colonies of Lophelia pertusa were collected in May 2018 (research cruise 2018613 RV K. Bonnevie) from 200 m depth on the Nakken CWC reef (59°49.83N, 05°33.44E) using the ROV Aglantha. On-board the colonies were transferred (submerged in water) from the biobox of the ROV to a large tank containing unfiltered water from 100 m depth. The corals were subsequently transported to the Institute of Marine Research’s laboratory in Bergen and into 300 L flow-through tanks receiving sand-filtered water from 120 m depth at a rate of 100 L h–1 for 5 months before the experiments started.
DNA Longevity – Microcosm Experiment 1
The experiment was carried out from 6 to 13 November 2018 in an 8°C climate-controlled room (Institute of Marine Research, Bergen). Four 10 L batch microcosms were each filled with 8 L of sand-filtered deep water (salinity 33‰). Lophelia pertusa colonies were transferred, one to each, of three of the microcosms and kept there for 24 h. The fourth microcosm served as a negative seawater control (control 1) and did not receive any coral. A fifth 10 L microcosm was filled with 8 L of distilled water (control 2). All microcosms were fitted with a submersible micropump operating at 100 ml min–1 to ensure water circulation and proper aeration of the microcosms throughout the experiment. The climate room utilized had no history of L. pertusa cultivation or handling and did not contain any animals other than the experimental colonies of L. pertusa during the experiment.
After 24 h, 1 L of water was carefully sampled from the center of each of the three microcosms containing L. pertusa, as well as from the two control microcosms (sampling time = T0) using a siphoning hose. One liter of tap water, also taken at T0, served as a third control. Water in the batch microcosms was not replaced after samplings hence the volume of water in the microcosms reduced over the course of the study. All water samples were individually filtered (using a vacuum pump) through sterile nitrocellulose membrane filters (Whatman, 0.45 μm pore size, 47 mm diameter, WHA7141004, Merck, Norway). Pressure in the pump was set so that filtering 1 liter of water took approximately 20 min. Filters were aseptically folded and placed in 2 mL sterile cryogenic vials to which 1.6 mL of tissue lysis buffer (Buffer ATL, 19076, Qiagen, Norway) was added before storage at −20°C. The filtration kit and forceps were sequentially cleaned with 10% (v/v) bleach followed by distilled water between samples in order to minimize cross-contamination.
After the first sets of water samples had been taken (T0), we removed the L. pertusa colonies from the microcosms and sampled the microcosm water after 5, 10, 24, 48, 96, and 168 h in order to check for the longevity of residual L. pertusa eDNA, following the same filtration procedure as described above. After removal from the tanks the coral colonies were weighed (wet weight, WW). Tissue samples for L. pertusa DNA were obtained by crushing 3–5 coral polyps in a mortar before transfer into a 2 mL cryovial containing 1.6 mL tissue lysis buffer (Qiagen, Norway). Samples were stored at −20°C until analysis.
DNA Shedding Rate – Microcosm Experiment 2
A second microcosm experiment was carried out from 5 to 13 March 2019 to assess the relationship between L. pertusa biomass and the concentration of L. pertusa eDNA in seawater. For this, three 80 L flow-through microcosms containing pieces of living coral colonies were set up. The microcosms received sand-filtered seawater from 120 m depth (salinity 33‰ and temperature 10°C) at a flow rate of 100 mL min–1 (turnover = 2 times day–1). The microcosms were allowed to equilibrate for 3 days before the first water samples of 2 L were taken from each of the microcosms. Inlet water to the microcosms and tap water (2 L) served as controls. All samples were filtered and preserved in the same manner as described for microcosm experiment 1. After the first water sampling round, 30–50% of the biomass in the microcosms was removed and the microcosms were allowed to equilibrate for 3 days before new water samples were taken and additional coral biomass was removed. The process of water sampling, coral removal and equilibration was repeated three times yielding a total of 5∗4 samples with varying coral biomass in the microcosms. Three days after complete removal of L. pertusa, the last set of water samples was collected. Coral biomass (g tissue dry weight) in each of the tanks at each time point was calculated by first drying the corals at 60°C to constant weight (roughly 14 days) and weighing them, thereafter burning them at 550°C for 6 h and re-weighing them. Tissue weight was calculated by subtracting ash weight from the dry weight for each individual coral.
DNA Extraction
Extraction of DNA from filters was carried out using the DNeasy Blood and Tissue kit (Qiagen, Norway) following Majaneva et al. (2018) with slight modifications. In short, 160 μL of Proteinase K (20 mg ml–1, Qiagen) was added to filters in 1.6 mL tissue lysis buffer, which were then incubated in a hybridization oven at 56°C with slight orbital rotation overnight. After lysis, total DNA was purified from one 200 μL subsample of lysate from each filter according to kit instructions. DNA was eluted in 100 μL and stored at −20°C. The resulting total DNA concentrations were measured using a Qubit fluorescence spectrophotometer and the dsDNA HS Assay kit (Invitrogen, Fisher Scientific, Norway). As positive control for DNA purification, we also extracted DNA from L. pertusa tissue using the identical protocol.
Primer Selection
Standard mitochondrial DNA (mtDNA) markers such as the cytochrome c oxidase I (COI) and cytochrome B (cytB) genes have proven to be very conserved in cnidarian species. Lophelia pertusa is nearly invariable in mtDNA throughout its range in the North East Atlantic (Flot et al., 2013). The solitary coral Desmophyllum dianthus is also closely related L. pertusa with little variation in the most commonly studied mtDNA regions (Addamo et al., 2016). In addition, there is limited data available in public sequence repositories reducing the number of possible target regions for construction of an assay specific to L. pertusa. However, based on an alignment of the rapidly evolving mtDNA control region from eleven D. dianthus and 7 L. pertusa sequences retrieved from the NCBI GenBank database, we identified a region unique to L. pertusa. The 7 L. pertusa individuals originated from a wide area, i.e., the Korallen reef (Northern Norway), Nord-Leksa reef (central Norway), Santa Maria de Leuca (Ionian Sea) and the Moria Mounds (Irish continental margin). Using NCBI primer Blast program for this region a pair of primers amplifying a 178 bp long fragment was identified and selected for further testing (Genbank Accessions: FR821799, KC875348, KC875349, KC875351, KC875353, KC875355, KM609293, KM609294, KX000882-KX000894) (Table 1). The L. pertusa assay was first tested in silico using GenBank, the results from which indicated it to be specific to L. pertusa. The assay readily amplified tissue samples of L. pertusa using a BioRad QX100 Droplet Digital PCR system (BioRad Inc., see below) and an annealing temperature of 62°C, further ensuring high assay specificity.
ddPCR Assay
Each 20 μL ddPCR reaction included 250 nM of each primer, 1× BioRad ddPCR EvaGreen Supermix, 5 μL template DNA, and PCR-grade water. Appropriate dilutions of DNA (i.e., 1 to 120 000 molecules per reaction) were used for ddPCR as necessary. The BioRad QX200 droplet generator partitioned each individual reaction mixture into nanodroplets according to manufacturer’s instructions (combining 20 μL of the reaction mixture with 70 μL of BioRad droplet oil). For amplification an annealing temperature of 62°C was used with a standard ddPCR program consisting of 40 amplification cycles after an initial denaturation at 95°C for 5 min. The 40 amplification cycles each consisted of 95°C for 30 s and 62°C for 1 min. Finishing steps were 4°C for 5 min and 90°C for 5 min. Ramp rate was set to +2°C s–1 for all steps. For each ddPCR analysis we used one positive control from a L. pertusa tissue extraction and two negative controls containing MilliQ water as template. Quantification of target DNA, in copies/μL in each reaction, is based on an assumption of a Poisson distribution of the target DNA among the slightly less than 20,000 droplets from a typical 20 μL reaction (Miotke et al., 2014). PCR products from positive control ddPCR reactions were sequenced and confirmed to be the correct gene fragments from L. pertusa.
Field Sampling
Seawater samples were collected during a research cruise on RV H. Brattström, 14–26 March 2019, at 10 sites in the Langenuen straight, western Norway, where earlier surveys had shown with a high degree of certainty either the lack of L. pertusa colonies, existence of single colonies, small or extensive reefs (Figure 1). The area was selected for the field campaign because this fjord system has been extensively scrutinized by marine zoologists from the academic institutions in the city of Bergen starting as early as the nineteen-fifties with dredge sampling campaigns (see e.g., Tambs-Lyche, 1958; Burdon-Jones and Tambs-Lyche, 1960) and with more that 200 ROV dives performed, by the Institute of Marine Research, between the years of 2011–2019 (for a full overview of surveys see the Fisken og Havet report series: Report on cruises and oceanographic data stations (nr 6/2011, 2/2012, 9/2013, 1/2014, 2/2015, 3/2016, 1/2017, 3/2018, and 1/2019). Here, water samples were collected at 160 m depth using Niskin bottles mounted on a CTD rosette, 50–100 m off three vertical walls. Two of these walls contain large wall reefs (site 3 and 6) and one contains scattered L. pertusa colonies (site 4) growing from 130 to 180 m depth. Samples were also collected 10 m above two different reefs growing on the flat seabed (site 1 and 5) and at five sites lacking corals. At all 10 sites, 5 L of seawater was sampled from two separate Niskin bottles and filtered immediately through Whatman nitrocellulose membrane filters following the procedure as described for microcosm experiment 1. Three liters of onboard tap water filtered, using the same equipment, served as sampling control for each site. Temperature and salinity were not measured when the water samples were collected, however, previous studies have shown temperature and salinity to be relatively stable at these depths, i.e., approximately 8°C and 34‰ (Skjelvan et al., 2016; Jones et al., 2018). Filters from field campaigns were stored and processed as described for microcosm experiment 1 (see above).
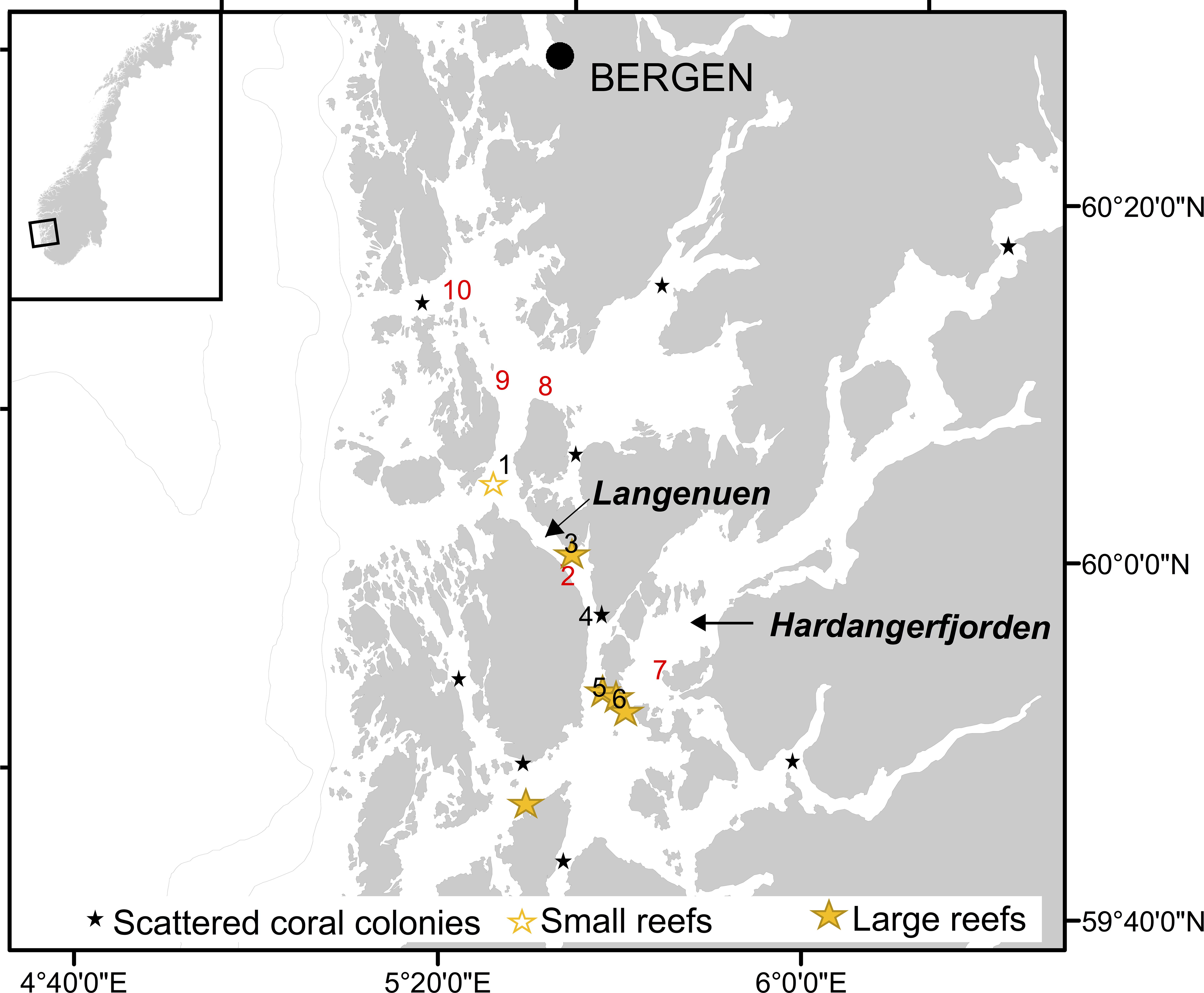
Figure 1. Map showing Hardangerfjorden and Langenuen straight (western Norway) and the locations where water samples were collected for quantification of Lophelia pertusa eDNA (nr. 1–10). Black numbers denote locations known to harbor cold-water corals (CWCs) and red number denote locations known not to have CWCs. Also shown (by stars) is the distribution of known CWC ecosystems in the region, classified into scattered coral colonies, small reefs and large reefs.
Dispersion Modeling
The predicted (i.e., simulated) concentration of genetic material (i.e., eDNA) from known CWC reefs in the 5600 km2 study area (Figure 1) was calculated using a hydrodynamic dispersion model. The current field was obtained from the Regional Ocean Model System (ROMS) with horizontal resolution of 160 m x 160 m and having 35 terrain following vertical layers (Shchepetkin and McWilliams, 2005; Albretsen et al., 2011;1). The simulated circulation has been shown to be in good accordance with field observations (Asplin et al., 2014; Espeland et al., 2015; Johnsen et al., 2016). The maximum currents at the seabed in the study area did not exceed 1.3 m s–1, and the average velocity was typically an order of magnitude smaller, hence CLF (Courant-Friedricks-Lewy) criteria was fulfilled for the study area. The particle tracking of genetic material from the reefs was conducted using the open source Lagrangian Advection and Diffusion Model (LADIM;2). The LADIM calculated the transport of particles using a 4th order Runge-Kutta scheme and a timestep of 120 s. 100 particles were released every hour from all sites where L. pertusa has been documented in the study area (Figure 1). The trajectories of neutrally buoyant particles, with no active behavior in the vertical or in the horizontal, were calculated. Non reflective boundaries were used along land, simply holding the particles fixed if the trajectory was directed onto land. To reflect the difference in size between the reefs the particles were parameterized as super individuals, reflecting the size of their origin reef grouped into three categories given as relative reef size in Figure 1 and Table 2. This was done because the reefs have not been mapped in sufficient detail to allow correct estimations of biomass of live L. pertusa. In the model the number of eDNA copies released is therefore only relative and does not represent the actual number released from each reef. The simulated concentration of genetic matter was calculated assuming an exponential decay of the particles in accordance to the observations in tank experiment 1 (Figure 2).
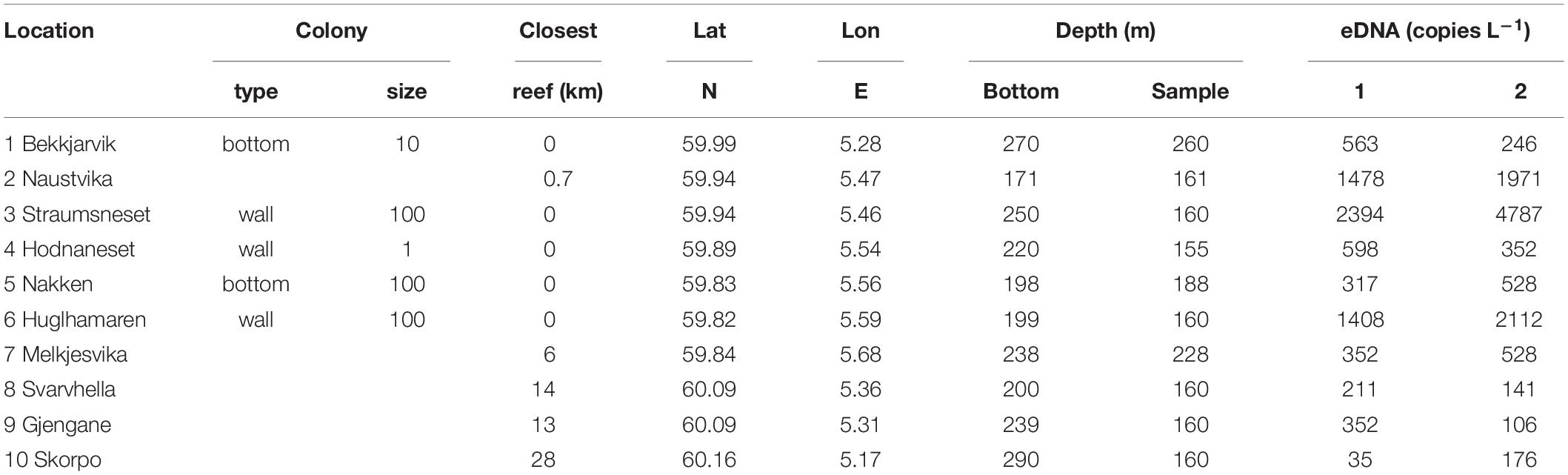
Table 2. Properties of the sampling sites, and eDNA concentrations (eDNA copies l–1) measured in two parallel Niskin bottles sampled at the 10 sites dispersed in the study region.
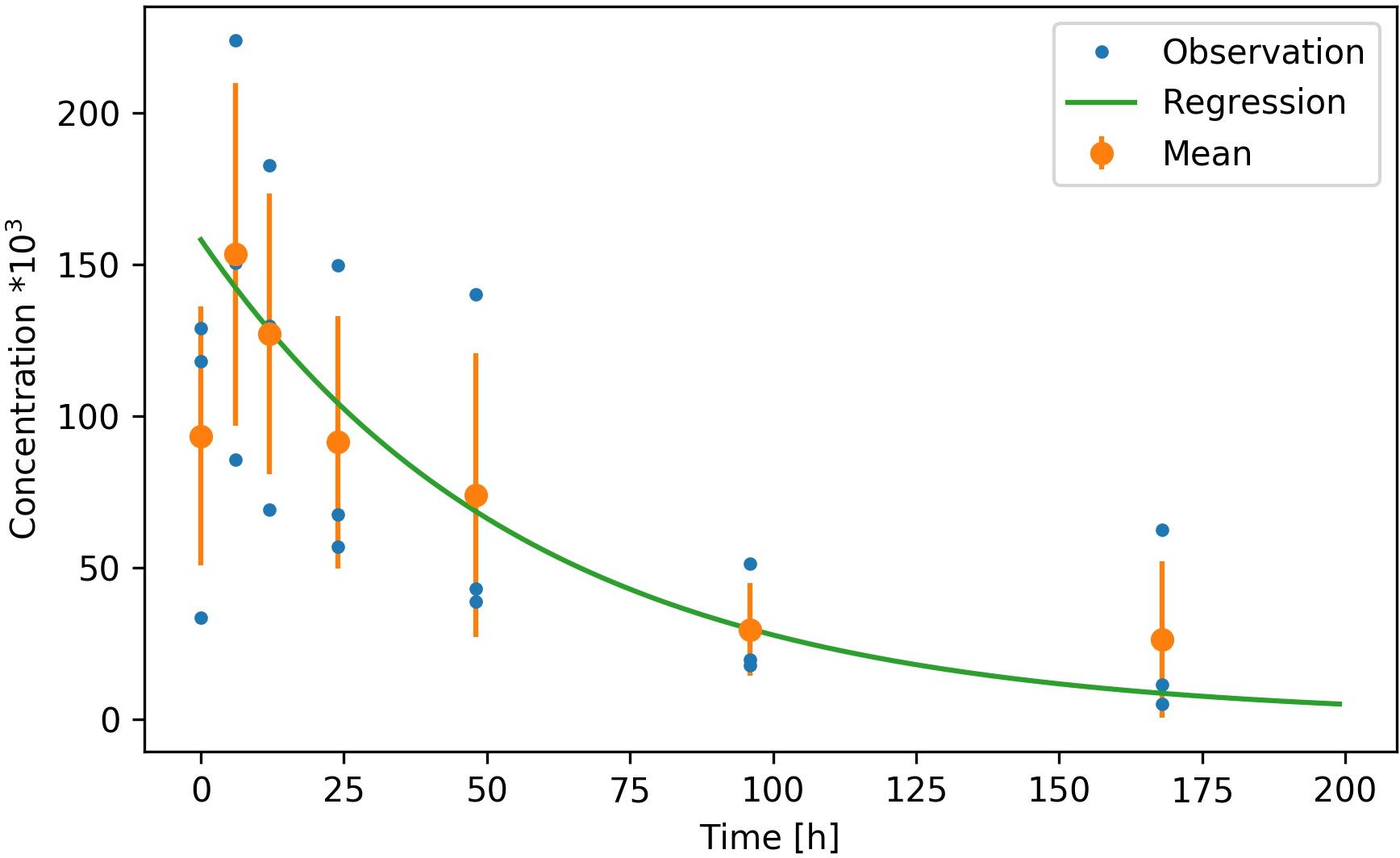
Figure 2. Decay rate for the Lophelia pertusa targeted eDNA fragment measured in microcosm experiments at ambient temperature (8°C) and salinity (33‰) over 168 h. Three replicate observations (attained by placing one coral colony into each of three separate microcosms) are given by blue dots, the mean and standard deviation of the observations are given by the orange dots and error bars and the exponential regression is given by the green line (excluding the initial 5 h of data).
Results
eDNA Decay and Shedding Rates
Our assay successfully amplified L. pertusa DNA from seawater. In experiment 1 the first sample (T0) showed a lower eDNA concentration compared to samples obtained after 5 and 10 h (Figure 2). We assumed that this was a result of the removal of the coral from the microcosms after the T0 sampling, which flushed the coral from surface DNA and increased DNA concentrations in the tank. T0 readings were therefore excluded from the regression. From T5 and onward the decay of eDNA at ambient temperature of 8°C and a salinity of 33‰ was exponential and followed the regression:
With C(t) being the concentration after time t in hours, C0 the concentration at t = 0 and the decay factorα = 0.017. The coefficient of determination (R2) of the regression to the mean observations was 0.97. The corresponding half-life for the 178 bp targeted fragment was 41 h (Figure 2).
eDNA shedding rates increased with coral biomass from 0.11 to 9.00 g dry tissue weight (Figure 3), following the linear regression: y = 19015x–2579 (Spearman rank correlation coefficient = 0.85, p < 0.001, n = 12, df = 10). Removing the data point generated by the largest L. pertusa fragment (i.e., that of 9.00 g) resulted in a slight reduction in the correlation, with a Spearman rank correlation coefficient of 0.80 and p = 0.003 (n = 11, df = 9).
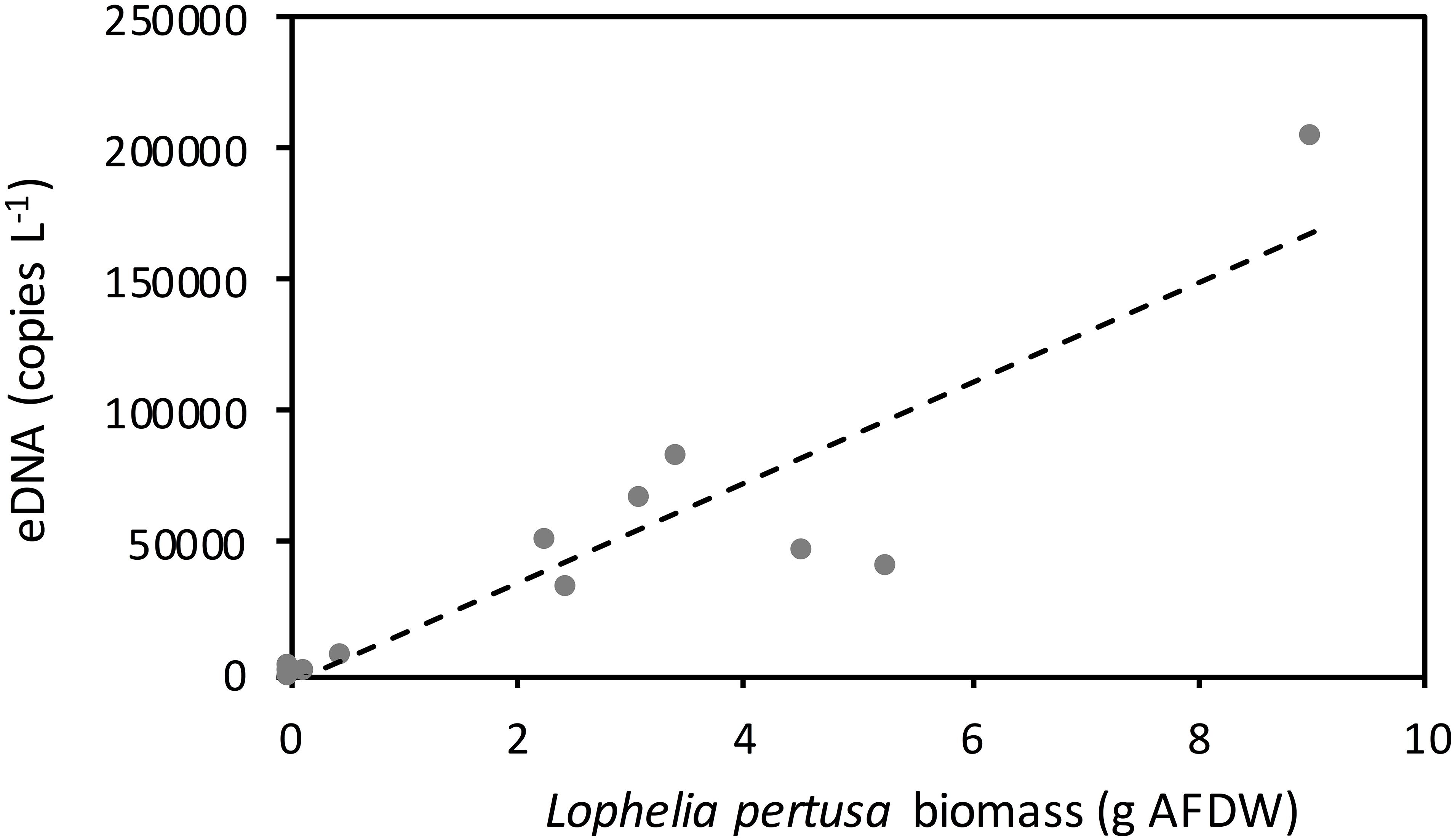
Figure 3. Concentration of the targeted Lophelia pertusa eDNA fragment in 80 liter flow through tanks containing decreasing biomass of corals. A linear regression is fitted for the concentration of eDNA in the water and biomass of corals in the tanks (y = 19015x–2579) (Spearman rank correlation coefficient = 0.85, p < 0.001). Temperature = 10oC and salinity = 33‰. Ash free dry weight ratio to dry weight ratio of whole living coral fragments was 0.03.
Field Study
Our field campaign, examining deep-water from five sites with known CWC occurrences and five sites without CWCs (i.e., where extensive video surveys had not documented any reefs, aggregations or single colonies of L. pertusa), showed the presence of L. pertusa eDNA at all examined sites and with no statistically significant difference between the two groups “reef” and “no reef” (Wilcoxon Rank Sum Test, p > 0.05). Instead, eDNA concentration seemed to be related to a combination of the factors colony/reef type, size and the proximity to nearest reef (Table 2). The community of small scattered colonies at the Hornaneset vertical wall did not produce an eDNA concentration above the mean background concentration, nor did the water samples collected 10 m above the sill reefs, Nakken and Bekkjarvik. L. pertusa eDNA concentrations in water sampled near the two large wall reefs (Straumsneset and Huglhamaren) were, however, 5 to 10 times higher than that of the background water (Table 2), as was the water collected at Naustvika 1 km south west of the wall reef at Straumsneset.
eDNA Dispersal Modeling
The simulated dispersal of genetic material shows the occurrence of L. pertusa eDNA in the whole region but with clearly elevated concentrations around the six large CWC reefs in the study area (depth integrated concentration field shown in Figure 4). Concentrations of genetic material from L. pertusa colonies rapidly decreased with distance from its source. For the 6 release points representing the large coral reefs only 5% of the original eDNA concentration remained at a distance of 12–31 km from the source (i.e., the reef). The simulated eDNA concentration field correlated well with the observed eDNA concentrations (shown log scaled in Figure 5) (Spearman rank correlation coefficient = 0.85, p < 0.01, y = 0.31x – 0.29, n = 10, df = 8).
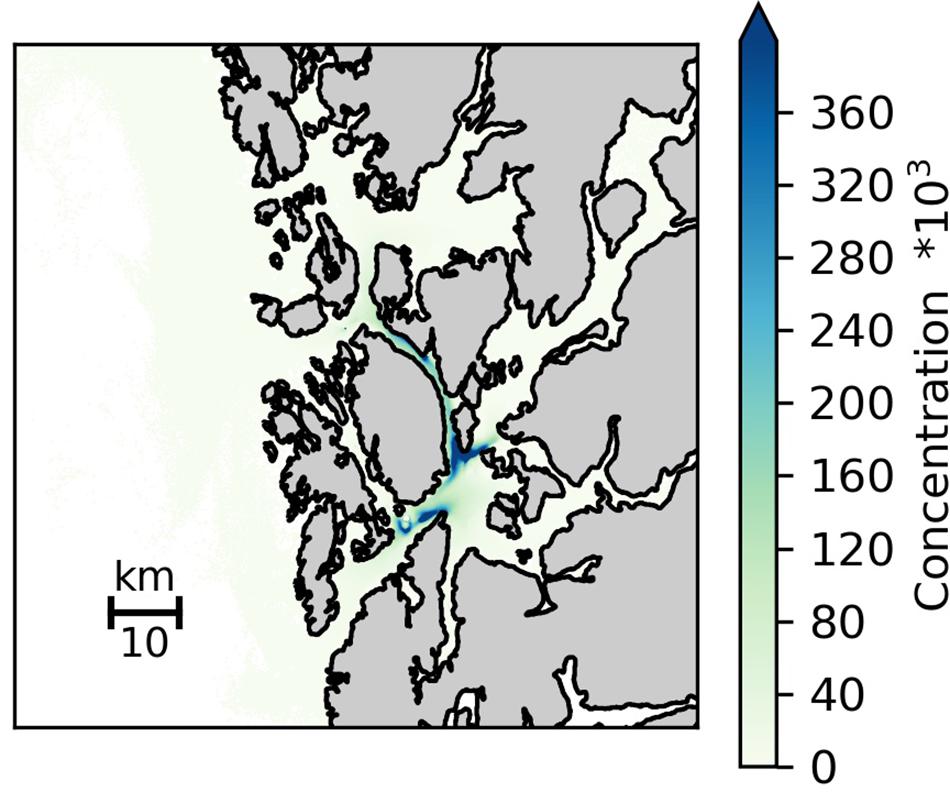
Figure 4. Simulated distribution of eDNA particles (averaged over all depths) dispersed from all known cold-water coral occurrences in the study area. Simulations were conducted for the time period 10–23 March 2019, and included releasing 100 particles every hour from each reefs with the trajectories of the particles calculated using passive drift, with no active behavior in the vertical nor in the horizontal. The particles were parameterized as super individuals, reflecting the size of its origin reef grouped into three categories given as relative reef size in Figure 1 and Table 2.
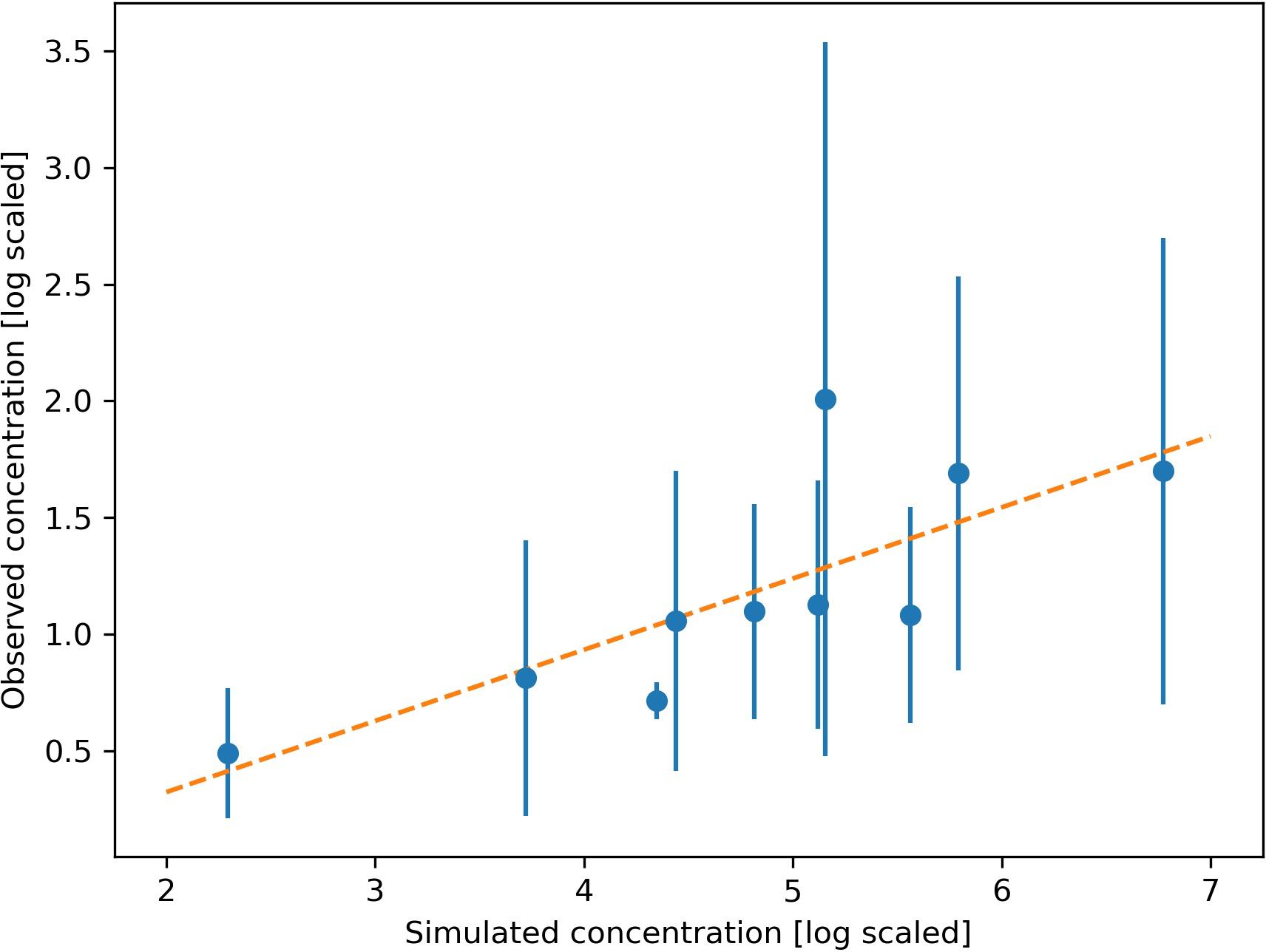
Figure 5. Relationship between the observed and simulated concentration of Lophelia pertusa eDNA at 10 sampling stations. Observed mean value and standard deviation from 2 replicate samples shown by dots and error bar. The linear regression model (y = 0.31x + 0.29) is shown by the orange line (Spearman rank correlation coefficient = 0.85, p < 0.01).
Discussion
Recent papers have demonstrated the applicability of environmental DNA (eDNA) in monitoring various aspects of tropical reef health; its usefulness for determining living coral biomass, ecological function of reefs and assessing the abundance of keystone predators (Uthicke et al., 2018; Carvalho et al., 2019; Nichols and Marko, 2019). Here, we demonstrate that quantifications of L. pertusa eDNA in deep water can also be used to determine the broad scale distribution of cold-water coral (CWC) reefs. It seems particularly successful for detecting CWC reefs on vertical surfaces (i.e., wall reefs), that are challenging to detect using traditional mapping techniques that rely heavily on multibeam bathymetry.
Shedding and Dispersal of eDNA
We calculated the release rate of eDNA from a L. pertusa colony (Figure 6A) composing 2 kg wet weight, 60 g tissue dry weight and being roughly the size of a football (following Uthicke et al., 2018) to 114 000 target gene copies min–1. This is in the same range as that shed by fish (250,000 target gene copies min–1 fish–1, Maruyama et al., 2014) but much less than measured rates for the crown of thorn sea-stars (i.e., 2,000,000 target gene copies min–1 individual–1, Uthicke et al., 2018). Corals are sessile suspension feeders known to release mucus at very high rates (Wild et al., 2004) to capture zooplankton prey (Murray et al., 2019) as well as to keep the body surface clean from settling particles (Larsson et al., 2013). It is thus plausible that the detected L. pertusa eDNA signal is largely derived from mucus production, as suggested also for gorgonian corals (Everett and Park, 2018). DNA yield may be low from mucous material (e.g., Winnepenninckx et al., 1993). It may therefore be important to use the same extraction method in shedding and degradation experiments as well as in the collection of field data (as done here) to attain consistent DNA copy numbers.
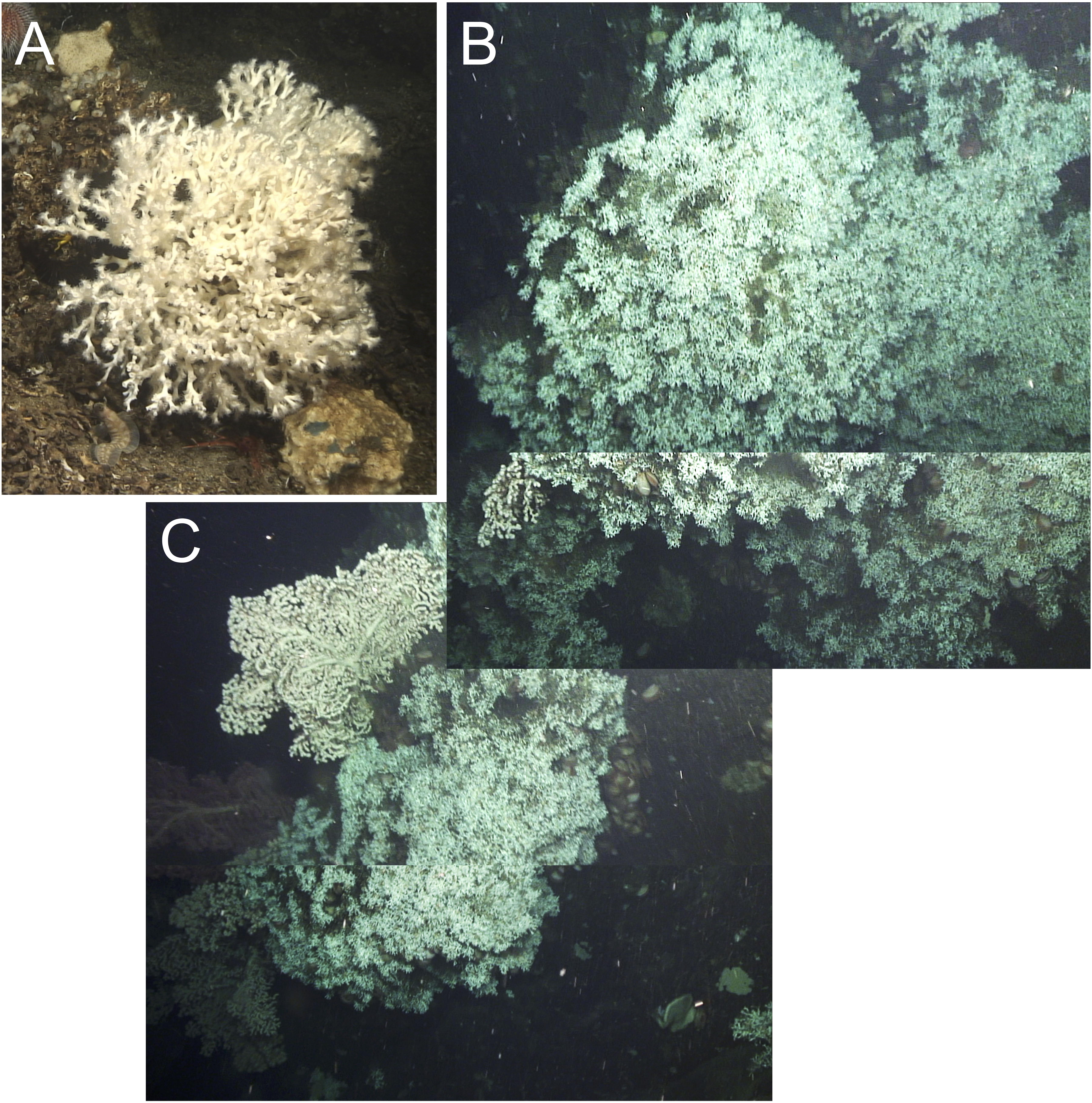
Figure 6. Lophelia pertusa colonies from the vertical wall at Straumsnes (A) small single colony 25 cm in diameter, (B) L. pertusa lobe 280 cm high and 350 cm wide and (C) L. pertusa lobe 300 cm high and 180 cm wide.
Despite the high incidence of L. pertusa in areas with complex hydrodynamics (Thiem et al., 2006; Davies et al., 2009), our study demonstrates that eDNA shedding from L. pertusa reefs can be detected using a low-precision, conventional water sampling cast carried out from a small survey vessel. Our dispersal model used experimentally derived values for L. pertusa eDNA decay rates and DNA shedding rates when exposed to realistic temperature and salinity (Harrison et al., 2019). Treating the DNA as a passive particle with no active vertical or horizontal movement, we successfully modeled the dispersal of genetic material from the known CWC reefs in the region and achieved a good fit with our measured eDNA concentrations (Figure 5). The modeled eDNA concentrations differed from the measured eDNA concentrations by about three orders in magnitude. However, it is important to note that the modeled concentration of eDNA around the reefs was based on our best knowledge of the reef size, not the actual weight of live coral tissue in each reef. The reefs were categorized according to their relative size (1, 10, and 100), hence an absolute fit could not have been achieved. Where large coral domes grow densely on vertical surfaces (Figures 6B,C), and there is an efficient horizontal dispersal of eDNA (Andruszkiewicz et al., 2019), the eDNA signal was clearly detectable in water samples taken 50 to 100 m off the reefs, in the same vertical depth layer as the live coral thicket.
Mapping Reefs in Complex Vertical Terrain
Steep, vertical and overhanging walls in fjords and deep canyons are known to provide habitats highly suitable for CWC growth (Huvenne et al., 2011; Gori et al., 2013; Järnegren and Kutti, 2014). Although likely frequent in both fjords and along the continental slope, these habitats are understudied and poorly known, mainly because they go undetected using traditional ship mounted downward-looking multibeam echo-sounders. Our field campaign near the two large wall reefs, Straumsneset and Huglhamaren, revealed that L. pertusa eDNA concentrations in seawater was 5 to 10 times higher than that of the background water. This shows the potential of quantitative eDNA analysis using ddPCR technology for a rapid assessment of the broad scale distribution of wall reefs within fjords or canyons, the precision in this system was 1–2.5 km. eDNA analysis could then guide targeted ROV video surveys, to verify L. pertusa occurrences and to describe reef composition and shape. Several studies have documented the ability of both qPCR and ddPCR to accurately determine DNA levels in target solutions but with ddPCR providing more precise and reliable results at low DNA concentrations (Zhao et al., 2013; Arvia et al., 2017; Taylor et al., 2017; Link-Lenczowska et al., 2018). While the assay developed in this study is likely to work also using a qPCR based approach, ddPCR might be better suited for the screening for small fjord reefs.
Recently, sideways-looking multibeam echo sounders on autonomous underwater vehicles (AUVs) have been introduced for the mapping of vertical walls (see e.g., Robert et al., 2017). Multibeams can provide detailed 3D maps of complex wall habitats but the presence of live coral colonies still needs to be verified with e.g., underwater video surveillance. eDNA analysis can provide a useful supplement to video verification of live L. pertusa colonies (this study) and associated gorgonian CWCs (Everett and Park, 2018) because eDNA concentrations are likely to be elevated several 100 m downstream of a reef while the field view of a ROV is often limited to 1-2 meters.
Mapping Reefs Close to the Seabed
While the signal of the eDNA shed from the large wall reefs was easily captured, the water sampling campaign failed in capturing the signal of eDNA shed from the reefs growing on the flat seabed. This may have been, at least partly, due to our inability in this study to sample bottom water very close to the sea-bed. The stratification of the water column normally observed in Norwegian fjords reduces the vertical movement of the water masses (Stigebrandt and Aure, 1989). Under normal conditions this mechanism will prevent rapid vertical transport of coral eDNA from the seabed and up to the seawater sampling depth 10 m above the seabed. Previous studies confirm little vertical exchange of eDNA between depth layers at the scale of tens of meters (Andruszkiewicz et al., 2017), with no or limited upward transport of eDNA (Andruszkiewicz et al., 2019). CWC reefs on the flat seabed are normally successfully mapped using ship mounted downward-looking multibeam echo-sounders. In large reef aggregations, however, only a small amount of what is suspected to be living CWC reefs can actually be confirmed using ROVs. In such instances, eDNA sampling could provide a useful way of ground-truthing whether CWC reefs identified by multibeam sonar are composed of living or dead coral colonies. Due to limited vertical mixing of genetic material on the flat seabed (Andruszkiewicz et al., 2019), water samples to confirm the presence of live L. pertusa colonies would have to be collected very close to the seabed using either ROVs or video-surveyed water sampling casts. In gorgonian corals, species presence observed by ROV underwater video footage matched well the diversity of coral eDNA from seawater collected using Niskin bottles mounted on the ROV (Everett and Park, 2018).
Limitations of the Protocol and Suggestions for Future Improvements
This study relied on the collection of single point water samples and the quantification of one 178 bp long mtDNA gene fragment from L. pertusa to locate CWC reefs and suggests that this is sufficient for detecting large reefs on vertical surfaces, with an accuracy of a few kilometers. We suggest two possible inclusions to the protocol which would profoundly increase the chances of accurately locating reefs and hence increase the ability of eDNA analysis to inform conservation and management programs. (1) As eDNA concentrations are diluted with increasing distance to the source (Taberlet et al., 2018), collecting clusters or transects of samples instead of single point samples would provide information on the heading to the DNA source and better so if fine scale local bathymetry and hydrodynamics are known. Furthermore, (2) eDNA studies often target fragments less than 200 bp in length because in situ DNA degradation precludes the persistence of longer eDNA fragments (i.e., 700–800 bp), resulting in reduced detection rates compared to shorter fragments (Jo et al., 2017; Taberlet et al., 2018; Nichols and Marko, 2019). However, primers targeting longer eDNA gene fragments would likely provide better estimates of recently shed eDNA as their decay rate is higher than for small eDNA fragments. In theory, long eDNA fragments are thus only present in high concentrations in samples collected very close to the eDNA source. While short DNA sequences may provide better estimates of fauna abundances on a regional scale, the distribution of single species on small spatial scales is probably better reflected in estimates based on detection of long eDNA fragments (Nichols and Marko, 2019). Moreover, by comparing the concentration of short (200 bp) and long (700 bp) eDNA fragments (with different decay rates) in a water sample, it is theoretically possible to determine the time since a given water mass came in contact with the coral reef eDNA source. With a better understanding of the direction of the reef relative to transect sampling the location of the reef could thereafter be modeled. In addition, differential target length detection could help in distinguishing, for example, whether an observed high eDNA concentration originates from a small but proximal reef (high concentration of both short and long fragments), or whether the signal originates from a large but distant reef (high concentration of short fragment but low concentration of long fragment).
Conclusion
We live in an era in which vulnerable marine ecosystems are increasingly subjected to stressors such as ocean warming and acidification, fisheries, offshore energy farms and aquaculture. In order to ensure effective management and conservation of vulnerable species and habitats, accurate spatial knowledge of these ecosystems is critical. Analysis of eDNA from keystone organisms shows great methodological promise for rapid and cost-efficient surveys of the broad scale distribution of CWC within fjord systems, that with time might be automated and carried out by AUVs (Yamahara et al., 2019). Results gained from this study may not be directly transferable to other areas as eDNA degradations and shedding rates will vary with environmental conditions. However, with further research and refinements of the method we believe that this technique will offer a useful complement to traditional sea-bed mapping. It seems particularly promising for detecting reefs in complex vertical terrain that cannot be imaged using ship mounted multi-beam echo-sounders and where other tools are needed to inform as to where targeted ROV surveys are needed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
TK, TD, JR, and VH conceived and designed the study. TK carried out the experiments and fieldwork, IJ performed the dispersal modeling, KS extracted DNA and analyzed all samples. TK took the lead in writing the manuscript. All authors provided critical feedback and helped in shaping the experiments, field study, analysis and manuscript.
Funding
The research reported in this publication was supported through funding from the Norwegian Ministry of Trade, Industry and Fisheries through IMR projects 14900, 15197, and 15598.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the personnel in the IMR laboratory in Bergen for logistical support as well as the crew of RV Kristine Bonnevie and Hans Brattström for assistance in the field. Dr. Aud Larsen, Dr. Sam Fredriksson and Dr. Nigel Keeley provided helpful comments and suggestions to improve the manuscript. This work was funded by the Institute of Marine Research trough the AkvaKyst (Nr 14900) and Vulnerable Benthic Habitats projects (Nr 15598). Additional funding to NORCE was provided by the Strategic Institute project «SIS-OMICS – et kapasitetsløft for analyze av digitale data» funded by the Norwegian Environment Agency.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00446/full#supplementary-material
Footnotes
References
Addamo, A. M., Vertino, A., Stolarski, J., García-Jiménez, R., Taviani, M., and Machordom, A. (2016). Merging scleractinian genera: the overwhelming genetic similarity between solitary desmophyllum and colonial Lophelia. BMC Evol. Biol. 16:149. doi: 10.1186/s12862-016-0654-8
Albretsen, J., Sperrevik, A. K., Staalstrøm, A., Sandvik, A. D., Vikebø, F., and Asplin, L. (2011). NorKyst-800 Report No. 1 User Manual and Technical Descriptions. Fisken og Havet 2/2011. Bergen: Institute of Marine Research.
Andruszkiewicz, E. A., Koseff, J. R., Fringer, O. B., Ouellette, N. T., Lowe, A. B., Edwards, C. A., et al. (2019). Modeling environmental DNA transport in the coastal ocean using lagrangian particle tracking. Front, Mar. Sci. 6:477. doi: 10.3389/fmars.2019.00477
Andruszkiewicz, E. A., Starks, H. A., Chavez, F. P., Sassoubre, L. M., Block, B. A., and Boehm, A. B. (2017). Biomonitoring of marine vertebrates in monterey bay using eDNA metabarcoding. PLoS One 12:e0176343. doi: 10.1371/journal.pone.0176343
Arvia, R., Sollai, M., Pierucci, F., Urso, C., Massi, D., and Zakrzewska, K. (2017). Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J. Virol. Methods 246, 15–20. doi: 10.1016/j.jviromet.2017.04.003
Asplin, L., Johnsen, I. A., Sandvik, A. D., Albretsen, J., Sundfjord, V., Aure, J., et al. (2014). Dispersion of salmon lice in the Hardangerfjord. Mar. Biol. Res. 10, 216–225. doi: 10.1080/17451000.2013.810755
Burdon-Jones, C., and Tambs-Lyche, H. (1960). Observations on the fauna of the North Brattholmen stone-coral reef near Bergen. Årbok for Universitetet i Bergen. Mat.Naturvitensk 1960, 1–24.
Burgos, J. M., Buhl-Mortensen, L., Buhl-Mortensen, P., Ólafsdóttir, S. H., Steingrund, P., Ragnarsson, S. Á., et al. (2020). Predicting the distribution of indicator taxa of vulnerable marine ecosystems in the arctic and sub-arctic waters of the nordic seas. Front. Mar. Sci. 7:131. doi: 10.3389/fmars.2020.00131
Carvalho, S., Aylagas, E., Villalobos, R., Kattan, Y., Berumen, M., and Pearman, J. K. (2019). Beyond the visual: using metabarcoding to characterize the hidden reef cryptobiome. Proc. R. Soc. B 286:20182697. doi: 10.1098/rspb.2018.2697
Cathalot, C., van Oevelen, D., Cox, T. J. S., Kutti, T., Lavaleye, M., Duineveld, G., et al. (2015). Cold-water coral reefs and adjacent sponge grounds: hotspots of benthic respiration and organic carbon cycling in the deep-sea. Front. Mar. Sci. 2:37. doi: 10.3389/fmars.2015.00037
Davies, A. J., Duineveld, G. C. A., Lavaleye, M. S. S., Bergman, M. J. N., van Haren, H., and Roberts, J. M. (2009). Downwelling and deep-water bottom currents as food supply mechanisms to the cold-water coral Lophelia pertusa (Scleractinia) at the mingulay reef complex. Limnol. Oceanogr. 54, 620–629. doi: 10.4319/lo.2009.54.2.0620
Doi, H., Uchii, K., Takahara, T., Matsuhashi, S., Yamanaka, H., and Minamoto, T. (2015). Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS One 10:e0122763. doi: 10.1371/journal.pone.0122763
Espeland, S. H., Albretsen, J., Olsen, E. M., and Bodvin, T. (2015). Modelling drift of pelagic offspring: the importance of egg surveys in providing a realistic model initialization. ICES J. of Mar. Sci. 72, 2578–2589. doi: 10.1093/icesjms/fsv134
Everett, M. V., and Park, L. K. (2018). Exploring deep-water coral communities using environmental DNA. Deep Sea Res. Part II. 150, 229–241. doi: 10.1016/j.dsr2.2017.09.008
Fisher, C. R., Hsing, P. Y., Kaiser, C. L., Yoerger, D. R., Roberts, H. H., Shedd, W. W., et al. (2014). Footprint of deep water horizon blowout impact to deep-water coral communities. Proc. Natl. Acad. Sci. U.S.A. 111, 11744–11749. doi: 10.1073/pnas.1403492111
Flot, J. F., Dahl, M., and André, C. (2013). Lophelia pertusa corals from the Ionian and Barents seas share identical nuclear ITS2 and near-identical mitochondrial genome sequences. BMC Res. Notes 6:144. doi: 10.1186/1756-0500-6-144
Fosså, J. H., Mortensen, P. B., and Furevik, D. M. (2002). The deep-water coral Lophelia pertusa in norwegian waters: distribution and fishery impact. Hydrobiologia 471, 1–12.
Freiwald, A. (2002). “Reef-forming cold-water corals,” in Ocean Margin Systems, eds G. Wefer, D. Billet, D. Hebbeln, B. B. Jorgensen, M. Schlüter, T. C. E. V. Weering, et al. (Berlin: Springer-Verlag), 365–385. doi: 10.1007/978-3-662-05127-6_23
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Gori, A., Orejas, C., Madurell, T., Bramanti, L., Martins, M., Quintanilla, E., et al. (2013). Bathymetrical distribution and size structure of cold-water coral populations in the cap de creus and lacaze-duthiers canyons (northwestern Mediterranean). Biogeosciences 10, 2049–2060. doi: 10.5194/bg-10-2049-2013
Hall-Spencer, J., Allain, V., and Fosså, J. H. (2002). Trawling damage to Northeast Atlantic ancient coral reefs. Proc. R. Soc. B 269, 507–511. doi: 10.1098/rspb.2001.1910
Harrison, J. B., Sunday, J. M., and Rogers, S. M. (2019). Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soci. B 286:20191409. doi: 10.1098/rspb.2019.1409
Heifetz, J., Stone, R. P., and Shotwell, S. K. (2009). Damage and disturbance to coral and sponge habitat of the Aleutian Archipelago. Mar. Ecol. Prog. Ser. 397, 295–303. doi: 10.3354/meps08304
Hoegh-Guldberg, O., Poloczanska, E. S., Skirving, W., and Dove, S. (2017). Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 4:158. doi: 10.3389/fmars.2017.00158
Holman, L. E., de Bruyn, M., Creer, S., Carvalho, G., Robidart, J., and Rius, M. (2019). Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci. Rep. 9:11559. doi: 10.1038/s41598-019-47899-7
Huvenne, V. A. I., Tyler, P. A., Masson, D. G., Fisher, E. H., Hauton, C., Hühnerbach, V., et al. (2011). A picture on the wall: innovative mapping reveals cold-water coral refuge in submarine canyons. PLoS One 6:e28755. doi: 10.1371/journal.pone.0028755
Järnegren, J., and Kutti, T. (2014). Lophelia Pertusa in Norwegian Waters. What Have We Learned Since 2008?. NINA Report 1028. Trondheim: Norwegian Institute for Nature Research, 40.
Jensen, S., Lynch, M. D., Ray, J. L., Neufeld, J. D., and Hovland, M. (2015). Norwegian deep-water coral reefs: cultivation and molecular analysis of planktonic microbial communities. Environ. Microbiol. 17, 3597–3609. doi: 10.1111/1462-2920.12531
Jo, T., Murakami, H., Masuda, R., Sakata, M. K., Yamamoto, S., and Minamoto, T. (2017). Rapid degradation of longer DNA fragments enables the improved estimation of distribution and biomass using environmental DNA. Mol. Ecol. Resour. 17, e25–e33. doi: 10.1111/1755-0998.12685
Johnsen, I. A., Asplin, L. C., Sandvik, A. D., and Serra-Llinares, R. M. (2016). Salmon lice dispersion in a northern Norwegian fjord system and the impact of vertical movements. Aquacult. Environ. Interact. 8, 99–116. doi: 10.3354/aei00162
Jones, E., Chierici, M., Skjelvan, I., Norli, M., Børsheim, K. Y., Lødemel, H. H., et al. (2018). Monitoring of Ocean Acidification in Norwegian seas in 2017. Monitoring Report M-1072 | 2018. Bergen: University of Bergen.
Kutti, T., Nordbø, K., Bannister, R. J., and Husa, V. (2015). Oppdrettsanlegg kan true korallrev i fjordane. Havforskningsrapporten 2015, 40–42.
Larsson, A. I., van Oevelen, D., Purser, A., and Thomsen, L. (2013). Tolerance to long-term exposure of suspended benthic sediments and drill cuttings in the cold-water coral Lophelia pertusa. Mar. Pollut. Bull. 70, 176–188. doi: 10.1016/j.marpolbul.2013.02.033
Link-Lenczowska, D., Pallisgaard, N., Cordua, S., Zawada, M., Czekalska, S., Krochmalczyk, D., et al. (2018). A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 97, 2299–2308. doi: 10.1007/s00277-018-3451-1
Majaneva, M., Diserud, O. H., Eagle, S. H. C., Boström, E., Hajibabaei, M., and Ekrem, T. (2018). Environmental DNA filtration techniques affect recovered biodiversity. Sci. Rep. 8:4682. doi: 10.1038/s41598-018-23052-8
Maruyama, A., Nakamura, K., Yamanaka, H., Kondoh, M., and Minamoto, T. (2014). The release rate of environmental DNA from juvenile and adult fish. PLoS One 9:e114639. doi: 10.1371/journal.pone.0114639
Miotke, L., Lau, B. T., Rumma, R. T., and Ji, H. P. (2014). High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal. Chem. 86, 2618–2624. doi: 10.1021/ac403843j
Mortensen, P. B., and Rapp, H. T. (1998). Oxygen and carbon isotope ratios related to growth line patterns in skeletons of Lophelia pertusa (L)(Anthozoa, Scleractinia): implications for determination of linear extension rates. Sarsia 83, 433–446. doi: 10.1080/00364827.1998.10413702
Murray, F., De Clippele, L. H., Hiley, A., Wicks, L., Roberts, J. M., and Hennige, S. (2019). Multiple feeding strategies observed in the cold-water coral Lophelia pertusa. J. Mar. Biol. Assoc. U. K. 99, 1281–1283. doi: 10.1017/S0025315419000298
Nichols, P. K., and Marko, P. B. (2019). Rapid assessment of coral cover from environmental DNA in Hawai’i. Environ. DNA 1, 40–53. doi: 10.1002/edn3.8
Port, J. A., O’Donnell, J. L., Romero-Maraccini, O. C., Leary, P. R., Litvin, S. Y., Nicklos, K. J., et al. (2016). Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol. Ecol. 25, 527–541. doi: 10.1111/mec.13481
Robert, K., Huvenne, V. A. I., Georgiopoulou, A., Jones, D. O. B., Marsh, L., Carters, G. D. O., et al. (2017). New approaches to high resolution mapping of marine vertical structures. Sci. Rep. 7:9005. doi: 10.1038/s41598-017-09382-z
Roberts, J. M., Wheeler, A., Freiwald, A., and Cairns, S. D. (2009). Cold-Water Corals: the Biology and Geology of Deep-Sea Coral Habitats. Cambridge: Cambridge University Press.
Rogers, A. D. (1999). The biology of Lophelia pertusa (Linnaeus 1758) and other deep-water reef-forming corals and impacts from human activities. Int. Rev. Hydrobiol. 84, 315–406. doi: 10.1002/iroh.199900032
Shchepetkin, A. F., and McWilliams, J. C. (2005). The regional oceanic modeling system (ROMS): a split-explicit, free-surface, topography-following-coordinate oceanic model. Ocean Model. 9, 347–404. doi: 10.1016/j.ocemod.2004.08.002
Skjelvan, I., Chierici, M., Sørensen, K., Jackson, K., Kutti, T., Lødemel, H. H., et al. (2016). Havforsuring i vestlandsfjorder og CO2-variabilitet i Lofoten. Rappo. Miljødirektoratet 642:2016.
Stigebrandt, A., and Aure, J. (1989). Vertical mixing in basin waters of fjords. J. Phys. Oceanogr. 19, 917–926. doi: 10.1175/1520-0485(1989)019<0917:vmibwo>2.0.co;2
Stone, R. P. (2006). Coral habitat in the Aleutian Islands of Alaska: depth distribution, fine-scale species associations, and fisheries interactions. Coral Reefs 25, 229–238. doi: 10.1007/s00338-006-0091-z
Sundahl, H., Buhl-Mortensen, P., and Buhl-Mortensen, L. (2020). Distribution and suitable habitat of the cold-water corals Lophelia pertusa, Paragorgia arborea, and Primnoa resedaeformis on the Norwegian Continental Shelf. Front. Mar. Sci. 7:213. doi: 10.3389/fmars.2020.00213
Taberlet, P., Bonin, A., Zinger, L., and Coissac, E. (2018). Environmental DNA: for Biodiversity Research and Monitoring. Oxford: Oxford University Press.
Tambs-Lyche, H. (1958). Zoogeographical and Faunistic Studies on West Norwegian Marine Animals. Naturvitenskapelig Rekke Nr. 7. Bergen: Universitetet i Bergen. 24.
Taylor, S. C., Laperriere, G., and Germain, H. (2017). Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 7, 1–8.
Thiem, Ø, Ravagnan, E., Fossa?, J. H., and Berntsen, J. (2006). Food supply mechanisms for cold-water corals along a continental shelf edge. J. Mar. Syst. 60, 207–219. doi: 10.1016/j.jmarsys.2005.12.004
Uthicke, S., Lamare, M., and Doyle, J. R. (2018). eDNA detection of corallivorous seastar (Acanthaster cf. solaris) outbreaks on the Great Barrier Reef using digital droplet PCR. Coral Reefs 37, 1–11. doi: 10.1007/s00338-018-1734-6
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y. M., and Jørgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Winnepenninckx, B., Backeljau, T., and De Wachter, R. (1993). Extraction of high molecular weight DNA from molluscs. Trends Genet. 9, 407–407. doi: 10.1016/0168-9525(93)90102-n
Yamahara, K. M., Preston, C. M., Birch, J., Walz, K., Marin, R., Jensen, S., et al. (2019). In situ autonomous acquisition and preservation of marine environmental DNA using an autonomous underwater vehicle. Front. Mar. Sci. 6:373. doi: 10.3389/fmars.2019.00373
Keywords: Lophelia pertusa, Desmophyllum pertusum, ddPCR, deep-sea, habitat mapping, vulnerable marine ecosystems
Citation: Kutti T, Johnsen IA, Skaar KS, Ray JL, Husa V and Dahlgren TG (2020) Quantification of eDNA to Map the Distribution of Cold-Water Coral Reefs. Front. Mar. Sci. 7:446. doi: 10.3389/fmars.2020.00446
Received: 21 February 2020; Accepted: 20 May 2020;
Published: 12 June 2020.
Edited by:
Iliana B. Baums, Pennsylvania State University (PSU), United StatesReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, United StatesErik Cordes, Temple University, United States
Copyright © 2020 Kutti, Johnsen, Skaar, Ray, Husa and Dahlgren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tina Kutti, dGluYS5rdXR0aUBoaS5ubw==; dGluYS5rdXR0aUBpbXIubm8=
 Tina Kutti
Tina Kutti Ingrid Askeland Johnsen2
Ingrid Askeland Johnsen2